
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 17 February 2017
Sec. Plant Development and EvoDevo
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.00153
This article is part of the Research TopicEvolution of Reproductive Organs in Land PlantsView all 16 articles
As a core regulatory gene of the anther development, DYSFUNCTIONAL TAPETUM1 (DYT1) was expressed in tapetum preferentially. Previous study had confirmed that a “CTCC” sequence within DYT1 promoter was indispensable for correct DYT1 expression. However, precise analysis on the function of each nucleotide of this sequence still lacks. Here we employed site mutation assay to identify the function roles of the nucleotides. As a result, the “T” and final “C” of “CTCC” were found essential for the temporal and spatial specificity of DYT1 expression, whereas the other two “C” nucleotides exhibited substitutable somewhat. The substitutes of two flanking nucleotides of “CTCC,” however, hardly affected the normal promoter function, suggesting that the “CTCC” sequence as a whole did meet the standard of a canonical cis-element by definition. In addition, it was found that as short as 497 bp DYT1 promoter was sufficient for tissue-specific expression, while longer 505 bp DYT1 promoter sequence was sufficient for species-specific expression.
Anther development is crucial for successful pollen production in flowering plants. The Arabidopsis anther during meiosis is a four-lobed structure comprised of concentric outer epidermis, endothecium, middle fibrous layer, tapetum and pollen mother cell (PMC; Goldberg et al., 1993; Yeung et al., 2011). The tapetum initially turns out as a single-cell layer surrounding PMC, and is the main nutrient tissue of PMC and pollen subsequently in the anther (Koltunow et al., 1990; Scott et al., 2004; Feng and Dickinson, 2007; Zhang et al., 2014; Li et al., 2015). A serial of regulatory genes have been identified to be essential for the tapetum function in Arabidopsis up to date, including DYSFUNCTIONAL TAPETUM1 (DYT1), DEFECTIVE IN TAPETAL DEVELOPMENT AND FUNCTION1 (TDF1), MYB103/MYB80, ABORTED MICROSPORE (AMS), MALE STERILITY1 (MS1), etc. (Zhang et al., 2006, 2007; Yang et al., 2007; Zhu et al., 2008; Phan et al., 2011; Wang et al., 2012; Fernández-Gómez and Wilson, 2014; Xu et al., 2014, 2015; Shumin et al., 2015; Yi et al., 2016). Among them, DYT1 as one of the earliest tapetum-preferential genes, initiates all aspects of tapetum function through regulating transcription of approximately 1,000 anther genes involved in callose synthesis and degradation, peptide and lipid transport, exine formation, etc. (Schiefthaler et al., 1999; Higginson et al., 2003; Sorensen et al., 2003; Ito et al., 2007; Liu et al., 2009; Feng et al., 2012; Phan et al., 2012; Li et al., 2013; Cui et al., 2016).
The expression profile of DYT1 is highly tissue-specific. Weak expression of DYT1 can be detected in the secondary parietal cell and sporogenous cell, the precursors of tapetum and PMC respectively at as early as anther stage 4 (Zhang et al., 2006; Shumin et al., 2015). Then DYT1 expression significantly enhances and culminates with maturation of tapetum at the anther stages 6, and exhibits as a tapetum-preferential pattern (Zhang et al., 2006; Shumin et al., 2015). With the end of meiosis of PMC, DYT1 expression declines rapidly, and disappears at stage 8 (Zhang et al., 2006; Gu et al., 2014; Shumin et al., 2015). The underlying mechanism(s) controlling DYT1 temporal and spatial expression pattern remains as a puzzle since DYT1 was firstly characterized one decade ago (Zhang et al., 2006). It has been known that at least two signal pathways are involved in initiation of DYT1 expression. The first one seems to be governed by transcription regulatory factors, including nuclear proteins NZZ/SPL and LFR, and SBP-domain transcription factor SPL8 (Yang et al., 1999; Xing et al., 2010; Wang et al., 2012). The second pathway is mediated by protein phosphorylation triggered by a series of receptor-like kinases, such as EXS/EMS1, SERK1 and SERK2, BAM1 and BAM2 (Zhao et al., 2002, 2008; Albrecht et al., 2005; Colcombet et al., 2005; Hord et al., 2006; Li et al., 2017). Both signal pathways are essential for normal DYT1 expression, though few details are known about how they crosstalk and activate DYT1 expression together (Zhang et al., 2006; Shumin et al., 2015).
In our previous study, it had been confirmed that as short as 513 bp sequence in front of the transcription start site (TSS) of DYT1 was essential and sufficient for proper temporal and spatial specificity of DYT1 expression. In addition, the deletion of a “CTCC” sequence at the position of −468 bp (i.e., 468 bp from the TSS) abolished DYT1 expression completely at the anther stage 6, suggesting that the “CTCC” sequence was indispensable for normal DYT1 expression (Shumin et al., 2015). Including our previous study, there have been only a couple of related reports about “CTCC” as a putative cis-element crucial for gene expression regulation in plants (Kano-Murakami et al., 1991; Ku et al., 2011). However, whether the “CTCC” sequence is a canonical cis-element in which the nucleotides are irreplaceable, remains to be addressed. In this study, we employed site mutation assay to characterize the function roles of the nucleotides, including the two flanking ones of the “CTCC” sequence to answer the question whether the “CTCC” sequence met the standard of a canonical cis-element or not. In addition, more truncation analysis was performed through using both transgenic Arabidopsis and tobacco bright yellow 2 (BY2) cell suspensions to identify which regions of DYT1 promoter were essential for tissue, and further species specificity of DYT1 expression.
Arabidopsis thaliana ecotype Col-0 was used in all of the transformation and promoter analysis in this study. The plants were cultivated under 16 h light/8 h dark photoperiod with 300 Es−1m−2 illumination intensity, at 22 ± 1°C. The seeds were stratified at 4°C for 4 days prior to growth.
The tobacco (Nicotiana tabacum L. cv Bright Yellow 2, BY2) was cultivated in a modified liquid Murashige and Skoog (MS) medium (Zhou et al., 2014) at 28°C with 120 rpm shaking avoiding light and maintained by weekly dilution (V/V = 1/10) of cell.
The pre-existing 513 bp DYT1 promoter-driven GFP expression construct, designated as DYT1513bp::GFP (Shumin et al., 2015), was used as PCR template in this study. The primers to generate site mutations of the constructsDYT1TTCC::GFP, DYT1CGCC::GFP, DYT1CTTC::GFP and DYT1CTCT::GFP; CTCC flanking site mutation constructDYT1TCTCCT::GFP were designed and synthesized respectively. Novel 5′ end primers of truncation constructs DYT1489bp::GFP, DYT1497bp::GFP and DYT1505bp::GFP were designed and synthesized, respectively (Figure 1). The PCR products were obtained and cloned into pCAMBIA1300 to make reporting constructs according to the report of Zhou (Shumin et al., 2015).
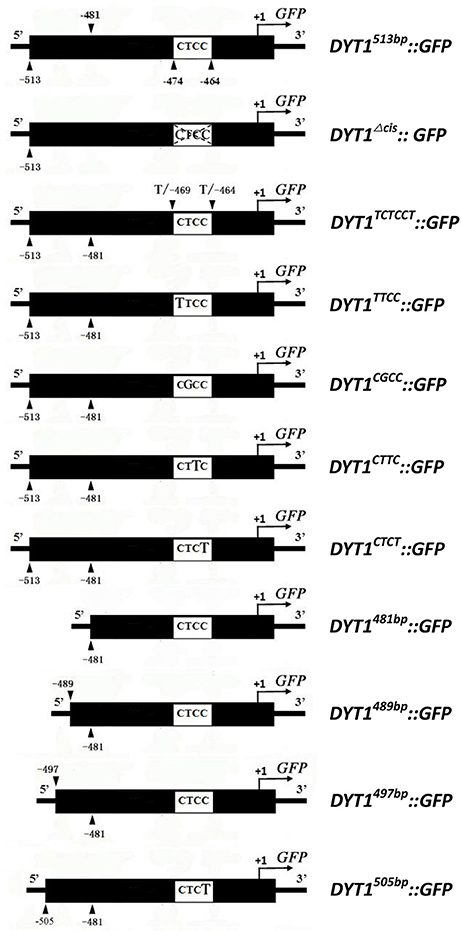
Figure 1. Promoter constructs used to drive GFP expression in transgenic plant anthers and BY2 cells.
Transgenic plants were generated via floral-dip transformation. The positive transgenic seedlings were screened on MS medium containing 25 mg/L hygromycin (Clough and Bent, 1998). At least 10 independent transgenic T1 generation lines for each construct were observed in this study.
The transformation of BY2 suspension was carried out according to the report of Zhou (Zhou et al., 2014). BY2 cell suspension was co-cultivated with the Agrobacterium GV3101 strain harboring transgenic construct in liquid medium without antibiotics avoiding light at 28°C for 48 h, so that the final concentration of cell suspension was approximately OD600 = 0.6. The resulted BY2 cell suspension was enriched by centrifuge and plated on MS solid medium containing 50 μg/ml hygromycin and 100 μg /ml vancomycin, and incubated at 28°C avoiding light. Two weeks later, grown-up antibiotics-resistant callus were subjected to amplified liquid cultivation, and the resulted BY2 cell suspension was used for genotyping and fluorescence observation. At least 10 independent original antibiotics-resistant callus were observed for each construct. The pre-existing transgenic callus of cauliflower mosaic leaf virus 35S promoter-driven GFP expression 35S::GFP was used as a positive control (Zhou et al., 2014).
Total RNA was extracted from the transgenic BY2 cell suspension and performed reverse transcription according to Zhou et al. (2014). Then GFP cDNA fragment was PCR amplified with GFP specific (GFP RT-F&R) primers with the sequence listed in Table 1.
Anthers were stripped and collected from transgenic plants flower bud just around male meiosis (anther stage 4–9) on a microscopy slides. Added one drop of sterile water on the anthers and covered a slide carefully without squeezing. Then the sample was observed and photographed under Zeiss LSM-710 confocal microscope (Zeiss, Germany) and Leica DM2500 fluorescence microscope. As for semi-quantification of the fluorescence intensity, randomly 10 sites on fluorescence images were selected and the intensity was measured and normalized by the SMART software. Statistics of at least 15 anthers per line, 10 independent T1 generation transgenic lines were counted for each construct transformation. As for BY2 cell suspension, at least 100 cells per callus ancestor were observed, and 10 callus were counted for each construct transformation.
Previous studies showed that the 513 bp length DYT1 promoter could faithfully regenerate the temporal and spatial profile of native DYT1 expression (Shumin et al., 2015). The GFP signal of transgenic DYT1513bp::GFP firstly appeared in the secondary parietal cell and microsporocyte of stage 4 Arabidopsis anther. Then the GFP expression increased significantly and reached its peak preferentially in the tapetum of stage 5 and 6 anthers. Subsequently, the GFP signal rapidly weakened at stage 7 and disappeared at stage 8 (Figures 2A,G). The “CTCC” cis-element locating at −468 bp from the TSS is particularly important for the correct expression of the DYT1 gene. The deletion of “CTCC” completely knocked out GFP expression (Shumin et al., 2015). To investigate the function of each nucleotide in the “CTCC” cis-element, a series of modifying constructs based on DYT1513bp::GFP with site mutations in or around the “CTCC” sequence were made, and transformed into Arabidopsis, respectively (Figure 1). The transgenic plants were identified by PCR using nucleotide specific primers (Table 1) and restriction endonuclease digestion assay (Supplementary Figure 1). The site mutations of the two flanking nucleotides of the “CTCC” cis-element (5′ end from “C” to “T,” and 3′ end from “C” to “T,” respectively), and the first and third nucleotide substitutes from “C” to “T” in the “CTCC” imposed no effect on the expression pattern of GFP (Figures 2B,C,E,G). On the contrast, however, “G” replacing “T” in the “CTCC” resulted in weak expression of GFP in the connective and epidermis tissues in addition to the tapetum and PMC (before stage 6, then microspore at stage 7 and 8; Figures 2D,H). Furthermore, “T” replacing the final “C” resulted in strong GFP expression in all tissues of stage 4–8 anthers (Figures 2F,H). Thus, the “T” and final “C” of the “CTCC” cis-element were suggested to play predominant roles in controlling the tissue specificity and appropriate intensity of the gene expression.
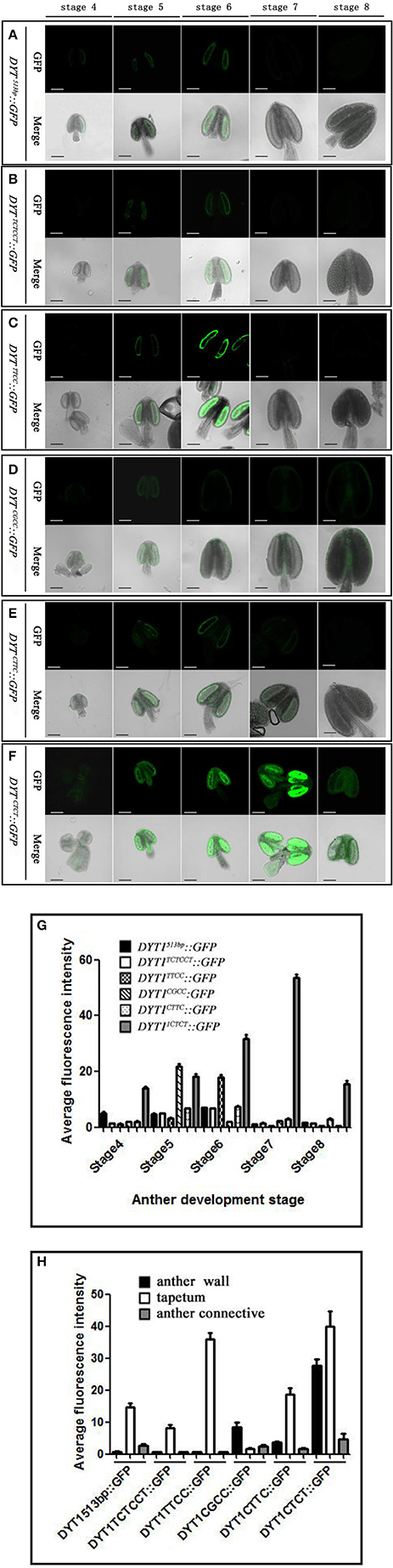
Figure 2. GFP expression in “CTCC” site mutation transgenic plant anthers. (A–F) Green fluorescence images of anthers at stage 4–8 in DYT1513bp ::GFP, DYT1TCTCCT::GFP, DYT1TTCC::GFP, DYT1CGCC::GFP, DYT1CTTC::GFP, and DYT1CTCT ::GFP in transgenic plants. (G,H) Semi-quantification of the average fluorescence intensity in stage 4–8 anthers and in different parts of stage 6 anthers of DYT1513bp::GFP, DYT1TCTCCT::GFP, DYT1TTCC::GFP, DYT1CGCC::GFP, DYT1CTTC::GFP, and DYT1CTCT ::GFP transgenic plantsthrough SMART software assay (n ≥ 30, ±SD; p < 0.1, Student's t-test), values were obtained from 3 independent lines of transgenic plants. Bar = 10 μm.
The previous study had elucidated that beside the core motif “CTCC,” the −481 to −513 bp region of DYT1 promoter was also indispensable for appropriate expression. To uncover finer structure within this region, in addition to original 481 and 513 bp truncated DYT1 promoter-driven GFP reporter constructs, 489, 497, and 505 bp truncated DYT1 promoter-driven GFP reporter constructs were made and transformed into Arabidopsis, respectively. As a result, both 505 and 497 bp DYT1 promoter gave rise of the identical expression pattern as the 513 bp DYT1 promoter (Figures 3A,B,E), suggesting as short as 497 bp DYT1 promoter was sufficient to recapitulate appropriate DYT1 expression in Arabidopsis anther. On the other side, in DYT489bp::GFP transgenic plants, GFP exhibited an ectopic and weaker expression losing the tapetum-preferential pattern, similar to that of the 481 bp DYT1 promoter. The detectable green fluorescence was distributed not only in the tapetum and PMC (before stage 6, then microspore at stage 7 and 8), but also in the connective and epidermis tissues (Figures 3C,D,F), suggesting that the sequence from −489 to −497 bp in DYT1 promoter was essential for tapetum-preferential expressing pattern, and as short as 497 bp DYT1 promoter sequence was sufficient for tissue-specific expression.
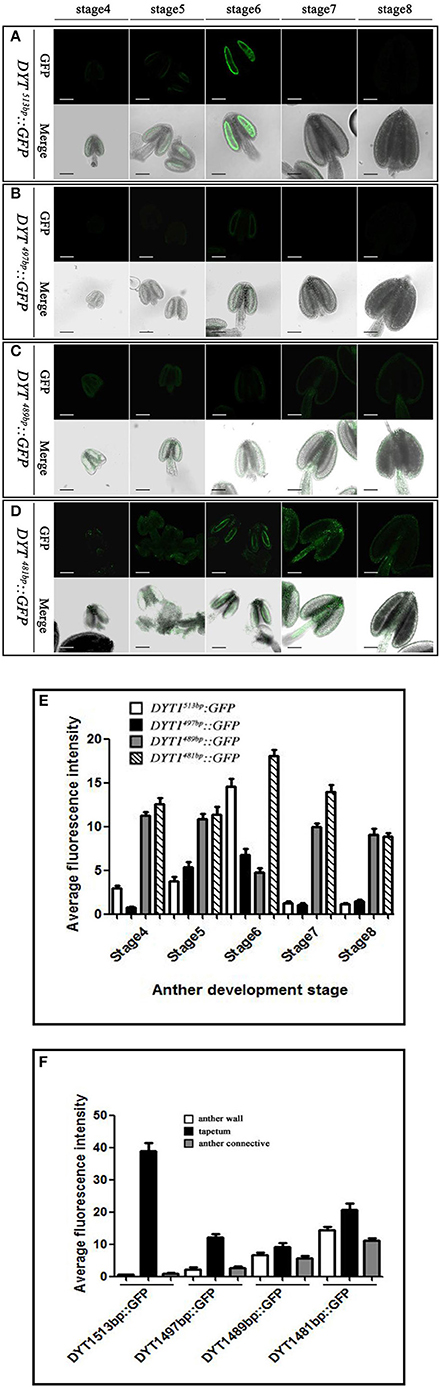
Figure 3. GFP expressing in DYT1513bp::GFP, DYT1489bp::GFP, DYT1497bp::GFP and DYT1481bp::GFP transgenic Arabidopsis anthers. (A) The green-fluorescence concentrates in the tapetal cells of transgenic plant anthers; (B–D) Obvious green-fluorescence displays both in the anther locules, tapetum and epidermis. The upper row are images of fluorescence, and the under row are merged images of light and fluorescence. (E,F) Semi-quantification of the average fluorescence intensity in stage4-8 anthers and in different parts of stage 6 anthers of DYT1513bp::GFP, DYT1497bp::GFP, DYT1489bp::GFP, and DYT1481bp::GFP transgenic plants through SMART software assay (n ≥ 30, ±SD; p < 0.1, Student's t-test), values were obtained from 3 to 5 independent lines of transgenic plants. Bar = 10 μm.
As mentioned before, the flanking −489 to −497 bp region seemed to play as a restriction element to limit DYT1 expression with certain spaces so that DYT1 expression exhibited as a specific spatial profile. Then one more question was brought up whether there was other region in 513 bp DYT1 promoter imparting the species specificity. In order to test such possibility, the series of truncated DYT1 promoter-driven GFP reporting constructs were transformed into tobacco BY2 cell suspension. In DYT1497bp::GFP, DYT1489bp::GFP and DYT1481bp::GFP transgenic BY2 cell suspension, weaker GFP expression comparing with that of 35S::GFP transgenic cells was found (Figures 4A,D–F,M). However, in DYT1513bp::GFP and DYT1505bp::GFP transformed cell lines, no GFP signal could be detected (Figures 4B,C,M). Thus, 505 bp DYT1 promoter sequence was sufficient for restricting the gene expression in A. thaliana rather than in other species such as tobacco BY2 cell suspension.
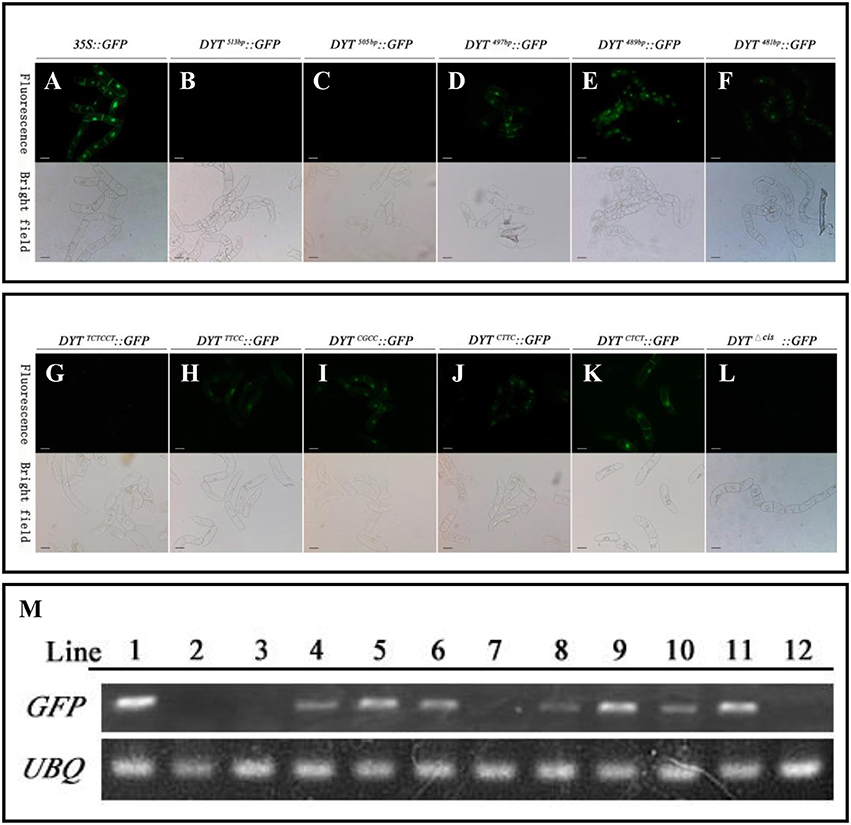
Figure 4. Functional segments assay of DYT1 promoterin transgenic BY2 cells. GFP fluorescence was detected under Confocal microscope. (A–F) The fluorescence and bright images of GFP driven by 35S promoter and different truncated DYT1 promoters. (G–L) The function assay of “CTCC” segment in DYT1 promoter driving GFP expression in BY2 cells. (M) RT-PCR analysis of GFP/ACTIN expression in different transgenic BY2 cells. Lane 1–12 represented 35S::GFP, DYT1513bp::GFP,DYT1505bp::GFP, DYT1497bp::GFP, DYT1489bp::GFP, DYT1481bp::GFP, DYT1TCTCCT ::GFP, DYT1TTCC::GFP, DYT1CGCC::GFP, DYT1CTTC::GFP, DYT1CTCT ::GFP, and DYT1Δcis::GFP transgenic cells. Bar = 20 μm.
Furthermore, all site mutations within “CTCC” based on DYT1513bp::GFP gave rise of ectopic GFP expression in BY2 cell suspension, suggesting that the “CTCC” cis-element participated in determining species specificity. However, the substitutes of the “T” and final “C” generated stronger ectopic expression than the other two nucleotides (Figures 4I,K), suggesting the “T” and final “C” also contributed in determining species specificity more than the other two “C” nucleotides (Figures 4H,J), though not so exclusively as in determining tissue specificity in Arabidopsis. Consistent to the results obtained from Arabidopsis study (Figure 2), the mutations of “CTCC” flanking nucleotides had no effect on the driven gene expression (Figure 4G), further supporting “CTCC” itself was a four-nucleotides motif. Unlike site mutations, the “CTCC” deletion DYT1513bpΔcis:: GFP generated little GFP fluorescence either in Arabidopsis anther (Shumin et al., 2015), or in BY2 cell suspension (Figures 4L,M), adding complexity to the function of intact “CTCC.” One explanation is that in addition to controlling spatial expression pattern of the driven gene, “CTCC” as a whole is also important for gene expression activation.
The findings of this study that the substitutes of the “T” and final “C” nucleotides in the “CTCC” sequence dramatically changed the driven gene (GFP here) expression profile, whereas the nucleotide replaces just out of “CTCC” imposed little effect on either tissue or species specificity, confirmed that the “CTCC” sequence did satisfy the definition of a canonical cis-element, and play as the core role in determining DYT1 expression profile. Furthermore, with more “CTCC” flanking sequences truncated from DYT1 promoter, the extent of gene expression specificity became weaker, reflected in the facts that the species specificity was lost firstly, then the expression region extended from the central locule to the connective tissue and epidermis in the Arabidopsis anther, resulting in a constitutive pattern at last. Thus it was suggested that DYT1 promoter was a functional unit comprised of multiple parts whose absence would lead to expression specificity attenuation, from both species and tissue-specific to only tissue-specific, and finally to constitutive. In other words, the core motif “CTCC” and its flanking sequences need work together to restrict the driven gene expressed precisely in specific tissues, and furthermore in specific species.
With the key cis-element identified, undoubtedly the main task of next stage work is to identify the trans-factor(s) which recognizes and binds to the “CTCC” cis-element, and finally activates DYT1 expression. As mentioned before, among the known DYT1 upstream regulatory factors which are involved in transcription regulation, SPL8 participates in the small RNA signaling in cell differentiation regulation in anther. As a SBP domain factor, the DNA-binding motif of SPL8 is zinc-binding motif rather than “CTCC” (Xing et al., 2010). Furthermore, both SPL/NZZ and LFR lack functional DNA-binding domain (Yang et al., 1999; Wang et al., 2012). Thus, it is proposed that the regulatory factor recognizing and binding to the “CTCC” cis-element of DYT1 promoter still needs to be characterized in future work. This unknown factor might be unable to activate DYT1 expression alone. Conversely it would associate with SPL/NZZ and/or LFR to form an active transcription complex to trigger DYT1 expression.
WZ and SZ designed all experiments, analyzed data, and wrote the manuscript. HZ performed experiments on transgenic expression assays. RL worked on the transgenic lines. QH performed experiments on construction of transformation vector. YL performed analysis of promoter function element. QX performed statistical analysis of fluorescence intensity.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was supported by grants from the Natural Science Foundation of Shanghai (Project Nos. 15ZR1416700), the National Natural Science Foundation of China (Project Nos. 30870225).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00153/full#supplementary-material
Albrecht, C., Russinova, E., Hecht, V., Baaijens, E., and de Vries, S. (2005). The Arabidopsis thaliana SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASES1 and 2 control male sporogenesis. Plant Cell 17, 3337–3349. doi: 10.1105/tpc.105.036814
Clough, S. J., and Bent, A. F. (1998). Floral dip: a simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743. doi: 10.1046/j.1365-313x.1998.00343.x
Colcombet, J., Boisson-Dernier, A., Ros-Palau, R., Vera, C. E., and Schroeder, J. I. (2005). Arabidopsis SOMATIC EMBRYOGENESIS RECEPTOR KINASES1 and 2 are essential for tapetum development and microspore maturation. Plant Cell 17, 3350–3361. doi: 10.1105/tpc.105.036731
Cui, J., You, C., Zhu, E., Huang, Q., Ma, H., and Chang, F. (2016). Feedback regulation of DYT1 by interactions with downstream bHLH factors promotes DYT1 nuclear localization and anther development. Plant Cell 28, 1078–1093. doi: 10.1105/tpc.15.00986
Feng, B., Lu, D., Ma, X., Peng, Y., Sun, Y., Ning, G., et al. (2012). Regulation of the Arabidopsis anther transcriptome by DYT1 for pollen development. Plant J. 72, 612–624. doi: 10.1111/j.1365-313X.2012.05104.x
Feng, X., and Dickinson, H. G. (2007). Packaging the male germline in plants. Trends Genet. 23, 503–510. doi: 10.1016/j.tig.2007.08.005
Fernández-Gómez, J., and Wilson, Z. A. (2014). A barley PHD finger transcription factor that confers male sterility by affecting tapetal development. Plant Biotechnol. J. 12, 765–777. doi: 10.1111/pbi.12181
Goldberg, R. B., Beals, T. P., and Sanders, P. M. (1993). Anther development: basic principles and practical applications. Plant Cell 5, 1217–1229. doi: 10.1105/tpc.5.10.1217
Gu, J. N., Zhu, J., Yu, Y., Teng, X. D., Lou, Y., Xu, X. F., et al. (2014). DYT1 directly regulates the expression of TDF1 for tapetum development and pollen wall formation in Arabidopsis. Plant J. 80, 1005–1013. doi: 10.1111/tpj.12694
Higginson, T., Li, S. F., and Parish, R. W. (2003). AtMYB103 regulates tapetum and trichome development in Arabidopsis thaliana. Plant J. 35, 177–192. doi: 10.1046/j.1365-313X.2003.01791.x
Hord, C. L., Chen, C., Deyoung, B. J., Clark, S. E., and Ma, H. (2006). The BAM1/BAM2 receptor-like kinases are important regulators of Arabidopsis early anther development. Plant Cell 18, 1667–1680. doi: 10.1105/tpc.105.036871
Ito, T., Nagata, N., Yoshiba, Y., Ohme-Takagi, M., Ma, H., and Shinozaki, K. (2007). Arabidopsis MALE STERILITY1 encodes a PHD-type transcription factor and regulates pollen and tapetum development. Plant Cell 19, 3549–3562. doi: 10.1105/tpc.107.054536
Kano-Murakami, Y, Suzuki, I., Sugiyama, T., and Matsuoka, M. (1991). Sequence-specific interactions of a maize factor with a GC-rich repeat in the phosphoenolpyruvate carboxylase gene. Mol. Gen. Genet. 225, 203–208. doi: 10.1007/BF00269849
Koltunow, A. M., Truettner, J., Cox, K. H., Wallroth, M., and Goldberg, R. B. (1990). Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2, 1201–1224. doi: 10.1105/tpc.2.12.1201
Ku, L., Wei, X., Zhang, S., Zhang, J., Guo, S., and Chen, Y. (2011). Cloning and characterization of a putative TAC1 ortholog associated with leaf angle in maize (Zea mays L.). PLoS ONE 6:e20621. doi: 10.1371/journal.pone.0020621
Li, J., Chen, X., Luo, L. Q., Yu, J., and Ming, F. (2013). Functions of ANAC092 involved in regulation of anther development in Arabidopsis thaliana. Yi Chuan 35, 913–922. doi: 10.3724/SP.J.1005.2013.00913
Li, L., Li, Y., Song, S., Deng, H., Li, N., Fu, X., et al. (2015). An anther development F-box (ADF) protein regulated by tapetum degeneration retardation (TDR) controls rice anther development. Planta 241, 157–166. doi: 10.1007/s00425-014-2160-9
Li, Z., Wang, Y., Huang, J., Ahsan, N., Biener, G., Paprocki, J., et al. (2017). Two SERK receptor-like kinases interact with EMS1 to control anther cell fate determination. Plant Physiol. 173, 326–337. doi: 10.1104/pp.16.01219
Liu, X., Huang, J., Parameswaran, S., Ito, T., Seubert, B., Auer, M., et al. (2009). The SPOROCYTELESS/NOZZLE gene is involved in controlling stamen identity in Arabidopsis. Plant Physiol. 151, 1401–1411. doi: 10.1104/pp.109.145896
Phan, H. A., Iacuone, S., Li, S. F., and Parish, R. W. (2011). The MYB80 transcription factor is required for pollen development and the regulation of tapetal programmed cell death in Arabidopsis thaliana. Plant Cell 23, 2209–2224. doi: 10.1105/tpc.110.082651
Phan, H. A., Li, S. F., and Parish, R. W. (2012). MYB80, a regulator of tapetal and pollen development, is functionally conserved in crops. Plant Mol. Biol. 78, 171–183. doi: 10.1007/s11103-011-9855-0
Schiefthaler, U., Balasubramanian, S., Sieber, P., Chevalier, D., Wisman, E., and Schneitz, K. (1999). Molecular analysis of NOZZLE, a gene involved in pattern formation and early sporogenesis during sex organ development in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 96, 11664–11669. doi: 10.1073/pnas.96.20.11664
Scott, R. J., Spielman, M., and Dickinson, H. G. (2004). Stamen structure and function. Plant Cell 16(Suppl.), 46–60. doi: 10.1105/tpc.017012
Shumin, Z., Yan, C., Hulin, S., Bang, Z., Licheng, S., and Wei, Z. (2015). One novel cis-element is essential for correct DYSFUNCTIONAL TAPETUM (DYT1) expression in Arabidopsis thaliana. Plant Cell Rep. 34, 1773–1780. doi: 10.1007/s00299-015-1823-8
Sorensen, A. M., Kröber, S., Unte, U. S., Huijser, P., Dekker, K., and Saedler, H. (2003). The Arabidopsis ABORTED MICROSPORES (AMS) gene encodes a MYC class transcription factor. Plant J. 33, 413–423. doi: 10.1046/j.1365-313X.2003.01644.x
Wang, X. T., Yuan, C., Yuan, T. T., and Cui, S. J. (2012). The Arabidopsis LFR gene is required for the formation of anther cell layers and normal expression of key regulatory genes. Mol. Plant 5, 993–1000. doi: 10.1093/mp/sss024
Xing, S. P., Salinas, M., Höhmann, S., Berndtgen, R., and Huijser, P. (2010). miR156-targeted and nontargeted SBP-box transcription factors act in concert to secure male fertility in Arabidopsis. Plant Cell 22, 3935–3950. doi: 10.1105/tpc.110.079343
Xu, X. F., Wang, B., Lou, Y., Han, W. J., Lu, J. Y., Li, D. D., et al. (2015). Magnesium transporter 5 plays an important role in Mg transport for male gametophyte development in Arabidopsis. Plant J. 84, 925–936. doi: 10.1111/tpj.13054
Xu, Y., Iacuone, S., Li, S. F., and Parish, R. W. (2014). MYB80 homologues in Arabidopsis cotton and Brassica: regulation and functional conservation in tapetal and pollen development. BMC Plant Biol. 14:278. doi: 10.1186/s12870-014-0278-3
Yang, C., Vizcay-Barrena, G., Conner, K., and Wilson, Z. A. (2007). MALE STERILITY1 is required for tapetal development and pollen wall biosynthesis. Plant Cell 19, 3530–3548. doi: 10.1105/tpc.107.054981
Yang, W. C., Ye, D., Xu, J., and Sundaresan, V. (1999). The SPOROCYTELESS gene of Arabidopsis is required for initiation of sporogenesis and encodes a novel nuclear protein. Genes Dev. 13, 2108–2117. doi: 10.1101/gad.13.16.2108
Yeung, E. C., Oinam, G. S., Yeung, S. S., and Harry, I. (2011). Anther, pollen and tapetum development in safflower, Carthamustinctorius L. Sex Plant Reprod. 24, 307–317. doi: 10.1007/s00497-011-0168-x
Yi, J., Moon, S., Lee, Y.-S., Zhu, L., Liang, W., Zhang, D., et al. (2016). Defective tapetum cell death 1 (DTC1) regulates ROS levels by binding to metallothionein during tapetum degeneration. Plant Physiol. 170, 1611–1623. doi: 10.1104/pp.15.01561
Zhang, D., Liu, D., Lv, X., Wang, Y., Xun, Z., Liu, Z., et al. (2014). The cysteine protease CEP1, a key executor involved in tapetal programmed cell death, regulates pollen development in Arabidopsis. Plant Cell 26, 2939–2961. doi: 10.1105/tpc.114.127282
Zhang, W., Sun, Y., Timofejeva, L., Chen, C., Grossniklaus, U., and Ma, H. (2006). Regulation of Arabidopsis tapetum development and function by DYSFUNCTIONAL TAPETUM1 (DYT1) encoding a putative bHLH transcription factor. Development 133, 3085–3095. doi: 10.1242/dev.02463
Zhang, Z. B., Zhu, J., Gao, J. F., Wang, C., Li, H., Li, H., et al. (2007). Transcription factor AtMYB103 is required for anther development by regulating tapetum development, callose dissolution and exine formation in Arabidopsis. Plant J. 52, 528–538. doi: 10.1111/j.1365-313X.2007.03254.x
Zhao, D. Z., Wang, G. F., Speal, B., and Ma, H. (2002). Theexcess microsporocytes1 gene encodes a putative leucine-rich repeat receptor protein kinase that controls somatic and reproductive cell fates in the Arabidopsis anther. Genes Dev. 16, 2021–2031. doi: 10.1101/gad.997902
Zhao, X., de-Palma, J., Oane, R., Gamuyao, R., Luo, M., Chaudhury, A., et al. (2008). Os TDL1A binds to the LRR domain of rice receptor kinase MSP1, and is required to limit sporocyte numbers. Plant J. 54, 375–387. doi: 10.1111/j.1365-313X.2008.03426.x
Zhou, S. M., Chu, Y. X., Zheng, B., and Zhang, W. (2014). Optimization of BY2 cell suspension as a stable transformable system. Not. Bot. HortiAgrobo. 42, 472–477.
Keywords: Arabidopsis, DYT1, cis-element, tissue specificity, tapetum
Key message: Through site mutation assay it was found that the “T” and final “C” nucleotides of key cis-element “CTCC” of Arabidopsis tapetum gene DYT1 promoter were irreplaceable for tissue specific gene expression.
Citation: Zhou S, Zhang H, Li R, Hong Q, Li Y, Xia Q and Zhang W (2017) Function Identification of the Nucleotides in Key cis-Element of DYSFUNCTIONAL TAPETUM1 (DYT1) Promoter. Front. Plant Sci. 8:153. doi: 10.3389/fpls.2017.00153
Received: 07 November 2016; Accepted: 25 January 2017;
Published: 17 February 2017.
Edited by:
Zhong-Jian Liu, The National Orchid Conservation Center of China, The Orchid Conservation and Research Center of Shenzhen, ChinaReviewed by:
Tatiana Arias, The Corporation for Biological Research, ColombiaCopyright © 2017 Zhou, Zhang, Li, Hong, Li, Xia and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Zhang, emh3NjIyMDdAc2h1LmVkdS5jbg==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.