- 1Laboratory of Plant Pathology, Department of Agriculture, Crop Production and Rural Environment, University of Thessaly, Volos, Greece
- 2Department of Electrical and Computer Engineering, Aristotle University of Thessaloniki, Thessaloniki, Greece
- 3Department of Ichthyology and Aquatic Environment, University of Thessaly, Volos, Greece
- 4Centre for Research and Technology Hellas, Institute of Applied Biosciences, Thessaloniki, Greece
- 5Department of Deciduous Fruit Trees in Naoussa, Institute of Plant Breeding and Plant Genetic Resources, Hellenic Agricultural Organization ‘Demeter’, Naoussa, Greece
Introduction
Botrytis cinerea is a ubiquitous fungus difficult to control because it possess a variety of attack modes, diverse hosts as inoculum sources, and it can survive as mycelia and/or conidia or for extended periods as sclerotia in crop debris. For these reasons the use of any single control measure is unlikely to succeed and a combination of cultural practices with the application of site-specific synthetic compounds provide the best protection for the crops (Williamson et al., 2007). However, the chemical control has been adversely affected by the development of fungicide resistance. The selection of resistant individuals in a fungal population subjected to selective pressure due to fungicides is an evolutionary mechanism that promotes advantageous genotypes (Walker et al., 2013). High levels of resistance to site-specific fungicides are commonly associated with point mutations. For example the mutations G143A, H272R, and F412S leading to changes in the target proteins CytB, SdhB, and Erg27 are conferring resistance of the pathogen to the chemical classes of QoIs, SDHIs, and hydroxyanilides, respectively (Leroux, 2007). Multidrug resistance is another mechanism associated with resistance in B. cinerea which involves mutations leading to overexpression of individual transporters such as ABC and MFS (Kretschmer et al., 2009). This mechanism is associated with low levels of resistance to multiple fungicides including the anilinopyrimidines and phenylpyrroles. However, a subdivision of gray mold populations was found to be more tolerant to these two classes of fungicides (Leroch et al., 2013).
Previous reports have clearly demonstrated that the resistance to anilinopyrimidines has a qualitative, disruptive pattern, and is monogenically controlled (Chapeland et al., 1999). In order to elucidate the mechanism of the resistance, the whole genome of three different samples (gene pools) was sequenced, each containing DNA of 10 selected strains of the same genotype regarding resistance to seven different classes of fungicides including anilinopyrimidines. This report presents the publicly available genomic data.
Materials and Methods
Isolation–Determination of Fungicide Resistance Profile
Pure cultures of B. cinerea were obtained on sterilized PDA media from infected lettuce plants by slight touching a flamed wire loop onto a freshly sporulating lesion. All isolates obtained from lettuce plants in a commercial lettuce glasshouse located at Krokion, Magnesia, Greece on February 26th, 2012. From each sample a single isolate was made. For all purposes single-spore isolates properly prepaired. The sensitivity of the isolates to the fungicides fenhexamid (class: Hydroxyanilides-Hyd), pyraclostrobin (class: Quinone outside Inhibitors-QoIs), boscalid (class: Succinate De-Hydrogenase Inhibitors-SDHIs; Bos), cyprodinil (class: Anilinopyrimidines-Ani), fludioxonil (class: Phenylpyrroles-Phen), carbendazim (class: Benzimidazoles-Ben), and iprodione (class: Dicarboximides-Dic) was determined by the point inoculation method using the discriminatory concentrations of each fungicide as defined by Chatzidimopoulos et al. (2013). The isolates were then classified in three major groups according to their respective fungicide resistance profile, i.e., (1) Wild type, sensitive to all seven fungicides reported previously; (2) Phenotype QoIRBosRAniRBenRDicR, resistant (R) to five fungicides; and (3) Phenotype HydRQoIRBosRAniRPhenRBenRDicR, resistant to all seven fungicides tested.
Nucleic Acid and Library Preparation—Sequencing
High quality genomic DNA was extracted from 10 selected single-spore strains of each group (30 in total) by applying a CTAB based protocol (Chatzidimopoulos et al., 2014). Then, three different gene pools were generated containing the DNA of the selected strains according to the fungicide resistance profile. The quantity of DNA was estimated by picogreen (Invitrogen) method using Victor 3 fluorometry. The condition of DNA was checked by a gel electrophoresis method and the purity was assessed on a NanoDrop instrument.
Library preparation performed with the TruSeq Nano DNA Kit (Illumina) with target insert size of 550 bp and read lengths of 101 bp using 200 ng input gDNA. Whole genome sequenced on Illumina HiSeq 2000 with paired-end libraries generated for each of the three fungal genomes (Macrogen INC, 10F, 254 Beotkkot-ro, Geumcheon-gu, Seoul, Rep. of Korea). Fragmented DNA was cleaned up and the overhangs converted into blunt ends using End Repair Mix 2 (Illumina). In order to verify the size of PCR enriched fragments, the template size distribution was checked on a Agilent Technologies 2100 Bioanalyzer using a DNA 1000 chip. The DNA sequence of each cluster on a flow cell determined with the TruSeq SBS Kit v3 (Illumina). The generation of raw data performed with the HiSeq Control Software v2.2 (Illumina).
Sequence Clean-Up, Alignment, and Variant Calling
In order to consistently apply quality and adapter trimming to the sequences, the Cutadapt and FastQC tools were applied through the Trim Galore! wrapper application (Andrews, 2010; Martin, 2011; Krueger, 2012). The sequences were consequently aligned on the B. cinerea B05.10 reference genome, as retrieved from Fungi Ensembl (http://fungi.ensembl.org/Botrytis_cinerea/Info/Index). After building the reference index files the reads were aligned to the reference genome by using Bowtie 2 (Langmead and Salzberg, 2012) and the produced alignments were parsed using SAMTools (Li et al., 2009). In order to identify potential SNPs and INDELs further analysis was performed using GATK's UnifiedGenotyper (DePristo et al., 2011) and SnpEff v 4.2 (Cingolani et al., 2012).
Results
A total of 12.8, 13, and 11.2 GB sequence data was obtained from the wild type (UTH.PPL.WT5), the 5-resistance variant (UTH.PPL.CR55), and the 7-resistance variant (UTH.PPL.MDR7) respectively. Low quality reads and adapter sequences were removed using FastQC and Cutadapt tools. After quality assessment, there was a loss of ~0.53, 0.45, and 0.46%, respectively (i.e., high quality sequences). Detailed information of the aligned reads is summarized in Table 1. The average coverage of the data across the reference genome was calculated using the samtools depth tool, and was found to be 95.9224, 108.681, and 87.2708 respectively. Moreover, the mapping rate of the three samples was approximately 80%, with the highest rate of 84.1% evident in UTH.PPL.CR55 and the lowest rate of 76.9 % in UTH.PPL.WT5. The datasets submitted to NCBI (raw reads) include the genome sequences of all three samples in FASTA format and can be accessed via the accession numbers reported in Table 1. Users can download and use the data freely for research purpose only with acknowledgment to us and quoting this paper as reference to the data.
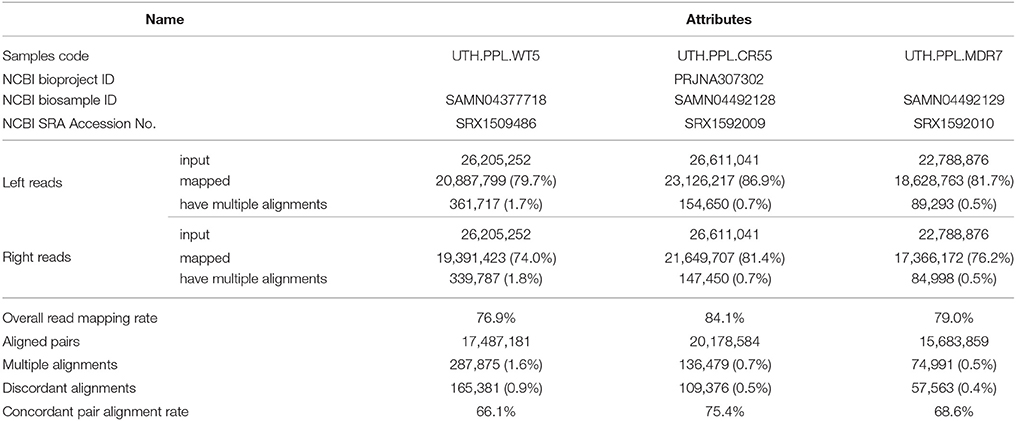
Table 1. Genome ID and overview of the three sample alignments against the Botrytis cinerea B05.10 reference genome.
Author Contributions
This project was planned and executed by MC under the supervision of PD. The data processing performed jointly by both FP and EM. IG, PM, and EV assisted substantially on the technical part of this work.
Funding
The research project was funded under the Action “Research & Technology Development Innovation projects (AgroETAK),” MIS 453350, in the framework of the Operational Program “Human Resources Development.” It was co-funded by the European Social Fund and by National Resources through the National Strategic Reference Framework 2007–2013 (NSRF 2007–2013) coordinated by the Hellenic Agricultural Organization “DEMETER”/Institute of Plant Breeding and Genetic Resources.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer LP and handling Editor declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
References
Andrews, S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Chapeland, F., Fritz, R., Lanen, C., Gredt, M., and Leroux, P. (1999). Inheritance and mechanisms of resistance to anilinopyrimidine fungicides in Botrytis cinerea (Botryotinia fuckeliana). Pestic. Biochem. Phys. 64, 85–100. doi: 10.1006/pest.1999.2414
Chatzidimopoulos, M., Ganopoulos, I., Madesis, P., Vellios, E., Tsaftaris, A., and Pappas, A. C. (2014). High-resolution melting (HRM) analysis for rapid detection and characterization of Botrytis cinerea phenotypes resistant to fenhexamid and boscalid. Plant Pathol. 63, 1336–1343. doi: 10.1111/ppa.12210
Chatzidimopoulos, M., Papaevaggelou, D., and Pappas, A. C. (2013). Detection and characterization of fungicide resistant phenotypes of Botrytis cinerea in lettuce crops in Greece. Eur. J. Plant Pathol. 137, 363–376. doi: 10.1007/s10658-013-0248-x
Cingolani, P., Platts, A., Wang, L. L., Coon, M., Nguyen, T., Wang, L., et al. (2012). A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w(1118); iso-2; iso-3. Fly 6, 80–92. doi: 10.4161/fly.19695
DePristo, M. A., Banks, E., Poplin, R. E., Garimella, K. V., Maguire, J. R., Hartl, C., et al. (2011). A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat. Genet. 43, 491–498. doi: 10.1038/ng.806
Kretschmer, M., Leroch, M., Mosbach, A., Walker, A. S., Fillinger, S., Mernke, D., et al. (2009). Fungicide-driven evolution and molecular basis of multidrug resistance in field populations of the grey mould fungus Botrytis cinerea. Plos Pathog. 5:e1000696. doi: 10.1371/journal.ppat.1000696
Krueger, F. (2012). Trim Galore!. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/trim_galore/
Langmead, B., and Salzberg, S. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Leroch, M., Plesken, C., Weber, R. W. S., Kauff, F., Scalliet, G., and Hahn, M. (2013). Gray mold populations in German strawberry fields are resistant to multiple fungicides and dominated by a novel clade closely related to Botrytis cinerea. Appl. Environ. Microb. 79, 159–167. doi: 10.1128/Aem.02655-12
Leroux, P. (2007). “Chemical control of Botrytis and its resistance to chemical fungicides,” in Botrytis: Biology, Pathology and Control, eds Y. Elad, B. Williamson, P. Tudzynski and N. Delen (Dordrecht: Springer). 195–222.
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet 17, 10–12. doi: 10.14806/ej.17.1.200
Walker, A. S., Micoud, A., Remuson, F., Grosman, J., Gredt, M., and Leroux, P. (2013). French vineyards provide information that opens ways for effective resistance management of Botrytis cinerea (grey mould). Pest. Manag. Sci. 69, 667–678. doi: 10.1002/Ps.3506
Keywords: Botrytis cinerea, anilinopyrimidines, whole genome sequencing, multi-drug resistance, next-generation sequencing (NGS)
Citation: Chatzidimopoulos M, Psomopoulos F, Malandrakis EE, Ganopoulos I, Madesis P, Vellios EK and Drogoudi P (2016) Comparative Genomics of Botrytis cinerea Strains with Differential Multi-Drug Resistance. Front. Plant Sci. 7:554. doi: 10.3389/fpls.2016.00554
Received: 17 March 2016; Accepted: 11 April 2016;
Published: 28 April 2016.
Edited by:
Alessio Mengoni, Università degli Studi di Firenze, ItalyReviewed by:
Jacob A. Tennessen, Oregon State University, USALuana Presta, University of Florence, Italy
Copyright © 2016 Chatzidimopoulos, Psomopoulos, Malandrakis, Ganopoulos, Madesis, Vellios and Drogoudi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Michael Chatzidimopoulos, mxatzid@agr.uth.gr
 Michael Chatzidimopoulos
Michael Chatzidimopoulos Fotis Psomopoulos
Fotis Psomopoulos Emmanouil E. Malandrakis
Emmanouil E. Malandrakis Ioannis Ganopoulos
Ioannis Ganopoulos Panagiotis Madesis
Panagiotis Madesis Evangelos K. Vellios1
Evangelos K. Vellios1 Pavlina Drogoudi
Pavlina Drogoudi