- 1Botany Department, Faculty of Science, Tanta University, Tanta, Egypt
- 2Commonwealth Scientific and Industrial Research Organization Agriculture, Queensland Bioscience Precinct, St Lucia, QLD, Australia
- 3Queensland Alliance for Agriculture & Food Innovation, The University of Queensland, Brisbane, QLD, Australia
- 4Plant-Microbe Interactions Laboratory, School of Agriculture and Food Sciences, The University of Queensland, Brisbane, QLD, Australia
Current technologies have changed biology into a data-intensive field and significantly increased our understanding of signal transduction pathways in plants. However, global defense signaling networks in plants have not been established yet. Considering the apparent intricate nature of signaling mechanisms in plants (due to their sessile nature), studying the points at which different signaling pathways converge, rather than the branches, represents a good start to unravel global plant signaling networks. In this regard, growing evidence shows that the generation of reactive oxygen species (ROS) is one of the most common plant responses to different stresses, representing a point at which various signaling pathways come together. In this review, the complex nature of plant stress signaling networks will be discussed. An emphasis on different signaling players with a specific attention to ROS as the primary source of the signaling battery in plants will be presented. The interactions between ROS and other signaling components, e.g., calcium, redox homeostasis, membranes, G-proteins, MAPKs, plant hormones, and transcription factors will be assessed. A better understanding of the vital roles ROS are playing in plant signaling would help innovate new strategies to improve plant productivity under the circumstances of the increasing severity of environmental conditions and the high demand of food and energy worldwide.
Introduction
Plants are increasingly subjected to a variety of environmental stresses which diminish the productivity of various economically important crops. Every year, the world loses a huge amount of crop production through scarcity of water, extreme temperatures, high soil salinity, herbivore attack, and pathogen infection.
The sessile nature of plants has resulted in the evolution of complicated protection mechanisms to survive different environmental challenges. One of the stress tolerance mechanisms is the ability to sense complex stress factors and respond appropriately. Activation of complex signaling pathways helps plants to achieve this. To better understand plant signaling pathways would enable us to modify plants to improve their adaptability. However, this requires reducing the complexity associated with signaling pathways. Focusing on the points at which different signaling pathways converge, rather than studying the branches of these pathways, would be helpful as a starting point. The rapid generation of reactive oxygen species (ROS) represents a common plant response to different biotic and abiotic stresses (Lamb and Dixon, 1997; Orozco-Cardenas and Ryan, 1999; Kovtun et al., 2000; Kotchoni and Gachomo, 2006; Mittler et al., 2011; Petrov and Van Breusegem, 2012; Noctor et al., 2014; Xia et al., 2015) and thus a basis to unify signaling events.
Recent genomic technologies, especially global gene expression tools, have not only produced new details about plant signaling pathways but also raised many historical questions including the followings: Is there a specific linear signaling pathway for each stress? If so, what about the observed cross-talk? Is there a big common signaling network from which many branches arise for specificity? If so, what about the different receptors? What do represent the points at which different branches of signaling pathways converge? If ROS are at the points of integrating signaling outputs from different signaling pathways, then what are the ROS receptors? What are the upstream and downstream signaling components of ROS? How do ROS set signaling specificity? What about the photosynthetic machinery that generates ROS; does it and its ROS and redox system represent a primary source of the plant signaling battery? As discussed throughout this review, most of these questions have been answered (or are being answered) while we will be in a better shape in providing more definite answers to the remaining ones in the near future.
This review presents a discussion about these historical questions by considering the so complex nature of plant stress signaling networks. A special attention will be given on reviewing signaling players and events such as receptors/sensors, secondary messengers, specificity, cross-talk, redundancy, feedback regulations, alternative promoter usage, alternative splicing, nucleo-cytoplasmic trafficking, and epigenetics. Here, an attempt will also be made to indicate the kinds of studies required to fill in the gaps. A specific thought to photosynthetic activities and ROS as the primary source of the signaling battery in plants will be presented. As ROS production represents a common plant response to almost all environmental challenges, a special emphasis will be devoted here to ROS production, scavenging, damaging effects, signaling roles and how they work upstream and downstream of other signaling components, e.g., calcium, redox homeostasis, membranes, G-proteins, MAPKs, plant hormones [such as salicylic acid (SA), jasmonic acid (JA), abscisic acid (ABA), and ethylene] and transcription factors (TFs). We hope to present a holistic summary of various signaling components and concepts that are important for a plant biologist to take into consideration when analyzing signaling events involved in plant response to the ever changing environment. This understanding would help construct comprehensive signaling networks which in turn innovate new strategies to improve plant productivity under the increasing severity of environmental stress conditions and the high global demand for food and energy.
Plant Signaling Networks
Signaling Networks are Complex
Recently, our knowledge about signaling mechanisms in plants starting from stimulus sensing to final response has increased. It is obvious that there is a large number of components underlying signaling mechanisms, including a high degree of interconnectivity, many spatio-temporal levels, and a complicated tune of signal transduction pathways. For example, the changes at the expression of certain genes under a definite environmental condition are not necessarily translated into metabolic and structural changes where the interactions between various aspects, including post-transcriptional and post-translational modifications, compartmentalization, metabolite stability, substrate availability may lead to an unexpected response (Krasensky and Jonak, 2012).
Moreover, it is becoming increasingly clear that signaling networks are not linear; rather they are part of a complicated and dynamic network with substantial overlap among their branches (Knight and Knight, 2001). Accordingly, rather than one sensor, there are many sensors that perceive certain stress conditions and control all downstream signals (Figure 1). Each sensor controls a branch of the signaling pathway activated by one aspect of the stress condition. For instance, temperature stress is well-known to change the physical state (fluidity) of membranes (Murata and Los, 1997; Königshofer et al., 2008), but this may not be the only condition that elicits signaling events under this stress. Changes of the conformation/activity of some intracellular proteins may also be involved in signaling to cold stress. Therefore, it is likely that the initial stress signal is perceived by multiple primary sensors, and then a cascade of signaling events is initiated by secondary signals such as plant hormones and calcium, which differ from the primary signal in time (coming late) and space (different compartments). Also, these secondary signals may differ in specificity from primary stimuli, may be shared by various stress pathways, and may underlie the interaction among signaling pathways for different insults and stress cross-protection (Figure 1). Consequently, multiple signaling mechanisms may be activated by one stimulus/stress initiating pathways differ in time, space, and outputs. Using shared signaling intermediates, such as phytohormones, these pathways may interconnect or interact with one another producing an intertwined signaling network (Xiong et al., 2002).
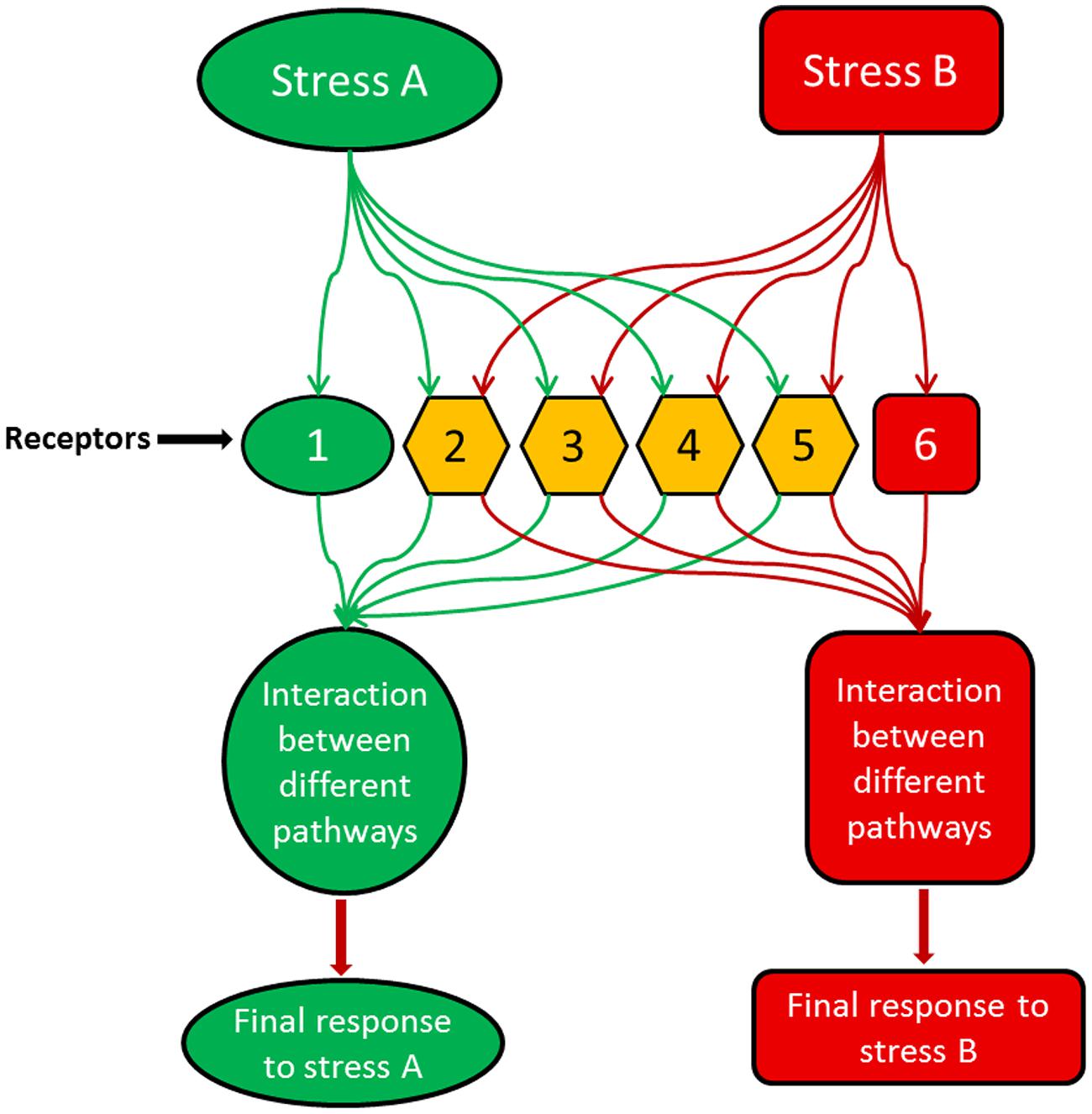
FIGURE 1. A model illustrating how different stresses or stimuli could activate overlapping receptors/sensors but produce distinct final outputs which are specific to each stimulus. In the model, stress A activates different receptors, e.g., 1, 2, 3, 4 and 5, while stress B is perceived by receptors 2, 3, 4, 5, and 6. Receptor 1 is activated only by stress A, while receptor 6 is activated only by stress B. The other receptors are shared between both stimuli representing the cross-talk between stress A and B. With stress A, the interaction between the downstream signaling events led by the receptor combination of 1, 2, 3, 4, and 5 produce a final output which can be completely different from the outcome of the receptor combination of 2, 3, 4, 5, and 6 with stress B.
The changes in gene expression do not represent end points for signaling pathways. There are many other aspects, including transcriptional, post-transcriptional and post-translational regulation, redundancy, alternative promoter usage, alternative splicing, protein trafficking, non-coding RNA, and epigenetic effects (Figure 2) that regulate signaling pathways. In the following sections, these latter aspects will be briefly discussed.
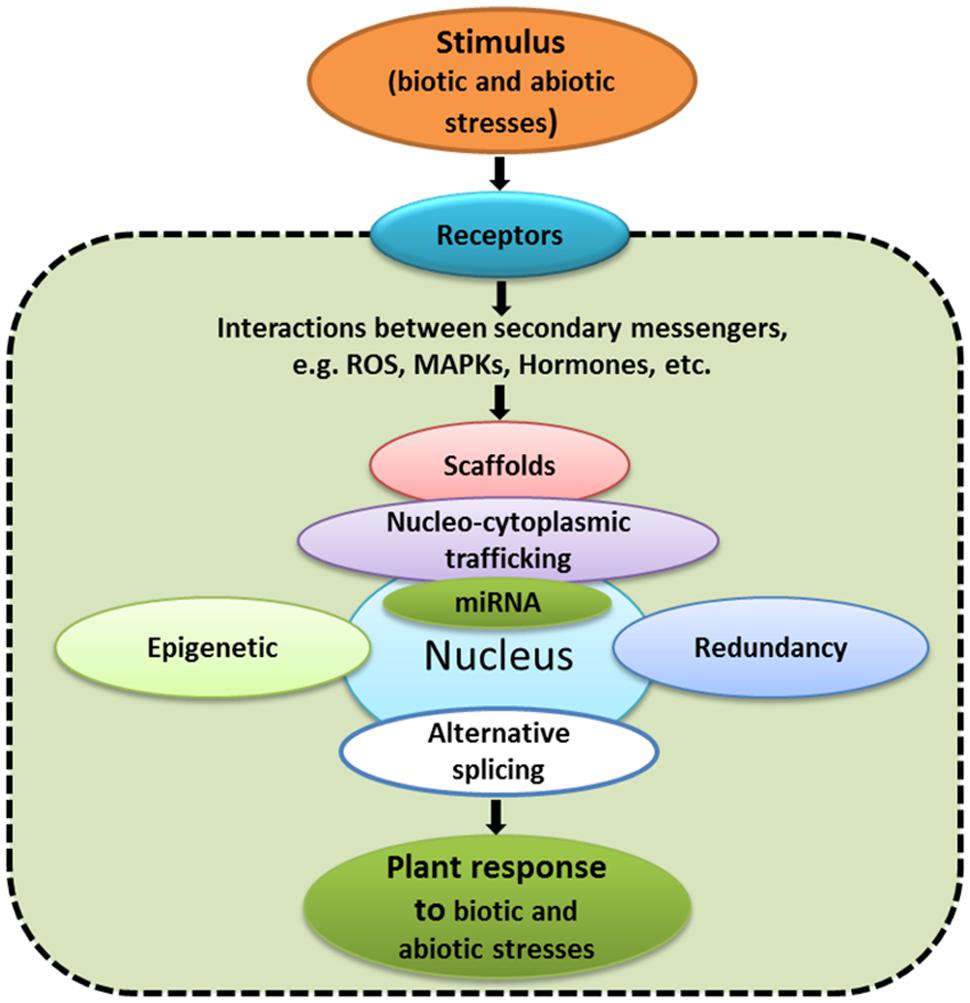
FIGURE 2. Examples of aspects that have to be taken into consideration while studying signaling networks.
Redundancy and Signaling
During stress, plant signaling networks have a high ability to compensate the effects of disturbances in neighboring nodes and related signaling pathways (Chiwocha et al., 2005). The simple explanation for functional redundancy may be brought about by duplicate genes that eukaryotic genomes contain (Stelling et al., 2004; De Smet and Van de Peer, 2012). It was reported that about one-quarter of functional redundancy in Saccharomyces cerevisiae can be explained by compensation by duplicate genes (Gu et al., 2003). However, while sequence redundancy explains some functional redundancy, the ability of networks to compensate the effects of perturbations in neighboring nodes and related pathways could be the main cause (Chiwocha et al., 2005). Of course, this redundancy represents a great advantage to the organism to cope with the ever changing environment. However, this represents a big problem to scientists who are studying signaling transduction pathways, especially when using knockout mutants. They usually prefer an altered phenotype after modulation of single genes (Chiwocha et al., 2005). Therefore, it is imperative, while studying a signaling pathway, to take into consideration the fact that many signaling components can be functionally redundant under a given experimental condition or current phenotypic methods are not sensitive enough to detect the consequences of knocking out single genes. Therefore, gene knockouts should be examined under a variety of experimental conditions, using genome-wide gene expression profiling and other omic technologies wherever possible.
Alternative Promoter Usage, Alternative Splicing, and Signaling
Many eukaryotic genes have multiple promoter elements. Each one is subjected to different regulatory factors under different situations. Alternative promoter usage is greatly linked to alternative splicing of internal exons and often has physiological implications (Kornblihtt, 2005). Alternative splicing produces multiple transcripts from the same gene and potentially different proteins. In turn, it represents a key post-transcriptional regulatory mechanism for expanding proteomic diversity and functional complexity in higher eukaryotes (Reddy, 2007; Carvalho et al., 2012). At the level of post-transcriptional mRNA processing, alternative splicing represents the primary mechanism to control the number of intracellular components (Bardo et al., 2002). There is substantial evidence that cellular signaling networks control the number and types of network components using alternative splicing. In the human genome, it was reported that 40–60% of the genes are subjected to alternative-splicing, with estimates of an average of 8 exons per gene (Thanaraj, 2004). In plants, alternative splicing has received less attention because this phenomenon was rare to be considered (Reddy, 2007). Others reported that, in plants, alternative splicing is ubiquitous and can mediate a bounty of transcriptome and proteome complexity (Kazan, 2003; Wang and Brendel, 2006). In the model plant Arabidopsis, 4,707 genes showed 8,264 alternative splicing events (Wang and Brendel, 2006). It was reported that alternative splicing of introns is involved in the regulation of the kinase activity of the MIK GCK-like MAP4K. Four different mature mRNAs of MIK were found to be accumulated with different expression profiles during maize development (Castells et al., 2006). Under stress conditions alternative splicing of pre-mRNAs dramatically increases (Reddy, 2007). Virus infection led to multiple novel intron-retaining splice variants in Brachypodium distachyon (Mandadi et al., 2015). So, while studying signaling networks, it is important to consider not only the signaling events leading to transcriptional changes, but also protein modifications (such as phosphorylation and glycosylation).
Protein Trafficking and Signaling
The nuclear envelope separates the nuclear compartment containing the genes from the cytoplasm where mRNA translation and protein synthesis occurs. Therefore, all nuclear proteins, including TFs, must be imported to the nucleus. This nucleo-cytoplasmic trafficking is under complex control. The Arabidopsis genome, for example, contains at least 17 genes encoding importin B-like nuclear transport receptors (Bollman et al., 2003). TFs and kinases are the main regulatory components in almost all signaling pathways. Hence, it is important, while studying signal transduction pathways, to consider not only the signaling events modulating the expression of regulatory genes and proteins and their downstream interactors, but also to think about how the access of these TFs to the target genes is regulated. In general, control of transcription on both the level of TF activity and the level of nucleo-cytoplasmic partitioning are combined to create a redundant network of regulatory switches to orchestrate different signaling mechanisms (Merkle, 2004; Parry, 2015). During pathogen infection, recent reports have suggested the involvement of the nucleo-cyctoplasmic trafficking of plant R proteins to achieve effector-triggered immunity and mediate disease resistance (Shen et al., 2007; Liu and Coaker, 2008). For more intensive discussion on nucleo-cytoplasmic trafficking and signaling, see Merkle (2004) and Parry (2015).
MicroRNA and Signaling
At the post-transcriptional level, microRNAs (miRNAs) are a class of small non-coding RNAs that are increasingly being recognized as key modulators of gene expression (Covarrubias and Reyes, 2010; van Rooij, 2011; Ding et al., 2013, 2015). miRNAs regulate the expression of relevant genes by binding to reverse complementary sequences, resulting in cleavage or translational inhibition of the target RNAs (Khraiwesh et al., 2012). miRNAs are reported to play important roles in biotic and abiotic stress responses in plants. Through repressing the expression of the respective target genes encoding regulatory and functional proteins, various miRNAs were reported to play crucial roles in drought stress responses, including ABA response, osmoprotection, and antioxidant defense (Ding et al., 2013). It was reported that H2O2 stress led to differential expression of seven miRNA families. The targets of these H2O2-responsive miRNAs were found to be involved in different cellular responses and metabolic processes including transcriptional regulation, nutrient transport, and programmed cell death (PCD; Li et al., 2011). The downregulation of miR398 was found to mediate post-transcriptional induction of two Cu/Zn superoxide dismutase (SOD) genes and be important for oxidative stress tolerance in Arabidopsis (Sunkar et al., 2006). During biotic stress, miRNAs were found to contribute to antibacterial resistance of Arabidopsis against Pseudomonas syringae via repressing auxin signaling (Navarro et al., 2006).
Epigenetic Effects and Signaling
Epigenetics (the study of heritable changes in gene expression that are not due to changes in DNA sequence; Bird, 2007) has become one of the hottest subjects of research in plant functional genomics since it plays an important role in developmental gene regulation, response to environmental stresses, and in natural variation of gene expression levels (Chinnusamy and Zhu, 2009; Sahu et al., 2013; Springer, 2013). Epigenetic effects are ascribed to a variety of molecular mechanisms including stable changes in protein structure, expression of small RNAs, and chromatin modifications. Chromatin modifications include DNA methylation, histone variants, remodeling of chromatin structure, and modification of histones including acetylation, methylation, ubiquitination, and phosphorylation (Springer, 2013). These mechanisms have the ability to regulate almost all genetic functions, including replication, DNA repair, gene transcription, gene transposition, and cell differentiation. For example, modifications in chromatin and generation of small RNAs have been shown to be involved in transcriptional and post-transcriptional control of gene expression during stress responses in plants (Madlung and Comai, 2004; Angers et al., 2010). These modifications are tissue-, species-, organelle-, and age-specific (Vanyushin and Ashapkin, 2011). The changes in hormonal levels that occur during biotic and abiotic stresses can control DNA methylation and other epigenetic effects (Zhang et al., 2012) resulting in plant adaptation (Mirouze and Paszkowski, 2011). Consequently, decoding how epigenetic mechanisms work in developmental gene regulation and during plant response to the environmental stresses is important. In turn, deciphering these mechanisms will also provide valuable information for potential applications, including genetic manipulation of plants toward enhanced tolerance to environmental stresses (Sahu et al., 2013; Springer, 2013).
Construction of Ever-Larger Signaling Networks is an Urgent Task
Indeed, cellular, genetic, genomic, proteomic, and metabolomic data platforms have resulted in increasingly more detailed descriptions of signaling mechanisms, which have raised the necessity for construction of ever-larger signaling networks (Papin et al., 2005; Baginsky et al., 2010). Understanding the function of these signaling networks through reconstructing the available data about signaling pathways is crucial for studying plant’s responses to different diseases and stresses.
Therefore, it is important to somewhat simplify this complexity. The start could be at the points at which the different signaling pathways converge, rather than studying the branches. Consequently, studying the phenomenon of cross-talk may represent a good point to start to unravel global signaling networks. Additionally, tolerance across different stresses is extremely important for agriculture where plants with tolerance to more than one stress can be produced through breeding as well as transformation (Sewelam et al., 2014b). It was stated that, although different environmental challenges use unique mechanisms to initiate their specific responses, all forms of stresses seem to induce a common set of responses (Levitt, 1972). More recently, it was reported that different stress-induced changes in gene and protein expression include similar fingerprints under various environmental insults in different organisms (Desikan et al., 2001; Scandalios, 2002; Laloi et al., 2004; Polidoros et al., 2005; Walley and Dehesh, 2009; Baena-González, 2010; Atkinson and Urwin, 2012). In this regard, it was found that the accelerated generation of ROS is a common plant response to different biotic and abiotic stresses (Allen et al., 1995; Goulet et al., 1997; Noctor, 1998; Orozco-Cardenas and Ryan, 1999; Asai et al., 2000; Miller et al., 2010; Petrov and Van Breusegem, 2012; Noctor et al., 2014; Perez and Brown, 2014; Hossain et al., 2015; Xia et al., 2015). The remainder of this review will be devoted for studying ROS signaling. ROS production, scavenging, damaging effects, signaling roles and how ROS work upstream or downstream of other signaling components will be discussed.
Reactive Oxygen Species at the Cross-Road
During normal growth and development, ROS are produced in different cellular compartments in living cells with increased production under biotic and abiotic challenges (Figure 3; Møller et al., 2007; Miller et al., 2010). The traditional notion that ROS are toxic by-products of plant metabolism has changed. Substantial experimental data are available assuring that ROS are highly controlled signaling molecules able to transfer the environmental signals, with other signaling intermediates, to the genetic machinery (Polidoros et al., 2005). Here, we present a summary about ROS chemistry and signaling that would help understanding of the next sections. For detailed descriptions, we suggest the following reviews; Mittler (2002), Apel and Hirt (2004), Laloi et al. (2004), Mittler et al. (2004, 2011), Asada (2006), Halliwell (2006), Møller et al. (2007), Heller and Tudzynski (2011), Wrzaczek et al. (2013), Baxter et al. (2014).
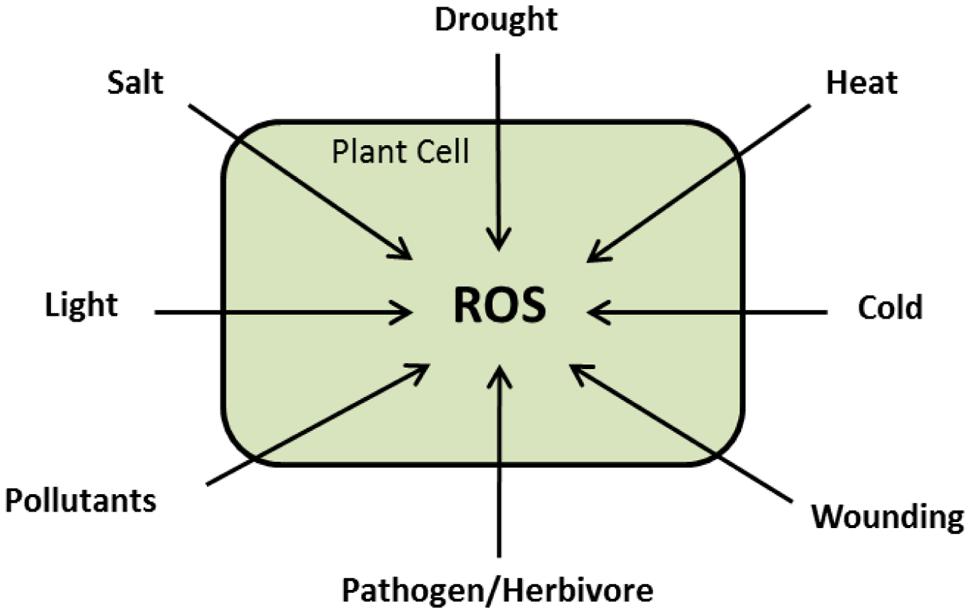
FIGURE 3. Schematic presentation showing that ROS are versatile signaling molecules during plant response to different stresses.
Definition and Chemistry of ROS
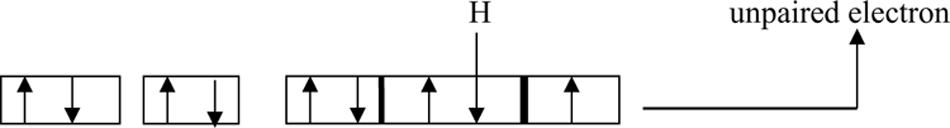
Molecular oxygen, in its ground state, is relatively unreactive. Nevertheless, during normal metabolic activity, and as a result of various environmental stresses, O2 is capable of giving rise to dangerous reactive states such as free radicals (Polidoros et al., 2005; Phaniendra et al., 2015). Reactive oxygen intermediates may result from the excitation of O2 to form singlet oxygen (1O2; Triantaphylidès and Havaux, 2009) or from the transfer of one, two, or three electrons to O2 to form, respectively, a superoxide radical (), H2O2 or a hydroxyl radical (OH∙; Mittler, 2002). The free radical might be defined as any species capable of an independent existence that contains one or more unpaired electrons; an unpaired electron being one that is alone in an orbital (Halliwell, 1991; Figure 4).
Radicals are generally more reactive than non-radicals because electrons are more stable when paired together in orbitals, but when an electron occupies an orbital by itself it has two possible directions of spin. On the other hand, H2O2 and singlet oxygen, themselves, can be quite toxic to cells although they are non-radicals. Accordingly, the term ROS has been introduced to describe collectively, not only free radicals but also other toxic non-radicals (Halliwell, 1991).
ROS Production In vivo
Reactive oxygen species are generated during normal metabolic processes. In addition, they are produced as an inevitable result of electron transport chains in chloroplast and mitochondria. As a result, electrons fall onto O2, generating different ROS. Furthermore, abiotic and biotic stresses can further exaggerate the production and accumulation of ROS (Bhattacharjee, 2005). Mittler (2002) mentioned ten sources for production of ROS in plant cells, including, in addition to photosynthetic and respiratory electron transport chains, NADPH oxidase, photorespiration, amine oxidase, and cell wall-bound peroxidases. In chloroplasts, for example, ROS can be produced at photosystem I (PSI) as well as at PSII. During stress conditions the absorbed light energy exceeds the capacity of photosynthesis to use it through photosynthetic electron transport. As a result, various ROS are formed, including singlet oxygen (1O2) at PSII and superoxide radicals () at PSI and PSII as byproducts (Pospisil et al., 2004; Asada, 2006; Schmitt et al., 2014). At PSII, the excess energy may be transferred from excited chlorophyll to molecular oxygen (energy is transferred not electrons) forming 1O2 as indicated below;
Reaction (1)
Under certain conditions (when the transport of photosynthetic products out of the chloroplast or the re-oxidation of NADPH is inhibited as occurs during different stresses) and O2 reduction (electron transfer) at PSI, superoxide radical formation takes place (Furbank et al., 1983). Then these radicals are dismutated into H2O2 spontaneously as well as through the action of SOD. Later on, inside the chloroplast, and H2O2 can react with each other in the presence of soluble metal ions, such as iron, to form the more reactive hydroxyl radicals according to the Haber–Weiss reaction (Bowler et al., 1992).
Reaction (2)
On the other hand, mitochondria represent a main source for ROS generation in aerobic organisms. It was estimated that from 1 to 5% of the oxygen taken up by isolated mitochondria is used in ROS production (Møller et al., 2007). The complete reduction of O2 to water through the respiratory electron transport chain requires four electrons.
Reaction (3)
But, as a consequence of spin restrictions, O2 cannot accept the four electrons at once, but one at a time. As a result, during O2 reduction, stable ROS intermediates such as , H2O2, and OH∙ are formed in a stepwise fashion as follow;
Reaction (4)
In peroxisomes, H2O2 is produced during the process of photorespiration via the action of the enzyme glycolate oxidase (Møller et al., 2007). Also, the plasma membrane-bound NADPH oxidases make a big contribution to ROS production in plant cells, especially during pathogen infections (Torres and Dangl, 2005).
Damaging Effects and Scavenging of ROS
Plants are well-adapted for minimizing the damage that could be induced by ROS under natural growth conditions. However, O2 toxicity emerges when the production of ROS exceeds the quenching capacity of the protective systems due to stress conditions (Bowler et al., 1992; Yuasa et al., 2001; Miller et al., 2009; Akter et al., 2015). As a consequence, different ROS, including , H2O2, OH∙, and singlet oxygen, are formed, leading to oxidizing and destroying lipids, proteins, and DNA in the stressed cells (for intense information on this topic see Scandalios, 2005; Møller et al., 2007; Vanderauwera et al., 2011). Thus, plant cells have evolved antioxidant mechanisms to combat the danger posed by the presence of ROS (Baxter et al., 2007; Gill and Tuteja, 2010; Miller et al., 2010; Heller and Tudzynski, 2011; Wrzaczek et al., 2013; Schmitt et al., 2014). Mittler (2002) has reported the presence of ten mechanisms to remove ROS, in addition to five ways to avoid ROS production in plant cells. These include several enzymatic and non-enzymatic mechanisms. The enzymatic mechanisms include antioxidant enzymes, such as SOD (which converts to H2O2), catalases and peroxidases (which remove H2O2). The non-enzymatic mechanisms of ROS removal include antioxidant molecules, such as ascorbic acid, glutathione, carotenoids, and α-tocopherol (Noctor, 1998; Asada, 1999; Mittler, 2002). It was reported that there is a network of 152 genes involved in managing the level of ROS in Arabidopsis (Mittler et al., 2004).
In addition to these antioxidant mechanisms which scavenge the already formed ROS, plants have evolved smart ways to avoid the production of toxic forms of oxygen. These avoiding mechanisms include anatomical adaptations, such as leaf movement and curling, C4 or CAM (Crassulacean Acid Metabolism), chlorophyll movement, suppression of photosynthesis, and photosystems and antenna modulators (Maxwell et al., 1999; Mittler, 2002).
ROS Signaling and Specificity
For a signaling molecule to be effective, it needs to be produced quickly and efficiently on demand, to induce distinct effects within the cell, and to be removed rapidly and efficiently when no longer required (Neill et al., 2003). ROS are produced instantly after the onset of the stress. In addition, ROS are very reactive; they can react with membrane lipids, carbohydrates, proteins and DNA. ROS such as H2O2 can defuse through the biological membranes through aquaporins (Bienert et al., 2007; D’Autreaux and Toledano, 2007; Dynowski et al., 2008; Mubarakshina et al., 2010; Borisova et al., 2012) leading to systemic responses. Moreover, living cells have very efficient antioxidant systems, including enzymatic and non-enzymatic mechanisms, to put ROS under a precise control (Foyer and Noctor, 2005). Collectively, all of these features of ROS render them ideal signaling components.
Levine et al. (1994) have suggested a signaling role for H2O2, controlling the hypersensitive response and promoting the expression of glutathione-S-transferase and glutathione peroxidase encoding genes. Many studies have suggested signaling roles for ROS in developmental processes as well as biotic and abiotic responses (Apel and Hirt, 2004; Foyer and Noctor, 2005; Gadjev et al., 2006; Miller et al., 2009; Mittler et al., 2011; Wrzaczek et al., 2013; Perez and Brown, 2014). In an early study, the genomic response of Escherichia coli cells to H2O2 treatment was examined with a DNA microarray composed of 4169 open reading frames (Li et al., 2001). In this study, the mRNA of 140 genes (in wild-type) was considerably induced after H2O2 treatment. On exposure of S. cerevisiae cells to H2O2, expression of about one-third of all yeast genes had changed suggesting that ROS can cause massive alterations in the biology of the oxidative-stressed cells (Gasch et al., 2000). Using cDNA microarray technology from a sample of 11,000 expressed sequence tags (ESTs), 175 non-redundant EST were identified that are regulated by H2O2 in Arabidopsis (Desikan et al., 2001).
To this end, it is quite evident that ROS operate as intracellular signaling molecules, but how they can set specific signaling duties is still controversial. This controversy arises from what seems to be a paradox between the reactive nature of ROS that renders them indiscriminate and the specificity that is required for signaling (D’Autreaux and Toledano, 2007). In general, the specificity in signaling pathways is mediated via the non-covalent binding of a ligand to its cognate receptor through a shape-complementary fit between macromolecules. On the other side, ROS deliver signaling events via chemical reactions with specific atoms, such as iron (Fe) and sulphur (S), of target proteins that lead to protein modifications (Nathan, 2003). ROS can also react with different target proteins whenever the chemical reaction is possible. The remaining question is how specificity in ROS signaling is managed? By looking into the chemical characteristics and the biological activities of each ROS, including , H2O2, OH∙ and singlet oxygen (1O2), an answer to this question could be revealed.
is a by-product of electron transport chains of photosynthesis and respiration and is produced by NADPH oxidases and cell wall peroxidases. In E. coli, the steady-state concentration of is very low (∼10-11 M; Halliwell and Gutteridge, 1999), which reflects its instability; this is mainly due to spontaneous and SOD-mediated dismutation to H2O2. The instability of and its inability to diffuse through membranes because of its negative charge make this ROS relatively poor signaling molecule. However, due to high attraction, oxidizes Fe–S clusters at a rate that is almost diffusion limited (Storz et al., 1990; Storz and Imlayt, 1999).
H2O2 is actually a poor oxidant and reacts mildly with [Fe–S] (rate constant of 102–103 M-1 s-1), loosely bound metals and, very slowly, with glutathione and free cysteine (Cys) (Imlay, 2003). By contrast, its reactivity toward Cys residues can significantly increase to 10–106 M-1 s-1. H2O2 is relatively stable (cellular half-life ∼1 ms, steady-state levels ∼10-7 M; D’Autreaux and Toledano, 2007), and can diffuse through biological membranes because it is not charged. Its selective reactivity, stability and diffusability make H2O2 fit for signaling. As a second messenger, H2O2 can mediate intracellular signal transduction through chemoselective oxidation of Cys residues in signaling proteins, such as glutathione, thioredoxins, and peroxiredoxins (Paulsen and Carroll, 2010).
OH∙ is the most highly toxic ROS. It has high indiscriminate reactivity, which limits its diffusion to sites of production (half-life 10-9 s; Halliwell and Gutteridge, 1999), even though OH∙ seems to operate in H2O2 sensing (D’Autreaux and Toledano, 2007).
Singlet oxygen (1O2) is an excited state molecule. The half-life time of 1O2 is very short (∼100 ns) and it can travel only a very short distance in cells (<100 nm; Moan, 1990; Niedre et al., 2002). This could be because it reacts very rapidly with amino acids, unsaturated lipids, and other cell constituents. As a result, 1O2 can react directly only with molecules in close proximity to its production location, i.e., in the chloroplast (Kochevar, 2004; Triantaphylidès and Havaux, 2009). This means that 1O2 could deliver specific signaling events mainly through spatial aspects of ROS production.
In addition to the previous chemical characteristics that render ROS able to set specificity as signaling molecules, a non-ROS intermediate in a ROS signaling pathway can regulate additional pathways that are physically non-adjacent to the pathway in which it was formed (Nathan, 2003). For instance, a ROS that is produced in a cellular compartment could specifically activate a secondary messenger such as a MAPK or a plant hormone, which in turn activate remote signaling pathways. In Arabidopsis, it was suggested that histidine kinase ethylene receptor ETR1 is important for H2O2 perception during stomatal closure (Bright et al., 2006). In addition, it was reported that indirect activation of TFs by ROS may be mediated by some members of MAPK cascades (Asai et al., 2000). Interestingly, it was shown that H2O2 originating in different subcellular sites induces different responses. H2O2 produced in chloroplasts was found to activate early signaling responses, including TFs and biosynthetic genes involved in production of secondary signaling messengers; while H2O2 produced in peroxisomes was found to induce transcripts involved in protein repair responses (Sewelam et al., 2014a). Moreover, ROS-mediated changes in the cellular redox homeostasis could set highly specific signaling roles for ROS. For example, different pathways could sense and weigh the change in cellular redox balance resulting from the change of intracellular ROS concentration, then translate these changes into highly specific cellular signals that direct the cell to produce a relevant adaptive response (Foyer and Noctor, 2005). In simple organisms, such as bacteria and yeast, the enhanced production of ROS is perceived by change in redox homeostasis which in turn is delivered to redox sensitive TFs (Costa and Moradas-Ferreira, 2001; Georgiou, 2002). In addition, it has been proposed that ROS may be perceived indirectly by sensing changes in the cellular redox potential (Price et al., 1994) or by detecting the products of ROS-inflicted damage (Evans et al., 2005). In addition, ROS can generate specific signaling effects through the peptides produced from proteolytic breakdown of oxidatively damaged proteins which act as secondary ROS messengers and contribute to a retrograde ROS signaling during different environmental challenges that generate oxidative stress (Moller and Sweetlove, 2010).
Components Involved in ROS Signaling
The perceived ROS signals work upstream as well as downstream from many other second messengers in addition to many feedback and feedforward regulations in an interwoven manner to establish specific responses to different developmental and environmental cues. Currently, a major gap exists in our understanding of how ROS induce large-scale and coordinated expression from many genes. In addition, the big challenge is to identify the upstream sensing and signaling events through which ROS are perceived and delivered to the ROS-induced TFs. Do ROS activate the expression of TFs directly or through another set of signaling intermediates? How could secondary messengers such as G proteins, MAPKs, Ca2+, JA, SA, and ABA mediate the ROS signals and which one is upstream or downstream from each other? Does ROS, produced passively during different stresses through their damaging effects on cellular structures, induce signaling events that differ from those signaling events produced actively through activation of cell membrane-bound enzymes, like NADPH oxidases? The discussion below is an overview of the interconnectivity between ROS and other individual components involved in plant signal transduction pathways (Figure 5).
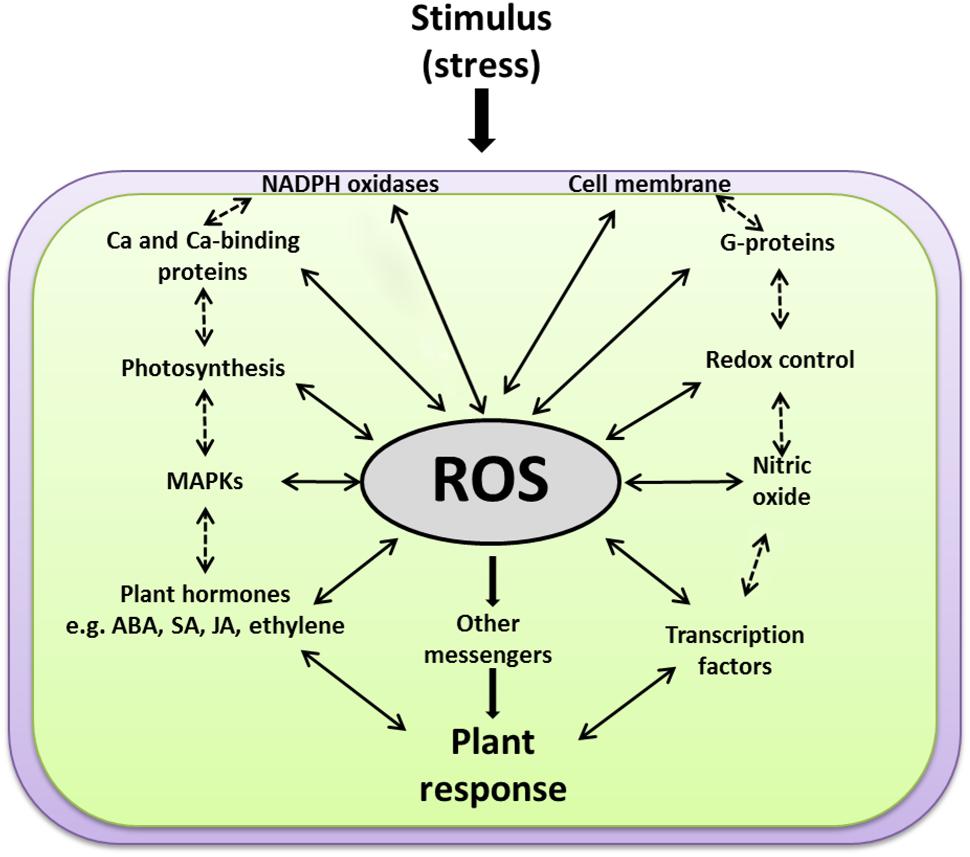
FIGURE 5. A scheme explaining how ROS function at the cross-road of various key signaling events. ROS work upstream and downstream of the other signaling components, e.g., membranes, NADPH oxidases, G-proteins, calcium, redox homeostasis, photosynthesis, MAPKs, plant hormones [such as salicylic acid (SA), jasmonic acid (JA), abscisic acid (ABA), and ethylene] and transcription factors. Solid arrows for direct ROS interactions with other signaling components, dashed arrows for expected indirect interactions.
Photosynthetic Activity
Photosynthesis represents the most peculiar feature that distinguishes plant and animal systems. In photosynthesis, through intersystem electron transport, the light energy captured by photosynthetic pigments is transformed into chemical energy which pumps reductants (NADPH) and ATP into the Calvin cycle (dark reactions) supplying carbon skeletons (sugars) for all major metabolic processes (Kromer, 1995). In addition to this role, plastids synthesize and store a large number of biomolecules, including carbohydrates, amino acids, fatty acids, and plant hormones (Buchanan et al., 2000). Therefore, it is self-evident that any change or imbalance in the function of the chloroplast will affect directly or/and indirectly the other cellular functions in plant cells. Consequently, environmental challenges can be primarily sensed via production of ROS and the concomitant changes of redox homeostasis of the chloroplast that act synergistically with other signaling pathways inducing then adaptive molecular and physiological responses (Huner et al., 1996). Earlier studies have suggested that the redox state of plastoquinone controls the rate of transcription of the chloroplast genes encoding reaction-center apoproteins of photosystem I and photosystem II (Pfannschmidt et al., 1999, 2009). Recently, it was reported that chloroplasts are able to sense light conditions and generate a remote control to modulate the nuclear gene expression to face the changing environment (Koussevitzky et al., 2007; Pogson et al., 2008; Pfannschmidt et al., 2009; Godoy Herz et al., 2014). As a retrograde signaling pathway, the redox state of components of the photosynthetic electron transport chain can sense the changes in photosysnthetic activity and in turn affect the nuclear gene expression (Fey et al., 2005; Nott et al., 2006; Godoy Herz et al., 2014). The plastidial metabolite, methylerythritol cyclodiphosphate (MEcPP), was found to regulate the expression of nuclear stress-response genes through a retrograde signal from the chloroplast to the nucleus (Xiao et al., 2012).
The thylakoid membranes and the involved redox complexes of the photosynthetic apparatus, especially the light energy harvesting PSII, are very sensitive targets to various environmental stress factors. During stress, excess of photosynthetically active light leads to the formation of ROS (e.g., 1O2 and OH∙; Glatz et al., 1999), which can induce membrane damage by attacking double bonds of unsaturated fatty acids. These effects are also expected to feedback signals for stress gene expression via the pathway which senses the physical state of the membrane (Glatz et al., 1999). This observation may reinforce the idea that receptors/sensors at the cell surface or the cell membranes could perceive stimuli, not directly from the stress, but indirectly through the chloroplast stress signals. It was reported that H2O2 from chloroplasts led to the induced expression of many genes coding for membrane-bound receptor proteins and signaling components (Sewelam et al., 2014a). In ozone-treated Arabidopsis leaves, Joo et al. (2005) reported that the chloroplastic ROS signal contributes to activating the membrane associated NADPH oxidases in intercellular signaling during the early component of the oxidative burst. Accordingly, they suggested that signaling from the chloroplast is central for oxidative stress induction by O3. Other studies also found that induction of light and stress response requires chloroplast signaling mediated by ROS (Allen et al., 1999; Fryer et al., 2003; Agrawal et al., 2004; Serrato et al., 2013).
Relatively little attention has been given to the role of photoproduced H2O2 and other ROS in defense signaling (Delledonne et al., 2001; Parker et al., 2001). This may be because the current models of signaling pathways controlling plant defense against pathogen infection are based mainly on animal models. Recently, new research has led to the development of models incorporating how the signaling pathways that are involved in light perception and in defense could operate and interact to form a complete defense signaling network, which includes systems to sense light and regulate gene expression. In this context, it was suggested that signals from the chloroplast and LESION SIMULATING DISEASE1 are integrated to mediate crosstalk between light acclimation and disease resistance in Arabidopsis (Mühlenbock et al., 2008). ROS produced from chloroplasts during the infection with Xanthomonas campestris play a major role in localized cell death in the non-host interaction between tobacco and this bacterial species (Zurbriggen et al., 2009). It was suggested that the chloroplast protein RPH1, a positive regulator of Phytophthora brassicae-induced oxidative burst, plays a role in the defense response of Arabidopsis and potato to P. brassicae (Belhaj et al., 2009). Based on these observations, it is clear that thylakoid membranes and hence photosynthesis, play a vital and very early role in stress sensing and signaling in plants, an idea that should be considered when constructing plant signaling networks.
Redox Homeostasis
In plants, the continuous energy conversions, in the chloroplast and mitochondria, and the optimal use of the available light energy are only guaranteed when all reduction–oxidation (redox) processes are under precise control. Information on the redox situation is generated and transferred by various redox components, including various ROS and different antioxidants, that are parts of a robust network that links metabolism with regulation and signaling. Under environmental challenges, the imbalance in the network is sensed, and transformed into redox signals that are transmitted in order to elicit specific responses at various levels of regulation and in different subcellular compartments (Scheibe and Dietz, 2012). Thus, ROS and redox cues, generated under stress conditions, are essential to control the main metabolic processes through which cells convert and distribute the energy and metabolic fluxes, optimize different cell functions, activate acclimation responses through retrograde signaling, and control whole-plant systemic signaling pathways (Noctor, 2006; Suzuki et al., 2012). Redox homeostasis in the plant cell is considered to be an “integrator” of information from the environment controlling plant growth and stress responses, as well as cell death events (Dietz, 2003; Foyer and Noctor, 2009; Potters et al., 2010). The antioxidants, ascorbate, glutathione, carotenoids, and tocopherol, are information-rich redox buffers that affect numerous cellular components. In addition to their vital roles in stress response and as enzyme cofactors, cellular redox components influence plant growth and development by orchestrating processes from cell division to senescence and cell death (de Pinto and De Gara, 2004; Potters et al., 2004; Tokunaga et al., 2005; Halliwell, 2006). Most importantly, antioxidants influence gene expression associated with responses to different environmental cues to maximize defense through tuning cellular ROS levels and redox state (Foyer and Noctor, 2005). Proteins with oxidisable thiols such as glutathione and thioredoxin-1 are crucial for many functions of cell nuclei, including transcription, chromatin stability, nucleo-cytoplasmic trafficking, and DNA replication and repair (Nose, 2005; Go and Jones, 2010; Lukosz et al., 2010). From bacteria to humans, the triplet peptide, glutathione, is involved in protein S-glutathionylation where it regulates a variety of cellular processes by modulating protein function and prevents irreversible oxidation of protein thiols under unfavorable conditions (Dalle-Donne et al., 2009).
As redox homeostasis is greatly influenced by most, if not all, conditions that affect plant growth and development, the changes of intracellular redox determine various signaling events through their interaction with many other secondary messengers, such as protein kinases and phosphatases, phytohormones and calcium. Intensive current research might confirm that ROS-antioxidant interactions act as a metabolic interface between environmental changes and the concomitant signaling responses (Foyer and Noctor, 2005, 2012). For example, the redox state determined by the ROS-antioxidant interactions could regulate, directly or indirectly, the work of TFs, such as TGA, Athb-9, and RAP2 and hence the regulation of the expression of their downstream genes (Dietz, 2008).
Membranes
The plasma membrane, as the selective barrier between living cells and their environments, plays a pivotal role in the perception of the changes in the surrounding environment (Guo et al., 2002). As a consequence of their rapid ability to modify their physical state, cellular membranes are not only the primary sites of stress damage, but also able to perceive environmental insults and activate remotely stress-defense genes (Glatz et al., 1999). The microdomain organization and physical state of cell membranes is known to be a very sensitive monitor of different environmental challenges (Horvath et al., 1998). It was stated that heat stress changes the membrane fluidity and H2O2 responds rapidly to this change, leading to the activation of small heat shock protein synthesis (Königshofer et al., 2008).
As a component of cell membranes, ion channels play a vital role in the transduction of environmental and internal signals (Binder et al., 2003). Ion channels are proteins forming hydrophilic pathways through the plasma membranes (Barnes et al., 1997). They function as permeation pores through which the electrically charged species can pass across biological membranes. It was reported that ion channels are directly involved in important cellular processes, such as plant defense responses induced by elicitors (Czempinski et al., 1997), light perception (Ermolayeva et al., 1997), and mechanical signals (Cosgrove and Hedrich, 1991). For example, it was reported that the efficiency of H2O2 signaling between cells is controlled by plasma membrane aquaporin pores where the expression of several plant plasma membrane aquaporins in yeast, such as Arabidopsis plasma membrane intrinsic protein PIP2.1, was found to enhance the toxicity of H2O2 when yeast cells were exposed to H2O2 treatment (Dynowski et al., 2008). It was found that the disruption of a cyclic nucleotide-gated calcium channel gene causes a hyper-thermosensitive phenotype in Arabidopsis and moss indicating that the plasma membrane cyclic nucleotide-gated calcium channels control plant thermal sensing and acquired thermotolerance (Saidi et al., 2009; Finka et al., 2012).
During exposure to stress, the major role of phospholipids, the backbone of cellular membranes, may be to serve as precursors for the generation of secondary messenger signaling molecules, such as phosphotidylinositol, inositol 1,4,5-tiphosphate (IP3), diacylglycerol, and jasmonates. IP3 and diacylglycerol are secondary messengers that can activate protein kinase and induce Ca2+ release, respectively. Additionally, IP3 itself is a signal and may be involved in several processes, such as the recruitment of signaling complexes to specific membrane location and their assembly (Guo et al., 2002). In addition, under oxidative stress, polyunsaturated fatty acids (PUFAs) are attacked by different ROS, specially 1O2 and OH∙. This causes production of lipid hydroperoxides, leading to a decrease of membrane fluidity (Møller et al., 2007). In turn, this change in membrane physical state could activate downstream signaling intermediates.
NADPH Oxidases
Membrane-bound NADPH oxidases are a group of enzymes that catalyze the production of superoxide radicals () in animals and plants (Sagi and Fluhr, 2006). In mammals, NADPH oxidases are also called respiratory burst oxidases (Rbo). Because of their functional homology with mammals, plant NADPH oxidases are known as respiratory burst oxidase homolog (Rboh; Torres and Dangl, 2005). In plants, Rboh enzymes are the source of ROS production under pathogen infection and in many of other processes (Torres and Dangl, 2005). The ability of Rboh enzymes to integrate various signaling players, such as calcium and protein phosphorylation with ROS production, suggests a crucial role for Rboh in many different biological processes in cells, and places them at the core of the ROS signaling network of cells, revealing their important functions in plants (Suzuki et al., 2012; Kadota et al., 2014).
In Arabidopsis, there are ten Rboh genes (Torres et al., 1998; Dangl and Jones, 2001). Many studies have reported the induction of Rboh gene expression by pathogens and fungal elicitors (Simon-Plas et al., 2002; Yoshioka et al., 2003; Wang et al., 2006). In addition, using mutant analysis, it was suggested that RbohD and RbohF are required for ROS production and cell death in Arabidopsis plants infected with P. syringae or Peronospora parasitica (Torres et al., 2002). The same group (Torres et al., 2005) reported that RbohD is required for ROS production but this ROS antagonizes cell death induced by Pseudomonas infection. In Nicotiana benthamiana, silencing of NbrbohA and NbrbohB led to reduction of ROS production and reduced resistance to Phytophthora infestans infection (Yoshioka et al., 2003). The Arabidopsis RbohF was suggested to be a vital player in defense-associated metabolism and a key factor in the interaction between oxidative stress and pathogen infection (Chaouch et al., 2012). Regarding the involvement of Rboh in abiotic interactions, it was reported that the Arabidopsis RbohD gene is involved in ROS-inducing a rapid systemic signal during various stress factors, such as heat, cold, high light, and salinity (Miller et al., 2009). The abiotic stress-mediating phytohormone ABA was reported to be regulated by the action of RbohD and RbohF in different ROS-ABA signaling pathways (Kwak et al., 2003; Joo et al., 2005; Xue and Seifert, 2015). In a microarray experiment, RbohD expression was downregulated by ABA treatment but upregulated by H2O2 treatment in Arabidopsis (Barraud et al., 2006). During salt stress, ROS produced by both AtrbohD and AtrbohF seem to function as signal molecules to regulate Na+/K+ homeostasis, where the two Arabidopsis double mutants atrbohD1/F1 and atrbohD2/F2 were found to produce less ROS and to be much more sensitive to NaCl treatments than wild-type (Ma et al., 2012). RbohD was found to contribute to the ROS-responsive expression of ERF6, a ROS regulator TF in Arabidopsis (Sewelam et al., 2013). As NADPH oxidases are physically located at the plasma membrane, they are proposed to play an early and vital signaling role and should be highly considered when constructing plant signaling networks.
G Proteins
GTP-binding proteins (G proteins) are found in almost all organisms from prokaryotes to humans (Assmann, 2004). G proteins mediate stimulus perception by G-protein-coupled receptors (GPCR), in addition to other regulatory proteins. In humans, there are about 1000 GPCR, representing the largest group of cell surface receptors encoded by mammalian genome (Nagarathnam et al., 2012). It is estimated that about 60% of all drugs currently available target G-protein-based pathways and G protein component disorders have been found to cause various genetic diseases (Assmann, 2004). G proteins are heterotrimeric proteins composed of three monomers; α, β, and γ. About 20 G protein α subunits (Gα), 6 Gβ subunits, and 20 Gγ subunits have been characterized in mammals (Gutkind, 2000). Controversially, in plants the situation seems to be much simpler than that in animal systems. For example, it was reported that the Arabidopsis genome encodes only single Gα and Gβ subunits, two Gγ subunits, just one GPCR protein, and one regulator of G protein signaling (Assmann, 2004).
The involvement of G proteins in plant stress signaling is evident, especially in plant-pathogen interactions (Assmann, 2005; Trusov et al., 2009; Maruta et al., 2015; Xu et al., 2015). Regarding ROS, many studies have suggested a tight relationship between ROS and G proteins in stress-mediated plant signaling. It was reported that, on exposure of Arabidopsis leaves to ozone, the first biphasic oxidative burst is greatly attenuated or completely absent in mutant plants lacking Gα protein or Gβ protein. This finding suggests that the ROS produced by ozone in the apoplastic fluid do not themselves enter cells to activate intracellular ROS-producing systems. Rather, the extracellular ROS activate the G protein either directly or indirectly (Joo et al., 2005). It is possible that G proteins themselves are directly activated by ROS. In this regard, it has been reported that two mammalian Gα proteins, Gαi and Gαo, are redox-controlled (Fujiki et al., 2000). The membrane-bound NADPH oxidases D and F were suggested to receive initial signals from G proteins to mediate ozone responses in Arabidopsis guard cells (Suharsono et al., 2002). The absence of the Gα subunit in the gpa1 mutant disrupts the interplay between ABA perception and ROS production, with a consequent inhibition of Ca2+-channel activation (Zhang et al., 2011). The membrane-bound ROS producing enzymes AtRbohD and AtRbohF work in the same pathway with the Gβ subunit of the heterotrimeric G protein for full disease resistance to different P. syringae strains (Torres et al., 2013). In plants, further studies are required to unravel the roles of G proteins and their signaling roles.
Calcium Signaling
The use of calcium ions as a secondary messenger represents an integral part in many signal transduction pathways in all life forms, from vertebrate animals to plants (Berridge et al., 2000; Stael et al., 2012). In contrast to other similar ions, such as Mn2+, the Ca2+ ion has many peculiar features, including a favorable ionic radius and hydration status, an irregular geometry, and flexible coordination chemistry (Jaiswal, 2001). The main calcium stores in plant cells are: the vacuole, the endoplasmic reticulum and the apoplast (Stael et al., 2012). Elevation in cytoplasmic Ca2+ represents an early response to many different biotic and abiotic stresses, including oxidative (McAinsh and Pittman, 2009; Dodd et al., 2010). As a second messenger in a wide range of signaling pathways in plants, calcium connects the perception of different stimuli and stresses to their downstream cellular responses (Evans et al., 2005). It has been stated that transient cellular calcium elevations are sensed by several Ca2+ sensors such as calmodulin (CAM), calmodulin-like (CML), calcium-dependent protein kinase (CDPK), and calcineurin B-like protein (CBL; Bouche et al., 2005; McCormack et al., 2005; Das and Pandey, 2010; Asai et al., 2013). A direct interconnection between CBL-CIPK-mediated Ca2+ and ROS signaling in plants was reported as evidence for a synergistic activation of the NADPH oxidase RbohF by direct Ca2+-binding to its EF-hands (Drerup et al., 2013). The Arabidopsis CPK5, an isoform of the plant CDPK family, was activated rapidly in response to infection with P. syringae, resulting in Rboh-mediated ROS production and enhanced SA-mediated resistance to this bacterial pathogen (Dubiella et al., 2013). In the same study, RbohD was reported to be an in vivo phosphorylation target of CPK5. Ca2+ ions also regulate long-distance root-to-shoot signaling and may also have roles in transmitting ROS signals (Choi et al., 2014).
The concentration of cytosolic Ca2+, the expression level of calmodulin 1 (CAM1) gene, the content of CAM proteins and the expression of many antioxidant genes in maize are increased after treatment with ABA or H2O2. Furthermore, pre-treating plants with CAM inhibitors almost completely blocked the upregulation of many antioxidant enzymes (Hu et al., 2007). These findings show that the increase in cytosolic Ca2+ requires CAM to deliver its signal to the downstream targets. Ca2+ elevations have been suggested, in some cases, to be upstream of ROS production; in other cases, Ca2+ elevations have been reported to be downstream of ROS production (Bowler and Fluhr, 2000). Several workers showed that oxidative stress results in increased cytosolic Ca2+. In tobacco seedlings, oxidative stress stimulates cytosolic Ca2+ increases (Bhattacharjee, 2005). The allelopathic toxin catechin was reported to cause rapid ROS production, followed by ROS-induced Ca2+ increases in diffuse knapweed (Centaurea diffusa) and Arabidopsis roots (Bais et al., 2003). It was reported that pre-treatment of Arabidopsis plants with the calcium channel blocker lanthanum chloride (LaCl3) attenuated the inducing effect of H2O2 on ERF6, suggesting that Ca2+ is playing a signaling role, which is downstream from ROS, in the induction of this TF by H2O2 (Sewelam et al., 2013). On the contrary, other research groups have reported that Ca2+ works upstream of ROS. For example, it was reported that inhibitors of Ca2+ fluxes inhibit both increase in cytosolic Ca2+ and H2O2, whereas inhibitors of NADPH oxidase blocks only the oxidative burst (Abuharbeid et al., 2004). Mechanical forces (e.g., touch) were found to trigger rapid and transient increases in cytosolic Ca2+ and to stimulate apoplastic ROS production. The production of ROS was inhibited by pre-treatment with Ca2+ channel blockers (Monshausen et al., 2009), suggesting a role for Ca2+ as a prerequisite of ROS production under mechanical stimuli. To avoid this ostensible contradiction, future studies should consider the presence of a large number of sources for ROS production as well as a plethora of Ca2+ subcellular sources, in addition to the expected feedback mechanisms. Nevertheless, these studies, at least, designate a crucial role for ROS-Ca2+ signaling during plant responses to stresses that should be considered when constructing global plant signaling networks.
Nitric Oxide (NO)
Nitric oxide (NO) is a small, water-, and lipid-soluble free radical gas with well-characterized signaling roles in mammalian systems (Furchgott, 1995; Neill et al., 2003; Moreau et al., 2010). Nitric oxide production by plants and its involvement in plant growth were described in the late 1970s (Anderson and Mansfield, 1979; Klepper, 1979). Research on the effects of NO in plants focused on atmospheric pollution by the oxides of nitrogen, NO and NO2 (nitrogen dioxide; Hufton et al., 1996). It was revealed that plants not only respond to atmospheric NO, but also produce considerable amounts of endogenous NO (Wildt et al., 1997). However, research on NO as a signaling molecule in plants started with the work done by Leshem and Haramaty (1996) and became well-established after the description of its role in plant defense signaling (Delledonne et al., 1998; Durner et al., 1998, 1999). Currently, it is well-known that NO plays an important signaling role in plant growth, development and defense responses (Besson-Bard et al., 2008; Moreau et al., 2010). It was reported that ROS and NO are produced concomitantly under various stresses and can interact with each other to induce a defense response (Neill et al., 2002; Yoshioka et al., 2009; Molassiotis and Fotopoulos, 2011). NO could have toxic or protective effects, depending on its concentration, combination with ROS compounds, and its subcellular localization (Correa-Aragunde et al., 2015).
Many reports have suggested interconnected signaling roles between ROS and NO during plant response to different stresses. Generation of NO at the same time as H2O2 in response to pathogen infection was found to mediate defense responses similar to those seen following H2O2 production (Delledonne et al., 1998; Durner et al., 1998; Asai and Yoshioka, 2009; Del Río, 2015). Delledonne et al. (1998) reported that treatment of soybean cultures with avirulent P. syringae induces rapid NO synthesis with kinetics similar to H2O2 generation, indicating an interaction between NO and H2O2 during plant response to pathogen attack. NO biosynthesis was reported to be regulated by H2O2-mediated activation of MAP Kinase 6 in Arabidopsis (Wang et al., 2010). A proteomic study on salt-stressed citrus plants pre-treated with H2O2 or NO has suggested an overlap between H2O2 and NO signaling pathways in acclimation to salinity (Tanou et al., 2009, 2010). Under drought stress, it was suggested that ROS and NO interact to induce ABA biosynthesis to affect stomatal closure (reviewed by Neill et al., 2003). Regarding the mechanisms by which NO exerts its effects, it is suggested that NO may deliver its signaling roles via modulating the activity of proteins through nitrosylation and probably tyrosine nitration, in addition to the role that NO can act as a Ca-mobilizing messenger (Besson-Bard et al., 2008). ABA signaling in guard cells was found to be negatively regulated by NO through S-nitrosylation-mediated inhibition of the open stomata 1 (OST1)/sucrose non-fermenting 1 (SNF1)-related protein kinase 2.6 (SnRK2.6; Wang et al., 2015). In fact, NO can interact with ROS in different ways and might work as an antioxidant molecule during various stresses (Beligni and Lamattina, 1999; Correa-Aragunde et al., 2015). Moreover, modulation of superoxide formation by NO (Caro and Puntarulo, 1998) and inhibition of lipid peroxidation (Boveris et al., 2000) could illustrate a potential antioxidant role for NO. The oxidative damage in sorghum embryos was found to be alleviated by pre-treatment with sodium nitroprusside and diethylenetriamine NONOate as sources of exogenous NO (Jasid et al., 2008). Alternatively, excess NO can result in nitrosative stress (Hausladen et al., 1998), so a positive balance of ROS/NO is essential.
Mitogen-Activated Protein Kinases
Mitogen (induces mitotic division)-activated protein kinases (MAPKs) are evolutionary conserved enzymes. In eukaryotes, signaling pathways arbitrated by MAPKs have been considered as a general signal transduction mechanism that links different receptors to their cellular and nuclear targets (Tena et al., 2001). The signaling events mediated by MAPKs are composed of three functionally interlinked protein kinases: a MAP kinase kinase kinase (MAPKKK), a MAP kinase kinase (MAPKK), and a MAP kinase (MAPK; Rodriguez et al., 2010; Sinha et al., 2011). In this phosphorylation module, a MAPKKK phosphorylates and activates a particular MAPKK, which in turn phosphorylates and activates a MAPK by phosphorylation of the tyrosine and therionine residues in the TXY motif (Qi and Elion, 2005).
In the Arabidopsis genome, 20 MAPK, 10 MAPKK, and 60 MAPKKK encoding genes were identified (Ichimura et al., 2002). The current functional analysis of MAPK cascades, mainly in Arabidopsis, revealed that plants have an overall of 24 MAPK pathways of which only a small set has been investigated so far (Wrzaczek and Hirt, 2001). This may reflect why MAPK signaling cascades are so complicated. The challenge ahead is to describe the elements of plant MAPK cascades and to specify roles of individual MAPK cascade genes, in particular signaling pathways (Wrzaczek and Hirt, 2001). The spatial and temporal expression and interaction characteristics of MAPKs are suggested to define their specificity in different signaling pathways (Dietz et al., 2010). The Arabidopsis mitogen-activated protein kinase 8 (MPK8) was reported to connect protein phosphorylation, Ca2+, and ROS in wound signaling pathways (Takahashi et al., 2011).
Some of the components of MAPK cascades are elicited by cold, drought, H2O2, heat, wounding, pathogens, elicitors, ABA, SA, and ethylene (reviewed by Bowler and Fluhr, 2000). In many eukaryotes, the transmission of oxidative signals is controlled by protein phosphorylation involving MAPKs (Kyriakis and Avruch, 1996; Gustin et al., 1998; Pitzschke and Hirt, 2006; Xing et al., 2008). On the one hand, MAPKs can be activated by accumulation of H2O2, on the other hand they can trigger an H2O2-induced oxidative burst (Nakagami et al., 2005; Pitzschke et al., 2009; reviewed by Petrov and Van Breusegem, 2012). In Nicotiana benthamiana, the MAPK cascades MEK2-SIPK/NTF4 and MEK1-NTF6 were reported to participate in the regulation of the radical burst induced by the oomycete pathogen P. infestans through NO and RbohB-dependant ROS generation (Asai et al., 2008). Using Arabidopsis protoplasts, a correlation was revealed between the activation of plant MAPK cascade and H2O2, which is generated by various stress factors. In this study, it was observed that H2O2 activates the MAPKKK, ANP1, which in turn phosphorylates the downstream kinases, AtMPK3 and AtMPK6 (Kovtun et al., 2000). Protein phosphorylation through MAPK cascades was suggested to trigger a positive feedback regulation of Ca2+ and ROS via the activation of RbohD and RbohF in Arabidopsis (Kimura et al., 2012). A maize MAPK, MAP65-1a, was reported to positively control H2O2 amplification and to enhance the antioxidant enzymes SOD and APX through the brassinosteroid signaling pathway (Zhu et al., 2013). The expression of the Arabidopsis OXI1 gene, encoding a serine/threonine kinase, was induced in response to a broad range of H2O2-producing stimuli and OXI1 kinase activity itself was also induced by H2O2 in vivo (Rentel et al., 2004). Application of bioinformatics and computational analysis would be required to illuminate how different MAPKs coordinate different plant signaling events.
Abscisic Acid (ABA)
Substantial evidence postulates that ABA plays a vital role in controlling downstream responses essential for adaptation to stress (Leung and Giraudat, 1998; Raghavendra et al., 2010). These responses include changes in stomatal conductance, growth, osmolyte accumulation, and gene expression (Chen et al., 2002; Verslues and Zhu, 2004; Krasensky and Jonak, 2012). In contrast to the positive role of ABA in abiotic stress response, ABA has been considered as a negative regulator of disease resistance. This negative effect appears to be due to the obstruction by ABA of biotic stress signaling pathways that are orchestrated by SA, JA, and ethylene (Coego et al., 2005). ABA can also improve disease resistance by modifying cell wall deposits, such as callose (Mauch-Mani and Mauch, 2005).
A simultaneous enhanced level of ROS and ABA in plant tissues has been monitored under different types of environmental stresses. The concomitant enhancement of ROS and ABA during stress has been suggested to be a node in cross-tolerance to multiple types of stresses (Verslues and Zhu, 2004). It has been indicated that ROS generated by NADPH oxidases work downstream of ABA in mediating stomatal closure during stress (reviewed by Verslues and Zhu, 2004). It was reported that the production of H2O2 in the chloroplasts, mitochondria and peroxisomes under water stress was abolished in the leaves of maize plants pre-treated with the ABA biosynthesis inhibitor (tungstate) or in an ABA mutant plants, indicating that ABA is required for H2O2 production in these compartments (Hu et al., 2006). It was demonstrated that a temporal-spatial interaction between ROS and ABA regulates rapid systemic acquired acclimation to environmental challenges in plants (Suzuki et al., 2013). In response to heat and oxidative stresses, it was reported that H2O2 mediates a crosstalk between the plant hormones; brassinosteroid and ABA, via a signaling pathway through which brassinosteroid induces a rapid and transient H2O2 production by NADPH oxidase. The process in turn activates increased ABA levels, leading to further increases in H2O2 production and improved stress tolerance in tomato plants (Zhou et al., 2014).
Salicylic Acid, Jasmonic Acid, and Ethylene
Various plant developmental and stress responses require a tuned coordination between the phytohormones SA, JA and ethylene. It is thought that, in Arabidopsis, a JA-ethylene signaling pathway is important to mediate resistance to necrotrophic pathogens (feed on dead tissues), such as Botrytis cinerea. On the other hand, the SA signaling pathway is supposed to mediate resistance to biotrophic pathogens (feed on living tissues), such as P. syringae (Thomma et al., 2001; Anderson et al., 2004). However, it has been suggested that many genes are co-regulated by these hormones and there is considerable genetic evidence for crosstalk between these signaling pathways (Schenk et al., 2000; Glazebrook et al., 2003; Leon-Reyes et al., 2009). Regarding ROS signaling, it was suggested that SA, JA, and ethylene work together with ROS and play crucial regulatory roles in plant defense responses (Mur et al., 2006; Mhamdi et al., 2010).
Salicylic acid (SA) is well-known to regulate both systemic acquired resistance (SAR) and local disease resistance mechanisms, including host cell death and defense gene expression (Park et al., 2007; Vlot et al., 2008). It was reported that SA elicits an oxidative burst, which in turn promotes SAR (Senaratna et al., 2000). One of the proposed modes of action of SA is the inhibition of catalase, a major enzyme scavenging H2O2, thereby increasing cellular concentrations of H2O2, which acts as a second messenger and activates defense-related genes (Ananieva et al., 2002). The extracellular production of ROS was found to enhance SA production inducing stomatal closure in Arabidopsis (Khokon et al., 2011). It was reported that SA accumulation in siz1 mutant plants enhances stomatal closure and drought tolerance through controlling ROS accumulation in Arabidopsis guard cells (Miura et al., 2013). It was suggested that SA accumulation and signaling is activated by increased H2O2 levels through changes in the glutathione pool in an Arabidopsis catalase 2 (cat2) mutant (Han et al., 2013a).
Jasmonic acid (JA) and methyl jasmonate (MeJA), are natural products regulating plant development, response to environmental challenges, and gene expression (Bell et al., 1995). A signaling role for JA in defense responses has been suggested in plants (Farmer and Ryan, 1992; Wasternack and Hause, 2013). Currently, there is accumulating evidence suggesting a strong relationship between ROS and JA signaling. For instance, it was suggested that MeJA pretreatment of Arabidopsis inhibited O3-induced H2O2 production and SA accumulation and completely abolished O3 induced cell death (Rao et al., 2000). It was reported that ROS generated by RbohD and RbohF enzymes are important for JA-induced expression of genes regulated by MYC2, a TF involved in the JA-mediated response, where treating RbohD and RbohF mutant plants with MeJA failed to increase the expression levels of various MYC2 downstream genes (Maruta et al., 2011). A dynamic interaction between JA and ROS was characterized to regulate lignin biosynthesis in response to cell wall damage where ROS produced by RbohD and JA-isoleucine generated by JASMONIC ACID RESISTANT1 were found to form a negative feedback loop that influence lignin accumulation (Denness et al., 2011). It was revealed that the intracellular ROS production in cat2 mutant Arabidopsis plants leads to activating the JA pathway and its related genes with glutathione accumulation as an intermediate (Han et al., 2013b).
Ethylene (C2H4) is one of the simplest organic molecules that have biological activity in plants (Zarembinski and Theologis, 1994). It is well-documented that ethylene is a main player in PCD, either during senescence (Orzaez and Granell, 1997), oxidative stress imposed by ozone (Overmyer et al., 2000), or plant pathogen interactions (Lund et al., 1998). In addition, it was suggested that ethylene is crucial for H2O2 production during PCD in tomato suspension cells (de Jong et al., 2002). It was reported that the ethylene receptor ETR1 mediates H2O2 signaling in guard cells in Arabidopsis (Desikan et al., 2005). Together, these findings suggest a cross-talk between ethylene and ROS in plant signaling. Treating Arabidopsis plants with the bacterial elicitor flagellin (flg22) enhanced an oxidative burst which was inhibited in ethylene-insensitive mutants, etr1 and ein2, indicating a requirement of ethylene signaling for ROS production (Mersmann et al., 2010). A synergistic biosynthesis of ethylene and ROS production, mediated by the plasma membrane bound enzymes RbohD and RbohF, was reported in tobacco plants infected with the hemibiotrophic Phytophthora parasitica (Wi et al., 2012).
Transcription Factors
Regulation of gene expression at the transcriptional level influences or controls many of the biological processes in a cell or organism, such as progression through the cell cycle, metabolic and physiological balance, and responses to environment (Riechmann et al., 2000). Plant stress responses are regulated by multiple signaling pathways that activate gene transcription and its downstream machinery. Using data from ROS-related microarray studies, Gadjev et al. (2006) examined the expression of 1,500 TFs in Arabidopsis in response to different ROS, including singlet oxygen, H2O2 and OH∙. They reported that different ROS induced or repressed the expression of about 500 of these annotated TFs in Arabidopsis. Nevertheless, the transcriptional regulation mechanisms mediating ROS signaling is not fully understood. It is suggested that the regulation of the TFs activity by the most important ROS, H2O2, is managed at several levels including: (1) upregulation of TF expression or increasing both mRNA stability and translation; (2) increasing the stability of the TF by decreasing its association with the protein degrading ubiquitin E3 ligase complex or by inhibiting this complex; (3) nucleo-cytoplasmic traffic by transferring or masking nuclear localization signals, or by releasing the TF from partners or from membrane anchors; and (4) DNA binding and nuclear transactivation by adapting TF affinity toward DNA, co-activators or repressors, and by targeting specific regions of chromatin to activate individual genes (Marinho et al., 2014). Many examples of TFs that are regulated by ROS have been revealed. Simple organisms, such as bacteria and yeast, sense the enhanced production of ROS using redox sensitive TFs (Mittler et al., 2004). In bacteria, OxyR (oxygen regulated) and PerR (Peroxide Regulon Repressor) are TFs that are directly activated by H2O2. The tetrameric OxyR protein is characterized as a regulatory protein that activates nine out of twelve early H2O2-induced proteins. The OxyR transcription activator exists in two forms, reduced and oxidized; only the oxidized state is able to initiate transcription (Storz et al., 1990; Storz and Imlayt, 1999). In yeast, four TFs, namely, Yap1, Maf1, Hsf1, and Msn2/4, were reported to be regulated by ROS. For example, Yap1 is regulated by H2O2 at the level of nucleo-cytoplasmic trafficking. Under oxidative stress, the export of Yap1 to the nucleus is decreased and Yap1 is kept longer in the nucleus where it regulates its target genes (Delaunay et al., 2000). Yap1 has a key role in the oxidative stress response, redox homeostasis and electrophilic response, regulating the transcription of genes encoding antioxidant and detoxification enzymes in yeast cells (Marinho et al., 2014). In multicellular organisms, nine different TFs, namely AP-1, NRF2, CREB, HSF1, HIF-1, TP53, NF- κB, NOTCH, SP1, and SCREB-1, are well-characterized to be regulated by ROS (Marinho et al., 2014).
Conclusion
As reviewed here, plants have evolved complicated protection mechanisms to survive different environmental challenges. The recent functional molecular and physiological studies have produced new details attempting to unravel the complexity of these signaling pathways. It is evident now that there is no a specific linear signaling pathway for each stress, instead, there are interconnected networks including common signaling events that are shared by various pathways represented by what we call cross-talk. From these big signaling networks many branches arise for specificity. As ROS are well-known to be produced by plants in response to different biotic and abiotic stresses, they are designated to work at the cross road within these complex signaling networks. ROS play this central signaling role through their evident interactions, whether upstream or downstream, with other key signaling components, including membranes, NADPH oxidases, G-proteins, calcium, redox homeostasis, MAPKs, plant hormones (such as SA, JA, ABA, and ethylene) and TFs. The recent research implies an early and vital role for photosynthesis in sensing various environmental insults, not only abiotic, but also biotic ones, a concept that needs to be taken into consideration when studying stress signaling pathways in plants. Despite all of these achievements, great efforts are still required to be able to reconstruct larger signaling networks that may include ROS at the convergent points. In this regard, bioinformatics and systems biology approaches are nominated to greatly help in constructing global signaling networks. As a result, these global networks would improve our understanding of plant biology and assist us to develop new strategies for higher plant productivity in the face of increasingly severe environmental conditions and the high demand for food, fiber, and energy crops.
Author Contributions
All authors wrote and approved the final version of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
We wish to thank the Australian Research Council (DP110104354), CSIRO and the Egyptian Government for financial support.
References
Abuharbeid, S., Apel, J., Sander, M., Fiedler, B., Langer, M., Zuzarte, M. L., et al. (2004). Cytotoxicity of the novel anti-cancer drug rViscumin depends on HER-2 levels in SKOV-3 cells. Biochem. Biophys. Res. Commun. 321, 403–412. doi: 10.1016/j.bbrc.2004.06.160
Agrawal, V., Zhang, C., Shapiro, A. D., and Dhurjati, P. S. (2004). A dynamic mathematical model to clarify signaling circuitry underlying programmed cell death control in Arabidopsis disease resistance. Biotechnol. Prog. 20, 426–442. doi: 10.1021/bp034226s
Akter, S., Huang, J., Waszczak, C., Jacques, S., Gevaert, K., Van Breusegem, F., et al. (2015). Cysteines under ROS attack in plants: a proteomics view. J. Exp. Bot. 66, 2935–2944. doi: 10.1093/jxb/erv044
Allen, G. J., Kwak, J. M., Chu, S. P., Llopis, J., Tsien, R. Y., Harper, J. F., et al. (1999). Cameleon calcium indicator reports cytoplasmic calcium dynamics in Arabidopsis guard cells. Plant J. 19, 735–747. doi: 10.1046/j.1365-313x.1999.00574.x
Allen, G. J., Muir, S. R., and Sanders, D. (1995). Release of Ca2+ from individual plant vacuoles by both InsP3 and cyclic ADP-ribose. Science 268, 735–737. doi: 10.1126/science.7732384
Ananieva, E. A., Alexieva, V. S., and Popova, L. P. (2002). Treatment with salicylic acid decreases the effects of paraquat on photosynthesis. J. Plant Physiol. 159, 685–693. doi: 10.1078/0176-1617-0706
Anderson, J. P., Badruzsaufari, E., Schenk, P. M., Manners, J. M., Desmond, O. J., Ehlert, C., et al. (2004). Antagonistic interaction between abscisic acid and jasmonate-ethylene signaling pathways modulates defense gene expression and disease resistance in Arabidopsis. Plant Cell 16, 3460–3479. doi: 10.1105/tpc.104.025833
Anderson, L. S., and Mansfield, T. A. (1979). The effects of nitric oxide pollution on the growth of tomato. Environ. Pollut. 20, 113–121. doi: 10.1016/j.jplph.2010.04.007
Angers, B., Castonguay, E., and Massicotte, R. (2010). Environmentally induced phenotypes and DNA methylation: how to deal with unpredictable conditions until the next generation and after. Mol. Ecol. 19, 1283–1295. doi: 10.1111/j.1365-294X.2010.04580.x
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Asada, K. (1999). The water-water cycle in chloroplasts: scavenging of active oxygens and dissipation of excess photons. Annu. Rev. Plant Physiol. Plant. Mol. Biol. 50, 601–639. doi: 10.1146/annurev.arplant.50.1.601
Asada, K. (2006). Production and scavenging of reactive oxygen species in chloroplasts and their functions. Plant Physiol. 141, 391–396. doi: 10.1104/pp.106.082040
Asai, S., Ichikawa, T., Nomura, H., Kobayashi, M., Kamiyoshihara, Y., Mori, H., et al. (2013). The variable domain of a plant calcium-dependent protein kinase (CDPK) confers subcellular localization and substrate recognition for NADPH oxidase. J. Biol. Chem. 288, 14332–14340. doi: 10.1074/jbc.M112.448910
Asai, S., Ohta, K., and Yoshioka, H. (2008). MAPK signaling regulates nitric oxide and NADPH oxidase-dependent oxidative bursts in Nicotiana benthamiana. Plant Cell 20, 1390–1406. doi: 10.1105/tpc.107.055855
Asai, S., and Yoshioka, H. (2009). Nitric oxide as a partner of reactive oxygen species participates in disease resistance to nectrotophic pathogen Botryis cinerea in Nicotiana benthamiana. Mol. Plant Microbe. Interact. 22, 619–629. doi: 10.1094/MPMI-22-6-0619
Asai, T., Stone, J. M., Heard, J. E., Kovtun, Y., Yorgey, P., Sheen, J., et al. (2000). Fumonisin B1-induced cell death in arabidopsis protoplasts requires jasmonate-, ethylene-, and salicylate-dependent signaling pathways. Plant Cell 12, 1823–1836. doi: 10.2307/3871195
Assmann, S. M. (2004). Plant G proteins, phytohormones, and plasticity: three questions and a speculation. Sci. STKE 2004:re20.
Assmann, S. M. (2005). G protein regulation of disease resistance during infection of rice with rice blast fungus. Sci. STKE 2005:cm13.
Atkinson, N. J., and Urwin, P. E. (2012). The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63, 3523–3543. doi: 10.1093/jxb/ers100
Baena-González, E. (2010). Energy signaling in the regulation of gene expression during stress. Mol. Plant 3, 300–313. doi: 10.1093/mp/ssp113
Baginsky, S., Hennig, L., Zimmermann, P., and Gruissem, W. (2010). Gene expression analysis, proteomics, and network discovery. Plant Physiol. 152, 402–410. doi: 10.1104/pp.109.150433
Bais, H. P., Vepachedu, R., Gilroy, S., Callaway, R. M., and Vivanco, J. M. (2003). Allelopathy and exotic plant invasion: from molecules and genes to species interactions. Science 301, 1377–1380. doi: 10.1126/science.1083245
Bardo, S., Robertson, B., and Stephens, G. J. (2002). Presynaptic internal Ca2+ stores contribute to inhibitory neurotransmitter release onto mouse cerebellar Purkinje cells. Br. J. Pharmacol. 137, 529–537. doi: 10.1038/sj.bjp.0704901
Barnes, S. A., McGrath, R. B., and Chua, N. H. (1997). Light signal transduction in plants. Trends Cell Biol. 7, 21–26. doi: 10.1016/S0962-8924(97)10043-5
Barraud, N., Hassett, D. J., Hwang, S. H., Rice, S. A., Kjelleberg, S., and Webb, J. S. (2006). Involvement of nitric oxide in biofilm dispersal of Pseudomonas aeruginosa. J. Bacteriol. 188, 7344–7353. doi: 10.1128/JB.00779-06
Baxter, A., Mittler, R., and Suzuki, N. (2014). ROS as key players in plant stress signaling. J. Exp. Bot. 65, 1229–1240. doi: 10.1093/jxb/ert375
Baxter, C. J., Redestig, H., Schauer, N., Repsilber, D., Patil, K. R., Nielsen, J., et al. (2007). The metabolic response of heterotrophic Arabidopsis cells to oxidative stress. Plant Physiol. 143, 312–325. doi: 10.1104/pp.106.090431
Belhaj, K., Lin, B., and Mauch, F. (2009). The chloroplast protein RPH1 plays a role in the immune response of Arabidopsis to Phytophthora brassicae. Plant J. 58, 287–298. doi: 10.1111/j.1365-313X.2008.03779.x
Beligni, M. V., and Lamattina, L. (1999). Nitric oxide protects against cellular damage produced by methylviologen herbicides in potato plants. Nitric Oxide 3, 199–208. doi: 10.1006/niox.1999.0222
Bell, E., Creelman, R. A., and Mullet, J. E. (1995). A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 92, 8675–8679. doi: 10.1073/pnas.92.19.8675
Berridge, M. J., Lipp, P., and Bootman, M. D. (2000). The versatility and universality of calcium signaling. Nat. Rev. Mol. Cell Biol. 1, 11–21. doi: 10.1038/35036191
Besson-Bard, A., Pugin, A., and Wendehenne, D. (2008). New insights into nitric oxide signaling in plants. Annu. Rev. Plant Biol. 59, 21–39. doi: 10.1146/annurev.arplant.59.032607.092830
Bhattacharjee, S. (2005). Reactive oxygen species and oxidative burst: roles in stress, senescence and signal transduction in plants. Curr. Sci. 89, 1113–1121.
Bienert, G. P., Moller, A. L., Kristiansen, K. A., Schulz, A., Moller, I. M., Schjoerring, J. K., et al. (2007). Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 282, 1183–1192. doi: 10.1074/jbc.M603761200
Binder, K. A., Wegner, L. H., Heidecker, M., and Zimmermann, U. (2003). Gating of Cl-currents in protoplasts from the marine alga Valonia utricularis depends on the transmembrane Cl-gradient and is affected by enzymatic cell wall degradation. J. Membr. Biol. 191, 165–178. doi: 10.1007/s00232-002-1052-2
Bollman, K. M., Aukerman, M. J., Park, M.-Y., Hunter, C., Berardini, T. Z., and Poethig, R. S. (2003). HASTY, the Arabidopsis ortholog of exportin 5/MSN5, regulates phase change and morphogenesis. Development 130, 1493–1504. doi: 10.1242/dev.00362
Borisova, M. M., Kozuleva, M. A., Rudenko, N. N., Naydov, I. A., Klenina, I. B., and Ivanov, B. N. (2012). Photosynthetic electron flow to oxygen and diffusion of hydrogen peroxide through the chloroplast envelope via aquaporins. Biochim. Biophys. Acta 1817, 1314–1321. doi: 10.1016/j.bbabio.2012.02.036
Bouche, N., Yellin, A., Snedden, W. A., and Fromm, H. (2005). Plant-specific calmodulin-binding proteins. Annu. Rev. Plant Biol. 56, 435–466. doi: 10.1146/annurev.arplant.56.032604.144224
Boveris, A., Costa, L. E., Poderoso, J. J., Carreras, M. C., and Cadenas, E. (2000). Regulation of mitochondrial respiration by oxygen and nitric oxide. Annu. N. Y. Acad. Sci. 899, 121–135. doi: 10.1111/j.1749-6632.2000.tb06181.x
Bowler, C., and Fluhr, R. (2000). The role of calcium and activated oxygens as signals for controlling cross-tolerance. Trends Plant Sci. 5, 241–246. doi: 10.1016/S1360-1385(00)01628-9
Bowler, C., Montagu, M. V., and Inze, D. (1992). Superoxide dismutase and stress tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 43, 83–116. doi: 10.1146/annurev.pp.43.060192.000503
Bright, J., Desikan, R., Hancock, J. T., Weir, I. S., and Neill, S. J. (2006). ABA-induced NO generation and stomatal closure in Arabidopsis are dependent on H2O2 synthesis. Plant J. 45, 113–122. doi: 10.1111/j.1365-313X.2005.02615.x
Buchanan, B. B., Gruissem, W., and Jones, R. L. (2000). Biochemistry and Molecular Biology of Plants. Rockville, MD: American Society of Plant Physiologists.
Caro, A., and Puntarulo, S. (1998). Nitric oxide decreases superoxide anion generation by microsomes from soybean embryonic axes. Physiol. Plant. 104, 357–364. doi: 10.1034/j.1399-3054.1998.1040310.x
Carvalho, A., Cunha, C., Almeida, A. J., Osório, N. S., Saraiva, M., Teixeira-Coelho, M., et al. (2012). The rs5743836 polymorphism in TLR9 confers a population-based increased risk of non-Hodgkin lymphoma. Genes Immun. 13, 197–201. doi: 10.1038/gene.2011.59
Castells, E., Puigdomènech, P., and Casacuberta, J. M. (2006). Regulation of the kinase activity of the MIK GCK-like MAP4K by alternative splicing. Plant. Mol. Biol. 61, 747–756. doi: 10.1007/s11103-006-0046-3
Chaouch, S., Queval, G., and Noctor, G. (2012). AtRbohF is a crucial modulator of defense-associated metabolism and a key factor in the interplay between intracellular oxidative stress and pathogenesis responses in Arabidopsis. Plant J. 69, 613–627. doi: 10.1111/j.1365-313X.2011.04816.x
Chen, W., Provart, N. J., Glazebrook, J., Katagiri, F., Chang, H. S., Eulgem, T., et al. (2002). Expression profile matrix of Arabidopsis transcription factor genes suggests their putative functions in response to environmental stresses. Plant Cell 14, 559–574. doi: 10.1105/tpc.010410
Chinnusamy, V., and Zhu, J. K. (2009). Epigenetic regulation of stress responses in plants. Curr. Opin. Plant Biol. 12, 133–139. doi: 10.1016/j.pbi.2008.12.006
Chiwocha, S. D., Cutler, A. J., Abrams, S. R., Ambrose, S. J., Yang, J., Ross, A. R., et al. (2005). The etr1-2 mutation in Arabidopsis thaliana affects the abscisic acid, auxin, cytokinin and gibberellin metabolic pathways during maintenance of seed dormancy, moist-chilling and germination. Plant J. 42, 35–48. doi: 10.1111/j.1365-313X.2005.02359.x
Choi, W. G., Toyota, M., Kim, S. H., Hilleary, R., and Gilroy, S. (2014). Salt stress-induced Ca2+ waves are associated with rapid, long-distance root-to-shoot signaling in plants. Proc. Natl. Acad. Sci. U.S.A. 111, 6497–6502. doi: 10.1073/pnas.1319955111
Coego, A., Ramirez, V., Gil, M. J., Flors, V., Mauch-Mani, B., and Vera, P. (2005). An Arabidopsis homeodomain transcription factor, OVEREXPRESSOR OF CATIONIC PEROXIDASE 3, mediates resistance to infection by necrotrophic pathogens. Plant Cell 17, 2123–2137. doi: 10.1105/tpc.105.032375
Correa-Aragunde, N., Foresi, N., and Lamattina, L. (2015). Nitric oxide is a ubiquitous signal for maintaining redox balance in plant cells: regulation of ascorbate peroxidase as a case study. J. Exp. Bot. 66, 2913–2921. doi: 10.1093/jxb/erv073
Cosgrove, D. J., and Hedrich, R. (1991). Stretch-activated chloride, potassium, and calcium channels coexisting in plasma membranes of guard cells of Vicia faba L. Planta 186, 143–153. doi: 10.1007/BF00201510
Costa, V., and Moradas-Ferreira, P. (2001). Oxidative stress and signal transduction in Saccharomyces cerevisiae: insights into ageing, apoptosis and diseases. Mol. Aspects Med. 22, 217–246. doi: 10.1016/S0098-2997(01)00012-7
Covarrubias, A. A., and Reyes, J. L. (2010). Post-transcriptional gene regulation of salinity and drought responses by plant microRNAs. Plant Cell Environ. 33, 481–489. doi: 10.1111/j.1365-3040.2009.02048.x
Czempinski, K., Zimmermann, S., Ehrhardt, T., and Muller-Rober, B. (1997). New structure and function in plant K+ channels: KCO1, an outward rectifier with a steep Ca2+ dependency. EMBO J. 16, 2565–2575. doi: 10.1093/emboj/16.10.2565
Dalle-Donne, I., Rossi, R., Colombo, G., Giustarini, D., and Milzani, A. (2009). Protein S-glutathionylation: a regulatory device from bacteria to humans. Trends Biochem. Sci. 34, 85–96. doi: 10.1016/j.tibs.2008.11.002
Dangl, J. L., and Jones, J. D. G. (2001). Plant pathogens and integrated defense responses to infection. Nature 411, 826–833. doi: 10.1038/35081161
Das, R., and Pandey, G. K. (2010). Expressional analysis and role of calcium regulated kinases in abiotic stress signaling. Curr. Genomics 11, 2–13. doi: 10.2174/138920210790217981
D’Autreaux, B., and Toledano, M. B. (2007). ROS as signaling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8, 813–824. doi: 10.1038/nrm2256
de Jong, A., Yakimova, E., Kapchina, V., and Woltering, E. (2002). A critical role for ethylene in hydrogen peroxide release during programmed cell death in tomato suspension cells. Planta 214, 537–545. doi: 10.1007/s004250100654
de Pinto, M. C., and De Gara, L. (2004). Changes in the ascorbate metabolism of apoplastic and symplastic spaces are associated with cell differentiation. J. Exp. Bot. 55, 2559–2569. doi: 10.1093/jxb/erh253
De Smet, R., and Van de Peer, Y. (2012). Redundancy and rewiring of genetic networks following genome-wide duplication events. Curr. Opin. Plant Biol. 15, 168–176. doi: 10.1016/j.pbi.2012.01.003
Del Río, L. A. (2015). ROS and RNS in plant physiology: an overview. J. Exp. Bot. 66, 2827–2837. doi: 10.1093/jxb/erv099
Delaunay, A., Isnard, A. D., and Toledano, M. B. (2000). H2O2 sensing through oxidation of the Yap1 transcription factor. EMBO J. 19, 5157–5166. doi: 10.1093/emboj/19.19.5157
Delledonne, M., Xia, Y., Dixon, R. A., and Lamb, C. (1998). Nitric oxide functions as a signal in plant disease resistance. Nature 394, 585–588. doi: 10.1038/29087
Delledonne, M., Zeier, J., Marocco, A., and Lamb, C. (2001). Signal interactions between nitric oxide and reactive oxygen intermediates in the plant hypersensitive disease resistance response. Proc. Natl. Acad. Sci. U.S.A. 98, 13454–13459. doi: 10.1073/pnas.231178298
Denness, L., McKenna, J. F., Segonzac, C., Wormit, A., Madhou, P., Bennett, M., et al. (2011). Cell wall damage-induced lignin biosynthesis is regulated by a reactive oxygen species- and jasmonic acid-dependent process in Arabidopsis. Plant Physiol. 156, 1364–1374. doi: 10.1104/pp.111.175737
Desikan, R., Hancock, J. T., Bright, J., Harrison, J., Weir, I., Hooley, R., et al. (2005). A role for ETR1 in hydrogen peroxide signaling in stomatal guard cells. Plant Physiol. 137, 831–834. doi: 10.1104/pp.104.056994
Desikan, R., Hancock, J. T., Ichimura, K., Shinozaki, K., and Neill, S. J. (2001). Harpin induces activation of the Arabidopsis mitogen-activated protein kinases AtMPK4 and AtMPK6. Plant Physiol. 126, 1579–1587. doi: 10.1104/pp.126.4.1579
Dietz, K. J. (2003). Redox control, redox signaling, and redox homeostasis in plant cells. Int. Rev. Cytol. 228, 141–193. doi: 10.1016/S0074-7696(03)28004-9
Dietz, K. J. (2008). Redox signal integration: from stimulus to networks and genes. Physiol. Plant. 133, 459–468. doi: 10.1111/j.1399-3054.2008.01120.x
Dietz, K. J., Jacquot, J. P., and Harris, G. (2010). Hubs and bottlenecks in plant molecular signaling networks. New Phytol. 188, 919–938. doi: 10.1111/j.1469-8137.2010.03502.x
Ding, J., Li, X., and Hu, H. (2015). MicroRNA modules prefer to bind weak and unconventional target sites. Bioinformatics 31, 1366–1374. doi: 10.1093/bioinformatics/btu833
Ding, Y., Tao, Y., and Zhu, C. (2013). Emerging roles of microRNAs in the mediation of drought stress response in plants. J. Exp. Bot. 64, 3077–3086. doi: 10.1093/jxb/ert164
Dodd, A. N., Kudla, J., and Sanders, D. (2010). The language of calcium signaling. Annu. Rev. Plant Biol. 61, 593–620. doi: 10.1146/annurev-arplant-070109-104628
Drerup, M. M., Schlücking, K., Hashimoto, K., Manishankar, P., Steinhorst, L., Kuchitsu, K., et al. (2013). The Calcineurin B-like calcium sensors CBL1 and CBL9 together with their interacting protein kinase CIPK26 regulate the Arabidopsis NADPH oxidase RBOHF. Mol. Plant. 6, 559–569. doi: 10.1093/mp/sst009
Dubiella, U., Seybold, H., Durian, G., Komander, E., Lassig, R., Witte, C. P., et al. (2013). Calcium-dependent protein kinase/NADPH oxidase activation circuit is required for rapid defense signal propagation. Proc. Natl. Acad. Sci. U.S.A. 110, 8744–8749. doi: 10.1073/pnas.1221294110
Durner, J., Gow, A. J., Stamler, J. S., and Glazebrook, J. (1999). Ancient origins of nitric oxide signaling in biological systems. Proc. Natl. Acad. Sci. U.S.A. 96, 14206–14207. doi: 10.1073/pnas.96.25.14206
Durner, J., Wendehenne, D., and Klessig, D. F. (1998). Defense gene induction in tobacco by nitric oxide, cyclic GMP, and cyclic ADP-ribose. Proc. Natl. Acad. Sci. U.S.A. 95, 10328–10333. doi: 10.1073/pnas.95.17.10328
Dynowski, M., Schaaf, G., Loque, D., Moran, O., and Ludewig, U. (2008). Plant plasma membrane water channels conduct the signaling molecule H2O2. Biochem. J. 414, 53–61. doi: 10.1042/BJ20080287
Ermolayeva, E., Sanders, D., and Johannes, E. (1997). Ionic mechanism and role of phytochrome-mediated membrane depolarisation in caulonemal side branch initial formation in the moss Physcomitrella patens. Planta 201, 109–118. doi: 10.1007/BF01007695
Evans, N. H., McAinsh, M. R., Hetherington, A. M., and Knight, M. R. (2005). ROS perception in Arabidopsis thaliana: the ozone-induced calcium response. Plant J. 41, 615–626. doi: 10.1111/j.1365-313X.2004.02325.x
Farmer, E. E., and Ryan, C. A. (1992). Octadecanoid precursors of jasmonic acid activate the synthesis of wound-inducible proteinase inhibitors. Plant Cell 4, 129–134. doi: 10.2307/3869566
Fey, V., Wagner, R., Brautigam, K., Wirtz, M., Hell, R., Dietzmann, A., et al. (2005). Retrograde plastid redox signals in the expression of nuclear genes for chloroplast proteins of Arabidopsis thaliana. J. Biol. Chem. 280, 5318–5328. doi: 10.1074/jbc.M406358200
Finka, A., Cuendet, A. F., Maathuis, F. J., Saidi, Y., and Goloubinoff, P. (2012). Plasma membrane cyclic nucleotide gated calcium channels control land plant thermal sensing and acquired thermotolerance. Plant Cell 24, 3333–3348. doi: 10.1105/tpc.112.095844
Foyer, C. H., and Noctor, G. (2005). Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17, 1866–1875. doi: 10.1105/tpc.105.033589
Foyer, C. H., and Noctor, G. (2009). Redox regulation in photosynthetic organisms: signaling, acclimation, and practical implications. Antioxid. Redox Signal. 11, 861–905. doi: 10.1089/ars.2008.2177
Foyer, C. H., and Noctor, G. (2012). Managing the cellular redox hub in photosynthetic organisms. Plant Cell Environ. 35, 199–201. doi: 10.1111/j.1365-3040.2011.02453.x
Fryer, M. J., Ball, L., Oxborough, K., Karpinski, S., Mullineaux, P. M., and Baker, N. R. (2003). Control of Ascorbate Peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J. 33, 691–705. doi: 10.1046/j.1365-313X.2003.01656.x
Fujiki, Y., Ito, M., Nishida, I., and Watanabe, A. (2000). Multiple signaling pathways in gene expression during sugar starvation. Pharmacological analysis of din gene expression in suspension-cultured cells of Arabidopsis. Plant Physiol. 124, 1139–1148. doi: 10.1104/pp.124.3.1139
Furbank, R. T., Badger, M. R., and Osmond, C. B. (1983). Photoreduction of oxygen in methophyll chloroplast of C4 plants. Plant Physiol. 37, 1038–1041. doi: 10.1104/pp.73.4.1038
Gadjev, I., Vanderauwera, S., Gechev, T. S., Laloi, C., Minkov, I. N., Shulaev, V., et al. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141, 436–445. doi: 10.1104/pp.106.078717
Gasch, A., Spellman, P., Kao, C., Harel, O., Eisen, M., Storz, G., et al. (2000). Genomic expression programs in the response of yeast cells to environmental changes. Mol. Biol. Cell 11, 4241–4257. doi: 10.1091/mbc.11.12.4241
Georgiou, G. (2002). How to Flip the (Redox) Switch. Cell 111, 607–610. doi: 10.1016/S0092-8674(02)01165-0
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Glatz, A., Vass, I., Los, D. A., and Vigh, L. (1999). The Synechocystis model of stress: from molecular chaperonesto membranes. Plant Physiol. Biochem. 37, 1–12. doi: 10.1016/S0981-9428(99)80061-8
Glazebrook, J., Chen, W., Estes, B., Chang, H. S., Nawrath, C., Metraux, J. P., et al. (2003). Topology of the network integrating salicylate and jasmonate signal transduction derived from global expression phenotyping. Plant J. 34, 217–228. doi: 10.1046/j.1365-313X.2003.01717.x
Go, Y. M., and Jones, D. P. (2010). Cysteine/cystine redox signaling in cardiovascular disease. Free Radic. Biol. Med. 50, 495–509. doi: 10.1016/j.freeradbiomed.2010.11.029
Godoy Herz, M. A., Kornblihtt, A. R., Barta, A., Kalyna, M., and Petrillo, E. (2014). Shedding light on the chloroplast as a remote control of nuclear gene expression. Plant Signal. Behav. 9:e976150. doi: 10.4161/15592324.2014.976150
Goulet, A. C., Goldmacher, V. S., Lambert, J. M., Baron, C., Roy, D. C., and Kouassi, E. (1997). Conjugation of blocked ricin to an anti-CD19 monoclonal antibody increases antibody-induced cell calcium mobilization and CD19 internalization. Blood 90, 2364–2375.
Gu, Z., Steinmetz, L. M., Gu, X., Scharfe, C., Davis, R. W., and Li, W.-H. (2003). Role of duplicate genes in genetic robustness against null mutations. Nature 421, 63–66. doi: 10.1038/nature01198
Guo, Y., Xiong, L., Ishitani, M., and Zhu, J. K. (2002). An Arabidopsis mutation in translation elongation factor 2 causes superinduction of CBF/DREB1 transcription factor genes but blocks the induction of their downstream targets under low temperatures. Proc. Natl. Acad. Sci. U.S.A. 99, 7786–7791. doi: 10.1073/pnas.112040099
Gustin, M. C., Albertyn, J., Alexander, M., and Davenport, K. (1998). MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62, 1264–1300.
Gutkind, J. S. (2000). Regulation of mitogen-activated protein kinase signaling networks by G protein-coupled receptors. Sci STKE 2000:re1.
Halliwell, B. (1991). Reactive oxygen species in living systems: source, biochemistry, and role in human disease. Am. J. Med. 91, 14S–22S. doi: 10.1016/0002-9343(91)90279-7
Halliwell, B. (2006). Reactive species and antioxidants. Redox biology is a fundamental theme of aerobic life. Plant Physiol. 141, 312–322. doi: 10.1104/pp.106.077073
Halliwell, B., and Gutteridge, J. M. C. (1999). Free Radicals in Biology and Medicine. Oxford: Oxford University Press.
Han, Y., Chaouch, S., Mhamdi, A., Queval, G., Zechmann, B., and Noctor, G. (2013a). Functional analysis of Arabidopsis mutants points to novel roles for glutathione in coupling H2O2 to activation of salicylic acid accumulation and signaling. Antioxid. Redox. Signal. 18, 2106–2121. doi: 10.1089/ars.2012.5052
Han, Y., Mhamdi, A., Chaouch, S., and Noctor, G. (2013b). Regulation of basal and oxidative stress-triggered jasmonic acid-related gene expression by glutathione. Plant Cell Environ. 36, 1135–1146. doi: 10.1111/pce.12048
Hausladen, A., Gow, A. J., and Stamler, J. S. (1998). Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc. Natl. Acad. Sci. U.S.A. 95, 14100–14105. doi: 10.1073/pnas.95.24.14100
Heller, J., and Tudzynski, P. (2011). Reactive oxygen species in phytopathogenic fungi: signaling, development, and disease. Annu. Rev. Phytopathol. 49, 369–390. doi: 10.1146/annurev-phyto-072910-095355
Horvath, I., Glatz, A., Varvasovszki, V., Torok, Z., Pali, T., Balogh, G., et al. (1998). Membrane physical state controls the signaling mechanism of the heat shock response in Synechocystis PCC 6803: identification of hsp17 as a ”fluidity gene”. Proc. Natl. Acad. Sci. U.S.A. 95, 3513–3518. doi: 10.1073/pnas.95.7.3513
Hossain, M. A., Bhattacharjee, S., Armin, S. M., Qian, P., Xin, W., Li, H. Y., et al. (2015). Hydrogen peroxide priming modulates abiotic oxidative stress tolerance: insights from ROS detoxification and scavenging. Front. Plant Sci. 6:420. doi: 10.3389/fpls.2015.00420
Hu, X., Jiang, M., Zhang, J., Zhang, A., Lin, F., and Tan, M. (2007). Calcium-calmodulin is required for abscisic acid-induced antioxidant defense and functions both upstream and downstream of H2O2 production in leaves of maize (Zea mays) plants. New Phytol. 173, 27–38. doi: 10.1088/1367-2630/9/2/027
Hu, X., Zhang, A., Zhang, J., and Jiang, M. (2006). Abscisic acid is a key inducer of hydrogen peroxide production in leaves of maize plants exposed to water stress. Plant Cell Physiol. 47, 1484–1495. doi: 10.1093/pcp/pcl014
Hufton, C. A., Besford, R. T., and Wellburn, A. R. (1996). Effects of NO (+ NO2) pollution on growth, nitrate reductase activities and associated protein contents in glasshouse lettuce grown hydroponically in winter with CO2 enrichment. New Phytol. 133, 495–501. doi: 10.1111/j.1469-8137.1996.tb01917.x
Huner, N. P. A., Maxwell, D. P., Gray, G. R., Savitch, L. V., Krol, M., Ivanov, A. G., et al. (1996). Sensing environmental temperature change through imbalances between energy supply and energy consumption: redox state of photosystem II. Physiol. Plant. 98, 358–364. doi: 10.1034/j.1399-3054.1996.980218.x
Ichimura, K., Shinozaki, K., Tena, G., Sheen, J., Henry, Y., Champion, A., et al. (2002). Mitogen-activated protein kinase cascades in plants: a new nomenclature. Trends Plant Sci. 7, 301–308. doi: 10.1016/S1360-1385(02)02302-6
Imlay, J. A. (2003). Pathways of oxidative damage. Annu. Rev. Microbiol. 57, 395–418. doi: 10.1146/annurev.micro.57.030502.090938
Jasid, S., Simontacchi, M., and Puntarulo, S. (2008). Exposure to nitric oxide protects against oxidative damage but increases the labile iron pool in sorghum embryonic axes. J. Exp. Bot. 59, 3953–3962. doi: 10.1093/jxb/ern235
Joo, J. H., Wang, S., Chen, J. G., Jones, A. M., and Fedoroff, N. V. (2005). Different signaling and cell death roles of heterotrimeric G protein α and β subunits in the Arabidopsis oxidative stress response to ozone. Plant Cell 17, 957–970. doi: 10.1105/tpc.104.029603
Kadota, Y., Sklenar, J., Derbyshire, P., Stransfeld, L., Asai, S., Ntoukakis, V., et al. (2014). Direct regulation of the NADPH Oxidase RBOHD by the PRR-associated kinase BIK1 during plant immunity. Mol. Cell. 54, 43–55. doi: 10.1016/j.molcel.2014.02.021
Kazan, K. (2003). Alternative splicing and proteome diversity in plants: the tip of the iceberg has just emerged. Trends Plant Sci. 8, 468–471. doi: 10.1016/j.tplants.2003.09.001
Khokon, A. R., Okuma, E., Hossain, M. A., Munemasa, S., Uraji, M., Nakamura, Y., et al. (2011). Involvement of extracellular oxidative burst in salicylic acid-induced stomatal closure in Arabidopsis. Plant Cell Environ. 34, 434–443. doi: 10.1111/j.1365-3040.2010.02253.x
Khraiwesh, B., Zhu, J. K., and Zhu, J. (2012). Role of miRNAs and siRNAs in biotic and abiotic stress responses of plants. Biochim. Biophys. Acta 1819, 137–148. doi: 10.1016/j.bbagrm.2011.05.001
Kimura, S., Kaya, H., Kawarazaki, T., Hiraoka, G., Senzaki, E., Michikawa, M., et al. (2012). Protein phosphorylation is a prerequisite for the Ca2+-dependent activation of Arabidopsis NADPH oxidases and may function as a trigger for the positive feedback regulation of Ca2+ and reactive oxygen species. Biochim. Biophys. Acta. 1823, 398–405. doi: 10.1016/j.bbamcr.2011.09.011
Klepper, L. (1979). Nitric oxide (NO) and nitrogen dioxide (NO2) emissions from herbicide-treated soybean plants. Atmos Environ. 13, 537–542. doi: 10.1016/0004-6981(79)90148-3
Knight, H., and Knight, M. R. (2001). Abiotic stress signaling pathways: specificity and cross-talk. Trends Plant Sci. 6, 262–267. doi: 10.1016/S1360-1385(01)01946-X
Königshofer, H., Tromballa, H. W., and Loppert, H. G. (2008). Early events in signaling high-temperature stress in tobacco BY2 cells involve alterations in membrane fluidity and enhanced hydrogen peroxide production. Plant Cell Environ. 31, 1771–1780. doi: 10.1111/j.1365-3040.2008.01880.x
Kornblihtt, A. R. (2005). Promoter usage and alternative splicing. Curr. Opin. Cell Biol. 17, 262–268. doi: 10.1016/j.ceb.2005.04.014
Kotchoni, S. O., and Gachomo, E. W. (2006). The reactive oxygen species network pathways: an essential prerequisite for perception of pathogen attack and disease resistance in plants. J. Biosci. 31, 389–404. doi: 10.1007/BF02704112
Koussevitzky, S., Nott, A., Mockler, T. C., Hong, F., Sachetto-Martins, G., Surpin, M., et al. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719. doi: 10.1126/science.%201140516
Kovtun, Y., Chiu, W. L., Tena, G., and Sheen, J. (2000). Functional analysis of oxidative stress-activated MAPK cascade in plants. Proc. Natl. Acad. Sci. U.S.A. 97, 2940–2945. doi: 10.1073/pnas.97.6.2940
Krasensky, J., and Jonak, C. (2012). Drought salt, and temperature stress-induced metabolic rearrangement and regulatory networks. J. Exp. Bot. 4, 1593–1608. doi: 10.1093/jxb/err460
Kromer, S. (1995). Respiration during photosynthesis. Annu. Rev. Plant Physiol. Plant Mol. Biol. 46, 45–70. doi: 10.1146/annurev.pp.46.060195.000401
Kwak, J. M., Mori, I. C., Pei, Z. M., Leonhardt, N., Torres, M. A., Dangl, J. L., et al. (2003). NADPH oxidase AtrbohD and AtrbohF genes function in ROS-dependent ABA signaling in Arabidopsis. EMBO J. 22, 2623–2633. doi: 10.1093/emboj/cdg277
Kyriakis, J. M., and Avruch, J. (1996). Sounding the alarm: protein kinase cascades activated by stress and inflammation. J. Biol. Chem. 271, 24313–24316. doi: 10.1074/jbc.271.40.24313
Laloi, C., Apel, K., and Danon, A. (2004). Reactive oxygen signaling: the latest news. Curr. Opin. Plant Biol. 7, 323–328. doi: 10.1016/j.pbi.2004.03.005
Lamb, C., and Dixon, R. A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant. Mol. Biol. 48, 251–275. doi: 10.1146/annurev.arplant.48.1.251
Leon-Reyes, A., Spoel, S. H., De Lange, E. S., Abe, H., Kobayashi, M., Tsuda, S., et al. (2009). Ethylene modulates the role of NONEXPRESSOR OF PATHOGENESIS-RELATED GENES1 in cross talk between salicylate and jasmonate signaling. Plant Physiol. 149, 1797–1809. doi: 10.1104/pp.108.133926
Leshem, Y. Y., and Haramaty, E. (1996). The characterization and contrasting effects of the nitric oxide free radical in vegetative stress and senescence of Pisum sativum Linn. Foliage. J. Plant Physiol. 148, 258–263. doi: 10.1016/S0176-1617(96)80251-3
Leung, J., and Giraudat, J. (1998). Abscisic acid signal transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 199–222. doi: 10.1146/annurev.arplant.49.1.199
Levine, A., Tenhaken, R., Dixon, R., and Lamb, C. (1994). H2O2 from the oxidative burst orchestrates the plant hypersensitive disease resistance response. Cell 79, 583–593. doi: 10.1016/0092-8674(94)90544-4
Li, H., Shen, J. J., Zheng, Z. L., Lin, Y., and Yang, Z. (2001). The Rop GTPase switch controls multiple developmental processes in Arabidopsis. Plant Physiol. 126, 670–684. doi: 10.1104/pp.126.2.670
Li, T., Li, H., Zhang, Y. X., and Liu, J. Y. (2011). Identification and analysis of seven H2O2-responsive miRNAs and 32 new miRNAs in the seedlings of rice (Oryza sativa L. ssp. indica). Nucleic Acids Res. 39, 2821–2833. doi: 10.1093/nar/gkq1047
Liu, J., and Coaker, G. (2008). Nuclear trafficking during plant innate immunity. Mol. Plant 1, 411–422. doi: 10.1093/mp/ssn010
Lukosz, M., Jakob, S., Büchner, N., Zschauer, T. C., Altschmied, J., and Haendeler, J. (2010). Nuclear redox signaling. Antioxid. Redox Signal 12, 713–742. doi: 10.1089/ars.2009.2609
Lund, S. T., Stall, R. E., and Klee, H. J. (1998). Ethylene regulates the susceptible response to pathogen infection in tomato. Plant Cell 10, 371–382. doi: 10.1105/tpc.10.3.371
Ma, L., Zhang, H., Sun, L., Jiao, Y., Zhang, G., Miao, C., et al. (2012). NADPH oxidase AtrbohD and AtrbohF function in ROS-dependent regulation of Na+/K+ homeostasis in Arabidopsis under salt stress. J. Exp. Bot. 63, 305–317. doi: 10.1093/jxb/err280
Madlung, A., and Comai, L. (2004). The effect of stress on genome regulation and structure. Ann. Bot. 94, 481–495. doi: 10.1093/aob/mch172
Mandadi, K. K., Pyle, J. D., and Scholthof, K. G. (2015). Characterization of SCL33 splicing patterns during diverse virus infections in Brachypodium distachyon. Plant Signal. Behav. 10:e1042641. doi: 10.1080/15592324.2015.1042641
Marinho, H. S., Real, C., Cyrne, L., Soares, H., and Antunes, F. (2014). Hydrogen peroxide sensing, signaling and regulation of transcription factors. Redox Biol. 2, 35–62. doi: 10.1016/j.redox.2014.02.006
Maruta, N., Trusov, Y., Brenya, E., Parekh, U., and Botella, J. R. (2015). Membrane-localized extra-large G proteins and Gbg of the heterotrimeric G proteins form functional complexes engaged in plant immunity in Arabidopsis. Plant Physiol. 167, 1004–1016. doi: 10.1104/pp.114.255703
Maruta, T., Inoue, T., Tamoi, M., Yabuta, Y., Yoshimura, K., Ishikawa, T., et al. (2011). Arabidopsis NADPH oxidases, AtrbohD and AtrbohF, are essential for jasmonic acid-induced expression of genes regulated by MYC2 transcription factor. Plant Sci. 180, 655–660. doi: 10.1016/j.plantsci.2011.01.014
Mauch-Mani, B., and Mauch, F. (2005). The role of abscisic acid in plant-pathogen interactions. Curr. Opin. Plant Biol. 8, 409–414. doi: 10.1016/j.pbi.2005.05.015
Maxwell, D. P., Wang, Y., and McIntosh, L. (1999). The alternative oxidase lowers mitochondrial reactive oxygen production in plant cells. Proc. Natl. Acad. Sci. U.S.A. 96, 8271–8276. doi: 10.1073/pnas.96.14.8271
McAinsh, M. R., and Pittman, J. K. (2009). Shaping the calcium signature. New Phytol. 181, 275–294. doi: 10.1111/j.1469-8137.2008.02682.x
McCormack, E., Tsai, Y. C., and Braam, J. (2005). Handling calcium signaling: Arabidopsis CaMs and CMLs. Trends Plant Sci. 10, 383–389. doi: 10.1016/j.tplants.2005.07.001
Merkle, T. (2004). Nucleo-cytoplasmic partitioning of proteins in plants: implications for the regulation of environmental and developmental signaling. Curr. Genet. 44, 231–260. doi: 10.1007/s00294-003-0444-x
Mersmann, S., Bourdais, G., Rietz, S., and Robatzek, S. (2010). Ethylene signaling regulates accumulation of the fls2 receptor and is required for the oxidative burst contributing to plant immunity. Plant Physiol. 54, 391–400. doi: 10.1104/pp.110.154567
Mhamdi, A., Hager, J., Chaouch, S., Queval, G., Han, Y., Taconnat, L., et al. (2010). Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 153, 1144–1160. doi: 10.1104/pp.110.153767
Miller, G., Schlauch, K., Tam, R., Cortes, D., Torres, M. A., Shulaev, V., et al. (2009). The plant NADPH oxidase RBOHD mediates rapid systemic signaling in response to diverse stimuli. Sci. Signal. 2:ra45. doi: 10.1126/scisignal.2000448
Miller, G., Suzuki, N., Ciftci-Yilmaz, S., and Mittler, R. (2010). Reactive oxygen species homeostasis and signaling during drought and salinity stresses. Plant Cell Environ. 33, 453–467. doi: 10.1111/j.1365-3040.2009.02041.x
Mirouze, M., and Paszkowski, J. (2011). Epigenetic contribution to stress adaptation in plants. Curr. Opin. Plant Biol. 14, 267–274. doi: 10.1016/j.pbi.2011.03.004
Mittler, R. (2002). Oxidative stress, antioxidants and stress tolerance. Trends Plant Sci. 7, 405–410. doi: 10.1016/S1360-1385(02)02312-9
Mittler, R., Vanderauwera, S., Gollery, M., and Van Breusegem, F. (2004). Reactive oxygen gene network of plants. Trends Plant Sci. 9, 490–498. doi: 10.1016/j.tplants.2004.08.009
Mittler, R., Vanderauwera, S., Suzuki, N., Miller, G., Tognetti, V. B., Vandepoele, K., et al. (2011). ROS signaling: the new wave? Trends Plant Sci. 16, 300–309. doi: 10.1016/j.tplants.2011.03.007
Miura, K., Okamoto, H., Okuma, E., Shiba, H., Kamada, H., Hasegawa, P. M., et al. (2013). SIZ1 deficiency causes reduced stomatal aperture and enhanced drought tolerance via controlling salicylic acid-induced accumulation of reactive oxygen species in Arabidopsis. Plant J. 73, 91–104. doi: 10.1111/tpj.12014
Moan, J. (1990). On the diffusion length of singlet oxygen in cells and tissues. J. Photochem. Photobiol. 6, 343–344. doi: 10.1016/1011-1344(90)85104-5
Molassiotis, A., and Fotopoulos, V. (2011). Oxidative and nitrosative signaling in plants: two branches in the same tree? Plant Signal. Behav. 6, 210–214. doi: 10.4161/psb.6.2.14878
Møller, I. M., Jensen, P. E., and Hansson, A. (2007). Oxidative modifications to cellular components in plants. Annu. Rev. Plant Biol. 58, 459–481. doi: 10.1146/annurev.arplant.58.032806.103946
Moller, I. M., and Sweetlove, L. J. (2010). ROS signaling - specificity is required. Trends Plant Sci. 15, 370–374. doi: 10.1016/j.tplants.2010.04.008
Monshausen, G. B., Bibikova, T. N., Weisenseel, M. H., and Gilroy, S. (2009). Ca2+ regulates reactive oxygen species production and pH during mechanosensing in Arabidopsis roots. Plant Cell 21, 2341–2356. doi: 10.1105/tpc.109.068395
Moreau, M., Lindermayr, C., Durner, J., and Klessig, D. F. (2010). NO synthesis and signaling in plants–where do we stand? Physiol. Plant. 138, 372–383. doi: 10.1111/j.1399-3054.2009.01308.x
Mubarakshina, M. M., Ivanov, B. N., Naydov, I. A., Hillier, W., Badger, M. R., and Krieger-Liszkay, A. (2010). Production and diffusion of chloroplastic H2O2 and its implication to signaling. J. Exp. Bot. 61, 3577–3587. doi: 10.1093/jxb/erq171
Mühlenbock, P., Szechynska-Hebda, M., Plaszczyca, M., Baudo, M., Mateo, A., Mullineaux, P. M., et al. (2008). Chloroplast signaling and LESION SIMULATING DISEASE1 regulate crosstalk between light acclimation and immunity in Arabidopsis. Plant Cell 20, 2339–2356. doi: 10.1105/tpc.108.059618
Mur, L. A., Kenton, P., Atzorn, R., Miersch, O., and Wasternack, C. (2006). The outcomes of concentration-specific interactions between salicylate and jasmonate signaling include synergy, antagonism, and oxidative stress leading to cell death. Plant Physiol. 140, 249–262. doi: 10.1104/pp.105.072348
Murata, N., and Los, D. A. (1997). Membrane fluidity and temperature perception. Plant Physiol. 115, 875–879.
Nagarathnam, B., Kalaimathy, S., Balakrishnan, V., and Sowdhamini, R. (2012). Cross-genome clustering of human and C. elegans G-Protein Coupled Receptors. Evol. Bioinform. 8, 229–259. doi: 10.4137/EBO.S9405
Nakagami, H., Pitzschke, A., and Hirt, H. (2005). Emerging MAP kinase pathways in plant stress signaling. Trends Plant Sci. 10, 339–346. doi: 10.1016/j.tplants.2005.05.009
Nathan, C. (2003). Specificity of a third kind: reactive oxygen and nitrogen intermediates in cell signaling. J. Clin. Invest. 111, 769–778. doi: 10.1172/JCI200318174
Navarro, L., Dunoyer, P., Jay, F., Arnold, B., Dharmasiri, N., Estelle, M., et al. (2006). A plant miRNA contributes to antibacterial resistance by repressing auxin signaling. Science 312, 436–439. doi: 10.1126/science.1126088
Neill, S. J., Desikan, R., Clarke, A., Hurst, R. D., and Hancock, J. T. (2002). Hydrogen peroxide and nitric oxide as signaling molecules in plants. J. Exp. Bot. 53, 1237–1247. doi: 10.1093/jexbot/53.372.1237
Neill, S. J., Desikan, R., and Hancock, J. T. (2003). Nitric oxide signaling in plants. New Phytol. 159, 11–35. doi: 10.1046/j.1469-8137.2003.00804.x
Niedre, M., Patterson, M. S., and Wilson, B. C. (2002). Direct near-infrared luminescence detection of singlet oxygen generated by photodynamic therapy in cells in vitro and tissues in vivo. Photochem. Photobiol. 75, 382–391. doi: 10.1562/0031-8655(2002)0750382DNILDO2.0.CO2
Noctor, G. (1998). Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 249–279. doi: 10.1146/annurev.arplant.49.1.249
Noctor, G. (2006). Metabolic signaling in defense and stress: the central roles of soluble redox couples. Plant Cell Environ. 29, 409–425. doi: 10.1111/j.1365-3040.2005.01476.x
Noctor, G., Mhamdi, A., and Foyer, C. H. (2014). The roles of reactive oxygen metabolism in drought: not so cut and dried. Plant Physiol. 164, 1636–1648. doi: 10.1104/pp.113.233478
Nose, K. (2005). Redox control of protein trafficking. Antioxid. Redox Signal. 7, 303–307. doi: 10.1089/ars.2005.7.303
Nott, A., Jung, H. S., Koussevitzky, S., and Chory, J. (2006). Plastid-to-nucleus retrograde signaling. Annu. Rev. Plant Biol. 57, 739–759. doi: 10.1146/annurev.arplant.57.032905.105310
Orozco-Cardenas, M., and Ryan, C. A. (1999). Hydrogen peroxide is generated systemically in plant leaves by wounding and systemin via the octadecanoid pathway. Proc. Natl. Acad. Sci. U.S.A. 96, 6553–6557. doi: 10.1073/pnas.96.11.6553
Orzaez, D., and Granell, A. (1997). DNA fragmentation is regulated by ethylene during carpel senescence in Pisum sativum. Plant J. 11, 137–144. doi: 10.1046/j.1365-313X.1997.11010137.x
Overmyer, K., Tuominen, H., Kettunen, R., Betz, C., Langebartels, C., Sandermann, H., et al. (2000). Ozone-sensitive Arabidopsis rcd1 mutant reveals opposite roles for ethylene and jasmonate signaling pathways in regulating superoxide-dependent cell death. Plant Cell 12, 1849–1862. doi: 10.1105/tpc.12.10.1849
Papin, J. A., Hunter, T., Palsson, B. O., and Subramaniam, S. (2005). Reconstruction of cellular signaling networks and analysis of their properties. Nat. Rev. Mol. Cell Biol. 6, 99–111. doi: 10.1038/nrm1570
Park, S.-W., Kaimoyo, E., Kumar, D., Mosher, S., and Klessig, D. F. (2007). Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318, 113–116. doi: 10.1126/science.1147113
Parker, J. E., Aarts, N., Austin, M. A., Feys, B. J., Moisan, L. J., Muskett, P., et al. (2001). Genetic analysis of plant disease resistance pathways. Novartis Found. Symp. 236, 153–161.
Parry, G. (2015). The plant nuclear envelope and regulation of gene expression. J. Exp. Bot. 66, 1673–1685. doi: 10.1093/jxb/erv023
Paulsen, C. E., and Carroll, K. S. (2010). Orchestrating redox signaling networks through regulatory cysteine switches. ACS Chem. Biol. 5, 47–62. doi: 10.1021/cb900258z
Perez, I. B., and Brown, P. J. (2014). The role of ROS signaling in cross-tolerance: from model to crop. Front Plant Sci. 5:754. doi: 10.3389/fpls.2014.00754
Petrov, V. D., and Van Breusegem, F. (2012). Hydrogen peroxide-a central hub for information flow in plant cells. AoB Plants 2012:ls014. doi: 10.1093/aobpla/pls014
Pfannschmidt, T., Bräutigam, K., Wagner, R., Dietzel, L., Schröter, Y., Steiner, S., et al. (2009). Potential regulation of gene expression in photosynthetic cells by redox and energy state: approaches towards better understanding. Ann. Bot. 103, 599–607. doi: 10.1093/aob/mcn081
Pfannschmidt, T., Nilsson, A., and Allen, J. F. (1999). Photosynthetic control of chloroplast gene expression. Nature 397, 625–628. doi: 10.1038/17624
Phaniendra, A., Jestadi, D. B., and Periyasamy, L. (2015). Free radicals: properties, sources, targets, and their implication in various diseases. Indian J. Clin. Biochem. 30, 11–26. doi: 10.1007/s12291-014-0446-0
Pitzschke, A., and Hirt, H. (2006). Mitogen-activated protein kinases and reactive oxygen species signaling in plants. Plant Physiol. 141, 351–356. doi: 10.1104/pp.106.079160
Pitzschke, A., Schikora, A., and Hirt, H. (2009). MAPK cascade signaling networks in plant defense. Curr. Opin. Plant Biol. 12, 421–426. doi: 10.1016/j.pbi.2009.06.008
Pogson, B. J., Woo, N. S., Förster, B., and Small, I. D. (2008). Plastid signaling to the nucleus and beyond. Trends Plant Sci. 13, 602–609. doi: 10.1016/j.tplants.2008.08.008
Polidoros, A. N., Mylona, P. V., Pasentsis, K., Scandalios, J. G., and Tsaftaris, A. S. (2005). The maize alternative oxidase 1a (Aox1a) gene is regulated by signals related to oxidative stress. Redox Rep. 10, 71–78. doi: 10.1179/135100005X21688
Pospisil, P., Arato, A., Krieger-Liszkay, A., and Rutherford, A. W. (2004). Hydroxyl radical generation by photosystem II. Biochemistry 43, 6783–6792. doi: 10.1021/bi036219i
Potters, G., Horemans, N., Bellone, S., Caubergs, R. J., Trost, P., Guisez, Y., et al. (2004). Dehydroascorbate influences the plant cell cycle through a glutathione-independent reduction mechanism. Plant Physiol. 134, 1479–1487. doi: 10.1104/pp.103.033548
Potters, G., Horemans, N., and Jansen, M. A. (2010). The cellular redox state in plant stress biology-a charging concept. Plant Physiol. Biochem. 48, 292–300. doi: 10.1016/j.plaphy.2009.12.007
Price, A. H., Taylor, A., Ripley, S. J., Griffiths, A., Trewavas, A. J., and Knight, M. R. (1994). Oxidative signals in tobacco increase cytosolic calcium. Plant Cell 6, 1301–1310. doi: 10.1105/tpc.6.9.1301
Qi, M., and Elion, E. A. (2005). MAP kinase pathways. J. Cell Sci. 118, 3569–3572. doi: 10.1242/jcs.02470
Raghavendra, A. S., Gonugunta, V. K., Christmann, A., and Grill, E. (2010). ABA perception and signaling. Trends Plant Sci. 15, 395–401. doi: 10.1016/j.tplants.2010.04.006
Rao, M. V., Lee, H., Creelman, R. A., Mullet, J. E., and Davis, K. R. (2000). Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12, 1633–1646. doi: 10.1105/tpc.12.9.1633
Reddy, A. S. N. (2007). Alternative splicing of pre-messenger RNAs in plants in the genomic era. Annu. Rev. Plant Biol. 58, 267–294. doi: 10.1146/annurev.arplant.58.032806.103754
Rentel, M. C., Lecourieux, D., Ouaked, F., Usher, S. L., Petersen, L., Okamoto, H., et al. (2004). OXI1 kinase is necessary for oxidative burst-mediated signaling in Arabidopsis. Nature 427, 858–861. doi: 10.1038/nature02353
Riechmann, J. L., Heard, J., Martin, G., Reuber, L., Jiang, C., Keddie, J., et al. (2000). Arabidopsis transcription factors: genome-wide comparative analysis among eukaryotes. Science 290, 2105–2110. doi: 10.1126/science.290.5499.2105
Rodriguez, M. C., Petersen, M., and Mundy, J. (2010). Mitogen-activated protein kinase signaling in plants. Annu. Rev. Plant. Biol. 61, 621–649. doi: 10.1146/annurev-arplant-042809-112252
Sagi, M., and Fluhr, R. (2006). Production of reactive oxygen species by plant NADPH oxidases. Plant Physiol. 141, 336–340. doi: 10.1104/pp.106.078089
Sahu, P. P., Pandey, G., Sharma, N., Puranik, S., Muthamilarasan, M., and Prasad, M. (2013). Epigenetic mechanisms of plant stress responses and adaptation. Plant Cell Rep. 32, 1151–1159. doi: 10.1007/s00299-013-1462-x
Saidi, Y., Finka, A., Muriset, M., Bromberg, Z., Weiss, Y. G., Maathuis, F. J., et al. (2009). The heat shock response in moss plants is regulated by specific calcium-permeable channels in the plasma membrane. Plant Cell 21, 2829–2843. doi: 10.1105/tpc.108.065318
Scandalios, J. G. (2002). Oxidative stress responses - what have genome-scale studies taught us? Genome Biol. 3:REVIEWS1019. doi: 10.1186/gb-2002-3-7-reviews1019
Scandalios, J. G. (2005). Oxidative stress: molecular perception and transduction of signals triggering antioxidant gene defenses. Braz. J. Med. Biol. Res. 38, 995–1014. doi: 10.1590/S0100-879X2005000700003
Scheibe, R., and Dietz, K. J. (2012). Reduction-oxidation network for flexible adjustment of cellular metabolism in photoautotrophic cells. Plant Cell Environ. 35, 202–216. doi: 10.1111/j.1365-3040.2011.02319.x
Schenk, P. M., Kazan, K., Wilson, I., Anderson, J. P., Richmond, T., Somerville, S. C., et al. (2000). Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc. Natl. Acad. Sci. U.S.A. 97, 11655–11660. doi: 10.1073/pnas.97.21.11655
Schmitt, F. J., Renger, G., Friedrich, T., Kreslavksi, V. D., Zharmukhadmedov, S. K., Los, D. A., et al. (2014). Reactive oxygen species: re-evaluation of generation, monitoring and role in stress-signaling in phototrophicorgan- isms. Biochim. Biophys. Acta 1837, 835–848. doi: 10.1016/j.bbabio.2014.02.005
Senaratna, T., Touchell, D., Bunn, E., and Dixon, K. (2000). Acetyl salicylic acid (Aspirin) and salicylic acid induce multiple stress tolerance in bean and tomato plants. Plant Growth Regulat. 30, 157–161. doi: 10.1023/A:1006386800974
Serrato, A. J., Fernández-Trijueque, J., Barajas-López, J. D., Chueca, A., and Sahrawy, M. (2013). Plastid thioredoxins: a “one-for-all” redox-signaling system in plants. Front. Plant Sci. 21:463. doi: 10.3389/fpls.2013.00463
Sewelam, N., Jaspert, N., Kelen, K. V. D., Schmitz, J., Frerigmann, H., Tognetti, V. B., et al. (2014a). Spatial H2O2 signaling specificity: H2O2 from chloroplasts and peroxisomes differentially modulates the plant transcriptome. Mol. Plant 7, 1191–1210. doi: 10.1093/mp/ssu070
Sewelam, N., Kazan, K., Thomas-Hall, S., Kidd, B. N., Manners, J. M., and Schenk, P. M. (2013). Ethylene response factor 6 is a regulator of reactive oxygen species signaling in Arabidopsis. PLoS ONE 8:e70289. doi: 10.1371/journal.pone.0070289
Sewelam, N., Oshima, Y., Mitsuda, N., and Ohme-Takagi, M. (2014b). A step towards understanding plant responses to multiple environmental stresses: a genome-wide study. Plant Cell Environ. 37, 2024–2035.
Shen, Q. H., Saijo, Y., Mauch, S., Biskup, C., Bieri, S., Keller, B., et al. (2007). Nuclear activity of MLA immune receptors links isolate-specific and basal disease-resistance responses. Science 315, 1098–1103. doi: 10.1126/science.1136372
Simon-Plas, F., Elmayan, T., and Blein, J.-P. (2002). The plasma membrane oxidase NtrbohD is responsible for AOS production in elicited tobacco cells. Plant J. 31, 137–147. doi: 10.1046/j.1365-313X.2002.01342.x
Sinha, A. K., Jaggi, M., Raghuram, B., and Tuteja, N. (2011). Mitogen-activated protein kinase signaling in plants under abiotic stress. Plant Signal. Behav. 6, 196–203. doi: 10.4161/psb.6.2.14701
Springer, N. M. (2013). Epigenetics and crop improvement. Trends Genet. 29, 241–247. doi: 10.1016/j.tig.2012.10.009
Stael, S., Wurzinger, B., Mair, A., Mehlmer, N., Vothknecht, U. C., and Teige, M. (2012). Plant organellar calcium signaling: an emerging field. J. Exp. Bot. 63, 1525–1542. doi: 10.1093/jxb/err394
Stelling, J., Sauer, U., Szallasi, Z., Doyle, F. J., and Doyle, J. (2004). Robustness of cellular functions. Cell 118, 675–685. doi: 10.1016/j.cell.2004.09.008
Storz, G., and Imlayt, J. A. (1999). Oxidative stress. Curr. Opin. Microbiol. 2, 188–194. doi: 10.1016/S1369-5274(99)80033-2
Storz, G., Tartaglia, L. A., and Ames, B. N. (1990). Transcriptional regulator of oxidative stress-inducible genes: direct activation by oxidation. Science 248, 189–194. doi: 10.1126/science.2183352
Suharsono, U., Fujisawa, Y., Kawasaki, T., Iwasaki, Y., Satoh, H., and Shimamoto, K. (2002). The heterotrimeric G protein alpha subunit acts upstream of the small GTPase Rac in disease resistance of rice. Proc. Natl. Acad. Sci. U.S.A. 99, 13307–13312. doi: 10.1073/pnas.192244099
Sunkar, R., Kapoor, A., and Zhu, J. K. (2006). Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 18, 2051–2065. doi: 10.1105/tpc.106.041673
Suzuki, N., Koussevitzky, S., Mittler, R., and Miller, G. (2012). ROS and redox signaling in the response of plants to abiotic stress. Plant Cell Environ. 35, 259–270. doi: 10.1111/j.1365-3040.2011.02336.x
Suzuki, N., Miller, G., Salazar, C., Mondal, H. A., Shulaev, E., Cortes, D. F., et al. (2013). Temporal-spatial interaction between reactive oxygen species and abscisic acid regulates rapid systemic acclimation in plants. Plant Cell 25, 3553–3569. doi: 10.1105/tpc.113.114595
Takahashi, F., Mizoguchi, T., Yoshida, R., Ichimura, K., and Shinozaki, K. (2011). Calmodulin-dependent activation of MAP kinase for ROS homeostasis in Arabidopsis. Mol. Cell 41, 649–660. doi: 10.1016/j.molcel.2011.02.029
Tanou, G., Job, C., Belghazi, M., Molassiotis, A., Diamantidis, G., and Job, D. (2010). Proteomic signatures uncover hydrogen peroxide and nitric oxide cross-talk signaling network in citrus plants. J. Proteome Res. 9, 5994–6006. doi: 10.1021/pr100782h
Tanou, G., Job, C., Rajjou, L., Arc, E., Belghazi, M., Diamantidis, G., et al. (2009). Proteomics reveals the overlapping roles of hydrogen peroxide and nitric oxide in the acclimation of citrus plants to salinity. Plant J. 60, 795–804. doi: 10.1111/j.1365-313X.2009.04000.x
Tena, G., Asai, T., Chiu, W. L., and Sheen, J. (2001). Plant mitogen-activated protein kinase signaling cascades. Curr. Opin. Plant Biol. 4, 392–400. doi: 10.1016/S1369-5266(00)00191-6
Thanaraj, T. A. (2004). ASD: the alternative splicing database. Nucleic Acids Res. 32, D64–D69. doi: 10.1093/nar/gkh030
Thomma, B. P., Penninckx, I. A., Broekaert, W. F., and Cammue, B. P. (2001). The complexity of disease signaling in Arabidopsis. Curr. Opin. Immunol. 13, 63–68. doi: 10.1016/S0952-7915(00)00183-7
Tokunaga, T., Miyahara, K., Tabata, K., and Esaka, M. (2005). Generation and properties of ascorbic acid-overproducing transgenic tobacco cells expressing sense RNA for l-galactono-1,4-lactone dehydrogenase. Planta 220, 854–863. doi: 10.1007/s00425-004-1406-3
Torres, M. A., and Dangl, J. L. (2005). Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr. Opin. Plant Biol. 8, 397–403. doi: 10.1016/j.pbi.2005.05.014
Torres, M. A., Dangl, J. L., and Jones, J. D. G. (2002). Arabidopsis gp91phox homologues AtrbohD and AtrbohF are required for accumulation of reactive oxygen intermediates in the plant defense response. Proc. Natl. Acad. Sci. U.S.A. 99, 517–522. doi: 10.1073/pnas.012452499
Torres, M. A., Jones, J. D., and Dangl, J. L. (2005). Pathogen-induced, NADPH oxidase-derived reactive oxygen intermediates suppress spread of cell death in Arabidopsis thaliana. Nat. Genet. 37, 1130–1134. doi: 10.1038/ng1639
Torres, M. A., Morales, J., Sánchez-Rodríguez, C., Molina, A., and Dangl, J. L. (2013). Functional interplay between Arabidopsis NADPH oxidases and heterotrimeric G protein. Mol. Plant Microbe Interact. 26, 686–694. doi: 10.1094/MPMI-10-12-0236-R
Torres, M. A., Onouchi, H., Hamada, S., Machida, C., Hammond-Kosack, K. E., and Jones, J. D. G. (1998). Six Arabidopsis thaliana homologues of the human respiratory burst oxidase (gp91phox). Plant J. 14, 365–370. doi: 10.1046/j.1365-313X.1998.00136.x
Triantaphylidès, C., and Havaux, M. (2009). Singlet oxygen in plants: production, detoxification and signaling. Trends Plant Sci. 14, 219–228. doi: 10.1016/j.tplants.2009.01.008
Trusov, Y., Sewelam, N., Rookes, J. E., Kunkel, M., Nowak, E., Schenk, P. M., et al. (2009). Heterotrimeric G proteins-mediated resistance to necrotrophic pathogens includes mechanisms independent of salicylic acid-, jasmonic acid/ethylene- and abscisic acid-mediated defense signaling. Plant J. 58, 69–81. doi: 10.1111/j.1365-313X.2008.03755.x
van Rooij, E. (2011). The art of microRNA research. Circ. Res. 108, 219–234. doi: 10.1161/CIRCRESAHA.110.227496
Vanderauwera, S., Suzuki, N., Miller, G., van de Cotte, B., Morsa, S., Ravanat, J. L., et al. (2011). Extranuclear protection of chromosomal DNA from oxidative stress. Proc. Natl. Acad. Sci. U.S.A. 108, 1711–1716. doi: 10.1073/pnas.1018359108
Vanyushin, B. F., and Ashapkin, V. V. (2011). DNA methylation in higher plants: past, present and future. Biochim. Biophys. Acta 1809, 360–368. doi: 10.1016/j.bbagrm.2011.04.006
Verslues, P. E., and Zhu, J. (2004). Before and beyond ABA: upstream sensing and internal signals that determine ABA accumulation and response under abiotic stress. Biochem. Soc. Trans. 33, 375–379. doi: 10.1042/BST0330375
Vlot, A. C., Liu, P.-P., Cameron, R. K., Park, S.-W., Yang, Y., Kumar, D., et al. (2008). Identification of likely orthologs of tobacco salicylic acid-binding protein 2 and their role in systemic acquired resistance in Arabidopsis thaliana. Plant J. 56, 445–456. doi: 10.1111/j.1365-313X.2008.03618.x
Walley, J. W., and Dehesh, K. (2009). Molecular mechanisms regulating rapid stress signaling networks in Arabidopsis. J. Integr. Plant Biol. 52, 354–359. doi: 10.1111/j.1744-7909.2010.00940.x
Wang, B.-B., and Brendel, V. (2006). Genomewide comparative analysis of alternative splicing in plants. Proc. Natl. Acad. Sci. U.S.A. 103, 7175–7180. doi: 10.1073/pnas.0602039103
Wang, P., Du, Y., Hou, Y. J., Zhao, Y., Hsu, C. C., Yuan, F., et al. (2015). Nitric oxide negatively regulates abscisic-acid signaling in guard cells by S-nitrosylation of OST1. Proc. Natl. Acad. Sci. U.S.A. 112, 613–618. doi: 10.1073/pnas.1423481112
Wang, P., Du, Y., Li, Y., Ren, D., and Song, C. P. (2010). Hydrogen peroxide–mediated activation of MAP kinase 6 modulates nitric oxide biosynthesis and signal transduction in Arabidopsis. Plant Cell 22, 2981–2998. doi: 10.1105/tpc.109.072959
Wang, P.-C., Du, Y.-Y., An, G.-Y., Zhou, Y., Miao, C., and Song, C.-P. (2006). Analysis of global expression profiles of Arabidopsis genes under abscisic acid and H2O2 applications. J. Integ. Plant Biol. 48, 62–74. doi: 10.1111/j.1744-7909.2006.00213.x
Wasternack, C., and Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Wi, S. J., Ji, N. R., and Park, K. Y. (2012). Synergistic biosynthesis of biphasic ethylene and reactive oxygen species in response to hemibiotrophic Phytophthora parasitica in tobacco plants. Plant Physiol. 159, 251–265. doi: 10.1104/pp.112.194654
Wildt, J., Kley, D., Rockel, A., Rockel, P., and Segschneider, H. J. (1997). Emission of NO from several higher plant species. J. Geophys. Res. 102, 5919–5927. doi: 10.1029/96JD02968
Wrzaczek, M., Brosché, M., and Kangasjärvi, J. (2013). ROS signaling loops — production, perception, regulation. Curr. Opin. Plant Biol. 16, 575–582. doi: 10.1016/j.pbi.2013.07.002
Wrzaczek, M., and Hirt, H. (2001). Plant MAP kinase pathways: how many and what for? Biol. Cell 93, 81–87. doi: 10.1016/S0248-4900(01)01121-2
Xia, X. J., Zhou, Y. H., Shi, K., Zhou, J., Foyer, C. H., and Yu, J. Q. (2015). Interplay between reactive oxygen species and hormones in the control of plant development and stress tolerance. J. Exp. Bot. 66, 2839–2856. doi: 10.1093/jxb/erv089
Xiao, Y., Savchenko, T., Baidoo, E. E., Chehab, W. E., Hayden, D. M., Tolstikov, V., et al. (2012). Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149, 1525–1535. doi: 10.1016/j.cell.2012.04.038
Xing, Y., Jia, W., and Zhang, J. (2008). AtMKK1 mediates ABA-induced CAT1 expression and H2O2 production via AtMPK6-coupled signaling in Arabidopsis. Plant J. 54, 440–451. doi: 10.1111/j.1365-313X.2008.03433.x
Xiong, L., Schumaker, K. S., and Zhu, J.-K. (2002). Cell Signaling during Cold, Drought, and Salt Stress. Plant Cell 14, s165–s183.
Xu, D. B., Chen, M., Ma, Y. N., Xu, Z. S., Li, L. C., Chen, Y. F., et al. (2015). A G-protein β subunit, AGB1, negatively regulates the ABA response and drought tolerance by down-regulating AtMPK6-related pathway in Arabidopsis. PLoS ONE 10:e0116385. doi: 10.1371/journal.pone.0116385
Xue, H., and Seifert, G. J. (2015). Fasciclin like arabinogalactan protein 4 and respiratory burst oxidase homolog d and F independently modulate abscisic acid signaling. Plant Signal. Behav. 10:e989064. doi: 10.4161/15592324.2014.989064
Yoshioka, H., Asai, S., Yoshioka, M., and Kobayashi, M. (2009). Molecular mechanisms of generation for nitric oxide and reactive oxygen species, and role of the radical burst in plant immunity. Mol. Cell 28, 321–329. doi: 10.1007/s10059-009-0156-2
Yoshioka, H., Numata, N., Nakajima, K., Katou, S., Kawakita, K., Rowland, O., et al. (2003). Nicotiana benthamiana gp91phox homologs NbrbohA and NbrbohB participate in H2O2 accumulation and resistance to Phytophthora infestans. Plant Cell 15, 706–718. doi: 10.1105/tpc.008680
Yuasa, T., Ichimura, K., Mizoguchi, T., and Shinozaki, K. (2001). Oxidative stress activates ATMPK6, an Arabidopsis homologue of MAP kinase. Plant Cell Physiol. 42, 1012–1016. doi: 10.1093/pcp/pce123
Zarembinski, T. I., and Theologis, A. (1994). Ethylene biosynthesis and action: a case of conservation. Plant Mol. Biol. 26, 1579–1597. doi: 10.1007/BF00016491
Zhang, L., Cheng, Z., Qin, R., Qiu, Y., Wang, J. L., Cui, X., et al. (2012). Identification and characterization of an epi-allele of fie1 reveals a regulatory linkage between two epigenetic marks in rice. Plant Cell 24, 4407–4421. doi: 10.1105/tpc.112.102269
Zhang, W., Jeon, B. W., and Assmann, S. M. (2011). Heterotrimeric G-protein regulation of ROS signaling and calcium currents in Arabidopsis guard cells. J. Exp. Bot. 62, 2371–2379. doi: 10.1093/jxb/erq424
Zhou, J., Wang, J., Li, X., Xia, X. J., Zhou, Y. H., Shi, K., et al. (2014). H2O2 mediates the crosstalk of brassinosteroid and abscisic acid in tomato responses to heat and oxidative stresses. J. Exp. Bot. 65, 4371–4383. doi: 10.1093/jxb/eru217
Zhu, Y., Zuo, M., Liang, Y., Jiang, M., Zhang, J., Scheller, H. V., et al. (2013). MAP65-1a positively regulates H2O2 amplification and enhances brassinosteroid-induced antioxidant defense in maize. J. Exp. Bot. 64, 3787–3802. doi: 10.1093/jxb/ert215
Zurbriggen, M. D., Carrillo, N., Tognetti, V. B., Melzer, M., Peisker, M., Hause, B., et al. (2009). Chloroplast-generated reactive oxygen species play a major role in localized cell death during the non-host interaction between tobacco and Xanthomonas campestrispv. vesicatoria. Plant J. 60, 962–973. doi: 10.1111/j.1365-313X.2009.04010.x
Keywords: abiotic stress, biotic stress, oxidative stress, plant defense, plant stress signaling, reactive oxygen species
Citation: Sewelam N, Kazan K and Schenk PM (2016) Global Plant Stress Signaling: Reactive Oxygen Species at the Cross-Road. Front. Plant Sci. 7:187. doi: 10.3389/fpls.2016.00187
Received: 24 September 2015; Accepted: 04 February 2016;
Published: 23 February 2016.
Edited by:
Woe Yeon Kim, Gyeongsang National University, South KoreaReviewed by:
Giridara Kumar Surabhi, Regional Plant Resource Centre, IndiaNobuhiro Suzuki, Sophia University, Japan
Copyright © 2016 Sewelam, Kazan and Schenk. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nasser Sewelam, c2V3ZWxhbUBzY2llbmNlLnRhbnRhLmVkdS5lZw==
 Nasser Sewelam
Nasser Sewelam Kemal Kazan
Kemal Kazan Peer M. Schenk
Peer M. Schenk