- 1Key Laboratory of Ministry of Education for Medicinal Plant Resource and Natural Pharmaceutical Chemistry, National Engineering Laboratory for Resource Developing of Endangered Chinese Crude Drugs in Northwest of China, College of Life Sciences, Shaanxi Normal University, Xi’an, China
- 2Laboratory of Phytopathology, Wageningen University, Wageningen, Netherlands
The CLAVATA2 (CLV2) gene encodes a leucine-rich repeat receptor-like protein, a class of cell surface receptors that lacks a cytoplasmic kinase domain. As such, CLV2 is capable of functioning in concert with additional receptor(s), possibly receptor-like kinase(s), to activate cellular responses upon ligand perception. Accumulating data indicate that CLV2 is implicated in distinct biological processes including plant growth and development as well as innate immunity to microbe and nematode infections. This article focuses on recent advances in our understanding of multiple signaling pathways mediated by multifunctional CLV2 that modulate various physiological processes. The challenges and future perspectives of elucidating the specificity of CLV2-mediated signaling pathways and identifying potential co-receptors and putative ligands for CLV2 are also discussed.
The Arabidopsis CLAVATA2 (CLV2) gene encodes a leucine-rich repeat (LRR) receptor-like protein (RLP) that lacks a cytoplasmic kinase domain, unlike receptor-like kinase (RLK) such as CLV1 which contains a cytoplasmic kinase domain (Clark et al., 1997; Jeong et al., 1999). It has been shown that, similar to CLV1 and CLV3, CLV2 is implicated primarily in the proper coordination between proliferation and differentiation of stem cells in the shoot apical meristem (SAM) (Clark et al., 1993, 1995, 1997; Kayes and Clark, 1998; Wang et al., 2008). However, unlike CLV1 and CLV3, CLV2 is expressed broadly not only in the SAM but also in many other tissues and is induced by multiple external stimuli, which suggests a wider role for CLV2 beyond SAM maintenance (Kayes and Clark, 1998; Wang et al., 2008, 2010b; Wu J. et al., 2016). Indeed, recent studies have indicated that CLV2 is involved in distinct physiological programs, including plant development and innate immunity to microbe and nematode infections (Wang et al., 2010a; Replogle et al., 2011; Hanemian et al., 2016). Identification of multiple functions of CLV2 raises the question of how the specificities of CLV2-mediated signaling are achieved. Other interesting aspects include the potential crosstalk of CLV2-mediated signaling pathways and how the formation of CLV2-associated receptor complex(es) is controlled. This article presents recent advances in our understanding of multiple functions of CLV2 in various biological processes.
CLV2 is Implicated in the Regulation of SAM Maintenance
Mutations in the CLV2 gene, similar to CLV1 and CLV3, resulted in enlarged meristems and abnormal organs, while the clv2 mutant exhibited weaker phenotypes than those of the clv1 and clv3 mutants (Clark et al., 1993, 1995, 1997; Kayes and Clark, 1998; Diévart et al., 2003; Wang et al., 2008). CLV3, one of the best studied CLAVATA3/EMBRYO SURROUNDING REGION-related (CLE) peptides (Wang et al., 2016), functions as an intercellular signaling molecule to repress expression of the stem cell-promoting gene WUSCHEL (WUS), thereby restricting the stem cell population (Fletcher et al., 1999; Brand et al., 2000; Schoof et al., 2000; Rojo et al., 2002; Lenhard and Laux, 2003). The CLV3 peptide is thought to be recognized in parallel by multiple receptor complexes (Figure 1), including CLV1 homomultimers, RECEPTOR-LIKE PROTEIN KINASE2/TOADSTOOL2 (RPK2/TOAD2) homodimers, heteromultimers of CLV2 and CORYNE/SUPPRESSOR OF OVEREXPRESSION OF LLP1-2 (CRN/SOL2), and heteromultimers of CLV1 with its close homologs BARELY ANY MERISTEM1 (BAM1) and BAM2 (Casamitjana-Martinez et al., 2003; Miwa et al., 2008; Müller et al., 2008; Bleckmann et al., 2010; Kinoshita et al., 2010; Zhu et al., 2010; Shinohara and Matsubayashi, 2015). Possibly, the CLV3 signal is also perceived by heteromultimers of CLV1 associated with CLV2-CRN/SOL2 (Figure 1), based on the result that CLV1 was found to weakly interact with the CLV2-CRN/SOL2 heterodimer (Bleckmann et al., 2010; Zhu et al., 2010). By applying the multiparameter fluorescence imaging spectroscopy (MFIS) method, it is further revealed that CLV homodimers, CLV2-CRN heteromultimers and CLV2-CRN-CLV1 multimers are organized in preformed complexes in the absence of CLV3. The addition of CLV3 stimulates the accumulation of CLV2-CRN-CLV1 multimers within specific domains along the plasma membrane (Somssich et al., 2015).

FIGURE 1. Schematic representation of different CLV2-associated receptor complexes that mediate CLE signaling in multiple biological processes. Particularly, the genetic and biochemical evidence are unknown for the physical interaction between CLV2 and CRN, both of which are potentially involved in R. solanacearum pathogenicity, and CLV2 associates with SOBIR1 that controls a yet uncharacterized physiological process (dash-line boxed). RLKa and RLKb represent unknown receptors that function together with CLV2 and CRN/SOL2 to form multimers and perceive CLE signals.
Specifically, CRN encodes a membrane-associated receptor-like cytoplasmic kinase (RLCK) that lacks a distinct extracellular domain (Miwa et al., 2008; Müller et al., 2008). It has demonstrated that CRN and CLV2 interact at the endoplasmic reticulum and relocalize to the plasma membrane (Bleckmann et al., 2010). Despite CRN kinase being catalytically inactive, the kinase domain of CRN is essential for CLV3 signaling perception in the SAM (Nimchuk et al., 2011; Somssich et al., 2016). Nevertheless, the CLV2-SOL2/CRN heterodimer likely functions together with an unknown RLK, independently of CLV1, to mediate CLV3 signal transduction in the SAM homeostasis (Figure 1).
Direct binding of CLV3 peptide with the extracellular LRR domains of CLV1 and BAM1 receptors has been reported (Ogawa et al., 2008; Shinohara and Matsubayashi, 2015). However, CLV2 did not exhibit direct binding to CLV3 peptide, albeit its ability to bind a number of CLE peptides (Guo et al., 2010; Shinohara and Matsubayashi, 2015). On one hand, these results suggest the need for identification of CLV2 ligands, and on the other the results further underscore the hypothesis that an unknown RLK controls SAM homeostasis together with the CLV2-CRN/SOL2 heterodimer (Figure 1).
CLV2 is Implicated in the Regulation of Root Apical Meristem (RAM) maintenance
A growing body of evidence suggests an important role for CLV2 in root meristem maintenance. Overexpression of many CLE genes, including CLV3, CLE14, CLE19, CLE20, and CLE40, results in an inhibition of root growth in a CLV2-dependent but CLV1-independent manner (Fiers et al., 2004, 2005; Strabala et al., 2006; Meng and Feldman, 2010). Consistently, the clv2 mutant is insensitive to chemically synthesized CLE peptides, indicating that CLV2 is required for CLE-induced consumption of the root meristem (Fiers et al., 2005; Ito et al., 2006; Kondo et al., 2006; Kinoshita et al., 2007). Similarly, the roots of crn/sol2 mutants are unaffected upon CLE peptide treatment, suggesting that CRN is also involved in transmitting CLE signals to regulate RAM homeostasis (Miwa et al., 2008; Müller et al., 2008). However, no visible phenotypic abnormalities were observed in roots of the clv2 mutant, the crn/sol2 mutant and the clv2 crn/sol2 double mutant under normal growth conditions (Wang et al., 2008; Miwa et al., 2008; Müller et al., 2008). Possibly, a closely related AtRLP gene could compensate for the loss of CLV2 function in the root (Wang et al., 2010b). It is also likely that the phenotype is very subtle and only become apparent at the microscopic level.
As has been shown in the SAM, CLV2 is also capable of forming a receptor complex with CRN/SOL2 to regulate RAM homoeostasis (Figure 1; Miwa et al., 2008; Müller et al., 2008; Bleckmann et al., 2010; Zhu et al., 2010). In addition to the previous observation that CRN lacked kinase activity (Nimchuk et al., 2011), it was reported recently that the CRN/SOL2 kinase domain was not essential for protein function in the root (Somssich et al., 2016). This finding indicates that an unknown receptor, probably a RLK, functions together with the CLV2-CRN heterodimer (Figure 1). As such, the root expressed RLKs, including BAMs and RPK2, might be the logical RLK candidates for the redundant role in root development, because of their pronounced expression in roots (DeYoung et al., 2006; Kinoshita et al., 2010). However, it has been shown that BAM1, independently of CLV2, functioned synergistically with RPK2 in CLE peptide-triggered root meristem arrest (Shimizu et al., 2015). An investigation of homozygous T-DNA insertion lines of root expressed RLKs for their sensitivity to CLV3 peptide treatment revealed none of the tested RLKs were involved in CLV3 perception (ten Hove et al., 2011). Thus it will be worthwhile in future to examine the homozygous lines with a wide range of CLE peptides (Qiang et al., 2013). A number of CLE genes are expressed in the RAM (Jun et al., 2010); however, it is still unclear which CLE peptide is responsible for RAM activity due to the lack of loss-of-function mutants. Nevertheless, it is suggested that CLV2-CRN/SOL2 may perceive the CLE14 and CLE20 ligands to trigger RAM termination, although biochemical evidence for this receptor-ligand interaction is missing (Meng and Feldman, 2010). Altogether, these studies argue that CLV2-CRN/SOL2 heteromers may act together with an unidentified RLK to modulate root meristem maintenance.
CLV2 is Implicated in the Regulation of Protoxylem Formation
It has been reported that ectopic protoxylem vessels were formed in clv2 roots (Kondo et al., 2011), suggesting that endogenous CLV2 functions in the inhibition of protoxylem vessel formation. Many chemically synthesized CLE peptides, including CLE10, suppresses protoxylem vessel formation in the root (Kondo et al., 2011). Consistently, transgenic plants overexpressing CLE10 under an estradiol-inducible promoter exhibited a protoxylem vessel defect similar to that of seedlings treated with CLE10 peptides (Kondo et al., 2011). By contrast, further investigation found that the protoxylem vessel defect is absent in CLE peptides treated clv2 mutants and crn/sol2 mutants (Kondo et al., 2011), indicating that CLV2 and CRN/SOL2 may function as receptors, presumably constituting a receptor complex as has been shown in the SAM and RAM, to perceive CLE10 signaling and modulate protoxylem vessel formation (Figure 1). Indeed, both CLV2 and CRN/SOL2 are strongly expressed in the root stele with greatly overlapping expression domains (Müller et al., 2008; Somssich et al., 2016). However, it remains unclear whether the kinase domain of CRN/SOL2 is involved in the transmission of CLE10 signaling to suppress protoxylem vessel formation. It is most likely that the kinase domain of CRN/SOL2 is dispensable for CRN/SOL2 function in the inhibition of protoxylem vessel formation similar to that in the RAM (Somssich et al., 2016). If this hypothesis holds true, it raises again the possibility that the interaction of CLV2-CRN/SOL2 with an unidentified RLK to form a functional receptor complex (Figure 1).
The CLV2 Mutant Affects Organ Development
Notably, the inactivation of CLV2 results in pleiotropic effects on Arabidopsis development in addition to enlarged meristem and increased organ number (Kayes and Clark, 1998; Wang et al., 2008, 2011), suggesting a multifactorial role for CLV2 that is verified by its broad expression pattern (Wang et al., 2008; Wang, 2009; Wu J. et al., 2016). Indeed, mutations in the CLV2 gene result in phenotypic alterations in the gynoecia, flower pedicels and stamens (Kayes and Clark, 1998). Particularly, it was found that the clv1, clv2, and crn mutants produced only extra fruit organs and generated floral meristems of similar dimensions to wild-type plants (Durbak and Tax, 2011). Characterization of gynoecium development in these mutants revealed increased cell proliferation and ectopic fruit organ initiation, which was marked by an expanded expression of cell proliferation-promoting gene SHOOTMERISTEMLESS (STM) (Durbak and Tax, 2011). In contrast to SAM development, in which CLV1 and CLV2-CRN2/SOL2 act in parallel, CLV1, CLV2 and CRN/SOL2 function together in a linear pathway during fruit development based on genetic analyses (Durbak and Tax, 2011).
The clv2 mutant developed reduced plant size with smaller and narrowed rosette leaves, which is consistent with the observation that CLV2 exhibited a constitutive expression in rosette leaves (Wang, 2009). Interestingly, overexpression of several CLE genes caused phenotypic abnormalities in leaf shape and size (Strabala et al., 2006). Furthermore, BAM1, BAM2, and BAM3 are also implicated in the leaf formation, as the loss of three BAM genes caused reduced leaf size (DeYoung et al., 2006). Moreover, PLL1, PLL4, and PLL5, three downstream genes involving in the CLV signaling pathway of the SAM, have also been shown to be functioned in leaf development (Song and Clark, 2005). Taken together, a CLV-like signaling pathway that includes CLE, CLV2, BAMs, and PLLs probably exists in the regulation of leaf development.
CLV2 Functions in Plant-Microbe Interactions
Beyond its roles in plant development, CLV2 is also implicated in plant-pathogen interactions. Overexpression of CLE-like genes from nematodes resulted in meristem termination and short roots, which mimic the overexpression phenotypes of plant CLE peptides (Replogle et al., 2011). Further studies have shown that CLV2, in conjunction with CLV1, RPK2, and CRN/SOL2, is required for perception of nematode CLE peptides allowing nematodes to successfully infect Arabidopsis roots (Replogle et al., 2011). Likewise, it has been shown that CLV1, CLV2, and CRN/SOL2 are required for full susceptibility to virulent strains of Ralstonia solanacearum (Hanemian et al., 2016). Interestingly, mutations in CLV1 and CLV2 provide resistance not only to R. solanacearum strains but also to the oomycete pathogen Hyaloperonospora arabidopsidis, suggesting that CLV1 and CLV2 may generally be involved in various plant-pathogen interactions (Hanemian et al., 2016). However, other CLV-signaling components involved in SAM stem cell homeostasis, e.g., CLV3, BAMs, WUS, POL, PLL1, and KAPP, are dispensable for plant susceptibility to R. solanacearum, indicating that the signaling pathways employed in R. solanacearum pathogenicity differ from those used in stem cell homoeostasis (Hanemian et al., 2016). As such, it is possible that CLV1, CLV2, and CRN/SOL2 control developmental programs that also provide plants to adapt to external stimuli through resource reallocation to balance the growth-immunity tradeoff. Notably, the clv2 mutant exhibited reduced growth (Wang et al., 2008), which makes biological sense because defense activation generally comes at the expense of plant growth.
The increased disease resistance mediated by clv1 and clv2 is independent of salicylic acid and ethylene (Hanemian et al., 2016). Gene expression analysis found that the nuclear transcription factor Y subunit alpha (NF-YA) genes were down-regulated in clv1 and clv2 mutants (Hanemian et al., 2016). Consistently, the accumulation of miR169, which is involved in the post-transcriptional regulation of NF-YA transcription factors, is drastically impaired in clv1 and clv2 mutants (Hanemian et al., 2016). Similar to the results reported in nematodes, it is speculated that R. solanacearum-derived peptides, either CLE-like peptides or unknown peptides, are recognized by CLV1 and CLV2, thereby promoting pathogenicity through the manipulation of intrinsic developmental CLV signaling. Alternatively, it is likely that an unidentified Arabidopsis CLE peptide, constituting a signaling pathway possibly with CLV1 and CLV2, may require for R. solanacearum pathogenicity.
Conclusions and Future Perspectives
Accumulating data indicate that CLV2 is capable of regulating various developmental and immunity signaling pathways (Figure 2). It is well-known that CLV2 needs to interact with additional component(s), possibly with RLK(s), and RLCK(s), to activate cellular responses upon ligand perception because of lacking the intracellular signaling domains (Figure 2). The association of CLV2 with different regulatory RLKs and RLCKs might result in activation of distinct biological responses, implying the diversity of CLV2-associated receptor complexes partially determine the specificity of CLV2-mediated signaling. It is also conceivable that ligand-dependent differential phosphorylations of regulatory RLKs and RLCKs could initiate a characteristic response (Figure 2). Nevertheless, potential interacting partners for CLV2 are unknown in most cases (Figure 1). Therefore, two available resources, the collection of homozygous T-DNA insertion lines for root expressed RLKs and the systematic expression atlas of GUS reporter lines for LRR-RLKs, are valuable for functional investigation of possible RLK candidates (ten Hove et al., 2011; Wu Y. et al., 2016). Furthermore, the organization and dynamics of different CLV2-associated receptor complexes could be further investigated using MFIS (Somssich et al., 2015). Notably, it has been shown that tomato SUPPRESSOR OF BIR1-1 (SlSOBIR1) associates with a broad range of tomato RLPs that are involved in either plant development or immunity (Liebrand et al., 2013). Specifically, SlSOBIR1 was found to interact with SlCLV2, the tomato homolog of Arabidopsis CLV2 (Liebrand et al., 2013). It is therefore hypothesized that, in Arabidopsis, SOBIR1 may also interact with CLV2. The speculation is in line with the fact that SOBIR is expressed in many tissues and organs overlapping with CLV2 (Wu Y. et al., 2016). However, the biological consequence of the interaction between SOBIR1 and CLV2 remains unclear.
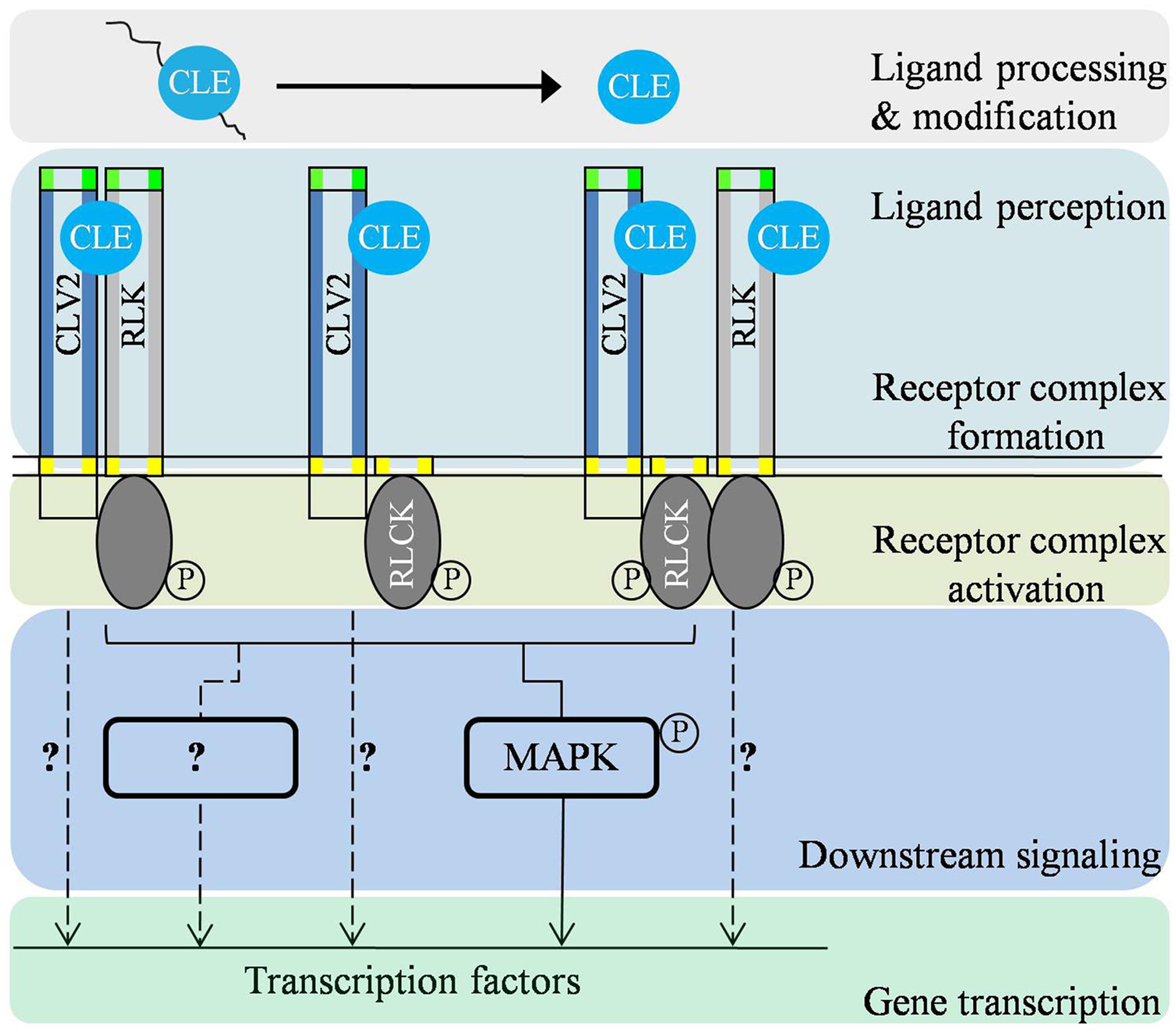
FIGURE 2. Predicted signal transduction cascades of CLV2-associated receptor complexes in modulating different biological processes. Firstly, CLE is proteolytically processed and/or post-translationally modified to be a functional CLE signaling peptide. On the plasma membrane, CLV2 forms receptor complexes with RLK(s)/RLCK(s) and perceive the putative ligand signal. In the cytoplasm, the ligand signal is modulated through regulation of phosphorylation status of the receptor complex. Upon activation, CLV2-associated receptor complexes subsequently transmit the signal to downstream signaling components such as mitogen-activated protein (MAP) kinases and possibly other yet-unidentified components (?). There also may be other mechanisms mediating the signaling output of CLV2-associated complexes (dotted lines with?).
Multiple studies have shown the physical interactions between RLKs and G-protein components. For instance, Arabidopsis Gβ protein AGB1 was found to interacts with ZAR1 to regulate cell division (Yu et al., 2016), and with ERECTA to control silique development and in response to pathogens (Lease et al., 2001; Llorente et al., 2005). Recently, the heterotrimeric G proteins composed of XLG2/3, AGB1, and AGG1/2 was found to interact with the FLS2-BIK1 complex and regulate immune signaling (Liang et al., 2016). Intriguingly, emerging evidence also exemplifies the interaction between CLV2-like proteins and G-protein components. FEA2, the maize ortholog of CLV2, is found to associate with Gα protein CT2 in coordinating maize SAM maintenance (Bommert et al., 2013). It leaves a question on how the FEA2-CT2 complex transmits the signal intracellularly. However, AGB1 was found to physically interact with RPK2, but not CLV2, to mediate CLV3 signaling (Ishida et al., 2014). The CLV2-dependent and CLV2-indpendent regulation of G-protein signaling potentially provide biochemical plasticity to diverse regulation of stem cell homeostasis. It remains elusive whether any canonical downstream regulators of G-protein signaling or any yet-unidentified components are involved in these processes (Figure 2). Altogether, there is little insight on how or whether CLV2 and CLV2-like proteins modulate the G-protein signaling.
Another aspect of future research will be to elucidate the putative ligand(s) for CLV2, which will be helpful in understanding the molecular mechanisms and the specificities of CLV2-mediated multiple biological processes. Thus far, no CLE peptide has been demonstrated as the ligand for CLV2, although it could potentially bind a variety of related CLE peptides (Guo et al., 2010; Shinohara and Matsubayashi, 2015). Inevitably, this raises again the question on how the specificity of CLV2-mediated signaling pathways is achieved to control plant development and innate immunity. Elucidating the potential ligands for CLV2 will also help to clarify the similarity and diversity of the signaling pathways mediated by CLV2.
Author Contributions
GW, JW, and LP conceived and wrote the manuscript; GW, LP, and SL contributed the figures; NY, YL, and ZL critically reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Work in our group is supported by the National Natural Science Foundation of China (31271575; 31200902), the Fundamental Research Funds for the Central Universities (GK201103005), the Specialized Research Fund for the Doctoral Program of Higher Education from the Ministry of Education of China (20120202120009), the Scientific Research Foundation for the Returned Overseas Chinese Scholars, State Education Ministry, and the Natural Science Basic Research Plan in Shaanxi Province of China (2014JM3064).
References
Bleckmann, A., Weidtkamp-Peters, S., Seidel, C., and Simon, R. (2010). Stem cell signalling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152, 166–176. doi: 10.1104/pp.109.149930
Bommert, P., Je, B. I., Goldshmidt, A., and Jackson, D. (2013). The maize Gα gene COMPACT PLANT2 functions in CLAVATA signalling to control shoot meristem size. Nature 502, 555–558. doi: 10.1038/nature12583
Brand, U., Fletcher, J. C., Hobe, M., Meyerowitz, E. M., and Simon, R. (2000). Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289, 617–619. doi: 10.1126/science.289.5479.617
Casamitjana-Martinez, E., Hofhuis, H. F., Xu, J., Liu, C. M., Heidstra, R., and Scheres, B. (2003). Root-specific CLE19 overexpression and the sol1/2 suppressors implicate a CLV-like pathway in the control of Arabidopsis root meristem maintenance. Curr. Biol. 13, 1435–1441. doi: 10.1016/S0960-9822(03)00533-5
Clark, S. E., Running, M. P., and Meyerowitz, E. M. (1993). CLAVATA1, a regulator of meristem and flower development in Arabidopsis. Development 119, 397–418.
Clark, S. E., Running, M. P., and Meyerowitz, E. M. (1995). CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121, 2057–2067.
Clark, S. E., Williams, R. W., and Meyerowitz, E. M. (1997). The CLAVATA1 gene encodes a putative receptor kinase that controls shoot and floral meristem size in Arabidopsis. Cell 89, 575–585. doi: 10.1016/S0092-8674(00)80239-1
DeYoung, B. J., Bickle, K. L., Schrage, K. J., Muskett, P., Patel, K., and Clark, S. E. (2006). The CLAVATA1-related BAM1, BAM2 and BAM3 receptor kinase-like proteins are required for meristem function in Arabidopsis. Plant J. 45, 1–16. doi: 10.1111/j.1365-313X.2005.02592.x
Diévart, A., Dalal, M., Tax, F. E., Lacey, A. D., Huttly, A., Li, J., et al. (2003). CLAVATA1 dominant-negative alleles reveal functional overlap between multiple receptor kinases that regulate meristem and organ development. Plant Cell 15, 1198–1211. doi: 10.1105/tpc.010504
Durbak, A. R., and Tax, F. E. (2011). CLAVATA signaling pathway receptors of Arabidopsis regulate cell proliferation in fruit organ formation as well as in meristems. Genetics 189, 177–194. doi: 10.1534/genetics.111.130930
Fiers, M., Golemiec, E., Xu, J., van der Geest, L., Heidstra, R., Stiekema, W., et al. (2005). The 14-amino acid CLV3, CLE19 and CLE40 peptides trigger consumption of the root meristem in Arabidopsis through a CLAVATA2-dependent pathway. Plant Cell 17, 2542–2553. doi: 10.1105/tpc.105.034009
Fiers, M., Hause, G., Boutilier, K., Casamitjana-Martinez, E., Weijers, D., Offringa, D., et al. (2004). Mis-expression of the CLV3/ESR-like gene CLE19 in Arabidopsis leads to a consumption of root meristem. Gene 327, 37–49. doi: 10.1016/j.gene.2003.11.014
Fletcher, J. C., Brand, U., Running, M. P., Simon, R., and Meyerowitz, E. M. (1999). Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283, 1911–1914. doi: 10.1126/science.283.5409.1911
Guo, Y., Han, L., Hymes, M., Denver, R., and Clark, S. E. (2010). CLAVATA2 forms a distinct CLE-binding receptor complex regulating Arabidopsis stem cell specification. Plant J. 63, 889–900. doi: 10.1111/j.1365-313X.2010.04295.x
Hanemian, M., Barlet, X., Sorin, C., Yadeta, K. A., Keller, H., Favery, B., et al. (2016). Arabidopsis CLAVATA1 and CLAVATA2 receptors contribute to Ralstonia solanacearum pathogenicity through a miR169-dependent pathway. New Phytol. 211, 502–515. doi: 10.1111/nph.13913
Ishida, T., Tabata, R., Yamada, M., Aida, M., Mitsumasu, K., Fujiwara, M., et al. (2014). Heterotrimeric G proteins control stem cell proliferation through CLAVATA signaling in Arabidopsis. EMBO Rep. 15, 1202–1209. doi: 10.15252/embr.201438660
Ito, Y., Nakanomyo, I., Motose, H., Iwamoto, K., Sawa, S., and Dohmae, N. (2006). Dodeca-CLE peptides as suppressors of plant stem cell differentiation. Science 313, 842–845. doi: 10.1126/science.1128436
Jeong, S., Trotochaud, A. E., and Clark, S. E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11, 1925–1934. doi: 10.2307/3871087
Jun, J., Fiume, E., Roeder, A. H., Meng, L., Sharma, V. K., Osmont, K. S., et al. (2010). Comprehensive analysis of CLE polypeptide signaling gene expression and overexpression activity in Arabidopsis. Plant Physiol. 154, 1721–1736. doi: 10.1104/pp.110.163683
Kayes, J. M., and Clark, S. E. (1998). CLAVATA2, a regulator of meristem and organ development in Arabidopsis. Development 125, 3843–3851.
Kinoshita, A., Betsuyaku, S., Osakabe, Y., Mizuno, S., Nagawa, S., Stahl, Y., et al. (2010). RPK2 is an essential receptor-like kinase that transmits the CLV3 signal in Arabidopsis. Development 137, 3911–3920. doi: 10.1242/dev.048199
Kinoshita, A., Nakamura, Y., Sasaki, E., Kyozuka, J., Fukuda, H., and Sawa, S. (2007). Gain-of-function phenotypes of chemically synthetic CLAVATA3/ESR-related (CLE) peptides in Arabidopsis thaliana and Oryza sativa. Plant Cell Physiol. 48, 1821–1825. doi: 10.1093/pcp/pcm154
Kondo, T., Sawa, S., Kinoshita, A., Mizuno, S., Kakimoto, T., Fukuda, H., et al. (2006). A plant peptide encoded by CLV3 identified by in Situ MALDI-TOF MS analysis. Science 313, 845–848. doi: 10.1126/science.1128439
Kondo, Y., Hirakawa, Y., Kieber, J. J., and Fukuda, H. (2011). CLE peptides can negatively regulate protoxylem vessel formation via cytokinin signaling. Plant Cell Physiol. 52, 37–48. doi: 10.1093/pcp/pcq129
Lease, K. A., Wen, J., Li, J., Doke, J. T., Liscum, E., and Walker, J. C. (2001). A mutant Arabidopsis heterotrimeric G-protein beta subunit affects leaf, flower, and fruit development. Plant Cell 13, 2631–2641. doi: 10.1105/tpc.13.12.2631
Lenhard, M., and Laux, T. (2003). Stem cell homeostasis in the Arabidopsis shoot meristem is regulated by intercellular movement of CLAVATA3 and its sequestration by CLAVATA1. Development 130, 3163–3173. doi: 10.1242/dev.00525
Liang, X., Ding, P., Lian, K., Wang, J., Ma, M., Li, L., et al. (2016). Arabidopsis heterotrimeric G proteins regulate immunity by directly coupling to the FLS2 receptor. Elife 5, e13568. doi: 10.7554/eLife.13568
Liebrand, T. W., van den Berg, G. C., Zhang, Z., Smit, P., Cordewener, J. H., America, A. H., et al. (2013). Receptor-like kinase SOBIR1/EVR interacts with receptor-like proteins in plant immunity against fungal infection. Proc. Natl. Acad. Sci. U.S.A. 110, 10010–10015. doi: 10.1073/pnas.1220015110
Llorente, F., Alonso-Blanco, C., Sanchez-Rodriguez, C., Jorda, L., and Molina, A. (2005). ERECTA receptor-like kinase and heterotrimeric G protein from Arabidopsis are required for resistance to the necrotrophic fungus Plectosphaerella cucumerina. Plant J. 43, 165–180. doi: 10.1111/j.1365-313X.2005.02440.x
Meng, L., and Feldman, L. J. (2010). CLE14/CLE20 peptides may interact with CLAVATA2/CORYNE receptor-like kinases to irreversibly inhibit cell division in the root meristem of Arabidopsis. Planta 232, 1061–1074. doi: 10.1007/s00425-010-1236-4
Miwa, H., Betsuyaku, S., Iwamoto, K., Kinoshita, A., Fukuda, H., and Sawa, S. (2008). The receptor-like kinase SOL2 mediates CLE signaling in Arabidopsis. Plant Cell Physiol. 49, 1752–1757. doi: 10.1093/pcp/pcn148
Müller, R., Bleckmann, A., and Simon, R. (2008). The receptor kinase CORYNE of Arabidopsis transmits the stem cell-limiting signal CLAVATA3 independently of CLAVATA1. Plant Cell 20, 934–946. doi: 10.1105/tpc.107.057547
Nimchuk, Z. L., Tarr, P. T., and Meyerowitz, E. M. (2011). An evolutionarily conserve pseudokinase mediates stem cell production in plants. Plant Cell 23, 851–854. doi: 10.1105/tpc.110.075622
Ogawa, M., Shinohara, H., Sakagami, Y., and Matsubayashi, Y. (2008). Arabidopsis CLV3 peptide directly binds CLV1 ectodomain. Science 319, 294. doi: 10.1126/science.1150083
Qiang, Y., Wu, J., Han, H., and Wang, G. (2013). CLE peptides in vascular development. J. Integr. Plant Biol. 55, 389–394. doi: 10.1111/jipb.12044
Replogle, A., Wang, J., Bleckmann, A., Hussey, R. S., Baum, T. J., Sawa, S., et al. (2011). Nematode CLE signaling in Arabidopsis requires CLAVATA2 and CORYNE. Plant J. 65, 430–440. doi: 10.1111/j.1365-313X.2010.04433.x
Rojo, E., Sharma, V. K., Kovaleva, V., Raikhel, N. V., and Fletcher, J. C. (2002). CLV3 is localized to the extracellular space, where it activates the Arabidopsis CLAVATA stem cell signaling pathway. Plant Cell 14, 969–977. doi: 10.1105/tpc.002196
Schoof, H., Lenhard, M., Haecker, A., Mayer, K. F., Jurgens, G., and Laux, T. (2000). The stem cell population of Arabidopsis shoot meristems in maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100, 635–644. doi: 10.1016/S0092-8674(00)80700-x
Shimizu, N., Ishida, T., Yamada, M., Shigenobu, S., Tabata, R., Kinoshita, A., et al. (2015). BAM 1 and RECEPTOR-LIKE PROTEIN KINASE 2 constitute a signaling pathway and modulate CLE peptide-triggered growth inhibition in Arabidopsis root. New Phytol. 208, 1104–1113. doi: 10.1111/nph.13520
Shinohara, H., and Matsubayashi, Y. (2015). Reevaluation of the CLV3-receptor interaction in the shoot apical meristem: dissection of the CLV3 signaling pathway from a direct ligand-binding point of view. Plant J. 82, 328–336. doi: 10.1111/tpj.12817
Somssich, M., Bleckmann, A., and Simon, R. (2016). Shared and distinct functions of the pseudokinase CORYNE (CRN) in shoot and root stem cell maintenance of Arabidopsis. J. Exp. Bot. 67, 4901–4915. doi: 10.1093/jxb/erw207
Somssich, M., Ma, Q., Weidtkamp-Peters, S., Stahl, Y., Felekyan, S., Bleckmann, A., et al. (2015). Real-time dynamics of peptide ligand-dependent receptor complex formation in planta. Sci. Signal. 8, ra76. doi: 10.1126/scisignal.aab0598
Song, S. K., and Clark, S. E. (2005). POL and related phosphatases are dosage-sensitive regulators of meristem and organ development in Arabidopsis. Dev. Biol. 285, 272–284. doi: 10.1016/j.ydbio.2005.06.020
Strabala, T. J., O’Donnell, P. J., Smit, A. M., Ampomah-Dwamena, C., JaneMartin, E., and Netzler, N. (2006). Gain-of-function phenotypes of many CLAVATA3/ESR genes, including four new family members, correlate with tandem variations in the conserved CLAVATA3/ESR domain. Plant Physiol. 140, 1331–1344. doi: 10.1104/pp.105.075515
ten Hove, C. A., Bochdanovits, Z., Jansweijer, V. M., Koning, F. G., Berke, L., Sanchez-Perez, G. F., et al. (2011). Probing the roles of LRR RLK genes in Arabidopsis thaliana roots using a custom T-DNA insertion set. Plant Mol. Biol. 76, 69–83. doi: 10.1007/s11103-011-9769-x
Wang, G. (2009). The Role of Receptor-like Proteins in Arabidopsis Development. Wageningen: Wageningen University.
Wang, G., Ellendorff, U., Kemp, B., Mansfield, J. W., Forsyth, A., Mitchell, K., et al. (2008). A genome-wide functional investigation into the roles of receptor-like proteins in Arabidopsis. Plant Physiol. 147, 503–517. doi: 10.1104/pp.108.119487
Wang, G., Fiers, M., Ellendorff, U., Wang, Z., de Wit, P. J., Angenent, C. G., et al. (2010a). The diverse roles of extracellular leucine-rich repeat-containing receptor-like proteins in Plants. Crit. Rev. Plant Sci. 29, 285–299. doi: 10.1080/07352689.2010.502082
Wang, G., Long, Y., Thomma, B. P., de Wit, P. J., Angenent, G. C., and Fiers, M. (2010b). Functional analyses of the CLAVATA2-like proteins and their domains that contribute to CLAVATA2 specificity. Plant Physiol. 152, 320–331. doi: 10.1104/pp.109.148197
Wang, G., Zhang, G., and Wu, M. (2016). CLE peptide signaling and crosstalk with phytohormones and environmental stimuli. Front. Plant Sci. 6:1211. doi: 10.3389/fpls.2015.01211
Wang, G., Zhang, Z., Angenent, G. C., and Fiers, M. (2011). New aspects of CLAVATA2, a versatile gene in the regulation of Arabidopsis development. J. Plant Physiol. 168, 403–407. doi: 10.1016/j.jplph.2010.08.015
Wu, J., Liu, Z., Zhang, Z., Lv, Y., Yang, N., Zhang, G., et al. (2016). Transcriptional regulation of receptor-like protein genes by environmental stresses and hormones and their overexpression activities in Arabidopsis thaliana. J. Exp. Bot. 67, 3339–3351. doi: 10.1093/jxb/erw152
Wu, Y., Xun, Q., Guo, Y., Zhang, J., Cheng, K., Shi, T., et al. (2016). Genome-wide expression pattern analyses of the Arabidopsis leucine-rich repeat receptor-like kinases. Mol. Plant 9, 289–300. doi: 10.1016/j.molp.2015.12.011
Yu, T. Y., Shi, D. Q., Jia, P. F., Tang, J., Li, H. J., Liu, J., et al. (2016). The Arabidopsis receptor kinase ZAR1 is required for zygote asymmetric division and its daughter cell fate. PLoS Genet 12:e1005933. doi: 10.1371/journal.pgen.1005933
Keywords: CLV2, receptor-like kinase, plant development, plant immunity, Arabidopsis
Citation: Pan L, Lv S, Yang N, Lv Y, Liu Z, Wu J and Wang G (2016) The Multifunction of CLAVATA2 in Plant Development and Immunity. Front. Plant Sci. 7:1573. doi: 10.3389/fpls.2016.01573
Received: 25 June 2016; Accepted: 06 October 2016;
Published: 24 October 2016.
Edited by:
Hideki Takahashi, Michigan State University, USACopyright © 2016 Pan, Lv, Yang, Lv, Liu, Wu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guodong Wang, Z3VvZG9uZ193YW5nQHNubnUuZWR1LmNu
 Lixia Pan1
Lixia Pan1 Guodong Wang
Guodong Wang