- 1National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan, China
- 2Microelement Research Center/Key Laboratory of Arable Land Conservation (Middle and Lower Reaches of Yangtze River), Ministry of Agriculture, Huazhong Agricultural University, Wuhan, China
- 3Southern Cross Plant Science, Southern Cross University, Lismore, NSW, Australia
Aquaporins (AQPs) are an abundant protein family and play important roles to facilitate small neutral molecule transport across membranes. Oilseed rape (Brassica napus L.) is an important oil crop in China and elsewhere in the world, and is very sensitive to low boron (B) stress. Several AQP family genes have been reported to be involved in B transport across plasma membranes in plants. In this study, a total of 121 full-length AQPs were identified and characterized in B. napus (AC genome), and could be classified into four sub-families, including 43 PIPs (plasma membrane intrinsic proteins), 35 TIPs (tonoplast intrinsic proteins), 32 NIPs (NOD26-like intrinsic proteins), and 11 SIPs (small basic intrinsic proteins). The gene characteristics of BnaAQPs were similar to those of BraAQPs (A genome) and BolAQPs (C genome) including the composition of each sub-family, gene structure, and substrate selectivity filters. The BnaNIP was the most complex AQP sub-family, reflecting the composition of substrate selectivity filter structures which affect the permeation of solution molecules. In this study, the seedlings of both B-efficient (QY10) and B-inefficient (W10) cultivars were treated with two boron (B) levels: deficient (0.25 μM B) and sufficient (25 μM B). The transcription of AQP genes in root (R), juvenile leaf (JL), and old leaf (OL) tissues of both cultivars was investigated under B deficient and sufficient conditions. Transcription of most BnaPIPs and BnaTIPs was significantly increased compared with other BnaAQPs in all the three tissues, especially in the roots, of both B-efficient and B-inefficient cultivars under both B conditions. With B deprivation, the expression of the majority of the BnaPIPs and BnaTIPs was down-regulated in the roots. However, the BnaNIPs were up-regulated. In addition, the BnaCnn_random.PIP1;4b, BnaPIP2;4s, BnaC04.TIP4;1a, BnaAnn_random.TIP1;1b, and BnaNIP5;1s (except for BnaA07.NIP5;1c and BnaC06.NIP5;1c) exhibited obvious differences at low B between B-efficient and B-inefficient cultivars. These results will help us to understand boron homeostasis in B. napus.
Introduction
Aquaporins (AQPs) are a superfamily of major intrinsic proteins (MIP) that selectively facilitate water and small neutral molecules across biological membranes (Maurel and Chrispeels, 2001). Recently, the AQP genes in the genomes of some plants have been identified, with 35 in Arabidopsis thaliana (A. thaliana), 60 in Brassica rapa (B. rapa), 67 in B. oleracea, 31 in maize, 33 in rice, 41 in common bean, and 30 in barley (Chaumont and Jung, 2001; Johanson et al., 2001; Sakurai et al., 2005; Tao et al., 2014; Ariani and Gepts, 2015; Diehn et al., 2015; Tombuloglu et al., 2015). Based on sequence similarity and subcellular localization, the AQPs are divided into five distinct sub-families in higher plants which consists of plasma membrane intrinsic proteins (PIPs), tonoplast intrinsic proteins (TIPs), NOD26-like intrinsic proteins (NIPs), small basic intrinsic proteins (SIPs), and uncategorized X intrinsic proteins (XIPs) (Ishikawa et al., 2005; Maurel et al., 2008; Wudick et al., 2009). AQP proteins of different sub-families share a conserved three-dimensional (3D) structure in cell membranes, although they vary in sequence, subcellular localization, and in planta physiological function, especially in substrate selectivity. Each AQP protein contains a six α-helical transmembrane structure (TM1-TM6), five loops (LA-LE), and two additional half-helices (HB, HE) (Daniels et al., 1999; Wallace et al., 2006). There are two selectivity filters within the protein structure that determine substrate specificity. One is composed of two highly conserved NPA motifs located in HB and HE, where HB and HE form a narrow central aqueous pore together, and the other is an ar/R (aromatic/arginine) selectivity filter consisting of four residues, one in transmembrane 2 (TM2), one in transmembrane 5 (TM5), and two in loop E (LE1 and LE2) (Wallace and Roberts, 2006; Hub and Groot, 2008; Mitaniueno et al., 2011).
Boron (B) is an essential micronutrient (Warington, 1923) for plant growth and development. Boron deficiency is a widespread problem for field crop production, causing large losses of crop yield annually both quantitatively (Wei et al., 1998) and qualitatively (Bell and Dell, 2008). AtNIP5;1 is involved in efficient B uptake in the roots of A. thaliana (Brassicaceae) under B deficiency (Takano et al., 2006) and further studies have indicated that the polar localization of NIP5;1 plays an important role in B absorption, which depends on threonine phosphorylation in the conserved three amino acid (ThrProGly, TPG) repeat of the N-terminal region, and is maintained by clathrin-mediated endocytosis (Wang S. et al., 2017). Moreover, the minimum open reading frame (AUG-stop, “AUGTAA”) in 5′UTR of AtNIP5;1 has been identified as the crucial element in response to cytoplasmic B concentration that regulates gene expression and induces ribosome stalling and mRNA degradation under high B conditions (Tanaka et al., 2016).
The Brassica genus diverged from a common ancestor with Arabidopsis around 18 MYA (Yang et al., 2006). B. napus (AnAnCnCn, 2n = 4x = 38) is an important oil crop species in the world, which most likely originated in domestication from the natural spontaneous hybridization between the diploid ancestors B. oleracea (CoCo, 2n = 2x = 18) and B. rapa (ArAr, 2n = 2x = 20) ~7,500 years ago, resulting in chromosome doubling (Chalhoub et al., 2014). In B. napus, we have previously identified six orthologous genes of NIP5;1, with BnaA03.NIP5;1b, and BnaC02.NIP5;1a proposed as candidate genes underlying the major-effect B efficiency QTLs qBEC-A3a and qBEC-C2a, respectively (Hua et al., 2016a,b). AtNIP6;1 is essential for preferential distribution of B into growing tissues under low B (Tanaka et al., 2008). Yeast complementation assays have confirmed that HvPIP1;3 and HvPIP1;4 can facilitate B transport across the membranes into yeast cells (Fitzpatrick and Reid, 2009). However, the overexpression of AtTIP5;1 and rice OsPIP2;4, OsPIP2;7, OsPIP1;3, and OsPIP2;6 in Arabidopsis notably increased its tolerance to high levels of B, and the latter two are involved the influx and efflux transport of B (Pang et al., 2010; Kumar et al., 2014; Mosa et al., 2015). In addition, AQP proteins can also facilitate transport of many other small molecule nutrients, such as Lis1 (OsNIP2;1) for silicon transport (Ma et al., 2006), AtTIP2;1 and AtTIP2;3 for NH3 transport (Loqué et al., 2005), AtNIP3;1 for arsenic uptake (Xu et al., 2015), and AtNIP1;2 facilitating Al-malate transport (Wang Y. et al., 2017), respectively.
The ancestral sub-genomes of Brassica species may be classified based on gene density and divergence, and have been nominated as LF (the least fractionized sub-genome), MF1 (the moderate gene fractionized sub-genome), and MF2 (the most gene fractionized sub-genome) (Cheng et al., 2012). A reference genome sequence of B. napus (cultivar Darmor-bzh) has been anchored to all 19 chromosomes (Chalhoub et al., 2014). In this study, we have identified and characterized a complete set of 121 full-length AQPs in the B. napus genome, which can be divided into four sub-families based on a phylogenetic tree that also incorporated A. thaliana orthologues. Phylogenetic relationships were established, and used as a context for investigating variation in gene structure, chromosomal and sub-genome distribution, and protein structural features of AQPs. The transcription of AQP genes in root (R), juvenile leaf (JL), and old leaf (OL) tissues of a B-efficient and a B-inefficient cultivar was also investigated under B deficient and sufficient conditions.
Materials and Methods
Identification of BnaAQP Genes in B. napus
AQP genes were identified in B. napus based on their homology similarity to Arabidopsis. The genome sequences and gene IDs of the 35 AQP genes were obtained from the TAIR10 database (http://www.arabidopsis.org/index.jsp) and used to perform a BLAST analysis and search for the AQP homologous genes in the CNS-Genoscope database (http://www.genoscope.cns.fr/brassicanapus/) (Chalhoub et al., 2014) and Brassica database (BRAD; http://brassicadb.org/brad/index.php), respectively. The genomic, cDNA, CDS, as well as protein sequences, of BnaAQPs were obtained from CNS-Genoscope database with inclusion of each AQP protein determined by checking the candidate genes using the Hidden Markov Model of the Pfam database (http://pfam.sanger.ac.uk/search), SMART database (http://smart.embl-heidelberg.de/), and NCBI Conserved Domain Search database (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi). Redundant sequences lacking either NPA motifs or ar/R selectivity filters were removed. Finally, the full-length BnaAQP family genes were identified. The BnaAQP genes were renamed according to the format: <genus (one letter)> <species (two letters)> <sub-genome (three letters)> <gene name (based on the orthologous gene name with Arabidopsis)> <allele (paralogous genes of each sub-genome according to their physical locations from the first chromosome to the next chromosome were named a, b, c, etc.)>, according to the standardized gene nomenclature for Brassica genus proposed by Østergaard and King (2008).
Multiple Alignment and Phylogenetic Analysis of BnAQP Family Genes
Multiple sequence alignment of protein sequences of B. napus and Arabidopsis was performed using ClustalW tool of MEGA 5.1 (Tamura et al., 2011). An unrooted phylogenetic tree of the 121 full-length AQP protein sequences was then constructed using MEGA 5.1 with the Neighbor Joining (NJ) method, and the analysis of bootstrap values was conducted using 1,000 replicates.
Chromosome Locations and Gene Structure Analysis of BnaAQP Genes
The chromosome location information of the BnAQP genes was obtained from BRAD. The MapInspect software was used to draw the gene chromosome location diagrams (Figure 2). The exon-intron structures of the BnAQP family genes were determined based on alignments of their coding sequences with the corresponding genomic sequences, and the diagram was drawn using GSDS (Gene structure display server: GSDS; http://gsds.cbi.pku.edu.cn/).
Conserved Motifs and Physicochemical Parameters Analysis of BnaAQP Proteins
Conserved motifs were identified by the MEME tool (Version 4.11.2) (http://meme-suite.org/tools/meme). The parameter settings were default values apart from: range of optimum width of each motif from 6 to 50 and maximum number of motifs to search: 15. The physicochemical parameters, including molecular weight (MW) and isoelectronic points (pI), of each BnaAQP protein were calculated using the compute pI/MW tool of ExPASy (http://www.expasy.org/tools/). GRAVY (grand average of hydropathy) values were calculated using the PROTPARAM tool (http://web.expasy.org/protparam/). Subcellular localization prediction was conducted using the WoLF PSORT (https://wolfpsort.hgc.jp/) server.
Syntenic Gene Analysis between A. thaliana and B. napus
Syntenic genes between A. thaliana and B. napus were searched by the online tool “syntenic gene analysis” (http://brassicadb.org/brad/searchSyntenytPCK.php) and each syntenic At-Bn ortholog in B. napus of AQP were obtained.
Transcriptional Profile of BnAQP Family Genes
The B. napus seedlings of the B-efficient cultivar Qingyou 10 (QY10) and the B-inefficient cultivar Westar10 (W10) were planted in Hoagland solution with 0.25 μM B (low B) and 25 μM B (high B) in an illuminated growth room (300–320 μmol m−2 s−1; 24°C daytime/22°C night; 16 h light/8 h dark) for 30 d. The roots (R), old leaves (OL, without petiole), and juvenile leaves (JL, without petiole) of QY10 and W10 were individually harvested, and each sample included three independent biological replicates. The total RNA of each sample was extracted using RNA extraction kit (BioTeke, Beijing, China) according to the manufacturer's recommendations. The RNA-seq sequencing libraries were subsequently sequenced using Illumina HiSeq™ 2000 (Illumina Inc., San Diego, CA, USA). All raw data were deposited in the NCBI Sequence Read Archive (Bioproject accession number, PRJNA393069). High-quality clean reads were mapped to the reference Darmor-bzh genome co-ordinates, and the RPKM (reads per kilo bases per million reads) measure was used to obtain the gene annotation and expression data. The RPKM values of genes were transformed to logarithmic values, which were used to produce a heat map using the program Multiviewer (MeV) (Saeed et al., 2003).
Results
Genome-Wide Identification and Phylogenetic Analysis of BnaAQP Gene Family in B. napus
The sequences and IDs of 35 AtAQP genes were used to carry out BLAST alignment against the B. napus reference genome, with annotation search used to identify 121 full-length BnaAQP genes as well as 15 non-full-length BnaAQP genes (Supplementary Table 1). The latter set lacked the transmembrane domains, resulting in the loss of the NPA motif and/or ar/R selectivity filter (Supplementary Table 1).
A phylogenetic tree was constructed based on the protein sequences of the 121 BnaAQP genes together with the 35 AtAQP genes (Figure 1). Four subgroups of BnaAQPs were readily identified, and coincided with the distribution of AtAQPs. All AtAQP genes had 1–7 corresponding orthologous genes in B. napus except from AtPIP2;8 and AtNIP1;1. Moreover, there were no XIP sub-family genes in either B. napus or A. thaliana. The B. napus sub-families included 43 PIPs, 35 TIPs, 32 NIPs, and 11 SIP members. The PIP sub-family had two sub-groups, 19 members in PIP1 and 24 in PIP2. The TIP sub-family was divided into five sub-groups (TIP1 to TIP5), nine members in TIP1, 13 in TIP2, 10 in TIP3, one in TIP4 and two in TIP5, respectively. SIPs were divided into the SIP1 (6 members) and SIP2 (5 members) sub-groups. The NIPs could be divided into seven sub-groups (NIP1 to NIP7), four members in NIP1, 2 and 6 sub-groups, six in NIP3, 4 and 5 sub-groups and two in NIP7 sub-group. Most (118 members) of the BnaAQPs, were clearly distinguished from each other and within the different sub-families and even different sub-groups of each sub-family. However, the BnaNIP4;1 and one BnaNIP4;2 appeared to have conserved amino acid sequences, distinguished not well each other. The orthologous genes of AtPIP1s, BnaA04.PIP1;1a, and BnaAnn_random.PIP1;1c appear to have undergone greater divergence from PIP1;2, although BnaA09.PIP1;1b and BnaC08.PIP1;1a, annotated as PIP1;2, were very close to PIP1;1 in the neighbor joining tree (Figure 1).
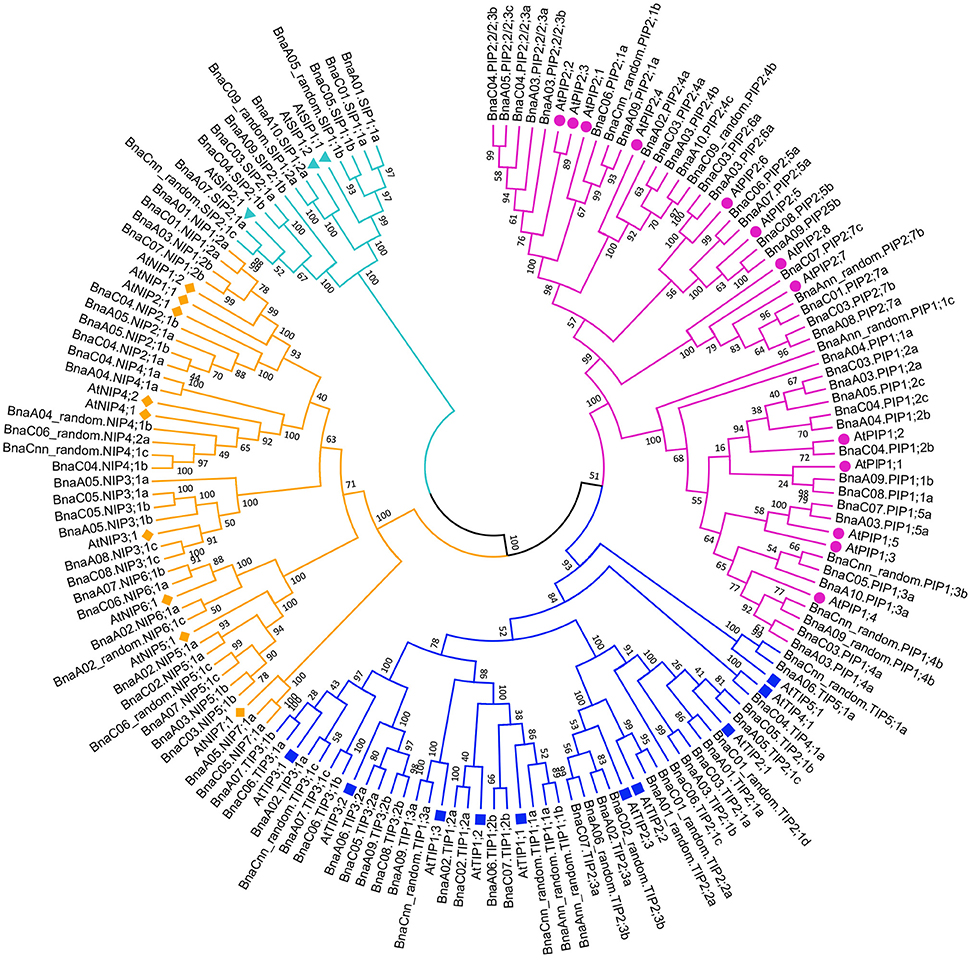
Figure 1. Phylogenetic tree of AQPs in Brassica crops. The AQPs phylogenetic tree was generated by MEGA 5.1 with Neighbor Joining (NJ) method and 1,000 replicates bootstraps, based on the amino acid sequences of 121 BnaAQP genes and 35 AtAQP genes. The gene name with purple, blue, yellow and cyan represented PIPs, TIPs, NIPs, and SIPs, respectively. AtAQPs, AtPIPs, AtTIPs, AtNIPs, and AtSIPs were marked with circle, square, diamond and triangle, respectively.
The Physicochemical Parameters of BnaAQPs
The length of BnaAQPs ranged from 206 aa (BnaAnn_random.PIP1;1c) to 323 aa (BnaC08.NIP3;1c), with BnaNIPs longer than the other BnaAQPs. The molecular weight (MW) ranged from 21.31 kDa (BnaA02.TIP2;3a) to 34.59 kDa (BnaA02_random.NIP6;1c), which was related to the protein length. All the BnaTIP proteins had a low pI (isoelectric point, pI < 7), except for BnaTIP5;1s. The majority of BnaPIPs and BnaNIPs, and all BnaSIPs proteins had pI > 7, while BnaPIP2;1s, BnaPIP2;2/2;3s, BnaPIP2;4s BnaC08.NIP3;1c, BnaC05.NIP7;1a, and BnaA05.NIP7;1a had a relatively low pI (pI < 7). The hydropathy value of all the amino acids was divided by the protein length, which was defined as grand average of hydropathicity (GRAVY). The GRAVY of all BnaAQPs were positive (ranging from 0.21 to 1.07; Table 1), indicating that all they were hydrophobic. Most proteins of each sub-family of BnaAQPs shared similar parameters, while different sub-families of BnaAQPs were distinguished from each other in the values of MW and pI (Table 1 and Supplementary Figure 1).
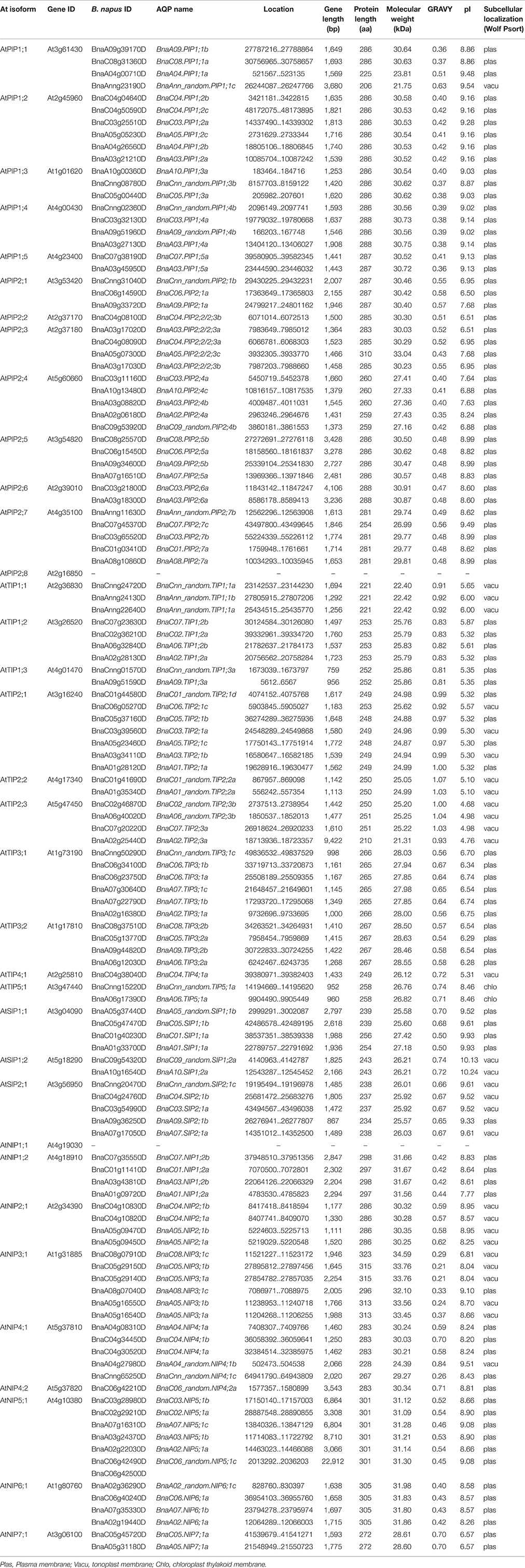
Table 1. Gene sequence characteristics of 121 BnaAQPs and their protein physicochemical parameters and putative subcellular localization.
The main types of subcellular localization for all 121 BnaAQPs were predicted to be plasma membrane (plas) and tonoplast membrane (vacu) (Table 1). 43% of TIPs were localized in the tonoplast membrane and almost all the BnaPIPs were localized in the plasma membrane. BnaNIPs were localized in plasma membrane except for BnaNIP2;1s and BnaNIP3;1s (tonoplast membrane). The members of BnaSIPs were equally localized in the plasma membrane and the tonoplast membrane.
Chromosomal Locations of BnaAQP Genes and Syntenic Block Analysis between A. thaliana and B. napus
Of the genes annotated in this paper as BnaAQPs, 95 full-length and 13 non-full-length had previously been incorporated in the genome scaffolds corresponding to 18 of the 19 psedochromosomes of B. napus (Figure 2), with the remainder located on scaffolds not yet anchored to a chromosome (Supplementary Table 2). The number of BnaAQPs located on each chromosome varied dramatically, with 12 on C04, two on A08 and none on C09. Genes within each BnaAQP subfamily were distributed unevenly on the chromosomes. Some clustering was observed at the top of chromosomes A05 and C04, the middle of A03 and C03, and the bottom of chromosome A07 and A09. There was an even distribution on each of the A (55) and C (53) genomes (Figure 2), with 76 full length BnaAQPs mapped to 17 syntenic blocks and three non-full-length BnaAQPs to two syntenic blocks (Table 2). Thirty three genes were located within the LF sub-genome with the relative proportion of 76 and 55% in MF1 (25) and MF2 (18), respectively, consistent with the total number of paralogues retained in these sub-genomes of both diploid ancestors (Liu et al., 2014).
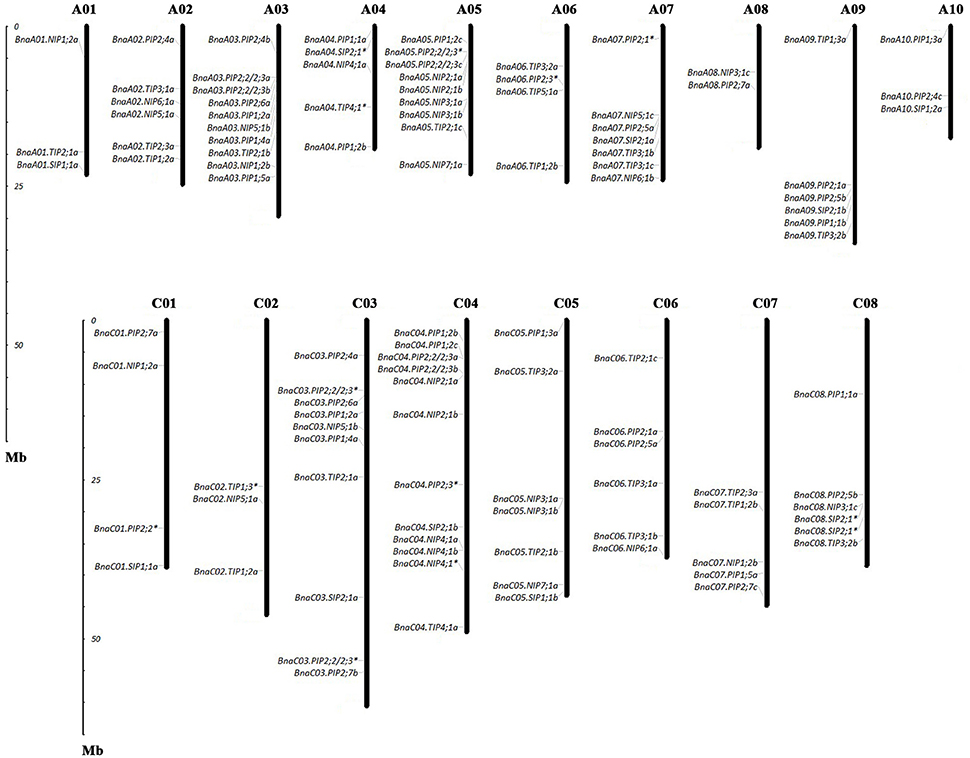
Figure 2. Chromosomal location of AQPs in Brassica napus. The gene chromosome location diagram was drawn using the MapInspect software. There were 95 full-length and 13 incomplete BnaAQPs located on 18 chromosomes. The genes marked with asterisk (*) represented the incomplete BnaAQPs.
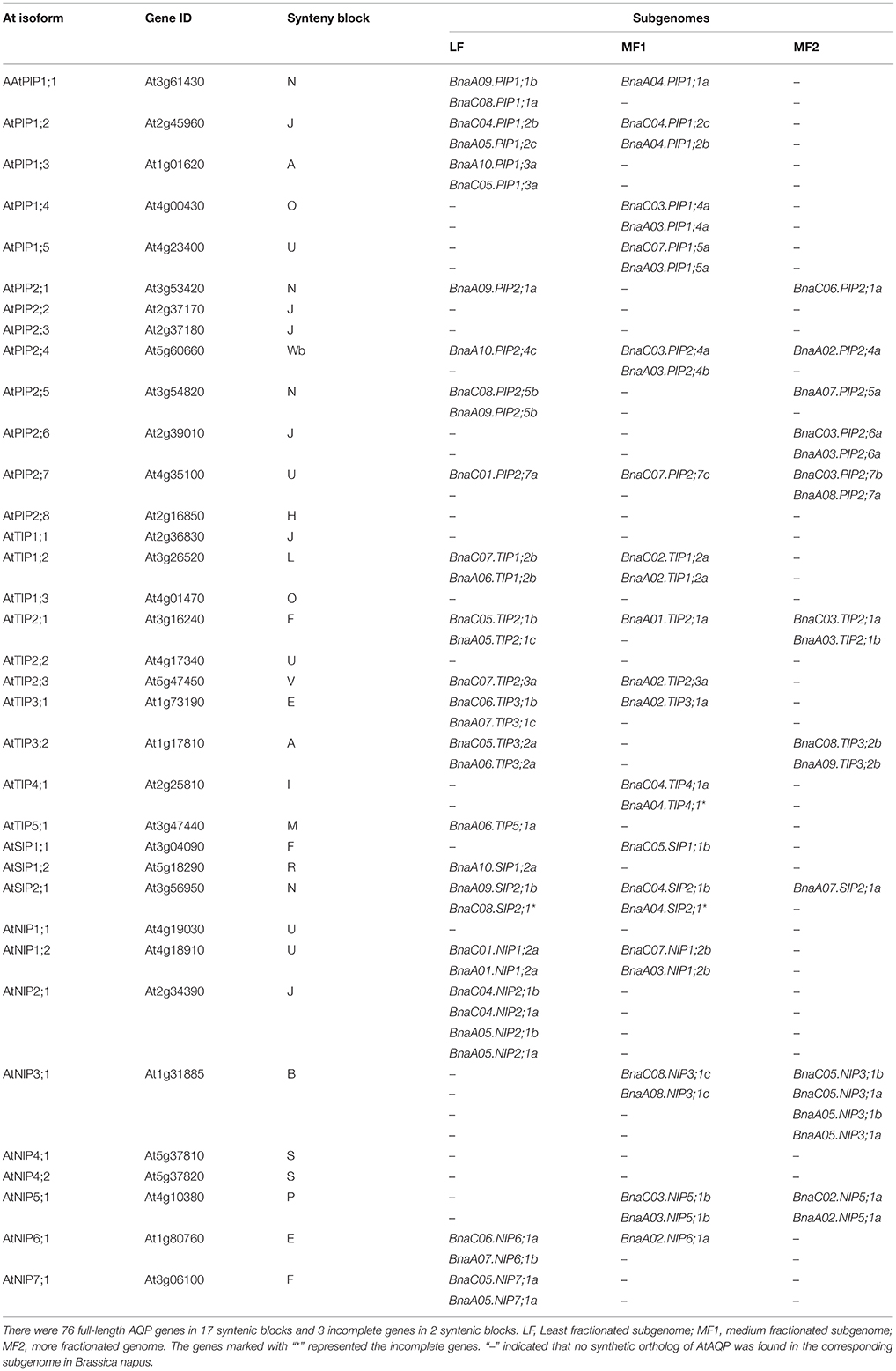
Table 2. Synteny block and distribution of aquaporin genes in the three sub-genomes of Brassica napus.
Gene Structure and Conserved Domains of BnaAQPs
The availability of the B. napus gene sequence enabled us to analyze and compare the gene structures of all 121 BnAQPs (Figure 3). As a whole, there was considerable variation in intron number between different BnaAQP sub-families. The majority of BnaPIPs had three introns, although BnaA10.PIP1;3a and BnaPIP1;5s had two, and BnaPIP2;4s, BnaA04.PIP1;1a and BnaC07.PIP2;7c had four. Within the BnaTIP sub-family 17 genes had two and 12 had one intron. All the BnaTIP3;2 genes had five introns, while two BnaTIP1;3s lacked an intron. Within the BnaNIP sub-family, 17 had four introns and 13, mostly BnaNIP2;1s and BnaNIP5;1s, had three introns. All SIP genes contained two introns. The gene structures of most BnaAQPs were conserved within a sub-family, and although there was some variation among homologous genes, they did not always show similarity amongst paralogous genes within the same sub-family. Additionally, the intron length changed frequently within the homologous genes of the isoforms of BnaPIP2;5, BnaPIP2;6, BnaNIP4;1, and BnaNIP5;1.
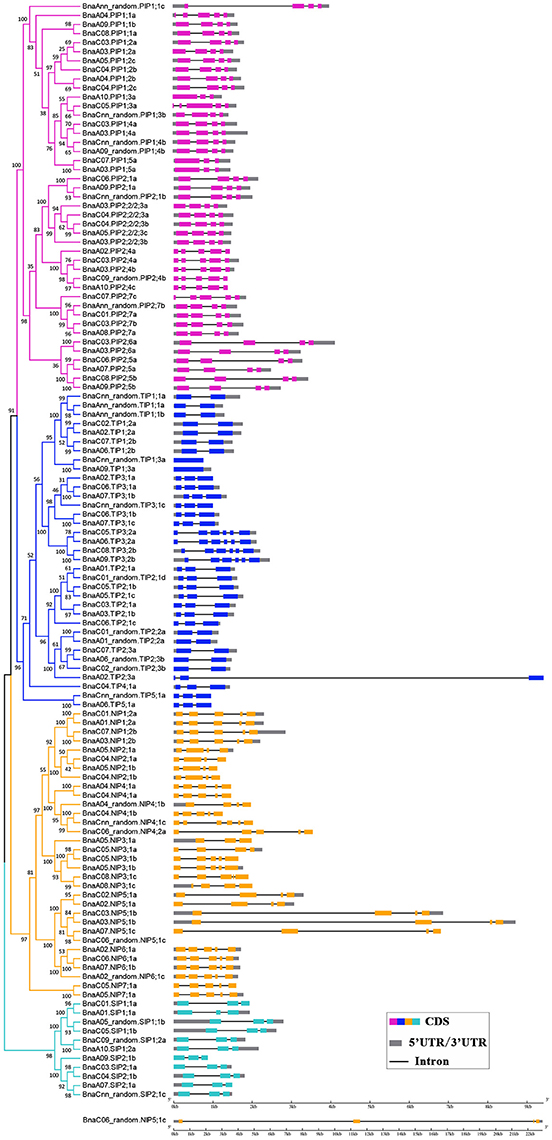
Figure 3. Gene structure of BnaAQPs. The exon-intron structures of the BnAQPs were determined by the alignments of coding sequences with corresponding genomic sequences, and the diagram was obtained using GSDS (Gene Structure Display Server 2.0) Web server. The purple, blue, yellow, and cyan rectangles represented the exons of BnaPIPs, BnaTIPs, BnaNIPs, and BnaSIPs, the gray rectangles represented the UTR and the black lines represented the introns, respectively. BnaC06_random.NIP5;1c was list alone because it had the longest gene length.
In addition, the conserved domains of all BnaAQPs were analyzed and fifteen motifs identified as conserved (Figure 4). Generally, the BnaAQPs within each sub-group shared similar motif compositions, but among different sub-groups their motif compositions varied. Motif 4 was common among all BnaAQPs, and was determined as a conserved region of BnaAQPs (Supplementary Figure 2C). Motif 5 was conserved in BnaPIPs, BnaTIPs and BnaNIPs (Supplementary Figure 2D). Two NPA motifs of all BnaAQPs were included in motif 1 and motif 2 (Supplementary Figures 2A,B). The BnaSIP family had the lowest number of motifs, and shared the lowest similarities with the proteins of other BnaAQP sub-families. The number and type of motifs of BnaNIPs changed frequently, which indicated that the protein diversity of this sub-family was high in B. napus.
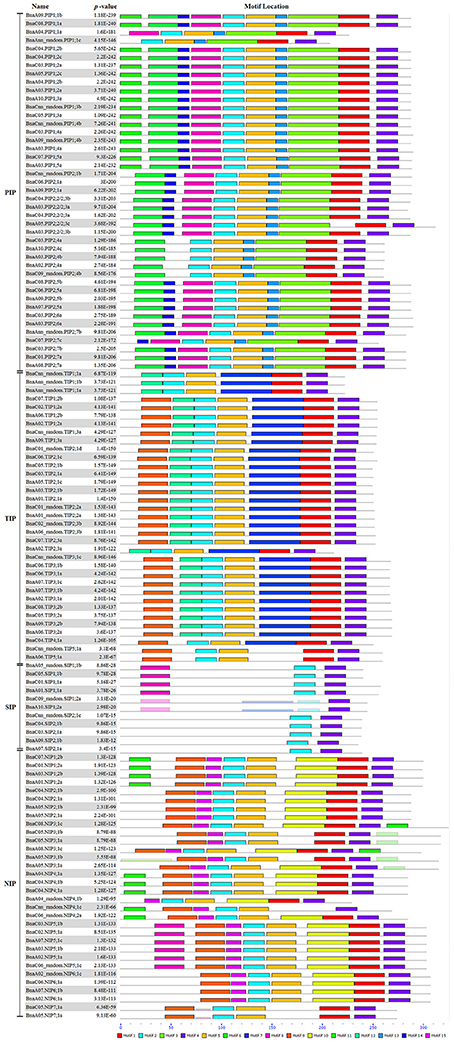
Figure 4. Distribution of conserved motifs in BnaAQPs. Conserved motifs of BnaAQPs were analyzed by MEME Web serve using the protein sequences of 121 full-length BnaAQPs. Fifteen conserved motifs were identified, and different motifs were distinguished by different colors.
The Comparison of NPA motifs and ar/R Selectivity Filters in BnaAQPs
NPA motifs and ar/R selectivity filters in BnaAQPs were identified since they are important for determining the permeation of solution molecules (Table 3). Two NPA motifs were identical in both BnaTIPs and BnaPIPs and all the NPA motifs in each sub-family were conserved. Additionally, the ar/R selectivity filters of BnaPIPs were also identical within different sub-groups, but that of BnaTIPs changed frequently. The second NPA motifs in BnaSIPs were more conserved than the first. The first NPA motifs of BnaSIP2;1s were present in two styles (NPV and NPL). The NPA motif and ar/R selectivity filters of BnaNIPs showed the greatest variation, particularly the ar/R selectivity filters of BnaNIP5;1s (Table 3). Moreover, the amino acids of H5 (BnaC06_random.NIP5;1c and BnaA07.NIP5;1c: A, N, A, R) and LE1 (BnaA03.NIP5;1b and BnaC03.NIP5;1b: A, I, A, R) differed from that of AtNIP5;1 (A, I, G, R) (Wallace and Roberts, 2004). In addition, we found that all the novel differences in the selectivity filters of BnaAQPs were paired in the corresponding A and C genomes (Table 3).
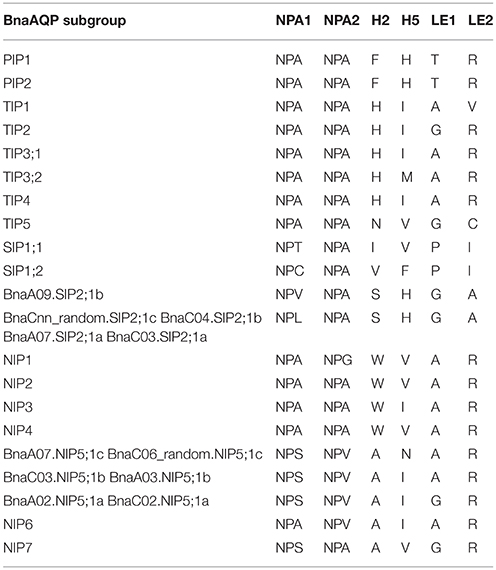
Table 3. Amino acid composition of the NPA motifs and ar/R selectivity filters of AQPs in Brassica napus.
Gene Expression of BnaAQPs in Response to Low B Stress
RNA-seq. was used to investigate the transcriptional profile of all the BnaAQPs in contrasting B-efficient (“Qingyou10”) and B-inefficient (“Westar10”) cultivar under B sufficient and deficient conditions. A total of 99 BnaAQPs (81%) were up-regulated or down-regulated in roots and/or leaves of both cultivars. The heat map displayed the transcript abundance pattern of BnaAQPs in roots (R), old leaves (OL), and juvenile leaves (JL) (Figure 5).
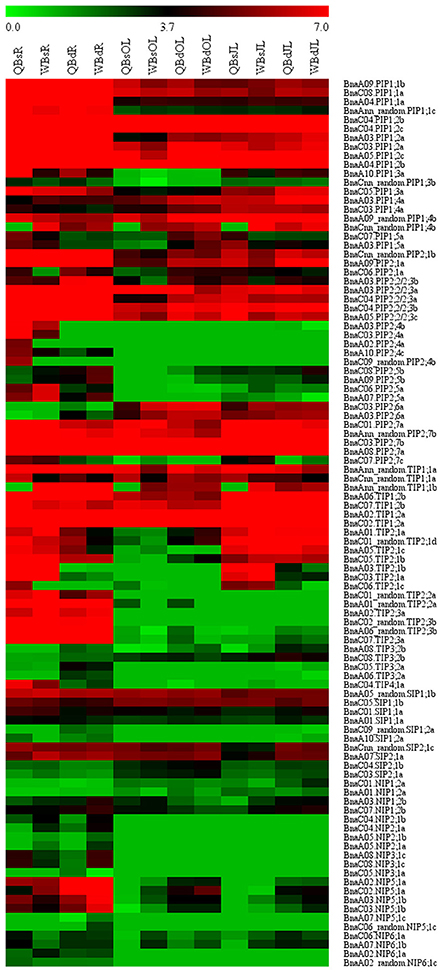
Figure 5. Expression profile of BnaAQPs in response to low boron stress in Brassica napus. The RPKM (reads per kilo bases per million reads) values of 99 BnaAQPs (non-expressed genes were not included) were obtained from the RNA-seq data, and then the RPKM values were converted into the logarithmic values. The heat map of expression profile of BnaAQPs in roots (R), old leaves (OL), and juvenile leaves (JL) was generated using the MeV program. The two cultivars of Brassica napus, QY10 (Q, B-efficient), and W10 (W, B-inefficient), were cultured in hydroponic solution with B deficient conditions (d, 0.25 μM) and B sufficient conditions (s, 25 μM) for 30 d.
Most of the BnaPIPs showed relatively higher expression levels in all the three tissues of B-efficient and B-inefficient cultivars compared with other sub-families of BnaAQPs. Moreover, approximately half of the BnaPIPs, e.g., BnaPIP1;1s, BnaPIP1;2s, and BnaPIP2;2/2;3s, had high expression levels in roots under the B sufficient condition; and they were dramatically down-regulated under low B stress in both cultivars (Figure 5 and Supplementary Table 3). A majority of BnaPIPs were expressed in all three tissues in both cultivars and both B levels, including BnaPIP1;1s, BnaPIP1;2s, BnaPIP1;4s (expect for BnaCnn_random.PIP1;4b), BnaPIP2;1s, BnaPIP2;2/2;3s, and BnaPIP2;7s (expect for BnaC07.PIP2;7c) (Figure 5 and Supplementary Table 3). However, several other BnaPIPs had tissue-preferential expression patterns in both cultivars, e.g., BnaPIP1;3s, BnaPIP2;4s, BnaPIP2;5s, and BnaPIP2;6s. Under B deprivation, the expression of BnaCnn_random.PIP1;4b was significantly up-regulated in all the three tissues of Qingyou10, but not in Westar10. However, the BnaPIP2;4 members were strikingly down-regulated in the roots of the two cultivars, particularly in Qingyou10. In addition, BnaC08.PIP2;5b and BnaA09.PIP2;5b showed up-regulation, while BnaC06.PIP2;5a and BnaA07.PIP2;5a were down-regulated with B deprivation.
BnaTIP1;1s and BnaTIP1;2s showed high expression levels in all tissues, whereas BnaTIP1;3s, BnaTIP3;1s, and BnaTIP5;1s were rarely expressed in any tissues. BnaTIP2;2s and BnaTIP2;3s showed an obvious root-preferential expression pattern in both B-efficient and B-inefficient cultivars at both B levels (Figure 5 and Supplementary Table 3). Under B deprivation, BnaC06.TIP2;1c, BnaC03.TIP2;1a, and BnaA03.TIP2;1b were distinctly down-regulated in roots and leaves of the two cultivars, while BnaC04.TIP4;1a was only down-regulated in roots. The degree of down-regulation for BnaC04.TIP4;1a in Qingyou10 was much greater than that for Westar10. BnaAnn_random.TIP1;1b only showed significantly up-regulated at low B in Qingyou10.
The expression levels of all the BnaSIPs in all tissues had slightly changed under B deprivation, and there was no significant difference between the B-efficient and B-inefficient cultivars. Moreover, transcription of all BnaSIPs varied in all tissues, apart from BnaA09.SIP2;1b which had no transcripts detected in any tissue, and BnaSIP1;2s which was not transcribed in leaves.
In comparison with other BnaAQPs, the expression levels of BnaNIPs were generally relatively lower in all the three tissues of B-efficient and B-inefficient cultivars under both B conditions. BnaA08.NIP3;1c, BnaC08.NIP3;1c, BnaA02_random.NIP6;1c, BnaA02.NIP6;1a as well as BnaNIP2;1s showed a root-specific expression pattern in both cultivars at low and high B levels (Figure 5 and Supplementary Table 3). The transcription of BnaC05.NIP3;1b, BnaA05.NIP3;1b, BnaNIP4;1s, BnaNIP4;2s, and BnaNIP7;1s was not detected in any tissue in both cultivars. In contrast, the transcription of BnaC03.NIP5;1b, BnaC02.NIP5;1a, BnaA03.NIP5;1b, and BnaA02.NIP5;1a was induced at low B in all three tissues of both cultivars, especially in the roots. Interestingly, the level of transcription for all the BnaNIP5;1s mentioned above in the B-inefficient cultivar (Westar10) was significantly higher than that in the B-efficient cultivar (Qingyou10).
Discussion
AQP Characterization in Brassica napus
Aquaporin is an abundant and high diversity protein family in plants, which facilitates transport of diverse small neutral molecules such as H2O, H2O2, boric acid, arsenic acid, silicic acid, urea and glycerol across membrane (Wallace et al., 2002; Ma et al., 2006, 2008; Takano et al., 2006; Hove and Bhave, 2011). Previously, AQP genes have been identified within the Brassicaceae, with 60 in the diploid A genome of B. rapa (Tao et al., 2014), 67 in the diploid C genome of B. oleracea (Diehn et al., 2015), and 35 in A. thaliana (Johanson et al., 2001). B. napus is a recent allopolyploid that originated by combining the intact genomes of B. oleracea and B. rapa. Gene replication and chromosome rearrangement in B. rapa and B. oleracea is therefore expected to result in a conserved gene distribution of AQP in the respective A and C chromosomes of B. napus. In this study, 121 full-length AQP family genes were identified in B. napus (Table 1), only slightly fewer than the sum of those reported in B. oleracea (Diehn et al., 2015) and B. rapa (Tao et al., 2014). Due to the origin and evolutionary independence of the two diploids (B. rapa and B. oleracea) over the past 4.6 MYA (Liu et al., 2014), the chromosomal location of BnaAQPs in the A genome are not completely conserved in homoeologous regions of the C genome (Figure 2).
In this study, we were able to allocate BnaAQPs according to the four well-established sub-families (43 PIPs, 35 TIPs, 32 NIPs, and 11 SIPs). As expected, no XIP sub-family was detected consistent with their absence in Arabidopsis, B. oleracea and B. rapa (Johanson et al., 2001; Tao et al., 2014; Diehn et al., 2015). There was also evidence of strong conservation within each BnaAQP sub-family since the allopolyploidisation of B. napus ~7,500 years ago, with membership closely matching the sum of B. rapa (23 PIPs, 16 TIPs, 15 NIPs, and 6 SIPs) and B. oleracea (25 PIPs, 19 TIPs, 17 NIPs, and 6 SIPs).
Comparison of collinearity between B. napus and orthologous Arabidopsis genes showed that 63% (76 of 121) of the AQP genes were present in 17 syntenic blocks, and these genes were distributed in proportion among the three ancestral Brassica sub-genomes of LF, MF1, and MF2 (Table 2). There was evidence of gene loss within the annotated sub-genomes of B. napus compared with the genomes of B. rapa and B. oleracea. The AQP gene membership of the LF, MF1, and MF2 sub-genomes in B. napus was 33, 25, and 18, however, there were 24, 16, and 13 AQP gene members in B. rapa and 30, 18, and 16 in B. oleracea, respectively (Tao et al., 2014; Diehn et al., 2015). The BnaAQPs density in the LF sub-genome was much higher than that of the other two MFs in B. napus, which is in proportion with the total number of all genes previously allocated to these categories for B. rapa and B. oleracea (Liu et al., 2014; Tao et al., 2014; Diehn et al., 2015). The two-step triplication that has been proposed for diploid Brassica species is based on evidence that a tetraploidization event was followed by substantial genome fractionation (MF1 and MF2), and subsequent hybridization with a third, less-fractionated sub-genome (LF) (Wang et al., 2011; Tang et al., 2012; Cheng et al., 2013). Most homoeologous AQP genes are represented in both A and C genomes of B. napus (Table 2). However, 17 AQPs were present only in the A or C genome, such as BnaA04.PIP1;1a. This may have arisen due to AQP gene loss or chromosome rearrangement either during the evolution of diploid Brassica species or subsequent to the formation of B. napus (Nicolas et al., 2007; Jeonghwan et al., 2009; Liu et al., 2014). Although 121 AQP genes were identified as full-length BnaAQPs (Table 1), 15 had an incomplete protein structure (Supplementary Table 1), mostly lacking fragments containing the conserved motifs corresponding to the NPA and ar/R selectivity filters.
The NPA motifs and ar/R selectivity filters of BnaAQPs showed variation among sub-families (Table 3). Within the PIP sub-families these were highly conserved in A. thaliana, B. rapa, B. oleracea, and B. napus. In addition, compared with those in barley (Tombuloglu et al., 2015), common bean (Ariani and Gepts, 2015) and rubber tree (Zhi et al., 2015), the NPA motifs and ar/R selectivity filters of BnaPIPs were also highly conserved, which suggested that the PIPs have been subject to strong selection in different plant taxonomic lineages. The Val (V) and Ile (I) residues near to P3 site have been considered as the conserved amino acids for water channel activity (Suga and Maeshima, 2004), and their differentiation can be used to distinguish PIP1s and PIP2s. In this study, all BnaPIP2s had the Val (V) and Ile (I) residues near P3 site, except for BnaPIP2;5s. However, all PIP1s had the Ile (I) and Ile (I) residues (Supplementary Data 1), which suggested that the BnaPIPs could be separated to BnaPIP1s and BnaPIP2s by the two amino acids.
All TIP sub-families of B. napus, B. rapa, B. oleracea, and A. thaliana shared two identical NPA motifs in HB and HE, although there still existed some differences in the ar/R selectivity filters. In Arabidopsis, the ar/R selectivity filters of TIPs were divided into 3 large subgroups (group I, group II and group III), with group II consisting of group IIA and group IIB (Wallace and Roberts, 2004). AtTIP3;2s belongs to group IIB, although all BnaTIP3;2s had a novel compositions of ar/R selectivity filters (H, M, A, R) (Table 3), which were the same as those of B. rapa and B. oleracea, but different from A. thaliana (H, I, A, R) (Tao et al., 2014; Diehn et al., 2015). The ar/R selectivity filters in TIP5s of the four species were considerably diverged from other TIPs (Table 3) (Tao et al., 2014; Diehn et al., 2015), indicating that TIP5s probably have a special function for solute permeation. Additionally, BnaTIP5;1s had a high pI and a distinct subcellular localization from the other BnaTIP members (Table 1). TIPs usually facilitate H2O, NH3 and urea transport (Wudick et al., 2009). Overexpression of AtTIP5;1 in A. thaliana significantly increases the tolerance to B toxicity (Pang et al., 2010), and the vacuolar compartmentation could explain the high B tolerance of Arabidopsis overexpression lines.
BnaNIPs and BnaSIPs showed larger variation in the third alanine of the NPA motifs, and a greater diversity in ar/R selectivity filters than BnaPIPs and BnaTIPs (Table 3), suggesting that they may have acquired a more specialized function for substrate absorption than the latter. In A. thaliana, the AtNIPs showed a large-scale permeation for substrates such as H2O, H2O2, As, B, Si, urea, and glycerol, which was consistent with their abundant amino acid constitutions of NPAs and ar/R selectivity filters of NIP sub-family (Mitaniueno et al., 2011).
The NPAs and ar/R selectivity filters of BnaAQPs were similar to those of the combination of BraAQPs and BolAQPs, but the specific ar/R selectivity filters in BraA.NIP4.c and BolC.NIP4.d (W, S, A, R) (Diehn et al., 2015) had been lost during the allopolyploidisation or subsequent domestication of B. napus (Table 3). The ar/R selectivity filters of BnaTIP3;2s and BnaNIP5;1s differed from those of their homologous genes in A. thaliana, which is consistent with the finding in B. oleracea (Tao et al., 2014). Moreover, all the novel differences in the selectivity filters of BnaAQPs were paired in the corresponding A and C genomes (Table 3), which suggested that the differentiation may have occurred before the species divergence of B. rapa and B. oleracea.
The majority of BnaAQPs were conserved both in the number and the position of introns among different paralogous genes in B. napus (Figure 3), although the intron lengths varied widely, such as BnaPIP2;5s, BnaPIP2;6s, BnaTIP2;3s, BnaNIP4;1s, and BnaNIP5;1s. The intron length of these paralogous genes had possibly undergone some changes since the relatively recent emergence of B. napus. In addition, the conserved domains of the four sub-families were significantly different, especially the SIP sub-family, which is in agreement with the result that the SIP sub-family was distant from the other three sub-families presented in the phylogenetic tree (Figure 1). However, the constitution of the predicted conserved domains of BnaAQPs within each sub-family was very similar apart from the NIP sub-family.
BnaAQPs and B Homeostasis in Brassica napus
Recent studies have shown that AtNIP5;1, AtNIP6;1, AtTIP5;1, OsPIP2;4, OsPIP2;7, OsPIP1;3, OsPIP2;6, and HvNIP2:1 play an important role in plant B homeostasis (Takano et al., 2006; Tanaka et al., 2008; Pang et al., 2010; Kumar et al., 2014; Mosa et al., 2015). In many cases, PIPs function as the water channels which mediate efficient water uptake in plant cells (Alexandersson et al., 2005; Boursiac et al., 2008; Mahdieh et al., 2008). In this study, the gene expression levels of most BnaPIPs were relatively higher than other BnaAQPs in all the tissues, especially in the roots under both B conditions (Figure 5 and Supplementary Table 3), which might be consistent with their functions as the water channel. Under B deficiency, the expression of most BnaPIPs was down-regulated in the roots of both cultivars (Figure 5 and Supplementary Table 3), indicating that they might be involved in the responses to the B deprivation in B. napus. Additionally, BnaPIP2;4s had high expression levels in the roots of both cultivars under the normal B condition, which was similar to their orthologues in Arabidopsis (Alexandersson et al., 2010), however, the gene expression of PIP2;4s in B. rapa and B. oleracea was not detected.
BnaTIP1;1s (except for BnaAnn_random.TIP1;1b) and BnaTIP1;2s displayed high expression levels in all tissues of both cultivars at two B levels and their expression was repressed in the roots with low B stress (Figure 5 and Supplementary Table 3), although recent studies report that the losses of AtTIP1;1 and AtTIP1;2 do not influence the growth of A. thaliana significantly (Schüssler et al., 2008; Beebo et al., 2009), thus suggesting that new functions for these genes in plants still need to be discovered. BnaTIP2;2s and BnaTIP2;3s clearly showed root-specific expression patterns, which was similar to that of B. oleracea (Diehn et al., 2015), B. rapa (Tao et al., 2014), and Arabidopsis (Alexandersson et al., 2010). Moreover, the BnaTIP2;2s and BnaTIP2;3s was strongly down-regulated at low B in both cultivars (Figure 5).
The gene expression levels of all the BnaSIPs were relatively low in all the tissues of both B-efficient and B-inefficient cultivars under low B condition (Figure 5). Hence, the function of BnaSIP genes was independent of the B supplement in this study. As a whole, at both B levels, the expression levels of BnaNIPs of both cultivars were lower than that of the other three sub-families in all tissues (Figure 5). Four members of BnaNIP5;1s (BnaA02.NIP5;1a, BnaC02.NIP5;1a, BnaA03.NIP5;1b, and BnaC03.NIP5;1b) exhibited significantly up-regulation whether in the root or in the leaves at low B stress, suggesting that they were not only involved in the B absorption in the root, but also in the response to low B in the shoot of B. napus.
Some AQP genes have tissue-specific expression characteristics. For instance, AtTIP1;3s, AtTIP5;1s, AtNIP4;1s, and AtNIP4;2s have been assigned as pollen-specific genes (Soto et al., 2008; Giorgio et al., 2016) and AtNIP7;1s showed an anther-specific expression pattern (Li et al., 2011). Thus, it is not surprising that transcription of BnaTIP1;3s, BnaTIP3;1s, BnaTIP5;1s, BnaNIP4;1s, BnaNIP4;2s, and BnaNIP7;1s was not detected in roots, old leaves and juvenile leaves of 30 d seedlings of QY10 and W10 under both B conditions in this study.
Significant differences in the transcription of several AQP genes between B-efficient cultivar QY10 and B-inefficient cultivar W10 were observed. This included BnaCnn_random.PIP1;4b, BnaC04.TIP4;1a, BnaAnn_random.TIP1;1b, BnaC03.NIP5;1b, BnaC02.NIP5;1a, BnaA03.NIP5;1b, and BnaA02.NIP5;1a (Figure 5 and Supplementary Table 3). Of these, we have previously mapped BnaA03.NIP5;1b as a B-efficient QTL qBEC-A3a detected in B. napus (Hua et al., 2016a). Moreover, the other member BnaC02.NIP5;1a, was also identified as a B-efficient QTL qBEC-C2a, and possibly facilitates efficient boron absorption under low B condition in B. napus (Hua et al., 2016b).
In summary, we identified and characterized 121 full-length AQP genes which belong to the PIP, TIP, SIP and NIP sub-families. The characteristics of gene structure, selectivity filters and transcriptional patterns for most AQP genes of B. napus were similar to those of B. rapa, B. oleracea, and Arabidopsis. We confirmed the identity and relationship of two candidate genes (BnaA03.NIP5;1b and BnaC02.NIP5;1a) underlying B-efficient QTL regions in B. napus. We found that most BnaPIPs and BnaTIPs had high expression levels in all the tissues of B-efficient and B-inefficient cultivars under both B conditions, especially in roots. Moreover, under low B condition, the transcription of these genes was down-regulated in roots although their responses to low B stress in leaves were only slight or with no change.
Author Contributions
Conceptual and experiment designs by LS and DY; Experiments were conducted by DY; RNA-seq data analysis performed by WL; Reagents/materials/analysis tools were contributed by LS, and the report was written by DY, YH, GK, FX, and LS. All the authors have commented, read and approved the final manuscript.
Funding
The authors acknowledge the financial support from the National Nature Science Foundation of China (Grant No. 31172018), Natural and Fundamental Research Funds for the Central Universities of China (Grant No. 2662015PY105), and Chutian scholarship from Hubei province.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01336/full#supplementary-material
Supplementary Figure 1. Putative MW and PI of BnaPIPs, BnaSIPs, BnaNIPs, and BnaTIPs in Brassica napus. The genes in the four sub-families were distinguished by different legends.
Supplementary Figure 2. WebLogo plot of the consensus motifs in BnaAQPs. The amino-acid residues of the consensus motifs in FASTA format were obtained from the MEME Web and WebLogo plot graphs were generated using the amino acid sequences, (A) Motif 1; (B) motif 2; (C) motif 4; (D) motif 5.
Supplementary Table 1. Gene location and description of incomplete BnaAQPs.
Supplementary Table 2. BnaAQP genes without chromosome localization information.
Supplementary Table 3. The RPKM values of the 121 BnaAQPs.
Supplementary Data 1. The multiple sequence alignment of 121 BnaAQPs. The yellow highlighted amino acids represent the residues near P3 site. The letters in a red font below P1 to P5 indicate the amino acid residues in the Froger's position.
References
Alexandersson, E., Danielson, J. Å. H., Råde, J., Moparthi, V. K., Fontes, M., Kjellbom, P., et al. (2010). Transcriptional regulation of aquaporins in accessions of Arabidopsis in response to drought stress. Plant J. 61, 650–660. doi: 10.1111/j.1365-313X.2009.04087.x
Alexandersson, E., Fraysse, L., Sjövalllarsen, S., Gustavsson, S., Fellert, M., Karlsson, M., et al. (2005). Whole gene family expression and drought stress regulation of aquaporins. Plant Mol. Biol. 59, 469–484. doi: 10.1007/s11103-005-0352-1
Ariani, A., and Gepts, P. (2015). Genome-wide identification and characterization of aquaporin gene family in common bean (Phaseolus vulgaris L.). Mol. Genet. Genom. 290, 1771–1785. doi: 10.1007/s00438-015-1038-2
Beebo, A., Thomas, D., Der, C., Sanchez, L., Leborgnecastel, N., Marty, F., et al. (2009). Life with and without AtTIP1;1, an Arabidopsis aquaporin preferentially localized in the apposing tonoplasts of adjacent vacuoles. Plant Mol. Biol. 70, 193–209. doi: 10.1007/s11103-009-9465-2
Bell, R. W., and Dell, B. (2008). Micronutrients for Sustainable Food, Feed, Fibre and Bioenergy Production. Paris: International Fertilizer Industry Association (IFA).
Boursiac, Y., Boudet, J., Postaire, O., Luu, D. T., Tournaire-Roux, C., and Maurel, C. (2008). Stimulus-induced downregulation of root water transport involves reactive oxygen species-activated cell signalling and plasma membrane intrinsic protein internalization. Plant J. 56, 207. doi: 10.1111/j.1365-313X.2008.03594.x
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953. doi: 10.1126/science.1253435
Chaumont, F., and Jung, R. (2001). Aquaporins constitute a large and highly divergent protein family in maize. Plant Physiol. 125, 1206–1215. doi: 10.1104/pp.125.3.1206
Cheng, F., Mandáková, T., Wu, J., Xie, Q., Lysak, M. A., and Wang, X. (2013). Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell 25, 1541–1554. doi: 10.1105/tpc.113.110486
Cheng, F., Wu, J., Fang, L., Sun, S., Liu, B., Lin, K., et al. (2012). Biased gene fractionation and dominant gene expression among the subgenomes of Brassica rapa. PLoS ONE 7:e36442. doi: 10.1371/journal.pone.0036442
Daniels, M. J., Chrispeels, M. J., and Yeager, M. (1999). Projection structure of a plant vacuole membrane aquaporin by electron cryo-crystallography. J. Mol. Biol. 294, 1337–1349. doi: 10.1006/jmbi.1999.3293
Diehn, T. A., Pommerrenig, B., Bernhardt, N., Hartmann, A., and Bienert, G. P. (2015). Genome-wide identification of aquaporin encoding genes in Brassica oleracea and their phylogenetic sequence comparison to Brassica crops and Arabidopsis. Front. Plant Sci. 6:166. doi: 10.3389/fpls.2015.00166
Fitzpatrick, K. L., and Reid, R. J. (2009). The involvement of aquaglyceroporins in transport of boron in barley roots. Plant Cell Environ. 32, 1357–1365. doi: 10.1111/j.1365-3040.2009.02003.x
Giorgio, J. P. D., Bienert, G. P., Ayub, N., Yaneff, A., Barberini, M. L., Mecchia, M. A., et al. (2016). Pollen-specific aquaporins NIP4;1 and NIP4;2 are required for pollen development and pollination in Arabidopsis thaliana. Plant Cell 28, 1053–1077. doi: 10.1105/tpc.15.00776
Hove, R. M., and Bhave, M. (2011). Plant aquaporins with non-aqua functions: deciphering the signature sequences. Plant Mol. Biol. 75, 413–430. doi: 10.1007/s11103-011-9737-5
Hua, Y., Zhang, D., Zhou, T., He, M., Ding, G., Shi, L., et al. (2016a). Transcriptomics-assisted quantitative trait locus fine mapping for the rapid identification of a nodulin 26-like intrinsic protein gene regulating boron efficiency in allotetraploid rapeseed. Plant Cell Environ. 39, 1601–1618. doi: 10.1111/pce.12731
Hua, Y., Zhou, T., Ding, G., Yang, Q., Shi, L., and Xu, F. (2016b). Physiological, genomic and transcriptional diversity in responses to boron deficiency in rapeseed genotypes. J. Exp. Bot. 67, 5769–5784. doi: 10.1093/jxb/erw342
Hub, J. S., and Groot, B. L. D. (2008). Mechanism of selectivity in aquaporins and aquaglyceroporins. Proc. Natl. Acad. Sci. U.S.A. 105, 1198–1203. doi: 10.1073/pnas.0707662104
Ishikawa, F., Suga, S., Uemura, T., Sato, M. H., and Maeshima, M. (2005). Novel type aquaporin SIPs are mainly localized to the ER membrane and show cell-specific expression in Arabidopsis thaliana. FEBS Lett. 579, 5814–5820. doi: 10.1016/j.febslet.2005.09.076
Jeonghwan, M., Soojin, K., Yang, T. J., Youngjoo, S., Jin, M., Jina, K., et al. (2009). Genome-wide comparative analysis of the Brassica rapa gene space reveals genome shrinkage and differential loss of duplicated genes after whole genome triplication. Genome Biol. 10:R111. doi: 10.1186/gb-2009-10-10-r111
Johanson, U., Karlsson, M., Johansson, I., Gustavsson, S., Sjövall, S., Fraysse, L., et al. (2001). The complete set of genes encoding major intrinsic proteins in Arabidopsis provides a framework for a new nomenclature for major intrinsic proteins in plants. Plant Physiol. 126, 1358–1369. doi: 10.1104/pp.126.4.1358
Kumar, K., Mosa, K. A., Chhikara, S., Musante, C., White, J. C., and Dhankher, O. P. (2014). Two rice plasma membrane intrinsic proteins, OsPIP2;4 and OsPIP2;7, are involved in transport and providing tolerance to boron toxicity. Planta 239, 187–198. doi: 10.1007/s00425-013-1969-y
Li, T., Choi, W. G., Wallace, I. S., Baudry, J., and Roberts, D. M. (2011). Arabidopsis thaliana NIP7;1: an anther-specific boric acid transporter of the aquaporin superfamily regulated by an unusual tyrosine in helix 2 of the transport pore. Biochemistry 50, 6633–6641. doi: 10.1021/bi2004476
Liu, S., Liu, Y., Yang, X., Tong, C., Edwards, D., Parkin, I. A., et al. (2014). The Brassica oleracea genome reveals the asymmetrical evolution of polyploid genomes. Nat. Commun. 5:3930. doi: 10.1038/ncomms4930
Loqué, D., Ludewig, U., Yuan, L., and Wirén, N. V. (2005). Tonoplast intrinsic proteins AtTIP2;1 and AtTIP2;3 facilitate NH3 transport into the vacuole. Plant Physiol. 137, 671–680. doi: 10.1104/pp.104.051268
Ma, J. F., Tamai, K., Yamaji, N., Mitani, N., Konishi, S., Katsuhara, M., et al. (2006). A silicon transporter in rice. Nature 440, 688–691. doi: 10.1038/nature04590
Ma, J. F., Yamaji, N., Mitani, N., Xu, X. Y., Su, Y. H., McGrath, S. P., et al. (2008). Transporters of arsenite in rice and their role in arsenic accumulation in rice grain. Proc. Natl. Acad. Sci. U.S.A. 105, 9931–9935. doi: 10.1073/pnas.0802361105
Mahdieh, M., Mostajeran, A., Horie, T., and Katsuhara, M. (2008). Drought stress alters water relations and expression of PIP-type aquaporin genes in Nicotiana tabacum plants. Plant Cell Physiol. 49, 801–813. doi: 10.1093/pcp/pcn054
Maurel, C., and Chrispeels, M. J. (2001). Aquaporins. A molecular entry into plant water relations. Plant Physiol. 125:135. doi: 10.1104/pp.125.1.135
Maurel, C., Verdoucq, L., Luu, D. T., and Santoni, V. (2008). Plant Aquaporins: Membrane channels with multiple integrated functions. Annu. Rev. Plant Biol. 59, 595–624. doi: 10.1146/annurev.arplant.59.032607.092734
Mitaniueno, N., Yamaji, N., Zhao, F. J., and Ma, J. F. (2011). The aromatic/arginine selectivity filter of NIP aquaporins plays a critical role in substrate selectivity for silicon, boron, and arsenic. J. Exp. Bot. 62, 4391–4398. doi: 10.1093/jxb/err158
Mosa, K. A., Kumar, K., Chhikara, S., Musante, C., White, J. C., and Dhankher, O. P. (2015). Enhanced boron tolerance in plants mediated by bidirectional transport through plasma membrane intrinsic proteins. Sci. Rep. 6:21640. doi: 10.1038/srep21640
Nicolas, S. D., Le, M. G., Eber, F., Coriton, O., Monod, H., Clouet, V., et al. (2007). Homeologous recombination plays a major role in chromosome rearrangements that occur during meiosis of Brassica napus haploids. Genetics 175, 487–503. doi: 10.1534/genetics.106.062968
Østergaard, L., and King, G. J. (2008). Standardized gene nomenclature for the Brassica genus. Plant Methods 4:10. doi: 10.1186/1746-4811-4-10
Pang, Y., Li, L., Ren, F., Lu, P., Wei, P., Cai, J., et al. (2010). Overexpression of the tonoplast aquaporin AtTIP5;1 conferred tolerance to boron toxicity in Arabidopsis. J. Genet. Genomics 37, 389–397. doi: 10.1016/S1673-8527(09)60057-6
Saeed, A. I., Sharov, V., White, J., Li, J., Liang, W., Bhagabati, N., et al. (2003). TM4: a free, open-source system for microarray data management and analysis. BioTechniques 34:374.
Sakurai, J., Ishikawa, F., Yamaguchi, T., Uemura, M., and Maeshima, M. (2005). Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 46, 1568–1577. doi: 10.1093/pcp/pci172
Schüssler, M. D., Alexandersson, E., Bienert, G. P., Kichey, T., Laursen, K. H., Johanson, U., et al. (2008). The effects of the loss of TIP1;1 and TIP1;2 aquaporins in Arabidopsis thaliana. Plant J. 56, 756–767. doi: 10.1111/j.1365-313X.2008.03632.x
Soto, G., Alleva, K., Mazzella, M. A., Amodeo, G., and Muschietti, J. P. (2008). AtTIP1;3 and AtTIP5;1, the only highly expressed Arabidopsis pollen-specific aquaporins, transport water and urea. FEBS Lett. 582, 4077–4082. doi: 10.1016/j.febslet.2008.11.002
Suga, S., and Maeshima, M. (2004). Water channel activity of radish plasma membrane aquaporins heterologously expressed in yeast and their modification by site-directed mutagenesis. Plant Cell Physiol. 45, 823–830. doi: 10.1093/pcp/pch120
Takano, J., Wada, M., Ludewig, U., Schaaf, G., Von, W. N., and Fujiwara, T. (2006). The Arabidopsis major intrinsic protein NIP5;1 is essential for efficient boron uptake and plant development under boron limitation. Plant Cell 18, 1498–1509. doi: 10.1105/tpc.106.041640
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tanaka, M., Sotta, N., Yamazumi, Y., Yamashita, Y., Miwa, K., Murota, K., et al. (2016). The minimum open reading frame, AUG-stop, induces boron-dependent ribosome stalling and mRNA degradation. Plant Cell 28, 2830–2849. doi: 10.1105/tpc.16.00481
Tanaka, M., Wallace, I. S., Takano, J., Roberts, D. M., and Fujiwara, T. (2008). NIP6;1 is a boric acid channel for preferential transport of boron to growing shoot tissues in Arabidopsis. Plant Cell 20, 2860–2875. doi: 10.1105/tpc.108.058628
Tang, H., Woodhouse, M. R., Cheng, F., Schnable, J. C., Pedersen, B. S., Conant, G., et al. (2012). Altered patterns of fractionation and exon deletions in Brassica rapa support a two-step model of paleohexaploidy. Genetics 190, 1563–1574. doi: 10.1534/genetics.111.137349
Tao, P., Zhong, X., Li, B., Wang, W., Yue, Z., Lei, J., et al. (2014). Genome-wide identification and characterization of aquaporin genes (AQPs) in Chinese cabbage (Brassica rapa ssp. pekinensis). Mol. Genet. Genom. 289, 1131–1145. doi: 10.1007/s00438-014-0874-9
Tombuloglu, H., Ozcan, I., Tombuloglu, G., Sakcali, S., and Unver, T. (2015). Aquaporins in boron-tolerant barley: identification, characterization, and expression analysis. Plant Mol. Biol. Rep. 34, 374–386. doi: 10.1007/s11105-015-0930-6
Wallace, I. S., and Roberts, D. M. (2004). Homology modeling of representative subfamilies of Arabidopsis major intrinsic proteins. Classification based on the aromatic/arginine selectivity filter. Plant Physiol. 135, 1059–1068. doi: 10.1104/pp.103.033415
Wallace, I. S., and Roberts, D. M. (2006). Distinct transport selectivity of two structural subclasses of the nodulin-like intrinsic protein family of plant aquaglyceroporin channels. Biochemistry 44, 16826–16834. doi: 10.1021/bi0511888
Wallace, I. S., Choi, W. G., and Roberts, D. M. (2006). The structure, function and regulation of the nodulin 26-like intrinsic protein family of plant aquaglyceroporins. Biochim. Biophys. Acta 1758, 1165–1175. doi: 10.1016/j.bbamem.2006.03.024
Wallace, I. S., Wills, D. M., Guenther, J. F., and Roberts, D. M. (2002). Functional selectivity for glycerol of the nodulin 26 subfamily of plant membrane intrinsic proteins. FEBS Lett. 523, 109–112. doi: 10.1016/S0014-5793(02)02955-1
Wang, S., Yoshinari, A., Shimada, T., Hara-Nishimura, I., Mitani-Ueno, N., Ma, J. F., et al. (2017). Polar localization of the NIP5;1 boric acid channel is maintained by endocytosis and facilitates boron transport in Arabidopsis roots. Plant Cell 29, 824–842. doi: 10.1105/tpc.16.00825
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Wang, Y., Li, R., Li, D., Jia, X., Zhou, D., Li, J., et al. (2017). NIP1;2 is a plasma membrane-localized transporter mediating aluminum uptake, translocation, and tolerance in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 114, 5047–5052. doi: 10.1073/pnas.1618557114
Warington, K. (1923). The effect of boric acid and borax on the broad bean and certain other plants. Ann. Bot. 37, 629–672. doi: 10.1093/oxfordjournals.aob.a089871
Wei, Y., Bell, R. W., Yang, Y., Ye, Z., Wang, K., and Huang, L. (1998). Prognosis of boron deficiency in oilseed rape (Brassica napus) by plant analysis. Aust. J. Agr. Res. 49, 867–874. doi: 10.1071/A97156
Wudick, M. M., Luu, D. T., and Maurel, C. (2009). A look inside: localization patterns and functions of intracellular plant aquaporins. New Phytol. 184, 289–302. doi: 10.1111/j.1469-8137.2009.02985.x
Xu, W., Dai, W., Yan, H., Li, S., Shen, H., Chen, Y., et al. (2015). Arabidopsis NIP3; 1 plays an important role in arsenic uptake and root-to-shoot translocation under arsenite stress conditions. Mol. Plant 8, 722–733. doi: 10.1016/j.molp.2015.01.005
Yang, T. J., Kim, J. S., Kwon, S. J., Lim, K. B., Choi, B. S., Kim, J. A., et al. (2006). Sequence-level analysis of the diploidization process in the triplicated FLOWERING LOCUS C region of Brassica rapa. Plant Cell 18, 1339–1347. doi: 10.1105/tpc.105.040535
Zhi, Z., Gong, J., Feng, A., Xie, G., Wang, J., Mo, Y., et al. (2015). Genome-wide identification of rubber tree (Hevea brasiliensis Muell. Arg.) aquaporin genes and their response to ethephon stimulation in the laticifer, a rubber-producing tissue. BMC Genomics 16:1001. doi: 10.1186/s12864-015-2152-6
Keywords: aquaporins (AQPs), Brassica napus, boron (B), transcriptional profile, boron homeostasis
Citation: Yuan D, Li W, Hua Y, King GJ, Xu F and Shi L (2017) Genome-Wide Identification and Characterization of the Aquaporin Gene Family and Transcriptional Responses to Boron Deficiency in Brassica napus. Front. Plant Sci. 8:1336. doi: 10.3389/fpls.2017.01336
Received: 22 May 2017; Accepted: 17 July 2017;
Published: 02 August 2017.
Edited by:
Gareth John Norton, University of Aberdeen, United KingdomReviewed by:
Rosario Vera-Estrella, National Autonomous University of Mexico, MexicoDoan Trung Luu, Centre National de la Recherche Scientifique (CNRS), France
Copyright © 2017 Yuan, Li, Hua, King, Xu and Shi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lei Shi, bGVpc2hAbWFpbC5oemF1LmVkdS5jbg==
 Dan Yuan
Dan Yuan Wei Li
Wei Li Yingpeng Hua
Yingpeng Hua Graham J. King
Graham J. King Fangsen Xu
Fangsen Xu Lei Shi
Lei Shi