
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 27 May 2016
Sec. Plant Breeding
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.00672
This article is part of the Research TopicCrop traits for defense against pests and disease: durability, breakdown and future prospectsView all 21 articles
 Pauline S. M. Van Weymers1†
Pauline S. M. Van Weymers1† Katie Baker2†
Katie Baker2† Xinwei Chen1†
Xinwei Chen1† Brian Harrower1
Brian Harrower1 David E. L. Cooke1
David E. L. Cooke1 Eleanor M. Gilroy1
Eleanor M. Gilroy1 Paul R. J. Birch1
Paul R. J. Birch1 Gaëtan J. A. Thilliez1
Gaëtan J. A. Thilliez1 Alison K. Lees1
Alison K. Lees1 James S. Lynott1
James S. Lynott1 Miles R. Armstrong1
Miles R. Armstrong1 Gaynor McKenzie1
Gaynor McKenzie1 Glenn J. Bryan1*
Glenn J. Bryan1* Ingo Hein1*
Ingo Hein1*The greatest threat to potato production world-wide is late blight, caused by the oomycete pathogen Phytophthora infestans. A screen of 126 wild diploid Solanum accessions from the Commonwealth Potato Collection (CPC) with P. infestans isolates belonging to the genotype 13-A2 identified resistances in the species S. bulbocastanum, S. capsicibaccatum, S. microdontum, S. mochiquense, S. okadae, S. pinnatisectum, S. polyadenium, S. tarijense, and S. verrucosum. Effector-omics, allele mining, and diagnostic RenSeq (dRenSeq) were utilized to investigate the nature of resistances in S. okadae accessions. dRenSeq in resistant S. okadae accessions 7129, 7625, 3762, and a bulk of 20 resistant progeny confirmed the presence of full-length Rpi-vnt1.1 under stringent mapping conditions and corroborated allele mining results in the accessions 7129 and 7625 as well as Avr-vnt1 recognition in transient expression assays. In contrast, susceptible S. okadae accession 3761 and a bulk of 20 susceptible progeny lacked sequence homology in the 5′ end compared to the functional Rpi-vnt1.1 gene. Further evaluation of S. okadae accessions with P. infestans isolates that have a broad spectrum of virulence demonstrated that, although S. okadae accessions 7129, 7625, and 7629 contain functional Rpi-vnt1.1, they also carry a novel resistance gene. We provide evidence that existing germplasm collections are important sources of novel resistances and that “omic” technologies such as dRenSeq-based genomics and effector-omics are efficacious tools to rapidly explore the diversity within these collections.
Potato is the most important non-cereal food crop worldwide and is consumed by more than a billion people (Birch et al., 2012). Global potato production between 1991 and 2007 has shown an increase of 21% that is driven by a 48% rise of potato production in the developing world, where the growing area has increased alongside yield. Pests and pathogens represent a serious and continuing threat to potato production, and the most widespread and economically significant of these is late blight, caused by the oomycete pathogen Phytophthora infestans. In agricultural systems major population changes of P. infestans lineages have been observed that often impact negatively on crop production. For example, in the European P. infestans population a new clonal lineage referred to as 13-A2 or “blue 13” was first detected in 2004 and, upon its arrival in Great Britain, came to dominate the population within 3 years (Cooke et al., 2012). Previously resistant potato cultivars such as Lady Balfour and Stirling were susceptible to the 13-A2 lineage and are consequently no longer suitable for the organically grown potato market. A conservative estimate of the chemical control costs and yield losses associated with late blight exceeds €6.7 Billion (Haverkort et al., 2009). In many parts of the world fungicide application is the only means to prevent disease. Predictions suggest that global potato production could exceed 400 Mt per year if diseases that reduce yields by ~25% could be controlled (Agrios, 1997).
The ability to withstand multiple biotic and abiotic stresses is critical for wild potato species, suggesting that many untapped, natural sources of resistance exist for exploitation in breeding programs. With the availability of extensive germplasm resources, including the Commonwealth Potato Collection (CPC) at the James Hutton Institute (Bradshaw et al., 2006), and improved genomics tools, the potential to exploit this natural biodiversity is considerable. Newly identified and deployed resistances could provide an environmentally benign opportunity to secure potatoes as a major food source in the future (Birch et al., 2012). Critical for the success of such disease control is, however, a detailed knowledge of the underlying mechanisms of defense to facilitate complementary deployment of resistances.
Inducible resistance responses in plants require the direct or indirect detection of pathogen molecules such as defense elicitors or effector molecules via plant receptors (Jones and Dangl, 2006; Wiesel et al., 2014). Effectors, once recognized, are known as avirulence (Avr) genes as their recognition often yields incompatibility for the pathogen on plants that carry the cognate resistance (R) protein. Genome-wide analysis of P. infestans and other oomycetes has shown that all identified Avr genes contain a canonical RXLR motif, which has led to coining of the term RXLR effectors (Armstrong et al., 2005; Hein et al., 2009; Raffaele et al., 2010; Cooke et al., 2012). Heterologous expression of these effectors is used as a novel tool for the identification of resistances and for disease resistance breeding (Birch et al., 2008; Vleeshouwers and Oliver, 2014; Lenman et al., 2016). The recognition of effectors is often dependent on R proteins that contain nucleotide binding (NB) and leucine-rich repeat (LRR) domains and are collectively known as NB-LRRs (Meyers et al., 1999). In the innate plant immune system this process is known as effector-triggered immunity (ETI; Jones and Dangl, 2006). NB-LRR genes are key to plant immunity and their presence, absence or allelic diversity is decisive for disease resistance. At least seven distinct potato NB-LRRs effective toward P. infestans have been cloned so far and their cognate effectors are well described (reviewed in Vleeshouwers and Oliver, 2014). Furthermore, allele mining for late blight resistance genes such as Rpi-blb1, Rpi-blb2, and Rpi-blb3 from the diploid Mexican species S. bulbocastanum has identified functional orthologs in other species (Lokossou et al., 2009, 2010). For example, Rpi-blb1 orthologous genes were identified in the Mexican diploid species S. cardiophyllum, the allopolyploid species S. papita and S. polytrichon as well as in S. stoloniferum amongst others (Wang et al., 2008; Lokossou et al., 2010). When seeking novel resistances in germplasm collections, it is thus imperative to exclude accessions that contain already characterized resistances as the sole means of defense against the pathogen in question.
Recent advances in genome sequencing technologies enable rapid analysis of entire crop genomes and have accelerated the identification of functional R genes. Indeed, 11 years since sequencing the model plant Arabidopsis thaliana, the genomes of two important Solanaceae crop plants, potato, and tomato, were reported (Potato Genome Sequencing Consortium (PGSC), 2011; Tomato Genome Consortium (TGC), 2012). These genomes provide a blueprint for identification of genes coding for important traits such as disease resistance. In the sequenced Solanum tuberosum group Phureja clone DM1-3 516 R44 (DM), 755 NB-LRR genes have been identified and their phylogenetic relationships as well as their physical locations in the 12 potato chromosomes described (Jupe et al., 2012, 2013). These studies formed the basis of a novel R gene enrichment and sequencing platform (RenSeq) that enables the improved annotation of resistance genes in sequenced genomes and facilitates rapid mapping and cloning of resistances via bulked-segregant analysis (Jupe et al., 2013).
In this study we utilized a combination of late blight infections, effector-omics, allele mining, and dRenSeq to identify and/or prioritize novel sources of resistance toward the P. infestans lineage 13-A2. As a proof of concept, dRenSeq was applied as a diagnostic tool to two accessions of the diploid potato species S. okadae and confirmed the presence of Rpi-vnt1.1 in this species.
Isolates of P. infestans were established in vivo on leaves of the late blight susceptible cultivar Craig's Royal and passaged through several generations according to Andrivon et al. (2011). Detached leaf tests were carried out as described by Whisson et al. (2007) and seedling and whole plant tests (two replicates) as described by Stewart et al. (1983) and Bradshaw et al. (2006), respectively. Disease was scored between 5 and 8 days post infection (dpi) on a scale of resistance ranging from 1 = very susceptible to 5 = very resistant for seedling and detached leaf tests and 1 = very susceptible to 9 = very resistant; symptomless plants, for whole plants according to the Malcolmson scale (Cruickshank et al., 1982).
P. infestans effectors were cloned into the binary vector pGRAB and transformed into the A. tumefaciens strain Agl1 with VirG and pSoup. An empty vector was used as a negative control. Infiltrations and analysis of infiltration sites were conducted as described previously (Gilroy et al., 2011).
Rpi-vnt1-like genes have been amplified from the S. okadae accessions 7129, 7625, and 7629 through PCRs with the Rpi-vnt1 specific primers Rpi-vnt1_F_full: 5′ATGAATTATTGTGTTTACAAGACTTGG3′ and Rpi-vnt1_R_full: 5′TTATAGTACCTGTGATATTCTCAACTTTGC3′. To assess the diversity of the Rpi-vnt1-like sequences PCR products were cloned into the vector pGEM-T easy for Sanger sequencing, according to the manufacturer's recommendations (pGEM®-T Easy Vector System—Promega). Recombinant clones were selected following transformation of the constructs into electro competent Escherichia coli DH10B and DH5α cells (Invitrogen) using colony PCR with the gene specific primers mentioned above. Sequencing products were subjected to a BLASTn analysis and compared to functional Rpi-vnt1 variants (Pel et al., 2009) using Geneious v5.6.3 (Biomatters).
RenSeq target enrichment and sequencing was performed according to Jupe et al. (2013, 2014) with minor modifications. The covaris sonicator M220 (Covaris), was used for the fragmentation of DNA to ~500 bp in length, with the following settings: 50 W Peak Incident Power, 20% Duty Factor, 200 cycles per burst, 60 s treatment time and 50 μL volume with 1 μg starting amount. The fragments sizes were checked using a Bioanalyser (Agilent) and no upper size selection was conducted. The samples were quantified using Qubit (Thermofisher) and the enrichment was started with 750 ng of indexed libraries. The Agilent SureSelect enrichment library utilized was designed to include all NB-LRRs identified by Jupe et al. (2013) and the sequences of the corresponding 46,220 probes can be accessed at http://solanum.hutton.ac.uk. Added to the hybridization was 1 μL of 1000 mM universal blocking primer, containing six inosines in place of the six nucleotide index sequence and a 3′ spacer C3 modification to prevent the primer from participating in any subsequent PCR amplification. The post capture amplification was performed with the Herculase II polymerase (Agilent). Sequencing was conducted on an Illumina MiSeq platform using the 2x 300 bp kit. The raw sequence reads were deposited at the European Nucleotide Archive under accession number PRJEB12834.
Paired-end Illumina MiSeq reads were first checked with FastQC (v0.10.0; Andrews, 2010) and then quality and adapter trimmed with cutadapt (v1.9; Martin, 2011) to a minimum length of 100 bp and minimum base quality of 20. The trimmed reads were then mapped to the potato DM reference genome (v4.03; Potato Genome Sequencing Consortium (PGSC), 2011; Sharma et al., 2013) or a FASTA file containing 12 cloned R genes using Bowtie2 (v2.2.1; Langmead and Salzberg, 2012) in very-sensitive end-to-end mode.
The known R genes comprise: R1 (GenBank: AF447489.1; Ballvora et al., 2002), R2 (GenBank: FJ536325.1; Lokossou et al., 2009), R2-like (GenBank: FJ536323.1; Lokossou et al., 2009), R3a (GenBank: AY849382.1; Huang et al., 2005), R3b (GenBank: JF900492.1; Li et al., 2011), Rpi-sto1 (GenBank: EU884421.1; Vleeshouwers et al., 2008), Rpi-pta1 (GenBank: EU884422.1; Vleeshouwers et al., 2008), Rpi-blb1 (GenBank: AY426259.1; van der Vossen et al., 2003), Rpi-blb2 (GenBank: DQ122125.1; van der Vossen et al., 2005, Rpi-blb3 (GenBank: FJ536346.1; Lokossou et al., 2010), Rpi-abpt (GenBank: FJ536324.1; Lokossou et al., 2009), and Rpi-vnt1.1 (GenBank: FJ423044.1; Foster et al., 2009).
For read mapping, discordant and mixed mappings were disabled and maximum insert was set to 1000 bp. Four score-min parameters were used in different mapping runs: “L,−0.03,−0.03,” “L,−0.06,−0.06,” “L,−0.3,−0.3,” and “L,−0.6,−0.6,” approximately equal to 0.5, 1, 5, and 10% mismatch rates, respectively. The resulting BAM files were sorted and indexed using SAMtools (v0.1.18; Li et al., 2009).
The percentage of mapped reads on target was calculated as the proportion of reads mapping to an annotated, targeted RenSeq region in the DM genome reference. Intersecting these RenSeq regions (plus 1000 bp up- and down-stream) against the mapped reads using BEDTools (v2.20.1; Quinlan and Hall, 2010) gave the number of on-target reads. The reads on target was then calculated as a proportion of the total number of mapped reads. Read coverage to on-target regions was estimated by dividing the number of base pairs mapped to the 704 R genes (plus 1000 bp up- and down-stream) on chromosomes 1–12 by their total length (plus 2000 bp per gene). Read coverage was also estimated for the 12 R gene reference set by dividing the total length of mapped reads by the total length of the reference set.
Seedlings and selected whole plants of 126 diploid CPC accessions belonging to 34 species (Supplementary Table S1) were tested with the P. infestans isolates 2006-3928A and/or 2009-7654A belonging to the P. infestans clonal lineage 13-A2. Resistance was observed within 29 of those accessions, belonging to the species S. bulbocastanum, S. capsicibaccatum, S. microdontum, S. mochiquense, S. okadae, S. pinnatisectum, S. polyadenium, S. tarijense, and S. verrucosum (Table 1). There was a strong correlation in the resistance phenotypes observed with both isolates and in the seedling vs. whole plant assays.
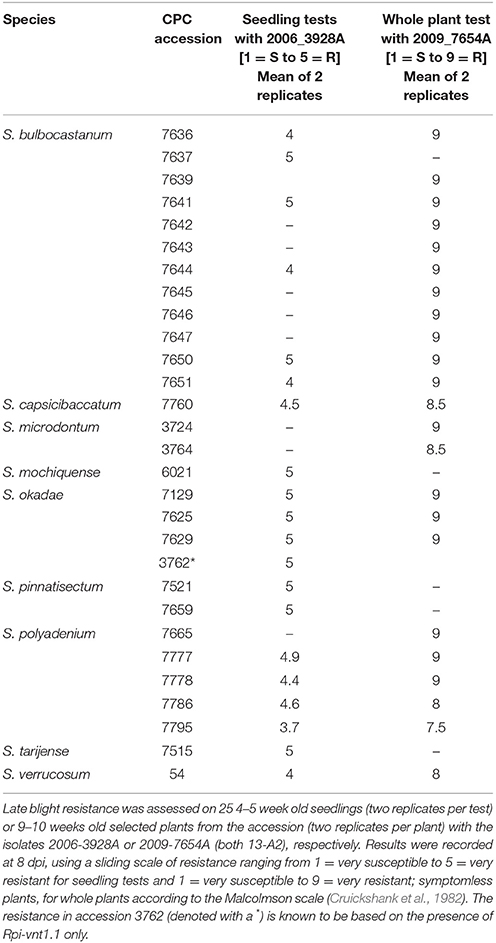
Table 1. Seedling and whole plant late blight resistance screening results for 29 diploid accessions from the CPC.
To determine if the resistances in these species are based on novel or already characterized resistance genes, a number of complementary assays were performed. In this study we report only on accessions of S. okadae and tested for the presence of Rpi-vnt1.1 amongst other characterized R genes. The resistance gene Rpi-vnt1.1 was initially cloned from S. venturii and S. okadae as well as S. phureja accessions and is a homolog of the tomato mosaic virus gene TM-2(2) (Foster et al., 2009).
A set of over 90 P. infestans RXLR effectors has been cloned into binary expressions systems to allow the heterologous expression via Agrobacterium tumefaciens. A subset of 82 effectors that includes known Avr genes (Supplementary Table S2) such as Avr-vnt1 (Pel, 2010) was screened on accessions of S. okadae including susceptible plants S. okadae 7775 and 3761. In at least seven independent replicates with more than 14 individual infiltration sites in total, Avr-vnt1 was recognized reproducibly in S. okadae accessions 7129, 7625, and 7629 but not in susceptible plants 7775 or 3761 (Figure 1). S. okadae accession 3762 was not responsive to Agrobacterium-based expression of effectors and controls (data not shown).
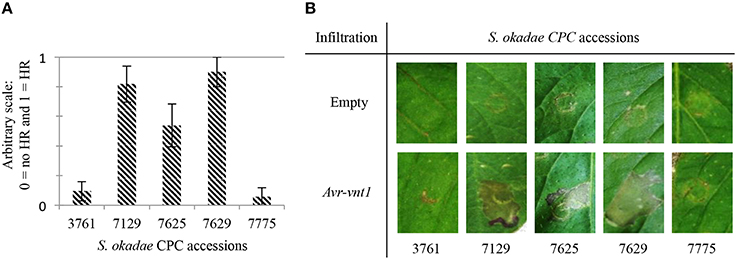
Figure 1. Recognition responses following transient, Agrobacterium tumefaciens-based expression of Avr-vnt1 in S. okadae. Accessions resistant to P. infestans genotype 13-A2, 7129, 7625, and 7629, yield a visible response whereas susceptible accessions 3761 and 7775 yield no specific response if compared to empty vector control. (A) Graph representing phenotypic response at the Avr-vnt1 infiltration sites from at least three independent replicates. Plants were scored at 5 dpi. A score of zero represent no HR and a score of one indicates that at least half the infiltrated leaf area responded with a cell death response. (B) Pictures of the infiltration sites of the empty vector control and Avr-vnt1 visualized under white light at 5 dpi. Transient expressions were performed by infiltration of A. tumefaciens strain Agl1, at an OD600 of 0.2.
Rpi-vnt1.1 gene specific PCR primers were designed and utilized to ascertain if the S. okadae accessions 7129, 7625, and 7629 contain the 2676 bp long gene Rpi-vnt1.1 (Foster et al., 2009) that is also present in S. okadae accession 3762 (Hein et al., unpublished). PCR products were cloned and Sanger sequenced to establish the sequences of individual clones. Alignment of PCR product sequences with Rpi-vnt1.1 indicates that all three accessions contain a sequence identical to Rpi-vnt1.1 alongside additional gene variations and truncated sequences (Figure 2).
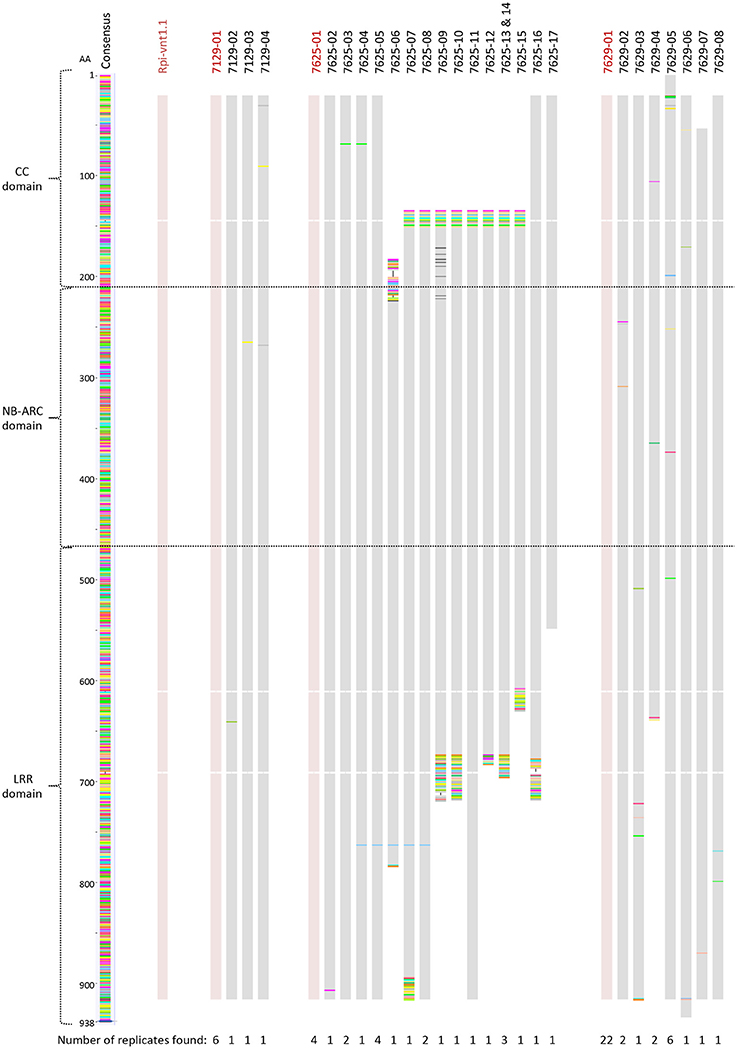
Figure 2. Rpi-vnt1.1 allele mining in S. okadae accessions. In total 9, 26, and 36 Rpi-vnt1-like genes have been amplified and sequenced from the S. okadae accessions 7129, 7625, and 7629, respectively. Nucleotide sequences were translated and the amino acid sequences aligned using Rpi-vnt1.1 as a reference. Sequences identical to Rpi-vnt1.1 are shown in red. The sequence redundancy for each clone is shown below the alignment.
RenSeq-based sequence analysis was conducted to corroborate the allele mining results and to establish whether RenSeq could be used as a diagnostic tool for validating the presence of functional NB-LRR genes. Genomic potato DNA samples from S. okadae accessions 7129 and 7625 were indexed, enriched for NB-LRR genes, and sequenced on a single lane of Illumina MiSeq. Each sample took a 12th of the MiSeq lane. Following quality control, 1,814,975 paired-end reads were obtained for S. okadae accession 7129 and 1,518,349 for 7625. Mapping against the sequenced potato clone DM, which has 704 NB-LRRs with known positions on chromosomes 1–12 (Jupe et al., 2013) was conducted at 0.5, 1, 5, and 10% mismatch rates. At 0.5 and 1% mismatch rates the systematic differences between S. okadae and S. phureja were apparent and a maximum of 6.49% of all reads could be mapped, of which more than 50% were on target. However, when allowing for a 5 or 10% mismatch rate, more than 46 or 70% of all reads could be mapped, respectively. Furthermore, the on-target rate increased to a maximum of 69.5% and mean coverage of NB-LRRs reached 108x (Table 2). Importantly, more of the 704 NB-LRR reference genes from DM were covered by reads from S. okadae accessions with conditions allowing for 5% or higher mismatch rates (Figure 3A, Supplementary Figure S1A, Table 3) indicating that the enrichment was successful.
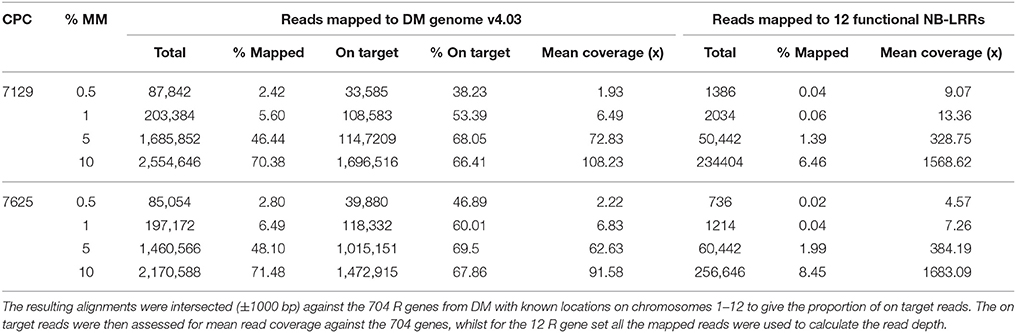
Table 2. RenSeq reads were mapped to DM genome v4.03 or a reference set of 12 R genes at various mismatch rates (% MM).
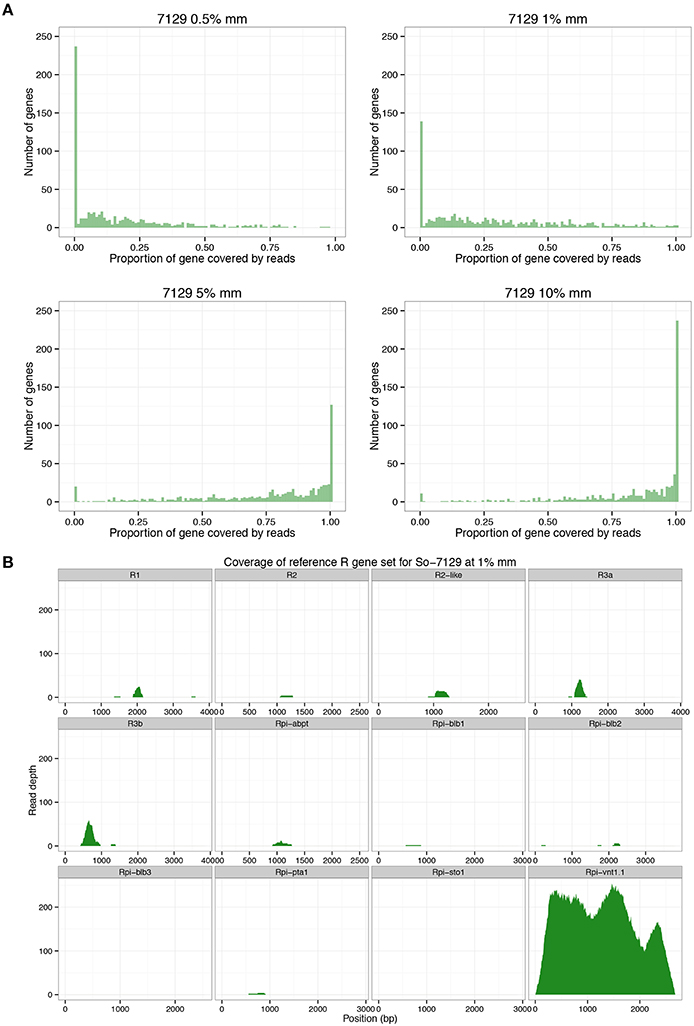
Figure 3. RenSeq analysis for S. okadae accession 7129. (A) The number of 704 R genes from DM with known locations on chromosomes 1–12 that are not covered (0.00), partially covered or fully covered (1.00) following RenSeq analysis in S. okadae accession 7129 is shown. Mismatch rates (%mm) ranging from stringent 0.5 or 1% to more relaxed 5 or 10% are displayed. (B) The read depth and coverage of 12 functional R genes with homologous sequences isolated from S. okadae accession 7129 following RenSeq analysis and mapping under stringent conditions (1% mismatch rate) are depicted.
Sequences derived from 7129 and 7625 were also mapped to a reference set of 12 characterized potato late blight NB-LRR sequences including R1, R2, R2-like, Rpi-abpt, Rpi-blb3, R3a, R3b, Rpi-blb1, Rpi-pta1, Rpi-sto1, Rpi-blb2, and Rpi-vnt1.1 in a dRenSeq analysis. At 1% mismatch rate, only functional Rpi-vnt1.1 was completely represented by dRenSeq reads (Figure 3B, Supplementary Figure S1B). Similar specific results were observed at 0.5% mismatch rate but not at 5 or 10% (Supplementary Figure S2). Indeed, at 5 and 10% mismatch rates, the mean read coverage of Rpi-vnt1.1 was comparable to other characterized R genes (Supplementary Figure S2).
Importantly, dRenSeq was also applied to resistant S. okadae accession 3762 (containing Rpi-vnt1.1) and susceptible S. okadae 3761 (without functional Rpi-vnt1.1) to validate the concept and to discern between resistant and susceptible plants from the same species. Included were also a pool of 20 resistant and 20 susceptible plants that are derived from a cross between both accessions (Figure 4, Supplementary Table S3). At a mismatch rate of either 0.5% (data not shown) or 1%, full-length Rpi-vnt1.1 was recovered from accession 3762 and the resistant pool. However, an Rpi-vnt1.1-like sequence with a truncated 5′ end, compared to the functional gene, was recovered from both the susceptible accession 3761 and the susceptible pool. Indeed, the lack of sequence conservation in this region was consistently detected in both susceptible samples (Figure 4).
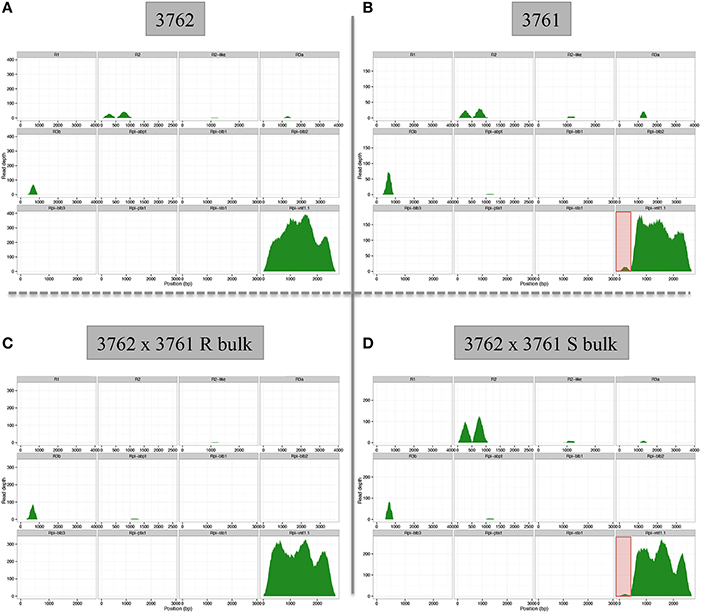
Figure 4. dRenSeq analysis for resistant and susceptible S. okadae accession and bulked progeny. The read depth and coverage of 12 functional R genes with homologous sequences isolated from S. okadae accession (A) 3762 carrying Rpi_vnt1.1, (B) 3761 (susceptible), (C) bulk of 20 resistant plants derived from a cross between 3762 and 3761, and (D) bulk of 20 susceptible plants derived from a cross between 3762 and 3761 following RenSeq analysis and mapping under stringent conditions (1% mismatch rate) are depicted.
Selected S. okadae accessions were screened with five additional P. infestans isolates that display broad race specificity (Supplementary Table S4). Importantly, the isolate EC1, which overcomes Rpi-vnt1.1 resistance, was included to discern between resistances that are exclusively based on the presence of Rpi-vnt1.1. The potato clone Rpi-vnt1.1_R6, which is an F1 clone derived from the cross between S. okadae accessions 3762 (containing Rpi-vnt1.1) and 3761 (susceptible), was used as a control.
In line with previous results, the clone Rpi-vnt1.1_R6 was resistant to the 13-A2 isolate 2009-7654A and other isolates but susceptible to EC1 (Table 4). The S. okadae accession 7775 was susceptible to the 13-A2 isolate but partially resistant to EC1. The three S. okadae accessions (7129, 7625, and 7629) recognizing Avr-vnt1 (Figure 1), however, were resistant to all isolates including EC1 (Figure 5, Table 4). This provides evidence that these accessions, unlike clone Rpi-vnt1.1_R6, carry at least one additional, novel resistance gene that functions independently of Rpi-vnt1.1.

Figure 5. Late blight screen of S. okadae accessions with EC1, a Rpi-vnt1.1 race specific isolate of P. infestans, and 13-A2. Isolates of P. infestans were drop-inoculated on detached leaves and symptoms assessed at 8 dpi. The S. okadae clone 3762-R6 has been independently characterized and only contains Rpi-vnt1.1, and was used as a control.
Potato production is constantly threatened by late blight. The risk of infection is further exacerbated by the rapidly evolving nature of the pathogen, marked by rapid expansion of population size through asexual multiplication or increased genetic diversity through sexual reproduction. Controlling late blight by host resistance requires the continuous development of cultivars by introgression of new resistance from wild species. Breeding strategies in the 1950s largely relied on the deployment of resistances from the hexaploid species S. demissum, which resulted in the release of cultivars carrying one or more resistance genes. Pentland Dell, for example, a potato cultivar released in Great Britain in 1963, contained three resistance genes R1, R2, and R3a (Bradshaw and Ramsay, 2005). However, these resistances proved to be short-lived and could be overcome quite easily within 4 years by adapted new genotypes of P. infestans (Malcolmson, 1969). Exploration of other wild species led to the identification and cloning of resistances conferred by Rpi-blb1 (van der Vossen et al., 2003) and Rpi-blb2 (van der Vossen et al., 2005) from S. bulbocastanum and Rpi-vnt1 from S. venturii (Foster et al., 2009). While these genes show a broad spectrum of resistance, there are some P. infestans isolates that can overcome individual R genes but not all three combined (Jones et al., 2014), showing the importance of pyramiding resistances. However, introgression of resistance genes is a long and laborious process. For example, Rpi-blb2 has been successfully introgressed into cultivars such as Toluca and Bionica that were developed after more than 30 years of breeding and selection efforts (Haverkort et al., 2009).
In light of these observations, the need for rapid and reliable diagnostic R gene tools is apparent. Effector-omics has proven useful for breeding and the identification of orthologous R gene in wild species (Vleeshouwers et al., 2008; Vleeshouwers and Oliver, 2014; Lenman et al., 2016). However, for this system to be successful, a detailed knowledge of the recognized effector is required alongside responsive plants that yield reproducible recognition response upon transient effector expression. We have obtained reproducible Avr-vnt1 recognition responses in S. okadae accessions 7129, 7625, and 7629 (Figure 1) but not for 3762 that contains the cognate R gene Rpi-vnt1.1. The latter proved non-responsive to the transient Agrobacterium-based expression system.
In line with the Avr-vnt1 recognition, PCR-based allele mining and Sanger sequencing confirmed the presence of Rpi-vnt1.1 in S. okadae accessions 7129, 7625, and 7629 (Figure 2). A similar approach has been utilized successfully to identify orthologous genes in wild potato species (Lokossou et al., 2009, 2010). A PCR-based screening for full-length R genes alone could, however, be prone to false-positives and/or false-negative results. Furthermore, the cloning and sequencing of PCR products, which is required to discriminate highly similar sequences (Figure 2), renders this process low to medium throughput.
This study has shown that mapping RenSeq reads with stringent mismatch rates against reference R genes, results in a quick and easy way to screen plants for the presence or absence of known R genes (Figures 3, 4, as well as Supplementary Figures S1, S2). Indeed, dRenSeq is specific enough that it could distinguish between functional Rpi-vnt1.1 in resistant accessions and its homologs in susceptible accessions as well as bulks. As such, dRenSeq could also be used for allele mining under various stringent mapping conditions and also aid evolutionary studies. Importantly, the obtained RenSeq sequence from plants that do contain novel resistances can subsequently be used as a reference in a bulked-segregant analysis if genetic crosses can be achieved (Jupe et al., 2013). Therefore, sequence data can be used to answer different biological questions.
Interestingly, the S. okadae accessions 7129, 7625, and 7629 all contain functional Rpi-vnt1.1 as demonstrated by effector recognition, allele mining and, in the case of 7129 and 7625, dRenSeq. However, they also contain a resistance that operates independent of Rpi-vnt1.1 as demonstrated by additional late blight screening (Figure 5). The clone Rpi-vnt1.1_R6 carries Rpi-vnt1.1 and is, as expected, resistant to blue 13 but susceptible to the isolate EC1 (Foster et al., 2009), whereas 7129, 7625, and 7629 were all resistant to both isolates (Figure 5). RenSeq-derived reads are of dual utility and the additional resistance(s) could be mapped via a bulked segregant RenSeq analysis as described in Jupe et al. (2013). In this case, the RenSeq reads that have been used for the dRenSeq analysis described here could be utilized to represent the resistant/susceptible parents. Using DM as a reference for the mapping, RenSeq reads are typically mapped at a 5% mismatch rate to allow for systematic differences between species which contrasts with dRenSeq where a 0.5 or 1% mismatch rate is used to establish the presence/absence of already known NB-LRRs.
Future efforts to identify resistances toward major pathogens in germplasm collection can quickly identify plants that contain novel resistances by taking advantage of target enrichment and sequencing technologies. For example, traditional allele mining based on PCR amplification, cloning of amplicons, and Sanger sequencing of individual clones can be omitted with dRenSeq application. Furthermore, a combination of late blight screening that includes isolates with a broad virulence spectrum followed by dRenSeq could be utilized to first prioritize plants that could subsequently be subjected to effector-omic analysis prior to a detailed genetic study. In breeding programs, dRenSeq (or similar enrichment strategies for additional genes) could be utilized to aid R gene pyramiding and/or to follow multiple important traits on a sequence-based level.
PV, XC, BH, GT, AL, JL were involved in late blight screening and effector recognition. DC characterised P. infestans isolates. GM provided high health CPC accessions for the experiments. PV conducted allele mining and RenSeq analysis with MA. EG and PB provided effectors and were involved in analysing effector recognition. KB conducted computational analysis of RenSeq and DRenSeq. PV, GB, XC, and IH wrote the manuscript. IH directed the work.
This work was funded by the Rural & Environment Science & Analytical Services Division of the Scottish Government, the BBSRC through the joint projects CRF/2009/SCRI/SOP 0929, BB/L008025/1, BB/ K018299/1 (IH) and the USDA NIFA grant 2011-68004-30154 (P SMVW). Additional funding was obtained through the James Hutton Institute SEEDCORN initiative.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We would like to acknowledge Pete Hedley for his input into the sequencing design, Micha Bayer and Florian Jupe for their contribution to the S. okadae sequence analysis pipeline and Shirley Gillard for her involvement in the effector library cloning. We also acknowledge Iain Milne and Linda Milne for generating the URL for the probe sets.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00672
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Andrivon, D., Avendano-Corcoles, J., Cameron, A. M., Carnegie, S. F., Cooke, L. R., Corbiere, R., et al. (2011). Stability and variability of virulence of Phytophthora infestans assessed in a ring test across European laboratories. Plant Pathol. 60, 556–565. doi: 10.1111/j.1365-3059.2010.02392.x
Armstrong, M. R., Whisson, S. C., Pritchard, L., Bos, J. I. B., Venter, E., Avrova, A. O., et al. (2005). An ancestral oomycete locus contains late blight avirulence gene Avr3a, encoding a protein that is recognized in the host cytoplasm. Proc. Natl. Acad. Sci. U.S.A. 102, 7766–7771. doi: 10.1073/pnas.0500113102
Ballvora, A., Ercolano, M. R., Weiû, J., Meksem, K., Bormann, C. A., Oberhagemann, P., et al. (2002). The R1 gene for potato resistance to late blight (Phytophthora infestans) belongs to the leucine zipper/NBS/LRR class of plant resistance genes. Plant J. 30, 361–371. doi: 10.1046/j.1365-313X.2001.01292.x
Birch, P. R. J., Boevink, P. C., Gilroy, E. M., Hein, I., Pritchard, L., and Whisson, S. C. (2008). Oomycete RxLR effectors: delivery, functional redundancy and durable disease resistance. Curr. Opin. Plant Biol. 11, 373–379. doi: 10.1016/j.pbi.2008.04.005
Birch, P. R. J., Bryan, G., Fenton, B., Gilroy, E. M., Hein, I., Jones, J. T., et al. (2012). Crops that feed the world 8: potato: are the trends of increased global production sustainable? Food Secur. 4, 477–508. doi: 10.1007/s12571-012-0220-1
Bradshaw, J. E., Bryan, G. J., and Ramsay, G. (2006). Genetic resources (Including Wild and Cultivated Solanum Species) and progress in their utilisation in potato breeding. Potato Res. 49, 49–65. doi: 10.1007/s11540-006-9002-5
Bradshaw, J. E., and Ramsay, G. (2005). Utilisation of the Commonwealth Potato Collection in potato breeding. Euphytica 146, 9–19. doi: 10.1007/s10681-005-3881-4
Cooke, D. E. L., Cano, L. M., Raffaele, S., Bain, R. A., Cooke, L. R., Etherington, G. J., et al. (2012). Genome analyses of an aggressive and invasive lineage of the Irish potato famine pathogen. PLOS Pathog. 8:e1002940. doi: 10.1371/journal.ppat.1002940
Cruickshank, G., Stewart, H. E., and Wastie, R. L. (1982). An illustrated assessment key for foliage blight of potatoes. Potato Res. 25, 213–214. doi: 10.1007/BF02359807
Foster, S. J., Park, T.-H., Pel, M., Brigneti, G., Sliwka, J., Jagger, L., et al. (2009). Rpi-vnt1.1, a Tm-22 Homolog from Solanum venturii, confers resistance to potato late blight. Mol. Plant Microbe Interact. 22, 598–600. doi: 10.1094/MPMI-22-5-0589
Gilroy, E. M., Breen, S., Whisson, S. C., Squires, J., Hein, I., Kaczmarek, M., et al. (2011). Presence/absence, differential expression and sequence polymorphisms between PiAVR2 and PiAVR2-like in Phytophthora infestans determine virulence on R2 plants. New Phytol. 191, 763–776. doi: 10.1111/j.1469-8137.2011.03736.x
Haverkort, A., Struik, P., Visser, R., and Jacobsen, E. (2009). Applied biotechnology to combat late blight in potato caused by Phytophthora infestans. Potato Res. 52, 249–264. doi: 10.1007/s11540-009-9136-3
Hein, I., Gilroy, E. M., Armstrong, M. R., and Birch, P. R. J. (2009). The zig-zag-zig in oomycete-plant interactions. Mol. Plant Pathol. 10, 547–562. doi: 10.1111/j.1364-3703.2009.00547.x
Huang, S., van der Vossen, E. A., Kuang, H., Vleeshouwers, V. G., Zhang, N., Borm, T. J., et al. (2005). Comparative genomics enabled the isolation of the R3a late blight resistance gene in potato. Plant J. 42, 251–261. doi: 10.1111/j.1365-313X.2005.02365.x
Jones, J. D. G., Witek, K., Verweij, W., Jupe, F., Cooke, D., Dorling, S., et al. (2014). Elevating crop disease resistance with cloned genes. Philos. Trans. R. Soc. B 369, 20130087. doi: 10.1098/rstb.2013.0087
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Jupe, F., Chen, X., Verweij, W., Witek, K., Jones, J. D., and Hein, I. (2014). Genomic DNA library preparation for resistance gene enrichment and sequencing (RenSeq) in plants. Methods Mol. Biol. 1127, 291–303. doi: 10.1007/978-1-62703-986-4_22
Jupe, F., Pritchard, L., Etherington, G. J., MacKenzie, K., Cock, P. J. A., Wright, F., et al. (2012). Identification and localisation of the NB-LRR gene family within the potato genome. BMC Genomics 13:75. doi: 10.1186/1471-2164-13-75
Jupe, F., Witek, K., Verweij, W., Sliwka, J., Pritchard, L., Etherington, G. J., et al. (2013). Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 76, 530–544. doi: 10.1111/tpj.12307
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Lenman, M., Ali, A., Mühlenbock, P., Carlson-Nilsson, U., Liljeroth, E., Champouret, N., et al. (2016). Effector-driven marker development and cloning of resistance genes against Phytophthora infestans in potato breeding clone SW93-1015. Theor. Appl. Genet. 129, 105–115. doi: 10.1007/s00122-015-2613-y
Li, G., Huang, S., Guo, X., Li, Y., Yang, Y., Guo, Z., et al. (2011). Cloning and characterization of R3b; members of the R3 superfamily of late blight resistance genes show sequence and functional divergence. Mol. Plant Microbe Interact. 24, 1132–1142. doi: 10.1094/MPMI-11-10-0276
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence alignment/map (SAM) format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Lokossou, A. A., Park, T. H., van Arkel, G., Arens, M., Ruyter-Spira, C., Morales, J., et al. (2009). Exploiting knowledge of R/Avr genes to rapidely clone a new LZ-NBS-LRR family of late blight resistance genes from potato linkage group IV. Mol. Plant-Microbe Interact. 22, 630–641. doi: 10.1094/MPMI-22-6-0630
Lokossou, A. A., Rietman, H., Wang, M., Krenek, P., van der Schoot, H., Henken, B., et al. (2010). Diversity, distribution, and evolution of Solanum bulbocastanum late blight resistance genes. Mol. Plant-Microbe Interact. 23, 1206–1216. doi: 10.1094/MPMI-23-9-1206
Malcolmson, J. F. (1969). Races of Phytophthora infestans occurring in Great Britain. Trans. Br. Mycol. Soc. 53, 417–423. doi: 10.1016/S0007-1536(69)80099-9
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 10–12. doi: 10.14806/ej.17.1.200
Meyers, B. C., Dickerman, A. W., Michelmore, R. W., Sivaramakrishnan, S., Sobral, B. W., and Young, N. D. (1999). Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 20, 317–332. doi: 10.1046/j.1365-313X.1999.t01-1-00606.x
Pel, M. A., Foster, S. J., Park, T. H., Rietman, H., van Arkel, G., Jones, J. D. G., et al. (2009). Mapping and cloning of late blight resistance genes from Solanum venturii using an interspecific candidate gene approach. Mol. Plant-Microbe Interact. 22, 601–615. doi: 10.1094/MPMI-22-5-0601
Pel, M. A. (2010). Mapping, Isolation and Characterization of Genes Responsible for Late Blight Resistance in Potato. Ph.D. thesis, Wageningen University, Wageningen, Netherlands.
Potato Genome Sequencing Consortium (2011). Genome sequence analysis of the tuber crop potato. Nature 475, 189–197. doi: 10.1038/nature10158
Quinlan, A. R., and Hall, I. M. (2010). BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842. doi: 10.1093/bioinformatics/btq033
Raffaele, S., Farrer, R. A., Cano, L. M., Studholme, D. J., MacLean, D., Thines, M., et al. (2010). Genome evolution following host jumps in the Irish potato famine pathogen lineage. Science 330, 1540–1543. doi: 10.1126/science.1193070
Sharma, S. K., Bolser, D., de Boer, J., Sønderkær, M., Amoros, W., Carboni, M. F., et al. (2013). Construction of reference chromosome-scale pseudomolecules for potato: integrating the potato genome with genetic and physical maps. G3 3, 2031–2047. doi: 10.1534/g3.113.007153
Stewart, H. E., Taylor, K., and Wastie, R. L. (1983). Resistance to late blight in foliage (Phytophthora infestans) of potatoes assessed as true seedlings and as adult plants in the glasshouse. Potato Res. 26, 363. doi: 10.1007/BF02356155
Tomato Genome Consortium (2012). The tomato genome sequence provides insights into fleshy fruit evolution. Nature 485, 635–641. doi: 10.1038/nature11119
van der Vossen, E. A. G., Sikkema, A., te Lintel Hekkert, B., Gros, J., Stevens, P., Muskens, M., et al. (2003). An ancient R gene from the wild potato species Solanum bulbocastanum confers broad-spectrum resistance to Phytophthora infestans in cultivated potato and tomato. Plant J. 36, 867–882. doi: 10.1046/j.1365-313X.2003.01934.x
van der Vossen, E. A. G., Gros, J., Sikkema, A., Muskens, M., Wouters, D., Wolters, P., et al. (2005). The Rpi-blb2 gene from Solanum bulbocastanum is an Mi-1 gene homolog conferring broad-spectrum late blight resistance in potato. Plant J. 44, 208–222. doi: 10.1111/j.1365-313X.2005.02527.x
Vleeshouwers, V. G. G. A., Rietman, H., Krenek, P., Champouret, N., Young, C., Oh, S.-K., et al. (2008). Effector genomics accelerates discovery and functional profiling of potato disease resistance and Phytophthora infestans avirulence genes. PLoS ONE 3:e2875. doi: 10.1371/journal.pone.0002875
Vleeshouwers, V. G. A. A., and Oliver, R. P. (2014). Effectors as tools in disease resistance breeding against biotrophic, hemibiotrophic, and necrotrophic plant pathogens. Mol. Plant Microbe Interact. 27, 196–206. doi: 10.1094/mpmi-10-13-0313-ia
Wang, M., Allefs, S., van den Berg, R. G., Vleeshouwers, V. G., van der Vossen, E. A., and Vosman, B. (2008). Allele mining in Solanum: conserved homologues of Rpi-blb1 are identified in Solanum stoloniferum. Theor. Appl. Genet. 116, 933–943. doi: 10.1007/s00122-008-0725-3
Whisson, S. C., Boevink, P. C., Moleleki, L., Avrova, A. O., Morales, J. G., Gilroy, E. M., et al. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450, 115–118. doi: 10.1038/nature06203
Keywords: germplasm collection, Commonwealth potato collection, diagnostic, RenSeq, Phytophthora infestans, oomycete, RXLR effectors
Citation: Van Weymers PSM, Baker K, Chen X, Harrower B, Cooke DEL, Gilroy EM, Birch PRJ, Thilliez GJA, Lees AK, Lynott JS, Armstrong MR, McKenzie G, Bryan GJ and Hein I (2016) Utilizing “Omic” Technologies to Identify and Prioritize Novel Sources of Resistance to the Oomycete Pathogen Phytophthora infestans in Potato Germplasm Collections. Front. Plant Sci. 7:672. doi: 10.3389/fpls.2016.00672
Received: 29 February 2016; Accepted: 02 May 2016;
Published: 27 May 2016.
Edited by:
Peter John Gregory, East Malling Research, UKReviewed by:
Matthew R. Willmann, Cornell University, USACopyright © 2016 Van Weymers, Baker, Chen, Harrower, Cooke, Gilroy, Birch, Thilliez, Lees, Lynott, Armstrong, McKenzie, Bryan and Hein. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Glenn J. Bryan, Z2xlbm4uQnJ5YW5AaHV0dG9uLmFjLnVr;
Ingo Hein, aW5nby5IZWluQGh1dHRvbi5hYy51aw==
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.