- 1Key Laboratory of Plant Molecular Physiology, Institute of Botany, Chinese Academy of Sciences, Beijing, China
- 2University of Chinese Academy of Sciences, Beijing, China
Phosphorus (P) is an essential mineral nutrient for plant growth and development. Low availability of inorganic phosphate (orthophosphate; Pi) in soil seriously restricts the crop production, while excessive fertilization has caused environmental pollution. Pi acquisition and homeostasis depend on transport processes controlled Pi transporters, which are grouped into five families so far: PHT1, PHT2, PHT3, PHT4, and PHT5. This review summarizes the current understanding on plant PHT families, including phylogenetic analysis, function, and regulation. The potential application of Pi transporters and the related regulatory factors for developing genetically modified crops with high phosphorus use efficiency (PUE) are also discussed in this review. At last, we provide some potential strategies for developing high PUE crops under salt or drought stress conditions, which can be valuable for improving crop yields challenged by global scarcity of water resources and increasing soil salinization.
Introduction
Phosphorus (P), one of the major plant macronutrients, is a structural component of nucleic acids and phospholipids and plays essential roles in energy transfer, signal transduction, and enzyme activation. P exists in two kinds of chemical forms in soil: organic form and inorganic form (orthophosphate, Pi). As the only form that can be assimilated by plant, Pi is commonly present at low concentration (less than 2 μM) in soil solution, even in fertile soil (10 μM), due to its uneven distribution, relative immobile and high fixation (Mimura, 1999; Raghothama, 1999). Low Pi availability has been a worldwide constraint for crop growth. Nowadays, phosphate fertilizer application is a conventional method to improve the Pi availability. However, only 10–20% of applied Pi can be absorbed by plants and overuse of phosphate fertilizer not only has been impeded by the non-renewable resource, phosphate rock, but also causes the water eutrophication (Holford, 1997; Oelkers and Valsami-Jones, 2008). Therefore, it is urgent and imperative to understand the plant Pi acquisition and distribution mechanism and enhance the crop phosphorus use efficiency (PUE). Plants have developed elaborate mechanisms to enhance Pi availability and assimilation. One adaptation to P deficiency commonly observed in most species is an increase in root surface area by formation of finer roots, aerenchyma, and root hairs, which would improve soil exploration (Lynch, 2007; Nestler et al., 2016). In addition, some plant species can secrete organic acids to release Pi through complexation reaction of organic acids with Al3+, Fe3+, Ca2+, or phosphatases to release Pi from the organic sources, thus to enhance the availability of Pi in soils (López-Arredondo et al., 2014). Plant uptake and translocation of available Pi occurs through the concerted action of various Pi transporters. Based on the sequence identity and their varied subcellular localization, plant Pi transporters are grouped into five phylogenetically distinct classes of families: PHT1, PHT2, PHT3, PHT4, and PHT5 (Mudge et al., 2002; Rausch and Bucher, 2002; Liu T.Y. et al., 2016; Mlodzińska and Zboińska, 2016). In this review, we summarize the current state of research on plant PHT families, including phylogenetic analysis, function, and regulation. The agricultural application aspects of Pi transporters and regulatory factors are also discussed here. We also provide some potential strategies for improving crop PUE under salt or drought stress conditions.
Phylogenetic Analysis and Roles of Plant Pi Transporters
Utilizing the identified Pi transporter amino acid sequences from Arabidopsis, rice, soybean, maize, wheat, poplar, Medicago truncatula as well as a halophyte E. salsugineum, we constructed a phylogenetic tree using the neighbor-joining method with MEGA 6 program. Phylogenetic analysis showed that all the divergence of five phosphate transporter families preceded divergence of monocots and dicots and the members of PHT1, PHT2, PHT3, PHT4, and PHT5 clustered separately (Figure 1). The putative localization, tissue expression and conserved sequence of Pi transporters in Arabidopsis were summarized in Table 1. In general, plasma membrane (PM) located PHT1s mainly function in Pi acquisition from soil, whereas Pi distribution within the plant such as translocation against chloroplasts, mitochondria, Golgi, and vacuole is mediated by PHT2, PHT3, PHT4, and PHT5 family members. The functions of these PHT families in Arabidopsis and rice were summarized in a recent review by Mlodzińska and Zboińska (2016). In this section, we draw more attention to the identification and function of Pi transporters in other plants, especially in some important crops, attempting to throw light on the underlying Pi uptake and distribution mechanisms in these crops and to find potential genetic target for crop improvement breeding.
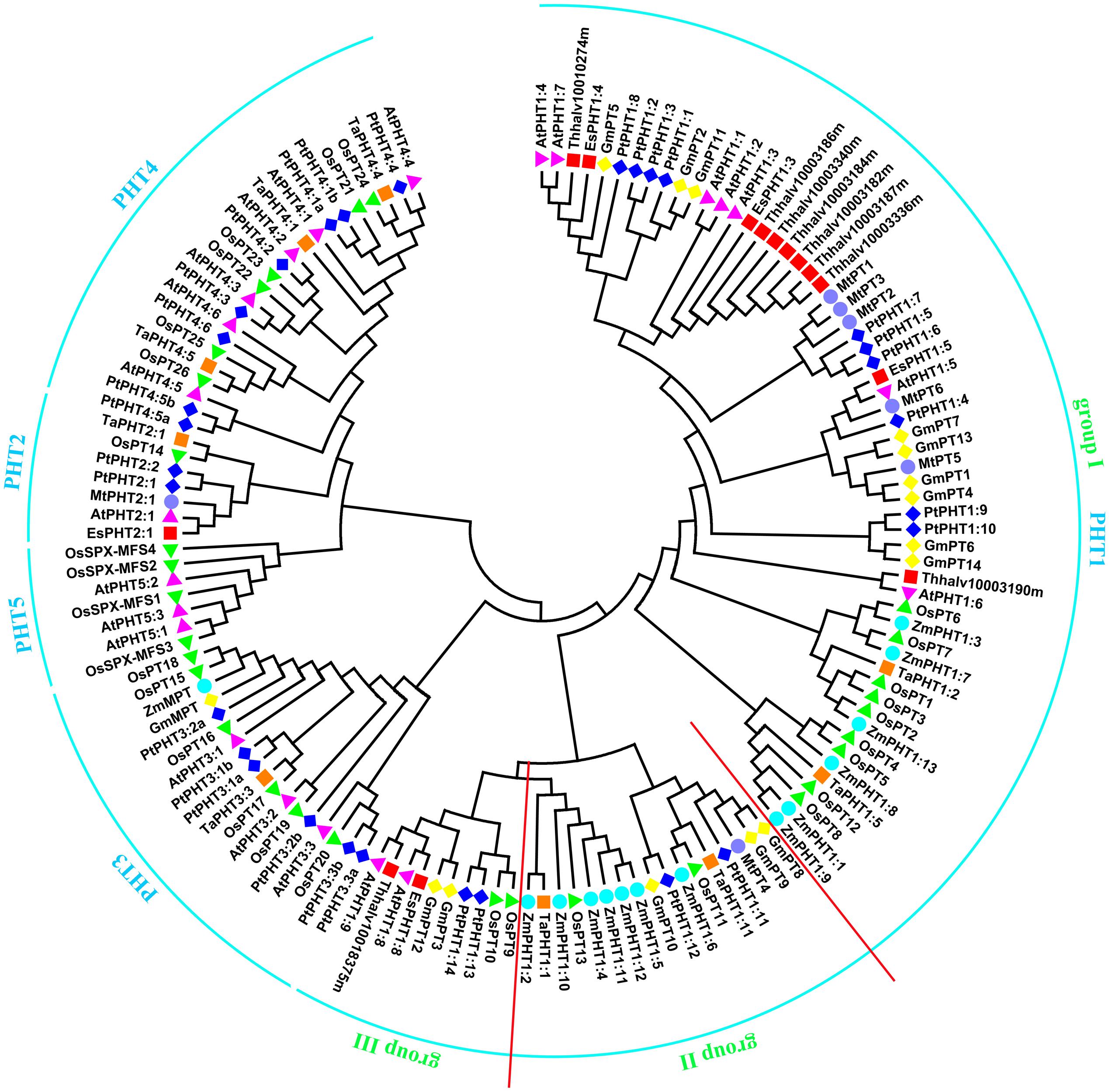
FIGURE 1. Phylogenetic analysis of Pi transporters. The phylogenetic tree was constructed by the Neighbor-Joining method with MEGA6.0, bootstrap values are from 1000 replicates. Accession numbers or locus IDs of all used proteins were given in Supplementary Table S1.
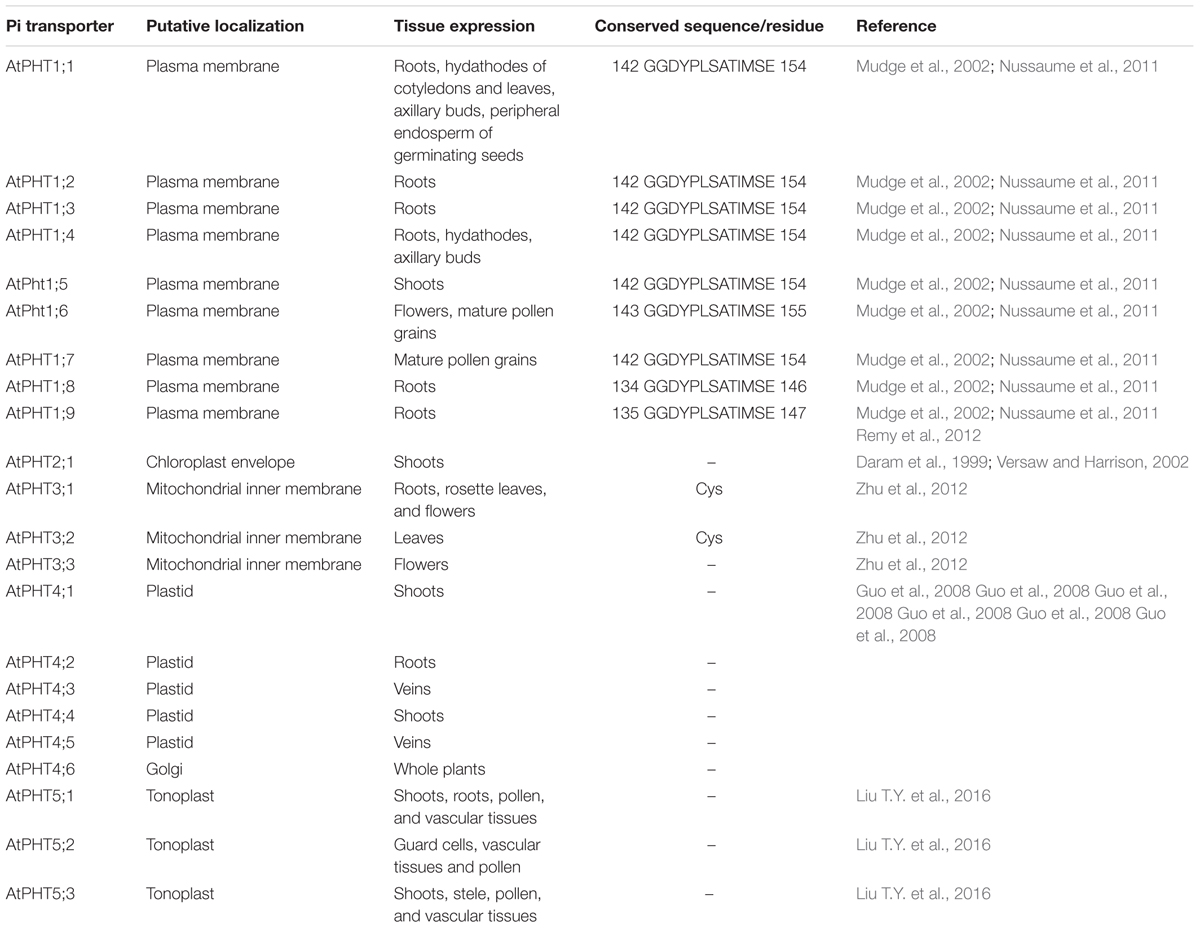
TABLE 1. Putative localization, tissue expression, and conserved sequence of Pi transporters in Arabidopsis.
PHT1s
PHT1 proteins are the best studied plant Pi transporters. These proteins have a conserved structure containing 12 transmembrane (TM) domains with a large hydrophilic loop between TM6 and TM7 and both hydrophilic N and C termini locate in the cytoplasm (Raghothama, 1999). All plant PHT1 proteins have the conserved PHT1 signature GGDYPLSATIxSE and in fungi that is GGDYPLSxxIxSE (Karandashov and Bucher, 2005). The first plant PHT1 gene was cloned from Arabidopsis (Muchhal et al., 1996) and exhibited similarities to genes encoding Pi transporters in Saccharomyces cerevisiae (PHO84; Bun-Ya et al., 1991). Based on protein sequence identity and conserved signature analysis, PHT1 members from several plant species have been identified, including tobacco (Baek et al., 2001), potato (Leggewie et al., 1997; Gordon-Weeks et al., 2003), rice (Paszkowski et al., 2002; Liu et al., 2011; Wang X. et al., 2014; Li et al., 2015), barley (Rae et al., 2003; Preuss et al., 2010, 2011), maize (Liu F. et al., 2016), Medicago truncatula (Javot et al., 2007a; Liu et al., 2008), wheat (Liu et al., 2013), soybean (Fan et al., 2013), Setaria italica (Ceasar et al., 2014), tomato (Chen et al., 2014), sorghum (Walder et al., 2015), flax (Walder et al., 2015), and poplar (Zhang et al., 2016). The numbers of PHT1 members identified in various plants were listed in Table 2. According to the phylogenetic tree, PHT1 proteins were well clustered into three groups. Group I contained most of the PHT1s from all species analyzed and those proteins from dicots were grouped separately with those from monocots. At least one member from all the species analyzed except Arabidopsis and E. salsugineum was included in group II. Group III contained AtPHT1;8 and AtPHT1;9 from Arabidopsis and their orthologs from E. salsugineum, OsPT9 and OsPT10 from rice, GmPT3 and GmPT12 from soybean, as well as PtPHT1;13 and PtPHT1;14 from poplar (Figure 1). Most of the group I members are reported to be involved in direct Pi uptake from soil such as AtPHT1;1 and AtPHT1;4 (Misson et al., 2004; Shin et al., 2004; Ayadi et al., 2015), OsPT1 (Sun et al., 2012), OsPT2 and OsPT6 (Ai et al., 2009), OsPT4 (Ye et al., 2015; Zhang et al., 2015). It should be pointed out that although AtPHT1;5 and OsPT8 were shown to function in Pi redistribution from source to sink organs (Nagarajan et al., 2011; Li et al., 2015), their roles in Pi uptake cannot be ruled out, considering the tolerance to arsenate (Pi structural analog) of the corresponding mutant plants (Nagarajan et al., 2011; Wang P. et al., 2016). TaPHT1;2 and TaPHT1;5 from wheat were strongly induced under Pi-limiting conditions in different cultivars, suggesting their roles in Pi uptake under limited Pi supply. Furthermore, their expression profiles showed genotype-dependent and correlated with plant phosphorus efficiency (Aziz et al., 2014). In poplar, PtPHT1;2, PtPHT1;3, PtPHT1;4, PtPHT1;6, PtPHT1;7, PtPHT1;9, and PtPHT1;10 clustered in group I were mainly expressed in roots, and PtPHT1;2, PtPHT1;4, PtPHT1;6, and PtPHT1;8 were greatly induced by Pi starvation (Zhang et al., 2016). Interestingly, PtPHT1;2 expression was remarkably induced by drought stress, irrespective of the Pi level (Zhang et al., 2016). In addition, seven orthologs of AtPHT1;3 are found in E. salsugineum, which was reported more acclimated to Pi limitation than Arabidopsis (Velasco et al., 2016). EsPHT1;4 (Thhalv10016497m) was up-regulated in Pi-deprived E. salsugineum plants, suggesting it roles in Pi uptake (Velasco et al., 2016).
Besides absorbing Pi directly from soil, most of vascular plants, including major crops, can obtain Pi from arbuscular mycorrhizal (AM) by exchange of carbohydrates through forming a mycorrhizal association (Javot et al., 2007b). All the PHT1 transporters included in group II (Figure 1) except PtPHT1;11 and PtPHT1;12 from poplar, ZmPHT1;10 and ZmPHT1;12 from maize have been reported to be involved in acquisition of Pi from AM (Glassop et al., 2005; Javot et al., 2007a; Tamura et al., 2012; Yang et al., 2012; Duan et al., 2015; Liu F. et al., 2016; Tian et al., 2017). MtPT4 was shown to be essential for Pi acquisition from AM fungus and loss of MtPT4 function even led to premature death of the arbuscules (Javot et al., 2007a). Interestingly, 7 of 13 ZmPHT1 members were clustered in group II (Figure 1), implying the importance of AM mediated Pi uptake in maize. ZmPHT1;6 was the best described AM-inducible Pi transporter in maize and its expression was significantly positively correlated with AM colonization rate, concentration of AM biomarkers in roots, Pi uptake and dry weight of shoot (Glassop et al., 2005; Tian et al., 2013). Recently, two studies reported other members including ZmPHT1;2, ZmPHT1;4, ZmPHT1;5, and ZmPHT1;11 were also up-regulated by AM and may participate in Pi uptake through mycorrhizal pathway (Liu F. et al., 2016; Sawers et al., 2017). Moreover, the expression of ZmPHT1;2, ZmPHT1;4, ZmPHT1;5, and ZmPHT1;6 was correlated with phosphorus acquisition efficiency in maize (Sawers et al., 2017). In fact, in most plant species forming mycorrhizal symbioses at least one mycorrhizal specific or up-regulated PHT1 gene has been discovered, such as LePT1 in Lycopersicon esculentum (Rosewarne et al., 1999), StPT3, StPT4, and StPT5 in potato (Rausch et al., 2001), AsPT1 in Astragalus sinicus (Xie et al., 2013), and BdPT3, BdPT7, BdPT12, and BdPT13 in Brachypodium distachyon (Hong et al., 2012). However, the molecular mechanisms of Pi and carbon transfer in the symbiosis are still largely unknown. In addition, Arabidopsis and E. salsugineum belong to the minority of plants that cannot associate with AM fungi and our analysis also showed that no Arabidopsis PHT1 proteins included in group II, which indicated a certain reliability of predicting unknown gene functions through phylogenetic analysis.
As for group III of PHT1 family, transporters may be involved in Pi uptake as well as Pi allocation from root to shoot, considering the well characterized AtPHT1;8 and AtPHT1;9 (Hamburger, 2002; Lapis-Gaza et al., 2014) and OsPT9 and OsPT10 (Wang X. et al., 2014) in this group. Also, the respective orthologs of AtPHT1;8 and AtPHT1;9 from E. salsugineum were included in this group. PtPHT1;13 and PtPHT1;14 were predominantly expressed in roots and induced by low Pi conditions, suggesting their roles in Pi uptake (Zhang et al., 2016). GmPT3 and GmPT12 were also predominantly expressed in roots. Interestingly, GmPT3 was up-regulated by N, P, or K deficiency simultaneously in both leaves and roots, suggesting it might be involved in a universal regulation network in response to multiple nutrient deficiencies (Qin et al., 2012).
PHT2s
PHT2 proteins have a putative topology of 12 TM domains interrupted by a large hydrophilic loop between TM8 and TM9. They have two boxes of eight and nine amino acids located in N- and C-terminal domains, respectively, conserved among eubacteria, archaea, fungi, plants, and animals (Daram et al., 1999). Most of the reported PHT2;1 genes were predominantly expressed in green tissue and the proteins were located in the chloroplast inner envelope membrane (Versaw and Harrison, 2002; Rausch et al., 2004; Loth-Pereda et al., 2011; Guo et al., 2013; Shi et al., 2013). The PHT2 family has long been thought to have only one member until two members were recently identified in poplar (Zhang et al., 2016). However, though the transcripts of PtPHT2;1 and PtPHT2;2 could be detected in various tissues, they were predominantly expressed in roots. Moreover, PtPHT2;2 significantly up-regulated under low-Pi conditions (Zhang et al., 2016), suggesting its different role from AtPHT2;1. Like AtPHT2;1, EsPHT2;1 (Thhalv10003891m) was mainly expressed in leaves and was not regulated by external Pi supply (Velasco et al., 2016). Contrast to AtPHT2;1 and EsPHT2;1, TaPHT2;1 and OsPHT2;1 (OsPT14) were regulated in response to external Pi concentrations (Guo et al., 2013; Shi et al., 2013). Knockdown of TaPHT2;1 significantly reduced Pi concentration in the chloroplast under both sufficient and limited Pi supply, suggesting that TaPHT2;1 is crucial in the mediation of Pi translocation from the cytosol to the chloroplast (Guo et al., 2013). Furthermore, TaPHT2;1 expression profile was genotype-dependent and correlated with plant phosphorus efficiency (Aziz et al., 2014). Therefore, it was proposed by Aziz et al. (2014) as a marker gene for screening high phosphorus efficiency genotypes. By contrast, overexpression of OsPHT2;1 (OsPT14) in rice increased Pi concentrations in leaves and the plant biomass, suggesting its roles in Pi accumulation in leaves and Pi translocation in plants (Shi et al., 2013).
PHT3s
PHT3 proteins have two TM α-helices separated by a hydrophilic extramembrane loop, which are conserved in all analyzed mitochondrial transporter proteins, being essential for mitochondrial targeting (Zhu et al., 2012). The first gene encoding mitochondrial Pi transporter (MPT) that belongs to PHT3 family was isolated from birch (Betula pendula Roth), named Mpt1 (Kiiskinen et al., 1997). Afterward, MPT cDNAs were continuously isolated and cloned from yeast, soybean, maize, rice, Arabidopsis, Lotus japonicas, and poplar (Takabatake et al., 1999; Nakamori et al., 2002; Hamel et al., 2004; Sha et al., 2014; Zhang et al., 2016). There are three members in Arabidopsis. Interestingly, six transporters from rice and poplar were clustered with AtPHT3 proteins in the phylogenetic tree (Figure 1). PHT3 proteins play a critical role in Pi exchange between cytoplasm and mitochondria matrix by Pi/H+ symport or Pi/OH- antiport, which is essential for the oxidative phosphorylation of ADP to ATP (Takabatake et al., 1999). Hamel et al. (2004) found that ectopic expression of AtMPT1 (AtPHT3;3) and AtMPT2 (AtPHT3;2) could complement the S. cerevisiae Δmir1Δpic2 mutant, which was not able to grow on non-fermentable medium at 28 and 36°C, and restored the Pi uptake into mitochondria. The different expression patterns of three AtMPT genes imply they play specific roles in various organs or developmental stages (Zhu et al., 2012). For example, overexpression of AtMPT3 (AtPHT3;1) caused multiple developmental defects including deformed leaves, dwarfed stature, and reduced fertility through regulating mitochondrial function (Jia et al., 2015). It is worth noting that PHT3s were suggested to be involved in wheat grain development, with high transcript abundance of TaPHT3;1 in embryo and rachis and TaPHT3;2 in aleurone (Shukla et al., 2016). Besides, PHT3 transporters were also found to be involved in response to salt and drought stress. AtMPTs were up-regulated by high salinity stress in Arabidopsis seedlings. And overexpression of AtMPTs increased plant sensitivity to salt stress compared with the wild-type plants, which might be through an ATP-dependent pathway and modulation of gibberellin homeostasis (Zhu et al., 2012). Recently, expression of PtPHT3;2b and PtPHT3;3b from poplar was found to be regulated by drought stress with a Pi level dependent manner (Zhang et al., 2016). To date, knowledge on the molecular mechanisms of PHT3s mediated biological functions in plants is still limited.
PHT4s
PHT4 family was first characterized and designated by Guo et al. (2008) in Arabidopsis. PHT4 proteins share similarity with SLC17/type I Pi transporters. There are six members in PHT4 family in Arabidopsis (Guo et al., 2008). The plastid and Golgi apparatus-located PHT4 members in Arabidopsis were found to participate in various processes including leaf development, plant defense, and salt response, which were well reviewed by Mlodzińska and Zboińska (2016). Recently, Hassler et al. (2016) found that pht4;6 mutant Arabidopsis plants displayed accelerated senescence induced by darkness. Further investigation indicated that the accelerated dark-induced senescence of pht4;6 mutant was caused by the reduced trans-zeatin concentration, which resulted from cellular Pi starvation (Hassler et al., 2016). Based on genome database searching and phylogenetic analysis, seven putative PHT4 members in rice (Mlodzińska and Zboińska, 2016), eight in poplar (Zhang et al., 2016) and six in wheat (Shukla et al., 2016) were identified recently. Phylogenetic analysis indicated that there are two orthologs of AtPHT4;6 in rice, and two of AtPHT4;1 and AtPHT4;5, respectively, in poplar (Figure 1). However, the functions of PHT4s in these plant species are largely unknown yet. TaPHT4;2 and TaPHT4;4 in wheat were found to be highly expressed in endosperm during grain development, suggesting their functions in the Pi-allocation within the seed compartments (Shukla et al., 2016). Similarly, all the PHT4 members in rice (OsPT21-26) were shown to be expressed in all seed tissues (husks, vascular bundle/aleurone, endosperm, and embryo), which suggested that their roles in grain P loading may be not specific (Wang F. et al., 2016). In poplar, PtPHT4;1a was up-regulated under low-Pi conditions, while PtPHT4;1b and PtPHT4;5b were induced under high-Pi (Zhang et al., 2016), suggesting their different roles in response to Pi supply. Interestingly, under high-Pi conditions, PtPHT4;4 and PtPHT4;6 were induced by drought stress (Zhang et al., 2016). Most recently, a chromoplast-localized ClPHT4;2 from watermelon was found to be necessary for flesh color development. It was reported that the ClPHT4;2 expression levels were closely correlated with flesh carotenoid contents among 198 watermelon accessions. Furthermore, down-regulation of ClPHT4;2 expression in transgenic watermelons decreased the fruit carotenoid accumulation (Zhang et al., 2017). Above all, plant PHT4s play various roles, further investigating their functions, especially in crops, will facilitate the development of crops with desired agronomic traits.
PHT5s
Plant vacuoles serve as the primary intracellular compartments for Pi storage. Recently, several studies shed light on the underlying molecular mechanisms for vacuolar Pi transport that had long remained unknown. Liu T.Y. et al. (2016) designated Arabidopsis SPX-MFS (SYG1/PHO81 /XPR1-Major Facility Superfamily) proteins as PHT5 family, which was also named Vacuolar Phosphate Transporter (VPT), function as vacuolar Pi transporters. In fact, OsSPX-MFS1, OsSPX-MFS2, and OsSPX-MFS3 localized on the tonoplast of rice protoplasts were already functionally characterized before (Wang et al., 2015). Another member OsSPX-MFS4 has not been characterized yet. The roles of PHT5s in Pi transport across the vacuole membrane in Arabidopsis and rice were well summarized by Mlodzińska and Zboińska (2016). Further identification and characterization of PHT5s in other crops will be helpful for our understanding on molecular mechanisms of vacuolar Pi transport.
Regulation of Pi Transporters
Great progress has been made also in deciphering the regulatory mechanism of plant Pi transporters, which has revealed a multi-layered network regulating gene expression, protein activity and protein turnover.
Transcriptional Regulation
To date, MYB-type, WRKY-type, bHLH-type and other kinds of transcription factors (TF) have been found involved in transcriptional regulation of Pi transporters. PHR1 (phosphate starvation response 1) is a member of the MYB superfamily (Rubio et al., 2001). It was reported that PHR1 regulated expression of PHT1 at low Pi concentrations through binding to the P1BS (PHR1-binding sequence) or P1BS-like domain, a cis-acting element in PHT1 gene promoters (Rubio et al., 2001; Schünmann et al., 2004a,b). Such interaction was validated in yeast cells where wheat TaPHR1 activated the expression of TaPHT1;2 through binding to P1BS (Wang et al., 2013). In fact, it has been reported that P1BS is an integrating cis-regulatory motif associated with genes that are highly induced by Pi starvation (Bustos et al., 2010). There are usually more than one P1BS in many PHT1 gene promoters, suggesting the important role of PHR1 in regulating PHT1 expression in response to Pi starvation. For example, P1BS was found in the promoters of 11 of 13 OsPHT1s, 8 of 13 ZmPHT1s, 10 of 14 PtPHT1s and all of the 8 LePHT1s in tomato with 1–6 copies (Liu et al., 2011; Chen et al., 2014; Liu F. et al., 2016; Zhang et al., 2016). However, some PHT1 genes containing P1BS in their promoters were not induced by Pi starvation. For example, OsPT11 and OsPT13 which have 6 and 4 P1BS motif, respectively, are AM-inducible Pi transporters but not involved in Pi starvation response (Yang et al., 2012). Similarly, ZmPHT1;1, LePT4 and LePT5 were not induced by Pi starvation (Chen et al., 2014; Liu F. et al., 2016). Another R2R3 type MYB TF, MYB62, is also induced by Pi starvation (Devaiah et al., 2009). However, the decreased expression of AtPHT1;1 and AtPHT1;4 in MYB62 overexpression lines suggested a negative regulation by MYB62, though the precise mechanism is unclear yet (Devaiah et al., 2009). As a plant-specific TF, WRKY has a conservative WRKYGQK domain at N-terminal and a zinc finger motif. WRKY specifically combines with W-box [TTTGAC(C/T)] in PHT1 promoters and regulates the expression of PHT1. Devaiah et al. (2007) proposed that when inhibiting the expression of WRKY75, a nucleus localized and roots expressed TF, the expression of AtPHT1;1 and AtPHT1;4 decreased, suggesting WRKY75 is a positive regulator of these two genes. A similar role was reported for WRKY45, which specifically regulated the expression of AtPHT1;1 (Wang H. et al., 2014; Ding et al., 2016). Recently, Yang et al. (2016) found that up-regulation of TabHLH1, a wheat bHLH-type TF, led to increased expression of NtPT1 in tobacco and improved plant tolerance to Pi deprivation. Additionally, TaZAT8, a wheat C2H2-ZFP-type TF, was also shown involved in regulation of Pi transporters and mediated low-Pi tolerance through regulating Pi acquisition, ROS homeostasis and root system establishment, however, the regulation mechanism is unclear yet (Ding et al., 2016). Besides, other regulatory elements were also found to regulate the expression of PHT1 genes, such as SPX3, histone H2A.Z and zinc finger TF ZAT6 (Devaiah et al., 2007; Duan et al., 2008; Smith et al., 2010).
In case of the transcriptional regulation of AM-induced Pi transporters, mycorrhiza transcription factor binding sequence (MYCS) were reported to be involved (Chen A. et al., 2011). In maize, though MYCS was found in the promoter of some AM-inducible ZmPHT1s, some ZmPHT1s without the MYCS element, such as ZmPHT1;2, ZmPHT1;4, ZmPHT1;7, and ZmPHT1;9, also upregulated by AM, implying that new regulation element(s) might exist in the promoter regions of these genes (Liu F. et al., 2016). Most recently, Zhang et al. (2017) identified two TFs (ClbZIP1 and ClbZIP2) from watermelon, binding to the promoter of ClPHT4;2, which shed light on our understanding of the regulatory mechanisms of PHT4s expression.
Post-transcriptional and Post-translational Regulation
MicroRNAs (miRNAs) have been reported to regulate gene expression by directing the cleavage of target gene transcript. Many studies have characterized the role of miRNA in regulating Pi homeostasis (Figure 2). Under Pi starvation conditions, up-regulated miR399 directed the cleavage of phosphate2 (PHO2) mRNA which could degrade PHT1 proteins in Arabidopsis (Fujii et al., 2005; Chiou et al., 2006; Kuo and Chiou, 2011). Up-regulation of miR399 in phloem sap was regarded as long-distance signal to regulate Pi homeostasis under Pi deficient conditions. miR399s from some crops including barley, sorghum, tea plant, and cotton were also identified and their predicted targets were PHO2 or UBC24 (Hackenberg et al., 2013; Djami-Tchatchou et al., 2017). miR827, which has been found to target NLA (Nitrogen Limitation Adaptation) gene, also regulates Pi homeostasis through proteolysis pathway (Kuo and Chiou, 2011). In addition, the PHT5 members OsSPX-MFS1 and OsSPX-MFS2 were also negatively regulated by osa-miR827 (Lin et al., 2010). Furthermore, similarly to the miR399-PHO2 pathway, this pathway is under the control of the central TF OsPHR2 (Wang et al., 2012).
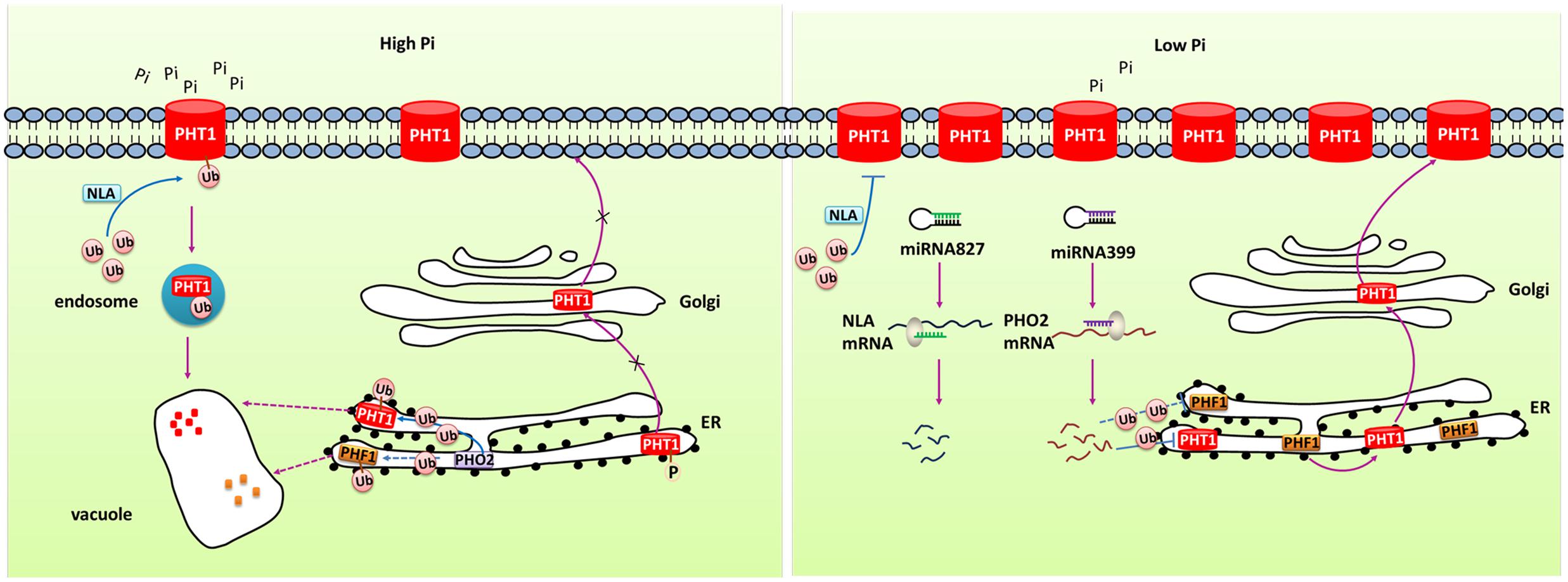
FIGURE 2. Simplified models for the post-transcriptional and post-translational regulation of PHT1 transporters. Under Pi-replete condition, phosphorylation of PHT1 transporters prevents their exit from ER and the subsequent targeting to the PM. NLA and PHO2 direct the ubiquitin-mediated degradation of PHT1 transporters at PM and ER, respectively, to avoid excessive accumulation of Pi. Under Pi deficiency condition, miR827 and miR399 mediate cleavage of NLA and PHO2 transcripts, respectively, thus to increase the amount of PHT1 transporters. PHF1 is accounted for the PHT1 transporters exit from ER and correct targeting to PM.
At the post-translational level, some PHT1 proteins can be modified by phosphorylation (Figure 2). Ser-514 phosphorylation of AtPHT1;1 C termini prevented its exit from ER suggesting phosphorylation regulated export of AtPHT1;1 from ER and its correct targeting to PM (Bayle et al., 2011). Furthermore, subcellular localization of several PHT1 proteins was revealed to be affected by PHF1 (phosphate transporter traffic facilitator1), a plant unique SCE12 protein (Figure 2). It was reported that AtPHT1;1, AtPHT1;2 and AtPHT1;4 transporters could not be targeted to the PM in phf1 mutant, and phosphate absorptivity of phf1 mutant was 80% lower than the wild type (González et al., 2005; Bayle et al., 2011). PHF1 was also shown to participate in PHT1 transport from endoplasm to Golgi (González et al., 2005; Bayle et al., 2011). In rice, OsPHF1 had a similar function in efficiently targeting low- and high-affinity Pi transporters to the PM (Chen J. et al., 2011). Hence, PHF1 plays an important role in post-translational regulation of PHT1. Another kind of post-translational regulation is degradation of PHT1 proteins localized at PM under Pi sufficient conditions (Figure 2). When Pi-starved seedlings were transferred to sufficient Pi conditions, the number of PHT1 transporters at PM decreased dramatically, indicating a rapid degradation mechanism specifically occurs in Pi-replete conditions (Bayle et al., 2011). An ubiquitin-mediated regulatory pathway has been found to be involved in the degradation of PHT1 members at PM through triggering the endocytosis and vacuolar degradation. The PM-localized NLA protein, a target of miR827, can negatively regulate PHT1 transporters to maintain Pi homeostasis in Arabidopsis. Under Pi-sufficient conditions, NLA had a putative ubiquitin ligase activity and mediated the ubiquitination of PHT1 transporters, thus triggered clathrin-dependent endocytosis and PHT1 transporters degradation (Lin et al., 2013). PHO2 is a membrane protein predominately associated with the endoplasmic reticulum (ER) and Golgi. As an ubiquitin-conjugating E2 enzyme, it shared similar mechanism with NLA in degradation of PHT1 proteins at ER including PHT1;1, PHT1;2, and PHT1;3, and likely PHF1 to prevent PHT1s exit from ER and their targeting to PM when the Pi supply is sufficient (Huang et al., 2013). Under Pi deficient condition, miR827 and miR399 mediate post-transcriptional cleavages on the transcripts of NLA and PHO2, thereby relieving the PHT1 transporter degradation and activating Pi uptake as well as root-to-shoot translocation (Figure 2). Rice OsNLA also directed the degradation of OsPT members (Lin et al., 2013). Most recently, OsNLA1 was reported to be involved in maintaining phosphate homeostasis in rice by mediating the degradation of OsPT2 and OsPT8. Most importantly, in contrast to the Arabidopsis ortholog, the expression of OsNLA1 was not responding to Pi limitation and also no changes were observed in OsmiR827 or OsPHR2-overexpressing lines. Furthermore, the authors concluded that OsNLA1 does not interact with OsPHO2, suggesting the requirement of additional unknown E2 conjugating enzyme for OsNLA1 mediated degradation of OsPTs (Yue et al., 2017).
Plants response to low phosphorus is a complex signal transduction process, including Pi sense, uptake, transport, translocation and remobilization. Pi transporters and regulatory factors play essential roles in this process. Although a few regulatory factors of PHT1 have been identified as mediators of plant adaptation to low-Pi stress, the accurate regulatory network of PHT1 transporters is still a great challenge to be deciphered. In addition, regulatory mechanisms of PHT2, PHT3, PHT4, and PHT5 family are still unclear as well. Further detailed analysis of these Pi transporter gene promoters is the basis of revelation of the complicated regulatory mechanism. A better understanding of regulation of phosphate transporters will provide novel avenues to improve crop yield with lower Pi inputs
Potential Applications and Perspectives in Agricultural Production
To Improve Crop PUE in Pi-Limited Soils
The concerted action of Pi transporters ensures Pi acquisition and distribution among tissues and cytosolic Pi homeostasis. Therefore, engineered alterations of the expression of Pi transporters provide an opportunity to optimize uptake and distribution of Pi in crops to improve yield. Several researches have been illustrated that modulating Pi transporters or regulatory factors could enhance Pi uptake, biomass or crop yields (Table 3). For instance, OsPT4 genetically modified (GM) rice showed increased Pi accumulation in roots and shoots and got significant increase in 1000-grain weight and grain yield per plant compared with wild type under various Pi concentrations (Zhang et al., 2015). Up-regulation of OsPT6 in soybean or rice also increased the plant biomass, grain weight and grain yield under low-Pi conditions or in field trials (Yan et al., 2014; Zhang et al., 2014). As for TFs, overexpressing OsWRKY74 in rice resulted in enhanced shoot and root biomass and increased grain yield under low-Pi conditions (Dai et al., 2016). Two regulatory factors (TaPHR1 and TaNFYA-B1) from wheat were also introduced in wheat plant to increase grain yield under low-Pi soils in field trials (Wang et al., 2013; Qu et al., 2015). Recently, knock out TaPHO2-A1 encoding the negative regulatory factor PHO2, which mediates PHT1 proteins degradation, improved Pi uptake and grain yield under low phosphorus conditions in wheat (Ouyang et al., 2016). These works provided useful candidates for breeding crops with high Pi use efficiency and improved yields. However, many Pi transporters or TFs were shown not to work as well as these proteins in improving plant growth and yields performance, which was usually due to Pi toxicity. For example, overexpression of OsPT8 in rice resulted in excessive Pi both in roots and shoots and significant growth suppression as well as other Pi toxic symptoms under the high-Pi conditions (Jia et al., 2011) (Table 3). Similar results were also observed in OsPT2, OsPT9, OsPT10, OsPHR2, and OsARF12 transgenic plants (Liu et al., 2010; Wang S. et al., 2014; Wang X. et al., 2014) (Table 3). Overexpression of miR399 in Arabidopsis and tomato also caused Pi toxicity and retarded growth (Fujii et al., 2005; Gao et al., 2009). However, a recent study demonstrated that monitoring expression of ath-miR399d by an abiotic stress inducible promoter (rd29A promoter) in tomato resulted in plant growth under low temperature and P deficiency conditions (Gao et al., 2015). Therefore, using low Pi or other environment inducible promoters, instead of the constitutive promoters, to control the expression of Pi transporters or their regulatory factors may be a good strategy to avoid Pi toxicity in GM crops.
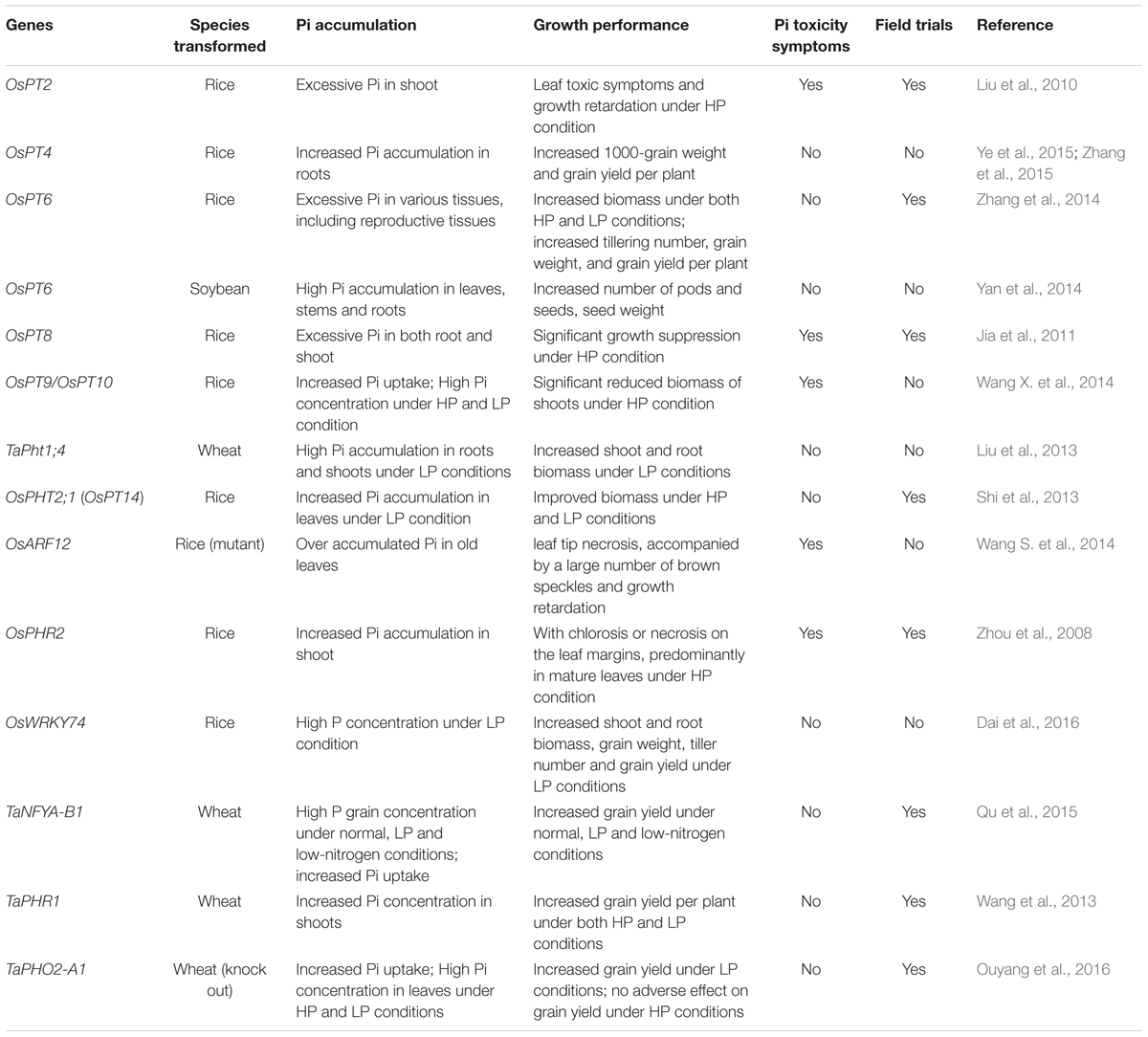
TABLE 3. Genes encoding Pi transporters and regulatory factors applied in improving Pi accumulation or crop yields.
All above target genes are involved in Pi uptake from soil, however, it should be pointed out that soil substrate (Pi) availability rather than transporter activity may be the limiting step in such soil conditions. P depletion experiments showed that the P inefficient genotype of rice was able to deplete Pi from nutrient solutions as rapidly and to an equally low level as a P deficiency tolerant genotype (Wissuwa, 2005), which suggested that transporter activity may be not limiting in inefficient genotype. Similarly, overexpressing a high-affinity Pi transporter in barley did not result in improved tolerance to P deficiency (Rae et al., 2004). In fact, several studies comparing gene expression under Pi limitation in different rice genotypes found higher expression of PHT1s in the inefficient genotype, which suggested that the Pi deficiency-induced expression of these Pi transporters may be part of the Pi starvation response, but not part of Pi starvation tolerance (Pariasca-Tanaka et al., 2009; Oono et al., 2013).
Taking into account the above problems in using PHT1s to improve crop yields under Pi-limited soils, more attention may be paid to genes encoding intracellular Pi transporters in the future, which play important roles in proper Pi distribution, Pi remobilization, and maintaining cytosolic Pi homeostasis, thus to improve the crop PUE instead of Pi uptake in Pi-limited soils. Consistent with this hypothesis, transgenic rice plants overexpressing OsPHT2;1 (OsPT14) exhibited more Pi accumulation in the top three leaves and panicle axis and significantly higher biomass than the wild type plants under various Pi conditions (Shi et al., 2013) (Table 3). In addition, Pi acquisition in crops via AM symbiosis is becoming increasingly important due to limited high grade rock Pi reserves and a demand for environmentally sustainable agriculture. The functional characterization of the interplay between direct and symbiotic Pi uptake, particularly under field conditions, mirroring agricultural practices, would be most valuable and relevant for crops.
To Improve Crop Growth under Salt and Drought Stress Conditions
Global scarcity of water resources and increasing soil salinity threatens crop productivity worldwide. High soil salinity is usually accompanied by the low availability of many mineral nutrients including P (Grattan and Grieve, 1999). Furthermore, salt and drought stress inhibit the uptake and translocation of mineral nutrients, and P fertilization can increase stress tolerance and dry weight in many plant species (Martinez and Lauchli, 1994; Jin et al., 2006; Waraich et al., 2011; Gao et al., 2015). Therefore, it is of great significance to understand the mechanism of Pi acquisition and utilization under salt and drought stress, thus to improve the crop PUE under abiotic stresses. Efforts can be made in the following three aspects.
Firstly, using identified Pi transporter genes involved in salt or drought response to develop GM crops. So far, several studies have found that Pi transporters are involved in response to salt and drought stress. Arabidopsis plants overexpressing AtPHT3;1, AtPHT3;2, and AtPHT3;3 were more sensitive to salt stress compared with wild type due to the energy status change and decreased expression of gibberellin (Zhu et al., 2012). In addition, protein N-glycosylation has been found associated with plant stress tolerance. AtPHT4;6 was shown to take part in recycling Pi released from glycosylation process and likely to relate to protein N-glycosylation. Interestingly, atpht4;6 mutant was more sensitive to salt stress compared with wild type (Cubero et al., 2009). Hence, intracellular Pi homeostasis is essential for plant salt stress tolerance. Further investigation of the regulatory mechanism of intracellular Pi transporters will be helpful for our understanding on plant Pi acquisition and utilization processes in response to salt stress. Recently, several genes encoding Pi transporters from poplar were found to be regulated by drought stress. PtPHT1;2 expression was remarkably induced by drought stress, irrespective of the Pi level, whereas changes of PtPHT1;3, PtPHT1;9, PtPHT1;11, PtPHT1;14, PtPHT2;2 PtPHT3;2b, PtPHT3;3b, PtPHT4;4, PtPHT4;6 expression under drought stress showed Pi level dependent. These genes especially those up-regulated by drought stress at low Pi level may contribute to drought tolerance of poplar plants in Pi-limited soils (Zhang et al., 2016).
Secondly, as discussed above, using salt or drought inducible promoters to control the expression of Pi transporters or their regulatory factors may be a potential strategy to improve crop PUE under salt or drought stress conditions.
Last but not least, characterization of Pi transporters in the halophytes will be valuable for our understanding on Pi acquisition and distribution under salinity. Some halophytes have evolved adaptive strategies to high salt and simultaneously low Pi habitat, whereas molecular mechanisms underlying the adaptability remain unclear yet. Comparative studies of the function and expression regulation of halophyte and glycophyte Pi transporters are essential to fully understand the plant Pi acquisition and utilization under salinity. E. salsugineum is well known as a model plant for the molecular elucidation of abiotic stress tolerance (Inan and Zhu, 2004; Amtmann et al., 2005; Kant et al., 2006). E. salsugineum is also found to be tolerant to nitrogen-limiting conditions by maintaining growth, nitrogen uptake and assimilation (Kant and Rothstein, 2008). Interestingly, it was reported most recently that without significant modification of root architecture, E. salsugineum was shown more acclimated to Pi limitation than Arabidopsis (Velasco et al., 2016). In this study, E. salsugineum and Arabidopsis grown either on medium or on soil were subjected to varying Pi treatments. Compared to the enormous changes in root architecture and shoot biomass under low-Pi treatment in Arabidopsis, E. salsugineum seedlings showed no difference in lateral root density and shoot biomass allocation, while adding NaCl increased lateral root density almost twofold. Furthermore, E. salsugineum seedlings had a higher Pi content than Arabidopsis. Pi deprived soil-grown Arabidopsis plants were stunted with senescing older leaves, whereas E. salsugineum plants were visually indistinguishable from Pi-supplemented plants (Velasco et al., 2016). These results suggested a higher Pi uptake and utilization capacity of E. salsugineum under low-Pi conditions. Interestingly, seven orthologs of AtPHT1;3 were found in E. salsugineum (Figure 1), which may be one of the mechanisms underlying the higher Pi uptake in E. salsugineum than Arabidopsis. Further investigations indicated that some Pi starvation associated genes including EsPHT1;4 were up-regulated in Pi-deprived E. salsugineum plants, while EsPHR1, EsWRKY75, and EsRNS1 showed constitutively higher expression levels relative to those in Arabidopsis regardless of external Pi (Velasco et al., 2016). These results suggested that specific regulatory mechanism of Pi transporters might be one of the adaptive strategies of E. salsugineum to high salt and simultaneously low Pi habitat. Further comparative expression patterns analysis of Pi transporters between E. salsugineum and Arabidopsis under salt stress and low-Pi conditions will help to identify members that are involved in Pi uptake and distribution under salinity conditions, which can be valuable for improving crop yields challenged by increasing soil salinization and shrinking farmland.
Conclusion
Plant Pi transporters play an essential role in Pi acquisition and distribution. PHT1 transporters are mainly involved in Pi uptake from soil and translocation, and they can be regulated at transcriptional, post-transcriptional and post-translational levels. PHT2/3/4/5 family participate in Pi transport within subcellular compartments, playing important roles in maintaining cytosolic Pi homeostasis, however, their regulatory mechanism is largely unknown yet. Although some PHT1 transporters and regulatory factors have been confirmed useful candidates for breeding crops with improved yields in Pi-limited soils, induced Pi toxicity as a result of PT overexpression is still a serious problem limiting the application of these proteins. Furthermore, many studies suggested that soil substrate (Pi) availability rather than transporter activity may be the limiting step in Pi-limited soil. In these respects, more attention should be paid to intracellular Pi transporters in the future to improve the crop PUE instead of Pi uptake in Pi-limited soils. In addition, three potential strategies for improving crop PUE under salt or drought stress conditions are provided, among which E. salsugineum is suggested as an appropriate system for studying plant Pi uptake and utilization under salinity and for identifying candidate genes that may be valuable for breeding crops with high PUE under salinized soils. Besides, Pi transporter may also be potential candidates for application in phytoremediation of arsenic contaminated soils, considering the roles of many PHT1s in plant AsV uptake.
Author Contributions
YL and SL conceived the manuscript. DW and SL wrote the manuscript. PJ and YL revised and checked the manuscript. All authors approved the final manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (grant no. 31270421).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00817/full#supplementary-material
References
Ai, P., Sun, S., Zhao, J., Fan, X., Xin, W., Guo, Q., et al. (2009). Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 57, 798–809. doi: 10.1111/j.1365-313X.2008.03726.x
Amtmann, A., Bohnert, H. J., and Bressan, R. A. (2005). Abiotic stress and plant genome evolution. Search for new models. Plant Physiol. 138, 127–130. doi: 10.1104/pp.105.059972
Ayadi, A., David, P., Arrighi, J. F., Chiarenza, S., Thibaud, M. C., Nussaume, L., et al. (2015). Reducing the genetic redundancy of Arabidopsis PHOSPHATE TRANSPORTER1 transporters to study phosphate uptake and signaling. Plant Physiol. 167, 1511–1526. doi: 10.1104/pp.114.252338
Aziz, T., Finnegan, P. M., Lambers, H., and Jost, R. (2014). Organ-specific phosphorus-allocation patterns and transcript profiles linked to phosphorus efficiency in two contrasting wheat genotypes. Plant Cell Environ. 37, 943–960. doi: 10.1111/pce.12210
Baek, S. H., Chung, I. M., and Yun, S. J. (2001). Molecular cloning and characterization of a tobacco leaf cDNA encoding a phosphate transporter. Mol. Cells 11, 1–6.
Bayle, V., Arrighi, J. F., Creff, A., Nespoulous, C., Vialaret, J., Rossignol, M., et al. (2011). Arabidopsis thaliana high-affinity phosphate transporters exhibit multiple levels of posttranslational regulation. Plant Cell 23, 1523–1535. doi: 10.1105/tpc.110.081067
Bun-Ya, M., Nishimura, M., Harashima, S., and Oshima, Y. (1991). The PHO84 gene of Saccharomyces cerevisiae encodes an inorganic phosphate transporter. Mol. Cell Biol. 11, 3229–3238. doi: 10.1128/mcb.11.6.3229
Bustos, R., Castrillo, G., Linhares, F., Puga, M. I., Rubio, V., Pérez-Pérezet, J., et al. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 6:e1001102. doi: 10.1371/journal.pgen.1001102
Ceasar, S. A., Hodge, A., Baker, A., and Baldwin, S. A. (2014). Phosphate concentration and arbuscular mycorrhizal colonisation influence the growth, yield and expression of twelve PHT1 family phosphate transporters in foxtail millet (Setaria italica). PLoS ONE 9:e108459. doi: 10.1371/journal.pone.0108459
Chen, A., Chen, X., Wang, H., Liao, D., Gu, M., Qu, H., et al. (2014). Genome-wide investigation and expression analysis suggest diverse roles and genetic redundancy of Pht1 family genes in response to Pi deficiency in tomato. BMC Plant Biol. 14:61. doi: 10.1186/1471-2229-14-61
Chen, A., Gu, M., Sun, S., Zhu, L., Hong, S., and Xu, G. (2011). Identification of two conserved cis-acting elements, MYCS and P1BS, involved in the regulation of mycorrhiza-activated phosphate transporters in eudicot species. New Phytol. 189, 1157–1169. doi: 10.1111/j.1469-8137.2010.03556.x
Chen, A., Hu, J., Sun, S., and Xu, G. (2007). Conservation and divergence of both phosphate- and mycorrhiza-regulated physiological responses and expression patterns of phosphate transporters in solanaceous species. New Phytol. 173, 817–831. doi: 10.1111/j.1469-8137.2006.01962.x
Chen, J., Liu, Y., Ni, J., Wang, Y., Bai, Y., Shi, J., et al. (2011). OsPHF1 regulates the plasma membrane localization of low- and high-affinity inorganic phosphate transporters and determines inorganic phosphate uptake and translocation in rice. Plant Physiol. 157, 269–278. doi: 10.1104/pp.111.181669
Chiou, T. J., Aung, K., Lin, S. I., Wu, C. C., Chiang, S. F., and Su, C. L. (2006). Regulation of phosphate homeostasis by microRNA in Arabidopsis. Plant Cell 18, 412–421. doi: 10.1105/tpc.105.038943
Cubero, B., Nakagawa, Y., Jiang, X. Y., Miura, K. J., Li, F., Raghothama, K. G., et al. (2009). The phosphate transporter PHT4;6 is a determinant of salt tolerance that is localized to the Golgi apparatus of Arabidopsis. Mol. Plant 2, 535–552. doi: 10.1093/mp/ssp013
Dai, X., Wang, Y., and Zhang, W. H. (2016). OsWRKY74, a WRKY transcription factor, modulates tolerance to phosphate starvation in rice. J. Exp. Bot. 67, 947–960. doi: 10.1093/jxb/erv515
Daram, P., Brunner, S., Rausch, C., Steiner, C., Amrhein, N., and Bucher, M. (1999). Pht2;1 encodes a low-affinity phosphate transporter from Arabidopsis. Plant Cell 11, 2153–2166. doi: 10.1105/tpc.11.11.2153
Devaiah, B. N., Karthikeyan, A. S., and Raghothama, K. G. (2007). WRKY75 transcription factor is a modulator of phosphate acquisition and root development in Arabidopsis. Plant Physiol. 143, 1789–1801. doi: 10.1104/pp.106.093971
Devaiah, B. N., Madhuvanthi, R., Karthikeyan, A. S., and Raghothama, K. G. (2009). Phosphate starvation responses and gibberellic acid biosynthesis are regulated by the MYB62 transcription factor in Arabidopsis. Mol. Plant 2, 43–58. doi: 10.1093/mp/ssn081
Davies, T. G. E., Ying, J., Xu, Q., Li, Z. S., Li, J., and Gordon-Weeks, R. (2002). Expression analysis of putative high-affinity phosphate transporters in Chinese winter wheats. Plant Cell Environ. 25, 1325–1339. doi: 10.1046/j.1365-3040.2002.00913.x
Ding, W., Wang, Y., Fang, W., Gao, S., Li, X., and Xiao, K. (2016). TaZAT8, a C2H2-ZFP type transcription factor gene in wheat, plays critical roles in mediating tolerance to Pi deprivation through regulating P acquisition, ROS homeostasis, and root system establishment. Physiol. Plant. 158, 297–311. doi: 10.1111/ppl.12467
Djami-Tchatchou, A. T., Sanan-Mishra, N., Ntushelo, K., and Dubery, I. A. (2017). Functional roles of microRNAs in agronomically important plants-potential as targets for crop improvement and protection. Front. Plant Sci 8:378. doi: 10.3389/fpls.2017.00378
Duan, J., Tian, H., Drijber, R. A., and Gao, Y. (2015). Systemic and local regulation of phosphate and nitrogen transporter genes by arbuscular mycorrhizal fungi in roots of winter wheat (Triticum aestivum L.). Plant Physiol. Biochem. 96, 199–208. doi: 10.1016/j.plaphy.2015.08.006
Duan, K., Yi, K., Dang, L., Huang, H., Wu, W., and Wu, P. (2008). Characterization of a sub-family of Arabidopsis genes with the SPX domain reveals their diverse functions in plant tolerance to phosphorus starvation. Plant J. 54, 965–975. doi: 10.1111/j.1365-313X.2008.03460.x
Fan, C., Wang, X., Hu, R., Wang, Y., Xiao, C., Jiang, Y., et al. (2013). The pattern of Phosphate transporter 1 genes evolutionary divergence in Glycine max L. BMC Plant Biol. 13:48. doi: 10.1186/1471-2229-13-48
Fujii, H., Chiou, T. J., Lin, S. I., Aung, K., and Zhu, J. K. (2005). A miRNA involved in phosphate-starvation response in Arabidopsis. Curr. Biol. 15, 2038–2043. doi: 10.1016/j.cub.2005.10.016
Gao, N., Qiang, X. M., Zhai, B. N., Min, J., and Shi, W. M. (2015). Transgenic tomato overexpressing ath-miR399d improves growth under abiotic stress conditions. Russ. J. Plant Physiol. 62, 360–366. doi: 10.1134/s1021443715030061
Gao, N., Su, Y., Min, J., Shen, W., and Shi, W. (2009). Transgenic tomato overexpressing ath-miR399d has enhanced phosphorus accumulation through increased acid phosphatase and proton secretion as well as phosphate transporters. Plant Soil 334, 123–136. doi: 10.1007/s11104-009-0219-3
Glassop, D., Smith, S. E., and Smith, F. W. (2005). Cereal phosphate transporters associated with the mycorrhizal pathway of phosphate uptake into roots. Planta 222, 688–698. doi: 10.1007/s00425-005-0015-0
González, E., Solano, R., Rubio, V., Leyva, A., and Paz-Ares, J. (2005). PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 is a plant-specific SEC12-related protein that enables the endoplasmic reticulum exit of a high-affinity phosphate transporter in Arabidopsis. Plant Cell 17, 3500–3512. doi: 10.1105/tpc.105.036640
Gordon-Weeks, R., Tong, Y., Davies, T. G., and Leggewie, G. (2003). Restricted spatial expression of a high-affinity phosphate transporter in potato roots. J. Cell Sci. 116, 3135–3144. doi: 10.1242/jcs.00615
Grattan, S. R., and Grieve, C. M. (1999). Salinity mineral nutrient relations in horticultural crops. Sci. Hortic. 78, 127–157.
Guo, B., Jin, Y., Wussler, C., Blancaflor, E. B., Motes, C. M., and Versaw, W. K. (2008). Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 177, 889–898. doi: 10.1111/j.1469-8137.2007.02331.x
Guo, C., Zhao, X., Liu, X., Zhang, L., Gu, J., Li, X., et al. (2013). Function of wheat phosphate transporter gene TaPHT2;1 in Pi translocation and plant growth regulation under replete and limited Pi supply conditions. Planta 237, 1163–1178. doi: 10.1007/s00425-012-1836-2
Hackenberg, M., Shi, B. J., Gustafson, P., and Langridge, P. (2013). Characterization of phosphorus-regulated miR399 and miR827 and their isomirs in barley under phosphorus-sufficient and phosphorus-deficient conditions. BMC Plant Biol. 13:214. doi: 10.1186/1471-2229-13-214
Hamburger, D. (2002). Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the Xylem. Plant Cell 14, 889–902. doi: 10.1105/tpc.000745
Hamel, P., Saint-Georges, Y., de Pinto, B., Lachacinski, N., Altamura, N., and Dujardin, G. (2004). Redundancy in the function of mitochondrial phosphate transport in Saccharomyces cerevisiae and Arabidopsis thaliana. Mol. Microbiol. 51, 307–317. doi: 10.1046/j.1365-2958.2003.03810.x
Hassler, S., Jung, B., Lemke, L., Novak, O., Strnad, M., Martinoia, E., et al. (2016). Function of the Golgi-located phosphate transporter PHT4;6 is critical for senescence-associated processes in Arabidopsis. J. Exp. Bot. 67, 4671–4684. doi: 10.1093/jxb/erw249
Holford, I. C. R. (1997). Soil phosphorus: its measurement, and its uptake by plants. Soil Res. 35, 227–239. doi: 10.1071/S96047
Hong, J. J., Park, Y.-S., Bravo, A., Bhattarai, K. K., Daniels, D. A., and Harrison, M. J. (2012). Diversity of morphology and function in arbuscular mycorrhizal symbioses in Brachypodium distachyon. Planta 236, 851–865. doi: 10.1007/s00425-012-1677-z
Huang, T. K., Han, C. L., Lin, S. I., Chen, Y. J., Tsai, Y. C., Chen, Y. R., et al. (2013). Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25, 4044–4060. doi: 10.1105/tpc.113.115998
Inan, G., and Zhu, J. K. (2004). Salt cress. A halophyte and cryophyte Arabidopsis relative model system and its applicability to molecular genetic analyses of growth and development of extremophiles. Plant Physiol. 135, 1718–1737. doi: 10.1104/pp.104.041723
Javot, H., Penmetsa, R. V., Terzaghi, N., Cook, D. R., and Harrison, M. J. (2007a). A Medicago truncatula phosphate transporter indispensable for the arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 104, 1720–1725. doi: 10.1073/pnas.0608136104
Javot, H., Pumplin, N., and Harrison, M. J. (2007b). Phosphate in the arbuscular mycorrhizal symbiosis: transport properties and regulatory roles. Plant Cell Environ. 30, 310–322. doi: 10.1111/j.1365-3040.2006.01617.x
Jia, F., Wan, X., Zhu, W., Sun, D., Zheng, C., Liu, P., et al. (2015). Overexpression of Mitochondrial Phosphate Transporter 3 severely hampers plant development through regulating mitochondrial function in Arabidopsis. PLoS ONE 10:e0129717. doi: 10.1371/journal.pone.0129717
Jia, H., Ren, H., Gu, M., Zhao, J., Sun, S., Zhang, X., et al. (2011). The phosphate transporter gene OsPht1;8 is involved in phosphate homeostasis in rice. Plant Physiol. 156, 1164–1175. doi: 10.1104/pp.111.175240
Jin, J., Wang, G., Liu, X., Pan, X., Herbert, S. J., and Tang, C. (2006). Interaction between phosphorus nutrition and drought on grain yield, and assimilation of phosphorus and nitrogen in two soybean cultivars differing in protein concentration in grains. J. Plant Nutr. 29, 1433–1449. doi: 10.1080/01904160600837089
Kai, M., Takazumi, K., Adachi, H., Wasaki, J., Shinano, T., and Osaki, M. (2002). Cloning and characterization of four phosphate transporter cDNAs in tobacco. Plant Sci. 163, 837–846. doi: 10.1016/S0168-9452(02)00233-9
Kant, S., Kant, P., Raveh, E., and Barak, S. (2006). Evidence that differential gene expression between the halophyte, Thellungiella halophila, and Arabidopsis thaliana is responsible for higher levels of the compatible osmolyte proline and tight control of Na+ uptake in T. halophila. Plant Cell Environ. 29, 1220–1234. doi: 10.1111/j.1365-3040.2006.01502.x
Kant, S., and Rothstein, S. J. (2008). The Arabidopsis halophytic relative Thellungiella halophila tolerates nitrogen-limiting conditions by maintaining growth, nitrogen uptake, and assimilation. Plant Physiol. 147, 1168–1180. doi: 10.1104/pp.108.118125
Karandashov, V., and Bucher, M. (2005). Symbiotic phosphate transport in arbuscular mycorrhizas. Trends Plant Sci. 10, 22–29. doi: 10.1016/j.tplants.2004.12.003
Kiiskinen, M., Korhonen, M., and Kangasjarvi, J. (1997). Isolation and characterization of cDNA for a plant mitochondrial phosphate translocator (Mpt1): ozone stress induces Mpt1 mRNA accumulation in birch (Betula pendula Roth). Plant Mol. Biol. 35, 271–279. doi: 10.1023/A:1005868715571
Kuo, H. F., and Chiou, T. J. (2011). The role of microRNAs in phosphorus deficiency signaling. Plant Physiol. 156, 1016–1024. doi: 10.1104/pp.111.175265
Lapis-Gaza, H. R., Jost, R., and Finnegan, P. M. (2014). Arabidopsis PHOSPHATE TRANSPORTER1 genes PHT1;8 and PHT1;9 are involved in root-to-shoot translocation of orthophosphate. BMC Plant Biol. 14:334. doi: 10.1186/s12870-014-0334-z
Leggewie, G., Willmitzer, L., and Riesmeier, J. W. (1997). Two cDNAs from potato are able to complement a phosphate uptake-deficient yeast mutant: identification of phosphate transporters from higher plants. Plant Cell 9, 381–392. doi: 10.1105/tpc.9.3.381
Li, Y., Zhang, J., Zhang, X., Fan, H., Gu, M., Qu, H., et al. (2015). Phosphate transporter OsPht1;8 in rice plays an important role in phosphorus redistribution from source to sink organs and allocation between embryo and endosperm of seeds. Plant Sci. 230, 23–32. doi: 10.1016/j.plantsci.2014.10.001
Lin, S. I., Santi, C., Jobet, E., Lacut, E., El Kholti, N., Karlowski, W. M., et al. (2010). Complex regulation of two target genes encoding SPX-MFS proteins by rice miR827 in response to phosphate starvation. Plant Cell Physiol. 51, 2119–2131. doi: 10.1093/pcp/pcq170
Lin, W. Y., Huang, T. K., and Chiou, T. J. (2013). Nitrogen limitation adaptation, a target of microRNA827, mediates degradation of plasma membrane-localized phosphate transporters to maintain phosphate homeostasis in Arabidopsis. Plant Cell 25, 4061–4074. doi: 10.1105/tpc.113.116012
Liu, F., Chang, X. J., Ye, Y., Xie, W. B., Wu, P., and Lian, X. M. (2011). Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice. Mol. Plant 4, 1105–1122. doi: 10.1093/mp/ssr058
Liu, F., Wang, Z., Ren, H., Shen, C., Li, Y., Ling, H. Q., et al. (2010). OsSPX1 suppresses the function of OsPHR2 in the regulation of expression of OsPT2 and phosphate homeostasis in shoots of rice. Plant J. 62, 508–517. doi: 10.1111/j.1365-313X.2010.04170.x
Liu, F., Xu, Y., Jiang, H., Jiang, C., Du, Y., Gong, C., et al. (2016). Systematic identification, evolution and expression analysis of the Zea mays PHT1 gene family reveals several new members involved in root colonization by arbuscular mycorrhizal fungi. Int. J. Mol. Sci. 17:930. doi: 10.3390/ijms17060930
Liu, T. Y., Huang, T. K., Yang, S. Y., Hong, Y. T., Huang, S. M., Wang, F. N., et al. (2016). Identification of plant vacuolar transporters mediating phosphate storage. Nat. Commun. 7:11095. doi: 10.1038/ncomms11095
Liu, J., Versaw, W. K., Pumplin, N., Gomez, S. K., Blaylock, L. A., and Harrison, M. J. (2008). Closely related members of the Medicago truncatula PHT1 phosphate transporter gene family encode phosphate transporters with distinct biochemical activities. J. Biol. Chem. 283, 24673–24681. doi: 10.1074/jbc.M802695200
Liu, X. M., Zhao, X. L., Zhang, L. J., Lu, W. J., Li, X. J., and Xiao, K. (2013). TaPht1;4, a high-affinity phosphate transporter gene in wheat (Triticum aestivum), plays an important role in plant phosphate acquisition under phosphorus deprivation. Funct. Plant Biol. 40, 329–341. doi: 10.1071/fp12242
López-Arredondo, D. L., Leyva-González, M. A., González-Morales, S. I., López-Bucio, J., and Herrera-Estrella, L. (2014). Phosphate nutrition: improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 65, 95–123. doi: 10.1146/annurev-arplant-050213-035949
Loth-Pereda, V., Orsini, E., Courty, P. E., Lota, F., Kohler, A., Diss, L., et al. (2011). Structure and expression profile of the phosphate Pht1 transporter gene family in mycorrhizal Populus trichocarpa. Plant Physiol. 156, 2141–2154. doi: 10.1104/pp.111.180646
Lynch, J. P. (2007). Roots of the second green revolution. Aust. J. Bot. 55, 493–512. doi: 10.1071/BT06118
Martinez, V., and Lauchli, A. (1994). Salt-induced inhibition of phosphate uptake in plants of cotton (Gossypium hirsutum L.). New Phytol. 126, 609–614. doi: 10.1111/j.1469-8137.1994.tb02955.x
Mimura, T. (1999). Regulation of phosphate transport and homeostasis in plant cells. Int. Rev. Cytol. 191, 149–200. doi: 10.1016/s0074-7696(08)60159-x
Ming, F., Mi, G. H., Lu, Q., Yin, S., Zhang, S. S., Guo, B., et al. (2005). Cloning and characterization of cDNA for the Oryza sativa phosphate transporter. Cell. Mol. Biol. Lett. 10, 401–411.
Misson, J., Thibaud, M. C., Bechtold, N., Raghothama, K., and Nussaume, L. (2004). Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 55, 727–741. doi: 10.1007/s11103-004-1965-5
Mlodzińska, E., and Zboińska, M. (2016). Phosphate uptake and allocation - a closer look at Arabidopsis thaliana L. and Oryza sativa L. Front. Plant Sci. 7:1198. doi: 10.3389/fpls.2016.01198
Muchhal, U. S., Pardo, J. M., and Raghothama, K. G. (1996). Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 93, 10519–10523. doi: 10.1073/pnas.93.19.10519
Mudge, S. R., Rae, A. L., Diatloff, E., and Smith, F. W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 31, 341–353. doi: 10.1046/j.1365-313X.2002.01356.x
Nagarajan, V. K., Jain, A., Poling, M. D., Lewis, A. J., Raghothama, K. G., and Smith, A. P. (2011). Arabidopsis Pht1;5 mobilizes phosphate between source and sink organs and influences the interaction between phosphate homeostasis and ethylene signaling. Plant Physiol. 156, 1149–1163. doi: 10.1104/pp.111.174805
Nagy, R., Karandashov, V., Chague, V., Kalinkevich, K., Tamasloukht, M., Xu, G., et al. (2005). The characterization of novel mycorrhiza-specific phosphate transporters from Lycopersicon esculentum and Solanum tuberosum uncovers functional redundancy in symbiotic phosphate transport in solanaceous species. Plant J. 42, 236–250. doi: 10.1111/j.1365-313X.2005.02364.x
Nakamori, K., Takabatake, R., Umehara, Y., Kouchi, H., Izui, K., and Hata, S. (2002). Cloning, functional expression, and mutational analysis of a cDNA for Lotus japonicus mitochondrial phosphate transporter. Plant Cell Physiol. 43, 1250–1253. doi: 10.1093/pcp/pcf141
Nestler, J., Keyes, S. D., and Wissuwa, M. (2016). Root hair formation in rice (Oryza sativa L.) differs between root types and is altered in artificial growth conditions. J. Exp. Bot. 67, 3699–3708. doi: 10.1093/jxb/erw115
Nussaume, L., Kanno, S., Javot, H., Marin, E., Pochon, N., Ayadi, A., et al. (2011). Phosphate import in plants: focus on the PHT1 transporters. Front. Plant Sci. 2:83. doi: 10.3389/fpls.2011.00083
Oelkers, E. H., and Valsami-Jones, E. (2008). Phosphate mineral reactivity and global sustainability. Elements 4, 83–87. doi: 10.2113/gselements.4.2.83
Oono, Y., Kawahara, Y., Yazawa, T., Kanamori, H., Kuramata, M., Yamagata, H., et al. (2013). Diversity in the complexity of phosphate starvation transcriptomes among rice cultivars based on RNA-Seq profiles. Plant Mol. Biol. 83, 523–537. doi: 10.1007/s11103-013-0106-4
Ouyang, X., Hong, X., Zhao, X., Zhang, W., He, X., Ma, W., et al. (2016). Knock out of the PHOSPHATE2 gene TaPHO2-A1 improves phosphorus uptake and grain yield under low phosphorus conditions in common wheat. Sci. Rep. 6:29850. doi: 10.1038/srep29850
Pariasca-Tanaka, J., Satoh, K., Rose, T., Mauleon, R., and Wissuwa, M. (2009). Stress response versus stress tolerance: a transcriptome analysis of two rice lines contrasting in tolerance to phosphorus deficiency. Rice 2, 167–185. doi: 10.1007/s12284-009-9032-0
Paszkowski, U., Kroken, S., Roux, C., and Briggs, S. P. (2002). Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 99, 13324–13329. doi: 10.1073/pnas.202474599
Preuss, C. P., Huang, C. Y., Gilliham, M., and Tyerman, S. D. (2010). Channel-like characteristics of the low-affinity barley phosphate transporter PHT1;6 when expressed in Xenopus oocytes. Plant Physiol. 152, 1431–1441. doi: 10.1104/pp.109.152009
Preuss, C. P., Huang, C. Y., and Tyerman, S. D. (2011). Proton-coupled high-affinity phosphate transport revealed from heterologous characterization in Xenopus of barley-root plasma membrane transporter, HvPHT1;1. Plant Cell Environ. 34, 681–689. doi: 10.1111/j.1365-3040.2010.02272.x
Qin, L., Guo, Y., Chen, L., Liang, R., Gu, M., Xu, G., et al. (2012). Functional characterization of 14 Pht1 family genes in yeast and their expressions in response to nutrient starvation in soybean. PLoS ONE 7:e47726. doi: 10.1371/journal.pone.0047726
Qu, B., He, X., Wang, J., Zhao, Y., Teng, W., Shao, A., et al. (2015). A wheat CCAAT box-binding transcription factor increases the grain yield of wheat with less fertilizer input. Plant Physiol. 167, 411–423. doi: 10.1104/pp.114.246959
Rae, A. L., Cybinski, D. H., Jarmey, J. M., and Smith, F. W. (2003). Characterization of two phosphate transporters from barley; evidence for diverse function and kinetic properties among members of the Pht1 family. Plant Mol. Biol. 53, 27–36. doi: 10.1023/B:PLAN.0000009259.75314.15
Rae, A. L., Jarmey, J. M., Mudge, S. R., and Smith, F. W. (2004). Over-expression of a high-affinity phosphate transporter in transgenic barley does not enhance phosphorus uptake rates. Funct. Plant Biol. 31, 141–148. doi: 10.1071/FP03159
Raghothama, K. G. (1999). Phosphate acquision. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 665–693. doi: 10.1146/annurev.arplant.50.1.665
Rausch, C., and Bucher, M. (2002). Molecular mechanisms of phosphate transport in plants. Planta 216, 23–37. doi: 10.1007/s00425-002-0921-3
Rausch, C., Daram, P., Brunner, S., Jansa, J., Laloi, M., Leggewie, G., et al. (2001). A phosphate transporter expressed in arbuscule-containing cells in potato. Nature 414, 462–470. doi: 10.1038/35106601
Rausch, C., Zimmermann, P., Amrhein, N., and Bucher, M. (2004). Expression analysis suggests novel roles for the plastidic phosphate transporter Pht2;1 in auto- and heterotrophic tissues in potato and Arabidopsis. Plant J. 39, 13–28. doi: 10.1111/j.1365-313X.2004.02106.x
Remy, E., Cabrito, T. R., Batista, R. A., Teixeira, M. C., Sá-Correia, I., and Duque, P. (2012). The Pht1;9 and Pht1;8 transporters mediate inorganic phosphate acquisition by the Arabidopsis thaliana root during phosphorus starvation. New Phytol. 195, 356–371. doi: 10.1111/j.1469-8137.2012.04167.x
Rosewarne, G. M., Barker, S. J., Smith, S. E., Smith, F. A., and Schachtman, D. P. (1999). A Lycopersicon esculentum phosphate transporter (LePT1) involved in phosphorus uptake from a vesicular-arbuscular mycorrhizal fungus. New Phytol. 144, 507–516. doi: 10.1046/j.1469-8137.1999.00536.x
Rubio, V., Linhares, F., Solano, R., Martin, A. C., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. doi: 10.1101/gad.204401
Sawers, R. J., Svane, S. F., Quan, C., Gronlund, M., Wozniak, B., Gebreselassie, M. N., et al. (2017). Phosphorus acquisition efficiency in arbuscular mycorrhizal maize is correlated with the abundance of root-external hyphae and the accumulation of transcripts encoding PHT1 phosphate transporters. New Phytol. 214, 632–643. doi: 10.1111/nph.14403
Schünmann, P. H., Richardson, A. E., Smith, F. W., and Delhaize, E. (2004a). Characterization of promoter expression patterns derived from the Pht1 phosphate transporter genes of barley (Hordeum vulgare L.). J. Exp. Bot. 55, 855–865. doi: 10.1093/jxb/erh103
Schünmann, P. H., Richardson, A. E., Vickers, C. E., and Delhaize, E. (2004b). Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiol. 136, 4205–4214. doi: 10.1104/pp.104.045823
Sha, A. H., Wu, H., Fu, X. M., Zhang, Q. L., Guo, Q. L., Chen, Y. H., et al. (2014). Isolation and expression analysis of the soybean GmPic gene. Genet. Mol. Res. 13, 4380–4391. doi: 10.4238/2014.June.11.2
Shi, S. L., Wang, D. F., Yan, Y., Zhang, F., Wang, H. D., Gu, M., et al. (2013). Function of phosphate transporter OsPHT2;1 in improving phosphate utilization in rice. Chin. J. Rice Sci. 27, 457–465.
Shin, H., Shin, H. S., Dewbre, G. R., and Harrison, M. J. (2004). Phosphate transport in Arabidopsis: Pht1;1 and Pht1;4 play a major role in phosphate acquisition from both low- and high-phosphate environments. Plant J. 39, 629–642. doi: 10.1111/j.1365-313X.2004.02161.x
Shukla, V., Kaur, M., Aggarwal, S., Bhati, K. K., Kaur, J., Mantri, S., et al. (2016). Tissue specific transcript profiling of wheat phosphate transporter genes and its association with phosphate allocation in grains. Sci. Rep. 6:39293. doi: 10.1038/srep39293
Sisaphaithong, T., Kondo, D., Matsunaga, H., Kobae, Y., and Hata, S. (2012). Expression of plant genes for arbuscular mycorrhiza-inducible phosphate transporters and fungal vesicle formation in sorghum, barley, and wheat roots. Biosci. Biotechnol. Biochem. 76, 2364–2367. doi: 10.1271/bbb.120782
Smith, A. P., Jain, A., Deal, R. B., Nagarajan, V. K., Poling, M. D., Raghothama, K. G., et al. (2010). Histone H2A.Z regulates the expression of several classes of phosphate starvation response genes but not as a transcriptional activator. Plant Physiol. 152, 217–225. doi: 10.1104/pp.109.145532
Sun, S., Gu, M., Cao, Y., Huang, X., Zhang, X., Ai, P., et al. (2012). A constitutive expressed phosphate transporter, OsPht1;1, modulates phosphate uptake and translocation in phosphate-replete rice. Plant Physiol. 159, 1571–1581. doi: 10.1104/pp.112.196345
Takabatake, R., Hata, S., Taniguchi, M., Kouchi, H., Sugiyama, T., and Izui, K. (1999). Isolation and characterization of cDNAs encoding mitochondrial phosphate transporters in soybean, maize, rice, and Arabidopis. Plant Mol. Biol. 40, 479–486. doi: 10.1023/A:1006285009435
Tamura, Y., Kobae, Y., Mizuno, T., and Hata, S. (2012). Identification and expression analysis of arbuscular mycorrhiza-inducible phosphate transporter genes of soybean. Biosci. Biotechnol. Biochem. 76, 309–313. doi: 10.1271/bbb.110684
Tian, H., Drijber, R. A., Li, X., Miller, D. N., and Wienhold, B. J. (2013). Arbuscular mycorrhizal fungi differ in their ability to regulate the expression of phosphate transporters in maize (Zea mays L.). Mycorrhiza 23, 507–514. doi: 10.1007/s00572-013-0491-1
Tian, H., Yuan, X., Duan, J., Li, W., Zhai, B., and Gao, Y. (2017). Influence of nutrient signals and carbon allocation on the expression of phosphate and nitrogen transporter genes in winter wheat (Triticum aestivum L.) roots colonized by arbuscular mycorrhizal fungi. PLoS ONE 12:e0172154. doi: 10.1371/journal.pone.0172154
Velasco, V. M. E., Mansbridge, J., Bremner, S., Carruthers, K., Summers, P. S., Sung, W. W. L., et al. (2016). Acclimation of the crucifer Eutrema salsugineum to phosphate limitation is associated with constitutively high expression of phosphate-starvation genes. Plant Cell Environ. 39, 1818–1834. doi: 10.1111/pce.12750
Versaw, W. K., and Harrison, M. J. (2002). A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14, 1751–1766. doi: 10.1105/tpc.002220
Walder, F., Brule, D., Koegel, S., Wiemken, A., Boller, T., and Courty, P. E. (2015). Plant phosphorus acquisition in a common mycorrhizal network: regulation of phosphate transporter genes of the Pht1 family in sorghum and flax. New Phytol. 205, 1632–1645. doi: 10.1111/nph.13292
Wang, C., Huang, W., Ying, Y., Li, S., Secco, D., Tyerman, S., et al. (2012). Functional characterization of the rice SPX-MFS family reveals a key role of OsSPX-MFS1 in controlling phosphate homeostasis in leaves. New Phytol. 196, 139–148. doi: 10.1111/j.1469-8137.2012.04227.x
Wang, C., Yue, W., Ying, Y., Wang, S., Secco, D., Liu, Y., et al. (2015). Rice SPX-Major Facility Superfamily3, a vacuolar phosphate efflux transporter, is involved in maintaining phosphate homeostasis in rice. Plant Physiol. 169, 2822–2831. doi: 10.1104/pp.15.01005
Wang, F., Rose, T., Jeong, K., Kretzschmar, T., and Wissuwa, M. (2016). The knowns and unknowns of phosphorus loading into grains, and implications for phosphorus efficiency in cropping systems. J. Exp. Bot. 67, 1221–1229. doi: 10.1093/jxb/erv517
Wang, H., Xu, Q., Kong, Y. H., Chen, Y., Duan, J. Y., Wu, W. H., et al. (2014). Arabidopsis WRKY45 transcription factor activates PHOSPHATE TRANSPORTER1;1 expression in response to phosphate starvation. Plant Physiol. 164, 2020–2029. doi: 10.1104/pp.113.235077
Wang, J., Sun, J., Miao, J., Guo, J., Shi, Z., He, M., et al. (2013). A phosphate starvation response regulator Ta-PHR1 is involved in phosphate signalling and increases grain yield in wheat. Ann. Bot. 111, 1139–1153. doi: 10.1093/aob/mct080
Wang, P., Zhang, W., Mao, C., Xu, G., and Zhao, F. J. (2016). The role of OsPT8 in arsenate uptake and varietal difference in arsenate tolerance in rice. J. Exp. Bot. 67, 6051–6059. doi: 10.1093/jxb/erw362
Wang, S., Zhang, S., Sun, C., Xu, Y., Chen, Y., Yu, C., et al. (2014). Auxin response factor (OsARF12), a novel regulator for phosphate homeostasis in rice (Oryza sativa). New Phytol. 201, 91–103. doi: 10.1111/nph.12499
Wang, X., Wang, Y., Pineros, M. A., Wang, Z., Wang, W., Li, C., et al. (2014). Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant Cell Environ. 37, 1159–1170. doi: 10.1111/pce.12224
Waraich, E. A., Ahmad, R., Saifullah, U., Ashraf, M. Y., and Ehsanullah. (2011). Role of mineral nutrition in alleviation of drought stress in plants. Aust. J. Crop Sci. 5, 764–777.
Wissuwa, M. (2005). Combining a modeling with a genetic approach in establishing associations between genetic and physiological effects in relation to phosphorus uptake. Plant Soil 269, 57–68. doi: 10.1007/s11104-004-2026-1
Xie, X., Huang, W., Liu, F., Tang, N., Liu, Y., Lin, H., et al. (2013). Functional analysis of the novel mycorrhiza-specific phosphate transporter AsPT1 and PHT1 family from Astragalus sinicus during the arbuscular mycorrhizal symbiosis. New Phytol. 198, 836–852. doi: 10.1111/nph.12188
Yan, W., Chen, G. H., Yang, L. F., Gai, J. Y., and Zhu, Y. L. (2014). Overexpression of the rice phosphate transporter gene OsPT6 enhances tolerance to low phosphorus stress in vegetable soybean. Sci. Hortic. 177, 71–76. doi: 10.1016/j.scienta.2014.07.037
Yang, S. Y., Gronlund, M., Jakobsen, I., Grotemeyer, M. S., Rentsch, D., Miyao, A., et al. (2012). Nonredundant regulation of rice arbuscular mycorrhizal symbiosis by two members of the phosphate transporter1 gene family. Plant Cell 24, 4236–4251. doi: 10.1105/tpc.112.104901
Yang, T., Hao, L., Yao, S., Zhao, Y., Lu, W., and Xiao, K. (2016). TabHLH1, a bHLH-type transcription factor gene in wheat, improves plant tolerance to Pi and N deprivation via regulation of nutrient transporter gene transcription and ROS homeostasis. Plant Physiol. Biochem. 104, 99–113. doi: 10.1016/j.plaphy.2016.03.023
Ye, Y., Yuan, J., Chang, X., Yang, M., Zhang, L., Lu, K., et al. (2015). The phosphate transporter gene OsPht1;4 is involved in phosphate homeostasis in rice. PLoS ONE 10:e0126186. doi: 10.1016/j.scienta.2014.07.037
Yue, W., Ying, Y., Wang, C., Zhao, Y., Dong, C., Whelan, J., et al. (2017). OsNLA1, a RING-type ubiquitin ligase, maintains phosphate homeostasis in Oryza sativa via degradation of phosphate transporters. Plant J. doi: 10.1111/tpj.13516 [Epub ahead of print].
Zhang, C., Meng, S., Li, M., and Zhao, Z. (2016). Genomic identification and expression analysis of the phosphate transporter gene family in poplar. Front. Plant Sci. 7:1398. doi: 10.3389/fpls.2016.01398
Zhang, F., Sun, Y., Pei, W., Jain, A., Sun, R., Cao, Y., et al. (2015). Involvement of OsPht1;4 in phosphate acquisition and mobilization facilitates embryo development in rice. Plant J. 82, 556–569. doi: 10.1111/tpj.12804
Zhang, F., Wu, X. N., Zhou, H. M., Wang, D. F., Jiang, T. T., Sun, Y. F., et al. (2014). Overexpression of rice phosphate transporter gene OsPT6 enhances phosphate uptake and accumulation in transgenic rice plants. Plant Soil 384, 259–270. doi: 10.1007/s11104-014-2168-8
Zhang, J., Guo, S. G., Ren, Y., Zhang, H. Y., Gong, G. Y., Zhou, M., et al. (2017). High-level expression of a novel chromoplast phosphate transporter ClPHT4;2 is required for flesh color development in watermelon. New Phytol. 213, 1208–1221. doi: 10.1111/nph.14257
Zhou, J., Jiao, F., Wu, Z., Li, Y., Wang, X., He, X., et al. (2008). OsPHR2 is involved in phosphate-starvation signaling and excessive phosphate accumulation in shoots of plants. Plant Physiol. 146, 1673–1686. doi: 10.1104/pp.107.111443
Keywords: crop, Eutrema salsugineum, drought, genetic modification, low phosphate, phosphate transporter, regulation, salinity
Citation: Wang D, Lv S, Jiang P and Li Y (2017) Roles, Regulation, and Agricultural Application of Plant Phosphate Transporters. Front. Plant Sci. 8:817. doi: 10.3389/fpls.2017.00817
Received: 21 January 2017; Accepted: 01 May 2017;
Published: 18 May 2017.
Edited by:
Matthias Wissuwa, Japan International Research Center for Agricultural Sciences, JapanReviewed by:
Marie-Christine Thibaud, Commissariat à l’énergie atomique et aux énergies alternatives, FranceFangsen Xu, Huazhong Agricultural University, China
Copyright © 2017 Wang, Lv, Jiang and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yinxin Li, eXhsaUBpYmNhcy5hYy5jbg==
†These authors have contributed equally to this work.
 Duoliya Wang1,2†
Duoliya Wang1,2† Yinxin Li
Yinxin Li