
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 21 November 2016
Sec. Plant Breeding
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.01718
This article is part of the Research Topic Plant Translational Research in Genomics Era View all 39 articles
 Hiroshi Mizuno1,2*†
Hiroshi Mizuno1,2*† Takayuki Yazawa1
Takayuki Yazawa1 Shigemitsu Kasuga3
Shigemitsu Kasuga3 Yuji Sawada4
Yuji Sawada4 Hiroyuki Kanamori1,2
Hiroyuki Kanamori1,2 Yuko Ogo1,2
Yuko Ogo1,2 Masami Yokota Hirai4
Masami Yokota Hirai4 Takashi Matsumoto1,2
Takashi Matsumoto1,2 Hiroyuki Kawahigashi1,2†
Hiroyuki Kawahigashi1,2†Sorghum (Sorghum bicolor L. Moench) exhibits various color changes in injured leaves in response to cutting stress. Here, we aimed to identify key genes for the light brown and dark brown color variations in tan-colored injured leaves of sorghum. For this purpose, sorghum M36001 (light brown injured leaves), Nakei-MS3B (purple), and a progeny, #7 (dark brown), from Nakei-MS3B × M36001, were used. Accumulated pigments were detected by using high-performance liquid chromatography: M36001 accumulated only apigenin in its light brown leaves; #7 accumulated both luteolin and a small amount of apigenin in its dark brown leaves, and Nakei-MS3B accumulated 3-deoxyanthocyanidins (apigeninidin and luteolinidin) in its purple leaves. Apigenin or luteolin glucoside derivatives were also accumulated, in different proportions. Differentially expressed genes before and after cutting stress were identified by using RNA sequencing (RNA-seq). Integration of our metabolic and RNA-seq analyses suggested that expression of only flavone synthase II (FNSII) led to the synthesis of apigenin in M36001, expression of both FNSII and flavonoid 3′-hydroxylase (F3′H) led to the synthesis of apigenin and luteolin in #7, and expression of both flavanone 4-reductase and F3′H led to the synthesis of 3-deoxyanthocyanidins in Nakei-MS3B. These results suggest that expression of FNSII is related to the synthesis of flavones (apigenin and luteolin) and the expression level of F3′H is related to the balance of apigenin and luteolin. Expression of FNSII and F3′H is thus associated with dark or light brown coloration in tan-colored injured leaves of sorghum.
Sorghum (Sorghum bicolor L. Moench) exhibits various leaf color changes in response to disease, injury by insects, or wounding (Doggett, 1988). Leaves can turn purple, red, orange, or tan (brown), or in-between colors. The color depends on the sorghum cultivar. For example, the leaves of Nakei-MS3B and JN43 turn purple; those of bmr-6 turn reddish orange; those of BTx623 turn orange (Mizuno et al., 2014); and those of M36001, JP501, JP43800, JP588, JP43764, and Greenleaf turn tan (brown; Kawahigashi et al., 2016). Moreover, it is possible to distinguish light or dark brown colorations in plants with tan colors (Doggett, 1988). The tan-colored sorghums accumulate relatively high levels of flavones (apigenin and luteolin) than red/purple colored sorghums (Siame et al., 1993; Dykes et al., 2009, 2011). Colored pigments are accumulated in response to infection with the fungi Bipolaris sorghicola (Kawahigashi et al., 2011; Mizuno et al., 2012), Colletotrichum sublineolum (Snyder and Nicholson, 1990), Cochliobolus heterostrophus (Aguero et al., 2002), Sporisorium reilianum (Zuther et al., 2012) or to wounding stress (Mizuno et al., 2014). Pigment accumulation is considered to enhance resistance to pathogen infection (Hipskind et al., 1990; Snyder and Nicholson, 1990; Lo et al., 1999; Clifford, 2000; Kawahigashi et al., 2011; Zuther et al., 2012).
The P locus is one of the loci responsible for sorghum coloration (purple, red, or orange) and is dominant to p (tan; Doggett, 1988). The protein encoded by the P gene is flavanone 4-reductase (FNR), which is responsible for the synthesis of 3-deoxyanthocyanidins (Kawahigashi et al., 2016). The variation of purple-red-orange is explained by the balance of two 3-deoxyanthocyanidins (luteolinidin and apigeninidin; Mizuno et al., 2014). The balance is controlled by the expression level of flavonoid 3′-hydroxylase (F3′H), which encodes a protein that hydroxylates the 3′ position of the B-ring of naringenin to produce a precursor of luteolinidin (Shih et al., 2006).
Sorghums with a tan injury response have non-functional FNR alleles (p). The FNR gene of JP501 and JP43800 has an insertion in the coding region; those of Greenleaf, JP588, and JP43764 have a Cys252Tyr amino acid substitution and encode unstable protein (Kawahigashi et al., 2016). Sorghums with a tan injury response have lower levels of flavan-4-ols (precursors of apigeninidin and luteolinidin—apiforol and luteoforol, respectively) than do sorghums with a purple or red injury response (Dykes et al., 2005); they are thus unable to synthesize 3-deoxyanthocyanidins. What genes are responsible for the coloration and color variation in sorghums with a tan (light or dark brown) injury response? These sorghums may show activation of an alternative pathway independent of 3-deoxyanthocyanidin synthesis.
Here, we aimed to identify key genes for the light brown and dark brown color variations in sorghum leaves with a tan injury response. For this purpose, we used sorghum Nakei-MS3B (purple response), M36001 (light brown response), and progeny #7 (dark brown response) derived from Nakei-MS3B × M36001. We performed a metabolic analysis to identify the accumulated pigments and an RNA sequencing (RNA-seq) analysis to identify the genes expressed in each line. Our data suggested that flavone synthase II (FNSII) pathways were activated to synthesize flavones (apigenin and luteolin) in sorghum with a tan response, and that levels of F3′H production changed the balance of the two flavones, thus resulting in their color response variation. We also discuss the differences in gene structure and gene expression networks determining each secondary metabolic pathway for coloration of sorghum leaves.
Sorghum Nakei-MS3B (purple response), M36001 (light brown response), and progeny #7 (dark brown response) derived from Nakei-MS3B × M36001 were used. For the plant color test, F2 populations were grown at Shinshu University in Nagano, Japan, in 2011. At Tsukuba, Ibaraki, Japan, in 2012, the F3 populations were grown and subjected to mRNA-seq analysis. Details of the materials used have been given previously (Mizuno et al., 2014). Accumulated pigments were quantified by using HPLC at an absorbance of 475 or 290 nm (Ogo et al., 2013). Metabolic analysis of the accumulated pigments in each line was performed by using LC-MS/MS and standard authentic compounds (Sawada et al., 2009).
Four days after cutting stress, the edges of sorghum leaf strips that exhibited color changes in response to injury were collected. As controls, leaf strips immediately after cutting stress were also collected. To extract RNA, five biological replicates were collected, immediately frozen in liquid nitrogen, and mixed to minimize the effect of transcriptome unevenness among plants. RNA quality was calculated with a Bioanalyzer 2100 algorithm (Agilent Technologies, Palo Alto, CA, USA); high-quality (RNA integrity number >8) RNA was used. The protocol used for extraction of RNA and sequencing with an Illumina GAIIx sequencer (Illumina, San Diego, CA, USA) has been described previously (Mizuno et al., 2010). Reads were deposited in the DDBJ (DNA Data Bank of Japan) Sequence Read Archive (Accession No. DRA001265).
The same RNA material was shared for use in the Illumina RNA-seq and Quantitative RT-PCR (qRT-PCR) analysis. First-strand cDNA was synthesized from RNA (1 μg) in a 20-μl reaction mixture with a TaKaRa RNA PCR kit (AMV) v. 3.0 (TaKaRa Bio, Inc.). PCR was performed with initial denaturation at 98° for 2 min; 42 cycles of 98° for 10 s, 60° for 10 s, and 68° 30 s. qRT-PCR was carried out using Mx3000P (Stratagene Products Division, Agilent Technologies) with KOD SYBR® qPCR Mix (Toyobo) according to the manufacturer’s recommendations and analyzed based on the delta-delta-Ct method. The gene transcripts were amplified with each specific primer pair (Supplementary Table S2). The value for each genes was normalized using SbActin (Sb03g040880) as an internal standard.
Trimming of low-quality nucleotides (<Q15) from both the 5′- and the 3′-ends and of adaptors was performed by using Cutadapt version 1.01. Bowtie 2 version 2.0.0 beta6 (Langmead and Salzberg, 2012) was used to align the reads against sorghum rRNA sequences (Ouyang and Buell, 2004); aligned reads were removed. Reads were aligned to the sorghum reference genome of BTx623 (Paterson et al., 2009) by using Bowtie 2, SAMtools version 0.1.18 (Li et al., 2009), and TopHat version 2.0.4 (Kim et al., 2013). RPKM (Reads Per Kilobase of exon model per Million mapped reads) values were calculated for each transcript annotated in Phytozome version 9 (Goodstein et al., 2012) or in non-annotated gene models constructed by using Cufflinks version 2.0.0 (Trapnell et al., 2010). Heatmaps were generated by using R2 package gplots version 2.10.1, with the RPKM of each gene model and the relative (after/before cutting stress) ratio of RPKM or that of the accumulated pigments determined by LC-MS/MS.
Sorghum leaves exhibited various colors in response to cutting stress. Nakei-MS3B leaves had a purple edge, M36001 had a light brown edge, and progeny #7 from a cross between Nakei-MS3B and M36001 had a dark brown edge (Figure 1). These accumulated pigments were analyzed by using high-performance liquid chromatography (HPLC). Nakei-MS3B accumulated the 3-deoxyanthocyanidins luteolinidin and (a small amount of) apigeninidin in its purple region after cutting stress, whereas #7 and M36001 did not (Figure 2, absorbance [A] 475 nm). #7 accumulated both luteolin and a small amount of apigenin in its dark brown region, whereas M36001 accumulated only apigenin in its light brown region (Figure 2, A 290 nm). In summary, Nakei-MS3B accumulated 3-deoxyanthocyanidins and M36001 and #7 accumulated flavones, with different proportions of luteolin and apigenin.
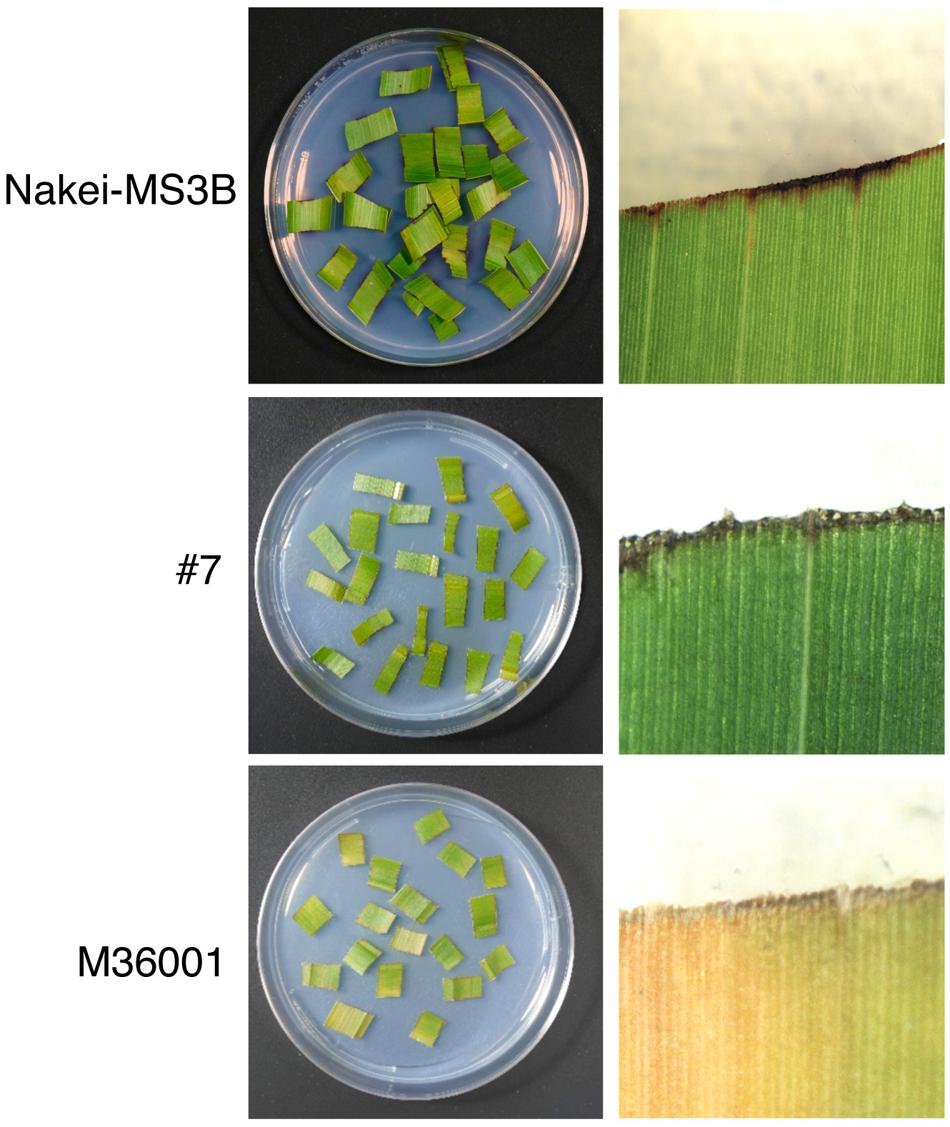
FIGURE 1. Coloration of sorghum leaves after cutting stress. Cut leaves on agarose gel (left) and enlarged images (right) are shown. Nakei-MS3B (Top) turns purple at the cut edge of each leaf fragment, but M36001 (Bottom) turns light brown. The F1 progeny #7 (Middle) of Nakei-MS3B × M36001 turns dark brown.
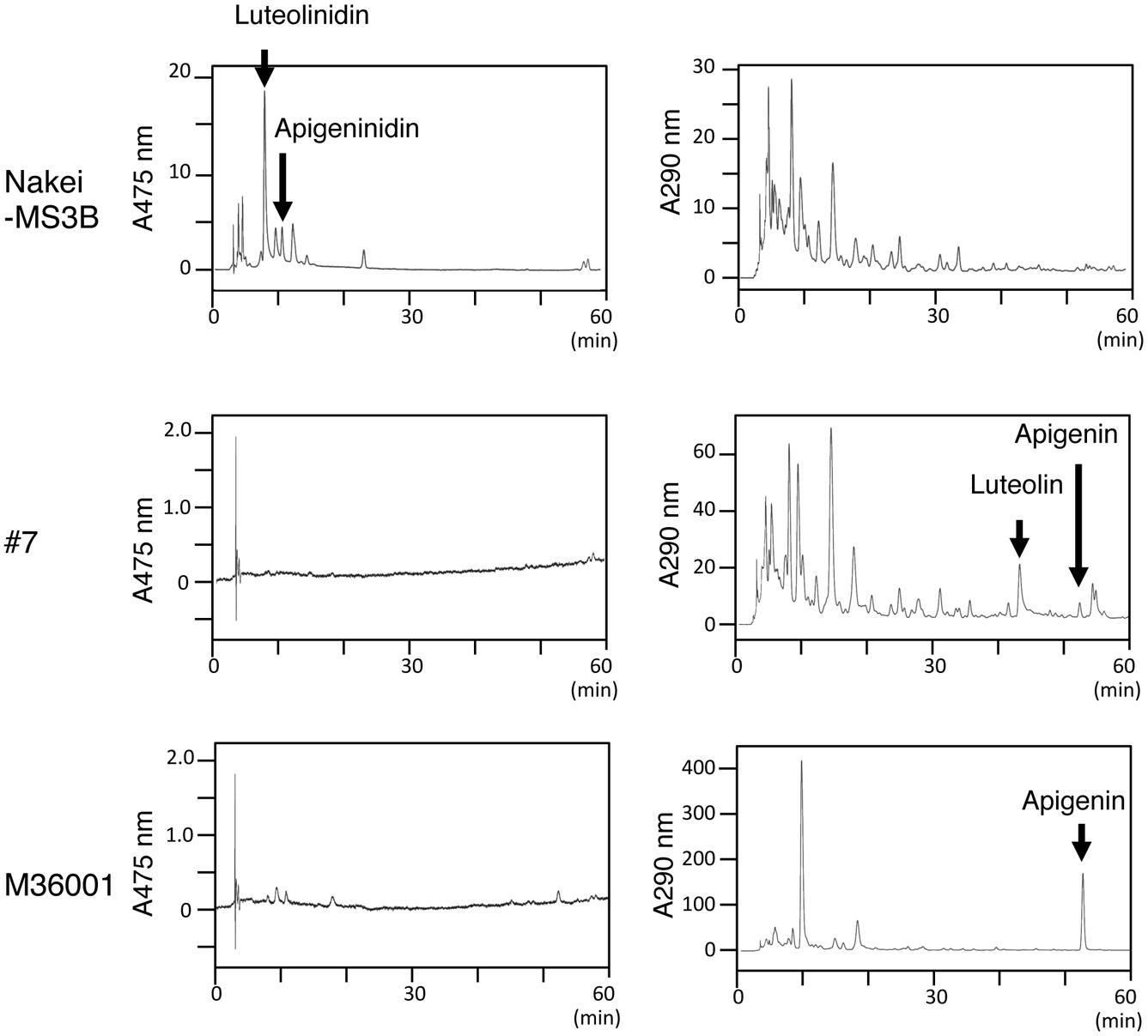
FIGURE 2. Pigments accumulated after cutting stress. Leaves were left for 4 days after cutting stress. Accumulated pigments were analyzed by HPLC at absorbances (A) of 475 or 290 nm. Nakei-MS3B accumulated 3-deoxyanthocyanidins (apigeninidin and luteolinidin), but #7 and M36001 did not. M36001 accumulated only apigenin, whereas line #7 accumulated both apigenin and luteolin.
We then performed a metabolic analysis of the color pigments in each line by using liquid chromatography – mass spectrometry/mass spectrometry (LC-MS/MS) and standard authentic compounds (Sawada et al., 2009). The proportions of each compound are shown in the heatmap (Figure 3). Nakei-MS3B accumulated 3-deoxyanthocyanidins (luteolinidin and apigeninidin) after cutting stress. M36001 and #7 accumulated apigenin, apigenin-8-C-glucoside, luteolin-3′, 7-di-O-glucoside, and luteolin-8-C-glucoside. Apigenin was accumulated at high levels in M36001, whereas luteolin-3′, 7-di-O-glucoside, and luteolin-8-C-glucoside were accumulated at high levels in #7 (Figure 3).
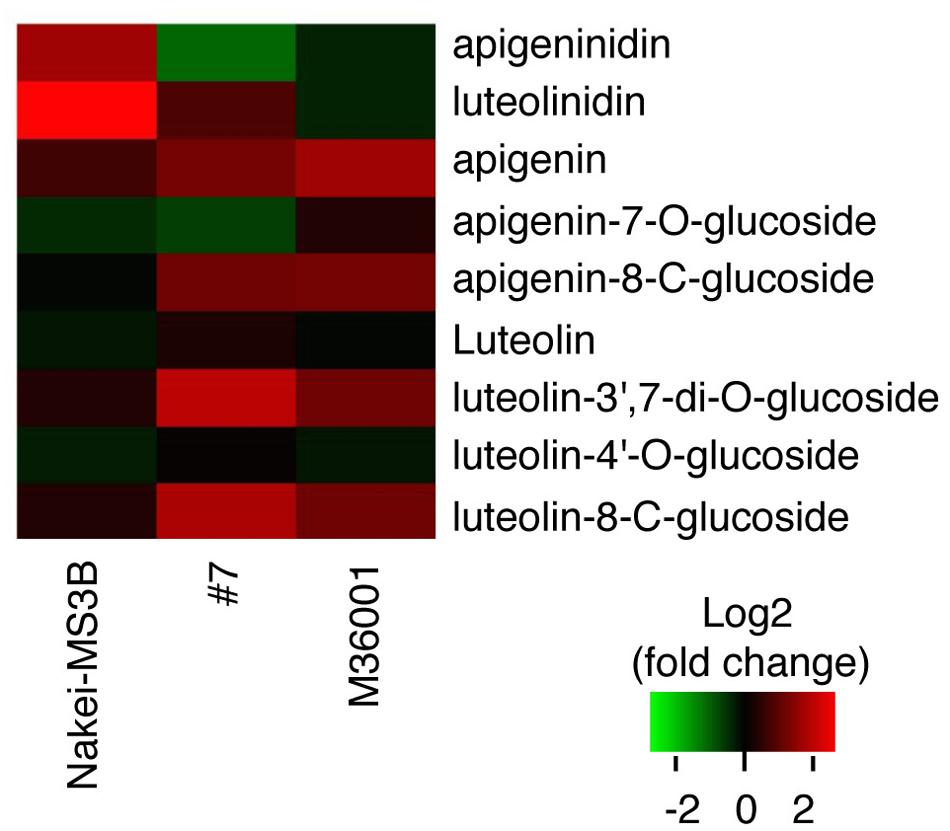
FIGURE 3. Accumulation of glycosylated flavones. Relative amount of glycosylated flavones is shown as a heatmap (red, upregulation; green, downregulation). Production of 3-deoxyanthocyanidins (luteolinidin and apigeninidin) was upregulated in Nakei-MS3B. Production of flavones and their derivatives was upregulated in M36001 and #7. Apigenin production was highly upregulated in M36001, and production of luteolin derivatives was highly upregulated in #7.
We constructed a metabolic map based on our data and on the previous reports on enzyme activity (Figure 4). We considered F3′H, FNR, and FNSII play pivotal roles in the synthesis of these pigments. Apigeninidin, apigenin, luteolinidin, and luteolin are synthesized from a common intermediate, naringenin, through sequential reactions. F3′H hydroxylates the 3′ position of the B-ring of naringenin to produce eriodictyol; both are called flavanones (Boddu et al., 2004; Shih et al., 2006). FNR converts flavanones (naringenin or eriodictyol) to flavan-4-ols (apiforol or luteoforol, respectively), and this is followed by the synthesis of 3-deoxyanthocyanidins (apigeninidin or luteolinidin, respectively; Kawahigashi et al., 2016). FNSII converts flavanones (naringenin or eriodictyol) to flavones (apigenin or luteolin, respectively; Du et al., 2010).
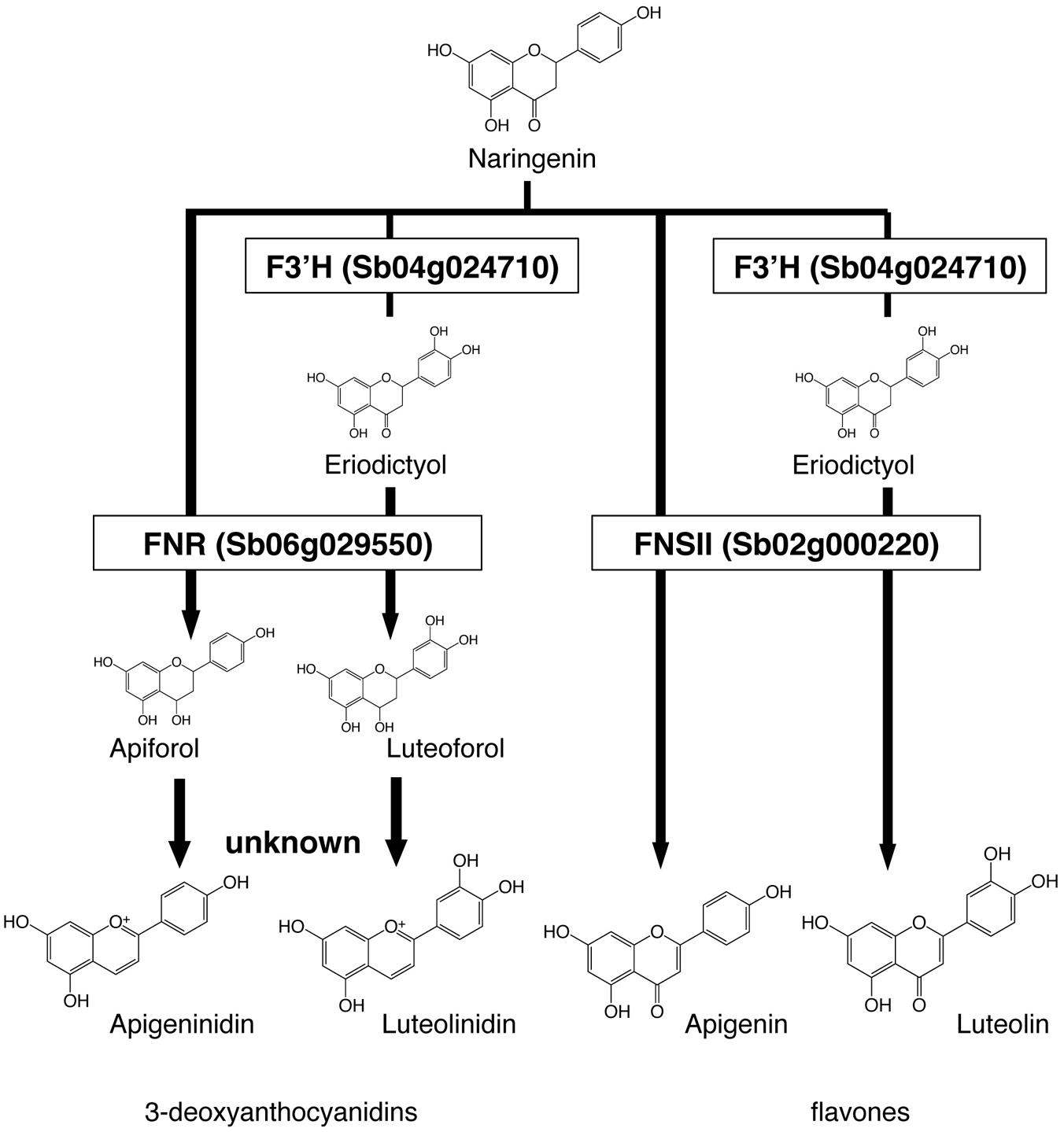
FIGURE 4. Roles of Flavanone 4-reductase (FNR), Flavone synthase II (FNSII), and Flavonoid 3′-hydroxylase (F3′H) in the metabolic pathways. Naringenin is the common intermediate of 3-deoxyanthocyanidins and flavones. FNR converts flavanones (naringenin or eriodictyol) to flavan-4-ols (apiforol, luteoforol). FNSII converts naringenin to apigenin or eriodictyol to luteolin. F3′H introduces a hydroxyl group at the 3′ position of ring B of naringenin, adding the step that leads to the production of luteolinidin or luteolin.
We assumed that the activation of each metabolic pathway (Figure 4) was controlled by the induction of gene expression. We performed RNA-seq to identify whole genes differentially expressed after cutting stress in each line (Supplementary Table S1) and made a schematic model of the relationship between the expressed genes and the pigments accumulated in each sorghum line. Here, we focused on FNR (Sb06g029550.1), FNSII (Sb02g000220.1), and F3′Hs (Sb04g024710.1, Sb04g024730.1, and Sb04g024750.1). The induction of each gene expression was confirmed by qRT-PCR.
(i) Nakei-MS3B (purple)
Nakei-MS3B accumulated apigeninidin and luteolinidin in the purple region of its leaves (Figures 1–3). RNA-seq indicated that FNR (Sb06g029550.1) was upregulated among tandemly duplicated similar genes (Sb06g029540.1 to Sb06g029630.1; Figure 5, Supplementary Figure S1), suggesting that FNR production leads to the synthesis of 3-deoxyanthocyanidin (apigeninidin from naringenin). In addition, F3′H genes (Sb04g024710.1, Sb04g024730.1, and Sb04g024750.1) were upregulated (Figures 5 and 6, Supplementary Figure S1C), suggesting the occurrence of the additional step of hydroxylation of the 3′ position of the B-ring of naringenin, followed by the synthesis of luteolinidin. Thus, expression of FNR and F3′H led to the synthesis of 3-deoxyanthocyanidins (apigeninidin and luteolinidin) in Nakei-MS3B (Figures 7 and 8). This was consistent with a previous report that Nakei-MS3B and #96, #62, #127, #3 accumulated 3-deoxyanthocyanidins and showed injury-responsive color variations (purple, red, or orange) that depended on the expression level of F3′H genes (Mizuno et al., 2014).
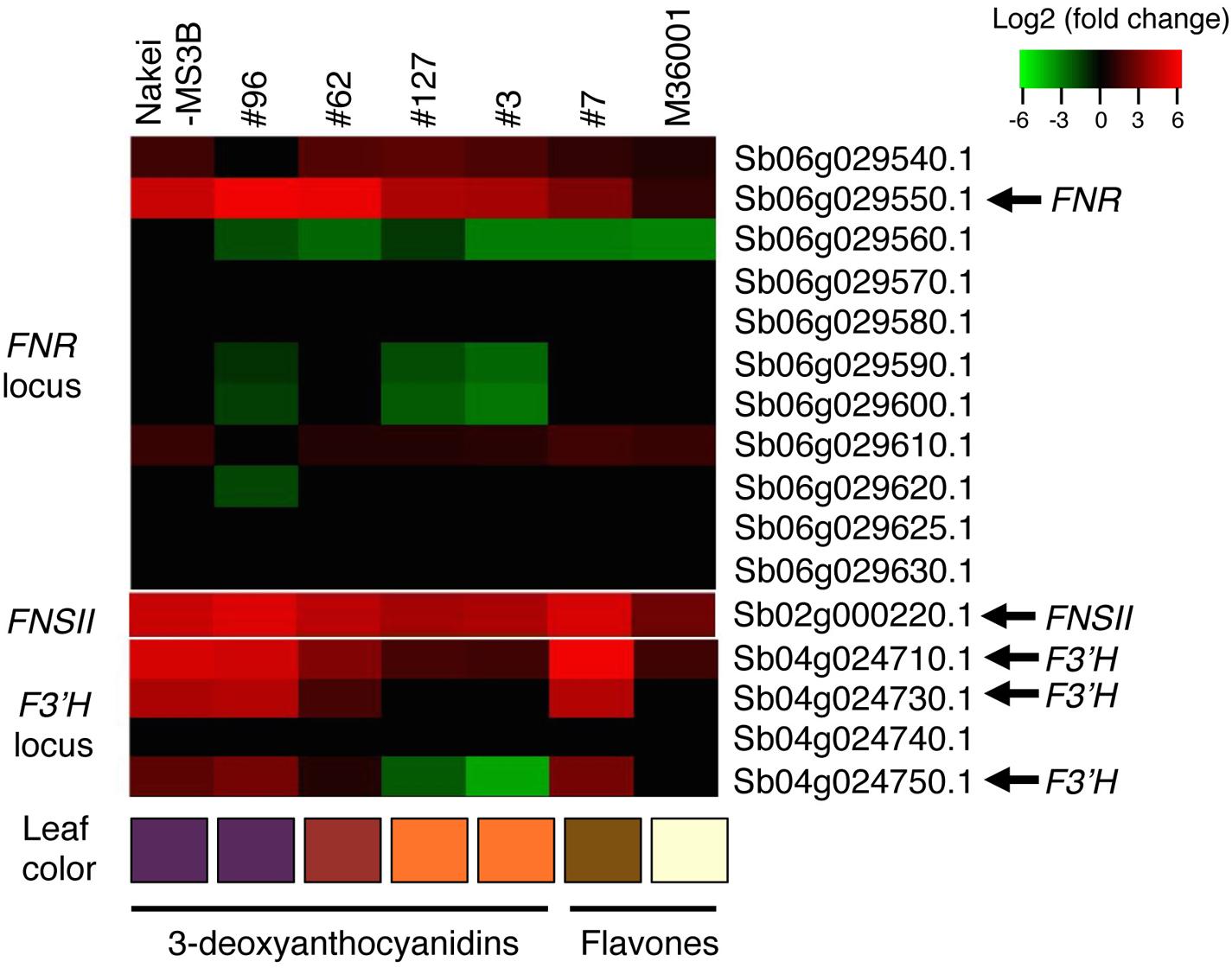
FIGURE 5. Differential expression of FNR, FNSII, and F3′H. Differentially expressed genes before and after cutting stress were identified by using RNA-seq. Log2 ratios of expression levels (4 days after cutting stress compared with immediately after cutting stress) of genes located at the FNR, FNSII, and F3′H loci are shown as a heatmap (red, upregulation; green, downregulation). Boxes at the bottom indicate colors of the cut edges of each sorghum leaf. FNR (Sb06g029550.1) expression suggests that this gene contributes to 3-deoxyanthocyanidin synthesis. Other tandemly arrayed homologous genes were similar to FNR, but their expression was not induced as strongly as that of FNR. Expression of FNSII was induced in all lines. The expression level of F3′H contributes to color variations determined by 3-deoxyanthocyanidins and flavones.
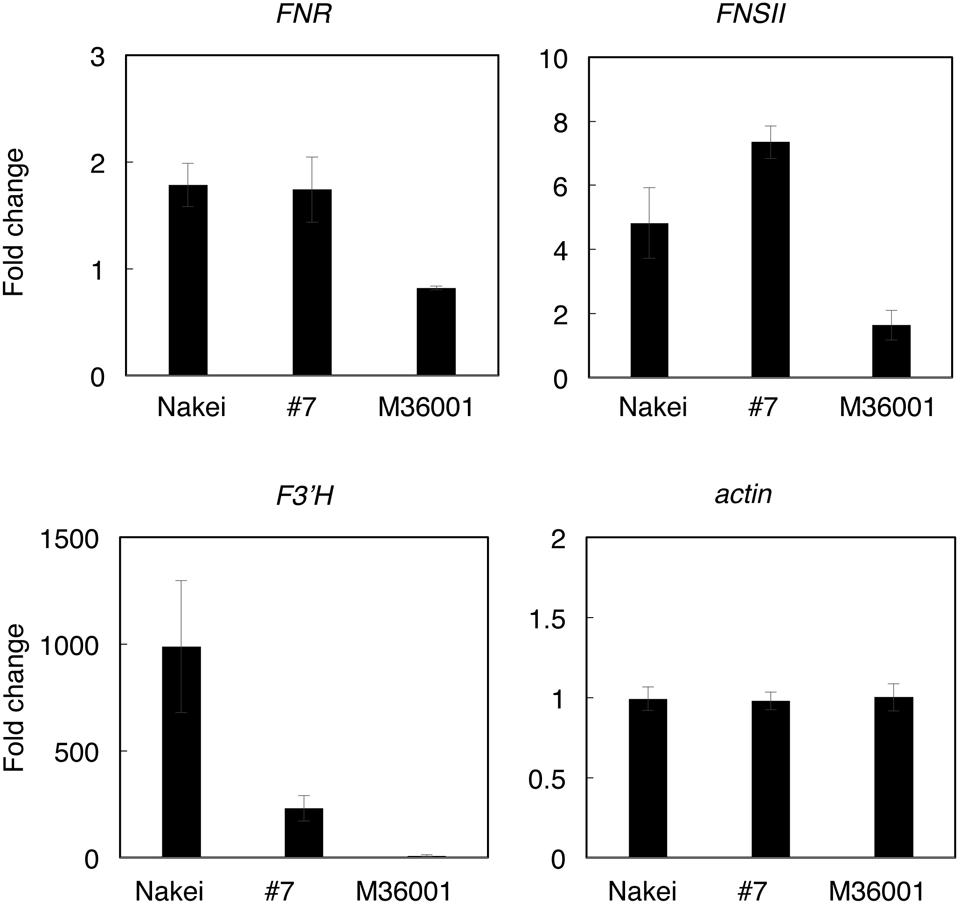
FIGURE 6. Quantitative RT-PCR (qRT-PCR) analysis of genes involved in synthesis of apigenin, luteolin, apigeninidin, and/or luteolinidine. Expression levels were quantified by qRT-PCR. Expression ratios (4 days after cutting stress compared with immediately after cutting stress) of genes for FNR (Sb06g029550), FNSII (Sb02g000220), F3′H (Sb04g024730), and actin (Sb03g040880) are shown. The expression level of each gene was normalized against that of the actin gene.
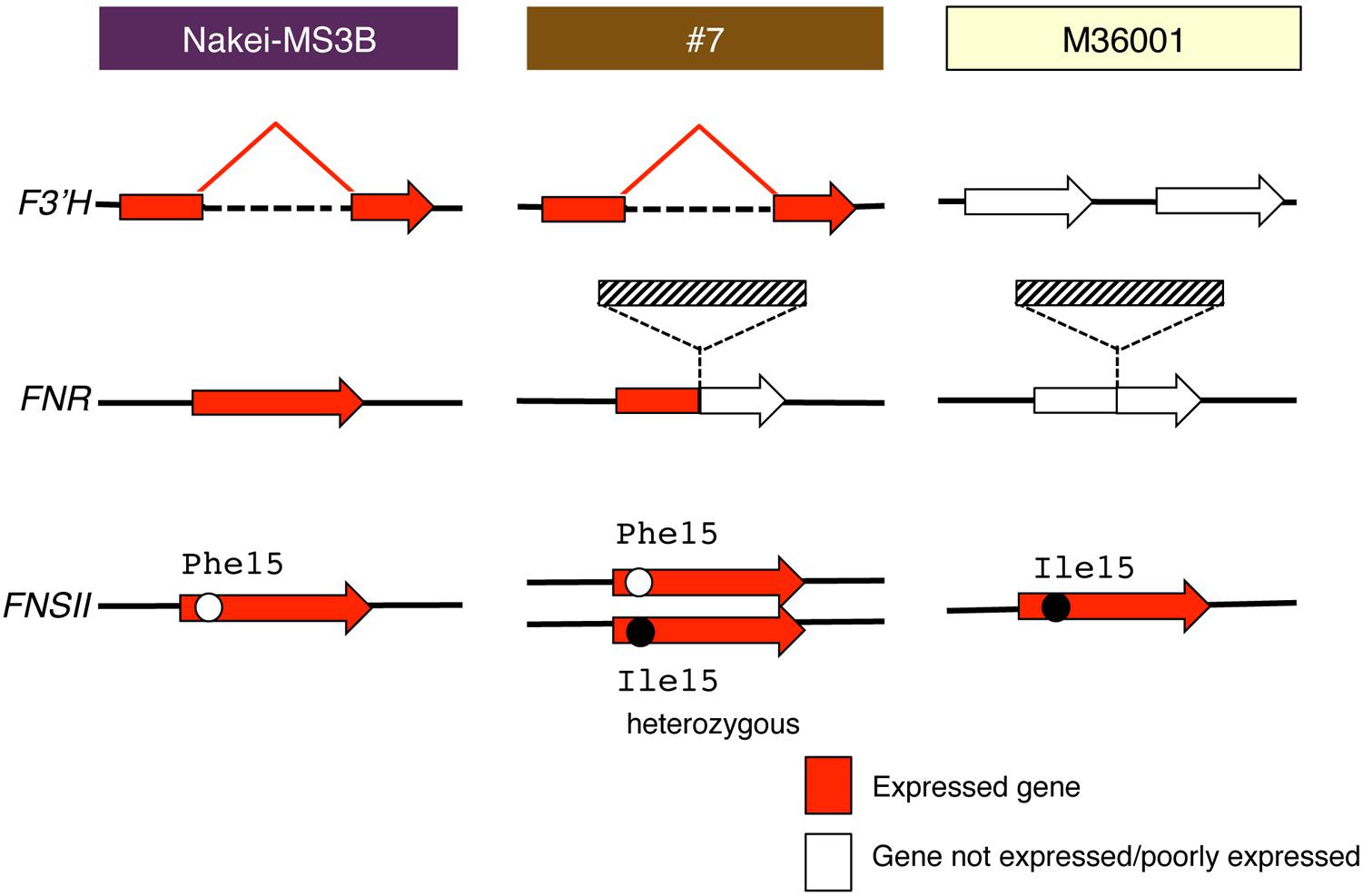
FIGURE 7. Schematic structure and expression of F3′H, FNR, and FNSII. Red arrows show gene expression; white arrows show that the gene is not expressed or is expressed very little. F3′H derived from Nakei-MS3B has a deletion and is highly expressed. FNR derived from M36001 has an insertion in the coding region; its expression is thus interrupted in #7. Hatched boxes show insertion in FNR. FNSII derived from Nakei-MS3B had Phe15, that from M36001 had Ile15, and that from #7 had heterozygous allele. The details of the gene structures and gene expression levels are shown in Supplementary Figure S1.
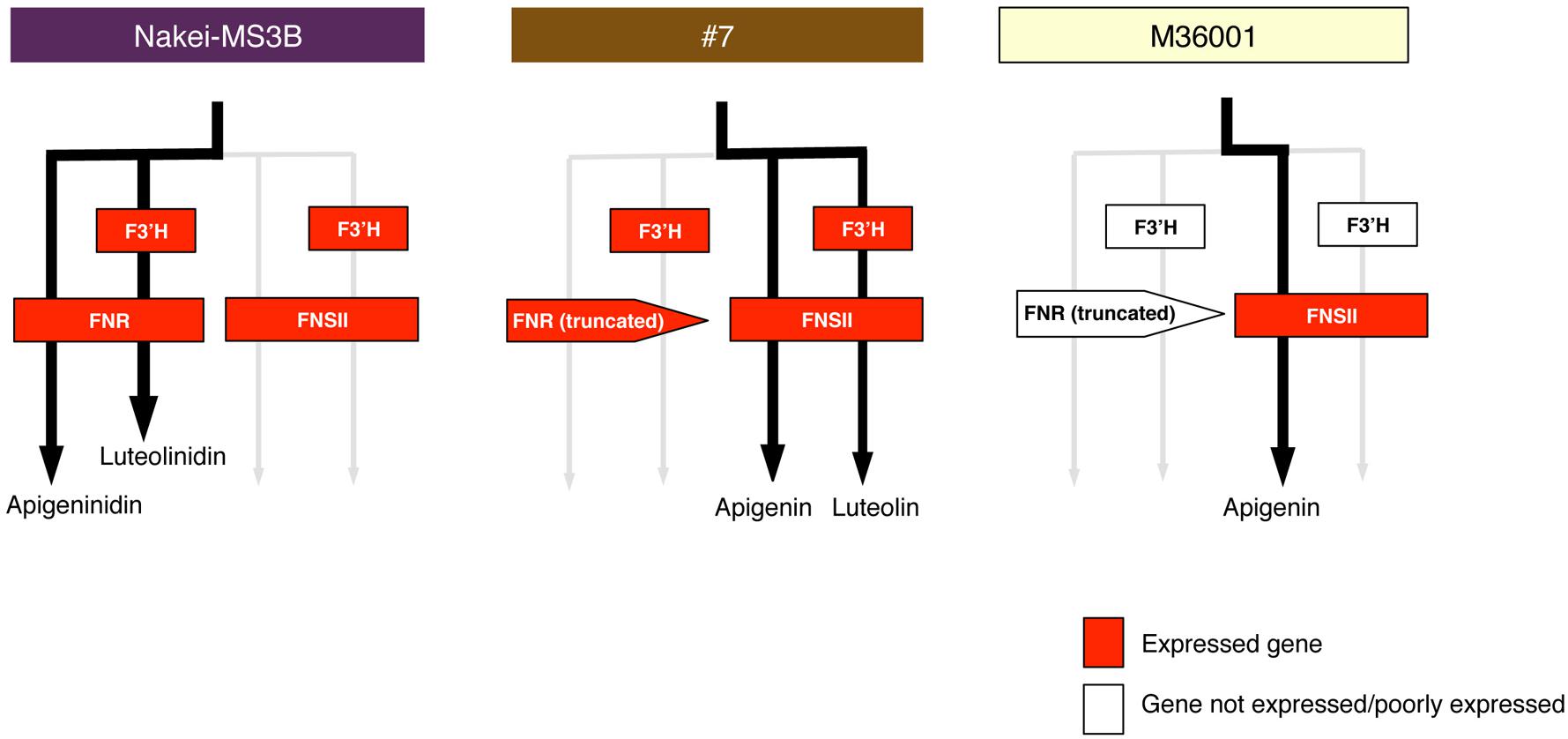
FIGURE 8. Schematic models of accumulated pigments and expressed genes. On the basis of the metabolome and transcriptome data, we show preferentially used metabolic pathways (black arrows) in Nakei-MS3B, M36001, and #7. Expressed genes are shown in red. Expression of FNR and F3′H led to synthesis of 3-deoxyanthocyanidins in Nakei-MS3B; expression of FNSII led to synthesis of apigenin (and its derivatives) in M36001; expression of FNSII and F3′H led to synthesis of apigenin and luteolin and their derivatives in #7.
(ii) M36001 (light brown)
M36001 accumulated apigenin and apigenin-8-C-glucoside in the light brown region of its leaves (Figures 1–3). FNSII was upregulated, but FNR and F3′H were not (Figures 5 and 6, Supplementary Figure S1), suggesting that expression of FNSII led to the synthesis of apigenin and its derivatives in M36001 (Figure 8).
(iii) #7 (dark brown, progeny of Nakei-MS3B and M36001)
#7 accumulated apigenin, luteolin, and their glucoside derivatives in the dark brown region of its leaves (Figures 1–3). Induction of the expression of FNSII and F3′H (Figures 5 and 6) thus likely led to the synthesis of these products in #7 (Figure 8). The FNSII allele was heterozygous: FNSII derived from Nakei-MS3B had Phe15, and that from M36001 had Ile15 (Figure 7). The F3′H gene was inherited from Nakei-MS3B, and highly expressed (Figure 7; Supplementary Figure S1C). The FNR gene in #7 inherited form M36001 had an insertion in the coding region at nucleotide 452 (Figure 7, Supplementary Figure S1A). Although the FNR gene was upregulated, this insertion divided the FNR gene and resulted in the production of truncated non-functional FNR protein (Figures 7 and 8).
Our metabolic data suggested that glycosylated flavones were also accumulated in the leaves of #7 and M36001 (Figure 3). We used the RNA-seq data to search for a putative sorghum glucosyltransferase responsible for glycosylated flavone synthesis. Sb03g032050.1 was upregulated among the putative glucosyltransferase genes (Supplementary Figure S2); the protein it encoded has 73.9% amino acid identity with the functionally validated rice OsCGT (ABC94602/LOC_Os06g18010.1; Brazier-Hicks et al., 2009). We thus consider that the protein encoded by Sb03g032050.1. is a candidate glucosyltransferase involved in the synthesis of glycosylated flavones after injury.
We aimed to elucidate the key genes for color variation in sorghum with leaves that turn tan upon injury. We identified accumulated pigments (Figures 1–4), determined the gene structure and expressed genes (Figures 5–7, Supplementary Table S1), and made a schematic model of a gene expression network for pigment synthesis (Figure 8). The expression of FNSII, F3′H, and/or FNR enables irreversible commitment to the metabolic pathway as they are located at the branch point from naringenin (Figure 4). We thus considered that the FNSII pathway was activated to synthesize flavones (apigenin and luteolin) and that the expression level of F3′H is associated with the balance of apigenin and luteolin, which in turn determined the dark or light brown variation in sorghums that respond to injury with tan pigmentation. Colors of extracted pigments from tan-colored sorghums were, however, different from those of the purified apigenin or luteolin monomers. One hypothesis is that dark brown or light brown pigments of tan-colored sorghum may be derived from polymers containing luteolin and/or apigenin, respectively. Moreover, glycosylation and other types of modifications of flavons (Figure 3) may change the colors of injured leaves of sorghum.
Here, among sorghum color variations (purple, red, orange, dark brown, and light brown), we focused on the color variation between dark brown (#7) and light brown (M36001). F3′H was highly expressed in #7; but not in M36001 and was consistent with luteolin accumulation (Figures 2, 5, 6, and Supplementary Figure S1). The high level of F3′H expression was likely due to the unique deletion in the F3′H locus inherited from Nakei-MS3B (Figure 7, Supplementary Figure S1C). We previously reported that other color variations (purple, red, and orange) are also determined by the level of F3′H expression, which results in differences in the proportions of 3-deoxyanthocyanidins (apigeninidin and luteolinidin; Mizuno et al., 2014). We therefore consider that expression of F3′H is associated with color variation in both the FNSII-dependent flavone pathway (light brown to dark brown) and the FNR-dependent 3-deoxyanthocyanidin pathway (purple to red to orange).
We speculate that at least two different transcription factors contributed to the expression of these color-related genes in sorghum: one for FNSII and the other for F3′H and FNR, because FNSII was upregulated in all lines used whereas F3′H and FNR was upregulated in Nakei-MS3B and #7, but not in M36001 (Figure 7; Supplementary Figures S1A–C). These results suggested M36001 lacks the functional transcription factor(s) that control injury-responsive expression of F3′H and FNR. As expression level of F3′H affected color variation in the FNR and FNSII pathways, the variation of these unknown transcription factors also have the potential to determine the injury-responsive color variation. As a candidate, Yellow seed1 (Y1), which encodes an R2R3-MYB-type regulatory protein (Boddu et al., 2006), had 66.9% amino acid identity with maize pericarp color1 (p1). P1 directly regulates the expression of ZmF3′H/pr1 in the 3-deoxyflavonoid biosynthesis in maize (Grotewold et al., 1994; Sharma et al., 2011, 2012). However, y1 (Sb01g037670) was not expressed with or without cutting stress in leaf (Supplementary Table S1). We therefore consider that regulation of color-related genes in sorghum differs between seed and leaf.
Naringenin is the branching point of the FNR-dependent 3-deoxyanthocyanidin pathway and the FNSII-dependent flavone pathway (Figure 4). If both FNR and FNSII were substantially upregulated in Nakei-MS3B (Figure 5), why were only the downstream products of the FNR pathways detected (Figure 2)? One hypothesis is that, irrespective of FNSII expression, the FNR, rather than the FNSII, preferentially catalyzes the metabolism of the naringenin substrate. Another hypothesis is that amino acid substitution decreases the enzyme activity of FNSII, because FNSII in Nakei-MS3B had an Ile15Phe amino acid substitution (Figure 7 and Supplementary Figure S1B). However, Ile15 was not conserved among rice, wheat, maize, and sorghum (Goodstein et al., 2012), suggesting that this substitution is not critical. In any case, we consider FNR-dependent pathway was dominant to FNSII-dependent pathway when both pathways were active. We thus consider that when the FNR-dependent pathway was not functional due to the insertion in FNR gene in #7 and M36001 (Figure 7), the FNSII-dependent pathway became dominant.
Naringenin is also the branching point of the anthocyanin pathway. The first product of the anthocyanin pathway from naringenin is synthesized by the action of flavanone 3-hydroxylase (F3H; Liu et al., 2010). However, in the tan-colored injury response we observed here, F3H (Sb06g031790.1) was not expressed (Supplementary Table S1), suggesting that the anthocyanin pathway was not activated; tan coloration was therefore independent of the production of anthocyanin metabolites. Our work expands scientific knowledge of the tan-colored injury response of sorghum cultivars—especially the gene expression behind this unique anthocyanin- and 3-deoxyanthocyanidin-independent coloration. Further comparative analysis will elucidate the diversity of the pigments accumulated and the gene expression network involved in the coloration of injured sorghums.
Sorghums accumulated flavones in their leaves (Figure 2 in this study), grains (Dykes and Rooney, 2006), and glumes and sheaths (Siame et al., 1993). One of the roles of flavones is to act as phytoalexins. Several plant flavones have activity against a variety of organisms, including other plants, nematodes, mollusks, fungi, oomycetes, and bacteria (Martens and Mithofer, 2005). Although flavones are phytoalexins with broad specificity for organisms, their disease resistance activity depends on their molecular structures. For example, luteolin in sorghum is a stronger spore germination inhibitor than apigenin in C. sublineolum (Du et al., 2010). Flavone glycosides function as antimicrobial agents in Thalassia testudinum (Harborne and Williams, 2000). Thus, regulation of the genes involved in flavone synthesis might control disease resistance in sorghum: overexpression of FNSII might enhance the amount of flavones; overexpression of F3′H might shift accumulation from apigenin to luteolin; and overexpression of glucosyltransferase genes might promote the synthesis of glycosylated derivatives. Flavones also have antioxidant, anti-inflammatory, and anti-allergic properties (Seelinger et al., 2008) and inhibitory effects on human cancer cell proliferation (Zhang et al., 2005). Thus, flavones have potential for agrobiological, physiological, and therapeutic application.
The dark or light brown color variation in injured leaves of sorghums is associated with the gene expression of FNSII and F3′H. We considered that FNSII pathway was activated to synthesize flavones (apigenin and luteolin) and that the expression level of F3′H changed the balance of apigenin and luteolin.
HKaw and SK prepared plant materials and performed cDNA synthesis; YS, MYH, and YO performed the metabolic analysis; HKan and TM performed the sequencing experiments; TY performed the data analysis; HM and HKaw designed the study; and HM wrote the manuscript. All authors read and approved the final manuscript.
This work was supported by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Genomics for Agricultural Innovation, QTL-5502 and QTL-5506).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank Mr. Muneo Sato, Mr. Yutaka Yamada, and Ms. Akane Sakata for their metabolic analysis and Mr. Jun Ogata for RNA preparation.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01718/full#supplementary-material
FIGURE S1 | Expression of Flavanone 4-reductase (FNR), Flavone synthase II (FNSII), and Flavonoid 3′-hydroxylase (F3′H), and occurrence of single nucleotide polymorphisms (SNPs). RNA-seq reads were mapped on the BTx623 reference genome and shown by using Integrative Genome Viewer. (A) FNR. FNR derived from M36001 has an insertion in the coding region; its expression is thus interrupted in #7. (B) FNSII. FNSII genes are expressed in all lines, but FNSII derived from Nakei-MS3B has two SNPs. One SNP results in amino I15F acid substitution (right arrowhead), but the other does not (left arrowhead). (C) F3′H. F3′H derived from Nakei-MS3B has a deletion and is highly expressed in Nakei-MS3B and #7.
FIGURE S2 | Differential expression of 10 putative C-glucosyl transferase genes. Gene expression is shown as a heatmap (red, upregulation). Strong expression of Sb03g032050.1 was induced after cutting stress.
Aguero, M. E., Gevens, A., and Nicholson, R. L. (2002). Interaction of Cochliobolus heterostrophus with phytoalexin inclusions in Sorghum bicolor. Physiol. Mol. Plant Pathol. 61, 267–271. doi: 10.1006/pmpp.2003.0440
Boddu, J., Jiang, C. H., Sangar, V., Olson, T., Peterson, T., and Chopra, S. (2006). Comparative structural and functional characterization of sorghum and maize duplications containing orthologous myb transcription regulators of 3-deoxyflavonoid biosynthesis. Plant Mol. Biol. 60, 185–199. doi: 10.1007/s11103-005-3568-1
Boddu, J., Svabek, C., Sekhon, R., Gevens, A., Nicholson, R. L., Jones, A. D., et al. (2004). Expression of a putative flavonoid 3′-hydroxylase in sorghum mesocotyls synthesizing 3-deoxyanthocyanidin phytoalexins. Physiol. Mol. Plant Pathol. 65, 101–113. doi: 10.1016/j.pmpp.2004.11.007
Brazier-Hicks, M., Evans, K. M., Gershater, M. C., Puschmann, H., Steel, P. G., and Edwards, R. (2009). The C-Glycosylation of flavonoids in Cereals. J. Biol. Chem. 284, 17926–17934. doi: 10.1074/jbc.M109.009258
Clifford, M. N. (2000). Anthocyanins - nature, occurrence and dietary burden. J. Sci. Food Agric. 80, 1063–1072. doi: 10.1002/(SICI)1097-0010(20000515)80:7<1126::AID-JSFA604>3.0.CO;2-0
Du, Y., Chu, H., Wang, M., Chu, I. K., and Lo, C. (2010). Identification of flavone phytoalexins and a pathogen-inducible flavone synthase II gene (SbFNSII) in sorghum. J. Exp. Bot. 61, 983–994. doi: 10.1093/jxb/erp364
Dykes, L., Peterson, G. C., Rooney, W. L., and Rooney, L. W. (2011). Flavonoid composition of lemon-yellow sorghum genotypes. Food Chem. 128, 173–179. doi: 10.1016/j.foodchem.2011.03.020
Dykes, L., and Rooney, L. W. (2006). Sorghum and millet phenols and antioxidants. J. Cereal Sci. 44, 236–251. doi: 10.1016/j.jcs.2006.06.007
Dykes, L., Rooney, L. W., Waniska, R. D., and Rooney, W. L. (2005). Phenolic compounds and antioxidant activity of sorghum grains of varying genotypes. J. Agric. Food Chem. 53, 6813–6818. doi: 10.1021/jf050419e
Dykes, L., Seitz, L. M., Rooney, W. L., and Rooney, L. W. (2009). Flavonoid composition of red sorghum genotypes. Food Chem. 116, 313–317. doi: 10.1016/j.foodchem.2009.02.052
Goodstein, D. M., Shu, S. Q., Howson, R., Neupane, R., Hayes, R. D., Fazo, J., et al. (2012). Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40, D1178–D1186. doi: 10.1093/nar/gkr944
Grotewold, E., Drummond, B. J., Bowen, B., and Peterson, T. (1994). The Myb-homologous P-Gene controls phlobaphene pigmentation in maize floral organs by directly activating a flavonoid biosynthetic gene subset. Cell 76, 543–553. doi: 10.1016/0092-8674(94)90117-1
Harborne, J. B., and Williams, C. A. (2000). Advances in flavonoid research since 1992. Phytochemistry 55, 481–504. doi: 10.1016/S0031-9422(00)00235-1
Hipskind, J. D., Hanau, R., Leite, B., and Nicholson, R. L. (1990). Phytoalexin accumulation in sorghum - identification of an apigeninidin acyl ester. Physiol. Mol. Plant Pathol. 36, 381–396. doi: 10.1016/0885-5765(90)90067-8
Kawahigashi, H., Kasuga, S., Ando, T., Kanamori, H., Wu, J., Yonemaru, J., et al. (2011). Positional cloning of ds1, the target leaf spot resistance gene against Bipolaris sorghicola in sorghum. Theor. Appl. Genet. 123, 131–142. doi: 10.1007/s00122-011-1572-1
Kawahigashi, H., Kasuga, S., Sawada, Y., Yonemaru, J. I., Ando, T., Kanamori, H., et al. (2016). The Sorghum gene for leaf color changes upon wounding (P) encodes a flavanone 4-reductase in the 3-deoxyanthocyanidin biosynthesis pathway. G3 (Bethesda) 6, 1439–1447. doi: 10.1534/g3.115.026104
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14:R36.
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Liu, H. J., Du, Y. G., Chu, H., Shih, C. H., Wong, Y. W., Wang, M. F., et al. (2010). Molecular dissection of the pathogen-inducible 3-deoxyanthocyanidin biosynthesis pathway in Sorghum. Plant Cell Physiol. 51, 1173–1185. doi: 10.1093/pcp/pcq080
Lo, S. C. C., De Verdier, K., and Nicholson, R. L. (1999). Accumulation of 3-deoxyanthocyanidin phytoalexins and resistance to Colletotrichum sublineolum in sorghum. Physiol. Mol. Plant Pathol. 55, 263–273. doi: 10.1006/pmpp.1999.0231
Martens, S., and Mithofer, A. (2005). Flavones and flavone synthases. Phytochemistry 66, 2399–2407. doi: 10.1016/j.phytochem.2005.07.013
Mizuno, H., Kawahara, Y., Sakai, H., Kanamori, H., Wakimoto, H., Yamagata, H., et al. (2010). Massive parallel sequencing of mRNA in identification of unannotated salinity stress-inducible transcripts in rice (Oryza sativa L.). BMC Genomics 11:683. doi: 10.1186/1471-2164-11-683
Mizuno, H., Kawahigashi, H., Kawahara, Y., Kanamori, H., Ogata, J., Minami, H., et al. (2012). Global transcriptome analysis reveals distinct expression among duplicated genes during sorghum-interaction. BMC Plant Biol. 12:121. doi: 10.1186/1471-2229-12-121
Mizuno, H., Yazawa, T., Kasuga, S., Sawada, Y., Ogata, J., Ando, T., et al. (2014). Expression level of a flavonoid 3’-hydroxylase gene determines pathogen-induced color variation in sorghum. BMC Res. Notes 7:761. doi: 10.1186/1756-0500-7-761
Ogo, Y., Ozawa, K., Ishimaru, T., Murayama, T., and Takaiwa, F. (2013). Transgenic rice seed synthesizing diverse flavonoids at high levels: a new platform for flavonoid production with associated health benefits. Plant Biotechnol. J. 11, 734–746. doi: 10.1111/pbi.12064
Ouyang, S., and Buell, C. R. (2004). The TIGR plant repeat databases: a collective resource for the identification of repetitive sequences in plants. Nucleic Acids Res. 32, D360–D363. doi: 10.1093/nar/gkh099
Paterson, A. H., Bowers, J. E., Bruggmann, R., Dubchak, I., Grimwood, J., Gundlach, H., et al. (2009). The Sorghum bicolor genome and the diversification of grasses. Nature 457, 551–556. doi: 10.1038/nature07723
Sawada, Y., Akiyama, K., Sakata, A., Kuwahara, A., Otsuki, H., Sakurai, T., et al. (2009). Widely targeted metabolomics based on large-scale MS/MS data for elucidating metabolite accumulation patterns in plants. Plant Cell Physiol. 50, 37–47. doi: 10.1093/pcp/pcn183
Seelinger, G., Merfort, I., and Schempp, C. M. (2008). Anti-oxidant, anti-inflammatory and anti-allergic activities of luteolin. Planta Med. 74, 1667–1677.
Sharma, M., Chai, C., Morohashi, K., Grotewold, E., Snook, M. E., and Chopra, S. (2012). Expression of flavonoid 3′-hydroxylase is controlled by P1, the regulator of 3-deoxyflavonoid biosynthesis in maize. BMC Plant Biol. 12:196. doi: 10.1186/1471-2229-12-196
Sharma, M., Cortes-Cruz, M., Ahern, K. R., McMullen, M., Brutnell, T. P., and Chopra, S. (2011). Identification of the Pr1 gene product completes the anthocyanin biosynthesis pathway of maize. Genetics 188, 69–79. doi: 10.1534/genetics.110.126136
Shih, C. H., Chu, I. K., Yip, W. K., and Lo, C. (2006). Differential expression of two flavonoid 3’-hydroxylase cDNAs involved in biosynthesis of anthocyanin pigments and 3-deoxyanthocyanidin phytoalexins in sorghum. Plant Cell Physiol. 47, 1412–1419. doi: 10.1093/pcp/pcl003
Siame, B. A., Ejeta, G., and Butler, L. G. (1993). Role of pigments and tannins in the reaction of tan and red near-isogenic sorghum check for this species in other resources lines to leaf diseases. Afr. Crop Sci. J. 1, 123–130.
Snyder, B. A., and Nicholson, R. L. (1990). Synthesis of phytoalexins in sorghum as a site-specific response to fungal ingress. Science 248, 1637–1639. doi: 10.1126/science.248.4963.1637
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–U174. doi: 10.1038/nbt.1621
Zhang, Y. J., Vareed, S. K., and Nair, M. G. (2005). Human tumor cell growth inhibition by nontoxic anthocyanidins, the pigments in fruits and vegetables. Life Sci. 76, 1465–1472. doi: 10.1016/j.lfs.2004.08.025
Keywords: apigenin, apigeninidin, FNR, FNSII, F3′H, luteolin, luteolinidin, RNA-seq
Citation: Mizuno H, Yazawa T, Kasuga S, Sawada Y, Kanamori H, Ogo Y, Hirai MY, Matsumoto T and Kawahigashi H (2016) Expression of Flavone Synthase II and Flavonoid 3′-Hydroxylase Is Associated with Color Variation in Tan-Colored Injured Leaves of Sorghum. Front. Plant Sci. 7:1718. doi: 10.3389/fpls.2016.01718
Received: 10 August 2016; Accepted: 01 November 2016;
Published: 21 November 2016.
Edited by:
Swarup Kumar Parida, National Institute of Plant Genome Research, IndiaReviewed by:
Yongzhong Xing, Huazhong Agricultural University, ChinaCopyright © 2016 Mizuno, Yazawa, Kasuga, Sawada, Kanamori, Ogo, Hirai, Matsumoto and Kawahigashi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hiroshi Mizuno, aG1penVub0BhZmZyYy5nby5qcA==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.