
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 14 August 2017
Sec. Plant Abiotic Stress
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01403
 Tao Lang1†
Tao Lang1† Shurong Deng2,3†
Shurong Deng2,3† Nan Zhao2†
Nan Zhao2† Chen Deng2
Chen Deng2 Yinan Zhang2
Yinan Zhang2 Yanli Zhang2
Yanli Zhang2 Huilong Zhang2
Huilong Zhang2 Gang Sa2
Gang Sa2 Jun Yao2
Jun Yao2 Caiwu Wu4
Caiwu Wu4 Yanhong Wu1
Yanhong Wu1 Qun Deng1
Qun Deng1 Shanzhi Lin2
Shanzhi Lin2 Jianxin Xia1*
Jianxin Xia1* Shaoliang Chen2*
Shaoliang Chen2*We investigated the effects of salt-sensitive signaling molecules on ionic fluxes and gene expression related to K+/Na+ homeostasis in a perennial herb, Glycyrrhiza uralensis, during short-term NaCl stress (100 mM, 24 h). Salt treatment caused more pronounced Na+ accumulation in root cells than in leaf cells. Na+ ions were mostly compartmentalized in vacuoles. Roots exposed to NaCl showed increased levels of extracellular ATP (eATP), cytosolic Ca2+, H2O2, and NO. Steady-state flux recordings revealed that these salt-sensitive signaling molecules enhanced NaCl-responsive Na+ efflux, due to the activated Na+/H+ antiport system in the plasma membrane (PM). Moreover, salt-elicited K+ efflux, which was mediated by depolarization-activated cation channels, was reduced with the addition of Ca2+, H2O2, NO, and eATP. The salt-adaptive effects of these molecules (Na+ extrusion and K+ maintenance) were reduced by pharmacological agents, including LaCl3 (a PM Ca2+ channel inhibitor), DMTU (a reactive oxygen species scavenger), cPTIO (an NO scavenger), or PPADS (an antagonist of animal PM purine P2 receptors). RT-qPCR data showed that the activation of the PM Na+/H+ antiport system in salinized roots most likely resulted from the upregulation of two genes, GuSOS1 and GuAHA, which encoded the PM Na+/H+ antiporter, salt overly sensitive 1 (SOS1), and H+-ATPase, respectively. Clear interactions occurred between these salt-sensitive agonists to accelerate transcription of salt-responsive signaling pathway genes in G. uralensis roots. For example, Ca2+, H2O2, NO, and eATP promoted transcription of GuSOS3 (salt overly sensitive 3) and/or GuCIPK (CBL-interacting protein kinase) to activate the predominant Ca2+-SOS signaling pathway in salinized liquorice roots. eATP, a novel player in the salt response of G. uralensis, increased the transcription of GuSOS3, GuCIPK, GuRbohD (respiratory burst oxidase homolog protein D), GuNIR (nitrate reductase), GuMAPK3, and GuMAPK6 (the mitogen-activated protein kinases 3 and 6). Moreover, GuMAPK3 and GuMAPK6 expression levels were enhanced by H2O2 in NaCl-stressed G. uralensis roots. Our results indicated that eATP triggered downstream components and interacted with Ca2+, H2O2, and NO signaling to maintain K+/Na+ homeostasis. We propose that a multiple signaling network regulated K+/Na+ homeostasis in NaCl-stressed G. uralensis roots.
Excess salts in the soil disrupts ion homeostasis in herbaceous and woody species (Munns and Tester, 2008; Polle and Chen, 2015). Maintaining cellular and whole-plant K+/Na+ homeostasis is required for plant adaptation to salt stress (Shabala et al., 2005; Sun et al., 2009a,b, 2010a,b; Chen and Polle, 2010; Chen et al., 2014). The plasma membrane (PM)-located H+-ATPase and Na+/H+ antiporter play crucial roles in maintaining K+/Na+ homeostasis in higher plants. The PM Na+/H+ antiporter, salt overly sensitive 1 (SOS1), prevents excessive Na+ accumulation in the cytoplasm (Zhu, 2001, 2016). The PM H+-ATPase sustains an H+ gradient to drive Na+ and H+ transport across the PM (Blumwald et al., 2000). Moreover, H+-pumps preserve a less-depolarized membrane potential, thus restricting K+ efflux through depolarization-activated outward rectifying K+ channels (DA-KORCs) and non-selective cation channels (DA-NSCCs, Sun et al., 2009b, 2012a; Zhang et al., 2015). A large body of evidence suggests that salt-sensitive signaling molecules, such as extracellular ATP (eATP), hydrogen peroxide (H2O2), calcium (Ca2+), nitric oxide (NO), and their crosstalk contribute to the regulation of the Na+/H+ antiport system (the H+-ATPase and Na+/H+ antiporter). This system contributes to K+/Na+ homeostasis in a variety of plant species (Zhang et al., 2007; Chen et al., 2010; Sun et al., 2010a,b, 2012a; Lu et al., 2013; Lang et al., 2014).
Salt-elicited cytosolic Ca2+ upregulates PM Na+/H+ antiporter activity via the SOS-signaling pathway in Arabidopsis (Qiu et al., 2002; Zhu, 2003), rice (Martínez-Atienza et al., 2007), and poplar (Tang et al., 2010). H2O2 induces the entry of Ca2+ through PM Ca2+-permeable channels (Pei et al., 2000; Mori and Schroeder, 2004), and this mechanism was suggested to trigger the Ca2+-SOS pathway (Sun et al., 2010b). NO functions as a gaseous signaling molecule, which induces resistance to salt injury by depleting the Na+ content, as previously shown in reed callus (Zhang et al., 2006) and in salt-secreting and non-secreting mangroves (Chen et al., 2010; Lu et al., 2013; Lang et al., 2014). Extracellular ATP acts as a signaling molecule and plays a significant role in protecting against NaCl stress (Kim et al., 2009; Sun et al., 2012a; Chen et al., 2014; Lang et al., 2014; Polle and Chen, 2015). It is suggested that eATP can be sensed by a purinergic ATP (P2) receptor in the PM, most likely P2K1 (Choi et al., 2014), and P2 receptor binding induces downstream signaling components, e.g., H2O2 and Ca2+ (Demidchik et al., 2009; Sueldo et al., 2010; Sun et al., 2010b, 2012a). Indeed, eATP interacted with H2O2 and Ca2+ to induce resistance to Na+ toxicity in mangrove roots (Lang et al., 2014). However, the effect of eATP signaling cascades on Na+ homeostasis remains to be elucidated in salt-resistant herbaceous species, e.g., Glycyrrhiza uralensis.
NaCl exposure caused membrane depolarization and net K+ efflux in Arabidopsis (Shabala et al., 2005, 2006), barley (Shabala et al., 2003; Chen et al., 2007), Populus euphratica (Sun et al., 2009b, 2010a,b, 2012a; Zhao et al., 2016), and mangrove species (Chen et al., 2010; Lu et al., 2013; Lang et al., 2014). Ca2+ blocked NaCl-induced K+ loss, which was mediated by depolarization-activated KORCs and NSCCs in Arabidopsis (Shabala et al., 2006) and in poplars (Sun et al., 2009b). This was mainly due to the activated PM H+-ATPase, which lowers the NaCl-depolarized membrane potential, thus restricting K+ loss through KORCs and NSCCs (Shabala et al., 2006; Sun et al., 2009b). H2O2, NO, and eATP were also shown to maintain K+ homeostasis by up-regulating PM proton pumps in poplar species (Zhang et al., 2007; Sun et al., 2010a,b, 2012a; Zhao et al., 2016) and mangroves (Chen et al., 2010; Lu et al., 2013; Lang et al., 2014). However, interactions between these stress signaling molecules in the regulation of K+ homeostasis remains to be established in liquorice plants.
Glycyrrhiza uralensis Fisch. (Licorice), a perennial herb of the genus Leguminosae, is naturally distributed in the arid and semi-arid areas of eastern Asia (Li et al., 2016). Licorice is frequently used as a crude therapeutic medicine to protect against multiple diseases in Asian populations (Mochida et al., 2017). Apart from its pharmaceutical functions, G. uralensis is ecologically important, both for conserving soil and water and for improving soil structure in semiarid ecosystems (Zhang and Ye, 2009). The deep-rooted nature of G. uralensis plants enables them to survive desert and semi-desert habitats in northwestern China. However, how G. uralensis sustains ionic homeostasis under saline conditions and whether salt-sensitive signals contribute to the demonstrated salt tolerance have not been investigated in this liquorice species.
In the present study, we aimed to characterize the importance of Ca2+, H2O2, NO, and eATP in mediating Na+/H+ transport in the salinized roots of G. uralensis. Flux measurements with non-invasive micro-test technology (NMT) revealed that these salt-induced signals were essential for restricting K+ efflux and enhancing Na+ exclusion in liquorice roots. We also screened for alterations in the transcription of genes involved in various salt-signaling pathways. We aimed to explore the network of multiple interactions among Ca2+, H2O2, NO, and eATP in the regulation of signaling and gene expression related to K+/Na+ homeostasis in G. uralensis roots.
Seeds of G. uralensis were obtained from the Mongolian Autonomous County of Hoboksar, Tarbagatay Prefecture, Xinjiang Uygur Autonomous Region (latitude 46°82′N, longitude 85°75′E). The seeds were planted in plastic pots (5 cm in diameter, 8 cm in height), containing a 2:1 mixture of sand and nursery soil, and placed in a growth chamber at Beijing Forestry University, Beijing, China. The potted G. uralensis were well irrigated, according to evaporation demand, and fertilized with one-quarter-strength Hoagland solution weekly. The temperature and relative humidity were maintained at 25–28°C and 60–70%, respectively. A photoperiod of 14 h (9:00–23:00) was applied, and photosynthetically active radiation varied from 280 to 350 μmol m-2s-1. After 2 weeks of culture, rooted liquorice seedlings were transferred to 300-ml pots containing one-quarter-strength Hoagland’s nutrient solution for hydroponic equilibration.
Hydroponic-equilibrated seedlings of G. uralensis were subjected to 0 or 100 mM NaCl for 24 h. Na+ concentrations in root and leaf cells were examined after 6, 12, and 24 h of treatment. Na+, K+, and H+ fluxes were measured along the root axes with the NMT technique. The effects of PM transporter/channel inhibitors were examined in NaCl-treated G. uralensis. A blocker of the Na+/H+ antiporter, amiloride (50 μM), and a specific inhibitor of the H+-ATPase, sodium orthovanadate (500 μM), were used to inhibit the Na+/H+ antiport system in the PM (Sun et al., 2009a). A typical K+ channel inhibitor, tetraethylammonium chloride (TEA, 50 μM), was used to reduce NaCl-elicited K+ efflux (Lu et al., 2013; Lang et al., 2014). In our study, control and NaCl-treated roots were treated with these inhibitors for 30 min before the flux recordings. In addition, two series of experiments (described below) were carried out to determine the involvement of Ca2+, H2O2, NO, and eATP in regulating Na+ and K+ fluxes and gene expression in NaCl-treated G. uralensis roots.
We added exogenous agonists, CaCl2 (10 mM), H2O2 (10 mM), the NO donor, sodium nitroprusside (SNP, 100 μM), and ATP-Na2 (300 μM), and measured the effects on NaCl-induced Na+ and K+ fluxes in young roots of G. uralensis. The chemicals were added to one-quarter-strength nutrient solution in the presence and absence of NaCl (100 mM). Control plants treated with or without salt were cultured in nutrient solution without the application of the chemicals mentioned above. The steady-state fluxes of K+ and Na+ were recorded along the root axis after 24-h NaCl treatments.
We also examined the expression levels of genes involved in salt transport and signaling after salt and agonist (Ca2+, H2O2, SNP, and ATP) treatments. Specifically, we examined expression of the PM H+-ATPase gene, GuAHA; the PM Na+/H+ antiporter gene, GuSOS1; the salt overly sensitive 3 gene, GuSOS3; the calcineurin B-like protein (CBL)-interacting protein kinase gene, GuCIPK; the respiratory burst oxidase homolog protein D gene, GuRbohD; the nitrate reductase gene, GuNIR; and the mitogen-activated protein kinases 3 and 6 genes, GuMAPK3 and GuMAPK6.
Control and NaCl (100 mM, 24 h)-stressed G. uralensis seedlings were treated with or without pharmacological agents for 30 min. These agents were: LaCl3, an inhibitor of the PM Ca2+ channel (5 mM); DMTU, a ROS scavenger (5 mM); cPTIO, a scavenger of NO (300 μM); and PPADS, an antagonist of animal PM P2 receptors (300 μM) (Sun et al., 2010a,b; Chen et al., 2013; Zhao et al., 2016). Next, young roots with apices of 2.0–3.0 cm were sampled and equilibrated in measuring solution for 30 min. Then, steady-state fluxes of K+ and Na+ along the root axes were recorded in plants after treating with NaCl and antagonist (LaCl3, DMTU, cPTIO, and PPADS). We also examined the abundances of GuAHA and GuSOS1 transcripts in these roots.
We used the NMT technique (NMT-YG-100, Younger United States LLC, Amherst, MA, United States) to measure the net Na+, K+, and H+ fluxes in G. uralensis roots. The microelectrodes were prepared and calibrated as previously described (Sun et al., 2009a,b; Lang et al., 2014).
After roots were exposed to NaCl treatment, with either an agonist (Ca2+, H2O2, SNP, and ATP) or an antagonist (amiloride, sodium orthovanadate, TEA, LaCl3, DMTU, cPTIO, and PPADS), root segments with 2.0–3.0 cm apices were selected and washed two or three times with redistilled water. When placed in a buffer with a lower Na+ concentration, the preloaded Na+ would diffuse from the surface of salt-stressed roots. To decrease the effect of this excess salt release on flux recordings, roots were equilibrated prior to flux recordings in a measuring solution (0.1 mM NaCl, 0.1 mM MgCl2, 0.1 mM CaCl2, and 0.5 mM KCl) for 30 min. The concentrations of Ca2+ and K+ in the measuring solution were set to 0.1 and 0.5 mM, respectively (Li et al., 2012), to reduce interference from Ca2+ and K+ on the Na+ electrodes (Cuin et al., 2011). The pH of the measuring solution was adjusted to 5.7 with HCl and KOH.
After equilibration, roots were immobilized on the bottom of a measuring chamber with 10 ml of fresh measuring solution. Flux measurements were started at 200 μm from the root apex and conducted along the root axis, up to 2700 μm from the root apex, at intervals of 200 or 300 μm (vigorous ion fluxes were typically observed at the apical regions; Lu et al., 2013; Lang et al., 2014). A 6–8 min continuous recording was performed at each measuring point in the apical zones. Five or six individual seedlings were measured from each treatment group.
To evaluate the NaCl-induced Na+ distribution in G. uralensis roots and leaves, we used a specific fluorescent probe, CoroNa-Green AM (Sun et al., 2012a). Two-week-old seedlings were exposed to 0 or 100 mM NaCl for 6, 12, or 24 h. Then, the roots and leaves were exposed to CoroNa-Green AM (20 μM) for 2 h in a 5 mM Mes-KCl loading buffer (pH 5.7). Cellular Na+ was visualized with a Leica SP5 confocal microscope (Leica Microsystems GmbH, Wetzlar, Germany). The confocal settings were as follows: excitation 488 nm, emission 510–530 nm, frame = 512 × 512.
In G. uralensis roots, we used specific fluorescent probes to detect cellular signal contents. We used Rhod-2 AM (Biotium) to detect cytosolic Ca2+ (Sun et al., 2012a; Zhang et al., 2015); H2DCF-DA (Eugene) to detect H2O2 (Sun et al., 2010a,b); and DAF-FM DA (Eugene) to detect NO (Sun et al., 2012a). Briefly, young roots were exposed to 0 or 100 mM NaCl for 30 min. Then, the roots were transferred to a 5 mM Mes-KCl loading buffer (pH 5.7) containing 2 μM Rhod-2 AM, 50 μM H2DCF-DA, or 10 μM DAF-FM DA. The staining was performed in the dark for 1 h at room temperature. Next, the roots were washed 4–5 times with Murashige and Skoog (MS) liquid medium prior to confocal microscope measurements. The confocal settings were as follows: excitation 488 nm, emission 510–530 nm for H2DCF-DA and DAF-FM DA; and excitation 543 nm, emission 570–590 nm for Rhod-2 AM (frame = 512 × 512).
Extracellular ATP levels were monitored with the Enlighten ATP assay system bioluminescence kit (Promega, Madison, WI, United States; Sun et al., 2012a; Deng et al., 2015). Briefly, G. uralensis roots were exposed to 0 or 100 mM NaCl at room temperature. The liquid culture medium of control and NaCl-treated roots was sampled at 0, 5, 10, 20, 40, 60, 120, and 240 min, then immediately frozen in liquid nitrogen. eATP was measured in an assay with luciferin-luciferase Turner Designs ModulusTM Microplate Multimode Reader (Promega Corp., Madison, WI, United States). The eATP levels were calculated, based on a standard curve created by measuring a linear range (0.01–100 nM) of standard eATP concentrations (Sun et al., 2012a; Deng et al., 2015).
The transcription levels of genes related to the PM Na+/H+ transport system and salt signaling were evaluated in salt-stressed plants. Quantitative real-time PCR assays were conducted according to Deng et al. (2015) with some modifications. Briefly, total RNA was isolated from G. uralensis roots with TRIzol reagent (Invitrogen). DNA was eliminated by treating for 0.5 h with DNase I (Promega). An aliquot of purified RNA (1 μg) was used as template for first strand cDNA synthesis with M-MLV reverse transcriptase (Promega) and oligo (dT) primers. Specific primers for GuAHA, GuSOS1, GuSOS3, GuCIPK, GuRbohD, GuNIR, GuMAPK3, and GuMAPK6 were designed, based on homologous sequences found in Populus trichocarpa or Arabidopsis. Forward and reverse primers are listed in Supplementary Table 1. Amplification was performed as described by Ding et al. (2010): 95°C for 5 min, followed by 32 cycles of 94°C for 30 s, 55°C for 30 s, and finally, 72°C for 30 s, with a final step of 72°C for 10 min. The transcripts of target genes were normalized to the expression level of the G. uralensis β-actin 2 gene (GuACT2), and relative expression was calculated with the 2-ΔΔCT method (Livak and Schmittgen, 2001). Each experiment was replicated at least three times, and mean values are shown.
Ion fluxes were evaluated with JCal V3.0, which was created by Yue Xu1. In the present study, positive values denote cation efflux and negative values denote cation influx. All experimental data were processed with SPSS 17.0 for statistical tests. Data were subjected to an Analysis of Variance (ANOVA), and comparisons between means were performed with Duncan’s multiple range test. P-values less than 0.05 were considered statistically significant.
The Na+ concentrations in roots and leaves of G. uralensis were detected with a Na+-sensitive fluorescent dye, CoroNa-Green AM. The Na+ fluorescence in roots and leaves increased with the duration of salt exposure (6, 12, and 24 h) (Figure 1). Intracellular Na+ was detected as a bright green fluorescence, which was typically observed in vacuoles (Figure 1). However, Na+ levels in roots were 1.69- to 2.40-fold higher than that in leaves over the observation period (Figure 1). This result indicated that G. uralensis roots could take up and accumulate high Na+ within a short period of salt treatment. Therefore, the roots were used to evaluate the effects of salt signaling molecules on ion fluxes and gene transcription.
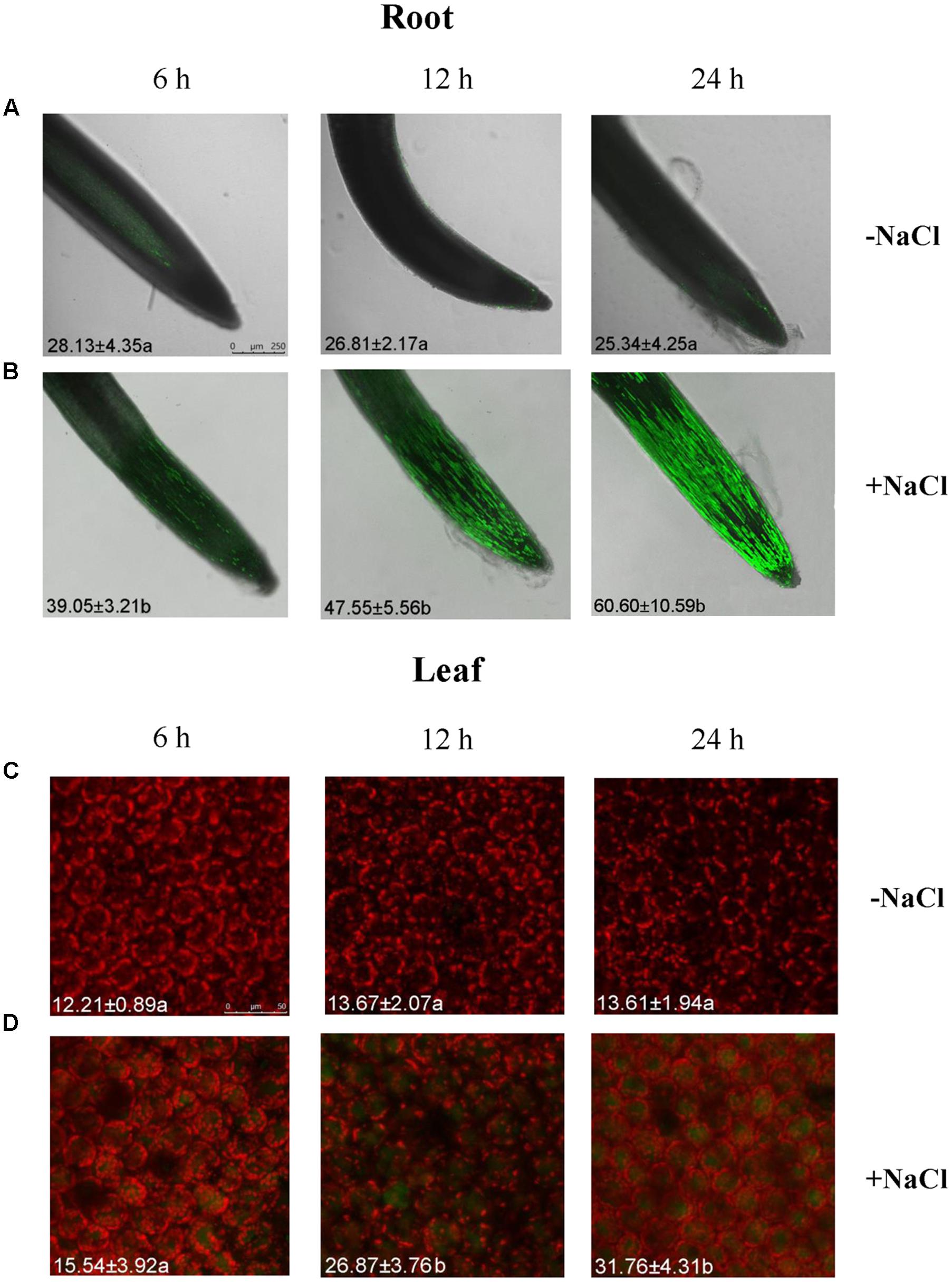
FIGURE 1. Na+ concentrations in root and leaf cells of NaCl-stressed Glycyrrhiza uralensis. Two-week-old G. uralensis seedlings were exposed to 0 or 100 mM NaCl for 6, 12, and 24 h in one-quarter-strength Hoagland solution, then stained with CoroNa-Green AM to detect Na+ concentrations. Representative confocal images of (A,B) roots (scale bar: 250 μm) and (C,D) leaves (scale bar: 50 μm) show the Na+ content (bright green fluorescence). The orange-red color is chlorophyll autofluorescence. The mean value (±SD) of 4–5 independent experiments is shown in the left bottom corner of each panel, and different letters (a and b) denote significant differences (P < 0.05) between control (–NaCl) and salt treatment (+NaCl).
Rhod-2 AM, H2DCF-DA, and DAF-FM DA, respectively, were used to detect cytosolic Ca2+, H2O2, and NO elicited by NaCl in G. uralensis roots (Sun et al., 2010a,b, 2012a). Confocal assays (Figure 2) revealed that cytosolic Ca2+ (color: pseudo-red), H2O2 (color: pseudo-green), and NO (color: pseudo-green) significantly increased by 71–111% after a 30 min salt shock. Similarly, in an ATP-bioluminescence assay, NaCl caused a marked rise in eATP after 20 min of stress, and the peak level occurred at 40 min of stress (Figure 3).
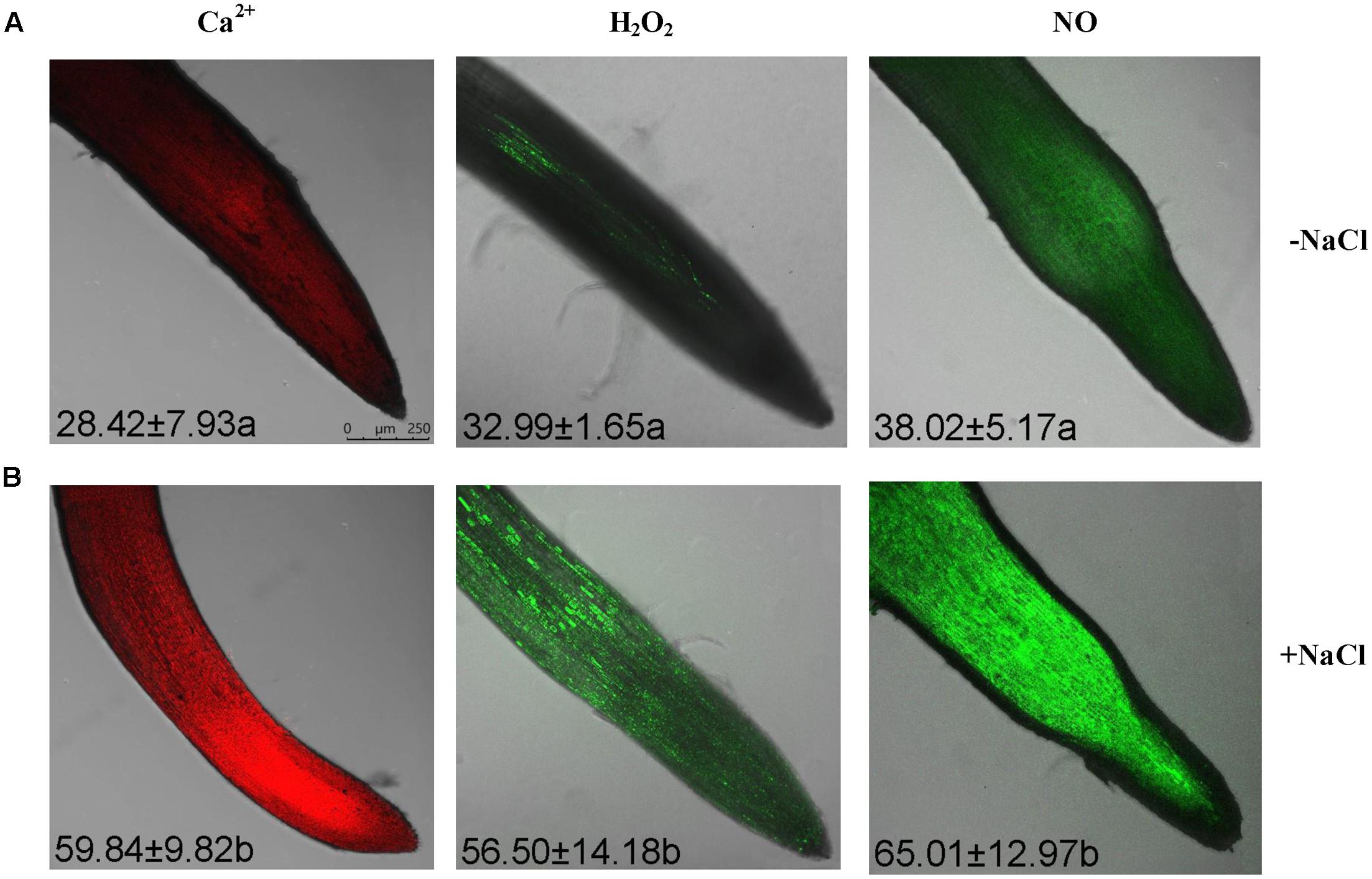
FIGURE 2. Cytosolic Ca2+, H2O2, and NO levels within root cells in NaCl-stressed G. uralensis. Two-week-old G. uralensis seedlings were exposed to 0 or 100 mM NaCl for 30 min in one-quarter-strength Hoagland solution, then stained with specific fluorescent probes for detecting Ca2+ (Rhod-2, orange-red), H2O2 (H2DCF, green), and NO (DAF-FM, green). Representative confocal images (scale bar: 250 μm) show (A) control and (B) NaCl-stressed roots. The bright green fluorescence corresponded to the detection of H2O2 and NO, while the orange-red color is the Ca2+ fluorescence. The mean value (±SD) of 4–5 independent experiments is shown in the left corner of each panel, and different letters (a and b) denote significant differences (P < 0.05) between control (–NaCl) and salt treatment (+NaCl).
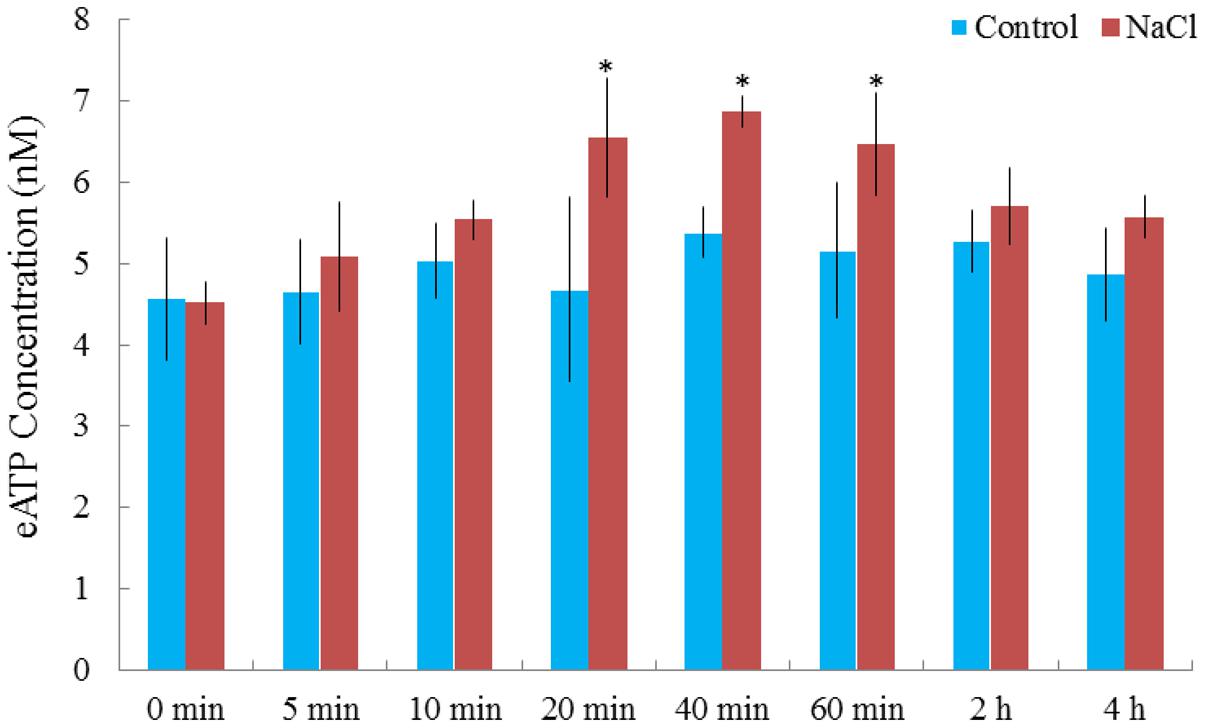
FIGURE 3. NaCl-induced alterations in extracellular ATP (eATP) in G. uralensis roots. Two-week-old G. uralensis seedlings were exposed to 0 or 100 mM NaCl for 4 h in one-quarter-strength Hoagland solution. Extracellular ATP was detected with the Enlighten ATP assay system bioluminescence kit. Each value (±SD) is the mean of 4–5 independent experiments. ∗P < 0.05, compared to no-salt control.
Under no-salt control conditions, G. uralensis roots exhibited stable, constant Na+ efflux along the root apex, with a mean value of 37.89 pmol cm-2 s-1 (Figure 4A). After exposure to NaCl (100 mM) for 24 h, Na+ efflux along the root tip significantly increased to 315.24 pmol cm-2 s-1 (Figure 4A). Of note, the maturation region (1700–2000 μm from the apex) displayed 10–20% higher Na+ efflux than the meristematic zone (200 μm from the apex).
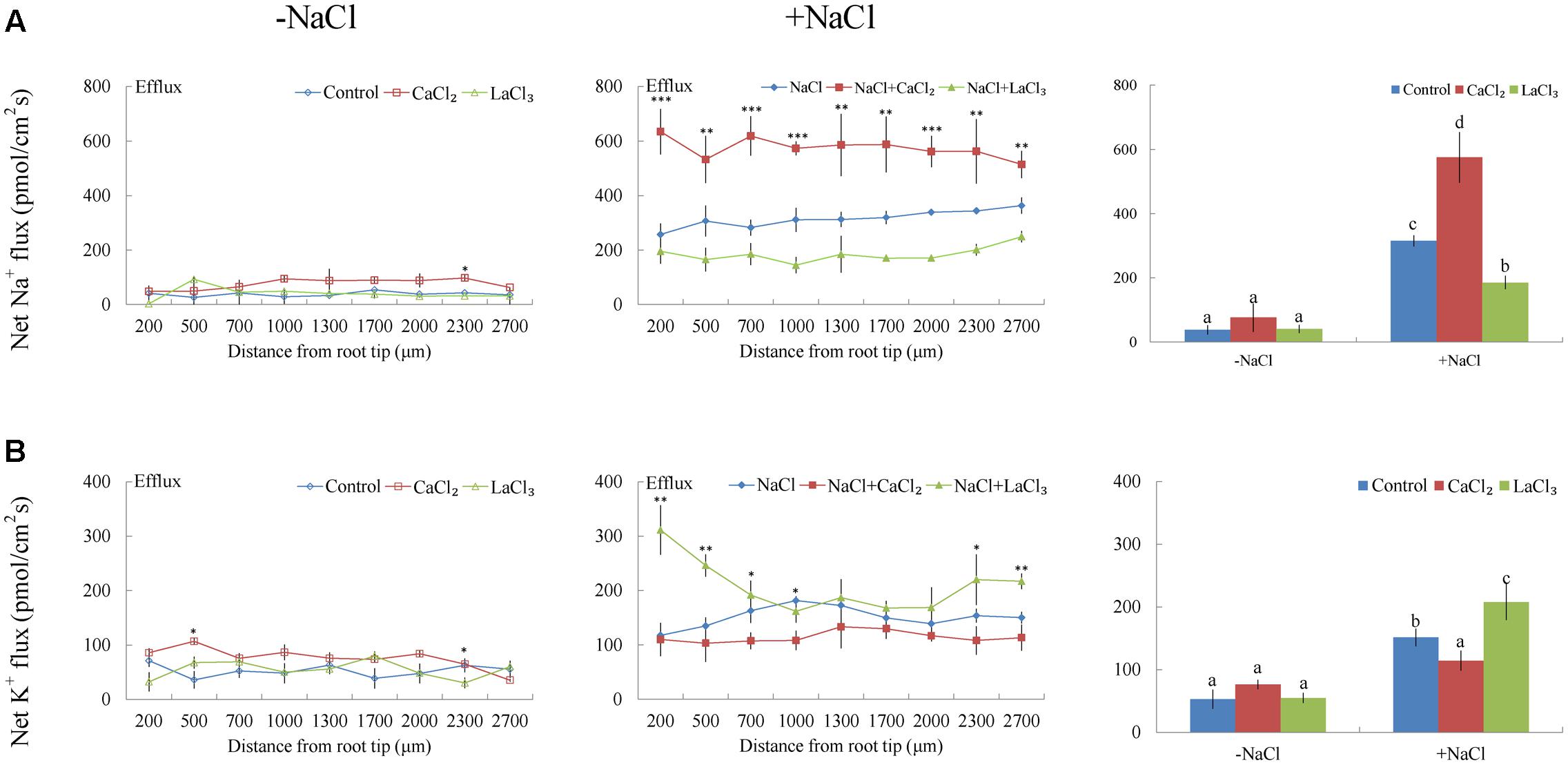
FIGURE 4. Effects of CaCl2 and LaCl3 on Na+ and K+ fluxes in G. uralensis roots under salt stress. Roots were untreated (control, blue) or exposed to CaCl2 (10 mM, red) for 24 h in the absence (–NaCl) and presence of NaCl (100 mM). For inhibitor treatment, control and NaCl-stressed roots were subjected to LaCl3 (5 mM, green) for 30 min. Steady-state flux profiles of (A) Na+ and (B) K+ were measured along the root axis at the apical zones (200–2700 μm from the root tip) in no-salt (left panels) and salt-stressed (center panels) conditions. Each point represents the mean of five to six individual plants. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, compared to controls. (Right panels) Means of Na+ and K+ fluxes at all measurement points, in no-salt (–NaCl) and salt-stressed (+NaCl) plants. Bars (±SD) represent the means of five to six individual plants; different letters (a, b, c, and d) indicate significant differences (P < 0.05) between treatments.
Under NaCl exposure, the addition of 10 mM Ca2+ markedly increased the Na+ efflux by 82% in the measured root regions (Figure 4A). However, the addition of LaCl3 (5 mM), an inhibitor of Ca2+-channels in the PM, markedly reduced the salt-elicited Na+ efflux (Figure 4A). Compared to NaCl treatment, in no-salt control conditions, exogenously applied CaCl2 or LaCl3 had no significant effect on root Na+ flux with the exception of a few measuring points (Figure 4A).
Pharmacological experiments revealed that salt-elicited Na+ efflux was significantly suppressed by amiloride (an inhibitor of the Na+/H+ antiporter) or sodium orthovanadate (a specific inhibitor of the PM H+-ATPase) (Figure 5A). Moreover, steady-state recordings showed that these inhibitors markedly decreased the H+ influx induced by salt treatment (Figure 5B). These results indicated that salt-stimulated Na+ efflux was due to active Na+ extrusion, i.e., Na+/H+ antiport across the PM, in this medicinal plant.
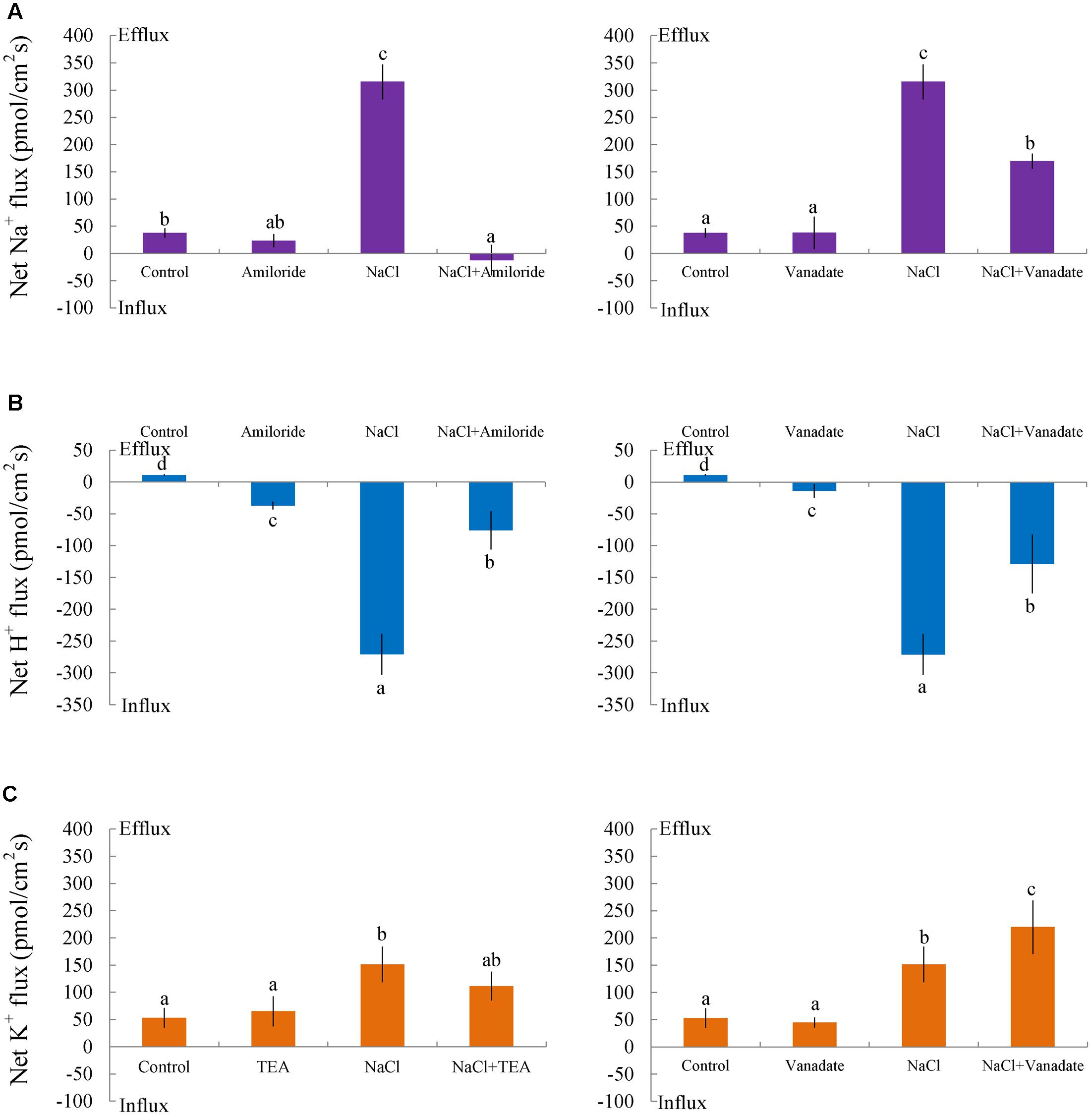
FIGURE 5. Effects of amiloride, sodium orthovanadate, and tetraethylammonium (TEA) on Na+, K+, and H+ fluxes in G. uralensis roots under salt stress. Roots were exposed to 0 (control) or 100 mM NaCl (NaCl) for 24 h, then exposed to transporter/channel inhibitors for 30 min. Steady-state fluxes were measured along the root axis at the apical zones (200–2700 μm from the root tip). Mean fluxes of (A) Na+ and (B) H+ were measured in the absence and presence of inhibitors, (left) amiloride (50 μM) and (right) sodium orthovanadate (500 μM). (C) The mean K+ flux was measured in the absence and presence of inhibitors, (left) TEA (50 μM) and (right) sodium orthovanadate (500 μM). Bars (±SD) represent the means of five to six individual plants; letters (a, b, c, and d) indicate significant differences between treatments (P < 0.05).
Under short-term NaCl stress, exogenously applied H2O2 (10 mM), SNP (a NO donor, 100 μM), or ATP (300 μM) produced an effect similar to that of CaCl2 (Figures 6A–8A). More pronounced effects were observed with ATP treatment, which induced a mean Na+ flux of 555.86 pmol cm-2 s-1, compared to fluxes of 458.84 pmol cm-2 s-1 with H2O2 and 469.56 pmol cm-2 s-1 with SNP treatments (Figures 6A–8A). Conversely, DMTU (a ROS scavenger, 5 mM), cPTIO (a NO scavenger, 300 μM), or PPADS (the antagonist of animal P2 receptors in the PM, 300 μM) significantly reduced NaCl-induced Na+ flux from G. uralensis roots (Figures 6A–8A). Our NMT data showed that the addition of agonists (H2O2, SNP, and eATP) or antagonists (DMTU, cPTIO, and PPADS) had no significant effect on Na+ flux in the absence of salt stress (Figures 6A–8A).
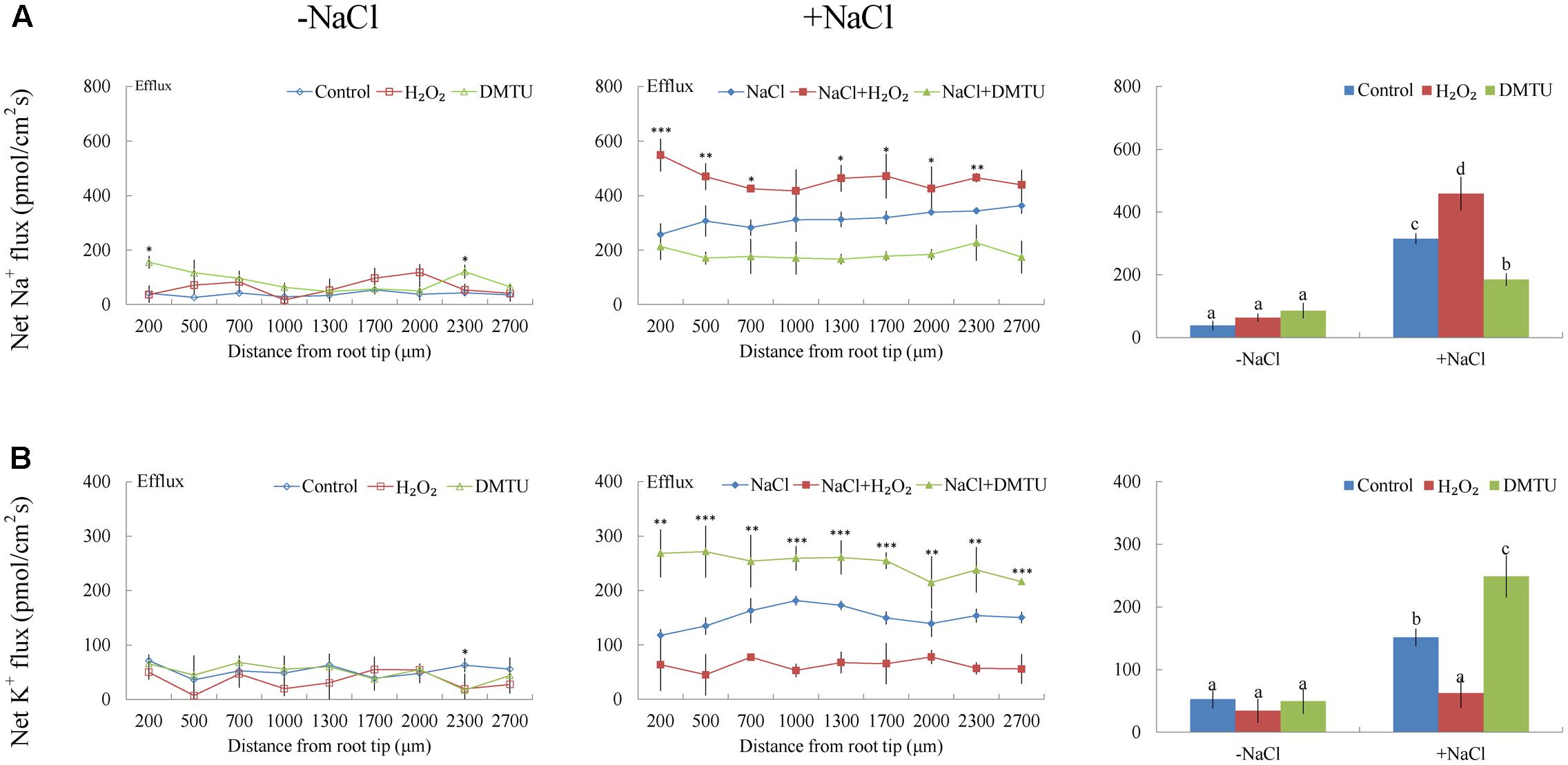
FIGURE 6. Effects of H2O2 and DMTU on Na+ and K+ fluxes in G. uralensis roots under salt stress. Roots were untreated (control, blue) or exposed to H2O2 (10 mM, red) for 24 h in the absence (–NaCl, no-salt) and presence of NaCl (100 mM). For inhibitor treatment, no-salt and NaCl-stressed roots were subjected to DMTU (5 mM, green) for 30 min. Steady-state flux profiles of (A) Na+ and (B) K+ were measured along the root axis at the apical zones (200–2700 μm from the root tip) in no-salt (left panels) and salt-stressed (center panels) conditions. Each point represents the mean of five to six individual plants. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001, compared to controls. (Right panels) Bars (±SD) represent the mean of five to six individual plants; letters (a, b, c, and d) indicate significant differences (P < 0.05) between treatments.
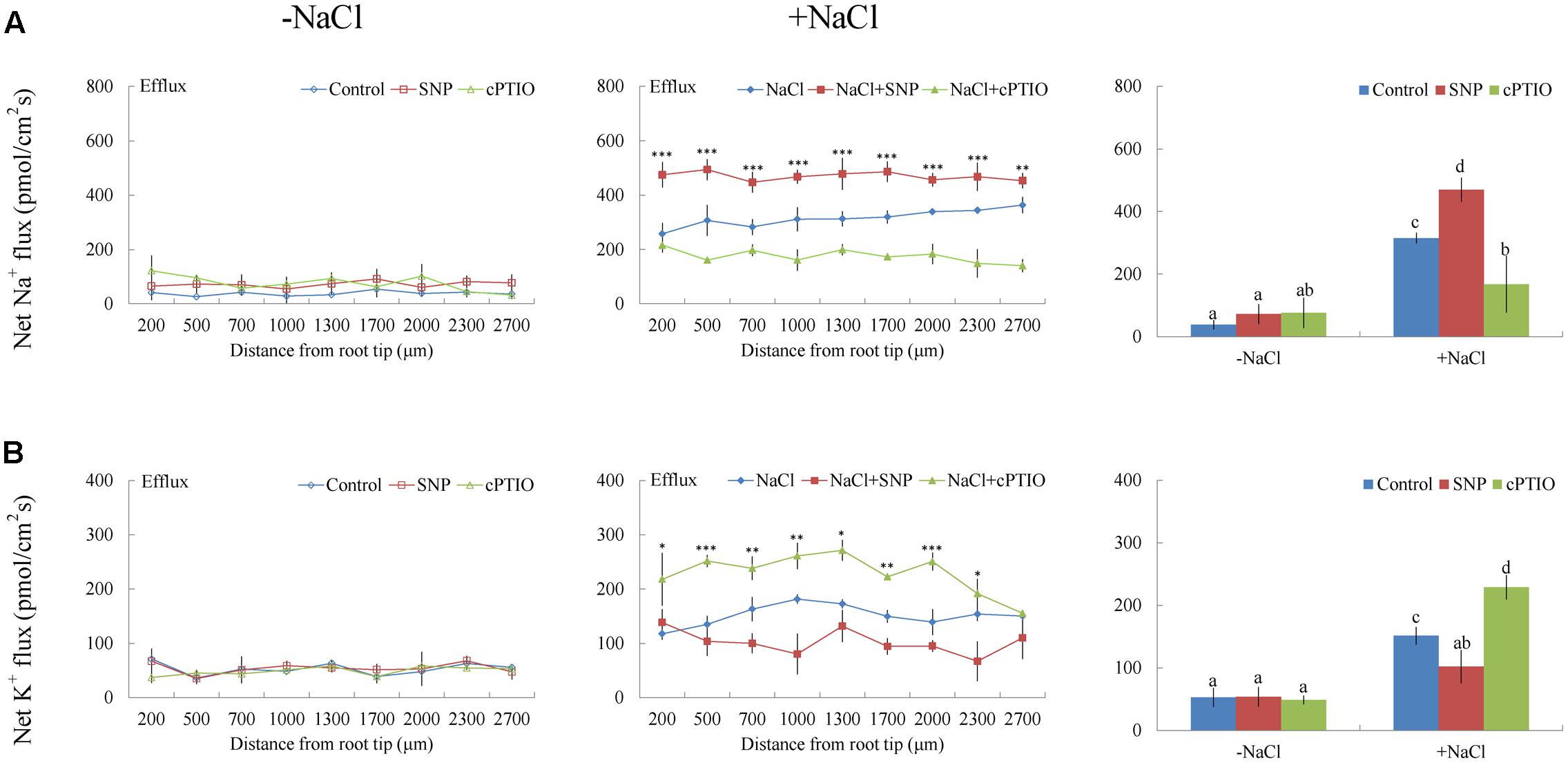
FIGURE 7. Effects of sodium nitroprusside (SNP) and cPTIO on Na+ and K+ fluxes in G. uralensis roots under salt stress. Roots were untreated (control, blue) or exposed to SNP (100 μM, red) for 24 h in the absence (–NaCl, no-salt) and presence of NaCl (100 mM). For inhibitor treatment, no-salt and NaCl-stressed roots were subjected to cPTIO (300 μM, green) for 30 min. Steady-state flux profiles of (A) Na+ and (B) K+ were measured along the root axis at the apical zones (200–2700 μm from the root tip) in no-salt (left panels) and salt-stressed (center panels) conditions. Each point represents the mean of five to six individual plants. ∗P < 0.05, ∗∗P < 0.01, ∗∗∗P < 0.001, compared to controls. (Right panels) Bars (±SD) represent the mean of five to six individual plants; letters (a, b, c, and d) indicate significant differences (P < 0.05) between treatments.
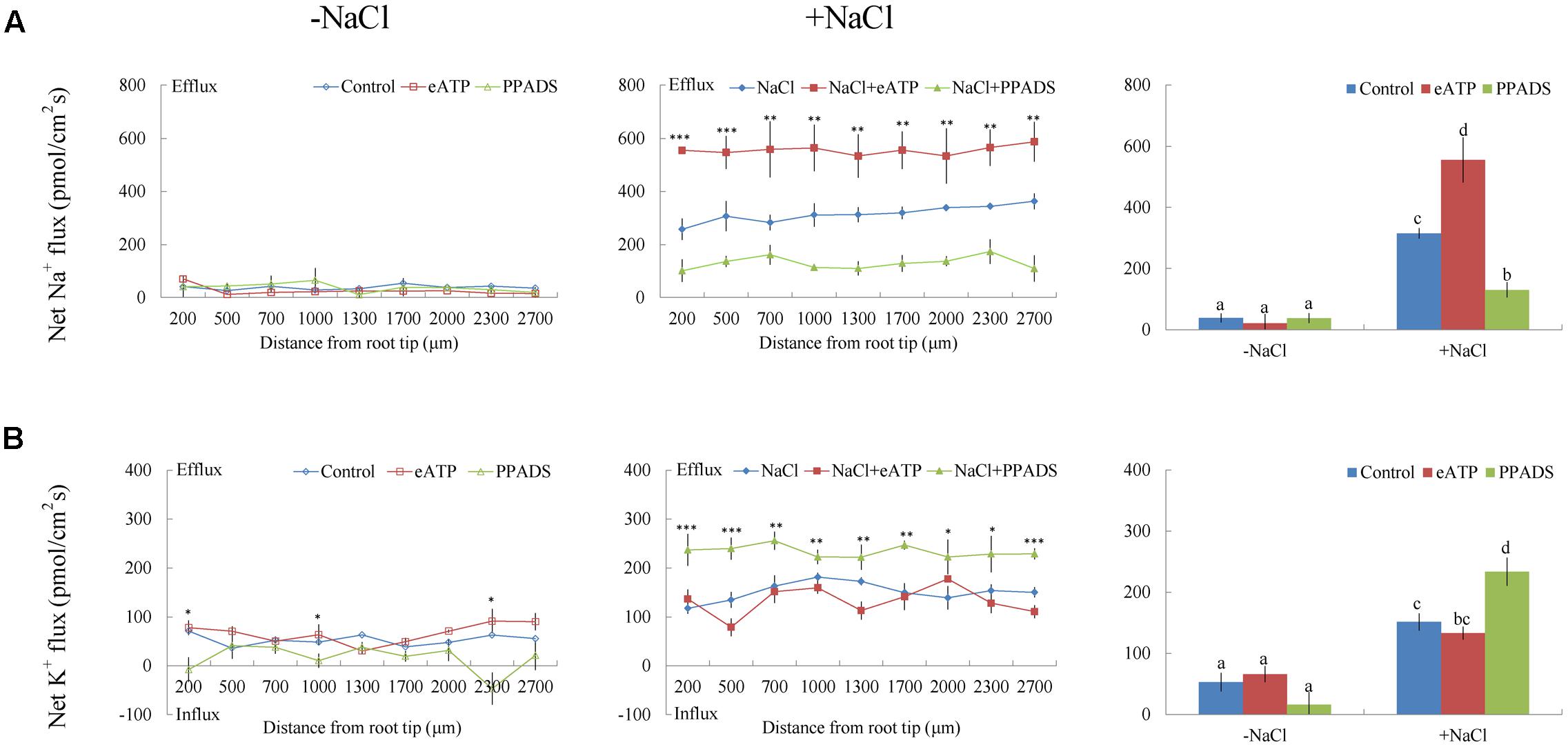
FIGURE 8. Effects of eATP and PPADS on Na+ and K+ fluxes in G. uralensis roots under salt stress. Roots were untreated (control, blue) or exposed to ATP-Na2 (300 μM, red) for 24 h in the absence (–NaCl, no-salt) and presence of NaCl (100 mM). For inhibitor treatment, no-salt and NaCl-stressed roots were subjected to PPADS (300 μM, green) for 30 min. Steady-state flux profiles of (A) Na+ and (B) K+ were measured along the root axis at the apical zones (200–2700 μm from the root tip) in no-salt (left panels) and salt-stressed (center panels) conditions. Each point represents the mean of five to six individual plants. ∗P < 0.05, ∗∗P < 0.01, and ∗∗∗P < 0.001, compared to controls. (Right panels) Bars (±SD) represent the mean of five to six individual plants; letters (a, b, c, and d) indicate significant differences (P < 0.05) between treatments.
Non-salinized G. uralensis roots displayed a stable, constant K+ efflux with a mean of 50.27 ± 8.49 pmol cm-2 s-1 (Figure 4B). Salt treatment markedly increased the K+ efflux, up to 151.35 pmol cm-2 s-1 in the measured regions (200–2700 μm from the apex) (Figure 4B). Inhibitor experiments showed that the salt-induced K+ loss was inhibited by a K+ channel blocker, TEA (Figure 5C). In contrast to TEA, sodium orthovanadate, the specific inhibitor of the PM H+-ATPase, markedly enhanced the salt-elicited K+ loss from liquorice roots (Figure 5C). This indicated that the K+ loss in salt-stressed roots was due to activation of DA-KORCs or NSCCs in the PM (Sun et al., 2009b; Zhang et al., 2015).
Of note, Ca2+, H2O2, SNP, or eATP reduced K+ efflux by 12–59% in salinized roots, although the effect H2O2 was more pronounced than that of the other agonists (Figures 4B, 6B–8B). In contrast, salt-induced K+ efflux was significantly enhanced by all the tested antagonists, LaCl3, DMTU, cPTIO, and PPADS (Figures 4B, 6B–8B). In general, none of the signaling molecules (Ca2+, H2O2, SNP, or eATP) or the inhibitors (LaCl3, DMTU, cPTIO, or PPADS) had a significant effect on K+ flux under no-salt control conditions (Figures 4B, 6B–8B).
NaCl treatment (100 mM, 24 h) induced significant increases in the expression of Na+/H+ antiport system genes, GuAHA (PM H+-ATPase gene) and GuSOS1 (PM Na+/H+ antiporter gene) (Figure 9). Interestingly, exogenously applied Ca2+, H2O2, SNP, or eATP increased the expression of GuAHA and/or GuSOS1 under NaCl stress (Figure 9). These data suggested that Ca2+, H2O2, SNP, and eATP were involved in regulating the transcription of the PM Na+/H+ antiport system. Accordingly, pharmacological data showed that the salt-elicited upregulation of GuAHA and GuSOS1 could be suppressed by DMTU, cPTIO, or PPADS (Figure 9). However, the Ca2+-channel inhibitor, LaCl3, did not block the salt-induced upregulation of GuAHA and GuSOS1 transcription (Figure 9). Moreover, we found that these salt signaling molecules and pharmacological agents had no obvious effects on gene expression in the absence of NaCl stress, with the exception of H2O2, which induced GuAHA expression in control conditions (Figure 9).
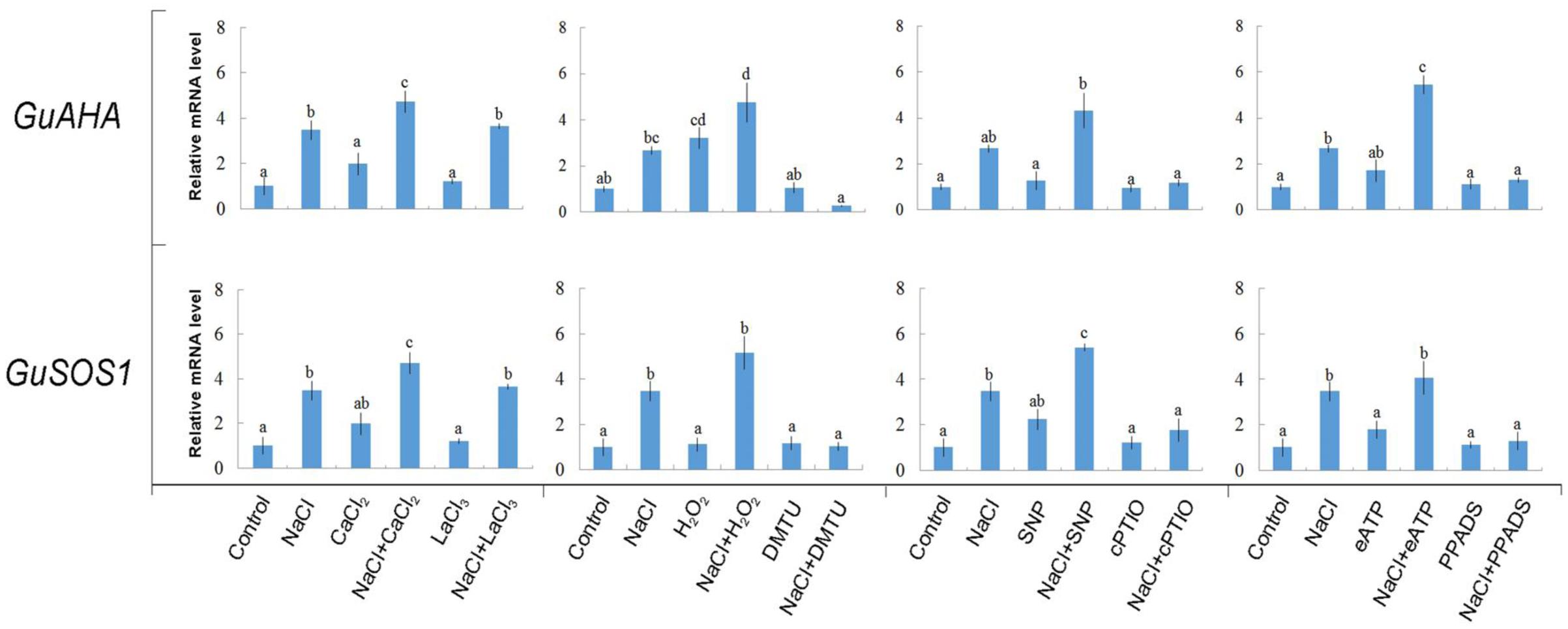
FIGURE 9. Effects of Ca2+, H2O2, SNP, eATP, and pharmacological agents on expression of GuAHA (PM H+-ATPase) and GuSOS1 (salt overly sensitive 1 Na+/H+ antiporter) in G. uralensis roots under salt stress. Roots were exposed for 24 h to 0 or 100 mM NaCl, supplemented with or without CaCl2 (10 mM), H2O2 (10 mM), SNP (a NO donor, 100 μM), or ATP-Na2 (300 μM). Then, control and NaCl-stressed roots were treated with LaCl3 (5 mM), DMTU (5 mM), cPTIO (300 μM), or PPADS (300 μM) for 30 min. Quantitative RT-PCR results show the relative transcript abundance of GuAHA and GuSOS1. GuActin2 served as the internal control for expression normalization. Forward and reverse primers for all tested genes are listed in Supplementary Table 1. Bars (±SD) represent the means of three to five individual plants; letters (a, b, c, and d) indicate significant differences between treatments (P < 0.05).
As shown in Figure 10, NaCl increased the transcription of a series of salt-responsive genes. GuSOS3 is important in Ca2+ signaling pathways (Zhu, 2001, 2003, 2016; Qiu et al., 2002; Yang et al., 2009; Ji et al., 2013); GuCIPK is important in Ca2+ signaling pathways (Xiang et al., 2007; Hu et al., 2015); GuRbohD is important in H2O2 signaling (Rejeb et al., 2015); GuNIR is important in NO signaling (Liu et al., 2007); and GuMAPK3 and GuMAPK6 are important in eATP signaling (Choi et al., 2014). We found that several signaling molecules changed the expression pattern of the selected salt-responsive genes under salt stress. For example, exposing NaCl-stressed plants to Ca2+, H2O2, or SNP enhanced transcription of GuSOS3 or GuCIPK (Figure 10). Of note, eATP produced a pronounced induction of Ca2+ signaling pathway genes; the expression levels of both GuSOS3 and GuCIPK were stimulated by eATP in NaCl-stressed roots (Figure 10). Also, GuRbohD transcription was enhanced by these signaling molecules, but Ca2+ and eATP produced more pronounced effects than H2O2 and SNP (Figure 10). GuNIR expression remained constant in NaCl-stressed roots, regardless of Ca2+, H2O2, or SNP treatment (Figure 10). However, GuNIR transcription was enhanced with eATP in salinized G. uralensis roots (Figure 10). The abundances of GuMAPK3 and/or GuMAPK6 transcripts increased in the presence of all signaling molecules, but H2O2 and eATP produced more pronounced effects on GuMAPK6 (Figure 10). We also noticed that, in general, Ca2+, H2O2, and eATP increased the expression of the tested salt-responsive genes under no-salt control conditions; in contrast, SNP had less of an effect (Figure 10).
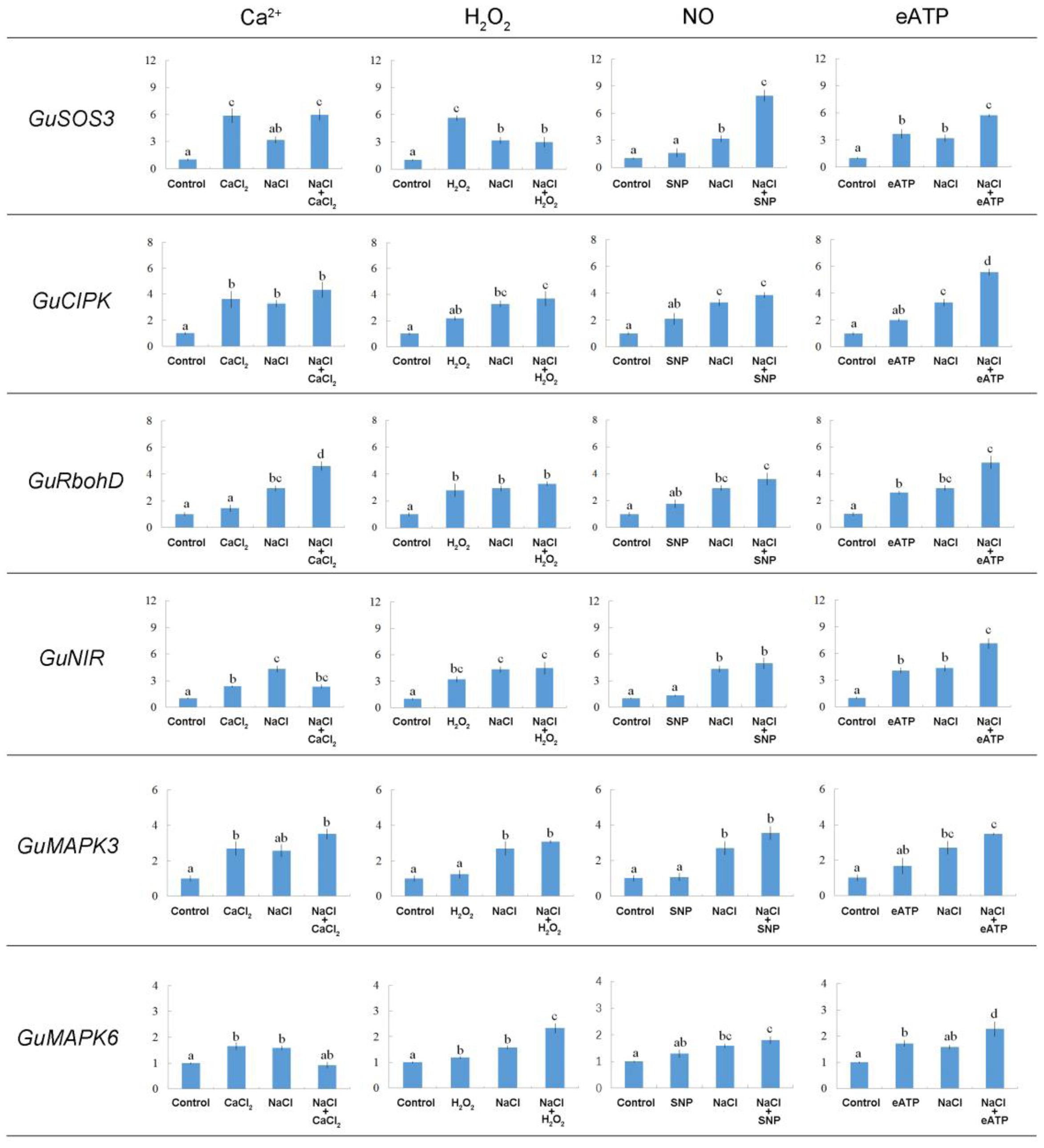
FIGURE 10. Effects of Ca2+, H2O2, SNP, and eATP on relative expression of salt-responsive genes in G. uralensis roots under salt stress. Roots were exposed for 24 h to 0 or 100 mM NaCl, supplemented with or without CaCl2 (10 mM), H2O2 (10 mM), SNP (a NO donor, 100 μM), or ATP-Na2 (300 μM). Quantitative RT-PCR results show the relative transcript abundance of homolog genes in G. uralensis, such as GuSOS3 (salt overly sensitive 3), GuCIPK (CBL-interacting protein kinase), GuRbohD (respiratory burst oxidase homolog protein D), GuNIR (nitrate reductase), GuMAPK3 (mitogen-activated protein kinase 3), and GuMAPK6 (mitogen-activated protein kinase 6). GuActin2 served as an internal control for expression normalization. Forward and reverse primers for all tested genes are listed in Supplementary Table 1. Bars (±SD) represent the means of three to five individual plants; letters (a, b, c, and d) indicate significant differences between treatments (P < 0.05).
A short period of NaCl exposure caused cellular Na+ accumulation, which was more pronounced in roots than in leaves (Figure 1). The buildup of Na+ in root cells resulted in remarkable increases in cytosolic Ca2+, H2O2, NO, and eATP (Figures 2, 3). The rapid increase of these signaling molecules indicated that G. uralensis roots could sense NaCl stress, and they set into motion a wide range of cellular processes required for salt adaptation (Chen and Polle, 2010; Chen et al., 2014; Polle and Chen, 2015). Accordingly, our NMT and RT-qPCR data revealed that eATP, Ca2+, H2O2, NO, and their interactions played crucial roles in regulating ion fluxes and gene transcription (Figures 4–10). These findings were similar to findings from our previous study in a salt-resistant poplar, P. euphratica (Sun et al., 2010a,b, 2012a).
To avoid toxicity, due to excessive Na+ in the cytosol, it is crucial for glycophyte plants to adapt to saline conditions (Shabala et al., 2005; Sun et al., 2009a,b, 2010a,b; Chen and Polle, 2010; Chen et al., 2014). The perennial species, G. uralensis, exhibited significant Na+ extrusion and a corresponding H+ uptake after exposure to 24-h NaCl treatments (Figures 4, 5). However, the salt-induced Na+ efflux and H+ influx were markedly blocked by amiloride (an inhibitor of Na+/H+ antiporters) or sodium orthovanadate (a specific inhibitor of the PM H+-ATPase) (Figure 5). These results suggested that salinized roots of G. uralensis extruded Na+ and took up H+ via the activated Na+/H+ antiport system in the PM (i.e., the H+-ATPase and Na+/H+ antiporter; Shabala et al., 2003, 2005; Sun et al., 2009a; Lu et al., 2013; Lang et al., 2014; Zhao et al., 2016). Notably, we found that the Na+ efflux was enhanced by Ca2+, H2O2, NO, and eATP (Figures 4, 6–8). Moreover, salt-induced Na+ extrusion could be reduced by pharmacological agents that blocked the pathways regulated by those molecules, i.e., LaCl3, DMTU, cPTIO, and PPADS, respectively (Figures 4, 6–8). These results indicated that the signaling molecules were required to activate the PM Na+/H+ antiport system in the presence of NaCl salinity.
Our RT-qPCR assays showed that the activated Na+/H+ antiport system in salinized roots presumably resulted from the upregulation of GuSOS1 and GuAHA genes (Figure 9). In a previous study, eATP was found to mediate the induction of PeSOS1 and PeAHA in the poplar, P. euphratica, during NaCl stress (Sun et al., 2012a; Zhang et al., 2015). Moreover, NO was found to enhance Na+ exclusion by increasing the expression of the PM H+-ATPase and Na+/H+ antiporter in a secretor mangrove, Avicennia marina, under high salinity (Chen et al., 2010). Our previous study revealed that NO most likely interacted with Ca2+ and H2O2 in Aegiceras corniculatum to up-regulate the PM Na+/H+ antiport system (Lang et al., 2014). Chung et al. (2008) found that reactive oxygen species mediated SOS1 mRNA stability in Na+-treated Arabidopsis.
In addition to our agonist findings, the pharmacological data also showed that the salt-induced transcription of GuSOS1 or GuAHA could be inhibited by DMTU, cPTIO, or PPADS in salt-stressed G. uralensis roots (Figure 9). These findings suggested that the endogenous salt-sensitive messengers, H2O2, NO, and eATP, contributed to the induction of G. uralensis Na+/H+ antiport genes during NaCl stress. However, the Ca2+-channel inhibitor, LaCl3, did not block the salt-responsive induction of GuAHA and GuSOS1 (Figure 9). This result implied that vacuolar Ca2+ release might facilitate cytosolic Ca2+ signaling in the salt response of G. uralensis (Zhang et al., 2015). Indeed, in a previous study, we showed that a vacuole-generated Ca2+ signaling pathway participated in the regulation of ionic homeostasis in NaCl-stressed P. euphratica cells (Zhang et al., 2015).
In G. uralensis roots, NaCl-induced K+ efflux was blocked by TEA (a specific inhibitor of K+ permeable channels), but enhanced by vanadate (Figure 5). These findings suggested that NaCl-induced K+ loss was mediated by depolarization-activated channels, e.g., KORCs and NSCCs (Shabala et al., 2005, 2006; Sun et al., 2009b; Lu et al., 2013; Lang et al., 2014; Zhao et al., 2016). The addition of Ca2+, H2O2, NO, and eATP reduced the salt-induced K+ efflux (Figures 4, 6–8). Presumably, this result was due to the inhibition of K+-channels by the activated PM H+-ATPase, because these signaling molecules upregulated GuAHA transcription in salinized roots (Figure 9). Previous studies have shown that NaCl-induced increases in PM H+-ATPase activity depended on H2O2 production, in P. euphratica (Zhang et al., 2007; Sun et al., 2010a,b) and in secretor and non-secretor mangroves (Lu et al., 2013; Lang et al., 2014). In A. marina leaves, NO remarkably enhanced PM H+-ATPase activity and AHA1 transcription, and conversely, these activities were reduced by NO synthesis inhibitors and NO scavengers (Chen et al., 2010). The maintenance of K+ homeostasis in P. euphratica cells was attributed to the eATP induction of AHA (Sun et al., 2012a). Moreover, in poplar cells, NaCl-induced K+ loss increased, when AHA transcription was inhibited by the glucose-hexokinase trap system or P2 receptor antagonists (suramin and PPADS) (Sun et al., 2012a; Zhao et al., 2016). In the present study, we also found that NaCl-induced K+ loss increased (Figures 4B, 6B–8B) and GuAHA expression was inhibited by the four tested antagonists, but LaCl3 produced less pronounced effects compared to DMTU, cPTIO, and PPADS (Figure 9). We concluded that salt-induced signaling molecules were required for upregulation of the PM H+-ATPase gene in G. uralensis roots. As a result, enhanced H+ pumping activity, on one hand, reduced K+ loss via depolarization-activated channels, and on the other hand, promoted Na+ extrusion via PM Na+/H+ antiporters (Chen and Polle, 2010; Chen et al., 2014; Polle and Chen, 2015).
Clear interactions occurred between these stress signals to accelerate the transcription of salt-adaptive signaling pathway genes in G. uralensis roots. Ca2+ increased the GuSOS3 expression (Figure 10), thus leading to enhanced Na+ extrusion via the SOS-signaling pathway (Zhu, 2001). In NaCl-treated roots of G. uralensis, H2O2, NO, or eATP, promoted the transcription of GuSOS3/GuCIPK (Figure 10), which indicated that these stress signals predominantly activated the Ca2+-SOS signaling pathway. A previous study in P. euphratica cells showed that exogenously applied H2O2 increased Ca2+ influx, which led to elevated cytosolic Ca2+ (Sun et al., 2010b). Based on our present results in G. uralensis roots, we suggest that H2O2 increased cytosolic Ca2+, which then mediated PM Na+/H+ antiport upregulation via the SOS-signaling pathway (Zhu, 2001, 2016). Furthermore, we found that NO enhanced the transcription of GuSOS3/GuCIPK in NaCl-stressed liquorice roots (Figure 10). Thus, NO-simulated Ca2+-SOS signaling would promote Na+ efflux and alleviate cellular Na+ toxicity in G. uralensis. Similarly, in the secretor mangrove, A. corniculatum, NO enhanced Na+ efflux elicited by Ca2+ (Lang et al., 2014).
Extracellular ATP signaling is a novel player in salt-stress acclimation. We found that eATP increased the expression of GuSOS3, GuCIPK, GuRbohD, GuNIR, GuMAPK3, and GuMAPK6 (Figure 10). Moreover, GuMAPK3 and GuMAPK6 expression levels were enhanced by H2O2 in salinized G. uralensis roots (Figure 10). This indicated that eATP interacted with H2O2 and Ca2+ signaling to maintain K+/Na+ homeostasis. Previously, eATP was shown to interact with H2O2 and Ca2+ to increase Na+ extrusion in two mangrove species, Kandelia obovata and A. corniculatum (Lang et al., 2014). In P. euphratica cells, eATP signaling was mediated by H2O2 and cytosolic Ca2+ in the salt response (Sun et al., 2012a). Accordingly, eATP is thought to bind P2-like receptors in the PM (Choi et al., 2014), which leads to an increase in H2O2 and a transient elevation in cytosolic Ca2+ (Jeter et al., 2004; Demidchik et al., 2009; Sun et al., 2012a). Thus, eATP could initiate the H2O2 and Ca2+ signaling cascades and cause an increase in Na+/H+ exchange across the PM of G. uralensis roots under NaCl stress.
Extracellular ATP also increased the expression of GuNIR (Figure 10). This finding indicated that NO was a downstream component of eATP signaling. Similarly, in P. euphratica cells, NO was triggered by eATP, although NO played a negligible role in eATP-stimulated cell death (Sun et al., 2012b). There are species–specific interactions between eATP and NO in the mediation of K+/Na+ homeostasis (Lang et al., 2014). In this study, eATP signaling appeared to be mediated by NO in G. uralensis roots (Figure 10). However, in the non-secretor, K. obovata, NO was redundant in the presence of eATP, because eATP alone exerted a pronounced effect on Na+/H+ antiporters (Lang et al., 2014).
Our findings suggested that salt exposure increased Ca2+, H2O2, NO, and eATP, which served as signaling molecules in mediating K+/Na+ balance by elevating Na+ efflux and restraining K+ loss in G. uralensis. Based on these results, we proposed a multiple signaling network for regulating ionic homeostasis in salinized G. uralensis (Figure 11). The NaCl-induced signaling molecules, Ca2+, H2O2, NO, and eATP, upregulated GuSOS1 and GuAHA expression, which increased the numbers of Na+/H+ antiporters and H+ pumps in the PM. The enhanced Na+/H+ antiport system promoted the SOS-signaling pathway. In addition, H+-pump activity preserved the membrane potential, which restricted K+ efflux through DA-KORCs and DA-NSCCs. Interestingly, we also found interactions between these stress signaling molecules and the expression of salt-responsive genes in G. uralensis roots. Ca2+, H2O2, NO, and eATP enhanced GuSOS3/GuCIPK genes, which are related to the Ca2+-SOS signaling pathway. Moreover, eATP exhibited novel interactions with Ca2+, H2O2, and NO signaling, which contributed to the upregulation of GuSOS3, GuCIPK, GuRbohD, and GuNIR. This crosstalk was thought to contribute to the upregulation of GuSOS1 and GuAHA expression in G. uralensis roots. Further investigations are needed to confirm these interactions.
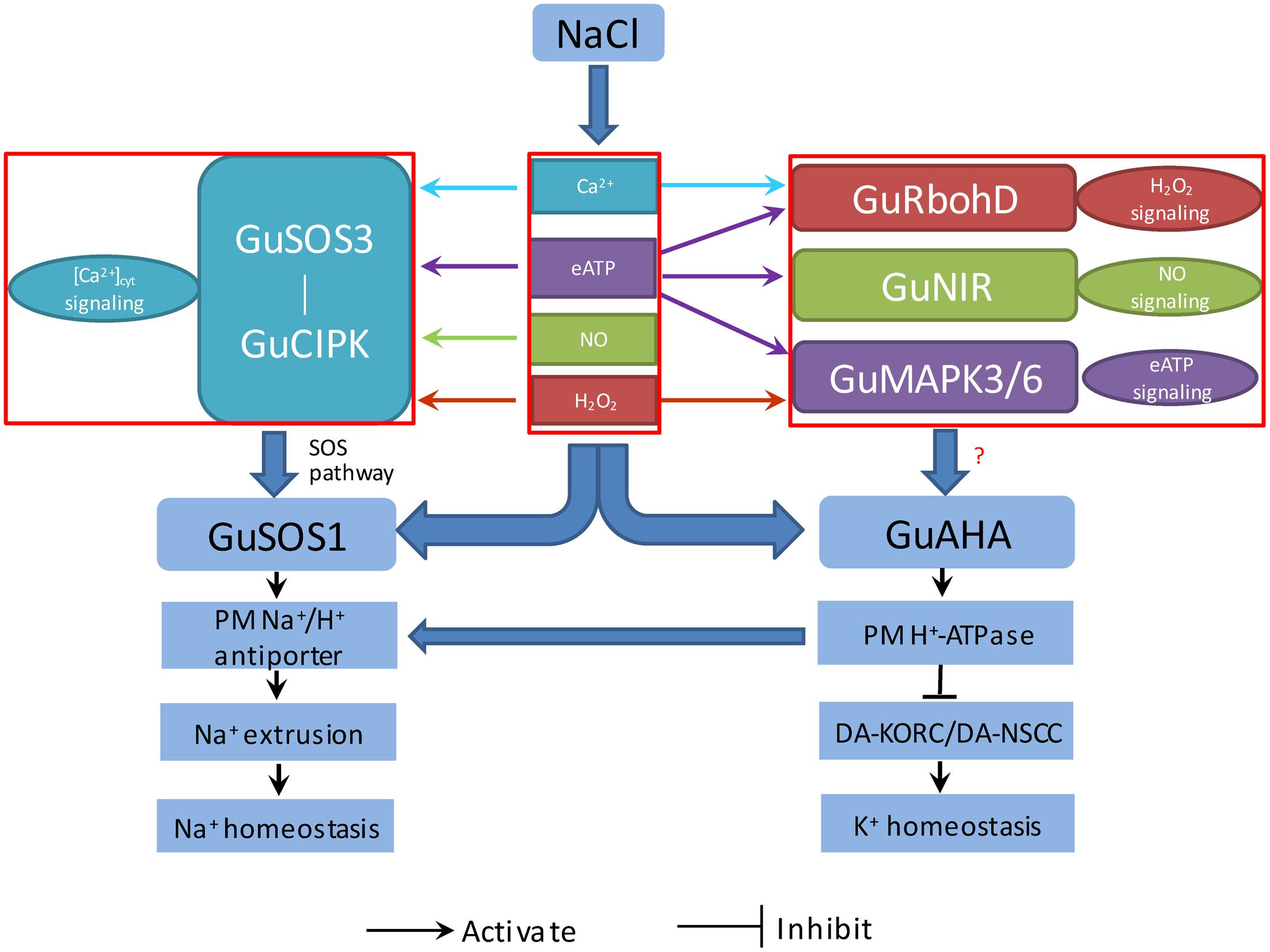
FIGURE 11. Proposed model of the signaling network (eATP, Ca2+, H2O2, and NO), which regulates salt-responsive genes, and their relationships to K+/Na+ homeostasis in G. uralensis roots. PM, plasma membrane; G. uralensis genes, GuAHA: PM H+-ATPase; GuSOS1, salt overly sensitive 1; GuSOS3, salt overly sensitive 3; GuCIPK, CBL-interacting protein kinase; GuRbohD, respiratory burst oxidase homolog protein D; GuNIR, nitrate reductase; GuMAPK3, mitogen-activated protein kinase 3; GuMAPK6, mitogen-activated protein kinase 6; Ion transporters: DA-KORCs, Depolarization-activated K+ outward rectifying channels; DA-NSCCs, Depolarization-activated non-selective cation channels.
TL, JX, and SC conceived of the original screening and research plans; SC supervised the experiments; TL, SD, NZ, CD, YnZ, YlZ, HZ, GS, and JY performed most of the experiments; CW, YW, QD, and SL provided technical assistance to TL, SD, and NZ; TL designed the experiments and analyzed the data; TL conceived of the project and wrote the article, with contributions from all the authors; SC supervised and complemented the writing. All authors have read and approved the manuscript.
The research was supported jointly by the National Science-technology Support Plan Projects of China (2014BAC15B04), the National Natural Science Foundation of China (grant nos. 31570587 and 31270654), the Research Project of the Chinese Ministry of Education (grant no. 113013A), the Key Project for Oversea Scholars by the Ministry of Human Resources and Social Security of PR China (grant no. 2012001), the Program for Changjiang Scholars and Innovative Research Teams in University (grant no. IRT_17R08), and the Program of Introducing Talents of Discipline to Universities (111 Project, grant no. B13007).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01403/full#supplementary-material
Blumwald, E., Aharon, G. S., and Apse, M. P. (2000). Sodium transport in plant cells. Biochim. Biophys. Acta 1465, 140–151. doi: 10.1016/S0005-2736(00)00135-8
Chen, J., Xiao, Q., Wu, F. H., Dong, X. J., He, J. X., Pei, Z. M., et al. (2010). Nitric oxide enhances salt secretion and Na+ sequestration in a mangrove plant, Avicennia marina, through increasing the expression of H+-ATPase and Na+/H+ antiporter under high salinity. Tree Physiol. 30, 1570–1585. doi: 10.1093/treephys/tpq086
Chen, J., Xiong, D. Y., Wang, W. H., Hu, W. J., Simon, M., Xiao, Q., et al. (2013). Nitric oxide mediates root K+/Na+ balance in a mangrove plant, Kandelia obovata, by enhancing the expression of AKT1-type K+ channel and Na+/H+ antiporter under high salinity. PLoS ONE 8:e71543. doi: 10.1371/journal.pone.0071543
Chen, S. L., Hawighorst, P., Sun, J., and Polle, A. (2014). Salt tolerance in Populus: significance of stress signaling networks, mycorrhization, and soil amendments for cellular and whole-plant nutrition. Environ. Exp. Bot. 107, 113–124. doi: 10.1016/j.envexpbot.2014.06.001
Chen, S. L., and Polle, A. (2010). Salinity tolerance of Populus. Plant Biol. 12, 317–333. doi: 10.1111/j.1438-8677.2009.00301.x
Chen, Z. H., Pottosin, I. I., Cuin, T. A., Fuglsang, A. T., Tester, M., Jha, D., et al. (2007). Root plasma membrane transporters controlling K+/Na+ homeostasis in salt stressed barley. Plant Physiol. 145, 1714–1725. doi: 10.1104/pp.107.110262
Choi, J., Tanaka, K., Cao, Y., Qi, Y., Qiu, J., Lang, Y., et al. (2014). Identification of a plant receptor for extracellular ATP. Science 343, 290–294. doi: 10.1126/science.1246609
Chung, J. S., Zhu, J. K., Bressan, R. A., Hasegawa, P. M., and Shi, H. Z. (2008). Reactive oxygen species mediate Na+-induced SOS1 mRNA stability in Arabidopsis. Plant J. 53, 554–565. doi: 10.1111/j.1365-313X.2007.03364.x
Cuin, T. A., Bose, J., Stefano, G., Jha, D., Tester, M., Mancuso, S., et al. (2011). Assessing the role of root plasma membrane and tonoplast Na+/H+ exchangers in salinity tolerance in wheat: in planta quantification methods. Plant Cell Environ. 34, 947–961. doi: 10.1111/j.1365-3040.2011.02296.x
Demidchik, V., Shang, Z., Shin, R., Tompson, E., Rubio, L., Laohavisit, A., et al. (2009). Plant extracellular ATP signalling by plasma membrane NADPH oxidase and Ca2+ channels. Plant J. 58, 903–913. doi: 10.1111/j.1365-313X.2009.03830.x
Deng, S. R., Sun, J., Zhao, R., Ding, M. Q., Zhang, Y. N., Sun, Y. L., et al. (2015). Populus euphratica APYRASE2 enhances cold tolerance by modulating vesicular trafficking and extracellular ATP in Arabidopsis plants. Plant Physiol. 169, 530–548. doi: 10.1104/pp.15.00581
Ding, M. Q., Hou, P. C., Shen, X., Wang, M. J., Deng, S. R., Sun, J., et al. (2010). Salt-induced expression of genes related to Na+/K+ and ROS homeostasis in leaves of salt-resistant and salt-sensitive poplar species. Plant Mol. Biol. 73, 251–269. doi: 10.1007/s11103-010-9612-9
Hu, D. G., Ma, Q. J., Sun, C. H., Sun, M. H., You, C. X., and Hao, Y. J. (2015). Overexpression of MdSOS2L1, a CIPK protein kinase, increases the antioxidant metabolites to enhance salt tolerance in apple and tomato. Physiol. Plant. 156, 201–214. doi: 10.1111/ppl.12354
Jeter, C. R., Tang, W., Henaff, E., Butterfield, T., and Roux, S. J. (2004). Evidence of a novelcell signaling role for extracellular adenosine triphosphates and diphosphates in Arabidopsis. Plant Cell 16, 2652–2664. doi: 10.1105/tpc.104.023945
Ji, H. T., Pardo, J. M., Batelli, G., Van Oosten, M. J., Bressan, R. A., and Li, X. (2013). The salt overly sensitive (SOS) pathway: established and emerging roles. Mol. Plant 6, 275–286. doi: 10.1093/mp/sst017
Kim, S. H., Yang, S. H., Kim, T. J., Han, J. S., and Suh, J. W. (2009). Hypertonic stress increased extracellular ATP levels and the expression of stress responsive genes in Arabidopsis thaliana seedlings. Biosci. Biotechnol. Biochem. 73, 1252–1256. doi: 10.1271/bbb.80660
Lang, T., Sun, H. M., Li, N. Y., Lu, Y. J., Shen, Z. D., Jing, X. S., et al. (2014). Multiple signaling networks of extracellular ATP, hydrogen peroxide, calcium, and nitric oxide in the mediation of root ion fluxes in secretor and non-secretor mangroves under salt stress. Aquat. Bot. 119, 33–43. doi: 10.1016/j.aquabot.2014.06.009
Li, J., Bao, S., Zhang, Y., Ma, X., Mishra-Knyrim, M., Sun, J., et al. (2012). Paxillus involutus strains MAJ and NAU mediate K+/Na+ homeostasis in ectomycorrhizal Populus × canescens under sodium chloride stress. Plant Physiol. 159, 1771–1786. doi: 10.1104/pp.112.195370
Li, Y., Zhang, W., Cui, J., Lang, D., Li, M., Zhao, Q., et al. (2016). Silicon nutrition alleviates the lipid peroxidation and ion imbalance of Glycyrrhiza uralensis seedlings under salt stress. Acta Physiol. Plant. 38:96. doi: 10.1007/s11738-016-2108-8
Liu, Y. G., Wu, R. R., Wan, Q., Xie, G. Q., and Bi, Y. R. (2007). Glucose-6-phosphate dehydrogenase plays a pivotal role in nitric oxide-involved defense against oxidative stress under salt stress in red kidney bean roots. Plant Cell Physiol. 48, 511–522. doi: 10.1093/pcp/pcm020
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT. Methods 24, 405–408. doi: 10.1006/meth.2001.1262
Lu, Y. J., Li, N. Y., Sun, J., Hou, P. C., Jing, X. S., Deng, S. R., et al. (2013). Exogenous hydrogen peroxide, nitric oxide and calcium mediate root ion fluxes in two non-secretor mangrove species subjected to NaCl stress. Tree Physiol. 33, 81–95. doi: 10.1093/treephys/tps119
Martínez-Atienza, J., Jiang, X. Y., Garciadeblas, B., Mendoza, I., Zhu, J. K., Pardo, J. M., et al. (2007). Conservation of the salt overly sensitive pathway in rice. Plant Physiol. 143, 1001–1012. doi: 10.1104/pp.106.092635
Mochida, K., Sakurai, T., Seki, H., Yoshida, T., Takahagi, K., Sawai, S., et al. (2017). Draft genome assembly and annotation of Glycyrrhiza uralensis, a medicinal legume. Plant J. 89, 181–194. doi: 10.1111/tpj.13385
Mori, I. C., and Schroeder, J. I. (2004). Reactive oxygen species activation of plant Ca2+ channels. A signaling mechanism in polar growth, hormone transduction, stress signaling, and hypothetically mechanotransduction. Plant Physiol. 135, 702–708. doi: 10.1104/pp.104.042069
Munns, R., and Tester, M. (2008). Mechanisms of salinity tolerance. Annu. Rev. Plant Biol. 59, 651–681. doi: 10.1146/annurev.arplant.59.032607.092911
Pei, Z. M., Murata, Y., Benning, G., Thomine, S., Klüsener, B., Allen, G. J., et al. (2000). Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 406, 731–734. doi: 10.1038/35021067
Polle, A., and Chen, S. L. (2015). On the salty side of life: molecular, physiological and anatomical adaptation and acclimation of trees to extreme habitats. Plant Cell Environ. 38, 1794–1816. doi: 10.1111/pce.12440
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., and Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441. doi: 10.1073/pnas.122224699
Rejeb, K. B., Vos, D. L. D., Disquet, I. L., Leprince, A. S., Bordenave, M., Maldiney, R., et al. (2015). Hydrogen peroxide produced by NADPH oxidases increases proline accumulation during salt or mannitol stress in Arabidopsis thaliana. New Phytol. 208, 1138–1148. doi: 10.1111/nph.13550
Shabala, L., Cuin, T. A., Newman, I. A., and Shabala, S. (2005). Salinity-induced ion flux patterns from the excised roots of Arabidopsis sos mutants. Planta 222, 1041–1050. doi: 10.1007/s00425-005-0074-2
Shabala, S., Demidchik, V., Shabala, L., Cuin, T. A., Smith, S. J., Miller, A. J., et al. (2006). Extracellular Ca2+ ameliorates NaCl-induced K+ loss from Arabidopsis root and leaf cells by controlling plasma membrane K+ permeable channels. Plant Physiol. 141, 1653–1665. doi: 10.1104/pp.106.082388
Shabala, S., Shabala, L., and Volkenburgh, E. V. (2003). Effect of calcium on root development and root ion fluxes in salinised barley seedlings. Funct. Plant Biol. 30, 507–514. doi: 10.1071/FP03016/1445-4408/03/050507
Sueldo, D. J., Foresi, N. P., Casalongue, C. A., Lamattina, L., and Laxalt, A. M. (2010). Phosphatidic acid formation is required for extracellular ATP-mediated nitric oxide production in suspension-cultured tomato cells. New Phytol. 185, 909–916. doi: 10.1111/j.1469-8137.2009.03165.x
Sun, J., Chen, S. L., Dai, S. X., Wang, R. G., Li, N. Y., Shen, X., et al. (2009a). NaCl-induced alternations of cellular and tissue ion fluxes in roots of salt-resistant and salt-sensitive poplar species. Plant Physiol. 149, 1141–1153. doi: 10.1104/pp.108.129494
Sun, J., Dai, S. X., Wang, R. G., Chen, S. L., Li, N. Y., Zhou, X. Y., et al. (2009b). Calcium mediates root K+/Na+ homeostasis in poplar species differing in salt tolerance. Tree Physiol. 29, 1175–1186. doi: 10.1093/treephys/tpp048
Sun, J., Li, L., Liu, M., Wang, M., Ding, M., Deng, S., et al. (2010a). Hydrogen peroxide and nitric oxide mediate K+/Na+ homeostasis and antioxidant defense in NaCl stressed callus cells of two contrasting poplars. Plant Cell Tissue Org. Cult. 103, 205–215. doi: 10.1007/s11240-010-9768-7
Sun, J., Wang, M. J., Ding, M. Q., Deng, S. R., Liu, M. Q., Lu, C. F., et al. (2010b). H2O2 and cytosolic Ca2+ signals triggered by the PM H+-coupled transport system mediate K+/Na+ homeostasis in NaCl-stressed Populus euphratica cells. Plant Cell Environ. 33, 943–958. doi: 10.1111/j.1365-3040.2010.02118.x
Sun, J., Zhang, C.-L., Deng, S.-R., Lu, C.-F., Shen, X., Zhou, X.-Y., et al. (2012b). An ATP signalling pathway in plant cells: extracellular ATP triggers programmed cell death in Populus euphratica. Plant Cell Environ. 35, 893–916. doi: 10.1111/j.1365-3040.2011.02461.x
Sun, J., Zhang, X., Deng, S., Zhang, C. L., Wang, M. J., Ding, M. Q., et al. (2012a). Extracellular ATP signaling is mediated by H2O2 and cytosolic Ca2+ in the salt response of Populus euphratica cells. PLoS ONE 7:e53136. doi: 10.1371/journal.pone.0053136
Tang, R. J., Liu, H., Bao, Y., Lv, Q. D., Yang, L., and Zhang, H. X. (2010). The woody plant poplar has a functionally conserved salt overly sensitive pathway in response to salinity stress. Plant Mol. Biol. 74, 367–380. doi: 10.1007/s11103-010-9680-x
Xiang, Y., Huang, Y., and Xiong, L. Z. (2007). Characterization of stress-responsive CIPK genes in rice for stress tolerance improvement. Plant Physiol. 144, 1416–1428. doi: 10.1104/pp.107.101295
Yang, Q., Chen, Z. Z., Zhou, X. F., Yin, H. B., Li, X., Xin, X. F., et al. (2009). Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol. Plant 2, 22–31. doi: 10.1093/mp/ssn058
Zhang, F., Wang, Y. P., Yang, Y. L., Wu, H., Wang, D., Liu, J. Q., et al. (2007). Involvement of hydrogen peroxide and nitric oxide in salt resistance in the calluses from Populus euphratica. Plant Cell Environ. 30, 775–785. doi: 10.1111/j.1365-3040.2007.01667.x
Zhang, Q., and Ye, M. (2009). Chemical analysis of the Chinese herbal medicine Gan-Cao (licorice). J. Chromatogr. A. 1216, 1954–1969. doi: 10.1016/j.chroma.2008.07.072
Zhang, X., Shen, Z. D., Sun, J., Yu, Y. C., Deng, S. R., Li, Z. Y., et al. (2015). NaCl-elicited, vacuolar Ca2+ release facilitates prolonged cytosolic Ca2+ signaling in the salt response of Populus euphratica cells. Cell Calcium 57, 348–365. doi: 10.1016/j.ceca.2015.03.0010
Zhang, Y. Y., Wang, L. L., Liu, Y. L., Zhang, Q., Wei, Q. P., and Zhang, W. H. (2006). Nitric oxide enhances salt tolerance in maize seedlings through increasing activities of proton-pump and Na+/H+ antiport in the tonoplast. Planta 224, 545–555. doi: 10.1007/s00425-006-0242-z
Zhao, N., Wang, S., Ma, X., Zhu, H., Sa, G., Sun, J., et al. (2016). Extracellular ATP mediates cellular K+/Na+ homeostasis in two contrasting poplar species under NaCl stress. Trees 30, 825–837. doi: 10.1007/s00468-015-1324-y
Zhu, J. K. (2001). Plant salt tolerance. Trends Plant Sci. 6, 66–71. doi: 10.1016/S1360-1385(00)01838-0
Zhu, J. K. (2003). Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 6, 441–445. doi: 10.1016/S1369-5266(03)00085-2
Keywords: liquorice, ion flux, eATP, H2O2, NO, NaCl, NMT, RT-qPCR
Citation: Lang T, Deng S, Zhao N, Deng C, Zhang Y, Zhang Y, Zhang H, Sa G, Yao J, Wu C, Wu Y, Deng Q, Lin S, Xia J and Chen S (2017) Salt-Sensitive Signaling Networks in the Mediation of K+/Na+ Homeostasis Gene Expression in Glycyrrhiza uralensis Roots. Front. Plant Sci. 8:1403. doi: 10.3389/fpls.2017.01403
Received: 06 April 2017; Accepted: 27 July 2017;
Published: 14 August 2017.
Edited by:
Sergey Shabala, University of Tasmania, AustraliaReviewed by:
Zhong-Hua Chen, Western Sydney University, AustraliaCopyright © 2017 Lang, Deng, Zhao, Deng, Zhang, Zhang, Zhang, Sa, Yao, Wu, Wu, Deng, Lin, Xia and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianxin Xia, anh4aWFAdmlwLnNpbmEuY29t Shaoliang Chen, bHNjaGVuQGJqZnUuZWR1LmNu
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.