
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 24 October 2016
Sec. Plant Proteomics and Protein Structural Biology
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.01482
This article is part of the Research Topic Biology of plant fruit, seed and tuberous tissues: An important topic in food security View all 21 articles
 Jianwei Gu1,2†
Jianwei Gu1,2† Hongbo Chao1†
Hongbo Chao1† Lu Gan1†
Lu Gan1† Liangxing Guo1
Liangxing Guo1 Kai Zhang1
Kai Zhang1 Yonghong Li3
Yonghong Li3 Hao Wang3
Hao Wang3 Nadia Raboanatahiry1
Nadia Raboanatahiry1 Maoteng Li1,2*
Maoteng Li1,2*The success of seed germination and establishment of a normal seedling are key determinants of plant species propagation. At present, only a few studies have focused on the genetic control of seed germination by using a proteomic approach in Brassica napus. In the present study, the protein expression pattern of seed germination was investigated using differential fluorescence two-dimensional gel electrophoresis in B. napus. One hundred and thirteen differentially expressed proteins (DEPs) that were mainly involved in storage (23.4%), energy metabolism (18.9%), protein metabolism (16.2%), defense/disease (12.6%), seed maturation (11.7%), carbohydrate metabolism (4.5%), lipid metabolism (4.5%), amino acids metabolism (3.6%), cell growth/division (3.6%), and some unclear functions (2.7%) were observed by proteomic analysis. Seventeen genes corresponding to 11 DEPs were identified within or near the associated linkage disequilibrium regions related to seed germination and vigor quantitative traits reported in B. napus in previous studies. The expression pattern of proteins showed that heterotrophic metabolism could be activated in the process of seed germination and that the onset of defense mechanisms might start during seed germination. These findings will help generate a more in-depth understanding of the mobilization of seed storage reserves and regulation mechanisms of the germination process in B. napus.
The success of seed germination and establishment of a normal seedling are determinant features for the propagation of plant species, which are of both economic and ecological importance (Rajjou et al., 2012). Seed germination and seedling establishment depend on the mobilization of storage reserves. For oilseed crops, effective degradation of storage lipid is essential for the success of seedling establishment, which is extremely important for plant adaptation to terrestrial environments (Bewley, 1997; Pritchard et al., 2002; Graham, 2008). In recent decades, many studies have been carried out on seed germination through physiological, proteomic, or transcriptomic analysis in Arabidopsis (Gallardo et al., 2001, 2002; Müller et al., 2006), rice (Oryza sativa; Yang et al., 2007), barley (Hordeum vulgare; Sreenivasulu et al., 2008), maize (Zea mays; Guo et al., 2013), soybean (Glycine max; Xu et al., 2011), Lepidium sativum (Müller et al., 2006), Jatropha curcas (Yang et al., 2009), Brassica napus (Li et al., 2005; Ge et al., 2013), and others. These studies have provided information about many aspects of the seed germination process, such as the roles of gibberellin and abscisic acid (Gallardo et al., 2002; Reyes and Chua, 2007), radicle emergence (Guo et al., 2013), defense (Rajjou et al., 2006; Xu et al., 2011), endosperm weakening (Zhang et al., 2014), and the mobilization of energy reserves (Kelly et al., 2011; Han et al., 2013), but the detailed regulatory mechanisms in the process of seed germination are still unclear.
Seed germination is a complex process controlled by many mechanisms. Plant hormones are the key regulation factors for breaking of seed dormancy and initiation of seed germination (Bewley, 1997; Gallardo et al., 2002; Rajjou et al., 2006; Reyes and Chua, 2007; Liu et al., 2013). Mobilization of storage reserves is essential for seed germination, but different storage reserves may have different roles during seed germination, such as seed storage oil mobilization, which was indicated as important but not essential for germination or seedling establishment in Arabidopsis (Pinfield-Wells et al., 2005; Kelly et al., 2011). Metabolic and regulatory network models in rice and Arabidopsis have been constructed in previous studies (Bassel et al., 2011; He et al., 2011), which showed that different crop species might have distinct mechanisms for reserve mobilization during germination (Han et al., 2013). Rapeseed is one of the most important oil crops in the world; rapeseed oil accounts for about 13–16% of the world vegetable oil production (Hajduch et al., 2006; Obermeier et al., 2013; Wang and Yin, 2014). In addition, rapeseed is also a potential bio-energy crop to alleviate the global energy shortage (Tsadilas and Shaheen, 2013). Seed germination and vigor in B. napus have been investigated in recent years, and it has been revealed that these are significantly influenced by many factors, such as salt (Srivastava et al., 2004; Bybordi and Tabatabaei, 2009), temperature (Kondra et al., 1983; Zhang et al., 2015), plant hormones (Schopfer and Plachy, 1985; Nguyen et al., 2016), and aging (Zhang et al., 2006; Janmohammadi et al., 2008), and can be enhanced by priming (Zheng et al., 1994; Mohammadi, 2009; Benincasa et al., 2013; Hatzig et al., 2014). In addition, many genes (Li et al., 2005; Ge et al., 2014; Kubala et al., 2015; Nguyen et al., 2016), proteins, (Srivastava et al., 2004; Kubala et al., 2015), or quantitative trait loci (QTLs; Nagel et al., 2011; Hatzig et al., 2015) have been shown to participate in the regulation of seed germination and vigor in B. napus. This research is consistent with previous research in other organisms, but is still not enough to clearly elucidate the regulation mechanisms of B. napus seed germination.
Important candidate genes can be easily identified in complex pathways to dissect genetic control mechanisms by combining the QTL mapping method with other “-omics” methods (Long et al., 2007; Gan et al., 2013; Schiessl et al., 2014). Gan et al. (2013) compared the QTLs for oil and total protein content in the “Tapidor” × “Ningyou7” cross doubled haploid (TNDH) mapping population with candidate genes that corresponded to differentially expressed proteins (DEPs) in two B. napus cultivars with high and low oil content. A total of 117 candidate genes were found located in the QTL confidence intervals for oil or protein content, which indicated that these DEPs might be involved in oil or protein formation. By using a similar approach, a dozen resistance gene loci for clubroot and blackleg have been identified in Brassica crops (Li and McVetty, 2013), and seven candidate genes that might be important for waterlogging tolerance and were co-localized with the QTL identified in maize were observed (Osman et al., 2013). These results have indicated the advantage of this candidate gene mapping approach in the dissection of complex pathways and regulation mechanisms.
In this study, differential fluorescence two-dimensional gel electrophoresis (2-D DIGE) was used to analyze the DEPs in different germination processes of B. napus, and a metabolism regulation pathway was constructed based on the DEPs. The results provide a more in-depth understanding of the mobilization of seed storage reserves and regulation mechanisms of the germination process in B. napus. With the aid of QTL analysis and genome-wide association mapping, new insights for seed germination in B. napus could be revealed.
The mature seed of B. napus (7792-95/772/772) was provided by Shaanxi Hybrid Rapeseed Research Center. The seeds were imbibed on two layers of moist filter paper (Whatman, GE healthcare, UK) at 28°C in the dark. The whole seeds were collected at 0, 6, 12, 18, 24, 36, 48, 60, and 72 h after imbibition (HAI) and stored at −70°C until further use.
The transmission electron microscopy (TEM) analysis followed the methods of Gan et al. (2013). Cotyledon tissues from dry and germinated seeds were fixed with 2.5% glutaraldehyde and postfixed in 1% OsO4, then dehydrated by a series of acetone. The samples were embedded in Spurr's epoxy resin and polymerized at 60°C for 24 h after infiltration through a series of acetone/Spurr's epoxy resin. The thin sections (70 nm) that were readily cut by an ultramicrotome (Leica MZ6, Germany) were collected onto copper grids, post-stained with supersaturated uranyl acetate and 0.4% lead citrate, respectively, rinsed six times, each for 15 s, with distilled water, and observed under a JEOL JEM-1230 transmission electron microscope.
The oil content during seed germination and post-germination growth was measured using a modified method of Wei et al. (2009). Each sample was extracted simultaneously in three replicates. For fatty acid (FA) analysis, the seeds or seedling tissues were milled and then vacuum freeze-dried overnight. The FAs were then extracted using the method of Browse et al. (1986). The absolute content of FAs was determined using gas chromatography (Shimadzu GC-2010, Japan, with DEGS-diethyl glycol succinate column) referring to Browse et al. (1986) and Rücker and Röbbelen (1996). Relative content of FAs was calculated from five independent biological replicates. The difference in FA contents between different processes of germination was evaluated by one-way analysis of variance (ANOVA) test.
Total protein was determined by using the Bradford method (Kruger, 2009). Between 10 and 100 μg protein in 100-μl total volume was collected into a test tube. The assay reagent (5 ml) was added into each tube and mixed well by inversion or gentle mixing on a vortex machine. The absorption value was then measured at 595 nm. γ-Globulin (Sigma) was used for the calibration curve, and 100 μl of distilled water was used as a blank.
To measure the total sugar content in germinating B. napus seed, the standard curve obtained by using glucose content was projected on the x-axis and the absorbance value on the y-axis. The sample (0.1 g) was weighed and then added to 1.5 ml water and 1 ml 6 M HCl and heated in a boiling water bath for 30 min. After the mixture was cooled down at room temperature, the pH was adjusted to neutral by using 10% NaOH solution. The filtrate was then filtered and diluted in 50 ml distilled water as sample solution. The sample solution was analyzed at an absorbance of 620 nm, and the total sugar content of samples was calculated according to the standard curve (Wen et al., 2005).
Proteins were extracted using a modified protocol according to Gan et al. (2013). For each sample, about 0.5 g seeds or seedlings was ground into fine powder. About 100 mg of each sample was homogenized in 750 μl Tris-saturated phenol (pH > 7.8) and 750 μl homogenization buffer [0.1 M Tris-HCl (pH 7.5), 0.9 M sucrose, 10 mM EDTA, 0.4% (g/ml) DTT] in an ice bath for 30 min. The homogenate was centrifuged at 5000 g for 15 min at 4°C. The supernatant was transferred to a new tube and precipitated using 1.5 ml 0.1 M ammonium acetate-methanol solution at −20°C overnight. The mixture was centrifuged at 5000 g for 10 min at 4°C, and the supernatant was discarded. The precipitation was washed twice with 0.1 M ammonium acetate-methanol solution and twice with acetone, then dried at room temperature for about 5 min. The dried powder was dissolved in a buffer solution containing 7 M urea, 2 M thiourea, and 4% CHAPS (pH 8.8). Three independent biological replicates were completed for each time-point of seed germination (0, 6, 12, 18, 24, and 36 HAI).
The concentration of proteins was measured using a 2-D Quant kit (GE Healthcare, UK) according the manufacturer's instructions, and then adjusted to 5 μg/μl. For labeling of the proteins, 50 pmol CyDye was mixed with 6 μl protein sample and incubated on ice for at least 2 h in the dark. The labeling reaction was terminated by adding 1 μl 10 mM lysine (Tang et al., 2008).
For 2-D DIGE analysis, a mixture of Cy2-, Cy3-, or Cy5-labeled protein was mixed with 2-D DIGE buffer (7 M urea, 2 M thiourea, 4% CHAPS, 0.4% DTT, 0.5% IPG buffer) and then loaded on an immobilized pH gradient (IPG; Amersham Biosciences, Uppsala, Sweden) for isoelectric focusing (Tang et al., 2008). The running conditions were as follows: rehydration for 14 h at 20°C, followed by holding at 100, 300, 600, and 1000 V for 1 h at each step; then the voltage was raised to 10,000 V linearly and held until reaching a total value of 120,000 V-h. Second-dimension electrophoresis was performed using 12.5% SDS-polyacrylamide gel (Gan et al., 2013).
The images of Cy2-, Cy3-, and Cy5-labeled proteins were acquired by a Typhoon 8600 scanner (GE Healthcare, UK) using difference wavelength and analyzed by DeCyder 6.5 software (GE Healthcare, UK). Spot detection was performed by the DIA (differential in-gel analysis) module. After removing the artifact spots by manual editing, the images were further analyzed by the DeCyder BVA (biological variation analysis) module. For each treatment, images from at least three biological replicates were used for statistical analysis.
The preparation of isoelectric focusing with 1 mg protein sample loaded into IPG strips for spot picking and second-dimension electrophoresis were performed as mentioned above. Gels were stained with CBB R-250 according to the protocol of Gan et al. (2013). The stained gels were scanned by a UMAX Power Look 2100XL scanner (UMAX, Inc., Taipei, China). These images were matched with the DIGE images to identify spots of interest. Protein spots were manually excised from the gels and cut into small pieces.
Protein spots were digested according to the method of Katayama et al. (2001). The digested protein samples were subjected to MS and MS/MS analysis by using an ABI 4800 MALDI-TOF/TOF Plus mass spectrometer (Shu et al., 2011). Both the MS and MS/MS data were integrated and processed by the GPS Explorer V3.6 software (Applied Biosystems) with default parameters. For protein identification, the acquired MS/MS spectra were automatically searched on the NCBInr green plants database, using the MASCOT V2.1 search engine (Matrix Science, London, UK). Search parameters were set as taxonomy: Rosids; enzyme specificity considered: trypsin; max missed cleavages: 1; fixed modifications: carbamidomethyl (C); variable modifications: Acetyl (Protein N-term), Deamidated (NQ), Dioxidation (W), and Oxidation (M); peptide mass tolerance: ±100 ppm; fragment mass tolerance: ±0.5 Da.
To minimize the inclusion of false positive hits, matches to peptides identified by SEQUEST were filtered according to their charge state, cross-correlation score (Xcorr), and normalized difference in correlation score (deltaCn). Peptide hits were accepted when singly, doubly, and triply charged peptides were with Xcorr >1.9, 2.2, and 3.75, respectively, and deltaCn >0.1 in all cases. After the peptide sequence raw data was searched using SEQUEST, a number of other criteria were considered in the final assignment of peptide and protein identifications: the number of matching peptides, the coverage, the Xcorr, and the molecular mass and isoelectric point of the protein.
Protein functional domains were predicted using the PSI and PHI-BLAST programs (http://www.ncbi.nlm.nih.gov/BLAST/). Proteins were classified into different categories by combining BLAST alignments with Gene Ontology and knowledge from the literature. A hierarchical cluster was constructed by SPSS using the K-means clustering approach in order to visualize the expression characteristics of the DEPs (Li et al., 2005).
A genome-wide association study to define genomic regions influencing seed germination and early seedling growth was carried out by Hatzig et al. (2015). Twenty-four associated linkage disequilibrium (LD) regions were identified to be associated with seven traits. Twenty associated LD regions related to seed germination and vigor were selected for comparative study. The related traits were as follows: volume increase within first 8 h (VI), total germination rate within 72 h after initiation of imbibition (GR72), first germination time (FG), radicle elongation speed (ES), time to reach 50% of germination (T50), and germination rate within 36 h after the initiation of imbibition (GR36). To identify the DEPs comprehensively and effectively, the genes within the flanking regions up to 1000 kbp on either side of the associated LD regions were considered as candidate regions for searching DEP genes.
Total RNA was extracted from 0.1 g of the frozen samples by using a RNA prep Pure Plant Kit (TOYOBO, DP441). The first chain was synthesized by ReverTra Ace qPCR RT Master Mix with gDNA Remover (TOYOBO, FSQ-301) according to the manufacturer's instructions. Expression analysis was assayed with a SYBR premix EX TaqTM kit (TaKaRa, Japan) on an ABI 7900HT Fast Real-Time PCR System. Relative gene expression was analyzed according to the method of Pfaffl et al. (2004). Actin 2.1 was used as a reference gene (Kubala et al., 2015), and other primers used in this study are listed in Table S1. Intra-assay variation was evaluated by calculating SD errors of arithmetic means of three sample replicates.
The seeds of B. napus were imbibed on moist filter paper for germination under darkness. It was revealed that the seed coat was ruptured, and hypocotyl elongation and radicle protrusion were observed at 18 HAI (Figure 1A); this process could be termed germination. Subsequently, the hypocotyl and radicle continued elongation to push the cotyledon out of the seed coat (Figure 1A). To obtain an overview, post-germination growth until 72 HAI was included in this work. The moisture content increased remarkably from 3.94 to 39% during the first 6 HAI, then experienced a stage of slow increase to 50.43% until 24 HAI, and increased rapidly again to 85.96% at 60 HAI (Figure 1B).
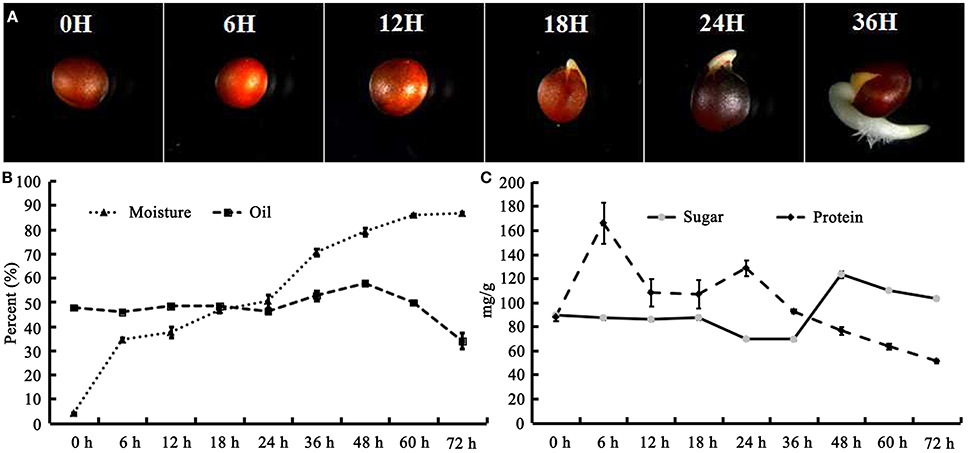
Figure 1. B. napus seed germination process and change of moisture, oil, protein, and sugar during seed germination and seedling establishment. (A) B. napus seed germination. The seeds were imbibed in moisture at 28°C under dark conditions. Photographs were taken at 0, 6, 12, 18, 24, and 36 HAI; (B) Change of moisture and oil during germination; (C) Change of protein and sugar during germination. Values are means of three biological replicates (±SD).
The oil content of B. napus seeds was stable at the germination stage and decreased sharply with more than 48 HAI (Figure 1B). Compared with 47.68% in dry mature seeds, the oil content was decreased to 33.86% at 72 HAI (Figure 1B), indicating that the oil might have been mobilized and consumed during the germination and post-germination processes. The FA compositions of the crude oil from different stages of germinating seeds were also determined (Table 1). It was revealed that the relative contents of arachidic acid (C20:0) and cis-11-eicosenoic acid (C20:1) were decreased from 0.27 and 0.5% to 0.14 and 0.39% of total FAs at 6 HAI, respectively (p < 0.01), and even lower at 72 HAI. No drastic change was observed for other FAs, while the absolute content of total FA increased to >250 μg/mg at 36 HAI and then decreased to about 200 μg/mg at 72 HAI. A similar pattern of change was observed in the absolute content of each FA except C20:0 and C20:1, which were significantly decreased at 6 HAI and remained lower even until 72 HAI (Figure S1).
The total protein and sugar in germinating B. napus seeds was also measured (Figure 1C). Generally, the total protein increased to its highest level at 6 HAI (166.02 mg/g) and then decreased continuously to 51.31 mg/g after 24 HAI. The total sugar content remained stable before 18 HAI and then decreased to 69.27 mg/g at 36 HAI. The total sugar content was observed to sharply increase to 123.35 mg/g at 48 HAI and then slowly decrease to 103.1 mg/g until 72 HAI. These results suggested that various physiological and biochemical processes occurred during post-germination.
TEM results allowed us to obtain additional data about the subcellular organization of the cotyledon cells during B. napus seed germination. In the mature dry seed, the cotyledon cells were completely filled by protein storage vacuoles (PBs) and oil bodies (OBs; Figure 2A). PBs of the cotyledon cells sh2owed different shapes (round and regular shape) with undulating contours and were formed by a homogenous and electron-dense matrix, and some electron-light, small, rounded areas were observed in the PB matrix. The rest of the cotyledon cell was filled up with smaller and electron-transparent bodies of rounded shape, corresponding to OBs (Figure 2A). No major changes were observed in cells examined before 12 HAI except for the different sizes of electron-light rounded areas in PBs, which obviously were all over the matrix (black stars; Figure 2B).
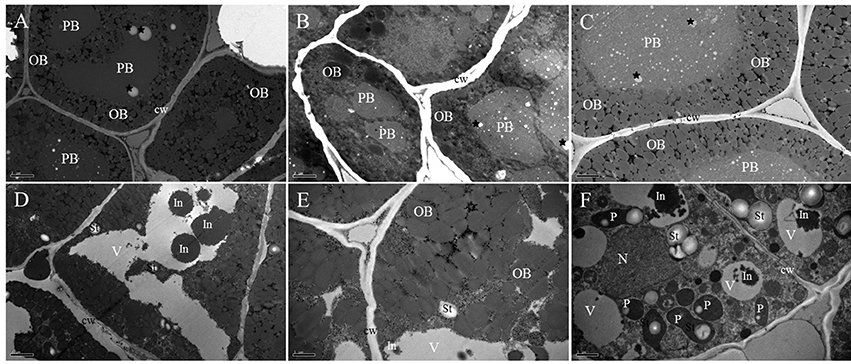
Figure 2. TEM study of cotyledons during imbibition and seedling establishment. (A) Dry mature seed. Electron-dense protein body (PB) with a homogenous matrix embedded in a cytoplasm, densely populated by small and less electron-dense OBs. Electron-light, small, rounded areas are presented in the PB matrix (black stars). (B) Twelve hours of germination. The cytoplasm is filled up with electron-dense PBs and numerous electron-light OBs. The different size electron-light, rounded area now is all over its matrix (black stars). (C) Twenty-four hours of germination. Enlarged PBs with different degrees of electron density are surrounded by OBs in close contact. The presence of electron-light, rounded area through its matrix (black stars) still can be seen. (D) Overall presentation of the 2 days of germination. The PB compartment is now similar to a vacuole (V) with a low electron density lumen that contains irregular electron-dense inclusions (In); in the surrounding cytoplasm, minute OBs and starch grains (St) are present. (E) Enlarged presentation of the 2 days of germination. The gap between the OBs is increased and boundaries of the OBs become blurred. Some cytoplasmic-like structures appear in the space between the OBs. (F) Three days of germination. The vacuolar compartment (V) with a low electron density lumen contains irregular, electron-dense inclusions (In); in the peripheral cytoplasm, developing plastids (P) with inner membrane system and starch grains (St) are present. A nearly rounded nucleus (N) with condensed chromatin is attached to the nuclear envelope. PB, protein body; OB, oil body; V, vacuole; St, starch grains; P, plastids; cw, cell wall. Bar (A–D) = 2 μm. Bar (E,F) = 1 μm.
By 24 HAI, the size of the PBs was increased, and they became rounded and less electron-dense (Figure 2C), indicating that the storage protein might begin to mobilize at this stage. The small, electron-transparent, rounded area inside the PBs was still present (Figure 2C, black stars). The OBs were observed surrounding the surface of the PBs and at the cell periphery. After 2 days of germination, the PB compartment was similar to a vacuole with a low electron density lumen that contained irregular, electron-dense inclusions. OBs as well as some starch grains were observed in the surrounding cytoplasm (Figure 2D). At the same time, the gap between the OBs was increased and the boundaries of the OBs became blurred, which indicated that the storage oil was also mobilized (Figure 2E). After 3 days of growth, developing plastids were easily identified in the cytoplasm, showing the presence of starch grains and an inner membrane system (Figure 2F). Some irregular, highly electron-dense inclusions located in the vacuole-like organelles were also observed. In the cytoplasm, a nearly rounded nucleus with condensed chromatin was attached to the nuclear envelope.
DEPs that existed during the five seed germination stages were detected by 2-D DIGE with a pH 4–7 IPG strip. The results revealed that approximately 1300 protein spots were detected on each gel (Figure 3; Figure S2). Among the DEPs, a total of 113 protein spots were identified through MS and MS/MS analysis and searching of the Mascot database in B. napus. Based on Gene Ontology, BLAST alignments, and information from the TAIR and Kyoto Encyclopedia of Genes and Genomes (KEGG) databases, the 113 protein spots were classified into 10 functional categories: storage proteins (23.4%), energy metabolism (18.9%), protein metabolism (16.2%), defense/disease (12.6%), seed maturation (11.7%), carbohydrate metabolism (4.5%), lipid metabolism (4.5%), amino acids metabolism (3.6%), cell growth/division (3.6%), and some unclear proteins (2.7%; Table 2; Figure 4B). These 113 protein spots only matched 68 unique proteins in Arabidopsis, and 20 proteins were matched with multiple protein spots, which were associated with defense/disease, storage proteins, protein metabolism, energy metabolism, lipid metabolism, seed maturation proteins, and carbohydrate metabolism (Table 2; Tables S2–S4). These multiply matched protein spots might be generated from alternative splicing, protein metabolism, and various post-translational modifications. There were 50, 23, 14, 11, and 55 DEPs showing significantly different expression at 6, 12, 18, 24, and 36 HAI, respectively (Figure S3; Table S2). The main metabolism pathways were quite different during seed germination and seedling establishment. For instance, most of the 50 DEPs were involved in energy metabolism (20%), protein metabolism (20%), storage proteins (14%), defense/disease (10%), and seed maturation (8%) at 6 HAI, which indicated the activity of protein synthesis, processing and selective degradation, storage reserve mobilization, and the onset of defense systems at the early germination stage. While at 36 HAI, storage proteins (27.3%), energy metabolism (18.2%), seed maturation (18.2%), and protein metabolism (16.4%) related DEPs were the major significantly differently expressed DEPs (Figure S3).
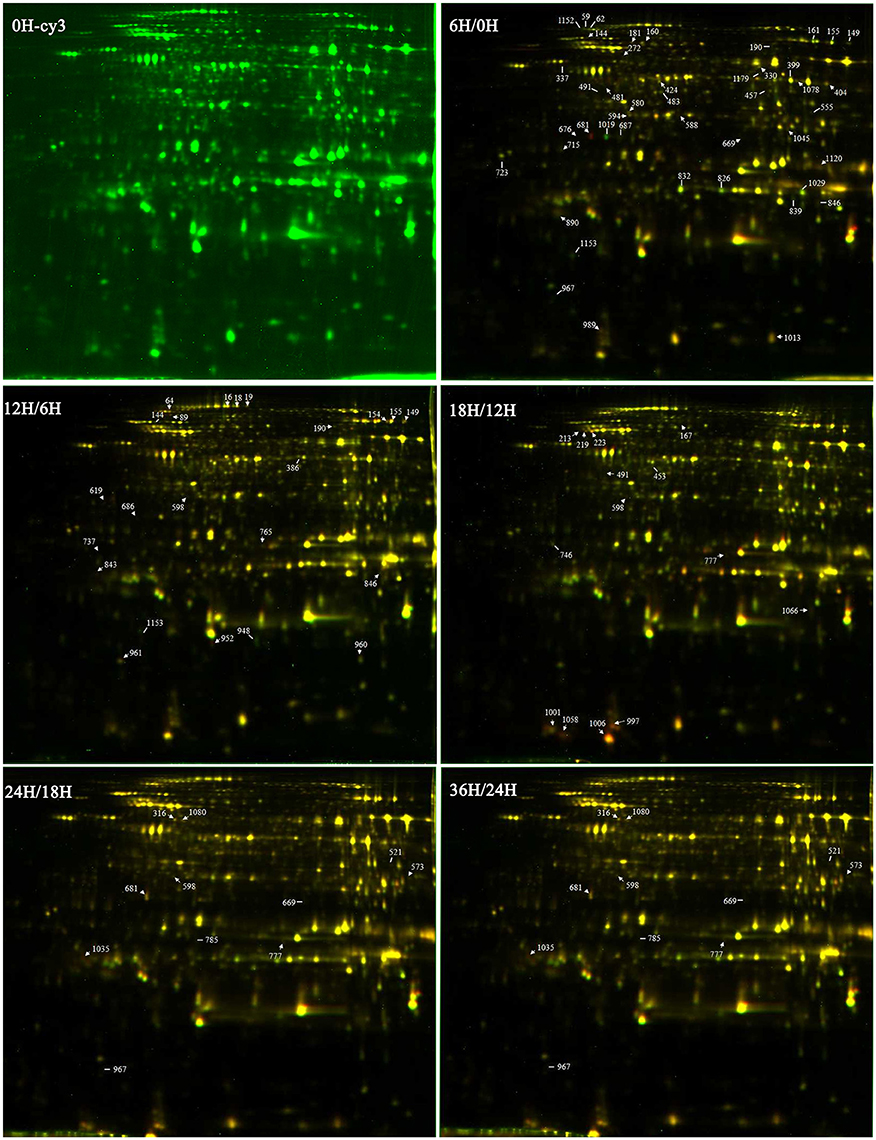
Figure 3. The 2-DIGE maps of different germination stages. 0H-cy3 represents the sample labeled with cy3. 6H/0H, 12H/6H, 18H/12H, 24H/18H, and 36H/24H represent comparisons between two samples.

Figure 4. Functional categorization and kinetic patterns of accumulation of DEPs detected in the germination process. (A) The kinetic patterns of accumulation of DEPs. Patterns I–V represents different accumulation patterns groups. (B) The functional categorization of DEPs. Patterns I–V correspond to the functional categorization of the DEPs in different accumulation patterns groups. All protein represents the functional categorization of all DEPs.
Eighteen DEPs involved in protein synthesis, folding, and degradation were also observed. Most of these proteins had up-regulated expression during seed germination, for example, translational initiation factor 4A-1 (spot 483), chaperonin 60 subunit beta 1 (spot 317), 60S acidic ribosomal protein P0-2 (spots 676 and 681), heat shock cognate protein 70-1 (spots 219 and 223), and T-complex protein 1 subunit alpha (spot 330) were all up regulated at the early stage of seed germination and retained relatively high expression levels during the following seed germination and post-germination seedling growth (Table 2; Table S2); this suggested that the activities of protein synthesis, processing, and selective degradation were essential for seed germination and post-germination seedling growth. Besides, 19 and 7 DEPs participated in energy and carbohydrate metabolism, such as glycolysis/gluconeogenesis, tricarboxylic acid (TCA) cycle, pentose phosphate pathway, ATP metabolism, and glycometabolism (Table 2; Table S2), most of which were also up-regulated in seed germination. For instance, malate dehydrogenase (spots 669, 692, and 1045) was found to be at low or undetectable levels in dry seeds but significantly increased in germinating seeds (Table 2; Table S2), indicating that mobilization and metabolism of the stored reserves took place in germinating seeds. Moreover, nine DEPs in other metabolism processes were observed, including lipid metabolism and amino acid metabolism. For instance, the acetyl-coenzyme A carboxylase carboxyl transferase subunit alpha (spot 160) was significantly induced from the beginning of germination, and the highest expression was observed at 36 HAI (Table 2; Table S2). Moreover, the expression of three amino acid metabolism-related DEPs, i.e., glutamine synthetase 1;1 (spot 598), spermidine synthase 1 (spot 634), and mercaptopyruvate sulfurtransferase 1 (spot 686), was stable at the beginning of germination, then increased from 12 HAI, and the highest expression level was observed at 36 HAI (Table 2; Table S2), indicating that other metabolic processes were also changed into a more active state after initiation of seed germination in addition to protein and energy-related metabolism. Fourteen DEPs involved in defense response or the oxidation-reduction process were also observed, including myrosinase-binding protein 2 (spots 16, 18, and 19), 1-Cys peroxiredoxin PER1 (spots 826 and 832), peptide methionine sulfoxide reductase B5 (spots 961 and 967), and others, indicating the onset of defense mechanisms during seed germination.
Hierarchical clustering analysis was performed for all the 113 DEPs, and five cluster patterns were revealed (Patterns I-V; Figure 4). Further analysis showed that Pattern II (29 DEPs) and pattern IV (18 DEPs) represented the abundance-decreased proteins in post-germination seedling growth and germinating seed, respectively. For instance, the expression of three storage proteins (cruciferin 2, 12S seed storage protein CRA1, and 12S seed storage protein CRD) was decreased during seed germination, indicating that these storage proteins might be mobilized and consumed as a nutrient reservoir in germinating seed. Pattern I (16 DEPs) was represented as the DEPs that kept increasing from 18 HAI. A defense-related protein, MBP2, which was involved in metabolizing glucosinolates and formed defense compounds to protect against herbivore attack, exhibited this pattern, implying that the myrosinase-glucosinolate defense system was activated at the beginning of post-germination seedling growth. In addition, pattern III (29 DEPs) and pattern V (17 DEPs) represented proteins that gradually changed during seed germination, and the highest protein abundance was detected at 36 HAI. These proteins reflected several active pathways (e.g., pentose phosphate pathway, glycolysis/gluconeogenesis, protein translation, and protein folding) during seed germination (Figure 4B). Thirty-three out of 113 DEPs fell into patterns I and V, which suggested that a number of DEPs might be de novo synthesized upon seed germination (Figure 4).
To predict the relationship among the DEPs, protein-protein interaction networks were constructed using STRING 10.0. The 113 DEPs were matched with 68 unique homologs in Arabidopsis by BLAST in the NCBI database (Table S3). Out of the 68 proteins, 66 representing 111 DEPs were depicted in the STRING database (Figure S4). The interaction networks were then re-visualized by Network Analyzer. As shown in Figure 5, nine tightly connected clusters were illuminated in the network based on the functional classification (Figure 5), for example, 13 unique homologous proteins were connected with each other in cluster E, and most of them were involved in protein synthesis, processing, and degradation. Besides, many members involved in the oxidation-reduction process, and energy and carbohydrate metabolism also appeared to be closely linked with these proteins, which indicated that redox homeostasis, and energy and carbohydrate metabolism were crucial for activating protein metabolism in germinating seed. Cluster H included multiple enzymes involved in glycolysis, gluconeogenesis, the pentose phosphate pathway, and the TCA cycle, implying the material and energy supply active upon seed germination (Figure 5). These DEPs also were linked with other DEPs involved in protein metabolism, amino metabolism, carbohydrate metabolism, and lipid metabolism, reflecting a crucial role in metabolism as the source of ATP, acetyl-CoA, NADH, and carbon skeleton (Figure 5).
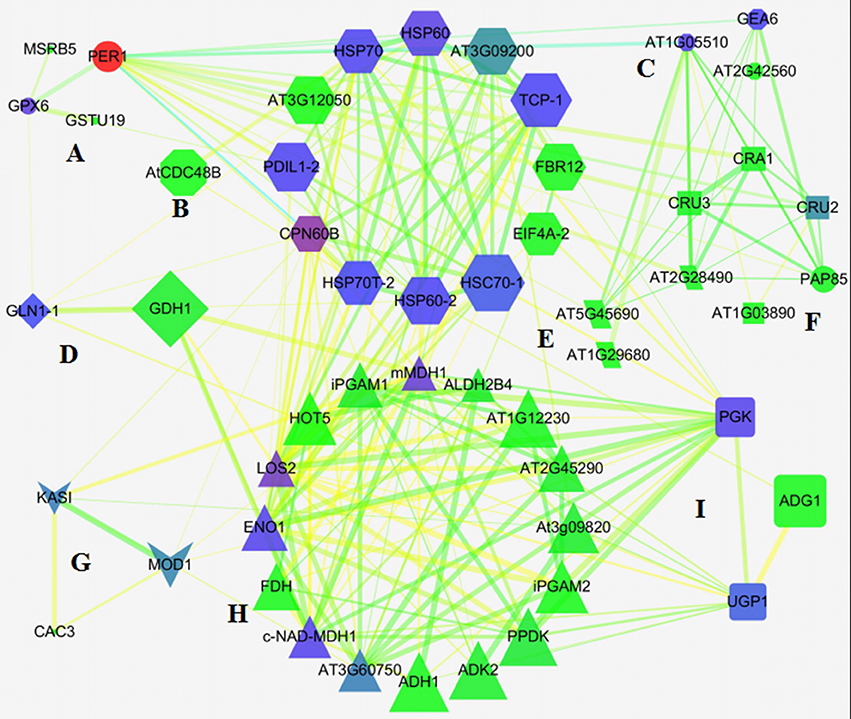
Figure 5. Network of DEPs in germinating seeds. (A) Defense/disease; (B) unknown; (C) seed maturation; (D) amino acids metabolism; (E) protein metabolism; (F) storage proteins; (G) lipid metabolism; (H) energy metabolism; (I) carbohydrate metabolism.
To further analyze the metabolism changes during seed germination, a potential metabolism pathway was constructed based on the DEPs during seed germination of B. napus by using the KEGG database (http://www.kegg.jp/) and previous research (Graham, 2008; He et al., 2011; Han et al., 2013; Figure 6). Here, we use a heatmap to show the expression patterns of the 33 DEPs that were constructed into the potential metabolism pathway according to a search of the KEGG database and previous research (Figure 6A; Graham, 2008; He et al., 2011; Han et al., 2013). These DEPs were located in several pathways, mainly involved in the TCA cycle, glyoxylate cycle, glycolysis/gluconeogenesis, pentose phosphate pathway, and amino acid metabolism (Figure 6B). Combination with the expression pattern shown in Figure 6A clearly revealed that the glycolysis/gluconeogenesis, pentose phosphate pathway, TCA cycle, and glyoxylate cycle might be activated earlier than amino acid metabolism during seed germination, while some sulfur amino acid metabolism-related DEPs, which were closely associated with plant redox homeostasis and defense systems (Rajjou et al., 2012), were also activated upon seed germination.
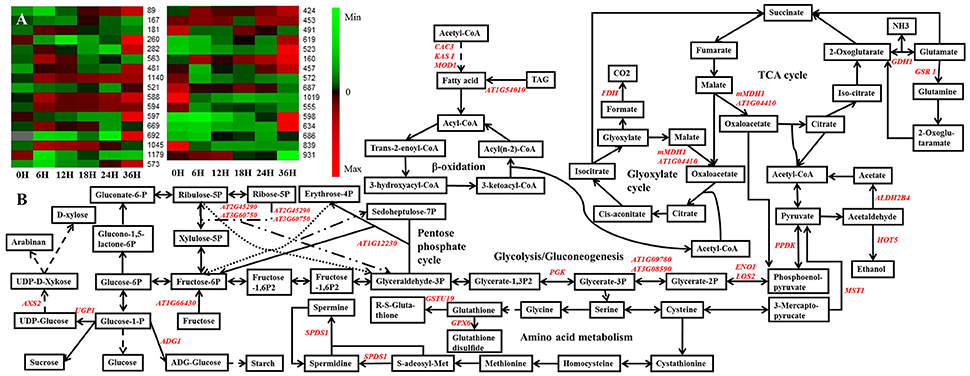
Figure 6. The metabolic pathways based on the proteomic data. (A) The expression pattern of DEPs showed in the metabolic pathways. (B) the metabolic pathways. Gene symbol and corresponding DEP spot numbers are as follows: ADG1, spot 491; ALDH2B4, spot 1179; AT1G04410, spot 1045; AT1G09780, spot 260; AT1G12230, spot 597; AT1G54010, spot 572; AT1G66430, spot 619; AT2G45290, spot 167; AT3G08590, spot 282; AT3G60750, spot 181; AXS2, spot 523; CAC3, spot 160; ENO1, spot 481; FDH, spot 573; GDH 1, spot 555; GPX6, spot 931; GSR 1, spot 598; GSTU19, spot 839; HOT5, spot 521 and 563; KAS I, spot 457; LOS2, spot 1140; mMDH1, spot 669 and 692; MOD1, spot 687 and 1019; MST1, spot 686; PGK, spot 588 and 594; PPDK, spot 89; SPDS1, spot 634; UGP1, spot 424 and 453.
To unravel the relationship between these DEPs and the genetic control of seed germination and vigor, the candidate genes for seed germination and vigor revealed through genome-wide association mapping performed by Hatzig et al. (2015) were used for further analysis. Through searching genes that were detected within and near 20 associated LD regions of six traits related to seed germination and vigor, 17 genes corresponding to 11 DEPs were identified in B. napus (Table 3). Among them, DEP AT4G25580, annotated as CAP160 protein, corresponding to BnaC07g39780D located near a LD region was associated with volume increase. Five DEPs (corresponding to BnaA03g15330D, BnaA03g16750D, BnaA03g32320D, BnaA03g33160D, and BnaC09g39680D) that were involved in protein and amino acid metabolism and the glycolysis pathway (Tables 2, 3) and located near three LD regions were associated with GR72 on chromosomes A03 and C09 (2 and 1, respectively) (Table 3). Four genes of three DEPs (APA1, AXS2, and AT1G03890) were observed to be close to a common LD region associated with T50 and GR36 on C08. Of them, the two copies (BnaC08g43570D and BnaC08g43590D) of AT1G03890 that encodes the 12S seed storage protein CRD could be found near this region. The other two DEPs (APA1 and AXS2) were identified as corresponding to BnaA09g47450D and BnaA09g48990D, and were close to another LD region, just associated with T50 on A09. APA1 (aspartic proteinase A1) is involved in proteolysis and response to salt stress during the seedling development stage in Arabidopsis (Huttlin et al., 2007; Mazorra-Manzano et al., 2010). AXS2, a UDP-D-apiose/UDP-D-xylose synthase, is involved in amino acid metabolism during the seedling development stage (Giavalisco et al., 2005). Near an ES-related LD region on chromosome A10, three copies (BnaA10g27060D, BnaA10g27070D, and BnaA10g27080D) of the gene that encodes HSC70-1 (a heat shock cognate protein) were identified. ADK2 (adenosine kinase 2) was also located near this region, and GDH1 (glutamate dehydrogenase 1) was identified adjacent to another LD region associated with ES on chromosome A10.
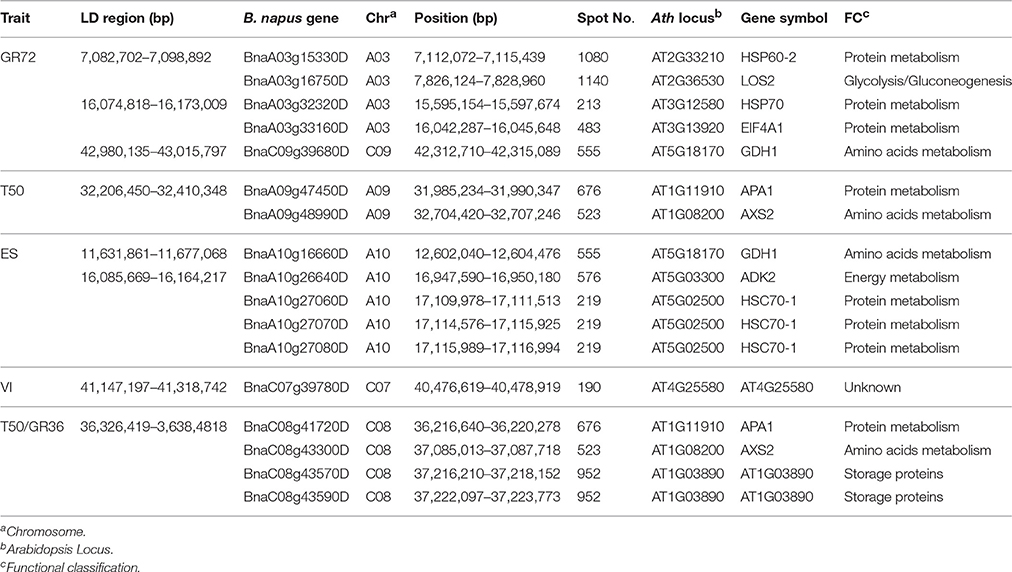
Table 3. DEPs verified within and near associated LD regions in seed germination and vigor-related genome-wide association mapping.
By combing our proteomic analysis results with genome-wide analysis results reported previously, we found 17 genes corresponding to 11 DEPs located in or near the confidence interval of QTLs for seed germination and vigor. The gene expression upon seed germination was analyzed by quantitative real-time (qRT)-PCR. The present results demonstrated that seven genes showed multiple expression patterns (Figure 7), and these were involved in glycolysis/gluconeogenesis, amino acid metabolism and protein metabolism (Table 2). Among them, glutamate dehydrogenase 1 (GDH1, spot 555) appeared to have similar expression patterns to the corresponding DEPs (Figure 7). In addition, three metabolism-related genes and one energy metabolism-related gene, Aha1 domain-containing protein (AT3G12050, spot 483), 60S acidic ribosomal protein P0-2 (APA 1, spot 676), heat shock protein 60-2 (HSP60-2, spot 1080), and adenosine kinase 2 (ADK2, spot 576), showed consistent trends of expression with the homologous proteins. Furthermore, gene expression of heat shock protein 70-4 (HSP70, spot 213) and bifunctional enolase 2/transcriptional activator (LOS2, spot 1140), which are involved in protein folding and energy metabolism, showed opposite trends to the expression of the homologous proteins, respectively (Figure 7). The results indicated that the aforementioned metabolic processes were modulated by post-transcriptional and/or post-translational regulation during seed germination. The inconsistent abundances of transcripts and proteins in germinating seed also supported the notion that pre-synthesized mRNA and proteins in mature dry seeds would function for seed germination (Sano et al., 2012).
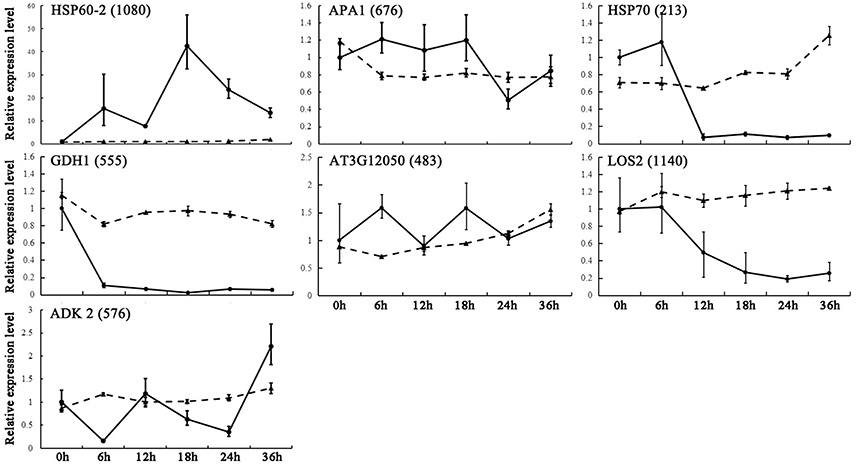
Figure 7. qRT-PCR analysis of gene expression patterns in germinating seeds. Relative expression levels of homologous genes of DEPs from B. napus seeds were determined by qRT-PCR analysis. The value were determined 0, 6, 12, 18, 24, and 36 HAI and are presented as means ± standard error (n = 3). Different lines show relative expression levels of homologous genes (solid line) and corresponding proteins (dotted line).
The physical and metabolic events occurring during germination and early seedling growth include respiration, DNA synthesis, transcription and translation of new mRNAs, radicle emergence, mobilization of reserves in storage tissues, and mobilization of major reserves (Graham, 2008; Nonogaki et al., 2010). Heterotrophic metabolism is suspected to contribute to seed germination, and the present TEM results revealed that the predominant reserve organs (OBs and PBs) decreased gradually during seed germination. Similarly, a clear decrease in the number of PBs and OBs after 3 and 4 days of olive (Olea europaea L.) cotyledon cell culture was also demonstrated (Zienkiewicz et al., 2011); an obvious decrease of OBs was also observed at 48 h of imbibition in the endosperm cells of germinating J. curcas seed (Yang et al., 2009). Consistently, it was observed that oil content remained stable at the early stage of germination and was rapidly consumed during seedling establishment in wild-type Arabidopsis WS and Col0 as well as the acyl-CoA oxidase gene mutants acx1-1, acx2-1, and acx1-1acx2-1 (Pinfield-Wells et al., 2005); the present results also showed a similar tendency in B. napus. This indicated that seed storage oil mobilization might be not essential for germination but is very important for seedling establishment.
By introduction of fluorescent cyanine dyes and an internal standard, the ability of 2-D DIGE to separate and resolve complex proteomic patterns was greatly improved (Van den Bergh and Arckens, 2004; Rozanas and Loyland, 2008). Although, 2-D DIGE has not totally overcome the shortfalls of gel-based proteomics technology, there is still not a method that can completely replace 2-D DIGE to simultaneously separate and display several thousand proteins from complex samples. Besides these above-mentioned physiology and biochemistry results, 46.9% DEPs involved in energy metabolism, carbohydrate metabolism, protein metabolism, amino acid metabolism, and lipid metabolism were also observed by 2-D DIGE combined with MS and MS/MS and a Mascot database search in B. napus. Similarly, 50 DEPs were identified during J. curcas seed germination, 34% (17) of which were oil mobilization-related proteins involved in the glyoxylate cycle, glycolysis, the citric acid cycle, gluconeogenesis, and the pentose phosphate pathway (Yang et al., 2009). With the assistance of KEGG analysis and previous research (Graham, 2008; He et al., 2011; Han et al., 2013), a metabolism pathway was constructed containing the DEPs involved during seed germination in B. napus. It was revealed that the pentose phosphate pathway (e.g., transketolase and transketolase, spots 167 and 181), TCA cycle (e.g., malate dehydrogenase, spots 669, 692, and 1045), glycometabolism (e.g., pfkB-like carbohydrate kinase family protein, spot 619), amino acid metabolism (e.g., mercaptopyruvate sulfurtransferase 1 and spermidine synthase 1, spots 686 and 634), and fatty acid metabolism (e.g., alcohol dehydrogenase 2B4, spot 1179) were actively altered upon seed germination (Figure 6; Table 2). This indicated that the mobilization and sequential hydrolysis of storage reserves (proteins, starch, and lipids) are tightly controlled temporally and spatially, which is important for energy metabolite biosynthesis during seed germination.
In B. napus, the necessary amino acids required for the de novo synthesis of proteins after germination is provided by cruciferin and napin (Nykiforuk and Johnson-Flanagan, 1999). The major storage protein, cruciferin, accounts for ~50–60% of the total seed protein (Crouch and Sussex, 1981; Höglund et al., 1992). Cruciferin was shown to be significantly decreased after 48 HAI at 22°C and was no longer detected at 72 HAI in B. napus (Nykiforuk and Johnson-Flanagan, 1999). The present results also revealed that the total protein content started to decrease sharply from 36 HAI, and 26 DPEs assigned as storage proteins during seed germination were identified, of which, 16 spots were dramatically decreased after 36 HAI. As storage proteins might have been generated by the initiation of seed storage mobilization, it is not surprising that 10 spots that might have been generated through the breakdown of storage proteins were significantly increased at 36 HAI (Table S2). Consistent with previous studies (Nykiforuk and Johnson-Flanagan, 1999; Fu et al., 2005; Yang et al., 2009), our results showed that heterotrophic metabolism is active in germinating B. napus seed in the dark. Storage sugar and protein, as the main energy and metabolite sources during B. napus seed germination sensu stricto, and the consumption of storage oil and protein are essential for seedling establishment.
Maturation usually happens during seed formation; however, Lopez-Molina et al. (2001) proposed that the formation of embryos also arrested in their progress toward germination based on the known function of the genes controlled by ABI3 during seed maturation, including those encoding LEA proteins. Seed germination of Arabidopsis was also shown to reset the maturation program, as the de novo synthesis of LEA proteins and storage proteins (cruciferins) was observed (Rajjou et al., 2004). The possibility of recapitulating the late maturation program at the transcriptional and translational levels would allow rapid adjustment of the response of imbibed seeds confronted with rapid fluctuations in environmental conditions (Lopez-Molina et al., 2001; Rajjou et al., 2004, 2006). A seed maturation-related protein (spot 1025, uncharacterized protein, corresponding gene: AT1G05510) induced during the B. napus seed germination was also observed; this showed that the maturation program might also be recapitulated during early stages of seed germination in B. napus.
It is well-known that plants have devised sophisticated mechanisms to cope with biotic and abiotic stresses imposed by their environment. In the plant life cycle, the seed and seedling stages are key developmental stages that are sensitive to stresses (Bewley and Black, 1994; Koornneef et al., 2002). The present results revealed that the expression of some sulfur amino acid metabolism-related proteins was increased during seed germination in B. napus. In both plants and animals, glutathione S-transferases are induced by diverse environmental stimuli and play direct roles in reducing oxidative damage or toxic products produced during xenobiotic metabolism (Dixon et al., 2002; Moons, 2005; Frova, 2006; Xu et al., 2016). The glutathione S-transferase TAU 19 (GSTU19; spot 839) was observed to have a relatively high expression level even at 36 HAI that was decreased upon seed germination. Glutathione peroxidases (spot 931) are a group of enzymes that protect cells against oxidative damage generated by reactive oxygen species, and play an important role in conversion of glutathione and glutathione disulfide, which is critical for the reduction of H2O2, organic hydroperoxides, and lipid peroxides (Sugimoto and Sakamoto, 1997; Milla et al., 2003); the expression of this enzyme was induced after imbibition but started to be repressed at 18 HAI. Methionine sulfoxide reductase B5 (spots 961 and 967) can reverse the reaction of methionine oxidation to methionine sulfoxide, which results in modification of activity and conformation for many proteins (Rouhier et al., 2006); these spots showed high expression levels during germination and post-germination growth and all were decreased at 36 HAI. The sulfur amino acid metabolism pathway represents a determinant biochemical key of the commitment of the seed to initiate its development toward germination (Rajjou et al., 2012), and inhibition of sulfur amino acid metabolism can strongly delay seed germination and seedling growth (Gallardo et al., 2002; Bassel et al., 2008; Fulneček et al., 2011). Methionine is a fundamental metabolite among the essential amino acids synthesized by plants; it does not only function as a building block for protein synthesis but also as the precursor of S-adenosylmethionine, the universal methyl-group donor, and the precursor of polyamines, ethylene, and the vitamin biotin (Ranocha et al., 2001). In Arabidopsis, some other sulfur amino acid metabolism-related proteins, such as methionine synthase and S-adenosylmethionine synthase, also showed accumulation at different stages of seed germination (Gallardo et al., 2001, 2002; Rajjou et al., 2012). These results highlight the important role of sulfur amino acid metabolism during seed germination.
Besides the sulfur amino acid metabolism pathway-related proteins, the myrosinase-glucosinolate system also is involved in plant development and defense, and could affect the behavior of herbivorous insects and pathogens (Rask et al., 2000; Capella et al., 2001). Myrosinase-binding proteins were identified as components of high-molecular-mass myrosinase complexes in extracts of B. napus seeds (Lenman et al., 1990; Falk et al., 1995; Taipalensuu et al., 1997). It was also shown that complex formation of myrosinase isoenzymes in oilseed rape seeds was dependent on the presence of myrosinase-binding proteins (Eriksson et al., 2002). Consistent with the results showed by Geshi and Brandt (1998), four myrosinase-binding protein spots were shown to be increased during seed germination and were highly expressed at 36 HAI. This implied the onset of defense mechanisms at the level of seed germination.
JG carried out the proteomic analysis and wrote the manuscript and Lu Gan participant in the proteomic experiments. HC built DEPs identification through the results of the genome-wide associated mapping and completed the manuscript. Liangxing Guo, NR, and KZ made helpful suggestions to the manuscript and participated in the qRT-PCR experiments. HW and YL provided plant materials. ML designed, led, and coordinated the overall study.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The National Science Foundation of China (31471532), the scientific research project of Hubei Collaborative Innovation Center for the Characteristic Resources Exploitation of Dabie Mountains (2015TD03), and the New Century Talents Support Program by the Ministry of Education of China (NCET110172).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01482
Table S1. Primer design for genes encoding key DEPs involved in seed germination.
Table S2. Protein expression patterns of DEPs during germination. 0H, 6H, 12H, 18H, 24H, and 36H represent the 0, 6, 12, 18, 24, and 36 HAI.
Table S3. The corresponding unique genes of DEPs. The unique genes that matched with each DEPs were searched by blast in NCBI database. achrome; bthe gi number of each deferentially expressed protein in the NCBI database; cthe name representing each DEPs in STRING.
Table S4. The peptide matches of DEPs. Peptide sequences identified by MS-MS sequencing and corresponding to DEPs in B. napus seeds identified in this work. % Cov, coverage.
Figure S1. The content of each fatty acid and total fatty acids determined during seed germination. (A) The change of total fatty acid content during B. napus seed germination; (B,C) The change of each fatty acid content during B. napus seed germination.
Figure S2. The significantly differently expressed DEPs during seed germination and seedling establishment. 6 h/0 h, 12 h/6 h, 18 h/12 h, 24 h/18 h, and 36 h/24 h represent the comparison among different germination stages.
Figure S3. The protein-protein interaction network in germinating seed revealed by STRING analysis. The name of each node is the same as the STRING symbol in Table S3.
Figure S4. The replicate DIGE gels of different germination stages. Green color shows that the sample was labeled by cy3. Red color shows the sample was labeled by cy5.
Bassel, G. W., Fung, P., Chow, T. F., Foong, J. A., Provart, N. J., Cutler, S. R., et al. (2008). Elucidating the germination transcriptional program using small molecules. Plant Physiol. 147, 143–155. doi: 10.1104/pp.107.110841
Bassel, G. W., Lan, H., Glaab, E., Gibbs, D. J., Gerjets, T., Krasnogor, N., et al. (2011). Genome-wide network model capturing seed germination reveals coordinated regulation of plant cellular phase transitions. Proc. Natl. Acad. Sci. U.S.A. 108, 9709–9714. doi: 10.1073/pnas.1100958108
Benincasa, P., Pace, R., Quinet, M., and Lutts, S. (2013). Effect of salinity and priming on seedling growth in rapeseed (Brassica napus var oleifera Del.). Acta Sci. Agron. 35, 479–486. doi: 10.4025/actasciagron.v35i4.17655
Bewley, J. D., and Black, M. (1994). Seeds: Physiology of Development and Germination. New York, NY: Springer US Press. doi: 10.1007/978-1-4899-1002-8
Browse, J., McCourt, P. J., and Somerville, C. R. (1986). Fatty acid composition of leaf lipids determined after combined digestion and fatty acid methyl ester formation from fresh tissue. Anal Biochem. 152, 141–145. doi: 10.1016/0003-2697(86)90132-6
Bybordi, A., and Tabatabaei, J. (2009). Effect of salinity stress on germination and seedling properties in canola cultivars (Brassica napus L.). Not. Bot. Hort. Agrobot. 37, 71. doi: 10.15835/nbha3723299
Capella, A. N., Menossi, M., Arruda, P., and Benedetti, C. E. (2001). COI1 affects myrosinase activity and controls the expression of two flower-specific myrosinase-binding protein homologues in Arabidopsis. Planta 213, 691–699. doi: 10.1007/s004250100548
Crouch, M. L., and Sussex, I. M. (1981). Development and storage-protein synthesis in Brassica napus L. embryos in vivo and in vitro. Planta 153, 64–74. doi: 10.1007/BF00385319
Dixon, R. A., Achnine, L., Kota, P., Liu, C. J., Reddy, M. S., and Wang, L. (2002). The phenylpropanoid pathway and plant defence-a genomics perspective. Mol. Plant Pathol. 3, 371–390. doi: 10.1046/j.1364-3703.2002.00131.x
Eriksson, S., Andréasson, E., Ekbom, B., Granér, G., Pontoppidan, B., Taipalensuu, J., et al. (2002). Complex formation of myrosinase isoenzymes in oilseed rape seeds are dependent on the presence of myrosinase-binding proteins. Plant Physiol. 129, 1592–1599. doi: 10.1104/pp.003285
Falk, A., Taipalensuu, J., Ek, B., Lenman, M., and Rask, L. (1995). Characterization of rapeseed myrosinase-binding protein. Planta 195, 387–395. doi: 10.1007/BF00202596
Frova, C. (2006). Glutathione transferases in the genomics era: new insights and perspectives. Biomol. Eng. 23, 149–169. doi: 10.1016/j.bioeng.2006.05.020
Fu, Q., Wang, B. C., Jin, X., Li, H. B., Han, P., Wei, K. H., et al. (2005). Proteomic analysis and extensive protein identification from dry, germinating Arabidopsis seeds and young seedlings. J. Biochem. Mol. Biol. 38:650. doi: 10.5483/BMBRep.2005.38.6.650
Fulneček, J., Matyášek, R., Votruba, I., Holý, A., Křížová, K., and Kovařík, A. (2011). Inhibition of SAH-hydrolase activity during seed germination leads to deregulation of flowering genes and altered flower morphology in tobacco. Mol. Genet. Genomics 285, 225–236. doi: 10.1007/s00438-011-0601-8
Gallardo, K., Job, C., Groot, S. P., Puype, M., Demol, H., Vandekerckhove, J., et al. (2001). Proteomic analysis of Arabidopsis seed germination and priming. Plant Physiol. 126, 835–848. doi: 10.1104/pp.126.2.835
Gallardo, K., Job, C., Groot, S. P., Puype, M., Demol, H., Vandekerckhove, J., et al. (2002). Proteomics of Arabidopsis seed germination. A comparative study of wild-type and gibberellin-deficient seeds. Plant Physiol. 129, 823–837. doi: 10.1104/pp.002816
Gan, L., Zhang, C. Y., Wang, X. D., Wang, H., Long, Y., Yin, Y. T., et al. (2013). Proteomic and comparative genomic analysis of two Brassica napus lines differing in oil content. J. Proteome Res. 12, 4965–4978. doi: 10.1021/pr4005635
Ge, F., Hu, Y., and Wang, J. (2013). Spatial and temporal gene expression during seed germination of Brassica napus. Acta Physiol. Plant. 35, 2939–2950. doi: 10.1007/s11738-013-1324-8
Ge, F. W., Tao, P., Zhang, Y., and Wang, J. B. (2014). Characterization of AQP gene expressions in Brassica napus during seed germination and in response to abiotic stresses. Biol. Plant. 58, 274–282. doi: 10.1007/s10535-013-0386-1
Geshi, N., and Brandt, A. (1998). Two jasmonate-inducible myrosinase-binding proteins from Brassica napus L. seedlings with homology to jacalin. Planta 204, 295–304. doi: 10.1007/s004250050259
Giavalisco, P., Nordhoff, E., Kreitler, T., Klöppel, K. D., Lehrach, H., Klose, J., et al. (2005). Proteome analysis of Arabidopsis thaliana by two-dimensional gel electrophoresis and matrix-assisted laser desorption/ionisation-time of flight mass spectrometry. Proteomics 5, 1902–1913. doi: 10.1002/pmic.200401062
Graham, I. A. (2008). Seed storage oil mobilization. Annu. Rev. Plant Biol. 59, 115–142. doi: 10.1146/annurev.arplant.59.032607.092938
Guo, B., Chen, Y., Zhang, G., Xing, J., Hu, Z., Feng, W., et al. (2013). Comparative proteomic analysis of embryos between a maize hybrid and its parental lines during early stages of seed germination. PLoS ONE 8:e65867. doi: 10.1371/journal.pone.0065867
Hajduch, M., Casteel, J. E., Hurrelmeyer, K. E., Song, Z., Agrawal, G. K., and Thelen, J. J. (2006). Proteomic analysis of seed filling in Brassica napus. Developmental characterization of metabolic isozymes using high-resolution two-dimensional gel ecectrophoresis. Plant Physiol. 141, 32–46. doi: 10.1104/pp.105.075390
Han, C., Yin, X., He, D., and Yang, P. (2013). Analysis of proteome profile in germinating soybean seed, and its comparison with rice showing the styles of reserves mobilization in different crops. PLoS ONE 8:e56947. doi: 10.1371/journal.pone.0056947
Hatzig, S., Zaharia, L. I., Abrams, S., Hohmann, M., Legoahec, L., Bouchereau, A., et al. (2014). Early osmotic adjustment responses in drought-resistant and drought-sensitive oilseed rape. J. Integr. Plant Biol. 56, 797–809. doi: 10.1111/jipb.12199
Hatzig, S. V., Frisch, M., Breuer, F., Nesi, N., Ducournau, S., Wagner, M. H., et al. (2015). Genome-wide association mapping unravels the genetic control of seed germination and vigor in Brassica napus. Front. Plant Sci. 6:221. doi: 10.3389/fpls.2015.00221
He, D., Han, C., Yao, J., Shen, S., and Yang, P. (2011). Constructing the metabolic and regulatory pathways in germinating rice seeds through proteomic approach. Proteomics 11, 2693–2713. doi: 10.1002/pmic.201000598
Höglund, A. S., Rödin, J., Larsson, E., and Rask, L. (1992). Distribution of napin and cruciferin in developing rape seed embryos. Plant Physiol. 98, 509–515. doi: 10.1104/pp.98.2.509
Huttlin, E. L., Hegeman, A. D., Harms, A. C., and Sussman, M. R. (2007). Comparison of full versus partial metabolic labeling for quantitative proteomics analysis in Arabidopsis thaliana. Mol. Cell. Proteomics 6, 860–881. doi: 10.1074/mcp.M600347-MCP200
Janmohammadi, M., Fallahnezhad, F., Golsha, M., and Mohammadi, H. (2008). Controlled ageing for storability assessment and predicting seedling early growth of canola cultivars (Brassica napus L.). ARPN J. Agric. Biol. Sci. 3, 22–26.
Katayama, H., Nagasu, T., and Oda, Y. (2001). Improvement of in-gel digestion protocol for peptide mass fingerprinting by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 15, 1416–1421. doi: 10.1002/rcm.379
Kelly, A. A., Quettier, A. L., Shaw, E., and Eastmond, P. J. (2011). Seed storage oil mobilization is important but not essential for germination or seedling establishment in Arabidopsis. Plant Physiol. 157, 866–875. doi: 10.1104/pp.111.181784
Kondra, Z. P., Campbell, D. C., and King, J. R. (1983). Temperature effects on germination of rapeseed (Brassica napus L. and B. campestris L.). Can. J. Plant Sci. 63, 1063–1065.
Koornneef, M., Bentsink, L., and Hilhorst, H. (2002). Seed dormancy and germination. Curr. Opin. Plant. Biol. 5, 33–36. doi: 10.1016/S1369-5266(01)00219-9
Kruger, N. J. (2009). “The Bradford method for protein quantitation,” in The Protein Protocols Handbook, ed J. M. Walker (New York, NY: Humana Press), 17–24. doi: 10.1007/978-1-59745-198-7_4
Kubala, S., Garnczarska, M., Wojtyla, Ł., Clippe, A., Kosmala, A., Żmieńko, A., et al. (2015). Deciphering priming-induced improvement of rapeseed (Brassica napus L.) germination through an integrated transcriptomic and proteomic approach. Plant Sci. 231, 94–113. doi: 10.1016/j.plantsci.2014.11.008
Lenman, M., Rödin, J., Josefsson, L. G., and and, Rask, L. (1990). Immunological characterization of rapeseed myrosinase. Eur. J. Biochem. 194, 747–753. doi: 10.1111/j.1432-1033.1990.tb19465.x
Li, F., Wu, X., Tsang, E., and Cutler, A. J. (2005). Transcriptional profiling of imbibed Brassica napus seed. Genomics 86, 718–730. doi: 10.1016/j.ygeno.2005.07.006
Li, G., and McVetty, P. B. (2013). “Genetics and gene mapping of disease resistance in Brassica,” in Translational Genomics for Crop Breeding: Biotic Stress, Vol. 1, eds R. K. Varshney and R. Tuberosa (New York, NY: John Wiley & Sons, Inc.), 327–344. doi: 10.1002/9781118728475.ch16
Liu, A., Gao, F., Kanno, Y., Jordan, M. C., Kamiya, Y., Seo, M., et al. (2013). Regulation of wheat seed dormancy by after-ripening is mediated by specific transcriptional switches that induce changes in seed hormone metabolism and signaling. PLoS ONE 8:e56570. doi: 10.1371/journal.pone.0056570
Long, Y., Shi, J., Qiu, D., Li, R., Zhang, C., Wang, J., et al. (2007). Flowering time quantitative trait loci analysis of oilseed Brassica in multiple environments and genomewide alignment with Arabidopsis. Genetics 177, 2433–2444. doi: 10.1534/genetics.107.080705
Lopez-Molina, L., Mongrand, S., and Chua, N. H. (2001). A postgermination developmental arrest checkpoint is mediated by abscisic acid and requires the ABI5 transcription factor in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 98, 4782–4787. doi: 10.1073/pnas.081594298
Mazorra-Manzano, M. A., Tanaka, T., Dee, D. R., and Yada, R. Y. (2010). Structure-function characterization of the recombinant aspartic proteinase A1 from Arabidopsis thaliana. Phytochemistry 71, 515–523. doi: 10.1016/j.phytochem.2009.12.005
Milla, M. A. R., Maurer, A., Huete, A. R., and Gustafson, J. P. (2003). Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 36, 602–615. doi: 10.1046/j.1365-313X.2003.01901.x
Mohammadi, G. R. (2009). The influence of NaCl priming on seed germination and seedling growth of canola (Brassica napus L.) under salinity conditions. Am. Euras. J. Agric. Environ. Sci. 5, 696–700.
Moons, A. (2005). Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases (GSTs). Vitam. Horm. 72, 155–202. doi: 10.1016/S0083-6729(05)72005-7
Müller, K., Tintelnot, S., and Leubner-Metzger, G. (2006). Endosperm-limited Brassicaceae seed germination: abscisic acid inhibits embryoinduced endosperm weakening of Lepidium sativum (cress) and endosperm rupture of cress and Arabidopsis thaliana. Plant Cell Physiol. 47, 864–877. doi: 10.1093/pcp/pcj059
Nagel, M., Rosenhauer, M., Willner, E., Snowdon, R. J., Friedt, W., and Börner, A. (2011). Seed longevity in oilseed rape (Brassica napus L.)–genetic variation and QTL mapping. Plant Genet. Resour. 9, 260–263. doi: 10.1017/S1479262111000372
Nguyen, T. C., Obermeier, C., Friedt, W., Abrams, S. R., and Snowdon, R. J. (2016). Disruption of germination and seedling development in Brassica napus by mutations causing severe seed hormonal imbalance. Front. Plant Sci. 7:322. doi: 10.3389/fpls.2016.00322
Nonogaki, H., Bassel, G. W., and Bewley, J. D. (2010). Germination—still a mystery. Plant Sci. 179, 574–581. doi: 10.1016/j.plantsci.2010.02.010
Nykiforuk, C. L., and Johnson-Flanagan, A. M. (1999). Storage reserve mobilization during low temperature germination and early seedling growth in Brassica napus. Plant Physiol. Biochem. 37, 939–947. doi: 10.1016/S0981-9428(99)00108-4
Obermeier, C., Hossain, M. A., Snowdon, R., Knüfer, J., von Tiedemann, A., and Friedt, W. (2013). Genetic analysis of phenylpropanoid metabolites associated with resistance against Verticillium longisporum in Brassica napus. Mol. Breed. 31, 347–361. doi: 10.1007/s11032-012-9794-8
Osman, K. A., Tang, B., Wang, Y., Chen, J., Yu, F., Li, L., et al. (2013). Dynamic QTL analysis and candidate gene mapping for waterlogging tolerance at maize seedling stage. PLoS ONE 8:e79305. doi: 10.1371/journal.pone.0079305
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnol Lett. 26, 509–515. doi: 10.1023/B:BILE.0000019559.84305.47
Pinfield-Wells, H., Rylott, E. L., Gilday, A. D., Graham, S., Job, K., Larson, T. R., et al. (2005). Sucrose rescues seedling establishment but not germination of Arabidopsis mutants disrupted in peroxisomal fatty acid catabolism. Plant J. 43, 861–872. doi: 10.1111/j.1365-313X.2005.02498.x
Pritchard, S. L., Charlton, W. L., Baker, A., and Graham, I. A. (2002). Germination and storage reserve mobilization are regulated independently in Arabidopsis. Plant J. 31, 639–647. doi: 10.1046/j.1365-313X.2002.01376.x
Rajjou, L., Belghazi, M., Huguet, R., Robin, C., Moreau, A., Job, C., et al. (2006). Proteomic investigation of the effect of salicylic acid on Arabidopsis seed germination and establishment of early defense mechanisms. Plant Physiol. 141, 910–923. doi: 10.1104/pp.106.082057
Rajjou, L., Duval, M., Gallardo, K., Catusse, J., Bally, J., Job, C., et al. (2012). Seed germination and vigor. Annu. Rev. Plant Biol. 63, 507–533. doi: 10.1146/annurev-arplant-042811-105550
Rajjou, L., Gallardo, K., Debeaujon, I., Vandekerckhove, J., Job, C., and Job, D. (2004). The effect of α-amanitin on the Arabidopsis seed proteome highlights the distinct roles of stored and neosynthesized mRNAs during germination. Plant Physiol. 134, 1598–1613. doi: 10.1104/pp.103.036293
Ranocha, P., McNeil, S. D., Ziemak, M. J., Li, C., Tarczynski, M. C., and Hanson, A. D. (2001). The S-methylmethionine cycle in angiosperms: ubiquity, antiquity and activity. Plant J. 25, 575–584. doi: 10.1046/j.1365-313x.2001.00988.x
Rask, L., Andréasson, E., Ekbom, B., Eriksson, S., Pontoppidan, B., and Meijer, J. (2000). “Myrosinase: gene family evolution and herbivore defense in Brassicaceae,” in Plant Molecular Evolution, eds J. J. Doyle and B. S. Gaut (Dordrecht: Springer Netherlands Press), 93–113. doi: 10.1007/978-94-011-4221-2_5
Reyes, J. L., and Chua, N. H. (2007). ABA induction of miR159 controls transcript levels of two MYB factors during Arabidopsis seed germination. Plant J. 49, 592–606. doi: 10.1111/j.1365-313X.2006.02980.x
Rouhier, N., Vieira Dos Santos, C., Tarrago, L., and Rey, P. (2006). Plant methionine sulfoxide reductase A and B multigenic families. Photosynth. Res. 89, 247–262. doi: 10.1007/s11120-006-9097-1
Rozanas, C. R., and Loyland, S. M. (2008). “Capabilities using 2-D DIGE in proteomics research,” in Tissue Proteomics, Vol. 441, eds B. C.-S. Liu and J. R. Ehrlich (New York, NY: Humana Press), 1–18. doi: 10.1007/978-1-60327-047-2_1
Rücker, B., and Röbbelen, G. (1996). Impact of low linolenic acid content on seed yield of winter oilseed rape (Brassica napus L.). Plant Breed. 115, 226–230. doi: 10.1111/j.1439-0523.1996.tb00908.x
Sano, N., Permana, H., Kumada, R., Shinozaki, Y., Tanabata, T., Yamada, T., et al. (2012). Proteomic analysis of embryonic proteins synthesized from long-lived mRNAs during germination of rice seeds. Plant Cell Physiol. 53, 687–698. doi: 10.1093/pcp/pcs024
Schiessl, S., Samans, B., Hüttel, B., Reinhard, R., and Snowdon, R. J. (2014). Capturing sequence variation among flowering-time regulatory gene homologs in the allopolyploid crop species Brassica napus. Front. Plant Sci. 5:404. doi: 10.3389/fpls.2014.00404
Schopfer, P., and Plachy, C. (1985). Control of Seed Germination by Abscisic Acid III. Effect on Embryo Growth Potential (Minimum Turgor Pressure) and Growth Coefficient (Cell Wall Extensibility) in Brassica napus L. Plant Physiol. 77, 676–686. doi: 10.1104/pp.77.3.676
Shu, L., Lou, Q., Ma, C., Ding, W., Zhou, J., Wu, J., et al. (2011). Genetic, proteomic and metabolic analysis of the regulation of energy storage in rice seedlings in response to drought. Proteomics 11, 4122–4138. doi: 10.1002/pmic.201000485
Sreenivasulu, N., Usadel, B., Winter, A., Radchuk, V., Scholz, U., Stein, N., et al. (2008). Barley grain maturation and germination: metabolicpathway and regulatory network commonalities and differences highlighted by new MapMan/PageMan Profiling Tools. Plant Physiol. 146, 1738–1758. doi: 10.1104/pp.107.111781
Srivastava, S., Fristensky, B., and Kav, N. N. (2004). Constitutive expression of a PR10 protein enhances the germination of Brassica napus under saline conditions. Plant Cell Physiol. 45, 1320–1324. doi: 10.1093/pcp/pch137
Sugimoto, M., and Sakamoto, W. (1997). Putative phospholipid hydroperoxide glutathione peroxidase gene from Arabidopsis thaliana induced by oxidative stress. Genes Genet. Syst. 72, 311–316. doi: 10.1266/ggs.72.311
Taipalensuu, J., Eriksson, S., and Rask, L. (1997). The Myrosinase-binding protein from Brassica napus seeds possesses lectin activity and has a highly similar vegetatively expressed wound-inducible counterpart. Eur. J. Biochem. 250, 680–688. doi: 10.1111/j.1432-1033.1997.00680.x
Tang, W., Deng, Z., Oses-Prieto, J. A., Suzuki, N., Zhu, S., Zhang, et al. (2008). Proteomics studies of brassinosteroid signal transduction using prefractionation and two-dimensional DIGE. Mol. Cell. Proteomics 7, 728–738. doi: 10.1074/mcp.M700358-MCP200
Tsadilas, C. D., and Shaheen, S. M. (2013). Utilization of biosolids in production of bioenergy crops II: impact of application rate on bioavailability and uptake of trace elements by canola. Commun. Soil Sci. Plant Anal. 44, 259–274. doi: 10.1080/00103624.2013.741773
Van den Bergh, G., and Arckens, L. (2004). Fluorescent two-dimensional difference gel electrophoresis unveils the potential of gel-based proteomics. Curr. Opin. Biotech. 15, 38–43. doi: 10.1016/j.copbio.2003.12.001
Wang, H. Z., and Yin, Y. (2014). Analysis and strategy for oil crop industry in China. Chin. J. Oil Crop Sci. 36, 414–421. doi: 10.7505/j.issn.1007-9084.2014.03.020
Wei, F., Yi, J. W., Wang, X. F., Dong, X. Y., Li, P. P., Li, Y. H., et al. (2009). A quick method for oil contents determination in small quantity of rape seed. Chin. J. Oil Crop Sci. 31, 517–521. doi: 10.3321/j.issn:1007-9084.2009.04.021
Wen, C.-f., Dong, A.-w., Li, G.-z., Lei, S., and Lei, Y. (2005). Determination of total sugar and reducing sugar in Viola Philippica ssp. Munda W. Becker by anthrone colorimetry. Mod. Food Sci. Technol. 21, 122–124. doi: 10.3969/j.issn.1673-9078.2005.03.044
Xu, J., Tian, Y. S., Xing, X. J., Peng, R. H., Zhu, B., Gao, J. J., et al. (2016). Over-expression of AtGSTU19 provides tolerance to salt, drought and methyl viologen stresses in Arabidopsis. Physiol. Plant. 156, 164–175. doi: 10.1111/ppl.12347
Xu, X. Y., Fan, R., Zheng, R., Li, C. M., and Yu, D. Y. (2011). Proteomic analysis of seed germination under salt stress in soybeans. J. Zhejiang Univ. Sci. B 12, 507–517. doi: 10.1631/jzus.B1100061
Yang, M. F., Liu, Y. J., Liu, Y., Chen, H., Chen, F., and Shen, S. H. (2009). Proteomic analysis of oil mobilization in seed germination and postgermination development of Jatropha curcas. J. Proteome Res. 8, 1441–1451. doi: 10.1021/pr800799s
Yang, P., Li, X., Wang, X., Chen, H., Chen, F., and Shen, S. (2007). Proteomic analysis of rice (Oryza sativa) seeds during germination. Proteomics 7, 3358–3368. doi: 10.1002/pmic.200700207
Zhang, J., Jiang, F., Yang, P., Li, J., Yan, G., and Hu, L. (2015). Responses of canola (Brassica napus L.) cultivars under contrasting temperature regimes during early seedling growth stage as revealed by multiple physiological criteria. Acta Physiol. Plant. 37, 1–10. doi: 10.1007/s11738-014-1748-9
Zhang, X. K., Yang, G. T., Chen, L., Yin, J. M., Tang, Z. L., and Li, J. N. (2006). Physiological differences between yellow-seeded and black-seeded rapeseed (Brassica napus L.) with different testa characteristics during artificial ageing. Seed Sci. Technol. 34, 373–381. doi: 10.15258/sst.2006.34.2.13
Zhang, Y., Chen, B., Xu, Z., Shi, Z., Chen, S., Huang, X., et al. (2014). Involvement of reactive oxygen species in endosperm cap weakening and embryo elongation growth during lettuce seed germination. J. Exp. Bot. 65, 3189–3200. doi: 10.1093/jxb/eru167
Zheng, G. H., Wilen, R. W., Slinkard, A. E., and Gusta, L. V. (1994). Enhancement of canola seed germination and seedling emergence at low temperature by priming. Crop Sci. 34, 1589–1593. doi: 10.2135/cropsci1994.0011183X003400060031x
Keywords: seed germination, Brassica napus, proteomic analysis, storage reserves
Citation: Gu J, Chao H, Gan L, Guo L, Zhang K, Li Y, Wang H, Raboanatahiry N and Li M (2016) Proteomic Dissection of Seed Germination and Seedling Establishment in Brassica napus. Front. Plant Sci. 7:1482. doi: 10.3389/fpls.2016.01482
Received: 31 May 2016; Accepted: 20 September 2016;
Published: 24 October 2016.
Edited by:
Sun Tae Kim, Pusan National University, South KoreaReviewed by:
Xiaoming Wu, The Chinese Academy of Agricultural Sciences, ChinaCopyright © 2016 Gu, Chao, Gan, Guo, Zhang, Li, Wang, Raboanatahiry and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Maoteng Li, bGltYW90ZW5nNDI2QG1haWwuaHVzdC5lZHUuY24=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.