
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 01 May 2017
Sec. Plant Abiotic Stress
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.00624
Acetyl-CoA carboxylase is an important enzyme, which catalyzes acetyl-CoA’s carboxylation to produce malonyl-CoA and to serve as a committed step for de novo fatty acid biosynthesis in plastids. In this study, 24 putative cotton BCCP genes were identified based on the lately published genome data in Gossypium. Among them, 4, 4, 8, and 8 BCCP homologs were identified in Gossypium raimondii, G. arboreum, G. hirsutum, and G. barbadense, respectively. These genes were divided into two classes based on a phylogenetic analysis. In each class, these homologs were relatively conserved in gene structure and motifs. The chromosomal distribution pattern revealed that all the BCCP genes were distributed equally on corresponding chromosomes or scaffold in the four cotton species. Segmental duplication was a predominant duplication event in both of G. hirsutum and G. barbadense. The analysis of the expression profile showed that 8 GhBCCP genes expressed in all the tested tissues with changed expression levels, and GhBCCP genes belonging to class II were predominantly expressed in developing ovules. Meanwhile, the expression analysis for the 16 cotton BCCP genes from G. raimondii, G. arboreum and G. hirsutum showed that they were induced or suppressed by cold or salt stress, and their expression patterns varied among different tissues. These findings will help to determine the functional and evolutionary characteristics of the BCCP genes in Gossypium species.
Polyploidy genomes have undergone rapid changes in genome structures and expression of duplicated genes (Li et al., 2011). The tetraploid cotton species originated from an inter-specific hybridization event between progenitors of A-genome species, G. arboreum (A2), and D-genome species, G. raimondii (D5) (Cronn et al., 1999; Senchina et al., 2003; Wendel et al., 2009). In tetraploid cotton (2n = 52), both Upland cotton (G. hirsutum, AD1) and Sea island cotton (G. barbadense, AD2), chromosome numbers 1 to 13 are reserved for the A subgenome (At), while chromosome numbers 14 to 26 have reserved for the D subgenome (Dt) (Kohel, 1973). Upland cotton accounts for more than 90% output of production, and cultivated in more than 130 countries due to its higher yield and fiber quality. Moreover, cotton ranks sixth among the world oil crops, providing cooking oil extracted from its oil-rich seeds which could be used for food industry either.
In plants, oil is one of the major components of seeds, stored in cottonseed as triacylglycerol (TAG). The accumulation of seed oil is mainly provided by TAG biosynthesis (Li et al., 2011; Troncosoponce et al., 2011; Hu et al., 2013; Sood and Chauhan, 2015). The pathway and related genes of lipid biosynthesis have been studied in many plants (Beisson et al., 2003; Bates et al., 2009; Yin et al., 2013; Jang et al., 2015). In Arabidopsis, at least 24 enzymes or subunits are involved in the plastid fatty acid synthetic pathway, which are encoded by 46 nuclear genes and one plastid gene (Beisson et al., 2003). Among them, acetyl-CoA carboxylase (ACCase) (E.C. 6.4.1.2) is an important enzyme, which catalyze acetyl-CoA to produce malonyl-CoA, and can be served as a committed step for de novo fatty acid biosynthesis in plastid (Sasaki and Nagano, 2004; Fukuda et al., 2013; Ran et al., 2015; Sood and Chauhan, 2015). In dicotyledon and non-graminaceous monocotyledon plants, plastidial ACCase is a multi-subunit complex comprised of four different polypeptides (biotin carboxyl carrier protein, BCCP; biotin carboxylase, BC; α- and β-carboxyltransferase subunits, α-CT and β-CT) with the exception of rapeseed plastid, which contains multifunctional ACCase comprised of a large multifunctional polypeptide (Elborough et al., 1996; Schulte et al., 1997; Sasaki and Nagano, 2004; Cui et al., 2017).
Among the subunits of heteromeric ACCase, BCCP subunit interacts with BC and accepts a bicarbonate ion when ATP is converted to ADP, thus it can be served as the donor of activated carboxyl group, then the complex (BCCP-biotin) transfers the ion to CT, and finally produces malonyl-CoA in fatty acid biosynthesis (Ke et al., 2000; Sasaki and Nagano, 2004; Chen et al., 2012; Jang et al., 2015). Previous studies revealed that BCCP subunits have variable protein lengths, and low similarity (Qiao and Liu, 2007), while each of BCCP subunits contains a biotinylated domain located at the C-terminal region, harboring a highly conserved motif (CIIEAMKLMNEIE) (Qiao and Liu, 2007; Xuan et al., 2015). Studies reported that BCCP subunit was redundant, and encoded by small gene family (Elborough et al., 1996; Reverdatto et al., 1999; Thelen et al., 2001). In rapeseed (Brassica napus), there were at least six copies of BCCP homolog, which can be divided into two distinct classes based on the comparison of amino acids (AAs) and nucleotide sequences (Elborough et al., 1996). In Arabidopsis, BCCP is encoded by two paralogous genes, AtBCCP1 and AtBCCP2 (Thelen et al., 2000, 2001), both share approximately 30% AA identity. AtBCCP1 was constitutively expressed in all the tissues, while AtBCCP2 was predominantly expressed in developing seeds (Thelen et al., 2000).
A BCCP homolog in Upland cotton, GhBCCP1, was constitutively expressed in all the tested tissues, indicating its indispensable role in cotton growth and metabolism. In addition, southern blotting result showed that there were several copies of BCCP genes in the cotton genome (Qiao and Liu, 2007), and overexpression of GhBCCP1 in cotton could increase cottonseed oil content (Cui et al., 2017). The recent availability of the completed genome sequences of G. raimondii (Paterson et al., 2012; Wang et al., 2012), G. arboreum (Li et al., 2014), G. hirsutum (Li et al., 2015; Zhang T. et al., 2015), and G. barbadense (Liu X. et al., 2015; Yuan et al., 2015) provide us new opportunity to identify and characterize BCCP genes in cotton.
In the present study, we performed a comprehensive analysis of BCCP genes in the four cotton species with the phylogenetic relationship, gene structure, conserved motifs, chromosomal distribution and expression profiling. Moreover, we assessed the paralogous and orthologous relationships of the four cotton species. The identification and comprehensive study of BCCP genes will provide valuable information for further study of the biological function and evolution in cotton.
The genome databases of G. arboreum (A2, BGI_V1.0), G. raimondii (D5, JGI_v2.1), G. hirsutum acc. TM-1 (NBI_V1.1), and G. barbadense acc.3-79 (NBI_V1.0) were downloaded from the CottonGen website1 (Yu et al., 2014).
The published AA sequences of BCCP for Arabidopsis, rapeseed, and soybean (Glycine max) were obtained from the NCBI (Supplementary Table 1). In order to identify all the candidate BCCP genes of four Gossypium species, several local BLAST searches (BlastP and tBlastN programs with default parameters) were performed using the BCCP protein sequences of Arabidopsis, rapeseed, and soybean as queries. Subsequently, the Pfam2 (Finn et al., 2014) and SMART3 (Letunic et al., 2015) databases were used to confirm each candidate of the BCCP gene family. Finally, in order to further verify the reliability of the initial results, all candidates were analyzed to confirm the presence of the conserved biotinyl domain using the InterProScan program4 (Quevillon et al., 2005). The online ExPASy tool5 was used to predict the theoretical MW (molecular weight) and pI (isoelectric point) of the BCCP proteins. Subcellular localization was predicted using the WoLF PSORT6 (Horton et al., 2007) and TargetP 1.17 (Emanuelsson et al., 2007), and the online Chlorop 1.1 Server8 was used to predict the presence of chloroplast transit peptides (cTP) in protein sequences. The 5′ upstream region, a 2-kb genomic DNA sequence of each gene was extracted from the genome database, and then subjected to the plantCARE database9 for a cis-element scan.
Multiple sequence alignment was performed using Clustal X version 2.0 program (Larkin et al., 2007) with default parameters. Subsequently, MEGA version 5.0 software (Tamura et al., 2011) were employed to construct an unrooted phylogenetic tree, using the method of Neighbor Joining with pairwise deletion option, poisson correction model and uniform rates (rates among sites). Bootstrap tests with 1000 replicates were carried out to evaluate the statistical reliability of phylogenetic tree. The gene structure of BCCP genes were obtained through comparing the genomic sequences and their predicted coding sequences using the Gene Structure Display Server (GSDS) tool10 (Hu et al., 2015). BCCP protein sequences in G. arboreum, G. raimondii, G. hirsutum, and G. barbadense were submitted to online MEME program11 (Bailey et al., 2009) for identification of conserved protein motifs. The MEME parameters were as follows: any number of repetitions, maximum number of motifs: 4, and optimum motif widths from 6 to 80 AA residues.
The chromosomal localization of each BCCP gene in G. arboreum, G. raimondii, G. hirsutum, and G. barbadense was deduced based on the available genomic information at the CottonGen database12 (Yu et al., 2014). Mapchart version 2.2 software (Voorrips, 2002) was used to visualize the distribution of BCCP genes on the chromosomes. Gene duplication events were defined when the following conditions were fulfilled: (1) the length of aligned sequence covered more than 80% of the longer gene, (2) the identity of the aligned regions was bigger than 80%, and (3) only one duplication event was taken into counted for tightly linked genes (Jiang et al., 2013; Wei et al., 2013; Dong et al., 2016). According to the chromosomal locations of BCCP genes, two types of gene duplications (tandem duplication and segmental duplication) were recognized.
The BCCP duplicated gene pairs of G. arboreum, G. raimondii, G. hirsutum, and G. barbadense were firstly aligned by Clustal X version 2.0 program (Larkin et al., 2007). Subsequently, synonymous substitution (Ks) and non-synonymous substitution (Ka) were calculated using the DnaSP version 5.0 software (DNA polymorphism analysis) (Rozas et al., 2003). Finally, the selection pressure for each gene pair was assessed by the Ka/Ks ratio.
For the expression analysis, the public expression data for various tissues (root, stem, and leaf), floral tissue (petal) and ovule tissues at different developmental stages (5, 10, 20, 25, and 35 DPA) in G. hirsutum TM-1 were obtained from Zhang T. et al. (2015), according to the identified GhBCCP ID. The expression data were gene-wise normalized and the heatmap for gene expression patterns was illustrated with the software MultiExperiment Viewer (MeV).
Cotton seedlings of G. arboreum L. Var. Shixiya 1, G. raimondii, and G. hirsutum L. acc TM-1 were grown in a temperature-controlled chamber with a photoperiod of 16 h light and 8 h darkness at 28°C. Three seedlings at trefoil stage were exposed to low temperature (4.0°C) for 24 h and salt (150 mM NaCl) for 24 h, respectively. Then the roots, stems, and leaves were sampled and frozen in liquid nitrogen immediately, and stored at -80°C for RNA isolation. Three biological repeats were performed for each treatment.
Approximately 1 μg RNA was used for first-strand cDNAs synthesis with the PrimeScript 1st Strand cDNA Synthesis Kit (TakaRa, Dalian, China) following the manufacturer’s instructions. Gene-specific primer pairs were designed using Primer version 5.0 software based on CDSs of the BCCP genes. The sequences of the primer pairs are listed in Supplementary Table 2. The qRT-PCR analysis was performed with the SYBR Premix Ex Taq (TakaRa, Dalian, China) following the manufacturer’s instructions. In all the qRT-PCR analyses, the cotton UBQ7 gene was used as an internal reference. Each sample was run with three biological replicates and three technical replicates on an ABI 7500 real-time PCR System (Applied Biosystems, Foster City, CA, USA). The thermal cycle applied was as follows: 95°C for 30 s, followed by 40 cycles of denaturation at 95°C for 5 s and annealing and elongation at 60°C for 35 s. The relative expression levels (RQ) were calculated according to the 2-ΔΔCt method (Livak and Schmittgen, 2001).
The genome-wide identification of BCCP genes have been performed on the basis of four cotton genome sequences, G. raimondii (Paterson et al., 2012), G. arboreum (Li et al., 2014), G. hirsutum (Zhang T. et al., 2015), and G. barbadense (Liu X. et al., 2015). BLASTP and BLASTN programs were used to search the candidate BCCP genes from the four cotton species genome databases with the query sequences of Arabidopsis (2), rapeseed (6), and soybean (2) BCCP proteins. Among the 6 BCCP genes in rapeseed (Elborough et al., 1996), only 2 (BnpBP4 and BnpBP6) coded completed proteins and were used. Subsequently, Interproscan (Quevillon et al., 2005) and SMART were used to verify the biotinyl domain (CIIEAMKLMNEIE) of the retrieved sequences. The results showed that a total of 24 BCCP genes, which contained biotinyl domain, were identified in the four cotton species genomes (Table 1). Among them, 4 were predicted in G. raimondii, 4 in G. arboreum, 8 in G. hirsutum, and 8 in G. barbadense. The predicted BCCP genes, GrBCCP1-GrBCCP4, GaBCCP1-GaBCCP4, GhBCCP2-GaBCCP8, and GbBCCP1-GaBCCP8 were numbered based on their chromosomal location. Though the size of G. arboreum genome was about twofold than the G. raimondii (Paterson et al., 2012; Wang et al., 2012; Li et al., 2014), each of the two diploid species has 4 BCCP genes. The length of 8 BCCP proteins from the two diploid cotton species varied from 244 to 295 AAs, and their predicted molecular weights and pI values were within the ranges of 25.50–31.57 kDa and 5.03–8.74, respectively. For G. hirsutum, the GhBCCP proteins were varied from 282 AA of GhBCCP1 to 313 AA of GhBCCP3, their molecular weights ranged between 29.43 kDa of GhBCCP5 to 33.41 kDa of GhBCCP2, and their pI values were distributed in a range from 4.91 of GhBCCP5 to 8.66 of GhBCCP3. For G. barbadense, the length of 8 GbBCCP proteins varied from 57 AA of GbBCCP4 to 515 AA of GbBCCP6, the MW ranged from 6.22 kDa of GbBCCP4 to 54.42 kDa of GbBCCP6, and the pI values were between 4.65 of GbBCCP4 and 8.90 of GbBCCP8. Compared with the length of BCCP homologs reported in other plants (Thelen et al., 2000; Gu et al., 2011), GbBCCP4, GbBCCP6, and GbBCCP8 were less than 200 AA or more than 350 AA.
Multiple sequence alignments of 24 BCCP proteins from the four cotton species showed that the C-terminal region was conserved, and a typical biotinyl domain was existed (Supplementary Figure 1). In addition, the result of multiple sequence alignments showed that GbBCCP4 only contains C-terminal sequence (Supplementary Figure 1). Due to the small protein length of GbBCCP4, we were unable to analyze it in the subsequent research.
Protein subcellular localization is important for understanding the function of genes (Chou and Shen, 2007). According to the Wolf PSORT assessment, the result of the signal peptide prediction showed that the N-terminal of 23 BCCP proteins of the four cotton species carried cTPs (Table 1). Meanwhile, the subcellular localization of 23 BCCP proteins was predicted again by TargetP software, the result showed that GaBCCP1, GbBCCP2, and GbBCCP8 proteins were located in the mitochondria, GbBCCP7 protein could not predict its subcellular location since it was not start with methionine (Supplementary Figure 1), the rest of 19 proteins were located in the chloroplast. Based on the predicted subcellular location results, the cTPs length of 20 BCCP proteins was predicted using the ChloroP 1.1. It was showed that their length ranged from 34 to 86 AA (Table 1), implying that those BCCP proteins are chloroplast-located proteins. The cTPs may facilitate the BCCP precursor entering from cytosol to chloroplast (Gu et al., 2011).
To assess the evolution of the BCCP homologs in G. arboreum, G. raimondii, G. hirsutum, and G. barbadense, 23 predicted full-length BCCP proteins were aligned to construct phylogenetic tree using a neighbor-joining (NJ) method (Figure 1A). Meanwhile, the phylogenetic trees were reconstructed with minimal evolution (ME) and maximum likelihood (ML) methods (Supplementary Figure 2). The trees produced by the two methods above showed less difference with the tree produced by NJ method, suggesting those three methods were largely consistent with each other, and the NJ tree was suitable for further analysis. According to the NJ tree, 23 BCCP proteins from the four cotton species were divided into two classes designated classes I and II (Figure 1A). This classification was consistent with BCCP genes in other plant species (Thelen et al., 2000; Li et al., 2010). Class I contained 12 members, composed of 2 members from G. raimondii, 2 from G. arboreum, 4 from G. hirsutum, and 4 from G. barbadense. Class II contained 11 members, 2, 2, 4, and 3 members in G. raimondii, G. arboreum, G. hirsutum, and G. barbadense, respectively.
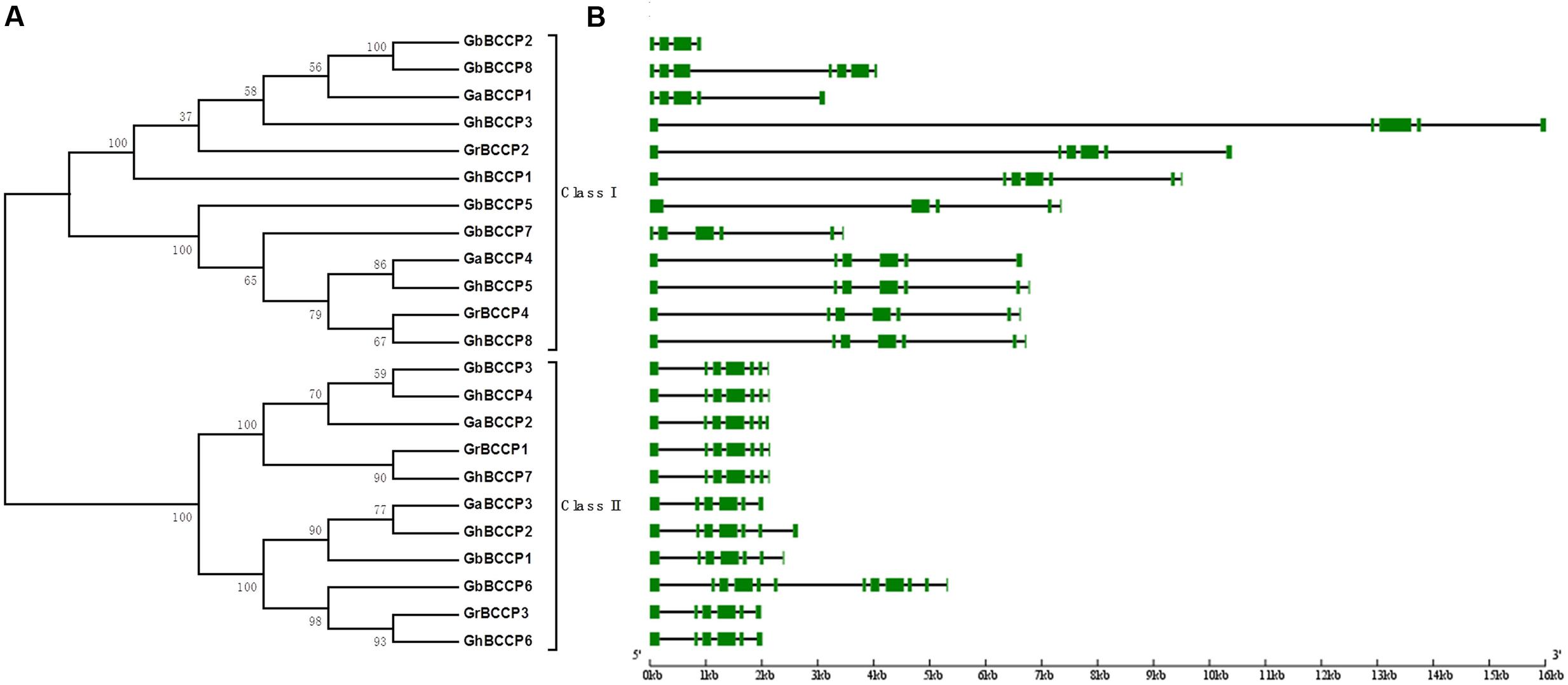
FIGURE 1. Phylogenetic tree and gene structure of the BCCP gene family in Gossypium. (A) The phylogenetic tree of all BCCP proteins in four Gossypium species was constructed using Neighbor-Joining method and the numbers at nodes represent bootstrap support values from 1000 replicates. (B) The exon/intron structure of BCCP genes in four Gossypium species. The green boxes represent exons and the black lines represent introns.
Gene structure analysis is a significant method to study genetic evolution. We calculated the number of exons and introns in BCCP family members by comparing the genomic sequences with the predicted coding sequences in the four cotton species, and created BCCP structure in cotton. As shown in Figure 1B, gene length in class I was longer than that in class II. The code length of gene members in class I ranged from 951 to 16,013 bp, while in class II ranged from 2,000 to 3,000 bp, except for GbBCCP6, which contained 5,502 bp. The exons/intron distribution patterns in class I genes were various. The exons numbers ranged from four of GbBCCP2 to nine of GbBCCP9. However, the exons/intron distribution patterns of class II genes were conserved except for GbBCCP6 which had 12 exons. For example, six exons were found in GrBCCP3, GaBCCP3, and GhBCCP6, and seven exons in each of GrBCCP1, GaBCCP2, GhBCCP2, GhBCCP4, GhBCCP7, GbBCCP1, and GbBCCP3. Four conserved motifs were identified from the 23 cotton BCCP proteins using the MEME motif research tool (Supplementary Figure 3), and Supplementary Table 3 listed the length and sequence information of these four motifs. Motif 1 was the biotinyl motif, and it was present in all BCCP genes. Although motifs 2–4 did not belong to any known functional domains based on the searches using interproscan database, motifs 2 and 3 were primarily present in the C-terminal regions. Moreover, motif 4 was only present in GrBCCP3, GaBCCP3, GhBCCP2, GhBCCP6, GbBCCP1, and GbBCCP6 proteins (Supplementary Figure 3A). The presence of the same type of conserved motifs might indicate similar function among cotton BCCP genes.
In order to reveal the orthologous relationships of BCCP genes between the four cotton species, the protein sequences of 4 GrBCCP genes, 4 GaBCCP genes, 8 GhBCCP genes, and 7 GbBCCP genes were applied to construct six unrooted phylogenetic trees (Figure 2). The results showed that there were 20 pairs of orthologous genes among the four cotton species, since they were in the terminal branches with high bootstrap values. Among them, four orthologous gene pairs in the two diploid cotton (G. raimondii and G. arboreum) (Figure 2A), and there were three pairs of orthologous genes in the two allotetraploid cotton (G. hirsutum and G. barbadense) (Figure 2B). Four pairs of orthologous genes in G. hirsutum and G. raimondii were identified (Figure 2C), and four pairs in G. hirsutum and G. arboreum were found (Figure 2D). While, there were three pairs of orthologous genes from G. barbadense and G. raimondii (Figure 2E), two pairs in G. barbadense and G. arboretum (Figure 2F). The orthologous relationships among the four cotton species were displayed in Supplementary Figure 4. As expected, gene structures of orthologous pairs were almost identical with only minor differences with the exception of GbBCCP6/GhBCCP6 and GbBCCP6/GrBCCP3 (Figures 2B,E). However, based on the others were divergent apparently, the orthologous relationships of them could not be confirmed. In addition, a total of eleven pairs of paralogous genes in the four cotton species were found (Supplementary Figure 5), since the BCCP genes from the same genome were in the terminal branches of the phylogenetic trees. Among them, two pairs of paralogous genes in respective genome of G. raimondii and G. arboreum, and there were four paralogous pairs in G. hirsutum and three in G. barbadense.
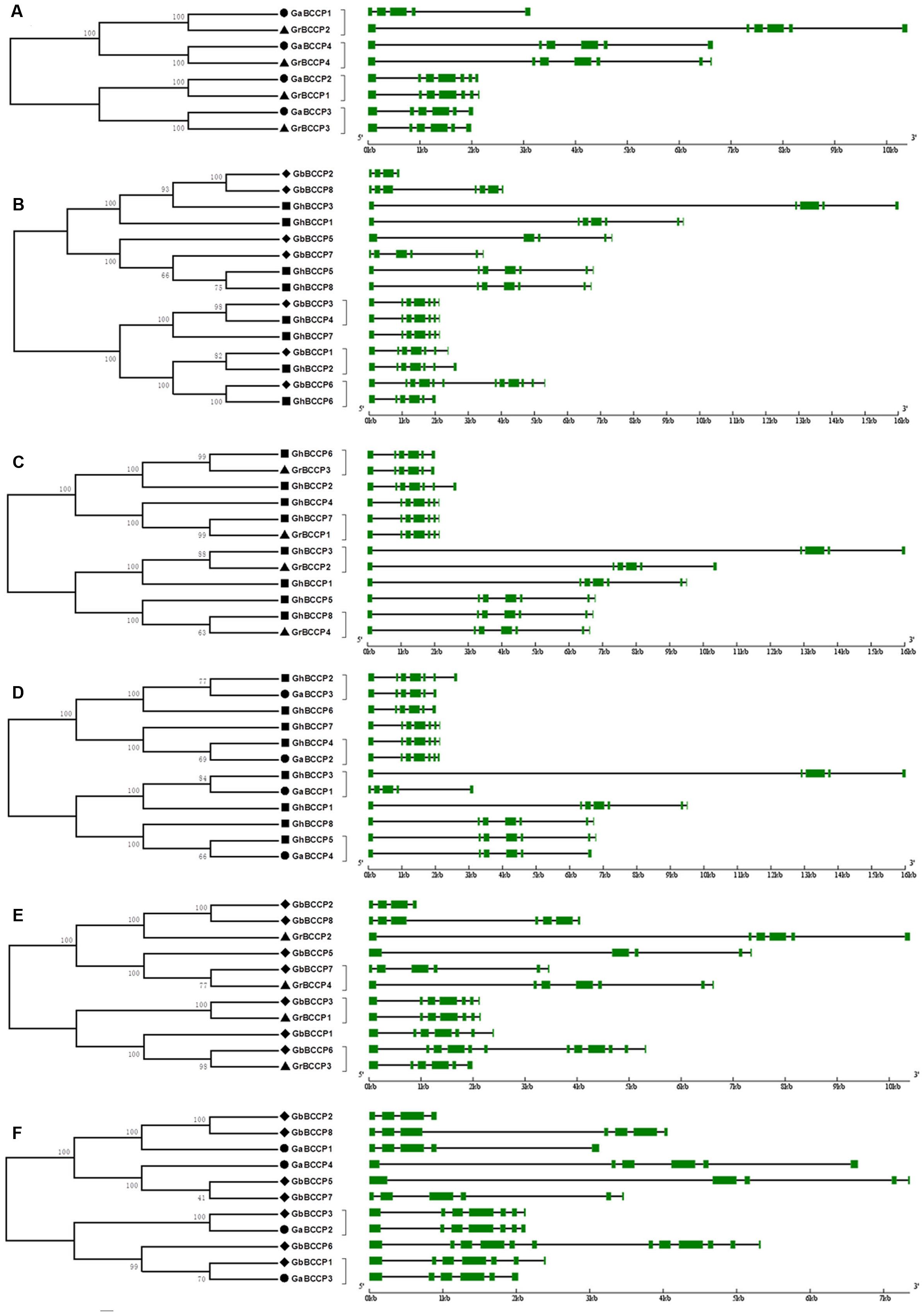
FIGURE 2. Phylogenetic relationships and gene structure of BCCP genes in and between G. raimondii, G. arboreum, G. hirsutum, and G. barbadense. (A) The phylogenetic tree and gene structure of BCCP genes in G. raimondii and G. arboreum. (B) The phylogenetic tree and gene structure of BCCP genes in G. hirsutum and G. barbadense. (C) The phylogenetic tree and gene structure of BCCP genes in G. hirsutum and G. raimondii. (D) The phylogenetic tree and gene structure of BCCP genes in G. hirsutum and G. arboreum. (E) The phylogenetic tree and gene structure of BCCP genes in G. barbadense and G. raimondii. (F) The phylogenetic tree and gene structure of BCCP genes in G. barbadense and G. arboreum. The BCCP genes from G. raimondii, G. arboreum, G. hirsutum and G. barbadense were marked with black triangles, black dots, black squares, and black rhombuses, respectively. Exons were represented by green boxes and introns by black lines.
Based on the coordinate of each BCCP gene on the chromosomes, the chromosomal distribution images of BCCP genes in G. raimondii, G. arboreum, G. hirsutum, and G. barbadense were generated. In the four cotton species, the BCCP genes were distributed uniformly, one gene on each chromosome or scaffold (Figure 3). In G. raimondii, one BCCP gene was found in each of chromosome 6, 10, 12, and 13 (Figure 3A). In G. arboreum, only one gene was in each of chromosome 8, 11, 12, and 13 (Figure 3B). There were 8 GhBCCP genes in G. hirsutum, 4 genes were assigned to A subgenome and 4 to D subgenome, respectively (Figure 3C). In A subgenome of G. hirsutum, each of chromosome 5, 6, 9, and 13 had only one gene, and only one in each of chromosome 4, 6, 9, and 13 in D subgenome of G. hirsutum. The 7 GbBCCP genes were mapped on six G. barbadense chromosomes and one scaffold (Figure 3D). Each only single BCCP gene was localized on chromosome 5, 6, 9, and 13 in At subgenome, and one gene on each of chromosomes 4 and 13 in Dt subgenome. GbBCCP8 was ambiguous and could not be assigned to either subgenome, but it merely anchored on unmapped scaffold.
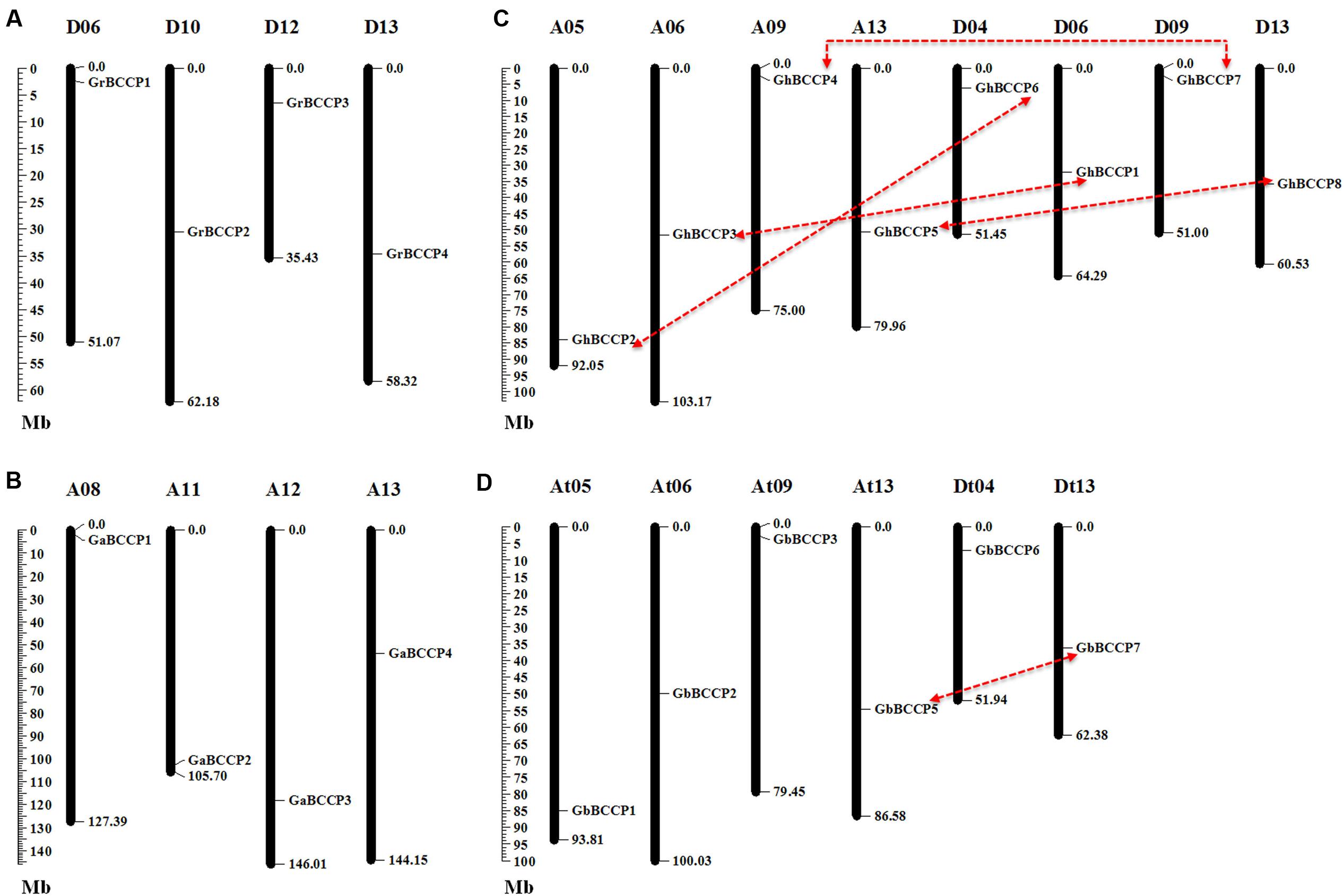
FIGURE 3. Localization of BCCP genes in the four cotton species. Twenty-two BCCP genes were mapped on different chromosomes in G. raimondii (A), G. arboreum (B), G. hirsutum (C), and G. barbadense (D). Only the chromosomes where BCCP genes were mapped are shown. Possible duplicated genes were connected by lines. The scale represents the megabases (Mb).
Genomic changes, including chromosomal rearrangement, gene duplication and expression change of genes, often occurred during the formation of polyploidy species (Cronn et al., 1999). And gene duplication events were considered to play an important role in the amplification of gene families (Cannon et al., 2004; Maere et al., 2005). We investigated the gene duplication events of BCCP genes in the four cotton species, respectively. Firstly, we used the following criteria (Zhou et al., 2004): the alignment length covered >70% of the longer aligned gene, and the AA identity between the sequences was >70%, to identify gene duplication events. A total of nine segmental duplication events were found (Supplementary Table 4). Among them, two duplication gene pairs were, respectively, found in genome of G. raimondii and G. arboretum, and four in G. hirsutum. In G. barbadense, only one gene pairs were found. All those duplicated gene pairs were located on different chromosomes, suggesting all of them were segmental duplication events. Subsequently, we employed a stringent criteria, the alignment length covered >80% of the longer gene, and the identity of the aligned regions >80%, (Jiang et al., 2013; Wei et al., 2013; Dong et al., 2016). Under this rule, only 4 and 1 segmental duplication events were found in G. hirsutum and G. barbadense, respectively (Table 2), comparable to the number generated with 70% cover length and AA identity criterion. These results suggested that segmental duplication played crucial roles in the expansion of the BCCP gene family in the two allotetraploid cotton species.
During the progress of evolution, the duplicated gene pairs might have undergone three alternative fates, i.e., non-functionalization, neofunctionalization, and subfunctionalization (Lynch and Conery, 2000). To explore different selective constrains on duplicated BCCP genes in G. hirsutum and G. barbadense, the Ka/Ks ratio for each pair of duplicates were calculated (Table 2). Generally, Ka/Ks > 1 indicates positive selection (accelerated evolution), Ka/Ks = 1 indicates neutral selection (the genes are pseudogenes), while Ka/Ks < 1 indicates negative or purifying selection (the functional constraint of the genes) (Liu et al., 2014; Dong et al., 2016). In this study, the Ka/Ks ratios for four duplicated BCCP gene pairs were less than 1, suggesting that the BCCP genes from G. hirsutum have mainly experienced purifying selection pressure. While in the case of G. barbadense, the one BCCP duplicated gene pair (GbBCCP5/GbBCCP7) with a ratio = 1, indicating neutral selection. Those results reflected that the function of the duplicated BCCP genes in the two cotton species did not diverge much during subsequent evolution, and the maintenance of function in G. hirsutum BCCP genes might contributed to purifying selection.
Phylogenetic tree was served as a common method to reveal homologous relationships and evolutionary root of BCCPs from species. To detect the evolutionary relationships of BCCP genes, a NJ phylogenetic tree was constructed with the alignments of BCCP protein sequences of four cotton species, Arabidopsis, rapeseed and soybean BCCP proteins (Figure 4). Compared with the other three species, BCCP proteins from the four cotton species had higher relative coefficient, suggesting a closer relationship. It also appeared that cotton BCCPs fell into two evolutionary classes defined by the two Arabidopsis BCCPs, which was consistent with BCCP genes in Brassicaceae oilseeds (Thelen et al., 2000). We surmised that they might have distinct roles in fatty acid biosynthesis. The phylogenetic tree clearly indicated that the BCCP genes (GrBCCP2 and GrBCCP4 of G. raimondii; GaBCCP1 and GaBCCP4 of G. arboreum; GhBCCP1, GhBCCP3, GhBCCP5, and GhBCCP8 of G. hirsutum; GbBCCP2, GbBCCP5, GbBCCP7, and GbBCCP8 of G. barbadense) were closely related to the AtBCCP1 and accB-1 of soybean (Figure 4), implying that they may have similar function(s).
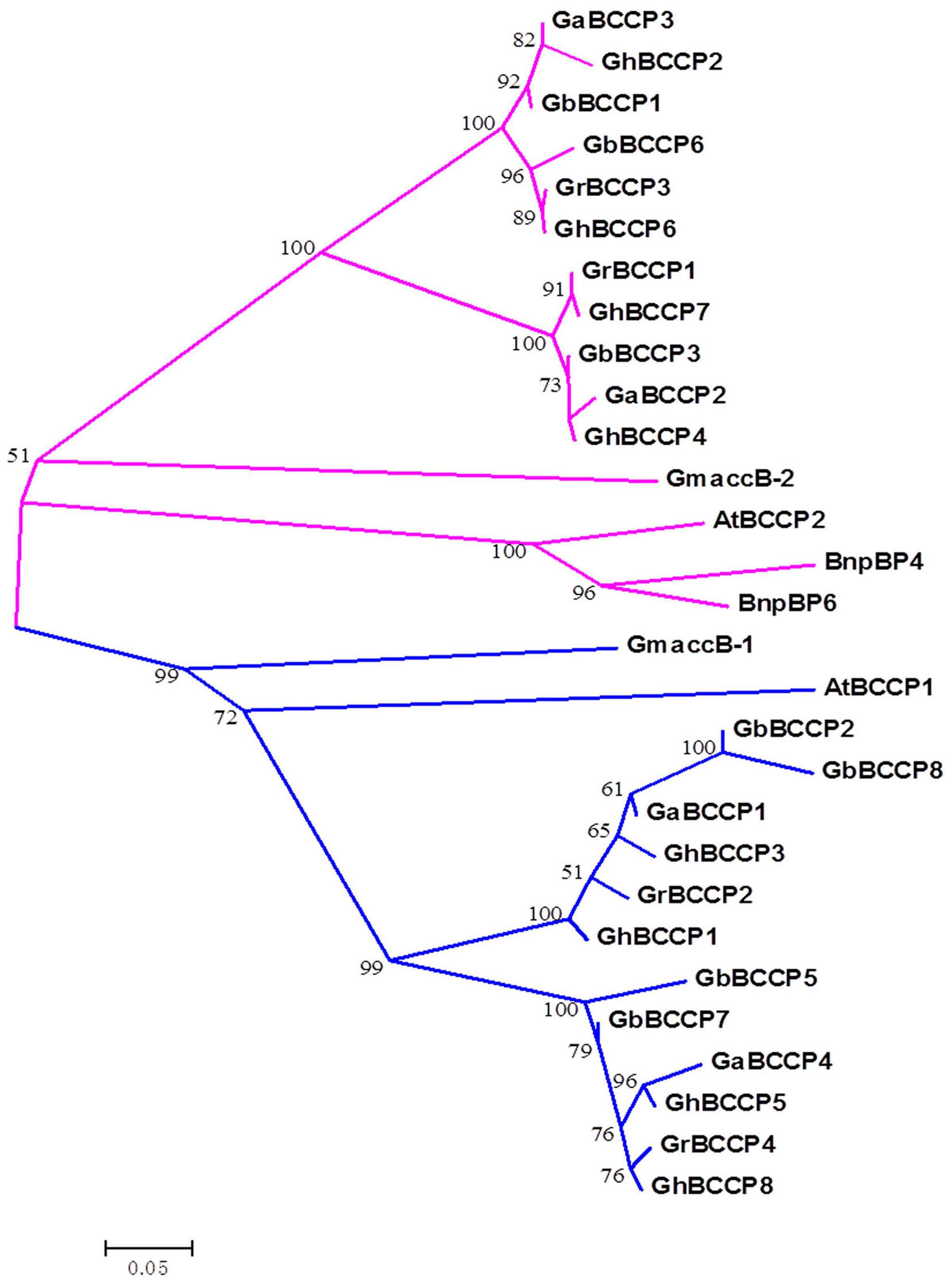
FIGURE 4. Phylogenetic analysis of BCCP genes in Gossypium and other plants. The unrooted phylogenetic tree containing 4 G. raimondii (GrBCCP), 4 G. arboreum (GaBCCP), 8 G. hirsutum (GhBCCP), 7 G. barbadense (GbBCCP), 2 Arabidopsis (AtBCCP), 2 soybean (GmaccB), and 2 rapeseed (BnpBP). BCCP genes were constructed using the neighbor-joining method with 1000 replicates.
In order to understand the role of GhBCCP genes better in G. hirsutum acc. TM-1, temporal expression patterns of these genes were analyzed using the public expression data, which contained vegetative tissues (root, stem, and leaf), floral tissue (petal), and ovule tissues at different developmental stages (5, 10, 20, 25, and 35 DPA) (Zhang T. et al., 2015). As shown in Figure 5, the expression of GhBCCP genes could be detected in all of the investigated tissues of TM-1, indicating that the GhBCCP genes were involved in multiple progresses during the development of the cotton. The heat map also revealed that the GhBCCP genes showed specific spatial expression patterns. Based on the hierarchical clustering analysis, the eight GhBCCP genes could be clustered into two classes (classes I and II) (Figure 5). Compared with the GhBCCP genes in class I, class II genes exhibited higher transcript abundance in the ovules at the five developmental stages. In addition, the expression levels of genes in class II were up-expressed in the ovules compared with that in vegetative and floral tissues (Figure 5). During the ovule developmental stages, GhBCCP genes in class II had higher expression in ovules at early and middle developmental stages (5–25 DPA), but relatively lower expressions in later developmental stages (35 DPA), indicating that they might contribute to fatty acids accumulation mainly occurring before the maturation stage in upland cotton seeds. Furthermore, the expression patterns of four duplicated gene pairs (Table 2) were slightly different in present research. Two duplicated gene pairs such as GhBCCP2/GhBCCP6 and GhBCCP4/GhBCCP7 were clustered together, and shared highly similar expression patterns in all the tested tissues (Figure 5). The expression of GhBCCP1/GhBCCP3 was similar in all investigated tissues except for in petals and 35 DPA ovules. However, the duplicated gene pairs of GhBCCP5/GhBCCP8 were divergent, which might be caused by the significant variation in gene regulation after the duplication events.
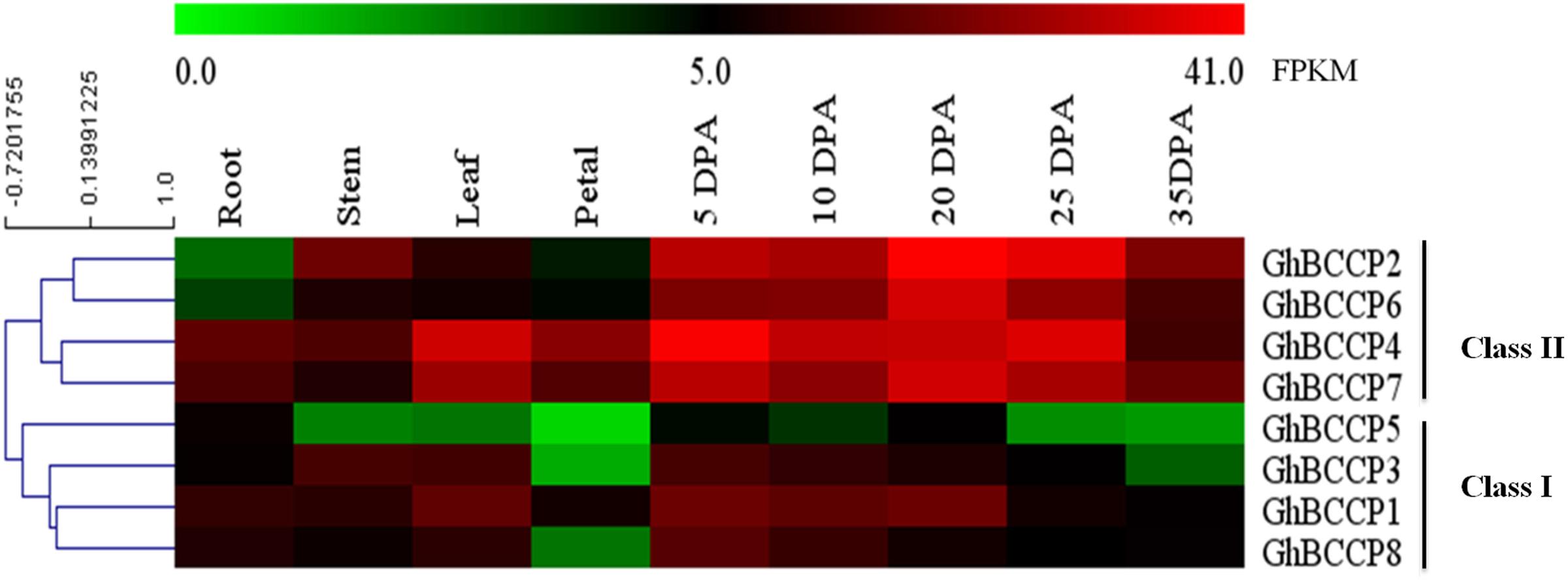
FIGURE 5. Expression analysis of ChBCCP genes in G. hirsutum acc TM-1 across 9 tissues. The RNA-Seq expression profiles of G. hirsutum acc. TM-1 (Zhang T. et al., 2015) were used to identify the expression levels of ChBCCP genes. FPKM represents fragments per kilobase of exon model per million mapped reads. DPA, days post anthesis.
Salt and cold stresses are the two serous environmental stresses that most plants might encounter during their growth and developmental processes. Precious study reported that GhBCCP1 gene has been induced by cold stress for 24 h (Cui et al., 2017). However, little is known about the function of cotton BCCP genes responsing to salt and cold stresses. Gene promoter is the control center of gene transcription, and the cis-elements in gene promoter regions could provide some evidence for dissection of gene functions in stress response (Zhou et al., 2013). In present study, we identified all the cis-regulatory elements in the promoter regions of 16 cotton BCCP genes from G. raimondii, G. arboreum, and G. hirsutum (Supplementary Table 5), and there were eight putative environmental stress-related elements in the promoter regions of the three cotton BCCP genes (Supplementary Table 6). Although there were no special items of salt responsive element and only one low temperature responsive cis-element existed in the PLACE database, some cis-elements might respond to multiple environment stimuli (Higo et al., 1999). The results showed that each BCCP gene in the three cotton species contained more than three environmental stress-related elements (Supplementary Table 6), indicating that these BCCP genes might the signal transduction of the cotton response to salt and cold stresses.
To investigate the expression patterns of BCCP genes in different tissues under salt and cold (4°C) stresses, roots, stems, and leaves in each of G. raimondii, G. arboreum var Shixiya 1, and G. hirsutum acc TM-1 at trefoil stage were treated and used for RNA extraction. The expression levels of BCCP genes responsive to salt and cold stresses were shown in Figure 6, and it showed that these BCCP genes from the three cotton species expressed diversely under both stresses. For the salt stress (Figure 6A), in roots, six BCCP genes in cotton showed up-regulated expression after salt treatment for 24 h, GrBCCP4 showed insignificantly up-regulated expression, while the rest genes expressed down-regulation. In stems, all the genes in G. raimondii and G. arboreum were up-regulated expression under salt stress, and seven of eight GhBCCP genes showed down-regulated. However, only a few up-regulated BCCP genes were found in leaves compared with roots and stems. Three GaBCCP genes showed down-regulated expression in leaves, GaBCCP4 and GhBCCP4 showed no significantly change compared with that control, others were down-regulated. For the cold stress (Figure 6B), the heat map showed that GrBCCP3, GaBCCP1, GaBCCP2, GaBCCP3, GaBCCP4, and GhBCCP7 were induced up-regulation in root. And only four genes were suppressed in roots compared with stem and leaves. In stem, only GrBCCP3, GaBCCP1, GaBCCP3, and GaBCCP4 were found up-regulated after cold stress for 24 h. In leaves, five genes were induced and the others were suppressed by cold stress for 24 h (Figure 6B). Notably, there were four BCCP genes (GrBCCP3, GaBCCP1, GaBCCP2, and GaBCCP4) up-regulated expression in roots under both salt and cold stresses. And also found four BCCP genes (GrBCCP3, GaBCCP1, GaBCCP3, and GaBCCP4) expressed up-regulated in stems under both salt and cold stresses. In leaves, two cotton BCCP genes (GaBCCP1 and GaBCCP3) were induced by both salt and cold treatment.
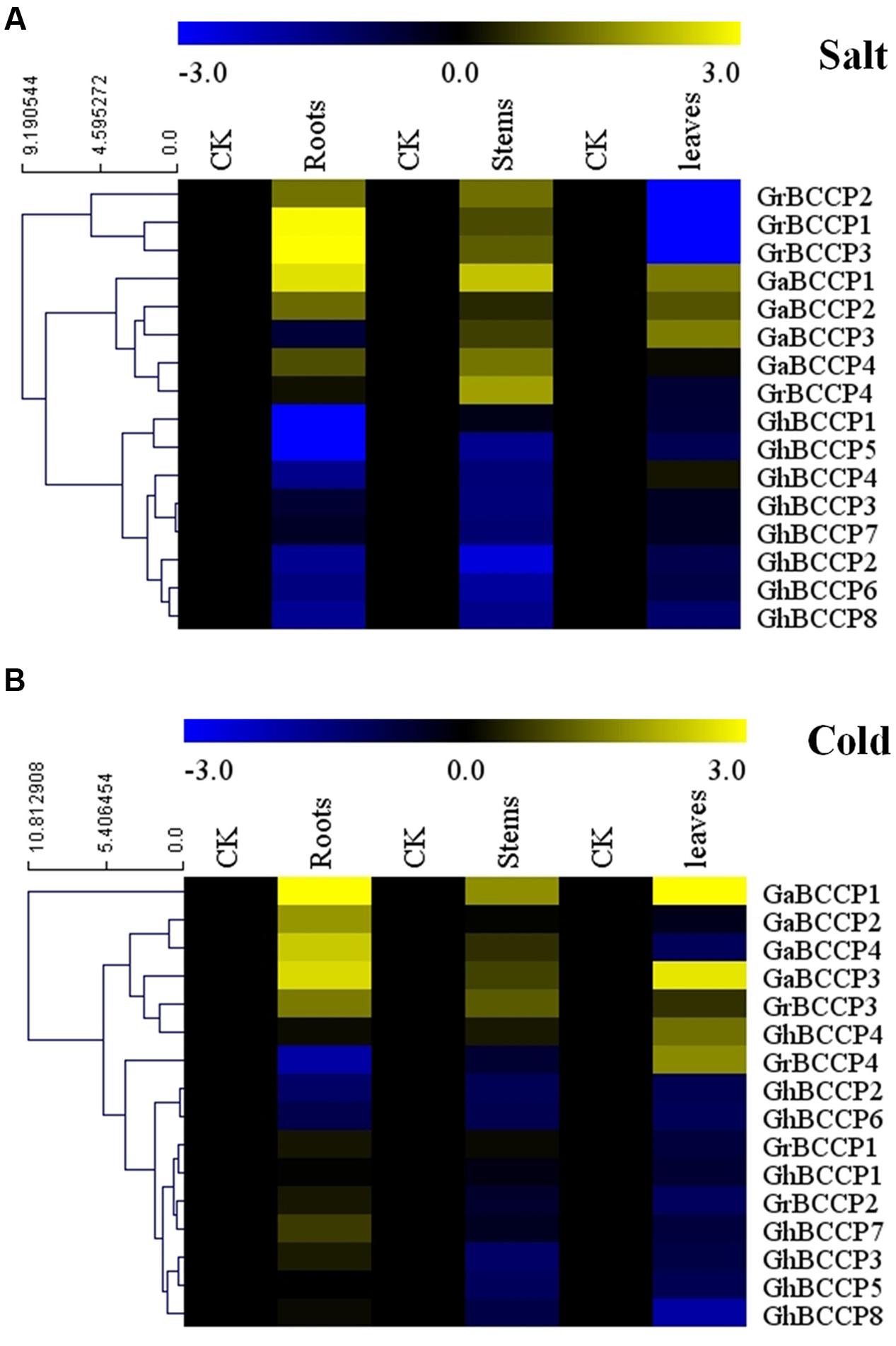
FIGURE 6. Expression patterns of 16 BCCP genes in three representative tissues of G. raimondii, G. arboreum, and G. hirsutum response to salt stress and cold stress. (A) Expression levels 16 cotton BCCP genes under salt stress, (B) Expression levels 16 cotton BCCP genes under cold stress. The color bar represents the relative signal intensity values.
The BCCP subunit is one of the four subunits of ACCase that catalyzes the irreversible carboxylation of acetyle-CoA to produce malonyl-CoA in fatty acid biosynthesis (Thelen et al., 2000; Fukuda et al., 2013; Jang et al., 2015). Many studies have indicated that modifying the BCCP genes could change the oil content of transgenic Arabidopsis (Thelen and Ohlrogge, 2002; Li et al., 2011). Cotton is a significant oilseed crop, and cottonseeds are important source of edible oil and potential industrial raw material (Cui et al., 2017). However, only GhBCCP1 gene has already been genetically manipulated for oil improvement in cottonseed (Cui et al., 2017). In present study, a comprehensive set of 24 BCCP genes was identified from the available genomes of the four cotton species. Undoubtedly, these identified cotton BCCP genes will provide candidate genes for the gene engineering of fatty acid biosynthesis in plants.
The results of present study revealed the details of 24 BCCP genes in the four cotton species. Among them, 4 were predicted in G. raimondii, 4 in G. arboreum, 8 in G. hirsutum, and 8 in G. barbadense, which indicated that BCCP genes in each of the two tetraploid cotton genomes were the sum of the two diploid cotton genomes. The predicted full-length 23 BCCP proteins were categorized into two classes, 12 genes in class I, and 11 in class II (Figure 1), and this classification was also presented in Figure 4. As shown in Figure 4, all the cotton BCCP genes could be divide into the same two classes according to the two AtBCCP genes in Arabidopsis (Figure 4), this was consistent with previous report in Brassicaceae oilseeds (Thelen et al., 2000). Each GrBCCP gene or GaBCCP gene in each of the diploid cotton species corresponded to two GhBCCP genes in the tetraploid cotton belonging to one homologous BCCP group, this was consistent with whole genome duplication events occurred during the evolution of Gossypium (Li et al., 2014). According to the distribution of intron/exon in BCCP genes, the gene in the same class shared the similar introns/exons structure and exon numbers (Figure 1B), but the gene length in class I were longer than class II, and the number of introns/exons in the terminal branch of phylogenetic tree were still different in some of the pairs. These findings indicated some introns loss, or introns gain, might have occurred during the BCCP structure evolution in the four cotton species. The prediction of motifs showed that all the BCCP proteins contained the biotinly domain (CIIEAMKLMNEIE) at C-terminal (Supplementary Figure 3A), but GbBCCP2 was one exception, which only harbored CIIEAMKLMNEIE sequence at C-terminal (Supplementary Figure 1) and could not presented motif 1 in Supplementary Figure 3A. Functional domains analysis indicated that the biotinyl domain of ACCasee is to transfer CO2 from one subsite to another allowing carboxylation reaction (Jitrapakdee and Wallace, 2003; Gu et al., 2011).
Gene duplication plays an important role in the process of plant genomic and organismal evolution, and gene duplication events contain tandem duplication, segmental duplication, transposition events and whole-genome duplication (Flagel and Wendel, 2009). In present study, we investigated gene duplicated events in order to further understand the expansion mechanism of BCCP genes in the four cotton species. Four duplicated gene pairs were identified in G. hirsutum, and one pair was found in G. barbadense. Among them, three segmental duplicated gene pairs, GhBCCP1/GhBCCP3, GhBCCP5/GhBCCP8, GbBCCP5/GbBCCP7, belonged to the class I, and the remaining two segmental duplicated gene pairs, GhBCCP2/GhBCCP6 and GhBCCP4/GhBCCP7, belonged to the class II. These results showed that the expansion of GhBCCP genes and GbBCCP genes in class I were mainly caused by the segmental duplication. Duplicated genes might have undergone three different fates, the result showed the Ka/Ks ratios for four duplicated GhBCCP gene pairs were less than 1, suggesting that these genes from G. hirsutum have mainly experienced purifying selection pressure. Gene expression patterns could provide useful clues for understanding these genes function. Based on these genes expression patterns in different tissues of TM-1 or response to salt and cold stresses performed in the study, the four GhBCCP duplicated gene pairs varied significantly. It was inferred that the functions of the four duplicated gene were different after duplication, and their fates could be described as neofunctionalization. These findings also further supported the assertion that expression divergence of duplicated genes is often the first step in the functional divergence, and this can increase the chance of duplicated genes being retained in a genome (Zhang, 2003).
Salt and cold stresses are the serious environmental stresses affecting the growth and yield of plants in many places of the world. Salt stress may increase the reactive oxygen species and damage the integrity of cell membrance (Zhu, 2002), and cold stress mainly alter the lipidic fluidity of membranes (Kargiotidou et al., 2008). Previous studies revealed that fatty acid synthesis related genes were induced or repressed by salt stress (Im et al., 2002; Zhang et al., 2009; Zeng et al., 2016) or cold stress (Liu et al., 2014; Liu W. et al., 2015; Zeng et al., 2016; Cui et al., 2017). In our study, the expression patterns of the 16 BCCP genes in the three cotton species (Figure 6) revealed that these genes were widely involved in responding to salt and cold stresses. Three of four GrBCCP genes (GrBCCP1, GrBCCP2, and GrBCCP3) were significantly up-regulated in response to salt stress in roots, suggesting these genes may be required to maintain certain activity of ACCase in cotton plants under salt treatment. Conversely, total of the four GrBCCP genes (GrBCCP1, GrBCCP2, GrBCCP3, and GrBCCP4) were down-regulated in leaves after salt treatment for 24 h (Figure 6A). The different expression patterns of GrBCCP genes responded to salt stress in roots and leaves might be associated with the fact that both tissues by themselves were distinct in structure and functions (Qing et al., 2009; Campo et al., 2014). Some cotton BCCP genes showed the same expression patterns in the same tissue after salt or cold stress, being either induced or suppressed, suggesting that these cotton BCCP genes were co-expressed in response to salt and cold stresses. However, a few of cotton BCCP genes in the same tissue responded to salt stress presented different expression patterns compared to cold stress. For instance, GaBCCP3 was up-regulated in root by cold stress, while it was down-regulated by salt stress. This indicated that two sets of cotton BCCP genes were separately involved in cold and salt stresses. All these results implied that the signaling network responded to abiotic stress in plants was complicated (Zhang L. et al., 2015).
In short, the BCCP gene family in G. raimondii, G. arboreum, G. hirsutum, and G. barbadense were identified and comprehensive analyzed using bioinformatics methods, and all of these results provided valuable clues in future efforts to identify specific gene functions for BCCP gene family and gene physiological roles among Gossypium species.
Conceived and designed the experiments: JH. Performed the experiments and analyzed the data: YC and YZ. Maintained the experimental platform and performed bench work: YW. Attended discussion and part of experiments: ZL, BI, and YH. Contributed reagents/materials/analysis tools: JH. Prepared the manuscript: YC. Edited and revised the manuscript: JH.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The work was supported by grants from National Natural Science Foundation of China (Grant Number 31371666) and National Key R & D Program for Crop Breeding (2016YFD0100203).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00624/full#supplementary-material
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37, W202–W208. doi: 10.1093/nar/gkp335
Bates, P. D., Durrett, T. P., Ohlrogge, J. B., and Pollard, M. (2009). Analysis of acyl fluxes through multiple pathways of triacylglycerol synthesis in developing soybean embryos. Plant Physiol. 150, 55–72. doi: 10.1104/pp.109.137737
Beisson, F., Koo, A. J. K., Ruuska, S., Schwender, J., Pollard, M., Thelen, J. J., et al. (2003). Arabidopsis genes involved in acyl lipid metabolism. A 2003 census of the candidates, a study of the distribution of expressed sequence tags in organs, and a web-based database. Plant Physiol. 132, 681–697. doi: 10.1104/pp.103.022988
Campo, S., Baldrich, P., Messeguer, J., Lalanne, E., Coca, M., and Segundo, B. S. (2014). Overexpression of a calcium-dependent protein kinase confers salt and drought tolerance in rice by preventing membrane lipid peroxidation. Plant Physiol. 165, 688–704. doi: 10.1104/pp.113.230268
Cannon, S. B., Mitra, A., Baumgarten, A., Young, N. D., and May, G. (2004). The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4:10. doi: 10.1186/1471-2229-4-10
Chen, Y., Elizondo-Noriega, A., Cantu, D. C., and Reilly, P. J. (2012). Structural classification of biotin carboxyl carrier proteins. Biotechnol. Lett. 34, 1869–1875. doi: 10.1007/s10529-012-0978-4
Chou, K. C., and Shen, H. B. (2007). Recent progress in protein subcellular location prediction. Anal. Biochem. 370, 1–16. doi: 10.1016/j.ab.2007.07.006
Cronn, R. C., Small, R. L., and Wendel, J. F. (1999). Duplicated genes evolve independently after polyploid formation in cotton. Proc. Natl. Acad. Sci. U.S.A. 96, 14406–14411. doi: 10.1073/pnas.96.25.14406
Cui, Y., Liu, Z., Zhao, Y., Wang, Y., Huang, Y., Li, L., et al. (2017). Overexpression of heteromeric GhACCase subunits enhanced oil accumulation in Upland cotton. Plant Mol. Biol. Rep. 35, 287–297. doi: 10.1007/s11105-016-1022-y
Dong, Y., Li, C., Zhang, Y., He, Q., Daud, M. K., Chen, J., et al. (2016). Glutathione s-transferase gene family in Gossypium raimondii and G. arboreum: comparative genomic study and their expression under salt stress. Front. Plant Sci. 7:139. doi: 10.3389/fpls.2016.00139
Elborough, K. M., Winz, R., Deka, R. K., Markham, J. E., White, A. J., Rawsthorne, S., et al. (1996). Biotin carboxyl carrier protein and carboxyltransferase subunits of the multi-subunit form of acetyl-CoA carboxylase from Brassica napus: cloning and analysis of expression during oilseed rape embryogenesis. Biochem. J. 315, 103–112. doi: 10.1042/bj3150103
Emanuelsson, O., Brunak, S., von Heijne, G., and Nielsen, H. (2007). Locating proteins in the cell using TargetP, SignalP and related tools. Nat. Protoc. 2, 953–971. doi: 10.1038/nprot.2007.131
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42, D222–D230. doi: 10.1093/nar/gkt1223
Flagel, L. E., and Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564. doi: 10.1111/j.1469-8137.2009.02923.x
Fukuda, N., Ikawa, Y., Aoyagi, T., and Kozaki, A. (2013). Expression of the genes coding for plastidic acetyl-CoA carboxylase subunits is regulated by a location-sensitive transcription factor binding site. Plant Mol. Biol. 82, 473–483. doi: 10.1007/s11103-013-0075-7
Gu, K., Chiam, H., Tian, D., and Yin, Z. (2011). Molecular cloning and expression of heteromeric ACCase subunit genes from Jatropha curcas. Plant Sci. 180, 642–649. doi: 10.1016/j.plantsci.2011.01.007
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Horton, P., Park, K. J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C. J., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35, W585–W587. doi: 10.1093/nar/gkm259
Hu, B., Jin, J., Guo, A. Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Hu, Z. Y., Hua, W., Zhang, L., Deng, L. B., Wang, X. F., Liu, G. H., et al. (2013). Seed structure characteristics to form ultrahigh oil content in rapeseed. PLoS ONE 8:e62099. doi: 10.1371/journal.pone.0062099
Im, Y. J., Han, O., Chung, G. C., and Cho, B. H. (2002). Antisense expression of an Arabidopsis omega-3 fatty acid desaturase gene reduces salt/drought tolerance in transgenic tobacco plants. Mol. Cells 13, 264–271.
Jang, Y. E., Kim, M. Y., Shim, S., Lee, J., and Lee, S.-H. (2015). Gene expression profiling for seed protein and oil synthesis during early seed development in soybean. Genes Genomics 37, 409–418. doi: 10.1007/s13258-015-0269-2
Jiang, H., Wu, Q., Jin, J., Sheng, L., Yan, H., Cheng, B., et al. (2013). Genome-wide identification and expression profiling of ankyrin-repeat gene family in maize. Dev. Genes Evol. 223, 303–318. doi: 10.1007/s00427-013-0447-7
Jitrapakdee, S., and Wallace, J. C. (2003). The biotin enzyme family: conserved structural motifs and domain rearrangements. Curr. Protein Pept. Sci. 4, 217–229. doi: 10.2174/1389203033487199
Kargiotidou, A., Deli, D., Galanopoulou, D., Tsaftaris, A., and Farmaki, T. (2008). Low temperature and light regulate delta 12 fatty acid desaturases (FAD2) at a transcriptional level in cotton (Gossypium hirsutum). J. Exp. Bot. 59, 2043–2056. doi: 10.1093/jxb/ern065
Ke, J., Wen, T. N., Nikolau, B. J., and Wurtele, E. S. (2000). Coordinate regulation of the nuclear and plastidic genes coding for the subunits of the heteromeric acetyl-coenzyme A carboxylase. Plant Physiol. 122, 1057–1071. doi: 10.1104/pp.122.4.1057
Kohel, R. J. (1973). Genetic nomenclature in cotton. J. Hered. 64, 291–295. doi: 10.1093/oxfordjournals.jhered.a108415
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Letunic, I., Doerks, T., and Bork, P. (2015). SMART: recent updates, new developments and status in 2015. Nucleic Acids Res. 43, D257–D260. doi: 10.1093/nar/gku949
Li, F., Fan, G., Lu, C., Xiao, G., Zou, C., Kohel, R. J., et al. (2015). Genome sequence of cultivated Upland cotton (Gossypium hirsutum TM-1) provides insights into genome evolution. Nat. Biotechnol. 33, 524–530. doi: 10.1038/nbt.3208
Li, F., Fan, G., Wang, K., Sun, F., Yuan, Y., Song, G., et al. (2014). Genome sequence of the cultivated cotton Gossypium arboreum. Nat. Genet. 46, 567–572. doi: 10.1038/ng.2987
Li, M. J., Xia, H., Zhao, C. Z., Li, A. Q., Li, C. S., Bi, Y. P., et al. (2010). Isolation and characterization of putative acetyl-CoA carboxylases in Arachis hypogaea L. Plant Mol. Biol. Rep. 28, 58–68. doi: 10.1007/s11105-009-0126-z
Li, Z. G., Yin, W. B., Song, L. Y., Chen, Y. H., Guan, R. Z., Wang, J. Q., et al. (2011). Genes encoding the biotin carboxylase subunit of acetyl-CoA carboxylase from Brassica napus and parental species: cloning, expression patterns, and evolution. Genome 54, 202–211. doi: 10.1139/G10-110
Liu, W., Li, W., He, Q., Daud, M. K., Chen, J., and Zhu, S. (2014). Genome-wide survey and expression analysis of calcium-dependent protein kinase in Gossypium raimondii. PLoS ONE 9:e98189. doi: 10.1371/journal.pone.0098189
Liu, W., Li, W., He, Q., Daud, M. K., Chen, J., and Zhu, S. (2015). Characterization of 19 genes encoding membrane-bound fatty acid desaturases and their expression profiles in Gossypium raimondii under low temperature. PLoS ONE 10:e0123281. doi: 10.1371/journal.pone.0123281
Liu, X., Zhao, B., Zheng, H. J., Hu, Y., Lu, G., Yang, C. Q., et al. (2015). Gossypium barbadense genome sequence provides insight into the evolution of extra-long staple fiber and specialized metabolites. Sci. Rep. 5:14139. doi: 10.1038/srep14139
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Lynch, M., and Conery, J. S. (2000). The evolutionary fate and consequences of duplicate genes. Science 290, 1151–1155. doi: 10.1126/science.290.5494.1151
Maere, S., Bodt, S. D., Raes, J., Casneuf, T., Montagu, M. V., Kuiper, M., et al. (2005). Modeling gene and genome duplications in eukaryotes. Proc. Natl. Acad. Sci. U.S.A. 102, 5454–5459. doi: 10.1126/science.290.5494.1151
Paterson, A. H., Wendel, J. F., Gundlach, H., Guo, H., Jenkins, J., Jin, D., et al. (2012). Repeated polyploidization of Gossypium genomes and the evolution of spinnable cotton fibres. Nature 492, 423–427. doi: 10.1038/nature11798
Qiao, Z. X., and Liu, J. Y. (2007). Cloning and characterization of cotton heteromeric acetyl-CoA carboxylase genes. Prog. Nat. Sci. 17, 1412–1418.
Qing, D. J., Lu, H. F., Li, N., Dong, H. T., Dong, D. F., and Li, Y. Z. (2009). Comparative profiles of gene expression in leaves and roots of maize seedlings under conditions of salt stress and the removal of salt stress. Plant Cell Physiol. 50, 889–903. doi: 10.1093/pcp/pcp038
Quevillon, E., Silventoinen, V., Pillai, S., Harte, N., Mulder, N., Apweiler, R., et al. (2005). InterProScan: protein domains identifier. Nucleic Acids Res. 33, W116–W120. doi: 10.1093/nar/gki442
Ran, H., Faigenboim-Doron, A., Kadmon, N., Hu, G., Zhang, X., Gallagher, J. P., et al. (2015). A transcriptome profile for developing seed of polyploid cotton. Plant Genome 8, 1–15. doi: 10.3835/plantgenome2014.08.0041
Reverdatto, S., Beilinson, V., and Nielsen, N. C. (1999). A multisubunit acetyl coenzyme A carboxylase from soybean. Plant Physiol. 119, 961–978. doi: 10.1104/pp.119.3.961
Rozas, J., Sánchezdelbarrio, J. C., Messeguer, X., and Rozas, R. (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics 19, 2496–2497. doi: 10.1093/bioinformatics/btg359
Sasaki, Y., and Nagano, Y. (2004). Plant acetyl-CoA carboxylase: structure, biosynthesis, regulation, and gene manipulation for plant breeding. Biosci. Biotechnol. Biochem. 68, 1175–1184. doi: 10.1271/bbb.68.1175
Schulte, W., Töpfer, R., Stracke, R., Schell, J., and Martini, N. (1997). Multifunctional acetyl-CoA carboxylase from Brassica napus is encoded by a multi-gene family: indication for plastidic localization of at least one isoform. Proc. Natl. Acad. Sci. U.S.A. 94, 3465–3470. doi: 10.1073/pnas.94.7.3465
Senchina, D. S., Alvarez, I., Cronn, R. C., Liu, B., Rong, J., Noyes, R. D., et al. (2003). Rate variation among nuclear genes and the age of polyploidy in Gossypium. Mol. Biol. Evol. 20, 633–643. doi: 10.1093/molbev/msg065
Sood, A., and Chauhan, R. S. (2015). Regulation of FA and TAG biosynthesis pathway genes in endosperms and embryos of high and low oil content genotypes of Jatropha curcas L. Plant Physiol. Biochem. 94, 253–267. doi: 10.1016/j.plaphy.2015.06.015
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Thelen, J. J., Mekhedov, S., and Ohlrogge, J. B. (2000). Biotin carboxyl carrier protein isoforms in Brassicaceae oilseeds. Biochem. Soc. Trans. 28, 595–598. doi: 10.1042/0300-5127:0280595
Thelen, J. J., Mekhedov, S., and Ohlrogge, J. B. (2001). Brassicaceae express multiple isoforms of biotin carboxyl carrier protein in a tissue-specific manner. Plant Physiol. 125, 2016–2028. doi: 10.1104/pp.125.4.2016
Thelen, J. J., and Ohlrogge, J. B. (2002). Both antisense and sense expression of biotin carboxyl carrier protein isoform 2 inactivates the plastid acetyl-coenzyme a carboxylase in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 32, 419–431. doi: 10.1046/j.1365-313X.2002.01435.x
Troncosoponce, M. A., Kilaru, A., Cao, X., Durrett, T. P., Fan, J., Jensen, J. K., et al. (2011). Comparative deep transcriptional profiling of four developing oilseeds. Plant J. 68, 1014–1027. doi: 10.1111/j.1365-313X.2011.04751.x
Voorrips, R. E. (2002). MapChart: software for the graphical presentation of linkage maps and QTLs. J. Hered. 93, 77–78. doi: 10.1093/jhered/93.1.77
Wang, K., Wang, Z., Li, F., Ye, W., Wang, J., Song, G., et al. (2012). The draft genome of a diploid cotton Gossypium raimondii. Nat. Genet. 44, 1098–1103. doi: 10.1038/ng.2371
Wei, H., Li, W., Sun, X., Zhu, S., and Zhu, J. (2013). Systematic analysis and comparison of nucleotide-binding site disease resistance genes in a diploid cotton Gossypium raimondii. PLoS ONE 8:e68435. doi: 10.1371/journal.pone.0068435
Wendel, J. F., Brubaker, C., Alvarez, I., Cronn, R., Stewart, J. M., and Paterson, A. H. (2009). “Evolution and natural history of the cotton genus,” in Genetics and Genomics of Cotton, ed. A. H. Paterson (New York, NY: Springer), 3–22. doi: 10.1007/978-0-387-70810-2_1
Xuan, W. Y., Zhang, Y., Liu, Z. Q., Feng, D., and Luo, M. Y. (2015). Molecular cloning and expression analysis of a novel BCCP subunit gene from Aleurites moluccana. Genet. Mol. Res. 14, 9922–9931. doi: 10.4238/2015.August.19.27
Yin, D., Wang, Y., Zhang, X., Li, H., Lu, X., Zhang, J., et al. (2013). De novo assembly of the peanut (Arachis hypogaea L.) seed transcriptome revealed candidate unigenes for oil accumulation pathways. PLoS ONE 8:e73767. doi: 10.1371/journal.pone.0073767
Yu, J., Jung, S., Cheng, C. H., Ficklin, S. P., Lee, T., Zheng, P., et al. (2014). CottonGen: a genomics, genetics and breeding database for cotton research. Nucleic Acids Res. 42, D1229–D1236. doi: 10.1093/nar/gkt1064
Yuan, D., Tang, Z., Wang, M., Gao, W., Tu, L., Xin, J., et al. (2015). The genome sequence of Sea-Island cotton (Gossypium barbadense) provides insights into the allopolyploidization and development of superior spinnable fibres. Sci. Rep. 5:17662. doi: 10.1038/srep17662
Zeng, L., Deng, R., Guo, Z., Yang, S., and Deng, X. (2016). Genome-wide identification and characterization of Glyceraldehyde-3-phosphate dehydrogenase genes family in wheat (Triticum aestivum). BMC Genomics 17:240. doi: 10.1186/s12864-016-2527-3
Zhang, J. (2003). Evolution by gene duplication: an update. Trends Ecol. Evol. 18, 292–298. doi: 10.1016/s0169-5347(03)00033-8
Zhang, J. T., Zhu, J. Q., Zhu, Q., Liu, H., Gao, X. S., and Zhang, H. X. (2009). Fatty acid desaturase-6 (Fad6) is required for salt tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 390, 469–474. doi: 10.1016/j.bbrc.2009.09.095
Zhang, L., Zhao, H. K., Dong, Q. L., Zhang, Y. Y., Wang, Y. M., Li, H. Y., et al. (2015). Genome-wide analysis and expression profiling under heat and drought treatments of HSP70 gene family in soybean (Glycine max L.). Front. Plant Sci. 6:773. doi: 10.3389/fpls.2015.00773
Zhang, T., Hu, Y., Jiang, W., Fang, L., Guan, X., Chen, J., et al. (2015). Sequencing of allotetraploid cotton (Gossypium hirsutum acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537. doi: 10.1038/nbt.3207
Zhou, M. L., Yang, X. B., Zhang, Q., Zhou, M., Zhao, E. Z., Tang, Y. X., et al. (2013). Induction of annexin by heavy metals and jasmonic acid in Zea mays. Funct. Integr. Genomics 13, 241–251. doi: 10.1007/s10142-013-0316-5
Zhou, T., Wang, Y., Chen, J. Q., Araki, H., Jing, Z., Jiang, K., et al. (2004). Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol. Genet. Genomics 271, 402–415. doi: 10.1007/s00438-004-0990-z
Keywords: biotin carboxyl carrier subunit (BCCP), gene family, expression profile, Gossypium, phylogenetic analysis
Citation: Cui Y, Zhao Y, Wang Y, Liu Z, Ijaz B, Huang Y and Hua J (2017) Genome-Wide Identification and Expression Analysis of the Biotin Carboxyl Carrier Subunits of Heteromeric Acetyl-CoA Carboxylase in Gossypium. Front. Plant Sci. 8:624. doi: 10.3389/fpls.2017.00624
Received: 24 January 2017; Accepted: 06 April 2017;
Published: 01 May 2017.
Edited by:
Takeshi Fukao, Virginia Tech, USAReviewed by:
Hak Soo Seo, Seoul National University, South KoreaCopyright © 2017 Cui, Zhao, Wang, Liu, Ijaz, Huang and Hua. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jinping Hua, amlucGluZ19odWFAY2F1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.