- 1School of Life Sciences, Anhui Agricultural University, Hefei, China
- 2State Key Laboratory of Tea Plant Biology and Utilization, Anhui Agricultural University, Hefei, China
Growth-regulating factors (GRFs) are plant-specific transcription factors that have important functions in regulating plant growth and development. Previous studies on GRF family members focused either on a single or a small set of genes. Here, a comparative genomic analysis of the GRF gene family was performed in poplar (a model tree species), Arabidopsis (a model plant for annual herbaceous dicots), grape (one model plant for perennial dicots), rice (a model plant for monocots) and Chinese pear (one of the economical fruit crops). In total, 58 GRF genes were identified, 12 genes in rice (Oryza sativa), 8 genes in grape (Vitis vinifera), 9 genes in Arabidopsis thaliana, 19 genes in poplar (Populus trichocarpa) and 10 genes in Chinese pear (Pyrus bretschneideri). The GRF genes were divided into five subfamilies based on the phylogenetic analysis, which was supported by their structural analysis. Furthermore, microsynteny analysis indicated that highly conserved regions of microsynteny were identified in all of the five species tested. And Ka/Ks analysis revealed that purifying selection plays an important role in the maintenance of GRF genes. Our results provide basic information on GRF genes in five plant species and lay the foundation for future research on the functions of these genes.
Introduction
Growth-regulating factors (GRFs) are plant-specific proteins. The first identified GRF, was rice OsGRF1 (van der Knaap et al., 2000). Subsequent studies found that GRF genes played a critical role in the regulation of plant growth and development (Kim et al., 2003; Horiguchi et al., 2005; Kuijt et al., 2014; Liang et al., 2014; Vercruyssen et al., 2015). In recent years, with the sequencing of tens of plant genomes, many GRF genes have been isolated and identified. The N-terminal of Arabidopsis GRF9 protein and Chinese cabbage GRF12 contain two WRC (Trp, Arg, Cys) structure domains (Kim et al., 2003; Wang et al., 2014), whereas the N-terminal of GRF proteins have one WRC and one QLQ (Gln, Leu, Gln) structure domain in the species studied (van der Knaap et al., 2000; Kim et al., 2003; Choi et al., 2004). The QLQ structure domain is similar to the N-terminal of SWI2/SNF2 in yeast, which could combine with SNF11 to form the chromatin remodeling complex (Treich et al., 1995). In addition, this structure domain could interact with the SNH structure domain in GIF (GRF-interacting factor) to form a functional complex to perform a transcriptional activation function (Kim and Kende, 2004). The WRC domain consists of one functional nuclear localization signal and one DNA binding motif (zinc finger structure), which is mainly involved in DNA binding. Moreover, the C-terminals of some GRF proteins also include the TQL (Thr, Gln, Leu), GGPL (Gly, Gly, Pro, Leu) and FFD structure domains (Kim et al., 2003; Choi et al., 2004; Zhang et al., 2008; Wang et al., 2014). The transcriptional expression of the GRF gene was found to be regulated by GA. For instance, after celery cabbage leaves were treated with GA3, the transcriptional expression levels of the BrGRF5, BrGRF8, BrGRF9, BrGRF11, BrGRF12, BrGRF13, BrGRF15, and BrGRF16 genes were increased by more than fivefolds and those of BrGRF2, BrGRF4, BrGRF7 were increased by more than 2–5 folds compared with controls (Wang et al., 2014). Moreover, miR396 is also involved in the regulation of GRF gene expression. The ath-miR396a gene of Arabidopsis thaliana was over-expressed and caused decreased GRF gene transcription levels (FG137771, FG165999, FG194560) in tobacco. Moreover, the petal, stamen and carpel of transgenic plants increased and fertility was reduced (Yang et al., 2009).
Although the GRF gene family has been reported in plants such as Chinese cabbage (Wang et al., 2014), Cucurbitaceae (Baloglu, 2014), Brachypodium distachyon (Filiz et al., 2014), and Zea mays (Zhang et al., 2008), both the mechanism of GRF gene expansion and specific evolutionary relationships remain elusive. Comparative genomic studies in plants would clarify the genome evolution by microsynteny analysis across different species. In our study, we analyzed the GRF genes from five flowering plant species including pear (Pyrus bretschneideri), poplar (Populus trichocarpa), Arabidopsis thaliana, grape (Vitis vinifera) and rice (Oryza sativa). By analysis of the phylogenetic relationship, intraspecies and interspecies differences in five plant species, gene duplication, origins and evolution were revealed. These results may contribute to the extrapolation of GRF gene function from one lineage to another.
Materials and Methods
Database Searches for Highly Conserved GRF Genes
In our study, the genomic data of pear, Arabidopsis, and Oryza sativa, were downloaded from the GigaDB database1, TAIR2, and the Rice Annotation Project3, respectively. And the genomic data of both poplar and grape were downloaded from the Phytozome database4. The WRC (PF08879.8) and QLQ (PF08880.9) domains were downloaded from the PFAM database5 (Punta et al., 2011) and were separately blasted against the corresponding plant genomes based on the hidden Markov model (HMM) using DNATools software. Subsequently, SMART software (Letunic et al., 2012) and the PFAM database (Punta et al., 2011) were used to identify the sequences that contain the WRC (Kim et al., 2003) and QLQ domains (Kim et al., 2003).
Phylogenetic Analysis of GRF Genes
To construct the phylogeny of the GRF genes from the five flowering plant species, multiple sequence alignments for all amino acid sequences of the full-length GRF proteins were conducted using ClustalX software with the default settings. A phylogenetic tree was generated using the neighbor-joining (NJ) method using MEGA 7.0 software (Kumar et al., 2016) with the following parameters: pairwise deletion mode, Poisson correction, and bootstrapping (1000 replicates).
Gene Structure Analysis and Motif Detection
The exon-intron structures of the GRF family were drawn using the GSDS website (Guo et al., 2007) (Gene Structure Display Server6) by comparing the coding sequences with their corresponding genomic sequences.
The conserved protein motifs were analyzed using the MEME online tool (Bailey et al., 2015) (Multiple Expectation Maximization for Motif Elicitation7 with the following parameters: maximum number of motifs of 20 and the optimum width from 6 to 200 residues. Additionally, we used the Pfam (Punta et al., 2011) and SMART databases (Letunic et al., 2012) to annotate the structural motif. All of the GRF gene functional annotations were obtained from Gene Ontology (GO8) using Blast2GO software (Conesa et al., 2005).
Microsynteny Analysis
Microsynteny analysis of chromosome segments containing GRF genes can identify and classify the expansion pattern of the GRF gene family. The physical locations of all GRF members on the chromosomes from pear, Populus, grape, Arabidopsis and rice were determined. If more than one gene family member was located in the same or neighboring regions of the genome, they were thought to be tandem duplications. If the two GRF genes were located on duplication blocks and their flanking protein-coding genes were highly similar at the amino acid level (Maher et al., 2006), they were thought to be large-scale duplication events. First, all of the GRF genes were located in the genome as the initial anchor point. Then, the protein-coding sequences 100 kb upstream and downstream of each anchor point were analyzed using the BLASTP program (Deleu et al., 2007) to identify whether the duplicated genes existed between the two independent regions. Then, the number of protein-coding genes exhibiting the highest non-self match (E-value < 10-10) (Sato et al., 2008) between the two flanking sequences of the anchor points was calculated. When four or more gene pairs with the synteny relationship were detected between the two regions, then these two regions were thought to originate from large-scale duplication events.
Environmental Selection Pressure and Duplication Event Dating Analysis
The Ks and the Ka/Ks ratios of gene pairs in the duplication blocks were calculated. The protein sequences of gene pairs were compared using MUSCLE software (Edgar, 2004). And results from protein sequence alignment were used to guide the comparison of nucleic acid sequence codons using the PAL2NAL program (Suyama et al., 2006). The comparison results of the codons were imported into DnaSP software (Librado and Rozas, 2009). Then, the Ka/Ks and Ks ratios were calculated. In addition, the parameters used in the sliding window analysis were as follows: window size 150 bp and step size 9 bp.
Functional Divergence Analysis
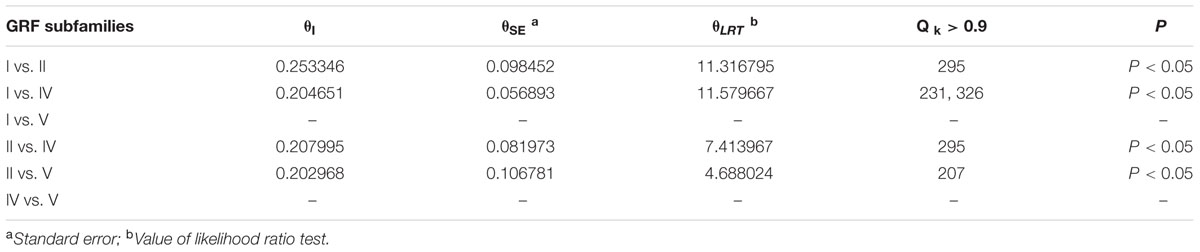
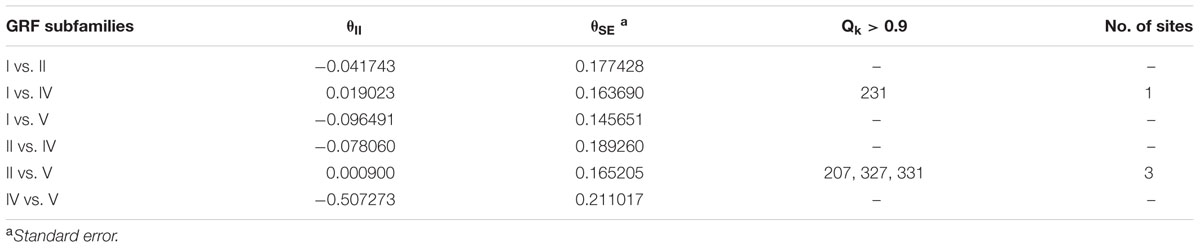
The functional divergence (I type and II type) between each GRF subfamily was calculated using V3.0B1 DIVERGE (Gu et al., 2013) software in accordance with the constructed phylogenetic tree. Type I functional divergence occurred after gene duplication, which usually led to a selectivity change in a specific amino acid, i.e., change in the evolution rate. The coefficient 𝜃I fluctuates from 0 to 1, reflecting weak to strong functional divergence between gene categories. Type II functional divergence occurred after gene duplication but only resulted in a change in the physical and chemical properties of amino acids (Gu, 1999, 2006). The evolutionary rate difference coefficient 𝜃I of each GRF subfamily and amino acid physico-chemical properties of the divergence coefficient 𝜃II and the corresponding posterior probability (Qk) were obtained. If Qk > 0.9, it was inferred that the amino acid site should have a functional differentiation after gene duplication.
Results
Identification and Chromosomal Location of GRF Genes in Five Genomes
A total of 58 GRF genes were identified from the five species studied, with 10 in pear (PbGRF01 to PbGRF10), 19 in poplar (PtGRF01 to PtGRF19), 9 in Arabidopsis (AtGRF01 to AtGRF09), 8 in grape (VvGRF01 to VvGRF08) and 12 in Oryza sativa (OsGRF01 to OsGRF12), respectively. In addition, we determined the physical location of GRF genes on the chromosomes according to the overall search in the complete genome sequences of the five plant species (Supplementary Table S1). The results showed that the distribution of the 58 GRF genes among the chromosomes of the five species was not even (Figure 1). In the Arabidopsis and Oryza sativa genomes, the GRF genes were mainly distributed on chromosome 2, chromosome 3 (2) and chromosome 4 (2). For grape, GRF genes were found on chromosomes 2, 8, 9, 11, 16, and 18. In poplar, GRF genes were distributed on chromosomes 1, 2, 3, 6, 7, 12, 13, 14, 15, 18, and 19. In pear, we found GRF genes on chromosomes 2, 6, 7, 9, and 15. Additionally, three GRF genes (VvGRF08, PbGRF08, and PbGRF09) could not be mapped to any chromosome in the grape or pear genomes (Supplementary Table S1).
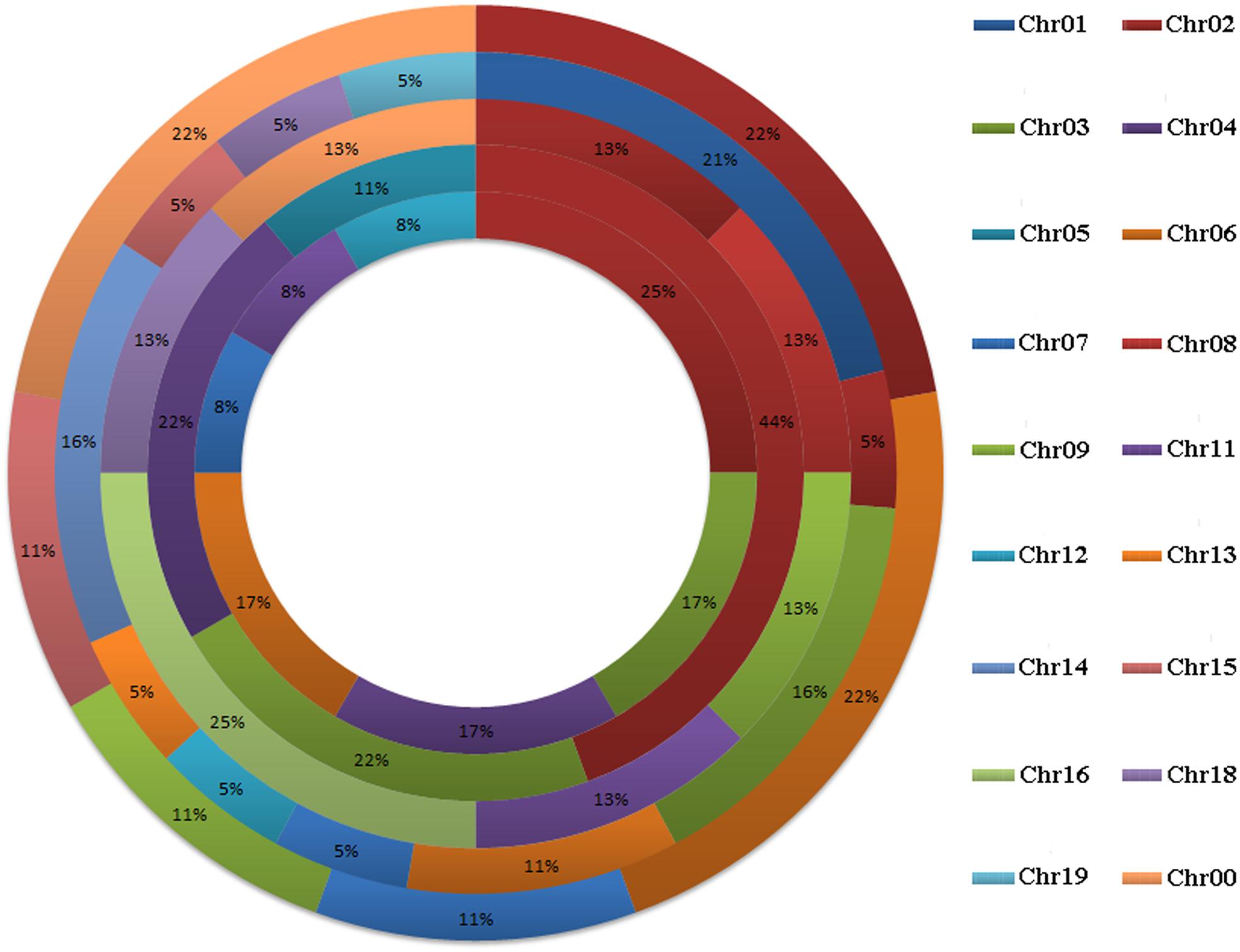
FIGURE 1. Chromosomal distribution and percentage share of GRF genes in pear, Populus, Arabidopsis, grape and Oryza sativa. The outermost ring represents chromosomes of pear, followed by Populus, grape, and Arabidopsis, and the innermost ring represents Oryza sativa.
Evolutionary Analysis of GRF Genes in Rice, Grape, Arabidopsis, Populus, and Pear
Using the well-described GRF proteins in representative plant species, including the monocots Brachypodium distachyon, Oryza sativa, Setaria italic, Zea mays, Sorghum bicolor and the dicots Arabidopsis thaliana, Populus trichocarpa, Glycine max, Citrus sinensis, Vitis vinifera, Cucumis sativus, Brassica rapa and Chinese pear, the evolutionary relationships between members of the GRF families proteins were evaluated through phylogenetic analysis. According to the nodes of the phylogenetic tree, the NJ tree could be divided into five subfamilies, designated as I, II, III, IV, and V, respectively (Supplementary Figure S1). Subsequently, to further understand the similarity and evolutionary history of the GRF genes in rice, grape, Arabidopsis, Populus and Chinese pear, we built an unrooted phylogenetic tree using the NJ method in the MEGA7 software (Kumar et al., 2016). The NJ tree showed that 58 GRF proteins were divided into five subfamilies (Figure 2), which was consistent with the result from phylogenetic analysis (Supplementary Figure S1). The topology of these two phylogenetic trees and the distribution of GRF gene in each subfamily were basically the same (Figure 2; Supplementary Figure S1). Therefore, we focused our research on the evolution of the GRF family members in rice, grape, Arabidopsis, Populus and pear. As shown in Figure 2, subfamily III contained the minimal GRF numbers (2), and subfamily I has the maximal GRF numbers (21), followed by subfamily V (16) and subfamily IV (12). Each of the five species (rice, grape, Arabidopsis, Populus and pear) contributed at least one GRF gene to subfamily I, subfamily II and subfamily V, whereas, the members of subfamily III and subfamily IV included one, two or three species. Subfamily III consisted of only rice (monocots) and subfamily IV consisted of grape, Arabidopsis, Populus, and pear (dicots). Therefore, we deduced that this phenomenon may correspond to a special gene expansion event (lost or obtained) during the evolutionary process (Supplementary Figure S1; Figure 2). In addition, according to the phylogenetic tree (Figure 2), we identified pairs of orthologous genes among the GRF genes: PbGRF01 and PtGRF16, and PbGRF06 and VvGRF06, and PbGRF04 and PtGRF01.
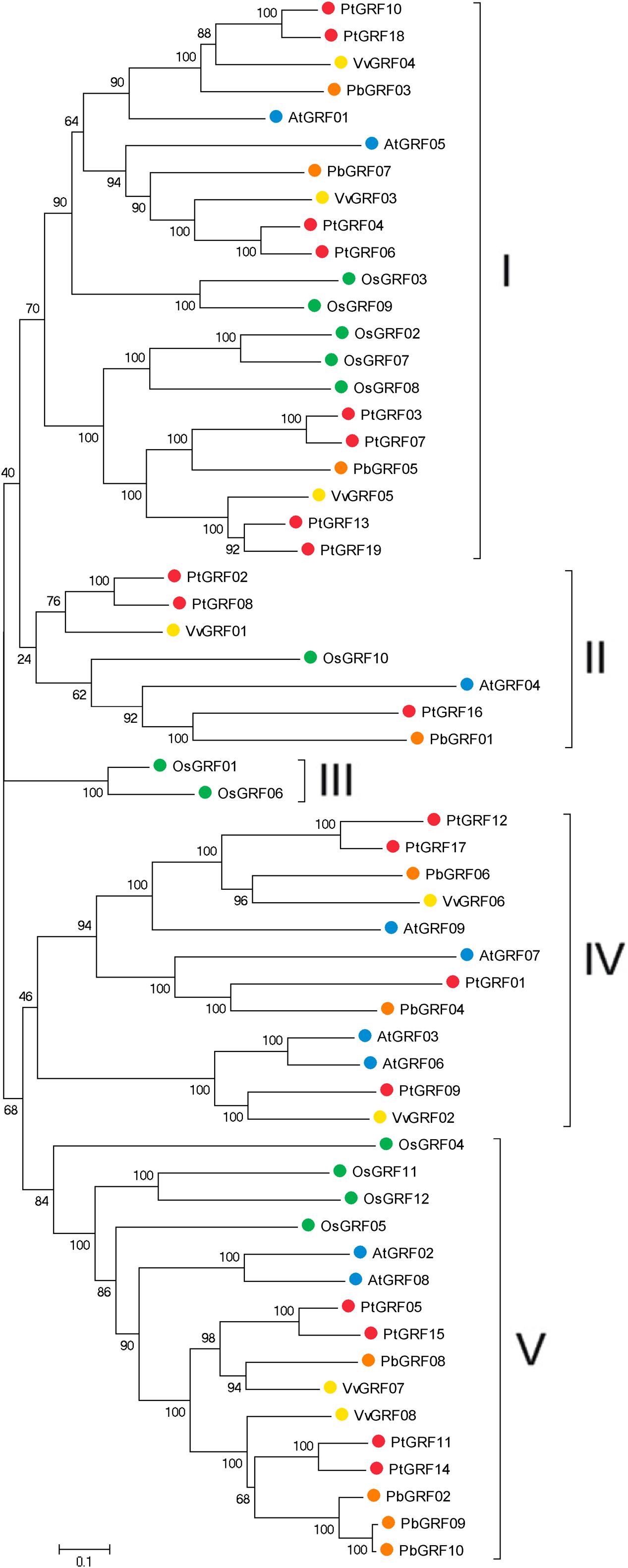
FIGURE 2. Phylogenetic analysis of GRF genes in pear, Populus, Arabidopsis, grape and Oryza sativa. The species background for each GRF protein is represented by different colors. Based on the bootstrap values and evolutionary distances, the tree was clustered into five subfamilies. Gene names are listed in Supplementary Table S1. The scale bar represents 0.1 amino acid changes per site.
Structural Analysis of GRF Genes from Pear, Poplar, Grape, Arabidopsis and Rice
To gain more insights into the structural diversity of GRF genes, exon-intron pattern maps of the individual GRF genes were generated. As shown in Figure 3A, the 58 GRF genes contained different numbers of exons, varying from 1 to 6. We found that 27 GRF genes had four introns and 25 had three exons, three genes had six introns and one had five exons, one gene had two exons, and vGRF06 had only one exon. These results implied that both of exon gain and loss occurred during the evolution of GRF genes, which may help to explain the functional diversity of closely related GRF genes. Exon-intron structures of the paralogous and orthologous GRF gene pairs were further analyzed. Among these gene pairs, the exon number of five gene pairs exhibits exon-intron gain or loss variations, including PtGRF04/PtGRF06, OsGRF02/OsGRF07, PbGRF06/VvGRF06, PtGRF01/PbGRF04, and PtGRF09/VvGRF02 (Figure 3A). In comparing the five gene pairs, PtGRF06, OsGRF02, PbGRF04 and PtGRF09 lost one exon whereas PtGRF04, OsGRF07, PtGRF01 and VvGRF02 gained one exon during the long evolutionary period. We speculated that these differences are possibly due to a single intron gain or loss event during the long evolutionary period.
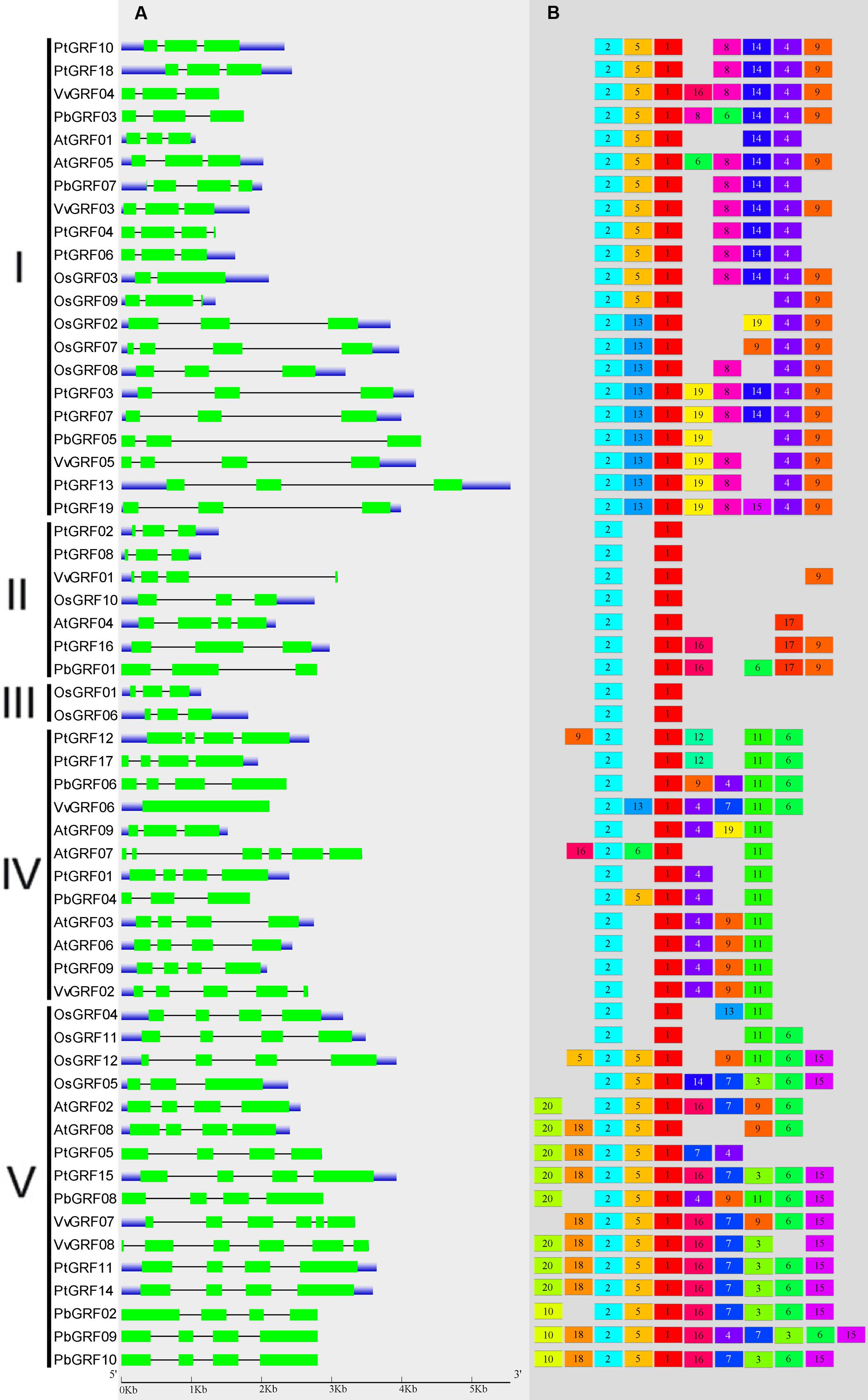
FIGURE 3. Exon-intron structure and motif compositions of GRF genes across five plant species. (A) Exon-intron structures of the GRF genes. Green rectangles: exons; thin lines: introns; blue boxes: untranslated regions (UTRs). (B) MEME motif search results. Note that different motifs are represented by different color boxes and that the length of each box does not show the true motif size.
Due to no high similarity among the 58 GRF genes, thus we used MEME software to identify the conserved motifs in the 58 GRF proteins. Twenty motifs were found on the GRF proteins (Figure 3B; Supplementary Table S2). Subsequently, the Pfam and SMART databases were used to annotate each of the putative motifs. Motif 1 and motif 17 were identified to encode the WRC domain (Figure 4), and motif 2 was found to encode the QLQ domain (Figure 4), while the remaining motifs did not have function annotation. As shown in Figure 3B, the most closely related members in each subfamily exhibit common motif compositions (e.g., PtGRF10 and PtGRF18, VvGRF05 and PtGRF13), implying functional similarities among GRF proteins. Both motif 1 and motif 2 were present in all 58 GRF proteins and thought as the most conserved motifs. In addition, some subfamily specific motifs, such as motif 18 and motif 20 in the subfamily V, were also found, implying that they might be important for the functions of GRF proteins in this subfamily. To further understand the function of different GRF genes, we searched the GO Database using Blast2GO software. The results show that the 58 GRF genes contain common functions such as regulation of metabolic process, biological process, organic cyclic compound binding, molecular function, intracellular organelles, and cellular components (Supplementary Table S3).
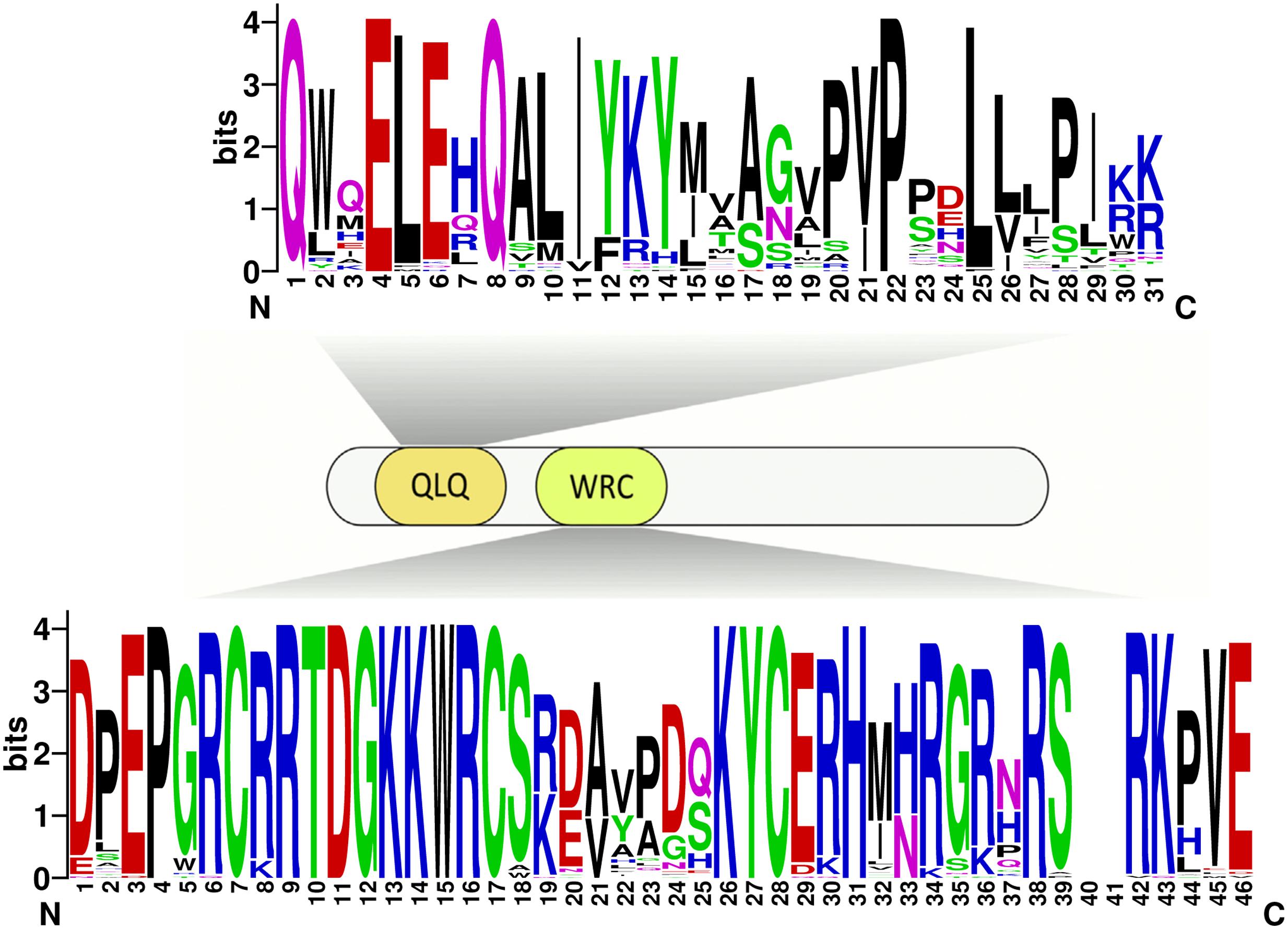
FIGURE 4. Domain composition of GRFs. Fifty-eight GRFs have both the characteristic WRC and QLQ domains. The sequences of the QLQ and WRC domains of the 58 GRFs were aligned using ClustalW software to analyze their sequence features. These domain diagrams were plotted using the online WebLogo tool.
Conserved Microsyntenies Were Found in Five Plant Species
In previous studies, microsynteny analysis of several plant species was performed to identify the location of homologous genes (Li et al., 2014; Lin et al., 2014; Wang et al., 2015; Cao et al., 2016b). In our research, microsynteny was investigated to infer the relationship of the GRF genes between eudicots (pear, Populus, grape and Arabidopsis) and monocots (rice). Additionally, since apple (Velasco et al., 2010) and pear (Wu et al., 2013) belong to the Rosaceae species, apple was also considered in the following analysis. The members of the GRF family of the six plant species (pear, apple, Populus, grape, Arabidopsis and rice) were used as anchor genes to clarify the molecular history of the surrounding chromosomal regions. Through pairwise comparisons and comparison of all of the proteins in the GRF gene flanking areas, the conserved microsyntenies were found in the six plant species (Figure 5).
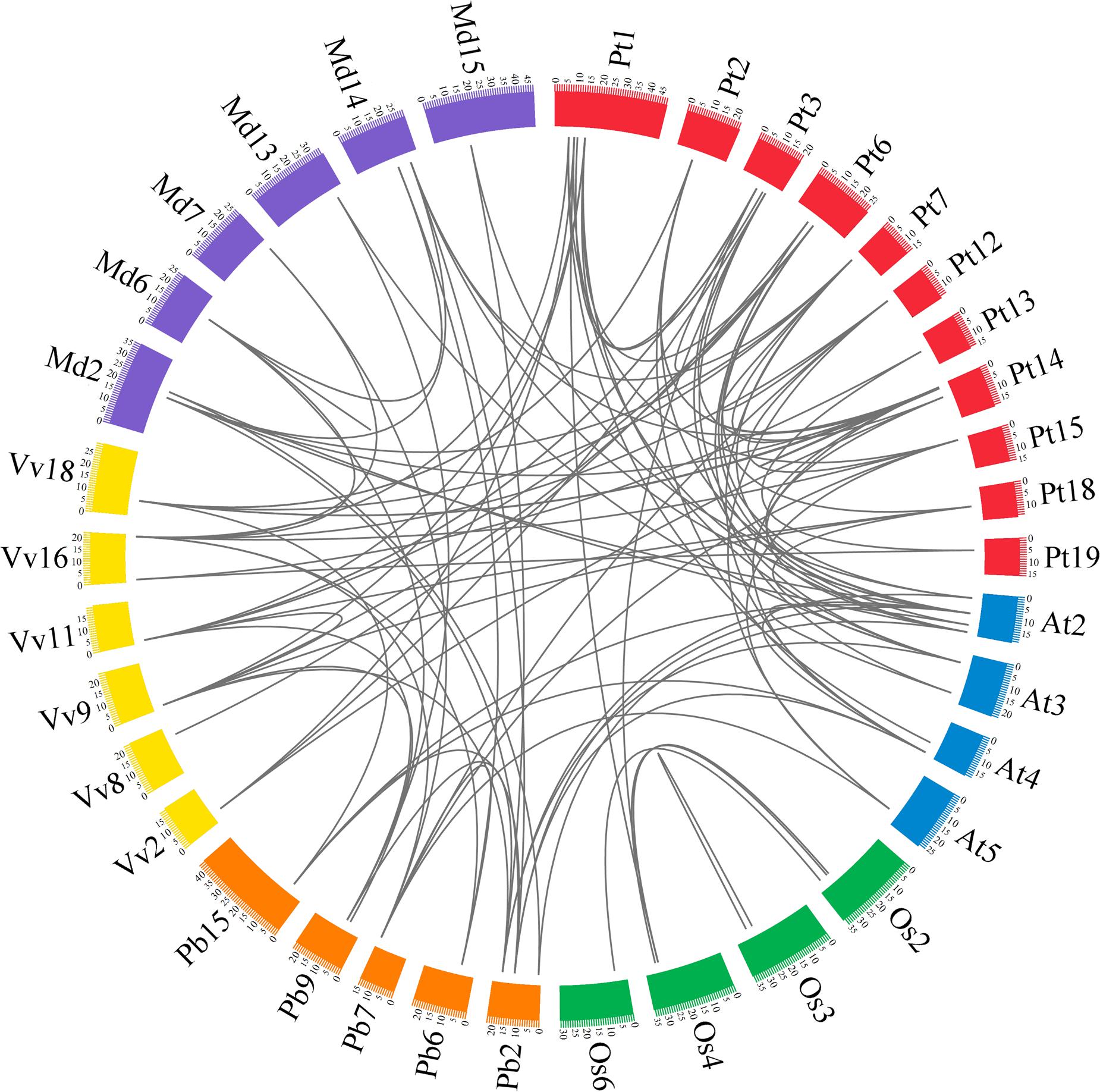
FIGURE 5. Extensive microsynteny of GRF regions across six plant species. The pear, apple, Populus, grape, Arabidopsis and rice chromosomes are depicted as different color boxes and labeled Pb, Md, Pt, Vv, At and Os, respectively. The scale on the circle is in megabases. Syntenic relationships between GRF regions are represented by black lines. The apple GRF gene family was obtained from The Apple Gene Function and Gene Family Database (Zhang et al., 2013).
Firstly, we identified intraspecies relationships of the GRF genes. A total of 14 collinear gene pairs in Populus, six in pear, four in apple, four in rice, two in Arabidopsis, and one in grape were found, respectively (Figure 5; Supplementary Table S4). Additionally, 15 GRF genes were not distributed in any microsynteny, such as PbGRF05 in pear and VvGRF01 in grape. These results revealed that there was not only a whole-genome duplication event but independent duplication events as well.
Subsequently, we analyzed the corresponding interspecies microsynteny in the six plant species. Twenty-five GRF genes were not present in the interspecies microsynteny analysis, including seven OsGRFs, six VvGRFs, five AtGRFs, three PtGRFs, two PbGRFs and two MdGRFs. By microsynteny analysis, 91 conserved syntenic segments were found (Figure 5; Supplementary Table S4). Among them, fourteen orthologous gene pairs were identified from pear and apple, 8 orthologous gene pairs were identified from pear and Arabidopsis, 7 orthologous gene pairs were identified from pear and grape, and 3 orthologous gene pairs were identified from pear and Populus. However, we did not find any orthologous gene pairs between pear and rice. These results may reflect that the relationship between pear and apple was closer than that between pear and grape/Arabidopsis/Populus/rice. Remarkably, some collinear gene pairs detected between pear and Arabidopsis/grape were not identified between pear and Populus, such as PbGRF07/VvGRF03, PbGRF08/AtGRF02, and PbGRF07/AtGRF05 (Figure 5; Supplementary Table S4), suggesting that these orthologous pairs were generated after Populus diverged from the common ancestor of pear and grape/Arabidopsis. In addition, we observed that two or more GRF genes from apple, Populus, Arabidopsis and grape matched one pear GRF gene (Supplementary Table S4), such as VvGRF03 and VvGRF04 orthologous to PbGRF07 and AtGRF02 and AtGRF08 orthologous to PbGRF08 (Supplementary Table S4), implying that these genes are probably paralogous gene pairs.
Gene Duplication of GRF Genes
The GRF gene family may have experienced many duplication processes, including whole-genome duplication, segmental duplication and tandem duplication, during evolution (Moore and Purugganan, 2003). To further understand the evolution of GRF genes, the gene duplication events of the GRF family were identified in five plant species (pear, Populus, grape, Arabidopsis and rice). The similarity of GRF flanking sequences was searched. If four or more genes in the upstream and downstream 100 kb of the two corresponding GRF genes obtained the best non-self match using the BLASTP program (E-value < 10-10), then these two regions were thought as the result of a large-scale duplication event. To avoid the GRF gene pairs that were located in the duplication region with the larger genetic difference, a set of relaxed criteria for gene gathering was defined according to a pair of flanking sequences of the GRF gene containing two or three conserved genes.
The pear genome contained nine GRF genes, eight of which (approximately 88.9%) were found in the duplication region of the genome (Figure 6E). In these gene pairs, six conserved genes were found in the flanking sequences of PbGRF02/PbGRF08; thus, this pair of genes was thought to have evolved from a large-scale duplication event. Nineteen GRF genes were included in the Populus genome, and 17 of these genes (approximately 89.5%) were found to be distributed on the duplicated segments of chromosomes (Figure 6A). As these genes (PtGRF03/PtGRF07, PtGRF05/PtGRF14, PtGRF05/PtGRF15, PtGRF04/PtGRF06, PtGRF11/PtGRF12, PtGRF11/PtGRF14, PtGRF12/PtGRF17, PtGRF13/PtGRF19, PtGRF14/PtGRF15, PtGRF08/PtGRF16) were located in high synteny regions (Figure 6A), their pairs were speculated to evolve from large-scale duplication events. In addition, the gene pair of PtGRF14 and PtGRF15 were located in the adjacent positions of chromosome 14 (Figure 6A), and therefore might be produced by tandem duplication. The Arabidopsis genome contained 9 GRF genes, four of which were found in the duplication regions of the genome. The flanking sequences of two gene pairs (AtGRF02/AtGRF08, AtGRF03/AtGRF06) had remarkable synteny (Figure 6C) and were inferred to have evolved from large-scale duplication events. Moreover, eight of 12 GRF genes were found in the duplication regions of the rice genome (Figure 6D). Conserved gene sequences were found in adjacent sequences of two gene pairs (OsGRF01/OsGRF06 and OsGRF02/OsGRF07), indicating that they were evolved from large-scale duplication events. In contrast, as OsGRF04 and OsGRF05 were located in adjacent positions on the same chromosome, the gene pair should be produced by tandem duplication. Only two of 8 GRF genes were found in the duplication regions of the grape genome. The synteny of the gene pair VvGRF03/VvGRF04 was weak in the duplicated region of the genome, and only two conserved genes were found on the flanking sequences (Figure 6B).
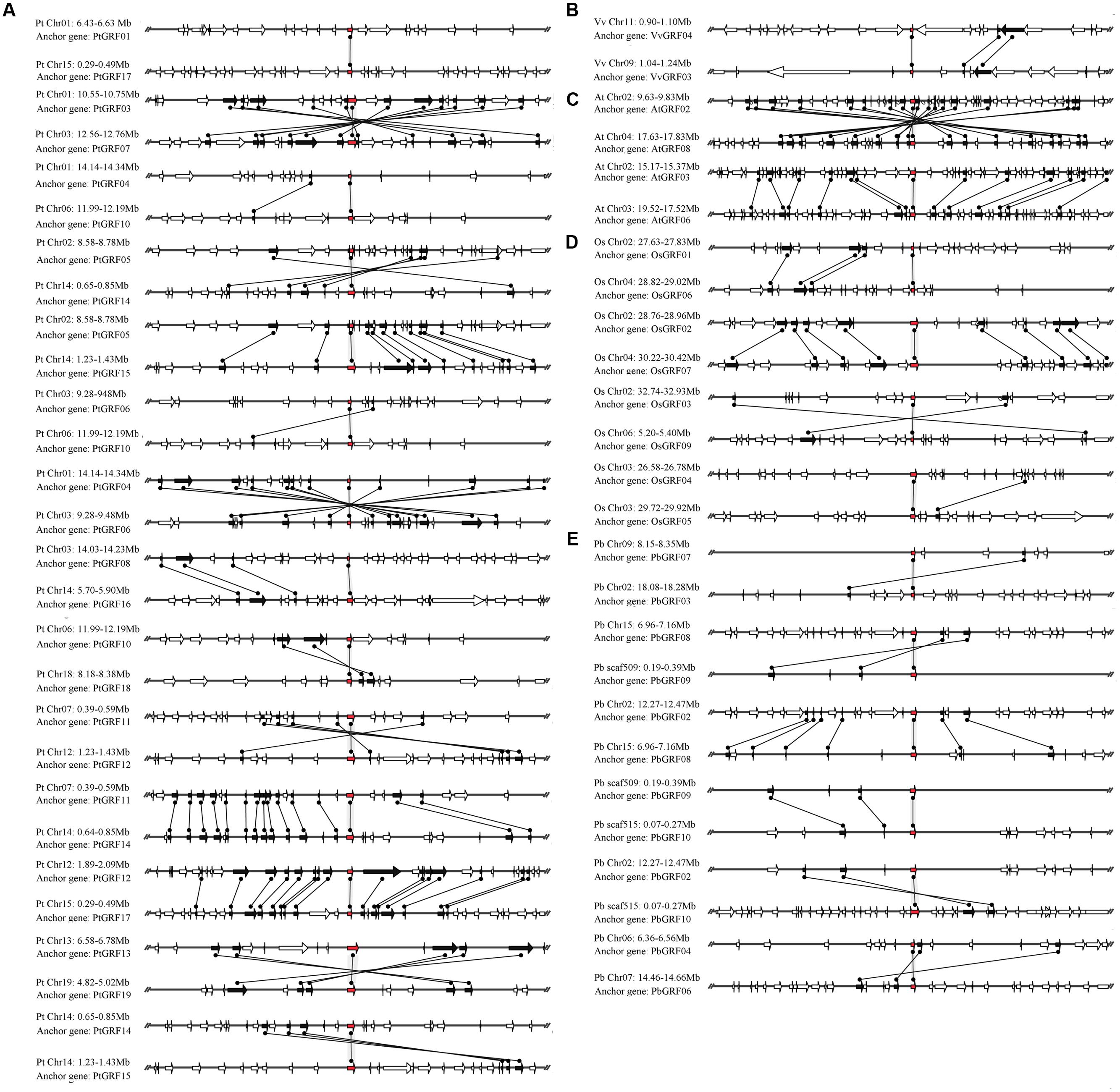
FIGURE 6. Microsynteny maps of GRF genes in Populus (A), grape (B), Arabidopsis (C), rice (D) and pear (E). The relative positions of all flanking protein-coding genes were defined by the anchored GRF genes, highlighted in red. Gray horizontal lines indicate the chromosome segments. Transcriptional orientations are represented by arrows. A gray line connects the conserved gene pairs among the segments.
Strong Purifying Selection for GRF Gene Pairs in Pear
To understand how gene duplications evolved into distinct GRF genes with different functions, we investigated the non-synonymous (Ka) and synonymous (Ks) substitutions and the ratio of Ka/Ks during the evolution of the GRF gene family in five plant species. In general, Ka/Ks < 1 indicates negative or purifying selection with functional constraint, Ka/Ks = 1 indicates neutral selection, and Ka/Ks > 1 indicates positive selection.
In our research, the Ka/Ks ratios of all pear GRF paralogous pairs were less than 0.2 (Supplementary Table S5), indicating that the GRF gene family evolved under strong purifying selection. Thus, we concluded that the GRF genes were slowly evolved at the protein level in pear. Remarkably, 27 GRF gene pairs appeared to be under purifying selection (Figure 7), because of their Ka/Ks ratios less than 1. Subsequently, we performed a sliding-window analysis of Ka/Ks between each pair of GRF paralogous and further clarified the selection pressures in pear. As shown in Figure 8, most Ka/Ks across coding regions were much less than one, with exception for one or several distinct peaks (Ka/Ks > 1). Compared with the other regions (peaks), the WRC and QLQ domains generally had lower Ka/Ks ratios (valleys), consistent with functional constraint being dominant in these domains. Together with the sliding window and Ka/Ks analysis (Figure 8), we deduced that strong purifying selections might have played a key role in the evolution of GRF genes, especially for the WRC and QLQ domains in pear.
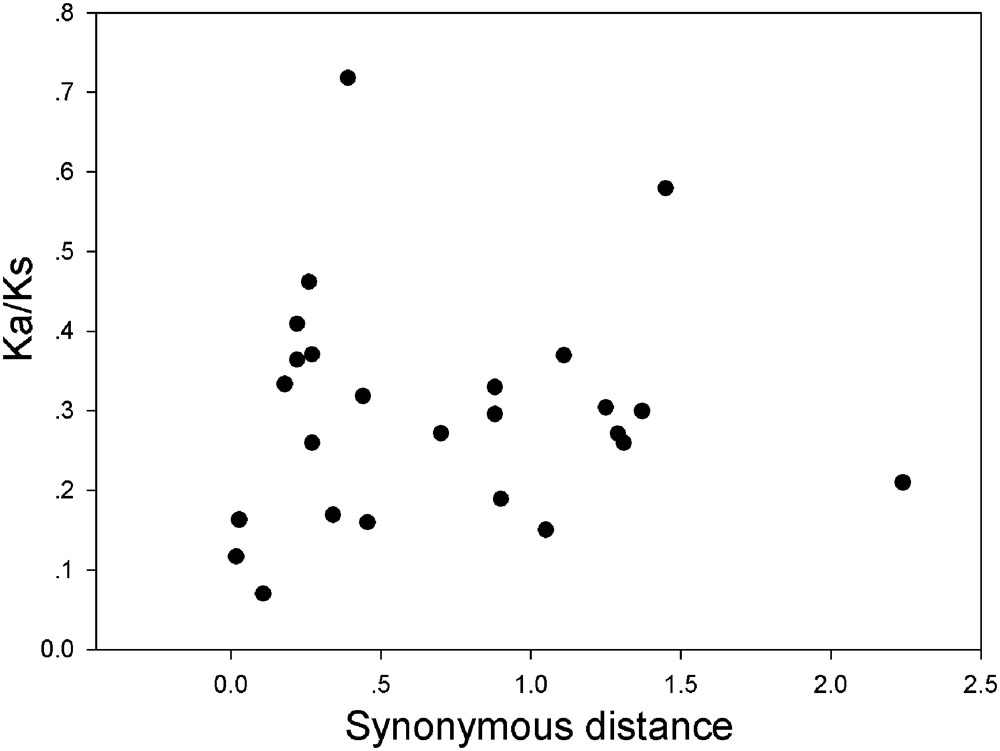
FIGURE 7. Scatter plots of the Ka/Ks ratios of duplicated GRF genes in pear, Populus, grape, Arabidopsis and rice. The X- and Y-axes denote the synonymous distance and Ka/Ks for each pair, respectively.
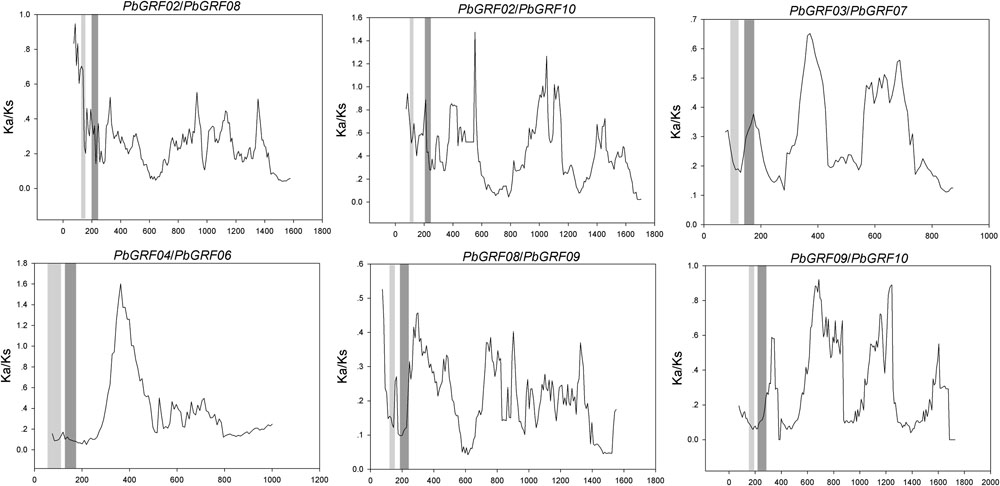
FIGURE 8. Sliding window plots of duplicated GRF genes in pear. The gray blocks, from light to dark, represent the positions of the WRC and QLQ domains, respectively. The window size is 150 bp, and the step size is 9 bp.
Functional Divergence in the GRF Gene Family
To understand whether amino acid substitutions in the GRF gene family caused adaptive functional diversification, DIVERGE 3.0B1 software (Gaucher et al., 2002; Gu et al., 2013) was used to estimate type I and type II functional divergence between gene subfamilies in the GRF gene family based on posterior analysis. In addition, because subfamily III contains only two sequences, which is less than 4 sequences required by the DIVERGE software analysis (Gaucher et al., 2002; Gu et al., 2013), we did not analyze subfamily III. Type I functional divergence usually results in a selective change in the specific amino acid, that is, the rate of evolution changes. The results revealed that 𝜃I was not gained between subfamily V and subfamily I or subfamily IV. This may be caused by an ML of less than 0 (Table 1). A card test (x 2) was performed for groups with a 𝜃I. The P-values of groups I and II, I and IV, II and IV, II and V were less than 0.05, reaching a significant level. To avoid the occurrence of false positives, we determined the sites of posterior probability (Qk) > 0.9 to be key amino acid sites leading to functional divergence according to previous research methods (Yin et al., 2013). The results showed there were significant type I functional differences in the 295th amino acid between I and II, in the 231st and 326th amino acid sites between I and IV, and in the 295th amino acid site between II and IV.
Type II functional divergence occurred after gene duplication, which only resulted in the change in the physical and chemical properties of amino acids. As shown in Table 2, the type II functional divergence coefficients between any two subfamilies are relatively small, some even negative (groups for which 𝜃II is negative were not included in the detailed analysis). Subfamilies II and V had three key amino acid sites (207, 327, and 331), and subfamilies I and IV had a critical amino acid site (231) at a locus that is a key site in the type I functional divergence analysis, suggesting that this locus may have a very close relationship with the change in GRF function (Table 2). Three key amino acid sites (207, 327, and 331) were detected in both subfamilies II and V. And another key amino acid site (231) in the type I functional divergence analysis, was detected in both subfamilies I and IV, suggesting that this site may have a very close relationship with the change in GRF gene function.
Discussion
Growth-regulating factors are plant-specific transcription factors that play key roles in plant growth and development. In our research, by searching local genome databases, 19, 12, 10, 9, and 8 GRF genes were identified in Populus, rice, pear, Arabidopsis, and grape, respectively. The GRF genes were divided into five classes, and orthologous pairs of pear and grape GRF proteins were more common according to the topology of phylogenetic tree, which revealed that some ancestor GRF genes existed before the divergence of pear and grape during evolution.
There exist functional differences of GRFs between the five plant species, which might be related to the diversity both of GRF genes’ exon-intron structures and motif components. In our study, the 58 GRF genes contain different numbers of introns/exons, implying that there is diversity in the GRF genes of the five plant species. For example, the GRF gene AtGRF07 contains five exons, while the VvGRF06 has only one exon. Nevertheless, the most closely related GRF genes shared similar exon-intron structure and motif composition in the same subfamily, either in their exon lengths or intron numbers. Furthermore, different conserved protein motifs were present in individual GRF proteins based on the MEME analysis. The differences in these features among the subfamilies revealed that the GRF members were functionally diversified. Interestingly, all known GRF proteins have motif 1 and motif 2, which encode a conserved WRC domain (containing a Trp-Arg-Cys structure) (Kim et al., 2003) and QLQ domain (containing a Gln, Leu, Gln structure) (Kim et al., 2003), respectively. Among these domains, the WRC domain is known as the zinc-finger structure (Rushton et al., 1995). As shown in Figure 4 and Supplementary Figure S2, zinc-finger structures are tightly connected in WRC motifs, implying that this domain functions in DNA binding.
Based on the comparative genome analyses, although the chromosome numbers and genome sizes of different plant species were diverse, gene orders among the related species were still highly conserved in the process of million years of evolution (Devos and Gale, 2000). Comparisons among the GRF genes across the five plant species’ genomic sequences implied the presence of one or more large-scale genome duplications during early evolution. Strong microsynteny was detected in the five dicot (pear, apple, Populus, Arabidopsis, and grape) genomes. In contrast, little or no microsynteny was detected between the five dicots (pear, apple, Populus, Arabidopsis, and grape) and one monocot (rice). For example, the low microsynteny (two pairs) of GRF genes from five dicots (pear, apple, Populus, grape, and Arabidopsis) and a monocot (rice) was probably because these plants are not closely related. Remarkably, the synteny blocks (14) in the Populus genome were much higher than the synteny blocks of the monocot (rice) and four other dicot (pear, apple, grape, and Arabidopsis) genomes, revealing that Populus GRF genes might have undergone large-scale duplication events during evolution, as shown in Figure 6A. Interestingly, we did not observe microsynteny relationships among OsGRF01-05, OsGRF07-12, PtGRF03, PtGRF07, PbGRF09, PbGRF10, MdGRF03, VvGRF03 and VvGRF04 and other 18 GRF gene members in these genomes studied, implying that these genes were either formed through complete transposition and loss of their primogenitors or ancient ones without detectable linkage to other GRF genes.
Gene duplications are of the major driving forces for generating novel genes, which would help organisms adapt to complex environments. Both events of tandem duplication and large-scale duplication are the main patterns of gene family expansion in plants, such as the MYB gene family in pear (Cao et al., 2016a), CHS gene family in maize (Han et al., 2016), or MYB gene family in Setaria italica (Muthamilarasan et al., 2014). In the present study, a high frequency of GRF genes was distributed in duplicated blocks, implying that large-scale duplications (whole-genome or segmental duplication) contributed to the expansion of the GRF gene family in plants. The Ka/Ks of the 27 paralogous gene pairs suggest that purifying selection may be largely responsible for maintaining the functions of GRF proteins from the four dicots (pear, Populus, Arabidopsis, and grape) and one monocot (rice). Furthermore, the Ka/Ks of pear GRF paralogous gene pairs were less than 0.2, suggesting that these genes underwent slow evolutionary non-diversification following duplication. In addition, we detected strong positive selection in coding regions in several pear GRF gene pairs, implying functional differentiation.
We used DIVERGE software to analyze subfamily I and subfamily IV. In the GRF sequence analysis, we detected 231 key functional divergences in sites, and the analysis of type I and type II functional divergence assayed important amino acid sites that may lead to functional differentiation of GRF decisive sites; thus, our study provides a reference for subsequent researchers exploring GRF functional divergence.
Conclusion
In the present study, 58 GRF gene members were analyzed, including their physical location, phylogenetic relationship, conserved microsynteny, gene duplication and Ka/Ks. By phylogenetic analysis, these GRF genes were divided into five subfamilies. In each subfamily, we found that gene structure and motif distribution features were relatively conserved. Based on genome sequences of the five species (pear, Populus, grape, Arabidopsis, and rice), a comprehensive analysis of GRF genes was performed and the results showed a wide range of synteny and the presence of one or more large-scale genome duplications during early evolution. Our results suggest that large-scale gene duplication was the major pattern of expansion for the vast majority of GRF genes. These genes were under strongly purifying selection and maintained their functional stability. The systematic analysis might contribute to the extrapolation of GRF gene function from one lineage to another.
Author Contributions
YuC conceived of and designed the experiments; YuC and YH performed the experiments; YuC analyzed the data; YuC, YH, QJ, YL, and YoC contributed reagents/materials/analysis tools; YuC and YaH wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Natural Science Foundation of China (grant 30771483, 31171944 and 31640068). We extend our thanks to the reviewers for their careful reading of and helpful comments on this manuscript.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01750/full#supplementary-material
FIGURE S1 | Phylogenetic tree of the GRF proteins in monocots Brachypodium distachyon, Oryza sativa, Setaria italic, Zea mays, Sorghum bicolor and the dicots Arabidopsis thaliana, Populus trichocarpa, Glycine max, Citrus sinensis, Vitis vinifera, Cucumis sativus, Brassica rapa and Chinese pear. For the thirteen-species GRF gene tree, the GRF proteins of Brachypodium distachyon, Setaria italic, Zea mays, Sorghum bicolor, Glycine max, Citrus sinensis, Cucumis sativus and Brassica rapa were obtained from PLAZA 3.0 database, The neighbor joining tree was constructed using MEGA7 software.
FIGURE S2 | Sequence alignment of GRF domains in GRF proteins from pear, Populus, grape, Arabidopsis and rice. The locations of the WRC and QLQ domains are represented by black dots.
TABLE S1 | Annotation of GRF genes in pear, Populus, grape, Arabidopsis and rice.
TABLE S2 | MEME motif sequences in GRF proteins.
TABLE S3 | Blast2GO annotation details of 68 GRF protein sequences.
TABLE S4 | Synteny data in pear, apple, Populus, grape, Arabidopsis, and rice. The apple GRF gene family was obtained from The Apple Gene Function and Gene Family Database (Zhang et al., 2013).
TABLE S5 | Estimates of the dates for the large-scale duplication events in pear.
Footnotes
- ^ http://gigadb.org/site/index
- ^ http://www.arabidopsis.org/
- ^ http://rapdb.dna.affrc.go.jp/
- ^ http://phytozome.jgi.doe.gov/pz/portal.html
- ^ http://pfam.xfam.org/
- ^ http://gsds.cbi.pku.edu.cn/
- ^ http://alternate.meme-suite.org/
- ^ http://www.geneontology.org
References
Bailey, T. L., Johnson, J., Grant, C. E., and Noble, W. S. (2015). The MEME suite. Nucleic Acids Res. 43(W1), W39–W49. doi: 10.1093/nar/gkv416
Baloglu, M. C. (2014). Genome-wide in silico identification and comparison of Growth Regulating Factor (GRF) genes in Cucurbitaceae family. Plant Omics 7, 260–270.
Cao, Y., Han, Y., Li, D., Lin, Y., and Cai, Y. (2016a). MYB transcription factors in Chinese pear (Pyrus bretschneideri Rehd.): genome-wide identification, classification, and expression profiling during fruit development. Front. Plant Sci. 7:577. doi: 10.3389/fpls.2016.00577
Cao, Y., Han, Y., Li, D., Lin, Y., and Cai, Y. (2016b). Systematic analysis of the 4-coumarate:coenzyme a ligase (4CL) related genes and expression profiling during fruit development in the Chinese pear. Genes 7:89. doi: 10.3390/genes7100089
Choi, D., Kim, J. H., and Kende, H. (2004). Whole genome analysis of the OsGRF gene family encoding plant-specific putative transcription activators in rice (Oryza sativa L.). Plant Cell Physiol. 45, 897–904. doi: 10.1093/pcp/pch098
Conesa, A., Götz, S., García-Gómez, J. M., Terol, J., Talón, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
Deleu, W., González, V., Monfort, A., Bendahmane, A., Puigdomènech, P., Arús, P., et al. (2007). Structure of two melon regions reveals high microsynteny with sequenced plant species. Mol. Genet. Genomics 278, 611–622. doi: 10.1007/s00438-007-0277-2
Devos, K. M., and Gale, M. D. (2000). Genome relationships: the grass model in current research. Plant Cell 12, 637–646. doi: 10.2307/3870991
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Filiz, E., Ko,Ý, and Tombuloðlu, H. (2014). Genome-wide identification and analysis of growth regulating factor genes in Brachypodium distachyon: in silico approaches. Turk. J. Biol. 38, 296–306. doi: 10.3906/biy-1308-57
Gaucher, E. A., Gu, X., Miyamoto, M. M., and Benner, S. A. (2002). Predicting functional divergence in protein evolution by site-specific rate shifts. Trends Biochem. Sci. 27, 315–321. doi: 10.1016/S0968-0004(02)02094-7
Gu, X. (1999). Statistical methods for testing functional divergence after gene duplication. Mol. Biol. Evol. 16, 1664–1674. doi: 10.1093/oxfordjournals.molbev.a026080
Gu, X. (2006). A simple statistical method for estimating type-II (Cluster-Specific) functional divergence of protein sequences. Mol. Biol. Evol. 23, 1937–1945. doi: 10.1093/molbev/msl056
Gu, X., Zou, Y., Su, Z., Huang, W., Zhou, Z., Arendsee, Z., et al. (2013). An update of DIVERGE software for functional divergence analysis of protein family. Mol. Biol. Evol. 30, 1713–1719. doi: 10.1093/molbev/mst069
Guo, A., Zhu, Q., Chen, X., and Luo, J. (2007). [GSDS: a gene structure display server]. Yi chuan 29, 1023–1026. doi: 10.1360/yc-007-1023
Han, Y., Ding, T., Su, B., and Jiang, H. (2016). Genome-wide identification, characterization and expression analysis of the chalcone synthase family in maize. Int. J. Mol. Sci. 17:161. doi: 10.3390/ijms17020161
Horiguchi, G., Kim, G. T., and Tsukaya, H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J. 43, 68–78. doi: 10.1111/j.1365-313X.2005.02429.x
Kim, J. H., Choi, D., and Kende, H. (2003). The AtGRF family of putative transcription factors is involved in leaf and cotyledon growth in Arabidopsis. Plant J. 36, 94–104. doi: 10.1046/j.1365-313X.2003.01862.x
Kim, J. H., and Kende, H. (2004). A transcriptional coactivator, AtGIF1, is involved in regulating leaf growth and morphology in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 13374–13379.
Kuijt, S. J. H., Greco, R., Agalou, A., Shao, J., Hoen, C. C. J. T., Overnäs, E., et al. (2014). Interaction between the GRF and KNOX families of transcription factors. Plant Physiol. 164, 1134–1147. doi: 10.1104/pp.113.222836
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Letunic, I., Doerks, T., and Bork, P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40, D302–D305. doi: 10.1093/nar/gkr931
Li, Z., Jiang, H., Zhou, L., Deng, L., Lin, Y., Peng, X., et al. (2014). Molecular evolution of the HD-ZIP I gene family in legume genomes. Gene 533, 218–228. doi: 10.1016/j.gene.2013.09.084
Liang, G., He, H., Li, Y., Wang, F., and Yu, D. (2014). Molecular mechanism of microRNA396 mediating pistil development in Arabidopsis. Plant Physiol. 164, 249–258. doi: 10.1104/pp.113.225144
Librado, P., and Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Lin, Y., Cheng, Y., Jin, J., Jin, X., Jiang, H., Yan, H., et al. (2014). Genome duplication and gene loss affect the evolution of heat shock transcription factor genes in legumes. PLoS ONE 9:e102825. doi: 10.1371/journal.pone.0102825
Maher, C., Stein, L., and Ware, D. (2006). Evolution of Arabidopsis microRNA families through duplication events. Genome Res. 16, 510–519. doi: 10.1101/gr.4680506
Moore, R. C., and Purugganan, M. D. (2003). The early stages of duplicate gene evolution. Proc. Natl. Acad. Sci. U.S.A. 100, 15682–15687. doi: 10.1073/pnas.2535513100
Muthamilarasan, M., Khandelwal, R., Yadav, C. B., Bonthala, V. S., Khan, Y., and Prasad, M. (2014). Identification and molecular characterization of MYB transcription factor superfamily in C 4 model plant foxtail millet (Setaria italica L. PLoS ONE 9:e109920. doi: 10.1371/journal.pone.0109920
Punta, M., Coggill, P. C., Eberhardt, R. Y., Mistry, J., Tate, J., Boursnell, C., et al. (2011). The Pfam protein families database. Nucleic Acids Res. 40, D290–D301.
Rushton, P. J., Macdonald, H., Huttly, A. K., Lazarus, C. M., and Hooley, R. (1995). Members of a new family of DNA-binding proteins bind to a conserved cis-element in the promoters of α-Amy2 genes. Plant Mol. Biol. 29, 691–702. doi: 10.1007/BF00041160
Sato, S., Nakamura, Y., Kaneko, T., Asamizu, E., Kato, T., Nakao, M., et al. (2008). Genome structure of the legume. Lotus japonicus. DNA Res. 15, 227–239. doi: 10.1093/dnares/dsn008
Suyama, M., Torrents, D., and Bork, P. (2006). PAL2NAL: robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612. doi: 10.1093/nar/gkl315
Treich, I., Cairns, B. R., De Los Santos, T., Brewster, E., and Carlson, M. (1995). SNF11, a new component of the yeast SNF-SWI complex that interacts with a conserved region of SNF2. Mol. Cell. Biol. 15, 4240–4248. doi: 10.1128/MCB.15.8.4240
van der Knaap, E., Kim, J. H., and Kende, H. (2000). A novel gibberellin-induced gene from rice and its potential regulatory role in stem growth. Plant Physiol. 122, 695–704. doi: 10.1104/pp.122.3.695
Velasco, R., Zharkikh, A., Affourtit, J., Dhingra, A., Cestaro, A., Kalyanaraman, A., et al. (2010). The genome of the domesticated apple (Malus [times] domestica Borkh.). Nat. Genet. 42, 833–839. doi: 10.1038/ng.654
Vercruyssen, L., Tognetti, V. B., Gonzalez, N., Van Dingenen, J., De Milde, L., Bielach, A., et al. (2015). GROWTH REGULATING FACTOR5 stimulates Arabidopsis chloroplast division, photosynthesis, and leaf longevity. Plant Physiol. 167, 817–832. doi: 10.1104/pp.114.256180
Wang, F., Qiu, N., Ding, Q., Li, J., Zhang, Y., Li, H., et al. (2014). Genome-wide identification and analysis of the growth-regulating factor family in Chinese cabbage (Brassica rapa L. ssp. pekinensis). BMC Genomics 15:807. doi: 10.1186/1471-2164-15-807
Wang, Y., Feng, L., Zhu, Y., Li, Y., Yan, H., and Xiang, Y. (2015). Comparative genomic analysis of the WRKY III gene family in populus, grape, arabidopsis and rice. Biol. Dir. 10:48. doi: 10.1186/s13062-015-0076-3
Wu, J., Wang, Z., Shi, Z., Zhang, S., Ming, R., Zhu, S., et al. (2013). The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 23, 396–408. doi: 10.1101/gr.144311.112
Yang, F., Liang, G., Liu, D., and Yu, D. (2009). Arabidopsis miR396 mediates the development of leaves and flowers in transgenic tobacco. J. Plant Biol. 52, 475–481. doi: 10.1007/s12374-009-9061-7
Yin, G., Xu, H., Xiao, S., Qin, Y., Li, Y., Yan, Y., et al. (2013). The large soybean (Glycine max) WRKY TF family expanded by segmental duplication events and subsequent divergent selection among subgroups. BMC Plant Biol. 13:148. doi: 10.1186/1471-2229-13-148
Zhang, D.-F., Li, B., Jia, G.-Q., Zhang, T.-F., Dai, J.-R., Li, J.-S., et al. (2008). Isolation and characterization of genes encoding GRF transcription factors and GIF transcriptional coactivators in Maize (Zea mays L.). Plant Sci. 175, 809–817. doi: 10.1016/j.plantsci.2008.08.002
Keywords: GRF, microsynteny, gene structure, molecular evolution, gene duplication
Citation: Cao Y, Han Y, Jin Q, Lin Y and Cai Y (2016) Comparative Genomic Analysis of the GRF Genes in Chinese Pear (Pyrus bretschneideri Rehd), Poplar (Populous), Grape (Vitis vinifera), Arabidopsis and rice (Oryza sativa). Front. Plant Sci. 7:1750. doi: 10.3389/fpls.2016.01750
Received: 24 August 2016; Accepted: 07 November 2016;
Published: 24 November 2016.
Edited by:
Karen M. Vasquez, University of Texas at Austin, USAReviewed by:
Xiyin Wang, North China Institute of Science and Technology, ChinaJuan Caballero, Autonomous University of Queretaro, Mexico
Copyright © 2016 Cao, Han, Jin, Lin and Cai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yongping Cai, c3dreDEyQGFoYXUuZWR1LmNu
 Yunpeng Cao
Yunpeng Cao Yahui Han2
Yahui Han2