- 1Unit of Molecular Signal Transduction in Inflammation, VIB-UGent Center for Inflammation Research, VIB, Ghent, Belgium
- 2Department of Biomedical Molecular Biology, Ghent University, Ghent, Belgium
- 3VIB-UGent Center for Plant Systems Biology, VIB, Ghent, Belgium
- 4Department of Plant Biotechnology and Bioinformatics, Ghent University, Ghent, Belgium
Abscisic acid (ABA) is a sesquiterpene signaling molecule produced in all kingdoms of life. To date, the best known functions of ABA are derived from its role as a major phytohormone in plant abiotic stress resistance. Different organisms have developed different biosynthesis and signal transduction pathways related to ABA. Despite this, there are also intriguing common themes where ABA often suppresses host immune responses and is utilized by pathogens as an effector molecule. ABA also seems to play an important role in compatible mutualistic interactions such as mycorrhiza and rhizosphere bacteria with plants, and possibly also the animal gut microbiome. The frequent use of ABA in inter-species communication could be a possible reason for the wide distribution and re-invention of ABA as a signaling molecule in different organisms. In humans and animal models, it has been shown that ABA treatment or nutrient-derived ABA is beneficial in inflammatory diseases like colitis and type 2 diabetes, which confer potential to ABA as an interesting nutraceutical or pharmacognostic drug. The anti-inflammatory activity, cellular metabolic reprogramming, and other beneficial physiological and psychological effects of ABA treatment in humans and animal models has sparked an interest in this molecule and its signaling pathway as a novel pharmacological target. In contrast to plants, however, very little is known about the ABA biosynthesis and signaling in other organisms. Genes, tools and knowledge about ABA from plant sciences and studies of phytopathogenic fungi might benefit biomedical studies on the physiological role of endogenously generated ABA in humans.
Introduction
Abscisic acid (ABA) is best known as a phytohormone regulating abiotic stress responses in plants, but ABA biosynthesis has been observed in a phylogenetically wide range of organisms (Hartung, 2010), ranging from cultured cyanobacteria (Maršálek et al., 1992) to human cells (Bruzzone et al., 2007). Although commonalities in ABA response between cells from diverse organism classes have been observed (Huddart et al., 1986), this evolutionary ancient signaling molecule shows several kingdom-specific features in both biosynthesis and signaling (Hirai et al., 2000; Hauser et al., 2011). It is not known why this particular molecule is produced by so many different types of organisms and why so many different types of organisms respond to ABA. Furthermore, ABA can exist in four different possible stereoisomeric forms, but naturally occurring ABA is predominantly of a single form [(+)cis, trans-ABA]. There are no special chemical properties that make ABA particularly useful as a signaling molecule, compared to all other potential alternative chemical structures. Despite the kingdom-specific signaling and biosynthesis, there are also commonalities between plants and animals where both plants and animals rely on intracellular free ABA for activation of their receptors (Klingler et al., 2010; Sturla et al., 2011). Intracellular ABA homeostasis is regulated by several different mechanisms: biosynthesis/catabolism, conjugation/deconjugation and export/import (Dong et al., 2015). Many of the mechanisms regulating intracellular free ABA homeostasis are however currently only known in plants. The vast majority of all studies on physiological effects of ABA to date are also focused on the role of ABA in plants. Most notably among studies of ABA in other organisms, several phytopathogenic fungal pathogens produce ABA as a virulence factor (Mbengue et al., 2016), and some mutualistic host-microbe interactions also rely on ABA (Stec et al., 2016). ABA influences immune responses in animals, and also animal pathogens can utilize this molecule as an effector molecule (Wang et al., 2009). Apart from direct effects on immunity, ABA also seems to influence metabolic control which might play a role in its protective activity against diabetes. In addition to endogenous biosynthesis, humans and other animals will also have a constant exposure of ABA from nutritional sources, and there are indications that a high ABA diet can have beneficial physiological effects (Magnone et al., 2015). One possible reason why ABA is so widely distributed among different types of organisms and why biosynthesis, perception and signaling for this molecule has been re-invented several times could be that ABA often is used as a signaling molecule for communication between different species. If that is the case, the role of ABA in host-pathogen and mutualistic interactions would be ancient and a universal theme among different organisms and organism interactions. The physiological effects of ABA in animals, especially related to immunity, inflammation and metabolic control, have sparked an interest in utilizing this pathway as a pharmacological target (Sakthivel et al., 2016), which has led to several recent papers on ABA functions in animals. However, the most famous plant hormone with pharmacological effects in humans is salicylic acid (SA). Just like with ABA, humans and other animals are constantly exposed to low doses of SA as part of their diet, which can have physiological effects (Klessig et al., 2016). Despite the well-documented effects of SA both in plants and animals, it is not until recently that several molecular targets have been identified (Manohar et al., 2014; Choi et al., 2015a,b, 2016; Dachineni et al., 2016). Earlier molecular targets of SA were often either only active at too high concentrations to be physiologically relevant, or only inhibited by acetyl-SA (i.e., Aspirin vs. COX-1 and -2). Interestingly, some of these new molecular targets are common between plants and animals (e.g., HMGB1 in humans and related plant homologs), and there are some indications of an endogenous SA biosynthesis pathway in humans (Klessig et al., 2016). This near-universal use of certain molecular structures in many different organisms and organism interactions, together with their interesting nutritional and pharmacological effects calls for a highly multidisciplinary approach in order to further elucidate the role and functions of ABA and other plant hormones in non-plant organisms. Here we will discuss what is known about the alternative “direct” biosynthesis pathway of ABA in fungi, and the role of ABA in immunity and inter-species communication. We will also explore the possibilities to utilize genes and tools from plants and phytopathogenic fungi for functional evaluation of the role of ABA in animals.
Different ABA Biosynthesis Pathways
ABA is a sesquiterpene (15 carbons) that can be found in many different kinds of organisms which produce it by different means (Hauser et al., 2011) (Figure 1). Plants synthesize their ABA via their plastids (an organelle that does not exist in fungi or animals) as a compound derived from large (40 carbons) carotenoid precursor molecules generated via the plastidial 2-C-methyl-D-erythritol 4-phosphate (MEP) pathway (Schwartz et al., 2003; Finkelstein, 2013). Rhizosphere and endophytic bacteria, such as Achromobacter, Bacillus and Pseudomonas, have all been shown to produce ABA in axenic cultures (Forchetti et al., 2007; Salomon et al., 2014). Also marine Streptomyces isolates have been shown to produce ABA and several other phytohormones without any obvious interaction with plants (Rashad et al., 2015). No ABA biosynthesis pathway is currently described in ABA-producing bacteria, but Achromobacter, Bacillus and Pseudomonas are all known to produce carotenoids, which makes it likely that also these bacteria depend on a carotenoid-dependent pathway for generation of ABA. Further underscoring this, in plants ABA synthesis from carotenoids takes place in the plastids, cell organelles thought to have originated from endosymbiotic cyanobacteria. It is however possible that different prokaryotic species utilize independently evolved carotenoid-dependent and -independent ABA biosynthesis pathways. Several phytopathogenic fungi, such as Cercospora rosicola (Assante et al., 1977), Botrytis cinerea (Hirai et al., 1986) and Magnaporthe oryzae (Spence et al., 2015) have been shown to produce ABA. In contrast to the plant carotenoid-dependent “indirect” pathway, the fungal ABA biosynthesis depends on a “direct” pathway with a 15 carbon (farnesyl diphosphate (FDP) or farnesyl pyrophosphate (FPP)) precursor molecule generated via the mevalonate (MVA) pathway (Hirai et al., 2000). FDP is a common and often rate-limiting precursor for several metabolites synthesized through the MVA pathway, including steroids, also in animals (Park et al., 2017). Several genes critical for this “direct” ABA biosynthesis have been identified in the broad host range phytopathogenic gray mold fungus B. cinerea: the P450 reductase CPR1 (Siewers et al., 2004) and a biosynthesis gene cluster (BcABA1-4) (Siewers et al., 2006). The enzyme catalyzing the first committed step in the fungal ABA biosynthesis pathway, the conversion of FDP to allofarnesene and ionylideneethane, is however not yet known (Siewers et al., 2006). Genetic and transcriptional studies of ABA over-expressing strains of B. cinerea (Ding et al., 2016) will hopefully result in the identification of genes for missing critical metabolic steps and a more complete general understanding of the direct ABA biosynthesis pathway in various phytopathogenic fungi. As a complementary strategy, defining the phylogenetic distribution of fungal ABA biosynthesis could further help to define and identify the genes responsible for critical biosynthetic steps. Other fungal pathogens, such as the rice blast pathogen M. oryzae, rely on the same biosynthesis genes as in B. cinerea (Spence et al., 2015). Furthermore, an NCBI BLASTp (Johnson et al., 2008) survey of homologs of the protein sequences from the B. cinerea ABA biosynthesis cluster easily identifies closely related proteins in many different endophytic or pathogenic fungi (e.g., Alternaria, Aspergillus, Aureobasidium, Colletotrichum, Dothistroma, Eutypa, Fusarium, Leptosphaeria, Magnaporthe, Pyrenophora, and Verticillium) and ABA has already been found in Aspergillus, Fusarium and many other kinds of saprophytic or parasitic fungi (Crocoll et al., 1991; Xu et al., 2008; Dörffling et al., 2014; Morrison et al., 2015; Uzor et al., 2017). Taken together, this might indicate that there is a conserved direct ABA biosynthesis pathway in fungi. It also indicates that ABA as a fungal virulence or compatibility strategy can be more common than currently appreciated. Fungi are part of the Opisthokont group together with animals (Steenkamp et al., 2006), which makes them closer related to animals than plants are and it is thus more likely that fungi and animals share a common ABA biosynthesis pathway. Of the four known genes in the B. cinerea ABA biosynthesis cluster, three (BcABA1, 2, and 4) have several homologs in the mammalian genomes and are thus candidate endogenous biosynthesis genes (Figure 2; Supplemental Texts 1, 2). In BLASTp surveys where fungi are excluded, animal homologs of BcABA2 are the top hits. BcABA3 homologs have a much narrower phylogenetic distribution (fungi, Amycolatopsis and Streptomyces), and this protein plays an unknown but critical biochemical role in B. cinerea ABA biosynthesis (Siewers et al., 2006). In contrast to what was previously reported (Spence et al., 2015), there is an M. oryzae BcABA3 homolog (Genbank: ELQ43177.1). Considering the long evolutionary distances between fungi and metazoa, it is possible that a conserved metabolic pathway will rely on unrelated proteins filling the same functions in animals and fungi through convergent evolution. Two of the critical ABA biosynthesis steps in Botrytis (BcABA1 and BcABA2) are represented by P450 proteins, and only CYP51 and CYP7 proteins are found in both fungi and metazoa (Nelson et al., 2013). It is however also possible that animals do not have a conserved biosynthetic pathway in common with fungi, and that they rely on carotenoid metabolites from nutritional sources to generate ABA in a manner similar to the retinoic acid biosynthesis pathway. Considering that humans lack the MEP pathway and that human cells are able to produce ABA in culture (Bruzzone et al., 2007), it is however more likely that animals rely on an endogenous “direct” (MVA) biosynthesis pathway similar to that of fungi. To distinguish between these two possibilities, it would be interesting to investigate this further in animal cells, for instance through isotope labeled metabolites, as previously done for fungi and plants (Hirai et al., 2000). Alternatively could ABA production from animal cells be investigated in presence of different types of inhibitors, such as statins and bisphosphonates, in order to determine the biosynthesis pathway.
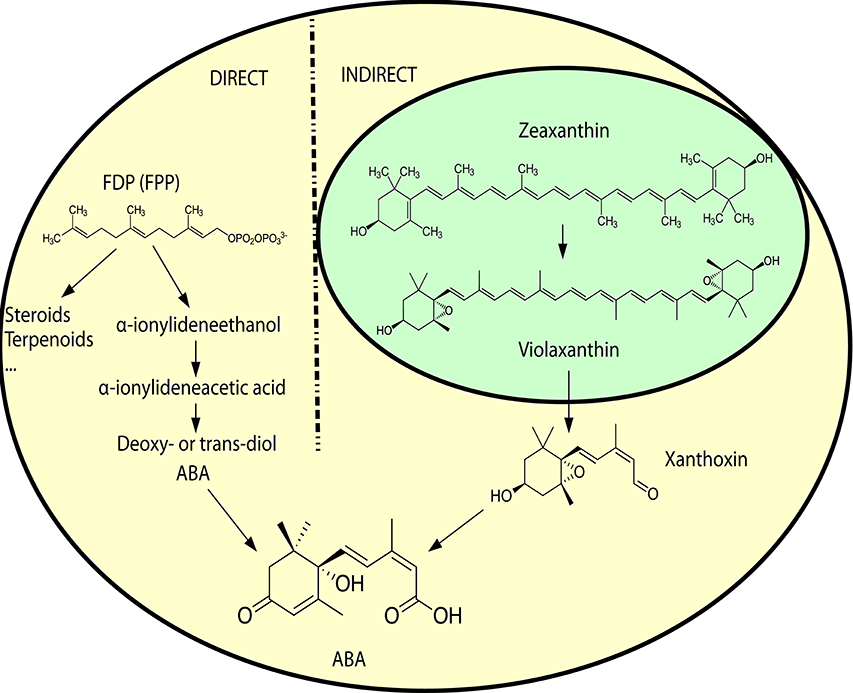
Figure 1. A simplified overview of the direct ABA biosynthesis in fungi and the indirect biosynthesis in plants. Yellow background indicates cytosolic compartment, and green background the chloroplast compartment. For more detailed biosynthetic pathways, see Siewers et al. (2006) for the direct pathway in fungi and Finkelstein (2013) for the indirect pathway in plants. Molecular structures in scalable vector graphics (SVG) format were obtained from Wikipedia (https://en.wikipedia.org) on 2017-02-08, and the images were licensed “public domain” by the original authors.
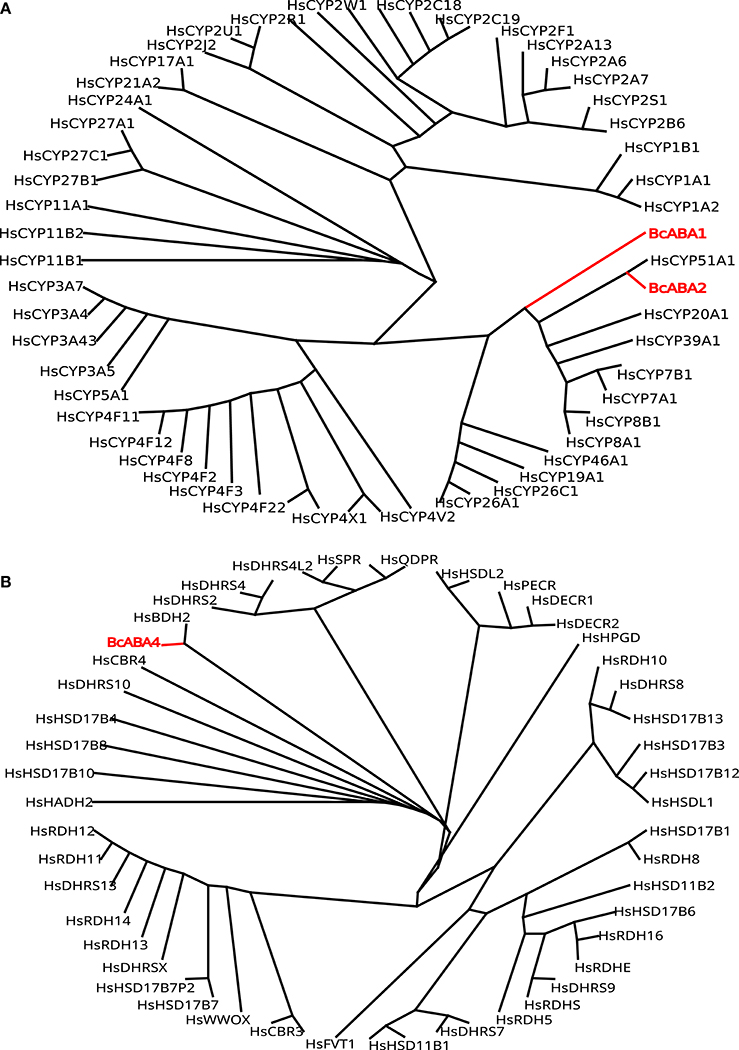
Figure 2. (A) Phylogeny of Botrytis P450 ABA biosynthesis proteins BcABA1 and BcABA2 (red) compared to all human P450 proteins (black). (B) Phylogeny of Botrytis BcABA4 biosynthesis protein (red) compared to all human carbonyl reductase family members (black).
ABA in Plant-Microbe Interactions
The effect of ABA on plant pathogen resistance is complex (Figure 3). Many pathosystems have demonstrated a negative effect from ABA on plant pathogen resistance, such as the Botrytis cinerea—tomato (Audenaert et al., 2002; Sivakumaran et al., 2016), Ralstonia solanacearum—tobacco (Zhou et al., 2008), Plectosphaerella cucumerina—Arabidopsis (Hernández-Blanco et al., 2007; Sánchez-Vallet et al., 2012), and Magnaporthe oryzae—barley (Ulferts et al., 2015) pathosystems. Emerging evidence also indicate that ABA plays an important role in compatible interactions in mutualistic plant-microbe interactions (Stec et al., 2016). For example, ABA is promoting infection and establishment of compatible interactions with arbuscular mycorrhizal fungi (Herrera-Medina et al., 2007; Charpentier et al., 2014; Fracetto et al., 2017) and several kinds of root-associated bacteria produce ABA in the rhizosphere (Salomon et al., 2014). Not only does ABA suppress immune responses, but immune responses are also able to suppress ABA signaling. For example can very early pathogen response signaling negatively regulate ABA responses (Kim et al., 2011; Desclos-Theveniau et al., 2012). In many diverse pathosystems, ABA acts antagonistically against the SA pathogen disease resistance hormone pathway (Audenaert et al., 2002; Jiang et al., 2010). Plants need to prioritize between many different environmental cues for an appropriate response, which could explain some of the antagonistic effects between different signaling pathways (Denancé et al., 2013; Vos et al., 2015; Kissoudis et al., 2016). The pathogen resistance responses can also be complex and multi-layered, where the effects of lesser responses only are apparent when the major responses have been disabled (Persson et al., 2009). Lately, complex interactions between multiple plant hormones and the traditional pathogen disease hormone [SA, jasmonic acid (JA) and ethylene (ET)] signaling pathways has been unraveled, which further highlights this inherent conflict between different plant responses (Shigenaga and Argueso, 2016; Verma et al., 2016). Also the different defense hormones play different mutually exclusive roles. In a simplified generalization, SA is involved in resistance against biotrophic pathogens and JA is involved in resistance against necrotrophic pathogens or insects. A vast majority of the studies of hormone interactions in disease resistance rely on the Arabidopsis model system, and there are indications that other plants may respond differently (De Vleesschauwer et al., 2014). As part of the antagonism between resistance against biotrophs and necrotrophs, SA suppresses JA responses at multiple levels (Caarls et al., 2015). ET, on the other hand, can enhance or influence both SA and JA responses (Broekgaarden et al., 2015). While the antagonism between ABA and SA is relatively clear in many models (de Torres Zabala et al., 2009; Moeder et al., 2010), the ABA influence on JA signaling is more complex where the JA/ABA branch regulates a different set of JA responses compared to the JA/ET branch, and these two branches are mutually exclusive (Anderson et al., 2004). The JA/ET branch responses rely on the ERF transcription factors, leading to expression of defensins and resistance against necrotrophs (Müller and Munné-Bosch, 2015). The JA/ABA branch responses, on the other hand, rely on the transcription factor MYC2, which regulates wounding responses, insect resistance and suppression of JA/ET-dependent innate immunity against necrotrophs (Kazan and Manners, 2013; Goossens et al., 2016). Interestingly, the transcription profile of genes downstream of MYC2 might be influenced directly by ABA via physical interactions with one of the intracellular ABA receptors (Aleman et al., 2016). In addition to the immune suppressive effects from ABA, it can also play a positive role in pathogen resistance. One of the first indications of a positive influence from plant ABA signaling on biotic stress resistance was the reliance of ABA for a β-aminobutyric acid (BABA)-induced priming for pathogen resistance (Ton and Mauch-Mani, 2004). Endogenous ABA was later shown to play a positive role directly in Brassica napus and Arabidopsis callose-dependent disease resistance to the hemibiotrophic fungal pathogen Leptosphaeria maculans (Kaliff et al., 2007). ABA is often a positive regulator of the callose-dependent disease resistance responses (Ton and Mauch-Mani, 2004; Flors et al., 2008; García-Andrade et al., 2011; Oide et al., 2013). Callose is a rapidly formed β-glucan barrier, which in turn also is in conflict with SA-dependent disease resistance responses (Nishimura et al., 2003; Oide et al., 2013). Also other kinds of physical barriers seem to interact with ABA and influence plant disease resistance, for example are Arabidopsis ABA-dependent resistance responses against R. solanacearum constituitively up-regulated in certain cellulose synthase mutants (Hernández-Blanco et al., 2007; Feng et al., 2012). In a more classical signaling sense, ABA has also been found to play a positive role together with JA in the resistance to Sclerotinia sclerotiorum in Arabidopsis (Perchepied et al., 2010), and the JA/ABA pathway has subsequently also been identified as important for resistance against insects (Verhage et al., 2011; Vos et al., 2013). An antagonistic action of ABA on ET-dependent infection has also been suggested as a mechanism in rice resistance against Cochliobolus miyabeanus (De Vleesschauwer et al., 2010). That ET can act as a virulence-promoting signal has been seen also in other pathosystems where ABA plays a positive role in resistance (Persson et al., 2009; Groen et al., 2013). In contrast, the ABA-ET antagonism is also important for establishment of arbuscular mycorrhiza where ET suppresses succesful colonization in ABA-deficient plants, most likely due to activation of JA/ET-dependent disease resistance responses against the invading fungus (Garrido et al., 2010; Fracetto et al., 2017). As an alternative mechanism, ABA can also positively influence disease resistance by regulating stomatal closure in order to deny pathogens entry into the plant (Lim et al., 2015). Interestingly, the JA-dependent pathway can sometimes antagonize this ABA-induced stomatal closure, which provides another example of opposing effects from ABA and other plant disease resistance hormones. Pathogens such as Pseudomonas syringae and various fungi can “hijack” the inherent antagonism between different resistance pathways in order to promote infection by using plant hormones or plant hormone-like compounds as effector molecules (Goossens et al., 2016; Toum et al., 2016).
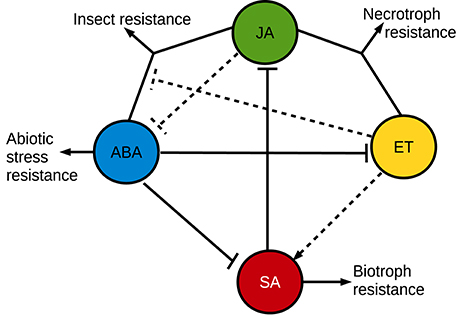
Figure 3. A simplified overview of synergistic and antagonistic interactions between plant stress resistance hormone signaling pathways. The simplified overview does not accurately describe the plant hormone involvement in all plant pathosystems, where variations may occur.
ABA as Pathogen Effector in Plant and Animal Hosts
Several phytopathogenic fungal pathogens produce ABA and other small molecule effectors in order to suppress plant immune responses (Mbengue et al., 2016), and there are indications that also human pathogens use ABA for a similar strategy (Wang et al., 2009). Some pathogens, like P. syringae, indirectly utilizes ABA as an effector molecule by modulating the endogenous ABA biosynthesis and response pathways in the host plant (de Torres-Zabala et al., 2007), which in turn indirectly modulates mutually exclusive pathogen responses, like the antagonistic relationship between SA- and callose-dependent responses (Oide et al., 2013). Other pathogens have their own ABA biosynthesis, most notably several phytopathogenic fungi such as B. cinerea. Most studies on B. cinerea ABA biosynthesis focus on its production in fermentation (Ding et al., 2015, 2016), and there are unfortunately no studies that have established the effect of altered B. cinerea ABA biosynthesis on virulence. The rice blast fungus M. oryzae, which depends on the same ABA biosynthesis pathway as B. cinerea (Spence et al., 2015), rely on ABA for virulence and respond to ABA in order to form appressoria for infection. The plastid-containing protozoan animal parasite Toxoplasma gondii also utilizes ABA as a virulence factor to suppress animal host responses by inducing autophagy (Wang et al., 2009). In contrast, the related malaria parasite Plasmodium produces SA to suppress animal host immunity, and ABA treatment seems to reduce the virulence of this pathogen (Matsubara et al., 2015; Glennon et al., 2016). Interpretations of the role of ABA in Toxoplasma virulence must however be made with caution, since Toxoplasma ABA production was inhibited using the herbicide Fluridone which also seems to affect ABA produced in mammalian cells (Magnone et al., 2013). Because of this, it would be interesting with transgenic ABA depletion studies by overexpression of an ABA catabolic enzyme in Toxoplasma, similar to how SA was depleted in Plasmodium by overexpression of a bacterial salicylate hydroxylase (Matsubara et al., 2015). Other pathogens, like the oomycete oyster parasite Perkinsus marinus, also produce ABA but it does not seem to influence its virulence (Sakamoto et al., 2016). ABA can also directly influence pathogen growth and development, such as the rice blast fungus M. oryzae (Spence et al., 2015) or the protist animal parasite T. gondii (Nagamune et al., 2008). In this context, the effect of ABA on pathogen development can fit into a much broader observation of complex hormone interactions between the host and the virulence of certain pathogens, like T. gondii (Galván-Ramírez et al., 2014). The use of ABA and other plant hormones as effector molecules or virulence factors on non-plant hosts is still a relatively unexplored area, and it is possible that more bacterial, protist, oomycete and fungal pathogens utilize this strategy.
Physiological and Pharmacological Effects of ABA in Animals
The presence of ABA in animal tissues has been known since the 1980s (Le Page-Degivry et al., 1986; Li et al., 2011), but has largely been ignored until recently. In early branching metazoans, ABA has been associated to abiotic stress responses (Zocchi et al., 2001) and nutrient-derived ABA has been suggested to promote innate immunity in honey bees (Negri et al., 2015). Administration of very high doses of ABA is well tolerated in mice without adverse effects (Li et al., 2011), which makes ABA treatments pharmacologically interesting. There are however reports that long-term ABA exposure might have adverse effects (Isik and Celik, 2015), which calls for caution. ABA treatment in humans and animal models has been suggested to be beneficial for type 2 diabetes (Guri et al., 2007; Bruzzone et al., 2015; Magnone et al., 2015), inflammatory bowel disease (IBD) (Guri et al., 2011; Viladomiu et al., 2013), atherosclerosis (Guri et al., 2010), systemic sclerosis (Bruzzone et al., 2012c), glioma (Zhou et al., 2016), depression (Qi et al., 2015b, 2016), and resistance against hepatitis C (Rakic et al., 2006), influenza (Hontecillas et al., 2013), malaria (Glennon et al., 2016), and mycobacteria (Clark et al., 2013). In addition to being administered either through nutritional sources or as a drug, there is also endogenous production of ABA in many different types of mammalian cells—such as stem cells, macrophages, microglia, granulocytes and keratinocytes (Bruzzone et al., 2007, 2012b; Scarfì et al., 2008; Bodrato et al., 2009; Magnone et al., 2009, 2012). Furthermore, endogenous induction of serum ABA in response to glucose challenge has been shown to stimulate insulin release and is impaired in patients suffering from type 2 diabetes (Bruzzone et al., 2008, 2012a; Ameri et al., 2015). ABA has recently attracted the attention of several research groups as a potential drug lead (Bassaganya-Riera et al., 2010; Sakthivel et al., 2016) because of these promising biological activities. One study has already shown that fruits and vegetables with high ABA contents (such as figs and apricots) are beneficial against hyperglycemia in both rats and humans (Magnone et al., 2015), which makes ABA a potential pharmacognostic drug or nutraceutical. Pharmaceutical research on ABA currently focuses on generation of ABA analogs that will be more stable or act as ABA antagonists (Grozio et al., 2011; Bellotti et al., 2015). Similar approaches are currently also pursued in the plant sciences where antagonistic ABA analogs have been designed (Takeuchi et al., 2014), and pyrabactin variants are utilized as more stable and chemically unrelated ABA agonists (Overtveldt et al., 2015). An alternative treatment approach is to manipulate ABA homeostasis indirectly by targeting the different mechanisms that influence the levels of free intracellular ABA (Todoroki and Ueno, 2010). A conceptually similar approach has already been tried in animal systems by treatments with a herbicide (Fluridone), which is known to influence ABA levels in plants (Magnone et al., 2013). At this moment, there are no reports describing an inhibition of the “direct” fungal ABA biosynthesis by Fluridone. How Fluoridone would influence ABA in mammalian cells is also unclear since the molecular target is the carotenoid biosynthesis protein phytoene desaturase (Chamovitz et al., 1993), which is absent in animals. More knowledge of the ABA homeostasis in humans and the molecular target of the herbicide is thus needed before it can be applied as a true drug strategy. There are however conflicting reports whether ABA plays a pro- or anti-inflammatory role in specific cell models (De Flora et al., 2014), but the over all pharmacological properties are anti-inflammatory.
The best studied proposed receptor for ABA in animals is the intracellular protein LANCL2, which is attached to the cell membrane by myristoylation (Sturla et al., 2009; Fresia et al., 2016). LANCL2 is homologous to the earlier proposed ABA receptor in plants: GCR2 (Liu et al., 2007; Chen and Ellis, 2008) and bacterial LanC lanthionine synthesis proteins. However, the mammalian LanC-like homologs do not show any evidence of having a conserved role in lanthionine biosynthesis (He et al., 2017). It is not yet known whether a currently unknown catalytic activity of LANCL2 could contribute to ABA signaling. GCR2 was proposed as a G-protein coupled receptor (GPCR) for ABA, but this was later challenged (Gao et al., 2007; Chen and Ellis, 2008) and an entirely different protein family (PYR/PYL/RCAR) of intracellular receptors is now thought to be the dominant ABA receptors in plants (Ma et al., 2009; Miyazono et al., 2009; Park et al., 2009). Signaling of ABA via the PYR/PYL/RCAR intracellular receptor family and its associated PP2C phosphatases is however a land plant-specific innovation with no animal homologs (Hauser et al., 2011). Despite the apparent low importance of GCR2 in plant ABA responses, the human LANCL2 seem to work as expected from an ABA receptor, since independent drugs targeting this protein result in effects similar to those seen by ABA treatment (Bissel et al., 2016). ABA is suggested to act through a LANCL2-PPARγ axis for its anti-inflammatory functions (Guri et al., 2008; Sturla et al., 2009, 2011) and through a GPCR—like signaling for pro-inflammatory responses (Bruzzone et al., 2007, 2012b; Sturla et al., 2009; Fresia et al., 2016). Interestingly, ABA worsened the inflammation in models of IBD when PPARγ was absent from T cells (Guri et al., 2011; Viladomiu et al., 2013), indicating that ABA indeed plays a dual role and acts both pro- and anti-inflammatory. Apart from its inflammation-modulating functions, ABA signals via LANCL2 have also been linked to metabolic reprogramming of adipocytes into brown fat cells (Sturla et al., 2017), which can be especially beneficial in the context of diabetes where a dual direct effect on immune cells and metabolism could contribute to reduced inflammation (Ray et al., 2016). One possible mechanism for the metabolic reprogramming is an effect of LANCL2 on the Akt/mTORC2 pathway, where LANCL2 influences insulin-dependent Akt phosphorylation (Zeng et al., 2014). The metabolic reprogramming and inflammation-modulating effects of ABA can however also be two sides of the same coin, since there is significant cross talk between inflammatory signaling pathways and metabolism in innate immune cells (Kelly and O'Neill, 2015). An increased oxidative metabolism through ABA signaling should promote the alternatively activated and often anti-inflammatory macrophages. Inspired by the protective effects of ABA, LANCL2 has been proposed as a drug target, especially for treatments of IBD (Lu et al., 2011, 2014; Basson et al., 2016). Apart from the proposed GPCR-like signaling and the PPARγ mediated signaling, LANCL2 has some interesting potential direct signaling effects. For example, binding of ABA causes nuclear translocation of LANCL2 (Fresia et al., 2016), and nuclear LANCL2 has already been shown to influence transcription factor activity (Park and James, 2003). On the other hand are there no spontaneous phenotypes reported for LANCL2 deficient mice (Leber et al., 2016). The lack of spontaneous phenotype in LancL2 knock-out mice is also not due to redundancy with the other mammalian LanC-like proteins, since the LancL1-3 triple knock-out mice still are viable and do not show any gross abnormalities (He et al., 2017). The downstream PPARγ is known to be required for alternative (M2) activation of macrophages (Odegaard et al., 2007). Macrophages can be activated (polarized) in two different ways: the classically activated pro-inflammatory M1 macrophages, and different kinds of alternatively activated M2 macrophages that often act anti-inflammatory and are important for tissue repair (Mantovani et al., 2002). The PPARγ–mediated M2 polarization could explain the anti-inflammatory role of ABA. Consistent with the proposed LANCL2-PPARγ ABA signaling pathway, LancL2 full-body or myeloid-specific knock-out mice show an impaired development of regulatory anti-inflammatory macrophages in response to Helicobacter pylori infection, which results in a stronger immune response but also greater tissue damage (Leber et al., 2016). It will be important to evaluate these LancL2 knock-out mice or the triple knock-out mice for ABA responses in models that have already shown clear effects from ABA—including models where other ABA receptors have been proposed. One such alternative pathway suggests that ABA acts as an agonist to the retinoic acid nuclear receptors (Zhou et al., 2016). ABA stimulation through cross-reactivity with the retinoic acid receptors could also cause anti-inflammatory effects, since retinoic acid has been shown to be protective in inflammatory models of rheumatoid arthritis (Kwok et al., 2012). Surveys of other ABA-binding human proteins have also identified HSP70 family members as ABA-binding (Kharenko et al., 2013), and ABA-mediated megakarocyte survival has been suggested to rely on both LANCL2 and HSP70 (GRP78) interactions with ABA (Malara et al., 2017). The possible role of HSP70 family members in ABA signaling is another intriguing parallel with plant ABA signaling, where overexpression of HSC70 caused an ABA-hypersensitive phenotype in Arabidopsis (Clément et al., 2011). ABA seems to bind many potential receptors (Figure 4) and seems to perform several physiological functions (Scarfì et al., 2009; Wang et al., 2009; Kharenko et al., 2013; Pydi et al., 2015; Qi et al., 2015a). Whether LANCL2 is the major ABA receptor in humans is thus still not entirely certain.
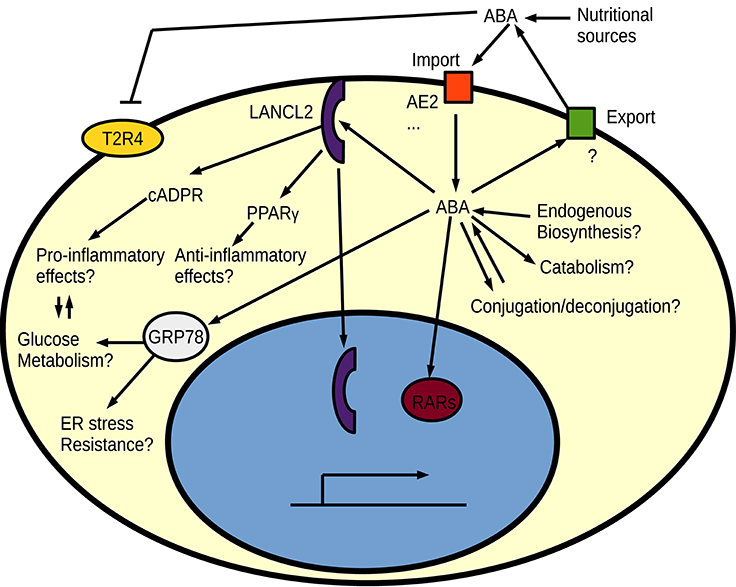
Figure 4. An overview of proposed signaling pathways for ABA in animal cells. Regulation of intracellular free ABA homeostasis by biosynthesis/catabolism, export/import and conjugation/deconjugation are currently unknown in animals. Different ABA receptors have been proposed for ABA, such as the bitter taste receptor (T2R4), LANCL2, retinoic acid receptors (RARs), and HSP70 (GRP78).
An intriguing indirect alternative effect of ABA is its role in inhibiting the bitter taste receptor, which is known to influence the gut microbiome and glucose responses (Dotson et al., 2008; Pydi et al., 2015; Latorre et al., 2016). This could explain many physiological effects of ABA, especially its protective effects in colitis and diabetes, and possibly also some psychological effects via the gut-brain axis. Gut bitter taste receptors would also be located at a primary site for nutrient-derived ABA, but it can also mean that microbe-generated ABA is an important signal between the gut microbiome and the host since gut microbes produce many complex metabolites (Lee and Hase, 2014; Palau-Rodriguez et al., 2015). Cultures of bacteria found in the animal gut (Escherichia coli, Klebsiella pneumonia and Proteus mirabilis) can produce low levels of ABA and other plant hormones (Karadeniz et al., 2006), but there are currently no reports on whether ABA or other plant hormones are present in the endogenous gut microbiome metabolome and if levels and presence of these compounds correlate with different host physiological states.
Being an endogenously generated anti-inflammatory secondary metabolite puts ABA in the same category as the glucocorticoids and other anti-inflammatory steroids, but with the important difference that we know very little about the function and regulation of ABA in humans. It is possible that the mammalian ABA receptor(s) are equally elusive as they used to be in plants prior to the discovery of the PYR/PYL/RCAR receptor family, with several ABA-binding proteins being proposed as receptor candidates (Klingler et al., 2010). It is also possible that many molecular targets of ABA will hit a gray zone where the question whether they should be considered receptors turns into a matter of semantics, similar to the current situation with SA (Klessig et al., 2016). It is however likely that there will be analogies between plant and animal ABA receptors. Intracellular receptors are for example likely, and since ABA cannot pass the cell membrane at physiological pH (Kuromori and Shinozaki, 2010), active transport is needed for signaling. Recently, two different ABA importers were identified in mammals (Vigliarolo et al., 2015, 2016), and this family of importers will most likely play an important role in regulating the ABA responsiveness of different cells. There could also be different receptor- and signal systems to respond to intracellular and extracellular ABA, which perhaps could explain some discrepancies between results suggesting an anti-inflammatory role of ABA and results where depletion of extracellular ABA with antibodies resulted in reduced activation of macrophages (Magnone et al., 2012). It is however also possible that ABA, just like in plants, plays a complex role in animal immunity where some immune responses are promoted whereas others are inhibited.
Perspectives: How Plant Science and Studies of Phytopathogenic Fungi can Help Biomedical Science
The near universal presence of ABA and its intriguing physiological effects in different organisms calls for a highly multidisciplinary approach to further understand its endogenous functions in the different organisms as well as its pharmacological potential (Bohlin et al., 2010). Similar approaches might also be fruitful for studies of other signaling molecules from plants and phytopathogenic fungi that have been found to show pharmacological effects in animals, like for example gibberellins (Hedden and Thomas, 2012; Annand et al., 2015; Bannon et al., 2015; Reihill et al., 2016) and SA (Klessig et al., 2016). Since no genes are known that regulate intracellular ABA homeostasis in animals, utilizing proteins and pathways known in plants and phytopathogenic fungi could help defining the physiological role of ABA in animals. The plant P450 (CYP707A) enzymes that degrades ABA to phaseic acid (Umezawa et al., 2006) could be used to deplete endogenous ABA (or intracellular ABA from other sources) in transgenic animals or animal cells. CYP707As are active in presence of animal P450 reductases, but expression of the Arabidopsis P450 reductase ATR1 (At4g24520) results in 70% higher activity (Saito et al., 2004). A transgenic approach comparing at least 2 different inactivation pathways is also important, since metabolic engineering carries a risk of phenotypic artifacts from the degradation products (van Wees and Glazebrook, 2003). Recently, the CYP707A ABA degradation product phaseic acid was found to be active in both mammalian and plant cells, which could cause such phenotypic artifacts (Hou et al., 2016; Rodriguez, 2016; Weng et al., 2016). This means that future ABA depletion studies relying on CYP707A activity should most likely also express the phaseic acid reductase (PAR, Arabidopsis ABH2: At4g27250) to avoid one potential side effect. One alternative ABA inactivation pathway is a UDP-dependent glycosyl transferase (UGT71C5, Arabidopsis: At1g07240), which inactivates free ABA by conjugating it to glucose (Liu et al., 2015). However, overexpression of this enzyme in plants gives a much lower level of ABA insensitivity compared to CYP707A overexpressing plants. Other alternative ABA inactivation strategies are overexpression of intracellular small single chain antibody (scFv) against ABA (Genbank: Z29480.1) (Artsaenko et al., 1995) or overexpression of an ABA exporter (ABCG25, Arabidopsis: At1g71960) protein (Park et al., 2016). The general concept of translating tools from plant sciences to animal models could also be applicable for other signaling molecules. For example, since there are indications that there is an endogenous SA biosynthesis pathway and basal SA levels from nutrition in animals (Paterson et al., 2008; Klessig et al., 2016), a bacterial salicylate hydroxylase nahG (NCBI: NC_007926.1) transgenic (Gaffney et al., 1993; Matsubara et al., 2015) animal model to deplete endogenous SA (and intracellular SA from other sources) could give interesting insights on the physiological role of the basal levels of SA in animals. As with the ABA depletions, keeping in mind that also the SA degradation product catechol can have biological functions (van Wees and Glazebrook, 2003). Because of this, an alternative inactivation mechanism, like for example overexpression of SA carboxyl methyltransferase (AtBSMT1: At3g11480) (Liu et al., 2009), would be advisable as a complementary approach. Apart from ABA depletion by using plant proteins, it might also be possible to enhance ABA production in animal cells using genes from phytopathogenic fungi if the biosynthetic pathway is conserved (Ding et al., 2015). Plant science has also provided us with several methods to measure ABA. ELISA assays for ABA originally developed for plant science have already been used in many studies of ABA produced in animals. Unfortunately, the commercial kits are quite expensive which might act as a barrier to entry for many biomedical research groups that are not specifically working on ABA. The recently described production of recombinant small single chain (scFv) anti-ABA antibodies (Badescu et al., 2016) and the recent development of anti-ABA aptamers (Grozio et al., 2013) might however significantly simplify the generation of in-house ELISAs or lower the prices of commercial kits in the near future, which would make it a viable standard assay for clinical samples. Another very exciting novel development is the ability to measure intracellular free ABA concentrations in real-time using ABA FRET biosensors (Jones et al., 2014; Waadt et al., 2014). Expression of such biosensors in mammalian cells might help us to understand under which signaling conditions free intracellular ABA is produced in the cell, and might help us to more accurately investigate the regulation and dynamics of free intracellular ABA in mammalian cells.
Author Contributions
JS came up with the original ideas which form the foundation of this manuscript and wrote the manuscript. LL performed preliminary experiments in mammalian cells under guidance of JS and RB. JP, RB, and AG were involved in further planning, discussions and comments on the manuscript.
Funding
Work in the Beyaert lab is financed by the Fund for Scientific Research Flanders (FWO), the Belgian Foundation Against Cancer, Interuniversity Attraction Poles, Concerted Research Actions (GOA) and the Group-ID Multidisciplinary Research Partnership of Ghent University.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00587/full#supplementary-material
Supplemental Text 1. FASTA sequences of Botrytis P450 BcABA1 and BcABA2 and all human P450 family member proteins.
Supplemental Text 2. FASTA sequences of Botrytis BcABA4 and all human carbonyl reductase family member proteins.
References
Aleman, F., Yazaki, J., Lee, M., Takahashi, Y., Kim, A. Y., Li, Z., et al. (2016). An ABA-increased interaction of the PYL6 ABA receptor with MYC2 Transcription Factor: a putative link of ABA and JA signaling. Sci. Rep. 6:28941. doi: 10.1038/srep28941
Ameri, P., Bruzzone, S., Mannino, E., Sociali, G., Andraghetti, G., Salis, A., et al. (2015). Impaired increase of plasma abscisic Acid in response to oral glucose load in type 2 diabetes and in gestational diabetes. PLoS ONE 10:e0115992. doi: 10.1371/journal.pone.0115992
Anderson, J. P., Badruzsaufari, E., Schenk, P. M., Manners, J. M., Desmond, O. J., Ehlert, C., et al. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16, 3460–3479. doi: 10.1105/tpc.104.025833
Annand, J. R., Bruno, P. A., Mapp, A. K., and Schindler, C. S. (2015). Synthesis and biological evaluation of pharbinilic acid and derivatives as NF-κB pathway inhibitors. Chem. Commun. Camb. Engl. 51, 8990–8993. doi: 10.1039/C5CC02918J
Artsaenko, O., Peisker, M., zur Nieden, U., Fiedler, U., Weiler, E. W., Müntz, K., et al. (1995). Expression of a single-chain Fv antibody against abscisic acid creates a wilty phenotype in transgenic tobacco. Plant J. Cell Mol. Biol. 8, 745–750. doi: 10.1046/j.1365-313X.1995.08050745.x
Assante, G., Merlini, L., and Nasini, G. (1977). (+)-Abscisic acid, a metabolite of the fungus Cercospora rosicola. Experientia 33, 1556–1557. doi: 10.1007/BF01933993
Audenaert, K., De Meyer, G. B., and Höfte, M. M. (2002). Abscisic acid determines basal susceptibility of tomato to Botrytis cinerea and suppresses salicylic acid-dependent signaling mechanisms. Plant Physiol. 128, 491–501. doi: 10.1104/pp.010605
Badescu, G. O., Marsh, A., Smith, T. R., Thompson, A. J., and Napier, R. M. (2016). Kinetic characterisation of a single chain antibody against the hormone abscisic acid: comparison with its parental monoclonal. PLoS ONE 11:e0152148. doi: 10.1371/journal.pone.0152148
Bannon, A., Zhang, S. D., Schock, B. C., and Ennis, M. (2015). Cystic fibrosis from laboratory to bedside: the role of A20 in NF-κB-mediated inflammation. Med. Princ. Pract. Int. J. Kuwait Univ. Health Sci. Cent. 24, 301–310. doi: 10.1159/000381423
Bassaganya-Riera, J., Skoneczka, J., Kingston, D. G., Krishnan, A., Misyak, S. A., Guri, A. J., et al. (2010). Mechanisms of action and medicinal applications of abscisic acid. Curr. Med. Chem. 17, 467–478. doi: 10.2174/092986710790226110
Basson, A., Trotter, A., Rodriguez-Palacios, A., and Cominelli, F. (2016). Mucosal interactions between genetics, diet, and microbiome in inflammatory bowel disease. Front. Immunol. 7:290. doi: 10.3389/fimmu.2016.00290
Bellotti, M., Salis, A., Grozio, A., Damonte, G., Vigliarolo, T., Galatini, A., et al. (2015). Synthesis, structural characterization and effect on human granulocyte intracellular cAMP levels of abscisic acid analogs. Bioorg. Med. Chem. 23, 22–32. doi: 10.1016/j.bmc.2014.11.035
Bissel, P., Boes, K., Hinckley, J., Jortner, B. S., Magnin-Bissel, G., Werre, S. R., et al. (2016). Exploratory studies with BT-11: a proposed orally active therapeutic for Crohn's disease. Int. J. Toxicol. 35, 521–529. doi: 10.1177/1091581816646356
Bodrato, N., Franco, L., Fresia, C., Guida, L., Usai, C., Salis, A., et al. (2009). Abscisic acid activates the murine microglial cell line N9 through the second messenger cyclic ADP-ribose. J. Biol. Chem. 284, 14777–14787. doi: 10.1074/jbc.M802604200
Bohlin, L., Göransson, U., Alsmark, C., Wedén, C., and Backlund, A. (2010). Natural products in modern life science. Phytochem. Rev. Proc. Phytochem. Soc. Eur. 9, 279–301. doi: 10.1007/s11101-009-9160-6
Broekgaarden, C., Caarls, L., Vos, I. A., Pieterse, C. M., and Van Wees, S. C. (2015). Ethylene: traffic controller on hormonal crossroads to defense. Plant Physiol. 169, 2371–2379. doi: 10.1104/pp.15.01020
Bruzzone, S., Ameri, P., Briatore, L., Mannino, E., Basile, G., Andraghetti, G., et al. (2012a). The plant hormone abscisic acid increases in human plasma after hyperglycemia and stimulates glucose consumption by adipocytes and myoblasts. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 26, 1251–1260. doi: 10.1096/fj.11-190140
Bruzzone, S., Basile, G., Mannino, E., Sturla, L., Magnone, M., Grozio, A., et al. (2012b). Autocrine abscisic acid mediates the UV-B-induced inflammatory response in human granulocytes and keratinocytes. J. Cell. Physiol. 227, 2502–2510. doi: 10.1002/jcp.22987
Bruzzone, S., Battaglia, F., Mannino, E., Parodi, A., Fruscione, F., Basile, G., et al. (2012c). Abscisic acid ameliorates the systemic sclerosis fibroblast phenotype in vitro. Biochem. Biophys. Res. Commun. 422, 70–74. doi: 10.1016/j.bbrc.2012.04.107
Bruzzone, S., Bodrato, N., Usai, C., Guida, L., Moreschi, I., Nano, R., et al. (2008). Abscisic acid is an endogenous stimulator of insulin release from human pancreatic islets with cyclic ADP ribose as second messenger. J. Biol. Chem. 283, 32188–32197. doi: 10.1074/jbc.M802603200
Bruzzone, S., Magnone, M., Mannino, E., Sociali, G., Sturla, L., Fresia, C., et al. (2015). Abscisic acid stimulates glucagon-like peptide-1 secretion from l-cells and its oral administration increases plasma glucagon-like peptide-1 levels in rats. PLoS ONE 10:e0140588. doi: 10.1371/journal.pone.0140588
Bruzzone, S., Moreschi, I., Usai, C., Guida, L., Damonte, G., Salis, A., et al. (2007). Abscisic acid is an endogenous cytokine in human granulocytes with cyclic ADP-ribose as second messenger. Proc. Natl. Acad. Sci. U.S.A. 104, 5759–5764. doi: 10.1073/pnas.0609379104
Caarls, L., Pieterse, C. M., and Van Wees, S. C. (2015). How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 6:170. doi: 10.3389/fpls.2015.00170
Chamovitz, D., Sandmann, G., and Hirschberg, J. (1993). Molecular and biochemical characterization of herbicide-resistant mutants of cyanobacteria reveals that phytoene desaturation is a rate-limiting step in carotenoid biosynthesis. J. Biol. Chem. 268, 17348–17353.
Charpentier, M., Sun, J., Wen, J., Mysore, K. S., and Oldroyd, G. E. (2014). Abscisic acid promotion of arbuscular mycorrhizal colonization requires a component of the PROTEIN PHOSPHATASE 2A complex. Plant Physiol. 166, 2077–2090. doi: 10.1104/pp.114.246371
Chen, J. G., and Ellis, B. E. (2008). GCR2 is a new member of the eukaryotic lanthionine synthetase component C-like protein family. Plant Signal. Behav. 3, 307–310. doi: 10.4161/psb.3.5.5292
Choi, H. W., Manohar, M., Manosalva, P., Tian, M., Moreau, M., and Klessig, D. F. (2016). Activation of plant innate immunity by extracellular high mobility group box 3 and its inhibition by salicylic acid. PLoS Pathog. 12:e1005518. doi: 10.1371/journal.ppat.1005518
Choi, H. W., Tian, M., Manohar, M., Harraz, M. M., Park, S. W., Schroeder, F. C., et al. (2015a). Human GAPDH Is a target of aspirin's primary metabolite salicylic acid and its derivatives. PLoS ONE 10:e0143447. doi: 10.1371/journal.pone.0143447
Choi, H. W., Tian, M., Song, F., Venereau, E., Preti, A., Park, S. W., et al. (2015b). Aspirin's active metabolite salicylic acid targets high mobility group box 1 to modulate inflammatory responses. Mol. Med. Camb. Mass 21, 526–535. doi: 10.2119/molmed.2015.00148
Clark, T. N., Ellsworth, K., Li, H., Johnson, J. A., and Gray, C. A. (2013). Isolation of the plant hormone (+)-abscisic acid as an antimycobacterial constituent of the medicinal plant endophyte Nigrospora sp. Nat. Prod. Commun. 8, 1673–1674. Available online at: www.naturalproduct.us
Clément, M., Leonhardt, N., Droillard, M. J., Reiter, I., Montillet, J. L., Genty, B., et al. (2011). The cytosolic/nuclear HSC70 and HSP90 molecular chaperones are important for stomatal closure and modulate abscisic acid-dependent physiological responses in Arabidopsis. Plant Physiol. 156, 1481–1492. doi: 10.1104/pp.111.174425
Crocoll, C., Kettner, J., and Dörffling, K. (1991). Abscisic acid in saprophytic and parasitic species of fungi. Phytochemistry 30, 1059–1060. doi: 10.1016/S0031-9422(00)95171-9
Dachineni, R., Ai, G., Kumar, D. R., Sadhu, S. S., Tummala, H., and Bhat, G. J. (2016). Cyclin A2 and CDK2 as novel targets of aspirin and salicylic acid: a potential role in cancer prevention. Mol. Cancer Res. 14, 241–252. doi: 10.1158/1541-7786.MCR-15-0360
De Flora, A., Bruzzone, S., Guida, L., Sturla, L., Magnone, M., Fresia, C., et al. (2014). Toward a medicine-oriented use of the human hormone/nutritional supplement abscisic acid. Messenger 3, 86–97. doi: 10.1166/msr.2014.1029
Denancé, N., Sánchez-Vallet, A., Goffner, D., and Molina, A. (2013). Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4:155. doi: 10.3389/fpls.2013.00155
Desclos-Theveniau, M., Arnaud, D., Huang, T. Y., Lin, G. J., Chen, W. Y., Lin, Y. C., et al. (2012). The Arabidopsis lectin receptor kinase LecRK-V.5 represses stomatal immunity induced by Pseudomonas syringae pv. tomato DC3000. PLoS Pathog. 8:e1002513. doi: 10.1371/journal.ppat.1002513
de Torres Zabala, M., Bennett, M. H., Truman, W. H., and Grant, M. R. (2009). Antagonism between salicylic and abscisic acid reflects early host-pathogen conflict and moulds plant defence responses. Plant J. Cell Mol. Biol. 59, 375–386. doi: 10.1111/j.1365-313X.2009.03875.x
de Torres-Zabala, M., Truman, W., Bennett, M. H., Lafforgue, G., Mansfield, J. W., Rodriguez Egea, P., et al. (2007). Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 26, 1434–1443. doi: 10.1038/sj.emboj.7601575
De Vleesschauwer, D., Xu, J., and Höfte, M. (2014). Making sense of hormone-mediated defense networking: from rice to Arabidopsis. Front. Plant Sci. 5:611. doi: 10.3389/fpls.2014.00611
De Vleesschauwer, D., Yang, Y., Cruz, C. V., and Höfte, M. (2010). Abscisic acid-induced resistance against the brown spot pathogen Cochliobolus miyabeanus in rice involves MAP kinase-mediated repression of ethylene signaling. Plant Physiol. 152, 2036–2052. doi: 10.1104/pp.109.152702
Ding, Z. T., Zhang, Z., Luo, D., Zhou, J. Y., Zhong, J., Yang, J., et al. (2015). Gene overexpression and RNA silencing tools for the genetic manipulation of the S-(+)-abscisic acid producing ascomycete Botrytis cinerea. Int. J. Mol. Sci. 16, 10301–10323. doi: 10.3390/ijms160510301
Ding, Z., Zhang, Z., Zhong, J., Luo, D., Zhou, J., Yang, J., et al. (2016). Comparative transcriptome analysis between an evolved abscisic acid-overproducing mutant Botrytis cinerea TBC-A and its ancestral strain Botrytis cinerea TBC-6. Sci. Rep. 6:37487. doi: 10.1038/srep37487
Dong, T., Park, Y., and Hwang, I. (2015). Abscisic acid: biosynthesis, inactivation, homoeostasis and signalling. Essays Biochem. 58, 29–48. doi: 10.1042/bse0580029
Dörffling, K., Petersen, W., Sprecher, E., Urbasch, I., and Hanssen, H.-P. (2014). Abscisic acid in phytopathogenic fungi of the genera botrytis, ceratocystis, fusarium, and rhizoctonia. Z. Für Naturforschung C 39, 683–684. doi: 10.1515/znc-1984-0626
Dotson, C. D., Zhang, L., Xu, H., Shin, Y. K., Vigues, S., Ott, S. H., et al. (2008). Bitter taste receptors influence glucose homeostasis. PLoS ONE 3:e3974. doi: 10.1371/journal.pone.0003974
Feng, D. X., Tasset, C., Hanemian, M., Barlet, X., Hu, J., Trémousaygue, D., et al. (2012). Biological control of bacterial wilt in Arabidopsis thaliana involves abscissic acid signalling. New Phytol. 194, 1035–1045. doi: 10.1111/j.1469-8137.2012.04113.x
Finkelstein, R. (2013). Abscisic acid synthesis and response. Arab. Book 11:e0166. doi: 10.1199/tab.0166
Flors, V., Ton, J., van Doorn, R., Jakab, G., García-Agustín, P., and Mauch-Mani, B. (2008). Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J. Cell Mol. Biol. 54, 81–92. doi: 10.1111/j.1365-313X.2007.03397.x
Forchetti, G., Masciarelli, O., Alemano, S., Alvarez, D., and Abdala, G. (2007). Endophytic bacteria in sunflower (Helianthus annuus L.): isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl. Microbiol. Biotechnol. 76, 1145–1152. doi: 10.1007/s00253-007-1077-7
Fracetto, G. G., Peres, L. E., and Lambais, M. R. (2017). Gene expression analyses in tomato near isogenic lines provide evidence for ethylene and abscisic acid biosynthesis fine-tuning during arbuscular mycorrhiza development. Arch. Microbiol. doi: 10.1007/s00203-017-1354-5. [Epub ahead of print].
Fresia, C., Vigliarolo, T., Guida, L., Booz, V., Bruzzone, S., Sturla, L., et al. (2016). G-protein coupling and nuclear translocation of the human abscisic acid receptor LANCL2. Sci. Rep. 6:26658. doi: 10.1038/srep26658
Gaffney, T., Friedrich, L., Vernooij, B., Negrotto, D., Nye, G., Uknes, S., et al. (1993). Requirement of salicylic Acid for the induction of systemic acquired resistance. Science 261, 754–756. doi: 10.1126/science.261.5122.754
Galván-Ramírez Mde, L., Gutiérrez-Maldonado, A. F., Verduzco-Grijalva, F., and Jiménez, J. M. (2014). The role of hormones on Toxoplasma gondii infection: a systematic review. Front. Microbiol. 5:503. doi: 10.3389/fmicb.2014.00503
Gao, Y., Zeng, Q., Guo, J., Cheng, J., Ellis, B. E., and Chen, J. G. (2007). Genetic characterization reveals no role for the reported ABA receptor, GCR2, in ABA control of seed germination and early seedling development in Arabidopsis. Plant J. Cell Mol. Biol. 52, 1001–1013. doi: 10.1111/j.1365-313X.2007.03291.x
García-Andrade, J., Ramírez, V., Flors, V., and Vera, P. (2011). Arabidopsis ocp3 mutant reveals a mechanism linking ABA and JA to pathogen-induced callose deposition. Plant J. Cell Mol. Biol. 67, 783–794. doi: 10.1111/j.1365-313X.2011.04633.x
Garrido, J. M., Morcillo, R. J., Rodríguez, J. A., and Bote, J. A. (2010). Variations in the mycorrhization characteristics in roots of wild-type and ABA-deficient tomato are accompanied by specific transcriptomic alterations. Mol. Plant Microbe Interact. 23, 651–664. doi: 10.1094/MPMI-23-5-0651
Glennon, E. K. K., Adams, L. G., Hicks, D. R., Dehesh, K., and Luckhart, S. (2016). Supplementation with abscisic acid reduces malaria disease severity and parasite transmission. Am. J. Trop. Med. Hyg. 94, 1266–1275. doi: 10.4269/ajtmh.15-0904
Goossens, J., Fernández-Calvo, P., Schweizer, F., and Goossens, A. (2016). Jasmonates: signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 91, 673–689. doi: 10.1007/s11103-016-0480-9
Groen, S. C., Whiteman, N. K., Bahrami, A. K., Wilczek, A. M., Cui, J., Russell, J. A., et al. (2013). Pathogen-triggered ethylene signaling mediates systemic-induced susceptibility to herbivory in Arabidopsis. Plant Cell 25, 4755–4766. doi: 10.1105/tpc.113.113415
Grozio, A., Gonzalez, V. M., Millo, E., Sturla, L., Vigliarolo, T., Bagnasco, L., et al. (2013). Selection and characterization of single stranded DNA aptamers for the hormone abscisic Acid. Nucleic Acid Ther. 23, 322–331. doi: 10.1089/nat.2013.0418
Grozio, A., Millo, E., Guida, L., Vigliarolo, T., Bellotti, M., Salis, A., et al. (2011). Functional characterization of a synthetic abscisic acid analog with anti-inflammatory activity on human granulocytes and monocytes. Biochem. Biophys. Res. Commun. 415, 696–701. doi: 10.1016/j.bbrc.2011.10.143
Guri, A. J., Evans, N. P., Hontecillas, R., and Bassaganya-Riera, J. (2011). T cell PPARγ is required for the anti-inflammatory efficacy of abscisic acid against experimental IBD. J. Nutr. Biochem. 22, 812–819. doi: 10.1016/j.jnutbio.2010.06.011
Guri, A. J., Hontecillas, R., Ferrer, G., Casagran, O., Wankhade, U., Noble, A. M., et al. (2008). Loss of PPAR gamma in immune cells impairs the ability of abscisic acid to improve insulin sensitivity by suppressing monocyte chemoattractant protein-1 expression and macrophage infiltration into white adipose tissue. J. Nutr. Biochem. 19, 216–228. doi: 10.1016/j.jnutbio.2007.02.010
Guri, A. J., Hontecillas, R., Si, H., Liu, D., and Bassaganya-Riera, J. (2007). Dietary abscisic acid ameliorates glucose tolerance and obesity-related inflammation in db/db mice fed high-fat diets. Clin. Nutr. Edinb. Scotl. 26, 107–116. doi: 10.1016/j.clnu.2006.07.008
Guri, A. J., Misyak, S. A., Hontecillas, R., Hasty, A., Liu, D., Si, H., et al. (2010). Abscisic acid ameliorates atherosclerosis by suppressing macrophage and CD4+ T cell recruitment into the aortic wall. J. Nutr. Biochem. 21, 1178–1185. doi: 10.1016/j.jnutbio.2009.10.003
Hartung, W. (2010). The evolution of abscisic acid (ABA) and ABA function in lower plants, fungi and lichen. Funct. Plant Biol. 37, 806–812. doi: 10.1071/FP10058
Hauser, F., Waadt, R., and Schroeder, J. I. (2011). Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. CB 21, R346–R355. doi: 10.1016/j.cub.2011.03.015
He, C., Zeng, M., Dutta, D., Koh, T. H., Chen, J., and van der Donk, W. A. (2017). LanCL proteins are not involved in lanthionine synthesis in mammals. Sci. Rep. 7:40980. doi: 10.1038/srep40980
Hedden, P., and Thomas, S. G. (2012). Gibberellin biosynthesis and its regulation. Biochem. J. 444, 11–25. doi: 10.1042/BJ20120245
Hernández-Blanco, C., Feng, D. X., Hu, J., Sánchez-Vallet, A., Deslandes, L., Llorente, F., et al. (2007). Impairment of cellulose synthases required for Arabidopsis secondary cell wall formation enhances disease resistance. Plant Cell 19, 890–903. doi: 10.1105/tpc.106.048058
Herrera-Medina, M. J., Steinkellner, S., Vierheilig, H., Ocampo Bote, J. A., and García Garrido, J. M. (2007). Abscisic acid determines arbuscule development and functionality in the tomato arbuscular mycorrhiza. New Phytol. 175, 554–564. doi: 10.1111/j.1469-8137.2007.02107.x
Hirai, N., Okamoto, M., and Koshimizu, K. (1986). The 1′,4′-trans-diol of abscisic acid, a possible precursor of abscisic acid in Botrytis cinerea. Phytochemistry 25, 1865–1868. doi: 10.1016/S0031-9422(00)81164-4
Hirai, N., Yoshida, R., Todoroki, Y., and Ohigashi, H. (2000). Biosynthesis of abscisic acid by the non-mevalonate pathway in plants, and by the mevalonate pathway in fungi. Biosci. Biotechnol. Biochem. 64, 1448–1458. doi: 10.1271/bbb.64.1448
Hontecillas, R., Roberts, P. C., Carbo, A., Vives, C., Horne, W. T., Genis, S., et al. (2013). Dietary abscisic acid ameliorates influenza-virus-associated disease and pulmonary immunopathology through a PPARγ-dependent mechanism. J. Nutr. Biochem. 24, 1019–1027. doi: 10.1016/j.jnutbio.2012.07.010
Hou, S. T., Jiang, S. X., Zaharia, L. I., Han, X., Benson, C. L., Slinn, J., et al. (2016). Phaseic acid, an endogenous and reversible inhibitor of glutamate receptors in mouse brain. J. Biol. Chem. 291, 27007–27022. doi: 10.1074/jbc.M116.756429
Huddart, H., Smith, R. J., Langton, P. D., Hetherington, A. M., and Mansfield, T. A. (1986). Is abscisic acid a universally active calcium agonist? New Phytol. 104, 161–173. doi: 10.1111/j.1469-8137.1986.tb00643.x
Isik, I., and Celik, I. (2015). Investigation of neurotoxic and immunotoxic effects of some plant growth regulators at subacute and subchronic applications on rats. Toxicol. Ind. Health 31, 1095–1105. doi: 10.1177/0748233713487247
Jiang, C. J., Shimono, M., Sugano, S., Kojima, M., Yazawa, K., Yoshida, R., et al. (2010). Abscisic acid interacts antagonistically with salicylic acid signaling pathway in rice-Magnaporthe grisea interaction. Mol. Plant Microbe Interact. 23, 791–798. doi: 10.1094/MPMI-23-6-0791
Johnson, M., Zaretskaya, I., Raytselis, Y., Merezhuk, Y., McGinnis, S., and Madden, T. L. (2008). NCBI BLAST: a better web interface. Nucleic Acids Res. 36, W5–W9. doi: 10.1093/nar/gkn201
Jones, A. M., Danielson, J. A., Manojkumar, S. N., Lanquar, V., Grossmann, G., and Frommer, W. B. (2014). Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife 3:e01741. doi: 10.7554/eLife.01741
Kaliff, M., Staal, J., Myrenås, M., and Dixelius, C. (2007). ABA is required for Leptosphaeria maculans resistance via ABI1- and ABI4-dependent signaling. Mol. Plant Microbe Interact. 20, 335–345. doi: 10.1094/MPMI-20-4-0335
Karadeniz, A., Topcuoğlu, Ş. F., and İnan, S. (2006). Auxin, gibberellin, cytokinin and abscisic acid production in some bacteria. World J. Microbiol. Biotechnol. 22, 1061–1064. doi: 10.1007/s11274-005-4561-1
Kazan, K., and Manners, J. M. (2013). MYC2: the master in action. Mol. Plant 6, 686–703. doi: 10.1093/mp/sss128
Kelly, B., and O'Neill, L. A. J. (2015). Metabolic reprogramming in macrophages and dendritic cells in innate immunity. Cell Res. 25, 771–784. doi: 10.1038/cr.2015.68
Kharenko, O. A., Polichuk, D., Nelson, K. M., Abrams, S. R., and Loewen, M. C. (2013). Identification and characterization of interactions between abscisic acid and human heat shock protein 70 family members. J. Biochem. (Tokyo) 154, 383–391. doi: 10.1093/jb/mvt067
Kim, T. H., Hauser, F., Ha, T., Xue, S., Böhmer, M., Nishimura, N., et al. (2011). Chemical genetics reveals negative regulation of abscisic acid signaling by a plant immune response pathway. Curr. Biol. 21, 990–997. doi: 10.1016/j.cub.2011.04.045
Kissoudis, C., Seifi, A., Yan, Z., Islam, A. T., van der Schoot, H., van de Wiel, C. C., et al. (2016). Ethylene and abscisic acid signaling pathways differentially influence tomato resistance to combined powdery mildew and salt stress. Front. Plant Sci. 7:2009. doi: 10.3389/fpls.2016.02009
Klessig, D. F., Tian, M., and Choi, H. W. (2016). Multiple targets of salicylic acid and its derivatives in plants and animals. Front. Immunol. 7:206. doi: 10.3389/fimmu.2016.00206
Klingler, J. P., Batelli, G., and Zhu, J. K. (2010). ABA receptors: the START of a new paradigm in phytohormone signalling. J. Exp. Bot. 61, 3199–3210. doi: 10.1093/jxb/erq151
Kuromori, T., and Shinozaki, K. (2010). ABA transport factors found in Arabidopsis ABC transporters. Plant Signal. Behav. 5, 1124–1126. doi: 10.4161/psb.5.9.12566
Kwok, S. K., Park, M. K., Cho, M. L., Oh, H. J., Park, E. M., Lee, D. G., et al. (2012). Retinoic acid attenuates rheumatoid inflammation in mice. J. Immunol. Baltim. 189, 1062–1071. doi: 10.4049/jimmunol.1102706
Latorre, R., Huynh, J., Mazzoni, M., Gupta, A., Bonora, E., Clavenzani, P., et al. (2016). Expression of the bitter taste receptor, T2R38, in enteroendocrine cells of the colonic mucosa of overweight/obese vs. lean subjects. PLoS ONE 11:e0147468. doi: 10.1371/journal.pone.0147468
Leber, A., Bassaganya-Riera, J., Tubau-Juni, N., Zoccoli-Rodriguez, V., Viladomiu, M., Abedi, V., et al. (2016). Modeling the role of lanthionine synthetase C-like 2 (LANCL2) in the modulation of immune responses to helicobacter pylori infection. PLoS ONE 11:e0167440. doi: 10.1371/journal.pone.0167440
Lee, W. J., and Hase, K. (2014). Gut microbiota-generated metabolites in animal health and disease. Nat. Chem. Biol. 10, 416–424. doi: 10.1038/nchembio.1535
Le Page-Degivry, M. T., Bidard, J. N., Rouvier, E., Bulard, C., and Lazdunski, M. (1986). Presence of abscisic acid, a phytohormone, in the mammalian brain. Proc. Natl. Acad. Sci. U.S.A. 83, 1155–1158. doi: 10.1073/pnas.83.4.1155
Li, H. H., Hao, R. L., Wu, S. S., Guo, P. C., Chen, C. J., Pan, L. P., et al. (2011). Occurrence, function and potential medicinal applications of the phytohormone abscisic acid in animals and humans. Biochem. Pharmacol. 82, 701–712. doi: 10.1016/j.bcp.2011.06.042
Lim, C. W., Baek, W., Jung, J., Kim, J. H., and Lee, S. C. (2015). Function of ABA in stomatal defense against biotic and drought stresses. Int. J. Mol. Sci. 16, 15251–15270. doi: 10.3390/ijms160715251
Liu, P. P., Yang, Y., Pichersky, E., and Klessig, D. F. (2009). Altering expression of benzoic acid/salicylic acid carboxyl methyltransferase 1 compromises systemic acquired resistance and PAMP-triggered immunity in Arabidopsis. Mol. Plant. Microbe Interact. 23, 82–90. doi: 10.1094/MPMI-23-1-0082
Liu, X., Yue, Y., Li, B., Nie, Y., Li, W., Wu, W. H., et al. (2007). A G protein-coupled receptor is a plasma membrane receptor for the plant hormone abscisic acid. Science 315, 1712–1716. doi: 10.1126/science.1135882
Liu, Z., Yan, J. P., Li, D. K., Luo, Q., Yan, Q., Liu, Z. B., et al. (2015). UDP-glucosyltransferase71c5, a major glucosyltransferase, mediates abscisic acid homeostasis in Arabidopsis. Plant Physiol. 167, 1659–1670. doi: 10.1104/pp.15.00053
Lu, P., Bevan, D. R., Lewis, S. N., Hontecillas, R., and Bassaganya-Riera, J. (2011). Molecular modeling of lanthionine synthetase component C-like protein 2: a potential target for the discovery of novel type 2 diabetes prophylactics and therapeutics. J. Mol. Model. 17, 543–553. doi: 10.1007/s00894-010-0748-y
Lu, P., Hontecillas, R., Philipson, C. W., and Bassaganya-Riera, J. (2014). Lanthionine synthetase component C-like protein 2: a new drug target for inflammatory diseases and diabetes. Curr. Drug Targets 15, 565–572. doi: 10.2174/1389450115666140313123714
Magnone, M., Ameri, P., Salis, A., Andraghetti, G., Emionite, L., Murialdo, G., et al. (2015). Microgram amounts of abscisic acid in fruit extracts improve glucose tolerance and reduce insulinemia in rats and in humans. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 29, 4783–4793. doi: 10.1096/fj.15-277731
Magnone, M., Bruzzone, S., Guida, L., Damonte, G., Millo, E., Scarfì, S., et al. (2009). Abscisic acid released by human monocytes activates monocytes and vascular smooth muscle cell responses involved in atherogenesis. J. Biol. Chem. 284, 17808–17818. doi: 10.1074/jbc.M809546200
Magnone, M., Scarfì, S., Sturla, L., Guida, L., Cuzzocrea, S., Di Paola, R., et al. (2013). Fluridone as a new anti-inflammatory drug. Eur. J. Pharmacol. 720, 7–15. doi: 10.1016/j.ejphar.2013.10.058
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Magnone, M., Sturla, L., Jacchetti, E., Scarfì, S., Bruzzone, S., Usai, C., et al. (2012). Autocrine abscisic acid plays a key role in quartz-induced macrophage activation. FASEB J. 26, 1261–1271. doi: 10.1096/fj.11-187351
Malara, A., Fresia, C., Di Buduo, C. A., Soprano, P. M., Moccia, F., Balduini, C., et al. (2017). The plant hormone abscisic acid is a pro-survival factor in human and murine megakaryocytes. J. Biol. Chem. 292, 3239–3251. doi: 10.1074/jbc.M116.751693
Manohar, M., Tian, M., Moreau, M., Park, S. W., Choi, H. W., Fei, Z., et al. (2014). Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front. Plant Sci. 5:777. doi: 10.3389/fpls.2014.00777
Mantovani, A., Sozzani, S., Locati, M., Allavena, P., and Sica, A. (2002). Macrophage polarization: tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 23, 549–555. doi: 10.1016/S1471-4906(02)02302-5
Maršálek, B., Zahradníčková, H., and Hronková, M. (1992). Extracellular abscisic acid produced by cyanobacteria under salt stress. J. Plant Physiol. 139, 506–508. doi: 10.1016/S0176-1617(11)80503-1
Matsubara, R., Aonuma, H., Kojima, M., Tahara, M., Andrabi, S. B., Sakakibara, H., et al. (2015). Plant hormone salicylic acid produced by a malaria parasite controls host immunity and cerebral malaria outcome. PLoS ONE 10:e0140559. doi: 10.1371/journal.pone.0140559
Mbengue, M., Navaud, O., Peyraud, R., Barascud, M., Badet, T., Vincent, R., et al. (2016). Emerging trends in molecular interactions between plants and the broad host range fungal pathogens Botrytis cinerea and Sclerotinia sclerotiorum. Front. Plant Sci. 7:422. doi: 10.3389/fpls.2016.00422
Miyazono, K., Miyakawa, T., Sawano, Y., Kubota, K., Kang, H. J., Asano, A., et al. (2009). Structural basis of abscisic acid signalling. Nature 462, 609–614. doi: 10.1038/nature08583
Moeder, W., Ung, H., Mosher, S., and Yoshioka, K. (2010). SA-ABA antagonism in defense responses. Plant Signal. Behav. 5, 1231–1233. doi: 10.4161/psb.5.10.12836
Morrison, E. N., Knowles, S., Hayward, A., Thorn, R. G., Saville, B. J., and Emery, R. J. (2015). Detection of phytohormones in temperate forest fungi predicts consistent abscisic acid production and a common pathway for cytokinin biosynthesis. Mycologia 107, 245–257. doi: 10.3852/14-157
Müller, M., and Munné-Bosch, S. (2015). Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 169, 32–41. doi: 10.1104/pp.15.00677
Nagamune, K., Hicks, L. M., Fux, B., Brossier, F., Chini, E. N., and Sibley, L. D. (2008). Abscisic acid controls calcium-dependent egress and development in Toxoplasma gondii. Nature 451, 207–210. doi: 10.1038/nature06478
Negri, P., Maggi, M. D., Ramirez, L., Feudis, L. D., Szwarski, N., Quintana, S., et al. (2015). Abscisic acid enhances the immune response in Apis mellifera and contributes to the colony fitness. Apidologie 46, 542–557. doi: 10.1007/s13592-014-0345-7
Nelson, D. R., Goldstone, J. V., and Stegeman, J. J. (2013). The cytochrome P450 genesis locus: the origin and evolution of animal cytochrome P450s. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 368:20120474. doi: 10.1098/rstb.2012.0474
Nishimura, M. T., Stein, M., Hou, B. H., Vogel, J. P., Edwards, H., and Somerville, S. C. (2003). Loss of a callose synthase results in salicylic acid-dependent disease resistance. Science 301, 969–972. doi: 10.1126/science.1086716
Odegaard, J. I., Ricardo-Gonzalez, R. R., Goforth, M. H., Morel, C. R., Subramanian, V., Mukundan, L., et al. (2007). Macrophage-specific PPARgamma controls alternative activation and improves insulin resistance. Nature 447, 1116–1120. doi: 10.1038/nature05894
Oide, S., Bejai, S., Staal, J., Guan, N., Kaliff, M., and Dixelius, C. (2013). A novel role of PR2 in abscisic acid (ABA) mediated, pathogen-induced callose deposition in Arabidopsis thaliana. New Phytol. 200, 1187–1199. doi: 10.1111/nph.12436
Palau-Rodriguez, M., Tulipani, S., Isabel Queipo-Ortuño, M., Urpi-Sarda, M., Tinahones, F. J., and Andres-Lacueva, C. (2015). Metabolomic insights into the intricate gut microbial-host interaction in the development of obesity and type 2 diabetes. Front. Microbiol. 6:1151. doi: 10.3389/fmicb.2015.01151
Park, J., Zielinski, M., Magder, A., Tsantrizos, Y. S., and Berghuis, A. M. (2017). Human farnesyl pyrophosphate synthase is allosterically inhibited by its own product. Nat. Commun. 8:14132. doi: 10.1038/ncomms14132
Park, S., and James, C. D. (2003). Lanthionine synthetase components C-like 2 increases cellular sensitivity to adriamycin by decreasing the expression of P-glycoprotein through a transcription-mediated mechanism. Cancer Res. 63, 723–727. Available online at: http://cancerres.aacrjournals.org/content/63/3/723
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Park, Y., Xu, Z. Y., Kim, S. Y., Lee, J., Choi, B., Lee, J., et al. (2016). Spatial regulation of ABCG25, an ABA Exporter, is an important component of the mechanism controlling cellular ABA levels. Plant Cell 28, 2528–2544. doi: 10.1105/tpc.16.00359
Paterson, J. R., Baxter, G., Dreyer, J. S., Halket, J. M., Flynn, R., and Lawrence, J. R. (2008). Salicylic acid sans aspirin in animals and man: persistence in fasting and biosynthesis from benzoic acid. J. Agric. Food Chem. 56, 11648–11652. doi: 10.1021/jf800974z
Perchepied, L., Balagué, C., Riou, C., Claudel-Renard, C., Rivière, N., Grezes-Besset, B., et al. (2010). Nitric oxide participates in the complex interplay of defense-related signaling pathways controlling disease resistance to Sclerotinia sclerotiorum in Arabidopsis thaliana. Mol. Plant Microbe Interact. 23, 846–860. doi: 10.1094/MPMI-23-7-0846
Persson, M., Staal, J., Oide, S., and Dixelius, C. (2009). Layers of defense responses to Leptosphaeria maculans below the RLM1- and camalexin-dependent resistances. New Phytol. 182, 470–482. doi: 10.1111/j.1469-8137.2009.02763.x
Pydi, S. P., Jaggupilli, A., Nelson, K. M., Abrams, S. R., Bhullar, R. P., Loewen, M. C., et al. (2015). Abscisic acid acts as a blocker of the bitter taste g protein-coupled receptor T2R4. Biochemistry (Mosc.) 54, 2622–2631. doi: 10.1021/acs.biochem.5b00265
Qi, C. C., Ge, J. F., and Zhou, J. N. (2015a). Preliminary evidence that abscisic acid improves spatial memory in rats. Physiol. Behav. 139, 231–239. doi: 10.1016/j.physbeh.2014.11.053
Qi, C. C., Shu, Y M., Chen, F. H., Ding, Y. Q., and Zhou, J. N. (2016). Sensitivity during the forced swim test is a key factor in evaluating the antidepressant effects of abscisic acid in mice. Behav. Brain Res. 300, 106–113. doi: 10.1016/j.bbr.2015.12.009
Qi, C. C., Zhang, Z., Fang, H., Liu, J., Zhou, N., Ge, J. F., et al. (2015b). Antidepressant effects of abscisic acid mediated by the downregulation of corticotrophin-releasing hormone gene expression in rats. Int. J. Neuropsychopharmacol. 18:pyu006. doi: 10.1093/ijnp/pyu006
Rakic, B., Clarke, J., Tremblay, T. L., Taylor, J., Schreiber, K., Nelson, K. M., et al. (2006). A small-molecule probe for hepatitis C virus replication that blocks protein folding. Chem. Biol. 13, 1051–1060. doi: 10.1016/j.chembiol.2006.08.010
Rashad, F. M., Fathy, H. M., El-Zayat, A. S., and Elghonaimy, A. M. (2015). Isolation and characterization of multifunctional Streptomyces species with antimicrobial, nematicidal and phytohormone activities from marine environments in Egypt. Microbiol. Res. 175, 34–47. doi: 10.1016/j.micres.2015.03.002
Ray, I., Mahata, S. K., and De, R. K. (2016). Obesity: an immunometabolic perspective. Front. Endocrinol. 7:157. doi: 10.3389/fendo.2016.00157
Reihill, J. A., Malcomson, B., Bertelsen, A., Cheung, S., Czerwiec, A., Barsden, R., et al. (2016). Induction of the inflammatory regulator A20 by gibberellic acid in airway epithelial cells. Br. J. Pharmacol. 173, 778–789. doi: 10.1111/bph.13200
Rodriguez, P. L. (2016). abscisic acid catabolism generates phaseic acid, a molecule able to activate a subset of ABA receptors. Mol. Plant 9, 1448–1450. doi: 10.1016/j.molp.2016.09.009
Saito, S., Hirai, N., Matsumoto, C., Ohigashi, H., Ohta, D., Sakata, K., et al. (2004). Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol. 134, 1439–1449. doi: 10.1104/pp.103.037614
Sakamoto, H., Suzuki, S., Nagamune, K., Kita, K., and Matsuzaki, M. (2016). Investigation into the physiological significance of the phytohormone abscisic acid in Perkinsus marinus, an oyster parasite harboring a nonphotosynthetic plastid. J. Eukaryot. Microbiol. doi: 10.1111/jeu.12379. [Epub ahead of print].
Sakthivel, P., Sharma, N., Klahn, P., Gereke, M., and Brude, D. (2016). Abscisic acid: a phytohormone and mammalian cytokine as novel pharmacon with potential for future development into clinical applications. Curr. Med. Chem. 23, 1549–1570. doi: 10.2174/0929867323666160405113129
Salomon, M. V., Bottini, R., de Souza Filho, G. A., Cohen, A. C., Moreno, D., Gil, M., et al. (2014). Bacteria isolated from roots and rhizosphere of Vitis vinifera retard water losses, induce abscisic acid accumulation and synthesis of defense-related terpenes in in vitro cultured grapevine. Physiol. Plant. 151, 359–374. doi: 10.1111/ppl.12117
Sánchez-Vallet, A., López, G., Ramos, B., Delgado-Cerezo, M., Riviere, M.-P., Llorente, F., et al. (2012). Disruption of abscisic acid signaling constitutively activates Arabidopsis resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant Physiol. 160, 2109–2124. doi: 10.1104/pp.112.200154
Scarfì, S., Ferraris, C., Fruscione, F., Fresia, C., Guida, L., Bruzzone, S., et al. (2008). Cyclic ADP-ribose-mediated expansion and stimulation of human mesenchymal stem cells by the plant hormone abscisic acid. Stem Cells Dayt. Ohio 26, 2855–2864. doi: 10.1634/stemcells.2008-0488
Scarfì, S., Fresia, C., Ferraris, C., Bruzzone, S., Fruscione, F., Usai, C., et al. (2009). The plant hormone abscisic acid stimulates the proliferation of human hemopoietic progenitors through the second messenger cyclic ADP-ribose. Stem Cells Dayt. Ohio 27, 2469–2477. doi: 10.1002/stem.173
Schwartz, S. H., Qin, X., and Zeevaart, J. A. (2003). Elucidation of the indirect pathway of abscisic acid biosynthesis by mutants, genes, and enzymes. Plant Physiol. 131, 1591–1601. doi: 10.1104/pp.102.017921
Shigenaga, A. M., and Argueso, C. T. (2016). No hormone to rule them all: Interactions of plant hormones during the responses of plants to pathogens. Semin. Cell Dev. Biol. 56, 174–189. doi: 10.1016/j.semcdb.2016.06.005
Siewers, V., Kokkelink, L., Smedsgaard, J., and Tudzynski, P. (2006). Identification of an abscisic acid gene cluster in the grey mold Botrytis cinerea. Appl. Environ. Microbiol. 72, 4619–4626. doi: 10.1128/AEM.02919-05
Siewers, V., Smedsgaard, J., and Tudzynski, P. (2004). The P450 monooxygenase BcABA1 is essential for abscisic acid biosynthesis in Botrytis cinerea. Appl. Environ. Microbiol. 70, 3868–3876. doi: 10.1128/AEM.70.7.3868-3876.2004
Sivakumaran, A., Akinyemi, A., Mandon, J., Cristescu, S. M., Hall, M. A., Harren, F. J., et al. (2016). ABA suppresses Botrytis cinerea Elicited NO production in tomato to influence H2O2 generation and increase host susceptibility. Front. Plant Sci. 7:709. doi: 10.3389/fpls.2016.00709
Spence, C. A., Lakshmanan, V., Donofrio, N., and Bais, H. P. (2015). Crucial roles of abscisic acid biogenesis in virulence of rice blast fungus Magnaporthe oryzae. Front. Plant Sci. 6:1082. doi: 10.3389/fpls.2015.01082
Stec, N., Banasiak, J., and Jasiński, M. (2016). Abscisic acid–an overlooked player in plant-microbe symbioses formation? Acta Biochim. Pol. 63, 53–58. doi: 10.18388/abp.2015_1210
Steenkamp, E. T., Wright, J., and Baldauf, S. L. (2006). The protistan origins of animals and fungi. Mol. Biol. Evol. 23, 93–106. doi: 10.1093/molbev/msj011
Sturla, L., Fresia, C., Guida, L., Bruzzone, S., Scarfì, S., Usai, C., et al. (2009). LANCL2 is necessary for abscisic acid binding and signaling in human granulocytes and in rat insulinoma cells. J. Biol. Chem. 284, 28045–28057. doi: 10.1074/jbc.M109.035329
Sturla, L., Fresia, C., Guida, L., Grozio, A., Vigliarolo, T., Mannino, E., et al. (2011). Binding of abscisic acid to human LANCL2. Biochem. Biophys. Res. Commun. 415, 390–395. doi: 10.1016/j.bbrc.2011.10.079
Sturla, L., Mannino, E., Scarfì, S., Bruzzone, S., Magnone, M., Sociali, G., et al. (2017). Abscisic acid enhances glucose disposal and induces brown fat activity in adipocytes in vitro and in vivo. Biochim. Biophys. Acta 1862, 131–144. doi: 10.1016/j.bbalip.2016.11.005
Takeuchi, J., Okamoto, M., Akiyama, T., Muto, T., Yajima, S., Sue, M., et al. (2014). Designed abscisic acid analogs as antagonists of PYL-PP2C receptor interactions. Nat. Chem. Biol. 10, 477–482. doi: 10.1038/nchembio.1524
Todoroki, Y., and Ueno, K. (2010). Development of specific inhibitors of CYP707A, a key enzyme in the catabolism of abscisic acid. Curr. Med. Chem. 17, 3230–3244. doi: 10.2174/092986710792231987
Ton, J., and Mauch-Mani, B. (2004). Beta-amino-butyric acid-induced resistance against necrotrophic pathogens is based on ABA-dependent priming for callose. Plant J. Cell Mol. Biol. 38, 119–130. doi: 10.1111/j.1365-313X.2004.02028.x
Toum, L., Torres, P. S., Gallego, S. M., Benavídes, M. P., Vojnov, A. A., and Gudesblat, G. E. (2016). Coronatine inhibits stomatal closure through guard cell-specific Inhibition of NADPH oxidase-dependent ROS production. Front. Plant Sci. 7:1851. doi: 10.3389/fpls.2016.01851
Ulferts, S., Delventhal, R., Splivallo, R., Karlovsky, P., and Schaffrath, U. (2015). Abscisic acid negatively interferes with basal defence of barley against Magnaporthe oryzae. BMC Plant Biol. 15:7. doi: 10.1186/s12870-014-0409-x
Umezawa, T., Okamoto, M., Kushiro, T., Nambara, E., Oono, Y., Seki, M., et al. (2006). CYP707A3, a major ABA 8′-hydroxylase involved in dehydration and rehydration response in Arabidopsis thaliana. Plant J. Cell Mol. Biol. 46, 171–182. doi: 10.1111/j.1365-313X.2006.02683.x
Uzor, P. F., Osadebe, P. O., and Nwodo, N. J. (2017). Antidiabetic activity of extract and compounds from an endophytic fungus Nigrospora oryzae. Drug Res. doi: 10.1055/s-0042-122777. [Epub ahead of print].
Van Overtveldt, M., Heugebaert, T. S., Verstraeten, I., Geelen, D., and Stevens, C. V. (2015). Phosphonamide pyrabactin analogues as abscisic acid agonists. Org. Biomol. Chem. 13, 5260–5264. doi: 10.1039/C5OB00137D
van Wees, S. C., and Glazebrook, J. (2003). Loss of non-host resistance of Arabidopsis NahG to Pseudomonas syringae pv. phaseolicola is due to degradation products of salicylic acid. Plant J. Cell Mol. Biol. 33, 733–742. doi: 10.1046/j.1365-313X.2003.01665.x
Verhage, A., Vlaardingerbroek, I., Raaymakers, C., Van Dam, N. M., Dicke, M., Van Wees, S. C. M., et al. (2011). Rewiring of the jasmonate signaling pathway in arabidopsis during insect herbivory. Front. Plant Sci. 2:47. doi: 10.3389/fpls.2011.00047
Verma, V., Ravindran, P., and Kumar, P. P. (2016). Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16:86. doi: 10.1186/s12870-016-0771-y
Vigliarolo, T., Guida, L., Millo, E., Fresia, C., Turco, E., De Flora, A., et al. (2015). Abscisic acid transport in human erythrocytes. J. Biol. Chem. 290, 13042–13052. doi: 10.1074/jbc.M114.629501
Vigliarolo, T., Zocchi, E., Fresia, C., Booz, V., and Guida, L. (2016). Abscisic acid influx into human nucleated cells occurs through the anion exchanger AE2. Int. J. Biochem. Cell Biol. 75, 99–103. doi: 10.1016/j.biocel.2016.03.006
Viladomiu, M., Hontecillas, R., Yuan, L., Lu, P., and Bassaganya-Riera, J. (2013). Nutritional protective mechanisms against gut inflammation. J. Nutr. Biochem. 24, 929–939. doi: 10.1016/j.jnutbio.2013.01.006
Vos, I. A., Moritz, L., Pieterse, C. M., and Van Wees, S. C. (2015). Impact of hormonal crosstalk on plant resistance and fitness under multi-attacker conditions. Front. Plant Sci. 6:639. doi: 10.3389/fpls.2015.00639
Vos, I. A., Verhage, A., Schuurink, R. C., Watt, L. G., Pieterse, C. M., and Van Wees, S. C. (2013). Onset of herbivore-induced resistance in systemic tissue primed for jasmonate-dependent defenses is activated by abscisic acid. Front. Plant Sci. 4:539. doi: 10.3389/fpls.2013.00539
Waadt, R., Hitomi, K., Nishimura, N., Hitomi, C., Adams, S. R., Getzoff, E. D., et al. (2014). FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. eLife 3:e01739. doi: 10.7554/eLife.01739
Wang, Y., Weiss, L. M., and Orlofsky, A. (2009). Host cell autophagy is induced by Toxoplasma gondii and contributes to parasite growth. J. Biol. Chem. 284, 1694–1701. doi: 10.1074/jbc.M807890200
Weng, J. K., Ye, M., Li, B., and Noel, J. P. (2016). Co-evolution of hormone metabolism and signaling networks expands plant adaptive plasticity. Cell 166, 881–893. doi: 10.1016/j.cell.2016.06.027
Xu, J., Nakazawa, T., Ukai, K., Kobayashi, H., Mangindaan, R. E., Wewengkang, D. S., et al. (2008). Tetrahydrobostrycin and 1-deoxytetrahydrobostrycin, two new hexahydroanthrone derivatives, from a marine-derived fungus Aspergillus sp. J. Antibiot. (Tokyo) 61, 415–419. doi: 10.1038/ja.2008.57
Zeng, M., van der Donk, W. A., and Chen, J. (2014). Lanthionine synthetase C-like protein 2 (LanCL2) is a novel regulator of Akt. Mol. Biol. Cell 25, 3954–3961. doi: 10.1091/mbc.E14-01-0004
Zhou, J., Zhang, H., Yang, Y., Zhang, Z., Zhang, H., Hu, X., et al. (2008). Abscisic acid regulates TSRF1-mediated resistance to Ralstonia solanacearum by modifying the expression of GCC box-containing genes in tobacco. J. Exp. Bot. 59, 645–652. doi: 10.1093/jxb/erm353
Zhou, N., Yao, Y., Ye, H., Zhu, W., Chen, L., and Mao, Y. (2016). Abscisic-acid-induced cellular apoptosis and differentiation in glioma via the retinoid acid signaling pathway. Int. J. Cancer 138, 1947–1958. doi: 10.1002/ijc.29935
Keywords: immunity, inflammation, host-microbe interactions, metabolic engineering, comparative biology, signal transduction, natural product chemistry, systems biology
Citation: Lievens L, Pollier J, Goossens A, Beyaert R and Staal J (2017) Abscisic Acid as Pathogen Effector and Immune Regulator. Front. Plant Sci. 8:587. doi: 10.3389/fpls.2017.00587
Received: 11 February 2017; Accepted: 31 March 2017;
Published: 19 April 2017.
Edited by:
Víctor Flors, Jaume I University, SpainReviewed by:
Gabor Jakab, University of Pécs, HungaryAntonio Molina, Universidad Politécnica de Madrid, Spain
Copyright © 2017 Lievens, Pollier, Goossens, Beyaert and Staal. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jens Staal, jens.staal@irc.vib-ugent.be
 Laurens Lievens
Laurens Lievens Jacob Pollier
Jacob Pollier Alain Goossens
Alain Goossens Rudi Beyaert
Rudi Beyaert Jens Staal
Jens Staal