
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
FOCUSED REVIEW article
Front. Plant Sci. , 03 February 2016
Volume 7 - 2016 | https://doi.org/10.3389/fpls.2016.00047
This article mentions parts of:
Genome-wide identification, isolation and expression analysis of auxin response factor (ARF) gene family in sweet orange (Citrus sinensis)
Auxin is a key regulator of virtually every aspect of plant growth and development from embryogenesis to senescence. Previous studies have indicated that auxin regulates these processes by controlling gene expression via a family of functionally distinct DNA-binding auxin response factors (ARFs). ARFs are likely components that confer specificity to auxin response through selection of target genes as transcription factors. They bind to auxin response DNA elements (AuxRE) in the promoters of auxin-regulated genes and either activate or repress transcription of these genes depending on a specific domain in the middle of the protein. Genetic studies have implicated various ARFs in distinct developmental processes through loss-of-function mutant analysis. Recent advances have provided information on the regulation of ARF gene expression, the role of ARFs in growth and developmental processes, protein–protein interactions of ARFs and target genes regulated by ARFs in plants. In particular, protein interaction and structural studies of ARF proteins have yielded novel insights into the molecular basis of auxin-regulated transcription. These results provide the foundation for predicting the contributions of ARF genes to the biology of other plants.
Auxins play a critical role in most major growth responses both throughout the different developmental stages of plants such as organogenesis, vascular tissue differentiation, apical dominance and root initiation, and tropism and on a cellular level cell in processes including extension, division, and differentiation (Guilfoyle and Hagen, 2007; Mockaitis and Estelle, 2008; Su et al., 2014). Three decades of studies have explored the rapid effects of auxin on gene expression and regulation (Di et al., 2015b). A large number of candidate genes that are potentially regulated by auxins and that may function in growth and developmental processes have been identified in Arabidopsis and other plant species (Rosado et al., 2012; Liu et al., 2014b; Di et al., 2015b; Guilfoyle, 2015). Among these genes, members of the auxin response factors (ARF) family have been suggested to play a key role in regulating the expression of auxin response genes (Liscum and Reed, 2002). To date, 22 ARF genes and one pseudogene have been isolated from Arabidopsis (Liscum and Reed, 2002; Guilfoyle and Hagen, 2007). ARF genes are expressed in dynamic and differential patterning during development, and genetic studies have shown that individual ARFs control distinct developmental processes (Rademacher et al., 2012). Members of the ARF family of proteins contain domains associated with DNA binding, transcriptional activation or repression, and protein-protein interactions during auxin perception and signaling processes (Guilfoyle and Hagen, 2007; Di et al., 2015a). Recently, the ARF gene family has been also investigated in several plants based on the recent release of the genome such as citrus, Medicago truncatula and Gossypium raimondii using both bioinformatics and molecular analyses (Li et al., 2015; Shen et al., 2015a; Sun et al., 2015). More importantly, a considerable amount of new information has been obtained regarding the mechanisms that control ARF protein activities, and gene expression profiles. This review will focus on recent advances that have provided insight into the roles played by ARFs in regulating a variety of plant growth and development processes and the mechanisms involved in this regulation in Arabidopsis and other plant species.
KEY CONCEPT 1. Organogenesis
An adult plant consists of many specialized cell organizations: tissues and organs. Tissues consist of cells of uniform shape and specialized function, such as meristem, cortex, and phloem. Several tissues are organized together to form an organ, such as leaves, roots, flowers, and fruit. The process of initiation and development of an organ is called organogenesis.
The plants response to auxin involves changes in gene regulation (Liscum and Reed, 2002). Genes that are up-regulated or down-regulated by auxin contain AuxRE in their promoters, which bind transcription factors of the ARF family (Guilfoyle and Hagen, 2007; Mockaitis and Estelle, 2008). The identification of the AuxRE sequence led to the isolation of Arabidopsis ARF1 (Ulmasov et al., 1997), and subsequent genetic, genomic, and molecular studies have identified 22 ARF genes in Arabidopsis (Liscum and Reed, 2002). The ARF gene family is a modular transcription factor family consisting of several domains that have remained conserved despite hundreds of millions of years of evolution (Finet et al., 2012). Most ARF proteins consist of an N-terminal B3-type DNA binding domain (DBD), a variable middle region that functions as an activation domain (AD) or repression domain (RD), and a carboxy-terminal dimerization domain (CTD: domain III/IV), which is involved in protein–protein interactions by dimerizing with auxin/indole-3-acetic acid (Aux/IAA) family genes as well as between ARFs (Kim et al., 1997; Guilfoyle and Hagen, 2007; Piya et al., 2014). The DBD is classified as a plant-specific B3-type protein domain, but requires additional amino-terminal and carboxyterminal amino acids for efficient in vitro binding to TGTCTC/GAGACA site (Tiwari et al., 2003). The first four bases of the recognition site are absolutely required for ARF binding, while more variation is tolerated in the last two bases (Boer et al., 2014). The AD and RD are located just carboxyterminal to the DBD and contain biased amino acid sequences (Ulmasov et al., 1999). The AD is enriched in glutamine along with leucine (L) and serine (S) residues, while the RD is enriched in glycine (Q), leucine (L), serine (S), and proline (P) residues (Ulmasov et al., 1999). The amino acid composition of the middle region is critical in determining ARF function, with S-rich ARFs acting as transcriptional repressors and Q-rich ARFs acting transcriptional activators by protoplast transfection assays (Tiwari et al., 2003; Guilfoyle and Hagen, 2007). Five ARF proteins (ARF5/ARF6/ARF7/ARF8/ARF19) were characterized as transcriptional activators based on transient assays in transfected protoplasts, the other ARFs were classified as repressors (Ulmasov et al., 1999; Tiwari et al., 2003). A recent crystallographic study revealed that two additional domains associate with the DBDs of some ARFs, and these are a dimerization domain (DD) and a Tudor-like ancillary domain within the C-terminal region of the flanking domain (FD). The DD facilitates cooperative binding of the B3 DBD to selected AuxREs. However, the function of Tudor-like ancillary domain has not been determined (Guilfoyle and Hagen, 2012; Boer et al., 2014; Guilfoyle, 2015; Korasick et al., 2015). Not all ARFs contain the five domains described above. In addition, the III–IV region of some ARF protein may form a type I/II Phox and Bem1p (PB1) protein–protein interaction domain, which provides both positive and negative electrostatic interfaces for directional protein interaction (Guilfoyle and Hagen, 2012; Guilfoyle, 2015; Korasick et al., 2015).
KEY CONCEPT 2. B3-type DNA binding domain (DBD)
It is a highly conserved domain found exclusively in transcription factors (TFs) of higher plants. It consists of 100–120 residues, includes seven beta strands and two alpha helices. There are three main families of TFs that contain B3 domain in Arabidopsis: ARF Auxin response factors (ARF), Abscisic acid insensitive 3 (ABI3), and Related to ABI3/VP1 (RAV).
Since, cloning of the first ARF gene from Arabidopsis (Ulmasov et al., 1997), ARF genes from 15 plant species have been identified based on genome-wide analysis studies (Table 1). For example, 22 genes from tomato (Zouine et al., 2014), 25 genes from rice (Wang et al., 2007), 19 genes from sweet orange (Li et al., 2015), 24 genes from Medicago truncatula (Shen et al., 2015a), 47 genes from banana (Hu et al., 2015), and 39 genes from Populus trichocarpa (Kalluri et al., 2007) were identified. Most ARF proteins from these plant species are nuclear proteins with described protein domains consistent with previous reports on the homologous genes from Arabidopsis (Table 1). However, some differences in the ARF protein family were also found between Arabidopsis and other plant species. For example, ARF3, ARF13, and ARF17 lack Domains III/IV, and ARF23 consists of a truncated DBD only in Arabidopsis (Guilfoyle and Hagen, 2007). Only one pseudogene (a truncated DBD) was found in citrus plants among these plant species (Li et al., 2015), whereas a large number of truncated proteins (lacking Domains III/IV) have been found in rice (Wang et al., 2007), maize (Liu et al., 2011), banana (Hu et al., 2015), and M. truncatula (Shen et al., 2015a) compared with Arabidopsis. Interestingly, some plant species contain more ARF genes than Arabidopsis (Table 1). One explanation for the higher number of ARF genes encoded in these genome could be that large-scale duplication event occurred early in the evolution of these plants (Blanc et al., 2003). It is noteworthy that most information about ARFs function, expression, and regulation comes from studies in annual herbaceous plants such as Arabidopsis, rice, and tomato (Guilfoyle and Hagen, 2007; Wang et al., 2007; Kumar et al., 2011), while relatively few reports focus on other plant species.
The ARF genes encode proteins with full-length DBDs that may recognize and compete for target sites in promoters of auxin response genes (Tiwari et al., 2003). Therefore, there has been increased interest to determining when and where these genes are expressed and what regulates their expression (Hagen and Guilfoyle, 2002; Guilfoyle and Hagen, 2007). It has long been recognized that ARFs directly bind to AuxREs in the promoters of auxin responsive genes through their DNA-binding domain (Wang and Estelle, 2014). ARF binding to AuxREs in particular requires C-terminal amino acids (Guilfoyle and Hagen, 2012). It has been proposed that the C-terminal domain enhances DNA binding by enabling ARF dimerization. Both ARF and Aux/IAA proteins contain conserved sequences near the C-terminus termed domains III and IV (Guilfoyle and Hagen, 2012). These domains mediate ARF-ARF, ARF-Aux/IAA, and Aux/IAA-Aux/IAA interactions as determined by yeast two-hybrid and bimolecular fluorescence complementation assays (Korasick et al., 2015). ARF regulation is well-studied, and a working model for ARF activation is now well-established (Figure 1) (Salehin et al., 2015). At low auxin levels, Aux/IAA proteins form dimers with ARFs to inhibit ARF activity by recruiting the co-repressor TOPLESS (TPL), which results in the repression of auxin-responsive genes (Figure 1A) (Szemenyei et al., 2008). At higher auxin levels, Aux/IAAs bind to the SCFTIR1∕AFB complex and subsequently become ubiquitinated and degraded by the 26S proteasome. The ARF is then released and can regulate the transcription of its target auxin response genes (Figure 1B) (Wang and Estelle, 2014). Recent structural studies of ARFs have led to exciting new insight into the molecular function of the ARF-Aux/IAA pathway. Crystal structures showed that the C-terminal domains of ARF5 and ARF7 conform to a well-known PB1 domain that confers protein-protein interactions with other PB1 domain proteins through electrostatic contacts (Boer et al., 2014; Guilfoyle, 2015). Further experiments confirmed the importance of these charged amino acids in conferring ARF and Aux/IAA interactions as proposed by the crystal structure of the PB1 domain (Korasick et al., 2015). In addition to the PB1 domain, a second protein-protein interaction module that functions in ARF-ARF dimerization and facilitates DNA binding has recently been revealed from structure-function analysis and saturating binding site selection on the ARF1 and ARF5 DNA binding domains (Boer et al., 2014). These studies provide an atomic-level explanation for DNA-binding specificity in the auxin pathway.
KEY CONCEPT 3. Bimolecular fluorescence complementation assays
It is a technology typically used to validate protein interactions. It is based on the association of fluorescent protein fragments that are attached to components of the same macromolecular complex. Through the Visualization and analysis of the intensity and distribution of fluorescence in live cells, one can identify both the location and interaction partners of proteins of interest.
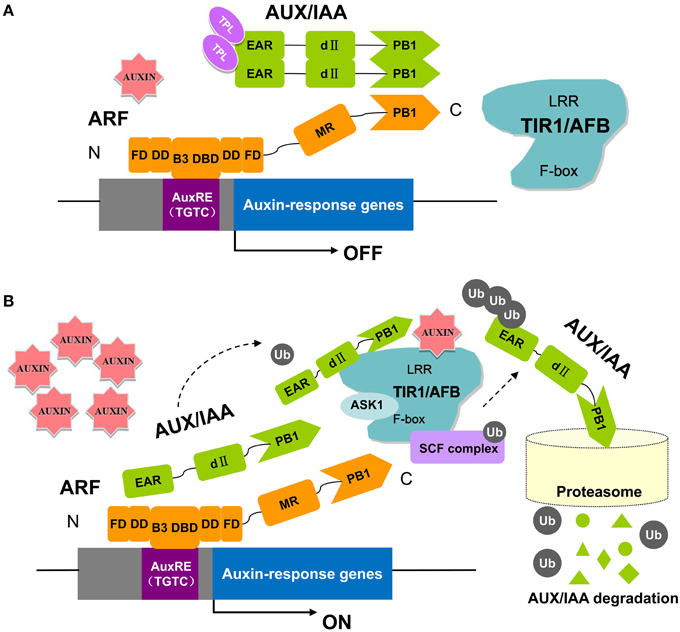
Figure 1. The key components in auxin perception and signaling in Arabidopsis. ARF proteins contain a non-conserved AD or RD flanked by an N-terminal DBD (composed of a B3 domain, a dimerization domain: DD, and a Tudor-like ancillary domain within the C-terminal region of the flanking domain: FD) and a C-terminal PB1 domain (previously referred to as domain III/IV). Parts of the DD and FD are found both N-terminal and C-terminal to the B3 domain. In this pathway, the TRANSPORT INHIBITOR RESPONSE 1/AUXIN SIGNALING F-BOX proteins (TIR1/AFBs) are F-box proteins that, together with other proteins (ASK1, CUL1, RBX), form the ubiquitin protein ligase complex, SCFTIR1. At low auxin levels (A), the Aux/IAA proteins form multimers with ARFs and recruit TPL to the chromatin. High levels of auxin (B) promote ubiquitination and degradation of Aux/IAAs through SCFTIR1∕AFB and the 26S proteasome (Kim et al., 1997; Guilfoyle and Hagen, 2007, 2012; Szemenyei et al., 2008; Boer et al., 2014; Guilfoyle, 2015; Korasick et al., 2015; Salehin et al., 2015).
In addition to the interaction between themselves, the ARFs have also been reported to regulate and be regulated by other transcription factors (Wang and Estelle, 2014). A recent study showed that a MYB transcription factor (MYB77) interacts with the ARF7 protein and that this interaction results in a strong reduction in lateral root numbers in Arabidopsis (Shin et al., 2007). Moreover, it has been shown that the bHLH transcription factor BIGPETALp (BPEp) interacts with ARF8 to effect petal growth. This interaction is mediated through the BPEp C-terminal domain and the C-terminal domain of ARF8 (Varaud et al., 2011). The Arabidopsis BREVIS RADIX (BRX) transcriptional co-regulator interacts with domain III/IV of ARF5 in yeast two-hybrid assays as well as in vitro pull-down assays, and this interaction enhances the transcriptional activation potential of this ARF (Guilfoyle and Hagen, 2012). In another recent report, HaIAA27 was shown to repress the transcriptional activation of the heat shock transcription factor HaHSFA9 in sunflower to repress its activity during seed development. As in the case of the ARFs, auxin also acts to relieve repression of the HaHSFA9 protein (Carranco et al., 2010). Recent data also suggest that post-translational modifications of ARFs may constitute another layer of regulation of auxin signaling outputs (Wang and Estelle, 2014; Hill, 2015). Phosphorylation of ARF7 and ARF19 by BRASSINOSTEROID-INSENSITIVE2 (BIN2) can potentiate auxin signaling output during lateral root organogenesis (Cho et al., 2014). Meanwhile, other previous report shows that BIN2 also phosphorylates ARF2 (Vert et al., 2008). These data suggest that ARF phosphorylation suppresses their interaction with Aux/IAAs, thus enhancing DNA binding and transcriptional activity. In addition, there is a growing body of evidence on the posttranscriptional regulation of ARF transcript abundance by miRNA and transacting-small interfering RNAs (ta-siRNA). While ARF6 and ARF8 are targets of miR167 and ARF10, ARF16, and ARF17 are targeted by miR160, ARF2, ARF3, and ARF4 are targets of TAS3 ta-siRNAs in Arabidopsis (Rhoades et al., 2002; Williams et al., 2005; Guilfoyle and Hagen, 2007; Lin et al., 2015).
KEY CONCEPT 4. Transacting-small interfering RNAs (Ta-siRNAs)
Ta-siRNAs are form of small interfering RNA (siRNA) that represses gene expression through post-transcriptional gene silencing in land plants. They are transcribed from the genome to form a polyadenylated, double-stranded segment of RNA that gets processed further, resulting in a segment of RNA that is 21-nucleotides long. These segments are incorporated into the RNA-induced Silencing Complex and direct the cleavage of target mRNA.
The Arabidopsis genome encodes 23 ARF proteins (Rademacher et al., 2011) and genetic analyses have shown that individual ARFs control distinct developmental processes based on their loss-of-function mutant phenotypes (Guilfoyle and Hagen, 2007; Rademacher et al., 2012). Although ARFs appear to have unique functions in some contexts, they display overlapping functions in others. For example, both ARF1 and ARF2 control leaf senescence and floral organ abscission in Arabidopsis (Ellis et al., 2005), while ARF3 interacts with KANADI proteins to form a functional complex essential for leaf polarity specification (Kelley et al., 2012). A recent study indicated that ARF3 integrates the functions of AGAMOUS (AG) and APETALA2 (AP2) in floral meristem determinacy (Liu et al., 2014b), while ARF4 has been studied primarily for its role in organ polarity (Hunter et al., 2006). However, the arf3arf4 double mutant plant has reduced abaxial identity in all lateral organs, including leaves (Pekker et al., 2005; Finet et al., 2010). ARF5 is critically required for embryonic root and flower formation (Hardtke and Berleth, 1998) and embryo patterning and vasculature defects observed in arf5 mutants are enhanced in arf5arf7 double mutants (Hardtke and Berleth, 1998). ARF8 is reported to regulate fertilization and fruit development (Goetz et al., 2006), and ARF6 and ARF8 act redundantly in flower maturation (Finet et al., 2010). ARF19 and ARF7 act redundantly with in controlling leaf expansion and lateral root growth (Wilmoth et al., 2005). While no phenotypic defects were reported for arf10 or arf16 single mutants (Okushima et al., 2005), arf10arf16 double mutants show a strong auxin phenotype that results in the absence of lateral root formation, which is not observed in neither the arf10 or arf16 single mutant (Wang et al., 2005).
In the case of tomato, genetic studies have shown that the mechanism of ARF signaling is different to that of Arabidopsis. A total of 21 putative functional SlARFs have been identified in tomato (Zouine et al., 2014). Although, SlARF3 RNAi lines do not display phenotypes such as floral organogenes or developmental timing changes (Sessions et al., 1997), SlARF3 plays multiple roles in tomato development and is involved in the formation of epidermal cells and trichomes (Zhang et al., 2015b). The functional analysis of SlARF9 indicated that it regulates cell division during early tomato fruit development (DeJong et al., 2015). A recent study confirmed that down-regulation of ARF6 and ARF8 by mi167 leads to floral development defects and female sterility in tomatoes. These results indicate that ARF6 and ARF8 have conserved roles in controlling growth and development of vegetative and flower organs in dicots (Liu et al., 2014a). SlARF7 acts as a negative regulator of fruit set until pollination and fertilization have taken place and moderates the auxin response during fruit growth in tomatoes (de Jong et al., 2009). Meanwhile, SlARF7 mediates cross-talk between auxin and gibberellin signaling during tomato fruit set and development (de Jong et al., 2011). Interestingly, SlARF4 is involved in the control of sugar metabolism during tomato fruit development (Sagar et al., 2013). In soybeans, the miR167-directed regulation of GmARF8a and GmARF8b is required for nodulation and lateral root development (Wang et al., 2015). In rice, OsARF16 and OsARF12 are required for iron deficiency response by regulating auxin redistribution (Wang et al., 2014; Shen et al., 2015b). OsARF3 mediates the auxin response during de novo shoot regeneration (Cheng et al., 2013). OsARF19 controls rice leaf angles through positively regulating OsGH3-5 and brassinosteroid insensitive 1 (OsBRI1) in rice (Zhang et al., 2015a).
During the last 10 years, our understanding of ARF regulatory mechanism and their role during model plant growth and development has been greatly improved by forward and reverse genetic approaches. Nonetheless, there are still many gaps in our knowledge and we lack a deep understanding of these regulatory processes. For example, it is still not clear how repressors of ARFs regulate gene repression and how other transcription factors and signaling proteins interact with ARF proteins. However, a larger number of candidate genes that are regulated by ARFs have been identified both experimentally and through bioinformatics analysis in recent years. Therefore, it will be interesting to understand the function of these candidate genes and regulatory mechanism of some important ARF proteins. In addition, our knowledge of ARFs in plant species beyond model plants (typically Arabidopsis) is very limited. The great challenge will be to integrate knowledge about ARF regulation of different developmental processes across in plants, and to understand how these processes work in different plant species.
JZ, SL, and ZX wrote the paper. CH provided some suggestions for the paper.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This research was supported financially by the National Natural Science Foundation of China (grant nos. 31130046, 31471863, 31360469, and 31372046), the Fundamental Research Funds for the Central Universities (2013PY083) and the International Foundation for Science No. C/5148-2.

Blanc, G., Hokamp, K., and Wolfe, K. H. (2003). A recent polyploidy superimposed on older large-scale duplications in the Arabidopsis genome. Genome Res. 13, 137–144. doi: 10.1101/gr.751803
Boer, D. R., Freire-Rios, A., van den Berg, W. A., Saaki, T., Manfield, I. W., Kepinski, S., et al. (2014). Structural basis for DNA binding specificity by the auxin-dependent ARF transcription factors. Cell 156, 577–589. doi: 10.1016/j.cell.2013.12.027
Carranco, R., Espinosa, J. M., Prieto-Dapena, P., Almoguera, C., and Jordano, J. (2010). Repression by an auxin/indole acetic acid protein connects auxin signaling with heat shock factor-mediated seed longevity. Proc. Natl. Acad. Sci. U.S.A. 107, 21908–21913. doi: 10.1073/pnas.1014856107
Cheng, Z. J., Wang, L., Sun, W., Zhang, Y., Zhou, C., Su, Y. H., et al. (2013). Pattern of auxin and cytokinin responses for shoot meristem induction results from the regulation of cytokinin biosynthesis by AUXIN RESPONSE FACTOR3. Plant Physiol. 161, 240–251. doi: 10.1104/pp.112.203166
Cho, H., Ryu, H., Rho, S., Hill, K., Smith, S., Audenaert, D., et al. (2014). A secreted peptide acts on BIN2-mediated phosphorylation of ARFs to potentiate auxin response during lateral root development. Nat. Cell Biol. 16, 66–76. doi: 10.1038/ncb2893
de Jong, M., Wolters-Arts, M., Feron, R., Mariani, C., and Vriezen, W. H. (2009). The Solanum lycopersicum auxin response factor 7 (SlARF7) regulates auxin signaling during tomato fruit set and development. Plant J. 57, 160–170. doi: 10.1111/j.1365-313X.2008.03671.x
de Jong, M., Wolters-Arts, M., García-Martínez, J. L., Mariani, C., and Vriezen, W. H. (2011). The Solanum lycopersicum AUXIN RESPONSE FACTOR 7 (SlARF7) mediates cross-talk between auxin and gibberellin signalling during tomato fruit set and development. J. Exp. Bot. 62, 617–626. doi: 10.1093/jxb/erq293
DeJong, M., Wolters-Arts, M., Schimmel, B. C., Stultiens, C. L., de Groot, P. F., Powers, S. J., et al. (2015). Solanum lycopersicum AUXIN RESPONSE FACTOR 9 regulates cell division activity during early tomato fruit development. J. Exp. Bot. 66, 3405–3416. doi: 10.1093/jxb/erv152
Di, D.-W., Zhang, C., and Guo, G.-Q. (2015a). Involvement of secondary messengers and small organic molecules in auxin perception and signaling. Plant Cell Rep. 34, 895–904. doi: 10.1007/s00299-015-1767-z
Di, D.-W., Zhang, C., Luo, P., An, C.-W., and Guo, G.-Q. (2015b). The biosynthesis of auxin: how many paths truly lead to IAA? Plant Growth Regul. 1–11. doi: 10.1007/s10725-015-0103-5. [Epub ahead of print].
Ellis, C. M., Nagpal, P., Young, J. C., Hagen, G., Guilfoyle, T. J., and Reed, J. W. (2005). AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132, 4563–4574. doi: 10.1242/dev.02012
Finet, C., Berne-Dedieu, A., Scutt, C. P., and Marlétaz, F. (2012). Evolution of the ARF gene family in land plants: old domains, new tricks. Mol. Biol. Evol. 30, 45–56. doi: 10.1093/molbev/mss220
Finet, C., Fourquin, C., Vinauger, M., Berne-Dedieu, A., Chambrier, P., Paindavoine, S., et al. (2010). Parallel structural evolution of auxin response factors in the angiosperms. Plant J. 63, 952–959. doi: 10.1111/j.1365-313X.2010.04292.x
Goetz, M., Vivian-Smith, A., Johnson, S. D., and Koltunow, A. M. (2006). AUXIN RESPONSE FACTOR8 is a negative regulator of fruit initiation in Arabidopsis. Plant Cell 18, 1873–1886. doi: 10.1105/tpc.105.037192
Guilfoyle, T. J. (2015). The PB1 domain in auxin response factor and Aux/IAA proteins: a versatile protein interaction module in the auxin response. Plant Cell 27, 33–43. doi: 10.1105/tpc.114.132753
Guilfoyle, T. J., and Hagen, G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10, 453–460. doi: 10.1016/j.pbi.2007.08.014
Guilfoyle, T. J., and Hagen, G. (2012). Getting a grasp on domain III/IV responsible for Auxin Response Factor–IAA protein interactions. Plant Sci. 190, 82–88. doi: 10.1016/j.plantsci.2012.04.003
Hagen, G., and Guilfoyle, T. (2002). Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol. Biol. 49, 373–385. doi: 10.1023/A:1015207114117
Hardtke, C. S., and Berleth, T. (1998). The Arabidopsis gene MONOPTEROS encodes a transcription factor mediating embryo axis formation and vascular development. EMBO J. 17, 1405–1411. doi: 10.1093/emboj/17.5.1405
Hill, K. (2015). Post-translational modifications of hormone-responsive transcription factors: the next level of regulation. J. Exp. Bot. 66, 4933–4945. doi: 10.1093/jxb/erv273
Hu, W., Zuo, J., Hou, X., Yan, Y., Wei, Y., Liu, J., et al. (2015). The auxin response factor gene family in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. Front. Plant Sci. 6:742. doi: 10.3389/fpls.2015.00742
Hunter, C., Willmann, M. R., Wu, G., Yoshikawa, M., de la Luz Gutiérrez-Nava, M., and Poethig, S. R. (2006). Trans-acting siRNA-mediated repression of ETTIN and ARF4 regulates heteroblasty in Arabidopsis. Development 133, 2973–2981. doi: 10.1242/dev.02491
Kalluri, U. C., Difazio, S. P., Brunner, A. M., and Tuskan, G. A. (2007). Genome-wide analysis of Aux/IAA and ARF gene families in Populus trichocarpa. BMC Plant Biol. 7:59. doi: 10.1186/1471-2229-7-59
Kelley, D. R., Arreola, A., Gallagher, T. L., and Gasser, C. S. (2012). ETTIN (ARF3) physically interacts with KANADI proteins to form a functional complex essential for integument development and polarity determination in Arabidopsis. Development 139, 1105–1109. doi: 10.1242/dev.067918
Kim, J., Harter, K., and Theologis, A. (1997). Protein–protein interactions among the Aux/IAA proteins. Proc. Natl. Acad. Sci. U.S.A. 94, 11786–11791. doi: 10.1073/pnas.94.22.11786
Korasick, D. A., Chatterjee, S., Tonelli, M., Dashti, H., Lee, S. G., Westfall, C. S., et al. (2015). Defining a two-pronged structural model for PB1 (Phox/Bem1p) domain interaction in plant auxin responses. J. Biol. Chem. 290, 12868–12878. doi: 10.1074/jbc.M115.648253
Kumar, R., Tyagi, A. K., and Sharma, A. K. (2011). Genome-wide analysis of auxin response factor (ARF) gene family from tomato and analysis of their role in flower and fruit development. Mol. Genet. Genomics 285, 245–260. doi: 10.1007/s00438-011-0602-7
Li, S.-B., OuYang, W.-Z., Hou, X.-J., Xie, L.-L., Hu, C.-G., and Zhang, J.-Z. (2015). Genome-wide identification, isolation and expression analysis of auxin response factor (ARF) gene family in sweet orange (Citrus sinensis). Front. Plant Sci. 6:119. doi: 10.3389/fpls.2015.00119
Lin, Y., Lai, Z., Tian, Q., Lin, L., Lai, R., Yang, M., et al. (2015). Endogenous target mimics down-regulate miR160 mediation of ARF10, -16 and -17 cleavage during somatic embryogenesis in Dimocarpus longan Lour. Front. Plant Sci. 6:956. doi: 10.3389/fpls.2015.00956
Liscum, E., and Reed, J. (2002). Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 49, 387–400. doi: 10.1023/A:1015255030047
Liu, K., Yuan, C., Li, H., Lin, W., Yang, Y., Shen, C., et al. (2015). Genome-wide identification and characterization of auxin response factor (ARF) family genes related to flower and fruit development in papaya (Carica papaya L.). BMC Genomics 16:901. doi: 10.1186/s12864-015-2182-0
Liu, N., Wu, S., Van Houten, J., Wang, Y., Ding, B., Fei, Z., et al. (2014a). Down-regulation of AUXIN RESPONSE FACTORS 6 and 8 by microRNA 167 leads to floral development defects and female sterility in tomato. J. Exp. Bot. 65, 2507–2520. doi: 10.1093/jxb/eru141
Liu, S. Q., and Hu, L. H. (2013). Genome-wide analysis of the auxin response factor gene family in cucumber. Genet. Mol. Res. 12, 4317–4331. doi: 10.4238/2013.April.2.1
Liu, X., Dinh, T. T., Li, D., Shi, B., Li, Y., Cao, X., et al. (2014b). AUXIN RESPONSE FACTOR 3 integrates the functions of AGAMOUS and APETALA2 in floral meristem determinacy. Plant J. 80, 629–641. doi: 10.1111/tpj.12658
Liu, Y., Jiang, H., Chen, W., Qian, Y., Ma, Q., Cheng, B., et al. (2011). Genome-wide analysis of the auxin response factor (ARF) gene family in maize (Zea mays). Plant Growth Regul. 63, 225–234. doi: 10.1007/s10725-010-9519-0
Luo, X.-C., Sun, M.-H., Xu, R. R., Shu, H.-R., Wang, J. W., and Zhang, S.-Z. (2014). Genomewide identification and expression analysis of the ARF gene family in apple. J. Genet. 93, 785–797. doi: 10.1007/s12041-014-0462-0
Mockaitis, K., and Estelle, M. (2008). Auxin receptors and plant development: a new signaling paradigm. Annu. Rev. Cell Dev. Biol. 24, 55–80. doi: 10.1146/annurev.cellbio.23.090506.123214
Mun, J.-H., Yu, H.-J., Shin, J. Y., Oh, M., Hwang, H.-J., and Chung, H. (2012). Auxin response factor gene family in Brassica rapa: genomic organization, divergence, expression, and evolution. Mol. Genet. Genomics 287, 765–784. doi: 10.1007/s00438-012-0718-4
Okushima, Y., Overvoorde, P. J., Arima, K., Alonso, J. M., Chan, A., Chang, C., et al. (2005). Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: unique and overlapping functions of ARF7 and ARF19. Plant Cell 17, 444–463. doi: 10.1105/tpc.104.028316
Pekker, I., Alvarez, J. P., and Eshed, Y. (2005). Auxin response factors mediate Arabidopsis organ asymmetry via modulation of KANADI activity. Plant Cell 17, 2899–2910. doi: 10.1105/tpc.105.034876
Piya, S., Shrestha, S. K., Binder, B., Stewart, C. N. Jr., and Hewezi, T. (2014). Protein-protein interaction and gene co-expression maps of ARFs and Aux/IAAs in Arabidopsis. Front. Plant Sci. 5:744. doi: 10.3389/fpls.2014.00744
Rademacher, E. H., Lokerse, A. S., Schlereth, A., Llavata-Peris, C. I., Bayer, M., Kientz, M., et al. (2012). Different auxin response machineries control distinct cell fates in the early plant embryo. Dev. Cell 22, 211–222. doi: 10.1016/j.devcel.2011.10.026
Rademacher, E. H., Möller, B., Lokerse, A. S., Llavata-Peris, C. I., van den Berg, W., and Weijers, D. (2011). A cellular expression map of the Arabidopsis AUXIN RESPONSE FACTOR gene family. Plant J. 68, 597–606. doi: 10.1111/j.1365-313X.2011.04710.x
Rhoades, M. W., Reinhart, B. J., Lim, L. P., Burge, C. B., Bartel, B., and Bartel, D. P. (2002). Prediction of plant microRNA targets. Cell 110, 513–520. doi: 10.1016/S0092-8674(02)00863-2
Rosado, A., Li, R., Van de Ven, W., Hsu, E., and Raikhel, N. V. (2012). Arabidopsis ribosomal proteins control developmental programs through translational regulation of auxin response factors. Proc. Natl. Acad. Sci. U.S.A. 109, 19537–19544. doi: 10.1073/pnas.1214774109
Sagar, M., Chervin, C., Mila, I., Hao, Y., Roustan, J.-P., Benichou, M., et al. (2013). SlARF4, an auxin response factor involved in the control of sugar metabolism during tomato fruit development. Plant Physiol. 161, 1362–1374. doi: 10.1104/pp.113.213843
Salehin, M., Bagchi, R., and Estelle, M. (2015). SCFTIR1/AFB-based auxin perception: mechanism and role in plant growth and development. Plant Cell 27, 9–19. doi: 10.1105/tpc.114.133744
Sessions, A., Nemhauser, J. L., McColl, A., Roe, J. L., Feldmann, K. A., and Zambryski, P. C. (1997). ETTIN patterns the Arabidopsis floral meristem and reproductive organs. Development 124, 4481–4491.
Shen, C., Yue, R., Sun, T., Zhang, L., Xu, L., Tie, S., et al. (2015a). Genome-wide identification and expression analysis of auxin response factor gene family in Medicago truncatula. Front. Plant Sci. 6:73. doi: 10.3389/fpls.2015.00073
Shen, C., Yue, R., Sun, T., Zhang, L., Yang, Y., and Wang, H. (2015b). OsARF16, a transcription factor regulating auxin redistribution, is required for iron deficiency response in rice (Oryza sativa L.). Plant Sci. 231, 148–158. doi: 10.1016/j.plantsci.2014.12.003
Shin, R., Burch, A. Y., Huppert, K. A., Tiwari, S. B., Murphy, A. S., Guilfoyle, T. J., et al. (2007). The Arabidopsis transcription factor MYB77 modulates auxin signal transduction. Plant Cell 19, 2440–2453. doi: 10.1105/tpc.107.050963
Su, Y. H., Liu, Y. B., Bai, B., and Zhang, X. S. (2014). Establishment of embryonic shoot–root axis is involved in auxin and cytokinin response during Arabidopsis somatic embryogenesis. Front. Plant Sci. 5:792. doi: 10.3389/fpls.2014.00792
Sun, R., Wang, K., Guo, T., Jones, D. C., Cobb, J., Zhang, B., et al. (2015). Genome-wide identification of auxin response factor (ARF) genes and its tissue-specific prominent expression in Gossypium raimondii. Funct. Integr. Genomics 15, 481–493. doi: 10.1007/s10142-015-0437-0
Szemenyei, H., Hannon, M., and Long, J. A. (2008). TOPLESS mediates auxin-dependent transcriptional repression during Arabidopsis embryogenesis. Science 319, 1384–1386. doi: 10.1126/science.1151461
Tiwari, S. B., Hagen, G., and Guilfoyle, T. (2003). The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15, 533–543. doi: 10.1105/tpc.008417
Ulmasov, T., Hagen, G., and Guilfoyle, T. J. (1997). ARF1, a transcription factor that binds to auxin response elements. Science 276, 1865–1868. doi: 10.1126/science.276.5320.1865
Ulmasov, T., Hagen, G., and Guilfoyle, T. J. (1999). Dimerization and DNA binding of auxin response factors. Plant J. 19, 309–319. doi: 10.1046/j.1365-313X.1999.00538.x
Van Ha, C., Le, D. T., Nishiyama, R., Watanabe, Y., Sulieman, S., Tran, U. T., et al. (2013). The auxin response factor transcription factor family in soybean: genome-wide identification and expression analyses during development and water stress. DNA Res. 20, 511–524. doi: 10.1093/dnares/dst027
Varaud, E., Brioudes, F., Szécsi, J., Leroux, J., Brown, S., Perrot-Rechenmann, C., et al. (2011). AUXIN RESPONSE FACTOR8 regulates Arabidopsis petal growth by interacting with the bHLH transcription factor BIGPETALp. Plant Cell 23, 973–983. doi: 10.1105/tpc.110.081653
Vert, G., Walcher, C. L., Chory, J., and Nemhauser, J. L. (2008). Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc. Natl. Acad. Sci. U.S.A. 105, 9829–9834. doi: 10.1073/pnas.0803996105
Wan, S., Li, W., Zhu, Y., Liu, Z., Huang, W., and Zhan, J. (2014). Genome-wide identification, characterization and expression analysis of the auxin response factor gene family in Vitis vinifera. Plant Cell Rep. 33, 1365–1375. doi: 10.1007/s00299-014-1622-7
Wang, D., Pei, K., Fu, Y., Sun, Z., Li, S., Liu, H., et al. (2007). Genome-wide analysis of the auxin response factors (ARF) gene family in rice (Oryza sativa). Gene 394, 13–24. doi: 10.1016/j.gene.2007.01.006
Wang, J.-W., Wang, L.-J., Mao, Y.-B., Cai, W.-J., Xue, H.-W., and Chen, X.-Y. (2005). Control of root cap formation by microRNA-targeted auxin response factors in Arabidopsis. Plant Cell 17, 2204–2216. doi: 10.1105/tpc.105.033076
Wang, R., and Estelle, M. (2014). Diversity and specificity: auxin perception and signaling through the TIR1/AFB pathway. Curr. Opin. Plant Biol. 21, 51–58. doi: 10.1016/j.pbi.2014.06.006
Wang, S., Zhang, S., Sun, C., Xu, Y., Chen, Y., Yu, C., et al. (2014). Auxin response factor (OsARF12), a novel regulator for phosphate homeostasis in rice (Oryza sativa). New Phytol. 201, 91–103. doi: 10.1111/nph.12499
Wang, Y., Li, K., Chen, L., Zou, Y., Liu, H., Tian, Y., et al. (2015). microrna167-directed regulation of the auxin response factors, GmARF8a and GmARF8b, is required for soybean (Glycine max L.) nodulation and lateral root development. Plant Physiol. 168, 984–999. doi: 10.1104/pp.15.00265
Williams, L., Carles, C. C., Osmont, K. S., and Fletcher, J. C. (2005). A database analysis method identifies an endogenous trans-acting short-interfering RNA that targets the Arabidopsis ARF2, ARF3, and ARF4 genes. Proc. Natl. Acad. Sci. U.S.A. 102, 9703–9708. doi: 10.1073/pnas.0504029102
Wilmoth, J. C., Wang, S., Tiwari, S. B., Joshi, A. D., Hagen, G., Guilfoyle, T. J., et al. (2005). NPH4/ARF7 and ARF19 promote leaf expansion and auxin-induced lateral root formation. Plant J. 43, 118–130. doi: 10.1111/j.1365-313X.2005.02432.x
Wu, J., Wang, F., Cheng, L., Kong, F., Peng, Z., Liu, S., et al. (2011). Identification, isolation and expression analysis of auxin response factor (ARF) genes in Solanum lycopersicum. Plant Cell Rep. 30, 2059–2073. doi: 10.1007/s00299-011-1113-z
Yu, H., Soler, M., Mila, I., San Clemente, H., Savelli, B., Dunand, C., et al. (2014). Genome-wide characterization and expression profiling of the AUXIN RESPONSE FACTOR (ARF) gene family in Eucalyptus grandis. PLoS ONE 9:e108906. doi: 10.1371/journal.pone.0108906
Zhang, S., Wang, S., Xu, Y., Yu, C., Shen, C., Qian, Q., et al. (2015a). The auxin response factor, OsARF19, controls rice leaf angles through positively regulating OsGH3−5 and OsBRI1. Plant Cell Environ. 38, 638–654. doi: 10.1111/pce.12397
Zhang, X., Yan, F., Tang, Y., Yuan, Y., Deng, W., and Li, Z. (2015b). Auxin response gene SlARF3 plays multiple roles in tomato development and is involved in the formation of epidermal cells and trichomes. Plant Cell Physiol. 56, 2110–2124. doi: 10.1093/pcp/pcv136
Keywords: auxin, ARF, auxin response DNA elements, Arabidopsis, DBD domain, a type I/II Phox and Bem1p (PB1)
Citation: Li S-B, Xie Z-Z, Hu C-G and Zhang J-Z (2016) A Review of Auxin Response Factors (ARFs) in Plants. Front. Plant Sci. 7:47. doi: 10.3389/fpls.2016.00047
Received: 04 November 2015; Accepted: 12 January 2016;
Published: 03 February 2016.
Edited by:
Elena Prats, Consejo Superior de Investigaciones Científicas, SpainReviewed by:
Lei Zhang, Washington State University, USACopyright © 2016 Li, Xie, Hu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: amluemhpemhhbmdAbWFpbC5oemF1LmVkdS5jbg==; amluemhpMzI3MzIwMDk0QDEyNi5jb20=
†These authors have contributed equally to this work.
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.