
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 05 April 2017
Sec. Plant Pathogen Interactions
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.00493
This article is part of the Research Topic Plant-Microbe-Insect Interaction: Source for Bio-fertilizers, Bio-medicines and Agent Research View all 43 articles
The TIFY domain contains approximately 36 conserved amino acids that form the core motif TIF[F/Y]XG, and they were reported to play important roles in plant growth, tissue development and defense regulation. Moreover, more and more evidence has shown that some members of the TIFY gene family perform their functions by modulating plant hormone signaling pathways. Poplar trees are found worldwide, and they comprise approximately 30 species. Benefit from the importance of poplar and its advanced platform, this tree is considered to be the model perennial plant. Here, we conducted a genome-wide identification of TIFY genes in poplar, and 24 TIFY genes were found. These 24 TIFY genes were assigned to different subfamilies according to the presence or absence of domains and motifs that they harbored. Careful analyses of their locations, structures, evolution and duplication patterns revealed an overview of this gene family in poplar. The expression profiles of these 24 TIFY genes were then analyzed in different tissues using publicly available expression data; their expression profiles following different JA/SA treatments and infection with leaf rust pathogen were also carefully examined by qRT-PCR assays. Based on their expression profiles, the functions of a number of TIFY genes could be predicted. By performing this study, we have provided valuable information for further functional characterisation of TIFY genes in poplar and candidate genes for the improvement of poplar disease resistance.
The TIFY domain contains approximately 36 conserved amino acids (AA) that form the core motif TIF[F/Y]XG (Vanholme et al., 2007; Bai et al., 2011). This conserved domain characterizes a plant-specific family of transcription factor (TF) genes called the TIFY gene family. TIFY genes were first characterized in Arabidopsis, and the gene AT4G24470 was reported to be a putative TF involved in inflorescence and flower development (Nishii et al., 2000). However, this gene was annotated as a ZIM gene in this study (Nishii et al., 2000). With a genome-wide survey of the Arabidopsis genome and because of the confusing use of the ZIM nomenclature, proteins containing TIFY domains were renamed as the TIFY gene family (Vanholme et al., 2007). The TIFY gene family can be classified into four subfamilies, TIFY, JAZ, ZML, and PPD, depending on whether they contain additional domains/motifs (Vanholme et al., 2007; Bai et al., 2011). Proteins with only the TIFY (PF06200) domain are classified as the TIFY subfamily; proteins with both the TIFY and jasmonate ZIM domains (JAZ, PF09425) are classified as the JAZ subfamily (Staswick, 2008); and proteins containing the TIFY domain and the CCT (PF06203) and/or ZML (PF00320) motif are classified as the ZML subfamily. In the PPD subfamily, the proteins contain the TIFY and PPD domains; proteins in this subfamily also sometimes contain a truncated JAZ domain (White, 2006).
To date, the functions of several TIFY genes have been fully investigated, and some of them have been found to play important roles in different biological processes. Of the four TIFY subfamilies, the functions of JAZ proteins are the most clear, and they have been found to be involved in the jasmonate (JA) signaling pathway. JAZ proteins act as JA repressors to inhibit TFs that regulate early JA-responsive genes (Pauwels and Goossens, 2011); in contrast, JA can also induce degradation of JAZ proteins, thereby allowing the expression of its response genes (Chung and Howe, 2009; Niu et al., 2011). Since JA is a key phytohormone in plant development, JAZ proteins were also found to play critical roles in the regulation of numerous aspects of plant development. In Astragalus sinicus, AsJAZ1 was found to interact with AsB2510 and participated in nodule development and nitrogen fixation (Li et al., 2015). In Arabidopsis, deletion of the two PPD genes (at the same locus) increased leaf lamina size and resulted in dome-shaped rather than flat leaves. Siliques were also altered in shape because of the additional lamina growth (White, 2006). JAZ and DELLA proteins were also found to bind to the WD-repeat/bHLH/MYB complex to modulate the synergistic effects of gibberellin and JA signaling; thus, these two types of proteins can integrate different hormonal signals to synergistically regulate plant development (Qi et al., 2014). Moreover, JAZ proteins were also found to play important roles in plant defense. In wild soybean, transcription of GsJAZ2 increased following exposure to different abiotic stresses including salt, alkali, cold and drought. Overexpression of GsJAZ2 in an Arabidopsis line resulted in enhanced tolerance to salt and alkali stresses (Zhu et al., 2012). In rice, OsJAZ8 was reported to regulate host immunity by modulation of JA-responsive volatile compounds (Taniguchi et al., 2014). In addition to the JAZ genes, other genes of the TIFY subfamilies were found to play roles in plant development and defense. In wild soybean, GsTIFY10, which could be induced by bicarbonate, salinity stress and JA, was isolated and overexpressed in Arabidopsis. The transgenic plants showed enhanced tolerance to bicarbonate stress during seed germination and during the early- and adult-seedling developmental stages (Zhu et al., 2011). Based on the preceding information, we conclude that the plant-specific TIFY proteins are very important in the regulation of plant development and defense.
Due to the importance of the TIFY gene family and benefitting from the accelerated release of genome data, a number of studies have focused on the genome-wide investigation and characterisation of TIFY genes in different plant species. In the respective dicot and monocot model plants, Arabidopsis and rice, 18 and 20 TIFY genes were reported, respectively (Ye et al., 2009; Bai et al., 2011). In addition, 18 TIFY family proteins were found in pigeonpea [Cajanus cajan (L.) Millsp.] (Sirhindi et al., 2016); 30 TIFY genes were found in apple (Malus × domestica Borkh.) (Li et al., 2014); 19 TIFY genes were found in grape (Vitis vinifera) (Zhang et al., 2012); 28 TIFY genes were found in the Gossypium raimondii genome (He et al., 2015); 21 TIFY genes were found in Brachypodium distachyon (Zhang et al., 2015); and 34 TIFY genes in found in wild soybean (Glycine soja) (Zhu et al., 2013). In these studies, the expression profiles of TIFY genes were investigated and their functions were predicted based on their preferentially transcriptional abundance in a given tissue or under a given stress condition. This information is very valuable for guiding further characterisation of the functions of TIFY genes. In poplar, 25 TIFY proteins were identified using the Populus trichocarpa genome version 1.0 annotations when global comparisons of the gene family were performed in a number of plant species (Bai et al., 2011). Recently, the P. trichocarpa genome annotation was updated to version 3.0, and the quality of the reference genomic sequences was also greatly improved. Therefore, it is necessary to conduct a genome-wide survey of TIFY genes in poplar using the P. trichocarpa genome annotation version 3.0, in addition to analyzing their expression profiles under different conditions.
Poplar (Populus spp.) is a tree that is found worldwide, and it comprises approximately 30 species. Due to its characteristics of being fast-growing, widely used and strong ability to adapt, poplar is grown in most of the Northern hemisphere. Most of the species in the genus can be used for wood, pulp, paper, and fuel. In some areas, poplar is also an important landscape tree. Moreover, poplar species usually have small genome sizes, and some species can easily be created transgenic lines. In 2006, the draft genomic sequence of P. trichocarpa was released (Tuskan et al., 2006). Subsequently, the genomic sequence and annotations have been updated several times and high quality genomic information has recently become available online1 (Wullschleger et al., 2013). Moreover, additional genomic information of value for the species is publicly available, including the expression database and the whole genomic sequence of P. euphratica (Ma et al., 2013; Sundell et al., 2015). According to the above information, poplar is considered to be a model plant in perennial tree species.
To our knowledge, no comprehensive genome-wide survey and characterisation of TIFY genes in poplar has been carried out. In this study, we mainly focused on the responses of TIFY genes to phytohormone treatments and biotic stresses, and we aimed to identify TIFY genes involved in poplar biotic defenses. To this end, we first conducted a genome-wide identification of TIFY genes using the P. trichocarpa genome annotation version 3.0; 24 TIFY genes were found. Based on the presence of certain domains/motifs within these 24 TIFY genes, they were assigned to different subfamilies. The duplication types of each of the TIFY genes were simulated and their phylogeny, gene structures and locations within the genome were also investigated. Based on the publicly available expression data, the expression profiles of the 24 TIFY genes in different tissues were subsequently analyzed; their expression profiles after different JA/SA treatments and M. larici-populina infection were also carefully examined by qRT-PCR assays. Based on their expression profiles, the functions of a number of TIFY genes could be predicted. By performing this study, we have provided valuable information for further functional characterisation of TIFY genes in poplar and candidate genes for the improvement of poplar disease resistance.
To identify members of the TIFY gene family in poplar, the Hidden Markov Model (HMM) profile of the TIFY domain (PF06200) was downloaded from the pfam database (Pfam2). The reference genome P. trichocarpa was downloaded from the Phytozome database3. Genome annotation version 3.0 of P. trichocarpa was used in this study. To search the 73,013 predicted proteins of the P. trichocarpa genome annotation version 3.0, 759 seed sequences in the PF06200 HMM profile were used. Two softwares, blastp and HMMER, were used to perform a search for proteins harboring the TIFY domain. Proteins of P. trichocarpa that showed E-values above 1e-6 in the search results of blastp or HMMER were considered to be candidate TIFY domain genes. Of the proteins from the same gene model, only the longest ones (the “0.1” gene model of TIFY genes in the P. trichocarpa genome annotation version 3.0) were kept for further study. To confirm the existence of the TIFY domain and the Jas, CCT and ZML motifs, candidate TIFY domain proteins were used to search the Pfam database. The Pfam accession numbers for the three motifs are PF09425, PF06203 and PF00320, respectively. We also checked for the existence of the PPD domain/motif in each of the candidate TIFY genes. We noted that there is no sequence information for the PPD domain/motif in the Pfam database. The previously annotated PPD genes in Arabidopsis and grape were used to build the conserved PPD sequence. Briefly, the previously annotated PPD sequences were first aligned by ClustX2.1, and an msf result was produced and imported into hmmbuild implemented in HMMER. The resulting PPD HMM profile was used to search all of the candidate TIFY genes.
The software MEGA5 was used to analyze the phylogeny of all of the TIFY genes (Tamura et al., 2011). The protein sequences of all of the identified poplar TIFY genes were imported into ClustalX2.1 to perform a complete alignment, and the resultant multiple alignment file was imported into MEGA5. Unrooted phylogenetic trees were constructed using the Neighbor-Joining (NJ) method, and the bootstrap test was carried out with 1,000 iterations. For constructing phylogenetic trees among the four species, the protein sequences of the three other species were obtained from Phytozome database4. The deduced TIFY protein information was obtained from Arabidopsis thaliana (Bai et al., 2011), Oryza sativa (Bai et al., 2011) and Vitis vinifera (Zhang et al., 2012).
Gene structures and their chromosomal locations were obtained from the P. trichocarpa genome annotation version 3.0. The gene structures were displayed using a custom R script. The duplication pattern for all of the poplar genes was analyzed using MCScanX software. Briefly, the 41,335 poplar gene models (the “0.1” gene model in the P. trichocarpa genome annotation version 3.0 represents all genes models if a gene has more than one alternative transcript) were extracted from the poplar genome. An all-vs.-all local blast for the 41,335 poplar gene models using the local blast software with E-values under 1e-4 was carried out. The blast output was imported into MCScanX software following the procedure. All of the 41,335 genes were classified into four types using default criteria, including segmental, tandem, proximal, and dispersed duplications. The duplication pattern for each poplar TIFY gene was then determined according to the results.
The PopGenExpress database was employed to investigate the expression levels of all of the poplar TIFY genes in different tissues5 (Wilkins et al., 2009). Briefly, the correspondence between microarray probes and poplar genes was obtained from the annotation files deposited for the microarray data. Because the annotation released for the poplar microarray used in the PopGenExpress database was based on P. trichocarpa genome annotation version 2.0, the relationship between the probes and poplar genes annotated in genome version 3.0 was transformed based on the correspondence between versions 2.0 and 3.0 of the P. trichocarpa genome annotation. For genes that had no corresponding probes in genome version 3.0, an additional reciprocal blast with released sequences for all of the probes was employed. If a given gene and probe pair were each other’s top hits in the reciprocal blast results, then the probe was considered to represent the gene. The relative expression levels of all of the TIFY genes in xylem, roots, mature and young leaves, male and female catkins were obtained from the PopGenExpress database. A heat map for the expression levels of all of the TIFY genes in the selected tissues was prepared with Cluster 3.0 software (de Hoon et al., 2004); the dendrogram was visualized using JavaTreeview6.
The hybrid poplar variety “NL895” (P. euramericana) was used in this study. The female parent of NL895 is P. deltoids cv. Lux” (I-69) and the male parent is also a hybrid variety “I-45” (P. × euramericana). Tissue culture seedlings of “NL895” were grown in 3-L pots containing a sand-peat (50:50, v/v) mixture, and the pots were placed in a greenhouse under a 16/8 h photoperiod. The light intensity in the greenhouse was set at 12,000 Lx. The temperatures were set at 28 and 25°C for day and night, respectively. The seedlings were watered twice per month with Hoagland’s complete nutrient solution. Additionally, the seedlings were watered with distilled water 2–3 times a week. Seedlings with 6–10 fully expanded leaves were used for the treatments.
For the plant hormone treatments, “NL895” seedlings were treated with 0.2 mM jasmonic acid (JA) and 0.5 mM salicylic acid (SA). Leaves with LPIs (leaf plastochron indices) ranging from 4 to 10 were sprayed with JA or SA solution. To ensure that the hormone treatments were uniform, leaves were sprayed on both sides just until the leaf surfaces began to form small drops. Leaf samples were collected at 0, 2, 6, 12, and 24 h after treatment. Sprayed leaves from each individual poplar plant were pooled into one biological sample. Three replicates were carried out for each treatment.
For the leaf rust pathogen infection treatments, a virulent M. larici-populina strain used as fungal material in this study was obtained from P. simonii Carr., which showed typical leaf rust symptoms (yellow bubbles on the lower surface of leaf) in the summer of 2015. This strain was also used as the fungal material in another study (Wang et al., 2017). The urediniospores of M. larici-populina isolates were allowed to multiply on 1-year-old potted P. simonii Carr. cuttings and diluted into an urediniospore suspension in agar-water (0.1% agar in distilled water). The concentrations of the urediniospore suspensions were 1–2 mg/ml. Fully expanded leaves from “NL895” with leaf plastochron indices (LPIs) of 4–10 from each branch were spray-inoculated on their abaxial surfaces with the prepared urediniospore suspension. Leaves from one separate cutting were considered to be one sample. Leaf samples were collected after 0, 2, 4, and 8 days treatments. The samples were immediately frozen in liquid nitrogen and stored at -80°C until required for further analysis.
Total RNA was isolated using an RNAprep Pure Kit (for Plants) according to the manufacturer’s protocol [TIANGEN Biotech (Beijing) Co., Ltd., Beijing, China]. The quality of all of the RNA samples was examined by performing agarose gel electrophoresis. The first strand cDNA was synthesized using the TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransBionovo Co., Ltd., Beijing, China). The primers used for the qRT-PCR assay of TIFY genes are listed in Supplementary Table S2. PCR products from cDNA or genomic DNA templates using these primer pairs were initially sent for sequencing to confirm that the correct targets had been amplified (data not shown). The correct primer pairs were used for subsequent qRT-PCR assays that were performed on the LightCycler 96 (Roche) platform by using the FastStart Essential DNA Green Master Mix (Roche). For each sample, two reference genes (ACTIN and 18S) were used to standardize the mRNA abundance, and three replications were performed. The 2-ΔΔCq method was used to calculate the relative gene expression based on the qRT-PCR data (Livak and Schmittgen, 2001).
A genome-wide search of TIFY family genes in poplar yielded a total of 24 non-redundant genes that were found to harbor the TIFY domain. The 24 non-redundant genes represent a total of 99 transcripts in the P. trichocarpa genome annotation version 3.0 (Table 1) and all the 99 transcripts have the TIFY domain. To avoid complexity in the subsequent study, only the “0.1” gene model for the 24 non-redundant genes were kept in further analyses. These genes were considered to be TIFY family genes (Table 1). Their phylogeny and gene structures are shown in Figures 1A,B, respectively. Of these 24 TIFY domain-containing genes, 12 of the predicted proteins contain both the TIFY domain and the Jas motif; these proteins were designated as PtJAZ1 through 12. Seven proteins have both a TIFY domain and CCT and ZML motifs, and one protein has both a TIFY domain and a CCT motif; thus, these eight proteins were designated as PtZML1 through 8. Two proteins contain the TIFY domain and the PPD motif, and they were designated as PtPPD1 and PtPPD2. Note that PtPPD1 also has a Jas motif in its protein structure. The remaining two proteins contain only the TIFY domain, and they were designated as PtTIFY1 and PtTIFY2. All of the domains or motifs located in the 24 TIFY genes are illustrated in Figure 1C. The nomenclatures of these genes were designated based on both their subfamilies and their locations on the poplar chromosomes (Figure 2). The correspondence of our nomenclatures for the identified TIFY family genes and their original gene IDs released by the P. trichocarpa genome annotation version 3.0 are shown in Table 1. In previous report, 25 TIFY proteins were identified using the P. trichocarpa genome version 1.0 annotations when global comparisons of the gene family were performed in a number of plant species (Bai et al., 2011). When comparing our results with theirs, we found PtZML4 and 5 were corresponding to 3 ZML proteins in Bai‘s results; while the other 22 TIFY genes are consistent in the two identifications.
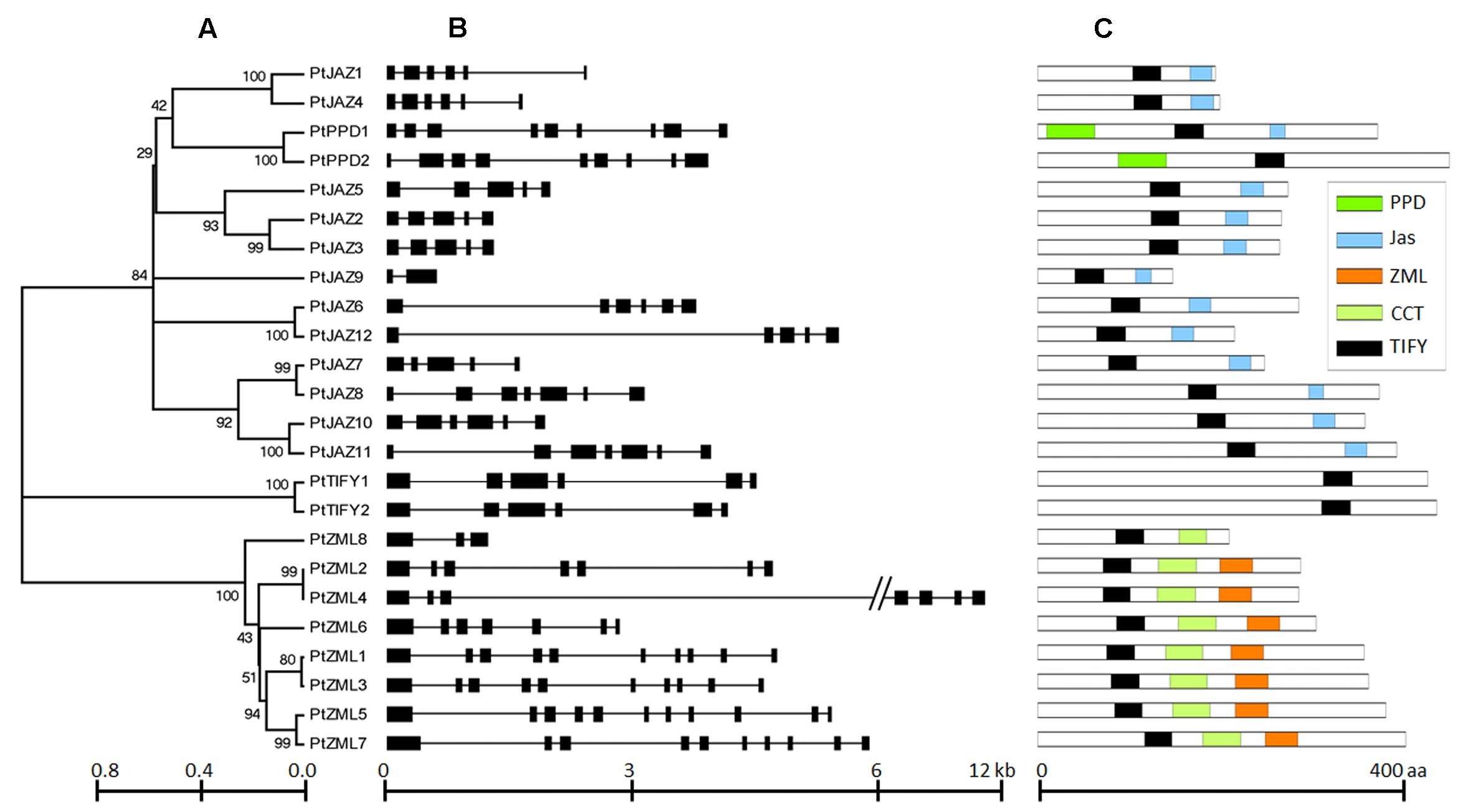
FIGURE 1. Phylogeny, structures and domains/motifs of the poplar TIFY gene family. The numbers labeled on the tree nodes in (A); represent bootstrap values in (C); black box represents exon and line represent intron in (B).
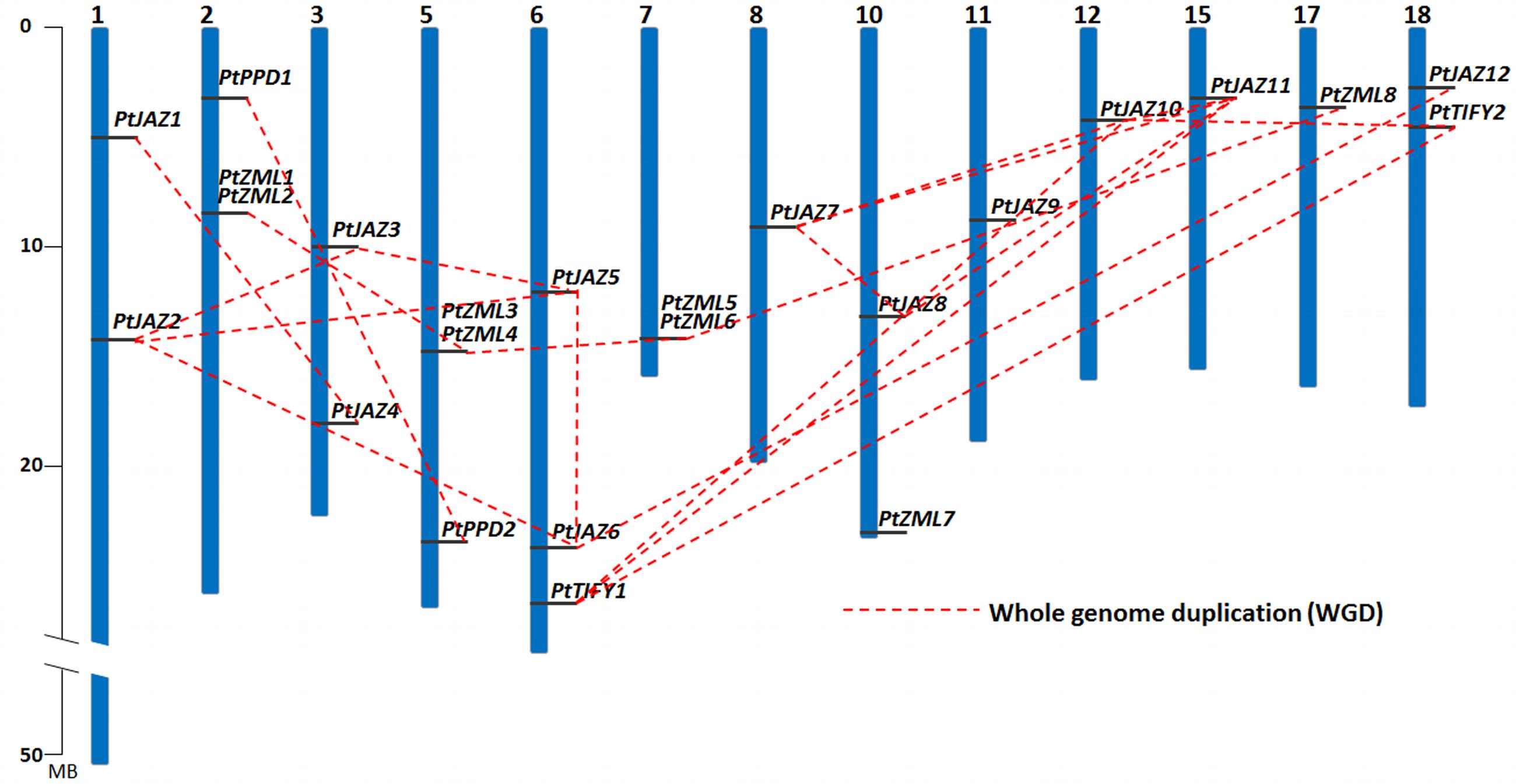
FIGURE 2. Chromosomal locations and WGD patterns of the poplar TIFY gene family. Blue bars represent 19 chromosomes and the gray lines on blue bars represent locations of TIFY genes. A dash line indicate a duplicated gene pair.
According to Figure 1B, all of the TIFY family genes have more than one exon. PtJAZ9 has the smallest number of exons at 2, whereas PtZML5 has the largest number of exons at 11. The genomic sequences of most of these genes are less than 6 kb in length, but PtZML4 has the longest genomic sequence of approximately 12 kb. The gene PtJAZ9 has the shortest open reading frame (ORF) of 149 AA, whereas PtPPD2 has the longest ORF of 454 AA. Insights into the phylogeny and gene structures of genes has indicated that proteins that are closely related phylogenetically tend to have similar gene structures. For example, PtJAZ1 and PtJAZ4 have the same number of exons in the same arrangement and tend to cluster together in the phylogenetic tree; this phenomenon was also observed for PtTIFY1 and PtTIFY2 and PtZML1 and PtZML3. In order to further insight into the phylogeny of TIFY proteins within plant, TIFY family proteins of three other representative species, including Arabidopsis, rice and grapes, were also employed to construct a phytogenic tree. Clearly, TIFY proteins in the four plants can be grouped into four clades (Supplementary Figure S1). According to this figure, proteins in the clade 1 and 2 tend to only have TIFY domain and have ZML motif, respectively; while proteins in the clade 3 and 4 have no clear difference when comparing with their domains or motifs.
The chromosomal locations of these 24 TIFY family genes is shown in Figure 2 and Table 1, and they are found on 13 of the 19 chromosomes. Chromosomes 4, 9, 13, 14, 16, and 19 contain no TIFY genes. Because duplication usually contributes to the expansion of gene families, we investigated the duplication patterns of each of the genes. In total, 19 genes were produced by whole genome duplication (WGD), and two genes were produced by tandem duplication. The other three genes did not show duplication patterns. The two tandemly duplicated genes, PtZML2 and Pt ZML6, were found to be tandemly copied from PtZML1 and PtZML5 (Figure 2), respectively, whereas both PtZML1 and PtZML5 were produced by WGD. This result suggested that PtZML1 and PtZML5 were the ancestors of PtZML2 and PtZML6, respectively. The gene duplication patterns for all 24 of the TIFY genes showed that WGD played large roles in this gene family in poplar.
Since the transcriptional abundance of a gene in different tissues is usually indicative of its function, we investigated the expression profiles of the 24 TIFY family genes in poplar. By analyzing of the publicly available expression data from the PopGenExpress database, we were able to obtain the relative expression levels of all 24 TIFY family genes in six tissues: xylem, roots, mature and young leaves and male and female catkins (Supplementary Table S1 and Figure 3). All 24 genes showed very high gene expression levels in female (7.11 ± 4.54) and male catkins (5.84 ± 1.63), whereas the lowest expression levels were found in roots (Supplementary Table S1). This result may suggest that the TIFY family of genes plays important roles in poplar catkin development.
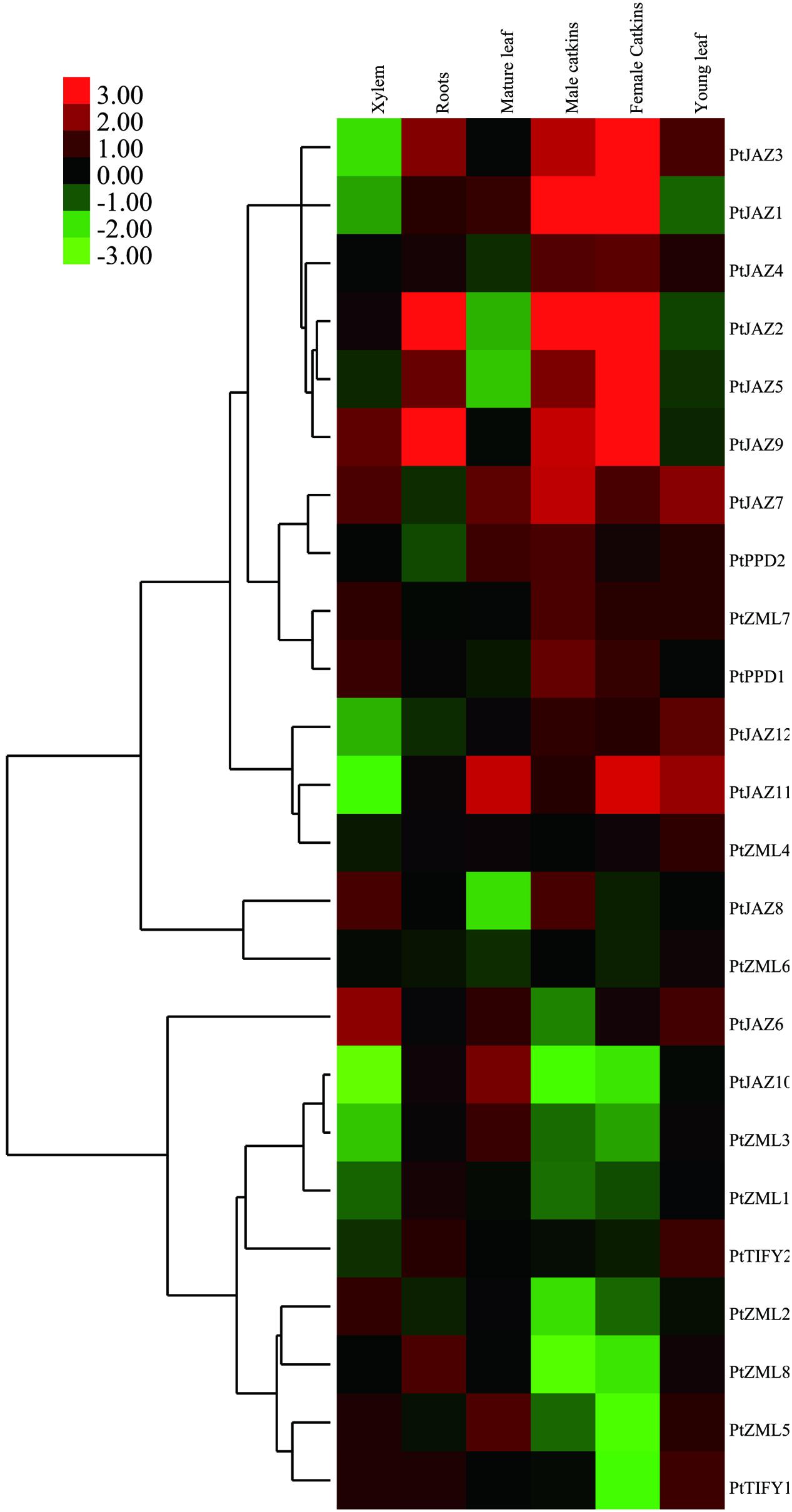
FIGURE 3. Hierarchical clustering of the expression profiles of the poplar TIFY gene family across different tissues. The relative expression levels of all of the TIFY genes in xylem, roots, mature and young leaves, male and female catkins were obtained from the PopGenExpress database. The expression of a mediate developmental stage of the plant material was set as control and all the gene expression at this stage was considered as “1” in the PopGenExpress database. A heat map for the expression levels of all of the TIFY genes in the selected tissues was prepared with Cluster 3.0 software.
Genes that are expressed abundantly in one or several specific tissues usually indicate their function related to the formation of these organs or the corresponding plant development. Further insights into the expression levels for each individual gene indicated that TIFY genes showed extremely high expression levels in some tissues. We filtered these types of genes when their expression was greater than two-fold the average expression level of all TIFY genes in the tissue. In total, seven TIFY genes were preferentially expressed in a given tissue, and some tissues had more than one preferentially expressed gene. PtJAZ6 and PtJAZ9 were preferentially expressed in xylem; PtJAZ2 and PtJAZ9 were preferentially expressed in roots and mature leaves; PtJAZ1 and PtJAZ2 were preferentially expressed in both male and female catkins, and PtJAZ9 was preferentially expressed in female catkins; PtJAZ7 and PtJAZ11 were preferentially expressed in young leaves (Supplementary Table S1 and Figure 3). These data suggest that PtJAZ1, PtJAZ2, PtJAZ6, PtJAZ7, and PtJAZ9 could be responsible for the formation of these tissues or contribute to the functions of these tissues. By summarizing these results, we were able to find that a large proportion of genes in the JAZ subfamily were filtered out by our method, although some genes in other subfamilies also showed relatively high abundance in the seven selected tissues. This suggests that most of the JAZ subfamily in the TIFY gene family plays important roles in poplar development. In addition, the expression values for the two reference genes, Actin and 18S, across the five developmental tissues were also obtained and listed in Supplementary Table S1. The data revealed these two genes were expressed similarly in different tissues and it suggested that these two genes were suitable for the subsequent qRT-PCR assay.
JA and SA are hormones that play important roles in signal transduction when plants are challenged with biotic and abiotic stresses. Treating plants with JA, SA or their derivatives MeJA and MeSA can modulate the symptoms of pathogenic or herbivory damage on plants (Yamada et al., 2012; Zhang et al., 2016). To investigate which genes in TIFY gene family can respond to these two plant hormones, we examined the expression profiles of TIFY genes in poplar leaves after treatment with MeJA and SA solutions. The relative expression levels were assayed by qRT-PCR. In the description to convenience, CK indicated untreated samples; JA-2 h, JA-6 h, JA-12 h, JA-24 h indicated samples collected from leaves after 2, 6, 12 and 24 h of MeJA treatment, respectively; and SA-2 h, SA-6 h, SA-12 h, and SA-24 h indicated samples collected from leaves after 2, 6, 12, and 24 h of SA treatment, respectively. In total, we were able to examine the expression profiles for 20 of the 24 TIFY genes; the expression profiles of PtJAZ8, PtPPD2, PtZML4 and PtZML5 were not examined due to unsuccessful primer design. The relative expression profiles of all 20 genes for all nine samples are shown in Figure 4.
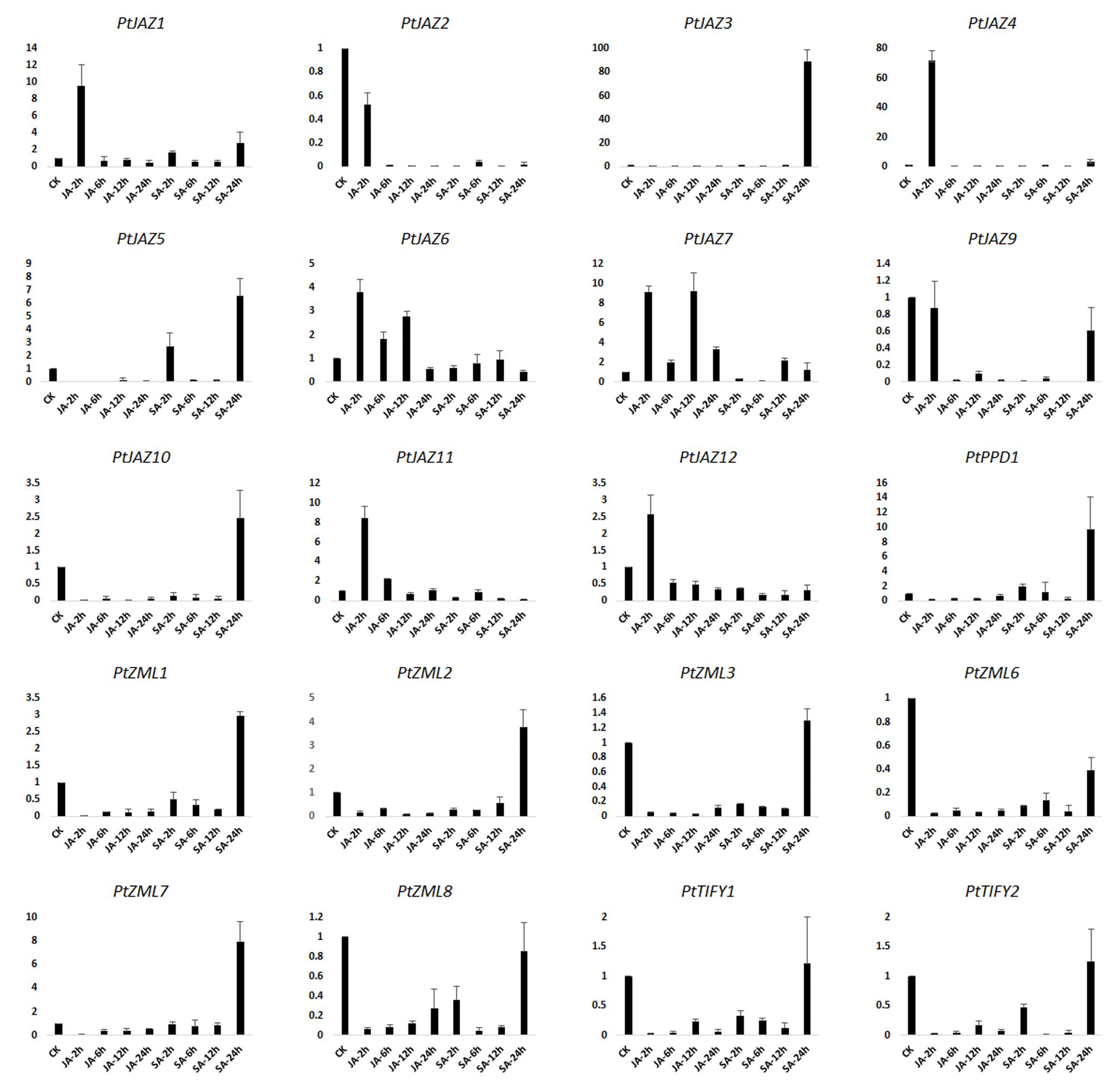
FIGURE 4. qRT-PCR assays of the expression profiles of the poplar TIFY gene family genes after MeJA and SA treatments. CK indicated untreated samples; JA-2 h, JA-6 h, JA-12 h, JA-24 h indicated samples collected from leaves after 2, 6, 12 and 24 h of MeJA treatment, respectively; and SA-2 h, SA-6 h, SA-12 h, and SA-24 h indicated samples collected from leaves after 2, 6, 12, and 24 h of SA treatment, respectively. Y-axis indicates relative expression folds when comparison with CK.
Fold-changes in expression larger than 2.0 or less than 0.5 and also with a p-value less than 0.05 between CK and time points after hormone treatment was considered that gene expression was influenced by MeJA or SA treatment. According to the data presented in Figure 4 and this criterion, gene expression patterns could be classified in four groups. The first group showed that both MeJA and SA increased gene expression at some time points but decreased it at other time points. This group included PtJAZ1, PtJAZ4 and PtJAZ7. The second group showed that both MeJA and SA decreased gene expression at all of the time points (except SA-24 h). This group included PtJAZ2, PtJAZ9, PtZML3, PtZML6, PtZML8, PtTIFY1 and PtTIFY2. Expression of most of the genes in this group was significantly decreased at JA-2 h, JA-6 h, JA-12 h, JA-24 h, SA-2 h, SA-6 h, and SA-12 h. In the third group, MeJA decreased gene expression at most time points, whereas SA increased it at some time points and decreased it at others. This group included PtJAZ3, PtJAZ5, PtJAZ10, PtPPD1, PtZML1, PtZML2 and PtZML7. In the fourth group, MeJA increased gene expression at some time points and decreased gene expression at the other time points, whereas SA decreased gene expression at all of the time points. This group included PtJAZ6, PtJAZ11 and PtJAZ12.
Previous reports suggested JAZ protein might act as TFs in response to rust fungi (Duplessis et al., 2009). To identify TIFY genes that respond to poplar leaf rust disease, we collected leaf samples from the poplar hybrid variety “NL895” that had been infected with M. larici-populina (see Materials and Methods); these samples were designated as CK, PI2, PI4, and PI8. The CK represented samples collected immediately after leaf rust infection, while PI2, PI4, and PI8 represented samples collected from leaves treated by urediniospore suspension of M. larici-populina after 2, 4, and 8 days, respectively. The symptom of leaf rust on leaves of “NL895” at these four selected time points were different. No visible symptom was observed at CK and 2 dpi; while very small white milk-white and plenty of uredinia could be observed at 4 and 8 dpi, respectively (data not shown). Previously, 48 hpi (equal to 2 dpi) was reported to be a key time point for the rust development (Rinaldi et al., 2007). Therefore, our plant material could be able to represent all key developmental stages of M. larici-populina on poplar leaves.
The relative expression of genes for PI2, PI4, and PI8 versus CK for all of the 24 TIFY genes was determined by qRT-PCR assays. We were able to test the expression of 18 of the 24 TIFY genes (Figure 5). According to the expression profiles of these 18 TIFY genes and using the criterion of fold changes less than 0.5 or larger than 2 and also with a p-value less than 0.05 when comparing CK with the other three treatments, we were able to classify them into four groups. The first group showed that gene expression was reduced by M. larici-populina infection at all the three stages of PI2, PI4, and PI8 when comparing with CK. This group included PtJAZ5, PtJAZ7, PtZML3, PtZML5 and PtZML6. The second group showed similar gene expression level of CK and PI8, while gene expression was largely reduced at PI2 and PI4 stages. This group had five genes and it included PtJAZ1, PtJAZ11, PtPPD1, PtTIFY1 and PtZML2. The third group showed that gene expression was similar at CK, PI2 and PI4 and highly increased at PI8. This group had six genes and it included PtJAZ2, PtJAZ6, PtJAZ9, PtJAZ12, PtTIFY2 and PtZML1. The fourth group showed that gene expression was reduced at PI2 and PI4 and highly increased at PI8. This group had two genes and it included PtJAZ3 and PtZML8. Based on these data, we could conclude that all tested TIFY genes respond to M. larici-populina infection and they also showed different responsive patterns.
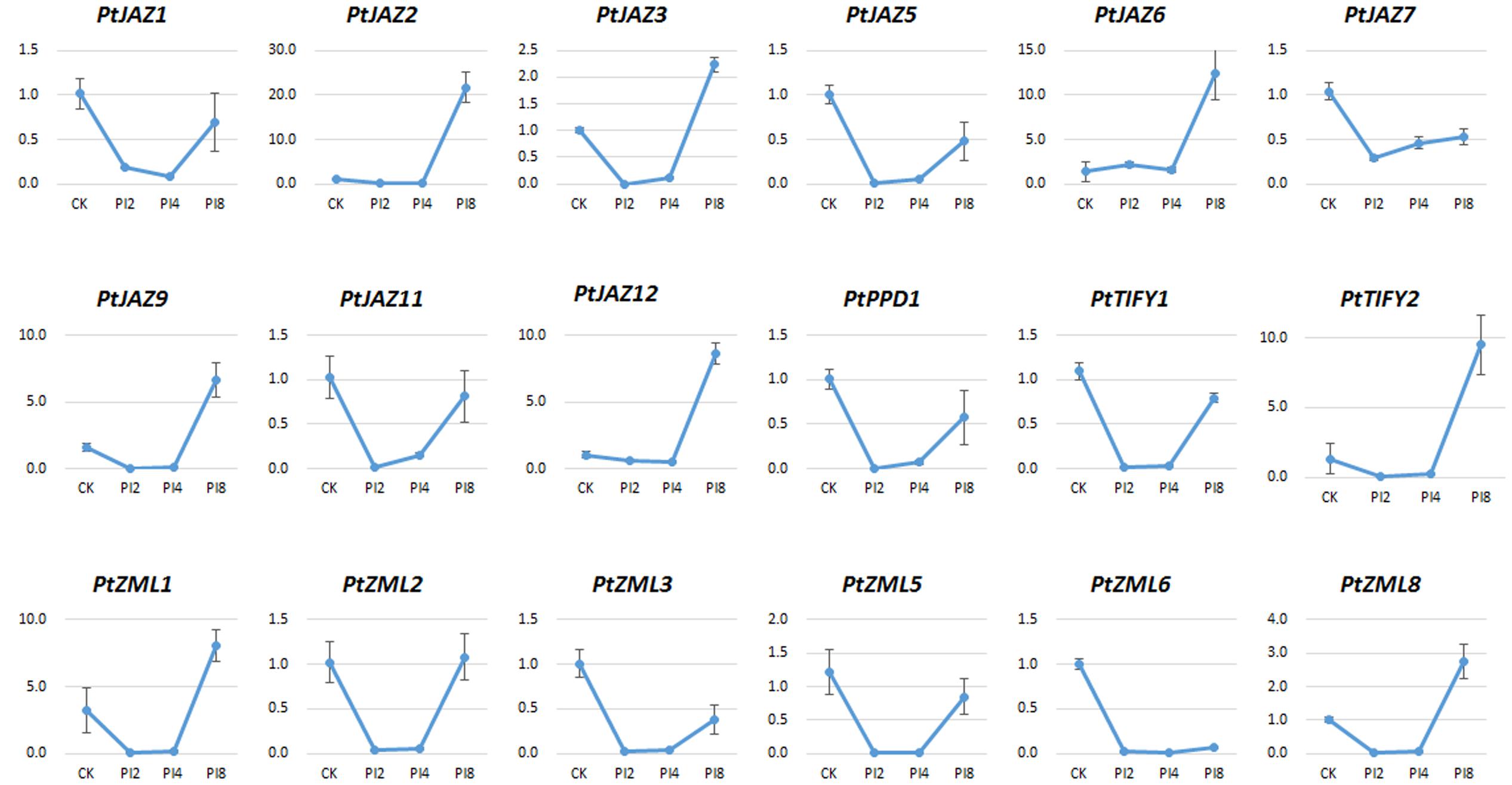
FIGURE 5. qRT-PCR assays of the expression profiles of the poplar TIFY gene family in response to M. larici-populina. The hybrid poplar variety “NL895” was used as plant material. CK indicated the control samples; 2, 4, and 8 dpi indicated samples collected from “NL895” leaves after 2, 4, and 8 days treatment by M. larici-populina inoculation. The virulent M. larici-populina strain used as fungal material in this qRT-PCR assay (see Materials and Methods). Y-axis indicates relative expression folds when comparison with CK.
In this study, we were able to identify 24 TIFY genes in the poplar genome using the P. trichocarpa genome annotation version 3.0. However, the similar total number of TIFY genes between the two identifications could suggest the accuracy of our results. In other plants (e.g., Arabidopsis, rice, apple, soybean, and grape), the number of TIFY genes ranges from 18 to 34 (Ye et al., 2009; Bai et al., 2011; Zhang et al., 2012; Li et al., 2014; Sirhindi et al., 2016). This result suggested that the number of TIFY genes in poplar is within the normal range. In a number of previous studies, the expansion of a given gene family usually indicates preferential roles that they played during the plant’s development. A largely expanded gene family in a plant genome sometimes results in unique characteristics for the plant. For example, when the genomes of P. trichocarpa and P. euphratica were compared, several gene families likely to be involved in tolerance to salt stress were found to be present in significantly greater gene copy numbers within the P. euphratica lineage (Ma et al., 2013). The expansion of TIFY genes in most plants did not show clear differences according to the information we obtained; this phenomenon might indicate that the function of TIFY genes in plants is essential and that they are involved in the basic plant processes.
The duplication pattern of a gene usually reveals how the gene was generated, how its function evolved and what roles it may played in plant growth and development (Wang et al., 2013). WGD is predicted to produce new genomic regions based on an ancestral genome, and this event occurs once or several times over many (sometimes hundreds of) millions of years (Wang et al., 2011). After WGD, the functions of the newly produced genes are usually modified to reduce genomic redundancy and could increase the plant’s ability to adapt to various growth environments (Flagel and Wendel, 2009; Magadum et al., 2013). In other words, gene families that were chiefly produced by WGD usually existed in the “ancestral genome” before duplication. This indicates that the functions of members of a gene family are indispensable for plant growth and development in a given species. In addition, genes in families that were mainly produced by WGD usually evolve slowly (Wang et al., 2013; Soltis and Soltis, 2016). In this study, we found that 19 of the 24 TIFY genes were produced by WGD. This revealed that WGD made a large contribution to the generation of the TIFY gene family in poplar and this result is consistent with reports from the poplar genome sequencing project (Tuskan et al., 2006). It also suggested that the TIFY genes are indispensable for poplar growth and development; this prediction is consistent with the prediction from gene numbers in TIFY families of most of the plants investigated.
Since JA and SA are important hormones for signal transduction when plants encounter pathogens, we chose to investigate whether there was commonality between the TIFY gene expression profiles in poplar leaves after JA/SA treatments and leaf rust pathogen infection. TIFY family genes could be arranged into four groups both after JA/SA treatments and leaf rust pathogen infection. However, we were not able to see a clear rule (data not shown) after analyzing the overlapping TIFY genes between the two classifications. This result suggests that although most of the TIFY genes can respond to a single elicitor, JA or SA treatment and leaf rust pathogen infection, the mechanisms underlying the response of the TIFY genes to each elicitor are different. This also indicates that gene regulation and signal transduction during plant responses to biotic stress are complicated.
Although there was no rule for the TIFY gene expression profiles in poplar leaves between JA/SA treatments and leaf rust pathogen infection, we were still able to obtain some valuable information by analyzing their expression profiles after phytohormone treatments and leaf rust pathogen infection in poplar. First, a large proportion of TIFY family genes can respond to JA/SA treatments and leaf rust pathogen infection. The expressions of all 20 tested TIFY genes were strongly influenced by JA/SA treatments (Figure 4), whereas all 18 tested TIFY genes were strongly influenced by leaf rust pathogen infection. Because TIFY family genes were reported to function in signal transduction when plants encountered various stresses and 65% of its members were found to respond to leaf rust pathogen infection, we speculated that the TIFY gene family in poplar also plays important roles in deploying the defense system against leaf rust via the JA/SA signal transduction pathway. Second, the TIFY genes which showed large changes in expression upon leaf rust pathogen infection could be used as a potential resource to increase resistance to the disease caused by M. larici-populina. The available transgenic platform that has been created in poplar allows overexpression or reduced expression transgenic lines to be generated. For example, the expressions of PtJAZ3, PtJAZ6, PtJAZ9, and PtJAZ15 were largely increased after leaf rust pathogen infection, and we can generate transgenic poplar lines that overexpress these genes (Figure 5). In contrast, the expressions of PtJAZ5, PtJAZ7, PtZML3, PtZML5 and PtZML6 were largely reduced after leaf rust pathogen infection, and we can generate transgenic poplar lines with reduced expression of these genes (Figure 5). Third, several previous studies showed a number of other genes could be able to respond to leaf rust infection and these genes would play important roles in defensive regulation in polar. For instance, a small number of pathogen-defense genes encoding PR-1, chitinases, and other pathogenesis-related proteins were found to be consistently upregulated throughout the whole leaf rust infection (Miranda et al., 2007); while 1,730 and 416 significantly differentially expressed transcripts were also found in the incompatible interaction between poplar and poplar rust (Rinaldi et al., 2007; Azaiez et al., 2009). Combining the previous reports with our study, we might be able to speculate that TIFY genes showed response to leaf rust infection could interact with genes reported in these studies. Finally, although TIFY family genes have been reported to function in signal transduction when plants encounter various stresses, the detailed mechanisms of how they perform their functions are still not clear, especially in the woody perennial plant poplar. Therefore, more work to understand how TIFY genes perform their functions in poplar should be performed in the future.
In summary, we conducted a genome-wide identification of TIFY genes in poplar. The characteristics of this gene family was carefully investigated. We also analyzed their expression profiles in response to phytohormone treatments and Melampsora larici-populina infection in poplar. The results of this study have provided valuable information for further functional characterisation of TIFY genes in poplar and have also provided candidate genes for the improvement of disease resistance in poplar.
NW, WX, HY, and PC organised the entire project and performed the experiments and data analysis. NW and JL wrote and edited this manuscript.
Financial support for this work was provided by the National Natural Science Foundation of China (NSFC accession No.: 31670651) and the Fundamental Research Funds for the Central Universities (No. 520.902-2678436).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful to AJE (American Journal Experts) for improving the English in this paper.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00493/full#supplementary-material
FIGURE S1 | Phylogeny of the TIFY proteins in the four representative species. The protein names start with “At” indicate TIFY genes from Arabidopsis; the protein names start with “Os” indicate TIFY genes from rice; the protein names start with “Vv” indicate TIFY genes from grapes; the protein names start with “Pt” indicate TIFY genes from poplar.
Azaiez, A., Boyle, B., Levee, V., and Seguin, A. (2009). Transcriptome profiling in hybrid poplar following interactions with Melampsora rust fungi. Mol. Plant Microbe Interact. 22, 190–200. doi: 10.1094/MPMI-22-2-0190
Bai, Y., Meng, Y., Huang, D., Qi, Y., and Chen, M. (2011). Origin and evolutionary analysis of the plant-specific TIFY transcription factor family. Genomics 98, 128–136. doi: 10.1016/j.ygeno.2011.05.002
Chung, H. S., and Howe, G. A. (2009). A critical role for the TIFY motif in repression of jasmonate signaling by a stabilized splice variant of the JASMONATE ZIM-domain protein JAZ10 in Arabidopsis. Plant Cell 21, 131–145. doi: 10.1105/tpc.108.064097
de Hoon, M. J., Imoto, S., Nolan, J., and Miyano, S. (2004). Open source clustering software. Bioinformatics 20, 1453–1454. doi: 10.1093/bioinformatics/bth078
Duplessis, S., Major, I., Martin, F., and Séguin, A. (2009). Poplar and pathogen interactions: insights from populus genome-wide analyses of resistance and defense gene families and gene expression profiling. Crit. Rev. Plant Sci. 28, 309–334. doi: 10.1080/07352680903241063
Flagel, L. E., and Wendel, J. F. (2009). Gene duplication and evolutionary novelty in plants. New Phytol. 183, 557–564. doi: 10.1111/j.1469-8137.2009.02923.x
He, D. H., Lei, Z. P., Tang, B. S., Xing, H. Y., Zhao, J. X., and Jing, Y. L. (2015). Identification and analysis of the TIFY gene family in Gossypium raimondii. Genet. Mol. Res. 14, 10119–10138. doi: 10.4238/2015.August.21.19
Li, X., Yin, X., Wang, H., Li, J., Guo, C., Gao, H., et al. (2014). Genome-wide identification and analysis of the apple (Malus (domestica Borkh.) TIFY gene family. Tree Genet. Genomes 11, 1–13. doi: 10.1016/j.plaphy.2013.07.017
Li, Y., Xu, M., Wang, N., and Li, Y. (2015). A JAZ protein in Astragalus sinicus interacts with a leghemoglobin through the TIFY domain and is involved in nodule development and nitrogen fixation. PLoS ONE 10:e0139964. doi: 10.1371/journal.pone.0139964
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Ma, T., Wang, J., Zhou, G., Yue, Z., Hu, Q., Chen, Y., et al. (2013). Genomic insights into salt adaptation in a desert poplar. Nat. Commun. 4:2797. doi: 10.1038/ncomms3797
Magadum, S., Banerjee, U., Murugan, P., Gangapur, D., and Ravikesavan, R. (2013). Gene duplication as a major force in evolution. J. Genet. 92, 155–161. doi: 10.1007/s12041-013-0212-8
Miranda, M., Ralph, S. G., Mellway, R., White, R., Heath, M. C., Bohlmann, J., et al. (2007). The transcriptional response of hybrid poplar (Populus trichocarpa x P. deltoides) to infection by Melampsora medusae leaf rust involves induction of flavonoid pathway genes leading to the accumulation of proanthocyanidins. Mol. Plant Microbe Interact. 20, 816–831. doi: 10.1094/MPMI-20-7-0816
Nishii, A., Takemura, M., Fujita, H., Shikata, M., Yokota, A., and Kohchi, T. (2000). Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 64, 1402–1409. doi: 10.1271/bbb.64.1402
Niu, Y., Figueroa, P., and Browse, J. (2011). Characterization of JAZ-interacting bHLH transcription factors that regulate jasmonate responses in Arabidopsis. J. Exp. Bot. 62, 2143–2154. doi: 10.1093/jxb/erq408
Pauwels, L., and Goossens, A. (2011). The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23, 3089–3100. doi: 10.1105/tpc.111.089300
Qi, T., Huang, H., Wu, D., Yan, J., Qi, Y., Song, S., et al. (2014). Arabidopsis DELLA and JAZ proteins bind the WD-repeat/bHLH/MYB complex to modulate gibberellin and jasmonate signaling synergy. Plant Cell 26, 1118–1133. doi: 10.1105/tpc.113.121731
Rinaldi, C., Kohler, A., Frey, P., Duchaussoy, F., Ningre, N., Couloux, A., et al. (2007). Transcript profiling of poplar leaves upon infection with compatible and incompatible strains of the foliar rust Melampsora larici-populina. Plant Physiol. 144, 347–366. doi: 10.1104/pp.106.094987
Sirhindi, G., Sharma, P., Arya, P., Goel, P., Kumar, G., Acharya, V., et al. (2016). Genome-wide characterization and expression profiling of TIFY gene family in pigeonpea (Cajanus cajan (L.) Millsp.) under copper stress. J. Plant Biochem. Biotechnol. 25, 301–310. doi: 10.1007/s13562-015-0342-6
Soltis, P. S., and Soltis, D. E. (2016). Ancient WGD events as drivers of key innovations in angiosperms. Curr. Opin. Plant Biol. 30, 159–165. doi: 10.1016/j.pbi.2016.03.015
Staswick, P. E. (2008). JAZing up jasmonate signaling. Trends Plant Sci. 13, 66–71. doi: 10.1016/j.tplants.2007.11.011
Sundell, D., Mannapperuma, C., Netotea, S., Delhomme, N., Lin, Y. C., Sjodin, A., et al. (2015). the plant genome integrative explorer resource: plantGenIE.org. New Phytol. 208, 1149–1156. doi: 10.1111/nph.13557
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Taniguchi, S., Hosokawa-Shinonaga, Y., Tamaoki, D., Yamada, S., Akimitsu, K., and Gomi, K. (2014). Jasmonate induction of the monoterpene linalool confers resistance to rice bacterial blight and its biosynthesis is regulated by JAZ protein in rice. Plant Cell Environ. 37, 451–461. doi: 10.1111/pce.12169
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Vanholme, B., Grunewald, W., Bateman, A., Kohchi, T., and Gheysen, G. (2007). The tify family previously known as ZIM. Trends Plant Sci. 12, 239–244. doi: 10.1016/j.tplants.2007.04.004
Wang, N., Cao, P., Xia, W., Fang, L. C., and Yu, H. Y. (2017). Identification and characterization of long non-coding RNAs in response to early infection by Melampsora larici-populina using genome-wide high-throughput RNA sequencing. Tree Genet. Genomes 13, 34. doi: 10.1007/s11295-017-1116-1
Wang, N., Xiang, Y., Fang, L., Wang, Y., Xin, H., and Li, S. (2013). Patterns of gene duplication and their contribution to expansion of gene families in grapevine. Plant Mol. Biol. Rep. 31, 852–861. doi: 10.1007/s11105-013-0556-5
Wang, Y., Wang, X., Tang, H., Tan, X., Ficklin, S. P., Feltus, F. A., et al. (2011). Modes of gene duplication contribute differently to genetic novelty and redundancy, but show parallels across divergent angiosperms. PLoS ONE 6:e28150. doi: 10.1371/journal.pone.0028150
White, D. W. (2006). PEAPOD regulates lamina size and curvature in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 13238–13243. doi: 10.1073/pnas.0604349103
Wilkins, O., Waldron, L., Nahal, H., Provart, N. J., and Campbell, M. M. (2009). Genotype and time of day shape the Populus drought response. Plant J. 60, 703–715. doi: 10.1111/j.1365-313X.2009.03993.x
Wullschleger, S. D., Weston, D. J., Difazio, S. P., and Tuskan, G. A. (2013). Revisiting the sequencing of the first tree genome: Populus trichocarpa. Tree Physiol. 33, 357–364. doi: 10.1093/treephys/tps081
Yamada, S., Kano, A., Tamaoki, D., Miyamoto, A., Shishido, H., Miyoshi, S., et al. (2012). Involvement of OsJAZ8 in jasmonate-induced resistance to bacterial blight in rice. Plant Cell Physiol. 53, 2060–2072. doi: 10.1093/pcp/pcs145
Ye, H., Du, H., Tang, N., Li, X., and Xiong, L. (2009). Identification and expression profiling analysis of TIFY family genes involved in stress and phytohormone responses in rice. Plant Mol. Biol. 71, 291–305. doi: 10.1007/s11103-009-9524-8
Zhang, L., You, J., and Chan, Z. (2015). Identification and characterization of TIFY family genes in Brachypodium distachyon. J. Plant Res. 128, 995–1005. doi: 10.1007/s10265-015-0755-2
Zhang, Y., Gao, M., Singer, S. D., Fei, Z., Wang, H., and Wang, X. (2012). Genome-wide identification and analysis of the TIFY gene family in grape. PLoS ONE 7:e44465. doi: 10.1371/journal.pone.0044465
Zhang, Y., Shi, X., Li, B., Zhang, Q., Liang, W., and Wang, C. (2016). Salicylic acid confers enhanced resistance to Glomerella leaf spot in apple. Plant Physiol. Biochem. 106, 64–72. doi: 10.1016/j.plaphy.2016.04.047
Zhu, D., Bai, X., Chen, C., Chen, Q., Cai, H., Li, Y., et al. (2011). GsTIFY10, a novel positive regulator of plant tolerance to bicarbonate stress and a repressor of jasmonate signaling. Plant Mol. Biol. 77, 285–297. doi: 10.1007/s11103-011-9810-0
Zhu, D., Bai, X., Luo, X., Chen, Q., Cai, H., Ji, W., et al. (2013). Identification of wild soybean (Glycine soja) TIFY family genes and their expression profiling analysis under bicarbonate stress. Plant Cell Rep. 32, 263–272. doi: 10.1007/s00299-012-1360-7
Keywords: TIFY domain gene family, poplar, jasmonic acid, salicylic acid, gene expression, Melampsora larici-populina
Citation: Xia W, Yu H, Cao P, Luo J and Wang N (2017) Identification of TIFY Family Genes and Analysis of Their Expression Profiles in Response to Phytohormone Treatments and Melampsora larici-populina Infection in Poplar. Front. Plant Sci. 8:493. doi: 10.3389/fpls.2017.00493
Received: 06 January 2017; Accepted: 21 March 2017;
Published: 05 April 2017.
Edited by:
Gero Benckiser, University of Giessen, GermanyReviewed by:
Daolong Dou, Nanjing Agricultural University, ChinaCopyright © 2017 Xia, Yu, Cao, Luo and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nian Wang, d2FuZ25AbWFpbC5oemF1LmVkdS5jbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.