- 1Department of Biology, Washington University in St. Louis, St. Louis, MO, USA
- 2Department of Chemistry, Washington University in St. Louis, St. Louis, MO, USA
Photosystem II (PSII) is a photosynthetic membrane-protein complex that undergoes an intricate, tightly regulated cycle of assembly, damage, and repair. The available crystal structures of cyanobacterial PSII are an essential foundation for understanding PSII function, but nonetheless provide a snapshot only of the active complex. To study aspects of the entire PSII life-cycle, mass spectrometry (MS) has emerged as a powerful tool that can be used in conjunction with biochemical techniques. In this article, we present the MS-based approaches that are used to study PSII composition, dynamics, and structure, and review the information about the PSII life-cycle that has been gained by these methods. This information includes the composition of PSII subcomplexes, discovery of accessory PSII proteins, identification of post-translational modifications and quantification of their changes under various conditions, determination of the binding site of proteins not observed in PSII crystal structures, conformational changes that underlie PSII functions, and identification of water and oxygen channels within PSII. We conclude with an outlook for the opportunity of future MS contributions to PSII research.
Introduction
Since the late 1990s, mass spectrometry (MS) has become a central tool for the study of proteins and their role in biology. The advent of electrospray ionization (ESI) and matrix-assisted laser desorption ionization (MALDI) permits the ionization of peptides and proteins and their introduction into the gas phase, enabling their analysis by MS. The typical “bottom-up” workflow that emerged in the wake of these breakthroughs involves: (1) enzymatic digestion (often by trypsin) of a protein to produce peptides of small enough size (typically 1–3 kDa) to be ionized and fragmented efficiently in a mass spectrometer; (2) liquid chromatographic (LC) separation of the peptides; and (3) online (or offline) injection of the separated peptides into a mass spectrometer. The “top-down” approach is an attractive alternative that eliminates the protein digestion step, but the subsequent steps are generally more difficult for intact proteins than peptides, and this approach is currently best-suited for small, soluble proteins. After injection of the peptides, a typical tandem MS analysis consists of: (1) ionization of the peptide sample by ESI or MALDI and introduction into the gas phase; (2) measurement of the mass-to-charge (m/z) ratio of the intact peptide (also referred to as “MS1” analysis); and (3) fragmentation of the precursor ion and measurement of its “product-ion” spectrum (“MS/MS” or “MS2” analysis), which provides information about the peptide's amino acid sequence. When genomic information is available to predict the sequence of all proteins in the organism, computer analysis of the peptide masses and product-ion spectra can determine the highest-scoring match for each peptide from the protein database. This highest-scoring match is taken as the identity of the peptide, assuming data quality meets certain statistical criteria. A given protein is then determined to have been present in the sample if the quality and number of its peptide hits meet an additional set of statistical criteria. The ability to identify many proteins in a sample at once by MS has become the cornerstone of the field of proteomics.
Protein identification is only the most basic application of MS-based proteomics, and it has traditionally been described as the first “pillar” of the field. The second pillar is characterization of the many proteoforms that exist for each protein, arising, e.g., from splice variants and post-translational modifications (PTMs). These two pillars address questions about the composition of a protein sample. The third pillar is quantification—either absolute or relative—of proteins using isotopic labeling or label-free approaches. This pillar is typically used to address questions about the dynamics of a system—how composition of proteins or proteoforms changes over time, space, or under different environmental conditions or perturbations. A proposed fourth pillar focuses on the emerging area of structural proteomics that uses MS-based techniques to address questions about the three-dimensional structure of proteins and protein complexes in a cell.
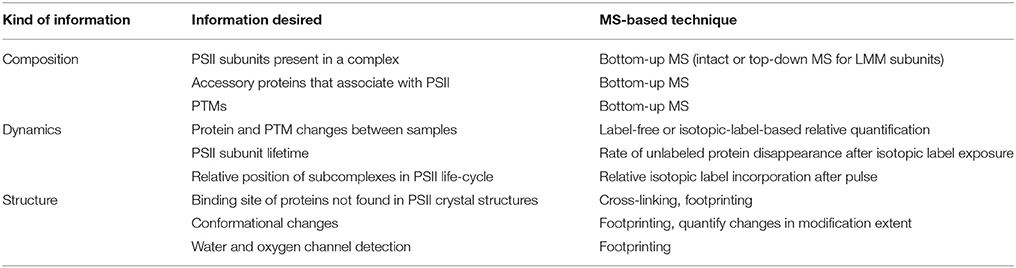
These four pillars of proteomics have each become indispensable tools for gleaning information about photosynthesis (Battchikova et al., 2015; Bricker et al., 2015; Heinz et al., 2016) and, in particular for this review, the life-cycle of PSII. A search for publications containing both “Photosystem II” and “mass spectrometry” in the article title, abstract, and/or keywords was performed on the Scopus database. The results, displayed in Figure 1, show that prior to the advent of ESI and MALDI in the late 1980s, publications were nearly zero per year. Starting in the early 1990s and continuing through 2015, publications have risen steadily, with around 20–30 publications per year in the last several years. The rise can be attributed to method and instrument development, and to increasing accessibility of MS instrumentation to biology researchers. An overview of how MS-based tools are typically applied to PSII life-cycle research is given in Table 1. This review focuses on MS of proteins. However, it should be noted that another widely used application of MS in PSII research is the analysis of the isotopic composition of evolved oxygen by membrane-inlet mass spectrometry. This technique has yielded significant insight into the mechanistic aspects of water oxidation by PSII (reviewed in Shevela and Messinger, 2013).
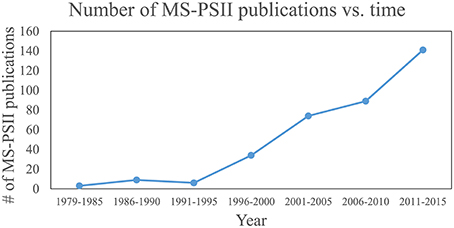
Figure 1. Plot of publications that use MS for PSII research over time. Publications that contain “Photosystem II” and “mass spectrometry” in their article title, abstract, or keywords were searched on the Scopus database. Each data point represents the total number of publications for that range of years.
In the sections that follow, we consider questions of PSII composition, dynamics, and structure separately. For each area, a brief overview of the relevant MS-based tools is given, followed by examples of several PSII life-cycle research areas that have benefitted from these techniques. In the final section, the outlook for future contributions of MS techniques to PSII life-cycle research is discussed.
Composition of PSII Complexes
MS-Based Methods to Study the Composition of PSII Complexes
PSII Subunits with Soluble Domains
The bottom-up MS workflow is highly effective at identifying soluble proteins or proteins with soluble domains. It is, therefore, the main MS strategy that has been used to detect the core PSII proteins D1, D2, CP43, and CP47, which are transmembrane proteins but have multiple soluble domains, the extrinsic (soluble) PSII proteins, or unknown PSII-bound proteins. Bottom-up MS analysis can be preceded by either in-gel or in-solution digestion of the protein, each with advantages. Gel electrophoresis serves as a one- or two-dimensional fractionation step, simplifying the mixture to be analyzed by MS. Using this approach to remove interferences can improve instrument sensitivity toward proteins in the band of interest. Native PAGE, either alone or followed by denaturing SDS-PAGE (2D-BN-PAGE), is a common choice for resolving multiple protein complexes in a thylakoid membrane or purified PSII preparation; unknown bands can be excised and analyzed by MS to identify components of specific complexes (Granvogl et al., 2008; Pagliano et al., 2014; Gao et al., 2015). However, targeted band excision can miss potentially important proteins that migrated at positions not selected for in-gel digestion. In theory, native PAGE can remove unbound proteins from complexes, simplifying MS analysis; however, disruption of certain relevant protein-protein interactions in complexes cannot ever be fully excluded. Alternatively, in-solution digestion allows a more comprehensive analysis of the protein components in a sample, but without the sample simplification or complex-specific resolution provided by prior SDS-PAGE or native PAGE.
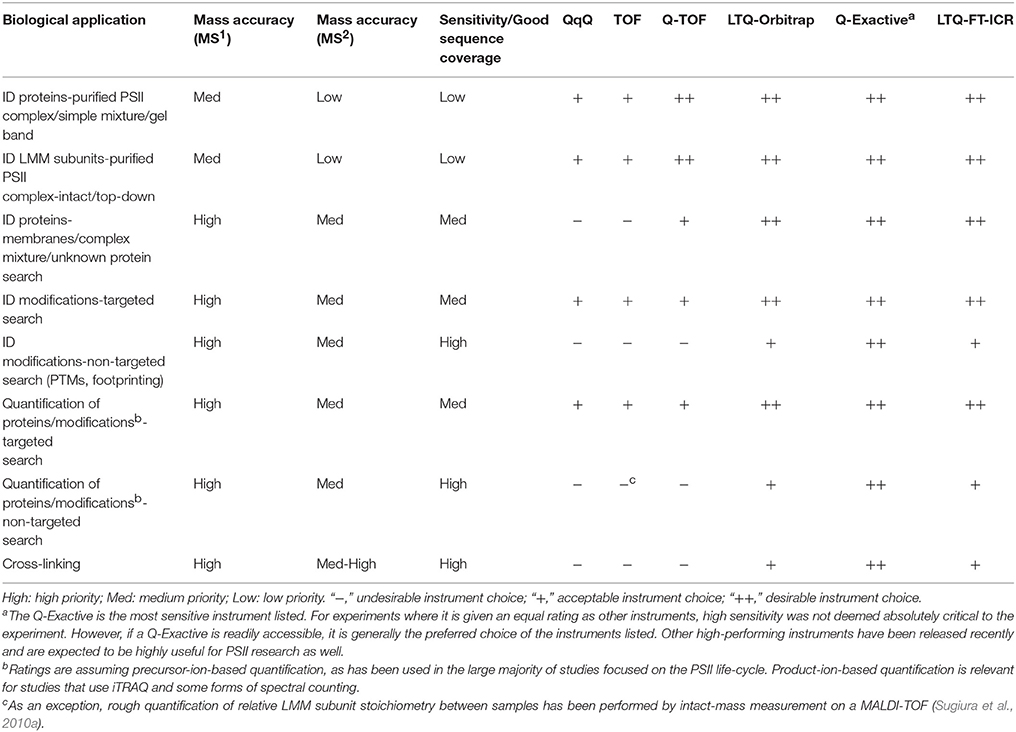
MS instrumentation, as well as membrane-protein sample preparation (Whitelegge, 2013; Battchikova et al., 2015; Heinz et al., 2016) and bioinformatics capabilities, has improved over the last two decades to facilitate PSII life-cycle research (Table 2 summarizes the kinds of experiments that have been performed and the main MS instrumentation and features that enable them). Early mass spectrometers that were applied to PSII research, especially triple-quadrupole (QqQ) and MALDI-time-of-flight (MALDI-TOF) instruments, had relatively low sensitivity, resolving power, and mass accuracy (on the order of 100-several hundred ppm; Michel et al., 1988; Sharma et al., 1997a,b,c; Frankel et al., 1999). Scarcity of genomic sequence data combined with low instrument sensitivity, mass accuracy, and fragmentation efficiency meant that sample analysis was mainly restricted to highly purified PSII complexes or individual subunits, with poor capability for novel protein identification. The mid-2000s saw the appearance of higher-performing instruments, especially the hybrid quadrupole-TOF (Q-TOF) and increasing availability of genomic sequence data for commonly studied photosynthetic organisms. These enabled routine bottom-up identification of the main subunits of PSII complexes (those with soluble domains) from more complex starting mixtures and identification of novel PSII-associated proteins (Kashino et al., 2002; Heinemeyer et al., 2004; Komenda et al., 2005). The fragmentation efficiency of the Q-TOF, however, still limited sequence coverage of proteins. The development and distribution of Fourier transform instruments (ion cyclotron resonance and orbitraps) sometimes interfaced with ion traps provided improved fragmentation efficiency and enabled analysis of highly complex mixtures with higher sequence coverage than ever before. These instruments allow proteome-wide experiments, enable routine confident PTM site identification, and have opened the door for bottom-up MS experiments on photosynthetic systems not before feasible (see Table 2 and sections below).
The Low-Molecular-Mass (LMM) Subunits
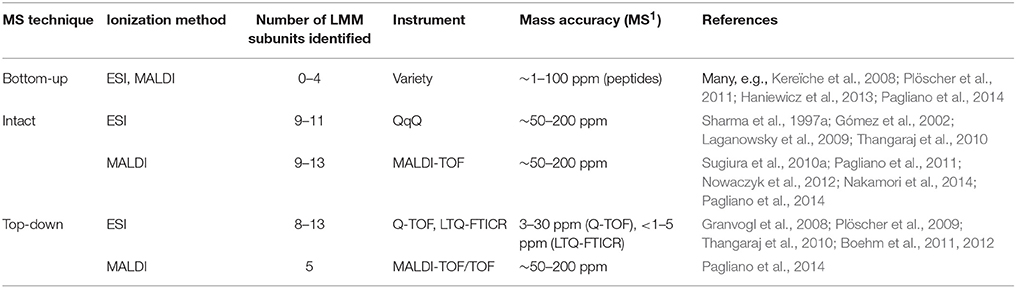
Fully assembled PSII contains around 13 low-molecular-mass (LMM) proteins (< 10 kDa) whose transmembrane domains account for around 40–85% of the sequence. Identification of these very hydrophobic proteins by bottom-up LC-MS/MS is challenging, with typically four or fewer LMM proteins detected (Granvogl et al., 2008; Haniewicz et al., 2013; Pagliano et al., 2014). Difficulties are associated with the proteins' hydrophobicity and lack of soluble domains, which lead to sample losses during preparation, poor tryptic digestion due to infrequent arginines and lysines, slow elution during chromatography, and poor ionization efficiency due to lack of abundant proton-accepting residues. Fractionation by gel electrophoresis carries the additional challenge of extracting the protein from the gel, made more difficult because tryptic digestion sites are infrequent (Granvogl et al., 2008).
To circumvent these difficulties, intact-mass measurement (no MS/MS fragmentation of the protein) and more recently top-down MS strategies have been employed, both of which avoid protein digestion and are able to identify nearly all the LMM subunits in a purified complex (summarized in Table 3). Intact-mass measurement of the LMM subunits was demonstrated by both ESI and MALDI methods, using QqQ and MALDI-TOF instruments (see references cited in Table 3). Both methods achieve roughly 50–200 ppm mass accuracy; especially without fragmentation data, this would typically not be enough for confident identification of an unknown protein. However, because there are only approximately 13 LMM subunits, predicted masses, which are available from genomic sequences in many organisms, are distinctive, and because the starting sample is typically a purified PSII complex, these intact-mass measurements are routinely accepted as confident identifications.
MS/MS fragmentation of intact LMM subunits, however, can be induced using both ESI and MALDI, although ESI has been more successful (see Table 3 and references cited therein). Whitelegge and co-workers (Thangaraj et al., 2010) identified 11 LMM proteins in purified PSII from G. sulphuraria with a linear ion trap quadrupole-Fourier transform ion cyclotron resonance (LTQ-FTICR) instrument after offline LC and confirmed several modifications. They employed both collisional-activated dissociation (CAD) and electron-capture dissociation (ECD) to fragment the proteins, but CAD gave better results for all LMM proteins. Eichacker and co-workers (Granvogl et al., 2008) demonstrated top-down analysis on a Q-TOF with sequence coverage ranging from 14 to 82%. This method has been used in several other recent studies (Plöscher et al., 2009; Boehm et al., 2011, 2012). Notably, Eichacker and co-workers (Granvogl et al., 2008) developed a protocol to perform in-gel extraction of intact LMM proteins prior to top-down analysis (capable of extracting all but the PsbZ protein from the gel matrix). This technique can be used to analyze individual BN-PAGE bands and, thus, identify the LMM components specific to individual types of PSII complexes in heterogeneous mixtures such as a thylakoid membrane proteome or affinity-tagged PSII complexes.
PSII Life-Cycle Application: Composition of Subcomplexes
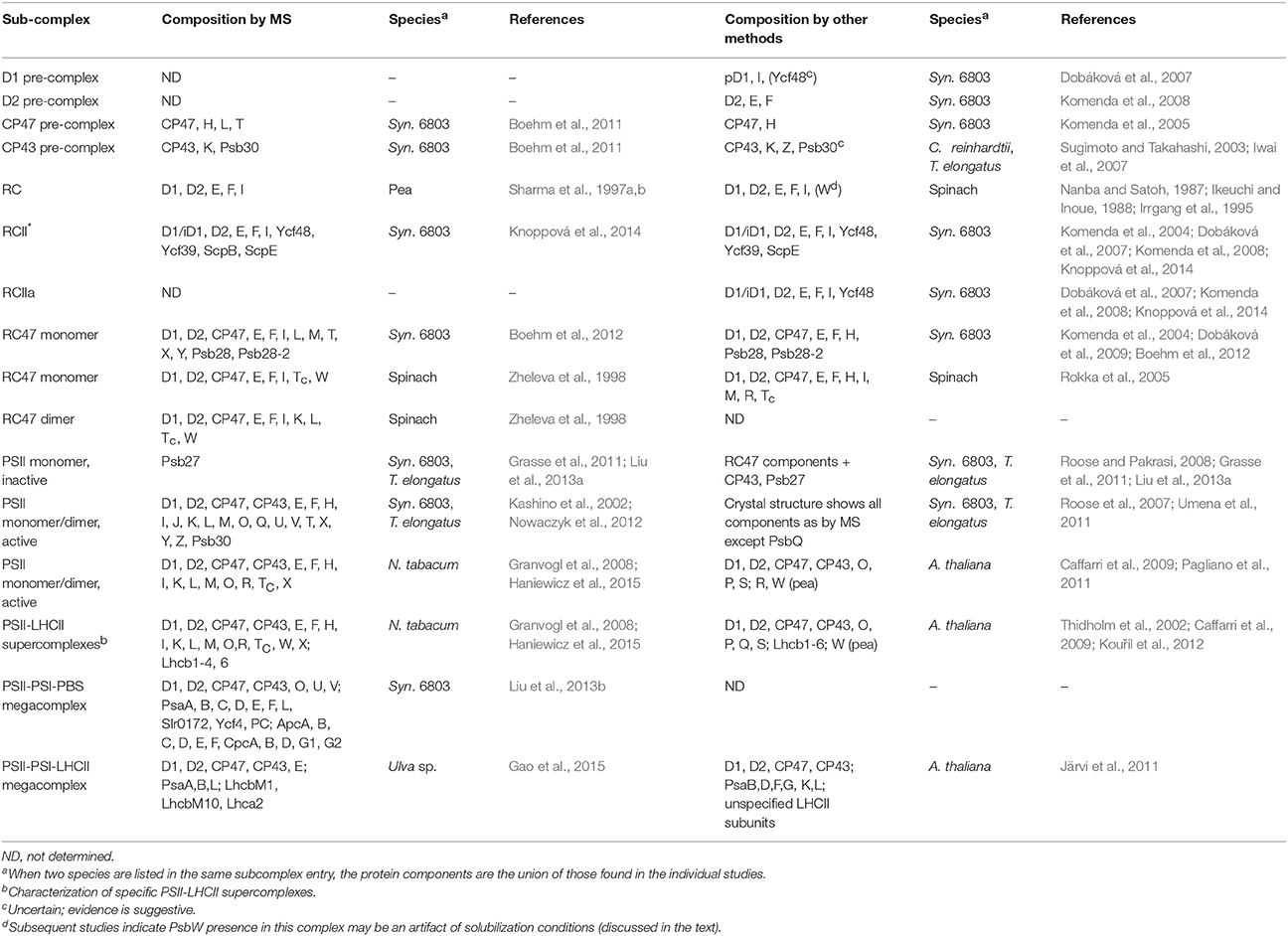
Many subcomplexes form during the PSII life-cycle, and MS has played a critical role, in combination with gel electrophoresis, immunoblotting, crystallography, electron microscopy and other biochemical techniques, in identifying their components (Heinz et al., 2016). A schematic of the life-cycle is shown in Figure 2 (for reviews of the life-cycle and the subcomplexes that form, see Baena-González and Aro, 2002; Aro et al., 2005; Nixon et al., 2010; Shi et al., 2012; Komenda et al., 2012b; Nickelsen and Rengstl, 2013; Järvi et al., 2015; Heinz et al., 2016). A summary of the main subcomplexes whose composition has been studied by MS is found in Table 4 (for completeness, several other subcomplexes are also included). MS analysis generally allows more rapid, comprehensive, and definitive profiling of PSII subunits than other methods, and is especially useful for the LMM subunits that tend to stain poorly on gels. However, owing to the high sensitivity of MS and because relative quantification by MS is not straightforward, it can be difficult to distinguish a trace component of a complex from one that is stoichiometric. Immunoblotting, therefore, complements MS for characterizing composition of subcomplexes.
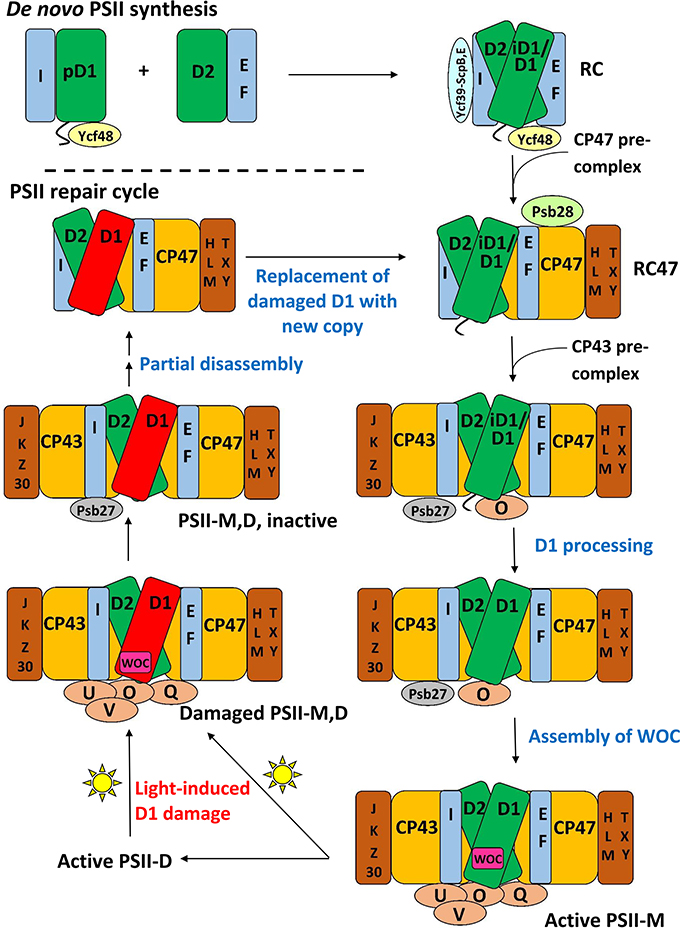
Figure 2. A schematic of the PSII life-cycle. Refer to the text for description of each step. This schematic represents the cyanobacterial PSII life-cycle. The subcomplex progression is similar in algae and higher plants, though several homologous subunits are named differently in these species than in cyanobacteria, and certain subunits are unique to each group (see Rokka et al., 2005; Shi et al., 2012; Nickelsen and Rengstl, 2013; Järvi et al., 2015; Heinz et al., 2016). In algae and higher plants, damaged complexes migrate from thylakoid grana to stromal lamellae for repair and the first steps of reassembly (Tikkanen and Aro, 2014; Järvi et al., 2015). In cyanobacteria, chloroplasts and such inter-thylakoid structure are absent, and repair is not believed to require spatial migration of damaged complexes. De novo PSII synthesis through RC formation appears to begin in specialized membrane subfractions in cyanobacteria, algae, and higher plants before PSII migration to the general thylakoid membrane space, though the details of this process in the various species classes remains to be resolved (Zak et al., 2001; Nickelsen et al., 2011; Nickelsen and Rengstl, 2013; Rast et al., 2015). E, F, H, I, J, K, L, M, O, Q, T, U, V, X, Y, Z, and 30 refer to the PsbE, PsbF, PsbH, etc. proteins, respectively. PSII-M, PSII monomer; PSII-D, PSII dimer.
At the start of de novo PSII assembly, each of the four core subunits D1, D2, CP47, and CP43, forms a pre-complex with specific LMM components. Using a ΔD1 mutant in Synechocystis sp. PCC 6803 (hereafter Synechocystis 6803) and top-down ESI-MS on a Q-TOF, Nixon and co-workers (Boehm et al., 2011) showed that the CP47 pre-complex contains the LMM subunits PsbH, PsbL, and PsbT, whereas the CP43 pre-complex contains the LMM subunits PsbK and Psb30. In this study, it was not possible by MS alone to demonstrate fully stoichiometric binding, just co-purification, of those LMM subunits to CP47 and CP43. However, these results are consistent with the PSII crystal structures and other non-MS-based results (Boehm et al., 2011 and references cited therein). Previous evidence implies PsbZ could also associate with the CP43 pre-complex (Iwai et al., 2007; Guskov et al., 2009; Takasaka et al., 2010), but it was not detected by MS in this study. As determined by affinity purification and immunoblotting, the D1 pre-complex contains PsbI and possibly Ycf48 (Dobáková et al., 2007). It was suggested that a Ycf39-ScpB-ScpE complex may also associate as early as this stage to insert chlorophyll into D1 (Knoppová et al., 2014). The D2 pre-complex contains PsbE and PsbF (Müller and Eichacker, 1999; Komenda et al., 2008).
The D1 and D2 pre-complexes merge to form the reaction center (RC) complex, the earliest subcomplex capable of charge separation (Baena-González and Aro, 2002; Dobáková et al., 2007). The RC complex, initially isolated from spinach and wheat by detergent solubilization of thylakoid membranes, was characterized by gel electrophoresis and immunoblotting to contain D1, D2, PsbE, PsbF, and PsbI (Nanba and Satoh, 1987; Ikeuchi and Inoue, 1988). Intact-mass and bottom-up MS studies later confirmed this composition (Sharma et al., 1997a,b,c). Several biochemical studies detected the 10-kDa PsbW subunit, which is found in green algae and higher plants but not in cyanobacteria, as an additional component (Irrgang et al., 1995; Lorković et al., 1995; Shi and Schröder, 1997). Subsequently, more specific studies (including an MS-based one, Granvogl et al., 2008) showed that PsbW associates later, to dimers during formation of PSII-Light-harvesting complex II (LHCII) supercomplexes (see below; Shi et al., 2000; Thidholm et al., 2002; Rokka et al., 2005; Granvogl et al., 2008). Despite attaching to PSII at a late stage of assembly, PsbW may bind tightly to the D1/D2 surface and, thus, remain partially attached to the RC complex during solubilization, while other peripheral PSII subunits are removed, explaining the controversy (Rokka et al., 2005). This case highlights that subcomplexes obtained from detergent solubilization, a technique used especially in early PSII subcomplex studies, do not necessarily represent subcomplexes that form in vivo. An alternative major method for isolating PSII subcomplexes is purifying them from mutant strains that are “blocked” at a particular stage of assembly. Such complexes are indeed formed in vivo, but it is possible that the altered relative quantity of PSII subunits in the thylakoid membrane arising from the mutation may lead to artefactual binding of certain subunits to some subcomplexes (Thidholm et al., 2002). In cyanobacteria, two slightly different forms of the RC complex were observed, labeled RCII* and RCIIa, which differ slightly in accessory protein content (see Table 3 and the section below). MS was critical in RCII* component characterization, and was indirectly used for RCIIa characterization as well by gel and immunoblot comparison (Knoppová et al., 2014).
The next complex formed during PSII assembly is the RC47 intermediate, also called the CP43-less core monomer in plants, formed by attachment of the CP47 pre-complex to the RC complex. In 1998, Barber and co-workers (Zheleva et al., 1998) showed by MS that the monomeric RC47 complex from spinach contains the D1, D2, CP47, PsbE, PsbF, PsbI, PsbTc, and PsbW proteins, and the dimeric form contains, in addition, PsbK and PsbL. From the later studies on PsbW cited above, PsbW presence may arise from a tight binding to the D1/D2 surface, not in vivo presence in the RC47 complex during assembly. Based on Nixon and co-workers' study (Boehm et al., 2011) on the CP47 pre-complex in Synechocystis 6803, it would be expected that RC47 also contains PsbH. Indeed, a more recent MS-based study of the RC47 complex from Synechocystis 6803 identified all the proteins found by Barber and co-workers (Zheleva et al., 1998) in their monomeric RC47 complex (except PsbW which is not found in cyanobacteria), plus PsbH, PsbM, PsbX, PsbY, and Psb28 (Boehm et al., 2012).
Attachment of the CP43 pre-complex to RC47 forms the inactive PSII monomer (Nickelsen and Rengstl, 2013). Active monomeric PSII is formed upon D1 processing (Liu et al., 2013a), dissociation of Psb27 (Liu et al., 2013a), assembly of the water-oxidizing manganese-calcium cluster and photoactivation (Dasgupta et al., 2008), and binding of PsbO, PsbU, and PsbV (cyanobacteria) or PsbO, PsbP, and PsbQ (algae and higher plants; Bricker et al., 2012). Active monomers dimerize and can attach to the phycobilisome antenna complex (cyanobacteria) (Mullineaux, 2008) or various oligomeric states of LHCII complexes (algae and higher plants) (Kouřil et al., 2012).
Although, crystal structures of active PSII dimers from cyanobacteria are available, several MS studies of fully-assembled cyanobacterial PSII have provided independent confirmation of the subunits present in purified complexes under more native conditions (Sugiura et al., 2010a; Nowaczyk et al., 2012). Using native conditions has even helped discover a component (PsbQ) that was lost during crystallization (Kashino et al., 2002; Roose et al., 2007). The majority of PSII from algae and higher plants is found in several PSII dimer-LHCII supercomplexes (for a review see Kouřil et al., 2012). MS studies (in concert with other techniques) have identified their subunit compositions, even in the absence of crystal structures of the complexes from these organisms. Eichacker and co-workers (Granvogl et al., 2008) showed that the four PSII-LHCII supercomplexes in Nicotiana tabacum contain identical PSII core and LMM subunits (of the eight LMM subunits identified), and that only PSII-LHCII supercomplexes contain the PsbW protein. These results support previous studies that suggest that PsbW may facilitate linkage of LHCII trimers to PSII (Shi et al., 2000; Thidholm et al., 2002; Rokka et al., 2005). Using both bottom-up and top-down MS techniques, Pagliano et al. (2014) found that the various supercomplexes in pea contain identical core and LMM subunits, but that the C2S2M2 supercomplex contains the PsbQ, PsbR, PsbP, Lhcb3, and Lhcb6 proteins whereas the C2S2 supercomplex does not. In light of the stabilizing effect of the PsbQ and PsbP proteins on oxygen evolution, this finding raises interesting questions about the role of the C2S2 supercomplex. Another recent study used MS to characterize PSII-LHCII supercomplexes in N. tabacum and found a few differences in subunit composition; in particular, the C2S2 supercomplex contained Lhcb1 isoform CB25, while the C2S2M2 supercomplex did not (Haniewicz et al., 2015).
Several studies indicate that PSII-PSI-antenna megacomplexes can form in both cyanobacteria and higher plants. Using in vivo cross-linking, Blankenship and co-workers (Liu et al., 2013b) captured a PSII-PSI-phycobilisome megacomplex in Synechocystis 6803. The authors used MS to demonstrate presence of subunits from each complex in the preparation (Tables 3, 5), and identified cross-links revealing specific inter-complex subunit interactions. Aro and co-workers (Tikkanen et al., 2008b, 2010) showed that LHCII can transfer excitation energy to PSI in grana margins of higher plants as a means of balancing energy flux under varying light conditions. In support of this hypothesis, two PSII-PSI-LHCII megacomplexes from Arabidopsis thaliana were observed by a novel large-pore BN-PAGE system (Järvi et al., 2011), and more recently, a PSII-PSI-LHCII megacomplex was identified by MS from the macroalga Ulva sp. under drought stress conditions (Gao et al., 2015).
PSII Life-Cycle Application: Identification of Accessory Proteins
Many accessory proteins bind transiently to PSII subcomplexes during the PSII life-cycle, serving key regulatory roles, but are not present in the crystal structure owing to their absence in fully assembled PSII. For reviews of the accessory proteins of PSII (see Shi et al., 2012; Komenda et al., 2012b; Nickelsen and Rengstl, 2013; Mabbitt et al., 2014; Järvi et al., 2015; Heinz et al., 2016). Bottom-up MS has played a key role in identifying some of the known ones, and others likely remain to be identified. Identifying a previously unknown PSII-associated protein in this manner, however, is not straightforward because the mass spectrometers used for bottom-up analysis are so sensitive that dozens of contaminant proteins are often detected even in “purified” complexes. Low signal intensity of a peptide compared to those of known PSII peptides does not necessarily indicate a contaminant at low abundance because different peptides have different intrinsic ionization efficiencies, and many accessory proteins bind sub-stoichiometrically to PSII. Certain contaminant proteins such as NDH-1 complex subunits (Nowaczyk et al., 2012), ATP synthase subunits (Komenda et al., 2005), phycobilisome subunits (Kufryk et al., 2008), certain ribosomal proteins (Liu et al., 2011b), and several carbon dioxide-concentrating mechanism proteins (Kufryk et al., 2008; Liu et al., 2011b) are frequently observed. Careful examination of the full list and consideration of the experimental conditions are needed to distinguish plausible PSII-interaction candidates from contaminant proteins (Kashino et al., 2002). Although, different MS search software packages use different algorithms for scoring protein hits, a strict statistical confidence threshold should be employed and reported. Overall, although a simple bottom-up experiment is a powerful tool to suggest new candidate proteins that associate with PSII, subsequent targeted experiments on each one are needed to confirm the interaction.
This strategy has proven successful many times for identifying new PSII interaction partners. An early example (Kashino et al., 2002) analyzed SDS-PAGE bands by MALDI-TOF MS from a highly purified PSII preparation and identified several novel proteins, Sll1638 (PsbQ), Sll1252, and Sll1398 (Psb32), that appeared to be plausible PSII interaction partners. Follow-up biochemical studies targeting these proteins confirmed their role in the PSII life-cycle and elucidated functional aspects of each (Inoue-Kashino et al., 2011; Wegener et al., 2011; Bricker et al., 2012). A later proteomic study of purified PSII complexes revealed that the Slr0144-Slr-0152 proteins, all part of one operon, associate with PSII, leading to further characterization of their role in PSII assembly (Wegener et al., 2008). In other cases, specific subcomplexes were isolated before MS analysis and identification of accessory proteins. For example, analysis of a gel band from ΔctpA-HT3-PSII revealed that the Psb27 protein binds specifically to a PSII subcomplex that accumulates before D1 processing (Roose and Pakrasi, 2004), initiating the studies that ultimately elucidated its role in PSII assembly (Nowaczyk et al., 2006; Roose and Pakrasi, 2008; Grasse et al., 2011; Liu et al., 2011a,b; Komenda et al., 2012a). MS analysis showed that the Ycf39, ScpB (HliC), and ScpE (HliD) proteins bind specifically to the RCII* form of the reaction center complex, but not the related RCIIa form (Knoppová et al., 2014). The specific binding of the accessory proteins Psb28 (Dobáková et al., 2009; Boehm et al., 2012) and Psb28-2 (Boehm et al., 2012) to the RC47 complex, and of Ycf48 to RCII* and RCIIa (Knoppová et al., 2014), was initially discovered by immunoblotting, but the proteins' presence was confirmed by MS, strengthening the finding.
PSII Life-Cycle Application: Identification of PTMs
Identification of Processing Events to Form Mature PSII Proteins
The D1 protein is synthesized as a precursor protein (pD1) with a C-terminal extension that gets cleaved during PSII assembly (Takahashi et al., 1988). An early study using peptide sequencing showed that in spinach, cleavage occurs after Ala-344, removing nine C-terminal residues (Takahashi et al., 1988). Several years later, it was found that, in Synechocystis 6803, cleavage also occurs after Ala-344, removing 16 C-terminal residues (Nixon et al., 1992). In this study, peptide sequencing as well as fast atom bombardment (FAB)-MS (a predecessor for ESI and MALDI) were used to pinpoint this cleavage site. Ala-344 serves as a ligand for a Mn ion in the water oxidation cluster (Umena et al., 2011) so that without cleavage, PSII remains incapable of oxygen evolution (Roose and Pakrasi, 2004). The extension, thus, protects early assembly intermediates from harmful premature water oxidation activity. Interestingly, although D1 in higher plants is cleaved in a single step, cyanobacterial D1 is cleaved in two steps, and an intermediate D1 (iD1) is formed transiently (Inagaki et al., 2001). Although, the iD1 cleavage site remained unknown for two decades, in 2007, MS and biochemical evidence demonstrated that the CtpA protease cleaves after Ala-352 to form iD1, which is then cleaved again after Ala-344 to form mature D1 (Komenda et al., 2007). The significance of the two-step cleavage remains unknown, although iD1 may serve as a signal for transferring an early PSII assembly intermediate from the cytoplasmic to the thylakoid membrane (Komenda et al., 2007).
The CP43 protein also appears to be cleaved before, or during an early stage of, PSII assembly. Tandem MS analysis identified a CP43 peptide in spinach starting with a modified form of Thr-15 (Michel et al., 1988). Based on the genomic sequence, the preceding residue is a leucine, so this peptide would not be a predicted trypsin cleavage product. It was also found that the N-terminus of CP43 is blocked from analysis by Edman degradation, likely owing to N-terminal modification. Taken together, these results show that the first 14 residues of CP43 are cleaved, leaving Thr-15 as the mature protein's N-terminus (Michel et al., 1988). Subsequent studies identified the corresponding CP43 peptide in A. thaliana (Vener et al., 2001) and Synechocystis 6803 (Wegener et al., 2008), suggesting that this cleavage is conserved. Crystal structures of cyanobacterial PSII were not able to resolve the most N-terminal portion of CP43, so those structures do not address this question of CP43 cleavage (Loll et al., 2005; Umena et al., 2011).
Cyanobacterial Psb27, PsbQ, and PsbP have unusually hydrophobic properties for soluble lumen-localized proteins and contain a lipoprotein signal motif and conserved cysteine in their N-terminal regions (Thornton et al., 2004; Nowaczyk et al., 2006; Fagerlund and Eaton-Rye, 2011). This led to the suggestion that they are N-terminally lipid-modified and, thus, anchored to the lumenal surface of the thylakoid membrane. Using lipase treatment and MALDI-TOF MS, Rögner and co-workers (Nowaczyk et al., 2006) showed that Psb27 from Thermosynechococcus elongatus does indeed contain such a modification. Also using MALDI-TOF MS, Wada and co-workers (Ujihara et al., 2008) confirmed this finding with Psb27 from Synechocystis 6803 and also found that Synechocystis 6803 PsbQ, recombinantly expressed in E. coli, is also N-terminally lipid modified. Notably, this group developed a method to extract lipid-modified peptides from a gel matrix after in-gel digestion, enabling downstream MS analysis (Ujihara et al., 2008). During PSII assembly, it is important that Psb27 binds to the lumenal surface before the other extrinsic proteins (Liu et al., 2013a), and the lipid anchor may facilitate this sequence by keeping Psb27 in close proximity at all times. A similar role for the lipid anchor of PsbQ was proposed recently (Liu et al., 2015). A lipid modification on PsbP has not yet been demonstrated although strong suggestive evidence indicates its presence (Fagerlund and Eaton-Rye, 2011).
Identification of Phosphorylation Sites
In the early 1980s, phosphorylation of the four PSII subunits that later came to be known as D1, D2, CP43, and PsbH, was observed. These studies were conducted in vivo and in vitro using 32P labeling of whole cells and thylakoid membranes from Chlamydomonas reinhardtii and pea, with detection of phosphoproteins by autoradiography (Owens and Ohad, 1982, 1983; Steinback et al., 1982). Immunoblotting with antibodies that recognize phosphorylated residues was introduced later and became another popular detection method (Rintamäki et al., 1997). Neither of these methods, however, reveal the modified residue. This information was first obtained by gas-phase sequencing using Edman degradation, which demonstrated that the PsbH phosphorylation site is Thr-2, its N-terminus, in spinach (Michel and Bennett, 1987) and C. reinhardtii (Dedner et al., 1988). Since then, MS analysis has replaced Edman degradation and become the dominant method for phosphorylation-site determination, as it is higher-throughput, more definitive, more sensitive, and not limited by N-terminal blockage (e.g., acetylation). The main sites identified are presented below (for reviews, see Vener, 2007; Pesaresi et al., 2011; Puthiyaveetil and Kirchhoff, 2013).
Tandem MS demonstrated phosphorylation of D1-Thr-2, D2-Thr-2, and CP43-Thr-15, the mature proteins' N-termini, in spinach (Michel et al., 1988), A. thaliana (Vener et al., 2001), and C. reinhardtii (Turkina et al., 2006). Phosphorylation of CP43 was also observed at Thr-20, Thr-22, and Thr-346 in spinach (Rinalducci et al., 2006), and at Thr-346 and Ser-468 in A. thaliana (Sugiyama et al., 2008; Reiland et al., 2009). MS analysis showed that PsbH is phosphorylated at its N-terminus in A. thaliana, supporting the Edman degradation data from spinach and C. reinhardtii, and additionally demonstrated phosphorylation of Thr-4 (Vener et al., 2001). Intact-mass MS evidence also indicates double PsbH phosphorylation in spinach and pea (Gómez et al., 1998, 2002). More recently, phosphorylation of the extrinsic proteins PsbP, PsbQ, and PsbR was observed in phosphoproteomic studies of A. thaliana (Sugiyama et al., 2008; Lohrig et al., 2009; Reiland et al., 2009). Although, not discussed here, phosphorylation of LHCII is well-documented, and it regulates state transitions in green algae and higher plants (for reviews see Lemeille and Rochaix, 2010; Minagawa, 2011; Schönberg and Baginsky, 2012; Tikkanen and Aro, 2014; Tikhonov, 2015).
Phosphorylation of PSII subunits is not absolutely required for PSII repair (Bonardi et al., 2005) but assists in transferring damaged PSII complexes from the stacked thylakoid grana to stromal lamellae, where repair occurs. Phosphorylation appears to induce architectural changes in the stacked grana and increase membrane fluidity in such a way as to promote mobility of damaged PSII centers to the stromal lamellae for repair (Tikkanen et al., 2008a; Fristedt et al., 2009, 2010; Herbstová et al., 2012; Järvi et al., 2015). For many of the PSII phosphorylation sites, light intensity and/or other environmental conditions affect the phosphorylation extent, with implications for the functional significance of these modifications. MS analysis has played a critical role in these quantitative studies, and methodology for such measurements is discussed in the dynamics section below. For reviews that discuss the role of PSII phosphorylation (see Pesaresi et al., 2011; Mulo et al., 2012; Schönberg and Baginsky, 2012; Järvi et al., 2015).
PSII phosphorylation may not be needed in cyanobacteria owing to the lack of spatial organization of thylakoids (Mulo et al., 2012). However, a recent global proteomics study of the cyanobacterium Synechococcus sp. PCC 7002 (hereafter Synechococcus 7002) found that a portion of D1 copies are phosphorylated at their N-terminus, Thr-2 (Yang et al., 2014), as in higher plants. This finding opens the possibility for a role of phosphorylation in PSII turnover in cyanobacteria.
Identification of Oxidative and Other Modifications
Light is necessary for PSII function, but even low light intensities can lead to PSII damage, particularly of the D1 protein. Damage triggers partial PSII disassembly, D1 degradation, insertion of a new D1 copy, and PSII re-assembly (Nickelsen and Rengstl, 2013). When the rate of damage exceeds that of repair, photosynthesis is inhibited, referred to as photoinhibition. Photodamage can be initiated in several ways, but a common result of each mechanism is production of highly oxidizing species (e.g., singlet O2, other reactive oxygen species (ROS), or radical PSII cofactors). These species rapidly oxidize PSII subunits, ultimately rendering the complex non-functional. For reviews of the photoinhibition process (see Barber and Andersson, 1992; Adir et al., 2003; Pospíšil, 2009; Allahverdiyeva and Aro, 2012; Tyystjärvi, 2013).
Although oxidative damage of PSII was long believed to be responsible for photoinhibition (Telfer et al., 1994), MS studies provided the first concrete evidence for specific oxidative modifications of PSII. Bottom-up MS analysis of the D1 and D2 subunits from pea PSII found up to three +16 oxidative modifications (each representing incorporation of an oxygen atom) on certain peptides (Sharma et al., 1997c). Interestingly, not all peptides were oxidized, but the oxidized ones were all located near the predicted D1 and D2 redox cofactor sites, supporting the idea that radical redox cofactors themselves, or ROS produced by reaction with them, cause oxidative damage to PSII. More recently, Bricker and co-workers (Frankel et al., 2012, 2013b) used tandem MS to identify oxidized residues on spinach D1, D2, and CP43 that are located near the QA, PheoD1, and manganese cluster sites, all reasonable sources of oxidizing species. Additionally, tryptophan oxidation products in spinach were identified on CP43-Trp-365 and D1-Trp-317, which are located near the manganese cluster (17 and 14 Å, respectively, in the crystal structure from T. elongatus; Anderson et al., 2002; Dreaden et al., 2011; Kasson et al., 2012). By monitoring the digested peptides' absorption at 350 nm, the authors found that these tryptophan oxidations are correlated with increased light intensity and decreased oxygen evolution. Other modifications to PSII subunits were also detected by MS (Gómez et al., 2002, 2003; Anderson et al., 2004; Rexroth et al., 2007; Sugiura et al., 2013). Notably, a recent global proteomics study of Synechococcus 7002 identified many new PSII PTMs (Yang et al., 2014), but the functional significance of these modifications remains to be determined.
Dynamics: Quantitative or Semi-Quantitative Changes in PSII Proteins and PTMs
MS-Based Methods to Study PSII Dynamics
Most MS-based quantification experiments seek the relative, not absolute quantity of a protein or PTM in one sample compared to another. We focus here on relative quantification methods because nearly all the work on PSII dynamics fell into that category.
Gel-Based Quantification
Perhaps the most basic MS-based semi-quantitative method is in-gel digestion at the same band in two different sample lanes, prompted by a significant staining-intensity difference between the two bands. This approach was used frequently when analyzing different purified PSII complexes (Liu et al., 2011b; Knoppová et al., 2014), yielding information about accessory proteins that bind specifically to certain subcomplexes. A proper loading control (typically equal chlorophyll) must be used to ensure a meaningful comparison. Multiple proteins are typically identified by MS in both bands, however, so it may not be immediately apparent which protein is the main component (Liu et al., 2011b). Confirmation may be necessary by western blotting or one of the more quantitative MS-based techniques described below.
The accuracy of gel-based quantification can be improved by introducing a second electrophoretic separation dimension before in-gel digestion and LC-MS/MS. Semi-quantitative two-dimensional denaturing gel electrophoresis (2DE) (distinct from 2D BN-PAGE described above), a popular technique especially in early proteomics studies, usually first separates proteins by size and then on the basis of pI (Rabilloud et al., 2010). The difference in staining intensity indicates the relative content of that protein in each sample. Because two proteins migrate less often together in two dimensions than in one, separation and quantification accuracy are improved. 2DE is useful for large-scale studies such as whole-cell or whole-organelle proteome profiling that require higher-resolution separation than a 1D gel provides. However, in recent years, 2DE has declined in popularity owing to its numerous drawbacks (reviewed in Rabilloud et al., 2010) and the improvements in other more versatile quantitative MS methods. Such large-scale proteomics studies have detected expression-level changes in several PSII proteins in response to a variety of stress conditions (e.g., Ingle et al., 2007; Aryal et al., 2011; Li et al., 2011; Guerreiro et al., 2014). However, insights into the PSII life-cycle have mainly emerged from more focused studies on purified PSII complexes.
Label-Free Quantification
Some MS-based relative quantification methods use a so-called label-free approach, but the better approach, when feasible, is to introduce a stable isotope into the sample. For label-free quantification, the samples to be compared are analyzed by LC-MS/MS separately. A variety of software tools can then be used to obtain an extracted ion chromatogram (EIC) of any peptide. The EIC displays the total intensity (peak area) of that peptide. Comparing the intensities of the same peptide from two different samples indicates the relative content of that peptide in those samples. Although the concept is simple, accurate label-free quantification depends on a number of factors: equal sample loading (on a relevant basis, e.g., chlorophyll concentration), reproducible LC runs, lack of ion suppression, and appropriate normalization during data analysis. For quantification of proteins, data from component peptides must be merged in a statistically sound way (Bantscheff et al., 2012; Nahnsen et al., 2013). Thorough mass spectral sampling of possible precursors—not as crucial in non-quantitative experiments—is necessary for accurate peak definition, but that typically diverts instrument time from obtaining product-ion spectra that give information for peptide identification and sequence coverage (Bantscheff et al., 2012). Various strategies have been designed to address this challenge (e.g., data-independent acquisition approaches such as MSE (Silva et al., 2006; Grossmann et al., 2010) and “all-ion fragmentation” (Geiger et al., 2010) especially when combined with Ultra-Performance LC (UPLC) (Bantscheff et al., 2012). Label-free quantification by spectral counting, which involves comparing the total number of product-ion (MS/MS) spectra obtained for a given peptide or protein, is a common approach (Lundgren et al., 2010), although that has been used in fewer PSII-related studies (Fristedt and Vener, 2011; Stöckel et al., 2011). Label-free quantification of intact proteins is more direct than comparing peptides, but best applied for small proteins. Intact-mass spectra (MALDI and ESI) of the LMM PSII proteins indeed have been used in a number of instances for label-free quantification between states (Laganowsky et al., 2009; Sugiura et al., 2010a).
Isotope Label-Based Quantification
The alternative to label-free quantification is introduction of a stable isotope label into one of the two samples being compared (certain methods also allow greater multiplexing, see below). In contrast to the label-free approach, the labeled and unlabeled samples (often called “heavy” and “light”) are mixed and analyzed in a single LC-MS/MS run. The mass spectra of the light and heavy peptide show two peaks shifted slightly in mass. Comparison of their peak areas, just as in label-free quantification, indicates the relative amount of that peptide in each sample (Bantscheff et al., 2012). Although, comparing peak areas from a single LC-MS/MS run eliminates the concerns of label-free LC reproducibility and ion suppression, labeling introduces additional sample preparation steps and often involves costly reagents.
Isotopic labeling (with 2H,13C, 15N, or 18O) of all proteins can be accomplished during cell growth (metabolic labeling), or by labeling a subset of proteins or peptides at various stages after cell lysis (chemical or enzymatic labeling). In the SILAC method (“stable isotope labeling by amino acids in cell culture”; reviewed in Chen et al., 2015), addition of labeled arginine or lysine to the growth medium results in incorporation of only the labeled form of that amino acid into all proteins. Hippler and co-workers (Naumann et al., 2007) used a SILAC-based method to measure changes in expression of PSII subunits and other proteins in C. reinhardtii under iron deficiency, and Jacobs and co-workers (Aryal et al., 2011) used this method to measure light-dark diurnal cycles in Cyanothece sp. ATCC 51142. A more common approach in PSII life-cycle research, however, has been 15N metabolic labeling (see “Measuring the temporal dynamics of life-cycle events using isotopic labeling” below), in which the growth medium is modified so that the only nitrogen source is a labeled salt such as potassium nitrate or ammonium chloride (Gouw et al., 2010).
Isotopic labeling at the peptide or protein level during downstream processing after cell lysis is an alternative to metabolic labeling. Tandem mass tags (TMT) (Thompson et al., 2003), isotope tags for relative and absolute quantification (iTRAQ) (Ross et al., 2004), enzymatic 18O labeling, and isotope-coded affinity tags (ICAT) can be used in proteomics experiments in photosynthetic organisms (Thelen and Peck, 2007). TMT and iTRAQ are related approaches that have become popular recently (Bantscheff et al., 2012). Both modify peptides with one of several possible isobaric tags that produce reporter ions during MS/MS fragmentation. Each sample is labeled with a different tag, but owing to the tags' isobaric nature, identical peptides from each sample are observed together chromatographically and as a single peak in a low-resolving power mass spectrum. Each tag, however, contains a unique reporter ion that appears as a distinct peak in the product-ion (MS/MS) spectra, and the ratio of these ions reveals the relative amounts of that peptide in each sample. The iTRAQ reagent modifies primary amines, and TMT tags are available that modify primary amines, thiols, or carbonyl groups. Advantages of these labeling approaches include the ability to multiplex up to 8 or 10 samples, greater than with metabolic and other chemical labeling methods, and the isobaric nature of the same peptide across all samples reduces both LC separation demands and MS data complexity (Bantscheff et al., 2012). Although, many proteomics studies on photosynthetic organisms have used these chemical labeling methods, most have not focused on PSII life-cycle issues (Thelen and Peck, 2007; Battchikova et al., 2015). Two relevant examples include the detection of elevated PsbO cysteine oxidation under DCMU and dark conditions (Guo et al., 2014), and intriguing evidence that PSII thermotolerance in Synechocystis 6803 may arise in part from antenna trimming and an increased rate of electron transfer to the cytochrome b6/f complex (Rowland et al., 2010).
PSII Life-Cycle Application: Measuring Changes in Phosphorylation Levels
As mentioned in the composition section above, phosphorylation of PSII subunits affects membrane fluidity and inter-thylakoid dynamics, thus playing a role in facilitating PSII turnover in green algae and higher plants (see the reviews cited in that section for in-depth treatment of this topic). Many of the studies that have contributed to our current understanding of this process used MS quantification techniques to compare phosphorylation levels between samples and under different environmental conditions.
When using peak-area-based label-free quantification to determine the change in a modified peptide between samples, it is crucial that the peak area of the unmodified peptide be taken into account as well, to distinguish a true change in modification extent from simply an increased level of protein expression in one of the states. This method is demonstrated in a study of phosphorylation and nitration in A. thaliana grown under low and high light regimes (Galetskiy et al., 2011b). The authors first normalized each modified-peptide peak area in each sample to that of its unmodified counterpart and then compared the modified peptides' normalized peak area to each other. This method can reveal fold-changes in modification extent between the two states, but not the absolute percentage of that peptide that contains the modification (the “modification stoichiometry”).
To find the modification stoichiometry, it is necessary to know in addition the relative “flyability” (ionization efficiency) of the modified and unmodified peptides. Vierstra and co-workers (Vener et al., 2001) showed that the relative flyabilities of six synthetic phosphopeptides and their non-phosphorylated counterparts are nearly identical. Suggesting this as a general phenomenon for phosphorylated peptides, they estimated the modification stoichiometry for the phosphorylated peptides of D1, D2, CP43, PsbH, and an LHCII protein. In 2010, Vener and co-workers (Fristedt et al., 2010) calculated the actual relative flyability ratio for these PSII peptide pairs, and reported reliable modification stoichiometry for these proteins for the first time under the various conditions in their study. Interestingly, the flyability ratios were indeed close to 1 for each pair (ranging from 0.89 to 1.23), supporting the earlier suggestion that this may be the case for most phosphorylated/non-phosphorylated peptide pairs (Vener et al., 2001). Other studies have since used those flyability ratios to determine changes in the modification stoichiometry of those same phosphorylation sites under other growth conditions (Fristedt and Vener, 2011; Romanowska et al., 2012; Samol et al., 2012). Knowledge of modification stoichiometry under different conditions is quite valuable; it enabled, for example, a greater level of confidence and detail in the model proposed for how phosphorylation affects thylakoid membrane stacking than would have been possible with fold-change data alone (Fristedt et al., 2010).
Chemical isotopic labeling of peptides has also been applied fruitfully to the study of PSII phosphorylation. Immobilized metal-ion affnity chromatography (IMAC) is a standard protocol for enrichment of phosphopeptides, taking advantage of the interaction between phosphoryl groups and a Fe3+-agarose matrix (Andersson and Porath, 1986). Given that free carboxyl groups can also interact with the resin, it has become common to convert free carboxylates to methyl esters after digestion and prior to IMAC, to avoid this interaction (Ficarro et al., 2002). Vener and co-workers (Vainonen et al., 2005) modified this approach by using deuterated methanol (CD3) as the esterification reagent for one sample, and unlabeled methanol for a second sample to quantify by “isotope encoding.” After mixing the samples and analyzing by LC-MS/MS, the relative amount of each phosphorylated peptide in the two samples is quantified by comparison of their mass spectral peak areas. It should be noted that this approach does not reveal the modification stoichiometry of any phosphorylation site; rather the techniques described above still need to be performed to gain that information. Instead, as with other isotope-labeling strategies, it enables more confident and straightforward comparisons of the level of any given peptide between samples. This labeling method was used to study phosphorylation of PSII under a variety of conditions and genetic backgrounds (Vainonen et al., 2005; Lemeille et al., 2010; Fristedt and Vener, 2011; Samol et al., 2012).
PSII Life-Cycle Application: Measuring Changes in Oxidation Levels
As discussed above, oxidation of PSII subunits is a well-documented phenomenon, and occurs, at least partially, from oxidizing species generated during the electron transfer reactions of PSII, especially under stress. However, relatively few studies have quantified changes in PSII subunit oxidation under different controlled conditions. Adamska and co-workers (Galetskiy et al., 2011a) used label-free quantification to compare oxidation and nitration (also associated with oxidative stress) levels of thylakoid membrane protein complexes from A. thaliana grown under low and high light. They found significantly more modified sites in PSII than in the PSI, cytochrome b6/f, and ATP synthase complexes. Interestingly, the modified D1, D2, and PsbO sites increased around 2-5-fold, whereas CP47, CP43, PsbE, and PsbR oxidation levels remained roughly constant. D1 and D2 bind most of the redox-active cofactors of PSII, so the increased oxidation especially of these two proteins is not surprising. Similarly, by measuring the increase in 350 nm absorption, Barry and co-workers (Dreaden et al., 2011; Kasson et al., 2012) found that two tryptophan oxidation products increase after exposure to high light, with a corresponding decrease in oxygen evolution activity. Adamska and co-workers (Galetskiy et al., 2011b) found that nitration levels in assembled PSII complexes decrease after exposure to high light, but increase in PSII subcomplexes. This may imply that once nitrated, PSII complexes are damaged and targeted for disassembly and repair.
PSII Life-Cycle Application: Measuring the Temporal Dynamics of Life-Cycle Events using Isotopic Labeling
Measurement of PSII subunit lifetimes has focused mainly on D1, using immunodetection following addition of a protein-synthesis inhibitor or by radioisotope pulse-chase labeling with detection by autoradiography or phosphorimaging (Aro et al., 1993; Mullet and Christopher, 1994; Ohnishi and Murata, 2006). Recently, several studies used 15N labeling pulses and quantified the disappearance of unlabeled PSII subunits using MS. This method enables simultaneous detection of a larger number of PSII subunits and eliminates any concern of overlapping signal from proteins with similar electrophoretic mobility (Yao et al., 2012b). From surveying nine PSII subunits from Synechocystis 6803, Vermaas and co-workers (Yao et al., 2012a) found that protein half-lives range from 1.5 to 33 h in a PSI-less mutant grown under low light (4 μmol m−2s−1 photon flux). In WT Synechocystis 6803 grown under 75 μmol m−2s−1 photon flux, half-lives of D1, D2, CP47, and CP43 ranged from < 1 to 11 h (Yao et al., 2012b). In both studies, D1 exhibited the shortest half-life. These studies highlight the wide range in PSII subunit lifetime and the tight regulation of protein synthesis and PSII assembly that must occur to ensure constant proper stoichiometric availability of all subunits. Interestingly, the chlorophyll half-life was several times longer than that of the core chlorophyll-binding proteins, but the half-life was reduced in the absence of the small CAB-like proteins (SCPs), implying that SCPs play a role in chlorophyll recycling during PSII turnover (Yao et al., 2012a).
Rögner, Nowaczyk, and co-workers demonstrated an elegant application of 15N labeling by purifying several subcomplexes in the PSII life-cycle after a pulse with 15N (from 15NH4Cl). Comparing extents of incorporation of 15N in different subcomplexes (e.g., monitoring D1 and D2 peptides) reveals the subcomplexes' position in the PSII life-cycle. Using this method, the authors demonstrated that in T. elongatus, Psb27 binds to a monomeric subcomplex early in the PSII assembly process (Nowaczyk et al., 2006), and that Psb27 binds again during disassembly to inactive dimers (Grasse et al., 2011). This information fits well with the current understanding of Psb27 as a gatekeeper preventing manganese cluster assembly in immature complexes (Liu et al., 2013a; Mabbitt et al., 2014).
Cyanobacteria contain multiple versions of the psbA gene, and the resulting versions of the D1 protein have some different properties and are expressed preferentially under different environmental conditions (for reviews see Mulo et al., 2009; Sugiura and Boussac, 2014). For example, the psbA1 gene product in T. elongatus is dominant under standard growth conditions, but expression of the psbA3 gene product, which differs from the PsbA1 copy by ~21 residues, increases under high light conditions (Clarke et al., 1993; Kós et al., 2008; Mulo et al., 2009). Characterization of PSII from mutants that express only specific versions of the gene has shown differences in electron-transfer properties, with the implication that PsbA3 assists in photoprotection of PSII under light stress conditions (Sander et al., 2010; Sugiura et al., 2010b). D1-copy expression was mainly monitored on the transcript level (Golden et al., 1986; Komenda et al., 2000; Kós et al., 2008; Sugiura et al., 2010b). However, using 15N labeling and MS-based quantification, Rögner and co-workers showed that PsbA3 incorporation on the protein level could be monitored unambiguously in T. elongatus under high light conditions (Sander et al., 2010) and in the ΔpsbJ mutant (Nowaczyk et al., 2012). Those studies used 15N-labeled PSII from a strain that only expresses the PsbA3 copy as a standard for 100% incorporation; relative peak area of the unlabeled PsbA3 peptides compared to this standard is a measure of the incorporation. Such definitive monitoring should allow further detailed studies of psbA gene incorporation dynamics.
Progress has also recently been made on the role of the PsbA4 D1 copy; an iTRAQ labeling study found elevated expression of PsbA4 in Cyanothece sp. PCC 7822 in the dark (Welkie et al., 2014), providing complementary evidence to that of Pakrasi and co-workers (Wegener et al., 2015) who found that PsbA4 incorporation into PSII renders the complex non-functional. PsbA4 replaces PsbA1 at night in cyanobacterial species that fix nitrogen during this time, protecting against even the trace levels of oxygen evolution that could occur and damage the nitrogenase enzyme (Wegener et al., 2015).
Structure: Determining Protein-Protein Interactions in PSII Complexes
MS-Based Methods to Study PSII Structure
X-ray crystallography remains the benchmark for determining the structure of protein complexes, but besides fully-assembled active PSII, many complexes that form during the PSII life-cycle are too transient and low in abundance to be easily amenable to crystallography. Valuable information about protein-protein interactions within PSII was obtained from immunogold labeling (Tsiotis et al., 1996; Promnares et al., 2006) and yeast two-hybrid assays (Schottkowski et al., 2009; Komenda et al., 2012a; Rengstl et al., 2013), but the former is primarily suitable for large PSII complexes (Dobáková et al., 2009), and the latter is time-consuming and low-throughput. Both provide relatively low-resolution structural information. Recently, advanced structural proteomics techniques bypass the limitations of the above techniques and offer higher-resolution structural data (although still lower than X-ray crystallography). Either chemical cross-linking or protein footprinting followed by MS detection of these modifications are enabled by MS instruments with high sensitivity, resolving power, and < 1–5 ppm mass accuracy on orbitrap- and FTICR-based instruments (Table 2). These methods allow not only identification of the binding partners of a specific protein but also a low-resolution mapping of the binding site.
Chemical Cross-Linking
Briefly, the chemical cross-linking technique (reviewed in Sinz, 2014) uses a small molecule with two functional groups on either end that can react with protein residues, separated by a spacer arm (typically less than 14 Å). Many types of cross-linkers are available (Paramelle et al., 2013). The ones most commonly used in PSII research (Bricker et al., 2015) can react with either the primary amine of a lysine and protein N-terminus (and under certain conditions, to a lesser extent with the hydroxyl group of a serine, threonine, or tyrosine, Mädler et al., 2009), or with the carboxylate of aspartate and glutamate side chains and protein C-termini. After both sides of the cross-linker react with neighboring proteins, digestion, LC-MS/MS, and specialized data analysis can identify cross-linked peptides. Inter-protein cross-linked peptides provide structural information about the complex because the two linked residues are constrained to the spacer arm-length distance from each other.
Cross-linking has been used for decades to study protein-protein interactions (Clegg and Hayes, 1974; Wetz and Habermehl, 1979; Walleczek et al., 1989; Back et al., 2003; Sinz, 2014), but its power was limited until modern MS instrumentation and the proteomics platform enabled high-throughput analysis and confident identification of linked peptides (Rappsilber, 2011). Identification of cross-linked peptides by MS is more challenging than for a typical protein digest, especially for large complexes, because the candidate peptide database increases roughly with the square of the number of peptides. As a result, false positives based on the mass spectrum are common even with high mass accuracy instruments, making high-quality product-ion spectra critical for a confident assignment. Despite powerful and constantly improving cross-link search algorithms (Rinner et al., 2008; Xu and Freitas, 2009; Petrotchenko and Borchers, 2010; Götze et al., 2012, 2015; Yang et al., 2012; Hoopmann et al., 2015), manual verification of the product-ion spectra of hits is highly recommended. Successful cross-linking requires high sequence coverage and high mass accuracy as is now practical with orbitrap- and FTICR-based instruments (Table 2).
Because cross-linked peptides give typically low-intensity signals compared to those of unlinked peptides, they are often not selected for fragmentation by the instrument's traditional “highest-abundance ion” selection criteria. Several strategies have been developed to improve cross-link selection and/or reduce false positives. They include various methods to enrich for cross-linked peptides before LC-MS/MS (Chu et al., 2006; Kang et al., 2009; Fritzsche et al., 2012; Leitner et al., 2012); use of isotope-coded linkers whose “fingerprint” increases confidence in an identification and can enable real-time guided selection of cross-links for fragmentation (Müller et al., 2001; Pearson et al., 2002; Seebacher et al., 2006; Petrotchenko et al., 2014); and MS-cleavable linkers that simplify data analysis by cleaving a cross-linked peptide into its component peptides before fragmentation (Kao et al., 2011; Petrotchenko et al., 2011; Weisbrod et al., 2013; Buncherd et al., 2014).
Protein Footprinting
Protein footprinting is another MS-based structural technique that has been used to study PSII. Its principle is that a protein residue's solvent accessibility determines its susceptibility to modification by a reagent in the solution; residues buried in a protein-protein interface are less susceptible to modification than surface-exposed residues. These modifications are then detected by MS. Instruments with high sensitivity, resulting in high sequence coverage, are critical so that footprinting experiments yield maximal information (Table 2). A common approach is hydroxyl radical footprinting using the well-established technique of synchrotron radiolysis of water to generate the radicals (Takamoto and Chance, 2006; Wang and Chance, 2011). Fast photochemical oxidation of proteins (FPOP) is a more recent hydroxyl radical fooptrinting technique that uses a laser pulse to generate the radicals and can probe protein dynamics that occur on a faster timescale, down to microseconds (Gau et al., 2011). Hydroxyl radical footprinting can modify 14 of the 20 amino acid side chains (Wang and Chance, 2011). Another technique, glycine ethyl ester (GEE) labeling, adapts a long-standing method for modifying and cross-linking carboxylate groups in proteins (Hoare and Koshland, 1967; Swaisgood and Natake, 1973) for protein footprinting (Wen et al., 2009; Gau et al., 2011). It is easier to implement than hydroxyl radical footprinting, and data interpretation is simpler, but it can only probe changes on aspartate, glutamate, and protein C-termini.
PSII Life-Cycle Application: Cross-Linking and Footprinting to Determine Interactions among PSII Subunits
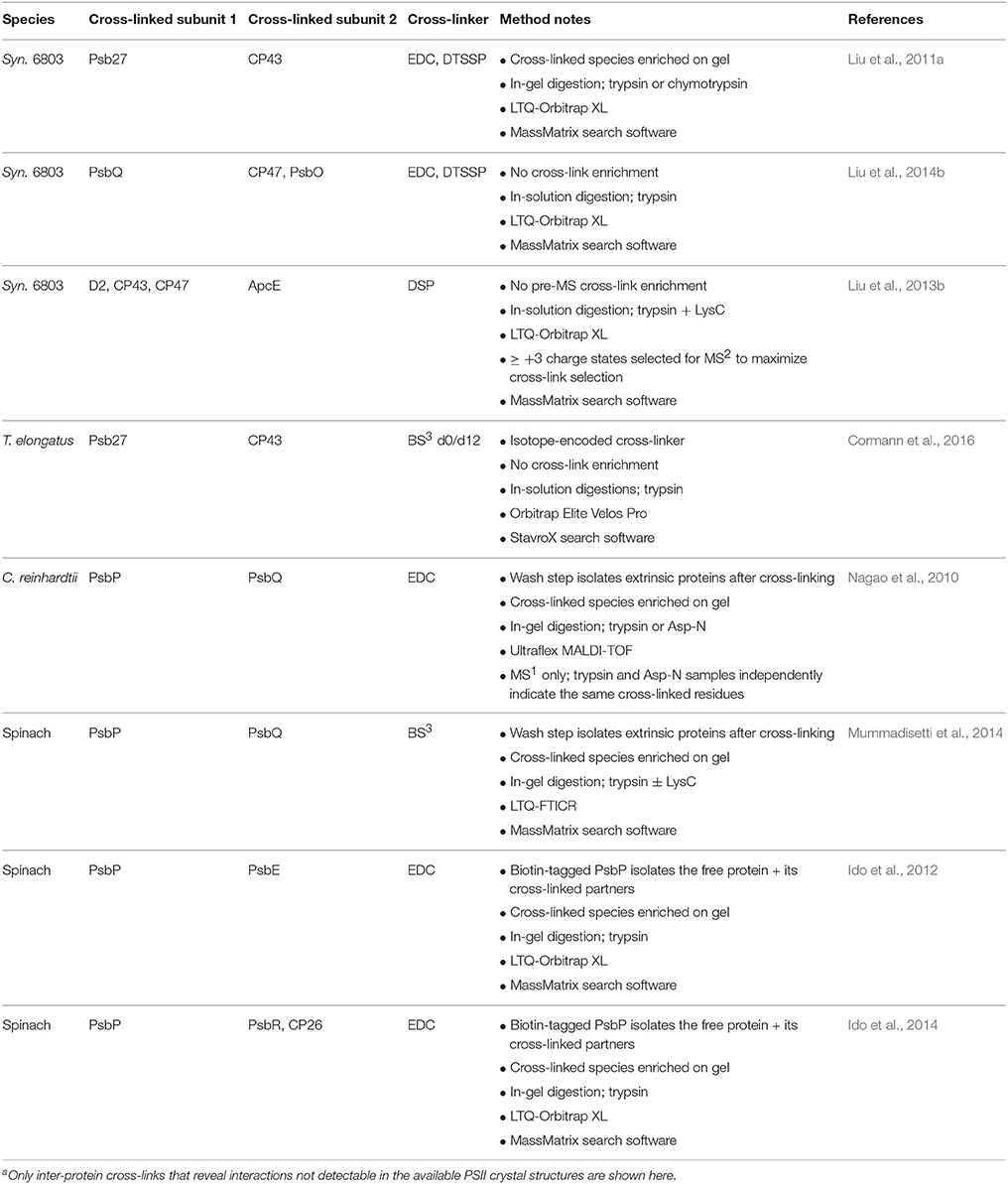
Early cross-linking studies on PSII provided information about subunit connectivity before PSII crystal structures were available. Many studies focused on the lumenal extrinsic proteins (Enami et al., 1987; Bricker et al., 1988; Odom and Bricker, 1992; Han et al., 1994), which are more easily accessible to soluble cross-linkers, but interactions involving the transmembrane subunits can also be detected (Tomo et al., 1993; Seidler, 1996; Harrer et al., 1998). In the absence of the MS-based platforms currently available, gel electrophoresis and immunoblotting identify cross-linked products and their likely component proteins. Those methods are still helpful today as confirmation and when cross-linked peptides are not detected by MS (Hansson et al., 2007; Nagao et al., 2010; Liu et al., 2011a, 2014b), but MS provides much greater confidence in the identification and pinpoints the exact cross-linked residues. Notably, Satoh and co-workers (Enami et al., 1998) used FAB-MS to identify intramolecular cross-linked peptides in PsbO, and deduced the linked residues even without MS/MS capability.
Since these early studies, crystal structures have elucidated the connectivity between the components of active cyanobacterial PSII. As a result, more recent cross-linking studies have focused on accessory proteins that bind only to subcomplexes and/or that are not found in the crystal structures, though work has continued on the lumenal extrinsic PSII subunits from algae and higher plants, PsbP and PsbQ, which differ significantly from their cyanobacterial counterparts (Bricker et al., 2012; results are summarized in Table 5). Cross-linking-MS has also been recently applied to study interactions within the phycobilisome (Tal et al., 2014) and between the phycobilisome and the photoprotective orange carotenoid protein (OCP) (Zhang et al., 2014; Liu et al., 2016), reviewed in Bricker et al. (2015).
With complementary use of the cross-linkers EDC and DTSSP, Pakrasi and co-workers (Liu et al., 2011a) demonstrated that the accessory protein Psb27 binds on the lumenal surface of CP43. Because this interaction is transient and occurs in only a small fraction of PSII centers in the cell at a given time, the authors purified PSII complexes from the ΔctpA mutant strain of Synechocystis 6803 that accumulates such complexes (Liu et al., 2011b), maximizing chances of capturing and observing Psb27 inter-protein cross-links. The two cross-linked species detected were used to map Psb27 onto the PSII crystal structure, showing how Psb27 accomplishes its role as a gatekeeper, protecting partially assembled PSII complexes from gaining premature harmful water oxidation activity (Roose and Pakrasi, 2008). Recently, Nowaczyk and co-workers (Cormann et al., 2016) identified a different cross-link between Psb27 and CP43 in T. elongatus using an isotope-encoded version of the BS3 cross-linker. Despite the different cyanobacterial species used in the two studies, and the different Psb27 residues that were cross-linked, both cross-links localize Psb27 to the same domain on CP43 (Liu et al., 2011a; Cormann et al., 2016).
Cyanobacterial PsbQ is a component of active PSII (Roose et al., 2007), but is not found in any of the crystal structures, presumably because it is destabilized under crystallization conditions. Pakrasi and co-workers (Liu et al., 2014b) again used EDC and DTSSP in parallel and detected a PsbQ-CP47 and two PsbQ-PsbO cross-links by MS. A PsbQ-PsbQ cross-link that appears to arise from two different copies of the protein was also detected. Taken together, these results position PsbQ along the lumenal PSII dimer interface, consistent with evidence that PsbQ stabilizes the PSII dimer (Liu et al., 2014b). In this study, in-solution digestion was used instead of in-gel digestion to avoid losses of large cross-linked peptides that are difficult to extract from the gel matrix.
Several recent studies have probed the binding sites of the higher plant lumenal extrinsic proteins PsbP and PsbQ, which help optimize Ca2+ and Cl− binding properties at the oxygen-evolving center (Bricker et al., 2012). Ifuku and co-workers (Ido et al., 2012, 2014) identified cross-links in spinach PSII between PsbP and PsbE, PsbR, and CP26 by MS and provided MS-based evidence for PsbP-CP43, PsbQ-CP43 and PsbQ-CP26 cross-links. The suggestive evidence arose from MS identification of CP43 or CP26 in individual cross-linked gel bands after affinity pull-downs using biotin-tagged PsbP or PsbQ (Ido et al., 2014). Their binding model for PsbP is different than that proposed by Bricker and co-workers (Mummadisetti et al., 2014), who identified nine intra-protein cross-links between the N-terminal and C-terminal regions of spinach PsbP that constrain significantly its binding conformation. The authors also identified a PsbP-PsbQ cross-link, consistent with that observed in C. reinhardtii by Enami and co-workers (Nagao et al., 2010).
The PsbQ-CP43 interaction in spinach PSII suggested by Ifuku and co-workers (Ido et al., 2014) contrasts with the PsbQ-CP47 cross-link identified in Synechocystis 6803 by Pakrasi and co-workers (Liu et al., 2014b) and their evidence for a PsbQ-PsbQ interaction at the PSII dimer interface. Significant sequence differences between cyanobacterial and plant PsbQ may explain this discrepancy. Bricker and co-workers (Mummadisetti et al., 2014) also found cross-linking evidence for a PsbQ-PsbQ interaction in spinach that may require a position at the dimer interface, consistent with the Pakrasi group's results in Synechocystis 6803. However, they suggest that that interaction could in theory arise from an inter-PSII-dimer interaction, and, thus, the results could alternatively be consistent with the Ifuku group's positioning of spinach PsbQ near CP43. Interestingly, the recently published crystal structure of PSII from the eukaryotic red alga Cyanidium caldarium indeed shows PsbQ′ binding to the lumenal surface of CP43 (Ago et al., 2016). PsbQ′ shares relatively low sequence homology to green algal or higher plant PsbQ; and though PsbQ′ can functionally replace PsbQ at least partially in C. reinhardtii, it cannot bind to spinach PSII (Ohta et al., 2003). Therefore, the red algal PsbQ′-CP43 interaction supports Ifuku and co-workers' (Ido et al., 2014) similar conclusion in spinach, but at the same time it does not necessarily contradict the alternate PsbQ-CP47 interaction observed by the other groups in spinach and Synechocystis 6803. The recent characterization of an active PSII complex from Synechocystis 6803 with multiple copies of the PsbQ protein (Liu et al., 2015) hints at one possible reconciliation of these findings, if such a complex is present in other species as well. Despite some discrepancies, these results begin to elucidate the binding orientation of the higher plant lumenal extrinsic proteins, suggesting a mechanism for stabilization of PSII-LHCII supercomplexes (Ido et al., 2014), and paving the road for further structural studies.
Although the advanced techniques for improving cross-link identification described in the methods section above have largely not yet been applied to PSII studies (with the exception of the recent use of isotope-encoded BS3 by Nowaczyk and co-workers, Cormann et al., 2016), several other creative approaches have been used. Enami and co-workers (Nagao et al., 2010) improved identification confidence by detecting the same cross-linked residues in peptides from two separate digestion experiments, one with trypsin and one with Asp-N. Pakrasi and co-workers (Liu et al., 2011a) provided strong evidence, using the thiol-cleavable cross-linker DTSSP and 2D gel electrophoresis, that Psb27 and CP43 cross-link to each other, allowing targeted data analysis and providing higher confidence in the subsequent MS cross-link identification. Ifuku and co-workers (Ido et al., 2012, 2014) used a biotin-tagged PsbP or PsbQ to purify only those cross-linked proteins. Although this method is not as efficient as purifying only cross-linked peptides by means of a tagged linker, because following digestion many non-linked peptides from the tagged protein will be present, it does simplify sample complexity and focuses on cross-links containing a particular protein of interest. Notably, Blankenship and co-workers (Liu et al., 2013b) demonstrated that in-vivo cross-linking of thylakoid membrane complexes is possible and can capture interactions between protein complexes that are otherwise difficult to preserve after cell lysis. Using the membrane-permeable cross-linker DSP, they captured a PSII-PSI-phycobilisome megacomplex and identified five cross-links between PSII subunits and the PBS, and five between PSI subunits and the PBS, providing the first molecular-level description of the interface of these complexes.
Like cross-linking, protein footprinting is a technique that has long been used in PSII structural studies but that has become significantly more powerful in combination with modern MS. Early studies using N-hydroxysuccinimidobiotin (NHS-biotin) and other modification reagents investigated the binding site of higher plant PsbO to PSII. In the absence of MS detection, specific modification sites could either not be identified (Bricker et al., 1988) or were localized to particular protein domains by N-terminal sequencing of peptides (Frankel and Bricker, 1992). With the rise of protein MS in the mid-1990s, MALDI-TOF and FAB-MS were used to identify modified peptides; lack of MS/MS capability, however, produced lower-confidence peptide identification than is achievable today, and meant that specific modified residues could only be pinpointed in favorable cases (Frankel and Bricker, 1995; Miura et al., 1997; Frankel et al., 1999). Nonetheless, these pioneering footprinting studies demonstrated, e.g., that PsbO interacts with Loop E of CP47 (Frankel and Bricker, 1992), and that charged residues on the surface of PsbO are involved in its interaction with PSII (Miura et al., 1997; Frankel et al., 1999).
Recently, hydroxyl radical footprinting using synchrotron radiolysis of water was used to study the binding surfaces of spinach PsbP and PsbQ to PSII, with detection of modified residues by MS (Mummadisetti et al., 2014). The results reveal buried regions on the surface of these proteins that complement the authors' cross-linking data and suggest these proteins' binding interfaces to other PSII subunits. The data also confirm and elaborate on the binding region identified by this group in a previous study using NHS-biotin as footprinting reagent (Meades et al., 2005).
Although the above footprinting studies detected whether or not a residue was modified in a given state, it is also possible to analyze footprinting data quantitatively to detect a conformational change in a complex in two different states. The label-free approaches described above can be used to monitor the relative change in modification, normalized to the unmodified peptide, in different PSII complexes. The utility of this approach was demonstrated in a study of the role of Psb27 in PSII assembly (Liu et al., 2013a) using GEE labeling. The authors monitored the relative changes in aspartate and glutamate modification of three PSII complexes representing different stages of PSII assembly, not only extending previous information about the Psb27 binding site (Liu et al., 2011a; Komenda et al., 2012a), but also demonstrating a conformational change upon D1 processing that prompts Psb27 dissociation and permits assembly of the oxygen evolving complex (Liu et al., 2013a). Blankenship and co-workers have also used quantitative GEE labeling to detect a light-dependent conformational change in the OCP protein that appears to underlie its photoprotective function (Liu et al., 2014a). The recent implementation of isotopically-labeled GEE (iGEE) footprinting (Zhang et al., 2016) will streamline, and increase confidence in, quantitative comparisons of modification extent between states.
Hydroxyl radical footprinting has also been used to identify putative water and oxygen channels in PSII (Frankel et al., 2013a), a topic that has been explored previously through computational studies (Murray and Barber, 2007; Ho and Styring, 2008; Gabdulkhakov et al., 2009; Vassiliev et al., 2012). This study provides general experimental support for the existence of such channels, confirms specific channel identifications from computational work (Ho and Styring, 2008; Vassiliev et al., 2012), and proposes a previously unidentified putative oxygen/ROS exit channel (see Bricker et al., 2015 for a discussion of the MS-based and computational results).
Future Directions
MS technology and associated sample preparation techniques are evolving rapidly. Increasing sensitivity and speed of instruments for bottom-up proteomics allows better coverage of transmembrane PSII proteins; for example, coverage of the core D1, D2, CP47, and CP43 proteins is routinely ~50–85% on a Thermo Q-Exactive Plus instrument, whereas ~20–40% coverage was reported on LTQ-Orbitrap, LTQ-FTICR, and MALDI-TOF instruments (Aro et al., 2005; Frankel et al., 2012; Liu et al., 2013a,b). This increased coverage will mean that more PTMs and cross-linked peptides can be identified, and a larger portion of the PSII complex can be mapped by footprinting. PTM analysis, especially using quantitative techniques to compare complexes exposed to different conditions, may help elucidate signals (largely unknown in cyanobacteria) that trigger D1 degradation. The increasing availability of high-sensitivity instruments that can achieve high sequence coverage is enabling detailed quantitative and non-quantitative global proteomic studies. The new challenge is to reduce the large amounts of information becoming available into specific testable hypotheses for targeted follow-up studies.
Improvements at all stages of the cross-linking workflow are occurring, from linker design to linked-peptide enrichment and software analysis. Specifically, isotope-labeled and MS-cleavable linkers are powerful tools that are just beginning to be applied to PSII research. In-vivo cross-linking is a promising approach to detect transient or unstable interactions that are difficult to capture after cell lysis. Cross-linking may enable binding site identification for at least some of the approximately 30 accessory proteins now known or believed to associate with PSII during its life-cycle (Nickelsen and Rengstl, 2013; Järvi et al., 2015). Detecting interactions between PSII subcomplexes and, e.g., D1 degradation proteases or proteins involved in chlorophyll loading would also be of prime interest.
Intact-mass measurements of the large core PSII proteins D1, D2, CP47, and CP43 were reported in several studies (Sharma et al., 1997b; Whitelegge et al., 1998; Huber et al., 2004; Thangaraj et al., 2010), with detection of the phosphorylated form of D1 as well in some cases (Whitelegge et al., 1998; Huber et al., 2004). However, their top-down analysis has not yet been achieved. Top-down technology is continuously developing, especially methods for increased product-ion sequence coverage (Frese et al., 2012; Shaw et al., 2013; Brunner et al., 2015) and analysis of larger integral membrane proteins and their PTMs (Ryan et al., 2010; Howery et al., 2012). Such analysis will make it easier to identify nearly-stoichiometric (and potentially important) PTM events from trace ones under different conditions, not an easy task using bottom-up MS. Native MS is capable of analyzing certain intact membrane protein complexes, although the technology is still developing, and no one approach works for all protein complexes (reviewed in Mehmood et al., 2015). Native MS analysis of PSII has not yet been demonstrated, but the technique could in theory serve as a complementary method to native gels to characterize the distribution of PSII subcomplexes under various conditions, and their components. This might be particularly useful to address the stoichiometry of accessory proteins and cofactors, and could add a new tool to address the long-standing question of chlorophyll loading in PSII.
Conclusion
The use of MS has been fueled by improvements in sample preparation methods for analysis of membrane proteins, increasing availability of MS instrumentation, and significant advances in instrument sensitivity, speed, and mass accuracy. Techniques from each of the four pillars of proteomics will continue to be employed to study the PSII life cycle. These techniques have addressed a wide range of questions regarding the composition of PSII complexes, the time-dependent dynamic changes of individual subunits and complexes under different environmental conditions, and the tertiary and quaternary structure of PSII complexes.
Modern MS techniques provide a higher level of detail and confidence than previous methods; examples are identification of a protein's phosphorylation site instead of mere detection of a phosphorylated protein, and identification of specific cross-linked residues instead of only suggestive evidence that two particular proteins are cross-linked to each other. For other applications, the use of MS permits entirely new questions to be asked (e.g., what proteins are present on a proteome-wide scale for a purified PSII complex).
The new information has opened up new questions about function. For example, what are the physiological roles of the many new PTMs that have been identified? What purpose does an accessory protein serve by binding at this particular location on a PSII complex? In some cases, the sensitivity of MS is a potential pitfall: identification of a protein in a PSII sample does not necessarily mean it is a stoichiometric component, or that it associates specifically with the complex at all. Thus, information from MS should be a starting point for more targeted genetic and biochemical studies, and MS is one component of an expanding toolbox for PSII life-cycle research. Rapidly developing MS technology promises continued contributions to this field, which has a wide range of fascinating questions about membrane protein complex composition, dynamics, and structure yet to be answered.
Author Contributions
HP, MG conceived of the article. DW collected data from the literature and drafted the manuscript. HP, MG, and DW revised the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Dr. Hao Zhang (Washington University) and Dr. Phil Jackson (University of Sheffield) for helpful discussions. This work was supported by the Photosynthetic Antenna Research Center, an Energy Frontier Research Center funded by the U.S. Department of Energy (DOE), Office of Basic Energy Sciences (Grant DE-SC 0001035 to MG and HP), by the Chemical Sciences, Geosciences, and Biosciences Division, Office of Basic Energy Sciences, Office of Science, U.S. Department of Energy (grant no. DE-FG02-99ER20350 to HP), and by the NIH (Grant P41GM103422) to MG.
Abbreviations
A.thaliana, Arabidopsis thaliana; BS3, (bis(sulfosuccinimidyl)suberate); C. reinhardtii, Chlamydomonas reinhardtii; DSP, dithiobis(succinimidyl propionate); DTSSP, 3,3′-dithiobis(sulfosuccinimidyl propionate); EDC, 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide; ESI, electrospray ionization; FAB, fast atom bombardment; FTICR, Fourier transform ion cyclotron resonance; GEE, glycine ethyl ester; LC, liquid chromatography; LHCII, Light-harvesting complex II; LMM, low-molecular-mass; LTQ, linear ion trap quadrupole; MALDI, matrix-assisted laser desorption ionization; MS, mass spectrometry; N. tabacum, Nicotiana tabacum; NHS, N-hydroxysuccinimide; OCP, orange carotenoid protein; PBS, phycobilisome; PSII, Photosystem II; PC, plastocyanin; PTM, post-translational modification; QqQ, triple-quadrupole; SCP, small CAB-like protein; Synechococcus 7002, Synechococcus sp. PCC 7002; Synechocystis 6803, Synechocystis sp. PCC 6803; T. elongatus, Thermosynechococcus elongatus; TOF, time-of-flight; WOC, water-oxidizing complex.
References
Adir, N., Zer, H., Shochat, S., and Ohad, I. (2003). Photoinhibition - a historical perspective. Photosynth. Res. 76, 343–370. doi: 10.1023/A:1024969518145
Ago, H., Adachi, H., Umena, Y., Tashiro, T., Kawakami, K., Kamiya, N., et al. (2016). Novel features of eukaryotic Photosystem II revealed by its crystal structure analysis from a red alga. J. Biol. Chem. 291, 5676–5687. doi: 10.1074/jbc.M115.711689
Allahverdiyeva, Y., and Aro, E. M. (2012). “Photosynthetic responses of plants to excess light: Mechanisms and conditions for photoinhibition, excess energy dissipation, and repair,” in Photosynthesis: Plastid Biology, Energy Conversion and Carbon Assimilation. Advances in Photosynthesis and Respiration, eds J. J. Eaton-Rye, B. C. Tripathy and T. D. Sharkey (Dordrecht: Springer), 275–297.
Anderson, L. B., Maderia, M., Ouellette, A. J. A., Putnam-Evans, C., Higgins, L., Krick, T., et al. (2002). Posttranslational modifications in the CP43 subunit of photosystem II. Proc. Natl. Acad. Sci. U.S.A. 99, 14676–14681. doi: 10.1073/pnas.232591599
Anderson, L. B., Ouellette, A. J. A., Eaton-Rye, J., Maderia, M., MacCoss, M. J., Yates, J. R., et al. (2004). Evidence for a post-translational modification, aspartyl aldehyde, in a photosynthetic membrane protein. J. Am. Chem. Soc. 126, 8399–8405. doi: 10.1021/ja0478781
Andersson, L., and Porath, J. (1986). Isolation of phosphoproteins by immobilized metal (Fe3+) affinity chromatography. Anal. Biochem. 154, 250–254. doi: 10.1016/0003-2697(86)90523-3
Aro, E. M., McCaffery, S., and Anderson, J. M. (1993). Photoinhibition and D1 protein degradation in peas acclimated to different growth irradiances. Plant Physiol. 103, 835–843.
Aro, E.-M., Suorsa, M., Rokka, A., Allahverdiyeva, Y., Paakkarinen, V., Saleem, A., et al. (2005). Dynamics of photosystem II: a proteomic approach to thylakoid protein complexes. J. Exp. Bot. 56, 347–356. doi: 10.1093/jxb/eri041
Aryal, U. K., Stöckel, J., Krovvidi, R. K., Gritsenko, M. A., Monroe, M. E., Moore, R. J., et al. (2011). Dynamic proteomic profiling of a unicellular cyanobacterium Cyanothece ATCC51142 across light-dark diurnal cycles. BMC Syst. Biol. 5:194. doi: 10.1186/1752-0509-5-194
Back, J. W., de Jong, L., Muijsers, A. O., and de Koster, C. G. (2003). Chemical cross-linking and mass spectrometry for protein structural modeling. J. Mol. Biol. 331, 303–313. doi: 10.1016/S0022-2836(03)00721-6
Baena-González, E., and Aro, E.-M. (2002). Biogenesis, assembly and turnover of photosystem II units. Phil. Trans. R. Soc. Lond. B. 357, 1451–1459. doi: 10.1098/rstb.2002.1141
Bantscheff, M., Lemeer, S., Savitski, M. M., and Kuster, B. (2012). Quantitative mass spectrometry in proteomics: critical review update from 2007 to the present. Anal. Bioanal. Chem. 404, 939–965. doi: 10.1007/s00216-012-6203-4
Barber, J., and Andersson, B. (1992). Too much of a good thing—light can be bad for photosynthesis. Trends Biochem. Sci. 17, 61–66. doi: 10.1016/0968-0004(92)90503-2
Battchikova, N., Angeleri, M., and Aro, E.-M. (2015). Proteomic approaches in research of cyanobacterial photosynthesis. Photosynth. Res. 126, 47–70. doi: 10.1007/s11120-014-0050-4
Boehm, M., Romero, E., Reisinger, V., Yu, J., Komenda, J., Eichacker, L. A., et al. (2011). Investigating the early stages of Photosystem II assembly in Synechocystis sp. PCC 6803: Isolation of CP47 and CP43 complexes. J. Biol. Chem. 286, 14812–14819. doi: 10.1074/jbc.M110.207944
Boehm, M., Yu, J., Reisinger, V., Beckova, M., Eichacker, L. A., Schlodder, E., et al. (2012). Subunit composition of CP43-less photosystem II complexes of Synechocystis sp. PCC 6803: implications for the assembly and repair of photosystem II. Phil. Trans. R. Soc. B 367, 3444–3454. doi: 10.1098/rstb.2012.0066
Bonardi, V., Pesaresi, P., Becker, T., Schleiff, E., Wagner, R., Pfannschmidt, T., et al. (2005). Photosystem II core phosphorylation and photosynthetic acclimation require two different protein kinases. Nature 437, 1179–1182. doi: 10.1038/nature04016
Bricker, T. M., Mummadisetti, M. P., and Frankel, L. K. (2015). Recent advances in the use of mass spectrometry to examine structure/function relationships in photosystem II. J. Photochem. Photobiol. B 152, 227–246. doi: 10.1016/j.jphotobiol.2015.08.031
Bricker, T. M., Odom, W. R., and Queirolo, C. B. (1988). Close association of the 33-kDa extrinsic protein with the apoprotein of CPa1 in photosystem II. FEBS Lett. 231, 111–117. doi: 10.1016/0014-5793(88)80713-0
Bricker, T. M., Roose, J. L., Fagerlund, R. D., Frankel, L. K., and Eaton-Rye, J. J. (2012). The extrinsic proteins of Photosystem II. Biochim. Biophys. Acta 1817, 121–142. doi: 10.1016/j.bbabio.2011.07.006
Brunner, A. M., Lossl, P., Liu, F., Huguet, R., Mullen, C., Yamashita, M., et al. (2015). Benchmarking multiple fragmentation methods on an Orbitrap Fusion for top-down phospho-proteoform characterization. Anal. Chem. 87, 4152–4158. doi: 10.1021/acs.analchem.5b00162
Buncherd, H., Roseboom, W., de Koning, L. J., de Koster, C. G., and de Jong, L. (2014). A gas phase cleavage reaction of cross-linked peptides for protein complex topology studies by peptide fragment fingerprinting from large sequence database. J. Proteomics 108, 65–77. doi: 10.1016/j.jprot.2014.05.003
Caffarri, S., Kouřil, R., Kereiche, S., Boekema, E. J., and Croce, R. (2009). Functional architecture of higher plant photosystem II supercomplexes. EMBO J. 28, 3052–3064. doi: 10.1038/emboj.2009.232
Chen, X., Wei, S., Ji, Y., Guo, X., and Yang, F. (2015). Quantitative proteomics using SILAC: principles, applications, and developments. Proteomics 15, 3175–3192. doi: 10.1002/pmic.201500108
Chu, F., Mahrus, S., Craik, C. S., and Burlingame, A. L. (2006). Isotope-coded and affinity-tagged cross-linking (ICATXL): An efficient strategy to probe protein interaction surfaces. J. Am. Chem. Soc. 128, 10362–10363. doi: 10.1021/ja0614159
Clarke, A. K., Soitamo, A., Gustafsson, P., and Öquist, G. (1993). Rapid interchange between two distinct forms of cyanobacterial photosystem II reaction center protein D1 in response to photoinhibition. Proc. Natl. Acad. Sci. U.S.A. 90, 9973–9977. doi: 10.1073/pnas.90.21.9973
Clegg, C., and Hayes, D. (1974). Identification of neighboring proteins in ribosomes of Escherichia coli- Topographical study with crosslinking reagent dimethyl suberimidate. Eur. J. Biochem. 42, 21–28. doi: 10.1111/j.1432-1033.1974.tb03309.x
Cormann, K. U., Möller, M., and Nowaczyk, M. M. (2016). Critical assessment of protein cross-linking and molecular docking: an updated model for the interaction between Photosystem II and Psb27. Front. Plant Sci. 7:157. doi: 10.3389/fpls.2016.00157
Dasgupta, J., Ananyev, G., and Dismukes, G. C. (2008). Photoassembly of the water-oxidizing complex in photosystem II. Coord. Chem. Rev. 252, 347–360. doi: 10.1016/j.ccr.2007.08.022
Dedner, N., Meyer, H. E., Ashton, C., and Wildner, G. F. (1988). N-terminal sequence analysis of the 8 kDa protein in Chlamydomonas reinhardii: localization of the phosphothreonine. FEBS Lett. 236, 77–82. doi: 10.1016/0014-5793(88)80288-6
Dobáková, M., Sobotka, R., Tichý, M., and Komenda, J. (2009). Psb28 protein is involved in the biogenesis of the Photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 149, 1076–1086. doi: 10.1104/pp.108.130039
Dobáková, M., Tichý, M., and Komenda, J. (2007). Role of the PsbI protein in photosystem II assembly and repair in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 145, 1681–1691. doi: 10.1104/pp.107.107805
Dreaden, T. M., Chen, J., Rexroth, S., and Barry, B. A. (2011). N-formylkynurenine as a marker of high light stress in photosynthesis. J. Biol. Chem. 286, 22632–22641. doi: 10.1074/jbc.M110.212928
Enami, I., Kamo, M., Ohta, H., Takahashi, S., Miura, T., Kusayanagi, M., et al. (1998). Intramolecular cross-linking of the extrinsic 33-kDa protein leads to loss of oxygen evolution but not its ability of binding to Photosystem II and stabilization of the manganese cluster. J. Biol. Chem. 273, 4629–4634. doi: 10.1074/jbc.273.8.4629
Enami, I., Satoh, K., and Katoh, S. (1987). Crosslinking between the 33 kDa extrinsic protein and the 47 kDa chlorophyll-carrying protein of the PSII reaction center core complex. FEBS Lett. 226, 161–165. doi: 10.1016/0014-5793(87)80571-9
Fagerlund, R. D., and Eaton-Rye, J. J. (2011). The lipoproteins of cyanobacterial photosystem II. J. Photochem. Photobiol. B 104, 191–203. doi: 10.1016/j.jphotobiol.2011.01.022
Ficarro, S. B., McCleland, M. L., Stukenberg, P. T., Burke, D. J., Ross, M. M., Shabanowitz, J., et al. (2002). Phosphoproteome analysis by mass spectrometry and its application to Saccharomyces cerevisiae. Nat. Biotechnol. 20, 301–305. doi: 10.1038/nbt0302-301
Frankel, L. K., and Bricker, T. M. (1992). Interaction of CPa-1 with the manganese-stabilizing protein of Photosystem II: identification of domains on CPa-1 which are shielded from N-hydroxysuccinimide biotinylation by the manganese-stabilizing protein. Biochemistry 31, 11059–11064. doi: 10.1021/bi00160a015
Frankel, L. K., and Bricker, T. M. (1995). Interaction of the 33-kDa extrinsic protein with Photosystem II: identification of domains on the 33-kDa protein that are shielded from NHS-biotinylation by Photosystem II. Biochemistry 34, 7492–7497. doi: 10.1021/bi00022a024
Frankel, L. K., Cruz, J. A., and Bricker, T. M. (1999). Carboxylate groups on the manganese-stabilizing protein are required for its efficient binding to Photosystem II. Biochemistry 38, 14271–14278. doi: 10.1021/bi991366v
Frankel, L. K., Sallans, L., Bellamy, H., Goettert, J. S., Limbach, P. A., and Bricker, T. M. (2013a). Radiolytic mapping of solvent-contact surfaces in Photosystem II of higher plants: experimental identification of putative water channels within the photosystem. J. Biol. Chem. 288, 23565–23572. doi: 10.1074/jbc.M113.487033
Frankel, L. K., Sallans, L., Limbach, P. A., and Bricker, T. M. (2013b). Oxidized amino acid residues in the vicinity of QA and PheoD1 of the Photosystem II reaction center: putative generation sites of reducing-side reactive oxygen species. PLoS ONE 8:e58042. doi: 10.1371/journal.pone.0058042
Frankel, L. K., Sallans, L., Limbach, P., and Bricker, T. M. (2012). Identification of oxidized amino acid residues in the vicinity of the Mn4CaO5 cluster of Photosystem II: implications for the identification of oxygen channels within the photosystem. Biochemistry 51, 6371–6377. doi: 10.1021/bi300650n
Frese, C. K., Altelaar, A. F. M., van den Toorn, H., Nolting, D., Griep-Raming, J., Heck, A. J. R., et al. (2012). Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal. Chem. 84, 9668–9673. doi: 10.1021/ac3025366
Fristedt, R., Granath, P., and Vener, A. V. (2010). A protein phosphorylation threshold for functional stacking of plant photosynthetic membranes. PLoS ONE 5:e10963. doi: 10.1371/journal.pone.0010963
Fristedt, R., and Vener, A. V. (2011). High light induced disassembly of Photosystem II supercomplexes in Arabidopsis requires STN7-dependent phosphorylation of CP29. PLoS ONE 6:e24565. doi: 10.1371/journal.pone.0024565
Fristedt, R., Willig, A., Granath, P., Crevecoeur, M., Rochaix, J.-D., and Vener, A. V. (2009). Phosphorylation of Photosystem II controls functional macroscopic folding of photosynthetic membranes in Arabidopsis. Plant Cell 21, 3950–3964. doi: 10.1105/tpc.109.069435
Fritzsche, R., Ihling, C. H., Götze, M., and Sinz, A. (2012). Optimizing the enrichment of cross-linked products for mass spectrometric protein analysis. Rapid Commun. Mass Spectrom. 26, 653–658. doi: 10.1002/rcm.6150
Gabdulkhakov, A., Guskov, A., Broser, M., Kern, J., Mueh, F., Saenger, W., et al. (2009). Probing the accessibility of the Mn4Ca cluster in Photosystem II: channels calculation, noble gas derivatization, and cocrystallization with DMSO. Structure 17, 1223–1234. doi: 10.1016/j.str.2009.07.010
Galetskiy, D., Lohscheider, J. N., Kononikhin, A. S., Popov, I. A., Nikolaev, E. N., and Adamska, I. (2011a). Mass spectrometric characterization of photooxidative protein modifications in Arabidopsis thaliana thylakoid membranes. Rapid Commun. Mass Spectrom. 25, 184–190. doi: 10.1002/rcm.4855
Galetskiy, D., Lohscheider, J. N., Kononikhin, A. S., Popov, I. A., Nikolaev, E. N., and Adamska, I. (2011b). Phosphorylation and nitration levels of photosynthetic proteins are conversely regulated by light stress. Plant Mol. Biol. 77, 461–473. doi: 10.1007/s11103-011-9824-7
Gao, S., Gu, W., Xiong, Q., Ge, F., Xie, X., Li, J., et al. (2015). Desiccation enhances phosphorylation of PSII and affects the distribution of protein complexes in the thylakoid membrane. Physiol. Plant. 153, 492–502. doi: 10.1111/ppl.12258
Gau, B., Garai, K., Frieden, C., and Gross, M. L. (2011). Mass spectrometry-based protein footprinting characterizes the structures of oligomeric apolipoprotein E2, E3, and E4. Biochemistry 50, 8117–8126. doi: 10.1021/bi200911c
Geiger, T., Cox, J., and Mann, M. (2010). Proteomics on an Orbitrap benchtop mass spectrometer using all-ion fragmentation. Mol. Cell. Proteomics 9, 2252–2261. doi: 10.1074/mcp.M110.001537
Golden, S. S., Brusslan, J., and Haselkorn, R. (1986). Expression of a family of psbA genes encoding a photosystem II polypeptide in the cyanobacterium Anacystis nidulans R2. EMBO J. 5, 2789–2798.
Gómez, S. M., Bil', K. Y., Aguilera, R., Nishio, J. N., Faull, K. F., and Whitelegge, J. P. (2003). Transit peptide cleavage sites of integral thylakoid membrane proteins. Mol. Cell. Proteomics 2, 1068–1085. doi: 10.1074/mcp.M300062-MCP200
Gómez, S. M., Nishio, J. N., Faull, K. F., and Whitelegge, J. P. (2002). The chloroplast grana proteome defined by intact mass measurements from liquid chromatography mass spectrometry. Mol. Cell. Proteomics 1, 46–59. doi: 10.1074/mcp.M100007-MCP200
Gómez, S. M., Park, J. J., Zhu, J., Whitelegge, J. P., and Thornber, J. P. (1998). “Isolation and characterization of a novel xanthophyll-rich pigment-protein complex from spinach,” in Photosynthesis: Mechanisms and Effects, Vol. 1,” ed G. Garab (Dordrecht: Kluwer Academic Publishers), 353–356.
Gouw, J. W., Krijgsveld, J., and Heck, A. J. R. (2010). Quantitative proteomics by metabolic labeling of model organisms. Mol. Cell. Proteomics 9, 11–24. doi: 10.1074/mcp.R900001-MCP200
Granvogl, B., Zoryan, M., and Plöscher, Eichacker, L. A. (2008). Localization of 13 one-helix integral membrane proteins in Photosystem II subcomplexes. Anal. Biochem. 383, 279–288. doi: 10.1016/j.ab.2008.08.038
Grasse, N., Mamedov, F., Becker, K., Styring, S., Rögner, M., and Nowaczyk, M. M. (2011). Role of novel dimeric Photosystem II (PSII)-Psb27 protein complex in PSII repair. J. Biol. Chem. 286, 29548–29555. doi: 10.1074/jbc.M111.238394
Grossmann, J., Roschitzki, B., Panse, C., Fortes, C., Barkow-Oesterreicher, S., Rutishauser, D., et al. (2010). Implementation and evaluation of relative and absolute quantification in shotgun proteomics with label-free methods. J. Proteomics 73, 1740–1746. doi: 10.1016/j.jprot.2010.05.011
Götze, M., Pettelkau, J., Fritzsche, R., Ihling, C. H., Schaefer, M., and Sinz, A. (2015). Automated assignment of MS/MS cleavable cross-links in protein 3D-structure analysis. J. Am. Soc. Mass. Spectrom. 26, 83–97. doi: 10.1007/s13361-014-1001-1
Götze, M., Pettelkau, J., Schaks, S., Bosse, K., Ihling, C., Krauth, F., et al. (2012). StavroX-A Software for analyzing crosslinked products in protein interaction studies. J. Am. Soc. Mass. Spectrom. 23, 76–87. doi: 10.1007/s13361-011-0261-2
Guerreiro, A. C. L., Benevento, M., Lehmann, R., van Breukelen, B., Post, H., Giansanti, P., et al. (2014). Daily rhythms in the cyanobacterium Synechococcus elongatus probed by high-resolution mass spectrometry-based proteomics reveals a small defined set of cyclic proteins. Mol. Cell. Proteomics 13, 2042–2055. doi: 10.1074/mcp.M113.035840
Guo, J., Nguyen, A. Y., Dai, Z., Su, D., Gaffrey, M. J., Moore, R. J., et al. (2014). Proteome-wide light/dark modulation of thiol oxidation in cyanobacteria revealed by quantitative site-specific redox proteomics. Mol. Cell. Proteomics 13, 3270–3285. doi: 10.1074/mcp.M114.041160
Guskov, A., Kern, J., Gabdulkhakov, A., Broser, M., Zouni, A., and Saenger, W. (2009). Cyanobacterial photosystem II at 2.9-Å resolution and the role of quinones, lipids, channels and chloride. Nat. Struct. Mol. Biol. 16, 334–342. doi: 10.1038/nsmb.1559
Han, K. C., Shen, J. R., Ikeuchi, M., and Inoue, Y. (1994). Chemical cross-linking studies of extrinsic proteins in cyanobacterial photosystem II. FEBS Lett. 355, 121–124. doi: 10.1016/0014-5793(94)01182-6
Haniewicz, P., De Sanctis, D., Büchel, C., Schröder, W. P., Loi, M. C., Kieselbach, T., et al. (2013). Isolation of monomeric photosystem II that retains the subunit PsbS. Photosynth. Res. 118, 199–207. doi: 10.1007/s11120-013-9914-2
Haniewicz, P., Floris, D., Farci, D., Kirkpatrick, J., Loi, M. C., and Büchel, C. (2015). Isolation of plant Photosystem II complexes by fractional solubilization. Front. Plant Sci. 6:1100. doi: 10.3389/fols.2015.01100
Hansson, M., Dupuis, T., Stromquist, R., Andersson, B., Vener, A. V., and Carlberg, I. (2007). The mobile thylakoid phosphoprotein TSP9 interacts with the light-harvesting complex II and the peripheries of both photosystems. J. Biol. Chem. 282, 16214–16222. doi: 10.1074/jbc.M605833200
Harrer, R., Bassi, R., Testi, M. G., and Schäfer, C. (1998). Nearest-neighbor analysis of a photosystem II complex from Marchantia polymorpha L. (liverwort), which contains reaction center and antenna proteins. Eur. J. Biochem. 255, 196–205. doi: 10.1046/j.1432-1327.1998.2550196.x
Heinemeyer, J., Eubel, H., Wehmhoner, D., Jansch, L., and Braun, H. P. (2004). Proteomic approach to characterize the supramolecular organization of photosystems in higher plants. Phytochemistry 65, 1683–1692. doi: 10.1016/j.phytochem.2004.04.022
Heinz, S., Liauw, P., Nickelsen, J., and Nowaczyk, M. (2016). Analysis of photosystem II biogenesis in cyanobacteria. Biochim. Biophys. Acta 1857, 274–287. doi: 10.1016/j.bbabio.2015.11.007
Herbstová, M., Tietz, S., Kinzel, C., Turkina, M., and Kirchhoff, H. (2012). Architectural switch in plant photosynthetic membranes induced by light stress. Proc. Natl. Acad. Sci. U.S.A. 109, 20130–20135. doi: 10.1073/pnas.1214265109
Ho, F. M., and Styring, S. (2008). Access channels and methanol binding site to the CaMn4 cluster in Photosystem II based on solvent accessibility simulations, with implications for substrate water access. Biochim. Biophys. Acta 1777, 140–153. doi: 10.1016/j.bbabio.2007.08.009
Hoare, D. G., and Koshland, D. E. (1967). A method for quantitative modification and estimation of carboxylic acid groups in proteins. J. Biol. Chem. 242, 2447–2453.
Hoopmann, M. R., Zelter, A., Johnson, R. S., Riffle, M., MacCoss, M. J., Davis, T. N., et al. (2015). Kojak: efficient analysis of chemically cross-linked protein complexes. J. Proteome Res. 14, 2190–2198. doi: 10.1021/pr501321h
Howery, A. E., Elvington, S., Abraham, S. J., Choi, K. H., Dworschak-Simpson, S., Phillips, S., et al. (2012). A designed inhibitor of a CLC antiporter blocks function through a unique binding mode. Chem. Biol. 19, 1460–1470. doi: 10.1016/j.chembiol.2012.09.017
Huber, C. G., Walcher, W., Timperio, A. M., Troiani, S., Porceddu, A., and Zolla, L. (2004). Multidimensional proteomic analysis of photosynthetic membrane proteins by liquid extraction-ultracentrifugation-liquid chromatography-mass spectrometry. Proteomics 4, 3909–3920. doi: 10.1002/pmic.200400823
Ido, K., Kakiuchi, S., Uno, C., Nishimura, T., Fukao, Y., Noguchi, T., et al. (2012). The conserved His-144 in the PsbP protein is important for the interaction between the PsbP N-terminus and the Cyt b559 subunit of Photosystem II. J. Biol. Chem. 287, 26377–26387. doi: 10.1074/jbc.M112.385286
Ido, K., Nield, J., Fukao, Y., Nishimura, T., Sato, F., and Ifuku, K. (2014). Cross-linking evidence for multiple interactions of the PsbP and PsbQ proteins in a higher plant Photosystem II supercomplex. J. Biol. Chem. 289, 20150–20157. doi: 10.1074/jbc.M114.574822
Ikeuchi, M., and Inoue, Y. (1988). A new 4.8-kDa polypeptide instrinsic to the PS II reaction center, as revealed by modified SDS-PAGE with improved resolution of low-molecular-weight proteins. Plant Cell Physiol. 29, 1233–1239.
Inagaki, N., Yamamoto, Y., and Satoh, K. (2001). A sequential two-step proteolytic process in the carboxyl-terminal truncation of precursor D1 protein in Synechocystis sp. PCC6803. FEBS Lett. 509, 197–201. doi: 10.1016/S0014-5793(01)03180-5
Ingle, R. A., Schmidt, U. G., Farrant, J. M., Thomson, J. A., and Mundree, S. G. (2007). Proteomic analysis of leaf proteins during dehydration of the resurrection plant Xerophyta viscosa. Plant Cell Environ. 30, 435–446. doi: 10.1111/j.1365-3040.2006.01631.x
Inoue-Kashino, N., Kashino, Y., Orii, H., Satoh, K., Terashima, I., and Pakrasi, H. B. (2011). S4 protein Sll1252 is necessary for energy balancing in photosynthetic electron transport in Synechocystis sp. PCC 6803. Biochemistry 50, 329–339. doi: 10.1021/bi101077e
Irrgang, K. D., Shi, L. X., Funk, C., and Schröder, W. P. (1995). A nuclear-encoded subunit of the Photosystem II reaction center. J. Biol. Chem. 270, 17588–17593.
Iwai, M., Suzuki, T., Dohmae, N., Inoue, Y., and Ikeuchi, M. (2007). Absence of the PsbZ subunit prevents association of PsbK and Ycf12 with the PSII complex in the thermophilic cyanobacterium Thermosynechococcus elongatus BP-1. Plant Cell Physiol. 48, 1758–1763. doi: 10.1093/pcp/pcm148
Järvi, S., Suorsa, M., and Aro, E.-M. (2015). Photosystem II repair in plant chloroplasts - Regulation, assisting proteins and shared components with photosystem II biogenesis. Biochim. Biophys. Acta 1847, 900–909. doi: 10.1016/j.bbabio.2015.01.006
Järvi, S., Suorsa, M., Paakkarinen, V., and Aro, E.-M. (2011). Optimized native gel systems for separation of thylakoid protein complexes: novel super- and mega-complexes. Biochem J. 439, 207–214. doi: 10.1042/BJ20102155
Kang, S., Mou, L., Lanman, J., Velu, S., Brouillette, W., Prevelige, P. E., et al. (2009). Synthesis of biotin-tagged chemical cross-linkers and their applications for mass spectrometry. Rapid Commun. Mass Spectrom. 23, 1719–1726. doi: 10.1002/rcm.4066
Kao, A., Chiu, C.-l., Vellucci, D., Yang, Y., Patel, V. R., Guan, S., et al. (2011). Development of a novel cross-linking strategy for fast and accurate identification of cross-linked peptides of protein complexes. Mol. Cell. Proteomics 10:002212. doi: 10.1074/mcp.M110.002212
Kashino, Y., Lauber, W. M., Carroll, J. A., Wang, Q. J., Whitmarsh, J., Satoh, K., et al. (2002). Proteomic analysis of a highly active photosystem II preparation from the cyanobacterium Synechocystis sp. PCC 6803 reveals the presence of novel polypeptides. Biochemistry 41, 8004–8012. doi: 10.1021/bi026012+
Kasson, T. M. D., Rexroth, S., and Barry, B. A. (2012). Light-induced oxidative stress, N-formylkynurenine, and oxygenic photosynthesis. PLoS ONE 7:42220. doi: 10.1371/journal.pone.0042220
Kereïche, S., Kouřil, R., Oostergetel, G. T., Fusetti, F., Boekema, E. J., Doust, A. B., et al. (2008). Association of chlorophyll a/c2 complexes to photosystem I and photosystem II in the cryptophyte Rhodomonas CS24. Biochim. Biophys. Acta 1777, 1122–1128. doi: 10.1016/j.bbabio.2008.04.045
Knoppová, J., Sobotka, R., Tichý, M., Yu, J., Konik, P., Halada, P., et al. (2014). Discovery of a chlorophyll binding protein complex involved in the early steps of Photosystem II assembly in Synechocystis. Plant Cell 26, 1200–1212. doi: 10.1105/tpc.114.123919
Komenda, J., Hassan, H. A. G., Diner, B. A., Debus, R. J., Barber, J., and Nixon, P. J. (2000). Degradation of the Photosystem II D1 and D2 proteins in different strains of the cyanobacterium Synechocystis PCC 6803 varying with respect to the type and level of psbA transcript. Plant Mol. Biol. 42, 635–645. doi: 10.1023/A:1006305308196
Komenda, J., Knoppová, J., Kopečná, J., Sobotka, R., Halada, P., Yu, J., et al. (2012a). The Psb27 assembly factor binds to the CP43 complex of Photosystem II in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 158, 476–486. doi: 10.1104/pp.111.184184
Komenda, J., Kuviková, S., Granvogl, B., Eichacker, L. A., Diner, B. A., and Nixon, P. J. (2007). Cleavage after residue Ala352 in the C-terminal extension is an early step in the maturation of the D1 subunit of Photosystem II in Synechocystis PCC 6803. Biochim. Biophys. Acta 1767, 829–837. doi: 10.1016/j.bbabio.2007.01.005
Komenda, J., Nickelsen, J., Tichý, M., Prášil, O., Eichacker, L., and Nixon, P. J. (2008). The cyanobacterial homologue of HCF136/YCF48 is a component of an early Photosystem II assembly complex and is important for both the efficient assembly and repair of Photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 283, 22390–22399. doi: 10.1074/jbc.M801917200
Komenda, J., Reisinger, V., Müller, B. C., Dobáková, M., Granvogl, B., and Eichacker, L. A. (2004). Accumulation of the D2 protein is a key regulatory step for assembly of the Photosystem II reaction center complex in Synechocystis PCC 6803. J. Biol. Chem. 279, 48620–48629. doi: 10.1074/jbc.M405725200
Komenda, J., Sobotka, R., and Nixon, P. J. (2012b). Assembling and maintaining the Photosystem II complex in chloroplasts and cyanobacteria. Curr. Opin. Plant Biol. 15, 245–251. doi: 10.1016/j.pbi.2012.01.017
Komenda, J., Tichý, M., and Eichacker, L. A. (2005). The PsbH protein is associated with the inner antenna CP47 and facilitates D1 processing and incorporation into PSII in the cyanobacterium Synechocystis PCC 6803. Plant Cell Physiol. 46, 1477–1483. doi: 10.1093/pcp/pci159
Kós, P. B., Deák, Z., Cheregi, O., and Vass, I. (2008). Differential regulation of psbA and psbD gene expression, and the role of the different D1 protein copies in the cyanobacterium Thermosynechococcus elongatus BP-1. Biochim. Biophys. Acta 1777, 74–83. doi: 10.1016/j.bbabio.2007.10.015
Kouřil, R., Dekker, J. P., and Boekema, E. J. (2012). Supramolecular organization of photosystem II in green plants. Biochim. Biophys. Acta 1817, 2–12. doi: 10.1016/j.bbabio.2011.05.024
Kufryk, G., Hernandez-Prieto, M. A., Kieselbach, T., Miranda, H., Vermaas, W., and Funk, C. (2008). Association of small CAB-like proteins (SCPs) of Synechocystis sp. PCC 6803 with Photosystem II. Photosynth. Res. 95, 135–145. doi: 10.1007/s11120-007-9244-3
Laganowsky, A., Gómez, S. M., Whitelegge, J. P., and Nishio, J. N. (2009). Hydroponics on a chip: analysis of the Fe deficient Arabidopsis thylakoid membrane proteome. J. Proteomics 72, 397–415. doi: 10.1016/j.jprot.2009.01.024
Leitner, A., Reischl, R., Walzthoeni, T., Herzog, F., Bohn, S., Förster, F., et al. (2012). Expanding the chemical cross-linking toolbox by the use of multiple proteases and enrichment by size exclusion chromatography. Mol. Cell. Proteomics 11:M111.014126. doi: 10.1074/mcp.M111.014126
Lemeille, S., and Rochaix, J.-D. (2010). State transitions at the crossroad of thylakoid signalling pathways. Photosynth. Res. 106, 33–46. doi: 10.1007/s11120-010-9538-8
Lemeille, S., Turkina, M., Vener, A. V., and Rochaix, J.-D. (2010). Stt7-dependent phosphorylation during state transitions in the green alga Chlamydomonas reinhardtii. Mol. Cell. Proteomics 9, 1281–1295. doi: 10.1074/mcp.M000020-MCP201
Li, X.-J., Yang, M.-F., Zhu, Y., Liang, Y., and Shen, S.-H. (2011). Proteomic analysis of salt stress responses in rice shoot. J. Plant Biol. 54, 384–395. doi: 10.1007/s12374-011-9173-8
Liu, H., Chen, J., Huang, R.-C., Weisz, D., Gross, M. L., and Pakrasi, H. B. (2013a). Mass spectrometry-based footprinting reveals structural dynamics of Loop E of the chlorophyll-binding protein CP43 during Photosystem II assembly in the cyanobacterium Synechocystis 6803. J. Biol. Chem. 288, 14212–14220. doi: 10.1074/jbc.M113.467613
Liu, H., Huang, R. Y.-C., Chen, J., Gross, M. L., and Pakrasi, H. B. (2011a). Psb27, a transiently associated protein, binds to the chlorophyll binding protein CP43 in photosystem II assembly intermediates. Proc. Natl. Acad. Sci. U.S.A. 108, 18536–18541. doi: 10.1073/pnas.1111597108
Liu, H., Roose, J. L., Cameron, J. C., and Pakrasi, H. B. (2011b). A genetically tagged Psb27 protein allows purification of two consecutive Photosystem II (PSII) assembly intermediates in Synechocystis 6803, a cyanobacterium. J. Biol. Chem. 286, 24865–24871. doi: 10.1074/jbc.M111.246231
Liu, H., Weisz, D. A., and Pakrasi, H. B. (2015). Multiple copies of the PsbQ protein in a cyanobacterial photosystem II assembly intermediate complex. Photosynth. Res. 126, 375–383. doi: 10.1007/s11120-015-0123-z
Liu, H., Zhang, H., King, J., Wolf, N., Prado, M., Gross, M. L., et al. (2014a). Mass spectrometry footprinting reveals the structural rearrangements of cyanobacterial orange carotenoid protein upon light activation. Biochim. Biophys. Acta 1837, 1955–1963. doi: 10.1016/j.bbabio.2014.09.004
Liu, H., Zhang, H., Niedzwiedzki, D., Prado, M., He, G., Gross, M. L., et al. (2013b). Phycobilisomes supply excitations to both photosystems in a megacomplex in cyanobacteria. Science 342, 1104–1107. doi: 10.1126/science.1242321
Liu, H., Zhang, H., Orf, G. S., Lu, Y., Jiang, J., King, J. D., et al. (2016). Dramatic domain rearrangements of the cyanobacterial orange carotenoid protein upon photoactivation. Biochemistry 55, 1003–1009. doi: 10.1021/acs.biochem.6b00013
Liu, H., Zhang, H., Weisz, D. A., Vidavsky, I., Gross, M. L., and Pakrasi, H. B. (2014b). MS-based cross-linking analysis reveals the location of the PsbQ protein in cyanobacterial photosystem II. Proc. Natl. Acad. Sci. U.S.A. 111, 4638–4643. doi: 10.1073/pnas.1323063111
Lohrig, K., Mueller, B., Davydova, J., Leister, D., and Wolters, D. A. (2009). Phosphorylation site mapping of soluble proteins: bioinformatical filtering reveals potential plastidic phosphoproteins in Arabidopsis thaliana. Planta 229, 1123–1134. doi: 10.1007/s00425-009-0901-y
Loll, B., Kern, J., Saenger, W., Zouni, A., and Biesiadka, J. (2005). Towards complete cofactor arrangement in the 3.0 Å resolution structure of photosystem II. Nature 438, 1040–1044. doi: 10.1038/nature04224
Lorković, Z. J., Schröder, W. P., Pakrasi, H. B., Irrgang, K. D., Herrmann, R. G., and Oelmüller, R. (1995). Molecular characterization of PsbW, a nuclear-encoded component of the Photosystem II reaction center complex in spinach. Proc. Natl. Acad. Sci. U.S.A. 92, 8930–8934. doi: 10.1073/pnas.92.19.8930
Lundgren, D. H., Hwang, S.-I., Wu, L., and Han, D. K. (2010). Role of spectral counting in quantitative proteomics. Expert Rev. Proteomics 7, 39–53. doi: 10.1586/EPR.09.69
Mabbitt, P. D., Wilbanks, S. M., and Eaton-Rye, J. J. (2014). Structure and function of the hydrophilic Photosystem II assembly proteins: Psb27, Psb28 and Ycf48. Plant Physiol. Bioch. 81, 96–107. doi: 10.1016/j.plaphy.2014.02.013
Mädler, S., Bich, C., Touboul, D., and Zenobi, R. (2009). Chemical cross-linking with NHS esters: a systematic study on amino acid reactivities. J. Mass Spectrom. 44, 694–706. doi: 10.1002/jms.1544
Meades, G. D., McLachlan, A., Sallans, L., Limbach, P. A., Frankel, L. K., and Bricker, T. M. (2005). Association of the 17-kDa extrinsic protein with Photosystem II in higher plants. Biochemistry 44, 15216–15221. doi: 10.1021/bi051704u
Mehmood, S., Allison, T. M., and Robinson, C. V. (2015). Mass spectrometry of protein complexes: from origins to applications. Annu. Rev. Phys. Chem. 66, 453–474. doi: 10.1146/annurev-physchem-040214-121732
Michel, H., Hunt, D. F., Shabanowitz, J., and Bennett, J. (1988). Tandem mass spectrometry reveals that three Photosystem II proteins of spinach chloroplasts contain N-acetyl-O-phosphothreonine at their NH2 termini. J. Biol. Chem. 263, 1123–1130.
Michel, H. P., and Bennett, J. (1987). Identification of the phosphorylation site of an 8.3 kDa protein from photosystem II of spinach. FEBS Lett. 212, 103–108. doi: 10.1016/0014-5793(87)81565-X
Minagawa, J. (2011). State transitions-The molecular remodeling of photosynthetic supercomplexes that controls energy flow in the chloroplast. Biochim. Biophys. Acta 1807, 897–905. doi: 10.1016/j.bbabio.2010.11.005
Miura, T., Shen, J. R., Takahashi, S., Kamo, M., Nakamura, E., Ohta, H., et al. (1997). Identification of domains on the extrinsic 33-kDa protein possibly involved in electrostatic interaction with Photosystem II complex by means of chemical modification. J. Biol. Chem. 272, 3788–3798.
Müller, B., and Eichacker, L. A. (1999). Assembly of the D1 precursor in monomeric Photosystem II reaction center precomplexes precedes Chlorophyll a-triggered accumulation of Reaction Center II in barley etioplasts. Plant Cell 11, 2365–2377. doi: 10.1105/tpc.11.12.2365
Müller, D. R., Schindler, P., Towbin, H., Wirth, U., Voshol, H., Hoving, S., et al. (2001). Isotope tagged cross linking reagents. A new tool in mass spectrometric protein interaction analysis. Anal. Chem. 73, 1927–1934. doi: 10.1021/ac001379a
Mullet, J. E., and Christopher, D. A. (1994). Separate photosensory pathways coregulate blue-light/ultraviolet-A-activated psbD-psbC transcription and light-induced D2 and CP43 degradation in barley (Hordeum vulgare) chloroplasts. Plant Physiol. 104, 1119–1129. doi: 10.1104/pp.104.4.1119
Mullineaux, C. W. (2008). Phycobilisome-reaction centre interaction in cyanobacteria. Photosynth. Res. 95, 175–182. doi: 10.1007/s11120-007-9249-y
Mulo, P., Sakurai, I., and Aro, E.-M. (2012). Strategies for psbA gene expression in cyanobacteria, green algae and higher plants: from transcription to PSII repair. Biochim. Biophys. Acta 1817, 247–257. doi: 10.1016/j.bbabio.2011.04.011
Mulo, P., Sicora, C., and Aro, E.-M. (2009). Cyanobacterial psbA gene family: optimization of oxygenic photosynthesis. Cell. Mol. Life Sci. 66, 3697–3710. doi: 10.1007/s00018-009-0103-6
Mummadisetti, M., Frankel, L. K., Bellamy, H. D., Sallans, L., Goettert, J. S., Brylinski, M., et al. (2014). Use of protein cross-linking and radiolytic footprinting to elucidate PsbP and PsbQ interactions within higher plant Photosystem II. Proc. Natl. Acad. Sci. U.S.A. 111, 16178–16183. doi: 10.1073/pnas.1415165111
Murray, J. W., and Barber, J. (2007). Structural characteristics of channels and pathways in photosystem II including the identification of an oxygen channel. J. Struct. Biol. 159, 228–237. doi: 10.1016/j.jsb.2007.01.f016
Nagao, R., Suzuki, T., Okumura, A., Niikura, A., Iwai, M., and Dohmae, N. (2010). Topological analysis of the extrinsic PsbO, PsbP and PsbQ proteins in a green algal PSII complex by cross-linking with a water-soluble carbodiimide. Plant Cell Physiol. 51, 718–727. doi: 10.1093/pcp/pcq042
Nahnsen, S., Bielow, C., Reinert, K., and Kohlbacher, O. (2013). Tools for label-free peptide quantification. Mol. Cell. Proteomics 12, 549–556. doi: 10.1074/mcp.R112.025163
Nakamori, H., Yatabe, T., Yoon, K.-S., and Ogo, S. (2014). Purification and characterization of an oxygen-evolving photosystem II from Leptolyngbya sp. strain O-77. J. Biosci. Bioeng. 118, 119–124. doi: 10.1016/j.jbiosc.2014.01.009
Nanba, O., and Satoh, K. (1987). Isolation of a photosystem II reaction center consisting of D-1 and D-2 polypeptides and cytochrome b-559. Proc. Natl. Acad. Sci. U.S.A. 84, 109–112. doi: 10.1073/pnas.84.1.109
Naumann, B., Busch, A., Allmer, J., Ostendorf, E., Zeller, M., Kirchhoff, H., et al. (2007). Comparative quantitative proteomics to investigate the remodeling of bioenergetic pathways under iron deficiency in Chlamydomonas reinhardtii. Proteomics 7, 3964–3979. doi: 10.1002/pmic.200700407
Nickelsen, J., and Rengstl, B. (2013). Photosystem II assembly: from cyanobacteria to plants. Annu. Rev. Plant Biol. 64, 609–635. doi: 10.1146/annurev-arplant-050312-120124
Nickelsen, J., Rengstl, B., Stengel, A., Schottkowski, M., Soll, J., and Ankele, E. (2011). Biogenesis of the cyanobacterial thylakoid membrane system - an update. FEMS Microbiol. Lett. 315, 1–5. doi: 10.1111/j.1574-6968.2010.02096.x
Nixon, P. J., Michoux, F., Yu, J., Boehm, M., and Komenda, J. (2010). Recent advances in understanding the assembly and repair of photosystem II. Ann. Bot. 106, 1–16. doi: 10.3389/fols.2075.07700
Nixon, P. J., Trost, J. T., and Diner, B. A. (1992). Role of the carboxy terminus of polypeptide D1 in the assembly of a functional water-oxidizing manganese cluster in Photosystem II of the cyanobacterium Synechocystis sp. PCC 6803: assembly requires a free carboxyl group at C-terminal position 344. Biochemistry 31, 10859–10871. doi: 10.1021/bi00159a029
Nowaczyk, M. M., Hebeler, R., Schlodder, E., Meyer, H. E., Warscheid, B., and Rögner, M. (2006). Psb27, a cyanobacterial lipoprotein, is involved in the repair cycle of Photosystem II. Plant Cell 18, 3121–3131. doi: 10.1105/tpc.106.042671
Nowaczyk, M. M., Krause, K., Mieseler, M., Sczibilanski, A., Ikeuchi, M., and Rögner, M. (2012). Deletion of psbJ leads to accumulation of Psb27-Psb28 photosystem II complexes in Thermosynechococcus elongatus. Biochim. Biophys. Acta 1817, 1339–1345. doi: 10.1016/j.bbabio.2012.02.017
Odom, W. R., and Bricker, T. M. (1992). Interaction of CPa-1 with the manganese-stabilizing protein of Photosystem II: identification of domains cross-linked by 1-ethyl-3-[3-(dimethylamino)propyl]carbodiimide. Biochemistry 31, 5616–5620. doi: 10.1021/bi00139a027
Ohnishi, N., and Murata, N. (2006). Glycinebetaine counteracts the inhibitory effects of salt stress on the degradation and synthesis of D1 protein during photoinhibition in Synechococcus sp. PCC 7942. Plant Physiol. 141, 758–765. doi: 10.1104/pp.106.076976
Ohta, H., Suzuki, T., Ueno, M., Okumura, A., Yoshihara, S., Shen, J. R., et al. (2003). Extrinsic proteins of photosystem II - An intermediate member of the PsbQ protein family in red algal PSII. Eur. J. Biochem. 270, 4156–4163. doi: 10.1046/j.1432-1033.2003.03810.x
Owens, G. C., and Ohad, I. (1982). Phosphorylation of Chlamydomonas reinhardi chloroplast membrane proteins in vivo and in vitro. J. Cell. Biol. 93, 712–718. doi: 10.1083/jcb.93.3.712
Owens, G. C., and Ohad, I. (1983). Changes in thylakoid polypeptide phosphorylation during membrane biogenesis in Chlamydomonas reinhardii y-1. Biochim. Biophys. Acta 722, 234–241. doi: 10.1016/0005-2728(83)90179-2
Pagliano, C., Chimirri, F., Saracco, G., Marsano, F., and Barber, J. (2011). One-step isolation and biochemical characterization of a highly active plant PSII monomeric core. Photosynth. Res. 108, 33–46. doi: 10.1007/s11120-011-9650-4
Pagliano, C., Nield, J., Marsano, F., Pape, T., Barera, S., Saracco, G., et al. (2014). Proteomic characterization and three-dimensional electron microscopy study of PSII-LHCII supercomplexes from higher plants. Biochim. Biophys. Acta 1837, 1454–1462. doi: 10.1016/j.bbabio.2013.11.004
Paramelle, D., Miralles, G., Subra, G., and Martinez, J. (2013). Chemical cross-linkers for protein structure studies by mass spectrometry. Proteomics 13, 438–456. doi: 10.1002/pmic.201200305
Pearson, K. M., Pannell, L. K., and Fales, H. M. (2002). Intramolecular cross-linking experiments on cytochrome c and ribonuclease A using an isotope multiplet method. Rapid Commun. Mass Spectrom. 16, 149–159. doi: 10.1002/rcm.554
Pesaresi, P., Pribil, M., Wunder, T., and Leister, D. (2011). Dynamics of reversible protein phosphorylation in thylakoids of flowering plants: the roles of STN7, STN8 and TAP38. Biochim. Biophys. Acta 1807, 887–896. doi: 10.1016/j.bbabio.2010.08.002
Petrotchenko, E. V., and Borchers, C. H. (2010). ICC-CLASS: isotopically-coded cleavable crosslinking analysis software suite. BMC Bioinformatics 11:64. doi: 10.1186/1471-2105-11-64
Petrotchenko, E. V., Makepeace, K. A. T., Serpa, J. J., and Borchers, C. H. (2014). Analysis of protein structure by cross-linking combined with mass spectrometry. Methods Mol. Biol. 1156, 447–463. doi: 10.1007/978-1-4939-0685-7_30
Petrotchenko, E. V., Serpa, J. J., and Borchers, C. H. (2011). An isotopically coded CID-cleavable biotinylated cross-linker for structural proteomics. Mol. Cell. Proteomics 10:M110.001420. doi: 10.1074/mcp.M110.001420
Plöscher, M., Granvogl, B., Zoryan, M., Reisinger, V., and Eichacker, L. A. (2009). Mass spectrometric characterization of membrane integral low molecular weight proteins from photosystem II in barley etioplasts. Proteomics 9, 625–635. doi: 10.1002/pmic.200800337
Plöscher, M., Reisinger, V., and Eichacker, L. A. (2011). Proteomic comparison of etioplast and chloroplast protein complexes. J. Proteomics 74, 1256–1265. doi: 10.1016/j.jprot.2011.03.020
Pospíšil, P. (2009). Production of reactive oxygen species by photosystem II. Biochim. Biophys. Acta 1787, 1151–1160. doi: 10.1016/j.bbabio.2009.05.005
Promnares, K., Komenda, J., Bumba, L., Nebesarova, J., Vacha, F., and Tichy, M. (2006). Cyanobacterial small chlorophyll-binding protein ScpD (HliB) is located on the periphery of Photosystem II in the vicinity of PsbH and CP47 subunits. J. Biol. Chem. 281, 32705–32713. doi: 10.1074/jbc.M606360200
Puthiyaveetil, S., and Kirchhoff, H. (2013). A phosphorylation map of the photosystem II supercomplex C2S2M2. Front. Plant Sci. 4:459. doi: 10.3389/fpls.2013.00459
Rabilloud, T., Chevallet, M., Luche, S., and Lelong, C. (2010). Two-dimensional gel electrophoresis in proteomics: Past, present and future. J. Proteomics 73, 2064–2077. doi: 10.1016/j.jprot.2010.05.016
Rappsilber, J. (2011). The beginning of a beautiful friendship: cross-linking/mass spectrometry and modelling of proteins and multi-protein complexes. J. Struct. Biol. 173, 530–540. doi: 10.1016/j.jsb.2010.10.014
Rast, A., Heinz, S., and Nickelsen, J. (2015). Biogenesis of thylakoid membranes. Biochim. Biophys. Acta 1847, 821–830. doi: 10.1016/j.bbabio.2015.01.007
Reiland, S., Messerli, G., Baerenfaller, K., Gerrits, B., Endler, A., Grossmann, J., et al. (2009). Large-scale Arabidopsis phosphoproteome profiling reveals novel chloroplast kinase substrates and phosphorylation networks. Plant Physiol. 150, 889–903. doi: 10.1104/pp.109.138677
Rengstl, B., Knoppová, J., Komenda, J., and Nickelsen, J. (2013). Characterization of a Synechocystis double mutant lacking the photosystem II assembly factors YCF48 and Sll0933. Planta 237, 471–480. doi: 10.1007/s00425-012-1720-0
Rexroth, S., Wong, C. C. L., Park, J. H., Yates, J. R. III, and Barry, B. A. (2007). An activated glutamate residue identified in Photosystem II at the interface between the manganese-stabilizing subunit and the D2 polypeptide. J. Biol. Chem. 282, 27802–27809. doi: 10.1074/jbc.M704394200
Rinalducci, S., Larsen, M. R., Mohammed, S., and Zolla, L. (2006). Novel protein phosphorylation site identification in spinach stroma membranes by titanium dioxide microcolumns and tandem mass spectrometry. J. Proteome Res. 5, 973–982. doi: 10.1021/pr050476n
Rinner, O., Seebacher, J., Walzthoeni, T., Mueller, L., Beck, M., Schmidt, A., et al. (2008). Identification of cross-linked peptides from large sequence databases. Nat. Methods 5, 315–318. doi: 10.1038/NMETH.1192
Rintamäki, E., Salonen, M., Suoranta, U. M., Carlberg, I., Andersson, B., and Aro, E.-M. (1997). Phosphorylation of Light-harvesting Complex II and Photosystem II core proteins shows different irradiance-dependent regulation in vivo: application of phosphothreonine antibodies to analysis of thylakoid phosphoproteins. J. Biol. Chem. 272, 30476–30482. doi: 10.1074/jbc.272.48.30476
Rokka, A., Suorsa, M., Saleem, A., Battchikova, N., and Aro, E. M. (2005). Synthesis and assembly of thylakoid protein complexes: multiple assembly steps of photosystem II. Biochem. J. 388, 159–168. doi: 10.1042/BJ20042098
Romanowska, E., Wasilewska, W., Fristedt, R., Vener, A. V., and Zienkiewicz, M. (2012). Phosphorylation of PSII proteins in maize thylakoids in the presence of Pb ions. J. Plant Physiol. 169, 345–352. doi: 10.1016/j.jplph.2011.10.006
Roose, J. L., Kashino, Y., and Pakrasi, H. B. (2007). The PsbQ protein defines cyanobacterial Photosystem II complexes with highest activity and stability. Proc. Natl. Acad. Sci. U.S.A. 104, 2548–2553. doi: 10.1073/pnas.0609337104
Roose, J. L., and Pakrasi, H. B. (2004). Evidence that D1 processing is required for manganese binding and extrinsic protein assembly into Photosystem II. J. Biol. Chem. 279, 45417–45422. doi: 10.1074/jbc.M408458200
Roose, J. L., and Pakrasi, H. B. (2008). The Psb27 protein facilitates manganese cluster assembly in Photosystem II. J. Biol. Chem. 283, 4044–4050. doi: 10.1074/jbc.M708960200
Ross, P. L., Huang, Y. L. N., Marchese, J. N., Williamson, B., Parker, K., Hattan, S., et al. (2004). Multiplexed protein quantitation in Saccharomyces cerevisiae using amine-reactive isobaric tagging reagents. Mol. Cell. Proteomics 3, 1154–1169. doi: 10.1074/mcp.M400129-MCP200
Rowland, J. G., Simon, W. J., Nishiyama, Y., and Slabas, A. R. (2010). Differential proteomic analysis using iTRAQ reveals changes in thylakoids associated with Photosystem II-acquired thermotolerance in Synechocystis sp. PCC 6803. Proteomics 10, 1917–1929. doi: 10.1002/pmic.200900337
Ryan, C. M., Souda, P., Bassilian, S., Ujwal, R., Zhang, J., Abramson, J., et al. (2010). Post-translational modifications of integral membrane proteins resolved by top-down Fourier transform mass spectrometry with collisionally activated dissociation. Mol. Cell. Proteomics 9, 791–803. doi: 10.1074/mcp.M900516-MCP200
Samol, I., Shapiguzov, A., Ingelsson, B., Fucile, G., Crevecoeur, M., Vener, A. V., et al. (2012). Identification of a Photosystem II phosphatase involved in light acclimation in Arabidopsis. Plant Cell 24, 2596–2609. doi: 10.1105/tpc.112.095703
Sander, J., Nowaczyk, M., Buchta, J., Dau, H., Vass, I., Deák, Z., et al. (2010). Functional characterization and quantification of the alternative PsbA copies in Thermosynechococcus elongatus and their role in photoprotection. J. Biol. Chem. 285, 29851–29856. doi: 10.1074/jbc.M110.127142
Schönberg, A., and Baginsky, S. (2012). Signal integration by chloroplast phosphorylation networks: an update. Front. Plant Sci. 3:256. doi: 10.3389/fpls.2012.00256
Schottkowski, M., Gkalympoudis, S., Tzekova, N., Stelljes, C., Schuenemann, D., Ankele, E., et al. (2009). Interaction of the periplasmic PratA factor and the PsbA (D1) protein during biogenesis of Photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 284, 1813–1819. doi: 10.1074/jbc.M806116200
Seebacher, J., Mallick, P., Zhang, N., Eddes, J., Aebersold, R., and Gelb, M. H. (2006). Protein cross-linking analysis using mass spectrometry, isotope-coded cross-linkers, and integrated computational data processing. J. Proteome Res. 5, 2270–2282. doi: 10.1021/pr060154z
Seidler, A. (1996). Intermolecular and intramolecular interactions of the 33-kDa protein in photosystem II. Eur. J. Biochem. 242, 485–490. doi: 10.1111/j.1432-1033.1996.0485r.x
Sharma, J., Panico, M., Barber, J., and Morris, H. R. (1997a). Characterization of the low molecular weight photosystem II reaction center subunits and their light-induced modifications by mass spectrometry. J. Biol. Chem. 272, 3935–3943.
Sharma, J., Panico, M., Barber, J., and Morris, H. R. (1997b). Purification and determination of intact molecular mass by electrospray ionization mass spectrometry of the photosystem II reaction center subunits. J. Biol. Chem. 272, 33153–33157.
Sharma, J., Panico, M., Shipton, C. A., Nilsson, F., Morris, H. R., and Barber, J. (1997c). Primary structure characterization of the photosystem II D1 and D2 subunits. J. Biol. Chem. 272, 33158–33166. doi: 10.1074/jbc.272.52.33158
Shaw, J. B., Li, W., Holden, D. D., Zhang, Y., Griep-Raming, J., Fellers, R. T., et al. (2013). Complete protein characterization using top-down mass spectrometry and ultraviolet photodissociation. J. Am. Chem. Soc. 135, 12646–12651. doi: 10.1021/ja4029654
Shevela, D., and Messinger, J. (2013). Studying the oxidation of water to molecular oxygen in photosynthetic and artificial systems by time-resolved membrane-inlet mass spectrometry. Front. Plant Sci. 4:473. doi: 10.3389/fpls.2013.00473
Shi, L.-X., Hall, M., Funk, C., and Schröder, W. (2012). Photosystem II, a growing complex: updates on newly discovered components and low molecular mass proteins. Biochim. Biophys. Acta 1817, 13–25. doi: 10.1016/j.bbabio.2011.08.008
Shi, L.-X., Lorković, Z. J., Oelmüller, R., and Schröder, W. P. (2000). The low molecular mass PsbW protein is involved in the stabilization of the dimeric Photosystem II complex in Arabidopsis thaliana. J. Biol. Chem. 275, 37945–37950. doi: 10.1074/jbc.M006300200
Shi, L.-X., and Schröder, W. P. (1997). Compositional and topological studies of the PsbW protein in spinach thylakoid membrane. Photosynth. Res. 53, 45–53. doi: 10.1023/A:1005830405809
Silva, J. C., Gorenstein, M. V., Li, G. Z., Vissers, J. P. C., and Geromanos, S. J. (2006). Absolute quantification of proteins by LCMSE - A virtue of parallel MS acquisition. Mol. Cell. Proteomics 5, 144–156. doi: 10.1074/mcp.M500230-MCP200
Sinz, A. (2014). The advancement of chemical cross-linking and mass spectrometry for structural proteomics: from single proteins to protein interaction networks. Expert Rev. Proteomics 11, 733–743. doi: 10.1586/14789450.2014.960852
Stöckel, J., Jacobs, J. M., Elvitigala, T. R., Liberton, M., Welsh, E. A., Polpitiya, A. D., et al. (2011). Diurnal rhythms result in significant changes in the cellular protein complement in the cyanobacterium Cyanothece 51142. PLoS ONE 6:e16680. doi: 10.1371/journal.pone.0016680
Steinback, K. E., Bose, S., and Kyle, D. J. (1982). Phosphorylation of the light-harvesting chlorophyll-protein regulates excitation energy distribution between Photosystem II and Photosystem I. Arch. Biochem. Biophys. 216, 356–361. doi: 10.1016/0003-9861(82)90221-1
Sugimoto, I., and Takahashi, Y. (2003). Evidence that the PsbK polypeptide is associated with the Photosystem II core antenna complex CP43. J. Biol. Chem. 278, 45004–45010. doi: 10.1074/jbc.M307537200
Sugiura, M., and Boussac, A. (2014). Some Photosystem II properties depending on the D1 protein variants in Thermosynechococcus elongatus. Biochim. Biophys. Acta 1837, 1427–1434. doi: 10.1016/j.bbabio.2013.12.011
Sugiura, M., Iwai, E., Hayashi, H., and Boussac, A. (2010a). Differences in the interactions between the subunits of Photosystem II dependent on D1 protein variants in the thermophilic cyanobacterium Thermosynechococcus elongatus. J. Biol. Chem. 285, 30008–30018. doi: 10.1074/jbc.M110.136945
Sugiura, M., Kato, Y., Takahashi, R., Suzuki, H., Watanabe, T., Noguchi, T., et al. (2010b). Energetics in Photosystem II from Thermosynechococcus elongatus with a D1 protein encoded by either the psbA1 or psbA3 gene. Biochim. Biophys. Acta 1797, 1491–1499. doi: 10.1016/j.bbabio.2010.03.022
Sugiura, M., Koyama, K., Umena, Y., Kawakami, K., Shen, J.-R., Kamiya, N., et al. (2013). Evidence for an unprecedented histidine hydroxyl modification on D2-His336 in Photosystem II of Thermosynechoccocus vulcanus and Thermosynechoccocus elongatus. Biochemistry 52, 9426–9431. doi: 10.1021/bi401213m
Sugiyama, N., Nakagami, H., Mochida, K., Daudi, A., Tomita, M., Shirasu, K., et al. (2008). Large-scale phosphorylation mapping reveals the extent of tyrosine phosphorylation in Arabidopsis. Mol. Syst. Biol. 4, 193. doi: 10.1038/msb.2008.32
Swaisgood, H., and Natake, M. (1973). Effect of carboxyl group modification on some of the enzymatic properties of L-glutamate dehydrogenase. J. Biochem. 74, 77–86.
Takahashi, M., Shiraishi, T., and Asada, K. (1988). COOH-terminal residues of D1 and the 44 kDa CPa-2 at spinach photosystem II core complex. FEBS Lett. 240, 6–8. doi: 10.1016/0014-5793(88)80330-2
Takamoto, K., and Chance, M. R. (2006). Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu. Rev. Biophys. Biomol. Struct. 35, 251–276. doi: 10.1146/annurev.biophys.35.040405.102050
Takasaka, K., Iwai, M., Umena, Y., Kawakami, K., Ohmori, Y., Ikeuchi, M., et al. (2010). Structural and functional studies on Ycf12 (Psb30) and PsbZ-deletion mutants from a thermophilic cyanobacterium. Biochim. Biophys. Acta 1797, 278–284. doi: 10.1016/j.bbabio.2009.11.001
Tal, O., Trabelcy, B., Gerchman, Y., and Adir, N. (2014). Investigation of phycobilisome subunit interaction interfaces by coupled cross-linking and mass spectrometry. J. Biol. Chem. 289, 33084–33097. doi: 10.1074/jbc.M114.595942
Telfer, A., Bishop, S. M., Phillips, D., and Barber, J. (1994). Isolated photosynthetic reaction center of Photosystem II as a sensitizer for the formation of singlet oxygen: detection and quantum yield determination using a chemical trapping technique. J. Biol. Chem. 269, 13244–13253.
Thangaraj, B., Ryan, C. M., Souda, P., Krause, K., Faull, K. F., Weber, A. P. M., et al. (2010). Data-directed top-down Fourier-transform mass spectrometry of a large integral membrane protein complex: Photosystem II from Galdieria sulphuraria. Proteomics 10, 3644–3656. doi: 10.1002/pmic.201000190
Thelen, J. J., and Peck, S. C. (2007). Quantitative proteomics in plants: choices in abundance. Plant Cell 19, 3339–3346. doi: 10.1105/tpc.107.053991
Thidholm, E., Lindström, V., Tissier, C., Robinson, C., Schröder, W. P., and Funk, C. (2002). Novel approach reveals localisation and assembly pathway of the PsbS and PsbW proteins into the photosystem II dimer. FEBS Lett. 513, 217–222. doi: 10.1016/S0014-5793(02)02314-1
Thompson, A., Schäfer, J., Kuhn, K., Kienle, S., Schwarz, J., Schmidt, G., et al. (2003). Tandem mass tags: a novel quantification strategy for comparative analysis of complex protein mixtures by MS/MS. Anal. Chem. 75, 1895–1904. doi: 10.1021/ac0262560
Thornton, L. E., Ohkawa, H., Roose, J. L., Kashino, Y., Keren, N., and Pakrasi, H. B. (2004). Homologs of plant PsbP and PsbQ proteins are necessary for regulation of Photosystem II activity in the cyanobacterium Synechopystis 6803. Plant Cell 16, 2164–2175. doi: 10.1105/tpc.104.023515
Tikhonov, A. N. (2015). Induction events and short-term regulation of electron transport in chloroplasts: an overview. Photosynth. Res. 125, 65–94. doi: 10.1007/s11120-015-0094-0
Tikkanen, M., and Aro, E.-M. (2014). Integrative regulatory network of plant thylakoid energy transduction. Trends Plant Sci. 19, 10–17. doi: 10.1016/j.tplants.2013.09.003
Tikkanen, M., Grieco, M., Kangasjarvi, S., and Aro, E.-M. (2010). Thylakoid protein phosphorylation in higher plant chloroplasts optimizes electron transfer under fluctuating light. Plant Physiol. 152, 723–735. doi: 10.1104/pp.109.150250
Tikkanen, M., Nurmi, M., Kangasjarvi, S., and Aro, E.-M. (2008a). Core protein phosphorylation facilitates the repair of photodamaged photosystem II at high light. Biochim. Biophys. Acta 1777, 1432–1437. doi: 10.1016/j.bbabio.2008.08.004
Tikkanen, M., Nurmi, M., Suorsa, M., Danielsson, R., Mamedov, F., Styring, S., et al. (2008b). Phosphorylation-dependent regulation of excitation energy distribution between the two photosystems in higher plants. Biochim. Biophys. Acta 1777, 425–432. doi: 10.1016/j.bbabio.2008.02.001
Tomo, T., Enami, I., and Satoh, K. (1993). Orientation and nearest-neighbor analysis of psbI gene product in the photosystem II reaction center complex using bifunctional cross-linkers. FEBS Lett. 323, 15–18. doi: 10.1016/0014-5793(93)81438-6
Tsiotis, G., Walz, T., Spyridaki, A., Lustig, A., Engel, A., and Ghanotakis, D. (1996). Tubular crystals of a Photosystem II core complex. J. Mol. Biol. 259, 241–248. doi: 10.1006/jmbi.1996.0316
Turkina, M. V., Kargul, J., Blanco-Rivero, A., Villarejo, A., Barber, J., and Vener, A. V. (2006). Environmentally modulated phosphoproteome of photosynthetic membranes in the green alga Chlamydomonas reinhardtii. Mol. Cell Proteomics 5, 1412–1425. doi: 10.1074/mcp.M600066-MCP200
Tyystjärvi, E. (2013). Photoinhibition of Photosystem II. Int. Rev. Cell. Mol. Biol. 300, 243–303. doi: 10.1016/B978-0-12-405210-9.00007-2
Ujihara, T., Sakurai, I., Mizusawa, N., and Wada, H. (2008). A method for analyzing lipid-modified proteins with mass spectrometry. Anal. Biochem. 374, 429–431. doi: 10.1016/j.ab.2007.11.014
Umena, Y., Kawakami, K., Shen, J., and Kamiya, N. (2011). Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å. Nature 473, 55–60. doi: 10.1038/nature09913
Vainonen, J. P., Hansson, M., and Vener, A. V. (2005). STN8 protein kinase in Arabidopsis thaliana is specific in phosphorylation of Photosystem II core proteins. J. Biol. Chem. 280, 33679–33686. doi: 10.1074/jbc.M505729200
Vassiliev, S., Zaraiskaya, T., and Bruce, D. (2012). Exploring the energetics of water permeation in photosystem II by multiple steered molecular dynamics simulations. Biochim. Biophys. Acta 1817, 1671–1678. doi: 10.1016/j.bbabio.2012.05.016
Vener, A. V. (2007). Environmentally modulated phosphorylation and dynamics of proteins in photosynthetic membranes. Biochim. Biophys. Acta 1767, 449–457. doi: 10.1016/j.bbabio.2007.11.007
Vener, A. V., Harms, A., Sussman, M. R., and Vierstra, R. D. (2001). Mass spectrometric resolution of reversible protein phosphorylation in photosynthetic membranes of Arabidopsis thaliana. J. Biol. Chem. 276, 6959–6966. doi: 10.1074/jbc.M009394200
Walleczek, J., Martin, T., Redl, B., Stofflermeilicke, M., and Stoffler, G. (1989). Comparative cross-linking study on the 50S ribosomal subunit from Escherichia coli. Biochemistry 28, 4099–4105. doi: 10.1021/bi00435a071
Wang, L. W., and Chance, M. R. (2011). Structural mass spectrometry of proteins using hydroxyl radical based protein footprinting. Anal. Chem. 83, 7234–7241. doi: 10.1021/ac200567u
Wegener, K. M., Bennewitz, S., Oelmüller, R., and Pakrasi, H. B. (2011). The Psb32 protein aids in repairing photodamaged Photosystem II in the cyanobacterium Synechocystis 6803. Mol. Plant 4, 1052–1061. doi: 10.1104/pp.114.253336
Wegener, K. M., Nagarajan, A., and Pakrasi, H. B. (2015). An atypical psbA gene encodes a sentinel D1 protein to form a physiologically relevant inactive Photosystem II complex in cyanobacteria. J. Biol. Chem. 290, 3764–3774. doi: 10.1074/jbc.M114.604124
Wegener, K. M., Welsh, E. A., Thornton, L. E., Keren, N., Jacobs, J. M., Hixson, K. K., et al. (2008). High sensitivity proteomics assisted discovery of a novel operon involved in the assembly of Photosystem II, a membrane protein complex. J. Biol. Chem. 283, 27829–27837. doi: 10.1074/jbc.M803918200
Weisbrod, C. R., Chavez, J. D., Eng, J. K., Yang, L., Zheng, C., and Bruce, J. E. (2013). In vivo protein interaction network identified with a novel real-time cross-linked peptide identification strategy. J. Proteome Res. 12, 1569–1579. doi: 10.1021/pr3011638
Welkie, D., Zhang, X., Markillie, M. L., Taylor, R., Orr, G., Jacobs, J., et al. (2014). Transcriptomic and proteomic dynamics in the metabolism of a diazotrophic cyanobacterium, Cyanothece sp. PCC 7822 during a diurnal light-dark cycle. BMC Genomics 15:1185. doi: 10.1186/1471-2164-15-1185
Wen, J. Z., Zhang, H., Gross, M. L., and Blankenship, R. E. (2009). Membrane orientation of the FMO antenna protein from Chlorobaculum tepidum as determined by mass spectrometry-based footprinting. Proc. Natl. Acad. Sci. U.S.A. 106, 6134–6139. doi: 10.1073/pnas.0901691106
Wetz, K., and Habermehl, K.-O. (1979). Topographical studies on poliovirus capsid proteins by chemical modification and cross-linking with bifunctional reagents. J. Gen. Virol. 44, 525–534. doi: 10.1099/0022-1317-44-2-525
Whitelegge, J. P. (2013). Integral membrane proteins and bilayer proteomics. Anal. Chem. 85, 2558–2568. doi: 10.1021/ac303064a
Whitelegge, J. P., Gundersen, C. B., and Faull, K. F. (1998). Electrospray-ionization mass spectrometry of intact intrinsic membrane proteins. Protein Sci. 7, 1423–1430.
Xu, H., and Freitas, M. A. (2009). MassMatrix: a database search program for rapid characterization of proteins and peptides from tandem mass spectrometry data. Proteomics 9, 1548–1555. doi: 10.1002/pmic.200700322
Yang, B., Wu, Y.-J., Zhu, M., Fan, S.-B., Lin, J., Zhang, K., et al. (2012). Identification of cross-linked peptides from complex samples. Nat. Methods 9, 904–909. doi: 10.1038/nmeth.2099
Yang, M.-K., Yang, Y.-H., Chen, Z., Zhang, J., Lin, Y., Wang, Y., et al. (2014). Proteogenomic analysis and global discovery of posttranslational modifications in prokaryotes. Proc. Natl. Acad. Sci. U.S.A. 111, E5633–E5642. doi: 10.1073/pnas.1412722111
Yao, D. C. I., Brune, D. C., Vavilin, D., and Vermaas, W. F. J. (2012a). Photosystem II component lifetimes in the cyanobacterium Synechocystis sp. strain PCC 6803: Small Cab-like proteins stabilize biosynthesis intermediates and affect early steps in chlorophyll synthesis. J. Biol. Chem. 287, 682–692. doi: 10.1074/jbc.M111.320994
Yao, D. C. I., Brune, D. C., and Vermaas, W. F. J. (2012b). Lifetimes of photosystem I and II proteins in the cyanobacterium Synechocystis sp. PCC 6803. FEBS Lett. 586, 169–173. doi: 10.1016/j.febslet.2011.12.010
Zak, E., Norling, B., Maitra, R., Huang, F., Andersson, B., and Pakrasi, H. B. (2001). The initial steps of biogenesis of cyanobacterial photosystems occur in plasma membranes. Proc. Natl. Acad. Sci. U.S.A. 98, 13443–13448. doi: 10.1073/pnas.241503898
Zhang, H., Liu, H., Blankenship, R. E., and Gross, M. L. (2016). Isotope-encoded carboxyl group footprinting for mass spectrometry-based protein conformational studies. J. Am. Soc. Mass Spectrom. 27, 178–181. doi: 10.1007/s13361-015-1260-5
Zhang, H., Liu, H., Niedzwiedzki, D. M., Prado, M., Jiang, J., Gross, M. L., et al. (2014). Molecular mechanism of photoactivation and structural location of the cyanobacterial orange carotenoid protein. Biochemistry 53, 13–19. doi: 10.1021/bi401539w
Keywords: Photosystem II, Photosystem II life-cycle, mass spectrometry, post-translational modification, chemical cross-linking, protein footprinting
Citation: Weisz DA, Gross ML and Pakrasi HB (2016) The Use of Advanced Mass Spectrometry to Dissect the Life-Cycle of Photosystem II. Front. Plant Sci. 7:617. doi: 10.3389/fpls.2016.00617
Received: 05 February 2016; Accepted: 22 April 2016;
Published: 10 May 2016.
Edited by:
Julian Eaton-Rye, University of Otago, New ZealandReviewed by:
Jian-Ren Shen, Okayama University, JapanJulian Whitelegge, University of California, Los Angeles, USA
Copyright © 2016 Weisz, Gross and Pakrasi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Himadri B. Pakrasi, cGFrcmFzaUB3dXN0bC5lZHU=
 Daniel A. Weisz
Daniel A. Weisz Michael L. Gross
Michael L. Gross Himadri B. Pakrasi
Himadri B. Pakrasi