
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 27 May 2016
Sec. Plant Physiology
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.00695
This article is part of the Research Topic Plant responses to biotic and abiotic stresses: lessons from cell signaling View all 24 articles
 Lingmin Dai1,2,3
Lingmin Dai1,2,3 Dan Wang1,2,3
Dan Wang1,2,3 Xiaoqing Xie1,2,3
Xiaoqing Xie1,2,3 Chaohong Zhang1,2,3
Chaohong Zhang1,2,3 Xiping Wang1,2,3
Xiping Wang1,2,3 Yan Xu1,2,3
Yan Xu1,2,3 Yuejin Wang1,2,3*
Yuejin Wang1,2,3* Jianxia Zhang1,2,3*
Jianxia Zhang1,2,3*Pathogenesis-related proteins (PRs) can lead to increased resistance of the whole plant to pathogen attack. Here, we isolate and characterize a PR-4 protein (VpPR4-1) from a wild Chinese grape Vitis pseudoreticulata which shows greatly elevated transcription following powdery mildew infection. Its expression profiles under a number of abiotic stresses were also investigated. Powdery mildew, salicylic acid, and jasmonic acid methyl ester significantly increased the VpPR4-1 induction while NaCl and heat treatments just slightly induced VpPR4-1 expression. Abscisic acid and cold treatment slightly affected the expression level of VpPR4-1. The VpPR4-1 gene was overexpressed in 30 regenerated V. vinifera cv. Red Globe via Agrobacterium tumefaciens-mediated transformation and verified by the Western blot. The 26 transgenic grapevines exhibited higher expression levels of PR-4 protein content than wild-type vines and six of them were inoculated with powdery mildew which showed that the growth of powdery mildew was repressed. The powdery mildew-resistance of Red Globe transformed with VpPR4-1 was enhanced inoculated with powdery mildew. Moreover, other powdery mildew resistant genes were associated with feedback regulation since VpPR4-1 is in abundance. This study demonstrates that PR-4 protein in grapes plays a vital role in defense against powdery mildew invasion.
Pathogenesis-related proteins (PRs) are involved in higher-plant responses to environment stress. They can be triggered by a large amount of pathogens including fungi, bacteria, and viruses (Van Loon, 1985; Lamb et al., 1989; Linthorst and Van Loon, 1991). Pathogenesis-related proteins were first detected in tobacco leaves under tobacco mosaic virus attack (Van Loon and Van Kammen, 1970). A large number of PRs have since been isolated from a range of other plant species (Sels et al., 2008). Currently, 17 families of PRs have been classified, based on their amino acid sequence similarities, enzymatic activities and other biological properties. These families have been numbered in sequence of discovery (Sels et al., 2008; Sinha et al., 2014). Usually, PRs possess enzymatic functions – so, for example, PR-2 is a 1,3-β-glucanase (Kauffmann et al., 1987), PR-7 has endoproteinase activity, PR-9 is a peroxidase, and PR-10 is a ribonuclease (Lagrimini et al., 1987; Vera and Conejero, 1988; Park et al., 2004; Xu et al., 2010). Moreover, a number of them, e.g., PR-1 and PR-6, have been identified as having antifungal and proteinase inhibitory properties (Green and Ryan, 1972; Niderman et al., 1995). In addition, chitinase activity has been detected in PR-3, PR-4, PR-8, and PR-11 (Van Loon, 1982; Métraux et al., 1988; Melchers et al., 1994), with PR-4 being referred to as chitin-binding proteins.
The PR-4 proteins were first described as wound-inducible proteins and were termed win-1 and win-2 in potato (Stanford et al., 1989; Friedrich et al., 1991). Since then, several PR-4 proteins have been isolated and characterized. PR-4 is not only induced by wounding but also by ethanol, abscisic acid (ABA), salicylic acid (SA), 2,6-dichloroisonicotinic acid, methyl jasmonate (MeJA), sugar starvation, viruses, and fungi (Broekaert et al., 1990; Boll, 1991; Friedrich et al., 1991; Linthorst and Van Loon, 1991; Potter et al., 1993; Melchers et al., 1994; Chevalier et al., 1995; Gregersen et al., 1997; Lee et al., 2001; Bravo et al., 2003; Hamada et al., 2005; Park et al., 2005; Guevara-Morato et al., 2010). The PR-4 proteins possess a Barwin domain in the C-terminal which comes from a barley seed protein related to the wound-induced proteins (Svensson et al., 1992). The secondary structure analysis of the barley seed protein shows the protein contains a large four-stranded antiparallel β-sheet and a small parallel β-sheet (Ludvigsen and Poulsen, 1992a). The binding site of the Barwin domain to the tetramer N-acetyl glucosamine was investigated by three-dimensional structural analysis (Ludvigsen and Poulsen, 1992b). The PR-4 proteins are classified into classes I and II based on whether they have an N-terminal Hevein domain or not (Neuhaus et al., 1996). Wheatwin1 and wheatwin2 belong to class II of the PR-4 proteins and show antifungal activity by inhibiting fungal hyphal growth (Caruso et al., 1996, 2001; Huet et al., 2013). Furthermore, wheatwin1 is able to digest RNA from wheat and Fusarium culmorum (Caporale et al., 2004). The ribonuclease activity of wheat PR-4 proteins contributes to their antifungal activity (Bertini et al., 2009). In addition, a PR-4 in Capsicum chinense has been shown to exhibit deoxyribonuclease and ribonuclease in in vitro assays (Guevara-Morato et al., 2010). AtHEL, a class I PR-4 protein of Arabidopsis, was found to interact with a fungal lectin having deoxyribonuclease and ribonuclease activities (Bertini et al., 2012; Huet et al., 2013).
Grape production is principally of Vitis vinifera cultivars and is also principally geared toward winemaking. Fungal disease is a critical factor in this major international industry, leading to heavy financial penalties due to reductions in both fruit yield and in fruit quality. When infected by fungi, a number of grape PRs are induced, including PR-4 (Kortekamp, 2006). V. pseudoreticulata accession Baihe 35-1 is a distinct accession of Chinese wild grape, which possesses high resistance to Erysiphe necator and powdery mildew-induced genes had been isolated from the cDNA library of the high powdery mildew resistant Baihe-35-1 inoculated by Erysiphe necator (Yu et al., 2013). Moreover, VpPR4 is different from the powdery mildew resistance gene of V. pseudoreticulata that we have already studied before (Yu et al., 2013; Dai et al., 2015) or other PR4 gene from similar species in sequence. Powdery mildew can induce abundant expression of PR-4 in V. vinifera (Fung et al., 2008). However, the expression of VpPR4 gene was significant higher than the PR4 detected in the European grapevine control by cDNA library analyzing in our previous study (Zhu et al., 2012) which suggested that the characterization and expression of VpPR4 is different compared with VvPR4. Moreover, the detailed function of PR-4 proteins in grape is unclear.
This study describes the detection and characterization of a PR-4 protein induced by powdery mildew (Erysiphe necator [Schw.] Burr) in the leaves of V. pseudoreticulata Baihe-35-1, and this gene expression profile following exposure to a range of plant hormones and abiotic stresses. The PR-4 gene was overexpressed in the powdery mildew susceptible variety Red Globe which shows enhanced resistance to powdery mildew and disarranged expression patterns of related defense genes.
Grapevines (wild Chinese V. pseudoreticulata accession Baihe 35-1) were propagated by tissue culture and plantlets were transplanted to pots grown in a culture room (Yu et al., 2013). The inoculation of vine leaves with powdery mildew was carried out as previously described (Guan et al., 2011). The young leaves were inoculated by touching the adaxial surface of the leaves with PM cv. NAFU1 (KJ539202) colonies maintained on the greenhouse-grown grapes. Young grapevine plantlets (V. pseudoreticulata Baihe 35-1) 20–25 cm in height were selected for hormone treatments and the leaves were allowed to expand in a greenhouse. Treatments with 100 μM SA, 50 μM MeJA, or 100 μM ABA were imposed by spraying these onto the fully expanded leaves. The solution of SA was 100 μM in distilled water (Wang and Li, 2006), solution of MeJA was 50 μM in 0.1% ethanol (Repka et al., 2004), and the solution of ABA was 100 μM in distilled water (Seong et al., 2007). For the environmental treatments, grapevine plantlets (V. pseudoreticulata Baihe 35-1) were incubated in 4°C (cold) or 40°C (hot). For NaCl treatment, the grapevines in pots were watered with 500 mM NaCl (Yang et al., 2012).
The well-known PR-4s were used to blast search the grape genome on http://www.genoscope.cns.fr/externe/GenomeBrowser/Vitis/. Three homologous sequences were isolated from the grape genome. One was a pseudogene and the other two were used as templates for homology-based cloning of PR-4s from V. pseudoreticulata. The amplified primers are listed in Table 1.
Protein sequences of VpPR4-1 and other well-known PR-4s were aligned by DNAman using default parameters. The phylogenetic tree was constructed by Neighbor-joining method using Mega 5.0 software (Tamura et al., 2011). Bootstrap analysis was carried out using 1000 replicates.
A modified SDS/phenol method was used to extract total RNA (Reid et al., 2006). First-strand cDNA synthesis and quantitative RT-PCR were carried out as Hou et al. (2013). The grape GAPDH gene (GenBank accession No. EF192466) was amplified as a normalized control. According to Dai et al. (2015), gene expression analysis was carried out by qRT-PCR with a Bio-Rad IQ5 Real-Time PCR Detection System (Hercules, CA, USA) using TaKaRa SYBR Premix Ex TaqTM II (Perfect Real Time). The qRT-PCR reaction was conducted in triplicate following parameters: 95°C for 10 s; 40 cycles of 94°C for 5 s and 60°C for 30 s. The normalized fold expression of RNA was calculated by comparison with the normalized control.
Binary vector construction was carried out as Yu et al. (2013). The VpPR4-1 gene was PCR cloned in-frame into plasmid pART-CAM-S (Xu et al., 2014) using Xho I and Xba I restriction enzymes to generate 35S::VpPR4-1, which contained a kanamycin resistance selective marker. The binary construct was introduced into Agrobacterium strain GV3101 using electroporation.
Proembryonic masses of V. vinifera L. Red Globe, initiated from immature stamens and maintained from a previous study, were used for genetic transformation with VpPR4-1. Transformation was carried out via the Agrobacterium-mediated transformation system as described previously (Zhou et al., 2014; Dai et al., 2015).
Total protein was extracted using phenol-based protocols as described previously (Vincent et al., 2006). The protein was fractionated by 5–10% SDS-PAGE and blotted to polyvinyl difluoride membranes (Roche) using a semi-dry blot apparatus as described by the manufacturer (Bio-Rad). The purified polyclonal antisera antibody of VpPR4-1 protein obtained from New Zealand rabbits immunized by purification of the prokaryotic expressed VpPR4-1 protein in our previously study (data not shown), and the polyclonal antisera was used at 1:2,000 dilution and secondary goat anti-rabbit IgG (TransGen Biotech) conjugated with alkaline phosphatase at 1:5,000 dilution. Detection was carried out using Pierce ECL Western Blotting Substrate (Thermo scientific) and imaged with Image Lab Software (Bio-Rad).
The trypan blue staining is used for observing powdery mildew hyphal development and carried out according to Xiao et al. (2003) with minor modification. Briefly, grape leaves were picked and stained with the Trypan blue staining solution in a boiling water bath for 15 min and then allowed to rest at room temperature for 24 h. The samples were then incubated in chloral hydrate solution for 2–6 h at room temperature with the chloral hydrate saturated solution changed once and then incubated for a further 8 h at room temperature. The chloral hydrate solution was then removed and the tissue immersed in 70% glycerol and examined using a compound light microscope.
Experimental results in Figures 2, 5, and 7 are mean values of triplicate experiments; error bars in Figures 2 and 5 indicate standard deviations (SD). Mean values and SD were calculated using SPASS 18.0 software (SPSS Inc, Chicago, IL, USA).
The full-length cDNA of VpPR4-1 was homologous cloned from the cDNA library of high powdery mildew resistant V. pseudoreticulata Baihe 35-1 inoculated by powdery mildew (Zhu et al., 2012). The VpPR4-1 gene is on the chromosome 14 of the whole genome of PN40024 (Jaillon et al., 2007), it is 432 base pairs (bp) long and codes for a unique polypeptide of 143 amino acids with two exons (Figure 1A) and possesses a Barwin domain (Figure 1B).
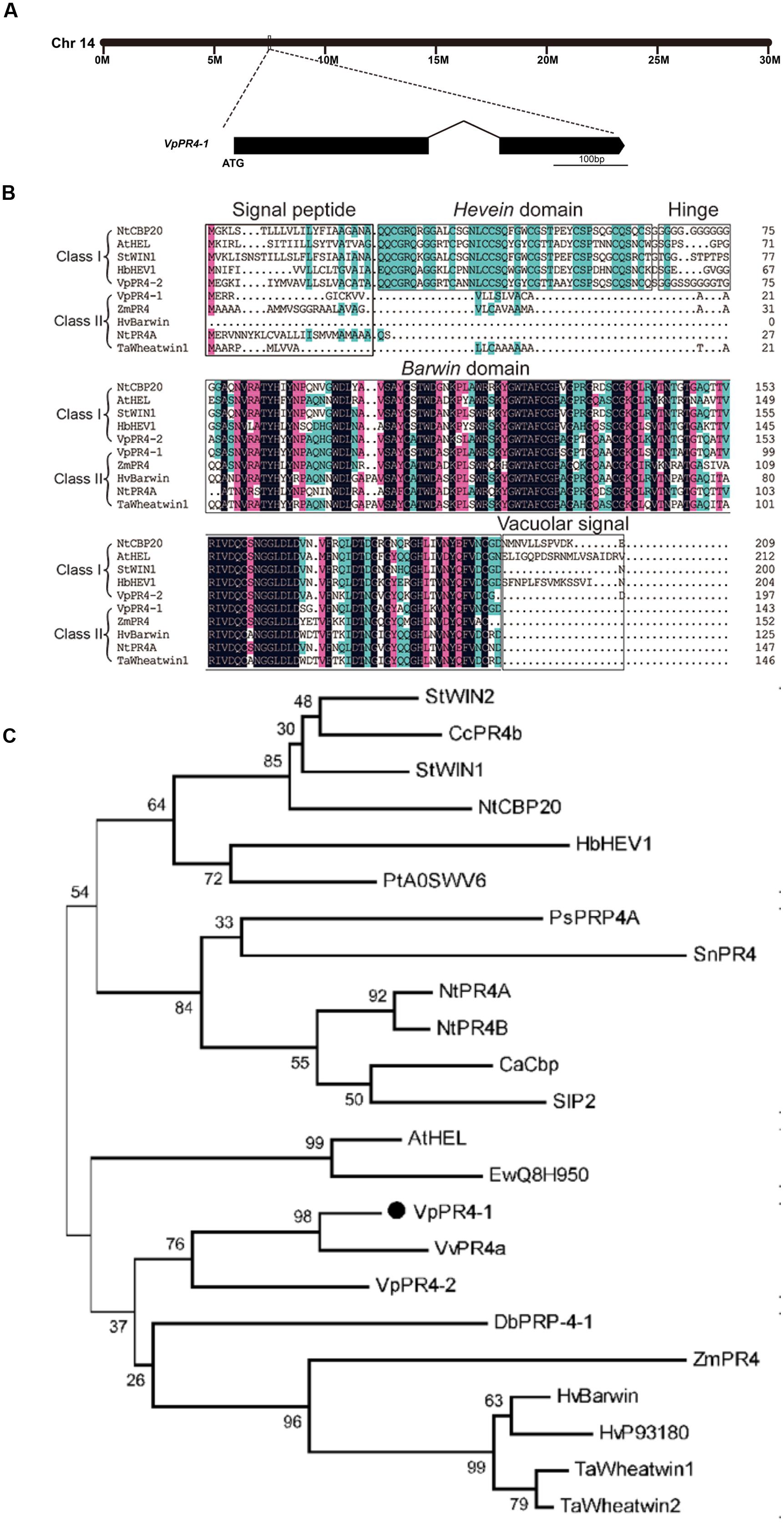
FIGURE 1. Phylogenetic analysis and alignment of PR-4 proteins. (A) Location on the chromosome and schematic representation of the VpPR4-1 gene. (B) Sequence alignment of VpPR4-1. The conserved domains are shown above the sequences. Identical amino acids are shaded with black, and similar amino acids are shaded with pink (<100 and ≥75%) or blue (<75 and ≥50%). (C) Phylogenetic analysis of various PR-4-1 proteins. Class I, subgroup of PR-4 proteins which contain a Hevein domain. Class II, subgroup of PR-4s which do not have a Hevein domain. Accession numbers are: VpPR4-1 (AEW12795; Vp, Vitis pseudoreticulata), VpPR4-2 (KP274873), StWIN2 (NP_001275628; St, Solanum tuberosum), CcPR4b (BAD11073; Cc, Capsicum chinense), StWIN1 (XP_006347743), NtCBP20 (AAB29960; Nt, Nicotiana tabacum), HbHEV1 (P02877; Hb, Hevea brasiliensis), PtA0SWV6 (ABK63195; Pt, Populus tremula × Populus alba), PsPRP4A (AAF61434; Ps, Pisum sativum), SnPR4 (CAA87070; Sn, Sambucus nigra), NtPR4A (XP_009763689), NtPR4B (XP_009614804), CaCbp (AFN21550; Capsicum annuum), SlP2 (NP_001234083; Sl, Solanum lycopersicum), AtHEL (NP_187123; At, Arabidopsis thaliana), EwQ8H950 (BAC16357; Ew, Eutrema wasabi), VvPR4a (AAC33732; Vv, Vitis vinifera), DbPRP-4-1 (AAB94514; Db, Dioscorea bulbifera), ZmPR4 (AFW60484; Zm, Zea mays), HvBarwin (BAK04328; Hv, Hordeum vulgare), HvP93180 (CAA71774), TaWheatwin1 (O64392; Ta, Triticum aestivum), and TaWheatwin2 (O64393).
Numerous PR-4 proteins have been detected from a variety of different species. To understand the evolutionary relationships between these PR-4 proteins, alignment and phylogenetic analyses were carried out. Alignment analysis of PR-4s showed that all contained a Barwin domain. However, PR-4s can be divided into two classes on the basis of whether they have a Hevein domain. VpPR4-1 fell into class II according to this classification scheme (Figure 1B). Phylogenetic analysis indicated that the PR-4s fell into two subgroups. The VpPR4-1 is closely referred to the PR-4 protein (BAK04328) from Hordeum vulgare subsp. at the amino acid level (Figure 1C). Vulgare sharing 79.3% of sequence identity (Matsumoto et al., 2011). For other PR-4 proteins used for alignment and phylogenetic analyses, the sequence identities ranged from 61.7 to 76.7%.
PRs are usually induced by pathogen attack. Expression of the VpPR4-1 induced by powdery mildew was demonstrated in the powdery mildew resistant genotype V. pseudoreticulata Baihe 35-1 (Figure 2A). Under normal conditions, the transcript level of VpPR4-1 was very low. Upon infection with powdery mildew VpPR4-1 transcript levels increased significantly, reaching a maximum 24 h post inoculation (hpi) and then declined. The induction increase of VpPR4-1 transcript level was very high, showing a nearly 800-fold increase compared with the normal level.
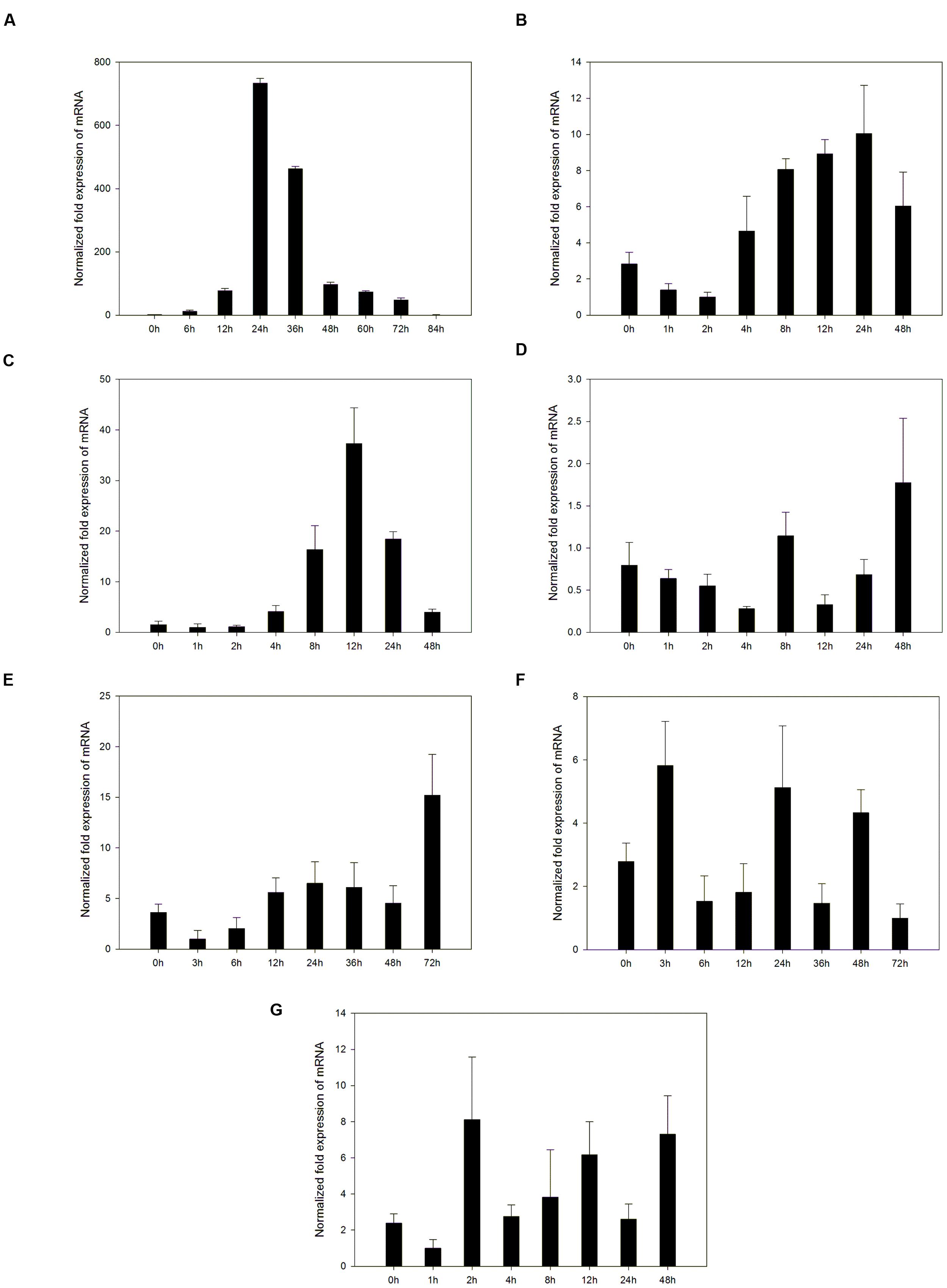
FIGURE 2. Accumulation of mRNA of VpPR4-1 was assessed by quantitative RT-PCR in leaves under a number of stresses. (A) Expression profiles of VpPR4-1 infection with powdery mildew. (B) Expression profiles of VpPR4-1 under SA treatment. (C) Expression profiles of VpPR4-1 under MeJA treatment. (D) Expression profiles of VpPR4-1 under ABA treatment. (E) Expression profiles of VpPR4-1 under NaCl treatment. (F) Expression profiles of VpPR4-1 under 4°C treatment. (G) Expression profiles of VpPR4-1 under 42°C treatment. Error bars indicate SD, from three independent experiments.
To determine the upstream pathway of VpPR4-1, exogenous hormones and abiotic stresses were imposed for RT-PCR. The VpPR4-1 responded differently to the applications of three exogenous hormones and three abiotic stresses (Figures 2B–G). The PR gene responds differently to different stresses in V. pseudoreticulata Baihe 35-1. ABA and cold treatment slightly affected the expression level of VpPR4-1. SA increased the level VpPR4-1 induction, which peaked at 24 hpi. MeJA increased VpPR4-1 induction strongly, with a peak at 12 hpi. This was about a 33-fold increase above normal. However, following NaCl and heat treatments, VpPR4-1 expression increased only about 3-fold.
To further understand the functional response of VpPR4-1 in grapevine, in relation to powdery mildew attack, functional analyses of VpPR4-1 in transgenic V. vinifera L. Red Globe were carried out with the overexpression construct VpPR4-1 (Figure 3) via Agrobacterium-media transformation (Supplementary Figure 1). A total of 30 independent transgenic Red Globe plantlets were obtained. Genomic DNA was extracted from the transgenic grapevines of VpPR4-1 and used to amplify the 520 bp fragment of NPTII. NPTII was detected in all transgenic lines and positive plasmid control, whereas no amplification was detected in the wild-type (WT) plant line (Figure 4A). The expression profiles of VpPR4-1 in transgenic plant lines were studied using the Western-blot method. Total proteins were extracted from the WT plant line and transgenic plant lines of VpPR4-1 under the same growth condition and blotting using polyclonal antibody. Twenty-six of VpPR4-1 transgenic lines showed higher expression levels of VpPR4-1 than the WT plant line (Figures 4B,C). However, expression levels of VpPR4-1 of transgenic lines 13, 14, 15, and 21 were lower than the WT grape. Transgenic lines VpPR4-1-4 and VpPR4-1-5 showed relatively higher levels of PR4 protein and were selected for further experiments.
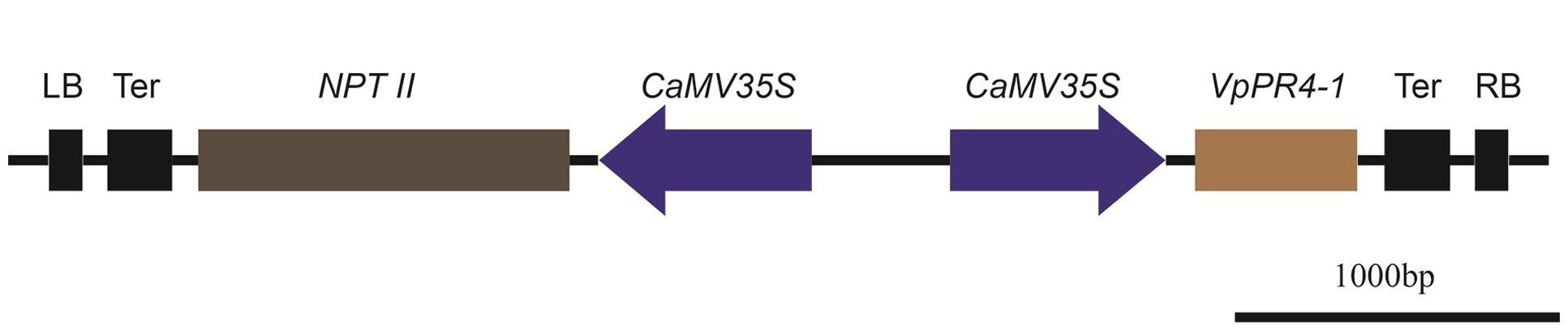
FIGURE 3. Physical maps of plant transformation vector VpPR4-1. LB, Left border of T-DNA; Ter, terminator; NPTII, neomycin phosphotransferase gene; CaMV35S, Cauliflower mosaic virus 35S promoter; VpPR4-1, pathogen-related protein gene from Vitis pseudoreticulata Baihe 35-1; RB, Right border of T-DNA.
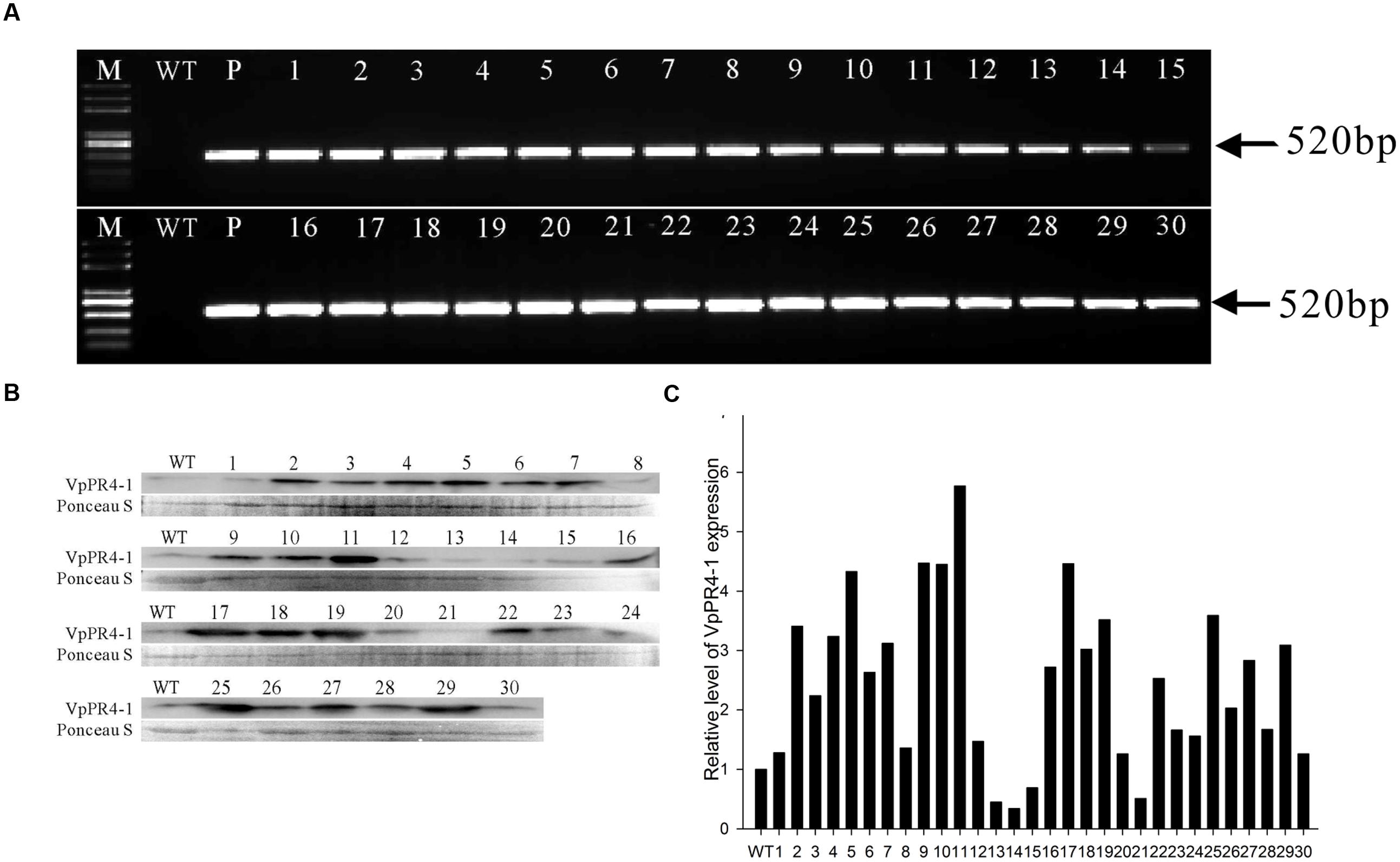
FIGURE 4. Molecular analysis of transgenic Vitis vinifera L. Red Globe by overexpression VpPR4-1 from V. pseudoreticulata. (A) PCR analysis of NPTII from transgenic V. vinifera L. Red Globe of VpPR4-1. (B) Western blotting analysis of transgenic V. vinifera L. Red Globe by overexpression VpPR4-1. VpPR4-1, showing the Western blotting of VpPR4-1 in transgenic lines; Ponceau S, showing the protein stained by Ponceau S. (C) Quantitative analysis for VpPR4-1 protein expression of Western blotting analysis of transgenic V. vinifera L. Red Globe by overexpression VpPR4-1 (B) were performed by Image lab 4.1 software. WT, wild-type line; 1–30, 30 transgenic lines of VpPR4-1.
To confirm that VpPR4-1 actually participates in antifungal activity, we compared the growth rates of powdery mildew hyphae between the six overexpression lines of the higher PR4 protein level and WT plants after powdery mildew infection. Hyphal growth during a 12-day period infection with powdery mildew revealed enhanced tolerance of transgenic plants to the fungus than of none transgenic plants. The VpPR4-1-2, VpPR4-1-3 showed better resistant than other transgenic lines (Figure 5). To investigate the growth of powdery mildew, light microscopy with trypan blue staining was used as previously described (Xiao et al., 2003). Images at 24, 72, 120, and 168 hpi are shown in Figure 6. In the WT lines, more hyphal growth was observed than in the transgenic plants, and the overexpression lines significant reduced powdery mildew hyphal growth not inhibited the fungal completely (Figure 6).
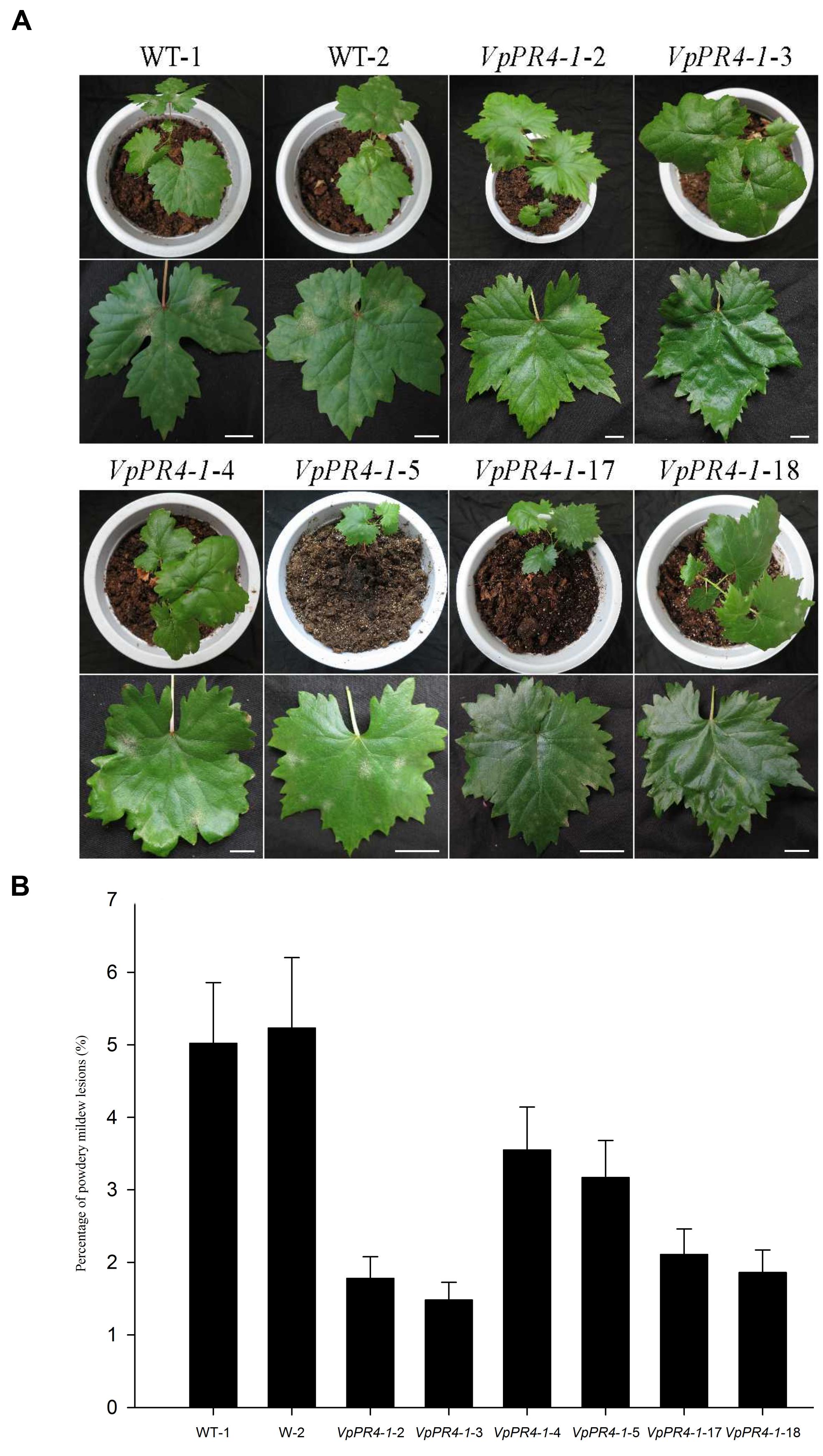
FIGURE 5. Powdery mildew-tolerance analysis for transgenic Vitis vinifera L. Red Globe by overexpression VpPR4-1 from V. pseudoreticulata. (A) Images were taken 12 days after inoculation. (B) Percentages of powdery mildew lesions were analyzed by ImageJ 1.43q software. Error bars indicate SD of three replications. WT-1 and WT-2, wild-type lines; VpPR4-1-2, -3, -4, -5, -17, and -18, transgenic lines of VpPR4-1. Scale bar = 2 cm.
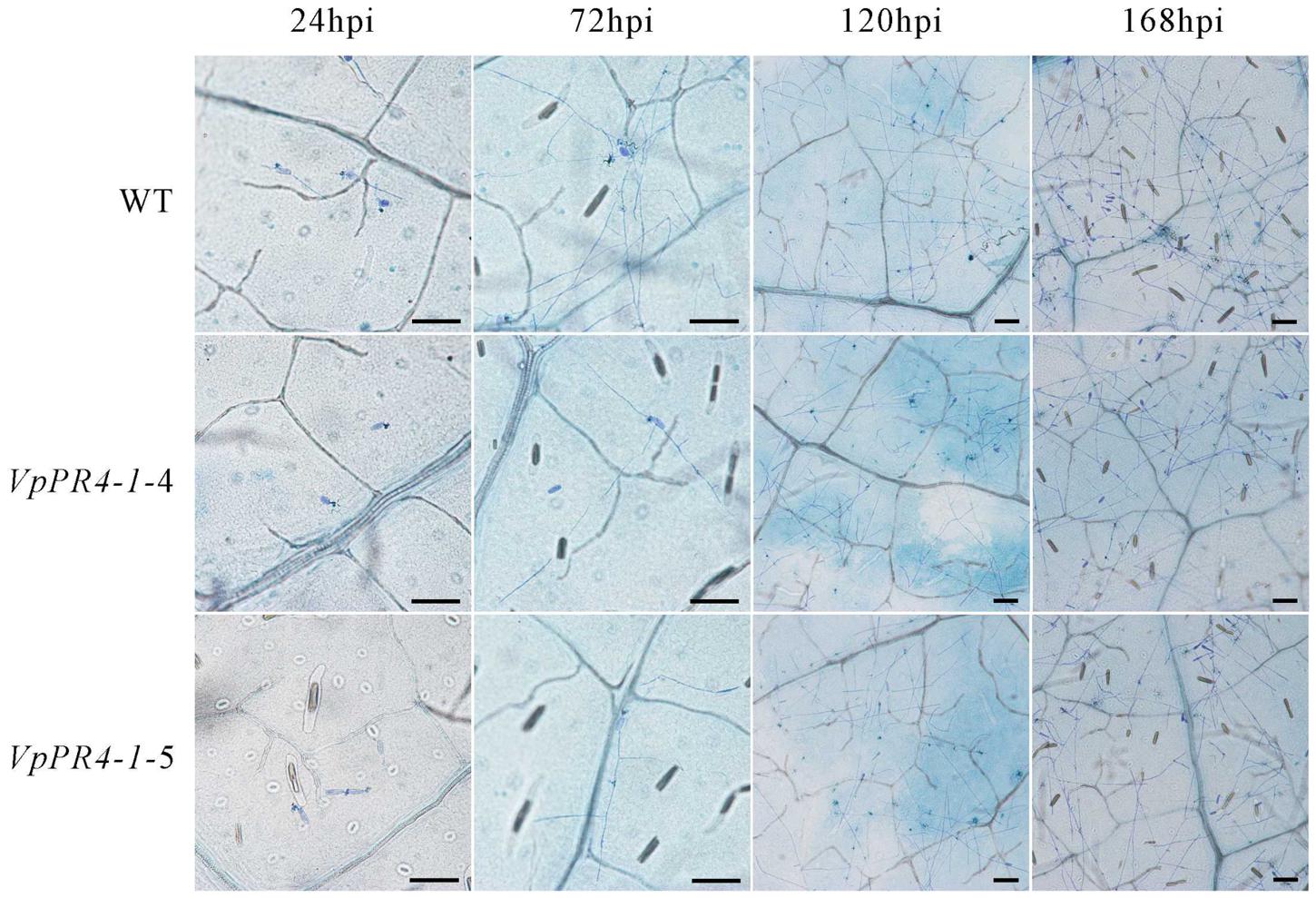
FIGURE 6. Powdery mildew invaded the leaves of transgenic Vitis vinifera L. Red Globe by overexpression VpPR4-1 from V. pseudoreticulata. WT, wild-type line; VpPR4-1-4 and VpPR4-1-5, transgenic line 4 and 5 of VpPR4-1, respectively. Scale bar = 100 μm.
To investigate expression changes of disease-resistance associated genes in the transgenic lines, NPR1, PR1, PR10, and PAL genes transcript levels were measured in the VpPR4-1-4, VpPR4-1-5, and WT lines inoculated with powdery mildew. These showed that the expression of disease-resistance associated genes was induced by powdery mildew and peaked between 12 and 48 hpi, before returning to the low baseline level in the WT line by 72 hpi. However, compared with WT grapes, the defense related genes were disturbed to different degrees by the VpPR4-1 overexpression when inoculated with powdery mildew. In transgenic lines, the expression of the NPR1, PR1, and PAL genes displayed a lower transcription level, and the peak of the PR10 gene transcript was delayed when inoculated with powdery mildew (Figure 7). Specifically, in the WT plants, NPR1 was induced and reached a 30-fold peak at 12 hpi but there was no significant change in the overexpression lines. Correspondingly, in WT, PR1 showed a double peak expression pattern while in the VpPR4-1 transgenic lines this was suppressed and showed a delayed response. However, PAL was not materially affected since both the WT and overexpression lines showed a tendency to rise at first but then decline later.
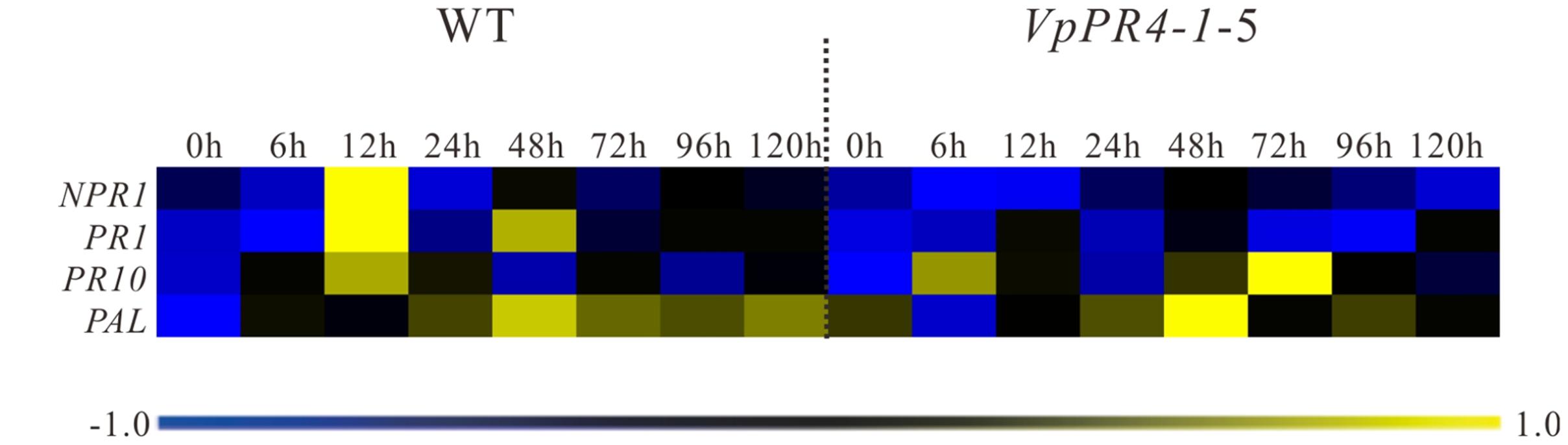
FIGURE 7. qPCR expression analysis for disease-resistance associated genes in leaves of transgenic Vitis vinifera L. Red Globe by overexpression VpPR4-1 from V. pseudoreticulata under powdery mildew treatment. WT, wild-type grapevines; VpPR4-1-5, transgenic line 5 containing VpPR4-1. The color heat map was created according to the standardized RT-PCR expression data by median normalization method using MeV 4.9.0 software, and the color coded showed the differences in gene expression (see key, bottom). The color scale represents relative expression levels: blue represents decreased transcript abundance and yellow represents increased transcript abundance.
Further gene function and the role of powdery mildew resistance of VpPR4-1 including the disease resistance tests in the vineyard will be studied in our future study based on the current work and the transgenic lines of VpPR4-1 obtained.
Powdery mildew is one of the most damaging fungal diseases of the European grapevines. Since powdery mildew resistance is regulated by multi-gene, grapevine is still recalcitrant to powdery mildew resistance and the genes from the cDNA library of the high powdery mildew resistant Baihe 35-1 is worth researching. In our study, detection and characterization of a PR-4 protein induced by powdery mildew in the leaves of V. pseudoreticulata Baihe 35-1 were detected, sequence analyses and expression patterns of VpPR4-1 were carried out. Finally, VpPR4-1 was overexpressed in the powdery mildew susceptible variety Red Globe to exhibit its resistance to powdery mildew.
Vitis pseudoreticulata shows high resistance to powdery mildew. The PR-4 proteins, VpPR4-1, were isolated from V. pseudoreticulata and characterized. VpPR4-1 has previously been cloned from expressed sequence tags (Shi et al., 2010). The PR-4 protein precursor has a short signal peptide and the mature protein format undergoes a strict cutoff. Based on the lack of a Hevein domain near the N terminal, VpPR4-1 belongs to class II. The class II PR-4 proteins are proposed to have evolved from the class I PR-4 proteins because of the highly conserved sequences in the C terminal and the deletion of the only Hevein domain (Friedrich et al., 1991; Ponstein et al., 1994). And sequence alignment and phylogenetic analysis indicate that VpPR4-1 belongs to class II and shows a high level of sequence conservation with these well characterized class II PR-4 proteins.
PR-4 mRNA expression level in grape increased during powdery mildew incubation. However, the induction patterns in susceptible and non-susceptible cultivars are quite different (Fung et al., 2008). Wild Chinese grape V. pseudoreticulata is an accession having high resistance to powdery mildew. The PR-4 gene expression levels are significantly higher in leaves of V. pseudoreticulata following inoculation with powdery mildew than that under normal conditions. Nevertheless, SA, MeJA, ABA, cold, heat, or NaCl stresses do not stimulate VpPR4-1 to a similar extent to powdery mildew attack although they do lead to PR-4 up-regulation. A possible explanation for the expression profile is that powdery mildew regulates VpPR4-1 in an independent or crossover pathway to these six abiotic stresses. It is well known that SA and JA have positive roles in disease resistance, while ABA has a negative role (Kunkel and Brooks, 2002; Mauch-Mani and Mauch, 2005). In this study, VpPR4-1 was induced by SA and MeJA (by 3- to 30-fold) but was insensitive to ABA. A different result was observed with ABA treatment in rice which shows threefold to ninefold change in the expression levels of the PR-4 genes. The expression profiles of VpPR4-1 were also induced by a number of environment stresses (including by salt, cold and heat shock) but the responses were slow. Together, these results suggest that VpPR4-1 has a positive role in stress responses in V. pseudoreticulata, which may be regulated by SA- and JA networks. Moreover, our results are consistent with Wang et al. (2011).
The Ribonuclease activity of PR-4 proteins has been reported a number of times and many experiments have confirmed that they can repress fungal growth in vitro (Bai et al., 2013; Vaghefi et al., 2013; Menezes et al., 2014). To expand our knowledge about PR-4 proteins and create resistant table-grape germplasm, we overexpressed the VpPR4-1 gene in V. vinifera cv. Red Globe. In spite of PR-4 proteins having been implicated in stress responses (Wang et al., 2011), no direct evidence has been reported previously showing that the PR-4 proteins participate in powdery mildew resistance in an important food crop such as grapevines. The VpPR4-1 gene was overexpressed in 30 regenerated V. vinifera cv. Red Globe via Agrobacterium tumefaciens-mediated transformation and confirmed by PCR of NPTII. The 26 transgenic grapevines exhibited higher expression levels of PR-4 protein content than WT vines verified by the Western blot. It is probably that the target gene between the right and left border of T-DNA in Agrobacterium was randomly transformed into the chromosome of host cell via Agrobacterium tumefaciens-mediated transformation and even multicopy of target genes transformation which could make gene expression different from each other or even gene silencing in transgenic lines (Bouquet et al., 2007), the expression of PR4 protein were not the same in all transgenic lines and the transgenic lines VpPR4-1-13, 14, 15, and 21 were lower than the WT grape, that is consistent with other gene transformation of grapevine (Zhou et al., 2014; Dai et al., 2015; Cheng et al., 2016). To confirm that VpPR4-1 actually participates in antifungal activity, we compared the growth rates of powdery mildew hyphae between the six overexpression lines of the higher PR4 protein level and WT plants after powdery mildew infection. The overexpression lines showed relatively high gene expression levels (Figures 4B,C) and significant reduced powdery mildew hyphal growth but not inhibited the fungal completely (Figure 6) which is probably due to powdery mildew resistance is regulated by multi-gene complex network and not by a dominant gene (Fung et al., 2008), and the result is similar with Bertini et al. (2012). Various related genes exhibit different expressions under normal or stress conditions when genes are overexpressed in vivo (Tillett et al., 2012; Xu et al., 2014). The PR-4 proteins are pathogenesis-related proteins involved in pathogen defense response. Since powdery mildew resistance is regulated by multi-gene of SA and/or MeJA pathways and other network (Fung et al., 2008), it is supposed that the significantly higher expression of PR4 protein may lead to affect the expression of other pathogen relative genes which also play important role in powdery mildew resistance network. Therefore, we detected the influences of overexpression of VpPR4-1 on related genes transcripts, including: NPR1, PR1, and PAL. In the overexpression lines, the expression level of NPR1 which plays a positive role in inducible plant disease resistance, was obviously down-regulated. The results may be associated with feedback regulation since PR-4 is in abundance. Accordingly, PR1 was also suppressed to some extent. It was different for PAL, this key enzyme catalyzes the rate-limiting step of phenylpropanoid biosynthesis in plants, that occurred negligibly during the long incubation period. And it is supposed that the higher expression of PR4 protein leading to the suppression expression of other disease resistant genes which resulted in that VpPR4-1-4 and VpPR4-1-5 with higher PR4 levels were not so resistant for powdery mildew as VpPR4-1-2 (Figure 5). Though numerous details for the regulation mechanism of VpPR4-1 in bestowing resistance to abiotic and biotic stress remains unclear (Wang et al., 2011), the twin facts that VpPR4-1 is responsive to powdery mildew inoculation and that the VpPR4-1 overexpression lines showed enhanced powdery mildew resistance in V. vinifera cv. Red Globe indicate that they do nevertheless play an important role in increasing fungal resistance.
LD had made the substantial contributions to the conception, design of the work, and the acquisition, analysis. DW had done interpretation of data for the work. XX did the revising manuscript critically for important intellectual content. JZ did drafting the work of the manuscript. YW gave final approval of the version to be published; XW, YX, and CZ made the agreement to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by the National Natural Science Foundation of China (grant no. 31171924) and the Program for Innovative Research Team of Grape Germplasm Resource and Breeding (2013KCT-25). The authors thank Dr. Alexander (Sandy) Lang from RESCRIPT Co. New Zealand for useful comments and language editing which have greatly improved the manuscript.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00695
Bai, S., Dong, C., Li, B., and Dai, H. (2013). A PR-4 gene identified from Malus domestica is involved in the defense responses against Botryosphaeria dothidea. Plant Physiol. Biochem. 62, 23–32. doi: 10.1016/j.plaphy.2012.10.016
Bertini, L., Caporale, C., Testa, M., Proietti, S., and Caruso, C. (2009). Structural basis of the antifungal activity of wheat PR4 proteins. FEBS Lett. 583, 2865–2871. doi: 10.1016/j.febslet.2009.07.045
Bertini, L., Proietti, S., Aleandri, M. P., Mondello, F., Sandini, S., Caporale, C., et al. (2012). Modular structure of HEL protein from Arabidopsis reveals new potential functions for PR-4 proteins. Biol. Chem. 393, 1533–1546. doi: 10.1515/hsz-2012-0225
Boll, J. F. (1991). Tobacco and tomato PR proteins homologous to win and Pro-hevein lack the “Hevein”. Domain. Mol. Plant Microbe Interact. 4, 586–592. doi: 10.1094/MPMI-4-586
Bouquet, A., Torregrosa, L., Iocco, P., and Thomas, M. R. (2007). Grapevine (Vitis vinifera L.) Agrobacterium Protocols, Vol. 2. (Berlin: Springer), 273–285.
Bravo, J., Campo, S., Murillo, I., Coca, M., and San Segundo, B. (2003). Fungus- and wound-induced accumulation of mRNA containing a class II chitinase of the pathogenesis-related protein 4 (PR-4) family of maize. Plant Mol. Biol. 52, 745–759. doi: 10.1023/A:1025016416951
Broekaert, I., Lee, H. I., Kush, A., Chua, N. H., and Raikhel, N. (1990). Wound-induced accumulation of mRNA containing a hevein sequence in laticifers of rubber tree (Hevea brasiliensis). Proc. Natl. Acad. Sci. U.S.A. 87, 7633–7637. doi: 10.1073/pnas.87.19.7633
Caporale, C., Di Berardino, I., Leonardi, L., Bertini, L., Cascone, A., Buonocore, V., et al. (2004). Wheat pathogenesis-related proteins of class 4 have ribonuclease activity. FEBS Lett. 575, 71–76. doi: 10.1016/j.febslet.2004.07.091
Caruso, C., Bertini, L., Tucci, M., Caporale, C., Nobile, M., Leonardi, L., et al. (2001). Recombinant wheat antifungal PR4 proteins expressed in Escherichia coli. Protein Exp. Purif. 23, 380–388. doi: 10.1006/prep.2001.1512
Caruso, C., Caporale, C., Chilosi, G., Vacca, F., Bertini, L., Magro, P., et al. (1996). Structural and antifungal properties of a pathogenesis-related protein from wheat kernel. J. Protein Chem. 15, 35–44. doi: 10.1007/BF01886809
Cheng, S., Xie, X., Xu, Y., Zhang, C., Wang, X., Zhang, J., et al. (2016). Genetic transformation of a fruit-specific, highly expressed stilbene synthase gene from Chinese wild Vitis quinquangularis. Planta 243, 1041–1053. doi: 10.1007/s00425-015-2459-1
Chevalier, C., Bourgeois, E., Pradet, A., and Raymond, P. (1995). Molecular cloning and characterization of six cDNAs expressed during glucose starvation in excised maize (Zea mays L.) root tips. Plant Mol. Biol. 28, 473–485. doi: 10.1007/BF00020395
Dai, L., Zhou, Q., Li, R., Du, Y., He, J., Wang, D., et al. (2015). Establishment of a picloram-induced somatic embryogenesis system in Vitis vinifera cv. chardonnay and genetic transformation of a stilbene synthase gene from wild-growing Vitis species. Plant Cell Tissue Organ. Cult. 121, 397–412. doi: 10.1007/s11240-015-0711-9
Friedrich, L., Moyer, M., Ward, E., and Ryals, J. (1991). Pathogenesis-related protein 4 is structurally homologous to the carboxy-terminal domains of hevein, Win-1 and Win-2. Mol. Gen. Genet. 230, 113–119. doi: 10.1007/BF00290658
Fung, R. W., Gonzalo, M., Fekete, C., Kovacs, L. G., He, Y., Marsh, E., et al. (2008). Powdery mildew induces defense-oriented reprogramming of the transcriptome in a susceptible but not in a resistant grapevine. Plant Physiol. 146, 236–249. doi: 10.1104/pp.107.108712
Green, T. R., and Ryan, C. A. (1972). Wound-induced proteinase inhibitor in plant leaves: a possible defense mechanism against insects. Science 175, 776–777. doi: 10.1126/science.175.4023.776
Gregersen, P. L., Thordal-Christensen, H., Förster, H., and Collinge, D. B. (1997). Differential gene transcript accumulation in barley leaf epidermis and mesophyll in response to attack by Blumeria graminisf sp. Hordei (syn. Erysiphe graminisf. sp. hordei). Physiol. Mol. Plant Pathol. 51, 85–97. doi: 10.1006/pmpp.1997.0108
Guan, X., Zhao, H., Xu, Y., and Wang, Y. (2011). Transient expression of glyoxal oxidase from the Chinese wild grape Vitis pseudoreticulata can suppress powdery mildew in a susceptible genotype. Protoplasma 248, 415–423. doi: 10.1007/s00709-010-0162-4
Guevara-Morato, M. Á., De Lacoba, M. G., García-Luque, I., and Serra, M. T. (2010). Characterization of a pathogenesis-related protein 4 (PR-4) induced in Capsicum chinense L3 plants with dual RNase and DNase activities. J. Exp. Bot. 61, 3259–3271. doi: 10.1093/jxb/erq148
Hamada, H., Takeuchi, S., Kiba, A., Tsuda, S., Suzuki, K., Hikichi, Y., et al. (2005). Timing and extent of hypersensitive response are critical to restrict local and systemic spread of Pepper mild mottle virus in pepper containing the L 3 gene. J. Gen. Plant Pathol. 71, 90–94. doi: 10.1007/s10327-004-0164-1
Hou, H., Li, J., Gao, M., Singer, S. D., Wang, H., Mao, L., et al. (2013). Genomic organization, phylogenetic comparison and differential expression of the SBP-box family genes in grape. PLoS ONE 8:e59358. doi: 10.1371/journal.pone.0059358
Huet, J., Teinkela Mbosso, E., Soror, S., Meyer, F., Looze, Y., Wintjens, R., et al. (2013). High-resolution structure of a papaya plant-defence barwin-like protein solved by in-house sulfur-SAD phasing. Acta Crystallogr. D Biol. Crystallogr. 69, 2017–2026. doi: 10.1107/S0907444913018015
Jaillon, O., Aury, J.-M., Noel, B., Policriti, A., Clepet, C., Casagrande, A., et al. (2007). The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 449, 463–467. doi: 10.1038/nature06148
Kauffmann, S., Legrand, M., Geoffroy, P., and Fritig, B. (1987). Biological function of ‘pathogenesis-related’ proteins: four PR proteins of tobacco have 1,3-β-glucanase activity. EMBO J. 6, 3209–3212.
Kortekamp, A. (2006). Expression analysis of defence-related genes in grapevine leaves after inoculation with a host and a non-host pathogen. Plant Physiol. Biochem. 44, 58–67. doi: 10.1016/j.plaphy.2006.01.008
Kunkel, B. N., and Brooks, D. M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. doi: 10.1016/S1369-5266(02)00275-3
Lagrimini, L. M., Burkhart, W., Moyer, M., and Rothstein, S. (1987). Molecular cloning of complementary DNA encoding the lignin-forming peroxidase from tobacco: molecular analysis and tissue-specific expression. Proc. Natl. Acad. Sci. U.S.A. 84, 7542–7546. doi: 10.1073/pnas.84.21.7542
Lamb, C. J., Lawton, M. A., Dron, M., and Dixon, R. A. (1989). Signals and transduction mechanisms for activation of plant defenses against microbial attack. Cell 56, 215–224. doi: 10.1016/0092-8674(89)90894-5
Lee, S. C., Kim, Y. J., and Hwang, B. K. (2001). A pathogen-induced chitin-binding protein gene from pepper: its isolation and differential expression in pepper tissues treated with pathogens, ethephon, methyl jasmonate or wounding. Plant Cell Physiol. 42, 1321–1330. doi: 10.1093/pcp/pce168
Linthorst, H. J. M., and Van Loon, L. C. (1991). Pathogenesis-related proteins of plants. Crit. Rev. Plant Sci. 10, 123–150. doi: 10.1080/07352689109382309
Ludvigsen, S., and Poulsen, F. M. (1992a). Secondary structure in solution of barwin from barley seed using proton nuclear magnetic resonance spectroscopy. Biochemistry 31, 8771–8782. doi: 10.1021/bi00152a013
Ludvigsen, S., and Poulsen, F. M. (1992b). Three-dimensional structure in solution of barwin, a protein from barley seed. Biochemistry 31, 8783–8789. doi: 10.1021/bi00152a013
Matsumoto, T., Tanaka, T., Sakai, H., Amano, N., Kanamori, H., Kurita, K., et al. (2011). Comprehensive sequence analysis of 24,783 barley full-length cDNAs derived from 12 clone libraries. Plant Physiol. 156, 20–28. doi: 10.1104/pp.110.171579
Mauch-Mani, B., and Mauch, F. (2005). The role of abscisic acid in plant–pathogen interactions. Curr. Opin. Plant Biol. 8, 409–414. doi: 10.1016/j.pbi.2005.05.015
Melchers, L. S., Groot, M. A.-D., Van Der Knaap, J. A., Ponstein, A. S., Sela-Buurlage, M. B., Bol, J. F., et al. (1994). A new class of tobacco chitinases homologous to bacterial exo-chitinases displays antifungal activity. Plant J. 5, 469–480. doi: 10.1046/j.1365-313X.1994.05040469.x
Menezes, S. P., de Andrade Silva, E. M., Lima, E. M., de Sousa, A. O., Andrade, B. S., Lemos, L. S. L., et al. (2014). The pathogenesis-related protein PR-4b from Theobroma cacao presents RNase activity, Ca2+ and Mg2+ dependent-DNase activity and antifungal action on Moniliophthora perniciosa. BMC Plant Biol. 14:161. doi: 10.1186/1471-2229-14-161
Métraux, J. P., Streit, L., and Staub, T. (1988). A pathogenesis-related protein in cucumber is a chitinase. Physiol. Mol. Plant Pathol. 33, 1–9. doi: 10.1016/0885-5765(88)90038-0
Neuhaus, J. M., Fritig, B., Linthorst, H. J. M., Meins, F., Mikkelsen, J. D., and Ryals, J. (1996). A revised nomenclature for chitinase genes. Plant Mol. Biol. Rep. 14, 102–104. doi: 10.1007/BF02684897
Niderman, T., Genetet, I., Bruyere, T., Gees, R., Stintzi, A., Legrand, M., et al. (1995). Pathogenesis-related PR-1 proteins are antifungal (Isolation and Characterization of Three 14-Kilodalton Proteins of Tomato and of a Basic PR-1 of Tobacco with Inhibitory Activity against Phytophthora infestans). Plant Physiol. 108, 17–27. doi: 10.1104/pp.108.1.17
Park, C.-J., Kim, K.-J., Shin, R., Park, J. M., Shin, Y.-C., and Paek, K.-H. (2004). Pathogenesis-related protein 10 isolated from hot pepper functions as a ribonuclease in an antiviral pathway. Plant J. 37, 186–198. doi: 10.1046/j.1365-313X.2003.01951.x
Park, Y., Jeon, M. H., Lee, S., Moon, J. S., Cha, J., Kim, H. Y., et al. (2005). Activation of defense responses in Chinese cabbage by a nonhost pathogen, Pseudomonas syringae pv. tomato. J. Biochem. Mol. Biol. 38:748. doi: 10.5483/BMBRep.2005.38.6.748
Ponstein, A. S., Bres-Vloemans, S. A., Sela-Buurlage, M. B., van den Elzen, P. J., Melchers, L. S., and Cornelissen, B. J. (1994). A novel pathogen-and wound-inducible tobacco (Nicotiana tabacum) protein with antifungal activity. Plant Physiol. 104, 109–118. doi: 10.1104/pp.104.1.109
Potter, S., Uknes, S., Lawton, K., Winter, A. M., Chandler, D., DiMaio, J., et al. (1993). Regulation of a hevein-like gene in Arabidopsis. Mol. Plant Microbe Interact. 6, 680–685. doi: 10.1094/MPMI-6-680
Reid, K. E., Olsson, N., Schlosser, J., Peng, F., and Lund, S. T. (2006). An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real-time RT-PCR during berry development. BMC Plant Biol. 6:27. doi: 10.1186/1471-2229-6-27
Repka, V., Ficherova, I., and Siharova, K. (2004). Methyl jasmonate is a potent elicitor of multiple defense responses in grapevine leaves and cell-suspension cultures. Biol. Plant. 48, 273–283. doi: 10.1023/B:BIOP.0000033456.27521.e5
Sels, J., Mathys, J., De Coninck, B. M. A., Cammue, B. P. A., and De Bolle, M. F. C. (2008). Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46, 941–950. doi: 10.1016/j.plaphy.2008.06.011
Seong, E., Choi, D., Cho, H., Lim, C., Cho, H., and Wang, M. (2007). Characterization of a stress-responsive ankyrin repeat-containing zinc finger protein of Capsicum annuum (CaKR1). J. Biochem. Mol. Biol. 40, 952–958. doi: 10.5483/BMBRep.2007.40.6.952
Shi, J.-L., Wang, Y.-J., Zhu, Z.-G., and Zhang, C.-H. (2010). The EST analysis of a suppressive subtraction cDNA Library of Chinese Wild Vitis pseudoreticulata Inoculated with Uncinula necator. Agric. Sci. China 9, 233–241. doi: 10.1016/S1671-2927(09)60088-2
Sinha, M., Singh, R. P., Kushwaha, G. S., Iqbal, N., Singh, A., Kaushik, S., et al. (2014). Current overview of allergens of plant pathogenesis related protein families. Sci. World J. 2014:19. doi: 10.1155/2014/543195
Stanford, A., Bevan, M., and Northcote, D. (1989). Differential expression within a family of novel wound-induced genes in potato. Mol. Gen. Genet. 215, 200–208. doi: 10.1007/BF00339718
Svensson, B., Svendsen, I., Hoejrup, P., Roepstorff, P., Ludvigsen, S., and Poulsen, F. M. (1992). Primary structure of barwin: a barley seed protein closely related to the C-terminal domain of proteins encoded by wound-induced plant genes. Biochemistry 31, 8767–8770. doi: 10.1021/bi00152a012
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Tillett, R. L., Wheatley, M. D., Tattersall, E. A., Schlauch, K. A., Cramer, G. R., and Cushman, J. C. (2012). The Vitis vinifera C-repeat binding protein 4 (VvCBF4) transcriptional factor enhances freezing tolerance in wine grape. Plant Biotechnol. J. 10, 105–124. doi: 10.1111/j.1467-7652.2011.00648.x
Vaghefi, N., Mustafa, B. M., Dulal, N., Selby-Pham, J., Taylor, P. W. J., and Ford, R. (2013). A novel pathogenesis-related protein (LcPR4a) from lentil, and its involvement in defence against Ascochyta lentis. Phytopathol. Mediter. 52, 192–201.
Van Loon, L. C. (1982). “Regulation of changes in proteins and enzymes associated with active defence against virus infection,” in Active Defense Mechanisms in Plants, ed. R. K. S. Wood (Berlin: Springer), 247–273.
Van Loon, L. C. (1985). Pathogenesis-related proteins. Plant Mol. Biol. 4, 111–116. doi: 10.1007/BF02418757
Van Loon, L. C., and Van Kammen, A. (1970). Polyacrylamide disc electrophoresis of the soluble leaf proteins from Nicotiana tabacum var. ‘Samsun’ and ‘Samsun NN’: II. Changes in protein constitution after infection with tobacco mosaic virus. Virology 40, 199–211. doi: 10.1016/0042-6822(70)90395-8
Vera, P., and Conejero, V. (1988). Pathogenesis-related proteins of tomato: P-69 as an alkaline endoproteinase. Plant Physiol. 87, 58–63. doi: 10.1104/pp.87.1.58
Vincent, D., Wheatley, M. D., and Cramer, G. R. (2006). Optimization of protein extraction and solubilization for mature grape berry clusters. Electrophoresis 27, 1853–1865. doi: 10.1002/elps.200500698
Wang, L., and Li, S. (2006). Thermotolerance and related antioxidant enzyme activities induced by heat acclimation and salicylic acid in grape (Vitis vinifera L.) leaves. Plant Growth Regul. 48, 137–144. doi: 10.1007/s10725-005-6146-2
Wang, N., Xiao, B., and Xiong, L. (2011). Identification of a cluster of PR4-like genes involved in stress responses in rice. J. Plant Physiol. 168, 2212–2224. doi: 10.1016/j.jplph.2011.07.013
Xiao, S., Brown, S., Patrick, E., Brearley, C., and Turner, J. G. (2003). Enhanced transcription of the Arabidopsis disease resistance genes rpw8. 1 and rpw8. 2 via a salicylic acid–dependent amplification circuit is required for hypersensitive cell death. Plant Cell 15, 33–45. doi: 10.1105/tpc.006940
Xu, W., Zhang, N., Jiao, Y., Li, R., Xiao, D., and Wang, Z. (2014). The grapevine basic helix-loop-helix (bHLH) transcription factor positively modulates CBF-pathway and confers tolerance to cold-stress in Arabidopsis. Mol. Biol. Rep. 41, 1–14. doi: 10.1007/s11033-014-3404-2
Xu, Y., Yu, H., He, M., Yang, Y., and Wang, Y. (2010). Isolation and expression analysis of a novel pathogenesis-related protein 10 gene from Chinese wild Vitis pseudoreticulata induced by Uncinula necator. Biologia 65, 653–659. doi: 10.2478/s11756-010-0056-0
Yang, Y., He, M., Zhou, Z., Li, S., Zhang, C., Singer, S., et al. (2012). Identification of the dehydrin gene family from grapevine species and analysis of their responsiveness to various forms of abiotc and biotic stress. BMC Plant Biol. 12:140. doi: 10.1186/1471-2229-12-140
Yu, Y., Xu, W., Wang, J., Wang, L., Yao, W., Yang, Y., et al. (2013). The Chinese wild grapevine (Vitis pseudoreticulata) E3 ubiquitin ligase Erysiphe necator-induced RING finger protein 1 (EIRP1) activates plant defense responses by inducing proteolysis of the VpWRKY11 transcription factor. New Phytol. 200, 834–846. doi: 10.1111/nph.12418
Zhou, Q., Dai, L., Cheng, S., He, J., Wang, D., Zhang, J., et al. (2014). A circulatory system useful both for long-term somatic embryogenesis and genetic transformation in Vitis vinifera L. cv. Thompson Seedless. Plant Cell Tissue Organ. Cult. 118, 157–168. doi: 10.1007/s11240-014-0471-y
Keywords: grapevine, pathogenesis-related protein-4 (PR-4), transformation, powdery mildew, qRT-PCR
Citation: Dai L, Wang D, Xie X, Zhang C, Wang X, Xu Y, Wang Y and Zhang J (2016) The Novel Gene VpPR4-1 from Vitis pseudoreticulata Increases Powdery Mildew Resistance in Transgenic Vitis vinifera L. Front. Plant Sci. 7:695. doi: 10.3389/fpls.2016.00695
Received: 24 December 2015; Accepted: 06 May 2016;
Published: 27 May 2016.
Edited by:
Sylvain Jeandroz, Agrosup Dijon, FranceReviewed by:
Anil Kumar Singh, ICAR-Indian Institute of Agricultural Biotechnology, IndiaCopyright © 2016 Dai, Wang, Xie, Zhang, Wang, Xu, Wang and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yuejin Wang, d2FuZ3lqQG53c3VhZi5lZHUuY24=; Jianxia Zhang, emhhbmdqeDY2NkAxMjYuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.