
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 21 February 2017
Sec. Plant Physiology
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.00150
This article is part of the Research Topic Phytohormones and the Regulation of Stress Tolerance in Plants: Current Status and Future Directions View all 28 articles
Multiple environmental stresses affect growth and development of plants. Plants try to adapt under these unfavorable condition through various evolutionary mechanisms like physiological and biochemical alterations connecting various network of regulatory processes. Transcription factors (TFs) like APETALA2/ETHYLENE RESPONSE FACTORS (AP2/ERFs) are an integral component of these signaling cascades because they regulate expression of a wide variety of down stream target genes related to stress response and development through different mechanism. This downstream regulation of transcript does not always positively or beneficially affect the plant but also they display some developmental defects like senescence and reduced growth under normal condition or sensitivity to stress condition. Therefore, tight auto/cross regulation of these TFs at transcriptional, translational and domain level is crucial to understand. The present manuscript discuss the multiple regulation and advantage of plasticity and specificity of these family of TFs to a wide or single downstream target(s) respectively. We have also discussed the concern which comes with the unwanted associated traits, which could only be averted by further study and exploration of these AP2/ERFs.
Plants are mostly affected by multiple stresses like salinity, toxicity, pathogen attack, and wounding. To survive and reproduce in adverse conditions, plants develop various adaptive traits governed through a complicated system. The interconnected and diverse networks operated during downstream signaling cascades are regulated by TFs which bind to the cis elements present in their promoters (Papdi et al., 2015). AP2/ERFs are one of the most important families of TF in plants which regulate various developmental and stress responsive pathways (Licausi et al., 2013; Li et al., 2015). The specific and/or differential binding affinity of these AP2/ERFs to multiple cis-elements and other proteins enable them to respond to different regulatory processes simultaneously (Shoji et al., 2013; Wang L. et al., 2014). In this review we will highlight the multiple roles of AP2/ERFs in regulating diverse plant processes, their mechanism of action, auto/cross-regulation properties and differential interaction at protein and DNA level. We will also discuss how AP2/ERF family of TFs are regulated by different plant growth regulators (PGRs) and their role in retrograde signaling. With multiple responses there comes both positive and negative regulation which needs proper concern before generation of recombinant plants. These unexplored detrimental effects need to be addressed first for commercialization of such plants. Our objective is to focus on the recent advancements made in this area to the best of our knowledge in order to make a comprehensive review on the roles of this family of proteins in plants.
AP2/ERFs are mainly plant-specific TFs with 122 and 139 ERF family genes in Arabidopsis and rice respectively, although it has been speculated that they might have been descended from non-plant organisms (Nakano et al., 2006). Number of AP2/ERFs present in different plants are: Triticum spp.- 117 (Zhuang et al., 2011), Populus spp.- 200 (Zhuang et al., 2008), Brassica spp.- 291 (Song et al., 2013, 2016), Vitis spp.- 149 (Ito et al., 2014), Citrus spp.- 108 (Licausi et al., 2010), Solanum spp.- 155 (Charfeddine et al., 2015), Phyllostachys spp.- 116 (Wu et al., 2015), Daucus spp.- 267 (Li et al., 2015), Eucalyptus spp.- 209 (Cao et al., 2015), Salix spp.- 173 (Rao et al., 2015), Cassava spp.- 147 (Fan et al., 2016), Barley spp.- 121 (Guo et al., 2016) Gossypium spp.- 271 (Lei et al., 2016) and Bamboo spp.- 142 (Huang Z. et al., 2016). His and Asn rich HNH class of homing endonucleases from Tetrahymena thermophile (Eukaryote ciliate), Trichodesmium erythraeum (cyanobacterium), Enterobacteria phage RB49 (virus), and Bacteriophage Felix 01 (Virus) are reported to have an AP2 domain (Magnani et al., 2004; Wuitschick et al., 2004). Homing endonucleases promote high-frequency lateral transfer of the intervening sequence, which might explain the origin and gene exchange theory between non-plant organisms and plants (Wessler, 2005). Most of the AP2/ERFs lack introns which further supports the hypothesis that they are supposed to be evolved from a prokaryote after horizontal gene transfer (Magnani et al., 2004). Based on DNA binding domain (DBD), AP2/ERFs are classified into AP2, RAV (related to Abscisic acid insensitive3/Viviparous1), DREB (subgroup A1–A6), ERF (subgroup B1-B6), and others (Sakuma et al., 2002). Due to their ability to bind with the multiple cis-elements with their highly conserved DBD this member of protein family is likely to be involved in multiple responses in plants. Earlier Dehydration Responsive Element Binding Proteins (DREB) were thought to impart tolerance to cold, dehydration and salt stress through DRE/CRT elements while ERF leads to biotic stress tolerance through GCC-box element (Shinozaki and Yamaguchi-Shinozaki, 2003; Guo and Ecker, 2004; Huang et al., 2008). But now many DREBs (CBF1, TINY, HARDY) and ERFs (RAP2.4, ERF1, HRE2, ThERF1, TdSHN1) have been identified that are activated by different interconnected pathways which binds to both DRE and GCC elements as shown in Table 1 (Sun et al., 2008; Yang et al., 2009; Cheng et al., 2013; Wang L. et al., 2014; Zhu et al., 2014; Djemal and Khoudi, 2015; Lee et al., 2015). One of the probable reasons for multiple regulatory responses are due to their differential binding to the promoters, they can mediate simultaneous regulation of multiple responses. But the mechanism of this complicated regulation is still far from being fully elucidated. The mechanism identified in a plant may vary for a homolog in another plant. Some recent reports suggest that AP2/ERFs can regulate both abiotic and biotic stress in plants (Li X. et al., 2014; Li Z. et al., 2014; Jisha et al., 2015; Tian et al., 2015; Zhang et al., 2015; Figueroa-Yanez et al., 2016; Romero et al., 2016; Wang X. et al., 2016). In one of our study an AP2/ERF (PsAP2) from Poppy which bind to both DRE as well as GCC box elements thus provide abiotic and biotic stress tolerance in tobacco (Mishra et al., 2015). The crosstalk in the signaling pathways or interaction/stimuli dependent activation or differential binding affinity could be the keys in regulational cascades of above mentioned ERFs (Eini et al., 2013; Pandey et al., 2015; Zeng et al., 2015).
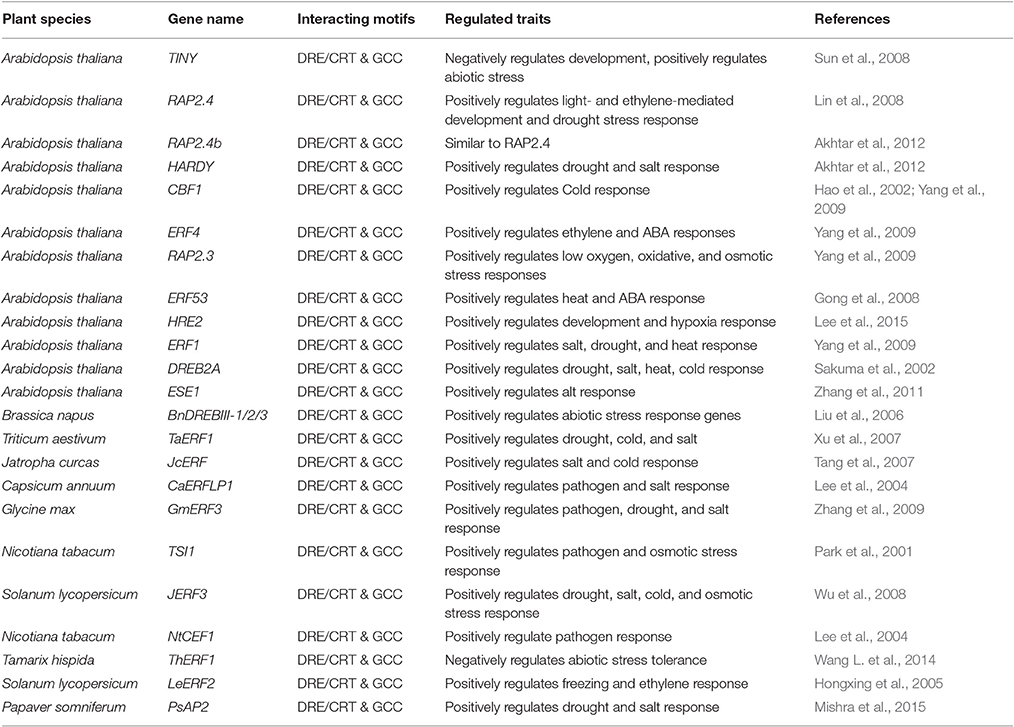
Table 1. List of AP2/ERFs that show multiple developmental and stress responses by binding to various cis-elements.
Expression of AP2/ERF and their downstream genes were tightly regulated in plants in order to maintain a perfect balance between stress and developmental responsive pathways. An example of complex transcriptional regulation was observed in DREBs under cold stress. Promoter of DREB1C/CBF2 contains several cold inducible motifs such as ICEr1 (Inducer of CBF Expression region 1), ICEr2 and CM2-Conserved DNA Motif 2 (Doherty et al., 2009; Zarka et al., 2003). bHLH (Basic Helix loop Helix) ICE1 (Inducer of CBF Expression 1) binds to ICEr1 motif under cold stress and activate transcription of DREB1C (Chinnusamy et al., 2003). Under control condition ICE1 remains unstable because of ubiquitination by a RING (Really Interesting New Gene) finger E3 ligase (HOS1) but under cold stress polyubiquitination of ICE1 is prevented by sumoylation by SIZ1-SUMO E3 ligase (Dong et al., 2006; Miura et al., 2007). ICE1 physically interacts with a R2R3 MYB transcriptional activator MYB15 to induce expression of DREB1/CBF and provide cold tolerance in Arabidopsis (Agarwal et al., 2006). Calmodulin-binding transcription activators CAMTA3 also binds to CM2 motif present in the promoter of DREB1/CBF and regulate their expression in calcium-dependent manner (Doherty et al., 2009). Another motif present in DREB1C/CBF2 promoter is G-box (CACGTG) to which a bHLH PIF7 (Phytochrome Interacting Factor) interacts and suppresses expression of DREB1C. PIF7 interacts with TOC1 (Timing of CAB expression 1) and PhyB (phytochrome B) to transcriptionally repress the expression (Kidokoro et al., 2009; Dong et al., 2011). DREB1C/CBF2 are also regulated by MYB-related TFs CCA1 (Circadian Clock Associated 1) and LHY (Late Elongated Hypocotyl) that bind to the EE (AAAATATCT) and CCA1 (AATCT) motifs of DREB1C/CBF2 promoters and positively regulate their expression (Alabadi et al., 2001; Nakamichi et al., 2009). These TFs are regulated so sophistically because under control conditions AtDREB2A leads to growth retardation and reduced reproduction as seen in over-expressed lines (Kim et al., 2012). Therefore, under non-stressed condition plants recruit GRF7 (Growth-Regulating Factor 7) that recognizes TGTCAGG element in the promoter of DREB2A and represses its expression. These complex regulation of DREBs through which they modulate different responses are mentioned in Table 2. Not only DREB2A but constitutive expression of AtDREB1A, AtERF1, OsERF1, and AtERF14 shows growth retardation which is not a desirable trait under normal condition (Onate-Sanchez et al., 2007). Another example where AP2/ERFs are tightly regulated is the regulation of ORCA 3 (Octadecanoid Responsive Catharanthus AP2-Domain Protein). ORCA3 has a regulatory role in primary and secondary metabolism in Catharanthus roseus, therefore it needs to be properly regulated at the transcriptional level. Its promoter carries a 74-bp long autonomous Jasmonate Responsive Element (JRE) having a quantitative sequence responsible for enhanced expression and a qualitative sequence that acts as an on/off switch in response to MeJA. CrMYC2 binds to T/G-box (CACGTG–G box with one mismatch) present in the qualitative sequence of JRE and mediate expression of ORCA3 that in turn regulate early jasmonate-responsive genes (Zhang et al., 2011). AT-Hook DNA-binding proteins also recognize JRE element and regulate expression of ORCA3 (Vom Endt et al., 2007). In Arabidopsis AtMYC2 regulate ORCA3 by binding with JRE element and regulate JA-responsiveness (Montiel et al., 2011). There are other reports in which these ERFs are transcriptionally regulated through different responses and complex mechanisms (Cole et al., 2009; Li et al., 2011; Ogata et al., 2015). So orchestration of these TFs (DREB2A, ORCA3, and most likely other ERFs) in plants through a single cascade is not possible because of multiple interacting and regulatory partners associated with them. One way could be the use of stress-inducible promoter rather than constitutive ones, but promoter and stimuli strength would greatly influence the expression of transgene under these conditions.
Apart from transcriptional regulation DREBs are regulated at the post-transcriptional level. Modifications through alternative splicing have been reported in rice OsDREB2A/2B (Matsukura et al., 2010), barley HvDRF1 (Xue and Loveridge, 2004), wheat WDREB2 (Egawa et al., 2006), and maize ZmDREB2A (Qin et al., 2007) to regulate proper functioning of them. Under normal conditions they produce an inactive transcript containing stop codon before the DNA-binding domain while under stress they produce an active transcript encoding a full-length protein. AP2/ERFs regulate multiple processes; therefore rDNA-technology mediated transcriptional inactivation through alternate splicing may interfere with the regulation of developmental responses. So exploration of the regulatory network involved in alternate splicing is necessary to modulate post-transcriptional changes in the genetically modified plants. Another mode of regulation involves non-coding small RNA or microRNA (miRNA). It is reported that miRNA172 regulate many protein-coding AP2s such as AT2G28550, AT5G60120, and AT5G67180 (Chen, 2004). They are involved in gene silencing and post-transcriptional gene regulation that occurs either via degradation of mRNA or silencing its translation. Mature miRNA often becomes a part of RNA-induced silencing complex (RISC) having Dicer and many other proteins to regulate RNA silencing. The miRNA172 regulates floral organ development and floral stem cell proliferation by acting as a potential translational repressor of AP2/ERF proteins. This regulation is very important for proper development of the reproductive organs and for the timely termination of floral stem cells. The putative miRNA172 binding sites are located within the coding regions of the genes but are outside of the conserved AP2 domains (Chen, 2004). Apart from miRNA172, other miRNAs such as miRNA156 and miRNA838 are reported to be probably involved in AP2/ERF regulation (Kavas et al., 2015). Now miRNA biogenesis is under tight temporal and spatial control, regulated at multiple levels like transcription, modification, processing in nucleus, and cytoplasm. So study of miRNA and its downstream regulation of AP2/ERF seem very important to further understand their role in different responses.
For proper regulation of several physiological processes in plants such as translational and post-translation modifications are key features of AP2/ERFs. Phosphorylation by kinases is one such event thereby which the AP2/ERFs are activated. Based on functional classification these kinases modify different substrates such as Serine/threonine protein kinases phosphorylate the OH group of serine or threonine and tyrosine-specific protein kinases phosphorylate tyrosine amino acid residues. Phosphorylation by different kinases usually results in a functional change of the target protein by changing activity, cellular location, or association with other proteins. For example PgDREB2A when phosphorylated at Thr residue(s) alters its binding affinity to DRE/CRT cis-element (Agarwal et al., 2007). When BWMK1 (blast and wound-induced MAP kinase1) through its TDY phosphorylation motif phosphorylates OsEREBP1 which enhances DNA-binding activity of OsEREBP1 which further activates several basic pathogenesis-related genes by binding to the GCC box element (AGCCGCC) present in their promoter (Cheong et al., 2003). Similarly JA-modulated MAPKK1 (JAM1) leads to increased transcriptional regulation of ORC1 through phosphorylation (De Boer et al., 2011). An example of kinase regulating cellular location is Histidine kinases and histidine-containing phosphotransfer proteins. Through phosphorylation it enables relocation of CRFs from the cytosol into the nucleus in response to cytokinin (Rashotte et al., 2006). Phosphorylation also affects protein-protein association as seen in the case of AtERF104. Pathogen-responsive mitogen-activated protein kinase MPK6 phosphorylates AtERF104, which is released as a substrate from MPK6 in response to FLG22 through ethylene signaling. This leads to reduced activation of AtERF104 and increased susceptibility to bacterial and fungal pathogen (Bethke et al., 2009). Hence, the phosphorylation is essential for activation of AP2/ERFs for different purposes, but limited information is available on the entire phosphorylation cascade. Both Up and downstream components of MAPKs; other interacting, inducing and inhibiting partners of MAPKs are unknown for most of the AP2/ERFs. Therefore, genetic alteration in plants through overexpression or silencing of MAPKs poses great uncertainty and so proper study is necessary to understand their role in activation of AP2/ERFs.
Many group VII hypoxia responsive AP2/ERFs are regulated by this unique N-end rule pathway. Under control condition these AP2/ERFs such as HRE1/2 except RAP2.12 are modified at the N-terminal by Pco1/2 (Plant Cysteine Oxidase 1) which are then recognized and degraded by N recognins ATE1/2 (Arginyl tRNA Protein Transferases) & PRT6 (Proteolysis 6) as shown in Figure 1. The N-end rule pathway is O2 and NO dependent in which N-terminal methionine is cleaved by methionine amino-peptidase and Cys which is oxidized by the process that depends upon availability of O2. This can be involved in the acidification of cytosol, change in redox state, ROS or NOS mode of action and enzymatic activities (Pucciariello and Perata, 2016). Under oxygen deprivation these TFs become stable and activate hypoxia responsive genes. RAP2.12 escape from N-end degradation by interacting with ankyrin-domain of plasma membrane linked ACBPs (Acyl-CoA binding protein). This interaction retains RAP2.12 at membrane site ensuring a minimum reservoir during the fast proteasomal degradation (Li et al., 2008; Licausi et al., 2011). Similarly ACBP4 is the interacting partner of RAP2.3 in the cytosol and nuclear membrane (Li et al., 2008). Under normal conditions Hypoxia Response Attenuator1 (HRA1) interacts with RAP2.12 to change and repress transcriptionally hypoxia-responsive genes (Giuntoli et al., 2014). N-end rule pathway can be targeted specifically to regulate certain conditions like hypoxia and submergence response. But Submergence-responsive (Sub1) is not regulated by N-end rule though it possesses the same N-terminal recognition sequence (Gibbs et al., 2011). It raises the question whether a single pathway can be applied universally to regulate a particular response due to N-end rule recognition. The further exploration of hypoxia-related group VII ERFs and role of N-end rule recognition will provide better understanding of role of this family of protein under oxygen deprived condition.
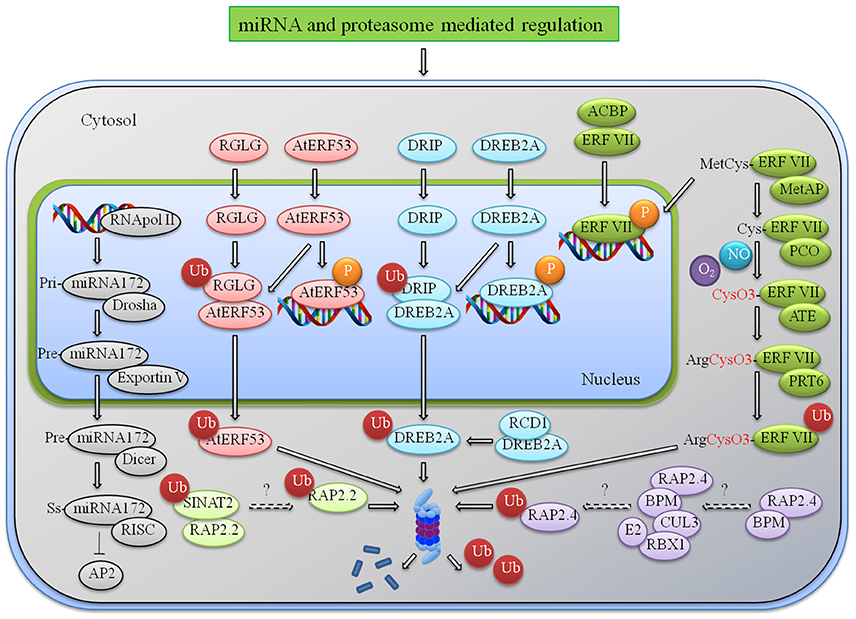
Figure 1. miRNA and proteasome mediated regulation of AP2/ERFs. An illustrative model representing the regulation of AP2/ERFs at various levels during multiple stress responses. After onset of stress multiple ERFs are induced and they in turn interact with the cis elements present in the promoter of stress-responsive genes and provide tolerance to various stress. Under normal condition they induce some negative effects like reduced height and senescence that needs to check by indegenous system. Degradation or translational repression by miRNA172 is one example. Other mechanisms include proteasome mediated degradation by different E3 ligases. RGLG interacts with AtERF53 and DRIP interacts with DREB to regulate their expression under normal condition. N-end rule mediated degradation of group VII ERFs under normal (no hypoxia) condition is another mechanism. RAP2.2 and RAP2.4 are also processed for proteasome degradation by SINAT2 and CUL3 ligases. Abbreviations: RGL, RalGDS-like; PRT6, Proteolysis6; SINAT, Seven in absentia 2 of Arabidopsis thaliana; DRIP, DREB2A-interacting protein; RCD1, Radical-induced cell death 1; BPM, BTB/POZ-MATH (bric-a-brac/POX virus and zinc finger—meprin and TRAF homology); RBX1, RING-box protein 1; CUL3, Cullin3; Met AP, methionine aminopeptidase; ATE, arginine transferase; PRT6, Proteolysis 6; RISC, RNA-induced silencing complex.
Ubiquitination is another post-translational mechanism through which AP2/ERFs are regulated. It may affect proteins in number of ways, either it process them for proteasomal-mediated degradation, or change cellular location, or alter their activity or enhance/prevent interaction of protein. Adding to the list of tight regulation of DREBs is the ubiquitination by DRIP1/2 (DREB2a-Interacting Protein1) under non-stressed condition. These are RING E3 ligases that interact with DREB2A in the nucleus and leads to its ubiquitination in the cytosol (Qin et al., 2008). RING domain containing ligase seems to majorly target AP2/ERFs for protein degradation as seen in Figure 1. For example SINAT2 (SEVEN IN ABSENTIA OF ARABIDOPSIS2) process AtRAP2.2 and RGL1/2 (RalGDS-like) process AtERF53 for proteasome-mediated degradation (Welsch et al., 2007; Cheng et al., 2012). Another cullin-based E3-ligases targets RAP2.4a to 26S proteasome by interacting with the MATH (Meprin and TRAF-tumor necrosis factor receptor-associated factor homology) domain of BTP/POZ (Broad complex, Tram track, bric-a-brac/POX virus and Zinc finger) proteins (Weber and Hellmann, 2009). Regulation of AP2/ERFs through proteasome-mediated degradation to obtain a balance in the stressed and control condition is not very well-understood. Also how protein turnover rate is maintained the under different environmental/developmental condition is a major area of future research.
AP2/ERFs physically interact with other proteins, which helps in localization, stability, abundance, transcriptional activity and target specificity. An example for protein-protein interaction includes DREB1A, DREB2A, and DREB2C proteins that specifically interact with the AREB1/ABF2 (ABRE-binding protein/ABRE-binding factor) and AREB2/ABF4 proteins to regulate ABA response (Lee et al., 2010). Huge number of interactions of other proteins with AP2/ ERF has been reported to mediate various responses (Buttner and Singh, 1997; Diaz-Martin et al., 2005; Song et al., 2005; Kagale and Rozwadowski, 2011; Wang X. H. et al., 2014). Some are involved in biotic responses (GmERF5 interact with GmbHLH and eukaryotic translation initiation factor GmEIF to provide Phytophthora sojae resistance in soyabean) while some are involved in development (OsERF3 and WOX11 interact with cytokinin-responsive gene RR2 to regulate crown root development in rice) (Dong et al., 2015; Zhao et al., 2015). Intensive complicated interaction is observed in certain AP2/ERFs such as AtERF5 physically interacts with AtERF6, AtERF8, SCL13, MPK3, and MPK6 like proteins to regulate wide array of responses. On one hand it positively regulate salicylic acid signaling and plant defense against the bacterial pathogen P. Syringae and fungal pathogen B. cinerea while on the other hand it negatively regulate chitin signaling and plant defense against the fungal pathogen Alternaria brassicicola (Moffat et al., 2012; Son et al., 2012). Yeast 2 hybrid, Co-immunoprecipitation and Bimolecular fluorescence complementation assays should be performed extensively not only in model plants but in other economically important plants to identify the vast network of AP2/ERF proteins and their interacting partners under different conditions. This is not an easy task but differential binding affinity in closely related species asks the need to perform these assays prior to genetic modifications in plants.
Approximately 60 amino acid DBD of AP2/ERFs consists of a three-stranded β-sheet and one α-helix running almost parallel to the β-sheet. The Arg and Trp residues of the β-sheet are necessary for the contact with DNA (Allen et al., 1998). Divergent DNA-binding specificities are seen in many ERFs affecting their affinity to different cis elements. Alignment of the DBD of AP2/ERFs based on their differential binding along with their phylogenetic analysis is shown in Figure 2. With our current knowledge based up on their characterization the AP2/ERFs that bind specifically to GCC motif carries an extra amino acid (basic polar) at 24th position of DBD accounting for 61 amino acid long DBD. While presence of Glu at 20th position and Ala at 49th position of DBD seems to favor DRE binding. Conserved Val (15th position) is also crucial in the regulation of the binding activity of DREB1A to the DRE cis-element (Cao et al., 2001). DRE binding AP2/ERFs consist of 2 different clades with one clade containing DREB2s and rest in other clade. Amino acid alignment of DREB2s showed more similarity with the AP2/ERFs that binds to GCC or GCC and DRE. At 18th, 38th, 43rd, and 60th position DREB2s showed similarity with GCC and GCC and DRE binding ERFs but not with other clade of DRE binding ERFs (Figure 2). Specific amino acid in DBD that regulate binding to both the cis elements is still not known but it is highly possible that motifs present outside DBD may regulate this differential binding. There are reports of modification in DBD which alters binding affinity of certain ERFs. Substitution of a conserved Arg to Lys in the first β-strand of AtERF189 which normally interacts with P-box (CCGCCCTCCA) was able to bind with the second GC pair of the GCC box cis element (AGAGCCGCCA). But an increased number of basic amino acids in the first two β-strands of ORCA3 allow recognition of more than one cis-element like GCC box, P box, and CS1 (TAGACCGCCT) presumably via increased electrostatic interactions with the negatively charged phosphate backbone of DNA (Shoji et al., 2013). In a recent report, it is shown that Arg in 156th position in DBD shows the highest binding affinity to the GCC box (Zhuang et al., 2016). Apart from DNA binding ability, AP2/ERF DBD is reported to be involved in protein-protein interactions. DBD of DRN and DRNL interact with PAS-like domain of HD-ZIP proteins Phavoluta, Phabulosa, Revoluta, and AtHB8. These complexes act as a transcriptional unit in the control of embryo patterning (Chandler et al., 2007). Though AP2 DBD is highly conserved but evolutionary modification is seen in different members of this family ranging from number of DBD (2/1) to linker domains. Other conserved motifs span the entire amino acid sequence of AP2/ERFs (Nakano et al., 2006). Therefore, alteration of DNA binding affinity/specificity of AP2/ERFs through modification of DBD may require attention from many amino acids and motifs present outside of DBD.
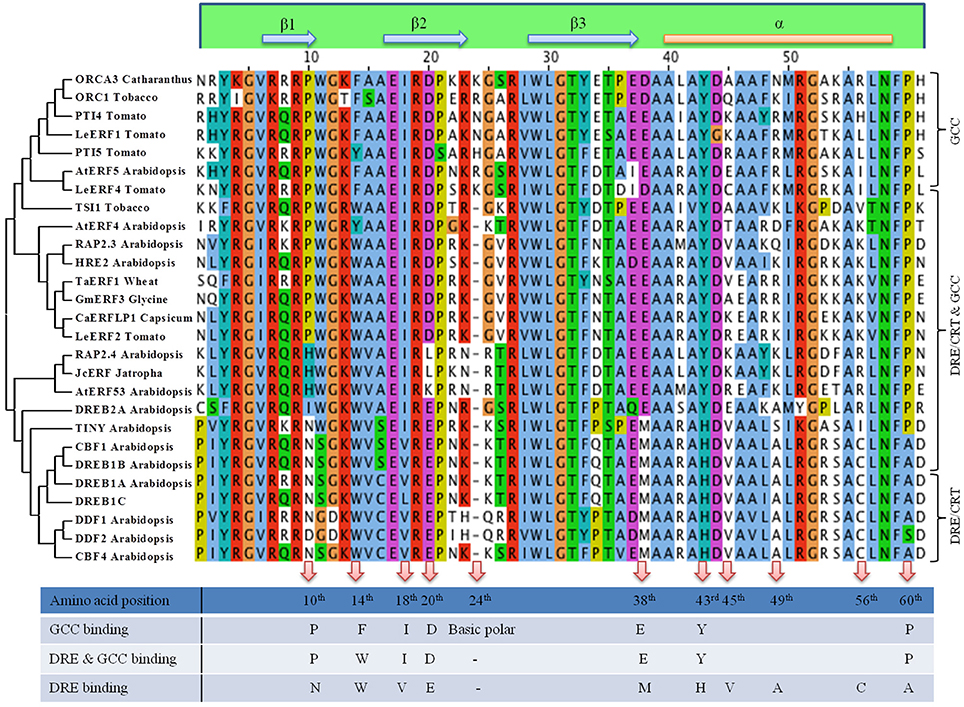
Figure 2. Clustal and Phylogenetic analysis of DNA binding domain of AP2/ERF. Based on the conserved amino acid sequences present in the DNA binding domain of AP2/ERFs Clustal Omega multiple alignment was generated. Three groups have been considered, one group that binds to both GCC box cis element, one group that binds to DRE cis element and one group that binds to both DRE and GCC cis element. Phylogenetic clustering was done using MEGA6 neighbor joining algorithm. Amino acids that may regulate specific or multiple binding to downstream cis element is also shown.
TFs interact with their DBD to the cis-motifs presents in the promoter of downstream genes and then activation or repressor domains further enhance or suppress expression of them. As studied earlier DREBs are extensively studied for their complex regulation. Another mechanism, through which DREB2A expression is regulated, is by hydrophilic Ser/Thr-rich negative regulatory domain (NRD) adjacent to DBD that makes DREB2A unstable under normal condition. Deletion of NRD converts DREB2A into its constitutively active form whose overexpression displays reduced growth but improved drought and salt tolerance (Sakuma et al., 2006). Tiwari et al. (2012) identified a potential transactivation domain in AtERF98 as EDLL motif (xxExxxxDxxxLxxxL) having acidic amino acids and hydrophobic leucines. EDLL motif of certain AP2/ERFs like ORA59, AtERF98, AtERF1, and AtERF15 physically interact with ACID domain of MED25 to regulate Jasmonate and stress signaling (Ou et al., 2011; Cevik et al., 2012). In another report, acidic C-terminal region of ERF2, as well as both N-terminal and C-terminal regions of ERF4 from tobacco, act as transactivation domains which regulate activation of downstream genes (Ohta et al., 2000). Apart from the activation domains, certain negative or repressor domains such as C-terminal EAR (L/FDLNL/F(x)P) motif are present in many AP2/ERFs (Ohta et al., 2001; Zeng et al., 2015; Park and Grabau, 2016). Another repression motif BRD (L/VR/KLFGVXM/V/L) is identified in B3 DBD containing TFs like AtRAV1 and AtRAV2 (Ikeda and Ohme-Takagi, 2009). EAR or BRD motif interacts with the transcriptional corepressors TOPLESS (TPL) and TOPLESS-RELATED (TPR) to suppress the transcription of downstream target genes (Causier et al., 2012). The DREB2A/2B/2C contains a RIM (RCD1-interacting motif-FDXXELLXXLN) motif that interacts with RST (RCD1–SRO–TAF4 in which TAF4 stands for TATA-box-binding protein-associated factor 4) domain of RCD1 (Radical- Induced Cell Death 1) to regulate proteasome-mediated degradation under normal conditions. Under heat stress expression of RCD1 is contained, leading to increased expression of DREBs. DREBs without RIM motif like DREB2E and alternative splice variant DREB2A.2 could not interact with RCD1 (Vainonen et al., 2012). Activation or repression of downstream genes through these motifs may positively or negatively influence different responses. Some of the AP2/ERFs (with a conserved C-terminal EAR repressor domain) like GmERF4 (enhanced salt & drought tolerance), GmERF5 (Enhanced P. sojae resistance), GmERF6 (enhanced drought tolerance), and NtERF3 (increased resistance to TMV) positively regulates stress response while AtERF7 (reduced sensitivity of guard cells to ABA and increased transpirational water loss) negatively regulates stress response (Fischer and Droge-Laser, 2004; Song et al., 2005; Zhang et al., 2010; Zhai et al., 2013; Dong et al., 2015). Therefore, identification of activation or repressor domain, downstream target genes, domain regulation of AP2/ERFs and their role in a particular response is necessary so that we could attain success in development of genetically modified crops with minimal unwanted effects.
Crosstalk between cellular organelles to regulate multiple responses is a quite unexplored field. Data analyzed through graphical Gaussian Models of gene networks and other studies reveal that many members of the AP2/ERF family are involved in integration of signals derived from organelles in retrograde feedback loops and in stress acclimation (Dietz et al., 2010). In this study, AT2G44940 was identified with the highest score for chloroplast targeting. Schwacke et al. (2007) conducted a bioinformatics search to identify more than 10 nuclear-encoded AP2/ERFs with potential targeting to plastids or mitochondria. Among them AT2G44940, AT1G77640, AT1G44830, AT1G21910 were showing highest potential to be involved in retrograde signaling. Generally there are 3 plastid to nucleus retrograde signaling pathways, first one includes accumulation of Mg–protoporphyrin IX and downregulation of many genes in Arabidopsis, second one represses Lhcb expression when plastid gene expression is inhibited and third one involves redox state of the photosynthetic electron transfer chain that affects both photosynthesis-related and stress-related genes (Strand et al., 2003; Nott et al., 2006). In all three pathways a chloroplast-localized PPR protein (pentatricopeptide repeat protein family) GUN1 (genomes uncoupled) and nuclear localized ERF ABI4 (ABA insensitive 4) plays an important role. In this process PTM4 (Plant homeodomain type TF with transmembrane domain) regulates ABI4 by moving out from chloroplast to the nucleus and helping in the transmission of retrograde singals from plastid to ABI4. ABI4 has also been identified as a sugar-insensitive mutant, and sugar signaling through ABI4 has been linked to chloroplast retrograde signaling (Oswald et al., 2001; Finkelstein et al., 2002). Aberrant plastid function in Arabidopsis leads to induced expression of GUN1 in plastids, finally leading to ABI4 regulated repression of nuclear-encoded genes (Figure 3). ABI4 binds to the promoter of Lhcb (Light Harvesting Complex B) carrying a CUF1 element (Cab Upstream Factor 1, a G-box element required for retrograde signaling) and inhibits light-induced expression of photosynthetic genes when chloroplast development is arrested (Koussevitzky et al., 2007). ABI4 also regulates CBFA (CCAAT binding factor A), which is a subunit of the Heme activator protein trimeric transcription complex (HAP2/HAP3/HAP5). During various environmental stresses under ABA and ROS accumulation, ABI4 down-regulates CBFA, and allow other TF subunits to enter the transcription complex and improves transcriptional efficiency of stress responsive genes instantaneously (Zhang et al., 2013). Characterization of GUN mutant (gun6) showed that increased accumulation of heme which is a positive regulator of the genes associated with photosynthesis by inhibiting the expression of ABI4. Heme not only have an important role in signaling in other organism but also known to be actively transported out of the plastids (Thomas and Weinstein, 1990; Von Gromoff et al., 2008). Apart from that, transcript of several AP2/ERFs like AtERF6, AtERF104, RRTF1, and AtERF105 were involved in fast retrograde response when low-light acclimated Arabidopsis plants were transferred to high light (Khandelwal et al., 2008; Vogel et al., 2014). It is proposed that this transfer accumulate higher levels of DHAP (dihydroxyacetone phosphate) that is transferred from chloroplast to cytosol which activate protein phosphorylation cascades regulated by MPK6. Phosphorylation of constitutive but inactive AP2/ERF initiates downstream HSPs (Heat Shock Proteins) and kinases for acclimation response (Vogel et al., 2014).
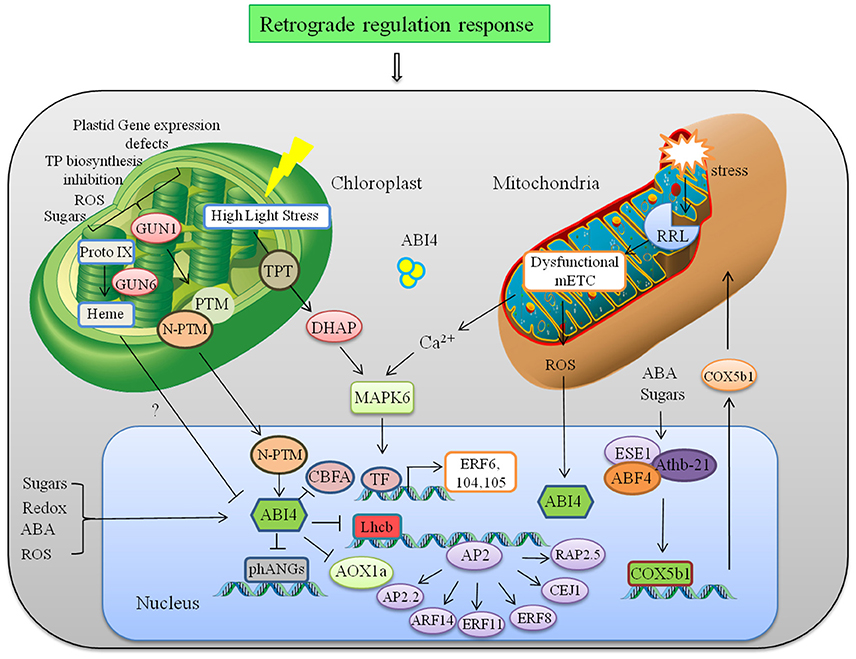
Figure 3. Retrograde regulation of AP2/ERFs. Abnormalities in plastid leads to induced expression of GUN1, which mediates retrograde signaling to regulate nuclear gene ABI4 and photosynthesis related genes like Lhcb. ABI4 is also regulated by mitochondrial retrograde signaling through mitochondria located RRL protein. RRL regulate expression of AOX1a to further to maintain ROS homeostasis. Another ERF ESE1 interacts physically with Athb21 and ABF4, all of which interact with the COX5b1 promoter to regulate respiratory chain complex. Abbreviations: GUN1, genomes uncoupled-1; PTM, Plant homeodomain type transcription factor with transmembrane domain; phANGs, Photosynthesis associated nuclear genes; CBFA, (CCAAT binding factor A;), ABI4, ABA-INSENTIVE 4; Lhcb, LIGHT HARVESTING COMPLEX B; AOX, Alternative oxidase; ESE1, Ethylene and salt inducible 1; ABF4, ABREbinding factor 4; COX5b1, cytochrome c oxidase subunit 5b-1.
Mitochondrial signal also regulates many AP2/ERFs through retrograde mechanism (Shaikhali et al., 2008; Mishra et al., 2015). ABI4 is not only involved in chloroplast retrograde signaling but also regulated transcriptionally through mitochondrial retrograde system (Yao et al., 2015). Mitochondrial stress leads to disruption of mitochondria-localized protein RRL (Retarded root growth-like) that causes impaired mitochondrial electron transport chain. It leads to reduced ABA-stimulated ROS production that induces expression of ABI4 in nucleus. ABI4 regulates expression of alternative oxidase (AOX1a) in the alternative respiratory pathway and is involved in seed germination and seedling growth (Figure 3). Another example of mitochondrial retrograde regulation involves expression of COX5b-1 (cytochrome c oxidase subunit 5b-1) gene that encodes an isoform of the cytochrome c oxidase zinc binding subunit involved in respiratory chain complex (Comelli et al., 2012). B-3 subgroup of AP2/ERF ESE1 (Early Somatic Embryogenesis 1) interacts with the distal B-like elements CCACTTG induced in response to ABA while HD-Zip protein Athb-21 interacts with ATCATT elements induced in response to carbohydrates in COX5b-1 promoter. ESE1 and Athb-21 interacts physically with AREB2/ABF4, which binds to the G-box which is required for expression of the COX5b-1 gene (Figure 3). Vogel et al. (2012) showed that many AP2/ERF such as ARF14, ERF11, ERF8, CEJ1, RAP2.2, and AP2-2 are likely to be involved in retrograde adjustment of nuclear gene expression. Retrograde and inter-organelle regulation is still an unexplored field and much information is required to fully understand the underlying mechanism. In context of the current knowledge photosynthesis-associated nuclear genes and nuclear respiratory complex genes are key components of this retrograde regulation.The net response of any stress is the cumulative interplay of plastid and mitochondria signaling toward nucleus and vice-versa. Therefore, understanding the role of AP2/ERFs in organelle signaling and their cross talk through retrograde signaling will further help us to manipulate the developmental, metabolic, and environmental need in different plants.
Ethylene signaling is an essential component involved in various development processes and stress responses. AP2/ERFs are highly responsive to these signaling cascades (Muller and Munne-Bosch, 2015). Receptors such as ETR1/2 (Ethylene Response1/2), ERS1/2 (Ethylene Response Sensor1/2), and EIN4 (Ethylene Insensitive4) located on the membrane of endoplasmic reticulum mainly perceive ethylene (Lacey and Binder, 2014). Downstream components of this signaling pathway includes CTR1 (Constitutive Triple Response1), EIN2/3 and ERFs (Kendrick and Chang, 2008; Stepanova and Alonso, 2009). Ethylene signaling regulates various ERFs such as AtERF1 (Cheng et al., 2013), AtERF4 (Yang et al., 2005), CarERF116 (Deokar et al., 2015), LchERF (Wu et al., 2014), and JcERF1 (Yang et al., 2014). CTR1and EIN2 both negatively and positively regulates ethylene signaling. Kinase activity of CTR1 is activated in absence of ethylene which phosphorylates the C-terminal of EIN2 preventing it from entering the nucleus (Kieber et al., 1993; Huang et al., 2003; Gao et al., 2008; Bisson and Groth, 2010; Ju et al., 2012). The absence of ethylene promotes interaction of EIN2 with F-box proteins ETP1/2 (Ethylene Insensitive2-Targeting Protein1/2) leading to reduced EIN2 level (Qiao et al., 2009). Under high ethylene level, CTR1 is inactivated and EIN2 is translocated to the nucleus where it directly or indirectly leads to the activation of EIN3 (Ju et al., 2012). Ethylene response is also enhanced by interaction of EIN2 with ECIP1 (EIN2 C-terminus Interacting Protein 1) an MA3 domain-containing protein (Lei et al., 2011). Ethylene also promotes accumulation of EIN3 that in turn initiates activation or repression of downstream stress responsive genes (An et al., 2010). EIN3 level is reduced in the absence of ethylene by interacting with F-box proteins EBF1/2 (Ethylene Insensitive3-Binding F-Box Protein1). Many waterlogging and submergence responsive genes are regulated by ethylene (Phukan et al., 2014, 2015; van Veen et al., 2014). EIN3 is reported to directly interact with SK1/2 (Snorkel1/2) that helps escape flooding by rapid ethylene and GA-mediated internode elongation (Hattori et al., 2009). AtRAP2.2 promotes activation of genes involved in ethylene biosynthesis such as ACS7 and ACO1 (Hinz et al., 2010). The Sub1A (Submergence 1) and Sub1C from rice are induced by ethylene but acts antagonistically to each other in response to flooding (Pena-Castro et al., 2011). Therefore, ethylene is an important upstream and downstream component of AP2/ERF mediated plant responsive pathways.
Many AP2/ERFs are responsive to ABA signaling. Apart from ABI4 that is involved in retrograde regulation as stated above, ABA-responsive AP2/ERFs regulate various responses. AtERF1/6 is negatively regulated by ABA (Cheng et al., 2013; Sewelam et al., 2013) while JERF1/3 (Wu et al., 2007, 2008), TaERF1 (Xu et al., 2007), GmERF3 (Zhang et al., 2009), CsERF (Ma et al., 2014), LchERF (Wu et al., 2014), and CarERF116 (Deokar et al., 2015) are positively regulated by ABA. It is reported that waterlogging leads to induced level of ABA that activates ERF RAP2.6 that provides enhanced oxidative stress tolerance in Arabidopsis (Liu et al., 2012). Importance of ABA-mediated regulation of ERF is highlighted from the fact that overexpression of AtERF4/7 inhibits ABA response and provide stress tolerance (Song et al., 2005; Yang et al., 2005). Although induction of AtERF1 is negatively regulated by ABA but its overexpression leads to increased ABA accumulation and proline which might be the one of the reason for enhanced drought, salt, and heat stress tolerance in transgenic lines (Cheng et al., 2013). JERF1 when overexpressed in tobacco activates ABA biosynthesis related genes such as NtSDR (short-chain dehydrogenase/reductase) and provide enhanced cold and salt stress tolerance (Wu et al., 2007). The overexpression of TSRF1 in tobacco increases the expression of NtSDR and ABA accumulation (Quan et al., 2010).
Various PGRs such as GA, SA, JA (Jasmonic acid) BR (brassinosteroid) also affect ERF mediated regulation. Hypoxia responsive HRE1 and HRE2 show ethylene response, but unlike RAP2.2 they lead to inhibition of ethylene signaling and provide GA response (Licausi et al., 2010). On the other hand SUB1A promotes activation of DWF1/4 (Dwarf1/4) involved in BR biosynthesis. DWF1/4 leads to accumulation of DELLA protein such as SLR1 (Slender Rice 1) and SLRL1 (SLR1 like) which inhibits GA responsiveness (Schmitz et al., 2013). AtERF6 is involved in complex crosstalk of ethylene and gibberellin/DELLA pathway. Under osmotic stress, AtERF6 activates Gibbereln2-Oxidase6 that leads to accumulation of DELLA protein which in turn suppresses GA responses. It was also found that AtERF6 induction was independent of EIN3 (Dubois et al., 2013). GA3 positively regulates the expression of MsERF8 and CaERF116 (Chen et al., 2012). It is reported that JA-mediated leaf senescence is promoted by ethylene. SUB1A delays the process of leaf senescence by inhibiting ethylene, JA and SA during stress. JA positively regulates expression of many ERFs such as Tsi1 (Park et al., 2001), OPBP1 (Guo and Ecker, 2004), JERF1 (Wu et al., 2007, 2008), AtERF1 (Cheng et al., 2013), GmERF3 (Zhang et al., 2009), AtERF6 (Sewelam et al., 2013), LcERF054 (Sun et al., 2014), and PsAP2 (Mishra et al., 2015). AtERF1 is considered to a key component in the ethylene/JA mediated defense response in Arabidopsis (Lorenzo et al., 2002). Activation of AtERF1under different stress conditions such as drought, salt, and heat which requires both ethylene and JA. JA-insensitive mutant failed to activate AtERF1 expression under stress conditions (Cheng et al., 2013). SA is an important plant defense hormone which positively regulates many ERFs such as TSRF1 (Huang et al., 2004), TaERF1 (Xu et al., 2007), GmERF3 (Zhang et al., 2009), MsERF8 (Chen et al., 2012), AtERF6 (Sewelam et al., 2013), and CarERF116 (Deokar et al., 2015) while it negatively regulates SodERF3 (Trujillo et al., 2008) and CsERF (Ma et al., 2014). JA cross-talk with SA to fine tune plant immune signaling network. It was reported that SA antagonizes certain JA-responsive genes, partly by ORA59 mediated transcriptional activation. The ERFs regulate the expression of ethylene and JA-responsive genes by targeting GCC box present in their promoters. It seems that GCC-box is sufficient for SA-mediated suppression of JA-responsive gene expression (Caarls et al., 2016).
Crop yield is severely affected because of various biotic and abiotic environmental factors. Over the last decade, AP2/ERF proteins have become the subject of intensive research activity in crops due to their involvement in a variety of biological processes. There are many reports in which significant results have been obtained through manipulation and regulation of AP2/ERF-like TFs. In the current scenario even commercialization of transgenics could be possible, but as stated above proper scrutiny at the every level is entirely necessary. As AP2/ERFs can regulate multiple responses in a simultaneous and cooperative manner they can be targeted for improved variety development which may deliver multiple tolerance traits to a single breed. Some of the recent findings are mentioned below.
It is one of the worldr's most important staple food crops. Generally half of the total rice cultivation is affected by environmental stress decreasing crop yield and quality. Also, it is a model plant where significant research has been done, which can be explored for generation of improved breeds with very limited unwanted traits. In a study using phylogenetic analysis of the rice genome, 170 AP2/ERF family genes were identified and divided into 11 groups, including four broad groups (AP2, ERF, DREB, and RAV), 10 subgroups, and two soloists (Rashid et al., 2012). SUB1A gene encoding an AP2 domain TF restricts GA functioning during sustained submergence in rice. SUB1A gene enhances accumulation of GA repressors SLR1 and SLRL1, thus limiting underwater internode elongation and augmenting submergence survival (Fukao and Bailey-Serres, 2008). Snorkel1 and Snorkel2 (SK1 and SK2) were identified to have an important role in internode elongation mainly post-mitotic expansion of differentiating cells separated from the region of intercalary meristem around node. SK1 and SK2 were suggested to trigger internode elongation via GA in response to rising water level (Hattori et al., 2009). By contrast, OsEATB was found to restrain GA responsiveness during the internode elongation process by down-regulating the expression of the GA biosynthetic gene OsCPS2 (Qi et al., 2011). Overexpression of OsAP37 showed enhanced tolerance to drought in rice plants under vegetative stage and improved grain yield (Oh et al., 2009). OsEREBP1overexpression activates the jasmonate and abscisic acid signaling pathways thus provide enhanced survival to rice plants under abiotic or biotic stress conditions (Jisha et al., 2015). Multi-Floret Spikelet1 (MFS1) gene was found to play an important role in regulating determinacy of spikelet meristem and floral organ identity. MFS1 is associated with unidentified function clade in the family of AP2/ERF which certainly regulates the expression pattern of Long Sterile Lemma (LSL) and the Indeterminate Spikelet1 (IDS1) like genes Supernumerary Bract and OsIDS1 (Ren et al., 2013). The auxin responsive CROWN ROOTLESS-5 (CRL5) has a function in crown root initiation in rice plants by inducing OsRR1 which act as type-A response regulator in cytokinin signaling. It was known that cytokinin has negative effects on de novo auxin-induced root formation (Kitomi et al., 2011). OsERF109 when overexpressed in rice negatively regulates ethylene biosynthesis and drought tolerance (Yu et al., 2017). OsERF71 overexpression in rice leads to root structure modifications, larger aerenchyma and radial root growth which provides improved drought tolerance of shoots. OsERF71 activate various stress responsive, cell wall-related and lignin biosynthesis associated genes causing root structural changes in rice (Lee et al., 2016). In a recent report OsERF115 target GCC box of OsNF-YB1 promoter to regulate endosperm development and grain filling in rice. OsNF-YB1 is specific to the aleurone layer of developing endosperms (Xu et al., 2016). CRISPR/Cas9-Targeted Mutagenesis has been used to generate rice lines with enhanced blast resistance through engineering OsERF922 mutation (Wang F. et al., 2016).
As rice, wheat is also important crop and cultivated all over the world. Several ERFs have been characterized from wheat whose regulation could potentially be used for improved variety development. TaPIE1 when over-expressed in wheat displayed significantly increased resistance to fungal pathogen Rhizoctonia cerealis and freezing stresses. TaPIE1 (Pathogen-Induced ERF1) transgenic wheat showed reduced electrolyte leakage and H2O2 content, while increased soluble sugar and proline content (Zhu et al., 2014). Another ERF, TaERF3 led to increased tolerance to salt and drought stresses. In TaERF3 overexpressing transgenic lines increased accumulation of proline was observed. These lines also inhibited chlorophyll degradation, H2O2 formation and stomatal conductance under both stresses. TaERF3 could interact specifically with the GCC-box cis-element present in the promoters of seven stress-responsive genes namely BG3 (β-glucans 3), Chit1 (Chitinase 1), RAB18 (ras-related protein 18), LEA3 (Late embryogenesis abundant 3), TIP2 (Delta tonoplast intrinsic protein 2), POX2 (Peroxidase 2), and GST6 (Glutathione S-Transferase 6) while could not bind with DRE cis-element (Rong et al., 2014). Overexpression of TaPIEP1 in wheat, which is induced in response to pathogen, confers increased resistance to fungal pathogen Bipolaris sorokiniana. The transgenic lines showed increased expression levels of certain defense-related genes in the ET/JA pathways, which might be responsible for enhanced disease resistance (Dong et al., 2010).
Tomatoes have been modified for slowing down the ripening process using genetic engineering like Flavr Savr, though none are commercially available now. But there are many recent developments made in this area till then. In transgenic tomato JREs, when overexpressed and silenced significantly affected Steroidal glycoalkaloids accumulation and expression of genes involved in its biosynthesis including the upstream mevalonate pathway. One of them, JRE4 binds with two structurally related elements, GCC box-like P box and the GCC box, which are also found on the promoters of biosynthetic genes like sterol reductase and glycoalkaloid metabolism 5 (Thagun et al., 2016). Another TF from tomato SlERF52 functions in flower pedicel abscission. Before the abscission stimulus, SlERF52 regulates the expression of pedicel abscission zone specific TFs, like tomato Wuschel Homolog, Goblet and Lateral Suppressor, which may lead to regulation of meristematic activities in pedicel abscission zones (Nakano et al., 2014). The tomato ERF Pti5 (Pto-interacting protein 5) contributes to potato aphid resistance in tomato as VIGS (Virus Induced Gene Silencing) of Pti5 enhanced aphid population growth (Wu et al., 2015). An extensive study of ERFs on ripening-impaired tomato mutants showed potential role of ERFs in Fruit Ripening in Tomato (Liu et al., 2016). When SlERF5 was overexpressed in tomato, plants showed increased tolerance to drought and salt stress and enhanced levels of relative water content (Pan et al., 2012). SlERF5 overexpressed tomato plants also accumulated higher levels of defense gene PR5 (Pathogenesis related 5) and displayed better tolerance to bacterial pathogen Ralstonia solanacearum. Constitutive expression of SlRAV2 in tomato also led to enhanced tolerance to R. solanacearum bacterial wilt and increased expression of SlERF5 and PR5 genes (Li et al., 2011). TERF2/LeERF2 when overexpressed in tomato, controls ethylene production and showed enhanced freezing tolerance (Zhang and Huang, 2010). Many ERFs from different cultivars of tomato have been identified which may be responsible for the defense mechanisms in response to Tomato yellow leaf curly virus (Huang Y. et al., 2016).
Lots of research and literature surrounding AP2/ERF are available for this model plant. Therefore, we are highlighting only some recent advances made in this area. AP2/ERFs positively/negatively regulate various processes to maintain proper homeostasis during stress and favorable conditions. AtERF019 overexpression delays flowering, plant growth and senescence in Arabidopsis thus impart drought tolerance. Genes involved in stress response such as BCAT3 (branched-chain-amino-acid aminotransferase3) and the zinc finger oxidative stress 2 are predicted targets of this ERF (Scarpeci et al., 2016). Recently reported AtERF105 provides cold tolerance by mediating CBF regulon in Arabidopsis (Bolt et al., 2017). AtERF014 is induced by both Pseudomonas syringae pv. tomato and Botrytis cinerea but it positively regulates P. syringae resistance while negatively regulates B. cinerea resistance in Arabidopsis. Overexpression of AtERF014 induces activation of SA-responsive AtPR1/5 genes while suppresses JA-ethylene responsive AtPDF1.2 (Zhang et al., 2016). Similarly AtERF15 is induced by P. syringae, B. cinerea, SA and JA but it provided enhanced resistance to both the pathogens in Arabidopsis (Zhang et al., 2015). Several JA/ethylene responsive ERFs also provide plant immunity in Arabidopsis such as AtERF1, AtERF6, ORA59, and AtERF96 (Huang P. Y. et al., 2016). AtERF96 provides increased resistance to B. cinerea and Pectobacterium carotovorum in Arabidopsis. It specifically interacts with the GCC box present in the promoters of defense responsive genes such as PDF1.2a, PR3/4, and ORA59 to modulate those responses (Catinot et al., 2016). AtERF96 has also been shown to be ABA-responsive. Overexpression lines significantly have elevated levels of ABA-responsive genes such as ABI5, RD29A, ABF4, ABF3, P5CS, and COR15A. AtERF96 overexpressed lines showed hypersensitivity to ABA and had reduced stomatal aperture (Wang X. et al., 2016). Group VII ERFs are responsive to hypoxia and are regulated through N-end rule pathway such as RAP2.2 and RAP2.12 (Gasch et al., 2016). But a recent report showed that under normoxia oxygen-dependent AtRAP2.12 stability plays a key role in central metabolic processes to maintain growth and development in Arabidopsis (Paul M. V. et al., 2016). Stem epidermis-specific WRI4 (Wrinkled4) regulates biosynthesis of cuticular wax in Arabidopsis stems by interacting promoters of various wax-biosynthesis related genes such as (LACS) 1long-chain acyl-CoA synthetase1, PAS2 (Pasticcino2), KCR1 (β-ketoacyl CoA reductase1), ECR (trans-2,3-enoyl-CoA reductase), and WSD1 (bifunctional wax synthase/acyl-CoA: Diacylglycerol acyltransferase). Wax layer in turn protects plants from high irradiance, desiccation and UV radiation (Park et al., 2016). AtERF11 leads to internode elongation by positively regulating both GA biosynthesis and GA signaling in Arabidopsis. Overexpression lines showed induced expression of GA3ox1 and GA20ox genes leading to elevated levels of GA while reduced ethylene levels (Zhou et al., 2016). AtERF11 also acts antagonistically to AtERF6 to regulate mannitol-induced growth inhibition in Arabidopsis. AtERF6 actually activates AtERF11, which in turn suppresses AtERF6 activated genes by directly competing for the promoters of downstream genes (Dubois et al., 2015).
Homologous gene expression is a difficult task as transformation and regeneration protocols differ from one species to other. Few examples of AP2/ERF have been successfully overexpressed in homologous system and found some promising results. GmERF5 is the first soybean EAR motif containing ERF proved to be involved against pathogen infection. GmERF5 overexpressing transgenic soybean enhanced resistance to P. sojae and positively regulates the expression of the pathogenesis-related (PR10, PR1-1, and PR10-1) genes (Dong et al., 2015). EjAP2-1 (AP2/ERF) from Eriobotrya japonica is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB (Zeng et al., 2015). Shi et al. (2014) characterized two tomato AP2/ERF genes, SlCRF1 and SlCRF2 (cytokinin response factor) under stress and found that SlCRF1 and SlCRF2 expressed prominently in vascular tissue during development and in response to cytokinin and specific stresses, indicating their importance in plant growth and environmental responses.
Environmental fluctuation mediated crop yield loss is a major problem around the world. Simultaneous impact of several other stresses increases the susceptibility of plants to many folds. The increasing population size of world also raises the continuous requirement of food crops to sustain their requirement. To overcome these adverse conditions without compromising the growth or yield new insight is required in the field of transgenic and improved variety development. In this review we sum up the current understanding and knowledge of regulations of AP2/ERFs under different conditions. These AP2/ERFs are tightly regulated at different transcriptional and translational level to attain a homeostasis during non-stressed conditions. AP2/ERFs are in prime focus in context of generating improved varities, as they are promising candidates for studying diverse network involved in development, metabolic and stress responses in plants. Because of their ability to bind with multiple cis-elements, allows them to regulate these diverse set of responses simultaneously. One important fact is that with so many positive effects, they also negatively influence many processes which are indeed and important for normal metabolism and growth of plants. So regulation of AP2/ERFs and downstream signaling has to be kept under proper check by different mechanisms for fruitful response. We believe that these crosstalks should be studied in detail and future research on AP2/ERFs and their integrated regulatory network should take following points into account:
1. MicroRNA and small RNA-mediated regulation are rarely discussed. MiR172 and MiR156 are involved in AP2 regulated intricate floral organogenesis. Recently legume nodulation during the rhizobia nitrogen-fixation symbiosis has been shown to be regulated by MiR172-AP2 complex. In order to identify additional transcriptional regulators associated with, genome-wide identification of miRNAs and their potential targets should be carried out. It seems highly probable that these do not play alone and other regulators need to be studied for the interpretation of the entire network to understand what regulates these regulators?
2. Many stress-responsive AP2/ERFs would impart growth abnormalities under normal condition if otherwise not regulated properly at post-transcriptional (capping, splicing, and histone modifications) and post-translational level (phosphorylation, ubiquitination, SUMOylation). Therefore, these aspects need to be exploited at a large level so that specific targets in the complex mechanism could be identified, which could be modulated for development of improved varieties.
3. AP2/ERFs activate or repress expression of a particular gene by interacting specifically to the cis-elements present in the promoter of that gene. We believe that this specificity to identify a single or multiple cis-elements is regulated not only by the rare anomalies in the DBD but also by the motifs flanking it. These bindings are also influenced by protein-protein interactions through different domains. So these domains and motifs, which lead to specificity or plasticity in the interaction of AP2/ERFs with their downstream targets to achieve multiple responses, should be thoroughly studied.
4. Also with recent findings, it is evident that retrograde signaling affects several AP2/ERF-mediated responses. Signaling from mitochondria or chloroplast to the nucleus and its feedback is not properly understood. Therefore, new insight is required in the field of retrograde regulation of AP2/ERFs.
5. Recent molecular tools like CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) and CRISPR-associated 9 (Cas9) genes could be applied to different non-model plants where functional mutants are not available. CRISPR/cas9 genome editing tool would have an edge over conventional tedious silencing approach (through pART vectors) or VIGS (through TRV vectors) in the coming era. Therefore, this technique could be exploited for functional characterization of AP2/ERFs from different plant species.
UP and GJ has compiled and written the review. VT and RS has edited and finalized the draft.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors acknowledge Director, CSIR-Central Institute of Medicinal & Aromatic Plants, Lucknow, India for providing the necessary facilities. RS acknowledge BSC107 CSIR-Network project for financial support. UP and GJ acknowledge UGC, New Delhi, India for providing fellowship.
Agarwal, P. K., Agarwal, P., Reddy, M. K., and Sopory, S. K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 25, 1263–1274. doi: 10.1007/s00299-006-0204-8
Agarwal, P., Agarwal, P. K., Nair, S., Sopory, S. K., and Reddy, M. K. (2007). Stress-inducible DREB2A transcription factor from Pennisetum glaucum is a phosphoprotein and its phosphorylation negatively regulates its DNA. Mol. Genet. Genomics 277, 189–198. doi: 10.1007/s00438-006-0183-z
Akhtar, M., Jaiswal, A., Taj, G., Jaiswal, J. P., Qureshi, M. I., and Singh, N. K. (2012). DREB1/CBF transcription factors: their structure, function and role in abiotic stress tolerance in plants. J. Genet. 91, 385–395. doi: 10.1007/s12041-012-0201-3
Alabadi, D., Oyama, T., Yanovsky, M. J., Harmon, F. G., Mas, P., and Kay, S. A. (2001). Reciprocal regulation between TOC1 and LHY/CCA1 within the Arabidopsis circadian clock. Science 293, 880–883. doi: 10.1126/science.1061320
Allen, M. D., Yamasaki, K., Ohme-Takagi, M., Tateno, M., and Suzuki, M. (1998). A novel mode of DNA recognition by a β-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA. EMBO J. 17, 5484–5496. doi: 10.1093/emboj/17.18.5484
An, F., Zhao, Q., Ji, Y., Li, W., Jiang, Z., Yu, X., et al. (2010). Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 22, 2384–2401. doi: 10.1105/tpc.110.076588
Bethke, G., Unthan, T., Uhrig, J. F., Poschl, Y., Gust, A. A., Scheel, D., et al. (2009). Flg22 regulates the release of an ethylene response factor substrate from MAP kinase 6 in Arabidopsis thaliana via ethylene signaling. Proc. Natl. Acad. Sci.U.S.A. 106, 8067–8072. doi: 10.1073/pnas.0810206106
Bisson, M. M., and Groth, G. (2010). New insight in ethylene signaling: autokinase activity of ETR1 modulates the interaction of receptors and EIN2. Mol. Plant 3, 882–889. doi: 10.1093/mp/ssq036
Bolt, S., Zuther, E., Zintl, S., Hincha, D. K., and Schmulling, T. (2017). ERF105 is a transcription factor gene of Arabidopsis thaliana required for freezing tolerance and cold acclimation. Plant Cell Environ. 40, 108–120. doi: 10.1111/pce.12838
Buttner, M., and Singh, K. B. (1997). Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc. Natl. Acad. Sci. U.S.A. 94, 5961–5966.
Caarls, L., van der Does, D., Hickman, R., Jansen, W., Van Verk, M. C., Proietti, S., et al. (2016). Assessing the Role of ETHYLENE RESPONSE FACTOR transcriptional repressors in salicylic acid-mediated suppression of jasmonic acid-responsive genes. Plant Cell Physiol. doi: 10.1093/pcp/pcw187. [Epub ahead of pront].
Cao, P. B., Azar, S., SanClemente, H., Mounet, F., Dunand, C., Marque, G., et al. (2015). Genome-wide analysis of the AP2/ERF family in Eucalyptus grandis: an intriguing over-representation of stress-responsive DREB1/CBF genes. PLoS ONE 10:e0121041. doi: 10.1371/journal.pone.0121041
Cao, Z. F., Li, J., Chen, F., Li, Y. Q., Zhou, H. M., and Liu, Q. (2001). Effect of two conserved amino-acid residues on DREB1A function. Biochemistry 66, 623–627. doi: 10.1023/A:1010251129429
Catinot, J., Huang, J. B., Huang, P. Y., Tseng, M. Y., Chen, Y. L., Gu, S. Y., et al. (2016). ETHYLENE RESPONSE FACTOR 96 positively regulates Arabidopsis resistance to necrotrophic pathogens by direct binding to GCC elements of jasmonate and ethylene-responsive defence genes. Plant Cell Environ. 38, 2721–2734. doi: 10.1111/pce.12583
Causier, B., Ashworth, M., Guo, W., and Davies, B. (2012). The TOPLESS Interactome: a framework for gene repression in Arabidopsis. Plant Physiol. 158, 423–438. doi: 10.1104/pp.111.186999
Cevik, V., Kidd, B. N., Zhang, P., Hill, C., Kiddle, S., Denby, K. J., et al. (2012). MEDIATOR25 acts as an integrative hub for the regulation of jasmonate-responsive gene expression in Arabidopsis. Plant Physiol. 160, 541–555. doi: 10.1104/pp.112.202697
Chandler, J. W., Cole, M., Flier, A., Grewe, B., and Werr, W. (2007). The AP2 transcription factors DORNROSCHEN and DORNROSCHEN-LIKE redundantly control Arabidopsis embryo patterning via interaction with PHAVOLUTA. Development 134, 1653–1662. doi: 10.1242/dev.001016
Charfeddine, M., Saidi, M. N., Charfeddine, S., Hammami, A., and Gargouri Bouzid, R. (2015). Genome-wide analysis and expression profiling of the ERF transcription factor family in potato (Solanum tuberosum L.) Mol. Biotechnol. 57, 348–358. doi: 10.1007/s12033-014-9828-z
Chen, T., Yang, Q., Gruber, M., Kang, J., Sun, Y., Ding, W., et al. (2012). Expression of an alfalfa (Medicago sativa L.) ethylene response factor gene MsERF8 in tobacco plants enhances resistance to salinity. Mol. Biol. Rep. 39, 6067–6075. doi: 10.1007/s11033-011-1421-y
Chen, X. (2004). A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 303, 2022–2025. doi: 10.1126/science.1088060
Cheng, M. C., Hsieh, E. J., Chen, J. H., Chen, H. Y., and Lin, T. P. (2012). Arabidopsis RGLG2, functioning as a RING E3 ligase, interacts with AtERF53 and negatively regulates the plant drought stress response. Plant Physiol. 158, 363–375. doi: 10.1104/pp.111.189738
Cheng, M. C., Liao, P. M., Kuo, W. W., and Lin, T. P. (2013). The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582. doi: 10.1104/pp.113.221911
Cheong, Y. H., Moon, B. C., Kim, J. K., Kim, C. Y., Kim, M. C., Kim, I. H., et al. (2003). BWMK1, a rice mitogen-activated protein kinase, locates in the nucleus and mediates pathogenesis-related gene expression by activation of a transcription factor. Plant Physiol. 132, 1961–1972. doi: 10.1104/pp.103.023176
Chinnusamy, V., Ohta, M., Kanrar, S., Lee, B. H., Hong, X., Agarwal, M., et al. (2003). ICE1: a regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 17, 1043–1054. doi: 10.1101/gad.1077503
Cole, M., Chandler, J., Weijers, D., Jacobs, B., Comelli, P., and Werr, W. (2009). DORNROSCHEN is a direct target of the auxin response factor MONOPTEROS in the Arabidopsis embryo. Development 136, 1643–1651. doi: 10.1242/dev.032177
Comelli, R. N., Welchen, E., Kim, H. J., Hong, J. C., and Gonzalez, D. H. (2012). Delta subclass HD-Zip proteins and a B-3 AP2/ERF transcription factor interact with promoter elements required for expression of the Arabidopsis cytochrome c oxidase 5b-1 gene. Plant Mol. Biol. 80, 157–167. doi: 10.1007/s11103-012-9935-9
De Boer, K., Tilleman, S., Pauwels, L., Bossche, R. V., De Sutter, V., Vanderhaeghen, R., et al. (2011). AP2/ERF and bHLH tobacco transcription factors cooperatively mediate jasmonate-elicited nicotine biosynthesis. Plant J. 66, 1053–1065. doi: 10.1111/j.1365-313X.2011.04566.x
Deokar, A. A., Kondawar, V., Kohli, D., Aslam, M., Jain, P. K., Karuppayil, S. M., et al. (2015). The CarERF genes in chickpea (Cicer arietinum L.) and the identification of CarERF116 as abiotic stress responsive transcription factor. Funct. Integr. Genomics 15, 27–46. doi: 10.1007/s10142-014-0399-7
Diaz-Martin, J., Almoguera, C., Prieto-Dapena, P., Espinosa, J. M., and Jordano, J. (2005). Functional interaction between two transcription factors involved in the developmental regulation of a small heat stress protein gene promoter. Plant Physiol. 139, 1483–1494. doi: 10.1104/pp.105.069963
Dietz, K. J., Vogel, M. O., and Viehhauser, A. (2010). AP2/EREBP transcription factors are part of gene regulatory networks and integrate metabolic, hormonal and environmental signals in stress acclimation and retrograde signalling. Protoplasma 245, 3–14. doi: 10.1007/s00709-010-0142-8
Djemal, R., and Khoudi, H. (2015). Isolation and molecular characterization of a novel WIN1/SHN1 ethylene-responsive transcription factor TdSHN1 from durum wheat (Triticum turgidum. L. subsp. durum). Protoplasma 252, 1461–1473. doi: 10.1007/s00709-015-0775-8
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., and Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. doi: 10.1105/tpc.108.063958
Dong, C. H., Agarwal, M., Zhang, Y., Xie, Q., and Zhu, J. K. (2006). The negative regulator of plant cold responses, HOS1, is a RING E3 ligase that mediates the ubiquitination and degradation of ICE1. Proc. Natl. Acad. Sci. U.S.A. 103, 8281–8286. doi: 10.1073/pnas.0602874103
Dong, L., Cheng, Y., Wu, J., Cheng, Q., Li, W., Fan, S., et al. (2015). Overexpression of GmERF5, a new member of the soybean EAR motif-containing ERF transcription factor, enhances resistance to Phytophthora sojae in soybean. J. Exp. Bot. 66, 2635–2647. doi: 10.1093/jxb/erv078
Dong, M. A., Farre, E. M., and Thomashow, M. F. (2011). Circadian clock-associated 1 and late elongated hypocotyl regulate expression of the C-repeat binding factor (CBF) pathway in Arabidopsis. Proc. Natl. Acad. Sci.U.S.A. 108, 7241–7246. doi: 10.1073/pnas.1103741108
Dong, N., Liu, X., Lu, Y., Du, L., Xu, H., Liu, H., et al. (2010). Overexpression of TaPIEP1, a pathogen-induced ERF gene of wheat, confers host-enhanced resistance to fungal pathogen Bipolaris sorokiniana. Funct. Integr. Genomics 10, 215–226. doi: 10.1007/s10142-009-0157-4
Dubois, M., Skirycz, A., Claeys, H., Maleux, K., Dhondt, S., De Bodt, S., et al. (2013). ETHYLENE RESPONSE FACTOR6 acts as a central regulator of leaf growth under waterlimiting conditions in Arabidopsis. Plant Physiol. 162, 319–332. doi: 10.1104/pp.113.216341
Dubois, M., Van den Broeck, L., Claeys, H., Van Vlierberghe, K., Matsui, M., and Inze, D. (2015). The ETHYLENE RESPONSE FACTORs ERF6 and ERF11 antagonistically regulate mannitol-induced growth inhibition in Arabidopsis. Plant Physiol. 169, 166–179. doi: 10.1104/pp.15.00335
Egawa, C., Kobayashi, F., Ishibashi, M., Nakamura, T., Nakamura, C., and Takumi, S. (2006). Differential regulation of transcript accumulation and alternative splicing of a DREB2 homolog under abiotic stress conditions in common wheat. Genes Genet. Syst. 81, 77–91.
Eini, O., Yang, N., Pyvovarenko, T., Pillman, K., Bazanova, N., Tikhomirov, N., et al. (2013). Complex regulation by Apetala2 domain-containing transcription factors revealed through analysis of the stress-responsive TdCor410b promoter from durum wheat. PLoS ONE 8:e58713. doi: 10.1371/journal.pone.0058713
Fan, W., Hai, M., Guo, Y., Ding, Z., Tie, W., Ding, X., et al. (2016). The ERF transcription factor family in cassava: genome-wide characterization and expression analyses against drought stress. Sci. Rep. 6:37379. doi: 10.1038/srep37379
Figueroa-Yanez, L., Pereira-Santana, A., Arroyo-Herrera, A., Rodriguez-Corona, U., Sanchez-Teyer, F., Espadas-Alcocer, J., et al. (2016). RAP2.4a is transported through the phloem to regulate cold and heat tolerance in papaya tree (Carica papaya cv. Maradol): implications for protection against abiotic stress. PLoS ONE 11:e0165030. doi: 10.1371/journal.pone.0165030
Finkelstein, R. R., Gampala, S. S., and Rock, C. D. (2002). Abscisic acid signalling in seeds and seedlings. Plant Cell 14, 15–45. doi: 10.1105/tpc.010441
Fischer, U., and Droge-Laser, W. (2004). Overexpression of NtERF5, a new member of the tobacco ethylene response transcription factor family enhances resistance to tobacco mosaic virus. Mol. Plant Microbe Interact. 17, 1162–1171. doi: 10.1094/MPMI.2004.17.10.1162
Fukao, T., and Bailey-Serres, J. (2008). Submergence tolerance conferred by Sub1A is mediated by SLR1 and SLRL1 restriction of gibberellin responses in rice. Proc. Natl. Acad. Sci.U.S.A. 105, 16814–16819. doi: 10.1073/pnas.0807821105
Gao, S., Zhang, H., Tian, Y., Li, F., Zhang, Z., and Lu, X. (2008). Expression of TERF1 in rice regulates expression of stress-responsive genes and enhances tolerance to drought and high-salinity. Plant Cell Rep. 27, 1787–1795. doi: 10.1007/s00299-008-0602-1
Gasch, P., Fundinger, M., Muller, J. T., Lee, T., Bailey-Serres, J., and Mustroph, A. (2016). Redundant ERF-VII transcription factors bind to an evolutionarily conserved cis-motif to regulate hypoxia-responsive gene expression in arabidopsis. Plant Cell. 28, 160–180. doi: 10.1105/tpc.15.00866
Gibbs, D. J., Lee, S. C., Isa, N. M., Gramuglia, S., Fukao, T., Bassel, G. W., et al. (2011). Homeostatic response to hypoxia is regulated by the N-end rule pathway in plants. Nature 479, 415–418. doi: 10.1038/nature10534
Giuntoli, B., Lee, S. C., Licausi, F., Kosmacz, M., Oosumi, T., Van Dongen, J. T., et al. (2014). A trihelix DNA binding protein counterbalances hypoxia-responsive transcriptional activation in Arabidopsis. PLoS Biol. 12:e1001950. doi: 10.1371/journal.pbio.100195
Gong, W., He, K., Covington, M., Dinesh-Kumar, S. P., Snyder, M., Harmer, S. L., et al. (2008). The development of protein microarrays and their applications in DNA-protein and protein-protein interaction analyses of Arabidopsis transcription factors. Mol. Plant 1, 27–41. doi: 10.1093/mp/ssm009
Guo, B., Wei, Y., Xu, R., Lin, S., Luan, H., Lv, C., et al. (2016). Genome-wide analysis of APETALA2/ethylene-responsive factor (AP2/ERF) gene family in barley (Hordeum vulgare L.). PLoS ONE 11:e0161322. doi: 10.1371/journal.pone.0161322
Guo, H., and Ecker, J. R. (2004). The ethylene signaling pathway: new insights. Curr. Opin. Plant Biol. 7, 40–49.
Hao, D., Yamasaki, K., Sarai, A., and Ohme-Takagi, M. (2002). Determinants in the sequence specific binding of two plant transcription factors, CBF1 and NtERF2, to the DRE and GCC motifs. Biochemistry 41, 4202–4208. doi: 10.1021/bi015979v
Hattori, Y., Nagai, K., Furukawa, S., Song, X. J., Kawano, R., Sakakibara, H. et al. (2009). The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460, 1026–1030. doi: 10.1038/nature08258
Hinz, M., Wilson, I. W., Yang, J., Buerstenbinder, K., Llewellyn, D., Dennis, E. S., et al. (2010). Arabidopsis RAP2.2: an ethylene response transcription factor that is important for hypoxia survival. Plant Physiol. 153, 757–772. doi: 10.1104/pp.110.155077
Hongxing, Z., Benzhong, Z., Bianyun, Y., Yanling, H., Daqi, F., Wentao, X., et al. (2005). Cloning and DNA-binding properties of ethylene response factor, LeERF1 and LeERF2, in tomato. Biotechnol. Lett. 27, 423–428. doi: 10.1007/s10529-005-1777-y
Huang, D., Wu, W., Abrams, S. R., and Cutler, A. J. (2008). The relationship of drought-related gene expression in Arabidopsis thaliana to hormonal and environmental factors. J. Exp. Bot. 59, 2991–3007. doi: 10.1093/jxb/ern155
Huang, P. Y., Catinot, J., and Zimmerli, L. (2016). Ethylene response factors in Arabidopsis immunity. J. Exp. Bot.. 67, 1231–1241. doi: 10.1093/jxb/erv518
Huang, Y., Li, H., Hutchison, C. E., Laskey, J., and Kieber, J. J. (2003). Biochemical and functional analysis of CTR1, a protein kinase that negatively regulates ethylene signaling in Arabidopsis. Plant J. 33, 221–233. doi: 10.1046/j.1365-313X.2003.01620.x
Huang, Y., Zhang, B. L., Sun, S., Xing, G. M., Wang, F., Li, M. Y., et al. (2016). AP2/ERF transcription factors involved in response to tomato yellow leaf curly virus in tomato. Plant Genome. 9, 1–15. doi: 10.3835/plantgenome2015.09.0082
Huang, Z., Zhang, Z., Zhang, X., Zhang, H., Huang, D., and Huang, R. (2004). Tomato TERF1 modulates ethylene response and enhances osmotic stress tolerance by activating expression of downstream genes. FEBS Lett. 573, 110–116. doi: 10.1016/j.febslet.2004.07.064
Huang, Z., Zhong, X. J., He, J., Jiang, M. Y., Yu, X. F., and Li, X. (2016). Identification and characterization of AP2/ERF transcription factors in moso bamboo (Phyllostachys edulis). Mol. Biol. 50, 785–796. doi: 10.7868/S0026898416050062
Ikeda, M., and Ohme-Takagi, M. (2009). A novel group of transcriptional repressors in Arabidopsis. Plant Cell Physiol. 50, 970–975. doi: 10.1093/pcp/pcp048
Ito, T. M., Polido, P. B., Rampim, M. C., Kaschuk, G., and Souza, S. G. (2014). Genome-wide identification and phylogenetic analysis of the AP2/ERF gene superfamily in sweet orange (Citrus sinensis). Genet. Mol. Res. 3, 7839–7851. doi: 10.4238/2014.September.26.22
Jisha, V., Dampanaboina, L., Vadassery, J., Mithofer, A., Kappara, S., and Ramanan, R. (2015). Overexpression of an AP2/ERF Type transcription factor OsEREBP1 confers biotic and abiotic stress tolerance in rice. PLoS ONE 10:e0127831. doi: 10.1371/journal.pone.0127831
Ju, C., Yoon, G. M., Shemansky, J. M., Lin, D. Y., Ying, Z. I., Chang, J., et al. (2012). CTR1 phosphorylates the central regulator EIN2 to control ethylene hormone signalling from the ER membrane to the nucleus in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 109, 19486–19491. doi: 10.1073/pnas.1214848109
Kagale, S., and Rozwadowski, K. (2011). EAR motif-mediated transcriptional repression in plants: an underlying mechanism for epigenetic regulation of gene expression. Epigenetics 6, 141–146. doi: 10.4161/epi.6.2.13627
Kavas, M., Kizildogan, A., Gokdemir, G., and Baloglu, M. C. (2015). Genome-wide investigation and expression analysis of AP2-ERF gene family in salt tolerant common bean. EXCLI J. 14, 1187–1206. doi: 10.17179/excli2015-600
Kendrick, M. D., and Chang, C. (2008). Ethylene signaling: new levels of complexity and regulation. Curr. Opin. Plant Biol. 11, 479–485. doi: 10.1016/j.pbi.2008.06.011
Khandelwal, A., Elvitigala, T., Ghosh, B., and Quatrano, R. S. (2008). Arabidopsis transcriptome reveals control circuits regulating redox homeostasis and the role of an AP2 transcription factor. Plant Physiol. 148, 2050–2058. doi: 10.1104/pp.108.128488
Kidokoro, S., Maruyama, K., Nakashima, K., Imura, Y., Narusaka, Y., Shinwari, Z. K., et al. (2009). The phytochrome-interacting factor PIF7 negatively regulates DREB1 expression under circadian control in Arabidopsis. Plant Physiol. 151, 12046–12057. doi: 10.1104/pp.109.147033
Kieber, J. J., Rothenberg, M., Roman, G., Feldmann, K. A., and Ecker, J. R. (1993). CTR1, a negative regulator of the ethylene response pathway in Arabidopsis, encodes a member of the raf family of protein kinases. Cell 72, 427–441.
Kim, J. S., Mizoi, J., Kidokoro, S., Maruyama, K., Nakajima, J., Nakashima, K., et al. (2012). Arabidopsis growth-regulating factor 7 functions as a transcriptional repressor of abscisic acid and osmotic stress-responsive genes, including DREB2A. Plant Cell 24, 3393–3405. doi: 10.1105/tpc.112.100933
Kitomi, Y., Ito, H., Hobo, T., Aya, K., Kitano, H., and Inukai, Y. (2011). The auxin responsive AP2/ERF transcription factor CROWN ROOTLESS5 is involved in crown root initiation in rice through the induction of OsRR1, a type-A response regulator of cytokinin signaling. Plant J. 67, 472–484. doi: 10.1111/j.1365-313X.2011.04610.x
Koussevitzky, S., Nott, A., Mockler, T. C., Hong, F., Sachetto-Martins, G., Surpin, M., et al. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719. doi: 10.1126/science.1140516
Lacey, R. F., and Binder, B. M. (2014). How plants sense ethylene gas–the ethylene receptors. J. Inorg. Biochem. 133, 58–62. doi: 10.1016/j.jinorgbio.2014.01.006
Lee, D. K., Yoon, S., Kim, Y. S., and Kim, J. K. (2016). Rice OsERF71-mediated root modification affects shoot drought tolerance. Plant Signal. Behav. 12:e1268311. doi: 10.1080/15592324.2016.1268311
Lee, J. H., Hong, J. P., Oh, S. K., Lee, S., Choi, D., and Kim, W. T. (2004). The ethylene-responsive factor like protein 1 (CaERFLP1) of hot pepper (Capsicum annuum L.) interacts in vitro with both GCC and DRE/CRT sequences with different binding affinities: possible biological roles of CaERFLP1in response to pathogen infection and high salinity conditions in transgenic tobacco plants. Plant Mol. Biol. 55, 61–81. doi: 10.1007/s11103-004-0417-6
Lee, S. Y., Hwang, E. Y., Seok, H. Y., Tarte, V. N., Jeong, M. S., Jang, S. B., et al. (2015). Arabidopsis AtERF71/HRE2 functions as transcriptional activator via cis-acting GCC box or DRE/CRT element and is involved in root development through regulation of root cell expansion. Plant Cell Rep. 34, 223–231. doi: 10.1007/s00299-014-1701-9
Lee, S. J., Park, J. H., Lee, M. H., Yu, J. H., and Kim, S. Y. (2010). Isolation and functional characterization of CE1 binding proteins. BMC Plant Biol. 10:277. doi: 10.1186/1471-2229-10-277
Lei, G., Shen, M., Li, Z. G., Zhang, B., Duan, K. X., Wang, N., et al. (2011). EIN2 regulates salt stress response and interacts with a MA3 domain-containing protein ECIP1 in Arabidopsis. Plant Cell Environ. 34, 1678–1692. doi: 10.1111/j.1365-3040.2011.02363.x
Lei, Z. P., He, D. H., Xing, H. Y., Tang, B. S., and Lu, B. X. (2016). Genome-wide comparison of AP2/ERF superfamily genes between Gossypium arboreum and G. raimondii. Genet. Mol. Res. doi: 10.4238/gmr.15038211. [Epub ahead of print].
Li, C. W., Su, R. C., Cheng, C. P., Sanjaya, You, S. J., Hsieh, T. H., et al. (2011). Tomato RAV transcription factor is a pivotal modulator involved in the AP2/EREBP-mediated defense pathway. Plant Physiol. 156, 213–227. doi: 10.1104/pp.111.174268
Li, H. Y., Xiao, S., and Chye, M. L. (2008). Ethylene and pathogen-inducible Arabidopsis acyl-CoA-binding protein 4 interacts with an ethylene-responsive element binding protein. J. Exp. Bot. 59, 3997–4006. doi: 10.1093/jxb/ern241
Li, M. Y., Xu, Z. S., Huang, Y., Tian, C., Wang, F., and Xiong, A. S. (2015). Genome-wide analysis of AP2/ERF transcription factors in carrot (Daucus carota L.) reveals evolution and expression profiles under abiotic stress. Mol. Genet. Genomics 290, 2049–2061. doi: 10.1007/s00438-015-1061-3
Li, X., Zhang, D., Li, H., Wang, Y., Zhang, Y., and Wood, A. J. (2014). EsDREB2B, a novel truncated DREB2-type transcription factor in the desert legume Eremosparton songoricum, enhances tolerance to multiple abiotic stresses in yeast and transgenic tobacco. BMC Plant Biol. 14:44. doi: 10.1186/1471-2229-14-44
Li, Z., Zhang, L., Li, J., Xu, X., Yao, Q., and Wang, A. (2014). Isolation and functional characterization of the ShCBF1 gene encoding a CRT/DRE-binding factor from the wild tomato species Solanum habrochaites. Plant Physiol. Biochem. 74, 294–303. doi: 10.1016/j.plaphy.2013.11.024
Licausi, F., Giorgi, F. M., Zenoni, S., Osti, F., Pezzotti, M., and Perata, P. (2010). Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genomics 11:719. doi: 10.1186/1471-2164-11-719
Licausi, F., Kosmacz, M., Weits, D. A., Giuntoli, B., Giorgi, F. M., Voesenek, L. A., et al. (2011). Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 479, 419–422. doi: 10.1038/nature10536
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Lin, R. C., Park, H. J., and Wang, H. Y. (2008). Role of Arabidopsis RAP2.4 in regulating light- and ethylene-mediated developmental processes and drought stress tolerance. Mol. Plant 1, 42–57. doi: 10.1093/mp/ssm004
Liu, D., Chen, X., Liu, J., Ye, J., and Guo, Z. (2012). The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 63, 3899–3911. doi: 10.1093/jxb/ers079
Liu, M., Gomes, B. L., Mila, I., Purgatto, E., Peres, L. E., Frasse, P., et al. (2016). Comprehensive profiling of ethylene response factor expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato. Plant Physiol. 170, 1732–1744. doi: 10.1104/pp.15.01859
Liu, Y., Zhao, T. J., Liu, J. M., Liu, W. Q., Liu, Q., Yan, Y. B., et al. (2006). The conserved Ala37 in the ERF/AP2 domain is essential for binding with the DRE element and the GCC box. FEBS Lett. 580, 1303–1308. doi: 10.1016/j.febslet.2006.01.048
Lorenzo, O., Piqueras, R., Sanchez-Serrano, J. J., and Solano, R. (2002). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 15, 165–178. doi: 10.1105/tpc.007468
Ma, Y., Zhang, L., Zhang, J., Chen, J., Wu, T., Zhu, S., et al. (2014). Expressing a Citrus ortholog of Arabidopsis ERF1 enhanced coldtolerance in tobacco. Sci. Hortic. 174, 64–76. doi: 10.1016/j.scienta.2014.05.009
Magnani, E., Sjolander, K., and Hake, S. (2004). From endonucleases to transcription factors: evolution of the AP2 DNA binding domain in plants. Plant Cell 16, 2265–2277. doi: 10.1105/tpc.104.023135
Matsukura, S., Mizoi, J., Yoshida, T., Todaka, D., Ito, Y., Maruyama, K., et al. (2010). Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genomics 283, 185–196. doi: 10.1007/s00438-009-0506-y
Mishra, S., Phukan, U. J., Tripathi, V., Singh, D. K., Luqman, S., and Shukla, R. K. (2015). PsAP2 an AP2/ERF family transcription factor from papaver somniferum enhances abiotic and biotic stress tolerance in transgenic tobacco. Plant Mol. Biol. 89, 173–186. doi: 10.1007/s11103-015-0361-7
Miura, K., Jin, J. B., Lee, J., Yoo, C. Y., Stirm, V., Miura, T., et al. (2007). SIZ1-mediated sumoylation of ICE1 controls CBF3/DREB1A expression and freezing tolerance in Arabidopsis. Plant Cell 19, 1403–1414. doi: 10.1105/tpc.106.048397
Moffat, C. S., Ingle, R. A., Wathugala, D. L., Saunders, N. J., Knight, H., and Knight, M. R. (2012). ERF5 and ERF6 play redundant roles as positive regulators of JA/ Et-mediated defense against Botrytis cinerea in Arabidopsis. PLoS ONE 7:e35995. doi: 10.1371/journal.pone.0035995
Montiel, G., Zarei, A., Korbes, A. P., and Memelink, J. (2011). The jasmonate-responsive element from the ORCA3 promoter from Catharanthus roseus is active in Arabidopsis and is controlled by the transcription factor AtMYC2. Plant Cell Physiol. 52, 578–587. doi: 10.1093/pcp/pcr016
Muller, M., and Munne-Bosch, S. (2015). Ethylene response factors: a key regulatory hub in hormone and stress signaling. Plant Physiol. 169, 32–41. doi: 10.1104/pp.15.00677
Nakamichi, N., Kusano, M., Fukushima, A., Kita, M., Ito, S., Yamashino, T., et al. (2009). Transcript profiling of an Arabidopsis PSEUDO RESPONSE REGULATOR arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 50, 447–462. doi: 10.1093/pcp/pcp004
Nakano, T., Fujisawa, M., Shima, Y., and Ito, Y. (2014). The AP2/ERF transcription factor SlERF52 functions in flower pedicel abscission in tomato. J. Exp. Bot. 65, 3111–3119. doi: 10.1093/jxb/eru154
Nakano, T., Suzuki, K., Fujimura, T., and Shinshi, H. (2006). Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol 140, 411–432. doi: 10.1104/pp.105.073783
Nott, A., Jung, H. S., Koussevitzky, S., and Chory, J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57, 739–759. doi: 10.1146/annurev.arplant.57.032905.105310
Ogata, T., Okada, H., Kawaide, H., Takahashi, H., Seo, S., Mitsuhara, I., et al. (2015). Involvement of NtERF3 in the cell death signalling pathway mediated by SIPK/WIPK and WRKY1 in tobacco plants. Plant Biol. 17, 962–972. doi: 10.1111/plb.12349
Oh, S. J., Kim, Y. S., Kwon, C. W., Park, H. Y., Jeong, J. S., and Kim, J. K. (2009). Overexpression of the transcription factor AP37 in rice improves grain yield under drought conditions. Plant Physiol. 150, 1368–1379. doi: 10.1104/pp.109.137554
Ohta, M., Matsui, K., Hiratsu, K., Shinshi, H., and Ohme-Takagi, M. (2001). Repression domains of class II ERF transcriptional repressors share an essential motif for active repression. Plant Cell 13, 1959–1968. doi: 10.1105/TPC.010127
Ohta, M., Ohme-Takagi, M., and Shinshi, H. (2000). Three ethylene-responsive transcription factors in tobacco with distinct transactivation functions. Plant J. 22, 29–38. doi: 10.1046/j.1365-313x.2000.00709.x
Onate-Sanchez, L., Anderson, J. P., Young, J., and Singh, K. B. (2007). AtERF14, a member of the ERF family of transcription factors, plays a nonredundant role in plant defense. Plant Physiol. 143, 400–409. doi: 10.1104/pp.106.086637
Oswald, O., Martin, T., Dominy, P. J., and Graham, I. A. (2001). Plastid redox state and sugars: interactive regulators of nuclear-encoded photosynthetic gene expression. Proc. Natl. Acad. Sci. U.S.A. 98, 2047–2052. doi: 10.1073/pnas.021449998
Ou, B., Yin, K. Q., Liu, S. N., Yang, Y., Gu, T., Wing, H., et al. (2011). A high-throughput screening system for Arabidopsis transcription factors and its application to Med25-dependent transcriptional regulation. Mol. Plant 4, 546–555. doi: 10.1093/mp/ssr002
Pan, Y., Seymour, G. B., Lu, C., Hu, Z., Chen, X., and Chen, G. (2012). An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 31, 349–360. doi: 10.1007/s00299-011-1170-3
Pandey, B., Sharma, P., Tyagi, C., Goyal, S., Grover, A., and Sharma, I. (2015). Structural modeling and molecular simulation analysis of HvAP2/EREBP from barley. J. Biomol. Struct. Dyn. 22, 1–44. doi: 10.1080/07391102.2015.1073630
Papdi, C., Perez-Salamo, I., Joseph, M. P., Giuntoli, B., Bogre, L., Koncz, C., et al. (2015). The low oxygen, oxidative and osmotic stress responses synergistically act through the ethylene response factor VII genes RAP2.12, RAP2.2 and RAP2.3. Plant J. 82, 772–784. doi: 10.1111/tpj.12848
Park, C. S., Go, Y. S., and Suh, M. C. (2016). Cuticular wax biosynthesis is positively regulated by WRINKLED4, an AP2/ERF-type transcription factor, in Arabidopsis stems. Plant J. 88, 257–270. doi: 10.1111/tpj.13248
Park, J. M., Park, C. J., Lee, S. B., Ham, B. K., Shin, R., and Paek, K. H. (2001). Overexpression of the tobacco tsi1 gene encoding an EREBP/AP2–type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell 13, 1035–1046. doi: 10.1105/tpc.13.5.1035
Park, S. Y., and Grabau, E. (2016). Differential isoform expression and protein localization from alternatively spliced Apetala2 in peanut under drought stress. J. Plant Physiol. 206, 98–102. doi: 10.1016/j.jplph.2016.09.007
Paul, M. V., Iyer, S., Amerhauser, C., Lehmann, M., van Dongen, J. T., and Geigenberger, P. (2016). Oxygen sensing via the ethylene response transcription factor RAP2.12 affects plant metabolism and performance under both normoxia and hypoxia. Plant Physiol. 172, 141–153. doi: 10.1104/pp.16.00460
Pena-Castro, J. M., van Zanten, M., Lee, S. C., Patel, M. R., Voesenek, L. A., Fukao, T., et al. (2011). Expression of rice SUB1A and SUB1C transcription factors in Arabidopsis uncovers flowering inhibition as a submergence tolerance mechanism. Plant J. 67, 434–446. doi: 10.1111/j.1365-313X.2011.04605.x
Phukan, U. J., Mishra, S., and Shukla, R. K. (2015). Waterlogging and submergence stress: affects and acclimation. Crit. Rev. Biotechnol. 36, 956–966. doi: 10.3109/0738855
Phukan, U. J., Mishra, S., Timbre, K., Luqman, S., and Shukla, R. K. (2014). Mentha arvensis exhibit better adaptive characters in contrast to Mentha piperita when subjugated to sustained waterlogging stress. Protoplasma 251, 603–614. doi: 10.1007/s00709-013-0561-4
Pucciariello, C., and Perata, P. (2016). New insights into reactive oxygen species and nitric oxide signalling under low oxygen in plants. Plant Cell Environ. doi: 10.1111/pce.12715. [Epub ahead of print].
Qi, W., Sun, F., Wang, Q., Chen, M., Huang, Y., Feng, Y. Q., et al. (2011). Rice ethylene-response AP2/ERF factor OsEATB restricts internode elongation by down-regulating a gibberellin biosynthetic gene. Plant Physiol. 157, 216–228. doi: 10.1104/pp.111.179945
Qiao, H., Chang, K. N., Yazaki, J., and Ecker, J. R. (2009). Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 23, 512–521. doi: 10.1101/gad.1765709
Qin, F., Kakimoto, M., Sakuma, Y., Maruyama, K., Osakabe, Y., Tran, L. S., et al. (2007). Regulation and functional analysis of ZmDREB2A in response to drought and heat stresses in Zea mays L. Plant J. 50, 54–69. doi: 10.1111/j.1365-313X.2007.03034.x
Qin, F., Sakuma, Y., Tran, L. S., Maruyama, K., Kidokoro, S., Fujita, Y., et al. (2008). Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 20, 1693–1707. doi: 10.1105/tpc.107.057380
Quan, R., Hu, S., Zhang, Z., Zhang, H., Zhang, Z., and Huang, R. (2010). Overexpression of an ERF transcription factor TSRF1 improves rice drought tolerance. Plant Biotechnol. J. 8, 476–488. doi: 10.1111/j.1467-7652.2009.00492.x
Rao, G., Sui, J., Zeng, Y., He, C., and Zhang, J. (2015). Genome-wide analysis of the AP2/ERF gene family in Salix arbutifolia. FEBS Open Biol. 5, 132–137. doi: 10.1016/j.fob.2015.02.002
Rashid, M., Guangyuan, H., Guangxiao, Y., Hussain, J., and Xu, Y. (2012). AP2/ERF transcription factor in rice: genome-wide canvas and syntenic relationships between monocots and eudicots. Evol. Bioinform. 8:321. doi: 10.4137/EBO.S9369
Rashotte, A., Mason, M., Hutchison, C., Ferreira, F., Schaller, G. E., and Kieber, J. J. (2006). A subset of Arabidopsis AP2 transcription factors mediate cytokinin responses in concert with a two-component pathway. Proc. Natl. Acad. Sci.U.S.A. 103, 11081–11085. doi: 10.1073/pnas.0602038103
Ren, D., Li, Y., Zhao, F., Sang, X., Shi, J., Wang, N., et al. (2013). MULTI-FLORET SPIKELET1, which encodes an AP2/ERF protein, determines spikelet meristem fate and sterile lemma identity in rice. Plant Physiol. 162, 872–884. doi: 10.1104/pp.113.216044
Romero, I., Vazquez-Hernandez, M., Escribano, M. I., Merodio, C., and Sanchez-Ballesta, M. T. (2016). Expression profiles and DNA-binding affinity of five ERF genes in bunches of Vitis vinifera cv. cardinal treated with high levels of CO2 at low temperature. Front. Plant Sci. 7:1748. doi: 10.3389/fpls.2016.01748
Rong, W., Qi, L., Wang, A., Ye, X., Du, L., Liang, H., et al. (2014). The ERF transcription factor TaERF3 promotes tolerance to salt and drought stresses in wheat. Plant Biotechnol. J. 12, 468–479. doi: 10.1111/pbi.12153
Sakuma, Y., Liu, Q., Dubouzet, J. G., Abe, H., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2002). DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 290, 998–1009. doi: 10.1006/bbrc.2001.6299
Sakuma, Y., Maruyama, K., Qin, F., Osakabe, Y., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2006). Dual function of an Arabidopsis transcription factor DREB2A in water-stressresponsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci.U.S.A. 103, 18822–18827. doi: 10.1073/pnas.0605639103
Scarpeci, T. E., Frea, V. S., Zanor, M. I., and Valle, E. M. (2016). Overexpression of AtERF019 delays plant growth and senescence and improves drought tolerance in Arabidopsis. J. Exp. Bot. doi: 10.1093/jxb/erw429. [Epub ahead of print].
Schmitz, A. J., Folsom, J. J., Jikamaru, Y., Ronald, P., and Walia, H. (2013). SUB1A-mediated submergence tolerance response in rice involves differential regulation of the brassinosteroid pathway. New Phytol. 198, 1060–1070. doi: 10.1111/nph.12202
Schwacke, R., Fischer, K., Ketelsen, B., Krupinska, K., and Krause, K. (2007). Comparative survey of plastid and mitochondrial targeting properties of transcription factors in Arabidopsis and rice. Mol. Gen. Genet. 277, 631–646. doi: 10.1007/s00438-007-0214-4
Sewelam, N., Kazan, K., Thomas-Hall, S. R., Kidd, B. N., Manners, J. M., and Schenk, P. M. (2013). Ethylene response factor 6 is a regulator of reactive oxygen species signaling in Arabidopsis. PLoS ONE 8:e70289. doi: 10.1371/journal.pone.0070289
Shaikhali, J., Heiber, I., Seidel, T., Stroher, E., Hiltscher, H., Birkmann, S., et al. (2008). The redox-sensitive transcription factor Rap2.4a controls nuclear expression of 2-Cys peroxiredoxin A and other chloroplast antioxidant enzymes. BMC Plant Biol. 8:48. doi: 10.1186/1471-2229-8-48
Shi, X., Gupta, S., and Rashotte, A. M. (2014). Characterization of two tomato AP2/ERF genes, SlCRF1 and SlCRF2 in hormone and stress responses. Plant Cell Rep. 33, 35–45. doi: 10.1007/s00299-013-1510-6
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2003). Molecular responses to dehydration and low temperature: differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 3, 217–223. doi: 10.1016/S1369-5266(00)80068-0
Shoji, T., Mishima, M., and Hashimoto, T. (2013). Divergent DNA-binding specificities of a group of ETHYLENE RESPONSE FACTOR transcription factors involved in plant defense. Plant Physiol. 162, 977–990. doi: 10.1104/pp.113.217455
Son, G. H., Wan, J., Kim, H. J., Nguyen, X. C., Chung, W. S., Hong, J. C., et al. (2012). Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Mol. Plant Microbe Interact. 25, 48–60. doi: 10.1094/MPMI-06-11-0165
Song, C. P., Agarwal, M., Ohta, M., Guo, Y., Halfter, U., Wang, P., et al. (2005). Role of an Arabidopsis AP2/EREBP-type transcriptional repressor in abscisic acid and drought stress responses. Plant Cell 17, 2384–2396. doi: 10.1105/tpc.105.033043
Song, X., Li, Y., and Hou, X. (2013). Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genomics 23:573. doi: 10.1186/1471-2164-14-573
Song, X., Wang, J., Ma, X., Li, Y., Lei, T., Wang, L., et al. (2016). Origination, expansion, evolutionary trajectory, and expression bias of AP2/ERF superfamily in Brassica napus. Front. Plant Sci. 7:1186. doi: 10.3389/fpls.2016.01186
Stepanova, A. N., and Alonso, J. M. (2009). Ethylene signaling and response: where different regulatory modules meet. Curr. Opin. Plant Biol. 12, 548–555. doi: 10.1016/j.pbi.2009.07.009
Strand, A., Asami, T., Alonso, J., Ecker, J. R., and Chory, J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421, 79–83. doi: 10.1038/nature01204
Sun, S., Yu, J. P., Chen, F., Zhao, T. J., Fang, X. H., Li, Y. Q., et al. (2008). TINY, a dehydration-responsive element (DRE)-binding protein-like transcription factor connecting the DRE- and ethylene-responsive element-mediated signaling pathways in Arabidopsis. J. Biol. Chem. 283, 6261–6271. doi: 10.1074/jbc.M706800200
Sun, Z. M., Zhou, M. L., Xiao, X. G., Tang, Y. X., and Wu, Y. M. (2014). Genome-wide analysis of AP2/ERF family genes from Lotus corniculatus shows LcERF054 enhances salt tolerance. Funct. Integr. Genomics 14, 453–466. doi: 10.1007/s10142-014-0372-5
Tang, M., Sun, J., Liu, Y., Chen, F., and Shen, S. (2007). Isolation and functional characterization of the JcERF gene, a putative AP2/EREBP domain-containing transcription factor, in the woody oil plant Jatropha curcas. Plant Mol. Biol. 63, 419–428. doi: 10.1007/s11103-006-9098-7
Thagun, C., Imanishi, S., Kudo, T., Nakabayashi, R., Ohyama, K., Mori, T., et al. (2016). Jasmonate-responsive ERF transcription factors regulate steroidal glycoalkaloid biosynthesis in tomato. Plant Cell Physiol. 57, 961–975. doi: 10.1093/pcp/pcw067
Thomas, J., and Weinstein, J. D. (1990). Measurement of heme efflux and heme content in isolated developing chloroplasts. Plant Physiol. 94, 1414–1423.
Tian, Z., He, Q., Wang, H., Liu, Y., Zhang, Y., Shao, F., et al. (2015). The potato ERF transcription factor StERF3 negatively regulates resistance to phytophthora infestans and salt tolerance in potato. Plant Cell Physiol. 56, 992–1005. doi: 10.1093/pcp/pcv025
Tiwari, S. B., Belachew, A., Ma, S. F., Young, M., Ade, J., Shen, Y., et al. (2012). The EDLL motif: a potent plant transcriptional activation domain from AP2/ERF transcription factors. Plant J. 70, 855–865. doi: 10.1111/j.1365-313X.2012.04935.x
Trujillo, L. E., Sotolongo, M., Menéndez, C., Ochogavía, M. E., Coll, Y., Hernández, I., et al. (2008). SodERF3, a novel sugarcane ethylene responsive factor (ERF), enhances salt and drought tolerance when overexpressed in tobacco plants. Plant Cell Physiol. 49, 512–525. doi: 10.1093/pcp/pcn025
Vainonen, J. P., Jaspers, P., Wrzaczek, M., Lamminmaki, A., Reddy, R. A., Vaahtera, L., et al. (2012). RCD1–DREB2A interaction in leaf senescence and stress responses in Arabidopsis thaliana. Biochem. J. 442, 573–581. doi: 10.1042/BJ20111739
van Veen, H., Akman, M., Jamar, D. C., Vreugdenhil, D., Kooiker, M., van Tienderen, P., et al. (2014). Group VII ethylene response factor diversification and regulation in four species from flood-prone environments. Plant Cell Environ. 37, 2421–2432. doi: 10.1111/pce.12302
Vogel, M. O., Gomez-Perez, D., Probst, N., and Dietz, K. J. (2012). Combinatorial signal integration by APETALA2/Ethylene Response Factor (ERF)-transcription factors and the involvement of AP2-2 in starvation response. Int. J. Mol. Sci. 13, 5933–5951. doi: 10.3390/ijms13055933
Vogel, M. O., Moore, M., Konig, K., Pecher, P., Alsharafa, K., Lee, J., et al. (2014). Fast retrograde signaling in response to high light involves metabolite Export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF Transcription factors in Arabidopsis. Plant Cell 26, 1151–1165. doi: 10.1105/tpc.113.121061
Vom Endt, D., Soares e Silva, M., Kijne, J. W., Pasquali, G., and Memelink, J. (2007). Identification of a bipartite jasmonate-responsive promoter element in the Catharanthus roseus ORCA3 transcription factor gene that interacts specifically with AT-Hook DNA-binding proteins. Plant Physiol. 144, 1680–1689. doi: 10.1104/pp.107.096115
Von Gromoff, E. D., Alawady, A., Meinecke, L., Grimm, B., and Beck, C. F. (2008). Heme, a plastid-derived regulator of nuclear gene expression in Chlamydomonas. Plant Cell 20, 552–567. doi: 10.1105/tpc.107.054650
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., et al. (2016). Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 11:e0154027. doi: 10.1371/journal.pone.0154027
Wang, L., Qin, L., Liu, W., Zhang, D., and Wang, Y. (2014). A novel ethylene-responsive factor from Tamarix hispida, ThERF1, is a GCC-box- and DRE-motif binding protein that negatively modulates abiotic stress tolerance in Arabidopsis. Physiol. Plant. 152, 84–97. doi: 10.1111/ppl.12159.
Wang, X. H., Li, Q. T., Chen, H. W., Zhang, W. K., Ma, B., Chen, S. Y., et al. (2014). Trihelix transcription factor GT-4 mediates salt tolerance via interaction with TEM2 in Arabidopsis. BMC Plant Biol. 14:339. doi: 10.1186/s12870-014-0339-7
Wang, X., Liu, S., Tian, H., Wang, S., and Chen, J. G. (2016). The Small Ethylene Response Factor ERF96 is Involved in the Regulation of the Abscisic Acid Response in Arabidopsis. Front. Plant Sci. 6:1064. doi: 10.3389/fpls.2015.01064
Weber, H., and Hellmann, H. (2009). Arabidopsis thaliana BTB/POZ-MATH proteins interact with members of the ERF/AP2 transcription factor family. FEBS J. 276, 6624–6635. doi: 10.1111/j.1742-4658.2009.07373.x
Welsch, R., Maass, D., Voegel, T., DellaPenna, D., and Beyer, P. (2007). Transcription factor RAP2.2 and its interacting partner SINAT2: stable elements in the carotenogenesis of Arabidopsis leaves. Plant Physiol. 145, 1073–1085. doi: 10.1104/pp.107.104828
Wessler, S. R. (2005). Homing into the origin of the AP2 DNA binding domain. Trends Plant Sci. 10, 54–56. doi: 10.1016/j.tplants.2004.12.007
Wu, C., Avila, C. A., and Goggin, F. L. (2015). The ethylene response factor Pti5 contributes to potato aphid resistance in tomato independent of ethylene signalling. J. Exp. Bot. 66, 559–570. doi: 10.1093/jxb/eru472
Wu, D., Ji, J., Wang, G., Guan, C., and Jin, C. (2014). LchERF, a novel ethyleneresponsive transcription factor from Lycium chinense, confers salt tolerance in transgenic tobacco. Plant Cell Rep. 33, 2033–2045. doi: 10.1007/s00299-014-1678-4
Wu, L., Chen, X., Ren, H., Zhang, Z., Zhang, H., Wang, J., et al. (2007). ERF protein JERF1 that transcriptionally modulates the expression of abscisic acid biosynthesis-related gene enhances the tolerance under salinity and cold in tobacco. Planta 226, 815–825. doi: 10.1007/s00425-007-0528-9
Wu, L., Zhang, Z., Zhang, H., Wang, X. C., and Huang, R. (2008). Transcriptional modulation of ethylene response factor protein JERF3 in the oxidative stress response enhances tolerance of tobacco seedlings to salt, drought, and freezing. Plant Physiol. 148, 1953–1963. doi: 10.1104/pp.108.126813
Wuitschick, J. D., Lindstrom, P. R., Meyer, A. E., and Karrer, K. M. (2004). Homing endonucleases encoded by germ line-limited genes in Tetrahymena thermophila have APETELA2 DNA binding domains. Eukaryotic Cell 3, 685–694. doi: 10.1128/EC.3.3.685-694.2004
Xu, J. J., Zhang, X. F., and Xue, H. W. (2016). Rice aleurone layer specific OsNF-YB1 regulates grain filling and endosperm development by interacting with an ERF transcription factor. J. Exp. Bot. 67, 6399–6411. doi: 10.1093/jxb/erw409
Xu, Z. S., Xia, L. Q., Chen, M., Cheng, X. G., Zhang, R. Y., Li, L. C., et al. (2007). Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol. Biol. 65, 719–732. doi: 10.1007/s11103-007-9237-9
Xue, G. P., and Loveridge, C. W. (2004). HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. Plant J. 37, 326–339. doi: 10.1046/j.1365-313X.2003.01963.x
Yang, H., Yu, C., Yan, J., Wang, X., Chen, F., Zhao, Y., et al. (2014). Overexpression of the Jatropha curcas JcERF1 gene coding an AP2/ERF-type transcription factor increase tolerance to salt in transgenic tobacco. Biochemistry 79, 1226–1236. doi: 10.1134/S0006297914110108
Yang, S., Wang, S., Liu, X., Yu, Y., Yue, L., Wang, X., et al. (2009). Four divergent Arabidopsis ethylene-responsive element-binding factor domains bind to a target DNA motif with a universal CG step core recognition and different flanking bases preference. FEBS J. 276, 7177–7186. doi: 10.1111/j.1742-4658.2009.07428.x
Yang, Z., Tian, L., Latoszek-Green, M., Brown, D., and Wu, K. (2005). Arabidopsis ERF4 is a transcriptional repressor capable of modulating ethylene and abscisic acid responses. Plant Mol. Biol. 58, 585–596. doi: 10.1007/s11103-005-7294-5
Yao, X., Li, J., Liu, J., and Liu, K. (2015). An Arabidopsis mitochondria-localized RRL protein mediates abscisic acid signal transduction through mitochondrial retrograde regulation involving ABI4. J. Exp. Bot. 66, 6431–6445. doi: 10.1093/jxb/erv356
Yu, Y., Yang, D., Zhou, S., Gu, J., Wang, F., Dong, J., et al. (2017). The ethylene response factor OsERF109 negatively affects ethylene biosynthesis and drought tolerance in rice. Protoplasma. 254, 401–408. doi: 10.1007/s00709-016-0960-4
Zarka, D. G., Vogel, J. T., Cook, D., and Thomashow, M. F. (2003). Cold induction of Arabidopsis CBF genes involves multiple ICE (inducer of CBF expression) promoter elements and a cold-regulatory circuit that is desensitized by low temperature. Plant Physiol. 133, 910–918. doi: 10.1104/pp.103.027169
Zeng, J. K., Li, X., Xu, Q., Chen, J. Y., Yin, X. R., Ferguson, I. B., et al. (2015). EjAP2-1, an AP2/ERF gene, is a novel regulator of fruit lignification induced by chilling injury, via interaction with EjMYB transcription factors. Plant Biotechnol. J. 13, 1325–1334. doi: 10.1111/pbi.12351
Zhai, Y., Wang, Y., Li, Y., Lei, T., Yan, F., Su, L., et al. (2013). Isolation and molecular characterization of GmERF7, a soybean ethylene-response factor that increases salt stress tolerance in tobacco. Gene 513, 174–183. doi: 10.1016/j.gene.2012.10.018
Zhang, G., Chen, M., Chen, X., Xu, Z., Li, L., Guo, J., et al. (2010). Isolation and characterization of a novel EAR-motif-containing gene GmERF4 from soybean (Glycine max L.) Mol. Biol. Rep. 37, 809–818. doi: 10.1007/s11033-009-9616-1
Zhang, G., Chen, M., Li, L., Xu, Z., Chen, X., Guo, J., et al. (2009). Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J. Exp. Bot. 60, 3781–3796. doi: 10.1093/jxb/erp214
Zhang, H., Hedhili, S., Montiel, G., Zhang, Y., Chatel, G., Pre, M., et al. (2011). The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 67, 61–71. doi: 10.1111/j.1365-313X.2011.04575.x
Zhang, H., Hong, Y., Huang, L., Li, D., and Song, F. (2016). Arabidopsis AtERF014 acts as a dual regulator that differentially modulates immunity against Pseudomonas syringae pv. tomato and Botrytis cinerea. Sci. Rep. 6:0251. doi: 10.1038/srep30251
Zhang, H., Huang, L., Dai, Y., Liu, S., Hong, Y., Tian, L., et al. (2015). Arabidopsis AtERF15 positively regulates immunity against Pseudomonas syringae pv. tomato DC3000 and Botrytis cinerea. Front. Plant Sci. 6:686. doi: 10.3389/fpls.2015.00686
Zhang, Z. W., Feng, L. Y., Cheng, J., Tang, H., Xu, F., Zhu, F., et al. (2013). The roles of two transcription factors, ABI4 and CBFA, in ABA and plastid signalling and stress responses. Plant Mol. Biol. 83, 455–458. doi: 10.1007/s11103-013-0102-8
Zhang, Z., and Huang, R. (2010). Enhanced tolerance to freezing in tobacco and tomato overexpressing transcription factor TERF2/LeERF2 is modulated by ethylene biosynthesis. Plant Mol. Biol. 73, 241–249. doi: 10.1007/s11103-010-9609-4
Zhao, Y., Cheng, S., Song, Y., Huang, Y., Zhou, S., Liu, X., et al. (2015). The interaction between Rice ERF3 and WOX11 promotes crown root development by regulating gene expression involved in cytokinin signaling. Plant Cell 27, 2469–2483. doi: 10.1105/tpc.15.00227
Zhou, X., Zhang, Z. L., Park, J., Tyler, L., Yusuke, J., Qiu, K., et al. (2016). The ERF11 transcription factor promotes internode elongation by activating gibberellin biosynthesis and signaling. Plant Physiol. 171, 2760–2770. doi: 10.1104/pp.16.00154
Zhu, X., Qi, L., Liu, X., Cai, S., Xu, H., Huang, R., et al. (2014). The wheat ethylene response factor transcription factor pathogen-induced ERF1 mediates host responses to both the necrotrophic pathogen Rhizoctonia cerealis and freezing stresses. Plant Physiol. 164, 1499–1514. doi: 10.1104/pp.113.229575
Zhuang, J., Chen, J. M., Yao, Q. H., Xiong, F., Sun, C. C., Zhou, X. R., et al. (2011). Discovery and expression profile analysis of AP2/ERF family genes from Triticum aestivum. Mol. Biol. Rep. 38, 745–753. doi: 10.1007/s11033-010-0162-7
Zhuang, J., Li, M. Y., Wu, B., Liu, Y. J., and Xiong, A. S. (2016). Arg156 in the AP2-domain exhibits the highest binding activity among the 20 individuals to the GCC Box in BnaERF-B3-hy15, a mutant ERF transcription factor from Brassica napus. Front. Plant Sci. 7:1603 doi: 10.3389/fpls.2016.01603
Keywords: AP2/ERF, ubiquitination, transcriptional regulation, translational regulation, retrograde regulation
Citation: Phukan UJ, Jeena GS, Tripathi V and Shukla RK (2017) Regulation of Apetala2/Ethylene Response Factors in Plants. Front. Plant Sci. 8:150. doi: 10.3389/fpls.2017.00150
Received: 24 October 2016; Accepted: 25 January 2017;
Published: 21 February 2017.
Edited by:
Sergi Munné-Bosch, University of Barcelona, SpainReviewed by:
Raffaella Cerana, University of Milano-Bicocca, ItalyCopyright © 2017 Phukan, Jeena, Tripathi and Shukla. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Rakesh K. Shukla, cmsuc2h1a2xhQGNpbWFwLnJlcy5pbg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.