
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
METHODS article
Front. Plant Sci. , 22 February 2016
Sec. Plant Biotechnology
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.00162
This article is part of the Research Topic Advances in microalgae biology and sustainable applications View all 12 articles
The fatty acid profiles of lipids from microalgae are unique. Polyunsaturated fatty acids are generally enriched in polar lipids, whereas saturated and monounsaturated fatty acids constitute the majority of fatty acids in triacylglycerols (TAG). Each species has characteristic fatty acids, and their content is positively or negatively correlated with TAGs. The marine oleaginous diatom Phaeodactylum tricornutum was used as the paradigm to determine the quantitative relationship between TAG and characteristic fatty acid content. Fatty acid profiles and TAG content of Phaeodactylum tricornutum were determined in a time course. C16:0/C16:1 and eicosapentaenoic acid (EPA, C20:5n3) were identified as characteristic fatty acids in TAGs and polar lipids, respectively. The percentage of those characteristic fatty acids in total fatty acids had a significant linear relationship with TAG content, and thus, the correlation coefficient presenting r2 were 0.96, 0.94, and 0.97, respectively. The fatty acid-based method for TAG quantification could also be applied to other microalgae such as Nannochloropsis oceanica in which the r2 of C16:0 and EPA were 0.94 and 0.97, respectively, and in Chlorella pyrenoidosa r2-values for C18:1 and C18:3 with TAG content were 0.91 and 0.99, repectively. This characteristic fatty acid-based method provided a distinct way to quantify TAGs in microalgae, by which TAGs could be measured precisely by immediate transesterification from wet biomass rather than using conventional methods. This procedure simplified the operation and required smaller samples than conventional methods.
Microalgae are potential triacylglycerol (TAG) resource for biofuel production (Chisti, 2007; Williams and Laurens, 2010). Despite analyses indicating that biofuels may be able to substitute for petroleum-derived transport fuels in the future (Chisti, 2008; Sander and Murthy, 2010), biofuels from microalgae have not yet been commercialized due to high cost (Williams and Laurens, 2010). Increasing lipid production, especially TAGs, is critical to reduce the cost of microalgae-based biofuels. Therefore, a suitable method to quantify the lipid content in microalgae is necessary.
Oil content in microalgae is commonly assessed using a gravimetric method. Oil in microalgae is extracted with organic solvents, using the general rule of “like dissolves like” (Bligh and Dyer, 1959; Ryckebosch et al., 2012; Axelsson and Gentili, 2014). However, many cellular components, such as pigments and unknown molecules, may be co-extracted with lipids due to their similar polarity, and these components may fluctuate during cultivation. Therefore, this method is unsuitable for assessing lipid production (Wang et al., 2009). Furthermore, this method requires a relatively large amount of sample, usually approximately 100 mg, and prevents conducting a time-course metabolic study of microalgae. Thin-layer chromatography (TLC) followed by capillary gas chromatography-flame ionization detection (GC-FID) is another method used to quantify lipids. This method separates lipids into individual classes by TLC after extraction from the cell using organic solvents. The lipids are then converted into fatty acid methyl esters (FAMEs) by GC-FID quantification. Due to the sensitivity of GC-FID, a few milligrams are sufficient for quantification. However, the method is time and labor intensive (Chen et al., 2009). High-performance liquid chromatography-mass spectrometry (HPLC-MS) has been used to detect lipids directly (MacDougall et al., 2011; Kind et al., 2012), but this approach is not routinely done due to the complex sample compositions. In addition, mutant screening is a powerful method to acquire an in-depth understanding of lipid metabolism (Doan and Obbard, 2012; Li et al., 2012; Manandhar-Shrestha and Hildebrand, 2013). Development of convenient and high-throughput methods to quantify lipids in microalgae has garnered increasing attention due to the labor-intensive nature of mutant screening (Terashima et al., 2015).
Lipids can be categorized into neutral and polar lipids, and neutral lipids are primarily referred to as TAGs. The fatty acid profiles in both microalgae and plants are unique to specific lipid classes. Neutral lipids mainly contain saturated (SFAs) and monounsaturated fatty acids (MUFAs), such as C16:0, C16:1, and C18:1, and more than 50% of C18:1 and C16:0 fatty acids that accumulate in the model algae Chlamydomonas reinhardtii are in the TAGs (Siaut et al., 2011). Contrarily, the neutral lipids in Chlamydomonas reinhardtii, polyunsaturated fatty acids (PUFAs), are enriched in polar lipids, such as C16:4, C18:3, and eicosapentaenoic acid (EPA, C20:5n3). C16:4 and C18:3 comprise the bulk of fatty acids in monogalactosyldiacylglycerols (MGDG), which account for more than 80% C16:4 and C18:3 of the total fatty acid content of Chlamydomonas reinhardtii (Zäuner et al., 2012). In the model plant Arabidopsis thaliana, C16:3 and C18:3 comprise the majority of MGDG and the main fatty acids in TAGs are C16:0, C18:0, and C18:1 (Fan et al., 2013). The total fatty acid profiles depend on the distinct fatty acid composition of the lipids. A strong correlation (r2 = 0.986) has been reported between the 16:0/16:4 ratio in FAMEs derived directly from algal cells and the TAG/total acyl group ratio in Chlamydomonas reinhardtii (Liu et al., 2013). More recently, we have reported that the neutral lipid content in Isochrysis zhangjiangensis can be quantified based on the C18:1 or C18:4 content (Wang et al., 2014). Therefore, we propose that TAG content can be quantified using specific fatty acids.
Here, the marine microalgae Phaeodactylum tricornutum, which was an ideal candidate for producing biodiesel, was studied as an example to support this hypothesis. First, the principles of characteristic fatty acid identification were discussed based on an analysis of the fatty acid profile of individual lipid in Phaeodactylum tricornutum. Subsequently, the relationship between the characteristic fatty acids and the TAG content was analyzed. This procedure was also applied to different microalgal species, such as Nannochloropsis oceanica and Chlorella pyrenoidosa, to expand its application.
The marine microalgae Phaeodactylum tricornutum was provided by Dr. Weidong Liu of the Liaoning Institute of Marine Fisheries. The marine microalgae Nannochloropsis oceanica IMET1 was provided by Dr. Jian Xu of the Qingdao Institute of Bioenergy and Bioprocess Technology, Chinese Academy of Sciences. Chlorella pyrenoidosa was obtained from the Freshwater Algae Culture Collection of the Institute of Hydrobiology, Chinese Academy of Sciences.
To identify the characteristic fatty acids, enriched f/2 medium, which contained three-fold the concentration of all ingredients, was inoculated with Phaeodactylum tricornutum as described by Feng et al. (2011). The medium was supplemented with sodium nitrate (N+), and the algae were stressed by omitting sodium nitrate (N−).
To correlate the characteristic fatty acids with neutral lipids, Phaeodactylum tricornutum and Nannochloropsis oceanica IMET1 were cultured under N− (Meng et al., 2015). Chlorella pyrenoidosa was cultured in BG11 medium without sodium nitrate to generate the N− condition.
Total fatty acid profiles and TAG content of three species were determined in a time course. The sampling time of the three species were 0, 6, and 18 h of Phaeodactylum tricornutum; 0, 2, 4, 7, 9, 12, and 14 h of Nannochloropsis oceanica IMET1; and 0, 1, 5, 6, and 8 days of Chlorella pyrenoidosa, respectively. Three replicates were taken for each time point. Each data point was a biological replicate (n = 3).
An average irradiance of 70 μmol photons m−2 s−1 was provided by continuous illumination with cool white fluorescent lamps. Temperature was maintained at 25 ± 2°C.
Cells were collected by centrifugation, and lipids were extracted according to the method described by Wang and Benning (2011) with slight modifications. A 300-μL aliquot of extraction solvent composed of methanol, chloroform, and formic acid (20:10:1, v/v/v) was added to approximately 5 mg biomass (dry weight). After shaking vigorously for 5 min, 150 μL of 0.2 M phosphoric acid and 1 M potassium chloride were added and vortexed for 1 min. Phase separation was achieved after centrifugation at 12,000 × g at room temperature for 5 min; the lipids dissolved in the lower chloroform phase were spotted onto TLC silica plates. Glycolipids (GL) and phospholipids (PL) were separated using a solvent system of acetone:toluene:water int the ration 91:30:7.5 mL, respectively. TAGs were separated with hexane:diethyl ether:acetic acid in the ratio 85:15:1, v/v/v, respectively.
The lipids separated by TLC or a microalgal biomass was added to methanol (2% H2SO4) and incubated for 1 h at 70°C to prepare the FAMEs. After methylation, deionized water and hexane were added to extract the FAMEs. C17:0-TAG was added as an internal standard for quantification. The FAMEs were identified qualitatively by mass spectroscopy and quantitatively by gas chromatography with a FID. The FAMEs of samples were detected on a 7890B gas chromatograph with a DB-23 capillary column (30 m × 0.32 mm × 0.25 μm; Agilent Technologies, Santa Clara, CA, USA) and a FID. The injector temperature was 260°C with a split ratio of 50:1. The initial column temperature was 130°C and was maintained for 1 min. The temperature was then increased to 170°C at a rate of 10°C min−1, followed by an increase to 215°C at a rate of 2.8°C min−1. The temperature was then maintained at 215°C for 1 min.
Correlation coefficients between characteristic fatty acids and TAG content were calculated using Origin Pro software, version 8.0 (Origin Lab, Hampton, Massachusetts, USA). Statistical significance in each result was calculated using the Two-tailed by SPSS 18.0 for windows (IBM, Armonk, New York City, USA), where the significance P < 0.01.
Characteristic fatty acids were present in different classes of lipids, and it was necessary to identify the characteristic fatty acids to accurately quantify TAGs. The fatty acid profiles of different lipid classes and TAG content were determined under both N+ and N− to identify the characteristic fatty acids. The total lipids (TLs) extracted from Phaeodactylum tricornutum cells were separated into GLs, PLs, and TAGs by TLC. All lipids except for TAGs were polar lipids. The fatty acid profile of individual lipid and TLs that were extracted from cells cultured under N+ and N− were analyzed (Table 1). PUFAs were the main fatty acids in GLs and PLs both under N+ and N−. EPA was the most abundant fatty acid in GLs and PLs and accounted for approximately 25% of the total fatty acids from these lipids. Therefore, EPA was the characteristic fatty acid of the polar lipids. TAGs were enriched with SFAs and MUFAs rather than PUFAs, such as C16:0, which was identified as a characteristic fatty acid. C16:1 almost evenly distributed across the aforementioned lipid classes and constituted 32–36% of the fatty acids in all lipid classes under N+. TAGs were specifically enriched with C16:1 under the N−, which was the most abundant fatty acid, accounting for almost 50% of fatty acid content. Although C16:1 content in PLs decreased dramatically, it remained largely unchanged in GLs and accounted for about 30% of its total fatty acids. Therefore, C16:1 was not specific to TAGs and could not serve as a characteristic fatty acid of TAGs.
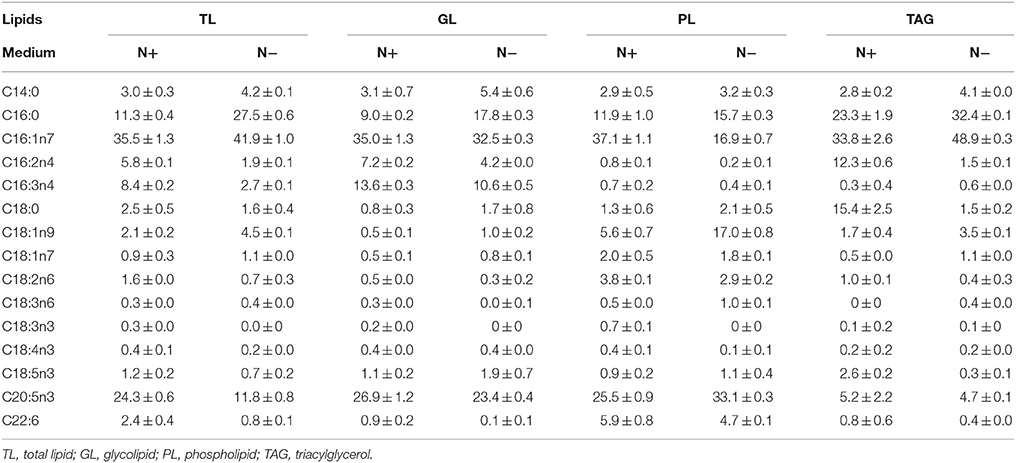
Table 1. Fatty acid profiles of different lipid components (means ± SD, n = 3) in Phaeodactylum tricornutum under nitrogen replete (N+) and nitrogen deplete (N−) were obtained by capillary FID-GC.
The fatty acid distributions were determined in different lipid classes to evaluate the contributions of each FAME to the total fatty acid profiles (Table 2). EPA was present in polar lipids under the N+, and more than 70% of the EPA was detected in polar lipids under the N−, despite the decreased amount of EPA compared with that detected under the N+. Only about 20% of the C16:0 was detected in TAGs under the N+, but this proportion increased to 80% under the N−. Because most of the EPA was found in polar lipids, the variation in polar lipids could be represented by the proportion of characteristic fatty acids of total fatty acids. Given the inverse relationship between polar lipids and TAGs, EPA was negatively correlated with TAG content. Similar to the characteristic fatty acids of polar lipids, the proportion of TAG-characteristic fatty acids also represented changes in TAG content.
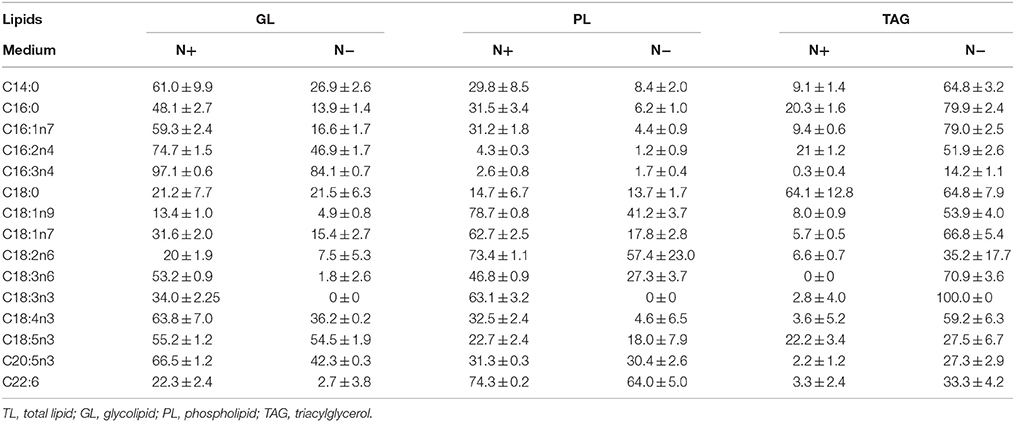
Table 2. Fatty acids distribution in different lipid classes (means ± SD, n = 3) in Phaeodactylum tricornutum under nitrogen replete (N+) and nitrogen deplete (N−).
Characteristic fatty acids had positive or negative correlations with TAGs in microalgae. The fatty acid profiles of TLs and TAG content were determined over time after nitrogen depletion in Phaeodactylum tricornutum to quantify the relationship between the characteristic fatty acids and TAG content. A correlation analysis revealed that fatty acid levels were highly and significantly correlated with TAG content (Figure 1). Specifically, the correlation coefficients of C16:0, C16:1, and EPA with the TAG content were 0.96, 0.94, and 0.97, respectively. The linear relationship between the characteristic fatty acids and the TAG content revealed that characteristic fatty acids could be used to quantify TAG content.
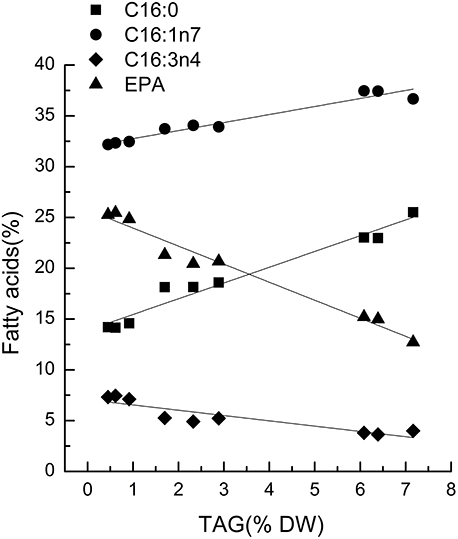
Figure 1. Correlations between characteristic fatty acids and triacylglycerol (TAG) content in Phaeodactylum tricornutum. The r2-values for C16:0, 16:1n7, C16:3n4, and eicosapentaenoic acid (EPA, C20:5n3) were 0.96, 0.94, 0.80, and 0.97, respectively. The sampling time of Phaeodactylum tricornutum was 0, 6, and 18 h. Each time point was made with three replicates, which represent biological replicates (n = 3). The points were drawn from individual result rather than the average of the three replicates. Statistical significance in each result was calculated using the Two-tailed, P < 0.01.
As some fatty acids were correlated with TAGs in microalgae, identifying the characteristic fatty acids was the first step to quantify TAG content. In order to quantify the characteristic fatty acid accurately, there was a threshold in the amount of characteristic fatty acids. Therefore, the proportion of fatty acids of TLs was a key rule for identifying characteristic fatty acids. Almost all C16:3 were found in GLs under the N− and N+ (Table 2). However, low total fatty acid content (Table 1) made it unsuitable to quantify TAGs. The correlation coefficient between the C16:3 content in total fatty acids and TAG content was only 0.798. We set 10% as the minimum content criterion for identifying characteristic fatty acids, therey considering a 10% variation in the percentage of each fatty acid determined by immediate transesterification of wet biomass (Liu et al., 2015), and C16:3 accounted for less than10% of total fatty acids in Phaeodactylum tricornutum. A significant change of cellular TAG accumulation was another key point when to determine the characteristic fatty acids, and the minimum fold-change was set to 1.5. Although C16:1 was increased in the TAGs under the N−, the ~1.2-fold change in C16:1 of the total fatty acids was insufficient for quantification. Taken together, 10% total fatty acid content and a 1.5-fold change were the thresholds for defining the characteristic fatty acids.
Chlorella pyrenoidosa (Chlorophyta) and Nannochloropsis oceanica (Eustigmatophyceae) were chosen as examples to expand the characteristic fatty acid-based method to other taxa. The fatty acid profiles of total lipids and TAG content were determined in a time course after nitrogen depletion in Nannochloropsis oceanica (Table 3) and Chlorella pyrenoidosa (Table 4), as described for Phaeodactylum tricornutum. The principles discussed above were used to identify the characteristic fatty acids in the two species. C16:0 and EPA were determined to be characteristic fatty acids in Nannochloropsis oceanica. The linear relationship between the characteristic fatty acids and TAG content was shown in Figure 2. The correlation coefficients of C16:0 and EPA with TAG content were 0.94 and 0.97 in Nannochloropsis oceanica, respectively. Similar to Nannochloropsis oceanica, C18:1 and C18:3 were determined to be characteristic fatty acids in Chlorella pyrenoidosa. The correlation coefficients of C18:1 and C18:3 with TAG content were 0.91 and 0.99 in Chlorella pyrenoidosa, respectively (Figure 3). The high correlation coefficients between the characteristic fatty acids and TAG content demonstrated that the characteristic fatty acid-based method could accurately measure TAG content. In addition, two types of characteristic fatty acids, such as EPA and C18:1, were detected that were negatively or positively correlated with TAGs, respectively, because they were from GLs or TAGs, which were different sources.
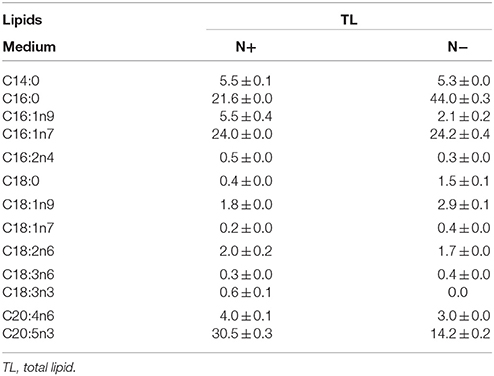
Table 3. Fatty acid profiles of total lipids (means ± SD, n = 3) in Nannochloropsis oceanica under nitrogen replete (N+) and nitrogen deplete (N−) were obtained by capillary FID-GC.
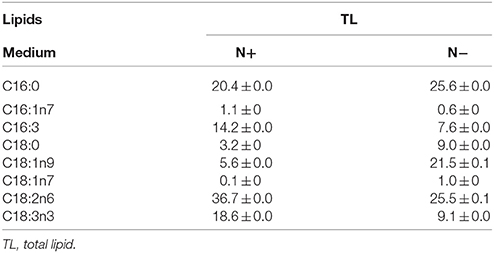
Table 4. Fatty acid profiles of total lipids (means ± SD, n = 3) in Chlorella pyrenoidosa under nitrogen replete (N+) and nitrogen deplete (N−) were obtained by capillary FID-GC.
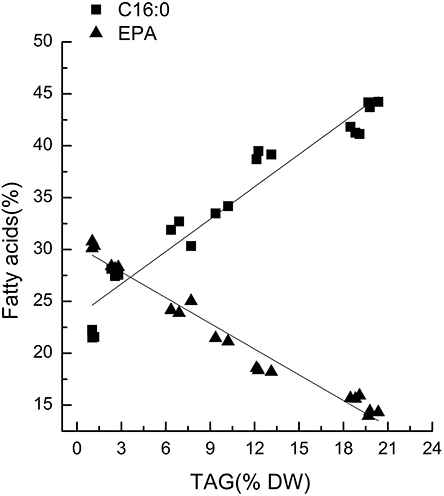
Figure 2. Correlations between characteristic fatty acids and triacylglycerol (TAG) content in Nannochloropsis oceanica IMET1. The r2-values for C16:0 and eicosapentaenoic acid (EPA, C20:5n3) were 0.94 and 0.97, respectively. The sampling time of Nannochloropsis oceanica IMET1 was 0, 2, 4, 7, 9, 12, and 14 h. Each time point was made with three replicates, which represent biological replicates (n = 3). The points were drawn from individual result rather than the average of the three replicates. Statistical significance in each result was calculated using the Two-tailed, P < 0.01.
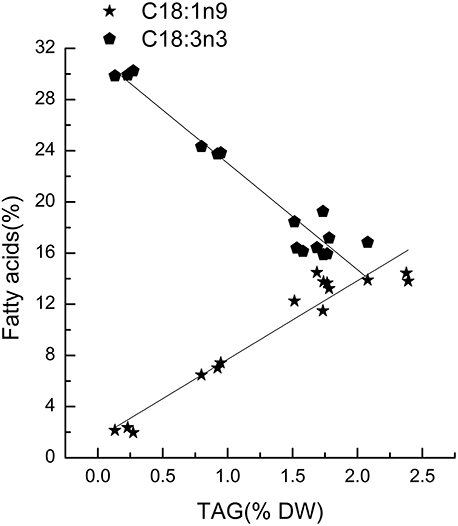
Figure 3. Correlations between characteristic fatty acids and triacylglycerol (TAG) content in Chlorella pyrenoidosa. The r2-values for C18:1n9 and C18:3n3 were 0.91 and 0.99, respectively. The sampling time of Chlorella pyrenoidosa was 0, 1, 5, 6, and 8 days. Each time point was made with three replicates, which represent biological replicates (n = 3). The points were drawn from individual result rather than the average of the three replicates. Statistical significance in each result was calculated using the Two-tailed, P < 0.01.
This idea was further supported by the statistical analysis of the microalgal fatty acid profiles under favorable and stressed conditions across eight phyla. The candidate characteristic fatty acids were shown in bold (Supplementary Material). Due to the diversity of microalgae, both fatty acid profiles and their response to the N− were species dependent. All species accumulated SFAs or MUFAs as TAG content increased in the same manner as in Phaeodactylum tricornutum, except in Porphyridium cruentum and Spirulina platensis, which accumulated C18:2n6. The level of PUFAs decreased in all microalgae following the accumulation of neutral lipids, such as C18:3n3 in Chlamydomonas sp. JSC4, C18:4n3 in Rhodomonas sp., and C18:5n3 in Gymnodinium sp.
The accumulation of SFAs or MUFAs coupled with the decrease in the level of PUFAs was also observed under high-light or salinity stress. Chlorella zofingiensis accumulated a large amount of C18:1 when exposed to high-light conditions, whereas the level of the PUFA C16:3 decreased by approximately 50% (Liu et al., 2012). The level of C18:1 increased from 17.8 to 39.4% in Sphenolithus obtusus XJ-15 under salinity stress (Xia et al., 2013). These results revealed that a linear relationship between specific fatty acids and TAG content may also be applicable to microalgae under stress conditions.
There were some microalgae of which PUFAs such as arachidonic acid (AA, C20:4n6), EPA, and docosahexaenoic (DHA, C22:6n3) were accumulated in TAGs under stress condition. For example, the unicellular green alga Parietochloris incise (Trebuxiophyceae, Chlorophyta) accumulated TAGs with over 90% of total AA under nitrogen deprivation (Cohen et al., 2000; Khozin-Goldberg et al., 2002; Merzlyak et al., 2007). In the marine Haptophyte Pavlova lutheri, 55 and 67% of the overall cellular EPA and DHA content were attributed to the cellular TAG accumulation triggered by bicarbonate addition under nitrogen starvation (Guiheneuf and Stengel, 2013). In the red microalgae Porphyridium cruentum (Cohen et al., 1988) and Gracilaria (Gracilariales, Rhodophyta) (Araki et al., 1990), TAGs were predominantly constructed of AA and EPA. In this case, TAG content still could be quantified with the corresponding PUFAs, firstly to determine the quantitative correlation between them.
The approach we have proposed differed from conventional approaches. All lipid quantification data presented previously relied on “like dissolves like” with many uncertain compounds, whereas quantification of TAGs in the proposed method relied on the characteristic fatty acids, which was much easier to quantify than the entire TAG class because only one compound needs to be determined. Furthermore, the characteristic fatty acid content in total fatty acids could be determined by immediate transesterification from wet biomass. Therefore, drying the biomass and extracting the lipids could be omitted. Hence, this approach simplified sample preparation and lipid determination procedure. Furthermore, less than 5 mg of biomass is necessary due to the high sensitivity of the capillary GC-FID analysis, which is 20-times less than the amount required for conventional gravimetric determination methods. Therefore, this procedure could be used to monitor variations in lipids throughout a culture. Our procedure saved time and labor when quantifying TAGs.
In conclusion, our procedure used characteristic fatty acids to obtain lipid content and provided a unique way to quantify TAGs. The significant feature of this procedure was indirect quantification of lipids by direct detection of the individual characteristic fatty acid. Total fatty acid content of 10% and a 1.5-fold change during TAG accumulation were set as criteria to define the characteristic fatty acids. A significant linear relationship was observed between the characteristic fatty acids and TAG content. This characteristic fatty acid-based method provided a new method to quantify TAG content in different microalgal species.
PS and HW designed the experiments. PS, HW, and YP performed the experiments and interpreted the data. PS and HW wrote the draft manuscript. YM established analysis methods. PW provided the subculture of the strains. SX made the critical revision of the manuscript. All the authors discussed the results and commented on the manuscript.
This work was supported by National Oceanic Administration public welfare fund project (Grant No. 201505030); National High Technology Research and Development Program “863” (2012AA052101).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00162
Araki, S., Sakurai, T., Oohusa, T., Kayama, M., and Nisizawa, K. (1990). Content of arachidonic and eicosapentaenoic acids in polarlipids from Gracilaria (Gracilariales, Rhodophyta). Hydrobiologia 204, 513–519. doi: 10.1007/BF00040279
Axelsson, M., and Gentili, F. (2014). A single-step method for rapid extraction of total lipids from Green Microalgae. PLoS ONE 9:e89643. doi: 10.1371/journal.pone.0089643
Bligh, E., and Dyer, W. (1959). A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/o59-099
Chen, W., Zhang, C., Song, L., Sommerfeld, M., and Hu, Q. (2009). A high throughput Nile red method for quantitative measurement of neutral lipids in microalgae. J. Microbiol. Methods 77, 41–47. doi: 10.1016/j.mimet.2009.01.001
Chisti, Y. (2007). Biodiesel from microalgae. Biotechnol. Adv. 25, 294–306. doi: 10.1016/j.biotechadv.2007.02.001
Chisti, Y. (2008). Biodiesel from microalgae beats bioethanol. Trends Biotechnol. 26, 126–131. doi: 10.1016/j.tibtech.2007.12.002
Cohen, Z., Khozin-Goldberg, I., Adlerstein, D., and Bigogno, C. (2000). The role of triacylglycerol as a reservoir of polyunsaturated fatty acids for the rapid production of chloroplastic lipids in certain microalgae. Biochem. Soc. Trans. 28, 740–743. doi: 10.1042/bst0280740
Cohen, Z., Vonshak, A., and Richmond, A. (1988). Effect of enviromental conditions on fatty acid composition of the red alga porphyridiumcruentum correlation to growth rate. J. Phycol. 24, 328–332.
Doan, T. T. Y., and Obbard, J. P. (2012). Enhanced intracellular lipid in Nannochloropsis sp. via random mutriglycerideenesis and flow cytometric cell sorting. Algal Res. 1, 17–21. doi: 10.1016/j.algal.2012.03.001
Fan, J., Yan, C., Zhang, X., and Xu, C. (2013). Dual role for phospholipid:diacylglycerol acyltransferase: enhancing fatty acid synthesis and diverting fatty acids from membrane lipids to triacylglycerol in Arabidopsis leaves. Plant Cell 25, 3506–3518. doi: 10.1105/tpc.113.117358
Feng, D., Chen, Z., Xue, S., and Zhang, W. (2011). Increased lipid production of the marine oleaginous microalagae Isochrysis zhangjiangensis (Chrysophyta) by nitrogen supplement. Bioresour. Technol. 102, 6710–6716. doi: 10.1016/j.biortech.2011.04.006
Guiheneuf, F., and Stengel, D. B. (2013). LC-PUFA-enriched oil production by microalgae: accumulation of lipid and triacylglycerols containing n-3 LC-PUFA is triggered by nitrogen limitation and inorganic carbon availability in the marine haptophyte Pavlova lutheri. Mar. Drugs 11, 4246–4266. doi: 10.3390/md11114246
Khozin-Goldberg, I., Bigogno, C., Shrestha, P., and Cohen, Z. (2002). Nitrogen starvation induces the accumulation of arachidonic acid in the freshwater green alga Parietochloris incisa (Trebuxiophyceae). J. Phycol. 38, 991–994. doi: 10.1046/j.1529-8817.2002.01160.x
Kind, T., Meissen, J. K., Yang, D., Nocito, F., Vaniya, A., Cheng, Y.-S., et al. (2012). Qualitative analysis of algal secretions with multiple mass spectrometric platforms. J. Chromatogr. A 1244, 139–147. doi: 10.1016/j.chroma.2012.04.074
Li, X., Moellering, E. R., Liu, B., Johnny, C., Fedewa, M., Sears, B. B., et al. (2012). A galactoglycerolipid lipase is required for triacylglycerol accumulation and survival following nitrogen deprivation in Chlamydomonas reinhardtii. Plant Cell 25, 3506–3518. doi: 10.1105/tpc.112.105106
Liu, B., Vieler, A., Li, C., Jones, A. D., and Benning, C. (2013). Triacylglycerol profiling of microalgae Chlamydomonas reinhardtii and Nannochloropsis oceanica. Bioresour. Technol. 146, 310–316. doi: 10.1016/j.biortech.2013.07.088
Liu, J., Liu, Y., Wang, H., and Xue, S. (2015). Direct transesterification of fresh microalgal cells. Bioresour. Technol. 176, 284–287. doi: 10.1016/j.biortech.2014.10.094
Liu, J., Sun, Z., Zhong, Y., Huang, J., Hu, Q., and Chen, F. (2012). Stearoyl-acyl carrier protein desaturase gene from the oleaginous microalga Chlorella zofingiensis: cloning, characterization and transcriptional analysis. Planta 236, 1665–1676. doi: 10.1007/s00425-012-1718-7
MacDougall, K., McNichol, J., McGinn, P., O'leary, S. B., and Melanson, J. (2011). Triacylglycerol profiling of microalgae strains for biofuel feedstock by liquid chromatography–high-resolution mass spectrometry. Anal. Bioanal. Chem. 401, 2609–2616. doi: 10.1007/s00216-011-5376-6
Manandhar-Shrestha, K., and Hildebrand, M. (2013). Development of flow cytometric procedures for the efficient isolation of improved lipid accumulation mutants in a Chlorella sp. microalga. J. Appl. Phycol. 25, 1643–1651. doi: 10.1007/s10811-013-0021-8
Meng, Y., Jiang, J., Wang, H., Cao, X., Xue, S., Yang, Q., et al. (2015). The characteristics of triglycerides and EPA accumulation in Nannochloropsis oceanica IMET1 under different nitrogen supply regimes. Bioresour. Technol. 179, 483–489. doi: 10.1016/j.biortech.2014.12.012
Merzlyak, M. N., Chivkunova, O. B., Gorelova, O. A., Reshetnikova, I. V., Solovchenko, A. E., Khozin-Goldberg, I., et al. (2007). Effect of nitrogen starvation on optical properties, pigments, and arachidonic acid content of the unicellular green alga Parietochloris incisa (Trebouxiophyceae, Chlorophyta). J. Phycol. 43, 833–843. doi: 10.1111/j.1529-8817.2007.00375.x
Ryckebosch, E., Muylaert, K., and Foubert, I. (2012). Optimization of an analytical procedure for extraction of lipids from microalgae. J. Am. Oil Chem. Soc. 89, 189–198. doi: 10.1007/s11746-011-1903-z
Sander, K., and Murthy, G. S. (2010). Life cycle analysis of algae biodiesel. Int. J. Life Cycle Assess. 15, 704–714. doi: 10.1007/s11367-010-0194-1
Siaut, M., Cuine, S., Cagnon, C., Fessler, B., Nguyen, M., and Carrier, P. (2011). Oil accumulation in the model green alga Chlamydomonas reinhardtii: characterization, variability between common laboratory strains and relationship with starch reserves. BMC Biotechnol. 11:7. doi: 10.1186/1472-6750-11-7
Terashima, M., Freeman, E. S., Jinkerson, R. E., and Jonikas, M. C. (2015). A fluorescence-activated cell sorting-based strategy for rapid isolation of high-lipid Chlamydomonas mutants. Plant J. 81, 147–159. doi: 10.1111/tpj.12682
Wang, H.-T., Yao, C.-H., Liu, Y.-N., Meng, Y.-Y., Wang, W.-L., Cao, X.-P., et al. (2014). Identification of fatty acid biomarkers for quantification of neutral lipids in marine microalgae Isochrysis zhangjiangensis. J. Appl. Phycol. 27, 1–7. doi: 10.1007/s10811-014-0300-z
Wang, Z. H., and Benning, C. (2011). Arabidopsis thalianapolar glycerolipid profiling by thin layer chromatography (TLC) coupled with gas-liquid chromatography (GLC). J. Vis. Exp. 49, 1–6. doi: 10.1167/11.6.1
Wang, Z., Ullrich, N., Joo, S., Waffenschmidt, S., and Goodenough, U. (2009). Algal lipid bodies: stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot. Cell 8, 1856–1868. doi: 10.1128/EC.00272-09
Williams, P. J. L. B., and Laurens, L. M. L. (2010). Microalgae as biodiesel & biomass feedstocks: review & analysis of the biochemistry, energetics & economics. Energy Environ. Sci. 3, 554–590. doi: 10.1039/b924978h
Xia, L., Ge, H., Zhou, X., Zhang, D., and Hu, C. (2013). Photoautotrophic outdoor two-striglyceridee cultivation for oleaginous microalgae Scenedesmus obtusus XJ-15. Bioresour. Technol. 144, 261–267. doi: 10.1016/j.biortech.2013.06.112
Keywords: microalgae, triacylglycerols, neutral lipids, characteristic fattyacids, biofuels
Citation: Shen P-L, Wang H-T, Pan Y-F, Meng Y-Y, Wu P-C and Xue S (2016) Identification of Characteristic Fatty Acids to Quantify Triacylglycerols in Microalgae. Front. Plant Sci. 7:162. doi: 10.3389/fpls.2016.00162
Received: 24 August 2015; Accepted: 30 January 2016;
Published: 22 February 2016.
Edited by:
Flavia Vischi Winck, University of São Paulo, BrazilReviewed by:
Surinder Singh, Commonwealth Scientific and Industrial Research Organisation, AustraliaCopyright © 2016 Shen, Wang, Pan, Meng, Wu and Xue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Song Xue, eHVlc29uZ0BkaWNwLmFjLmNu
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.