- National Institute of Plant Genome Research, New Delhi, India
14-3-3s are highly conserved, multigene family proteins that have been implicated in modulating various biological processes. The presence of inherent polyploidy and genome complexity has limited the identification and characterization of 14-3-3 proteins from globally important Brassica crops. Through data mining of Brassica rapa, the model Brassica genome, we identified 21 members encoding 14-3-3 proteins namely, BraA.GRF14.a to BraA.GRF14.u. Phylogenetic analysis indicated that B. rapa contains both ε (epsilon) and non-ε 14-3-3 isoforms, having distinct intron-exon structural organization patterns. The non-ε isoforms showed lower divergence rate (Ks < 0.45) compared to ε protein isoforms (Ks > 0.48), suggesting class-specific divergence pattern. Synteny analysis revealed that mesohexaploid B. rapa genome has retained 1–5 orthologs of each Arabidopsis 14-3-3 gene, interspersed across its three fragmented sub-genomes. qRT-PCR analysis showed that 14 of the 21 BraA.GRF14 were expressed, wherein a higher abundance of non-ε transcripts was observed compared to the ε genes, indicating class-specific transcriptional bias. The BraA.GRF14 genes showed distinct expression pattern during plant developmental stages and in response to abiotic stress, phytohormone treatments, and nutrient deprivation conditions. Together, the distinct expression pattern and differential regulation of BraA.GRF14 genes indicated the occurrence of functional divergence of B. rapa 14-3-3 proteins during plant development and stress responses.
Introduction
14-3-3 proteins derived their unique name from the studies of fractionation of bovine brain proteins on DEAE cellulose and their electrophoretic mobility on starch gel electrophoresis (Moore and Perez, 1967). These regulatory proteins are present in all eukaryotes and involved in protein interactions mediated signal transduction pathways. In plants, 14-3-3 proteins function by binding to numerous “client” proteins in a phosphorylation-dependent manner to modulate their activities, degradation, or sub-cellular localization (Rosenquist et al., 2001; Paul et al., 2012). So far, more than 300 putative 14-3-3 interacting client proteins have been reported in plants out of which nitrate reductase, sucrose-phosphate synthase, plasma membrane H+-ATPase, EmBP1, and VP1 transcription factors, the RSG transcription activator, a lipoxygenase from barley, a membrane bound ascorbate peroxidase and a outward-rectifying K+ channel in tomato are well-characterized (reviewed in Oecking and Jaspert, 2009; Denison et al., 2011; de Boer et al., 2013). This high number reflects the potential role of the 14-3-3s in controlling various signaling and developmental processes in plants. Current literatures clearly suggest the involvement of 14-3-3 proteins in various physiological processes including primary carbon and nitrogen metabolism (Comparot et al., 2003), abiotic and biotic stress responses (Roberts et al., 2002; Umezawa et al., 2004; Yan et al., 2004; Chen et al., 2006; Yang et al., 2013; Catalá et al., 2014; Zhou et al., 2014; He et al., 2015; Li et al., 2015), signaling pathways of phytohormones like ABA, GA, and BR (Testerink et al., 1999; Igarashi et al., 2001; Ryu et al., 2007; Kim et al., 2009; Zhou et al., 2015), and also during plant growth and development (Radwan et al., 2012; de Boer et al., 2013; Sun et al., 2014; van Kleeff et al., 2014).
To carry such diverse roles, almost all eukaryotes harbor multiple isoforms of 14-3-3 genes, with two present in yeast, seven in humans, and more than a dozen in vascular plants. The model monocot (Oryza sativa) and dicot (Arabidopsis thaliana) plant genomes encode 8 and 13 expressed 14-3-3 genes, respectively (DeLille et al., 2001; Rosenquist et al., 2001; Chen et al., 2006; Yao et al., 2007). Data mining of sequenced plant genomes has led to the identification of much higher number of 14-3-3 genes, particularly from polyploid genomes. A total of thirty-one 14-3-3 cDNAs encoding 25 unique proteins were identified from allotetraploid cotton (Sun et al., 2011), whereas the diploidized tetraploid soybean genome has eighteen 14-3-3 gene homologs, of which 16 are expressed (Li and Dhaubhadel, 2011). The chromosomal/segmental duplications and the evolutionary diversification are largely known to shape the quantitative variability of functional 14-3-3 proteins across plant species. Even though high sequence similarity exists among multiple copies of 14-3-3s, the specificity of these protein isoforms harboring definite subcellular localization, tissue-specific expression, and dynamic regulation in response to environmental changes is well-reviewed (Kjarland et al., 2006). Also each isoform, expressing in different subcellular location inside the cell, interacts with different client partners and relay the downstream signaling. This partly explains the versatility of so many isoforms in a plant species regulating a wide range of biological processes and functions.
Brassica species, the closest crop relatives to Arabidopsis, play an important role in global agriculture and horticulture. Brassica rapa (field mustard) is one of the globally important Brassica crop because of its enormous genetic and morphological diversity, and being utilized as leafy vegetables, vegetable oils, turnips roots, turnip greens, turnip tops, and fodder turnip. Besides, it is one of the diploid progenitor species (n = 10) which contributed the “A” genome to the important oilseed crops, Brassica juncea (n = 18, AABB) and Brassica napus (n = 19, AACC). Because of its pivotal position among the Brassica species, the recent sequencing of B. rapa genome offers an excellent opportunity to study the structural and functional evolution of candidate genes (The B. rapa Genome Sequencing Project Consortium; Wang et al., 2011). Sequence level studies although reflect high similarity in functional genes present between Arabidopsis and B. rapa, quantitative variation and evolutionary divergence in members of gene families present in polyploid Brassica genome, however, may contribute to the remarkable phenotypic plasticity and environmental adaptability of economically important Brassica species.
To investigate the important roles played by 14-3-3 protein isoforms in Brassica crops, comprehensive analysis of 14-3-3 gene homologs was undertaken from B. rapa. Present study through data mining of the recently sequenced B. rapa genome identified 21 divergent 14-3-3 genes providing their chromosomal and sub-genomic localization, phylogenetic relationship, and divergence analysis. We further carried out comprehensive expression profiling of 14 expressed 14-3-3 gene family members in B. rapa across plant development stages, abiotic stress conditions, phytohormone treatments and under nutrient deprivation conditions. We observed highly coordinated tissue- and condition-specific expression of B. rapa 14-3-3 transcripts, suggesting their multifarious roles across plant growth, development, and environmental cues. The study provides an excellent base for conducting further in-depth research on various signaling pathways regulated by B. rapa 14-3-3 proteins, which could be utilized for agricultural improvements of the mustard crop.
Materials and Methods
Plant Materials and Growth Condition
B. rapa genotype YID1 was grown in a controlled growth conditions set as day (22°C, 10 h)/night (15°C, 14 h) cycle and 70% relative humidity. Different developmental stages namely, seedling, root, stem, leaf, and silique (20 days post anthesis), were harvested and immediately frozen in liquid nitrogen and stored at −80°C.
Identification of the 14-3-3 Isoforms and Phylogenetic Comparisons
Arabidopsis 14-3-3 gene sequences were used to search against the B. rapa genome database (http://brassicadb.org/brad/; Cheng et al., 2011). The retrieved sequences were then named according to the existing nomenclature as BraA.GRF14.a to BraA.GRF14.u. Multiple sequence alignments of the encoded proteins were done using Clustal W and the phylogenetic trees were constructed using the Neighborhood-joining method of MEGA5 (Tamura et al., 2011). The human 14-3-3 theta isoform (NP_006817) was used as an out-group protein.
RNA Isolation and cDNA Synthesis
RNA was extracted from plant tissues using the Spectrum Plant Total RNA Kit (Sigma Life Sciences, USA) according to manufacturer's instructions. The quantity and quality of RNA sample was checked using Nano spectrophotometer (ND-1000 Thermo scientific); and RNA samples with 260/280 ratio (1.9–2.1) and 260/230 ratio (2.0–2.5) were used for further analysis. First strand cDNA was synthesized by reverse transcribing 2 μg of total RNA with random primers of high-capacity cDNA Reverse Transcription kit (Applied Biosystems, USA) in a 20 μl reaction according to manufacturer's instructions. Diluted cDNA (1:50) was used for the real-time qRT-PCR reaction.
Expression Profiling of B. rapa 14-3-3 Genes
Real-Time PCR was performed using standard cycling conditions (95°C for 10 min, 40 cycles of 15 s at 95 and 60°C for 60 s) in final volume of 20 μl in a 7900 HT real time PCR machine (Applied Biosystems). Reaction mixture contained SYBR Green Master Mix (Kapa Biosystems), 10 pmol of gene-specific forward and reverse primers, and 2 μl of the diluted cDNA (~200 pg). To check for the specificity of PCR amplification dissociation curve was generated (Figure S3). The Ct-values were determined for each reaction using SDS version 2.3 and RQ manager version 1.2 (Applied Biosciences) software with default parameters. GAPDH and ACT2 genes of Brassica origin were used as endogenous control (Chandna et al., 2012). Three independent sets of experiments were conducted with two technical replicates each to confirm results. Primers used for qRT-PCR analysis are tabulated in Table S4.
Elicitor and Stress Treatments
For elicitor and stress treatments, seedlings were grown initially for 5 days on agar plates containing one-half strength Murashige and Skoog (MS) medium. Before elicitor induction, seedlings were adapted to liquid MS culture containing 1% sucrose for 24 h. For hormones induction, methyl jasmonate (MeJA, 0.2 mM), salicylic acid (SA, 0.2 mM), indole-3-acetic acid (IAA, 0.1 mM), and abscisic acid (ABA, 0.1 mM) were independently added to the medium (Chandna et al., 2012). Seedlings were also subjected to different stress conditions such as heat shock (37°C), cold (4°C), salinity (300 mM NaCl), and dehydration (between folds of tissue paper). The plants were harvested at 15 min, 30 min, 3 h, and 6 h durations and the mock treated seedling for same time interval served as control. For elicitor treatment experiments, the expression cut-off of 1.5-fold change (w.r.t. corresponding control) was used to identify the up- and down-regulated transcripts.
Nutrient Deprivation Experiments
Plants were initially grown in MS agar medium for 3 days at 22°C with 10 h daylight at 200 μmol m−2 s−1. Seedlings were then transferred and adapted for 2 days on hydroponic nutrient solution containing 5 mM KNO3, 2.5 mM KH2PO4, 2.5 mM Fe-EDTA, 2 mM Ca(NO3)2, 2 mM MgSO4, and the following micronutrients: 1 mM NaCl, 1.4 mM MgCl2, 0.001 mM CaCl2, 7 mM H3BO3, 0.1 mM ZnSO4, 0.05 mM CuSO4, and 0.002 mM Na2MoO4. For nutrient deprivation experiments, the 2 days old hydroponically adapted seedlings were then transferred to nutrient solution lacking either Phosphorus (P), or Potassium (K), or Nitrogen (N). For the “P” deprivation, 2.5 mM KH2PO4 was replaced with 2.5 mM K2SO4. For the “K” deprivation, 5 mM KNO3 was replaced with 3 mM Ca(NO3)2; whereas 2.5 mM KH2PO4 was replaced with 0.25 mM K2SO4. For the “N” deprivation, 5 mM KNO3, and 2 mM Ca(NO3)2 were replaced with 5 mM KCl and 2 mM CaCl2, respectively. The untreated control and nutrient deprived seedlings were harvested at 1, 6, 24, and 48 h after treatments, immediately frozen in liquid nitrogen, and stored at −80°C until RNA extraction.
Results
Identification and Sequence Analysis of 14-3-3 Gene Family from B. rapa
The availability of initial draft of B. rapa genome project (http://brassicadb.org/brad/) led us to perform the comprehensive search of 14-3-3 gene sequences. Using Arabidopsis GRF (General Regulatory Factor) cDNA sequences as query, BLAST search in B. rapa genome database resulted in the identification of 21 putative gene sequences encoding 14-3-3 proteins (Table 1). The 21 BraA.GRF14 genes were located on eight out of 10 linkage groups (LG) of B. rapa genome, with A07 LG containing up to six BraA.GRF14 genes (Figure S1, Table 1). Adopting the standard gene nomenclature for Brassica species (Ostergaard and King, 2008), the genes were named as BraA.GRF14.a to BraA.GRF14.u, in order of their identification. The 21 BraA.GRF14 genes were variable in their sizes ranging from 998 to 1857 bp. The coding DNA sequences (CDS) of BraA.GRF14 genes ranged from 738 to 918 bp and shared 41.8–92.6% sequence identity among them (Table 1, Table S1).
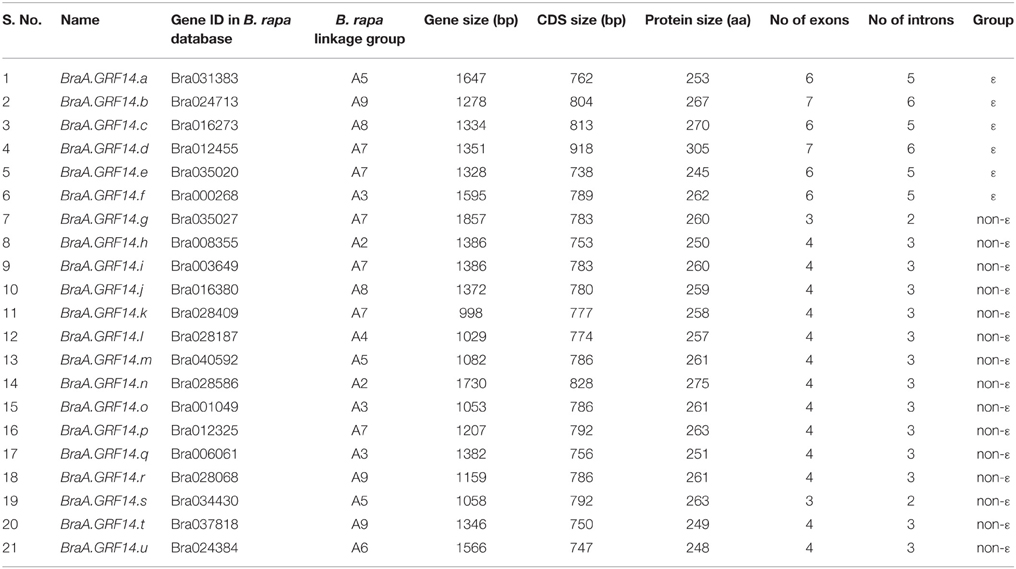
Table 1. Summary of gene structure attributes of the twenty-one 14-3-3 proteins identified in B. rapa genome (http://brassicadb.org/brad/).
The BraA.GRF14 genes encoded proteins ranging from 245 to 305 amino acids, with their calculated molecular masses and pI that ranged from 27.79 to 35.28 kDa and 4.34 to 4.77, respectively. Amino-acid sequence alignment of the deduced BraA.GRF14 proteins indicated that these are highly conserved proteins (Figure 1) sharing a high level of sequence identity (45.4–96.8%) among them (Table S2). Further, when we queried the deduced BraA.GRF14 proteins at ExPASy proteomics tool (www.expasy.org) all proteins, except for BraA.GRF14.e (Bra035020), showed the presence of highly conserved 14-3-3 signature motifs RNL(L/V)SV(G/A)YKNV and YKDSTLIMQLLRDNLTLWTS, thereby confirming their identity as 14-3-3 proteins. The divergence observed in the deduced Bra035020 (BraA.GRF14.e) protein sequence might be an evolutionary consequence; although the possibility of its mis-annotation in the current version of B. rapa genome assembly cannot be ruled out completely. In addition, the conserved phosphorylation sites reported earlier for plant's 14-3-3 proteins were also present in BraA.GRF14 proteins. The pY137 of maize GF14-6 known to decreases binding of the H+-ATPase (Giacometti et al., 2004); and pS216, pS220, pT221 reported to be phosphorylated during the seed development in oilseed rape (Agrawal and Thelen, 2006), were all found to be conserved across BraA.GRF14 proteins. The pS93/95 residue, identified as being phosphorylated by SnRK2.8 in roots of the three Arabidopsis GRFs (χ, κ, ψ; Shin et al., 2007), was however found to be present in 7 of the 21 BraA.GRF14 proteins, thereby suggesting isoform-specific phosphorylation pattern and functional specificity of 14-3-3 proteins in B. rapa.

Figure 1. Amino acid alignment of the deduced 14-3-3 proteins of B. rapa (BraA.GRF14). The sequence alignment of the 21 BraA.GRF14 proteins was performed using ClustalW. The 14-3-3 signature motifs RNL(L/V)SV(G/A)YKNV and YKDSTLIMQLLRDNLTLWTS, are underlined. The phosphorylation sites reported earlier for plant 14-3-3 proteins (Paul et al., 2012) are marked with asterisk.
Genomic Structure and Phylogenetic Relationships of B. rapa 14-3-3 Genes
The presence of multiple 14-3-3 type sequences in B. rapa having significant sequence divergence led us to investigate the evolution of BraA.GRF14 gene family. We therefore, analyzed the genomic structure and phylogenetic relationship of BraA.GRF14 gene family members. Analysis of genomic structure of 21 BraA.GRF14 genes showed that the members of 14-3-3 gene in B. rapa contain 3–7 exons, interspersed by highly divergent introns (Figure 2). On the basis of organization of exons and introns, the BraA.GRF14 genes were broadly categorized into two distinct sub-groups namely, the ε (epsilon) and non-ε (non-epsilon) groups. The six genes belonging to ε group contained 6–7 exons each, whereas 3–4 exons were present in the 15 non-ε group genes. Furthermore, the length of the exons was highly conserved among most of the 15 non-ε group genes; whereas genes belonging to ε group showed comparably higher divergence in their exon length. The extreme conservation of the exon organization in both ε and non-ε BraA.GRF14 genes possibly suggest independent evolution and expansion of the ε and non-ε groups genes in B. rapa. In comparison, the introns in both ε and non-ε groups genes of B. rapa were highly divergent in their sizes and sequences.
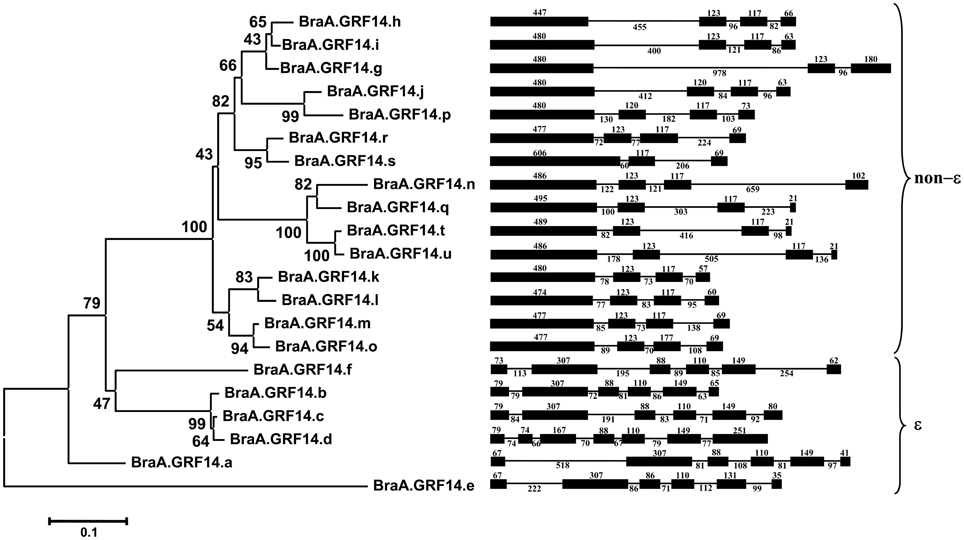
Figure 2. Phylogenetic analysis and gene structures of the deduced 14-3-3 proteins of B. rapa (BraA.GRF14). The evolutionary history was inferred by using the Maximum Likelihood method based on the JTT matrix-based model conducted in MEGA5 (Tamura et al., 2011). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) is shown next to the branches. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site. The sizes (in bp) and organization of exons (dark boxes) and introns (lines) of BraA.GRF14 genes are marked along with.
To get a better insight into expansion of B. rapa 14-3-3 gene family, a phylogenetic tree was constructed based on the multiple sequence alignment of full-length protein sequences from B. rapa (21), Arabidopsis (13) and Oryza sativa (7). Phylogenetic analysis showed that 14-3-3 protein isoforms from the three plant genomes were clustered into distinct ε and non-ε groups (Figure 2, Figure S2). Within each class, the 14-3-3 proteins from Arabidopsis and B. rapa genomes were grouped together, whereas the rice 14-3-3 proteins formed separate branches. This indicates that the 14-3-3 proteins of each class existed before the divergence of monocots and dicots, and have expanded independently in species-specific manner, as also observed for other gene families in plants (Zhang et al., 2005; Jain et al., 2007).
Sub-Genomic Location and Divergence Analysis of B. rapa 14-3-3 Genes
It is well-known that the whole genome triplication (WGT) event in Brassica lineage had formed multiple homologs (paralogs) in each Brassica species. To analyze the degree of expansion of 14-3-3 genes between B. rapa and its nearest model plant Arabidopsis, we identify the sub-genomic location of the 21 BraA.GRF14 genes on B. rapa genome (http://brassicadb.org/brad/). The BraA.GRF14 genes were identified in all the three sub-genomes of B. rapa, with both least fractionated (LF) and most-fractionated (MF2) sub-genomes containing eight genes each, and the moderately fractionated (MF1) sub-genome having five genes only (Table 2).
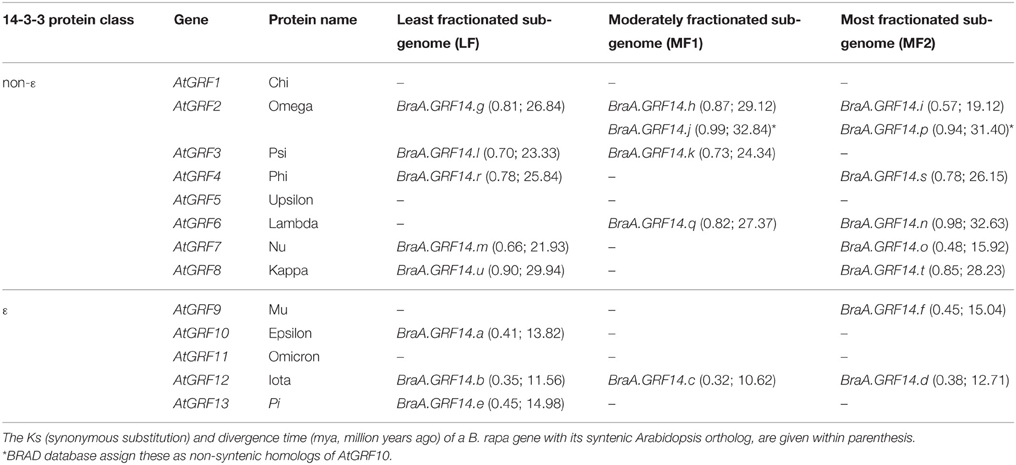
Table 2. Gene fractionation and divergence analysis of syntenic 14-3-3 genes identified in the three sub-genomes of B. rapa (http://brassicadb.org/brad/) with their corresponding Arabidopsis orthologs.
Of the 13 AtGRF14 genes, syntenic homologs for 10 could be identified in the B. rapa, with each Arabidopsis gene having variable number of B. rapa paralogs (Table 2; Figure S2). A total of five and three B. rapa paralogs was identified for AtGRF2 (omega) and AtGRF12 (iota) genes, respectively; whereas two paralogs were identified for five Arabidopsis 14-3-3 genes in B. rapa genome. Only one B. rapa homologs was identified for three AtGRF genes. The gene-fractionation events in Brassica lineage in all possibility have led to the presence of variable number of 14-3-3 gene orthologs in B. rapa. Surprisingly, for three 14-3-3 genes of Arabidopsis, namely AtGRF1, AtGRF5, and AtGRF11 no syntenic homolog could be identified in the B. rapa genome. Phylogenetic analysis showed that AtGRF1/AtGRF4, AtGRF5/AtGRF7, and AtGRF11/AtGRF12 gene pairs formed individual branches, and could have arisen as the result of Arabidopsis specific At-α WGD event, dating around 24–40 mya (Franzke et al., 2011). Divergence analysis of these proteins pairs also showed that at least AtGRF1/AtGRF4, and AtGRF5/AtGRF7 gene pairs might have duplicated recently in Arabidopsis lineage around 20.92 and 24.76 mya, respectively, after the Arabidopsis–Brassica split.
We further estimated the divergence of BraA.GRF14 genes retained in the extant B. rapa genome by performing the pairwise comparisons to estimate the synonymous base substitution (Ks)-values between the duplicated B. rapa and Arabidopsis genes. Divergence times were calculated assuming a mutation rate of 1.5 × 10−8 synonymous substitutions per year (Koch et al., 2000). The Ka/Ks ratios of Arabidopsis–B. rapa 14-3-3 orthologs were less than 1, suggesting purifying selection on these duplicated pairs (Table 2; Table S3). The Ks-values of BraA.GRF14 genes ranged from 0.32 to 0.99, which indicated that the BraA.GRF14 genes might have diverged somewhere between 10.62 and 32.84 million years ago (mya). Interestingly, the ε protein orthologs shared between the Arabidopsis–B. rapa genomes showed lower range of Ks-values (0.32–0.45) compared to the non-ε protein orthologs with higher range of Ks-values (0.48–0.99). This class-specific divergence pattern indicated that the non-ε proteins might have diverge recently (10.62–15.04 mya) compared to the ε proteins (>15.92 mya) during the evolution of extant B. rapa genome.
Tissue Specific Expression of BraA.GRF14 Genes
The study on gene expression patterns of all the members of a gene family provides insight about their functional diversification. The multiplicity of 14-3-3 genes, therefore led us to investigate if all the 21 BraA.GRF14 genes are expressed in B. rapa. Gene specific primers (Table S4) were designed for each BraA.GRF14 genes and real time quantitative PCR (qRT-PCR) was performed using the cDNA samples prepared from different tissue types representing various stages of plant development.
It is interesting to note that the genes belonging to ε (epsilon) group, in general, showed lower levels of transcripts abundance in all the tissue type tested. For example, only two genes namely BraA.GRF14.a and BraA.GRF14.f showed moderate transcript abundance, when compared to the expression of BraGAPDH, whereas four genes namely BraA.GRF14.b, BraA.GRF14.c, BraA.GRF14.d, and BraA.GRF14.r showed almost negligible transcript abundance across different tissue types tested. In contrast, among the 15 non-ε group genes, only three genes (BraA.GRF14.n, BraA.GRF14.s, and BraA.GRF14.u) showed lower transcript accumulation across plant development. This observation was confirmed using multiple primer pairs from different regions of the representative genes (Table S4). The quantitatively higher transcript abundance of non-ε group genes compared to the ε group genes obtained for B. rapa 14-3-3 gene family, is somewhat similar to the expression observed for AtGRF genes in Arabidopsis (Table S5).
The BraA.GRF14 genes also exhibited a high degree of tissue specificity (Figure 3). Hierarchal clustering based on gene expression profile suggested that BraA.GRF14 genes, in general, are abundantly expressed in root, stem and seedling stages, compared to leaf and silique stages where moderate level of transcript abundance was detected. In contrast, two of the genes namely, BraA.GRF14.k and BraA.GRF14.l showed a high level of transcript abundance in the developing leaf stages only, thereby suggesting that these members may play a specialized role in these tissue types.
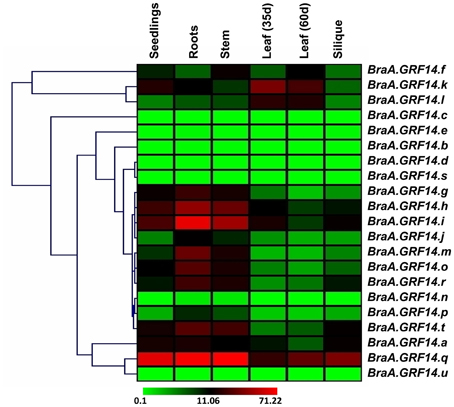
Figure 3. Expression profile of B. rapa 14-3-3 genes (BraA.GRF14) across plant developmental stages. A heat map representing hierarchical clustering of average fold values of BraA.GRF14 genes expression (w.r.t. GAPDH, set at 100) in various developmental tissues (mentioned at the top of each lane) is shown. The color scale representing average signal is shown at the bottom of the heat map.
We also examined the expression pattern of multiple paralogs of each GRF gene, resulted from WGT event in Brassica lineage. Hierarchical clustering of three paralogs of GRF2 namely BraA.GRF14.g, BraA.GRF14.h, and BraA.GRF14.i showed almost similar expression levels and tissue specificity when tested across different tissue types of B. rapa (Figure 3). The B. rapa paralogs of GRF3 (BraA.GRF14.k and BraA.GRF14.l) and GRF7 (BraA.GRF14.m and BraA.GRF14.o) also showed similar expression patterns, thereby suggesting that these B. rapa paralogs, resulted from polyploidization, had conserved their expression levels and tissue-specificity during the evolution of Brassica species. However, in other cases the paralogs of GRF4 (BraA.GRF14.r and BraA.GRF14.s), GRF6 (BraA.GRF14.q and BraA.GRF14.n), and GRF8 (BraA.GRF14.u and BraA.GRF14.t), showed contrasting variation in their transcript abundance in different tissue type tested, suggesting tissue-specific transcriptional sub-functionalization of B. rapa 14-3-3 paralogs.
Expression Analysis of 14-3-3 Genes in Response to Abiotic Stresses and Hormone Treatments in B. rapa
To investigate the effects of various abiotic stress conditions and hormone treatments on the expression of BraA.GRF14 genes, B. rapa seedlings were treated with different abiotic stress conditions (dehydration, cold, heat, salt) and exogenously supplied hormones (IAA, MeJA, SA, and ABA) for different time points (15 min, 30 min, 3 h, and 6 h). The qRT-PCR expression analysis was performed to detect the transcriptional regulation of 14 BraA.GRF14 genes, having detectable transcripts levels, compared to the untreated control seedlings.
The expression of BraA.GRF14 genes were altered differentially in response to different abiotic stress treatments (Figure 4A). The expression of BraA.GRF14 genes was found to be unaltered or down-regulated, particularly during the early stages (15 and 30 min) of dehydration and heat stress treatments compared to higher induction observed during later time points (3 and 6 h; Figures 4B,C). Most of the BraA.GRF14 genes were found to be highly induced by salt treatment at all tested time points, thereby suggesting their crucial roles during salt stress conditions (Figure 4A). In response to cold, BraA.GRF14.t was up-regulated within 15 min, whereas transcript accumulation of eight BraA.GRF14 genes was found to be induced after delayed cold treatment. In response to dehydration, none of the BraA.GRF14 genes were up-regulated during early time points, whereas three BraA.GRF14 genes namely, BraA.GRF14.k, BraA.GRF14.l, and BraA.GRF14.m showed up-regulation in their transcripts during later time points (Figures 4B,C). Under cold and salt treatments, five BraA.GRF14 genes namely, BraA.GRF14.h, BraA.GRF14.i, BraA.GRF14.k, BraA.GRF14.l, and BraA.GRF14.m, were found to be commonly induced during late time points (Figures 4A,C). Similarly, the three common genes namely BraA.GRF14.k, BraA.GRF14.l, and BraA.GRF14.m were also found to be up-regulated during the later time points of cold, dehydration and salt stresses. However, using the stringent criterion, expression of only one BraA.GRF14 gene i.e., BraA.GRF14.m was found to be up-regulated during late time points under all the four tested abiotic stress conditions (Figure 4C).
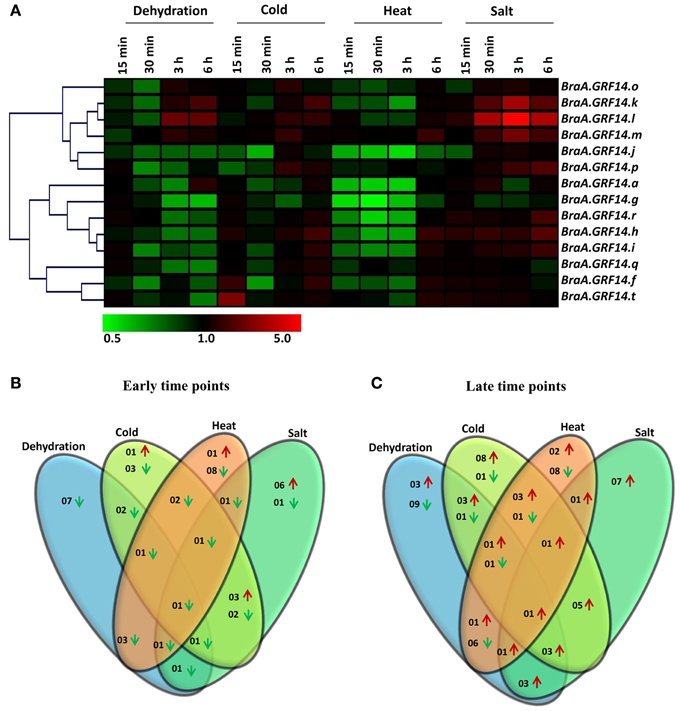
Figure 4. Expression profile of BraA.GRF14 genes in 6 days old B. rapa seedlings in response to abiotic stress conditions. (A) A heat map representing hierarchical clustering of average fold change of BraA.GRF14 genes expression in response to dehydration, cold (4°C), heat (37°C), and salt (300 mM NaCl) treatments at different time points (15 min, 30 min, 3 h, and 6 h) is shown (mentioned at the top of each lane). The mock treated seedlings of same time interval served as control (set at 1) and expression was normalized with ACT2 gene. The color scale representing average signal is shown at the bottom of the heat map. The Venn diagrams represent the total number of BraA.GRF14 genes which were upregulated (red upward arrow) and down-regulated (green downward arrow) during (B) early (15 and 30 min), and (C) late (3 and 6 h) abiotic stress conditions.
The transcriptional regulation of BraA.GRF14 genes in response to exogenously supplied phytohormones was also analyzed. Our results showed that most of the BraA.GRF14 genes were differentially expressed and showed up-regulation in their transcript particularly during later time points (3 and 6 h; Figures 5A–C). Of 14 BraA.GRF14 genes, a total of 13, 5, 13, and 13 genes were found to be up-regulated during later time points on IAA, MeJA, SA, and ABA treatments, respectively, thereby suggesting that B. rapa 14-3-3 proteins could mediate various plant responses via phytohormone sensing and signaling. In response to IAA treatment, the BraA.GRF14 genes showed up-regulation of their transcripts at a later time point (6 h); whereas the exogenous treatment of ABA showed a pronounced up-regulation of almost all the BraA.GRF14 genes except BraA.GRF14.f within few minutes, suggesting differential transcriptional response of B. rapa 14-3-3 genes during abiotic stress. In response to SA treatment, a hormone mimicking the biotic stress condition, the expression of most of the BraA.GRF14 genes was found to be induced within 15 min and up to 6 h of treatment. On contrary to this, with the application of MeJA on B. rapa seedlings, the expression of only two BraA.GRF14 genes namely, BraA.GRF14.j and BraA.GRF14.k showed up-regulation in their transcript during early time points (Figures 5A,B). In response to all the tested phytohormone treatments, although two genes (BraA.GRF14.j and BraA.GRF14.k) were found to be up-regulated during early time points; a total of five genes (BraA.GRF14r; BraA.GRF14.i, BraA.GRF14.j, BraA.GRF14.k, and BraA.GRF14.l) were up-regulated during later time points. None of the single BraA.GRF14 gene was found to be commonly down-regulated in response to all the phytohormone treatments, during both early and late time points.
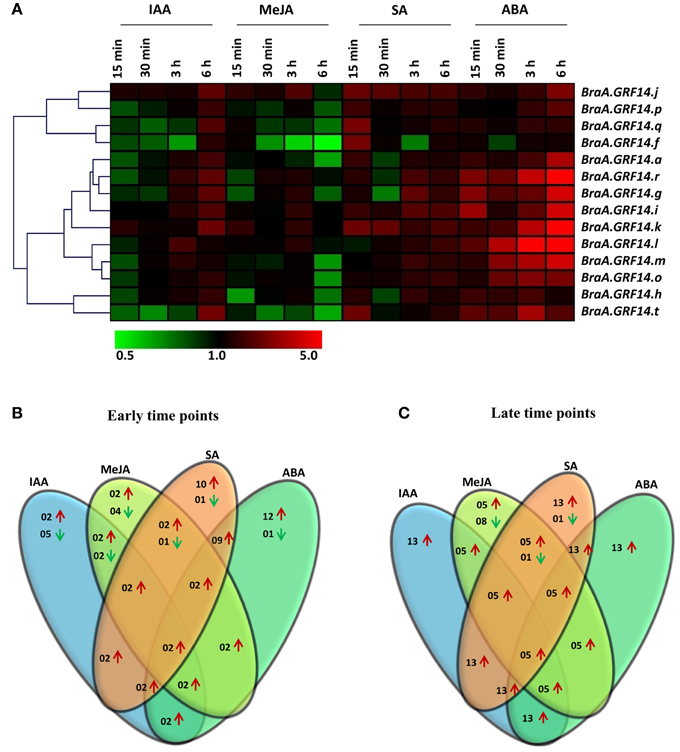
Figure 5. Expression profile of BraA.GRF14 genes in 6 days old B. rapa seedlings in response to phytohormone treatments. (A) A heat map representing hierarchical clustering of average fold change of BraA.GRF14 genes expression in response to methyl jasmonate (MeJA, 0.2 mM), salicylic acid (SA, 0.2 mM), 1-aminocyclopropane carboxylate (ACC, 0.1 mM), indole-3-acetic acid (IAA, 0.1 mM), and abscisic acid (ABA, 0.1 mM) at different time points (15, 30 min, 3 and 6 h) is shown (mentioned at the top of each lane). The mock treated seedlings of same time interval served as control (set at 1) and expression was normalized with ACT2 gene. The color scale representing average signal is shown at the bottom of the heat map. The Venn diagrams represent the total number of BraA.GRF14 genes which were upregulated (red upward arrow) and down-regulated (green downward arrow) during (B) early (15 and 30 min), and (C) late (3 and 6 h) phytohormone treatments.
Expression Analysis of 14-3-3 Genes during Nutrient Deprivation Conditions in B. rapa
Previous findings suggest a connection of 14-3-3 isoforms with plant nutrient metabolism and signaling (Xu and Shi, 2006; Shin et al., 2011). To better understand the transcriptional regulation of 14-3-3 genes toward changes in plant nutrient status, B. rapa seedlings were deprived of nitrogen (N), phosphorus (P), and potassium (K) from early (1 and 6 h) to late (24 and 48 h) time intervals, and the expression of BraA.GRF14 genes was analyzed using qRT-PCR.
The BraA.GRF14 genes showed differential transcriptional variation in response to the tested nutrient deprivation conditions. It was observed in case of nitrogen deprivation condition (-N), the transcript abundance of most of the BraA.GRF14 isoforms was found to be down-regulated by less than two-folds at early time points (Figures 6A–C). Interestingly, during delayed nitrogen deprivation (24 and 48 h), the expression of almost all BraA.GRF14 genes showed pronounced up-regulation. In response to phosphorus deprivation condition (-P), the expression of 12 BraA.GRF14 genes were found to be down-regulated during early time points, whereas after delayed phosphorus deficiency only BraA.GRF14.g was found to be up-regulated (Figures 6B,C). In contrast, the potassium deficient (-K) B. rapa seedlings showed a profound up-regulation of most of the BraA.GRF14 genes within 6 h of treatment (Figure 6A). For example, expression of BraA.GRF14.g, BraA.GRF14.j, BraA.GRF14.m, BraA.GRF14.o, and BraA.GRF14.r genes were significantly up-regulated during early time points. During the prolonged K deprivation condition, the expression of seven BraA.GRF14 was also found to be up-regulated, suggesting a profound yet differential transcriptional response of B. rapa 14-3-3 genes in response to -K condition (Figures 6B,C). Overall, in response to all the tested nutrient deprivation conditions, two genes namely, BraA.GRF14.f and BraA.GRF14.h were commonly down regulated during early time points; whereas BraA.GRF14.g was only found to be up-regulated during later time points (Figures 6B,C).
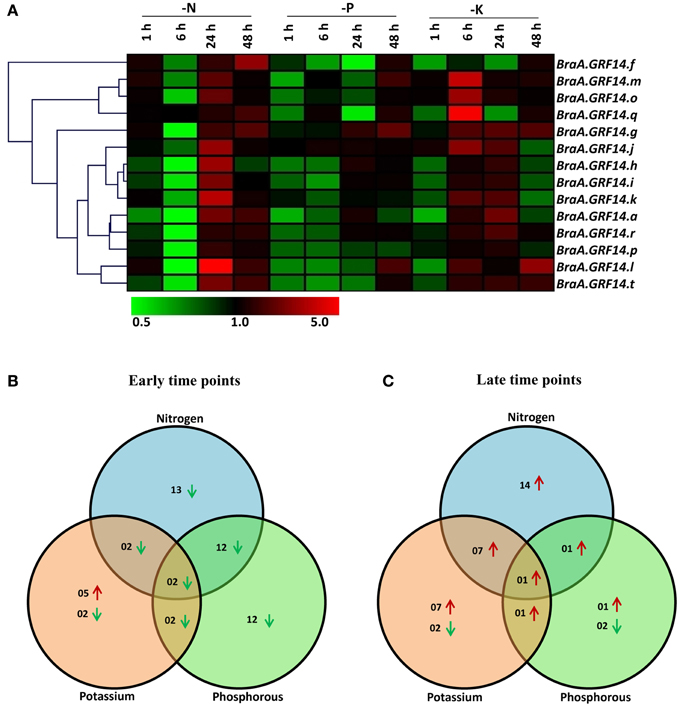
Figure 6. Expression profile of BraA.GRF14 genes during nutrient deprivation conditions in 5 days old B. rapa seedlings. (A) A heat map representing hierarchical clustering of average fold change of BraA.GRF14 gene expression in response to nutrient deprived conditions at different time points (mentioned at the top of each lane). The 5 days old B. rapa seedlings were harvested at different time points (1, 6, 24, and 48 h) after they were exposed to nutrient solution lacking Nitrogen (N), Phosphorus (P), and Potassium (K). The mock treated seedlings of same time interval served as control (set at 1) and expression was normalized with ACT2 gene. The color scale representing average signal is shown at the bottom of the heat map. The Venn diagrams represent the total number of BraA.GRF14 genes which were upregulated (red upward arrow) and down-regulated (green downward arrow) during (B) early (1 and 6 h), and (C) late (24 and 48 h) nutrient deprived conditions.
Analysis of cis-Regulatory Divergence of 5′ Upstream Sequences of B. rapa 14-3-3 Genes
The differential transcriptional regulation of BraA.GRF14 genes during plant growth and developmental stages and in response to various elicitor treatments tested in this study could be attributed to sequence divergence and cis-regulatory elements present in their 5′ upstream regulatory sequences. Approximately 1.5 kb sequence upstream of transcription start site of BraA.GRF14 genes were obtained from the BRAD database and analyzed using ClustalW. The 5′ upstream sequence of BraA.GRF14 genes were highly divergent showing a low level of sequence identity ranging from 18.5 to 31.7% (Table S6), which is quite in agreement with the differential expression pattern obtained among BraA.GRF14 genes.
To identify various cis-acting regulatory elements present in the 5′ upstream sequences, the BraA.GRF14 genes were further analyzed using the PLACE database (http://www.dna.affrc.go.jp/PLACE/). In-silico analysis revealed the presence of various motifs involved in phytohormones, abiotic and biotic stress responses (Table S7). In general, the BraA.GRF14 genes, which were found to be up-regulated under ABA treatment in current study, showed abundance of ABA responsive elements like ABRELATERD1, DRE1COREZMRAB17, and ABREZMRAB28, thereby indicating the involvement of B. rapa 14-3-3 gene family members in various ABA mediated cellular responses. Similarly, upstream sequence of B. rapa 14-3-3 genes like BraA.GRF14.g, BraA.GRF14.h, BraA.GRF14.p, and BraA.GRF14.m, which were significantly up-regulated under salt stress, were found to have GT1GMSCAM4 response element known to be involved in salt stress and plant defense. Various pathogen and elicitor response elements like ELRECOREPCRP1, GCCCORE, T/GBOXATPIN2, and TATCCACHVAL21 were also present, which confirm up-regulation of few BraA.GRF14 genes under MeJA and SA treatments. A significant up-regulation of most of the BraA.GRF14 genes during late time point of IAA treatment (6 h) can also be correlated with the presence of different types of auxin inducible cis-acting elements like ARFAT, CATATGGMSAUR, D3GMAUX28, and NTBBF1ARROLB in their 5′ upstream sequences. Expression of all the B. rapa 14-3-3 genes were either unaltered or down-regulated during different time point of heat stress, which could be due to the presence of only few heat stress related cis-acting elements. Although the upstream sequence of B. rapa 14-3-3 genes showed the presence of few cis-acting elements for nutrient deprivation conditions, but interestingly all the B. rapa 14-3-3 genes were highly expressed during late time of nutrient deprivation, which could be due to the presence of still unknown regulatory motifs involved in the nutrient sensing in B. rapa. Nonetheless, the differential transcriptional alteration observed in the expression of B. rapa 14-3-3 genes could be nicely attributed to the presence of various cis-regulatory elements and their quantitative variability, across multiple BraA.GRF14 genes.
Discussion
The 14-3-3 proteins are a family of highly conserved regulatory proteins present across phyla, which function by binding to the phosphorylated target proteins (effectors) to play vital roles in many biological processes in plants, including primary metabolism and hormone signaling as well as in response to the abiotic and biotic stresses. In this study, through data mining we identified 21 genes encoding 14-3-3 like proteins from the recently sequenced B. rapa, the model Brassica genome.
Evolutionary Expansion of B. rapa 14-3-3 Multigene Family
It is quite expected that the inherent polyploidy in plants has shaped the expansion of 14-3-3 gene family. For example, Li and Dhaubhadel (2011) identified 18 genes encoding 14-3-3 proteins in soybean, an allotetraploid genome (ca. 1115 Mb), having undergone two whole genome duplication events (ca. 14 and 42 mya). Comparative genomics study in cotton, an allotetraploid crop species (ca. 2500 Mb), identified the highest thirty-one 14-3-3 cDNAs encoding 25 unique proteins, resulting from a recent duplication event after the divergence of cotton from its progenitor species (Sun et al., 2011).
It is interesting that given the higher size (haploid genome ca. 500 Mb) and ploidy (mesohexaploidy) level of the B. rapa genome compared to that of the diploid A. thaliana (ca. 120 Mb), the B. rapa has most likely only 21 isoforms of 14-3-3 proteins, in comparison with 13 expressed isoforms reported in the closest dicot model (Figure 1). Various comparative genomics studies have clearly suggested that the Arabidopsis and the cultivable Brassica species had split from a common ancestral Brassicaceae around 13–17 million years ago (mya; Lysak et al., 2007; Panjabi et al., 2008; Franzke et al., 2011; Wang et al., 2011; Cheng et al., 2013). The Brassica lineage has further undergone whole genome triplication (WGT) event after the Arabidopsis–Brassica split, as a result of which the so called diploid Brassica species, including B. rapa, are paleohexaploid containing three sub-genomes. Sequence level studies in recent year strongly suggested that among these three sub-genomes, biased gene-fractionation (gene-loss) phenomenon has occurred, which resulted in the formation of least fractionized (LF), moderate gene fractionized (MF1), and the most gene fractionized (MF2) sub-genomes in B. rapa (Cheng et al., 2013). Thus, all Brassica species analyzed to date are supposed to contain multiple copies of orthologous genomic regions of A. thaliana (Lysak et al., 2007; Panjabi et al., 2008). We presume that the variable copies (1–5) of each 14-3-3 Arabidopsis genes identified in B. rapa could be the consequence of differential level of gene-fractionation (or gene-loss) phenomenon that occurred after the WGT event in the extant B. rapa genome (Table 2). As extreme cases, for few Arabidopsis 14-3-3 genes no syntenic ortholog were observed in B. rapa. Nonetheless, polyploidy coupled with genomic shrinkage and rearrangements have caused noteworthy expansion of 14-3-3 gene family members (21 isoforms) in mesohexaploid B. rapa, compared to the A. thaliana, rice, soybean, and cotton having 13, 8, 18, and 25 members, respectively.
Sequence alignment revealed that the orthologous genes shared between Arabidopsis–B. rapa genomes have a high level of amino-acid similarity, suggesting possibility of functional conservation of the 14-3-3 proteins across the two genomes. The B. rapa 14-3-3 proteins could be classified into two groups namely epsilon (ε) and non-epsilon (non-ε), with each sub-group showing extreme conservation of the intron-exon organization, and a distinct divergence pattern (Ks-values) between the two sub-groups, which in all possibility suggest independent evolution and expansion of the ε and non-ε groups 14-3-3 genes in B. rapa. Such class specific divergence pattern is also reported recently for the three types of plant G-gamma (Gγ) subunits (Type-A, -B, and -C) of the heterotrimeric G-proteins (Trusov et al., 2012; Arya et al., 2014; Kumar et al., 2014), an important class of signaling proteins, wherein the divergent Gγ subunits are known to provide functional selectivity to G-proteins in plants. We presume that the quantitative variation as well as the divergent residues present across 14-3-3 isoforms could shape some degree of specificity with regard to their expression profiles and the target protein(s) with which they interact, thereby contributing to the remarkable phenotypic plasticity and environmental adaptability of B. rapa.
B. rapa 14-3-3 Genes Exhibit Redundant and Divergent Expression Patterns
Gene duplication although raises the functional redundancy of duplicated genes, it is known to serve as a mechanism to increase the functional diversity. The duplicated genes often evolved cis-regulatory divergence in their regulatory regions; as a consequence there exist both immediate and long-term alterations in the expression of genes arising from polyploidy, such as differential expression, transcriptional bias, or gene silencing (Adams, 2007; Chaudhary et al., 2009). As a result, the duplicated genes may undergo diversification of gene function(s) such as neo-, sub- or, non-functionalization.
The BraA.GRF14 genes are ubiquitously expressed during B. rapa growth and developmental stages (Figure 3). Since 14-3-3 proteins represent one of the key components of the plant signaling cascade (reviewed in Sehnke et al., 2002; de Boer et al., 2013), the ubiquitous activity of BraA.GRF14 proteins vis-à-vis their interaction with their client proteins, are quite necessary for regulating a wide variety of biological processes in B. rapa. Interestingly, the class specific transcription abundance of the B. rapa non-ε group genes compared to the ε group genes observed in this study, is somewhat similar to that of the Arabidopsis 14-3-3 genes (Paul et al., 2012). Three of the five ε group genes of Arabidopsis have significantly low expression intensities compared to the highly expressed non-ε group genes (Table S4). This evolutionary conservation of expression pattern of the Arabidopsis–Brassica orthologs in all possibility suggests that non-ε group 14-3-3 genes might have retained functional dominance in regulating various growth and development processes across Brassicaceae.
Recent studies in polyploid Brassica species have functionally demonstrated that members of a multigene family can be expressed at different levels and can respond differentially to polyploidy in various organs of the plant or in response to various environmental stimuli (Higgins et al., 2012; Augustine et al., 2013; Meenu et al., 2015). Our study also suggest that multiple paralogs of few GRF genes, resulted from WGT event in Brassica lineage, although show almost similar expression patterns and tissue specificity; whereas in other cases the paralogs have contrasting variation in their transcript abundance and pattern across plant developmental stages. The overlapping and divergent expression patterns of BraA.GRF14 genes suggest that the multiple members have evolved to perform redundant yet tissue-specific functions during plant growth and development in B. rapa.
B. rapa 14-3-3 Genes are Differentially Regulated in Response to Various Stimuli
There are evidences that plant 14-3-3 proteins show changes in their gene expression in response to various environmental stresses (Roberts et al., 2002). These stresses often have variable effects on different isoforms in terms of change in expression level and the time period. Our study also demonstrates that the expressed B. rapa 14-3-3 genes behave differentially in response to the tested abiotic stress conditions, exogenously supplied phytohormones, and nutrient deprivation conditions indicating that different members of this gene family might have undergone differential transcriptional regulation to play specific roles during altered environmental conditions.
In general, most of the BraA.GRF14 genes were significantly induced during high salt treatment both during early and late time points (Figure 4). The rice OsGF14f was originally identified as the 14-3-3 transcript that accumulated in the callus and seedling of rice when exposed to high salt or cold temperature (Kidou et al., 1993). Similarly, the up-regulation of 14-3-3 genes has been reported earlier under NaCl treatment in tomato, cotton and rice (Chen et al., 2006; Xu and Shi, 2006; Yao et al., 2007; Sun et al., 2011). In Arabidopsis, two 14-3-3 genes namely, RCI1A and RCI1B were shown to be involved in cold and freezing stress tolerance (Jarillo et al., 1994; Catalá et al., 2014). Under cold stress condition, eight BraA.GRF14 genes were up-regulated during later time points (Figure 4C). Likewise, the expression of few members of Phaseolus vulgaris 14-3-3 gene family has been recently reported to be up-regulated by cold stress (Li et al., 2015). Over-expression of the Arabidopsis 14-3-3 “lambda” gene into cotton has been shown to impart enhanced tolerance to drought in transgenic lines, as determined by less wilting and visible damage to the leaves (Yan et al., 2004). Interestingly, we found only few BraA.GRF14 genes were up-regulated under dehydration and heat stress both during early and later time points (Figure 4), suggesting that BraA.GRF14 gene members have evolved to perform condition-specific functions.
It has been widely accepted that 14-3-3 genes act as key components in regulating phytohormone mediated plants responses (reviewed in Denison et al., 2011). Our study shows a profound up-regulation of BraA.GRF14 genes under MeJA and SA treatments during later time points (Figure 5), indicating that B. rapa 14-3-3 multigene family plays regulatory roles in response to biotic stress. Earlier, it has been shown that 14-3-3 genes are involved in plant defense response in poplar (Lapointe et al., 2001). Similarly, altered expression pattern of 14-3-3 genes under various biotic stress conditions has been reported in rice (Chen et al., 2006; Yao et al., 2007). The constitutive up-regulation of BraA.GRF14 genes under ABA treatment, coupled with the presence of high number of ABA responsive cis-regulatory elements in their 5′ upstream region, suggests that the B. rapa 14-3-3 proteins play a vital role in ABA mediated cellular responses. The 14-3-3 proteins are also known to interact with the ABA signaling pathway in barley, cotton and rice, by interacting with the AREB-like transcription factors (ABF1, ABF2, ABF3, and ABI5), that binds to ABA-responsive elements (Schoonheim et al., 2007; Zhang et al., 2010; Hong et al., 2011). The differential transcriptional regulation of BraA.GRF14 genes in response to the tested phytohormones suggests their complex cross-talk with phytohormone signaling components to regulate wide range of physiological processes.
In higher plants, 14-3-3 proteins play a significant role in response to nutrient sensing. The phosphorus deprivation (-P) condition, in general, causes a significant down-regulation of B. rapa 14-3-3 gene expression (Figure 6). This is in agreement with that observed for Arabidopsis GRF orthologs (Cao et al., 2007), thereby suggesting that P deficiency potentially affects transcript levels of 14-3-3 isoforms and their dependent processes in different cellular compartments. In response to nitrogen deprivation (-N) condition, the expression of B. rapa 14-3-3 genes was induced at later time points. In contrast, potassium deprivation (- K) treatment seems to up-regulate the expression of most of the 14-3-3 isoforms as also reported for tomato 14-3-3 proteins (Wang et al., 2002; Xu and Shi, 2006). The expression of BraA.GRF14.t gene was increased under potassium deprivation (-K) condition during the late time points. Similarly, the expression level of Arabidopsis 14-3-3 κ increased after “K” deprivation in leaves (Shin et al., 2011). The plant's 14-3-3 proteins are known to interact with regulatory enzymes involved in nutrient sensing, metabolism and transport in plants. Glutamate synthase (GS) and nitrate reductase, key enzymes that regulate N metabolism, were identified as a 14-3-3 interacting proteins (Bachmann et al., 1996; Shin et al., 2011). Various signaling proteins including 14-3-3 interact with phosphorus deficiency response factors including protein kinases, phosphatases (Baldwin et al., 2008; Xu et al., 2012). Regulatory evidences of 14-3-3 genes were also observed of plant K+ channels, which are known to play a role in potassium homeostasis in the plants (Bunney et al., 2002; Wijngaard et al., 2005). The differential transcriptional response of 14-3-3 genes in response to abiotic stress conditions, phytohormones treatments, and nutrient deprivation conditions indicated that each member of this family participate for condition specific function in multiple signaling pathways.
It is well-known that various post-translational modifications of proteins increase the functional diversity of the proteome by the covalent addition of functional groups or proteins, proteolytic cleavage of regulatory subunits or degradation of the entire protein. Recent studies in Arabidopsis and other plants suggested evidences for phosphorylation of several isoforms at conserved as well as divergent serine (S) and tyrosine (Y) residues, providing the potential for phosphorylation to affect 14-3-3 proteins in an isoform-specific manner (reviewed by Paul et al., 2012). Thus, in addition to the transcriptional regulation observed in the current study, the isoform-specific phosphorylation patterns could also provide a key post-translational regulation of the multiple BraGRF14 proteins toward controlling their binding with “client” proteins and the functional diversity in B. rapa.
In conclusion, our study provides a comprehensive classification together with a structural and evolutionary analysis of the BraA.GRF14 gene family in B. rapa. A total of 21 isoforms of BraA.GRF14s are identified from B. rapa, of which 14 isoforms were found to be expressed throughout the plant developmental tissues. The BraA.GRF14 genes, on the basis of introns and exons, were broadly categorized into two distinct sub-groups namely the ε (epsilon) and non-ε (non-epsilon) groups. It was also observed that multiple orthologs for most of the Arabidopsis 14-3-3 genes existed in B. rapa. Expression analysis of the 14 BraA.GRF14 genes in response to abiotic stress, hormone stress and nutrient deficiency in the B. rapa seedlings suggest that B. rapa 14-3-3s are either directly or indirectly involved in the regulation of majority of physiological and metabolic pathways. A systemic and comparative analysis of the target proteins of the B. rapa 14-3-3 isoforms, in future, would contribute to fundamental understanding of the conservation and divergence of biological processes controlled by these key signaling proteins.
Author Contributions
RC, RA conducted the real time PCR experiments and data analysis. Kanchupati P, RK, Kumar P, GA carried out identification and in-silico analysis of candidate genes. RC, RK, and NB compiled the data and writing of manuscript. All authors read and approved the final manuscript.
Funding
This work was supported by the core research grant from the NIPGR, India. RC and RA were supported from NIPGR short-term research fellowships; Junior Research Fellowships of CSIR (to GCA), UGC (to Kanchupati P, RK), and DBT (to Kumar P) are also acknowledged.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The reviewer Yashwanti Mudgil and handling Editor Girdhar Kumar Pandey declared their shared affiliation, and the handling Editor states that the process nevertheless met the standards of a fair and objective review.
Acknowledgments
The central instrumentation and plant growth facility at NIPGR are highly acknowledged.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2016.00012
References
Adams, K. L. (2007). Evolution of duplicate gene expression in polyploid and hybrid plants. J. Hered. 98, 136–141. doi: 10.1093/jhered/esl061
Agrawal, G. K., and Thelen, J. J. (2006). Large scale identification and quantitative profiling of phosphoproteins expressed during seed filling in oilseed rape. Mol. Cell Proteomics 5, 2044–2059. doi: 10.1074/mcp.m600084-mcp200
Arya, G. C., Kumar, R., and Bisht, N. C. (2014). Evolution, expression differentiation and interaction specificity of heterotrimeric G-protein subunit gene family in the mesohexaploid Brassica rapa. PLoS ONE 9:e105771. doi: 10.1371/journal.pone.0105771
Augustine, R., Majee, M., Gershenzon, J., and Bisht, N. C. (2013). Four genes encoding MYB28, a major transcriptional regulator of the aliphatic glucosinolate pathway, are differentially expressed in the allopolyploid Brassica juncea. J. Exp. Bot. 64, 4907–4921. doi: 10.1093/jxb/ert280
Bachmann, M., Huber, J. L., Athwal, G. S., Wu, K., Ferl, R. J., and Huber, S. C. (1996). 14-3-3 proteins associate with the regulatory phosphorylation site of spinach leaf nitrate reductase in an isoform-specific manner and reduce dephosphorylation of Ser-543 by endogenous protein phosphatases. FEBS Lett. 398, 26–30. doi: 10.1016/s0014-5793(96)01188-x
Baldwin, J. C., Karthikeyan, A. S., Cao, A., and Raghothama, K. G. (2008). Biochemical and molecular analysis of LePS2;1: a phosphate starvation induced protein phosphatase gene from tomato. Planta 228, 272–280. doi: 10.1007/s00425-008-0736-y
Bunney, T. D., van den Wijngaard, P. W., and De Boer, A. H. (2002). 14-3-3 protein regulation of proton pumps and ion channels. Plant Mol. Biol. 50, 1041–1051. doi: 10.1023/A:1021231805697
Cao, A., Jain, A., Baldwin, J. C., and Raghothama, K. G. (2007). Phosphate differentially regulates 14-3-3 family members and GRF9 plays a role in Pi-starvation induced responses. Planta 226, 1219–1230. doi: 10.1007/s00425-007-0569-0
Catalá, R., López-Cobollo, R., Mar Castellano, M., Angosto, T., Alonso, J. M., Ecker, J. R., et al. (2014). The Arabidopsis 14-3-3 protein RARE COLD INDUCIBLE 1A links low-temperature response and ethylene biosynthesis to regulate freezing tolerance and cold acclimation. Plant Cell 26, 3326–3342. doi: 10.1105/tpc.114.127605
Chandna, R., Augustine, R., and Bisht, N. C. (2012). Evaluation of candidate reference genes for gene expression normalization in Brassica juncea using real time quantitative RT-PCR. PLoS ONE 7:e36918. doi: 10.1371/journal.pone.0036918
Chaudhary, B., Flagel, L., Stupar, R. M., Udall, J. A., Verma, N., Springer, N. M., et al. (2009). Reciprocal silencing, transcriptional bias and functional divergence of homeologs in polyploid cotton (Gossypium). Genetics 182, 503–517. doi: 10.1534/genetics.109.102608
Chen, F., Li, Q., Sun, L., and He, Z. (2006). The rice 14-3-3 gene family and its involvement in responses to biotic and abiotic stress. DNA Res. 13, 53–63. doi: 10.1093/dnares/dsl001
Cheng, F., Liu, S., Wu, J., Fang, L., Sun, S., Liu, B., et al. (2011). BRAD, the genetics and genomics database for Brassica plants. BMC Plant Biol. 11:136. doi: 10.1186/1471-2229-11-136
Cheng, F., Mandáková, T., Wu, J., Xie, Q., Lysak, M. A., and Wang, X. (2013). Deciphering the diploid ancestral genome of the mesohexaploid Brassica rapa. Plant Cell 25, 1541–1554. doi: 10.1105/tpc.113.110486
Comparot, S., Lingiah, G., and Martin, T. (2003). Function and specificity of 14-3-3 proteins in the regulation of carbohydrate and nitrogen metabolism. J. Exp. Bot. 382, 595–604. doi: 10.1093/jxb/erg057
de Boer, A. H., van Kleeff, P. J., and Gao, J. (2013). Plant 14-3-3 proteins as spiders in a web of phosphorylation. Protoplasma 250, 425–440. doi: 10.1007/s00709-012-0437-z
DeLille, J. M., Sehnke, P. C., and Ferl, R. J. (2001). The Arabidopsis 14-3-3 family of signaling regulators. Plant Physiol. 126, 35–38. doi: 10.1104/pp.126.1.35
Denison, F. C., Paul, A. L., Zupanska, A. K., and Ferl, R. J. (2011). 14-3-3 proteins in plant physiology. Semin. Cell Dev. Biol. 22, 720–727. doi: 10.1016/j.semcdb.2011.08.006
Franzke, A., Lysak, M. A., Al-Shehbaz, I. A., Koch, M. A., and Mummenhoff, K. (2011). Cabbage family affairs: the evolutionary history of Brassicaceae. Trends Plant Sci. 16, 108–116. doi: 10.1016/j.tplants.2010.11.005
Giacometti, S., Camoni, L., Albumi, C., Visconti, S., De Michelis, M. I., and Aducci, P. (2004). Tyrosine phosphorylation inhibits the interaction of 14-3-3 proteins with the plant plasma membrane H+-ATPase. Plant Biol. 6, 422–431. doi: 10.1055/s-2004-820933
He, Y., Wu, J., Lv, B., Li, J., Gao, Z., Xu, W., et al. (2015). Involvement of 14-3-3 protein GRF9 in root growth and response under polyethylene glycol-induced water stress. J. Exp. Bot. 66, 2271–2281. doi: 10.1093/jxb/erv149
Higgins, J., Magusin, A., Trick, M., Fraser, F., and Bancroft, I. (2012). Use of mRNA-seq to discriminate contributions to the transcriptome from the constituent genomes of the polyploid crop species Brassica napus. BMC Genomics 13:247. doi: 10.1186/1471-2164-13-247
Hong, J. Y., Chae, M. J., Lee, I. S., Lee, Y. N., Nam, M. H., Kim, D. Y., et al. (2011). Phosphorylation-mediated regulation of a rice ABA responsive element binding factor. Phytochemistry 72, 27–36. doi: 10.1016/j.phytochem.2010.10.005
Igarashi, D., Ishida, S., Fukazawa, J., and Takahashi, Y. (2001). 14-3-3 proteins regulate intracellular localization of the bZIP transcriptional activator RSG. Plant Cell 13, 2483–2497. doi: 10.1105/tpc.13.11.2483
Jain, M., Nijhawan, A., Arora, R., Agarwal, P., Ray, S., Sharma, P., et al. (2007). F-box proteins in rice: genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiol. 143, 1467–1483. doi: 10.1104/pp.106.091900
Jarillo, J. A., Capel, J., Leyva, A., Martínez-Zapater, J. M., and Salinas, J. (1994). Two related low-temperature-inducible genes of Arabidopsis encode proteins showing high homology to 14-3-3 proteins, a family of putative kinase regulators. Plant Mol. Biol. 25, 693–704. doi: 10.1007/BF00029607
Kidou, S., Umeda, M., Kato, A., and Uchimiya, H. (1993). Isolation and characterization of a rice cDNA similar to the bovine brain-specific 14-3-3 protein gene. Plant Mol. Biol. 21, 191–194. doi: 10.1007/bf00039631
Kim, T. W., Guan, S., Sun, Y., Deng, Z., Tang, W., Shang, J. X., et al. (2009). Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat. Cell Biol. 11, 1254–1260. doi: 10.1038/ncb1970
Kjarland, E., Keen, T. J., and Kleppe, R. (2006). Does isoform diversity explain functional differences in the 14-3-3 protein family? Curr. Pharm. Biotechnol. 7, 217–223. doi: 10.2174/138920106777549777
Koch, M. A., Haubold, B., and Mitchell-Olds, T. (2000). Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 17, 1483–1498. doi: 10.1093/oxfordjournals.molbev.a026248
Kumar, R., Arya, G. C., and Bisht, N. C. (2014). Differential expression and interaction specificity of the heterotrimeric G-protein family in Brassica nigra reveal their developmental- and condition-specific roles. Plant Cell Physiol. 55, 1954–1968. doi: 10.1093/pcp/pcu126
Lapointe, G., Luckevich, M. D., Cloutier, M., and Séguin, A. (2001). 14-3-3 gene family in hybrid poplar and its involvement in tree defence against pathogens. J. Exp. Bot. 52, 1331–1338. doi: 10.1093/jexbot/52.359.1331
Li, R., Jiang, X., Jin, D., Dhaubhadel, S., Bian, S., and Li, X. (2015). Identification of 14-3-3 family in common bean and their response to abiotic stress. PLoS ONE 10:e0143280. doi: 10.1371/journal.pone.0143280
Li, X., and Dhaubhadel, S. (2011). Soybean 14-3-3 gene family: identification and molecular characterization. Planta 233, 569–582. doi: 10.1007/s00425-010-1315-6
Lysak, M. A., Cheung, K., Kitschke, M., and Bures, P. (2007). Ancestral chromosomal blocks are triplicated in Brassiceae species with varying chromosome number and genome size. Plant Physiol. 145, 402–410. doi: 10.1104/pp.107.104380
Meenu, Augustine, R., Majee, M., Pradhan, A. K., and Bisht, N. C. (2015). Genomic origin, expression differentiation and regulation of multiple genes encoding CYP83A1, a key enzyme for core glucosinolate biosynthesis, from the allotetraploid Brassica juncea. Planta 241, 651–665. doi: 10.1007/s00425-014-2205-0
Moore, B., and Perez, V. (1967). “Specific acidic proteins of the nervous system,” in Physiological and Biochemical Aspects of Nervous Integration, ed F. Carlson (Woods Hole, MA: Prentice–Hall), 343–359.
Oecking, C., and Jaspert, N. (2009). Plant 14-3-3 proteins catch up with their mammalian orthologs. Curr. Opin. Plant Biol. 12, 760–765. doi: 10.1016/j.pbi.2009.08.003
Ostergaard, L., and King, G. J. (2008). Standardized gene nomenclature for the Brassica genus. Plant Methods 4:10. doi: 10.1186/1746-4811-4-10
Panjabi, P., Jagannath, A., Bisht, N., Padmaja, K. L., Sharma, S., Gupta, V. C., et al. (2008). Comparative mapping of Brassica juncea and Arabidopsis thaliana using intron polymorphism (IP) markers: homeologous relationships, diversification and evolution of the A, B and C Brassica genomes. BMC Genomics 9:113. doi: 10.1186/1471-2164-9-113
Paul, A. L., Denison, F. C., Schultz, E. R., Zupanska, A. K., and Ferl, R. J. (2012). 14-3-3 phosphoprotein interaction networks-does isoform diversity present functional interaction specification? Front. Plant. Sci. 3:190. doi: 10.3389/fpls.2012.00190
Radwan, O., Wu, X., Govindarajulu, M., Libault, M., Neece, D. J., Oh, M. H., et al. (2012). 14-3-3 proteins SGF14c and SGF14l play critical roles during soybean nodulation. Plant Physiol. 160, 2125–2136. doi: 10.1104/pp.112.207027
Roberts, M. R., Salinas, J., and Collinge, D. B. (2002). 14-3-3 proteins and the response to abiotic and biotic stress. Plant Mol. Biol. 50, 1031–1039. doi: 10.1023/A:1021261614491
Rosenquist, M., Alsterfjord, M., Larsson, C., and Sommarin, M. (2001). Data mining the Arabidopsis genome reveals fifteen 14-3-3 genes. Expression is demonstrated for two out of five novel genes. Plant Physiol. 127, 142–149. doi: 10.1104/pp.127.1.142
Ryu, H., Kim, K., Cho, H., Park, J., Choe, S., and Hwang, I. (2007). Nucleocytoplasmic shuttling of BZR1 mediated by phosphorylation is essential in Arabidopsis brassinosteroid signaling. Plant Cell 19, 2749–2762. doi: 10.1105/tpc.107.053728
Schoonheim, P. J., Sinnige, M. P., Casaretto, J. A., Veiga, H., Bunney, T. D., Quatranoet, R. S., et al. (2007). 14-3-3 Adapter proteins are intermediates in ABA signal transduction during barley seed germination. Plant J. 49, 289–301. doi: 10.1111/j.1365-313x.2006.02955.x
Sehnke, P. C., Rosenquist, M., Alsterfjord, M., DeLille, J., Sommarin, M., Larsson, C., et al. (2002). Evolution and isoform specificity of plant 14-3-3 proteins. Plant Mol. Biol. 50, 1011–1018. doi: 10.1023/A:1021289127519
Shin, R., Alvarez, S., Burch, A. Y., Jez, J. M., and Schachtman, D. P. (2007). Phosphoproteomic identification of targets of the Arabidopsis sucrose nonfermenting-like kinase SnRK2.8 reveals a connection to metabolic processes. Proc. Natl. Acad. Sci. U.S.A. 104, 6460–6465. doi: 10.1073/pnas.0610208104
Shin, R., Jez, J. M., Basra, A., Zhang, B., and Schachtman, D. P. (2011). 14-3-3 proteins fine-tune plant nutrient metabolism. FEBS Lett. 585, 143–147. doi: 10.1016/j.febslet.2010.11.025
Sun, G., Xie, F., and Zhang, B. (2011). Transcriptome-wide identification and stress properties of the 14-3-3 gene family in cotton (Gossypium hirsutum L.). Funct. Integr. Genome 11, 627–636. doi: 10.1007/s10142-011-0242-3
Sun, X., Luo, X., Sun, M., Chen, C., Ding, X., Wang, X., et al. (2014). A Glycine soja 14-3-3 protein GsGF14o participates in stomatal and root hair development and drought tolerance in Arabidopsis thaliana. Plant Cell Physiol. 55, 99–118. doi: 10.1093/pcp/pct161
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739. doi: 10.1093/molbev/msr121
Testerink, C., van der Meulen, R. M., Oppedijk, B. J., de Boer, A. H., Heimovaara-Dijkstra, S., Kijne, J. W., et al. (1999). Differences in spatial expression between 14-3-3 isoforms in germinating barley embryos. Plant Physiol. 121, 81–88. doi: 10.1104/pp.121.1.81
Trusov, Y., Chakravorty, D., and Botella, J. R. (2012). Diversity of heterotrimeric G-protein γ subunits in plants. BMC Res. Notes 5:608. doi: 10.1186/1756-0500-5-608
Umezawa, T., Yoshida, R., Maruyama, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2004). SRK2C, a SNF1-related protein kinase 2, improves drought tolerance by controlling stress-responsive gene expression in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 101, 17306–17311. doi: 10.1073/pnas.0407758101
van Kleeff, P. J., Jaspert, N., Li, K. W., Rauch, S., Oecking, C., and de Boer, A. H. (2014). Higher order Arabidopsis 14-3-3 mutants show 14-3-3 involvement in primary root growth both under control and abiotic stress conditions. J. Exp. Bot. 65, 5877–5888. doi: 10.1093/jxb/eru338
Wang, X., Wang, H., Wang, J., Sun, R., Wu, J., Liu, S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039. doi: 10.1038/ng.919
Wang, Y. H., Garvin, D. F., and Kochian, L. V. (2002). Rapid induction of regulatory and transporter genes in response to phosphorus, potassium, and iron deficiencies in tomato roots: evidence for cross-talk and root/rhizosphere-mediated signal. Plant Physiol. 130, 1361–1370. doi: 10.1104/pp.008854
Wijngaard, P. W. J., Sinnige, M. P., Roobeek, L., Reumer, A., Schoonheim, P. J., Mol, J. N., et al. (2005). Abscisic acid and 14-3-3 proteins control K+ channel activity in barley embryonic root. Plant J. 41, 43–55. doi: 10.1104/pp.008854
Xu, W. F., and Shi, W. M. (2006). Expression profiling of the 14-3-3 gene family in response to salt stress and potassium and iron deficiencies in young tomato (Solanum lycopersicum) roots: Analysis by real-time RT–PCR. Ann. Bot. 98, 965–974. doi: 10.1093/aob/mcl189
Xu, W. F., Shi, W. M., Jia, L. G., Liang, J. S., and Zhang, J. H. (2012). TFT6 and TFT7, two different members of tomato 14-3-3 gene family, play distinct roles in plant adaption to low phosphorus stress. Plant Cell Environ. 35, 1393–1406. doi: 10.1111/j.1365-3040.2012.02497.x
Yan, J., He, C., Wang, J., Mao, Z., Holaday, S. A., Allen, R. D., et al. (2004). Overexpression of the Arabidopsis 14-3-3 protein GF14 lambda in cotton leads to a “staygreen” phenotype and improves stress tolerance under moderate drought conditions. Plant Cell Physiol. 45, 1007–1014. doi: 10.1093/pcp/pch115
Yang, J. L., Chen, W. W., Chen, L. Q., Qin, C., Jin, C. W., Shi, Y. Z., et al (2013.). The 14-3-3 protein GENERAL REGULATORY FACTOR11 (GRF11) acts downstream of nitric oxide to regulate iron acquisition in Arabidopsis thaliana. New Phytol. 197, 815–824. doi: 10.1111/nph.12057
Yao, Y., Du, Y., Jiang, L., and Liu, J. Y. (2007). Molecular analysis and expression patterns of the 14-3-3 gene family from Oryza sativa. J. Biochem. Mol. Biol. 40, 349–357. doi: 10.5483/BMBRep.2007.40.3.349
Zhang, S., Chen, C., Li, L., Meng, L., Singh, J., Jiang, N., et al. (2005). Evolutionary expansion, gene structure, and expression of the rice wall-associated kinase gene family. Plant Physiol. 139, 1107–2114. doi: 10.1104/pp.105.069005
Zhang, Z. T., Zhou, Y., Li, Y., Shao, S. Q., Li, B. Y., Shi, H. Y., et al. (2010). Interactome analysis of the six cotton 14-3-3s that are preferentially expressed in fibres and involved in cell elongation. J. Exp. Bot. 61, 3331–3344. doi: 10.1093/jxb/erq155
Zhou, H., Lin, H., Chen, S., Becker, K., Yang, Y., Zhao, J., et al. (2014). Inhibition of the Arabidopsis salt overly sensitive pathway by 14-3-3 proteins. Plant Cell 26, 1166–1182. doi: 10.1105/tpc.113.117069
Keywords: 14-3-3, Brassica rapa, expression differentiation, gene divergence, polyploidy
Citation: Chandna R, Augustine R, Kanchupati P, Kumar R, Kumar P, Arya GC and Bisht NC (2016) Class-Specific Evolution and Transcriptional Differentiation of 14-3-3 Family Members in Mesohexaploid Brassica rapa. Front. Plant Sci. 7:12. doi: 10.3389/fpls.2016.00012
Received: 15 August 2015; Accepted: 07 January 2016;
Published: 26 January 2016.
Edited by:
Girdhar Kumar Pandey, University of Delhi, IndiaReviewed by:
Yueyun Hong, Huazhong Agricultural University, ChinaYashwanti Mudgil, University of Delhi, India
Copyright © 2016 Chandna, Augustine, Kanchupati, Kumar, Kumar, Arya and Bisht. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Naveen C. Bisht, bmNiaXNodEBuaXBnci5hYy5pbg==
†These authors have contributed equally to this work.
 Ruby Chandna
Ruby Chandna Rehna Augustine†
Rehna Augustine† Praveena Kanchupati
Praveena Kanchupati Roshan Kumar
Roshan Kumar Pawan Kumar
Pawan Kumar Gulab C. Arya
Gulab C. Arya Naveen C. Bisht
Naveen C. Bisht