
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 12 July 2017
Sec. Plant Breeding
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.01223
 Ningfei Jiang1,2†
Ningfei Jiang1,2† Shilai Shi1,2†
Shilai Shi1,2† Huan Shi1,2†
Huan Shi1,2† Hira Khanzada1
Hira Khanzada1 Ghulam M. Wassan1
Ghulam M. Wassan1 Changlan Zhu1,2,3
Changlan Zhu1,2,3 Xiaosong Peng1,2
Xiaosong Peng1,2 Qiuying Yu1
Qiuying Yu1 Xiaorong Chen1,2
Xiaorong Chen1,2 Xiaopeng He1,2
Xiaopeng He1,2 Junru Fu1,2
Junru Fu1,2 Lifang Hu1,2,3
Lifang Hu1,2,3 Jie Xu1,2
Jie Xu1,2 Linjuan Ouyang1,2
Linjuan Ouyang1,2 Xiaotang Sun1,2,3
Xiaotang Sun1,2,3 Dahu Zhou1,2
Dahu Zhou1,2 Haohua He1,2,3*
Haohua He1,2,3* Jianmin Bian1,2,3*
Jianmin Bian1,2,3*Mapping major quantitative trait loci (QTL) responsible for rice seed germinability under low temperature (GULT) can provide valuable genetic source for improving cold tolerance in rice breeding. In this study, 124 rice backcross recombinant inbred lines (BRILs) derived from a cross indica cv. Changhui 891 and japonica cv. 02428 were genotyped through re-sequencing technology. A bin map was generated which includes 3057 bins covering distance of 1266.5 cM with an average of 0.41 cM between markers. On the basis of newly constructed high-density genetic map, six QTL were detected ranging from 40 to 140 kb on Nipponbare genome. Among these, two QTL qCGR8 and qGRR11 alleles shared by 02428 could increase GULT and seed germination recovery rate after cold stress, respectively. However, qNGR1 and qNGR4 may be two major QTL affecting indica Changhui 891germination under normal condition. QTL qGRR1 and qGRR8 affected the seed germination recovery rate after cold stress and the alleles with increasing effects were shared by the Changhui 891 could improve seed germination rate after cold stress dramatically. These QTL could be a highly valuable genetic factors for cold tolerance improvement in rice lines. Moreover, the BRILs developed in this study will serve as an appropriate choice for mapping and studying genetic basis of rice complex traits.
Low temperature germination is a major determinant for stable establishing of the cultivated rice in tropical or subtropical area where optimum temperatures for rice growth during germination and seedling stages are from 25 to 35°C (Fujino et al., 2004; Xu et al., 2015). The low temperature existing during seed germination stage results poor plants survival rate and alternatively cause reductions in the final yield. Cold tolerance improvement at germination stage is therefore necessary for yield stability and an increase in crop productivity (Fujino et al., 2008). Moreover, paddy field with direct seeding become one of the choice with low labor and cost to overcome the damage of low temperature in rice cultivars (Mao et al., 2015). Improving cold tolerance in rice varieties with conventional breeding methods is labor-and time consuming as cold tolerance is complicated and controlled by quantitative trait loci (QTL). Thus, better understanding of the complexity of QTL underlying cold tolerance at the seed germination stage is estimated to accelerate the progress of cold-tolerant cultivars development by using molecular assisted breeding technology (Koseki et al., 2010).
Rice has been divided into two main subspecies, indica and japonica. In general, indica rice cultivars are cold sensitive and are distributed mainly in tropical and subtropical regions, while japonica cultivars have tolerance to low temperatures and well adapt to temperate climates or higher altitudes (Oka, 1958; Mao et al., 2015). In other words, optimization of these two subspecies become necessary to improve cold-tolerant of rice cultivars.
To find out the genetic basis of cold at germination stage for indica and japonica, several independent studies have been reported and some low-temperature germinability related QTL have been identified in different populations (Miura et al., 2001; Zhang et al., 2005; Jiang et al., 2006; Ji et al., 2008; Wang Z. et al., 2011; Li et al., 2013; Satoh et al., 2016). For example, Miura et al. (2001) detected five low temperature germinability related QTL on chromosomes 2, 4, 5, and 11. Zhang et al. (2005) identified 34 QTL associated with seedling vigor traits (including germination rate, shoot length, root length, and dry weight) by using three temperature regimes (15, 20, and 25°C) (Borjas et al., 2016). Jiang et al. (2006) reported eleven putative QTL for low temperature germination ability on chromosomes 3–5, 7, 9–11 in an indica × japonica cross. Ji et al. (2008) carried out a low-temperature germinability genetic analysis and a total of 11 QTL on chromosomes 2, 5, 7, 8, 11, and 12 were detected. Wang Z. et al. (2011) studied cold tolerance at germination stage in a japonica × indica cross, two minor QTL responsible for cold tolerance at germination were observed. Li et al. (2013) found three QTL (qLTG-7, qLTG-9, and qLTG-12) associated with low-temperature germination, and qLTG-9 was fine mapped to a 72.3-kb region in chromosome 9. Among these QTL, only one QTL (qLTG3-1) has been cloned (Fujino et al., 2008). This is because all these low-temperature germinability related QTL were identified based on traditional markers such as SSRs (simple sequence repeats), which were always sparsely distributed on rice 12 chromosomes; thus it is very difficult to get precise and complete information for these low-temperature germinability QTL (Mao et al., 2015). These uncertain results hindered the mapping of these QTL and limit our understanding of the cold-tolerant presence for indica and japonica. Therefore, the development of new type makers is necessary to facilitate fine mapping and cloning of QTL.
Recently, advances in whole-genome sequencing approach have provided an effective platform for direct identification of million of single nucleotide polymorphisms (SNPs) across the whole genome. The adjacent SNPs with the same genotype in an internal are combined into bins via sliding-window approach (Huang et al., 2009); these bins could demarcate recombination events across the whole population and be used as an effective type of genetic markers for QTL analysis (Wang L. et al., 2011). The high density map developed by the bin-markers has successfully accelerated the genetic studies for quantitative traits in a variety of species (Xie et al., 2010; Xu et al., 2010; Poland et al., 2012; Byrne et al., 2013; Sonah et al., 2013; Wang et al., 2015).
In this study, high-resolution QTL mapping have been reported through sequencing-based genotyping of 124 rice backcross recombinant inbred lines (BRILs). The population was generated from the crossing Oryza sativa spp. indica Changhui 891 with Oryza sativa ssp. japonica 02428. A total of six QTL within relatively small genomic regions were identified for rice germinability under low temperature (GULT). The high-resolution QTL mapping method thus greatly improved the precision and resolution of QTL mapping, meanwhile, the QTL within relatively small genomic regions for rice germination will facilitate the development of novel cold-tolerant cultivars using molecular breeding strategies (Koseki et al., 2010).
Two rice (Oryza sativa L.) varieties; Changhui 891, an excellent indica restorer line in south China, and 02428, a leading japonica wide compatibility variety, were used to develop this mapping population. A BRILs (BC1F6) population consisting of 124 individuals derived from Changhui 891/02428//02428 was developed for subsequently analysis. The population was developed in the experimental field at Jiangxi Agricultural University in Nanchang, Jiangxi Province, and Linwang, Hainan Province. After continuous of six generations of self-fertilization, the genomic DNA from each line of the BC1F6 BRILs was extracted and subjected for genotyping.
The seeds with high-quality were selected by removal of shriveled and unfilled seeds. The seeds were surface-sterilized by dipping 15 min in a 10% solution of sodium hypochlorite and were rinsed for three times with fresh clean tap water (Satoh et al., 2016). Each sample with 30 seeds was placed in a Petri dish containing 5 ml of tap water and incubated at 15°C for 1 week in a growth chamber. Tap water was added every day according to requirement to avoid desiccation, germinated seeds were counted on the seventh day.
The germination potential of seeds that did not germinate at low temperature were evaluated by further incubation of these seeds at 25°C for 24 h and the germinated seeds were counted on the eighth day. The germination test was repeated two times for each line and the germination rate (%) was calculated as: Germination rate (%) = (number of germinated seeds during the test period/total number of germinated seeds) × 100; Seed germination recovery rate = [(number of germinated seeds on eighth day - number of germinated seeds on seventh day)/number of total germinated seeds] × 100.
Healthy and young fresh leaves were collected from 124 BRILs along with their parental lines and subjected to DNA extraction using cetyltrimethylammonium bromide (CTAB) method with minor modifications. Each DNA was cut by an enzyme Ecor I and the adapters were ligated to barcode the DNA of each line. Size of each sample was selected by gel electrophoresis and DNA fragments were consisted of 300–500 bp in length. Finally, the libraries were enriched by PCR amplification and sequenced was performed using Illumina HiSeq 4000 instrument using paired-end reads (100 bp). The raw reads were then filtered and sorted according to indices and the high-quality SNPs between parents were termed by alignment with Nipponbare reference genome1.
Due to the high linkage disequilibrium, high-density genetic map of populations often contain many redundant markers that provides no any new information. Otherwise, a small number of genotypes are falsely termed due to sequencing error. To overcome these issues, a modified sliding-window approach was recommended (Huang et al., 2009). According to the method of Chen et al. (2014), consecutive 40-kb intervals that lack a recombination event in the population were combined into bins via sliding-window approach (Huang et al., 2009); these bins could demarcate recombination events across the whole population and be used as a genetic marker for linkage map construction using IciMapping v4.1 Mapping software2.
To explore the gene expression for seed germination for the BRIL population, QTL IciMapping v4.1 software was used for QTL analysis, threshold LOD of 2.5 was applied (Bian et al., 2015). QTL nomenclature was followed by the method of McCouch et al. (2008).
For each BRIL individual, the reads of the 100-bp sequences were sort out on indices basis. A total of ∼890.09 million reads with average of ∼7.18 million reads per BRIL individual were generated (Supplementary Table S1). The ratio of Q20 for each sample was above 90%, thus the quality of the data is very high and meet the requirements for further analysis. The sequencing reads were then aligned to reference genome using SOAP2 software (Li et al., 2009b) and SNPs were identified with the SOAPsnp software package (Li et al., 2009a). The SNP numbers of parental lines 02428 and Changhui 891 are 22,358 and 85,605 as compared to the Nipponbare rice reference. Finally, a total of 70,480 high-quality SNPs were detected in two parents were selected (Figure 1). According to the method as described above, a total of 3057 bin markers were obtained to construct the recombination map.
A linkage map was developed using the 3057 recombination bins which were generated from whole-genome re-sequencing of the 124 BRILs. Total genetic distance of linkage map was 1266.5 cM with an average interval of 0.41 cM between adjacent bins (Supplementary Data Sheet 1 and Figure 2). The genetic distance between adjacent bins was ranging from 0 to 9.07 cM. The 12 rice linkage groups varied in the number of markers and marker density (Figures 2, 3). Total marker density (0.24 cM/marker) was highest in linkage group on chromosome 8. The lowest marker density (0.54 cM/marker) was in linkage group on chromosome 10.
Overall, 41.7% of the parental line 02428 seeds were germinated at 15°C at 7 days after sowing, whereas most of the Changhui 891 seeds had not germinated (Table 1 and Figure 4). While after incubation at 25°C for 24 h after the germination test at low temperature, the germinate rate of 02428 line was 75.0% while Changhui 891 had germinated rate of 70.0%. The seed germinate recovery rate of Changhui 891 (68.3%) is higher than that of 02428 (33.3%) (Figure 4).

FIGURE 4. The germination of the two parents: Changhui 891 (left) and 02428 (right) on seventh day (A) and eighth day (B).
The average number of seed germination % per each BRIL individual was 21.7% ranging from 0 to 73.3% under 15°C at 7 days after sowing (Table 1). After incubation at 25°C for 24 h of the germination test at low temperature, the average germinate rate was 76.4% with a range of 18.3 to 96.7%, and in the end, seed germinate recovery rate was 54.7% with a range of 6.7 to 96.7% in the BRIL population.
In total, six QTL were identified by interval mapping with IciMapping v4.1 software using high-density map, One QTL for seed germination on seventh day, two QTL for seed germination on eighth day and three QTL for seed germination rate recovery after cold stress were identified in BRIL population (Table 2 and Figure 5).
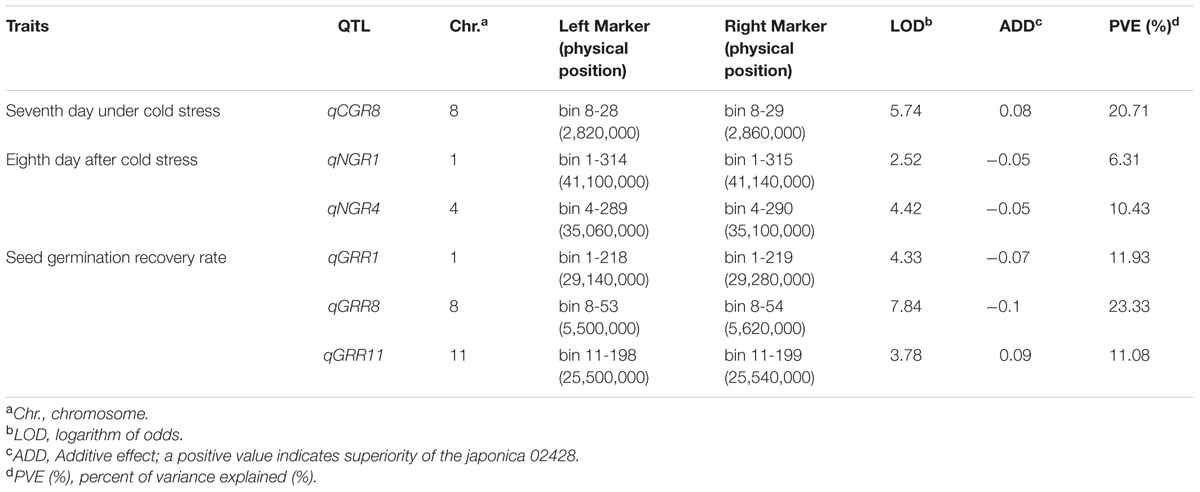
TABLE 2. Putative QTL associated with seed germination under cold stress in the BRIL population in rice.
Only one QTL (qCGR8) for seed germination on seventh day was detected with phenotypic variation explained (PVE) of 20.71% and mapped on a region between bin 8-28 and bin 8-29 on chromosome 8. The genetic distance was 0.2 cM and the physical distance was 40 kb in Nipponbare genome. The allele with increasing effects on the seed germination was shared by japonica 02428.
Two QTL for seed germination on eighth day were detected. QTL (qNGR1) was harbored on chromosome 1 on a region between bin 1-314 and bin 1-315 and covered the genetic distance was about 1.15 cM, and physical distance was about 40 kb in Nipponbare genome. QTL qNGR4 was identified on chromosome 4 and covered genetic distance of about 0.2 cM between bin 4-289 and bin 4-290, and the physical distance was about 40 kb in Nipponbare genome. The alleles with increasing effects for seed germination rate were shared by indica Changhui 891 line.
Among the three QTL detected for seed germination rate recovery after cold stress, QTL (qGRR8) was mapped on chromosome 8 on the region of bin 8-53 and bin 8-54. It contributed the largest PVE value (23.33%). The genetic distance was 1.96 cM, and a physical distance was about 120 kb in Nipponbare genome. The QTL (qGRR1) is located between bin 1-218 and bin1-219 on chromosome 1, the genetic distance covered is 0.2 cM, and a physical distance of about 140 kb to corresponding genome. While QTL (qGRR11) is detected on chromosome 11 between bin 11-198 and bin11-199, the genetic distance was about 0.62 cM, and a physical distance of about 40 kb in Nipponbare genome. The alleles with the increasing additive effects on seed germination recovery rate after cold stress for QTL qGRR1 and qGRR8 were from Changhui 891 while for QTL qGRR11 were shared by parental line 02428.
An appropriate mapping population is effective in QTL analysis and gene cloning. Due to genetic background noise, the standard segregating populations cannot give an precise information (the location and the size) for each individual QTL, particularly small effect QTL. Near-isogenic lines (NILs) (CSSLs) and chromosome segment substitution lines can overcome this issue and therefore ideal for QTL detection (Kubo et al., 1999; Bian et al., 2010), but they are time consuming and labor-intensive which limited the acceleration of gene cloning. BRILs being a permanent population are easy to develop compare to CSSLs and NILs. Moreover, because of the high percentage of recurrent parent genome, it can be rapidly crossed with the recurrent parent to construct NILs for the target QTL cloning. Therefore, it become necessary to develop BRIL population for the genetic analysis of quantitative traits in rice (Wang B. et al., 2011; Hosseini et al., 2012). In this study, a BRIL population consisting of 124 individuals derived from Changhui 891/02428//02428 was developed, hence we assume this population will serve as better choice for mapping and studying important quantitative traits of rice.
Quantitative trait loci mapping resolution depended on the marker density and the size of confidence interval of QTL (Visscher et al., 1996; Da et al., 2000). Generally speaking, a gain of information by more markers density resulted in smaller intervals, therefore, the application of additional markers is an effective way to increase QTL mapping resolution (Liu et al., 2008). The traditional markers such as SSRs and restriction fragment length polymorphism (RFLP) are not suitable when their linkage distance are zero and cannot meet the requirements of the high QTL mapping resolution. Development of SNP markers by next-generation sequencing technologies will increase the feasibility to increase QTL mapping resolution. In this study, an ultra-high density bin-map for the BRIL population was constructed through re-sequence strategy and used for QTL mapping in rice for seed germination. Compared with previous QTL associated with rice seed germination, the QTL interval size in this study was significantly narrow and ranged from 40 to 140 kb in Nipponbare genome. These results confirmed that genotyping by sequencing has significantly increased the QTL mapping resolution compared with traditional markers.
The position of qCGR8 on chromosome 8 is near to the major QTL qCTS8.1 which is responsible for increasing seedling cold tolerance in japonica rice variety Daugandao (Wang Z. et al., 2011). These varieties may thus share the QTL for cold tolerance. In this study, qCGR8 was identified on a region about 0.2 cM, and the physical distance was about 40 kb. Six candidate genes were predicted, which provide a good clue for the cold tolerance gene isolation (Table 3). Otherwise, chromosome 11 locus qGRR11 maps to a similar location as does by qLTG11-2 (Jiang et al., 2006) and qLTG11-2 (Wang Z. et al., 2011). It has been noticed that the region (marker by bin 11-198–bin 11-199) may play an important role in seedling germination under low temperature (Satoh et al., 2016). Identification of the gene from different varieties allow us to understand the roles of this region in seedling germination (Satoh et al., 2016) but the accurate location for this QTL is still not clear. Luckily, qGRR11 was mapped between bin11-198 and bin11-199 on chromosome 11 in this study covering the genetic distance of 0.62 cM, corresponding to a physical distance of about 40 kb. Six candidate genes were predicted in this region (Table 3), it would be only a matter of time before the isolation of qNGR11. Four new QTL; qNGR1, qNGR4, qGRR1, and qGRR8 are newly detected QTL and never reported before, these all QTL were located on small genomic regions (40–140 kb). This result contributes to the fine mapping of these QTL and provides a foundation for understanding the mechanism of rice germination and seedling growth under low-temperature conditions (Li et al., 2013). Moreover, the BRILs harboring these QTL will serve as an excellent materials and enable us to map and study genetic basis of GULT.
Vigorous GULT is an important agricultural trait for direct seeding in rice especially at high altitudes in subtropical and tropical regions, and in areas with cold irrigated water (Fujino et al., 2004; Li et al., 2013; Mao et al., 2015; Satoh et al., 2016). Generally, GULT of japonica varieties was higher than that of indica varieties, cold resistant genes/QTL from japonica rice have been used as gene resource to improve the cold resistance in indica rice (Mao et al., 2015). However, only few of these QTL are considered appropriate for improving cold tolerance in indica because of the ambiguous chromosome position. In present study, six QTL ranged from 40 to 140 kb in Nipponbare genome were fine-mapped, further small genomic region can be transferred directly by molecular assisted selection (MAS). Among these, two QTL, qCGR8 and qGRR11 whose alleles from 02428 could increase GULT and seed germination recovery rate after cold stress, respectively. These QTL will be available in cold-tolerant rice breeding programs especially for cold-tolerant indica rice breeding program. For qNGR1 and qNGR4, the increasing effect of alleles were detected from Changhui 891 for seed germination and may be two major QTL from Changhui 891 affecting germination under normal condition. However, two QTL qGRR1 and qGRR8 affecting the seed germination recovery rate after cold stress, the alleles from Changhui 891 could improve seed germination rate dramatically after cold stress. This also suggest that alleles shared by Changhui 891 for QTL qGRR1 and qGRR8 would be a highly valuable genetic factors for improving cold tolerant in rice lines. In other words, the rice lines harboring Changhui 891 alleles for qGRR1 and qGRR8 could significantly improve the seed germinability even after a period of cold stress.
JB and HH designed the research work, annotated the data and drafted the manuscript. NJ, HS, SS, CZ, XP, and QY performed the experiments. XC, XH, JF, LH, LO, JX, XS, and DZ developed the population. HK and GW revised the manuscript for the language.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank the referees for their critical comments on this manuscript. This research was supported by grant (2016YFD0101801) from The National Key Research and Development Program of China and Technology Department and grants (31560386 and 31360326) from National Nature Science Foundation of China.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01223/full#supplementary-material
DATA SHEET 1 | The bin marker information of the high density map.
Bian, J., Zhu, G., Zhu, C., Peng, X., Li, C., He, X., et al. (2015). Molecular dissection of developmental behavior of tiller number and the relationship with effective panicle using indica–japonica introgression lines in rice. Mol. Breed. 35:91. doi: 10.1007/s11032-015-0269-6
Bian, J. M., Jiang, L., Liu, L. L., Wei, X. J., Xiao, Y. H., Zhang, L. J., et al. (2010). Construction of a new set of rice chromosome segment substitution lines and identification of grain weight and related traits QTLs. Breed. Sci. 60, 305–313. doi: 10.1270/jsbbs.60.305
Borjas, A. H., Teresa, B. D. L., and Prasanta, K. S. (2016). Genetic analysis of germinating ability and seedling vigor under cold stress in US weedy rice. Euphytica 208, 251–264. doi: 10.1007/s10681-015-1584-z
Byrne, S., Czaban, A., Studer, B., Panitz, F., Bendixen, C., and Asp, T. (2013). Genome wide allele frequency fingerprints (GWAFFs) of populations via genotyping by sequencing. PLoS ONE 8:e57438. doi: 10.1371/journal.pone.0057438
Chen, Z., Wang, B., Dong, X., Liu, H., Ren, L., Chen, J., et al. (2014). An ultra-high density bin-map for rapid QTL mapping for tassel and ear architecture in a large F2 maize population. BMC Genomics 15:433. doi: 10.1186/1471-2164-15-433
Da, Y., Vanraden, P. M., and Schook, L. B. (2000). Detection and parameter estimation for quantitative trait loci using regression models and multiple markers. Genet. Sel. Evol. 32, 357–381. doi: 10.1186/1297-9686-32-4-357
Fujino, K., Sekiguchi, H., Matsuda, Y., Sugimoto, K., Ono, K., and Yano, M. (2008). Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. U.S.A. 105, 12623–12628. doi: 10.1073/pnas.0805303105
Fujino, K., Sekiguchi, H., Sato, T., Kiuchi, H., Nonoue, Y., Takeuchi, Y., et al. (2004). Mapping of quantitative trait loci controlling low-temperature germinability in rice (Oryza sativa L.). Theor. Appl. Genet. 108, 794–799. doi: 10.1007/s00122-003-1509-4
Hosseini, M., Houshmand, S., Mohamadi, S., Tarang, A., Khodambashi, M., and Rahimsoroush, H. (2012). Detection of QTLs with main, epistatic and QTL × environment interaction effects for rice grain appearance quality traits using two populations of backcross inbred lines (BILs). Field Crops Res. 135, 97–106. doi: 10.1016/j.fcr.2012.07.009
Huang, X., Feng, Q., Qian, Q., Zhao, Q., Wang, L., Wang, A., et al. (2009). High-throughput genotyping by whole-genome resequencing. Genome Res. 19, 1068–1076. doi: 10.1101/gr.089516.108
Ji, S. L., Jiang, L., Wang, Y. H., Liu, S. J., Liu, X., Zhai, H. Q., et al. (2008). QTL and epistasis for low temperature germinability in rice. Acta Agron. Sin. 34, 551–556. doi: 10.3724/SP.J.1006.2008.00551
Jiang, L., Liu, S., Hou, M., Tang, J., Chen, L., Zhai, H., et al. (2006). Analysis of QTLs for seed low temperature germinability and anoxia germinability in rice (Oryza sativa L.). Field Crop Res. 98, 68–75. doi: 10.1016/j.fcr.2005.12.015
Koseki, M., Kitazawa, N., Yonebayashi, S., Maehara, Y., Wang, Z., and Minobe, Y. (2010). Identification and fine mapping of a major quantitative trait locus originating from wild rice, controlling cold tolerance at the seedling stage. Mol. Genet. Genomics 284, 45–54. doi: 10.1007/s00438-010-0548-1
Kubo, T., Nakamura, K., and Yoshimura, A. (1999). Development of a series of Indica chromosome segment substitution lines in Japonica background of rice. Rice Genet. Newsl. 16, 104–106.
Li, L., Liu, X., Xie, K., Wang, Y., Liu, F., Lin, Q., et al. (2013). qLTG-9, a stable quantitative trait locus for low temperature germination in rice (Oryza sativa L.). Theor. Appl. Genet. 126, 2313–2322. doi: 10.1007/s00122-013-2137-2
Li, R., Li, Y., Fang, X., Yang, H., Wang, J., Kristiansen, K., et al. (2009a). SNP detection for massively parallel whole-genome resequencing. Genome Res. 19, 1124–1132. doi: 10.1101/gr.088013.108
Li, R., Yu, C., Li, Y., Lam, T. W., Yiu, S. M., Kristiansen, K., et al. (2009b). SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics 25, 1966–1967. doi: 10.1093/bioinformatics/btp336
Liu, X., Zhang, H., Li, H., Li, N., Zhang, Y., Zhang, Q., et al. (2008). Fine-mapping quantitative trait loci for body weight and abdominal fat traits: effects of marker density and sample size. Poult. Sci. 87, 1314–1319. doi: 10.3382/ps.2007-00512
Mao, D., Yu, L., Chen, D., Li, L., Zhu, Y., Xiao, Y., et al. (2015). Multiple cold resistance loci confer the high cold tolerance adaptation of Dongxiang wild rice (Oryza rufipogon) to its high-latitude habitat. Theor. Appl. Genet. 128, 1359–1371. doi: 10.1007/s00122-015-2511-3
McCouch, S. R., and Committee on Gene Symbolization, Nomenclature, and Linkage [CGSNL], Rice Genetics Cooperative. (2008). Gene nomenclature system for rice. Rice 1, 72–84. doi: 10.1007/s12284-008-9004-9
Miura, K., Lin, S. Y., Yano, M., and Nagamine, T. (2001). Mapping quantitative trait loci controlling low temperature germinability in rice (Oryza sativa L.). Breed. Sci. 51, 293–299. doi: 10.1270/jsbbs.51.293
Oka, H. I. (1958). Intervarietal variation and classification of cultivated rice. Indian J. Genet. Plant Breed. 18, 79–89.
Poland, J. A., Brown, P. J., Sorrells, M. E., and Jannink, J. L. (2012). Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS ONE 7:e32253. doi: 10.1371/journal.pone.0032253
Satoh, T., Tezuka, K., Kawamoto, T., Matsumoto, S., Satoh-Nagasawa, N., Ueda, K., et al. (2016). Identification of QTLs controlling low-temperature germination of the East European rice (Oryza sativa L.) variety Maratteli. Euphytica 207, 245–254. doi: 10.1007/s10681-015-1531-z
Sonah, H., Bastien, M., Iquira, E., Tardivel, A., Légaré, G., Boyle, B., et al. (2013). An improved genotyping by sequencing (GBS) approach offering increased versatility and efficiency of SNP discovery and genotyping. PLoS ONE 8:e54603. doi: 10.1371/journal.pone.0054603
Visscher, P. M., Thompson, R., and Haley, C. S. (1996). Confidence intervals in QTL mapping by bootstrapping. Genetics 143, 1013–1020.
Wang, B., Zhu, C., Liu, X., Wang, W., Ding, H., Jiang, M., et al. (2011). Fine mapping of qHD4-1, a QTL controlling the heading date, to a 20.7-kb DNA fragment in rice (Oryza sativa L.). Plant Mol. Biol. Rep. 29, 702–713. doi: 10.1007/s11105-010-0278-x
Wang, L., Wang, A., Huang, X., Zhao, Q., Dong, G., Qian, Q., et al. (2011). Mapping 49 quantitative trait loci at high resolution through sequencing-based genotyping of rice recombinant inbred lines. Theor. Appl. Genet. 122, 327–340. doi: 10.1007/s00122-010-1449-8
Wang, X., Yu, K., Li, H., Peng, Q., Chen, F., Zhang, W., et al. (2015). High-density SNP map construction and QTL identification for the apetalous character in Brassica napus L. Front. Plant Sci. 6:1164. doi: 10.3389/fpls.2015.01164
Wang, Z., Wang, F., Zhou, R., Wang, J., and Zhang, H. (2011). Identification of quantitative trait loci for cold tolerance during the germination and seedling stages in rice (Oryza sativa L.). Euphytica 181, 405–413. doi: 10.1007/s10681-011-0469-z
Xie, W., Feng, Q., Yu, H., Huang, X., Zhao, Q., Xing, Y., et al. (2010). Parent-independent genotyping for constructing an ultrahigh-density linkage map based on population sequencing. Proc. Natl. Acad. Sci. U.S.A. 107, 10578–10583. doi: 10.1073/pnas.1005931107
Xu, J., Zhao, Q., Du, P., Xu, C., Wang, B., Feng, Q., et al. (2010). Developing high throughput genotyped chromosome segment substitution lines based on population whole-genome re-sequencing in rice (Oryza sativa L.). BMC Genomics 11:656. doi: 10.1186/1471-2164-11-656
Xu, M., Ye, X., Wang, W., Wei, C., Zhang, J., and Tu, J. (2015). Genetic analysis and molecular mapping of a high threshold and low temperature-sensitive mutant in rice (Oryza sativa L.) at the seedling stage. Euphytica 203, 71–82. doi: 10.1007/s10681-014-1260-8
Keywords: rice, low temperature, seed germinability, high-density genetic map, fine mapping
Citation: Jiang N, Shi S, Shi H, Khanzada H, Wassan GM, Zhu C, Peng X, Yu Q, Chen X, He X, Fu J, Hu L, Xu J, Ouyang L, Sun X, Zhou D, He H and Bian J (2017) Mapping QTL for Seed Germinability under Low Temperature Using a New High-Density Genetic Map of Rice. Front. Plant Sci. 8:1223. doi: 10.3389/fpls.2017.01223
Received: 04 April 2017; Accepted: 28 June 2017;
Published: 12 July 2017.
Edited by:
Chengdao Li, Murdoch University, AustraliaReviewed by:
Liezhao Liu, Southwest University, ChinaCopyright © 2017 Jiang, Shi, Shi, Khanzada, Wassan, Zhu, Peng, Yu, Chen, He, Fu, Hu, Xu, Ouyang, Sun, Zhou, He and Bian. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jianmin Bian, am1iaWFuODFAMTI2LmNvbQ== Haohua He, aGhodWE2NEAxNjMuY29t
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.