
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 May 2017
Sec. Plant Abiotic Stress
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.00877
Calcium ion (Ca2+) is a universal second messenger that plays a critical role in plant responses to diverse physiological and environmental stimuli. The stimulus-specific signals are perceived and decoded by a series of Ca2+ binding proteins serving as Ca2+ sensors. The majority of Ca2+ sensors possess the EF-hand motif, a helix-loop-helix structure which forms a turn-loop structure. Although EF-hand proteins in model plant such as Arabidopsis have been well described, the identification, classification, and the physiological functions of EF-hand-containing proteins from soybean are not systemically reported. In this study, a total of at least 262 genes possibly encoding proteins containing one to six EF-hand motifs were identified in soybean genome. These genes include 6 calmodulins (CaMs), 144 calmodulin-like proteins (CMLs), 15 calcineurin B-like proteins, 50 calcium-dependent protein kinases (CDPKs), 13 CDPK-related protein kinases, 2 Ca2+- and CaM-dependent protein kinases, 17 respiratory burst oxidase homologs, and 15 unclassified EF-hand proteins. Most of these genes (87.8%) contain at least one kind of hormonal signaling- and/or stress response-related cis-elements in their -1500 bp promoter regions. Expression analyses by exploring the published microarray and Illumina transcriptome sequencing data revealed that the expression of these EF-hand genes were widely detected in different organs of soybean, and nearly half of the total EF-hand genes were responsive to various environmental or nutritional stresses. Quantitative RT-PCR was used to confirm their responsiveness to several stress treatments. To confirm the Ca2+-binding ability of these EF-hand proteins, four CMLs (CML1, CML13, CML39, and CML95) were randomly selected for SDS–PAGE mobility-shift assay in the presence and absence of Ca2+. Results showed that all of them have the ability to bind Ca2+. This study provided the first comprehensive analyses of genes encoding for EF-hand proteins in soybean. Information on the classification, phylogenetic relationships and expression profiles of soybean EF-hand genes in different tissues and under various environmental and nutritional stresses will be helpful for identifying candidates with potential roles in Ca2+ signal-mediated physiological processes including growth and development, plant-microbe interactions and responses to biotic and abiotic stresses.
During their life cycles, plants encounter a variety of external stresses such as heat, cold, drought, flooding, salinity, nutrient deficiency, and attacks from insects and pathogens. Calcium (Ca2+) is one of the most abundant elements on earth, and serves as a universal second intracellular messenger that plays a critical role in plant responses to environmental stresses, as well as hormonal and developmental cues (Sanders et al., 2002). Ca2+ concentration in the cytoplasm and nucleus is kept low (∼100–200 nM), since high levels of cytosolic Ca2+ are toxic to cells by forming insoluble compounds with phosphate derivatives and complex with macromolecules. A steep Ca2+ gradient across the plasma membrane as well as the inner membrane system enclosing the cellular Ca2+ storages is established under the controls of stimulus responsive Ca2+ channels, pumps and transporters (McAinsh and Pittman, 2009; Kudla et al., 2010). It has been well-documented that various external biotic or abiotic stimuli can quickly trigger specific and distinct spatial-temporal patterns of changes in cytosolic Ca2+ concentration, and “Ca2+ signatures" have been coined to describe the specificities of calcium signals triggered by the causative stimuli (Webb et al., 1996; McAinsh and Pittman, 2009). The stimulus-specific signals are perceived and decoded by a series of Ca2+ binding proteins which function as Ca2+ sensors to generate specific or overlapping responses (DeFalco et al., 2010; Batistic and Kudla, 2012). Upon Ca2+ binding, Ca2+ sensors undergo conformational changes that either promote their interactions with target proteins or alter their own enzymatic activities, and therefore transmit these stimulus-specific signals into biochemical, molecular, cellular or physiological events, which help plants survive the unfavorable environmental conditions.
The majority of Ca2+ sensors possess the EF-hand motif, which is composed of a 29-amino acid helix-loop-helix structure, with the central 12 residues forming a turn-loop structure that is responsible for the coordination of Ca2+ ion (Gifford et al., 2007). Within the coordination loop, ligating residues are found at positions 1, 3, 5, 7, 9, and 12 (X∗Y∗Z∗-Y∗-X∗∗-Z, “∗” represents an intervening residue). The loop is enriched in negatively charged residues, and a Gly at position six allows the loop to wrap around the Ca2+ ion, a feature critical for high-affinity Ca2+ binding (Gifford et al., 2007). Structural properties of the EF-hand motif allow for rapid on/off Ca2+ binding, which permits quick responses to changes in cytosolic Ca2+ concentration. Typically, EF-hands occur in pairs and facilitate high-affinity cooperative binding of Ca2+ (DeFalco et al., 2010). Calmodulins (CaMs) and CaM-like proteins (CMLs), calcineurin B-like proteins (CBLs), and calcium-dependent protein kinases (CDPKs) are the three largest groups of Ca2+ sensors in plants (Luan et al., 2002; DeFalco et al., 2010; Zeng et al., 2015). CaMs are ubiquitous Ca2+-binding proteins existing in all eukaryotes, whereas CMLs, CDPKs, and CBLs are restricted to plant taxa and certain protist groups (Poovaiah et al., 2013; Hamel et al., 2014; Kleist et al., 2014; Zhu et al., 2015). In Arabidopsis thaliana genome, there are at least 250 genes encoding for EF-hand proteins which include 7 CaM genes, 50 CML genes, 10 CBL genes, 34 CDPK genes (Day et al., 2002; Hrabak et al., 2003; McCormack and Braam, 2003; Kolukisaoglu et al., 2004). However, the identification, classification and the physiological functions of EF-hand-containing proteins in other plants are largely unaddressed.
Soybean is an important economic crop which provides an oil- and protein-rich food for human around the world, and also an important crop for the production of renewable fuel of biodiesel. However, global productivity of soybean is frequently affected by various environmental stresses, such as drought, salt, and nutrient deficiency (Tran and Mochida, 2010; Zeng et al., 2010; Deshmukh et al., 2014). With the availability of whole-genome sequences of soybean, genes encoding proteins with specific domains like EF-hand motif can be identified at the whole-genome scale (Schmutz et al., 2010). In this study, we carried out a genome-wide search and identified and analyzed all the available EF-hand motif-containing proteins in soybean genome.
Proteins containing an EF-hand domain or in the family of Ca2+-binding proteins which included domains PF00036, PTHR10891, PS00018, PS50222, SM00054, and SSF47473, were collected by keyword searching from the Phytozome’s soybean database1. Furthermore, “EF-hand,” “calmodulin,” “calmodulin-like,” “calcineurin B-like,” and “calcium-dependent protein kinase” were each used as a key word to perform homolog searches. In addition, the amino acid sequences of Arabidopsis CaM2, CBL1, CDPK1, CRK1, and RbohD were used as queries to blast against the soybean genome database using the BlastP program. The nucleotide and protein sequences were obtained and analyzed for EF-hands and other domains using InterPro database2 (Hunter et al., 2009). Proteins not showing EF-hand domains were eliminated from the list.
Sequences of EF-hand proteins of Arabidopsis and soybean were obtained from TAIR3 and Phytozome4, respectively. The amino acid sequences of EF-hand proteins were aligned using ClustalW, and a phylogenetic tree was constructed by the neighbor-joining method using the software MEGA6 (Tamura et al., 2013). The location of soybean genes encoding EF-hand proteins was determined based on their physical positions on chromosomes corresponding to their locus number in the JBrowse genome browser5. If the distance between two neighboring paralogous genes were less than 100 kb and separated by five or less genes, they were considered to be tandemly duplicated genes (Zhu et al., 2016).
Conserved domains other than EF-hand motif were predicted using InterPro6. Myristoylation sites were predicted using PlantsP7. Promoter sequences of 1.5 kb upstream to the transcription start site of the genes encoding EF-hand proteins were retrieved from JBrowse, and the location of stress-related cis-acting regulatory elements was analyzed using Regulatory Sequence Analysis Tools8 (Medina-Rivera et al., 2015).
Genome-wide public RNA-seq datasets [Reads/Kb/Million (RPKM) normalized data] in different tissues of soybean were downloaded from soybean transcriptome atlas (Libault et al., 2010b). Microarray or high-throughput sequencing datasets for soybean in responses to cold, drought, phosphorus deficiency, and symbiotic bacteria, were also retrieved from previously published data (Libault et al., 2010a; Le et al., 2012; Maruyama et al., 2012; Chen et al., 2016; Zeng et al., 2016b).
Soybean seeds (Glycine max var. Williams 82) were soaked in sterilized water for 4 h, and then germinated at room temperature in the dark between two layers of filter paper moistened with sterilized water (Wang et al., 2015). After 4 days, seedlings were grown hydroponically in half-strength modified Hoagland nutrient solution, the pH of the nutrient solution was adjusted to 5.6, and the nutrient solution was changed every 2 days. The seedlings were grown in a growth chamber under controlled conditions (photoperiod 16-h-light/8-h-dark at 26/22°C, light intensity 150 μmol m-2 s-1). After cultivating for 8 days, soybean seedlings were used for various stress treatments. For salt stress, the roots of seedlings were immersed in nutrient solution containing a relative high concentration of NaCl (200 mM) (Zhang et al., 2008). Plant responses to high salinity stress may be obvious and significant relative to moderate salinity stress (e.g., 100 mM). For dehydration treatment, seedlings were treated with 10% polyethylene glycol (PEG) 6000; for nutrient deficiency treatment, seedlings were transferred into normal nutrient solution (control) and nutrient-deficient solution (without phosphate, iron, or zinc). Roots of the stress-treated and non-treated plants were collected at time intervals of 0, 2, 8, and 24 h. After collection, the samples were immediately frozen in liquid nitrogen and stored at -80°C until RNA extraction.
Gene-specific primers were designed using NCBI Primer-BLAST9. Primer specificity was then confirmed by blasting each primer sequence against the soybean genome. Total RNA was extracted from soybean tissues using RNApure Plant Kit (with DNase I) (CoWin Biotech, Beijing, China) and digested with DNaseI to eliminate genomic DNA contamination according to the manufacturer’s instruction. cDNA was synthesized from 1.0 μg total RNA by SuperRT Reverse Transcriptase (CoWin Biotech, Beijing, China) using oligo(dT) primers in a 20 μL reaction system. Quantitative RT-PCR (qRT-PCR) was performed on a real-time PCR system (CFX96 system, Bio-Rad) as described previously (Zeng et al., 2016a). Amplifications were run in triplicate together with controls that contained no template and no reverse transcription for each of the examined gene. Relative expression levels were normalized to that of an internal control GmACTIN11 (Gm18g290800). The gene-specific primers are listed in Supplemental Table S11.
Gene-specific primers were used to clone the coding sequences of the genes GmCML1, GmCML13, GmCML39, and GmCML95 by PCR from cDNA of soybean seedlings. AtCaM7 were cloned from Arabidopsis cDNA. The gene-specific primers are listed in Supplemental Table S11. Recombinant proteins were expressed and purified according to (Routray et al., 2013). cDNAs were subcloned into the multiple cloning sites of the pET28a expression vector (Novagen) and confirmed by DNA sequencing. Recombinant proteins were expressed in Escherichia coli BL21(DE3) pLysS (Stratagene). Bacterial cultures were grown under antibiotic selection in Luria-Bertani medium with agitation at 37°C to an OD600 of 0.4–0.5 and protein expression was induced over the course of 3 h by the addition of 0.5 mM isopropyl-β-D-thiogalactopyranoside (IPTG). His-tagged recombinant proteins were purified using Ni-NTA agarose affinity beads (Qiagen) as described by the manufacturer. The electrophoresis mobility-shift assay was performed as described (Garrigos et al., 1991) using 1–2 μg of denatured protein supplemented with either 1.0 mM CaCl2, 1.0 mM EGTA, or 1.0 mM MgCl2. Each sample was electrophoresed on a 12% SDS–PAGE containing either 1.0 mM CaCl2, 1.0 mM EGTA, or 1.0 mM MgCl2, respectively.
A total of at least 262 putative EF-hand-containing proteins were found in soybean genome by using the methods described in the “Materials and Methods” section. These proteins were checked for the presence of EF-hands by using InterPro database (Hunter et al., 2009) (Supplemental Table S1). The number of EF-hand motifs in each protein varied from one to six; among these EF-hand proteins, nearly half (47%) contained four EF-hands, and 24% contained two EF-hands (Figure 1).
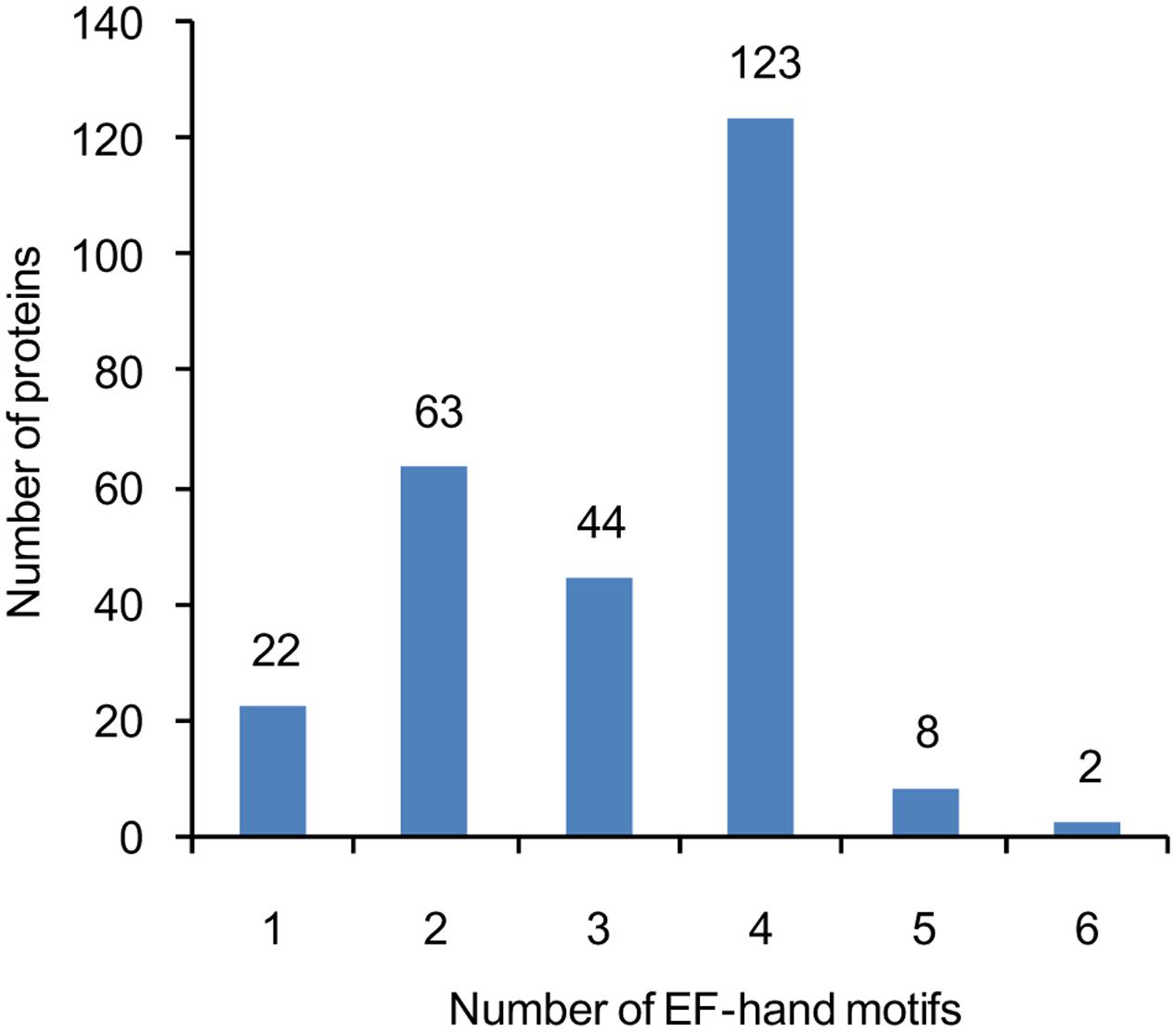
FIGURE 1. The number of genes encoding proteins containing 1, 2, 3, 4, 5, or 6 EF-hand motifs. The number of EF-hand motifs in each protein was predicted using InterPro database (http://www.ebi.ac.uk/interpro/).
The 262 EF-hand proteins identified in this study were distributed on all of the 20 soybean chromosomes. However, the number of EF-hand proteins on each chromosome appeared to be uneven, ranging from 6 to 17 (Figure 2). For example, chromosome 2, 5, and 17 contained the highest number of 17 EF-hand proteins, whereas only 6 EF-hand proteins were found on chromosome 9 and 15. Tandem duplication is a factor contributing to the evolution of many gene families (Cannon et al., 2004). Genes located within 100 kb from each other and separated by five or less genes were marked with a light green box to indicate possible tandem duplications. There were 13 possible tandem duplicated sites among these EF-hand proteins (Figure 2).
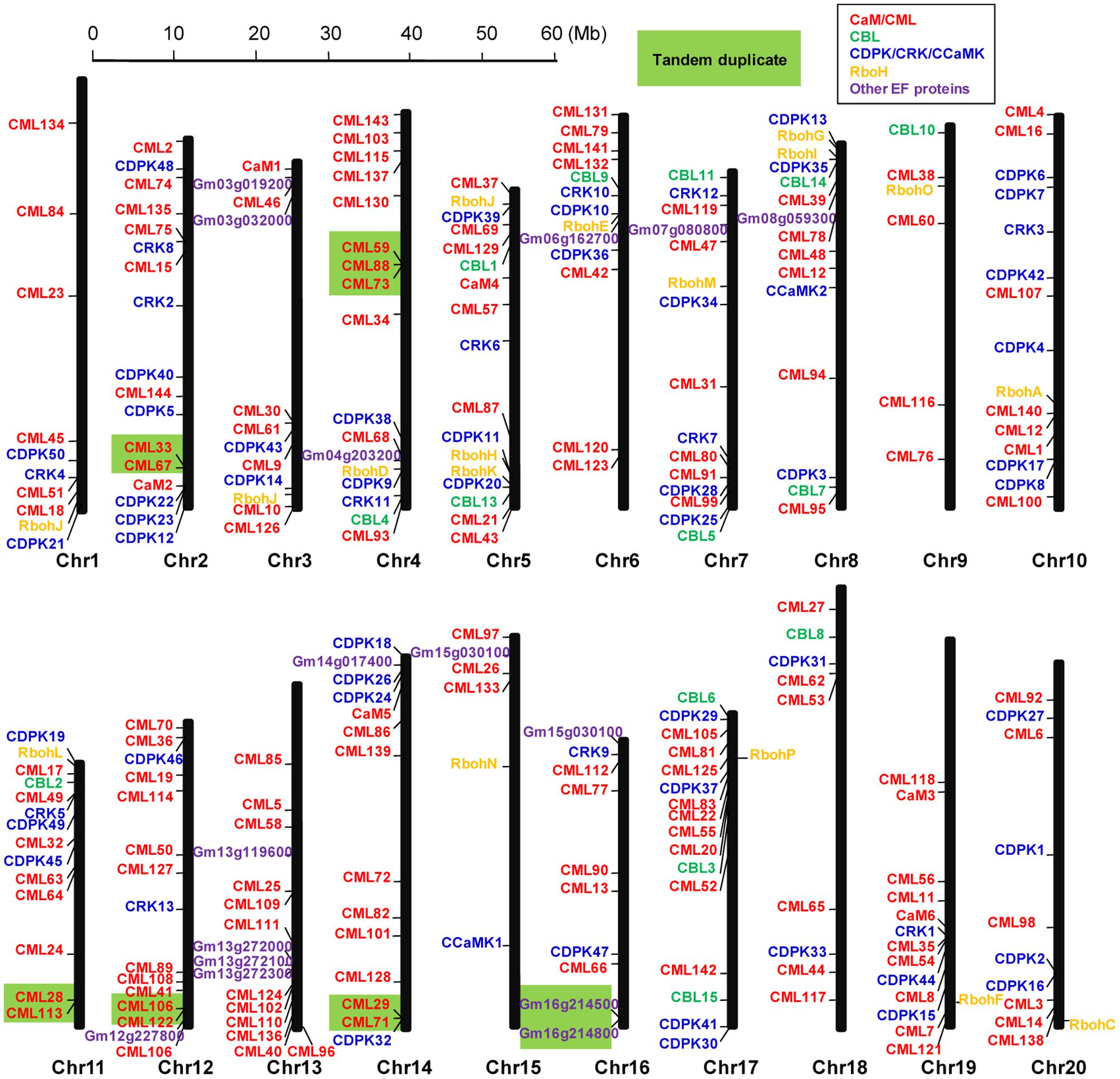
FIGURE 2. Graphical representation of locations for putative EF-hand protein-encoding genes on each soybean chromosome. CaM/CMLs are marked in red, CBLs are marked in green, CDPK/CRK/CCaMKs are marked in blue, respiratory burst oxidase homologs (Rbohs) are marked in orange, and the other unclassified EF-hand proteins (UEPs) are marked in purple. Genes within a light green box are putatively tandemly duplicated.
Sequences of all the identified EF-hand proteins were aligned using ClustalW and a phylogenetic tree was constructed using the neighbor-joining method. The phylogenetic tree of the overall 262 EF-hand proteins was shown in Figure 3. According to the presence of predicted functional domains other than EF-hands, and their similarity to classical Ca2+ sensors, such as CaM/CML, CBL, and CDPK, these proteins were classified into five groups, CaM/CMLs, CBLs, CDPK/CRK/CCaMKs, Rbohs, and UEPs (Supplemental Tables S2–S7 and Figure 3). The schematic diagrams of representative EF-hand proteins of various groups containing different number of EF-hands were shown in Figure 4. As can be seen, the CaMs, CMLs, and CBLs have no functional domain other than EF-hands, while the other proteins have additional domains, such as protein kinase domain, TIR domain, mitochondrial carrier domain, actin-binding domain, and FAD-binding domain (Figure 4).

FIGURE 3. Unrooted phylogenetic tree of EF-hand proteins in soybean (Glycine max). The alignment for the phylogenetic tree was performed with ClustalW using full-length amino acid sequences. The phylogenetic tree was created with the MEGA6 software and the neighbor-joining method with 1,000 bootstrap replications. All the 150 CaM/CML proteins are marked with red circles, 15 CBL proteins are marked with green squares, 65 CDPK/CRK/CCaMK proteins are marked with purple triangles, 17 Rboh proteins are marked with pink inverted triangles, and 15 UEPs are marked with blue diamonds. The bar indicates the relative divergence of the sequences examined.
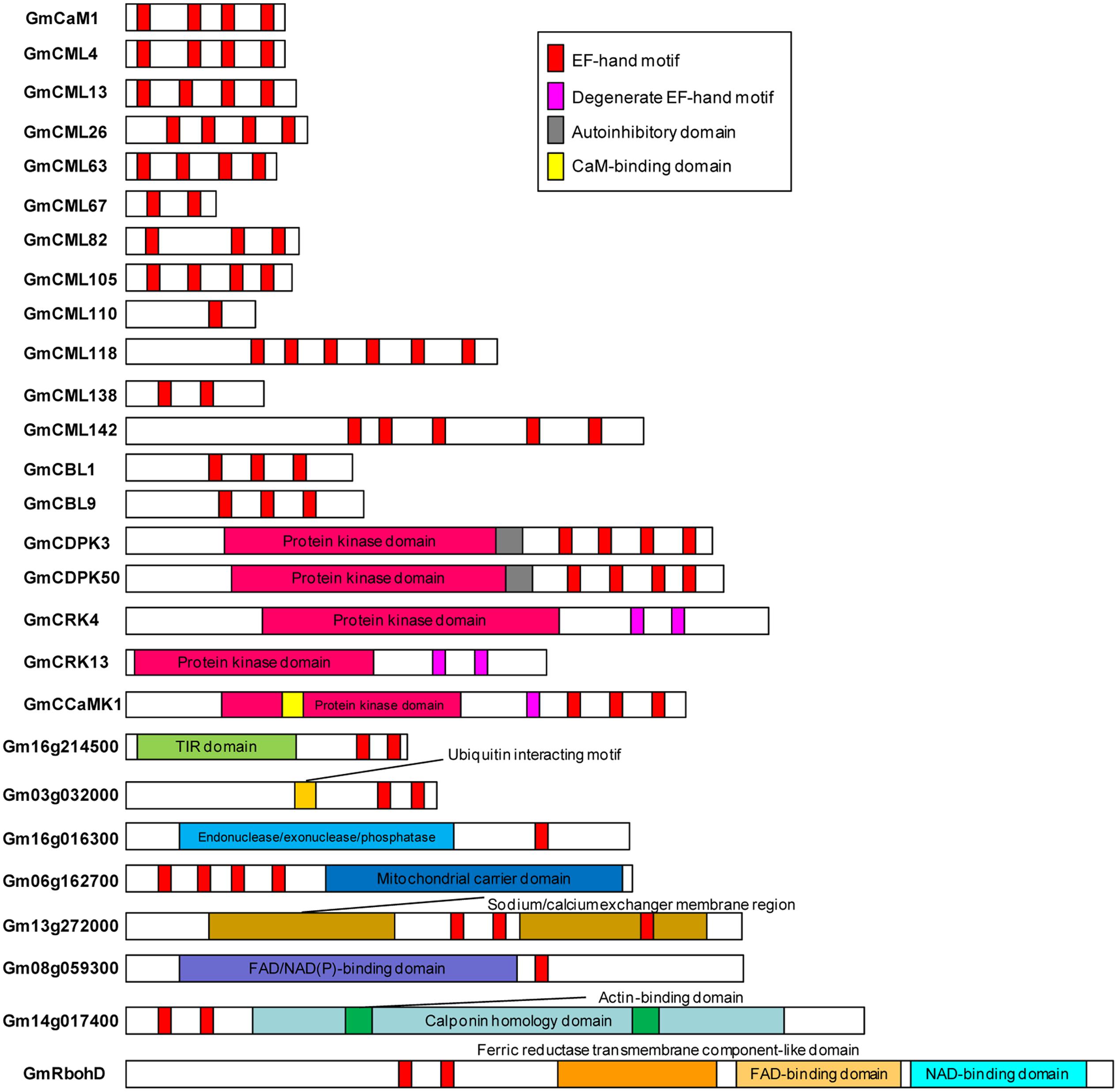
FIGURE 4. Schematic diagrams of representative soybean EF-hand proteins. EF-hands and other domains were predicted using InterPro database (http://www.ebi.ac.uk/interpro/).
Based on the extremely high amino acid identity to Arabidopsis thaliana CaM2 (AtCaM2) (98.0–98.7%), six genes were found to encode two isoforms of CaMs in soybean (CaM1/2/3/4/5, CaM6), both are 149 amino acid (AA) in length. In addition, there were at least 144 genes encoding for CML proteins; these CMLs share at least 12.8% overall amino acid identity with AtCaM2, and have no other known functional domains except the EF-hands (Supplemental Table S2). The length of soybean CMLs ranged from 80 to 501 AA. Half of these CMLs (72/144) have four EF-hands, and only two CMLs (CML101 and CML118) have six EF-hands. CML1 and CML2 have previously been known as SCaM4 and SCaM5, respectively (Lee et al., 1995). However, because the encoded proteins share only 79% amino acid identity with AtCaM2, they are likely to have distinct functions from the conserved CaMs. Protein sequences of CaM/CMLs from soybean and Arabidopsis were retrieved for phylogenetic analysis, and the results showed that these CaM/CMLs can be classified into nine subgroups, with each subgroup containing different number of CaM/CML proteins (Figure 5).
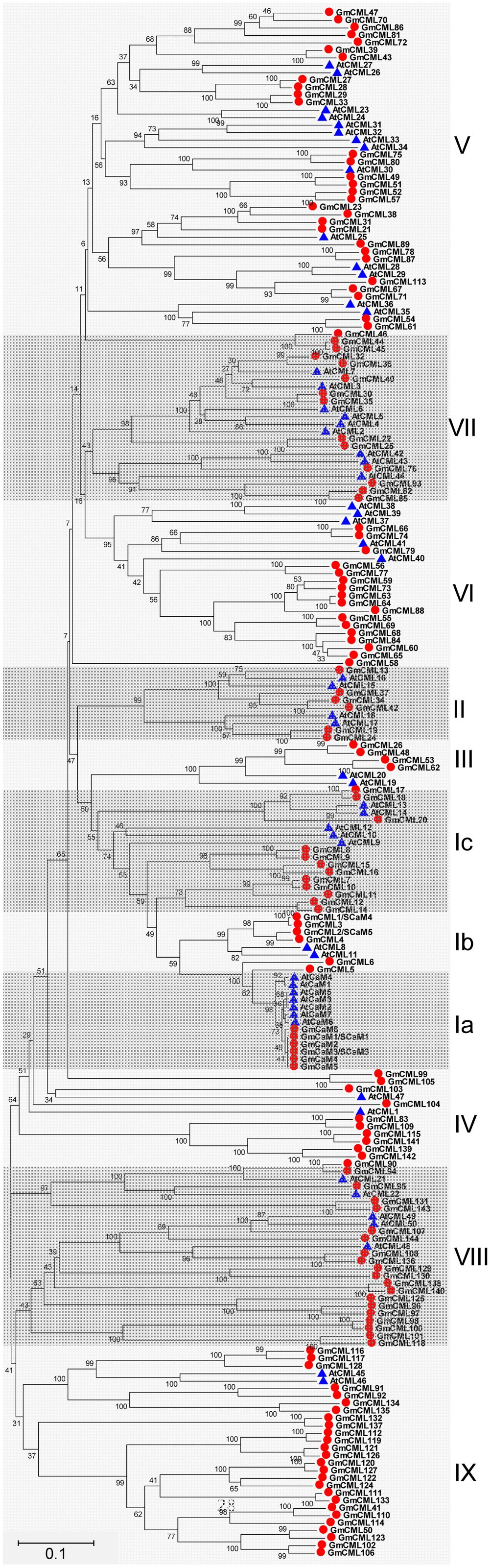
FIGURE 5. Unrooted phylogenetic tree of the CaM and CML proteins from soybean (G. max) and Arabidopsis thaliana. The alignment for the phylogenetic tree was performed with ClustalW using full-length amino acid sequences. The phylogenetic tree was created with the MEGA6 software and the neighbor-joining method with 1,000 bootstrap replications. All the 150 CaM/CML proteins from soybean are marked with red circles, while 57 CaM/CML proteins from Arabidopsis are marked with blue triangles. Roman numerals designate the subfamilies. The bar indicates the relative divergence of the sequences examined and bootstrap values are displayed next to the branch.
A total of 15 genes encoding putative CBLs were found in soybean genome, and they showed high sequence identity with AtCBL1 (48.8–77.5%). These genes were sequentially named GmCBL1-GmCBL15, based on their amino acid identity to AtCBL1 (Supplemental Table S3). Similar to their counterparts in other plants, GmCBLs are relatively small proteins with lengths ranging from 212 to 265 AA, and all proteins contain three putative EF-hands (Supplemental Table S3). Phylogenetic analysis indicated that GmCBLs are closely related to the 10 AtCBLs (Figure 6).

FIGURE 6. Unrooted phylogenetic tree of the total CBL proteins from soybean (G. max) and A. thaliana. The alignment for the phylogenetic tree was performed with ClustalW using full-length amino acid sequences. The phylogenetic tree was created with the MEGA6 software and the neighbor-joining method with 1,000 bootstrap replications. The 15 CBLs from soybean are marked with green squares, while the 10 CBLs from Arabidopsis are not marked. Locus name of each gene is shown in brackets. The bar indicates the relative divergence of the sequences examined and bootstrap values are displayed next to the branch.
The soybean genome contains 50 CDPKs, 2 CCaMKs, and 13 CRKs. The detailed characteristics are shown in Supplemental Tables S4, S5. Most GmCDPKs contain four predicted EF-hands except for GmCDPK16, GmCDPK17, and GmCDPK35, which contain five, five, and one EF-hands, respectively (Supplemental Table S4). GmCDPK1, GmCDPK2, and GmCDPK3 have previously been defined as GmCDPKα, GmCDPKβ, and GmCDPKγ, respectively (Lee et al., 1998). Both the GmCCaMK1 and GmCCaMK2 contain three predicted EF-hands, and their amino acid identities to MtCCaMK are very high (87 and 88%). The CRKs possess the CaM-like domain with poorly conserved EF-hands (Hrabak et al., 2003). All GmCRKs were predicted to contain two degenerated EF-hands (Supplemental Table S5). In addition, 80% (40/50) of GmCDPKs and 77% (9/13) of GmCRKs have predicted myristoylation motifs at their N-termini (Supplemental Tables S4, S5), a feature observed in their orthologs from Arabidopsis (Hrabak et al., 2003). Myristoylation was reported to be an important mechanism for the membrane attachment of plant CDPKs (Martín and Busconi, 2000; Rutschmann et al., 2002). Figure 7 shows the unrooted phylogenetic tree for CDPKs/CRKs from soybean and Arabidopsis, and also CCaMKs from soybean, Medicago truncatula and Lotus japonicus, based on the comparisons of their amino acid sequences. These GmCDPK/CRK/CCaMK proteins are closely related to their orthologs in Arabidopsis and other plants.
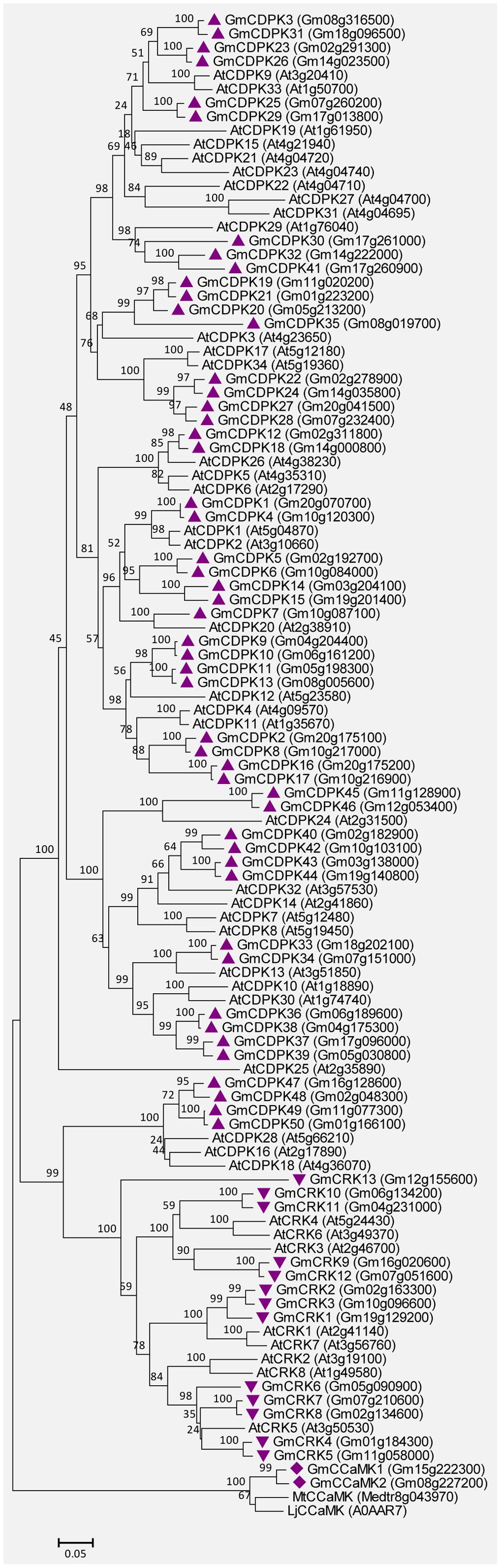
FIGURE 7. Unrooted phylogenetic tree of the GmCDPK, GmCRK, and GmCCaMK proteins. The alignment for the phylogenetic tree was performed with ClustalW using full-length amino acid sequences. The phylogenetic tree was created with the MEGA6 software and the neighbor-joining method with 1,000 bootstrap replications. All the 50 GmCDPKs are marked with triangles, 13 GmCRKs are marked with inverted triangles, and 2 GmCCaMKs are marked with diamonds, while proteins from Arabidopsis (34 CDPKs and 8 CRKs), and CCaMKs from Medicago truncatula (MtCCaMK) or Lotus japonicus (LjCCaMK) are not marked. Locus name of each gene is shown in brackets. The bar indicates the relative divergence of the sequences examined and bootstrap values are displayed next to the branch.
A total of 17 Rboh genes were found in the soybean genome. They were successively named GmRbohA-Q based on their homology to AtRbohD (Supplemental Table S6). The GmRboh proteins are 820–941 AA long and share 42–67% identity to AtRbohD. The intron numbers contained in GmRboh genes vary from 10 to 13 (Supplemental Table S6). Similar to their counterparts in Arabidopsis and other plants, all GmRboh proteins contain two putative EF-hand motifs. In addition, the evolutionary relationships between GmRbohs and 10 AtRboh members are closely related (Figure 8).
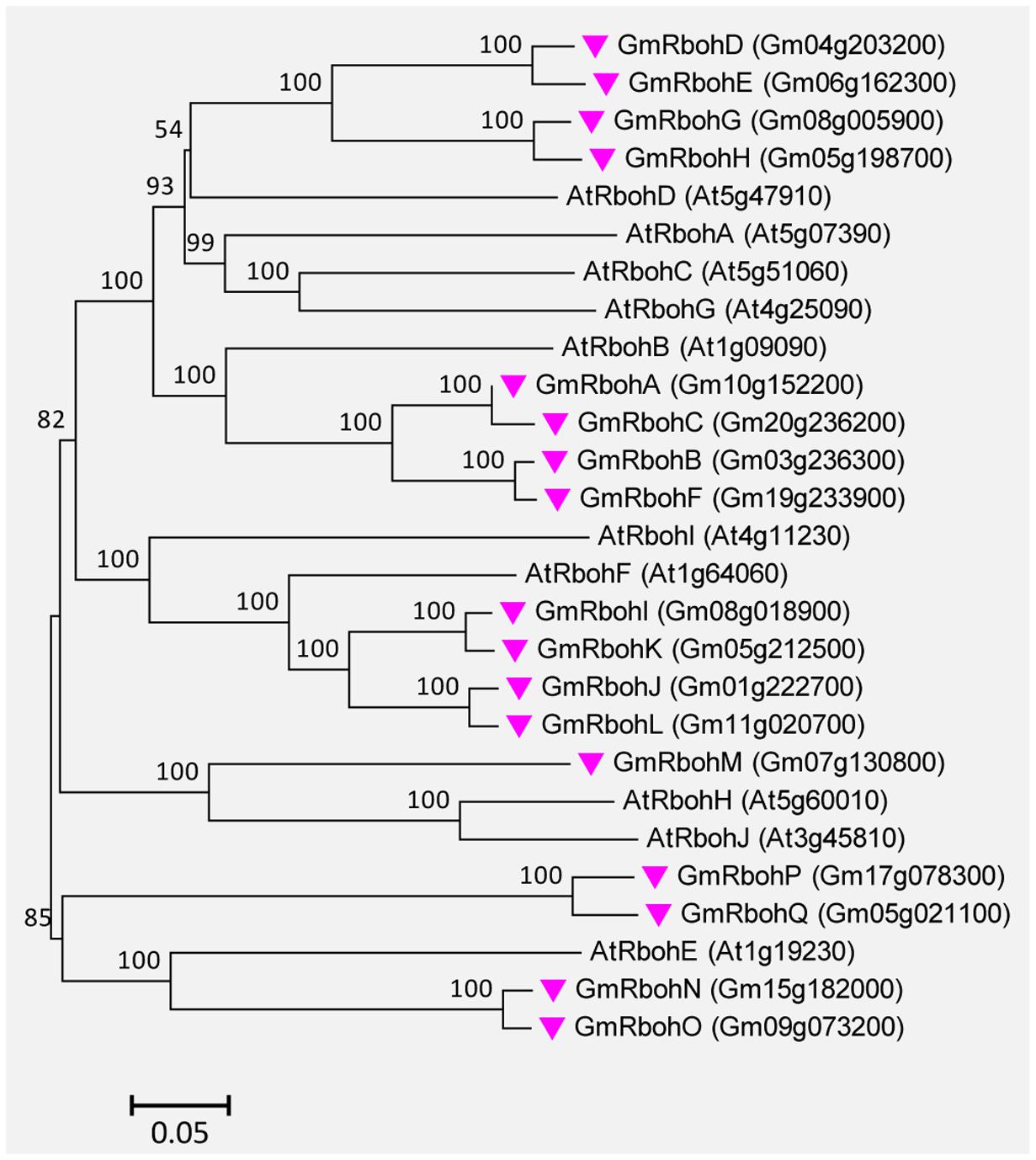
FIGURE 8. Unrooted phylogenetic tree of the Rboh proteins from soybean (G. max) and A. thaliana. The alignment for the phylogenetic tree was performed with ClustalW using full-length amino acid sequences. The phylogenetic tree was created with the MEGA6 software and the neighbor-joining method with 1,000 bootstrap replications. Seventeen GmRbohs are marked with pink inverted triangles, while 10 AtRboh proteins are not marked. Locus name of each gene is shown in brackets. The bar indicates the relative divergence of the sequences examined and bootstrap values are displayed next to the branch.
In addition, there are genes in soybean genome encoding 15 EF-hand proteins which are not belonging to CaM/CMLs, CBLs, CDPK/CRK/CCaMKs, and Rbohs, and they are named UEPs, (Supplemental Table S7). For example, UEP8 (Gm13g272000), UEP9 (Gm13g272100), and UEP10 (Gm13g272300) are tandemly duplicated genes encoding EF-hand proteins with putative sodium/calcium exchanger function (Figures 2, 4).
It is well known that transcription of a gene in plants is usually altered when an appropriate transcription factor recognizes and binds to a specific DNA motif (cis-element). Here we surveyed the presence of eight classes of cis-elements (Supplemental Table S8) related to hormonal signal and/or stress responses, in the -1500 bp promoter regions upstream to the predicted transcription start sites of these EF-hand genes (Williams et al., 1992; Abe et al., 1997; Ulmasov et al., 1997; Fujimoto et al., 2000; Sakuma et al., 2002; Yang and Poovaiah, 2002; Osakabe et al., 2014; Ruan et al., 2015). Of these 262 EF-hand genes, 39 contain dehydration- and cold-responsive element DRE/CRT, 78 contain ABA-responsive element ABRE, 112 contain auxin-responsive element AuxRE, 17 contain ethylene-responsive element GCC-box, 45 contain environmental response-related element G-box, 81 contain signal response-related element CG-box, 28 contain drought and ABA signaling-related element MYB2-BS, and 66 contain phosphate starvation response-related element P1BS (Supplemental Table S9). However, 32 genes did not contain any of these elements (Supplemental Table S9). Interestingly, more than one type of these cis-elements exist in the promoters of 142 EF-hand genes. For example, there are five types of cis-elements in the promoters of CML15, CML40, RbohB, RbohE, UEP3 (Gm06g162700), and UEP12 (Gm15g030100) (Supplemental Table S9). The enrichment of hormone-/stress-responsive cis-elements in the promoters of these EF-hand genes suggests that they are likely to be involved in plant responses to various hormone signals and stresses.
Transcriptome data derived from Illumina sequencing were used to assess the expression patterns of EF-hand genes in different tissues of soybean. The transcriptome atlas provided expression data for 55,616 putative soybean genes in eight types of tissues and organs, including root tips, roots, root hairs, nodules, leaves, shoot apical meristems, flowers and green pods (Libault et al., 2010b). The expression data of almost all EF-hand genes can be found in the transcriptome atlas, with the exception of five genes (CML56, CML65, CML120, CML144, and CRK6) (Figures 9, 10). However, 17 genes were found to have no expression in any of these tissues; these genes are CML11, CML15, CML16, CML23, CML42, CML47, CML54, CML60, CML70, CML72, CML81, CML86, CML130, CML139, CML142, CBL2, and CBL3. These genes could be pseudogenes or could be expressed only under specific developmental stages or environmental conditions which were not met in these studies. As shown in the heat maps, some genes were ubiquitous in various tissues, while some genes are tissue-specific (Figures 9, 10). For example, CML6, CML18, CML26, CML48, CML61, CBL6, CBL11, CDPK18, CDPK19, and CDPK20 were constitutive expressed in various tissues; CML20, CML30, CML71, CML78, CML87, CML113, CML118, and CDPK28 were specifically expressed in flower tissue; CaM2, CML2, CML9, CML19, CML24, CML55, CML77, CML137, CDPK6, CDPK14, CCaMK1, CCaMK2, and RbohN were preferentially expressed in roots/root tips/root hairs (Figures 9, 10).
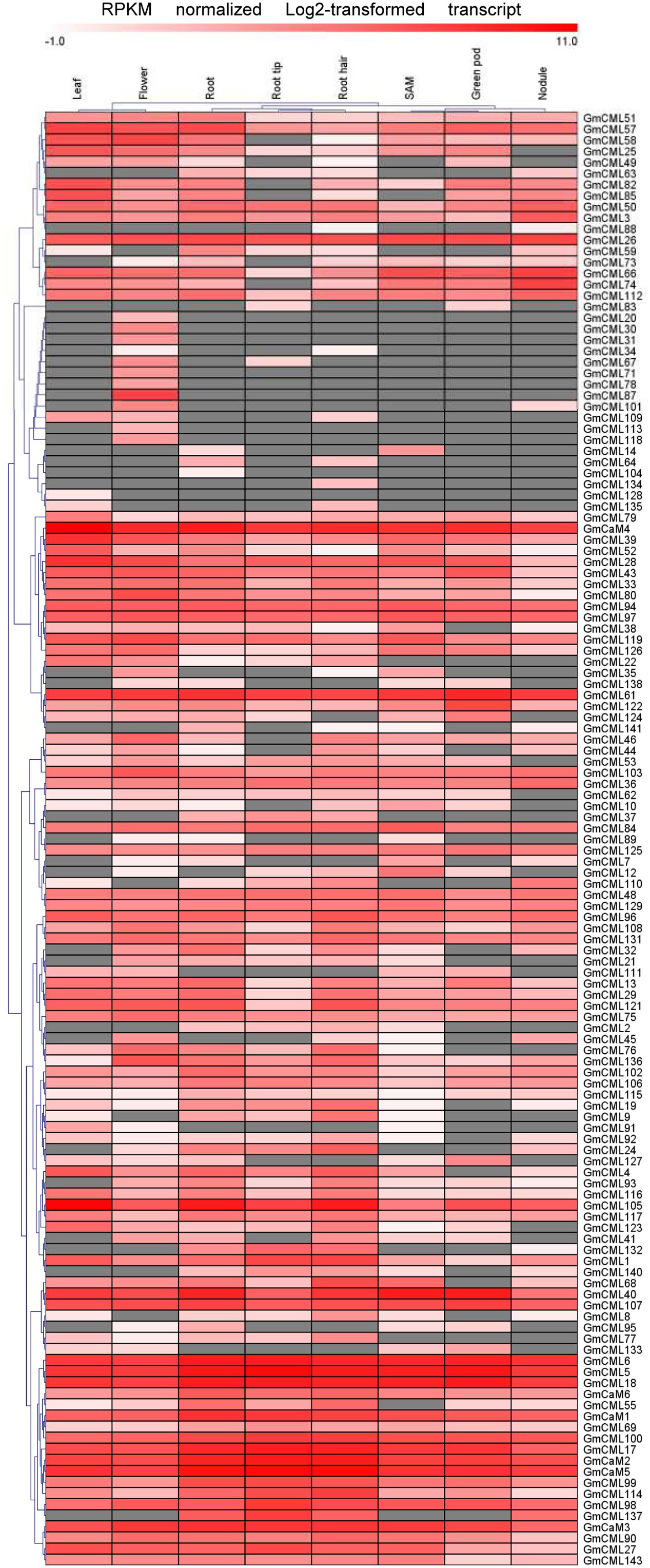
FIGURE 9. Heat map representation for tissue-specific expression patterns of the predicted GmCaM/CML genes according to Illumina transcriptome data. The Reads/Kb/Million (RPKM) normalized log2 transformed counts were visualized in the heat map. The red colors indicate expression intensity, gray color indicates no expression.
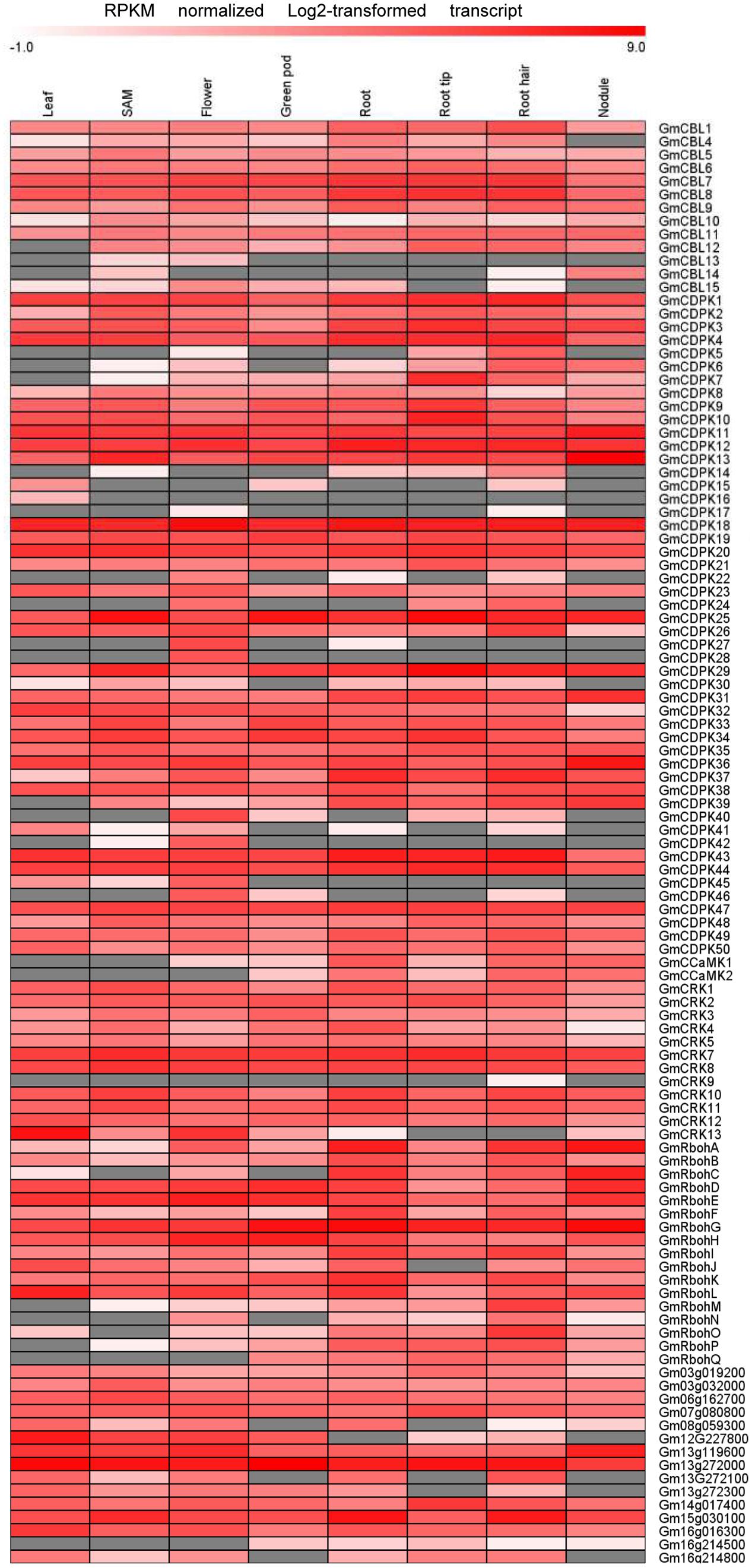
FIGURE 10. Heat map representation for tissue-specific expression patterns of predicted EF-hand protein-coding genes with the exception of GmCaM/CMLs according to Illumina transcriptome data. The RPKM normalized log2 transformed counts were visualized in the heat map. The red colors indicate expression intensity, gray color indicates no expression detected.
In order to investigate whether the EF-hand genes are responsive to environmental stresses, we retrieved the published soybean microarray and/or deep sequencing data reflecting soybean responses to various stresses including cold, drought, flooding, phosphate deficiency, and Bradyrhizobium japonicum inoculation (Libault et al., 2010a; Le et al., 2012; Maruyama et al., 2012; Chen et al., 2016; Zeng et al., 2016b). Of all the EF-hand genes, 113 (43.1%) genes were shown to be differentially expressed under one or multiple stresses (Figure 11). For example, CML59, CML102, CML117, and RbohB were induced by cold, drought, and flooding; CML109 and UEP6 (Gm12g227800) were repressed by cold and flooding; CML82 and UEP11 (Gm14g017400) were induced by cold but repressed by flooding; CML28, CDPK45, and UEP10 (Gm13g272300) were only responsive to cold stress.
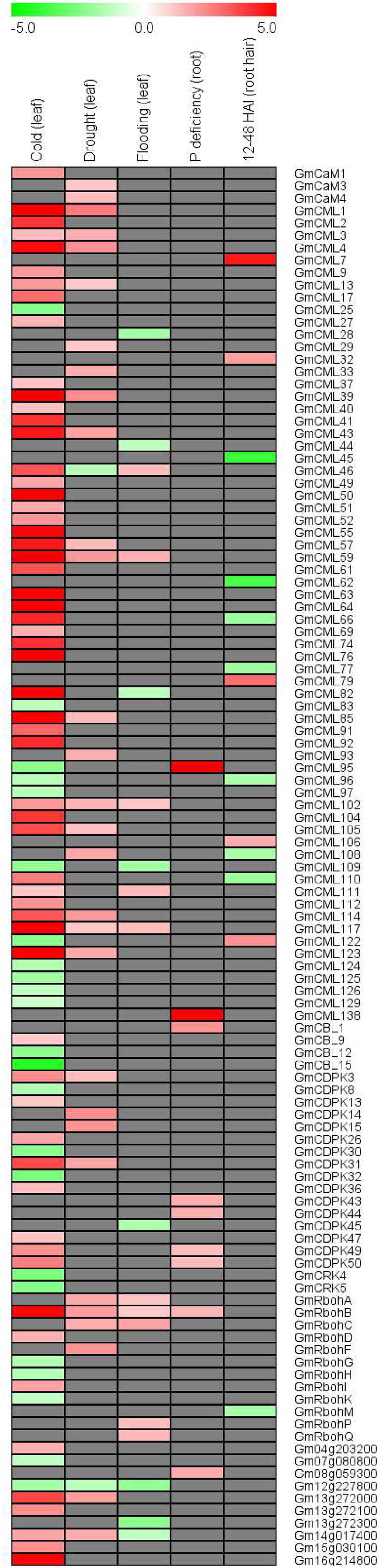
FIGURE 11. Heatmap representation for expression profiles of EF-hand protein-encoding genes in response to cold, drought, flooding, phosphorus deficiency, and Bradyrhizobium japonicum inoculation (24–48 HAI). The intensities of the color represent the relative magnitude of fold changes in log2 values according to microarray or high-throughput sequencing data. Red color indicates induction, green color indicates repression, gray color means there is no significant expressional change.
In addition, we also selected 14 EF-hand genes to confirm their transcriptional responses to environmental stresses including salt, dehydration, phosphate deficiency, iron deficiency, and zinc deficiency by qRT-PCR (Figure 12). Nearly all of these genes were responsive to diverse stresses. Most of them were induced by salt stress, but were repressed by various nutrient deficiencies, and eight genes of them were responsive to PEG treatment (dehydration), four were induced (CML82, CML122, CRK8, and CBL15) and four [CML1, CML39, CML109, and UEP12 (Gm15g030100)] were repressed (Figure 12).
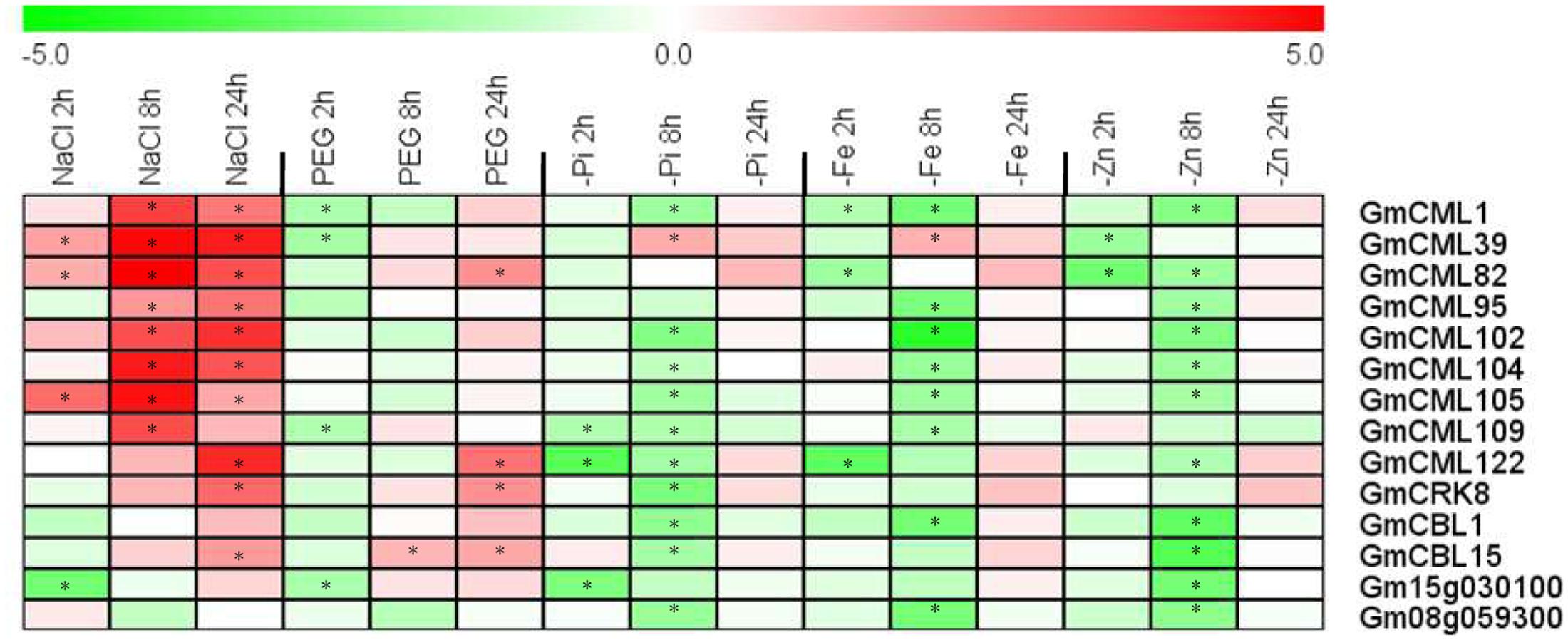
FIGURE 12. Heat map representation for expression profiles of some selected EF-hand genes in response to stresses. The expression of EF-hand genes were analyzed by qRT-PCR in roots of 12-day-old seedling, after treatment with 200 mM NaCl (salt), 10% PEG 6000, phosphate deficiency (-Pi), iron deficiency (-Fe), and zinc deficiency (-Zn) for 2, 8, and 24 h. The intensities of the color represent the relative magnitude of fold changes (treatment/control) in log2 values of three technical replicates. The asterisks indicate an absolute fold change ≥ 2 and P-value < 0.05 by Student’s t-test. Red color indicates induction, green color indicates repression.
Based on the gene expression analysis, we further examined four representative soybean CMLs (CML1, CML13, CML39, and CML95) to confirm their Ca2+-binding ability by using SDS–PAGE mobility-shift assay. The accelerated electrophoretic migration of CaM/CMLs in the presence of Ca2+ is a well-documented phenomenon (Garrigos et al., 1991; Vanderbeld and Snedden, 2007). As shown in Figure 13, like the conserved CaM, the CML1, CML13, and CML39 exhibit the characteristic Ca2+ shift, and CML95 exhibit a small Ca2+ shift. Interestingly, a detectable increased shift in the mobility of CML39 was observed in the presence of MgCl2. We used IPD3, a non-Ca2+-binding protein (Routray et al., 2013) for negative control, and no shift was observed in the presence of CaCl2 or MgCl2 (Supplemental Figure S1). Mg2+ binding to some particular EF-hand proteins was reported for affecting EF-hand protein stability and functions (Wingard et al., 2005; Gifford et al., 2007; Vanderbeld and Snedden, 2007).
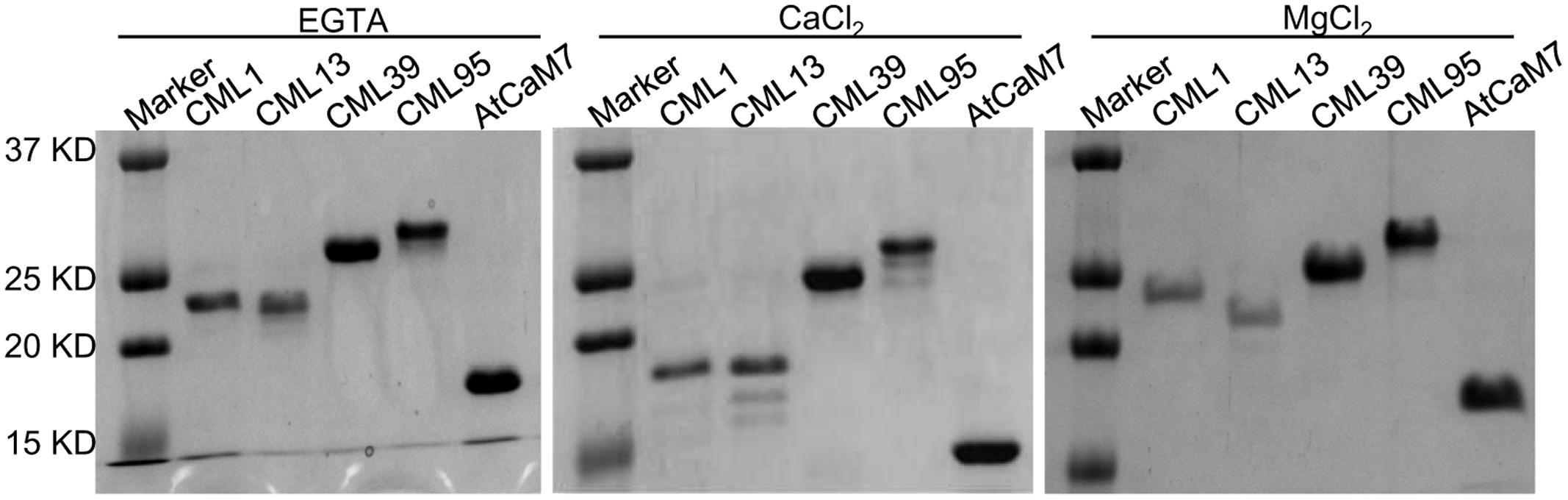
FIGURE 13. Effect of Ca2+ on the electrophoretic mobility of GmCML1, GmCML13, GmCML39, and GmCML95 proteins. Samples (1–2 μg) of purified, recombinant AtCaM7 (positive control), GmCML1, GmCML13, GmCML39, and GmCML95 containing 1.0 mM CaCl2, 1.0 mM EGTA, or 1.0 mM MgCl2 were separated on SDS–PAGE gels supplemented with 1.0 mM CaCl2, 1.0 mM EGTA, or 1.0 mM MgCl2, respectively, and stained with Coomassie Brilliant Blue. The positions of the molecular weight markers (kDa) are indicated on the left.
Ca2+ mediated signaling is involved in various developmental processes and responses to a variety of abiotic and biotic stresses. The Ca2+ binding EF-hand motif is highly conserved in most of the Ca2+ sensors. CaMs/CMLs, CBLs, and CDPKs are the major subfamilies of EF-hand Ca2+ sensors (Weinl and Kudla, 2009; DeFalco et al., 2010; Poovaiah et al., 2013). Although subfamilies of Ca2+ sensors (e.g., CaM/CML, CBL, and CDPK) have been analyzed in model plants and some major crops, no detailed information about these Ca2+ sensors in soybean is systemically reported so far (Hrabak et al., 2003; McCormack and Braam, 2003; Kolukisaoglu et al., 2004; Asano et al., 2005; Boonburapong and Buaboocha, 2007; Chen et al., 2013; Kong et al., 2013; Zhao et al., 2013; Kleist et al., 2014; Meena et al., 2015; Munir et al., 2016). Using domain prediction programs and BLAST searches against known EF-hand proteins (e.g., AtCaM2, AtCBL1, AtCDPK1, AtCRK1, and AtRbohD) at the genomic level in soybean, at least 262 genes encoding proteins with varying number of EF-hand motifs were identified (Figures 1–3). The EF-hand genes in soybean include 6 CaM genes, 144 CML genes, 15 CBL genes, 50 CDPK genes, 13 CRK genes, 2 CCaMK genes, 17 Rboh genes, and 15 UEP genes (Supplemental Tables S1–S7). EF-hand motifs tend to occur in pairs, which can increase stability as well as affinity of binding between the EF-hand proteins and Ca2+ (Gifford et al., 2007). Consistent with the EF-hand family members in other plants, the majority of the soybean EF-hand proteins (71.8%) have pairs of EF-hand motifs (two, four, or six) (Figure 1) (Day et al., 2002; McCormack and Braam, 2003; Boonburapong and Buaboocha, 2007; Munir et al., 2016).
The number of total EF-hand proteins in Arabidopsis and rice plants were previously found to be 250 and 243, respectively (Day et al., 2002; Boonburapong and Buaboocha, 2007). The number of EF-hand proteins in soybean is similar to that found in Arabidopsis and rice. However, the number of CMLs in soybean (144) is much more than that in Arabidopsis (50), rice (32), and tomato (52) (McCormack and Braam, 2003; Boonburapong and Buaboocha, 2007; Munir et al., 2016). In addition, the number of CBLs, CDPKs, CRKs, and Rbohs in soybean is at least 1.5-folds as much as that in Arabidopsis and soybean (Supplemental Table S10). The large size of the CML/CBL/CDPK/CRK/Rboh gene families in soybean could be attributed to the whole-genome duplication events occurred approximately 59 and 13 million years ago (Schmutz et al., 2010). However, the EF-hand proteins other than CaMs/CMLs/CBLs/CDPKs/CRKs/CCaMKs in soybean are much less than that in Arabidopsis and rice plants (Supplemental Table S10). This may be caused by reasons that some loci encoding putative EF-hand proteins are not annotated in the current version of soybean genome (version 2.0), or the composition of different subfamilies of EF-hand Ca2+ sensor may be a little different between legume and non-legume plants.
The expression patterns of EF-hand genes in different tissues revealed that these genes were diversely expressed in various tissues of soybean (Figures 9, 10). Ca2+ sensor subfamilies, such as CaM/CMLs and CDPKs also show tissue and developmentally specific expression patterns (McCormack et al., 2005; Kong et al., 2013; Munir et al., 2016). It is clear that Ca2+ signaling is highly complex and plays an important role during plant growth and development. For example, Ca2+/CaM signaling is critical for brassinosteroid biosynthesis and plant growth by regulating the function of DWARF1 (Du and Poovaiah, 2005); Arabidopsis CML42 regulates trichome branching by interacting with KIC (kinesin interacting Ca2+-binding protein) (Dobney et al., 2009); CBL-interacting protein kinase CIPK6 is involved in root development (Tripathi et al., 2009); CDPK28 functions as a regulatory component controlling stem elongation and vascular development (Matschi et al., 2013); Arabidopsis RbohC regulates root cell expansion through the activation of Ca2+ channels (Foreman et al., 2003). Ca2+ signaling-components, such as DMI1 (does not make infections 1), Ca2+-dependent ATPase MCA8, the cyclic nucleotide-gated channels (CNGCs) and CCaMK are required for the establishment of root symbiosis in legumes (Oldroyd, 2013; Poovaiah et al., 2013; Charpentier et al., 2016). Interestingly, some EF-protein-coding genes were preferentially expressed in root nodule of soybean, such as CML45, CML63, CML73, CML110, CDPK6, CDPK14, CCaMK1, and CCaMK2 (Figures 9, 10). Notably, CML45 and CML110 were responsive to the inoculation of rhizobia (B. japonicum) (Figure 11). Whether these genes are possibly associated with symbiosis establishment or nodule development should deserve further researches.
A variety of stimuli including hormonal signals, abiotic and biotic stresses, regulate the expression of diverse subfamilies of EF-hand-containing proteins (McCormack et al., 2005; Boudsocq and Sheen, 2013; Chen et al., 2013; Kong et al., 2013; Zhao et al., 2013; Meena et al., 2015; Mohanta et al., 2015; Munir et al., 2016). Nowadays, a lot of EF-hand proteins have been documented to mediate plant response and tolerance to various environmental stresses (Weinl and Kudla, 2009; Xi et al., 2012; Bender and Snedden, 2013; Boudsocq and Sheen, 2013; Zhang et al., 2014; Zeng et al., 2015). For example, Arabidopsis CaM3 is involved in heat shock signal transduction because cam3 mutant was more sensitive to heat stress (Zhang et al., 2009); Arabidopsis CML9 knockout mutant plants express more tolerance to salinity and drought (Magnan et al., 2008); overexpression of GmCaM4 confers soybean enhanced resistances to pathogens and salt stress (Rao et al., 2014); overexpression of rice CML4 enhances drought tolerance in transgenic rice (Yin et al., 2015); Arabidopsis cml42 mutant plants are more resistant to herbivory than the wild type plants (Vadassery et al., 2012); Arabidopsis CBL1, CBL2, CBL3, CBL4, CBL9, and CBL10 are involved in cellular ion homeostasis regulation (e.g., Na+, K+, Mg2+, and NO3-) (Weinl and Kudla, 2009; Tang et al., 2015); overexpression of Arabidopsis CBL5 and soybean CBL1 improves salt and drought tolerance in transgenic Arabidopsis plants (Cheong et al., 2010; Li et al., 2012); Arabidopsis CDPK8 functions in ABA-mediated stomatal regulation in response to drought by phosphorylating CATALASE3 (Zou et al., 2015); rice CPK21 positively regulates salt stress tolerance (Asano et al., 2011); overexpression of a Medicago EF-hand protein gene MtCaMP1 enhances drought and salt stress tolerance (Wang et al., 2013). In the present study, 43.1% of all the EF-hand genes were found to be differentially expressed under one or multiple stresses including cold, drought, salinity, flooding, rhizobia inoculation, and nutrient deficiencies of phosphate, iron, or zinc (Figures 11, 12). It is notable that most of these genes (85%) contain at least one type of stress-related cis-elements in their promoters (Supplemental Table S9). In this study, we did not analyzed the physiological role of any soybean EF-hand proteins. But in the future, researches through the combination of biochemical, molecular, and genetic approaches are required to dissect the exact physiological roles of these EF-hand proteins considering the important functions of their counterparts in model plants and their responsiveness to one or multiple stresses.
In this study, a total of at least 262 genes encoding proteins with varying number of EF-hand motifs were identified in soybean genome, including 6 CaM genes, 144 CML genes, 15 CBL genes, 50 CDPK genes, 13 CRK genes, 2 CCaMK genes, 17 Rboh genes, and 15 UEP genes. Most of these genes (87.8%) contain one or multiple hormonal signal and/or stress-responsive cis-elements in the -1500 bp promoter regions. Expression profiling revealed that these EF-hand genes were broadly expressed in different organs of soybean. Expression pattern analyses also revealed that nearly half of these genes (43.1%) could be induced or repressed under various environmental or nutritional stresses, indicating their potential role in stress responses. Further in-depth functional characterization will enhance our understanding of Ca2+-mediated signaling underling plant responses to environmental and nutritional stresses and facilitate the development of stress resistant crops.
HZ conceived and designed the study, and wrote the manuscript. HZ, YxZ, and XZ performed the bioinformatic analysis and the experiments. EP and YyZ participated in the data analysis. All authors read and approved the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Prof. Liqun Du (Hangzhou Normal University) for his critical reading and revision of the manuscript. This work was supported by grants from the National Natural Science Foundation of China (31201679 and U1130304) and Zhejiang Provincial Natural Science Foundation of China (LY15C020006).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00877/full#supplementary-material
Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of Arabidopsis MYC and MYB homologs in drought-and abscisic acid-regulated gene expression. Plant Cell 9, 1859–1868. doi: 10.1105/tpc.9.10.1859
Asano, T., Hakata, M., Nakamura, H., Aoki, N., Komatsu, S., Ichikawa, H., et al. (2011). Functional characterisation of OsCPK21, a calcium-dependent protein kinase that confers salt tolerance in rice. Plant Mol. Biol. 75, 179–191. doi: 10.1007/s11103-010-9717-1
Asano, T., Tanaka, N., Yang, G., Hayashi, N., and Komatsu, S. (2005). Genome-wide identification of the rice calcium-dependent protein kinase and its closely related kinase gene families: comprehensive analysis of the CDPKs gene family in rice. Plant Cell Physiol. 46, 356–366. doi: 10.1093/pcp/pci035
Batistic, O., and Kudla, J. (2012). Analysis of calcium signaling pathways in plants. Biochim. Biophys. Acta 1820, 1283–1293. doi: 10.1016/j.bbagen.2011.10.012
Bender, K. W., and Snedden, W. A. (2013). Calmodulin-related proteins step out from the shadow of their namesake. Plant Physiol. 163, 486–495. doi: 10.1104/pp.113.221069
Boonburapong, B., and Buaboocha, T. (2007). Genome-wide identification and analyses of the rice calmodulin and related potential calcium sensor proteins. BMC Plant Biol. 7:4. doi: 10.1186/1471-2229-7-4
Boudsocq, M., and Sheen, J. (2013). CDPKs in immune and stress signaling. Trends Plant Sci. 18, 30–40. doi: 10.1016/j.tplants.2012.08.008
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Charpentier, M., Sun, J., Vaz Martins, T., Radhakrishnan, G. V., Findlay, K., Soumpourou, E., et al. (2016). Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352, 1102–1105. doi: 10.1126/science.aae0109
Chen, F., Fasoli, M., Tornielli, G. B., Dal Santo, S., Pezzotti, M., Zhang, L., et al. (2013). The evolutionary history and diverse physiological roles of the grapevine calcium-dependent protein kinase gene family. PLoS ONE 8:e80818. doi: 10.1371/journal.pone.0080818
Chen, W., Yao, Q., Patil, G. B., Agarwal, G., Deshmukh, R. K., Lin, L., et al. (2016). Identification and comparative analysis of differential gene expression in soybean leaf tissue under drought and flooding stress revealed by RNA-Seq. Front. Plant Sci. 7:1044. doi: 10.3389/fpls.2016.01044
Cheong, Y. H., Sung, S. J., Kim, B. G., Pandey, G. K., Cho, J. S., Kim, K. N., et al. (2010). Constitutive overexpression of the calcium sensor CBL5 confers osmotic or drought stress tolerance in Arabidopsis. Mol. Cells 29, 159–165. doi: 10.1007/s10059-010-0025-z
Day, I. S., Reddy, V. S., Ali, G. S., and Reddy, A. S. (2002). Analysis of EF-hand-containing proteins in Arabidopsis. Genome Biol. 3:0056. doi: 10.1186/gb-2002-3-10-research0056
DeFalco, T. A., Bender, K. W., and Snedden, W. A. (2010). Breaking the code: Ca2+ sensors in plant signalling. Biochem. J. 425, 27–40. doi: 10.1042/BJ20091147
Deshmukh, R., Sonah, H., Patil, G., Chen, W., Prince, S., Mutava, R., et al. (2014). Integrating omic approaches for abiotic stress tolerance in soybean. Front. Plant Sci. 5:244. doi: 10.3389/fpls.2014.00244
Dobney, S., Chiasson, D., Lam, P., Smith, S. P., and Snedden, W. A. (2009). The calmodulin-related calcium sensor CML42 plays a role in trichome branching. J. Biol. Chem. 284, 31647–31657. doi: 10.1074/jbc.M109.056770
Du, L., and Poovaiah, B. W. (2005). Ca2+/calmodulin is critical for brassinosteroid biosynthesis and plant growth. Nature 437, 741–745. doi: 10.1038/nature03973
Foreman, J., Demidchik, V., Bothwell, J. H. F., Mylona, P., Miedema, H., Torres, M. A., et al. (2003). Reactive oxygen species produced by NADPH oxidase regulate plant cell growth. Nature 422, 442–446. doi: 10.1038/nature01485
Fujimoto, S. Y., Ohta, M., Usui, A., Shinshi, H., and Ohme-Takagi, M. (2000). Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box–mediated gene expression. Plant Cell 12, 393–404. doi: 10.1105/tpc.12.3.393
Garrigos, M., Deschamps, S., Viel, A., Lund, S., Champeil, P., Møller, J. V., et al. (1991). Detection of Ca2+-binding proteins by electrophoretic migration in the presence of Ca2+ combined with 45Ca2+ overlay of protein blots. Anal. Biochem. 194, 82–88. doi: 10.1016/0003-2697(91)90154-L
Gifford, J. L., Walsh, M. P., and Vogel, H. J. (2007). Structures and metal-ion-binding properties of the Ca2+-binding helix-loop-helix EF-hand motifs. Biochem. J. 405, 199–221. doi: 10.1042/BJ20070255
Hamel, L. P., Sheen, J., and Séguin, A. (2014). Ancient signals: comparative genomics of green plant CDPKs. Trends Plant Sci. 19, 79–89. doi: 10.1016/j.tplants.2013.10.009
Hrabak, E. M., Chan, C. W., Gribskov, M., Harper, J. F., Choi, J. H., Halford, N., et al. (2003). The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 132, 666–680. doi: 10.1104/pp.102.011999
Hunter, S., Apweiler, R., Attwood, T. K., Bairoch, A., Bateman, A., Binns, D., et al. (2009). InterPro: the integrative protein signature database. Nucleic Acids Res. 37, D211–D215. doi: 10.1093/nar/gkn785
Kleist, T. J., Spencley, A. L., and Luan, S. (2014). Comparative phylogenomics of the CBL-CIPK calcium-decoding network in the moss Physcomitrella, Arabidopsis, and other green lineages. Front. Plant Sci. 5:187. doi: 10.3389/fpls.2014.00187
Kolukisaoglu,Ü, Weinl, S., Blazevic, D., Batistic, O., and Kudla, J. (2004). Calcium sensors and their interacting protein kinases: genomics of the Arabidopsis and rice CBL-CIPK signaling networks. Plant Physiol. 134, 43–58. doi: 10.1104/pp.103.033068
Kong, X., Lv, W., Jiang, S., Zhang, D., Cai, G., Pan, J., et al. (2013). Genome-wide identification and expression analysis of calcium-dependent protein kinase in maize. BMC Genomics 14:433. doi: 10.1186/1471-2164-14-433
Kudla, J., Batistic, O., and Hashimoto, K. (2010). Calcium signals: the lead currency of plant information processing. Plant Cell 22, 541–563. doi: 10.1105/tpc.109.072686
Le, D. T., Nishiyama, R., Watanabe, Y., Tanaka, M., Seki, M., Ham, L. H., et al. (2012). Differential gene expression in soybean leaf tissues at late developmental stages under drought stress revealed by genome-wide transcriptome analysis. PLoS ONE 7:e49522. doi: 10.1371/journal.pone.0049522
Lee, J. Y., Yoo, B. C., and Harmon, A. C. (1998). Kinetic and calcium-binding properties of three calcium-dependent protein kinase isoenzymes from soybean. Biochemistry 37, 6801–6809. doi: 10.1021/bi980062q
Lee, S. H., Kim, J. C., Lee, M. S., Heo, W. D., Seo, H. Y., Yoon, H. W., et al. (1995). Identification of a novel divergent calmodulin isoform from soybean which has differential ability to activate calmodulin-dependent enzymes. J. Biol. Chem. 270, 21806–21812. doi: 10.1074/jbc.270.37.21806
Li, Z. Y., Xu, Z. S., He, G. Y., Yang, G. X., Chen, M., Li, L. C., et al. (2012). Overexpression of soybean GmCBL1 enhances abiotic stress tolerance and promotes hypocotyl elongation in Arabidopsis. Biochem. Biophys. Res. Commun. 427, 731–736. doi: 10.1016/j.bbrc.2012.09.128
Libault, M., Farmer, A., Brechenmacher, L., Drnevich, J., Langley, R. J., Bilgin, D. D., et al. (2010a). Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 152, 541–552. doi: 10.1104/pp.109.148379
Libault, M., Farmer, A., Joshi, T., Takahashi, K., Langley, R. J., Franklin, L. D., et al. (2010b). An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 63, 86–99. doi: 10.1111/j.1365-313X.2010.04222.x
Luan, S., Kudla, J., Rodriguez-Concepcion, M., Yalovsky, S., and Gruissem, W. (2002). Calmodulins and calcineurin B-like proteins: calcium sensors for specific signal response coupling in plants. Plant Cell 14(Suppl.), S389–S400.
Magnan, F., Ranty, B., Charpenteau, M., Sotta, B., Galaud, J. P., and Aldon, D. (2008). Mutations in AtCML9, a calmodulin-like protein from Arabidopsis thaliana, alter plant responses to abiotic stress and abscisic acid. Plant J. 56, 575–589. doi: 10.1111/j.1365-313X.2008.03622.x
Martín, M. L., and Busconi, L. (2000). Membrane localization of a rice calcium-dependent protein kinase (CDPK) is mediated by myristoylation and palmitoylation. Plant J. 24, 429–435. doi: 10.1046/j.1365-313x.2000.00889.x
Maruyama, K., Todaka, D., Mizoi, J., Yoshida, T., Kidokoro, S., Matsukura, S., et al. (2012). Identification of cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in Arabidopsis, rice, and soybean. DNA Res. 19, 37–49. doi: 10.1093/dnares/dsr040
Matschi, S., Werner, S., Schulze, W. X., Legen, J., Hilger, H. H., and Romeis, T. (2013). Function of calcium-dependent protein kinase CPK28 of Arabidopsis thaliana in plant stem elongation and vascular development. Plant J. 73, 883–896. doi: 10.1111/tpj.12090
McAinsh, M. R., and Pittman, J. K. (2009). Shaping the calcium signature. New Phytol. 181, 275–294. doi: 10.1111/j.1469-8137.2008.02682.x
McCormack, E., and Braam, J. (2003). Calmodulins and related potential calcium sensors of Arabidopsis. New Phytol. 159, 585–598. doi: 10.1046/j.1469-8137.2003.00845.x
McCormack, E., Tsai, Y. C., and Braam, J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389. doi: 10.1016/j.tplants.2005.07.001
Medina-Rivera, A., Defrance, M., Sand, O., Herrmann, C., Castro-Mondragon, J. A., Delerce, J., et al. (2015). RSAT 2015: regulatory sequence analysis tools. Nucleic Acids Res. 43, W50–W56. doi: 10.1093/nar/gkv362
Meena, M. K., Ghawana, S., Sardar, A., Dwivedi, V., Khandal, H., Roy, R., et al. (2015). Investigation of genes encoding calcineurin B-like protein family in legumes and their expression analyses in chickpea (Cicer arietinum L.). PLoS ONE 10:e0123640. doi: 10.1371/journal.pone.0123640
Mohanta, T. K., Mohanta, N., Mohanta, Y. K., Parida, P., and Bae, H. (2015). Genome-wide identification of calcineurin B-Like (CBL) gene family of plants reveals novel conserved motifs and evolutionary aspects in calcium signaling events. BMC Plant Biol. 15:189. doi: 10.1186/s12870-015-0543-0
Munir, S., Khan, M. R., Song, J., Munir, S., Zhang, Y., Ye, Z., et al. (2016). Genome-wide identification, characterization and expression analysis of calmodulin-like (CML) proteins in tomato (Solanum lycopersicum). Plant Physiol. Biochem. 102, 167–179. doi: 10.1016/j.plaphy.2016.02.020
Oldroyd, G. E. D. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Osakabe, Y., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. S. (2014). ABA control of plant macroelement membrane transport systems in response to water deficit and high salinity. New Phytol. 202, 35–49. doi: 10.1111/nph.12613
Poovaiah, B. W., Du, L., Wang, H., and Yang, T. (2013). Recent advances in calcium/calmodulin-mediated signaling with an emphasis on plant-microbe interactions. Plant Physiol. 163, 531–542. doi: 10.1104/pp.113.220780
Rao, S. S., El-Habbak, M. H., Havens, W. M., Singh, A., Zheng, D., Vaughn, L., et al. (2014). Overexpression of GmCaM4 in soybean enhances resistance to pathogens and tolerance to salt stress. Mol. Plant Pathol. 15, 145–160. doi: 10.1111/mpp.12075
Routray, P., Miller, J. B., Du, L., Oldroyd, G., and Poovaiah, B. W. (2013). Phosphorylation of S344 in the calmodulin-binding domain negatively affects CCaMK function during bacterial and fungal symbioses. Plant J. 76, 287–296. doi: 10.1111/tpj.12288
Ruan, W., Guo, M., Cai, L., Hu, H., Li, C., Liu, Y., et al. (2015). Genetic manipulation of a high-affinity PHR1 target cis-element to improve phosphorous uptake in Oryza sativa L. Plant Mol. Biol. 87, 429–440. doi: 10.1007/s11103-015-0289-y
Rutschmann, F., Stalder, U., Piotrowski, M., Oecking, C., and Schaller, A. (2002). LeCPK1, a calcium-dependent protein kinase from tomato. Plasma membrane targeting and biochemical characterization. Plant Physiol. 129, 156–168. doi: 10.1104/pp.000869
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-Binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Sanders, D., Pelloux, J., Brownlee, C., and Harper, J. F. (2002). Calcium at the crossroads of signaling. Plant Cell 14(Suppl.), S401–S417.
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30,2725–2729. doi: 10.1093/molbev/mst197
Tang, R. J., Zhao, F. G., Garcia, V. J., Kleist, T. J., Yang, L., Zhang, H. X., et al. (2015). Tonoplast CBL-CIPK calcium signaling network regulates magnesium homeostasis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 112, 3134–3139. doi: 10.1073/pnas.1420944112
Tran, L. S. P., and Mochida, K. (2010). Functional genomics of soybean for improvement of productivity in adverse conditions. Funct. Integr. Genomics 10, 447–462. doi: 10.1007/s10142-010-0178-z
Tripathi, V., Parasuraman, B., Laxmi, A., and Chattopadhyay, D. (2009). CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J. 58, 778–790. doi: 10.1111/j.1365-313X.2009.03812.x
Ulmasov, T., Hagen, G., and Guilfoyle, T. J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. doi: 10.1126/science.276.5320.1865
Vadassery, J., Reichelt, M., Hause, B., Gershenzon, J., Boland, W., and Mithofer, A. (2012). CML42-mediated calcium signaling coordinates responses to Spodoptera herbivory and abiotic stresses in Arabidopsis. Plant Physiol. 159, 1159–1175. doi: 10.1104/pp.112.198150
Vanderbeld, B., and Snedden, W. A. (2007). Developmental and stimulus-induced expression patterns of Arabidopsis calmodulin-like genes CML37, CML38 and CML39. Plant Mol. Biol. 64, 683–697. doi: 10.1007/s11103-007-9189-0
Wang, G., Zeng, H., Hu, X., Zhu, Y., Chen, Y., Shen, C., et al. (2015). Identification and expression analyses of calmodulin-binding transcription activator genes in soybean. Plant Soil 386, 205–221. doi: 10.1007/s11104-014-2267-6
Wang, T. Z., Zhang, J. L., Tian, Q. Y., Zhao, M. G., and Zhang, W. H. (2013). A Medicago truncatula EF-hand family gene, MtCaMP1, is involved in drought and salt stress tolerance. PLoS ONE 8:e58952. doi: 10.1371/journal.pone.0058952
Webb, A. A., McAinsh, M. R., Taylor, J. E., and Hetherington, A. M. (1996). “Calciumions as intracellular second messengers in higher plants,” in Advances in Botanical Research, ed. J. A. Callow (Waltham, MA: Academic Press), 45–96.
Weinl, S., and Kudla, J. (2009). The CBL-CIPK Ca2+-decoding signaling network: function and perspectives. New Phytol. 184, 517–528. doi: 10.1111/j.1469-8137.2009.02938.x
Williams, M. E., Foster, R., and Chua, N. H. (1992). Sequences flanking the hexameric G-box core CACGTG affect the specificity of protein binding. Plant Cell 4, 485–496. doi: 10.1105/tpc.4.4.485
Wingard, J. N., Chan, J., Bosanac, I., Haeseleer, F., Palczewski, K., Ikura, M., et al. (2005). Structural analysis of Mg2+ and Ca2+ binding to CaBP1, a neuron-specific regulator of calcium channels. J. Biol. Chem. 280, 37461–37470. doi: 10.1074/jbc.M508541200
Xi, J., Qiu, Y., Du, L., and Poovaiah, B. W. (2012). Plant-specific trihelix transcription factor AtGT2L interacts with calcium/calmodulin and responds to cold and salt stresses. Plant Sci. 185, 274–280. doi: 10.1016/j.plantsci.2011.11.013
Yang, T., and Poovaiah, B. W. (2002). A calmodulin-binding/CGCG box DNA-binding protein family involved in multiple signaling pathways in plants. J. Biol. Chem. 277, 45049–45058. doi: 10.1074/jbc.M207941200
Yin, X. M., Huang, L. F., Zhang, X., Wang, M. L., Xu, G. Y., and Xia, X. J. (2015). OsCML4 improves drought tolerance through scavenging of reactive oxygen species in rice. J. Plant Biol. 58, 68–73. doi: 10.1016/j.plantsci.2012.08.003
Zeng, H. Q., Di, T. J., Zhu, Y. Y., and Subbarao, G. V. (2016a). Transcriptional response of plasma membrane H+-ATPase genes to ammonium nutrition and its functional link to the release of biological nitrification inhibitors from sorghum roots. Plant Soil 398, 301–312. doi: 10.1007/s11104-015-2675-2
Zeng, H. Q., Wang, G. P., Zhang, Y. Q., Hu, X. Y., Pi, E. X., Zhu, Y. Y., et al. (2016b). Genome-wide identification of phosphate-deficiency-responsive genes in soybean roots by high-throughput sequencing. Plant Soil 398, 207–227. doi: 10.1007/s11104-015-2657-4
Zeng, H. Q., Xu, L. Q., Singh, A., Wang, H. Z., Du, L. Q., and Poovaiah, B. W. (2015). Involvement of calmodulin and calmodulin-like proteins in plant responses to abiotic stresses. Front. Plant Sci. 6:600. doi: 10.3389/fpls.2015.00600
Zeng, H. Q., Zhu, Y. Y., Huang, S. Q., and Yang, Z. M. (2010). Analysis of phosphorus-deficient responsive miRNAs and cis-elements from soybean (Glycine max L.). J. Plant Physiol. 167, 1289–1297. doi: 10.1016/j.jplph.2010.04.017
Zhang, G., Chen, M., Chen, X., Xu, Z., Guan, S., Li, L. C., et al. (2008). Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J. Exp. Bot. 59, 4095–4107. doi: 10.1093/jxb/ern248
Zhang, L., Du, L., Shen, C., Yang, Y., and Poovaiah, B. W. (2014). Regulation of plant immunity through ubiquitin-mediated modulation of Ca2+-calmodulin-AtSR1/CAMTA3 signaling. Plant J. 78, 269–281. doi: 10.1111/tpj.12473
Zhang, W., Zhou, R. G., Gao, Y. J., Zheng, S. Z., Xu, P., Zhang, S. Q., et al. (2009). Molecular and genetic evidence for the key role of AtCaM3 in heat-shock signal transduction in Arabidopsis. Plant Physiol. 149, 1773–1784. doi: 10.1104/pp.108.133744
Zhao, Y., Liu, W., Xu, Y. P., Cao, J. Y., Braam, J., and Cai, X. Z. (2013). Genome-wide identification and functional analyses of calmodulin genes in Solanaceous species. BMC Plant Biol. 13:70. doi: 10.1186/1471-2229-13-70
Zhu, K., Chen, F., Liu, J., Chen, X., Hewezi, T., and Cheng, Z. M. (2016). Evolution of an intron-poor cluster of the CIPK gene family and expression in response to drought stress in soybean. Sci Rep. 6:28225. doi: 10.1038/srep28225
Zhu, X., Dunand, C., Snedden, W., and Galaud, J. P. (2015). CaM and CML emergence in the green lineage. Trends Plant Sci. 20, 483–489. doi: 10.1038/srep28225
Zou, J. J., Li, X. D., Ratnasekera, D., Wang, C., Liu, W. X., Song, L. F., et al. (2015). Arabidopsis CALCIUM-DEPENDENT PROTEIN KINASE8 and CATALASE3 function in abscisic acid-mediated signaling and H2O2 homeostasis in stomatal guard cells under drought stress. Plant Cell 27, 1445–1460. doi: 10.1105/tpc.15.00144
Keywords: soybean (Glycine max), EF-hand motif, calcium signal, calmodulin, calmodulin-like protein (CML), calcineurin B-like protein (CBL), calcium-dependent protein kinase (CDPK), Rboh (respiratory burst oxidase homolog)
Citation: Zeng H, Zhang Y, Zhang X, Pi E and Zhu Y (2017) Analysis of EF-Hand Proteins in Soybean Genome Suggests Their Potential Roles in Environmental and Nutritional Stress Signaling. Front. Plant Sci. 8:877. doi: 10.3389/fpls.2017.00877
Received: 15 March 2017; Accepted: 10 May 2017;
Published: 24 May 2017.
Edited by:
Viswanathan Chinnusamy, Indian Agricultural Research Institute (ICAR), IndiaReviewed by:
Lee Jeong Hwan, Chonbuk National University, South KoreaCopyright © 2017 Zeng, Zhang, Zhang, Pi and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Houqing Zeng, emVuZ2hxQGh6bnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.