- 1Graduate School of Agriculture, Kyoto University, Kyoto, Japan
- 2Crop Production Systems Division, NARO Agricultural Research Center, Tsukuba, Japan
- 3Faculty of Life and Environmental Sciences, University of Tsukuba, Tsukuba, Japan
- 4Plant Genome Engineering Research Unit, Institute of Agrobiological Sciences, National Agriculture and Food Research Organization, Tsukuba, Japan
- 5National Institute for Agro-Environmental Sciences, Tsukuba, Japan
- 6Central Region Agricultural Research Center, National Agriculture and Food Research Organization, Tsu, Japan
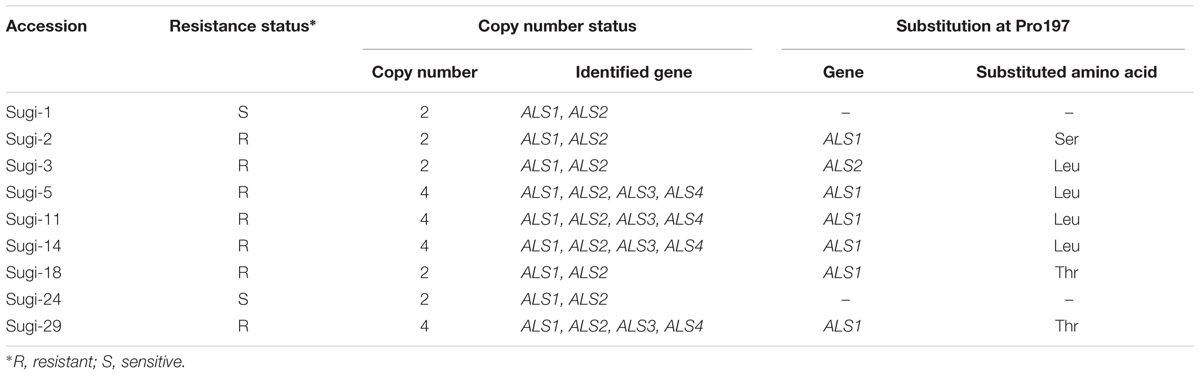
Severe infestations of Alopecurus aequalis (shortawn foxtail), a noxious weed in wheat and barley cropping systems in Japan, can occur even after application of thifensulfuron-methyl, a sulfonylurea (SU) herbicide. In the present study, nine accessions of A. aequalis growing in a single wheat field were tested for sensitivity to thifensulfuron-methyl. Seven of the nine accessions survived application of standard field rates of thifensulfuron-methyl, indicating that severe infestations likely result from herbicide resistance. Acetolactate synthase (ALS) is the target enzyme of SU herbicides. Full-length genes encoding ALS were therefore isolated to determine the mechanism of SU resistance. As a result, differences in ALS gene copy numbers among accessions were revealed. Two copies, ALS1 and ALS2, were conserved in all accessions, while some carried two additional copies, ALS3 and ALS4. A single-base deletion in ALS3 and ALS4 further indicated that they represent pseudogenes. No differences in ploidy level were observed between accessions with two or four copies of the ALS gene, suggesting that copy number varies. Resistant plants were found to carry a mutation in either the ALS1 or ALS2 gene, with all mutations causing an amino acid substitution at the Pro197 residue, which is known to confer SU resistance. Transcription of each ALS gene copy was confirmed by reverse transcription PCR, supporting involvement of these mutations in SU resistance. The information on the copy number and full-length sequences of ALS genes in A. aequalis will aid future analysis of the mechanism of resistance.
Introduction
Herbicides that inhibit acetolactate synthase (ALS) cause depletion of branched chain amino acids such as valine, leucine and isoleucine, leading to plant death (Duggleby et al., 2008; Yu and Powles, 2014). However, recurrent use of these herbicides has resulted in the rapid evolution of herbicide resistance in weeds. Globally, resistance to ALS inhibitors has been reported in 159 weed species (Heap, 2016). ALS inhibitors are categorized into five groups based on their chemical structure: sulfonylurea (SU), triazolopyrimidine (TP), pyrimidinylthiobenzoate (PTB), imidazolinone (IMI), and sulfonylaminocarbonyltriazolinone (SCT). In Japan, SUs are predominantly used due to their excellent crop safety, broad spectrum of weed control and low toxicity to animals. Thus, most cases of resistance to ALS-inhibiting herbicides reported in Japan, such as Monochoria vaginalis (Ohsako and Tominaga, 2007), Schoenoplectus juncoides (Uchino et al., 2007), and Sagittaria trifolia (Iwakami et al., 2014b), have evolved under SU selection (Uchino et al., 2016).
Resistance of weeds to herbicides is caused by target-site and /or non-target-site resistance mechanisms (Powles and Yu, 2010). Target-site resistance involves alterations to the target site such as overproduction and amino acid substitution of the target protein, while non-target-site resistance includes all other mechanisms such as enhanced herbicide metabolism, restricted herbicide translocation and reduced herbicide uptake. In the case of ALS inhibitor resistance, target-site resistance resulting from amino acid substitution frequently occurs (Yu and Powles, 2014). A substitution at one of eight amino acid residues in the ALS protein sequence of Arabidopsis thaliana, Ala122, Pro197, Ala205, Asp376, Arg377, Trp574, Ser653, or Gly654, was found to cause resistance to ALS inhibitors (Yu and Powles, 2014).
Accumulating evidence further suggests that copy numbers of genes encoding herbicide targets also has an effect on the evolution of herbicide resistance. For example, in a polyploid, a single mutation in one homoeologous copy encoding a target enzyme tends to confer lower levels of herbicide resistance compared to diploid plants carrying the same mutation in a single-copy gene (Yu et al., 2013). The locus of a mutated copy among multiple homoeologous copies of genes encoding herbicide targets could also influence the degree of herbicide resistance (Ostlie et al., 2015). In addition, within-species copy number variation (CNV) was previously observed in the gene encoding the glyphosate target enzyme, 5-enolpyruvoylshikimate-3-phosphate synthase (EPSPS), of which a higher copy number was associated with glyphosate resistance (Sammons and Gaines, 2014). It is therefore important to determine copy numbers of genes encoding target enzymes and identify the particular copy carrying a resistance-conferring mutation.
Alopecurus aequalis Sobol. (shortawn foxtail), family Poaceae, is a diploid species distributed throughout Europe, temperate Asia and North America (Cope, 1982). Its strong tillering capacity allows it to out-compete wheat seedlings, causing yield losses of more than 50 % (Guo et al., 2015a). In Japan, A. aequalis is a major weed in barley and wheat fields. Since the early 1990s, management has relied on postemergence application of thifensulfuron-methyl, a SU herbicide. However, resistance to thifensulfuron-methyl was confirmed in 2004, after seven consecutive years of herbicide treatment (Uchikawa et al., 2005). Resistance of A. aequalis to ALS inhibitors was also recently reported in China (Guo et al., 2015b, 2016; Xia et al., 2015). Although mutations in ALS gene causing Pro197Arg, Pro197Thr, or Trp574Leu have been reported, it remains unknown whether these populations are homozygous or heterozygous at the ALS loci and which copies of the ALS gene carry a mutation. In this study, we therefore determined the full-length sequences and copy numbers of ALS genes in A. aequalis, and identified mutations in resistant plants thought to be responsible for resistance to thifensulfuron-methyl. CNV in ALS genes in A. aequalis was also examined.
Materials and Methods
Plant Materials
Seeds of nine A. aequalis plants (hereafter referred to as accessions) were collected in May 2012 from a ∼1-ha wheat field in Kumamoto City, Kumamoto Prefecture, Japan (Table 1). The field was under a rice–wheat cropping system and was severely infested with A. aequalis after thifensulfuron-methyl treatment. Single plants of each accession were grown in a greenhouse and self-pollinated once and the seeds assayed for thifensulfuron-methyl sensitivity. Nucleic acids were also extracted from the seedlings for gene cloning and Southern blot analysis. Individual seedlings from each accession were self-pollinated and used for gene expression and genotyping analyses. An A. japonicus accession was also used in this study, and was collected in May 2012 from a wheat field in Mifune, Kumamoto Prefecture, Japan.
Thifensulfuron-Methyl Dose-Response Assay
Seeds were germinated on 0.6% agar plates in a growth chamber at 25/15°C (day / night) with a 12 h photoperiod. After germination, six seedlings per accession were transplanted in a cell tray filled with soil and kept at an ambient temperature in a vinyl greenhouse at Kyoto University during winter 2016 (January to March). At the 3–4 leaf stage, plants were treated with a commercial formulation of thifensulfuron-methyl (Harmony, DuPont, Tokyo, Japan) at 0, 1/3, 1 and 3× the recommended rate (75 g a.i. ha-1), respectively. Three weeks after thifensulfuron-methyl application, the dry weights of shoots were measured to compare relative growth among the accessions. The experiment was repeated twice with three replications. Results of a single experiment are shown since they were similar between experiments.
Statistical analyses were performed using square root transformed data. One-way ANOVA with Dunnett’s post-test was performed using R version 3.3.1 (R core team, 2016) to determine differences in sensitivity of the Sugi-1 accession with all other accessions.
Isolation and Sequencing of ALS Genes
Isolation and sequencing analysis of ALS genes was carried out using plants of all nine accessions grown in a greenhouse. Green leaves were harvested at the heading stage, snap-frozen and stored at –80°C until use. RNA was isolated using the RNeasy Plant Mini Kit (Qiagen, CA, USA) and genomic DNA removed using the TURBO DNA-free Kit (Life Technologies, CA, USA). Complementary DNA (cDNA) was synthesized from the RNA using the SMART RACE cDNA Amplification Kit (TaKaRa, Otsu, Japan), and extracted using the DNeasy Plant Mini Kit (Qiagen).
Partial ALS genes of all accessions were amplified from genomic DNA samples using KOD FX (Toyobo, Osaka, Japan) with primers designed based on ALS genes from other grass species: forward primer, 5′-AAGGGCGCSGACATCCT-3′; reverse primer, 5′-ATCTGCTGYTGGATGTCCTT-3′. Amplicons were subjected to direct sequencing using BigDye Terminator V3.1 on a 3130xl Genetic Analyzer (Applied Biosystems, CA, USA). Fragments of ALS genes amplified from accessions Sugi-5 and Sugi-24 were cloned into pGEM-T Easy (Promega, Madison, WI, USA) and the inserts sequenced.
The 5′ and 3′ regions of each ALS gene fragment from Sugi-5 were determined by Rapid Amplification of cDNA Ends (RACE) according to either the manufacturer’s instructions for the SMART RACE cDNA Amplification Kit, Thermal Asymmetric Interlaced (TAIL) PCR (Liu and Whittier, 1995) or fusion primer and nested integrated (FPNI) PCR (Wang et al., 2011). The gene-specific primers used for PCR are listed in Table 2. The resulting amplicons were then subjected to direct sequencing or cloned and sequenced as described above. Full-length ALS gene sequences were amplified using KOD FX with the primers listed in Table 3 and the sequences confirmed by direct sequencing.
Phylogenetic Analysis of ALS Protein Sequences
Amino acid sequences of plant ALS genes were obtained from GenBank, the MSU Rice Genome Annotation Project1 or Phytozome v11.02, and a phylogenetic tree generated using MEGA6 (Tamura et al., 2013). Sequences were aligned using ClustalW, and the JTT matrix-based method used to compute evolutionary distances.
Analysis of Genetic Inheritance of the ALS3 and ALS 4 Genes
Seedlings of accession Sugi-5 were self-pollinated twice using the single-seed descent method in the greenhouse at Kyoto University. DNA was then extracted from shoot tissues using the One Step Method according to instructions provided by the manufacturer of KOD FX polymerase. PCR was conducted using KOD FX with primers designed from conserved regions of the ALS3 and ALS4 genes (Table 3). The amplicons were then subjected to direct sequencing to determine which sequence is carried in the progeny.
Southern Blot Analysis
Genomic DNA was extracted from shoots of individual plants of single-self-pollinated Sugi-5 and Sugi-24 seedlings using a Plant Genomic DNA Kit (Tiangen, Beijing, China) according to the manufacturer’s instructions. Ten microgram of DNA was digested with EcoRV and HindIII (New England BioLabs Japan, Tokyo, Japan), then separated electrophoretically on 1% agarose gels and transferred to positively charged nylon membranes (Roche, Basel, Switzerland). A probe for the ALS gene was prepared by PCR using the PCR DIG Probe Synthesis Kit (Roche) with primers (forward, 5′-CTTTTGCAAGCAGGTCCAAG-3′; reverse, 5′-ATCCCCATCAATGTCAACAAC-3′) and the ALS1 PCR product as a template. Hybridization was conducted according to the DIG Application Manual (Roche) and the signals detected using CDP-Star (Roche) on ChemiDoc Touch (Bio-Rad, Irvine, CA, USA).
Flow Cytometric Analysis
Ploidy levels were analyzed using an Attune Flow Cytometer (Applied Biosystems). A. aequalis leaves were chopped using a razor blade in extraction buffer (Galbraith et al., 1983). After filtration using a CellTrics® 20-μm filter (Sysmex, Kobe, Japan), propidium iodide (25 mg/l) was added to stain the DNA. The nuclei suspension was then analyzed by laser excitation at 488 nm.
Transcription of ALS Genes
Plants were grown in a growth chamber at 25°C with a 12 h photoperiod for 20 days after germination. Shoot and root RNA was extracted as described above from three Sugi-5 and Sugi-24 plants, respectively. RNA samples (1 μg) were reverse-transcribed using ReverTra Ace (Toyobo) and PCR amplification conducted using rTaq DNA polymerase (Toyobo) with primers listed in Table 4. PCR conditions were as follows: 94 °C for 2 min followed by 35 cycles of 94°C for 30 s, 64°C for 30 s, 72°C for 30 s, and ending with 72°C for 5 min. The PCR products were run on 1% agarose gel and stained with ethidium bromide. Experiments were conducted twice using RNA samples from distinct individuals. Similar results were obtained in each experiment.
Results
Thifensulfuron-Methyl Sensitivity
Figure 1 shows the results of the thifensulfuron-methyl sensitivity assay in nine A. aequalis accessions collected from a severely infested wheat field. Growth of two accessions, Sugi-1 and Sugi-24, was severely affected by application of 1/3× the recommended field rate, with dry weights reaching only 35% that of the untreated control. Moreover, plants of both accessions died under treatment with 1 and 3× the recommended field rate. These accessions were therefore considered thifensulfuron-methyl susceptible. In comparison, the remaining seven accessions survived application of 1× the recommended field rate, achieving dry weights of >60% that of the untreated control. Even under 3× the field rate, these accessions exhibited dry weights ≥50% that of the untreated control, indicating thifensulfuron-methyl resistance. All plants from each accession responded to each dose of thifensulfuron-methyl in a similar manner, with no segregation of resistance.
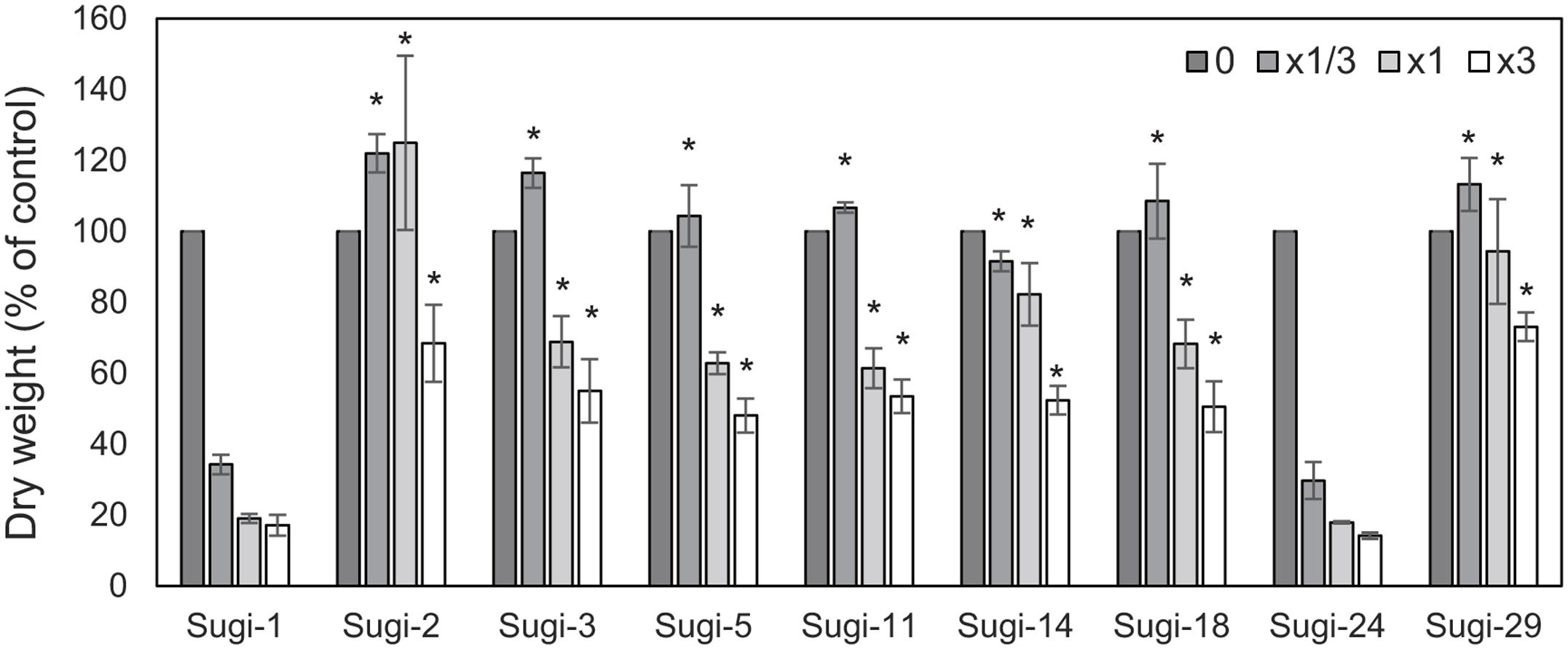
FIGURE 1. Thifensulfuron-methyl responses of the Alopecurus aequalis accessions. Each accession was treated with 0, 1/3, 1, or 3× the recommended field application rate of thifensulfuron-methyl. Shoot dry weights are shown as percentages of untreated controls. Bars indicate the standard error (n = 3). Dry weights of accession Sugi-1 were compared with remaining accessions using Dunnett’s test. ∗p < 0.01.
Isolation and Analysis of ALS Genes
Partial fragments of ALS genes were amplified from genomic DNA of each accession and subjected to direct sequencing. More than one peak was observed at some nucleotide positions in chromatograms of each accession, suggesting the existence of multiple copies of the ALS gene. Interestingly, the chromatogram patterns differed among accessions, with double peaks observed in some accessions, and very sharp single peaks in others (Figure 2A), suggesting CNV of the ALS genes.
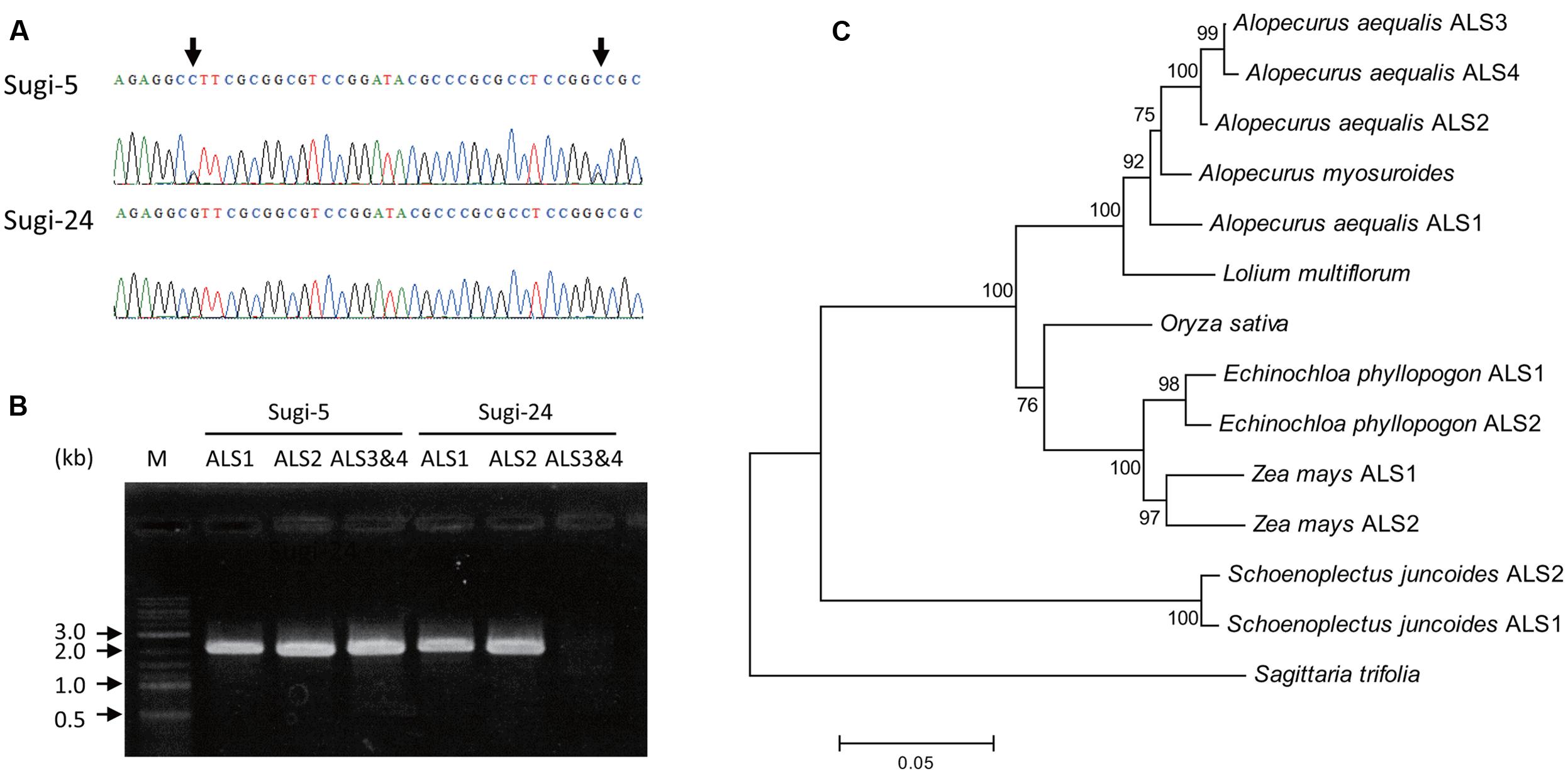
FIGURE 2. ALS genes in A. aequalis. (A) Chromatograms of DNA sequences of an ALS gene fragment. Arrows indicate double peaks. (B) Amplification of full-length ALS genes from accessions Sugi-5 and Sugi-24. M, DNA size standard. (C) Phylogenetic tree of the ALS. Numbers at each node indicate the reliability of branches estimated by bootstrap analysis with 1,000 replicates. Accession numbers for each sequence are as follows: Alopecurus myosuroides, AJ437300; Lolium multiflorum, AAG30931.1; Oryza sativa, LOC_Os02g30630.2; Echinochloa phyllopogon ALS1, AB636580.1; Echinochloa phyllopogon ALS2, AB636581.1; Zea mays ALS1, GRMZM2G143008_T01; Zea mays ALS2, GRMZM2G143357_T01; Schoenoplectus juncoides ALS1, BAE97675.1; Schoenoplectus juncoides ALS2, BAE97677.1; Sagittaria trifolia, AB301496.1. In the case of rice ALS, an additional ALS-like sequence, LOC_Os04g32010.1, was found in the MUS database; however, this sequence was excluded due to its low level of expression and the large number of unconserved residues among grass ALSs.
PCR amplification of the 3′ and 5′ regions of the partial sequences resulted in the isolation of two copies of the ALS gene (ALS1 and ALS2) from accession Sugi-24 and four copies (ALS1, ALS2, ALS3, and ALS4) from accession Sugi-5 (accession numbers: LC200800 to LC200803) (Figures 2, 3). The double peaks observed in direct sequencing of the Sugi-5 PCR product (Figure 2A) therefore represented ALS3 and ALS4. The ALS2 gene sequence was identical to that of the A. aequalis ALS gene sequence previously deposited in GenBank (JQ743908.1). The ALS3 and ALS4 genes each contained a single base deletion in a region containing four consecutive Cs at positions +146 to +149 (Figure 3), respectively, which would result in aberrant protein sequences. Thus, ALS3 and ALS4 are thought to be pseudogenes that share 99.5% identity with only three single nucleotide polymorphisms (SNPs) in the coding sequence (Figures 2C, 3). Polymorphisms between ALS3 and ALS4 were limited even in untranslated regions (UTRs) and the promoter region; no polymorphisms were observed in the 3′-UTR (157 bp), while four SNPs and three bp deletions were revealed in the promoter and 5′-UTR, respectively (603 and 600 bp for ALS3 and ALS4, respectively) (Supplementary Figure S1). This discovery of a limited number of polymorphisms between ALS3 and ALS4 precluded the design of gene-specific primers. None of the A. aequalis ALS genes contained introns, as previously observed in ALS genes in Poaceae (Iwakami et al., 2012). The predicted protein sequences of ALS1 and ALS4 exhibited high homology with those of ALS proteins from A. myosuroides (AJ437300) at 96.2 to 97.5% identity (Figure 2C). Furthermore, ALS3, which shared 99.5 % identity with ALS4, exhibited 95.8 and 98.9% identity with ALS1 and ALS2, respectively.

FIGURE 3. Sequence alignment of coding sequences of ALS genes in A. aequalis. The gray box (+339 to +380) represents the region shown in the chromatogram in Figure 2A. Red arrows indicate the positions of SNPs between ALS3 and ALS4, and the blue arrow indicates the deletion that caused a frameshift in ALS3 and ALS4.
The possibility that ALS3 and ALS4 represent alleles of a single gene, rather than two different genes, was subsequently investigated since the two were found to be highly homologous. To do so, sequences were investigated in the self-pollinated progeny of next-generation plants carrying both sequences. Full-length sequences of ALS3 and ALS4 were amplified and the DNA fragments directly sequenced. A chromatogram of all the progeny (more than 30 individuals) showed a double peak at the three polymorphic sites between the ALS3 and ALS4 sequences only. This finding confirms that all the progeny carried both the ALS3 and ALS4 sequences, contradicting the suggestion that they represent alleles and providing strong evidence that they represent two different genes.
The results of Southern blot analysis supported the differences in copy number among accessions. In Sugi-24, which carried two copies (ALS1 and ALS2), two signals were detected both in the EcoRV and HindIII digests (Supplementary Figure S2). In contrast, three signals were detected in the HindIII digest from Sugi-5, in which an additional two copies were found, although no differences in hybridization patterns of the EcoRV digests were observed. In both accessions, lower molecular weight signals in the EcoRV digest were thought to represent ALS2, considering the restriction sites of EcoRV (Supplementary Figure S2C).
The full-length sequences of ALS genes amplified from all nine accessions were subsequently sequenced. ALS1 and ALS2 were observed in all accessions analyzed so far (48 accessions from eight fields, data not shown), while some were found to carry additional ALS3 and ALS4 copies (Table 1). All sequences of each gene were identical, except at the codon corresponding to Pro197, mutations of which are known to confer resistance to several SU herbicides. Here, all of the thifensulfuron-methyl resistant accessions carried mutations at Pro197, with substitutions of Ser, Leu or Thr identified in ALS1 (Table 1). However, in Sugi-3, a Leu substitution was observed in the Pro197 of ALS2.
Ploidy Levels
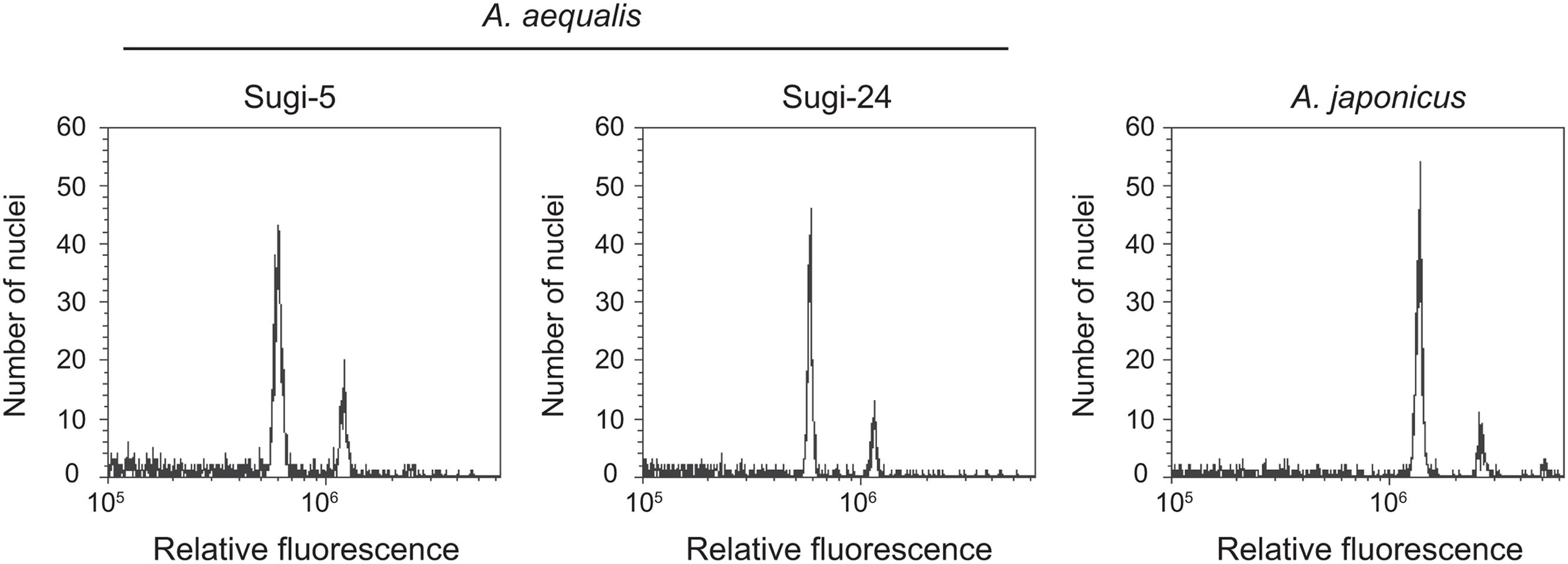
Two nuclei peaks were detected in flow cytometric analyses of two accessions of A. aequalis and one accession of A. japonicus (Figure 4). The fluorescence intensities of the first and second peaks were the same in the A. aequalis accessions, with two and four copies of the ALS genes. In contrast, peak intensities of the closely related tetraploid A. japonicus (Xu et al., 2014) were double those of the A. aequalis accessions, suggesting that DNA contents of single nuclei from two- and four-copy A. aequalis accessions are equal and approximately half that of A. japonicus.
Transcription of the ALS Genes
To confirm transcription of the ALS genes in seedlings from accessions carrying two or four copies, reverse transcription (RT)-PCR was conducted. PCR amplification could not distinguish ALS3 and ALS4 due to the lack of nucleotide polymorphisms. Thus, primers targeting the conserved region between the two genes were designed in order to amplify the area containing the polymorphisms. The resulting PCR product was subjected to direct sequencing to confirm which genes were transcribed.
ALS1 and ALS2 were transcribed both in the shoots and roots in accessions carrying two or four copies of the ALS genes; namely, Sugi-24 and Sugi-5, respectively (Figure 5). ALS3 and ALS4 DNA fragments were subsequently amplified from Sugi-5 shoot and root template cDNA. Sequencing of the PCR products revealed transcription of both the ALS3 and ALS4 genes. No DNA fragments of any of the ALS genes were detected in controls without reverse transcriptase (data not shown).
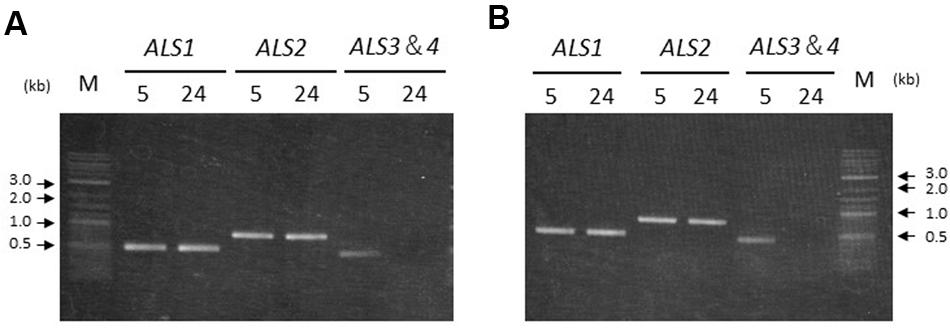
FIGURE 5. Transcription of ALS genes in two A. aequalis accessions. Labels 5 and 24 represent the Sugi-5 and Sugi-24 accessions, respectively. The primers do not discriminate between the ALS3 and ALS4 genes. M, DNA size standard. (A) Seedling shoots. (B) Seedling roots.
Discussion
In the present study, thifensulfuron-methyl resistant A. aequalis accessions were found to carry a mutation at the Pro197 residue in either the ALS1 or ALS2 gene. All amino acid substitutions identified in this study are known to confer SU resistance (Yu and Powles, 2014). Moreover, both the ALS1 and ALS2 genes were found to be transcribed in seedlings of A. aequalis, supporting involvement of this mutation in thifensulfuron-methyl resistance. However, it remains to be determined whether the mutation site or number of mutated loci influences the degree of resistance. More detailed characterization of these accessions will therefore be carried out in the future.
The number of ALS gene sequences differed among the nine accessions. Differences in the copy number of genes can result from CNV or from different ploidy levels within a species. In the present study, analysis of ploidy level revealed no differences in the amount of nuclear DNA among accessions. It is therefore unlikely that the higher-copy number accessions are tetraploid, although ploidy level variation within a single species has been reported in Alopecurus spp. including A. aequalis (Sieber and Murray, 1980; Koul and Gohil, 1990). The two highly similar DNA sequences of the higher–copy number accessions (ALS3 and ALS4 sequences) suggested that they might represent alleles of a single gene rather than two different genes. However, analysis of self-pollinated progeny revealed that all offspring carried both sequences, confirming that they are not alleles. Meanwhile, Southern blot analysis indicated two or three bands rather than four, which is consistent with different restriction enzymes (data not shown). This discrepancy can be explained by the hypothesis that ALS3 and ALS4 are tandemly duplicated and tightly linked on the same chromosome, supporting the observation that no three-copy individuals were present among 48 accessions in eight fields (data not shown). The occurrence of CNV in ALS genes has also been suggested in the tetraploid A. japonicus (Feng et al., 2016), with three copies identified in one population and an additional copy in another. This additional copy might represent an ortholog of ALS3 and ALS4 identified in this study, since one of the subgenomes of the hexaploid A. japonicus is thought to have been derived from A. aequalis (Matumura, 1968).
The CNV revealed in this study is unlikely to influence the evolution or level of resistance since both additional copies were thought to be pseudogenes without functions. However, copy number amplification of functional ALS genes could have resulted in ALS inhibitor resistance as previously observed in cultured cells (Odell et al., 1990), although resistance levels would be lower than that of target-site resistance resulting from amino acid substitution. Activity of plant ALS is subject to feedback inhibition from branched chain amino acids (Duggleby and Pang, 2000) and is not necessarily positively correlated with protein levels. In line with this, overproduction of ALS was found to result in only a slight decrease in ALS inhibitor sensitivity (Odell et al., 1990; Whaley et al., 2007).
The discovery of CNV in the ALS gene warrants further analysis of copy numbers in genes encoding herbicide targets, especially EPSPS. ALS genes are frequently used as an internal control in real-time PCR analysis of copy numbers of EPSPS genes, since ALS copy number was previously thought to be stable among individuals. However, this study revealed that the ALS copy number is, in fact, unstable. Selection of genes for use as an internal control should therefore be made carefully, and copy number amplification of genes of interest validated by other methods such as Southern blotting or fluorescence in situ hybridization as in Gaines et al. (2010), Jugulam et al. (2014), and Dillon et al. (2016). Alternatively, additional real-time PCR using a second internal control gene such as the cinnamoyl-CoA reductase gene (Salas et al., 2012, 2015) should also be performed for validation.
One interesting finding of this study was the presence of various mutation patterns in accessions randomly collected from a small field. Since A. aequalis is a self-fertile species, one might expect resistant plants in a single field to be derived from a single progenitor, as previously seen in resistant populations of M. vaginalis in Japan (Imaizumi et al., 2008). However, the discovery of four resistant genotypes suggests that the evolution of resistance occurred multiple independent times. Further research is now needed to determine how these resistance alleles evolved.
Despite the findings, the present results do not exclude the possible involvement of a non-target-site resistance mechanism. Non-target-site resistance is a major mechanism in A. myosuroides, which is a close outcrossing relative of A. aequalis (Délye et al., 2011). Non-target-site resistance to ALS inhibitors is often associated with enhanced activity of cytochrome P450 (Yuan et al., 2007). Recently, we discovered that overexpression of herbicide-metabolizing P450 genes is associated with resistance to ALS inhibitors in Echinochloa phyllopogon, a self-pollinating Poaceae species (Iwakami et al., 2014a). It is therefore possible that resistance evolved via a similar mechanism in A. aequalis.
Author Contributions
SI, YS, YM, AU, and TT designed the experiments; SI, YS, YM, ME, and HS performed the experiments and analyzed the results; and SI, YS, AU, and TT wrote and approved the manuscript.
Funding
This work was partly supported by a Grant-in-Aid for Scientific Research, No. 25292013 to TT.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00254/full#supplementary-material
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer PTFM and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Footnotes
References
Cope, T. A. (1982). “Poaceae,” in Flora of Pakistan, eds S. Ali and E. Nasir (Rawalpindi: Gordon College), 1–678.
Délye, C., Gardin, J. A. C., Boucansaud, K., Chauvel, B., and Petit, C. (2011). Non-target-site-based resistance should be the centre of attention for herbicide resistance research: Alopecurus myosuroides as an illustration. Weed Res. 51, 433–437. doi: 10.1111/j.1365-3180.2011.00864.x
Dillon, A. J., Varanasi, V. K., Danilova, T., Koo, D.-H., Nakka, S., Peterson, D., et al. (2016). Physical mapping of amplified 5-enolpyruvylshikimate-3-phosphate synthase gene copies in glyphosate-resistant Amaranthus tuberculatus. Plant Physiol. 173, 1226–1234. doi: 10.1104/pp.16.01427
Duggleby, R. G., McCourt, J. A., and Guddat, L. W. (2008). Structure and mechanism of inhibition of plant acetohydroxyacid synthase. Plant Physiol. Biochem. 46, 309–324. doi: 10.1016/j.plaphy.2007.12.004
Duggleby, R. G., and Pang, S. S. (2000). Acetohydroxyacid synthase. J. Biochem. Mol. Biol. 33, 1–36.
Feng, Y., Gao, Y., Zhang, Y., Dong, L., and Li, J. (2016). Mechanisms of resistance to pyroxsulam and ACCase inhibitors in Japanese foxtail (Alopecurus japonicus). Weed Sci. 64, 695–704. doi: 10.1614/Ws-D-16-00042.1
Gaines, T. A., Zhang, W., Wang, D., Bukun, B., Chisholm, S. T., Shaner, D. L., et al. (2010). Gene amplification confers glyphosate resistance in Amaranthus palmeri. Proc. Natl. Acad. Sci. U.S.A. 107, 1029–1034. doi: 10.1073/pnas.0906649107
Galbraith, D. W., Harkins, K. R., Maddox, J. M., Ayres, N. M., Sharma, D. P., and Firoozabady, E. (1983). Rapid flow cytometric analysis of the cell cycle in intact plant tissues. Science 220, 1049–1051. doi: 10.1126/science.220.4601.1049
Guo, W., Liu, W., Li, L., Yuan, G., Du, L., and Wang, J. (2015a). Molecular basis for resistance to fenoxaprop in shortawn foxtail (Alopecurus aequalis) from China. Weed Sci. 63, 416–424. doi: 10.1614/WS-D-14-00105.1
Guo, W., Lv, L., Zhang, L., Li, Q., Wu, C., Lu, X., et al. (2016). Herbicides cross resistance of a multiple resistant short-awn foxtail (Alopecurus aequalis Sobol.) population in wheat field. Chil. J. Agr. Res. 76, 163–169. doi: 10.1614/WS-D-14-00105.1
Guo, W., Yuan, G., Liu, W., Bi, Y., Du, L., Zhang, C., et al. (2015b). Multiple resistance to ACCase and AHAS-inhibiting herbicides in shortawn foxtail (Alopecurus aequalis Sobol.) from China. Pestic. Biochem. Physiol. 124, 66–72. doi: 10.1016/j.pestbp.2015.04.006
Heap, I. (2016). The International Survey of Herbicide Resistant Weeds. Available at: www.weedscience.com [accessed October 15, 2016].
Imaizumi, T., Wang, G., Ohsako, T., and Tominaga, T. (2008). Genetic diversity of sulfonylurea-resistant and -susceptible Monochoria vaginalis populations in Japan. Weed Res. 48, 187–196. doi: 10.1111/j.1365-3180.2008.00622.x
Iwakami, S., Endo, M., Saika, H., Okuno, J., Nakamura, N., Yokoyama, M., et al. (2014a). Cytochrome P450 CYP81A12 and CYP81A21 are associated with resistance to two acetolactate synthase inhibitors in Echinochloa phyllopogon. Plant Physiol. 165, 618–629. doi: 10.1104/pp.113.232843
Iwakami, S., Uchino, A., Watanabe, H., Yamasue, Y., and Inamura, T. (2012). Isolation and expression of genes for acetolactate synthase and acetyl-CoA carboxylase in Echinochloa phyllopogon, a polyploid weed species. Pest Manag. Sci. 68, 1098–1106. doi: 10.1002/ps.3287
Iwakami, S., Watanabe, H., Miura, T., Matsumoto, H., and Uchino, A. (2014b). Occurrence of sulfonylurea resistance in Sagittaria trifolia L., a basal monocot species, based on target-site and non-target-site resistance. Weed Biol. Manag. 14, 43–49. doi: 10.1111/wbm.12031
Jugulam, M., Niehues, K., Godar, A. S., Koo, D. H., Danilova, T., Friebe, B., et al. (2014). Tandem amplification of a chromosomal segment harboring 5-enolpyruvylshikimate-3-phosphate synthase locus confers glyphosate resistance in Kochia scoparia. Plant Physiol. 166, 1200–1207. doi: 10.1104/pp.114.242826
Koul, K. K., and Gohil, R. (1990). Cytological studies on some Kashmir grasses. VI. Cytomorphological polymorphism in Alopecurus aequalis Sobol. Cytologia 55, 217–223. doi: 10.1508/cytologia.55.217
Liu, Y. G., and Whittier, R. F. (1995). Thermal asymmetric interlaced PCR: automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25, 674–681. doi: 10.1016/0888-7543(95)80010-J
Matumura, M. (1968). Some observations on the F1 hybrids of Alopecurus japonicus and up- and low-land types of A. aequalis. Gifu Univ. Fac. Agric. Res. Bull. 26, 195–203.
Odell, J. T., Caimi, P. G., Yadav, N. S., and Mauvais, C. J. (1990). Comparison of increased expression of wild-type and herbicide-resistant acetolactate synthase genes in transgenic plants, and indication of posttranscriptional limitation on enzyme activity. Plant Physiol. 94, 1647–1654. doi: 10.1104/pp.94.4.1647
Ohsako, T., and Tominaga, T. (2007). Nucleotide substitutions in the acetolactate synthase genes of sulfonylurea-resistant biotypes of Monochoria vaginalis (Pontederiaceae). Genes Genet. Syst. 82, 207–215. doi: 10.1266/ggs.82.207
Ostlie, M., Haley, S. D., Anderson, V., Shaner, D., Manmathan, H., Beil, C., et al. (2015). Development and characterization of mutant winter wheat (Triticum aestivum L.) accessions resistant to the herbicide quizalofop. Theor. Appl. Genet. 128, 343–351. doi: 10.1007/s00122-014-2434-4
Powles, S. B., and Yu, Q. (2010). Evolution in action: plants resistant to herbicides. Annu. Rev. Plant Biol. 61, 317–347. doi: 10.1146/annurev-arplant-042809-112119
R core team (2016). R: A Language and Environment for Statistical Computing. Vienna: R Foundation for Statistical Computing.
Salas, R. A., Dayan, F. E., Pan, Z., Watson, S. B., Dickson, J. W., Scott, R. C., et al. (2012). EPSPS gene amplification in glyphosate-resistant Italian ryegrass (Lolium perenne ssp. multiflorum) from Arkansas. Pest Manag. Sci. 68, 1223–1230. doi: 10.1002/ps.3342
Salas, R. A., Scott, R. C., Dayan, F. E., and Burgos, N. R. (2015). EPSPS gene amplification in glyphosate-resistant Italian ryegrass (Lolium perenne ssp. multiflorum) populations from Arkansas, USA. J. Agric. Food Chem. 63, 5885–5893. doi: 10.1021/acs.jafc.5b00018
Sammons, R. D., and Gaines, T. A. (2014). Glyphosate resistance: state of knowledge. Pest Manag. Sci. 70, 1367–1377. doi: 10.1002/ps.3743
Sieber, V. K., and Murray, B. (1980). Spontaneous polyploids in marginal populations of Alopecurus bulbosus Gouan (Poaceae). Bot. J. Linnean Soc. 81, 293–300. doi: 10.1111/j.1095-8339.1980.tb01680.x
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Uchikawa, O., Miyazaki, M., Tanaka, K., and Ohdan, H. (2005). Occurrence of low sensitive biotype of some herbicides of water foxtail (Alopecurus aequalis) in wheat fields in Fukuoka prefecture, and effects of some herbicides to its biotype. J. Weed Sci. Technol. 50(Suppl.), 68–69. doi: 10.3719/weed.50.Supplement_68
Uchino, A., Iwakami, S., Tominaga, T., and Itoh, K. (2016). Herbicide Resistant Weeds in Japan. Available at: http://www.wssj.jp/~hr/
Uchino, A., Ogata, S., Kohara, H., Yoshida, S., Yoshioka, T., and Watanabe, H. (2007). Molecular basis of diverse responses to acetolactate synthase-inhibiting herbicides in sulfonylurea-resistant biotypes of Schoenoplectus juncoides. Weed Biol. Manag. 7, 89–96. doi: 10.1111/j.1445-6664.2007.00240.x
Wang, Z., Ye, S., Li, J., Zheng, B., Bao, M., and Ning, G. (2011). Fusion primer and nested integrated PCR (FPNI-PCR): a new high-efficiency strategy for rapid chromosome walking or flanking sequence cloning. BMC Biotechnol. 11:109. doi: 10.1186/1472-6750-11-109
Whaley, C. M., Wilson, H. P., and Westwood, J. H. (2007). A new mutation in plant Als confers resistance to five classes of Als-inhibiting herbicides. Weed Sci. 55, 83–90. doi: 10.1614/WS-06-082.1
Xia, W., Pan, L., Li, J., Wang, Q., Feng, Y., and Dong, L. (2015). Molecular basis of ALS- and/or ACCase-inhibitor resistance in shortawn foxtail (Alopecurus aequalis Sobol.). Pestic. Biochem. Physiol. 122, 76–80. doi: 10.1016/j.pestbp.2014.12.019
Xu, H., Zhang, W., Zhang, T., Li, J., Wu, X., and Dong, L. (2014). Determination of ploidy level and isolation of genes encoding acetyl-CoA carboxylase in Japanese Foxtail (Alopecurus japonicus). PLoS ONE 9:e114712. doi: 10.1371/journal.pone.0114712
Yu, Q., Ahmad-Hamdani, M. S., Han, H., Christoffers, M. J., and Powles, S. B. (2013). Herbicide resistance-endowing ACCase gene mutations in hexaploid wild oat (Avena fatua): insights into resistance evolution in a hexaploid species. Heredity 110, 220–231. doi: 10.1038/hdy.2012.69
Yu, Q., and Powles, S. B. (2014). Resistance to AHAS inhibitor herbicides: current understanding. Pest Manag. Sci. 70, 1340–1350. doi: 10.1002/ps.3710
Keywords: shortawn foxtail, ALS inhibitor, herbicide resistance, target-site resistance, copy number variation
Citation: Iwakami S, Shimono Y, Manabe Y, Endo M, Shibaike H, Uchino A and Tominaga T (2017) Copy Number Variation in Acetolactate Synthase Genes of Thifensulfuron-Methyl Resistant Alopecurus aequalis (Shortawn Foxtail) Accessions in Japan. Front. Plant Sci. 8:254. doi: 10.3389/fpls.2017.00254
Received: 21 December 2016; Accepted: 09 February 2017;
Published: 02 March 2017.
Edited by:
Rafael De Prado, University of Córdoba, SpainReviewed by:
Todd Gaines, Colorado State University, USAPablo Tomás Fernández-Moreno, University of Córdoba, Spain
Copyright © 2017 Iwakami, Shimono, Manabe, Endo, Shibaike, Uchino and Tominaga. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Satoshi Iwakami, aXdha2FtaS5zYXRvc2hpLjJ2QGt5b3RvLXUuYWMuanA=
 Satoshi Iwakami
Satoshi Iwakami Yoshiko Shimono
Yoshiko Shimono Yohei Manabe1
Yohei Manabe1 Masaki Endo
Masaki Endo Hiroyuki Shibaike
Hiroyuki Shibaike Akira Uchino
Akira Uchino Tohru Tominaga
Tohru Tominaga