- Department of Agricultural Sciences, University of Naples ‘Federico II’, Portici, Italy
One of the greatest challenges for agricultural science in the 21st century is to improve yield stability through the progressive development of superior cultivars. The increasing numbers of infectious plant diseases that are caused by plant-pathogens make it ever more necessary to develop new strategies for plant disease resistance breeding. Targeted genome engineering allows the introduction of precise modifications directly into a commercial variety, offering a viable alternative to traditional breeding methods. Genome editing is a powerful tool for modifying crucial players in the plant immunity system. In this work, we propose and discuss genome-editing strategies and targets for improving resistance to phytopathogens. First of all, we present the opportunities to rewrite the effector-target sequence for avoiding effector-target molecular interaction and also to modify effector-target promoters for increasing the expression of target genes involved in the resistance process. In addition, we describe potential approaches for obtaining synthetic R-genes through genome-editing technologies (GETs). Finally, we illustrate a genome editing flowchart to modify the pathogen recognition sites and engineer an R-gene that mounts resistance to some phylogenetically divergent pathogens. GETs potentially mark the beginning of a new era, in which synthetic biology affords a basis for obtaining a reinforced plant defense system. Nowadays it is conceivable that by modulating the function of the major plant immunity players, we will be able to improve crop performance for a sustainable agriculture.
An Important Reason for Enhancing the Plant Immune System
The principal aim of sustainable intensification of agriculture is to increase food production while minimizing pressure on the environment. Phytopathogens limit crop yields and pose a threat to food sustainability worldwide. In the absence of genetic resistance, crop production relies heavily on chemical control of pathogens. Reducing the dependence of food production on chemical control is a key goal for avoiding negative environmental impacts caused by current practices (Tilman et al., 2002) and taking significant global climate change into account (IPCC, 2007). Plant domestication and breeding processes allow crops to be obtained with improved performance and tailored traits. The most renewable strategy to manage plant disease is to develop resistant plants, thereby obtaining environmental, economic, and social benefits. Therefore, the genes for resistance to pests and diseases can be rightfully considered essential resources to meet human food requirements (Mundt, 1994).
Plant Immunity Components
Plants have developed a plethora of defense mechanisms underlying disease suppression to ward off damage caused by pathogens. The response of plants to pathogen attack relies on pathogen recognition at the cellular level, which then triggers complex signaling pathways (Jones and Dangl, 2006; Andolfo and Ercolano, 2015). When the plant perceives the signals of danger as pathogen-associated molecular patterns (PAMPs) or damage-associated molecular patterns (DAMPs), effectors, prompt the stereotypical defense program (Wise et al., 2007). The plant innate immune system is based on two distinct but interconnected components, namely the immunity activation component (IAC) and the immunity modulation component (IMC). The IAC is based on a large number of surveillance receptors: pattern-recognition receptors (PRRs) and Nibblers (NB-LRR receptors) that recognize the presence of pathogens and convey the message of invasion. The second component (IMC) is based on the phytohormones that play a fundamental role in regulating plant immune response (Shah, 2003; von Essen et al., 2010).
In an exemplified model, three distinct stages (1: interaction, 2: activation/modulation, and 3: effective resistance/immunity) can be identified in a generic plant-pathogen interaction (Andolfo and Ercolano, 2015). During the first stage, the conformation of virulence factor targets is modified and several alterations of primary plant metabolism are detected. In the second stage, modification of virulence factor targets induces the Nibblers/PRR-triggered signaling (NTS and PTS). Furthermore, a feedback regulation of primary metabolisms, mediated by the metabolic alterations, induced a hormone-tempered resistance (HTR). In the effective resistance/immunity stage, the NTS/PTS, and the HTR converge to confer a pathogen lifestyle-specific resistance (PSR).
Rapid adaptation to threats is orchestrated by a complex regulatory network of interconnected signaling pathways. The newly emerging picture indicates that complex crosstalk among different classes of hormones might modulate disease resistance, with outcomes dependent on pathogen lifestyles and the genetic background of the host (Andolfo and Ercolano, 2015). Plant defense systems have been extensively investigated in the last decades, but exactly how they recognize pathogens and how IAC and IMC are regulated remains unknown. During the plant immunity process several gene networks are established following signaling cascades, in which regulators must fine-tune their activity to cooperate with or antagonize other regulators. Although pathogens have evolved to hijack this highly interconnected network of regulators to promote their virulence (Grant and Jones, 2009), emerging evidence suggests that crosstalk between immunity regulators offers the potential to fine-tune plant defense responses. Knowledge of host receptors variation should be combined with complementary knowledge of PMAPs/Effectors variation in the pathogen in order to provide effective new resistance genes. The large number of genes that are thought to be involved in the resistance process complicates our understanding of the biological molecules and pathways involved. Naturally variable alleles in pathogen receptor genes and downstream components of the resistance process, such as mitogen-activated protein kinases, transcription factors, and proteases/lipases, have been shown to contribute to disease resistance (Druka et al., 2008; Chen et al., 2010; Moscou et al., 2011; Camañes et al., 2012; Corwin et al., 2016). Identification of such variants may help engineer long-lasting and broad-spectrum disease resistance in crops with both durable resistances to pathogens and increased yields (Dangl et al., 2013).
Following classical breeding methodologies, we can introgress the resistance traits, making use of natural genetic variation, through several rounds of genetic recombination. New alleles can be introduced by random mutagenesis, although this is usually followed by the time-consuming screening of large populations to identify mutants (Parry et al., 2009; Sikora et al., 2011). Genome-editing technologies (GETs) allow site-specific mutagenesis to be achieved, overcoming the limits imposed by previous methods. Indeed, plant disease resistance can be increased by targeting suitable actors of plant defense machinery. However, to extend GET applicability to the ever-increasing number of crops some bottlenecks should be solved. In this work we describe suitable methods with potential pitfalls and crucial targets for engineering plant immunity.
Targeted Genome Engineering Techniques
Site-directed mutagenesis relies on the introduction of targeted DNA double-strand breaks (DSBs) by action of programmable nucleases. Small deletions, targeted insertions, and multiplex genome modifications can result both from non-homologous end joining (NHEJ) and homologous recombination (HR) cellular DNA repair mechanisms. Artificial zinc-finger nucleases (ZFNs; Kim et al., 1996) and transcription activator-like effector nucleases (TALENs; Christian et al., 2010) contain a DNA cleavage domain from the restriction enzyme Fok I fused to an engineered DNA-binding domain. The CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/Cas9(CRISPR-associated protein-9 nuclease) is based on RNA-guided engineered nucleases. Such genome-editing technology holds great promise due to its simplicity, efficiency and versatility (Jinek et al., 2012). CRISPR/Cas9 cleavage coupled with homology-directed repair (HDR) has the potential to enable engineering of new alleles of endogenous genes or the sequential insertion of transgenes at the same locus (Feng et al., 2014). Moreover, the CRISPR/Cas9 system is advantageous over ZFNs and TALENs since it allows simultaneous editing at multiple sites across the genome (Cong et al., 2013).
The applicability of GETs in the field of plant biology was already demonstrated in the model species Arabidopsis thaliana (Christian et al., 2010; Osakabe et al., 2010; Cermak et al., 2011; Li et al., 2013) and Nicotiana benthamiana (Nekrasov et al., 2013; Gao et al., 2015) as well as in other crops including rice, sorghum, wheat, corn, soybean, tobacco, potato, petunia, sweet orange, liver worth, and poplar (Shan et al., 2014; Luo et al., 2016; Rani et al., 2016). Stable inheritance of homozygous mutations induced by GETs and segregation of the mutation in the off springs was reported in several species (Maeder et al., 2008; Christian et al., 2010; Zhang et al., 2010; Qi et al., 2013; Brooks et al., 2014; Fauser et al., 2014; Feng et al., 2014; Jia et al., 2014; Schiml et al., 2014; Zhang et al., 2014; Zhou et al., 2014; Forner et al., 2015). GETs have been shown to be excellent tools to engineer metabolic circuits for synthetic biology applications (Bortesi and Fischer, 2015) since gene expression could be modulated by an inactive nuclease fused with either transcriptional activation or repression domains (Gilbert et al., 2013; Maeder et al., 2013; Bortesi and Fischer, 2015). Indeed, the transcription of a PDS gene of N. benthamiana was modified by Piatek et al. (2014). With the availability of an increasing amount of genomic data, genome engineering is developing increasingly precise methodologies. There is no general strategy for obtaining successful modifications. In silico approaches can help to predict candidates for resistance (Sanseverino and Ercolano, 2012) and to select target site, minimizing the occurrence of off-targets (Peng et al., 2016; Rani et al., 2016). Identification of amino acid residues under selective pressure can also provide valuable support (Iovieno et al., 2015). In order to reduce off-target activity two separate sgRNA target sequences can be used to guide a Cas9 nickase variant to two adjacent positions in the genome. Indeed, the enzyme induces a single-strand break (SSB) in each of the two DNA strands, increasing cleavage specificity (Schiml and Puchta, 2016). Optimization of delivery methods, HDR incidence and enzyme activity could also increase editing efficiency and specificity (Peng et al., 2016). Finally, genetic transformation and plant regeneration are bottlenecks in plant editing of several species, in particular tree species and need to be improved to extend the use of GETs in important crops (Luo et al., 2016).
Genome-Editing Applications for Plant Disease Resistance
The growing need for crop yield stability has prompted breeding research to design plants able to respond to pathogen attacks without fitness penalties. GETs have been employed to modify major players of plant immunity at different levels in several crops. Host susceptibility genes (S-genes) have been successfully manipulated to promote resistance to key pathogens. TALEN and CRISPR/Cas9 technologies were both used to target the mildew-resistance locus O (MLO) in wheat (Wang et al., 2014), generating plants resistant to powdery mildew disease. GETs were used to generate plants resistant to bacterial leaf blight, caused by Xanthomonas oryzae pv. oryzae, impairing down the transcriptional regulation of S-genes by the effector. Indeed, the plants stably edited in the OsSWEET14 promoter were resistant to bacterial strains since the effector was unable to activate the transcription of its target (Li et al., 2012). The metabolic pathways that regulate hormonal balance can be modified to enhance the IMC component of plant immunity. This goal was achieved by using GETs to cause the down-regulation of ethylene-responsive factors (ERF). In particular, the ethylene pathway in rice was successfully modified to increase resistance to Magnaporthe oryzae (Liu et al., 2012), using CRISPR/Cas9 technology to target a mutation in OsERF922 (Wang et al., 2016). In addition, the utility of GETs to introduce resistance was also demonstrated by the deletion of a host factor not directly involved in IMC but strictly required for pathogen survival (Pyott et al., 2016).
Furthermore, a new player can be introduced into plant immunity by using GETs. Indeed, the portability of the CRISPR/Cas9 system was demonstrated for introducing a new source of resistance to the geminivirus, circular single-stranded DNA (ssDNA) viruses that replicate within the nuclei of plant cells, causing serious damage to many dicotyledonous crop plants such as beet severe curly top virus in Arabidopsis and N. benthamiana (Ji et al., 2015) and bean yellow dwarf and tomato yellow leaf curl virus in N. benthamiana (Ali et al., 2015; Baltes et al., 2015). When plant immunity can target the D/S DNA replicative form, using CRISPR/Cas9 in a similar way to its endogenous role in Archaea, it is possible to trigger mutations and interfere with the copy number of freely replicating viruses. CRISPR/Cas9 was also used to knock down the eIF(iso)4E gene encoding for a translation complex in cucumber (Cucumis sativus). The induced mutation in the host eIF(iso)4E, confers resistance to cucumber vein yellowing virus (CVYV), zucchini yellow mosaic virus (ZYMV), and papaya ringspot virus-type W (PRSV-W) (Chandrasekaran et al., 2016). A novel approach to developing therapies for infectious diseases is to block bacteria, without killing them. In Phytophthora sojae, the possibility of editing a pathogen gene (Avr4/6) involved in the immunity activation was efficiently proved (Fang and Tyler, 2016).
Opportunity to Obtain a Synthetic R-Gene for Single or Multi-Resistance
Traditional resistance breeding is based on the introgression of resistance traits, such as NLR (nucleotide-binding, leucine-rich repeat) genes, from wild species into elite varieties (Ercolano et al., 2012; Andolfo et al., 2014). Genetic variation for disease resistance within a plant is most often explained by allelic variation in the receptor encoding genes. Unfortunately, R-gene-mediated resistance is based on recognition of a single elicitor and the frequency of resistance breakdown is typically high. Therefore, a continuous influx of novel resistance genes in breeding programs is required. Recent transgenic strategies also allow the efficient transfer of R-genes between plant species (Faino et al., 2010; Horvath et al., 2012; Narusaka et al., 2013). However, the deployment of novel resistance genes through both conventional breeding and transgenic approaches is hampered by the low occurrence of R-genes with the useful response specificities. Specific R-gene targets could be edited since one or few polymorphic amino acids in the coiled-coil (CC) and/or nucleotide-binding (NB) domain are known to be responsible for recognition specificity (Ashikawa, 2012). In addition, it was observed that a double aminoacidic mutation enhanced the ability of the R-protein to trigger cell death (Stirnweis et al., 2014).
A recent work demonstrated that synthetic immune receptors (I2) can be engineered to confer resistance to phylogenetically divergent pathogens (Giannakopoulou et al., 2015). GETs could be very useful for designing and engineering R-genes with novel activities, where mutants identified in one gene could be transferred to homologs (Figure 1A). As shown in Figure 1B, genome editing could be useful also exploited to combine several pathogen recognition sites (PRSs) into a novel engineered R-gene able to mount resistance to some conserved pathogen effectors and/or PAMPs. Indeed, several works have shown that certain motifs are sufficient to determine resistance in the plant host. The highly conserved EDVID motif of the CC domain has been shown to be important for the function of the R proteins (Rairdan et al., 2008). Other studies revealed that overexpression of the isolated Toll/interleukin-1 receptor (TIR) domains of several Nibbler proteins is sufficient to trigger a hypersensitive reaction (Zhang et al., 2004; Swiderski et al., 2009; Bernoux et al., 2011; Collier et al., 2011; Maekawa et al., 2011). Furthermore, several studies showed that modular assembly of subdomains from different PRRs is used to form functional receptors. Indeed, the extracellular-leucine-rich repeat (eLRR) receptor kinases of the EFR receptor were replaced by corresponding parts from different families or species (for example from FLS2 or XA21), broadening its spectrum against diverse pathogens (Albert et al., 2010; De Lorenzo et al., 2011; Schwessinger et al., 2015). Knowledge gathered from one R-gene could be exploited to improve the candidates from other plant species to rapidly deliver agronomically useful resistance genes. A promising approach to improving disease resistance could be achieved by combining engineered R-genes in the same cultivar for conferring resistance to different pathogens (Piquerez et al., 2014).
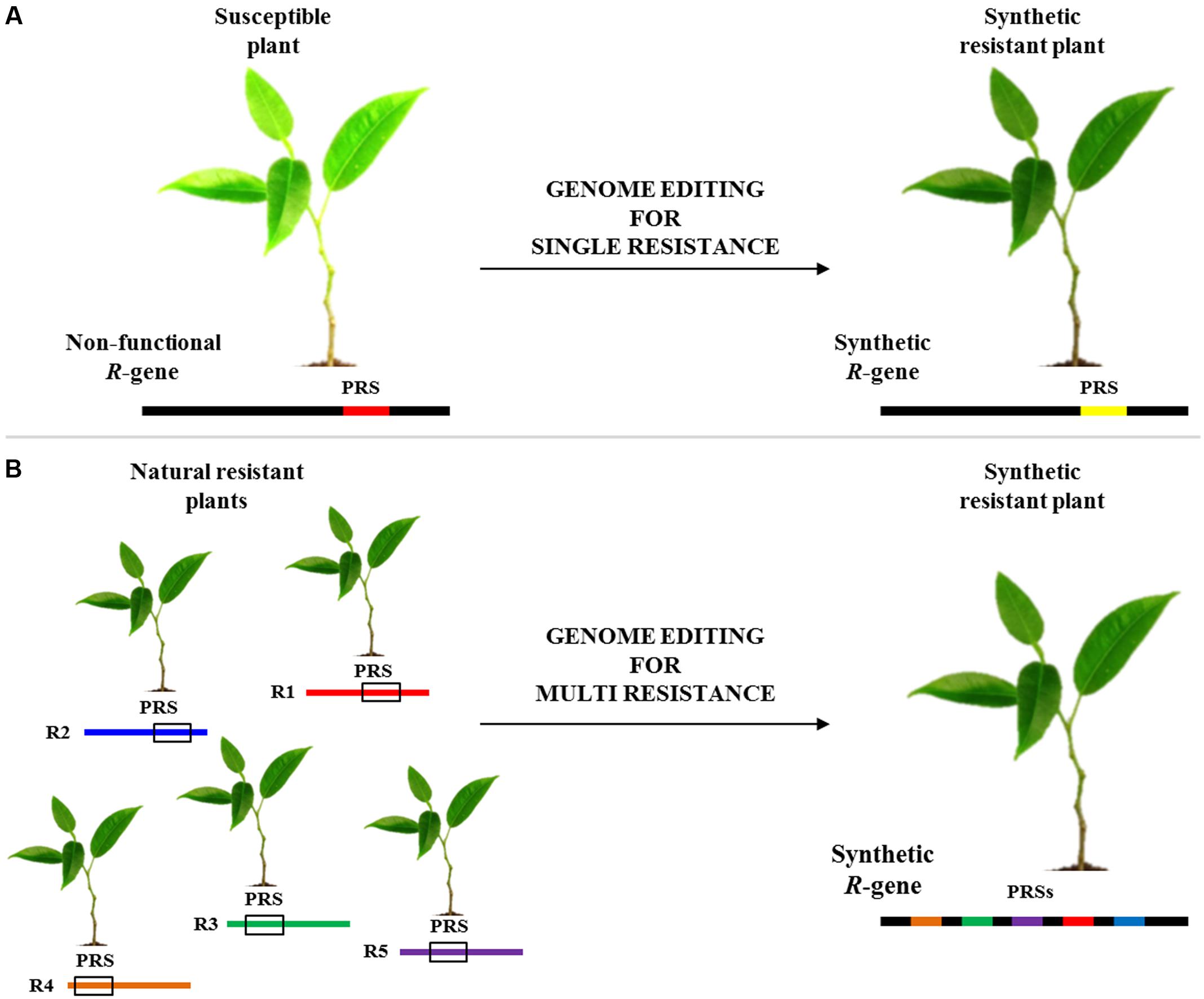
FIGURE 1. Flowchart of disease resistance genes editing. (A) Modification of a non-functional pathogen recognition site (PRS; red line) to obtain a synthetic functional R-gene (yellow line). (B) Another potential use of editing technology is the engineering of a novel synthetic R-gene able to mount resistance to several pathogens by combining PRS from different R-genes (R1-R5).
Additional Genome-Editing Targets for Disease Resistance in Crop Plants
One way to achieve broader spectrum resistance is to make use of PRRs. As with the identification of NLRomes, efforts have been made to identify PRRomes (Tang et al., 2010; Andolfo et al., 2013). In addition, detailed knowledge of plant immunity signaling will enable the construction of a novel, resilient immune response network in plants. It is well known that phytopathogens secrete effector proteins that suppress plant immunity (Figure 2A). Effector-target genes have great potential in breeding for plant disease resistance (Gawehns et al., 2013). GETs could be used to rewrite the effector-target sequence to avoid their molecular interaction (Figure 2A) and to modify the interaction during IAC. Indeed, it was highlighted that a single amino acid change in the effectors (such as in Phytophthora infestans EPIC1 and in Phytophthora mirabilis PmEPIC) and in their corresponding targets (in tomato PLCP and potato RCR3) impairs interaction (Dong et al., 2014). Moreover, the sequence variations in effector targets may cause quantitative variations in resistance phenotypes (Niks et al., 2015). Some important components of immunity, such as RIN4, are targets of effectors and might be successfully manipulated by GETs. A rin4 mutant exhibits increased resistance to the oomycete Peronospora parasitica and the bacteria Pseudomonas syringae (Luo et al., 2009) in A. thaliana. The effector may suppress immunity through attenuation ofHTR, acting on hormone synthesis routes, which are required for resistance to many pathogens. GETs might be used to target the negative regulators of HTR. Indeed, impaired function of negative regulators of the salicylic acid (SA) response (such as the MAP kinase MPK4) leads to increased resistance in A. thaliana against Pseudomonas syringae and Peronospora parasitica (Petersen et al., 2000). Some pathogen effectors could target the host cell physiology through ubiquitination. Since this process contributes crucially to plant immunity, it could be engineered by genome editing. Several studies showed that the knockout of host ubiquitin ligase increased resistance to biotrophic pathogens in A. thaliana (Trujillo et al., 2008) and to Phytophthora infestans in potato (Bos et al., 2010). The effectors not only interfere with IAC surveillance system but can also modify the plant defense transcriptome more directly. DNA target fragments within the regulatory sequences upstream of the genes that determine resistance to pathogens could be modified as shown in Figure 2B. Indeed, early works demonstrated that insertions of short donor sequences can be achieved through the CRISPR/Cas9 system (Li et al., 2013; Shan et al., 2013). It would thus be possible to reprogram the promoter, inserting cis-regulatory elements (CREs) to enhance transcription (Figure 2B). Recently, it was demonstrated that the overexpression of a mutant of the phosphatase catalytic subunits (PP1c-1) attenuates infection from Phytophthora infestans, interacting with the RXLR effector (Boevink et al., 2016).
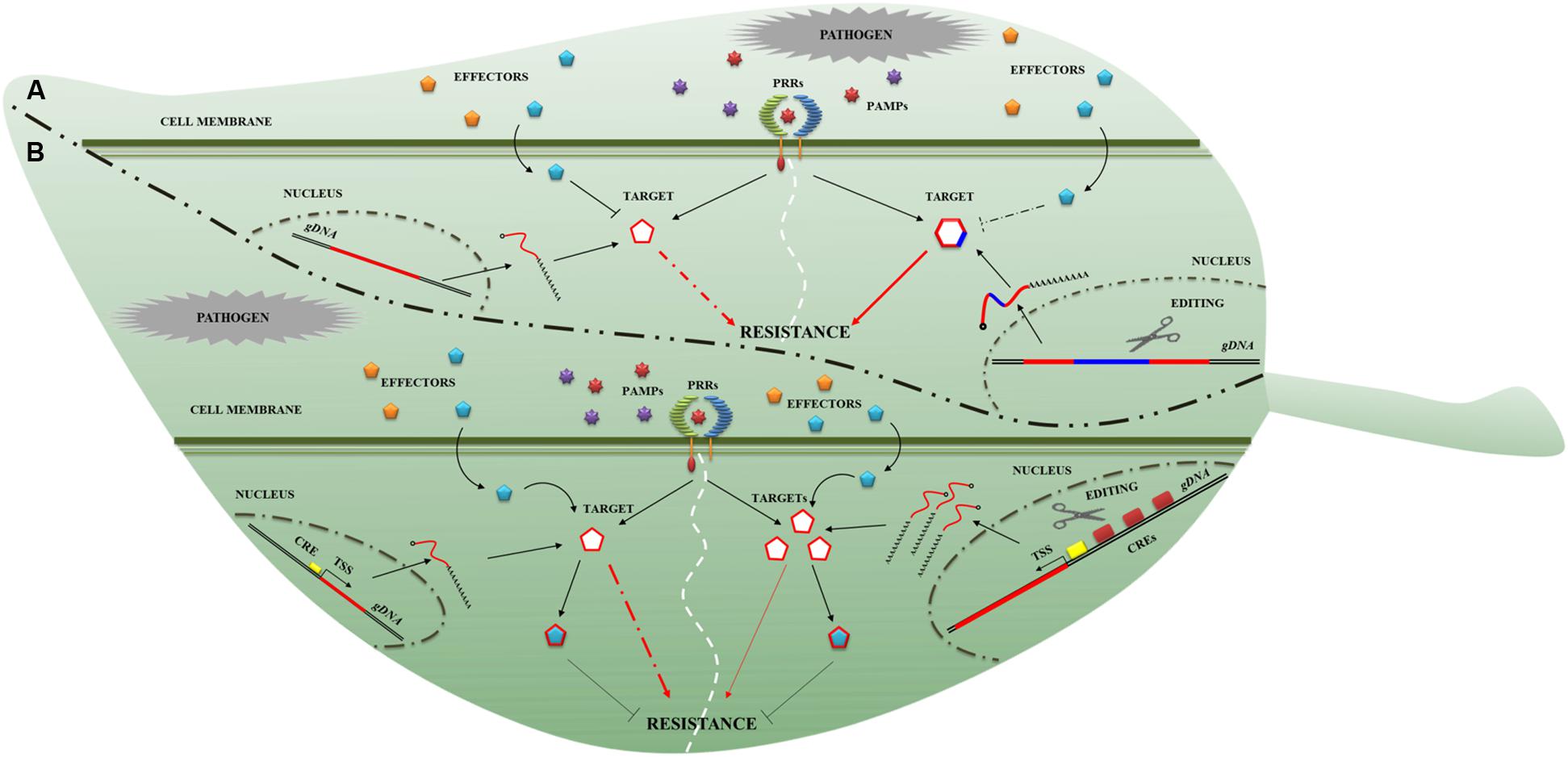
FIGURE 2. Novel genome editing targets for plant disease resistance breeding. Two genome editing applications (A,B) to obtain resistance plant are presented. (A) In the first example, the modification (blue line) of the effector-target to restore resistance is proposed. The target gene sequence (red line on the left side) is modified (red-blue line on the right side) to impede the effector- (cyan pentagon) target (red-blue hexagon) interaction. (B) The second example describes the possibility of modifying the effector-target promoter. In particular, it is depicted a cis-acting regulatory element (CRE; yellow rectangle on the left side) that controls the gene-expression of effector-target (curve red line) and product release (cyan pentagon). On the right, the increase of CREs (red rectangles on the right side) enhances the effector-target expression (curve red lines) and product release (cyan pentagons) conferring a partial resistance (thin red arrows). The dashed red arrows in (A,B) panels and dashed black line in (A) indicate an interrupted connection between components involved in the resistance process.
In general, GETs allow plant resistance to be modulated by acting on the immunity players, improving the performance of important crops for a sustainable agriculture. Such technologies allow specific mutations to be introduced into effector targets, reducing the pleiotropic effects of complete gene deletion and help to bring about gain-of-function mutations that may promote the use of a quantitative grading of resistance as a valuable approach to protecting crops.
Author Contributions
GA and PI were primarily involved in drafting the manuscript and producing the figures. LF critically read the manuscript and improved the text. ME conceived the study, drafted and edited the text and coordinated the work. All of the authors read and approved the final manuscript.
Funding
This research was carried out within GenoPOM-Pro and GenHORT projects funded by the Italian Ministry of Education, University and Research.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer CM and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgment
We thank Mark Walters for language editing and the reviewers for their constructive comments that contributed to improving the final version of the paper.
References
Albert, M., Jehle, A. K., Mueller, K., Eisele, C., Lipschis, M., and Felix, G. (2010). Arabidopsis thaliana pattern recognition receptors for bacterial elongation factor tu and flagellin can be combined to form functional chimeric receptors. J. Biol. Chem. 285, 19035–19042. doi: 10.1074/jbc.M110.124800
Ali, Z., Abulfaraj, A., Idris, A., Ali, S., Tashkandi, M., and Mahfouz, M. M. (2015). CRISPR/Cas9-mediated viral interference in plants. Genome Biol. 16, 238. doi: 10.1186/s13059-015-0799-6
Andolfo, G., and Ercolano, M. R. (2015). Plant innate immunity multicomponent model. Front. Plant Sci. 6:987. doi: 10.3389/fpls.2015.00987
Andolfo, G., Jupe, F., Witek, K., Etherington, G. J., Ercolano, M. R., and Jones, J. D. (2014). Defining the full tomato NB-LRR resistance gene repertoire using genomic and cDNA RenSeq. BMC Plant Biol. 14:120. doi: 10.1186/1471-2229-14-120
Andolfo, G., Sanseverino, W., Rombauts, S., Van de Peer, Y., Bradeen, J. M., Carputo, D., et al. (2013). Overview of tomato (Solanum lycopersicum) candidate pathogen recognition genes reveals important Solanum R locus dynamics. New Phytol. 197, 1. doi: 10.1111/j.1469-8137.2012.04380.x
Ashikawa, I. (2012). Regions outside the leucine-rich repeat domain determine the distinct resistance specificities of the rice blast resistance genes Pik and Pik-m. Mol. Breed. 30, 1531–1535. doi: 10.1007/s11032-012-9732-9
Baltes, N. J., Hummel, A. W., Konecna, E., Cegan, R., Bruns, A. N., Bisaro, D. M., et al. (2015). Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat. Plants 1, 15145. doi: 10.1038/nplants.2015.145
Bernoux, M., Ve, T., Williams, S., Warren, C., Hatters, D., Valkov, E., et al. (2011). Structural and functional analysis of a plant resistance protein TIR domain reveals interfaces for self-association, signaling, and autoregulation. Cell Host Microbe 9, 200–211. doi: 10.1016/j.chom.2011.02.009
Boevink, P. C., Wang, X., McLellan, H., He, Q., Naqvi, S., Armstrong, M. R., et al. (2016). A Phytophthora infestans RXLR effector targets plant PP1c isoforms that promote late blight disease. Nat. Commun. 7, 10311. doi: 10.1038/ncomms10311
Bortesi, L., and Fischer, R. (2015). The CRISPR/Cas9 system for plant genome editing and beyond. Biotechnol. Adv. 33, 1. doi: 10.1016/j.biotechadv.2014.12.006
Bos, J. I, Armstrong, M. R., Gilroy, E. M., Boevink, P. C., Hein, I., Taylor, R. M., et al. (2010). Phytophthorainfestans effector AVR3a is essential for virulence and manipulates plant immunity by stabilizing host E3 ligase CMPG1. Proc. Natl. Acad. Sci. U.S.A. 107, 9909–9914. doi: 10.1073/pnas.0914408107
Brooks, C., Nekrasov, V., Lippman, Z. B., and Van Eck, J. (2014). Efficient gene editing in tomato in the first generation using the CRISPR/Cas9 system. Plant Physiol. 166, 1292–1297. doi: 10.1104/pp.114.247577
Camañes, G., Pastor, V., Cerezo, M., García-Andrade, J., Vicedo, B., García-Agustín, P., et al. (2012). A deletion in NRT2.1 attenuates Pseudomonas syringae-induced hormonal perturbation, resulting in primed plant defenses. Plant Physiol. 158, 1054–1066. doi: 10.1104/pp.111.184424
Cermak, T., Doyle, E. L., Christian, M., Wang, L., Zhang, Y., Schmidt, C., et al. (2011). Efficient design and assembly of custom TALEN and other TAL effector-based constructs for DNA targeting. Nucleic Acids Res. 39, e82. doi: 10.1093/nar/gkr218
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 7. doi: 10.1111/mpp.12375
Chen, X., Hackett, C. A., Niks, R. E., Hedley, P. E., Booth, C., Druka, A., et al. (2010). An eQTL analysis of partial resistance to Puccinia hordei in barley. PLoS ONE 5:e8598. doi: 10.1371/journal.pone.0008598
Christian, M., Cermak, T., Doyle, E. L., Schmidt, C., Zhang, F., Hummel, A., et al. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761. doi: 10.1534/genetics.110.120717
Collier, S. M., Hamel, L. P., and Moffett, P. (2011). Cell death mediated by the N-terminal domains of a unique and highly conserved class of NB-LRR protein.Mol. Plant Microbe Interact. 24, 918–931. doi: 10.1094/MPMI-03-11-0050
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Corwin, J. A., Copeland, D., Feusier, J., Subedy, A., Eshbaugh, R., Palmer, C., et al. (2016). The quantitative basis of the Arabidopsis innate immune system to endemic pathogens depends on pathogen genetics. PLoS Genet. 12:e1005789. doi: 10.1371/journal.pgen.1005789
Dangl, J. L., Horvath, D. M., and Staskawicz, B. J. (2013). Pivoting the plant immune system from dissection to deployment. Science 34, 746–751. doi: 10.1126/science.1236011
De Lorenzo, G., Brutus, A., Savatin, D. V., Sicilia, F., and Cervone, F. (2011). Engineering plant resistance by constructing chimeric receptors that recognize damage-associated molecular patterns (DAMPs). FEBS Lett. 585, 1521–1528. doi: 10.1016/j.febslet.2011.04.043
Dong, S., Stam, R., Cano, L. M., Song, J., Sklenar, J., Yoshida, K., et al. (2014). Effector specialization in a lineage of the Irish potato famine pathogen. Science 343, 552–555. doi: 10.1126/science.1246300
Druka, A., Potokina, E., Luo, Z., Bonar, N., Druka, I., Zhang, L., et al. (2008). Exploiting regulatory variation to identify genes underlying quantitative resistance to the wheat stem rust pathogen Puccinia gramini sf. sptritici in barley. Theor. Appl. Genet. 117, 261–272. doi: 10.1007/s00122-008-0771-x
Ercolano, M. R., Sanseverino, W., Carli, P., Ferriello, F., and Frusciante, L. (2012). Genetic and genomic approaches for R-gene mediated disease resistance in tomato: retrospects and prospects. Plant Cell Rep. 31, 973–985. doi: 10.1007/s00299-012-1234-z
Faino, L., Carli, P., Testa, A., Cristinzio, G., Frusciante, L., and Ercolano, M. R. (2010). Potato R1 resistance gene confers resistance against Phytophthora infestans in transgenic tomato plants. Eur. J. Plant Pathol. 128, 233. doi: 10.1007/s10658-010-9649-2
Fang, Y., and Tyler, B. M. (2016). Efficient disruption and replacement of an effector gene in the oomycete Phytophthora sojae using CRISPR/Cas9. Mol. Plant Pathol. 17, 127–139. doi: 10.1111/mpp.12318
Fauser, F., Schiml, S., and Puchta, H. (2014). Both CRISPR/Cas-based nucleases and nickases can be used efficiently for genome engineering in Arabidopsis thaliana. Plant J. 79, 348–359. doi: 10.1111/tpj.12554
Feng, Z., Mao, Y., Xu, N., Zhang, B., Wei, P., Yang, D. L., et al. (2014). Multigeneration analysis reveals the inheritance, specificity, and patterns of CRISPR/Cas-induced gene modifications in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 111, 4632–4637. doi: 10.1073/pnas.1400822111
Forner, J., Pfeiffer, A., Langenecker, T., Manavella, P., and Lohmann, J. U. (2015). Germline-transmitted genome editing in Arabidopsis thaliana using TAL-effector-nucleases. PLoS ONE 10:e0121056. doi: 10.1371/journal.pone.0121056
Gao, J., Wang, G., Ma, S., Xie, X., Wu, X., Zhang, X., et al. (2015). CRISPR/Cas9-mediated targeted mutagenesis in Nicotiana tabacum. Plant Mol. Biol. 87, 1. doi: 10.1007/s11103-014-0263-0
Gawehns, F., Cornelissen, B. J. C., and Takken, F. L. W. (2013). The potential of effector-target genes in breeding for plant innate immunity. Microb. Biotech. 6, 223–229. doi: 10.1111/1751-7915.12023
Giannakopoulou, A., Steele, J. F. C., Segretin, M. E., Bozkurt, T. O., Zhou, J., Robatzek, S., et al. (2015). Tomato I2 immune receptor can be engineered to confer partial resistance to the oomycete Phytophthora infestans in addition to the fungus Fusarium oxysporum. Mol. Plant Microbe Interact. 28, 1316–1329. doi: 10.1094/MPMI-07-15-0147-R
Gilbert, L. A., Larson, M. H., Morsut, L., Liu, Z., Brar, G. A., Torres, S. E., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. doi: 10.1016/j.cell.2013.06.044
Grant, M., and Jones, J. G. (2009). Hormone (dis) harmony moulds plant health and disease. Science 324, 750–752. doi: 10.1126/science.1173771
Horvath, D. M., Stall, R. E., Jones, J. B., Pauly, M. H., Vallad, G. E., Dahlbeck, D., et al. (2012). Transgenic resistance confers effective field level control of bacterial spot disease in tomato. PLoS ONE 7:e42036. doi: 10.1371/journal.pone.0042036
Iovieno, P., Andolfo, G., Schiavulli, A., Catalano, D., Ricciardi, L., Frusciante, L., et al. (2015). Structure, evolution and functional inference on the MildewLocusO (MLO) gene family in three cultivated Cucurbitaceae. BMC Genomics 16:1112. doi: 10.1186/s12864-015-2325-3
IPCC (2007). “Climate change 2007: synthesis report,” in Contribution of Working Groups I, II, and III to the Fourth Assessment Report of the Intergovernmental Panel on Climate Change, eds Core Writing Team, R. K. Pachauri, and A. Reisinger (Geneva: IPCC), 104.
Ji, X., Zhang, H., Zhang, Y., Wang, Y., and Gao, C. (2015). Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 1, 15144. doi: 10.1038/nplants.2015.144
Jia, W., Yang, B., and Weeks, D. P. (2014). Efficient CRISPR/Cas9-mediated gene editing in Arabidopsis thaliana and inheritance of modified genes in the T2 and T3 generations. PLoS ONE 9:6. doi: 10.1371/journal.pone.0099225
Jinek, M., Chylinski, K., Fonfara, I., Hauer, M., Doudna, J. A., and Charpentier, E. (2012). A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337, 816–821. doi: 10.1126/science.1225829
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kim, Y. G., Cha, J., and Chandrasegaran, S. (1996). Hybrid restriction enzymes: zinc finger fusions to FokI cleavage domain. Proc. Natl. Acad. Sci. U.S.A. 93, 1156–1160. doi: 10.1073/pnas.93.3.1156
Li, J. F., Norville, J. E., Aach, J., McCormack, M., Zhang, D., Bush, J., et al. (2013). Multiplex and homologous recombination-mediated genome editing in Arabidopsis and Nicotiana benthamiana using guide RNA and Cas9. Nat. Biotechnol. 31, 688–691.
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., and Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392. doi: 10.1038/nbt.2199
Liu, D., Chen, X., Liu, J., Ye, J., and Guo, Z. (2012). The rice ERF transcription factor OsERF922 negatively regulates resistance to Magnaporthe oryzae and salt tolerance. J. Exp. Bot. 63, 3899–3912. doi: 10.1093/jxb/ers079
Luo, M., Gilbert, B., and Ayliffe, M. (2016). Applications of CRISPR/Cas9 technology for targeted mutagenesis, gene replacement and stacking of genes in higher plants. Plant Cell Rep. 35, 7. doi: 10.1007/s00299-016-1989-8
Luo, Y., Caldwell, K. S., Wroblewski, T., Wright, M. E., and Michelmore, R. W. (2009). Proteolysis of a negative regulator of innate immunity is dependent on resistance genes in tomato and Nicotiana benthamiana and induced by multiple bacterial effectors. Plant Cell 21, 2458–2472. doi: 10.1105/tpc.107.056044
Maeder, M. L., Linder, S. J., Cascio, V. M., Fu, Y., Ho, Q. H., and Joung, J. K. (2013). CRISPR RNA-guided activation of endogenous human genes. Nat. Methods 10, 977–979. doi: 10.1038/nmeth.2598
Maeder, M. L., Thibodeau-Beganny, S., Osiak, A., Wright, D. A., Anthony, R. M., Eichtinger, M., et al. (2008). Rapid “open-source” engineering of customized zinc-finger nucleases for highly efficient gene modification. Mol. Cell 31, 294–301. doi: 10.1016/j.molcel.2008.06.016
Maekawa, T., Cheng, W., Spiridon, L. N., Toller, A., Lukasik, E., Saijo, Y., et al. (2011). Coiled-coil domain-dependent homodimerization of intracellular barley immune receptors defines a minimal functional module for triggering cell death. Cell Host Microbe 9, 187–199. doi: 10.1016/j.chom.2011.02.008
Moscou, M. J., Lauter, N., Steffenson, B., and Wise, R. P. (2011). Quantitative and qualitative stem rust resistance factors in barley are associated with transcriptional suppression of defense regulons. PLoS Genet. 7:e1002208. doi: 10.1371/journal.pgen.1002208
Mundt, C. C. (1994). “Use of genetic diversity to control cereal diseases: implications for rice blast,” in RiceBlast Disease, eds R. S. Zeigler, S. A. Leong, and P. S. Teng (Corvallis, OR: Oregon State University), 293–308.
Narusaka, M., Kubo, Y., Hatakeyama, K., Imamura, J., Ezura, H., Nanasato, Y., et al. (2013). Interfamily transfer of dual NB-LRR Genes confers resistance to multiple pathogens. PLoS ONE 8:e55954. doi: 10.1371/journal.pone.0055954
Nekrasov, V., Staskawicz, B., Weigel, D., Jones, J. D., and Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol 31, 691–693. doi: 10.1038/nbt.2655
Niks, R. E., Qi, X., and Marcel, T. C. (2015). Quantitative resistance to biotrophic filamentous plant pathogens: concepts, misconceptions, and mechanisms. Annu. Rev. Phytopathol. 53, 1. doi: 10.1146/annurev-phyto-080614-115928
Osakabe, K., Osakabe, Y., and Toki, S. (2010). Site-directed mutagenesis in Arabidopsis using custom-designed zinc finger nucleases. Proc. Nat.l Acad. Sci U.S.A. 107, 12034–12039. doi: 10.1073/pnas.1000234107
Parry, M. A. J., Madgwick, P. J., Bayon, C., Tearall, K., Hernandez-Lopez, A., Baudo, M., et al. (2009). Mutation discovery for crop improvement. J. Exp. Bot. 60, 2817–2825. doi: 10.1093/jxb/erp189
Peng, R., Lin, G., and Li, J. (2016). Potential pitfalls of CRISPR/Cas9-mediated genome editing. FEBS J. 283, 7. doi: 10.1111/febs.13586
Petersen, M., Brodersen, P., Naested, H., Andreasson, E., Lindhart, U., Johansen, B., et al. (2000). Arabidopsis MAP kinase 4 negatively regulates systemic acquired resistance. Cell 103, 1111–1120. doi: 10.1016/S0092-8674(00)00213-0
Piatek, A., Ali, Z., Baazim, H., Li, L., Abulfaraj, A., Al-Shareef, S., et al. (2014). RNA-guided transcriptional regulation in planta via synthetic dCas9-based transcription factors. Plant Biotechnol. J. 13, 578–589. doi: 10.1111/pbi.12284
Piquerez, S. J. M., Harvey, S. E., Beynon, J. L., and Ntoukakis, V. (2014). Improving crop disease resistance: lessons from research on Arabidopsis and tomato. Front. Plant Sci. 5:671. doi: 10.3389/fpls.2014.00671
Pyott, D. E., Sheehan, E., and Molnar, A. (2016). Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol. Plant Pathol. 4, 1–13. doi: 10.1111/mpp.12417
Qi, Y., Li, X., Zhang, Y., Starker, C. G., Baltes, N. J., Zhang, F., et al. (2013). Targeted deletion and inversion of tandemly arrayed genes in Arabidopsis thaliana using zinc finger nucleases. G3 (Bethesda). 3, 1707–1715. doi: 10.1534/g3.113.006270
Rairdan, G. J., Collier, S. M., Sacco, M. A., Baldwin, T. T., Boettrich, T., and Moffett, P. (2008). The coiled-coil and nucleotide binding domains of the potato Rx disease resistance protein function in pathogen recognition and signaling. Plant Cell 20, 739–751. doi: 10.1105/tpc.107.056036
Rani, R., Yadav, P., Barbadikar, K. M., Baliyan, N., Malhotra, E. V., Singh, B. K., et al. (2016). CRISPR/Cas9: a promising way to exploit genetic variation in plants. Biotechnol. Lett. 38, 12. doi: 10.1007/s10529-016-2195-z
Sanseverino, W., and Ercolano, M. R. (2012). In silicoapproach to predict candidate R proteins and to define their domain architecture. BMC Res. Notes 5:678. doi: 10.1186/1756-0500-5-678
Schiml, S., Fauser, F., and Puchta, H. (2014). The CRISPR/Cas system can be used as nuclease for in planta gene targeting and as paired nickases for directed mutagenesis in Arabidopsis resulting in heritable progeny. Plant J. 80, 6. doi: 10.1111/tpj.12704
Schiml, S., and Puchta, H. (2016). Revolutionizing plant biology: multiple ways of genome engineering by CRISPR/Cas. Plant methods 12, 8. doi: 10.1186/s13007-016-0103-0
Schwessinger, B., Bahar, O., Thomas, N., Holton, N., Nekrasov, V., Ruan, D., et al. (2015). Transgenic expression of the dicotyledonous pattern recognition receptor EFR in rice leads to ligand-dependent activation of defense responses. PLoS Pathog. 11:e1004809. doi: 10.1371/journal.ppat.1004809
Shah, J. (2003). The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 6, 365–371. doi: 10.1016/S1369-5266(03)00058-X
Shan, Q., Wang, Y., Li, J., and Gao, C. (2014). Genome editing in rice and wheat using the CRISPR/Cas system. Nat. Protoc. 9, 2395–2410.
Shan, Q., Wang, Y., Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR-Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Sikora, P., Chawade, A., Larsson, M., Olsson, J., and Olsson, O. (2011). Mutagenesis as a tool in plant genetics, functional genomics, and breeding. Int. J. Plant Genomics 2011, 314829. doi: 10.1155/2011/314829
Stirnweis, D., Milani, S. D., Jordan, T., Keller, B., and Brunner, S. (2014). Substitutions of two amino acids in the nucleotide-binding site domain of a resistance protein enhance the hypersensitive response and enlarge the PM3F resistance spectrum in wheat. Mol. Plant Microbe Interact. 27, 265–276. doi: 10.1094/MPMI-10-13-0297-FI
Swiderski, M. R., Birker, D., and Jones, J. D. G. (2009). The TIR domain of TIR-NB-LRR resistance proteins is a signaling domain involved in cell death induction. Mol. Plant Microbe Interact. 22, 157–165. doi: 10.1094/MPMI-22-2-0157
Tang, P., Zhang, Y., Sunc, X., Tian, D., Yang, S., and Ding, J. (2010). Disease resistance signature of the leucine-rich repeat receptor-like kinase genes in four plant species. Plant Sci. 179, 4. doi: 10.1016/j.plantsci.2010.06.017
Tilman, D., Cassman, K. G., Matson, P. A., Naylor, R., and Polasky, S. (2002). Agricultural sustainability and intensive production practices. Nature 418, 671–677. doi: 10.1038/nature01014
Trujillo, M., Ichimura, K., Casais, C., and Shirasu, K. (2008). Negative regulation of PAMP-triggered immunity by an E3 ubiquitin ligase triplet in Arabidopsis. Curr. Biol. 18, 1396–1401. doi: 10.1016/j.cub.2008.07.085
von Essen, M. R., Kongsbak, M., Schjerling, P., Olgaard, K., Odum, N., and Geisler, C. (2010). Vitamin D controls T cell antigen receptor signaling and activation of human T cells. Nat. Immunol. 11, 344–349. doi: 10.1038/ni.1851
Wang, F., Wang, C., Liu, P., Lei, C., Hao, W., Gao, Y., et al. (2016). Enhanced rice blast resistance by CRISPR/Cas9-targeted mutagenesis of the ERF transcription factor gene OsERF922. PLoS ONE 11:e0154027. doi: 10.1371/journal.pone.0154027
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–952. doi: 10.1038/nbt.2969
Wise, R. P., Moscou, M. J., Bogdanove, A. J., and Whitham, S. A. (2007). Transcript profiling in host-pathogen interactions. Annu. Rev. Phytopathol. 45, 329–369. doi: 10.1146/annurev.phyto.45.011107.143944
Zhang, F., Maeder, M. L., Unger-Wallace, E., Hoshaw, J. P., Reyon, D., Christian, M., et al. (2010). High frequency targeted mutagenesis in Arabidopsis thaliana using zinc finger nucleases. Proc. Natl. Acad. Sci. U.S.A. 107, 12028–12033. doi: 10.1073/pnas.0914991107
Zhang, H., Zhang, J., Wei, P., Zhang, B., Gou, F., Feng, Z., et al. (2014). The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol. J. 12, 797–807. doi: 10.1111/pbi.12200
Zhang, Y., Dorey, S., Swiderski, M., and Jones, J. D. (2004). Expression of RPS4 in tobacco induces an AvrRps4-independent HR that requires EDS1, SGT1 and HSP90. Plant J. 40, 213–224. doi: 10.1111/j.1365-313X.2004.02201.x
Keywords: defense system, plant immunity, IAC, IMC, genome editing, effector, R-gene
Citation: Andolfo G, Iovieno P, Frusciante L and Ercolano MR (2016) Genome-Editing Technologies for Enhancing Plant Disease Resistance. Front. Plant Sci. 7:1813. doi: 10.3389/fpls.2016.01813
Received: 01 September 2016; Accepted: 16 November 2016;
Published: 01 December 2016.
Edited by:
Chidananda Nagamangala Kanchiswamy, Fondazione Edmund Mach, ItalyReviewed by:
Claudio Moser, Fondazione Edmund Mach, ItalyHanne Volpin, Danziger Innovations, Israel
Copyright © 2016 Andolfo, Iovieno, Frusciante and Ercolano. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maria R. Ercolano, ZXJjb2xhbm9AdW5pbmEuaXQ=
†These authors have contributed equally to this work.
 Giuseppe Andolfo
Giuseppe Andolfo Paolo Iovieno
Paolo Iovieno Luigi Frusciante
Luigi Frusciante Maria R. Ercolano
Maria R. Ercolano