- Key Laboratory of Sivilculture in Shandong Province, College of Forestry, Shandong Agriculture University, Tai'an, China
In plants, sex determination is a comprehensive process of correlated events, which involves genes that are differentially and/or specifically expressed in distinct developmental phases. Exploring gene expression profiles from different sex types will contribute to fully understanding sex determination in plants. In this study, we conducted RNA-sequencing of female and male buds (FB and MB) as well as ovulate strobilus and staminate strobilus (OS and SS) of Ginkgo biloba to gain insights into the genes potentially related to sex determination in this species. Approximately 60 Gb of clean reads were obtained from eight cDNA libraries. De novo assembly of the clean reads generated 108,307 unigenes with an average length of 796 bp. Among these unigenes, 51,953 (47.97%) had at least one significant match with a gene sequence in the public databases searched. A total of 4709 and 9802 differentially expressed genes (DEGs) were identified in MB vs. FB and SS vs. OS, respectively. Genes involved in plant hormone signal and transduction as well as those encoding DNA methyltransferase were found to be differentially expressed between different sex types. Their potential roles in sex determination of G. biloba were discussed. Pistil-related genes were expressed in male buds while anther-specific genes were identified in female buds, suggesting that dioecism in G. biloba was resulted from the selective arrest of reproductive primordia. High correlation of expression level was found between the RNA-Seq and quantitative real-time PCR results. The transcriptome resources that we generated allowed us to characterize gene expression profiles and examine differential expression profiles, which provided foundations for identifying functional genes associated with sex determination in G. biloba.
Introduction
Sexual phenotypes in plants are exceedingly diverse and understanding the mechanisms underlying sex determination will require data from ecology, developmental biology, and genetics (Ming et al., 2011). Hermaphroditic or monoecious species make up 94% of all flowering plants, while dioecious species make up only 4% (Renner and Ricklefs, 1995). Dioecious species are ideal models for studying sex determination. Species such as Silene latifolia have been extensively investigated and several sex-linked genes have been identified and isolated (Matsunaga et al., 1996; Guttman and Charlesworth, 1998; Moore et al., 2003; Wu et al., 2010). Nevertheless, sex determination in plants is a complex process that involves various genes that are differentially and/or specifically expressed in different developmental phases, the identification and characterization of only a scant handful of involved genes will not provide a full understanding of the mechanism of sex determination (Charlesworth, 2002; Charlesworth and Mank, 2010). Previously used transcriptome techniques, such as expressed sequence tags (ESTs) and microarray analysis (Irizarry et al., 2005; Terefe and Tatlioglu, 2005; Akao et al., 2007; Andersen et al., 2008), suffered from a limited depth of coverage and sensitivity as well as background or cross-hybridization problems that restricted their applications in fully elucidating the functional complexity of plant sex determination. Furthermore, it is more difficult to identify candidate genes in non-model plant species that the total genome sequences are unknown. The introduction of high-throughput RNA-sequencing (RNA-Seq) technology for transcriptome analysis has provided a more powerful and cost-efficient approach (Wang et al., 2009, 2010). RNA-Seq has been used extensively and successfully in transcriptome analyses of willow (Liu et al., 2013), wheat (Yang et al., 2015) and cucumber (Wu et al., 2010) to greatly accelerate the understanding of the complexity of gene expression, regulation, and networks in plant sex determination.
Ginkgo biloba L. (also referred as the “golden living fossil”) is a typical dioecious gymnosperm species with great economic and ecological values (Zhang et al., 2015). Like other gymnosperms such as species of Cycadopsida and Gnetales, the reproductive organ of male ginkgo individual is staminate strobilus (male cone) while female one bears ovulate strobilus (female cone) (Wu, 1999) (Figures 1A,B). The morphological traits, growth habits, and physiological characteristics are significantly different between mature female and male individuals, which directly influence the practical uses of this plant (Huang et al., 2013). Male individuals are mainly used in ornamental horticulture and wood industry while female ones are used for seed production. These differences are directly derived from sex determination and the following sex differentiation in G. biloba, however, sex determination has not yet been explored in this species. Furthermore, to our knowledge, previous and recent genomic studies on G. biloba mainly focused on genes involved in ginkgolides and/or bilobalide biosynthesis (Han et al., 2015; He et al., 2015), no genome-wide analyses to systematically characterize gene expression levels and to compare differentially expressed genes between male and female reproductive organs has been conducted so far. Such information is essential to understand the mechanism of sex determination in this species. Considering that sex determination involves genes expressed in different developmental stages, in the present study we used RNA-Seq to investigate and compare the transcriptomes of buds (Figures 1C,D) and strobilus of G. biloba. Genes related to phytohormone signal and transduction, DNA methylation were found to be differentially expressed between male and female organs. Our results provided valuable transcriptome resources for a non-model gymnosperm species and may help to reveal clues for further investigations on plant sex determination.
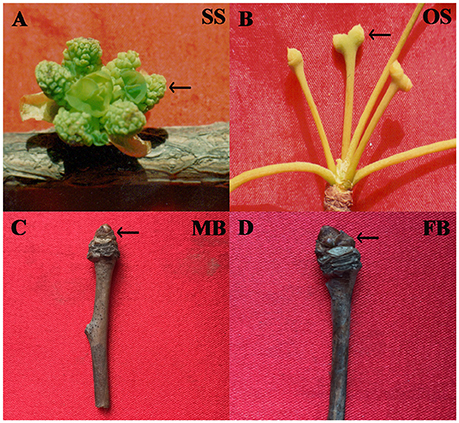
Figure 1. Samples used in this study. (A) Staminate strobilus (SS); (B) Ovulate strobilus (OS); (C) male bud (MB); (D) female bud (FB).
Materials and Methods
Plant Materials and RNA Extraction
The G. biloba plants used in this study were planted in the Forestry Experimental Field of Shandong Agriculture University located in Taian, Shandong Province, China (35°38′−36°28′ N, 116°02′−117°59′ E). Five male trees and five female ones of 25-year-old from the same family were chosen to exclude stochastic error caused by different genetic background. For female and male buds as well as ovulate strobilus and staminate strobilus, two samples were collected from each tree on 4th and 28th March 2015, respectively, based on the development stages categorized previously (Shi et al., 1998; Zhang W. P. et al., 2001). For the buds, the bud bracts were removed immediately after sampling. All the materials were frozen in liquid nitrogen and stored at −80°C till used.
For each group of materials, five samples from different trees of the same gender were pooled to form one biological sample [two biological replicates were performed for each group as in other studies (Frey et al., 2015; Yang et al., 2015)] for total RNA extraction using TRIZOL reagent according to the manufacturer's instructions (Invitrogen, USA). The quality and quantity of the total RNA were assessed using 1% agarose gels and a NanoPhotometer® spectrophotometer (Implen, CA, USA). Subsequently, genomic DNA was digested by treatment with DNase I and mRNA was isolated using Dynabeads Oligo (dT) (Invitrogen, USA). Then, the concentration and integrity of the mRNA were quantified using a Qubit® RNA Assay Kit in Qubit 2.0 Fluorometer (Life Technologies, CA, USA) and RNA Nano 6000 Assay Kit in an Agilent Bioanalyzer 2100 system (Agilent Technologies, CA, USA).
Libraries Construction, Deep Sequencing, and De novo Assembly
Eight transcriptome sequencing libraries were generated using a NEB Next® Ultra™ RNA Library Prep Kit for Illumina® (NEB, USA) following the manufacturer's recommendations. The libraries were sequenced on an Illumina HiSeq 2500 platform. To obtain high-quality clean reads for assembly, the raw reads were filtered through in-house perl scripts by removing adaptor sequences, reads containing poly-N sequences, and low quality reads. All the downstream analyses were conducted using the clean read sequences. All the clean reads were pooled and assembled using the Trinity de novo assembly program (Grabherr et al., 2011) with the minimum kmer_cov set to 2 as the default, and all other parameters set to their default values (He et al., 2015).
Functional Annotation
The assembled unigenes were searched against the Nr and Nt databases, and the Swiss-Prot protein and COG/KOG databases using BLAST with an cutoff E-value of 1e−5. To assign functional annotations, the unigenes were searched against Pfam using HMMER 3.0 (Finn et al., 2011) with E-value of 1e−2, the KEGG database using KAAS (KEGG Automatic Annotation Server) (Moriya et al., 2007) with E-value of 1e−10, and the GO database using Blast2GO (Gotz, 2008) with an E-value of 1e−6.
Identification of Differentially Expressed Genes (DEGs)
For all the comparisons, read counts were normalized by calculating the FPKM value (Trapnell et al., 2010) to obtain relative expression levels. An FPKM value >0.3 was defined as the threshold of significant gene expression. Differential expression analysis was performed using the DESeq R package 1.10.1 (Anders, 2010), which provides statistical routines for determining differential expression in digital gene expression data using a model based on the negative binomial distribution. The resulting P-values were adjusted using the Benjamin and Hochberg's approach for controlling the false discovery rate (Storey and Tibshirani, 2003). Unigenes with an adjusted P < 0.05 determined by DESeq were assigned as DEGs. GO enrichment analysis of the DEGs was performed using the GOseq R packages based on the Wallenius' non-central hypergeometric distribution (Young et al., 2010), which can adjust for gene length bias in DEGs. We also used the KOBAS software (Mao et al., 2005) to test the statistical enrichment of DEGs in the KEGG pathways.
qRT-PCR Validation
Based on the classification of genes and/or pathways revealed in this study (see below), we selected 26 genes from different categories generated by RNA-Seq for validation. We designed specific primers that corresponded to the conserved region of each cDNA in the sequenced database (Table S1). Real-time assays were performed with SYBR Green Dye (Takara, Dalian, China) using a Bio-Rad CFX96 real-time PCR platform (BioRad Laboratories, Hercules, CA, USA) with the following cycle conditions: 95°C for 5 min, followed by 45 cycles of 95°C for 10 s, 60°C for 10 s, and 72°C for 20 s. Three biological replicates were used for each gene. RNA transcript fold changes were calculated using the 2−ΔΔCt method (Livak and Schmittgen, 2001) with GAPDH as the internal control (Wang et al., 2012).
Results
Sequencing Analysis and De novo Assembly
Eight cDNA libraries were constructed using female and male buds (FB and MB) as well as ovulate strobilus and staminate strobilus (OS and SS) of 25 year-old G. biloba trees. The libraries were sequenced on an Illumina Hiseq 2500 platform. The sequences of the raw reads have been deposited in NCBI Sequence Read Archive (SRA) under accession numbers SRR2147720 (SS), SRR2147715 (OS), SRR2147717 (FB), and SRR2147721 (MB). After the raw reads were filtered, 119,494,172 (MB), 115,958,434 (FB), 121,730,470 (SS), and 116,504,860 (OS) clean reads were obtained (Table S2). All the clean reads were pooled together and assembled de novo using Trinity (Grabherr et al., 2011). A total of 108,307 unigenes with an average length of 796 bp and a N50 of 1648 bp were obtained, among which 23,624 unigenes (21.81%) were longer than 1000 bp. The length distribution of the unigenes is shown in Figure S1. The fasta file of the unigene set is available from the Dryad Digital Repository (http://dx.doi.org/10.5061/dryad.nb028). To evaluate the efficiency of short-read usage during the de novo assembly, the clean reads were mapped back onto the unigenes using RSEM (Bo and Dewey, 2011). A total of 100,682,078 (MB), 98,967,076 (FB), 104,142,618 (SS), and 99,124,608 (OS) sequences were mapped (~85%), which indicated that the assembled unigenes could be used for the subsequent analysis.
Functional Annotation of the Unigenes
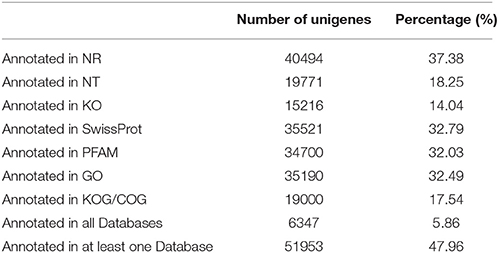
The unigenes were matched against sequences in the Nr, Nt, Swiss-Prot, COG/KOG, Pfam databases to assign functional annotations. A total of 6347 (5.86%) unigenes found matches in all the searched databases and 51,953 (47.97%) unigenes matched sequences in at least one of the databases; 56,354 (52.03%) unigenes did not match any of the known sequences in the public databases (Table 1 and Table S3). The number of unigenes assembled and annotated varied between the present study and other transcriptomic studies of G. biloba (Han et al., 2015; He et al., 2015), which may result from the novel genes specially expressed in different organs, or they may be the result of technical or biological biases (Rao et al., 2014).
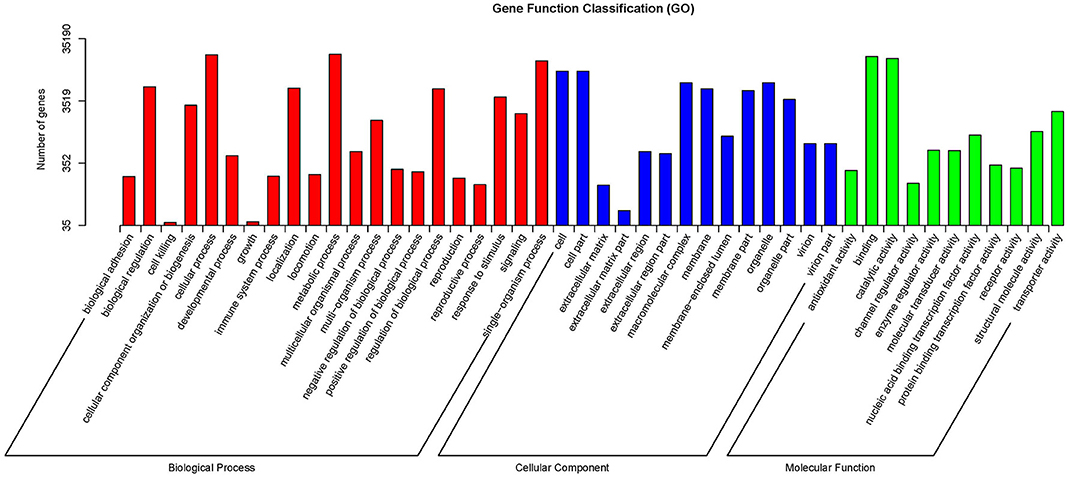
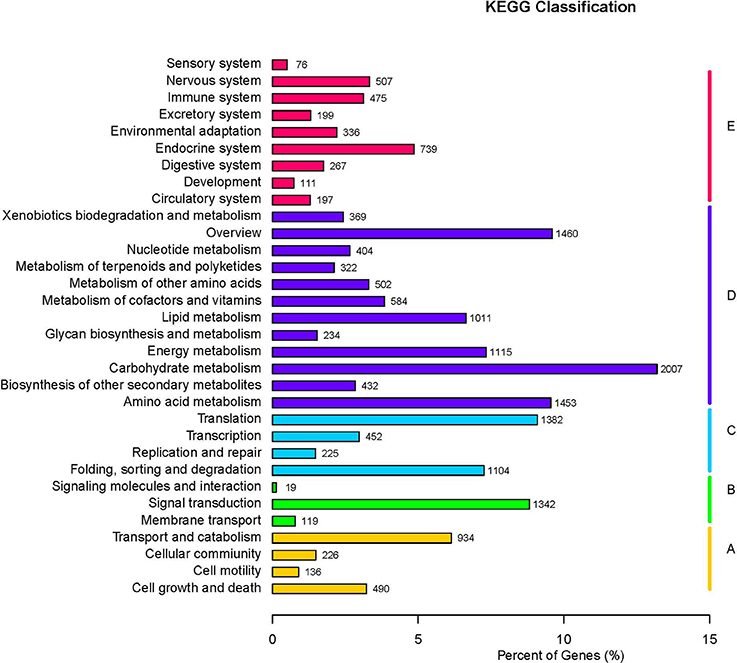
The GO database was searched using Blast2GO (Conesa et al., 2005) to functionally classify the unigenes. In several cases, multiple categories were assigned to the same unigene. A total of 180,424 terms were assigned under 23 subcategories of biological process, 14 subcategories of molecular function, and 18 subcategories of cellular component (Figure 2). The KEGG pathway database was used to predict the biological functions and the interactions of the gene products. Overall, 15,216 unigenes were assigned to 32 KEGG pathways (Figure 3).
Analysis of DEGs
To identify unigenes potentially involved in sex determination of G. biloba, we compared the transcriptome profiles between FB and MB, and between OS and SS. The total numbers of differentially expressed genes (DEGs) in MB vs. FB and SS vs. OS were 4709 and 9802 respectively, in which 1944 unigenes were commonly expressed (Figure 4), with an adjusted P < 0.05 as the threshold. In MB vs. FB, 2235 genes were up-regulated and 2474 were down-regulated, while in SS vs. OS, 5849 genes were up-regulated and 3953 were down-regulated.
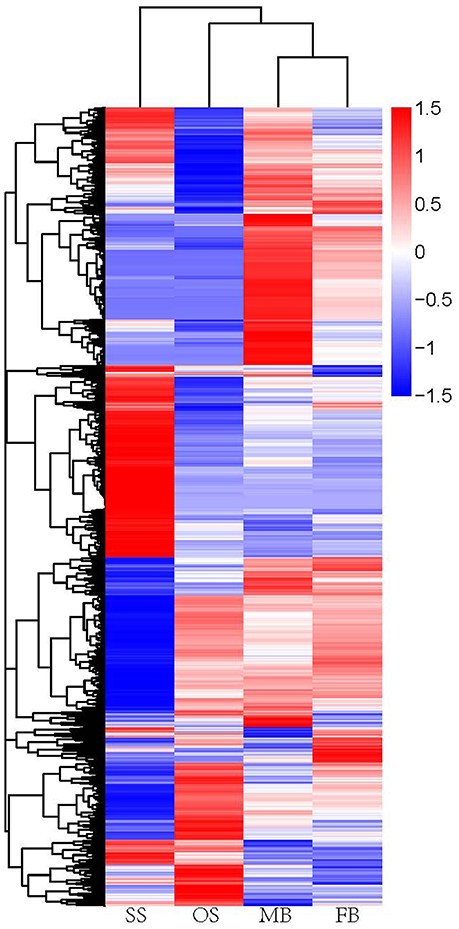
Figure 4. Heatmap of hierarchical clustering of DEGs. Each column represens a sample, each row represents a unigene. The differences in expression level were shown in distinct colors. Positive numbers indicate up-regulation DEGs and negative number indicates down-regulated DEGs. SS, staminate strobilus; OS, ovulate strobilus; MB, male bud; FB, female bud.
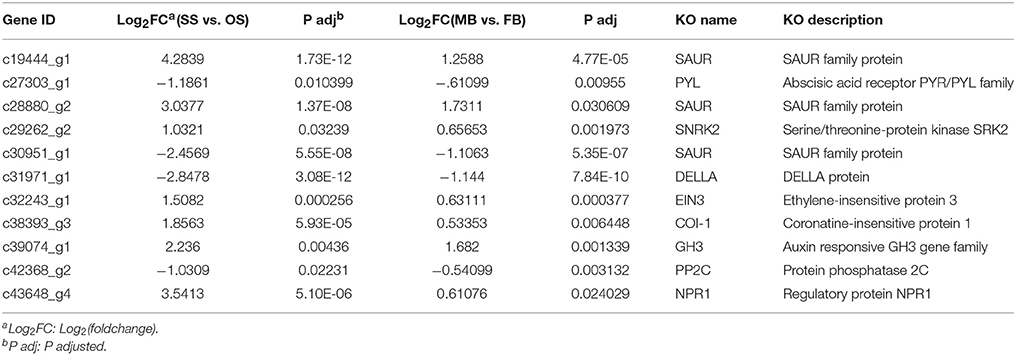
GO enrichment analysis of the DEGs in MB vs. FB showed that they were enriched in 36 categories with metabolic process (GO:0008152; 2154 unigenes) and cellular metabolic process (GO:0044237; 1678 unigenes) representing the most abundant categories, followed by cell part (GO:0044464; 1241 unigenes) and cellular macromolecule metabolic process (GO:0044260; 1222 unigenes) (Figure S2). Among the 9802 DEGs in SS vs. OS, 3115 were enriched in 18 categories. Cellular component biogenesis (GO:0044085; 427 unigenes) and ribonucleoprotein complex biogenesis (GO:0022613; 272 unigenes) were the most abundant categories, followed by ribosome biogenesis (GO: 0042254; 268 unigenes) and hydrolase activity, acting on glycosyl bonds (GO:0016798; 233 unigenes) (Figure S3). The most enriched KEGG pathways in SS vs. OS were ribosome (143 unigenes) and starch and sucrose metabolism (97 unigenes), followed by plant hormone signal transduction (76 unigenes) and phenylpropanoid biosynthesis (67 unigenes) (Figure S4). Among the 76 DEGs annotated to be involved in plant hormone signal and transduction (Table S4), 11 DEGs were commonly differentially expressed in MB vs. FB and SS vs. OS (Table 2, Data sheet 1), which might be involved in sex determination of G. biloba. In MB vs. FB, ribosome (190 unigenes) and phenylpropanoid biosynthesis (33 unigenes) were the most enriched pathways (Figure S5).
qRT-PCR Analysis
qRT-PCR was performed on 26 unigenes including 5 out of the 11 commonly differentially expressed unigenes involved in plant hormone signal and transduction (c29262_g2, c30951_g1, c31971_g1, c32243_g1, c38393_g3) to valid the expression profiling results obtained from the RNA-Seq data (Figure 5). The qRT-PCR and RNA-Seq results in the different organs were compared and the correlation between them was determined by calculating the correlation coefficient (R2). High correlation (R2 >0.9) was found between the RNA-Seq and qRT-PCR results, which indicated that the measured changes in gene expression detected by RNA-Seq reflected the actual transcriptome differences between the different libraries (Figure S6).
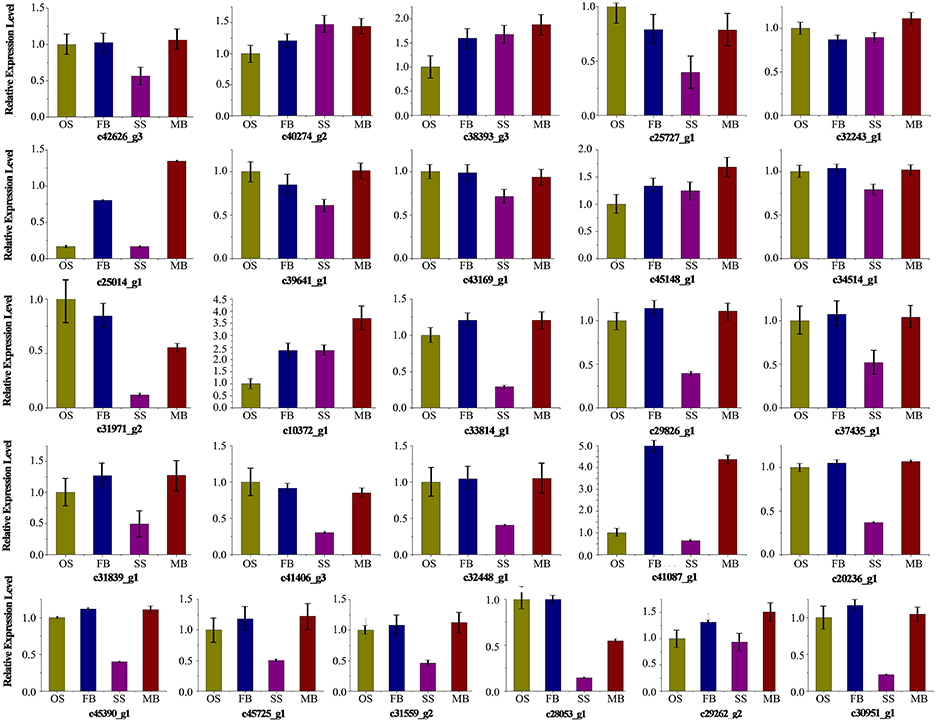
Figure 5. Expression analysis of 26 genes in different samples. Relative expression level in ovulate strobilus was set to 1.0. Error bars indicate the calculated maximum and minimum expression quantity of replicates. SS, staminate strobilus; OS, ovulate strobilus; MB, male bud; FB, female bud.
Discussion
Sex determination is regulated at several points in the intricate network of involved genes in plants (Aryal and Ming, 2014). Moreover, genes known to be implicated in floral development (e.g., ABC model genes) do not have direct roles in sex determination (Hardenack et al., 1994; Ainsworth et al., 1995). It is well-appreciated that plant sex determination is mainly regulated by genes involved in phytohormone synthesis and transduction (Guo et al., 2010; Aryal and Ming, 2014; Zhang et al., 2014). As shown in Table 2, abscisic acid (ABA)-related genes encoding PP2C (protein phosphatase 2C, c42368_g2), and PYL (abscisic acid receptor 2, c27303_g1) that negatively regulated ABA transduction were down-regulated while one SNF1-related protein kinase subfamily 2 (SNRK2) gene that positively regulating ABA signaling was up-regulated in male organs. Antisense expression of SNRK gene in Hordeum vulgare interfered with pollen development, which caused male sterility (Zhang Y. et al., 2001). This may suggest that these proteins involved in ABA signaling in G. biloba is critical for reproduction development (Fujii and Zhu, 2009; Park et al., 2009). An auxin-reduced SAUR (small auxin up RNA) gene (c30951_g1) was down-regulated in male, two other SAUR genes (c19444_g1 and c28880_g2) and one GH3 (Gretchen Hagen3) gene (c39074_g1) involved in auxin response showed higher expression level in male, which indicated that auxin related genes possessed diverse functions as in other plants (Kong et al., 2013). Coronatine insensitive 1 (COI1), which involved in jasmonate (JA) perception and transduction in plants, showed higher expression level in male. Mutations of COI1 in Arabidopsis displayed various degrees of male sterility (Ellis and Turner, 2002; Turner et al., 2002). The higher expression of COI1 in male indicated that JA may play a crucial role in male reproductive development and sterility in G. biloba. Higher expression of NPR1 (nonexpressor of PR gene) implicated in salicylic acid (SA) signaling in male may directly correlate with immunity to pathogens in G. biloba (Mou et al., 2003). Furthermore, interaction of these phytohormones may play important roles in G. biloba sex determination as in other developmental processes (Domagalska and Leyser, 2011; de Jong et al., 2014), further studies are needed to testify this hypothesis.
It is well-appreciated that transcription factors play important roles in regulating gene expression by temporarily and spatially regulating the transcription of their target genes (Hobert, 2008). Transcription factors such as a maize DELLA protein D8 and a melon zinc finger protein (CmW1P1) (Peng et al., 1999; Martin et al., 2009) have been shown to be associated with the sex determination process. Several transcription factors that belong to the C2C2-GATA, VOZ, and BZR1-BES1 families may play important roles in sex determination of cucumber (Guo et al., 2010). In the present study, a DELLA protein involved in gibberellin (GA) transduction (c31971_g1) and ethylene-insensitive protein 3 (EIN3) (c32243_g1) showed simultaneously high expression level in male (Table 2). Ethylene plays a significant role in sex determination of plant species and can induce femaleness (Sisler, 1994). In cucumber, genes involved in ethylene biosynthesis and perception, as well as some ethylene-induced genes, have been found to be involved in sex determination (Boualem, 2009; Wu et al., 2010). Arabidopsis insensitive mutant, ein3, was defective in its ability to perceive or respond to ethylene (Solano et al., 1998). The higher expression of EIN3 in male may inhibit the perceive and response to ethylene, which may result in male differentiation.
There is increasing evidence that DNA methylation is an important regulatory factor in plant sex determination. Furthermore, DNA methylation is hypothesized to be the first step toward the evolution of dioecy (Gorelick, 2003). Exploration of the methylation status in various species will help to understand the evolution of sex determination and sex chromosomes in plants (Aryal and Ming, 2014). In maize, sex determination was regulated by various small non-coding RNAs through DNA methylation (Parkinson et al., 2007; Hultquist and Dorweiler, 2008). Female sex suppression in S. latifolia males was depended on methylation of specific DNA sequences in the Y chromosome (Janoušek et al., 1996). In Populus, which has a ZW sex chromosome system, two genes related to DNA methylation were reported to be localized in the sex determining region of chromosome 19 (Song et al., 2013). At least 200 unigenes related to methyltransferase activity in GO enrichment analysis (e.g., GO0032259, GO0008168, GO0008649, see Table S5) showed higher expression level in female than those in male, which included genes that have been reported previously to be involved in plant sex determination, such as MET1 and COMT1 (Do et al., 2007; Schmidt et al., 2013). This is similar to the findings in Populus where more methylation sites were detected in female flowers compared with that in male flowers (Song et al., 2012). We suggested that the differentially expressed unigenes that encode proteins involved in DNA methylation may be related to sex determination in G. biloba. This result may help provide clues to the role of primary DNA methylation in sex determination in this species.
In plants, the formation of unisexual flowers can be classified mainly into two types. The first type includes Asparagus officinalis (Lazarte and Palser, 1979), cucumber (Bai et al., 2004), Ceratonia siliqua (Tucker, 1992), and S. latifolia ssp. alba (Ye et al., 1991) in which, at early developmental stages, male and female floral primordia are initially bisexual and contain primordial anthers and pistils. Sex determination appears to be the result of the selective arrest of pistillate primordial in male and of staminate primordial in female (Lazarte and Palser, 1979). Thus, sex determination genes do not become active at the moment of flower initiation when the different organ primordia appear, but at a crucial time during a later developmental stage (Caporali et al., 1994). The second type includes species such as Spinacia oleracea and Mercurialis annua (Sherry et al., 1993; Pannell, 1997), in which there are no traces of gynoecia in male flowers or of stamens in female flowers. Thus, sex determination occurs before inflorescence development, prior to or at evocation (Sherry et al., 1993). In MB, unigenes encoding pistil-specific extensin-like proteins were detected and expressed at a relatively high level (FPKM = 203.76). Similarly, in FB, unigenes encoding the pollen-specific protein SF21 and anther development proteins were detected (FPKM = 19.64 and 7.7 respectively). Furthermore, unigenes encoding apoptosis regulator proteins (e.g., metacaspase involved in regulation of apoptosis and apoptosis-related protein) and hydroxysteroid dehydrogenase (e.g., 3-beta hydroxysteroid dehydrogenase) were detected in both MB and FB (Table S6), which involved in triggering the organ death program and contributed to the selective abortion of the development of either the staminate or the pistillate primordia (DeLong et al., 1993). Based on these findings, we suggested that the formation of dioecism in G. biloba may resulted from the selective arrest of reproductive primordia and the unigenes implicated in the apoptosis pathway may play specific roles.
Conclusions
In the present study, we sequenced and characterized the transcriptomes of G. biloba female and male buds and strobilus. The transcriptome resources that we obtained provided foundations for identifying functional genes related to sex determination in G. biloba. Based on these resources, we found that genes involved in plant hormone signal and those encoding DNA methyltransferase were differentially expressed between different sex types and dioecism in G. biloba was caused by the selective abortion of reproductive primordia. Concerted efforts with various model and non-model systems and studies over a pool of putative regulatory elements are necessary to further understand the complex mechanism of sex determination in plants.
Author Contributions
SX, YS designed the work, SD, XL, YS performed the experiment, analyzed the data and wrote the paper. HT, JL, LS collected the samples and revised it critically for important intellectual content.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Zhaoshan Wang and Dr. Jianguo Zhang from the Research Institute of Forestry, Chinese Academy of Forestry, Beijing, China for suggestions and constructive criticism of this manuscript. We also thank Dr. Yu Wendong from School of Horticulture and Plant Protection, Yangzhou University and Mr. Zhao Pengcheng from Jinan Urban Construction Co., Ltd., for their help in figure modification. This work was supported by China Postdoctoral Science Foundation (2015M582124), Shandong Agriculture Breeding Project: Collection and Evaluation of Valuable Native Tree Germplasm Resources (Shandong Nongliang No. [2011]7) and Research Award Fund for Outstanding Young and Middle-aged Scientists of Shandong Province (BS2012NY006).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00871
Figure S1. Length distribution of the unigene sequences.
Figure S2. GO enrichment analysis of differentially expressed unigenes in MB vs. FB. BP, Biological Process; CC, Cellular Component; MF, Molecular Function); MB, male bud; FB, female bud.
Figure S3. GO enrichment analysis of differentially expressed unigenes in SS vs. OS. BP, Biological Process; CC, Cellular Component; MF, Molecular Function; SS, staminate strobilus; OS, ovulate strobilus.
Figure S4. KEGG enrichment analysis of differentially expressed unigenes in SS vs. OS. SS, staminate strobilus; OS, ovulate strobilus.
Figure S5. KEGG enrichment analysis of differentially expressed unigenes in MB vs. FB. MB, male bud; FB, female bud.
Figure S6. Correlation of the expression levels of 26 genes measured by qRT-PCR and RNA-seq. SS: staminate strobilus; OS, ovulate strobilus; MB, male bud; FB, female bud.
Table S1. Summary of RNA-Seq and do novo assembly results.
Table S2. Summary of the 26 genes in qRT-PCR analysis.
Table S3. Annotation results of the unigenes in public databases.
Table S4. Differentially expressed unigenes related to plant hormone signal and transduction.
Table S5. Differentially expressed unigenes related to DNA methyltransferase.
Table S6. Differentially expressed unigenes related to apoptosis regulator proteins and hydroxysteroid dehydrogenase. MB, male bud; FB, female bud.
Data Sheet 1. Unigenes sequences of the 11 commenly DEGs.
References
Ainsworth, C., Crossley, S., Buchanan-Wollaston, V., Thangavelu, M., and Parker, J. (1995). Male and female flowers of the dioecious plant sorrel show different patterns of MADS Box gene expression. Plant Cell 7, 1583–1598. doi: 10.1105/tpc.7.10.1583
Akao, T., Sano, M., Yamada, O., Akeno, T., Fujii, K., Goto, K., et al. (2007). Analysis of expressed sequence tags from the fungus Aspergillus oryzae cultured under different conditions. DNA Res. 14, 47–57. doi: 10.1093/dnares/dsm008
Anders, S. (2010). Differential expression analysis for sequence count data. Genome Biol. 11, 79–82. doi: 10.1186/gb-2010-11-10-r106
Andersen, M. R., Vongsangnak, W., Panagiotou, G., Salazar, M. P., Lehmann, L., and Nielsen, J. (2008). A trispecies Aspergillus microarray: comparative transcriptomics of three Aspergillus species. Proc. Natl. Acad. Sci. U.S.A. 105, 4387–4392. doi: 10.1073/pnas.0709964105
Aryal, R., and Ming, R. (2014). Sex determination in flowering plants: papaya as a model system. Plant Sci. 217, 56–62. doi: 10.1016/j.plantsci.2013.10.018
Bai, S. L., Peng, Y. B., Cui, J. X., Gu, H. T., Xu, L. Y., Li, Y. Q., et al. (2004). Developmental analyses reveal early arrests of the spore-bearing parts of reproductive organs in unisexual flowers of cucumber (Cucumis sativus L.). Planta 220, 230–240. doi: 10.1007/s00425-004-1342-2
Bo, L., and Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323. doi: 10.1186/1471-2105-12-323
Boualem, A. (2009). A conserved ethylene biosynthesis enzyme leads to andromonoecy in two Cucumis species. PLoS ONE 4:e6144. doi: 10.1371/journal.pone.0006144
Caporali, E., Carboni, A., Galli, M., Rossi, G., Spada, A., and Longo, G. M. (1994). Development of male and female flower in Asparagus officinalis. Search for point of transition from hermaphroditic to unisexual developmental pathway. Sex. Plant Reprod. 7, 239–249. doi: 10.1007/BF00232743
Charlesworth, D., and Mank, J. E. (2010). The birds and the bees and the flowers and the trees: lessons from genetic mapping of sex determination in plants and animals. Genetics 186, 9–31. doi: 10.1534/genetics.110.117697
Charlesworth, D. (2002). Plant sex determination and sex chromosomes. Heredity (Edinb). 88, 94–101. doi: 10.1038/sj.hdy.6800016
Conesa, A., Gotz, S., Garcia, J. M., Terol, J., Talon, M., and Robles, M. (2005). Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21, 3674–3676. doi: 10.1093/bioinformatics/bti610
de Jong, M., George, G., Ongaro, V., Williamson, L., Willetts, B., Ljung, K., et al. (2014). Auxin and strigolactone signaling are required for modulation of Arabidopsis shoot branching by nitrogen supply. Plant Physiol. 166, 384–395. doi: 10.1104/pp.114.242388
DeLong, A., Calderon-Urrea, A., and Dellaporta, S. L. (1993). Sex determination gene TASSELSEED2 of maize encodes a short-chain alcohol dehydrogenase required for stage-specific floral organ abortion. Cell 74, 757–768. doi: 10.1016/0092-8674(93)90522-R
Do, C. T., Pollet, B., Thévenin, J., Sibout, R., Denoue, D., Barrière, Y., et al. (2007). Both caffeoyl Coenzyme A 3-O-methyltransferase 1 and caffeic acid O-methyltransferase 1 are involved in redundant functions for lignin, flavonoids and sinapoyl malate biosynthesis in Arabidopsis. Planta 226, 1117–1129. doi: 10.1007/s00425-007-0558-3
Domagalska, M. A., and Leyser, O. (2011). Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 12, 211–221. doi: 10.1038/nrm3088
Ellis, C., and Turner, J. G. (2002). A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215, 549–556. doi: 10.1007/s00425-002-0787-4
Finn, R. D., Clements, J., and Eddy, S. R. (2011). HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39, 29–37. doi: 10.1093/nar/gkr367
Frey, F. P., Urbany, C., Hüttel, B., Reinhardt, R., and Stich, B. (2015). Genome-wide expression profiling and phenotypic evaluation of European maize inbreds at seedling stage in response to heat stress. BMC Genomics 16:123. doi: 10.1186/s12864-015-1282-1
Fujii, H., and Zhu, J. K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 106, 8380–8385. doi: 10.1073/pnas.0903144106
Gorelick, R. (2003). Evolution of dioecy and sex chromosomes via methylation driving Muller's ratchet. Biol. J. Linn. Soc. 80, 353–368. doi: 10.1046/j.1095-8312.2003.00244.x
Gotz, S. (2008). High-throughput functional annotation and data mining with the Blast2GO suite. Nucleic Acids Res. 36, 3420–3435. doi: 10.1093/nar/gkn176
Grabherr, M. G., Haas, B. J., Yassour, M., Levin, J. Z., Thompson, D. A., Amit, I., et al. (2011). Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652. doi: 10.1038/nbt.1883
Guo, S., Zheng, Y., Joung, J.-G., Liu, S., Zhang, Z., Crasta, O. R., et al. (2010). Transcriptome sequencing and comparative analysis of cucumber flowers with different sex types. BMC Genomics 11:384. doi: 10.1186/1471-2164-11-384
Guttman, D. S., and Charlesworth, D. (1998). An X-linked gene with a degenerate Y-linked homologue in a dioecious plant. Nature 393, 263–266. doi: 10.1038/30492
Han, S., Wu, Z., Jin, Y., Yang, W., and Shi, H. (2015). RNA-Seq analysis for transcriptome assembly, gene identification, and SSR mining in ginkgo (Ginkgo biloba L.). Tree Genet. Genomes 11, 1–10. doi: 10.1007/s11295-015-0868-8
Hardenack, S., Ye, D., Saedler, H., and Grant, S. (1994). Comparison of MADS box gene expression in developing male and female flowers of the dioecious plant white campion. Plant Cell 6, 1775–1787. doi: 10.1105/tpc.6.12.1775
He, B., Gu, Y., Xu, M., Wang, J., Cao, F., and Xu, L.-A. (2015). Transcriptome analysis of Ginkgo biloba kernels. Front. Plant Sci. 6:819. doi: 10.3389/fpls.2015.00819
Hobert, O. (2008). Gene regulation by transcription factors and microRNAs. Science 319, 1785–1786. doi: 10.1126/science.1151651
Huang, Q., Liu, Q. Y., Cao, M., and Yang, S. S. (2013). Advance of gender characteristic expression and identification of Ginkgo biloba. J. Fruit Sci. 30, 1065–1071. doi: 10.13925/j.cnki.gsxb.2013.06.026
Hultquist, J., and Dorweiler, J. (2008). Feminized tassels of maize mop1 and ts1 mutants exhibit altered levels of miR156 and specific SBP-box genes. Planta 229, 99–113. doi: 10.1007/s00425-008-0813-2
Irizarry, R. A., Warren, D., Spencer, F., Kim, I. F., Biswal, S., Frank, B. C., et al. (2005). Multiple-laboratory comparison of microarray platforms. Nat. Methods 2, 345–350. doi: 10.1038/nmeth756
Janoušek, B., Široký, J., and Vyskot, B. (1996). Epigenetic control of sexual phenotype in a dioecious plant, Melandrium album. Mol. Gen. Genet. 250, 483–490. doi: 10.1007/BF02174037
Kong, Y., Zhu, Y., Gao, C., She, W., Lin, W., Chen, Y., et al. (2013). Tissue-specific expression of SMALL AUXIN UP RNA41 differentially regulates cell expansion and root meristem patterning in Arabidopsis. Plant Cell Physiol. 54, 609–621. doi: 10.1093/pcp/pct028
Lazarte, J. E., and Palser, B. F. (1979). Morphology, vascular anatomy and embryology of pistillate and staminate flowers of Asparagus officinalis. Am. J. Bot. 66, 753–764. doi: 10.2307/2442462
Liu, J., Yin, T., Ye, N., Chen, Y., Yin, T., Liu, M., et al. (2013). Transcriptome analysis of the differentially expressed genes in the male and female shrub willows (Salix suchowensis). PLoS ONE 8:e60181. doi: 10.1371/journal.pone.0060181
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using Real-time quantitative PCR and the 2−ΔΔCT Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mao, X., Tao, C. J. G. O., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Martin, A., Troadec, C., Boualem, A., Rajab, M., Fernandez, R., Morin, H., et al. (2009). A transposon-induced epigenetic change leads to sex determination in melon. Nature 461, 1135–1138. doi: 10.1038/nature08498
Matsunaga, S., Kawano, S., Takano, H., Uchida, H., Sakai, A., and Kuroiwa, T. (1996). Isolation and developmental expression of male reproductive organ-specific genes in a dioecious campion, Melandrium album (Silene latifolia). Plant J. 10, 679–689. doi: 10.1046/j.1365-313X.1996.10040679.x
Ming, R., Bendahmane, A., and Renner, S. S. (2011). Sex chromosomes in land plants. Annu. Rev. Plant Biol. 62, 485–514. doi: 10.1146/annurev-arplant-042110-103914
Moore, R. C., Kozyreva, O., Lebel-Hardenack, S., Siroky, J., Hobza, R., Vyskot, B., et al. (2003). Genetic and functional analysis of DD44, a sex-linked gene from the dioecious plant Silene latifolia, provides clues to early events in sex chromosome evolution. Genetics 163, 321–334.
Moriya, Y., Itoh, M., Okuda, S., Yoshizawa, A. C., and Kanehisa, M. (2007). KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 35, 182–185. doi: 10.1093/nar/gkm321
Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. doi: 10.1016/S0092-8674(03)00429-X
Pannell, J. (1997). Mixed genetic and environmental sex determination in an androdioecious population of Mercurialis annua. Heredity 78, 50–56. doi: 10.1038/hdy.1997.6
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Parkinson, S. E., Gross, S. M., and Hollick, J. B. (2007). Maize sex determination and abaxial leaf fates are canalized by a factor that maintains repressed epigenetic states. Dev. Biol. 308, 462–473. doi: 10.1016/j.ydbio.2007.06.004
Peng, J., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). 'Green revolution' genes encode mutant gibberellin response modulators. Nature 400, 256–261. doi: 10.1038/22307
Rao, G., Sui, J., Zeng, Y., He, C., Duan, A., and Zhang, J. (2014). De novo transcriptome and small RNA analysis of two Chinese willow cultivars reveals stress response genes in Salix matsudana. PLoS ONE 9:e109122. doi: 10.1371/journal.pone.0109122
Renner, S. S., and Ricklefs, R. E. (1995). Dioecy and its correlates in the flowering plants. Am. J. Bot. 596–606. doi: 10.2307/2445418
Schmidt, A., Wöhrmann, H. J. P., Raissig, M. T., Arand, J., Gheyselinck, J., Gagliardini, V., et al. (2013). The Polycomb group protein MEDEA and the DNA methyltransferase MET1 interact to repress autonomous endosperm development in Arabidopsis. Plant J. 73, 776–787. doi: 10.1111/tpj.12070
Sherry, R. A., Eckard, K. J., and Lord, E. M. (1993). Flower development in dioecious Spinacia oleracea (Chenopodiaceae). Am. J. Bot. 283–291. doi: 10.2307/2445351
Shi, J. K., Fan, W. G., and Wen, X. P. (1998). Female flower bud differentiation of Ginkgo biloba. Acta Hortic. Sin. 1, 33–36.
Solano, R., Stepanova, A., Chao, Q., and Ecker, J. R. (1998). Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev. 12, 3703–3714. doi: 10.1101/gad.12.23.3703
Song, Y., Ma, K., Bo, W., Zhang, Z., and Zhang, D. (2012). Sex-specific DNA methylation and gene expression in andromonoecious poplar. Plant Cell Rep. 31, 1393–1405. doi: 10.1007/s00299-012-1255-7
Song, Y., Ma, K., Ci, D., Chen, Q., Tian, J., and Zhang, D. (2013). Sexual dimorphic floral development in dioecious plants revealed by transcriptome, phytohormone, and DNA methylation analysis in Populus tomentosa. Plant Mol. Biol. 83, 559–576. doi: 10.1007/s11103-013-0108-2
Storey, J. D., and Tibshirani, R. (2003). Statistical significance for genomewide studies. Proc. Natl. Acad. Sci. U.S.A. 100, 9440–9445. doi: 10.1073/pnas.1530509100
Terefe, D., and Tatlioglu, T. (2005). Isolation of a partial sequence of a putative nucleotide sugar epimerase, which may involve in stamen development in cucumber (Cucumis sativus L.). Theor. Appl. Genet. 111, 1300–1307. doi: 10.1007/s00122-005-0058-4
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Tucker, S. C. (1992). The developmental basis for sexual expression in Ceratonia siliqua (Leguminosae: Caesalpinioideae: Cassieae). Am. J. Bot. 318–327. doi: 10.2307/2445022
Turner, J. G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14, 153–164.
Wang, B., Guo, G., Wang, C., Lin, Y., Wang, X., Zhao, M., et al. (2010). Survey of the transcriptome of Aspergillus oryzae via massively parallel mRNA sequencing. Nucleic Acids Res. 38, 5075–5087. doi: 10.1093/nar/gkq256
Wang, H., Cao, F., Zhang, W., Wang, G., and Yu, W. (2012). Cloning and Expression of Stearoyl-ACP Desaturase and Two Oleate Desaturases Genes from Ginkgo biloba L. Plant Mol. Biol. Rep. 31, 633–648. doi: 10.1007/s11105-012-0525-4
Wang, Z., Gerstein, M., and Snyder, M. (2009). RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 10, 57–63. doi: 10.1038/nrg2484
Wu, T., Qin, Z., Zhou, X., Feng, Z., and Du, Y. (2010). Transcriptome profile analysis of floral sex determination in cucumber. J. Plant Physiol. 167, 905–913. doi: 10.1016/j.jplph.2010.02.004
Yang, Z., Peng, Z., Wei, S., Liao, M., Yu, Y., and Jang, Z. (2015). Pistillody mutant reveals key insights into stamen and pistil development in wheat (Triticum aestivum L.). BMC Genomics 16:211. doi: 10.1186/s12864-015-1453-0
Ye, D., Oliveira, M., Veuskens, J., Wu, Y., Installe, P., Hinnisdaels, S., et al. (1991). Sex determination in the dioecious Melandrium. The X/Y chromosome system allows complementary cloning strategies. Plant Sci. 80, 93–106. doi: 10.1016/0168-9452(91)90275-D
Young, M. D., Wakefield, M. J., Smyth, G. K., and Oshlack, A. (2010). Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11:R14. doi: 10.1186/gb-2010-11-2-r14
Zhang, J., Boualem, A., Bendahmane, A., and Ming, R. (2014). Genomics of sex determination. Curr. Opin. Plant Biol. 18, 110–116. doi: 10.1016/j.pbi.2014.02.012
Zhang, Q., Li, J., Sang, Y., Xing, S., Wu, Q., and Liu, X. (2015). Identification and characterization of microRNAs in Ginkgo biloba var. epiphylla Mak. PLoS ONE 10:e0127184. doi: 10.1371/journal.pone.0127184
Zhang, W. P., Shi, J. K., Fan, W. G., and Wen, X. P. (2001). Male flower bud differentiation of Ginkgo biloba. Acta Hortic. Sin. 28, 255–258.
Keywords: Ginkgo biloba, sex determination, RNA-sequencing, plant hormone signal and transduction, DNA methyltransferase, hydroxysteroid dehydrogenase
Citation: Du S, Sang Y, Liu X, Xing S, Li J, Tang H and Sun L (2016) Transcriptome Profile Analysis from Different Sex Types of Ginkgo biloba L. Front. Plant Sci. 7:871. doi: 10.3389/fpls.2016.00871
Received: 22 January 2016; Accepted: 02 June 2016;
Published: 16 June 2016.
Edited by:
Daniel Pinero, Universidad Nacional Autónoma de México, MexicoReviewed by:
Stefan De Folter, Centro de Investigación y de Estudios Avanzados del Instituto Politécnico Nacional, MexicoRishi Aryal, Western Sydney University, Australia
Copyright © 2016 Du, Sang, Liu, Xing, Li, Tang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shiyan Xing, eGluZ3N5QHNkYXUuZWR1LmNu
†These authors have contributed equally to this work.
 Shuhui Du
Shuhui Du Yalin Sang†
Yalin Sang†