
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 01 June 2016
Sec. Plant Physiology
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.00713
This article is part of the Research TopicMechanisms of abiotic stress responses and tolerance in plants: physiological, biochemical and molecular interventionsView all 121 articles
 Elena Gottardini1
Elena Gottardini1 Antonella Cristofori1
Antonella Cristofori1 Elisa Pellegrini2*
Elisa Pellegrini2* Nicola La Porta1,3,4
Nicola La Porta1,3,4 Cristina Nali2
Cristina Nali2 Paolo Baldi5
Paolo Baldi5 Gaurav Sablok1,6
Gaurav Sablok1,6Tropospheric ozone (O3) is a global air pollutant that causes high economic damages by decreasing plant productivity. It enters the leaves through the stomata, generates reactive oxygen species, which subsequent decrease in photosynthesis, plant growth, and biomass accumulation. In order to identify genes that are important for conferring O3 tolerance or sensitivity to plants, a suppression subtractive hybridization analysis was performed on the very sensitive woody shrub, Viburnum lantana, exposed to chronic O3 treatment (60 ppb, 5 h d−1 for 45 consecutive days). Transcript profiling and relative expression assessment were carried out in asymptomatic leaves, after 15 days of O3 exposure. At the end of the experiment symptoms were observed on all treated leaves and plants, with an injured leaf area per plant accounting for 16.7% of the total surface. Cloned genes were sequenced by 454-pyrosequencing and transcript profiling and relative expression assessment were carried out on sequenced reads. A total of 38,800 and 12,495 high quality reads obtained in control and O3-treated libraries, respectively (average length of 319 ± 156.7 and 255 ± 107.4 bp). The Ensembl transcriptome yielded a total of 1241 unigenes with a total sequence length of 389,126 bp and an average length size of 389 bp (guanine-cytosine content = 49.9%). mRNA abundance was measured by reads per kilobase per million and 41 and 37 ensembl unigenes showed up- and down-regulation respectively. Unigenes functionally associated to photosynthesis and carbon utilization were repressed, demonstrating the deleterious effect of O3 exposure. Unigenes functionally associated to heat-shock proteins and glutathione were concurrently induced, suggesting the role of thylakoid-localized proteins and antioxidant-detoxification pathways as an effective strategy for responding to O3. Gene Ontology analysis documented a differential expression of co-regulated transcripts for several functional categories, including specific transcription factors (MYB and WRKY). This study demonstrates that a complex sequence of events takes place in the cells at intracellular and membrane level following O3 exposure and elucidates the effects of this oxidative stress on the transcriptional machinery of the non-model plant species V. lantana, with the final aim to provide the molecular supportive knowledge for the use of this plant as O3-bioindicator.
Ozone (O3) is a gas naturally present in both troposphere and stratosphere. Particularly, tropospheric O3 results from a series of complex photochemical reactions involving primary pollutants, such as nitrogen oxides (NOx), volatile organic compounds (VOC) and carbon monoxide (CO) mainly generated by human activities (Jenkin and Clemitshaw, 2000). O3-producing photochemical reactions are favored by high temperatures and elevated light intensities (Cristofanelli and Bonasoni, 2009). During summertime, the Mediterranean basin is characterized by specific meteorological conditions (i.e., sunny, hot, and dry climate) that enhance photochemical O3 formation (Millàn et al., 2000). At the mid-latitudes of the Northern Hemisphere, O3 concentrations have more than doubled over the last decades (Monks et al., 2015). Especially in Europe, a general trend toward a decline in peak concentrations has been observed, taking into account the implementation of European air pollution policies reducing precursor emissions. On the other hand, Dawnay and Mills (2009) documented a concomitant increase in O3 background concentrations, due to the rise in anthropogenic emissions on a global scale. The ambient O3 concentrations have a marked impact not only on human health (Yang and Omaye, 2009; Norval et al., 2011), but also on natural ecosystems, crop productivity (yield and quality), manufactures and works of art (Cass et al., 1989). For these reasons, it is important to understand the regulatory behavior of O3 induced stress in plants (e.g., Braun et al., 2014; Doring et al., 2014a; González-Fernández et al., 2014).
After entering into the leaf via the open stomata, O3 interacts immediately with biological molecules (like bio-membranes and enzymes) and releases reactive oxygen species (ROS), thereby triggering an oxidative burst (Jaspers et al., 2005). Plants deploy several response mechanisms, some of which are universally conserved among species (Whaley et al., 2015). Through a signaling cascade (Vainonen and Kangasjärvi, 2015), O3 affects primarily biological processes involved in plant productivity, such as regulation of structural and chemical components of photosynthesis (Pellegrini et al., 2015), respiration and transpiration (Heath, 2008). The toxicology of this pollutant is complex: many factors such as species, provenance, genotype and leaf age together with environmental, nutritional, and health conditions, play a key role in determining the overall plant response (Manninen et al., 2009). Moreover, contrasting results may be caused by different O3 concentrations as well as by different spatial and temporal scales of this pollutant. In this respect, it is important to categorize acute vs. chronic exposures, respectively in short- and long-time treatments (Miller, 2011).
Taking into account the literature, a bio-molecular approach might enable a better understanding of oxidative stress-plant interactions. Current knowledge concerning specific molecular alterations caused by O3 at the transcriptomic level is limited (Heath, 2008; Kanter et al., 2013) and has been primarily elucidated (i) in model plants, like Arabidopsis thaliana (Hirayama and Shinozaki, 2010) and Medicago truncatula (Puckette et al., 2009, 2012); (ii) in crops, like Pisum sativum (Savenstrand et al., 2000), Oryza sativa (Frei, 2015; Sarkar et al., 2015); and (iii) in herbaceous annual plants, like Centaurea jacea (Francini et al., 2008), Melissa officinalis (Doring et al., 2014b). Moreover, interactions occurring between O3 exposure and changes in the expression profile of several genes have been described in some woody species.
Olbrich et al. (2010) studied transcriptional responses in juvenile Fagus sylvatica saplings fumigated with a double concentration of ambient O3 over 3 years by conducting microarray hybridization. Recently, the effects of O3 (twice ambient concentration) on F. sylvatica were investigated performing a large-scale protein analysis based on 2-D Fluorescence Difference Gel Electrophoresis (2-DE DIGE) (Kerner et al., 2014). Rizzo et al. (2007) identified the differential expression of genes induced by an episodic O3 treatment (150 ppb for 5 h) in two poplar clones with different O3-sensitivity, performing Suppression Subtractive Hybridization (SSH). Nathaniel et al. (2011) investigated the transcriptional and genetics O3 responsiveness (chronic and acute treatment) in two divergent Populus species. Using an inbred F2 mapping population derived from these two species, they mapped quantitative trait loci (QTLs) associated with O3 response, and examined segregation of the transcriptional response to O3 and co-localized genes showing divergent responses between tolerant and sensitive genotypes. Tuomainen et al. (1996) studied the acute O3-induced reactions at biochemical and transcriptomic levels in two Betula clones differing in O3-sensitivity, whereas Zinser et al. (2000) focalized on Pinus sylvestris. Similarly, Kontunen-Soppela et al. (2010) revealed the patterns of gene expression in Betula papyrifera leaves exposed to twice ambient O3 concentration using microarray analyses.
In this study, we focused on wayfaring tree (Viburnum lantana L.), a common deciduous shrub species, widespread in most part of Europe, North Africa, North America and temperate Asia, and well-known for its sensitivity to O3 (Novak et al., 2003; Calatayud et al., 2010). Wayfaring tree sensitivity has been already assessed in terms of morphological (i.e. foliar symptoms) and physiolgical traits in order to evaluate its potential as bio-indicator (Gottardini et al., 2010, 2014a,b). Although a series of studies has been conducted on Viburnum spp. (Clement and Donoghue, 2012), this report represents the first attempt to assess the O3 sensitivity of this species at molecular level. Specifically, paired SSH and 454-pyrosequencing analyses were performed in plants exposed to near-ambient O3 concentrations in controlled environmental conditions. Data collected in this study may be useful to better understand results obtained in natural field conditions.
One-year-old agamically reproduced saplings of V. lantana were grown for 1 month in plastic pots containing a mix of steam sterilized medium soil and peat (1:1) in a controlled environment facility (steady temperature of 20 ± 1°C, relative humidity (RH) of 85 ± 5% and photon flux density at plant height of 530 μmol photon m−2 s−1 provided by incandescent lamps, following a 14 h photoperiod). A sub-sample of 18 uniform plants were selected when they were ca. 35 cm tall (ca. 30 fully expanded leaves), and were placed in a controlled fumigation environment facility under the same climatic conditions as the growth chamber. Nine plants were exposed to 60 ± 13 ppb of O3 (1 ppb = 1.96 μg m−3, at 20°C and 101.325 kPa) for 45 consecutive days (5 h d−1, in form of a square wave between 9:00 a.m. and 2:00 p.m.). At the same time, nine control plants were exposed to charcoal-filtered air. The entire methodology was performed according to Lorenzini et al. (1994). Leaf samples (n = 5) were collected from three treated and three control plants after 15 days of fumigation (before the onset of foliar symptoms), immediately frozen in liquid nitrogen (N2) and kept at −80°C for RNA extraction.
Each marked leaf was scored in percent of O3-damaged surface (5%-classes) by in-hand examination with a 10x hand lens and symptoms identified as reported by Gottardini et al. (2014a). For each plant, the mean percentage of injured leaf area of the marked leaves per date was calculated.
Total RNA was extracted from leaves using the protocol described by Gambino et al. (2008). Frozen leaves were ground to fine powder using a pre-chilled mortar and a pestle. Five milliliter of extraction buffer [2% CTAB, 2.5% PVP-40, 2 M NaCl, 100 mM Tris-HCl (pH 8.0), 25 mM EDTA (pH 8.0), and 2% β-mercaptoethanol, added just before use] were heated at 65°C and added to 1 g of ground tissue. After 10 min of incubation at 65°C, two independent extractions were performed, using chloroform:isoamyl alcohol (24:1 v/v). The supernatant was transferred to a new tube and LiCl (3 M final concentration) was added. After 30 min on ice, the RNA was selectively pelleted by centrifugation (21,000 × g for 20 min at 4°C) and re-suspended in 500 μl of SSTE buffer [10 mM Tris-HCl (pH 8.0), 1 mM EDTA (pH 8), 1% SDS, 1 M NaCl], pre-heated at 65°C. An equal volume of chloroform:isoamyl alcohol (24:1 v/v) was added, centrifuged (11,000 × g for 10 min at 4°C) and the supernatant was transferred to a new tube and precipitated with one volume of cold isopropanol. The pellet was washed with ethanol (70%), air-dried and re-suspended in DEPC-treated water. Messenger RNA was isolated using GenElute™ mRNA Miniprep kit (Sigma-Aldrich, St. Louis, MO, USA) according to the manufacturer's instructions. SSH was performed using PCR-select™cDNA subtraction kit (Clontech Laboratories, Mountain View, CA, USA) following the procedure described in the user manual. Forward and reverse subtractions were performed using control and O3-treated leaves and PCR products were subsequently sequenced using 454-pyrosequencing (GS FLX+ System, Roche Diagnostics GmbH, Penzberg, Germany). Raw reads obtained from the present study are recorded in the European Nucleotide Archive (ENA) under the project number PRJEB9317.
Sequencing reads, obtained from the two libraries (induced and repressed), were cleaned, removing adapter and primer sequences and trimming low-quality ends. We further removed reads shorter than 100 bp and having an average Phred equivalent quality score lower than 15 bp. After read cleaning, homo-polymer stretches (polyA/T) were estimated and were subsequently masked using an in-house PERL script. Masking of the polyA/T was done to increase the sensitivity and specificity in the assembly. For the creation of the Ensembl transcriptome, all the cleaned reads of the induced and repressed libraries were concatenated and a single Ensembl transcriptome was built using MIRA (Chevreux et al., 2004) and CAP3, an overlap layout consensus assembly approach (Huang and Madan, 1999) with an overlap percentage identity cut-off of 97%, to avoid the formation of the spurious assemblies. Resulting contigs and singletons were clustered into representative set of unigenes for each library. All the repetitive reads that were falling into the mega-hub during the MIRA assembly were discarded so as to ensure the correctness and the accuracy of the assembled unigenes.
Following unigene assembly, unigenes in each library were subjected to BLASTx with an E-value cut off of 1E-5 against NCBI database available from http://www.ncbi.nlm.nih.gov. All the unigenes were translated into six possible translational frames using the sixpack package of the EMBOSS available from http://emboss.sourceforge.net/ and the putative open-reading frames (ORF) were extracted using the Getorf of the EMBOSS package. All the translated frames were queried for the identification of the InterPro domains and the longest frame with no internal stop codon and with assigned InterPro domain was kept as an assigned functional domain to that respective unigene. FastAnnotator (Chen T. W. et al., 2012) and PLAZA version 2.5 (Van Bel et al., 2013) were run to identify functional annotations associated with the unigenes. Gene Ontology (GO) was derived for each unigenes, slimmed using the plant GO slim (available from http://www.geneontology.org) and classified according to biological and molecular functions and cellular localization. Transcription factors were identified using the PlantTFcat (Dai et al., 2013).
To evaluate the expression levels of the unigenes in induced/repressed libraries, we mapped the individual library reads back to the ensembl transcriptome and reads per kilobase per million (RPKM) was calculated as expression estimate: RPKM (A) = 1,000,000 × C × 1000)/(N × L), where A is defined as the expression of the unigene, C corresponds to the reads that align uniquely to the unigene, N refers to total number of reads that uniquely aligned to all genes, and L refers to the length of gene A. Transcript mapping was performed using BWA-SW algorithm, as implemented in Burrows-Wheeler Aligner [BWA, available from http://bio-bwa.sourceforge.net/; Li and Durbin (2009)]. Expression values were further analyzed to identify the transcripts whose expression was significantly up- or down- regulated during the O3 treatment, using the log2 fold change (RPKM induced/RPKM repressed), as previously described in Kanter et al. (2013). To identify the functionally and statistically enriched biological pathways and GOs in the up- and down-regulated unigenes, those showing up- and down-regulation in Ensembl unigenes were analyzed using KOBAS, with A. thaliana as a background dataset. All the identified biological pathways and GOs were statistically evaluated using the hypergeometric test/Fisher's exact test followed by Benjamini and Hochberg FDR correction [P < 0.001 (Mao et al., 2005; Wu et al., 2006; Xie et al., 2011)].
After 15 days of O3 fumigation leaves did not show any visible symptoms. Thirty days from the beginning of the exposure [AOT40 = 3000 ppb h; AOT40: ozone Accumulated Over a Threshold of 40 ppb, sensu de Leeuw and van Zantwoort (1997)], fully expanded leaves from O3-fumigated plants showed several minute (Ø 1-2 mm) roundish dark-blackish necrosis located in the interveinal area of the adaxial surface. Symptoms were observed on all the examined leaves (n = 30) and plants (n = 6), with an injured leaf area per plant accounting for 4.2% (SE 1.4%; range 1–20%). At the end of the experiment (AOT40 4500 ppb h, 45 days from the beginning of exposure), the injured area was 16.7% (SE 2.47%; range 5–60%) of the total surface. Visible foliar injury has been used in many field experiments as an indicator of the response of V. lantana to O3 exposure (e.g., Gottardini et al., 2010). Fully expanded leaves showed symptoms similar to those previously reported in seedlings from OTC experiments (Novak et al., 2005, 2008) and in plants grown in natural conditions (Gottardini et al., 2014a,b).
A total of 43,815 and 13,610 high quality reads were generated in induced and repressed libraries, respectively (Table 1). Following sequencing, reads were filtered as described in the Materials and Methods section, resulting in a total of 38,800 and 12,495 clean reads (Table 1). The mean read length for the induced and repressed library was 319 ± 156.7 and 255 ± 107.4 bp, respectively (data not shown). 454-assembly using MIRA of the Ensembl transcriptome yielded a total of 1238 unigenes with a total sequence length of 389,126 bp and an average length size of 389 bp (GC = 49.9%, Table 1). ORF predictions revealed a total of 161 (13.0%) sequences with a proper start codon and a total of 574 sequences (46.3%) with proper stop codons. Interestingly, a low number of unigenes (73, 5.9%) with predicted ORF contain frameshifts, which indicates a good quality of the assembly. Identified frameshifts were corrected using FrameDP (Gouzy et al., 2009).
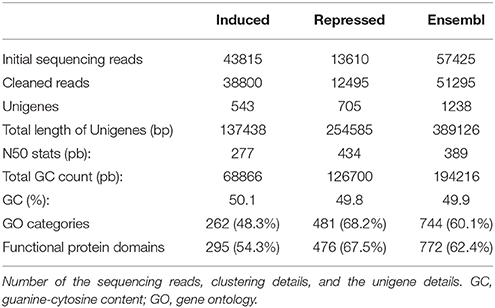
Table 1. Summary of RNA sequencing and de novo assembly to construct the gene set of Viburnum lantana plants exposed to O3 treatment (60 ppb of O3, 5 h d−1 for 15 consecutive days).
Functional classification of the assembled unigenes indicates the putative functional changes occurring at gene level in plants subjected to O3. The Ensembl transcriptome was annotated by performing stringent BLASTx searches (E-value threshold, 1E-5) against NCBI and PLAZA version 2.5. Functional annotation of the Ensembl transcriptome (1,238 unigenes) revealed a total of 744 transcript sequences (60.1%), with an assigned GO category and a total of 772 transcript sequences (62.4%), with an assigned InterPro domain, respectively (Table 1 and Supplementary Table 1 and Supplementary Datasheet 1). The results of BLASTx (E-value threshold, 1E-5) searches against NCBI database and PLAZA version 2.5 (Van Bel et al., 2013) resembled those reported for whole genome expressed genes and tissue-specific cDNA extracts in other plant species (Legrand et al., 2007, 2010; Hao et al., 2015). Based on GO-slim annotations, Ensembl unigenes were classified into three ontological categories: cellular component, biological process, and molecular function (Figure 1). Within the cellular component category, 21 GO slims were identified including cell, membrane, thylakoid, and other apparatus (Figure 1A). Cell, cellular component, intracellular, and cytoplasm were the most represented slims in terms of number of genes (393, 393, 323, and 231, respectively). On the other hand, 42 GO slims were recognized within the biological process category, such as translation, reproduction, transport, photosynthesis, cell death, and other functions (Figure 1B). Among all these slims, metabolic and cellular processes were the most frequent ones (502 and 419 genes, respectively). Aside from GO cellular processes, the response to stress was the next most abundant GO slim (110 gene numbers) after biological and biosynthetic process (400 and 154, respectively). It comprised responses to different stresses, including abiotic and endogenous stimuli (93 and 46 genes, respectively, Figure 1B). Within the molecular function category, 24 GO slims were observed including protein binding, transcription regulator activity, carbohydrate binding, hydrolase activity, receptor activity, and other enzymatic function (Figure 1C). Among all of these slims, binding and catalytic activity appeared more frequently (415 and 380, respectively).
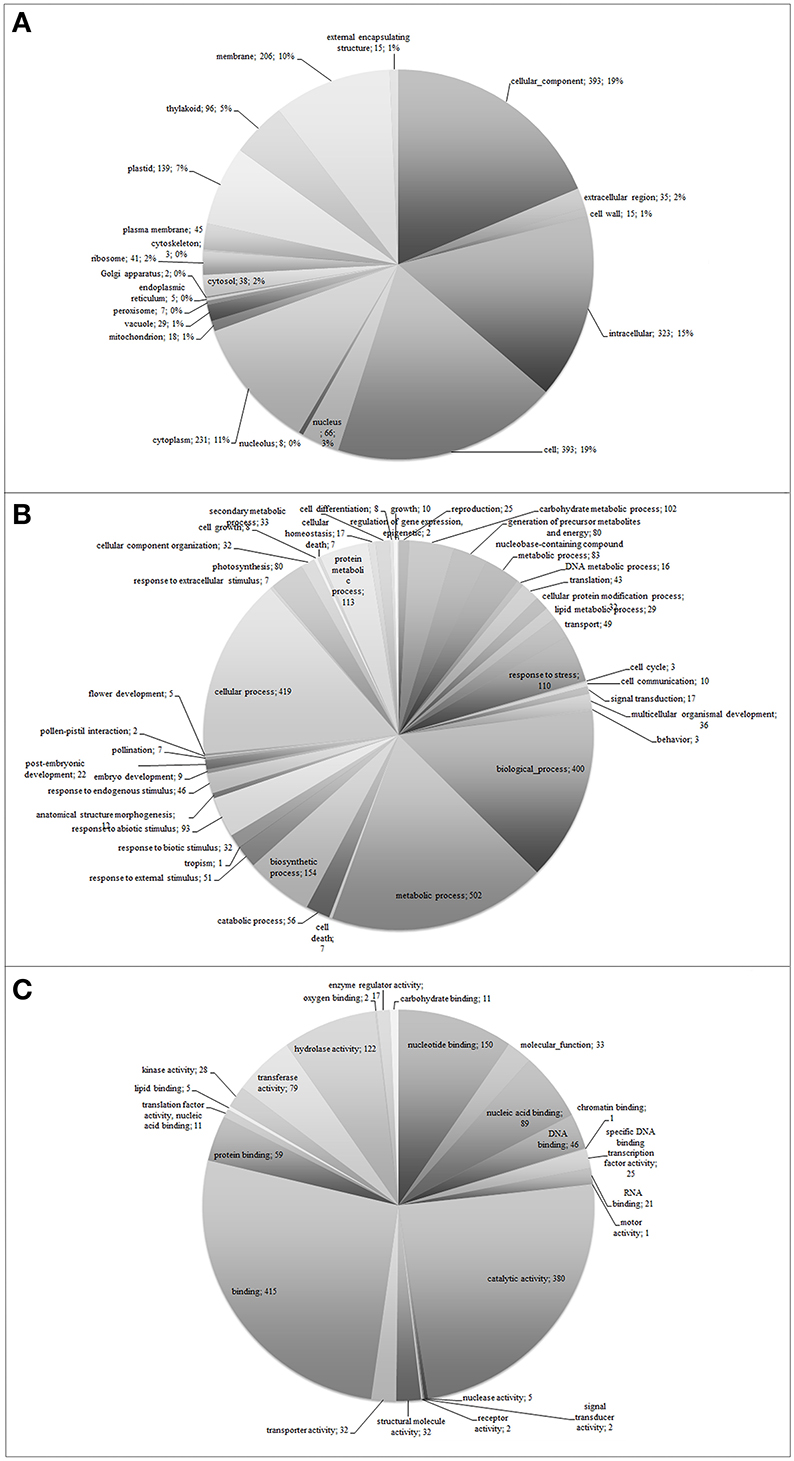
Figure 1. Functional classification of Viburnum lantana ensembl unigenes within cellular component (A), biological process (B), and molecular function (C) categories.
Expression abundance for the unigenes was estimated by mapping the reads from the individual library to the assembled Ensemble transcriptome and was assessed as number of reads per RPKM. According to Kanter et al. (2013), prior to linking the expression estimates to biological functions, unigenes having a RPKM lower than 7 were discarded to avoid the false interpretation of the log2 fold change values. A total of 78 (6.3%) Ensembl unigenes showed a variation in expression values after O3-treatment. In particular, 41 ensembl unigenes showed an up-regulation average [log2 (RPKM induced/RPKM repressed) of 3.7 (2.1 SE) ranged between 0.4 and 8.7 (Table 2)] and the other 37 showed a down-regulation [log2 (RPKM induced/RPKM repressed) ranged between –0.1 and –8.7 (Table 3)]. Functional ontology and the assigned InterPro domain of the up- and down-regulated unigenes (30 and 29, respectively) are tabulated in the Supplementary Table 2. We observed induction of unigenes (EnsVib0416 and EnsVib0041), which are functionally associated to heat-shock proteins (IPR008978, HSP20-like chaperone; IPR002068, Heat shock protein Hsp20; Supplementary Table 2). The expression levels of HSPs (IPR008978 and IPR002068) in terms of RPKM for the induced library were 58,328.8 and 12,510.56 (vs. 140.4 and 212.3, for the repressed library respectively), which clearly indicate a strong induction of HSPs in O3-treated plants [log2 fold 8.7 and 5.9, respectively (Table 2)]. The observed results are in accordance with previous observations reported by Eckey-Kaltenbach et al. (1997) in Petroselinum crispum seedlings exposed to O3 (200 ppb, 10 h) and post-cultivated in pollutant-free air, using a Northern blot analysis. Recently, Al-Whaibi (2011) reviewed the role of HSPs demonstrating their function as molecular chaperones, regulating the (i) localization, (ii) degradation, (iii) accumulation, and (iv) folding of proteins during their synthesis. Taking into account these studies, HSPs can be considered as the first line of defense against O3 (Haslbeck and Vierling, 2015).
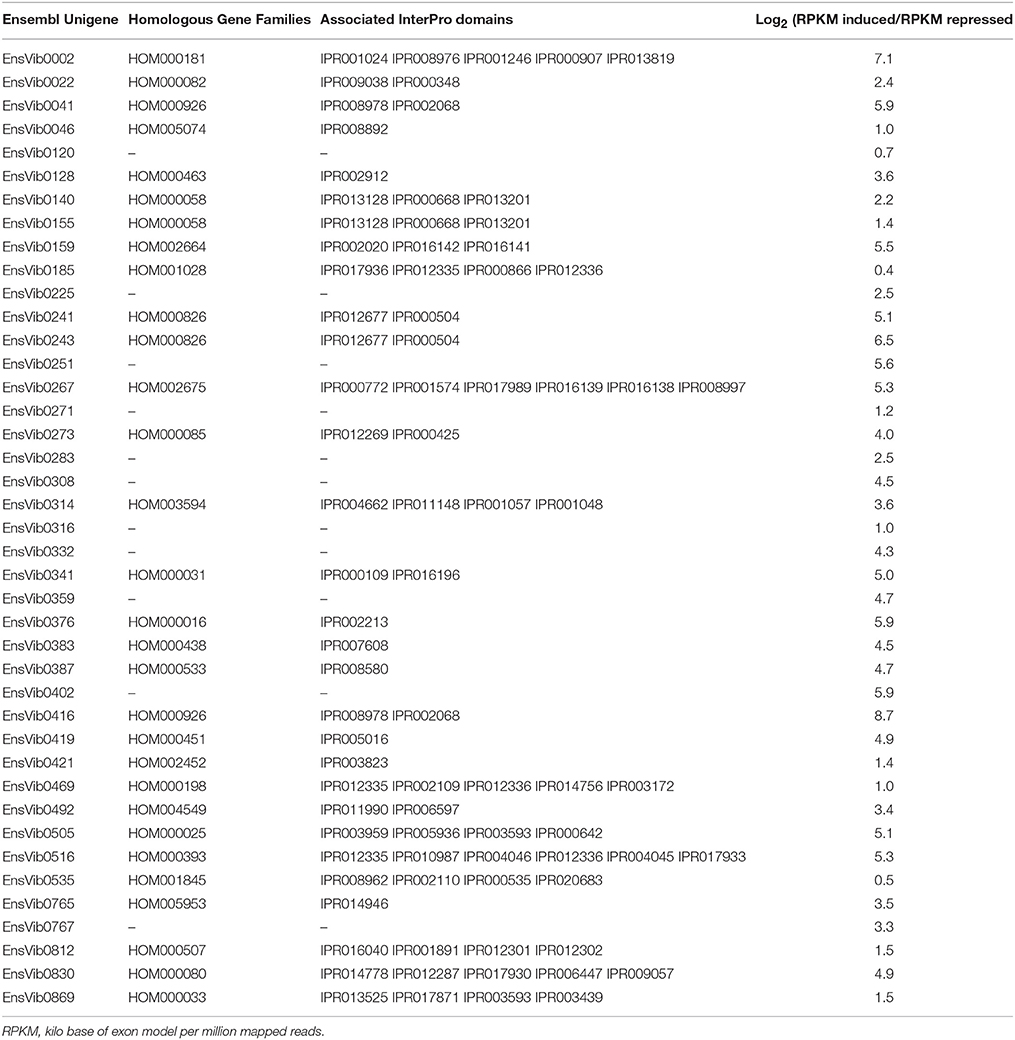
Table 2. Expression log2(RPKM induced/RPKM repressed) of the up-regulated Ensembl unigenes having homologous gene families according to PLAZA version 2.5 and associated Interpro domains.
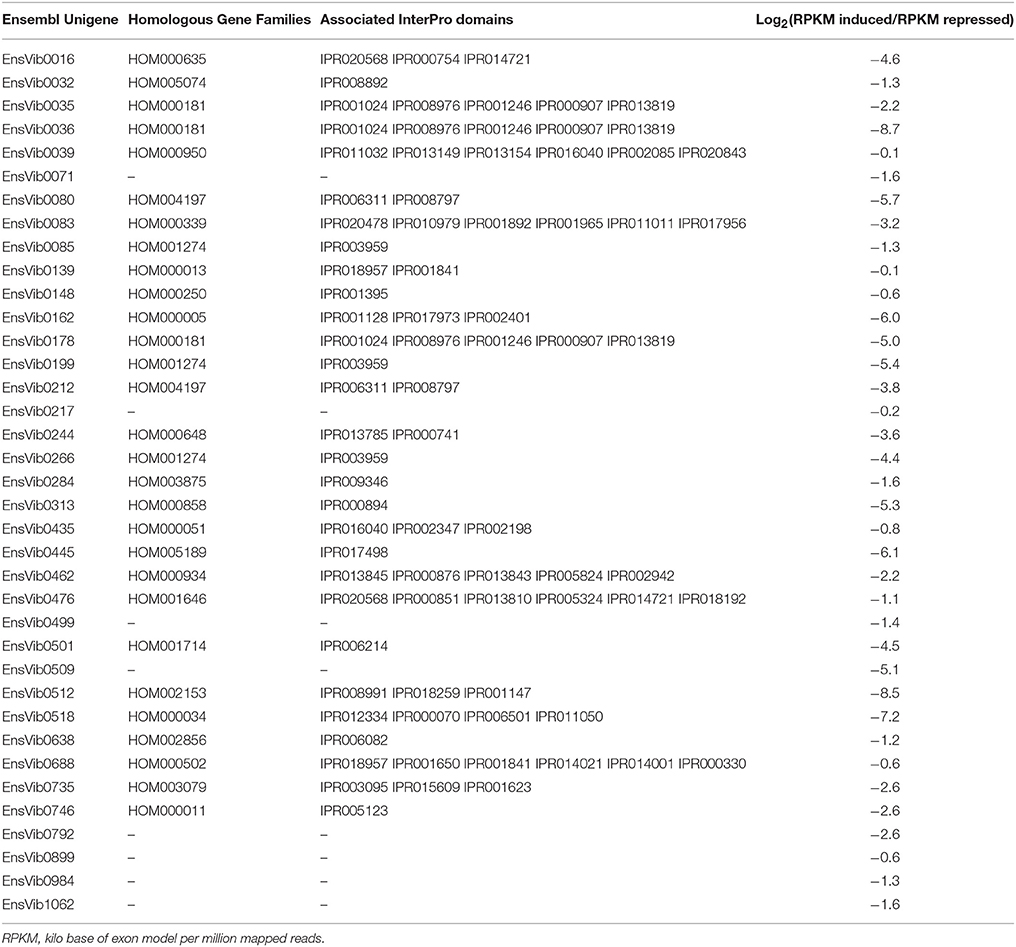
Table 3. Expression log2(RPKM induced/RPKM repressed) of the down-regulated Ensembl unigenes having homologous gene families according to PLAZA version 2.5 and associated Interpro domains.
Using a proteomic approach, Torres et al. (2007) documented that expression levels of HSPs were strongly increased in O3-stressed young leaves of maize (200 ppb, 3 h) and that their functions were correlated with glycolysis, photosynthesis, antioxidant-, and pathogen-related defense. It is worthwhile to mention that HSPs (especially HSP20 superfamily) play a pivotal role in protecting the photosynthetic machinery against damage caused by photo-oxidative stress (Lee et al., 2000). Although this property has been demonstrated during heat stress, we can suppose that the induction of HSPs by O3 treatment could (i) increase the resistance of photosynthetic machinery to photoinhibition, and (ii) affect stress tolerance.
We observed a log2 fold up-regulation (RPKM induced/RPKM repressed = 7.1) of a unigene (EnsVib0002) functionally associated to lipoxygenase activity (GO:0016165, Supplementary Table 2). This is supported by similar findings in Lens culinaris seedlings exposed to O3 flux after 30 min from the beginning of the treatment (Maccarrone et al., 1997). In soybean seedlings, O3 up-regulated the lipoxygenase gene and its activity, with a concomitant enhanced membrane lipid peroxidation (Maccarrone et al., 1992). Recent RNA-seq based transcriptomics indicated an increased expression of genes involved in lipid metabolic process in two soybean varieties exposed to an episodic O3 treatment [25–75 ppb, 4 h (Whaley et al., 2015)]. Plant lipoxygenases, which use molecular oxygen to produce hydroperoxides from unsaturated fatty acids, play a key role in (i) growth and development, (ii) senescence, and (iii) responses to biotic and abiotic stresses. In particular, lipoxygenase activity has been implicated in membrane alteration and could mediate the O3-effect. It is worthwhile to mention that lipid oxidation is a double-edged event due to (i) its damaging effects on lipids and membrane components and (ii) a putative beneficial role in the signaling pathway [e.g., jasmonic acid formation, Vaultier and Jolivet (2015)].
An induction of unigenes (EnsVib0516) functionally associated with glutathione (GSH) was observed with GO GSH binding (GO:0043295), GSH transferase (GO:0004364), and GSH peroxidase activities (GO:0004602, Supplementary Table 2), according with the earlier reports by Tosti et al. (2006) in A. thaliana ecotype Columbia (Col-0) plants exposed to O3 (300 ppb, 6 h) after 3 h from the beginning of the treatment. Antioxidant enzymatic activities were expected to rise during a situation that leaded to increased oxidative stress. Results indicate that GSH-dependent detoxification pathways were induced by O3. Specifically, the expression level of GSH in terms of RPKM for the induced library was 1,198.5 (vs. 312.6 for the repressed library), which suggests an involvement of disulphide bridges in redox-control process. This is in agreement with the results reported by D'Haese et al. (2006) for A. thaliana plants exposed to an episodic O3 treatment (150 ppb, 8 h).
A repression of unigenes (EnsVib0080 and EnsVib0212) functionally associated to photosynthetic process with GO: photosystem (GO:0009521) and photosynthesis (GO:0015979, Supplementary Table 2) was found. The expression levels of photosynthetic process in terms of RPKM for the induced library were 95.8 and 368.2 (vs. 4,909.0 and 5,013.0, for the repressed library respectively), clearly suggesting a decrease in photosynthetic performance in O3-treated plants [log2 (RPKM induced/RPKM repressed) = −5.8 and −3.8, respectively (Table 3)]. Similarly, Kontunen-Soppela et al. (2010) reported a decreased expression of photosynthesis- and carbon fixation-related genes in B. papyrifera plants exposed to O3 alone (2x ambient O3 concentration) or in combination with CO2 (target 550 ppm), during the growing season since 1998. A previous microarray analysis indicated that many genes involved in photosynthesis were down-regulated in Populus tremuloides plants (clone 216) subjected to chronic O3 fumigation (1.5x ambient O3 concentration for 5 consecutive years; Gupta et al., 2005).
Oxidation reactions would be expected to reduce net photosynthesis predisposing plants to the inhibition of PSII electron transport (Pellegrini, 2014) and possibly, accelerating the onset of cell senescence (Pellegrini et al., 2015). In the present study, impaired photosynthesis was seen as a down-regulation of PSII oxygen-evolving complex (OEC) PsbQ genes (regulators for the biogenesis of optically active PSII). According to Gururani et al. (2015), the down-regulation of OEC is an efficient and dynamic feedback mechanism to (i) reduce/regulate the generation of reactive oxygen radicals in PSII (favoring electron donation by non-water electron donors with a high rate constant) and (ii) to provide protection against photodamage in response to abiotic stress. In our case, the easy accessibility of non-water electrons from antioxidant molecules and the concomitant increase of GSH-related genes confirm that plants use a specific set of active mechanisms for ROS scavenging during O3 exposure.
To identify the functional enriched terms and pathways and to approximate the coverage of sequenced and assembled unigenes, we compared the identified 78 unigenes, whose expression altered during the O3 stress in V. lantana to A. thaliana genome predicted coding sequences (CDS) using KOBAS (E-value threshold, 1E-08; Supplementary Figure 1). Interestingly, 45 out of 78 unigenes showed putative functional orthologs in A. thaliana (Table 4). GO enrichment was observed according to the observed functionally enriched linoleate 13S-lipoxygenase activity (GO:0016165, P = 1.86E-07, corrected P = 1.59E-05), which might indicate the conversion of the linoleate into 13-HPODE, responsible for the activation of the lipid peroxidation process. This suggests that O3 treatments may induce deleterious effects on (i) integrity, (ii) conformation, and (iii) transport capacity of membranes in V. lantana, as reported in other species (Yan et al., 2010; Pellegrini et al., 2011). Dynamics of gas exchange results strongly altered as confirmed by the enrichment of GO categories related to thylakoid (GO:0009579, GO:0009535, GO:0055035, GO:0044436, GO:0009534, GO:0031976). It confirms that O3 exposure, by free-radical production, induces alterations in the (i) photosynthetic apparatus, (ii) content/pattern of thylakoid, and (iii) functional state of chloroplast membranes. Particularly, O3-derivative molecular species induce changes in gene expression responsible for rearrangements in the thylakoid architecture of V. lantana leaves limiting the damage of PSII activity (as a compensatory mechanism for the inhibiting photosynthetic effects) and, generally, counteracting the oxidative stress.
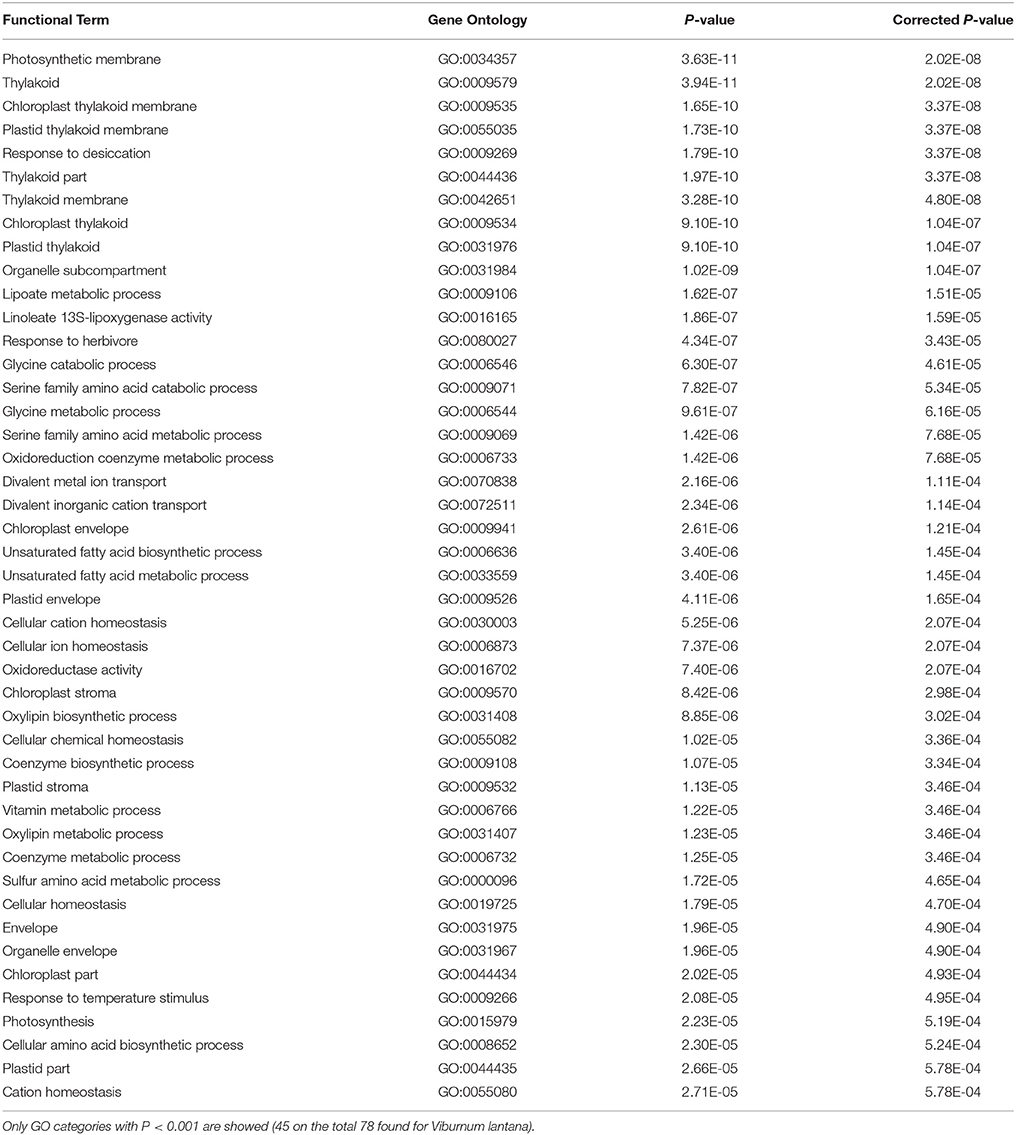
Table 4. Statistical analysis on functional homologs (GO categories) between Viburnum lantana and Arabidopsis thaliana ontology assignment and enrichment analysis using hypergeometric test/Fisher's exact test and corrected values after Benjamini and Hochberg FDR correction.
Using RNA-seq, Liu et al. (2015) indicated the abundance and enrichment of the differentially expressed genes involved in the thylakoid, plastid part, chloroplast, and plastids envelope in Reaumuria soongorica leaves subjected to UV-B radiation. Additionally, previous immunological studies documented that O3 treatment could affect energy transfer processes by inducing alterations in thylakoid membrane proteins (Tognini et al., 1997; Ranieri et al., 2000). Following O3 treatment, a complex sequence of events takes place in the cells of V. lantana (at intracellular and membrane level), altering key biological processes [such as metabolism, protein fate (folding, modification, and destination) and transports] and molecular functions (catalytic and hydrolase activities). This suggests that plants react to O3 changing metabolic processes (for example lipid catabolism, sugar and amino acid metabolism) that are both used (i) directly, as alternative sources of energy, (nitrogen and carbon skeletons) and (ii) indirectly, as substrates of secondary metabolite modifications. According to Heath (2008), we can conclude that O3 induces deep changes in the expression of genes responsible for biochemical adjustments and metabolic shifts.
Thirty-four families of transcription factors were observed for Ensembl unigenes (Supplementary Table 3). Myeloblast (MYB)-factors were previously described to be associated with a diverse array of cellular responses, including plant secondary metabolism as well as biotic and abiotic tolerance (Kwon et al., 2013) and were observed in the induced library. MYB proteins responded at the transcriptional level to O3 stress in V. lantana (EnsVib0607_ORF+3, IPR001005, and IPR009057 domains). MYB transcription factors could have repressing effects on genes involved in the biosynthesis of phenylpropanoids (Bender and Fink, 1998), flavonoids (Borevitz et al., 2000), auxin and consequently have an effect on the maintenance of cell wall development, cuticle formation, and lipid metabolism. Few studies based on the evaluation of plant responses to drought, salt, and UV stress (Hemm et al., 2001; Golldack et al., 2014) have reported the functional role of MYBs.
In our study, WRKY-factors were found, which have been previously reported to be important components in the complex signaling processes during plant stress responses (Dong et al., 2003; Zhang et al., 2015). Far less information is available to understand the function of WRKY proteins in abiotic stress. Some studies demonstrated that the expression of many WRKY genes is greatly and rapidly induced in response to wounding, temperature, nutrient deficiency, drought, and salinity (Chen L. et al., 2012). WRKY proteins respond to O3 stress at the transcriptional level in V. lantana plants (EnsVib0769_ORF-1, IPR003657 domain) and WRKY transcription factors could have inducing effects on genes involved in O3 perception/signal transduction pathway and in redox regulation. This result suggests that WRKY-factors could act as redox-responsive sequences and, consequently, as promoter elements specific for redox regulation (since they possess a redox-sensitive zinc-finger DNA binding domain). Similar findings were reported by Tosti et al. (2006) in Col-0 Arabidopsis plants 3 h after the beginning of O3 treatment. SSH analysis indicated that WRKY genes may be involved in redox regulation in two poplar hybrid clones exposed to an episodic O3 treatment (Rizzo et al., 2007). Similarly, Mahalingam et al. (2003) documented an over-representation of WRKY motifs in the promoter region of genes up-regulated by an episodic O3 exposure (350 ppb, 6 h) in Col-0 Arabidopsis plants. Xu et al. (2015) observed that four WRKY-transcriptional factors genes were highly induced by O3 treatment (350 ppb, 2 h) in Col-0 Arabidopsis plants. Furthermore, the expression gene profile after O3 fumigation was similar to that of tomato after Botrytis cinerea (a fungus) (Journot-Catalino et al., 2006) and Pseudomonas syringae (a bacterium) infections (Birkenbihl et al., 2012), suggesting that (i) O3 resembles a biotic elicitor and (ii) stress-regulated genes represent a general stress response.
This is the first study on differentially expressed genes after O3 treatment in V. lantana plants. A large number of genes involved in signaling/transcription, stress/defense, and protein metabolism showed significant differences in expression of plants exposed to chronic O3 treatment, suggesting that complex molecular alterations occurred. By GO slims and pathways enrichment of the co-regulated genes, it could be demonstrated that following O3 exposure, a complex sequence of events takes place in the cells at intracellular and membrane level, altering a series of biological processes [such as metabolism, protein fate (folding, post-translational modification, and destination) and transport] and molecular functions (catalytic and hydrolase activities). Specifically, the down-regulation of genes associated to photosynthesis demonstrates the deleterious effects of O3. Up-regulation of genes involved in antioxidant-detoxification pathway and thylakoid-localized proteins may be an effective strategy of defense against O3. This research can be considered as an useful basis to (i) generate the functional resources for the putative characterization of identified unigenes in V. lantana and (ii) better understand the response to O3 exposure in a non-model species.
The work presented here was carried out in collaboration among all authors. NL and CN defined the research theme and obtained funding. EG, AC, EP, PB, and GS designed methods, carried out laboratory experiments, and analyzed the data. AC, EP, and GS co-designed experiments, discussed analyses, interpreted the results, and wrote the paper. All authors have contributed to discuss the results and implications of the work and to comment on the manuscript at all stages before approvation.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00713
Supplementary Figure 1. Statistical pathway enrichment of the differentially expressed genes against Arabidopsis thaliana.
Supplementary Table 1. Table showing the GO and associated InterPro annotations with ensembl transcripts showing the up- and down-regulations during the ozone treatment.
Supplementary Table 2. Detailed classification of expression log2(RPKM induced/RPKM repressed) of the up- and down-regulated Ensembl unigenes having homologous gene families according to PLAZA version 2.5 and associated InterPro domains. Abbreviations: RPKM: kilo base of exon model per million mapped reads. Detailed Classification of the associated functional categories associated with the expressed transcripts in ozone treatment.
Supplementary Table 3. Table showing the family and associated InterPro domains of ensembl transcription factors.
Al-Whaibi, M. H. (2011). Plant heat-shock proteins: a mini review. J. King Saud Univ. Sci. 23, 139–150. doi: 10.1016/j.jksus.2010.06.022
Bender, J., and Fink, G. R. (1998). A myb homologue, ATR1, activates tryptophan gene expression in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 95, 5655–5660. doi: 10.1073/pnas.95.10.5655
Birkenbihl, R. P., Diezel, C., and Somssich, I. E. (2012). Arabidopsis WRKY33 is key transcriptional regulator of hormonal and metabolic responses toward Botrytis cinerea infection. Plant Physiol. 159, 266–285. doi: 10.1104/pp.111.192641
Borevitz, J. O., Xia, Y., Blount, J., Dixon, R. A., and Lamb, C. (2000). Activation tagging identifies a conserved myb regulator of phenylpropanoid biosynthesis. Plant Cell 12, 2383–2394. doi: 10.1105/tpc.12.12.2383
Braun, S., Schindler, C., and Rihm, B. (2014). Growth losses in Swiss forests caused by ozone: epidemiological data analysis of stem increment of Fagus sylvatica L. and Picea abies karst. Environ. Pollut. 192, 129–138. doi: 10.1016/j.envpol.2014.05.016
Calatayud, V., Marco, F., Cervero, J., Sanchez-Pena, G., and Jose Sanz, M. (2010). Contrasting ozone sensitivity in related evergreen and deciduous shrubs. Environ. Pollut. 158, 3580–3587. doi: 10.1016/j.envpol.2010.08.013
Cass, G. R., Druzik, J. R., Grosjean, D., Nazaroff, W. W., Whitmore, P. M., and Wittman, C. L. (1989). Protection of Works of Art from Photochemical Smog. Final Report Submitted to the Getty Conservation Institute, Environmental Quality Laboratory, Pasadena, CA, California Institute of Technology.
Chen, L., Song, Y., Li, S., Zhang, L., Zou, C., and Yu, D. (2012). The role of WRKY transcription factors in plant abiotic stresses. Biochim. Biophys. Acta 1819, 120–128. doi: 10.1016/j.bbagrm.2011.09.002
Chen, T. W., Gan, R. C., Wu, T. H., Huang, P. J., Lee, C. Y., Chen, Y. Y., et al. (2012). FastAnnotator: an efficient transcript annotation web tool. BMC Genomics 13:S9. doi: 10.1186/1471-2164-13-S7-S9
Chevreux, B., Pfisterer, T., Drescher, B., Driesel, A. J., Müller, W. E., Wetter, T., et al. (2004). Using the miraEST assembler for reliable and automated mRNA transcript assembly and SNP detection in sequenced ESTs. Genome Res. 14, 1147–1159. doi: 10.1101/gr.1917404
Clement, W. L., and Donoghue, M. J. (2012). Barcoding success as a function of phylogenetic relatedness in Viburnum, a clade of woody angiosperms. BMC Evol. Biol. 12:73. doi: 10.1186/1471-2148-12-73
Cristofanelli, P., and Bonasoni, P. (2009). Background ozone in the southern Europe and Mediterranean area: influence of the transport processes. Environ. Pollut. 157, 1399–1406. doi: 10.1016/j.envpol.2008.09.017
Dai, X., Sinharoy, S., Udvardi, M. K., and Zhao, P. X. (2013). PlantTFcat: an online plant transcription factor and transcriptional regulator categorization and analysis tool. BMC Bioinformatics 14:321. doi: 10.1186/1471-2105-14-321
Dawnay, L., and Mills, G. (2009). Relative effects of elevated background ozone concentrations and peak episodes on senescence and above-ground growth in four populations of Anthoxanthum odoratum L. Environ. Pollut. 157, 503–510. doi: 10.1016/j.envpol.2008.09.003
D'Haese, D., Horemans, N., De Coen, W., and Guisez, Y. (2006). Identification of late O3-responsive genes in Arabidopsis thaliana by cDNA microarray analysis. Physiol. Plant. 128, 70–79. doi: 10.1111/j.1399-3054.2006.00711.x
de Leeuw, F. A. A. M., and van Zantwoort, E. D. G. (1997). Mapping of exceedances of ozone critical levels for crops and forest trees in the Netherlands. Environ. Pollut. 96, 89–98. doi: 10.1016/S0269-7491(96)00141-8
Dong, J., Chen, C., and Chen, Z. (2003). Expression profiles of the Arabidopsis WRKY genes superfamily during plant defense response. Plant Mol. Biol. 51, 21–37. doi: 10.1023/A:1020780022549
Doring, A. S., Pellegrini, E., Campanella, A., Trivellini, A., Gennai, C., Petersen, M., et al. (2014a). How sensitive is Melissa officinalis to realistic ozone concentrations? Plant Physiol. Biochem. 74, 156–164. doi: 10.1016/j.plaphy.2013.11.006
Doring, A. S., Pellegrini, E., Della Bartola, M., Nali, C., Lorenzini, G., and Petersen, M. (2014b). How do backgroung ozone concentrations affect the biosynthesis of rosmarinic acid in Melissa officinalis? J. Plant Physiol. 171, 35–41. doi: 10.1016/j.jplph.2013.11.005
Eckey-Kaltenbach, H., Kiefer, E., Grosskopf, E., Ernst, D., and Sandermann, H. Jr. (1997). Differential transcript induction of parsley pathogenesis-related proteins and of a small shock protein by ozone and heat shock. Plant Mol. Biol. 33, 343–350. doi: 10.1023/A:1005786317975
Francini, A., Nali, C., Pellegrini, E., and Lorenzini, G. (2008). Characterization and isolation of some genes of the shikimate pathway in sensitive and resistant Centaurea jacea plants after ozone exposure. Environ. Pollut. 151, 272–279. doi: 10.1016/j.envpol.2007.07.007
Frei, M. (2015). Breeding of ozone resistant rice: relevance, approaches and challenges. Environ. Pollut. 197, 144–155. doi: 10.1016/j.envpol.2014.12.011
Gambino, G., Perrone, I., and Gribaudo, I. (2008). A rapid and effective method for RNA extraction from different tissues of grapevine and other woody plants. Phytochem. Anal. 19, 520–525. doi: 10.1002/pca.1078
Golldack, D., Li, C., Mohan, H., and Probst, N. (2014). Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front. Plant Sci. 5:151. doi: 10.3389/fpls.2014.00151
González-Fernández, I., Calvo, E., Gerosa, G., Bermejo, V., Marzuoli, R., Calatayud, V., et al. (2014). Setting ozone critical levels for protecting horticultural Mediterranean crops: case study of tomato. Environ. Pollut. 10, 178–187. doi: 10.1016/j.envpol.2013.10.033
Gottardini, E., Cristofolini, F., Cristofori, A., and Ferretti, M. (2014b). Ozone risk and foliar injury on Viburnum lantana L.: a meso-scale epidemiological study. Sci. Total Environ. 493, 954–960. doi: 10.1016/j.scitotenv.2014.06.041
Gottardini, E., Cristofori, A., Cristofolini, F., Bussotti, F., and Ferretti, M. (2010). Responsiveness of Viburnum lantana L. to tropospheric ozone: field evidence under contrasting site conditions in Trentino, northern Italy. J. Environ. Monit. 12, 2237–2243. doi: 10.1039/c0em00299b
Gottardini, E., Cristofori, A., Cristofolini, F., Nali, C., Pellegrini, E., Bussotti, F., et al. (2014a). Chlorophyll-related indicators are linked to visible ozone symptoms: evidence from a field study on native Viburnum lantana L. plants in Northern Italy. Ecol. Indic. 39, 65–74. doi: 10.1016/j.ecolind.2013.11.021
Gouzy, J., Carrere, S., and Schiex, T. (2009). FrameDP: sensitive peptide detection on noisy matured sequences. Bioinformatics 25, 670–671. doi: 10.1093/bioinformatics/btp024
Gupta, P., Duplessis, S., White, H., Karnosky, D. F., Martin, F., and Podila, G. K. (2005). Gene expression patterns of trembling aspen trees following long-term exposure to interacting elevated CO2 and tropospheric O3. New Phytol. 167, 129–142. doi: 10.1111/j.1469-8137.2005.01422.x
Gururani, M. A., Benkatesh, J., and Tran, L. S. P. (2015). Regulation of photosynthesis during abiotic stress-induced photoinhibition. Int. J. Mol. Sci. 16, 19055–19085. doi: 10.3390/ijms160819055
Hao, D. C., Chen, S. L., Osbourn, A., Kontogianni, V. G., Liu, L. W., and Jordán, M. J. (2015). Temporal transcriptome changes induced by methyl jasmonate in Salvia sclarea. Gene 558, 41–53. doi: 10.1016/j.gene.2014.12.043
Haslbeck, M., and Vierling, E. (2015). A first line of stress defence: small heat shock proteins and their function in protein homeostasis. J. Mol. Biol. 427, 1537–1548. doi: 10.1016/j.jmb.2015.02.002
Heath, R. L. (2008). Modification of the biochemical pathways of plants induced by ozone: what are the varied routes to change? Environ. Pollut. 155, 453–463. doi: 10.1016/j.envpol.2008.03.010
Hemm, M. R., Herrmann, K. M., and Chapple, C. (2001). AtMYB4: a transcription factor general in the battle against UV. Trends Plant. Sci. 6, 135–136. doi: 10.1016/S1360-1385(01)01915-X
Hirayama, T., and Shinozaki, K. (2010). Research on plant abiotic stress responses in the post-genome era: past, present and future. Plant J. 61, 1041–1052. doi: 10.1111/j.1365-313X.2010.04124.x
Huang, X., and Madan, A. (1999). CAP3: A DNA sequence assembly program. Genome Res. 9, 868–877. doi: 10.1101/gr.9.9.868
Jaspers, P., Kollist, H., Langebartels, C., and Kangasjärvi, J. (2005). “Plant responses to ozone”, in Antioxidants and Reactive Oxygen Species in Plants, ed N. Smirnoff (Oxford: Blackwell), 268–292.
Jenkin, M., and Clemitshaw, K. (2000). Ozone and other secondary photochemical pollutants: chemical processes governing their formation in the planetary boundary layer. Atmos. Environ. 34, 2499–2527. doi: 10.1016/S1352-2310(99)00478-1
Journot-Catalino, N., Somssich, I. E., Roby, D., and Kroj, T. (2006). The transcription factors WRKY11 and WRKY17 act as negative regulators of basal resistance in Arabidopsis thaliana. Plant Cell 18, 3289–3302. doi: 10.1105/tpc.106.044149
Kanter, U., Heller, W., Durner, J., Winkler, J. B., Engel, M., Behrendt, H., et al. (2013). Molecular and immunological characterization of ragweed (Ambrosia artemisiifolia L.) pollen after exposure of the plants to elevated ozone over a whole growing season. PLoS ONE 8:e61518. doi: 10.1371/journal.pone.0061518
Kerner, R., Delgado-Eckert, E., Ernst, D., Dupuy, J.-W., Grams, T. E. E., Winkler, J. B., et al. (2014). Large-scale protein analysis of European beech trees following four vegetation periods of twice ambient ozone exposure. J. Proteomics 109, 417–435. doi: 10.1016/j.jprot.2014.05.021
Kontunen-Soppela, S., Parviainen, J., Ruhanen, H., Brosché, M., Keinänen, M., Thakur, R. C., et al. (2010). Gene expression responses of paper birch (Betula papyrifera) to elevated CO2 and O3 during leaf maturation and senescence. Environ. Pollut. 158, 959–968. doi: 10.1016/j.envpol.2009.10.008
Kwon, Y., Kim, J. H., Nguyen, H. N., Jikumaru, Y., Kamiya, Y., Hong, S.-W., et al. (2013). A novel Arabidopsis MYB-like transcription factor, MYBH, regulates hypocotyl elongation by enhancing auxin accumulation. J. Exp. Bot. 64, 3911–3922. doi: 10.1093/jxb/ert223
Lee, B. H., Won, S. H., Lee, H. S., Miyao, M., Chung, W. I., Kim, I. J., et al. (2000). Expression of the chloroplast-localized small heat shock protein by oxidative stress in rice. Gene 245, 283–290. doi: 10.1016/S0378-1119(00)00043-3
Legrand, S., Hendriks, T., Hilbert, J. L., and Quillet, M. C. (2007). Characterization of expressed sequence tags obtained by SSH during somatic embryogenesis in Cichorium intybus L. BMC Plant Biol. 7:27. doi: 10.1186/1471-2229-7-27
Legrand, S., Valot, N., Nicolé, F., Moja, S., Baudino, S., Jullien, F., et al. (2010). One-step identification of conserved miRNAs, their targets, potential transcription factors and effector genes of complete secondary metabolism pathways after 454 pyrosequencing of calyx cDNAs form the Labiate Salvia sclarea L. Gene 450, 55–62. doi: 10.1016/j.gene.2009.10.004
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows-wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Liu, M., Li, X., Liu, Y., Shi, Y., and Ma, X. (2015). Analysis of differentially expressed genes under UV-B radiation in the desert plant Reaumuria soongorica. Gene 15, 265–272. doi: 10.1016/j.gene.2015.08.026
Lorenzini, G., Medeghini Bonatti, P., Nali, C., and Baroni Fornasiero, R. (1994). The protective effect of rust infection against ozone, sulphur dioxide and paraquat toxicity symptoms in broad bean. Physiol. Mol. Plant Path. 45, 263–279. doi: 10.1016/S0885-5765(05)80058-X
Maccarrone, M., Veldink, G. A., and Vliegenthart, F. G. (1992). Thermal injury and ozone stress affect soybean lipoxygenases expression. FEBS Lett. 309, 225–230. doi: 10.1016/0014-5793(92)80778-F
Maccarrone, M., Veldink, G. A., Vliegenthart, F. G., and Agrò, A. F. (1997). Ozone stress modulates amine oxidase and lipoxygenase expression in lentil (Lens culinaris) seedlings. FEBS Lett. 408, 241–244. doi: 10.1016/S0014-5793(97)00433-X
Mahalingam, R., Gomez-Buitrago, A., Eckardt, N., Shah, N., Guevara-Garcia, A., Day, P., et al. (2003). Characterizing the stress/defense transcriptome of Arabidopsis. Genome Biol. 4:R20. doi: 10.1186/gb-2003-4-3-r20
Manninen, S., Huttunen, S., Vanhatalo, M., Pakonen, T., and Hämäläinen, A. (2009). Inter- and intra-specific responses to elevated ozone and chamber climate in northern birches. Environ. Pollut. 157, 1679–1688. doi: 10.1016/j.envpol.2008.12.008
Mao, X., Cai, T., Olyarchuk, J. G., and Wei, L. (2005). Automated genome annotation and pathway identification using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 21, 3787–3793. doi: 10.1093/bioinformatics/bti430
Millàn, M. M., Mantilla, E., Salvador, R., Carratalà, A., and Sanz, M. J. (2000). Ozone cycles in the Western Mediterranean basin: interpretation of monitoring data in complex coastal terrain. J. Appl. Meteorol. 39, 487–508. doi: 10.1175/1520-0450(2000)039<0487:OCITWM>2.0.CO;2
Miller, P. (2011). The long and short of it: a national ozone standard for farm and forest. Environ. Manag. Air Waste Manag. 6–9.
Monks, P. S., Archibald, A. T., Colette, A., Cooper, O., Coyle, M., Derwent, R., et al. (2015). Tropospheric ozone and its precursors from the urban to the global scale from air quality to short-lived climate forcer. Atmos. Chem. Phys. 15, 8889–8973. doi: 10.5194/acp-15-8889-2015
Nathaniel, R. S., Tallis, M. J., Tucker, J., Brosché, M., Kangasjärvi, J., Broadmeadow, M., et al. (2011). The physiological, transcriptional and genetic responses of an ozone- sensitive and an ozone tolerant poplar and selected extremes of their F2 progeny. Environ. Pollut. 159, 45–54. doi: 10.1016/j.envpol.2010.09.027
Norval, M., Lucas, R. M., Cullen, A. P., de Gruijl, F. R., Longstreth, J., Takizawa, Y., et al. (2011). The human health effects of ozone depletion and interactions with climate change. Photochem. Photobiol. Sci. 10, 199–225. doi: 10.1039/c0pp90044c
Novak, K., Schaub, M., Fuhrer, J., Skelly, J. M., Frey, B., and Kräuchi, N. (2008). Ozone effects on visible foliar injury and growth of Fagus sylvatica and Viburnum lantana seedlings grown in monoculture or in mixture. Environ. Exp. Bot. 62, 212–200. doi: 10.1016/j.envexpbot.2007.08.008
Novak, K., Schaub, M., Fuhrer, J., Skelly, J. M., Hug, C., Landolt, W., et al. (2005). Seasonal trends in reduced leaf gas exchange and ozone-induced foliar injury in three ozone sensitive woody plant species. Environ. Pollut. 136, 33–45. doi: 10.1016/j.envpol.2004.12.018
Novak, K., Skelly, J. M., Schaub, M., Kräuchi, N., Hug, C., Landolt, W., et al. (2003). Ozone air pollution and foliar injury development on native plants of Switzerland. Environ. Pollut. 125, 41–52. doi: 10.1016/S0269-7491(03)00085-X
Olbrich, M., Gerstner, E., Bahnweg, G., Häberle, K.-H., Welzl, G., Heller, W., et al. (2010). Transcriptional signatures in leaves of adult European beech trees (Fagus sylvatica L.) in an experimentally enhanced free air ozone setting. Environ. Pollut. 158, 977–982. doi: 10.1016/j.envpol.2009.08.001
Pellegrini, E. (2014). PSII photochemistry is the primary target of oxidative stress imposed by ozone in Tilia americana. Urban For. Urban Green. 13, 94–102. doi: 10.1016/j.ufug.2013.10.006
Pellegrini, E., Francini, A., Lorenzini, G., and Nali, C. (2011). PSII photochemistry and carboxylation efficiency in Liriodendron tulipifera under ozone exposure. Environ. Exp. Bot. 70, 217–226. doi: 10.1016/j.envexpbot.2010.09.012
Pellegrini, E., Francini, A., Lorenzini, G., and Nali, C. (2015). Ecophysiological and antioxidant traits of Salvia officinalis under ozone stress. Environ. Sci. Pollut. Res. 22, 13083–13093. doi: 10.1007/s11356-015-4569-5
Puckette, M., Iyer, N. J., Tanh, Y., Dai, X.-B., Zhao, P., and Mahalingam, R. (2012). Differential mRNA translation in Medicago truncatula accessions with contrasting responses to ozone-induced oxidative stress. Mol. Plant 5, 187–204. doi: 10.1093/mp/ssr069
Puckette, M., Peal, L., Steele, J., Tang, Y., and Mahalingam, R. (2009). Ozone responsive genes in Medicago truncatula: analysis by suppression subtraction hybridization. J. Plant Physiol. 166, 1284–1295. doi: 10.1016/j.jplph.2009.01.009
Ranieri, A., Serini, R., Castagna, A., Nali, C., Baldan, B., Lorenzini, G., et al. (2000). Differential sensitivity to ozone in two poplar clones. Analysis of thylakoid pigment-protein complexes. Physiol. Plant. 110, 181–188. doi: 10.1034/j.1399-3054.2000.110206.x
Rizzo, M., Bernardi, R., Salvini, M., Nali, C., Lorenzini, G., and Durante, M. (2007). Identification of differentially expressed genes induced by ozone stress in sensitive and tolerant poplar hybrids. J. Plant Physiol. 164, 945–949. doi: 10.1016/j.jplph.2006.07.012
Sarkar, A., Singh, A. A., Agrawal, S. B., Ahmad, A., and Rai, S. P. (2015). Cultivar specific variations in antioxidative defense system, genome and proteome of two tropical rice cultivars against ambient and elevated ozone. Ecotox. Environ. Safe. 115, 101–111. doi: 10.1016/j.ecoenv.2015.02.010
Savenstrand, H., Brosche, M., Angehagen, M., and Strid, A. (2000). Molecular markers for ozone stress isolated by suppression subtractive hybridization: specificity of gene expression and identification of a novel stress-regulated gene. Plant Cell Environ. 23, 689–700. doi: 10.1046/j.1365-3040.2000.00586.x
Tognini, M., Ranieri, A., Castagna, A., Nali, C., Lorenzini, G., and Soldatini, G. F. (1997). Ozone-induced alterations in thylakoid protein patterns in pumpkin leaves of different age. Phyton 37, 277–282.
Torres, N. L., Cho, K., Shibato, J., Hirano, M., Kubo, A., Masuo, Y., et al. (2007). Gel-based proteomics reveals potential novel protein markers of ozone stress in leaves of cultivated bean and maize species of Panama. Electrophoresis 28, 4369–4381. doi: 10.1002/elps.200700219
Tosti, N., Pasqualini, S., Borgogni, A., Ederli, L., Falistocco, E., Crispi, S., et al. (2006). Gene expression profiles of O3-treated Arabidopsis plants. Plant Cell Environ. 29, 1686–1702. doi: 10.1111/j.1365-3040.2006.01542.x
Tuomainen, J., Pellinen, R., Roy, S., Kiiskinen, M., Eloranta, R., Karjalainen, R., et al. (1996). Ozone affects birch (Betula pendula Roth) phenylpropanoid, polyamine and active oxygen detoxifying pathways at biochemical and gene expression level. J. Plant Physiol. 148, 179–188. doi: 10.1016/S0176-1617(96)80312-9
Vainonen, J. P., and Kangasjärvi, J. (2015). Plant signalling in acute ozone exposure. Plant Cell Environ. 38, 240–252. doi: 10.1111/pce.12273
Van Bel, M., Proost, S., Van Neste, C., Deforce, D., Van de Peer, Y., and Vandepoele, K. (2013). TRAPID: an efficient online tool for the functional and comparative analysis of de novo RNA-Seq transcriptomes. Genome Biol. 14:R134. doi: 10.1186/gb-2013-14-12-r134
Vaultier, M. N., and Jolivet, Y. (2015). Ozone sensing and early signaling in plants: an outline from the cloud. Environ. Exp. Bot. 114, 144–152. doi: 10.1016/j.envexpbot.2014.11.012
Whaley, A., Sheridan, J., Safari, S., Burton, A., Burkey, K., and Schlueter, J. (2015). RNA-seq analysis reveals genetic response and tolerance mechanisms to ozone exposure in soybean. BMC Genomics 16:7. doi: 10.1186/s12864-015-1637-7
Wu, J., Mao, X., Cai, T., Luo, J., and Wei, L. (2006). KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 34, 720–724. doi: 10.1093/nar/gkl167
Xie, C., Mao, X., Huang, J., Ding, Y., Wu, J., Dong, S., et al. (2011). KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, 316–322. doi: 10.1093/nar/gkr483
Xu, E., Vaahtera, L., and Brosché, M. (2015). The role of defense hormones in regulation of ozone-induced changes in gene expression and cell death. Mol. Plant 8, 1776–1794. doi: 10.1016/j.molp.2015.08.008
Yan, K., Chen, W., He, X., Zhang, G., Xu, S., and Wang, L. (2010). Responses of photosynthesis, lipid peroxidation and antioxidant system in leaves of Quercus mongolica to elevated O3. Environ. Exp. Bot. 69, 198–204. doi: 10.1016/j.envexpbot.2010.03.008
Yang, W., and Omaye, S. T. (2009). Air pollutants, oxidative stress and human health. Mutat. Res. 674, 45–54. doi: 10.1016/j.mrgentox.2008.10.005
Zhang, L., Gu, L., Ringler, P., Smith, S., Rushton, P. J., and Shen, Q. J. (2015). Three WRKY transcription factors additively repress abscisic acid and gibberellin signaling in aleurone cells. Plant Sci. 236, 214–222. doi: 10.1016/j.plantsci.2015.04.014
Zinser, C., Jungblut, T., Heller, W., Seidlitz, H. K., Schnitzler, J.-P., Ernst, D., et al. (2000). The effect of ozone in Scots pine (Pinus sylvestris L.): gene expression, biochemical changes and interactions with UV-B radiation. Plant Cell Environ. 23, 975–982. doi: 10.1046/j.1365-3040.2000.00613.x
Keywords: SSH, gene ontology, photosynthesis, detoxification, HSP20-like chaperone, PCR-select™, lipoxygenase activity
Citation: Gottardini E, Cristofori A, Pellegrini E, La Porta N, Nali C, Baldi P and Sablok G (2016) Suppression Substractive Hybridization and NGS Reveal Differential Transcriptome Expression Profiles in Wayfaring Tree (Viburnum lantana L.) Treated with Ozone. Front. Plant Sci. 7:713. doi: 10.3389/fpls.2016.00713
Received: 18 December 2015; Accepted: 09 May 2016;
Published: 01 June 2016.
Edited by:
Mohammad Anwar Hossain, Bangladesh Agricultural University, BangladeshReviewed by:
Golam Jalal Ahammed, Zhejiang University, ChinaCopyright © 2016 Gottardini, Cristofori, Pellegrini, La Porta, Nali, Baldi and Sablok. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elisa Pellegrini, ZWxpc2EucGVsbGVncmluaUBmb3IudW5pcGkuaXQ=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.