- 1The Institute of Agriculture, The University of Western Australia, Perth, WA, Australia
- 2Department of Biochemistry, Cell and Molecular Biology of Plants, Estacion Experimental del Zaidin, Spanish National Research Council, Granada, Spain
- 3Centre for Environment and Life Sciences, Agriculture and Food, Commonwealth Scientific and Industrial Research Organisation, Floreat, WA, Australia
Vicilins (7S globulins) are seed storage proteins and constitute the main protein family in legume seeds, particularly in narrow-leafed lupin (Lupinus angustifolius L.; NLL), where seven vicilin genes, called β1- to β7-conglutin have been identified. Vicilins are involved in germination processes supplying amino acids for seedling growth and plant development, as well as in some cases roles in plant defense and protection against pathogens. The roles of NLL β-conglutins in plant defense are unknown. Here the potential role of five NLL β-conglutin family members in protection against necrotrophic fungal pathogens was investigated and it was demonstrated that recombinant purified 6xHis-tagged β1- and β6-conglutin proteins exhibited the strongest in vitro growth inhibitory activity against a range of necrotrophic fungal pathogens compared to β2, β3, and β4 conglutins. To examine activity in vivo, two representative necrotrophic pathogens, the fungus Sclerotinia sclerotiorum and oomycete Phytophthora nicotianae were used. Transient expression of β1- and β6-conglutin proteins in Nicotiana benthamiana leaves demonstrated in vivo growth suppression of both of these pathogens, resulting in low percentages of hyphal growth and elongation in comparison to control treated leaves. Cellular studies using β1- and β6-GFP fusion proteins showed these conglutins localized to the cell surface including plasmodesmata. Analysis of cellular death following S. sclerotiorum or P. nicotianae revealed both β1- and β6-conglutins suppressed pathogen induced cell death in planta and prevented pathogen induced suppression of the plant oxidative burst as determined by protein oxidation in infected compared to mock-inoculated leaves.
Introduction
Plants are under constant exposure to potential microbial pathogens. One of the mechanisms they employ to defend themselves is via the production of bioactive antimicrobial proteins (AMPs). In addition to plants, other organisms may produce a diverse array of AMPs for defense purposes and these can confer a high level of antimicrobial activity against competing microorganisms such as bacteria, viruses, protozoa, filamentous fungi and yeasts (Niyonsaba et al., 2009; Tam et al., 2015). In plants AMPs can play a role in constitutive immunity or can be induced upon pathogen attack. Inducible responses can include the expression of pathogen-related (PR) proteins such as the enzymes (1-3)-β-glucanases (PR-2), chitinases (PR-3, -4, -8, and -11), peroxidases (PR-9) and oxalate oxidases (PR-16 and -17; Thatcher et al., 2005; Tam et al., 2015). In addition, it has been proposed that proteins involved in the delivery of storage and energy requirements to plant embryos during germination may also be involved in defense responses (Chrispeels and Raikhel, 1991; Marcus et al., 1999; Gábrišová et al., 2016). For example, members of the following storage protein families 2S albumins, Kunitz proteinase inhibitors, plant lectins and vicilins or vicilin-like proteins (including 7S globulins and β-conglutins; De Souza Candido et al., 2011).
Plant storage proteins can be classified into vegetative storage proteins and seed storage proteins, where the latter can represent a significant proportion of seed composition (Gomes et al., 2014). Storage proteins perform essential roles in plant survival. They provide a source of amino acids that can be mobilized and utilized for maintenance and growth during both seed embryonic developmental, and germination stages (Zienkiewicz et al., 2011; Tan-Wilson and Wilson, 2012; Jimenez-Lopez et al., 2016). These proteins accumulate in cellular storage vacuoles of seeds, nuts, and kernels; stem parenchyma of trees; grains and legumes; and some roots and tubers. The vicilins, also called conglutins in some legume species, constitute a class of proteins abundantly found as reserves in seeds of leguminous and non-leguminous plants, representing as much as 70 to 80% of total protein in the seeds of these plants (Duranti and Gius, 1997). Their structure consists of a trimeric organization, and unlike most plant storage proteins individual subunits with molecular masses typically around 15–70 kDa (Melo et al., 1994), NLL’s individual subunits are larger and range from 150 to 170 kDa in size (Argos et al., 1985).
Vicilins appear to play multifunctional roles, acting as an energy source and providing amino acids during the germination process, while in some cases, also being involved in defense responses against fungi and insects (Yunes et al., 1998). This includes for example, vicilins from the legumes Vigna unguiculata (cowpea), V. radiata (mung bean), Phaseolus vulgaris (common bean) and Canavalia ensiformis (jack bean; Gomes et al., 1997, 1998; Oliveira et al., 1999; Coda et al., 2008). The insecticidal activity of vicilins relates to their capacity to bind chitinous structures, thereby interfering with insect development, as shown for cowpea and the cowpea seed beetle (Callosobruchus maculatus; Sales et al., 2001). This chitin-binding activity can also inhibit yeast and fungal growth (Gomes et al., 1998). The potency of vicilin antifungal activity varies among plant species. For example, Gomes et al. (Gomes et al., 1998) extracted a vicilin from V. unguiculata showing inhibitory activity between 90 and 100% against the yeast S. cerevisiae, in addition to interfering with spore germination of the fungi Fusarium solani, F. oxysporum, Colletotrichum musae, Phytophthora capsici, Neurospora crassa and Ustilago maydis sporidia. Vicilin extracted from V. radiata seeds showed 65% inhibitory activity against Candida albicans (Gomes et al., 1998), whereas vicilin isolated from the non-legume Malva parviflora Malva (an annual or perennial herb) showed inhibitory activity against Phytophthora infestans (Wang et al., 2001).
Narrow-leafed lupin (Lupinus angustifolius L.; NLL) is a recently domesticated important pulse crop, and increasingly popular due to its wide range of agricultural and health benefits (Berger et al., 2013). The NLL grain constitutes an important source of protein for humans and animals with low starch content and free of gluten (reviewed in Foley et al., 2011). In NLL the seed storage proteins are collectively called conglutins and fall into four sub-families called α, β, γ, and δ-conglutins (Foley et al., 2011, 2015). In addition to dissection of lupin-based health benefits, the identification of lupin seed storage proteins playing roles in resistance against pathogens is of interest. Recently, antifungal activity from a multifunctional glyco-oligomer with 210 kDa, mainly composed by BLAD (banda de Lupinus albus doce), a 20 kDa polypeptide, a stable intermediary product of β-conglutin catabolism, was demonstrated and found to exclusively accumulate in the cotyledons of Lupinus species (Monteiro et al., 2015).
The recent development of a reference NLL genome assembly (Hane et al., 2016)1 and extensive RNA expression analysis from various tissues including seeds (Foley et al., 2015; Kamphuis et al., 2015) facilitated the identification of 16 conglutin genes, where the β-conglutin family was the most abundant, representing 56% of the total seed storage protein RNA expression levels (Foley et al., 2011). The NLL β-conglutin family comprises seven members, namely β1- to β7-conglutin (Foley et al., 2011). These β-conglutins share sequence identities ranging from 77.4 to 94.7%, reflected presumably in differential structure-functionality between some of them (Jin et al., 2014), and are highly expressed in the seeds compared to other NLL tissues (Foley et al., 2015).
Pathogenic fungi of lupins, as is the case for many other grain legume crops, cause substantial annual crop losses and are of major economic significance (Rubiales et al., 2015). For example, Sclerotinia stem rot, Rhizoctonia barepatch, Phytophthora root rot, and anthracnose stem and pod blights caused by Colletotrichum lupini causes several million dollars of losses in Australia, the largest producer of NLL globally (Sinden et al., 2004; Murray and Brennan, 2012). Considering the demonstrated antifungal activity of some seed storage proteins from several legume species, it was of interest to determine if seed storage proteins such as β-conglutins from NLL may also have roles in protection against fungal pathogens. Therefore the antifungal activity of NLL β-conglutins was examined using both in vitro and in planta assays for protection against fungal and oomycete pathogen growth known to induce necrotic host tissue damage. Furthermore, insight into the potential inhibitory mechanisms by which these proteins act against pathogens was obtained through an assessment of their subcellular localization and impact on plant oxidative processes.
Materials and Methods
Plant Material and Growth Conditions
Plant experiments were conducted with Nicotiana benthamiana accession “lab” (Bally et al., 2015) in temperature controlled growth rooms as described by Petrie et al. (2010). Plants were grown under a 16-h light/8-h dark cycle at 22°C.
Fungal Isolates
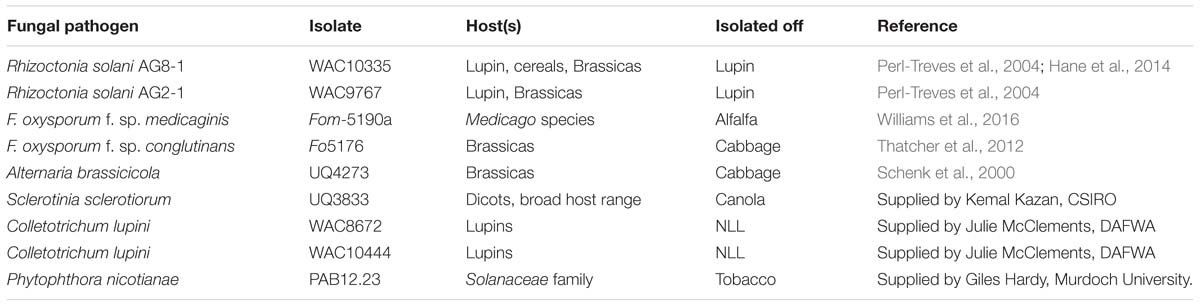
Details of fungal isolates are listed in Table 1 and were maintained as pure cultures with Rhizoctonia solani, Alternaria brassicicola, F. oxysporum, Phytophthora nicotianae and C. lupini isolates grown on 1/2 strength Potato Dextrose Agar (PDA), and S. sclerotiorum on 1/8 strength PDA. Spores, mycelia or sclerotia were inoculated on PDA plates, which were placed at room temperature in the dark until plates were fully covered by the pathogen. Mycelial plugs from these plates were used for subsequent experiments.
Construction of Expression Plasmids
β1- and β6-conglutins were overexpressed using the pET28b construct (Novogen)2 that contains an N-terminal polyhistidine (6xHis) tag. pUC57 vectors carrying synthesized β1, β2, β3, β4, or β6 conglutin sequences based on Genbank HQ670409 (β1), HQ670410 (β2), HQ670411 (β3), HQ670412 (β4) and HQ670414 (β6) sequences but altered for optimum bacterial codon usage with NcoI/XhoI restriction enzyme linkers were synthesized and constructed by GenScript3 (Supplementary Figure S1). The bacterial expression vectors for β-conglutins were obtained via NcoI/XhoI digestion of respective pUC57-β-conglutin constructs followed by ligation of the β-conglutin fragments into the pET28b vector.
Overexpression and Purification of NLL β-Conglutin Proteins
All β-conglutin proteins were expressed in RosettaTM 2(DE3) pLysS SinglesTM Competent Cells (Novogen). Protein expression was performed using an auto-induction method (Studier, 2005). Briefly, a single clone containing the expression construct was isolated and grown for 20 h in LB plus kanamycin at 50 μg/mL at 37°C and continuous shaking (200 rpm). The culture was diluted 1:150 in ZYM-5052 medium and grown for a further 5 h until the cell density reached an OD600 of 0.7. The cells were then induced to overexpress the proteins by adjusting the temperature to 19°C for another 20 h. Cells were collected by centrifugation at 5000 × g at 4°C. The bacterial cell pellet was rinsed two times with phosphate buffered saline (PBS), pH 7.5, removing the supernatant, then flash frozen in liquid nitrogen and stored at -80°C until further use.
Purification of Recombinant β1- and β6-Conglutin Proteins
Protein purification from bacterial pellets was performed following the manufacturers’ recommendations for His-tagged proteins (Qiagen)4. Briefly, the steps consisted of lysing cells followed by nickel affinity chromatography using Ni-NTA spin columns, and histidine (6xHis) tags at the N-terminal part of the β-conglutin proteins. After elution of 6xHis-tagged proteins from the column with an increasing imidazole concentration gradient (10–300 mM), 2.5 mL fractions were collected. Fractions containing protein were analyzed using SDS-PAGE and fractions showing a single band corresponding to the expected molecular weight were pooled, and dialyzed five times against Tris-HCl 100 mM, pH 7.5, 150 mM NaCl to eliminate the imidazole reagent. The protein was concentrated using a 30 kDa Amicon centrifuge filter (Millipore)5. The aliquots were flash-frozen in liquid nitrogen and kept at -80°C until further use. Protein purities were >95% as determined by densitometry analysis of the SDS-PAGE gel image. An aliquot of each protein was used to measure their concentration using Bradford assays (BioRad, Hercules, CA, USA) using bovine serum albumin (BSA) as a standard. The β-conglutins purifications yields ranged between 10–15 mg/mL.
β-Conglutin Antibody Production
The peptide sequence Nt – VDEGEGNYELVGIR – Ct, was chosen as this region was 100% homologous among all NLL β-conglutins and did not share any significant homology to other known lupin sequences. This peptide was generated by Agrisera6 and was used to immunize rabbits and to produce polyclonal antiserum (Agrisera). The rabbit immune serum was affinity-purified against the same synthetic peptide.
SDS-PAGE and Immunoblotting
SDS-PAGE and Immunoblotting were performed as previously described (Foley et al., 2015).
In vitro Assays for Fungal Growth Inhibition
A disk diffusion method was performed on 90 mm petri dishes containing PDA to test the sensitivity of different fungi strains toward the β-conglutin proteins. Fungal isolates were initially grown on PDA plates as described previously at 21°C until mycelial growth had developed. A mycelial plug was then taken from the growing edge of the colony and placed in the center of a new full-strength PDA plate in which sterile blank paper disks (12.7 mm diameter) containing 800 μg of purified β-conglutin protein (dissolved in BSA buffer) or buffer only control were placed 30 mm away. For in vitro assay of C. lupini, 1 mg of purified β6-conglutin protein was used. The plates were incubated in the dark at 21°C and the zone of fungal inhibition around the disks recorded over 30 days. Assays were performed in triplicate.
Antifungal activity of β-conglutin proteins were expressed as the IC50 (μM) values for the fungi tested in Table 1. The mycelial growth inhibition assays were used to determine the concentration required for 50% growth inhibition (IC50), using a β-conglutin protein concentration range from 5 to 125 μM (5, 10, 15, 20, 25, 35, 50, 75, and 125 μM) for each β-conglutin. Results were expressed as mean ± standard deviation (SD). To determine the statistical significant differences of the β-conglutins antifungal activity on the growth of these fungal pathogens, the data was analyzed using statistical package SPSS 15.0 (SPSS, Inc., Chicago, IL, USA). Significant differences between the mean values of each cohort were determined using Tukey Kramer HSD test (p < 0.05). To characterize the antifungal activity of β-conglutins further, a subsequent experiment with the necrotrophic fungal pathogen of NLL causing anthracnose disease (C. lupini isolates WAC8672 and WAC10444) was conducted, where the antifungal activity of β6-conglutin was determined by placing a mycelial plug in the center of a PDA plate and radial outgrowth determined over 14 days. For each time point significant differences were determined using a student’s t-test using the JMP software v7.0 (SAS Institute).
Agroinfiltration of Nicotiana benthamiana Leaves
Four-week-old N. benthamiana plants grown at 22–24°C in culture rooms were used for Agrobacterium tumefaciens-mediated transient expression as described previously (Sparkes et al., 2006). The β1- and β6-conglutin coding sequence were cloned into the vector pMDC83 to generate C-terminal GFP fusion proteins and transformed into A. tumefaciens AGL1. Transformed A. tumefaciens AGL1 were cultured at 28°C until stationary phase (∼24 h), washed and resuspended in infiltration medium (50 mM MES, 0.5% (w/v) glucose, 100 μM acetosyringone (Sigma-Aldrich7 pH 5.6). The bacterial suspension was inoculated using a 1-mL syringe without a needle by gentle pressure through a <1 mm hole punched on the lower epidermal surface of the upper leaves of N. benthamiana plants. Following infiltration, plants were incubated under normal growth conditions at 22–24°C. This protocol was used for in vivo fungal growth inhibition assays, oxyblot assays, and subcellular localization studies.
Trypan Blue Staining for Fungal Hyphae and Dead Plant Cells
Forty hours after Agrobacterium infiltration of β1- or β6-conglutin protein constructs into N. benthamiana plants, agar plugs containing hyphae of S. sclerotiorum or P. nicotianae were placed on the infiltrated leaf areas (control and β1- or β6-conlgutin overexpression) and plants were incubated in a growth chamber at 18°C for 3 days under a 16 h long-day light regime. Leaves were assessed for visible necrotic disease progression, then detached and further visualized after lactophenol trypan blue staining based on Keogh et al. (1980). Briefly, mature fourth leaves of N. benthamiana containing visible infection sites (tissue necrosis) were cleared with acetic acid: ethanol (1:1 v/v) then stained for 1–2 h in lactophenol (10 mL phenol; 10 mL lactic acid, 10 mL water) with 0.05% (w/v) trypan blue at 60°C. Excess staining was removed with lactic acid: water (1:1 v/v) until leaves were clear. Leaves were examined by light microscopy on a Nikon N400-M light microscope (Nikon, Tokyo, Japan). Control areas were infiltrated with GFP-only vectors. The experiment was repeated three times. In each experiment, leaves from 4–6 plants were analyzed for each treatment.
Subcellular Localization of β1- and β6-Conglutin in Plant Cells
Fusion proteins were expressed in 3-week-old N. benthamiana via Agrobacterium infiltration of leaves as previously described (Sparkes et al., 2006). Leaves were excised 3 days following infiltration and mounted with water under a 0.17 mm coverslip and imaged using a Nikon A1Si confocal microscope (Nikon Plan Apo VC 60x NA1.2 water-immersion objective). For GFP imaging, the 488 nm laser line and a 521/50 nm band pass filter was utilized, while a 561 nm laser line and 595/50 nm filter was used for RFP imaging.
Images were analyzed using ImageJ software (Schneider et al., 2012). Images were converted to 8-bit grayscale and the intensity correlation analysis (ICA) method was used for determine the levels of colocalization and by using the JACoP plugin according to (Bolte and Cordelières, 2006). As a control, empty vector was used to transform leaf cells expressing 35S::GFP alone as described previously by Thatcher et al. (2007).
Oxyblot Assays
Proteins were extracted from N. bentamiana leaves infiltrated with GFP, β1-GFP or β6-GFP fusion expression constructs following either control or S. sclerotiorum or P. nicotianae treatments [extraction buffer: 25 mM Tris–HCl, pH 7.0, 0.05% Triton X-100, 1 mM dithiothreitol (DTT), and protease inhibitors (Roche, Basel, Switzerland)]. 25 μg of total proteins were loaded onto 12% polyacrylamide gels for protein separation. Proteins separated by SDS-PAGE were electrotransferred to PVDF membranes. The OxyBlotTM Protein Oxidation Detection Kit (EMD Millipore) was used according to the manufacturer’s instructions for immunoblot detection of carbonyl groups introduced into proteins by reaction with reactive oxygen species (ROS).
Results
In vitro Inhibition of Fungal Growth by β-Conglutin Recombinant Proteins
To assess the potential for antifungal activity in NLL seed storage proteins, we focussed on the most abundant seed storage proteins in the NLL grain, the β-conglutins (Foley et al., 2011). The 6xHis-tag recombinant β-conglutin proteins were expressed in E. coli and purified using nickel affinity chromatography. To confirm the identity of purified β-conglutins, SDS–PAGE analysis of the purified proteins was performed, which indicated a single protein band of approximately 65 kDa, which is the predicted size of β-conglutin (Supplementary Figure S2A). This was followed by immunoblotting using an anti-β-conglutin antibody which confirmed the identity of the recombinant proteins as β-conglutin (Supplementary Figure S2B). We were successful in expressing and purifying the β1, β2, β3, β4 and β6 recombinant proteins but not β5 and β7.
Subsequently, we examined the effect of the purified β-conglutin proteins on the growth rate of a range of phytopathogenic necrotrophic fungi using in vitro bioassays. The fungal pathogens selected included the legume pathogen F. oxysporum forma specialis (f. sp.) medicaginis (Fom-5190a, a root pathogen), the broad host range pathogens R. solani AG8-1 (isolated from lupin) and S. sclerotiorum (isolated from canola) and the brassica-specific pathogens F. oxysporum f. sp. conglutinans (Fo-5176), R. solani AG2-1 and A. brassicicola (Brassicaceae hosts; details of these pathogens are listed in Table 1). The IC50 values (μM) for each of these fungal isolates was determined (Table 2). Overall the β1 and β6 conglutins showed significantly stronger mycelium growth inhibition when compared to β2-, β3-, and β4-conglutin proteins for both R. solani isolates and A. brassicicola by Tukey–Kramer honestly significant difference (HSD) test (P < 0.05). The β1-conglutin showed a significantly stronger growth inhibition to S. sclerotiorum and the two F. oxysporum isolates, compared to β2-, β3-, and β4-conglutin, where β6-conglutin was not significantly different from β1 (Table 2). Overall β1-, and β6-conglutin showed the strongest mycelial growth inhibition to the various pathogens tested, but interestingly, a sequence alignment of the seven β-conglutin proteins showed β6 exhibits the highest sequence identity to other β-conglutin protein isoforms (78–98%) while β1 had an amino acid sequence with the lowest identity (77–81%; Supplementary Figure S3). Control treatment of filter disks with BSA buffer showed no fungal growth inhibition against any of the isolates tested.
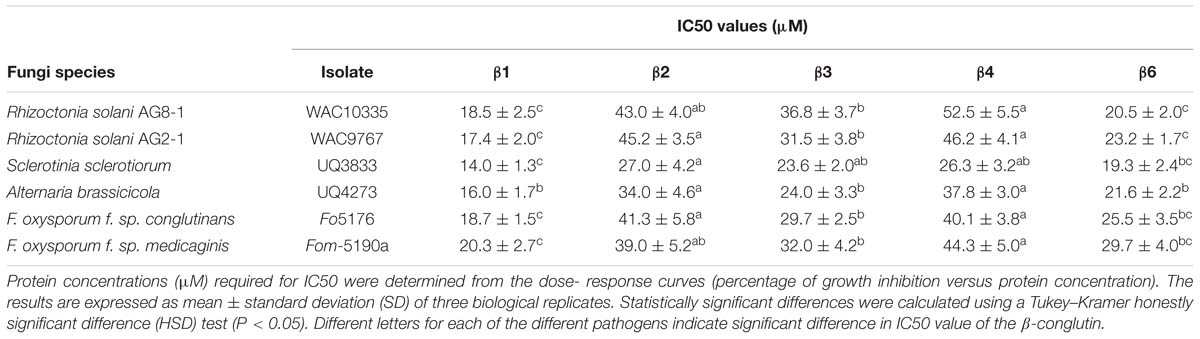
TABLE 2. Antifungal activity of the NLL recombinant purified β1- to β4-conglutins and β6-conglutins.
Based on the β-conglutin protein alignments and IC50 data, we decided to focus on β6-conglutin as a representative member of the β-conglutin family. Further characterization of the in vitro anti-fungal properties of β6-conglutin was performed in a detailed time course experiment against two isolates (WAC8672 and WAC10444) of a major fungal pathogen of lupins, C. lupini which causes anthracnose disease (Fischer et al., 2015). Radial outgrowth of C. lupini mycelium on PDA plates toward Whatman filter disks containing control protein (BSA), β6-conglutin or no protein was recorded. Radial growth inhibition was only observed toward filter disks containing β6-conglutin protein and this occurred from as early as 6 days post-inoculation with the mycelial plug for isolate WAC10444 and 8 days post-inoculation for WAC8672 (Figure 1). Combined with the IC50 data in Table 2, these results indicate recombinant NLL β-conglutins exhibit antifungal activity in vitro against both leaf and root-infecting pathogens of legumes and non-legume hosts.
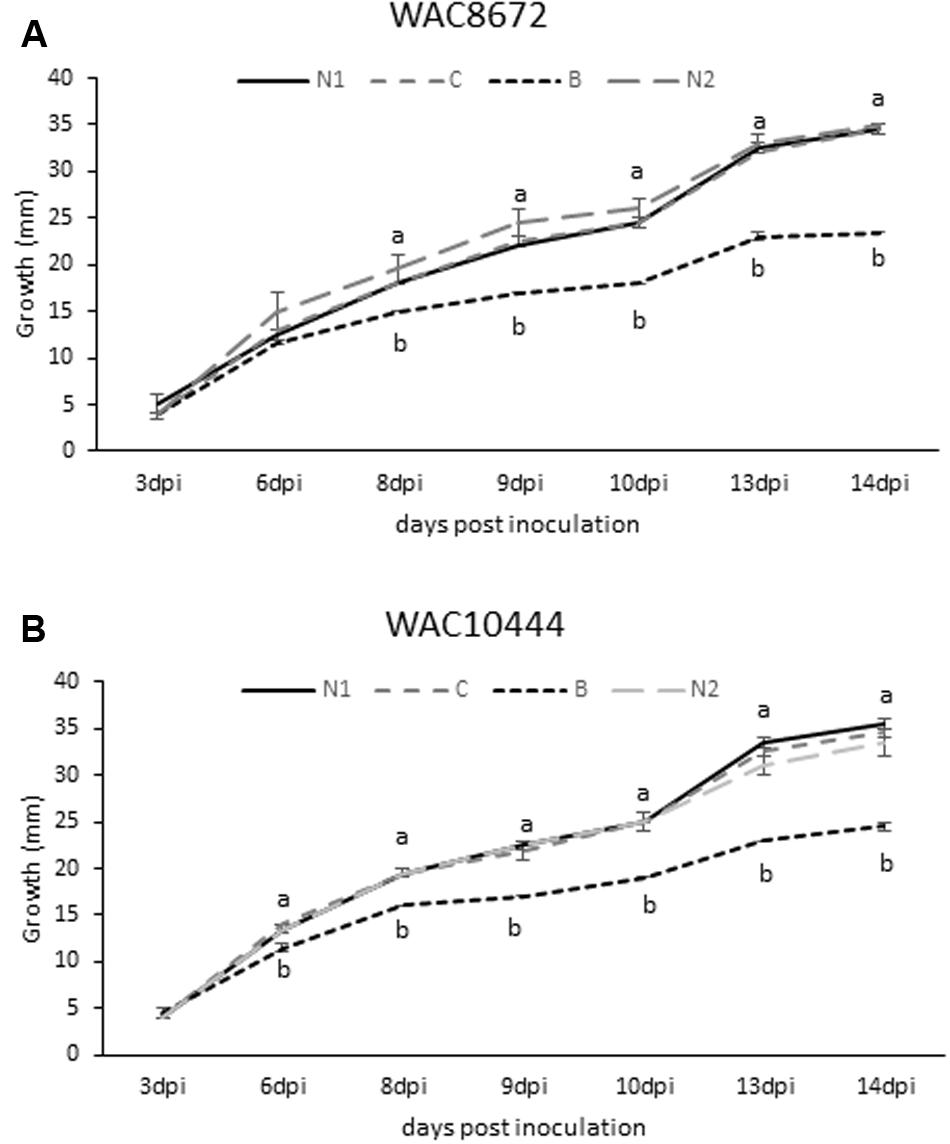
FIGURE 1. Antifungal activity of recombinant β6-conglutin in in vitro bioassays against the lupin pathogen Colletotrichum lupini. Significant growth inhibition toward β6-conglutin was observed from as early as (A) 8 dpi for isolate WAC8672 and (B) 6 dpi for isolate WAC10444. Means and standard error of three biological replicates. Letters indicate significant differences for a given day after inoculation (dpi) with the mycelial plug by student’s t-test. C, control containing 1 mg BSA; N1, no protein; B, 1 mg β6-conglutin; N2, no protein.
β1- and β6-Conglutins Exhibit in planta Anti-fungal and Oomycete Activity
To examine the effect of NLL β6-conglutin in planta, we selected the N. benthamiana infiltration system as a model for assessing the functionality of proteins against various phytopathogens (Ma et al., 2012). This involved Agrobacterium-mediated infiltration into leaves of N. benthamiana plants followed by assessment of antifungal activity in disease assays. The broad host range leaf pathogen S. sclerotiorum was chosen which is readily amenable to N. benthamiana leaf disease assays and secretes the non-host selective toxin oxalic acid to induce disease symptom development (Kim et al., 2008; Williams et al., 2011). In addition, β6-conglutin showed strong inhibition of this pathogen’s growth as compared to other fungal pathogens tested in our in vitro assays (Table 2). The Agrobacterium-infiltration system was used to transiently express β6-GFP or a GFP-only control. The GFP control was infiltrated into one half of the leaf with β6-GFP infiltrated into the other leaf half. Forty-eight hours after infiltration, leaves were inoculated with S. sclerotiorum and progression of lesions was observed over 72 h (Figure 2). In the GFP-only control, necrosis was apparent within 24 h of S. sclerotiorum inoculation with the size of the necrotic lesions progressing rapidly to engulf half of the leaf by 72 h. In stark contrast, there was only limited necrotic damage in the leaves expressing the β6-GFP protein. As β1-conglutin demonstrated strong antifungal activity in vitro yet exhibits the least amino acid identity amongst the NLL β-conglutins, we also assayed β1-GFP in the above experiments. As with β6-GFP, β1-GFP infiltrated leaves also exhibited limited necrotic damage after S. sclerotiorum inoculation (Supplementary Figure S4).
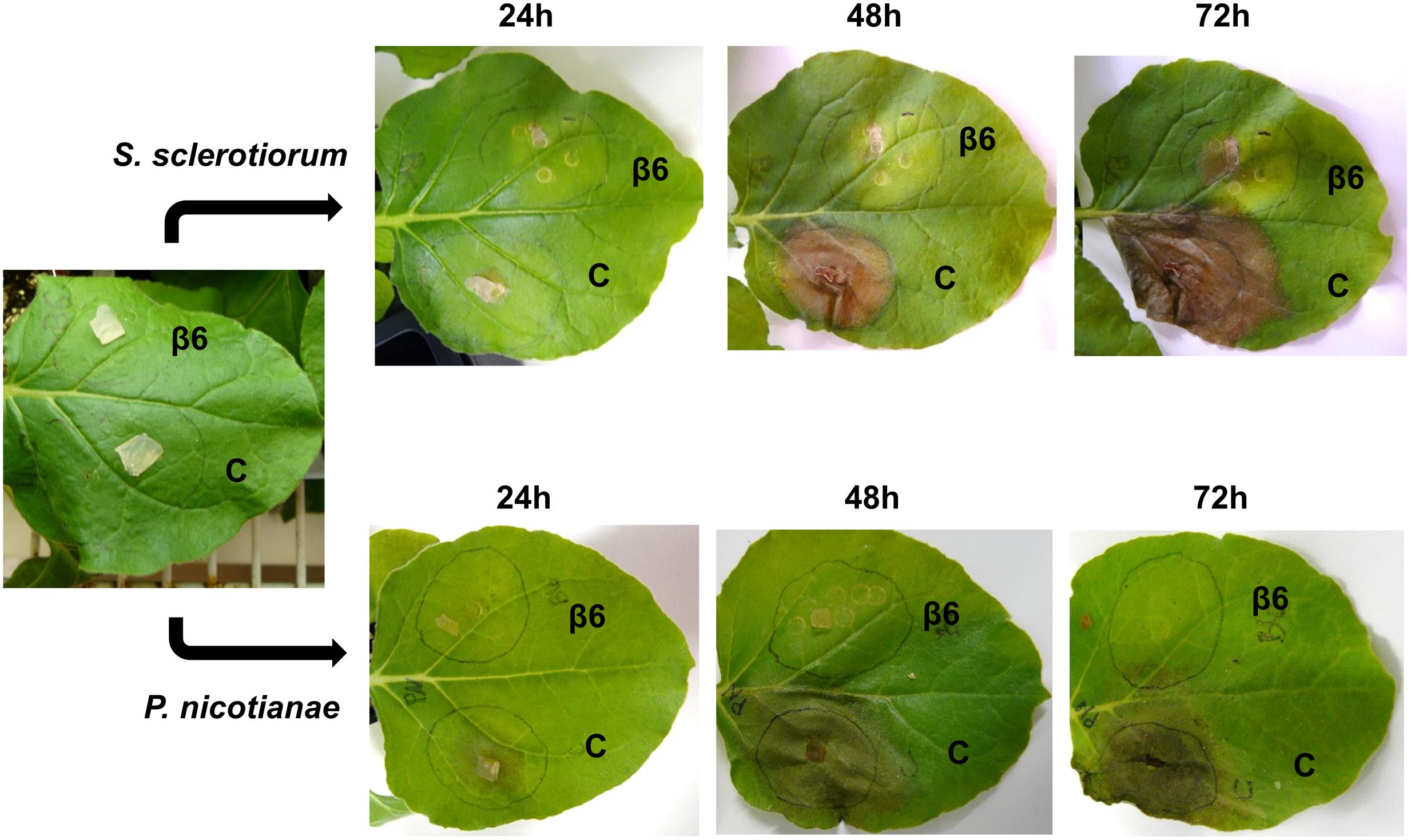
FIGURE 2. Recombinant β6-conglutin exhibits in planta anti-fungal and oomycete activity. Shown are representative images of Agrobacterium infiltrated N. benthamiana leaves expressing recombinant β6-conglutin proteins and subsequently inoculated with either S. sclerotiorum or P. nicotianae. The experiment was repeated three times with similar results. C, control agroinfiltration of the leaf area with Agrobacterium expressing GFP only; β6, agroinfiltration of the leaf area with Agrobacterium expressing GFP tagged β6-conglutin.
The soil-borne oomycete pathogen of N. benthamiana, P. nicotianae, was also assayed. P. nicotianae is a hemibiotrophic pathogen that causes root rot, leaf necrosis and stem lesions (Liu et al., 2016). As with the S. sclerotiorum assays, both β6- and β1-conglutin strongly inhibited lesion development by P. nicotianae in our N. benthamiana Agrobacterium-infiltration disease assays (Figure 2 and Supplementary Figure S4). At 72 h post S. sclerotiorum or P. nicotianae inoculation, β6-conglutin infiltrated leaf zones exhibited a 71.7 and 85.7% reduction in lesion size, respectively, relative to control treated leaf zones. Similar reductions were recorded for β1-conglutin infiltrated leaf zones (94.2 and 90.3%).
β1- and β6-Conglutin Reduce Pathogen Growth and Pathogen Induced Cell Death In planta
The striking inhibition of S. sclerotiorum and P. nicotianae induced lesions on N. benthamiana leaves expressing β1- or β6-conglutin suggests these pathogens are unable to grow or their growth is severely impaired by these β-conglutins. To examine fungal/oomycete growth we challenged N. benthamiana β-conglutin expressing leaves with pathogen and allowed 72 h for disease symptom development in controls, then assessed the leaves and mycelial growth microscopically after staining with trypan blue, which stains dead plant cells (van Wees, 2008). In leaves expressing β6-GFP or β1-GFP we observed strong inhibition of the S. sclerotiorum and P. nicotianae mycelial growth compared to controls (Figure 3 and Supplementary Figure S5). There was evidence of aggregated cells with short pseudohyphae, particularly at the site of inoculation, however, these were few and sparse in comparison to controls. Combined, our results suggest β1- and β6-conglutins from NLL inhibit hyphal growth of a range of phytopathogenic oomycete and fungi, both in vitro and in vivo.
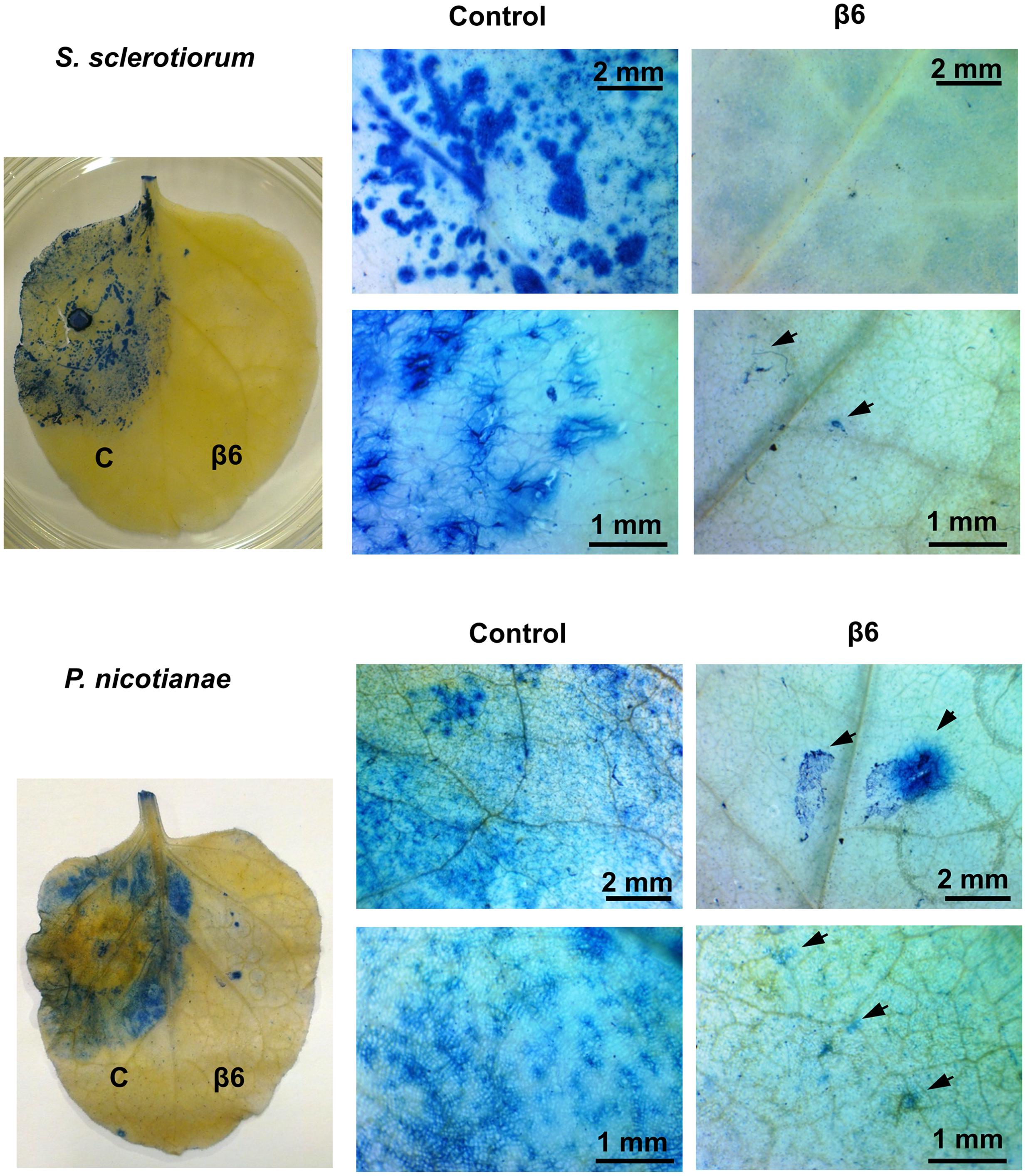
FIGURE 3. Recombinant β6-conglutin reduces pathogen growth and pathogen induced cell death in planta. Shown are representative images of Agrobacterium infiltrated N. benthamiana leaves expressing recombinant β6-conglutin proteins and subsequently inoculated either S. sclerotiorum or P. nicotianae. Trypan blue staining was performed to visualize hyphal growth and cell death. Arrows point to hyphae and hyphal damage.
Subcellular Localization of β-Conglutin in N. benthamiana Leaves
The effect of β-conglutins on the growth of fungal and oomycete pathogens tested led to the hypothesis that β1- and β6-conglutins might localize at the cell surface, at sites closest to initial pathogen attack. Therefore, the subcellular localization of the β-conglutin proteins using Agrobacterium-mediated transient expression of β-GFP constructs in N. benthamiana leaves was examined. Two-to-three days following Agrobacterium infiltration, the localisation of β-GFP in N. benthamiana leaf epidermal cells was examined by confocal microscopy. Both β6-GFP and β1-GFP were expressed in punctate structures close to the cell surface (plasma membrane; Figure 4, Supplementary Figure S6). To determine the nature of the β6-GFP- and β1-GFP-positive structures, which resembled the pattern described for plasmodesmata (Lee and Lu, 2011), we co-expressed the constructs with the known plasmodesmata marker plasmodesmata-located protein1 or AtPDLP1-mCherry (Thomas et al., 2008). β6-GFP and β1-GFP partially overlapped the expression pattern of AtPDLP1-mCherry (R = 0.0725–0.755), indicating that β6-GFP and β1-GFP were partially located at plasmodesmata (Figure 5, and Supplementary Figure S6).
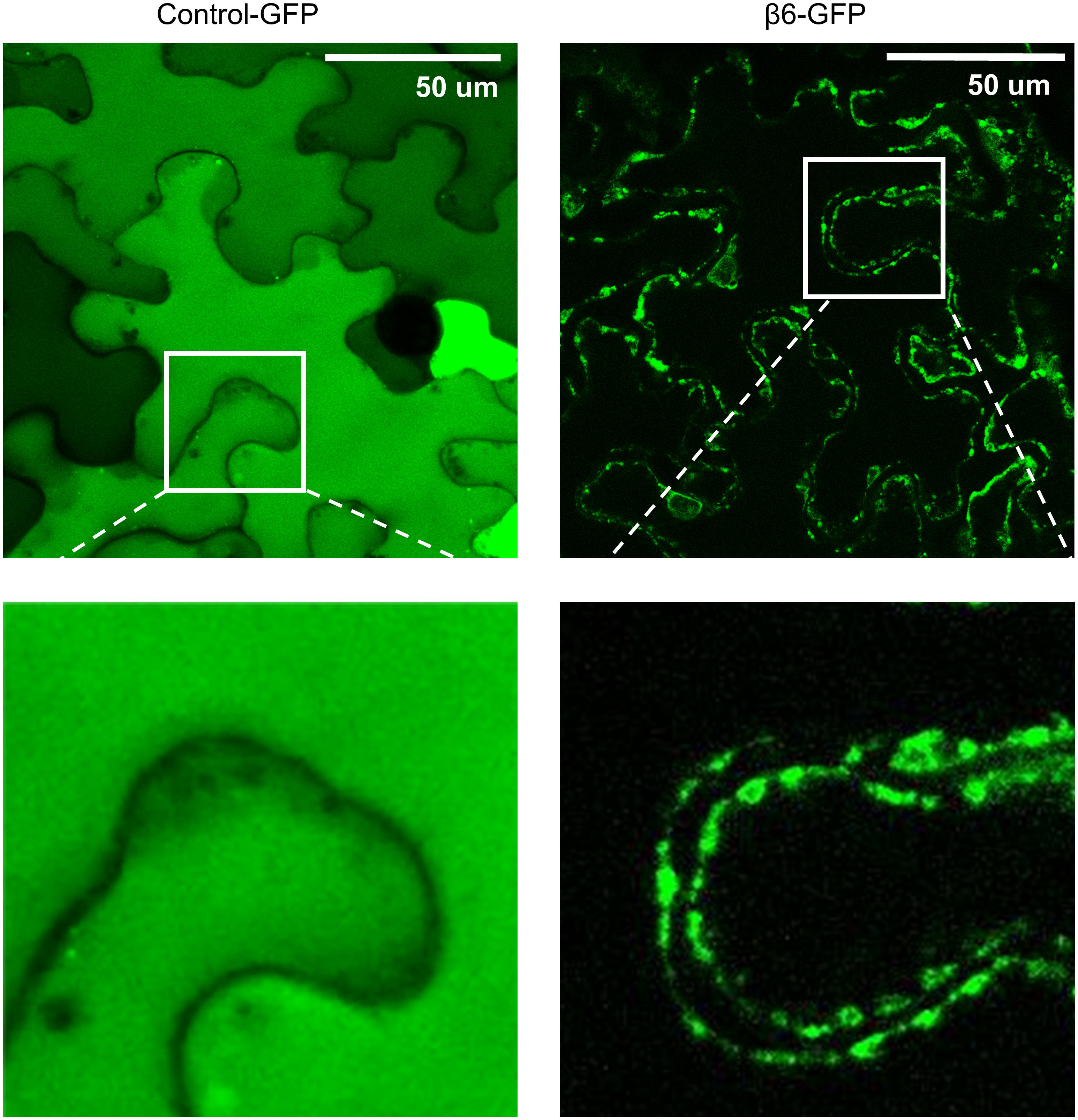
FIGURE 4. β6-conglutin is localized to the cell surface. Confocal images of tobacco epidermis cell expressing GFP alone or β6-GFP shows GFP alone homogenously expressed throughout the cytoplasm while β6-conglutin localizes to the plasma membrane through the whole cell, with no expression in cytosol or intracellular organelles. Insert: β6-GFP shows punctate labeling at the cell surface.
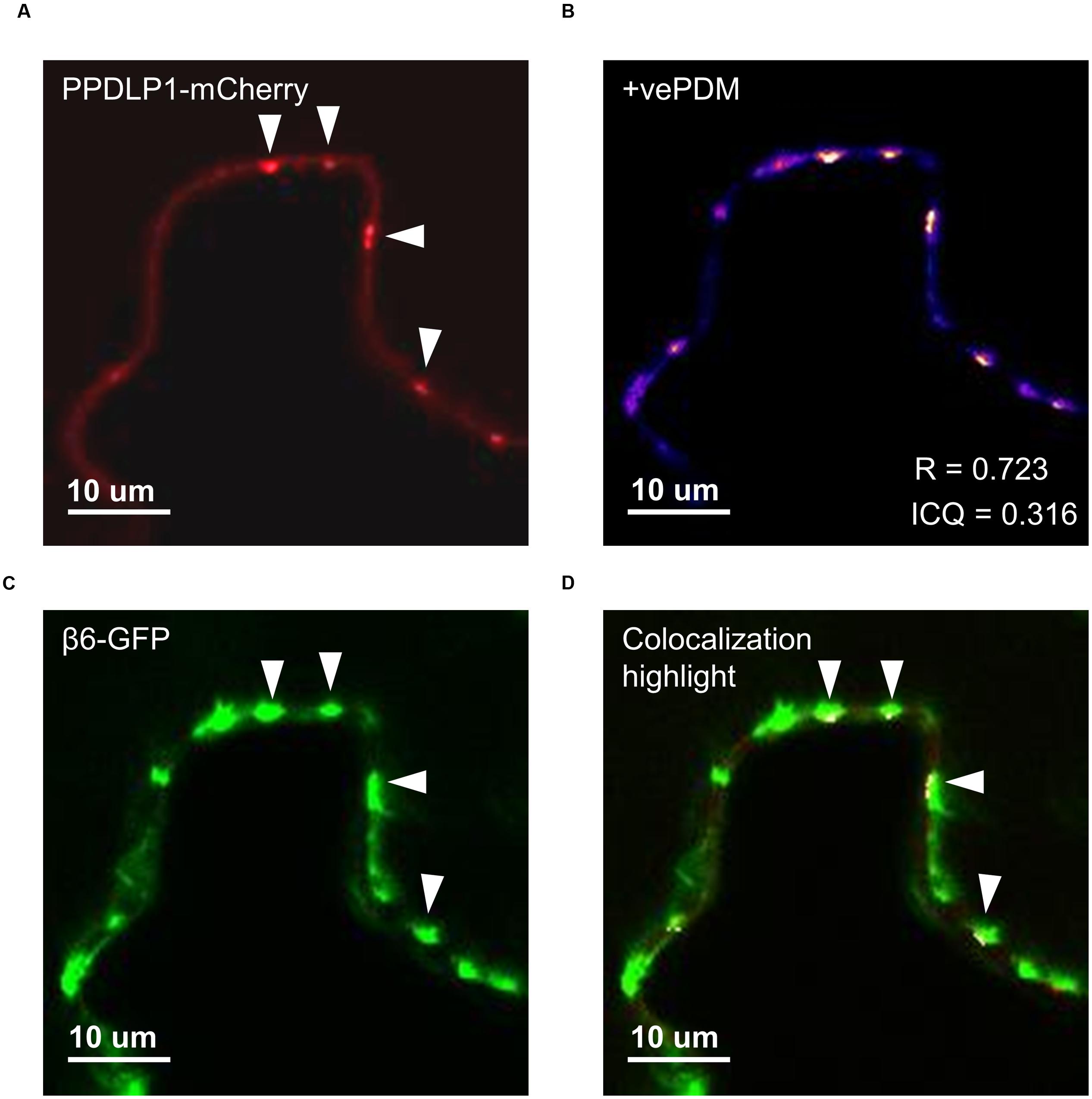
FIGURE 5. β6-conglutin localizes to the plasmodesmata. Single-slice confocal images of co-expression GFP-β6 with the plasmodesmata marker PDLP1-mCherry after transient expression in N. benthamiana; (A) PPDLP1-mCherry, (B) β6-GFP, (C) Image showing pixel pairs that have a positive PDM value equal to the value (intensity of A- mean A intensity) ∗ (intensity of B-mean B intensity) as described in Li et al. (2004), (D) merge of (A,B) with highlighted co-localized pixels. ICQ, Intensity correlation quotient; R, Mandel’s overlap coefficient. 60× immersion objective.
Protein Oxidation Levels in N. benthamiana Leaves Expressing the β1- and β6-Conglutin Proteins Following Infection with S. sclerotiorum and P. nicotianae
One of the mechanisms employed by S. sclerotiorum and P. nicotianae during infection of a compatible host is to initially suppress the plant second-phase oxidative burst that occurs 3–6 h after pathogen contact (Levine et al., 1994; Cessna et al., 2000), thereby compromising the capacity of the plant to activate downstream defense pathways (Doke, 1985; Levine et al., 1994; Williams et al., 2011). Therefore the effect of β-conglutin protein on the capacity of S. sclerotiorum and P. nicotianae to suppress the plant oxidative burst was examined. Leaves were infiltrated and allowed to transiently express GFP or β6-GFP conglutin for 48 h before being infected with S. sclerotiorum or P. nicotianae. As soon as hyphae and lesions became visible (within 24 h) leaves were collected for analysis (as shown in Figure 6A). We estimated production of ROS and oxidative burst capacity by examining the level of protein carbonylation in infected compared to mock-inoculated leaves using an OxyBlot Protein Oxidation Detection and immunoassay (Rinalducci et al., 2008). Protein oxidation is one of the covalent modification of proteins induced by ROS such as H2O2 or other products of oxidative stress, and carbonylation is one of the most commonly occurring oxidative modifications of proteins, which may be responsible for the alteration in protein activity, for example, signaling (Oracz et al., 2007). Carbonylated proteins have been identified in many plant species at different stage of growth and development (Barba-Espín et al., 2011; Morscher et al., 2015).
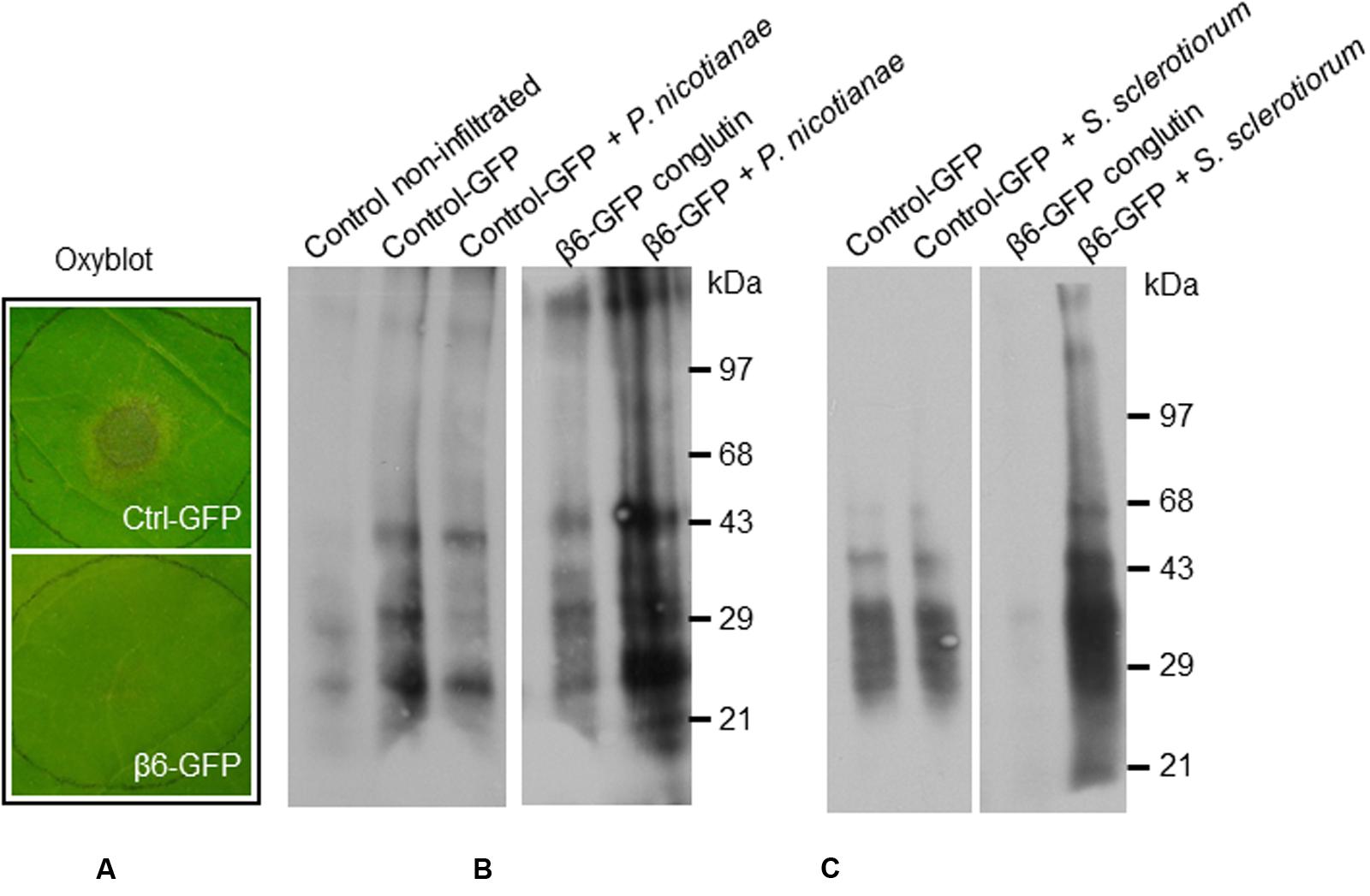
FIGURE 6. β6-conglutin oxyblots assayes. Protein carbonyl formation in tobacco leaves 48 h after agroinfiltration with indicated constructs and 24 h after inoculation with pathogens. Protein carbonyls were assessed using an OxyBlot TM kit. (A) Typical levels of pathogen growth 24 h after inoculation of leaf samples used for assay. (B) Representative blot showing basal carbonylation levels in non-transformed leaves, control leaves expressing GFP mock-inoculated and after infection with P. nicotianae, and leaves expressing β6-GFP mock-inoculated and after infection. (C) Protein carbonylation levels after infection with S. sclerotiorum, same constructs as in (B).
Basal levels of protein oxidation, as generated through normal metabolic activity (Alscher et al., 1997; Rinalducci et al., 2008) were observed in the mock-inoculated control leaves expressing GFP-only, as well as in mock-inoculated leaves expressing β6-GFP (Figures 6B–C). Following inoculation with S. sclerotiorum or P. nicotianae protein oxidation remained at similar levels in the GFP-only control leaves (Figures 6B–C). In contrast, we observed a marked increase in the levels of protein oxidation in leaves expressing the β6-GFP following infection with S. sclerotiorum or P. nicotianae when compared to the respective mock-inoculated β6-GFP or the infected GFP-only leaves. The inoculated leaves expressing β6-GFP were nevertheless healthy, as expected (Figure 6A). This suggests that the over-expression of β-conglutin proteins effectively circumvents the initial suppression of the plant oxidative burst by S. sclerotiorum or P. nicotianae.
Discussion
β-conglutins are the most abundant seed storage proteins in NLL (Foley et al., 2011) and while in other plant species these vicilin-like proteins may have roles in plant defense, the functional roles of β-conglutins in this aspect remain largely unknown (Khuri et al., 2001; Dunwell et al., 2004). In this study we identified two NLL β-conglutin proteins that strongly inhibited the growth of a range of necrotrophic fungal or oomycete pathogens, both in vitro and in vivo when transiently expressed in N. benthamiana leaves. Reduced in planta fungal growth was associated with a significant reduction in pathogen-induced host cell death and interestingly the NLL β-conglutins examined were localized near the plant cell surface. These results provide the first demonstration for any NLL β-conglutin in protection against pathogen attack, and add to the growing list of vicilin-like proteins that accumulate during seed development and have roles in plant defense (Gomes et al., 1998; Marcus et al., 1999; Rietz et al., 2012; Monteiro et al., 2015).
Vicilin-like proteins are members of the cupin superfamily which is extremely diverse, encompassing 18 different functional classes including the vicilins and similar germin-like seed storage proteins, as well as single-barrel isomerases, epimerases, and auxin-binding proteins (Dunwell et al., 2001). Given the varying antifungal potency of vicilin and vicilin-like proteins from various plant species (Gomes et al., 1998), to determine and compare and contrast the ability of NLL β-conglutins to inhibit fungal growth we assayed each of the five synthesizable NLL β-conglutins against a range of necrotrophic pathogens. Of the five NLL β-conglutins, β1 and β6 exhibited the strongest activity in vitro. Sequence comparisons among the β-conglutins does not reveal any motif common between β1 and β6 but not in the other tested β-conglutins (Supplementary Figure S3) so at this stage we are unable to hypothesize why β1 and β6 have stronger fungal inhibition activity than the other β-conglutins. Furthermore, we demonstrated β-conglutin antifungal activity in planta against S. sclerotiorum as well as against a hemibiotrophic oomycete pathogen, P. nicotianae. Necrotrophic fungal pathogens actively kill host tissue, while hemibiotrophic pathogens switch to this attack mode during later stages of their infection cycle (Glazebrook, 2005).
Although plant defense responses against pathogen attack are the result of various integrated preformed and induced mechanisms, one of the most prominent is the hypersensitivity response (HR) resulting from the generation of host ROS (Mittler, 2002). While the HR response is a type of programmed cell death that can limit the growth of biotrophic pathogens, it is favorable to necrotrophic pathogens that thrive off the dead host cells (Glazebrook, 2005; Laluk and Mengiste, 2010). Both S. sclerotiorum and P. nicotianae are capable of inciting necrotic lesions on a broad range of host plants (Agrios, 2005; Gallup et al., 2006) where S. sclerotiorum produces the major pathogenicity factor oxalic acid (Cessna et al., 2000), the primary determinant contributing to its pathogenic success (Kim et al., 2011). In compatible interactions, oxalic acid initially dampens the plant oxidative burst (Williams et al., 2011). However, once the pathogen is established, oxalic acid induces apoptotic-like programmed cell death in plant hosts, triggered by the generation of ROS at detrimental levels (Kim et al., 2008). We found in planta expression of NLL β1- and β6-conglutins effectively impaired host cell death induced by both S. sclerotiorum and P. nicotianae, evident within 24 h of pathogen challenge and lasting over the 72 h assayed. In planta expression of NLL β1 and β6-conglutins also increased levels of pathogen (S. sclerotiorum, P. nicotianae) induced protein oxidation whilst maintaining leaf health, suggesting overexpression of these two β-conglutins inhibits pathogen induced suppression of the early phase plant oxidative burst.
To dissect how NLL β-conglutins inhibit pathogen growth and host cell death in planta, we utilized GFP-tagged versions of these proteins to visualize their sub-cellular localisation. The vacuolar localisation of vicilin-like proteins (to supply amino acids during seed germination and seedling growth) has been extensively reported, however, almost no studies have been conducted for these protein classes in organs other than seeds (Overvoorde et al., 1997). Germin-like proteins from peanut localize to both the cytoplasm and the cell surface (cell membrane or cell wall) when transiently expressed within onion epidermal cells (Wang et al., 2013). Here we observed both the NLL β1- and β6-conglutin proteins localizing to the cell surface in distinct structures that included plasmodesmata when expressed in N. benthamiana leaf epidermal cells. Many studies have demonstrated that ROS are produced at the plant cell wall in a highly regulated manner (Wojtaszek, 1997; Sewelam et al., 2016), where they play key signaling roles in the control of physiological processes such as cellular growth and development (Gapper and Dolan, 2006; Kärkönen and Kuchitsu, 2015), as well as adaptation to environmental changes and pathogen attack (Wu et al., 1997; Sewelam et al., 2016). In plants one of the major contributors to ROS production during pathogen infection are the plasma membrane localized NADPH oxidases (Torres et al., 2006; reviewed in Schopfer and Liszkay, 2006). It is possible that NLL β-conglutins facilitate/mediate the production of ROS directed to the oxidative burst (Bolwell et al., 1995; Bolwell and Wojtaszek, 1997), which is known to induce structural reinforcement of the cell wall through lignin crosslinking. This has been reported for some cupins (germin and germin-like proteins) from wheat (Schweizer et al., 1999). Alternatively, ROS such as H2O2 could play direct antimicrobial roles or act as a signaling molecule in defense response pathways (reviewed in Shetty et al., 2008).
Structurally, the β-conglutin proteins are similar to germins, germin-like proteins, and vicilin-like glucose binding proteins, which are also glycoproteins characterized by a beta-barrel core structure that can be associated with the cell wall (Lane et al., 1992), the plasma membrane (Overvoorde et al., 1997; Kukavica et al., 2005) and/or plasmodesmata (Ham et al., 2012). The structure of β-conglutin is unique as it possesses two cupin domains forming a Rossmann fold reminiscent of enzymes that use molecular oxygen as a substrate (Jimenez-Lopez et al., 2015). Germins and germin-like proteins have been shown to play dual roles in seed germination and also in pathogen defense (Cândido et al., 2011). Identified from germinating wheat embryos, the wheat germin protein exhibits oxalate oxidase activity, catalyzing the conversion of oxalates (the conjugate base of oxalic acid) into CO2 and H2O2 (Woo et al., 2000; Pan et al., 2007). Other enzymatic properties of germins or germin-like proteins include superoxide dismutase (SOD) activity, ADP glucose pyrophosphatase/phosphodiesterase activity or polyphenol oxidase (PPO) activity (reviewed in Barman and Banerjee, 2015). Over-expression of germin in several plant species can lead to increased resistance to fungal pathogens such as S. sclerotiorum (Donaldson et al., 2001; Dong et al., 2008; Walz et al., 2008), and the over-expression of a germin-like oxalate oxidase in rice or sunflowers lead to increased resistance, respectively, against R. solani (Molla et al., 2013) or both R. solani and S. sclerotiorum (Beracochea et al., 2015). Moreover, overexpression of the sunflower germin-like protein in Arabidopsis altered host redox and increased endogenous ROS levels (Beracochea et al., 2015). Germin-like proteins from Brassica napus have also been linked to the initiation of an oxidative burst that impedes pathogenesis of S. sclerotiorum (Rietz et al., 2012).
A role for β-conglutin in pathogen resistance has also been proposed based on cleavage and secretion of a β-conglutin peptide (BLAD) upon germination in L. albus (Monteiro et al., 2015). BLAD, a 20 kDa polypeptide, accumulates exclusively in the cotyledon between days 4 and 12 after the onset of germination. BLAD forms a 120 kDA oligomeric structure which exhibits lectin-like activity, catalytic activities of β-N-acetyl-D-glucosaminidase and chitin-binding activity, and provides effective antifungal activity against a range of plant pathogens (Monteiro et al., 2015). Whilst the results presented in our current study indicate the involvement of the NLL β-conglutin proteins in facilitating the production of ROS following pathogen infection in planta, it remains possible that some of the effects observed, particularly those obtained with the in vitro plate assays, may be partially linked to anti-fungal activities similar to those observed with the BLAD peptide. Protein exudates of germinating L. albus seeds showed fungal growth inhibition to five of six pathogens tested (Scarafoni et al., 2013). This protein exudate contains a range of different proteins including both β- and γ-conglutins. Our research presented herein has shown that β-conglutins have antifungal activity and the β-conglutins from L. albus in the protein exudate could thus be a good candidate for contributing to the causal antifungal activity observed. It is therefore possible that lupins secrete β-conglutins during the vulnerable initial seedling germination stage as a means to protect itself from plant pathogens. As the BLAD peptide has been processed from β-conglutin, it remains to be determined if β1 and β6 would have altered antifungal properties if these proteins were also processed similar to that of BLAD.
The results presented herein suggest that NLL β-conglutins may be more versatile in their physiological roles than previously thought. While a clear causal connection cannot be given at present, our results show that several NLL β-conglutins inhibit fungal growth in vitro and that expression of at least two of these in planta enhances plant resistance to fungal/oomycete necrotrophic pathogens.
Author Contributions
Conceived and designed the experiments: JJ-L, SM, KD, and LT. Performed the experiments: JJ-L, SM, KD, LT, and LK. Analyzed the data: JJ-L, SM, KD, LT, LK, and KS. Contributed reagents/materials/analysis tools: RF, JJ-L, and KS. Wrote the paper: JJ-L, SM, KD, LT, RF, LK, and KS.
Funding
This work was supported by the European Research Program MARIE CURIE (FP7-PEOPLE-2011-IOF) for the grant ref. number PIOF-GA-2011-301550 to JJ-L and KS. JJ-L thanks the Spanish Ministry of Economy and Competitiveness for the grant ref. number RYC-2014-16536 (Ramon y Cajal Research Program). SM was supported by a CSIRO OCE Postdoctoral Fellowship.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer SF and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
We thank Roger Shivas for the F. oxysporum f. sp. conglutinans strain Fo-5176, John Irwin for the F. oxysporum f. sp. medicaginis strain Fom-5190a (BRIP 5190a), Kemal Kazan (CSIRO) for the A. brassicicola (UQ4273) and S. sclerotiorum (UQ3833) isolates, Mark Sweetingham (DAFWA) for the R. solani isolates AG8-1 and AG2-1, and Prof. Giles Hardy (Murdoch University) for the P. nicotianae (PAB12.23). We thank the Department of Agriculture and Food of Western Australia for supplying the C. lupini isolates WAC8672 and WAC10444. We acknowledge the facilities, the scientific and technical assistance of the Australian Microscopy and Microanalysis Research Facility at the Centre for Microscopy, Characterisation and Analysis, The University of Western Australia, a facility funded by the University, State and Commonwealth Governments. We also thank Nicholas Pain for excellent technical assistance and TJ Higgins for helpful comments on the manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01856/full#supplementary-material
FIGURE S1 | Sequences of synthetic β1, β2, β3, β4, and β6 conglutins that were cloned into the expression vector, pET28b.
FIGURE S2 | Purification and confirmation of recombinant β1- to β4- and β6-conglutins. (A) Purified proteins (10 μg per sample) were separated by SDS–PAGE analyses to indicate a single protein band (6xHis-tag) of approximately 65 kDa at high purity level (>95%). (B) Immunoblot confirmation using anti-β-conglutin protein antibody. The arrow indicates the correct sized band.
FIGURE S3 | Comparison of the NLL β-conglutin protein sequences. Alignment of the seven β-conglutin proteins identified in NLL, and comparison of the level of variability among them. Similarity percentages for each compared-pair of sequences are described in the table below. The lowest and the highest percentages of similarity are highlighted with gray color.
FIGURE S4 | Recombinant β1-conglutin exhibits in planta anti-fungal and oomycete activity. Shown are representative images of Agrobacterium infiltrated N. benthamiana leaves expressing recombinant β1-conglutin proteins and subsequently inoculated with either S. sclerotorium or P. nicotianae. The experiment was repeated three times with similar results.
FIGURE S5 | Recombinant β1-conglutin reduces pathogen growth and pathogen induced cell death in planta. Shown are representative images of Agrobacterium infiltrated N. benthamiana leaves expressing recombinant β1-conglutin proteins and subsequently inoculated either S. sclerotorium or P. nicotianae. Trypan blue staining was performed to visualize hyphal growth and cell death. Arrows point to hyphae, asterisk marks inoculation site.
FIGURE S6 | β1-conglutin is localized to the cell surface and plasmodesmata. (A) Confocal images of tobacco epidermis cell expressing GFP alone or β1-GFP. Insert: β1-GFP shows punctate labeling at the cell surface. (B–E) Single-slice confocal images of co-expression β1-GFP with the plasmodesmata marker PPDLP1-mCherry after transient expression in N. benthamiana; (B) PDLP1-mCherry, (C) β1-GFP, (D) Image showing pixel pairs that have a positive PDM value equal to the value (intensity of B- mean B intensity) ∗ (intensity of C-mean C intensity) as described in Li et al. (2004), (E) merge of (B,C) with highlighted co-localized pixels. ICQ, Intensity correlation quotient; R = Mandel’s overlap coefficient. 60× immersion objective.
Footnotes
- ^ http://www.lupinexpress.org/
- ^ www.novogen.com
- ^ www.genscript.com
- ^ www.qiagen.com
- ^ http://www.emdmillipore.com
- ^ http://www.agrisera.com/
- ^ www.sigmaaldrich.com
References
Agrios, G. N. (2005). “Plant diseases caused by fungi,” in Plant Pathology, 5th Edn, ed. G. N. Agrios (San Diego, CA: Academic Press), 385–614.
Alscher, R. G., Donahue, J. L., and Cramer, C. L. (1997). Reactive oxygen species and antioxidants: Relationships in green cells. Physiol. Plant. 100, 224–233. doi: 10.1111/j.1399-3054.1997.tb04778.x
Argos, P., Narayana, S. V., and Nielsen, N. C. (1985). Structural similarity between legumin and vicilin storage proteins from legumes. EMBO J. 4, 1111–1117.
Bally, J., Nakasugi, K., Jia, F., Hung, H., Ho, S. Y. W., Wong, M., et al. (2015). The extremophile Nicotiana benthamiana has traded viral defence for early vigour. Nat. Plants 1:15165. doi: 10.1038/nplants.2015.165
Barba-Espín, G., Diaz-Vivancos, P., Job, D., Belghazi, M., Job, C., and Hernández, J. A. (2011). Understanding the role of H2O2 during pea seed germination: a combined proteomic and hormone profiling approach. Plant Cell Environ. 34, 1907–1919. doi: 10.1111/j.1365-3040.2011.02386.x
Barman, A. R., and Banerjee, J. (2015). Versatility of germin-like proteins in their sequences, expressions, and functions. Funct. Integr. Genomics 15, 533–548. doi: 10.1007/s10142-015-0454-z
Beracochea, V. C., Almasia, N. I., Peluffo, L., Nahirnak, V., Hopp, E. H., Paniego, N., et al. (2015). Sunflower germin-like protein HaGLP1 promotes ROS accumulation and enhances protection against fungal pathogens in transgenic Arabidopsis thaliana. Plant Cell Rep. 34, 1717–1733. doi: 10.1007/s00299-015-1819-4
Berger, J. D., Clements, J. C., Nelson, M. N., Kamphuis, L. G., Singh, K. B., and Buirchell, B. (2013). The essential role of genetic resources in narrow-leafed lupin improvement. Crop Pasture Sci. 64, 361–373. doi: 10.1071/CP13092
Bolte, S., and Cordelières, F. P. (2006). A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 224, 213–232. doi: 10.1111/j.1365-2818.2006.01706.x
Bolwell, G. P., Butt, V. S., Davies, D. R., and Zimmerlin, A. (1995). The origin of the oxidative burst in plants. Free Radic. Res. 23, 517–532. doi: 10.3109/10715769509065273
Bolwell, G. P., and Wojtaszek, P. (1997). Mechanisms for the generation of reactive oxygen species in plant defence: a broad prespective. Physiol. Mol. Plant Pathol. 51, 347–366. doi: 10.1006/pmpp.1997.0129
Cândido, E. S., Pinto, M. F. S., Pelegrini, P. B., Lima, T. B., Silva, O. N., Pogue, R., et al. (2011). Plant storage proteins with antimicrobial activity: novel insights into plant defense mechanisms. FASEB J. 25, 3290–3305. doi: 10.1096/fj.11-184291
Cessna, S. G., Sears, V. E., Dickman, M. B., and Low, P. S. (2000). Oxalic acid, a pathogenicity factor for Sclerotinia sclerotiorum, suppresses the oxidative burst of the host plant. Plant Cell 12, 2191–2200. doi: 10.2307/3871114
Chrispeels, M. J., and Raikhel, N. V. (1991). Lectins, lectin genes, and their role in plant defense. Plant Cell 3, 1–9. doi: 10.2307/3869195
Coda, R., Rizzello, C. G., Nigro, F., De Angelis, M., Arnault, P., and Gobbetti, M. (2008). Long-term fungal inhibitory activity of water-soluble extracts of Phaseolus vulgaris cv. Pinto and sourdough lactic acid bacteria during bread storage. Appl. Environ. Microbiol. 74, 7391–7398. doi: 10.1128/AEM.01420-08
De Souza Candido, E., Pinto, M., Barbosa Pelegrini, P., Bergamin Lima, T., Nascimento Silva, O., Pogue, R., et al. (2011). Plant storage proteins with antimicrobial activity: novel insights into plant defense mechanisms. FASEB J. 25, 3290–3305. doi: 10.1096/fj.11-184291
Doke, N. (1985). NADPH-dependent O2- generation in membrane fractions isolated from wounded potato tubers inoculated with Phytophthora infestans. Physiol. Plant Pathol. 27, 311–322. doi: 10.1016/0048-4059(85)90044-X
Donaldson, P. A., Anderson, T., Lane, B. G., Davidson, A. L., and Simmonds, D. H. (2001). Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf-2.8 (germin) gene are resistant to the oxalate-secreting pathogen Sclerotinia sclerotiorum. Physiol. Mol. Plant Pathol. 59, 297–307. doi: 10.1006/pmpp.2001.0369
Dong, X., Ji, R., Guo, X., Foster, S. J., Chen, H., Dong, C., et al. (2008). Expressing a gene encoding wheat oxalate oxidase enhances resistance to Sclerotinia sclerotiorum in oilseed rape (Brassica napus). Planta 228, 331–340. doi: 10.1007/s00425-008-0740-2
Dunwell, J. M., Culham, A., Carter, C. E., Sosa-Aguirre, C. R., and Goodenough, P. W. (2001). Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 26, 740–746. doi: 10.1016/S0968-0004(01)01981-8
Dunwell, J. M., Purvis, A., and Khuri, S. (2004). Cupins: the most functionally diverse protein superfamily? Phytochemistry 65, 7–17. doi: 10.1016/j.phytochem.2003.08.016
Duranti, M., and Gius, C. (1997). Legume seeds: protein content and nutritional value. Field Crops Res. 53, 31–45. doi: 10.1016/S0378-4290(97)00021-X
Fischer, K., Dieterich, R., Nelson, M. N., Kamphuis, L. G., Singh, K. B., Rotter, B., et al. (2015). Characterization and mapping of LanrBo: a locus conferring anthracnose resistance in narrow-leafed lupin (Lupinus angustifolius L.). Theor. Appl. Genet. 128, 2121–2130. doi: 10.1007/s00122-015-2572-3
Foley, R. C., Gao, L. L., Spriggs, A., Soo, L. Y., Goggin, D. E., Smith, P. M., et al. (2011). Identification and characterisation of seed storage protein transcripts from Lupinus angustifolius. BMC Plant Biol. 11:59. doi: 10.1186/1471-2229-11-59
Foley, R. C., Jimenez-Lopez, J. C., Kamphuis, L. G., Hane, J. K., Melser, S., and Singh, K. B. (2015). Analysis of conglutin seed storage proteins across lupin species using transcriptomic, protein and comparative genomic approaches. BMC Plant Biol. 15:106. doi: 10.1186/s12870-015-0485-6
Gábrišová, D., Klubicová, K., Danchenko, M., Gömöry, D., Berezhna, V. V., Skultety, L., et al. (2016). Do cupins have a function beyond being seed storage proteins? Front. Plant Sci. 6:1215. doi: 10.3389/fpls.2015.01215
Gallup, C. A., Sullivan, M. J., and Shew, H. D. (2006). Black shank of tobacco. Plant Health Instructor doi: 10.1094/PHI-I-2006-0717-01
Gapper, C., and Dolan, L. (2006). Control of plant development by reactive oxygen species. Plant Physiol. 141, 341–345. doi: 10.1104/pp.106.079079
Glazebrook, J. (2005). Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43, 205–227. doi: 10.1146/annurev.phyto.43.040204.135923
Gomes, L. S., Senna, R., Sandim, V., Silva-Neto, M. A. C., Perales, J. E. A., Zingali, R. B., et al. (2014). Four conventional soybean [Glycine max (L.) Merrill] seeds exhibit different protein profiles as revealed by proteomic analysis. J. Agric. Food Chem. 62, 1283–1293. doi: 10.1021/jf404351g
Gomes, V. M., Mosqueda, M.-I., Blanco-Labra, A., Sales, M. P., Fernandes, K. V. S., Cordeiro, R. A., et al. (1997). Vicilin storage proteins from Vigna unguiculata (Legume) seeds inhibit fungal growth. J. Agric. Food Chem. 45, 4110–4115. doi: 10.1021/jf960942g
Gomes, V. M., Okorokov, L. A., Rose, T. L., Fernandes, K. V., and Xavier-Filho, J. (1998). Legume vicilins (7S storage globulins) inhibit yeast growth and glucose stimulated acidification of the medium by yeast cells. Biochim. Biophys. Acta 1379, 207–216. doi: 10.1016/S0304-4165(97)00100-1
Ham, B. K., Li, G., Kang, B. H., Zeng, F., and Lucas, W. J. (2012). Overexpression of Arabidopsis plasmodesmata germin-like proteins disrupts root growth and development. Plant Cell 24, 3630–3648. doi: 10.1105/tpc.112.101063
Hane, J. K., Anderson, J. P., Williams, A. H., Sperschneider, J., and Singh, K. B. (2014). Genome sequencing and comparative genomics of the broad host-range pathogen Rhizoctonia solani AG8. PLoS Genet. 10:e1004281. doi: 10.1371/journal.pgen.1004281
Hane, J. K., Ming, Y., Kamphuis, L. G., Nelson, M. N., Garg, G., Atkins, C. A., et al. (2016). A comprehensive draft genome sequence for lupin (Lupinus angustifolius), an emerging health food: insights into plant–microbe interactions and legume evolution. Plant Biotechnol. J. doi: 10.1111/pbi.12615 [Epub ahead of print]
Jimenez-Lopez, J. C., Lima-Cabello, E., Melser, S., Foley, R. C., Singh, K. B., and Alche, J. D. (2015). Lupin allergy: uncovering structural features and epitopes of β-conglutin proteins in Lupinus angustifolius L. with a focus on cross-allergenic reactivity to peanut and other legumes. Lect. Notes Comput. Sci. 9043, 96–107. doi: 10.1007/978-3-319-16483-0_10
Jimenez-Lopez, J. C., Zienkiewicz, A., Zienkiewicz, K., Alche, J. D., and Rodriguez-Garcia, M. I. (2016). Biogenesis of protein bodies during legumin accumulation in Olea europaea L. seed development and differentiation. Protoplasma 253, 517–530. doi: 10.1007/s00709-015-0830-5
Jin, T., Wang, Y., Chen, Y. W., Fu, T. J., Kothary, M. H., McHugh, T. H., et al. (2014). Crystal structure of Korean pine (Pinus koraiensis) 7S seed storage protein with copper ligands. J. Agric. Food Chem. 62, 222–228. doi: 10.1021/jf4039887
Kamphuis, L. G., Hane, J. K., Nelson, M. N., Gao, L., Atkins, C. A., and Singh, K. B. (2015). Transcriptome sequencing of different narrow-leafed lupin tissue types provides a comprehensive uni-gene assembly and extensive gene-based molecular markers. Plant Biotechnol. J. 13, 14–25. doi: 10.1111/pbi.12229
Kärkönen, A., and Kuchitsu, K. (2015). Reactive oxygen species in cell wall metabolism and development in plants. Phytochemistry 112, 22–32. doi: 10.1016/j.phytochem.2014.09.016
Keogh, R. C., Deverall, B. J., and McLeod, S. (1980). Comparison of histological and physiological responses to Phakopsora pachyrhizi in resistant and susceptible soybean. Trans. Br. Mycol. Soc. 74, 329–333. doi: 10.1016/S0007-1536(80)80163-X
Khuri, S., Bakker, F. T., and Dunwell, J. M. (2001). Phylogeny, function, and evolution of the cupins, a structurally conserved, functionally diverse superfamily of proteins. Mol. Biol. Evol. 18, 593–605. doi: 10.1093/oxfordjournals.molbev.a003840
Kim, H. J., Chen, C., Kabbage, M., and Dickman, M. B. (2011). Identification and characterization of Sclerotinia sclerotiorum NADPH oxidases. Appl. Environ. Microbiol. 77, 7721–7729. doi: 10.1128/AEM.05472-11
Kim, K. S., Min, J. Y., and Dickman, M. B. (2008). Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant Microbe Interact. 21, 605–612. doi: 10.1094/MPMI-21-5-0605
Kukavica, B., Vucinic, Z., and Vuletic, M. (2005). Superoxide dismutase, peroxidase, and germin-like protein activity in plasma membranes and apoplast of maize roots. Protoplasma 226, 191–197. doi: 10.1007/s00709-005-0112-8
Laluk, K., and Mengiste, T. (2010). Necrotroph attacks on plants: wanton destruction or covert extortion? The Arabidopsis Book 8:e0136. doi: 10.1199/tab.0136
Lane, B. G., Cuming, A. C., Frégeau, J., Carpita, N. C., Hurkman, W. J., Bernier, F., et al. (1992). Germin isoforms are discrete temporal markers of wheat development. Pseudogermin is a uniquely thermostable water-soluble oligomeric protein in ungerminated embryos and like germin in germinated embryos, it is incorporated into cell walls. Eur. J. Biochem. 209, 961–969. doi: 10.1111/j.1432-1033.1992.tb17369.x
Lee, J. Y., and Lu, H. (2011). Plasmodesmata: the battleground against intruders. Trends Plant Sci. 16, 201–210. doi: 10.1016/j.tplants.2011.01.004
Levine, A., Tenhaken, R., Dixon, R., and Lamb, C. (1994). H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79, 583–593. doi: 10.1016/0092-8674(94)90544-4
Li, Q., Lau, A., Morris, T. J., Guo, L., Fordyce, C. B., and Stanley, E. F. (2004). A syntaxin 1, Galpha(o), and N-type calcium channel complex at a presynaptic nerve terminal: analysis by quantitative immunocolocalization. J. Neurosci. 24, 4070–4081. doi: 10.1523/JNEUROSCI.0346-04.2004
Liu, H., Ma, X., Yu, H., Fang, D., Li, Y., Wang, X., et al. (2016). Genomes and virulence difference between two physiological races of Phytophthora nicotianae. Gigascience 5:3. doi: 10.1186/s13742-016-0108-7
Ma, L., Lukasik, E., Gawehns, F., and Takken, F. L. W. (2012). The use of agroinfiltration for transient expression of plant resistance and fungal effector proteins in Nicotiana benthamiana leaves. Methods Mol. Biol. 835, 61–74. doi: 10.1007/978-1-61779-501-5_4
Marcus, J. P., Green, J. L., Goulter, K. C., and Manners, J. M. (1999). A family of antimicrobial peptides is produced by processing of a 7S globulin protein in Macadamia integrifolia kernels. Plant J. 19, 699–710. doi: 10.1046/j.1365-313x.1999.00569.x
Melo, T. S., Ferreira, R. B., and Teixeira, A. N. (1994). The seed storage proteins from Lupinus albus. Phytochemistry 37, 641–648. doi: 10.1016/S0031-9422(00)90331-5
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. doi: 10.1016/S1360-1385(02)02312-9
Molla, K. A., Karmakar, S., Chanda, P. K., Ghosh, S., Sarkar, S. N., Dattam, S. K., et al. (2013). Rice oxalate oxidase gene driven by green tissue-specific promoter increases tolerance to sheath blight pathogen (Rhizoctonia solani) in transgenic rice. Mol. Plant Pathol. 14, 910–922. doi: 10.1111/mpp.12055
Monteiro, S., Carreira, A., Freitas, R., Pinheiro, A. M., and Ferreira, R. B. (2015). A nontoxic polypeptide oligomer with a fungicide potency under agricultural conditions which is equal or greater than that of their chemical counterparts. PLoS ONE 10:e0122095. doi: 10.1371/journal.pone.0122095
Morscher, F., Kranner, I., Arc, E., Bailly, C., and Roach, T. (2015). Glutathione redox state, tocochromanols, fatty acids, antioxidant enzymes and protein carbonylation in sunflower seed embryos associated with after-ripening and ageing. Ann. Bot. 116, 669–678. doi: 10.1093/aob/mcv108
Murray, G. M., and Brennan, J. P. (2012). The Current and Potential Costs from Diseases of Pulse Crops in Australia: GRDC Research Code: CER00002. Barton, ACT: Grains Research and Development Corporation.
Niyonsaba, F., Nagaoka, I., Ogawa, H., and Okumura, K. (2009). Multifunctional antimicrobial proteins and peptides: natural activators of immune systems. Curr. Pharm. Des. 15, 2393–2413. doi: 10.2174/138161209788682271
Oliveira, A. E. A., Gomes, V. M., Sales, M. P., Fernandes, K. V. S., Carlini, C. R., and Xavier-Filho, J. (1999). The toxicity of jack bean [Canavalia ensiformis (L.) dc.] canatoxin to plant pathogenic fungi. Rev. Bras. Biol. 59, 59–62. doi: 10.1590/S0034-71081999000100008
Oracz, K., El-Maarouf Bouteau, H., Farrant, J. M., Cooper, K., Belghazi, M., Job, C., et al. (2007). ROS production and protein oxidation as a novel mechanism for seed dormancy alleviation. Plant J. 50, 452–465. doi: 10.1111/j.1365-313X.2007.03063.x
Overvoorde, P. J., Chao, W. S., and Grimes, H. D. (1997). A plasma membrane sucrose-binding protein that mediates sucrose uptake shares structural and sequence similarity with seed storage proteins but remains functionally distinct. J. Biol. Chem. 272, 15898–15904. doi: 10.1074/jbc.272.25.15898
Pan, H. Y., Whittaker, M. M., Bouveret, R., Berna, A., Bernier, F., and Whittaker, J. W. (2007). Characterization of wheat germin (oxalate oxidase) expressed by Pichia pastoris. Biochem. Biophys. Res. Commun. 356, 925–929. doi: 10.1016/j.bbrc.2007.03.097
Perl-Treves, R., Foley, R. C., Chen, W., and Singh, K. B. (2004). Early induction of the Arabidopsis GSTF8 promoter by specific strains of the fungal pathogen Rhizoctonia solani. Mol. Plant Microbe Interact. 17, 70–80. doi: 10.1094/MPMI.2004.17.1.70
Petrie, J. R., Shrestha, P., Liu, Q., Mansour, M. P., Wood, C. C., Zhou, X.-R., et al. (2010). Rapid expression of transgenes driven by seed-specific constructs in leaf tissue: DHA production. Plant Methods 6, 1–6. doi: 10.1186/1746-4811-6-8
Rietz, S., Bernsdorff, F. E., and Cai, D. (2012). Members of the germin-like protein family in Brassica napus are candidates for the initiation of an oxidative burst that impedes pathogenesis of Sclerotinia sclerotiorum. J. Exp. Bot. 63, 5507–5519. doi: 10.1093/jxb/ers203
Rinalducci, S., Murgiano, L., and Zolla, L. (2008). Redox proteomics: basic principles and future perspectives for the detection of protein oxidation in plants. J. Exp. Bot. 59, 3781–3801. doi: 10.1093/jxb/ern252
Rubiales, D., Fondevilla, S., Chen, W., Gentzbittel, L., Higgins, T. J. V., Castillejo, M. A., et al. (2015). Achievements and challenges in legume breeding for pest and disease resistance. CRC Crit. Rev. Plant Sci. 34, 1–3. doi: 10.1080/07352689.2014.898445
Sales, M. P., Pimenta, P. P., Paes, N. S., Grossi-de-Sá, M. F., and Xavier-Filho, J. (2001). Vicilins (7S storage globulins) of cowpea (Vigna unguiculata) seeds bind to chitinous structures of the midgut of Callosobruchus maculatus (Coleoptera: Bruchidae) larvae. Braz. J. Med. Biol. Res. 34, 27–34. doi: 10.1590/S0100-879X2001000100003
Scarafoni, A., Ronchi, A., Prinsi, B., Espen, L., Assante, G., Venturini, G., et al. (2013). The proteome of exudates from germinating Lupinus albus seeds is secreted through a selective dual-step process and contains proteins involved in plant defence. FEBS J. 280, 1443–1459. doi: 10.1111/febs.12140
Schenk, P. M., Kazan, K., Wilson, I., Anderson, J. P., Richmond, T., Somerville, S. C., et al. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. U.S.A. 97, 11655–11660. doi: 10.1073/pnas.97.21.11655
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Schopfer, P., and Liszkay, A. (2006). Plasma membrane-generated reactive oxygen intermediates and their role in cell growth of plants. Biofactors 28, 73–81. doi: 10.1002/biof.5520280202
Schweizer, P., Christoffel, A., and Dudler, R. (1999). Transient expression of members of the germin-like gene family in epidermal cells of wheat confers disease resistance. Plant J. 20, 541–552. doi: 10.1046/j.1365-313X.1999.00624.x
Sewelam, N., Kazan, K., and Schenk, P. M. (2016). Global plant stress signaling: reactive oxygen species at the cross-road. Front. Plant Sci. 7:187. doi: 10.3389/fpls.2016.00187
Shetty, N. P., Jorgensen, H. J. L., Jensen, J. D., Collinge, D. B., and Shetty, H. S. (2008). Roles of reactive oxygen species in interactions between plants and pathogens. Eur. J. Plant Pathol. 121, 267–280. doi: 10.1007/s10658-008-9302-5
Sinden, J. A., Jones, R., Hester, S., Odom, D., Kalisch, C., James, R., et al. (2004). The Economic Impact of Weeds in Australia: Report to the CRC for Australian Weed Management. Glen Osmond, SA: CRC for Australian Weed Management.
Sparkes, I. A., Runions, J., Kearns, A., and Hawes, C. (2006). Rapid, transient expression of fluorescent fusion proteins in tobacco plants and generation of stably transformed plants. Nat. Protoc. 1, 2019–2025. doi: 10.1038/nprot.2006.286
Studier, F. W. (2005). Protein production by auto-induction in high density shaking cultures. Prot. Exp. Pur. 41, 207–234. doi: 10.1016/j.pep.2005.01.016
Tam, J. P., Wang, S., Wong, K. H., and Tan, W. L. (2015). Antimicrobial peptides from plants. Pharmaceuticals 8, 711–757. doi: 10.3390/ph8040711
Tan-Wilson, A. L., and Wilson, K. A. (2012). Mobilization of seed protein reserves. Physiol. Plant. 145, 140–153. doi: 10.1111/j.1399-3054.2011.01535.x
Thatcher, L. F., Anderson, J. P., and Singh, K. B. (2005). Plant defence responses: what have we learnt from Arabidopsis? Funct. Plant Biol. 32, 1–19. doi: 10.1071/FP04135
Thatcher, L. F., Carrie, C., Andersson, C. R., Sivasithamparam, K., Whelan, J., and Singh, K. B. (2007). Differential gene expression and subcellular targeting of Arabidopsis glutathione S-transferase F8 is achieved through alternative transcription start sites. J. Biol. Chem. 282, 28915–28928. doi: 10.1074/jbc.M702207200
Thatcher, L. F., Gardiner, D. M., Kazan, K., and Manners, J. M. (2012). A highly conserved effector in Fusarium oxysporum is required for full virulence on Arabidopsis. Mol. Plant Microbe Interact. 25, 180–190. doi: 10.1094/MPMI-08-11-0212
Thomas, C. L., Bayer, E. M., Ritzenthaler, C., Fernandez-Calvino, L., and Maule, A. J. (2008). Specific targeting of a plasmodesmal protein affecting cell-to-cell communication. PLoS Biol. 6:e7. doi: 10.1371/journal.pbio.0060007
Torres, M. A., Jones, J. D. G., and Dangl, J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. doi: 10.1104/pp.106.079467
van Wees, S. (2008). Phenotypic analysis of Arabidopsis mutants: trypan blue stain for fungi, oomycetes, and dead plant cells. CSH Protoc. 2008:db.rot4982. doi: 10.1101/pdb.prot4982
Walz, A., Zingen-Sell, I., Loeffler, M., and Sauer, M. (2008). Expression of an oxalate oxidase gene in tomato and severity of disease caused by Botrytis cinerea and Sclerotinia sclerotiorum. Plant Pathol. 57, 453–458. doi: 10.1111/j.1365-3059.2007.01815.x
Wang, T., Chen, X., Zhu, F., Li, H., Li, L., Yang, Q., et al. (2013). Characterization of peanut germin-like proteins, AhGLPs in plant development and defense. PLoS ONE 8:e61722. doi: 10.1371/journal.pone.0061722
Wang, X., Bunkers, G. J., Walters, M. R., and Thoma, R. S. (2001). Purification and characterization of three antifungal proteins from cheeseweed (Malva parviflora). Biochem. Biophys. Res. Commun. 282, 1224–1228. doi: 10.1006/bbrc.2001.4716
Williams, A. H., Sharma, M., Thatcher, L. F., Azam, S., Hane, J. K., Sperschneider, J., et al. (2016). Comparative genomics and prediction of conditionally dispensable sequences in legume–infecting Fusarium oxysporum formae speciales facilitates identification of candidate effectors. BMC Genomics 17:191. doi: 10.1186/s12864-016-2486-8
Williams, B., Kabbage, M., Kim, H.-J., Britt, R., and Dickman, M. B. (2011). Tipping the balance: Sclerotinia sclerotiorum secreted oxalic acid suppresses host defenses by manipulating the host redox environment. PLoS Pathog. 7:e1002107. doi: 10.1371/journal.ppat.1002107
Wojtaszek, P. (1997). Oxidative burst: an early plant response to pathogen infection. Biochem. J. 322, 681–692. doi: 10.1042/bj3220681
Woo, E. J., Dunwell, J. M., Goodenough, P. W., Marvier, A. C., and Pickersgill, R. W. (2000). Germin is a manganese containing homohexamer with oxalate oxidase and superoxide dismutase activities. Nat. Struct. Mol. Biol. 7, 1036–1040. doi: 10.1038/80954
Wu, G., Shortt, B. J., Lawrence, E. B., Leon, J., Fitzsimmons, K. C., Levine, E. B., et al. (1997). Activation of host defence mechanisms by elevated production of H2O2 in transgenic plants. Plant Physiol. 115, 427–435. doi: 10.1104/pp.115.2.427
Yunes, A. N. A., de Andrade, M. T., Sales, M. P., Morais, R. A., Fernandes, K. V. S., Gomes, V. M., et al. (1998). Legume seed vicilins (7S storage proteins) interfere with the development of the cowpea weevil (Callosobruchus maculatus (F)). J. Sci. Food Agric. 76, 111–116. doi: 10.1002/(SICI)1097-0010(199801)76:1<111::AID-JSFA932>3.0.CO;2-4
Keywords: 7S globulins, fungal pathogen, legume, oxidative stress, plant defense, seed storage protein, vicilins
Citation: Jimenez-Lopez JC, Melser S, DeBoer K, Thatcher LF, Kamphuis LG, Foley RC and Singh KB (2016) Narrow-Leafed Lupin (Lupinus angustifolius) β1- and β6-Conglutin Proteins Exhibit Antifungal Activity, Protecting Plants against Necrotrophic Pathogen Induced Damage from Sclerotinia sclerotiorum and Phytophthora nicotianae. Front. Plant Sci. 7:1856. doi: 10.3389/fpls.2016.01856
Received: 07 October 2016; Accepted: 24 November 2016;
Published: 09 December 2016.
Edited by:
Diego Rubiales, Spanish National Research Council, SpainReviewed by:
Sara Fondevilla, Spanish National Research Council, SpainMarcello Duranti, University of Milan, Italy
Rebecca Ford, Griffith University, Australia
Copyright © 2016 Jimenez-Lopez, Melser, DeBoer, Thatcher, Kamphuis, Foley and Singh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Karam B. Singh, Karam.Singh@csiro.au
†Present address: Su Melser, INSERM U1215, NeuroCentre Magendie, Group Endocannabinoids and Neuroadaptation, Bordeaux, France; Université de Bordeaux, NeuroCentre Magendie, Bordeaux, France
‡These authors have contributed equally to this work.
 Jose C. Jimenez-Lopez
Jose C. Jimenez-Lopez Su Melser
Su Melser Kathleen DeBoer1,3
Kathleen DeBoer1,3 Louise F. Thatcher
Louise F. Thatcher Lars G. Kamphuis
Lars G. Kamphuis Karam B. Singh
Karam B. Singh