
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 26 October 2016
Sec. Plant Biotechnology
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.01539
This article is part of the Research TopicMechanisms of abiotic stress responses and tolerance in plants: physiological, biochemical and molecular interventionsView all 121 articles
 Jubin N. Shah1†
Jubin N. Shah1† Olga Kirioukhova1,2†
Olga Kirioukhova1,2† Pallavi Pawar1
Pallavi Pawar1 Muhammad Tayyab1
Muhammad Tayyab1 Juan L. Mateo3*
Juan L. Mateo3* Amal J. Johnston1,2*
Amal J. Johnston1,2*Molecular dissection of apomixis – an asexual reproductive mode – is anticipated to solve the enigma of loss of meiotic sex, and to help fixing elite agronomic traits. The Brassicaceae genus Boechera comprises of both sexual and apomictic species, permitting comparative analyses of meiotic circumvention (apomeiosis) and parthenogenesis. Whereas previous studies reported local transcriptome changes during these events, it remained unclear whether global changes associated with hybridization, polyploidy and environmental adaptation that arose during evolution of Boechera might serve as (epi)genetic regulators of early development prior apomictic initiation. To identify these signatures during vegetative stages, we compared seedling RNA-seq transcriptomes of an obligate triploid apomict and a diploid sexual, both isolated from a drought-prone habitat. Uncovered were several genes differentially expressed between sexual and apomictic seedlings, including homologs of meiotic genes ASYNAPTIC 1 (ASY1) and MULTIPOLAR SPINDLE 1 (MPS1) that were down-regulated in apomicts. An intriguing class of apomict-specific deregulated genes included several NAC transcription factors, homologs of which are known to be transcriptionally reprogrammed during abiotic stress in other plants. Deregulation of both meiotic and stress-response genes during seedling stages might possibly be important in preparation for meiotic circumvention, as similar transcriptional alteration was discernible in apomeiotic floral buds too. Furthermore, we noted that the apomict showed better tolerance to osmotic stress in vitro than the sexual, in conjunction with significant upregulation of a subset of NAC genes. In support of the current model that DNA methylation epigenetically regulates stress, ploidy, hybridization and apomixis, we noted that ASY1, MPS1 and NAC019 homologs were deregulated in Boechera seedlings upon DNA demethylation, and ASY1 in particular seems to be repressed by global DNA methylation exclusively in the apomicts. Variability in stress and transcriptional response in a diploid apomict, which is geographically distinct from the triploid apomict, pinpoints both common and independent features of apomixis evolution. Our study provides a molecular frame-work to investigate how the adaptive traits associated with the evolutionary history of apomicts co-adapted with meiotic gene deregulation at early developmental stage, in order to predate meiotic recombination, which otherwise is thought to be favorable in stress and low-fitness conditions.
Sexual reproduction, otherwise known as amphimixis, is the most prevalent mode of reproduction found across phylogenetic boundaries of multi-cellular organisms over two billion years. Sex leads to genetic recombination, allowing natural selection to act upon individual traits and segregation of mutations from the natural population (Smith, 1971). The evolutionary advantages of meiotic recombination have long been debated in biology. The deterministic mutation hypothesis sees recombination as a mean to efficiently purge deleterious mutations (Kondrashov, 1988). The Red Queen hypothesis favors recombination as an optimal way of adaptation in arms races with parasites (Jaenike, 1978). Fisher suggested that recombination might accelerate species adaptation to a changing environment by bringing beneficial allele combinations together (Fisher, 1930). Whereas Fisher’s proposal is fully supported by a recent molecular experiment in yeast (McDonald et al., 2016), it was also extensively debated by others. For instance, there are cases where recombination breaks fitter-combinations more than it does so to create novel beneficial mutation sets (Eshel and Feldman, 1970). A compromising “fitness-associated recombination (FAR)” theory proposes increase in recombination as a consequence of stress-associated DNA damage in individuals with lower fitness thus less tolerant to stress, in stark contrast to better adapted organisms bearing beneficial genetic combinations. Therefore, FAR would favor preserving the fitter genotypes, and enhance segregation of alleles in progenies of the less fit ones thus enabling new genetic combinations to occur and to be exposed to selective pressure (Hadany and Beker, 2003a,b). Following the “restoration” view of Bernstein and co-workers that sex evolved for the repair of DNA lesions, a refined hypothesis proposes that meiosis has selectively been retained in evolution mainly for its role in repair of DNA damage caused by oxidative stress and reactive oxygen species (ROS), instead of its role in generation of recombination [summarized in (Horandl and Hadacek, 2013)]. The latter view advocates that redox status between oxidized DNA and a key meiotic protein could be necessary for generating double-strand breaks (DSB), and could possibly be considered as a consequence of genome-wide epimutations caused by DNA or histone methylation changes that restrict deleterious transposable elements (de Massy, 2013; Zamudio et al., 2015). Together, the paradoxical view of sex in biology has seen several changes over the past decades, and the most recent ones address stress, fitness, DNA repair, epigenetics and trans-generational responses as key underlying determinants.
Asexual reproduction, on the other end, represents offspring propagation from somatic cells, including deviation of the sexual pathway by avoidance of meiotic recombination. The latter has been infrequently observed in some Eukaryotes, and is considered to be disadvantageous in a population in the long run impeding adaptation, since deleterious mutations may accumulate in the absence of meiotic recombination due to Muller’s Ratchet (Muller, 1964; Charlesworth, 2006), as experimentally documented in yeast (McDonald et al., 2016). Despite the popular views of Fisher (1930) and Muller (1964), the dynamics of adaptation in asexual populations has constantly been debated. For instance, ‘clonal interference’ theory postulates that many single beneficial mutations of varying effect contend for fixation in a two-loci model in asexuals (Gerrish and Lenski, 1998). Kim and Orr (2005) could not see that sex could be advantageous over asexuality, when multiple beneficial mutations are involved in a finite population. Multiple-mutations model of (Desai et al., 2007) assumes that all clonal mutations are of the same beneficial effect and that clonal competition between clones bearing different numbers of beneficial mutations might occur among genotypes. The problem here is that superior clonal competitors might not emerge under the assumption that beneficial mutations are of the same effect. An addendum to the latter (Sniegowski and Gerrish, 2010) discussed that asexual populations might harbor mutations of different effects, and that competitions might arise between clones that bear multiple beneficial mutations. Adaptation rate in these populations should, however, assess both beneficial and deleterious mutations, population bottlenecks, and clonal interference (Campos and Wahl, 2010). Summarizing the old and new population theories, evolution of asexuality could often be associated with genetic conflicts deviating from a faithful sexual meiotic program, and asexuals could suffer from reduced fitness and an increase in genetic drift, but the population size and the number of mutations and their interactions might play a lead role in deciding whether or not clonal populations are advantageous.
Asexual organisms do not switch ploidy cycles between generations, and asexuality is predominantly found in polyploid organisms; therefore, asexuality and polyploidy seem to be positively correlated. Meiosis fails to a large extent in polyploids, specifically in organisms with odd numbered genomes, potentially due to difficulty in pairing of greater than two chromosomes of each type (William and Barraclough, 2009). Increased ploidy level, particularly in conjunction with hybridization and resulting heterosis, enhance heterozygosity of an individual organism hence genetic variation within the population (reviewed in Horandl, 2009). All the above may lead to better tolerance of deleterious mutations due to higher number of homologous chromosomes and therefore the functional gene copies in asexuals. This view is further supported by a broader epigenetic landscape and narrow epigenetic resetting in polyploids and asexuals, for instance involving DNA methylation and small RNA pathways, and the enhanced epigenetic stability in clonal organisms might confer adaptive advantages (reviewed in Verhoeven and Preite, 2014). In addition, asexual reproduction provides short-term advantages for organisms primarily because the costs underlying the energy-consuming process of meiosis and involvement of two parents (twofold cost) are avoided (Dawson, 1995). Clonal reproduction may also be an advantage by fixing beneficial allelic combination in one organism. Extant ancient asexual species might confirm the evolutionary stability of this reproductive mode (Judson and Normark, 1996; Martens et al., 2003); however, the recent work uncovered that their evolution is based on horizontal gene transfer instead of meiotic sex (Eyres et al., 2015). It is equally important to note that most theories above utilized population or generation of microbes or fungus and/or simulations; the rate of mutation accumulation in clonal populations could as well be extremely slow in plastic organisms like plants (discussed later) across time scale and might provide advantage over sexuals in the wake of changing climatic conditions, as evident from the stable and wide-spread inter-continental occurrence of apomicts such as Taraxacum and Hieracium. Regulation of oxidative stress machinery was proposed as a switch between meiotic program as DNA repair and ameiotic reproduction in organisms with mixed (facultative) reproductive mode (Horandl and Hadacek, 2013). Nonetheless, selection of sexual versus asexual reproduction in organisms has remained an enigma and a stimulating research topic.
Gametophytic apomixis refers to asexual reproduction in plants, during which the sexual development is modified via avoidance of meiosis and fertilization (Winkler, 1908; Koltunow, 1993). Apomixis has been noted in about 0.1% of the flowering plants, leading to clonal reproduction through seeds sensu stricto (Mogie, 1992). Apomixis comprises four developmental processes: (a) alteration of meiosis known as apomeiosis, during which a diploid somatic cell termed as the Megaspore Mother Cell (MMC) undergoes only one cell division and ultimately yields an unreduced diploid egg cell (diplospory); or (b) avoidance of meiosis by a diploid somatic cell in the vicinity of MMC so that the former surpasses meiosis and mitotically develops a diploid egg cell (apospory); (c) formation of embryo from unfertilized egg cell, i.e., by parthenogenesis; and (d) autonomous (in the absence of fertilization) or sexual development of endosperm, which nourishes the parthenogenetic embryo (Koltunow, 1993; Hojsgaard et al., 2014b). These four developmental steps seem to be regulated independently, and genetic studies support both dominant and/or additive genetic effects underlying these traits. Apomixis generally occurs outside of the major cultivated plants, for instance, aposporous Hieracium and diplosporous Taraxacum; and several model systems have been established both in dicots and monocots, such that apomictic traits could possibly be transferred into the closest sexual relatives (Koltunow, 1993). Deciphering apomixis mechanisms in plants is a high priority research not only for evolutionary and developmental biologists, but also it is a long quest for plant breeders as it would allow propagation of elite clonal genotypes like F1 hybrids with superior breeding traits such as grain yield, biomass or timber, if apomictic traits are incorporated into the traditional breeding programs.
Following an initial observation in a wild apomict that led to a proposal that apomixis can be a mere deregulation of sexual developmental program, one of the widely accepted hypothesis is that the expression of genes controlling sexual reproduction may occur at the wrong place and/or the wrong time in apomicts (Nogler, 1984; Koltunow and Grossniklaus, 2003). In support of this hypothesis, transcriptome-wide changes of gene expression during apomixis events in the ovules or specific cell-types have been documented in various apomictic species (Hand and Koltunow, 2014). Expression of apomixis-specific gene fragments, for instance like those of BABY BOOM of apomictic Pennisetum (Conner et al., 2015) inducing partial parthenogenesis in sexual plants, or triple Arabidopsis meiotic mutants mimicking apomeiosis (d’Erfurth et al., 2009) lend support to a mutational model in which apomixis can be seen as mutation(s) modifying the sexual pathway.
In evolutionary terms, apomixis was proposed to have arisen due to hybridization between species that are inter-related, given that most apomicts are polyploids and maintain high degree of heterozygosity as means of buffering deleterious mutations (Ernst, 1918). Both hybridity and polyploidy presumably might have led to genome-wide effects, and might induce apomixis (Carman, 1997; Madlung et al., 2002; Bicknell and Koltunow, 2004; Wang et al., 2004). Although it is extremely rare, apomixis could also be sustained in diploid conditions, particularly when the apomictic populations represent diploid-aneuploids, or hybrids between two unrelated ecotypes or related species with differences in reproductive characters (Carman, 1997; Lovell et al., 2013). Apomictic polyploids were likely derived from initially unstable diploid apomictic hybrids, and they seem to feature high heterozygosity necessary for sustenance of apomixis due to dosage-dependent gene regulation leading to efficient management of deleterious mutations (Archetti, 2004). In addition, ploidy shifts lead to changes in epigenetic landscape, for instance in DNA methylation within several loci, and concomitant transcriptional reprogramming wired by the RNA-dependent DNA methylation (RdDM) pathways required for initiation and maintenance of apomictic traits (Verhoeven et al., 2010; Hand and Koltunow, 2014; Podio et al., 2014; Zappacosta et al., 2014). Thus, hybridization-derived polyploidy could sustain apomixis in plant populations. This view had lent support for the hybridization theory for evolution of apomicts (Carman, 1997), which suggests that apomixis might occur when two unrelated ecotypes or related species with differences in reproductive characters hybridized. The two distinct sets of genes from these hybrids would be asynchronously expressed, which might lead to precocious embryo sac initiation and parthenogenesis. Therefore, this theory is based on additive expression of sets of genes involved in reproduction, rather than mutations in genes involved in sexual reproduction, as evident in some apomictic species (Carman, 1997; Hojsgaard et al., 2014a). In summary, genetic control underlying apomixis in diploids versus polyploids remains largely unresolved, but it is conceivable that the molecular mechansims of evolutionary history traits related to hybridization, and/or ploidy and the environment (discussed below) would be indispensable for apomixis.
Environment is one of the important aspects that control organismal reproduction and fitness trade-offs. Due to their sessile nature, plants frequently encounter unfavorable abiotic conditions such as severe drought, extreme temperatures, contamination of soil by heavy metals and/or high salt concentration, and the same is true for a plethora of biotic conditions. Variation in parameters like temperature, light, water and nutrient levels act as stress agents and influence reproduction in plants (Hedhly et al., 2009; Thakur et al., 2010). Sexual reproduction is sensitive to stressful environmental conditions, and stress tolerance starting as early as in the vegetative phase and continuing into the reproductive phase is a key factor in sustainable plant productivity (Herrero and Johnson, 1980; De Storme and Geelen, 2014). The reproductive phase also gives the plant a chance to acclimatize to environmental variations. Genetic and epigenetic stress mechanisms have been extensively studied mainly in sexual plants more so during the sporophytic phase; data suggest that both vegetative and reproductive stress responses are inter-linked. For instance, under certain salinity thresholds, the female germline tissues of the sexual plant Arabidopsis thaliana seem to buffer stress much better than the male tissues, possibly due to a sink mechanism from the female tissues that are tightly dependent on the plant sporophyte (Sun et al., 2004). This proposition is in line with the role of maternal sporophytic tissue that could lend support for epigenetic, metabolic and genetic reprogramming in order to sustain apomictic events upon stress. It is important to note that apomictic hybrids and polyploids generally arose in stressful environments. Polyploids are generally known for their greater efficiency to tolerate abiotic stress such as osmotic stress, drought, light stress and to scavenge ROS species than the corresponding diploid individuals (Chandra and Dubey, 2010; Coate et al., 2013; Hao et al., 2013; Syamaladevi et al., 2016), and admittedly might offer a genomic haven to sustain apomixis in challenging environmental conditions (Lynch, 1984). Taken together, response to environmental factors such as abiotic stress acclimation is an important life-history trait, and might influence sexual and possibly apomictic reproduction.
Boechera, a close relative of the sexual model plant Arabidopsis, is a unique model system to study apomixis; the genus comprises of both diploid sexual, diploid and polypoid apomictic species, giving an opportunity to conduct comparative analyses of apomixis versus sex in the wild (Böcher, 1947, 1951; Roy, 1995; Naumova et al., 2001; Al-Shehbaz et al., 2006). Most apomictic Boechera are reproductively isolated, showing primary distribution in the western USA and Canada (Windsor et al., 2006). B. stricta (Graham) Al-Shehbaz (commonly known as Drummond’s Rockcress; hereafter referred to as line Sex-1 in this work; 2n = 14) seems to be the most widespread sexual Boechera; repeated hybridization of this species likely paved way to reproductively isolated hybrids that ultimately resulted in the birth of apomictic species. Ploidy and zygosity are important for apomixis in Boechera, similar to the situation in most apomictic plants (Hand and Koltunow, 2014). The majority of the apomictic Boechera are triploids, and aneuploids, tetraploids and very rare diploids have also been identified (Roy, 1995; Schranz et al., 2006). In contrast to the high degree of inbreeding and homozygosity of sexual diploids, Boechera apomicts are highly heterozygous for most loci (Roy, 1995; Rushworth et al., 2011). Most apomictic Boechera are diplosporous, but rare mixed occurrence of apospory and diplospory have also been noted; parthenogenesis accompanies diplospory or apospory, and endosperm development is predominantly sexual (Böcher, 1947, 1951; Roy, 1995; Carman, 1997; Naumova et al., 2001; Lovell et al., 2013). Boechera genomes seem to have over 80% nucleotide sequence similarity to the genome of A. thaliana, making comparative analyses amenable. Recent work on Boechera uncovered genome-wide alterations of the transcriptomes from the reproductive lineage organs such as ovules or specific cell types such as the MMC, in diploid or triploid apomicts (Sharbel et al., 2010; Schmidt et al., 2014). These data, together with the unpublished data from our lab, support an unprecedented wave of transcriptional changes in the apomictic organ or cell-types of Boechera, hinting alterations in several transcription factors, meiotic and mitotic cell cycle machinery, hormonal regulation, DNA or histone methylation pathway etc., but a defined pattern of regulation has not been discernible. One of the intriguing classes of transcriptional changes noted in these transcriptome datasets is the abiotic and biotic stress category. It is interesting to note that in its native range varying between high altitude to coastal regions and deserts, Boechera species are often challenged by recurrent stress such as drought, heat, light, freezing etc.; thus highly prone to natural selection on stress tolerance traits (Roy, 1995; Franzke et al., 2011). Hence, Boechera has become a model plant to study particularly abiotic stress tolerance mechanisms (Rushworth et al., 2011; Gallas and Waters, 2015).
It is important to note that taxonomic revision within the Boechera genus is one of the daunting exercises for systematic botanists, as several Boechera species are known to repeatedly inter-hybridize, leading to novel species that are either sexuals or mostly facultative apomicts (Rushworth et al., 2011; Alexander et al., 2013). Several apomictic Boechera (diploid or polyploid) had been classified under B. holboellii clade and/or several others previously belong to this clade, e.g., B. divaricarpa (used in this study) had been reclassified into “trashcan” hybrids involving sexuals like B. stricta and B. sparsifolia (Rushworth et al., 2011). Diploids and polyploids (e.g., triploids) can be tractable within these “holboeleii and hybrid” clades (Naumova et al., 2001; Aliyu et al., 2010; Voigt-Zielinski et al., 2012), and current studies in several labs focus on the genome evolution of the identified apomictic diploids and their sexual parents by genome sequencing. Although exhaustive comparative studies have not yet been conducted, Roy concluded that B. holboellii apomictic populations are likely polyphyletic with substantial allelic variation thus greater fixed heterozygosity than what were observed for B. gunnisoniana, a stand-alone monophyletic apomictic triploid (Roy, 1995). We and others have used the latter for characterization of apomixis events (current study; Roy, 1995; Taskin et al., 2004; Schmidt et al., 2014; Kirioukhova et al., in preparation). B. gunnisoniana is not genome-sequenced, and no diploid individuals could be traceable in the wild. Unfortunately, the influence of hybridity but polyploidy on apomeiotic expressivity cannot be separated in this case, therefore, omic-datasets like Schmidt et al. (2014) or this study will have to be carefully compared and interpreted. As apomixis is also independent in evolution, independent apomictic Boechera species or clades might exhibit independent molecular signatures.
Given the importance of Boechera genus in evolutionary point of view, with its intrinsic characteristics to give rise to a number of independent populations with apomictic mode of reproduction and its origin from a habitat with elevated environmental stress, the main questions, which we address in this work are: (1) If there are specific gene expression alterations at the seedling stage, i.e., way in advance of the apomeiotic germline transition as a preparatory mechanism for apomixis-specific gene expression program, which could likely be continued at the reproductive stage of development. (2) In particular, we hypothesize that apomict-specific transcriptional shifts already in the seedling perhaps in light of genome-wide changes underlie (a) regulation of components of meiotic machinery, since the chosen diplosporous Boechera accessions exhibit a shortcut of meiotic division, and (b) stress-related regulatory networks as a response to harsh environmental conditions and hybrid genomic collisions predicted to have occurred during their evolution.
Using Sex-1 as a baseline, we compared the seedling transcriptome of the obligate triploid apomict B. gunnisoniana (Rollins) W. A. Weber (commonly known as Gunnison’s Rockcress; hereafter referred to as line Apo-1). Both these strains were originally collected from a severe drought-prone gold mine high-elevation habitat in Colorado, USA (Roy, 1995) (Supplementary Figure S1). For further validations, we used a diploid apomict B. divaricarpa (A. Nelson) A. Löve and D. Löve of the holboellii hybrid clade; hereafter referred to as line Apo-2, which is evolutionarily distinct from Apo-1, and derived from a less-disturbed high-elevation habitat in Montana, USA (Schranz et al., 2005) (Supplementary Figure S1). Our RNA-seq approach revealed that, similar to apomictic ovules, apomictic seedlings show an array of transcriptional changes in several genes including down-regulation of two important meiotic regulators ASYNAPTIC 1 (ASY1) and MULTIPOLAR SPINDLE 1 (MPS1), accompanied by activation of stress-responsive genes particularly belonging to the NAC-DOMAIN CONTAINING (NAC) transcription factor family and LATE EMBRYOGENESIS ABUNDANT (LEA) family. Furthermore, we show evidence that osmotic stress, as an environmental factor; and DNA methylation, as an epigenetic modifier, seems to be involved in gene regulation during apomictic seedling development. Together, we propose a model integrating stress response machinery as a developmental buffer from early seedling development and simultaneously co-evolved apomeiosis/apomixis-specific regulatory network under polyploidy. In line with hypothesis of stress-related background of sex (Hadany and Beker, 2003a,b; Horandl and Hadacek, 2013), our findings suggest that the shift in the stress-response regulation might have created stress tolerant genomic environment for successful apomeiotic transition in Boechera.
Boechera seeds (sexual and asexual) were generous donations of Bitty Roy (University of Oregon, USA) and Eric Schranz (Wageningen University, USA); seeds were originally isolated from the native habitat of Boechera in the USA (Supplementary Figure S1), and they represent green-house or growth-chamber grown bulked seeds when donated. The progeny were further screened by bulked and single seed flow cytometry to remove odd genotypes (4n, 6n, aneuploids), and 3n and 2n plants of Apo-1 and Apo-2 were identified and characterized by cytology (unpublished data). Subsequent generation of seeds were surface-sterilized, stratified at 4°C and grown on half-strength MS agar plates for 17–18 days. For reproductive development, they were transferred subsequently to soil (Profi-substrate, Einheitserde, Germany) and quartz sand in a 4:1 combination. Seedlings and plants were grown in dedicated plant growth chambers with long day photoperiod (16-h light/8-h darkness cycle), under cool white light with ca. 120 μmol m-2s-1 intensity, 60–65% air humidity and 22°C. Plants were ploidy genotyped either in a Partec flow cytometer (Sysmex, Germany) or in a Guava® easyCyte flow cytometer (Millipore, USA).
Seventeen to eighteen days old seedlings from Sex-1 and Apo-1 accessions were subjected to mRNA sequencing. Seedlings were snap-frozen and kept at -80°C until RNA extraction; tissue was ground using a tissue lyser (Retsch, Germany). RNA extraction was performed using QIAGEN RNeasy Mini Kit and DNase–treated according to manufacturer’s instructions (Qiagen, USA). The RNA quality was further validated in a Bioanalyzer (Life Technologies, USA). mRNA transcripts purification, cDNA synthesis, library preparation and NGS sequencing were performed as per the routine pipeline established at the facility of Fasteris, Geneva, Switzerland. Illumina HiSeq2000 was used to carry out paired-end sequencing generating a minimum of 100 bp per read.
All reads from Illumina sequencing (ArrayExpress accession: E-MTAB-4972) were first quality-assessed using FastQC (Andrews, 2010) and mapped to Arabidopsis (TAIR-10) genome using Tophat-2.0.9 (Kim et al., 2013). The percentage of reads mapped from Sex-1 library were 33.0% and from Apo-1 library 34.9%, respectively. To quantify transcript abundances and differential expression analysis, Cufflinks-2.1.1 and Cuffdiff-2.1.1 tools were used with upper quartile normalization and multi-read correction was applied to procure fragments per kilobase of transcript per million mapped reads (FPKM) values (Trapnell et al., 2010, 2012). In parallel to obtaining a list of differentially expressed genes based on corrected p-values from cuffdiff (N = 87), FPKM values from cufflinks file were used to generate the second list of differentially expressed candidate genes based on twofold changes between expression values across both genotypes. It must be noted that mapping Boechera reads to Arabidopsis genome (80% similarity) is of heterologous nature, therefore, we relaxed the filtering criteria and categorized genes showing mis-regulation by twofold change. Based on this list, we found significant number of genes (∼4100) to be differentially expressed across both genotypes. The list of differentially expressed genes was then subjected to GO enrichment analysis using BiNGO tool from Cytoscape after Benjamini and Hochberg false discovery rate (FDR) correction with p-value cut-off of ≤0.05 (Maere et al., 2005). Gene ontology and annotations were improved with data acquired from the following websites: www.arabidopsis.org; http://planttfdb.cbi.pku.edu.cn; http://ahd.cbi.pku.edu.cn, where appropriate.
In vitro osmotic stress treatment was carried out using half-strength Murashige and Skoog (½MS) agar plates infused with PEG-8000 (polyethylene glycol). Once the MS media was solidified, PEG was overlaid and allowed to equilibrate overnight. Two concentrations of PEG (400 and 550 g/L) were tested on Boechera seedlings. Prior to transferring seedlings, PEG solution was poured according to (Verslues et al., 2006). Since 550 g/L PEG almost completely inhibited the root growth, 400 g/L PEG was used for final experiments. For salt (osmotic + ionic) stress, ½MS agar plates supplemented with 50 mM NaCl were used as this concentration causes a similar effect on primary root elongation in Arabidopsis (Verslues et al., 2006). Individual seedlings (n = 20 seedlings each for mock and PEG stress treatment in four replicates, and n = 8 for salt stress treatment) grown for 11–12 days on mock ½MS agar plates in vertical position were then transferred to both stress treatment and mock plates, arranged in a manner to have the same start point for further growth of roots and grown for next 7 days. Each plate contained seedlings from all three genotypes, and the experiments were replicated at least four times to avoid plate effect. Following the seventh day of treatment, images were captured and primary root length measurements were carried out using the software ImageJ (Schneider et al., 2012). Drought treatment of flowering plants pre-grown at control conditions was performed by stopping watering for 9 days until soil in pots was almost dry and some plants showed first signs of wilting, and then resuming watering to maintain the acquired weight of the pots and prevent plants drying out.
Zebularine, a chemical analog of the cytosine nucleoside that efficiently inhibit DNA methyltransferase activity (Baubec et al., 2009) was used as a DNA methylation inhibitor. Different concentrations of zebularine (50–175 μM) were first tested for their growth effect on Boechera seedlings in vitro. For final experiments, we used 175 μM zebularine treatment, as the seedlings still retained chlorophyll but their growth was inhibited. Seedlings (n = 10 each for control and for treated) grown on ½MS agar plates for 11–12 days were transferred to ½MS mock agar plates or supplemented with 175 μM zebularine. Arrangement of seedlings in the plates was similar to that of PEG treatment. Images were recorded and samples were snap-frozen for RNA allowing 7 days of growth upon exposure to the nucleoside.
Seedlings grown on vertical ½MS agar plates, and premeiotic buds from plants grown on soil in control conditions were collected and immediately snap-frozen for RNA extraction. Developmental staging of pre-meiotic flower buds used for gene expression validation was determined based on clearing of ovule samples by DIC microscopy. The bud size and images of corresponding ovule development for each genotype are furnished in Supplementary Figure S3. RNA isolation and cDNA synthesis were performed as described previously (Johnston et al., 2008, 2010). In brief, frozen tissues were ground similarly to the RNA-seq experiment; RNA was extracted using Trizol method (Chomczynski, 1993) and treated with DNase I; reverse transcription was done with SuperScript III First-Strand Synthesis System, according to the manufacturer instructions (Thermofisher, USA). To validate RNA-seq results, qRT-PCR of selected genes with strongly increased or decreased expression in the apomict was carried out. Three biological replicates of three seedlings each in three technical replicates were used for all control seedlings; similar experimental setup was also used for qRT experiments on premeiotic buds and zebularine-treated seedling experiments. The list of candidates included genes from meiotic and stress machinery. Primer sequences are listed in Supplementary Table S5. SYBR Green assays were performed with StepOnePlus Real-Time-PCR System (Applied Biosystems, USA) and normalized as described in (Pellino et al., 2011). Calculations of relative expression levels and generation of graphs was carried out using Microsoft Excel 2010.
Pre- and post-meiotic ovules were fixed in ice-cold 9:1 ethanol:acetic acid fixative and incubated overnight at 4°C, then re-hydrated in 90, 80, and 70% ethanol series. Ethanol was replaced with clearing solution (8:2:1 chloralhydrate:glycerol:water) for 12–16 h at 4°C, and ovules were dissected out on microscopy slides with help of insulin needles in a drop of clearing solution, and mounted under cover slips. Differential interference contrast (DIC) imaging was performed on a Leica DMI6000 microscope.
Venn diagrams were made with online Venn diagram generator available at http://bioinformatics.psb.ugent.be/webtools/Venn. Images were treated in Adobe suite of software such as Illustrator or Photoshop CS for quality of representation only.
Quantitative data were analyzed using one-tailed unpaired Student’s t-test for comparisons between groups. In addition, significance of differential gene expression response to treatments was evaluated using two-way ANOVA. Significance levels used were α ≤ 0.01 (∗∗) and α ≤ 0.05 (∗). Calculations were carried out using Microsoft Excel 2010 and http://vassarstats.net/anova2u.html.
We conducted a transcriptome-wide differential gene expression analyses between a sexual diploid, and an apomictic triploid Boechera that originated presumably through hybridization and polyploidization in the same stress-prone habitat (Figure 1, Supplementary Figure S1). We performed comparative RNA-seq analyses between the apomictic seedlings versus the sexual seedlings, in order to uncover the molecular signatures associated with the evolutionary history that might have formed the basis of evolution and maintenance of apomixis. A little over 3000 Boechera genes were found differentially expressed between Apo-1 and Sex-1 seedlings, accounting for about 15% of the total number of corresponding Arabidopsis loci qualified for differential analyses upon heterologous mapping. From the analysis of gene ontology (GO) enrichment of these genes we noted that response to stimulus and stress categories predominated, ranging up to 23% of the total (Figure 1, Supplementary Table S1). Both biotic and abiotic GO stress categories were present; and abiotic stress categories were frequented by response to osmotic, drought and salt stress categories. Other primary categories included response to wounding, hormones, ethylene, oxidative stress, metabolic processes etc. Regardless of lists of genes upregulated or down-regulated in the apomictic seedlings, the overall annotation enrichment helped us to pinpoint the essence of stress being an important category to distinguish the vegetative transcriptome of the apomict over the corresponding sexual (see the bar chart in Figure 1; Supplementary Figures S1 and S2). Intriguingly, a total of 12 genes associated with meiosis were found differentially expressed in apomictic seedlings (Table 1). We validated our transcriptome data by a two-fold approach: (a) comparison of real-time qPCR-determined expression levels of representative candidate genes not only between Apo-1 and Sex-1, but also in another apomict, Apo-2, which is distinct from Apo-1 in terms of ploidy and evolutionary history, and (b) testing deregulation of meiotic and stress response genes also in reproductive organs at onset of (apo)meiosis.
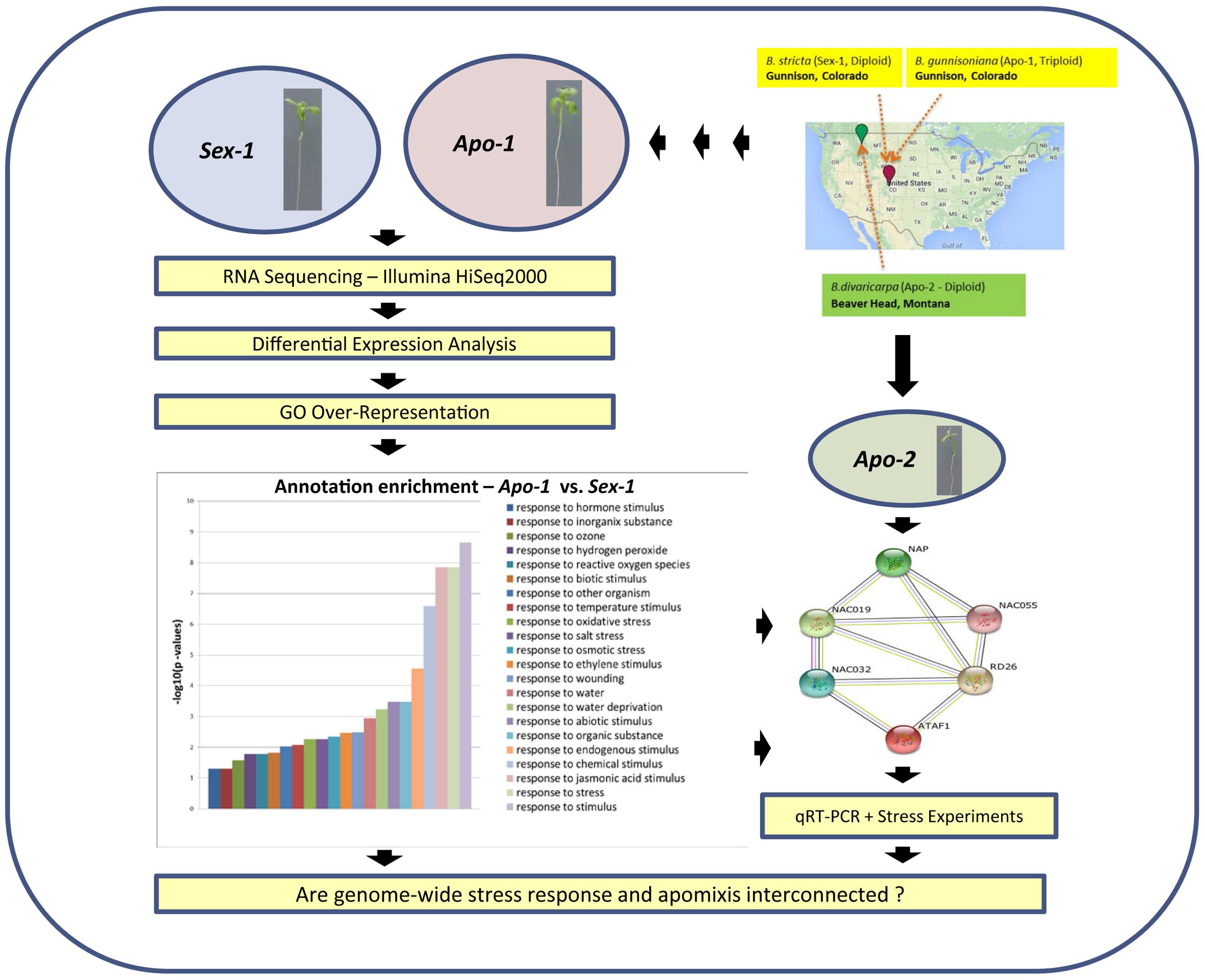
FIGURE 1. A flow chart describing the transcriptome approach and validation in order to identify stress-specific changes underlying apomixis at seedling stage. Seedlings of Sex-1 and Apo-1 Boechera strains were subjected to mRNA sequencing followed by differential gene expression analysis and identification of GO (Gene Ontology) enrichment for stress. The bar chart represents –log10 values of corrected p-values for stress-related annotations obtained after GO enrichment using Bingo tool from Cytoscape (http://www.cytoscape.org). Apo-2 accession was used as a secondary apomictic genotype for real-time qRT-PCR-based validations and stress experiments.
Consistent with our RNA-seq data (Table 1, Supplementary Table S3, Figures 2A,D), we could confirm that homologs of ASYNAPTIC 1 (ASY1) and MULTI-POLAR SPINDLE 1 (MPS1) were significantly down-regulated in seedlings of Apo-1 compared to Sex-1 (Figures 2B,E) as well as at pre-meiotic stage in flower buds (Figures 2C,F, Supplementary Figure S3). Between Apo-1 and Apo-2 seedlings, ASY1 level was lower in Apo-1 than Apo-2 while MPS1 showed insignificant differences between the two apomicts. Similar trend was also observed in premeiotic flower buds. In order to subtract ploidy from reproduction mode-related effects, we compared the gene expression in groups of the diploids with triploid (2n vs. 3n) and the sexual with apomicts (Sex vs. Apo). When mean seedling expression value of both apomicts was compared to that of the sexual, MPS1 expression was found significantly decreased, and ASY1 displayed a similar downregulation trend. However, ploidy groups also exhibited significant difference in gene expression, and MPS1 expression in seedlings was tested as ploidy-independent (Figure 2E). Meiotic gene deregulation was further accompanied by upregulation of gene homolog of an abiotic (light) stress response GATA TRANSCRIPTION FACTOR 24 (GATA24) (Supplementary Table S2), otherwise annotated as ZIM-LIKE 1 (ZML1) in Arabidopsis, both in apomictic seedling tissues and flower buds of Apo-1 and Apo-2 (Figures 2G–I). However, ZML1 levels were much higher in Apo-1 compared to Apo-2 in both the stages and probably reflected ploidy-specific gene expression pattern. Similarly, a biotic stress-specific gene JASMONATE-ZIM-DOMAIN PROTEIN 9 (JAZ9) was found activated in Apo-1 only (Figures 2J,K, Supplementary Table S2). ZML1 showed both significant effect between reproductive and ploidy groups, but JAZ9 expression seemed to be rather ploidy-dependent. Together, a subset of meiotic and stress-specific genes was deregulated in the apomictic seedlings and premeiotic buds, thus confirming our RNA-seq data and a possible association with the reproductive stage. This finding let us to propose that the identified transcriptional modifications in an early stage could likely follow throughout apomictic development. Accession-specific differences between the two apomicts could also be noticed pointing at the role of ploidy and/or evolutionary history in gene expression, yet suggesting their role preferentially in the apomicts. The above validations support that our dataset of about 700 stress- and stimulus-associated genes differentially expressed in the asexual over sexual seedlings could be resourceful for connecting stress, ploidy and reproductive nodes of gene regulatory networks in development.
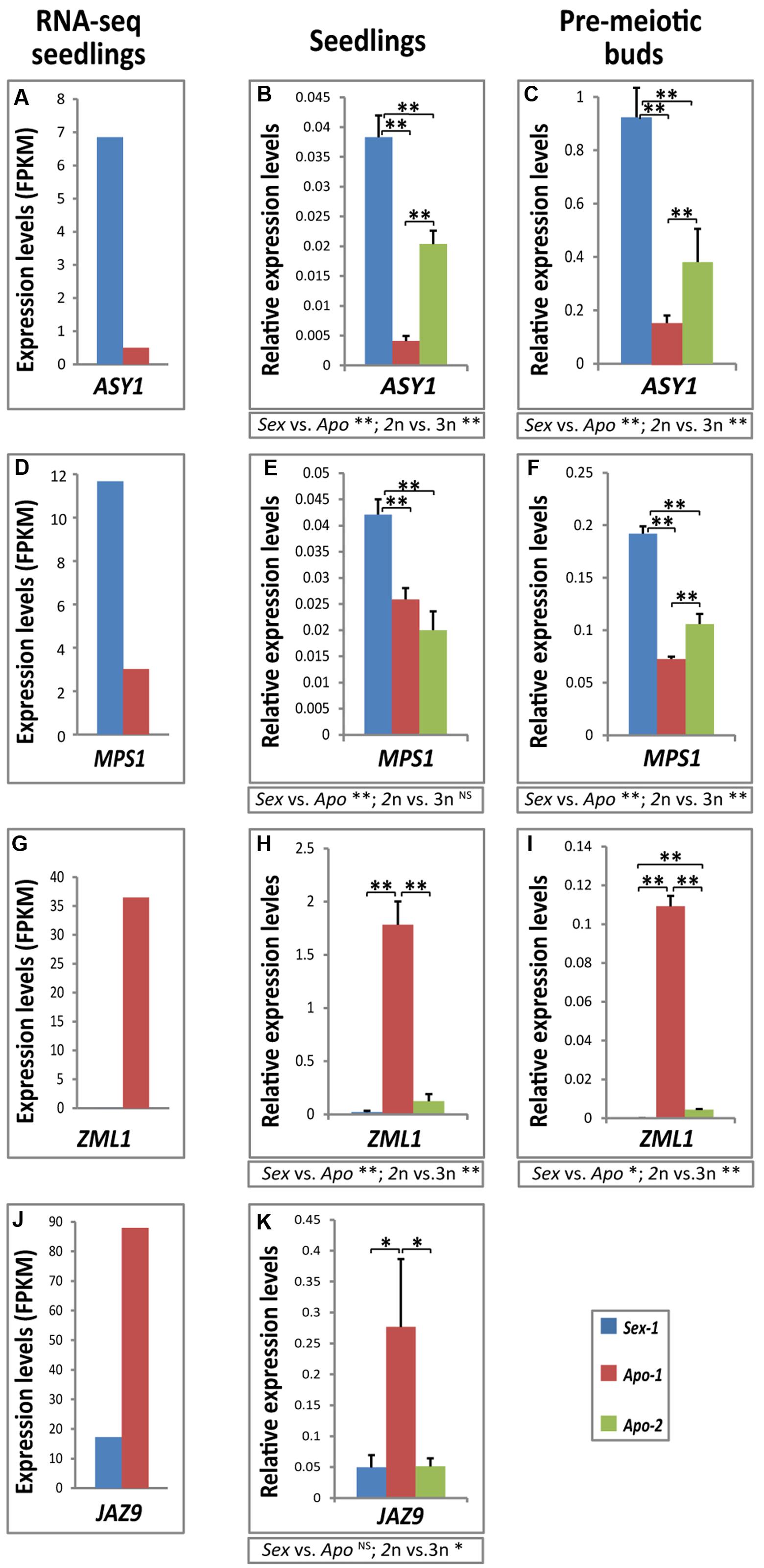
FIGURE 2. Differential real-time quantification of gene expression validates our transcriptome approach. Demonstrated here is down-regulation of homologs of meiotic genes ASY1 (A–C) and MPS1 (D–F), accompanied by upregulation of stress-response genes, a GATA type (ZML1) transcription factor (G–I) and JASMONATE-ZIM-DOMAIN PROTEIN 9 (JAZ9) (J,K). Sex-1 versus Apo-1 seedlings-derived RNA-seq differential expression data (first column) were validated by real-time qRT-PCR (second column), and the expression analyses were further assayed by qRT-PCR in corresponding floral buds of the pre-meiotic MMC-stage (Supplementary Figure S3) (third column). Apo-2 was used as an additional apomictic genotype for qRT-PCR comparisons. FPKM – fragments per kilobase per million. Statistical significance was calculated using t-test; significance levels: ∗∗α ≤ 0.01; ∗α ≤ 0.05; NS, not significant. t-test results for comparisons between groups for reproductive mode and ploidy (Sex vs. Apo, and diploid vs. triploid) are indicated below the respective bar chart.
A careful examination of our dataset indicated that transcription factor deregulation is indeed one of the major components of the apomict/stress-specific transcriptome accounting for about 15% (Supplementary Tables S1 and S2). Whereas members of the AP2/EREBP (APETALA2/ethylene-responsive element binding proteins) transcription factor family predominated equally in both upregulated and down-regulated genes in apomictic seedlings, transcription factors belonging to the NAC [NAC domain: NAM (for NO APICAL MERISTEM), ATAF 1 and 2, and CUC2 (for CUP-SHAPED COTYLEDON 2)], bZIP (BASIC LEUCINE ZIPPER) and MYB (R2R3-type) families were enriched in the upregulated gene lists (Table 2, Supplementary Tables S2 and S4). bHLH (BASIC HELIX-LOOP-HELIX), WRKY (WRKY DNA-BINDING PROTEIN) and C2H2-type Zn-Finger transcription factor families were exclusively abundant in the down-regulated category (Supplementary Table S2). Since the stimulus, hormonal control and transcription factor regulation categories are tightly inter-connected; we examined if stress-related hormonal regulatory features could be identified as specific for Apo-1 seedlings transcriptome. We noticed that nearly one-third of differentially expressed genes (26 and 36% of down-regulated and upregulated, respectively) respond to hormones such as abscisic acid (ABA), auxin, jasmonic acid (JA), ethylene, salicylic acid (SA), gibberellic acid (GA) and cytokinin (Supplementary Table S2). It is interesting to note that genes of the auxin biosynthesis and response [e.g., YUCCA8, CYTOCHROME P450 CYP79B2, INDOLE-3-ACETIC ACID INDUCIBLE 5 (IAA5), IAA19, IAA32], cytokinin response [e.g., CYTOKININ INDEPENDENT 2 (CKI2), TYPE-A RESPONSE REGULATOR (ARR4), ARR5, Arabidopsis HISTIDINE-CONTAINING PHOSPHOTRANSFER FACTOR 5 (AHP5)] regulatory members are generally down-regulated, in contrast to those that belong to biosynthesis and response of ABA [e.g., NINE-CIS-EPOXYCAROTENOID DIOXYGENASE 3 (NCED3); RESPONSIVE TO ABA 18 (RAB18; Figures 2C,D); ABSCISIC ACID RESPONSIVE ELEMENTS-BINDING FACTOR 2 (ABF2), DRE-BINDING PROTEIN 2A (DREB2A), RD26 NAC] and the JA pathway [LIPOXYGENASE 2 (LOX2), FATTY ACID DESATURASE 8 (FAD8), ALLENE OXIDE SYNTHASE (AOS), JAZ9] were upregulated in the apomictic Boechera seedlings (Table 2, Supplementary Table S2). The antagonistic situation between different hormone responses, enrichment of specific hormone regulation categories of ABA and JA, and upregulation of several genes coding for the LATE EMBRYOGENESIS ABUNDANT (LEA)-type storage proteins [e.g., LEA30, RAB18 and LEA28 (Figures 3J–N)] suggest that the apomictic seedlings likely have better stress acclimation than the sexual seedlings, perhaps in a ploidy-dependent manner.
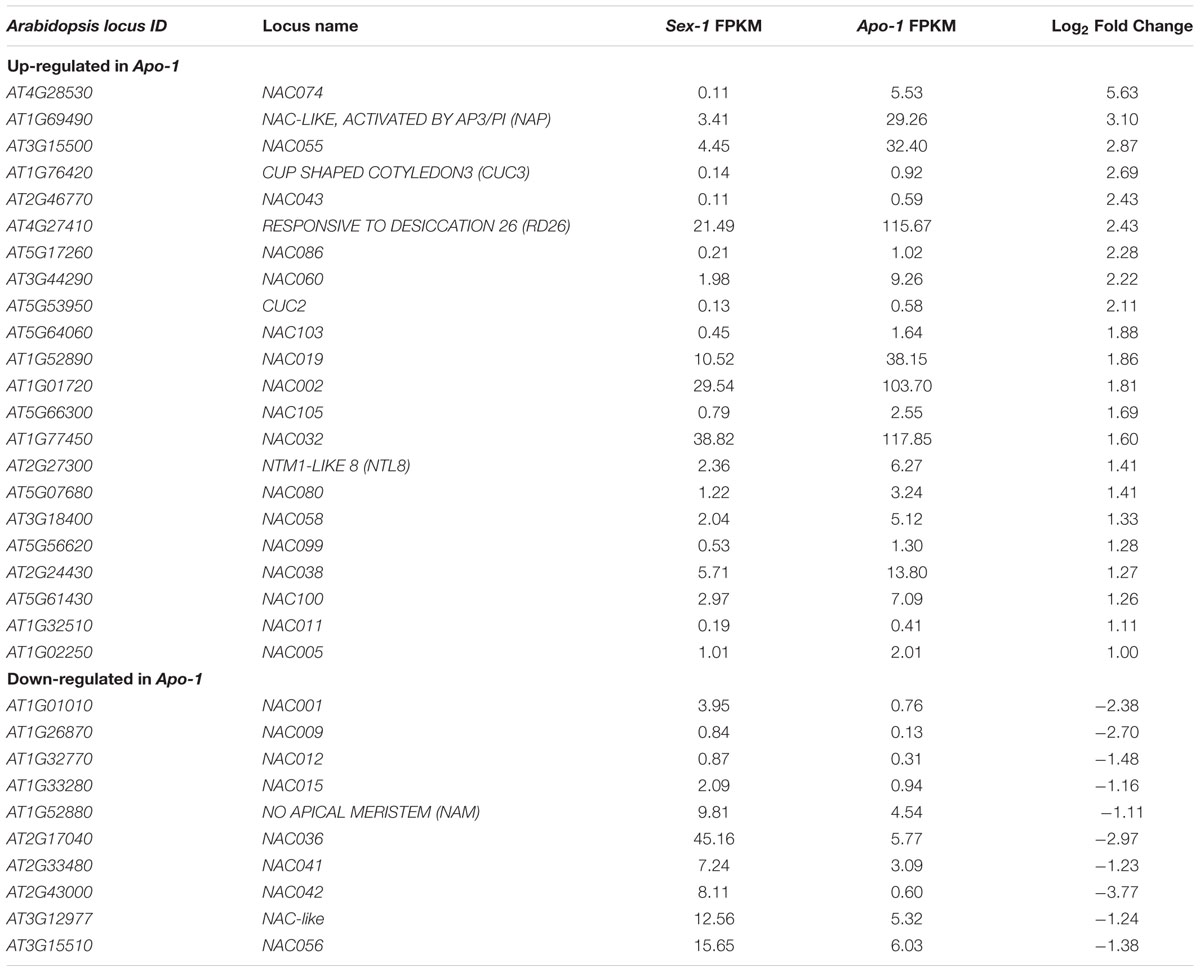
TABLE 2. NAC-DOMAIN CONTAINING (NAC) transcription factor homologs deregulated in apomictic seedlings of Boechera.
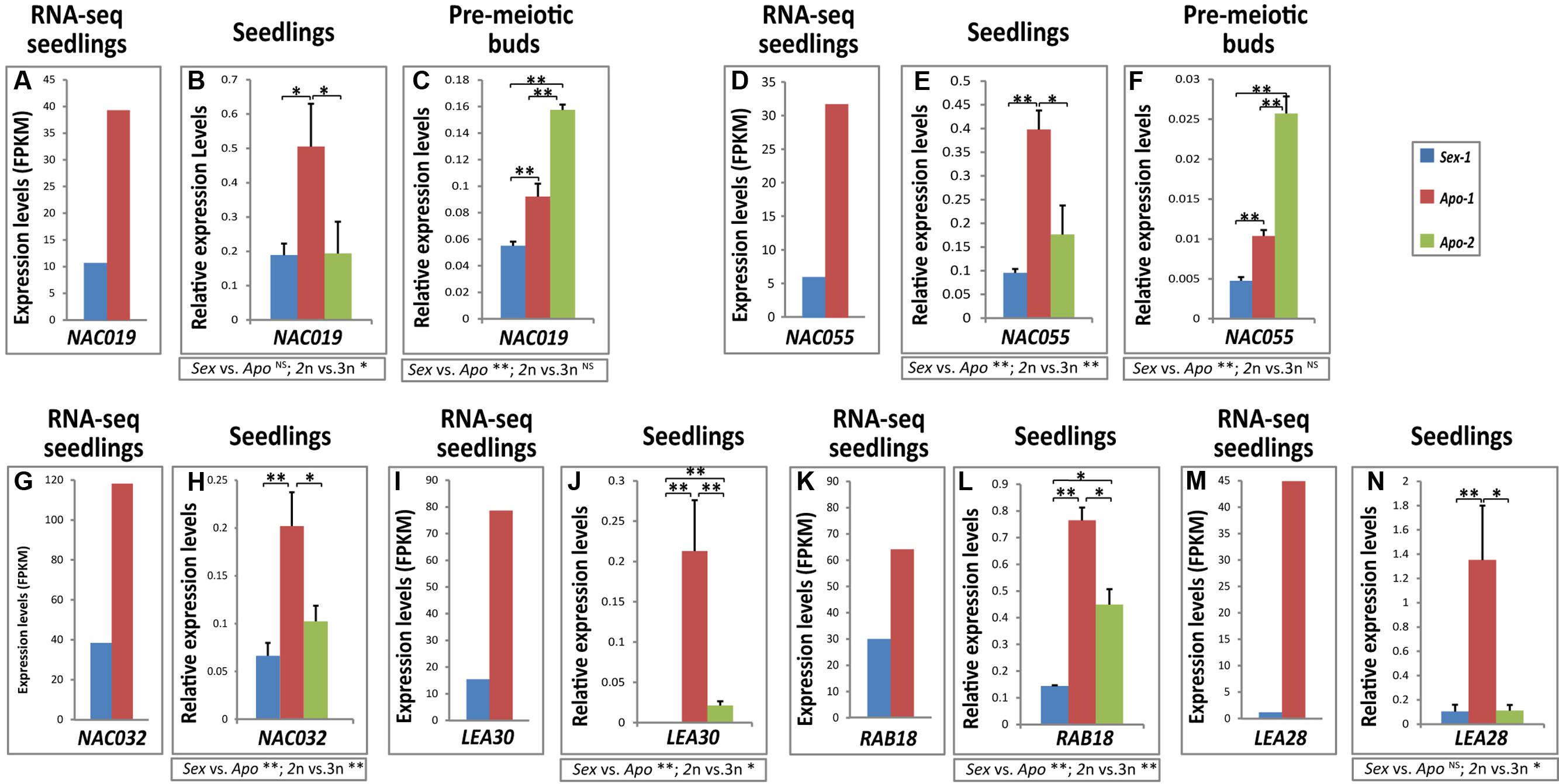
FIGURE 3. Transcripts of stress-related NAC transcription factors and LEA-type storage protein homologs are upregulated in the apomict. Histograms showing real-time qRT-PCR results for (A–C) NAC019, (D–F) NAC055, (G,H) NAC032, (I,J) LEA30, (K,L) RESPONSIVE TO ABA 18 (RAB18) and (M,N) LEA28. The initial RNA-seq data used in validation are graphically represented in (A,D,G,I,K,M). Statistical significance calculated by t-tests is indicated by asterisks. t-test results for comparisons between groups for reproductive mode and ploidy (Sex vs. Apo, and diploid vs. triploid) are indicated below the respective bar chart. Statistical significance levels: ∗∗α ≤ 0.01; ∗α ≤ 0.05; NSnot significant.
Among the several stress-responsive transcription factors identified to be deregulated in the apomict (discussed above), we chose to examine the NAC transcription factors further. Table 2 enlists all the NAC factors uncovered by our RNA-seq approach. Particularly six Boechera protein homologs of the Arabidopsis NACs, NAC019, NAC032, NAC055, RD026, NAP and ATAF1 (Table 2, Supplementary Table S4) that presumably might function as well during apomeiotic transition, could be mapped within a probable regulatory network in Arabidopsis, as pinpointed by the String database (Figure 1)1. We examined if some of these NAC homologs were indeed differentially expressed. Boechera homologs of NAC019 and NAC055 showed significant upregulation both in the apomictic seedlings and in the apomeiotic flower buds of Apo-1 (Figures 3A–F). NAC032 confirmed the trend from transcriptome data and showed upregulation in Apo-1 seedlings as well. Although these genes did not show significant upregulation in the seedlings of Apo-2, the corresponding apomeiotic floral buds of Apo-2 showed stronger enrichment than Sex-1 and Apo-1 (Figures 3C,F) exhibiting a significant expression increase of the average between apomicts. The expression changes for NAC019, NAC055 and NAC032 between Apo-1 and Apo-2 were also found to be significantly different. Although there were certain differences in NAC expression profiles between the apomicts at early developmental stages, which could concern rather difference in ploidy level or genetic background, NAC055 and NAC019 expression in flower buds was not different between ploidy groups but only significantly reproductive-mode-dependent. Together, NAC gene family upregulation may represent a key unifying background for early stress response, in apomictic Boechera species.
To understand stress sensitivity of Boechera species, we carried out in vitro osmotic stress treatments in line with the output from transcriptome analysis of Boechera seedlings featuring stress signatures such as drought, salinity and osmotic stress tolerance. We treated Boechera seedlings with PEG-8000 (polyethylene glycol 8000), using plate assays, generating changes in osmotic balance and therefore decreasing water availability as a major component of drought stress (Verslues et al., 2006). We tested multiple concentrations of PEG for Boechera growth and found that PEG 400 g/L would impose moderate osmotic stress; PEG 550 g/L seemed to be too harsh for the seedlings as most of the exposed plants did not survive well, regardless of accessions (data not shown). Alteration in osmotic conditions had a noticeable root growth effect in Sex-1 roots but not in the apomicts. Primary root elongation of the Sex-1 seedlings treated with PEG 400g/L was found to be smaller by ∼25% in comparison to mock treated Sex-1 seedlings (Figures 4A–C). However, for both Apo-1 and Apo-2 seedlings, no significant difference was observed in primary root lengths in either of the treatments, suggesting the prevalence of a better osmotic stress tolerance mechanism in both apomicts than in the sexual. This finding was further corroborated by the differential response of the sexual and apomicts to salt concentrations that cause both osmotic and ionic stress using 50 mM NaCl which caused a very similar effect decreasing the primary root growth exclusively in the sexual (Figure 4C). Whereas primary root extension was similar in the control treatment across all Boechera accessions, only the sexual exhibited stress-induced effect, and both osmotic and salt stress caused significantly different effect between reproductive groups but that was not ploidy-dependent. In addition to the in vitro assays, we enforced moderate drought on soil grown plants in standard growth room conditions at 22°C to understand if osmotic stress induced by drought impact the overall growth and reproduction of the three genotypes. We analyzed the ratio of reproductive modes in ovules upon drought stress, asking if stress impacted meiosis versus apomeiosis. The proportion of rare sexual-like triads and the routine meiotic tetrads in relation to apomeiotic dyads did not detectably change upon drought exposure in both apomicts (Figure 4). In sum, apomictic seedlings seem to be better acclimated to osmotic and ionic stress in seedlings, but stress does not significantly shift obligate apomeiotic mode to sexual mode during the reproductive phase.
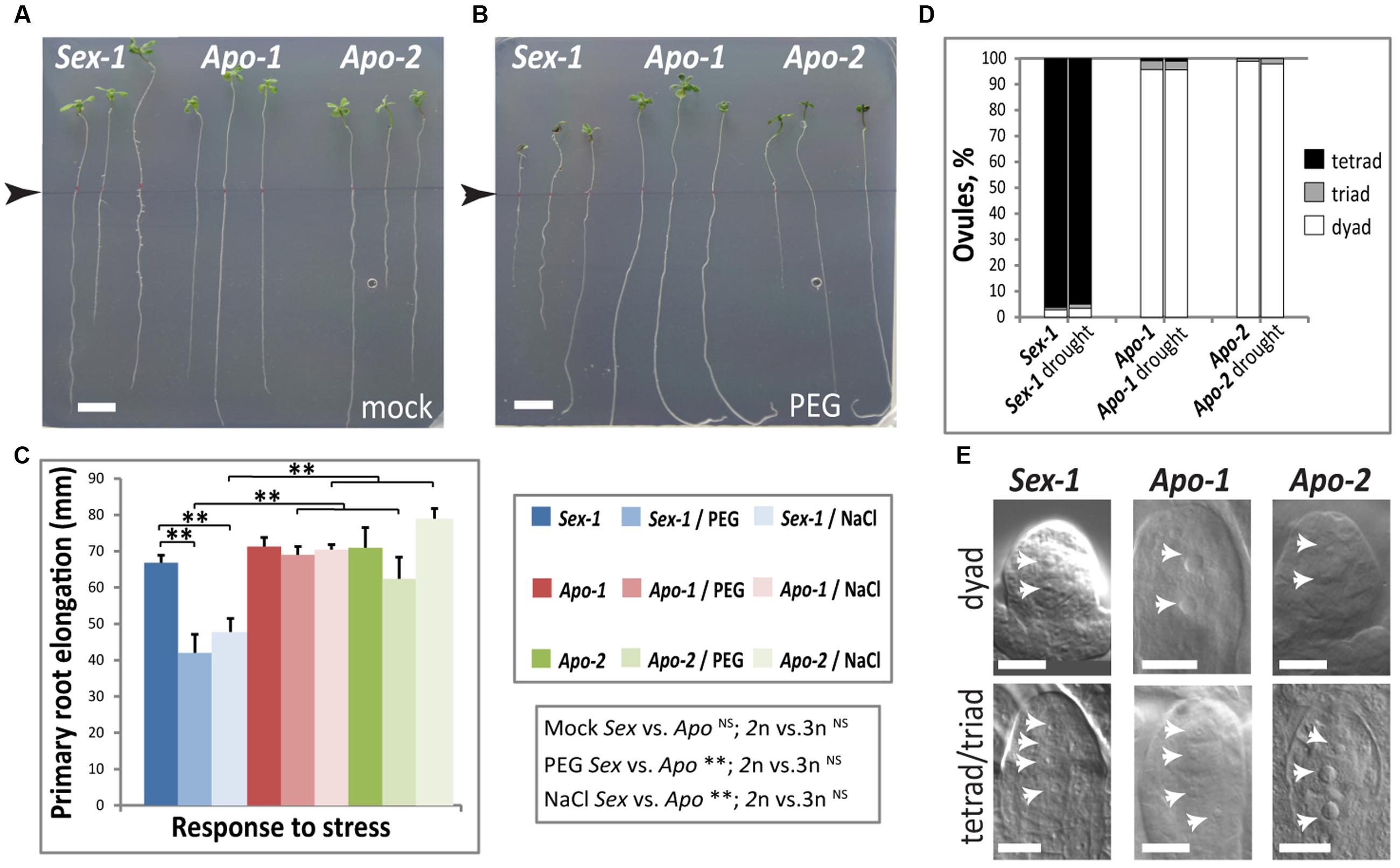
FIGURE 4. Osmotic stress effects on early seedling and reproductive stages of development in Boechera. (A–C) An in vitro primary root elongation assay demonstrates that the apomicts buffer osmotic stress better than the corresponding sexual. Representative photographs of Boechera seedlings grown without (A) and with osmotic stress imposed by an osmoticum, polyethylene glycol (PEG8000, 400 g/L) (B). Root length measurement start points are marked by arrow-heads in both images. (C) Quantification of root elongation in the presence of PEG, and an additional osmoticum NaCl (50 mM). Statistical significance verified by t-test is indicated by asterisks. t-test results for comparisons between groups for reproductive mode and ploidy (Sex vs. Apo, and diploid vs. triploid) are indicated below the respective bar chart. (D) Quantification of meiotic versus apomeiotic events (tetrads and dyads, respectively) in post-meiotic/apo-meiotic ovules of plants that were well-watered (N = 107, 117, 95, respectively) versus those that underwent moderate drought stress (N = 59, 91, 68, respectively), did not reveal obvious differences (E). Representative differential interference contrast (DIC) images of cleared ovules quantified in (D). Scale bars in (A,B) 10 mm, in (E) 10 μm. Statistical significance levels: ∗∗α ≤ 0.01; NSnot significant.
Our transcriptome approach uncovered the parallel alterations between stress and meiosis regulation in the apomictic Boechera. Unpublished data from our lab and meta-analysis of our transcriptome data to large scale databases indicated definite role of methylation machinery in apomixis specific gene expression changes. We indeed observed that the Arabidopsis homologs of several of our stress-specific candidate transcription factors and meiotic genes are likely epigenetically regulated in seedlings via DNA methylation and/or small RNAs, or repressive histone methylation marks (Supplementary Tables S2–S4). In the absence of full genome and methylome sequences of all three species studied here, we conducted a DNA methylation inhibitor assay using a demethylation agent zebularine (Baubec et al., 2009), to identify gene expression changes associated with global DNA methylation. Zebularine exposure resulted in significant effects on all genotypes causing strong decrease in root length (Figure 5A, compare to Figure 4A), due to the genome-wide demethylation effect of this nucleoside combined with its redox-active nature, similar to growth retardation of Arabidopsis when applied in vitro. It must be noted, however, that there was small but significant difference in response upon zebularine treatment; both apomicts retained a slightly better root elongation than the sexual (Figure 5B). We examined the effect of this DNA methylation inhibitor on selected candidate genes by real-time qRT-PCR comparing zebularine-treated versus mock-treated seedling gene expression of meiotic genes: ASY1 and MPS1; and a subset of stress-responsive transcription factors: ZML1, NAC019, NAC032 and NAC055. The first four candidates seem to be regulated by DNA methylation as their expression was affected by zebularine exposure (Figures 5C–E), and both NAC032 and NAC055 did not show similar regulation (data not shown). The qRT-PCR results showed some interesting changes in expression of the candidate genes. MPS1 seems to be repressed by DNA methylation in all species regardless of the reproductive mode, as there was no significant difference in response to zebularine application between the Boechera accessions used (Figure 5B). ASY1, ZML1 and NAC019 expression upon zebularine treatment was either unaltered or down-regulated in the sexual seedlings but they were strongly activated in Apo-1, and ASY1 upregulation seem to be common in both apomicts and different from the sexual (Figures 5C–F). ASY1 and NAC019 expression responses to zebularine were significantly different between species based on two-way ANOVA test, while ZML1 exhibited rather ploidy-dependent effect. Our data suggest locus-specific gene expression changes upon induced global DNA demethylation coupled with cellular stress distinguish between both ploidy groups and apomictic and sexual genotypes.
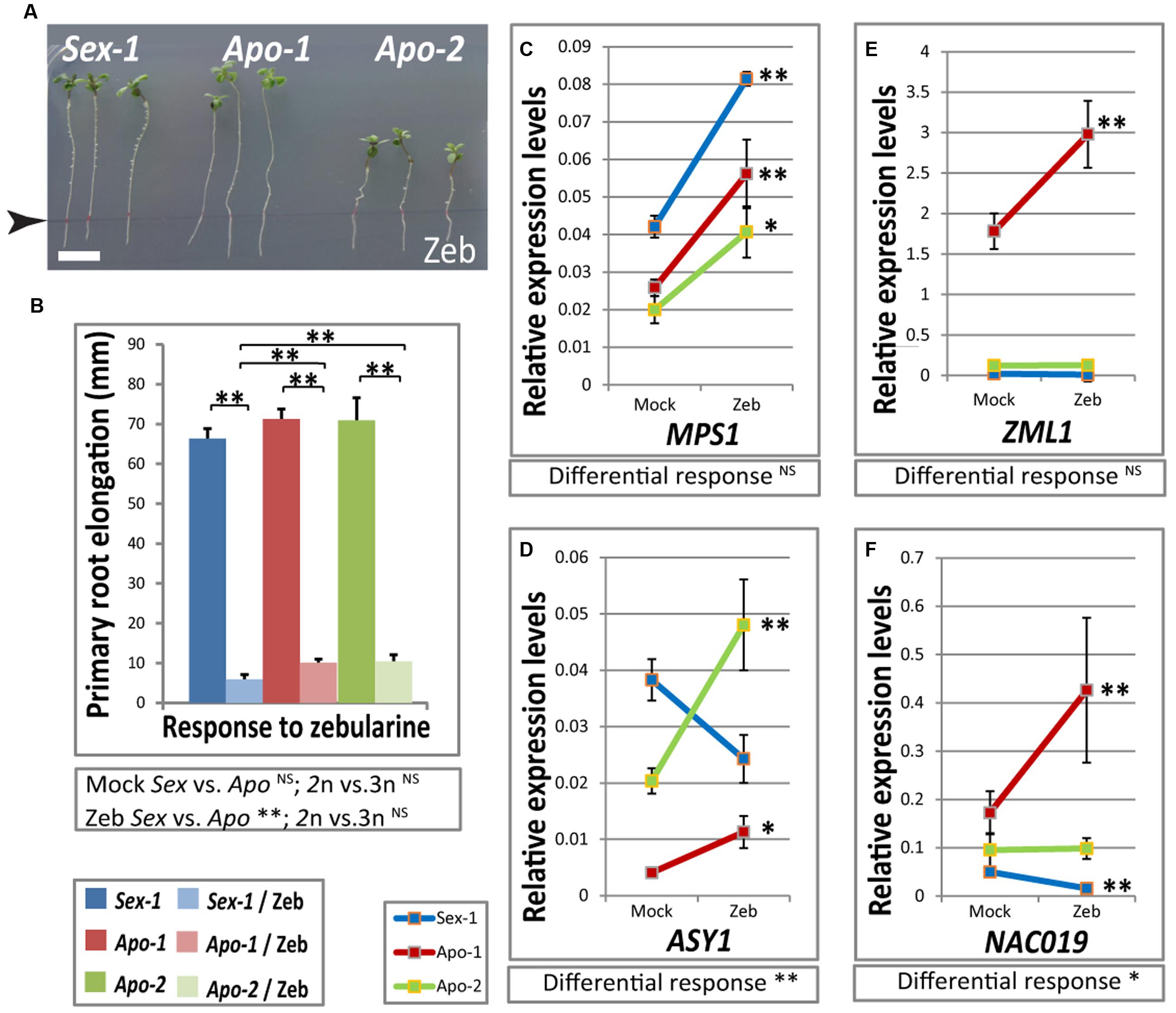
FIGURE 5. DNA de-methylation agent, zebularine, has impact on primary root growth, regulation of meiotic genes and abiotic stress factors in Boechera. (A) A representative photograph of Boechera seedlings upon in vitro application of a DNA methylation inhibitor, zebularine, exhibiting strong root growth retardation (for a comparable representative control, see Figure 4A). Note that zebularine (175 μM) inhibited growth in all three genotypes. (B) Zebularine-treated sexual plants had significantly shorter primary root elongation when compared to that of both apomicts. Statistical significance verified by t-test is indicated by asterisks. t-test results for comparisons between groups for reproductive mode and ploidy (Sex vs. Apo, and diploid vs. triploid) are indicated below the respective bar chart. (C–F) Real-time qRT-PCR-based gene expression of MPS1 (C), ASY1 (D), ZML1 (E) and NAC019 (F) homologs in three Boechera genotypes upon zebularine treatment. Significance of differential statistical response between genotypes verified by two-way ANOVA is given below each graph. Scale bar in (A) 10 mm. Statistical significance levels: ∗∗α ≤ 0.01; ∗α ≤ 0.05; NSnot significant.
Boechera is one of the primary plant model systems to study evolution and regulation of apomixis associated traits such as meiotic circumvention (apomeiosis), apospory, parthenogenesis, polyploidy, hybridity and environment (Roy, 1995; Schranz et al., 2005; Rushworth et al., 2011). Boechera genus in particular comprises of numerous independently emerged asexual (sub)species indicating that this genus may harbor a specifically pronounced (epi)genetic background favorable for evolution of apomixis. Along with polyploid apomicts, Boechera genus also includes diploid apomictic populations that were presumably sired from apomictic polyploids or aneuploids, or occurred via hybridization (Kantama et al., 2007; Lovell et al., 2013). Apomixis in diploid condition is extremely rare, and polyploidy and hybridity seems to have been the springboard for evolution and maintenance of apomixis (Comai, 2005; Hand and Koltunow, 2014; Hojsgaard et al., 2014b). Unlike the polyploid apomictic parents, the rare apomict diploids might display segregation of apomictic traits (Noyes and Wagner, 2014) and fitness trade-offs, hence apomictic diploids do not seem to survive in most apomictic genera. In triploids, where meiotic synapsis of chromosomes fails causing so-called “triploid block,” apomixis remains the only possible mean of reproduction through seeds; to some extent this concerns other polyploids too. However, as most diploid apomicts in Boechera seem to represent products of hybridization (Beck et al., 2012), a similar “genomic block” for meiotic bivalent formation may take place. Yet another route for evolution of a triploid apomict might have been endo-duplication due to triggered stress response following exposure to harsh environmental conditions (Scholes and Paige, 2015). This scenario might have led to diploid (sexual) gamete in one genotype, which was then fertilized by a haploid sexual gamete, thus resulting in triploid genotype. We reported similar endo-duplication events in an Arabidopsis mutant (Johnston et al., 2010), and we believe it was due to genomic stress in the female gamete mediated by loss of a crucial cell cycle regulator. Especially triploids could have paved way for evolution of apomixis by fixing and maintaining heterozygosity. Spontaneous apospory reported in Ranunculus triploids (Hojsgaard et al., 2014a) might lend additional support along this line. In addition, asynchronous gene expression from different genomes is thought to facilitate asexual re-programming in hybrids (Carman, 1997; Hojsgaard et al., 2014a). As hybridity leads to “genomic shock” thus “genomic stress,” we may not be able to tease apart cause and consequence.
Understanding apomixis traits has been mostly restricted to transcriptome analyses of apomixis-specific organs or cell-types, and population genetic studies. For instance, Sharbel et al. (2010) reported heterochronic changes in gene expression during initiation of apomixis along with decrease of their transcript levels in the apomict by comparing expression tags across four stages of ovule development between diploid apomictic and diploid sexual Boechera species. Another study analyzed apomictic cell type-specific (e.g., MMC) gene expression profiles of a triploid apomictic Boechera similar to the one we used in this work, and compared to the corresponding profiles of a distantly related (10 million years of divergence) sexual species A. thaliana (Schmidt et al., 2014). Although number of interesting gene expression changes related to meiotic and mitotic cell cycle regulation, hormonal pathways, signal transduction, stress and epigenetic regulatory pathways were alluded to be specific for the apomeiotic MMC, comparative analyses seem to be constrained due to the lack of appropriate sexual control. It was also noted that all major core meiotic genes were expressed in the apomictic MMC in this triploid apomict, and the authors proposed that diplosporous apomeiosis likely result from modification of the core meiotic and cell division pathway in the MMC. Although some candidate loci underlying apomixis have been isolated or reported, functional analyses of apomixis in Boechera are limited primarily owing to the inherent difficulties in plant transformation of these species.
Environment has always remained a crucial factor with the highest impact on homeostasis and plant life cycle. Whereas most accessions of the model plant A. thaliana are drought-sensitive, the Boechera clade in general and a number of Boechera species in particular seem to have acquired an asset of biotic and abiotic stress-tolerance mechanisms, perhaps due to their predominant ability to frequently hybridize, forming novel species that occupy distinct ecological niches primarily in their native habitat ranges; and due to their repeated exposure to extreme and fluctuating environmental conditions involving major abiotic stress factors like drought, cold and several biotic factors as depicted in Figure 6 (Rushworth et al., 2011). Apomixis is absent in the Arabidopsis clade but prevalent in the Boechera; therefore, it is possible that general stress acclimation could also be an inherent characteristic background for evolution of apomicts, as it might likely provide an efficient (epi)genomic constitution to efficiently tolerate environmental hazards and to buffer mutation loads in the absence of recombination. In parallel, several stress-response genes have been deregulated in transcriptomes of apomictic organs and cell-types of Boechera (our unpublished data; Sharbel et al., 2010; Schmidt et al., 2014). In sum, Boechera represents an excellent model system for identifying molecular cues to gain in-depth understanding of apomixis regulation and its correlation with environmental change and stress response in a diverse genetic background linked to hybridity and polyploidy.
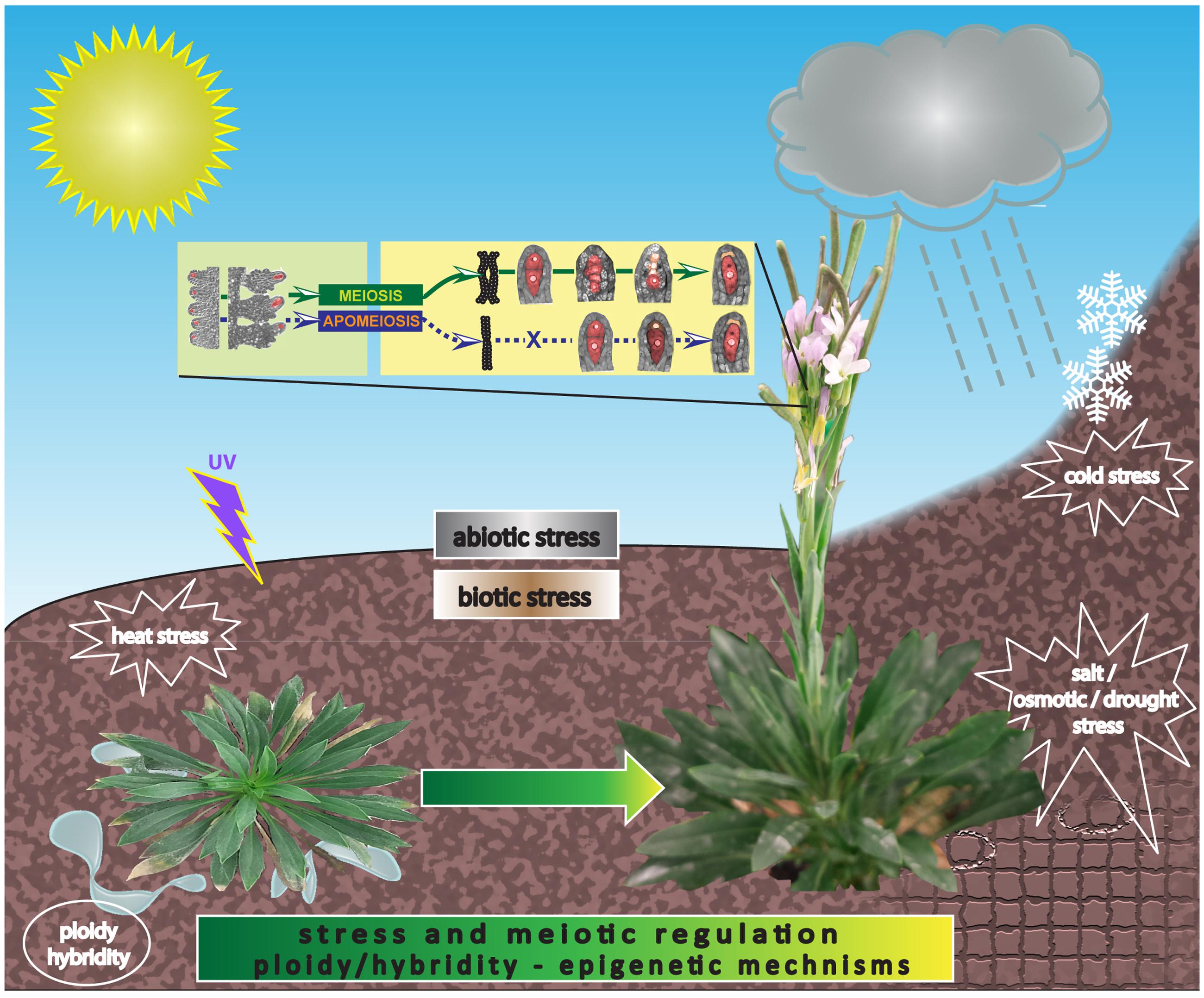
FIGURE 6. An artist’s view illustrating how apomictic Boechera presumably sired from sexual ancestors, were and are exposed to extreme abiotic and biotic conditions in the natural habitat where they evolved. Both genetic and epigenetic control of stress and apomeiotic regulation throughout plant development could be viewed as co-adapted gene responses during stress amelioration acting as an efficient buffer to tolerate the mutation load in the absence of meiotic recombination.
Apomictic Boechera in general seem to be more stress tolerant, at least in terms of osmotic stress acclimation, than the corresponding sexual. The apomicts grown in normal conditions exhibit elevated levels of a considerable number of stress-response genes when compared to the sexual (Supplementary Tables S1 and S4, Table 2, Figure 3). These genes include prominent drought and ABA response factors such as transcription factors NACs, bZIPs, DREB2A, MYBs, and LEA proteins (Nuruzzaman et al., 2013). For example, proper function of bZIP transcription factors AREB1, AREB2 and AREB3 has been shown to be important for drought tolerance (Amir Hossain et al., 2010; Golldack et al., 2011). When overexpressed, Arabidopsis NAC019, NAC055, and NAC072 elevated drought tolerance (Tran et al., 2004).
We noticed though that expression levels of some genes tested were significantly different between the two apomictic accessions used in this study. Both Apo-1 and Apo-2 are obligate diplosporous apomicts, and the main difference between them is their evolutionary history including ploidy level (Apo-1 being triploid and Apo-2 diploid), ecological habitats with different abiotic stress severity, distant geographical location and, therefore, almost certainly distinct parental genotypes. In order to disentangle the ploidy effect on the gene expression, in the absence of available diploid versus polyploid Boechera transcriptome data, we compared our results with published datasets of the Arabidopsis clade. About 4% of the Apo-1-specific upregulated genes showed the same behavior in Arabidopsis allo-tetraploids in an inter-specific hybridization study (Wang et al., 2004), and for stress-related genes it was only 1.8%. Here, three genes NAC019, NAC055 and ZML1 from our validated list were upregulated. Similar changes are not discernible in Arabidopsis auto-tetraploids compared to the corresponding diploid transcriptomes (Pignatta et al., 2010). Therefore, ploidy might not have been the sole cause of genotype-specific transcription differences and shaped up the entire transcriptome of the Apo-1. As Boechera apomicts were presumably sexual hybrids by origin that were eventually fixed by apomixis, hybridity between different genomes, thus genome collisions could have played a role for activation of stress-response genes and initiation of apomixis, as evident from upregulation of genes like NAC019 and NAC055 homologs in both apomictic floral buds. Upregulation of the homologs of stress-related genes in drought-tolerant apomictic Boechera correlates with their crucial function in stress adaptation shown in other plants and support the hypothesis that this constitutive overexpression confers better stress amelioration, in particular osmotic stress tolerance. Our transcriptome data show an intricate network between hormonal regulation and stress response; for instance, preferential activation of several ABA and JA-specific biosynthesis genes and responsive transcription factors in Apo-1 pinpoints that the apomict has an ability to withstand multiple stresses including osmotic stress and drought (Nguyen et al., 2016). Activation of mobile mRNAs of ascorbic peroxidases (APX1 and APX2) that are pivotal for maintaining cellular redox homeostasis by scavenging hydrogen peroxide and ROS, consequent down-regulation of genes of the auxin biosynthesis and response genes, giving thus a possibility to perturb auxin homeostasis, collectively suggest that the apomict Apo-1 has an efficient auxin-dependent growth control in order to cope-up with stress in comparison to Sex-1 (Tognetti et al., 2012; Naser and Shani, 2016), and in turn might activate ABA-specific responses required for stress acclimation. Intriguingly, whereas specific auxin-response factors (ARFs) seem to be down-regulated in seedlings (this study) and many ARFs down-regulated in ovules (Shah et al., unpublished data), they were reported to be enriched in the MMC cells of Apo-1 (Schmidt et al., 2014), perhaps due to the underlying hormonal homeostasis shifts between the seedling, nucellus stages, and the MMC prior apomeiosis. The latter data, however, is based on comparison between MMC cells of Arabidopsis versus Boechera, therefore, further validation would be required. The expression of most genes tested showed significant difference between the sexual and apomicts, with ploidy effect being significant at many instances; yet the effect of stress on in vitro growth was clearly dependent on the reproductive mode of the Boechera species but not the ploidy level. In sum, whether it is Apo-1, or the evolutionarily distinct Apo-2, there is a clear trend that the Boechera apomictic seedlings confer higher level of stress acclimation and underlying gene response in seedlings than the sexual counterpart, and likely play a pivotal role in sustaining apomixis through to apomeiotic transition.
Apomeiotic switch has been perceived as a developmental reprogramming event connected to meiotic shunt, stress, epigenetic and transcriptional changes, transposon control etc. (reviewed in Grimanelli, 2012; Horandl and Hadacek, 2013; Mercier et al., 2015). Previous work in Arabidopsis has indicated the importance of meiotic mutations in producing unreduced gametes by apomeiosis-like program. For instance, DYAD/SWITCH1 (SWI1) is an important candidate gene in Arabidopsis reported for switching the meiosis program toward apomeiosis (Ravi et al., 2008). In MiMe-1 and MiMe-2 triple mutants, a diplospory-like division also leads to the formation of unreduced gametes. By combining different meiotic mutant combinations [(MiMe-1: sporulation11-1 (spo11-1), omission of second division1 (osd1) and recombination8 (rec8); MiMe-2: spo11-1, osd1 and cyc1;2/tardy asynchronous meiosis (tam)], diplospory-like meiotic restitution have been successfully recreated in Arabidopsis (d’Erfurth et al., 2009, 2010). In maize, a mutation in an effector of small RNA pathway has been shown to be important for recreating diplospory-like event (Singh et al., 2011). Whereas role of these genes or mutations underlying apomixis in a natural apomict like Boechera has not yet been documented, Arabidopsis versus Boechera (Apo-1) MMC comparisons suggested possible down-regulation of certain meiotic genes: PARTING DANCERS, SPO11-2, TAM, SWI1, SOLO DANCERS and RAD50 (Schmidt et al., 2014). Whereas this data need real-time validation, none of these genes were deregulated in our transcriptome of Apo-1 versus Sex-1 seedlings, but deregulation of two other meiotic genes were noteworthy. Acute down-regulation of core meiotic homologs such as ASY1 and MPS1 both in seedlings and apomeiotic buds gives a hint that a global alteration in expression of these genes are likely necessary in order to make the apomeiotic switch. ASY1 is a Horma-domain containing DNA-binding protein component of the meiotic axis involved in synapsis and crossover formation, and it is required for DMC1-mediated interhomolog recombination during female and male meiosis (Armstrong and Jones, 2001; Sanchez-Moran et al., 2007). MPS1 is a coiled-coil protein involved in meiotic spindle formation and it is required for faithful chromosome segregation during female and male meiosis (Jiang et al., 2009). Interestingly, none of the meiotic genes we identified in our study (Table 1, Supplementary Table S3) showed transcriptional changes in transcriptomes of Arabidopsis polyploids or hybrids (Wang et al., 2004; Yu et al., 2010). Could evolutionary history traits in Boechera different from that of Arabidopsis influence expression of these genes? It is also likely that this transcriptional homeostasis reflects a genome-wide change in apomictic Boechera and might involve heritable epigenetic silencing.
Abiotic stress has been shown to affect meiotic progression, whereas different type, severity and duration of the stress factors may have different effects on reproduction (reviewed in De Storme and Geelen, 2014). In addition, how stress affects meiosis could be different between male and female tissues. As female tissues are more somatically dependent than the male organs, for instance, female meiosis seems to progress during water deficit but male meiosis abort in Arabidopsis (Saini, 1997). In barley, heat stress leads to precocious onset of meiotic chromosome condensation during male meiosis and a concomitant ASY1 upregulation (Oshino et al., 2007). In Arabidopsis seedlings, ASY1 seems to be marginally down-regulated during drought, and ASY1 expression levels seem to fluctuate inconsistently between time points during prolonged stress condition (Kilian et al., 2007). The same is true for MPS1 upon light stress, and for DYAD during cold, drought, and heat; therefore, a link between stress response and meiotic machinery may exist much earlier than onset of reproduction itself. However, direct connections between stress response and regulation of meiotic genes in apomicts have not been elaborated earlier. The stress-co-adapted and highly heterozygous apomictic genomes, similarly to polyploids (Chandra and Dubey, 2010; Coate et al., 2013; Hao et al., 2013; Syamaladevi et al., 2016), generally are considered to be a haven for buffering mutation load in the absence of meiotic recombination (Lynch, 1984). Therefore, not only those apomictic plants are well prepared for stress, they also feature “apomeiotic” mutations or down-regulation of genes such as MPS1 and ASY1 right from the very growth of the apomictic plant. The broadly adapted apomicts may more efficiently buffer mild abiotic stress than the sexuals, thus limiting cellular oxidative stress and formation of ROS and resulting DNA damage, hence, there is no need for meiosis and recombination in the apomict [sensu. FAR theory (Hadany and Beker, 2003a,b) and oxidative damage initiation hypothesis or meiosis (Bernstein and Bernstein, 2013; Horandl and Hadacek, 2013)]. In agreement with this, in our experimental setting, we observed neither reproducible transcriptional changes of Boechera homologs of MPS1 and ASY1 in seedlings upon osmotic stress (data not shown) nor reversion of apomeiosis to sexual-like meiosis. Recent preliminary reports indicate that a facultative apomictic Boechera species might incline toward sexual meiosis in a substantial fraction of ovules under drought and heat (Carman et al., 2015). These results are also reminiscent of similar observations in a facultative apomict Ranunculus under extended photoperiod (Klatt et al., 2016), and collectively hint that residual sexuality contained in the facultative apomicts could be triggered upon environmental cues. In light of our observation that obligate apomictic Boechera accessions exposed to mild drought did not exhibit any significant reversion to sexuality in our conditions, we propose that additional stress may not impinge on the reproductive mode of obligate apomicts that were already co-adapted for stress acclimation and meiotic circumvention.
Among several transcription factor groups that are predominantly transcriptionally activated in the stress-tolerant apomict, we have noticed upregulation of the family of NAC transcription factors, which have previously been reported to work in tandem with each other (Hickman et al., 2013). This reproduction mode-specific activation extends beyond seedling to reproductive phase, proposing the prime importance of this regulatory network during apomixis. Stress-response and developmental role of NACs have previously been discussed in various plant systems (Meng et al., 2009; Nuruzzaman et al., 2013; You et al., 2015). It is interesting to note that NAC genes chosen as candidates from our RNA-seq (NAC019, NAC032, NAC055) are also known to be expressed in specific cell-types and ovules in Arabidopsis (Yu et al., 2005; Johnston et al., 2007; Wuest et al., 2010; Schmidt et al., 2011; Schmid et al., 2012). Although the micro-dissected RNA-seq did not give consistent presence calls for several NACs in laser-captured single cell transcriptomes presumably due to experimental noise, NAC032, NAC002 and NAC103 homologs of Boechera were consistently present in Apo-1 apomeiotic cell types such as the MMC and surrounding ovular cells (Schmidt et al., 2014) (Supplementary Table S4). A recent work identified novel role of NAC family members in ovule separation (Goncalves et al., 2015), thus in addition to stress regulation, NACs might govern early reproductive development. A number of NACs are also known for their non-cell-autonomous movement via movement of their mobile mRNA (Thieme et al., 2015); therefore, one possibility may be that they maternally control decisions on sexual or apomictic reproductive development.
Epigenetic regulation involving DNA methylation and histone modifications has long been recognized as a potent regulator of stress-response in plants (reviewed in Probst and Mittelsten Scheid, 2015; Bilichak and Kovalchuk, 2016). Whereas the underlying regulation is extremely complex, a two-step model proposed for transcriptional activation of stress-response genes includes first DNA methylation in CG context, which then directs H3K9 methylation recruiting non-CG methylation (reviewed in Du et al., 2015; Probst and Mittelsten Scheid, 2015). Whereas DNA hypermethylation of stress-inducible genes might lead to enrichment of H3K9me2 and a concomitant depletion of histone acetylation, as shown for Arabidopsis seedlings under saline stress (Bilichak et al., 2012), drought activated genes are associated with elevation of H3K4 trimethylation and H3K9 acetylation (Kim et al., 2008). However, epigenetic specificity between DNA and histone methylation under stress might vary between genes, as shown for salt-responsive transcription factors in soybean (Song et al., 2012). A review of the previous data showing locus-specific DNA and histone methylation marks, and small RNAs in Arabidopsis (Supplementary Table S4) suggests that the corresponding Boechera loci might be under complex epigenetic control. Specifically, NACs, MYBs, and several other genes coding for stress-related transcription factors in different plant species have been shown to be targets of miRNAs (Rhoades et al., 2002; Guo et al., 2005), a class of small RNAs involved in post-transcriptional gene silencing and, in many of these cases, induced by stress. In light of these previous findings, our data of Boechera NAC019 being significantly deregulated in response to global demethylation, supports an underlying epigenetic regulation in the apomicts. The Arabidopsis NAC019 was previously shown to be activated upon allo-polyploidization in large-scale transcriptomic datasets (Wang et al., 2004), supporting the hybridization effect also in Boechera. As Boechera apomicts studied here likely represent allopolyploid and diploid hybrids, it is important to consider the importance of epigenetic landscape of the entire genome to be successful apomicts. Global changes in DNA methylation, RdDM-dependent transcriptional changes etc. would be important for the hybrid/polyploid genome to perpetuate apomixis in plant populations (reviewed in Verhoeven and Preite, 2014). Besides, the heterozygous genome and epigenetic landscape instigated by hybridity or polyploidy is compatible with the efficient stress ameliorative advantages in the apomicts, as discussed above.
In addition to the previous data that apomixis is largely epigenetically controlled via pathways of DNA methylation in several apomictic plant systems (Garcia-Aguilar et al., 2010; Zappacosta et al., 2014; reviewed in Hand and Koltunow, 2014), and also by observing the fact that DNA methylation pathway members are deregulated in Boechera apomictic cell types (Schmidt et al., 2014) and generally at different developmental stages of Boechera apomicts (unpublished data), our data indicating control of MPS1 and ASY1 by DNA methylation highlights the importance of an epigenetic angle underlying meiotic deregulation. In this work, we have provided indirect evidence that global DNA methylation might possibly control the apomict-specific transcriptional regulation of stress and meiotic genes in the seedlings in a ploidy-independent manner, though this effect could be coupled to a rather marginal DNA-damaging effect of zebularine (Liu et al., 2015). It is important to note that correlation of DNA methylation status with expression is complex and modifications specific to each cytosine will have to be mapped in the future by locus-specific bisulfite sequencing. Nonetheless, our work provides a base to further investigate the connection between genome-wide and locus-specific DNA methylation and gene expression alterations in Boechera species and its possible correlation with the apomixis program. It will also be interesting to examine how epigenetics, the redox status and DNA breaks play a role in regulation of meiotic genes like ASY1. Are there connection between apomict-dependent ASY1 regulation, and SPO11 (Horandl and Hadacek, 2013), which are critical for double strand break repair and chromosome pairing, and may their expression be induced by DSB-dependent signaling network at different developmental stages? The decreased levels of Boechera MPS1 and ASY1 expression starting from seedling stage along with constitutive stress response in the apomicts allow us to propose that apomictic initiation is pre-programmed from early plant development onward, perhaps due to global (epi)genome-wide deregulation and resulting changes in the transcriptome. Taken together, our work forms one of the first glimpses to view apomixis as a possible read-out of stress-related genome- and transcriptome-wide alterations. Whereas previous studies in Boechera have given importance to gene expression changes that occurred during the apomeiotic shift or parthenogenetic reprogramming during the reproductive phase, our work for the first time examines the earliest molecular events during the vegetative phase of the apomicts, and provides a transcriptomic resource for targeted work. The dynamic changes connecting these factors in evolutionary timeline have possibly resulted in genome-wide transcriptome adjustments uncovered in Boechera. The work has hinted the importance of the evolutionary history, constitutive stress response elevation, meiotic gene deregulation for priming apomixis in the apomictic species, and we anticipate that this phenomenon will likely be a common background for most, if not all apomicts, similar to the proposals in the apomeiotic dandelion (Verhoeven and van Gurp, 2012). Not only in plants, stress and/or epigenetics are important denominators that impinge on asexual reproductive mode in animals too, for instance in insects and lizards (Kearney et al., 2006; Jedlicka et al., 2015) and in mice (Kono et al., 2004). Nature must have dictated co-evolution of these mechanisms across kingdoms.
AJ conceived of the project, and both AJ and JM supervised the wet lab and bioinformatics work, respectively. Lab experiments described here were performed by: JS, OK, PP, and MT; NGS sequencing was performed by Fasteris, Switzerland. Bioinformatic analyses were conducted by JS and JM; annotations, data comparisons, figures and tables were prepared by JS and OK. The manuscript was drafted by JS, OK, and AJ, and all authors read and approved of the content.
We are grateful for funding from German Science Foundation (DFG) and University of Heidelberg to AJ, and financial support to JM by J. Wittbrodt (University of Heidelberg). Acknowledged are additional support from Alexander von Humbodt Foundation to AJ, a DAAD fellowship to P. Sarhanova, a Landesgraduiertenförderung (LGF) fellowship to JS, and general support from IPK-Gatersleben. We acknowledge the financial support of the Deutsche Forschungsgemeinschaft and Ruprecht-Karls-Universität Heidelberg within the funding programme Open Access Publishing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Sincere thanks to B. Roy and E. Schranz for seed donations. P. Sarhanova and A. Busching are thanked for bulking of Boechera accessions and technical help with flow-cytometry-based screening (IPK); T. Schmutzer for advice on data analyses, H. Baeumlein for sharing lab facilities (IPK), F. Blattner and J. Kumlehn for additional interactions. AJ thanks, J. Wittbrodt and J. Lohmann for a dedicated growth facility and their support, T. Holstein for sharing lab facilities (Heidelberg). Additional thanks are due to D. Larsen for various help, A. Cornean and K. Gross for assistance with RNA isolation, S. Schmitt (DKFZ, Heidelberg) for the flow-cytometry application and his helpful suggestions, and K. Piiper for advices on growth room management (Heidelberg). We thank the reviewers for insightful comments that helped to improve the manuscript.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01539
Alexander, P. J., Windham, M. D., Beck, J. B., and Bailey, C. D. (2013). Molecular phylogenetics and taxonomy of the genus Boechera and related genera (Brassicaceae: Boechereae). Syst. Bot. 38, 192–209. doi: 10.1600/036364413X661917
Aliyu, O. M., Schranz, M. E., and Sharbel, T. F. (2010). Quantitative variation for apomictic reproduction in the genus Boechera (Brassicaceae). Am. J. Bot. 97, 1719–1731. doi: 10.3732/ajb.1000188
Al-Shehbaz, I. A., Beilstein, M. A., and Kellogg, E. A. (2006). Systematics and phylogeny of the Brassicaceae (Cruciferae): an overview. Plant Syst. Evol. 259, 89–120. doi: 10.1007/s00606-006-0415-z
Amir Hossain, M., Lee, Y., Cho, J. I., Ahn, C. H., Lee, S. K., Jeon, J. S., et al. (2010). The bZIP transcription factor OsABF1 is an ABA responsive element binding factor that enhances abiotic stress signaling in rice. Plant Mol. Biol. 72, 557–566. doi: 10.1007/s11103-009-9592-9
Andrews, S. (2010) FastQC: A Quality Control Tool for High Throughput Sequence Data. Available at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Archetti, M. (2004). Recombination and loss of complementation: a more than two-fold cost for parthenogenesis. J. Evol. Biol. 17, 1084–1097. doi: 10.1111/j.1420-9101.2004.00745.x
Armstrong, J. S., and Jones, H. G. (2001). Female meiosis in wild-type Arabidopsis thaliana and in two meiotic mutants. Sex. Plant Reprod. 13, 177–183. doi: 10.1007/s004970000050
Baubec, T., Pecinka, A., Rozhon, W., and Mittelsten Scheid, O. (2009). Effective, homogeneous and transient interference with cytosine methylation in plant genomic DNA by zebularine. Plant J. 57, 542–554. doi: 10.1111/j.1365-313X.2008.03699.x
Beck, J. B., Alexander, P. J., Allphin, L., Al-Shehbaz, I. A., Rushworth, C., Bailey, C. D., et al. (2012). Does hybridization drive the transition to asexuality in diploid Boechera? Evolution 66, 985–995. doi: 10.1111/j.1558-5646.2011.01507.x
Bernstein, H., and Bernstein, C. (2013) Evolutionary origin and adaptive function of meiosis. in Meiosis, ed. C. Bernstein (Rijeka: InTech). doi: 10.5772/56557
Bicknell, R. A., and Koltunow, A. M. (2004). Understanding apomixis: recent advances and remaining conundrums. Plant Cell 16: S228-S245. doi: 10.1105/tpc.017921
Bilichak, A., Ilnystkyy, Y., Hollunder, J., and Kovalchuk, I. (2012). The progeny of Arabidopsis thaliana plants exposed to salt exhibit changes in DNA methylation, histone modifications and gene expression. PLoS ONE 7:e30515. doi: 10.1371/journal.pone.0030515
Bilichak, A., and Kovalchuk, I. (2016). Transgenerational response to stress in plants and its application for breeding. J. Exp. Bot. 67, 2081–2092. doi: 10.1093/jxb/erw066
Böcher, T. W. (1951). Cytological and embryological studies in the amphi-apomictic Arabis holboellii complex. Kong Dansk Vidensk. Selsk. Biol. Skr. 6, 1–57.
Campos, P. R., and Wahl, L. M. (2010). The adaptation rate of asexuals: deleterious mutations, clonal interference and population bottlenecks. Evolution 64, 1973–1983. doi: 10.1111/j.1558-5646.2010.00981.x
Carman, J. G. (1997). Asynchronous expression of duplicate genes in angiosperms may cause apomixis, bispory, tetraspory, and polyembryony. Biol. J. Linn. Soc. 61, 51–94. doi: 10.1111/j.1095-8312.1997.tb01778.x
Carman, J. G., de Arias, M. M., Nelson, S. M., Zhao, X., Gao, L., Srivastava, M., et al. (2015). “Hot on the trail of the sex-apomixis switch in Boechera (Brassicaceae),” in Proceedings of the Plant and Animal Genome Conference (PAG XXIV), San Diego, CA.
Chandra, A., and Dubey, A. (2010). Effect of ploidy levels on the activities of delta(1)-pyrroline-5-carboxylate synthetase, superoxide dismutase and peroxidase in Cenchrus species grown under water stress. Plant Physiol. Biochem. 48, 27–34. doi: 10.1016/j.plaphy.2009.10.003
Charlesworth, B. (2006). The evolutionary biology of sex. Curr. Biol. 16: R693–R695. doi: 10.1016/j.cub.2006.08.023
Chomczynski, P. (1993). A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15: 532–534, 536–537.
Coate, J. E., Powell, A. F., Owens, T. G., and Doyle, J. J. (2013). Transgressive physiological and transcriptomic responses to light stress in allopolyploid Glycine dolichocarpa (Leguminosae). Heredity (Edinb) 110, 160–170. doi: 10.1038/hdy.2012.77
Comai, L. (2005). The advantages and disadvantages of being polyploid. Nat. Rev. Genet. 6, 836–846. doi: 10.1038/nrg1711
Conner, J. A., Mookkan, M., Huo, H., Chae, K., and Ozias-Akins, P. (2015). A parthenogenesis gene of apomict origin elicits embryo formation from unfertilized eggs in a sexual plant. Proc. Natl. Acad. Sci. U.S.A. 112, 11205–11210. doi: 10.1073/pnas.1505856112
Dawson, K. J. (1995). The advantage of asexual reproduction – when is it 2-fold. J. Theor. Biol. 176, 341–347. doi: 10.1006/jtbi.1995.0203
de Massy, B. (2013). Initiation of meiotic recombination: how and where? Conservation and specificities among eukaryotes. Annu. Rev. Genet. 47, 563–599. doi: 10.1146/annurev-genet-110711-155423
De Storme, N., and Geelen, D. (2014). The impact of environmental stress on male reproductive development in plants: biological processes and molecular mechanisms. Plant Cell Environ. 37, 1–18. doi: 10.1111/pce.12142
d’Erfurth, I., Cromer, L., Jolivet, S., Girard, C., Horlow, C., Sun, Y., et al. (2010). The cyclin-A CYCA1;2/TAM is required for the meiosis I to meiosis II transition and cooperates with OSD1 for the prophase to first meiotic division transition. PLoS Genet. 6:e1000989. doi: 10.1371/journal.pgen.1000989
d’Erfurth, I., Jolivet, S., Froger, N., Catrice, O., Novatchkova, M., and Mercier, R. (2009). Turning meiosis into mitosis. PLoS Biol. 7:e1000124. doi: 10.1371/journal.pbio.1000124
Desai, M. M., Fisher, D. S., and Murray, A. W. (2007). The speed of evolution and maintenance of variation in asexual populations. Curr. Biol. 17, 385–394. doi: 10.1016/j.cub.2007.01.072
Du, J., Johnson, L. M., Jacobsen, S. E., and Patel, D. J. (2015). DNA methylation pathways and their crosstalk with histone methylation. Nat. Rev. Mol. Cell Biol. 16, 519–532. doi: 10.1038/nrm4043
Eshel, I., and Feldman, M. W. (1970). On the evolutionary effect of recombination. Theor. Popul. Biol. 1, 88–100. doi: 10.1016/0040-5809(70)90043-2
Eyres, I., Boschetti, C., Crisp, A., Smith, T. P., Fontaneto, D., Tunnacliffe, A., et al. (2015). Horizontal gene transfer in bdelloid rotifers is ancient, ongoing and more frequent in species from desiccating habitats. BMC Biol. 13:90. doi: 10.1186/s12915-015-0202-9
Fisher, R. A. (1930). The Genetical Theory of Natural Selection. Oxford: Clarendon Press. doi: 10.5962/bhl.title.27468
Franzke, A., Lysak, M. A., Al-Shehbaz, I. A., Koch, M. A., and Mummenhoff, K. (2011). Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci. 16, 108–116. doi: 10.1016/j.tplants.2010.11.005
Gallas, G., and Waters, E. R. (2015). Boechera species exhibit species-specific responses to combined heat and high light stress. PLoS ONE 10:e0129041. doi: 10.1371/journal.pone.0129041
Garcia-Aguilar, M., Michaud, C., Leblanc, O., and Grimanelli, D. (2010). Inactivation of a DNA methylation pathway in maize reproductive organs results in apomixis-like phenotypes. Plant Cell 22, 3249–3267. doi: 10.1105/tpc.109.072181
Gerrish, P. J., and Lenski, R. E. (1998). The fate of competing beneficial mutations in an asexual population. Genetica 102-103, 127–144. doi: 10.1023/A:1017067816551
Golldack, D., Luking, I., and Yang, O. (2011). Plant tolerance to drought and salinity: stress regulating transcription factors and their functional significance in the cellular transcriptional network. Plant Cell Rep. 30, 1383–1391. doi: 10.1007/s00299-011-1068-0
Goncalves, B., Hasson, A., Belcram, K., Cortizo, M., Morin, H., Nikovics, K., et al. (2015). A conserved role for CUP-SHAPED COTYLEDON genes during ovule development. Plant J. 83, 732–742. doi: 10.1111/tpj.12923
Grimanelli, D. (2012). Epigenetic regulation of reproductive development and the emergence of apomixis in angiosperms. Curr. Opin. Plant Biol. 15, 57–62. doi: 10.1016/j.pbi.2011.10.002
Guo, H. S., Xie, Q., Fei, J. F., and Chua, N. H. (2005). MicroRNA directs mRNA cleavage of the transcription factor NAC1 to downregulate auxin signals for arabidopsis lateral root development. Plant Cell 17, 1376–1386. doi: 10.1105/tpc.105.030841
Hadany, L., and Beker, T. (2003a). Fitness-associated recombination on rugged adaptive landscapes. J. Evol. Biol. 16, 862–870. doi: 10.1046/j.1420-9101.2003.00586.x
Hadany, L., and Beker, T. (2003b). On the evolutionary advantage of fitness-associated recombination. Genetics 165, 2167–2179.
Hand, M. L., and Koltunow, A. M. (2014). The genetic control of apomixis: asexual seed formation. Genetics 197, 441–450. doi: 10.1534/genetics.114.163105
Hao, G. Y., Lucero, M. E., Sanderson, S. C., Zacharias, E. H., and Holbrook, N. M. (2013). Polyploidy enhances the occupation of heterogeneous environments through hydraulic related trade-offs in Atriplex canescens (Chenopodiaceae). New Phytol. 197, 970–978. doi: 10.1111/nph.12051
Hedhly, A., Hormaza, J. I., and Herrero, M. (2009). Global warming and sexual plant reproduction. Trends Plant Sci. 14, 30–36. doi: 10.1016/j.tplants.2008.11.001
Herrero, M. P., and Johnson, R. R. (1980). High-temperature stress and pollen viability of maize. Crop Sci. 20, 796–800. doi: 10.2135/cropsci1980.0011183X002000060030x
Hickman, R., Hill, C., Penfold, C. A., Breeze, E., Bowden, L., Moore, J. D., et al. (2013). A local regulatory network around three NAC transcription factors in stress responses and senescence in Arabidopsis leaves. Plant J. 75, 26–39. doi: 10.1111/tpj.12194
Hojsgaard, D., Greilhuber, J., Pellino, M., Paun, O., Sharbel, T. F., and Horandl, E. (2014a). Emergence of apospory and bypass of meiosis via apomixis after sexual hybridisation and polyploidisation. New Phytol. 204, 1000–1012. doi: 10.1111/nph.12954
Hojsgaard, D., Klatt, S., Baier, R., Carman, J. G., and Horandl, E. (2014b). Taxonomy and biogeography of apomixis in angiosperms and associated biodiversity characteristics. CRC Crit. Rev. Plant Sci. 33, 414–427. doi: 10.1080/07352689.2014.898488
Horandl, E. (2009). A combinational theory for maintenance of sex. Heredity (Edinb) 103, 445–457. doi: 10.1038/hdy.2009.85
Horandl, E., and Hadacek, F. (2013). The oxidative damage initiation hypothesis for meiosis. Plant Reprod. 26, 351–367. doi: 10.1007/s00497-013-0234-7
Jaenike, J. (1978). An hypothesis to account for the maintenance of sex within populations. Evol. Theory 3, 191–194.
Jedlicka, P., Jedlickova, V., and Lee, H. J. (2015). Expression of stress-related genes in the parthenogenetic forms of the pea aphid, Acyrthosiphon pisum. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 180, 32–37. doi: 10.1016/j.cbpa.2014.11.009
Jiang, H., Wang, F. F., Wu, Y. T., Zhou, X., Huang, X. Y., Zhu, J., et al. (2009). MULTIPOLAR SPINDLE 1 (MPS1), a novel coiled-coil protein of Arabidopsis thaliana, is required for meiotic spindle organization. Plant J. 59, 1001–1010. doi: 10.1111/j.1365-313X.2009.03929.x
Johnston, A. J., Kirioukhova, O., Barrell, P. J., Rutten, T., Moore, J. M., Baskar, R., et al. (2010). Dosage-sensitive function of retinoblastoma related and convergent epigenetic control are required during the Arabidopsis life cycle. PLoS Genet. 6:e1000988. doi: 10.1371/journal.pgen.1000988
Johnston, A. J., Matveeva, E., Kirioukhova, O., Grossniklaus, U., and Gruissem, W. (2008). A dynamic reciprocal RBR-PRC2 regulatory circuit controls Arabidopsis gametophyte development. Curr. Biol. 18, 1680–1686. doi: 10.1016/j.cub.2008.09.026
Johnston, A. J., Meier, P., Gheyselinck, J., Wuest, S. E., Federer, M., Schlagenhauf, E., et al. (2007). Genetic subtraction profiling identifies genes essential for Arabidopsis reproduction and reveals interaction between the female gametophyte and the maternal sporophyte. Genome Biol. 8: R204. doi: 10.1186/gb-2007-8-10-r204
Judson, O. P., and Normark, B. B. (1996). Ancient asexual scandals. Trends Ecol. Evol. 11, 41–46. doi: 10.1016/0169-5347(96)81040-8
Kantama, L., Sharbel, T. F., Schranz, M. E., Mitchell-Olds, T., de Vries, S., and de Jong, H. (2007). Diploid apomicts of the Boechera holboellii complex display large-scale chromosome substitutions and aberrant chromosomes. Proc. Natl. Acad. Sci. U.S.A. 104, 14026–14031. doi: 10.1073/pnas.0706647104
Kearney, M., Blacket, M. J., Strasburg, J. L., and Moritz, C. (2006). Waves of parthenogenesis in the desert: evidence for the parallel loss of sex in a grasshopper and a gecko from Australia. Mol. Ecol. 15, 1743–1748. doi: 10.1111/j.1365-294X.2006.02898.x
Kilian, J., Whitehead, D., Horak, J., Wanke, D., Weinl, S., Batistic, O., et al. (2007). The AtGenExpress global stress expression data set: protocols, evaluation and model data analysis of UV-B light, drought and cold stress responses. Plant J. 50, 347–363. doi: 10.1111/j.1365-313X.2007.03052.x
Kim, D., Pertea, G., Trapnell, C., Pimentel, H., Kelley, R., and Salzberg, S. L. (2013). TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 14: R36. doi: 10.1186/gb-2013-14-4-r36
Kim, J. M., To, T. K., Ishida, J., Morosawa, T., Kawashima, M., Matsui, A., et al. (2008). Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 49, 1580–1588. doi: 10.1093/pcp/pcn133
Kim, Y., and Orr, H. A. (2005). Adaptation in sexuals vs. asexuals: clonal interference and the Fisher-Muller model. Genetics 171, 1377–1386. doi: 10.1534/genetics.105.045252
Klatt, S., Hadacek, F., Hodac, L., Brinkmann, G., Eilerts, M., Hojsgaard, D., et al. (2016). Photoperiod extension enhances sexual megaspore formation and triggers metabolic reprogramming in facultative apomictic Ranunculus auricomus. Front. Plant Sci. 7: 278. doi: 10.3389/fpls.2016.00278
Koltunow, A. M. (1993). Apomixis – embryo sacs and embryos formed without meiosis or fertilization in ovules. Plant Cell 5, 1425–1437. doi: 10.1105/tpc.5.10.1425
Koltunow, A. M., and Grossniklaus, U. (2003). Apomixis: a developmental perspective. Annu. Rev. Plant Biol. 54, 547–574. doi: 10.1146/annurev.arplant.54.110901.160842
Kondrashov, A. S. (1988). Deleterious mutations and the evolution of sexual reproduction. Nature 336, 435–440. doi: 10.1038/336435a0
Kono, T., Obata, Y., Wu, Q., Niwa, K., Ono, Y., Yamamoto, Y., et al. (2004). Birth of parthenogenetic mice that can develop to adulthood. Nature 428, 860–864. doi: 10.1038/nature02402
Liu, C. H., Finke, A., Diaz, M., Rozhon, W., Poppenberger, B., Baubec, T., et al. (2015). Repair of DNA damage induced by the cytidine analog zebularine requires ATR and ATM in Arabidopsis. Plant Cell 27, 1788–1800. doi: 10.1105/tpc.114.135467
Lovell, J. T., Aliyu, O. M., Mau, M., Schranz, M. E., Koch, M., Kiefer, C., et al. (2013). On the origin and evolution of apomixis in Boechera. Plant Reprod. 26, 309–315. doi: 10.1007/s00497-013-0218-7
Lynch, M. (1984). Destabilizing hybridization, general-purpose genotypes and geographic parthenogenesis. Q. Rev. Biol. 59, 257–290. doi: 10.1086/413902
Madlung, A., Masuelli, R. W., Watson, B., Reynolds, S. H., Davison, J., and Comai, L. (2002). Remodeling of DNA methylation and phenotypic and transcriptional changes in synthetic Arabidopsis allotetraploids. Plant Physiol. 129, 733–746. doi: 10.1104/pp.003095
Maere, S., Heymans, K., and Kuiper, M. (2005). BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449. doi: 10.1093/bioinformatics/bti551
Martens, K., Rossetti, G., and Horne, D. J. (2003). How ancient are ancient asexuals? Proc. Biol. Sci. 270, 723–729. doi: 10.1098/rspb.2002.2270
McDonald, M. J., Rice, D. P., and Desai, M. M. (2016). Sex speeds adaptation by altering the dynamics of molecular evolution. Nature 531, 233–236. doi: 10.1038/nature17143
Meng, C. M., Cai, C. P., Zhang, T. Z., and Guo, W. Z. (2009). Characterization of six novel NAC genes and their responses to abiotic stresses in Gossypium hirsutum L. Plant Sci. 176, 352–359. doi: 10.1016/j.plantsci.2008.12.003
Mercier, R., Mezard, C., Jenczewski, E., Macaisne, N., and Grelon, M. (2015). The molecular biology of meiosis in plants. Annu. Rev. Plant Biol. 66, 297–327. doi: 10.1146/annurev-arplant-050213-035923
Muller, H. J. (1964). The relation of recombination to mutational advance. Mutat. Res. 106: 2–9. doi: 10.1016/0027-5107(64)90047-8
Naser, V., and Shani, E. (2016). Auxin response under osmotic stress. Plant Mol. Biol. 91, 661–672. doi: 10.1007/s11103-016-0476-5
Naumova, T. N., van der Laak, J., Osadtchiy, J., Matzk, F., Kravtchenko, A., Bergervoet, J., et al. (2001). Reproductive development in apomictic populations of Arabis holboellii (Brassicaceae). Sex Plant Reprod. 14, 195–200. doi: 10.1007/s00497-001-0118-0
Nguyen, D., Rieu, I., Mariani, C., and van Dam, N. M. (2016). How plants handle multiple stresses: hormonal interactions underlying responses to abiotic stress and insect herbivory. Plant Mol. Biol. 91, 727–740. doi: 10.1007/s11103-016-0481-8
Nogler, G. A. (1984). Genetics of apospory in apomictic Ranuncuclus auricomus. V. Conclusion. Bot. Helv. 94, 411–422.
Noyes, R. D., and Wagner, J. D. (2014). Dihaploidy yields diploid apomicts and parthenogens in Erigeron (Asteraceae). Am. J. Bot. 101, 865–874. doi: 10.3732/ajb.1400008
Nuruzzaman, M., Sharoni, A. M., and Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Front. Microbiol. 4:248. doi: 10.3389/fmicb.2013.00248
Oshino, T., Abiko, M., Saito, R., Ichiishi, E., Endo, M., Kawagishi-Kobayashi, M., et al. (2007). Premature progression of anther early developmental programs accompanied by comprehensive alterations in transcription during high-temperature injury in barley plants. Mol. Genet. Genomics 278, 31–42. doi: 10.1007/s00438-007-0229-x
Pellino, M., Sharbel, T. F., Mau, M., Amiteye, S., and Corral, J. M. (2011). Selection of reference genes for quantitative real-time PCR expression studies of microdissected reproductive tissues in apomictic and sexual Boechera. BMC Res. Notes 4:303. doi: 10.1186/1756-0500-4-303
Pignatta, D., Dilkes, B. P., Yoo, S. Y., Henry, I. M., Madlung, A., Doerge, R. W., et al. (2010). Differential sensitivity of the Arabidopsis thaliana transcriptome and enhancers to the effects of genome doubling. New Phytol. 186, 194–206. doi: 10.1111/j.1469-8137.2010.03198.x
Podio, M., Caceres, M. E., Samoluk, S. S., Seijo, J. G., Pessino, S. C., Ortiz, J. P., et al. (2014). A methylation status analysis of the apomixis-specific region in Paspalum spp. suggests an epigenetic control of parthenogenesis. J. Exp. Bot. 65, 6411–6424. doi: 10.1093/jxb/eru354
Probst, A. V., and Mittelsten Scheid, O. (2015). Stress-induced structural changes in plant chromatin. Curr. Opin. Plant Biol. 27, 8–16. doi: 10.1016/j.pbi.2015.05.011
Ravi, M., Marimuthu, M. P., and Siddiqi, I. (2008). Gamete formation without meiosis in Arabidopsis. Nature 451, 1121–1124. doi: 10.1038/nature06557
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., and Bartel, D. P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. doi: 10.1016/S0092-8674(02)00863-2
Roy, B. A. (1995). The breeding systems of six species of Arabis (Brassicaceae). Am. J. Bot. 82, 869–877. doi: 10.2307/2445973
Rushworth, C. A., Song, B. H., Lee, C. R., and Mitchell-Olds, T. (2011). Boechera, a model system for ecological genomics. Mol. Ecol. 20, 4843–4857. doi: 10.1111/j.1365-294X.2011.05340.x
Saini, S. H. (1997). Effects of water stress on male gametophyte development in plants. Sex. Plant Reprod. 10, 67–73. doi: 10.1007/s004970050069
Sanchez-Moran, E., Santos, J. L., Jones, G. H., and Franklin, F. C. (2007). ASY1 mediates AtDMC1-dependent interhomolog recombination during meiosis in Arabidopsis. Genes Dev. 21, 2220–2233. doi: 10.1101/gad.439007
Schmid, M. W., Schmidt, A., Klostermeier, U. C., Barann, M., Rosenstiel, P., and Grossniklaus, U. (2012). A powerful method for transcriptional profiling of specific cell types in eukaryotes: laser-assisted microdissection and RNA sequencing. PLoS ONE 7:e29685. doi: 10.1371/journal.pone.0029685
Schmidt, A., Schmid, M. W., Klostermeier, U. C., Qi, W., Guthorl, D., Sailer, C., et al. (2014). Apomictic and sexual germline development differ with respect to cell cycle, transcriptional, hormonal and epigenetic regulation. PLoS Genet. 10:e1004476. doi: 10.1371/journal.pgen.1004476
Schmidt, A., Wuest, S. E., Vijverberg, K., Baroux, C., Kleen, D., and Grossniklaus, U. (2011). Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development. PLoS Biol. 9:e1001155. doi: 10.1371/journal.pbio.1001155
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675. doi: 10.1038/nmeth.2089
Scholes, D. R., and Paige, K. N. (2015). Plasticity in ploidy: a generalized response to stress. Trends Plant Sci. 20, 165–175. doi: 10.1016/j.tplants.2014.11.007
Schranz, M. E., Dobes, C., Koch, M. A., and Mitchell-Olds, T. (2005). Sexual reproduction, hybridization, apomixis, and polyploidization in the genus Boechera (Brassicaceae). Am. J. Bot. 92, 1797–1810. doi: 10.3732/ajb.92.11.1797
Schranz, M. E., Kantama, L., de Jong, H., and Mitchell-Olds, T. (2006). Asexual reproduction in a close relative of Arabidopsis: a genetic investigation of apomixis in Boechera (Brassicaceae). New Phytol. 171, 425–438. doi: 10.1111/j.1469-8137.2006.01765.x
Sharbel, T. F., Voigt, M. L., Corral, J. M., Galla, G., Kumlehn, J., Klukas, C., et al. (2010). Apomictic and sexual ovules of Boechera display heterochronic global gene expression patterns. Plant Cell 22, 655–671. doi: 10.1105/tpc.109.072223
Singh, M., Goel, S., Meeley, R. B., Dantec, C., Parrinello, H., Michaud, C., et al. (2011). Production of viable gametes without meiosis in maize deficient for an ARGONAUTE protein. Plant Cell 23, 443–458. doi: 10.1105/tpc.110.079020
Smith, J. M. (1971). What use is sex?. J. Theor. Biol. 30, 319–335. doi: 10.1016/0022-5193(71)90058-0
Sniegowski, P. D., and Gerrish, P. J. (2010). Beneficial mutations and the dynamics of adaptation in asexual populations. Philos. Trans. R Soc. Lond B Biol. Sci. 365, 1255–1263. doi: 10.1098/rstb.2009.0290
Song, Y., Ji, D., Li, S., Wang, P., Li, Q., and Xiang, F. (2012). The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS ONE 7:e41274. doi: 10.1371/journal.pone.0041274
Sun, K., Hunt, K., and Hauser, B. A. (2004). Ovule abortion in Arabidopsis triggered by stress. Plant Physiol. 135, 2358–2367. doi: 10.1104/pp.104.043091
Syamaladevi, D. P., Meena, S. S., and Nagar, R. P. (2016). Molecular understandings on ‘the never thirsty’ and apomictic Cenchrus grass. Biotechnol. Lett. 38, 369–376. doi: 10.1007/s10529-015-2004-0
Taskin, K. M., Turgut, K., and Scott, R. J. (2004). Apomictic development in Arabis gunnisoniana. Israel J. Plant Sci. 52, 155–160.
Thakur, P., Kumar, S., Malik, J. A., Berger, J. D., and Nayyar, H. (2010). Cold stress effects on reproductive development in grain crops: an overview. Environ. Exp. Bot. 67, 429–443. doi: 10.1016/j.envexpbot.2009.09.004
Thieme, C. J., Rojas-Triana, M., Stecyk, E., Schudoma, C., Zhang, W., Yang, L., et al. (2015). Endogenous Arabidopsis messenger RNAs transported to distant tissues. Nat. Plants 1: 15025. doi: 10.1038/nplants.2015.25
Tognetti, V. B., Muhlenbock, P., and Van Breusegem, F. (2012). Stress homeostasis – the redox and auxin perspective. Plant Cell Environ. 35, 321–333. doi: 10.1111/j.1365-3040.2011.02324.x
Tran, L. S., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Trapnell, C., Roberts, A., Goff, L., Pertea, G., Kim, D., Kelley, D. R., et al. (2012). Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578. doi: 10.1038/nprot.2012.016
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Verhoeven, K. J., Jansen, J. J., van Dijk, P. J., and Biere, A. (2010). Stress-induced DNA methylation changes and their heritability in asexual dandelions. New Phytol. 185, 1108–1118. doi: 10.1111/j.1469-8137.2009.03121.x
Verhoeven, K. J., and Preite, V. (2014). Epigenetic variation in asexually reproducing organisms. Evolution 68, 644–655. doi: 10.1111/evo.12320
Verhoeven, K. J., and van Gurp, T. P. (2012). Transgenerational effects of stress exposure on offspring phenotypes in apomictic dandelion. PLoS ONE 7:e38605. doi: 10.1371/journal.pone.0038605
Verslues, P. E., Agarwal, M., Katiyar-Agarwal, S., Zhu, J. H., and Zhu, J. K. (2006). Methods and concepts in quantifying resistance to drought, salt and freezing, abiotic stresses that affect plant water status. Plant J. 45, 523–539. doi: 10.1111/j.1365-313X.2005.02593.x
Voigt-Zielinski, M. L., Piwczynski, M., and Sharbel, T. F. (2012). Differential effects of polyploidy and diploidy on fitness of apomictic Boechera. Sex. Plant Reprod. 25, 97–109. doi: 10.1007/s00497-012-0181-8
Wang, J., Tian, L., Madlung, A., Lee, H. S., Chen, M., Lee, J. J., et al. (2004). Stochastic and epigenetic changes of gene expression in Arabidopsis polyploids. Genetics 167, 1961–1973. doi: 10.1534/genetics.104.027896
William, B. C., and Barraclough, T. G. (2009) “Asexual speciation,” in Lost Sex: The Evolutionary Biology of Parthenogenesis, eds I. Schön K. Martens and P. van Dijk (Berlin: Springer).
Windsor, A. J., Schranz, M. E., Formanova, N., Gebauer-Jung, S., Bishop, J. G., Schnabelrauch, D., et al. (2006). Partial shotgun sequencing of the Boechera stricta genome reveals extensive microsynteny and promoter conservation with Arabidopsis. Plant Physiol. 140, 1169–1182. doi: 10.1104/pp.105.073981
Winkler, H. (1908). Über parthenogenesis und apogamie im pflanzenreiche. Progr. Rei. Bot. 2, 293–454.
Wuest, S. E., Vijverberg, K., Schmidt, A., Weiss, M., Gheyselinck, J., Lohr, M., et al. (2010). Arabidopsis female gametophyte gene expression map reveals similarities between plant and animal gametes. Curr. Biol. 20, 506–512. doi: 10.1016/j.cub.2010.01.051
You, J., Zhang, L. H., Song, B., Qi, X. Q., and Chan, Z. L. (2015). Systematic analysis and identification of stress-responsive genes of the NAC gene family in Brachypodium distachyon. PLoS ONE 10:e0122027. doi: 10.1371/journal.pone.0122027
Yu, H. J., Hogan, P., and Sundaresan, V. (2005). Analysis of the female gametophyte transcriptome of Arabidopsis by comparative expression profiling. Plant Physiol. 139, 1853–1869. doi: 10.1104/pp.105.067314
Yu, Z., Haberer, G., Matthes, M., Rattei, T., Mayer, K. F., Gierl, A., et al. (2010). Impact of natural genetic variation on the transcriptome of autotetraploid Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 17809–17814. doi: 10.1073/pnas.1000852107
Zamudio, N., Barau, J., Teissandier, A., Walter, M., Borsos, M., Servant, N., et al. (2015). DNA methylation restrains transposons from adopting a chromatin signature permissive for meiotic recombination. Genes Dev. 29, 1256–1270. doi: 10.1101/gad.257840.114
Keywords: stress response, asexual reproduction, apomixis, apomeiosis, NAC transcription factors, gene regulatory network, DNA methylation, Boechera
Citation: Shah JN, Kirioukhova O, Pawar P, Tayyab M, Mateo JL and Johnston AJ (2016) Depletion of Key Meiotic Genes and Transcriptome-Wide Abiotic Stress Reprogramming Mark Early Preparatory Events Ahead of Apomeiotic Transition. Front. Plant Sci. 7:1539. doi: 10.3389/fpls.2016.01539
Received: 02 May 2016; Accepted: 30 September 2016;
Published: 26 October 2016.
Edited by:
Mohammad Anwar Hossain, Bangladesh Agricultural University, BangladeshReviewed by:
Franz Hadacek, University of Göttingen, GermanyCopyright © 2016 Shah, Kirioukhova, Pawar, Tayyab, Mateo and Johnston. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Amal J. Johnston, YW1hbC5qb2huc3RvbkBncmVlbnRlY2hsYWIubmV0; YW1hbC5qb2huc3RvbkBjb3MudW5pLWhlaWRlbGJlcmcuZGU= Juan L. Mateo, anVhbi5tYXRlb0Bjb3MudW5pLWhlaWRlbGJlcmcuZGU=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.