- 1College of Crop Science, Fujian Agriculture and Forestry University, Fuzhou, China
- 2Department of Botany, Government College University, Faisalabad, Pakistan
- 3Department of Botany, Government College Women University, Faisalabad, Pakistan
- 4Department of Botany, Government College Women University, Sialkot, Pakistan
- 5Institute of Pure and Applied Biology, Bahauddin Zakariya University, Multan, Pakistan
- 6Division of Science & Technology, Department of Botany, University of Education, Lahore, Pakistan
- 7Department of Botany, University of Lahore, Sargodha, Pakistan
- 8State Key Laboratory of Grassland Agro-Ecosystems, School of Life Science, Lanzhou University, Lanzhou, China
- 9National Education Minister, Key Laboratory of Plant Genetic Improvement and Comprehensive Utilization, Fujian Agriculture and Forestry University, Fuzhou, China
Plants respond to cold stress by modulating biochemical pathways and array of molecular events. Plant morphology is also affected by the onset of cold conditions culminating at repression in growth as well as yield reduction. As a preventive measure, cascades of complex signal transduction pathways are employed that permit plants to endure freezing or chilling periods. The signaling pathways and related events are regulated by the plant hormonal activity. Recent investigations have provided a prospective understanding about plant response to cold stress by means of developmental pathways e.g., moderate growth involved in cold tolerance. Cold acclimation assays and bioinformatics analyses have revealed the role of potential transcription factors and expression of genes like CBF, COR in response to low temperature stress. Capsella bursa-pastoris is a considerable model plant system for evolutionary and developmental studies. On different occasions it has been proved that C. bursa-pastoris is more capable of tolerating cold than A. thaliana. But, the mechanism for enhanced low or freezing temperature tolerance is still not clear and demands intensive research. Additionally, identification and validation of cold responsive genes in this candidate plant species is imperative for plant stress physiology and molecular breeding studies to improve cold tolerance in crops. We have analyzed the role of different genes and hormones in regulating plant cold resistance with special reference to C. bursa-pastoris. Review of collected data displays potential ability of Capsella as model plant for improvement in cold stress regulation. Information is summarized on cold stress signaling by hormonal control which highlights the substantial achievements and designate gaps that still happen in our understanding.
Introduction
Environmental stresses hamper seed germination, plant growth, development and productivity (Chinnusamy et al., 2006; Zaynab et al., 2017). During chilling or freezing stress, plants adjust a repertoire of metabolic pathways to tolerate the conditions (Xin and Browse, 2000; Tang et al., 2006). Temperate plants acquire low temperature tolerance by means of cold acclimation (Wang et al., 2004). During cold acclimation process, precise regulation of various genes i.e., cold-regulated (COR) and transcription factors (TFs) is carried out (Gong et al., 2002; Zhou et al., 2014). In the recent years, many reports have supplemented the knowledge regarding different genes and their associated machinery for cold stress tolerance (Barrero-Gil and Salinas, 2013; Peng et al., 2015). Several transcriptional, post-transcriptional, and post-translational regulators have been recognized for chilling or freezing temperature-induced expression of COR genes (Lin et al., 2016).
Changes in membrane fluidity are vital contributors in response to temperature fluctuations (Ruelland et al., 2009). It is considered that cold stimulus is transduced by unspecified modes to the nucleus. The degree of cold tolerance in plants can be linked to their differential capabilities for acclimation to cold (Knight and Knight, 2012). In angiosperms, the C-repeat binding factors (CBF), dehydration responsive element (DRE) (Noman et al., 2016) or cold responsive genes (COR) are key players in cold acclimation (Yamaguchi-Shinozaki and Shinozaki, 1994). Plant growth inhibition during cold stress is a commonly observed phenomenon. But the signals for this growth retardation are mostly unknown and mechanisms involved are largely unexplored (Zhou et al., 2014).
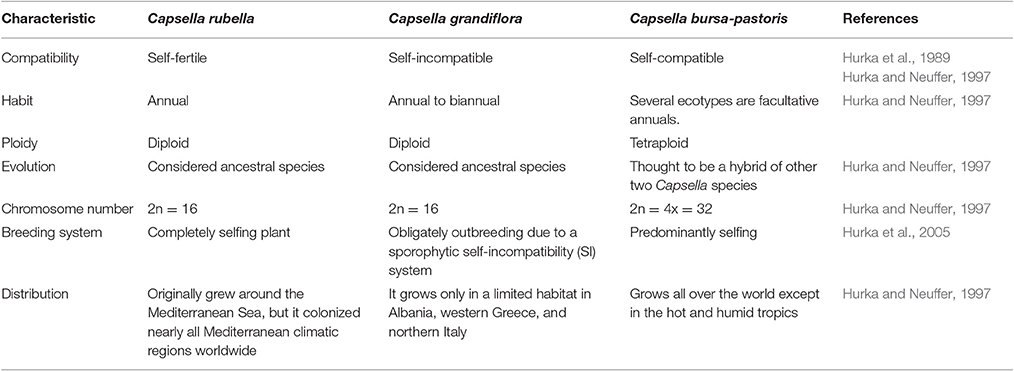
Other than generally used model plants such as Arabidopsis and rice, the addition of further plants in stress biology research would offer new systems of molecular investigations. This can provide novel insights into the genomics and genetic engineering. The genus Capsella is closely related to Arabidopsis and has three species (Table 1). Capsella bursa-pastoris is well adapted to varied environmental conditions particularly low temperatures (Ceplitis et al., 2005). This plant can grow and set seeds normally at low temperatures, suggesting that it has a strong cold-acclimation pathway. Capsella possesses strongly ability of tolerating cold by modulating its metabolism and accumulation of numerous cold prompted transcripts (Wang et al., 2004; Lin et al., 2007). The expressional characterization of different genes and their subsequent products from Capsella has presented this plant as a model to study plant resistance to low temperature (Zhou et al., 2016). The mechanism for high cold resistance has yet not been clearly understood and requires exhaustive study. Identification and validation of C repeat binding factors (CBF), COR (cold regulated) genes and other signaling components in C. bursa-pastoris has a non-conventional and broad range perspective in the fields of plant stress physiology and crop breeding for improving tolerance to low temperature.
Because of close affiliation between Arabidopsis and Capsella (Table 2), abundant experimental strategies are available and additional are being developed. The resembling gene orientation and sequences between these two plants will escalate identification of genes and expose novel regulatory, dogmatic and evolutionary modes through inter-species comparison. However, we still need more information related to different transcriptional regulators and networks involved in cold acclimation. Keeping in view the immense significance of cold tolerant Capsella plant, we tried to sum up topical research in cold stress responsive elements and associated pathways. This review highlights the prospective research and substantial functioning of crucial components for low temperature tolerance in C. bursa-pastoris. Moreover, we discussed the role of plant growth regulators as key players in determining plant responses to low or freezing temperature.
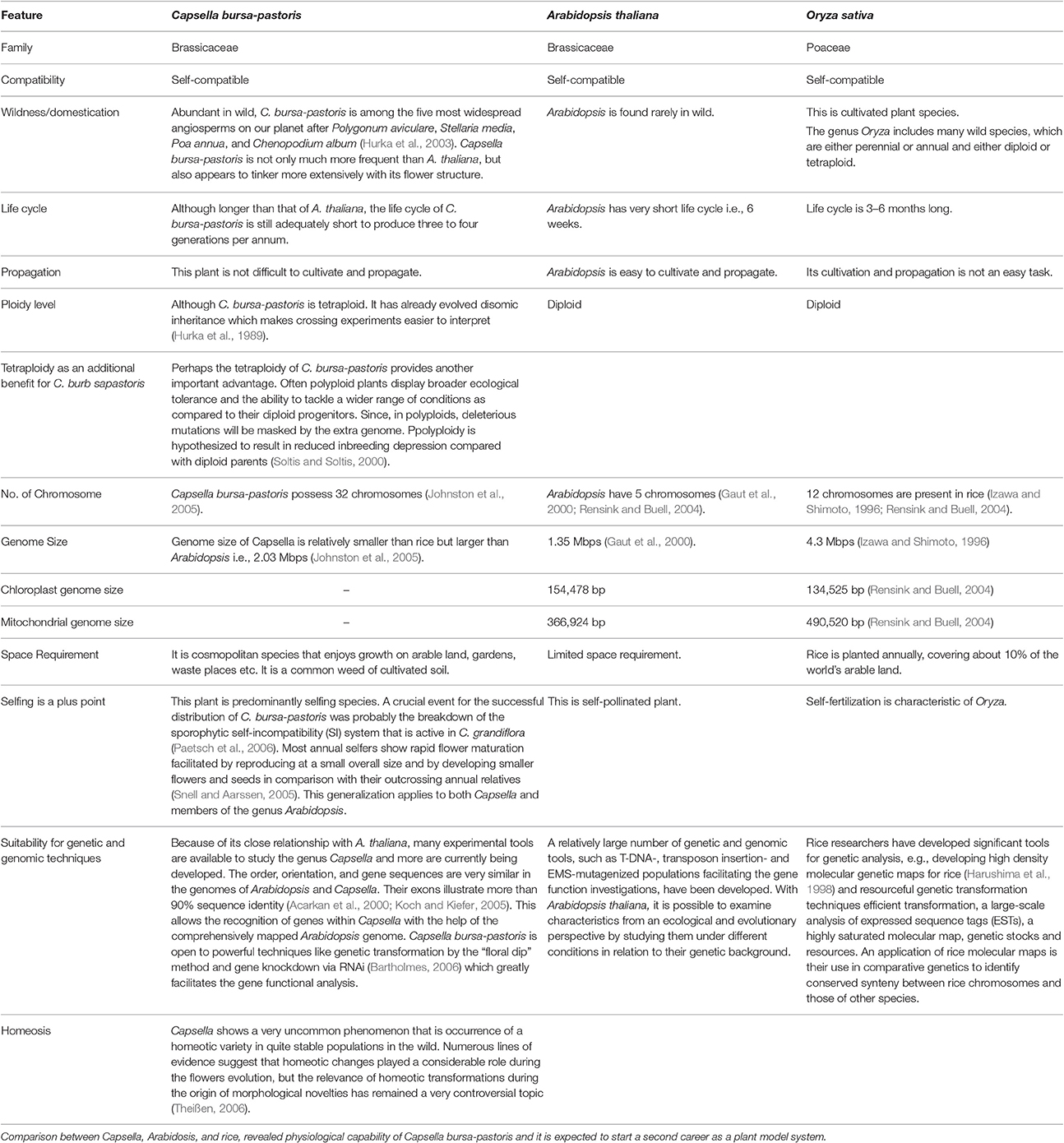
Table 2. Why Capsella is gaining importance as model plant in presence of Arabidopsis and Rice? Due to its interesting biology and close relationship with Arabidopsis, Capsella bursa-pastoris is appearing as model plant for studying abiotic stress tolerance.
PGRs Are Front Line Players in Cold Stress Tolerance
Temperate plants adopt a variety of mechanisms such as germination or developmental modulations to avoid stress damages (Ali et al., 2017; Noman et al., 2017b). In addition to physio-biochemical adjustments (Cramer et al., 2011), cold adaptation includes modified expression of various genes and associated machinery of extensive biological significance (Xin and Browse, 2000; Fowler and Thomashow, 2002; Noman et al., 2017a). The frequently occurring events like altered membranes composition or structure help in reducing the cellular injuries triggered by freezing or very low temperatures (Figure 1; Ruelland et al., 2009). Plant growth regulators (PGRs) are front line players in controlling these molecular trails during cold stress. Moreover, the hormonal signaling serves to stimulate stress response pathways. This hormonal signaling network integrates exterior information from environment into endogenous developmental programs to activate plants stress response pathways. It is not astonishing that PGRs are very important features for cold stress responses (Eremina et al., 2016). However, our comprehension about the molecular modes responsible for stress needs extensive investigations.
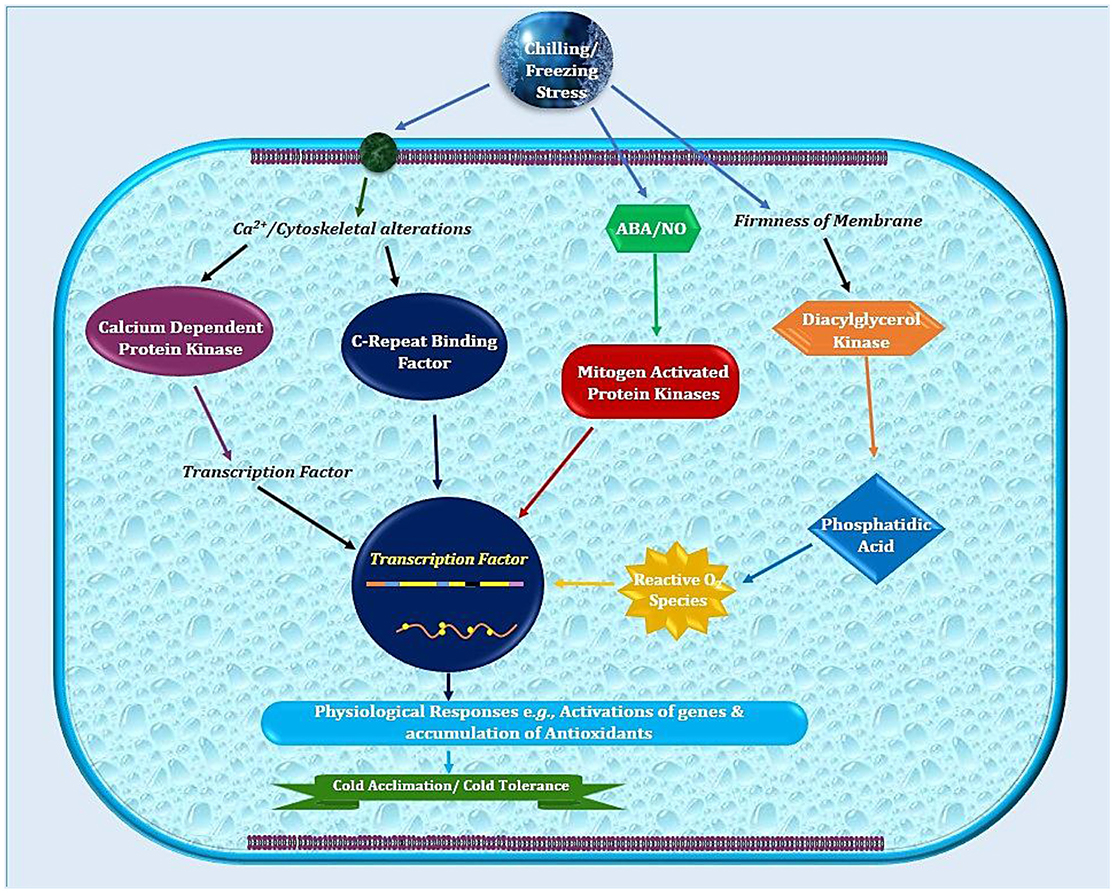
Figure 1. Schematic illustration of sub-cellular events in plant cell after exposure to low temperature. Plasma membrane lipids in cold sensitive plants possess high degree of saturated fatty acids that contribute in higher freezing tolerance. Later on, combination of physio-biochemical and molecular changes leads to cold stress tolerance.
The modes of hormonal activity are usually species dependent that complicate the research in this field. Abscisic acid (ABA) is regarded as chief contributor in tolerating freezing or low temperature stress. Of note, increase in ABA level is directly proportional to increased cold tolerance (Nakashima et al., 2014). Similarly, OSTI (open stomata 1) protein is activated by ABA. The activity of OSTI is induced after implication of cold stress. Interaction of OST1 and ICE1 stabilize this protein and enhance its transcriptional activity (Ding et al., 2015).
Investigations have highlighted that cold stress affects auxin levels differentially depending on plant type, developmental stage and physiology (Eremina et al., 2016). For example, low temperature treatment for many days substantially augmented IAA levels in spring wheat crown tissues only. Besides, IAA concentration was significantly increased in winter wheat after 12 days of cold stress (Majlath et al., 2012). Some contradictions have also been recorded in case of other plants such as rice facing low temperature treatment (Maruyama et al., 2014).
In mutant Arabidopsis and rice plants, it has been established that GA signaling components can modify the plant responses to chilling conditions (Richter et al., 2013). PIF4 may have involvement in mediating CBF control by GAs. PIF4 and other likely factors may appear as central nodes for integrating different environmental stimuli for growth. Quite opposite to GAs, brassinosteroid (BR) enhance cold tolerance in several plants including chilling-sensitive plants like Zea mays, Cucumis sativus etc. (Jiang et al., 2013). BR receptor BRI1 mutant, bri1-116 exhibited increased ion leakage due to cold stress, thereby attested the role of BR signaling in promoting cold stress acclimation (Qu et al., 2011). But some studies have also presented opposite results. Therefore, extensive experimentation and verification of results is needed. It is generally agreed that cytokinins (CK) application can improve chilling tolerance in plants. In CK-deficient mutant of Arbidopsis, application of cytokinins also enhanced tolerance to low temperature in CBF1-independent manner (Jeon et al., 2010). Low CK levels in response to chilling have been reported in model plant like rice (Maruyama et al., 2014).
There are still some questions regarding the positive or negative regulatory role of ethylene in plant tolerance to cold. Some reports mentioned boost in ethylene levels against cold in various plant species e.g., Medicago sp. (Guo et al., 2014). Increased ethylene concentration during cold stress was linked to amplified expression of enzymes in Arabidopsis. On the other hand, decreased ethylene level in response to low temperature would fit well to its suggested function as negative regulator of chilling tolerance in some plants (Shi et al., 2012; Zhao et al., 2014).
Is Concept of Plant Cold Acclimation Incomplete Without CBFs?
To respond against cold stress, the signaling pathway of C-repeat binding factor (CBF) is essential in angiosperms (Chinnusamy et al., 2007; Welling and Palva, 2008). In robust system Capsella, CbCBF expression is apparently feasible strategy for studying chilling stress tolerance (Zhou et al., 2011a). In model plant tobacco, over expression of CbCBF improved delayed flowering, dwarfism as well as tolerance to freezing and chilling (Zhou et al., 2012b). Consistently, in tobacco the reduced bioactive GA content coupled with impaired GA metabolism was due to CbCBF over-expression (Kasuga et al., 2004). So, we can build an opinion that by interacting with cell cycle pathways, CbCBF confers ultimate resistance to cold seemingly through downstream target genes stimulation in tobacco cells.
Thomashow (1999) has described CBF as master switches to increase cold tolerance. Interestingly, the COR genes expression is activated by CBF genes in Arabidopsis (Figures 1, 2). Comparative account of CBF from model plant Arabidopsis and Capsella presents interesting facts. In spite of the dissimilarities, the resembling genome sequences for many genes and TFs reveal high level functional similarity among both relatives. AtCBF1 and AtCBF4 play a more substantial role than the CBF3 under chilling stress (Wang and Hua, 2009) while AtCBF2 exhibits dissimilar expression pattern from AtCBF1and AtCBF3 (Novillo et al., 2007). In comparison with Arabidopsis, CbCBF have a great impact in both chilling and freezing tolerance in cold sensitive N. tabacum plants. This indicates that both severe and moderate cold responses are regulated by the participation of CbCBF. Stimulation, activation and transcription of CbCBF promoters have been recognized in shoot as well as root system (Zhou et al., 2014). On the whole, in Capsella a stronger cold responsive signaling cascade may be induced by CbCBF during cold exposure as compared to species sensitive to low temperature.
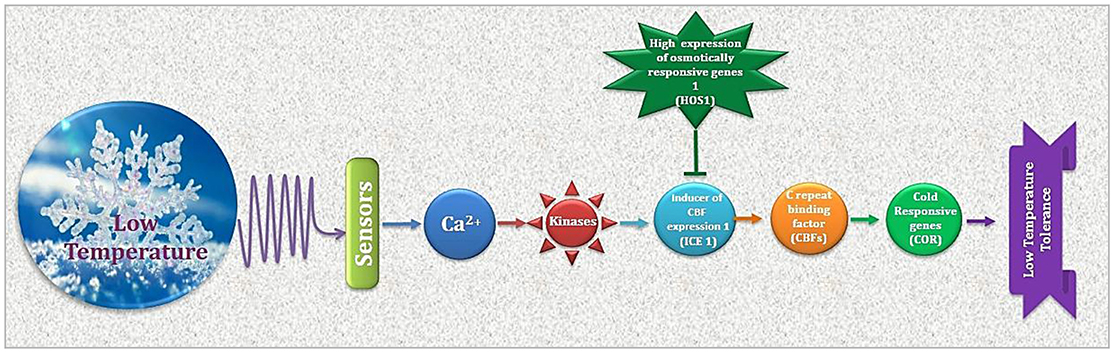
Figure 2. Cold stress perception and ultimate plant response is determined by regulation of CBFs and CORs. As a premier regulator of cold acclimation, CBF controls COR gene expression. Products of CORs i.e., regulatory and functional proteins result in physiological adjustments for appropriate plant response to low temperature.
Besides, slow growth rate, inhibited growth, stunted appearance and delayed flowering was also exhibited by 35S::CbCBF in tobacco plants. Already in Arabidopsis, over-expression of CYCD genes has been considered responsible for the shortening of G1 in cells (Menges et al., 2006). In Capsella CBF-ox plants, both the reduction in mRNA levels for CYCD genes and increased number of G1 phase cells supports the involvement of CYCDs as rate-limiting factors for the G1-S transition. The hypothesis that CbCBF inhibits the G1-S transition is also checked by the contrasting properties of CbCBF over-expression to that of 35S::AtCYCD3;1 (Menges et al., 2006). However, Guo and Wang (2008) reported that cold stress causes reduction in NtCYCD genes expression. Consequently, CbCBF may possibly hinder the G1-S transition by suppressing the manifestation of CYCD genes in response to cold stress. These findings are in agreement with the previous study on rice that over-expression of OsCYCB1;1 enhanced cold stress tolerance (Ma et al., 2009). From all above findings, we agree that CbCBF contributes in regulation of cell cycle progression.
Yamaguchi (2008) reported considerably lower GA1 and GA3 level in plants harboring 35S::CbCBF. The most GA deprived site was young leaves and few leaves from the apical nodes. Having said all this, CbCBF is responsible for the reduction in bioactive GA levels (Achard et al., 2006) in new growing leaves and have slight influence in old leaves. Contrarily, the dwarfism of CbCBF transgenic plants may be reversed by exogenous application of GA. Therefore, the growth retardation is likely to be due to suppression of bioactive GA. It has been reported in studies on rice as well as Capsella that CDK and some cyclin genes expression can be stimulated by exposure to GA. This stimulation can occur at both G1-S and G2-M transitions (Lorbiecke and Sauter, 1998; Zhou et al., 2014). The decreased GA levels may also be partially responsible for the delayed G1-S transition conferred by CbCBF. The specific suppression of the G1-S transition might be based upon altered transcription of CYCD.
In view of these findings and the CbCBF suppression by GA exposure under cold stress, we speculate that CbCBF-dependent regulatory pathway interacts with the GA signaling pathway. This interaction regulates plant growth especially in growing tissues and ultimately modulates cold tolerance. Meanwhile, there seems to be other pathway(s) downstream of CbCBF but independent of GA in cold response. In summary, CbCBF is strongly induced by cold and participates in a regulatory network of cold acclimation.
The inverse relationship between PGRs i.e., GA and CBF can be a crucial output of cold induced gene regulation in Capsella. In addition, CbCBF is also involved in cell cycle control by interfering CYCD genes expression. Based on the above analysis, CbCBF is an excellent candidate for application in breeding of plants with dwarf forms and lawn grasses. Further investigation on the exact target nodes of CbCBF on growth reduction will contribute to the production of CbCBF transgenic plants with stronger cold tolerance but without growth retardation.
Expression of COR Gene Along with Identification of cis- Elements is Imperative for Low Temperature (LT) Tolerance
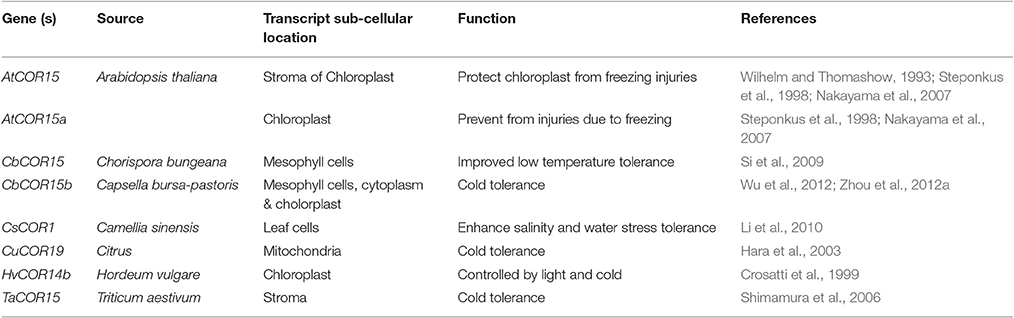
The cryoprotective proteins are special product of cold-regulated (COR) genes. These proteins function by increasing membrane expandability during melting and reduce permeability of membranes upon exposure to freezing. Transcription factors (TFs) coupled with definite nuclear events directly regulate COR gene expression. Success is evident in exploiting TFs among transgenic plants. Different genes and transcription factors appeared helpful for cold tolerance in different plant species (Table 3). But, the up-stream regulatory modes that control these activities are still indefinable. Various COR genes have been defined from different spermatophytes comprising of COR15a from C. bursa-pastoris, BN19, BN115, and BN26 from B. napus, CbCOR15 from C. bungeana and COR14b gene from H. vulgare (Table 4; Figure 2). Array of expression patterns of these genes have been exposed after low temperature treatment (Si et al., 2009; Chen et al., 2011). In A. thaliana and Capsella the arbitrate expression of COR genes have been validated by cis-acting elements of putative COR15 promoter (Stockinger et al., 1997; Lin et al., 2016).
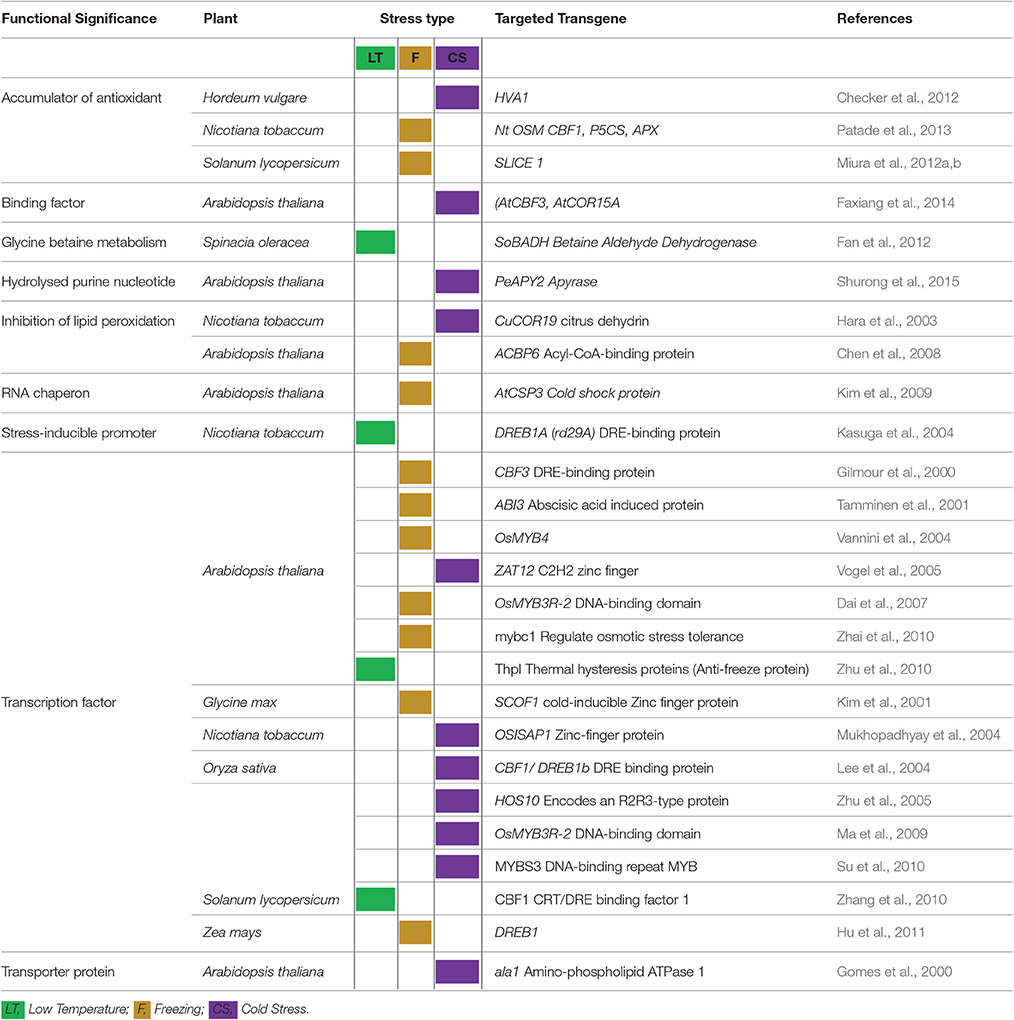
Table 3. Success of transgenic plants and different transgenes in enhancing plant tolerance to varied temperature ranges.
In Arabidopsis thaliana COR15a/b (Lin et al., 2016), COR78 (Thomashow et al., 2001), RAB18 (Lang and Palva, 1992), and KIN1/2 (Kurkela and Borg-Franck, 1992) are different promoters of COR genes. These encompass extremely conserved cis-elements such as CRT, DRE, or LTRE. ICEs are the upstream regulators and inducers of CBF expression as well. They work as positive regulator of CBFs, though COR genes are regulated by CBFs by attaching to the CRT/DRE element (Lissarre et al., 2010). Promoter fusion revealed that COR gene expression can be initiated by two cis-acting CRT/DRE elements of CbCOR15a gene and one cis-acting CRT/DRE element of CbCOR15b. It had already been illustrated that under non-acclimated situation the expression of AtRD29a-GUS (Yamaguchi-Shinozaki and Shinozaki, 1993) and AtCOR78-GUS (Horvath et al., 1993) gene was either imperceptible or very low in almost all plants tissues.
Different patterns of gene expression indicate completely different role of COR genes in different plant species (Bajji et al., 2002; Kang et al., 2009). The data presented by Wu et al. (2012) affirmed that CbCOR15b expressed primarily in leaves and stems. However, in response to cold, it may additionally play a function in roots. We are convinced that the CbCOR15b protein localization within the plastid and cold inductive activity of CbCOR15b in leaves (Wilhelm and Thomashow, 1993) confirms the presence of preserved chloroplast-targeting signal peptide in its N-terminal as a member of late embryogenesis abundant (LEA) proteins. Cleavage of this protein might occurr and also the residual peptide could be targeted into the stroma of chloroplast (Liu et al., 2004). CbCOR15b localization in epidermal cells, stele, and endodermis revealed its alternative functions in roots as compared to the leaves. Its function may be the protection of chloroplast from freezing and thawing damage in leaves. This functional disparity between CbCOR15b and CbCOR15a indicates a different function of CbCOR15b in plants with respect to temperature variation and organ type. From the former researches it has been revealed that cold regulated genes contained 3 gene pairs which had been isolated from A. thaliana, such as KIN1/KIN2, COR15a/COR15b, and RD29a/RD29b. In every case, totally different regulation was observed by the members of the sequence pairs of genes. However, expression patterns of these genes were differed spatially as well as temporally in response to chilling stress and ABA treatment. However, accumulation of COR15b was in high levels under drought stress conditions (Wilhelm and Thomashow, 1993). Whereas, in C. bursa-pastoris, gene pairs like CbCOR15a/CbCOR15b expressed diverse characteristics with treatments of different phytohormones such as SA, GA3, IAA, and MeJA. Treatment with ABA exhibited the similar expression trends in same plant. So, CbCOR15b expression was only distinguished under ABA application (Zhou et al., 2010, 2011b).
Enhanced chilling tolerance in transgenic plants could be due to the constitutive expression of the COR genes (Artus et al., 1996; Grossi et al., 1998; Zhou et al., 2012b). The three physiological indices i.e., electrolyte leakage, glucose contents and the relative water content are significant indicators of plant freezing resistance (Campos et al., 2003; Nakashima et al., 2006). These physiological modulations positively correlate with improved cold tolerance in plant cells. Transgenic tobacco lines revealed amplified freezing resistance with the anticipated CbCOR15b functions after crystallization of cellular components and chloroplast preventing water loss (Si et al., 2009). On the basis of presented information, we attribute cytoplasm and chloroplast-targeted CbCOR15b for maintaining the cytoplasmic homeostasis by triggering the reactions. It also protects the membrane-targeted and cellular active proteins under freezing stress.
In some plant tissues the molecular reason for absence of expression of the COR promoter is not well defined at normal growth temperature (Lin et al., 2016). Besides, cold stress dependent COR gene up-regulation is tissue specific in various plants. Experiments have affirmed the least promoter activity of AtCOR15a in roots but significantly enhanced in flowers, leaves and siliques after exposure to 4°C (Baker et al., 1994). Whereas, in cold treated leaves, flower sepals, stems and roots, the activity of AtCOR78 promoter was significantly enhanced. However, no activity was observed in other plant parts such as anthers, styles, stigmas, or ovaries of the flowers (Horvath et al., 1993). It is evident that in all plant tissues including cauline leaves, rosette leaves, inflorescence, seedlings and siliques the action of CbCOR15 promoter was encouraged significantly. The presented data clearly elucidate that various expression patterns are controlled by different cis-acting elements under low or normal temperature conditions.
Furthermore, the control of constitutive CAMV35S by the over expression of CbCOR15b resulted in no dwarf phenotype. The use of endogenous environment-inducible promoter of plant is better to circumvent hazardous effects on growth and development by driving the expression of those genes causing dwarf phenotype than the 35S promoter. The non-dwarfism of transformant plants and cold inductive activity of CbCOR15b (Shimamura et al., 2006; Wu et al., 2012) provide a potential evidence and it can be used in transgenic crops for the improvement of cold resistance. Studying the interaction between different COR genes would endow us a much better understanding of the freezing or chilling stress responsive pathway and also offer a pragmatic tool for increasing or inducing plant resistance to low temperature.
ROS Homeostasis and Cold Stress
Low temperature obstructs plant development by affecting the cellular metabolism and gene regulating networks. In response to various abiotic stresses, antioxidant enzymes e.g., superoxide dismutase, peroxidase, catalase play a great role in controlling and regulating the ROS production and accumulation (Noman and Aqeel, 2017; Noman et al., 2017b). In Capsella, belonging to type III peroxidase family the CbRCI35 (Rare Cold-Inducible 35) gene has been reported as cold responsive gene. Heterologous expression tests unravel the fact that cold responsive endogenous signaling and low temperature resistance in tobacco is conferred by CbRCI35 (Zhou et al., 2016). Conversely, a moderate increase in ROS accumulation was noticed under normal conditions and CbRCI53 linked superoxide dismutase activity was enhanced in transgenic plants after exposure to chilling stress. A consequent alteration was reported in the gene expression related to ROS metabolism.
Different scientists used Arabidopsis cDNA library screening for identifying the cold responsive RCI genes e.g., AtRCI1A/B or AtRCI2A/B (Kim et al., 2010; Sivankalyani et al., 2015). CbRCI35 gene displayed comparatively high transcription level and obvious cold-inducible expressions in roots as well as great resemblance to Arabidopsis RCI3. Another interesting fact about these genes is their differential responses to various conditions. During cold, AtRCI3 respond by gradually elevating its transcription and then reach its maximal level after 24 h of 4°C exposure (Llorente et al., 2002). CbRCI35 expressed gradually at different temperature levels. High expression level was noted after 8 h of treatment and then returned to a low level at the 24 h exposure to similar temperature. This behavior indicates its potential role and quick activation to low temperature at the earlier stage of response. As far as organs specified expressions are concerned, in roots CbRCI35 displayed high expression level. However, it can also encourage expression in stems and leaves. Quite opposite to earlier described RCIs, AtRCI3 exhibits a root specific transcription. Moreover, AtRCI3 expressed itself in the cortex and stele. Contrarily, transcription expression of CbRCI35 was restricted to the root cortex only (Llorente et al., 2002). The data collected from different plant species reveal that type III peroxidase genes exhibit a variety of expressional regulation. Subsequently, the AtRCI3 protein is localized in the endoplasmic reticulum (ER) and can be secreted to the cell wall while the CbRCI35 protein is restricted in the cytoplasm (Kim et al., 2010). The protein localization and distinct transcription level entails that CbRCI35 from C. bursa-pastoris might have more distinct function as compared to Arabidopsis RCI3. This opinion provides innovative insight to understand the cold tolerance regulation in plants by AtRCI3-like type III peroxidases.
The production and scavenging mechanism of ROS exhibit a key role in plant stress acclimation (Noman et al., 2015; Ali et al., 2016; Zafar et al., 2016). The signal transduction and ROS level are jointly controlled by adequate amount of ROS homeostasis regulators (Mangano et al., 2016; Noman et al., 2017b). In plants during stress responses a wide range of ROS scavengers along with cold resistance positive modulators have been reported (Table 5; Kim et al., 2010). Our analysis of the available information recommends resemblance in over-expression of CbRCI35 and AtRCI3 which contribute in managing ROS under normal circumstances.
According to Zhou et al. (2016), in CbRCI35-ox seedlings, enhanced NtSOD expression is positively correlated with CbRCI35 gene function against ROS accumulation. Due to feedback mechanism the NtSOD transcripts can increase. Most of the genes were negatively controlled in transgenic tobacco as compared to control during chilling treatment. As a result, the level of ROS was identical to the control describing the transcriptional control of ROS metabolic genes transformed by the CbRCI35 for ROS homeostatic mechanism (Zhou et al., 2016). Evidence supports the SOD activity was complex in transgenic plants under both the chilling and warm environments (Table 3). This attribute reflect that CbRCI35 gene might contribute in the protection of bioactive enzymes during chilling stress. Although, the total level of ROS was not dropped, electrolyte leakage and malondialdehyde (MDA) content indicated the alleviated membrane injury in CbRCI35-ox tobacco plants during chilling temperatures (Zhou et al., 2016). Therefore, we can infer that CbRCI35 significantly contribute in enhancing freezing resistance as well as in plant cold acclimation by activating the COR genes and regulating ROS homeostasis. Moreover, application of CbRCI35 has comprehensive prospects for crop improvements in plant breeding.
Take a Pause, Some Links are Missing
As a model plant, Capsella bursa-pastoris is a viable system for inquiring plant stress responses and adaptation. But with reference to angiosperms generally and Capsella particularly, several important links in cold acclimation are missing. So many questions arise that are crucially linked with low temperature tolerance and acclimation process. For example, still there are question marks upon the molecular identity of the cold-regulated Ca2+ channel(s) in plants. Over the years, Ca2+ channel activities have been intensively studied for their electro-physiological aid in tolerating low temperature stress (Carpaneto et al., 2007). But contrary to animals (Karashima et al., 2009), point to ponder is ignorance to clone plant genes encoding the proteins accountable for cold tolerance activities. With particular reference to Capsella sp., we do not have answers about identification of Ca2+ channel. Similarly, different mutant screens for expression of cold-induced gene and cold tolerance have yet not been able to recognize any plant Ca2+ channels. On the other hand, functional cloning by using a heterologous system can be very good approach for identifying Ca2+-sensing receptor like in Arabidopsis (Han et al., 2003) and could be employed in case of Capsella.
Another missing link is deciphering of the cold Ca2+ response and role of different Ca2+-binding signaling proteins. As a response to different degrees of temperature reductions, even before the temperature needed for triggering cold acclimation, Ca2+ levels increases (Larkindale and Knight, 2002). At the moment, we need to investigate the Capsella's capacity to make a distinction between changes in modulated Ca2+ levels especially when the temperature falls below 5°C (Knight and Knight, 2000). For presenting C. bursa-pastoris as a model plant to unravel molecular basis of cold acclimation, it will be beneficial to quantify the in vitro activity of Ca2+-responsive proteins like CaM in response to various Ca2+ signatures. It is speculated that Ca2+ alterations update the cell only about temperature reduction while other signaling mechanisms independent of Ca2+ cover information regarding absolute temperature to inform the plant for type of response needed. Future identification of Ca2+ channels would make possible a genetic advance to be made and applied to the questions relating to Ca2+ encoded information.
In plants presence of Ca2+-dependent and MAPK dependent pathways for mediating the gene expression regulation propose different messages conveyed during temperature variations (Knight and Knight, 2012). It is motivating to find out whether mutation in regulatory authority of each pathway awards differential sensitivity or augment the response to particular temperature range because different gene groups are regulated by each sensing system. Winfield et al. (2009, 2010) have presented the wheat CBF gene selective response against cold shock and slow cooling. It will be very enticing to verify which gene group is regulated by different mechanisms. This analysis would differentiate between the genes taking part in chilling, freezing temperature or vernalization.
It is very clear that response to chilling and freezing temperature requires different gene operations. The question is how we can differentiate between the cold-regulated genes involved in chilling and freezing. In plants with capacity to acclimatize in cold, genes expression against low temperatures i.e., 5°C or below is seemingly a pre-requisite for the acquisition of freezing tolerance or gain/maintenance of chilling tolerance. So far with the help of transcriptomic profiling in model species like Arabidopsis and non-model plants such as Solanum tuberosum, we are unable to differentiate between chilling and freezing tolerance. Therefore, this situation has necessitated additional studies for gene identification specifically involved in chilling tolerance. We speculate that transcriptomic comparison among chilling-sensitive and freezing-tolerant plants may grant support here. Investigations by using A. thaliana have not only substantiated the effectiveness of this approach for identification of genes linked with one exact form of cold tolerance. From the data, it can easily be inferred that components of chilling and freezing tolerance pathways display high level of conservation with modifications in the target genes expression only (Narsai et al., 2010).
In continuation of above quoted discussion, the question arises if plants are responsive to temperature fluctuations, how do they discriminate between those that are worthy and those that should be overlooked? It looks very important that a plant does not employ a full cold acclimation response whenever it experience temperature fluctuations. Hence, a competent system of checks is expected to discriminate between injurious and harmless fluctuations with the help of warning signals. This may engage sensing the duration of low temperature and incite acclimation response after independent confirmation of low temperature. Knocking out of individual cold signaling pathways can assist us to solve such issues. Unequivocally the circadian clock as well as light quality signals makes available significant contextual data regarding relevance of low-temperature alterations to the plant. It is agreed that any abrupt drop in temperature to 5°C at noon entails a very different explanation to the same degree of temperature drop experienced by plant at dusk or night. The recognition of manifold temperature parameters by means of parallel signal transduction systems may appear as an aegis against wasteful or inapt responses.
Conclusion
Knowledge about low temperature responses in plants has been enriched by genetic as well as molecular techniques. However, the existing information can be augmented by exploring involvement of numerous transcriptional regulators and interaction between the signaling pathways operating in the process of low temperature acclimation. Scientific advances in the fields like metabolite profiling have highlighted the contribution of cellular metabolic signals in freezing or chilling stress tolerance. Many genes taking part in RNA splicing, export or remodeling of chromatin proteins have been recognized for their significant functions during plant acclimation to low temperature. But accurate details and the exact mechanism of the whole complex network remain to be elucidated. Today, we need mandatory studies such as mutation analysis and identification of regulation cascades for ambiguous metabolic signals. On the other hand for the studied genes, obviously intensive effects in entire network and homologous analysis in novel and candidate model plant species are required. Forward and reverse genetic research in concert with physio-biochemical and bioinformatics analyses will offer more inspiring discoveries for LT acclimation in plants. Moreover, for highly efficient utilization of the elements in the CBF or COR-dependent signaling pathways, novel approach and techniques need to be established. In near future, it will become essential to have these systems more enthusiastically adopted and optimized to study plant responses to low temperature. This would accelerate the progress made in this field during recent years.
Author Contributions
AN and MA had major and equal contribution in overall preparation of manuscript. AN has collected research data and compiled manuscript. MA has made all the figures and tables and made corrections. HK and NK contributed in writing the different sections. AT and AM has checked and corrected the grammar and TS have compiled table. SS corrected the references as well as DOI. SH has provided technical guidance and critically read this manuscript and suggested for improvement and corrected mistakes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Acarkan, A., Roβberg, M., Koch, M., and Schimdt, R. (2000). Comparative genome analysis reveals extensive conservation of genome organization for Arabidopsis thaliana and Capsella rubella. Plant J. 23, 55–62. doi: 10.1046/j.1365-313x.2000.00790.x
Achard, P., Cheng, H., De Grauwe, L., Decat, J., Schoutteten, H., Moritz, T., et al. (2006). Integration of plant responses to environmentally activated phytohormonal signals. Science 311, 91–94. doi: 10.1126/science.1118642
Ali, Q., Haider, M. Z., Iftikhar, W., Jamil, S., Javed, M. T., Noman, A., et al. (2016). Drought tolerance potential of Vigna mungo L. lines as deciphered by modulated growth, antioxidant defense, and nutrient acquisition patterns. Braz. J. Bot. 39, 801–812. doi: 10.1007/s40415-016-0282-y
Ali, Q., Javed, M. T., Noman, A., Haider, M. Z., Waseem, M., Iqbal, N., et al. (2017). Assessment of drought tolerance in Mung-bean cultivars/lines as depicted by the activities of germination enzymes, seedling's antioxidative potential and nutrient acquisition. Arch. Agron. Soil Sci. doi: 10.1080/03650340.2017.1335393
Ao, P. X., Li, Z. G., Fan, D. M., and Gong, M. (2013). Involvement of antioxidant defense system in chill hardening-induced chilling tolerance in Jatropha curcas seedlings. Acta Physiol. Plant. 35, 153–160. doi: 10.1007/s11738-012-1058-z
Artus, N. N., Uemura, M., Steponkus, P. L., Gilmour, S. J., Lin, C., and Thomashow, M. F. (1996). Constitutive expression of the cold regulated Arabidopsis thaliana COR15a gene affects both chloroplast and protoplast freezing tolerance. Proc. Natl. Acad. Sci. U.S.A. 93, 13404–13409. doi: 10.1073/pnas.93.23.13404
Bajji, M., Kinet, J. M., and Lutts, S. (2002). The use of the electrolyte leakage method for assessing cell membrane stability as a water stress tolerance test in durum wheat. Plant Growth Regul. 36, 61–70. doi: 10.1023/A:1014732714549
Baker, S. S., Wilhelm, K. S., and Thomashow, M. F. (1994). The 5'-region of Arabidopsis thaliana COR15a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. doi: 10.1007/bf00029852
Barrero-Gil, J., and Salinas, J. (2013). Post-translational regulation of cold acclimation response. Plant Sci. 205–206, 48–54. doi: 10.1016/j.plantsci.2013.01.008
Bartholmes, C. (2006). Versuch der gezielten Manipulation der Genexpression von Capsella bursa-pastoris (L.) Medik. Mittels Agrobacterium-vermittelter Transformation. Diploma Thesis, Friedrich-Schiller-Universitat Jena.
Campos, P. S., Quartin, V., Ramalho, J. C., and Nunes, M. A. (2003). Electrolyte leakage and lipid degradation account for cold sensitivity in leaves of Coffee sp. plants. J. Plant Physiol. 160, 283–292. doi: 10.1078/0176-1617-00833
Carpaneto, A., Ivashikina, N., Levchenko, V., Krol, E., Jeworutzki, E., Zhu, J. K., et al. (2007). Cold transiently activates calcium-permeable channels in Arabidopsis mesophyll cells. Plant Physiol. 143, 487–494. doi: 10.1104/pp.106.090928
Ceplitis, A., Su, Y. T., and Lascoux, M. (2005). Bayesian inference of evolutionary history from chloroplast microsatellites in the cosmopolitan weed Capsella bursa-pastoris (Brassicaceae). Mol. Ecol. 5, 4221–4233. doi: 10.1111/j.1365-294x.2005.02743.x
Checker, V. G., Chhibbar., A. K., and Khurana, P. (2012). Stress-inducible expression of barley Hva1 gene in transgenic mulberry displays enhanced tolerance against drought, salinity and cold stress. Trans. Res. 21, 939–957. doi: 10.1007/s11248-011-9577-8
Chen, L., Zhong, H., Ren, F., Guo, Q. Q., Hu, X. P., and Li, X. B. (2011). A novel cold-regulated gene, COR25, of Brassica napus is involved in plant response and tolerance to cold stress. Plant Cell Rep. 30, 463–471. doi: 10.1007/s00299-010-0952-3
Chen, Q. F., Xiao, S., and Chye, M. L. (2008). Overexpression of the Arabidopsis 10-kDa acyl-CoA-binding proteinACBP6 enhances freezing tolerance. Plant Physiol. 148, 304–315. doi: 10.1104/pp.108.123331
Chinnusamy, V., Zhu, J., and Zhu, J. K. (2006). Gene regulation during cold acclimation in plants. Physiol. Plant. 126, 52–61. doi: 10.1111/j.1399-3054.2006.00596.x
Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi: 10.1016/j.tplants.2007.07.002
Chu, J., Yao, X., and Zhang, Z. (2010). Responses of wheat seedlings to exogenous selenium supply under cold stress. Biol. Trace Element Res. 136, 355–363. doi: 10.1007/s12011-009-8542-3
Cramer, G. R., Urano, K., Delrot, S., Pezzotti, M., and Shinozaki, K. (2011). Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 11:163. doi: 10.1186/1471-2229-11-163
Crosatti, C., Polverino de Laureto, P., Bassi, R., and Cattivelli, L. (1999). The interaction between cold and light controls the expression of the cold-regulated barley gene COR14b and the accumulation of the corresponding protein. Plant Physiol. 119, 671–680. doi: 10.1104/pp.119.2.671
Dai, X., Xu, Y., Ma, Q., Xu, W., Wang, T., Xue, Y., et al. (2007). Overexpression of an R1R2R3 MYB gene, OsMYB3R-2, increases tolerance to freezing, drought and salt stress in transgenic Arabidopsis. Plant Physiol. 143, 1739–1751. doi: 10.1104/pp.106.094532
Ding, Y., Li, H., Zhang, X., Xie, Q., Gong, Z., and Yang, S. (2015). OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell. 32, 278–289. doi: 10.1016/j.devcel.2014.12.023
Eremina, M., Rozhan, W., and Poppenberger, B. (2016). Hormonal control of cold stress responses in plants. Cell. Mol. Life Sci. 73, 797–810. doi: 10.1007/s00018-015-2089-6
Fan, W., Zhang, M., Zhang, H., and Zhang, P. (2012). Improved tolerance to various abiotic stresses in transgenic sweet potato (Ipomoea batatas) expressing spinach betaine aldehyde dehydrogenase. PLoS ONE 7:e37344. doi: 10.1371/journal.pone.0037344
Faxiang, W., Yu, P., Jinghua, L., Xiangfu, C., Yanglu, P., Yongqing, W., et al. (2014). Heterologous expression of Arabidopsis C -repeat binding factor 3 (AtCBF3) and cold -regulated 15A (AtCOR15A) enhanced chilling tolerance in transgenic eggplant (Solanum melongena L.). Plant Cell Rep. 33:1670. doi: 10.1007/s00299-014-1670-z
Fortunato, A. S., Lidon, F. C., Batista-santos, P., Leitào, A. E., Pais, I. P., Ribeiro, A. I., et al. (2010). Biochemical and molecular characterization of the antioxidative system of Coffea sp. Under cold conditions in genotypes with contrasting tolerance. J. Plant Physiol. 167, 333–342. doi: 10.1016/j.jplph.2009.10.013
Fowler, S., and Thomashow, M. F. (2002). Arabidopsis transcriptome profiling indicates that multiple regulatory pathways are activated during cold acclimation in addition to the CBF cold response pathway. Plant Cell. 14, 1675–1690. doi: 10.1105/tpc.003483
Gaut, B. S., Maud, L. T., Andrew, S. P., and Mark, C. S. (2000). Maize as a model for the evolution of plant nuclear genomes. Proc. Natl. Acad. Sci. USA 97, 7008–7015. doi: 10.1073/pnas.97.13.7008
Gilmour, S. J., Sebolt, A. M., Salazar, M. P., Everard, J. D., and Thomashow, M. F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 124, 1854–1865. doi: 10.1104/pp.124.4.1854
Gomes, E., Jakobsen, M. K., Axelsen, K. B., Geisler, M., and Palmgreen, M. G. (2000). Chilling tolerance in Arabidopsis involves ALA1, a member of a new family of putative aminophospholipid translocases. Plant Cell 12, 2441–2454. doi: 10.1105/tpc.12.12.2441
Gong, Z., Lee, H., Xiong, L., Jagendorf, A., Stevenson, B., and Zhu, J. K. (2002). RNA helicase-like protein as an early regulator of transcription factors for plant chilling and freezing tolerance. Proc. Natl. Acad. Sci. USA 99, 11507–11512. doi: 10.1073/pnas.172399299
Grossi, M., Giorni, E., Rizza, F., Stanca, A. M., and Cattivelli, L. (1998). Wild and cultivated barleys show differences in the expression pattern of a cold-regulated gene family under different light and temperature conditions. Plant Mol. Biol. 38, 1061–1069. doi: 10.1023/A:1006079916917
Guo, J., and Wang, M. H. (2008). Transgenic tobacco plants overexpressing the Nicta; CycD3; 4 genes demonstrate accelerated growth rates. BMB Rep. 7, 542–547. doi: 10.5483/BMBRep.2008.41
Guo, Z., Tan, J., Zhuo, C., Wang, C., Xiang, B., and Wang, Z. (2014). Abscisic acid, H2O2 and nitric oxide interactions mediated cold induced S-adenosyl methionine synthetase in Medicago sativa sub sp. falcata that confers cold tolerance through up-regulating polyamine oxidation. Plant Biotechnol. J. 12, 601–612. doi: 10.1111/pbi.12166
Han, S. C., Tang, R. H., Anderson, L. K., Woerner, T. E., and Pei, Z. M. (2003). A cell surface receptor mediates extracellular Ca2+ sensing in guard cells. Nature 425, 196–200. doi: 10.1038/nature01932
Hara, M., Terashima, S., Fukaya, T., and Kuboi, T. (2003). Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 217, 290–298. doi: 10.1007/s00425-003-0986-7
Harushima, Y., Yano, M., Shomura, A., Sato, M., Shimano, T., Kuboki, Y., et al. (1998). A high-density rice genetic linkage map with 2275 markers using a single F2 population. Genetics 148, 479–494.
Horvath, D. P., Mclarney, B. K., and Thomashow, M. F. (1993). Regulation of Arabidopsis thaliana L. (Heyn) COR78 in response to low temperature. Plant Physiol. 103, 1047–1053. doi: 10.1104/pp.103.4.1047
Hu, Y., Zhang, L., Zhao, L., Li, J., He, S., Zhou, K., et al. (2011). Trichostatin A selectively suppresses the cold-induced transcription of the ZmDREB1 gene in maize. PLoS ONE 6:e22132. doi: 10.1371/journal.pone.0022132
Hurka, F., Freuedener, S., Brown, H. D., and Plantholt, U. (1989). Aspartate aminotransferase isozyme in genus Capsella (Brassicacae), subcellular location, gene duplication and polymorphism. Biochem. Gen. 27, 77–90. doi: 10.1007/bf00563019
Hurka, H., and Neuffer, B. (1997). Evolutionary processes in the genus Capsella (Brassicaceae). Plant Sys. Evol. 206, 295–316. doi: 10.1007/BF00987954
Hurka, H., Bleeker, W., and Neuffer, B. (2003). Evolutionary process associated with biological invasions in the Brassicaceae. Biol. Invas. 5, 281–292. doi: 10.1023/b:binv.0000005571.19401.81
Hurka, H., Paetsch, M., Bleeker, W., and Neuffer, B. (2005). Evolution within the Brassicaceae. Nova Acta Leopoldina 92, 113–127.
Izawa, T., and Shimoto, K. (1996). Becoming a model plant: the importance of rice to plant science. Trends Plant Sci. 1, 95–99. doi: 10.1016/s1360-1385(96)80041-0
Jeon, J., Kim, N. Y., Kim, S., Kang, N. Y., Nova'k, O., Ku, S. J., et al. (2010). A subset of cytokinin two component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 285, 23371–23386. doi: 10.1074/jbc.M109.096644
Jiang, Y. P., Huang, L. F., Cheng, F., Zhou, Y. H., Xia, X. J., Mao, W. H., et al. (2013). Brassinosteroids accelerate recovery of photosynthetic apparatus from cold stress by balancing the electron partitioning, carboxylation and redox homeostasis in cucumber. Physiol. Plant. 148, 133–145. doi: 10.1111/j.1399-3054.2012.01696.x
Johnston, J. S., Pepper, A. E., Hall, A. E., Chen, Z. J., Hodnett, G., Drabek, J., et al. (2005). Evolution of genome size in Brassicaceae. Ann. Bot. 95, 229–235. doi: 10.1093/aob/mci016
Kang, G. Z., Zhu, Z. H., Guo, T. C., and Ren, J. P. (2009). Isolation and expression pattern of COR15b and KIN1 genes in watermelon and pumpkin. Afr. J. Biotechnol. 8, 5666–5672. doi: 10.5897/AJB09.1225
Karashima, Y., Talavera, K., Everaerts, W., Janssens, A., Kwan, K. Y., Vennekens, R., et al. (2009). TRPA1 acts as a cold sensor in vitro and in vivo. Proc. Nat. Acad. Sci. U.S.A. 106, 1273–1278. doi: 10.1073/pnas.0808487106
Kasuga, M., Miura, S., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). A combination of the Arabidopsis DREB1a gene and stress-inducible RD29a promoter improved drought- and low-temperature stress tolerance in tobacco by gene transfer. Plant Cell Physiol. 45, 346–350. doi: 10.1093/pcp/pch037
Kazemi-Shahandashti, S. S., Maali Amiri, R., Zeinali, H., and Ramezanpour, S. S. (2013). Change in membrane fatty acid compositions and cold-induced responses in chickpea. Mol. Biol. Rep. 40, 893–903. doi: 10.1007/s11033-012-2130-x
Kim, J. C., Lee, S. H., Cheong, Y. H., Yoo, C. M., Lee, S. I., Chun, H. J., et al. (2001). A novel cold-inducible zinc finger protein from soybean, SCOF-1, enhances cold tolerance in transgenic plants. Plant J. 25, 247–259. doi: 10.1046/j.1365-313x.2001.00947.x
Kim, M. H., Sasaki, K., and Imai, R. (2009). Cold shock domain protein 3 regulates freezing tolerance in Arabidopsis thaliana. J. Biol. Chem. 284, 23454–23460. doi: 10.1074/jbc.m109.025791
Kim, M. J., Ciani, S., and Schachtman, D. P. (2010). A peroxidase contributes to ROS production during Arabidopsis root response to potassium deficiency. Mol. Plant 3, 420–427. doi: 10.1093/mp/ssp121
Knight, H., and Knight, M. R. (2000). Imaging spatial and cellular characteristics of low temperature calcium signature after cold acclimation in Arabidopsis. J. Exp. Bot. 51, 1679–1686. doi: 10.1093/jexbot/51.351.1679
Knight, M. R., and Knight, H. (2012). Low-temperature perception leading to gene expression and cold tolerance in higher plants. New Phytol. 195, 737–751. doi: 10.1111/j.1469-8137.2012.04239.x
Koch, M. A., and Kiefer, M. (2005). Genome evolution among Cruciferous plants: a lecture from the comparison of the genetic maps of three diploid species- Capsella rubella, Arabidopsis lyrata ssp. petraea, and A. thaliana. Amer. J. Bot. 92, 761–767. doi: 10.3732/ajb.92.4.761
Kurkela, S., and Borg-Franck, M. (1992). Structure and expression of kin2, one of two cold- and ABA-induced genes of Arabidopsis thaliana. Plant Mol. Biol. 19, 689–692. doi: 10.1007/bf00026794
Lang, V., and Palva, E. T. (1992). The expression of a rab-related gene, rab 18, is induced by abscisic acid during the cold acclimation process of Arabidopsis thaliana (L.) Heyn. Plant Mol. Biol. 20, 951–962. doi: 10.1007/BF00027165
Larkindale, J., and Knight, M. R. (2002). Protection against heat stress-induced oxidative damage in Arabidopsis involves calcium, abscisic acid, ethylene, and salicylic acid. Plant Physiol. 128, 682–695. doi: 10.1104/pp.010320
Lee, S. C., Won, H. K., An, K., An, G., and Kim, S. R. (2004). Ectopic expression of a cold- inducible transcription factor, CBF1/DREB1b, in transgenic rice (Oryza sativa L.). Mol. Cells 18, 107–14.
Li, X. W., Feng, Z. G., Yang, H. M., Zhu, X. P., Liu, J., and Yuan, H. Y. (2010). A novel cold-regulated gene from Camellia sinensis. CsCOR1, enhances salt- and dehydration-tolerance in tobacco. Biochem. Biophys. Res. Commun. 394, 354–359. doi: 10.1016/j.bbrc.2010.03.011
Lin, J., Zhang, W., Zhou, X. W., Wang, X. L., Shi, M. Z., Sun, X. F., et al. (2007). Molecular cloning and characterization of cold-responsive gene Cbrci35 from Capsella bursa-pastoris. Biologia 62, 690–696. doi: 10.2478/s11756-007-0145-x
Lin, P., Wu, L., Wei, D., Chen, H., Zhou, M., Yao, X., et al. (2016). Promoter analysis of cold-responsive (COR) gene from Capsella bursa-pastoris and expression character in response to low temperature. Int. J. Agric. Biol. 18, 346–352. doi: 10.17957/ijab/15.0093
Lissarre, M., Ohta, M., Sato, A., and Miura, K. (2010). Cold-responsive gene regulation during cold acclimation in plants. Plant Signal Behav. 5, 948–952. doi: 10.4161/psb.5.8.12135
Liu, S. X., Wang, X. L., Fan, Z. Q., Pang, Y. Z., Sun, X. F., Wang, X. R., et al. (2004). Molecular cloning and characterization of a novel cold-regulated gene from Capsella bursa-pastoris. DNA Seq. 15, 262–268. doi: 10.1080/10425170400002421
Liu, W., Kenming, Y., Tengfei, H., Feifei, L., Dongxu, Z., and Jianxia, L. (2013). The Low Temperature Induced Physiological Responses of Avena nuda L., a Cold-Tolerant Plant Species. Sci. World J. 7:658793. doi: 10.1155/2013/658793
Liu, X., Wang, L., Liu, L., Guo, Y., and Ren, H. (2011). Alleviating effect of exogenous nitric oxide in cucumber seedling against chilling stress. Afr. J. Biotech. 10, 4380–4386. doi: 10.5897/AJB10.81
Llorente, F., Lopez-Cobollo, R. M., Catala, R., Martinez-Zapater, J. M., and Salinas, J. (2002). An ovel cold-inducible gene from Arabidopsis, RCI3, encodes a peroxidase that constitutes a component for stress tolerance. Plant J. 32, 13–24. doi: 10.1046/j.1365-313X.2002.01398.x
Lorbiecke, R., and Sauter, M. (1998). Induction of cell growth and cell division in the intercalary meristem of submerged deepwater rice (Oryza sativa L.). Planta 204, 140–145. doi: 10.1007/s004250050240
Ma, Q. B., Dai, X. Y., Xu, Y. Y., Guo, J., Liu, Y. J., Chen, N., et al. (2009). Enhanced tolerance to chilling stress in OsMYB3R-2transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 150, 244–256. doi: 10.1104/pp.108.133454
Majlath, I., Szalai, G., Soo's, V., Sebestye'n, E., Bala'zs, E., Vankova, R., et al. (2012). Effect of light on the gene expression and hormonal status of winter and spring wheat plants during cold hardening. Physiol. Plant 145, 296–314. doi: 10.1111/j.1399-3054.2012.01579.x
Mangano, S., Juarez, S. P., and Estevez, J. M. (2016). ROS regulation of polar growth in plant cells. Plant Physiol. 171, 1593–1605. doi: 10.1104/pp.16.00191
Maruyama, K., Urano, K., Yoshiwara, K., Morishita, Y., Sakurai, N., Suzuki, H., et al. (2014). Integrated analysis of the effects of cold and dehydration on rice metabolites, phytohormones, and gene transcripts. Plant Physiol. 164, 1759–1771. doi: 10.1104/pp.113.231720
Menges, M., Samland, A. K., Planchais, S., and Murray, J. (2006). The D-Type cyclin CYCD3; 1 is limiting for the G1-to-S-phase transition in Arabidopsis. Plant Cell. 18, 893–906. doi: 10.1105/tpc.105.039636
Miura, K., Sato, A., Shiba, H., Kang, S. W., Kamada, H., and Ezura, H. (2012a). Accumulation of antioxidants and antioxidant activity in tomato, Solanum lycopersicum, are enhanced by the transcription factor SlICE1. Plant Biotechnol. 29, 261–269. doi: 10.5511/plantbiotechnology.12.0303b
Miura, K., Shiba, H., Ohta, M., Kang, S. W., Sato, A., Yuasa, T., et al. (2012b). SlICE1 encoding a MYC-type transcription factor controls cold tolerance in tomato, Solanum lycopersicum. Plant Biotechnol. 29, 253–260. doi: 10.5511/plantbiotechnology.12.0303a
Mukhopadhyay, A., Vij, S., and Tyagi, A. K. (2004). Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. U.S.A. 101, 6309–6314. doi: 10.1073/pnas.0401572101
Nakashima, K., Fujita, Y., Katsura, K., Maruyama, K., Narusaka, Y., Seki, M., et al. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60, 51–68. doi: 10.1007/s11103-005-2418-5
Nakashima, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5:170. doi: 10.3389/fpls.2014.00170
Nakayama, K., Okawa, K., Kakizaki, T., Honma, T., Itoh, H., and Inaba, T. (2007). Arabidopsis Cor15am is a chloroplast stromal protein that has cryoprotective activity and forms oligomers. Plant Physiol. 144, 513–523. doi: 10.1104/pp.106.094581
Narsai, R., Castleden, I., and Whelan, J. (2010). Common and distinct organ and stress responsive transcriptomic patterns in Oryza sativa and Arabidopsis thaliana. BMC Plant Biol. 10:262. doi: 10.1186/1471-2229-10-262
Noman, A., Ali, Q., Naheed, F., Rizwan, M., Ali, S., and Irshad, K. (2015). Foliar application of asCORbate enhances the physiological and biochemical attributes of maize (Zea mays L.) cultivars under drought stress. Arch. Agron. Soil Sci. 61, 1659–1672. doi: 10.1080/03650340.2015.1028379
Noman, A., and Aqeel, M. (2017). miRNA-based heavy metal homeostasis and plant growth. Environ. Sci. Poll. Res. 24, 10068–10082. doi: 10.1007/s11356-017-8593-5
Noman, A., Aqeel, M., Deng, J., Khalid, N., Sanaullah, T., and Shuilin, H. (2017a). Biotechnological advancements for improving floral attributes in ornamental plants. Front. Plant Sci. 8:530. doi: 10.3389/fpls.2017.00530
Noman, A., Bashir, R., Aqeel, M., Anwer, S., Iftikhar, W., Zainab, M., et al. (2016). Success of transgenic cotton (Gossypium hirsutum L.): fiction or reality? Cogent Food Agric. 2:1207844. doi: 10.1080/23311932.2016.1207844
Noman, A., Fahad, S., Aqeel, M., Ali, U., Ullah, A., Anwer, S., et al. (2017b). miRNAs: major modulators for crop growth and development under abiotic stresses. Biotechnol. Lett. 39, 685–700. doi: 10.1007/s10529-017-2302-9
Novillo, F., Medina, J., and Salinas, J. (2007). Arabidopsis CBF1 and CBF3 have a different function than CBF2 in cold acclimation and define different gene classes in the CBF regulon. Proc. Nat. Acad. Sci. U.S.A. 104, 21002–21007. doi: 10.1073/pnas.0705639105
Paetsch, M., Mayland-Quellhorst, S., and Neuffer, B. (2006). Evolution of the self-incompatibility system in the Brassicaceae: identification of S-locus receptor kinase (SRK) in self-incompatible Capsella grandiflora. Heredity 97, 283–290. doi: 10.1038/sj.hdy.6800854
Patade, V. Y., Khatri, D., Kumari, M., Grover, A., Gupta, S. M., and Ahmed, Z. (2013). Cold tolerance inOsmotin transgenic tomato (Solanum lycopersicum L.) is associated with modulation in transcript abundance of stress responsive genes. Springer Plus 2:117. doi: 10.1186/2193-1801-2-117
Peng, X., Wu, Q., Teng, L., Tang, F., Pi, Z., and Shen, S. (2015). Transcriptional regulation of the paper mulberry under cold stress as revealed by a comprehensive analysis of transcription factors. BMC Plant Biol. 15:108. doi: 10.1186/s12870-015-0489-2
Qu, T., Liu, R., Wang, W., An, L., Chen, T., Liu, G., et al. (2011). Brassinosteroids regulate pectin methylesterase activity and AtPME41 expression in Arabidopsis under chilling stress. Cryobiology 63, 111–117. doi: 10.1042/BCJ20160238
Rensink, W. A., and Buell, C. R. (2004). Arabidopsis to rice. Applying knowledge from a weed to enhance our understanding of a crop species. Plant Physiol. 135, 622–629. doi: 10.1104/pp.104.040170
Richter, R., Bastakis, E., and Schwechheimer, C. (2013).Cross-repressive interactions between SOC1 and the GATAs GNC and GNL/CGA1 in the control of greening, cold tolerance, and flowering time in Arabidopsis. Plant Physiol. 162, 1992–2004. doi: 10.1104/pp.113.219238
Ruelland, E., Vaultier, M. N., Zachowski, A., and Hurry, V. (2009). Cold signaling and cold acclimation in plants. Adv. Bot. Res. 49, 35–150. doi: 10.1016/s0065-2296(08)00602-2
Shi, Y., Tian, S., Hou, L., Huang, X., Zhang, X., Guo, H., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell. 24, 2578–2595. doi: 10.1105/tpc.112.098640
Shimamura, C., Ohno, R., Nakamura, C., and Takumi, S. (2006). Improvement of freezing tolerance in tobacco plants expressing a cold responsive and chloroplast-targeting protein WCOR15 of wheat. J. Plant Physiol. 163, 213–219. doi: 10.1016/j.jplph.2005.06.008
Shurong, D., Jian, S., Zhao, R., Ding, M., Zhang, Y., Sun, Y., et al. (2015). Populus euphratica APYRASE2 enhances cold tolerance by modulating vesicular trafficking and extracellular ATP in Arabidopsis plants. Plant Physiol. 169, 530–548. doi: 10.1104/pp.15.00581
Si, J., Wang, J. H., Zhang, L. J., Zhang, H., Liu, Y. J., and An, L. Z. (2009). CbCOR15, a cold-regulated gene from alpine Chorispora bungeana, confers cold tolerance in transgenic tobacco. J. Plant Biol. 52, 593–601. doi: 10.1007/s12374-009-9077-z
Sivankalyani, V., Geetha, M., Subramanyam, K., and Girija, S. (2015). Ectopic expression of Arabidopsis RCI2A gene contributes to cold tolerance in tomato. Trans. Res. 24, 237–251. doi: 10.1007/s11248-014-9840-x
Snell, R., and Aarssen, L. W. (2005). Life history traits in selfing versus outcrossing annuals: exploring the ‘time-limitation’ hypothesis for the fitness benefit of self-pollination. BMC Ecol. 5:2. doi: 10.1186/1472-6785-5-2
Soltis, P. S., and Soltis, D. E. (2000). The role of genetic and genomic attributes in the success of polyploids. Proc. Nat. Acad. Sci. U.S.A. 97, 7051–7057. doi: 10.1073/pnas.97.13.7051
Steponkus, P. L., Uemura, M., Joseph, R. A., and Gilmour, S. J. (1998). Mode of action of the COR15a gene on the freezing tolerance of Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 95, 14570–14575. doi: 10.1073/pnas.95.24.14570
Stockinger, E. J., Gilmour, S. J., and Thomashow, M. F. (1997). Arabidopsis thaliana CBF1 encodes an AP2 domain-containing transcriptional activator that binds to the C-repeat/DRE, a cis-acting DNA regulatory element that stimulates transcription in response to low temperature and water deficit. Proc. Natl. Acad. Sci. U.S.A. 94, 1035–1040. doi: 10.1073/pnas.94.3.1035
Su, C. F., Wang, Y. C., Hsieh, T. H., Lu, C. A., Tseng, T. H., and Yu, S. M. (2010). A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 153, 145–158. doi: 10.1104/pp.110.153015
Tamminen, I., MaÈkelaÈ, P., Heino, P., and Palva, E. T. (2001). Ectopic expression of ABI3 gene enhances freezing tolerance in response to abscisic acid and low temperature in Arabidopsis thaliana. Plant J. 25, 1–8. doi: 10.1111/j.1365-313x.2001.00927.x
Tang, K. X., Wang, X. L., Sun, X. F., and Deng, Z. X. (2006). “Cold: a double-edged sword to plants,” in Floriculture, Ornamental and Plant Biotechnology, Vol. 3, ed J. A. Teixeira da Silva (Global Science Books), 102–107.
TheiSSen, G. (2006). The proper place of hopeful monsters in evolutionary biology. Theor. Biosci. 124, 349–369. doi: 10.1016/j.thbio.2005.11.002
Thomashow, M. F. (1999). Plant cold acclimation: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Thomashow, M. F., Gilmour, S. J., Stockinger, E. J., Jaglo-Ottosen, K. R., and Zarka, D. G. (2001). Role of the Arabidopsis CBF transcriptional activators in cold acclimation. Physiol. Plant. 112, 171–175. doi: 10.1034/j.1399-3054.2001.1120204.x
Turan, O., and Ekmekçi, Y. (2011). Activities of photosystem II and antioxidant enzymes in chickpea (Cicer arietinum L.) cultivars exposed to chilling temperatures. Acta Physiol. Plant. 33, 67–78. doi: 10.1007/s11738-010-0517-7
Vannini, C., Locatelli, F., Bracale, M., Magnani, E., Marsoni, M., Osnato, M., et al. (2004). Overexpression of the rice Osmyb4 gene increases chilling and freezing tolerance of Arabidopsis thaliana plants. Plant J. 37, 115–127. doi: 10.1046/j.1365-313X.2003.01938.x
Vogel, J. T., Zarka, D. G., Van Anbuskirk, H. A., Fowler, S. G., and Thomashow, M. F. (2005). Roles of the CBF2 andZAT12 transcription factors in configuring the low temperature transcriptome of Arabidopsis. Plant J. 41, 195–211. doi: 10.1111/j.1365-313x.2004.02288.x
Wang, L. J., and Li, S. H. (2006). Salicylic acid-induced heat or cold tolerance in relation to Ca2+ homeostasis and antioxidant systems in young grape plants. Plant Sci. 170, 685–694. doi: 10.1016/j.plantsci.2005.09.005
Wang, X., Liu, L., Liu, S., Sun, X., Deng, Z., Pi, Y., et al. (2004). Isolation and molecular characterization of a new CRT binding factor gene from Capsella bursa-pastoris. J. Biochem. Mol. Biol. 37, 538–545. doi: 10.5483/BMBRep.2004.37.5.538
Wang, Y., and Hua, J. (2009). A moderate decrease in temperature induces COR15a expression through the CBF signaling cascade and enhances freezing tolerance. Plant J. 60, 340–349. doi: 10.1111/j.1365-313X.2009.03959.x
Welling, A., and Palva, E. T. (2008). Involvement of CBF transcription factors in winter hardiness in birch. Plant Physiol. 147, 1199–1211. doi: 10.1104/pp.108.117812
Wilhelm, K., and Thomashow, M. F. (1993). Arabidopsis thaliana CORl5b, an apparent homologue of CORl5a, is strongly responsive to cold and ABA, but not drought. Plant Mol. Biol. 23, 1073–1077. doi: 10.1007/bf00021822
Winfield, M. O., Lu, C., Wilson, I. D., Coghill, J. A., and Edwards, K. J. (2009). Cold- and light-induced changes in the transcriptome of wheat leading to phase transition from vegetative to reproductive growth. BMC Plant Biol. 9:55. doi: 10.1186/1471-2229-9-55
Winfield, M. O., Lu, C., Wilson, I. D., Coghill, J. A., and Edwards, K. J. (2010). Plant responses to cold: transcriptome analysis of wheat. Plant Biotech. J. 8, 749–771. doi: 10.1111/j.1467-7652.2010.00536.x
Wu, L., Zhou, M., Shen, C., Liang, J., and Lin, J. (2012). Transgenic tobacco plants over expressing cold regulated protein CbCOR15b from Capsella bursa-pastoris exhibit enhanced cold tolerance. J. Plant Physiol. 169, 1408–1416. doi: 10.1016/j.jplph.2012.05.016
Xin, Z., and Browse, J. (2000). Cold comfort farm: the acclimation of plants to freezing temperatures. Plant Cell Environ. 23, 1365–3040. doi: 10.1046/j.1365-3040.2000.00611.x
Xu, J., Jun, Y., Xiaoguang, D., Yueming, J., and Peng, Z. (2014). Increased expression of native cytosolic Cu/Zn superoxide dismutase and ascorbate peroxidase improves tolerance to oxidative and chilling stresses in cassava Manihot esculenta Crantz. BMC Plant Biol. 14:208. doi: 10.1186/s12870-014-0208-4
Xu, J., Li, Y., Sun, J., Du, L., Zhang, Y., Yu, Q., et al. (2013). Comparative physiological and proteomic response to abrupt low temperature stress between two winter wheat cultivars differing in low temperature tolerance. Plant Biol. 15, 292–303. doi: 10.1111/j.1438-8677.2012.00639.x
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Ann. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1993). Characterization of the expression of a desiccation-responsive rd29 gene of Arabidopsis thaliana and analysis of its promoter in transgenic plants. Mol. Gen. Genet. 236, 331–340. doi: 10.1007/bf00277130
Yamaguchi-Shinozaki, K., and Shinozaki, K. (1994). A novel cis-acting element in an Arabidopsis gene is involved in responsiveness to drought, low temperature, or high-salt stress. Plant Cell 6, 251–264. doi: 10.2307/3869643
Yang, H., Wu, F., and Cheng, J. (2011). Reduced chilling injury in cucumber by nitric oxide and the antioxidant response. Food Chem. 127, 1237–1242. doi: 10.1016/j.foodchem.2011.02.011
Zafar, S., Ashraf, M. Y., Ali, Q., Ashraf, A., Anwer, S., Iqbal, N., et al. (2016). Antioxidant activity and secondary metabolites in selected vegetables irrigated with sewage water. App. Ecol. Environ. Res. 14, 35–48. doi: 10.15666/aeer/1405_035048
Zaynab, M., Kanwal, S., Furqan, M., Islam, W., Noman, A., Ali, G. M., et al. (2017). Proteomic approach to address low seed germination in Cyclobalnopsis gilva. Biotechnol. Let. doi: 10.1007/s10529-017-2393-3. [Epub ahead of print].
Zhai, H., Bai, X., Zhu, Y., Li, Y., Cai, H., Ji, W., et al. (2010). A single-repeat R3-MYB transcription factor MYBC1 negatively regulates freezing tolerance in Arabidopsis. Biochem. Biophys. Res. Commun. 394, 1018–1023. doi: 10.1016/j.bbrc.2010.03.114
Zhang, Y. J., Yang, J. S., Guo, S. J., Meng, J. J., Zhang, L., Wan, S. B., et al. (2010). Over-expression of the Arabidopsis CBF1 gene improves resistance of tomato leaves to low temperature under lowirradiance. Plant Biol. 13, 362–367. doi: 10.1111/j.1438-8677.2010.00365.x
Zhang, Z., Zhang, Q., Wu, J., Zheng, X., Zheng, S., Sun, X., et al. (2013). Gene knockout study reveals that cytosolic ascorbate peroxidase 2(OsAPX2) plays a critical role in growth and reproduction in rice under drought, salt and cold stresses. PLoS ONE 8:e57472. doi: 10.1371/journal.pone.0057472
Zhao, D. Y., Shen, L., Yu, M. M., Zheng, Y., and Sheng, J. P. (2009). Relationship between activities of antioxidant enzymes and cold tolerance of postharvest tomato fruits. Food Sci.14, 309–313.
Zhao, M., Liu, W., Xia, X., Wang, T., and Zhang, W. H. (2014). Cold acclimation-induced freezing tolerance of Medicago truncatula seedlings is negatively regulated by ethylene. Physiol. Plant. 152, 115–129. doi: 10.1111/ppl.12161
Zhou, M. Q., Shen, C., Wu, L. H., Tang, K. X., and Lin., J. (2011a). CBF-dependent signaling pathway: a key responder to low temperature stress in plants. Crit. Rev. Biotechnol. 31, 186–192. doi: 10.3109/07388551.2010.505910
Zhou, M. Q., Wu, L. H., Liang, J., Shen, C., and Lin, J. (2012a). Expression analysis and functional characterization of a novel cold-responsive gene CbCOR15a from Capsella bursa-pastoris. Mol. Biol. Rep. 39, 5169–5179. doi: 10.1007/s11033-011-1313-1
Zhou, M. Q., Wu, L. H., Liang, J., Shen, C., and Lin, J. (2012b). Cold induced modulation of CbICE53 gene activates endogenous cold responsive genes to enhance cold acclimation in transgenic tobacco. Mol. Breed. 30, 1611–1620. doi: 10.1007/s11032-012-9744-5
Zhou, M. Q., Wu, L. H., Shen, C., and Lin, J. (2010). Regulation of cold-responsive genes in CBF signaling pathway from Capsella bursa-pastoric induced by ABA, Me JA and SA. J. Agric. Sci. Technol. 12, 75–80.
Zhou, M. Q., Wu, L. H., Shen, C., and Lin, J. (2011b). A study on the regulation of the expression of cold responsive genes in CBF signaling pathway from Capsella bursa-pastoric induced by IAA and GA3. J. Sichuan Univ. (Nat. Sci. Ed.) 48, 201–205.
Zhou, M. Q., Xu, M., Wu, L., Shen, C., Ma, H., and Lin, J. (2014). CbCBF from Capsella bursa-pastoris enhances cold tolerance and restrains growth in Nicotiana tabacum by antagonizing with gibberellin and affecting cell cycle signaling. Plant Mol. Biol. 85, 259–275. doi: 10.1007/s11103-014-0181-1
Zhou, M., Li, W., Zheng, Y., Lin, P., Yao, X., and Lin, J. (2016). CbRCI35, a cold responsive peroxidase from Capsella bursa-pastoris regulates reactive oxygen species homeostasis and enhances cold tolerance in tobacco. Front. Plant Sci. 7:1599. doi: 10.3389/fpls.2016.01599
Zhu, B., Xiong, A. S., Peng, R. H., Xu, J., Jin, X. F., Memg, X. R., et al. (2010). Over-expression of ThpI from Choristoneura fumiferana enhances tolerance to cold in Arabidopsis. Mol. Biol. Rep. 37, 961–966. doi: 10.1007/s11033-009-9759-0
Keywords: Capsella, CBF, COR, cold tolerance, plant breeding, physiology
Citation: Noman A, Kanwal H, Khalid N, Sanaullah T, Tufail A, Masood A, Sabir S, Aqeel M and He S (2017) Perspective Research Progress in Cold Responses of Capsella bursa-pastoris. Front. Plant Sci. 8:1388. doi: 10.3389/fpls.2017.01388
Received: 11 April 2017; Accepted: 25 July 2017;
Published: 14 August 2017.
Edited by:
Mingqi Zhou, University of Florida, United StatesReviewed by:
Mirza Hasanuzzaman, Sher-e-Bangla Agricultural University, BangladeshGang Yu, Shanghai Center for Plant Stress Biology (PSC), China
Copyright © 2017 Noman, Kanwal, Khalid, Sanaullah, Tufail, Masood, Sabir, Aqeel and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Aqeel, YXFlZWxidXR0OTlAZ21haWwuY29t
Shuilin He, c2hsaGUyMDEzMDRAYWxpeXVuLmNvbQ==
 Ali Noman
Ali Noman Hina Kanwal
Hina Kanwal Noreen Khalid
Noreen Khalid Tayyaba Sanaullah
Tayyaba Sanaullah Aasma Tufail
Aasma Tufail Atifa Masood
Atifa Masood Sabeeh-ur-Rasool Sabir
Sabeeh-ur-Rasool Sabir Muhammad Aqeel
Muhammad Aqeel Shuilin He
Shuilin He