- Institute of Plant and Microbial Biology, University of Zurich, Zurich, Switzerland
The gene-for-gene hypothesis states that for each gene controlling resistance in the host, there is a corresponding, specific gene controlling avirulence in the pathogen. Allelic series of the cereal mildew resistance genes Pm3 and Mla provide an excellent system for genetic and molecular analysis of resistance specificity. Despite this opportunity for molecular research, avirulence genes in mildews remain underexplored. Earlier work in barley powdery mildew (B.g. hordei) has shown that the reaction to some Mla resistance alleles is controlled by multiple genes. Similarly, several genes are involved in the specific interaction of wheat mildew (B.g. tritici) with the Pm3 allelic series. We found that two mildew genes control avirulence on Pm3f: one gene is involved in recognition by the resistance protein as demonstrated by functional studies in wheat and the heterologous host Nicotiana benthamiana. A second gene is a suppressor, and resistance is only observed in mildew genotypes combining the inactive suppressor and the recognized Avr. We propose that such suppressor/avirulence gene combinations provide the basis of specificity in mildews. Depending on the particular gene combinations in a mildew race, different genes will be genetically identified as the “avirulence” gene. Additionally, the observation of two LINE retrotransposon-encoded avirulence genes in B.g. hordei further suggests that the control of avirulence in mildew is more complex than a canonical gene-for-gene interaction. To fully understand the mildew–cereal interactions, more knowledge on avirulence determinants is needed and we propose ways how this can be achieved based on recent advances in the field.
Avirulence Genes in Fungal Plant Pathogens
The identification of avirulence (Avr) genes in plant pathogenic fungi has been accelerating in recent years due to rapid advances in ‘omics’ technologies. At present, at least 35 Avrs have been cloned from filamentous fungi infecting a wide variety of agronomically important crops. Examples of Avrs fitting the gene-for-gene model are found in Cladosporium fulvum and Leptosphaeria maculans, where single Avrs are recognized by their cognate resistance (R) genes (Wulff et al., 2009; Hayward et al., 2012). However, in Magnaporthe oryzae and Melampsora lini, there are cases of single Avrs recognized by multiple R genes, as it was found for Avr-Pik/km/kp (Yoshida et al., 2009), AvrL567, and AvrP123 (Barrett et al., 2009; Ravensdale et al., 2012). Another level of complexity has been described in Fusarium oxysporum f. sp. lycopersici, which encodes Avr1 that is recognized by the tomato R gene I-1, but also acts as a suppressor of the recognition of Avr2 and Avr3 by I-2 and I-3, respectively (Houterman et al., 2008).
Thus far, most cloned avirulence genes encode a typical effector protein (63–314 amino acids) with a predicted signal peptide for secretion, with Ace1 (4034 amino acids) from Magnaporthe grisea being one of three exceptions (the other two are AvrMla genes, see below). Ace1 encodes a hybrid polyketide synthase non-ribosomal peptide synthetase involved in the biosynthesis of the actual avirulence factor (Boehnert et al., 2004). In contrast to AVR proteins from Oomycetes that contain few or no cysteines, fungal AVRs are often cysteine-rich, which confers stability in the leaf apoplast. Most AVRs cloned from fungal pathogens forming specialized infection structures (e.g., Blumeria graminis, M. lini, M. grisea) appear to contain fewer cysteines than AVRs from fungal pathogens that invade exclusively with hyphae (for a summary, see Rouxel and Balesdent, 2010). Thus, differences in cysteine content might reflect the mechanistic differences between these two infection strategies.
Avirulence in Cereal Mildews: Genetic Analysis in Segregating Populations
The genetic basis of Avr–R interactions in cereal powdery mildews was investigated using classical genetic approaches in eleven crosses of B.g. hordei (see Skamnioti et al., 2008, for a summary), two of B.g. tritici (Bourras et al., 2015; Parlange et al., 2015) and at least in one cross of B.g. secalis and B.g. tritici hybrids (Tosa, 1994). In the resulting haploid F1 progeny, each genetic locus from the parental genotypes is expected to segregate in a 1:1 ratio, so that only one of the two parental alleles is present per locus and per individual. Deviations of Avr segregation from the classical 1:1 single gene model were frequently observed, indicating the involvement in avirulence of at least two or three genes (see Table 1 for a summary and Figures 1B–D). Thus, in addition to the single gene control of avirulence that is commonly observed in plant pathogenic fungi, there are more complex genetic situations in cereal powdery mildews. Furthermore, classical genetic studies have often reported the emergence of additional phenotypic classes in the segregating progeny, where a quantitative variation in virulence contrasting with the parental phenotypes was observed (Table 1; Brown and Jessop, 1995; Bourras et al., 2015; Parlange et al., 2015). For instance, in the cross between the avirulent B.g. tritici isolate 96224, and the virulent isolate JIW2, 33 progeny showed consistent intermediate phenotype on wheat lines containing the mildew resistance gene Pm3c (Table 1). Similar examples were reported in segregating progeny growing on the mildew resistance genes Pm3b, Pm3d, and Mla7 (Table 1). Thus, in cereal powdery mildews there is a quantitative component in race-specific Avr–R interactions that is reminiscent of a polygenic, quantitative trait.
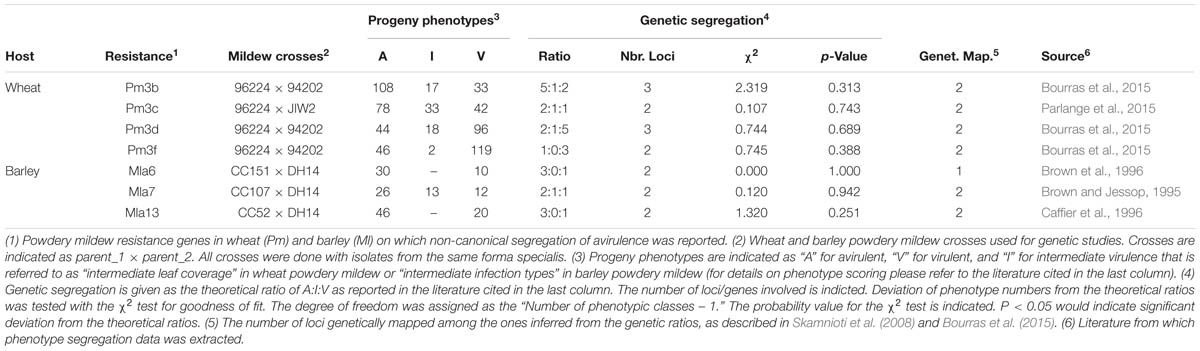
TABLE 1. Non-canonical segregation of race-specific avirulence in the F1 progeny of wheat and barley powdery mildew crosses.
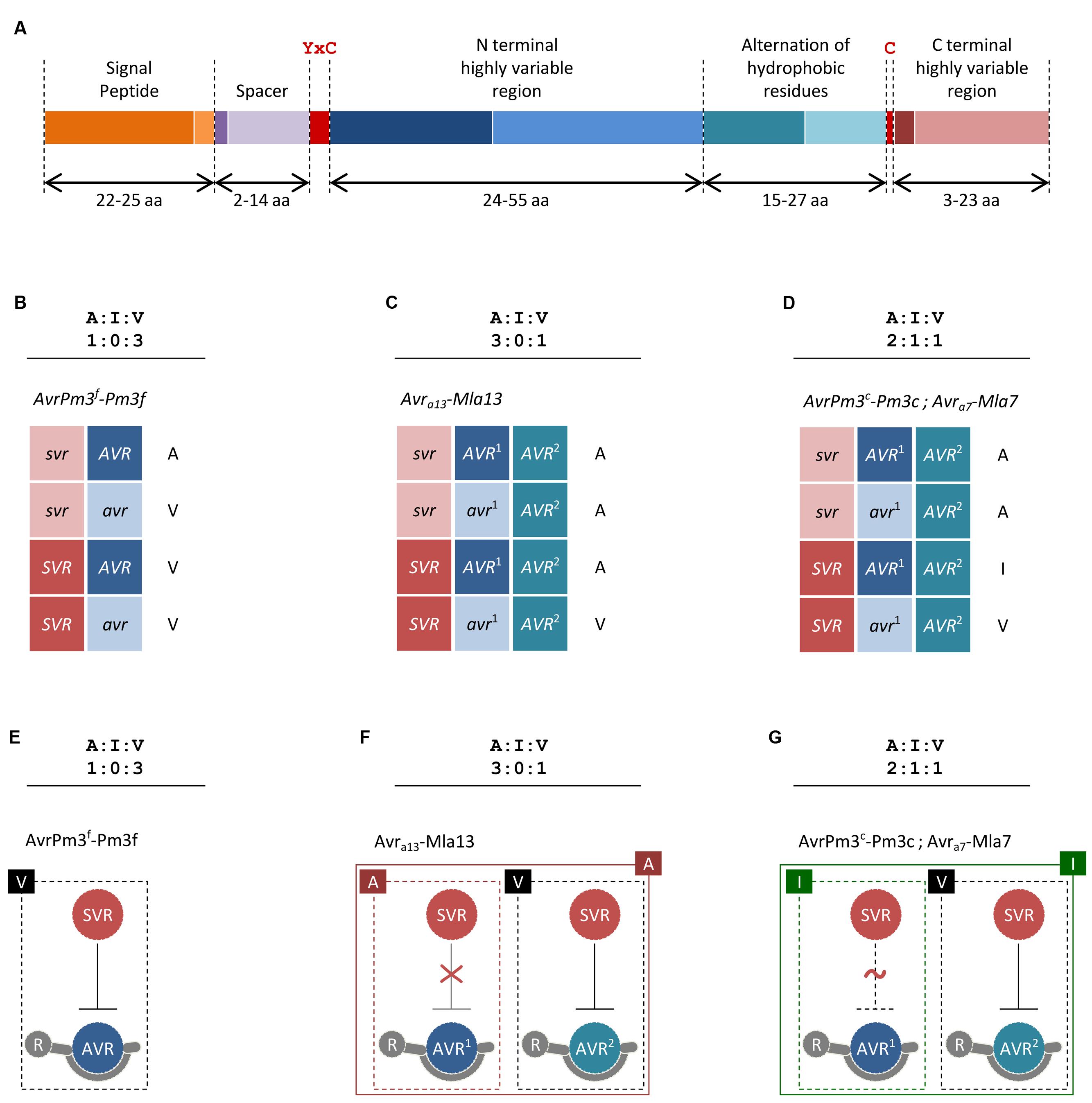
FIGURE 1. Proposed models for Avr–R–Svr interactions in cereal powdery mildews. (A) Structure of the AvrPm3a2/f2 effector family. Minimum and maximum sizes of each region as observed among family members are given in amino acids (aa) and differentiated with dark and light color shades, respectively. The YxC motif and the conserved cysteines are indicated in red. (B–D) Proposed Avr-R-Svr genetic models for the interpretation of genetic segregation ratios deviating from the canonical 1:1 single gene hypothesis. For readability, active and inactive alleles are distinguished with upper and lower case. Here, we have only considered examples of mildew Avr–R interactions where genetic segregation ratios and mapping data were consistent with at least two genetically independent loci being involved in avirulence. (B) Considering the Avr-R-Svr model and a genetic segregation ratio of 1:0:3, avirulence results from a combination of an inactive suppressor allele (svr) and an active avirulence allele (AVR). In the presence of the active SVR and the active AVR, the interaction result in virulence. A molecular model for this suppression scenario is depicted in (E). (C) Considering the Avr-R-Svr model, a genetic segregation ratio of 3:0:1 can be explained by a model involving one suppressor locus and two loci for avirulence (AVR1 and AVR2). Importantly, the second locus for avirulence (AVR2) is not polymorphic in this cross, thus only the active allele (AVR2) is present. In this model, the active SVR is only effective in suppressing AVR2 but not AVR1. A molecular model for this suppression scenario is depicted in (F). (D) Considering the Avr-R-Svr model, a genetic segregation ratio of 2:1:1 can be explained by a model involving one suppressor locus and two loci for avirulence (AVR1 and AVR2). Importantly, the second locus for avirulence (AVR2) is not polymorphic in this cross, thus only the active allele (AVR2) is present. In this model, the active SVR can fully suppress AVR2 but is only partially effective on AVR1, thus resulting in avirulence in the first case and intermediate virulence in the second. A molecular model for this suppression scenario is depicted in (G). (E–G) Proposed suppression scenarios based on the genetic models described in (B–D) and resulting in three major phenotypic classes of avirulence ‘A,’ virulence ‘V,’ and intermediate virulence ‘I.’ Only active suppressor and avirulence proteins are shown. For simplicity, AVR–R interactions are represented as physical binding. Absence of suppression is depicted as a red “X” sign. Partial suppression is depicted as a dotted line and a red “∼” sign. The phenotype resulting from single and combined interactions is indicated.
In addition to studies of inheritance, linkage between different Avr encoding loci was resolved for 15 Avr–R interactions in B.g. hordei and six in B.g. tritici (Skamnioti et al., 2008; Bourras et al., 2015). In both formae speciales, two genetically unlinked regions in the genome that commonly control several Avr specificities were reported (see Table 1 for some examples). In B.g. hordei there is the AVRa10 locus controlling the AVRa10, AVRk1, AVRa22, AVRa9, AVRa13-1, AVRg and possibly AVRa6 and AVRa7-2 specificities. There is also the AVRa12 locus comprised of AVRa6, AVRa12, AVRP17, and AVRLa. Similarly in B.g. tritici there is locus_1 controlling the AvrPm3a, AvrPm3b1, AvrPm3c1, AvrPm3d1, AvrPm3f1, and AvrPm3e specificities, and locus_3 for AvrPm3b2, AvrPm3c2, and AvrPm3d2. However, while in B.g. hordei the AVRa10 and the AVRa12 loci were described as clusters of linked but recombining Avr genes (Skamnioti et al., 2008), locus_1 in B.g. tritici was characterized as encoding for a single Avr factor commonly involved in the interaction with six alleles of the Pm3 resistance genes (Bourras et al., 2015; Parlange et al., 2015). Considering the AvrPm3a/f-Pm3a/f interaction involving locus_1 and locus_2 in B.g. tritici, it was found that specificity is only encoded by locus_2, while locus_1 encodes a factor acting unspecifically on several AvrPm3–Pm3 interactions. Assuming this genetic model stands true for B.g. hordei, it is possible that the AVRa10 and AVRa12 loci encode for general factors that are reminiscent of the B.g. tritici locus_1, while the actual avirulence proteins are encoded within specific loci outside of these clusters.
Genes Encoding CSEPs in Cereal Mildews: The Primary Candidates for Avirulence Genes
Genes encoding so-called candidate secreted effector proteins (CSEPs; Spanu et al., 2010) have been suggested as major determinants of the mildew–host interaction and as good candidates for avirulence proteins. Refined analyses of the CSEP complement in the Blumeria genomes indicated there are close to 500 of such genes in B.g. hordei (Pedersen et al., 2012) and over 600 in B.g. tritici (Fabrizio Menardo, personal communication), hence much more than initially predicted (Spanu et al., 2010; Wicker et al., 2013). Pedersen et al. (2012) showed that many CSEPs are highly expressed in haustoria, and thus expected to be released from the haustorial membrane into the extrahaustorial matrix. How effector proteins subsequently enter the plant cell is still unclear, although data from site directed mutagenesis suggest that specific conserved protein motifs such as the HRxxH motif in the CSEP BEC1019 may mediate translocation of the effector through the plant membrane (Whigham et al., 2015).
Beyond the common features described above, CSEPs have very limited sequence similarity and only a few common protein motifs are found (e.g., the YxC motif, Figure 1A), suggesting they may target different proteins in the host and fulfill different functions (Pedersen et al., 2012; Wicker et al., 2013). One group of predicted CSEP proteins share structural similarities to ribonucleases (Pedersen et al., 2012), which is why they are suspected to mimic and compete with plant proteins involved in pathogen defense. Furthermore, several CSEPs have homology to enzymes that could be involved in processes of plant–pathogen interaction such as cell-wall remodeling or protein degradation (Pliego et al., 2013). Indeed, a B.g. hordei CSEP of the latter type (BEC1019) was recently shown to suppress plant cell death (Whigham et al., 2015). In summary, our understanding of the actual ‘effects’ of CSEPs is very limited, but we can expect that data from comparative analyses and functional assays will soon shed more light on this large class of fungal proteins.
The Identification of Avra10 and Avrk1: A Novel Class of Avirulence Genes Encoded by Line Retrotransposons
The two avirulence genes Avra10 and Avrk1 were isolated by map-based cloning from B.g. hordei by Ridout et al. (2006). Both encode unusual avirulence proteins as they lack signal peptides. Avrk1 was functionally validated by bombardment assays that demonstrated induction of cell death in a Mlk-resistance gene dependent manner, as well as enhanced infection on susceptible varieties after transient expression. It was found that recognition of AVRa10 by MLA10 induced nuclear associations between the immune receptor and WRKY transcription factors (Shen et al., 2007). Furthermore, Nowara et al. (2010) described that silencing of Avra10 resulted in reduced fungal development in the absence but not in the presence of the cognate resistance gene Mla10.
In B.g. hordei Avra10 and Avrk1 were found to map only 0.7 cM from Avra22 (Skamnioti et al., 2008), and the evolutionary origin of these Avr genes has been subject of intense debate. Recent genome-wide analysis and comparisons with LINE elements from animals and plants made clear that Avra10 and Avrk1 are derived from non-LTR retrotransposons (also known as Long Interspersed Nuclear Elements or LINEs). These retro-elements usually consist of two open reading frames (ORFs), where ORF2 encodes a reverse transcriptase and RNaseH (RT/RH), while ORF1 is thought to encode a protein that mediates the transfer of retrovirus-like particles into the nucleus, at least in the human L1 element. Avra10 and Avrk1 are both derivatives of ORF1, but their homology at the DNA and protein level is very limited, thus indicating that both genes arose independently from distantly related LINE families (Amselem et al., 2015). Since LINEs are among the most abundant types of transposable elements in the Blumeria genomes (Parlange et al., 2011), it is not surprising that more than a thousand Avra10 and Avrk1 homologs were originally interpreted as a very large gene family that happened to co-evolve with the RT/RH domain of LINEs (Sacristán et al., 2009). The evolutionary origin of Avra10 and Avrk1 from LINEs has the intriguing implication that the large amount of repetitive DNA found in powdery mildew genomes serves as raw material from which new proteins and regulatory regions can emerge (Amselem et al., 2015).
The Identification of SvrPm3a1/f1 and AvrPm3a2/f2 in B.g. tritici: Two CSEPs Involved in Pm3-Mediated Avirulence
The identification of the first Avr gene in wheat powdery mildews was enabled by a combination of new genomic tools, next generation sequencing and high-throughput genotyping technologies (Bourras et al., 2015). Analysis of the two loci controlling the AvrPm3a/f–Pm3a/f interaction resulted in the cloning of AvrPm3a2/f2, a typical CSEP encoded within locus_2 (Figure 1A). The avirulent allele is specifically recognized by Pm3a and Pm3f, whereas the virulent allele differing by two amino acid polymorphisms escapes recognition. Similarly, SvrPm3a1/f1, also a typical CSEP gene encoded within the general locus_1 was cloned (Parlange et al., 2015). Most likely, this gene acts as a suppressor (Svr) of the AvrPm3–Pm3 mediated avirulence (Bourras et al., 2015). Thus, in the AvrPm3–Pm3 interaction model, a second layer of regulation is provided by a suppression mechanism. Here, in the presence of an active SVR suppressor, primary recognition of the avirulence effector by the cognate resistance protein is not sufficient to induce a resistance response. In this model, it is possible for the pathogen to maintain an unaltered and active AVR effector while still escaping R gene recognition which represents an evolutionary advantage on the long term.
It was shown that both Avr and Svr genes have the same kinetics of expression with a peak at 2 days after mildew inoculation (Bourras et al., 2015). This time point coincides with the formation of the haustorium, a fungal feeding structure involved in effector delivery, and a milestone for successful infection. It was also shown that Avr and Svr regulation is inherited in a parent-of-origin-specific manner in progeny from a cross between the Pm3a/f avirulent isolate 96224 and the Pm3a/f virulent isolate 94202 (Bourras et al., 2015). Considering these parental isolates and the critical time point of 2 days, it was found that the Avr gene is upregulated in the avirulent parent while the Svr suppressor is downregulated, and the exact opposite situation was found in the virulent parent. This situation can be explained by mechanisms such as epiallelic variation, mutations in cis-elements, alteration of trans-acting factors, or epigenetic modulation, all of which can affect effector protein accumulation without altering protein sequence (Bakkeren and Valent, 2014; Gijzen et al., 2014). Thus, in addition to sequence polymorphism distinguishing Avr and avr alleles, and the presence of an active (Svr) or inactive (svr) suppressor, there is a third layer of regulation of the AvrPm3–Pm3 interaction at the gene expression level.
Based on these results, an extension of Flor’s gene-for-gene model that accounts for a suppressor locus was proposed as the Avr-R-Svr genetic model (Figures 1B–D). Here, resistance is mediated by an interaction involving an allele-specific avirulence effector (Avr), a resistance gene allele (R), and an allele-unspecific pathogen-encoded suppressor of avirulence (Svr). At the molecular level, recognition and suppression of recognition are determined by sequence polymorphism as well as gene expression levels differentiating Avr vs. avr and Svr vs. svr alleles. Thus, resistance can only occur if the suppressor is inactive and the AVR protein is produced in sufficient amounts for R gene activation.
How is Resistance Specificity to Allelic Series of Resistance Genes Controlled by Powdery Mildew?
The recent studies on the Pm3a and Pm3f resistance genes (Stirnweis et al., 2014) on the host side, as well as the identification of AvrPm3a2/f2 on the pathogen side (Bourras et al., 2015) have given a first insight into the control of molecular specificity. Considering these two Pm3 alleles, Pm3a is a stronger form of Pm3f as Pm3a recognizes all isolates which are recognized by Pm3f, plus some additional ones (Brunner et al., 2010). It was found that activation efficiency through the ARC2 domain controls the difference in the recognition spectrum of these two alleles, and there is no difference in recognition specificity (Stirnweis et al., 2014). Furthermore, there is strong evidence that specificity in the allelic interactions is based on the recognition of different AvrPm3 or AvrMla genes. AvrPm3a2/f2 is not recognized by any of the other Pm3 alleles and genetic mapping (Bourras et al., 2015) and mutant analysis (Parlange et al., 2015) have revealed at least three additional genetic loci controlling recognition specificity toward Pm3 alleles in B.g. tritici. In B.g. hordei also, the complex genetic ratios of avirulence inheritance indicate the involvement of many genes (Table 1). Thus, the corresponding avirulence genes seem to be quite different from each other, a surprising finding given that some allelic PM3 protein variants differ only by two amino acids in the LRR domain. Finally, resistance specificity also involves the pathogen encoded SvrPm3a1/f1 suppressor of the Pm3 resistance (Figures 1B–G) (Bourras et al., 2015). In B.g. hordei, the molecular basis of recognition specificity of the Avra10 and Avrk1 alleles is not yet understood. As discussed below, there are several ways to further explore the function of these genes. In addition, it will be essential to characterize at the protein level the determinants of recognition, and also to isolate more AvrMla genes for the many members of the Mla allelic series (Seeholzer et al., 2010) to get further insight into this interaction.
Clearly, genetic studies in mildew are greatly simplified by haploid inheritance, but haploid genetics has the disadvantage that dominant and recessive traits cannot easily be distinguished. Thus, based on genetics only it is not possible to predict whether an “avirulence gene” actually encodes for an AVR that is recognized by the cognate resistance protein, or an inactive suppressor that is ineffective in inhibiting AVR-R recognition and/or resistance signaling. In addition to this first layer of complexity, gene expression possibly plays an important regulatory role in the determination of specificity (Bourras et al., 2015). Therefore, differences in gene expression levels between different B.g. tritici or B.g. hordei isolates might actually give important hints on functional avirulence and suppressor genes. In this context, we propose that comparative RNAseq studies in many isolates might be useful to detect such diagnostic expression differences.
Next Steps in the Characterization of Cereal Powdery Mildew Avirulence
Despite the progress in the characterization of mildew avirulence genetics and genomics described above, there are large remaining gaps in our knowledge. Considering the identified components of avirulence in mildew, the specific interactions leading to AvrPm3a2/f2 recognition/suppression need to be studied at the molecular and biochemical level. In addition, the role of the LINE encoded B.g. hordei avirulence genes Avra10 and Avrk1 needs to be clarified. It is also possible that these two genes are not recognized by the resistance genes (in a situation similar to SvrPm3a1/f1 in the case of B.g. tritici) but rather play a different, molecularly unknown role as activators or suppressors of avirulence. In this context, it will be important to isolate more AvrPm3 and AvrMla genes to understand the molecular basis of specificity in cereal mildews.
So far, all known Avr loci in Blumeria were isolated by map-based cloning approaches which are solid but very time-consuming. The B.g. tritici and B.g. hordei reference genomes should be improved by new sequencing technologies such as PacBio (Eid et al., 2009) which would greatly support map-based cloning and pave the way for genome-wide association studies (GWAS), an attractive alternative for Avr identification. The decreased costs of next-generation sequencing allow us to sequence a large number of isolates which is the basis for GWAS. However, it remains to be seen if GWAS approaches will be capable of identifying mildew Avrs, where avirulence seems to result from complex genetic networks and epistatic interactions.
A largely unexplored area of research is the identification of specific mutants in avirulence genes. Such mutants should theoretically be easy to identify by mutagenizing avirulent races and selecting for growth on a barley or wheat genotype containing the cognate resistance gene. Two such mutants at new and still uncharacterized genetic loci have been identified for loss-of-recognition by Pm3a and Pm3f, with no impact on the other Pm3 alleles (Parlange et al., 2015). Based on whole genome sequencing of pools from segregants of mapping populations, and considering the haploid nature of the mildew genome, it should be straightforward to identify the causative mutations and we suggest that larger screens for many different Avr mutants should be performed. Given that there are relatively few gene families in mildew (with the exception of rapidly diverging CSEP families, Wicker et al., 2013) genetic redundancy should be a minor problem for mutant identification. AvrPm3a2/f2 belongs to a family of 36 members identified in silico. Twenty-three of these members, including the most homologous paralogs by protein sequence, PU_24 (61%) and PU_23 (59%), were functionally tested, but none of them was recognized by Pm3a/f (Bourras et al., 2015; our unpublished data).
Finally, there is a need for additional systems to study gene function. Particle bombardment has been a successful system to study Avr gene function, either directly or by host-induced gene silencing (Ridout et al., 2006; Nowara et al., 2010; Bourras et al., 2015), but it is time-consuming and needs practice and optimization in individual labs. In the case of the AvrPm3a2/f2 avirulence gene, transient expression studies after infiltration of the heterologous species N. benthamiana have been successful (Bourras et al., 2015). However, it remains to be seen how broadly this system is suitable for mildew avirulence research. Therefore, alternative delivery approaches for functional studies, e.g., by bacterial vectors (Upadhyaya et al., 2014) should be developed for mildew. In addition, further work should be invested into the development of a highly reproducible mildew transformation system. This would then allow us to fully exploit the current rapid development in mildew avirulence biology and fully understand the interaction of this fascinating obligate biotroph and the co-evolution with its host.
In a more applied perspective, knowledge on the molecular interactions between R and Avr genes could be highly productive to develop pathogen-informed strategies for resistance improvement in the host. On the pathogen side, information on the allelic diversity of Avrs and Svrs in mildew populations can guide the spatio-temporal deployment of R genes in the field. On the molecular level, the identification of novel sources of resistance should be facilitated by the characterization of the genes and gene networks involved in resistance activation by AVR proteins vs. resistance suppression by the pathogen encoded suppressor. In particular, synthetic modification of wheat proteins co-opted by mildew suppressors might provide a strategy to achieve durable resistance.
Author Contributions
All authors reviewed literature and contributed to writing the manuscript. SB and BK coordinated the manuscript.
Funding
This work was supported by a grant from the Swiss National Science Foundation: 310030_163260.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We acknowledge the technical support of Gerhard Herren, Helen Zbinden, Gabriele Buchmann, Linda Lüthi, and Simon Flückiger.
References
Amselem, J., Vigouroux, M., Oberhaensli, S., Brown, J. K., Bindschedler, L. V., Skamnioti, P., et al. (2015). Evolution of the EKA family of powdery mildew avirulence-effector genes from the ORF 1 of a LINE retrotransposon. BMC Genomics 16:917. doi: 10.1186/s12864-015-2185-x
Bakkeren, G., and Valent, B. (2014). Do pathogen effectors play peek-a-boo? Front. Plant Sci. 5:731. doi: 10.3389/fpls.2014.00731
Barrett, L. G., Thrall, P. H., Dodds, P. N., van der Merwe, M., Linde, C. C., Lawrence, G. J., et al. (2009). Diversity and evolution of effector loci in natural populations of the plant pathogen Melampsora lini. Mol. Biol. Evol. 26, 2499–2513. doi: 10.1093/molbev/msp166
Boehnert, H. U., Fudal, I., and Dioh, W., Tharreau, D., Notteghem, J.-L., and Lebrun, M.-H. (2004). A putative polyketide synthase peptide synthetase from Magnaporthe grisea signals pathogen attack to resistant rice. Plant Cell 16, 2499–2513. doi: 10.1105/tpc.104.022715
Bourras, S., McNally, K. E., Ben-David, R., Parlange, F., Roffler, S., Praz, C. R., et al. (2015). Multiple avirulence loci and allele-specific effector recognition control the pm3 race-specific resistance of wheat to powdery mildew. Plant Cell 27, 2991–3012.
Brown, J. K. M., and Jessop, A. C. (1995). Genetics of avirulences in Erysiphe graminis f. sp. hordei. Plant Pathol. 44, 1039–1049. doi: 10.1111/j.1365-3059.1995.tb02663.x
Brown, J. K. M., Le Boulaire, S., and Evans, N. (1996). Genetics of responses to morpholine-type fungicides and of avirulences in Erysiphe graminis f. sp. hordei. Eur. J. Plant Pathol. 102, 479–490. doi: 10.1007/BF01877142
Brunner, S., Hurni, S., Streckeisen, P., Mayr, G., Albrecht, M., Yahiaoui, N., et al. (2010). Intragenic allele pyramiding combines different specificities of wheat Pm3 resistance alleles. Plant J. 64, 433–445. doi: 10.1111/j.1365-313X.2010.04342.x
Caffier, V., de Vallavieille-Pope, C., and Brown, J. K. M. (1996). Segregation of avirulences and genetic basis of infection types in Erysiphe graminis f. sp. hordei. Phytopathology 86, 1112–1121. doi: 10.1094/Phyto-86-1112
Eid, J., Fehr, A., Gray, J., Luong, K., Lyle, J., Otto, G., et al. (2009). Real-time DNA sequencing from single polymerase molecules. Science 323, 133–138. doi: 10.1126/science.1162986
Gijzen, M., Ishmael, C., and Shrestha, S. D. (2014). Epigenetic control of effectors in plant pathogens. Front. Plant Sci. 5, 1–4. doi: 10.3389/fpls.2014.00638
Hayward, A., McLanders, J., Campbell, E., Edwards, D., and Batley, J. (2012). Genomic advances will herald new insights into the Brassica: Leptosphaeria maculans pathosystem. Plant Biol. 14, 1–10. doi: 10.1111/j.1438-8677.2011.00481.x
Houterman, P. M., Cornelissen, B. J. C., and Rep, M. (2008). Suppression of plant resistance gene-based immunity by a fungal effector. PLoS Pathog. 4:e1000061. doi: 10.1371/journal.ppat.1000061
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Parlange, F., Oberhaensli, S., Breen, J., Platzer, M., Taudien, S., Simková, H., et al. (2011). A major invasion of transposable elements accounts for the large size of the Blumeria graminis f.sp. tritici genome. Funct. Integr. Genomics. 11, 671–677. doi: 10.1007/s10142-011-0240-5
Parlange, F., Roffler, S., Menardo, F., Ben-David, R., Bourras, S., McNally, K. E., et al. (2015). Genetic and molecular characterization of a locus involved in avirulence of Blumeria graminis f. sp. tritici on wheat Pm3 resistance alleles. Fungal Genet. Biol. 82, 181–192. doi: 10.1016/j.fgb.2015.06.009
Pedersen, C., Ver Loren van Themaat, E., McGuffin, L. J., Abbott, J. C., Burgis, T. A., Barton, G., et al. (2012). Structure and evolution of barley powdery mildew effector candidates. BMC Genomics 13:694. doi: 10.1371/journal.ppat.1000061
Pliego, C., Nowara, D., Bonciani, G., Gheorghe, D. M., Xu, R., Surana, P., et al. (2013). Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. Mol. Plant Microbe Interact. 26, 633–642. doi: 10.1094/MPMI-01-13-0005-R
Ravensdale, M., Bernoux, M., Ve, T., Kobe, B., Thrall, P. H., Ellis, J. G., et al. (2012). Intramolecular interaction influences binding of the flax L5 and L6 resistance proteins to their AvrL567 ligands. PLoS Pathog. 8:e1003004. doi: 10.1371/journal.ppat.1003004
Ridout, C. J, Skamnioti, P., Porritt, O., Sacristan, S., Jones, J. D. G., and Brown, J. M. K. (2006). Multiple avirulence paralogous in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell 18, 2402–2414. doi: 10.1105/tpc.106.043307
Rouxel, T., and Balesdent, M.-H. (2010). “Avirulence Genes,” in Encyclopedia of Life Sciences (ELS), (Chichester: John Wiley & Sons, Ltd). doi: 10.1002/9780470015902.a0021267
Sacristán, S., Vigouroux, M., Pedersen, C., Skamnioti, P., Thordal-Christensen, H., Micali, C., et al. (2009). Coevolution between a family of parasite virulence effectors and a class of LINE-1 retrotransposons. PLoS ONE 4:e7463. doi: 10.1371/journal.pone.0007463
Seeholzer, S., Tsuchimatsu, T., Jordan, T., Bieri, S., Pajonk, S., Yang, W., et al. (2010). Diversity at the Mla powdery mildew resistance locus from cultivated barley reveals sites of positive selection. Mol. Plant. Microbe. Interact. 23, 497–509. doi: 10.1094/MPMI-23-4-0497
Shen, Q.-H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., et al. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315, 1098–1103. doi: 10.1126/science.1136372
Skamnioti, P., Pedersen, C., Al-Chaarani, G. R., Holefors, A., Thordal-Christensen, H., Brown, J. K., et al. (2008). Genetics of avirulence genes in Blumeria graminis f.sp. hordei and physical mapping of AVR(a22) and AVR(a12). Fungal Genet. Biol. 45, 243–252. doi: 10.1016/j.fgb.2007.09.011
Spanu, P. D., Abbott, J. C., Amselem, J., Burgis, T. A., Soanes, D. M., Stüber, K., et al. (2010). Genome expansion and gene loss in powdery mildew fungi reveal tradeoffs in extreme parasitism. Science 330, 1543–1546. doi: 10.1126/science.1194573
Stirnweis, D., Milani, S. D., Jordan, T., Keller, B., and Brunner, S. (2014). Substitutions of two amino acids in the nucleotide-binding site domain of a resistance protein enhance the hypersensitive response and enlarge the PM3F resistance spectrum in wheat. Mol. Plant. Microbe. Interact. 27, 265–276. doi: 10.1094/MPMI-10-13-0297-FI
Tosa, Y. (1994). Gene-for-gene interactions between the rye mildew fungus and wheat cultivars. Genome. 37, 758–762. doi: 10.1139/g94-108
Upadhyaya, N. M., Mago, R., Staskawicz, B. J., Ayliffe, M. A., Ellis, J. G., and Dodds, P. N. (2014). A bacterial type III secretion assay for delivery of fungal effector proteins into wheat. Mol. Plant Microbe Interact. 27, 255–264. doi: 10.1094/MPMI-07-13-0187-FI
Whigham, E., Qi, S., Mistry, D., Surana, P., Xu, R., Fuerst, G., et al. (2015). Broadly conserved fungal effector bec1019 suppresses host cell death and enhances pathogen virulence in powdery mildew of barley (Hordeum vulgare L.). Mol. Plant Microbe Interact. 28, 968–983. doi: 10.1094/MPMI-02-15-0027-FI
Wicker, T., Oberhaensli, S., Parlange, F., Buchmann, J. P., Shatalina, M., Roffler, S., et al. (2013). The wheat powdery mildew genome shows the unique evolution of an obligate biotroph. Nat. Genet. 45, 1092–1096. doi: 10.1038/ng.2704
Wulff, B. B. H., Chakrabarti, A., and Jones, D. A. (2009). Recognitional specificity and evolution in the tomato-Cladosporium fulvum pathosystem. Mol. Plant. Microbe. Interact. 22, 1191–1202. doi: 10.1094/MPMI-22-10-1191
Keywords: wheat, powdery mildew, resistance gene, avirulence gene, barley
Citation: Bourras S, McNally KE, Müller MC, Wicker T and Keller B (2016) Avirulence Genes in Cereal Powdery Mildews: The Gene-for-Gene Hypothesis 2.0. Front. Plant Sci. 7:241. doi: 10.3389/fpls.2016.00241
Received: 29 December 2015; Accepted: 12 February 2016;
Published: 01 March 2016.
Edited by:
Pietro Daniele Spanu, Imperial College London, UKReviewed by:
Guus Bakkeren, Agriculture and Agri-Food Canada, CanadaGuido Van Den Ackerveken, Utrecht University, Netherlands
Copyright © 2016 Bourras, McNally, Müller, Wicker and Keller. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Beat Keller, YmtlbGxlckBib3RpbnN0LnV6aC5jaA==
 Salim Bourras
Salim Bourras Kaitlin E. McNally
Kaitlin E. McNally Marion C. Müller
Marion C. Müller Thomas Wicker
Thomas Wicker Beat Keller
Beat Keller