
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 20 January 2017
Sec. Plant Abiotic Stress
Volume 8 - 2017 | https://doi.org/10.3389/fpls.2017.00016
 Mi Young Byun1†
Mi Young Byun1† Li Hua Cui1†
Li Hua Cui1† Tae Kyung Oh1
Tae Kyung Oh1 Ye-Jin Jung1
Ye-Jin Jung1 Andosung Lee1
Andosung Lee1 Ki Youl Park1
Ki Youl Park1 Bin Goo Kang2
Bin Goo Kang2 Woo Taek Kim1*
Woo Taek Kim1*Rice U-box E3 Ub ligases (OsPUBs) are implicated in biotic stress responses. However, their cellular roles in response to abiotic stress are poorly understood. In this study, we performed functional analyses of two homologous OsPUB2 and OsPUB3 in response to cold stress (4°C). OsPUB2 was up-regulated by high salinity, drought, and cold, whereas OsPUB3 was constitutively expressed. A subcellular localization assay revealed that OsPUB2 and OsPUB3 were localized to the exocyst positive organelle (EXPO)-like punctate structures. OsPUB2 was also localized to the nuclei. OsPUB2 and OsPUB3 formed a hetero-dimeric complex as well as homo-dimers in yeast cells and in vitro. OsPUB2/OsPUB3 exhibited self-ubiquitination activities in vitro and were rapidly degraded in the cell-free extracts with apparent half-lives of 150–160 min. This rapid degradation of OsPUB2/OsPUB3 was delayed in the presence of the crude extracts of cold-treated seedlings (apparent half-lives of 200–280 min). Moreover, a hetero-dimeric form of OsPUB2/OsPUB3 was more stable than the homo-dimers. These results suggested that OsPUB2 and OsPUB3 function coordinately in response to cold stress. OsPUB2- and OsPUB3-overexpressing transgenic rice plants showed markedly better tolerance to cold stress than did the wild-type plants in terms of survival rates, chlorophyll content, ion leakage, and expression levels of cold stress-inducible marker genes. Taken together, these results suggested that the two homologous rice U-box E3 Ub ligases OsPUB2 and OsPUB3 are positive regulators of the response to cold stress.
Higher plants are constantly subjected to adverse environmental conditions owing to either biotic factors, such as pathogens and herbivores, or abiotic factors, such as extreme temperature, water availability, and high salinity. Being sessile organisms, plants are unable to move to more favorable places, and thus have developed the ability to sense and adjust to stressful conditions. Rice, a monocot model plant and the food source of more than half of the world’s population, is normally grown in tropical and temperate climate zones and is sensitive to chilling stress (Mukhopadhyay et al., 2004). Cold stress affects germination and reduces fertility, which are the key factors responsible for the decline in crop yields (Andaya and Tai, 2006; Suzuki et al., 2008; Ma et al., 2009). Thus, plants with enhanced tolerance to cold stress are able to show better growth (Cabello et al., 2014).
Ubiquitination is a post-translational modification of cellular proteins, which mainly identifies them for degradation via the 26S proteasome complex (Vierstra, 2009; Sadanandom et al., 2012). The ubiquitin (Ub)-proteasome system (UPS) regulates the stability and activity of many proteins and influences diverse cellular processes, including signal transduction, cell division, and response to biotic and abiotic stresses, in higher plants (Santner and Estelle, 2010; Lyzenga and Stone, 2012; Stone, 2014; Zhang et al., 2015; Yu et al., 2016). The UPS is conducted via successive reactions catalyzed by three enzymes (E1 Ub-activating enzymes, E2 Ub-conjugating enzymes, and E3 Ub ligases) that stimulate the tethering of poly-ubiquitin chain to a target protein for its degradation. In general, E3 Ub ligases play a crucial role in the specific recognition of appropriate target proteins and attachment of a poly-ubiquitin chain (Chen and Hellmann, 2013). E3 Ub ligases are divided into two groups based on their structures: single-subunit and multi-subunit E3 ligases (Lee and Kim, 2011; Guerra and Callis, 2012; Sharma et al., 2016). The former group consists of RING (for Really Interesting New Gene)/U-box and HECT (for Homology to E6-AP Carboxyl Terminus) E3 Ub ligases. The latter group includes SCF (for Skp1-Cullin-F-box) and APC (for Anaphase-Promoting Complex) E3 ligases.
The U-box E3 Ub ligases contain a modified RING domain and widely exist in eukaryotic organisms. While yeasts and humans contain 2 and 21 U-box E3 genes, respectively (Koegl et al., 1999; Hatakeyama et al., 2001; Ohi et al., 2003), at least 64 and 77 U-box E3 Ub ligases are predicted to be present in Arabidopsis and rice genomes, respectively (Mudgil et al., 2004; Zeng et al., 2008). Increased number of U-box proteins (PUBs) in higher plants might indicate their important roles in the adjustment of diverse cellular processes that are specific to plants (Yee and Goring, 2009). U-box E3 Ub ligases were recently implicated in biotic and abiotic stress responses in higher plants (Trujillo and Shirasu, 2010; Lyzenga and Stone, 2012; Duplan and Rivas, 2014; Stone, 2014; Zhang et al., 2015; Yu et al., 2016).
Rice PUB proteins have been reported to play roles in biotic stress responses. For example, SPL11 is known to ubiquitinate Rho GTPase-activating protein (RhoGAP) SPIN6 and negatively regulate innate immunity in rice (Zeng et al., 2004; Liu et al., 2015). OsPUB15 is involved in reducing cellular oxidative stress during seedling establishment (Park et al., 2011). OsPUB15 interacts with the receptor-like kinase PID2 and regulates cell death and immunity (Wang et al., 2015). OsPUB44 was found to be positively involved in PAMP-triggered immunity (Ishikawa et al., 2014). In addition, rice PUBs are known to participate in various cellular aspects, including brassinosteroid hormone signaling and phosphate starvation response (Hu et al., 2013; Hur et al., 2014; Ren et al., 2014). Nevertheless, the cellular roles of OsPUBs in response to abiotic stress are largely unknown in rice.
In this study, we identified two homologous U-box-type E3 Ub ligases, OsPUB2 and OsPUB3, in rice (Oryza sativa L.). The OsPUB2 gene was up-regulated by low temperature (4°C), whereas the transcript level of OsPUB3 remained unchanged after 48 h of cold treatment. Subcellular localization assay revealed that OsPUB2 and OsPUB3 were localized to the exocyst positive organelle (EXPO)-like punctate structures that were closely overlapped with Exo70E2 proteins. OsPUB2 was also localized to the nuclei. Yeast-two hybrid and in vitro pull-down assays indicated that OsPUB2 and OsPUB3 formed a hetero-dimeric complex as well as homo-dimers. Cell-free protein degradation assay indicated that OsPUB2 and OsPUB3 were more stable when they formed a hetero-dimer than when they formed homo-dimers. Both OsPUB2- and OsPUB3-overexpressing transgenic rice plants exhibited markedly enhanced tolerance to cold stress compared to wild-type rice plants. These results suggest that OsPUB2 and OsPUB3 are positively involved in the response to cold stress in rice.
Rice (Oryza sativa L.) japonica variety ‘Dong-jin’ was used in this study. Dry rice seeds were washed with 70% ethanol and subsequently with distilled water. They were then sterilized with 0.4% NaClO solution for 30 min and washed extensively with sterilized water until the NaClO solution was washed off. Sterilized seeds were germinated and grown on half-strength Murashige and Skoog (MS) medium containing vitamins (Duchefa Biochemie, Haarlem, The Netherlands), 3% sucrose, and 0.7% phytoagar for 8–10 days. Seedlings were transplanted to soil and grown at 28°C under long-day (16-h light and 8-h dark) conditions in a green house.
Total RNA was extracted from various tissues of wild-type and Ubi:sGFP-OsPUB2, Ubi:sGFP-OsPUB3, Ubi:RNAi-OsPUB2, and Ubi:RNAi-OsPUB3 transgenic rice plants by using Easy Spin Plants Total RNA Extraction kit (iNtRON Biotechnology, South Korea) according to the manufacturer’s protocol. RNA was quantified using a spectrophotometer (NanoDrop1000; Thermo Scientific, USA). Total RNA (2 μg) was used to synthesize cDNA by using TOPscript Reverse Transcriptase (Enzynomics, South Korea) and oligo (dT) primers.
Reverse transcription polymerase chain reaction (RT-PCR) was conducted as described previously (Seo et al., 2012). PCR products were separated on a 2% agarose gel and visualized under UV light. Real-time quantitative RT-PCR (qRT-PCR) was conducted on an IQ5 light cycler (Bio-Rad, USA) in 20 μL reaction mixtures by using SYBR Premix Ex Taq II (TAKARA, Japan). The amplification procedures were as follows: 5 min of denaturation and enzyme activation at 95°C, followed by 50 cycles of 5 s at 95°C, 10 s at 55°C, and 10 s at 72°C. The Actin (Os11g06390) gene was used as an internal control, and DREB1B/CBF1 (Os09g35010; CCAAT-binding factor) was used as a positive control for cold stress treatment. GAD (Os03g13300; glutamate decarboxylase), WRKY77 (Os01g40260), MRP4 (Os01g50100; multidrug resistance protein 4), MYBS3 (Os01g50100), TPP2 (Os10g40550; trehalose-6-phosphate phosphatase 2), and DREB1B/CBF1 were cold stress-induced genes (Jain et al., 2007; Su et al., 2010). Gene-specific primers used for PCR are listed in Supplementary Table S1.
In vitro self-ubiquitination assay was performed according to the established protocol described in a previous study (Bae et al., 2011). The Myc-OsPUB2, Myc-OsPUB3, Myc-OsPUB2C281A, and Myc-OsPUB3C280A fusion genes were cloned to pProEx hta vectors (Invitrogen, USA). Briefly, bacterially expressed Myc-OsPUB2, Myc-OsPUB2C281A, Myc-OsPUB3, and Myc-OsPUB3C280A recombinant fusion proteins (500 ng) were incubated for 2 h in the presence or absence of 100 ng E1 (Arabidopsis UBA1), 100 ng E2 (Arabidopsis UBC8), 10 mM ATP, and 0.1 μg/mL Ub with ubiquitination reaction buffer (50 mM Tris-HCl, pH 7.5, 2.5 mM MgCl2, and 0.5 mM DTT) at 30°C. The reaction products were subjected to immuno-blotting by using anti-Myc (Applied Biological Materials, Canada) and anti-Ub (Santa Cruz Biotechnology, USA) antibodies.
The 3′ ends of OsPUB2 and OsPUB3 coding regions were tagged with synthetic green fluorescent protein (sGFP) or monomeric red fluorescent protein (mRFP) in-frame and inserted into pEarleyGate (pEG) 100 binary vectors that contains the 35S CaMV promoter. The vector was then transformed into Agrobacterium tumefaciens strain LBA4404 by electroporation. The 35S:OsPUB2-sGFP, 35S:OsPUB2-mRFP, 35S:OsPUB3-sGFP, 35S:OsPUB3-mRFP, 35S:NLS-mRFP, and 35S:AtExo70E2-sGFP constructs were expressed in tobacco (Nicotiana benthamiana) leaves by using Agrobacterium-mediated method as described in a previous study (Kim and Kim, 2013). Two days after infection, protoplasts were extracted from the tobacco leaves, and fluorescent protein signals were visualized by fluorescence microscopy (BX51, Olympus, Japan) as described by Byun et al. (2015). NLS-mRFP and AtExo70E2-sGFP were used as nucleus and EXPO marker proteins, respectively.
Rice protoplasts were obtained from 11-day-old seedlings of Ubi:sGFP-OsPUB2 and Ubi:sGFP-OsPUB3 transgenic rice plants before and after 48 h cold treatment and were used for subcellular localization analysis of OsPUB2 and OsPUB3. Fluorescent signals were visualized by fluorescence microscopy in the presence of 50 μM MG132.
Transgenic rice plants were produced by transforming the pGA2897 binary vector plasmids that contained the maize ubiquitin promoter (Ubi) and OsPUB2/OsPUB3 (Ubi:sGFP-OsPUB2, Ubi:sGFP-OsPUB3, Ubi:RNAi-OsPUB2, and Ubi:RNAi-OsPUB3) into the A. tumefaciens strain LBA4404 via electroporation. Callus was generated by germinating wild-type rice (Oryza sativa L. japonica variety called ‘Dong-Jin’) seeds on the callus induction medium (2 mg L-1 2,4-D, 0.003% casein hydrolysate, 0.4% CHU stock containing vitamins [Duchefa Biochemie], 0.2% gelite, 0.03% L-proline, and 3% sucrose, pH 5.8) and used for Agrobacterium-mediated rice transformation. Transformed callus was selected on hygromycin B (40 mg L-1) and carbenicillin (250 mg L-1) containing medium and then transferred to the regeneration medium (2 mg L-1 kinetin, 4% MS medium containing B5 vitamins, 1 mg L-1 NAA, 1.2% phytoagar, 2% sorbitol, and 5% sucrose, pH 5.8). All processes during rice transformation were followed as described previously (Byun and Kim, 2014). Transgenic T0 plants were transplanted to soil and independent T4 overexpressing (lines #1 and #2) and T3 RNAi knock-down (lines #1 and #2) transgenic rice plants were used for phenotypic analysis.
Total genomic DNA was extracted from developing leaves of wild-type and transgenic rice plants by using the CTAB (2% CTAB, 2% PVP-40, 1.4 M NaCl, 100 mM Tris-HCl, pH 8.0, and 20 mM EDTA) method as described by Byun and Kim (2014). Total genomic DNA (10 μg) was digested with BamHI or EcoRI restriction enzyme (Thermo Scientific, USA) and separated by electrophoresis on 0.7% agarose gel. Separated DNA on the gel was transferred to Hybond-N nylon membrane, and the blot was hybridized with 32P-labeled hygromycin B phosphotransferase (Hph) probes under high stringency conditions (65°C) as described by Byun and Kim (2014). The autoradiography signals were visualized using the Bio-Imaging Analyzer (BAS2500; Fuji Film, Japan).
Yeast two-hybrid assays were performed according to the method of Bae and Kim (2013) with modifications. The full-length OsPUB2 and OsPUB3 coding regions were inserted into pGAD T7 and pGBK T7 vectors (Clontech, USA), respectively. These constructs or empty vector were co-transformed into AH109 yeast cells by using the LiAc/single-stranded carrier DNA/PEG method. Transformed yeast cells were serially diluted, plated onto four-minus (-Leu/-Trp/-His/-Ade) medium and grown at 30°C for 2 days (Bae and Kim, 2014).
OsPUB2 and OsPUB3 were cloned into pProEx hta vector and pMAL c2X vector (New England BioLabs, UK) plasmids, respectively. Maltose binding protein (MBP) and (His)6-OsPUB2, (His)6-OsPUB3, MBP-OsPUB2, and MBP-OsPUB3 recombinant proteins were expressed in Escherichia coli BL21 (DE3). Expressed proteins were purified by affinity chromatography by using Ni-NTA agarose (Qiagen, Germany) for (His)6-tagged proteins and MBP Excellose (TAKARA, Japan) for MBP fused proteins, respectively. The recombinant proteins were co-incubated with His resin for 3 h at 4°C in the affinity precipitation (AP) buffer (50 mM Tris-HCl, pH 7.5, 1 mM EDTA, 1 mM MgCl2, 0.5× proteinase inhibitor cocktail, and 0.3% Triton X-100) and captured proteins were washed five times with AP buffer. The bound proteins were eluted, resolved on 10% SDS-PAGE, transferred to PVDF membrane (Millipore, Germany), and subjected to immuno-blot analysis by using anti-MBP antibody (Applied Biological Materials) or anti-His antibody (Applied Biological Materials).
Cell-free degradation assays were conducted as described by Kim and Kim (2013). Cell-free protein crude extracts were prepared from 8-day-old mock or cold-treated rice seedlings by using protein extraction buffer (50 mM Tris-HCl, pH 7.2, 100 mM NaCl, and protease inhibitor cocktail [Roche, Switzerland]). Arabidopsis RGA, which was tagged with Flag, was cloned into pProEx hta vector. Bacterially expressed MBP-OsPUB2, MBP-OsPUB3, and Flag-RGA recombinant proteins were incubated with cell-free extracts for 2, 4, and 6 h at 30°C. Each sample was harvested, boiled, separated by 10% SDS-PAGE, and analyzed by immunoblotting by using the anti-MBP antibody and anti-Flag antibody (Applied Biological Materials). Rubisco was used as a loading control.
For cold stress treatment, 5-week-old wild-type and transgenic rice plants were transferred to cold room at 4°C for 6 days, after which plants were recovered at 28°C and their growth patterns were monitored as described previously (Byun et al., 2015). Total chlorophyll (chlorophyll a + chlorophyll b) was extracted from wild-type and transgenic leaves before and after cold treatment according to Lichtenthaler (1987) with modifications as described by Min et al. (2016). The amounts of chlorophyll a and chlorophyll b were measured at 664.2 and 648.6 nm, respectively, by ELISA microplate reader (VERSAmax, Molecular Devices, USA).
Electrolyte leakage analysis was conducted using 8-day-old rice seedlings before and after cold stress treatments in different time points (0, 6, and 11 days) at 4°C. Seedlings of wild-type and transgenic plants were soaked in a test tube containing 35 mL of distilled water on an orbital shaker (200 rpm) at 28°C for overnight. The electrolyte conductivity of each sample was determined before and after autoclaving by using conductivity meter (Orion Star A212, Thermo Scientific, USA) by the method of Min et al. (2016).
OsPUB2 and OsPUB3 are homologous genes that encode putative U-box E3 Ub ligases with 75% amino-acid sequence identity in rice (Figure 1A). Both predicted proteins contained a single U-box motif and Armadilo (ARM) repeats in their central and C-terminal regions, respectively. OsPUB2 and OsPUB3 are 71–82% identical to putative U-box proteins from monocot plants, such as millet (Setaria italica), false brome (Brachypodium distachyon), and maize (Zea mays), whereas they share 42% sequence identity with AtPUB17 from dicot Arabidopsis thaliana (Supplementary Figure S1). The U-box and ARM domains are well-conserved in both monocot and dicot U-box proteins.
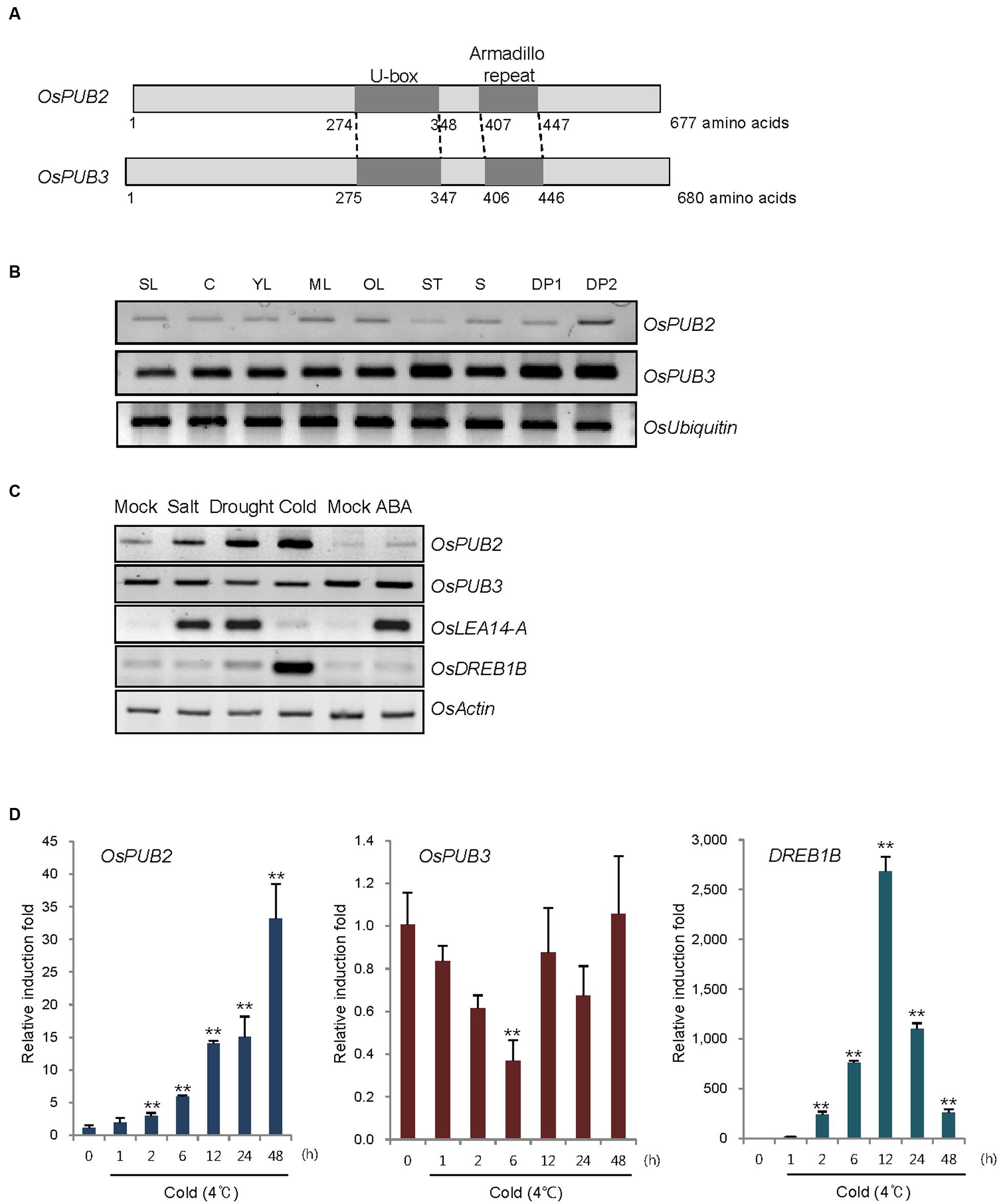
FIGURE 1. Identification and expression of OsPUB2 and OsPUB3 in rice. (A) Schematic structures of predicted rice U-box E3 Ub ligases OsPUB2 and OsPUB3. Gray bars depict the coding regions. The U-box motif and Armadillo repeat are indicated by dark gray bars. (B) Spatial expression patterns of OsPUB2 and OsPUB3 in rice. Total RNA was extracted from various tissues and analyzed by RT-PCR. OsUbiquitin is a loading control. SL, 10-day-old whole seedlings; C, callus; YL, young leaves; ML, mature leaves; OL, old leaves; ST, stems; S, seeds; DP1, 3–5 cm-long developing panicles; DP2, 5–8 cm-long developing panicles. (C) (Left panel) Expression profiles of OsPUB2 and OsPUB3 in response to the different abiotic stress conditions. Light-grown, 10-day-old wild-type rice seedlings were subjected to high salt (200 mM NaCl for 3 h), dry (40% reduction of fresh weight), low temperature (4°C for 8 h), and ABA (150 μM for 3 h). Total RNA was prepared from the treated tissues and analyzed by RT-PCR using gene-specific primer sets. OsLEA14-A served as a positive control for drought, salt, and ABA treatments, whereas OsDREB1B was used as a cold stress marker gene. OsActin is a loading control. (Right panel) Time course inductions of OsPUB2 and OsPUB3 in response to cold (4°C for 4 and 8 h) and drought (15, 30, and 45% reduction of fresh weight) treatments. (D) Real-time qRT-PCR analysis of OsPUB2 and OsPUB3. Total RNA was isolated from the leaves treated with cold stress (4°C) for different time periods (0, 1, 2, 6, 12, 24, and 48 h) and used for qRT-PCR. Fold induction of OsPUB2 and OsPUB3 was normalized to the level of OsActin mRNA, which was used as an internal control. Data represent mean ± SE (∗∗P < 0.01, Student’s t-test) from three independent experiments. Nucleotide sequences of primers used for RT-PCR are shown in Supplementary Table S1.
Reverse transcription polymerase chain reaction analysis showed that transcripts of OsPUB2 and OsPUB3 were detected in all tissues examined, including early seedlings, developing and mature leaves, stems, developing seeds, and panicles, with the expression level of OsPUB3 being higher than that of OsPUB2 (Figure 1B). The transcript level of OsPUB2 was elevated in response to high salt (200 mM NaCl for 3 h), dry (15–45% reduction of fresh weight), and low temperature (4°C for 4–8 h) treatments in 10-day-old rice seedlings (Figure 1C). In contrast, the transcript level of OsPUB3 was not affected by stress treatments. Neither of the genes was induced by ABA (150 μM for 3 h). Real-time qRT-PCR assay indicated that the amount of OsPUB2 mRNA began to increase at 1 h after exposure to cold stress and was continuously elevated up to 35-fold after 48 h (Figure 1D). The induction pattern of OsPUB2 was different from that of DREB1B, a cold stress marker gene, the expression of which was maximum at 12 h after cold treatment. The transcript level of OsPUB3 was reduced at 6 h after cold treatment and restored to normal levels thereafter.
Whether OsPUB2 and OsPUB3 possess E3 Ub ligase enzymatic activity was determined by performing in vitro self-ubiquitination assay. Bacterially expressed Myc-tagged OsPUB2 or OsPUB3 recombinant proteins were incubated at 30°C for 2 h in the presence or absence of E1(Arabidopsis UBA1), E2 (Arabidopsis UBC8), Ub, and ATP. Reaction mixtures were separated by SDS-PAGE and subjected to immuno-blot analysis by using anti-Myc and anti-Ub antibodies. As shown in Figure 2, Myc-OsPUB2 and Myc-OsPUB3 yielded high-molecular-mass ubiquitinated bands detected by both anti-Myc and anti-Ub antibodies. In contrast, the exclusion of E1, E2, ATP, or Ub from the incubation mixture abrogated ubiquitinated smear bands. Myc-OsPUB2C281A and Myc-OsPUB3C280A, in which the conserved Cys residue in the U-box motif was replaced by Ala, failed to exhibit the E3 ligase activity even in the presence of all reaction components (Figure 2). Overall, the results presented in Figures 1 and 2 indicated that OsPUB2 and OsPUB3 are homologous U-box E3 Ub ligases in rice.
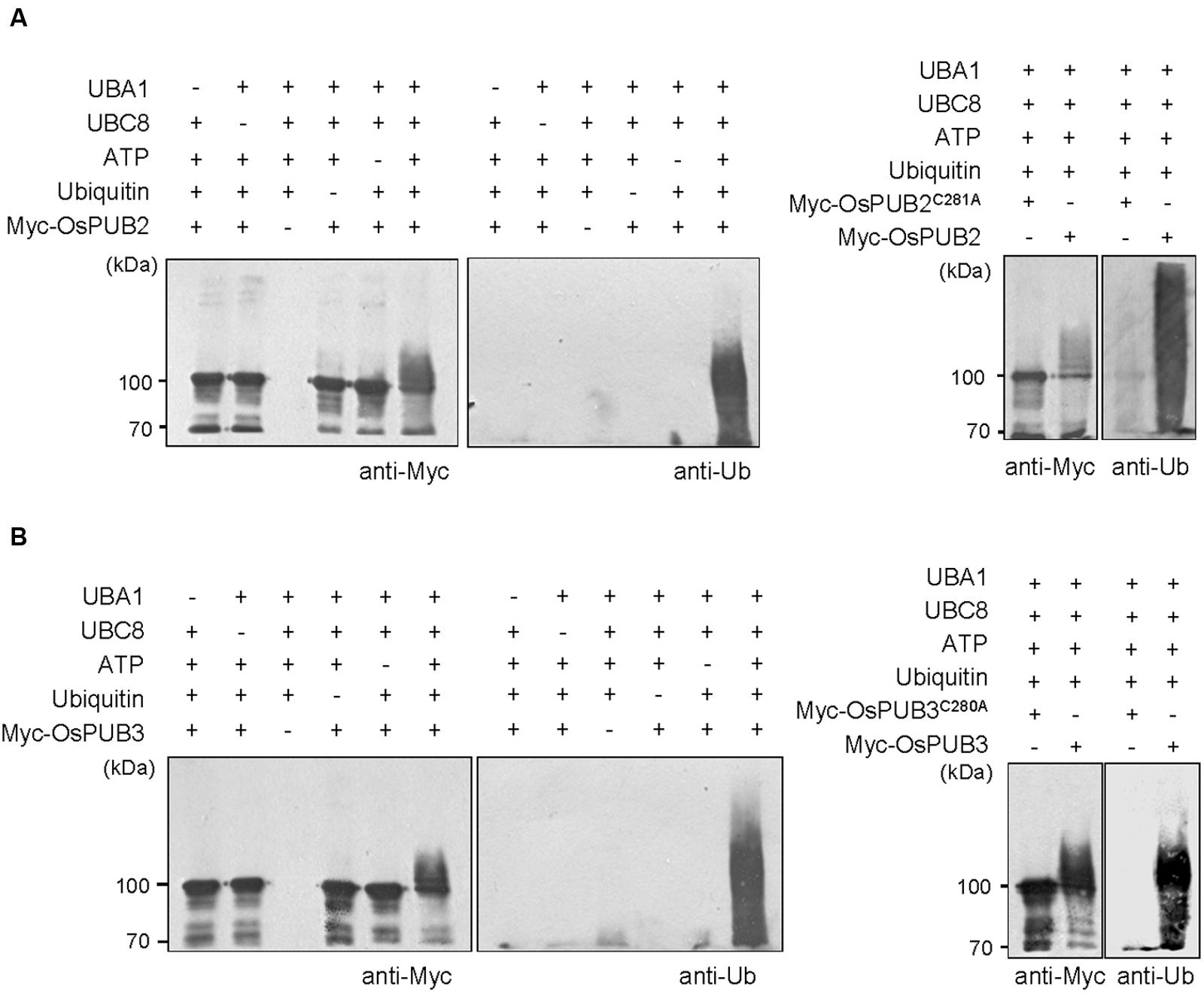
FIGURE 2. In vitro self-ubiquitination assays of OsPUB2 and OsPUB3. (A) (Left panel) Bacterially expressed Myc-tagged OsPUB2 fusion protein was purified and incubated at 30°C for 2 h in the presence or absence of E1 (Arabidopsis UBA1), E2 (Arabidopsis UBC8), Ub, and ATP. Samples were resolved by 8% SDS-PAGE and subjected to immuno-blot analysis by using anti-Myc and anti-Ub antibodies. (Right panel) Wild-type Myc-OsPUB2 and a single amino acid substitution mutant Myc-OsPUB2C281A were incubated at 30°C for 2 h with E1, E2, Ub, and ATP, and analyzed by immuno-blotting by using anti-Myc and anti-Ub antibodies. (B) Purified recombinant Myc-OsPUB3 (Left panel) and a single amino acid substitution mutant Myc-OsPUB3C280A (Right panel) proteins were used for in vitro self-ubiquitination assay by using anti-Myc and anti-Ub antibodies as described above.
The subcellular localization of OsPUB2 and OsPUB3 was investigated by conducting an in vivo protein targeting experiment. The 35S:OsPUB2-sGFP or 35S:OsPUB3-sGFP chimeric construct was co-expressed with 35S:NLS-mRFP in tobacco (N. benthamiana) leaves by using Agrobacterium-mediated infiltration method. The NLS-mRFP was used as a nuclear marker protein. Protoplasts were prepared from tobacco leaves, and expressed fusion proteins were visualized by fluorescence microscopy. The results revealed that the fluorescence signal of OsPUB2-sGFP was exhibited as small cytosolic punctate bodies and was also found in the nucleus, where it merged with the NLS-mRFP signal (Figure 3Aa). In the case of OsPUB3-sGFP, the fluorescence signals were only exhibited in the cytosolic punctae. When OsPUB2-mRFP and OsPUB3-sGFP constructs were co-expressed in tobacco leaves, the cytosolic punctate fluorescence signals were closely merged, indicating the co-localization of OsPUB2 and OsPUB3 (Figure 3Ab). These punctate localization patterns of OsPUB2/OsPUB3 were reminiscent of EXPOs (Wang et al., 2010; Ding et al., 2014). Indeed, the localization signals of OsPUB2-mRFP and OsPUB3-mRFP closely overlapped with those of AtExo70E2-sGFP, a marker protein of EXPO, in tobacco leaf protoplasts (Figure 3B). We detected very weak punctate localization signals of both OsPUB2 and OsPUB3 in the protoplasts prepared from Ubi:OsPUB2-sGFP and Ubi:OsPUB3-sGFP transgenic rice plants (Supplementary Figure S3). These localization patterns were unchanged in response to cold treatment (Supplementary Figure S3). Thus, OsPUB2 appeared to be localized to the EXPO-like punctate bodies and nuclei, whereas OsPUB3 was predominantly localized to the EXPO-like structure.
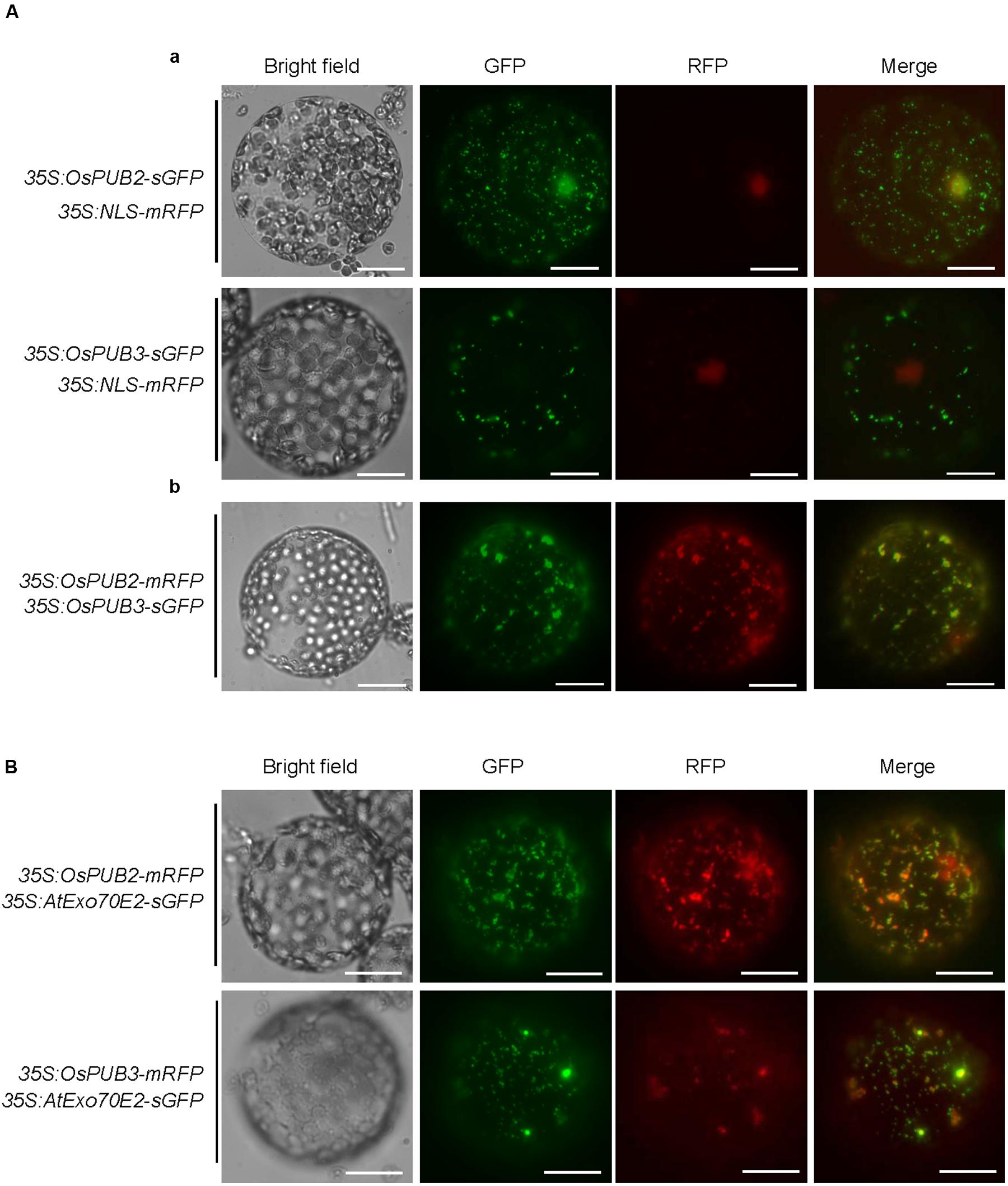
FIGURE 3. Subcellular localization of OsPUB2 and OsPUB3. (A-a) The 35S:OsPUB2-sGFP or 35S:OsPUB3-sGFP fusion construct was co-expressed with 35S:NLS-mRFP in tobacco (N. benthamiana) leaf epidermal cells by using Agrobacterium-mediated infiltration method. After 2 days of infiltration, protoplasts were prepared from the leaves, and fluorescent signals were visualized by fluorescence microscopy under dark-field conditions. The NLS-mRFP was used as a nucleus-localized marker protein. Bars = 10 μm. (A-b) The 35S:OsPUB2-mRFP and 35S:OsPUB3-sGFP fusion constructs were co-expressed in tobacco leaves. The co-localization signals of OsPUB2-mRFP and OsPUB3-sGFP in the protoplasts were detected by fluorescence microscopy under dark-field conditions. Bars = 10 μm. (B) The 35S:OsPUB2-mRFP+ 35S:AtExo70E2-sGFP and 35S:OsPUB3-mRFP+ 35S:AtExo70E2-sGFP constructs were infiltrated into tobacco leaves, and their co-localization signals were visualized by fluorescence microscopy. The AtExo70E2-sGFP served as a marker protein of exocyst positive organelles (EXPOs). Bars = 10 μm.
The RING/U-box E3 Ub ligases often form stable dimeric complexes, which is critical for their activity (Nikolay et al., 2004; Berndsen and Wolberger, 2014). To determine whether OsPUB2 and OsPUB3 form dimers, we performed a yeast two-hybrid assay. The full-length OsPUB2 and OsPUB3 coding regions were inserted into pGAD T7 and pGBK T7 vectors, respectively, and co-transformed into yeast cells. Transformed yeast cells were serially diluted and plated onto three-minus (-Leu/-Trp/-Ade) and four-minus (-Leu/-Trp/-His/-Ade) growth media. The results showed that both OsPUB2 and OsPUB3 could interact with themselves and formed homo-dimers in the yeast cells (Figure 4A). Considering the high sequence identity (75%) (Figure 1A; Supplementary Figure S1A) and identical subcellular localization patterns (Figure 3) of OsPUB2 and OsPUB3, we speculated that they formed a hetero-dimeric complex. As expected, OsPUB2 and OsPUB3 bound each other to form hetero-dimers in the yeast cells (Figure 4A).
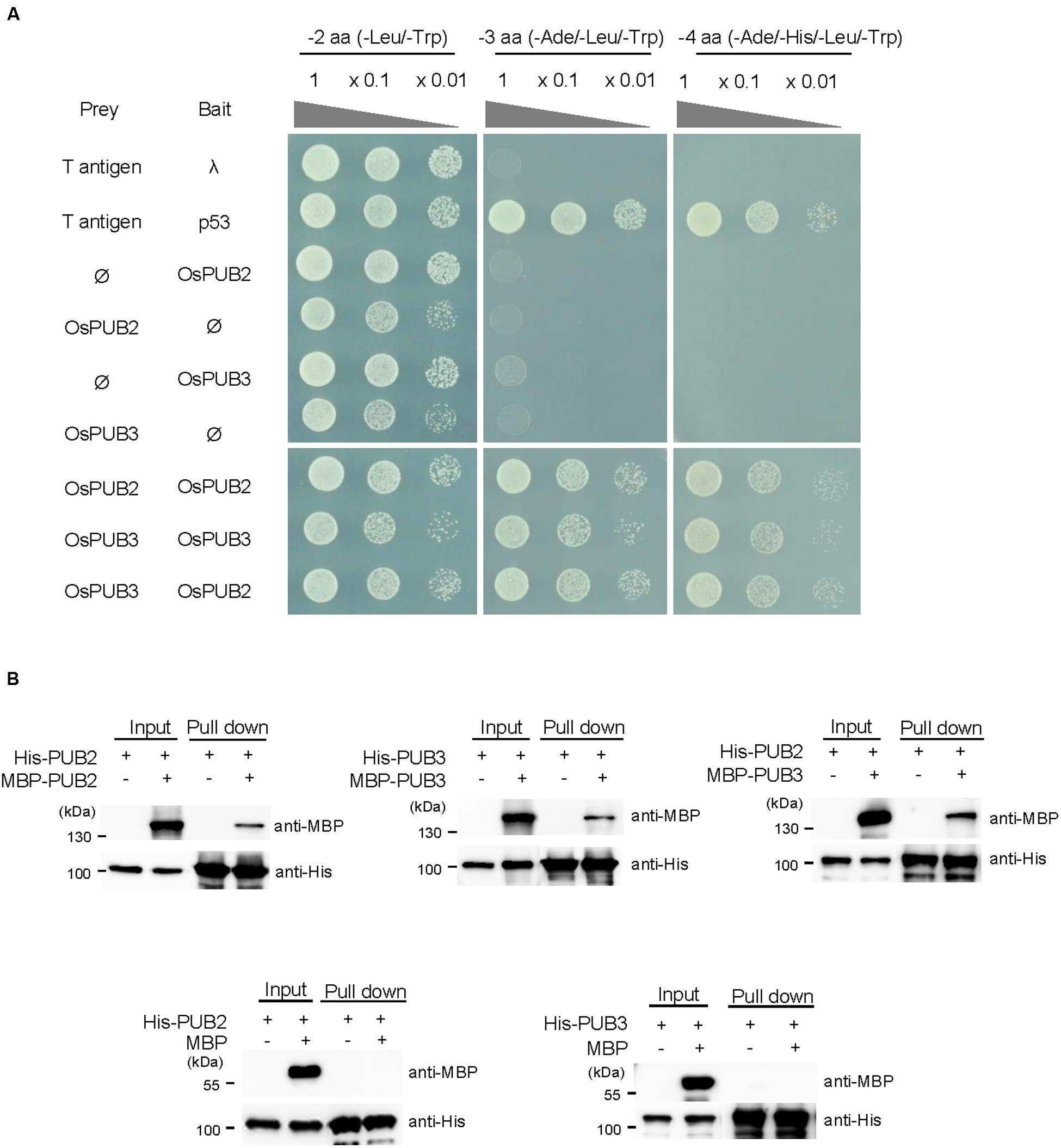
FIGURE 4. Homo- and hetero-dimeric complex formation of OsPUB2 and OsPUB3. (A) Yeast two-hybrid assay. The full-length coding regions of OsPUB2 and OsPUB3 were cloned into pGAD T7 and pGBK T7 vectors, respectively, and co-transformed into yeast AH109 cells with a combination of the indicated plasmids. Transformed yeast cells were serially diluted, plated onto two-minus (-Leu/-Trp), three-minus (-Leu/-Trp/-Ade), and four-minus (-Leu/-Trp/-His/-Ade) growth media, and incubated at 30°C for 2 days. The p53 + T-antigen were used as a positive control, whereas lambda + T-antigen were used as a negative control. (B) In vitro pull down assay. Bacterially expressed MBP-OsPUB2, MBP-OsPUB3, or MBP recombinant protein was co-incubated with (His)6-OsPUB2 or (His)6-OsPUB3 as indicated in the presence of a Ni-NTA agarose affinity matrix. The bound protein was eluted, resolved by 10% SDS-PAGE, and immuno-blotted with anti-MBP and anti-His antibodies.
To further confirm the dimeric-complex formation of OsPUB2 and OsPUB3, we performed in vitro pull-down assay. Bacterially expressed (His)6-OsPUB2 + MBP-OsPUB2, (His)6-OsPUB3 + MBP-OsPUB3, and (His)6-OsPUB2 + MBP-OsPUB3 protein mixtures were co-incubated with a Ni-NTA resin. The bound proteins were eluted from the resin and analyzed by immuno-blotting by using anti-MBP and anti-His antibodies. As indicated in Figure 4B, the MBP-tagged OsPUB2 and OsPUB3 proteins were pull-downed from the Ni-NTA resin by the (His)6-tagged OsPUB2 and OsPUB3, indicating that OsPUB2 and OsPUB3 formed both homo- and hetero-dimeric complexes in vitro.
We next considered the possibility that the formation of a hetero-dimeric complex of OsPUB2 and OsPUB3 might affect their stability. To test this hypothesis, we performed an in vitro cell-free degradation assay. MBP-OsPUB2 and MBP-OsPUB3 recombinant proteins were incubated for different time periods (0, 2, 4, and 6 h) with the crude protein extracts prepared from 8-day-old rice seedlings grown under normal conditions. The levels of MBP-OsPUB2 and MBP-OsPUB3 rapidly decreased over time (0–6 h) in the cell-free extracts. At 4 h of incubation, 76.8 ± 2.0 and 80.2 ± 2.2% of OsPUB2 and OsPUB3 were degraded, respectively (Figure 5A). After 6 h, only 6.0 ± 0.9 and 6.7 ± 3.0% of OsPUB2 and OsPUB3 proteins were detected, respectively. Under our experimental conditions, the apparent half-lives of MBP-OsPUB2 and MBP-OsPUB3 were approximately 150 min and 160 min, respectively, in the mock-treated cell-free extracts (Figure 5B). However, the degradation of MBP-OsPUB2 and MBP-OsPUB3 appeared to be slower in the cell-free extracts prepared from the cold-treated seedlings than in the mock-treated seedlings: apparent half-lives of MBP-OsPUB2 and MBP-OsPUB3 were about 200 min and 280 min, respectively, in the cold-treated cell-free extracts (Figure 5B). We repeated identical sets of experiments using MBP as a negative control. As shown in Figure 5C, the level of MBP was consistent in mock- and cold-treated crude extracts. In addition, RGA, which was degraded by the 26S proteasome complex (Lee et al., 2010; Kim and Kim, 2013), was degraded in a cold-independent manner (Figure 5D). These results suggested that OsPUB2 and OsPUB3 are more stable in cold-treated cell-free extracts than in mock-treated extracts, with the half-life of OsPUB3 being more apparently increased.
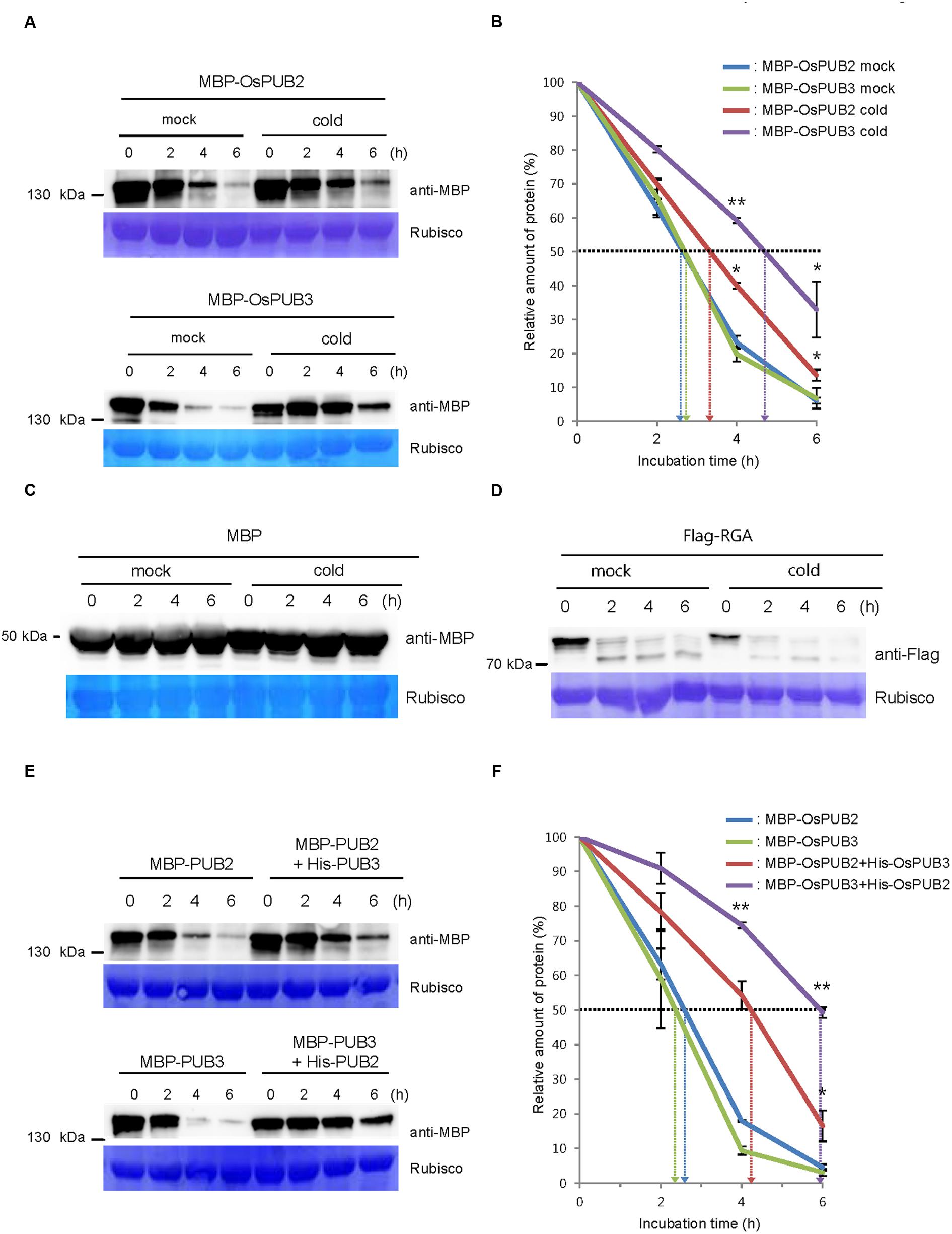
FIGURE 5. Regulation of OsPUB2 and OsPUB3 stability in cell-free protein crude extracts. (A) In vitro cell-free degradation assays of OsPUB2 and OsPUB3 under mock and cold stress conditions. The MBP-OsPUB2 or MBP-OsPUB3 protein was incubated for different time periods (0, 2, 4, and 6 h) with the protein crude extracts prepared from light-grown 8-day-old wild-type rice seedlings before and after cold treatment. The time-dependent protein levels were analyzed by immuno-blotting by using anti-MBP antibody. Rubisco detected by Coomassie blue served as a loading control. (B) Apparent half-lives of OsPUB2 and OsPUB3 in the cell-free crude extracts. The graph shows the decrease in the relative amounts of proteins in the cell-free degradation assay. The levels of proteins were calculated by quantification of band intensities by using ImageJ software. Bars represent mean ± SE (∗P < 0.05, ∗∗P < 0.01, Student’s t-test) from three independent experiments. (C,D) In vitro cell-free degradation assays of MBP and RGA under mock and cold stress conditions. The MBP (C) or Flag-RGA (D) protein was incubated for different time periods (0, 2, 4, and 6 h) with the protein crude extracts prepared from light-grown 8-day-old wild-type rice seedlings before and after cold treatment. The time-dependent protein levels were analyzed by immuno-blotting by using anti-MBP (C) and anti-Flag (D) antibodies. Rubisco detected by Coomassie blue served as a loading control. (E) In vitro cell-free degradation assay of homo- and hetero-dimeric complexes of OsPUB2 and OsPUB3. The MBP-OsPUB2 and MBP-OsPUB3 were incubated for different time periods (0, 2, 4, and 6 h) in the presence or absence of (His)6-OsPUB3 and (His)6-OsPUB2, respectively, with mock-treated crude extracts. The levels of proteins were detected by immuno-blotting with anti-MBP antibody. Rubisco was used as a loading control. (F) Apparent half-lives of OsPUB2 and OsPUB3 in the cell-free crude extracts when they formed homo- and hetero-dimers. The graph shows the decrease in the relative amounts of proteins in the cell-free degradation assay. Bars indicate mean ± SE (∗P < 0.05, ∗∗P < 0.01, Student’s t-test) from three independent experiments.
Furthermore, when OsPUB2 and OsPUB3 were mixed together in the cell-free extracts, their degradation was markedly delayed (Figure 5E); thus, their apparent half-lives were increased from 150 min and 140 min to 250 min and 350 min, respectively (Figure 5F). On the other hand, both OsPUB2 and OsPUB3 were rapidly degraded in the presence of equal amount of RGA (Supplementary Figures S2A,B), suggesting that slower degradation of OsPUB2 + OsPUB3 hetero-dimer was not due to the increased amounts of proteins in the reaction mixture. This indicated that a hetero-dimeric form of OsPUB2 and OsPUB3 was more stable than the homo-dimers in a cell-free system. Taken together, these results raised the possibility that the stability of OsPUB2 and OsPUB3 are ameliorated in response to cold stress (Figures 5A,B) and by the formation of a hetero-dimeric complex (Figures 5E,F).
To address the cellular roles of OsPUB2 and OsPUB3, we generated transgenic rice plants (Ubi:sGFP-OsPUB2 and Ubi:sGFP-OsPUB3), in which the sGFP-OsPUB2 and sGFP-OsPUB3 genes were ectopically expressed under the control of the maize ubiquitin promoter (Ubi) (Figure 6A). The qRT-PCR analysis results showed markedly increased amounts of OsPUB2 and OsPUB3 transcripts in two different T4 transgenic lines (#1 and #2) of Ubi:sGFP-OsPUB2 and Ubi:sGFP-OsPUB3 plants, respectively, under normal growth conditions (Figure 6B). The overexpression of sGFP-OsPUB2 and sGFP-OsPUB3 proteins was confirmed by immuno-blot analysis by using anti-GFP antibody (Figure 6C). DNA gel-blot analysis showed that these over-expressing transgenic lines were independent (Supplementary Figure S4). In addition, the independent transgenic rice plants (Ubi:RNAi-OsPUB2 and Ubi:RNAi-OsPUB3), in which OsPUB2 and OsPUB3 were suppressed, were constructed using the RNA interference (RNAi) method (Figure 6A; Supplementary Figure S4). The results of qRT-PCR analysis indicated that the expression of OsPUB2 was reduced approximately by 40% in Ubi:RNAi-OsPUB2 (transgenic lines #1 and #2), whereas that of OsPUB3 was decreased by 50% (line #1) to 70% (line #2) in Ubi:RNAi-OsPUB3 (Figure 6D). The transcript levels of OsPUB2 and OsPUB3 were slightly increased (1.1–1.5 times), in Ubi:RNAi-OsPUB3 and Ubi:RNAi-OsPUB2 plants, respectively (Figure 6D). These overexpressing and RNAi-mediated knock-down transgenic rice plants were used for phenotypic analysis in response to cold stress.
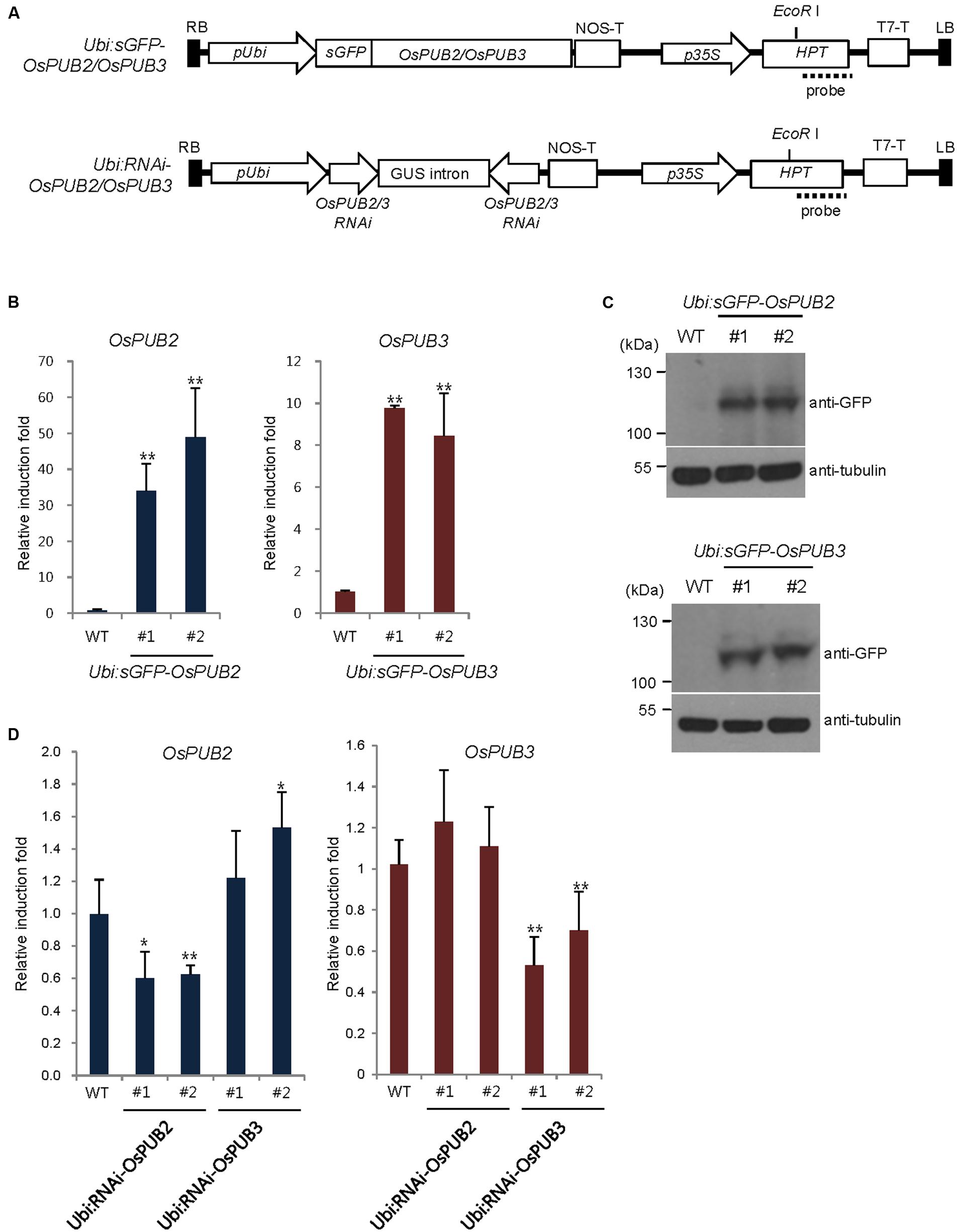
FIGURE 6. Generation and molecular characterization of OsPUB2- and OsPUB3-overexpressing and RNAi-mediated knock-down transgenic rice plants. (A) Schematic structures of the OsPUB2 and OsPUB3 overexpression and RNAi-mediated knock-down binary vector constructs. RB, right border; pUbi, maize ubiquitin promoter; NOS-T, NOS terminator; p35S, 35S CaMV promoter; Hpt, hygromycin phosphotransferase; T7-T, T7 terminator; LB, left border. Dash lines indicate DNA probes for genomic Southern blot analysis in Supplementary Figure S4. (B) qRT-PCR analysis of wild-type (WT) and T4 Ubi:sGFP-OsPUB2 (independent transgenic lines #1 and #2) and Ubi:sGFP-OsPUB3 (lines #1 and #2) transgenic rice plants. Data represent the fold induction of OsPUB2 and OsPUB3 in overexpressing transgenic rice plants relative to wild-type plants. The relative expression levels of OsPUB2 and OsPUB3 transcripts were normalized to the level of OsActin mRNA, which was used as an internal reference gene. Data represent mean ± SE (∗∗P < 0.01, Student’s t-test) from three independent experiments. (C) Immuno-blot analysis of wild-type (WT) and T4 Ubi:sGFP-OsPUB2 (independent transgenic lines #1 and #2) and Ubi:sGFP-OsPUB3 (lines #1 and #2) transgenic rice plants by using anti-GFP antibody. Tubulin was used as the loading control. (D) qRT-PCR analysis of wild-type (WT) and T3 Ubi:RNAi-OsPUB2 (independent transgenic lines #1 and #2), and Ubi:RNAi-OsPUB3 (lines #1 and #2) transgenic rice plants. Data represent the fold reduction of OsPUB2 and OsPUB3 in RNAi-knock-down transgenic progeny relative to wild-type (WT) plants. The relative expression levels of OsPUB2 and OsPUB3 mRNAs were normalized to that of OsActin mRNA, an internal control. Data represent mean ± SE (∗P < 0.05, ∗∗P < 0.01, Student’s t-test) from three independent experiments.
Wild-type, T4 Ubi:sGFP-OsPUB2 (lines #1 and #2), and T4 Ubi:sGFP-OsPUB3 (lines #1 and #2) rice plants were grown at 28°C under long day (16-h light and 8-h dark) conditions for 5 weeks. These plants were then subjected to cold stress by transferring them to a cold room at 4°C. After 6 days of low temperature treatment, plants were retransferred to the growth room at 28°C, and their growth patterns were monitored. As shown in Figure 7, most of the wild-type plants exhibited pale green and yellowish leaves after cold stress. They were unable to grow and eventually died (survival rate = 1.7 ± 1.4–7.4 ± 6.4%). In contrast, OsPUB2- and OsPUB3-overexpressing plants showed markedly increased tolerance to cold temperature compared to the wild-type rice plant. The survival rates of Ubi:sGFP-OsPUB2 was 79.1 ± 5.7% (line #1) and 32.5 ± 6.0% (line #2), whereas those of Ubi:sGFP-OsPUB3 were 60.7 ± 9.7% (line #1) and 50.7 ± 4.4% (line #2) (Figure 7).
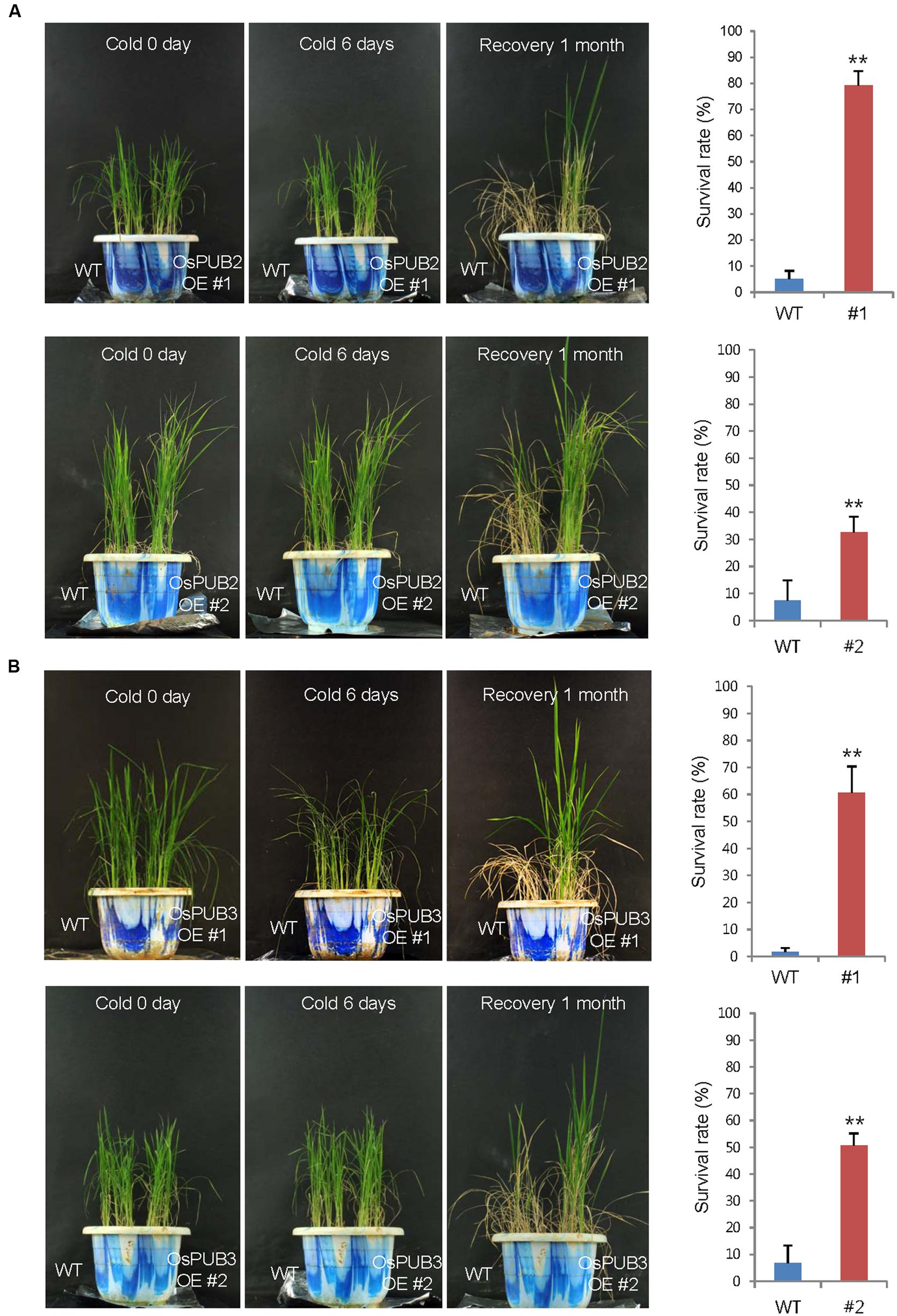
FIGURE 7. Cold stress tolerant phenotypes of OsPUB2- and OsPUB3-overexpressing transgenic rice plants. Overexpression of OsPUB2 (A) and OsPUB3 (B) conferred enhanced tolerance to cold stress compared to that in wild-type rice plants. Wild-type and T4 Ubi:sGFP-OsPUB2 (lines #1 and #2) and Ubi:sGFP-OsPUB3 (lines #1 and #2) transgenic rice plants were grown for 5 weeks under normal condition (28°C). These plants were transferred to cold room at 4°C for 6 days and recovered at 28°C. The survival rates of cold-treated plants were monitored. Data represent mean ± SE (n ≥ 5 independent experiments; more than 100 plants were used in each assay, ∗∗P < 0.01, Student’s t-test). OE, overexpressing transgenic plants.
In addition, 5-week-old wild-type and OsPUB2/OsPUB3-overexpressing plants exhibited similar leaf chlorophyll content (chlorophyll a + chlorophyll b) under normal condition (Figure 8A). Consistent with their tolerance, OsPUB2- and OsPUB3-overexpressing progeny contained higher amount of chlorophyll compared to that in the wild-type plants in response to cold stress. At 1 month recovery of cold treatment (4°C), the leaf chlorophyll content of wild-type plants was 3.5 ± 1.6 mg/g DW, whereas those of Ubi:sGFP-OsPUB2 and Ubi:sGFP-OsPUB3 was 12.5 ± 2.2–13.5 ± 1.1 mg/g DW and 12.6 ± 0.5–16.2 ± 1.7 mg/g DW, respectively (Figure 8A). As a next experiment, electrolyte leakage from 8-day-old cold-stressed seedlings was determined. Wild-type and OsPUB2/OsPUB3-overexpressing plants were grown at 4°C for 0, 6, and 11 days, and whole seedlings were soaked in distilled water for measuring the rates of electrolyte leakage. The data showed that both Ubi:sGFP-OsPUB2 and Ubi:sGFP-OsPUB3 seedlings showed lower ion leakage (9.7 ± 0.2–10.3 ± 1.0% at 6 days and 17.8 ± 2.1–19.5 ± 1.5% at 11 days) than the wild-type (13.9 ± 0.4% at 6 days and 27.0 ± 1.5% at 11 days) plants in response to low temperature (Figure 8B). Overall, phenotypic analyses in Figures 7 and 8 indicated that OsPUB2- and OsPUB3-overexpressors were more tolerant to severe cold stress than the wild-type plants, suggesting that OsPUB2 and OsPUB3 have positive roles in cold stress response in rice plants.
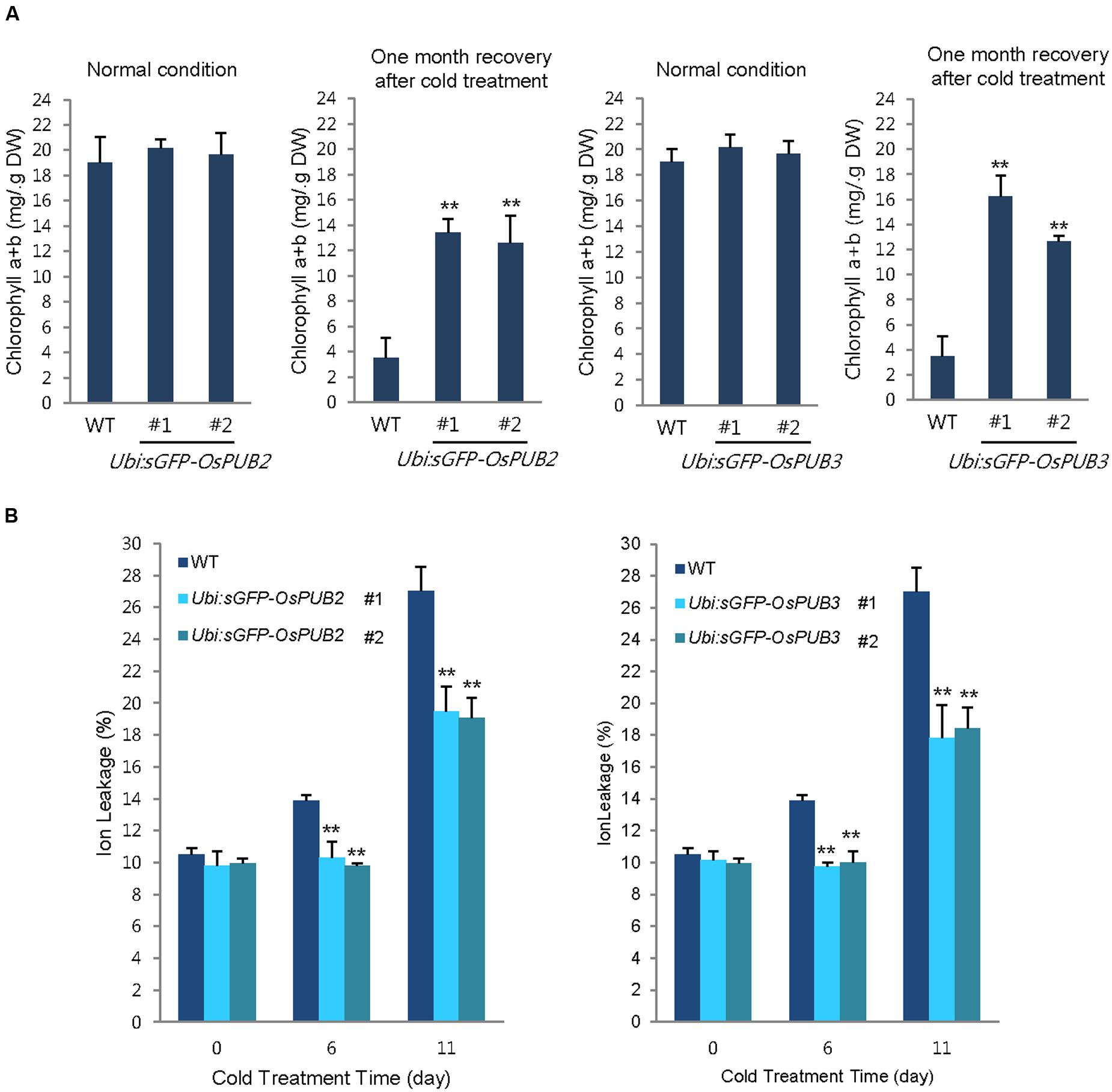
FIGURE 8. Effects of OsPUB2 and OsPUB3 overexpression on leaf chlorophyll content and electrolyte leakage in response to cold stress. (A) Total chlorophyll content of wild-type and T4 Ubi:sGFP-OsPUB2 (lines #1 and #2) and Ubi:sGFP-OsPUB3 (lines #1 and #2) transgenic rice plants. Light-grown, 5-week-old wild-type and transgenic plants were grown for 6 days under normal (28°C) or cold (4°C) condition. The amount of total leaf chlorophyll (chlorophyll a + chlorophyll b) was determined at normal growth condition and 1 month recovery after cold (4°C) treatment. Data indicate the mean ± SE (9 ≥ n ≥ 3 independent experiments; 10 plants were used in each experiment, ∗∗P < 0.01, Student’s t-test). (B) Electrolyte leakage analysis was performed using 8-day-old seedlings of wild-type and T4 Ubi:sGFP-OsPUB2 (lines #1 and #2) and Ubi:sGFP-OsPUB3 (lines #1 and #2) plants before and after cold (4°C) treatment in different time points (0, 6, and 11 days). Data represent ± SE (8 ≥ n ≥ 3 independent experiments; three plants were used in each experiment, ∗∗P < 0.01, Student’s t-test).
Next, we examined the phenotype of Ubi:RNAi-OsPUB2 (lines #1 and #2) and Ubi:RNAi-OsPUB3 (lines #1 and #2) knock-down transgenic lines. Unlike the overexpressing lines, RNAi-knock-down plants were very similar to wild-type plants in terms of tolerance to cold temperature (Supplementary Figure S5). The total chlorophyll content and electrolyte leakage rate of RNAi-knock-down plants were also indistinguishable from those of the wild-type rice plants (Supplementary Figure S6). These results led us to hypothesize that the basal levels of OsPUB2 and OsPUB3 might be considerably high, and hence, their partial suppression resulted in undetectable effects on the Ubi:RNAi-OsPUB2 and Ubi:RNAi-OsPUB3 knock-down transgenic lines. Alternatively, the suppression of OsPUB2 could be complemented by OsPUB3 and, conversely, the knock-down of OsPUB3 could be rescued by OsPUB2. This assumption seems to be reasonable, since OsPUB2 and OsPUB3 shared high degree of sequence identity (75%), had a similar structure (Figure 1A), and formed a hetero-dimeric complex (Figure 4).
Because the ectopic expression of OsPUB2 and OsPUB3 conferred increased tolerance to cold stress in rice plants, we next determined whether OsPUB2 or OsPUB3 affected the expression profiles of cold-stress responsive genes. Light-grown, 8-day-old wild-type, Ubi:sGFP-OsPUB2, and Ubi:sGFP-OsPUB3 transgenic seedlings were subjected to cold stress (4°C) for 24 h. Total RNA was isolated from the shoot tissue and analyzed using real-time qRT-PCR by using gene-specific primer sets (Supplementary Table S1). The expressions of GAD (Os03g13300; glutamate decarboxylase), WRKY77 (Os01g40260), and MRP4 (Os01g50100; multidrug resistance protein 4), which are cold stress-inducible genes (Jain et al., 2007; Su et al., 2010), were markedly upregulated in Ubi:sGFP-OsPUB2 and Ubi:sGFP-OsPUB3 transgenic plants than in the wild-type plants under both normal and cold-stressed conditions (Figure 9). Although, TPP2 (Os10g40550; trehalose-6-phosphate phosphatase 2) was not induced by cold treatment in the wild-type rice plant, it was 2–6-fold upregulated in OsPUB2/3-overexpressing transgenic plants relative to wild-type plants after 24 h of cold treatment. The transcript level of MYBS3 (Os01g50100) was similar in wild-type and OsPUB2/OsPUB3-overexpressing plants under normal condition, but was higher in the overexpressors after cold treatment (Figure 9). Expression of DREB1B/CBF1 (CCAAT-binding factor) was higher in Ubi:sGFP-OsPUB2/3 plants in normal condition relative to that of wild-type plant, but it became lower after cold treatment. These results suggested that the cold stress-tolerant phenotypes of Ubi:sGFP-OsPUB2 and Ubi:sGFP-OsPUB3 plants were correlated with increased expression levels of cold stress-related genes before or after low temperature treatment.
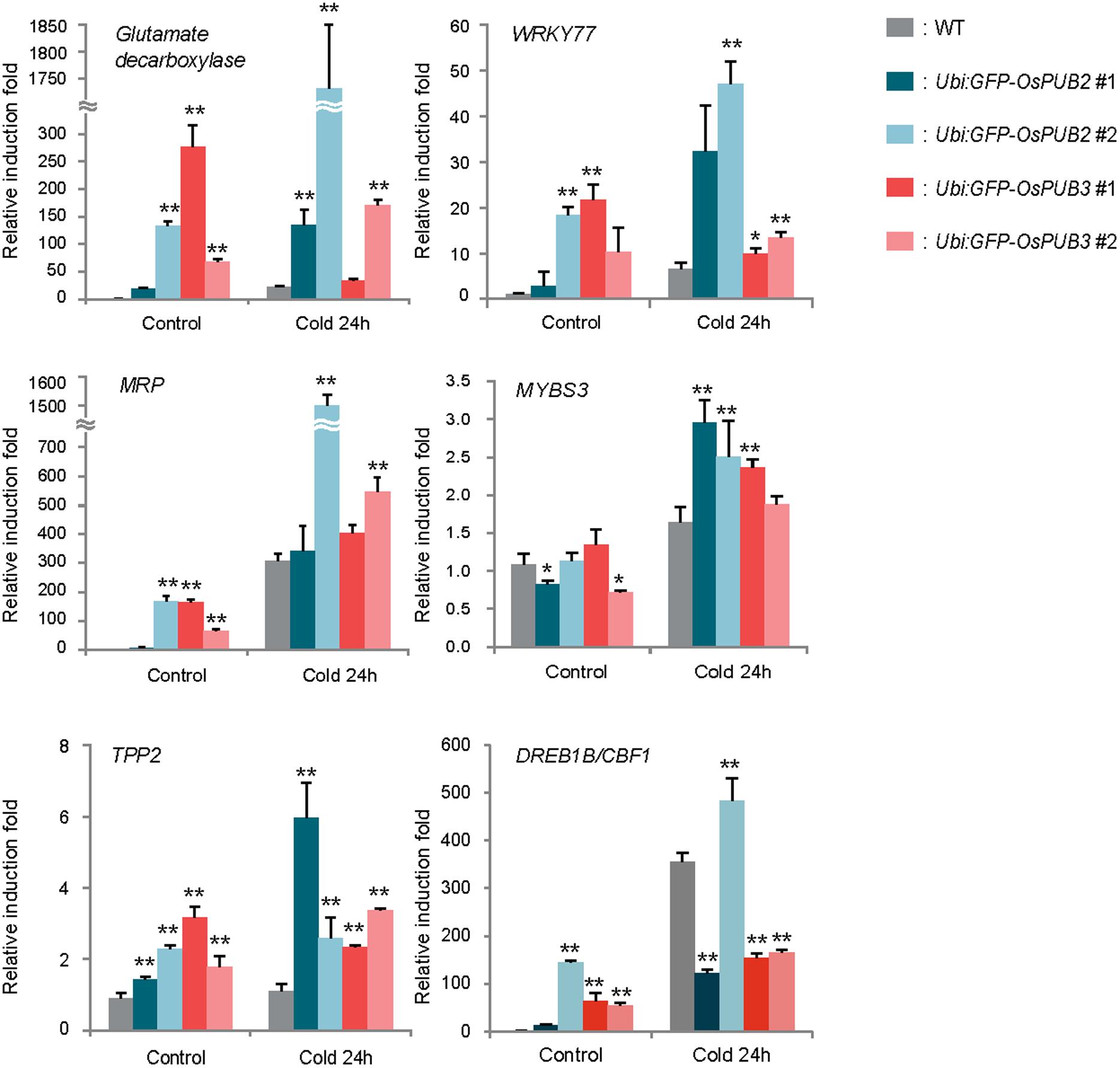
FIGURE 9. Expression analysis of cold stress-inducible genes in wild-type and OsPUB2/OsPUB3-overexpressing transgenic rice plants. Light-grown, 8-day-old wild-type and T4 Ubi:sGFP-OsPUB2 and Ubi:sGFP-OsPUB3 transgenic plants were subjected to cold (4°C) stress for 24 h. Induction patterns of five different cold-responsive genes were analyzed by real-time qRT-PCR by using the gene-specific primers listed in Supplementary Table S1. Data represent the fold inductions of GAD, WRKY77, MRP4, MYBS3, TPP2, and DREB1B/CBF1 in response to cold stress relative to those in the control treatment. The relative expression level of each gene was normalized to that of OsActin, an internal reference gene. Data indicate mean ± SE (∗P < 0.05, ∗∗P < 0.01, Student’s t-test) from three independent experiments.
Rice U-box E3 Ub ligases are implicated in biotic stress responses such as innate immunity and PAMP-triggered cell death (Zeng et al., 2004; Ishikawa et al., 2014; Liu et al., 2015; Wang et al., 2015). Recent genome-wide expression analysis showed that OsPUBs, which contain the ARM repeat motif, are induced by a broad spectrum of abiotic stress, suggesting their roles in the response to environmental stimuli in rice plants (Sharma et al., 2014). Nevertheless, cellular roles of rice OsPUBs in response to abiotic stress are only beginning to be understood. In this study, we conducted functional analyses of two homologous rice U-box E3 Ub ligases, OsPUB2 and OsPUB3 (Figure 1A; Supplementary Figure S1A). Although, the transcripts of OsPUB2 and OsPUB3 were detected in various tissues of developing rice plants (Figure 1B), their expression profiles were different: OsPUB2 was up-regulated by abiotic stresses, including low temperature, drought, and high salinity, whereas OsPUB3 was constitutively expressed (Figures 1C,D). Consistent with their close overall sequence identity (75%) and conserved functional motifs, such as the U-box domain and ARM repeats, OsPUB2 and OsPUB3 showed an in vitro E3 Ub ligase activity (Figure 2) and similar subcellular localization patterns in the EXPO-like punctate bodies (Figure 3). Considering these features of OsPUB2 and OsPUB3, we speculated that they might play a cellular role in an inter-connected manner. This view was further supported by the results that OsPUB2 and OsPUB3 form a hetero-dimeric complex in addition to homo-dimers in yeast cells and in vitro (Figure 4).
E3 Ub ligases can catalyze their own ubiquitination, thereby targeting themselves in a form of negative feedback loop (Ryan et al., 2006; de Bie and Ciechanover, 2011). Both OsPUB2 and OsPUB3 exhibited self-ubiquitination activities in vitro (Figure 2) and were rapidly degraded in the cell-free extracts prepared from developing rice seedlings; they had apparent half-lives of 150–160 min (Figure 5). This rapid degradation of OsPUB2/OsPUB3 was delayed when the crude extracts of cold-treated seedlings were used (apparent half-lives of 200–280 min). Thus, the stability of OsPUB2/OsPUB3 might be regulated by low temperature stress. Moreover, a hetero-dimeric form of OsPUB2/OsPUB3 was more stable than homo-dimers in a cell-free degradation system (apparent half-lives of 250–350 min). These results are in agreement with the notion that OsPUB2 and OsPUB3 function coordinately in response to cold stress. Notably, the stability of OsPUB3 was more evidently increased than that of OsPUB2 in the cold-treated cell-free extracts and when they formed a hetero-dimer (Figure 5). OsPUB3 was constitutively expressed in all the tissues examined in rice plants, whereas the basal level of OsPUB2 was low, but rapidly increased by cold stress (Figure 1). With these results in mind, we hypothesized that (1) under the normal growth conditions, constitutively expressed OsPUB3 was self-ubiquitinated and rapidly degraded; (2) in response to cold stress, OsPUB2 was induced and interacted with OsPUB3, which resulted in the formation of a more stable hetero-dimer as well as homo-dimers; and (3) homo- and hetero-dimeric complexes of OsPUB2/OsPUB3 play a positive role in the cold stress tolerance mechanism in rice plants (Figure 10). In this scenario, however, it remains to be clarified how the OsPUB2/OsPUB3 hetero-dimeric form is more resistant to self-ubiquitination and subsequent degradation. Possible involvement of EXPO-localized OsPUB2/OsPUB3 in the exocytosis process should also be investigated.
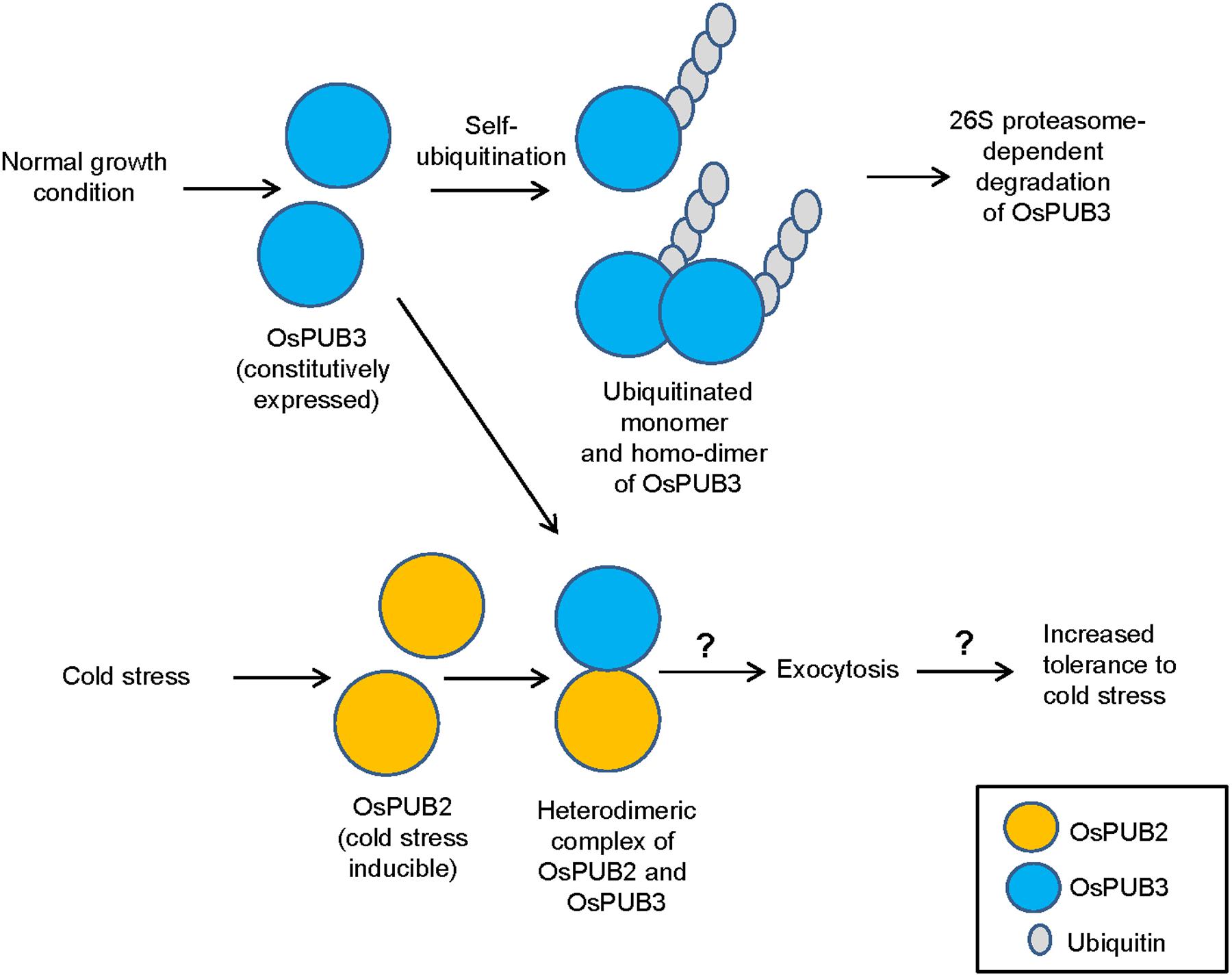
FIGURE 10. A simplified working model of EXPO-localized OsPUB2 and OsPUB3 in response to cold stress in rice plants. Under the normal growth conditions, constitutively expressed OsPUB3 is self-ubiquitinated and rapidly degraded. OsPUB2 is induced by cold stress and interacts with OsPUB3, which results in the formation of a more stable hetero-dimer. Hetero-dimeric complex of OsPUB2/OsPUB3 plays a positive role in the cold stress tolerance mechanism in rice plants. Possible involvement of OsPUB2/OsPUB3 hetero-dimeric form in the exocytosis process remains to be investigated.
Transgenic rice plants, in which OsPUB2 and OsPUB3 were over-expressed (Ubi:sGFP-OsPUB2 and Ubi:sGFP-OsPUB3) or suppressed (Ubi:RNAi-OsPUB2 and Ubi:RNAi-OsPUB3), were morphologically normal (Figure 7; Supplementary Figure S5). Their development and seed yields were indistinguishable from those of wild-type rice plants. Thus, OsPUB2 and OsPUB3 might not be involved in the normal cellular processes, but could specifically participate in the response to abiotic stress. Our results showed that OsPUB2/OsPUB3-overexpressors were markedly tolerant to low temperature treatment (4°C) with regard to survival rates (Figure 7), chlorophyll content and ion leakage (Figure 8), and expression levels of cold stress-inducible marker genes (Figure 9) compared to that in wild-type rice plants. These results strongly suggested that OsPUB2 and OsPUB3 are positive factors in response to cold stress. However, we failed to detect opposite phenotypes by using Ubi:RNAi-OsPUB2 and Ubi:RNAi-OsPUB3 knock-down transgenic plants (Supplementary Figures S5 and S6). These results could be reconciled by the interpretation that partial suppression of individual OsPUB2 and OsPUB3 was reciprocally complemented by OsPUB3 and OsPUB2, respectively, since they play a role in an inter-connected or coordinated fashion. In addition, the basal levels of OsPUB2 and OsPUB3 might be sufficiently high; hence, their partial suppression resulted in undetectable effects on the RNAi-mediated knock-down transgenic lines. We obtained the T-DNA inserted loss-of-function knock-out mutant line (PFG_1B-02809.L) for OsPUB2 from the rice T-DNA insertion sequence database1. However, this mutant was turned out to be an activation tagging line rather than a knock-out line. In addition, functional seeds of ospub3 knock-out mutant line (PFG_K-04006.L) were unavailable. Identification and characterization of additional knock-out mutant lines will be necessary.
The Exo70 (exocyst component of 70 kDa) protein is one of the core components of an evolutionarily conserved exocyst vesicle-tethering complex (Chong et al., 2010). OsPUB2 and OsPUB3 are co-localized with the Exo70E2 subunit to EXPO-like cytosolic punctae (Figure 3). In Arabidopsis, an EXPO-localized U-box E3 Ub ligase PUB22 ubiquitinates Exo70B2 to regulate the PAMP-triggered responses (Stegmann et al., 2012). Most recently, Seo et al. (2016) reported that Arabidopsis PUB18 negatively regulates ABA-mediated stomatal movements by ubiquitinating Exo70B1. At this moment, however, it is unknown how EXPO-localized OsPUB2 and OsPUB3 regulate cold stress response in rice plants (Figure 10). Thus, the target proteins that are ubiquitinated by OsPUB2/OsPUB3 need to be identified to decipher the detailed modes of action of these rice U-box E3 Ub ligases. In addition to EXPO-like structure, OsPUB2 was found in the nuclei (Figure 3). This raised the possibility that OsPUB2 plays an additional role, unlike OsPUB3, which is predominantly present in the EXPO. In this respect, it should be noted that OsPUB2, in contrast to OsPUB3, is induced by not only low temperature but also drought and high salinity (Figure 1C). Thus, as a next experiment, we intend to perform phenotypic analysis to elucidate whether OsPUB2-overexpressors are tolerant to drought and salt stress.
Our data indicate that two homologous U-box E3 Ub ligases OsPUB2 and OsPUB3 are the positive factors in response to low temperature stress in rice plants.
MB, LC, TO, Y-JJ, AL, and KP performed the experiments. MB, LC, BK, and WK analyzed the data. MB, LC, and WK designed the project and drafted the manuscript. WK supervised the project and complemented the writing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer CL and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
This work was supported by grants from the Next-Generation BioGreen 21 Program for Agriculture and Technology Development (Project No. PJ01113801 funded by the Rural Development Administration) and the National Research Foundation (Project No. 2014R1A2A2A01003891), Republic of Korea, to WK and from the Korea Institute of Science and Technology Information to BK.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2017.00016/full#supplementary-material
Andaya, V. C., and Tai, T. H. (2006). Fine mapping of the qCTS12 locus, a major QTL for seedling cold tolerance in rice. Theor. Appl. Genet. 113, 467–475. doi: 10.1007/s00122-006-0311-5
Bae, H., Kim, S. K., Cho, S. K., Kang, B. G., and Kim, W. T. (2011). Overexpression of OsRDCP1, a rice RING domain-containing E3 ubiquitin ligase, increased tolerance to drought stress in rice (Oryza sativa L.). Plant Sci. 180, 775–782. doi: 10.1016/j.plantsci.2011.02.008
Bae, H., and Kim, W. T. (2013). The N-terminal tetra-peptide (IPDE) short extension of the U-box motif in rice SPL11 E3 is essential for the interaction with E2 and ubiquitin-ligase activity. Biochem. Biophys. Res. Commun. 433, 266–271. doi: 10.1016/j.bbrc.2013.03.005
Bae, H., and Kim, W. T. (2014). Classification and interaction modes of 40 rice E2 ubiquitin-conjugating enzymes with 17 rice ARM-U-box E3 ubiquitin ligases. Biochem. Biophys. Res. Commun. 21, 575–580. doi: 10.1016/j.bbrc.2014.01.098
Berndsen, C. E., and Wolberger, C. (2014). New insights into ubiquitin E3 ligase mechanism. Nature Struct. Mol. Biol. 21, 301–307. doi: 10.1038/nsmb.2780
Byun, M. Y., and Kim, W. T. (2014). Suppression of OsRAD51D results in defects in reproductive development in rice (Oryza sativa L.). Plant J. 79, 256–269. doi: 10.1111/tpj.12558
Byun, M. Y., Lee, J., Cui, L. H., Kang, Y., Oh, T. K., Park, H., et al. (2015). Constitutive expression of DaCBF7, an Antarctic vascular plant, Deschampsia antarctica CBF homolog, resulted in improved cold tolerance in transgenic rice plants. Plant Sci. 236, 61–74. doi: 10.1016/j.plantsci.2015.03.020
Cabello, J. V., Lodeyro, A. F., and Zurbriggen, M. D. (2014). Novel perspectives for the engineering of abiotic stress tolerance in plants. Curr. Opin. Biotechnol. 26, 62–70. doi: 10.1016/j.copbio.2013.09.011
Chen, L., and Hellmann, H. (2013). Plant E3 ligases: flexible enzymes in a sessile world. Mol. Plant 6, 1388–1404. doi: 10.1093/mp/sst005
Chong, Y. T., Gidda, S. K., Sanford, C., Parkinson, J., Mullen, R. T., and Goring, D. R. (2010). Characterization of the Arabidopsis thaliana exocyst complex gene families by phylogenetic, expression profiling, and subcellular localization studies. New Phytol. 185, 401–419. doi: 10.1111/j.1469-8137.2009.03070.x
de Bie, P., and Ciechanover, A. (2011). Ubiquitination of E3 ligases: self-regulation of the ubiquitin system via proteolytic and non-proteolytic mechanisms. Cell Death. Differ. 18, 1393–1402. doi: 10.1038/cdd.2011.16
Ding, Y., Wang, J., Lai, J., Chan, V., Wang, X., Cai, Y., et al. (2014). Exo70E2 is essential for exocyst subunit recruitment and EXPO formation in both plants and animals. Mol. Biol. Cell 25, 412–426. doi: 10.1091/mbc.E13-10-0586
Duplan, V., and Rivas, S. (2014). E3 ubiquitin-ligases and their target proteins during the regulation of plant innate immunity. Front. Plant Sci. 5:42. doi: 10.3389/fpls.2014.00042
Guerra, D. D., and Callis, J. (2012). Ubiquitin on the move: the ubiquitin modification pathway plays diverse roles in the regulation of ER- and membrane-localized proteins. Plant Physiol. 160, 56–64. doi: 10.1104/pp.112.199869
Hatakeyama, S., Yada, M., Matsumoto, M., Ishida, N., and Nakayama, K. I. (2001). U box proteins as a new family of ubiquitin–protein ligases. J. Biol. Chem. 276, 33111–33120. doi: 10.1074/jbc.M102755200
Hu, X., Qian, Q., Xu, T., Zhang, Y., Dong, G., Gao, T., et al. (2013). The U-box E3 ubiquitin ligase TUD1 functions with a heterotrimeric G α subunit to regulate brassinosteroid - mediated growth in rice. PLoS Genet. 9:e1003391. doi: 10.1371/journal.pgen.1003391
Hur, Y. J., Yi, Y. B., Lee, J. H., Chung, Y. S., Jung, H. W., Yun, D. J., et al. (2014). Molecular cloning and characterization of OsUPS, a U-box containing E3 ligase gene that respond to phosphate starvation in rice (Oryza sativa). Mol. Biol. Rep. 39, 5883–5888. doi: 10.1007/s11033-011-1399-5
Ishikawa, K., Yamaguchi, K., Sakamoto, K., Yoshimura, S., Inoue, K., Tsuge, S., et al. (2014). Bacterial effector modulation of host E3 ligase activity suppresses PAMP-triggered immunity in rice. Nat. Commun. 5:5430. doi: 10.1038/ncomms6430
Jain, M., Nijhawan, A., Arora, R., Agarwal, P., Ray, S., Sharma, P., et al. (2007). F-box proteins in rice: genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143, 1467–1483. doi: 10.1104/pp.106.091900
Kim, J. H., and Kim, W. T. (2013). The Arabidopsis RING E3 ubiquitin ligase AtAIRP3/LOG2 participates in positive regulation of high-salt and drought stress. Plant Physiol. 162, 1733–1749. doi: 10.1104/pp.113.220103
Koegl, M., Hoppe, T., Schlenker, S., Ulrich, H. D., Mayer, T. U., and Jentsch, S. (1999). A novel ubiquitination factor, E4, is involved in multiubiquitin chain assembly. Cell 96, 635–644. doi: 10.1016/S0092-8674(00)80574
Lee, J. H., and Kim, W. T. (2011). Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol. Cells 31, 201–208. doi: 10.1007/s10059-011-0031-9
Lee, J. H., Yoon, H. J., Terzaghi, W., Martinez, C., Dai, M., Li, J., et al. (2010). DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22, 1716–1732. doi: 10.1105/tpc.109.073783
Lichtenthaler, H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148, 350–382. doi: 10.1016/0076-6879(87)48036-1
Liu, J., Park, C. H., He, F., Nagano, M., Wang, M., Bellizzi, M., et al. (2015). The RhoGAP SPIN6 associates with SPL11 and OsRac1 and negatively regulates programmed cell death and innate immunity in rice. PLoS Pathog. 11:e1004629. doi: 10.1371/journal.ppat.1004629
Lyzenga, W. J., and Stone, S. L. (2012). Abiotic stress tolerance mediated by protein ubiquitination. J. Exp. Bot. 63, 599–616. doi: 10.1093/jxb/err310
Ma, Q., Dai, X., Xu, Y., Guo, J., Liu, Y., Chen, N., et al. (2009). Enhanced tolerance to chilling stress in OsMYB3R-2 transgenic rice is mediated by alteration in cell cycle and ectopic expression of stress genes. Plant Physiol. 150, 244–256. doi: 10.1104/pp.108.133454
Min, H. J., Jung, Y. J., Kang, B. G., and Kim, W. T. (2016). CaPUB1, a hot pepper U-box E3 ubiquitin ligase, confers enhanced cold stress tolerance and decreased drought stress tolerance in transgenic rice (Oryza sativa L.). Mol. Cells 39, 250–257. doi: 10.14348/molcells.2016.2290
Mudgil, Y., Shiu, S. H., Stone, S. L., Salt, J. N., and Goring, D. R. (2004). A large complement of the predicted Arabidopsis ARM repeat proteins are members of the U-Box E3 ubiquitin ligase family. Plant Physiol. 134, 59–66. doi: 10.1104/pp.103.029553
Mukhopadhyay, A., Vij, S., and Tyagi, A. K. (2004). Overexpression of a zinc-finger protein gene from rice confers tolerance to cold, dehydration, and salt stress in transgenic tobacco. Proc. Natl. Acad. Sci. U.S.A. 101, 6309–6314. doi: 10.1073/pnas.0401572101
Nikolay, R., Wiederkehr, T., Rist, W., Kramer, G., Mayer, M., and Bukau, B. (2004). Dimerization of the human E3 ligase CHIP via a coiled-coil domain is essential for its activity. J. Biol. Chem. 279, 2673–2678. doi: 10.1074/jbc.M311112200
Ohi, M. D., Vander Kooi, C. W., Rosenberg, J. A., Chazin, W. J., and Gould, K. L. (2003). Structural insights into the U-box, a domain associated with multi-ubiquitination. Nat. Struct. Mol. Biol. 10, 250–255. doi: 10.1038/nsb906
Park, J. J., Yi, J., Yoon, J., Cho, L. H., Ping, J., Jeong, H. J., et al. (2011). OsPUB15, an E3 ubiquitin ligase, functions to reduce cellular oxidative stress during seedling establishment. Plant J. 65, 194–205. doi: 10.1111/j.1365-313X.2010.04416.x
Ren, Y., Tang, Y., Xie, K., Li, W., Ye, S., Gao, F., et al. (2014). Mutation of a U-box E3 ubiquitin ligase results in brassinosteroid insensitivity in rice. Mol. Breed. 34, 115–125. doi: 10.1007/s11032-014-0021-7
Ryan, P. E., Davies, G. C., Nau, M. M., and Lipkowitz, S. (2006). Regulating the regulator: negative regulation of CBL ubiquitin ligases. Trends Biochem. Sci. 31, 79–88. doi: 10.1016/j.tibs.2005.12.004
Sadanandom, A., Bailey, M., Ewan, R., Lee, J., and Nelis, S. (2012). The ubiquitin-proteasome system: central modifier of plant signalling. New Phytol. 196, 13–28. doi: 10.1111/j.1469-8137.2012.04266
Santner, A., and Estelle, M. (2010). The ubiquitin-proteasome system regulates plant hormone signaling. Plant J. 61, 1029–1040. doi: 10.1111/j.1365-313X.2010.04112.x
Seo, D. H., Ahn, M. Y., Park, K. Y., Kim, E. Y., and Kim, W. T. (2016). The N-terminal UND motif of the Arabidopsis U-box E3 ligase PUB18 is critical for the negative regulation of ABA-mediated stomatal movement and determines its ubiquitination specificity for exocyst subunit Exo70B1. Plant Cell doi: 10.1105/tpc.16.00347
Seo, D. H., Ryu, M. Y., Jammes, F., Hwang, J. H., Turek, M., Kang, B. G., et al. (2012). Roles of four Arabidopsis U-box E3 ubiquitin ligases in negative regulation of abscisic acid-mediated drought stress responses. Plant Physiol. 160, 556–568. doi: 10.1104/pp.112.202143
Sharma, B., Joshi, D., Yadav, P. K., Gupta, A. K., and Bhatt, T. K. (2016). Role of ubiquitin-mediated degradation system in plant biology. Front. Plant Sci. 7:806. doi: 10.3389/fpls.2016.00806
Sharma, M., Singh, A., Shankar, A., Pandey, A., Baranwal, V., Kapoor, S., et al. (2014). Comprehensive expression analysis of rice Armadillo gene family during abiotic stress and development. DNA Res. 21, 267–283. doi: 10.1093/dnares/dst056
Stegmann, M., Anderson, R. G., Ichimura, K., Pecenkova, T., Reuter, P., Žársky, V., et al. (2012). The ubiquitin ligase PUB22 targets a subunit of the exocyst complex required for PAMP-triggered responses in Arabidopsis. Plant Cell 24, 4703–4716. doi: 10.1105/tpc.112.104463
Stone, S. L. (2014). The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front. Plant Sci. 5:135. doi: 10.3389/fpls.2014.00135
Su, C., Wang, Y., Hsieh, T., Lu, C., Tseng, T., and Yu, S. (2010). A novel MYBS3-dependent pathway confers cold tolerance in rice. Plant Physiol. 153, 145–158. doi: 10.1104/pp.110.153015
Suzuki, K., Nagasuga, K., and Okada, M. (2008). The chilling injury induced by high root temperature in the leaves of rice seedlings. Plant Cell Physiol. 49, 433–442. doi: 10.1093/pcp/pcn020
Trujillo, M., and Shirasu, K. (2010). Ubiquitination in plant immunity. Curr. Opin. Plant Biol. 13, 402–408. doi: 10.1016/j.pbi.2010.04.002
Vierstra, R. D. (2009). The ubiquitin-26s proteasome system at the nexus of plant biology. Nat. Rev. Mol. Cell Biol. 10, 385–397. doi: 10.1038/nrm2688
Wang, J., Ding, Y., Wang, J., Hillmer, S., Miao, Y., Lo, S., et al. (2010). EXPO, an exocyst-positive organelle distinct from multivesicular endosomes and autophagosomes, mediates cytosol to cell wall exocytosis in Arabidopsis and tobacco cells. Plant Cell 22, 4009–4030. doi: 10.1105/tpc.110.080697
Wang, J., Qu, B., Dou, S., Li, L., Yin, D., Pang, Z., et al. (2015). The E3 ligase OsPUB15 interacts with the receptor-like kinase PID2 and regulates plant cell death and innate immunity. BMC Plant Biol. 15:49. doi: 10.1186/s12870-015-0442-4
Yee, D., and Goring, D. R. (2009). The diversity of plant U-box E3 ubiquitin ligases: from upstream activators to downstream target substrates. J. Exp. Bot. 60, 1109–1121. doi: 10.1093/jxb/ern369
Yu, F., Wu, Y., and Xie, Q. (2016). Ubiquitin-proteasome system in ABA signaling: from perception to action. Mol. Plant 9, 21–33. doi: 10.1016/j.molp.2015.09.015
Zeng, L. R., Park, C. H., Venu, R. C., Gough, J., and Wang, G. L. (2008). Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol. Plant 1, 800–815. doi: 10.1093/mp/ssn044
Zeng, L. R., Qu, S., Bordeos, A., Yang, C., Baraoidan, M., Yan, H., et al. (2004). Spotted leaf11, a negative regulator of plant cell death and defense, encodes a U-box/armadillo repeat protein endowed with E3 ubiquitin ligase activity. Plant Cell 16, 2795–2808. doi: 10.1105/tpc.104.025171
Keywords: cold stress, dimerization, E3 Ub ligase, rice (Oryza sativa), U-box motif
Citation: Byun MY, Cui LH, Oh TK, Jung Y - J, Lee A, Park KY, Kang BG and Kim WT (2017) Homologous U-box E3 Ubiquitin Ligases OsPUB2 and OsPUB3 Are Involved in the Positive Regulation of Low Temperature Stress Response in Rice (Oryza sativa L.). Front. Plant Sci. 8:16. doi: 10.3389/fpls.2017.00016
Received: 17 October 2016; Accepted: 04 January 2017;
Published: 20 January 2017.
Edited by:
Sung Chul Lee, Chung-Ang University, South KoreaReviewed by:
Cheol Soo Kim, Chonnam National University, South KoreaCopyright © 2017 Byun, Cui, Oh, Jung, Lee, Park, Kang and Kim. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Woo Taek Kim, d3RraW1AeW9uc2VpLmFjLmty
†These authors are co-first authors.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.