- 1Key Laboratory for Plant Diversity and Biogeography of East Asia, Kunming Institute of Botany, Chinese Academy of Science, Kunming, China
- 2Plant Germplasm and Genomics Center, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 3Institute of Tibetan Plateau Research at Kunming, Kunming Institute of Botany, Chinese Academy of Sciences, Kunming, China
- 4University of Chinese Academy of Sciences, Beijing, China
- 5School of Life Sciences, Yunnan University, Kunming, China
The calcineurin B-like protein (CBL)–CBL-interacting protein kinase (CIPK) complex has been identified as a primary component in calcium sensors that perceives various stress signals. Turnip (Brassica rapa var. rapa) has been widely cultivated in the Qinghai–Tibet Plateau for a century as a food crop of worldwide economic significance. These CBL–CIPK complexes have been demonstrated to play crucial roles in plant response to various environmental stresses. However, no report is available on the genome-wide characterization of these two gene families in turnip. In the present study, 19 and 51 members of the BrrCBL and BrrCIPK genes, respectively, are first identified in turnip and phylogenetically grouped into three and two distinct clusters, respectively. The expansion of these two gene families is mainly attributable to segmental duplication. Moreover, the differences in expression patterns in quantitative real-time PCR, as well as interaction profiles in the yeast two-hybrid assay, suggest the functional divergence of paralog genes during long-term evolution in turnip. Overexpressing and complement lines in Arabidopsis reveal that BrrCBL9.2 improves, but BrrCBL9.1 does not affect, salt tolerance in Arabidopsis. Thus, the expansion of the BrrCBL and BrrCIPK gene families enables the functional differentiation and evolution of some new gene functions of paralog genes. These paralog genes then play prominent roles in turnip's adaptation to the adverse environment of the Qinghai–Tibet Plateau. Overall, the study results contribute to our understanding of the functions of the CBL–CIPK complex and provide basis for selecting appropriate genes for the in-depth functional studies of BrrCBL–BrrCIPK in turnip.
Introduction
Plants are exposed to various adverse stress conditions during their growth and development process. To cope with numerous environmental stimuli, plants form a series of complex signal transduction mechanisms to perceive, transduce, and respond to different stresses. These responses can minimize the injury derived from adversity. Calcium is a ubiquitous intracellular secondary messenger in plants. This element regulates various plant growth and development processes, as well as abiotic and biotic stress responses (Xiong et al., 2002). In calcium regulatory networks, calcium-binding proteins that function as sensor molecules specifically receive cellular calcium signals and transmit these signals to downstream pathways (Roberts and Harmon, 1992; Li et al., 2016). The calcineurin B-like protein (CBL) belongs to a unique group of calcium sensors in plants. Hence, the calcium signature can bind to the elongation factor (EF) hand domains of the CBL proteins. Then, the CBL proteins bind to the NAF/FISL domain of the C-terminal of the CBL-interacting protein kinases (CIPKs). As such, the CBL proteins control the affinities and activities of numerous ion transporters (Kolukisaoglu et al., 2004; Luan, 2009). The CBL–CIPK network represents an example of a significantly divergent Ca2+-decoding paradigm unique to plants. The N-terminal MGCXXS/T motif of the CBL protein that mediates lipid modification by myristoylation and palmitoylation directs the CBL–CIPK system to an exact cellular target region. As a result, the CIPK is stimulated to phosphorylate the proper target proteins (Batistic et al., 2008; Batistic and Kudla, 2009). CBL1, -4, -5, -8, and -9 contained the shortest N-termini (Batistic et al., 2010; Sanchez-Barrena et al., 2013). Most of the genes contained the conserved MGCXXS/T motif that helped the CBLs to anchor in the membrane (Batistic et al., 2008; Weinl and Kudla, 2009). Arabidopsis CBL1, -4, -5, and -9 localize to the plasma membrane (Batistic et al., 2010).
The CBL and CIPK genes have been identified in many species, such as Arabidopsis thaliana (Kolukisaoglu et al., 2004), Populus (Zhang et al., 2008), Oryza sativa (Chen X. F. et al., 2011), Zea mays (Chen X. F. et al., 2011), Brassica napus L. (Zhang et al., 2014), Triticum aestivum L. (Sun et al., 2015), Solanum melongena L. (Li et al., 2016), Physcomitrella patens (Weinl and Kudla, 2009), and Selaginella moellendorffii (Weinl and Kudla, 2009). These studies have extended the analysis of CBL–CIPK interactions to the entire family of CBLs and CIPKs to discover functional pairs of CBLs and CIPKs. Previous studies reported that CBL–CIPK complexes are involved in mediating Ca2+ signals elicited by many kinds of stresses, such as low potassium content (CBL1/CBL9–CIPK23 pathway) (Li et al., 2006; Xu et al., 2006; Cheong et al., 2007; Lee et al., 2007), high salt level (CBL4/CBL10–CIPK24 pathway) (Qiu et al., 2002; Cheng et al., 2004), abscisic acid (ABA) exposure (CBL9–CIPK3 pathway) (Pandey et al., 2004), and cold (CBL1–CIPK7 pathway) (Huang et al., 2011). Studies on the CBL–CIPK network extensively demonstrated the interaction specificity and overlap among various members of the CBL and CIPK family. This finding reflects both the functional specificity and redundancy of the CBL and CIPK genes. For example, Arabidopsis CBL1 and CBL9 are similar in their amino-acid sequences and serve overlapping and specific functions (Albrecht et al., 2003; Pandey et al., 2004; Li et al., 2006; Xu et al., 2006; Cheong et al., 2007). This observation suggests that calcium sensors with high sequence similarities or a close evolutionary relationship may perform very distinct functions, and duplicated genes usually evolve some new functions. The overexpression and mutant analysis of CBLs and CIPKs have greatly enriched our understanding of the gene's functions (Thapa et al., 2011). These analyses have shown that the genes might develop tolerance to different stresses; thus, the plant harboring these genes can adapt to unfavorable conditions. For instance, CBL1 expression is induced by wounding, high-salt conditions, cold, ABA exposure, and drought (Kudla et al., 1999; Cheong et al., 2003). The cbl1 mutant is hypersensitive to drought, high-salt conditions, and hyperosmotic stress (Albrecht et al., 2003; Cheong et al., 2003). Meanwhile, CBL9 is involved in ABA signaling and stress-induced ABA biosynthetic pathways (Pandey et al., 2004). ThCBL9 overexpression conferred salt tolerance to transgenic A. thaliana (Sun et al., 2008). cbl9 mutant plants are hypersensitive to ABA or high salt and mannitol (Pandey et al., 2004; Sun et al., 2008). Moreover, the double mutant cbl1cbl9 has been generated to investigate the functions of the corresponding genes as that in the study of the CBL1/9–CIPK23 complex involved in the regulation of potassium absorption. cbl1cbl9 double mutants display defects in low-K conditions (Li et al., 2006; Xu et al., 2006; Lee et al., 2007), whereas cbl1 and cbl9 signaling mutants did not show phenotypic changes in K uptake (Cheong et al., 2003; Pandey et al., 2004).
Turnip (Brassica rapa var. rapa) belongs to a well-known plant family called Brassicaceae. The plant is a traditional crop highly adapted to the extreme environments of the Qinghai–Tibet Plateau and hold edible, feeding, and pharmaceutical values (Liang et al., 2006). Turnip undergoes a range of environmental stresses in its natural environment and has evolved a wide range of mechanisms to cope with these stresses. Accordingly, in this study, we explore the mechanisms and functions of the CBL–CIPK signaling system involved turnip's response to extreme environments. The functions of some members of CBL and CIPK have been studied in some species. However, the functions of many CBLs and CIPKs remain to be characterized to date. Moreover, the CBL–CIPK network in other plants remains fragmentary. To date, no systematic analysis of CBL or CIPK family genes has been reported in turnip. The manner by which specific CBL–CIPK complexes participate in turnip growth and development, as well as in response to abiotic and jasmonic acid (stress hormone) treatments, remains to be revealed. Thus, to analyze the mechanisms and functions of the CBL–CIPK signaling system in turnip in response to extreme environments is of great significance. In this present study, we carried out a genome-wide analysis and identified 19 BrrCBL and 51 BrrCIPK genes in turnip. We determined the genomic information; phylogenetic relationships; chromosome localization; and expression analysis for different tissues (root, stem, leaf, and flower), different development stages (early stage [ES], cortex-splitting stage [CSS], and root-thickening stage [RTS]), and stress treatments (drought, cold, NaCl, and methyl jasmonate [MeJA]) related to the two families of genes and proteins. We also investigated the BrrCBL–BrrCIPK interactions, determined their effects on salt tolerance, and performed overexpression and complementation analyses of BrrCBL9.1 and -9.2. Functional divergence was determined between paralog genes in the two families of genes and proteins. The results provide a solid foundation and may help in further elucidating how the turnip CBL–CIPK network enables the integration of multiple signals from the plant's environment and coordinate downstream responses to stresses in the Qinghai–Tibet Plateau. The findings also offer a useful framework for additional functional studies in this area.
Materials and Methods
Identification and Structure Analyses of the CBL and CIPK Gene Family in Turnip
The 10 Arabidopsis CBL and 26 CIPK genes were downloaded from TAIR (http://www.arabidopsis.org) as queries to search against turnip genomes (http://www.bioinformatics.nl/brassica/index.html?data=bras_tp%2Fdata&loc=A03%3A22821424..22821483&tracks=DNA%2CTranscripts%2CGenes&highlight=). The domains and functional sites in each protein were examined with ps_scan.pl. All CBL protein sequences containing the EF-hand calcium-binding domains (PS50222), as well as all CIPK protein sequences with protein kinase domains (PS50011) and the NAF/FISL motif (PS50816) were extracted as candidates. All candidates were used to search against the GenBank nonredundant protein database. Homology analysis between turnip and Arabidopsis was conducted using DNAMAN software. Physicochemical parameters including the MW, theoretical PI, grand average of hydropathicity, and the number of amino acids were calculated using the ProtParam tool of ExPaSy (http://web.expasy.org/protparam/; Gasteiger et al., 2005). Putative EF-hand was predicated using a simple modular architecture research tool (http://smart.embl-heidelberg.de/). Myristoylaton and palmitoylation motifs were predicted using PlantsP (http://mendel.imp.ac.at/myristate/SUPLpredictor.htm) and CSS-Palm 3.0 software (Ren et al., 2008), respectively. The diagram of the intron/exon structures of BrrCBL and BrrCIPK was analyzed using the online Gene Structure Display Server (http://gsds.cbi.pku.edu.cn/; Hu et al., 2015). Subsequently, the MEME program was used to search for conserved motifs in the turnip BrrCBL and BrrCIPK protein sequences (Bailey et al., 2006).
Phylogenetic Analysis and Chromosomal Location, Divergence Time Estimation
The BrrCBL and BrrCIPK protein sequences were aligned using the MAFFT version 7 program, and phylogenetic trees were constructed using the MEGA 5.0 software with the neighbor-joining method and the 1,000 bootstrap test replicates (Tamura et al., 2011).
To map the locations of BrrCBL and BrrCIPK genes in turnip, the chromosomal distribution of turnip genomic sequences were generated by circus software (http://www.circos.ca/software/download/circos/). Then, two types of gene duplications were recognized, namely, segment duplication and tandem duplication (Flagel and Wendel, 2009). The synonymous (Ks) and nonsynonymous (Ka) substitution rates were estimated by the codeml program of PAML4 (Yang, 2007). The divergence time (T) of BrrCBL and BrrCIPK gene pairs was calculated as T = Ks/2λ (divergence rates λ = 1.5 × 10−8 for Arabidopsis; Cao et al., 2011).
Subcellular Localization and Confocal Laser Scanning Microscopy
The coding region (CDS) of BrrCBL genes was cloned and fused to a binary vector pRI101-AN DNA, with a green fluorescent protein (GFP) and driven by the Cauliflower mosaic virus (CaMV) 35S promoter, forming a 35S:BrrCBL-GFP construct. After confirmation by sequencing, these constructs were separately transferred into Agrobacterium tumefaciens EHA105 by electroporation for infiltration into leaves of Nicotiana benthamiana (Wydro et al., 2006). For the control, we tested the subcellular localization of GFP alone in the leaf cells of N. benthamiana. Freshly made transformed Agrobacteria cell cultures were resuspended in media containing 10 mM MES–KOH (pH 5.6), 10 mM MgCl2·6H2O, and 100 μM/L acetosyringone (AS, Sigma), adjusted to an OD600 of 0.6–0.8. Next, 3 mL of culture was obtained by sterile single-use syringes to inject into the abaxial air space of N. benthamiana leaves. The leaf section near the injection site was squashed 3 days after infiltration. Fluorescence images were detected using a laser-scanning confocal microscope (Olympus Optical Co. Ltd., Tokyo, Japan). GFP was excited with a 488 nm laser line, and emissions were captured using a 505–530 band-pass filter.
Different Expression Profile of BrrCBL and BrrCIPK Genes
The estimated expression profiles expressed in fragments per kilobase of exon model per million mapped reads values for each BrrCBLs and BrrCIPKs from different developmental stages were obtained from RNA-Seq data in the reanalyzed turnip (Li et al., 2015). Samples were obtained in the early stage before cortex splitting (ES1 and ES2), cortex-splitting stage (CSS1 and CSS2), and secondary root-thickening stage (RTS1 and RTS2) in turnip. These stages were described in detail in a previous study (Li et al., 2015). The normalization and hierarchical clustering analysis of gene expression patterns were performed on the basis of Pearson coefficients with average linkage using the Genesis software (version 1.7.1; Sturn et al., 2002).
Plant Material, Growth Condition, and Stress Treatments
Seeds of turnip were grown in soil pots and 1/4 Hoagland's nutrient solution (pH 5.5) under controlled conditions (28°C day/25°C night cycle, relative humidity of 75–80%, 200 mmol photons m−2 s−1 light intensity). After 3 weeks of germination, drought stress was executed by withholding water for 7 days. For MeJA, high salinity, and cold stress, seeding was exposed to MeJA (1 mM), NaCl (100 mM), and cold temperature (4°C). The roots and leaves of seedlings were harvested after treatment for 3 h, with 0 h as the control. The samples, including the root, stem, leaf, and flower, were harvested at the flowering stage. All samples were immediately frozen in liquid nitrogen and then stored at −80°C for RNA extraction.
RNA Isolation and Quantitative Real-Time PCR (qRT-PCR) Analysis
Total RNA samples were isolated using the Eastep® Super Total RNA Extraction Kit (Promega, Madison, WI, USA). RNA was quantified by NanoDrop1000 (NanoDrop Technologies, Inc.) with integrity checked on 0.8% agarose gel. Approximately 5 μg RNA was reverse transcribed using the GoScript Reverse Transcription System (Promega, Madison, WI) to generate cDNA. qRT-PCR was conducted in triplicate with different cDNAs synthesized from three biological replicates of different tissues and treatments using FastStart Universal SYBR Green Master (Rox, Roche, Indianapolis, IN, USA) and a 7,500 Sequence Detection System (Applied Biosystems, USA). The reaction parameters for thermal cycling were as follows: 95°C for 10 min, followed by 40 cycles of 94°C for 5 s, and 60°C for 15 s. B. rapa tubulin beta-2 chain-like (LOC103873913) was amplified as an internal control. The primers used for qRT-PCR are listed in Supplementary Table 1. The relative gene expression levels were obtained by dividing the extrapolated transcript levels of the target genes by the levels of the internal control from the same sample. The results were obtained from the comparison of the treatment with the control using independent-samples T-test. Statistical analysis was performed using the software IBM SPSS Statistics 20.0.
Yeast Two-Hybrid (Y2H) and Bimolecular Fluorescence Complementation (BiFC) Assays
Y2H assays were conducted using the MatchMaker Y2H system (http://www.clontech.com/). The CDS regions of BrrCBL and BrrCIPK genes were first subcloned into pGADT7 and pGBKT7 vectors, respectively. The primers used for vector construction are listed in Supplementary Table 1. Then, the plasmids of BrrCBL and BrrCIPK were sequentially transformed into yeast strain AH109 through the lithium acetate method following the protocol described in the Yeast Protocols Handbook (Clontech). The transformed yeast cells were grown on medium as follows: (1) lacking leucine and tryptophan SD−Trp−Leu to serve as a positive control for transformation and loading; (2) lacking leucine, tryptophan, and histidine SD−Trp−Leu−His to test for PPIs under low stringency, and (3) lacking leucine, tryptophan, histidine, and adenine SD−Trp−Leu−His−Ade to test for interactions under stringent conditions. Cell growth was recorded at 48 h intervals over the course of 6 days. The method of assessment of the interaction strength between sets of proteins was as follows (Kleist et al., 2014). Red boxes indicate vigorous growth on -LTHA plates. Orange boxes indicate weak growth on -LTHA plates. Yellow boxes indicate robust growth on -LTH plates but no growth on -LTHA plates. Pale yellow boxes indicate weak growth on -LTH plates but with each CBL–CIPK interaction conferring superior growth to that in the empty vector control. Gray boxes indicate only growth on -LT plates.
For BiFC assays, the CDS regions of selected BrrCBL and BrrCIPK genes were sub-cloned into 35s-SPYCE and 35s-SPYNE vectors, respectively. The primers used for vector construction are listed in Supplementary Table 1. The fusion construct was transferred into A. tumefaciens strain EHA105, then into tobacco leaves by Agrobacterium infiltration (Wydro et al., 2006). Infiltrated leaves were observed with a laser scanning confocal microscope (Olympus Optical Co. Ltd., Tokyo, Japan).
Arabidopsis Transformation and Treatments
The full-length coding sequences of BrrCBL9.1 and BrrCBL9.2 were each inserted into a binary vector pRI101-AN DNA fused with a GFP and driven by Cauliflower mosaic virus (CaMV) 35S promoter. Consequently, a 35S: BrrCBL-GFP construct was obtained. Arabidopsis plants were transformed using the floral-dip method with the A. tumefaciens strain EHA105 (Clough and Bent, 1998). The assayed plants (overexpression, cbl9 mutant, complemented lines, and WT) were grown on 1/2 solid Murashige and Skoog (MS) medium containing NaCl (100 mM) for stress analysis. After 8 days, the seedling root lengths were measured. For expression analysis of corresponding BrrCIPK genes specially interacted with BrrCBL9.1 and BrrCBL9.2 in overexpression, cbl9 mutant, and complemented lines, the assayed plants were grown on MS medium for about 10 days. Then they were transferred to soil in a growth chamber (photoperiod of 16-h light/8 h darkness) at 21°C. After 2 weeks of germination, seedings were exposed to NaCl (100 mM) and were harvested after treatment for 3 h, with 0 h as the control. All samples were immediately frozen in liquid nitrogen and then stored at −80°C for RNA extraction.
Results
Identification and Characterization of BrrCBL and BrrCIPK Genes
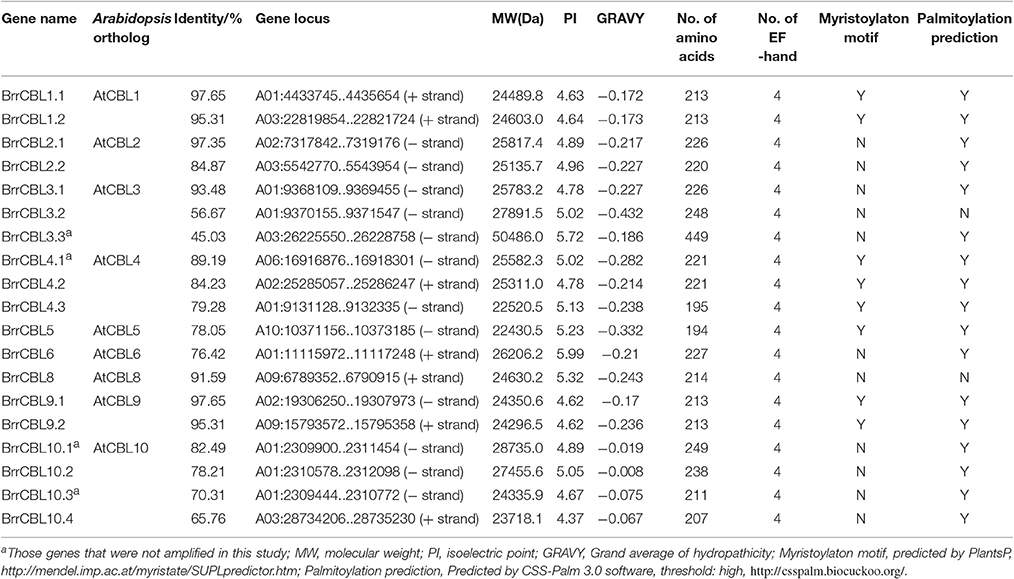
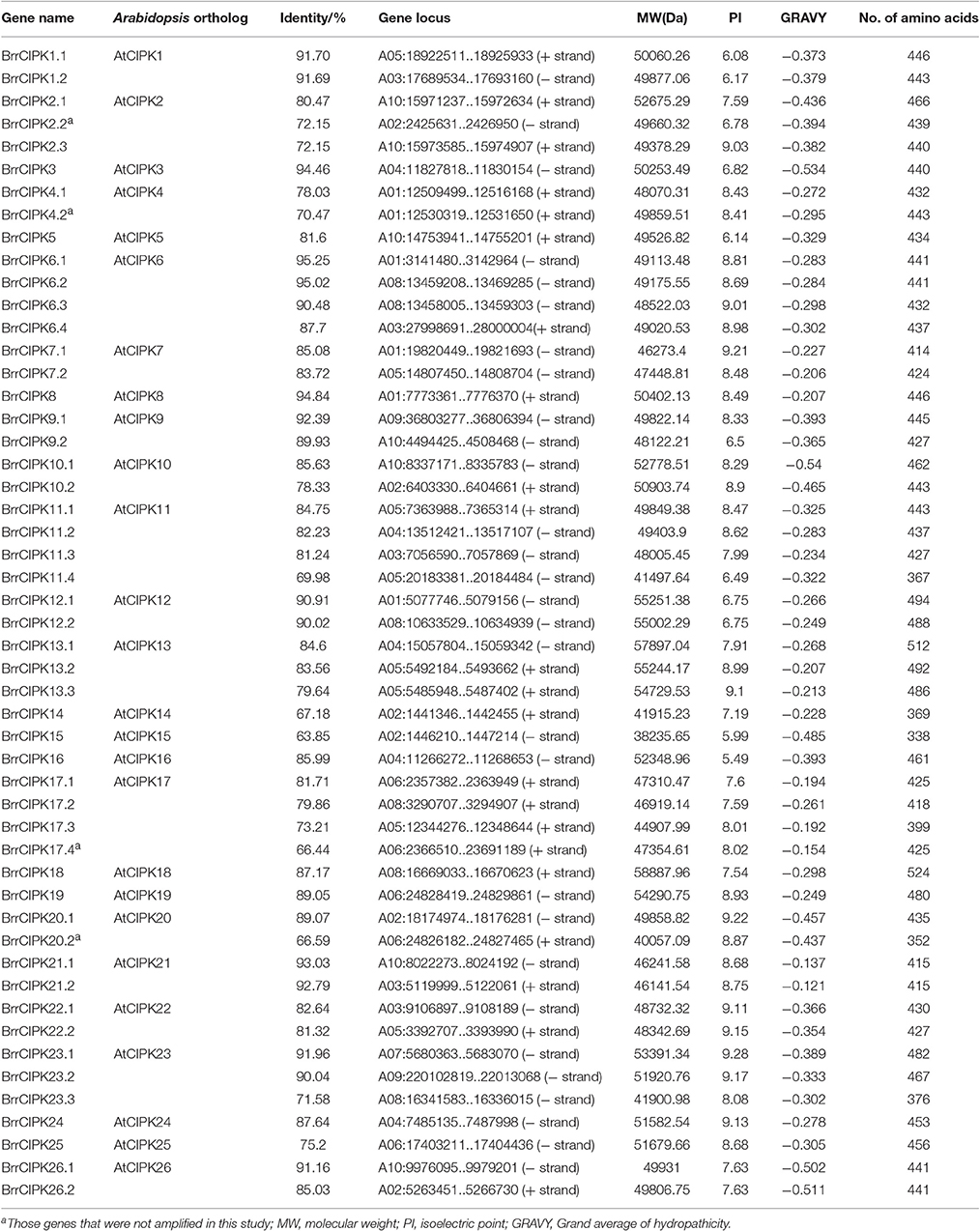
We conducted a genome-wide identification of CBLs and CIPKs in turnip. Accordingly, we used Arabidopsis 10 CBL and 26 CIPK CDS sequences (Zhang et al., 2014) as queries to search against the published genome of turnip. After the search, we identified 19 CBLs and 51 CIPKs, the names assigned to these genes, and their phylogenies and sequence similarities to corresponding individual AtCBLs and AtCIPKs (Tables 1, 2). The full-length turnip CBL proteins were 194 (BrrCBL5) to 249 (BrrCBL10.1) amino acids long, except BrrCBL3.3 with 449 amino acids (Table 1). All the BrrCBLs contained four EF-hand motifs, which provide the structural basis for calcium binding. BrrCBL1.1, -1.2, -4.1, -4.2, -4.3, -5, -9.1, and -9.2 proteins each harbored a conserved N-myristoylation and palmitoylation motif (MGCXXS/T) that may be responsible for membrane association (Batistic et al., 2008). The BrrCIPKs identified in our study ranged in isoelectric points (PIs) from 5.49 (BrrCIPK16) to 9.28 (BrrCIPK23.1), with coding sequences of 352–524 amino acids (Table 2). These ranges suggest the diversity of the biochemical properties of BrrCIPKs. Among the identified BrrCBL and BrrCIPK genes, the CDS of 15 BrrCBLs and 47 BrrCIPKs were successfully amplified by PCR from cDNA prepared from turnip as the template using available primers (Supplementary Table 1). We further predicted the localization of 15 BrrCBL proteins by confocal laser microscopy (Supplementary Table 1). The BrrCBL1.1, -1.2, -2.2, -3.2, -4.3, -5, -8, -9.1, and -9.2–GFR fusion proteins each emitted a green fluorescent signal in the plasma membranes and nuclei of the epidermal cells of N. benthamiana leaves. By contrast, BrrCBL2.1, -3.1, -4.2, -6, -10.2, and -10.4–GFR were detected in plasma membranes. Although, lacking myristoylation and palmitoylation motifs, BrrCBL3.2 and BrrCBL8 localized in the membrane. The dynamic localization of CIPKs was determined by their specific CBL partners (Batistic et al., 2010; Batistic and Kudla, 2012). We did not analyze the subcellular localization of the CIPK protein.
Phylogenetic Relationships and Gene Structural Analyses of the BrrCBLs and BrrCIPKs
To gain insights into the phylogenetic analysis of both CBL and CIPK families, we generated two phylogenetic trees for each family. The BrrCBL proteins were divided into three groups, namely, groups I, II, and III (Figure 1). We also compared the evolutionary relationship between turnip and Arabidopsis in terms of the CBLs (Supplementary Figure 2A). All BrrCBLs were clustered closely with AtCBL orthologs. However, CBL7 was present in Arabidopsis but absent in turnip. To analyze the structural characteristics of the BrrCBLs, we mapped gene structures, including exons and introns, on the basis of the turnip genome sequence. The results showed that all the BrrCBLs were intron rich, with six to eight introns (except BrrCBL3.3 with the 15 introns). Hence, the exon/intron numbers of the CBL gene potentially vary. Moreover, we analyzed the EF-hand motifs for the BrrCBL proteins (Supplementary Figure 3). BrrCBL1, -3, -4, -5, and -8 possessed no canonical EF hand, whereas BrrCBL6, -7, and -10 possessed one and BrrCBL1 and -9 harbored two canonical EF hands. The space of each EF-hand motif was absolutely conserved in the BrrCBL proteins. Up to 23 amino acids were observed to lie between EF1 and EF2, 25 amino acids between EF2 and EF3, and 32 amino acids between EF3 and EF4.
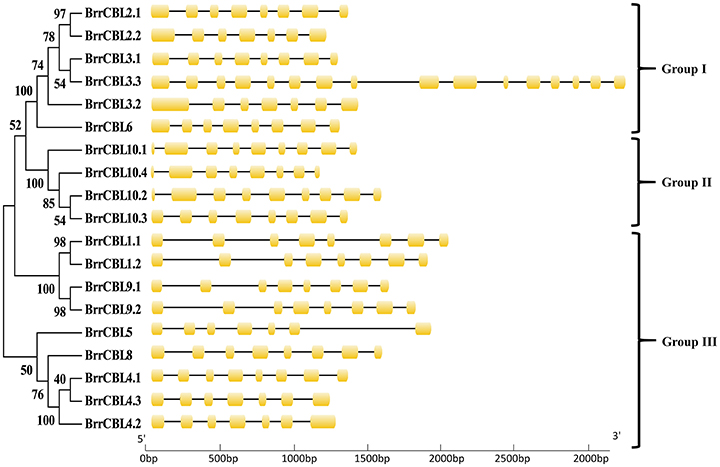
Figure 1. Phylogenetic relationship of Turnip CBL proteins. The BrrCBL protein sequences were aligned using the MAFFT version 7 program, and phylogenetic trees were constructed using the MEGA 5.0 software with the neighbor-joining method and the 1,000 bootstrap test replicates. The tree can be divided into three major clades (Group I – Group III).
As expected, the phylogenetic tree divided the BrrCIPKs into two groups, namely, group A (intron-rich) (18 BrrCIPKs) and group B (intron-less) (33 BrrCIPKs); this finding was consistent with previous evolutionary analyses (Kolukisaoglu et al., 2004) (Figure 2). The structural differences in the BrrCIPKs may allow BrrCIPK genes to function differently because the functional structural gene domains determine the gene's function (Albrecht et al., 2001). All of the 51 BrrCIPKs were clustered closely with AtCIPK orthologs (Supplementary Figure 2B). In the Arabidopsis CIPKs (Batistic and Kudla, 2004), all BrrCIPKs contained domain structures similar to that of AtCIPK24, including an N-terminal protein kinase catalytic (PKC) domain and a C-terminal regulatory domain (NAF/FISL motif) was identified (Supplementary Figure 4A). Previous studies showed that the PKC domain in the N-terminal and a short motif called NAF/FISL motif located in the C-terminal regulatory domain were necessary and sufficient for mediating interactions with CBLs (Albrecht et al., 2001; Guo et al., 2001; Hashimoto et al., 2012). Thus, amino-acid sequences were produced to gain insight into the BrrCIPK functional domains within each residue position.
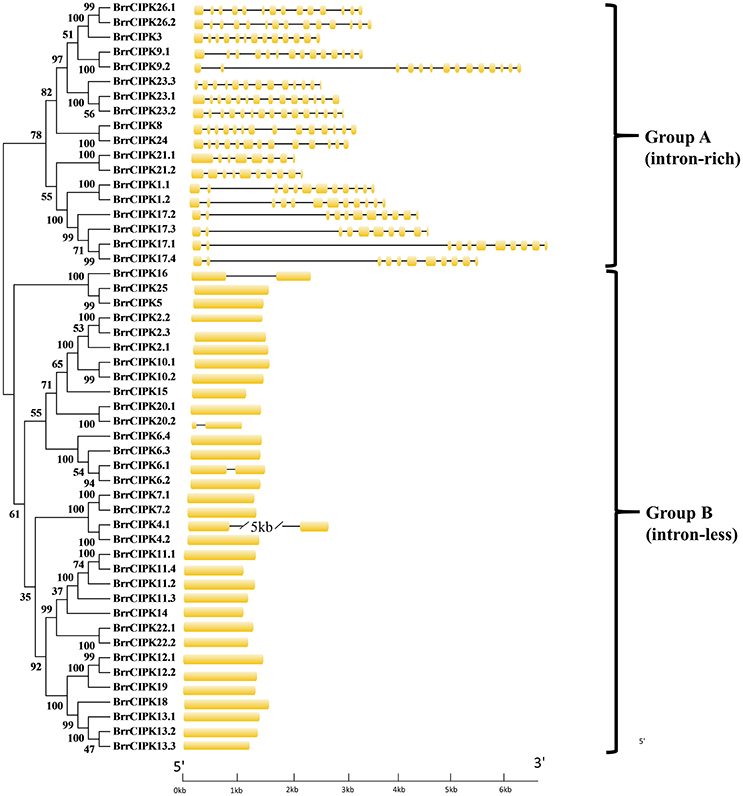
Figure 2. Phylogenetic relationship of Turnip CIPK proteins. The BrrCIPK protein sequences were aligned using the MAFFT version 7 program, and phylogenetic trees were constructed using the MEGA 5.0 software with the neighbor-joining method and the 1,000 bootstrap test replicates. The tree can be divided into two major clades (Group A and Group B).
We further searched for the conserved motifs in both BrrCBL and BrrCIPK proteins using the MEME program to analyze the diversity of motif compositions. A total of 15 conserved motifs designated as motif 1 to motif 15, respectively, were identified within the genes (Supplementary Figures 5, 6). These motifs may also help predict the genes' functions. Our analysis revealed that motif 10 contained the N-myristoylation motif of 12 BrrCBL proteins (Supplementary Figure 5). For the BrrCIPKs, motifs 1 and 2 were found within the PKC domain. Motif 8 in the protein–protein interaction (PPI) domain was shared by all BrrCIPKs. Motifs 7 and 11, also shared by all the BrrCIPK members, were found within the region of the NAF/FISL domain and showed relatively conserved amino acids (N, A, F, I, S, and L; Supplementary Figure 6). The amino acid residues at the 2nd, 3rd, 4th, 7th, and 10th sites of the NAF/FISL motif were rather conserved (Supplementary Figure 4B). The phylogenetic analysis together with the structure analysis presented herein may facilitate the functional annotation and study of CBLs and CIPKs in turnip.
Chromosomal Location Analysis and Evolutionary History of the Turnip BrrCBL and BrrCIPK Gene Families
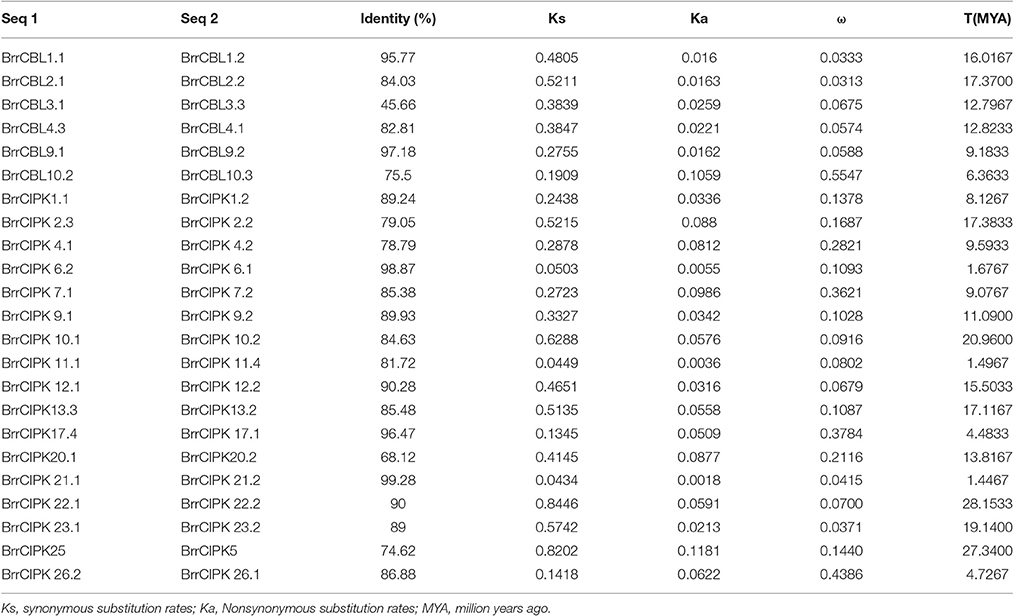
To determine the relationship between gene duplication and genetic divergence in the CBL or CIPK gene family in turnip, we investigated the chromosomal location of BrrCBL and BrrCIPK (Figure 3). All the BrrCBLs and BrrCIPKs were located at the top sections of chromosomes. Among the 10 chromosomes, chromosome 1 contained the maximum number of seven BrrCBL genes. Chromosomes 4, 5, and 7 carried no BrrCBL. Chromosomes 6, 8, and 10 each contained only one BrrCBL gene, and all other chromosomes contained 2–4 BrrCBLs (Figure 3A). Given the phylogenetic analyses and an entire genome analysis of gene duplications, we observed that six pairs of BrrCBL were putative paralogs, including five segmental duplications (BrrCBL1.1/1.2, BrrCBL2.1/2.2, BrrCBL3.1/3.3, BrrCBL4.1/4.3, and BrrCBL9.1/9.2) and one tandem duplication (BrrCBL10.2/10.3) (Table 3). Furthermore, we found that some paralog genes were located on different chromosomes or at different locations on the same chromosome (Figure 3B). Two pairs of BrrCBL3.1/3.3 and BrrCBL10.2/10.3 were located on the same chromosomes at different locations. Meanwhile, other paralogs were located on different chromosomes. Among all the genes studied, the BrrCIPK genes were found variably distributed on the turnip chromosomes. Chromosomes 5 and 9 contained the maximum and the minimum numbers, respectively, of BrrCIPK genes. A total of 17 pairs of putative BrrCIPK paralog genes were produced by segmental duplication, including 14 duplication events between chromosomes (BrrCIPK1.1/1.2, BrrCIPK2.2/2.3, BrrCIPKV6.1/6.2, BrrCIPK7.1/7.2, BrrCIPK9.1/9.2, BrrCIPK10.1/10.2, BrrCIPK12.1/12.2, BrrCIPK13.2/13.3, BrrCIPK20.1/20.2, BrrCIPK21.1/21.2, BrrCIPK22.1/22.2, BrrCIPK23.1/23.2, BrrCIPK5/25, and BrrCIPK26.1/26.2), as well as three duplication events within the same chromosome (BrrCIPK4.1/4.2, BrrCIPK11.1/11.4, and BrrCIPK17.1/17.4; Figure 3B, Table 3). These observations suggest the large-scale segmental duplication events involved in the expansion of BrrCBL and BrrCIPK gene families in turnip.
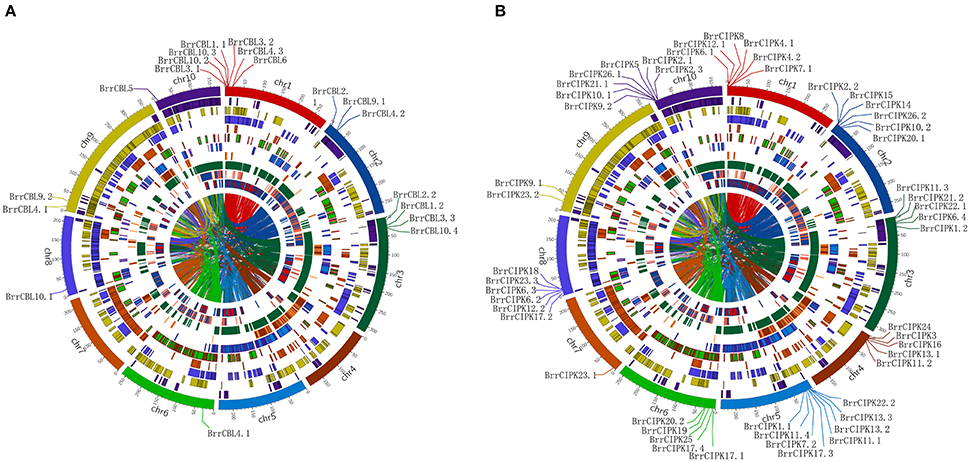
Figure 3. Turnip CBLs and CIPKs Chromosome distributions. Ten kinds of color in the outer circle represented 10 turnip chromosomes. Within the circle were numerous duplications between and within chromosomes. The distribution of BrrCBL and BrrCIPK on 10 chromosomes were given in the outer circle, where the numbers represent the chromosome length in 100 kb. (A) The distribution of BrrCBL on 10 chromosomes (Chromosome 4, 5, 7 containing no BrrCBLs). (B) BrrCIPKs variably distributed among all on the turnip chromosomes.
We estimated the divergence time (T) of six pairs of turnip putative BrrCBL paralog proteins by measuring the synonymous (Ks) and nonsynonymous (Ka) mutation rates (Table 3). In the process, we used a divergence rate of 1.5 × 10−8 mutations per synonymous site per year. The estimated divergence time (T) for the turnip CBL paralogs was approximately between 6.3633 and 17.3700 million years ago (MYA), with an average duplication time of ~12.4256 MYA. Ka/ks (ω) was calculated for each pair of CBL paralog genes. The ω-values for all of the putative CBL paralogs with mean value of 0.134 were less than one. Thus, the five pairs of turnip CBL proteins were potentially under strong purifying selection pressure. However, one pair of turnip CBL genes (BrrCBL10.2/BrrCBL10.3, ω = 0.5547) attained relatively large ω-values. This result implied that the corresponding genes may have evolved rapidly from the last common ancestor. We also estimated the divergence time of 17 pairs of turnip putative CIPK paralog genes. Moreover, the estimated T for BrrCIPK paralogs was between 1.4467 and 28.1533 MYA, with an average duplication time of approximately 12.4194 MYA. Interestingly, the divergence time of three BrrCIPK paralogs (BrrCIPK6.1/6.2, BrrCIPK11.1/11.4, and BrrCIPK5/25) were estimated to have occurred recently (only approximately one). Schranz and Mitchell–Olds (Schranz et al., 2006) estimated the time of very early radiation of Brassicaceae at 34 MYA. Other authors proposed that the conserved linkage arrangements between B. rapa and A. thaliana diverged from a common ancestor at ~14.5–20.4 MYA (Ferguson et al., 2003). However, the divergence time of three pairs of BrrCIPK paralogs (BrrCIPK10.1/10.2, BrrCIPK22.1/22.2, and BrrCIPK5/25; 20.9600–28.1533 MYA) occurred precedent to the period of the origin of the B. rapa. The ω in all the putative BrrCIPK paralogs with mean value of 0.167 were less than one. Hence, the 17 pairs of turnip BrrCIPK genes were possibly under strong purifying selection pressure. By contrast, three pairs of BrrCIPK genes, namely, BrrCIPK7.1/7.2 (ω = 0.3621), BrrCIPK17.1/17.2 (ω = 0.3784), and BrrCIPK26.1/26.2 (ω = 0.4386), achieved relatively large ω-values, which indicate that they may have evolved rapidly from those of the last common ancestor.
Expression Profiles of Turnip BrrCBL and BrrCIPK Genes at Different Tissues and Different Developmental Stages
We determined the spatial and temporal expression profiles of 19 BrrCBL and 51 BrrCIPK genes in different tissues of turnip (root, stem, leaf, and flower) by quantitative real-time PCR (qRT-PCR; Figure 4A). A heat map was created through the hierarchical clustering of the gene expression profiles of the BrrCBL and BrrCIPK genes. These profiles can be classified into two clusters by expression pattern. Cluster I consisted of 16 BrrCBLs and 28 BrrCIPKs with different expression patterns in different tissues. Some of these genes were highly expressed in certain tissue tested. BrrCBL1.1, -1.2, -4.2, and -4.3, as well BrrCIPK6.3, -10.2, and -17.1, almost showed peak transcript levels in the root. BrrCIPK13.2 and 23.3 were highly transcribed in the stem, whereas BrrCIPK3, -9.1, -15, -17.2, and -26.1 were highly transcribed in the leaf. These transcriptional patterns indicate that these genes may be involved in organ development and growth. By contrast, some genes, especially BrrCBL10.2, showed no transcript levels in all tissues. Cluster II included three BrrCBL and 23 BrrCIPK genes with high transcript abundance in the flowers. Hence, the genes in Cluster II may play important roles in flower development. Additionally, some paralogs showed opposite expression patterns. Examples include 4 out of 6 BrrCBL paralogs (BrrCBL2.1/2.2, BrrCBL4.1/4.3, BrrCBL10.2/10.3, and BrrCBL12.1/12.2) and 4 out of 17 BrrCIPK paralogs (BrrCIPK1.1/1.2, BrrCIPK2.2/2.3, BrrCIPK17.1/17.4, and BrrCIPK26.1/26.2) in different tissues (Figure 4A).
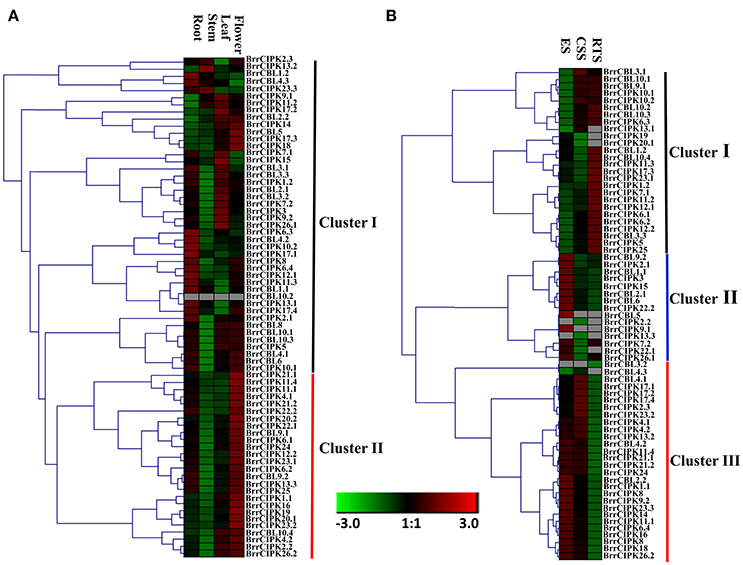
Figure 4. Expression profiles of turnip CBL and CIPK genes in different tissues and different development stages. (A) Heat map of real-time quantitative PCR (qRT-PCR) analysis of BrrCBL and BrrCIPK genes in the four tissues of roots, stems, leaves, flowers, with three biological and technical replicates. (B) Heat map of expression profiles (in log2-based FPKM) for BrrCBL and BrrCIPK in early stage before cortex splitting (ES), cortex splitting stage (CSS), and secondary root thickening stage (RTS). The expression levels were represented by the color bar.
The expression levels of BrrCBL and BrrCIPK genes were reanalyzed using publicly available RNA-sequence data from three different developmental stages (Li et al., 2015). These stages include the average of the early stage before cortex splitting (ES1 and ES2), the cortex-splitting stage (CSS1 and CSS2), and the secondary root-thickening stage (RTS1 and RTS2) in turnip corresponding to ES, CSS, and RTS, respectively (Figure 4B). Likewise, the transcripts of the BrrCBL and BrrCIPK genes can be detected in different developmental stages and can be divided into three clusters by expression pattern. The eight BrrCBL and 18 BrrCIPK genes were assigned to Cluster I, except BrrCIPK13.1, -19, and -20.1, which were relative highly expressed in the secondary RTS. Five BrrCBL and 10 BrrCIPK genes were found in Cluster II, and they were almost exclusively expressed in the early stage before cortex splitting (ES), except for BrrCIPK2.2 and BrrCIPK13.3. Among the Cluster III genes, 6 BrrCBL and 23 BrrCIPK genes were highly expressed in the early stage before cortex splitting (ES) and CSS. By contrast, the expression of BrrCBL3.2 was not detected and BrrCBL4.3 was expressed at a very low level. These results show that some of the genes were modestly expressed in developmental stages. Meanwhile, some genes were specifically and abundantly expressed in certain developmental stages. Therefore, such genes may play important role during developmental stages. We also found that a subset of paralogous genes were differentially expressed in the three developmental stages (Figure 4B). BrrCBL9.1 was expressed abundantly in the CSS and RTS stages, whereas BrrCBL9.2 was specifically expressed in the ES. The expression patterns of two BrrCBL paralogous pairs (BrrCBL1.1/1.2 and BrrCBL4.1/4.3) and six BrrCIPK paralogous pairs (BrrCIPK1.1/1.2, BrrCIPK2.2/2.3, BrrCIPK7.1/7.2, BrrCIPK13.2/13.3, BrrCIPK7.1/7.2, and BrrCIPK23.1/23.2) also diverged dramatically in developmental stages. Overall, the expression patterns of BrrCBL and BrrCIPK genes at different tissues and different developmental stages may indicate the functional divergence between paralogs.
Expression Profiles of BrrCBL and BrrCIPK Genes during Stress Treatment
We then further investigated the expression profiles of BrrCBL and BrrCIPK genes under stress conditions. We also evaluated the possible functional divergence of paralog genes. To achieve these goals, we conducted qRT-PCR analysis. The transcript level of the BrrCBL1.1 gene showed the most significant increases among those of other genes after various stress treatments (Figure 5). By contrast, BrrCBL2.1, -4.2, and -4.3 expression was remarkably downregulated in four stress treatments. Under NaCl treatment, the BrrCBL1.1, -3.2, -4.1, -5, -6, and -10.2 transcript levels significantly increased. Meanwhile, BrrCBL1.2, -3.3, -9.2, and -10.4 expression was still slightly upregulated after NaCl treatment. Under the drought treatment, BrrCBL1.1, -1.2, -2.2, -4.1, -6, -9.1, and -10.3 expression significantly induced. In addition, cold stress significantly upregulated the expression levels of BrrCBL1.1, -2.2, -8, -9.2, -10.1, and -10.4. In response to MeJA treatment, BrrCBL1.1, -3.2, -5, -6, -8, -9.1, -9.2, and -10.2 were highly upregulated. Additionally, we investigated the expression profiles of BrrCBL paralog gene pairs and found that they exhibited different expression patterns under different stress conditions. BrrCBL9.1 expression was downregulated, whereas BrrCBL9.2 expression was upregulated by NaCl and cold treatments. The results suggest the functional divergence of the BrrCBL paralog pairs.
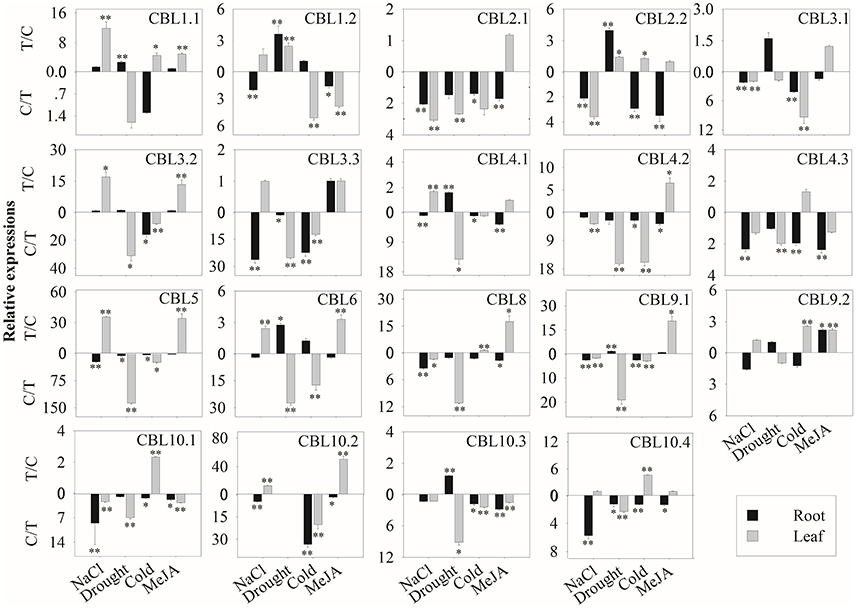
Figure 5. Expression analysis of BrrCBL in leaves and roots responding to various treatments, including NaCl (100 mM), drought, cold (4°C), and MeJA (1 mM). Two time points (0 and 3 h) were used to detect the gene expressions, with three biological and technical replicates. Error bars represented standard errors of repeats. The stress treatments with significant change of expression level compared to the controls with P < 0.05 and P < 0.01 were marked by “*” and “**,” respectively.
For the 51 BrrCIPK genes assayed under four stress treatments, the most striking induction was observed for 34 BrrCIPK genes after MeJA treatment, followed by 30 BrrCIPK genes after NaCl treatment. Among these genes, BrrCIPK2.2, -3.2, -5, -8, -9.1, -10.2, -18, -19, and -20.1 exhibited remarkable MeJA-induced upregulation. BrrCIPK10.2, -20.2, -23.2, and -26.1 also exhibited remarkable NaCl-induced upregulation. Drought treatment induced the expression of 24 BrrCIPKs as that under cold treatment. Strikingly, BrrCIPK17.4 was markedly upregulated in response to drought. The transcript levels of BrrCIPK9.1 and BrrCIPK21.1 showed remarkable increases after cold treatment. Conversely, the transcript levels of BrrCIPK17.1 gene were significantly decreased under the four treatments (Figure 6). Several paralog gene pairs (such as BrrCIPK1.1/1.2, BrrCIPK4.1/4.2, and BrrCIPK10.1/10.2) demonstrated divergent expression patterns, that is, one gene is induced, whereas the other is suppressed by the treatment. The results reveal that the BrrCIPK paralogs exhibit distinct expression patterns under different stress conditions. This occurrence also suggests the functional divergence of the BrrCIPK-duplicated genes.
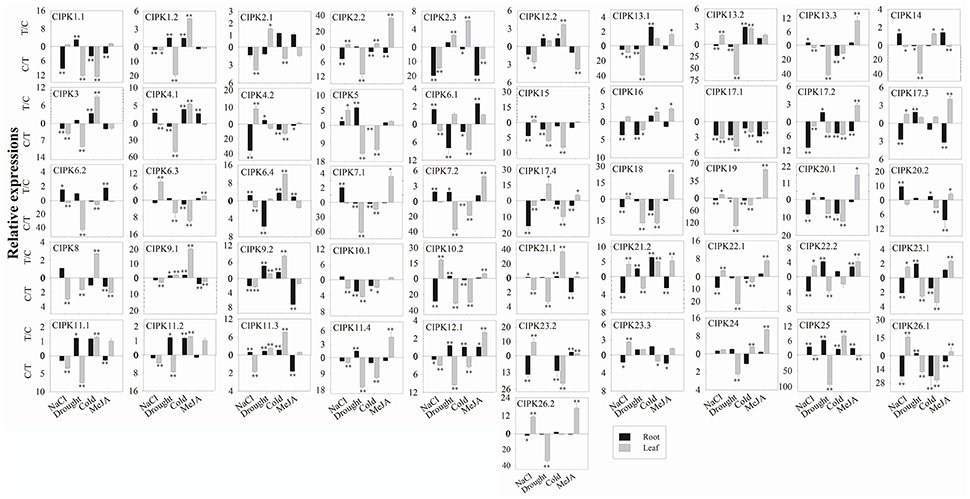
Figure 6. Expression analysis of BrrCIPK in leaves and roots responding to various treatments, including NaCl (100 mM), drought, cold (4°C), and MeJA (1 mM). Two time points (0 and 3 h) were used to detect the gene expressions, with three biological and technical replicates. Error bars represented standard errors of repeats. The stress treatments with significant change of expression level compared to the controls with P < 0.05 and P < 0.01 were marked by “*” and “**,” respectively.
According to previous reports, CBL–CIPK complexes play highly important roles in response to external stimuli (Manik et al., 2015; Sun et al., 2015). We wish to determine whether the same stress responses of BrrCBL and BrrCIPK transcripts can interact with each other. To investigate this aspect in the turnip expression profiles, we performed coexpression analysis, which can predict the interaction of the BrrCBL–BrrCIPK complex in response to stress. A number of BrrCBLs and BrrCIPKs were upregulated and downregulated in response to one or more stress treatments (Supplementary Figure 7). For example, the transcript abundances of BrrCBL1.1, -8, -9.2, and -10.4 as well as BrrCIPK2.3, -8, -14, and -24 were increased by cold. We therefore speculate that potential interactions may exist between these BrrCBL and BrrCIPK genes.
Protein Interaction Analysis between BrrCBLs and BrrCIPKs
To further elucidate the interaction preferences between CBLs and CIPKs in turnip, we used a yeast two-hybrid (Y2H) system. Up to 15 BrrCBL and 47 BrrCIPK genes were cloned and used in Y2H assays. In the current study, we demonstrated 166 interactions between BrrCBLs and BrrCIPKs; the interactions displayed varying strengths and specificities (Figure 7). Notably, BrrCBL9.1 and -9.2 were paralog proteins and showed strong interactions with 22 and 25 BrrCIPKs, respectively. Some specific BrrCIPKs were identified, and these genes interacted specifically with either of the others. BrrCBL9.1 specifically interacted with BrrCIPK6.1, -25, and -26.2, whereas BrrCBL9.2 interacted specifically with BrrCIPK6.2, -6.4, -7.1, -18, -22.1, and -23.3. Thus, BrrCBL9.1 and -9.2 were involved in more interactions with BrrCIPKs than those of the other BrrCBLs. However, the two genes displayed a fairly different interaction with its paralogs, and hence may be appropriate for further in-depth research. Overall, this type of interaction analysis enabled the deduction of potential gene functions to guide future functional studies. We then selected 10 of the interactions (BrrCBL3.2-BrrCIPK16, BrrCBL4.2-BrrCIPK7.1, BrrCBL9.1-BrrCIPK16, -20.1 and -23.2, BrrCBL9.2-BrrCIPK7.1, -16, -20.1, -23.2, -23.3) detected between BrrCBL and BrrCIPK proteins using the Y2H assay for further analyses using the BiFC method. We found that green fluorescence signals were observed when BrrCBLs and BrrCIPKs were co-expressed in epidermal cells of tobacco leaves (Figure 8). It seemed that the fluorescent signal were consistent with the localization signal of corresponding BrrCBL proteins (Supplementary Figure 1). In contrast, in controls in which BrrCBL3.2, -4.2, -9.1, or -9.2 was expressed together with YFPN only, as well BrrCIPK7.1, -16, -20.1, -23.2, or -23.3 with YFPC only, no signal of reconstructed YFP appeared.
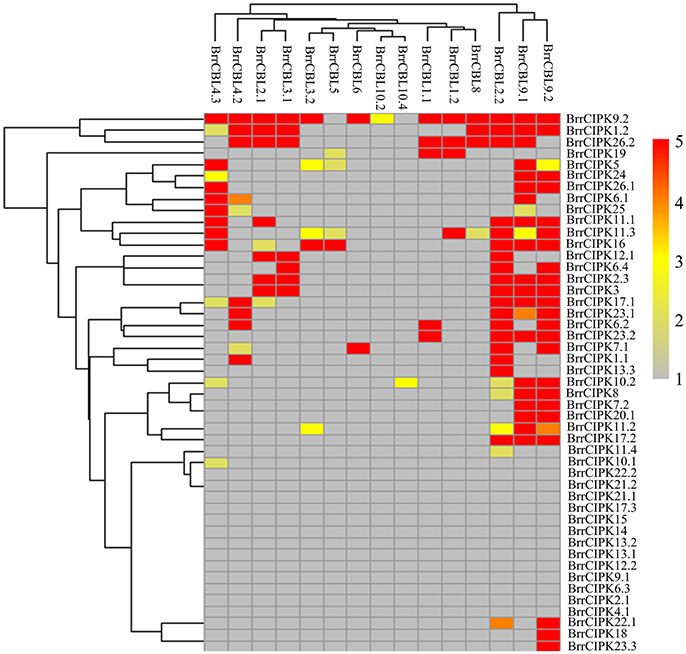
Figure 7. Heat map summarizing yeast two-hybrid (Y2H) results for all interactions between turnip CBL and CIPK proteins. Interaction strength was inferred by serial growth dilutions on selective media lacking one or two auxotrophic markers and summarized qualitatively by heat map. Red boxes indicate vigorous growth on -LTHA plates; Orange boxes indicate weaker growth on -LTHA plates; Yellow boxes indicate robust growth on -LTH plates but no growth on -LTHA plates; Pale yellow boxes indicate weak growth on -LTH plates, but each CBL-CIPK interaction conferred better growth than the empty vector control; Gray boxes indicate only growth on -LT plates. The phylogenetic relationships of both BrrCBLs and BrrCIPKs family were inferred by cladogram.
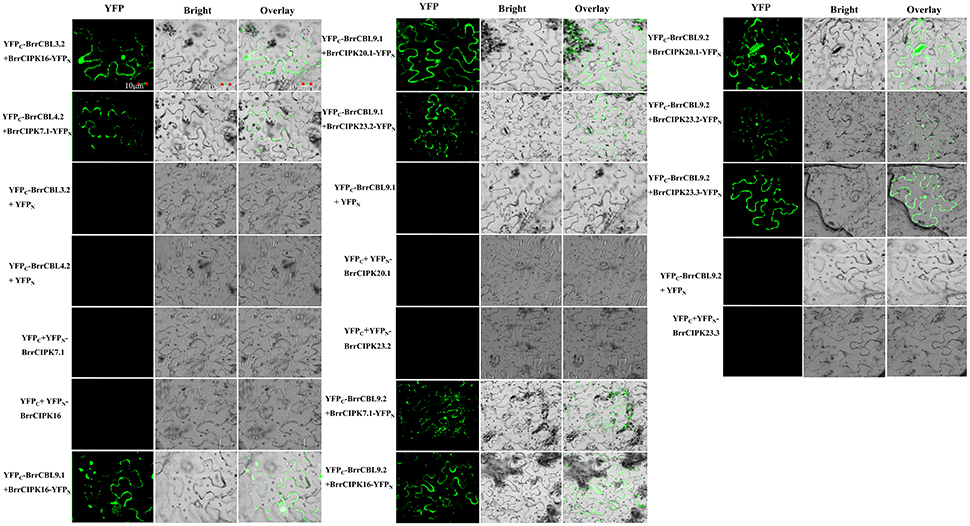
Figure 8. The bimolecular fluorescence complementation (BiFC) experiments in N. benthamiana leaf cells. The CDS regions of BrrCBL and BrrCIPK genes were sub-cloned into 35s-SPYCE and 35s-SPYNE vectors, respectively. Bar = 10 μm.
Functional Analysis of BrrCBL9.1 and BrrCBL9.2 under Salt Stress
The AtCBL9 gene in Arabidopsis possesses two counterparts, namely, BrrCBL9.1, and -9.2, in turnip. The differences between these genes were discovered through expression and interaction profiles. Previous studies revealed that the Arabidopsis cbl9 mutant is hypersensitive to ABA, salt, drought, and glucose stress (Pandey et al., 2004). To dissect the biological function of turnip CBL9.1 and -9.2, we overexpressed each gene in Arabidopsis using a CaMV 35S promoter (OE-BrrCBL9.1 and OE-BrrCBL9.2, respectively). To examine whether BrrCBL9.1 and -9.2 can rescue the cbl9 mutant phenotype, we transformed a cbl9 mutant plant with BrrCBL9.1 and -9.2 using the CaMV 35S promoter (cbl9/BrrCBL9.1 and cbl9/BrrCBL9.2, respectively). Seeds from wild-type (WT), cbl9 mutant, complemented, and overexpressing plants were germinated on 1/2 Murashige and Skoog (MS) and 1/2 MS with 100 mM NaCl (Figures 9A,B). On the 1/2 MS medium plates, the WT, cbl9 mutant, complemented, and overexpressing lines displayed relatively similar growth phenotypes. The latter three plants showed no significant difference in primary root length from the WT. After 100 mM NaCl treatment, no significance difference in the primary roots was observed between cbl9 and cbl9/BrrCBL9.1, as well as between the OE-BrrCBL9.1 and WT under salt-stress conditions. However, the primary roots of cbl9/BrrCBL9.2 were restored to WT background levels, and the primary roots of OE-BrrCBL9.2 were much longer than those of WT plants. Therefore, our data indicate that the BrrCBL9.2 gene can promote root elongation in both transgenic and complementary Arabidopsis under salt stress. We also detected the expression level of corresponding BrrCIPK genes specially interacted with BrrCBL9.1 and -9.2 in complemented and overexpressing transgenic plants (Figure 9C). As a result, BrrCIPK6.4 and -18 showed the most significant increase in complemented and overexpressing transgenic Arabidopsis of BrrCBL9.2 under NaCl treatment, but BrrCBL9.1 does not affect. This indicated that the interaction between BrrCBL9.2 and BrrCIPK6.4 and -18 may play a role in salt responses.
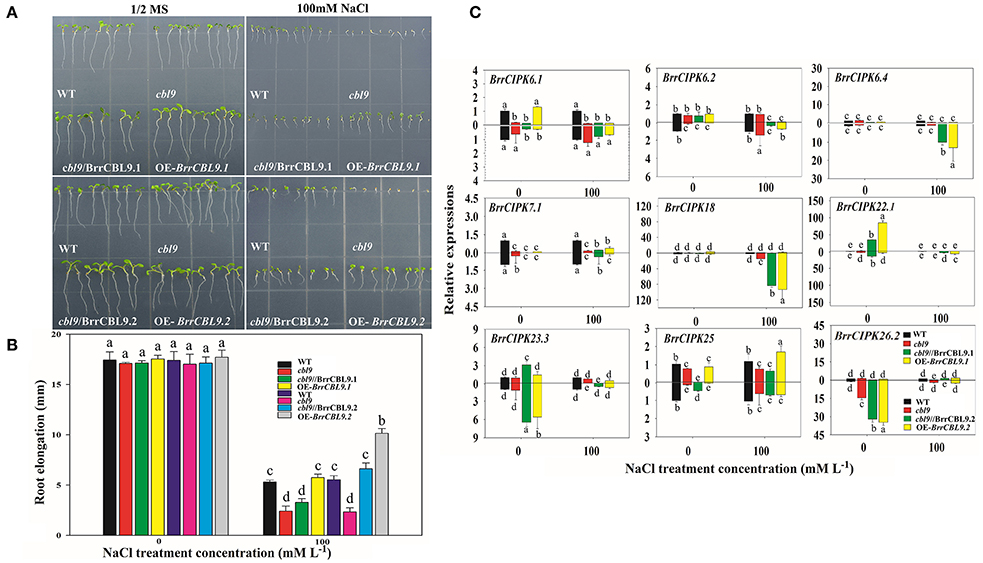
Figure 9. Overexpression of BrrCBL9.1 and -9.2 in Arabidopsis and functional complementation of Arabidopsis cbl9 mutant by BrrCBL9.1 and -9.2 gene. (A) The plants grown on control (1/2MS medium) and stress condition (1/2MS + 100 mM NaCl), respectively. (B) Statistical analysis of the changing root length between 1/2MS control and salt stress condition. (C) Expression analysis of corresponding BrrCIPK genes specially interacted with BrrCBL9.1 and 9.2 in WT, cbl9 mutant, complemented, and overexpressing plants responding to NaCl (100 mM). Two time points (0 and 3 h) were used to detect the gene expressions, with three biological and technical replicates. Error bar denoted standard deviation, and different letters within a column indicate a significant difference (P < 0.05; Tukey's test).
Discussion
The signaling system composed of CBLs and CIPKs is a key regulatory node in various stress signaling pathways in plants. Previously, CBL and CIPK members were comprehensively analyzed in many species, such as P. patens (four CBLs and seven CIPKs), S. moellendorffii (four CBLs and five CIPKs), Arabidopsis (10 CBLs and 26 CIPKs) (Luan et al., 2002; Kolukisaoglu et al., 2004; Zhang et al., 2014), O. sativa (10 CBLs and 31 CIPKs) (Kolukisaoglu et al., 2004), Populus trichocarpa (10 CBLs and 27 CIPKs) (Yu et al., 2007; Zhang et al., 2008), Z. mays (Chen X. F. et al., 2011) (eight CBLs and 43 CIPKs), B. napus L. (seven CBLs and 23 CIPKs) (Zhang et al., 2014), and S. melongena L. (five CBLs and 15 CIPKs) (Li et al., 2016). The studies suggested that the CBLs and CIPKs developed during early plant evolution. However, no CBL and CIPK gene has been found in turnip (B. rapa var. rapa). In our study, a genome-wide database search based on conserved domains and sequence similarities to known Arabidopsis CBLs and CIPKs revealed 19 BrrCBL and 51 BrrCIPK genes in the turnip genome (Tables 1,2). The phylogenetic analysis indicated that BrrCIPK family can be divided into intron-rich and intron-less clades, similar to those in Arabidopsis and rice (Kolukisaoglu et al., 2004; Chen X. F. et al., 2011; Mao et al., 2016). And an alternative mRNA splicing of OsCIPK3 and OsCIPK24 were detected, as observed in AtCIPK3 and AtCIPK9 (Kolukisaoglu et al., 2004; Mao et al., 2016). Moreover, BrrCIPK harbored 7 to 15 introns and exhibited a high degree of intron conservation with their relative Arabidopsis gene family members (Kolukisaoglu et al., 2004). Thus, we speculated that the clustering of BrrCIPK was possibly due to evolutionarily intron retention and alternative splicing mechanism in turnip. This structural differences in BrrCIPK indicated they may be involved in different stress response. Previous studies revealed that B. rapa underwent a polyploidization event, such as the γ triplication (135 MYA), β (90–100 MYA), and α (24–40 MYA) duplications (Wang and Kole, 2015). These three polyploidization events occurred in the evolutionary history of turnip. These events were followed by chromosomal reduction and rearrangement as well as numerous gene losses. As a result, highly complex gene families were obtained. In our study, 19 BrrCBL and 51 BrrCIPK members were always clustered closely with AtCBL and AtCIPK orthologs. However, CBL7 presented in Arabidopsis (Kolukisaoglu et al., 2004) but was absent in turnip mainly because of gene loss. Thus, we conclude that some of the CBL family members were conserved, whereas others were lost after the divergence. Many CBL and CIPK genes in Arabidopsis also held two or more counterparts in turnip. This observation suggests that the expansion of the CBL and CIPK families in turnip may be caused by genome duplication processes, including multiple segmental duplication, tandem duplication, transposition events, and entire-genome duplication (Cannon et al., 2004; Flagel and Wendel, 2009). We found evidence for five (26.3%) segmental duplication and one (0.5%) tandem duplication in 19 BrrCBLs and 17 (33.3%) segmental duplication in 51 BrrCIPK genes (Figure 3, Table 3). This finding indicates that segmental duplication predominantly contributed to the expansion of both BrrCBL and BrrCIPK in turnip. The increase in the number of CBL and CIPK genes during plant evolution was likely related to the functional evolution of the CBL–CIPK signaling system (Weinl and Kudla, 2009).
The polyploidization event also caused structural and functional domain diversification. As first found, BrrCBL3.3 contained the largest number of introns (15 introns) in its coding regions relative to those in AtCBL3 (Mahajan et al., 2006), OsCBL3 (Kanwar et al., 2014), ZmCBL3 (Zhang et al., 2016), and PsCBL3 (Mahajan et al., 2006). The large number of introns may have been produced by structural diversification (Figure 1). Likewise, BrrCBL3.3 attained the highest molecular weight (MW) (50.486 kDa; Table 1). By contrast, CBL3 proteins in other plants were relatively conserved in size (MW ranged from 23 to 29 kDa) (Shen et al., 2014; Zhang et al., 2014). BrrCBL proteins possessed variable N-terminal, which play essential roles in the localization of CBL proteins (Supplementary Figure 3). The two segmental duplication pairs (BrrCBL2.1/2.2 and BrrCBL3.1/3.2) differed in the N-terminal. Confocal fluorescence microscopy analysis showed that BrrCBL2.1 and -3.1 localized in the plasma membrane. By contrast, their segmental duplication partners BrrCBL2.2 and -3.2 localized in the plasma membrane and nuclei (Supplementary Figure 1). This result differed from that of the vacuolar membrane-localized proteins AtCBL2 and -3 in Arabidopsis (Batistic et al., 2008). However, both BrrCBL1.1/1.2 and BrrCBL9.1/9.2 (harboring the same myristoylation motif in the N-terminal) localized in the plasma membrane and nuclei (Supplementary Figure 1). AtCBL1 and -9 localized to the plasma membrane (Batistic et al., 2010). Even the intronless BrrCIPKs were highly conserved in the intron phase. Variations in intron lengths also existed in the BrrCIPKs (Figure 2). The longest intron length was found in BrrCIPK4.1 (5.37 kb) relative to those of AtCIPK4 (Li et al., 2009; Mao et al., 2016) and TaCIPK4 (Sun et al., 2015). Previous studies showed that the NAF/FISL domain itself was sufficient to mediate CBL interaction (Albrecht et al., 2001; Guo et al., 2001). Even at the conserved sites of this motif, variations existed in the turnip CIPK proteins (Additional file 4). The second amino-acid residue of the NAF/FISL motif in BrrCIPK7.1, -7.2, -4.1, and -4.2 was T (threonine) instead of N (asparagine), similar to that in BnaCIPK7 in canola (Zhang et al., 2014), AtCIPK4, and AtCIPK7 in Arabidopsis (Li et al., 2009). This difference in amino-acid residue of a BrrCIPK protein may influence its ability to interact with BrrCBLs. Changes in structure may result in the functional divergence between paralog genes. Polyploidization events that occurred in B. rapa played important roles in the evolution of the plant species. This even afforded the plant with the ability to be diversified and to respond to changing habitats. The genes generated from polyploidization events developed new functions (Wang and Kole, 2015).
The large size of these two gene family members in turnip indicated their importance in the control of turnip-specific processes. In our study, the expression profiles of 19 CBLs and 51 CIPKs in different tissues and different developmental stages revealed that the majority of BrrCBL and BrrCIPK genes are expressed at different levels in all organs and at all developmental stages (Figure 4). By contrast, the expression patterns of some paralogous pairs, as such BrrCBL9.1/9.2, differed dramatically. Differences were also noted in the tandemly duplicated pair (OsCIPK12/30) and segmentally duplicated pairs (OsCIPK6/27, OsCIPK1/17, and OsCIPK3/31) in rice. Meanwhile, considering all the results for abiotic stresses and phytohormones, we noted that the transcript levels of all the tested BrrCBL and BrrCIPK genes either increased or decreased in response to many of the treatments (Figures 5, 6). Conversely, the responses of BrrCBL9.2 were significantly upregulated, whereas BrrCBL9.1 was downregulated by NaCl stress. Similar to paralog pairs OsCIPK3 and OsCIPK31 in rice and ZmCIPK8 and -31 in maize under stress treatments (Chen X. et al., 2011; Kanwar et al., 2014). Hence, this type of differential expression in paralog genes reflects that CBL and CIPK genes evolved some new functions for turnip adaptation to the adverse environmental factors in the Qinghai–Tibet Plateau. Moreover, the CBL and CIPK contribute to plant stress-response mechanisms usually by forming a CBL–CIPK signaling component (Manik et al., 2015; Mao et al., 2016). Among the interactions, we found that the paralog pair BrrCBL9.1 and -9.2 can interact with 22 and 25 BrrCIPKs, respectively (Figure 7). They had the common and specific interactors with BrrCIPKs. In this regard, they may serve functional roles in different signaling pathways and choose different interacting partners to propagate signals downstream. Along with the differences between BrrCBL9.1 and -9.2 in expression patterns and interactions, we speculated that the two genes serve dissimilar functions.
The functional divergence of homologous genes may play a vital role in a plant's adaptation to adverse environments. In Arabidopsis, AtCBL1 functions as a positive regulator of salt and drought responses and as a negative regulator of cold responses in plants (Cheong et al., 2003). Meanwhile, AtCBL9 acts as a negative regulator of ABA signaling and is involved in ABA biosynthesis under stress (Pandey et al., 2004). Similar findings were obtained for Arabidopsis CIPK8 (Hu et al., 2009) and CIPK24 (Liu et al., 2000). Our transgenic/complement Arabidopsis assay indicated that BrrCBL9.2 play roles in salt responses. This result suggests that the gene is not functionally redundant (Figure 9). Previous studies of CBL9 in other species demonstrated that overexpressing ZmCBL9 enhances the resistance or tolerance to ABA, glucose, salt, and osmotic stress. The studies also showed that the process complements the hypersensitive phenotype of the Arabidopsis cbl9 mutant in response to ABA and abiotic stress (Pandey et al., 2004; Zhang et al., 2016). Overexpressing Thellungiella halophila ThCBL9 increases salt and osmotic stress tolerance in transgenic Arabidopsis (Sun et al., 2008). These results indicated that BrrCBL9.2 may play a similar molecular function to those of AtCBL9, ZmCBL9, and ThCBL9 in salt response. Thus, an evolutionary conserved function of BrrCBL9.2 genes may exist between dicots and monocots. However, BrrCBL9.1 may serve some new functions in other stress responses. Thus, the functional divergence of the paralog pair BrrCBL9.1 and -9.2 may play a prominent role in turnip's adaptation to the environment of the Qinghai–Tibet Plateau.
In conclusion, we provided a systematic genome-wide analysis of the CBL and CIPK gene families in turnip. Up to 19 and 51 members of the BrrCBL and BrrCIPK genes were identified, respectively. Phylogenetic and chromosomal distribution analyses revealed that the main reason for the expansion of these two gene families was a segmental duplication event. Different expression patterns showed that the BrrCBL and BrrCIPK involved various stresses and some paralog genes functionally diverged during long-term evolution. Y2H assay demonstrated the interaction complex between BrrCBLs and BrrCIPKs and the different interactions between some paralog proteins. Further functional analysis suggested that BrrCBL9.2 was upregulated by salt stress. Overall, this study provides insight into the characterization of CBLs and CIPKs in turnip. These insights then contributes to the understanding of the mechanisms and functions of CBL–CIPK complexes and offers basis for selecting appropriate genes for the in-depth functional studies of CBL–CIPK in turnip.
Author Contributions
Designed the experiments: YoY, YuY, and XY. Performed the experiments: XY, QC, and NX. Analyzed the data: YuY and XY. Contributed reagents/materials/analysis tools: XY, QC, NX, and QW. Wrote the paper: XY.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported financially by the Major Projects of National Natural Science Foundation of China (No. 31590823) and the National Natural Science Foundation of China (No. 31601999).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.01191/full#supplementary-material
Supplementary Table 1. Forward and reverse primers used in gene cloning, expression analysis, Y2H and BiFC vector constructions.
Supplementary Figure 1. Subcellular localization of 15 35S: BrrCBL-GFP fusion protein in N. benthamiana leaf cells. Bar = 10 μm.
Supplementary Figure 2. Phylogenetic relationships of BrrCBL and BrrCIPK proteins in Arabidopsis and Brassica rapa var. rapa. (A) Phylogenetic relationships of BrrCBL proteins in Arabidopsis and Brassica rapa var. rapa. (B) Phylogenetic relationships of BrrCIPK proteins in Arabidopsis and Brassica rapa var. rapa. The protein sequences were aligned using the MAFFT version 7 program, and phylogenetic trees were constructed using the MEGA 5.0 software with the neighbour-joining method and the 1,000 bootstrap test replicates.
Supplementary Figure 3. Multiple sequence alignments of the amino acids of 19 BrrCBLs and 10 Arabidopsis CBLs. The myristoylation and palmitoylation site was the red rectangle. The four EF-hand conserved domains were all indicated with straight lines.
Supplementary Figure 4. Multiple sequence alignments of the amino acids of 51 turnip BrrCIPKs and Arabidopsis CIPK24. (A) CIPK proteins consist of a conserved N-terminal kinase domain, and a C-terminal regulatory domain. The activation loop was denoted by straight lines. The conserved NAF/FISL motif within the rather divergent C-terminal regulatory domain was denoted by dashed lines. The protein-phosphatase interaction (PPI) motif within the C-terminus of these kinases was marked by dots above the sequences. (B) An MEME analysis of the NAF/FISL motif in 51 BrrCIPKs.
Supplementary Figure 5. An MEME analysis of motif compositions of BrrCBLs. A total of 15 conserved motifs designated as motif 1 to motif 15 were identified within the genes.
Supplementary Figure 6. An MEME analysis of motif compositions of BrrCIPKs. A total of 15 conserved motifs designated as motif 1 to motif 15 were identified within the genes.
Supplementary Figure 7. The co-expression analysis among BrrCBLs and BrrCIPKs under NaCl, drought, cold, and MeJA treatment, respectively.
References
Albrecht, V., Ritz, O., Linder, S., Harter, K., and Kudla, J. (2001). The NAF domain defines a novel protein-protein interaction module conserved in Ca2+-regulated kinases. EMBO J. 20, 1051–1063. doi: 10.1093/emboj/20.5.1051
Albrecht, V., Weinl, S., Blazevic, D., D'angelo, C., Batistic, O., Kolukisaoglu, Ü., et al. (2003). The calcium sensor CBL1 integrates plant responses to abiotic stresses. Plant J. 36, 457–470. doi: 10.1046/j.1365-313X.2003.01892.x
Bailey, T. L., Williams, N., Misleh, C., and Li, W. W. (2006). MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 34, W369–W373. doi: 10.1093/nar/gkl198
Batistic, O., and Kudla, J. (2004). Integration and channeling of calcium signaling through the CBL calcium sensor/CIPK protein kinase network. Planta 219, 915–924. doi: 10.1007/s00425-004-1333-3
Batistic, O., and Kudla, J. (2009). Plant calcineurin B-like proteins and their interacting protein kinases. Biochim. Biophys. Acta 1793, 985–992. doi: 10.1016/j.bbamcr.2008.10.006
Batistic, O., and Kudla, J. (2012). Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta 1820, 1283–1293. doi: 10.1016/j.bbagen.2011.10.012
Batistic, O., Sorek, N., Schultke, S., Yalovsky, S., and Kudla, J. (2008). Dual fatty acyl modification determines the localization and plasma membrane targeting of CBL/CIPK Ca2+ signaling complexes in Arabidopsis. Plant Cell 20, 1346–1362. doi: 10.1105/tpc.108.058123
Batistic, O., Waadt, R., Steinhorst, L., Held, K., and Kudla, J. (2010). CBL-mediated targeting of CIPKs facilitates the decoding of calcium signals emanating from distinct cellular stores. Plant J. 61, 211–222. doi: 10.1111/j.1365-313X.2009.04045.x
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Cao, J., Huang, J., Yang, Y., and Hu, X. (2011). Analyses of the oligopeptide transporter gene family in poplar and grape. BMC Genomics 12:465. doi: 10.1186/1471-2164-12-465
Chen, X., Gu, Z., Xin, D., Hao, L., Liu, C., Huang, J., et al. (2011). Identification and characterization of putative CIPK genes in maize. J. Genet. Genomics 38, 77–87. doi: 10.1016/j.jcg.2011.01.005
Chen, X. F., Gu, Z. M., Liu, F., Ma, B. J., and Zhang, H. S. (2011). Molecular analysis of rice CIPKs involved in both biotic and abiotic stress responses. Rice Sci. 18, 1–9. doi: 10.1016/S1672-6308(11)60001-2
Cheng, N. H., Pittman, J. K., Zhu, J. K., and Hirschi, K. D. (2004). The protein kinase SOS2 activates the Arabidopsis H+/Ca2+ antiporter CAX1 to integrate calcium transport and salt tolerance. J. Biol. Chem. 279, 2922–2926. doi: 10.1074/jbc.M309084200
Cheong, Y. H., Kim, K. N., Pandey, G. K., Gupta, R., Grant, J. J., and Luan, S. (2003). CBL1, a calcium sensor that differentially regulates salt, drought, and cold responses in Arabidopsis. Plant Cell 15, 1833–1845. doi: 10.1105/tpc.012393
Cheong, Y. H., Pandey, G. K., Grant, J. J., Batistic, O., Li, L., Kim, B. G., et al. (2007). Two calcineurin B-like calcium sensors, interacting with protein kinase CIPK23, regulate leaf transpiration and root potassium uptake in Arabidopsis. Plant J. 52, 223–239. doi: 10.1111/j.1365-313X.2007.03236.x
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Ferguson, N. M., Galvani, A. P., and Bush, R. M. (2003). Unravelling angiosperm genome evolution by phylogenetic analysis of chromosomal duplication events. Nature 422, 428–433. doi: 10.1038/nature01509
Flagel, L. E., and Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564. doi: 10.1111/j.1469-8137.2009.02923.x
Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S. E., Wilkins, M. R., Appel, R. D., and Bairoch, A. (2005). “Protein identification and analysis tools on the ExPASy server,” in The Proteomics Protocols Handbook, ed. J. M. Walker (Totowa, NJ: Humana Press), 571–607.
Guo, Y., Halfter, U., Ishitani, M., and Zhu, J. K. (2001). Molecular characterization of functional domains in the protein kinase SOS2 that is required for plant salt tolerance. Plant Cell 13, 1383–1400. doi: 10.1105/tpc.13.6.1383
Hashimoto, K., Eckert, C., Anschutz, U., Scholz, M., Held, K., Waadt, R., et al. (2012). Phosphorylation of calcineurin B-like (CBL) calcium sensor proteins by their CBL-interacting protein kinases (CIPKs) is required for full activity of CBL-CIPK complexes toward their target proteins. J. Biol. Chem. 287, 7956–7968. doi: 10.1074/jbc.M111.279331
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, H. C., Wang, Y. Y., and Tsay, Y. F. (2009). AtCIPK8, a CBL-interacting protein kinase, regulates the low-affinity phase of the primary nitrate response. Plant J. 57, 264–278. doi: 10.1111/j.1365-313X.2008.03685.x
Huang, C., Ding, S., Zhang, H., Du, H., and An, L. (2011). CIPK7 is involved in cold response by interacting with CBL1 in Arabidopsis thaliana. Plant Sci. 181, 57–64. doi: 10.1016/j.plantsci.2011.03.011
Kanwar, P., Sanyal, S. K., Tokas, I., Yadav, A. K., Pandey, A., Kapoor, S., et al. (2014). Comprehensive structural, interaction and expression analysis of CBL and CIPK complement during abiotic stresses and development in rice. Cell Calcium 56, 81–95. doi: 10.1016/j.ceca.2014.05.003
Kleist, T. J., Spencley, A. L., and Luan, S. (2014). Comparative phylogenomics of the CBL-CIPK calcium-decoding network in the moss Physcomitrella, Arabidopsis, and other green lineages. Front. Plant Sci. 5:187. doi: 10.3389/fpls.2014.00187
Kolukisaoglu, U., Weinl, S., Blazevic, D., Batistic, O., and Kudla, J. (2004). Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134, 43–58. doi: 10.1104/pp.103.033068
Kudla, J., Xu, Q., Harter, K., Gruissem, W., and Luan, S. (1999). Genes for calcineurin B-like proteins in Arabidopsis are differentially regulated by stress signals. Proc. Natl. Acad. Sci. U.S.A. 96, 4718–4723. doi: 10.1073/pnas.96.8.4718
Lee, S. C., Lan, W. Z., Kim, B. G., Li, L., Cheong, Y. H., Pandey, G. K., et al. (2007). A protein phosphorylation/dephosphorylation network regulates a plant potassium channel. Proc. Natl. Acad. Sci. U.S.A. 104, 15959–15964. doi: 10.1073/pnas.0707912104
Li, J., Ding, Q., Wang, F., Zhang, Y., Li, H., and Gao, J. (2015). Integrative analysis of mRNA and miRNA expression profiles of the tuberous root development at seedling stages in turnips. PLoS ONE 10:e0137983. doi: 10.1371/journal.pone.0137983
Li, J., Jiang, M. M., Ren, L., Liu, Y., and Chen, H. Y. (2016). Identification and characterization of CBL and CIPK gene families in eggplant (Solanum melongena L.). Mol. Genet. Genomics 291, 1769–1781. doi: 10.1007/s00438-016-1218-8
Li, L., Kim, B. G., Cheong, Y. H., Pandey, G. K., and Luan, S. (2006). A Ca2+ signaling pathway regulates a K+ channel for low-K response in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 12625–12630. doi: 10.1073/pnas.0605129103
Li, R., Zhang, J., Wei, J., Wang, H., Wang, Y., and Ma, R. (2009). Functions and mechanisms of the CBL–CIPK signaling system in plant response to abiotic stress. Prog. Nat. Sci. 19, 667–676. doi: 10.1016/j.pnsc.2008.06.030
Liang, Y. S., Kim, H. K., Lefeber, A. W., Erkelens, C., Choi, Y. H., and Verpoorte, R. (2006). Identification of phenylpropanoids in methyl jasmonate treated Brassica rapa leaves using two-dimensional nuclear magnetic resonance spectroscopy. J. Chromatogr. A 1112, 148–155. doi: 10.1016/j.chroma.2005.11.114
Liu, J., Ishitani, M., Halfter, U., Kim, C. S., and Zhu, J. K. (2000). The Arabidopsis thaliana SOS2 gene encodes a protein kinase that is required for salt tolerance. Proc. Natl. Acad. Sci. U.S.A. 97, 3730–3734. doi: 10.1073/pnas.97.7.3730
Luan, S. (2009). The CBL-CIPK network in plant calcium signaling. Trends Plant Sci. 14, 37–42. doi: 10.1016/j.tplants.2008.10.005
Luan, S., Kudla, J., Rodriguez-Concepcion, M., Yalovsky, S., and Gruissem, W. (2002). Calmodulins and calcineurin B–like proteins calcium sensors for specific signal response coupling in plants. Plant Cell 14, S389–S400. doi: 10.1105/tpc.001115
Mahajan, S., Sopory, S. K., and Tuteja, N. (2006). Cloning and characterization of CBL-CIPK signalling components from a legume (Pisum sativum). FEBS J. 273, 907–925. doi: 10.1111/j.1742-4658.2006.05111.x
Manik, S. M., Shi, S., Mao, J., Dong, L., Su, Y., Wang, Q., et al. (2015). The calcium sensor CBL-CIPK is involved in plant's response to abiotic stresses. Int. J. Genomics 2015, 10. doi: 10.1155/2015/493191
Mao, J., Manik, S. M., Shi, S., Chao, J., Jin, Y., Wang, Q., et al. (2016). Mechanisms and physiological roles of the CBL-CIPK networking system in Arabidopsis thaliana. Genes 7:62. doi: 10.3390/genes7090062
Pandey, G. K., Cheong, Y. H., Kim, K. N., Grant, J. J., Li, L., Hung, W., et al. (2004). The calcium sensor calcineurin B-like 9 modulates abscisic acid sensitivity and biosynthesis in Arabidopsis. Plant Cell 16, 1912–1924. doi: 10.1105/tpc.021311
Qiu, Q. S., Guo, Y., Dietrich, M. A., Schumaker, K. S., and Zhu, J. K. (2002). Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc. Natl. Acad. Sci. U.S.A. 99, 8436–8441. doi: 10.1073/pnas.122224699
Ren, J., Wen, L., Gao, X., Jin, C., Xue, Y., and Yao, X. (2008). CSS-Palm 2.0: an updated software for palmitoylation sites prediction. Protein Eng. Des. Sel. 21, 639–644. doi: 10.1093/protein/gzn039
Roberts, D. M., and Harmon, A. C. (1992). Calcium-modulated proteins: targets of intracellular calcium signals in higher plants. Annu. Rev. Plant Biol. 43, 375–414. doi: 10.1146/annurev.pp.43.060192.002111
Sanchez-Barrena, M. J., Martinez-Ripoll, M., and Albert, A. (2013). Structural biology of a major signaling network that regulates plant abiotic stress: the CBL-CIPK Mediated Pathway. Int. J. Mol. Sci. 14, 5734–5749. doi: 10.3390/ijms14035734
Schranz, M. E., Lysak, M. A., and Mitchell-Olds, T. (2006). The ABC's of comparative genomics in the Brassicaceae: building blocks of crucifer genomes. Trends Plant Sci. 11, 535–542. doi: 10.1016/j.tplants.2006.09.002
Shen, J. Q., Zhang, Z. Z., P, W. H., and Pan, J. W. (2014). Functions and action mechanisms of CBL-CIPK signaling system in plants. Plant Physiol. J. 50, 641–650.
Sturn, A., Quackenbush, J., and Trajanoski, Z. (2002). Genesis: cluster analysis of microarray data. Bioinformatics 18, 207–208. doi: 10.1093/bioinformatics/18.1.207
Sun, T., Wang, Y., Wang, M., Li, T., Zhou, Y., Wang, X., et al. (2015). Identification and comprehensive analyses of the CBL and CIPK gene families in wheat (Triticum aestivum L.). BMC Plant Biol. 15:269. doi: 10.1186/s12870-015-0657-4
Sun, Z., Qi, X., Li, P., Wu, C., Zhao, Y., Zhang, H., et al. (2008). Overexpression of a Thellungiella halophila CBl9 homolog, ThCBL9, confers salt and osmotic tolerances in transgenic Arabidopsis thaliana. J. Plant Biol. 51, 25–34. doi: 10.1007/BF03030737
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thapa, G. D., Dey, M., Sahoo, L., and Panda, S. K. (2011). An insight into the drought stress induced alterations in plants. Biol. Plant 55, 603–613 doi: 10.1007/s10535-011-0158-8
Weinl, S., and Kudla, J. (2009). The CBL–CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol. 184, 517–528. doi: 10.1111/j.1469-8137.2009.02938.x
Wydro, M., Kozubek, E., and Lehmann, P. (2006). Optimization of transient Agrobacterium-mediated gene expression system in leaves of Nicotiana benthamiana. Acta Biochim. Pol. 53, 289.
Xiong, L., Schumaker, K. S., and Zhu, J. K. (2002). Cell signaling during cold, drought, and salt stress. Plant Cell 14, S165–S183. doi: 10.1105/tpc.000596
Xu, J., Li, H. D., Chen, L. Q., Wang, Y., Liu, L. L., He, L., et al. (2006). A protein kinase, interacting with two calcineurin B-like proteins, regulates K+ transporter AKT1 in Arabidopsis. Cell 125, 1347–1360. doi: 10.1016/j.cell.2006.06.011
Yang, Z. (2007). PAML 4: phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591. doi: 10.1093/molbev/msm088
Yu, Y., Xia, X., Yin, W., and Zhang, H. (2007). Comparative genomic analysis of CIPK gene family in Arabidopsis and Populus. Plant Growth Regul. 52, 101–110. doi: 10.1007/s10725-007-9165-3
Zhang, F., Li, L., Jiao, Z., Chen, Y., Liu, H., Chen, X., et al. (2016). Characterization of the calcineurin B-Like (CBL) gene family in maize and functional analysis of ZmCBL9 under abscisic acid and abiotic stress treatments. Plant Sci. 253, 118–129. doi: 10.1016/j.plantsci.2016.09.011
Zhang, H., Yang, B., Liu, W. Z., Li, H., Wang, L., Wang, B., et al. (2014). Identification and characterization of CBL and CIPK gene families in canola (Brassica napus L.). BMC Plant Biol. 14:8. doi: 10.1186/1471-2229-14-8
Keywords: BrrCBL–BrrCIPK, expression profiles, preferential interactions, functional differentiation, turnip
Citation: Yin X, Wang Q, Chen Q, Xiang N, Yang Y and Yang Y (2017) Genome-Wide Identification and Functional Analysis of the Calcineurin B-like Protein and Calcineurin B-like Protein-Interacting Protein Kinase Gene Families in Turnip (Brassica rapa var. rapa). Front. Plant Sci. 8:1191. doi: 10.3389/fpls.2017.01191
Received: 24 March 2017; Accepted: 22 June 2017;
Published: 07 July 2017.
Edited by:
Xiaowu Wang, Biotechnology Research Institute (CAAS), ChinaReviewed by:
Xiaolin Yu, Zhejiang University, ChinaXiaonan Li, Chungnam National University, South Korea
Copyright © 2017 Yin, Wang, Chen, Xiang, Yang and Yang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yunqiang Yang, eWFuZ3l1bnFpYW5nQG1haWwua2liLmFjLmNu
Yongping Yang, eWFuZ3lwQG1haWwua2liLmFjLmNu
 Xin Yin
Xin Yin Qiuli Wang1,2,3,5
Qiuli Wang1,2,3,5 Yunqiang Yang
Yunqiang Yang Yongping Yang
Yongping Yang