
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 09 August 2016
Sec. Plant Genetics and Genomics
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.01175
In the current context of food security, increase of plant protein production in a sustainable manner represents one of the major challenges of agronomic research, which could be partially resolved by increased cultivation of legume crops. Medicago truncatula is now a well-established model for legume genomic and genetic studies. With the establishment of genomics tools and mutant populations in M. truncatula, it has become an important resource to answer some of the basic biological questions related to plant development and stress tolerance. This review has an objective to overview a decade of genetic studies in this model plant from generation of mutant populations to nowadays. To date, the three biological fields, which have been extensively studied in M. truncatula, are the symbiotic nitrogen fixation, the seed development, and the abiotic stress tolerance, due to their significant agronomic impacts. In this review, we summarize functional genetic studies related to these three major biological fields. We integrated analyses of a nearly exhaustive list of genes into their biological contexts in order to provide an overview of the forefront research advances in this important legume model plant.
Medicago, a Mediterranean origin species, has been intensively used as a legume pasture plants worldwide. Nowadays, Alfalfa (Medicago sativa) is the most cultivated forage plants in the USA and represents the most economically valuable forage for animal feed. In the past 20 years, the increasing number of research projects on legumes allowed the emergence of model plants for legume species. Crop and pasture legumes are generally poor model systems for genetic and genomic research. Some cultivated legumes are tetraploid (e.g., peanut), many have large genomes (e.g., pea and faba beans) and many are recalcitrant to transformation or difficult to regenerate (e.g., common bean, pea, and soybean). Most grain legumes have large but relatively few seeds per plant, and large seedlings, which prevents high-density culture (e.g., chickpea, black-eyed pea, mung bean, pea, bean, and soybean). Some legumes, such as soybean, have genome duplications, and some are self-incompatible or have a long generation time. As a result, two species, Medicago truncatula and Lotus japonicus have been proposed as models for legume research (Barker et al., 1990; Handberg and Stougaard, 1992). M. truncatula was first proposed as a model by Barker et al. (1990) to study the rhizobia-legume symbiosis. Now, it is internationally recognized as a model legume for all the legume studies. M. truncatula has several advantages for plant genomic research: diploid genome (2n = 16), autogamous, relatively small genome (~375 Mbp), which was sequenced and annotated (Young et al., 2011), and a relatively short generation time (around 4 months seed to seed).
Dedicated meetings and workshops on Medicago have allowed a rapid and coordinated development of genetic and genomic tools. For instance, transcriptomics studies have been facilitated by the development of microarray chips such as the 16k microarray of 70-mer oligos used in studies such as Hohnjec et al. (2005) or Gallardo et al. (2007) and Affymetrix GeneChip used in studies such as Benedito et al. (2008), Verdier et al. (2013b), and Zhang et al. (2014). Most of the data obtained from the Affymetrix GeneChip experiments have been stored and publicly shared on a dedicated webserver to provide a M. truncatula Gene Expression atlas (MtGEA, www.mtgea.noble.org; He et al., 2009). Transcriptomics tools also comprise a high-throughput quantitative PCR platform to profile all known transcription factors used in studies such as Verdier et al. (2008). Recently, the development of RNA-seq technologies has allowed a comprehensive identification and quantification of transcripts in M. truncatula such those responding to different stresses (e.g., Gruber et al., 2009; Li et al., 2009; Zhang et al., 2014). In parallel to transcriptomics tools, M. truncatula also has libraries for metabolomics studies (Broeckling et al., 2004) and reference maps for proteomics studies (Mathesius et al., 2001; Gallardo et al., 2003; Watson et al., 2003). Recently, 330 M. truncatula accessions from a germplasm collection were sequenced and have been used for genome-wide association studies such as Stanton-Geddes et al. (2013) and Kang et al. (2015).
Numerous bioinformatics resources are also available for Medicago, some have been developed specifically for Medicago (and legumes) such as the Medicago Gbrowser (http://gb.sc.noble.org/cgi-bin/gb2/gbrowse), LegumeGRN (Wang M. et al., 2013), legumeIP (Li et al., 2012), and Legoo (http://www.legoo.org); and others have been adapted from Arabidopsis to Medicago such as PathExpress (Goffard and Weiller, 2007) and AgriGO (Du et al., 2010). Another key step in the adoption of M. truncatula as a model plant for legume studies was the possibility of Agrobacterium-mediated transformation of the whole plant via somatic embryogenesis using Agrobacterium tumefaciens (Thomas et al., 1992), transformation of seedling using Agrobacterium tumefaciens (Trieu et al., 2000) or more specifically transformation of roots to generate transient hairy root transformants using Agrobacterium rhizogenes (Boisson-Dernier et al., 2001). The emergence of functional genetics in M. truncatula has been possible due to its capacity to be transformed and more recently due to the generation of different mutant populations.
Mutant populations play a central role in functional genomics analyses and are used in both forward and reverse genetic studies. To date, the three largest mutant populations of M. truncatula have been produced by three different approaches: chemical mutagenesis using Ethyl Methane Sulfonate (EMS population), fast neutron bombardment (FNB population), and finally transposon tagging with the introduction of the Tnt1 transposon of Tobacco within the M. truncatula genome (Tnt1 population). Despite that these populations are the largest and the most popular according to the number of mutated plants and the number of published papers, smaller collections exist and have been used in functional studies such as a gamma-rays induced mutations (Sagan et al., 1995) and activation-tagging population (Porceddu et al., 2008).
An EMS population was generated by treating seeds using EMS, a chemical mutagen, inducing point mutations throughout the genome by C/G to A/T substitutions. EMS mutagenesis is very popular to generate mutant populations because of its ability in introducing high-density mutations. This population in M. truncatula genotype A17 has been extensively used in both forward and reverse genetic screens. It comprises almost 9000 M2 plants derived from 4500 M2 plants obtained from 500 M1 (not using single seed descent) and from 4350 M2 derived from 4350 M1 (using single seed descent; Le Signor et al., 2009). For reverse genetic screening, the population is screened using a Target Induced Local Lesion IN Genomes (TILLING) approach. This technique permits to localize point mutations in pooled genomic DNA sequences from various mutagenized plants. It requires the availability of genomic sequences and relies on a specific digestion enzyme (e.g., Cell), which is able to cleave heteroduplexes formed by the association of wild-type and mutated PCR products at the site of mismatch (Till et al., 2004). Two websites are associated to this population: the REVGENUK web-server (http://revgenuk.jic.ac.uk/order.htm) to request a reverse screen of your candidate gene and the phenotypic database (http://www.inra.fr/legumbase) to facilitate the identification of mutant line phenotypes and to request specific mutated lines.
Fast neutron bombardment (FNB) is another typical approach to mutate plant genomes. Unlike EMS, it results in DNA deletions ranging from a few bases to more than 30 Kb, and possible chromosomal rearrangements. This technique generates a high proportion of knockout mutations (i.e., KO mutations) but low mutation densities (Tadege et al., 2005). Thus, it requires large population sets to achieve saturation mutagenesis. For instance, M. truncatula FNB population consists of more than 80,000 M1 lines. Deletion-based TILLING (DeTILLING) has been established to identify the deletions within a large population (Rogers et al., 2009). The method adopts a three-dimensional pooling strategy together with PCR-based screening to enhance the efficiency of mutant recovery. All the information related to request a screen or to access to the phenotypes of mutated lines is available at the FNB dedicated webserver (http://bioinfo4.noble.org/mutant/).
Insertional mutagenesis is one of the most powerful approaches to obtain null mutants (i.e., KO) as demonstrated by the success of Arabidopsis T-DNA populations. In M. truncatula, the well-studied tobacco retro-transposon Tnt1 was introduced into R108 genome, another genotype of M. truncatula. A study of its transposition indicated that Tnt1 transposes actively during tissue culture (4–50 transpositions per genome) and that insertions are stable (D'Erfurth et al., 2003). Furthermore, Tnt1 inserts were proven to preferentially target exons making a perfect tool to preferentially knocking out genes (D'Erfurth et al., 2003). Because of these characteristics, saturation mutagenesis of the Tnt1 population requires a relatively small population. To date, the M. truncatula population comprises ~22,000 mutant lines and is near saturation with insertions in around 90% of all genes (Cheng et al., 2014). Tnt1 mutant population has been used in forward genetic screens using inverse-PCR or thermal asymmetric interlaced-PCR (TAIL-PCR) to recover the flanking sequences of the insertion (Benlloch et al., 2006; Cheng et al., 2014). The use of this population in forward genetic screens was recently facilitated by the development of an algorithm, ITIS, which retrieves Tnt1 insertion sites from mutant line genomes using low coverage genome sequencing data (Jiang et al., 2015). Meanwhile reverse genetic screening has been developed, including PCR-based DNA pool screening for candidate genes, and more recently using a BLAST database containing flanking sequence tags (FST; i.e., flanking regions of the Tnt1 insertions) (Cheng et al., 2014). All the information regarding the Tnt1 population such as request of mutated lines, description of mutant line phenotypes and a BLAST-able database of FSTs are available at http://medicago-mutant.noble.org/.
Besides the mutant populations described previously, other tools have shown their efficiency to functionally characterize multiple genes such as RNA interference approaches (RNAi) or the use of MERE1 (Medicago RetroElement1), a low-copy retro-element naturally present in M. truncatula that showed transposition events preferentially in genic regions occurring during tissue culture (Rakocevic et al., 2009). Other strategies have emerged for functional characterization of genes and are under construction, such as the activation tag population, which consists of the introduction of a T-DNA together with the cauliflower mosaic virus (CaMV) 35S promoter into genome with the potential to generate gain-of-function phenotypes.
In this review, we will provide a non-exhaustive list of M. truncatula genes, which have been functionally characterized using one of the previously described mutant populations. To demonstrate that M. truncatula is a valuable model legume in broad research fields of plant biology, we decided to divide the functional studies into three important biological areas based on the number of published studies: (i) Rhizobia-legume symbiosis, (ii) seed biology, and (iii) abiotic stress biology.
Plant root system is crucial for anchorage to the soil and acquisition of nutrients. In dicotyledonous plants, taproot architecture is mainly composed of an embryonically derived primary root and post-embryonically generated lateral roots. Both root nodules (an organ formed after the invasion by diazotrophs, see below for details) and lateral roots (LR) originate by dedifferentiation of a few cells in the pericycle and cortex of main root (Herrbach et al., 2014). LR development is influenced by symbiotic bacterial inoculation and nodule formation in legumes. Several plant and bacterial mutants have demonstrated abnormal nodule organogenesis featuring a central vascular bundle, like lateral roots (Guan et al., 2013). Together, these observations suggest that evolution of nodule development involved extensive recruitment and reengagement of pre-existing LR developmental pathways. Extensive functional genetic studies have been carried out to understand root nodule development in Medicago (Table 1).
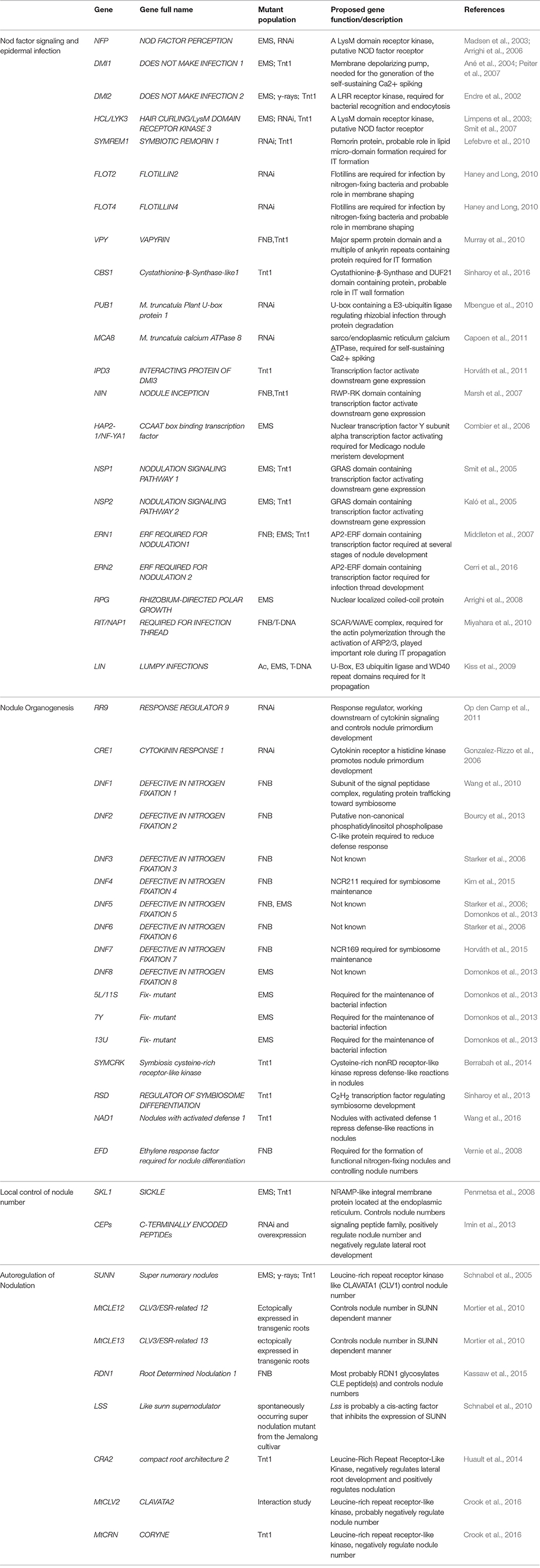
Table 1. List of nodule development related genes that have been functionally characterized in M. truncatula.
Development of nodules is initiated by an exchange of chemical signals between plant cells and soil bacteria called rhizobia (Oldroyd, 2013). In most of the legumes including Medicago, rhizobia produce and release lipochitooligosaccharides called Nod factors (NF) in response to flavonoids secreted by the host. NF are perceived by the root hair cells. Perception of NF leads to the induction of a calcium (Ca+2) influx and is followed by Ca+2 oscillations around the nucleus of the root hair cell. All these molecular changes lead to asymmetric growth (curling) of the root hair cells and division of cortical cells (Limpens et al., 2003; Radutoiu et al., 2003). Rhizobia enter and traverse epidermal and underlying cortical cells via a plant cell wall and plasma membrane invagination called the infection thread (ITs). Bacteria are eventually released into the cytoplasm of nodule primordial cells by endocytosis. Rhizobial endocytosis results in bacteria surrounded by a plant membrane, called the symbiosome membrane (SM), creating a novel organelle called symbiosome (Goodchild and Bergersen, 1966; Robertson and Lyttleton, 1984; Roth and Stacey, 1989). Finally, rhizobia (now called the bacteroids) undergo a process of differentiation that involves induction of nitrogen fixation genes and repression of ammonium assimilation genes, turning symbiosomes into an ammonium-exporting organelle (Udvardi and Day, 1997; Udvardi and Poole, 2013).
Nodules in M. truncatula always retain the meristem and keep on growing (indeterminate type). A mature nodule is organized into five well-defined developmental zones: Zone I (meristem), Zone II (invasion zone), Zone II-III (interzone), Zone III (nitrogen fixation zone), and Zone IV (senescent zone, only present in older nodules). Bacteroid genomes in M. truncatula undergo several rounds of replication without any cell division (endoreduplication), which leads to polyploidy of the bacteroids. Endoreduplication leads to the formation of elongated or Y-shaped bacteroids. Both bacterial cell division and elongated or Y-shaped bacteroid formation take place in the invasion zone (Oldroyd, 2013). Several parallel approaches have been undertaken to understand nodule development in M. truncatula. Two main methods have been forward genetic approach and transcriptomic analyses followed by reverse genetic approach. Extensive forward genetic and biochemical studies have been performed to understand the molecular mechanism behind nodule development. In the last decade, a rapid advancement of functional genomic technology helped us better understand the process of nodule development.
A schematic description of nodule formation, nodule zonation and symbiotic plant cell of the invasion zone is provided in Figure 1.
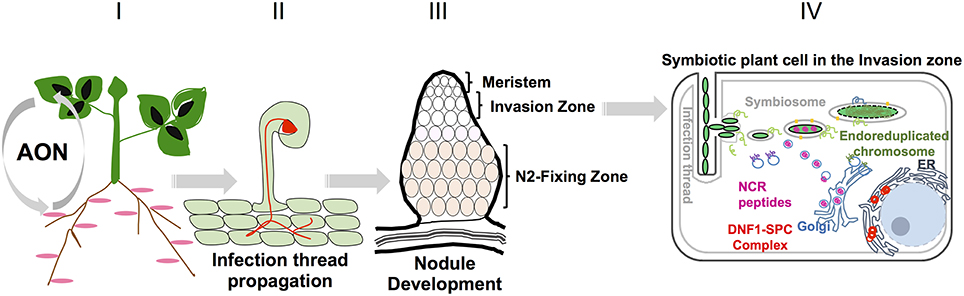
Figure 1. Different nodule developmental stages in Medicago: I Nodule number is controlled systemically by long distance signaling from root to shoot and back again to root, called autoregulation of nodulation (AON). II Rhizobia enter the root epidermis through the formation of infection threads. III A fully mature Medicago nodule with different nodule zones. Bacteroid maturation takes place in the invasion zone and nitrogen fixation takes place in nitrogen fixation zone. IV Schematic representation of one infected cell in the invasion zone. Bacterial endocytosis, differentiation to functional bacteroids takes place in this specific zone.
A systematic mutagenesis approach has been undertaken in M. truncatula Jemalong A17 using EMS (Penmetsa and Cook, 2000) to unravel the major players behind nodule development. Mutation in LysM domain containing receptor-like kinase NOD FACTOR PERCEPTION (NFP; Amor et al., 2003) eliminates any response in the root hair cells in presence of rhizobia/NF. Two other genes involved in nuclear-associated Ca+2 spiking response are DOES NOT MAKE INFECTION (DMI) genes DMI1 and DMI2 (Endre et al., 2002; Ané et al., 2004). dmi1 and dmi2 respond to NFs by causing swelling of the root hair in the absence of Ca+2 spiking (Ané et al., 2002). dmi2 encodes a leucine rich receptor (LRR) domain containing receptor-like kinase and acts in parallel with NFP. Another membrane-localized LysM domain containing receptor-like kinase has been implicated in NF recognition. This gene is named HAIR CURLING (HCL), and it encodes a LysM DOMAIN RECEPTOR KINASE 3 (LYK3; Smit et al., 2007). Excessive root hair curling and cortical cell division occurs in the hcl/lyk3 mutant, but rhizobia are not entrapped into the root curls.
In a reverse genetic approach, SYMBIOTIC REMORIN 1 (MtSYMREM1) has been identified. The above-mentioned three membrane-bound kinases (DMI2, NFP, and LYK3) interact with MtSYMREM1. The mutant of MtSYMREM1 was isolated from Tnt1 mutant population, and its phenotype indicated its involvement during bacterial endocytosis rather than IT formation (Lefebvre et al., 2010). Another gene family that has been implicated in M. truncatula IT development by reverse genetic studies is “FLOTILLIN.” Flotillins are peripheral membrane proteins known to be required for endocytosis and membrane shaping. Two flotillin genes FLOT2 and FLOT4 are induced downstream of the NF signaling pathway. Silencing of FLOT2 and FLOT4 expression revealed a non-redundant requirement for both genes in IT initiation and nodule development (Haney and Long, 2010). The VAPYRIN (VPY) gene encodes a protein with a major sperm protein domain and a series of ankyrin repeats. Ankyrin repeats are typically found in proteins involved in membrane trafficking and biogenesis. The vpy mutant showed abnormal IT formation and fewer underdeveloped nodules (Murray et al., 2010). Mutation in Cystathionine-β-Synthase-like1 (CBS1) causes a defect in the infection thread propagation. cbs1 plants have more micro-colonies but less propagating infection threads. CBS1 encodes for a putative membrane-localized domain of unknown function (DUF21) and a cystathionine-β-synthase domain, and is has been speculated that CBS1 participate in infection thread cell wall development (Sinharoy et al., 2016).
In summary, the three receptor-like kinases (DMI2, NFP, and LYK3) and FLOT4 probably localize at the tip of the root hair cells. They change their localization and organize themselves into a microdomain after the application of NFs (Haney et al., 2011). These, in turn, activate a nuclear Ca+2 spiking that acts as a secondary messenger to activate gene expression in the nucleus. Interaction of LYK3 with an M. truncatula Plant U-box protein 1 (PUB1, a E3-ubiquitin ligase) suggests that the initial signaling involves protein degradation (Mbengue et al., 2010). Recently, it has been shown that PUB1 interacts and is phosphorylated by DMI2, which supports the findings that the concerted action of the receptor kinases controls rhizobial entry (Vernié et al., 2016). The actual function of VAPYRIN and CBS1 in this context is not clear.
DMI1 is an inner-membrane-localized channel protein. DMI1 localizes to the ER membrane, and the ER is one of the major calcium storages in the cell (Ané et al., 2004). A Ca2+ pump (MCA8, sarco/endoplasmic reticulum calcium ATPase), three cyclic nucleotide-gated channels (CNGC15a, CNGC15b, CNGC15c) and potassium-permeable channel (DMI1) are needed for the generation of the self-sustaining Ca2+ spiking. The three cyclic nucleotide-gated channels form a complex with the DMI1 in nuclear envelope, which modulates nuclear Ca(2+) release (Charpentier et al., 2016). All the three CNGC15a-c and MCA8 has been identified by a reverse genetic approach (Capoen et al., 2011).
A Ca2+/calmodulin dependent protein kinase, named DMI3, acts downstream to the symbiotic Ca2+ spiking. DMI3 has an exceptional ability to bind free Ca2+ both directly using EF-hand domains and indirectly through a calmodulin (CaM) binding domain. It has been hypothesized that DMI3 decodes the calcium spiking signal that leads to nodule development. Indeed, the dmi3 mutant has normal Ca2+ spiking responses but does not form any nodules. Several transcription factors acting downstream of DMI3 have been identified (Oldroyd, 2013). Among them, INTERACTING PROTEIN OF DMI3 (IPD3) represents a unique class of transcription factor family. DMI3 phosphorylates IPD3, leading to the activation of the latter, and eventually activation of its downstream targets like NODULE INCEPTION (NIN) and a CCAAT-box binding transcription factor (HAP2-1; Oldroyd, 2013; Singh et al., 2014).
Several other transcription factors have been implicated in nodule development. Plants showing mutation(s) in these genes either develop small bump-like nodules without bacterial colonization or no nodules. NODULATION SIGNALING PATHWAY genes (NSP1 and NSP2) are plant specific GRAS transcription factor/regulators. nsp1 and nsp2 mutants show root hair deformation but the induction of cortical cells in response to NF is blocked (Oldroyd and Long, 2003; Kaló et al., 2005; Smit et al., 2005). Three more TFs have been implicated in this signaling process. Among them, ERF REQUIRED FOR NODULATION (ERN1) contains a highly conserved AP2-DNA binding domain. ern1 initiate the development of ITs but still form small bumps (Middleton et al., 2007). Medicago ERN1 has a close homolog, ERN2. ERN1/ERN2 act in concert in the root epidermis and the ern1/ern2 double mutant displays a severe phenotype where the initiation of infection is completely abolished (Cerri et al., 2016). MtNF-YA1/HAP2-1 is a Medicago CCAAT box-binding TF. The nf-ya1 was isolated from an EMS mutant population. This gene is required to initiate IT formation and to maintain the persistent meristem activity in mature nodules (Laporte et al., 2014). Probably, the most vital transcription factor that controls epidermal infection, cortical cell division and nodule number is NIN. nin mutants undergo excessive root hair curling in response to inoculation by Sinorhizobium, but are impaired in infection and do not show any cortical cell division (Marsh et al., 2007; Soyano et al., 2014; Yoro et al., 2014). RHIZOBIUM-DIRECTED POLAR GROWTH (RPG) gene of M. truncatula is also worth mentioning. Nitrogen-fixing nodules are rarely formed in rpg mutants. ITs are abnormally thick and progress slowly; moreover, root hair curling is abnormal. This gene encodes for a yet uncharacterized putative long coiled-coil protein. This protein has a nuclear localization signal (NLS) and was found to actually localize to the nucleus in Nicotiana cells (Arrighi et al., 2008). The actual role of this gene is not clear to date.
The first thing that is apparent from the complexity of IT development is the need of cytoskeleton rearrangement. In a forward genetic approach, M. truncatula REQUIRED FOR INFECTION THREAD (rit/nap1) mutant has been isolated, which encodes for a component of the SCAR/WAVE (suppressor of cAMP receptor/WASP-family verprolin homologous protein) complex. This complex regulates actin polymerization through the activation of ARP2/3, suggesting that actin cytoskeleton rearrangement is crucial for IT propagation (Miyahara et al., 2010). LUMPY INFECTIONS (LIN) is another gene required for the growth of ITs in the root hair cells. It encodes for a protein with U-Box, E3 ubiquitin ligase and WD40 repeat domains. This also suggests that protein degradation is a crucial mechanism downstream of NF signaling in the root hair cells (Kiss et al., 2009).
Although the last two decades have brought significant increase in our knowledge of the early infection process, a lot of unanswered questions still remain. A transcriptional profiling of M. truncatula root hairs prior to and during the initial stages of rhizobial infection has been undertaken (Breakspear et al., 2014). A Systems biology approach of this single cell model revealed (a) activation of plant cell cycle, (b) involvement of several hormone related pathways like auxin, gibberellin, strigolactone, brassinosteroid, jasmonic acid and salicylic acid, and (c) expression of infection specific flavonoid synthesis genes and bacterial NF degradation genes. From this study, it appears that at the onset of infection, the plant cell cycle is reactivated, and along with that, hormone and flavonoid biosynthesis is necessary for rhizobial infection. It also suggests that both positive and negative feedback loops control levels of NF during rhizobial infection. Further genetic studies are needed to find out unique and redundant pathways that are required during bacterial infection (Breakspear et al., 2014).
The first genetic evidence that highlighted the fact that bacterial infection and nodule development are two independent phenomena came from Lotus japonicus. In Lotus, snf1 and snf2 mutants initiated nodule development independently of any rhizobial inoculation (Tirichine et al., 2006, 2007). Lotus snf1 encodes for CCaMK, which is orthologous to M. truncatula DMI3. Deregulation of the kinase activity of DMI3 leads to auto-nodule development in M. truncatula as well (Gleason et al., 2006). DMI3 has a dual sensing module, which is able to perceive the Ca2+ spiking and to promote the downstream NF signaling pathway. DMI3 controls bacterial infection in the epidermis and promotes nodule organogenesis. If the tuning of the kinase activity is hampered, it would lead to activation of the NF signaling pathway (Miller et al., 2013). The second report of the cortical cell division in absence of rhizobia came from the overexpression of RESPONSE REGULATOR 9 (RR9) in M. truncatula. RR9 is a response regulator that acts downstream of cytokinin signaling. Overexpression of RR9 leads to cortical cell division and generation of a bulge like structure in roots (Op den Camp et al., 2011). Involvement of cytokinin in spontaneous nodule development was reported earlier in Lotus. snf2 is a Lotus cytokinin receptor and have a histidine kinase domain. A point mutation in the cytokinin perceiving domain leads to auto-activation of its kinase activity, causing spontaneous nodule development in Lotus (Tirichine et al., 2007). The orthologous gene of Lotus snf2 has been identified in M. truncatula and named CYTOKININ RESPONSE 1 (CRE1; Gonzalez-Rizzo et al., 2006). cre1 mutants show an early inhibition of cortical cell divisions during nodule initiation (Gonzalez-Rizzo et al., 2006). The third report that shows spontaneous nodulation in M. truncatula came from the overexpression of the intracellular kinase-domain DMI2. Deregulation of DMI2 activity leads to hyper-activation of the nodule organogenesis program (Saha et al., 2014).
The overexpression of RWP-RK transcription factor (i.e., NIN) in Lotus is sufficient to induce spontaneous nodule-like structures (Soyano et al., 2013). Further, specific overexpression of NIN separately in epidermis (using an epidermal specific promoter) and in cortex (using a cortical specific promoter) promotes spontaneous nodules in Medicago. Additionally, NIN directly binds to the promoter of CRE1 gene and induces cytokinin signaling. These results suggest that cytokinin is the best described downstream factor involved in spontaneous nodule organogenesis (Vernié et al., 2015).
In summary, nodule organogenesis is spatio-temporally separated into two sophistically regulated events, an epidermal bacterial infection and cortical cell division. Disruption of this regulation leads to spontaneous nodule development in both Medicago and Lotus. It appears that phosphorylation of the downstream targets induces activation of series of transcription factors, including NIN, which directly activates the expression of a cytokinin receptor (CRE1) in the cortex followed by the downstream cytokinin signaling inducing nodule development.
In a wild type context, nodule development, and bacterial endocytosis are tightly regulated. ITs start to penetrate the dividing cortical cells after ~33–35 h of bacterial inoculation. Eventually, bacterial endocytosis takes place (Xiao et al., 2014). NF signaling module that operates in the epidermis during bacterial entrapment is also functional during the bacterial release process in the host cell cytoplasm. Specific knock-down of the NFP gene inside the nodule highlights its requirement for the release of the bacteria from the ITs. Knocking-down DMI2 specifically inside the nodule prevents bacterial endocytosis. Rice CCaMK can restore the epidermal block in the dmi3 mutant, but cannot rescue bacterial endocytosis (Godfroy et al., 2006). In the ipd3-1, bacteria are entrapped inside the ITs and nearly never released into the nodule primordium (Horváth et al., 2011). Thus, NFP-DMI2-DMI3 and IPD3 act in nodule epidermis and cortex during the plant-bacteria recognition phase.
Rhizobia are released inside the host cytoplasm and form symbiosomes after about 80 h of inoculation (Xiao et al., 2014). Symbiosomes undergo several rounds of division followed by endoreduplication. Endoreduplication of the bacteroid genome in M. truncatula leads to elongated or Y-shaped bacteroids (called terminally differentiated). Simultaneously, host cortical cells undergo several rounds of endoreduplication (Oldroyd, 2013). A transcriptomic approach has been undertaken to understand the connection between the cellular differentiation and the transcriptional activation during nodule development (Maunoury et al., 2010). This study unraveled two waves of transcriptional activation. The first wave starts with the repression of plant defense-related genes and simultaneous activation of cell cycle and protein synthesis genes. The second wave is marked by the induction of secretory pathways along with a large number of small peptides, secretory and transmembrane proteins.
Several forward genetic studies also support the above-mentioned hypothesis of transcription reprogramming. Mutant lines with normal bacterial infection in the epidermis but impaired in nitrogen fixation are called DEFECTIVE IN NITROGEN FIXATION (dnf mutants). Eleven mutants belonging to the dnf category have been described, to date, namely dnf1 to dnf8, 5L/11S, 7Y, and 13U (Starker et al., 2006; Domonkos et al., 2013). Among them, the genes responsible for dnf1, dnf2 phenotypes have been identified. Bacteroids in dnf1 remain small like their free-living counterparts, unlike the Y-shaped bacteroids in wild type. DNF1 mutated gene encodes a subunit of the signal peptidase complex without the signal peptide of secretory proteins. DNF1 is involved in protein trafficking to symbiosomes (Wang et al., 2010). Around 600 small peptide genes (NCR peptides) are responsible for the elongation of bacteroids and are present in M. truncatula but not in L. japonicus (Mergaert et al., 2003). Two NCR peptides, NCR169 and NCR211 are required for symbiotic nitrogen fixation in M. truncatula nodules (Horváth et al., 2015; Kim et al., 2015). At least, few NCR peptides are transported inside the symbiosome and DNF1 is required for their transport (Van de Velde et al., 2010). dnf2 nodules have a narrow zone with infected cells. Nonetheless, improper differentiation of bacteroids results in rapidly prematurely senescing nodules. DNF2 is a putative non-canonical phosphatidylinositol phospholipase C-like protein, which lacks its Y domain. DNF2 has N-terminal signal peptide, indicating its probable entry to the secretory pathway. It is proposed that DNF2 is required for the repression of defense response, but the actual role of this protein in nodule development remains elusive (Bourcy et al., 2013).
Two more crucial genes for nodule development have been identified in a reverse genetic approach [based on Mt Gene Expression Atlas Benedito et al., 2008]. One of them is symCRK. symcrk mutant plants develops non-functional nodules. symcrk nodules show defense-like reactions and early senescence. The predicted protein encodes for a membrane-bound kinase, which contains two extracellular cysteine-rich domains. The exact role of this protein in nodule development has not been understood, but it is predicted that symCRK may be involved in suppression of host defense response (Berrabah et al., 2014). The second one is a transcription factor, REGULATOR OF SYMBIOSOME DIFFERENTIATION (RSD). RSD encodes for a Cysteine-2/Histidine-2 (C2H2) zinc-finger family of plant TFs. In rsd-1 mutant nodules, bacteroids fail to differentiate normally. The endoreduplication and the viability of the bacteroids are also compromised. MtRSD is a transcriptional regulator of plant secretory pathway genes VAMP721a and thereby controls symbiosome development (Sinharoy et al., 2013). RSD is also implicated in the repression of plant defenses in nodules (Berrabah et al., 2015) and predicted to work similarly as symCRK in defense suppression. Recently, another M. truncatula mutant has been described via reverse genetics, called NODULE WITH ACTIVATED DEFENSE 1 (NAD1). nad1 nodules shows very early defense response. The rhizobia, along with their symbiotic plant cells, become necrotic immediately after the bacteria are released from infection threads into the nodule symbiotic cells. NAD1 encodes a small and uncharacterized protein with two predicted transmembrane helices and is localized at the endoplasmic reticulum (Wang et al., 2016). Presence of high load of NCR peptides and their involvement in nodule development confer to M. truncatula a unique situation. The coherent action of DNF2, RSD, symCRK, NAD1, NCR peptides, and most probably several hundred uncharacterized genes, act together to repress the defense response and enhance efficient symbiotic nitrogen fixation after bacterial endocytosis. Detailed studies should be performed in order to unravel the molecular mechanism. Another transcription factor, EFD (Ethylene response factor required for nodule differentiation) controls nodule number by negatively regulating nodule development. This gene has been proposed to be involved in the bacteroid differentiation processes by activating MtRR4, a type-A cytokinin primary response regulator (Vernie et al., 2008).
Functional nodule development is a highly energy consuming process. The host plant spends up to 25% of a legume's net photosynthate for root nodule development (Oono and Denison, 2010). To efficiently manage the distribution of energy, plants developed a mechanism to control nodule number by terminating nodule development. This mechanism is called autoregulation of nodulation (AON; Kosslak and Bohlool, 1984). AON involves through long distance signaling, from root to shoot and back again to root. AON was discovered by the classical split root inoculation experiments, where one part of the split root was inoculated inducing a systemic suppression of nodule development on the other part of the split root (Kassaw et al., 2015).
More recently, genetic studies have been valuable to dissect the AON pathway. M. truncatula the sickle mutant lines display a drastically increased underdeveloped number of nodules. SICKLE gene encodes an integral membrane protein comprising an N-terminal segment that shows similarity with a family of metal transporters and a unique C-terminal segment (CEND, the C-terminal end of EIN2) that does not contain a known and characterized motif (Alonso et al., 1999; Penmetsa et al., 2008). The EIN2 (ETHYLENE INSENSITIVE2) protein was previously identified in Arabidopsis. Studies from Arabidopsis and several other plants highlighted that EIN2 plays a central role in ethylene signaling. After, the first phase of infection, the competency for further root hair mediated infection is controlled locally by SICKLE/EIN2-dependent pathway. Transcriptional profiling of the Mtein2/sickle mutant highlighted a SICKLE/EIN2 dependent pathway negatively controls NF-mediated nodule organogenesis (Lauressergues et al., 2015). The AON pathway is shoot-controlled and work systemically instead of locally like SICKLE/EIN2. The first AON gene that was cloned is SUPER NUMERARY NODULES (SUNN), encoding a leucine-rich repeat receptor kinase like CLAVATA1 (CLV1) of Arabidopsis thaliana (Schnabel et al., 2005). The sunn mutants display a super-nodulation phenotype (van Noorden et al., 2006). Another shoot controlled super-nodulation mutant was identified and called like sunn supernodulator (lss). Transcriptomics studies highlighted that the main cause behind the lss phenotype is reduced SUNN expression. The prevailing hypothesis suggests that lss is a cis-acting factor that inhibits the expression of SUNN (Schnabel et al., 2010).
As mentioned earlier, AON involves long distance signaling from root to shoot with feedback signal to root. The root-derived signal is perceived by SUNN receptor kinase (Leucine-Rich Repeat Receptor-Like Kinase [LRR-RLK]). In Arabidopsis CLV1 receptor perceives short secreted CLE peptides (CLAVATA3/Endosperm surrounding region-related) and controls several aspects of plant development. Arabidopsis CLV1 functions within a larger receptor complex. AtCLV1-AtCLV2 heterodimer perceives AtCLV3 peptide (Jeong et al., 1999). Again, AtCLV2 forms a stable heterodimer with a membrane bound pseudokinase, CORYNE (CRN). AtCLV2-AtCRN pathway is able to function independently of AtCLV1 (Bleckmann et al., 2010). Over-expression of two CLE peptides (MtCLE12/MtCLE13 and LjCLE-RS1/LjCLE-RS2) systemically reduces nodule number in a MtCLV1-like receptor kinase dependent manner (Okamoto et al., 2009; Mortier et al., 2010). LjCLE-RS1 and LjCLE-RS2 were shown as direct targets of LjNIN in L. japonicus (Soyano et al., 2014). Expression of NIN is significantly reduced as a systemic effect of a CLE peptide over-expression in Medicago. It is still not clear whether or not CLE peptide genes are direct target of NIN in Medicago (Mortier et al., 2010). In a recent study, it has been shown that MtSUNN form homomers, and heteromers (with both MtCRN and MtCLV2). Further, in Medicago the crn mutant, shoot controlled AON is destroyed and it forms increased number of nodules. Indicate, that the same trio that control Arabidopsis shoot development, recruited to controls nodule number in Medicago. Another gene that has been identified in M. truncatula is ROOT DETERMINED NODULATION 1 (RDN1; Schnabel et al., 2011). RDN1 belongs to the HPAT gene family of Arabidopsis. In Arabidopsis, HPAT genes are able to glycosylate some of the CLE peptide (Ogawa-Ohnishi et al., 2013).
Conversely, C-TERMINALLY ENCODED PEPTIDEs (CEPs) positively regulate nodule number and negatively regulate lateral root emergence locally (Imin et al., 2013). Again, compact root architecture (cra2) mutant have more lateral roots and reduced number of nodules. The CRA2 gene encodes another a LRR-RLK. Like CEPs, CRA2 negatively regulates lateral root formation and positively regulates symbiotic nodulation (Huault et al., 2014). In a recent study it has been shown that unlike in wild-type, application of MtCEP1 is unable to increase nodule number in cra2 mutant (Mohd-Radzman et al., 2016). CRA2 is very closely related to A. thaliana XIP1 (XYLEM INTERMIXED IN PHLOEM1) receptor (Bryan et al., 2012) which specifically binds CEPs. Taking together the knowledge from Medicago and Arabidopsis it appears that MtCEPs can bind to MtCRA2 receptor and regulate nodule number. MtCEP1 treatment can increase nodule number in the sunn mutant but failed to increase nodule number in the sickle background, suggest there is an overlap between CEP medicated and EIN2 dependent nodule number controlling pathway (Crook et al., 2016).
Double mutant analysis using sickle and sunn mutants highlighted that SICKLE and SUNN participate in distinct genetic pathways (Penmetsa et al., 2003). Ectopic overexpression of the cytoplasmic kinase domain of DMI2 in sickle background increases the hyper nodulation phenotype, but it completely abolishes nodule development in sunn background. This confirms, SUNN-mediated systemic AON signaling pathway and SICKLE-mediated local nodule number control pathway acts through separate pathways (Saha and DasGupta, 2015). This result also suggests that DMI2 can be another determinant controlling nodule number.
To summarize, combined negative action of SICKLE and SUNN mediated pathways and positive action CRA2 and CEPs medicated induction of nodule development, tightly control nodule number. The AON mechanism starts very early during nodule development, probably MtNIN activates CLE peptides gene expression. CLE peptide repress NIN expression in a negative feedback loop. Most likely, SUNN receptor kinase act as a receptor of RDN1 glycosylate CLE peptide, CRA2 and CEPs medicated positive regulation of nodule number work through SICKLE and positively regulate nodule number.
Given the economical and nutritional importance of legume seeds, seed development of M. truncatula has been intensively studied and various reports have been published regarding the accumulation of transcripts (Gallardo et al., 2007; Benedito et al., 2008; Verdier et al., 2008), proteins (Gallardo et al., 2003), lipids and sugars (Djemel et al., 2005), and metabolites (Verdier et al., 2013b) during the development of the seeds. Seed development is usually divided into three phases: the embryogenesis marked by the development of the embryo, the early maturation or seed filling marked by the accumulation of storage macromolecules in cotyledons, and the late maturation marked by the desiccation of the seed and its entry into dormancy. In M. truncatula, several reports (Gallardo et al., 2007; Verdier et al., 2008) described the timing of seed development with embryogenesis starting at the double fertilization until 12–13 days after pollination (dap), then the early maturation until around 20 dap, and finally the late maturation until the seed become mature at ~48 dap. The seed is also characterized according to its tissues: the embryo, the endosperm and the seed coat. The formation of these tissues is derived from the double fertilization (Goldberg et al., 1994). The integument of the ovule develops into seed coat, and the inside of the ovule progresses to embryo sac. Within the sac, the central cell (2n) fuses with one sperm cell (1n) to form the triploid endosperm. In the meanwhile, the egg cell (1n) together with another sperm cell (1n) will form the diploid zygote, which will go through the embryogenesis. A schematic description of the seed tissues and seed developmental stages with the timing of M. truncatula seed development is provided in Figure 2.
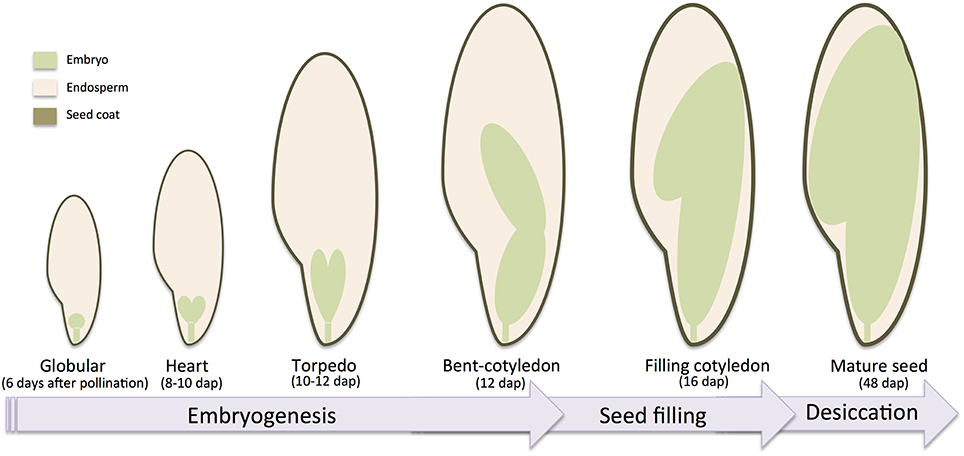
Figure 2. Schematic development of Medicago truncatula seed. Major seed tissues and developmental seed and embryo stages are indicated with corresponding days after pollination.
In this section, we classified the gene functional studies into three subfields related to their role during seed development: genes controlling embryogenesis, seed composition and desiccation tolerance. All the genes proposed in the following section are described in Table 2.
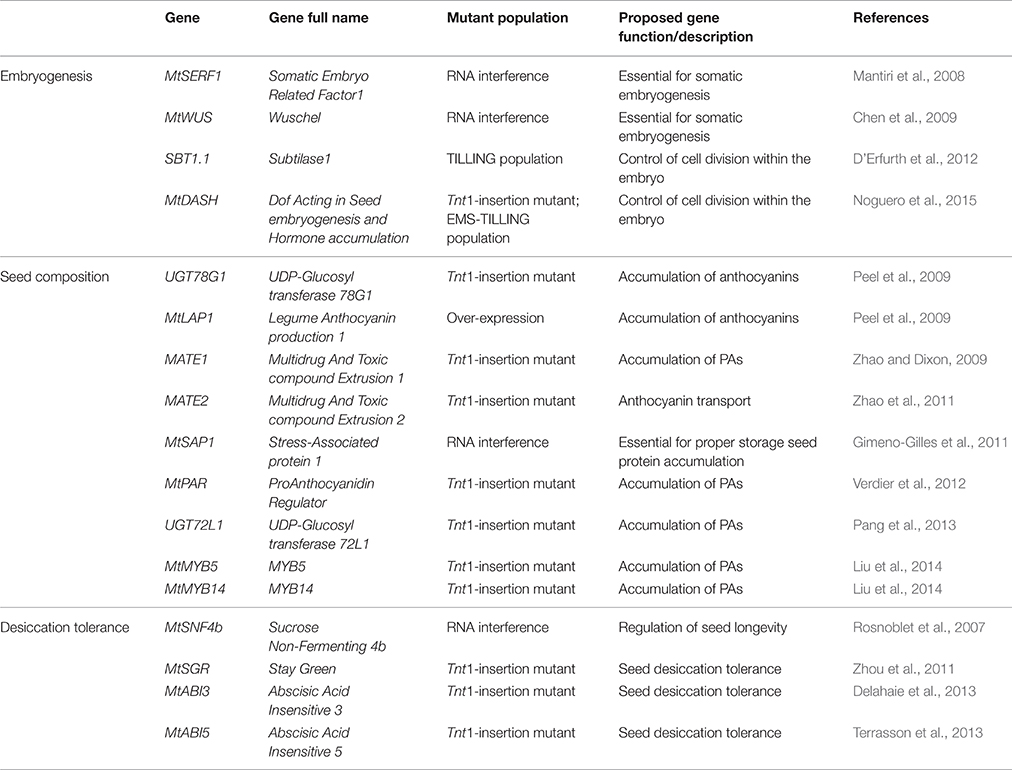
Table 2. List of seed development related genes that have been functionally characterized in M. truncatula.
Embryogenesis is a very well-studied process in plants and many genes are necessary for the acquisition of the polarity and the morphogenesis. For instance, it has been shown in Arabidopsis that more than 400 genes are required for the proper development of the embryo (Muralla et al., 2011). In M. truncatula, the formation of the embryo starts after the double fertilization event and the embryo goes through the different developmental stages known as globular (around 6 dap), heart (around 8–10 dap), torpedo (around 10–11 dap), and reaches the bent cotyledon stage around 12 DAP marking the end of the embryogenesis (Verdier et al., 2013a; Noguero et al., 2015; Figure 2). Some of the essential genes required for M. truncatula embryo development have been transcriptionally profiled (Kurdyukov et al., 2014). Two of them expressed in zygotic embryos have been functionally characterized showing their essential roles during somatic embryogenesis. The first one from the ERF subfamily transcription factors, SOMATIC EMBRYO RELATED FACTOR1 (MtSERF1), was showed to be induced by ethylene, in combination with other phytohormones, to control the initiation of somatic embryogenesis. Indeed, the serf1 knock-down (KD) lines, obtained using RNAi, resulted in a strong suppression of somatic embryogenesis (Mantiri et al., 2008). Some evidence supported that MtSERF could be regulated by WUSCHEL gene (MtWUS). Indeed, wus RNAi lines displayed similar phenotype as serf1, with no formation of callus and somatic embryos, and WUS binding sites were identified in the SERF1 promoter (Chen et al., 2009).
Apart from the regulation of the morphogenesis, embryogenesis is also marked by active cell divisions within the embryo, which is crucial for the final determination of the mature seed size. Indeed, a positive correlation was observed between the number of cotyledon cells and the final seed size in pea and soybean (Munier-Jolain and Ney, 1998). Two genes expressed in the endosperm of M. truncatula have been described as regulators of embryo cell division thereby impacting the seed size. The loss-of-function mutation in the SBT1.1 gene, a subtilase, was obtained from an EMS population by TILLING screening and its phenotype showed reduced cell number and a decrease of seed size compared with wild type plants. This suggests that SBT1.1 can regulate the cell number of cotyledon during the seed development, probably by providing molecules that act as signals to control cell division within the embryo (D'Erfurth et al., 2012). Moreover, a DOF transcription factor, DASH (DOF Acting in Seed embryogenesis and Hormone accumulation) was demonstrated to regulate the cell division rate of the embryo by its impact on auxin homeostasis leading to final seed size determination (Noguero et al., 2015). Three alleles were identified for MtDASH, one weak allele (i.e., Tnt1 insertion in the promoter region) showing a moderate phenotype on seed size and two strong alleles (i.e., Tnt1 insertion just before the starting codon and an EMS substitution inducing premature stop codon) displaying embryo lethal phenotypes and defect in auxin homeostasis.
The seed composition represents a nutritionally important trait. Indeed, the nature and amount of stored macromolecules will define the final quality of seeds for human and animal diets. In M. truncatula, as in the dicot seeds, most of these storage macromolecules are stored in the embryo during the seed filling phase from 14 dap to 36 dap. To our knowledge, only one gene, MtSAP1, has been shown to directly regulate the nature and amount of the major storage molecules in M. truncatula. Using RNAi, sap1 seeds displayed reduction of the globulin content (i.e., legumin and vicillin storage proteins). This phenotype affecting the seed composition was associated with reduced seed size and decrease of germination rate (Gimeno-Gilles et al., 2011). Other molecules, called secondary metabolites, are accumulated during the seed development, in another seed tissue, the seed coat. These molecules are not required for the proper seed development and are proposed to participate in defense-related processes. Amongst these compounds, the flavonoids such as proanthocyanindins (or PA) and anthocyanins have recently received a lot of attention because of their potential beneficial effects on human and animal health. To date, regarding the regulation of the PA biosynthetic pathway, several genes have been identified in M. truncatula such as MtWD40-1 (Pang et al., 2009), MtMYB5 and MtMYB14 (Liu et al., 2014), MtPAR (PROANTHOCYANIDIN REGULATOR, a regulator of PA accumulation by its action on MtWD40-1; Verdier et al., 2012). All these regulator genes have been shown to directly or indirectly control one of the major enzymes leading to PA production, the ANTHOCYANIDIN REDUCTASE (ANR). Others genes with involvement in PA accumulation have been revealed, such as a glycosyltransferase gene (UGT72L1) involved in modification and assembly of PA precursors (Pang et al., 2013) and MATE1, a PA transporter (Zhao and Dixon, 2009). In parallel to the PA pathway, the anthocyanin biosynthetic pathway also received some attention. MATE2 has been shown to transport the anthocyanin by redirecting the flavonoid precursors to the anthocyanin pathway at the expense of the PAs (Zhao et al., 2011). Moreover, the glycosyltransferase gene (UGT78G1) was found to be essential to anthocyanin modification and accumulation (Peel et al., 2009). Most of the advances in the PA/anthocyanin gene regulation have become possible using Tnt1 insertion mutants of the previously mentioned genes.
Desiccation tolerance represents an important seed quality trait that allows mature seeds to survive in a dry state for long periods of time. This characteristic is acquired at the later stages of maturation, after 20–24 dap in M. truncatula (Figure 2). Several protective molecular processes have been described to acquire the desiccation tolerance including accumulation of non-reducing sugars such as raffinose family oligosaccharides (RFO), late embryogenesis abundant (LEA) proteins and other stress-related proteins. In M. truncatula, several regulators of these processes have been identified. Knock-down of the MtSNF4b gene, which encodes the γ-subunit of SnRK1 (sucrose non-fermenting-related kinase complex), resulted in impaired accumulation of RFO in seeds leading to a decrease of desiccation tolerance and seed longevity (Rosnoblet et al., 2007). Loss-of-function mutants of two other key regulators of seed development, ABSCISIC ACID INSENSITIVE3 (MtABI3) and ABSCISIC ACID INSENSITIVE5 (MtABI5), have been functionally characterized and revealed a loss of desiccation tolerance in mutant lines with impaired RFO content, decrease of LEA protein expression and changes in stress-related protein expression (Delahaie et al., 2013; Terrasson et al., 2013). Finally, STAY GREEN gene (MtSGR) have been identified and loss-of-function sgr mutants displayed a delayed senescence of the whole plant (Zhou et al., 2011). Even if the sgr mutant seed phenotype has not been investigated in the study, sgr mutant seed remained green at maturity, which mimics the loss of ability to desiccate of the abi3 mutant seeds (Delahaie et al., 2013).
In conclusion to this section, functional genetic studies of seed-related genes in M. truncatula are facing the same challenges than in other species: (i) the embryo and/or seed lethality phenotypes and (ii) the complexity of regulation between seed tissues with the seed coat (diploid and maternal origin, 2n♀), the endosperm (triploid and hybrid origin, 2n ♀ + 1n♂), and the embryo (1n♀ + 1n♂). Regarding the first challenge, M. truncatula mutant populations are kept and distributed at the segregating population status, which allows the conservation of most of the lethal mutations at the heterozygote status, when the mutation is non-dominant. However, this advantage of keeping living seeds containing 1 allele of lethal mutation, could turn as a time consuming process when it comes to identify homozygote mutant plants to reveal putative seed phenotype due to the M. truncatula generation time (~4 months). So far, most of the functional genetic studies have focused on the seed coat composition and flavonoid accumulation because it is a non-lethal mutation and easy to phenotypically visualize (i.e., change of seed coat color). This aspect represents an economical interest because of the beneficial effects of tannins on plant defense, flower color, forage quality and human health (see review, Dixon et al., 2013). The second focus of M. truncatula seed studies was the genes related to desiccation tolerance because we know how to rescue seeds non-tolerant to desiccation. At the opposite, to date, there is no efficient protocol in M. truncatula for embryo-rescue, which limits the functional studies of genes essential for proper embryogenesis. Regarding the second challenge, most of the genetic studies related to the complexity of regulation of seed development have been using A. thaliana as model plant. As demonstrated in the current section, M. truncatula seed genetic studies have so far focused on more applied aspects of seed biology such as yield, seed composition or desiccation/longevity.
Being sessile, plants have acquired the ability to respond promptly to extreme environmental conditions in order to survive. Among all the factors that affect plant growth, abiotic stresses probably have the largest effect. The primary abiotic stresses are drought, salinity, low and high temperature, nutrient deficiency, and flooding. Plants respond to each abiotic stress specifically but crosstalks among different stresses at physiological, biochemical, cellular, and molecular levels are common (Araujo et al., 2015). For example, both drought and salinity induce osmotic stress and therefore plants respond similarly to both stresses in osmotic-stress correlated pathways.
On large scales, microarray and RNA-seq experiments have been widely used to perform whole genome transcript profiling in M. truncatula. For example, identification of microRNAs responsive to drought (Wang et al., 2011), salinity (Long et al., 2015), and heavy metals (Zhou et al., 2011; Chen et al., 2012), and identification of gene expression changes under phosphate and nitrogen limitation (Bonneau et al., 2013), salinity stress (Gruber et al., 2009; Li et al., 2009), gradual drought stress (Zhang et al., 2014), as well as ozone stress (Puckette et al., 2008). At single gene level, functional studies of abiotic stress-related genes covered all the major aspects including gene expression (transcription factors), signaling transduction, osmolyte regulation, as well as antioxidant biosynthesis and DNA stability. Because of the crosstalk among stresses, researchers often identify plants' responses to multiple abiotic stresses in gene functional studies. Therefore, we classified the abiotic stress-related genes that have been functionally studied in M. truncatula based on functional category rather than the type(s) of abiotic stress (Table 3).
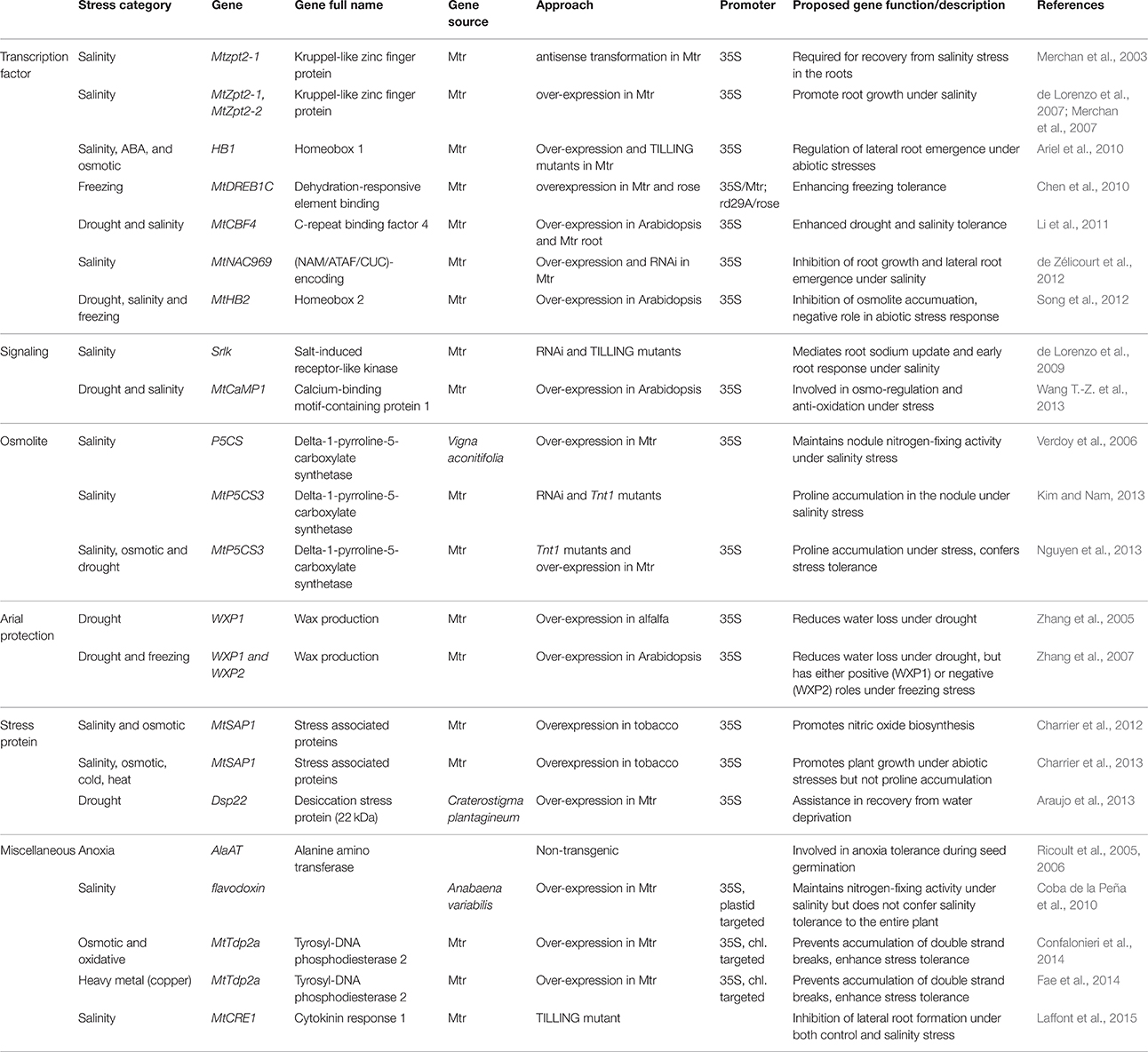
Table 3. List of abiotic-stress related genes that have been functionally characterized in M. truncatula.
Among all the transcription factors that have been characterized in M. truncatula, Mtzpt2 (encoding a putative Krüppel-like Cys-2/His-2 zinc finger protein) has been extensively characterized. A homolog of zpt2 was first identified in alfalfa (Mscp17 and renamed Mszpt2-1) for its strong induction during nodulation (Frugier et al., 1998). Ectopic expression of Mszpt2-1 and its Arabidopsis homolog, Stz, in yeast was able to induce salinity tolerance (Lippuner et al., 1996; Frugier et al., 2000). In M. truncatula, Mtzpt2-1 is rapidly induced by salt treatment in both the nodules and the roots. Notably, antisense Mtzpt2-1 transgenic M. truncatula plants had a significantly slower recovery compared to the control plants after salinity stress, although the root growth was similar before and during salinity stress between the transgenic and the control plants (Merchan et al., 2003). In an independent study, Mtzpt2-1 and Mtzpt2-2 were identified as among the most differentially regulated genes between M. truncatula genotypes Jemalong A17 and R108 under salinity stress, where Jemalong A17 was more tolerant to salinity than R108 (de Lorenzo et al., 2007). Further, overexpression of Mtzpt2-1 and Mtzpt2-2 in the salinity-sensitive genotype R108, but not in Jemalong A17, promoted root growth under salt stress (de Lorenzo et al., 2007). These results suggest a potential role of Mtzpt2 in adaptation of M. truncatula to saline soils. Interestingly, at the same time period, Merchan et al. (2007) identified Mtzpt2-1 and Mtzpt2-2 as the top regulated genes in the R108 root during salt stress and recovery with a subtractive hybridization approach, and they revealed that overexpression of Mtzpt2-1 in roots conferred plant salt tolerance by maintaining root growth. Taken together, Mtzpt2 appears to be a strongly salinity-induced gene in the root and may confer protection to the root under salinity stress conditions.
Besides Mtzpt2, which has been studied in detail, HB1, HB2, DREB1C, CBF4, and NAC969 have all been shown to be involved in gene expression regulation under various stress conditions in M. truncatula (Table 3). It is interesting to note that overexpression of MtHB2 in Arabidopsis inhibited osmolyte accumulation rendering plants more sensitive to abiotic stresses (Song et al., 2012), while overexpression of MtHB1 in M. truncatula altered root structure without negative effect on stress tolerance (Ariel et al., 2010).
Compared to transcription factors, genes involved in stress signaling transduction are less studied. In the same experiment as previously mentioned, Merchan et al. (2007) identified a SRLK (for Salt-induced Receptor-Like Kinase) gene. They showed that expression of SRLK was rapidly induced after salt stress in roots, especially in the root epidermis and root apex. srlk-TILLING mutants were able to maintain root growth under salt stress and also accumulated less sodium ions than the control plants in both root and shoot (de Lorenzo et al., 2009). Several early salt-regulated genes including Mtzpt2-1 were down-regulated in the srlk mutant lines under salt stress, indicating that SRLK may function in the early steps of a salinity stress signal transduction pathway.
Osmolyte accumulation in plants under abiotic stress has been widely observed (reviewed in Ashraf and Foolad, 2007; Kavi Kishor and Sreenivasulu, 2014). The primary forms of osmolytes are proline, glycine betaine, sugars, and polyols. In M. truncatula, proline biosynthesis has been the focus of manipulation under abiotic stress. P5CS (delta-1-pyrroline-5-carboxylate synthetase), a gene encoding the major proline biosynthetic enzyme, has been studied with both loss- and gain-of function approaches. It was shown that under salinity stress, overexpression of P5CS gene in M. truncatula could maintain nodule nitrogen fixation activity under salinity stress (Verdoy et al., 2006) and the transgenic plant displayed increased tolerance to salinity and osmotic stresses (Nguyen et al., 2013). On the other hand, a loss-of-function mutant of MtP5CS3 formed fewer nodules, and had lower nitrogen-fixation efficiency than the wild type under salinity (Kim and Nam, 2013). The non-nodulated MtP5CS3 insertion-mutant plant (Tnt1-insertion mutant) accumulated less proline and showed increased sensitivity to drought, salinity, and osmotic stresses, resulting in decreased seedling growth and leaf chlorophyll content (Nguyen et al., 2013).
In addition to the above three major categories of abiotic stress-regulated genes, genes that participate in other processes of plant abiotic stress responses have also been explored in M. truncatula, though not extensively. WXP1 and WXP2 (wax production protein) are two homologous genes involved in the wax production pathway in M. truncatula. Overexpression of WXP1 or WXP2 in alfalfa and Arabidopsis significantly reduced water loss under drought and generated drought-resistant transgenic plants (Zhang et al., 2005, 2007). Interestingly, WXP1 overexpression also promoted freezing tolerance, but WXP2 overexpression had the opposite effect; though, the underlying mechanism has not been explained (Zhang et al., 2007). In addition, two other stress-related proteins were characterized. MtSAP1 (stress associated protein 1) was over-expressed in tobacco and the transgenic plants performed better under multiple stresses with more nitric oxide production independently of enhanced proline accumulation (Charrier et al., 2012, 2013). Moreover, DSP22 gene (desiccation stress protein 22 kDa) identified from the resurrection plant Craterostigma plantagineum was over-expressed in M. truncatula and the transgenic plants were able to recover from drought stress better than the wild-type plants (Araujo et al., 2013). Finally, other genes involved in amino acid production, ROS scavenging, DNA stability, as well as hormone signaling were functionally studied and are listed in Table 3.
In conclusion to this section, if we analyze together the abiotic stress-related genes that have been functionally studied in M. truncatula, we see clear trends. First, salinity stress, rather than drought stress, has been the top focus among all abiotic stresses, due to the simplicity of controlling specific amount of salt in petri dishes experiments. Second, the majority of the functional genomics studies used over-expression approach, either alone or in combination with loss-of-function mutants. To generate overexpressing transgenic plants, the constitutive cauliflower mosaic virus (CaMV) 35S promoter was used in almost all studies except one (Chen et al., 2010). This potentially complicates the interpretation of transformed plant phenotypes considering the tissue-specific expression manner of the target genes. Finally, transgenic plants showing abiotic-stress tolerant phenotypes were only tested in the petri dish/growth chamber conditions, but not under soil and/or field conditions.
In this review, we presented a non-exhaustive list of M. truncatula genes that have been functionally characterized in three primary research areas of legume biology. With the recent completion of the M. truncatula genome, the development of several genomics tools and the new technological advances, we have no doubt that we are only at the onset of functional genetics and genomics in this species and many other gene functions will be investigated and revealed in the coming years. For instance, transcriptomics studies have estimated that more than 20,000 plant genes are expressed during nodule development and more than 19,000 are differentially expressed during seed development in M. truncatula. Among them, only ~100 genes have been characterized functionally in different plant species. Extensive use of next generation technology coupled with genomics and systems biology approaches is needed for the better understanding of the functional aspects of the gene pool mentioned above. Finally, a technical breakthrough with the recent development of genome editing technologies, such as TALENs, ZINC-FINGER nucleases, and CRISPR-CAS9, provide new tools to precisely mutate gene sequences in order to advance functional genomics studies.
All authors contributed equally to the work. YK, ML, SS and JV wrote the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Alonso, J. M., Hirayama, T., Roman, G., Nourizadeh, S., and Ecker, J. R. (1999). EIN2, a bifunctional transducer of ethylene and stress responses in Arabidopsis. Science 284, 2148–2152. doi: 10.1126/science.284.5423.2148
Amor, B. B., Shaw, S. L., Oldroyd, G. E., Maillet, F., Penmetsa, R. V., Cook, D., et al. (2003). The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 34, 495–506. doi: 10.1046/j.1365-313X.2003.01743.x
Ané, J. M., Kiss, G. B., Riely, B. K., Penmetsa, R. V., Oldroyd, G. E., Ayax, C., et al. (2004). Medicago truncatula DMI1 required for bacterial and fungal symbioses in legumes. Science 303, 1364–1367. doi: 10.1126/science.1092986
Ané, J. M., Levy, J., Thoquet, P., Kulikova, O., de Billy, F., Penmetsa, V., et al. (2002). Genetic and cytogenetic mapping of DMI1, DMI2, and DMI3 genes of Medicago truncatula involved in Nod factor transduction, nodulation, and mycorrhization. Mol. Plant Microbe Interact. 15, 1108–1118. doi: 10.1094/MPMI.2002.15.11.1108
Araujo, S. S., Beebe, S., Crespi, M., Delbreil, B., Gonzalez, E. M., Gruber, V., et al. (2015). Abiotic stress responses in legumes: strategies used tocope with environmental challenges. Crit. Rev. Plant Sci. 34, 237–280. doi: 10.1080/07352689.2014.898450
Araujo, S. S., Duque, A. S., Silva, J. M., Santos, D., Silva, A. B., and Fevereiro, P. (2013). Water deficit and recovery response of Medicago truncatula plants expressing the ELIP-like DSP22. Biol. Plant. 57, 159–163. doi: 10.1007/s10535-012-0235-7
Ariel, F., Diet, A., Verdenaud, M., Gruber, V., Frugier, F., Chan, R., et al. (2010). Environmental regulation of lateral root emergence in Medicago truncatula requires the HD-Zip I transcription factor HB1. Plant Cell 22, 2171–2183. doi: 10.1105/tpc.110.074823
Arrighi, J.-F., Barre, A., Ben Amor, B., Bersoult, A., Soriano, L. C., Mirabella, R., et al. (2006). The Medicago truncatula lysine motif-receptor-like kinase gene family includes NFP and new nodule-expressed genes. Plant Physiol. 142, 265–279. doi: 10.1104/pp.106.084657
Arrighi, J. F., Godfroy, O., de Billy, F., Saurat, O., Jauneau, A., and Gough, C. (2008). The RPG gene of Medicago truncatula controls Rhizobium-directed polar growth during infection. Proc. Natl. Acad. Sci. U.S.A. 105, 9817–9822. doi: 10.1073/pnas.0710273105
Ashraf, M., and Foolad, M. R. (2007). Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot. 59, 206–216. doi: 10.1016/j.envexpbot.2005.12.006
Barker, D., Bianchi, S., Blondon, F., Dattée, Y., Duc, G., Essad, S., et al. (1990). Medicago truncatula, a model plant for studying the molecular genetics of the Rhizobium-legume symbiosis. Plant Mol. Biol. Report. 8, 40–49. doi: 10.1007/BF02668879
Benedito, V. A., Torres-Jerez, I., Murray, J. D., Andriankaja, A., Allen, S., Kakar, K., et al. (2008). A gene expression atlas of the model legume Medicago truncatula. Plant J. 55, 504–513. doi: 10.1111/j.1365-313X.2008.03519.x
Benlloch, R., d'Erfurth, I., Ferrandiz, C., Cosson, V., Beltrán, J. P., Cañas, L. A., et al. (2006). Isolation of mtpim proves Tnt1 a useful reverse genetics tool in Medicago truncatula and uncovers new aspects of AP1-like functions in legumes. Plant Physiol. 142, 972–983. doi: 10.1104/pp.106.083543
Berrabah, F., Bourcy, M., Eschstruth, A., Cayrel, A., Guefrachi, I., Mergaert, P., et al. (2014). A nonRD receptor-like kinase prevents nodule early senescence and defense-like reactions during symbiosis. New Phytol. 203, 1305–1314. doi: 10.1111/nph.12881
Berrabah, F., Ratet, P., and Gourion, B. (2015). Multiple steps control immunity during the intracellular accommodation of rhizobia. J. Exp. Bot. 66, 1977–1985. doi: 10.1093/jxb/eru545
Bleckmann, A., Weidtkamp-Peters, S., Seidel, C. A. M., and Simon, R. (2010). Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152, 166–176. doi: 10.1104/pp.109.149930
Boisson-Dernier, A., Chabaud, M., Garcia, F., Bécard, G., Rosenberg, C., and Barker, D. G. (2001). Agrobacterium rhizogenes-transformed roots of Medicago truncatula for the study of nitrogen-fixing and endomycorrhizal symbiotic associations. Mol. Plant Microbe Interact. 14, 695–700. doi: 10.1094/MPMI.2001.14.6.695
Bonneau, L., Huguet, S., Wipf, D., Pauly, N., and Truong, H. N. (2013). Combined phosphate and nitrogen limitation generates a nutrient stress transcriptome favorable for arbuscular mycorrhizal symbiosis in Medicago truncatula. New Phytol. 199, 188–202. doi: 10.1111/nph.12234
Bourcy, M., Brocard, L., Pislariu, C. I., Cosson, V., Mergaert, P., Tadege, M., et al. (2013). Medicago truncatula DNF2 is a PI-PLC-XD-containing protein required for bacteroid persistence and prevention of nodule early senescence and defense-like reactions. New Phytol. 197, 1250–1261. doi: 10.1111/nph.12091
Breakspear, A., Liu, C., Roy, S., Stacey, N., Rogers, C., Trick, M., et al. (2014). The root hair “infectome” of Medicago truncatula uncovers changes in cell cycle genes and reveals a requirement for Auxin signaling in rhizobial infection. Plant Cell 26, 4680–4701. doi: 10.1105/tpc.114.133496
Broeckling, C. D., Huhman, D. V., Farag, M. A., Smith, J. T., May, G. D., Mendes, P., et al. (2004). Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J. Exp. Bot. 56, 323–336. doi: 10.1093/jxb/eri058
Bryan, A. C., Obaidi, A., Wierzba, M., and Tax, F. E. (2012). XYLEM INTERMIXED WITH PHLOEM1, a leucine-rich repeat receptor-like kinase required for stem growth and vascular development in Arabidopsis thaliana. Planta 235, 111–122. doi: 10.1007/s00425-011-1489-6
Capoen, W., Sun, J., Wysham, D., Otegui, M. S., Venkateshwaran, M., Hirsch, S., et al. (2011). Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. U.S.A. 108, 14348–14353. doi: 10.1073/pnas.1107912108
Cerri, M. R., Frances, L., Kelner, A., Fournier, J., Middleton, P. H., Auriac, M. C., et al. (2016). The symbiosis-related ERN transcription factors act in concert to coordinate rhizobial host root infection. Plant Physiol. 171, 1037–1054. doi: 10.1104/pp.16.00230
Charpentier, M., Sun, J., Martins, T. V., Radhakrishnan, G. V., Findlay, K., Soumpourou, E., et al. (2016). Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352, 1102–1105. doi: 10.1126/science.aae0109
Charrier, A., Lelièvre, E., Limami, A. M., and Planchet, E. (2013). Medicago truncatula stress associated protein 1 gene (MtSAP1) overexpression confers tolerance to abiotic stress and impacts proline accumulation in transgenic tobacco. J. Plant Physiol. 170, 874–877. doi: 10.1016/j.jplph.2013.01.008
Charrier, A., Planchet, E., Cerveau, D., Gimeno-Gilles, C., Verdu, I., Limami, A. M., et al. (2012). Overexpression of a Medicago truncatula stress-associated protein gene (MtSAP1) leads to nitric oxide accumulation and confers osmotic and salt stress tolerance in transgenic tobacco. Planta 236, 567–577. doi: 10.1007/s00425-012-1635-9
Chen, J.-R., Lue, J.-J., Liu, R., Xiong, X.-Y., Wang, T.-X., Chen, S.-Y., et al. (2010). DREB1C from Medicago truncatula enhances freezing tolerance in transgenic M. truncatula and China Rose (Rosa chinensis Jacq.). Plant Growth Regul. 60, 199–211. doi: 10.1007/s10725-009-9434-4
Chen, L., Wang, T., Zhao, M., Tian, Q., and Zhang, W.-H. (2012). Identification of aluminum-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. Planta 235, 375–386. doi: 10.1007/s00425-011-1514-9
Chen, S. K., Kurdyukov, S., Kereszt, A., Wang, X. D., Gresshoff, P. M., and Rose, R. J. (2009). The association of homeobox gene expression with stem cell formation and morphogenesis in cultured Medicago truncatula. Planta 230, 827–840. doi: 10.1007/s00425-009-0988-1
Cheng, X., Wang, M., Lee, H., Tadege, M., Ratet, P., Udvardi, M., et al. (2014). An efficient reverse genetics platform in the model legume Medicago truncatula. New Phytol. 201, 1065–1076. doi: 10.1111/nph.12575
Coba de la Pena, T., Redondo, F. J., Manrique, E., Lucas, M. M., and Pueyo, J. J. (2010). Nitrogen fixation persists under conditions of salt stress in transgenic Medicago truncatula plants expressing a cyanobacterial flavodoxin. Plant Biotechnol. J. 8, 954–965. doi: 10.1111/j.1467-7652.2010.00519.x
Combier, J.-P., Frugier, F., de Billy, F., Boualem, A., El-Yahyaoui, F., Moreau, S., et al. (2006). MtHAP2-1 is a key transcriptional regulator of symbiotic nodule development regulated by microRNA169 in Medicago truncatula. Genes Dev. 20, 3084–3088. doi: 10.1101/gad.402806
Confalonieri, M., Fae, M., Balestrazzi, A., Dona, M., Macovei, A., Valassi, A., et al. (2014). Enhanced osmotic stress tolerance in Medicago truncatula plants overexpressing the DNA repair gene MtTdp2 alpha (tyrosyl-DNA phosphodiesterase 2). Plant Cell Tissue Organ Cult. 116, 187–203. doi: 10.1007/s11240-013-0395-y
Crook, A. D., Schnabel, E. L., and Frugoli, J. A. (2016). The systemic nodule number regulation kinase SUNN in Medicago truncatula interacts with MtCLV2 and MtCRN. Plant J. doi: 10.1111/tpj.13234. [Epub ahead of print].
D'Erfurth, I., Cosson, V., Eschstruth, A., Lucas, H., Kondorosi, A., and Ratet, P. (2003). Efficient transposition of the Tnt1 tobacco retrotransposon in the model legume Medicago truncatula. Plant J. 34, 95–106. doi: 10.1046/j.1365-313X.2003.01701.x
D'Erfurth, I., Le Signor, C., Aubert, G., Sanchez, M., Vernoud, V., Darchy, B., et al. (2012). A role for an endosperm-localized subtilase in the control of seed size in legumes. New Phytol. 196, 738–751. doi: 10.1111/j.1469-8137.2012.04296.x
Delahaie, J., Hundertmark, M., Bove, J., Leprince, O., Rogniaux, H., and Buitink, J. (2013). LEA polypeptide profiling of recalcitrant and orthodox legume seeds reveals ABI3-regulated LEA protein abundance linked to desiccation tolerance. J. Exp. Bot. 64, 4559–4573. doi: 10.1093/jxb/ert274
de Lorenzo, L., Merchan, F., Blanchet, S., Megias, M., Frugier, F., Crespi, M., et al. (2007). Differential expression of the TFIIIA regulatory pathway in response to salt stress between Medicago truncatula genotypes(1 W). Plant Physiol. 145, 1521–1532. doi: 10.1104/pp.107.106146
de Lorenzo, L., Merchan, F., Laporte, P., Thompson, R., Clarke, J., Sousa, C., et al. (2009). A novel plant leucine-rich repeat receptor kinase regulates the response of Medicago truncatula roots to salt stress. Plant Cell 21, 668–680. doi: 10.1105/tpc.108.059576
de Zélicourt, A., Diet, A., Marion, J., Laffont, C., Ariel, F., Moison, M. L., et al. (2012). Dual involvement of a Medicago truncatula NAC transcription factor in root abiotic stress response and symbiotic nodule senescence. Plant J. 70, 220–230. doi: 10.1111/j.1365-313X.2011.04859.x
Dixon, R., Liu, C., and Jun, J. H. (2013). Metabolic engineering of anthocyanins and condensed tannins in plants. Curr. Opin. Biotechnol. 24, 329–335. doi: 10.1016/j.copbio.2012.07.004
Djemel, N., Guedon, D., Lechevalier, A., Salon, C., Miquel, M., Prosperi, J. M., et al. (2005). Development and composition of the seeds of nine genotypes of the Medicago truncatula species complex. Plant Physiol. Biochem. 43, 557–566. doi: 10.1016/j.plaphy.2005.04.005
Domonkos, A., Horvath, B., Marsh, J. F., Halasz, G., Ayaydin, F., Oldroyd, G. E., et al. (2013). The identification of novel loci required for appropriate nodule development in Medicago truncatula. BMC Plant Biol. 13:157. doi: 10.1186/1471-2229-13-157
Du, Z., Zhou, X., Ling, Y., Zhang, Z., and Su, Z. (2010). agriGO: A GO analysis toolkit for the agricultural community. Nucleic Acids Res. 38, 64–70. doi: 10.1093/nar/gkq310
Endre, G., Kereszt, A., Kevei, Z., Mihacea, S., Kalo, P., and Kiss, G. B. (2002). A receptor kinase gene regulating symbiotic nodule development. Nature 417, 962–966. doi: 10.1038/nature00842
Fae, M., Balestrazzi, A., Confalonieri, M., Dona, M., Macovei, A., Valassi, A., et al. (2014). Copper-mediated genotoxic stress is attenuated by the overexpression of the DNA repair gene MtTdp2 alpha (tyrosyl-DNA phosphodiesterase 2) in Medicago truncatula plants. Plant Cell Rep. 33, 1071–1080. doi: 10.1007/s00299-014-1595-6
Frugier, F., Kondorosi, A., and Crespi, M. (1998). Identification of novel putative regulatory genes induced during alfalfa nodule development with a cold-plaque screening procedure. Mol. Plant-Microbe Interact. 11, 358–366. doi: 10.1094/MPMI.1998.11.5.358
Frugier, F., Poirier, S., Satiat-Jeunemaitre, B., Kondorosi, A., and Crespi, M. (2000). A Kruppel-like zinc finger protein is involved in nitrogen-fixing root nodule organogenesis. Genes Dev. 14, 475–482.
Gallardo, K., Firnhaber, C., Zuber, H., Hericher, D., Belghazi, M., Henry, C., et al. (2007). A combined proteome and transcriptome analysis of developing Medicago truncatula seeds: evidence for metabolic specialization of maternal and filial tissues. Mol. Cell. Proteomics 6, 2165–2179. doi: 10.1074/mcp.M700171-MCP200
Gallardo, K., Le Signor, C., Vandekerckhove, J., Thompson, R. D., and Burstin, J. (2003). Proteomics of Medicago truncatula seed development establishes the time frame of diverse metabolic processes related to reserve accumulation. Plant Physiol. 133, 664–682. doi: 10.1104/pp.103.025254
Gimeno-Gilles, C., Gervais, M. L., Planchet, E., Satour, P., Limami, A. M., and Lelievre, E. (2011). A stress-associated protein containing A20/AN1 zing-finger domains expressed in Medicago truncatula seeds. Plant Physiol. Biochem. 49, 303–310. doi: 10.1016/j.plaphy.2011.01.004
Gleason, C., Chaudhuri, S., Yang, T., Munoz, A., Poovaiah, B. W., and Oldroyd, G. E. (2006). Nodulation independent of rhizobia induced by a calcium-activated kinase lacking autoinhibition. Nature 441, 1149–1152. doi: 10.1038/nature04812
Godfroy, O., Debellé, F., Timmers, T., and Rosenberg, C. (2006). A rice calcium- and calmodulin-dependent protein kinase restores nodulation to a legume mutant. Mol. Plant Microbe Interact. 19, 495–501. doi: 10.1094/MPMI-19-0495
Goffard, N., and Weiller, G. (2007). PathExpress: a web-based tool to identify relevant pathways in gene expression data. Nucleic Acids Res. 35, W176–W181. doi: 10.1093/nar/gkm261
Goldberg, R. B., de Paiva, G., and Yadegari, R. (1994). Plant embryogenesis: zygote to seed. Science 266, 605–614. doi: 10.1126/science.266.5185.605
Gonzalez-Rizzo, S., Crespi, M., and Frugier, F. (2006). The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18, 2680–2693. doi: 10.1105/tpc.106.043778
Goodchild, D. J., and Bergersen, F. J. (1966). Electron microscopy of the infection and subsequent development of soybean nodule cells. J. Bacteriol. 92, 204–213.
Gruber, V., Blanchet, S., Diet, A., Zahaf, O., Boualem, A., Kakar, K., et al. (2009). Identification of transcription factors involved in root apex responses to salt stress in Medicago truncatula. Mol. Genet. Genomics 281, 55–66. doi: 10.1007/s00438-008-0392-8
Guan, D., Stacey, N., Liu, C., Mysore, J. W. K. S., Torres-Jerez, I., Vernié, T., et al. (2013). Rhizobial infection is associated with the development of peripheral vasculature in nodules of Medicago truncatula. Plant Physiol. 162, 107–116. doi: 10.1104/pp.113.215111
Handberg, K., and Stougaard, J. (1992). Lotus japonicus, an autogamous, diploid legume species for classical and molecular-genetics. Plant J. 2, 487–496. doi: 10.1111/j.1365-313X.1992.00487.x
Haney, C. H., and Long, S. R. (2010). Plant flotillins are required for infection by nitrogen-fixing bacteria. Proc. Natl. Acad. Sci. U.S.A. 107, 478–483. doi: 10.1073/pnas.0910081107
Haney, C. H., Riely, B. K., Tricoli, D. M., Cook, D. R., Ehrhardt, D. W., and Long, S. R. (2011). Symbiotic rhizobia bacteria trigger a change in localization and dynamics of the Medicago truncatula receptor kinase LYK3. Plant Cell 23, 2774–2787. doi: 10.1105/tpc.111.086389
He, J., Benedito, V. A., Wang, M., Murray, J. D., Zhao, P. X., Tang, Y., et al. (2009). The Medicago truncatula gene expression atlas web server. BMC Bioinform. 10:441. doi: 10.1186/1471-2105-10-441
Herrbach, V., Remblière, C., Gough, C., and Bensmihen, S. (2014). Lateral root formation and patterning in Medicago truncatula. J. Plant Physiol. 171, 301–310. doi: 10.1016/j.jplph.2013.09.006
Hohnjec, N., Vieweg, M. F., Pühler, A., Becker, A., and Küster, H. (2005). Overlaps in the transcriptional profiles of Medicago truncatula roots inoculated with two different Glomus fungi provide insights into the genetic program activated during arbuscular mycorrhiza. Plant Physiol. 137, 1283–1301. doi: 10.1104/pp.104.056572
Horváth, B., Domonkos, A., Kereszt, A., Szucs, A., Abraham, E., Ayaydin, F., et al. (2015). Loss of the nodule-specific cysteine rich peptide, NCR169, abolishes symbiotic nitrogen fixation in the Medicago truncatula dnf7 mutant. Proc. Natl. Acad. Sci. U.S.A. 112, 15232–15237. doi: 10.1073/pnas.1500777112
Horváth, B., Yeun, L. H., Domonkos, A., Halasz, G., Gobbato, E., Ayaydin, F., et al. (2011). Medicago truncatula IPD3 is a member of the common symbiotic signaling pathway required for rhizobial and mycorrhizal symbioses. Mol. Plant Microbe Interact. 24, 1345–1358. doi: 10.1094/MPMI-01-11-0015
Huault, E., Laffont, C., Wen, J., Mysore, K. S., Ratet, P., Duc, G., et al. (2014). Local and systemic regulation of plant root system architecture and symbiotic nodulation by a receptor-like kinase. PLoS Genet. 10:e1004891. doi: 10.1371/journal.pgen.1004891
Imin, N., Mohd-Radzman, N. A., Ogilvie, H. A., and Djordjevic, M. A. (2013). The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J. Exp. Bot. 64, 5395–5409. doi: 10.1093/jxb/ert369
Jeong, S., Trotochaud, A. E., and Clark, S. E. (1999). The Arabidopsis CLAVATA2 gene encodes a receptor-like protein required for the stability of the CLAVATA1 receptor-like kinase. Plant Cell 11, 1925–1934.
Jiang, C., Chen, C., Huang, Z., Liu, R., and Verdier, J. (2015). ITIS, a bioinformatics tool for accurate identification of transposon insertion sites using next-generation sequencing data. BMC Bioinform. 16:72. doi: 10.1186/s12859-015-0507-2
Kaló, P., Gleason, C., Edwards, A., Marsh, J., Mitra, R. M., Hirsch, S., et al. (2005). Nodulation signaling in legumes requires NSP2, a member of the GRAS family of transcriptional regulators. Science 308, 1786–1789. doi: 10.1126/science.1110951
Kang, Y., Sakiroglu, M., Krom, N., Stanton-Geddes, J., Wang, M., Lee, Y.-C., et al. (2015). Genome-wide association of drought-related and biomass traits with HapMap SNPs in Medicago truncatula. Plant. Cell Environ. 38, 1997–2011. doi: 10.1111/pce.12520
Kassaw, T., Bridges, W. Jr., and Frugoli, J. (2015). Multiple Autoregulation of Nodulation (AON) signals identified through split root analysis of Medicago truncatula sunn and rdn1 mutants. Plants (Basel). 4, 209–224. doi: 10.3390/plants4020209
Kavi Kishor, P. B., and Sreenivasulu, N. (2014). Is proline accumulation per se correlated with stress tolerance or is proline homeostasis a more critical issue? Plant Cell Environ. 37, 300–311. doi: 10.1111/pce.12157
Kim, G.-B., and Nam, Y.-W. (2013). A novel Delta(1)-pyrroline-5-carboxylate synthetase gene of Medicago truncatula plays a predominant role in stress-induced proline accumulation during symbiotic nitrogen fixation. J. Plant Physiol. 170, 291–302. doi: 10.1016/j.jplph.2012.10.004
Kim, M., Chen, Y., Xi, J., Waters, C., Chen, R., and Wang, D. (2015). An antimicrobial peptide essential for bacterial survival in the nitrogen-fixing symbiosis. Proc. Natl. Acad. Sci. U.S.A. 112, 15238–15243. doi: 10.1073/pnas.1500123112
Kiss, E., Oláh, B., Kalo, P., Morales, M., Heckmann, A. B., Borbola, A., et al. (2009). LIN, a novel type of U-box/WD40 protein, controls early infection by rhizobia in legumes. Plant Physiol. 151, 1239–1249. doi: 10.1104/pp.109.143933
Kosslak, R. M., and Bohlool, B. B. (1984). Suppression of nodule development of one side of a split-root system of soybeans caused by prior inoculation of the other side. Plant Physiol. 75, 125–130.
Kurdyukov, S., Song, Y., Sheahan, M. B., and Rose, R. J. (2014). Transcriptional regulation of early embryo development in the model legume Medicago truncatula. Plant Cell Rep. 33, 349–362. doi: 10.1007/s00299-013-1535-x
Laffont, C., Rey, T., Andre, O., Novero, M., Kazmierczak, T., Debelle, F., et al. (2015). The CRE1 cytokinin pathway is differentially recruited depending on Medicago truncatula root environments and negatively regulates resistance to a pathogen. PLoS ONE 10:e0116819. doi: 10.1371/journal.pone.0116819
Laporte, P., Lepage, A., Fournier, J., Catrice, O., Moreau, S., Jardinaud, M. F., et al. (2014). The CCAAT box-binding transcription factor NF-YA1 controls rhizobial infection. J. Exp. Bot. 65, 481–494. doi: 10.1093/jxb/ert392
Lauressergues, D., Couzigou, J.-M., Clemente, H. S., Martinez, Y., Dunand, C., Bécard, G., et al. (2015). Primary transcripts of microRNAs encode regulatory peptides. Nature, 520, 90–93. doi: 10.1038/nature14346
Lefebvre, B., Timmers, T., Mbengue, M., Moreau, S., Herve, C., Toth, K., et al. (2010). A remorin protein interacts with symbiotic receptors and regulates bacterial infection. Proc. Natl. Acad. Sci. U.S.A. 107, 2343–2348. doi: 10.1073/pnas.0913320107
Le Signor, C., Savois, V., Aubert, G., Verdier, J., Nicolas, M., Pagny, G., et al. (2009). Optimizing TILLING populations for reverse genetics in Medicago truncatula. Plant Biotechnol. J. 7, 430–441. doi: 10.1111/j.1467-7652.2009.00410.x
Li, D., Su, Z., Dong, J., and Wang, T. (2009). An expression database for roots of the model legume Medicago truncatula under salt stress. BMC Genomics 10:517. doi: 10.1186/1471-2164-10-517
Li, D., Zhang, Y., Hu, X., Shen, X., Ma, L., Su, Z., et al. (2011). Transcriptional profiling of Medicago truncatula under salt stress identified a novel CBF transcription factor MtCBF4 that plays an important role in abiotic stress responses. BMC Plant Biol. 11:109. doi: 10.1186/1471-2229-11-109
Li, J., Dai, X., Liu, T., and Zhao, P. X. (2012). LegumeIP: an integrative database for comparative genomics and transcriptomics of model legumes. Nucleic Acids Res. 40, D1221–D1229. doi: 10.1093/nar/gkr939
Limpens, E., Franken, C., Smit, P., Willemse, J., Bisseling, T., and Geurts, R. (2003). LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 302, 630–633. doi: 10.1126/science.1090074
Lippuner, V., Cyert, M. S., and Gasser, C. S. (1996). Two classes of plant cDNA clones differentially complement yeast calcineurin mutants and increase salt tolerance of wild-type yeast. J. Biol. Chem. 271, 12859–12866. doi: 10.1074/jbc.271.22.12859
Liu, C., Jun, J. H., and Dixon, R. A. (2014). MYB5 and MYB14 play pivotal roles in seed coat polymer biosynthesis in Medicago truncatula. Plant Physiol. 165, 1424–1439. doi: 10.1104/pp.114.241877
Long, R. C., Li, M. N., Kang, J. M., Zhang, T. J., Sun, Y., and Yang, Q. C. (2015). Small RNA deep sequencing identifies novel and salt−stress−regulated microRNAs from roots of Medicago sativa and Medicago truncatula. Physiol. Plant. 154, 13–27. doi: 10.1111/ppl.12266
Madsen, E. B., Madsen, L. H., Radutoiu, S., Olbryt, M., Rakwalska, M., Szczyglowski, K., et al. (2003). A receptor kinase gene of the LysM type is involved in legume perception of rhizobial signals. Nature 425, 637–640. doi: 10.1038/nature02045
Mantiri, F. R., Kurdyukov, S., Chen, S.-K., and Rose, R. J. (2008). The transcription factor MtSERF1 may function as a nexus between stress and development in somatic embryogenesis in Medicago truncatula. Plant Signal. Behav. 3, 498–500. doi: 10.4161/psb.3.7.6049
Marsh, J. F., Rakocevic, A., Mitra, R. M., Brocard, L., Sun, J., Eschstruth, A., et al. (2007). Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 144, 324–335. doi: 10.1104/pp.106.093021
Mathesius, U., Keijzers, G., Natera, S. H., Weinman, J. J., Djordjevic, M. A., and Rolfe, B. G. (2001). Establishment of a root proteome reference map for the model legume Medicago truncatula using the expressed sequence tag database for peptide mass fingerprinting. Proteomics 1, 1424–1440. doi: 10.1002/1615-9861(200111)1:11<1424::AID-PROT1424>3.0.CO;2-J
Maunoury, N., Redondo-Nieto, M., Bourcy, M., Van de Velde, W., Alunni, B., Laporte, P., et al. (2010). Differentiation of symbiotic cells and endosymbionts in Medicago truncatula nodulation are coupled to two transcriptome-switches. PLoS ONE 5:e9519. doi: 10.1371/journal.pone.0009519
Mbengue, M., Camut, S., de Carvalho-Niebel, F., Deslandes, L., Froidure, S., Klaus-Heisen, D., et al. (2010). The Medicago truncatula E3 ubiquitin ligase PUB1 interacts with the LYK3 symbiotic receptor and negatively regulates infection and nodulation. Plant Cell 22, 3474–3488. doi: 10.1105/tpc.110.075861
Merchan, F., Breda, C., Hormaeche, J. P., Sousa, C., Kondorosi, A., Aguilar, O. M., et al. (2003). A Kruppel-like transcription factor gene is involved in salt stress responses in Medicago spp. Plant Soil 257, 1–9. doi: 10.1023/A:1026256415556
Merchan, F., de Lorenzo, L., Gonzalez Rizzo, S., Niebel, A., Manyani, H., Frugier, F., et al. (2007). Identification of regulatory pathways involved in the reacquisition of root growth after salt stress in Medicago truncatula. Plant J. 51, 1–17. doi: 10.1111/j.1365-313X.2007.03117.x
Mergaert, P., Nikovics, K., Kelemen, Z., Maunoury, N., Vaubert, D., Kondorosi, A., et al. (2003). A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 132, 161–173. doi: 10.1104/pp.102.018192
Middleton, P. H., Jakab, J., Penmetsa, R. V., Starker, C. G., Doll, J., Kalo, P., et al. (2007). An ERF transcription factor in Medicago truncatula that is essential for Nod factor signal transduction. Plant Cell 19, 1221–1234. doi: 10.1105/tpc.106.048264
Miller, J. B., Pratap, A., Miyahara, A., Zhou, L., Bornemann, S., Morris, R. J., et al. (2013). Calcium/Calmodulin-dependent protein kinase is negatively and positively regulated by calcium, providing a mechanism for decoding calcium responses during symbiosis signaling. Plant Cell 25, 5053–5066. doi: 10.1105/tpc.113.116921
Miyahara, A., Richens, J., Starker, C., Morieri, G., Smith, L., Long, S., et al. (2010). Conservation in function of a SCAR/WAVE component during infection thread and root hair growth in Medicago truncatula. Mol. Plant Microbe Interact. 23, 1553–1562. doi: 10.1094/MPMI-06-10-0144
Mohd-Radzman, N. A., Laffont, C., Ivanovici, A., Patel, N., Reid, D., Stougaard, J., et al. (2016). Different pathways act downstream of the peptide receptor CRA2 to regulate lateral root and nodule development. Plant Physiol. 171, 2536–2548. doi: 10.1104/pp.16.00113
Mortier, V., Den Herder, G., Whitford, R., Van de Velde, W., Rombauts, S., D'Haeseleer, K., et al. (2010). CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol. 153, 222–237. doi: 10.1104/pp.110.153718
Munier-Jolain, N. G., and Ney, B. (1998). Seed growth rate in grain legumes II. Seed growth rate depends on cotyledon cell number. J. Exp. Bot. 49, 1971–1976. doi: 10.1093/jxb/49.329.1971
Muralla, R., Lloyd, J., and Meinke, D. (2011). Molecular foundations of reproductive lethality in Arabidopsis thaliana. PLoS ONE 6:e28398. doi: 10.1371/journal.pone.0028398
Murray, J. D., Muni, R. R., Torres-Jerez, I., Tang, Y., Allen, S., Andriankaja, M., et al. (2010). Vapyrin, a gene essential for intracellular progression of arbuscular mycorrhizal symbiosis, is also essential for infection by rhizobia in the nodule symbiosis of Medicago truncatula. Plant J. 65, 244–252. doi: 10.1111/j.1365-313X.2010.04415.x
Nguyen, M., Kim, G.-B., Hyun, S.-H., Lee, S.-Y., Lee, C.-Y., Choi, H.-K., et al. (2013). Physiological and metabolomic analysis of a knockout mutant suggests a critical role of MtP5CS3 gene in osmotic stress tolerance of Medicago truncatula. Euphytica 193, 101–120. doi: 10.1007/s10681-013-0957-4
Noguero, M., Le Signor, C., Vernoud, V., Bandyopadhyay, K., Sanchez, M., Fu, C., et al. (2015). DASH transcription factor impacts Medicago truncatula seed size by its action on embryo morphogenesis and auxin homeostasis. Plant J. 81, 453–466. doi: 10.1111/tpj.12742
Ogawa-Ohnishi, M., Matsushita, W., and Matsubayashi, Y. (2013). Identification of three hydroxyproline O-arabinosyltransferases in Arabidopsis thaliana. Nat Chem Biol. 9, 726–730. doi: 10.1038/nchembio.1351
Okamoto, S., Ohnishi, E., Sato, S., Takahashi, H., Nakazono, M., Tabata, S., et al. (2009). Nod factor/nitrate-induced CLE genes that drive HAR1-mediated systemic regulation of nodulation. Plant Cell Physiol. 50, 67–77. doi: 10.1093/pcp/pcn194
Oldroyd, G. E. (2013). Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat. Rev. Microbiol. 11, 252–263. doi: 10.1038/nrmicro2990
Oldroyd, G. E., and Long, S. R. (2003). Identification and characterization of nodulation-signaling pathway 2, a gene of Medicago truncatula involved in Nod actor signaling. Plant Physiol. 131, 1027–1032. doi: 10.1104/pp.102.010710
Oono, R., and Denison, R. F. (2010). Comparing symbiotic efficiency between swollen versus nonswollen rhizobial bacteroids. Plant Physiol. 154, 1541–1548. doi: 10.1104/pp.110.163436
Op den Camp, R. H., De Mita, S., Lillo, A., Cao, Q., Limpens, E., Bisseling, T., et al. (2011). A phylogenetic strategy based on a legume-specific whole genome duplication yields symbiotic cytokinin type-A response regulators. Plant Physiol. 157, 2013–2022. doi: 10.1104/pp.111.187526
Pang, Y., Cheng, X., Huhman, D., Ma, J., Peel, G., Yonekura-Sakakibara, K., et al. (2013). Medicago glucosyltransferase UGT72L1: potential roles in proanthocyanidin biosynthesis. Planta 238, 139–154. doi: 10.1007/s00425-013-1879-z
Pang, Y., Wenger, J. P., Saathoff, K., Peel, G. J., Wen, J., Huhman, D., et al. (2009). A WD40 repeat protein from Medicago truncatula is necessary for tissue-specific anthocyanin and proanthocyanidin biosynthesis but not for trichome development. Plant Physiol. 151, 1114–1129. doi: 10.1104/pp.109.144022
Peel, G. J., Pang, Y., Modolo, L. V., and Dixon, R. A. (2009). The LAP1 MYB transcription factor orchestrates anthocyanidin biosynthesis and glycosylation in Medicago. Plant J. 59, 136–149. doi: 10.1111/j.1365-313X.2009.03885.x
Peiter, E., Sun, J., Heckmann, A. B., Venkateshwaran, M., Riely, B. K., Otegui, M. S., et al. (2007). The Medicago truncatula DMI1 protein modulates cytosolic calcium signaling. Plant Physiol. 145, 192–203. doi: 10.1104/pp.107.097261
Penmetsa, R. V., and Cook, D. R. (2000). Production and characterization of diverse developmental mutants of Medicago truncatula. Plant Physiol. 123, 1387–1398. doi: 10.1104/pp.123.4.1387
Penmetsa, R. V., Frugoli, J. A., Smith, L. S., Long, S. R., and Cook, D. R. (2003). Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol. 131, 998–1008. doi: 10.1104/pp.015677
Penmetsa, R. V., Uribe, P., Anderson, J., Lichtenzveig, J., Gish, J. C., Nam, Y. W., et al. (2008). The Medicago truncatula ortholog of Arabidopsis EIN2, sickle, is a negative regulator of symbiotic and pathogenic microbial associations. Plant J. 55, 580–595. doi: 10.1111/j.1365-313X.2008.03531
Porceddu, A., Panara, F., Calderini, O., Molinari, L., Taviani, P., Lanfaloni, L., et al. (2008). An Italian functional genomic resource for Medicago truncatula. BMC Res. Notes 1:129. doi: 10.1186/1756-0500-1-129
Puckette, M. C., Tang, Y., and Mahalingam, R. (2008). Transcriptomic changes induced by acute ozone in resistant and sensitive Medicago truncatula accessions. BMC Plant Biol. 8:46. doi: 10.1186/1471-2229-8-46
Radutoiu, S., Madsen, L. H., Madsen, E. B., Felle, H. H., Umehara, Y., Gronlund, M., et al. (2003). Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 425, 585–592. doi: 10.1038/nature02039
Rakocevic, A., Mondy, S., Tirichine, L., Cosson, V., Brocard, L., Iantcheva, A., et al. (2009). MERE1, a low-copy-number copia-type retroelement in Medicago truncatula active during tissue culture. Plant Physiol. 151, 1250–1263. doi: 10.1104/pp.109.138024
Ricoult, C., Cliquet, J. B., and Limami, A. M. (2005). Stimulation of alanine amino transferase (AlaAT) gene expression and alanine accumulation in embryo axis of the model legume Medicago truncatula contribute to anoxia stress tolerance. Physiol. Plant. 123, 30–39. doi: 10.1111/j.1399-3054.2005.00449.x
Ricoult, C., Echeverria, L. O., Cliquet, J.-B., and Limami, A. M. (2006). Characterization of alanine aminotransferase (AlaAT) multigene family and hypoxic response in young seedlings of the model legume Medicago truncatula. J. Exp. Bot. 57, 3079–3089. doi: 10.1093/jxb/erl069
Robertson, J. G., and Lyttleton, P. (1984). Division of peribacteroid membranes in root nodules of white clover. J. Cell Sci. 69, 147–157.
Rogers, C., Wen, J., Chen, R., and Oldroyd, G. (2009). Deletion-based reverse genetics in Medicago truncatula. Plant Physiol. 151, 1077–1086. doi: 10.1104/pp.109.142919
Rosnoblet, C., Aubry, C., Leprince, O., Vu, B. L., Rogniaux, H., and Buitink, J. (2007). The regulatory gamma subunit SNF4b of the sucrose non-fermenting-related kinase complex is involved in longevity and stachyose accumulation during maturation of Medicago truncatula seeds. Plant J. 51, 47–59. doi: 10.1111/j.1365-313X.2007.03116.x
Roth, L. E., and Stacey, G. (1989). Bacterium release into host cells of nitrogen-fixing soybean nodules: the symbiosome membrane comes from three sources. Eur. J. Cell Biol. 49, 13–23.
Sagan, M., Morandi, D., Tarenghi, E., and Duc, G. (1995). Selection of nodulation and mycorrhizal mutants in the model plant Medicago truncatula (Gaertn.) after gamma-ray mutagenesis. Plant Sci. 111, 63–71. doi: 10.1016/0168-9452(95)04229-N
Saha, S., and DasGupta, M. (2015). Does SUNN-SYMRK Crosstalk occur in Medicago truncatula for regulating nodule organogenesis? Plant Signal Behav. 10:e1028703. doi: 10.1080/15592324.2015.1028703
Saha, S., Dutta, A., Bhattacharya, A., and DasGupta, M. (2014). Intracellular catalytic domain of symbiosis receptor kinase hyperactivates spontaneous nodulation in absence of rhizobia. Plant Physiol. 166, 1699–1708. doi: 10.1104/pp.114.250084
Schnabel, E., Journet, E. P., de Carvalho-Niebel, F., Duc, G., and Frugoli, J. (2005). The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol. Biol. 58, 809–822. doi: 10.1007/s11103-005-8102-y
Schnabel, E. L., Kassaw, T. K., Smith, L. S., Marsh, J. F., Oldroyd, G. E., Long, S. R., et al. (2011). The ROOT DETERMINED NODULATION1 gene regulates nodule number in roots of Medicago truncatula and defines a highly conserved, uncharacterized plant gene family. Plant Physiol. 157, 328–340. doi: 10.1104/pp.111.178756
Schnabel, E., Mukherjee, A., Smith, L., Kassaw, T., Long, S., and Frugoli, J. (2010). The lss supernodulation mutant of Medicago truncatula reduces expression of the SUNN gene. Plant Physiol. 154, 1390–1402. doi: 10.1104/pp.110.164889
Singh, S., Katzer, K., Lambert, J., Cerri, M., and Parniske, M. (2014). CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15, 139–152. doi: 10.1016/j.chom.2014.01.011
Sinharoy, S., Liu, C., Breakspear, A., Guan, D., Shailes, S., Nakashima, J., et al. (2016). A Medicago truncatula Cystathionine-β-synthase-like domain-containing protein is required for rhizobial infection and symbiotic nitrogen fixation. Plant Physiol. 170, 2204–2217. doi: 10.1104/pp.15.01853
Sinharoy, S., Torres-Jerez, I., Bandyopadhyay, K., Kereszt, A., Pislariu, C. I., Nakashima, J., et al. (2013). The C2H2 transcription factor regulator of symbiosome differentiation represses transcription of the secretory pathway gene VAMP721a and promotes symbiosome development in Medicago truncatula. Plant Cell 25, 3584–3601. doi: 10.1105/tpc.113.114017
Smit, P., Limpens, E., Geurts, R., Fedorova, E., Dolgikh, E., Gough, C., et al. (2007). Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 145, 183–191. doi: 10.1104/pp.107.100495
Smit, P., Raedts, J., Portyanko, V., Debelle, F., Gough, C., Bisseling, T., et al. (2005). NSP1 of the GRAS protein family is essential for rhizobial Nod factor-induced transcription. Science 308, 1789–1791. doi: 10.1126/science.1111025
Song, S., Chen, Y., Zhao, M., and Zhang, W.-H. (2012). A novel Medicago truncatula HD-Zip gene, MtHB2, is involved in abiotic stress responses. Environ. Exp. Bot. 80, 1–9. doi: 10.1016/j.envexpbot.2012.02.001
Soyano, T., Hirakawa, H., Sato, S., Hayashi, M., and Kawaguchi, M. (2014). Nodule inception creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc Natl Acad Sci U.S.A. 111, 14607–14612. doi: 10.1073/pnas.1412716111
Soyano, T., Kouchi, H., Hirota, A., and Hayashi, M. (2013). NODULE INCEPTION Directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 9:e1003352. doi: 10.1371/journal.pgen.1003352
Stanton-Geddes, J., Paape, T., Epstein, B., Briskine, R., Yoder, J., Mudge, J., et al. (2013). Candidate genes and genetic architecture of symbiotic and agronomic traits revealed by whole-genome, sequence-based association genetics in Medicago truncatula. PLoS ONE 8:e65688. doi: 10.1371/journal.pone.0065688
Starker, C. G., Parra-Colmenares, A. L., Smith, L., Mitra, R. M., and Long, S. R. (2006). Nitrogen fixation mutants of Medicago truncatula fail to support plant and bacterial symbiotic gene expression. Plant Physiol. 140, 671–680. doi: 10.1104/pp.105.072132
Tadege, M., Ratet, P., and Mysore, K. S. (2005). Insertional mutagenesis: a Swiss Army knife for functional genomics of Medicago truncatula. Trends Plant Sci. 10, 229–235. doi: 10.1016/j.tplants.2005.03.009
Terrasson, E., Buitink, J., Righetti, K., Ly Vu, B., Pelletier, S., Zinsmeister, J., et al. (2013). An emerging picture of the seed desiccome: confirmed regulators and newcomers identified using transcriptome comparison. Front. Plant Sci. 4:497. doi: 10.3389/fpls.2013.00497
Thomas, M. R., Rose, R. J., and Nolan, K. E. (1992). Genetic transformation of Medicago truncatula using Agrobacterium with genetically modified Ri and disarmed Ti plasmids. Plant Cell Rep. 11, 113–117. doi: 10.1007/BF00232161
Till, B. J., Burtner, C., Comai, L., and Henikoff, S. (2004). Mismatch cleavage by single-strand specific nucleases. Nucleic Acids Res. 32, 2632–2641. doi: 10.1093/nar/gkh599
Tirichine, L., Imaizumi-Anraku, H., Yoshida, S., Murakami, Y., Madsen, L. H., Miwa, H., et al. (2006). Deregulation of a Ca2+/calmodulin-dependent kinase leads to spontaneous nodule development. Nature 441, 1153–1156. doi: 10.1038/nature04862
Tirichine, L., Sandal, N., Madsen, L. H., Radutoiu, S., Albrektsen, A. S., Sato, S., et al. (2007). A gain-of-function mutation in a cytokinin receptor triggers spontaneous root nodule organogenesis. Science 315, 104–107. doi: 10.1126/science.1132397
Trieu, A. T., Burleigh, S. H., Kardailsky, I. V., Maldonado-Mendoza, I. E, Versaw, W. K, Blaylock, L. A., et al. (2000). Transformation of Medicago truncatula via infiltration of seedlings or flowering plants with Agrobacterium. Plant J. 22, 531–541. doi: 10.1046/j.1365-313x.2000.00757.x
Udvardi, M. K., and Day, D. A. (1997). Metabolite transport across symbiotic membranes of legume nodules. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 493–523. doi: 10.1146/annurev.arplant.48.1.493
Udvardi, M., and Poole, P. S. (2013). Transport and metabolism in legume-rhizobia symbioses. Annu. Rev. Plant Biol. 64, 781–805. doi: 10.1146/annurev-arplant-050312-120235
Van de Velde, W., Zehirov, G., Szatmari, A., Debreczeny, M., Ishihara, H., Kevei, Z., et al. (2010). Plant peptides govern terminal differentiation of bacteria in symbiosis. Science 327, 1122–1126. doi: 10.1126/science.1184057
van Noorden, G. E., Ross, J. J., Reid, J. B., Rolfe, B. G., and Mathesius, U. (2006). Defective long-distance auxin transport regulation in the Medicago truncatula super numeric nodules mutant. Plant Physiol. 140, 1494–1506. doi: 10.1104/pp.105.075879
Verdier, J., Dessaint, F., Schneider, C., and Abirached-Darmency, M. (2013a). A combined histology and transcriptome analysis unravels novel questions on Medicago truncatula seed coat. J. Exp. Bot. 64, 459–470. doi: 10.1093/jxb/ers304
Verdier, J., Kakar, K., Gallardo, K., Le Signor, C., Aubert, G., Schlereth, A., et al. (2008). Gene expression profiling of M-truncatula transcription factors identifies putative regulators of grain legume seed filling. Plant Mol. Biol. 67, 567–580. doi: 10.1007/s11103-008-9320-x
Verdier, J., Lalanne, D., Pelletier, S., Torres-Jerez, I., Righetti, K., Bandyopadhyay, K., et al. (2013b). A regulatory network based approach dissects late maturation processes related to the acquisition of desiccation tolerance and longevity of Medicago truncatula seeds. Plant Physiol. 163, 757–774. doi: 10.1104/pp.113.222380
Verdier, J., Zhao, J., Torres-Jerez, I., Ge, S., Liu, C., He, X., et al. (2012). MtPAR MYB transcription factor acts as an on switch for proanthocyanidin biosynthesis in Medicago truncatula. Proc Natl Acad Sci U.S.A. 109, 1766–1771. doi: 10.1073/pnas.1120916109
Verdoy, D., De la Pena, T. C., Redondo, F. J., Lucas, M. M., and Pueyo, J. J. (2006). Transgenic Medicago truncatula plants that accumulate proline display nitrogen-fixing activity with enhanced tolerance to osmotic stress. Plant Cell Environ. 29, 1913–1923. doi: 10.1111/j.1365-3040.2006.01567.x
Vernié, T., Camut, S., Camps, C., Rembliere, C., de Carvalho-Niebel, F., Mbengue, M., et al. (2016). PUB1 interacts with the receptor kinase DMI2 and negatively regulates rhizobial and arbuscular mycorrhizal symbioses through its ubiquitination activity in Medicago truncatula. Plant Physiol. 170, 2312–2324. doi: 10.1104/pp.15.01694
Vernié, T., Kim, J., Frances, L., Ding, Y., Sun, J., Guan, D., et al. (2015). The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell 27, 3410–3424. doi: 10.1105/tpc.15.00461
Vernie, T., Moreau, S., de Billy, F., Plet, J., Combier, J. P., Rogers, C., et al. (2008). EFD Is an ERF transcription factor involved in the control of nodule number and differentiation in Medicago truncatula. Plant Cell 20, 2696–2713. doi: 10.1105/tpc.108.059857
Wang, T., Chen, L., Zhao, M., Tian, Q., and Zhang, W.-H. (2011). Identification of drought-responsive microRNAs in Medicago truncatula by genome-wide high-throughput sequencing. BMC Genomics 12:367. doi: 10.1186/1471-2164-12-367
Wang, C., Yu, H., Luo, L., Duan, L., Cai, L., He, X., et al. (2016). NODULES WITH ACTIVATED DEFENSE 1 is required for maintenance of rhizobial endosymbiosis in Medicago truncatula. New Phytol. doi: 10.1111/nph.14017. [Epub ahead of print].
Wang, D., Griffitts, J., Starker, C., Fedorova, E., Limpens, E., Ivanov, S., et al. (2010). A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science 327, 1126–1129. doi: 10.1126/science.1184096
Wang, M., Verdier, J., Benedito, V., a, Tang, Y., Murray, J. D., Ge, Y., et al. (2013). LegumeGRN: a gene regulatory network prediction server for functional and comparative studies. PLoS ONE 8:e67434. doi: 10.1371/journal.pone.0067434
Wang, T.-Z., Zhang, J.-L., Tian, Q.-Y., Zhao, M.-G., and Zhang, W.-H. (2013). A Medicago truncatula EF-Hand family gene, MtCaMP1, is involved in drought and salt stress tolerance. Plos ONE 8:58952. doi: 10.1371/journal.pone.0058952
Watson, B. S., Asirvatham, V. S., Wang, L., and Sumner, L. W. (2003). Mapping the proteome of barrel medic (Medicago truncatula). Plant Physiol. 131, 1104–1123. doi: 10.1104/pp.102.019034
Xiao, T. T., Schilderink, S., Moling, S., Deinum, E. E., Kondorosi, E., Franssen, H., et al. (2014). Fate map of Medicago truncatula root nodules. Development 141, 3517–3528. doi: 10.1242/dev.110775
Yoro, E., Suzaki, T., Toyokura, K., Miyazawa, H., Fukaki, H., and Kawaguchi, M. (2014). A positive regulator of nodule organogenesis, NODULE INCEPTION, acts as a negative regulator of rhizobial infection in Lotus japonicus. Plant Physiol. 165, 747–758. doi: 10.1104/pp.113.233379
Young, N. D., Debellé, F., Oldroyd, G. E. D., Geurts, R., Cannon, S. B., Udvardi, M. K., et al. (2011). The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature 480, 520–524. doi: 10.1038/nature10625
Zhang, J. Y., Broeckling, C. D., Blancaflor, E. B., Sledge, M. K., Sumner, L. W., and Wang, Z. Y. (2005). Overexpression of WXP1, a putative Medicago truncatula AP2 domain-containing transcription factor gene, increases cuticular wax accumulation and enhances drought tolerance in transgenic alfalfa (Medicago sativa). Plant J. 42, 689–707. doi: 10.1111/j.1365-313X.2005.02405.x
Zhang, J.-Y., Broeckling, C. D., Sumner, L. W., and Wang, Z.-Y. (2007). Heterologous expression of two Medicago truncatula putative ERF transcription factor genes, WXP1 and WXP2, in Arabidopsis led to increased leaf wax accumulation and improved drought tolerance, but differential response in freezing tolerance. Plant Mol. Biol. 64, 265–278. doi: 10.1007/s11103-007-9150-2
Zhang, J. Y., Cruz DE Carvalho, M. H., Torres-Jerez, I., Kang, Y., Allen, S. N., Huhman, D. V., et al. (2014). Global reprogramming of transcription and metabolism in Medicago truncatula during progressive drought and after rewatering. Plant. Cell Environ. 37, 2553–2576. doi: 10.1111/pce.12328
Zhao, J., and Dixon, R. A. (2009). MATE Transporters Facilitate Vacuolar Uptake of Epicatechin 39-O-Glucoside for Proanthocyanidin Biosynthesis in Medicago truncatula and Arabidopsis. Plant Cell 21, 2323–2340. doi: 10.1105/tpc.109.067819
Zhao, J., Huhman, D., Shadle, G., He, X.-Z., Sumner, L. W., Tang, Y., et al. (2011). MATE2 mediates vacuolar sequestration of flavonoid glycosides and glycoside malonates in Medicago truncatula. Plant Cell 23, 1536–1555. doi: 10.1105/tpc.110.080804
Keywords: Medicago truncatula, functional genomics, seed, symbiosis, abiotic stresses
Citation: Kang Y, Li M, Sinharoy S and Verdier J (2016) A Snapshot of Functional Genetic Studies in Medicago truncatula. Front. Plant Sci. 7:1175. doi: 10.3389/fpls.2016.01175
Received: 08 May 2016; Accepted: 21 July 2016;
Published: 09 August 2016.
Edited by:
Georgina Hernandez, National Autonomous University of Mexico, MexicoReviewed by:
Ulrike Mathesius, Australian National University, AustraliaCopyright © 2016 Kang, Li, Sinharoy and Verdier. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jerome Verdier, amF2ZXJkaWVyQGljbG91ZC5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.