
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 18 July 2017
Sec. Plant Proteomics and Protein Structural Biology
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01267
Legume plants are key elements in sustainable agriculture and represent a significant source of plant-based protein for humans and animal feed worldwide. One specific feature of the family is the ability to establish nitrogen-fixing symbiosis with Rhizobium bacteria. Additionally, like most vascular flowering plants, legumes are able to form a mutualistic endosymbiosis with arbuscular mycorrhizal (AM) fungi. These beneficial associations can enhance the plant resistance to biotic and abiotic stresses. Understanding how symbiotic interactions influence and increase plant stress tolerance are relevant questions toward maintaining crop yield and food safety in the scope of climate change. Proteomics offers numerous tools for the identification of proteins involved in such responses, allowing the study of sub-cellular localization and turnover regulation, as well as the discovery of post-translational modifications (PTMs). The current work reviews the progress made during the last decades in the field of proteomics applied to the study of the legume-Rhizobium and -AM symbioses, and highlights their influence on the plant responses to pathogens and abiotic stresses. We further discuss future perspectives and new experimental approaches that are likely to have a significant impact on the field including peptidomics, mass spectrometric imaging, and quantitative proteomics.
The Fabaceae or Leguminosae family, commonly referred to as “legumes,” is the third largest family of flowering plants, second only to cereals in terms of agricultural importance. Some of the most widely studied plants in the family include crops such as soybean (Glycine max L. Merr.), common bean (Phaseolus vulgaris L.), chickpea (Cicer arietinum L.), lentil (Lens culinaris Medik.), pea (Pisum sativum L.), or alfalfa (Medicago sativa L.). However, given the large size and genome complexity of these major crops, the scientific community has focused its efforts in the development of tools and protocols for other legume plants, commonly referred to as model legumes, namely Medicago truncatula Gaertn. and Lotus japonicus L. For many of these legume species there is genomic sequence information available, which greatly facilitates protein identification using mass spectrometry-based proteomic approaches. Many species within the family are able to establish endosymbiotic relationships with nitrogen-fixing Rhizobium bacteria and arbuscular mycorrhizal (AM) fungi. These symbiotic interactions involve complex signal exchanges between both symbionts and an intimate communication to allow the establishment of the bacteria or fungi inside root cells. These interactions are considered mutualistic associations, with Rhizobium bacteria providing a source of reduced nitrogen inside specialized root organs named nodules, and AM fungi facilitating the capture of important nutrients for the plant such as phosphorous or sulfur, and even improving the plant responses to biotic and abiotic stress conditions (Ruiz-Lozano et al., 2001; Dimkpa et al., 2009; Pieterse et al., 2014). In order to understand the biological processes governing these symbiotic interactions and their effects on plant fitness, scientists have employed an array of methodologies, ranging from gene expression to proteomic analysis. Proteomics is a powerful tool for the study of subcellular compartmentalization important to understand nodule formation, symbiosome function and to unravel the molecular mechanisms involved in the enhanced stress tolerance of legumes under symbiotic conditions. The current work reviews and summarizes the progress made during the last decades in the field of proteomics applied to the study of legume-Rhizobium and -AM symbioses, and their interactions with biotic and abiotic stresses, discussing future perspectives and new experimental approaches.
During the last decades a considerable effort has been made to characterize the diversity of proteins expressed in different tissues under a variety of conditions at the international level. This effort was initiated using 2D-PAGE-based approaches, generating reference maps for various organs in different legume species. Taking M. truncatula as an example, Mathesius et al. (2001) were the first to establish a root reference map, which included ~2,500 protein spots, from which 179 were identified. It was remarkable that close to half of them were present as protein isoforms, including key metabolic enzymes such as S-adenosyl-L-methionine synthase, malate dehydrogenase, or ascorbate peroxidase. Gallardo et al. (2003) reported the characterization of the seed proteome during seed filling, with the identification of 84 proteins including proteins belonging to the main storage protein families as well as proteins involved in carbon and sulfur metabolism, among others. Subsequently, works from the Sumner laboratory published a comprehensive analysis of the M. truncatula proteome at the different organ level, including cell cultures, with the high-confidence identification of close to 2,000 proteins, the largest proteomic identification reported so far (Watson et al., 2003; Lei et al., 2005).
Focusing on the symbiotic perspective, several comparative proteomics works have been devoted to the analysis of the differential root proteome of a number of legumes when inoculated with their corresponding microbial partners. Depending on the focus of the study, works in the field of symbiosis can be divided in two major groups: (i) studies focused on the characterization of the legume plant proteome, and (ii) works focused on the Rhizobium partner (Table 1). In the latter case, a classical comparison is the analysis of the proteome of free-living Rhizobium cells vs. their differentiated nitrogen-fixing forms, named bacteroids. This strategy has been applied to identify symbiosis-specific proteins synthesized in the symbionts of the main legume species. Comparison of the proteomic profiles of cultured cells vs. Sinorhizobium meliloti nodule bacteroids suggested that nodule bacteria do not express sugar transporters or enzymes involved in the early steps of glycolysis, while containing multiple transporters for nitrogen compounds including amino acids and oligopeptides (Djordjevic et al., 2003; Djordjevic, 2004). The high adaptability of symbiotic bacteria depending on the carbon source was also reported in proteomic works carried out in Bradyrhizobium japonicum (Sarma and Emerich, 2005, 2006). Authors reported the unusually low levels in proteins related to fatty acid and nucleic acid metabolism in bacteroids, suggesting that bacteroids and cell cultured bacteria may present differential mechanisms to regulate the levels of ribonucleotides. Subsequent work in B. japonicum using more powerful mass spectrometry techniques, however, did not observe such repression in nucleotide metabolism, identifying almost the full set of enzymes involved in de novo nucleoside and nucleotide biosynthesis expressed at the gen and/or protein level (Delmotte et al., 2010). Regarding the symbiont of L. japonicus, Tatsukami et al. (2013) identified 722 proteins commonly found under the free-living and symbiotic conditions, while 125 proteins were uniquely identified under symbiotic conditions. Interestingly, proteins involved in peptidoglycan biosynthesis and proteins related to the flagellum were uniquely detected under free-living conditions, suggesting that once within the symbiosomes, bacteroids simplify their cell surface by losing their cell wall and motility structures (Tatsukami et al., 2013). Furthermore, the quantitative time-course proteomic analysis of M. loti suggested that bacteroids experience nitrogen-deficiency at early stages of nodule development, while at intermediate stages high levels of nitrogenase protein lead to nitrogen-rich conditions in the symbiosome (Nambu et al., 2015).
In the case of plant-oriented studies, it is common to compare differences in the proteome of uninoculated vs. inoculated plants. One of the pioneer works in this line was carried out by Krause and Broughton (1992), reporting 12 symbiosis-specific proteins potentially involved in root-hair deformation in Vigna unguiculata, although at the time the lack of genomic sequence information limited protein identification. The availability of expressed sequence tags (ESTs) and, subsequently, genomic sequences of several legume plants have led to greatly improve the number of identified proteins. For instance, the model legume M. truncatula has been subjected to detailed proteomic characterization in terms of symbiotic responses. One of the first studies in nodules identified two leghemoglobin isoforms and one enolase protein as some of the proteins that most accumulated in roots upon inoculation with symbiotic bacteria (Bestel-Corre et al., 2002). The protein profiling of the plant fraction of M. truncatula root nodules led to the identification of 377 unique proteins, most of them with roles in amino acid metabolism and protein synthesis and degradation (Larrainzar et al., 2007). Proteomics is also a valuable tool for the analysis of local and systemic responses upon inoculation. Through the analysis of the proteomic changes occurring in shoots and roots of inoculated M. truncatula plants, 18 proteins were found to accumulate in roots including sucrose synthase 1, a fructose-bisphosphate aldolase, and an alcohol dehydrogenase, while in shoots the content of several proteins involved in defense responses or abiotic stress responses was found to increase (Molesini et al., 2014). Proteomics has been also applied to investigate the effects of the addition of the ethylene precursor aminocyclopropane carboxylic acid (ACC) on nodule development using the supernodulating, ethylene-insensitive mutant sickle (Penmetsa and Cook, 1997) during the early stages of the symbiotic interaction (Prayitno et al., 2006). Among other findings, authors observed that Sinorhizobium inoculation increased the abundance of one ACC oxidase isoform in wild-type roots but not in sickle roots, suggesting that a feedback mechanism regulates the expression this gene. Nevertheless, subsequent work using RNA-seq techniques has shown that at least three genes of the ACC oxidase family are induced in sickle upon inoculation (Larrainzar et al., 2015), which highlights the usefulness of combining data at the proteomic and transcriptomic level.
Further characterization of specific metabolic pathways using absolute quantification techniques has been also applied to this model legume, including the detailed analysis of the nitrogen assimilation and ethylene biosynthesis pathways in root nodules (Larrainzar et al., 2009, 2014). Similarly, the symbiotic proteome of soybean, a crop of major economical importance, has been also extensively studied. A time-course proteomic analysis of wild type and the soybean mutant SS2-2, which lacks autoregulation of nodulation, has revealed that there is a protein-mediated suppression of defense-related responses in root cells upon inoculation with Rhizobium bacteria (Lim et al., 2010). A similar observation was done when comparing the proteomic changes associated to inoculation of soybean plants with differential nodulation capacities (Salavati et al., 2012). In this work, a correlation between the levels of a peroxidase isoform and levels of nodulation was found, although the regulation did not occur at the transcript level. The plant fraction of soybean nodules has been also subjected to proteomic analysis, leading to the identification of 69 proteins mainly related to carbon and nitrogen metabolic activities (Oehrle et al., 2008), similarly to previous observations in M. truncatula.
Since root hairs are most frequently the main entry point for Rhizobium bacteria, several works have been devoted to identify the proteomic changes occurring in this specialized root cell upon inoculation. This work has the obvious technical limitation that collecting sufficient amount of plant material is challenging and requires a large number of plants per proteomic sample, with estimations of around 1,500 soybean roots and 4,000 soybean seedlings (Wan et al., 2005; Brechenmacher et al., 2009). A time course of the proteomic changes occurring in root hairs revealed that there is a specific induction of phospholipases and phosphoglucomutases, as well as a lectin and an actin isoform upon inoculation (Wan et al., 2005). Under uninoculated conditions, Brechenmacher et al. (2009) combined traditional 2D-PAGE and shotgun methods for the identification of 1,492 proteins present in root hairs, establishing a reference map for future work.
Several recent studies have provided broad insights into the systemic effects the legume-Rhizobium symbiotic interaction at the root and, especially, leaf metabolic level (Staudinger et al., 2012, 2016; Desalegn et al., 2016; Turetschek et al., 2016). In all these studies, one of the major response found was related to a significantly induction of the plant translational apparatus and an accumulation of plant proteins involved in stress responses.
Proteomic approaches are particularly suited to gain information about protein subcellular localizations. Analyses of enriched fractions or, ideally, purified organelles, or sub-organelle compartments allow the validation of protein compartmentalization and isoform localization data. Furthermore, subcellular fractionation provides valuable information on the specific changes in the proteome of organelles in response to various stresses, allowing for the development of accurate proteomic pathways and networks (Hooper et al., 2014). Legume plants have been also subjected to this type of analysis. Most of work, however, has been done under non-symbiotic conditions. Comprehensive reviews in the field of subcellular proteomics in legumes have been published elsewhere (Lee et al., 2013; Wang and Komatsu, 2016; Yin and Komatsu, 2016). Thus, in the current review we will discuss subcellular proteomic works with a symbiotic focus.
Nodules are complex structures containing a combination of infected and non-infected plant cells. Infected cells are filled with nitrogen-fixing bacteroids arranged in symbiosomes surrounded by a specialized plant membrane named peribacteroid membrane (PBM). The identification of proteins present in this specialized membrane is of key relevance, since it represents the direct interface where nutrient and signal exchange occurs between the legume host plant and Rhizobium bacteroids. In order to identify which proteins are localized at the PBM, extensive proteomic work has been carried out in this membrane fraction. Panter et al. (2000) carried out one of the first studies, with the 2D-PAGE analysis of the PBM of soybean nodules. The proteomic characterization of the pea PBM and peribacteroid space, a much more challenging approach, has also been studied (Saalbach et al., 2002). In this work, proteins of the Coatomer-coated vesicles like V-ATPase, BIP, were found in the PBM fraction, supporting the role of the endomembrane system in PBM biogenesis. These studies were followed by more comprehensive LC-MS/MS-based analyses of the PBM in legumes such as L. japonicus (Wienkoop and Saalbach, 2003), M. truncatula (Catalano et al., 2004), and more recently, soybean (Clarke et al., 2015). Identification of protein components in these membranes has been shown an essential first step for increasing our knowledge on the metabolic exchange processes between plant and bacteroid. For instance, the proteomic analysis of the PBM in L. japonicus led to the identification of a symbiosis-specific sulfate transporter 1 (SST1) and around 80 other abundant membrane or membrane-associated proteins such as the hypersensitive response protein and remorin (SYMREM1; Wienkoop and Saalbach, 2003). Both SST1 and SYMREM1 have subsequently been characterized in detail and shown to be relevant for nodule development and functioning (Krusell et al., 2005; Lefebvre et al., 2010; Toth et al., 2012; Domonkos et al., 2013; Kalloniati et al., 2015).
One of the strengths of mass spectrometry-based proteomic approaches is that it allows the identification of post-translational modification (PTM) sites in proteins, something that cannot be accurately predicted with genomic information alone. PTMs have a huge impact on plant signaling and metabolism, contributing to the regulation of protein activity, stability/degradation, interactions, and ultimately gene expression. In recent years, the number of studies focused on the identification of PTMs in plants has considerable grown (for an extensive review, see Friso and van Wijk, 2015). Particular interest has received the large-scale identification of phosphoproteins at early stages of the legume-Rhizobium symbiotic interaction. The reason behind this interest is that a number of protein kinases have been shown essential for rhizobial infection and/or nodule development. These include LysM-domain-containing receptor kinases, implicated in the binding of Nod factors, as well as other membrane receptor-like kinases and calcium/cadmodulin-dependent protein kinases (Antolín-Llovera et al., 2014). Analysis of the phosphoproteome at early symbiotic stages has been carried out in L. japonicus roots (Serna-Sanz et al., 2011), soybean root hairs (Nguyen et al., 2012), and M. truncatula roots both applying discovery proteomics (Rose et al., 2012) and targeted approaches (van Ness et al., 2016). These studies are important first steps toward understanding how Nod-factor signaling is transmitted from the plasma membrane and decoded to activate the developmental reprogramming root cells undergo to allow nodule formation. Previous studies identified several phosphoproteins present in mature nitrogen-fixing nodules, including several phosphopeptides in sucrose synthase 1 and alkaline invertase, the main sucrose-degrading enzymes in this specialized organ (Wienkoop et al., 2008). Similarly, large-scale analyses of M. truncatula roots (Grimsrud et al., 2010) or during nodulation (Marx et al., 2016) have allowed the identification of a repertoire of in vivo phosphorylated peptides and phosphorylation motifs, which can be queried online (http://www.phospho.medicago.wisc.edu and http://compendium.medicago.wisc.edu, respectively).
Ubiquitination is another PTM that plays important roles for rhizobial infection and nodule organogenesis in legumes. For instance, proteins such as the receptor kinase M. truncatula LYK3 has been found ubiquitinated in vitro by the E3 ubiquitin ligase PUB1 (Mbengue et al., 2010) and a deubiquitinating enzyme named AMSH1 has been shown to be required for the establishment of an effective symbiosis in L. japonicus (Małolepszy et al., 2015). However, to our knowledge, the large-scale identification of ubiquitinated proteins under symbiotic conditions has not been carried out to date.
The role of the signaling molecule nitric oxide and its associated protein modifications, nitrosylation and nitration, has also drawn considerable attention in legume studies. At the level of root nodules, two proteins have been identified as targets of Tyr nitration: the ammonium-assimilating enzyme glutamine synthetase (GS; Melo et al., 2011; Blanquet et al., 2015) and the hemeprotein leghemoglobin (Sainz et al., 2015). In the latter work it was found that leghemoglobin not only plays an important role as an O2 transporter but may also act as sink of toxic peroxynitrite and thus be part of a protective mechanism in symbiosis.
Other PTM studies in the field include the identification of sulfenylated proteins in the M. truncatula-S. meliloti symbiosis (Oger et al., 2012). Interestingly, both in the plant and bacterial partners a large proportion of the identified proteins were related to the glycolytic pathway, tricarboxylic acid cycle and amino-acid metabolism, including one of the nodule-enhanced sucrose synthase isoforms. Additionally, a cytosolic isoform of glutamine synthetase has been found as a target of sulfoxidation, although this modification did not alter the activity of the enzyme (Matamoros et al., 2013).
The plant-AM fungi (Glomeromycota) is the most extensively observed association with roots of land plants (>80%; Schüβler et al., 2001). The symbiotic interaction includes formation of appressoria on the root surface, the entrance into the root epidermis, proliferation within the cortical parenchyma and the formation of arbuscule structures (Giovannetti et al., 1996). These hyphal branches are surrounded by a plant-derived plasma membrane, called periarbuscular membrane (Alexander et al., 1989; Harrison, 1999). Similarly to the PBM, this membrane is the actual site of the plant-microbe interaction. AM symbiosis is described as a bilateral nutritional beneficial association whereby AM fungi supply plants primarily with phosphorus and also nitrogen, while plants provide corporate fungi with carbohydrates (Harrison, 1999). At the gene level, the induction of phosphate transporter genes during AM symbiosis has been reported in several plant species (Rausch et al., 2001; Harrison et al., 2002; Paszkowski et al., 2002). By analyzing the protein profiles of the periarbuscular membrane, the localization of such transporters can be confirmed, as demonstrated in the model legume M. truncatula (Harrison et al., 2002). Further proteomic works in this legume have identified changes in the levels of H1-ATPase and a predicted glycosylphosphatidylinositol-anchored blue copper-binding protein in response to AM-association (Gianinazzi-Pearson et al., 2000; Bestel-Corre et al., 2004; Valot et al., 2006). The high H1-ATPase activity was described to support the existence of an active nutrient transport between the partners (Ferrol et al., 2002).
Recently, Abdallah et al. (2014) examined the profile of the M. truncatula root membrane proteome after microsomal enrichment. The most abundant organelle components that were retrieved encompassed the plastid, the nucleus and the plasma membrane. Comparing AM-colonized vs. non-mycorrhized plants, they found a lysine/histidine transporter, as well as a differential abundance of about 100 other proteins upon mycorrhization, including known sulfate transporters, the above described H1-ATPase and blue copper protein. Comparison between mutants with contrasting AM-colonization genotypes showed differences at the level of appressorium-responsive proteins (Amiour et al., 2006). Another proteomic study on M. truncatula roots colonized with two different Glomus species identified a conserved plant responses to mycorrhizal colonization, which include proteins related to redox homeostasis, carbon metabolism, and energy generation (Recorbet et al., 2010). In this regard, a closer look into the root plastid proteome revealed that arbuscule development was potentially slowing down the hosts anabolic reactions such as N assimilation, fatty acid biosynthesis, glycolysis, and pentose phosphate pathway (Daher et al., 2016). Authors also proposed that the reduced C and N assimilation was concurrent with the reallocation of other molecules, possibly to be stored as N-rich compounds. In addition and in accordance with investigations of the leaf proteome of P. sativum (Desalegn et al., 2016), they also found an AM symbiosis-induced oxidative stress signature.
All in all, a strong influence of the AM symbiosis on the plant metabolism has been evidenced by these proteomic studies, a response that may be comparable to the previously described systemic resistance induced by rhizosphere bacteria (van Loon et al., 1998).
Although a number of proteomic studies have been published on the response of legumes to abiotic stresses, in most studies, however, plants under study have not been grown under symbiotic conditions. For instance, analyses of peanut (Arachis hypogaea L.) varieties with contrasting tolerance to water deficit have shown a relation between plant tolerance to the stress and the abundance of proteins involved in stress signaling and wax biosynthesis in leaves (Kottapalli et al., 2009). A similar approach was taken by Katam et al. (2016) who identified proteins related to nitrogen metabolism, defense and cellular biogenesis exclusively in the tolerant peanut cultivar. Abiotic stress responses in a major legume crop like soybean have been reviewed elsewhere (Hossain et al., 2012, 2013; Hossain and Komatsu, 2014; Komatsu et al., 2015). Here we will focus on the proteomic works analyzing the effects of abiotic stress of legumes grown under symbiotic conditions.
Regarding the legume-Rhizobium symbiosis, the effects of drought stress on the M. truncatula nodule proteome have been characterized in most detailed. Proteomic analysis of the plant protein fraction of drought-stressed nodules showed a general decline in enzymes involved in symbiotic nitrogen fixation and N assimilation (Larrainzar et al., 2007). In contrast, the microsymbiont showed an accumulation of chaperonins and heat-shock proteins, along with enzymes involved in energy metabolism. In a later study, the application of an integrative proteomic and metabolic approach to M. truncatula nodules allowed the identification of the main effects of drought followed by a recovery treatment (Larrainzar et al., 2009). Both works highlighted the role of enzymes related to sulfur assimilation in nodules, with the identification of a specific, nodule-enhanced methionine synthase isoform responding to water deficit conditions. Thus, in a subsequent work, Larrainzar et al. (2014) analyzed in detail the involvement of sulfur metabolism in the nodule response to drought. Gene expression and absolute quantification of proteins in the methionine and ethylene biosynthesis pathways were found strongly down-regulated during drought, demonstrating the contribution of nodule sulfur metabolism and the phytohormone ethylene for nodule functioning under water deficit conditions. Using a split-root system and proteomics, local inhibition of nitrogen fixation and nodule metabolism was evidenced in M. truncatula and soybean plants partially exposed to drought stress (Gil-Quintana et al., 2013). More recently, the drought response of two legume species was compared, finding a similar molecular response to drought but a higher drought tolerance in the tropical legume (Gil-Quintana et al., 2015). In this study, protein isoforms that share same function and location could be identified across species.
Induced systemic resistance and the positive effect of microbes in the plant immune system has received considerable in recent years (van Wees et al., 2008). In this line, the effect of Rhizobium in priming plant metabolism in response to salt and drought stress has been analyzed in M. truncatula using a combination of proteomic and metabolomic techniques (Staudinger et al., 2012, 2016). These works highlighted the involvement of another group of nodule proteins in the plant response to drought; the family of lipoxygenases, enzymes related to lipid metabolism and jasmonate biosynthesis. Additionally, authors found an influence of the Rhizobium symbiosis on the abundance levels of stress responsive proteins prior application of the stress treatment. In subsequent works, plants grown under symbiotic conditions were found to present reduced levels of leaf senescence under drought stress, regardless the efficiency of the Rhizobium strain used (Staudinger et al., 2016). Authors concluded that, besides increased jasmonate biosynthesis and potassium ion concentrations, allocation of reserves to osmolytes and a shift in carbon partitioning from starch to sugar are key responses for the observed symbiont-induced stay-green effect.
The plant-AM symbiosis has been shown to have a positive impact on the plant nutritional status, thus, improving growth and crop productivity. In legumes, an overall enhanced tolerance to abiotic stress has been also reported (Ruiz-Lozano et al., 2001). However, specifically in the field of proteomics applied to the legume-AM symbiosis, works describing this positive effect are scarce. One of the few examples are the analyses by Aloui and colleagues focused on the changes in the proteome of M. truncatula roots (Aloui et al., 2009) and shoots (Aloui et al., 2011) when colonized with the AM fungus G. intraradices in cadmium (Cd)-free and Cd-contaminated substrates. Authors identified nine out of 15 proteins changing under Cd treatment in non-mycorrhizal roots that were not observed in Cd-treated, colonized roots. In shoots Cd induced the accumulation of proteins related to photosynthesis in plants under symbiotic conditions. As a conclusion, when exposed to Cd, symbiotically grown plants partially escaped metal toxicity through a concerted increase in shoot biomass and thanks to allocation plasticity strategies compared to non-symbiotic plants.
Collectively, in the last decade proteomics research has provided a significant contribution to the understanding of the legume symbiotic interactions in response to biotic and abiotic stress. A summary of the main conclusions drawn can be found in Figure 1.
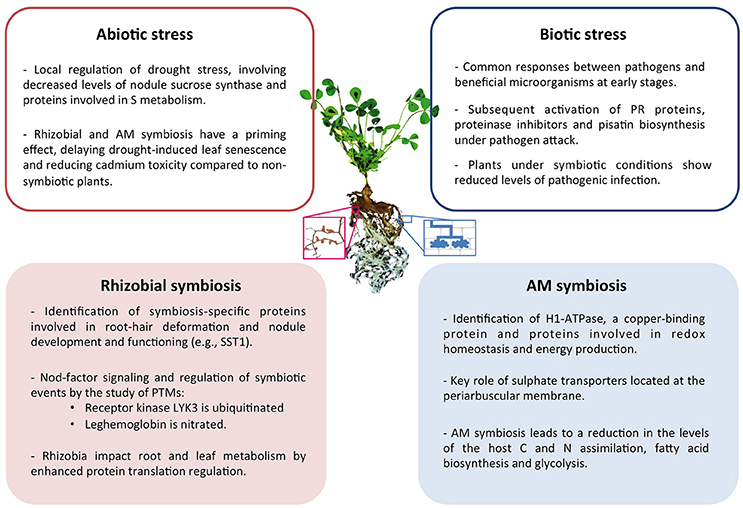
Figure 1. Summary of the main conclusions drawn from proteomic studies of symbiotic legume plants and their interactions under abiotic and biotic stress conditions. Center image represents a M. truncatula plant and a magnified image of nodulated roots (left) and schematic representation of cells containing AM fungi (right).
Symbiotic interactions and responses to a pathogen attack share some features especially at the early stage of the interaction. Legume hosts initially recognize their symbiotic partners as potential threats, activating plant defense responses that are subsequently down-regulated at later stages (Zamioudis and Pieterse, 2012). Since plants under symbiotic conditions appear to show an improved tolerance to abiotic stresses, it could be hypothesized that a similar pattern would be observed when challenged by pathogens. In terms of proteomic analysis, few are the works that have analyzed this question and most studies have been carried out on legumes grown under non-symbiotic conditions. One of the few works on the topic analyzed the proteomic changes occurring during the tripartite interactions with S. meliloti, mycorrhizal fungi, and the pathogenic oomycota Aphanomyces euteiches in M. truncatula roots (Schenkluhn et al., 2010). Among the proteins detected after inoculation either with G. intraradices and/or S. meliloti the highest increase in relative content was observed for a calmodulin-2, which could be related to the activation of Nod- and Myc-factor signaling cascades. In agreement with previous studies, authors observed the activation of proteins involved in antioxidant defense and/or scavenging of reactive oxygen species. At later stages the pathogen induced the expression of a different set of proteins including pathogen response (PR) proteins, Kunitz-type proteinase inhibitors, a lectin, and proteins related to primary carbohydrate metabolism. Interestingly, the induction of these pathogen responses occurred to a lesser extent in plants under mixed infections (i.e., symbiotic plants infected with A. euteiches), which indeed reinforces the idea of the beneficial effects of symbiosis toward pathogenic infections. A similar observation was made when analyzing the leaf proteome and metabolome of pea plants after the co-inoculation with R. leguminosarum and AM fungi, or either of those combined with the pathogenic fungi Didymella pinodes (Desalegn et al., 2016). The Rhizobium symbiosis considerably increased the levels of proteins involved in the pisatin pathway upon pathogen attack compared to mycorrhized, co-inoculated plants or plants under non-symbiotic conditions. Hence, it was concluded that proteins and pathways involved in symbiotic interactions might indirectly be linked to specific pathogen-response pathways. However, the regulatory link between the different molecular responses induced by symbionts and pathogens remains unknown. In fact, when in a subsequent work the effect of the symbionts and pathogen disease levels were compared on two pea cultivars with differential susceptibility, the influence of the microsymbiont was found superimposed by genotypic resistance traits (Turetschek et al., 2016).
The storage and public availability of large proteomic data resources is of great interest for the wider distribution of proteomic data in the scientific community. Several databases give either access to proteome sequence information of fully sequenced genomes and their functional annotation such as the Universal Protein Resource (UniProt, http://www.uniprot.org) or even of mass spectrometry spectral identification like ProMEX (Hummel et al., 2007; Wienkoop et al., 2012). Nevertheless, to date genomic databases for most plant model species, including legumes, lack proteome subcellular localization annotation (Hooper et al., 2016). One of the best-characterized subcellular proteome is that of the non-legume model plant A. thaliana, which is stored in the SUBA3 database (Tanz et al., 2013). Recently, subcellular localization data based on gene co-expression have been extended for agriculturally relevant plants including soybean (Obayashi et al., 2014). An extension of this database integrating proteomic data of those species has been also made publically available (Hooper et al., 2016).
Although a summary of the main databases specific for legume plants has been published elsewhere (González et al., 2015; Ramalingam et al., 2015), the current section reviews some additional databases that were not included above. The platform for integrative legume biology (LEGOO; https://www.legoo.org) offers a tool for the automatic annotation of several legume proteomes including these of M. truncatula, G. max and L. japonicus. One of the strengths of this database is that it allows for the rapid identification of orthologous genes/proteins across different legume species, simplifying the task of tracking down proteins with multiple IDs. Recently, Lotus Base an integral information portal providing genomic, proteomic, and expression resources for the model legume L. japonicus has been also made available (https://lotus.au.dk; Mun et al., 2016). Regarding data on post-translational modification, Schulze et al. (2015) have summarized the online databases available in the field of phosphoproteomics for a range of plant species, including the model legume M. truncatula.
In terms of databases focused on the microsymbionts, one useful platform is the RhizoGATE (http://www.cebitec.uni-bielefeld.de/CeBiTec/rhizogate; Becker et al., 2009), which collects several databases containing genomic and transcriptomic information of a number of Rhizobium species. Similarly, the RhizoBase (http://genome.microbedb.jp/rhizobase; Fujisawa et al., 2014) is a manually-curated genome annotation database containing genome data for Rhizobium, including the option of downloading data on several microsymbiont proteomes. However, to our knowledge, the only database containing proteomic spectral and experimental metadata on Rhizobium is ProMEX, including spectral peptide information on S. meliloti (955 entries) and B. japonicum (157 entries).
One major challenge of proteomic data is that the vast amount of information generated is difficult to present in a convenient, accessible way, thus mostly ending up in huge lists of metadata that are difficult to access for further work. Additionally, transfer of knowledge from one organism to another is very restricted; even if protein functions can be estimated based on sequence similarity, their localization and specific responses to stress varies depending on the plant species. One example of such comparative approach can be found in Gil-Quintana et al. (2015), who compared the M. truncatula and soybean root nodule proteome in response to drought. Authors found that the nodule proteome in response to stress in grain and forage legumes was very similar, suggesting that proteome research conducted on the model legume might be extended to other economically relevant crop legumes.
Taken together, there are several legume-specific proteomic databases available. The development of a platform gathering and allowing the analysis of this proteomic information in legumes, similarly to the MASCP gator for the visualization and extraction of proteomic information in A. thaliana (Joshi et al., 2011), might be a future goal in the legume-symbiosis community.
This work reviews some of the most important outcomes of proteomic analysis of the legume-Rhizobium and -AM symbiosis and their interactions with abiotic and biotic stresses. It has become evident that symbiotic interactions have a significant, positive impact on the plant fitness levels, improving their tolerance to stress conditions and pathogen attack. Although, supplementation based on nitrogen fertilizers under optimal conditions may still lead to higher crop yield in global terms, the positive influence of the microsymbiont should be taken into consideration for future breeding programmes, particularly now under the predicted climatic change scenarios.
During the last decade, improvements in terms of speed and accuracy of mass spectrometric instruments has led to about one order of magnitude increase in the amount of high-throughput proteomic data obtained, with the identification of >1,000 different proteins per experiment. However, these improvements have not yet been fully exploited in the field of the legume-microbial symbiosis. There is, for instance, growing interest in extending the work on endogenous plant peptides described to act as signaling molecules and shown to be involved in plant-microbe communication (van de Velde et al., 2010). Hence, application of peptidomic techniques may significantly contribute to unravel the plant control mechanisms for both beneficial and pathogenic interactions in the root microbiome. One example of potential applications is the use of MALDI mass spectrometric imaging, a technique that allows for the visualization of spatial protein distributions, recently applied to analyze changes in the levels of endogenous peptides and proteins at different developmental stages in M. truncatula (Ye et al., 2013; Gemperline et al., 2016). Similarly, other relatively novel proteomic techniques are slowly being introduced in the field, including, isotopic labeling strategies for protein turnover (Lyon et al., 2014, 2016), or absolute quantification studies (Lehmann et al., 2008; Larrainzar et al., 2009). Combinations of spatial and temporal experiments are rare and there is a need of performing phenotyping of protein abundance at the organelle level to improve our understanding of the legume symbiotic interactions. In this regard, it would be of great interest to connecting protein localization with organelle abundance profiling (Parsons and Heazlewood, 2015). This can be done through the combination of (i) unbiased approachs using e.g., spectral count or LFQs (Maxquant; Hoehenwarter and Wienkoop, 2010; Wienkoop, 2013a) for all proteins belonging to a target organelle (provided that subcellular localization is known) and (ii) selective reaction monitoring (Wienkoop, 2013b; Recuenco-Munoz et al., 2015) of proteotypic peptides selected from abundant and organelle-specific proteins. Also, non-aqueous fractionation, a technique that has been demonstrated to be useful for the integrative subcellular analysis of metabolites and proteins (Arrivault et al., 2014), has thus far not been applied for legume symbiosis research.
Taking into account the large number of proteins involved in response to abiotic and biotic stresses that have been identified, it is now necessary to integrate and compile this information in an accessible way so that this proteomic data and the numerous potential markers identified are analyzed in terms of their contribution to symbiotic and pathogen signaling, nodule formation and nitrogen-fixation efficiency, as well as in improved stress tolerance. The growing application of CRISPR/Cas9 technology and the availability of mutant lines for several model legumes will most likely contribute to this end. Interactions between plant genomic and proteomic specialists and integration of their techniques are key aspects that need to be strengthened in the future.
EL and SW performed the literature review, analysis, drew conclusions, and wrote the article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the Spanish Ministry of Economy and Competitiveness (AGL2014-56561-P and the corresponding FEDER funding, and “Juan de la Cierva” JCI-2012-13175 postdoctoral contract to EL) and SW gratefully acknowledges the Austrian Science Fund (FWF) [P24870-B22] and the COST Action FA1306 for support. Reinhard Turetschek helped with figure design.
2D-PAGE, two-dimensional polyacrylamide gel electrophoresis; AM, arbuscular mycorrhizal; CRISPR, clustered regularly interspaced short palindromic repeats; LC-MS/MS, liquid chromatography coupled to tandem mass spectrometry; MALDI-TOF, matrix-assisted laser desorption/ionization-time of flight mass spectrometry; PBM, peribacteroid membrane; PR, pathogen response; PTM, post-translational modification; SST1, symbiosis-specific sulfate transporter 1.
Abdallah, C., Valot, B., Guillier, C., Mounier, A., Balliau, T., Zivy, M., et al. (2014). The membrane proteome of Medicago truncatula roots displays qualitative and quantitative changes in response to arbuscular mycorrhizal symbiosis. J. Proteomics 108, 354–368. doi: 10.1016/j.jprot.2014.05.028
Alexander, T., Toth, R., Meier, R., and Weber, H. C. (1989). Dynamics of arbuscule development and degeneration in onion, bean, and tomato with reference to vesicular–arbuscular mycorrhizae in grasses. Canad. J. Bot. 67, 2505–2513. doi: 10.1139/b89-320
Aloui, A., Recorbet, G., Gollotte, A., Robert, F., Valot, B., Gianinazzi-Pearson, V., et al. (2009). On the mechanisms of cadmium stress alleviation in Medicago truncatula by arbuscular mycorrhizal symbiosis: a root proteomic study. Proteomics 9, 420–433. doi: 10.1002/pmic.200800336
Aloui, A., Recorbet, G., Robert, F., Schoefs, B., Bertrand, M., Henry, C., et al. (2011). Arbuscular mycorrhizal symbiosis elicits shoot proteome changes that are modified during cadmium stress alleviation in Medicago truncatula. BMC Plant Biol. 11:75. doi: 10.1186/1471-2229-11-75
Amiour, N., Recorbet, G., Robert, F., Gianinazzi, S., and Dumas-Gaudot, E. (2006). Mutations in DMI3 and SUNN modify the appressorium-responsive root proteome in arbuscular mycorrhiza. Mol. Plant Microbe Interact. 19, 988–997. doi: 10.1094/MPMI-19-0988
Antolín-Llovera, M., Petutsching, E. K., Ried, M. K., Lipka, V., Nürnberger, T., Robatzek, S., et al. (2014). Knowing your friends and foes–plant receptor-like kinases as initiators of symbiosis or defence. New. Phytol. 204, 791–802. doi: 10.1111/nph.13117
Arrivault, S., Guenther, M., Florian, A., Encke, B., Feil, R., Vosloh, D., et al. (2014). Dissecting the subcellular compartmentation of proteins and metabolites in arabidopsis leaves using non-aqueous fractionation. Mol. Cell Proteomics 13, 2246–2259. doi: 10.1074/mcp.M114.038190
Becker, A., Barnett, M. J., Capela, D., Dondrup, M., Kamp, P.-B., Krol, E., et al. (2009). A portal for rhizobial genomes: RhizoGATE integrates a Sinorhizobium meliloti genome annotation update with postgenome data. J. Biotechnol. 140, 45–50. doi: 10.1016/j.jbiotec.2008.11.006
Bestel-Corre, G., Dumas-Gaudot, E., and Gianinazzi, S. (2004). Proteomics as a tool to monitor plant-microbe endosymbioses in the rhizosphere. Mycorrhiza 14, 1–10. doi: 10.1007/s00572-003-0280-3
Bestel-Corre, G., Dumas-Gaudot, E., Poinsot, V., Dieu, M., Dierick, J. F., van, T. D., et al. (2002). Proteome analysis and identification of symbiosis-related proteins from Medicago truncatula Gaertn. by two-dimensional electrophoresis and mass spectrometry. Electrophoresis. 23, 122–137. doi: 10.1002/1522-2683(200201)23:1<122::AID-ELPS122>3.0.CO;2-4
Blanquet, P., Silva, L., Catrice, O., Bruand, C., Carvalho, H., and Meilhoc, E. (2015). Sinorhizobium meliloti controls nitric oxide-mediated post-translational modification of a Medicago truncatula nodule protein. Mol. Plant Microbe Interact. 28, 1353–1363. doi: 10.1094/MPMI-05-15-0118-R
Brechenmacher, L., Lee, J., Sachdev, S., Song, Z., Nguyen, T. H., Joshi, T., et al. (2009). Establishment of a protein reference map for soybean root hair cells. Plant Physiol. 149, 670–682. doi: 10.1104/pp.108.131649
Catalano, C. M., Lane, W. S., and Sherrier, D. J. (2004). Biochemical characterization of symbiosome membrane proteins from Medicago truncatula root nodules. Electrophoresis 25, 519–531. doi: 10.1002/elps.200305711
Clarke, V. C., Loughlin, P. C., Gavrin, A., Chen, C., Brear, E. M., Day, D. A., et al. (2015). Proteomic analysis of the soybean symbiosome identifies new symbiotic proteins. Mol. Cell. Proteomics 14, 1301–1322. doi: 10.1074/mcp.M114.043166
Daher, Z., Recorbet, G., Solymosi, K., Wienkoop, S., Mounier, A., Morandi, D., et al. (2016). Changes in plastid proteome and structure in arbuscular mycorrhizal roots display a nutrient starvation signature. Physiol. Plant. 159, 13–29. doi: 10.1111/ppl.12505
Dam, S., Dyrlund, T. F., Ussatjuk, A., Jochimsen, B., Nielsen, K., Goffard, N., et al. (2014). Proteome reference maps of the Lotus japonicus nodule and root. Proteomics 14, 230–240. doi: 10.1002/pmic.201300353
Delmotte, N., Ahrens, C. H., Knief, C., Qeli, E., Koch, M., Fischer, H. M., et al. (2010). An integrated proteomics and transcriptomics reference data set provides new insights into the Bradyrhizobium japonicum bacteroid metabolism in soybean root nodules. Proteomics 10, 1391–1400. doi: 10.1002/pmic.200900710
Delmotte, N., Mondy, S., Alunni, B., Fardoux, J., Chaintreuil, C., Vorholt, J. A., et al. (2014). A proteomic approach of Bradyrhizobium/Aeschynomene root and stem symbioses reveals the importance of the fixA locus for symbiosis. Int. J. Mol. Sci. 15, 3660–3670. doi: 10.3390/ijms15033660
Desalegn, G., Turetschek, R., Kaul, H.-P., and Wienkoop, S. (2016). Microbial symbionts affect Pisum sativum proteome and metabolome under Didymella pinodes infection. J. Proteomics 143, 173–187. doi: 10.1016/j.jprot.2016.03.018
Dimkpa, C., Weinand, T., and Asch, F. (2009). Plant-rhizobacteria interactions alleviate abiotic stress conditions. Plant Cell Environ. 32, 1682–1694. doi: 10.1111/j.1365-3040.2009.02028.x
Djordjevic, M. A. (2004). Sinorhizobium meliloti metabolism in the root nodule: a proteomic perspective. Proteomics 4, 1859–1872. doi: 10.1002/pmic.200300802
Djordjevic, M. A., Chen, H. C., Natera, S., Van Noorden, G., Menzel, C., Taylor, S., et al. (2003). A global analysis of protein expression profiles in Sinorhizobium meliloti: discovery of new genes for nodule occupancy and stress adaptation. Mol. Plant Microbe Interact. 16, 508–524. doi: 10.1094/MPMI.2003.16.6.508
Domonkos, A., Horvath, B., Marsh, J. F., Halasz, G., Ayaydin, F., Oldroyd, G. E. D., et al. (2013). The identification of novel loci required for appropriate nodule development in Medicago truncatula. BMC Plant Biol. 13:157. doi: 10.1186/1471-2229-13-157
Ferrol, N., Pozo, M. J., Antelo, M., and Azcón-Aguilar, C. (2002). Arbuscular mycorrhizal symbiosis regulates plasma membrane H+-ATPase gene expression in tomato plants. J. Exp. Bot. 53, 1683–1687. doi: 10.1093/jxb/erf014
Friso, G., and van Wijk, K. J. (2015). Posttranslational protein modifications in plant metabolism. Plant Physiol. 169, 1469–1487. doi: 10.1104/pp.15.01378
Fujisawa, T., Okamoto, S., Katayama, T., Nakao, M., Yoshimura, H., Kajiya-Kanegae, H., et al. (2014). CyanoBase and RhizoBase: databases of manually curated annotations for cyanobacterial and rhizobial genomes. Nucleic Acids Res. 42, D666–D670. doi: 10.1093/nar/gkt1145
Gallardo, K., Le Signor, C., Vandekerckhove, J., Thompson, R. D., and Burstin, J. (2003). Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiol. 133, 664–682. doi: 10.1104/pp.103.025254
Gemperline, E., Keller, C., Jayaraman, D., Maeda, J., Sussman, M. R., Ané, J.-M., et al. (2016). Examination of endogenous peptides in Medicago truncatula using mass spectrometry imaging. J. Proteome Res. 15, 4403–4411. doi: 10.1021/acs.jproteome.6b00471
Gianinazzi-Pearson, V., Arnould, C., Oufattole, M., Arango, M., and Gianinazzi, S. (2000). Differential activation of H+-ATPase genes by an arbuscular mycorrhizal fungus in root cells of transgenic tobacco. Planta 211, 609–613. doi: 10.1007/s004250000323
Gil-Quintana, E., Larrainzar, E., Seminario, A., Díaz-Leal, J. L., Alamillo, J. M., Pineda, M., et al. (2013). Local inhibition of nitrogen fixation and nodule metabolism in drought-stressed soybean. J. Exp. Bot. 64, 2171–2182. doi: 10.1093/jxb/ert074
Gil-Quintana, E., Lyon, D., Staudinger, C., Wienkoop, S., and González, E. M. (2015). Medicago truncatula and Glycine max : different drought tolerance and similar local response of the root nodule proteome. J. Proteome Res. 14, 5240–5251. doi: 10.1021/acs.jproteome.5b00617
Giovannetti, M., Sbrana, C., Silvia, A., and Avio, L. (1996). Analysis of factors involved in fungal recognition response to host-derived signals by arbuscular mycorrhizal fungi. New Phytol. 133, 65–71. doi: 10.1111/j.1469-8137.1996.tb04342.x
González, E. M., Larrainzar, E., Marino, D., Wienkoop, S., Gil-Quintana, E., and Arrese-Igor, C. (2015). “Physiological responses of N2-fixing legumes to water limitation,” in Legume Nitrogen Fixation in a Changing Environment: Achievements and Challenges, eds S. Sulieman and L. S. P. Tran (Cham: Springer International Publishing), 1–133.
Grimsrud, P. A., den Os, D., Wenger, C. D., Swaney, D. L., Schwartz, D., Sussman, M. R., et al. (2010). Large-scale phosphoprotein analysis in Medicago truncatula roots provides insight into in vivo kinase activity in legumes. Plant Physiol. 152, 19–28. doi: 10.1104/pp.109.149625
Harrison, M. J. (1999). Molecular and cellular aspects of the arbuscular mycorrhizal symbiosis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 361–389. doi: 10.1146/annurev.arplant.50.1.361
Harrison, M. J., Dewbre, G. R., and Liu, J. (2002). A phosphate transporter from Medicago truncatula involved in the acquisition of phosphate released by arbuscular mycorrhizal fungi. Plant Cell 14, 2413–2429. doi: 10.1105/tpc.004861
Hoehenwarter, W., and Wienkoop, S. (2010). Spectral counting robust on high mass accuracy mass spectrometers. Rapid Commun. Mass Spectrom. 24, 3609–3614. doi: 10.1002/rcm.4818
Hooper, C. M., Castleden, I. R., Aryamanesh, N., Jacoby, R. P., and Millar, A. H. (2016). Finding the subcellular location of barley, wheat, rice and maize proteins: the compendium of crop proteins with annotated locations (cropPAL). Plant Cell Physiol. 57:e9. doi: 10.1093/pcp/pcv170
Hooper, C. M., Tanz, S. K., Castleden, I. R., Vacher, M. A., Small, I. D., and Millar, A. H. (2014). SUBAcon: a consensus algorithm for unifying the subcellular localization data of the Arabidopsis proteome. Bioinformatics 30, 3356–3364. doi: 10.1093/bioinformatics/btu550
Hossain, Z., and Komatsu, S. (2014). Potentiality of soybean proteomics in untying the mechanism of flood and drought stress tolerance. Proteomes 2, 107–127. doi: 10.3390/proteomes2010107
Hossain, Z., Khatoon, A., and Komatsu, S. (2013). Soybean proteomics for unraveling abiotic stress response mechanism. J. Proteome. Res. 12, 4670–4684. doi: 10.1021/pr400604b
Hossain, Z., Nouri, M. Z., and Komatsu, S. (2012). Plant cell organelle proteomics in response to abiotic stress. J. Proteome. Res. 11, 37–48. doi: 10.1021/pr200863r
Hummel, J., Niemann, M., Wienkoop, S., Schulze, W., Steinhauser, D., Selbig, J., et al. (2007). ProMEX: a mass spectral reference database for proteins and protein phosphorylation sites. BMC Bioinform. 8:216. doi: 10.1186/1471-2105-8-216
Irar, S., González, E. M., Arrese-Igor, C., and Marino, D. (2014). A proteomic approach reveals new actors of nodule response to drought in split-root grown pea plants. Physiol Plant 152, 634–645. doi: 10.1111/ppl.12214
Joshi, H. J., Hirsch-Hoffmann, M., Baerenfaller, K., Gruissem, W., Baginsky, S., Schmidt, R., et al. (2011). MASCP Gator: an aggregation portal for the visualization of Arabidopsis proteomics data. Plant Physiol. 155, 259–270. doi: 10.1104/pp.110.168195
Kalloniati, C., Krompas, P., Karalias, G., Udvardi, M. K., Rennenberg, H., Herschbach, C., et al. (2015). Nitrogen-fixing nodules are an important source of reduced sulfur, which triggers global changes in sulfur metabolism in Lotus japonicus. Plant Cell 27, 2384–2400. doi: 10.1105/tpc.15.00108
Katam, R., Sakata, K., Suravajhala, P., Pechan, T., Kambiranda, D. M., Naik, K. S., et al. (2016). Comparative leaf proteomics of drought-tolerant and -susceptible peanut in response to water stress. J. Proteomics 143, 209–226. doi: 10.1016/j.jprot.2016.05.031
Komatsu, S., Tougou, M., and Nanjo, Y. (2015). Proteomic techniques and management of flooding tolerance in soybean. J. Proteome Res. 14, 3768–3778. doi: 10.1021/acs.jproteome.5b00389
Kottapalli, K. R., Rakwal, R., Shibato, J., Burow, G., Tissue, D., Burke, J., et al. (2009). Physiology and proteomics of the water-deficit stress response in three contrasting peanut genotypes. Plant Cell Environ. 32, 380–407. doi: 10.1111/j.1365-3040.2009.01933.x
Krause, A., and Broughton, W. J. (1992). Proteins associated with root-hair deformation and nodule initiation in Vigna unguiculata. Mol. Plant Microbe Interact. 5, 96–103. doi: 10.1094/MPMI-5-096
Krusell, L., Krause, K., Ott, T., Desbrosses, G., Kramer, U., Sato, S., et al. (2005). The sulfate transporter SST1 is crucial for symbiotic nitrogen fixation in Lotus japonicus root nodules. Plant Cell 17, 1625–1636. doi: 10.1105/tpc.104.030106
Larrainzar, E., Molenaar, J., Wienkoop, S., Gil-Quintana, E., Alibert, B., Limami, A., et al. (2014). Drought stress provokes the down-regulation of methionine and ethylene biosynthesis pathways in Medicago truncatula roots and nodules. Plant Cell Environ. 37, 2051–2063. doi: 10.1111/pce.12285
Larrainzar, E., Riely, B. K., Kim, S. C., Carrasquilla-Garcia, N., Yu, H. J., Hwang, H. J., et al. (2015). Deep sequencing of the Medicago truncatula root transcriptome reveals a massive and early interaction between nodulation factor and ethylene signals. Plant. Physiol. 169, 233–265. doi: 10.1104/pp.15.00350
Larrainzar, E., Wienkoop, S., Scherling, C., Kempa, S., Ladrera, R., Arrese-Igor, C., et al. (2009). Carbon metabolism and bacteroid functioning are involved in the regulation of nitrogen fixation in Medicago truncatula under drought and recovery. Mol. Plant Microbe Interact. 22, 1565–1576. doi: 10.1094/MPMI-22-12-1565
Larrainzar, E., Wienkoop, S., Weckwerth, W., Ladrera, R., Arrese-Igor, C., and Gonzalez, E. M. (2007). Medicago truncatula root nodule proteome analysis reveals differential plant and bacteroid responses to drought stress. Plant Physiol. 144, 1495–1507. doi: 10.1104/pp.107.101618
Lee, J., Lei, Z., Watson, B. S., and Sumner, L. W. (2013). Sub-cellular proteomics of Medicago truncatula. Front. Plant Sci. 4:112. doi: 10.3389/fpls.2013.00112
Lefebvre, B., Timmers, T., Mbengue, M., Moreau, S., Hervé, C., Tóth, K., et al. (2010). A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 107, 2343–2348. doi: 10.1073/pnas.0913320107
Lei, Z., Elmer, A. M., Watson, B. S., Dixon, R. A., Mendes, P. J., and Sumner, L. W. (2005). A two-dimensional electrophoresis proteomic reference map and systematic identification of 1367 proteins from a cell suspension culture of the model legume Medicago truncatula. Mol. Cell. Proteomics 4, 1812–1825. doi: 10.1074/mcp.D500005-MCP200
Lehmann, U., Wienkoop, S., Tschoep, H., and Weckwerth, W. (2008). If the antibody fails–a mass western approach. Plant J. 55, 1039–1046. doi: 10.1111/j.1365-313X.2008.03554.x
Lim, C. W., Park, J. Y., Lee, S. H., and Hwang, C. H. (2010). Comparative proteomic analysis of soybean nodulation using a supernodulation mutant, SS2-2. Biosci. Biotechnol. Biochem. 74, 2396–2404. doi: 10.1271/bbb.100421
Lyon, D., Castillejo, M. A., Mehmeti-Tershani, V., Staudinger, C., Kleemaier, C., and Wienkoop, S. (2016). Drought and recovery: independently regulated processes highlighting the importance of protein turnover dynamics and translational regulation. Mol. Cell. Proteomics 15, 1921–1937. doi: 10.1074/mcp.M115.049205
Lyon, D., Castillejo, M. A., Staudinger, C., Weckwerth, W., Wienkoop, S., and Egelhofer, V. (2014). Automated protein turnover calculations from 15N partial metabolic labeling LC/MS shotgun proteomics data. PLoS ONE 9:e94692. doi: 10.1371/journal.pone.0094692
Małolepszy, A., Urbański, D. F., James, E. K., Sandal, N., Isono, E., Stougaard, J., et al. (2015). The deubiquitinating enzyme AMSH1 is required for rhizobial infection and nodule organogenesis in Lotus japonicus. Plant J. 83, 719–731. doi: 10.1111/tpj.12922
Marx, H., Minogue, C. E., Jayaraman, D., Richards, A. L., Kwiecien, N. W., Siahpirani, A. F., et al. (2016). A proteomic atlas of the legume Medicago truncatula and its nitrogen-fixing endosymbiont Sinorhizobium meliloti. Nat. Biotechnol. 3, 1198–1205. doi: 10.1038/nbt.3681
Matamoros, M. A., Fernández-García, N., Wienkoop, S., Loscos, J., Saiz, A., and Becana, M. (2013). Mitochondria are an early target of oxidative modifications in senescing legume nodules. New Phytol. 197, 873–885. doi: 10.1111/nph.12049
Mathesius, U., Keijzers, G., Natera, S. H. A., Weinman, J. J., Djordjevic, M. A., and Rolfe, B. G. (2001). Establishment of a root proteome reference map for the model legume Medicago truncatula using the expressed sequence tag database for peptide mass fingerprinting. Proteomics 1, 1424–1440. doi: 10.1002/1615-9861(200111)1:11<1424::AID-PROT1424>3.0.CO;2-J
Mbengue, M., Camut, S., de Carvalho-Niebel, F., Deslandes, L., Froidure, S., Klaus-Heisen, D., et al. (2010). The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22, 3474–3488. doi: 10.1105/tpc.110.075861
Melo, P. M., Silva, L. S., Ribeiro, I., Seabra, A. R., and Carvalho, H. G. (2011). Glutamine synthetase is a molecular target of nitric oxide in root nodules of Medicago truncatula and is regulated by tyrosine nitration. Plant Physiol. 157, 1505–1517. doi: 10.1104/pp.111.186056
Molesini, B., Cecconi, D., Pii, Y., and Pandolfini, T. (2014). Local and systemic proteomic changes in Medicago truncatula at an early phase of Sinorhizobium meliloti infection. J. Proteome Res. 13, 408–421. doi: 10.1021/pr4009942
Mun, T., Bachmann, A., Gupta, V., Stougaard, J., and Andersen, S. U. (2016). Lotus Base: an integrated information portal for the model legume Lotus japonicus. Sci. Rep. 6:39447. doi: 10.1038/srep39447
Nambu, M., Tatsukami, Y., Morisaka, H., Kuroda, K., and Ueda, M. (2015). Quantitative time-course proteome analysis of Mesorhizobium loti during nodule maturation. J. Proteomics 125, 112–120. doi: 10.1016/j.jprot.2015.04.034
Natera, S. H., Guerreiro, N., and Djordjevic, M. A. (2000). Proteome analysis of differentially displayed proteins as a tool for the investigation of symbiosis. Mol. Plant. Microbe. Interact. 13, 995–1009. doi: 10.1094/MPMI.2000.13.9.995
Nguyen, T. H., Brechenmacher, L., Aldrich, J. T., Clauss, T. R., Gritsenko, M. A., Hixson, K. K., et al. (2012). Quantitative phosphoproteomic analysis of soybean root hairs inoculated with Bradyrhizobium japonicum. Mol. Cell Proteomics 11, 1140–1155. doi: 10.1074/mcp.M112.018028
Obayashi, T., Okamura, Y., Ito, S., Tadaka, S., Aoki, Y., Shirota, M., et al. (2014). ATTED-II in 2014: evaluation of gene coexpression in agriculturally important plants. Plant Cell Physiol. 55, 1–7. doi: 10.1093/pcp/pct193
Oehrle, N. W., Sarma, A. D., Waters, J. K., and Emerich, D. W. (2008). Proteomic analysis of soybean nodule cytosol. Phytochemistry 69, 2426–2438. doi: 10.1016/j.phytochem.2008.07.004
Oger, E., Marino, D., Guigonis, J. M., Pauly, N., and Puppo, A. (2012). Sulfenylated proteins in the Medicago truncatula-Sinorhizobium meliloti symbiosis. J. Proteomics 75, 4102–4113. doi: 10.1016/j.jprot.2012.05.024
Panter, S., Thomson, R., de Bruxelles, G., Laver, D., Trevaskis, B., and Udvardi, M. (2000). Identification with proteomics of novel proteins associated with the peribacteroid membrane of soybean root nodules. Mol. Plant Microbe Interact. 13, 325–333. doi: 10.1094/MPMI.2000.13.3.325
Parsons, H. T., and Heazlewood, J. L. (2015). Beyond the Western front: targeted proteomics and organelle abundance profiling. Front. Plant Sci. 6:301. doi: 10.3389/fpls.2015.00301
Paszkowski, U., Kroken, S., Roux, C., and Briggs, S. P. (2002). Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 99, 13324–13329. doi: 10.1073/pnas.202474599
Penmetsa, R. V., and Cook, D. R. (1997). A legume ethylene-insensitive mutant hyperinfected by its rhizobial symbiont. Science 275, 527–530. doi: 10.1126/science.275.5299.527
Pieterse, C. M., Zamioudis, C., Berendsen, R. L., Weller, D. M., Van Wees, S. C., and Bakker, P. A. (2014). Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 52, 347–375. doi: 10.1146/annurev-phyto-082712-102340
Prayitno, J., Imin, N., Rolfe, B. G., and Mathesius, U. (2006). Identification of ethylene-mediated protein changes during nodulation in Medicago truncatula using proteome analysis. J. Proteome. Res. 5, 3084–3095. doi: 10.1021/pr0602646
Ramalingam, A., Kudapa, H., Pazhamala, L. T., Weckwerth, W., and Varshney, R. K. (2015). Proteomics and metabolomics: two emerging areas for legume improvement. Front. Plant Sci. 6:1116. doi: 10.3389/fpls.2015.01116
Rausch, C., Daram, P., Brunner, S., Jansa, J., Laloi, M., Leggewie, G., et al. (2001). A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 414, 462–470. doi: 10.1038/35106601
Recorbet, G., Valot, B., Robert, F., Gianinazzi-Pearson, V., and Dumas-Gaudot, E. (2010). Identification of in planta-expressed arbuscular mycorrhizal fungal proteins upon comparison of the root proteomes of Medicago truncatula colonised with two Glomus species. Fungal Genet. Biol. 47, 608–618. doi: 10.1016/j.fgb.2010.03.003
Recuenco-Munoz, L., Offre, P., Valledor, L., Lyon, D., Weckwerth, W., and Wienkoop, S. (2015). Targeted quantitative analysis of a diurnal RuBisCO subunit expression and translation profile in Chlamydomonas reinhardtii introducing a novel Mass Western approach. J. Proteomics 113, 143–153. doi: 10.1016/j.jprot.2014.09.026
Rose, C. M., Venkateshwaran, M., Volkening, J. D., Grimsrud, P. A., Maeda, J., Bailey, D. J., et al. (2012). Rapid phosphoproteomic and transcriptomic changes in the rhizobia-legume symbiosis. Mol. Cell. Proteomics 11, 724–744. doi: 10.1074/mcp.M112.019208
Ruiz-Lozano, J. M., Collados, C., Barea, J. M., and Azcón, R. (2001). Arbuscular mycorrhizal symbiosis can alleviate drought-induced nodule senescence in soybean plants. New Phytologist. 151, 493–502. doi: 10.1046/j.0028-646x.2001.00196.x
Saalbach, G., Erik, P., and Wienkoop, S. (2002). Characterisation by proteomics of peribacteroid space and peribacteroid membrane preparations from pea (Pisum sativum) symbiosomes. Proteomics 2, 325–337. doi: 10.1002/1615-9861(200203)2:3<325::AID-PROT325>3.0.CO;2-W
Sainz, M., Calvo-Begueria, L., Pérez-Rontomé, C., Wienkoop, S., Abián, J., Staudinger, C., et al. (2015). Leghemoglobin is nitrated in functional legume nodules in a tyrosine residue within the heme cavity by a nitrite/peroxide-dependent mechanism. Plant J. 81, 723–735. doi: 10.1111/tpj.12762
Salavati, A., Bushehri, A. A., Taleei, A., Hiraga, S., and Komatsu, S. (2012). A comparative proteomic analysis of the early response to compatible symbiotic bacteria in the roots of a supernodulating soybean variety. J. Proteomics 75, 819–832. doi: 10.1016/j.jprot.2011.09.022
Sarma, A. D., and Emerich, D. W. (2005). Global protein expression pattern of Bradyrhizobium japonicum bacteroids: a prelude to functional proteomics. Proteomics 5, 4170–4184. doi: 10.1002/pmic.200401296
Sarma, A. D., and Emerich, D. W. (2006). A comparative proteomic evaluation of culture grown vs. nodule isolated Bradyrhizobium japonicum. Proteomics 6, 3008–3028. doi: 10.1002/pmic.200500783
Schenkluhn, L., Hohnjec, N., Niehaus, K., Schmitz, U., and Colditz, F. (2010). Differential gel electrophoresis (DIGE) to quantitatively monitor early symbiosis- and pathogenesis-induced changes of the Medicago truncatula root proteome. J. Proteomics 73, 753–768. doi: 10.1016/j.jprot.2009.10.009
Schulze, W. X., Yao, Q., and Xu, D. (2015). Databases for plant phosphoproteomics. Methods Mol. Biol. 1306, 207–216. doi: 10.1007/978-1-4939-2648-0_16
Schüβler, A., Schwarzott, D., and Walker, C. (2001). A new fungal phylum, the Glomeromycota: phylogeny and evolution. Mycol. Res. 105, 1413–1421. doi: 10.1017/S0953756201005196
Serna-Sanz, A., Parniske, M., and Peck, S. C. (2011). Phosphoproteome analysis of Lotus japonicus roots reveals shared and distinct components of symbiosis and defense. Mol. Plant Microbe Interact. 24, 932–937. doi: 10.1094/MPMI-09-10-0222
Staudinger, C., Mehmeti, V., Turetschek, R., Lyon, D., Egelhofer, V., and Wienkoop, S. (2012). possible role of nutritional priming for early salt and drought stress responses in Medicago truncatula. Front. Plant Sci. 3:285. doi: 10.3389/fpls.2012.00285
Staudinger, C., Mehmeti-Tershani, V., Gil-Quintana, E., Gonzalez, E. M., Hofhansl, F., Bachmann, G., et al. (2016). Evidence for a rhizobia-induced drought stress response strategy in Medicago truncatula. J. Proteomics 136, 202–213. doi: 10.1016/j.jprot.2016.01.006
Tanz, S. K., Castleden, I., Hooper, C. M., Vacher, M., Small, I., and Millar, H. A. (2013). SUBA3: a database for integrating experimentation and prediction to define the SUBcellular location of proteins in Arabidopsis. Nucleic Acids Res. 41, 1185–1191. doi: 10.1093/nar/gks1151
Tatsukami, Y., Nambu, M., Morisaka, H., Kuroda, K., and Ueda, M. (2013). Disclosure of the differences of Mesorhizobium loti under the free-living and symbiotic conditions by comparative proteome analysis without bacteroid isolation. BMC. Microbiol. 13:180. doi: 10.1186/1471-2180-13-180
Toth, K., Stratil, T. F., Madsen, E. B., Ye, J., Popp, C., Antolín-Llovera, M., et al. (2012). Functional domain analysis of the remorin protein LjSYMREM1 in Lotus japonicus. PLoS ONE 7:817. doi: 10.1371/journal.pone.0030817
Turetschek, R., Lyon, D., Desalegn, G., Kaul, H.-P., and Wienkoop, S. (2016). “A Proteomic workflow using high-throughput de novo sequencing towards complementation of genome information for improved comparative crop science,” in Proteomis in Systems Biology SE - 17 Methods in Molecular Biology, ed J. Reinders (New York, NY: Springer), 233–243.
Valot, B., Negroni, L., Zivy, M., Gianinazzi, S., and Dumas-Gaudot, E. (2006). A mass spectrometric approach to identify arbuscular mycorrhiza-related proteins in root plasma membrane fractions. Proteomics 6(Suppl. 1), S145–S155. doi: 10.1002/pmic.200500403
van de Velde, W., Zehirov, G., Szatmari, A., Debreczeny, M., Ishihara, H., Kevei, Z., et al. (2010). Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126. doi: 10.1126/science.1184057
van Loon, L. C., Bakker, P. A., and Pieterse, C. M. (1998). Systemic resistance induced by rhizosphere bacteria. Annu. Rev. Phytopathol. 36, 453–483. doi: 10.1146/annurev.phyto.36.1.453
van Ness, L. K., Jayaraman, D., Maeda, J., Barrett-Wilt, G. A., Sussman, M. R., and Ané, J. M. (2016). Mass spectrometric-based selected reaction monitoring of protein phosphorylation during symbiotic signaling in the model legume, Medicago truncatula. PLoS ONE 11:e0155460. doi: 10.1371/journal.pone.0155460
van Wees, S. C., Van der Ent, S., and Pieterse, C. M. (2008). Plant immune responses triggered by beneficial microbes. Curr. Opin. Plant Biol. 11, 443–448. doi: 10.1016/j.pbi.2008.05.005
Wan, J. R., Torres, M., Ganapathy, A., Thelen, J., DaGue, B. B., Mooney, B., et al. (2005). Proteomic analysis of soybean root hairs after infection by Bradyrhizobium japonicum. Mol. Plant-Microbe Interact. 18, 458–467. doi: 10.1094/MPMI-18-0458
Wang, X., and Komatsu, S. (2016). Gel-free/label-free proteomic analysis of endoplasmic reticulum proteins in soybean root tips under flooding and drought stresses. J. Proteome Res. 15, 2211–2227. doi: 10.1021/acs.jproteome.6b00190
Watson, B. S., Asirvatham, V. S., Wang, L. J., and Sumner, L. W. (2003). Mapping the proteome of barrel medic (Medicago truncatula). Plant Physiol. 131, 1104–1123. doi: 10.1104/pp.102.019034
Wienkoop, S. (2013a). “Spectral count,” in Encyclopedia of Systems Biology, eds W. Dubitzky, O. Wolkenhauer, K.-H. Cho, and H. Yokota (New York, NY: Springer), 1967.
Wienkoop, S. (2013b). “Selective reaction monitoring,” in Encyclopedia of Systems Biology, eds W. Dubitzky, O. Wolkenhauer, K.-H. Cho, and H. Yokota (New York, NY: Springer), 1914.
Wienkoop, S., and Saalbach, G. (2003). Proteome analysis. Novel proteins identified at the peribacteroid membrane from Lotus japonicus root nodules. Plant Physiol. 131, 1080–1090. doi: 10.1104/pp.102.015362
Wienkoop, S., Larrainzar, E., Glinski, M., Gonzalez, E. M., Arrese-Igor, C., and Weckwerth, W. (2008). Absolute quantification of Medicago truncatula sucrose synthase isoforms and N-metabolism enzymes in symbiotic root nodules and the detection of novel nodule phosphoproteins by mass spectrometry. J. Exp. Bot. 59, 3307–3315. doi: 10.1093/jxb/ern182
Wienkoop, S., Staudinger, C., Hoehenwarter, W., Weckwerth, W., and Egelhofer, V. (2012). ProMEX - a mass spectral reference database for plant proteomics. Front. Plant Sci. 3:125. doi: 10.3389/fpls.2012.00125
Ye, H., Gemperline, E., Venkateshwaran, M., Chen, R., Delaux, P. M., Howes-Podoll, M., et al. (2013). MALDI mass spectrometry-assisted molecular imaging of metabolites during nitrogen fixation in the Medicago truncatula-Sinorhizobium meliloti symbiosis. Plant J. 75, 130–145. doi: 10.1111/tpj.12191
Yin, X., and Komatsu, S. (2016). Plant nuclear proteomics for unraveling physiological function. N. Biotechnol. 33, 644–654. doi: 10.1016/j.nbt.2016.03.001
Keywords: proteomics, legume, Rhizobium, arbuscular mycorrhizal fungi, pathogen, abiotic stress, drought
Citation: Larrainzar E and Wienkoop S (2017) A Proteomic View on the Role of Legume Symbiotic Interactions. Front. Plant Sci. 8:1267. doi: 10.3389/fpls.2017.01267
Received: 04 April 2017; Accepted: 05 July 2017;
Published: 18 July 2017.
Edited by:
Wei Wang, Henan Agricultural University, ChinaReviewed by:
Ramesh Katam, Florida A&M University, United StatesCopyright © 2017 Larrainzar and Wienkoop. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Estíbaliz Larrainzar, ZXN0aWJhbGl6LmxhcnJhaW56YXJAdW5hdmFycmEuZXM=
Stefanie Wienkoop, c3RlZmFuaWUud2llbmtvb3BAdW5pdmllLmFjLmF0
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.