
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 16 September 2016
Sec. Plant Nutrition
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.01398
This article is part of the Research TopicNutrient sensing, signaling and transport in plantsView all 12 articles
Inorganic phosphate is one of key macronutrients essential for plant growth. The acquisition and distribution of phosphate are mediated by phosphate transporters functioning in various physiological and biochemical processes. In the present study, we comprehensively evaluated the phosphate transporter (PHT) gene family in the latest release of the Populus trichocarpa genome (version 3.0; Phytozome 11.0) and a total of 42 PHT genes were identified which formed five clusters: PHT1, PHT2, PHT3, PHT4, and PHO. Among the 42 PHT genes, 41 were localized to 15 Populus chromosomes. Analysis of these genes led to identification of 5–14 transmembrane segments, most of which were conserved within the same cluster. We identified 234 putative cis elements in the 2-kb upstream regions of the 42 PHT genes, many of which are related to development, stress, or hormone. Tissue-specific expression analysis of the 42 PtPHT genes revealed that 25 were highly expressed in the roots of P. tremula, suggesting that most of them might be involved in Pi uptake. Some PtPHT genes were highly expressed in more than six of the twelve investigated tissues of P. tremula, while the expression of a few of them was very low in all investigated tissues. In addition, the expression of the PtPHT genes was verified by quantitative real-time PCR in four tissues of P. simonii. Transcripts of 7 PtPHT genes were detected in all four tested tissues of P. simonii. Most PtPHT genes were expressed in the roots of P. simonii at high levels. Further, PtPHT1.2 and PtPHO9 expression was increased under drought conditions, irrespective of the phosphate levels. In particular, PtPHT1.2 expression was significantly induced by approximately 90-fold. However, the transcriptional changes of some PtPHT genes under drought stress were highly dependent on the phosphate levels. These results will aid in elucidation of the functions of PtPHT in the growth, development, and stress response of the poplar plant.
Phosphorus is a major macronutrient required for plant growth and development. It constitutes up to 0.2% of the dry weight of plant cells and consequently is required in significant quantities (Schachtman et al., 1998). However, it is often a limiting factor for plant growth because the soil inorganic phosphate (Pi) concentration is usually lower than 10 μM (Shen et al., 2011). Plants have adopted several strategies to increase the acquisition of poorly available P source (Bucher, 2007). For example, plants could produce root exudates such as organic acids, sugars, and phenolic acids in the rhizosphere to improve the acquisition of poorly available P from soil (Cesco et al., 2010; Valentinuzzi et al., 2015). Additionally, plants have evolved specific transport systems for the uptake and partitioning of Pi (Poirier and Bucher, 2002). Both low- and high-affinity Pi-transporter systems have been identified in plants (Clark et al., 2000; Misson et al., 2004). The genes encoding for low-affinity Pi transport are constitutively expressed and function at relatively high Pi in the millimolar range, whereas high-affinity Pi transporters are transcriptionally induced at low Pi levels in the micromolar range (Raghothama, 2000).
Generally, the phosphate transporters are classified into PHT1, PHT2, PHT3, and PHT4 families (Rausch and Bucher, 2002; Liu et al., 2011). PHT1s belonging to the major facilitator superfamily are homologs of the yeast PHO84 Pi transporter (Pao et al., 1998). Since the initial identification of PHT1 genes in Arabidopsis (Muchhal et al., 1996), homologs of PHT1 transporters have been characterized in many species, such as tomato (Chen et al., 2014), rice (Liu et al., 2011), and poplar (Loth-Pereda et al., 2011). The PHT1 family, as H+/Pi symporters, has been reported to include proteins involved in the Pi uptake from the soil and distribution within plants (Nussaume et al., 2011). Regarding the PHT2 family of Pi transporters, one member (PHT2;1) has been functionally studied in Arabidopsis (Daram et al., 1999). PHT2 family proteins function as H+/Pi cotransporters in the plastids of plants (Versaw and Harrison, 2002). In addition, the PHT3 family was reported after the cloning of the first putative mitochondrial Pi transporter gene in plants (Takabatake et al., 1999). The PHT3 family, which are supposed to act as a Pi/H+ symporter Pi/OH− antiport, catalyze Pi/Pi exchange between the matrix and cytosol (Stappen and Krämer, 1994). This family included at least three members in Arabidopsis to date, and all of them show high conservation within the mitochondrial transporter family (Rausch and Bucher, 2002). The PHT4 gene family, which includes six members in Arabidopsis, has been reported to be involved in the Pi transport in plastids and the Golgi apparatus (Guo et al., 2008). Recently, the PHO1 gene in Arabidopsis was identified by a map-based positional cloning strategy (Hamburger et al., 2002). The members of PHO1 family were mainly expressed in root stellar cells and in the lower part of the hypocotyl, which is consistent with the role of PHO1 in the loading of Pi to the xylem (Poirier and Bucher, 2002). The expression patterns of 11 PHO1 homologs in the Arabidopsis genome suggest that PHO1 proteins likely play a role in transferring Pi to the vascular cylinders in various tissues, as well as in acquiring Pi into cells, such as pollen, root epidermal cells, and cortical cells (Wang et al., 2004). Most studies of PHT have focused on a single cluster of the PHT family, such as the PHT1 family (Loth-Pereda et al., 2011; Nussaume et al., 2011); however, to our knowledge, there is no report of the whole PHT family, including PHT1, PHT2, PHT3, PHT4, and PHO, in any plant species. In addition, there are few reports of coordinated expression of the entire set of PHTs on a global scale in woody tree species.
Poplar is a model woody tree species used in functional genomic studies due to its small genome, easy transformation, fast growth, and vegetative propagation (Brunner et al., 2004a). Populus simonii is an important poplar species in the northwestern area of China with a wide distribution, and it is capable of growing under low- or high-Pi conditions (Weisgerber and Han, 2001). In this study, we conducted comprehensive genome-wide analysis to identify phosphate transporter gene family in poplar, including PHT1, PHT2, PHT3, PHT4, and PHO, in the P. trichocarpa genome (version 3.0; Phytozome 11.0); in addition, we analyzed the phylogeny, gene structures, genomic locations, and cis elements of these genes. We also investigated the expression patterns of all of the PHT genes in various tissues, as well as their expression profiles at different Pi levels (Pi starvation, low and high Pi levels) in P. simonii. Moreover, we examined the transcriptional profiles of PHT genes in P. simonii roots under drought conditions. The results of this study may provide a basic foundation for future functional genomic studies of the PHT family in poplar.
The cuttings of P. simonii (ca. 15 cm in length, 2 cm in diameter) were rooted in pots (10 L) filled with fine sand. The plants were cultivated in a glasshouse (natural light, 75% relative humidity) and irrigated with 50 ml nutrient solution (Hoagland and Arnon, 1950) (10 μM EDTA·FeNa, 5 μM MnSO4·H2O, 1 μM ZnSO4·7H2O, 1 μM CuSO4·5H2O, 30 μM H3BO3, 0.5 μM H2MoO4, 1000 μM MgSO4·7H2O, 1000 μM CaCl2, 1000 μM Na2SO4, and 1000 μM NH4NO3, pH 6.5) containing Pi at one of three different levels (0 μM, 1000 μM, and 3000 μM KH2PO4) separately every other day. Each Pi treatment was applied to 12 plants for a period of 8 weeks. In addition, a phosphate-free solution was prepared by replacing KH2PO4 with K2SO4. Six plants for each Pi treatment were subjected to drought stress for 2 weeks before harvesting, and the other six plants for each treatment were not subjected to drought stress and served as controls. After the treatments, whole plantlets were harvested and separated into roots, stems, new leaves, and old leaves. Then, samples from the same tissues were pooled for each treatment, frozen in liquid nitrogen for further analysis.
To identify phosphate transporters in poplar, the reported protein sequences from both Arabidopsis and Oryza sativa were used as query sequence with E-value cutoff set as 1e−10 to perform a local BLASTP against P. trichocarpa JGI gene catalog (phytozome v11.0, https://phytozome.jgi.doe.gov/pz/portal.html), respectively. The choice of candidate phosphate transporters was based on the E-value, the sequence homology value (>60%) and the value of score (>450). All corresponding DNA and protein sequences of poplar phosphate transporters were obtained from Phytozome 11.0 (http://phytozome.jgi.doe.gov/pz/portal.html) and also verified with the Pfam database (http://pfam.xfam.org/). Various splicing variants of one gene and incomplete genes with short length were discarded. In addition, redundant sequences were removed, and the remaining sequences were used in further analyses.
For each putative protein, the grand average of hydropathicity (GRAVY) was calculated using ProtParam (http://web.expasy.org/protparam/). Further, TMpred (http://www.ch.embnet.org/software/TMPRED_form.html) was used to predict the transmembrane domains (TMDs) of each PHT protein. In addition, the subcellular location of each PHT was predicted using WoLF PSORT (http://www.genscript.com/wolf-psort.html). Multiple amino acid alignments of the PHTs were generated using ClustalW (Larkin et al., 2007). Phylogenetic tree was constructed using the neighbor-joining method with the 1000 bootstrap in MEGA 6 (http://www.megasoftware.net/).
The chromosomal locations and gene structures of the PHT genes were obtained from Phytozome 11.0. The chromosomal locations of the PHT genes were mapped with MapDraw program (Liu and Meng, 2003). The exon/intron organization of the individual PHT gene was drawn using Gene Structure Display Server (GSDS) program (Hu et al., 2015).
Promoter sequences located 2 kb upstream of the transcription start sites (ATG codons) of the Populus PHT genes were retrieved from the Populus trichocarpa genome v3.0 in Phytozome 11.0 (http://phytozome.jgi.doe.gov/pz/portal.html). The cis-regulatory elements were scanned in PLACE (Higo et al., 1999).
Total RNA was isolated from roots, stems, new leaves, and old leaves using a plant RNA extraction kit (R6827, Omega Bio-Tek, GA, USA) according to the manufacturer's instructions. The first-strand cDNA was synthesized using a PrimeScript RT Reagent Kit (DRR037S, Takara, Dalian, China) following the removal of trace genomic DNA using DNase I (E1091, Omega Bio-Tek). The specific primers (Table S1) for quantitative real time PCR analysis were designed by Primer Premier 6.0 (Premier Biosoft, Palo Alto, CA, USA). We performed PCR in a 20 μl reaction mixture containing 10 μl of 2X SYBR Green Premix Ex Taq II (Bioteke, China), 2 μl of cDNA, and 1 μl of 20 mM of each primer (Table S1) using a Roche LightCycle 96 machine (Roche, Germany). PCR amplification was performed under the following conditions: one cycle of 2 min at 95°C, followed by 45 cycles at 95°C for 10 s, 55°C for 20 s, and 72°C for 20 s. Actin2/7 was used as a reference gene (Brunner et al., 2004b). Three biological replicates with three technical replicates were assayed for each sample. Reactions for the reference gene were included in each plate. The relative expression levels of all the PtPHT genes were calculated using the 2−ΔΔCT method (Livak and Schmittgen, 2001).
We identified a total of 42 putative PtPHT genes, including 12 PHO, 14 PtPHT1, 2 PtPHT2, 6 PtPHT3, and 8 PtPHT4 genes from the genome of P. trichocarpa according to the full AtPHT family in Arabidopsisf (Table 1). The lengths of the encoded proteins ranged from 314 amino acids to 802 amino acids, and their sequences contained 5 to 14 TMDs. The molecular weight (MW) of 42 putative proteins ranged from 33.5 to 93.5 kDa. The GRAVY value of putative phosphate transporter protein (PHT family) was positive and ranged from 0.128 to 0.795, while that of PHO family was negative ranging from −0.255 to −0.064.
The amino acid sequences of all of the PHT genes of P. trichocarpa were aligned with those of A. thaliana. The resulting phylogenetic tree contained five clusters with PHO, PHT1, PHT2, PHT3, and PHT4, respectively (Figure 1). Based on the phylogenetic analysis results, the P. trichocarpa PHT genes were named after the A. thaliana PHT genes (Figure 1, Table 1).
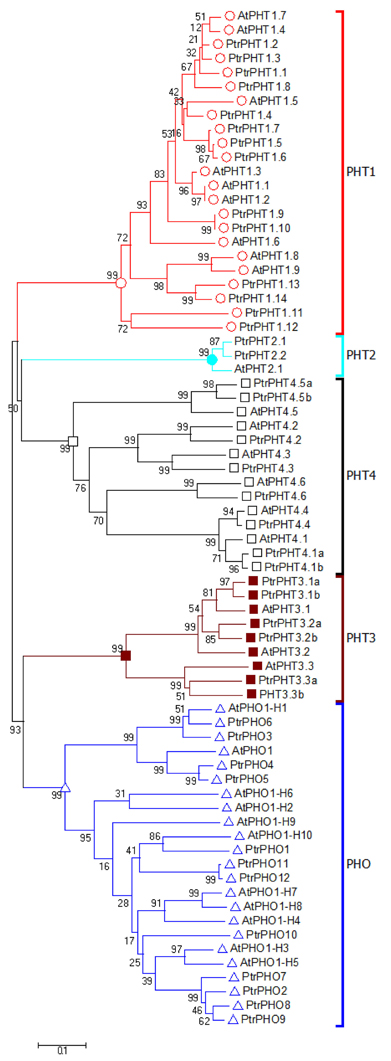
Figure 1. Phylogenetic tree of P. trichocarpa and A. thaliana PHT proteins. The phylogenetic tree was constructed using the neighbor-joining method with the 1000 bootstrap in MEGA 6 (http://www.megasoftware.net/).
The PtPHT genes were marked on a physical map of Populus linkage groups based on the location information for PHTs in Phytozome 11.0. The Populus PtPHT genes showed a heterogeneous distribution pattern among the chromosomes (Figure 2). A total of 41 of the 42 PtPHT genes were localized to 15 of the 19 linkage groups of Populus (Figure 2). Only PtPHO12 was localized to unattributed scaffold fragments.
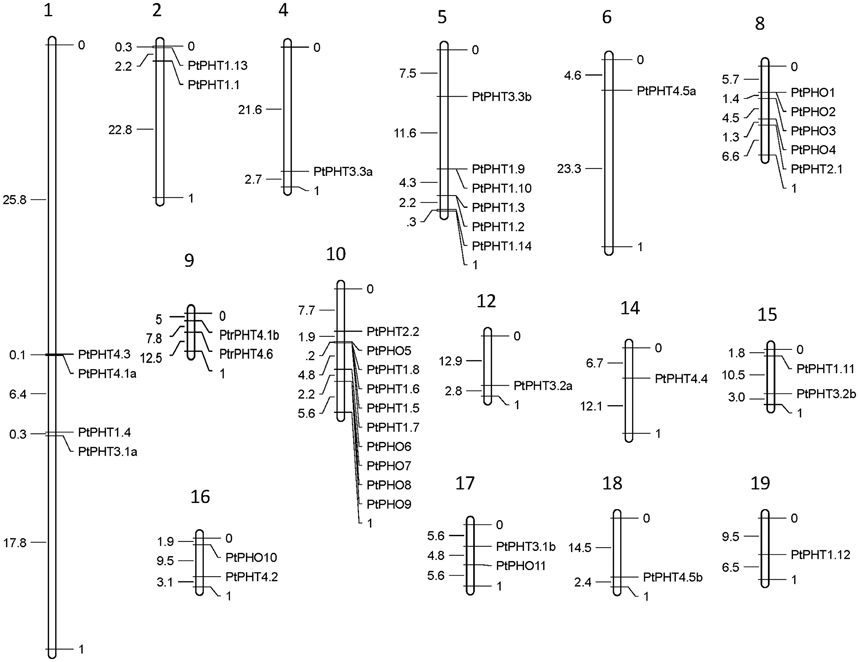
Figure 2. Chromosomal locations of Populus PHT family members. The chromosomal locations of the PHT genes were mapped with MapDraw program (Liu and Meng, 2003).
The lengths of the 42 PtPHT genes ranged from 945 to 2409 bp, and they contained 1 to 16 exons (Table 1). These genes were distributed across 15 of the 19 poplar chromosomes excluding chromosomes 3, 7, 11, and 13 (Table 1, Figure 2). In addition, the lengths of the 12 PtPHO genes ranged from 1890 to 2409 bp, and they contained 11 to 14 exons (Table 1). Most of these genes were localized to chromosomes 8 and 10 (Table 1). Further, the lengths of the 14 PtPHT1 genes ranged from 1311 to 1620 bp, and they contained 1 to 3 exons. Most of these genes were located on chromosomes 5 and 10 (Table 1). Two members of the PHT2 family, PtPHT2.1 and PtPHT2.2, had lengths of 963 and 1773 bp and contained 2 and 3 exons, respectively. The PHT3 family included six PtPHT genes in poplar, with lengths ranging from 945 to 1125 bp and containing 6 exons. Further, the PHT4 family included nine PtPHT genes, with lengths ranging from 864 to 1662 bp and containing 1 to 11 exons.
We further analyzed the exon/intron structures of the 42 PtPHT genes. The genes belonging to the same PHT family, with the exception of the PHT2 family, were well conserved in terms of their exon/intron structures, with similar exon lengths (Figure 3). Unexpectedly, compared with other members of the PHT4 family, which contained 7–16 exons, PtPHT4.6 possessed only one long exon, and its coding sequence was shorter. These substantial differences in gene structure are due to differences in the numbers of exons and introns among the various genes.
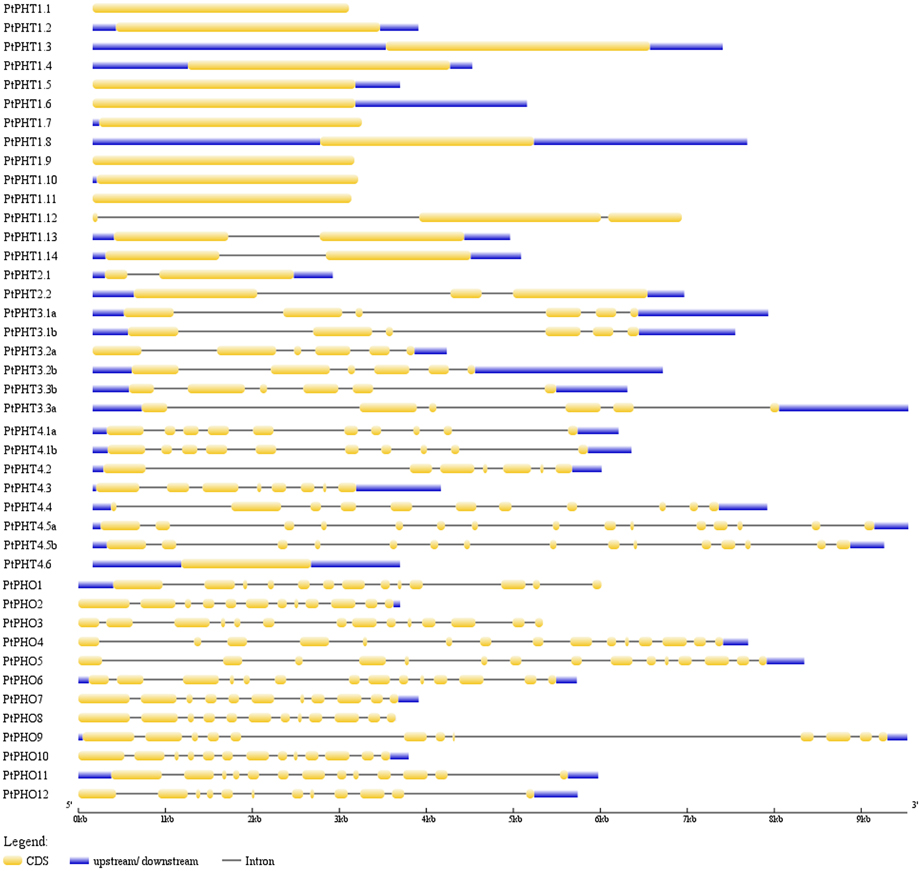
Figure 3. Exon/intron structures of PHT genes in Populus. The exon/intron organization of the individual PHT gene was drawn using Gene Structure Display Server (GSDS) program (Hu et al., 2015).
The 2 kb upstream regions of the 42 PtPHT genes were analyzed using PLACE signal scan program. A total of 234 putative cis elements were detected, many of which are involved in development, stress, or hormone activity (Table S2). Twenty-six of the detected cis elements were found in all of the PtPHT genes involved in energy, light, hormone, defense, carbon metabolism, and tissue-specific expression. For example, GATABOX, GT1CONSENSUS, and IBOXCORE are light-responsive cis elements, and CACTFTPPCAL is carbon metabolism related cis element (Gowik et al., 2004).
Some Pi-response and other stress-related cis regulatory elements including ABRE elements, helix–loop–helix elements, PHO-like, PHR1, TATA-box-like, and WRKY1 were detected in the upstream regions of PtPHT genes. AACACOREOSGLUB1, a WRKY1 element, and ACGTABREMOTIFA2OSEM, an ABRE element, were present in 22 and 7 PtPHT genes, respectively. In addition, A PHO-like element (AACACOREOSGLUB1) and a TATA-box-like element (TATABOX2) was detected in 22 and 34PtPHT genes, respectively; A PHR1 element (P1SB) was observed in 20 PtPHT genes; and a helix–loop–helix element, (CATATGGMSAUR) was present in 11 PtPHT genes. Furthermore, 17 PtPHT genes had typical cis elements that were present especially in the respective genes, and the total number of these cis elements was 32.
The specificities of tissues or organs may allow for the detailed elucidation of their functions. We measured the transcript levels of all of the PtPHT genes in 12 various tissues of Populus tremula using a public database, PopGenIE (http://popgenie.org/) (Figure 4). These tissues included roots, dormant cambium-phloem, mature petiole, freshly expanded leaves, expanding young leaves, mature leaves, suckers, dormant buds, dormant flowers, expanded flowers, expanding flowers, and mature seeds. Twenty-five PtPHT genes were highly expressed in the roots of P. tremula, suggesting that most PtPHT genes might be involved in Pi uptake. Six PtPHT genes, including PtPHO3, PtPHO4, PtPHO5, PtPHO6, PtPHT1.4, and PtPHT1.14, were highly expressed in more than six of the twelve investigated tissues. For example, PtPHO6 was highly expressed in nine tissues, including cambium, petiole, expanded freshly leaves, expanding young leaves, mature leaves, dormant buds, dormant flowers, expanding flowers, and mature seeds. In contrast, the expression levels of several PtPHT genes, including PtPHT4.3 and PtPHT3.3b, were very low in all of the investigated tissues. In particular, PtPHT1.9 and PtPHT1.10 were not expressed in any of the 12 tissues examined; this finding is not shown in Figure 4. Further, some PtPHT genes were only highly expressed in a few specific tissues. For example, PtPHT1.6 was exclusively expressed in the roots of P. tremula, while PtPHT1.5 and PtPHO9 were only highly expressed in the roots and suckers of P. tremula.
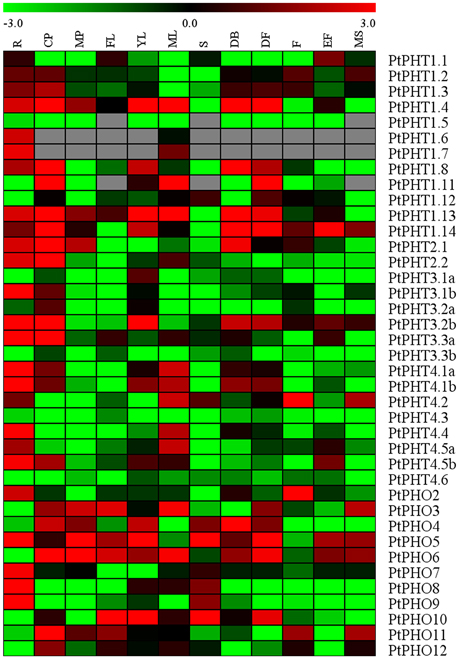
Figure 4. The expression profiles of PHT genes in different tissues of Populus tremula. Heatmap shows PHT gene expression across 12 various tissues including roots (R), dormant cambium-phloem (CP), mature petiole (MP), freshly expanded leaves (FL), expanding young leaves (YL), mature leaves (ML), suckers (S), dormant buds (DB), dormant flowers (DF), expanded flowers (F), expanding flowers (EF), and mature seeds (MS). The data were obtained from PopGenIE (http://popgenie.org/) (Sjodin et al., 2009).
Real-time quantitative RT-PCR was also performed to analyze the transcript levels of all 42 PtPHT genes in the roots, stems, young leaves, and mature leaves of P. simonii. Transcripts of 35 PtPHT genes were detected in the examined tissues (Figure 5), but some of them were present at relatively low levels (such as PtPHT1.5/1.6, PtPHO1, and PtPHO2) (Figures 5A,D). Most of the PtPHT genes were confirmed to be expressed in the roots of poplar. Transcripts of 7 PtPHT genes (such as PtPHT1.3, PtPHT2.2, and PtPHT4.1a) were detected in all four tested tissues (Figures 5A–D). Most PtPHT genes were expressed in the roots of P. simonii at high levels. PtPHT1.8 and PtPHO7 exhibited increased transcript accumulation in the leaves compared to the roots (Figures 5A,D). In addition, PtPHT3.1a/b expression was similar in all four investigated tissues (Figure 5B).
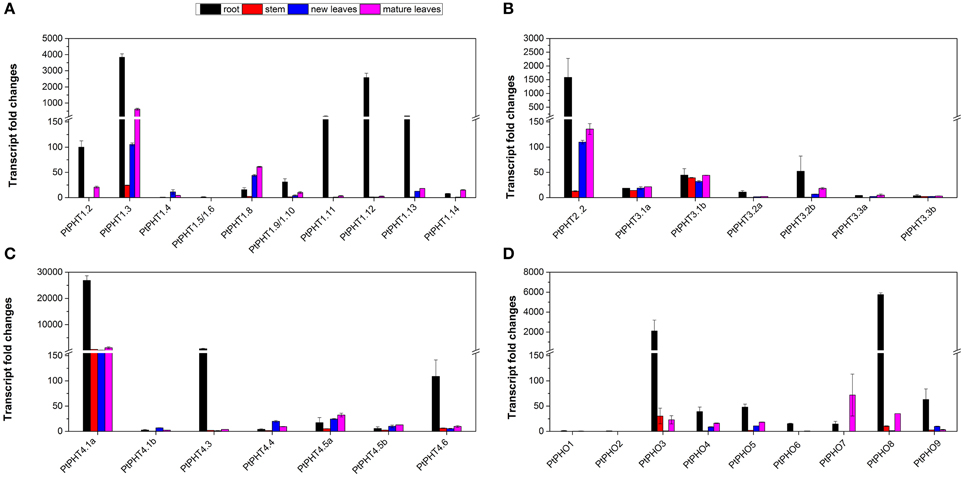
Figure 5. The expression of PtPHT genes (A: PHT1; B: PHT2 and PHT3; C: PHT4; D: PHO) in different tissues of Populus simonii. Total RNA was isolated using a plant RNA extraction kit (R6827, Omega Bio-Tek, GA, USA) according to the manufacturer's instructions. The relative expression of PHT1.2 was set as a standard (100). The RNA was used for qRT-PCR with PHT-specific primers. The signal intensities were calibrated based on a constitutively expressed poplar actin gene (Brunner et al., 2004b). The data are presented as the mean ± SD of three separate measurements.
To better understand the functions of PtPHT genes in relation to phosphate uptake, the transcription patterns of PtPHT genes in the roots of P. simonii were examined at varying phosphate levels. There were 33 PtPHT genes in total expressed in roots (Figure 6), but the expression of a few PtPHTs, such as PtPHT3.2a/b and PtPHO1/2/3, was very low (Figures 6B,D). In the roots, the expression of PtPHT1.12 was up-regulated and that of PtPHT1.5 was only slightly altered under phosphate starvation conditions; in addition, the expression of PtPHT1.2, PtPHT1.3, PtPHT2.2, PtPHT3.1b, and PtPHT4.1a was markedly up-regulated under low-phosphate conditions, and that of PtPHT3.3a, PtPHT4.1b, PtPHT4.5b, PtPHO8, and PtPHO9 was up-regulated under high-phosphate conditions (Figures 6A–D).
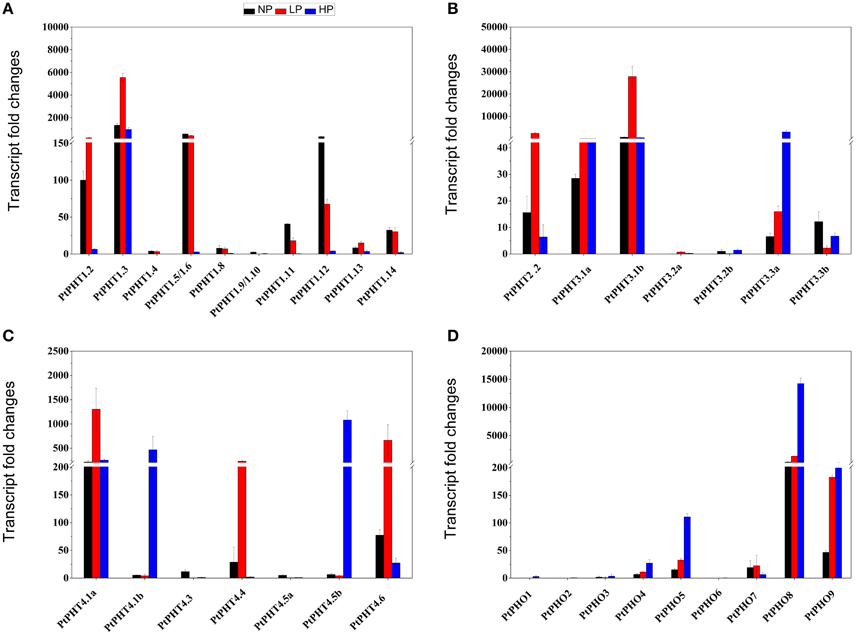
Figure 6. The expression of PtPHT genes (A: PHT1; B: PHT2 and PHT3; C: PHT4; D: PHO) under different phosphate levels in roots of P. simonii seedlings. The relative expression of PHT1.2 was set as a standard (100). Total RNA was isolated using a plant RNA extraction kit (R6827, Omega Bio-Tek, GA, USA) according to the manufacturer's instructions. The RNA was used for qRT-PCR with PHT-specific primers. The signal intensities were calibrated based on a constitutively expressed poplar actin gene (Brunner et al., 2004b). The data are presented as the mean ± SD of three separate measurements.
Most of the PtPHT genes were expressed at high levels in the roots of P. simonii in the presence of low phosphate levels, indicating that they are high-affinity genes. The accumulation of transcripts of many PtPHT genes was reduced when the level of H2 was increased in the roots of P. simonii, with the exception of PtPHO8 and PtPHO9.
To understand the transcriptional changes of PtPHT genes induced by drought, P. simonii plants supplied with different phosphate levels were subjected to drought stress for 2 weeks. PtPHT1.2 and PtPHO9 expression was found to be increased under drought conditions, irrespective of the phosphate level. In particular, PtPHT1.2 expression was significantly induced by approximately 90-fold. However, changes in the transcription of some PtPHT genes under drought stress were largely dependent on the phosphate level. For example, after 14 days of drought stress, the expression of PtPHT3.3b, PtPHT4.3, and PtPHT4.5a was significantly down-regulated, while that of PtPHT1.9, PtPHO4, and PtPHO9 was only slightly increased when no phosphate was supplied (Figure 7). However, the expression of these genes was up-regulated under drought stress in the presence of a low level of phosphate. Further, in the presence of a high level of phosphate, the expression of PtPHT1.9, PtPHT1.11, PtPHT1.3, PtPHT1.14, PtPHT2.2, PtPHT4.4, PtPHT4.6, PtPHO1, and PtPHO8 was increased under drought conditions. These data suggested that the changes in the transcription of some PtPHT genes under drought conditions were strongly influenced by the phosphate level.
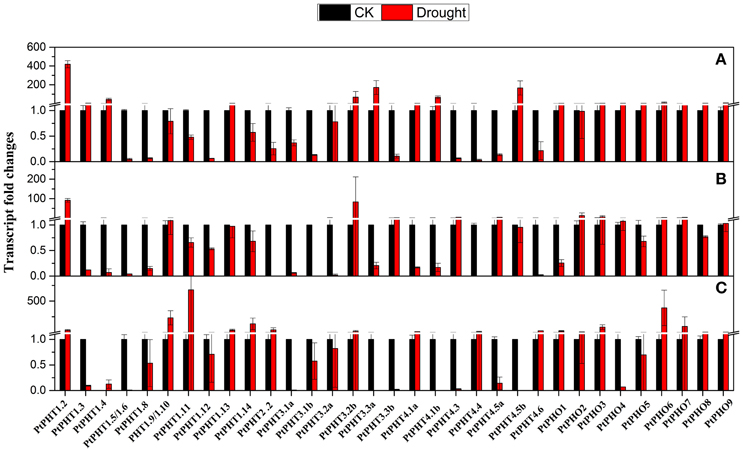
Figure 7. The expression of PHT genes affected by drought at different phosphate levels (A: No P; B: low P; C: high P) in roots of P. simonii seedlings. The control is defined as 1 in the figures. Total RNA was isolated using a plant RNA extraction kit (R6827, Omega Bio-Tek, GA, USA) according to the manufacturer's instructions. The RNA was used for qRT-PCR with PHT-specific primers. The signal intensities were calibrated based on a constitutively expressed poplar actin gene (Brunner et al., 2004b). The data are presented as the mean ± SD of three separate measurements.
Phosphorus is a mineral nutrient that is essential for plant growth and development. Pi was assimilated in roots via PHT transporters. In this study, we identified the whole PHT gene family in poplar and examined the expression patterns of these genes in different plant tissues and in response to differing Pi levels and drought. The PHT family, the cis elements of the PHT genes, and transcriptional changes in the PHT genes in response to different Pi levels and drought are discussed below.
In the current study, we conducted comprehensive analysis of PHT genes in poplar. A total of 42 PtPHT genes were retrieved from Populus genome (Phytozome 11.0, P. trichocarpa v3.0) with improved annotation, and they were classified into five clusters: PHT1, PHT2, PHT3, PHT4, and PHO. In a subsequent study, Loth-Pereda et al. (2011) reported the identification of 12 PHT1 family members in an earlier version of the Populus genome (v1), and we expanded upon their findings with the identification of 14 PHT1 family members in the latest version of Populus genome (v3). Notably, the PHT1 family is larger in poplar (14 genes) than in Arabidopsis (9 genes), barley (8 genes), rice (13 genes), and tomato (8 genes) (Muchhal et al., 1996; Rae et al., 2003; Liu et al., 2011; Chen et al., 2014).
The PHT2, PHT3, PHT4, and PHO families contained more members of PtPHT genes in poplar than that in Arabidopsis. One PHT2 family gene has been identified in Arabidopsis (Daram et al., 1999), while two (PtPHT2.1 and PtPHT2.2) were detected in poplar. These two genes are located on different chromosomes (Chr8 and Chr10), and the lengths of their encoded proteins greatly differ which is constituent with the previous report that AtPHT2.1 cDNA encodes a 61-kD protein that is structurally similar but different from the PHT1 family proteins in Arabidopsis (Daram et al., 1999). However, phylogenetic analysis showed that PtPHT2.1 and PtPHT2.2 share 100% similarity (Table S3). At least three AtPHT3 genes have been identified in Arabidopsis to date (Rausch and Bucher, 2002), while we found six PHT3 family genes in poplar. Further, six PHT4 family genes have been characterized in Arabidopsis (Guo et al., 2008), while 8 PtPHT4 genes were detected in the present study. Ten genes with homology to AtPHO1 are present in the Arabidopsis genome (Hamburger et al., 2002), and in this study, 12 PHO family genes were detected with similarities to the PHO1 protein in Arabidopsis.
The presence of a large number of genes within a family reflects the successive expansion and rearrangement of the genome by the extensive duplication and diversification that frequently occur over the course of evolution (Wang et al., 2007). The P. trichocarpa genome is duplicated and contains more protein-coding genes than the Arabidopsis genome; on average, there are 1.4 to 1.6 putative Populus genes for every Arabidopsis gene (Tuskan et al., 2006). Whole-genome duplication increases gene gains and losses, which greatly affects the amplification of members of gene families in the genome (Guo, 2013). The expanded genes in the PHT family, including PHT1, PHT2, PHT3, PHT4, and PHO, in poplar compared with Arabidopsis might be the result of gene duplication. Gene duplication has been reported in P. trichocarpa in many studies, including studies of the glutamine synthetase family, nitrate transporter gene family, and CCCH zinc finger family (Castro-Rodríguez et al., 2011; Chai et al., 2012; Bai et al., 2013). In the present study gene expansion and loss might also occur in the PHT family of P. trichocarpa similar to that of Arabidopsis. For example, two closely related orthologs of Arabidopsis, AtPHT4.1, AtPHT4.5, and AtPHO1, were found in Populus suggesting that duplications might be a partial reason for the expansion of PHT gene family in Populus. Forty-three PtPHT genes were localized to 15 poplar chromosomes, and some PHT genes were found to be clustered together as closely related genes (Figure 2). The presence of these closely related clusters suggests that these genes might evolve from regional duplications (e.g., PtPHO6, PtPHO7, PtPHO8, and PtPHO9).
Cis elements may control the efficiency of promoters and thus regulate the expression of the genes that they control by interacting with the corresponding trans-regulatory factors (Liu et al., 2011). Studies of cis elements might play a crucial role in dissecting the functions of genes. A previous study has described the cis elements of PHT1 family members in poplar (Loth-Pereda et al., 2011), but the cis elements of all PHT gene family members in poplar have not yet been identified. In this study, the upstream regions of all the PtPHTs contain cis elements affecting energy, light, hormone, defense, carbon metabolism, and tissue-specific expression. These findings suggest that the expression of these PtPHT genes might be regulated by many factors, such as light-, hormone-, and defense-related factors. For instance, GATABOX and GT1CONSENSUS are cis elements required for light regulation (Lam and Chua, 1989; Terzaghi and Cashmore, 1995), and they have also been identified in the upstream regions of all OsPHT genes in rice (Liu et al., 2011). Further, putative PHO-like (CGCGTGGG) and TATA-box-like (TATAAATA) cis regulatory elements have been identified in the promoter regions of Arabidopsis genes (Hammond et al., 2003), and they were observed in 22 and 34 of the 42 PtPHT promoters, respectively. Alternatively, because the core sequence of the Arabidopsis PHO-like element (CACGTG) is also similar to that of the ABRE element (ABREOSRAB21), which is present in 6 of the 42 PtPHT promoters, proteins in the bZIP class of transcription factors might bind to them (Schindler et al., 1992). The presence of these cis elements in poplar PtPHT genes and their likely roles in regulating gene expression suggest that PtPHT genes may be involved in various stress responses in poplar.
Some cis elements function under phosphate deficiency, for example, the P1BS element can be bound to PHR1 (a MYB transcription factor) to potentially regulate the phosphate starvation response in Arabidopsis (Rubio et al., 2001), PHR1 has been suggested as a central integrator to role in the transcriptional regulation of phosphate starvation responses (Bustos et al., 2010). In the current study, P1BS-like elements were found in 20 of the 42 PtPHT promoters examined suggesting that expression of these genes might be influenced by Pi starvation. Similarly, P1BS-like elements have also been detected in OsPHT genes in rice (Liu et al., 2011). These elements are found to be in the promoters of phosphate regulated genes, not only in Arabidopsis but also in other plant species such as barley and rice (Schünmann et al., 2004; Liu et al., 2011), which suggests that they may participate in a conserved signaling pathway for the phosphate starvation response in plants. The existence of P1BS-like elements in poplar, rice, and Arabidopsis suggests that they may serve a similar role in both woody species and herbs.
The number of Pi transporters present in roots may reflect the complexity and significance of the process of Pi transport. In the current study, most PHT family members were expressed in the roots of P. tremula and P. simonii. In particular, most PHT1 family members were highly expressed in the roots of poplar, consistent with a previous study conducted by Loth-Pereda et al. (2011) suggesting that they may function in Pi uptake. Notably, most PHT1 genes in other plant species are also expressed in the roots. For example, eight of the nine PHT1 family genes in Arabidopsis are expressed in the roots (Bucher et al., 2001; Mudge et al., 2002), in addition, 8 PHT1 genes in tomato (Chen et al., 2014) and 10 of the 13 PHT1 genes in rice are expressed in the roots (Paszkowski et al., 2002). These results indicate that PHT1 family genes may play similar roles in Pi uptake in different species.
Homologous PHT1, PHT2, PHT3, PHT4, and PHO genes showed substantial differences in their regulation in different tissues. Nine PHT1 family genes were highly expressed in the roots of P. tremula, but the expression of the other five genes was very low, and two of these genes (PtPHT1.9 and PtPHT1.10) were not expressed in any tissues examined. PtPHT2.1 was highly expressed in the roots, cambium-phloem, mature petiole, and dormant buds, while PtPHT2.2 was mainly expressed in the roots, cambium-phloem, and mature leaves. Six homologous PtPHT3 genes exhibited substantial differences in their expression in the various tissues examined. Three of them (PtPHT3.1b, PtPHT3.2b, and PtPHT3.3a) were highly expressed in the roots of P. tremula, while the others (PtPHT3.1a, PtPHT3.2a, and PtPHT3.3b) were expressed at very low levels. Although most PHT4 family members, excluding PtPHT4.3 and PtPHT4.6, were highly expressed in the roots and leaves, PtPHT4.1 was also highly expressed in the cambium-phloem, dormant buds, and dormant flowers. Further, PtPHO5 and PtPHO6 were highly expressed in nine tissues examined, while PtPHO7 and PtPHO8 were exclusively highly expressed in only a few of the tissues. Expression of the PtPHT genes varied among the different tissues examined, suggesting that these genes may have different functions in Pi uptake, transport, and storage. These results suggest that the PHT genes may have different functions since they expressed in a wide range of tissues, which may be partly crucial for the growth of perennial species.
The differential expression of PtPHT genes in different tissues might be related to their subcellular locations (Table 1). For example, two PHT2 family members were expressed in the leaves of P. simonii, in agreement with a previous study demonstrating that the AtPHT2.1 gene, which is located in the chloroplast, is predominantly expressed in green tissues in Arabidopsis (Liu et al., 2011). In addition, the expression patterns of PtPHT4 genes in both roots and leaves, and the phosphate transport activities as well as their subcellular locations, indicate that PHT4 proteins play roles in transferring Pi between the cytosol and chloroplasts, heterotrophic plastids, and the Golgi apparatus (Guo et al., 2008). For example, AtPHT4.6 transports Pi out of the Golgi lumenal space to recycle Pi released from glycosylation processes in Arabidopsis (Cubero et al., 2009). In addition, the PHO1 protein has been reported to be localized to stellar cells of the roots as well as the lower part of the hypocotyl in Arabidopsis (Ribot et al., 2008). In the present study, PHO family members were found to be highly expressed in many of the tissues examined, suggesting that they have diverse functions in loading Pi to different parts of plants.
The transcription of PtPHT genes differed at different Pi levels. Upon Pi starvation, PtPHT1.9, PtPHT1.11, PtPHT1.12, PtPHT3.3b, PtPHT4.3, and PtPHT4.5a expression was up-regulated in P. simonii (Figure 5). The up-regulation of some PtPHTs, such as PtPHT9 and PtPHT11, under Pi starvation is consistent with the findings of a previous study of P. trichocarpa conducted by Loth-Pereda et al. (2011). Most PHT1 family genes expressed in roots show up-regulation in phosphate deprived Arabidopsis plants; in particular, PtPHT9 and PtPHT11 in poplar are highly homologous to AtPHT8 and AtPHT9, which have been shown to be strongly up-regulated during Pi starvation (Mudge et al., 2002). In rice, high-affinity Km values of OsPHT9 and OsPHT10 for Pi transport have been reported, as well as specific induction of their expression by Pi starvation (Wang et al., 2014). Therefore, some of these PHT transporters likely play roles in Pi scavenging under conditions of low soil Pi availability. PtPHT1.2, PtPHT1.3, PtPHT2.2, PtPHT3.1b, and PtPHT4.1a expression was markedly up-regulated under low-Pi conditions, suggesting that their expression might be induced under these conditions. The transcript levels of a few PHT genes in Arabidopsis, such as AtPHO1, AtPHO;H1, and AtPHO;H10, were increased by Pi deficiency (Ribot et al., 2008); however, the expression of most PHO family members was not induced by Pi deficiency in poplar, suggesting that PHT genes might have different functions in woody species and herbs. PtPHT3.3a, PtPHT4.1b, PtPHT4.5b, PtPHO8, and PtPHO9 expression was up-regulated under high-phosphate conditions, indicating that these proteins might be low-affinity transporters. In addition, some PHT4 family members might be low-affinity transporters, consistent with the relatively high Pi levels that have been reported in the cytosol and other subcellular compartments (Mimura, 1999).
The expression of PHT genes varied at different Pi levels, suggesting that PHT family members are likely to encode both high- and low-affinity transporters. For example, OsPHT2 shows low-affinity characteristics on the mM scale in oocytes (Ai et al., 2009). By contrast, a Km value of 19 μM has been reported for HvPHT1 (barley high-affinity transporter, Preuss et al., 2010). AtPHT1.1 has been demonstrated to be a high-affinity transporter in a gene over-expression study (Km value of 3.1 μM, Mitsukawa et al., 1997), in addition to HvPHT1.1 (9.06 μM) and HvPHT1;6 (385 μM) in barley (Rae et al., 2003). The affinities of PHT genes in poplar require further study, including assessment of the heterologous transformation of Xenopus oocytes and gene over-expression studies using plant cell cultures.
The expression levels of some PtPHT genes, such as PtPHT1.2 and PtPHO9, were up-regulated under drought conditions, irrespective of the Pi level, suggesting that their expression was not affected by the Pi level under these conditions. However, the changes in expression of the other PtPHT genes, including PtPHT3.3b, PtPHT4.3, and PtPHT4.5a, under drought stress were largely dependent on the phosphate level (Figure 6). Especially, the upregulation of PtPHT genes induced by a low level of Pi may contribute to drought tolerance of poplar plants. These results suggest that under drought conditions, Pi uptake in plants likely changes in association with alterations in PHT gene expression. This altered uptake in the presence of different levels of phosphate might be related to the drought resistance of plants.
CZ and ZZ conceived and designed the experiments. SM and CZ performed the experiments. CZ, ML, and SM analyzed the data. CZ, ML, and ZZ wrote the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was sponsored by the State Key Basic Research Development Program (2012CB416902), the National Natural Science Foundation of China (31500543), and the Natural Basic Research Plan in Shaanxi Province of China (2015JQ3082).
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.01398
Table S1. The primers used for quantitative real time PCR.
Table S2. Cis elements of PHT genes in Populus.
Table S3. The sequence similarity of PHT genes in Populus.
Ai, P., Sun, S., Zhao, J. N., Fan, X. R., Xin, W. J., Guo, Q., et al. (2009). Two rice phosphate transporters, OsPht1;2 and OsPht1;6, have different functions and kinetic properties in uptake and translocation. Plant J. 57, 798–809. doi: 10.1111/j.1365-313X.2008.03726.x
Bai, H., Euring, D., Volmer, K., Janz, D., and Polle, A. (2013). The nitrate transporter (NRT) gene family in poplar. PLoS ONE 8:e72126. doi: 10.1371/journal.pone.0072126
Brunner, A. M., Busov, V. B., and Strauss, S. H. (2004a). Poplar genome sequence: functional genomics in an ecologically dominant plant species. Trends Plant Sci. 9, 49–56. doi: 10.1016/j.tplants.2003.11.006
Brunner, A. M., Yakovlev, I. A., and Strauss, S. H. (2004b). Validating internal controls for quantitative plant gene expression studies. BMC Plant Biol. 4:14.
Bucher, M. (2007). Functional biology of plant phosphate uptake at root and mycorrhiza interfaces. New Phytol. 173, 11–26. doi: 10.1111/j.1469-8137.2006.01935.x
Bucher, M., Rausch, C., and Daram, P. (2001). Molecular and biochemical mechanisms of phosphorus uptake into plants. J. Plant Nutr. Soil Sci. 164, 209–217. doi: 10.1002/1522-2624(200104)164:2<209::AID-JPLN209>3.0.CO;2-F
Bustos, R., Castrillo, G., Linhares, F., Puga, M. I., Rubio, V., Perez-Perez, J., et al. (2010). A central regulatory system largely controls transcriptional activation and repression responses to phosphate starvation in Arabidopsis. PLoS Genet. 6:e1001102. doi: 10.1371/journal.pgen.1001102
Castro-Rodríguez, V., García-Gutiérrez, A., Canales, J., Avila, C., Kirby, E. G., and Cánovas, F. M. (2011). The glutamine synthetase gene family in Populus. BMC Plant Biol. 11:119. doi: 10.1186/1471-2229-11-119
Cesco, S., Neumann, G., Tomasi, N., Pinton, R., and Weisskopf, L. (2010). Release of plant-borne flavonoids into the rhizosphere and their role in plant nutrition. Plant Soil 329, 1–25. doi: 10.1007/s11104-009-0266-9
Chai, G., Hu, R., Zhang, D., Qi, G., Zuo, R., Cao, Y., et al. (2012). Comprehensive analysis of CCCH zinc finger family in poplar (Populus trichocarpa). BMC Genomics 13:253. doi: 10.1186/1471-2164-13-253
Chen, A., Chen, X., Wang, H., Liao, D., Gu, M., Qu, H., et al. (2014). Genome-wide investigation and expression analysis suggest diverse roles and genetic redundancy of Pht1 family genes in response to Pi deficiency in tomato. BMC Plant Biol. 14:61. doi: 10.1186/1471-2229-14-61
Clark, G. T., Dunlop, J., and Phung, H. T. (2000). Phosphate absorption by Arabidopsis thaliana: interactions between phosphorus status and inhibition by arsenate. Aust. J. Plant Physiol. 27, 959–965. doi: 10.1071/PP99108
Cubero, B., Nakagawa, Y., Jiang, X. Y., Miura, K. J., Li, F., Raghothama, K. G., et al. (2009). The phosphate transporter PHT4; 6 is a determinant of salt tolerance that is localized to the golgi apparatus of Arabidopsis. Mol. Plant 2, 535–552. doi: 10.1093/mp/ssp013
Daram, P., Brunner, S., Rausch, C., Steiner, C., Amrhein, N., and Bucher, M. (1999). Pht2;1 encodes a low-affinity phosphate transporter from Arabidopsis. Plant Cell 11, 2153–2166. doi: 10.1105/tpc.11.11.2153
Gowik, U., Burscheidt, J., Akyildiz, M., Schlue, U., Koczor, M., Streubel, M., et al. (2004). Cis-regulatory elements for mesophyll-specific gene expression in the C4 plant flaveria trinervia, the promoter of the C4 phosphoenolpyruvate carboxylase gene. Plant Cell 16, 1077–1090. doi: 10.1105/tpc.019729
Guo, B., Jin, Y., Wussler, C., Blancaflor, E. B., Motes, C. M., and Versaw, W. K. (2008). Functional analysis of the Arabidopsis PHT4 family of intracellular phosphate transporters. New Phytol. 177, 889–898. doi: 10.1111/j.1469-8137.2007.02331.x
Guo, Y. L. (2013). Gene family evolution in green plants with emphasis on the origination and evolution of Arabidopsis thaliana genes. Plant J. 73, 941–951. doi: 10.1111/tpj.12089
Hamburger, D., Rezzonico, E., Petetot, J. M. C., Somerville, C., and Poirier, Y. (2002). Identification and characterization of the Arabidopsis PHO1 gene involved in phosphate loading to the xylem. Plant Cell 14, 889–902. doi: 10.1105/tpc.000745
Hammond, J. P., Bennett, M. J., Bowen, H. C., Broadley, M. R., Eastwood, D. C., May, S. T., et al. (2003). Changes in gene expression in Arabidopsis shoots during phosphate starvation and the potential for developing smart plants. Plant Physiol. 132, 578–596. doi: 10.1104/pp.103.020941
Higo, K., Ugawa, Y., Iwamoto, M., and Korenaga, T. (1999). Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucl. Acids Res. 27, 297–300. doi: 10.1093/nar/27.1.297
Hoagland, D. R., and Arnon, D. I. (1950). The Water-Culture Method for Growing Plants Without Soil. Circular. Berkeley: California Agricultural Experiment Station.
Hu, B., Jin, J., Guo, A.-Y., Zhang, H., Luo, J., and Gao, G. (2015). GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics 31, 1296–1297. doi: 10.1093/bioinformatics/btu817
Lam, E., and Chua, N. H. (1989). ASF-2: a factor that binds to the cauliflower mosaic virus 35S promoter and a conserved GATA motif in Cab promoters. Plant Cell 1, 1147–1156. doi: 10.1105/tpc.1.12.1147
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., Mcgettigan, P. A., Mcwilliam, H., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics 23, 2947–2948. doi: 10.1093/bioinformatics/btm404
Liu, F., Chang, X.-J., Ye, Y., Xie, W.-B., Wu, P., and Lian, X.-M. (2011). Comprehensive sequence and whole-life-cycle expression profile analysis of the phosphate transporter gene family in rice. Mol. Plant 4, 1105–1122. doi: 10.1093/mp/ssr058
Liu, R. H., and Meng, J. L. (2003). MapDraw: a microsoft excel macro for drawing genetic linkage maps based on given genetic linkage data. Hereditas (Beijing) 25, 317–321. doi: 10.3321/j.issn:0253-9772.2003.03.019
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Loth-Pereda, V., Orsini, E., Courty, P. E., Lota, F., Kohler, A., Diss, L., et al. (2011). Structure and expression profile of the phosphate Pht1 transporter gene family in mycorrhizal Populus trichocarpa. Plant Physiol. 156, 2141–2154. doi: 10.1104/pp.111.180646
Mimura, T. (1999). Regulation of phosphate transport and homeostasis in plant cells. Int. Rev. Cytol. 191, 149–200. doi: 10.1016/S0074-7696(08)60159-X
Misson, J., Thibaud, M. C., Bechtold, N., Raghothama, K., and Nussaume, L. (2004). Transcriptional regulation and functional properties of Arabidopsis Pht1;4, a high affinity transporter contributing greatly to phosphate uptake in phosphate deprived plants. Plant Mol. Biol. 55, 727–741. doi: 10.1007/s11103-004-1965-5
Mitsukawa, N., Okumura, S., Shirano, Y., Sato, S., Kato, T., Harashima, S., et al. (1997). Overexpression of an Arabidopsis thaliana high-affinity phosphate transporter gene in tobacco cultured cells enhances cell growth under phosphate-limited conditions. Proc. Natl. Acad. Sci. U.S.A. 94, 7098–7102. doi: 10.1073/pnas.94.13.7098
Muchhal, U., Pardo, J., and Raghothama, K. (1996). Phosphate transporters from the higher plant Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 93, 10519–10523. doi: 10.1073/pnas.93.19.10519
Mudge, S. R., Rae, A. L., Diatloff, E., and Smith, F. W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 31, 341–353. doi: 10.1046/j.1365-313X.2002.01356.x
Nussaume, L., Kanno, S., Javot, H., Marin, E., Pochon, N., Ayadi, A., et al. (2011). Phosphate import in plants: focus on the PHT1 transporters. Front. Plant Sci. 2:83. doi: 10.3389/fpls.2011.00083
Pao, S. S., Paulsen, I. T., and Saier, M. H. (1998). Major facilitator superfamily. Microbiol. Mol. Biol. Rev. 62, 1–34.
Paszkowski, U., Kroken, S., Roux, C., and Briggs, S. P. (2002). Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proc. Natl. Acad. Sci. U.S.A. 99, 13324–13329. doi: 10.1073/pnas.202474599
Poirier, Y., and Bucher, M. (2002). Phosphate transport and homeostasis in arabidopsis. Arabidopsis Book 1:e0024. doi: 10.1199/tab.0024
Preuss, C. P., Huang, C. Y., Gilliham, M., and Tyerman, S. D. (2010). Channel-Like characteristics of the low-affinity barley phosphate transporter PHT1;6 when expressed in xenopus oocytes. Plant Physiol. 152, 1431–1441. doi: 10.1104/pp.109.152009
Rae, A., Cybinski, D., Jarmey, J., and Smith, F. (2003). Characterization of two phosphate transporters from barley; evidence for diverse function and kinetic properties among members of the Pht1 family. Plant Mol. Biol. 53, 27–36. doi: 10.1023/B:PLAN.0000009259.75314.15
Raghothama, K. (2000). Phosphate transport and signaling. Curr. Opin. Plant Biol. 3, 182–187. doi: 10.1016/S1369-5266(00)00062-5
Rausch, C., and Bucher, M. (2002). Molecular mechanisms of phosphate transport in plants. Planta 216, 23–37. doi: 10.1007/s00425-002-0921-3
Ribot, C., Wang, Y., and Poirier, Y. (2008). Expression analyses of three members of the AtPHO1 family reveal differential interactions between signaling pathways involved in phosphate deficiency and the responses to auxin, cytokinin, and abscisic acid. Planta 227, 1025–1036. doi: 10.1007/s00425-007-0677-x
Rubio, V., Linhares, F., Solano, R., Martín, A. C., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. doi: 10.1101/gad.204401
Schachtman, D. P., Reid, R. J., and Ayling, S. M. (1998). Phosphorus uptake by plants: From soil to cell. Plant Physiol. 116, 447–453. doi: 10.1104/pp.116.2.447
Schindler, U., Beckmann, H., and Cashmore, A. R. (1992). TGA1 and G-box binding factors: two distinct classes of Arabidopsis leucine zipper proteins compete for the G-box-like element TGACGTGG. Plant Cell 4, 1309–1319. doi: 10.1105/tpc.4.10.1309
Schünmann, P. H. D., Richardson, A. E., Vickers, C. E., and Delhaize, E. (2004). Promoter analysis of the barley Pht1;1 phosphate transporter gene identifies regions controlling root expression and responsiveness to phosphate deprivation. Plant Physiol. 136, 4205–4214. doi: 10.1104/pp.104.045823
Shen, J., Yuan, L., Zhang, J., Li, H., Bai, Z., Chen, X., et al. (2011). Phosphorus dynamics: from soil to plant. Plant Physiol. 156, 997–1005. doi: 10.1104/pp.111.175232
Sjodin, A., Street, N. R., Sandberg, G., Gustafsson, P., and Jansson, S. (2009). The Populus Genome Integrative Explorer (PopGenIE): a new resource for exploring the Populus genome. New Phytol. 182, 1013–1025. doi: 10.1111/j.1469-8137.2009.02807.x
Stappen, R., and Krämer, R. (1994). Kinetic mechanism of phosphate/phosphate and phosphate/OH− antiports catalyzed by reconstituted phosphate carrier from beef heart mitochondria. J. Biol. Chem. 269, 11240–11246.
Takabatake, R., Hata, S., Taniguchi, M., Kouchi, H., Sugiyama, T., and Izui, K. (1999). Isolation and characterization of cDNAs encoding mitochondrial phosphate transporters in soybean, maize, rice, and Arabidopsis. Plant Mol. Biol. 40, 479–486. doi: 10.1023/A:1006285009435
Terzaghi, W. B., and Cashmore, A. R. (1995). Light-regulated transcription. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 445–474. doi: 10.1146/annurev.pp.46.060195.002305
Tuskan, G. A., Difazio, S., Jansson, S., Bohlmann, J., Grigoriev, I., Hellsten, U., et al. (2006). The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science 313, 1596–1604. doi: 10.1126/science.1128691
Valentinuzzi, F., Pii, Y., Vigani, G., Lehmann, M., Cesco, S., and Mimmo, T. (2015). Phosphorus and iron deficiencies induce a metabolic reprogramming and affect the exudation traits of the woody plant Fragaria×ananassa. J. Exp. Bot. 66, 6483–6495. doi: 10.1093/jxb/erv364
Versaw, W. K., and Harrison, M. J. (2002). A chloroplast phosphate transporter, PHT2;1, influences allocation of phosphate within the plant and phosphate-starvation responses. Plant Cell 14, 1751–1766. doi: 10.1105/tpc.002220
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24. doi: 10.1016/j.gene.2007.01.006
Wang, X. F., Wang, Y. F., Pineros, M. A., Wang, Z. Y., Wang, W. X., Li, C. Y., et al. (2014). Phosphate transporters OsPHT1;9 and OsPHT1;10 are involved in phosphate uptake in rice. Plant Cell Environ. 37, 1159–1170. doi: 10.1111/pce.12224
Wang, Y., Ribot, C., Rezzonico, E., and Poirier, Y. (2004). Structure and expression profile of the Arabidopsis PHO1 gene family indicates a broad role in inorganic phosphate homeostasis. Plant Physiol. 135, 400–411. doi: 10.1104/pp.103.037945
Keywords: phosphate transporter, poplar, genome-wide analysis, expression profile, phosphate levels, drought
Citation: Zhang C, Meng S, Li M and Zhao Z (2016) Genomic Identification and Expression Analysis of the Phosphate Transporter Gene Family in Poplar. Front. Plant Sci. 7:1398. doi: 10.3389/fpls.2016.01398
Received: 11 July 2016; Accepted: 01 September 2016;
Published: 16 September 2016.
Edited by:
Karabi Datta, University of Calcutta, IndiaReviewed by:
Stefano Cesco, Free University of Bozen-Bolzano, ItalyCopyright © 2016 Zhang, Meng, Li and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunxia Zhang, Y3poYW5nMjAxMkAxNjMuY29t
Zhong Zhao, emhhb3poQG53c3VhZi5lZHUuY24=
†These authors have contributed equally to these work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.