
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 12 May 2017
Sec. Plant Physiology
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.00652
This article is part of the Research TopicMolecular and metabolic mechanisms associated with fleshy fruit qualityView all 30 articles
Improving fruit quality has become a major goal in plant breeding. Direct approaches to tackling fruit quality traits specifically linked to consumer preferences and environmental friendliness, such as improved flavor, nutraceutical compounds, and sustainability, have slowly been added to a breeder priority list that already includes traits like productivity, efficiency, and, especially, pest and disease control. Breeders already use molecular genetic tools to improve fruit quality although most advances have been made in producer and industrial quality standards. Furthermore, progress has largely been limited to simple agronomic traits easy-to-observe, whereas the vast majority of quality attributes, specifically those relating to flavor and nutrition, are complex and have mostly been neglected. Fortunately, wild germplasm, which is used for resistance against/tolerance of environmental stresses (including pathogens), is still available and harbors significant genetic variation for taste and health-promoting traits. Similarly, heirloom/traditional varieties could be used to identify which genes contribute to flavor and health quality and, at the same time, serve as a good source of the best alleles for organoleptic quality improvement. Grape (Vitis vinifera L.) and tomato (Solanum lycopersicum L.) produce fleshy, berry-type fruits, among the most consumed in the world. Both have undergone important domestication and selection processes, that have dramatically reduced their genetic variability, and strongly standardized fruit traits. Moreover, more and more consumers are asking for sustainable production, incompatible with the wide range of chemical inputs. In the present paper, we review the genetic resources available to tomato/grape breeders, and the recent technological progresses that facilitate the identification of genes/alleles of interest within the natural or generated variability gene pool. These technologies include omics, high-throughput phenotyping/phenomics, and biotech approaches. Our review also covers a range of technologies used to transfer to tomato and grape those alleles considered of interest for fruit quality. These include traditional breeding, TILLING (Targeting Induced Local Lesions in Genomes), genetic engineering, or NPBT (New Plant Breeding Technologies). Altogether, the combined exploitation of genetic variability and innovative biotechnological tools may facilitate breeders to improve fruit quality tacking more into account the consumer standards and the needs to move forward into more sustainable farming practices.
Since the dawn of agriculture in Neolithic communities some 12,000–10,000 years ago, the selection of plants exhibiting the most desirable traits has never ceased. This, so-called, domestication process appears to have been instrumental in our ancestors' transition from a hunter-gatherer to an agricultural lifestyle (Gepts, 2014), and was characterized by the low number of plant species to succeed as widely-grown crops in modern societies. Initially an intuitive process, selection was made on a few easy-to-observe desirable traits (e.g., fruit size, shape and color, or seed quality; Chalhoub et al., 2014; Vogel, 2014). As in species reduction, only a few genes exercising large phenotypic effects within this limited number of species were selected (Tang et al., 2010).
In fruit crops, initial selection was probably based on nutritious, non-toxic, and palatable features. Hedonic and culinary qualities, including flavor, succulence, juiciness, and other consumer-desirable characteristics were added later (Table 1). However, since the 1930s breeders, including tomato breeders, have centered their efforts on productivity and have basically neglected fruit quality, including traits of interest to consumers (e.g., flavor or nutritious). This can be explained in many ways: one is the fact that it is difficult to breed for complex multigene traits such as flavor; another is our lack of understanding of the molecular genetic basis of fruit quality (Klee, 2010; Lim et al., 2014). Together with changes in consumer habits, this has led to lower fruit quality and loss of flavor, which indirectly have a negative impact on fruit consumption (Klee, 2010; Orzaez et al., 2010). Hence, scientists and breeders are faced with a real challenge to improve grapes and tomatoes so that they meet the needs both of producers, i.e., productivity, and consumers, i.e., taste and healthiness (Handa et al., 2014). The relevance of this goal lies in the importance of nutrition (i.e., vitamins, antioxidants, and minerals) to remedy physiological disorders and reduce the incidence of human diseases (Klee, 2010). Today, regarding what quality parameters are crucial to improve, yield, and sustainability are the first, because of their role to ensure food security and healthiness. So, we need to maintain the yield per hectare, reducing fertilizers, and pesticides and increasing resilience to biotic and abiotic stresses in a global climate change scenario. The next objective should be increasing nutritional content, especially for crops that will be cultivated in poor areas. Enable crop diversification in poor areas could be a solution. Moreover, depending on the crop, different nutritional contents will be easier to increase. In the case of tomato, carotenoid related compounds are a clear target. For grapes, polyphenols are the main topic of studies. Finally, consumer preferences and taste should be taking into account.
Grape (Vitis vinifera L.) and tomato (Solanum lycopersicum L.) are the focus of the present review. Both produce fleshy, berry-type fruit, and have undergone important domestication and selection processes that have dramatically reduced their genetic variability. Tomato and grapevine have been selected to satisfy the quality standards required by humans. This has entailed a preference for varieties that were more productive, gave larger fruits or displayed defined organoleptic characteristics. In grapevine, despite the thousands of cultivars available, the market is dominated by a few and these are classified as a function of the final product: table grapes or raisins, or their use in winemaking (This et al., 2006). In tomato, there has also been a progressive/dramatic reduction in variability during the domestication process in the original centers of diversification and, later, when introduced into Europe, and then reintroduced into North America (Blanca et al., 2015). Initially, selection was performed by farmers; later, breeders and researchers became involved. Ultimately, this has led to the development of tomato cultivars yielding fruits of the shape, color, and size of choice. For a long time, tomatoes have been used both as a fresh product and as a processed commodity in soups, juices, sauce, pastes, powders, or concentrates, all of which require different characteristics (Bai and Lindhout, 2007; Bergougnoux, 2014). While grape and tomato share a past history of reduced variability, important differences exist: loss of flavor has more dramatically affected tomato, in part, due to more active breeding for productivity than in grapevine. Knowledge of the molecular genetic basis of fruit quality traits and of environmental impact on these traits will facilitate the maintenance of and/or an increase in production while enabling us to improve or change flavor at will.
Despite the biotechnological advances of recent decades, breeding programs often fail when dealing with complex quality traits (Handa et al., 2014). Progress in biotechnology and omics technologies applied to the variability available are likely to help us decode the underlying genetic basis of complex traits. Best alleles could subsequently be transferred into cultivars by crossing, genetic engineering, or NPBT (New Plant Breeding Technologies), to improve the quality of tomato or grape fruit. The present review is based on four fundamental approaches to increase fruit quality: (i) to enhance/maintain germplasm diversity as the source for best alleles; (ii) to understand the biochemical and genetic basis of fruit quality traits using this genomic and phenotypic diversity; (iii) to develop and use tools to dissect fruit quality traits, including improved computational technologies and network analysis; and (iv) to conduct functional studies of cultivar improvement. In conclusion, we will present an up-to-date view of the genetic resources and technologies that can improve fruit quality.
Sources of germplasm, here defined as the collection of genes and their alleles available for plant improvement, include cultivated species and sexually-compatible wild species but could also include sexually-incompatible species harboring genes that can impact on fruit quality and be transferred through genetic engineering. Only a minimal part of the wide variability present in wild germplasm was domesticated and resulted in selective gain of phenotypical or physiological traits of interest for humans. Similarly, the domestication process also resulted in a loss of genes that were left behind in non-selected wild relatives, but were needed to improve crop adaptation to environmental changes. Modern plant breeding programs are based on a process of human selection which differs dramatically from that of natural evolution: selective pressure is no longer defined primarily by a multifactorial changing environment but by narrow human standards that focus on a few traits. Hence, even if the number and nature of genes under selection may vary across the different domesticated species, phenotypic, and genetic diversity are more heavily reduced in “domesticated” germplasm than in their wild relatives. These so-called bottlenecks occurred during domestication and cultivar development, and have recently been confirmed by sequencing (Tang et al., 2010; Abbo et al., 2014; Amini et al., 2014; Andersen et al., 2015). This reduction in genetic variability is particularly evident in cultivated grapevine, in part, as a consequence of its vegetative propagation (Roby et al., 2014), but it also occurs in tomato. On the whole, as the (agronomical) traits selected by humans differed from those oriented toward optimal adaptation to the natural environment, a clear dichotomy arose between crops and their wild progenitors (Gepts, 2014). This particular genetic bottleneck, known as genetic erosion, could compromise modern cultivars as they may be unable to cope with global warming or newly emerging diseases (Prada, 2009; Chen et al., 2013; Bai et al., 2016). For instance, the wild North American grapevine species Muscadinia rotundifolia is known to be resistant to both powdery and downy mildew (Feechan et al., 2013). This resistance was mapped to a single locus that contains a family of seven TIR-NB-LRR genes known to be involved in effector-triggered immunity. Therefore, these wild species could constitute a source of resistance-related genes to be introgressed into susceptible cultivars.
In light of the consequences of genetic erosion and the importance of preserving sources of genetic and phenotypic diversity in crops, the scientific community has developed germplasm banks (Prada, 2009). Nowadays, there are more than a thousand seed banks distributed all over the world. Tomato genetic resources in gene banks have been reviewed by (Bai and Lindhout, 2007; Di Matteo et al., 2011) (Table 2) and altogether may account for over 20,000 accessions. Grapevine germplasm also exhibits great diversity with up to 10,000 cultivars predicted (Laucou et al., 2011). In this context, many seed centers have been dedicated specifically to grapevine species—especially in countries with a tradition of viticulture (Table 2). Furthermore, the Svalbard Global Seed Vault conserves in permafrost the seeds of over four thousand plant species (>774,601 accessions, of which 7,382 correspond to tomato or wild relatives of tomato clade) (www.seedvault.no) (Fowler, 2008; Westengen et al., 2013).
Genetic resources include wild, landraces (heirlooms and old cultivars of local importance), modern cultivars, and synthetic populations, and constitute the ground material for breeders. Populations of wild relatives offer breeders untapped genetic and phenotypic diversity that has evolved over millions of years to adapt to a wide range of environmental niches (Honnay et al., 2012). It is very much in our interest to study this in depth (Khan et al., 2012). Landraces/heirlooms or traditional varieties represent old cultivars that may be of more or less local importance and were developed/selected by traditional farmers over hundreds or a few thousand years to best fit their needs. Landraces (local varieties) generally display greater diversity than modern cultivars as they have been selected to adapt to local, sometimes hostile environments, at a time when agronomic technology (i.e., irrigation, fertilizers, pesticides) was not yet widely available. Cultivar uniformity was not desirable when varieties had to successfully adapt to a range of environmental conditions (Fernie et al., 2006; Cebolla-Cornejo et al., 2013). Modern agronomic practices often result in more homogeneous environmental conditions: tomato cultivation in greenhouses entails controlled watering, facilitating the selection/development of genetically uniform cultivars to enhance yield performance. Hence, landraces constitute a source of allelic variants lost to modern breeding (i.e., over the last 80 years) but potentially available for variety improvement (Mazzucato et al., 2008; Prada, 2009; Leida et al., 2015). Because of their greater proximity to modern cultivars than their wild relatives, landrace cultivars with the desired phenotypes hold great potential for cultivar improvement (Zhu et al., 2008; Prada, 2009; Biasi and Brunori, 2015). For example, Corrado et al. (2013), studied variability in a set of 214 tomato accessions which included wild relatives, cultivated landraces, and commercial varieties. They identified a number of loci which were under strong positive selection among landrace and commercial cultivars. Although the diversity present in wild and landrace populations makes them useful for the identification of genotypes carrying genes of agronomic importance, they are of less use when we attempt to accurately dissect the underlying genetic basis. To overcome these difficulties, researchers and breeders have developed a wide range of cross populations such as Recombinant Inbred Lines (RILs), Near Isogenic Lines (NILs) or Introgression Lines (ILs), Double Haploid Lines (DHLs), Induced Mutant Lines (IMLs), TILLING (Targeting Induced Local Lesions in Genomes) Lines (TLs) (Varshney et al., 2014; Henikoff et al., 2004), Multiparent Advanced Generation Intercross (MAGIC, Cavanagh et al., 2008) and Nested Association Mapping (NAM, McMullen et al., 2009; Table 3). In grape, as in other perennial/long generation time and/or self-incompatible species, for which it is difficult or impossible to generate inbred lines, F1 segregating populations (also termed Cross-Pollinators, CP) have been developed for genetic mapping (Grattapaglia and Sederoff, 1994) and propagated by grafting. Finally, germplasm collection can also be used directly as a mapping population in Genome-Wide Association Studies (GWAS; Rosenberg et al., 2010).
One way to unravel the genetic basis of fruit quality traits is by analyzing spontaneous/natural or induced mutant lines (Di Matteo et al., 2011; Bauchet and Causse, 2012). For tomato, several natural mutants have been identified but these resources are limited in comparison with induced mutant collections (Bauchet and Causse, 2012; and Table 1). The carotenoid pathway, for example, is one of the best elucidated metabolism in tomato fruit due to the availability of a series of well-characterized mutations (Figure 1). These mutants provide distinct berry color phenotypes: apricot, at, loss of function in the isopentenyl diphosphate 1 (ID11) gene (Pankratov et al., 2016); yellow flesh, r, knockout of the phytoene synthase 1 (PSY1) gene (Fray and Grierson, 1993); tangerine, t, loss of function in the carotenoid isomerase 1 (CrtISO1) enzymatic activity (Isaacson et al., 2002); Beta, B, and Delta, Del, gain of function in the lycopene β- and ε-cyclase (CYC-b; LCY-e) genes (Gil et al., 1999; Ronen et al., 2000); high-pigment 3, hp3, loss of function in the transcript coding for the zeaxanthin epoxydase (ZEP) (Galpaz et al., 2008); neoxanthin deficient 1, nxd1, defected in the neoxathin synthase (NXS) enzymatic activity (Neuman et al., 2014). In this context, the only known exception of a carotenoid structural gene which, if mutated, does not affect the berry color is represented by the β-carotene hydroxylase 2 (CHY2), whose knock out produce the, so called, white flower (wf) mutant, displaying, respectively, regular and not pigmented fruits and flowers (Galpaz et al., 2006). Additionally, a series of well-known mutants in ABA biosynthesis are also available thanks to the studies carried out by the german scientist Hans Stubbe: notabilis, not, loss of function in the 9-cis-epoxycarotenoid dioxygenase (NCED) gene (Burbidge et al., 1999); flacca, flc, knockout of the gene coding for a molybdenum cofactor (MoCo) (Sagi et al., 2002); sitiens, sit, deficient in the aldehyde oxidase (AAO) enzymatic activity (Harrison et al., 2011). More recently, the first mutant in the strigolactone pathway (ORT1) has been identified, although the source of the mutation has not yet been elucidated (Kohlen et al., 2012) (Figure 1). In addition, The Solanaceae genome network (SGN) and the Tomato Genetic Resource Center (TGRC) host large collections of tomato genotypes and mutants, which are available to researchers (Di Matteo et al., 2011; Saito et al., 2011; Bauchet and Causse, 2012; Sacco et al., 2013). More recently, a collection of ethyl methanesulfonate (EMS) and γ-ray-derived tomato mutants in the Micro-Tom dwarf background has been generated (Saito et al., 2011; Shikata et al., 2016). To date, it comprises over a thousand genotypes which have been used to create the TOMATOMA database, representing an interesting resource to research scored traits/phenotypes easily. Other EMS tomato mutant collections include the M82 processing tomato collection (http://zamir.sgn.cornell.edu/mutants/) and the Red Setter collection (http://www.agrobios.it/tilling/). These monogenic mutant populations could be directly screened to identify the genes responsible for a specific function (Menda et al., 2004; Long et al., 2006), or individual mutant lines could be analyzed to confirm the function of a gene previously identified by other means, such as QTL analysis (Goldsbrough et al., 1994).
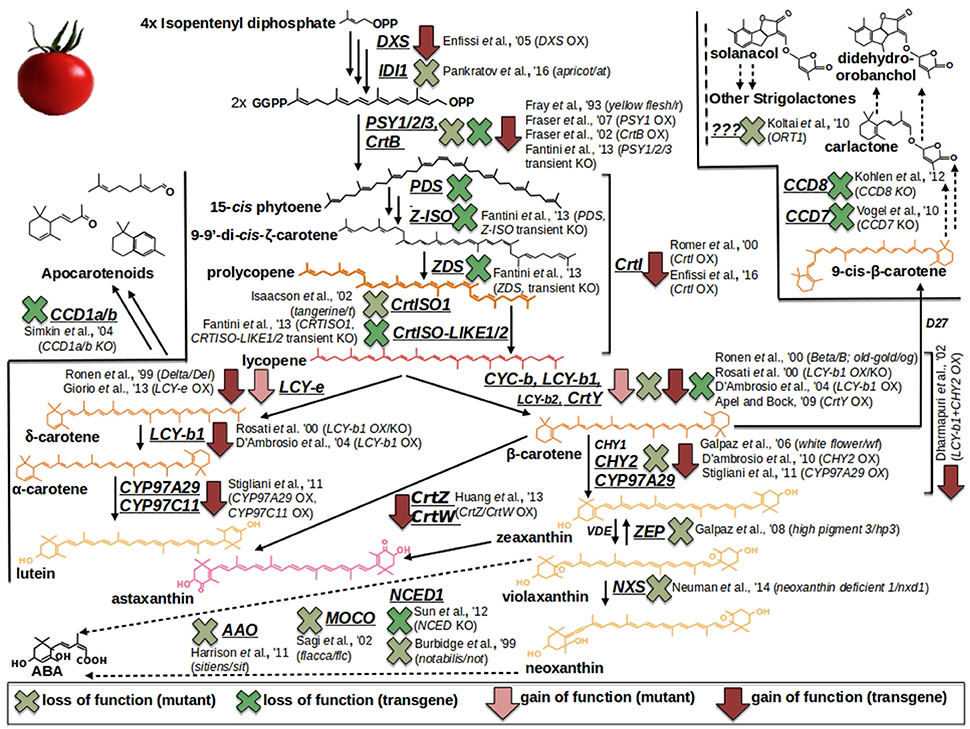
Figure 1. Carotenoid/Apocarotenoid (Volatiles, VOCs; Abscisic acid, ABA; Strigolactones, SL) biosynthesis and overview of tomato natural (mutants) and metabolically engineered (ME) resources. Light red arrows and light green crosses refer to, respectively, gain and loss of function mutants. Dark red arrows and dark green crosses pinpoint overexpression and knockout ME interventions, respectively.
Unlike tomato, collections of grapevine-induced mutants are quite rare (Fortes et al., 2015). Consequently, almost all studies in grape aimed at deciphering the molecular basis of traits use natural mutants (This et al., 2006). The FAO/IAEA Mutant Variety Database (MVD) maintains a wide range of plant mutant cultivars including tomato and grapevine. In grape, the counterpart of the conspicuous tomato/carotenoid system is represented by the phenylpropanoid pathway and, more specifically, by the synthesis of high-value sub-classes of phenypropanoid compounds (anthocyanins, stilbenes etc). An overview of grape genes and genetic resources for important phenylpropanoids affecting fruit quality is shown in Figure 2. While, contrary to the situation in tomato, it is not possible to clearly define grapevine monogenic mutants, several studies have unraveled the genetic basis of the difference between red and white cultivars, which is mainly due to a group of MYB transcription factors (MYBA1-1/2, MYBA2, MYB5a/b), mutated in the latter and, thus, preventing anthocyanin synthesis (Kobayashi et al., 2002; Deluc et al., 2006, 2008; Walker et al., 2007; Rinaldo et al., 2015; Figure 2). Similarly, Rinaldo et al. (2015) have reported that the acylated-anthocyanin phenotype is associated to the expression of the 3AT gene, coding for an ANTHOCYANIN 3-O-GLUCOSIDE-6″-O-ACYLTRANSFERASE, which is lacking in white cultivars, as well in some red varities as Pinot-Noir, that do not accumulate acylated anthocyanins. TILLING was also used to screen the tomato mutant database (Kurowska et al., 2011) for validation of gene function and as a source/tool for crop improvement (Minoia et al., 2010). Furthermore, it can also be applied to the identification of SNPs in spontaneous mutants (EcoTILLING) making it, thus, extremely useful in characterizing the variability present in germplasm banks (Mba, 2013).
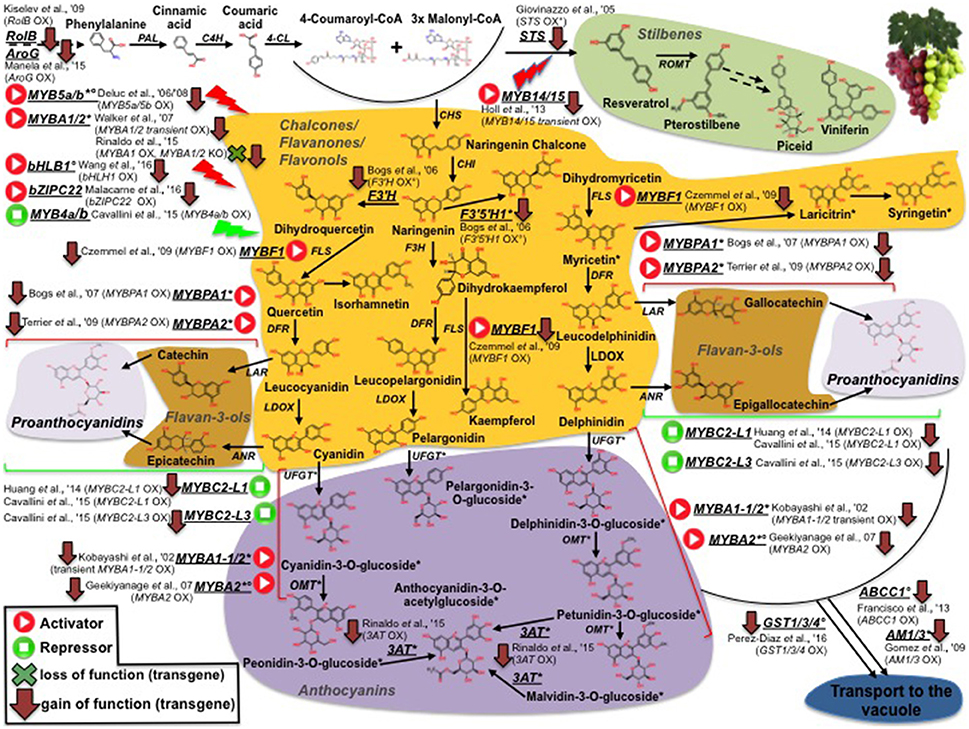
Figure 2. Phenylpropanoid biosynthesis in grapevine and survey of structural and regulative genes engineered by genetic modification (GM). Transcription factors positively (activators) or negatively (repressors) affecting phenylpropanoid pathway are represented by “Play” and “Stop” symbols, respectively. Red arrows and green crosses pinpoint overexpression and knockout GM interventions, respectively. Asterisks indicate genes and metabolites previously reported to be not/low expressed and accumulated, respectively, in white genotypes. Degrees refer to ME studies in which grapevine genes were ectopically expressed only in heterologous systems.
Genomic variations can be the result of SNPs, insertions/deletions (Indels), copy number variations (CNV), and presence absence variations (PAV); they are responsible for crop evolution and domestication (Xu and Bai, 2015). Historically, to decipher genomic diversity, two types of molecular markers were developed (reviewed by Yang et al., 2015). The first were generated before the genomic era and were able to identify genetic diversity in a wide range of genotypes (and different conditions) without the need for DNA or genome sequences. For example, the first markers developed in the 1980s were the restriction fragment length polymorphism (RFLP). Anonymous PCR-based markers such as Random Amplified Polymorphic DNA (RAPD) markers and Amplified fragment length polymorphism (AFLP) were developed later. Single Sequence Repeat (SSR) or microsatellite markers were more popular during the 1990s and the early 2000s, when a large source of reliable medium-throughput markers was generated. However, even with these markers, molecular mapping remained time-consuming, expensive, and yielded relatively low mapping resolution (Xu and Bai, 2015). While several QTLs were identified on large genomic regions, few have been used in breeding programs (Bernardo, 2008).
Three generations of sequencing technologies resulting in three “waves” of genome sequencing facilitated the study of germplasm diversity and, thus, the production of new markers and high-throughput genotyping technologies that impact on breeding methods (Bolger et al., 2014; Varshney et al., 2014; Xu and Bai, 2015; Yang et al., 2015). In 2007, the genomes of an inbred line (PN40024) derived from Pinot Noir (Jaillon et al., 2007) and a heterozygous genotype nowadays used by winemakers (Velasco et al., 2007), were published by two groups independently. Both studies used whole genome shotgun (WGS) methods and predicted around 30 k protein-coding genes (Jaillon et al., 2007; Velasco et al., 2007) distributed around 38 chromosomes (n = 19). On the other side, a high quality, well-annotated reference genome is available for tomato (Sato et al., 2012). From this genome (around 900 Mb divided up to 12 chromosomes), 34,727 protein-coding genes were identified and 30,855 of these were confirmed by RNA sequencing. Moreover, using comparative genomics with grape and A. thaliana genomes, this study highlighted that two consecutive genome triplication events might have occurred during its evolution (Sato et al., 2012). The use of NGS methods is not limited to sequencing and de novo assembly but, thanks to an increase in high-throughput read lengths, single-base accuracy, reduced costs, and assembling methods, NGS enables whole-genome resequencing to identify genetic variations on a genome-wide basis (Xu and Bai, 2015). A number of resequencing projects have already identified genomic variations by resequencing and identifying a huge number of DNA markers (cited above). Divergence between the wild (S. pimpinellifolium) and domesticated tomato genomes was estimated at around 0.6%, representing 5.4 million SNPs, distributed along the chromosomes mostly in the gene-poor regions (Sato et al., 2012). Despite this, more than 12,500 genes carry non-synonymous changes. Another study has revealed that the Micro-Tom genome presents about 1,230,000 SNPs and 190,000 indels, by comparison with the “Heinz 1706” genome (Aoki et al., 2013). Using a high-density polymorphism array (7,720 SNPs, also known as the SolCAP array), Sim et al. (2012) genotyped a collection of 426 tomato accessions, which revealed that over 97% of the markers in the collection were polymorphic. Currently, several hundred resequenced genomes of tomato varieties, S. lycopersicum vr cerasiformes, and S. pimpinellifolium are available for marker and variability studies at https://solgenomics.net/jbrowse_solgenomics/. They are being used to gain an understanding of genetic base domestication and improvement, and for GWAS (Lin et al., 2014). WGS of induced tomato mutants reveals many DNA markers, such as SNPs (Menda et al., 2004; Saito et al., 2011; Xu and Bai, 2015). In some cases, NGS can be applied to a limited number of sites in the genome and the throughput can be increased using Genotyping By Sequencing (GBS) (Kumar and Khurana, 2014; Xu and Bai, 2015). For example, Víquez-Zamora et al. (2014) used GBS on a RIL population of a cross between S. lycopersicum cv. Moneymaker and S. pimpinellifolium G1.1554 to develop a linkage map of 715 unique genetic loci from 1,974 SNPs. These results were subsequently used to map QTL responsible for TYLCV (Tomato yellow leaf curl virus) resistance. A similar strategy based on the SolCAP was used by Rambla et al. (2017a) to define a volatile QTL map in an RIL population derived from the cross between S. lycopersicum (Money maker) and the TO-937 accession of S. pimpinellifolium.
Recent studies have shown the differential regulation of genes encoding epigenetic regulators as well as local chromatin and DNA methylation changes in response to a variety of abiotic stresses including cold, salinity, drought, osmolality, or mineral nutrition (reviewed by Fortes and Gallusci, 2017). Epigenetics constitutes another process that greatly influences gene expression and, therefore, contributes to genetic plasticity. DNA methylation represents a layer of regulatory complexity beyond that encoded in the basic structure of the plant genome (Harrigan et al., 2007). Using techniques such as bisulfite Sanger sequencing, whole-genome bisulfite sequencing, and chromatin immunoprecipitation sequencing (ChIP-seq), Zhong et al. (2013) have shown that tomato ripening involves specific epigenetic remodeling. They found that binding sites for RIN, one of the key ripening transcription factors, were frequently localized in the demethylated regions of the promoters of numerous ripening genes. This binding process occurred in concert with demethylation. The binding of RIN to regulate fruit ripening genes is attenuated in the cnr ripening mutant. In addition, they found that DNA regions near the 5′ ends of genes were hypermethylated in the cnr mutant (Zhong et al., 2013). In a more recent study (Liu et al., 2015), a direct relationship between DNA demethylase (SlDML2) activity and tomato fruit ripening was reported. Briefly, silencing SlDML2 caused ripening inhibition via hypermethylation. Simultaneously, a drastic reduction in the expression of both transcription factors controlling fruit ripening and of down-stream pathways (e.g., carotenoids) occurred. Consequently, crop-improvement strategies should take account of both DNA sequence variation between plant lines and information encoded in the epigenome. In this context, the grape was recently proposed as an essential model for epigenetic and epigenomic studies in agriculturally-important, woody perennials to enable so-called epigenetic breeding (Fortes and Gallusci, 2017). Currently, a Tomato Epigenome database (http://ted.bti.cornell.edu/epigenome/1196099620) is available to investigate the presence of DNA methylation phenomena for each tomato gene. Epigenetic mechanisms have also been reported as being involved in defining the levels of Vitamin E accumulation in tomato fruits (Quadrana et al., 2014). Epigenetic marks may participate in the priming mechanisms to better withstand biotic and abiotic stresses, a topic that deserves attention in order to moderate stress susceptibility and increase climate change resilience in grapevine and tomato (reviewed by Fortes and Gallusci, 2017).
While sequencing and genotyping technologies have leaped forward significantly, limited progress in the throughput and price affordability of phenotyping technologies has slowed the identification of genetic-phenotypic associations (Fiorani and Schurr, 2013; Bolger et al., 2014).
Phenotype-based selection came long before DNA discovery and the use of genotyping technologies. However, sequencing and molecular biotechnologies made rapid progress while phenotyping biotechnologies still need to be improved. Indeed, while the sequence of genonic DNA gives a comprehensive view of genetic capacity, the information it contains is cryptic and does not directly explain the differences between cells and all plant phenotypes (Angel et al., 2012). When it comes to fruit such as tomato or grape, it is the phenotype that is directly linked to our interest. Until now, plant phenotyping mainly focused on a single scale (molecule, cell, organ, plant, field, or canopy) depending on the organ of interest (shoots or roots) and the technology used. However, Rousseau et al. (2015) insist on the importance of multi-scale analysis. Indeed, genome expression can be observed at multiple microscopic and macroscopic levels including proteomics, metabolomics, physiological traits, and others that are visible/invisible to the naked eye. Hence, phenotypic traits provide more direct information about plant production and health than genomic data do. Nevertheless, because few technologies are available, phenotyping methods have traditionally been restricted to macroscopic traits.
Fortunately, the recent improvement in phenotyping methods (reviewed by Fiorani and Schurr, 2013) enable us to broaden the concept of phenotyping and include both molecular mechanisms (proteomic and metabolomic) and all intermediate layers that result in macroscopic physiological and phenological traits (architecture, yield, taste). Progress is mainly related to the development of a wide range of sensors, their automatization and adaptation to both indoor and outdoor conditions. Hence, advances in phenotyping technologies, including cost reductions and time gains, facilitate an increase in throughput phenotyping for multi-level traits under control or field conditions (reviewed by Fiorani and Schurr, 2013; Araus and Cairns, 2014). In fact, the global phenotype can be considered the result of all the measurable traits, influenced in a complex and dynamic manner (time and space) by both genome expression and environmental effects.
Macroscopic shoot phenotyping improvements have mainly been due to the development of new sensors (Table 4) (Araus and Cairns, 2014; Fahlgren et al., 2015). For root phenotyping, new technologies were recently established (Wasson et al., 2012; Fiorani and Schurr, 2013; Kuijken et al., 2015) by easily accessing the roots (artificial growth medium and dynamic 2D or 3D imaging), and by indirect methods which phenotype roots in the soil (Table 5). For example, using a time-lapse scanning system, Dresbøll et al. (2013) demonstrated that the growth rate of tomato roots decreased under waterlogging. More recently, a series of platforms that integrate morphological parameters and, in some cases, gene expression have been developed. Among these, for example, MorphoGraphX is able to quantify several morphogenetic processes in 4D (Barbier de Reuille et al., 2015). New sensors were also developed to improve post harvested practices such as shelf life (Abano and Buah, 2014). For example, NIR spectroscopy was used to optimize the storage time of apple lots (Giovanelli et al., 2014).
On the other hand, automated facilities have evolved into high-throughput phenotyping platforms providing a powerful tool to fundamental research that can be conducted at growth chamber, greenhouse or field levels. In order to reduce error variance under field conditions, most of the sensors described above could be adapted to allow high-throughput measures, thus increasing the number of samples under analysis (reviewed by Araus and Cairns, 2014). Ground vehicles equipped with sensors were used in several studies (Andrade-Sanchez et al., 2014), while aerial vehicles with dedicated instruments facilitate rapid plant characterization in many plots, notably for phenotyping canopy traits (Araus and Cairns, 2014; Sankaran et al., 2015). Among them, due to their reduced cost, user-friendly flying control, and high autonomy, polycopters also called Unmanned Aerial Platforms (UAPs) could constitute the future of field phenotyping. The laboratory of plant-microbe interactions (INRA, Toulouse, France) set up a low cost phenotyping platform so called “Heliaphen,” which allows the growth and the high throughput phenotyping of 1,300 plants in outdoor semi-natural conditions (https://www.youtube.com/watch?v=VZSvgeWuhlw). The development of plants in high capacity pots (15 L) makes possible the study of crops during their entire life cycle. In this way, the effect of soil heterogeneity is reduced compared to field conditions. The use of a mobile robot, which phenotypes and monitors hydric conditions for each plant, is one of the original aspects of this platform (personal communication from N. Langlade).
In microscopic imaging technologies, improvements in time acquisition, automatization, and user-friendly interface make high-throughput phenotyping possible on a microscopic scale (Sozzani et al., 2014; Rousseau et al., 2015). In a recent study, Legland et al. (2012) coupled microscopic and macroscopic approaches to create a synthetic representation of cell morphology variations at the whole fruit level. The complexity and the high volume of data produced by high-throughput phenotyping platforms require computing power and robust bioinformatic tools (Araus and Cairns, 2014). Furthermore, to date, phenotyping data are still dispersed in different file types, programs, and databases and, therefore, efforts to comply with defined standards, which enable comparison and information exchange between phenotyping experiments and conditions, are needed (Krajewski et al., 2015).
The proteome integrates environmental and genetic information and is, therefore, fundamental. Knowledge of the proteome permits a more direct connection between proteins and the corresponding phenotypes (Boggess et al., 2013). Nowadays, significant improvements have been achieved in this field (reviewed by Angel et al., 2012). For example, coupling liquid chromatography (LC) separations with mass spectrometry (MS)-based technologies that enable the characterization of a protein at the proteome and sub-proteome levels, such as post-translational modifications (PTMs) of proteins like, for instance, lysine succinylation (Jin and Wu, 2016). Hence, many studies have used proteomic analyses to highlight the link between proteomic and phenotypic variations (Tanou et al., 2009; Zhao et al., 2013; Kumar and Khurana, 2014). Several studies of tomato proteome have provided both qualitative and quantitative data (reviewed by Kumar and Khurana, 2014). For example, using shotgun proteomic analysis of fruit tissues, Shah et al. (2012) presented data about the interaction between tomato fruit and Botrytis cinerea showing that the proteins produced by the fungus include those that facilitate the pathogen's penetration and growth on the plant tissue, those that inhibit resistance responses by the plant, and those that enable the pathogen to use the nutrient resources within the plant. On the other hand, the proteins produced by the plant include those that limit pathogenic infection and protect the plant tissue from additional damage.
A similar study by (Parker et al., 2013) analyzed the interaction between tomato and the Pseudomonas syringae bacteria through an iTRAQ (isobaric tags for relative and absolute quantification) quantitative proteomic approach. Proteomic data could also be used as biomarkers to facilitate the rapid identification of biotic or abiotic stress before it becomes visible through diagnostic tools (Angel et al., 2012). An interesting, novel approach involves the use of combined genomic-proteomic data to predict DNA-binding proteins (like transcription factors), integrated through computational models which can greatly promote functional annotation of tomato or other plant genomes (Motion et al., 2015). However, in contrast to the genomic data common to all cells of the same organism, proteomic data could be highly tissue-, cell-, or compartment-specific, making it more difficult to access the overview offered by plant proteome. In this context, another important issue is represented by the characterization of the protein fraction at sub-cellular level, like those specifically synthesized in plastids (Barsan et al., 2012), which can significantly influence a series of physiological processes such as fruit ripening. In another example, the characterization of proteomic changes induced during ripening processes into grape fruit skin provided important information to determine the skin parameters which could impact on wine quality (Deytieux et al., 2007). Furthermore, alterations in sugar and phenylpropanoid metabolism due to thermal stress were revealed by a quantitative proteomic study of Cabernet Sauvignon grape cells (George et al., 2015).
Metabolomics has played a remarkable role in assessing genotypic and phenotypic diversity in plants, in defining biochemical changes associated with developmental changes during plant growth and, increasingly, in compositional comparisons. Furthermore, metabolic information is often viewed as a more accurate reflection of biological endpoints than transcript or protein analysis (Harrigan et al., 2007). Therefore, metabolomic data may strongly support breeding and selection of novel yield-enhanced and nutritionally improved crops (Harrigan et al., 2007). It also seems that metabolite composition, although genetically based, is heavily influenced by environmental factors, much more, even, than enzyme activity (Biais et al., 2014). Reassuring results have proved that the hereditability of the tomato fruit metabolome, including that part of the metabolome affecting flavor, in terms of mQTL, was relatively high, in both primary metabolites (sugars and acids) (Schauer et al., 2008) and volatiles (Rambla et al., 2016). Obviously, flavor-related traits have attracted much attention. The combination of a taste panel and other omics technologies have facilitated the definition of sugars, organic acids, and volatile compounds underlying flavor and consumer preferences (Mathieu et al., 2009). Furthermore, the robustness of the mQTL and the release of flavor compounds often depend on enzymatic activities that cleave the chemical bond between the flavor compound and a glycosyl moiety. One example is represented by the non-smoky glycosyltransferase1 (NSGT1) gene, that takes part in the phenylpropanoid pathway, which was shown to prevent the “smoky” aroma attribute (Tikunov et al., 2013). Similar glycosylation/glycosidation mechanisms operate in grape varieties that usually accumulate large amounts of volatile precursors as conjugated compounds that are released following tissue maceration (Rambla et al., 2016, 2017b). Using target approaches based on knowledge of metabolic pathways has led to the characterization of several genes involved in the biosynthesis of phenylpropanoids (Tieman et al., 2010; Mageroy et al., 2012), fatty acid-derived volatiles (Speirs et al., 1998; Chen et al., 2004; Matsui et al., 2007; Shen et al., 2014), apocarotenoids (Simkin et al., 2004), esters (Goulet et al., 2015), and other phenylalanine-derived volatile compounds (Tieman et al., 2010), and in the conjugation and/or deconjugation and emission of volatiles (Tikunov et al., 2013). Moreover, Schauer et al. (2005) performed one of the first GC–MS-based surveys of the relative metabolic levels of leaves and fruits of S. lycopersicum and five sexually-compatible wild tomato species (S. pimpinellifolium, S. neorickii, S. chmielewskii, S. habrochaites, and S. pennellii). Interestingly, several biochemical markers associated with the desired traits (stress resilience, nutritional quality) were identified in the wild species. A series of robust LC–MS-based protocols for tomato metabolome have been developed at WUR (De Vos et al., 2007) and KAZUSA (Iijima et al., 2008), and exploited in several studies of fruit development and physiology (Yin et al., 2010; Mounet et al., 2012), and stress response (Etalo et al., 2013; Lucatti et al., 2013). In a recent study (D'Esposito et al., 2017), genotype × environment interaction, particularly related to sensorial attributes, was investigated in three tomato varieties using a combination of genomic, transcriptomic and metabolomic technologies. The varieties in question included the “cosmopolitan” Heinz 1706—which showed high resilience in the different environments tested—and two Italian Protected Designation of Origin (DOP) ecotypes—San Marzano and Vesuviano—which displayed high plasticity to environmental variations.
In grape, studies focusing on ripening and using complementary platforms such as NMR and GC–MS to identify metabolic markers of pre-ripening and ripening stages, are available (Fortes et al., 2011; Agudelo-Romero et al., 2013). Using an integrated transcriptomic/metabolomic approach, Agudelo-Romero et al. (2013) provided hints about how the development of a grape cultivar-specific aroma is controlled at transcriptional level. In the same context, the distinctive processes regulating the accumulation of polyphenols in berry skins of Cabernet Sauvignon and Shiraz cultivars were investigated at gene expression and metabolite levels (Degu et al., 2014).
One important phenological aspect, the terroir (i.e., the complex of all environmental factors responsible for the qualities of a grapevine cultivar grown in a specific habitat), was studied for the Corvina variety using volatile/non-volatile metabolomics, and transcriptomics. On the whole, a strong terroir-specific effect was revealed in clones grown in different vineyards—an effect that persists over several vintages (Anesi et al., 2015). The primary aromatic profile of a wine is mainly due to the genotype × environment-derived relationship between volatile metabolites and their precursors. Volatiles have been extensively studied in grape (reviewed in: González-Barreiro et al., 2015), whereas volatile precursors have scarcely been investigated (Martin et al., 2012). Recently, Rambla et al. (2016) performed an in-depth analysis of volatile and precursor metabolites in white (Airén) and red (Tempranillo) grape variety berries at different developmental stages. The use of a series of bioinformatic approaches—such as correlation networks—proved the existence of complex metabolite-metabolite patterns that were more complex in Airén, as would be expected given the enriched aroma bouquet typical of white varieties. Metabolomics has contributed much to our increased understanding of the molecular basis of biotic stress resistance. A series of metabolites, including quercetin-3-O-glucoside and a trans-feruloyl derivative, have been shown to underlie cultivar resistance to downy mildew infection (Kashif et al., 2009). More recently, Agudelo-Romero et al. (2015) concluded that berries infected with B. cinerea, reprogram carbohydrate and lipid metabolisms toward increased synthesis of secondary metabolites like trans-resveratrol and gallic acid, which are involved in plant defense.
Furthermore, metabolomic approaches have been used to assess the impact on the metabolome and fruit quality traits of mutations or genetically engineered approaches in structural/regulatory genes. Of special significance are the metabolic boost identified in tomato fruit by the light-hyperresponsive high-pigment (hp) gene (Bino et al., 2005). The authors concluded that fruits from hp plants overproduced many metabolites with antioxidant or photoprotective activities. A number of additional tomato fruit color mutants that affect the metabolite profile have been identified (list available at http://kdcomm.net/~tomato/Tomato/color.html). However, not all of these them resulted in the accumulation of quality molecules (with positive health or organoleptic effects) in the fruit. Among these mutants are the B (Beta) and Bc/Bog mutants, yielding high amounts in β-carotene and lycopene, respectively, due to a gain or loss of function in chromoplast-specific lycopene β-cyclase (Cyc-B) activity (Ronen et al., 2000, and Figure 1). Similarly, the Abg (Aubergine), Aft (Anthocyanin fruit) and Atv (Atroviolaceum) loci result in anthocyanin-accumulating fruits (Mes et al., 2008; Schreiber et al., 2012), phenotypes associated with a perturbation in the expression of the transcription factors controlling anthocyanin synthesis, such as ANTHOCYANIN 1 (ANT1) and ANTHOCYANIN 2 (AN2). In contrast to classical mutants, metabolic engineering overcomes a number of classic breeding constraints, including a limited gene-pool, time consuming processes, etc. Against this broader scenario, tomato fruits have been engineered to accumulate large amounts of many high-value nutrients (in an approach known as metabolic engineering, ME): vitamins such as folate (Díaz de la Garza et al., 2007) and ascorbate (Nunes-Nesi et al., 2005); secondary metabolites such as carotenoids, for which tomato represents a model system. An overview of ME studies of carotenoids in tomato is shown in Figure 1: so far, transgenic fruits enriched in lycopene [(Fraser et al., 2002, 2007); (ectopic expression of the bacterial (CrtB) or the tomato (PSY1) phytoene synthase genes); (Rosati et al., 2000) (down-regulation by antisense technology, of the lycopene-b-cyclase 1 (LCY-b1) gene)], β-carotene [(Apel and Bock, 2009); transplastomic expression of the bacterial lycopene-β-cyclase (CrtY) activity]; (D'Ambrosio et al., 2004, 2011) (stable transgenics for the tomato LCY-b1 gene); (Römer et al., 2000) [ectopic expression of the bacterial carotenoid isomerase (CrtI); (Rosati et al., 2000) (stable transformants espressing the arabidopsis LCY-b1 gene), lutein (Giorio et al., 2013; over-expression of the endogenous lycopene ε-cyclase (LCY-ε-) activity)], and β–xanthophylls [Dharmapuri et al., 2002; simultaneous expression of the arabidopsis LCY-b1 and of a pepper β-carotene hydroxylase 1 (CHY1)]; (D'Ambrosio et al., 2011) [overexpression of the tomato β-carotene hydroxylase 2 (CHY2)] have been achieved. Furthermore, ME tomatoes accumulating high-value ketocarotenoids (e.g., astaxanthin) have been obtained by the simultaneous expression of the β-carotene hydroxylase (CrtZ) from Haematococcus pluvialis and the algal β-carotene ketolase (CrtW) from Chlamydomonas reinhardtii (Huang et al., 2013) (Figure 1). In some cases, it is not possible to achieve stable silenced transgenic plants for a specific activity, likely due to the occurrence of a lethal phenotype in the transformant cells; in this context, an useful alternative is represented by virus induced gene silencing (VIGS), which allows to study a specific enzymatic step by transient transformation assays. In tomato fruits, this tool has been exploited to investigate the functions of all the genes involved in lycopene biosynthesis (PSY1, 2, 3; phytoene desaturase, PDS; 15-cis-ζ-carotene isomerase, Z-ISO; ζ-carotene desaturase, ZDS; carotenoid isomerase 1, like-1, like-2,CrtISO1, CrtISO-LIKE1, CrtISO-LIKE2), and the presence of three functional units, comprising PSY1, PDS/ZISO, and ZDS/CrtISO has been found (Fantini et al., 2013). ME has also been used to elucidate enzymatic activities taking place in carotenoid catabolism: with this purpose, apocarotenoid emission has been strongly reduced by the down-regulation, via RNAi technology, of the carotenoid cleavage dioxygenase 1b (CCD1b) gene (Simkin et al., 2004). Similarly, ABA biosynthesis has been investigated by through the production of RNAi plants for the 9-cis-epoxycarotenoid dioxygenase (NCED1) gene (Sun et al., 2012); and two CCD (CCD7 and CCD8) transcripts, involved in strigolactone pathway, have been characterized by tomato stable transformants, in which the two enzymatic functions had been knocked out (Vogel et al., 2010; Kohlen et al., 2012). Engineering tomatoes for high flavonoids in the fruit is a biotechnology goal as theise types of healthy metabolites are deficient in the fruit. To this end, successful efforts for flavonoid increase (Schijlen et al., 2006) and de novo anthocyanin accumulation (Zhang et al., 2013) have been reported; in a recent study, Zhang et al. (2015) used the AtMYB12 transcription factor to engineer high levels of novel phenylpropanoids in tomato. This up-regulation of specific branches of phenylpropanoid metabolism was disclosed by a combination of RNA sequencing and LC–MS analyses. Phenylpropanoids have also been the target molecules of the few ME attempts reported in grape (illustrated in Figure 2): interestingly, while only limited studies have modified the expression of structural genes, most efforts have focused on the identification of biosynthetic transcriptional regulators. Within the formers, flavonoid 3′-hydroxylase (F3′H) and flavonoid 3′,5′-hydroxylase (F3′5′H), key genes for flavonoid hydroxylation (and, thus, for their stability, color and antioxidant capacity) have been cloned in red grapevine, cv Shiraz, and their functionality has been proved by ectopic expression in Petunia hybrida (Bogs et al., 2006); in another study, Giovinazzo et al. (2005) have achieved stilbene accumulation in tomato fruits by expressing a grape stilbene synthase (STS). In the latter, a vast range of MYB transcription factors acting as activators or repressors of the pathway have has been described: interestingly, some of them have been found to perturb the whole biosynthesis [positively: MYBA1-1/2, MYBA2, MYB5a/b (Kobayashi et al., 2002; Deluc et al., 2006, 2008; Walker et al., 2007; Rinaldo et al., 2015); negatively: MYB4a/b (Cavallini et al., 2015)], while another group looks to affect distinct phenylpropanoid sub-classes [MYB14/15, directly activating STS genes (STSs)] (Höll et al., 2013); MYBF1, positively regulating flavonol synthase (FLS) expression (Czemmel et al., 2009); MYBPA1/2 and MYBC2-L1/3, respectively boosting or repressing flavan-3-ols/ proanthocyanidin synthesis (Bogs et al., 2007; Cavallini et al., 2015; Figure 2). Besides MYBs, additional transcription factors affecting phenylpropanoid metabolite pool have been isolated and characterized in grape: Wang et al. (2016), for instance, have identified a VvbHLH1 factor, whose ectopic expression in Arabidopsis resulted in increased flavonoid content, although this factor looks to be also associated to ABA-related processes, like drought and salt stresses; similarly, Malacarne et al. (2016) have recently described a new bZIP factor, named VvibZIPC22, whose ectopic expression in tobacco has proved its role in triggering flavonoid synthesis and accumulation (Figure 2). Once synthesized, flavonoids and anthocyanins are rapidly transported to the vacuole: basically, three mechanisms including vesicle trafficking, membrane transporters and glutathione S-transferase (GST)-mediated transport have been described. In grape, in particular, two kinds of anthocyanin active transporters, and localized to the tonoplast, have been discovered: two belonging to the Multidrug And Toxic Extrusion (MATE) family and called anthoMATE1-3 (AM1 and AM3), which can bind acylated anthocyanins and translocate them to the vacuole in the presence of MgATP (Gomez et al., 2009); and an ABC-type transporter, ABCC1, shown to perform the transport of glucosylated anthocyanidins (Francisco et al., 2013). More recently, three GSTs (VviGST1, VviGST3, VviGST4) have been tested for their ability to bind glutathione and monomers of different phenylpropanoids (anthocyanin, PAs, and flavonols): interestingly, all the three genes displayed the binding activity, although with distinct specificity according the phenylpropanoid class (Pérez-Díaz et al., 2016).
Over the last 25 years, a number of papers have started to dissect the genetic basis of fruit quality traits by means of QTL analysis (Duchêne et al., 2012; Klee and Tieman, 2013). In tomato, fruit morphology, yield, fruit color, and soluble solid concentration were the major focus of attention during the early QTL mapping years but recently, more complex traits such as primary metabolites, nutritional, antioxidant, and volatile compounds have received more attention (reviewed by Grandillo et al., 2013; see Table 6). The translation of those early studies into gene discovery and/or application to breeding programs remains slow. This low impact can be explained in several ways, including the limited accuracy of QTL mapping experiments due to the lack of sufficient markers; the accuracy of phenotypic evaluations; or the limitations or poor suitability of mapping population designs (Collard et al., 2005), among others.
In spite of these shortcomings, genes involved in tomato fruit morphology and sugar content QTLs have been isolated (Fridman et al., 2000; Monforte et al., 2014). Recent advances in sequencing, genotyping, and phenotyping technologies, combined with the development of a wide range of plant germplasm collections and populations, facilitate more accurate QTL detection (Chen et al., 2015; Li and Sillanpää, 2015). Today, these technologies permit the fine mapping of QTLs and candidate genes for a wide range of complex traits such as seed characteristics (Doligez et al., 2013), developmental stages (Duchêne et al., 2012), or tolerance to root chilling (Arms et al., 2015). In this last study, Arms et al. (2015) took advantage of a sub-NILs population in order to identify and functionally test candidate genes. Recently, Houel et al. (2015) worked on QTLs related to leaf area and berry quality using high-throughput genotyping technology from the Illumina® 18 K SNP chip and a mapping population of 129 microvines derived from Picovine × Ugni Blanc flb. The compact size, early flowering, and continuous production of reproductive organs make the Microvine or Dwarf and Rapid Cycling and Flowering (DRCF) mutant a valuable tool for QTL mapping (Houel et al., 2015). Combined with the 6,000 SNP markers given by the 18 K SNP chip, this microvine population has facilitated the identification of 10 QTLs of the 43 traits analyzed simultaneously (Houel et al., 2015). In tomato, the development of the Illumina® 8 K SNP chip (Sim et al., 2012) gave the research community access to affordable high-throughput genotyping. The combination of bulk segregant analysis with whole genome sequencing (i.e., QTL-seq) is another approach that has proved a cost-effective method of identifying QTLs involved in tomato fruit morphology (Illa-Berenguer et al., 2015).
Hence, several studies insist on the importance of the populations used to permit QTL fine mapping (Nicolas et al., 2016). Indeed, the choice of an appropriate genotype panel from the vast germplasm available is particularly relevant for QTL identification either in the case of using a segregating mapping population (Table 2) or in GWASenome Wide Association Studies (GWAS). Take, for example, one of the biggest collection of grapevine cultivars: that of the Institut National de la Recherche Agronomique (France). The 2,486 unique grapevine cultivars in this collection can be used to identify new QTLs (Nicolas et al., 2016). From this huge population, Nicolas et al. (2016) designed a diversity core panel of 247 grapevine cultivars with limited relatedness to use in identifying new QTLs with the GWAS approach as it captures most of the genetic and phenotypic diversity present in the original collection. Even though GWAS is a very promising strategy, the development of bi-/multi-parent populations is still highly relevant (Pascual et al., 2016) when comparing QTL detection in tomato RIL, MAGIC populations and GWAS, to find significant differences between the populations. RILs and MAGICs are especially powerful tools for rare allele mappings, whereas GWAS provides a more general view of common variants. An integration of different populations and mega QTL analysis (Monforte et al., 2014), would help detect an increasing number of small effect loci. High-throughput genotyping methods also help speed up the construction of time-consuming populations as IL collections (Barrantes et al., 2014). We would encourage the development of a larger number of these populations (especially ILs and MAGICs/NAMs) in the near future, to allow easy access to a wide range of germplasm resources.
One critical issue following QTL identification is to determine the stability and robustness of their genetic basis in different backgrounds and environments. Several studies have addressed the stability of QTLs over time and generation, as well as across environments (Monforte et al., 2001; Gur and Zamir, 2004; Chaïb et al., 2006; Doligez et al., 2013; Arms et al., 2015; Houel et al., 2015). These authors have shown that selecting stable QTLs to introgress into agronomic cultivars is feasible, a finding that must especially be taken into account considering issues relating to global warming. Introgression lines have been proved to be a highly suitable population design to address these questions (Monforte et al., 2001; Gur and Zamir, 2004).
Quantitative trait loci maps have been published for most descriptors of tomato fruit quality (color, texture, flavor) and also for specific metabolites associated with these quality descriptors. For these tomato fruit volatiles, QTLs have been identified in experimental populations obtained from crosses between tomato cultivars and different germplasm sources used as donor parents—e.g., cherry tomato (Saliba-Colombani et al., 2001; Zanor et al., 2009) or the distantly related, green-fruited, wild tomato species Solanum pennellii (Tadmor et al., 2002; Tieman et al., 2006) and Solanum habrochaites (Mathieu et al., 2009). In some cases, QTL validity (Zanor et al., 2009; Rambla et al., 2016, 2017a) has been confirmed in other populations which are, therefore, useful for breeding. Genomics has been successfully used in a limited number of cases to narrow down the regions of several hundreds of genes to a plausible candidate gene, as in the aforementioned case of the “smoky” aroma (Tikunov et al., 2013), and the gene for Brix (Zanor et al., 2009). In most cases, however, the gene underlying the QLT has yet to be identified.
Over the past 10 years, the introduction of so-called, new plant breeding techniques (NPBT) has constituted a breakthrough in the field of crop improvement. A number of technologies have been developed to produce new plants with desired traits, in which the main bottlenecks to standard genetic modification (i.e., the presence of foreign DNA in the modified food plant) are no longer an issue. In this context, several different strategies, based on the exploitation of chimeric nucleases have been applied. Overall, they rely on a system composed of sequence-specific DNA-binding domains coupled to a non-specific DNA cleavage module (reviewed in: Gaj, 2014; Sprink et al., 2015; Schaart et al., 2016) that expedite efficient genomic modifications through the introduction of sequenced specific/targeted DNA double-strand breaks (DSBs), which boost all the DNA repair components, like error-prone non-homologous end joining (NHEJ), and homology-directed repair (HDR). To date, the most widely utilized NPBTs are: zinc finger nucleases, ZFNs; transcription activator-like effector nucleases, TALENs; and Clustered Regulatory Interspaced Short Palindromic Repeats (CRISPR)/CRISPR-associated (Cas) system, CRISPR/Cas. Each strategy has its own advantages and disadvantages, as illustrated in Table 7. To date, no TALEN and ZNF studies of grape are available, whereas two proof-of-concept trials have been described in tomato: Lor et al. (2014) knocked out the PROCERA (PRO) gene involved in the negative regulation of gibberellin signaling; in contrast, Hilioti et al. (2016) have shown the effectiveness of the ZFN approach by targeting the expression of the LEAFY-COTYLEDON1-LIKE4 (L1L4) transcription factor, coding for the β subunit of nuclear factor Y and severely affecting plant development.
Currently, the most promising NPBT is based on the exploitation of the CRISPR/Cas9 system. Involved in the immune response processes of the prokaryotes (Barrangou et al., 2007), CRISPRs have been identified in 90% of sequenced archaea (Grissa et al., 2007). A simplified CRISPR system, relying on a single protein (Cas9), has been shown capable of modulating expression of specific one-by-one targets in human cells, insects and plants (Shalem et al., 2014; Konermann et al., 2015). More recently, a powerful tool for multi-modular expression of several plant genes in a single construct (with so-called “Goldenbraid” technology; Sarrion-Perdigones et al., 2011, 2013) has been adapted to CRISPR/Cas9 technology to build constructs able to modify the expression of a series of targets of interest (Vazquez-Vilar et al., 2016). Examples of efficient modifications of specific target genes have been reported both for tomato and grape: by using the CRISPR/Cas9 system. In fact, the ripening inhibitor (RIN) gene, encoding a MADS-box transcription factor regulating ethylene synthesis and, thus, fruit ripening, has been successfully mutagenized (Ito et al., 2015); simultaneously, the efficient knockout of the L-idonate dehydrogenase gene (IdnDH), involved in the tartaric acid pathway, has been achieved in both grape cell suspension and plants (Ren et al., 2016). Additionally, still in grape, a computational survey of all the CRISPR/Cas9 sites available in the genome has been performed. This has revealed the presence of 35,767,960 potential CRISPR/Cas9 target sites, distributed across all chromosomes with a preferential localization at the coding region level (Wang et al., 2016). A Grape-CRISPR website of all possible protospacers and target sites has been created and made available to the public (http://biodb.sdau.edu.cn/gc/index.html).
New plant breeding techniques have already proved successful in the potential improvement of apple and citrus fruit quality (Jia and Nian, 2014; Nishitani et al., 2016), although the feasibility of the technology has been exploited as proof-of-concept by the knockout of the PDS gene, acting on carotenoid biosynthesis at vegetative and reproductive levels. In contrast, to date, only two advanced studies in tomato have been described: precise targeting of the pectate lyase (PL) gene, which resulted in delayed fruit softening without perturbing other ripening-related parameters (Uluisik et al., 2016); and editing the SlAGAMOUS-LIKE6 (SlAGL6), a MADS-box transcription factor which provides tolerance to heat stress conditions and results in parthenocarpic fruits (Klap et al., 2016).
Taking into consideration the potential of these technologies, a more precise metabolic refinery is expected to come by selecting specific targets for nutritional and anti-nutritional molecules. This would imply the loss (knock out) and/or gain of function (activation) of selected enzymatic activities, respectively. Overall, these technologies potentially represent a powerful, innovative opportunity to introduce fine modifications in specific target genes. However, although the effect of knocking out genes has already proved successful, more work is needed for other kinds of gene remodeling (e.g., activation, production of allelic variants, etc.). To this end, significant contributions are likely to be provided by combining the CRISPR systems with additional enzymatic activities acting on DNA, such as recombinases, transposases, and DNA histone methyltransferases/acetyltransferases. These additional editing capabilities could potentially enable a vaster array of gene changes that, in the case of the fruit quality trait, may lead to a revolution in efficiency and respond better to consumer interests.
Three elements required to identify the genetic basis responsible for suitable phenotypes, and to use them to improve fruit quality produced in fields, have experienced huge technological progresses in the recent years. The first one is the constitution of germplasm banks in order to conserve the existing genetic diversity, including both natural and artificialy-induced variability. The second one is the ability to identify suitable phenotypes, notably innovations from wild genotypes, and to decipher their genetic basis. Finally, the third element is represented by the capacity to introduce the genetic elements into agronomic germplasm, remarkably through NPBT or selection assisted by markers. Altogether, the important advances in plant biotechnologies described in this review could last for long time, further facilitating plant breeding.
Indeed, biotechnologies are often praised for assuring food security to a growing Human population, through their impact on crop yield, and de facto, hunger has diminished drastically. Nevertheless, malnutrition still remains a global health problem, which also concerns developed countries (e.g., obesity) (FAO, 2015 hunger report; Steiber et al., 2004), suggesting that access to balanced and quality food is a combination of multiple factors besides agronomic yield as food allocation, waste and nutritional quality (Foley et al., 2011; Tilman and Clark, 2015). Hence, the responsibility of plant scientist is to develop solution in order to try to solve the society concerns. This could be achieved by a wide range of biotechnologies, dedicated to setting up the best suited genotypes, and producing knowledge that enables the optimization of agronomic practices (Chappell and LaValle, 2011; Amini et al., 2014).
However, in the context of recent societal mistrust about biotechnologies, sustainability of fruit production is becoming a quality trait more and more demanded by consumers, and awareness by research institutes. If one wants biotechnologies to be synonym of sustainability, improving yields and fruit quality in a long run on diverse field conditions, the notion of cost-benefits should be weighted ensuring that (i) Human and environmental health are not threatened, (ii) scientist and farmer self-reliance is not jeopardized by monopoles hold by international conglomerates including seed, chemical, and processing companies (Francis et al., 2003; Altieri and Nicholls, 2005; Chappell and LaValle, 2011; Guillemaud et al., 2016), and (iii) biotechnologies bring real benefits compared to existing processes (Temple et al., 2011; Abbo et al., 2014; Amini et al., 2014; Andersen et al., 2015; Reganold and Wachter, 2016). This debate around biotechnology use is well-illustrated by the debate around GMOs whose use could be more problematic than genetic manipulation itself (Altieri and Rosset, 1999; Chappell and LaValle, 2011; Amini et al., 2014; Guillemaud et al., 2016).
Therein, biotechnologies have their place within agroecology which bases the design of agricultural systems on the valorization of ecosystemic services to set up agri-food system economically viable, socially fair, and sustainable for the environment (Francis et al., 2003; Altieri and Nicholls, 2005; Wezel et al., 2009, 2014; García et al., 2013; Kershen, 2013). In this frame, evaluation of of biotechnologies relevance taking into account their global impact on all components of our societies, could be considered as a sustainable way to integrate biotechnologies to agriculture.
QG, AF, and AG designed the perspective and all the authors wrote the manuscript.
AF was provided by the Portuguese Foundation for Science and Technology (SFRH/BPD/100928/2014, FCT Investigator IF/00169/2015, PEst-OE/BIA/UI4046/2014), and to AG by the EC H2020 program (TRADITOM project 634561). QG benefited of the support of the Sunrise project ANR-11-BTBR-0005 funded by the ANR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors would like to thank the COST (European Cooperation in Science and Technology) Action FA1106 “Quality fruit” and Action CA15136 “EUROCAROTEN.”
Abano, E. E., and Buah, J. N. (2014). Biotechnological approaches to improve nutritional quality and shelf life of fruits and vegetables. Int. J. Eng. Technol. 4.
Abbo, S., Pinhasi van-Oss, R., Gopher, A., Saranga, Y., Ofner, I., and Peleg, Z. (2014). Plant domestication versus crop evolution: a conceptual framework for cereals and grain legumes. Trends Plant Sci. 19, 351–360. doi: 10.1016/j.tplants.2013.12.002
Agudelo-Romero, P., Erban, A., Rego, C., Carbonell-Bejerano, P., Nascimento, T., Sousa, L., et al. (2015). Transcriptome and metabolome reprogramming in Vitis vinifera cv. Trincadeira berries upon infection with Botrytis cinerea. J. Exp. Bot. 66, 1769–1785. doi: 10.1093/jxb/eru517
Agudelo-Romero, P., Erban, A., Sousa, L., Pais, M. S., Kopka, J., and Fortes, A. M. (2013). Search for transcriptional and metabolic markers of grape pre-ripening and ripening and insights into specific aroma development in three portuguese cultivars. PLoS ONE 8:e60422. doi: 10.1371/journal.pone.0060422
Altieri, M. A., and Nicholls, C. I. (2005). Agroecology and the Search for a Truly Sustainable Agriculture. United Nations Environmental Programme, Environmental Training Network for Latin America and the Caribbean.
Altieri, M. A., and Rosset, P. (1999). Ten reasons why biotechnology will not ensure food security, protect the environment and reduce poverty in the developing world. AgBioForum 2, 155–162.
Amini, S., Sharaf, S., Komeili, H. R., and Tabaee, N. A. (2014). Effect of biotechnology on biodiversity. Int. J. Farm. Allied Sci. 3, 910–915.
Andersen, M. M., Landes, X., Xiang, W., Anyshchenko, A., Falhof, J., Østerberg, J. T., et al. (2015). Feasibility of new breeding techniques for organic farming. Trends Plant Sci. 20, 426–434. doi: 10.1016/j.tplants.2015.04.011
Andrade-Sanchez, P., Gore, M. A., Heun, J. T., Thorp, K. R., Carmo-Silva, A. E., French, A., et al. (2014). Development and evaluation of a field-based, high-thoughput phenotyping platform. Funct. Plant Biol. 41, 68–79. doi: 10.1071/FP13126
Anesi, A., Stocchero, M., Dal Santo, S., Commisso, M., Zenoni, S., Ceoldo, S., et al. (2015). Towards a scientific interpretation of the terroir concept: plasticity of the grape berry metabolome. BMC Plant Biol. 15:191. doi: 10.1186/s12870-015-0584-4
Angel, T. E., Aryal, U. K., Hengel, S. M., Baker, E. S., Kelly, R. T., Robinson, W., et al. (2012). Mass spectrometry-based proteomics: Existing capabilities and future directions. Chem. Soc. Rev. 41, 3912–3928. doi: 10.1039/c2cs15331a.Mass
Aoki, K., Ogata, Y., Igarashi, K., Yano, K., Nagasaki, H., Kaminuma, E., et al. (2013). Functional genomics of tomato in a post-genome-sequencing phase. Breed. Sci. 63, 14–20. doi: 10.1270/jsbbs.63.14
Apel, W., and Bock, R. (2009). Enhancement of carotenoid biosynthesis in transplastomic tomatoes by induced lycopene-to-provitamin A conversion. Plant Physiol. 151, 59–66. doi: 10.1104/pp.109.140533
Araus, J. L., and Cairns, J. E. (2014). Field high-throughput phenotyping: the new crop breeding frontier. Trends Plant Sci. 19, 52–61. doi: 10.1016/j.tplants.2013.09.008
Arms, E. M., Bloom, A. J., and St. Clair, D. A. (2015). High-resolution mapping of a major effect QTL from wild tomato Solanum habrochaites that influences water relations under root chilling. Theor. Appl. Genet. 128, 1713–1724. doi: 10.1007/s00122-015-2540-y
Bai, H., Tao, F., Xiao, D., Liu, F., and Zhang, H. (2016). Attribution of yield change for rice-wheat rotation system in China to climate change, cultivars and agronomic management in the past three decades. Clim. Change 135, 539–553. doi: 10.1007/s10584-015-1579-8
Bai, Y., and Lindhout, P. (2007). Domestication and breeding of tomatoes: what have we gained and what can we gain in the future? Ann. Bot. 100, 1085–1094. doi: 10.1093/aob/mcm150
Barba, P., Cadle-Davidson, L., Harriman, J., Glaubitz, J. C., Brooks, S., Hyma, K., et al. (2014). Grapevine powdery mildew resistance and susceptibility loci identified on a high-resolution SNP map. Theor. Appl. Genet. 127, 73–84. doi: 10.1007/s00122-013-2202-x
Barbier de Reuille, P., Routier-Kierzkowska, A.-L., Kierzkowski, D., Bassel, G. W., Schüpbach, T., Tauriello, G., et al. (2015). MorphoGraphX: a platform for quantifying morphogenesis in 4D. Elife 4:5864. doi: 10.7554/eLife.05864
Barker, C. L., Donald, T., Pauquet, J., Ratnaparkhe, M. B., Bouquet, A., Adam-Blondon, A. F., et al. (2005). Genetic and physical mapping of the grapevine powdery mildew resistance gene, Run1, using a bacterial artificial chromosome library. Theor. Appl. Genet. 111, 370–377. doi: 10.1007/s00122-005-2030-8
Barrangou, R., Fremaux, C., Deveau, H., Richards, M., Boyaval, P., Moineau, S., et al. (2007). CRISPR provides acquired resistance against viruses in prokaryotes. Science 315, 1709–1712. doi: 10.1126/science.1138140
Barrantes, W., Fernández-del-Carmen, A., López-Casado, G., González-Sánchez, M. Á., Fernández-Muñoz, R., Granell, A., et al. (2014). Highly efficient genomics-assisted development of a library of introgression lines of Solanum pimpinellifolium. Mol. Breed. 34, 1817–1831. doi: 10.1007/s11032-014-0141-0
Barsan, C., Zouine, M., Maza, E., Bian, W., Egea, I., Rossignol, M., et al. (2012). Proteomic analysis of chloroplast-to-chromoplast transition in tomato reveals metabolic shifts coupled with disrupted thylakoid biogenesis machinery and elevated energy-production components. Plant Physiol. 160, 708–725. doi: 10.1104/pp.112.203679
Bauchet, G., and Causse, M. (2012). “Genetic diversity in tomato (Solanum lycopersicum) and its wild relatives,” in Genetic Diversity in Tomato (Solanum lycopersicum) and Its Wild Relatives, Genetic Diversity in Plants, ed M. Caliskan (InTech), 133–162. Available online at: http://www.intechopen.com/books/genetic-diversity-in-plants/genetic-diversity-in-tomato-solanum-lycopersicum-and-its-wild-relatives
Bélanger, M. C., Roger, J. M., Cartolaro, P., Viau, A. A., and Bellon-Maurel, V. (2008). Detection of powdery mildew in grapevine using remotely sensed UV-induced fluorescence. Int. J. Remote Sens. 29, 1707–1724. doi: 10.1080/01431160701395245
Berger, S., Papadopoulos, M., Schreiber, U., Kaiser, W., and Roitsch, T. (2004). Complex regulation of gene expression, photosynthesis and sugar levels by pathogen infection in tomato. Physiol. Plant. 122, 419–428. doi: 10.1111/j.1399-3054.2004.00433.x
Bergougnoux, V. (2014). The history of tomato: from domestication to biopharming. Biotechnol. Adv. 32, 170–189. doi: 10.1016/j.biotechadv.2013.11.003
Bernacchi, D., Beck-Bunn, T., Eshed, Y., Lopez, J., Petiard, V., Uhlig, J., et al. (1998). Advanced backcross QTL analysis in tomato. I. Identification of QTLs for traits of agronomic importance from Lycopersicon hirsutum. Theor. Appl. Genet. 97, 381–397. doi: 10.1007/s001220050908
Bernardo, R. (2008). Molecular markers and selection for complex traits in plants: learning from the last 20 years. Crop Sci. 48, 1649–1664. doi: 10.2135/cropsci2008.03.0131
Biais, B., Bénard, C., Beauvoit, B., Colombié, S., Prodhomme, D., Ménard, G., et al. (2014). Remarkable reproducibility of enzyme activity profiles in tomato fruits grown under contrasting environments provides a roadmap for studies of fruit metabolism. Plant Physiol. 164, 1204–1221. doi: 10.1104/pp.113.231241
Biasi, R., and Brunori, E. (2015). The on-farm conservation of grapevine (Vitis vinifera L.) landraces assures the habitat diversity in the viticultural agro-ecosystem. Vitis J. Grapevine Res. 54, 265–269.
Bino, R. J., De Vos, C. H. R., Lieberman, M., Hall, R. D., Bovy, A., Jonker, H. H., et al. (2005). The light-hyperresponsive high pigment-2dg mutation of tomato: alterations in the fruit metabolome. New Phytol. 166, 427–438. doi: 10.1111/j.1469-8137.2005.01362.x
Blanca, J., Montero-Pau, J., Sauvage, C., Bauchet, G., Illa, E., Díez, M. J., et al. (2015). Genomic variation in tomato, from wild ancestors to contemporary breeding accessions. BMC Genomics 16:257. doi: 10.1186/s12864-015-1444-1
Boggess, M. V., Lippolis, J. D., Hurkman, W. J., Fagerquist, C. K., Briggs, S. P., Gomes, A. V., et al. (2013). The need for agriculture phenotyping: moving from genotype to phenotype. J. Proteomics 93, 20–39. doi: 10.1016/j.jprot.2013.03.021
Bogs, J., Ebadi, A., McDavid, D., and Robinson, S. P. (2006). Identification of the flavonoid hydroxylases from grapevine and their regulation during fruit development. Plant Physiol. 140, 279–291. doi: 10.1104/pp.105.073262
Bogs, J., Jaffe, F. W., Takos, A. M., Walker, A. R., and Robinson, S. P. (2007). The grapevine transcription factor VvMYBPA1 regulates proanthocyanidin synthesis during fruit development. Plant Physiol. 143, 1347–1361. doi: 10.1104/pp.106.093203
Bolger, M. E., Weisshaar, B., Scholz, U., Stein, N., Usadel, B., and Mayer, K. F. X. (2014). Plant genome sequencing - applications for crop improvement. Curr. Opin. Biotechnol. 26, 31–37. doi: 10.1016/j.copbio.2013.08.019
Broman, K. W. (2005). The genomes of recombinant inbred lines. Genetics 169, 1133–1146. doi: 10.1534/genetics.104.035212
Brooks, C., Nekrasov, V., Lippman, Z. B., and Van Eck, J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166, 1292–1297. doi: 10.1104/pp.114.247577
Burbidge, A., Grieve, T. M., Jackson, A., Thompson, A., McCarty, D. R., and Taylor, I. B. (1999). Characterization of the ABA-deficient tomato mutant notabilis and its relationship with maizeVp14. Plant J. 17, 427–431. doi: 10.1046/j.1365-313X.1999.00386.x
Calafiore, R., Ruggieri, V., Raiola, A., Rigano, M. M., Sacco, A., Hassan, M. I., et al. (2016). Exploiting genomics resources to identify candidate genes underlying antioxidants content in tomato fruit. Front. Plant Sci. 7:397. doi: 10.3389/fpls.2016.00397
Calderón, R., Castro, O., Arauz, F., and Bonatti, J. (2014). “Use of infrared sensors for early detection of bacterial wilt caused by Ralstonia solanacearum in tomato plants,” in CIGR Proceedings, Vol. 1 (San Jose, CL: Costa Rica).
Capel, C., Del Carmen, A. F., Alba, J. M., Lima-Silva, V., Hernández-Gras, F., Salinas, M., et al. (2015). Wide-genome QTL mapping of fruit quality traits in a tomato RIL population derived from the wild-relative species Solanum pimpinellifolium L. Theor. Appl. Genet. 128, 2019–2035. doi: 10.1007/s00122-015-2563-4
Carrera, J., Fernández del Carmen, A., Fernández-Muñoz, R., Rambla, J. L., Pons, C., Jaramillo, A., et al. (2012). Fine-tuning tomato agronomic properties by computational genome redesign. PLoS Comput. Biol. 8:e1002528. doi: 10.1371/journal.pcbi.1002528
Causse, M., Duffe, P., Gomez, M. C., Buret, M., Damidaux, R., Zamir, D., et al. (2004). A genetic map of candidate genes and QTLs involved in tomato fruit size and composition. J. Exp. Bot. 55, 1671–1685. doi: 10.1093/jxb/erh207
Cavallini, E., Matus, J. T., Finezzo, L., Zenoni, S., Loyola, R., Guzzo, F., et al. (2015). The phenylpropanoid pathway is controlled at different branches by a set of R2R3-MYB C2 repressors in grapevine. Plant Physiol. 167, 1448–1470. doi: 10.1104/pp.114.256172
Cavanagh, C., Morell, M., Mackay, I., and Powell, W. (2008). From mutations to MAGIC: resources for gene discovery, validation and delivery in crop plants. Curr. Opin. Plant Biol. 11, 215–221 doi: 10.1016/j.pbi.2008.01.002
Cebolla-Cornejo, J., Roselló, S., and Nuez, F. (2013). Phenotypic and genetic diversity of Spanish tomato landraces. Sci. Hortic. 162, 150–164. doi: 10.1016/j.scienta.2013.07.044
Čermák, T., Baltes, N. J., Čegan, R., Zhang, Y., and Voytas, D. F. (2015). High-frequency, precise modification of the tomato genome. BMC Genomics 16:232. doi: 10.1186/s13059-015-0796-9
Chaerle, L., Lenk, S., Leinonen, I., Jones, H. G., Van Der Straeten, D., and Buschmann, C. (2009). Multi-sensor plant imaging: towards the development of a stress-catalogue. Biotechnol. J. 4, 1152–1167. doi: 10.1002/biot.200800242
Chaïb, J., Lecomte, L., Buret, M., and Causse, M. (2006). Stability over genetic backgrounds, generations and years of quantitative trait locus (QTLs) for organoleptic quality in tomato. Theor. Appl. Genet. 112, 934–944. doi: 10.1007/s00122-005-0197-7
Chalhoub, B., Denoeud, F., Liu, S., Parkin, I. A., Tang, H., Wang, X., et al. (2014). Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345, 950–953.
Chappell, M. J., and LaValle, L. A. (2011). Food security and biodiversity: can we have both? An agroecological analysis. Agric. Hum. Values 28, 3–26. doi: 10.1007/s10460-009-9251-4
Chen, G., Hackett, R., Walker, D., Taylor, A., Lin, Z., and Grierson, D. (2004). Identification of a specific isoform of tomato lipoxygenase (TomloxC) involved in the generation of fatty acid-derived flavor compounds. Plant Physiol. 136, 2641–2651. doi: 10.1104/pp.104.041608
Chen, J., Wang, N., Fang, L.-C., Liang, Z.-C., Li, S.-H., and Wu, B.-H. (2015). Construction of a high-density genetic map and QTLs mapping for sugars and acids in grape berries. BMC Plant Biol. 15:28. doi: 10.1186/s12870-015-0428-2
Chen, X., Chen, F., Chen, Y., Gao, Q., Yang, X., Yuan, L., et al. (2013). Modern maize hybrids in Northeast China exhibit increased yield potential and resource use efficiency despite adverse climate change. Glob. Chang. Biol. 19, 923–936. doi: 10.1111/gcb.12093
Coleman, C., Copetti, D., Cipriani, G., Hoffmann, S., Kozma, P., Kovács, L., et al. (2009). The powdery mildew resistance gene REN1 co-segregates with an NBS-LRR gene cluster in two Central Asian grapevines. BMC Genet. 10:89. doi: 10.1186/1471-2156-10-89
Collard, B. C. Y., Jahufer, M. Z. Z., Brouwer, J. B., and Pang, E. C. K. (2005). An introduction to markers, quantitative trait loci (QTL) mapping and marker-assisted selection for crop improvement: the basic concepts. Euphytica 142, 169–196. doi: 10.1007/s10681-005-1681-5
Corrado, G., Piffanelli, P., Caramante, M., Coppola, M., and Rao, R. (2013). SNP genotyping reveals genetic diversity between cultivated landraces and contemporary varieties of tomato. BMC Genomics 14:835. doi: 10.1186/1471-2164-14-835
Czemmel, S., Stracke, R., Weisshaar, B., Cordon, N., Harris, N. N., Walker, A. R., et al. (2009). The Grapevine R2R3-MYB transcription factor VvMYBF1 regulates flavonol synthesis in developing grape berries. Plant Physiol. 151, 1513–1530. doi: 10.1104/pp.109.142059
D'Ambrosio, C., Giorio, G., Marino, I., Merendino, A., Petrozza, A., Salfi, L., et al. (2004). Virtually complete conversion of lycopene into β-carotene in fruits of tomato plants transformed with the tomato lycopene β-cyclase (tlcy-b) cDNA. Plant Sci. 166, 207–214. doi: 10.1016/j.plantsci.2003.09.015
D'Ambrosio, C., Stigliani, A. L., and Giorio, G. (2011). Overexpression of CrtR-b2 (carotene beta hydroxylase 2) from S. lycopersicum L. differentially affects xanthophyll synthesis and accumulation in transgenic tomato plants. Transgenic Res. 20, 47–60. doi: 10.1007/s11248-010-9387-4
D'Esposito, D., Ferriello, F., Dal Molin, A., Diretto, G., Sacco, A., Minio, A., et al. (2017). Unraveling the complexity of transcriptomic, metabolomic and quality environmental response of tomato fruit. BMC Plant Biol. 17:66. doi: 10.1186/s12870-017-1008-4
de Toledo Thomazella, D. P., Brail, Q., Dahlbeck, D., and Staskawicz, B. J. (2016). CRISPR-Cas9 mediated mutagenesis of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. bioRxiv, 064824. doi: 10.1101/064824
De Vos, R. C., Moco, S., Lommen, A., Keurentjes, J. J., Bino, R. J., and Hall, R. D. (2007). Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2, 778–791. doi: 10.1038/nprot.2007.95
Deery, D., Jimenez-Berni, J., Jones, H., Sirault, X., and Furbank, R. (2014). Proximal remote sensing buggies and potential applications for field-based phenotyping. Agronomy 4, 349–379. doi: 10.3390/agronomy4030349
Degu, A., Hochberg, U., Sikron, N., Venturini, L., Buson, G., Ghan, R., et al. (2014). Metabolite and transcript profiling of berry skin during fruit development elucidates differential regulation between Cabernet Sauvignon and Shiraz cultivars at branching points in the polyphenol pathway. BMC Plant Biol. 14:188. doi: 10.1186/s12870-014-0188-4
Deluc, L., Barrieu, F. C., Marchive, C., Lauvergeat, V., Decendit, A., Richard, T., et al. (2006). Characterization of a grapevine R2R3-MYB transcription factor that regulates the phenylpropanoid pathway. Plant Physiol. 140:499. doi: 10.1104/pp.105.067231
Deluc, L., Bogs, J., Walker, A. R., Ferrier, T., Decendit, A., Merillon, J.-M., et al. (2008). The transcription factor VvMYB5b contributes to the regulation of anthocyanin and proanthocyanidin biosynthesis in developing grape berries. Plant Physiol. 147, 2041–2053. doi: 10.1104/pp.108.118919
Deytieux, C., Geny, L., Lapaillerie, D., Claverol, S., Bonneu, M., and Donèche, B. (2007). Proteome analysis of grape skins during ripening. J. Exp. Bot. 58, 1851–1862. doi: 10.1093/jxb/erm049
Dharmapuri, S., Rosati, C., Pallara, P., Aquilani, R., Bouvier, F., Camara, B., et al. (2002). Metabolic engineering of xanthophyll content in tomato fruits. FEBS Lett. 519, 30–34. doi: 10.1016/S0014-5793(02)02699-6
Di Matteo, A., Rigano, M. M., and Sacco, A. (2011). Genetic transformation in tomato: novel tools to improve fruit quality and pharmaceutical production,” in Genetic Transformation. Available online at: http://www.intechopen.com/source/pdfs/18813/InTech-Genetic_transformation_in_tomato_novel_tools_to_improve_fruit_quality_and_pharmaceutical_production.pdf
Díaz de la Garza, R. I., Gregory, J. F., and Hanson, A. D. (2007). Folate biofortification of tomato fruit. Proc. Natl. Acad. Sci. U.S.A. 104, 4218–4222. doi: 10.1073/pnas.0700409104
Doganlar, S., Frary, A., Daunay, M. C., Lester, R. N., and Tanksley, S. D. (2002). Conservation of gene function in the Solanaceae as revealed by comparative mapping of domestication traits in eggplant. Genetics 161, 1713–1726.
Doligez, A., Bertrand, Y., Farnos, M., Grolier, M., Romieu, C., Esnault, F., et al. (2013). New stable QTLs for berry weight do not colocalize with QTLs for seed traits in cultivated grapevine (Vitis vinifera L.). BMC Plant Biol. 13:217. doi: 10.1186/1471-2229-13-217
Doucleff, M., Jin, Y., Gao, F., Riaz, S., Krivanek, A. F., and Walker, M. A. (2004). A genetic linkage map of grape, utilizing Vitis rupestris and Vitis arizonica. Theor. Appl. Genet. 109, 1178–1187. doi: 10.1007/s00122-004-1728-3
Dresbøll, D. B., Thorup-Kristensen, K., McKenzie, B. M., Dupuy, L. X., and Bengough, A. G. (2013). Timelapse scanning reveals spatial variation in tomato (Solanum lycopersicum L.) root elongation rates during partial waterlogging. Plant Soil 369, 467–477. doi: 10.1007/s11104-013-1592-5
Duchêne, E., Butterlin, G., Dumas, V., and Merdinoglu, D. (2012). Towards the adaptation of grapevine varieties to climate change: QTLs and candidate genes for developmental stages. Theor. Appl. Genet. 124, 623–635. doi: 10.1007/s00122-011-1734-1
Eitel, J. U., Vierling, L. A., Long, D. S., and Hunt, E. R. (2011). Early season remote sensing of wheat nitrogen status using a green scanning laser. Agric. For. Meteorol. 151, 1338–1345. doi: 10.1016/j.agrformet.2011.05.015
Elizondo, R., and Oyanedel, E. (2011). Field testing of tomato chilling tolerance under varying light and temperature conditions. Chilean J. Agric. Res. 70, 552–558. doi: 10.4067/S0718-58392010000400004
Eshed, Y., and Zamir, D. (1995). An introgression line population of Lycopersicon pennellii in the cultivated tomato enables the identification and fine mapping of yield-associated QTL. Genetics 141, 1147–1162.
Etalo, D. W., Stulemeijer, I. J. E., Esse, H. P., van Vos, R. C. H., de Bouwmeester, H. J., and Joosten, M. H. A. J. (2013). System-wide hypersensitive response-associated transcriptome and metabolome reprogramming in tomato. Plant Physiol. 162, 1599–1617. doi: 10.1104/pp.113.217471
Fahlgren, N., Gehan, M. A., and Baxter, I. (2015). Lights, camera, action: high-throughput plant phenotyping is ready for a close-up. Curr. Opin. Plant Biol. 24, 93–99. doi: 10.1016/j.pbi.2015.02.006
Fantini, E., Falcone, G., Frusciante, S., Giliberto, L., and Giuliano, G. (2013). Dissection of tomato lycopene biosynthesis through virus-induced gene silencing. Plant Physiol. 163, 986–998. doi: 10.1104/pp.113.224733
FAO IFAD, WFP (2015). “The State of Food Insecurity in the World 2015,” in Meeting the 2015 International Hunger Targets: Taking Stock of Uneven Progress. Rome: Food and Agriculture Organization Publications.
Feechan, A., Anderson, C., Torregrosa, L., Jermakow, A., Mestre, P., Wiedemann-Merdinoglu, S., et al. (2013). Genetic dissection of a TIR-NB-LRR locus from the wild North American grapevine species Muscadinia rotundifolia identifies paralogous genes conferring resistance to major fungal and oomycete pathogens in cultivated grapevine. Plant J. 76, 661–674. doi: 10.1111/tpj.12327
Fernie, A. R., Tadmor, Y., and Zamir, D. (2006). Natural genetic variation for improving crop quality. Curr. Opin. Plant Biol. 9, 196–202. doi: 10.1016/j.pbi.2006.01.010
Fiorani, F., and Schurr, U. (2013). Future scenarios for plant phenotyping. Annu. Rev. Plant Biol. 64, 267–291. doi: 10.1146/annurev-arplant-050312-120137
Fiorani, F., Rascher, U., Jahnke, S., and Schurr, U. (2012). Imaging plants dynamics in heterogenic environments. Curr. Opin. Biotechnol. 23, 227–235. doi: 10.1016/j.copbio.2011.12.010
Fischer, B. M., Salakhutdinov, I., Akkurt, M., Eibach, R., Edwards, K. J., Töpfer, R., et al. (2004). Quantitative trait locus analysis of fungal disease resistance factors on a molecular map of grapevine. Theor. Appl. Genet. 108, 501–515. doi: 10.1007/s00122-003-1445-3
Fodor, A., Segura, V., Denis, M., Neuenschwander, S., Fournier-Level, A., Chatelet, P., et al. (2014). Genome-wide prediction methods in highly diverse and heterozygous species: proof-of-concept through simulation in grapevine. PLoS ONE 9:e110436. doi: 10.1371/journal.pone.0110436
Foley, J. A., Ramankutty, N., Brauman, K. A., Cassidy, E. S., Gerber, J. S., Johnston, M., et al. (2011). Solutions for a cultivated planet. Nature 478, 337–342. doi: 10.1038/nature10452
Fortes, A. M., Agudelo-Romero, P., Silva, M. S., Ali, K., Sousa, L., Maltese, F., et al. (2011). Transcript and metabolite analysis in Trincadeira cultivar reveals novel information regarding the dynamics of grape ripening. BMC Plant Biol. 11:149. doi: 10.1186/1471-2229-11-149
Fortes, A. M., and Gallusci, P. (2017). Plant stress responses and phenotypic plasticity in the epigenomics era: perspectives on the grapevine scenario, a model for perennial crop plants. Front. Plant Sci. 8:82. doi: 10.3389/fpls.2017.00082
Fortes, A. M., Teixeira, R. T., and Agudelo-Romero, P. (2015). Complex interplay of hormonal signals during grape berry ripening. Molecules 20, 9326–9343. doi: 10.3390/molecules20059326
Fowler, C. (2008). The Svalbard seed vault and crop security. Bioscience 58:190. doi: 10.1641/B580302
Francis, C., Lieblein, G., Gliessman, S., Breland, T. A., Creamer, N., Harwood, R., et al. (2003). Agroecology : the ecology of food systems agroecology : the ecology of food systems. J. Sustain. Agric. 22, 99–118. doi: 10.1300/J064v22n03_10
Francisco, R. M., Regalado, A., Ageorges, A., Burla, B. J., Bassin, B., Eisenach, C., et al. (2013). ABCC1, an ATP binding cassette protein from grape berry, transports anthocyanidin 3-O-glucosides. Plant Cell 25, 1840–1854. doi: 10.1105/tpc.112.102152
Fraser, P. D., Enfissi, E. M. A., Halket, J. M., Truesdale, M. R., Yu, D., Gerrish, C., et al. (2007). Manipulation of phytoene levels in tomato fruit: effects on isoprenoids, plastids, and intermediary metabolism. Plant Cell 19, 3194–3211. doi: 10.1105/tpc.106.049817
Fraser, P. D., Romer, S., Shipton, C. A., Mills, P. B., Kiano, J. W., Misawa, N., et al. (2002). Evaluation of transgenic tomato plants expressing an additional phytoene synthase in a fruit-specific manner. Proc. Natl. Acad. Sci. U.S.A. 99, 1092–1097. doi: 10.1073/pnas.241374598
Fray, R. G., and Grierson, D. (1993). Identification and genetic analysis of normal and mutant phytoene synthase genes of tomato by sequencing, complementation and co-suppression. Plant Mol. Biol. 22, 589–602. doi: 10.1007/BF00047400
Fridman, E., Pleban, T., and Zamir, D. (2000). A recombination hotspot delimits a wild-species quantitative trait locus for tomato sugar content to 484 bp within an invertase gene. Proc. Natl. Acad. Sci. U.S.A. 97, 4718–4723. doi: 10.1073/pnas.97.9.4718
Fuentes, S., De Bei, R., Pech, J., and Tyerman, S. (2012). Computational water stress indices obtained from thermal image analysis of grapevine canopies. Irrigat. Sci. 30, 523–536. doi: 10.1007/s00271-012-0375-8
Fulton, T. M., Beck-Bunn, T., Emmatty, D., Eshed, Y., Lopez, J., Petiard, V., et al. (1997). QTL analysis of an advanced backcross of Lycopersicon peruvianum to the cultivated tomato and comparisons with QTLs found in other wild species. Theor. Appl. Genet. 95, 881–894. doi: 10.1007/s001220050639
Gaj, T. (2014). ZFN, TALEN and CRISPR/Cas based methods for genome engineering. 31, 397–405. doi: 10.1016/j.tibtech.2013.04.004.ZFN
Galpaz, N., Ronen, G., Khalfa, Z., Zamir, D., and Hirschberg, J. (2006). A chromoplast-specific carotenoid biosynthesis pathway is revealed by cloning of the tomato white-flower locus. Plant Cell 18, 1947–1960. doi: 10.1105/tpc.105.039966
Galpaz, N., Wang, Q., Menda, N., Zamir, D., and Hirschberg, J. (2008). Abscisic acid deficiency in the tomato mutant high-pigment 3 leading to increased plastid number and higher fruit lycopene content. Plant J. 53, 717–730. doi: 10.1111/j.1365-313X.2007.03362.x
García, A. C., Izquierdo, F. G., Hernández, O. L., Margarita, M., Armas, D., De López, R. H., et al. (2013). Biotechnology of humified materials obtained from vermicomposts for sustainable agroecological purposes. Afr. J. Biotechnol. 12, 625–634. doi: 10.5897/AJBX12.014
George, I. S., Pascovici, D., Mirzaei, M., and Haynes, P. A. (2015). Quantitative proteomic analysis of cabernet sauvignon grape cells exposed to thermal stresses reveals alterations in sugar and phenylpropanoid metabolism. Proteomics 15, 3048–3060. doi: 10.1002/pmic.201400541
Gepts, P. (2014). The contribution of genetic and genomic approaches to plant domestication studies. Curr. Opin. Plant Biol. 18, 51–59. doi: 10.1016/j.pbi.2014.02.001
Gil, R., Merav, C., Dani, Z., and Joseph, H. (1999). Regulation of carotenoid biosynthesis during tomato fruit development: expression of the gene for lycopene epsilon-cyclase is down-regulated during ripening and is elevated in the mutant Delta. Plant J. 17, 341–351. doi: 10.1046/j.1365-313X.1999.00381.x
Giorio, G., Yildirim, A., Stigliani, A. L., and D'Ambrosio, C. (2013). Elevation of lutein content in tomato: a biochemical tug-of-war between lycopene cyclases. Metab. Eng. 20, 167–176. doi: 10.1016/j.ymben.2013.10.007
Giovanelli, G., Sinelli, N., Beghi, R., Guidetti, R., and Casiraghi, E. (2014). NIR spectroscopy for the optimization of postharvest apple management. Postharvest Biol. Technol. 87, 13–20. doi: 10.1016/j.postharvbio.2013.07.041
Giovinazzo, G., D'Amico, L., Paradiso, A., Bollini, R., Sparvoli, F., and DeGara, L. (2005). Antioxidant metabolite profiles in tomato fruit constitutively expressing the grapevine stilbene synthase gene. Plant Biotechnol. J. 3, 57–69. doi: 10.1111/j.1467-7652.2004.00099.x
Goldsbrough, A., Belzile, F., and Yoder, J. (1994). Complementation of the Tomato anthocyanin without (aw) mutant using the Dihydroflavonol 4-Reductase Gene. Plant Physiol. 105, 491–496.
Gomez, C., Terrier, N., Torregrosa, L., Vialet, S., Fournier-Level, A., Verries, C., et al. (2009). Grapevine MATE-type proteins act as vacuolar H+-dependent acylated anthocyanin transporters. Plant Physiol. 150, 402–415. doi: 10.1104/pp.109.135624
González-Barreiro, C., Rial-Otero, R., Cancho-Grande, B., and Simal-Gándara, J. (2015). Wine aroma compounds in grapes: a critical review. Crit. Rev. Food Sci. Nutr. 55, 202–218. doi: 10.1080/10408398.2011.650336
Goulet, C., Kamiyoshihara, Y., Lam, N. B., Richard, T., Taylor, M. G., Tieman, D. M., et al. (2015). Divergence in the enzymatic activities of a tomato and Solanum pennellii alcohol acyltransferase impacts fruit volatile ester composition. Mol. Plant 8, 153–162. doi: 10.1016/j.molp.2014.11.007
Grandillo, S., Termolino, P., and van der Knaap, E. (2013). Molecular mapping of complex traits in tomato. Genet. Genomics Breed. Tomato 150–227. doi: 10.1201/b14578-7
Grattapaglia, D., and Sederoff, R. (1994). Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics 137, 1121–1137. doi: 10.1007/s11033-010-0612-2
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). The CRISPRdb database and tools to display CRISPRs and to generate dictionaries of spacers and repeats. BMC Bioinformatics 8:172. doi: 10.1186/1471-2105-8-172
Guillemaud, T., Lombaert, E., and Bourguet, D. (2016). Conflicts of interest in GM Bt crop efficacy and durability studies. PLoS ONE 11:e0167777. doi: 10.1371/journal.pone.0167777
Gur, A., and Zamir, D. (2004). Unused natural variation can lift yield barriers in plant breeding. PLoS Biol. 2:245. doi: 10.1371/journal.pbio.0020245
Handa, A. K., Anwar, R., and Mattoo, A. K. (2014). Biotechnology of fruit quality. Fruit Ripening Physiol. Signal. Genomics 259–290. doi: 10.1079/9781845939625.0259
Harrigan, G. G., Martino-Catt, S., and Glenn, K. C. (2007). Metabolomics, metabolic diversity and genetic variation in crops. Metabolomics 3, 259–272. doi: 10.1007/s11306-007-0076-0
Harrison, E., Burbidge, A., Okyere, J. P., Thompson, A. J., and Taylor, I. B. (2011). Identification of the tomato ABA-deficient mutant sitiens as a member of the ABA-aldehyde oxidase gene family using genetic and genomic analysis. Plant Growth Regul. 64, 301–309. doi: 10.1007/s10725-010-9550-1
Henikoff, S., Till, B. J., Comai, L., Division, B. S., Hutchinson, F., and Washington, S. H. (2004). TILLING. Traditional mutagenesis meets functional genomics. Plant Physiol. 135, 630–636. doi: 10.1104/pp.104.041061.630
Herzog, K., Wind, R., and Töpfer, R. (2015). Impedance of the grape berry cuticle as a novel phenotypic trait to estimate resistance to Botrytis cinerea. Sensors 15, 12498–12512. doi: 10.3390/s150612498
Hilioti, Z., Ganopoulos, I., Ajith, S., Bossis, I., and Tsaftaris, A. (2016). A novel arrangement of zinc finger nuclease system for in vivo targeted genome engineering: the tomato LEC1-LIKE4 gene case. Plant Cell Rep. 35, 1–15. doi: 10.1007/s00299-016-2031-x
Höll, J., Vannozzi, A., Czemmel, S., D'Onofrio, C., Walker, A. R., Rausch, T., et al. (2013). The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 25, 4135–4149. doi: 10.1105/tpc.113.117127
Honnay, O., Jacquemyn, H., and Aerts, R. (2012). Crop wild relatives: more common ground for breeders and ecologists. Front. Ecol. Environ. 10:121. doi: 10.1890/12.WB.007
Hosoi, F., and Omasa, K. (2012). Estimation of vertical plant area density profiles in a rice canopy at different growth stages by high-resolution portable scanning lidar with a lightweight mirror. ISPRS J. Photogramm. Remote Sens. 74, 11–19. doi: 10.1016/j.isprsjprs.2012.08.001
Hosoi, F., Nakabayashi, K., and Omasa, K. (2011). 3-D modeling of tomato canopies using a high-resolution portable scanning lidar for extracting structural information. Sensors 11, 2166–2174. doi: 10.3390/s110202166
Houel, C., Chatbanyong, R., Doligez, A., Rienth, M., Foria, S., Luchaire, N., et al. (2015). Identification of stable QTLs for vegetative and reproductive traits in the microvine (Vitis vinifera L.) using the 18 K Infinium chip. BMC Plant Biol. 15, 205. doi: 10.1186/s12870-015-0588-0
Huang, J. C., Zhong, Y. J., Liu, J., Sandmann, G., and Chen, F. (2013). Metabolic engineering of tomato for high-yield production of astaxanthin. Metab. Eng. 17, 59–67. doi: 10.1016/j.ymben.2013.02.005
Huang, Z., and van der Knaap, E. (2011). Tomato fruit weight 11.3 maps close to fasciated on the bottom of chromosome 11. Theor. Appl.Genetics 123, 465–474. doi: 10.1007/s00122-011-1599-3
Iijima, Y., Nakamura, Y., Ogata, Y., Tanaka, K., Sakurai, N., Suda, K., et al. (2008). Metabolite annotations based on the integration of mass spectral information. Plant J. 54, 949–962. doi: 10.1111/j.1365-313X.2008.03434.x
Illa-Berenguer, E., Van Houten, J., Huang, Z., and van der Knaap, E. (2015). Rapid and reliable identification of tomato fruit weight and locule number loci by QTL-seq. Theor. Appl. Genet. 128, 1329–1342. doi: 10.1007/s00122-015-2509-x
Isaacson, T., Ronen, G., Zamir, D., and Hirschberg, J. (2002). Cloning of tangerine from tomato reveals a carotenoid isomerase essential for the production of beta-carotene and xanthophylls in plants. Plant Cell 14, 333–342. doi: 10.1105/tpc.010303.2001
Ishimwe, R., Abutaleb, K., and Ahmed, F. (2014). Applications of thermal imaging in agriculture—A review. Adv. Remote Sens. 3, 128. doi: 10.4236/ars.2014.33011
Ito, Y., Nishizawa-Yokoi, A., Endo, M., Mikami, M., and Toki, S. (2015). CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem. Biophys. Res. Commun. 467, 76–82. doi: 10.1016/j.bbrc.2015.09.117
Jacobs, T. B., and Martin, G. B. (2016). High-throughput CRISPR vector construction and characterization of DNA modifications by generation of tomato hairy roots. J. Vis. Exp. e53843–e53843. doi: 10.3791/53843
Jaillon, O., Aury, J.-M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi: 10.1038/nature06148
Jia, H., and Nian, W. (2014). Targeted genome editing of sweet orange using Cas9/sgRNA. PLoS ONE 9:93806. doi: 10.1371/journal.pone.0093806
Jiménez-Gómez, J. M., Alonso-Blanco, C., Borja, A., Anastasio, G., Angosto, T., Lozano, R., et al. (2007). Quantitative genetic analysis of flowering time in tomato. Genome 50, 303–315. doi: 10.1139/G07-009
Jin, W., and Wu, F. (2016). Proteome-wide identification of lysine succinylation in the proteins of tomato (Solanum lycopersicum). PLoS ONE 11:0147586. doi: 10.1371/journal.pone.0147586
Kashif, A., Federica, M., Eva, Z., Martina, R., Young, H. C., and Robert, V. (2009). NMR metabolic fingerprinting based identification of grapevine metabolites associated with downy mildew resistance. J. Agric. Food Chem. 57, 9599–9606. doi: 10.1021/jf902069f
Kershen, D. L. (2013). The contested vision for agriculture's future: sustainable intensive agriculture and agroecology. Creighton Law Rev. 46, 1–36.
Khan, N., Kazmi, R. H., Willems, L. A. J., van Heusden, A. W., Ligterink, W., and Hilhorst, H. W. M. (2012). Exploring the natural variation for seedling traits and their link with seed dimensions in tomato. PLoS ONE 7:e43991. doi: 10.1371/journal.pone.0043991
Klap, C., Yeshayahou, E., Bolger, A. M., Arazi, T., Gupta, S. K., Shabtai, S., et al. (2016). Tomato facultative parthenocarpy results from Sl AGAMOUS-LIKE 6 loss of function. Plant Biotechnol. J. 15, 634–647. doi: 10.1111/pbi.12662
Klee, H. J. (2010). Improving the flavor of fresh fruits: genomics, biochemistry, and biotechnology. New Phytol. 187, 44–56. doi: 10.1111/j.1469-8137.2010.03281.x
Klee, H. J., and Tieman, D. M. (2013). Genetic challenges of flavor improvement in tomato. Trends Genet. 29, 257–262. doi: 10.1016/j.tig.2012.12.003
Kobayashi, S., Ishimaru, M., Hiraoka, K., and Honda, C. (2002). Myb-related genes of the Kyoho grape (Vitis labruscana) regulate anthocyanin biosynthesis. Planta 215, 924–933. doi: 10.1007/s00425-002-0830-5
Kohlen, W., Charnikhova, T., Lammers, M., Pollina, T., Tóth, P., Haider, I., et al. (2012). The tomato carotenoid cleavage dioxygenase8 (SlCCD8) regulates rhizosphere signaling, plant architecture and affects reproductive development through strigolactone biosynthesis. New Phytol. 196, 535–547. doi: 10.1111/j.1469-8137.2012.04265.x
Konermann, S., Brigham, M. D., Trevino, A. E., Abudayyeh, O. O., Barcena, C., Hsu, P. D., et al. (2015). Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 61422–61427. doi: 10.1038/nature14136.Genome-scale
Krajewski, P., Chen, D., Ćwiek, H., Van Dijk, A. D. J., Fiorani, F., Kersey, P., et al. (2015). Towards recommendations for metadata and data handling in plant phenotyping. J. Exp. Bot. 66, 5417–5427. doi: 10.1093/jxb/erv271
Krivanek, A. F., Riaz, S., and Walker, M. A. (2006). Identification and molecular mapping of PdR1, a primary resistance gene to Pierce's disease in Vitis. Theor. Appl. Genet. 112, 1125–1131. doi: 10.1007/s00122-006-0214-5
Kuijken, R. C. P., Van Eeuwijk, F. A., Marcelis, L. F. M., and Bouwmeester, H. J. (2015). Root phenotyping: from component trait in the lab to breeding. J. Exp. Bot. 66, 5389–5401. doi: 10.1093/jxb/erv239
Kumar, R., and Khurana, A. (2014). Functional genomics of tomato: opportunities and challenges in post-genome NGS era. J. Biosci. 39, 917–929. doi: 10.1007/s12038-014-9480-6
Kurowska, M., Daszkowska-Golec, A., Gruszka, D., Marzec, M., Szurman, M., Szarejko, I., et al. (2011). TILLING - a shortcut in functional genomics. J. Appl. Genet. 52, 371–390. doi: 10.1007/s13353-011-0061-1
Laucou, V., Lacombe, T., Dechesne, F., Siret, R., Bruno, J. P., Dessup, M., et al. (2011). High throughput analysis of grape genetic diversity as a tool for germplasm collection management. Theor. Appl. Genet. 122, 1233–1245. doi: 10.1007/s00122-010-1527-y
Legland, D., Devaux, M. F., Bouchet, B., Guillon, F., and Lahaye, M. (2012). Cartography of cell morphology in tomato pericarp at the fruit scale. J. Microsc. 247, 78–93. doi: 10.1111/j.1365-2818.2012.03623.x
Leida, C., Moser, C., Esteras, C., Sulpice, R., Lunn, J. E., de Langen, F., et al. (2015). Variability of candidate genes, genetic structure and association with sugar accumulation and climacteric behavior in a broad germplasm collection of melon (Cucumis melo L.). BMC Genet. 16:28. doi: 10.1186/s12863-015-0183-2
Li, Z., and Sillanpää, M. J. (2015). Dynamic quantitative trait locus analysis of plant phenomic data. Trends Plant Sci. 20, 822–833. doi: 10.1016/j.tplants.2015.08.012
Lim, W., Miller, R., Park, J., and Park, S. (2014). Consumer sensory analysis of high flavonoid transgenic tomatoes. J. Food Sci. 79, S1212–S1217. doi: 10.1111/1750-3841.12478
Lin, T., Zhu, G., Zhang, J., Xu, X., Yu, Q., Zheng, Z., et al. (2014). Genomic analyses provide insights into the history of tomato breeding. Nat. Genet., 46, 1220–1226.
Liu, R., How-Kit, A., Stammitti, L., Teyssier, E., Rolin, D., Mortain-Bertrand, A., et al. (2015). A DEMETER-like DNA demethylase governs tomato fruit ripening. Proc. Natl. Acad. Sci. U.S.A. 112, 10804–10809. doi: 10.1073/pnas.1503362112
Liu, Y. S., Gur, A., Ronen, G., Causse, M., Damidaux, R., Buret, M., et al. (2003). There is more to tomato fruit colour than candidate carotenoid genes. Plant Biotechnol. J. 1, 195–207. doi: 10.1046/j.1467-7652.2003.00018.x
Llorens, J., Gil, E., and Llop, J. (2011). Ultrasonic and LIDAR sensors for electronic canopy characterization in vineyards: advances to improve pesticide application methods. Sensors 11, 2177–2194. doi: 10.3390/s110202177
Long, M., Millar, D. J., Kimura, Y., Donovan, G., Rees, J., Fraser, P. D., et al. (2006). Metabolite profiling of carotenoid and phenolic pathways in mutant and transgenic lines of tomato: identification of a high antioxidant fruit line. Phytochemistry 67, 1750–1757. doi: 10.1016/j.phytochem.2006.02.022
Lor, V. S., Starker, C. G., Voytas, D. F., Weiss, D., and Olszewski, N. E. (2014). Targeted mutagenesis of the tomato PROCERA gene using TALENs. Plant Physiol. 166, 1288–1291. doi: 10.1104/pp.114.247593
Lucatti, A. F., van Heusden, A. W., de Vos, R. C., Visser, R. G., and Vosman, B. (2013). Differences in insect resistance between tomato species endemic to the Galapagos Islands. BMC Evol. Biol. 13:175. doi: 10.1186/1471-2148-13-175
Mageroy, M. H., Tieman, D. M., Floystad, A., Taylor, M. G., and Klee, H. J. (2012). A Solanum lycopersicum catechol-O-methyltransferase involved in synthesis of the flavor molecule guaiacol. Plant J. 69, 1043–1051. doi: 10.1111/j.1365-313X.2011.04854.x
Malacarne, G., Coller, E., Czemmel, S., Vrhovsek, U., Engelen, K., Goremykin, V., et al. (2016). The grapevine VvibZIPC22 transcription factor is involved in the regulation of flavonoid biosynthesis. J. Exp. Bot. 67, 3509–3522. doi: 10.1093/jxb/erw181
Malacarne, G., Costantini, L., Coller, E., Battilana, J., Velasco, R., Vrhovsek, U., et al. (2015). Regulation of flavonol content and composition in (Syrah × Pinot Noir) mature grapes: integration of transcriptional profiling and metabolic quantitative trait locus analyses. J. Exp. Bot. 66, 4441–4453. doi: 10.1093/jxb/erv243
Marguerit, E., Boury, C., Manicki, A., Donnart, M., Butterlin, G., Némorin, A., et al. (2009). Genetic dissection of sex determinism, inflorescence morphology and downy mildew resistance in grapevine. Theor. Appl. Genet. 118, 1261–1278. doi: 10.1007/s00122-009-0979-4
Martin, D. M., Chiang, A., Lund, S. T., and Bohlmann, J. (2012). Biosynthesis of wine aroma: transcript profiles of hydroxymethylbutenyl diphosphate reductase, geranyl diphosphate synthase, and linalool/nerolidol synthase parallel monoterpenol glycoside accumulation in Gewürztraminer grapes. Planta 236, 919–929. doi: 10.1007/s00425-012-1704-0
Mathieu, S., Cin, V. D., Fei, Z., Li, H., Bliss, P., Taylor, M. G., et al. (2009). Flavour compounds in tomato fruits: identification of loci and potential pathways affecting volatile composition. J. Exp. Bot. 60, 325–337. doi: 10.1093/jxb/ern294
Matsui, K., Ishii, M., Sasaki, M., Rabinowitch, H. D., and Ben-Oliel, G. (2007). Identification of an allele attributable to formation of cucumber-like flavor in wild tomato species (Solanum pennellii) that was inactivated during domestication. J. Agric. Food Chem. 55, 4080–4086. doi: 10.1021/jf063756b
Maxwell, K., and Johnson, G. N. (2000). Chlorophyll fluorescence—a practical guide. J. Exp. Bot. 51, 659–668.
Mazzucato, A., Papa, R., Bitocchi, E., Mosconi, P., Nanni, L., Negri, V., et al. (2008). Genetic diversity, structure and marker-trait associations in a collection of Italian tomato (Solanum lycopersicum L.) landraces. Theor. Appl. Genet. 116, 657–669. doi: 10.1007/s00122-007-0699-6
Mba, C. (2013). Induced mutations unleash the potentials of plant genetic resources for food and agriculture. Agronomy 3, 200–231. doi: 10.3390/agronomy3010200
McMullen, M. D., Kresovich, S., Villeda, H. S., Bradbury, P., Li, H., Sun, Q., et al. (2009). Supporting online material for: genetic properties of the maize nested association mapping population. Science 325, 737–741. doi: 10.1126/science.1174320
Menda, N., Semel, Y., Peled, D., Eshed, Y., and Zamir, D. (2004). In silico screening of a saturated mutation library of tomato. Plant J. 38, 861–872. doi: 10.1111/j.1365-313X.2004.02088.x
Menzel, M. I., Tittmann, S., Buehler, J., Preis, S., Wolters, N., Jahnke, S., et al. (2009). Non-invasive determination of plant biomass with microwave resonators. Plant Cell Environ. 32, 368–379. doi: 10.1111/j.1365-3040.2009.01931.x
Meron, M., Sprintsin, M., Tsipris, J., Alchanatis, V., and Cohen, Y. (2013). Foliage temperature extraction from thermal imagery for crop water stress determination. Precis. Agric. 14, 467–477. doi: 10.1007/s11119-013-9310-0
Mes, P. J., Boches, P., Myers, J. R., and Durst, R. (2008). Characterization of tomatoes expressing anthocyanin in the fruit. J. Am. Soc. Hort. Sci. 133, 262–269.
Minoia, S., Petrozza, A., D'Onofrio, O., Piron, F., Mosca, G., Sozio, G., et al. (2010). A new mutant genetic resource for tomato crop improvement by TILLING technology. BMC Res. Notes 3:69. doi: 10.1186/1756-0500-3-69
Mishra, K. B., Iannacone, R., Petrozza, A., Mishra, A., Armentano, N., La Vecchia, G., et al. (2012). Engineered drought tolerance in tomato plants is reflected in chlorophyll fluorescence emission. Plant Sci. 182, 79–86. doi: 10.1016/j.plantsci.2011.03.022
Monforte, A. J., and Tanksley, S. D. (2000). Development of a set of near isogenic and backcross recombinant inbred lines containing most of the Lycopersicon hirsutum genome in a L. esculentum genetic background: a tool for gene mapping and gene discovery. Genome 43, 803–813. doi: 10.1139/gen-43-5-803
Monforte, A. J., Diaz, A., Caño-Delgado, A., and Van Der Knaap, E. (2014). The genetic basis of fruit morphology in horticultural crops: lessons from tomato and melon. J. Exp. Bot. 65, 4625–4637. doi: 10.1093/jxb/eru017
Monforte, A. J., Friedman, E., Zamir, D., and Tanksley, S. D. (2001). Comparison of a set of allelic QTL-NILs for chromosome 4 of tomato: deductions about natural variation and implications for germplasm utilization. Theor. Appl. Genet. 102, 572–590. doi: 10.1007/s001220051684
Motion, G. B., Howden, A. J. M., Huitema, E., and Jones, S. (2015). DNA-binding protein prediction using plant specific support vector machines: validation and application of a new genome annotation tool. Nucleic Acids Res. 43:e158. doi: 10.1093/nar/gkv805
Mounet, F., Moing, A., Kowalczyk, M., Rohrmann, J., Petit, J., Garcia, V., et al. (2012). Down-regulation of a single auxin efflux transport protein in tomato induces precocious fruit development. J. Exp. Bot. 63, 4901–4917. doi: 10.1093/jxb/ers167
Nakano, H., Kobayashi, N., Takahata, K., Mine, Y., and Sugiyama, N. (2016). Quantitative trait loci analysis of the time of floral initiation in tomato. Sci. Hortic. 201, 199–210. doi: 10.1016/j.scienta.2016.02.009
Neuman, H., Galpaz, N., Cunningham, F. X., Zamir, D., and Hirschberg, J. (2014). The tomato mutation nxd1 reveals a gene necessary for neoxanthin biosynthesis and demonstrates that violaxanthin is a sufficient precursor for abscisic acid biosynthesis. Plant J. 78, 80–93. doi: 10.1111/tpj.12451
Nicolas, S. D., Péros, J.-P., Lacombe, T., Launay, A., Le Paslier, M.-C., Bérard, A., et al. (2016). Genetic diversity, linkage disequilibrium and power of a large grapevine (Vitis vinifera L) diversity panel newly designed for association studies. BMC Plant Biol. 16:74. doi: 10.1186/s12870-016-0754-z
Nishitani, C., Hirai, N., Komori, S., Wada, M., Okada, K., Osakabe, K., et al. (2016). Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6:31481. doi: 10.1038/srep31481
Nunes-Nesi, A., Carrari, F., Lytovchenko, A., Smith, A. M. O., Loureiro, M. E., Ratcliffe, R. G., et al. (2005). Enhanced photosynthetic performance and growth as a consequence of decreasing mitochondrial malate dehydrogenase activity in transgenic tomato plants. Plant Physiol. 137, 611–622. doi: 10.1104/pp.104.055566
Orzaez, D., Monforte, A. J., and Granell, A. (2010). Using genetic variability available in the breeder's pool to engineer fruit quality. GM Crops 1, 120–127. doi: 10.4161/gmcr.1.3.12327
Oyanedel, E., Wolfe, D. W., Monforte, A. J., Tanksley, S. D., and Owens, T. G. (2001). Using Lycopersicon hirsutum as a source of cold tolerance in processing tomato breeding. Acta Hortic. doi: 10.17660/ActaHortic.2001.542.51
Pan, C., Ye, L., Qin, L., Liu, X., He, Y., Wang, J., et al. (2016). CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci. Rep. 6:24765. doi: 10.1038/srep24765
Pankratov, I., McQuinn, R., Schwartz, J., Bar, E., Fei, Z., Lewinsohn, E., et al. (2016). Fruit carotenoid-deficient mutants in tomato reveal a function of the plastidial isopentenyl diphosphate isomerase (IDI1) in carotenoid biosynthesis. Plant J. 88, 82–94. doi: 10.1111/tpj.13232
Parker, J., Koh, J., Yoo, M.-J., Zhu, N., Feole, M., Yi, S., et al. (2013). Quantitative proteomics of tomato defense against Pseudomonas syringae infection. Proteomics 13, 1934–1946. doi: 10.1002/pmic.201200402
Pascual, L., Albert, E., Sauvage, C., Duangjit, J., Bouchet, J. P., Bitton, F., et al. (2016). Dissecting quantitative trait variation in the resequencing era: complementarity of bi-parental, multi-parental and association panels. Plant Sci. 242, 120–130. doi: 10.1016/j.plantsci.2015.06.017
Pascual, L., Desplat, N., Huang, B. E., Desgroux, A., Bruguier, L., Bouchet, J.-P., et al. (2015). Potential of a tomato MAGIC population to decipher the genetic control of quantitative traits and detect causal variants in the resequencing era. Plant Biotechnol. J. 13, 565–577. doi: 10.1111/pbi.12282
Pérez-Díaz, R., Madrid-Espinoza, J., Salinas-Cornejo, J., González-Villanueva, E., and Ruiz-Lara, S. (2016). Differential roles for VviGST1, VviGST3, and VviGST4 in proanthocyanidin and anthocyanin transport in Vitis vinífera. Front. Plant Sci. 7:1166. doi: 10.3389/fpls.2016.01166
Prada, D. (2009). Molecular population genetics and agronomic alleles in seed banks: searching for a needle in a haystack? J. Exp. Bot. 60, 2541–2552. doi: 10.1093/jxb/erp130
Prashar, A., and Jones, H. G. (2014). Infra-red thermography as a high-throughput tool for field phenotyping. Agronomy 4, 397–417. doi: 10.3390/agronomy4030397
Quadrana, L., Almeida, J., Asís, R., Duffy, T., Dominguez, P. G., Bermúdez, L., et al. (2014). Natural occurring epialleles determine vitamin E accumulation in tomato fruits. Nat. Commun. 5, 3027. doi: 10.1038/ncomms5027
Rambla, J. L., Gianfranco, D., Angela, R.-M., Giovanni, G., Antonio, G., Lourdes, G.-G., et al. (2017b). Evolution of volatile compounds and their relationship with their precursors and the expression profiles of some genes involved in their release during maturation of Airén and Tempranillo grape varieties. Front. Plant Sci. 7:1619. doi: 10.3389/fpls.2016.01619
Rambla, J. L., Medina, A., Fernández-del-Carmen, A., Barrantes, W., Garndillo, S., Cammareri, M., et al. (2017a). Identification, validation and introgression of fruit volatile QTLs from a red-fruited wild tomato species J. Exp. Bot. 68, 4269–4442. doi: 10.1093/jxb/erw455
Rambla, J. L., Trapero-Mozos, A., Diretto, G., Rubio-Moraga, A., Granell, A., Gómez-Gómez, L., et al. (2016). Gene-metabolite networks of volatile metabolism in airen and Tempranillo grape cultivars revealed a distinct mechanism of aroma bouquet production. Front. Plant Sci. 7:1691. doi: 10.3389/fpls.2016.01619
Raza, S. E., Prince, G., Clarkson, J. P., and Rajpoot, N. M. (2015). Automatic detection of diseased tomato plants using thermal and stereo visible light images. PLOS ONE 10:e0123262. doi: 10.1371/journal.pone.0123262
Reganold, J. P., and Wachter, J. M. (2016). Organic agriculture in the twenty-first century. Nat. Plants 2, 1–8. doi: 10.1038/nplants.2015.221
Ren, C., Liu, X., Zhang, Z., Wang, Y., Duan, W., Li, S., et al. (2016). CRISPR/Cas9-mediated efficient targeted mutagenesis in Chardonnay (Vitis vinifera L.). Sci. Rep. 6:32289. doi: 10.1038/srep32289
Riaz, S., Krivanek, A. F., Xu, K., and Walker, M. A. (2006). Refined mapping of the Pierce's disease resistance locus, PdR1, and Sex on an extended genetic map of Vitis rupestris× V. arizonica. Theor. Appl.Genet. 113, 1317–1329. doi: 10.1007/s00122-006-0385-0
Riaz, S., Tenscher, A. C., Ramming, D. W., and Walker, M. A. (2011). Using a limited mapping strategy to identify major QTLs for resistance to grapevine powdery mildew (Erysiphe necator) and their use in marker-assisted breeding. Theor. Appl. Genet. 122, 1059–1073. doi: 10.1007/s00122-010-1511-6
Rinaldi, M., Llorens, J., and Gil, E. (2013). “Electronic characterization of the phenological stages of grapevine using a LIDAR sensor,” in Precision Agriculture, Vol. 13. (Wageningen: Academic Publishers), 603–609.
Rinaldo, A. R., Cavallini, E., Jia, Y., Moss, S. M. A., McDavid, D. A. J., Hooper, L. C., et al. (2015). A grapevine anthocyanin acyltransferase, transcriptionally regulated by VvMYBA, can produce most acylated anthocyanins present in grape skins. Plant Physiol. 169, 1897–1916. doi: 10.1104/pp.15.01255
Roby, J. P., van Leeuwen, C., Gonçalves, E., Graça, A., and Martins, A. (2014). “The preservation of genetic resources of the vine requires cohabitation between institutional clonal selection, mass selection and private clonal selection,” in BIO Web of Conferences, Vol. 3, (EDP Sciences).
Römer, S., Fraser, P. D., Kiano, J. W., Shipton, C. A., Misawa, N., Schuch, W., et al. (2000). Elevation of the provitamin A content of transgenic tomato plants. Nat. Biotechnol. 18, 666–669. doi: 10.1038/76523
Ron, M., Kajala, K., Pauluzzi, G., Wang, D., Reynoso, M. A., Zumstein, K., et al. (2014). Hairy root transformation using Agrobacterium rhizogenes as a tool for exploring cell type-specific gene expression and function using tomato as a model. Plant Physiol. 166, 455–469. doi: 10.1104/pp.114.239392
Ronen, G., Carmel-Goren, L., Zamir, D., and Hirschberg, J. (2000). An alternative pathway to β-carotene formation in plant chromoplasts discovered by map-based cloning of Beta and old-gold color mutations in tomato. Proc. Natl. Acad. Sci. U.S.A. 97, 11102–11107. doi: 10.1073/pnas.190177497
Rosati, C., Aquilani, R., Dharmapuri, S., Pallara, P., Marusic, C., Tavazza, R., et al. (2000). Metabolic engineering of beta-carotene and lycopene content in tomato fruit. Plant J. 24, 413–419. doi: 10.1046/j.1365-313x.2000.00880.x
Rosenberg, N. A., Huang, L., Jewett, E. M., Szpiech, Z. A., Jankovic, I., and Boehnke, M. (2010).Genome-wide association studies in diverse populations. Nat. Rev. Genet. 11, 356–366 doi: 10.1038/nrg2760
Rousseau, D., Chéné, Y., Belin, E., Semaan, G., Trigui, G., Boudehri, K., et al. (2015). Multiscale imaging of plants: current approaches and challenges. Plant Methods 11, 6. doi: 10.1186/s13007-015-0050-1
Rousseaux, M. C., Jones, C. M., Adams, D., Chetelat, R., Bennett, A., and Powell, A. (2005). QTL analysis of fruit antioxidants in tomato using Lycopersicon pennellii introgression lines. Theor. Appl. Genet. 111, 1396–1408. doi: 10.1007/s00122-005-0071-7
Ruggieri, V., Francese, G., Sacco, A., D'Alessandro, A., Rigano, M. M., Parisi, M., et al. (2014). An association mapping approach to identify favourable alleles for tomato fruit quality breeding. BMC Plant Biol. 14:337. doi: 10.1186/s12870-014-0337-9
Sacco, A., Ruggieri, V., Molisso, M., and Barone, A. (2013). “Omics” approaches in tomato aimed at identifying candidate genes for ascorbic acid accumulation in the fruit. Afr. J. Biotechnol. 12, 6791–6800. doi: 10.5897/AJBX12.007
Sagi, M., Scazzocchio, C., and Fluhr, R. (2002). The absence of molybdenum cofactor sulfuration is the primary cause of the flacca phenotype in tomato plants. Plant J. 31, 305–317. doi: 10.1046/j.1365-313X.2002.01363.x
Saito, T., Ariizumi, T., Okabe, Y., Asamizu, E., Hiwasa-Tanase, K., Fukuda, N., et al. (2011). TOMATOMA: a novel tomato mutant database distributing micro-tom mutant collections. Plant Cell Physiol. 52, 283–296. doi: 10.1093/pcp/pcr004
Saliba-Colombani, V., Causse, M., Langlois, D., Philouze, J., and Buret, M. (2001). Genetic analysis of organoleptic quality in fresh market tomato. 1. Mapping QTLs for physical and chemical traits. Theor. Appl. Genet. 102, 259–272. doi: 10.1007/s001220051643
Sankaran, S., Khot, L. R., Espinoza, C. Z., Jarolmasjed, S., Sathuvalli, V. R., Vandemark, G. J., et al. (2015). Low-altitude, high-resolution aerial imaging systems for row and field crop phenotyping: a review. Eur. J. Agron. 70, 112–123. doi: 10.1016/j.eja.2015.07.004
Sanz, R., Rosell, J. R., Llorens, J., Gil, E., and Planas, S. (2013). Relationship between tree row LIDAR-volume and leaf area density for fruit orchards and vineyards obtained with a LIDAR 3D dynamic measurement system. Agric. For. Meteorol. 171, 153–162. doi: 10.1016/j.agrformet.2012.11.013
Sarrion-Perdigones, A., Falconi, E. E., Zandalinas, S. I., Juárez, P., Fernández-del-Carmen, A., Granell, A., et al. (2011). GoldenBraid: An iterative cloning system for standardized assembly of reusable genetic modules. PLoS ONE 6:e21622. doi: 10.1371/journal.pone.0021622
Sarrion-Perdigones, A., Vazquez-Vilar, M., Palaci, J., Castelijns, B., Forment, J., Ziarsolo, P., et al. (2013). GoldenBraid 2.0: a comprehensive DNA assembly framework for plant synthetic biology. Plant Physiol. 162, 1618–1631. doi: 10.1104/pp.113.217661
Sato, S., Tabata, S., Hirakawa, H., Asamizu, E., Shirasawa, K., Isobe, S., et al. (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
Schaart, J. G., van de Wiel, C. C. M., Lotz, L. A. P., and Smulders, M. J. M. (2016). Opportunities for products of new plant breeding techniques. Trends Plant Sci. 21, 438–449. doi: 10.1016/j.tplants.2015.11.006
Schauer, N., Semel, Y., Balbo, I., Steinfath, M., Repsilber, D., Selbig, J., et al. (2008). Mode of inheritance of primary metabolic traits in tomato. Plant Cell 20, 509–523. doi: 10.1105/tpc.107.056523
Schauer, N., Semel, Y., Roessner, U., Gur, A., Balbo, I., Carrari, F., et al. (2006). Comprehensive metabolic profiling and phenotyping of interspecific introgression lines for tomato improvement. Nat. Biotechnol. 24, 447–454. doi: 10.1038/nbt1192
Schauer, N., Zamir, D., and Fernie, A. R. (2005). Metabolic profiling of leaves and fruit of wild species tomato: a survey of the Solanum lycopersicum complex. J. Exp. Bot. 56, 297–307. doi: 10.1093/jxb/eri057
Schijlen, E., Ric de Vos, C. H., Jonker, H., van den Broeck, H., Molthoff, J., van Tunen, A., et al. (2006). Pathway engineering for healthy phytochemicals leading to the production of novel flavonoids in tomato fruit. Plant Biotechnol. J. 4, 433–444. doi: 10.1111/j.1467-7652.2006.00192.x
Schreiber, G., Reuveni, M., Evenor, D., Oren-Shamir, M., Ovadia, R., Sapir-Mir, M., et al. (2012). ANTHOCYANIN1 from Solanum chilense is more efficient in accumulating anthocyanin metabolites than its Solanum lycopersicum counterpart in association with the ANTHOCYANIN FRUIT phenotype of tomato. Theor. Appl. Genet. 124, 295–307. doi: 10.1007/s00122-011-1705-6
Shah, P., Powell, A. L., Orlando, R., Bergmann, C., and Gutierrez Sanchez, G. (2012). Proteomic analysis of ripening tomato fruit infected by Botrytis cinerea. J. Proteome Res. 11, 2178–2192. doi: 10.1021/pr200965c
Shalem, O., Sanjana, E. N., Hartenian, E., and Zhang, F. (2014). Genome-scale CRISPR-Cas9 knockout. Science 343, 84–88. doi: 10.1038/nbt.2647
Shen, J., Tieman, D., Jones, J. B., Taylor, M. G., Schmelz, E., Huffaker, A., et al. (2014). A 13-lipoxygenase, TomloxC, is essential for synthesis of C5 flavour volatiles in tomato. J. Exp. Bot. 65, 419–428. doi: 10.1093/jxb/ert382
Shikata, M., Hoshikawa, K., Ariizumi, T., Fukuda, N., Yamazaki, Y., and Ezura, H. (2016). TOMATOMA update: phenotypic and metabolite information in the micro-tom mutant resource. Plant Cell Physiol. 57, e11. doi: 10.1093/pcp/pcv194
Sim, S. C., Durstewitz, G., Plieske, J., Wieseke, R., Ganal, M. W., van Deynze, A., et al. (2012). Development of a large snp genotyping array and generation of high-density genetic maps in tomato. PLoS ONE 7:e40563. doi: 10.1371/journal.pone.0040563
Simkin, A. J., Schwartz, S. H., Auldridge, M., Taylor, M. G., and Klee, H. J. (2004). The tomato carotenoid cleavage dioxygenase 1 genes contribute to the formation of the flavor volatiles β-ionone, pseudoionone, and geranylacetone. Plant J. 40, 882–892. doi: 10.1111/j.1365-313X.2004.02263.x
Sozzani, R., Busch, W., Spalding, E., and Benfey, P. (2014). Advanced imaging technique for the study of plant growth and development. Trends Plant Sci. 19, 304–310. doi: 10.1016/j.biotechadv.2011.08.021.Secreted
Speirs, J., Lee, E., Holt, K., Yong-Duk, K., Steelen Scott, N., Loveys, B., et al. (1998). Genetic manipulation of alcohol dehydrogenase levels in ripening tomato fruit affects the balance of some flavor aldehydes and alcohols1. Plant Physiol. 117, 1047–1058. doi: 10.1104/pp.117.3.1047
Sprink, T., Metje, J., and Hartung, F. (2015). Plant genome editing by novel tools: TALEN and other sequence specific nucleases. Curr. Opin. Biotechnol. 32, 47–53. doi: 10.1016/j.copbio.2014.11.010
Steiber, A., Hegazi, R., Herrera, M., Landy Zamor, M., Chimanya, K., Pekcan, A. G., et al. (2004). Spotlight on global malnutrition: a continuing challenge in the 21st century. J. Acad. Nutr. Diet. 115, 1335–1341. doi: 10.1016/j.jand.2015.05.015
Sun, Y. D., Liang, Y., Wu, J. M., Li, Y. Z., Cui, X., and Qin, L. (2012). Dynamic QTL analysis for fruit lycopene content and total soluble solid content in a Solanum lycopersicum x S. pimpinellifolium cross. Genet. Mol. Res. 11, 3696–3710. doi: 10.4238/2012.August.17.8
Tadmor, Y., Fridman, E., Gur, A., Larkov, O., Lastochkin, E., Ravid, U., et al. (2002). Identification of malodorous, a wild species allele affecting tomato aroma that was selected against during domestication. J. Agric. Food Chem. 50, 2005–2009. doi: 10.1021/jf011237x
Tang, H., Sezen, U., and Paterson, A. H. (2010). Domestication and plant genomes. Curr. Opin. Plant Biol. 13, 160–166. doi: 10.1016/j.pbi.2009.10.008
Tanksley, S. D., Grandillo, S., Fulton, T. M., Zamir, D., Eshed, Y., Petiard, V., et al. (1996). Advanced backcross QTL analysis in a cross between an elite processing line of tomato and its wild relative L. pimpinellifolium. Theor. Appl.Genet. 92, 213–224. doi: 10.1007/BF00223378
Tanou, G., Job, C., Rajjou, L., Arc, E., Belghazi, M., Diamantidis, G., et al. (2009). Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J. 60, 795–804. doi: 10.1111/j.1365-313X.2009.04000.x
Temple, L., Kwa, M., Tetang, J., and Bikoi, A. (2011). Organizational determinant of technological innovation in food agriculture and impacts on sustainable development. Agron. Sustain. Dev. 31, 745–755. doi: 10.1007/s13593-011-0017-1
Thapa, S. P., Miyao, E. M., Davis, R. M., and Coaker, G. (2015). Identification of QTLs controlling resistance to Pseudomonas syringae pv. tomato race 1 strains from the wild tomato, Solanum habrochaites LA1777. Theor. Appl. Genet. 128, 681–692.
This, P., Lacombe, T., and Thomas, M. R. (2006). Historical origins and genetic diversity of wine grapes. Trends Genet. 22, 511–519. doi: 10.1016/j.tig.2006.07.008
Tieman, D. M., Zeigler, M., Schmelz, E. A., Taylor, M. G., Bliss, P., Kirst, M., et al. (2006). Identification of loci affecting flavour volatile emissions in tomato fruits. J. Exp. Bot. 57, 887–896. doi: 10.1093/jxb/erj074
Tieman, D., Zeigler, M., Schmelz, E., Taylor, M. G., Rushing, S., Jones, J. B., et al. (2010). Functional analysis of a tomato salicylic acid methyl transferase and its role in synthesis of the flavor volatile methyl salicylate. Plant J. 62, 113–123. doi: 10.1111/j.1365-313X.2010.04128.x
Tikunov, Y. M., Molthoff, J., de Vos, R. C. H., Beekwilder, J., van Houwelingen, A., van der Hooft, J. J. J., et al. (2013). Non-smoky glycosyltransferase1 prevents the release of smoky aroma from tomato fruit. Plant Cell 25, 3067–3078. doi: 10.1105/tpc.113.114231
Tilman, D., and Clark, M. (2015). Food, agriculture and the environment: can we feed the world and save the earth? Daedalus 144, 8–23. doi: 10.1162/DAED_a_00350
Truco, M. J., Randall, L. B., Bloom, A. J., and St. Clair, D. A. (2000). Detection of QTLs associated with shoot wilting and root ammonium uptake under chilling temperatures in an interspecific backcross population from Lycopersicon esculentum× L. hirsutum. Theor. Appl.Genet. 101, 1082–1092. doi: 10.1007/s001220051583
Uluisik, S., Chapman, N. H., Smith, R., Poole, M., Adams, G., Gillis, R. B., et al. (2016). Genetic improvement of tomato by targeted control of fruit softening. Nat. Biotechnol. 1, 1–11. doi: 10.1038/nbt.3602
Vadivambal, R., and Jayas, D. S. (2011). Applications of thermal imaging in agriculture and food industry—a review. Food Bioprocess. Technol. 4, 186–199. doi: 10.1007/s11947-010-0333-5
Van der Knaap, E., Lippman, Z. B., and Tanksley, S. D. (2002). Extremely elongated tomato fruit controlled by four quantitative trait loci with epistatic interactions. Theor. Appl.Genet. 104, 241–247. doi: 10.1007/s00122-001-0776-1
Varshney, R. K., Terauchi, R., and McCouch, S. R. (2014). Harvesting the promising fruits of genomics: applying genome sequencing technologies to crop breeding. PLoS Biol. 12:e1001883. doi: 10.1371/journal.pbio.1001883
Vazquez-Vilar, M., Bernabé-Orts, J. M., Fernandez-Del-Carmen, A., Ziarsolo, P., Blanca, J., Granell, A., et al. (2016). A modular toolbox for gRNA-Cas9 genome engineering in plants based on the GoldenBraid standard. Plant Methods 12, 10. doi: 10.1186/s13007-016-0101-2
Velasco, R., Zharkikh, A., Troggio, M., Cartwright, D. A., Cestaro, A., Pruss, D., et al. (2007). A high quality draft consensus sequence of the genome of a heterozygous grapevine variety. PLoS ONE 2:e1326. doi: 10.1371/journal.pone.0001326
Víquez-Zamora, M., Caro, M., Finkers, R., Tikunov, Y., Bovy, A., Visser, R. G., et al. (2014). Mapping in the era of sequencing: high density genotyping and its application for mapping TYLCV resistance in Solanum pimpinellifolium. BMC Genomics 15:1152. doi: 10.1186/1471-2164-15-1152
Vogel, B. (2014). Marker assited selection: a biotechnology for plant breeding without genetic. Greenpeace Res. Lab. 59.
Vogel, J. T., Walter, M. H., Giavalisco, P., Lytovchenko, A., Kohlen, W., Charnikhova, T., et al. (2010). SlCCD7 controls strigolactone biosynthesis, shoot branching and mycorrhiza-induced apocarotenoid formation in tomato. Plant J. 61, 300–311. doi: 10.1111/j.1365-313X.2009.04056.x
Walker, A. R., Lee, E., Bogs, J., McDavid, D. A. J., Thomas, M. R., and Robinson, S. P. (2007). White grapes arose through the mutation of two similar and adjacent regulatory genes. Plant J. 49, 772–785. doi: 10.1111/j.1365-313X.2006.02997.x
Wang, Y., Liu, X., Ren, C., Zhong, G.-Y., Yang, L., Li, S., et al. (2016). Identification of genomic sites for CRISPR/Cas9-based genome editing in the Vitis vinifera genome. BMC Plant Biol. 16:96. doi: 10.1186/s12870-016-0787-3
Wasson, A. P., Richards, R. A., Chatrath, R., Misra, S. C., Prasad, S. V. S., Rebetzke, G. J., et al. (2012). Traits and selection strategies to improve root systems and water uptake in water-limited wheat crops. J. Exp. Bot. 63, 3485–3498. doi: 10.1093/jxb/ers111
Westengen, O. T., Jeppson, S., and Guarino, L. (2013). Global Ex-Situ crop diversity conservation and the svalbard global seed vault: assessing the current status. PLoS ONE 8:e64146. doi: 10.1371/journal.pone.0064146
Wezel, A., Bellon, S., Doré, T., Francis, C., Vallod, D., and David, C. (2009). Agroecology as a science, a movement and a practice. Sustain. Agric. 2, 27–43. doi: 10.1007/978-94-007-0394-0_3
Wezel, A., Casagrande, M., Celette, F., Vian, J.-F., Ferrer, A., and Peigné, J. (2014). Agroecological practices for sustainable agriculture. A review. Agron. Sustain. Dev. 34, 1–20. doi: 10.1007/s13593-013-0180-7
Xu, K., Riaz, S., Roncoroni, N. C., Jin, Y., Hu, R., Zhou, R., et al. (2008). Genetic and QTL analysis of resistance to Xiphinema index in a grapevine cross. Theor. Appl.Genet. 116, 305–311. doi: 10.1007/s00122-007-0670-6
Xu, X., and Bai, G. (2015). Whole-genome resequencing: changing the paradigms of SNP detection, molecular mapping and gene discovery. Mol. Breed. 35, 33. doi: 10.1007/s11032-015-0240-6
Xu, C., Park, S. J., Van Eck, J., and Lippman, Z. B. (2016). Control of inflorescence architecture in tomato by BTB/POZ transcriptional regulators. Genes Dev. 30, 2048–2061. doi: 10.1101/gad.288415.116
Yang, H., Li, C., Lam, H. M., Clements, J., Yan, G., and Zhao, S. (2015). Sequencing consolidates molecular markers with plant breeding practice. Theor. Appl. Genet. 128, 779–795. doi: 10.1007/s00122-015-2499-8
Yin, Y. G., Kobayashi, Y., Sanuki, A., Kondo, S., Fukuda, N., Ezura, H., et al. (2010). Salinity induces carbohydrate accumulation and sugar-regulated starch biosynthetic genes in tomato (Solanum lycopersicum L. cv. “Micro-Tom”) fruits in an ABA-and osmotic stress-independent manner. J. Exp. Bot. 61, 563–574. doi: 10.1093/jxb/erp333
Zanor, M. I., Rambla, J. L., Chaïb, J., Steppa, A., Medina, A., Granell, A., et al. (2009). Metabolic characterization of loci affecting sensory attributes in tomato allows an assessment of the influence of the levels of primary metabolites and volatile organic contents. J. Exp. Bot. 60, 2139–2154. doi: 10.1093/jxb/erp086
Zhang, J., Zhao, J., Xu, Y., Liang, J., Chang, P., Yan, F., et al. (2015). Genome-wide association mapping for tomato volatiles positively contributing to tomato flavor. Front. Plant Sci. 6:1042. doi: 10.3389/fpls.2015.01042
Zhang, Y., Butelli, E., De Stefano, R., Schoonbeek, H. J., Magusin, A., Pagliarani, C., et al. (2013). Anthocyanins double the shelf life of tomatoes by delaying overripening and reducing susceptibility to gray mold. Curr. Biol. 23, 1094–1100. doi: 10.1016/j.cub.2013.04.072
Zhao, Q., Zhang, H., Wang, T., Chen, S., and Dai, S. (2013). Proteomics-based investigation of salt-responsive mechanisms in plant roots. J. Proteomics 82, 230–253. doi: 10.1016/j.jprot.2013.01.024
Zhong, S., Fei, Z., Chen, Y., Zheng, Y., Huang, M., Vrebalov, J., et al. (2013). Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 31, 154–159. doi: 10.1038/nbt.2462
Zhu, C., Gore, M., Buckler, E. S., and Yu, J. (2008). Status and prospects of association mapping in plants. Plant Genome J. 1, 5–20. doi: 10.3835/plantgenome2008.02.0089
Keywords: fruit quality, germplasm, grape, omics, new plant breeding techniques, tomato, QTLs
Citation: Gascuel Q, Diretto G, Monforte AJ, Fortes AM and Granell A (2017) Use of Natural Diversity and Biotechnology to Increase the Quality and Nutritional Content of Tomato and Grape. Front. Plant Sci. 8:652. doi: 10.3389/fpls.2017.00652
Received: 21 December 2016; Accepted: 10 April 2017;
Published: 12 May 2017.
Edited by:
Irene Murgia, Università degli Studi di Milano, ItalyReviewed by:
Golam Jalal Ahammed, Zhejiang University, ChinaCopyright © 2017 Gascuel, Diretto, Monforte, Fortes and Granell. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quentin Gascuel, cWdhc2N1ZWxAcXVlbnRpbmdhc2N1ZWwud2ViNG1lLmZy
Antonio Granell, YWdyYW5lbGxAaWJtY3AudXB2LmVz
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.