- School of Chemical and Biotechnology, SASTRA University, Thanjavur, India
Plant specialized metabolites are being used worldwide as therapeutic agents against several diseases. Since the precursors for specialized metabolites come through primary metabolism, extensive investigations have been carried out to understand the detailed connection between primary and specialized metabolism at various levels. Stress regulates the expression of primary and specialized metabolism genes at the transcriptional level via transcription factors binding to specific cis-elements. The presence of varied cis-element signatures upstream to different stress-responsive genes and their transcription factor binding patterns provide a prospective molecular link among diverse metabolic pathways. The pattern of occurrence of these cis-elements (overrepresentation/common) decipher the mechanism of stress-responsive upregulation of downstream genes, simultaneously forming a molecular bridge between primary and specialized metabolisms. Though many studies have been conducted on the transcriptional regulation of stress-mediated primary or specialized metabolism genes, but not much data is available with regard to cis-element signatures and transcription factors that simultaneously modulate both pathway genes. Hence, our major focus would be to present a comprehensive analysis of the stress-mediated interconnection between primary and specialized metabolism genes via the interaction between different transcription factors and their corresponding cis-elements. In future, this study could be further utilized for the overexpression of the specific transcription factors that upregulate both primary and specialized metabolism, thereby simultaneously improving the yield and therapeutic content of plants.
Introduction
Plants produce a wide array of biomolecules through metabolic pathways that are essential for sustenance of life. All the processes involved in plant primary metabolism are essential for maintenance of plant life and growth, whereas compounds resulting from specialized metabolism (specialized metabolites) have a role in plant defense and are also used as therapeutics in human disease treatment. Although primary and specialized metabolic processes are intimately interconnected, with the former providing precursors to the latter, yet most of the specialized metabolism processes have been studied largely in isolation and relatively little is known about their integration with primary metabolism (Tohge et al., 2013; Caretto et al., 2015).
The extensive interrelationship between primary and specialized metabolism is a combined consequence of metabolite partitioning, energy donation and molecular signaling (Ibrahim and Jaafar, 2012). Principal primary metabolic pathways like Pentose Phosphate Pathway, TCA cycle, Photosynthesis, Glycolysis, etc. contribute to these intermediate metabolites, which act as precursors for specialized metabolic processes (Caretto et al., 2015; KEGG Map01100, Figure 1). The levels of these intermediates in their respective pools is governed by various physiological and genetic factors, like environmental stress, location of the system, inherited mutations, etc. (Tohge et al., 2013). Among all, environmental stress acts as a common mediator toward simultaneous upregulation of many primary and specialized metabolic pathways in plants (Bhargava and Sawant, 2013; Schlüter et al., 2013). It is also known to influence primary metabolic pathways like Carbon, Nitrogen and Phosphorous metabolism, as well as specialized metabolic pathways like Phenylpropanoid and Indole Alkaloid biosynthesis (Bhargava and Sawant, 2013; Schlüter et al., 2013; Rejeb et al., 2014), thereby causing upregulation of cascade of stress-responsive genes which impart stress-tolerance to the plants (Gao et al., 1998; Shulze et al., 2005; Ramakrishna and Ravishankar, 2011; Bhargava and Sawant, 2013; Schlüter et al., 2013; Gujjar et al., 2014; Caretto et al., 2015; Le Gall et al., 2015). Under stressed conditions, molecular level changes occurring in plants are principally brought about by transcription factors (TF) binding to their specific recognition sequences upstream to the stress-responsive genes (called as cis-elements). Although exhaustive data is available pertaining to the broad effects of the stress mediated primary and specialized metabolism (Bhargava and Sawant, 2013; Caretto et al., 2015), not many reports highlight the plausible role of cis-element and TF interactions in simultaneous regulation of primary and specialized metabolism genes.
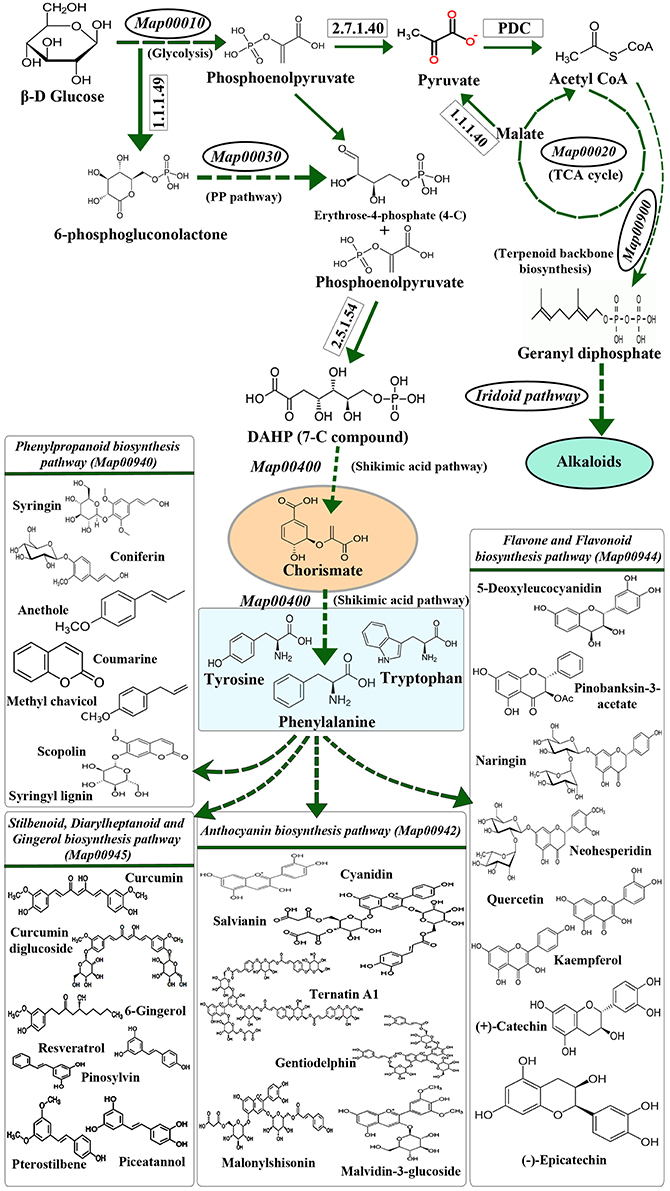
Figure 1. The biochemical link between primary and specialized metabolism. Primary and major specialized metabolisms (example, phenylpropanoid metabolism) are interconnected through intermediates like chorismate. The precursors for the synthesis of amino acids phenylalanine, tyrosine and tryptophan are derived through the Shikimic acid pathway and utilized in the biosynthesis of various specialized metabolites via the Phenylpropanoid biosynthesis pathway. (Map numbers indicate the KEGG pathway ID; PP pathway refers to Pentose phosphate pathway; PDC refers to Pyruvate Dehydrogenase Complex).
The aim of this article is to unravel the interconnection between primary and specialized metabolism under various stress conditions, especially at the transcriptional level. As a part of our study, we have shown the effects of different stress on metabolites and genes specific to primary and specialized metabolic pathways. Further, we present an in-depth analysis of the stress-mediated primary and specialized metabolism genes with regard to their cis-element and TF interactions. To conclude, a detailed study is presented on the TFs that might play a role in simultaneous upregulation of primary and specialized metabolism genes.
Stress Conditions Regulating Primary and Specialized Metabolism Genes
Plant systems are prone to a wide spectrum of stress conditions, like drought, salinity, temperature extremities, heavy metals, biotic (pathogen attacks) and human factors (herbicides, pesticides, weedicides, pollution, loss of gene pool) (Yadav, 2010). As a consequence, an estimated average global crop loss of 50% is caused due to varied stress conditions (Grover et al., 1998; Peleg et al., 2011; Haggag et al., 2015). Farmers additionally face numerous problems, including erratic and scanty rainfall, saline/alkaline soils, flash floods, water logging and global warming, which basically act as environmental stress, thereby hampering the overall productivity (Jenks and Hasegawa, 2005).
Plant stress has been one of the most widely studied areas of biological research, wherein scientific efforts are involved in studying its effects and devising techniques toward its mitigation. As a direct consequence of stress, plants undergo gross biochemical, physiological and molecular changes (as depicted in Figure 2). Due to variations in the metabolic profile of plants under stressed conditions (as described in Supplementary Table 1), the natural requirement of free energy toward maintenance of homeostasis and growth-associated processes get lowered, thereby causing growth-retardation and reduction in the overall plant productivity (Caretto et al., 2015). Plants inherently possess various systems to protect themselves from different forms of stress. This exercise is a combination of a complex array of regulations that occur at various levels, i.e., at whole plant, tissue, cellular, sub-cellular, genetic and molecular levels (Shulze et al., 2005; Prasad et al., 2008; Yadav, 2010; Qados, 2011; Ramakrishna and Ravishankar, 2011; Rejeb et al., 2014). Primarily, plants combat stress by redirecting the metabolic machinery to overproduce certain defense-associated primary and specialized metabolites (Caretto et al., 2015). As seen in Supplementary Table 1, distinct forms of stress display similar metabolite profiles, belonging to primary and specialized metabolism. The elevated levels of diverse metabolites under similar conditions of stress may arise due to coregulation of biochemical pathways at the molecular level. For example, literature evidence points toward an increased accumulation of at least 15 amino acids (belonging to the primary metabolism) together with volatile organic compounds (VOCs) under drought stress (Joshi and Jander, 2009; Fraire-Velázquez et al., 2011; Gill and Tuteja, 2011; Álvarez et al., 2012; Du and Wang, 2012; Hayat et al., 2012; Kendziorek et al., 2012; Griesser et al., 2015; Hudson, 2015; Niinemets, 2015; Weldegergis et al., 2015). Additionally, it was noted that abiotic stresses like temperature and salinity could regulate the levels of other primary (sugar alcohols and sugars) and specialized (phenylpropanoids, alkaloids, etc.) metabolites. (Flores and Galston, 1982; Smith, 1984; Cho et al., 1999; Streeter et al., 2001; Weise et al., 2006; Cuevas et al., 2008; Rosa et al., 2009; Gill and Tuteja, 2010; Hochberg et al., 2013, 2015; Zhao et al., 2013; Alam et al., 2014; Mouradov and Spangenberg, 2014; Zhang et al., 2014; Le Gall et al., 2015; Saleh and Madany, 2015; Sheshadri et al., 2015; Wei et al., 2015). Furthermore, biotic stress like herbivory also displayed a remarkably similar metabolite profile in plants; while primary metabolites like phenylalanine and allantoin were found to be elevated, the levels of VOCs (specialized metabolites) were also enhanced (Fraire-Velázquez et al., 2011; Du and Wang, 2012; Hayat et al., 2012; Kendziorek et al., 2012; Griesser et al., 2015; Hudson, 2015; Weldegergis et al., 2015; Takagi et al., 2016). Thus, the trend of overproduction of primary and specialized metabolites arising from diverse pathways under similar conditions of stress, further confirms the predominant role of stress as a possible link to elucidate the crosstalk between primary and specialized metabolism (Tuteja, 2007; Bolton, 2009; Qados, 2011; Bhargava and Sawant, 2013; Chamoli and Verma, 2014).
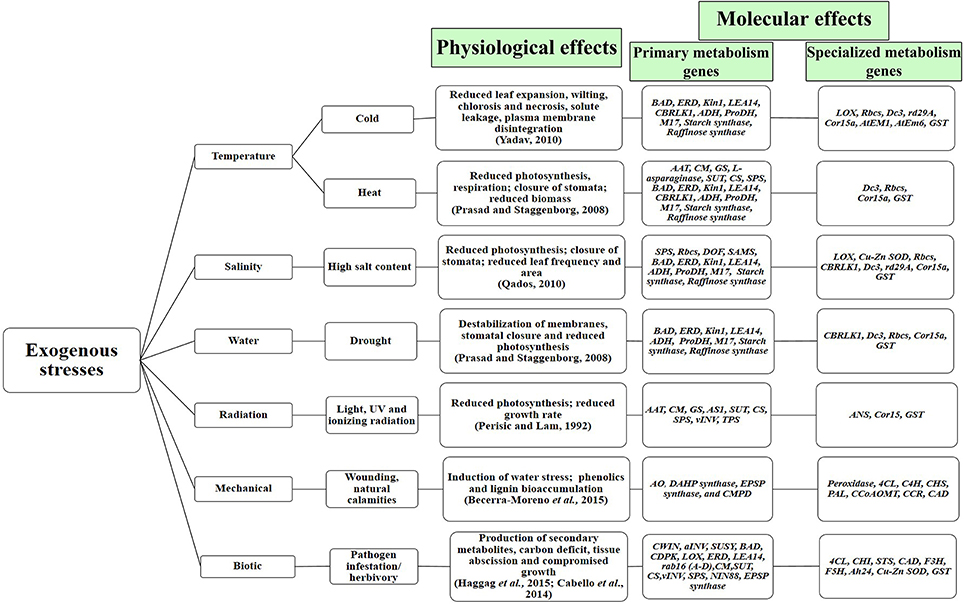
Figure 2. Physiological and molecular effects of phyto-stress. Temperature, salinity and drought stress have similar physiological and molecular footprints. The depicted primary and specialized metabolism genes show synchronized upregulation under abiotic and biotic stress (Full forms, the detailed list of stress-regulated genes and their references have been given in Table 1).
The process of stress tolerance in plants principally involves the regulation of stress-responsive genes that encode for primary metabolites, specialized metabolites and TFs (Davuluri et al., 2003; Floris et al., 2009; Osakabe et al., 2014). This advantage combined with various inherent signaling mechanisms (like pH, metal ions, symbionts, etc.) causes the upregulation of several cascade of genes in plant systems (Tuteja, 2007; Palmieri et al., 2008; Rushton et al., 2012). The mechanism adopted by these genes in bringing about stress tolerance depends on their inherent function, type of stress and the plant system (Davuluri et al., 2003; Shinozaki and Yamaguchi-Shinozaki, 2007; Floris et al., 2009; Osakabe et al., 2014). Functionally, majority of these genes are involved directly in stress mitigation by regulating physiological parameters like water homeostasis and osmoregulation via endogenous signaling (Tuteja, 2007). Table 1 illustrates the predominantly studied primary, specialized and TF genes that are coregulated under different stress conditions.
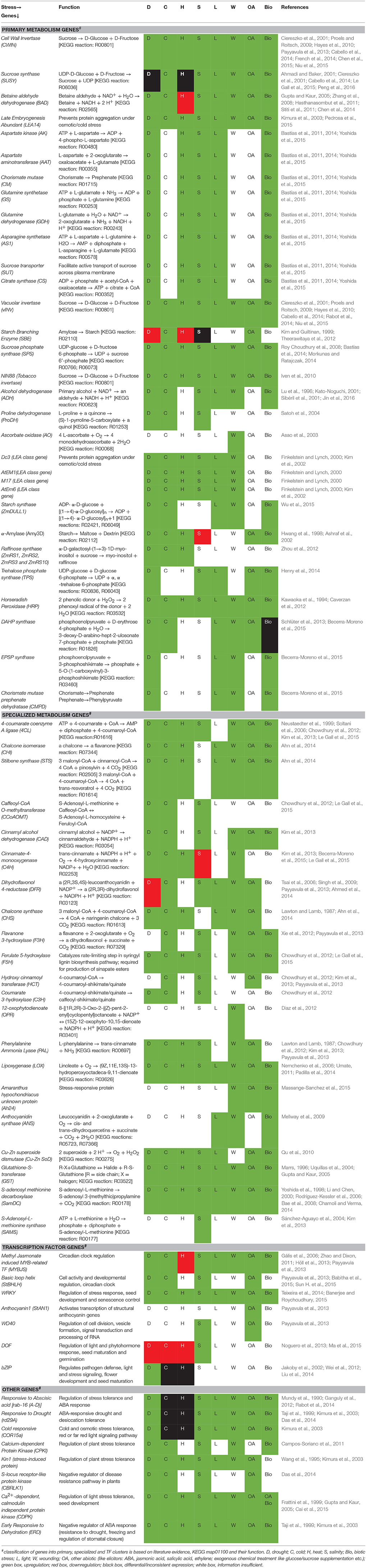
Table 1. Predominant primary metabolism, specialized metabolism and TF genes coregulated under similar stress conditions.
The regulation of a wide spectrum of genes under stress occurs principally at the transcriptional level (Shinozaki and Yamaguchi-Shinozaki, 2007). This is especially brought about by TF binding to their specific cis-elements present in the 5′ flanking regions of gene(s) (Passricha et al., 2016). Moreover, the patterns of cis-elements present among the promoter and intronic regions decide the levels of gene expression (Rombauts et al., 2003; Brown et al., 2007; Zou et al., 2011; Hernandez-Garcia and Finer, 2014), and any mutation(s) occurring in this region can greatly influence the stress-responsiveness of the coded genes (Wittkopp and Kalay, 2012). Most notably, diverse forms of stress may activate similar cis-element TF regulatory networks (Faktor et al., 1996; Kim et al., 2006; Soltani et al., 2006; Mellway et al., 2009; Cao et al., 2012; Payyavula et al., 2013; Ahn et al., 2014; Chen et al., 2015; Zhu et al., 2015). Despite having a great depth of understanding on plant stress and its significance in connecting diverse pathways, not many reports are available that connect the primary and specialized metabolism at the transcriptional level. The forthcoming sections present an in-depth understanding of the molecular level regulation of stress-responsive genes, with an insight into the transcriptional regulation mediated by cis-element and TF interactions.
As observed from Table 1, the expression profile of genes show remarkable similarity among primary and specialized metabolism. Under similar stressed conditions, it could be observed that most of the primary, specialized and TF/other genes show enhanced expression. Literature evidences indicate that coexpression of wide spectrum of genes is a resultant of coregulation at the transcriptional level, primarily via cis-elements and TF interactions (Brown et al., 2007; Shinozaki and Yamaguchi-Shinozaki, 2007; Floris et al., 2009; Nakashima et al., 2009; Lata et al., 2011; Zou et al., 2011; Basu et al., 2014). The study involving cis-elements has gained impetus in the recent years, especially in elucidating the link between pathways which are known to be dependent on each other, but whose genetic inter-dependency is not much known. Bioinformatics has enabled researchers to elucidate and forecast the type of stress-responsive transcriptional regulation of genes by studying the pattern of cis-elements present in the upstream regions of these genes (Ibraheem et al., 2010). Several tools have been made available for this purpose, like Plant Cis-Acting Regulatory DNA Elements Database (http://www.dna.affrc.go.jp/PLACE; Lescot et al., 2002), Genomatix (http://www.genomatix.de; Cartharius et al., 2005) and Arabidopsis Gene Regulatory Information Server (AGRIS; http://arabidopsis.med.ohio-state.edu/; Davuluri et al., 2003). Based on a highly efficient Hidden Markov Model, a database of probable TF binding sites in the promoters of stress–responsive genes of A. thaliana is also available for further research (Malhotra and Sowdhamini, 2014). Cis-elements are further involved in imparting several auxiliary functions to the plant systems, like developmental regulation of growth associated processes, morphological modifications, regulating senescence, DNA damage repair mechanisms, etc. (Floris et al., 2009; Nakashima et al., 2009; Zou et al., 2011).
Among the stress-responsive genes enlisted in Table 1, the promoter analysis data was available for 33 genes, and their characteristic cis-elements have been shown in Supplementary Table 2. It can be inferred that there are several cis-elements that are commonly overrepresented in the promoter regions of various primary and specialized metabolism genes. Such elements can present a plausible molecular link between diverse pathways. As discussed earlier, these elements possess additional roles (like developmental regulation, controlling circadian cycle, etc.) apart from their characteristic role of stress-responsive transcriptional regulation. The promoter regions of most of the primary and specialized metabolism genes possessed following cis-elements: ABRE, G-box, W-box and MYB-recognizing elements. ABRE and G-box elements are favorable binding sites of bZIP TFs that regulate stress responses (Heinekamp et al., 2002, 2004). Studies have shown that one of the bZIP TFs, BZI-1 is involved in imparting auxin responsiveness and regulating pollen development via carbohydrate allocation (Heinekamp et al., 2004; Iven et al., 2010). BZI-1 TFs bind specifically to the ACEs (ACGT core elements; example: G-box, GT-box, etc.), thereby controlling stress-specific regulation of primary (NIN88, Adh, α-amylase, AtEM6, ProDH, Dc3, LEA, Kin1, BAD, GST and SbeI; Lu et al., 1996; Finkelstein and Lynch, 2000; Kim et al., 2002; Satoh et al., 2004; Wobbes, 2004; Iven et al., 2010; Bastías et al., 2011, 2014) and specialized metabolism genes (CHI, CHS, Ah24, DFR and PAL; Strathmann et al., 2001; Heinekamp et al., 2002, 2004; Fujita et al., 2005; Iven et al., 2010; Yoshida et al., 2015). It can therefore be inferred that the ACEs and bZIP TFs interactions can play a central role in coregulating primary and specialized metabolism in plants. In addition, the WRKY binding sites (W-box) were also found to be present in the promoter regions of some primary (GST, ANS, SUSY, vINV and CWIN) and specialized metabolism genes (HCT, CHS, C3H, F3H, PAL and DFR). Physiologically, the WRKY TFs binding to W-boxes regulate various developmental activities (trichome development and controlling senescence) and defense associated processes (like regulating responses to pathogen infestation and other abiotic stresses) (Aken et al., 2013; Llorca et al., 2014). This mechanism of coregulating diverse genes under stressed conditions indicates at WRKY-W-box interactions as a prospective link between primary and specialized metabolism. Similarly, the MYB-binding sites are present in the promoters of several primary (LEA14, CWIN, vInv 1 and SUSY) and specialized metabolism genes (CHI, HCT, ANS, DFR, F3H, PAL, C3H and GST). Moreover, it is known that MYB TFs binding to their respective cis-elements regulate changes in various processes like hormonal signaling, specialized metabolism (phenylpropanoid and anthocyanin biosynthesis), cellular morphogenesis, and formation of meristem (Cao et al., 2013; Höll et al., 2013). This striking similarity observed in the promoter regions of functionally distinct genes provides ample scope to draw a link between diverse pathways at the transcriptional level via cis-element-TF interactions serving as the bridges.
TF Families Regulating Stress-Mediated Link between Primary and Specialized Metabolism
In plants, the transcriptional mode of gene regulation is mediated synergistically by a combination of TFs acting in tandem to bring about different expression patterns. Studies have revealed that in Arabidopsis, about 5–10% of the functional genes are TFs, which regulate diverse genes under different environmental conditions (Mitsuda and Ohme-Takagi, 2009). The most studied stress-responsive TFs principally belong to six families, namely bZIP, WRKY, MYB, APETALA2 (AP2 family), NAC and Zinc finger family (ZnF) (Saibo et al., 2009; Gujjar et al., 2014; Malhotra and Sowdhamini, 2014). However, the largest TF families- bZIP, WRKY, MYB and AP2 are more extensively involved in regulating diverse metabolic pathways in plants under stress (Heinekamp et al., 2002; Jakoby et al., 2002; Katiyar et al., 2012; Rushton et al., 2012; Wei et al., 2012; Alves et al., 2013; Llorca et al., 2014; Liu et al., 2015; Wang et al., 2015). Also, most TFs can recognize secondary motifs (apart from their primary recognition sequences), which allow them to bind to distinct sites in the promoters. Further, TFs having upto 79% amino acid similarity in their recognition domain have shown distinct DNA binding profiles. Several other TFs like ERFs, bZIPs, etc. also demonstrated their ability to recognize and bind to secondary motifs which partially differ from their respective primary motifs (Franco-Zorrilla et al., 2014). The forthcoming sections describe the mode of action of the above four predominant TF family proteins and their role in simultaneously regulating primary and specialized metabolism genes.
The bZIP Family
The bZIP family (basic leucine zipper) is one of the largest TF families in plants, which is involved in diverse regulatory functions, like abiotic and biotic stress tolerance, hormone signaled gene regulation, sugar signaling, nitrogen, carbon and energy metabolism, light responsiveness and developmental regulation (like cell elongation, differentiation, flowering, senescence and maturation of seedlings, Chuang et al., 1999; Wei et al., 2012; Bastías et al., 2014; Llorca et al., 2014; Zhao et al., 2016). The bZIP TFs have a widespread presence among eukaryotes (17 in S. cerevisiae, 27 in Drosophila, 75 in A. thaliana, 89 in rice, 125 in maize, 131 in soybean, 69 in tomato and 585 among six leguminous plants: G. max, M. truncatula, P. vulgaris, C. arietinum, C. cajan, and L. japonicus, Fassler et al., 2002; Wei et al., 2012; Llorca et al., 2014; Li D. et al., 2015; Wang et al., 2015). These TFs possess a binding affinity toward the core motif–ACGT- (ACEs), which is found in G-Box, A-Box, C-Box and ABRE.
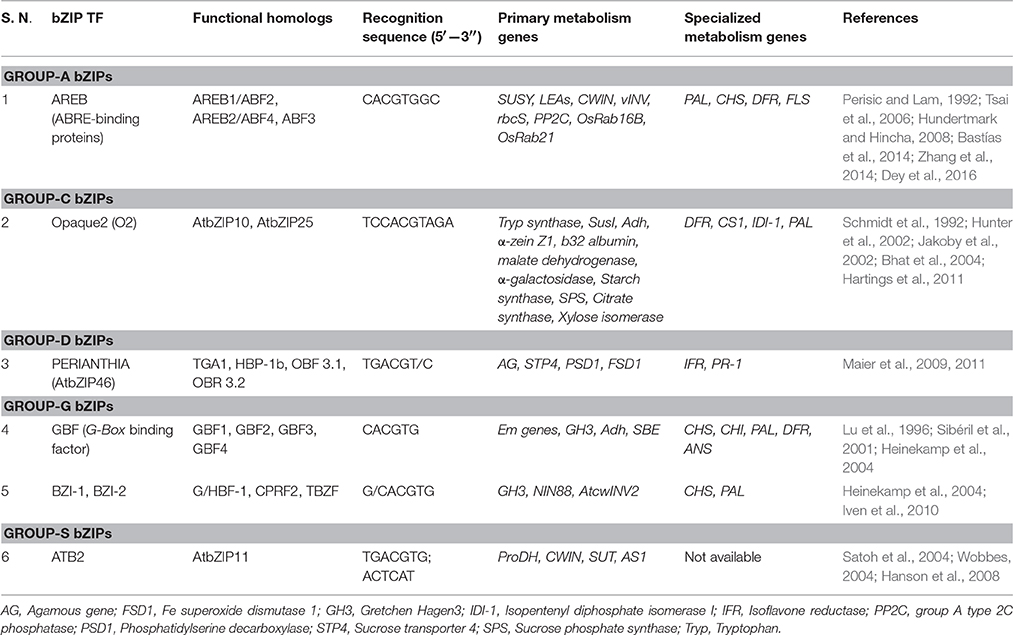
bZIP TFs are comprised of a short basic region linked to a DNA recognition domain followed by a leucine repeat region that imparts amphipathic nature to the protein (Jakoby et al., 2002; Alves et al., 2013; Llorca et al., 2014). The leucine zipper region of the protein binds to the bZIP recognition sequences in a chopstick fashion (Sibéril et al., 2001; Iven et al., 2010; Alves et al., 2013; Llorca et al., 2014). bZIP TFs can be sub-classified into 10 groups, out of which groups A, C, D, G and S have been studied extensively (Jakoby et al., 2002; Dey et al., 2016). Table 2 describes these groups with special emphasis on its relevance in simultaneously regulating primary and specialized metabolism genes.
Among the bZIPs, varied cis-element and TF binding patterns bring about differential expression of diverse stress-responsive genes. For example, studies have indicated that the expression of primary (SUSY, LEAs, CWIN, vINV, PP2C) and specialized metabolism genes (PAL, CHS, DFR, FLS) is regulated by AREB-ABRE interactions via ABA signaling (Narusaka et al., 2003; Gómes-Porras et al., 2007; Bastías et al., 2011, 2014; Basu et al., 2014; Yoshida et al., 2015). Similarly, Opaque 2 (O2) is an endosperm-specific TF that was found to enhance the expression of several primary and specialized metabolism genes (Table 2). O2 TF binding to its recognition sites is mediated by certain transcriptional coactivators (like GCN5 and ADA2), which acetylate histone residues and thereby reinforce the binding. Further, a loss-of-function O2 mutant severely impaired several developmental activities (like seed storage by downregulating storage protein coding genes; Schmidt et al., 1992; Schmitz et al., 1997; Zhang et al., 2015) and defense processes (Hunter et al., 2002; Bhat et al., 2004; Hartings et al., 2011). Further, one of the group-D bZIP TFs, PERIANTHIA was found to regulate developmental processes like controlling floral organ number (via regulation of the MADS domain TF gene Agamous; Maier et al., 2009), shoot meristem regulation (via FEA4, an ortholog of PERIANTHIA, Pautler et al., 2015) and regulating pathogen defense responses (Maier et al., 2011). It is associated with TGA regulators which is known to act upstream to the PR (Pathogenesis-related) gene, thereby conferring pathogen responsiveness to the plant systems. It was further observed that PERIANTHIA TF binds to the promoter regions of several primary and specialized metabolism genes, thereby causing their simultaneous regulation (Table 2). This pattern of simultaneous regulation of primary and specialized metabolism genes is demonstrated by several other bZIP TFs as well (GBFs, BZI, etc.; Lu et al., 1996; Sibéril et al., 2001; Heinekamp et al., 2002, 2004; Iven et al., 2010). HY5, a bZIP TF known to induce chlorophyll and carotenoid genes in plants, acts as a bridge between ABA and GA signaling pathways (owing to the fact that GA and ABA share the common precursor, Geranyl geranyl diphosphate; Mohanty et al., 2016). Despite being associated largely with phytohormone mediated stress responses, the bZIP family is involved in regulating growth and developmental activities like flowering, senescence, seed storage regulation, etc. (Rook et al., 1998; Hunter et al., 2002; Hanson et al., 2008; Hartings et al., 2011). The similarity in the pattern of occurrence of bZIP recognition sites (ACEs) among the promoters of primary and specialized metabolism genes depict bZIP-cis-elements interactions as a credible link to bridge diverse metabolic pathways in planta.
Although several reports indicated positive regulation of downstream genes by bZIP TFs, it has been observed that overexpressing BZI-4 (a bZIP TF that possesses strong affinity toward G-Box element) caused significant reduction in the expression of NIN88 (Iven et al., 2010). Evidence suggests that BZI subfamily possesses conflicting roles in regulating developmental processes. Although BZI-1 and BZI-2 are involved in the transcriptional activation of NIN88 gene, homo-dimerized BZI-4 acts as a repressor (Iven et al., 2010). Furthermore, despite the S-group being the largest bZIP subgroup in Arabidopsis (Jakoby et al., 2002), its significance in regulating specialized metabolism processes have not been studied much. Therefore, an in-depth research into this area needs to be conducted to understand the finer details on bZIP TFs and their involvement in linking the primary and specialized metabolism in planta.
The WRKY Family
WRKY protein family in model plants A. thaliana and N. benthamiana is one of the largest TF families, which majorly bind to the W-box, a 6-bp region (C/TTGACC/T) present in the promoters of various primary and specialized metabolism genes. This has been known to bring about tolerance to abiotic and biotic stresses and regulate developmental processes in plants (trichome development and senescence; Basu et al., 2014; Llorca et al., 2014). W-boxes are present in the upstream regions of several genes like PR1, isochorismate synthase 1 and ABA responsive genes: SamDC, RD29A, COR47, iso1, etc. (Supplementary Table 2; Sun et al., 2003; Rushton et al., 2012; Aken et al., 2013; Basu et al., 2014; Llorca et al., 2014; Singh and Laxmi, 2015). Although every WRKY TF has an affinity toward W-Box, they also possess additional DNA sequence affinities (like SUSIBA2 and SURE, sucrose responsive elements, core motif TGGACGG; Sun et al., 2003; Bi et al., 2016). The nucleotide sequences flanking the core W-Box element also decide the binding specificity of WRKYs. Most WRKYs are induced by plant hormones, like SA, ABA, etc. However, research reports highlight that SA induction and subsequent binding is more evident for extended W-boxes (Franco-Zorrilla et al., 2014). Since majority of WRKYs bind to W-Boxes to bring about transcriptional and posttranscriptional regulation of diverse genes, plants have developed an extrinsic mechanism to eliminate non-specific binding of repressor WRKYs to cause the activator WRKY to fit in and perform the stress-responsive gene regulation (Llorca et al., 2014).
One of the mechanisms involved in upregulation of stress-responsive genes is via ABA, which triggers the removal of repressor WRKYs from the promoter regions of ABA responsive genes (ABF4, ABI4, DREB1a, MYB2, RAB18; Rushton et al., 2012; Aken et al., 2013). According to studies, some WRKY proteins (AtWRKY40, AtWRKY18 and AtWRKY60) disallow the transcription of ABA responsive genes upon binding to the W-box sequence [(C/T)TGAC(T/C)] in the promoter region. To ensure successful ABA-mediated stress mitigation, these WRKY TFs are translocated from the nucleus to the cytosol by making use of the affinity between the C-terminus of ABA-bound ABA receptor and WRKY proteins (Rushton et al., 2012; Aken et al., 2013). ABAR (or Mg-chelatase H-subunit/putative ABA receptor) is a chloroplast membrane-localized receptor which exposes its N and C-termini to the cytoplasm. ABA, upon binding to the C-terminal of ABAR, promotes the exit of WRKY proteins (WRKY40, WRKY18 and WRKY60) from the nucleus to cytosol, thereby facilitating the enhanced expression of ABA-responsive genes via binding of other activator WRKY TFs (example, WRKY63) to the W-box in the promoter regions (Figure 3; Rushton et al., 2012; Aken et al., 2013).
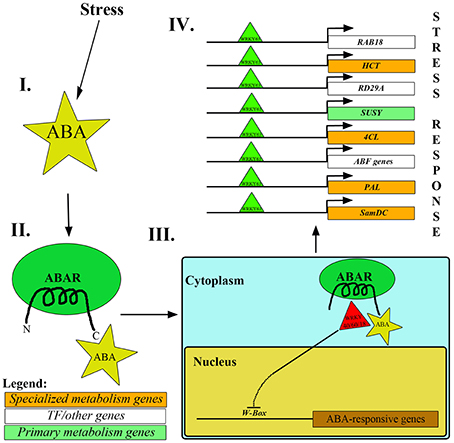
Figure 3. Mechanism of repressor WRKYs removal from nucleus mediated by ABA. (I, II). Under stress conditions, ABA binds to the C-terminus of ABAR. (III) Consequently, it leads to the transport of AtWRKY40/18/60 from the nucleus to the cytoplasm. (IV) Subsequently, AtWRKY63 binds to the promoter regions of stress-responsive genes like RAB18, RD29A, HCT, SUSY, 4CL, PAL, SamDC and ABF genes, thereby enhancing their expression and mitigating stress.
ABA-mediated WRKY-W box binding can form a crucial link between primary/growth-associated metabolic processes and stress-responsive/defense pathways. ABA helps mitigate drought through closure of guard cells of the stomata (Tuteja, 2007; Yoshida et al., 2015), simultaneously regulating the expression of several drought associated and cold stress responsive genes (ABI genes, MYB2, RAB18, RD29A, ABF4, AOX1, DREB2, etc.; Rushton et al., 2012; Qin et al., 2015). The occurrence of ABRE and W-box elements upstream to the coding regions of specialized metabolism genes further determines the plant response to ABA under stress (Fujita et al., 2005; Gómes-Porras et al., 2007; Yoshida et al., 2015). Additionally, reports suggest that in O. sativa, the presence of W-box elements upstream to the polyamine synthesis gene (SamDC) plays a key role in its upregulation (Basu et al., 2014). Furthermore, TaWRKY93 (WRKY protein from wheat) was found to enhance the levels of Pyrroline-5-carboxylate synthase (P5CS; Qin et al., 2015) involved in proline biosynthesis (Proline is known to be directly involved in drought mitigation as osmoticum; Chamoli and Verma, 2014). It was also noted that WRKYs have a significant role in upregulating several other stress-responsive genes (like ABF3, ABIs, DREB2A, RDs, etc.; Qin et al., 2015). The synergistic binding of WRKY to W-box; ABF to ABRE; MYB TFs to MYB recognition elements; CBFs to LTRE and GBFs to GATA was found to upregulate SamDC gene in O. sativa (Basu et al., 2014). Under P. infestans infection, StWRKY1 was found to regulate the levels of 4CL and HCT by binding to the W-boxes present in their promoters (Yogendra et al., 2015).
Scientific evidence suggests that WRKY operation is often synergistically linked to the occurrence of ABA responsive bZIP TFs (Llorca et al., 2014). The pattern of occurrence of similar cis-elements among the upstream sequences of genes constituting diverse pathways (primary metabolism: Invertases, SUSY, SUT; specialized metabolism: DREB1a, PAL, SamDC; Yogendra et al., 2015) leads to a substantial hypothesis that these TF-cis-element interactions can form a regulatory bridge between primary and specialized metabolism. However, the detailed mechanism of WRKY as a plausible link between primary and specialized metabolism genes yet needs to be unraveled.
The MYB Family
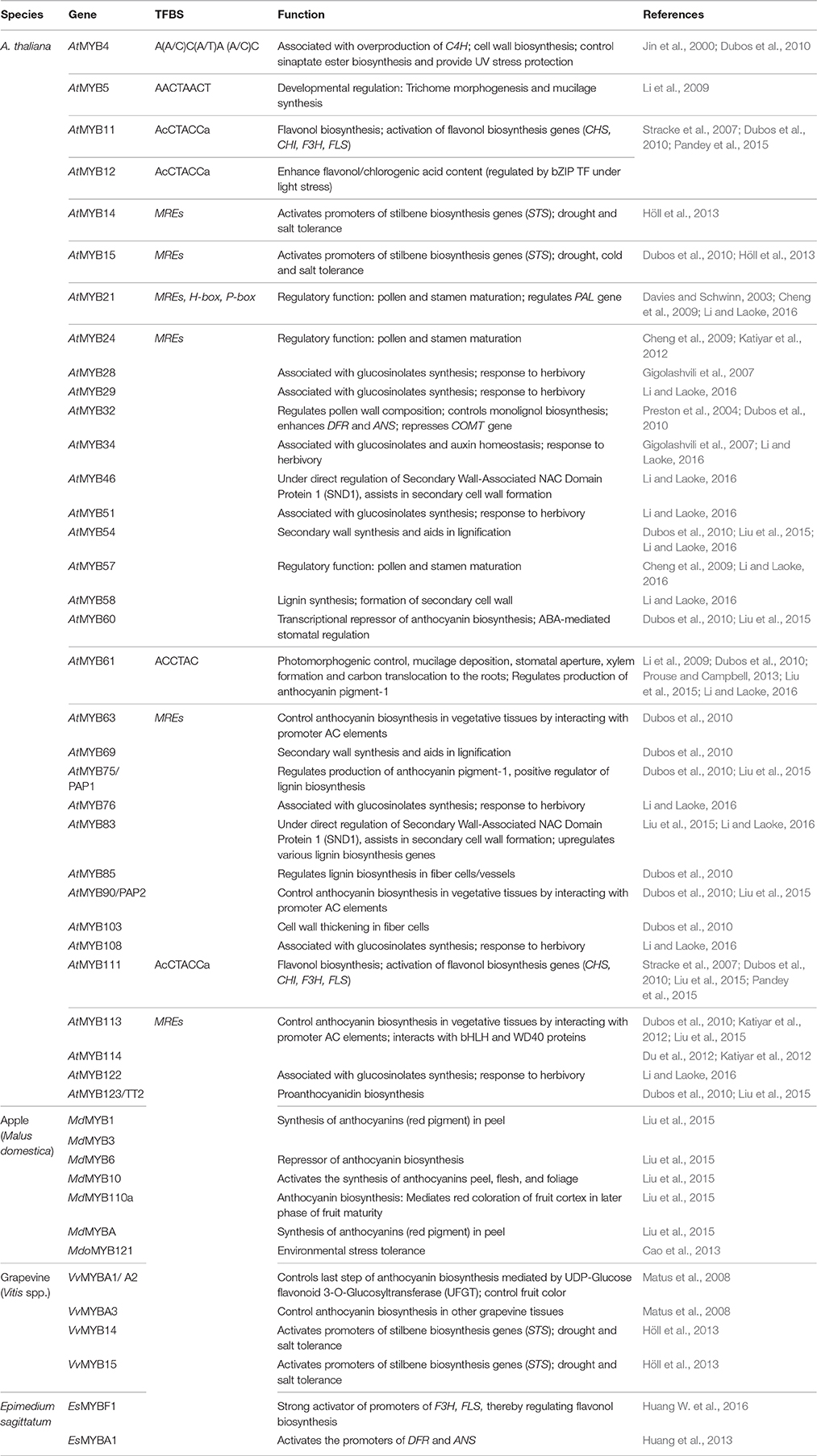
The MYB TF family is also one of the largest TF families in plants. As many as 125 in A. thaliana (Stracke et al., 2007), 205 in G. raimondii (He et al., 2016) and 559 in S. lycopersicum (Gates et al., 2016). Based on the number of MYB domains they contain, MYB TF family can be subdivided into four sub-families, namely 1R (R1/2/3), 2R (R2R3), 3R (R1R2R3) and 4R (R1R2R2R1/2), among which the R2R3-MYBs form the largest population (56.77% in O. sativa; 70.05% in A. thaliana). Literature evidence strongly suggests the involvement of 2R (R2R3-MYBs) in regulating several diverse metabolic processes.
This subfamily of MYB proteins is known to bind to the MYB-recognizing elements (MREs having the consensus sequence ANCNNCC, as demonstrated in MBSI, MBSII and MBSIIG; Franco-Zorrilla et al., 2014; Zhu et al., 2015), regulate Phenylpropanoid metabolism in plants (Liu et al., 2015). Additionally, R2R3-MYBs have been associated with several pleiotropic roles, like cell wall synthesis, regulation of pollen wall composition, glucosinolate biosynthesis, developmental processes, responses to physiological stress and determination of cell fate and identity (Lu et al., 2002; Du et al., 2012; Cao et al., 2013; Höll et al., 2013; He et al., 2016; Gates et al., 2016).
The R2R3-MYB TFs that play a crucial role in transcriptional regulation of primary and specialized metabolism have been enlisted in Table 3. From the table, it can be inferred that although AtMYB32, AtMYB3, AtMYB4, AtMYB26/MS35, AtMYB28, AtMYB29, AtMYB76, AtMYB103, AtMYB34, AtMYB51 and AtMYB122 have been associated largely with primary and developmental processes; AtMYB58, AtMYB63, AtMYB75, AtMYB85, AtMYB68, AtMYB111, AtMYB114 and AtMYB123 are involved much into regulating Phenylpropanoid pathway (lignin/anthocyanin biosynthesis processes).
Interestingly, several MYB TFs have dual roles, like AtMYB 52, AtMYB 54, and AtMYB 69 regulate lignin biosynthesis (specialized metabolism), simultaneously regulating xylan and cellulose biosynthesis (primary metabolism). Similarly, AtMYB46 is also associated with lignification in fibers and vessel tissues, simultaneously regulating xylan and cellulose deposition in A. thaliana. Most notably, research evidence pointed that the cis-element MBSIIG was bound favorably by MYB59 as well as MYB111. While MYB59 has been known to regulate cell cycle and root growth, MYB111 binding to MBSIIG was found to regulate flavonoid biosynthesis along with MYB11 and MYB12. Further, this element was found to be highly overrepresented in the promoter regions of several genes belonging to primary and specialized metabolism (Franco-Zorrilla et al., 2014). It is also known that some MYBs (AtMYB63, AtMYB90, AtMYB113 and AtMYB114) bring about the transcriptional regulation via binding to the AC elements present upstream to the stress-responsive genes through synergistic interaction with bHLH and WD40 TFs. The promoter regions of several primary (LEA14, CWIN, vInv 1, SUSY) and specialized metabolism genes (CHI, HCT, ANS, DFR, F3H, PAL, C3H, GST) have characteristic presence of AC-rich elements in their promoter regions. It thus makes it evident that MYB TFs play a bridging role to link primary and specialized metabolism in plants. Table 3 presents a comprehensive overview of the MYB TFs and their role in regulating metabolic processes in various plant genera.
The AP2/ERF Superfamily
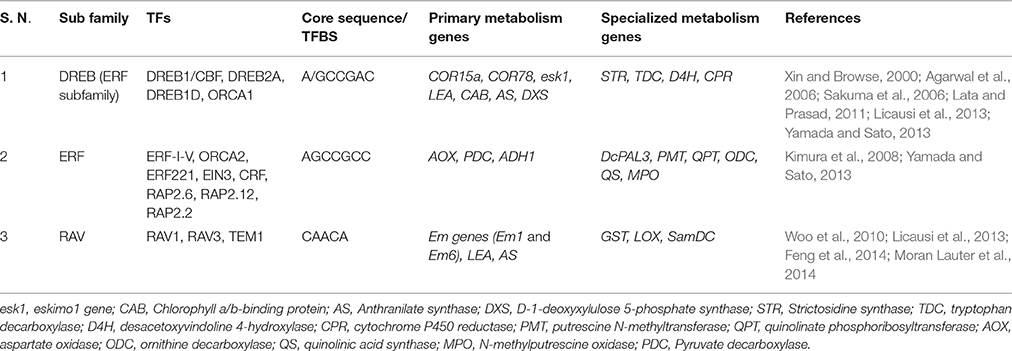
The APETALA2 TF family was initially linked to developmental regulation in plants, like floral development, seed germination and yield regulation. This TF family is associated with a few other pleiotropic roles, like regulating stress tolerance via expression of genes involved in abiotic stress response, disease resistance and ethylene/jasmonic acid/salicylic acid response (Cui et al., 2016; Guo et al., 2016). Based on the number of AP2/ERF DNA binding domains they possess, the AP2/ERF family is further classified into four subfamilies, namely ERF, DREB (one AP2/ERF domain); AP2 (two AP2/ERF domains) and RAV (one AP2 and an additional B3 DNA binding domain; Licausi et al., 2013; Guo et al., 2016; Huang Z. et al., 2016). The ERF subfamily in Arabidopsis is regulated either via a phytohormone dependent (like Ethylene, JA, ABA, auxin, cytokinin and SA; Guo and Ecker, 2004; Arora, 2005; Cheng et al., 2013; Dey and Vlot, 2015) or independent manner (via Ethylene Insensitive or EIN genes, stress like wounding, etc., Guo and Ecker, 2004; Arora, 2005; Dey and Vlot, 2015). ERFs have the ability to distinctly bind to the GCC box and DRE elements (under abiotic and biotic stress; Cheng et al., 2013; Guo et al., 2016) and upregulate downstream genes, thus forming a crucial component of stress mitigation mechanisms in plants. The DREB subfamily also plays a crucial role in abiotic stress mitigation by binding to the DRE/CRT elements (Dehydration Responsive Element/ C-Repeat Element) present upstream to stress responsive genes (like RD29A, COR15a, etc.), leading to plant responses to abiotic stresses like cold, drought and salinity (Chinnusamy et al., 2010; Basu et al., 2014). Similar cis-element LTRE (Low temperature Responsive Element) was found to be involved in mitigating cold stress. The promoter regions of Arabidopsis Cor15A gene (encoding cold-regulated chloroplastic protein, principally involved in cold stress regulation) showed the characteristic presence of DRE elements, while polyamine synthesis gene, SamDC (specialized metabolism) in rice showed the presence of both DREs as well as LTREs in their promoters (Basu et al., 2014). Promoter analysis of principal abiotic stress responsive genes in A. thaliana (COR15A, COR15B, KIN1, KIN2 RD29B, RD29A, RD29B, RD22, RAB18 and COR47) demonstrated an overrepresentation of DREs, which is favorably bound by DREB1A and DREB2A (Sakuma et al., 2006). Research reports highlight the involvement of ERF and DREB subfamily in simultaneously upregulating genes belonging to the primary (esk1, LEA, CAB, AS, DXS) and specialized metabolism (DcPAL3, STR, TDC, D4H, CPR) by binding to the GCC boxes in their promoters. It can therefore be inferred that the AP2 family TFs are involved not only in imparting stress tolerance to plants, but also form a crucial molecular link among diverse metabolic pathways.
The RAV subfamily TFs are more involved in imparting biotic stress tolerance to the plants via activation of the PR genes (Woo et al., 2010; Fu et al., 2014). One of the RAV proteins, RAV1 is known to be involved in ABA signaling, where it increased ABA insensitivity of seeds during germination (Feng et al., 2014). Scientific reports indicate that the RAV TFBS are overrepresented in the promoter regions of primary (Em genes (Em1 and Em6), LEA, AS) and specialized metabolism genes (GST, LOX, SamDC, Feng et al., 2014; Moran Lauter et al., 2014). The RAV family TFs are also involved in regulating several other allied processes, like regulating metal starvation tolerance and controlling senescence-related gene expression. In A. thaliana, the promoter regions of principal cold responsive genes COL1 (CONSTANS-like 1) and COR27 demonstrated the presence of certain sequences called as “Evening elements (EE) and EE-like (EEL) elements” which were amplified in the presence of ABRE-like (ABREL) motif. Three ABREL motifs, along with four EE motifs could induce the expression of cold-responsive genes COL1 and COR27 (Mikkelsen and Thomashow, 2009). AP2/ERF family TFs can therefore serve as the missing molecular link between primary and specialized metabolism in plants. Table 4 presents a detailed account of the AP2/ERF TF family. However, among the AP2/ERF TFs, not many reports highlight the role of AP2 subfamily in regulating crucial genes under stressed conditions and futuristic research needs to highlight more in this aspect.
There are several other additional TFBS, which bring about cold stress mitigation, like the MYC binding sites, G-box and ABRE (Maruyama et al., 2012). Reports also suggest that AP2/ERF TFs work in tandem with bZIPs and MYBs to bring about synergistic regulation of cold stress tolerance by controlling ABA mediated gene expression in Arabidopsis (Pandey et al., 2005; Xu et al., 2011). Therefore, it can be suggested that a network of TFs is involved in coregulating diverse stress-responsive genes, which potentially form the missing molecular link between primary and specialized metabolism genes under stressed conditions. Although the active role of AP2 TFs subfamily in upregulating primary and specialized metabolism genes is not fully uncovered, deeper insights into this area would present a promising prospective in interconnecting diverse metabolic pathways.
Future Prospective
Newer insights into the interrelationships among multiple metabolic pathways are important to realize the subtle interplay of biomolecules within plants, as well as between plants and their environment. Primary and specialized metabolism serve as the backbone for the production of several therapeutically significant metabolites in planta (Tohge et al., 2013). Though under stress conditions plants overproduce certain key therapeutic metabolites, however it might have a negative impact on plant yield and productivity (Caretto et al., 2015). Therefore, an attempt to study coregulation of primary and specialized metabolism genes under certain stress conditions could pave a way to enhance both plant productivity and plant-derived therapeutic compounds.
Conventional stress-mitigation programmes either focus on breeding to develop robust, stress-tolerant plants or using plant growth regulators (like salicylic acid, ascorbic acid, brassinolides, etc.) to provide momentary stress-mitigation effects. Genetic engineering techniques to impart abiotic stress-tolerance to plants have focused on engineering stress-responsive TF genes in order to bring about effective stress response (Mickelbart et al., 2015). The current knowledge of cis-elements and TF interactions that bring about simultaneous upregulation of primary and specialized metabolism genes would help in developing tolerance to wide range of environmental stresses. Therefore, by adopting cis-element and TF engineering, scientists can develop robust crop varieties with high therapeutic potential. However, exhaustive research needs to be carried out before putting this technology to practice.
Conclusion
The link between primary and specialized metabolic pathways in plants has been an area of extensive research in the recent years with prime focus being laid on stress mitigation, increased plant yield and enhanced production of specialized metabolites. In the recent decades, rising population has contributed to increased levels of environmental stress, thereby plunging the overall crop productivity. However, it is also known that stress alters the biochemical fingerprint of plants, thereby enhancing the production of therapeutic metabolites like alkaloids, flavonoids, stillbenoids and phenylpropanoids. Scientific studies aimed at imparting stress tolerance focus mainly on enhancing production of specialized metabolites through genetic engineering approaches. However, not many studies highlight the significance of the molecular interface connecting primary and specialized metabolites under stress conditions. Since the precursors for the specialized metabolites originate from primary metabolism, much efforts are needed to unravel the cross talk between the two pathways at the molecular level.
In our review, we have presented a comprehensive analysis of the interplay between primary and specialized metabolism in plants under stressed conditions. Phyto-stress brings about a remarkable change in the metabolic profile of plants, wherein diverse primary and specialized metabolites are overproduced. Among the primary metabolites, the levels of sugars, sugar alcohols and amino acids were predominantly enhanced. On the other hand, VOCs, phenylpropanoids and alkaloids were mainly overproduced from among the specialized metabolites. The striking observation that diverse forms of stress leaves behind similar biomolecular patterns points at the molecular level regulation occurring between these metabolic processes. In the process of stress mitigation, plants are known to concurrently induce the expression of diverse stress-responsive genes belonging to primary metabolism, specialized metabolism and TFs. Since the principal mode of regulation of various genes occurs at the transcriptional level, the prime focus was laid on the cis-element and TF interactions that can simultaneously regulate primary and specialized metabolism genes. Upon spanning the immense literature available on cis-element profiles in the promoter regions of these genes in different plant systems, it could be inferred that a predictable pattern of cis-element-TF interactions (like bZIP TFs which recognize and bind to ABRE, AREB, G-box, ABI elements; WRKY TFs binding to their recognition sites W-box; MYB TFs binding to the MREs and AP2 TFs binding to DRE and GCC boxes) could be seen among primary, specialized as well as TF genes. Many of these TFs possessed pleiotropic roles, like developmental regulation, controlling senescence, physiological functioning, phenylpropanoid metabolism regulation, etc. Moreover, this pattern was observed among genes belonging to diverse metabolic pathways in different plant species also (V. vinifera, S. tuberosum, S. lycopersicum, E. haichowensis. O. sativa, N. tabacum, C. sativus, R. hybrida, Populus spp., H. vulgare, Z. mays, S. liaotungensis, M. acuminate, C. melo, etc.) This pattern of cis-element-TF interactions holds the key toward simultaneous upregulation of diverse genes. However, despite the immense genomic data available for several plants (genome sequence available for more than 60 plant species), scientific reports discretely attempt at elucidating the transcriptional regulatory mechanisms of either primary metabolism or specialized metabolism or TF genes. The immense prospective offered by simultaneous transcriptional regulation of primary and specialized metabolism genes toward achieving a two-tier objective of stress-tolerance as well as improved therapeutic values needs to be harnessed at a full potential, as it is still in a nascent stage.
Author Contributions
The authors have equally contributed to the manuscript. SS and MN wrote the article, while BS conceptualized the manuscript and corrected the same.
Funding
This work was supported by Department of Biotechnology, Ministry of Science and Technology, Govt. of India [grant number BT/Bio-CARe/02/10078/2013-14].
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to Department of Biotechnology, Govt. of India for funding the research and SASTRA University for providing the requisite infrastructure facilities.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01725/full#supplementary-material
References
Agarwal, P. K., Agarwal, P., Reddy, M. K., and Sopory, S. K. (2006). Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep. 25, 1263–1274. doi: 10.1007/s00299-006-0204-8
Ahmadi, A., and Baker, D. A. (2001). The effect of water stress on the activities of key regulatory enzymes of the sucrose to starch pathway in wheat. Plant. Growth. Regulation. 35, 81–91. doi: 10.1023/A:1013827600528
Ahmed, N. U., Park, J. I., Jung, H. J., Yang, T. J., Hur, Y., and Nou, I. S. (2014). Characterization of dihydroflavonol 4-reductase (DFR) genes and their association with cold and freezing stress in Brassica rapa. Gene 550, 46–55. doi: 10.1016/j.gene.2014.08.013
Ahn, S. Y., Kim, S. A., Cho, K. S., and Yun, H. K. (2014). Expression of genes related to flavonoid and stilbene synthesis as affected by signaling chemicals and Botrytis cinerea in grapevines. Biol. Plant. 58, 758–767. doi: 10.1007/s10535-014-0437-2
Akladious, S. A., and Abbas, S. M. (2013). Alleviation of sea water stress on tomato plants by foliar application of aspartic acid and glutathione. Bangladesh J. Bot. 42, 31–43. doi: 10.3329/bjb.v42i1.15822
Alam, M. M., Nahar, K., Hasanuzzaman, M., and Fujita, M. (2014). Trehalose-induced drought stress tolerance: a comparative study among different Brassica species. Plant Omics J. 7, 271–283.
Álvarez, C., Bermúdez, M. Á., Romero, L. C., Gotor, C., and García, I. (2012). Cysteine homeostasis plays an essential role in plant immunity. New Phytol. 193, 165–177. doi: 10.1111/j.1469-8137.2011.03889.x
Alves, M. S., Dadalto, S. P., Goncalves, A. B., De Souza, G. B., Barros, V. A., and Fietto, L. G. (2013). Plant bZIP transcription factors responsive to pathogens: a review. Int. J. Mol. Sci. 14, 7815–7828. doi: 10.3390/ijms14047815
Arora, A. (2005). Ethylene receptors and molecular mechanism of ethylene sensitivity in plants. Curr. Sci. 89, 1348–1361.
Asao, H., Yoshida, K., Nishi, Y., and Shinmyo, A. (2003). Wound-responsive cis-element in the 5′-upstream region of cucumber ascorbate oxidase gene. Biosci. Biotechnol. Biochem. 67, 271–277. doi: 10.1271/bbb.67.271
Ashraf, M. Y., Sarwar, G., Ashraf, M., Afaf, R., and Sattar, A. (2002). Salinity induced changes in α-amylase activity during germination and early cotton seedling growth. Biologia Plantarum. 45, 589–591. doi: 10.1023/A:1022338900818
Babitha, K. C., Vemanna, R. S., Nataraja, K. N., and Udayakumar, M. (2015). Overexpression of EcbHLH57 transcription factor from Eleusine coracana L. in tobacco confers tolerance to salt, oxidative and drought stress. PLoS ONE 10:e0137098. doi: 10.1371/journal.pone.0137098
Bae, H., Kim, S. H., Kim, M. S., Sicher, R. C., Lary, D., Strem, M. D., et al. (2008). The drought response of Theobroma cacao (cacao) and the regulation of genes involved in polyamine biosynthesis by drought and other stresses. Plant Physiol. Biochem. 46, 174–188. doi: 10.1016/j.plaphy.2007.10.014
Baker, S. S., Wilhelm, K. S., and Thomashow, M. F. (1994). The 5′-region of Arabidopsis thaliana corl5a has cis-acting elements that confer cold-, drought- and ABA-regulated gene expression. Plant Mol. Biol. 24, 701–713. doi: 10.1007/BF00029852
Banerjee, A., and Roychoudhury, A. (2015). WRKY proteins: signaling and regulation of expression during abiotic stress responses. Scientific. World. J. 2015:807560. doi: 10.1155/2015/807560
Bastías, A., López-Climent, M., Valcárcel, M., Rosello, S., Gómez-Cadenas, A., and Casaretto, J. A. (2011). Modulation of organic acids and sugar content in tomato fruits by an abscisic acid-regulated transcription factor. Physiol. Plant. 141, 215–226. doi: 10.1111/j.1399-3054.2010.01435.x
Bastías, A., Yañez, M., Osorio, S., Arbona, V., Gómez-Cadenas, A., Fernie, A. R., et al. (2014). The transcription factor AREB1 regulates primary metabolic pathways in tomato fruits. J. Exp. Bot. 65, 2351–2363. doi: 10.1093/jxb/eru114
Basu, S., Roychoudhury, A., and Sengupta, D. N. (2014). Deciphering the role of various cis-acting regulatory elements in controlling SamDC gene expression in Rice. Plant Signal. Behav. 9:e28391. doi: 10.4161/psb.28391
Becerra-Moreno, A., Redondo-Gil, M., Benavides, J., Nair, V., Cisneros-Zevallos, L., and Jacobo-Velázquez, D. A. (2015). Combined effect of water loss and wounding stress on gene activation of metabolic pathways associated with phenolic biosynthesis in carrot. Front. Plant Sci. 6:837. doi: 10.3389/fpls.2015.00837
Bhargava, S., and Sawant, K. (2013). Drought stress adaptation: metabolic adjustment and regulation of gene expression. Plant Breed. 132, 21–32. doi: 10.1111/pbr.12004
Bhat, R. A., Borst, J. W., Riehl, M., and Thompson, R. D. (2004). Interaction of maize Opaque-2 and the transcriptional co-activators GCN5 and ADA2, in the modulation of transcriptional activity. Plant Mol. Biol. 55, 239–252. doi: 10.1007/s11103-004-0553-z
Bi, C., Xu, Y., Ye, Q., Yin, T., and Ye, N. (2016). Genome-wide identification and characterization of WRKY gene family in Salix suchowensis. PeerJ. 4:e2437. doi: 10.7717/peerj.2437
Bolton, M. D. (2009). Primary metabolism and plant defense—fuel for the fire. Mol. Plant Microbe Interact. 22, 487–497. doi: 10.1094/MPMI-22-5-0487
Borsani, O., Valpuesta, V., and Botella, M. A. (2001). Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiol. 126, 1024–1030. doi: 10.1104/pp.126.3.1024
Brown, C. D., Johnson, D. S., and Sidow, A. (2007). Functional architecture and evolution of transcriptional elements that drive gene. Science 317, 1557–1560. doi: 10.1126/science.1145893
Cabello, S., Lorenz, C., Crespo, S., Cabrera, J., Ludwig, R., Escobar, C., et al. (2014). Altered sucrose synthase and invertase expression affects the local and systemic sugar metabolism of nematode infected Arabidopsis thaliana plants. J. Exp. Bot. 65, 201–212. doi: 10.1093/jxb/ert359
Cai, H., Cheng, J., Yan, Y., Xiao, Z., Li, J., Mou, S., et al. (2015). Genome-wide identification and expression analysis of calcium-dependent protein kinase and its closely related kinase genesin Capsicum annuum. Front. Plant Sci. 6:737. doi: 10.3389/fpls.2015.00737
Campos-Soriano, L., Gómez-Ariza, J., Bonfante, P., and San Segundo, B. (2011). A rice calcium-dependent protein kinase is expressed in cortical root cells during the presymbiotic phase of the arbuscular mycorrhizal symbiosis. BMC Plant Biol. 11:90. doi: 10.1186/1471-2229-11-90
Cao, Y., Hu, S. L., Huang, S. X., Ren, P., and Lu, X. (2012). Molecular cloning, expression pattern, and putative cis-acting elements of a 4-coumarate:CoA ligase gene in bamboo (Neosinocalamus affinis). Electron. J. Biotechnol. 15:5. doi: 10.2225/vol15-issue5-fulltext-10
Cao, Z. H., Zhang, S. Z., Wang, R. K., Zhang, R. F., and Hao, Y. J. (2013). Genome wide analysis of the apple MYB transcription factor family allows the identification of MdoMYB121 gene confering abiotic stress tolerance in plants. PLoS ONE 8:e69955. doi: 10.1371/journal.pone.0069955
Caretto, S., Linsalata, V., Colella, G., Mita, G., and Lattanzio, V. (2015). Carbon fluxes between primary metabolism and phenolic pathway in plant tissues under stress. Int. J. Mol. Sci. 16, 26378–26394. doi: 10.3390/ijms161125967
Cartharius, K., Frech, K., Grote, K., Klocke, B., Haltmeier, M., Klingenhoff, A., et al. (2005). MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics 21, 2933–2942. doi: 10.1093/bioinformatics/bti473
Caverzan, A., Passaia, G., Rosa, S. B., Ribeiro, C. W., Lazzarotto, F., and Margis-Pinheiro, M. (2012). Plant responses to stresses: role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 35, 1011–1019. doi: 10.1590/S1415-47572012000600016
Chamoli, S., and Verma, A. K. (2014). “Targeting of metabolic pathways for genetic engineering to combat abiotic stress tolerance in crop plants,” in Approaches to Plant Stress and Their Management, eds R. K. Gaur and P. Sharma (New Delhi: Springer), 23–38.
Chen, Z., Chen, M., Xu, Z. S., Li, L. C., Chen, X. P., and Ma, Y. Z. (2014). Characteristics and expression patterns of the aldehyde dehydrogenase (ALDH) gene superfamily of foxtail millet (Setaria italica L.). PLoS ONE 9:e101136. doi: 10.1371/journal.pone.0101136
Chen, Z., Gao, K., Su, X., Rao, P., and An, X. (2015). Genome-wide identification of the invertase gene family in populus. PLoS ONE 10:e0138540. doi: 10.1371/journal.pone.0138540
Cheng, H., Song, S., Xiao, L., Soo, H. M., Cheng, Z., Xie, D., et al. (2009). gibberellin acts through jasmonate to control the expression of MYB21, MYB24, and MYB57 to promote stamen filament growth in Arabidopsis. PLoS Genet. 5:e1000440. doi: 10.1371/journal.pgen.1000440
Cheng, M. C., Liao, P. M., Kuo, W. W., and Lin, T. P. (2013). The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 162, 1566–1582. doi: 10.1104/pp.113.221911
Chinnusamy, V., Zhu, J., and Sunkar, R. (2010). Gene regulation during cold stress acclimation in plants. Methods Mol. Biol. 639, 39–55. doi: 10.1007/978-1-60761-702-0_3
Cho, Y., Lightfoot, D. A., and Wood, A. J. (1999). Trigonelline concentrations in salt stressed leaves of cultivated Glycine max. Phytochemistry 52, 1235–1238. doi: 10.1016/S0031-9422(99)00410-0
Chowdhury, E. M., Choi, B. S., Park, S. U., Lim, H. S., and Bae, H. (2012). Transcriptional analysis of hydroxycinnamoyl transferase (HCT) in various tissues of Hibiscus cannabinus in response to abiotic stress conditions. POJ 5, 305–313.
Chuang, C. F., Running, M. P., Williams, R. W., and Meyerowitz, E. M. (1999). The PERIANTHIA gene encodes a bZIP protein involved in the determination of floral organ number in Arabidopsis thaliana. Genes Dev. 13, 334–344. doi: 10.1101/gad.13.3.334
Ciereszko, I., Johansson, H., and Kleczkowski, L. A. (2001). Sucrose and light regulation of a cold-inducible UDP-glucose pyrophosphorylase gene via a hexokinase-independent and abscisic acid-insensitive pathway in Arabidopsis. Biochem. J. 354, 67–72. doi: 10.1042/bj3540067
Creelman, R. A., and Mullet, J. E. (1995). Jasmonic acid distribution and action in plants: Regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. U.S.A. 92, 4114–4119. doi: 10.1073/pnas.92.10.4114
Cuevas, J. C., López-Cobollo, R., Alcázar, R., Zarza, X., Koncz, C., Altabella, T., et al. (2008). Putrescine is involved in Arabidopsis freezing tolerance and cold acclimation by regulating abscisic acid levels in response to low temperature. Plant Physiol. 148, 1094–1105. doi: 10.1104/pp.108.122945
Cui, L., Feng, K., Wang, M., Wang, M., Deng, P., Song, W., et al. (2016). Genome-wide identification, phylogeny and expression analysis of AP2/ERF transcription factors family in Brachypodium distachyon. BMC Genomics 17:636. doi: 10.1186/s12864-016-2968-8
Das, R., Pandey, A., and Pandey, G. K. (2014). “Role of calcium/calmodulin in plant stress response and signalling,” in Approaches to Plant Stress and Their Management, eds R. K. Gaur and P. Sharma (New Delhi: Springer), 53–84. doi: 10.1007/978-81-322-1620-9_4
Davies, K. M., and Schwinn, K. E. (2003). Transcriptional regulation of secondary metabolism. Funct. Plant Biol. 30, 913–925. doi: 10.1071/FP03062
Davuluri, R. V., Sun, H., Palaniswamy, S. K., Matthews, N., Molina, C., Kurtz, M., et al. (2003). AGRIS: Arabidopsis gene regulatory information server, an information resource of Arabidopsis cis-regulatory elements and transcription factors. BMC Bioinformatics 4:25. doi: 10.1186/1471-2105-4-25
de Meaux, J., Pop, A., and Michell-Olds, T. (2006). Cis-regulatory evolution of Chalcone-synthase expression in the genus Arabidopsis. Genetics 174, 2181–2202. doi: 10.1534/genetics.106.064543
Dey, A., Samanta, M. K., Gayen, S., Sen, S. K., and Maiti, M. K. (2016). Enhanced gene expression rather than natural polymorphism in coding sequence of the OsbZIP23 determines drought tolerance and yield improvement in rice genotypes. PLoS ONE 11:e0150763. doi: 10.1371/journal.pone.0150763
Dey, S., and Vlot, A. C. (2015). Ethylene responsive factors in the orchestration of stress responses in monocotyledonous plants. Front. Plant Sci. 6:640. doi: 10.3389/fpls.2015.00640
Diaz, M., Polanco, V., Ramirez, I., and Pena-Cortes, H. (2012). Molecular cloning and expression analysis of 12-oxophytodienoate reductase cDNA by wounding in Solanum tuberosum. Electron. J. Biotechnol. 15:1. doi: 10.2225/vol15-issue1-fulltext-3
Du, H., Yang, S.-S., Liang, Z., Feng, B.-R., Liu, L., Huang, Y.-B., et al. (2012). Genome-wide analysis of the MYB transcription factor superfamily in soybean. BMC Plant Biol. 12:106. doi: 10.1186/1471-2229-12-106
Du, H., and Wang, Z. (2012). Metabolic responses of hybrid bermudagrass to short-term and long-term drought stress. J. Amer. Soc. Hort. Sci. 137, 411–420.
Dubos, C., Stracke, R., Grotewold, E., Weisshaar, B., Martin, C., and Lepiniec, L. (2010). MYB transcription factors in Arabidopsis. Trends Plant. Sci. 15, 573–581. doi: 10.1016/j.tplants.2010.06.005
Du Fall, L. A., and Solomon, P. S. (2011). Role of cereal secondary metabolites involved in mediating the outcome of plant-pathogen interactions. Metabolites 1, 64–78. doi: 10.3390/metabo1010064
Faktor, O., Kooter, J. M., Dixon, R. A., and Lamb, C. J. (1996). Functional dissection of a bean chalcone synthase gene promoter in transgenic tobacco plants reveals sequence motifs essential for floral expression. Plant Mol. Biol. 32, 849–859. doi: 10.1007/BF00020482
Fassler, J., Landsman, D., Acharya, A., Moll, J. R., Bonovich, M., and Vinson, C. (2002). B-ZIP proteins encoded by the drosophila genome: evaluation of potential dimerization partners. Genome Res. 12, 1190–1200. doi: 10.1101/gr.67902
Feng, C. Z., Chen, Y., Wang, C., Kong, Y. H., Wu, W. H., and Chen, Y. F. (2014). Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 80, 654–668. doi: 10.1111/tpj.12670
Finkelstein, R. R., and Lynch, T. J. (2000). The Arabidopsis abscisic acid response gene ABI5 encodes a basic leucine zipper transcription factor. Plant Cell 12, 599–609. doi: 10.1105/tpc.12.4.599
Flores, H. E., and Galston, A. W. (1982). Polyamines and plant stress: activation of putrescine biosynthesis by osmotic shock. Science 217, 1259–1260. doi: 10.1126/science.217.4566.1259
Floris, M., Mahgoub, H., Lanet, E., and Robaglia, C., Menand, B. (2009). Posttranscriptional regulation of gene expression in plants during abiotic stress. Int. J. Mol. Sci. 10, 3168–3185. doi: 10.3390/ijms10073168
Fraire-Velázquez, S., Rodriguez-Guerra, R., and Sanchez-Calderon, L. (2011). “Abiotic and biotic stress response crosstalk in plants,” in Abiotic Stress Response in Plants - Physiological, Biochemical and Genetic Perspectives, eds A. Shankar and B. Venkateswarlu (InTech Publication), 3–26.
Frattini, M., Morello, L., and Breviario, D. (1999). Rice calcium-dependent protein kinase isoforms OsCDPK2 and OsCDPK11 show different responses to light and different expression patterns during seed development. Plant Mol. Biol. 41, 753–764. doi: 10.1023/A:1006316422400
Franco-Zorrilla, J. M., López-Vidriero, I., Carrasco, J. L., Godoy, M., Vera, P., and Solano, R. (2014). DNA-binding specificities of plant transcription factors and their potential to define target genes. Proc. Natl. Acad. Sci. U.S.A. 111, 2367–2372. doi: 10.1073/pnas.1316278111
French, S. R., Abu-Zaitoon, Y., Uddin, M. M., Bennett, K., and Nonhebel, H. M. (2014). Auxin and cell wall invertase related signaling during rice grain development. Plants 3, 95–112. doi: 10.3390/plants3010095
Fu, M., Kang, H. K., Son, S.-H., Kim, S.-K., and Nam, K. H. (2014). A subset of Arabidopsis RAV transcription factors modulates drought and salt stress responses independent of ABA. Plant Cell Physiol. 55, 1892–1904. doi: 10.1093/pcp/pcu118
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell Online 17, 3470–3488. doi: 10.1105/tpc.105.035659
Gális, I., Simek, P., Narisawa, T., Sasaki, M., Horiguchi, T., Fukuda, H., et al. (2006). A novel R2R3 MYB transcription factor NtMYBJS1 is a methyl jasmonate-dependent regulator of phenylpropanoid conjugate biosynthesis in tobacco. Plant J. 46, 573–592. doi: 10.1111/j.1365-313X.2006.02719.x
Ganguly, M., Datta, K., Roychoudhury, A., Gayen, D., Sengupta, D. N., and Datta, S. K. (2012). Overexpression of Rab16A gene in indica rice variety for generating enhanced salt tolerance. Plant Signal. Behav. 7, 502–509. doi: 10.4161/psb.19646
Gao, Z., Sagi, M., and Lips, S. H. (1998). Carbohydrate metabolism in leaves and assimilate partitioning in fruits of tomato (Lycopersicon esculentum L.) as affected by salinity. Plant Sci. 135, 149–159. doi: 10.1016/S0168-9452(98)00085-5
Gates, D. J., Strickler, S. R., Mueller, L. A., Olson, B. J., and Smith, S. D. (2016). Diversification of R2R3-MYB transcription factors in the tomato family solanaceae. J. Mol. Evol. 83, 26–37. doi: 10.1007/s00239-016-9750-z
Gigolashvili, T., Yatusevich, R., Berger, B., Müller, C., and Flügge, U. I. (2007). The R2R3-MYB transcription factor HAG1/MYB28 is a regulator of methionine-derived glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 51, 247–261. doi: 10.1111/j.1365-313X.2007.03133.x
Gill, S. S., and Tuteja, N. (2010). Polyamines and abiotic stress tolerance in plants. Plant Signal. Behav. 5, 26–33. doi: 10.4161/psb.5.1.10291
Gill, S. S., and Tuteja, N. (2011). Cadmium stress tolerance in crop plants. Probing the role of sulfur. Plant Signal. Behav. 6, 215–222. doi: 10.4161/psb.6.2.14880
Gómes-Porras, J. L., Riano-Pachón, D. M., Dryer, I., Mayer, J. E., and Mueller-Roeber, B. (2007). Genome-wide analysis of ABA-responsive elements ABRE and CE3 reveals divergent patterns in Arabidopsis and rice. BMC Genomics 8:260. doi: 10.1186/1471-2164-8-260
Grace, S. C., Logan, B. A., and Adams, W. W. III. (1998). Seasonal differences in foliar content of chlorogenic acid, a phenylpropanoid antioxidant, in Mahonia repens. Plant Cell Environ. 21, 513–521. doi: 10.1046/j.1365-3040.1998.00282.x
Griesser, M., Weingart, G., Schoedl-Hummel, K., Neumann, N., Becker, M., Varmuza, K., et al. (2015). Severe drought stress is affecting selected primary metabolites, polyphenols, and volatile metabolites in grapevine leaves (Vitis vinifera cv. Pinot noir). Plant Physiol. Biochem. 88, 17–26. doi: 10.1016/j.plaphy.2015.01.004
Grover, A., Pareek, A., Singla, S. L., Minhas, D., Katiyar, S., Ghawana, S., et al. (1998). Engineering crops for tolerance against abiotic stress through gene manipulation. Curr. Sci. 75, 689–696.
Gujjar, R. S., Akhtar, M., and Singh, M. (2014). Transcription factors in abiotic stress tolerance. Ind. J. Plant Physiol. 19:306. doi: 10.1007/s40502-014-0121-8
Guo, B., Wei, Y., Xu, R., Lin, S., Luan, H., Lv, C., et al. (2016). Genome-wide analysis of APETALA2/Ethylene-responsive factor (AP2/ERF) gene family in barley (Hordeum vulgare L.). PLoS ONE 11:e0161322. doi: 10.1371/journal.pone.0161322
Guo, H., and Ecker, J. R. (2004). The ethylene signaling pathway: new insights. Curr. Op. Plant Biol. 7, 40–49. doi: 10.1016/j.pbi.2003.11.011
Gupta, A. K., and Kaur, N. (2005). Sugar signaling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants. J. Biosci. 30, 101–116. doi: 10.1007/BF02703574
Haggag, W. H., Abouziena, H. F., Abd-El-Kreem, F., and El Habbasha, S. (2015). Agriculture biotechnology for management of multiple biotic and abiotic environmental stress in crops. J. Chem. Pharm. Res. 7, 882–889.
Hanson, J., Hanssen, M., Weise, A., Hendriks, M. M., and Smeekens, S. (2008). The sucrose regulated transcription factor bZIP11 affects amino acid metabolism by regulating the expression of ASPARAGINE SYNTHETASE1 and PROLINE DEHYDROGENASE2. Plant J. 53, 935–949. doi: 10.1111/j.1365-313X.2007.03385.x
Hartings, H., Lauria, M., Lazzaroni, N., Pirona, R., and Motto, M. (2011). The Zea mays mutants opaque-2 and opaque-7 disclose extensive changes in endosperm metabolism as revealed by protein, amino acid, and transcriptome-wide analyses. BMC Genomics 12:41. doi: 10.1186/1471-2164-12-41
Hasthanasombut, S., Paisarnwipatpong, N., Triwitayakorn, K., Kirdmanee, C., and Supaibulwatana, K. (2011). Expression of OsBADH1 gene in Indica rice (Oryza sativa L.) in correlation with salt, plasmolysis, temperature and light stresses. POJ 4, 400–407.
Hayat, S., Hayat, Q., Alyemeni, M. N., Wani, A. S., Pichtel, J., and Ahmad, A. (2012). Role of proline under changing environments: a review. Plant. Signal. Behav. 7, 1456–1466. doi: 10.4161/psb.21949
Hayes, M. A., Feechan, A., and Dry, I. B. (2010). Involvement of abscisic acid in the coordinated regulation of a stress-inducible hexose transporter (VvHT5) and a cell wall invertase in grapevine in response to biotrophic fungal infection. Plant. Physiol. 153, 211–221. doi: 10.1104/pp.110.154765
He, Q., Jones, D. C., Li, W., Xie, F., Ma, J., Sun, R., et al. (2016). Genome-wide identification of R2R3-MYB genes and expression analyses during abiotic stress in Gossypium raimondii. Nat. Sci. Rep. 6:22980. doi: 10.1038/srep22980
Heinekamp, T., Kuhlmann, M., Lenk, A., Strathmann, A., and Droge-Laser, W. (2002). The tobacco bZIP transcription factor BZI-1 binds to G-box elements in the promoters of phenylpropanoid pathway genes in vitro, but it is not involved in their regulation in vivo. Mol. Genet. Genomics 267, 16–26. doi: 10.1007/s00438-001-0636-3
Heinekamp, T., Strathmann, A., Kuhlmann, M., Froissard, M., Müller, A., Perrot-Rechenmann, C., et al. (2004). The tobacco bZIP transcription factor BZI-1 binds the GH3 promoter in vivo and modulates auxin-induced transcription. Plant J. 38, 298–309. doi: 10.1111/j.1365-313X.2004.02043.x
Henry, C., Bledsoe, S. W., Siekman, A., Kollman, A., Waters, B. M., Feil, R., et al. (2014). The trehalose pathway in maize: conservation and gene regulation in response to the diurnal cycle and extended darkness. J. Exp. Bot. 65, 5959–5973. doi: 10.1093/jxb/eru335
Hernandez-Garcia, C. M., and Finer, J. J. (2014). Identification and validation of promoters and cis-acting regulatory elements. Plant. Sci. 217–218, 109–119. doi: 10.1016/j.plantsci.2013.12.007
Hettenhausen, C., Sun, G., He, Y., Zhuang, H., Sun, T., Qi, J., et al. (2016). Genome-wide identification of calcium-dependent protein kinases in soybean and analyses of their transcriptional responses to insect herbivory and drought stress. Sci. Rep. 6:18973. doi: 10.1038/srep18973
Himi, E., and Taketa, S. (2015). Barley Ant17, encoding flavanone 3-hydroxylase (F3H), is a promising target locus for attaining anthocyanin/proanthocyanidin-free plants without pleiotropic reduction of grain dormancy. Genome 58, 1–11. doi: 10.1139/gen-2014-0189
Hochberg, U., Batushansky, A., Degu, A., Rachmilevitch, S., and Fait, A. (2015). Metabolic and physiological responses of shiraz and cabernet sauvignon (Vitis vinifera L.) to near optimal temperatures of 25 and 35°C. Int. J. Mol. Sci. 16, 24276–24294. doi: 10.3390/ijms161024276
Hochberg, U., Degu, A., Toubiana, D., Gendler, T., Nikoloski, Z., Rachmilevitch, S., et al. (2013). Metabolite profiling and network analysis reveal coordinated changes in grapevine water stress response. BMC Plant Biol. 13:184. doi: 10.1186/1471-2229-13-184
Höll, J., Vannozzi, A., Czemmel, S., D'Onofrio, C., Walker, A. R., Rausch, T., et al. (2013). The R2R3-MYB transcription factors MYB14 and MYB15 regulate stilbene biosynthesis in Vitis vinifera. Plant Cell 25, 4135–4149.
Holopainen, J. K., and Blande, J. D. (2013). Where do herbivore-induced plant volatiles go? Front. Plant Sci. 4:185. doi: 10.3389/fpls.2013.00185
Horvath, E., Szalai, G., and Janda, T. (2007). Induction of abiotic stress tolerance by salicylic acid signaling. J. Plant Growth Regul. 26, 290–300. doi: 10.1007/s00344-007-9017-4
Hu, T., He, S., Yang, G., Zeng, H., Wang, G., Chen, Z., et al. (2011). Isolation and characterization of a rice glutathione S-transferase gene promoter regulated by herbicides and hormones. Plant Cell Rep. 30, 539–549. doi: 10.1007/s00299-010-0964-z
Huang, W., Khaldun, A. B., Chen, J., Zhang, C., Lv, H., Yuan, L., et al. (2016). A R2R3-MYB Transcription Factor Regulates the Flavonol Biosynthetic Pathway in a Traditional Chinese Medicinal Plant, Epimedium sagittatum. Front. Plant Sci. 7:1089. doi: 10.3389/fpls.2016.01089
Huang, W., Sun, W., Lv, H., Luo, M., Zeng, S., Pattanayak, S., et al. (2013). A R2R3-MYB transcription factor from Epimedium sagittatum regulates the flavonoid biosynthetic pathway. PLoS ONE 8:e70778. doi: 10.1371/journal.pone.0070778
Huang, Z., He, J., Zhong, X.-J., Guo, H.-D., Jin, S.-H., Li, X., et al. (2016). Molecular cloning and characterization of a novel freezing-inducible DREB1/CBF transcription factor gene in boreal plant Iceland poppy (Papaver nudicaule). Genet. Mol. Biol. doi: 10.1590/1678-4685-gmb-2015-0228. [Epub ahead of print].
Hudson, A. O. (2015). “Tyrosine aminotransferase,” in Amino Acids in Higher Plants, ed J. P. F. D'Mello (Edinburgh: CAB International), 68–81.
Hundertmark, M., and Hincha, D. K. (2008). LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9:118. doi: 10.1186/1471-2164-9-118
Hunter, B. G., Beatty, M. K., Singletary, G. W., Hamaker, B. R., Dilkes, B. P., Larkins, B. A., et al. (2002). Maize opaque endosperm mutations create extensive changes in patterns of gene expression. Plant Cell 14, 2591–2612. doi: 10.1105/tpc.003905
Hwang, Y. S., Karrer, E. E., Thomas, B. R., Chen, L., and Rodriguez, R. L. (1998). Three cis-elements required for rice amylase Amy3D expression during sugar starvation. Plant Mol. Biol. 36, 331–341. doi: 10.1023/A:1005956104636
Ibraheem, O., Botha, C. E., and Bradley, G. (2010). In silico analysis of cis-acting regulatory elements in 5_ regulatory regions of sucrose transporter gene families in rice (Oryza sativa Japonica) and Arabidopsis thaliana. Comput. Biol. Chem. 34, 268–283. doi: 10.1016/j.compbiolchem.2010.09.003
Ibrahim, M. H., and Jaafar, H. Z. E. (2012). Primary, secondary metabolites, H2O2, malondialdehyde and photosynthetic responses of Orthosiphon stimaneus Benth. to different irradiance levels. Molecules 17, 1159–1176. doi: 10.3390/molecules17021159
Iven, T., Strathmann, A., Böttner, S., Zwafink, T., Heinekamp, T., Guivarch, A., et al. (2010). Homo- and heterodimers of tobacco bZIP proteins counteract as positive or negative regulators of transcription during pollen development. Plant J. 63, 155–166. doi: 10.1111/j.1365-313x.2010.04230.x
Jakoby, M., Weisshaar, B., Dröge-Laser, W., Vicente-Carbajosa, J., Tiedemann, J., Kroj, T., et al. (2002). bZIP transcription factors in Arabidopsis. Trends Plant. Sci. 7, 106–111. doi: 10.1016/S1360-1385(01)02223-3
Jamalian, S., Gholami, M., and Esna-Ashari, M. (2013). Abscisic acid-mediated leaf phenolic compounds, plant growth and yield is strawberry under different salt stress regimes. Theor. Exp. Plant Physiol. 25, 291–299.
Jin, H., Cominelli, E., Bailey, P., Parr, A., Mehrtens, F., Jones, J., et al. (2000). Transcriptional repression by AtMYB4 controls production of UV-protecting sunscreens in Arabidopsis. EMBO J. 19, 6150–6161. doi: 10.1093/emboj/19.22.6150
Jin, Y., Zhang, C., Liu, W., Tang, Y. F., Qi, H., Chen, H., et al. (2016). The alcohol dehydrogenase gene family in melon (Cucumis melo L.): bioinformatic analysis and expression patterns. Front. Plant Sci. 7:670. doi: 10.3389/fpls.2016.00670
Jing, Z., Shen-Wen, C., Zhong-Rui, Z., and Zhi-Ting, X. (2016). Isolation and activity analysis of cell wall invertase gene promoter (EhcwINVP) from Elsholtzia haichowensis Sun. Plant Sci. J. 34, 420–429.
Joshi, V., and Jander, G. (2009). Arabidopsis methionine g-lyase is regulated according to isoleucine biosynthesis needs but plays a subordinate role to threonine deaminase. Plant Physiol. 151, 367–378. doi: 10.1104/pp.109.138651
Katiyar, A., Smita, S., Lenka, S. K., Rajwanshi, R., Chinnusamy, V., and Bansal, K. C. (2012). Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genomics 13:544. doi: 10.1186/1471-2164-13-544
Kato-Noguchi, H. (2001). Wounding stress induces alcohol dehydrogenase in maize and lettuce seedlings. Plant Growth Regul. 35, 285–288. doi: 10.1023/A:1014489922792
Kawaoka, A., Kawamoto, T., Sekine, M., Yoshida, K., Takano, M., and Shinmyo, A. (1994). A cis-acting element and a trans-acting factor involved in the wound-induced expression of a horseradish peroxidase gene. Plant J. 6, 87–97. doi: 10.1046/j.1365-313X.1994.6010087.x
Kaya, C., Sonmez, O., Aydemir, S., Ashraf, M., and Dikilitas, M. (2013). Exogenous application of mannitol and thiourea regulates plant growth and oxidative stress responses in salt-stressed maize (Zea mays L.). J. Plant Interact. 8, 234–241. doi: 10.1080/17429145.2012.725480
Kendziorek, M., Paszkowski, A., and Zagdanska, B. (2012). Differential regulation of alanine aminotransferase homologues by abiotic stresses in wheat (Triticum aestivum L.) seedlings. Plant. Cell. Rep. 31, 1105–1117. doi: 10.1007/s00299-012-1231-2
Khairy, A. I. H., and Roh, K. S. (2016). Effect of salicylic acid, benzoic acid, and p-Coumaric acid on growth, chlorophyll, proline, and vitamin C of salinity-stressed tobacco (Nicotiana tabacum). Int. J. Plant. Soil Sci. 9, 1–10. doi: 10.9734/IJPSS/2016/22396
Khan, M. I., Fatma, M., Per, T. S., Anjum, N. A., and Khan, N. A. (2015). Salicylic acid-induced abiotic stress tolerance and underlying mechanisms in plants. Front. Plant Sci. 6:462. doi: 10.3389/fpls.2015.00462
Kim, J., Choi, B., Natarajan, S., and Bae, H. (2013). Expression analysis of kenaf cinnamate 4-hydroxylase (C4H) ortholog during developmental and stress responses. POJ 6, 65–72.
Kim, K. N., and Guiltinan, M. J. (1999). Identification of cis-acting elements important for expression of the starch-branching enzyme I gene in maize endosperm. Plant Physiol. 121, 225–236. doi: 10.1104/pp.121.1.225
Kim, S. H., Lee, J. R., and Kim, S. R. (2006). Characterization of an apple anthocyanidin synthase gene in transgenic tobacco plants. J. Plant Biol. 49, 326–330. doi: 10.1007/BF03031164
Kim, S. Y., Ma, J., Perret, P., Li, Z., and Thomas, T. L. (2002). Arabidopsis ABI5 subfamily members have distinct DNA-binding and transcriptional activities. Plant Physiol. 130, 688–697. doi: 10.1104/pp.003566
Kimura, M., Yamamoto, Y. Y., Seki, M., Sakurai, T., Sato, M., Abe, T., et al. (2003). Identification of Arabidopsis genes regulated by high light–stress using cDNA microarray. Phytochem. Photobiol. 77, 226–233. doi: 10.1562/0031-8655(2003)0770226IOAGRB2.0.CO2
Kimura, S., Chikagawa, Y., Kato, M., Maeda, K., and Ozeki, Y. (2008). Upregulation of the promoter activity of the carrot (Daucus carota) phenylalanine ammonia-lyase gene (DcPAL3) is caused by new members of the transcriptional regulatory proteins, DcERF1 and DcERF2, which bind to the GCC-box homolog and act as an activator to the DcPAL3 promoter. J. Plant Res. 121, 499–508. doi: 10.1007/s10265-008-0170-z
Lata, C., and Prasad, M. (2011). Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 62, 4731–4748. doi: 10.1093/jxb/err210
Lata, C., Yadav, A., and Prasad, M. (2011). “Role of plant transcription factors in abiotic stress tolerance,” in Abiotic Stress Response in Plants - Physiological, Biochemical and Genetic Perspectives, eds A. Shankar and B. Venkateswarlu (InTech Publication), 269–296.
Lavinsky, A. O., Magalhães, P. C., Ávila, R. G., Diniz, M. M., and de Souza, T. C. (2015). Partitioning between primary and secondary metabolism of carbon allocated to roots in four maize genotypes under water deficit and its effects on productivity. Crop J. 3, 379–386. doi: 10.1016/j.cj.2015.04.008
Lawton, M. A., and Lamb, C. J. (1987). transcriptional activation of plant defense genes by fungal elicitor, wounding, and infection. Mol. Cell. Biol. 7, 335–341. doi: 10.1128/MCB.7.1.335
Le Gall, H., Philippe, F., Domon, J. M., Gillet, F., Pelloux, J., and Rayon, C. (2015). Cell wall metabolism in response to abiotic stress. Plants 4, 112–166. doi: 10.3390/plants4010112
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., de Peer, Y. V., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in-silico analysis of promoter sequences. Nucleic Acids Res. 30, 325–327. doi: 10.1093/nar/30.1.325
Li, D., Fu, F., Zhang, H., and Song, F. (2015). Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genomics. 16:771. doi: 10.1186/s12864-015-1990-6
Li, D. M., Nie, Y. X., Zhang, J., Yin, J. S., Li, Q., Wang, X. J., et al. (2013). Ferulic acid pretreatment enhances dehydration-stress tolerance of cucumber seedlings. Biol. Plant. 57, 711–717. doi: 10.1007/s10535-013-0326-0
Li, F., Hao, C., Yan, L., Wu, B., Qin, X., Lai, J., et al. (2015). Gene structure, phylogeny and expression profile of the sucrose synthase gene family in cacao (Theobroma cacao L.). J. Genet. 94, 461–472. doi: 10.1007/s12041-015-0558-1
Li, S. F., Milliken, O. N., Pham, H., Seyit, R., Napoli, R., Preston, J., et al. (2009). The Arabidopsis MYB5 transcription factor regulates mucilage synthesis, seed coat development, and trichome morphogenesis. Plant Cell 21, 72–89. doi: 10.1105/tpc.108.063503
Li, Y., and Laoke, G. J. (2016). “Chapter 24 – redox-regulated plant transcription factors,” in Plant Transcription Factors: Evolutionary, Structural and Functional Aspects, ed D. H. Gonzales (Santa Fe, NM: Elsevier Publishers), 373–384.
Li, Z. Y., and Chen, S. Y. (2000). Differential accumulation of the S-adenosylmethionine decarboxylase transcript in rice seedlings in response to salt and drought stresses. Theor. Appl. Genet. 100, 782–788. doi: 10.1007/s001220051352
Licausi, F., Ohme-Takagi, M., and Perata, P. (2013). APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: mediators of stress responses and developmental programs. New Phytol. 199, 639–649. doi: 10.1111/nph.12291
Liu, J., Chen, N., Chen, F., Cai, B., Dal Santo, S., Tornielli, G. B., et al. (2014). Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genomics 15:281. doi: 10.1186/1471-2164-15-281
Liu, J., Osbourn, A., and Ma, P. (2015). MYB transcription factors as regulators of phenylpropanoid metabolism in plants. Mol. Plant. 8, 689–708. doi: 10.1016/j.molp.2015.03.012
Llorca, C. M., Potschin, M., and Zentgraf, U. (2014). bZIPs and WRKYs: two large transcription factor families executing two different functional strategies. Front. Plant Sci. 5:169. doi: 10.3389/fpls.2014.00169
Logemann, E., Parniske, M., and Hahlbrock, K. (1995). Modes of expression and common structural features of the complete phenylalanine ammonia-lyase gene family in parsley. Proc. Natl. Acad. Sci. U.S.A. 92, 5905–5909. doi: 10.1073/pnas.92.13.5905
Lu, C. A., Ho, T. D., Ho, S. L., and Yu, S. M. (2002). Three novel MYB proteins with one DNA binding repeat mediate sugar and hormone regulation of α-amylase gene expression. Plant Cell 14, 1963–1980. doi: 10.1105/tpc.001735
Lu, G., Paul, A. L., McCarty, D. R., and Ferl, R. J. (1996). Transcription factor veracity: 1s GBF3 responsible for ABA-regulated expression of Arabidopsis Adh? Plant Cell 8, 847–857. doi: 10.1105/tpc.8.5.847
Ma, J., Li, M. Y., Wang, F., Tang, J., and Xiong, A. S. (2015). Genome-wide analysis of Dof family transcription factors and their responses to abiotic stresses in Chinese cabbage. BMC Genomics 16:33. doi: 10.1186/s12864-015-1242-9
Maier, A. T., Stehling-Sun, S., Offenburger, S. L., and Lohmann, J. U. (2011). The bZIP transcription factor PERIANTHIA: a multifunctional hub for meristem control. Front. Plant Sci. 2, 1–17. doi: 10.3389/fpls.2011.00079
Maier, A. T., Stehling-Sun, S., Wollmann, H., Demar, M., Hong, R. L., Haubei, S., et al. (2009). Dual roles of the bZIP transcription factor PERIANTHIA in the control of floral architecture and homeotic gene expression. Development 136, 1613–1620. doi: 10.1242/dev.033647
Maiti, S., Ghosh, N., Mandal, C., Das, K., Dey, N., and Adak, M. K. (2012). Responses of the maize plant to chromium stress with reference to antioxidation activity. Brazilian Soc. Plant Physiol. 24, 203–212. doi: 10.1590/S1677-04202012000300007
Malhotra, S., and Sowdhamini, R. (2014). Interactions among plant transcription factors regulating expression of stress-responsive genes. Bioinform. Biol. Insights 8, 193–198. doi: 10.4137/BBI.S16313
Marrs, K. A. (1996). The functions and regulation of glutathione s-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 127–158. doi: 10.1146/annurev.arplant.47.1.127
Maruyama, K., Todaka, D., Mizoi, J., Yoshida, T., Kidokoro, S., Matsukura, S., et al. (2012). Identification of Cis-acting promoter elements in cold- and dehydration-induced transcriptional pathways in arabidopsis, rice, and soybean. DNA Res. 19, 37–49. doi: 10.1093/dnares/dsr040
Massange-Sanchez, J. A., Palmeros-Suarez, P. A., Martinez-Gallardo, N. A., Castrillon-Arbelaez, P. A., Avilés-Arnaut, H., Alatorre-Cobos, F., et al. (2015). The novel and taxonomically restricted Ah24 gene from grain amaranth (Amaranthus hypochondriacus) has a dual role in development and defense. Front. Plant Sci. 6:602. doi: 10.3389/fpls.2015.00602
Matus, J. T., Aquea, F., and Arce-Johnson, P. (2008). Analysis of the grape MYB R2R3 subfamily reveals expanded wine quality-related clades and conserved gene structure organization across Vitis and Arabidopsis genomes. BMC Plant Biol. 8:83. doi: 10.1186/1471-2229-8-83
Medici, L. O., Nazareno, A. C., Gaziola, S. A., Schmidt, D., and Azevedo, R. A. (2015). “Lysine metabolism,” in Amino Acids in Higher Plants, ed J. P. F. D'Mello (Edinburgh: CAB International), 234–250.
Mellway, R. D., Tran, L. T., Prouse, M. B., Campbell, M. M., and Constabe, C. P. (2009). The Wound-, pathogen-, and ultraviolet B-responsive MYB134 gene encodes an R2R3 MYB transcription factor that regulates proanthocyanidin synthesis in poplar. Plant Physol. 150, 924–941. doi: 10.1104/pp.109.139071
Mickelbart, M. V., Hasegawa, P. M., and Bailey-Serres, J. (2015). Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 16, 237–251. doi: 10.1038/nrg3901
Mikkelsen, M. D., and Thomashow, M. F. (2009). A role for circadian evening elements in cold-regulated gene expression in Arabidopsis. Plant J. 60, 328–339. doi: 10.1111/j.1365-313X.2009.03957.x
Mitsuda, N., and Ohme-Takagi, M. (2009). Functional analysis of transcription factors in Arabidopsis. Plant Cell Physiol. 50, 1232–1248. doi: 10.1093/pcp/pcp075
Miura, K., and Tada, Y. (2014). Regulation of water, salinity, and cold stress responses by salicylic acid. Front. Plant Sci. 5:4. doi: 10.3389/fpls.2014.00004
Mohanty, B., Lakshmanan, M., Lim, S. H., Kim, J. K., Ha, S. H., and Lee, D. Y. (2016). Light-specific transcriptional regulation of the accumulation of carotenoids and phenolic compounds in rice leaves. Plant Signal. Behav. 11:e1184808. doi: 10.1080/15592324.2016.1184808
Moran Lauter, A. N., Peiffer, G. A., Yin, T., Whitham, S. A., Cook, D., Shoemaker, R. C., et al. (2014). Identification of candidate genes involved in early iron deficiency chlorosis signaling in soybean (Glycine max) roots and leaves. BMC Genomics 15:702. doi: 10.1186/1471-2164-15-702
Morkunas, I., and Ratajczak, L. (2014). The role of sugar signaling in plant defense responses against fungal pathogens. Acta. Physiol. Plant. 36, 1607–1619. doi: 10.1007/s11738-014-1559-z
Mouradov, A., and Spangenberg, G. (2014). Flavonoids: a metabolic network mediating plants adaptation to their real estate. Front. Plant. Sci. 5:620. doi: 10.3389/fpls.2014.00620
Mundy, J., Yamaguchi-Shinozaki, K., and Chua, N. (1990). Nuclear proteins bind conserved elements in the abscisic acid-responsive promoter of a rice rab gene. Proc. Natl. Acad. Sci. U.S.A. 87, 1406–1410. doi: 10.1073/pnas.87.4.1406
Nakashima, K., Fujita, Y., Kanamori, N., Katagiri, T., Umezawa, T., Kidokoro, S., et al. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2,SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50, 1345–1363. doi: 10.1093/pcp/pcp083
Nakashima, K., Fujita, Y., Katsura, K., Maruyama, K., Narusaka, Y., Seki, M., et al. (2006). Transcriptional regulation of ABI3- and ABA-responsive genes including RD29B and RD29A in seeds, germinating embryos, and seedlings of Arabidopsis. Plant Mol. Biol. 60, 51–68. doi: 10.1007/s11103-005-2418-5
Narusaka, Y., Nakashima, K., Shinwari, Z. K., Sakuma, Y., Furihata, T., Abe, H., et al. (2003). Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stress. Plant J. 34, 137–148. doi: 10.1046/j.1365-313X.2003.01708.x
Nemchenko, A., Kunze, S., Feussner, I., and Kolomiets, M. (2006). Duplicate maize 13-lipoxygenase genes are differentially regulated by circadian rhythm, cold stress, wounding, pathogen infection, and hormonal treatments. J. Exp. Bot. 57, 3767–3779. doi: 10.1093/jxb/erl137
Neustaedter, D. A., Lee, S. P., and Douglas, C. J. (1999). A novel parsley 4CL1 cis-element is required for developmentally regulated expression and protein-DNA complex formation. Plant J. 18, 77–88. doi: 10.1046/j.1365-313X.1999.00430.x
Niinemets, Ü. (2015). Uncovering the hidden facets of drought stress: secondary metabolites make the difference. Tree. Physiol. 36, 129–132. doi: 10.1093/treephys/tpv128
Niu, J. Q., Wang, A. Q., Huang, J. L., Yang, L. T., and Li, Y. R. (2015). Isolation, characterization and promoter analysis of cell wall invertase gene SoCIN1 from sugarcane (Saccharum spp.). Sugar Tech 17, 65–76. doi: 10.1007/s12355-014-0348-8
Noguero, M., Atif, R. M., Ochatt, S., and Thompson, R. D. (2013). The role of the DNA-binding One Zinc Finger (DOF) transcription factor family in plants. Plant Sci. 209, 32–45. doi: 10.1016/j.plantsci.2013.03.016
Osakabe, Y., Osakabe, K., Shinozaki, K., and Tran, L. S. (2014). Response of plants to water stress. Front. Plant Sci. 5:86. doi: 10.3389/fpls.2014.00086
Ou, Y., Song, B., Liu, X., Xie, C., Li, M., Lin, Y., et al. (2013). Promoter regions of potato vacuolar invertase gene in response to sugars and hormones. Plant Physiol. Biochem. 69, 9–16. doi: 10.1016/j.plaphy.2013.04.015
Padilla, M. N., Hernández, M. L., Sanz, C., and Martínez-Rivas, J. M. (2014). Stress-dependent regulation of 13-lipoxygenases and 13-hydroperoxide lyase in olive fruit mesocarp. Phytochemistry 102, 80–88. doi: 10.1016/j.phytochem.2014.01.024
Palmieri, M. C., Sell, S., Huang, X., Scherf, M., Werner, T., Durner, J., et al. (2008). Nitric oxide-responsive genes and promoters in Arabidopsis thaliana: a bioinformatics approach. J. Exp. Bot. 59, 177–186. doi: 10.1093/jxb/erm345
Pandey, A., Misra, P., and Trivedi, P. K. (2015). Constitutive expression of Arabidopsis MYB transcription factor, AtMYB11, in tobacco modulates flavonoid biosynthesis in favor of flavonol accumulation. Plant Cell Rep. 34, 1515–1528. doi: 10.1007/s00299-015-1803-z
Pandey, G. K., Grant, J. J., Cheong, Y. H., Kim, B. G., Li, L., and Luan, S. (2005). ABR1, an APETALA2-domain transcription factor that functions as a repressor of ABA response in Arabidopsis. Plant Physiol. 139, 1185–1193. doi: 10.1104/pp.105.066324
Passricha, N., Saifi, S., Ansari, M. W., and Tuteja, N. (2016). Prediction and validation of cis-regulatory elements in 5′ upstream regulatory regions of lectin receptor-like kinase gene family in rice. Protoplasma. doi: 10.1007/s00709-016-0979-6. [Epub ahead of print].
Pautler, M., Eveland, A. L., LaRue, T., Yang, F., Weeks, R., Lunde, C., et al. (2015). FASCIATED EAR4 encodes a bZIP transcription factor that regulates shoot meristem size in maize. Plant Cell 27, 104–120. doi: 10.1105/tpc.114.132506
Payyavula, R. S., Singh, R. K., and Navarre, D. A. (2013). Transcription factors, sucrose, and sucrose metabolic genes interact to regulate potato phenylpropanoid metabolism. J. Exp Bot. 64, 5115–5131. doi: 10.1093/jxb/ert303
Pedrosa, A. M., Martins Cde, P., Gonçalves, L. P., and Costa, M. G. (2015). Late embryogenesis abundant (LEA) constitutes a large and diverse family of proteins involved in development and abiotic stress responses in sweet orange (Citrus sinensis L. Osb.). PLoS ONE 10:e0145785. doi: 10.1371/journal.pone.0145785
Peleg, Z., Apse, M. P., and Blumwald, E. (2011). Engineering salinity and water-stress tolerance in crop plants: getting closer to the field. Adv. Bot. Res. 57, 405–443. doi: 10.1016/B978-0-12-387692-8.00012-6
Peng, J., Liu, J., Zhang, L., Luo, J., Dong, H., Ma, Y., et al. (2016). Effects of soil salinity on sucrose metabolism in cotton leaves. PLoS ONE 11:e0156241. doi: 10.1371/journal.pone.0156241
Perisic, O., and Lam, E. (1992). A tobacco DNA binding protein that lnteracts with a light-responsive box II element. Plant Cell 4, 831–838. doi: 10.1105/tpc.4.7.831
Planchet, E., and Limami, A. M. (2015). “Amino acid synthesis under abiotic stress,” in Amino Acids in Higher Plants, ed J. P. F. D'Mello (Edinburgh: CAB International), 262–276.
Prasad, P. V. V., Staggenborg, S. A., and Ristic, Z. (2008). “Impacts of drought and/or heat stress on physiological, developmental, growth, and yield processes of crop plants,” in Response of Crops to Limited Water: Understanding and Modeling Water Stress Effects on Plant Growth Processes, eds L. R. Ahuja, V. R. Reddy, S. A. Saseendran, and Q. Yu (Madison, WI), 301–355.
Preston, J., Wheeler, J., Heazlewood, J., Li, S. F., and Parish, R. W. (2004). AtMYB32 is required for normal pollen development in Arabidopsis thaliana. Plant J. 40, 979–995. doi: 10.1111/j.1365-313X.2004.02280.x
Proels, R. K., and Roitsch, T. (2009). Extracellular invertase LIN6 of tomato: a pivotal enzyme for integration of metabolic, hormonal, and stress signals is regulated by a diurnal rhythm. J. Exp. Bot. 60, 1555–1567. doi: 10.1093/jxb/erp027
Prouse, M. B., and Campbell, M. M. (2013). Interactions between the R2R3-MYB transcription factor, AtMYB61, and target DNA binding sites. PLoS ONE 8:e65132. doi: 10.1371/journal.pone.0065132
Qados, A. M. S. A. (2011). Effect of salt stress on plant growth and metabolism of bean plant Vicia faba (L.). J. Saudi Soc. Agri. Sci. 10, 7–15. doi: 10.1016/j.jssas.2010.06.002
Qin, Y., Tian, Y., and Liu, X. (2015). A wheat salinity-induced WRKY transcription factor TaWRKY93 confers multiple abiotic stress tolerance in Arabidopsis thaliana. Biochem. Biophys. Res. Comm. 464, 428–433. doi: 10.1016/j.bbrc.2015.06.128
Qu, C. P., Xu, Z. R., Liu, G. J., Liu, C., Li, Y., Wei, Z. G., et al. (2010). Differential expression of copper-zinc superoxide dismutase gene of Polygonum sibiricum leaves, stems and underground stems, subjected to high-salt stress. Int. J. Mol. Sci. 11, 5234–5245. doi: 10.3390/ijms11125234
Rabot, A., Portemer, V., Péron, T., Mortreau, E., Leduc, N., Hamama, L., et al. (2014). Interplay of sugar, light and gibberellins in expression of Rosa hybrida Vacuolar Invertase 1 regulation. Plant Cell Physiol. 55, 1734–1748. doi: 10.1093/pcp/pcu106
Rai, V. K. (2002). Role of amino acids in plant responses to stresses. Biol. Plant. 45, 481–487. doi: 10.1023/A:1022308229759
Ramakrishna, A., and Ravishankar, G. A. (2011). Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 6, 1720–1731 doi: 10.4161/psb.6.11.17613
Rasmussen, T. B., and Donaldson, I. A. (2006). Investigation of the endosperm-specific sucrose synthase promoter from rice using transient expression of reporter genes in guar seed tissue. Plant Cell Rep. 25, 1035–1042. doi: 10.1007/s00299-006-0158-x
Rejeb, I. B., Pastor, V., and Mauch-Mani, B. (2014). Plant responses to simultaneous biotic and abiotic stress: molecular mechanisms. Plants 3, 458–475. doi: 10.3390/plants3040458
Rivas-San Vicente, M., and Plasencia, J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62, 3321–3338, doi: 10.1093/jxb/err031
Rodriguez-Kessler, M., Alpuche-Solis, A. G., Ruiz, O. A., and Jimenex-Bremont, J. F. (2006). Effect of salt stress on the regulation of maize (Zea mays L.) genes involved in polyamine biosynthesis. Plant Growth Regul. 48, 175–185. doi: 10.1007/s10725-005-5990-4
Rombauts, S., Florquin, K., Lescot, M., Marchal, K., Rouzé, R., and van de Peer, Y. (2003). Computational approaches to identify promoters and cis- regulatory elements in plant genomes. Plant Physiol. 132, 1162–1176. doi: 10.1104/pp.102.017715
Rook, F., Weisbeek, P., and Smeekens, S. (1998). The light-regulated Arabidopsis bZIP transcription factor gene ATB2 encodes a protein with an unusually long leucine zipper domain. Plant Mol. Biol. 37, 171–178. doi: 10.1023/A:1005964327725
Rosa, M., Prado, C., Podazza, G., Interdonato, R., González, J. A., Hilal, M., et al. (2009). Soluble sugars-Metabolism, sensing and abiotic stress. A complex network in the life of plants. Plant. Signal. Behav. 4, 388–393. doi: 10.4161/psb.4.5.8294
Roy Choudhury, S., Roy, S., Das, R., and Sengupta, D. N. (2008). Differential transcriptional regulation of banana sucrose phosphate synthase gene in response to ethylene, auxin, wounding, low temperature and different photoperiods during fruit ripening and functional analysis of banana SPS gene promoter. Planta 229, 207–223. doi: 10.1007/s00425-008-0821-2
Rushton, D. L., Tripathi, P., Rabara, R. C., Lin, J., Ringler, P., Boken, A. K., et al. (2012). WRKY transcription factors: key components in abscisic acid signalling. Plant Biotech. J. 10, 2–11. doi: 10.1111/j.1467-7652.2011.00634.x
Saibo, N. J., Lourenço, T., and Oliveira, M. M. (2009). Transcription factors and regulation of photosynthetic and related metabolism under environmental stresses. Ann. Bot. 103, 609–623. doi: 10.1093/aob/mcn227
Sakuma, Y., Maruyama, K., Osakabe, Y., Qin, F., Seki, M., Shinozaki, K., et al. (2006). Functional analysis of an Arabidopsis transcription factor, DREB2A, involved in drought-responsive gene expression. Plant Cell 18, 1292–1309. doi: 10.1105/tpc.105.035881
Sakuragi, Y., Maeda, H., Dellapenna, D., and Bryant, D. A. (2006). α-tocopherol plays a role in photosynthesis and macronutrient homeostasis of the Cyanobacterium Synechocystis sp. PCC 6803 that is independent of its antioxidant function. Plant Physiol. 141, 508–521. doi: 10.1104/pp.105.074765
Saleh, A. M., and Madany, M. M. Y. (2015). Coumarin pretreatment alleviates salinity stress in wheat seedlings. Plant Physiol. Biochem. 88, 27–35. doi: 10.1016/j.plaphy.2015.01.005
Sánchez-Aguayo, I., Rodriguez-Galan, J. M., Garcia, R., Torreblanca, J., and Pardo, J. M. (2004). Salt stress enhances xylem development and expression of S-adenosyl-l-methionine synthase in lignifying tissues of tomato plants. Planta 220, 278–285. doi: 10.1007/s00425-004-1350-2
Satoh, R., Fujita, Y., Nakashima, K., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2004). Novel subgroup of bZIP proteins functions as transcriptional activators in hypoosmolarity-responsive expression of the ProDH gene in Arabidopsis. Plant Cell Physiol. 45, 309–317. doi: 10.1093/pcp/pch036
Schlüter, U., Colmsee, C., Scholz, U., Bräutigam, A., Weber, A. P., Zellerhoff, N., et al. (2013). Adaptation of maize source leaf metabolism to stress related disturbances in carbon, nitrogen and phosphorus balance. BMC Genomics 14:442. doi: 10.1186/1471-2164-14-442
Schlüter, U., Colmsee, C., Scholz, U., Bräutigam, A., Weber, A. P., Zellerhoff, N., et al. (2013). Adaptation of maize source leaf metabolism to stress related disturbances in carbon, nitrogen and phosphorus balance. BMC Genomics 14:442. doi: 10.1186/1471-2164-14-442
Schmidt, R. J., Ketudat, M., Aukerman, M. J., and Hoschek, G. (1992). Opaque-2 1s a transcriptional activator that recognizes a specific target site in 22-kD zein genes. Plant Cell 4, 689–700. doi: 10.1105/tpc.4.6.689
Schmitz, D., Lohmer, S., Salamini, F., and Thompson, R. D. (1997). The activation domain of the maize transcription factor Opaque-2 resides in a single acidic region. Nucleic Acids Res. 25, 756–763. doi: 10.1093/nar/25.4.756
Sengupta-Gopalan, C., and Ortega, J. L. (2015). “An insight into the role and regulation of glutamine synthetase in plants,” in Amino Acids in Higher Plants, ed J. P. F. D'Mello (Edinburgh: CAB International), 82–99.
Sheshadri, S. A., Sriram, S., Balamurugan, P., Anupriya, R., Princy, S. A., Brindha, P., et al. (2015). Melatonin improves bioreductant capacity and silver nanoparticles synthesis using Catharanthus roseus leaves. RSC Adv. 5, 47548–47554. doi: 10.1039/C5RA01848J
Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 58, 221–227. doi: 10.1093/jxb/erl164
Shulaev, V., Silverman, P., and Raskin, I. (1997). Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 385, 718–721. doi: 10.1038/385718a0
Shulze, E. D., Beck, E., and Muller-Hohenstein, K. (2005). “Environment as stress factor: stress physiology of plants,” in Plant Ecology (Springer-Verlag), 9–11.
Sibéril, Y., Dioreau, P., and Gantet, P. (2001). Plant bZIP G-box binding factors. Eur. J. Biochem. 268, 5655–5666. doi: 10.1046/j.0014-2956.2001.02552.x
Singh, B., and Sharma, R. A. (2015). Plant terpenes: defense responses, phylogenetic analysis, regulation and clinical applications. 3 Biotech 5, 129–519. doi: 10.1007/s13205-014-0220-2
Singh, D., and Laxmi, A. (2015). Transcriptional regulation of drought response: a tortuous network of transcriptional factors. Front. Plant Sci. 6:895. doi: 10.3389/fpls.2015.00895
Singh, K., Kumar, S., Kumar, S., and Ahuja, P. S. (2009). Characterization of dihydroflavonol 4-reductase cDNA in tea [Camellia sinensis (L.) O. Kuntze]. Plant. Biotechno. Rep. 3, 95–101. doi: 10.1007/s11816-008-0079-y
Singh, N., and Bhardwaj, R. D. (2016). Ascorbic acid alleviates water deficit induced growth inhibition in wheat seedlings by modulating levels of endogenous antioxidants. Biologia 71, 402–413. doi: 10.1515/biolog-2016-0050
Singh, P. K., Singh, R., and Singh, S. (2013). Cinnamic acid induced changes in reactive oxygen species scavenging enzymes and protein profile in maize (Zea mays L.) plants grown under salt stress. Physiol. Mol. Biol. Plants 19, 53–59. doi: 10.1007/s12298-012-0126-6
Smith, T. A. (1984). “Putrescine and inorganic ions,” in Phytochemical Adaptations to Stress, eds B. N. Timmerman, C. Steelink, and F. A. Loewus (Washington, DC: Springer), 7–54. doi: 10.1007/978-1-4684-1206-2_2
Soltani, B. M., Ehlting, J., Hamberger, B., and Douglas, C. J. (2006). Multiple cis-regulatory elements regulate distinct and complex patterns of developmental and wound-induced expression of Arabidopsis thaliana 4CL gene family members. Planta 224, 1226–1238. doi: 10.1007/s00425-006-0296-y
Stintzi, A., Weber, H., Reymond, P., Browse, J., and Farmer, E. E. (2001). Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc. Natl. Acad. Sci. U.S.A. 98, 12837–12842. doi: 10.1073/pnas.211311098
Stiti, N., Missihoun, T. D., Kotchoni, S. O., Kirch, H. H., and Bartels, D. (2011). Aldehyde dehydrogenases in Arabidopsis thaliana: biochemical requirements, metabolic pathways, and functional analysis. Front. Plant Sci. 2:65. doi: 10.3389/fpls.2011.00065
Stracke, R., Ishihara, H., Huep, G., Barsch, A., Mehrtens, F., Niehaus, K., et al. (2007). Differential regulation of closely related R2R3-MYB transcription factors controls flavonol accumulation in different parts of the Arabidopsis thaliana seedling. Plant J. 50, 660–677. doi: 10.1111/j.1365-313X.2007.03078.x
Strathmann, A., Kuhlmann, M., Heinekamp, T., and Dröge-Laser, W. (2001). BZI-1 specifically heterodimerises with the tobacco bZIP transcription factors BZI-2, BZI-3/TBZF and BZI-4, and is functionally involved in flower development. Plant J. 28, 397–408. doi: 10.1046/j.1365-313X.2001.01164.x
Streeter, J. G., Lohnes, D. G., and Fioritto, R. J. (2001). Patterns of pinitol accumulation in soybean plants and relationships to drought tolerance. Plant. Cell. Environ. 24, 429–438. doi: 10.1046/j.1365-3040.2001.00690.x
Sun, C., Palmqvist, S., Olsson, H., Borén, M., Ahlandsberg, S., and Jansson, C. (2003). A novel WRKY transcription factor, SUSIBA2, participates in sugar signaling in barley by binding to the sugar-responsive elements of the iso1 promoter. Plant Cell 15, 2076–2092. doi: 10.1105/tpc.014597
Sun, H., Fan, H. J., and Ling, H. Q. (2015). Genome-wide identification and characterization of the bHLH gene family in tomato. BMC Genomics 16:9. doi: 10.1186/s12864-014-1209-2
Sun, N., Liu, M., Zhang, W., Yang, W., Bei, X., Ma, H., et al. (2015). Bean metal-responsive element-binding transcription factor confers cadmium resistance in tobacco. Plant Phys. 167, 1136–1148. doi: 10.1104/pp.114.253096
Taji, T., Seki, M., Yamaguchi-Shinozaki, K., Kamada, H., Giraudat, J., and Shinozaki, K. (1999). Mapping of 25 drought-inducible genes, RD and ERD, in Arabidopsis thaliana. Plant Cell Physiol. 40, 119–123. doi: 10.1093/oxfordjournals.pcp.a029469
Takagi, H., Ishiga, Y., Watanabe, S., Konishi, T., Egusa, M., Akiyoshi, N., et al. (2016). Allantoin, a stress-related purine metabolite, can activate jasmonate signaling in a MYC2-regulated and abscisic acid-dependent manner. J. Exp. Bot. 67, 2519–2532. doi: 10.1093/jxb/erw071
Tanaka, K., Taniguchi, S., Tamaoki, D., Yoshitomi, K., Akimitsu, K., and Gomi, K. (2014). Multiple roles of plant volatiles in jasmonate-induced defense response in rice. Plant Signal. Behav. 9:e29247. doi: 10.4161/psb.29247
Tari, I., Kiss, G., Deer, A. K., Csiszar, J., Erdei, L., Galle, A., et al. (2010). Salicylic acid increased aldose reductase activity and sorbitol accumulation in tomato plants under salt stress. Biol. Plant. 54, 677–683. doi: 10.1007/s10535-010-0120-1
Teixeira, P. J., Thomazella, D. P., Reis, O., do Prado, P. F., do Rio, M. C., Fiorin, G. L., et al. (2014). High-resolution transcript profiling of the atypical biotrophic interaction between Theobroma cacao and the fungal pathogen Moniliophthora perniciosa. Plant Cell 26, 4245–4269. doi: 10.1105/tpc.114.130807
Theerawitaya, C., Boriboonkaset, T., Cha-Um, S., Supaibulwatana, K., and Kirdmanee, C. (2012). Transcriptional regulations of the genes of starch metabolism and physiological changes in response to salt stress rice (Oryza sativa L.) seedlings. Physiol. Mol. Biol. Plants 18:197. doi: 10.1007/s12298-012-0114-x
Timpa, J. D., Burke, J. J., Quisenberry, J. E., and Wendt, C. W. (1986). Effects of water stress on the organic acid and carbohydrate compositions of cotton plants. Plant Physiol. 82, 724–728. doi: 10.1104/pp.82.3.724
Tiwari, V., Patel, M. K., Chaturvedi, A. K., Mishra, A., and Jha, B. (2016). Functional characterization of the tau class glutathione-S-transferases gene (SbGSTU) promoter of Salicornia brachiata under salinity and osmotic stress. PLoS ONE 11:e0148494. doi: 10.1371/journal.pone.0148494
Tohge, T., Watanabe, M., Hoefgen, R., and Fernie, A. R. (2013). Shikimate and phenylalanine biosynthesis in the green lineage. Front. Plant Sci. 4:62. doi: 10.3389/fpls.2013.00062
Tsai, C., Kayal, W. E., and Harding, S. A. (2006). Populus, the new model system for investigating phenylpropanoid complexity. Int. J. Appl. Sci. Eng. 4, 221–233.
Tuteja, N. (2007). Abscisic acid and abiotic stress signaling. Plant Signal. Behav. 2, 135–138. doi: 10.4161/psb.2.3.4156
Umate, P. (2011). Genome-wide analysis of lipoxygenase gene family in Arabidopsis and rice. Plant Signal. Behav. 6, 335–338. doi: 10.4161/psb.6.3.13546
Uquillas, C., Letelier, I., Blanco, F., Jordana, X., and Holuigue, L. (2004). NPR1-independent activation of immediate early salicylic acid-responsive genes in Arabidopsis. Mol. Plant Microbe Interact. 17, 34–42. doi: 10.1094/MPMI.2004.17.1.34
Van Aken, O., Zhang, B., Law, S., Narsai, R., and Whelan, J. (2013). AtWRKY40 and AtWRKY63 modulate the expression of stress-responsive nuclear genes encoding mitochondrial and chloroplast proteins. Plant Physiol. 162, 254–271. doi: 10.1104/pp.113.215996
Vasquez-Robinet, C., Mane, S. P., Ulanov, A. V., Watkinson, J. I., Stromberg, V. K., De Koeyer, D., et al. (2008). Physiological and molecular adaptations to drought in Andean potato genotypes. J. Exp. Bot. 59, 2109–2123. doi: 10.1093/jxb/ern073
Wang, H., Datla, R., Georges, F., Loewen, M., and Cutler, A. J. (1995). Promoters from kinl and cor6.6, two homologous Arabidopsis thaliana genes: transcriptional regulation and gene expression induced by low temperature, ABA, osmoticum and dehydration. Plant Mol. Biol. 28, 605–617. doi: 10.1007/BF00021187
Wang, Z., Cheng, K., Wan, L., Yan, L., Jiang, H., Liu, S., et al. (2015). Genome-wide analysis of the basic leucine zipper (bZIP) transcription factor gene family in six legume genomes. BMC Genomics 16:1053. doi: 10.1186/s12864-015-2258-x
Wei, K., Chen, J., Wang, Y., Chen, Y., Chen, S., Lin, Y., et al. (2012). Genome-wide analysis of bZIP-encoding genes in maize. DNA Res. 19, 463–476. doi: 10.1093/dnares/dss026
Wei, W., Li, Q.-T., Chu, Y.-N., Reiter, R. J., Yu, X.-M., Zhu, D.-H., et al. (2015). Melatonin enhances plant growth and abiotic stress tolerance in soybean plants. J. Exp. Bot. 66, 695–707. doi: 10.1093/jxb/eru392
Weise, S. E., Schrader, S. M., Kleinbeck, K. R., and Sharkey, T. D. (2006). Carbon balance and circadian regulation of hydrolytic and phosphorolytic breakdown of transitory starch. Plant. Physiol. 141, 879–886. doi: 10.1104/pp.106.081174
Weldegergis, B. T., Zhu, F., Poelman, E. H., and Dicke, M. (2015). Drought stress affects plant metabolites and herbivore preference but not host location by its parasitoids. Oecologia 177, 701–713. doi: 10.1007/s00442-014-3129-x
Wittkopp, P. J., and Kalay, G. (2012). Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69. doi: 10.1038/nrg3095
Wobbes, B. (2004). Control of Plant Carbohydrate Partitioning by the Arabidopsis thaliana ATB2 bZIP Transcription Factor Gene. dissertation/doctoral thesis, University of Utrecht.
Woo, H. R., Kim, J. H., Kim, J., Kim, J., Lee, U., Song, I. J., et al. (2010). The RAV1 transcription factor positively regulates leaf senescence in Arabidopsis. J. Exp. Bot. 61, 3947–3957. doi: 10.1093/jxb/erq206
Wu, J. D., Jiang, C. P., Zhu, H. S., Jiang, H. Y., Cheng, B. J., and Zhu, S. W. (2015). Cloning and functional analysis of the promoter of a maize starch synthase III gene (ZmDULL1). Genet. Mol. Res. 14, 5468–5479. doi: 10.4238/2015.May.22.17
Xie, C., Wang, D., Kou, F., Kang, D., and Yang, X. (2012). The pathogen and wound induces expression of genes related to proanthocyanidins (PAs) synthesis in cotton leaves. AJPS 3, 228–234. doi: 10.4236/ajps.2012.32027
Xin, Z., and Browse, J. (2000). Cold comfort farm: the acclimation of plants to freezing temperatures. Plant Cell Environ. 23, 893–902. doi: 10.1046/j.1365-3040.2000.00611.x
Xu, X., Liu, M., Lu, L., He, M., Qu, W., Xu, Q., et al. (2015). Genome-wide analysis and expression of the calcium-dependent protein kinase gene family in cucumber. Mol. Genet. Genom. 290, 1403–1414. doi: 10.1007/s00438-015-1002-1
Xu, Z. S., Chen, M., Li, L. C., and Ma, Y. Z. (2011). Functions and application of the AP2/ERF transcription factor family in crop improvement. J. Integr. Plant Biol. 53, 570–585. doi: 10.1111/j.1744-7909.2011.01062.x
Yadav, S. K. (2010). Cold stress tolerance mechanisms in plants. A review. Agron. Sustain. Dev. 30, 515–527. doi: 10.1051/agro/2009050
Yamada, Y., and Sato, F. (2013). Transcription factors in alkaloid biosynthesis. Int. Rev. Cell Mol. Biol. 305, 339–382. doi: 10.1016/B978-0-12-407695-2.00008-1
Yogendra, K. N., Kumar, A., Sarkar, K., Li, Y., Pushpa, D., Mosa, K. A., et al. (2015). Transcription factor StWRKY1 regulates phenylpropanoid metabolites conferring late blight resistance in potato. J. Exp. Bot. 66, 7377–7389. doi: 10.1093/jxb/erv434
Yoshida, I., Yamagata, H., and Hirasawa, E. (1998). Light-regulated gene expression of S-adenosylmethionine decarboxylase in Pharbitis nil. J. Exp. Bot. 49, 617–620. doi: 10.1093/jxb/49.320.617
Yoshida, T., Fujita, Y., Maruyama, K., Mogami, J., Todaka, D., Shinozaki, K., et al. (2015). Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 38, 35–49. doi: 10.1111/pce.12351
Zhang, W.-W., Li, J.-B., Xu, F., Tang, Y., Cheng, S.-Y., and Cao, F.-L. (2014). Isolation and characterization of a phenylalanine ammonia-lyase gene (PAL) promoter from Ginkgo biloba and its regulation of gene expression in transgenic tobacco plants. Plant Omics J. 7, 353–360.
Zhang, Y., Yin, H., Li, D., Zhu, W., and Li, Q. (2008). Functional analysis of BADH gene promoter from Suaeda liaotungensis K. Plant Cell Rep. 27, 585–592. doi: 10.1007/s00299-007-0459-8
Zhang, Z., Yang, J., and Wu, Y. (2015). Transcriptional regulation of zein gene expression in maize through the additive and synergistic action of opaque2, prolamine-box binding factor, and O2 heterodimerizing proteins. Plant Cell 27, 1162–1172. doi: 10.1105/tpc.15.00035
Zhao, J., Guo, R., Guo, C., Hou, H., Wang, X., and Gao, H. (2016). Evolutionary and expression analyses of the apple basic leucine zipper transcription factor family. Front. Plant Sci. 7:376. doi: 10.3389/fpls.2016.00376
Zhao, Q., and Dixon, R. A. (2011). Transcriptional networks for lignin biosynthesis: more complex than we thought? Trends Plant Sci. 16, 227–233. doi: 10.1016/j.tplants.2010.12.005
Zhao, X. Q., Wang, W. S., Zhang, F., Zhang, T., Zhao, W., Fu, B. Y., et al. (2013). Temporal profiling of primary metabolites under chilling stress and its association with seedling chilling tolerance of rice (Oryza sativa L.). Rice 6:23. doi: 10.1186/1939-8433-6-23
Zhou, J., Yang, Y., Yu, J., Wang, L., Yu, X., Ohtani, M., et al. (2014). Responses of Populus trichocarpa galactinol synthase genes to abiotic stresses. J. Plant Res. 127, 347–358. doi: 10.1007/s10265-013-0597-8
Zhou, M. L., Zhang, Q., Zhou, M., Sun, Z. M., Zhu, X. M., Shao, J. R., et al. (2012). Genome-wide identification of genes involved in raffinose metabolism in Maize. Glycobiol. 22, 1775–1785. doi: 10.1093/glycob/cws121
Zhu, J., Xu, J., Chang, W., and Zhang, Z. (2015). Isolation and molecular characterization of 1-aminocyclopropane-1-carboxylic acid synthase genes in Hevea brasiliensis. Int. J. Mol. Sci. 16, 4136–4149. doi: 10.3390/ijms16024136
Keywords: stress, cis-elements, primary metabolism, specialized metabolism, transcriptional regulation
Citation: Sheshadri SA, Nishanth MJ and Simon B (2016) Stress-Mediated cis-Element Transcription Factor Interactions Interconnecting Primary and Specialized Metabolism in planta. Front. Plant Sci. 7:1725. doi: 10.3389/fpls.2016.01725
Received: 07 June 2016; Accepted: 02 November 2016;
Published: 25 November 2016.
Edited by:
Basil J. Nikolau, Iowa State University, USAReviewed by:
Thomas D. Sharkey, Michigan State University, USATimothy Patrick Durrett, Kansas State University, USA
Copyright © 2016 Sheshadri, Nishanth and Simon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bindu Simon, YmluZHVzaW1vbkBzY2J0LnNhc3RyYS5lZHU=
 S. A. Sheshadri
S. A. Sheshadri M. J. Nishanth
M. J. Nishanth