
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 01 March 2017
Sec. Plant Breeding
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.00269
This article is part of the Research Topic Advances in Parasitic Weed research View all 28 articles
 Lilach Iasur Kruh1,2
Lilach Iasur Kruh1,2 Tamar Lahav3
Tamar Lahav3 Jacline Abu-Nassar1
Jacline Abu-Nassar1 Guy Achdari1
Guy Achdari1 Raghda Salami2
Raghda Salami2 Shiri Freilich3
Shiri Freilich3 Radi Aly1*
Radi Aly1*
Broomrapes (Phelipanche/Orobanche spp.) are holoparasitic plants that subsist on the roots of a variety of agricultural crops, establishing direct connections with the host vascular system. This connection allows for the exchange of various substances and a possible exchange of endophytic microorganisms that inhabit the internal tissues of both plants. To shed some light on bacterial interactions occurring between the parasitic Phelipanche aegyptiaca and its host tomato, we characterized the endophytic composition in the parasite during the parasitization process and ascertained if these changes were accompanied by changes to endophytes in the host root. Endophyte communities of the parasitic weed were significantly different from that of the non-parasitized tomato root but no significant differences were observed between the parasite and its host after parasitization, suggesting the occurrence of bacterial exchange between these two plants. Moreover, the P. aegyptiaca endophytic community composition showed a clear shift from gram negative to gram-positive bacteria at different developmental stages of the parasite life cycle. To examine possible functions of the endophytic bacteria in both the host and the parasite plants, a number of unique bacterial candidates were isolated and characterized. Results showed that a Pseudomonas strain PhelS10, originating from the tomato roots, suppressed approximately 80% of P. aegyptiaca seed germination and significantly reduced P. aegyptiaca parasitism. The information gleaned in the present study regarding the endophytic microbial communities in this unique ecological system of two plants connected by their vascular system, highlights the potential of exploiting alternative environmentally friendly approaches for parasitic weed control.
Phelipanche aegyptiaca (broomrapes) is an obligate holoparasite that attacks the roots of almost all economically important crops in the Solanaceae, Fabaceae, Compositae, Brassicaceae, and Umbelliferae plant families (Parker and Riches, 1993; Westwood et al., 2010). This parasitic weed invades host plants by using a highly specialized detection system for strigolactones (hormones secreted by the host roots), whose presence is essential for the germination of parasite seeds (Yoder, 1999; Bouwmeester et al., 2003; Joel et al., 2006; Cardoso et al., 2011). Lacking functional roots and a photosynthetic system, the parasite develops special intrusive organs (haustorium) that directly connect to the vascular system of the host plants (Westwood et al., 2010; Joel et al., 2013). By developing a metabolic sink stronger than that of the host, the parasite channels the flow of water and nutrients from the host, thereby damaging crop (its host plant) development. Following successful attachment to the host root, the adjacent broomrape tissue grows into a bulbous structure called a tubercle (spider stage) from which short root-like organs arise that can form secondary attachments to neighboring host roots. After approximately 4 weeks of growth, a floral meristem is produced (shoot stage), which emerges aboveground to flower and disseminate seeds.
This parasite is the main limiting factor in processing-tomato production in numerous Middle East countries (Dor et al., 2014). A wide variety of parasitic weed control methods have been applied in attempts to control broomrape (Joel et al., 2006; Aly, 2007; Aly et al., 2009; Cochavi et al., 2016), most of which are based on chemical sprays that can be windborne and toxic to non-target plants. Therefore there is a need to find alternative solutions to reduce plant–plant parasitization.
One such alternative is to harness endophytic bacteria that naturally inhabit the internal tissues of most plants (Hallmann, 2001). These bacteria often play important beneficial roles in numerous aspects of their host plant’s biology, including enhanced host growth rate, acceleration of seed germination, increased stress tolerance, and the provision of critical nutrients to the host. Endophytic bacteria may also contribute resistance to their hosts by suppressing pathogens and enhancing the plant’s immune system (Azevedo et al., 2000; Rosenblueth and Martínez-Romero, 2006; Ryan et al., 2008; East, 2013). Furthermore, as recently reviewed by Joel et al. (2013), plant endophytes may influence the interaction of their hosts with parasitic weeds, For example the bacterium Azospirillum brasilense inhibits seed germination and radical elongation in the broomrape P. aegyptiaca; Pseudomonas fluorescens reduces both the quantity and biomass of the broomrape Orobanche foetida (Zermane et al., 2007); and the bacterium Rhizobium spp. reduces not only seed germination of O. foetida but also the number of tubercles on its host’s (chickpea) roots (Hemissi et al., 2013). Despite these reports, to date, there is no published information regarding endophytic bacteria inhabiting the parasitic weed during the establishment of parasitism. Furthermore, the parasite and host connect through their vascular system forming a unique ecological system that potentially enables movement of bacteria from one plant to another, resulting in a host-parasite-endophyte triangle. Therefore, the first step in clarifying the role of these bacteria in the parasitism is to investigate the endophytic communities during the phases of parasitization and compared them with parasitized and non- parasitized host tissues.
To do so, we used tomato (Solanum lycopersicum) as a host and P. aegyptiaca as the parasitic weed.
The parasitic weed and its host were grown as was previously described (Eizenberg et al., 2004): Solanum lycopersicum L. ‘MP-1’ plants were served as hosts for P. aegyptiaca parasitization. The parasitic seeds were collected from an infested tomato field near Qiryat Shemona (northern Israel), dried and kept at 8°C until use. The host plant was planted into 4 L pots filled with soil (light-medium clay with 63% sand, 12% silt, and 22% clay) and grown in a greenhouse under natural lighting with an average 14 h of daylight and a temperature of 20 ± 6°C. These plants were watered and fertilized as needed. Four developmental stages of P. aegyptiaca were sampled: seeds, pre-haustorium stage, tubercle (spider stage) and shoot. To ensure that only endophytic bacteria from the plant tissue were being examined, samples were surface-sterilized twice by 2-min incubation in 70% ethanol and 10 min in 6% sodium hypochlorite, followed by a double rinsing in sterile double-distilled water (DDW) (10 min each). To deprive contamination by external bacteria, 100 μl of DDW from the second wash was plated on nutrient agar by Drigalski spatula. In addition, we examined the plant surface by general bacterial probe using FISH analysis (Supplementary Figure S1). No external contamination was detected.
DNA was extracted from plant tissue using the cetyltrimethylammonium bromide (CTAB) method (Chen and Ronald, 1999).
DNA from sterilized tissues of non-parasitized tomato roots, parasitized tomato roots (collected a few cm from parasite-attachment point) and from the tubercle of the parasite (spider stage). PCR was performed using general 16S rRNA bacterial primers, which reduce plastid amplification (63F+1401R) (Supplementary Table S1). The PCR products were sent for sequencing to the DNA Services Facility (Chicago, IL, USA) and 10 μl of each reaction was kept at −20°C as a reference. Sequencing was performed by high-throughput amplicon sequencing using the Illumina MiSeq platform at Research and Testing Laboratory (Lubbock, TX, USA) with the primers 515F+806R (Supplementary Table S1). We used five replicates for each treatment.
To identify the bacterial community associated with P. aegyptiaca, DNA was extracted at different stages of the parasite development (pre-haustorium, tubercles and shoots) and PCR analysis was performed using general 16S rRNA bacterial primers, which reduce plastid amplification (27F+783R). PCR products were isolated with AgencountAMPure® XP beads (Beckman Coulter, Nyon, Switzerland) and next generation sequencing was performed by Ion TorrentTM sequencer (Life Technologies, Grand Island, NY, USA) using primers 27F+338R (Supplementary Table S1). We used three replicates for each treatment.
Two mass-sequencing methods and primer sets were applied to the parasitic weeds at the spider stage and the resultant community composition data were similar, indicating that both can be used interchangeably. Still, we treated these two dataset independently (Figures 1, 2).
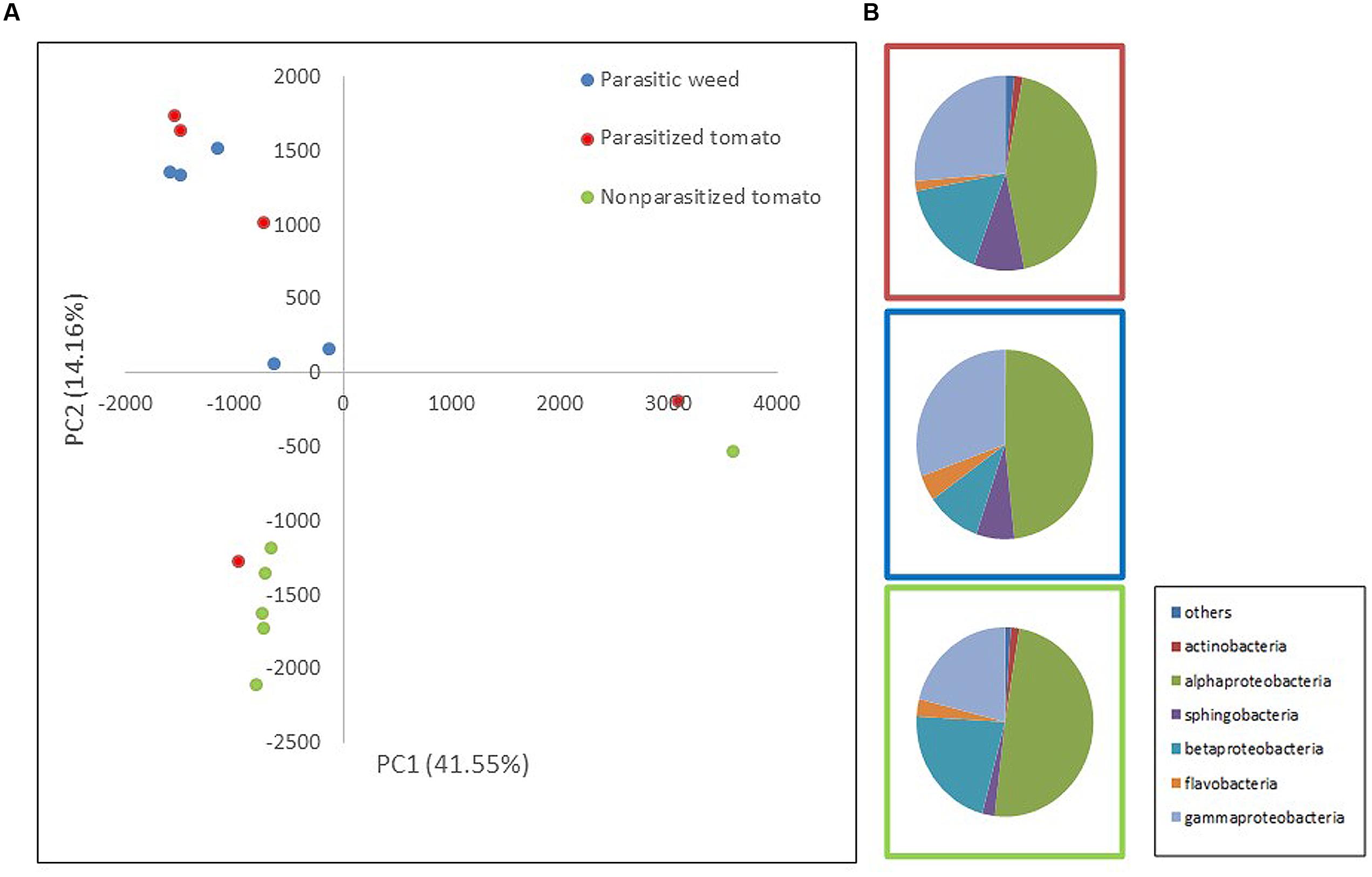
FIGURE 1. Comparison of endophytic communities based on mass sequencing of ribosomal 16S rRNA gene. The bacterial community composition was determined for parasitized tomato root (red), parasite (Phelipanche aegyptiaca – spider stage) (blue) and non-parasitized tomato root (green) in five repeats each. Sequences were classified into operational taxonomic units using a 97% similarity threshold. (A) The first and second dimensions of PCA analysis. (B) Relative abundance of bacterial classes among treatments (an average of all five repeats).
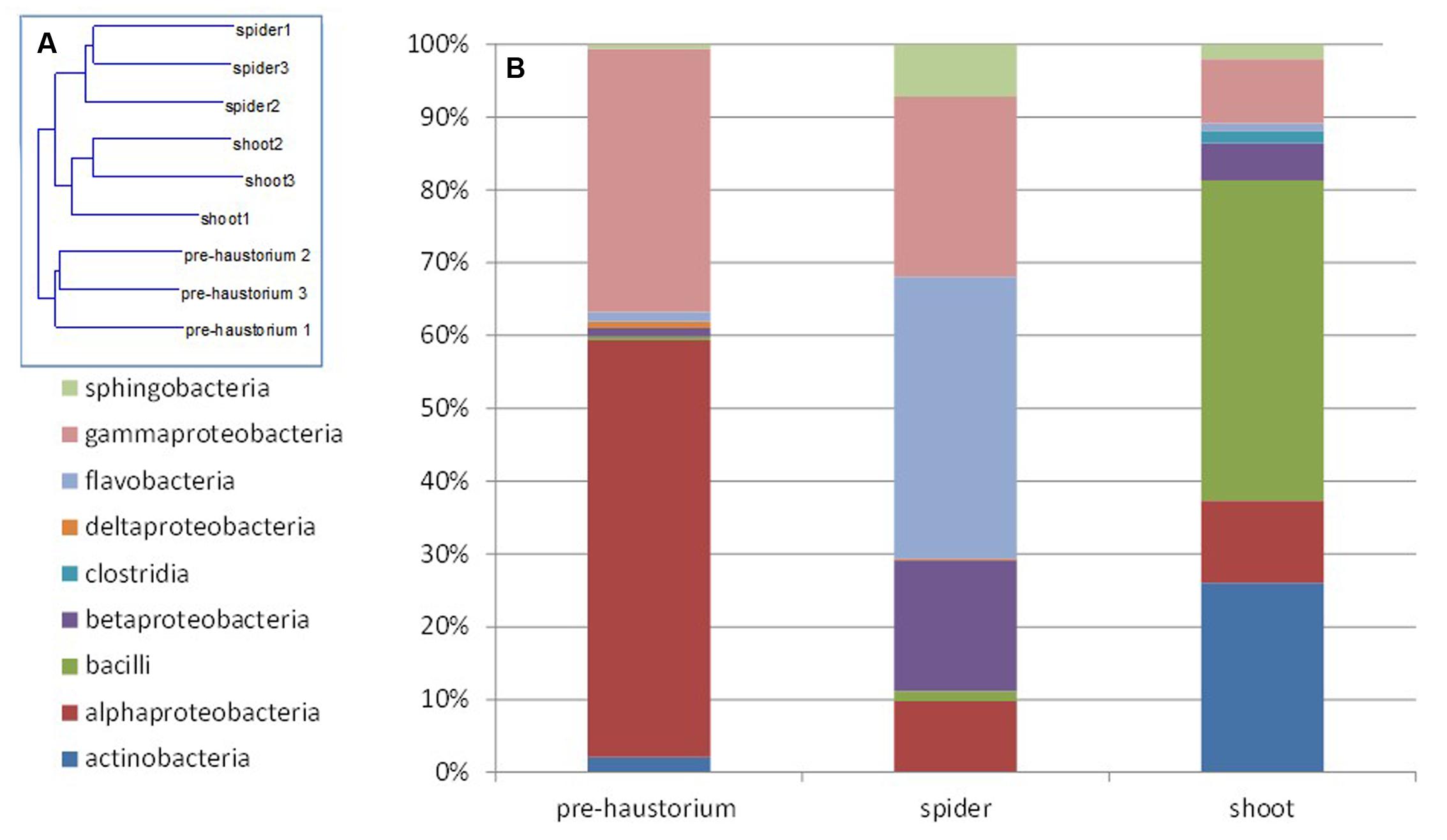
FIGURE 2. Endophytic community composition at different stages of the parasite development as obtained from 16S rRNA gene mass sequencing. The bacterial community composition was determined for P. aegyptiaca germinated seeds before attachment to the host root (pre-haustorium), P. aegyptiaca tubercles (spider stage) and shoot, after attachment to the host root. Sequences were classified into operational taxonomic units using a 97% similarity threshold. (A) Neighbor-joining tree calculated by Rho. (B) Relative abundance of bacterial class at different parasite developmental stages.
Retrieved sequences were analyzed using MOTHUR software (Schloss et al., 2009). Sequences shorter than 150 bp, as well as those of low quality (multiple N, chimeras, etc.), were omitted. Bacterial sequences were aligned using the Silva-compatible alignment database and a distance matrix was calculated. Sequences were grouped into OTUs at a 97% sequence similarity threshold (i.e., sequences that differed by 3% were clustered in the same OTU). A “sub.sample” command was performed and all samples were adjusted, by random selection, to the sample with the lowest number of sequences (13000 and 1100 sequences per sample for Illumina and ion torrent data, respectively). “Indicator” command was performed in order to identify the specific OTUs (indicator species) of each treatment. Diversity indices, principal component analysis (PCA) and ANOVA statistics (Bray–Curtis distance measure) were calculated using PAST software (Hammer et al., 2001). Diversity indices (Supplementary Tables S2, S3) showing observed OTUs, Chao-1 index of richness – representing the predicted number of OTUs in each sample, Dominance – representing relative abundance distribution among species within each sample and Shannon index of diversity.
In order to make sure that the bacteria in the current study are endophytes, we sterilized the examined plant tissue twice as specified above. Surface-sterilized tissue (100 mg) was homogenized with 1 ml of sterile saline solution (9% NaCl) by sterilized pestle. Lysate (100 μL) was spread on NA plates and kept at 28°C in the dark. Three days after incubation, bacterial isolates were counted and classified according to different colony morphologies. Five repeats were carried out for each treatment.
To determine the phylogenetic affiliation of the bacterial isolates, single colonies were collected and subjected to PCR analysis using 27F+1513R primers targeting the 16S rRNA gene by direct colony PCR method as was previously described (Iasur-Kruh et al., 2011) with primers 27F and 1513R (Supplementary Table S1). The PCR procedure was as follows: DNA was denatured at 95°C for 5 min, followed by 30 cycles at 95°C for 30 s each, 57°C for 30 s and 72°C for 1 min, followed by 5 min at 72°C. The PCR product was sequenced by 3130xl Genetic Analyzer (Applied Biosystems). Phylogenetic affiliation of the isolates was determined by comparing with sequences obtained from the NCBI GenBank database. These strains are kept in glycerol at −20°C in our laboratory collection.
Parasite seeds were surface-sterilized, dispersed on Whatman GF/A glass-fiber filters (0.7 cm diameter), covered with the same filter and placed on a Petri dish. After 1 week GR24 (1 mg L-1) was added to the disks. To examine the effect of different isolates on the germination, each isolate was grown in LB (Luria broth- Life Technologies, Israel) overnight at 28°C, centrifuged, the pellet was washed with DDW and the culture was adapted to an optical density at 600 nm (OD600) of 0.3. The culture was added to the disks with the parasite seeds. After incubation in the dark at 26°C for 7 days, the seeds were rated for seed germination in comparison to seed germination control (without bacteria). The experiment was conducted in five repeats for each isolate.
We used an in planta system in which tomato plants were transplanted into polyethylene bags and kept moist with half-strength nutrient solution (Hoagland and Arnon, 1950). Plant growth conditions were 25°C, with 14 h light at 100 μE/s m2. Surface-sterilized P. aegyptiaca seeds were applied on and around the host roots with care taken to achieve even inoculation of all plant roots. After allowing 7 days for parasite seed preconditioning, 10 mL of 2 mg L−1 GR24, a germination stimulant (Mangnus et al., 1992), was added to each bag to synchronize the germination of the P. aegyptiaca seeds. To examine the effect of chosen isolates on parasite development, it was grown in LB overnight at 28°C, centrifuged, the pellet was washed with DDW and the culture was adapted to an OD600 of 0.4 (equivalent to 1.00E+08 cfu∖ml). The culture was added to the polyethylene bags. After 2–3 weeks, parasitism was evaluated by counting the number of live and dead tubercles on each plant using a binocular microscope. The experiment was conducted in five repeats for each treatment (control and endophytic isolate).
Sequences obtained from mass sequencing were deposited in the European Nucleotide Archive (ENA) (study accession number: PRJEB7137). Sequences from isolated endophytes were deposited in GenBank (NCBI) (KP219403-KP219415).
To assess whether bacterial community composition changes following attachment of the parasite (P. aegyptiaca) to its host (tomato), we compared the endophytic communities in parasitized and non-parasitized host roots with those found in the parasite during ongoing parasitization (i.e., occurring during the spider stage, the stage of attachment to the host). The results showed that endophytic communities of the parasite and the non-parasitized host differed significantly (t-test; p = 0.0048). Furthermore, the overall community of the parasitized host root did not differ from that of the parasite on the one hand or from that of the non-parasitized host root (t-test; p > 0.05) (Figure 1A) on the other. A greater number of sphingobacterial sequences were found in the parasitized tomato roots and in the parasite than in the non-parasitized tomato roots (t-test; p = 0.03) (Figure 1B). We observed higher numbers of beta-Proteobacteria and fewer gamma-Proteobacteria in the tomato roots (both non-parasitized and parasitized tomato) in comparison to endophytic community structure of the parasitic weed. Furthermore, there was an introduction of additional taxonomic groups such as actinomycetes in the parasitic weed.
The most abundant OTUs, belonging to the proteobacteria genera; Rhizobium, Pseudomonas, Comamonadaceae sp., Sphingomonas and Burkholderia, are similarly represented in all three “treatments” (parasitized and non-parasitized tomato roots and parasitic weed) with no significant differences between them. However, examination of specific indicator genera showed that Novosphingobium and Methylophilus (representing 1 and 0.8% of the total sequence dataset, respectively) were specific for the parasitic weed and Devosia (representing 0.5% of the total sequence dataset) was significantly associated with the parasitized tomato root.
Next, we characterized the dynamics of the endophyte communities in the parasitic weed across its life cycle, that is, before and after attachment to the host. FISH analysis clearly showed that endophytic bacteria inhabit the inner tissue of all developmental stages of the parasitic weed (Supplementary Figure S1). The endophyte communities of the two stages of the parasite at the post-attachment stages (spider and shoot) were more similar to each other than to those detected in the parasite prior to attachment to the host (pre-haustorium stage) (Figure 2A). In the pre-attachment stage the dominant classes found in the parasite were alpha- and gamma-Proteobacteria (mostly represented by Sphingomonas and Acinetobacter species), comprising 55 and 35% of the bacterial community composition, respectively (Figure 2B). Just after attachment, at the spider stage, there was a decrease in the proportions of these classes and an increase in the fraction of Flavobacteria and beta-Proteobacteria (mostly representative of the Flavobacterium and Methylophilus genera). An increased community of Bacilli and Actinobacteria (40 and 25%, respectively), mostly representative of Jeotgalibacillus and Propionibacterium, were found at the shoot stage (Figure 2B).
In addition, we isolated bacteria from surface-sterilized host tomato roots and from different surface-sterilized developmental stages of P. aegyptiaca. Similarly, to the mass sequencing results the endophytic bacteria isolated from pre-haustorium stage belonged to alpha- and gamma-Proteobacteria genera; Acinetobacter and Sphingobium as well as Roseomonas and Pseudomonas. Gram positive bacteria were isolated from both the parasite tubercle (spider stage) and its host belonging to Agrococcus and Bacillus as well as Roseomonas and Pseudomonas which are gram negative. Unique isolates were identified for the parasite: Bacillus sp. and Rhizobium sp. and for its host: Pseudomonas sp. The diversity of the isolated endophytes was reduced, containing only one Bacillus species in the shoot stage and in its host (Table 1).
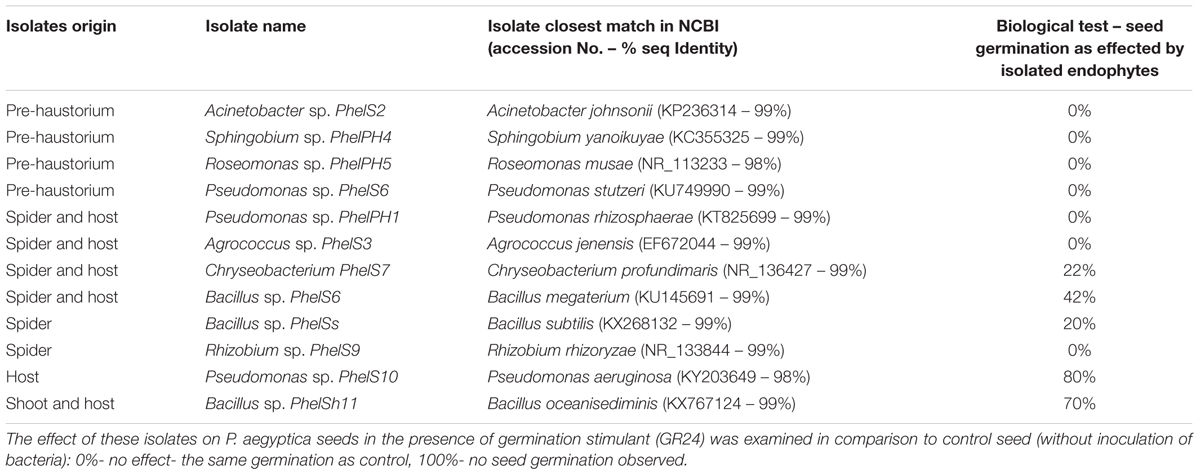
TABLE 1. Phylogenetic identification of bacteria isolated from internal tissues of different developmental stages (pre-haustorium, spider and shoot) of Phelipanche aegyptica and from host (tomato roots).
To examine the effect of different endophytic isolates on the interactions between the parasitic weed and its host, we then examined the effect of the isolates on development of the parasitic weed. We screened for the ability of parasite’s seeds to germinate in vitro in the presence of each isolate (Table 1) and found that two isolates (from the host) reduced the germination of the seeds. Pseudomonas PhelS10 reduced germination by 80% and Bacillus sp. PhelSh11 reduced it by 70%. These bacteria were then chosen for examination in planta using the polyethylene bag test (Figure 3). This test showed that the Bacillus strain dramatically reduced the viability of tomato roots coincident with the reduction of the parasite tubercles, while the Pseudomonas strain reduced the number of tubercles without harming the tomato roots (Figure 3).
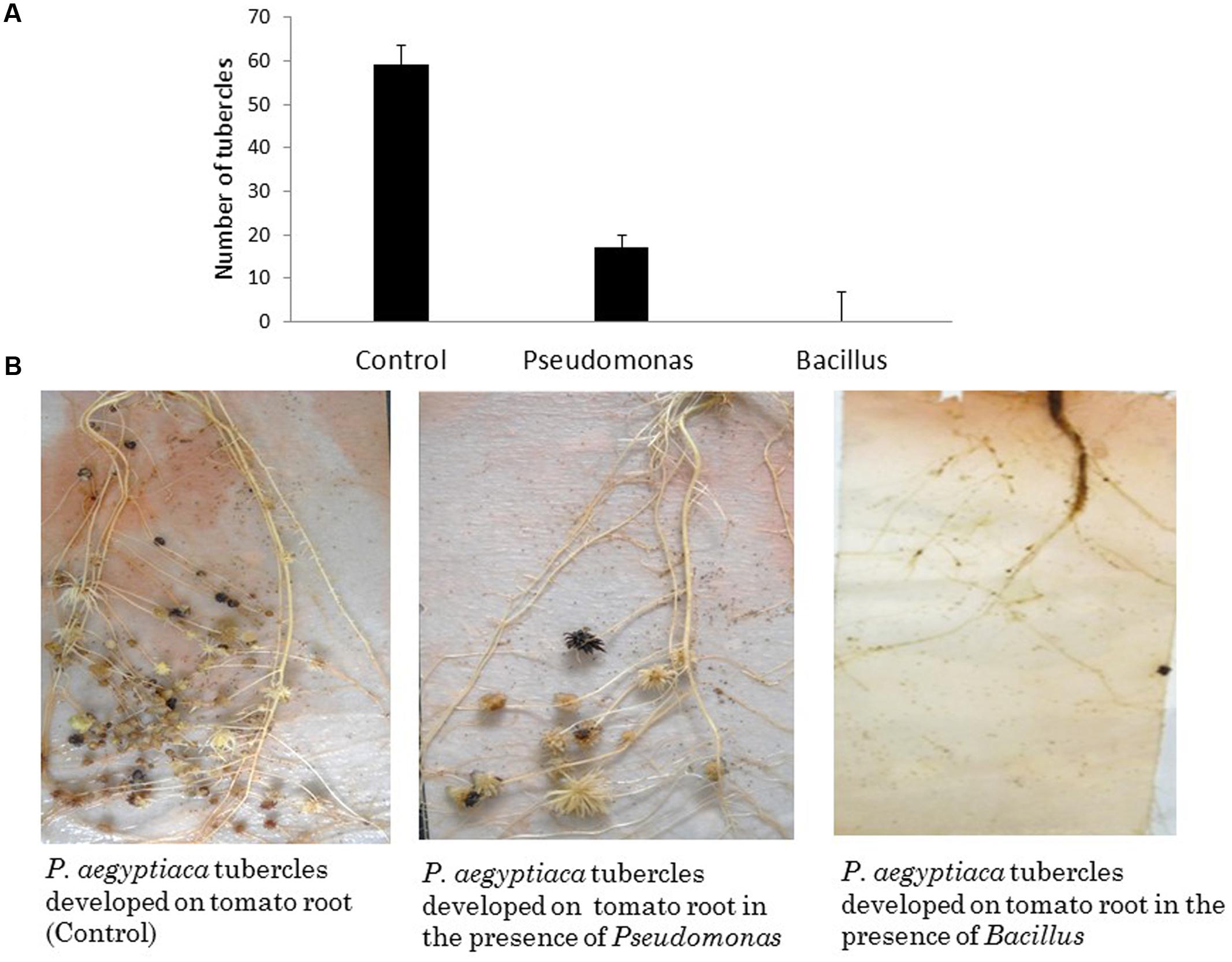
FIGURE 3. The effect of in planta test of two isolated endophytes (approximately 1.00E+08 cfu∖ml each) on parasitic weed development. (A) The number of P. aegyptica tubercles developing in the presence of selected isolates. (B) The effects of the bacterial isolates on P. aegyptica tubercles and on tomato roots in PE system. While Pseudomonas PhelS10 reduced the number of tubercles without harming the host root, the Bacillus sp. PhelSh11 destroyed the host root making the parasitic weed unable to develop. The results are an average of five repeats.
Endophytes are microorganisms (bacteria, fungi, etc.) inhabiting the inner tissue of plants without causing apparent disease. Tomato is hosting both endophytic fungi and endophytic bacteria, some of them showing the ability to promote seed germination and protect tomato plants from pathogens (Larran et al., 2001; Pozo et al., 2002; Shittu et al., 2009; Xu et al., 2014). Furthermore, since endophytic mycorrhizal fungi contain a large amount of bacteria in the ectomycorrhizal root tips (Sbrana et al., 2002), it might spread endophytic bacteria while colonizing the tomato. de Vega et al. (2010) demonstrated tripartite association among a holoparasitic weed (Cytinus), its host Cistaceae species, and mycorrhizal fungi. However, studies on endophytic bacteria in parasitic weed have not been reported yet. In the current work we show that attachment of a parasitic weed to its host has an effect on the bacterial endophytic community composition of both plants. Parasitic weeds are considered ecosystem engineers, due to their ability to induce changes in the abundance and diversity of plants. Such changes may lead to up-stream effects on herbivores, pollinators and seed vectors (Press and Phoenix, 2005). Therefore changes in parasite endophytic communities may affect parasitism and as a consequence plant community makeup. In the current study, we showed for the first time, that bacterial endophytic communities changed with different stages of parasitism, and that they also affect the host’s endophytic composition. The communities inhabiting the root of the host were examined in parallel to the parasite tubercle because this is the stage that the parasite is well-established on the host and the endophytic exchange may occur.
Endophytic bacteria are located in planta, where they are protected from the outside environment on the one hand, but are easily affected by changes occurring in the plant tissue on the other (Ryan et al., 2008). The fact that both the parasitic weed and its host endophytic communities changed following parasitization interaction and become more similar to each other can be explained by the following: the parasite affects the environment of the inner tissue of the host plant. Such changes may be exemplified by the reported changes in metabolic profile of the tomato host that occur following parasitization (Hacham et al., 2016). Thus, the changes in endophytic community occurring in the host plant may be affected by the parasite in an indirect manner.
Alternatively, the endophytic community composition is affected in direct manner by an exchange of endophytic bacteria between the P. aegyptiaca and its host. Indeed, in the present study the endophytic community of the parasitic weed did not differ from that of the parasitized tomato host but was significantly different from the non-parasitized host. However, the fact that bacterial community composition in the parasitized tomato root is not significantly different from either the non-parasitized tomato root or the parasitic weed itself, makes it difficult to determine the direction(s) in which the bacteria are being transferred. In general endophytic bacterial movement can be either apolplasitc via intercellular space (Maheshwari et al., 2013) or may be facilitated through xylem tubes (Rosenblueth and Martínez-Romero, 2006; Compant et al., 2010). Since the P. aegyptiaca tubercle is composed mostly of parenchyma cells which are traversed by both xylem and phloem (Joel et al., 2013), both movement types can serve as potential transmission routes between the parasitic weed and its host through the haustorium bridge.
To date, most of the data in the literature on endophytes and their biological impact on their host plants were established by classical microbiological methods resulting in the loss of much valuable information regarding uncultured endophytes (Hallmann, 2001; Zermane et al., 2007; Hemissi et al., 2013). By using molecular tools we were able to show that even though the dominant endophyte taxonomy in the parasite is similar to that in other known plant endophytes (Lodewyckx et al., 2002), the microbial ecology of these endophytic communities in P. aegyptiaca is greatly affected by its connection to the host.
The greater similarity between the endophyte communities of the parasitic weed in the different post-attachment stages to each other, than to those detected prior to attachment to the host (pre-haustorium stage) indicate that connection to the host is affecting the endophytic community composition of the parasite as well as the host. Isolation of and further investigation of indicator bacterial species from both the host and parasite may therefore be of interest. Indeed investigating indicator bacteria such as Novosphingobium and Methylophilus, from the host P. aegyptiaca will enable the understanding of their unique interaction with the parasitic weed.
Endophytes may be equipped with a rich arsenal of metabolites involved in defense, as well as in interaction with the plant (Bouwmeester et al., 2003; Brader et al., 2014), supporting their host plant in different aspects. Therefore, we further examined specific endophytic bacteria isolated from the tomato-parasitic weed system and assessed their effect on P. aegyptiaca development. In agreement to previous studies (reviewed by Joel et al., 2013), our results showed that two isolates, associated with tomato-plant, reduced the germination of the parasite’s seeds. Similar mechanisms were shown in a number of studies (Azevedo et al., 2000; Eljounaidi et al., 2016), where bacteria obtained from different crops were shown to protect their hosts against pests and diseases. Furthermore, Pseudomonas sp. PhelS10, which originated from the inner tissue of the host tomato root, was able to suppress both seed germination (in vitro) and parasitism (in vivo) of P. aegyptiaca, suggesting it may be used by the host for protection against parasitic weed attack. This protection may be through secreting different substances that suppress the pest and∖or enhancing the plant’s immune system (ISR), enabling higher resistance against the parasite as was demonstrated by other beneficial endophytes against other pathogens (Berg et al., 2005; Compant et al., 2005; Innerebner et al., 2011). Therefore it may be possible to harness these naturally occurring microbial partners as possible bio-controls agents against parasitization, reducing the need for harmful chemical pesticide agents.
In this study, we showed that attachment to the host had a major effect on all tested bacterial parameters of the parasitic weed, suggesting an exchange of endophytes between the host and the parasite. Considering the impact of parasitic weeds on agriculture and the difficulty in establishing efficient control methods further research is required to characterize additional endophytic isolates and to fully optimize this mechanism of resistance. Moreover, a deeper understanding of the relationships in the host-parasite-endophyte triangle may lead to new weed control methods and help alleviate weed-related ecological, agricultural, and economic issues.
RA conceived, planned and supervised the work. LI designed and performed the experiments and analyzed the data. SF analyzed the data. TL, JA-N, GA, and RS contributed in data production.
Agricultural Research Organization, The Volcani Center, Bet-Dagan, Israel, number 566/17.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Thanks to Dr. Stefan Green, Dr. Asaf Sade, and Prof. Harry Paris for their suggestions and scientific support.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00269/full#supplementary-material
Aly, R. (2007). Conventional and biotechnological approaches for control of parasitic weeds. In Vitro Cell. Dev. Biol. Plant 43, 304–317. doi: 10.3109/07388551.2012.743502
Aly, R., Cholakh, H., Joel, D. M., Leibman, D., Steinitz, B., Zelcer, A., et al. (2009). Gene silencing of mannose 6-phosphate reductase in the parasitic weed Orobanche aegyptiaca through the production of homologous dsRNA sequences in the host plant. Plant Biotechnol. 7, 487–498. doi: 10.1111/j.1467-7652.2009.00418.x
Azevedo, J. L., Maccheroni, W. Jr., Pereira, J. O., and De Araújo, W. L. (2000). Endophytic microorganisms: a review on insect control and recent advances on tropical plants. Electron. J. Biotechnol. 3, 15–16. doi: 10.2225/vol3-issue1-fulltext-4
Berg, G., Krechel, A., Ditz, M., Sikora, R. A., Ulrich, A., and Hallmann, J. (2005). Endophytic and ectophytic potato-associated bacterial communities differ in structure and antagonistic function against plant pathogenic fungi. FEMS Microbiol. Ecol. 51, 215–229. doi: 10.1016/j.femsec.2004.08.006
Bouwmeester, H. J., Matusova, R., Zhongkui, S., and Beale, M. H. (2003). Secondary metabolite signalling in host–parasitic plant interactions. Curr. Opin. Plant Biol. 6, 358–364. doi: 10.1016/S1369-5266(03)00065-7
Brader, G., Compant, S., Mitter, B., Trognitz, F., and Sessitsch, A. (2014). Metabolic potential of endophytic bacteria. Curr. Opin. Biotechnol. 27, 30–37. doi: 10.1016/j.copbio.2013.09.012
Cardoso, C., Ruyter-Spira, C., and Bouwmeester, H. J. (2011). Strigolactones and root infestation by plant-parasitic Striga, Orobanche and Phelipanche spp. Plant Sci. 180, 414–420. doi: 10.1016/j.plantsci.2010.11.007
Chen, D., and Ronald, P. (1999). A rapid DNA minipreparation method suitable for AFLP and other PCR applications. Plant Mol. Biol. Rep. 17, 53–57. doi: 10.1023/A:1007585532036
Cochavi, A., Rubin, B., Achdari, G., and Eizenberg, H. (2016). Thermal time model for Egyptian broomrape (Phelipanche aegyptiaca) parasitism dynamics in carrot (Daucus carota L.): field validation. Front. Plant Sci. 7:1807. doi: 10.3389/fpls.2016.01807
Compant, S., Clément, C., and Sessitsch, A. (2010). Plant growth-promoting bacteria in the rhizo- and endosphere of plants: their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 42, 669–678. doi: 10.1016/j.soilbio.2009.11.024
Compant, S., Duffy, B., Nowak, J., and Cle, C. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005
Daims, H., Ramsing, N. B., Schleifer, K. H., and Wagner, M. (2001). Cultivation-independent, semiautomatic determination of absolute bacterial cell numbers in environmental samples by fluorescent in situ hybridization. Appl. Environ. Microbiol. 67, 5810–5818. doi: 10.1128/AEM.67.12.5810-5818.2001
de Vega, C., Arista, M., Ortiz, P. L., and Talavera, S. (2010). Anatomical relations among endophytic holoparasitic angiosperms, autotrophic host plants and mycorrhizal fungi: a novel tripartite interaction. Am. J. Bot. 97, 730–737. doi: 10.3732/ajb.0900147
Dor, E., Eizenberg, H., Joel, D. M., Smirnov, E., Achdari, G., and Hershenhorn, J. (2014). Orobanche palaestina: a potential threat to agricultural crops in Israel. Phytoparasitica 42, 285–291. doi: 10.1007/s12600-013-0368-z
Dowd, S. E., Callaway, T. R., Wolcott, R. D., Sun, Y., McKeehan, T., Hagevoort, R. G., et al. (2008). Evaluation of the bacterial diversity in the feces of cattle using 16S rDNA bacterial tag-encoded FLX amplicon pyrosequencing (bTEFAP). BMC Microbiol. 8:125. doi: 10.1186/1471-2180-8-125
Eizenberg, H., Goldwasser, Y., Golan, S., Plakhine, D., and Hershenhorn, J. (2004). Egyptian broomrape (Orobanche aegyptiaca) control in tomato with sulfonylurea herbicides-greenhouse studies 1. Weed Technol. 18, 490–496. doi: 10.1614/WT-03-023R3
Eljounaidi, K., Lee, S. K., and Bae, H. (2016). Bacterial endophytes as potential biocontrol agents of vascular wilt diseases–review and future prospects. Biol. Control 103, 62–68. doi: 10.1016/j.biocontrol.2016.07.013
Hacham, Y., Hershenhorn, J., Dor, E., and Amir, R. (2016). Primary metabolic profiling of Egyptian broomrape (Phelipanche aegyptiaca) compared to its host tomato roots. J. Plant Physiol. 205, 11–19. doi: 10.1016/j.jplph.2016.08.005
Hallmann, J. (2001). Plant Interactions with Endophytic Bacteria. New York, NY: CABI Publishing, 87–119. doi: 10.1079/9780851995120.0087
Hammer, H. Q., Harper, D. A. T., and Ryan, P. D. (2001). PAST: saleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 9.
Harasawa, R., Pitcher, D. G., Ramírez, A. S., and Bradbury, J. M. (2004). A putative transposase gene in the 16S–23S rRNA intergenic spacer region of Mycoplasma imitans. Microbiology 150, 1023–1029. doi: 10.1099/mic.0.26629-0
Hemissi, I., Mabrouk, Y., Abdi, N., Bouraoui, M., Saidi, M., and Sifi, B. (2013). Growth promotion and protection against Orobanche foetida of chickpea (Cicer aerietinum) by two Rhizobium strains under greenhouse conditions. Afr. J. Biotechnol. 12, 1371–1377.
Hoagland, D. R., and Arnon, D. I. (1950). The Water-Culture Method for Growing Plants without Soil. Berkeley, CA: College of Agriculture, University of California.
Iasur-Kruh, L., Hadar, Y., and Minz, D. (2011). Isolation and bioaugmentation of an estradiol-degrading bacterium and its integration into a mature biofilm. Appl. Environ. Microbiol. 77, 3734–3740. doi: 10.1128/AEM.00691-11
Innerebner, G., Knief, C., and Vorholt, J. A. (2011). Protection of Arabidopsis thaliana against leaf-pathogenic Pseudomonas syringae by Sphingomonas strains in a controlled model system. Appl. Environ. Microbiol. 77, 3202–3210. doi: 10.1128/AEM.00133-11
Joel, D. M., Gressel, J., and Musselman, L. J. (2013). Parasitic Orobanchaceae: Parasitic Mechanisms and Control Strategies. Berlin: Springer-Verlag. doi: 10.1007/978-3-642-38146-1
Joel, D. M., Hershenhorn, Y., Eizenberg, H., Aly, R., Ejeta, G., Rich, P. J., et al. (2006). “Biology and management of weedy root parasites,” in Horticultural Reviews, ed. J. Janick (Hoboken, NJ: John Wiley & Sons), 267–349.
Larran, S., Monaco, C., and Alippi, H. E. (2001). Endophytic fungi in leaves of Lycopersicon esculentum Mill. World J. Microbiol. Biotechnol. 17, 181–184. doi: 10.1023/A:1016670000288
Lodewyckx, C., Vangronsveld, J., Porteous, F., Moore, E., Taghavi, S., Mezgeay, M., et al. (2002). Endophytic bacteria and their potential applications. CRC Crit. Rev. Plant Sci. 21, 583–606. doi: 10.1080/0735-260291044377
Maheshwari, D. K., Meenu, S., and Abhinav, A. (2013). Bacteria in Agrobiology: Crop Productivity. Berlin: Springer Science & Business Media.
Mangnus, E. M., Van Vliet, L. A., Vandenput, D. A., and Zwanenburg, B. (1992). Structural modifications of strigol analogs. Influence of the B and C rings on the bioactivity of the germination stimulant GR24. J. Agric. Food Chem. 40, 1222–1229. doi: 10.1021/jf00019a030
Parker, C., and Riches, C. R. (1993). Parasitic Weeds of the World: Biology and Control. Wallingford, CT: CAB Int.
Pozo, M. J., Cordier, C., Dumas-Gaudot, E., Gianinazzi, S., Barea, J. M., and Azcón-Aguilar, C. (2002). Localized versus systemic effect of arbuscular mycorrhizal fungi on defence responses to Phytophthora infection in tomato plants. J. Exp. Bot. 53, 525–534. doi: 10.1093/jexbot/53.368.525
Press, M. C., and Phoenix, G. K. (2005). Impacts of parasitic plants on natural communities. New phytol. 166, 737–751. doi: 10.1111/j.1469-8137.2005.01358.x
Rosenblueth, M., and Martínez-Romero, E. (2006). Bacterial endophytes and their interactions with hosts. Mol. Plant Microbe Interact. 19, 827–837. doi: 10.1094/MPMI-19-0827
Ryan, R. P., Germaine, K., Franks, A., Ryan, D. J., and Dowling, D. N. (2008). Bacterial endophytes: recent developments and applications. FEMS Microbiol. Lett. 278, 1–9. doi: 10.1111/j.1574-6968.2007.00918.x
Sakai, M., Matsuka, A., Komura, T., and Kanazawa, S. (2004). Application of a new PCR primer for terminal restriction fragment length polymorphism analysis of the bacterial communities in plant roots. J. Microbiol. Methods 59, 81–89. doi: 10.1016/j.mimet.2004.06.005
Sbrana, C., Agnolucci, M., Bedini, S., Lepera, A., Toffanin, A., Giovannetti, M., et al. (2002). Diversity of culturable bacterial populations associated to Tuber borchii ectomycorrhizas and their activity on T. borchii mycelial growth. FEMS Microbiol. Lett. 211, 195–201. doi: 10.1111/j.1574-6968.2002.tb11224.x
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Shittu, H. O., Castroverde, D. C. M., Nazar, R. N., and Robb, J. (2009). Plant-endophyte interplay protects tomato against a virulent Verticillium. Planta 229, 415–426. doi: 10.1007/s00425-008-0840-z
Weisburg, W. G., Barns, S. M., Pelletier, D. A., and Lane, D. J. (1991). 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703. doi: 10.1128/jb.173.2.697-703.1991
Westwood, J. H., Yoder, J. I., and Timko, M. P. (2010). The evolution of parasitism in plants. Trends Plant Sci. 15, 227–235. doi: 10.1016/j.tplants.2010.01.004
Xu, M., Sheng, J., Chen, L., Men, Y., Gan, L., Guo, S., et al. (2014). Bacterial community compositions of tomato (Lycopersicum esculentum Mill.) seeds and plant growth promoting activity of ACC deaminase producing Bacillus subtilis (HYT-12-1) on tomato seedlings. World J. Microbiol. Biotechnol. 30, 835–845. doi: 10.1007/s11274-013-1486-y
Yoder, J. I. (1999). A species-specific recognition system directs haustorium development in the parasitic plant Triphysaria (Scrophulariaceae). Planta 202, 407–413. doi: 10.1007/s004250050144
Keywords: broomrapes, parasitic weed, endophytic bacteria, tomato (Solanum lycopersicum), biocontrol
Citation: Iasur Kruh L, Lahav T, Abu-Nassar J, Achdari G, Salami R, Freilich S and Aly R (2017) Host-Parasite-Bacteria Triangle: The Microbiome of the Parasitic Weed Phelipanche aegyptiaca and Tomato-Solanum lycopersicum (Mill.) as a Host. Front. Plant Sci. 8:269. doi: 10.3389/fpls.2017.00269
Received: 28 December 2016; Accepted: 14 February 2017;
Published: 01 March 2017.
Edited by:
Maurizio Vurro, National Research Council, ItalyReviewed by:
Grama Nanjappa Dhanapal, University of Agricultural Sciences, Bengaluru, IndiaCopyright © 2017 Iasur Kruh, Lahav, Abu-Nassar, Achdari, Salami, Freilich and Aly. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Radi Aly, cmFkaUB2b2xjYW5pLmFncmkuZ292Lmls
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.