- 1Soybean Research Institute, Key Laboratory of Soybean Biology of Chinese Education Ministry, Northeast Agricultural University, Harbin, China
- 2Center for Plant Biotechnology, College of Agronomy, Jilin Agricultural University, Changchun, China
- 3First Affiliated Hospital of Harbin Medical University, Harbin, China
- 4Soybean Research Institute, Key Laboratory of Soybean Cultivation of Ministry of Agriculture, Heilongjiang Academy of Agricultural Sciences, Harbin, China
- 5Heihe Branch of Heilongjiang Academy of Agricultural Sciences, Heihe, China
Phytophthora root and stem rot of soybean [Glycine max (L.) Merr.] caused by the oomycete Phytophthora sojae, is a destructive disease worldwide. The molecular mechanism of the soybean response to P. sojae is largely unclear. We report a novel WRKY transcription factor (TF) in soybean, GmWRKY31, in the host response to P. sojae. Overexpression and RNA interference analysis demonstrated that GmWRKY31 enhanced resistance to P. sojae in transgenic soybean plants. GmWRKY31 was targeted to the nucleus, where it bound to the W-box and acted as an activator of gene transcription. Moreover, we determined that GmWRKY31 physically interacted with GmHDL56, which improved resistance to P. sojae in transgenic soybean roots. GmWRKY31 and GmHDL56 shared a common target GmNPR1 which was induced by P. sojae. Overexpression and RNA interference analysis demonstrated that GmNPR1 enhanced resistance to P. sojae in transgenic soybean plants. Several pathogenesis-related (PR) genes were constitutively activated, including GmPR1a, GmPR2, GmPR3, GmPR4, GmPR5a, and GmPR10, in soybean plants overexpressing GmNPR1 transcripts. By contrast, the induction of PR genes was compromised in transgenic GmNPR1-RNAi lines. Taken together, these findings suggested that the interaction between GmWRKY31 and GmHDL56 enhances resistance to P. sojae by regulating defense-related gene expression in soybean.
Introduction
Phytophthora root and stem rot (PRR), which is caused by the oomycete Phytophthora sojae, is one of the most destructive diseases of soybean [Glycine max (L.) Merr.] and is responsible for $1–2 billion losses per year worldwide (Tyler, 2007). Since the first identification of PRR in Indiana in 1948, PRR has been observed in all major soybean-growing regions all around the world (Schmitthenner, 1985; Anderson and Buzzell, 1992; Su and Shen, 1993; Jee et al., 1998; Dorrance and Grunwald, 2009). Resistant cultivars carrying major resistance (R) genes against P. sojae (Rps genes) have been a cornerstone for the management of the pathogen for 50 years (Schmitthenner, 1985; Buzzell and Anderson, 1992). Rps gene resistance is race-specific, qualitatively inherited and confers an immune type of response to infection by P. sojae. However, this qualitative resistance tends to be short-lived because R-genes are neutralized by adaptation of P. sojae populations (Schmitthenner, 1985).
WRKY transcription factors (TFs) are one of the largest families of TFs in plants (Eulgem et al., 2000; Rushton et al., 2010). WRKY TFs form complex webs to modulate a great number of processes in plants, including senescence, seed development, seed dormancy and germination, and stress responses, particularly in response to biotic and abiotic stresses (Ulker and Somssich, 2004; Zhou et al., 2009; Ren et al., 2010; Rushton et al., 2010; Tao et al., 2011; Jiang et al., 2012; Vanderauwera et al., 2012; Lin et al., 2014). WRKY TFs feature a DNA-binding domain containing the conserved residues WRKYGQK and a novel zinc finger motif (Eulgem et al., 2000). WRKY TFs modulate target gene expression by specifically binding to the cis-acting element known as the W-box (C/T)TGAC(T/C) (Rushton et al., 1995; de Pater et al., 1996), which exists in the promoters of many defense-related genes (Maleck et al., 2000; Chen and Chen, 2002; Ulker and Somssich, 2004). GmWRKY27 inhibits the expression of the downstream gene GmNAC29 by binding to the W-boxes in the promoter region of GmNAC29, leading to abiotic stress tolerance (Wang F. et al., 2015), and OsWRKY6 binds directly to WLE1 and W-boxes in the promoters of defense-related genes and regulates pathogen-defense responses (Choi et al., 2015).
Seventy-four WRKY TFs have been reported in Arabidopsis (Pandey and Somssich, 2009; Bakshi and Oelmüller, 2014), and 109 WRKY TFs have been identified in rice, including four with incomplete WRKY domains (Zhang and Wang, 2005). In Arabidopsis, WRKY TFs play a crucial role in the response to pathogen infection (Abuqamar et al., 2006; Journot-Catalino et al., 2006; Li et al., 2006; Xu et al., 2006; Knoth et al., 2007; Lippok et al., 2007; Murray et al., 2007; Zheng et al., 2007; Hu et al., 2008; Mao et al., 2011). Overexpression of AtWRKY70 leads to the up-regulation of PR genes and the down-regulation of PDF1.2, leading to enhanced resistance against biotrophic pathogens and increased susceptibility to constitutive necrotrophic pathogens (Li et al., 2004). In addition, mutations of the Arabidopsis WRKY33 gene enhance susceptibility to the necrotrophic fungal pathogens Botrytis cinerea and Alternaria brassicicola. By contrast, ectopic over-expression of WRKY33 increases resistance to these two necrotrophic fungal pathogens (Zheng et al., 2006). Arabidopsis plants overexpressing AtWRKY28 and AtWRK75 exhibit enhanced resistance to Sclerotinia sclerotiorum (Chen et al., 2013). Several studies have also suggested the importance of specific OsWRKYs in the transcriptional regulation of defense-related genes in the rice response to pathogens (Hwang et al., 2011; Lee et al., 2013; Choi et al., 2015). For example, OsWRKY55 (OsWRKY31 according to the nomenclature of Wu et al. (2005) is induced by the rice blast fungus, and the transgenic rice plants over-expressing OsWRKY22 are more resistant to M. oryzae (Zhang et al., 2008). OsWRKY6 plays a role in the pathogen- or salicylic acid (SA)-inducible expression of OsPR1a. Indeed, OsWRKY6 is induced by Xanthomonas oryzae pv. oryzae (Xoo) and SA and activates the OsPR1a promoter in rice (Hwang et al., 2011). OsWRKY22 is a regulator of rice resistance to M. oryzae and is also involved in the rice response to non-host barley powdery mildew (Abbruscato et al., 2012). A total of 197 WRKY genes have been identified in soybean [G. max (L.) Merr.] (Schmutz et al., 2010), but only four members of the WRKY family in this species have been functionally characterized. Over-expressing GmWRKY21 or GmWRKY54 enhances cold tolerance and salt and drought tolerance in Arabidopsis. By contrast, over- expression of GmWRKY13 leads to increased sensitivity to salt and mannitol stress (Zhou et al., 2008). Transgenic soybean hairy roots overexpressing the GmWRKY27 gene exhibit enhanced tolerance to drought and salt stresses, whereas GmWRKY27 RNAi roots exhibit hypersensitivity to these stresses (Wang F. et al., 2015). Although a role of WRKY TFs in abiotic stress responses in soybean has been identified, the potential function of these TFs in biotic stress responses remains unclear.
Homeodomain leucine-zipper (HD-ZIP) TFs, several are known to be rapidly induced in response to altered environmental conditions and to integrate hormonal signals (Brandt et al., 2014). Members of the HD-ZIP proteins play roles related to drought stress and ABA-signaling in different plant species (Dezar et al., 2005; Deng et al., 2006; Agalou et al., 2008). Expression of the HD-ZIP genes AtHB6 and AtHB7 is induced by ABA application or water deficiency (Söderman et al., 1996; Himmelbach et al., 2002; Lechner et al., 2011). Recently, it was shown that both AtHB7 and AtHB12 act to repress the transcription of genes encoding the ABA receptors PYL5 and PYL8 in response to an ABA stimulus (Valdés et al., 2012). In soybean, it has been demonstrated that GmHDL56 and GmHDL57 could interact with the -611 to -451 portion of the VspB promoter (Tang et al., 2001). However, the molecular mechanism of HD-ZIP proteins in response to pathogen infection still remains elusive.
In a previous study, a cDNA library of mRNAs encoding expressed sequence tags (ESTs) exhibiting increased expression during P. sojae infection was constructed using suppression subtractive hybridization (SSH) from the leaf tissues of the highly resistant soybean ‘Suinong 10’. A novel EST homologous to the WRKY TF AtWRKY31 (GenBank accession no. NM_118328.3) was significantly induced after infection with P. sojae (Xu et al., 2012). In this study, we isolated this WRKY TF, designated GmWRKY31 (GenBank accession no. XM_003546112.3), from soybean ‘Suinong 10’ and studied the expression and function of this gene. Transgenic soybean plants overexpressing GmWRKY31 exhibited enhanced resistance to P. sojae, whereas RNA interference (RNAi) in transgenic soybean plants increased susceptibility to P. sojae. Further analysis indicated that GmWRKY31 physically interacted with GmHDL56 which improved resistance to P. sojae in transgenic soybean roots. GmWRKY31 and GmHDL56 shared a common target GmNPR1 (GenBank accession no. NM_001251745.1). GmWRKY31 and GmHDL56 coregulated the expression of GmNPR1 gene, leading to improved soybean resistance to P. sojae. These data suggested that GmWRKY31 interacted with GmHDL56 in response to P. sojae infection via the activation of GmNPR1 expression.
Materials and Methods
Plant Materials and Stress Treatments
‘Suinong 10,’ a popular soybean cultivar with high resistance against the predominant race 1 of P. sojae in Heilongjiang, China (Zhang et al., 2010), was used for gene isolation. Seeds of ‘Suinong 10’ were planted in pots filled with sterile vermiculite in a growth chamber with a 14-h photoperiod (at a light intensity of 350 μmol m-2s-1) at 22°C/18°C day/night temperatures and relative humidity of 70 ± 10%. Fourteen days after planting, the seedlings at the first-node stage (V1) (Fehr et al., 1971) were used for various treatments.
Phytophthora sojae race 1, PSR01, which was isolated from infected soybean plants in Heilongjiang (Zhang et al., 2010), was cultivated at 25°C for 7 days on V8 juice agar in a polystyrene dish. For P. sojae treatment, the soybean plants were infected with zoospores according to the method of Ward et al. (1979) with minor modifications for incubation of 48 h. The unifoliate leaves were also harvested at 0, 6, 12, 24, 36, 48, and 72 h after the treatment, immediately frozen in liquid nitrogen, and stored at -80°C until RNA extraction and cDNA analysis.
Cloning of the Full-Length GmWRKY31 cDNA
The cDNA library of mRNAs encoding ESTs with increased expression during P. sojae infection was constructed previously using SSH from the leaf tissues of the highly resistant soybean cultivar ‘Suinong 10’ (Xu et al., 2012). A novel up-regulated EST encoding a putative WRKY TF was identified by searching the phytozome database1. In the present study, the full-length cDNA (termed GmWRKY31) was isolated by RT-PCR from the cDNA of ‘Suinong 10’ using the primers GmWRKY31-F/R (Supplementary Table S1). The PCR procedure was performed as follows: 94°C for 5 min, followed by 30 cycles of 94°C for 30 s, 58°C for 30 s, and 72°C for 2 min, with a final extension at 72°C for 10 min. The amplification products were cloned into pMD18-T vector (TaKaRa, Dalian, China) for sequencing.
Quantitative Real-Time PCR Analysis
Quantitative real-time PCR (qRT-PCR) analysis was performed to determine the transcript abundance of the GmWRKY31 gene using a SYBR® Green Real Time PCR Master Mix Plus kit according to the manufacturer’s instructions (TOYOBO, Japan) on a CFX96 TouchTM Real-Time PCR Detection System (Bio-Rad, USA). The primers was GmWRKY31-qF/R (Supplementary Table S1). Total RNA was isolated from soybean leaves using TRIzol reagent (Invitrogen, Shanghai, China) according to the manufacturer’s protocol. Reverse transcription was performed using 1 μg of total RNA and a ReverTra Ace® qPCR RT Kit (TOYOBO, Japan). The PCR protocol was 95°C for 1 min, followed by 40 cycles of 95°C for 15 s, 60°C for 15 s, and 72°C for 45 s. The amplification product was confirmed by melting curve analysis in one-degree intervals from 95 to 60°C. The relative expression value was calculated by the 2-ΔΔCT method using the soybean internal control gene GmEF1b (GenBank accession no. NM_001248778) with the primers GmEF1b-F/R (Supplementary Table S1). Each qRT-PCR was performed on three biological replicates with three technical replicates each.
Generation and Resistance Identification of Transgenic Plants
To obtain the 35S:GmWRKY31-overexpression construct, the coding region of GmWRKY31 was amplified by RT-PCR using the primers GmWRKY31-F/R and ligated into the plant overexpression vector pCAMBIA33012 with the bar gene as the selective marker. To obtain the 35S:GmWRKY31-RNAi silencing construct, a 381 bp fragment of the GmWRKY31 was amplified by RT-PCR using the primers GmWRKY31-rF/R and inserted into the RNAi transformation vector pJawohl8 as an inverted-repeat construct with the pat gene as the selective marker. To construct the GmNPR1 overexpression and RNAi vector, the full-length cDNA and a specific cDNA fragment of GmNPR1 were amplified using the primers GmNPR1-F/R and GmNPR1-rF/R and inserted into the pCAMBIA3301 and pJawohl8 vectors, respectively. The overexpression construct and RNAi silencing construct were transferred into Agrobacterium tumefaciens LBA4404 via tri-parental mating. The cotyledonary nodes were used as explants for soybean transformation using the Agrobacterium-mediated transformation method described by Paz et al. (2004). Homozygous T2 plants were identified by glyphosate spraying, PCR amplification, qRT-PCR analysis and southern blot hybridization using a DIG High Prime DNA Labeling and Detection Starter Kit II (Roche, Germany). The full-length cDNA of GmHDL56 was amplified using the primers GmHDL56-F/R. The PCR products were inserted into the pCAMBIA3301 vectors. GmHDL56 transgenic soybean hairy roots were generated by A. rhizogenes-mediated transformation according to the method described by Kereszt et al. (2007). The transgenic soybean hairy roots were verified by LibertyLink strip and qRT-PCR analysis. LibertyLink strips (Envirologix, Portland, OR, USA) were used to determinate genetically modified plants containing the phosphinothricin N-acetyltransferase protein following the manufacturer’s instruction. All the primers for genotyping and vector construction were listed in Supplementary Table S1.
The transgenic soybean seeds were planted in soil and maintained under greenhouse conditions. For disease resistance analysis, the living cotyledons at the first-node stage (V1) and the trifoliate leaf from the top of each plant at the third-node stage (V3) (Fehr et al., 1971) were treated with P. sojae as described by Ward et al. (1979) with minor modifications for the inoculation site and investigated according to the methods described by Dou et al. (2003) with minor modifications for the live plants. The living cotyledons and trifoliate leaf were incubated in a mist chamber at 25°C with 90% relative humidity under a 14-h photoperiod at a light intensity of 350 μmol m-2 s-1. Non-transformed cotyledons and leaves were used as controls. The disease symptoms on each leaf were observed and photographed using a Nikon D700 camera after inoculation.
Subcellular Localization of GmWRKY31
The coding sequence of GmWRKY31 was amplified by RT-PCR using the primers GmWRKY31-gF/R (Supplementary Table S1). Then, the coding sequence was ligated into the N-terminus of green fluorescence protein (GFP) under the control of the constitutive CaMV35S promoter. The resulting expression plasmid, 35S:GmWRKY31-GFP, was transformed into Arabidopsis protoplasts cells via polyethylene glycol (PEG)-mediated transfection as described by Yoo et al. (2007). The fluorescence signal was imaged using a TCS SP2 confocal spectral microscope imaging system (Leica, Germany). The vector 35S:GFP was used as a control.
Expression and Purification of Fusion Proteins
The full-length cDNA of GmWRKY31 was amplified by RT-PCR using the primers GmWRKY31-yF/R (Supplementary Table S1). The product was inserted at the BamHɪ/EcoRɪ site of the pET29b(+) vector (Novagen, Germany) to generate pET29b(+)-GmWRKY31. The recombinant fusion plasmid was transformed into Escherichia coli BL21(DE3) cells. His-tagged proteins were induced with 0.5 mM isopropyl-β-D-thiogalactoside at 37°C for 5 h. The fusion protein was purified at room temperature and quantified according to the pET System Manual (Novagen).
Electrophoretic Mobility Shift Assay (EMSA)
The DNA binding activity of GmWRKY31 was examined using digoxigenin- ddUTP-labeled double-stranded oligonucleotide W-box or GmNPR1 promoter DNA N1P (including the W-box domain TTGACC or TTGACT) probes. EMSA was performed as described by Garner and Revzin (1981). The signal was detected by chemiluminescence and recorded on X-ray film (Kodak).
Transactivation Assays
For transactivation assay, the GUS gene in pCAMBIA3301 was replaced by GmWRKY31 as the effector plasmid. The W-box and sequence from the GmNPR1 gene promoter was multimerized three times and inserted upstream of the CaMV35S promoter (-42 to +8) containing a TATA box. This construct was inserted into pXGUS-P (Chen et al., 2009) and fused to the GUS gene as the reporter plasmid. The transactivation assay was performed by PEG transfection of Arabidopsis protoplasts as described by Yoo et al. (2007). Twenty micrograms of reporter plasmid and 20 μg of effector plasmid or control plasmid (pXGUS-P-35Smini) were co-transfected into 4 × 104 protoplasts. The transfected cells were incubated at 22°C in light for 18–20 h. GUS activity was determined following the methods described by Lu et al. (1998).
Yeast One-Hybrid Experiment
GmWRKY31 and GmHDL56 coding regions were amplified and cloned into the pGADT7 prey vector (Clontech)3, which created a translational fusion between the GAL4 activation domain and the TF. A specific DNA fragment of the promoter of GmNPR1 was amplified using the primers N1-F/R (Supplementary Table S1) and cloned into pHIS2 vector at the EcoRɪ/Sacɪ site. Competent yeast cells were prepared according to the Clontech Yeast Protocols Handbook using the Y187 yeast strain. For yeast transformation, 50 μl of competent yeast cells were incubated with 100 ng of pHIS2 bait vector and 100 ng of pGADT7 prey vector, 50 μg of salmon sperm carrier DNA and 0.5 ml of PEG/LiAc solution. Cells were transformed according to the manufacturer’s instructions. Transformations were plated onto SD media (-Leu, -Trp) to select co-transformed cells and incubated at 28°C for 4 days. Transformed yeast cells were subsequently grown in SD media (-Leu, -Trp) liquid media to an OD600 of 0.1 and diluted in a 10 × dilution series. From each dilution, 5 μl was spotted on SD media (-Leu, -Trp) and on SD media (-His, -Leu, -Trp) media plates supplemented with 100 mM 3-amino-1, 2, 4-triazole (3AT; Sigma-Aldrich). The plates were then incubated for 3 days at 28°C
Library Screening
The coding sequence of GmWRKY31 was amplified by RT-PCR using the primers GmWRKY31-tF and GmWRKY31-tR. Then, the purified PCR product was cloned into the bait vector pGBKT7 (Clontech, USA). The pGBKT7-GmWRKY31 plasmids were transformed into Y2H Gold yeast cells. Yeast two-hybrid transformation screens were performed by transforming the pGBKT7-GmWRKY31 expressing strain with the soybean cDNA library, which was constructed using the prey vector pGADT7-rec and mRNA isolated from seedlings of the soybean cultivar ‘Suinong 10’ (Dong et al., 2015) according to the manufacturer’s protocols (Clontech, USA). Cells were screened for growth on SD/-Leu-Trp-His-Ade media for 3–5 days at 30°C. The colonies were transferred to selective media containing X-α-gal (20 μg ml-1) and AbA (125 μg ml-1). The blue colonies were sequenced and characterized using homology BLAST at NCBI4.
Yeast Two-Hybrid Experiment
The coding region of GmHDL56 was amplified and cloned into pGADT7 (Clontech, USA). The plasmids pGBKT7- GmWRKY31 and pGADT7-GmHDL56 were co-transferred into yeast Y2H Gold cells, and interactions were detected by growth on several types of media: SD/-Trp/-Leu media; SD/-Trp/-Leu/-His/-Ade media; and SD/-Trp/-Leu/-His/-Ade/X-α-gal media. Two independent clones for each transformation were tested. The interaction between mammalian pGBKT7-53 and pGADT7-SV40 served as a positive control, whereas the coexpression of pGBKT7-Lam and pGADT7-SV40 served as a negative control (Clontech, USA).
Bimolecular Fluorescence Complementation (BiFC) Assay
To further confirm the interactions between GmWRKY31 and GmHDL56, a BiFC assay based on yellow fluorescence protein (YFP) was performed. For the constructs, the coding sequence of GmWRKY31 was amplified by RT-PCR using the primers GmWRKY31-bF/R and cloned into pSAT6-nEYFP-N1, and the coding sequence of GmHDL56 was amplified by RT-PCR using the primers GmHDL56-bF/R and cloned into pSAT6-cEYFP-N1. The resulting constructs were transformed into Arabidopsis protoplasts cells via polyethylene glycol (PEG)-mediated transfection as described by Yoo et al. (2007). Empty vectors of pSAT6-nEYFP-N1 and pSAT6-cEYFP-N1 were used as controls. Transfected cells were imaged using the TCS SP5 confocal spectral microscope imaging system.
Pull-Down Assays
Glutathione S-transferase-GmWRKY31 and His-GmHDL56 proteins were separately expressed in E. coli BL21(DE3) cells. To purify the recombinant protein, the bacterial cells were pelleted after induction, resuspended in 10 ml of ice-cold 1× binding buffer, and sonicated on ice for 10 min (30-s pulse/min) until the sample was no longer viscous. Following centrifugation at 1,200 × g for 15 min at 4°C, the supernatant was harvested and loaded onto a GST- or His-bind Resin column (EMD Millipore, USA). The recombinant protein in the elutes was analyzed by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE).
Glutathione S-transferase-GmWRKY31 was incubated with GST-sepharose beads for 2 h and washed with 1× binding buffer three times. The beads containing GST-GmWRKY31 were incubated with His-GmHDL56 protein for 1 h. Then, the sepharose beads with associated protein were spun down, washed three times with 1× binding buffer, and boiled in Laemmli sample buffer for 10 min. The pull-down assays were performed at 4°C. The inputs and pull-down pellets of these experiments and the supernatants in the competitive pull-down experiments were subjected to standard SDS-PAGE and immunoblotting using an anti-His antibody.
Detection of Luciferase Activity in Tobacco Leaf
This assay was performed as previously described (Shang et al., 2010; Song et al., 2013). Briefly, the 1.5 kb promoter sequence of GmNPR1 (GmNPR1P) was cloned using the primers GmNPR1-pF/R, and linked to pBI121-LUC (GUS gene of pBI121 was replaced luciferase gene). The reporter construct GmNPR1P-LUC and the effector constructs 35S: GmWRKY31 and 35S: GmHDL56 were transformed into A. tumefaciens strain GV3101, and transfected into tobacco leaves by agroinfiltration as described previously (Liu et al., 2012). The activity of luciferase was observed using a CCD camera (Berthold Technologies) 72 h after infiltration. All the primers for genotyping and vector construction were listed in Supplementary Table S1.
SA Measurement
To test whether GmNPR1 could regulate SA accumulation, the content of SA was analyzed in T4 GmNPR1 transgenic plants. SA was extracted and measured from soybean plant leaves, as described previously by Aboul-Soud et al. (2004). Leaf tissues (0.5 g) were extracted in 1 mL of 90 % methanol following homogenization in liquid nitrogen. 3-Hydroxybenzoic acid (Sigma) was used as an internal standard. The SA extracts were analyzed automatically using a fluorescence detector (excitation at 305 nm and emission at 405 nm) with reversed-phase high-performance liquid chromatography on a Waters 515 system (Waters, Milford, MA, USA) with a C18 column.
Results
GmWRKY31 Expression Is Significantly Induced by P. sojae
The full-length cDNA sequence of GmWRKY31 was cloned from ‘Suinong10’ total RNA by RT-PCR. Sequence analysis indicated that GmWRKY31 encoded a protein with a deduced polypeptide sequence of 557 amino acids. A WRKY domain was identified at amino acid positions 159–219 (Supplementary Figure S1A). Sequence alignment revealed that GmWRKY31 shares 49, 42, 36, and 33% amino acid identity with AtWRKY31, CaWRKY31, GmWRKY23, and GmWRKY6, respectively (Supplementary Figure S1B), and the WRKY domains were highly conserved (Supplementary Figure S1C).
To study GmWRKY31 expression, we first examined the accumulation of GmWRKY31 mRNA in various tissues by qRT-PCR. GmWRKY31 was constitutively and highly expressed in the leaves, followed by the roots and the stems (Supplementary Figure S2A). We further explored the expression pattern of GmWRKY31 in soybean ‘Suinong 10’ during P. sojae infection. GmWRKY31 expression was significantly induced by P. sojae, and the accumulation of GmWRKY31 mRNA reached a peak at 24 h under P. sojae stress, followed by a decline (Supplementary Figure S2B). The expression of GmWRKY31 was higher in resistant cultivars (‘Suinong 10,’ ‘Williams 82,’ and ‘Dongnong 1’) than that in susceptible cultivars (‘Dongnong 50,’ ‘Hefeng 35,’ and ‘Hefeng 25’) (Supplementary Figure S2C).
GmWRKY31 Is a W-Box DNA Binding Transcriptional Activator
To determine whether GmWRKY31 functions as a TF, we determined the subcellular localization of the GmWRKY31 protein. We transformed the GmWRKY31-GFP construct under the control of the CaMV35S promoter into Arabidopsis mesophyll protoplasts. A strong fluorescent signal derived from GFP alone was observed in the cytoplasm and nuclei, whereas the transformed cells carrying GmWRKY31-GFP displayed a strong green fluorescent signal in the nucleus (Figure 1A), just like GmERF5-GFP did (Dong et al., 2015), indicating nuclear localization of GmWRKY31 (Figure 1A). We then examined whether GmWRKY31 acts as a transcription regulator in plant cells. We performed a transactivation assay in Arabidopsis mesophyll protoplasts using a reporter gene with three tandem copies of the W-box or mW-box and effector plasmids with GmWRKY31 (Figure 1B). As shown in Figure 1C, constitutive expression of GmWRKY31 clearly activated the expression of the W-box-driven reporter gene and did not activate the expression of the mW-box-driven reporter gene. The relative GUS activity driven by both the W-box and GmWRKY31 was approximately 1.7 times that of the control (driven by the 35Sm and GmWRKY31), which indicated that GmWRKY31 was able to bind to the W-box and trans-activate reporter gene expression in plants. Overall, these results suggest that GmWRKY31 can act as a transcriptional activator in plant cells.
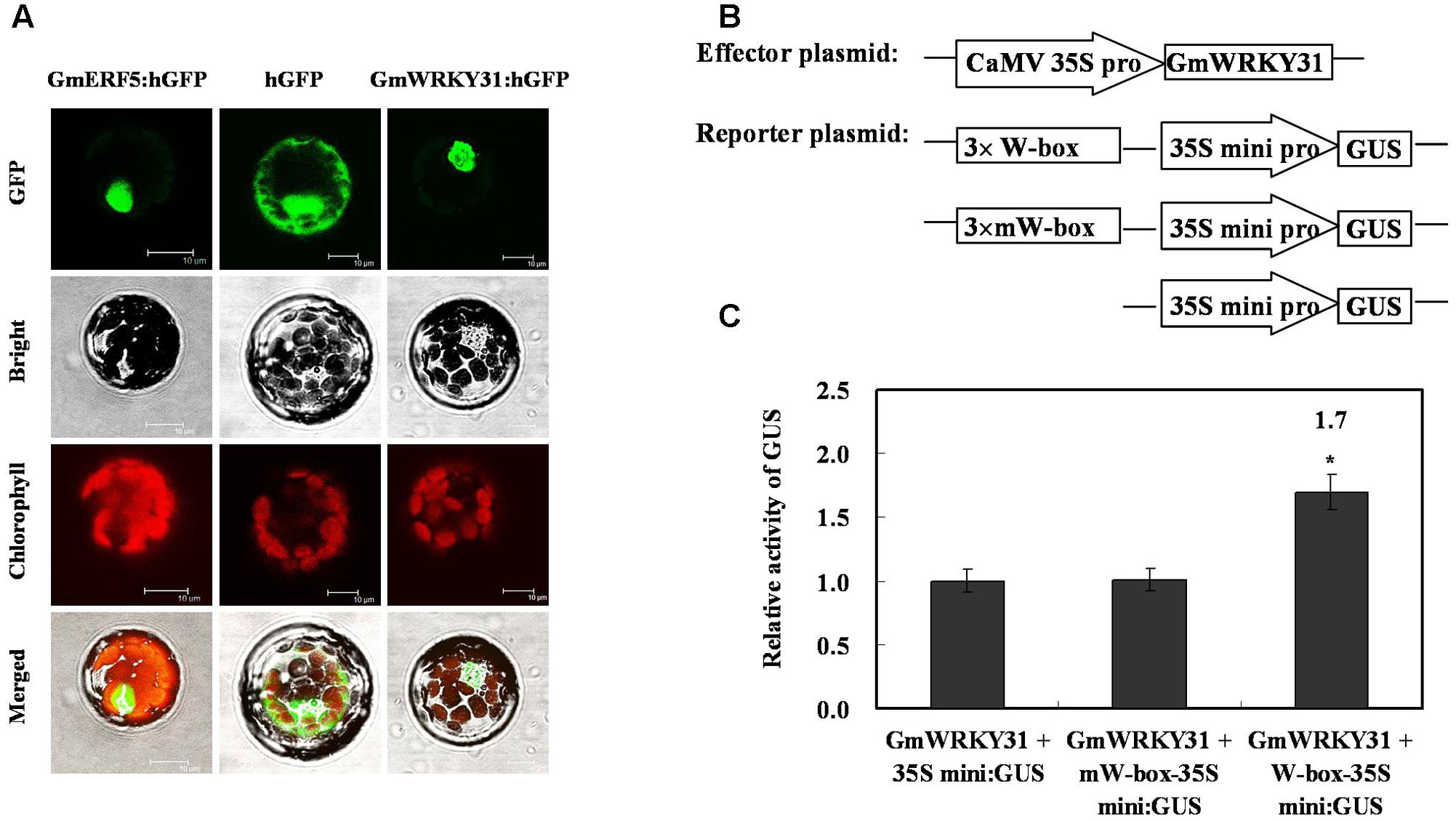
FIGURE 1. Subcellular localization of GmWRKY31 protein and transactivation assays of GmWRKY31 to the W-box element. (A) Subcellular localization was investigated in Arabidopsis mesophyll protoplasts under a confocal microscope. The fluorescent distribution of humanized hGFP, the fusion protein GmERF5-hGFP and GmWRKY31-hGFP were observed under white light, UV light, and red light, respectively. Bars, 10 μm. (B) Schematic diagram of effector, reporter and internal control plasmid constructs. Effector plasmids encoded GmWRKY31, under the control of the CaMV35S promoter. Reporter plasmids contained three repeats of the wild type or mutant W-box and 35Smini. (C) Relative GUS activities in transactivation assays. The effector plasmid encoding GmWRKY31 and the reporter plasmid were co-transfected into Arabidopsis mesophyll protoplasts with internal control plasmid by polyethylene glycol-mediated transformation. The numbers showed the fold increase in GUS activity compared with the vector GmWRKY31+35S mini. Results were averages of three replicates ± standard deviation. Statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01).
GmWRKY31 Enhances Resistance to P. sojae in Transgenic Soybean Plants
To study the function of GmWRKY31 in response to P. sojae, the plant over-expressing vector 35S:GmWRKY31 and RNA interference vector 35S:GmWRKY31-RNAi were constructed and transformed into soybean plants by the Agrobacterium-mediated transformation system. Southern blot results showed that the T2 GmWRKY31-overexpressing transgenic lines (GmWRKY31-OE) and T2 GmWRKY31-RNAi transgenic soybean lines were integrated into the genomes of the three transgenic lines with one copy, respectively (Figure 2A). These transgenic lines were developed into T3 transgenic soybean plants. Expression analysis of the transgenic population led to the identification of lines with significantly increased GmWRKY31 mRNA levels compared with wild-type plants (WT), and expression analysis of T3 GmWRKY31-RNAi transgenic soybean plants led to the identification of lines with remarkably reduced GmWRKY31 mRNA levels compared with WT (Figure 2B).
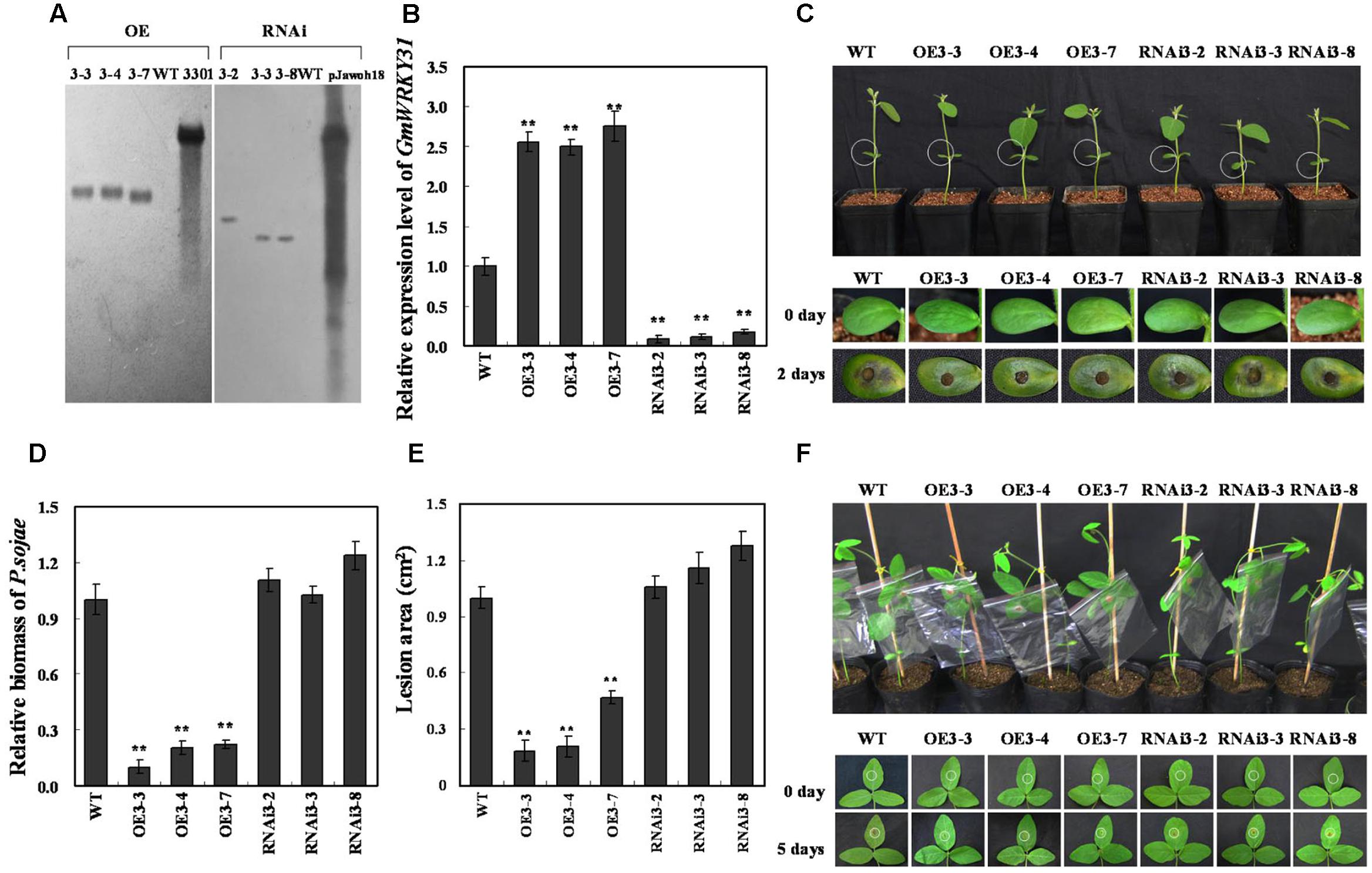
FIGURE 2. Response of GmWRKY31 transgenic soybean plants to P. sojae. (A) Southern-blot assay of the T2 GmWRKY31-OE, GmWRKY31-RNAi and wild-type plants (WT). Twenty micrograms of genomic DNA digested by the restriction enzyme Hind III was hybridized with the probe derived from the bar gene. (B) Quantitative real-time PCR of the T3 GmWRKY31-overexpression, GmWRKY31-RNAi transgenic soybean plants and WT. GmEF1b gene (NM_001248778) was used as an internal control to normalize all data. The experiments were performed on three biological replicates with their respective three technical replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicated standard error of the mean. (C) Disease symptoms on the living cotyledons of the transgenic lines and WT treated with zoospores of P. sojae at 0 and 2 days. (D) Accumulation of P. sojae biomass in the living cotyledons of the transgenic soybean lines and WT. Transcript levels of P. sojae TEF1 (EU079791) in infected living cotyledons (2 days) were plotted relative to soybean GmEF1b expression levels as determined by qRT-PCR. The experiments were performed on three biological replicates with their respective three technical replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicated standard error of the mean. (E) Disease symptoms on the living leaves of the transgenic lines and WT treated with a P. sojae race 1 inoculum at 0 and 5 days. The experiments were performed on three biological replicates. (F) The lesion area of the transgenic lines and WT were detected after 5 days of incubation with P. sojae. The experiment was performed on three biological replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicated standard error of the mean.
The living cotyledons of those transgenic soybean plants were selected to investigate resistance to P. sojae. After 2 days of incubation with zoospores of P. sojae, a remarkable difference in the development of disease symptoms was observed (Figure 2C). In GmWRKY31-RNAi lines, the cotyledons became soft, emitted a foul odor and exhibited clear water-soaked lesions compared with those of the GmWRKY31-OE lines, and there were nearly no visible lesions in GmWRKY31-OE lines and the cotyledons keep hard compared with those of the GmWRKY31-RNAi lines or WT. Meanwhile, tthe relative biomass of P. sojae in infected living cotyledons after 2 days of incubation with zoospores of P. sojae was also analyzed. The biomass of P. sojae based on the transcript level of the P. sojae TEF1 gene (GenBank accession no. EU079791) was significantly (P < 0.01) lower in GmWRKY31-OE lines than that in WT, and it was higher in GmWRKY31-RNAi lines than that in WT but did not reach a significant level (Figure 2D). Similar results were obtained after 5 days of incubation with P. sojae, and the living leaves of the WT and GmWRKY31-RNAi soybean plants exhibited clear, large lesions compared with those of the GmWRKY31-OE lines (Figure 2E). The lesion area of the GmWRKY31-OE lines was significantly (P < 0.01) smaller than that of WT soybean plants after 5 days of incubation with P. sojae (Figure 2F). These results indicate that overexpression of GmWRKY31 in soybean plants improves resistance to P. sojae and that GmWRKY31-RNAi transgenic soybean plants have increased susceptibility to P. sojae.
GmWRKY31 Interacts with GmHDL56
To determine which protein is responsible for the interaction with GmWRKY31, we performed yeast two-hybrid screening to identify WRKY31-interacting partners using a soybean cDNA library (Dong et al., 2015). We identified 19 putative GmWRKY31-interacting proteins (Supplementary Table S2), of which a cDNA corresponding to the homeodomain-leucine zipper transcript factor GmHDL56 (LOC100796213) was selected for further study. Yeast two-hybrid assays revealed that GmWRKY31 interacts strongly with GmHDL56 (Figure 3A). Bimolecular fluorescence complementation (BiFC) analyses revealed that GmWRKY31 interacts with GmHDL56 in the nuclei of Arabidopsis cells (Figure 3B). Consistent with the results of the BiFC assay, an in vitro glutathione S-transferase (GST) pull-down assay was performed using a recombinant N-terminal GST-tagged GmWRKY31 protein and a recombinant C-terminal His-tagged GmHDL56 protein (Figure 3C). As shown in Figure 3D, the His-tagged GmHDL56 protein was pulled down by GST-GmWRKY31 but not by GST alone, indicating that GmWRKY31 interacts with GmHDL56 in vitro. These results suggest that GmWRKY31 directly interacts with GmHDL56.
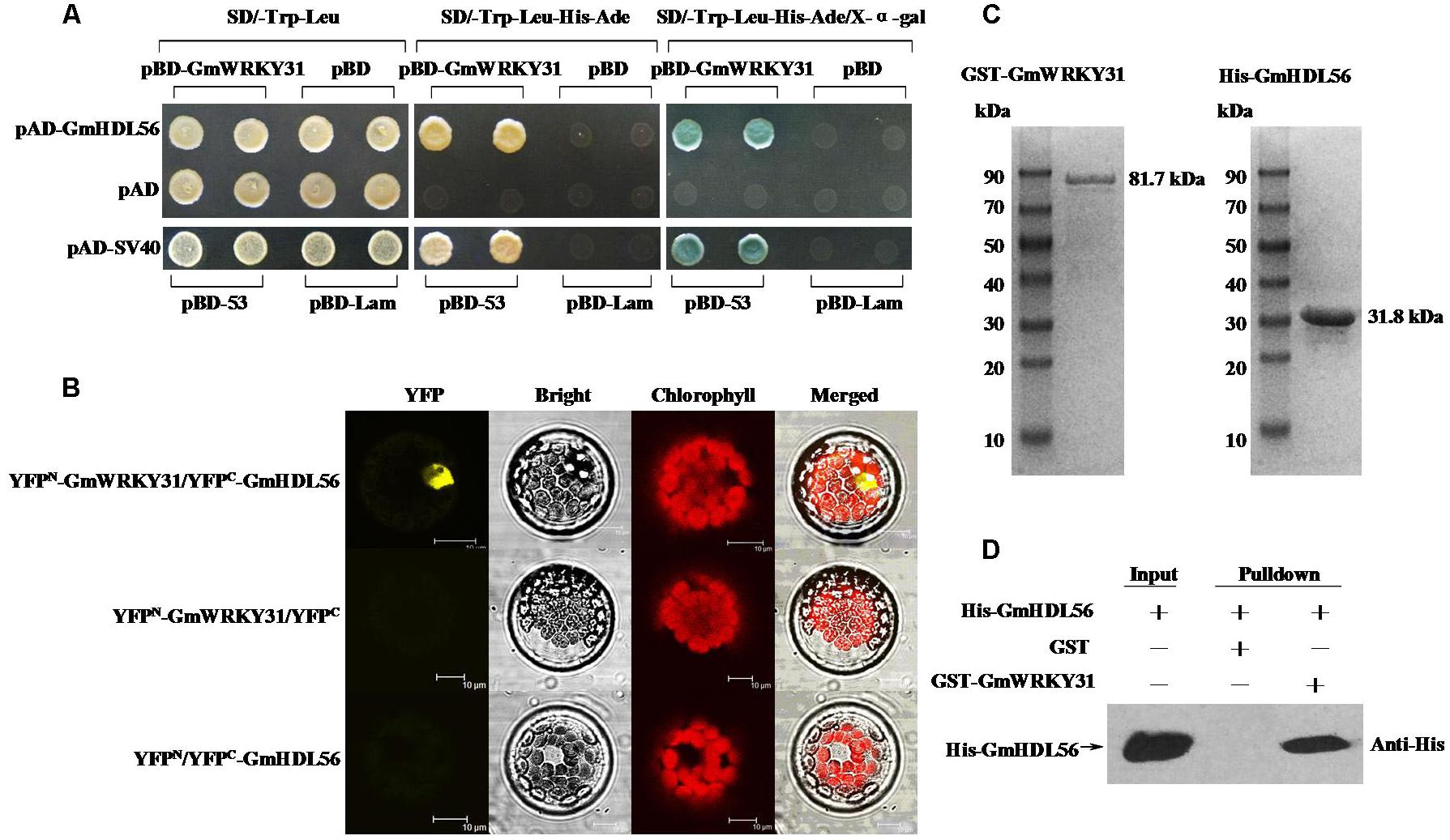
FIGURE 3. Interaction of GmWRKY31 with GmHDL56 in vitro and in vivo. (A) Analysis of interactions between GmWRKY31 and GmHDL56 protein in yeast cells. The yeast cells of strain Y2H harboring pBD-GmWRKY31 and pAD-GmHDL56 plasmid combinations were grown on either SD/-Trp/-Leu media or SD/-Trp/-Leu/-His/-Ade media, followed by β-galactosidase assay (SD/-Trp/-Leu/-His/-Ade/X-α-gal media). (B) Bimolecular fluorescence complementation (BiFC) analysis of interaction between GmWRKY31 and GmHDL56 in Arabidopsis protoplast cells. The plasmid combinations are indicated on top. The fluorescence of YFP was observed by confocal laser microscopy 16 h after transfection. Bars = 10 μm. (C) SDS-PAGE analysis of the purified recombinant proteins GST-GmWRKY31 and His-GmHDL56 using GST- or His-Bind Kits. (D) Pull-down assay of GmWRKY31 interaction with GmHDL56. His-GmHDL56 protein was incubated with immobilized GST or GST-GmWRKY31 proteins, and immunoprecipitated fractions were detected by anti-His antibody.
GmHDL56 Enhances Resistance to P. sojae in Transgenic Soybean Hairy Roots
The interaction between GmWRKY31 and GmHDL56 raised the question of whether GmHDL56 was also required for P. sojae responses in soybean. To this end, we examined the phenotype of GmHDL56 in response to P. sojae infection. The plant over-expressing vector 35S:GmHDL56 was constructed and transformed into soybean hairy roots by high-efficiency Agrobacterium rhizogenes-mediated transformation (Kereszt et al., 2007). The transgenic soybean hairy roots were tested using LibertyLink strips (Figure 4A) and qRT-PCR (Figure 4B). Three overexpression hairy roots (OE101, OE106, and OE108) were selected to investigate resistance to P. sojae. After 5 days of incubation with zoospores of P. sojae, three GmHDL56-overexpressing lines (OE101, OE106, and OE108) displayed significantly enhanced resistance compared with the wild-type plants (Figure 4C). Moreover, we analyzed the relative biomass of P. sojae in soybean root based on the transcript level of the P. sojae TEF1 gene. The results indicated that the P. sojae biomass was lower in GmHDL56-overexpressing lines than that in wild-type plants (Figure 4D). These data suggest that GmHDL56 positively regulates the soybean defense in response to P. sojae infection.
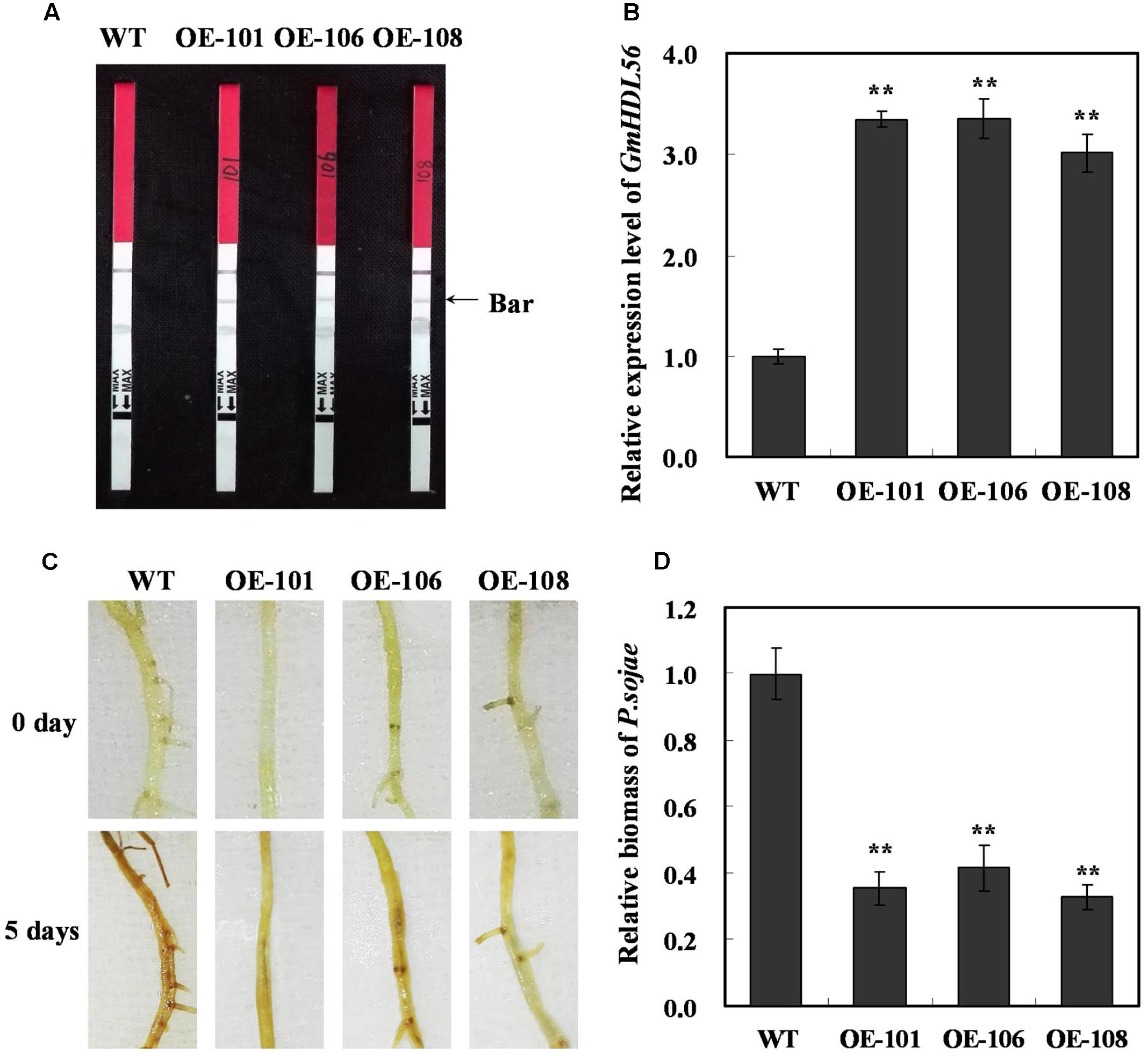
FIGURE 4. GmHDL56 enhances resistance to P. sojae in transgenic soybean hairy roots. (A) Transgenic soybean plants were tested using Liberty Link strips. (B) Quantitative real-time PCR of transgenic soybean hairy roots overexpressing GmHDL56 and WT. GmEF1b gene (NM_001248778) was used as an internal control to normalize all the data. The experiments were performed on three biological replicates with their respective three technical replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate standard error of the mean. (C) Disease symptoms on the hairy roots of the transgenic lines and WT treated with zoospores of P. sojae at 0 and 5 days. (D) Accumulation of P. sojae biomass in the hairy roots of the transgenic soybean lines and WT. Transcript levels of P. sojae TEF1 (EU079791) in infected hairy roots (5 days) were plotted relative to soybean GmEF1b expression levels as determined by qRT-PCR. The experiments were performed on three biological replicates with their respective three technical replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicated standard error of the mean.
GmWRKY31 Positively Regulates GmNPR1 Expression via Directly Binds to the W-Box on the Promoter of GmNPR1
WRKY proteins act upstream of NPR1 and positively regulate its expression during the activation of the plant defense response (Yu et al., 2001; Liu et al., 2005; Choi et al., 2015). We previously reported that GmNPR1 was induced after infection with the oomycete P. sojae (Xu et al., 2012). To determine whether GmNPR1 is involved in GmWRKY31-overexpression-mediated defense pathways in soybean, we analyzed GmNPR1 mRNA levels in GmWRKY31 transgenic soybean plants by qRT-PCR. GmNPR1 was highly induced in GmWRKY31-overexpressing lines and repressed in GmWRKY31-RNAi lines (Figure 5A), which suggested that GmWRKY31 might be directly or indirectly involved in the regulation of GmNPR1. We then analyzed the GmNPR1 genomic sequence and identified two W-box elements in the -766 to -638 region of the GmNPR1 promoter (N1P) (Figure 5B). To determine whether the GmWRKY31 protein directly binds to these element sequences, EMSAs using DNA fragments corresponding to the N1P region and the GmWRKY31 protein produced in E. coli was performed. As shown in Figure 5C, the recombinant GmWRKY31 bound only to N1P, and not mN1P. In addition, increasing molar excesses of unlabeled N1P fragment (competitor) inhibited binding (Figure 5C). These results indicate that GmWRKY31 specifically binds to the -766 to -638 region of the GmNPR1 promoter in vitro.
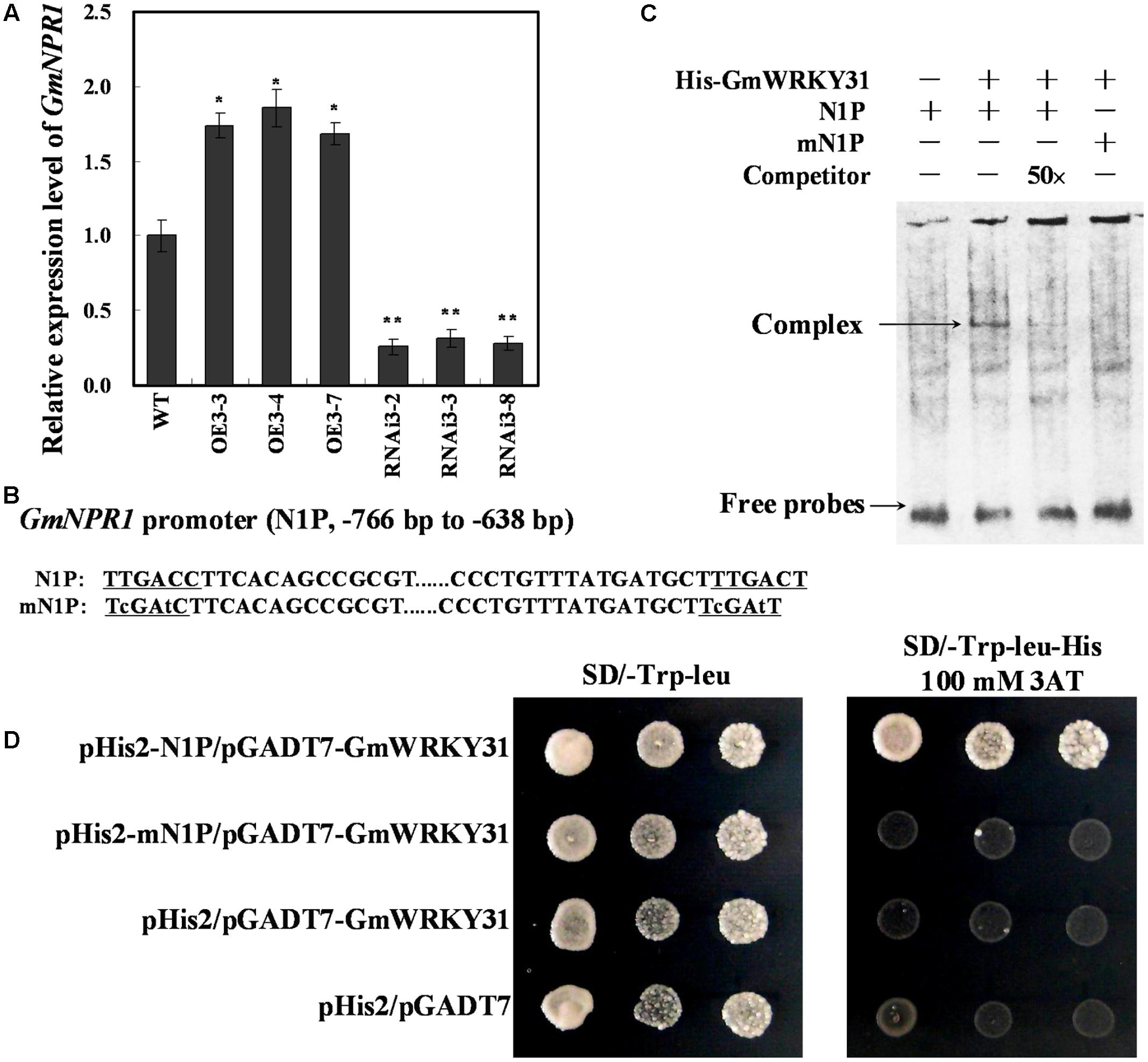
FIGURE 5. GmWRKY31 positively regulate GmNPR1 expression and bound to the W-box on the –766 to –638 region of the GmNPR1 promoter. (A) Relative expression level of GmNPR1 gene determined by qRT-PCR. GmEF1b gene (NM_001248778) was used as an internal control to normalize all data. The experiment was performed on three biological replicates with their respective three technical replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicated standard error of the mean. (B) Nucleotide sequences of the N1P and mN1P probes. (C) EMSA analysis of binding of GmWRKY31 protein to the W-box. A 129 bp GmNPR1 promoter fragment containing the W-box or mW-box was labeled with digoxigenin-ddUTP and used as probe. An unlabeled probe was used as a cold competitor. (D) The result of yeast-one-hybrid assay for interaction between GmWRKY31 and W-box. The –766 to –638 region of the GmNPR1 promoter containing two W-box (N1P) or two mutated W-box (mN1P) were used as bait. Yeast cells were dropped in 10-fold dilutions onto selective media (SD/-Leu-Trp-His) containing 100 mM 3-AT (3-amino-1, 2, 4-triazole). pHIS2 co-transformed with pGAD-GmWRKY31 was used as negative control. Each transformation used three different colonies of each pairwise interaction test.
To further confirm that GmWRKY31 binds to the -766 to -638 region of the GmNPR1 promoter in vivo, a yeast one-hybrid assay was performed. As shown in Figure 5D, GmWRKY31 bound to N1P but not mN1P in the Y1H assay. These results indicate that GmWRKY31 directly binds to the -766 to -638 region of the GmNPR1 promoter in vivo.
GmHDL56 and GmWRKY31 Coregulates GmNPR1 Expression
To determine whether GmHDL56 regulates GmNPR1 gene expression, we analyzed GmNPR1 mRNA levels in the GmHDL56 transgenic hairy roots. RT-qPCR results showed that GmNPR1 was highly induced in GmHDL56 overexpression hairy roots (Figure 6A). GmHDL56 could bind to the phosphate response domain ATTAATTA of the soybean VspB tripartite promoter (Tang et al., 2001). To determine whether GmHDL56 directly binds to GmNPR1 promoter, we analyzed the nucleotide sequence of GmNPR1 promoter and identified an ATTAATTA element in the -1263 to -1205 region of the GmNPR1 promoter (N2P) (Figure 6B). To determine whether GmHDL56 directly binds to the element sequences, we performed EMSA using DNA fragments corresponding to the N2P region. As shown in Figure 6C, recombinant GmHDL56 bound to N2P but not mN2P. In addition, increasing molar excesses of the unlabeled N2P fragment (competitor) inhibited the binding. These results indicated that GmHDL56 binds specifically to the ATTAATTA element (-1263 to -1205) present in the promoter of region of GmNPR1 in vitro. To further confirm that GmHDL56 binds to the -1263 to -1205 region of the GmNPR1 promoter in vivo, a yeast one-hybrid assay was performed. As shown in Figure 6D, GmHDL56 bound to N2P but not mN2P in the Y1H assay. These results indicated that GmHDL56 directly binds to the -1263 to -1205 region of the GmNPR1 promoter in vivo.
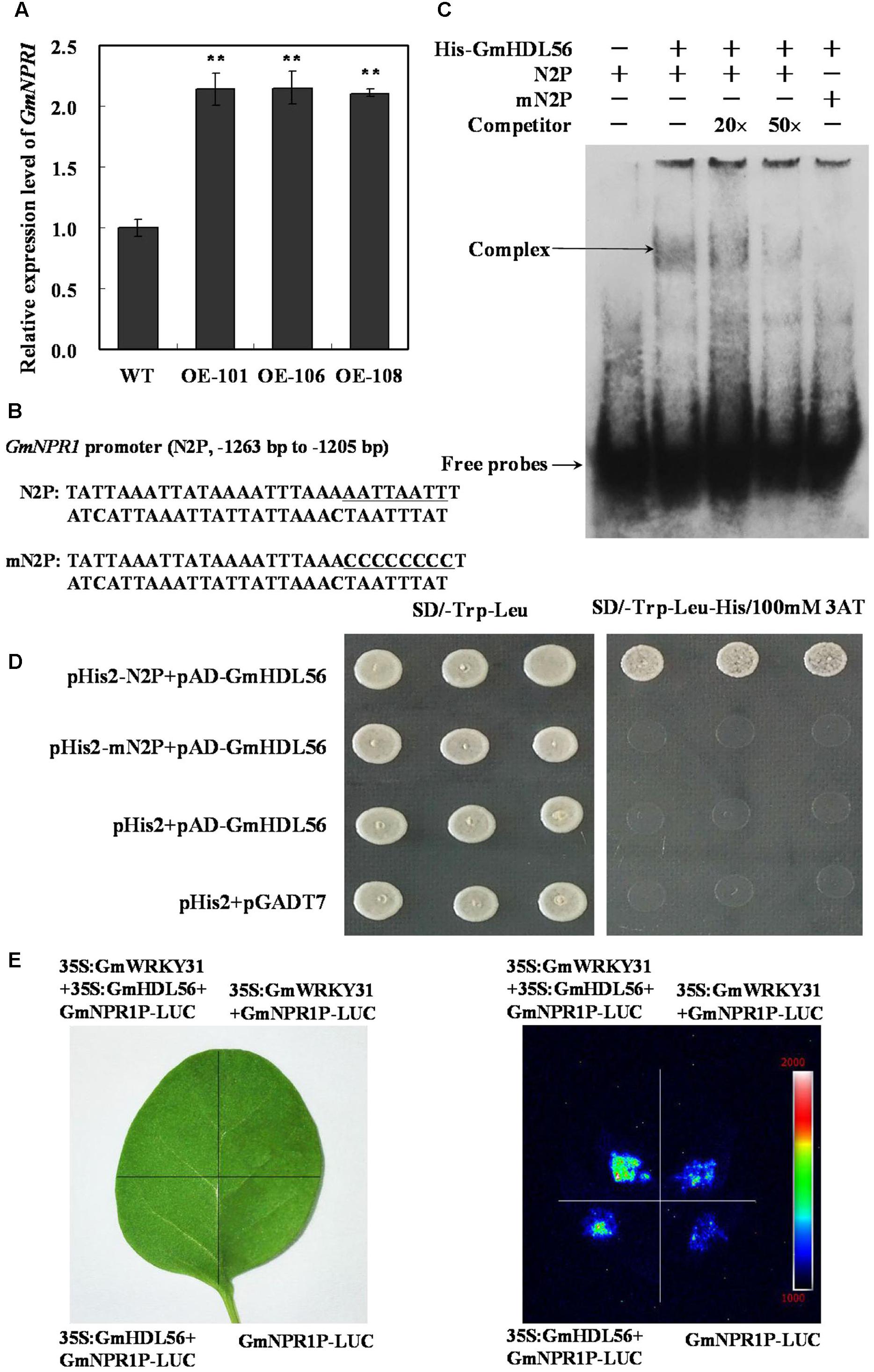
FIGURE 6. GmWRKY31 and GmHDL56 coregulated GmNPR1 expression. (A) Relative expression level of GmNPR1 gene determined by qRT-PCR. GmEF1b gene (NM_001248778) was used as an internal control to normalize all the data. The experiments were performed on three biological replicates with their respective three technical replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate standard error of the mean. (B) Nucleotide sequences of the N2P and mN2P probes. (C) EMSA analysis of binding of GmHDL56 protein to the ATTAATTA element present at –1263 to –1205 region of the GmNPR1 promoter. A 59 bp GmNPR1 promoter fragment containing the element was labeled with digoxigenin-ddUTP and used as probe. An unlabeled probe was used as a cold competitor. (D) The result of yeast-one-hybrid assay for interaction between GmHDL56 and ATTAATTA element. The –1263 to –1205 region of the GmNPR1 promoter containing ATTAATTA element (N2P) or mutated W-box (mN2P) were used as bait. Yeast cells were dropped in 10-fold dilutions onto selective media (SD/-Leu-Trp-His) containing 100 mM 3-AT (3-amino-1, 2, 4-triazole). pHIS2 co-transformed with pGAD-GmHDL56 was used as negative control. Each transformation used three different colonies of each pairwise interaction test. (E) GmWRKY31 and Gm56HDL activate GmNPR1 promoter activity in tobacco leaves. A. tumefaciens GV3101 strains harboring GmNPR1P-LUC and 35S: GmWRKY31 and/or GmHDL56 were transfected into tobacco leaves. Luciferase imaging was performed 72 h after injection.
To learn how the interaction between GmWRKY31 and GmHDL56 regulates the expression of GmNPR1 gene. We examined whether the GmNPR1 promoter activity was activated by the two proteins using the luciferase expression system in vivo. The luciferase under the control of the GmNPR1 promoter was injected into tobacco leaves simultaneously with 35S:GmWRKY31and/or35S:GmHDL56. Our results showed that both GmWRKY31 and GmHDL56 improved GmNPR1 promoter activity, and the activation was stronger when the two proteins were simultaneously expressed (Figure 6E). The above findings suggest that both GmWRKY31 and GmHDL56 are transcriptional activators of GmNPR1, and they work together to improve transcriptional activity.
GmNPR1 Enhances Resistance to P. sojae and Positively Regulated the Expression of the Pathogenesis-Related (PR) Genes in Transgenic Soybean Plants
To evaluate the responsiveness of GmNPR1 to P. sojae, qRT-PCR was performed to determine the expression patterns of GmNPR1 in ‘Suinong 10’ plants. The examination of tissue-specific transcript abundance in ‘Suinong 10’ demonstrated that GmNPR1 was constitutively and highly expressed in the roots, followed by the leaves and stems (Supplementary Figure S3A). Under P. sojae stress, GmNPR1 mRNA rapidly increased and reached a maximum level at 72 h after the treatment (Supplementary Figure S3B). The expression of GmNPR1 was higher in resistant cultivars (‘Suinong 10,’ ‘Williams 82,’ ‘Dongnong 1’) than that that in susceptible cultivars (‘Dongnong 50,’ ‘Hefeng 35,’ and ‘Hefeng 25’) (Supplementary Figure S3C).
To determine whether the resistance to P. sojae phenotypes in GmWRKY31 transgenic soybean plants is related to GmNPR1 expression, we produced GmNPR1-overexpressing (GmNPR1-OE) and RNAi (GmNPR1-RNAi) transgenic plants. Southern blot results showed that three T2 GmNPR1-overexpressing transgenic lines (GmNPR1-OE) and three T2 GmNPR1-RNAi transgenic soybean lines were integrated into the genomes of the three transgenic lines with one copy, respectively (Figure 7A). These transgenic lines were developed into T3 transgenic soybean plants. Expression analysis of the transgenic population led to the identification of lines with remarkably increased GmNPR1 mRNA levels, and the expression analysis of T3 GmWRKY31-RNAi transgenic soybean plants led to the identification of lines with significantly reduced GmNPR1 mRNA levels (Figure 7B).
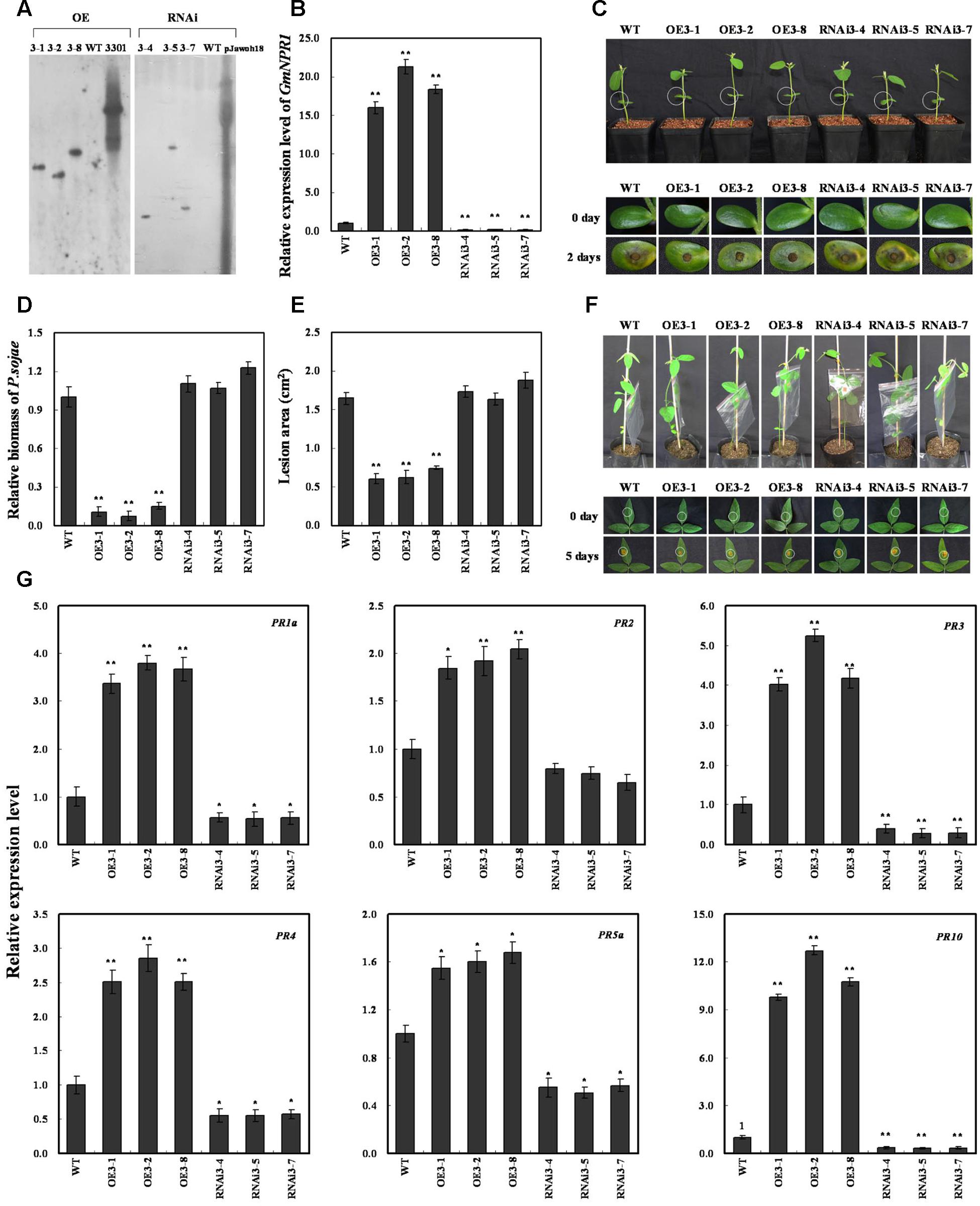
FIGURE 7. Response of GmNPR1 to P. sojae in transgenic soybean plants. (A) Southern-blot assay of the T2 Gm NPR1-OE, Gm NPR1-RNAi and wild-type plants (WT). Twenty micrograms of genomic DNA digested by the restriction enzyme Hind III was hybridized with the probe derived from the bar gene. (B) Quantitative real-time PCR of the T3 GmNPR1-overexpression, GmNPR1-RNAi transgenic soybean plants and WT. GmEF1b gene (NM_001248778) was used as an internal control to normalize all data. The experiments were performed on three biological replicates with their respective three technical replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate standard error of the mean. (C) Disease symptoms on the living cotyledons of the transgenic lines and WT treated with zoospores of P. sojae at 0 and 2 days. The experiments were performed on three biological replicates with their respective three technical replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate standard error of the mean. (D) Accumulation of P. sojae biomass in the living cotyledons of the transgenic soybean lines and WT. Transcript levels of P. sojae TEF1 (EU079791) in infected living cotyledons (2 days) were plotted relative to soybean GmEF1b expression levels as determined by qRT-PCR. The experiments were performed on three biological replicates with their respective three technical replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate standard error of the mean. (E) Disease symptoms on the living leaves of the transgenic lines and WT treated with a P. sojae race 1 inoculum at 0 and 5 days. The experiments were performed on three biological replicates with their respective three technical replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate standard error of the mean. (F) The lesion area of the transgenic lines and (WT) were detected after 5 days of incubation with P. sojae. The experiments were performed on three biological replicates with their respective three technical replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate standard error of the mean. (G) The relative transcript abundance of GmPR1a (AF136636), GmPR2 (M37753), GmPR3 (AF202731), GmPR4 (BT090788), GmPR5a (M21297), GmPR10 (FJ960440) in GmNPR1 overexpression transgenic lines (GmNPR1-OE) and RNA-interference transgenic lines (GmNPR1-RNAi) were compared with that in WT. The amplification of GmEF1b gene (NM_001248778) was used as an internal control to normalize all the data. Statistically significant differences were performed between the transgenic lines and WT. The experiment was performed on three biological replicates with their respective three technical replicates and statistically analyzed using Student’s t-test (∗P < 0.05, ∗∗P < 0.01). Bars indicate standard error of the mean.
The living cotyledons of the GmNPR1-OE and GmNPR1-RNAi transgenic soybean plants were selected to investigate the resistance to P. sojae. After 2 days of incubation with zoospores of P. sojae, the remarkable differences in the development of disease symptoms were observed (Figure 7C). In GmNPR1-RNAi lines, the cotyledons became soft, emitted a foul odor and exhibited clear water-soaked lesions compared with those of the GmNPR1-OE lines. However, there were nearly no visible lesions in GmNPR1-OE lines, and the cotyledons keep hard compared with those of the GmNPR1-RNAi lines or WT. Meanwhile, the relative biomass of P. sojae in infected living cotyledons after 2 days of incubation with zoospores of P. sojae was also analyzed. The biomass of P. sojae based on the transcript level of the P. sojae TEF1 gene was significantly (P < 0.01) lower in GmNPR1-OE lines than that in WT, and it was higher in GmNPR1-RNAi lines than that in WT but did not reach a significant level (Figure 7D). Similar results were obtained after 5 days of incubation with P. sojae, and the ability of living leaves to resist P. sojae significantly increased in the GmNPR1-OE lines and decreased in the GmNPR1-RNAi lines compared with WT (Figure 7E). The lesion area of the GmNPR1-OE lines was significantly (P < 0.01) smaller and the lesion area of the GmNPR1-RNAi lines was larger than that of WT after 5 days infection with P. sojae (Figure 7F). These results indicate that the overexpression of GmNPR1 in soybean plants improved resistance to P. sojae, whereas susceptibility to P. sojae was enhanced in GmNPR1-RNAi transgenic soybean plants.
To determine whether GmNPR1 regulates PR gene expression, we analyzed mRNA levels in GmNPR1 transgenic plants by qRT-PCR. GmPR1a (GenBank accession no. AF136636), GmPR2 (GenBank accession no. M37753), GmPR3 (GenBank accession no. AF202731), GmPR4 (GenBank accession no. BT090788), GmPR5a (GenBank accession no. M21297), and GmPR10 (GenBank accession no. FJ960440) were highly induced in GmNPR1-overexpressing lines, and the transcript abundance of GmPR10 increased up to 12-fold compared to WT (Figure 7G). In GmNPR1-RNAi transgenic soybean plants, the transcript abundances of GmPR1a, GmPR3, GmPR4, GmPR5a and GmPR10 were significantly lower than those of WT (Figure 7G). These data indicate that GmNPR1 directly or indirectly regulates these PR genes.
The T4 GmNPR1 transgenic plants were identified by qRT-PCR (Supplementary Figure S4A), and the content of SA was analyzed in T4 GmNPR1 transgenic plants. The results showed that the accumulation of SA in GmNPR1-OE lines was significantly (P < 0.05) higher than that in WT or GmNPR1-RNAi lines, and it was lower in GmNPR1-RNAi lines than that in WT but did not reach a significant level (Supplementary Figure S4B). These results indicated that overexpression of GmNPR1 in soybean leads to higher SA level.
Discussion
Since the first WRKY protein (SPF1) was identified in sweet potato (Rushton and Somssich, 1998), much progress has been made in studying the functions of WRKY family TFs. In model plants, the functional roles and underlying mechanisms of many WRKY TFs in pathogen responses have been studied (Chen and Chen, 2002; Kim et al., 2006; Chen et al., 2010; Abbruscato et al., 2012), but little is known about the functional roles of WRKYs and their mechanisms in soybean. In this study, we determined that GmWRKY31, a new member of the soybean WRKY family of TFs, plays a crucial role in soybean during infection with P. sojae. To cope with pathogenic challenge, plants rapidly activate defense responses regulated by the major signaling molecules SA, JA, and ET (Reymond and Farmer, 1998; Kunkel and Brooks, 2002; Spoel et al., 2003; Robert-Seilaniantz et al., 2011). WRKY genes are rapidly induced by pathogens, pathogen elicitors, or SA treatment in a number of plant species (Eulgem et al., 1999; Chen and Chen, 2000). Arabidopsis WRKY70 is a common regulatory component of SA- and JA- dependent defense responses and is also required for R gene-mediated resistance (Li et al., 2004, 2006; Knoth et al., 2007). In rice, OsWRKY13 functions as an activator of the SA-dependent defense response and a suppressor of the JA-dependent response, which mediates disease resistance to bacterial blight and fungal blast (Qiu et al., 2007). Here, we demonstrated that mRNA transcripts of GmWRKY31 are remarkably increased by P. sojae, suggesting an important role of GmWRKY31 in soybean resistance to P. sojae. To understand the molecular basis of GmWRKY31 in response to P. sojae, we further demonstrated that overexpression of GmWRKY31 significantly increases resistance to P. sojae in transgenic soybean plants, whereas resistance to P. sojae was compromised in GmWRKY31 RNA interference transgenic soybean plants. These results indicate that GmWRKY31 is required for soybean defense responses to P. sojae.
WRKY TFs are mainly involved in the response to biotic and abiotic stresses by binding to the W-box present in the promoter of defense-related genes (Maleck et al., 2000; Wang H.H. et al., 2015). OsWRKY4 specifically binds the promoter regions of OsPR1b and OsPR5, which contain W-box or W-box-like cis-elements that mediate resistance to rice sheath blight fungus (Wang H.H. et al., 2015). In this study, we demonstrated that GmWRKY31 is localized in the nucleus. We also demonstrated that GmWRKY31 activates basal transcription levels of a reporter gene in Arabidopsis cells. These findings suggest that GmWRKY31 acts as a W-box-mediated transcriptional activator. WRKY genes act upstream of NPR1 and positively regulate its expression during the activation of plant defense responses (Yu et al., 2001). Liu et al. (2005) demonstrated that OsNPR1 expression is constitutively induced in OsWRKY03 transgenic plants but not in control plants, suggesting that OsWRKY03 functions upstream of OsNPR1 in the defense signal pathway. Accordingly, Choi et al. (2015) determined that OsWRKY6 positively regulates OsNPR1 expression in transient expression assays but did not detect any amplified PCR bands by ChIP-PCR assay in OsWRKY6-ox lines, suggesting that OsWRKY6 does not directly bind to the OsNPR1 promoter. We previously reported differential abundance of GmNPR1 in response to the oomycete P. sojae by SSH coupled with cDNA microarrays (Xu et al., 2012). In this work, GmWRKY31 overexpression activated GmNPR1 gene expression, and GmWRKY31 RNA interference transgenic soybean plants reduced GmNPR1 gene expression. In addition, we also demonstrated that GmWRKY31 directly binds to the W-box within the -766 to -638 region of the GmNPR1 promoter using EMSA and yeast-one-hybrid assays. Taken together, these results suggest that GmWRKY31 promotes GmNPR1 expression by directly binding to the W-box within the -766 to -638 region of the GmNPR1 promoter in vivo and in vitro.
WRKY TFs perform their diverse functions in various stress signaling pathways through physical interactions with different proteins, such as the VQ protein, MAP kinases, His deacetylases, and calmodulin (Park et al., 2005; Kim et al., 2008; Qiu et al., 2008; Ishihama et al., 2011). In Arabidopsis, AtWRKY33, which interacts with multiple VQ proteins, is critical not only for resistance to necrotrophic pathogens but also for tolerance to important abiotic stresses (Zheng et al., 2006; Jiang and Deyholos, 2009; Li et al., 2011). In rice, OsWRKY30 interacts with several rice MAPKs and is phosphorylated by some interacting MAPKs (Shen et al., 2012). More recent research indicates that GmWRKY27 interacts with GmMYB174, which cooperatively suppresses GmNAC29 expression and enhances drought stress tolerance in soybean plants (Wang F. et al., 2015). The present data demonstrate that GmWRKY31 interacts with GmHDL56 in vivo and in vitro and responds to P. sojae infection.
Homeodomain leucine-zipper (HD-ZIP) TFs are rapidly induced in response to altered environmental conditions (Brandt et al., 2014). GmHDL56 belongs to the HD-ZIP TF family and binds to the ATTAATTA sequence present in the promoter of GmVspB (Scott et al., 1989; Tang et al., 2001). The WRKY protein-binding elements and HD-ZIP protein-binding elements are located very close to each other in the promoter region of GmNPR1. We determined that GmHDL56 overexpression activated GmNPR1 gene expression and GmHDL56 also specifically bound to the ATTAATTA element (-1263 to -1205) present in the promoter of region of GmNPR1. We also demonstrated that GmWRKY31 and GmHDL56 cooperatively improved the expression of GmNPR1. These results suggested that the interaction between GmWRKY31 and GmHDL56 might contribute to the spatial and temporal patterns of expression of GmNPR1.
NPR1 was first identified by screening for mutants blocked in SA-mediated PR gene expression and resistance (Cao et al., 1994; Delaney et al., 1995; Wang et al., 2006). NPR1 is constitutively expressed in plants, and its expression level is elevated upon SA treatment or pathogen infection (Malnoy et al., 2007; Shi et al., 2010; Zhang et al., 2012). In addition, the overexpression of the AtNPR1 gene or its orthologs enhances resistance to biotrophic and necrotrophic fungal, viral and bacterial pathogens in a number of plant species, including rice, wheat, tomato and tobacco (Lin et al., 2004; Chern et al., 2005; Makandar et al., 2006; Yuan et al., 2007; Meur et al., 2008). In the present study, the expression of GmNPR1 was significantly induced by P. sojae. Further transgenic analysis revealed that the overexpression of GmNPR1 in soybean improved resistance to P. sojae and enhanced susceptibility to P. sojae infection in GmNPR1-RNAi lines, and the accumulation of SA in the GmNPR1-OE lines was significantly higher than that of WT and GmNPR1-RNAi lines. In Arabidopsis, the TF NPR1 is essential for the induction of defense genes such as PR genes (Zhou et al., 2000; Fan and Dong, 2002). Here, we observed that GmPR1a, GmPR2, GmPR3, GmPR4, GmPR5a, and GmPR10 were highly induced in GmNPR1-OE lines and that GmPR1a, GmPR3, GmPR4, GmPR5a and GmPR10 were significantly repressed in GmNPR1-RNAi lines. Taken together, these results indicated that overexpression of GmNPR1 in soybean leads to higher SA level. Previous studies with SA in other plants showed that SA-induced defense responses are mediated by NPR1 (Cao et al., 1997; Spoel et al., 2003) and the accumulation of SA leads to up-regulation of defense-related genes including the PR genes PR1, PR2 and PR5, and results in enhanced disease resistance against biotrophic pathogens (Gaffney et al., 1993; Delaney et al., 1994). So, we speculate a model that GmWRKY31 and GmHDL56 interact with each other and directly activate the expression of GmNPR1, then GmNPR1 positively regulates the content of the SA, and accumulation of SA leads to up-regulation of GmPR1a, GmPR2, GmPR3, GmPR4, GmPR5a, and GmPR10 genes, which ultimately leads to enhanced resistance to P. sojae in soybean (Figure 8).
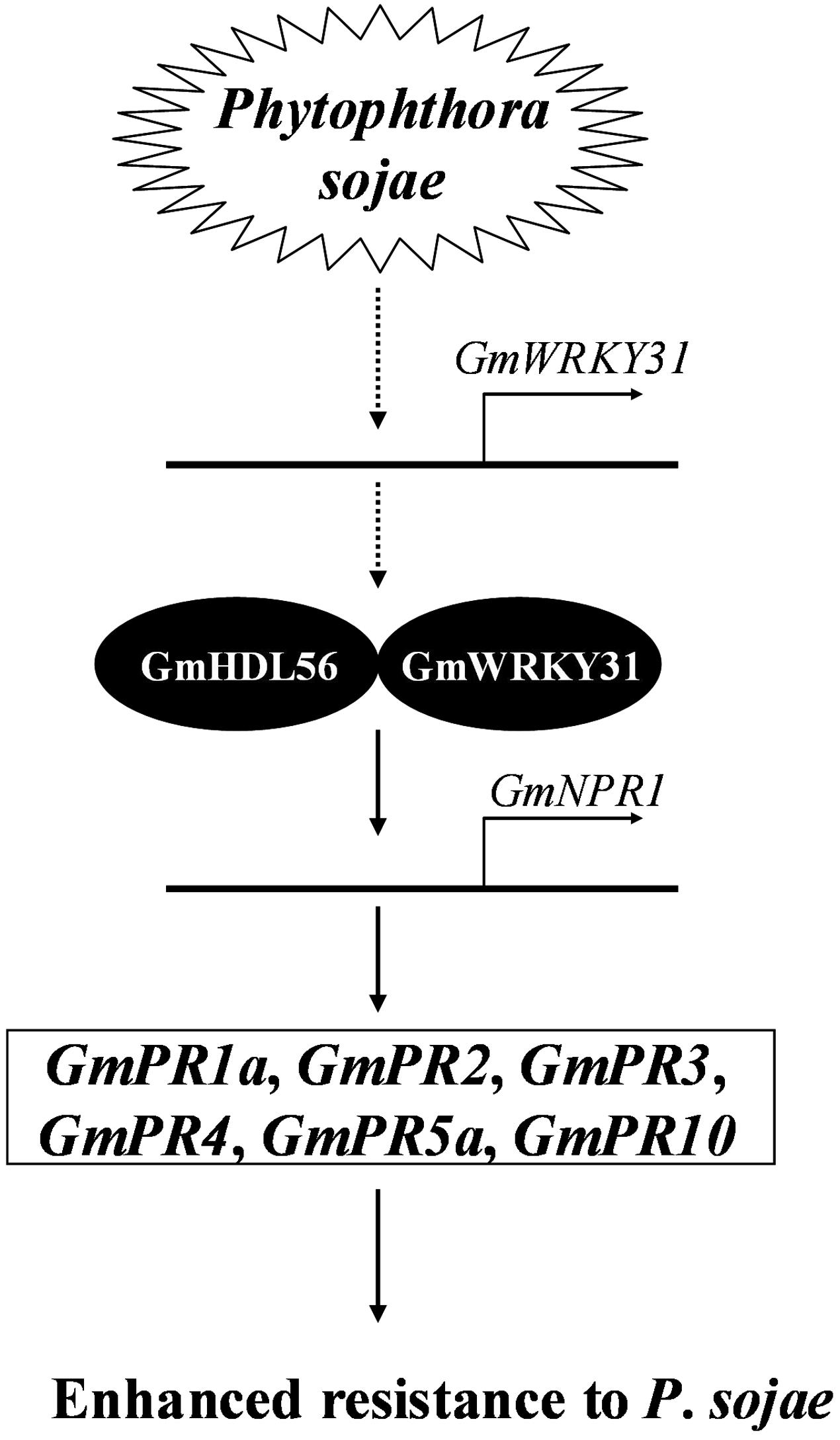
FIGURE 8. Proposed model of GmWRKY31 function in response to Phytophthora sojae in soybean. Expression of the GmWRKY31 gene is induced by P. sojae. GmWRKY31 interacts with GmHDL56, and cooperatively activate the expression of GmNPR1 by binding to the core sequences in its promoter. GmNPR1 positively regulates the expression of the GmPR1a (AF136636), GmPR2 (M37753), GmPR3 (AF202731), GmPR4 (BT090788), GmPR5a (M21297), GmPR10 (FJ960440) genes, finally leading to enhance resistance to P. sojae in soybean.
In addition, the GmNPR1-OE lines show a 15–20 fold increase in transcripts for GmNPR1, but the GmWRKY31 or GmHDL56 transactivation assays show a rather weak increase in GmNPR1 (<2-fold). In previous studies, there were reports that a few of genes were induced by P. sojae, such as N-rich protein (Ludwig and Tenhaken, 2001), GmPR10 (Xu et al., 2014), GmPRP (Jiang et al., 2015), and Gly m 4l (Fan et al., 2015), then we speculate that there maybe P. sojae induced genes changed in their expression when the expression of GmWRKY31 or GmHDL56 is modified, which might affect the expression of GmNPR1. So, in the follow-up work, it is valuable to make a deeper research on other P. sojae induced genes, defensive barriers and phytoalexins in transgenic soybean plants, and it would improve the interpretation of the findings that the mechanism of the GmWRKY31 and GmHDL56 enhances resistance. This study provides new insights into the mechanism by which WRKY proteins regulate biotic stress responses in soybean and presents GmWRKY31 as an ideal candidate target to improve P. sojae resistance in soybean and other crops.
Author Contributions
Conceived and designed the experiments: PX and SZ. Performed the experiments: SF, LD, DH, and FZ. Analyzed the data: JW, LJ, WL, QC, RL, and FM. Contributed reagents/materials/analysis tools: SZ and PX.
Funding
This work was supported by NSFC Projects (31171577, 31671719), Outstanding Talents and Innovative Team of Agricultural Scientific Research, Young and middle-aged scientific and technological innovation leader (MOST), Special Fund for Agro-scientific Research in the Public Interest (201303018), Natural Science Foundation of Heilongjiang Province (JC201308, C2015010), Changjiang Scholar Candidates Program for Provincial Universities in Heilongjiang (2013CJHB003).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00781/full#supplementary-material
FIGURE S1 | Sequence and sequence alignment of GmWRKY31 gene. (A) The open reading frame sequence and deduced polypeptide sequence of GmWRKY31. (B) Phylogenetic relationship between GmWRKY31 and the related WRKY proteins in other species. (C) Alignment of the conserved WRKY domain amino acid sequences of GmWRKY31 and nearby 7 WRKY proteins in (B).
FIGURE S2 | Expression patterns of GmWRKY31 in soybean. (A) GmWRKY31 mRNA levels in various tissues of soybean. (B) GmWRKY31 expression in soybean leaves infected with Phytophthora sojae. (C) GmWRKY31 expression in soybean resistant cultivars and susceptible cultivars.
FIGURE S3 | Expression patterns of GmNPR1 in soybean. (A) GmNPR1 mRNA levels in various tissues of soybean. (B) GmNPR1 expression in soybean leaves infected with Phytophthora sojae. (C) GmNPR1 expression in soybean resistant cultivars and susceptible cultivars.
FIGURE S4 | The content of SA in T4 GmNPR1 transgenic plants. (A) Quantitative real-time PCR of the T4 GmNPR1-overexpression. (B) The content of SA in T4 GmNPR1 transgenic plants.
Footnotes
- ^http://www.phytozome.net/
- ^http://www.cambia.org
- ^http://www.clontech.com
- ^http://blast.ncbi.nlm.nih.gov/Blast.cgi
References
Abbruscato, P., Nepusz, T., Mizzi, L., Corv, M. D., Morandin, P., Fumasoni, I., et al. (2012). OsWRKY22, a monocot WRKY gene, plays a role in the resistance response to blast. Mol. Plant Pathol. 13, 828–841. doi: 10.1111/j.1364-3703.2012.00795.x
Aboul-Soud, M. A. M., Cook, K., and Loake, G. J. (2004). Measurement of salicylic acid by a high-performance liquid chromatography procedure based on ion-exchange. Chromatographia 59, 129–133. doi: 10.1365/s10337-003-0136-1
Abuqamar, S., Chen, X., Dhawan, R., Bluhm, B., Salmeron, J., Lam, S., et al. (2006). Expression profiling and mutant analysis reveals complex regulatory networks involved in Arabidopsis response to Botrytis infection. Plant. J. 48, 28–44. doi: 10.1111/j.1365-313X.2006.02849.x
Agalou, A., Purwantomo, S., Överäns, E., Johannesson, H., Zhu, X., Estiati, A., et al. (2008). A genome-wide survey of HD-Zip genes in rice and analysis of drought-responsive family members. Plant Mol. Biol. 66, 87–103. doi: 10.1007/s11103-007-9255-7
Anderson, T. R., and Buzzell, R. I. (1992). Diversity and frequency of races of Phytophthora megasperma f. sp. glycinea in soybean fields in Essex County, Ontario, 1980–1989. Plant Dis. 76, 587–589. doi: 10.1094/PD-76-0587
Bakshi, M., and Oelmüller, R. (2014). WRKY transcription factors: jack of many trades in plants. Plant Signal. Behav. 9:e27700. doi: 10.4161/psb.27700
Brandt, R., Cabedo, M., Xie, Y., and Wenkel, S. (2014). Homeodomain leucine-zipper proteins and their role in synchronizing growth and development with the environment. J. Integr. Plant Biol. 56, 518–526. doi: 10.1111/jipb.12185
Buzzell, R. I., and Anderson, T. R. (1992). Inheritance and race reaction of a new soybean Rps1 allele. Plant Dis. 76, 600–601. doi: 10.1094/PD-76-0600
Cao, H., Bowling, S. A., Gordon, A. S., and Dong, X. (1994). Characterization of an Arabidopsis mutant that is nonresponsive to inducers of systemic acquired resistance. Plant Cell 6, 1583–1592. doi: 10.1105/tpc.6.11.1583
Cao, H., Glazebrook, J., Clarke, J. D., Volko, S., and Dong, X. N. (1997). The Arabidopsis NPR1 gene that controls systemic acquired resistance encodes a novel protein containing ankyrin repeats. Cell 88, 57–63. doi: 10.1016/S0092-8674(00)81858-9
Chen, C., and Chen, Z. (2000). Isolation and characterization of two pathogen- and salicylic acid-induced genes encoding WRKY DNA-binding proteins from tobacco. Plant Mol. Biol. 42, 387–396. doi: 10.1023/A:1006399311615
Chen, C. H., and Chen, Z. X. (2002). Potentiation of developmentally regulated plant defense response by AtWRKY18, a pathogen-induced Arabidopsis transcription factor. Plant Physiol. 129, 706–716. doi: 10.1104/pp.001057
Chen, L., Zhang, L., and Yu, D. (2010). Wounding-induced WRKY8 is involved in basal defense in Arabidopsis. Mol. Plant Microbe Interact. 23, 558–565. doi: 10.1094/MPMI-23-5-0558
Chen, S. B., Songkumarn, P., Liu, J. L., and Wang, G. L. (2009). A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol. 150, 1111–1121. doi: 10.1104/pp.109.137125
Chen, X. T., Liu, J., Lin, G. F., Wang, A., Wang, Z. H., and Lu, G. D. (2013). Overexpression of AtWRKY28 and AtWRKY75 in Arabidopsis enhances resistance to oxalic acid and Sclerotinia sclerotiorum. Plant Cell Rep. 32, 1589–1599. doi: 10.1007/s00299-013-1469-3
Chern, M., Fitzgerald, H. A., Canlas, P. E., Navarre, D. A., and Ronald, P. C. (2005). Overexpression of a rice NPR1 homolog leads to constitutive activation of defense response and hypersensitivity to light. Mol. Plant Microbe Interact. 18, 511–520. doi: 10.1094/MPMI-18-0511
Choi, C., Hwang, S. H., Fang, I. R., Kwon, S. I., Park, S. R., Ahn, I., et al. (2015). Molecular characterization of Oryza sativa WRKY6, which binds to W-box-like element 1 of the Oryza sativa pathogenesis related (PR) 10a promoter and confers reduced susceptibility to pathogens. New Phytol. 208, 846–859. doi: 10.1111/nph.13516
de Pater, S., Greco, V., Pham, K., Memelink, J., and Kijne, J. (1996). Characterization of a zinc-dependent transcriptional activator from Arabidopsis. Nucleic Acids Res. 24, 4624–4631. doi: 10.1093/nar/24.23.4624
Delaney, T. P., Friedrich, L., and Ryals, J. A. (1995). Arabidopsis signal transduction mutant defective in chemically and biologically induced disease resistance. Proc. Natl. Acad. Sci. U.S.A. 92, 6602–6606. doi: 10.1073/pnas.92.14.6602
Delaney, T. P., Uknes, S., Bernooij, B., Friedrich, L., Weymann, K., Negrotto, D., et al. (1994). A central role of salicylic acid in plant disease resistance. Science 266, 1247–1250. doi: 10.1126/science.266.5188.1247
Deng, X., Phillips, J., Bräutigam, A., Engström, P., Johannesson, H., Ouwerkerk, P. B. F., et al. (2006). A homeodomain leucine zipper gene from Craterostigma plantagineum regulates abscisic acid responsive gene expression and physiological responses. Plant Mol. Biol. 61, 469–489. doi: 10.1007/s11103-006-0023-x
Dezar, C. A., Gago, G. M., González, D. H., and Chan, R. L. (2005). Hahb-4, a sunflower homeobox-leucine zipper gene, is a developmental regulator and confers drought tolerance to Arabidopsis thaliana plants. Transgenic Res. 14, 429–440. doi: 10.1007/s11248-005-5076-0
Dong, L. D., Cheng, Y. X., Wu, J. J., Cheng, Q., Li, W. B., Fan, S. J., et al. (2015). Overexpression of GmERF5, a new member of the soybean EAR-motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 66, 2635–2647. doi: 10.1093/jxb/erv078
Dorrance, A. E., and Grunwald, N. (2009). “Phytophthora sojae: diversity among and within populations,” in Oomycetes Genetics and Genomics: Diversity, Interactions, and Research Tools, eds K. Lamour and S. Kamoun (Hoboken, NJ: Wiley).
Dou, D. L., Wang, B. S., Zhu, S. W., Tang, Y. X., Wang, Z. X., Sun, J. S., et al. (2003). Transgenic tobacco with NDR1 gene improved its resistance to two fungal diseases. Sci. Agric. Sin. 36, 1120–1124.
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Eulgem, T., Rushton, P. J., Schmelzer, E., Hahlbrock, K., and Somssich, I. E. (1999). Early nuclear events in plant defence signalling, Rapid gene activation by WRKY transcription factors. EMBO. J. 18, 4689–4699. doi: 10.1093/emboj/18.17.4689
Fan, S. J., Jiang, L. Y., Wu, J. J., Dong, L. D., Cheng, Q., Xu, P. F., et al. (2015). A novel pathogenesis-related class 10 protein Gly m 4l, increases resistance upon Phytophthora sojae infection in soybean (Glycine max [L.] Merr.). PLOS ONE 10:e014036. doi: 10.1371/journal.pone.0140364
Fan, W. H., and Dong, X. N. (2002). In vivo interaction between NPR1 and transcription factor TGA2 leads to salicylic acid-mediated gene activation in Arabidopsis. Plant Cell 14, 1377–1389. doi: 10.1105/tpc.001628
Fehr, W. R., Caviness, C. E., Burmood, D. T., and Pennington, J. (1971). Stage of development descriptions for soybeans, Glycine max (L.) Merrill. Crop Sci. 11, 929–931. doi: 10.1007/BF02197963
Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., et al. (1993). Requirement of salicylic acid for the induction of systemic acquired resistance. Science 261, 754–756. doi: 10.1126/science.261.5122.754
Garner, M. M., and Revzin, A. (1981). A gel electrophoresis method for quantifying the binding of proteins to specific DNA regions: application to components of the Escherichia coli lactose operon regulatory system. Nucleic Acids Res. 9, 3047–3060. doi: 10.1093/nar/9.13.3047
Himmelbach, A., Hoffmann, T., Leube, M., Höhener, B., and Grill, E. (2002). Homeodomain protein ATHB6 is a target of the protein phosphatase ABI1 and regulates hormone responses in Arabidopsis. EMBO. J. 21, 3029–3038. doi: 10.1093/emboj/cdf316
Hu, J., Barlet, X., Deslandes, L., Hirsch, J., Feng, D. X., Somssich, I., et al. (2008). Transcriptional responses of Arabidopsis thaliana during wilt disease caused by the soil-borne phytopathogenic bacterium Ralstonia solanacearum. PLoS ONE 3:e2589. doi: 10.1371/journal.pone.0002589
Hwang, S. H., Won Yie, S., and Hwang, D. J. (2011). Heterologous expression of OsWRKY6 gene in Arabidopsis activates the expression of defense related genes and enhances resistance to pathogens. Plant Sci. 181, 316–323. doi: 10.1016/j.plantsci.2011.06.007
Ishihama, N., Yamada, R., Yoshioka, M., Katou, S., and Yoshioka, H. (2011). Phosphorylation of the Nicotiana benthamiana WRKY8 transcription factor by MAPK functions in the defense response. Plant Cell 23, 1153–1170. doi: 10.1105/tpc.110.081794
Jee, H., Kim, W., and Cho, W. (1998). Occurance of Phytophthora root rot on soybean (Glycine max) and identification of the causal fungus. Crop Prot. 40, 16–22.
Jiang, L. Y., Wu, J. J., Fan, S. J., Li, W. B., Dong, L. D., Cheng, Q., et al. (2015). Isolation and characterization of a novel pathogenesis-related protein gene (GmPRP) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. PLoS ONE 10:e0129932. doi: 10.1371/journal.pone.0129932
Jiang, Y., and Deyholos, M. K. (2009). Functional characterization of Arabidopsis NaCl-inducible WRKY25 and WRKY33 transcription factors in abiotic stresses. Plant Mol. Biol. 69, 91–105. doi: 10.1007/s11103-008-9408-3
Jiang, Y., Lianga, G., and Yu, D. (2012). Activated expression of WRKY57 confers drought tolerance in Arabidopsis. Mol. Plant 5, 1375–1388. doi: 10.1093/mp/sss080
Journot-Catalino, N., Somssich, I. E., Roby, D., and Kroj, T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18, 3289–3302. doi: 10.1105/tpc.106.044149
Kereszt, A., Li, D., Indrasumunar, A., Nguyen, C. D., Nontachaiyapoom, S., Kinkema, M., et al. (2007). Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2, 948–952. doi: 10.1038/nprot.2007.141
Kim, K. C., Fan, B., and Chen, Z. (2006). Pathogen-induced Arabidopsis WRKY7 is a transcriptional repressor and enhances plant susceptibility to Pseudomonas syringae. Plant Physiol. 142, 1180–1192. doi: 10.1104/pp.106.082487
Kim, K. C., Lai, Z., Fan, B., and Chen, Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20, 2357–2371. doi: 10.1105/tpc.107.055566
Knoth, C., Ringler, J., Dangl, J., and Eulgem, T. (2007). Arabidopsis WRKY70 is required for full RPP4-mediated disease resistance and basal defense against Hyaloperonospora parasitica. Mol. Plant Microbe Interact. 20, 120–128. doi: 10.1094/MPMI-20-2-0120
Kunkel, B. N., and Brooks, D. M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. doi: 10.1016/S1369-5266(02)00275-3
Lechner, E., Leonhardt, N., Eisler, H., Alioua, M., Jacquet, H., Leung, J., et al. (2011). MATH/BTB CRL3 receptors target the homeodomain-leucine zipper ATHB6 to modulate abscisic acid signalling. Dev. Cell 21, 1116–1128. doi: 10.1016/j.devcel.2011.10.018
Lee, H., Ko, Y. J., Cha, J. Y., Park, S. R., Ahn, I., and Hwang, D. J. (2013). The C-terminal region of OsWRKY30 is sufficient to confer enhanced resistance to pathogen and activate the expression of defense-related genes. Plant Biotechnol. Rep. 7, 221–230. doi: 10.1007/s11816-012-0252-1
Li, J., Brader, G., Kariola, T., and Palva, E. T. (2006). WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 46, 477–491. doi: 10.1111/j.1365-313X.2006.02712.x
Li, J., Brader, G., and Palva, E. T. (2004). The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell 16, 319–331. doi: 10.1105/tpc.016980
Li, S., Fu, Q., Chen, L., Huang, W., and Yu, D. (2011). Arabidopsis thaliana WRKY25, WRKY26, and WRKY33 coordinate induction of plant thermotolerance. Planta 233, 1237–1252. doi: 10.1007/s00425-011-1375-2
Lin, F., Zhao, M. X., Baumann, D. D., Ping, J. Q., Sun, L. J., Liu, Y. F., et al. (2014). Molecular response to the pathogen Phytophthora sojae among ten soybean near isogenic lines revealed by comparative transcriptomics. BMC Genomics 15:18. doi: 10.1186/1471-2164-15-18
Lin, W. C., Lu, C. F., Wu, J. W., Cheng, M. L., Lin, Y. M., Yang, N. S., et al. (2004). Transgenic tomato plants expressing the Arabidopsis NPR1 gene display enhanced resistance to a spectrum of fungal and bacterial diseases. Trans. Res. 13, 567–581. doi: 10.1007/s11248-004-2375-9
Lippok, E., Birkenbihl, R. P., Rivory, G., Brümmer, J., Schmelzer, E., Logemann, E., et al. (2007). Expression of AtWRKY33 encoding a pathogen or PAMP responsive WRKY transcription factor is regulated by a composite DNA motif containing Wbox elements. Mol. Plant Microbe Interact. 20, 420–429. doi: 10.1094/MPMI-20-4-0420
Liu, X. Q., Bai, X. Q., Qian, Q., Wang, J. X., Chen, M. S., and Chu, C. C. (2005). OsWRKY03, a rice transcriptional activator that functions in defense signaling pathway upstream of OsNPR1. Cell Res. 15, 593–603. doi: 10.1038/sj.cr.7290329
Liu, Z. Q., Yan, L., Wu, Z., Mei, C., Lu, K., Yu, Y. T., et al. (2012). Cooperation of three WRKY-domain transcription factors WRKY18, WRKY40, and WRKY60 in repressing two ABA-responsive genes ABI4 and ABI5 in Arabidopsis. J. Exp. Bot. 63, 6371–6392. doi: 10.1093/jxb/ers293
Lu, C. A., Lim, E. K., and Yu, S. M. (1998). Sugar response sequence in the promoter of a rice α-amylase gene serves as a transcriptional enhancer. J. Biol. Chem. 273, 10120–10131. doi: 10.1074/jbc.273.17.10120
Ludwig, A. A., and Tenhaken, R. (2001). A new cell wall located N-rich protein is strongly induced during the hypersensitive response in Glycine Max L. Eur. J. Plant Pathol. 107, 323–336. doi: 10.1023/A:1011202225323
Makandar, R., Essig, J. S., Schapaugh, M. A., Trick, H. N., and Shah, J. (2006). Genetically engineered resistance to fusarium head blight in wheat by expression of Arabidopsis NPR1. Mol. Plant Microbe Interact. 19, 123–129. doi: 10.1094/MPMI-19-0123
Maleck, K., Levine, A., Eulgem, T., Morgan, A., Schmid, J., Lawton, K. A., et al. (2000). The transcriptome of Arabidopsis thaliana during systemic acquired resistance. Nat. Genet. 26, 403–410. doi: 10.1038/82521
Malnoy, M., Jin, Q., Borejsza-Wysocka, E. E., He, S. Y., and Aldwinckle, H. S. (2007). Over-expression of the apple MpNPR1 gene confers increased disease resistance in Malus x domestica. Mol. Plant Microbe Interact. 20, 1568–1580. doi: 10.1094/MPMI-20-12-1568
Mao, G., Meng, X., Liu, Y., Zheng, Z., Chen, Z., and Zhang, S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639–1653. doi: 10.1105/tpc.111.084996
Meur, G., Budatha, M., Srinivasan, T., Rajesh Kumar, K. R., Dutta Gupta, A., and Kirti, P. B. (2008). Constitutive expression of Arabidopsis NPR1 confers enhanced resistance to the early instars of Spodoptera litura in transgenic tobacco. Plant Physiol. 133, 765–775. doi: 10.1111/j.1399-3054.2008.01098.x
Murray, S. L., Ingle, R. A., Petersen, L. N., and Denby, K. J. (2007). Basal resistance against Pseudomonas syringae in Arabidopsis involves WRKY53 and a protein with homology to a nematode resistance protein. Mol. Plant Microbe Interact. 20, 1431–1438. doi: 10.1094/MPMI-20-11-1431
Pandey, S. P., and Somssich, I. E. (2009). The role of WRKY transcription factors in plant immunity. Plant Physiol. 150, 1648–1655. doi: 10.1104/pp.109.138990
Park, C. Y., Lee, J. H., Yoo, J. H., Moon, B. C., Choi, M. S., Kang, Y. H., et al. (2005). WRKY Group IId transcription factors interact with calmodulin. FEBS. Lett. 579, 1545–1550. doi: 10.1016/j.febslet.2005.01.057
Paz, M. M., Shou, H., Guo, Z., Zhang, Z., Banerjee, A. K., and Wang, K. (2004). Assessment of conditions affecting Agrobacterium-mediated soybean transformation using the cotyledonary node explants. Euphytica 136, 167–179. doi: 10.1023/B:EUPH.0000030670.36730.a4
Qiu, D., Xiao, J., Ding, X., Xiong, M., Cai, M., Cao, Y., et al. (2007). OsWRKY13 mediates rice disease resistance by regulating defense-related genes in salicylate- and jasmonate-dependent signaling. Mol. Plant Microbe Interact. 20, 492–499. doi: 10.1094/MPMI-20-5-0492
Qiu, J. L., Fiil, B. K., Petersen, K., Nielsen, H. B., Botanga, C. J., Thorgrimsen, S., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO. J. 27, 2214–2221. doi: 10.1038/emboj.2008.147
Ren, X., Chen, Z., Liu, Y., Zhang, H., Zhang, M., Liu, Q., et al. (2010). ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 63, 417–429. doi: 10.1111/j.1365-313X.2010.04248.x
Reymond, P., and Farmer, E. E. (1998). Jasmonate and salicylate as global signals for defense gene expression. Curr. Opin. Plant Biol. 1, 404–411. doi: 10.1016/S1369-5266(98)80264-1
Robert-Seilaniantz, A., Grant, M., and Jones, J. D. G. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate–salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. doi: 10.1146/annurev-phyto-073009-114447
Rushton, P. J., Macdonald, H., Huttly, A. K., Lazarus, C. M., and Hooley, R. (1995). Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of alpha-Amy2 genes. Plant Mol. Biol. 29, 691–702. doi: 10.1007/BF00041160
Rushton, P. J., and Somssich, I. E. (1998). Transcriptional control of plant genes responsive to pathogens. Curr. Opin. Plant Biol. 1, 311–315. doi: 10.1016/j.tplants.2010.02.006
Rushton, P. J., Somssich, I. E., Ringler, P., and Shen, Q. J. (2010). WRKY transcription factors. Trends Plant Sci. 15, 247–258. doi: 10.1016/j.tplants.2010.02.006
Schmitthenner, A. F. (1985). Problems and progress in control of Phytophthora root rot of soybean. Plant Dis. 69, 362–368.
Schmutz, J., Cannon, S. B., Schlueter, J., Ma, J., Mitros, T., Nelson, W., et al. (2010). Genome sequence of the palaeopolyploid soybean. Nature 463, 178–183. doi: 10.1038/nature08670
Scott, M. P., Tamkun, J. W., and Hartzell, G. W. (1989). The structure and function of the homeodomain. Biochim. Biophys. Acta 989, 25–48. doi: 10.1016/0304-419X(89)90033-4
Shang, Y., Yan, L., Liu, Z. Q., Cao, Z., Mei, C., Xin, Q., et al. (2010). The Mg-chelatase H subunit of Arabidopsis antagonizes a group of WRKY transcription repressors to relieve ABA-responsive genes of inhibition. Plant Cell 22, 1909–1935. doi: 10.1105/tpc.110.073874
Shen, H., Liu, C., Zhang, Y., Meng, X., Zhou, X., Chu, C., et al. (2012). OsWRKY30 is activated by MAP kinases to confer drought tolerance in rice. Plant Mol. Biol. 80, 241–253. doi: 10.1007/s11103-012-9941-y
Shi, Z., Maximova, S., Liu, Y., Verica, J., and Guiltinan, M. (2010). Functional analysis of the Theobroma cacao NPR1 gene in Arabidopsis. BMC Plant Biol. 10:248. doi: 10.1186/1471-2229-10-248
Söderman, E., Mattsson, J., and Engström, P. (1996). The Arabidopsis homeobox gene ATHB-7 is induced by water deficit and by abscisic acid. Plant J. 10, 375–381. doi: 10.1046/j.1365-313X.1996.10020375.x
Song, Q. X., Li, Q. T., Liu, Y. F., Zhang, F. X., Ma, B., Zhang, W. K., et al. (2013). Soybean GmbZIP123 gene enhances lipid content in the seeds of transgenic Arabidopsis plants. J. Exp. Bot. 64, 4329–4341. doi: 10.1093/jxb/ert238
Spoel, S. H., Koornneef, A., Claessens, S. M., Korzelius, J. P., Van Pelt, J. A., Mueller, M. J., et al. (2003). NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15, 760–770.
Su, Y. C., and Shen, C. Y. (1993). The discovery and biological characteristics studies of Phytophthora megasperma f. sp. glycinea on soybean in China. Acta Phytopathol. Sin. 23, 341–347.
Tang, Z. J., Sadka, A., Motishige, D. T., and Mullet, J. E. (2001). Homeodomain leucine zipper proteins bind to the phosphate response domain of the Soybean VspB Tripartite Promoter. Plant Physiol. 125, 797–809. doi: 10.1104/pp.125.2.797
Tao, Z., Kou, Y., Liu, H., Li, X., Xiao, J., and Wang, S. (2011). OsWRKY45 alleles play different roles in abscisic acid signaling and salt stress tolerance but similar roles in drought and cold tolerance in rice. J. Exp. Bot. 62, 4863–4874. doi: 10.1093/jxb/err144
Tyler, B. M. (2007). Phytophthora sojae: root rot pathogen of soybean and model oomycete. Mol. Plant Pathol. 8, 1–8. doi: 10.1111/j.1364-3703.2006.00373.x
Ulker, B., and Somssich, I. E. (2004). WRKY transcription factors: from DNA binding towards biological function. Curr. Opin. Plant Biol. 7, 491–498. doi: 10.1016/j.pbi.2004.07.012
Valdés, A. E., Övernäs, E., Johansson, H., Rada-Iglesias, A., and Engström, P. (2012). The homeodomain-leucine zipper (HD-Zip) class I transcription factors ATHB7 and ATHB12 modulate abscisic acid signalling by regulating protein phosphatase 2C and abscisic acid receptor gene activities. Plant Mol. Biol. 80, 405–418. doi: 10.1007/s11103-012-9956-4
Vanderauwera, S., Vandenbroucke, K., Inze, A., Cotte, B., Muhlenbock, P., Rycke, R., et al. (2012). AtWRKY15 perturbation abolishes the mitochondrial stress response thatsteers osmotic stress tolerance in Arabidopsis. Proc. Natl Acad. Sci. U.S.A. 109, 20113–20118. doi: 10.1073/pnas.1217516109
Wang, D., Amornsiripanitch, N., and Dong, X. (2006). A genomic approach to identify regulatory nodes in the transcriptional network of systemic acquired resistance in plants. PLoS Pathog. 2:e123. doi: 10.1371/journal.ppat.0020123
Wang, F., Chen, H. W., Li, Q. T., Wei, W., Li, W., Zhang, W. K., et al. (2015). GmWRKY27 interacts with GmMYB174 to reduce expression of GmNAC29 for stress tolerance in soybean plants. Plant J. 83, 224–236. doi: 10.1111/tpj.12879
Wang, H. H., Meng, J., Peng, X. X., Tang, X. K., Zhou, P. L., Xiang, J. H., et al. (2015). Rice WRKY4 acts as a transcriptional activator mediating defense responses toward Rhizoctonia solani, the causing agent of rice sheath blight. Plant Mol. Biol. 89, 157–171. doi: 10.1007/s11103-015-0360-8
Ward, E. W. B., Lazarovits, G., Unwin, C. H., and Buzzell, R. I. (1979). Hypocotyl reactions and glyceollin in soybeans inoculated with zoospores of Phytophthora megasperma var. sojae. Phytopathology 69, 951–955. doi: 10.1094/Phyto-69-951
Wu, K., Guo, Z., Wang, H., and Li, J. (2005). The WRKY family of transcription factors in rice and Arabidopsis and their origins. DNA Res. 12, 9–26. doi: 10.1093/dnares/12.1.9
Xu, P. F., Chen, W. Y., Lv, H. Y., Fan, S. J., Wang, X., Jiang, L. Y., et al. (2012). Differentially expressed genes of soybean during infection by Phytophthora sojae. J. Integr. Agric. 11, 368–377.
Xu, P. F., Jiang, L. Y., Wu, J. J., Li, W. B., Fan, S. J., and Zhang, S. Z. (2014). Isolation and characterization of a pathogenesis-related protein 10 gene (GmPR10) with induced expression in soybean (Glycine max) during infection with Phytophthora sojae. Mol. Biol. Rep. 41, 4899–4909. doi: 10.1007/s11033-014-3356-6
Xu, X., Chen, C., Fan, B., and Chen, Z. (2006). Physical and functional interactions between pathogen-induced Arabidopsis WRKY18, WRKY40, and WRKY60 transcription factors. Plant Cell 18, 1310–1326. doi: 10.1016/S2095-3119(12)60021-5
Yoo, S. D., Cho, Y. H., and Sheen, J. (2007). Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572. doi: 10.1105/tpc.105.037523
Yu, D., Chen, C., and Chen, Z. (2001). Evidence for an important role of WRKY DNA binding proteins in the regulation of NPR1 gene expression. Plant Cell 13, 1527–1540. doi: 10.1105/tpc.13.7.1527
Yuan, Y., Zhong, S., Li, Q., Zhu, Z., Lou, Y., Wang, L., et al. (2007). Functional analysis of rice NPR1-like genes reveals that OsNPR1/NH1 is the rice orthologue conferring disease resistance with enhanced herbivore susceptibility. Plant Biol. J. 5, 313–324. doi: 10.1111/j.1467-7652.2007.00243.x
Zhang, J., Peng, Y., and Guo, Z. (2008). Constitutive expression of pathogen inducible OsWRKY31 enhances disease resistance and affects root growth and auxin response in transgenic rice plants. Cell Res. 18, 508–521. doi: 10.1038/cr.2007.104
Zhang, J. Y., Qiao, Y. S., Lv, D., Gao, Z. H., Qu, S. C., and Zhang, Z. (2012). Malus hupehensis NPR1 induces pathogenesis-related protein gene expression in transgenic tobacco. Plant Biol. 14, 46–56. doi: 10.1111/j.1438-8677.2011.00483.x
Zhang, S. Z., Xu, P. F., Wu, J. J., Allen, X., Zhang, J. X., Li, W. B., et al. (2010). Races of Phytophthora sojae and their virulences on commonly grown soybean varieties in Heilongjiang, China. Plant Dis. 94, 87–91. doi: 10.1094/PDIS-94-1-0087
Zhang, Y., and Wang, L. (2005). The WRKY transcription factor superfamily: its origin in eukaryotes and expansion in plants. BMC Evol. Biol. 5:1. doi: 10.1186/1471-2148-5-1
Zheng, Z., Mosher, S. L., Fan, B., Klessig, D. F., and Chen, Z. (2007). Functional analysis of Arabidopsis WRKY25 transcription factor in plant defense against Pseudomonas syringae. BMC Plant Biol. 7:2. doi: 10.1186/1471-2229-7-2
Zheng, Z., Qamar, S. A., Chen, Z., and Mengiste, T. (2006). Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 48, 592–605. doi: 10.1111/j.1365-313X.2006.02901.x
Zhou, J. M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., et al. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13, 191–202. doi: 10.1094/MPMI.2000.13.2.191
Zhou, L. C., Mideros, S. X., Bao, L., Hanlon, R., Arredondo, F. D., Tripathy, S., et al. (2009). Infection and genotype remodel the entire soybean transcriptome. BMC Genomics 10:49. doi: 10.1186/1471-2164-10-49
Zhou, Q. Y., Tian, A. G., Zou, H. F., Xie, Z. M., Lei, G., Huang, J., et al. (2008). Soybean WRKY-type transcription factor genes, GmWRKY13, GmWRKY21, and GmWRKY54, confer differential tolerance to abiotic stresses in transgenic Arabidopsis plants. Plant Biotechnol. J. 6, 486–503. doi: 10.1111/j.1467-7652.2008.00336.x
Keywords: Glycine max, GmWRKY31, GmHDL56, Phytophthora sojae, response selection
Citation: Fan S, Dong L, Han D, Zhang F, Wu J, Jiang L, Cheng Q, Li R, Lu W, Meng F, Zhang S and Xu P (2017) GmWRKY31 and GmHDL56 Enhances Resistance to Phytophthora sojae by Regulating Defense-Related Gene Expression in Soybean. Front. Plant Sci. 8:781. doi: 10.3389/fpls.2017.00781
Received: 22 February 2017; Accepted: 25 April 2017;
Published: 12 May 2017.
Edited by:
Stanislav Kopriva, University of Cologne, GermanyReviewed by:
Raimund Tenhaken, University of Salzburg, AustriaMark A. Bernards, University of Western Ontario, Canada
Copyright © 2017 Fan, Dong, Han, Zhang, Wu, Jiang, Cheng, Li, Lu, Meng, Zhang and Xu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Pengfei Xu, eHVwZW5nZmVpQG5lYXUuZWR1LmNu Shuzhen Zhang, emhhbmdzaHV6aGVuQG5lYXUuZWR1LmNu
†These authors have contributed equally to this work.
 Sujie Fan
Sujie Fan Lidong Dong1†
Lidong Dong1† Shuzhen Zhang
Shuzhen Zhang Pengfei Xu
Pengfei Xu