- 1National Botanical Research Institute (CSIR), Lucknow, India
- 2Academy of Scientific and Innovative Research, New Delhi, India
Pearl millet is one of the most important small-grained C4 Panicoid crops with a large genome size (∼2352 Mb), short life cycle and outbreeding nature. It is highly resilient to areas with scanty rain and high temperature. Pearl millet is a nutritionally superior staple crop for people inhabiting hot, drought-prone arid and semi-arid regions of South Asia and Africa where it is widely grown and used for food, hay, silage, bird feed, building material, and fuel. Having excellent nutrient composition and exceptional buffering capacity against variable climatic conditions and pathogen attack makes pearl millet a wonderful model crop for stress tolerance studies. Pearl millet germplasm show a large range of genotypic and phenotypic variations including tolerance to abiotic and biotic stresses. Conventional breeding for enhancing abiotic and biotic stress resistance in pearl millet have met with considerable success, however, in last few years various novel approaches including functional genomics and molecular breeding have been attempted in this crop for augmenting yield under adverse environmental conditions, and there is still a lot of scope for further improvement using genomic tools. Discovery and use of various DNA-based markers such as EST-SSRs, DArT, CISP, and SSCP-SNP in pearl millet not only help in determining population structure and genetic diversity but also prove to be important for developing strategies for crop improvement at a faster rate and greater precision. Molecular marker-based genetic linkage maps and identification of genomic regions determining yield under abiotic stresses particularly terminal drought have paved way for marker-assisted selection and breeding of pearl millet cultivars. Reference collections and marker-assisted backcrossing have also been used to improve biotic stress resistance in pearl millet specifically to downy mildew. Whole genome sequencing of pearl millet genome will give new insights for processing of functional genes and assist in crop improvement programs through molecular breeding approaches. This review thus summarizes the exploration of pearl millet genetic and genomic resources for improving abiotic and biotic stress resistance and development of cultivars superior in stress tolerance.
Introduction
Pearl millet [Pennisetum glaucum (L.) R. Br.] (2n = 2x = 14) is an important tropical C4 small-grained cereal crop that belongs to family Poaceae and sub family Panicoideae. It is a highly tillering, cross-pollinating millet crop with a short life cycle and a large genome size (∼2352 Mbp) (Bennett et al., 2000). It is usually grown in marginal environments of arid and semi-arid regions of Sub-Saharan Africa and the Indian subcontinent characterized by scanty and erratic rainfall, poor soil conditions and high temperature where staple cereals such as rice, wheat, maize, and even sorghum are likely to fail (Vadez et al., 2012). It is the sixth most economically important and promising cereal crop for providing food security to almost 90 million poor people inhabiting across the high temperate regions of Africa and Asia1. It is a dry land crop and can withstand poor environmental conditions such as low moisture, high temperature, and nutrient deficient soil where other cereal crops could not hold up their growth. It is cultivated principally for grain production but it also cultivated by marginal farmers for hay, bird feed, fuel, and forage materials. Globally, it is cultivated on approximately 31 million ha of land and contributes around 50% of total global millet production2. In India, it is cultivated in an area of approximately 7.8 million ha with an average of about 9.25 million tons of grain production3. However, in the year 2013–2014, India was the largest producer of pearl millet (10 mt) followed by Nigeria (5 mt) (Malik, 2015). Pearl millet grains are high in nutrient composition and are considered an inexpensive source of energy as compared to staple cereals such as wheat, rice and maize in terms of micronutrients (Zinc and Iron) (Kumar et al., 2016), protein content and amino acid composition for the resource-poor farmers (Kanatti et al., 2014). In fact, pearl millet is superior in protein content, quality, protein energy ratio, and metabolizable energy levels as compared to sorghum (Vadez et al., 2012). Thus millions of people depend on this crop for their dietary needs and livelihood2. Furthermore, pearl millet has also been long known as a potential biofuel crop (Wu et al., 2005; Lata, 2015).
Pearl millet accessions exhibit a wide range of phenotypic and genotypic variations for various agronomic and stress tolerance traits owing to its cultivation in varied agro-climatic conditions and soil types across the world. Despite having a wide and diverse germplasm collection and genetic resources, pearl millet was until recently considered an orphan crop and only a narrow range of its germplasm could be exploited for improving its valuable agronomic traits, stress tolerance, and productivity (Passot et al., 2016). In the past few years most of the allele mining studies in this crop for improvement of various agronomic traits such as yield and stover quality as well as for abiotic and biotic stress resistance such as drought tolerance (DT) and downy mildew resistance have been achieved by generating and using bi-parental mapping populations (Vengadessan et al., 2013). Genetic maps based on molecular markers and identified quantitative trait loci (QTLs) have been used for developmental and breeding programs in pearl millet to enhance its productivity under the present scenario of global climatic change. Molecular markers like restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), expressed sequence – simple sequence repeats (EST-SSRs), conserved intron spanning primer (CISP), single strand conformational polymorphism-single nucleotide polymorphism (SSCP-SNP) and diversity array technology (DArT) are dominant and proven to be useful in the evaluation of QTLs associated with various traits (Jones et al., 1995, 2002; Yadav R.S. et al., 2003; Bidinger et al., 2007), terminal drought stress tolerance and adaptation (Yadav R.S. et al., 2004; Kholova et al., 2012) and disease resistance (Hash et al., 2006a; Jogaiah et al., 2014). Along with genetic markers, core or mini core and reference collections are also helpful to identify new sources of abiotic and biotic stress resistance in pearl millet.
Though, conventional linkage mapping has helped in identifying different quantitative traits associated with stress tolerance in pearl millet but these are limited by the resolution provided by two parent-derived mapping populations (Dwivedi et al., 2012). Association mapping, also known as linkage disequilibrium (LD) mapping opens a new platform for allele mining with the help of ancestral recombination events in natural populations or germplasm collections to make marker-phenotype associations. It has the advantage over QTLs associated linkage mapping in terms of less laborious, time efficient and generation of 1000s of recombinants with large and diverse gene pool. It has been widely used in many crops including maize, barley, sorghum, rice, and common wheat to detect important markers or genes associated with abiotic and biotic stress resistance (Rajaram et al., 2010, 2013; Senthilvel et al., 2010; Sehgal et al., 2012, 2015; Jogaiah et al., 2014). Recently a pearl millet inbred germplasm association panel (PMiGAP) of about 346 lines was generated that represented a collection of approximately 1000 diverse cultivars, landraces and parents of mapping population from various regions of Africa and Asia, out of which 250 lines were used for association mapping of DT traits (Sehgal et al., 2015). This PMiGAP is thought to provide new insights for fine mapping of QTLs and allele mining of favorable genes for important agronomic traits.
Apart from genomics tools, various advanced technologies such as high throughput sequencing, insertional mutagenesis, targeted induced local lesion in genomes (TILLING), gene silencing, genome editing, and transgenics may also play important role in improving our understanding of complex biological mechanisms of stress regulation, adaptation, and resistance. Huge datasets obtained from various omics technologies could be useful for identifying candidate genes/proteins that can be incorporated in crop improvement programs for developing cultivars superior in stress tolerance. Until now there have been limited reports on identification and exploitation of potential genes for abiotic tolerance or biotic stress resistance in pearl millet (Girgi et al., 2006; Latha et al., 2006; Mishra et al., 2007; Agarwal et al., 2008). It is anticipated that the ongoing pearl millet genome sequencing project will be very useful for exploring its genetic and genomic resources for crop improvement programs directed toward improving important agronomic traits such as yield and stress resistance. This review thus primarily focuses on the sources of tolerance to abiotic and biotic stresses, agronomic traits of importance, as well as efforts to identify new sources of variation in pearl millet germplasm collections and to promote these resources for genomics/marker- assisted breeding program(s) for product development related to food, feed, and bioenergy traits in pearl millet.
Morphology, Origin, and Domestication
Pearl millet is a robust erect highly cross-pollinated annual C4 grass with slender culms and profuse root system and the plant may reach up to a height of 4 m. However, improved open-pollinated varieties (OPVs) and hybrids are reportedly shorter1 (Manga and Kumar, 2011). Its leaves are minutely serrated alternate, simple, flat, green with linear blades that can be up to 1.5 m long and 8 cm wide while its inflorescence is a 12–30 cm long panicle with a circumference of 7–9 cm that bears the seeds4. Involucres composed of bristles each of which enclosing 1–9 spikelets is one of the most characteristic features of Pennisetum glaucum (de Wet et al., 1992). There could be around 500–3000 spikelets on a panicle depending upon the variety. Pearl millet seeds are small (0.5–3 mm long and each could weigh between 0.003 and 0.02 g), wedge-shaped to spherical and can be of various colors ranging from white or yellow, brown or even purple5. Seeds could attain physiological maturity within 27–30 days after 50% flowering6. It is well-adapted to sandy or light loam well-drained soils and has a good DT capacity. It could be cultivated from late May to September and high temperature is a prerequisite for its optimum growth5. Pearl millet could broadly be divided into two morphotypes namely, tall and dwarf. Dwarf types are usually leafier and are commonly used for grazing.
Pearl millet is thought to be originated in Dhar Tichitt, a Saharan site in Mauritania of West Africa about 3500 B.C. ago (Amblard and Pernès, 1989). The findings of an archeologist Birimi, confirms the Sahara and Sahel hypothesis of origin and their widespread distribution and cultivation of wild and cultivated pearl millet across sub-Saharan Africa (Amblard and Pernès, 1989; D’Andrea and Casey, 2002). de Wet et al. (1992) reported pearl millet to be a native along the southern fringes of Sahara with a distribution across semi-arid tropics of Africa, and also suggested its introduction as a cereal crop into the South Asian continent. It has been introduced in India since 2000 B.C., in the USA in the 1850s and in Brazil in the 1960s (Garí, 2002; Oumar et al., 2008; Rai et al., 2013). Across different regions of the world, pearl millet is called by various common names including geohatsii. mahangu, sanio, babala, etc. in different areas of Africa; milheto kaustubh in Brazil; bajri, sajje, kambu, bajra in different states of India; miglio in Italian, grano in Span, Type de graine in France, etc. (Malik, 2015). With approximately 140 species, Pennisetum is the largest genera in the Paniaceae tribe with diverse ploidy and reproductive behavior7 (Table 1). Brunken et al. (1977) categorized Pennisetum into two reproductively isolated species, namely P. purpureum Schumach (Napier grass) (2n = 4x = 28) and P. americanum (syn. P. glaucum) (2n = 2x = 14). Napier grass is perennial and is found across wet tropics of the world while P. americanum is annual and is a native of semi-arid tropics of Africa and India. P. americanum has been classified into three distinct morphological units or subspecies (i) wild plants Subsp., violaceum (monodii) – that neither show any characteristic of a cultivated plant nor are dependent upon man for their survival; (ii) weedy plants (Subsp., stenostachyum) – that exhibit intermediate morphology and are exclusively associated with pearl millet cultivation and are also indirectly dependent upon man; and (iii) cultivated plants (Subsp., glaucum) – that show distinctly stalked and involucres persistent at maturity, and are directly dependent upon agricultural activities taken up by man for their survival. Together these three subspecies displayed three distinct adaptive strategies in response to the selective pressures thrust upon them during domestication. P. glaucum is the only cultivated species used for seed purpose while P. purpureum and its hybrids with pearl millet are usually cultivated for fodder in different parts of the world7.
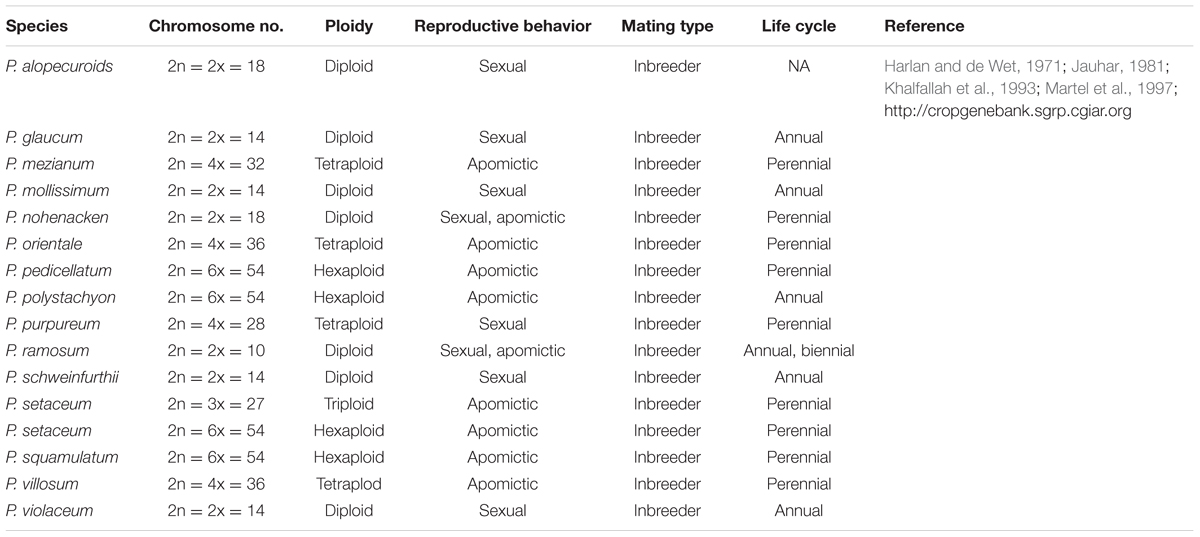
TABLE 1. Details of genome organization and other characteristics of significant cultivated and wild Pennisetum species.
Nutritional Value and Medicinal Uses
Pearl millet has an excellent nutritional composition and is a rich source of energy (361 Kcal/100 g) as compared to staple cereals like rice (345 Kcal/100 g) and wheat (346 Kcal/100 g) (Malik, 2015). Further with regard to nutritional quality, it is generally equivalent to maize and superior to sorghum in terms of protein content, quality, efficiency ratio, and metabolizable energy (Vadez et al., 2012). Although it is deficient in essential amino acids, it contains 35% more lysine as compared to sorghum (Rooney and McDonough, 1987). Additionally, it also does not contain condensed polyphenols which are a major cause of decreased digestibility, for example, tannins in sorghum. Pearl millet grains contain 5–6% oil and are also rich in important micronutrients like iron and zinc (Jambunathan and Subramanian, 1988; Malik, 2015). Hence, pearl millet significantly contributes toward protein, iron, and zinc uptake as well as serve as the cheapest source of energy to low-income group consumers of semi-arid tropics including India.
Furthermore, pearl millet has plenty of health-promoting attributes owing to its nutritional composition and hence has an immense potential toward medicinal uses. As for example, its high fiber content can make it a potential component in the diets of patients suffering from constipation, obesity, and gallstones (Malik, 2015). Also since pearl millet grains are gluten free and have low glycemic index, they can be very beneficial for persons with celiac disease and/or diabetes. Further due to its anti- or hypo-allergic properties, it can be safely incorporated in the diets of pregnant women, infants, lactating mothers, elderly and convalescents. Its flour can be conveniently used for making chapattis, porridge or can be used in the boiled or roasted form or even as weaning mixtures. Pearl millet grains are also locally brewed in Africa and Asia to produce alcoholic or non-alcoholic beverages (Dwivedi et al., 2012). Thus pearl millet grains have immense medicinal value and should be aggressively promoted by dieticians and nutritionists so that a large section of society could be benefited.
Germplasm Collections and Diversity Assessment
Ex situ seed storage is one of the most extensively used methods for conserving pearl millet genetic resources (Dwivedi et al., 2012). Germplasm collections help in maintaining the genetic resources so that they can be used for crop development and improvement programs globally. Pearl millet germplasm have been preserved in various national and international gene banks. India with 11243 accessions has the largest collection of pearl millet germplasm followed by Brazil (7225) and Canada (3764) (Dwivedi et al., 2012). The initiative for the conservation of pearl millet genetic resources in India was taken up by International Crop Research Institute for Semi-Arid Tropics (ICRISAT), Patancheru in collaboration with different national and international organizations including several national organizations such as National Bureau of Plant Genetic Resources (NBPGR), All India Coordinated Pearl Millet Improvement Project (AICPMIP), National Agricultural Research Systems (NARS), Agricultural Universities, and other ICAR Institutes etc. About 65 organizations had contributed approximately 10,764 accessions of pearl millet in different disciplines of ICRISAT, in past few years. Major international contributors for pearl millet germplasm are Institute Francais de Recherche Scientifique pour le development en Cooperation (ORSTOM), France (2,178); Rockefeller Foundation, New Delhi, India (2,022); and International Bureau for Plant Genetic Resources (IBPGR), Rome, Italy (974) at ICRISAT. ICRISAT has a total of 21,594 pearl millet accessions from 51 countries, thus becoming the largest center for pearl millet germplasm collection in the world (Upadhyaya et al., 2007; Upadhyaya et al., 2015) (Table 2). ICRISAT also maintains some of the pearl millet wild relatives in an ex situ field gene bank at its Patancheru campus (Dwivedi et al., 2012). Conservation of wild relatives is also very important as they could contribute beneficial traits to the cultivated gene pool, for example, a few accessions of P. glaucum subsp. monodii namely PS# 202, 637, 639, and 727 are good sources of resistance to a cereal parasitic weed, Striga hermonthica, in sub-Saharan West Africa. PS 202 is also a source of resistance to one of the most devastating fungal diseases of pearl millet, downy mildew (Wilson et al., 2004). A few wild accessions could serve as a source for rust resistance (Hanna and Dujardin, 1985) or also as alternative systems for cytoplasmic male sterility (Hanna, 1989). Thus a more detailed exploration for useful traits from pearl millet wild relatives is required to be carried out.
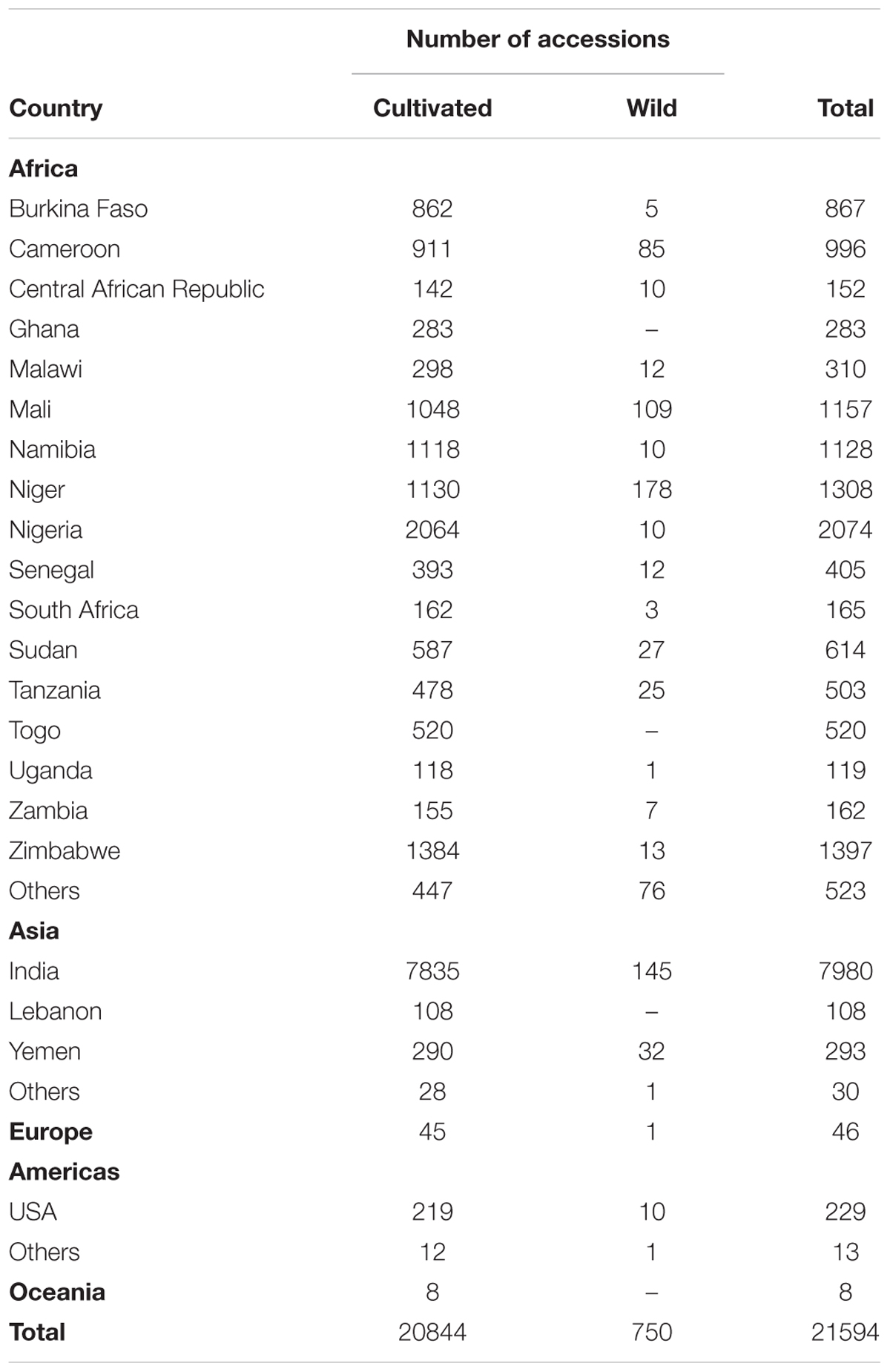
TABLE 2. Geographical distribution of pearl millet germplasm accessions assembled at ICRISAT gene bank (as on 1 January 2007; Upadhyaya et al., 2015).
Descriptor list has also been developed and used to characterize pearl millet (IBPGR and ICRISAT, 1993) germplasm for various morphological and agronomic traits (Dwivedi et al., 2012). The information so generated along with passport data has been used to assess patterns of diversity in germplasm collection, as a result of which several interesting information regarding their utility in pearl millet genetics and breeding could be obtained. As for example, Stich et al. (2010) reported patterns of diversity in flowering, photoperiod response, panicle length and population structure differentiation in 145 inbreds derived from 122 Western and Central African landraces. These landraces also exhibited exceptional buffering capacity against variable environmental conditions while the landraces from Cameroon, Togo, and Ghana were found to be good sources of earliness and bigger seeds. On the other hand, landraces from Yemen were found to be potential sources of variation for early maturity, short stature, large seeds, and cold tolerance. Similarly, Yadav and Bidinger (2008) evaluated 169 landraces from India for their grain and stover yield and discerned significant differences among them for the examined parameters such as early flowering, profuse tillering, more panicles and larger seeds. In fact, several landraces also outperformed controls in terms of both grain and stover yield and were thus considered as potential resources for developing dual-purpose hybrids adapted for arid climatic conditions. Upadhyaya et al. (2007) evaluated 20,844 accessions from 51 countries for 23 traits including number of flowering days, plant height, tiller number, 100 seed-weight, panicle length and shape, seed shape and color, and reported significant variations among the accessions studied. Therefore, germplasm collection (both cultivated and wild) and their characterization using both conventional and advanced techniques such as TILLING need to be done for expansion of its cultivated gene pool.
Developing Core, Mini-Core, and Reference Sets
Pearl millet exhibits a vast range of genetic diversity in its global germplasm collection. Core and mini-core collections of pearl millet germplasm could serve as wonderful cost effective resources for allele mining and identification of genotypic variants for resistance to biotic and abiotic stresses as well as other important agronomic traits. A core collection comprises of about 10% of the entire germplasm collection while a mini core is comprised of ∼1% of the total collection. Upadhyaya et al. (2009) reported a core collection of 2094 pearl millet accessions which was huge in itself to be evaluated by breeders for crop improvement. Hence, a mini-core collection of 238 pearl millet accessions representing all accessions of the core collection was constituted using data on 10 qualitative and eight quantitative traits (Upadhyaya et al., 2011). This mini-core collection with its greatly reduced size could be effectively utilized for accurate evaluation of desirable traits including tolerance to abiotic and biotic stresses as well as for mapping with molecular markers and identification of trait-specific germplasm and discovery of candidate genes (Upadhyaya et al., 2011; Manga, 2015). Other than the core and mini- core collections, a genotype based reference set comprised of 300 accessions is also available in pearl millet (Upadhyaya et al., 2011). A global composite collection of accessions in pearl millet based on SSRs and high-throughput assays has also been developed at ICRISAT prior to the formation of reference sets for assessing population structure and diversity (Dwivedi et al., 2012). This reference set reportedly capture 87–95% of the allelic diversity of the composite collections (ICRISAT, 2008/2009). However, a persistent research needs to be directed in order to maintain up-to-date pearl millet core and mini- core collections.
Identification of Pearl Millet Germplasm for Stress Tolerance
Tolerance to Abiotic Stresses
Abiotic stresses adversely affect all crops by negatively impacting their growth and yield potential, and pearl millet is also no exception to it. In fact, it is also vulnerable to various abiotic stresses like other crops despite the fact it is generally considered to be well-adapted to drought, salinity, high temperature, lodging, and poor soil conditions (Zegada-Lizarazu and Iijima, 2005). Therefore, it is very imperative that identification and utilization of genetic variations for abiotic stress tolerance in pearl millet could be helpful in enhancing its adaptation to various stresses. Considering a large collection of pearl millet germplasm (both cultivated and landraces) in various national and international gene banks, there is a huge opportunity for researchers to identify and characterize the hidden DNA stretches responsible for abiotic stress tolerance which would eventually be beneficial in speeding up the crop improvement programs. Further accurate phenotyping along with improved understanding of the physiological and molecular basis of abiotic stress tolerance in this crop will be helpful in identifying and utilizing new potential candidate genes for developing varieties with superior stress tolerance. Among various millets, pearl millet has received comparatively more attention toward the identification of sources of tolerance to drought, salinity, and high temperature (Dwivedi et al., 2012). ICRISAT even has developed phenotypic screens for various abiotic stresses for identifying tolerant germplasm (Krishnamurthy et al., 2007; ICRISAT, 2008/2009). Identification and exploitation of pearl millet germplasm for abiotic stress tolerance is discussed in following sections.
Drought
Pearl millet is a DT crop and substantial work has been done to understand its response and adaptation to water deficit conditions at different developmental growth stages. Further impact of low water stress conditions depends on growth stages of the crop. Water stress during germination stage or seedling emergence stages cause seedling death resulting in poor crop establishment (Farooq et al., 2009; Lata et al., 2015). Severe moisture stress at seedling stage is the major cause of low yield of pearl millet in the semi-arid regions (Carberry et al., 1985; Soman and Peacock, 1985; Soman et al., 1987). It has been found that after the seedling establishment, the impact of the drought stress has little effect on pearl millet grain yield (Lahiri and Kumar, 1966). Effect of drought stress on the germination of seedling depends on the availability of water and it has been shown that impact of low water stress is closely related to leaf formation and secondary root development (Gregory, 1983). The impact of moisture stress has also been analyzed using varying concentrations of polyethylene glycol (PEG) 6000 and found that it significantly affected various seedling parameters such as germination percentage, root and shoot length, and root/shoot ratio (Govindaraj et al., 2010). Use of PEG 6000 for screening pearl millet germplasm at germination and early seedling growth stages was also established as a simple, rapid and cost-effective method. The study revealed TNBH 0538, TNBH 0642, and ICMV-221 genotypes as better performers under all four imposed moisture stress regime while PT6034 was found to be least resistant to moisture stress. Drought stress at vegetative phase exhibit little or almost negligible reduction in crop growth and yield owing to its asynchronous tillering and rapid growth rate which allow it to recover quickly (Bidinger et al., 1987; Mahalakshmi et al., 1987). Further drought stress during the vegetative phase delays the flowering time of the main shoot. This phenological plasticity gives it a chance to escape from the most sensitive flowering phase until the stress has been relieved (Henson and Mahalakshmi, 1985). However, post-flowering drought stress or terminal drought stress impose the most drastic impact on pearl millet grain and stover yield as well as on yield stability (Mahalakshmi et al., 1987; Winkel et al., 1997; Bidinger and Hash, 2004; Kholová and Vadez, 2013). It has been reported that early flowering pearl millet cultivars having few but effective basal tillers, low biomass, and high harvest index (including panicle harvest index) can effectively overcome the effects of terminal drought stress over other genotypes (Yadav O.P. et al., 2003; Bidinger et al., 2005). Further in order to understand whether control of water loss under non-limiting water conditions is implicated in terminal DT of pearl millet, a study was carried out involving two contrasting pair of parents of mapping population (PRLT2/88-33 X H77/833-2 and 863B-P2 X ICMB 841-P3) and near-isogenic lines (NILs) generated from introgression of a major DT-QTL from PRLT2/88-33 (donor) into H77/833-2 (NILs-QTL) under water deficit conditions. Tolerant varieties and NILs-QTL showed low transpiration rate as compared to sensitive genotypes in well-watered conditions (Kholova et al., 2010a). Kholova et al. (2010b) established that pearl millet genotypes carrying a DT-QTL for terminal DT, including tolerant and NIL-QTLs, have higher ABA levels and lower transpiration rate (Tr) under well-watered conditions as compared to sensitive genotypes at all vapor pressure deficit (VPD) levels, ultimately contributing to absolute water saving during grain-filling. Furthermore, F7 recombinant inbred lines (RILs) from a cross between 863B X ICMB 841, were used to identify four QTLs that contributed to increased Tr under high VPD conditions, out of which the major QTL was mapped on LG 6 (Aparna et al., 2015). This study also provided an additional evidence for a link between DT and water savings from lower Tr under high VPD.
Across different regions of West Africa, Ghana, and North Western India, a number of landraces have been identified which could serve as a rich source of diversity for abiotic stress tolerance in pearl millet (Table 3). These landraces because of their high grain and stover yield as compared to the conventionally bred lines are preferentially cultivated in arid zones of northwestern India (Gujarat, Rajasthan, and Haryana) where drought is a common phenomenon (Bidinger et al., 2009). As for example, CZP9802, the first open pollen pearl millet variety derived from the landraces of Rajasthan is not only highly adapted to drought but is also high yielding (14–33% higher grain yield and 18–36% higher stover yield) as compared to the controls Pusa 266 and ICTP 8203 (Yadav, 2004; Dwivedi et al., 2012). CZP9802 is endowed with the property to escape terminal drought stress owing to its exceptional attributes of flowering within 48 days and maturity in 75 days making it a suitable cultivar for arid zones of India (Yadav O.P. et al., 2004). Further Okashana 1, another early maturing variety is very popular in Namibia (Daisuke, 2005). Similarly, a relatively photoperiod insensitive early maturing West African landrace Iniadi with compact, conical panicles and bold grains has significantly contributed toward the development of several pearl millet cultivars worldwide including ICMV 88904 (released as ICMV 221) (Andrews and Kumar, 1996; Witcombe et al., 1997). ICMV 221 which is being cultivated in India and several African countries is terminal DT, downy mildew resistant and has improved grain yield potential.
Salinity
Salinity is also a major abiotic stress constraint for crops of arid and semi-arid regions of Africa and Asia where high surface evaporation, low precipitation, and poor irrigation habits result in the enhancement in the levels of soluble salts that make the ground water unavailable to plants (Minitzer, 1993; Goyal, 2004). Salinization becomes more severe in areas affected by intense drought and high-temperature stress resulting in increased upward movement of capillary water and water soluble salts to the root zone of plants (Szabolcs, 1990; Varallyay, 1994).
Though pearl millet is a crop with inbuilt capacity to withstand soil salinity and therefore is capable of cultivation in saline lands for grain and forage production, salinity also acts as a significant abiotic constraint for its cultivation in several areas of Africa and India, with more intense effects in the West Asia and North Asia (WANA) zones of Central Asia. As compared to other cereal crops only limited information has been available on response to soil salinity in pearl millet. Reduced shoot N content and increased K+ and Na+ content is usually associated with salinity tolerance in pearl millet (Dwivedi et al., 2012). According to Krishnamurthy et al. (2007), shoot biomass ratio associated with salt tolerance and shoot Na+ concentration could be used as potential selection criteria for screening of pearl millet germplasm at vegetative stage. ICRISAT has carried out some basic research on salinity tolerance of pearl millet in collaboration with the International Centre for Biosaline Agriculture (ICAB) along with NARS partners in both India and WANA region (Dwivedi et al., 2012). For the screening of salinity tolerant breeding materials of pearl millet, pot culture method has been followed in the salinity affected fields. Screening of pearl millet germplasm has resulted in the development of advanced breeding materials, improved population including OPVs, gene pools and composites, parental lines of potential hybrids, and germplasm accessions with high grain and forage yield presumably with a high degree of salinity tolerance (Table 3). In the short-to-medium terms, some of these materials can be released for cultivation after extensive validation of their yield performances at on-farm trials. Working on these lines, a pearl millet variety “HASHAKI I” has been identified for release in Uzbekistan in 2012 as a high-forage variety for salt-affected areas. The identified salinity-tolerant pearl millet lines should be utilized in breeding programs to develop salinity-tolerant locally adapted cultivars (both OPVs and hybrids). This will enable farmers in salt-affected areas to adopt and grow a new crop such as pearl millet in lands that otherwise are fallow most of the years (Yadav et al., 2012).
High Temperature
Most of the growth processes of pearl millet like rate of seed germination, coleoptile elongation, rate of photosynthesis, etc. requires optimum temperature of about 35°C indicating that it is adapted to the hot arid zones of Sahel and many parts of India (Gracia-Huidobro et al., 1985) where mid-day surface temperature could exceed 45°C (Singh et al., 1993). While temperature beyond 35°C could be detrimental to the development of most of the other cereal crops, pearl millet could thrive in hot environmental conditions and maintain their optimal growth and yield potential (Yadav et al., 2010).
In order to understand the effects of temperature on seed germination, supra-optimal effects of temperature have been studied (Yadav et al., 2010). It was found that seed germination in pearl millet usually occurs at 35°C till 45°C but decreases at 47°C and almost stop at 50°C (Garcia-Huidobro et al., 1982). Further, the high temperature of seedbed has been established as a major cause of poor plant stand in pearl millet (Ong, 1983a,b). It was also indicated from several field studies in Sahel region that pearl millet seedlings are most vulnerable to high temperatures during the first 10 days of sowing (Stomph, 1990). Since it is difficult to control high soil surface temperature by cultural methods, it becomes essential to identify genetic variations in pearl millet germplasm for tolerance to high-temperature seedbeds. H 77/833-2, an elite pearl millet inbred line, hybrids of which are widely used by farmers in north-western India, is sensitive to terminal drought stress but tolerant to high-temperature stress (Yadav et al., 2014). This cultivar has been used as one of the parents (recurrent) to generate a set of mapping population (NILs) to map QTLs for terminal DT of grain and stover yield as well as their component traits in pearl millet (Yadav R.S. et al., 2003, 2004; Hash et al., 2006b).
Resistance to Biotic Stresses
Like other cereal crops, pearl millet is also prone to various biotic stresses including fungal, viral, and bacterial infections. Fungal infections including downy mildew, rust, smut, blast, and ergot are considered to impact pearl millet production and yield more severely than any other plant disease causing pathogens. Downy mildew or ‘green ear’ caused by Sclerospora graminicola (Sacc. Schroet.) is one of the most devastating diseases of pearl millet causing maximum yield loss in India and Africa (Kumar et al., 2012). Downy mildew attacks panicles and their effects may range from mild symptoms to catastrophes when large fields have been destroyed. The disease was first reported in India and was considered of minor importance till 1970 when HB3, a popular Indian pearl millet hybrid (Singh, 1995) suffered severe yield loss from approximately 8.2 million metric tons in 1970–1971 to 4.6 million metric tons in 1971–1972 due to downy mildew epidemic (Dwivedi et al., 2012; Kumar et al., 2012). Blast (Pyricularia grisea) and rust (Puccinia substriata var. indica) are other common fungal diseases affecting pearl millet fodder and grain production. Blast is a foliar disease while ergot (Claviceps spp.) and smut (Moesziomyces penicillariae) are tissue-specific diseases and mainly infect ovary part of the plant (Thakur and Williams, 1980). A combination of various disease management practices can be helpful in combating fungal disease including cultural methods, chemical methods, and host–plant resistance (Williams et al., 1981; Singh and Gopinath, 1985). Further identification and utilization of new genes for host plant resistance for developing varieties resistant to biotic stresses are very important. It can be achieved by developing precise phenotyping methods, exploiting natural genetic variations among germplasm, and with a detailed understanding of pathogen variability, as well as molecular and genetic mechanisms of resistance. At different centers of ICRISAT various techniques and resources have been refined for the identification and screening of virulent traits for downy mildew, rust, smut, and blast and ergot diseases. Furthermore effective phenotypic screens have been developed for downy mildew (Singh et al., 1997; Jones et al., 2002; Thakur et al., 2008), ergot (Thakur et al., 1982), rust (Singh et al., 1997), and smut (Thakur and King, 1988b) in pearl millet by ICRISAT and other research organizations. Pearl millet germplasm have also been extensively evaluated for resistance to major fungal diseases and several sources of resistance to downy mildew, blast, rust, smut and ergot have been identified. These sources of resistance have also been transferred into improved genetic backgrounds for developing resistant varieties (Table 3). Four widely cultivated OPVs from India namely, WC-C75, ICMS 7703, ICTP 8203, and ICMV 155 are resistant to downy mildew8 while ICMH 451 and Pusa 23 are popular downy mildew resistant hybrids (Singh, 1995). ICRISAT has also produced a top cross hybrid ICMH 88088 that possess a high level of downy mildew resistance and is also high yielding (Singh, 1995). A few downy mildew resistant varieties have also been developed by ICRISAT for Western Africa such as ICMV1 and ICMV2 for Senegal and IKMP2, IKMP3, and IKMV 8201 for Burkina Faso (Singh et al., 1993). Researchers have also gained some success toward identification of stable sources of disease resistance for smut (Thakur and King, 1988b), ergot (Thakur and King, 1988a), and rust (Singh et al., 1997) and some of these have also been used in breeding programs for enhanced disease resistance (Hash et al., 1999, 2006a).
However, in the last 10 years, the scenario has been changed for downy mildew as well as for other fungal diseases globally. Development and commercialization of new hybrid cultivars have also given rise to new strains of downy mildew pathogens (about 20 new virulent pathotypes of S. graminicola have been screened so far) (Thakur et al., 2011) and in coming years more new virulent strains could be identified. Further host-directed evolution of pathogenic variation has rendered downy mildew resistance rather elusive in Indian pearl millet inbred lines and hybrid cultivars (Thakur et al., 1992). Similarly, in the last few years, blast and rust disease outbreaks have been severe in some states of India. Therefore, it becomes essential to understand the pathogen variability, its mechanism of action and genetics of host plant resistance for the formulation of effective strategies for crop improvement against biotic stresses. Resistance to downy mildew shows dominance over susceptibility, additive or recessive traits, however, partial host plant resistance is controlled by one or more genes along with some modifiers (Hash and Witcombe, 2001; Breese et al., 2002; Dwivedi et al., 2012). Six major downy mildew pathotypes have been reported in India (Thakur et al., 2006). Sudisha et al. (2009) used Inter-simple sequence repeat (ISSR) primers to characterize pathogen variability; while 27 downy mildew isolates were grouped into six major pathotypes using 20 rapid amplified polymorphic DNA (RAPD) and 19 ISSR markers (Jogaiah et al., 2008). Similarly, 46 isolates of downy mildew could be classified into 21 pathotypes on the basis of disease incidence, latent period and virulence index, with pathotype P11 found to be most virulent (Sharma et al., 2010). In another study, 48 pearl millet inbred lines were evaluated against nine diverse S. graminicola isolates obtained from five different geographical locations in India which confirmed that gene pyramiding could enhance resistance to diverse isolates of downy mildew (Hash et al., 2006a). In a very recent study 39 native endophytic actinomycetes isolates from pearl millet roots were evaluated for their proteolytic action against downy mildew, of which seven strains could directly suppress sporangium formation in S. graminicola suggesting them as potential effective biocontrol agents (Jogaiah et al., 2016). Hence, despite the impressive progress toward managing various fungal diseases in pearl millet, there is still a lot of scope toward developing strategies for better management of diseases with the focus primarily on virulence monitoring, identification and characterization of newer isolates and developing resistant hybrids/cultivars for commercial cultivation.
Genetic and Genomic Resources
Markers, Genetic Linkage Maps, and Trait-Genetics for Marker-Assisted Breeding
DNA-based molecular markers, genetic linkage maps, and sequence information are essential genomic resources to carry out genetic studies or marker-aided breeding in any crop. Though the discovery of molecular markers and construction of genetic maps in millets lag far behind those of staple cereals, pearl millet reportedly has comparatively substantial stock of genetic and genomic resources in the form of DNA-based markers, mapping populations or linkage maps (Lata et al., 2013; Lata, 2015). In pearl millet various DNA-based molecular markers such as AFLP, RFLP, RAPD, expressed sequence tag-based (EST) markers, sequence-tagged sites (STSs), simple sequence repeat (SSRs/microsatellites), DArTs, CISP and SNP have been developed to distinguish genetic variability, linkage map analysis and marker assisted screening to expedite the breeding programs (Liu et al., 1994; Devos et al., 1995; Allouis et al., 2001; Qi et al., 2004; Supriya et al., 2011; Sehgal et al., 2012; Lata, 2015) (Table 4). Molecular markers help in evaluating genetic differences in germplasm collections for appropriate selection of mating parents for hybrid breeding, studying population structure, and analysis of QTLs for abiotic and biotic stress resistance. Pearl millet exhibits a very high level of DNA marker polymorphism even between elite inbred parental lines of popular hybrids (Vadez et al., 2012).

TABLE 4. Summery of DNA based markers available in pearl millet related abiotic and biotic stress tolerance.
RFLP-based molecular markers were developed in pearl millet by Liu et al. (1992, 1994) and were mapped over 180 loci ranged approximately 350 cM in seven linkage groups, one extra couplet, and other was a free-floating point and later was used to map QTLs for resistance to downy mildew in pearl millet (Jones et al., 1995). A set of 21 polymorphic EST-based SSRs markers and six genomic markers were developed in pearl millet and were tested to detect polymorphism across 11 pairs of pearl millet mapping population parental lines (Senthilvel et al., 2010). These EST-based markers are also used for marker assisted breeding and crop improvement programs in pearl millet at ICRISAT. Senthilvel et al. (2010) also reported the development of an array of about 6900 DArT clones using PstI/BanII complexity reduction, and identification of a total of 256–277 polymorphic DArT markers in three RILs of pearl millet. A total of 574 polymorphic DArT markers were identified in a set of 24 genetically diverse pearl millet inbreds from 7000 DArT clones obtained from 95 diverse genotypes using PstI/BanII complexity reduction (Supriya et al., 2011). With the help of DArT markers, comparative genomics and genome organization studies can be easily done and the cost of marker-assisted backcrossing (MABC) is also low as compared to others. Sehgal et al. (2012) used available pearl millet ESTs and developed 75 SNP and CISP markers and demonstrated their use in identifying candidate genes underlying a major DT-QTL using four genotypes namely, H 77/833-2, PRLT 2/89-33, ICMR 01029 and ICMR 01004, representing parents of two mapping populations. There is another recent research on identification of 83,875 SNP markers using genotyping by sequencing (GBS) of PstI-MspI reduced representation libraries in 500 genotypes of pearl millet, comprised of 252 global accessions and 248 Senegalese landraces, which exhibited high genetic diversity relative to other genotypes of Africa and Asia (Hu et al., 2015). Further ISSR-derived sequence characterized amplified region (SCAR) markers were developed to screen out polymorphism in a pair of pearl millet genotypes ICMR-01007 (P1) and ICMR-01004 (P2) and their population for downy mildew resistance. A polymorphic band of 1.4 kb size was produced in ISMR-01004 variety only from a single primer set ISSR-22 and further PCR amplification of these polymorphic band was found to be tightly linked to downy mildew resistant linkage group (χ2 3:1 = 0.86, P < 0.01) with a genetic distance of 0.72 cM. This SCAR marker was further validated in different Asian and African pearl millet cultivars and the result showed that the marker was associated with downy mildew disease resistant genotypes only (Jogaiah et al., 2014). Further, Ambawat et al. (2016) constructed a linkage map comprising 286 loci (229 DArT and 57 SSR markers) through the genotyping of 256 DArT and 70 SSR markers on 168 F7 RILs derived from the cross between 81B-P6 and ICMP 451-P8. Using these markers, QTL for rust resistance have been mapped on LG1 with LOD score of 27.
These molecular markers have been used to construct a pearl millet genetic linkage map (Liu et al., 1994) and to identify and map QTLs for terminal drought (Yadav et al., 2002), reduced salt uptake (Sharma et al., 2010, 2014), grain and stover yield (Yadav et al., 2002; Yadav R.S. et al., 2003, 2004), and for downy mildew resistance (Jones et al., 1995, 2002; Breese et al., 2002; Gulia, 2004; Gulia et al., 2007), rust and blast resistance (Morgan et al., 1998). These genetic tools have also been used for diversity assessment (Liu et al., 1992; Bhattacharjee et al., 2002), studying recombination rates (Busso et al., 1995; Liu et al., 1996), analyzing domestication syndrome (Poncet et al., 2000, 2002) and comparative genetics (Devos et al., 1998; Devos and Gale, 2000). The present pearl millet genetic linkage map covers approximately 700 cM distributed across the seven linkage groups as expected, and at least one pair of a free-floating RFLP-linked marker, however, telomeric regions of chromosomes have yet not been mapped (Vadez et al., 2012). Intriguingly these DNA marker-based linkage groups could not be distinctly linked with the pearl millet chromosome map (Kaul and Sidhu, 1997) developed using morphological markers (Kumar and Andrews, 1993) and conventional cytogenetic methods (Jauhar and Hanna, 1998) suggesting the scope of more focused research in this direction. Further identification of minor QTLs using conventional mapping population is thought to be a complex process partly due to the interaction(s) between QTLs and/or due to the strong overshadowing effects of major QTLs. Kumari et al. (2014) developed a set of chromosome segment substitution lines (CSSLs) for the identification of minor QTLs by introgressing overlapping chromosome segments from 863B into ICMB 841 background for use in QTL detection, fine mapping, and complex trait mechanism studies. Advanced generation backcross progenies (1492) that were expected to provide coverage across each of the seven pearl millet linkage groups, were genotyped at 74 marker loci involving SSRS, SSCP-SNP, and STSs markers, identifying 124 segment introgression homozygotes. Recently GBS approach is gaining popularity for the development of genetic markers and high density linkage map for species lacking reference genome (Punnuri et al., 2016). The authors have developed 96-plex ApeKI GBS library from the 186 RILs and from their parents (‘Tift 99D2B1’ and ‘Tift 454’) and F1 population. DNA of these populations was sequenced and the results were used for the development of reference genetic map using 150 RILs that contained a total of 16,650 SNPs and 333,567 sequence tags spread across all seven pearl millet chromosomes. The final map has a genetic distance of 716.7 cM with 23.23/cM overall average density of SNPs and 1.66 unique linkage bins per cM. This map was further used in mapping QTLs for flowering and resistance to Pyricularia leaf spot disease caused by [Pyricularia grisea (Cke.) Sacc.] in pearl millet.
It has also been proposed that pearl millet genome has undergone several structural rearrangements (Devos and Gale, 2000) that could be possibly associated with its evolution and adaptation in severe abiotic stresses and nutrient deficient soils although until now marker relationships are almost collinear across all the 10 mapping populations developed and mapped (Liu et al., 1996; Azhaguvel, 2001; Kolesnikova, 2001; Vadez et al., 2012).
Identification and Functional Validation of Genes Associated with Abiotic and Biotic Stress Tolerance
In last few years, the scientific community has gained some success toward understanding the physiological basis of stress tolerance and open new avenues for the development of more drought- tolerant pearl millet inbred lines and hybrids using MABC (Serraj et al., 2005). In pearl millet several QTLs have been identified and mapped to downy mildew, DT and yield components (Table 5), however linkage analysis in most of these studies suggest presence of genes/QTLs at a distance of 10–40 cM from the closest markers making it difficult for MAB or functional validation of candidate genes (Dwivedi et al., 2012). Until now only genes associated with a major DT-QTL have been identified in this crop (Sehgal et al., 2012). Moreover, it is apparent that in comparison to other cereal crops, little attention has been given toward identification and characterization of candidate genes for various abiotic and biotic stress resistances in pearl millet. There have been only a few reports on transcriptome analysis in pearl millet for delineating the mechanism of abiotic stress tolerance till date (Mishra et al., 2007; Choudhary et al., 2015). Both studies were done using suppression subtractive hybridization (SSH) technique. Mishra et al. (2007) reported the identification of about 2,494 differentially regulated transcripts in response to drought, salinity, and cold stress indicating the existence of a complex gene regulatory network in this stress tolerant crop. While Choudhary et al. (2015) reported transcript profiling of 22-days old seedlings of P. glaucum subjected to 30% PEG for different time periods ranging from 0.5 to 48 h that led to the identification of 745 ESTs in response to drought stress which could be helpful in unraveling the molecular basis underlying tolerance to drought stress. In another study, transcriptome analysis using 454 Roche NGS tool was done to understand the mechanisms underlying resistance to downy mildew in pearl millet (Kulkarni et al., 2016). A total of 1396 and 936 up- and down- regulated transcripts in resistant inoculated/resistant control, and 1000 and 1591 transcripts in susceptible inoculated/susceptible control were identified. The study revealed the up-regulation of phenylpropanoid pathway genes in resistant genotype and also evoked potentials of hypersensitive response and systemic acquired resistance as possible defense mechanisms operating against downy mildew infection in pearl millet. A comprehensive pearl millet transcriptome assembly (∼43 Mbp) has also been developed by integrating data from three independent investigations carried out by Zeng et al. (2011) and Rajaram et al. (2013) and is comprised of about 69, 398 tentative assembly contigs9. This transcriptome assembly also includes pearl millet ovule transcriptome data. Recently three studies have been published regarding identification and selection of suitable reference genes for assessment of gene expression through quantitative real-time (qRT)-PCR analysis under individual as well as multiple stress conditions (Saha and Blumwald, 2014; Reddy et al., 2015; Shivhare and Lata, 2016).
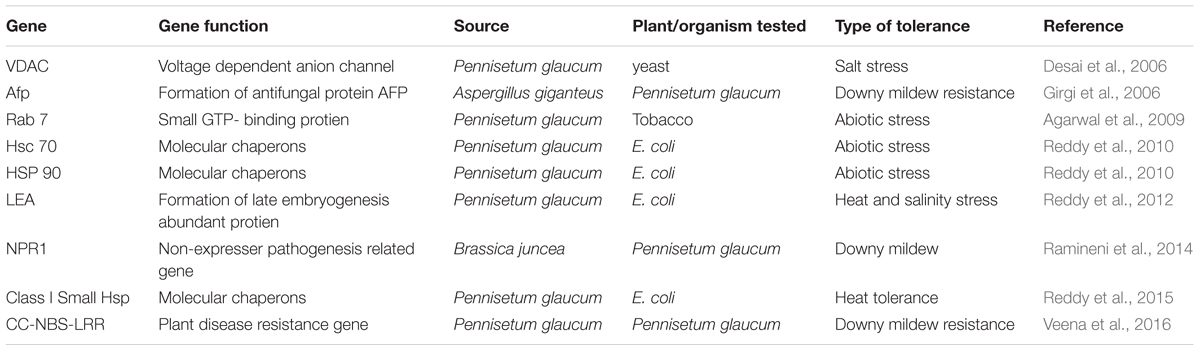
TABLE 5. Functional validation of important genes linked with biotic and abiotic stress response in pearl millet.
Improved transformation of millets linked to significant gene targets which may offer resistance to various biotic and abiotic stresses will play important role in crop improvement. Of late development of cultivars using genetic engineering approaches is gaining significance in plant biology and stress physiology. The understanding of mechanisms that regulate gene expression and the capacity to transfer important genes from other organisms into plants will expand the ways in which plants can be utilized. The use of newer approaches combining physiological, biochemical, molecular and genetic techniques should provide excellent results in near future. The task of generating transgenic cultivars is not only limited to the success of the transformation process but also the proper emphasis on incorporation of stress tolerance. Functional analysis in vivo is fundamental to transgenic technology which helps to investigate molecular mechanisms of biotic and abiotic stress regulation. Important methods to functionally validate candidate gene(s) for stress tolerance include overexpression, gene silencing or genome editing in a plant system. In pearl millet, the biolistic method for gene delivery has been frequently used for transformation (Ceasar and Ignacimuthu, 2009; Lata, 2015). There have been only a few reports on achieving stress tolerance through transgenic approaches in pearl millet (Table 6) as for example, Girgi et al. (2006), introduced an afp gene from the mold Aspergillus giganteus, encoding an antifungal protein AFP into pearl millet through particle bombardment. The transgenic plants showed a significant reduction in disease symptoms against rust and downy mildew infection as compared to control plants with an increase in disease resistance by up to 90%. Similarly, a Brassica juncea non-expressor of pathogenesis-related gene 1 (BjNPR1) has been stably integrated and expressed into a pearl millet male fertility restorer line ICMP451 through Agrobacterium-mediated transformation (Ramineni et al., 2014). However, until now there has been no report on the generation of abiotic stress tolerant transgenic pearl millet line. Therefore, it can be inferred that development of genetically engineered pearl millet varieties is still in nascent stage despite its economic importance and hence calls for a concerted effort for generation and evaluation of transgenic lines under various stress conditions.
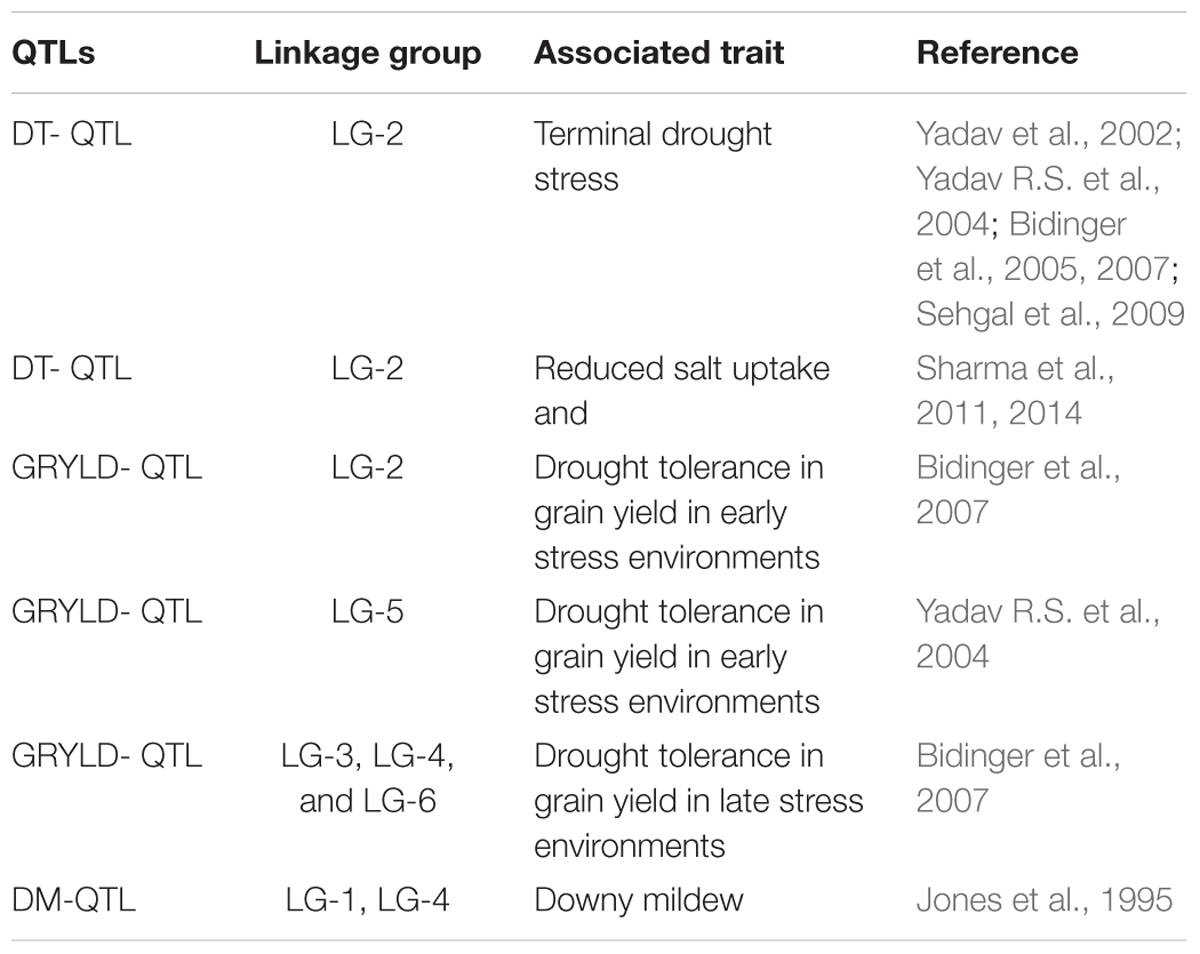
TABLE 6. Quantitative trait loci (QTLs) associated with important traits under abiotic and biotic stresses in pearl millet.
Pearl Millet Genome Sequencing
Though, in past few years crucial efforts have been made toward generating genomic resources in the form of molecular markers and genetic linkage maps in pearl millet but the collection is still not up to the mark as compared to other cereal crops. Hence, with the aim to generate sufficient genomic resources for augmenting breeding programs in pearl millet by sequencing pearl millet genome, a consortium of various international organizations namely ICRISAT; L’Institut de Recherché pour le Développement (IRD), France; the Indian Council of Agricultural Research (ICAR), India; the University of Georgia, USA; Cornell University, USA; University of Florida USA; L’Institut Sénégalais de Recherches Agricoles (ISRA); and Pioneer Overseas Corporation, India has been formed10. A global pearl millet reference genotype, Tift 23DB2B1 has been chosen to develop its draft genome sequence through whole genome shotgun and Bacterial artificial chromosome (BAC) sequence approaches. There have also been efforts to resequence around 993 pearl millet germplasm lines for validation. Hence, the rigorous sequencing efforts will be definitely helpful to the researchers working in crop genetics and breeding particularly pearl millet breeding.
Conclusion and Future Perspectives
In summary, pearl millet could serve as an excellent model for abiotic and biotic stress research owing to its hardy nature. Therefore, efforts should be focused on collection and characterization of pearl millet germplasm so that potential sources of genetic variation could be identified and incorporated in crop improvement programs. A concerted effort is also needed to develop and upgrade phenotypic screens for abiotic stress tolerance particularly high temperature, and biotic stress resistance such as downy mildew. There is a necessity for continuous monitoring of pathogen variability in pearl millet against various fungal pathogens. There is also a dire need to expedite the genetic and genomic resources of pearl millet for the development of more efficient genomic and sequenced based molecular markers for carrying out trait genetics and association mapping. This will help in identification and validation of candidate genes associated with important agronomic traits including abiotic stress tolerance and biotic stress resistance. The potential candidate genes identified through various transcriptomic and proteomic studies could be utilized for the development of cultivars with improved adaptability to survive harsh environmental conditions either through transgenic technology or MAB (Figure 1). It is hoped that with the completion and release of pearl millet genome sequence, the genetic and molecular studies in this otherwise naturally stress tolerant crop will progress swiftly. The advanced high throughput assays and next-generation sequencing could be used to resequence a large number of pearl millet germplasm. It will not only help in identification and tracking of natural genetic variations present within the germplasm collection but will also help in the discovery of genes and molecular markers associated with important agronomic traits, leading to newer opportunities for crop improvement in pearl millet and related crops.
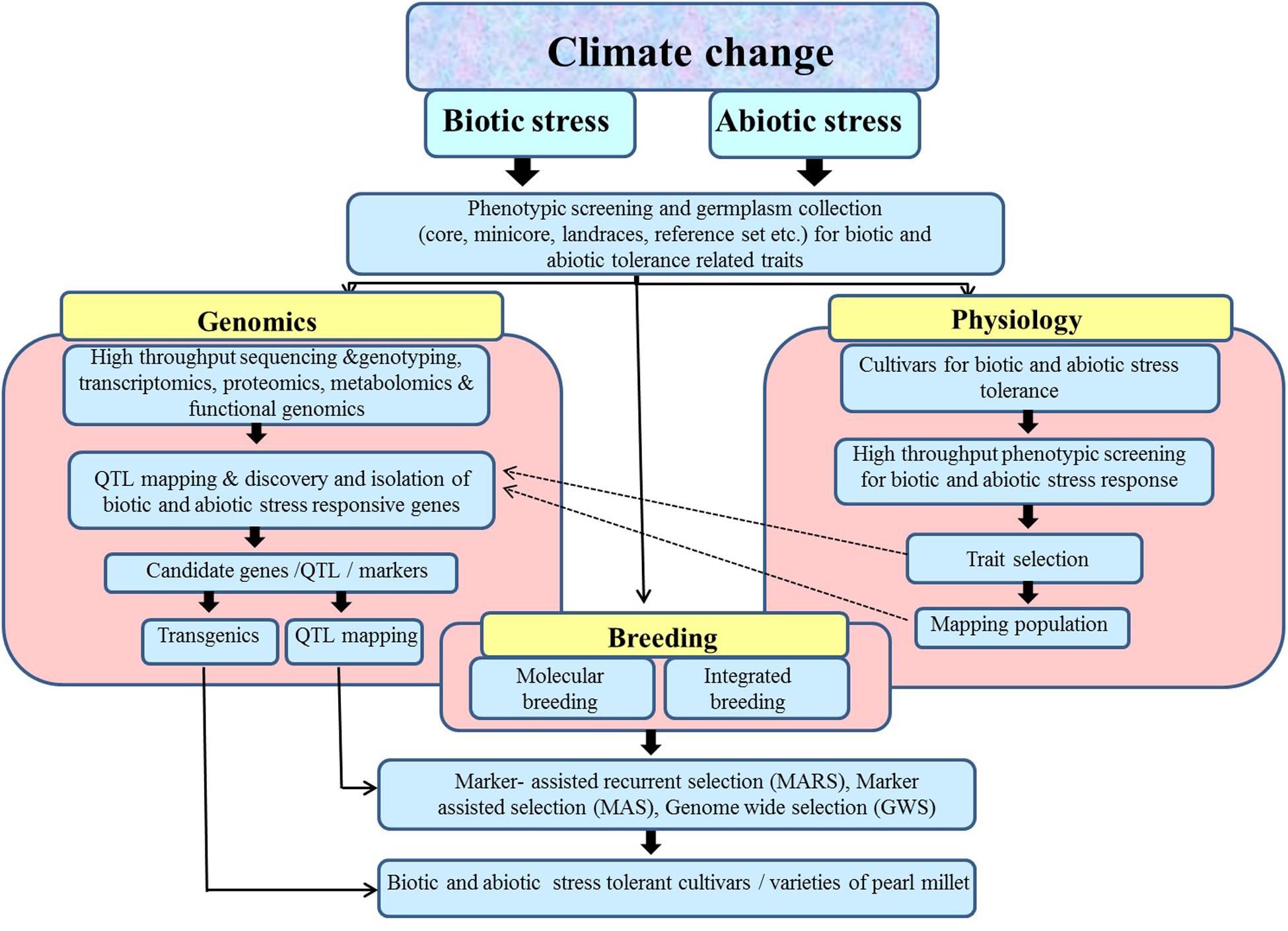
FIGURE 1. A combined approach for integrating genomics, physiology, and breeding techniques for developing improved cultivars with enhanced abiotic and biotic stress resistance in pearl millet.
Author Contributions
CL and RS wrote and reviewed the manuscript. Both authors contributed equally.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
CL acknowledges the research grant from INSPIRE Faculty Award [IFA-11LSPA-01] by Department of Science and Technology (DST), GoI. Authors are also thankful to the Director, CSIR-National Botanical Research Institute, Lucknow, India for providing facilities and support for the study.
Footnotes
- ^http://exploreit.icrisat.org/page/pearl_millet/680/273
- ^http://exploreit.icrisat.org/page/pearl_millet/680
- ^http://www.aicpmip.res.in/pmnews2016.pdf
- ^http://www.feedipedia.org/node/399
- ^http://edis.ifas.ufl.edu/ag347
- ^http://agritech.tnau.ac.in/seed_certification/seed_cm_pearl\%20miilets.html
- ^http://www.aicpmip.res.in/pmbiology.pdf
- ^http://agropedia.iitk.ac.in/content/downy-mildew-pearl-millet
- ^http://www.ceg.icrisat.org/transcriptome.html
- ^http://www.ceg.icrisat.org/ipgsc.html
References
Agarwal, P., Agarwal, P. K., Joshi, A. J., Sporoy, S. K., and Reddy, M. K. (2009). Overexpression of PgDREB2A transcription factor enhances abiotic stress tolerance and activates downstream stress-responsive genes. Mol. Biol. Rep. 37, 1125–1135. doi: 10.1007/s11033-009-9885-8
Agarwal, P. K., Agarwal, P., Jain, P., Jha, B., Reddy, M. K., and Sopory, S. K. (2008). Constitutive overexpression of a stress-inducible small GTP-binding protein PgRab7 from Pennisetum glaucum enhances abiotic stress tolerance in transgenic tobacco. Plant Cell Rep. 27, 105–115. doi: 10.1007/s00299-007-0446-0
Ali, G. M., Khan, N. M., Hazara, R., and McNeilly, T. (2004). Variability in the response of pearl millet (Pennisetum americanum (L.) Leeke) accessions to salinity. Acta Agron. Hung. 52, 277–286. doi: 10.1556/AAgr.52.2004.3.9
Allouis, S., Qi, X., Lindup, S., Gale, M. D., and Devos, K. M. (2001). Construction of a BAC library of pearl millet, Pennisetum glaucum. Theor. Appl. Genet. 102, 1200–1205. doi: 10.1007/s001220100559
Ambawat, S., Senthivel, S., Hash, C. T., Nepolean, T., Rajaram, V., Eshwar, K., et al. (2016). QTL mapping for rust resistance in pearl millet using an integrated DArT- and SSR-based linkage map. Euphytica 209, 461–476. doi: 10.1007/s10681-016-1671-9
Amblard, S., and Pernès, J. (1989). The identification of cultivated pearl millet (Pennisetum) amongst plant impressions on pottery from Oued Chebbi (Dhar Oualata, Mauritania). Afr. Archaeol. Rev. 7, 117–126. doi: 10.1007/BF01116840
Andrews, D. J., and Kumar, K. A. (1996). Use of the West African pearl millet landrace Iniadi in cultivar development. Plant Genet. Resour. Newsl. 105, 15–22.
Aparna, K., Nepolean, T., Srivastava, R. K., Kholova, J., Rajaram, V., Kumar, S., et al. (2015). Quantitative trait loci associated with constitutive traits controlling water use in pearl millet [Pennisetum glaucum (L.) R. Br.]. Plant Biol. 5, 1073–1084. doi: 10.1111/plb.12343
Arya, L., Verma, M., Gupta, P. K., and Karihaloo, J. (2009). Development of EST-SSRs in finger millet (Eleusine coracana ssp coracana) and their transferability to pearl millet (Pennisetum glaucum). J. Plan Biochem. Biotech. 18, 97–100. doi: 10.1007/BF03263303
Azhaguvel, P. (2001). Linkage Map Construction and Identification of QTLs for Downy Mildew (Sclerospora Graminicola) Resistance in Pearl Millet (Pennisetum glaucum (L) R Br). Ph.D. thesis, Agricultural University, Madurai.
Bennett, M. D., Bhandol, P., and Leitch, I. J. (2000). Nuclear DNA amounts in angiosperms and their modern uses-807 new estimates. Ann. Bot. (Lond.) 86, 859–909. doi: 10.1111/nph.13264
Bertin, I., Zhu, J. H., and Gale, M. D. (2005). SSCP-SNP in pearl millet—a new marker system for comparative genetics. Theor. Appl. Genet. 110, 1467–1472. doi: 10.1007/s00122-005-1981-0
Bhattacharjee, R., Bramel, P. J., Hash, C. T., Kolesnikova-Allen, M. A., and Khairwal, I. S. (2002). Assessment of genetic diversity within and between pearl millet landraces. Theor. Appl. Genet. 105, 666–673. doi: 10.1007/s00122-002-0917-1
Bidinger, F. R., and Hash, C. T. (2004). “Pearl millet,” in Physiology and Biotechnology Integration for Plant Breeding, eds H. T. Nguyen and A. Blum (New York, NY: Marcel Dekker), 225–270.
Bidinger, F. R., Mahalakshmi, V., and Rao, G. D. P. (1987). Assessment of drought resistance in pearl millet [Pennisetum americanum (L) Leeke]. Factors affecting yields under stress. Aust. J. Agric. Res. 38, 37–48. doi: 10.1071/AR9870037
Bidinger, F. R., Nepolean, T., Hash, C. T., Yadav, R. S., and Howarth, C. J. (2007). Identification of QTL for grain yield of pearl millet [Pennisetum glaucum (L.) R. Br.] in environments with variable moisture during grain filling. Crop Sci. 47, 969–980. doi: 10.2135/cropsci2006.07.0465
Bidinger, F. R., Serraj, R., Rizvi, S. M. H., Howarth, C. J., Yadav, R. S., and Hash, C. T. (2005). Field evaluation of drought tolerance QTL effects on phenotype and adaptation in pearl millet [Pennisetum glaucum (L.) R. Br.] top cross hybrids. Field Crops Res. 94, 14–32. doi: 10.1016/j.fcr.2004.11.006
Bidinger, F. R., Yadav, O. P., and Weltzien, R. E. (2009). Genetic improvement of pearl millet for the arid zone of northern India: lessons from two decades of collaborative ICRISAT-ICAR research. Expt. Agric. 45, 107–115. doi: 10.1017/S0014479708007059
Bourland, F. M. (1987). Registration of ICML11 rust resistant pearl millet germplasm. Crop Sci. 27: 367. doi: 10.2135/cropsci1987.0011183X002700020061x
Breese, W. A., Hash, C. T., Devos, K. M., and Howarth, C. J. (2002). “Pearl millet genomics—an overview with respect to breeding for resistance to downy mildew,” in Sorghum and Millets Pathology, ed. J. F. Leslie (Ames, IA: Iowa State Press), 243–246.
Brunken, J., de Wet, J. M. J., and Harlan, J. R. (1977). The morphology and domestication of pearl millet. Econ. Bot. 31, 163–174. doi: 10.1371/journal.pone.0022404
Budak, H., Cregan, P. B., Baenziger, P. S., and Dweikat, I. (2003). Development and utilization of SSRs to estimate the degree of genetic relationships in a collection of pearl millet germplasm. Crop Sci. 43, 2284–2290. doi: 10.2135/cropsci2003.2284
Burton, G. W., and Wilson, J. P. (1995). Registration of Tift 65 parental inbred line of pearl millet. Crop Sci. 35: 1244. doi: 10.2135/cropsci1995.0011183X003500040111x
Busso, C. S., Liu, C. J., Hash, C. T., Witcombe, J. T., Devos, K. M., de Wet, J. M. J., et al. (1995). Analysis of recombination rate in female and male gametogenesis in pearl millet (Pennisetum glaucum) using RFLP markers. Theor. Appl. Genet. 90, 253–257. doi: 10.1007/BF00222208
Carberry, P. S., Cambell, L. E., and Bidinger, F. R. (1985). The growth and development of pearl millet as affected by plant population. Field Crops Res. 11, 193–220. doi: 10.1016/0378-4290(85)90102-9
Ceasar, S. A., and Ignacimuthu, E. S. (2009). Genetic engineering of millets: current status and future prospects. Biotechnol. Lett. 31, 779–788. doi: 10.1007/s10529-009-9933-4
Choudhary, M., Jayanand, and Padaria, J. C. (2015). Transcriptional profiling in pearl millet (Pennisetum glaucum L.R. Br.) for identification of differentially expressed drought responsive genes. Physiol. Mol. Biol. Plants 21, 187–196. doi: 10.1007/s12298-015-0287-1
Daisuke, U. N. O. (2005). Farmer’s selection of local and improved pearl millet varieties in Ovamboland, Northern Namibia. Afr. Study Monogr. 30, 107–117.
D’Andrea, A. C., and Casey, J. (2002). Pearl millet and Kintampo subsistence. Afr. Archaeol. Rev. 19, 147–173. doi: 10.1023/A:1016518919072
de Wet, J. M. J., Bidinger, F. R., and Peacock, J. M. (1992). “Pearl millet (Pennisetum glaucum) – a cereal of the Sahel,” in Desertified Grasslands, their Biology, and Management, ed. G. P. Chapman (London: Academic Press),k259–267.
Desai, M. K., Mishra, R. N., Verma, D., Nair, S., Sopory, S. K., and Reddy, M. K. (2006). Structural and functional analysis of a salt stress inducible gene encoding voltage dependent anion channel (VDAC) from pearl millet (Pennisetum glaucum). Plant Physiol. Biochem. 44, 483–493. doi: 10.1016/j.plaphy.2006.08.008
Devos, K. M., and Gale, M. D. (2000). Genome relationships: the grass model in current research. Plant Cell 12, 637–646. doi: 10.1105/tpc.12.5.637
Devos, K. M., Pittaway, T. S., Busso, C. S., Gale, M. D., Witcombe, J. R., and Hash, C. T. (1995). Molecular tools for the pearl millet nuclear genome. Int. Sorghum Millets Newslett. 36, 64–66.
Devos, K. M., Wang, Z. M., Beales, J., Sasaki, T., and Gale, M. D. (1998). Comparative genetic maps of foxtail millet (Setaria italica) and rice (Oryza sativa). Theor. Appl. Genet. 96, 63–68. doi: 10.1007/s001220050709
Dwivedi, S., Upadhyaya, H., Senthilvel, S., and Hash, C. (2012). “Millets: Genetic, and Genomic Resources,” in Plant Breeding Reviews, ed. J. Janick (Hoboken, NJ: John Wiley, and Sons, Inc.), 247–375.
Dwivedi, S., Upadhyaya, H., Subudhi, P., Gehring, C., Bajic, V., and Ortiz, R. (2010). Enhancing abiotic stress tolerance in cereals through breeding and transgenic interventions. Plant Breed. Rev. 33, 31–114.
Farooq, M., Wahid, A., Kobayashi, N., Fujita, D., and Basra, S. M. A. (2009). Plant drought stress; effects, mechanisms and management. Agron. Sustain. Dev. 29, 185–212. doi: 10.1051/agro:2008021
Garcia-Huidobro, J., Monteith, J. L., and Squire, G. R. (1985). Time, temperature and germination of pearl millet (Pennisetum typhoides S. & H.): III. Inhibition of germination by short exposure to high temperature. J. Exp. Bot. 36, 338–343. doi: 10.1093/jxb/36.2.338
Garcia-Huidobro, T., Monteith, T., and Squire, G. R. (1982). Time temperature and germination of pearl millet (Pennisetum typhoides S. & H.): I. Constant temperature. J. Exp. Bot. 33, 288–296. doi: 10.1093/jxb/33.2.288
Garí, A. J. (2002). “Review of the African millet diversity,” in Proceedings of the International Workshop on Fonio, Food Security and Livelihood Among the Rural Poor in West Africa (Rome: International Plant Genetic Resources Institute), 1–18.
Girgi, M., Breese, W. A., Lörz, H., and Oldach, K. H. (2006). Rust and downy mildew resistance in pearl millet (Pennisetum glaucum) mediated by heterologous expression of the afp gene from Aspergillus giganteus. Trans. Res. 15, 313–324. doi: 10.1007/s11248-006-0001-8
Govindaraj, M., Shanmugasundaram, P., Sumathi, P., and Muthiah, A. R. (2010). Simple, rapid and cost effective screening method for drought resistant breeding in pearl millet. Electron. J. Plant Breed. 1, 590–599.
Goyal, R. K. (2004). Sensitivity of evapotranspiration to global warming: a ase study of arid zone of Rajasthan India. Agric. Water Manag. 69, 1–11. doi: 10.1016/j.agwat.2004.03.014
Gracia-Huidobro, J., Monteith, J. L., and Squire, G. R. (1985). Time, temperature(and)germination of pearl millet (Pennisetum typhoides S & H). III. Inhibition of germination by short exposure to high temperature. J. Exp. Bot. 36, 338–343. doi: 10.1093/jxb/36.2.338
Gregory, P. J. (1983). Response to temperature in a stand of pearl millet (Pennisetum typhoides S. & H.): III. Root development.]. J. Exp. Bot. 34, 744–756. doi: 10.1093/jxb/34.6.744
Gulia, S. K. (2004). QTL Mapping for Improvement of Downy Mildew [Sclerospora graminicola (Sacc.) J. Schroet.] Resistance (DMR) in Pearl Millet [Pennisetum glaucum (L.) R. Br.] Hybrid Parental Line ICMB 89111. Ph.D. thesis, Chaudhary Charan Singh Haryana Agricultural University, Hisar.
Gulia, S. K., Hash, C. T., Thakur, R. P., Breese, W. A., and Sangwan, R. S. (2007). “Mapping new QTLs for downy mildew [Sclerospora graminicola (Sacc.) J. Schroet.] resistance in pearl millet [Pennisetum glaucum (L.) R. Br.],” in Crop Production in Stress Environments— Genetic and Management Options, eds D. S. Singh, V. S. Tomar, R. K. Behl, S. D. Upadhyaya, M. S. Bhale, and D. Khare (Jodhpur: Agrobios), 373–386.
Hanna, W. W. (1989). Characteristics and stability of a new cytoplasmic – nuclear male-sterile source in pearl millet. Crop Sci. 29, 1457–1459. doi: 10.2135/cropsci1989.0011183X002900060026x
Hanna, W. W., and Dujardin, M. (1985). “Interspecific transfer of apomixis in Pennisetum,” in Proceedings. of the XV International Grassland Congress, Kyoto, 249–250.
Hanna, W. W., Hill, G. M., Gates, R. N., Wilson, J. P., and Burton, G. W. (1997). Registration of tif leaf 3 pearl millet. Crop Sci. 37:1388. doi: 10.2135/cropsci1997.0011183X003700040095x
Harlan, J. R., and de Wet, J. M. J. (1971). Toward a rational classification of cultivated plants. Taxon 20, 509–517. doi: 10.2307/1218252
Hash, C. T., Sharma, A., Kolesnikova-Allen, M. A., Singh, S. D., Thakur, R. P., Raj, A. G. B., et al. (2006a). Teamwork delivers biotechnology products to Indian small-holder crop-livestock products: Pearl millet hybrid ‘HHB 67 Improved’ enters seed delivery pipeline. J. SAT Agric. Res. 2, 1–3.
Hash, C. T., Thakur, R. P., Rao, V. P., and Raj, A. G. B. (2006b). Evidence for Enhanced Resistance to Diverse Isolates of Pearl Millet Downy Mildew through Gene Pyramiding, Vol. 47. Patancheru: International Crops Research Institute for the Semi-Arid Tropics, 134–138.
Hash, C. T., and Witcombe, J. R. (2001). Pearl millet molecular marker research. Int. Sorghum Millets Newsl. 42, 8–15.
Hash, C. T., Yadav, R. S., Cavan, G. P., Howarth, C. J., Liu, H., Xiaoquan, Q., et al. (1999). “Marker-assisted backcrossing to improve terminal drought tolerance in pearl millet,” in Proceedings of a Strategic Planning Workshop on Molecular Approaches for the Genetic Improvement of Cereals for Stable Production in Water-Limited Environments, Improve Terminal drought Tolerance in Pearl Millet, eds J.-M. Ribaut and D. Poland (El Batan: International Maize and Wheat Improvement Center(CIMMYT)), 114–119.
Henson, I. E., and Mahalakshmi, V. (1985). Evidence for panicle control of stomatal behaviour in water stressed plants of pearl millet. Field Crops Res. 11, 281–290. doi: 10.1016/0378-4290(85)90109-1
Hu, Z., Mbacké, B., Perumal, R., Guèye, M. C., Sy, O., Bouchet, S., et al. (2015). Population genomics of pearl millet [Pennisetum glaucum (L.) R. Br.]: comparative analysis of global accessions and Senegalese landraces. BMC Genomics 16:1048. doi: 10.1186/s12864-015-2255-0
IBPGR and ICRISAT (1993). Descriptors for Pearl Millet [Pennisetum glaucum (L.) R. Br.]. Rome: International Board for Plant Genetic Resources.
ICRISAT (2008/2009). MTP Project 5: Producing More and Better Food at Lower Cost of Staple Cereal and Legume Hybrids in the Asian SAT (Sorghum, Pearl Millet, and Pigeonpea) through Genetic Improvement. Patancheru: ICRISAT, 158–159.
Jambunathan, R., and Subramanian, V. (1988). “Grain quality and utilization of sorghum and pearl millet,” in Proceedings of the International Biotechnology Workshop: Biotechnology in tropical crop improvement, (Patancheru: ICRISAT), 133–139.
Jauhar, P. P. (1981). Cytogenetics and Breeding of Pearl Millet and Related Species. New York, NY: Alan R. Liss.
Jauhar, P. P., and Hanna, W. W. (1998). Cytogenetics andgenetics of pearl millet. Adv. Agron. 64, 1–26. doi: 10.1016/S0065-2113(08)60501-5
Jogaiah, S., Kurjogi, M., Govind, S. R., Huntrike, S. S., Basappa, V. A., and Tran, L. P. (2016). Isolation and evaluation of proteolytic actinomycete isolates as novel inducers of pearl millet downy mildew disease protection. Sci. Rep. 6:30789. doi: 10.1038/srep30789
Jogaiah, S., Sharathchandra, R. G., and Raj, N. (2014). Development of SCAR marker associated with downy mildew disease resistance in pearl millet (Pennisetum glaucum L.). Mol. Biol. Rep. 41, 7815. doi: 10.1007/s11033-014-3675-7
Jogaiah, S., Soshee, A. K., and Hunthrike, S. S. (2008). Characterization of downy mildew isolates of Sclerospora graminicola by using differential cultivars and molecular markers. J. Cell Mol. Biol. 7, 41–55. doi: 10.1094/PHYTO.1998.88.4.292
Jones, E. S., Breese, W. A., Liu, C. J., Singh, S. D., Shaw, D. S., and Witcombe, J. R. (2002). Mapping quantitative trait loci for resistance to downy mildew in pearl millet: field and glasshouse detect the same QTL. Crop Sci. 42, 1316–1323. doi: 10.2135/cropsci2002.1316
Jones, E. S., Liu, C. J., Gale, M. D., Hash, C. T., and Witcombe, R. J. (1995). Mapping quantitative trait loci for downy mildew resistance in pearl millet. Theor. Appl. Genet. 91, 448–456. doi: 10.1007/BF00222972
Kanatti, A., Rai, K. N., Radhika, K., Govindaraj, M., Sahrawat, K. L., and Rao, A. S. (2014). Grain iron and zinc density in pearl millet: combining ability, heterosis and association with grain yield and grain size. Springerplus 3:763. doi: 10.1186/2193-1801-3-763
Kaul, J., and Sidhu, J. S. (1997). Establishment of chromosome map of pearl millet through the use of interchanges. J. Cytol. Genet. 32, 113–123.
Khairwal, I. S., and Yadav, O. P. (2005). Pearl millet (Pennisetum glaucum) improvement in India—retrospect and prospects. Indian J. Agric. Sci. 75, 183–191.
Khalfallah, N., Sarr, A., and Siljak-Yakovlev, S. (1993). Kary-ological study of some cultivated and wild stocks of pearl millet from Africa (Pennisetum typhoides Stapf and Hubb. and P. violaceum Rich). Caryologia 46, 127–138. doi: 10.1080/00087114.1993.10797254
Kholova, J., Hash, C. T., Kakkera, A., Kocova, M., and Vadez, V. (2010a). Constitutive water-conserving mechanisms are correlated with the drought tolerance of pearl millet [Pennisetum glaucum (L.) R. Br.]. J. Exp. Bot. 61, 369–377. doi: 10.1093/jxb/erp314
Kholova, J., Hash, C. T., Kakkera, A., Kocova, M., and Vadez, V. (2010b). Terminal drought- tolerant pearl millet [Pennisetum glaucum (L.) R. Br.] have high leaf ABA and limit transpiration at high vapour pressure deficit. J. Exp. Bot. 61, 1431–1440. doi: 10.1093/jxb/erq013
Kholova, J., Nepolean, T., Hash, C. T., Supriya, A., Rajaram, V., Senthilvel, S., et al. (2012). Water saving traits co-map with a major terminal drought tolerance quantitative trait locus in pearl millet [Pennisetum glaucum (L.) R. Br.]. Mol. Breed. 30, 1337–1353. doi: 10.1007/s11032-012-9720-0
Kholová, J., and Vadez, V. (2013). Water extraction under terminal drought explains the genotypic differences in yield, not the anti-oxidant changes in leaves of pearl millet (Pennisetum glaucum). Funct. Plant Biol. 40, 44–53.
Kolesnikova, M. (2001). Mapping New Quantitative Trait Loci (QTL) for Downy Mildew Resistance in Pearl Millet. Ph.D. thesis, Russia Academy of Science, Moscow.
Krishnamurthy, L., Serraj, R., Rai, K. N., Hash, C. T., and Dakheel, A. J. (2007). Identification of pearl millet [Pennisetum glaucum (L.) R. Br.] lines tolerant to soil salinity. Euphytica 158, 179–188. doi: 10.1007/s10681-007-9441-3
Kulkarni, K. S., Zala, H. N., Bosamia, T. C., Shukla, Y. M., Kumar, S., Fougat, R., et al. (2016). De novo transcriptome sequencing to dissect candidate genes associated with pearl millet-downy mildew (Sclerospora graminicola Sacc.) interaction. Front. Plant Sci. 7:847. doi: 10.3389/fpls.2016.00847
Kumar, A., and Andrews, D. J. (1993). Genetics of quantitative traits in pearl millet: a review. Crop Sci. 33, 1–20. doi: 10.2135/cropsci1993.0011183X003300010001x
Kumar, A., Sharma, N., Panwar, P., and Gupta, A. K. (2012). Use of SSR, RAPD markers and protein profiles based analysis to differentiate Eleusine coracana genotypes differing in their protein content. Mol. Biol. Rep. 39, 4949–4960. doi: 10.1007/s11033-011-1291-3
Kumar, S., Hash, C. T., Thirunavukkarasu, N., Singh, G., Rajaram, V., Rathore, A., et al. (2016). Mapping quantitave trait loci controlling high iron and zinc in self and open pollinated grains of pearl millet [Pennisetum glaucum (L) R. Br.]. Front. Plant Sci. 7:1636. doi: 10.3389/fpls.2016.01636
Kumari, B. R., Kolesnikova-Allen, M. A., Hash, C. T., Senthivel, S., Nepolean, T., Kavi Kishor, P. B., et al. (2014). Development of a set of chromosome segment substitution lines in pearl millet [Pennisetum glaucum (L.) R. Br.]. Crop Sci. 54, 2175–2182. doi: 10.2135/cropsci2013.09.0589
Lahiri, A. N., and Kumar, V. (1966). Studies on plant-water relationship III: further studies on the drought mediated alterations in the performance of bulrush millet. Proc. Natl. Inst. Sci. India B. 32, 116–129.
Lata, C. (2015). Advances in omics for enhancing abiotic stress tolerance in millets. Proc. Indian Natl. Sci. Acad. 81, 397–417.
Lata, C., Gupta, S., and Prasad, M. (2013). Foxtail millet: a model crop for genetic and genomic studies in bioenergy grasses Crit. Rev. Biotechnol. 33, 328–343. doi: 10.3109/07388551.2012.716809
Lata, C., Muthamilarasan, M., and Prasad, M. (2015). “Drought stress responses and signal transduction in plants,” in The Elucidation of Abiotic Stress Signaling in Plants, ed. G. K. Pandey (New York, NY: Springer), 195–225.
Latha, A. M., Rao, K. V., Reddy, T. P., and Reddy, V. D. (2006). Development of transgenic pearl millet (Pennisetum glaucum (L.) R. Br.) plants resistant to downy mildew. Plant Cell. Rep. 25, 927–935. doi: 10.1007/s00299-006-0141-6
Liu, C. J., Devos, K. M., Witcombe, J. R., Pittaway, T. S., and Gale, M. D. (1996). The effect of genome and sex on recombination rates in Pennisetum species. Theor. Appl. Genet. 93, 902–908. doi: 10.1007/BF00224092
Liu, C. J., Witcombe, J. R., Pittaway, T. S., Nash, M., Hash, C. T., Busso, C. S., et al. (1994). An RFLP-based genetic map of pearl millet (Pennisetum glaucum). Theor. Appl. Genet. 89, 481–487. doi: 10.1007/BF00225384
Liu, C. J., Witcombe, J. R., Pittaway, T. S., Nash, M., Hash, C. T., and Gale, M. D. (1992). “Restriction fragment length polymorphism in pearl millet, Pennisetum glaucum,” in Complexes D’espèces, Flux de Gènes et Ressources Génétiques des Plantes. Colloque International, eds J. C. Mounolou and A. Sarr (Paris: Bureau des Ressources Génétiques), 233–241.
Mahalakshmi, V., Bidinger, F. R., and Raju, D. S. (1987). Effect of timing of water deficit on pearl millet (Pennisetum americanum). Field Crops Res. 15, 327–339. doi: 10.1016/0378-4290(87)90020-7
Malik, S. (2015). Pearl millet- nutritional value and medicinal use. Int. J. Adv. Res. Innov. Ideas Educ. 1, 414–418.
Manga, V. K. (2015). Diversity in pearl millet [Pennisetum Glaucum (L.) R. BR.]. Indian J. Plant Sci. 4, 38–51.
Manga, V. K., and Kumar, A. (2011). Cultivar options for increasing pearl millet productivity in arid. Indian J. Fundam. Appl. Life Sci. 1, 200–208.
Mariac, C., Luong, V., Kapran, I., Mamadou, A., Sagnard, F., Deu, M., et al. (2006). Diversity of wild and cultivated pearl millet accessions (Pennisetum glaucum [L.] R. Br.) in Niger assessed by microsatellite markers. Theor. Appl. Genet. 114, 49–58. doi: 10.1007/s00122-006-0409-9
Martel, E., Ney, D. D., Siljak-Yakovlev, S., Brown, S., and Sarr, A. (1997). Genome size variation and basic chromosome number in pearl millet and fourteen Pennisetum species. J. Hered. 88, 139–143. doi: 10.1093/oxfordjournals.jhered.a023072
Minitzer, I. M. (1993). Confronting Climate Change: Risk, Implications and Responses. Cambridge: Cambridge University Press.
Mishra, R. N., Reddy, P. S., Nair, S., Markandeya, G., Reddy, A. R., Sopory, S., et al. (2007). Isolation and characterization of expressed sequence tags (ESTs) from subtracted cDNA libraries of Pennisetum glaucum seedlings. Plant Mol. Biol. 64, 713–732.
Morgan, R. N., Wilson, J. P., Hanna, W. W., and Ozias-Akins, P. (1998). Molecular markers for rust and pyricularia leaf spot disease resistance in pearl millet. Theor. Appl. Genet. 96, 413–420. doi: 10.1007/s001220050757
Ong, C. K. (1983a). Response to temperature in a stand of pearl millet (Pennisetium typhoides S&H): I. Vegetative development. J. Exp. Bot. 34, 322–336. doi: 10.1093/jxb/34.3.322
Ong, C. K. (1983b). Response to temperature in a stand of pearl millet (Pennisetium typhoides S&H): II. Reproductive development. J. Exp. Bot. 34, 337–348. doi: 10.1093/jxb/34.3.337
Oumar, I., Mariac, C., Pham, J. L., and Vigouroux, Y. (2008). Phylogeny and origin of pearl millet (Pennisetum glaucum [L.] R. Br) as revealed by microsatellite loci. Theor. Appl. Genet. 117, 489–497. doi: 10.1007/s00122-008-0793-4
Passot, S., Gnacko, F., Moukouanga, D., Lucas, M., Guyomarc’h, S., Ortega, B. M., et al. (2016). Characterization of pearl millet root architecture and anatomy reveals three types of lateral roots. Front. Plant Sci. 7:829. doi: 10.3389/fpls.2016.00829
Poncet, V., Lamy, F., Devos, K. M., Gale, M. D., Sarr, A., and Robert, T. (2000). Genetic control of domestication traits in pearl millet (Pennisetum glaucum L., Poaceae). Theor. Appl. Genet. 100, 147–159. doi: 10.1007/s001220050020
Poncet, V., Martel, E., Allouis, S., Devos, K. M., Lamy, F., Sarr, A., et al. (2002). Comparative analysis of QTLs affecting domestication traits between two domesticated _ wild pearl millet (Pennisetum glaucum L., Poaceae) crosses. Theor. Appl. Genet. 104, 965–975. doi: 10.1007/s00122-002-0889-1
Punnuri, S. M., Wallace, J. G., Knoll, J. E., Hyma, K. E., Mitchell, S. E., Buckler, E. S., et al. (2016). Development of high density linkage map and tagging leaf spot resistance in pearl millet using genotyping-by-sequencing markers. Plant Genome 9, 1–13. doi: 10.3835/plantgenome2015.10.0106
Qi, X., Pittaway, T. S., Lindup, S., Liu, H., Wateran, E., Padi, F. K., et al. (2004). An integrated genetic map and new set of simple sequence repeat markers for pearl millet, Pennisetum glaucum. Theor. Appl. Genet. 109, 1485–1493. doi: 10.1007/s00122-004-1765-y
Rai, K. N., Talukdar, B. S., and Rao, A. S. (1998a). Registration of pearl millet parental lines ICMA 92666 and ICMB 92666 with multiple disease resistance. Crop Sci. 38: 575. doi: 10.2135/cropsci1998.0011183X003800020103x
Rai, K. N., Thakur, R. P., and Rao, A. S. (1998b). Registration of pearl millet parental linesICMA 88006 and ICMB 88006. Crop Sci. 38, 575–576. doi: 10.2135/cropsci1998.0011183X003800020103x
Rai, K. N., Yadav, O. P., Rajpurohit, B. S., Patil, H. T., Govindaraj, M., Khairwal, I. S., et al. (2013). Breeding pearl millet cultivars for high iron density with zinc density as an associated trait. J. SAT Agric. Res. 11, 1–7.
Rajaram, V., Nepolean, T., Senthilvel, S., Varshney, R. K., Vadez, V., Srivastava, R. K., et al. (2013). Pearl millet [Pennisetum glaucum (L.) R. Br.] consensus linkage map constructed using four RIL mapping populations and newly developed EST-SSRs. BMC Genomics 14:159. doi: 10.1186/1471-2164-14-159
Rajaram, V., Varshney, R. K., Vadez, V., Nepolean, T., Senthilvel, S., Kholova, J., et al. (2010). Development of EST resources in pearl millet and their use in development and mapping of EST-SSRs in four RIL populations. Plant Anim. Gen. 18, 9–13.
Ramineni, R., Sadumpati, V., Khareedu, V. R., and Vudem, D. R. (2014). Transgenic pearl millet male fertility restorer line (ICMP451) and hybrid (ICMH451) expressing Brassica juncea nonexpressor of pathogenesis related genes 1 (BjNPR1) exhibit resistance to downy mildew disease. PLoS ONE 3:e90839. doi: 10.1371/journal.pone.0090839
Reddy, P. S., Mallikarjuna, G., Kaul, T., Chakradhar, T., Sopory, S. K., and Reddy, M. K. (2010). Molecular cloning and characterization of gene encoding for cytoplasmic Hsc70 from Pennisetum glaucum may play a protective role against abiotic stresses. Mol. Genet. Genomic. 283, 243–254. doi: 10.1007/s00438-010-0518-7
Reddy, P. S., Reddy, D. S., Sharma, Kiran, K., Bhatnagar-Mathur, P., and Vadez, V. (2015). Cloning and validation of reference genes for normalization of gene expression studies in pearl millet [Pennisetum glaucum (L.) R. Br.] by quantitative real-time PCR. Plant Gene. 1, 35–42. doi: 10.1016/j.plgene.2015.02.001
Reddy, P. S., Reddy, G. M., Pandey, P., Chandrasekhar, K., and Reddy, M. K. (2012). Cloning and molecular characterization of a gene encoding late embryogenesis abundant protein from Pennisetum glaucum: protection against abiotic stresses. Mol. Biol. Rep. 39, 7163–7174. doi: 10.1007/s11033-012-1548-5
Rooney, L. W., and McDonough, C. M. (1987). “Food quality and consumer acceptance of pearl millet,” in Proceedings of the lnternaUonal Pearl Millet Workshop, (Patancheru: ICRISAT).
Saha, P., and Blumwald, E. (2014). Assessing reference genes for accurate transcript normalization using quantitative real-time PCR in pearl millet [Pennisetum glaucum (L.) R. Br]. PLoS ONE 9:e106308. doi: 10.1371/journal.pone.0106308
Sehgal, D., Rajaram, V., Armstead, I. P., Vadez, V., Yadav, Y. P., Hash, C. T., et al. (2012). Integration of gene-based markers in a pearl millet genetic map for identification of candidate genes underlying drought tolerance quantitative trait loci. BMC Plant Biol. 12:9. doi: 10.1186/1471-2229-12-9
Sehgal, D., Rajpal, V. R., Raina, S. N., Sasanuma, T., and Sasakuma, T. (2009). Assaying polymorphism at DNA level for genetic diversity diagnostics of the safflower (Carthamus tinctorius L.) world germplasm resources. Genetica 135, 457–470. doi: 10.1007/s10709-008-9292-4
Sehgal, D., Skot, L., Singh, R., Srivastava, R. K., Das, S. P., Taunk, J., et al. (2015). Exploring potential of pearl millet germplasm association panel for association mapping of drought tolerance traits. PLoS ONE 10:e0122165. doi: 10.1371/journal.pone.0122165
Senthilvel, S., Jayashree, B., Mahalakshmi, V., Sathish Kumar, P., Nakka, S., Nepolean, T., et al. (2008). Development and mapping of simple sequence repeat markers for pearl millet from data mining of expressed sequence tags. BMC Plant Biol. 8:119. doi: 10.1186/1471-2229-8-119
Senthilvel, S., Nepolean, T., Supriya, A., Rajaram, V., Kumar, S., Hash, C. T., et al. (2010). “Development of a molecular linkage map of pearl millet integrating DArT and SSR markers,” in Proceedings of the Plant and Animal Genome 18 Conference, San Diego, CA, 9–13.
Serraj, R., Hash, C. T., Rizvi, S. M. H., Sharma, A., Yadav, R. S., and Bidinger, F. R. (2005). Recent advances in marker-assisted selection for drought tolerance in pearl millet. Plant Prod. Sci. 8, 334–337. doi: 10.1626/pps.8.334
Sharma, P. C., Sehgal, D., Singh, D., Singh, G., and Yadav, R. S. (2011). A major terminal drought tolerance QTL of pearl millet is also associated with reduced salt uptake and enhanced growth under salt stress. Mol. Breed. 27, 207–222. doi: 10.1007/s11032-010-9423-3
Sharma, P. C., Singh, D., Sehgal, D., Singh, G., Hash, C. T., and Yadav, R. S. (2014). Further evidence that terminal drought tolerance QTL of pearl millet is associated with reduced salt uptake. Environ. Exp. Bot. 102, 48–57. doi: 10.1016/j.envexpbot.2014.01.013
Sharma, R., Rao, V. P., Varshney, R. K., Prasanth, V. P., Kannan, S., and Thakur, R. P. (2010). Characterization of pathogenic and molecular diversity in Sclerospora graminicola, the causal agent of pearl millet downy mildew. Arch. Phytopathol. Plant Protect 43, 538–551. doi: 10.1080/03235400801939896
Sharma, Y. K., Yadav, S. K., and Khairwal, I. S. (2007). Evaluation of pearl millet germplasm lines against downy mildew incited by Sclerospora graminicola in western Rajasthan. J. SAT Agric. 3: 2.
Shivhare, R., and Lata, C. (2016). Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Sci. Rep. 6: 23036. doi: 10.1038/srep23036
Singh, S. D., and Gopinath, R. (1985). A seedling inoculation technique for detecting downy mildew resistance in pearl millet. Plant Dis. 69, 582–584. doi: 10.1094/PD-69-582
Singh, S. D., King, S. B., and Werder, J. (1993). Downy Mildew Disease of Pearl Millet. Patancheru: ICRISAT.
Singh, S. D., Wilson, J. P., Navi, S. S., Talukdar, B. S., Hess, D. E., and Reddy, K. N. (1997). Screening Techniques and Sources of Resistance to Downy Mildew and Rust in Pearl Millet. Patancheru: ICRISAT.
Soman, P., Jayachandran, R., and Bidinger, F. R. (1987). Uneven variation in plant to plant spacing in pearl millet. Agron. J. 79, 891–895. doi: 10.2134/agronj1987.00021962007900050027x
Soman, P., and Peacock, J. M. (1985). A laboratory technique to screen seedling emergence of sorghum and pearl millet at high soil temperature. Exp. Agric. 21, 335–341. doi: 10.1017/S0014479700013168
Stich, B., Haussmann, B. I. G., Pasam, R., Bhosale, S., Hash, C. T., Melchinger, A. E., et al. (2010). Patterns of molecular and phenotypic diversity in pearl millet [Pennisetum glaucum (L.) R. Br.] from West and Central Africa and their geographical and environmental parameters. BMC Plant Biol. 10: 216. doi: 10.1186/1471-2229-10-216
Stomph, T. J. (1990). Seedling Establishment in Pearl Millet [Pennisetum glaucum (L.) R. Br.]: the Influence of Genotype, Physiological Seed Quality Soil Temperature and Soil Water. Ph.D. thesis, University of Reading, Reading.
Sudisha, J., Kumar, S. A., Thakur, R. P., Rao, V. P., and Shetty, H. K. (2009). Molecular characterization of Sclerospora graminicola, the incitant of pearl millet downy mildew using ISSR markers. J. Phytopathol. 157, 748–755. doi: 10.1111/j.1439-0434.2009.01547.x
Supriya, A., Senthilvel, S., Nepolean, T., Eshwar, K., Rajaram, V., Shaw, R., et al. (2011). Development of a molecular linkage map of pearl millet integrating DArT and SSR markers. Theor. Appl. Genet. 123, 239–250. doi: 10.1007/s00122-011-1580-1
Szabolcs, I. (1990). “Impact of climate change on soil attributes: influence on salinization and alkalinization,” in Soils on a Warmer Earth, eds H. W. Scharpenseel, M. Schomaker, and A. Ayoub (Amsterdam: Elsevier), 61–69.
Thakur, R. P., and King, S. B. (1988a). Registration of four ergot resistant germplasms of pearl millet. Crop Sci. 28: 382. doi: 10.2135/cropsci1988.0011183X002800020057x
Thakur, R. P., and King, S. B. (1988b). Registration of six smut resistant germplasms of pearl millet. Crop Sci. 28, 382–383. doi: 10.2135/cropsci1988.0011183X002800020058x
Thakur, R. P., King, S. B., King, Rai, K. N., and Rao, V. P. (1992). Identification and Utilization of Smut Resistance in Pearl Millet. Patancheru: ICRISAT.
Thakur, R. P., Rai, K. N., Khairwal, I. S., and Mahala, R. S. (2008). Strategy for downy mildew resistance breeding in pearl millet in India. J. SAT Agric. Res. 6, 1–11.
Thakur, R. P., Sharm, R., and Rao, V. P. (2011). Screening Techniques for Pearl Millet Diseases. Patancheru: ICRISAT, 56.
Thakur, R. P., Shetty, H. S., and Khairwal, I. S. (2006). Pearl millet downy mildew research in India: progress and perspectives. J. SAT Agric. 2: 6.
Thakur, R. P., Subba Rao, K. V., and Williams, R. J. (1983). Evaluation of new field screening technique for smut resistance in pearl millet. Phytopathology 73, 1255–1258. doi: 10.1094/Phyto-73-1255
Thakur, R. P., and Williams, R. J. (1980). Pollination effects on pearl millet ergot. Phytopathology 70, 80–84. doi: 10.1094/Phyto-70-80
Thakur, R. P., Williams, R. J., and Rao, V. P. (1982). Development of resistance to ergot in pearl millet. Phytopathology 72, 406–408. doi: 10.1094/Phyto-72-406
Upadhyaya, H. D., Gowda, C. L. L., Reddy, K. N., and Singh, S. (2009). Augmenting the pearl millet core collection for enhancing germplasm utilization in crop improvement. Crop Sci. 49, 573–580. doi: 10.2135/cropsci2008.06.0378
Upadhyaya, H. D., Reddy, K. N., Ahmed, M. I., Kumar, V., Gumma, M. K., and Ramachandran, S. (2015). Geographical distribution of traits and diversity in the world collection of pearl millet [Pennisetum glaucum (L.) R. Br.] landraces conserved at ICRISAT genebank. Genet. Res. Crop Evol. 1–17. doi: 10.1007/s10722-016-0442-8
Upadhyaya, H. D., Reddy, K. N., and Gowda, C. L. L. (2007). Pearl millet germplasm at ICRISAT gene bank-status and impact. J. SAT Agric. 3:1.
Upadhyaya, H. D., Yadav, D., Reddy, K. N., Gowda, C. L. L., and Singh, S. (2011). Development of pearl millet minicore collection for enhanced utilization of germplasm. Crop Sci. 51, 217–223. doi: 10.2135/cropsci2010.06.0336
Vadez, V., Hash, T., Bidinger, F. R., and Kholova, J. (2012). Phenotyping pearl millet for adaptation to drought. Front. Physiol. 3:386. doi: 10.3389/fphys.2012.00386
Varallyay, G. (1994). Climate Change, Soil Salinity and Alkalinity, NATO ASI Series, Vol. 23. London: Springer, 3–11.
Veena, M., Melvin, P., Prabhu, S. A., Shailashree, S., Shetty, S. H., and Kini, K. R. (2016). Molecular cloning of a coiled- coil- nucleotide-binding- site leucine- rich repeat gene from pearl millet and its expression pattern in response to the downy mildew pathogen. Mol. Biol. Rep. 43, 117–128. doi: 10.1007/s11033-016-3944-8
Vengadessan, V., Rai, K. N., Bapu, J. R. K., Hash, C. T., Bhattacharjee, R., Senthilvel, S., et al. (2013). Construction of genetic linkage map and QTL analysis of sink-size traits in pearl millet (Pennisetum glaucum). ISRN Genet. 2013:1–14. doi: 10.5402/2013/471632
Williams, R. J., Singh, S. D., and Pawar, M. N. (1981). An improved field screening technique for downy mildew resistance in pearl millet. Plant Dis. 65, 239–241. doi: 10.1094/PD-65-239
Willingale, J., Mantle, P. G., and Thakur, R. P. (1986). Post-pollination stigmatic constriction,the basis of ergot resistance in selected lines of pearl millet. Phytopathology 76, 536–539. doi: 10.1094/Phyto-76-536
Wilson, J. P., and Burton, G. W. (1991). Registration of Tift 3 and Tift 4 rust resistant pearl millet germplasms. Crop Sci. 31:1713. doi: 10.2135/cropsci1991.0011183X003100060090x
Wilson, J. P., Hess, D. E., Hanna, W. W., Kumar, K. A., and Gupta, S. C. (2004). Pennisetum glaucum subsp. monodii accessions with Striga resistance in West Africa. Crop Prot. 23, 865–870. doi: 10.1016/j.cropro.2004.01.006
Winkel, T., Renno, J. F., and Payne, W. A. (1997). Effect of the timing of water deficit on growth, phenology and yield of pearl millet (Pennisetum glaucum (L.) R. Br.) grown in Sahelian conditions. J. Exp. Bot. 48, 1001–1009. doi: 10.1093/jxb/48.5.1001
Witcombe, J. R., Rao, M. N. V. R., Raj, A. G. B., and Hash, C. T. (1997). Registration of ‘ICMV 88904’ pearl millet. Crop Sci. 37:10221023. doi: 10.2135/cropsci1997.0011183X0037
Wu, X., Wang, D., Bean, S. R., and Wilson, J. P. (2005). Ethanol production from pearl millet using Saccharomyces cerevisiae. Cereal Chem. 83, 127–131. doi: 10.1094/CC-83-0127
Yadav, A. K., Arya, R. K., and Narwal, M. S. (2014). Screening of pearl millet F1 hybrids for heat tolerance at early seedling stage. Adv. Agric. 2014: 17.
Yadav, M. S., and Duhan, J. C. (1996). Screening of pearl millet genotypes for resistance to smut. Plant Dis. Res. 11, 95–96.
Yadav, O. P. (2004). CZP 9802—a new drought-tolerant cultivar of pearl millet. Indian Farming 54, 15–17.
Yadav, O. P., and Bidinger, F. R. (2008). Dual-purpose landraces of pearl millet (Pennisetum glaucum) as sources of high stover and grain yield for arid zone environments. Plant Genet. Res. 6, 73–78. doi: 10.1017/S1479262108993084
Yadav, O. P., Mitchell, S. E., Fulton, T. M., and Kresovich, S. (2008). Transferring molecular markers from sorghum, rice and other cereals to pearl millet and identifying polymorphic markers. J. SAT Agric. Res. 6, 1–4.
Yadav, O. P., Mitchell, S. E., Zamora, A., Fulton, T. M., and Kresovich, S. (2007). Development of new simple sequence repeat markers for pearl millet. SAT eJournal 3, 34–37.
Yadav, O. P., Rai, K. N., and Gupta, S. K. (2012). “Improving crop productivity in sustainable agriculture,” in Pearl Millet: Genetic improvement for tolerance to abiotic stresses, eds N. Tuteja, S. S. Gill, and R. Tuteja (Weinheim: Wl1ey⋅VCH Verlag GmbH & Co. KGaA publisher), 261–288.
Yadav, O. P., Weltzien-Rattunde, E., and Bidinger, F. R. (2003). Genetic variation in drought response of landraces of pearl millet (Pennisetum glaucum (L.) R. Br.). Indian J. Genet. 63, 37–40.
Yadav, O. P., Weltzien-Rattunde, E., and Bidinger, F. R. (2004). Diversity among pearl millet landraces collected in north-western. India Ann. Arid Zone 43, 45–53.
Yadav, R. S., Bidinger, F. R., Hash, C. T., Yadav, Y. P., Bhatnagar, S. K., and Howarth, C. J. (2003). Mapping and characterization of QTL × E interactions for traits determining grain and stover yield in pearl millet. Theor. Appl. Genet. 106, 512–520. doi: 10.1007/s00122-002-1081-3
Yadav, R. S., Hash, C. T., Bidinger, F. R., Cavan, G. P., and Howarth, C. L. (2002). Quantitative trait loci associated with traits determining grain and stover yield in pearl millet under terminal drought stress conditions. Theor. Appl. Genet. 104, 67–83. doi: 10.1007/s001220200008
Yadav, R. S., Hash, C. T., Bidinger, F. R., Devos, K. M., and Howarth, C. J. (2004). Genomic regions associated with grain yield and aspects of post-flowering drought tolerance in pearl millet across stress environments and testers backgrounds. Euphytica 136, 265–277. doi: 10.1023/B:EUPH.0000032711.34599.3a
Yadav, R. S., Sehgal, D., and Vadez, V. (2010). Using genetic mapping and genomics approaches in understanding and improving drought tolerance in pearl millet. J. Exp. Bot. 62, 397–408. doi: 10.1093/jxb/erq265
Zegada-Lizarazu, W., and Iijima, M. (2005). Deep root water uptake ability and water use efficiency of pearl millet in comparison to other millet species. Plant Prod. Sci. 8, 454–460. doi: 10.1626/pps.8.454
Keywords: diversity, downy mildew, germplasm, marker-assisted breeding, panicoid, quantitative trait loci, stress tolerance, terminal drought
Citation: Shivhare R and Lata C (2017) Exploration of Genetic and Genomic Resources for Abiotic and Biotic Stress Tolerance in Pearl Millet. Front. Plant Sci. 7:2069. doi: 10.3389/fpls.2016.02069
Received: 30 September 2016; Accepted: 27 December 2016;
Published: 23 January 2017.
Edited by:
Rattan Yadav, Aberystwyth University, UKReviewed by:
Andrew Doust, Oklahoma State University-Stillwater, USARakesh Srivastava, International Crops Research Institute for the Semi-Arid Tropics (ICRISAT), India
Desalegn D. Serba, Kansas State University, USA
Copyright © 2017 Shivhare and Lata. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Charu Lata, Y2hhcnVsYXRhQG5icmkucmVzLmlu; Y2hhcnVsYXRhMTRAZ21haWwuY29t
†These authors have contributed equally to this work.
 Radha Shivhare
Radha Shivhare Charu Lata
Charu Lata