- National Institute of Plant Genome Research, New Delhi, India
The sedentary life of plants has forced them to live in an environment that is characterized by the presence of numerous challenges in terms of biotic and abiotic stresses. Phytohormones play essential roles in mediating plant physiology and alleviating various environmental perturbations. Jasmonates are a group of oxylipin compounds occurring ubiquitously in the plant kingdom that play pivotal roles in response to developmental and environmental cues. Jasmonates (JAs) have been shown to participate in unison with key factors of other signal transduction pathway, including those involved in response to abiotic stress. Recent findings have furnished large body of information suggesting the role of jasmonates in cold and heat stress. JAs have been shown to regulate C-repeat binding factor (CBF) pathway during cold stress. The interaction between the integrants of JA signaling and components of CBF pathway demonstrates a complex relationship between the two. JAs have also been shown to counteract chilling stress by inducing ROS avoidance enzymes. In addition, several lines of evidence suggest the positive regulation of thermotolerance by JA. The present review provides insights into biosynthesis, signal transduction pathway of jasmonic acid and their role in response to temperature stress.
Introduction
Adequate perception, amalgamation and transduction of signals are obligatory for the growth and development of an organism. Plant hormones are a group of structurally diverse signal molecules that organize all cellular processes, consequently ensuring an effectual developmental plan and rationalized use of resources. Plant hormones therefore act as middlemen in the transmittance of information from the environment to the organism. The study of plant hormones is centuries old when the 5 classical hormones auxin, cytokinin, ABA, GA, and ethylene were described. However, over the last decade many “non-traditional” plant growth regulators have been described. These include highly diverse group of oxidized compounds, collectively known as oxylipins. Oxylipins execute diverse functions ranging from developmental processes to stress responses in plants (Andersson et al., 2006). Plant oxylipins can be produced either enzymatically by LIPOXYGENASES (LOXs) or α-DIOXYGENASES (α-DOXsas) or non-enzymatically by autoxidation of polyunsaturated fatty acids (Göbel and Feussner, 2009). One of the well characterized examples of oxylipins is Jasmonates (JAs).
The history of jasmonates is very old and dates back to 1960 when Demole successfully characterized methyl jasmonate (MeJA) from jasmine flower Jasminum grandiflorum. (Demole et al., 1962). Like many small esters, MeJA is volatile and has a sweet fragrance. Jasmonic acid (JA), on the other hand was isolated from fungal culture filtrate of Lasiodiplodia theobromae (Aldridge et al., 1971). JAs modulate many essential roles in plant development ranging from germination to vegetative growth to senescence. The role of JAs in dicotyledons such as tomato and Arabidopsis is well known, they are directly entailed in a number of physiological processes like stamen and trichome development, vegetative growth, cell cycle regulation, senescence, anthocyanin biosynthesis regulation, fruit ripening, cell cycle regulation (Parthier, 1991; Koda et al., 1992; Sembdner and Parthier, 1993; Creelman and Mullet, 1995, 1997; Koda, 1997; Wasternack and Hause, 2002; Browse, 2005; Wasternack, 2007; Balbi and Devoto, 2008; Pauwels et al., 2008; Zhang and Turner, 2008; Reinbothe et al., 2009; Yoshida et al., 2009). In addition, JAs activate plant defense mechanisms in response to insect-driven wounding, pathogen attack, and environmental stress, such as low temperature, salinity, heavy metal toxicity (Creelman and Mullet, 1997; Wasternack, 2007; Howe and Jander, 2008; Browse, 2009; Pauwels and Goossens, 2011). Studies in monocots have also confirmed the indispensible role of JAs in reproductive bud initiation and elongation, sex determination, leaf senescence and responses to the attack by pathogens and insects (Engelberth et al., 2004; Tani et al., 2008; Acosta et al., 2009; Yan et al., 2012).
Biosynthesis of Jasmonates
JAs are biosynthesized by the sequential action of enzymes present in plastid, peroxisome and cytoplasm (Feussner and Wasternack, 2002). JA biosynthesis is initiated by the release of α-LINOLENIC ACID (α-LeA) (18:3) from chloroplast membranes by PHOSPHOLIPASE1 (PLA1) to generate JA substrate (Vick and Zimmerman, 1983). α-LeA liberation is followed by the incorporation of molecular oxygen by the lipoxygenase family enzyme, LINOLEATE OXYGEN OXIDOREDUCTASE (13-LOX) at carbon atom 13 of the substrate forming 13S-HYDROPEROXY-(9Z,11E,15)-OCTADECATRIENOIC ACID (13-HPOT). 13-HPOT undergoes dehydration by the ALLENE OXIDE SYNTHASE (AOS) to form cis (+)12-OXO-PHYTODIENOIC ACID (OPDA) (Turner et al., 2002; Devoto and Turner, 2003; Wasternack, 2007). Similarly, LOX, AOS and AOC together catalyze hexadecatrienonic acid (C16:3) to form dinor-OPDA (dnOPDA). OPDA and dnOPDA are further transported to peroxisome via transporter COMATOSE (CTS1) (Theodoulou et al., 2005), wherein, they are reduced to OXOPHYTOENIC ACID (OPC-8) and 12-OXOPHYTOENIC ACID (OPC-6), respectively by OPDA reductase 3 (OPR3). OPDA, dnOPDA, OPC8 and OPC6 are activated by the ACYL-COENZYME A SYNTHETASES to form CoA esters, so that the carboxylic acid side chains can be shorted by two or three rounds of β-oxidation by ACYL-COA OXIDASE (ACX), a MULTIFUNCTIONAL PROTEIN (MFP), and L-3-KETOACYL COA THIOLASE (KAT) (Schneider et al., 2005). Jasmonoyl-CoA, the final product of the β-oxidation reactions, is cleaved by THIOESTERASE (TE) to form cis-7-iso-jasmonic acid [(+)-7-iso-JA]. It is then catabolized further by JA CARBOXYL METHYLTRANSFERASE (JMT) to form volatile counterpart MeJA. MeJA ESTERASE (MJE) in turn converts MeJA back to JA. The reversible conversion between JA and Jasmonoyl-isoleucine (JA-Ile) is catalyzed by a JASMONATE AMINO ACID SYNTHETASE (JAR1).
Metabolic Fate of JA
There are plethora of jasmonate compounds. JA goes through several biochemical modifications (Sembdner and Parthier, 1993; Koch et al., 1997; Seo et al., 2001; Staswick and Tiryaki, 2004; Swiatek et al., 2004). JA, Cis Jasmone (CJ), MeJA and JA-Ile have some biological activity in plants (Wasternack, 2007; Fonseca et al., 2009a). CJ, a volatile counterpart of JA is biologically active and is released in response to herbivory and insect driven attack. Transcriptome analysis data of CJ treated Arabidopsis plants provided insights into a COI1-independent CJ signaling (Matthes et al., 2010).
Miersch and co-workers in 2008 reported the presence of high levels of 12-OH-JA, 12-HSO4-JA, and 12-O-Glc-JA in immature seeds and leaves of Glycine max and Zea mays (Miersch et al., 2008). The role of jasmonates (JAs) 12-OH-JA, 12-HSO4-JA, and 12-O-Glc-JA in sex determination has been studied in Zea mays (Acosta et al., 2009; Browse, 2009). JA, MeJA, and CJ are considered useful tools in anti-cancer therapy as they are known to induce cell death by mitochondria perturbation and subsequent release of cytochrome oxidase (Kim et al., 2004; Rotem et al., 2005). Michelet et al. (2012) reported the anti-aging potential of tetra-hydro-jasmonic acid in humans. Tetra-hydro-jasmonic acid is known to increase the synthesis of hyaluronic acid by increasing the expression of hyaluronase synthase 2 and hyaluronase synthase 3.
Jasmonic Acid Perception and Signaling
JA signal perception and transduction involve numerous TFs, repressors and members of ubiquitin proteasomal pathway. The section gives information of different signaling components. The current model of JA signal transduction is given in Figure 1.
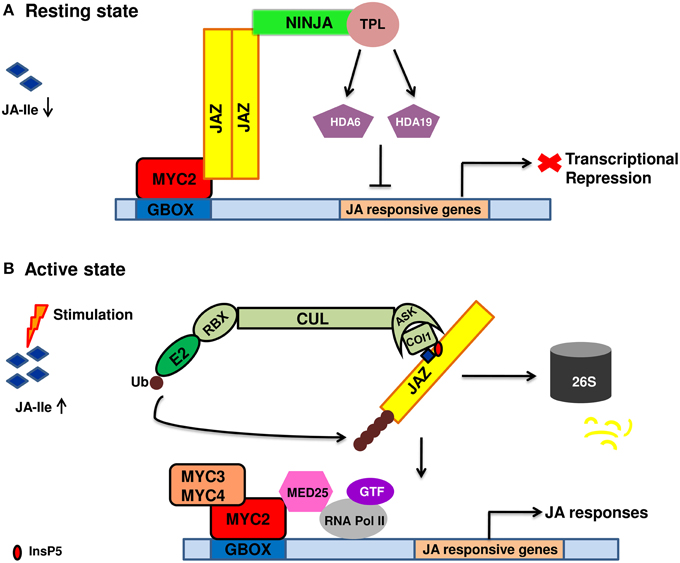
Figure 1. Diagrammatic representation of jasmonic acid perception and signal transduction pathway. (A) In the absence of a stimuli, jasmonic acid is not synthesized. As a result, JA mediated gene expression is inhibited due to the binding of JAZ repressors to the transcriptional activator MYC2. JAZ proteins recruit TPL and adaptor protein NINJA. Together, JAZ-NINJA-TPL form an active transcriptional repression complex that inhibit jasmonate responses by converting an open complex to a closed complex by recruiting HDA6, HDA19. (B) Upon stimulation by pathogen/insect/wounding, JA is rapidly synthesized and readily epimerizes to JA-Ile. It then binds to COI1-JAZ-InsP5 co-receptor complex causing ubiquitination and proteasomal degradation of JAZ. This frees MYC2 and its homologs from repression which then bind to G-box element present downstream of JA-responsive genes upon homo/heterodimerization. This is followed by the recruitment of MED25 that in turn bring RNAPol II and general transcription factors hence, causing diverse jasmonate responses. JA, jasmonic acid; JA-Ile, Jasmonate–isoleucine; JAZ, Jasmonate ZIM domain; NINJA, Novel adaptor of JAZ; TPL, topless; COI1, coronatine insensitive; Cul, Cullin1; RBX1, ring box1; Ub, ubiquitin; ASK1, Arabidopsis skp1 homolog 1; InsP5, inositol pentakisphosphate; GTF, general transcription factor; HDA6, HDA19, histone deacetylase 6,19; MED25, mediator25; RNAPol II, RNA polymerase II.
Bioactive Ligand
Fonseca and co-workers in 2009 provided evidences that (+)-7-iso-JA-L-Ile is the sole natural ligand of A. thaliana as revealed by detailed GC-MS and HPLC analyses. Also, experiments carried out by Thines et al. (2007) revealed that only JA-Ile out of MeJA, OPDA, and JA can promote COI1-JAZ binding, thus confirming JA-Ile to be the direct JA signaling ligand in plants.
SCF Complex
The ubiquitin-proteasome comprises of Skp1/Cullin/F-box (SCF). Earlier, researchers believed that screening Arabidopsis mutants insensitive to growth inhibition with bacterial coronatine, a structural and functional homolog of JA-Ile, would result in discovering JA receptor in plants (Feys et al., 1994; Fonseca et al., 2009b). Exhaustive genetic screens identified the allele of coronatine insensitive 1 (coi1), suggesting COI1 functions in JA perception in plants. It was considered as the receptor from two lines of evidences- first, coi1 mutant exhibits male sterility, defective responses to JA-treatment and wounding and susceptibility to necrotrophic pathogens and insects; secondly, COI1 locus encodes an F-box protein that associates with its other counterparts SKP1, Cullin, and Rbx proteins to form an E3 ubiquitin ligase (Xie et al., 1998). COI1 show approximately 33% sequence similarity with the auxin receptor TIR1 in amino acid sequence having leucine-rich-repeats and F-box motif (Yan et al., 2009).
JAZ Proteins
After the discovery of the receptor the most fascinating question was to find out the substrate for SCFCOI1 E3 ubiquitin ligase complex. This substrate was anticipated to be the key negative regulator of JA signaling. In 2007, three independent research groups discovered a new family of protein in Arabidopsis called JASMONATE ZIM DOMAIN (JAZ) proteins (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007). The JAZ proteins belong to the larger plant specific TIFY family, consisting of a core TIF[F/Y]XG motif within the ZN-FINGER PROTEIN EXPRESSED IN INFLORESCENCE MERISTEM (ZIM) protein domain. A. thaliana consists of 12 JAZ proteins (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007; Chung et al., 2009) that are differentiated from other TIFY family proteins by the presence of C-terminally located Jas motif, SLX2FX2KRX2RX5PY (Nishii et al., 2000; Vanholme et al., 2007; Yan et al., 2007). They contain N-terminal domain, a highly conserved C-terminal Jas domain that mediates the interaction with the COI1 and several transcription factors, and the conserved protein- protein interaction domain, the ZIM (TIFY) domain that helps in JAZ dimerization and interaction with NINJA (Vanholme et al., 2007; Chung et al., 2009; Pauwels and Goossens, 2011; Wasternack and Hause, 2013). The Jas domain is exclusively required to repress downstream targets of JAZ proteins (Chini et al., 2007; Thines et al., 2007; Yan et al., 2007).
The initial clue about the role of JAZ proteins in JA-signaling came from the jasmonate-insensitive 3 mutant (jai3), which is a mutant of JAI3/JAZ3 gene. In jai3-1 mutant, the JAZ3 protein lacks the C-terminal portion which perturbs its binding and degradation via SCFCOI1 complex. This resulted in accumulation of truncated JAI3/JAZ3 proteins in the mutant which blocked the JA-induced degradation of other JAZ proteins and hence dominant JA-insensitive phenotype (Chini et al., 2007).
Co-receptor Complex
The co-receptor complex is formed by the physical interaction of COI1 with the Jas domain of JAZ proteins in the presence of JA-Ile (Yan et al., 2009; Sheard et al., 2010). More recently, the role of inositol pentakisphosphate (IP5) as a cofactor in the formation of co-receptor complex has been substantiated (Sheard et al., 2010; Mosblech et al., 2011). JA, OPDA, MeJA, and JA-Ile were tested for affinity in COI1 JAZ1 binding. Surprisingly, only JA-Ile functioned as ligand for COI1-JAZ interaction (Thines et al., 2007). Based on the information available hitherto, the true jasmonates receptor is a co-receptor complex, consisting of the SCFCOI1 E3 ubiquitin ligase complex, JAZ degrons (JAZ1 to JAZ12) and IP5 (Sheard et al., 2010).
Co-repressors
Co-repressors are transcriptional regulators that inhibit transcription initiation. One such example is the group of Groucho/Tup1 corepressor family comprising of TOPLESS (TPL) and TPL-related proteins (TPRs). TPL and TPR mediate repression by recruiting histone deacetylases and demethylases that cause chromatin modification (Macrae and Long, 2011). TPL interacts with JAZ proteins via ETHYLENE RESPONSE FACTOR (ERF)-ASSOCIATED AMPHIPHILIC REPRESSION (EAR) motif. Those JAZ proteins that do not have the repression motif recruit TPL through an adapter protein called NOVEL INTERACTOR OF JAZ (NINJA) (Pauwels et al., 2010). NINJA was first identified by Tandem affinity purification as an interactor of JAZ1 (Pauwels et al., 2010).
JAZ Targets
The role of bHLH transcription factor MYC2 in mediating the transcriptional regulation of JA is well defined and thus has been considered the master regulator of many biological processes (Lorenzo et al., 2004; Dombrecht et al., 2007). The role of MYC2 in JA mediated responses is revealed by the study of its mutant jasmonate-insensitive1 (jin1). Microarray analysis of wild type and the mutant myc2/jin1 exposed the role of MYC2 in JA-dependent transcriptional regulation. MYC2 has twin function of an activator of JA-induced root growth inhibition, anthocyanin biosynthesis and oxidative stress tolerance and a repressor in mediating resistance to necrotrophic pathogens, insects and biosynthesis of tryptophan and indol glucosinolates (Lorenzo et al., 2004; Dombrecht et al., 2007). Besides MYC2, several other TFs control diverse JA response. These TFs are MYC3, MYC4, MYB, GL3, EGL3 AP, GL1 etc. (Cheng et al., 2011). MYC2 forms homo or heterodimers with its close homologs MYC3 and MYC4 to regulate the transcription of downstream targets (Fernández-Calvo et al., 2011) by binding to the G-box (5′-CAC GTG-3′) and G-box related hexamers (Abe et al., 1997; Boter et al., 2004; Yadav et al., 2005).
Dissecting the Role of Jasmonates in Abiotic Stress
Out of 13 billion hectares of total land, only 1.6 billion is under farmland production accounting to only 12% of arable land (Syngenta, 2014)1. Agriculture must evolve in order to meet the demands of the increasing population. However, every year some part of the world suffers from drought, global increase in temperature, variable precipitation that eventually hampers the quality and quantity of crops. All these visible warning signs can have erratic production patterns all over the world. Plants encounter numerous challenges in terms of competition from other plants, organisms and because of the complex environment. All these provocations have made the plants tougher and more flexible. The morphological flexibility has given them the advantage to counteract, inhabit and endure biotic and abiotic challenges. Rapid changes in the plant biochemistry and physiology are mediated by the action of several phytohormones. By tradition cytokinin, auxins, brassinosteroids, and giberallins have always been associated to regulate developmental processes of plants, whereas, salicylic acid, JA and ethylene associate with plant defense and ABA regulates plant's response to abiotic stress. Now, it has been quite evident from many reports that all hormones affect multiple plant functions. Thus, one can say that hormones not only participate in plant developmental processes but also have a say in plant's response to abiotic stresses like drought, osmotic stress, chilling injury, heavy metal toxicity etc. These adversities have forced the plants to either employ avoidance as a mechanism in order to surmount the stress or choose defense over growth (Band et al., 2012; Murray et al., 2012; Petricka et al., 2012; Wasternack and Hause, 2013). Thus, stress activates signal transduction of hormones which may promote specific protective mechanisms.
Cold Stress
Among various environmental perturbations, cold stress or low temperature stress limits plant performance and geographical distribution. Cold stress can be categorized into chilling (0–15°C) and freezing that causes mayhem in tropical and subtropical plants by inducing chlorosis, necrosis, membrane damage, changes in cytoplasm viscosity, changes in enzyme activities (Ruelland and Zachowski, 2010) and ultimately death. All these physiological and biochemical changes elicit a cascade of events that cause changes in gene expression pattern and protein products and thus in due course induce plant species to adopt stratagems to tolerate low non-freezing temperatures and complete their life cycle, an experience known as cold acclimation response.
ICE-DREB1/CBF regulon plays an imperative role in cold response pathway in model plant Arabidopsis thaliana (Thomashow, 1999; Chinnusamy et al., 2007). Inducer of CBF EXPRESSION 1 (ICE1) is a MYC-type transcription factor that acts as a master regulator and controls CBF/DREB1 pathway. In Arabidopsis, three CBF/DREB1 are involved in the regulation of COLD REGULATED (COR) gene expression and tolerance to cold stress (Gilmour et al., 2000, 2004). In this pathway, ICE1 positively regulates and activates CBF/DREB1 genes that encode AP2/ ERF type TF family. CBFs, by binding to C-repeat (CRT) element induce COR genes leading to tolerance to cold stress (Thomashow, 2010).
Tropical and subtropical fruits like mango, avocado, papaya etc. exhibit symptoms due to chilling injuries such as browning discoloration and off-flavor in the fruit. Previous studies have specified the role of MeJA in alleviating chilling injury by inducing the production of cryo-protective agents, proteinase inhibitors, polyamines, ABA, lower activity of LOXs, and antioxidants (Wang and Buta, 1994; González-Aguilar et al., 2000; Cao et al., 2009; Zhao et al., 2013). Role of MeJA in response to freezing tolerance has also been studied in rice seedlings. It has been reported that rather than treating rice with MeJA during or after stress imposition, MeJA treatment before chilling remarkably enhances the survival ratio of chilled rice seedlings (Lee et al., 1997). Furthermore, it was observed that MeJA maintained the well-watered status of chilled plants by preventing stomatal opening and enhancing hydrolytic conductivity.
Du et al. (2013) reported an increase in the level of endogenous JA on exposure to cold stress. Microarray analysis of cold treated rice seedlings revealed upregulation of JA biosynthesis genes OsDAD1, OsLOX2, OsAOC, OsAOS1, OsAOS2, OsOPR1, and OsOPR7. Besides this, JA signaling genes such as OsJAR1, OsbHLH148, and OsCOI1a showed up regulation upon cold exposure. Furthermore, positive role of jasmonate in enhancing constitutive and cold acclimation–induced freezing tolerance of Arabidopsis has been reported by Hu et al. (2013). Treatment of WT Arabidopsis seedlings with exogenous methyl jasmonate improved the endurance and plant freezing tolerance. On the contrary, blocking JA biosynthesis and signaling pathway rendered the plant hypersensitive to freezing stress.
Of lately, role of JAZ repressors in controlling cold stress tolerance has also been investigated. Under normal growth conditions Arabidopsis JAZ1 and JAZ4 interact and suppress cold TFs ICE1 and ICE2, thus quelling ICE1-CBF/DREB1 pathway (Hu et al., 2013). Also, over expression of JAZ1 or JAZ4 repressed the cold-induced expression of CBF/DREB1, in that way contributing the transgenic plants sensitive to freezing. However, over expression of ICE1 was able to salvage freezing sensitive phenotype of coi1-1 mutant plants.
Recent studies have suggested the role of downstream transcription factors in the regulation of cold responses. Homologs of Arabidopsis MYC2 TF have been isolated and characterized in Musa accuminata (Peng et al., 2013; Zhao et al., 2013). MaMYC2a and MaMYC2b have been shown to be rapidly induced by MeJA treatment upon cold exposure. Expression profiles of CBF cold responsive pathway genes, including MaCBF1, MaCBF2, MaCOR1, MaKIN2, MaRD2, and MaRD5 demonstrated the induction of CBF genes by MeJA upon cold stress. Also, they have been accounted to physically interact with MaICE1, therefore suggesting a potential cross talk between two signal transduction pathways.
Arabidopsis SENSITIVE TO FREEZING 6 (SFR6) controls cold regulated gene expression and is well known to act post-translationally on the CBF module (Knight et al., 1999, 2009; Boyce et al., 2003). Additionally, it is involved in regulating JA responses (Wathugala et al., 2012; Zhang et al., 2012). Very recently, SFR6 has been identified as the MEDIATOR16 of the plant mediator complex that is involved in recruiting RNA polymerase II to promoters carrying CRT/DREB motif (Hemsley et al., 2014).
Similar to JA, salicylic acid (SA) is a powerful tool in regulating cold stress tolerance. Exogenous application of SA on H. vulgare genotypes resulted in cold tolerance by enhancing antioxidant enzymes, ice nucleation activity and the patterns of apoplastic proteins (Mutlu et al., 2013). Accumulation of endogenous free SA and glucosyl SA has been reported during chilling in Arabidopsis shoots, wheat and grape berry (Scott et al., 2004; Wan et al., 2009; Kosová et al., 2012). However, it has been observed that concentration and duration of applied SA greatly influence its utility. High concentration and continual application of SA decreases the cold tolerance capacity of plants as observed in some Arabidopsis mutants, such as CONSTITUTIVE EXPRESSER OF PATHOGENESIS-RELATED GENE1 (CPR1) AND ACCELERATED CELL DEATH6 (ACD6), in which SA is over-accumulated, exhibit a dwarf phenotype and freezing sensitivity (Scott et al., 2004; Miura et al., 2010).
Fung et al. (2004) reported an alleviation of chilling injury in freshly harvested green bell pepper (Capsicum annuum) by methyl SA (MeSA) and MeJA vapors. This reduction of chilling injury in the green bell pepper was related with an increase in the expression of the ALTERNATIVE OXIDASE (AOX) gene induced by MeSA and MeJA vapors. Feng et al. (2008) later reported that the expression of AOX was enhanced under chilling stress. It has been earlier reported that AOX expression increased in response to low temperature stresses in rice (Ito et al., 1997). All these observations suggest that AOX is involved in combating cold stress. Siboza et al. (2014) reported that combined treatment of MeJA and SA reduced ROS accumulation, lipid peroxidation and increased chilling tolerance in lemon during cold storage by increasing the synthesis of total phenolics and phenylalanine ammonia lyase (PAL) and inhibiting the activity of polyphenol oxidase (PPO) and peroxidase (POD). Induction of PAL activity has been considered a good marker of CI (Martínez-Téllez and Lafuente, 1993; Sanchez-Ballesta et al., 2000).
Findings by Miura and Ohta (2010) have indicated ICE1 to be an essential integrator of SA signaling and cold response pathway. ice1 mutant also showed an up-regulation of SA-inducible genes. Additionally, CALMODULIN BINDING TRANSCRIPTION ACTIVATOR 3 CAMTA3/AtSR1 has been shown to participate in enhancing cold tolerance by binding to the promoter of CBF1 and CBF2/DREB1C (Doherty et al., 2009). Recently, Kim et al. (2013) extended the findings by showing the up-regulation of 15% cold inducible genes by CAMTA TFs. It has been established that CAMTA3 behaves as a repressor of SA biosynthesis at warm temperature under non-stressed condition (Du et al., 2009). But, under cold conditions this repression is overcome as reported by Kim and co-workers in 2013, leading to an increase in the level of SA and up regulation of SA responsive genes. However, the results indicate that SA does not contribute to freezing tolerance but genes induced by SA at low temperature increase the resistance to pathogen attack. The above findings raise the possibility that CAMTA3 does not only cause changes in gene expression at low temperature but also plays a role in regulating genes involved in SA biosynthesis at low temperature.
Unlike the close interaction between components of JA signaling and cold regulated transcription factors and JA functioning as a crucial upstream signal to ICE-CBF/DREB1 pathway (Figure 2), very few reports are available on the molecular mechanisms underlying SA-mediated improved plant tolerance to cold temperature. However, the above reports shed light upon the relatedness between cold signaling and SA signaling. Moreover, the participation of ICE1 in both JA and SA signaling pathway, ROS avoidance mechanism employed both by SA and JA points out the possible crosstalk between JA and SA signal transduction pathway to fight cold stress.
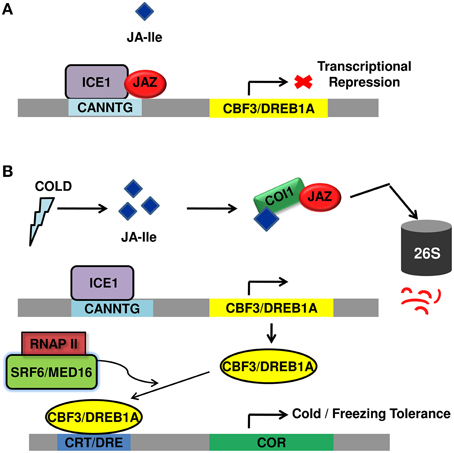
Figure 2. Diagrammatic representation of regulation of cold stress tolerance by JA signal transduction pathway. (A) Under normal growth conditions, JAZ repressor proteins physically interact and suppress cold TF ICE1, thus repressing ICE/CBF-DREB1 pathway and rendering plants sensitive to freezing. (B) Upon cold induction, JA is synthesized that rapidly isomerizes to JA-Ile and lead to proteasomal degradation of JAZ. This frees ICE1 that binds to CBF3 responsive element leading to its expression. The CBF proteins bind to CRT/DRE element causing the expression of COR genes that participate in cold/freezing tolerance. JAZ, jasmonate ZIM domain protein; TF, transcription factor; ICE1, inducer of CBF expression; CBF-DREB1, C repeat binding factor 1-dehydartion responsive element binding factor1B; COR, cold regulated; DRE, dehydration responsive element; CRT, C-repeat; SFR6/MED16, sensitive to freezing 6/Mediator 16; RNAPII, RNA polymerase II; COI1, coronatine insensitive.
Earlier work conducted by Wilen et al. (1993) pointed out synergism between ABA and JA in inducing freezing tolerance in bromegrass cell cultures. The role of JAs in imparting freezing stress tolerance via CBF1/DREB1 pathway is well known. To further investigate their role in imparting freezing tolerance via CBF/DREB1 independent pathway, Hu et al. (2013) carried out microarray and real time analysis in WT and coi1 mutant plants upon cold treatment. They observed that many cold responsive genes that do not fall in CBF1/DREB1 pathway were down-regulated in coi1 mutant plants, thus indicating that jasmonate might play a role in imparting freezing tolerance via CBF/DREB1 independent pathway. To further assess whether JAs affect all cold regulated expression of genes, Hu and co-workers carried out expression analysis of cold responsive genes falling in ABA pathway in WT and coi1 plants and it was observed that the transcript level in coi1 mutant plants changed similar to WT upon cold treatment. Hence, cold tolerance may be provided independently via ABA and JA signaling pathways.
Like jasmonates, ethylene (ET) has been demonstrated to modulate various abiotic stress responses. Based on findings from Arabidopsis, ET signaling pathway has been shown to negatively regulate freezing stress responses (Shi et al., 2012). EIN3, a positive regulator of ET signaling pathway transcriptionally represses the CBF pathway and thus acts antagonist to JA. Furthermore, Zhu and co-workers in 2011 revealed a crosstalk between JA and ET signaling pathways through the interaction between JAZ and their targets EIN3/EIL1 (Zhu et al., 2011). According to their model, ET is required for stabilization of EIN3/EIL1 and JA is needed for their release from JAZ degradation, as speculated by them. This dual regulation balances the detrimental effects on plant growth and development and establishes appropriate stress tolerance, as speculated by them. Thus, one can say that the CBF signaling pathway is regulated at multiple levels by JAs as it positively regulates freezing tolerance and at the same time represses the CBF/DREB1 pathway via EIN3/EIL1.
The role of JAs and its interplay with other phytohormones to modulate cold stress responses has been demonstrated in this section. The interaction between different components for example JAZ with EIN3/EIL1 or JAZ with ICE1/ICE2 mediates ET, JA, cold signal transduction pathways. Identifying the novel components involved in crosstalk between JA and other hormones in regulating CBF cold response pathway would help in dissecting the exact molecular mechanism.
Heat Stress
Temperature above the optimum for growth can be detrimental, causing injury or irreversible damage to plant growth and development. IPCC (2014) reports suggest an approximate increase of 4–5°C in the average temperature by the end of the twenty-first century. High temperature stress negatively influences plant processes and cellular machinery, thereby impairing cell homeostasis (Bokszczanin et al., 2013). Plants have the inherent ability to ameliorate the adverse effects of heat shock by the phenomenon of basal thermotolerance whereas, acquired thermotolerance is achieved when plants are pre-exposed to high, non-lethal temperature (Bokszczanin et al., 2013). In response to high temperature, plants synthesize HEAT SHOCK PROTEINS (HSPs) that prevent denaturation and assist refolding of damaged proteins (Boston et al., 1996).
The role of JA signaling in contributing to thermotolerance has been recently established in WT Arabidopsis (Clarke et al., 2009). Exogenous application of low concentration of MeJA maintained cell viability in heat stressed plants as demonstrated by electrolyte leakage assays. Moreover, heating WT Arabidopsis led to the accumulation of several jasmonates including OPDA, MeJA, JA, and JA-Ile. But, no evidence was found that thermotolerance conferred by MeJA elicited HSP gene expression. Expression level of jasmonate inducible gene PDF1.2 was found to be high upon heat stress exposure. The final proof of the role of jasmonates in imparting thermotolerance was confirmed by mutant analysis wherein JA and SA signaling mutants coi1-1, opr3, and jar1-1cpr5-1 were found to be sensitive to heat stress (Clarke et al., 2009). Thus, establishing the fact that both SA and JA provide basal thermotolerance.
SUPPRESSOR OF G2 ALLELE OF SKP1 (SGT1) protein operates as a cofactor of HEAT SHOCK PROTEIN 90 (HSP90) in both plants and mammals forming functional complexes and providing thermotolerance. Intensive genetic and biochemical screening confirmed the role of SGT1 in plant hormone signaling pathways that involve F-box proteins and ubiquitin ligases such as COI1 of JA signaling. SGT1 maintains the steady state of COI1 (Zhang et al., 2015). Reduced transcript levels of JA marker genes in HSP90 RNAi lines confirmed the crucial role played by HSP90 and HSP70 in JA-COR (coronatine) responses. Additionally, pre-treating WT Arabidopsis with HSP inhibitor attenuated COR (coronatine) triggered gene expression. Given the facts that HSP proteins have numerous substrates including transcription factors, E3 ligases, kinases and the above data suggested that COI1 is a client protein of SGT1b–HSP70–HSP90 chaperone complexes. This widens the functional capacity of SGT1b–HSP70–HSP90 chaperone complexes in regulating JA responses.
WRKY super family consists of 74 members in Arabidopsis thaliana (Eulgem and Somssich, 2007) and is subdivided into three groups based on the number of WRKY domains and the features of their Zn finger like motifs (Eulgem et al., 2000). They are a group of regulatory proteins that participate in plant developmental processes but notably in a plethora of biotic and abiotic challenges. There are sufficient evidences that many WRKY genes participate in abiotic stresses, including heat stress. Several WRKY TFs have been revealed to impart thermotolerance like AtWRKY25 (Zhu et al., 2009), AtWRKY39 (Li et al., 2010), and OsWRKY11 (Wu et al., 2009). Dang et al. (2013) showed that CaWRKY40 was involved in heat stress and was transcriptionally induced by exogenous application of JA. Moreover, over expression lines of CaWRKY40 derepressed JA biosynthesis NtLOX1 by heat stress. Together, the findings suggest that JA mediates the expression of CaWRKY40 leading to the expression of downstream thermotolerance-related genes.
JA and ET act as antagonists in regulating heat stress responses. Studies by Clarke et al. (2009) showed that despite being produced in response to heat stress, ET negatively regulates heat stress tolerance. They found out that ein2 mutant displayed thermotolerance, hence suggesting that EIN2 mediated pathway negatively regulates thermotolerance. They also demonstrated that ET production was augmented by JA from studies carried out in WT and opr3 mutant.
Concluding Remarks
The daunting issue of nutritional and food security has resulted in a quest among researchers to elucidate the action of phytohormones in stress related responses. Tolerance to (a)biotic stresses is a challenge of agro-economic impact. For this, a model eudicot, Arabidopsis has emerged as a quintessential system to study plant stress tolerance. However, crop plants are exposed to complex environmental perturbations in the field. Therefore, thorough research will be required to understand how crops respond to multiple abiotic stresses in order to develop new varieties that can withstand global climate changes.
The role of jasmonates in plant development is very well established (Wasternack and Hause, 2013). Nevertheless, a significant body of research suggests the role of jasmonic acid in plant responses to abiotic stresses. Growing sagacity of crosstalk between JAs and other hormones and with different components of abiotic signal transduction may allow us to dissect key factors involved in the crosstalk. Recent reports suggest interplay between cold regulated transcription factors and components of JA biosynthesis and signaling. The interaction of JAZ repressors with transcriptional activators ICE1 and ICE2 and falling upstream to the cold response pathway (Hu et al., 2013) as well as the induction of JA genes upon cold stimulus (Du et al., 2013; Hu et al., 2013) demonstrates the sharing of common signaling components and hence a closed interaction between the two signal transduction pathway. Additionally, several lines of evidences suggest the role of JAs in imparting thermotolerance. In addition, mutant analyses have also confirmed the role of JAs in heat tolerance responses. Modifying different upstream/downstream signaling components by genetic engineering can improve the adaptability of plants in response to temperature stress. However, there is still obscurity in the crosstalk among different signaling pathways. The application of forward and reverse genetic analysis in model plants along with the genomics and proteomics tools will help us in the discovery of new regulatory components and in dissecting the complex interactions between different signaling pathways to elucidate hormone action in stress related context.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are thankful to National Institute of Plant Genome Research core grant. The authors are also thankful to University Grants Commission for research fellowship to MS.
Footnotes
1. ^Syngenta (2014). Our industry 2014.
References
Abe, H., Yamaguchi-Shinozaki, K., Urao, T., Iwasaki, T., Hosokawa, D., and Shinozaki, K. (1997). Role of arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 9, 1859–1868.
Acosta, I. F., Laparra, H., Romero, S. P., Schmelz, E., Hamberg, M., Mottinger, J. P., et al. (2009). Tasselseed1 is a lipoxygenase affecting jasmonic acid signaling in sex determination of maize. Science 323, 262–265. doi: 10.1126/science.1164645
Aldridge, D. C., Galt, S., Giles, D., and Turner, W. D. (1971). Metabolites of Lasiodiplodia theobromae. J. Chem. Soc. C 1971, 1623–1627. doi: 10.1039/j39710001623
Andersson, M. X., Hamberg, M., Kourtchenko, O., Brunnström, A., McPhail, K. L., Gerwick, W. H., et al. (2006). Oxylipin profiling of the hypersensitive response in Arabidopsis thaliana. Formation of a novel oxo-phytodienoicacid-containing galactolipid, arabidopside E. J. Biol. Chem. 281, 31528–31537. doi: 10.1074/jbc.M604820200
Balbi, V., and Devoto, A. (2008). Jasmonate signalling network in Arabidopsis thaliana: crucial regulatory nodes and new physiological scenarios. New Phytol. 177, 301–318. doi: 10.1111/j.1469-8137.2007.02292.x
Band, L. R., Fozard, J. A., Godin, C., Jensen, O. E., Pridmore, T., Bennett, M. J., et al. (2012). Multiscale systems analysis of root growth and development: modeling beyond the network and cellular scales. Plant Cell 24, 3892–3906. doi: 10.1105/tpc.112.101550
Bokszczanin, K. L., and Solanaceae Pollen Thermotolerance Initial Training Network (SPOT ITN) Consortium, and Fragkostefanakis, S. (2013). Perspectives on deciphering mechanisms underlying plant heat stress response and thermotolerance. Front. Plant. Sci. 4:315. doi: 10.3389/fpls.2013.00315
Boston, R. S., Viitanen, P. V., and Vierling, E. (1996). Molecular chaperones and protein folding in plants. Plant Mol. Biol. 32, 191–222. doi: 10.1007/BF00039383
Boter, M., Ruíz-Rivero, O., Abdeen, A., and Prat, S. (2004). Conserved MYC transcription factors play a key role in jasmonate signaling both in tomato and Arabidopsis. Genes Dev. 18, 1577–1591. doi: 10.1101/gad.297704
Boyce, J. M., Knight, H., Deyholos, M., Openshaw, M. R., Galbraith, D. W., Warren, G., et al. (2003). The sfr6 mutant of Arabidopsis is defective in transcriptional activation via CBF/DREB1 and DREB2 and shows sensitivity to osmotic stress. Plant J. 34, 395–406. doi: 10.1046/j.1365-313X.2003.01734.x
Browse, J. (2005). Jasmonate: an oxylipin signal with many roles in plants. Vitam. Horm. 72, 431–456. doi: 10.1016/S0083-6729(05)72012-4
Browse, J. (2009). Jasmonate: preventing the maize tassel from getting in touch with his feminine side. Sci. Signal. 2:pe9. doi: 10.1126/scisignal.259pe9
Cao, S., Zheng, Y., Wang, K., Jin, P., and Rui, H. (2009). Methyl jasmonate reduces chilling and enhances antioxidant enzyme activity in postharvest loquat fruit. Food Chem. 115, 1458–1463. doi: 10.1016/j.foodchem.2009.01.082
Cheng, Z., Sun, L., Qi, T., Zhang, B., Peng, W., Liu, Y., et al. (2011). The bHLH transcription factor MYC3 interacts with the Jasmonate ZIM-domain proteins to mediate jasmonate response in Arabidopsis. Mol. Plant. 4, 279–288. doi: 10.1093/mp/ssq073
Chini, A., Fonseca, S., Fernández, G., Adie, B., Chico, J. M., Lorenzo, O., et al. (2007). The JAZ family of repressors is the missing link in jasmonate signalling. Nature 448, 666–671. doi: 10.1038/nature06006
Chinnusamy, V., Zhu, J., and Zhu, J. K. (2007). Cold stress regulation of gene expression in plants. Trends Plant Sci. 12, 444–451. doi: 10.1016/j.tplants.2007.07.002
Chung, H. S., Niu, Y., Browse, J., and Howe, G. A. (2009). Top hits in contemporary JAZ: an update on jasmonate signaling. Phytochemistry 70, 1547–1559. doi: 10.1016/j.phytochem.2009.08.022
Clarke, S. M., Cristescu, S. M., Miersch, O., Harren, F. J., Wasternack, C., and Mur, L. A. (2009). Jasmonates act with salicylic acid to confer basal thermotolerance in Arabidopsis thaliana. New Phytol. 182, 175–187. doi: 10.1111/j.1469-8137.2008.02735.x
Creelman, R. A., and Mullet, J. E. (1995). Jasmonic acid distribution and action in plants: regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. U.S.A. 92, 4114–4119. doi: 10.1073/pnas.92.10.4114
Creelman, R. A., and Mullet, J. E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 355–381. doi: 10.1146/annurev.arplant.48.1.355
Dang, F. F., Wang, Y. N., Yu, L., Eulgem, T., Lai, Y., Liu, Z. Q., et al. (2013). CaWRKY40, a WRKY protein of pepper, plays an important role in the regulation of tolerance to heat stress and resistance to Ralstonia solanacearum infection. Plant Cell Environ. 36, 757–774. doi: 10.1111/pce.12011
Demole, E., Lederer, E., and Mercier, D. (1962). Isolement et determination de la structure du jasmonate de methyle, constituant odorant caracteristique de l'essence de jasmin. Helv. Chim. Acta 45, 675–685. doi: 10.1002/hlca.19620450233
Devoto, A., and Turner, J. G. (2003). Regulation of jasmonate-mediated plant responses in Arabidopsis. Ann. Bot. 92, 329–337. doi: 10.1093/aob/mcg151
Doherty, C. J., Van Buskirk, H. A., Myers, S. J., and Thomashow, M. F. (2009). Roles for Arabidopsis CAMTA transcription factors in cold-regulated gene expression and freezing tolerance. Plant Cell 21, 972–984. doi: 10.1105/tpc.108.063958
Dombrecht, B., Xue, G. P., Sprague, S. J., Kirkegaard, J. A., Ross, J. J., Reid, J. B., et al. (2007). MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 19, 2225–2245. doi: 10.1105/tpc.106.048017
Du, H., Liu, H., and Xiong, L. (2013). Endogenous auxin and jasmonic acid levels are differentially modulated by abiotic stresses in rice. Front. Plant Sci. 4:397. doi: 10.3389/fpls.2013.00397
Du, L., Ali, G. S., Simons, K. A., Hou, J., Yang, T., Reddy, A. S., et al. (2009). Ca(2+)/calmodulin regulates salicylic-acid-mediated plant immunity. Nature 457, 1154–1158. doi: 10.1038/nature07612
Engelberth, J., Alborn, H. T., Schmelz, E. A., and Tumlinson, J. H. (2004). Airborne signals primeplants against insect herbivore attack. Proc. Natl. Acad. Sci. U.S.A. 101, 1781–1785. doi: 10.1073/pnas.0308037100
Eulgem, T., Rushton, P. J., Robatzek, S., and Somssich, I. E. (2000). The WRKY superfamily of plant transcription factors. Trends Plant Sci. 5, 199–206. doi: 10.1016/S1360-1385(00)01600-9
Eulgem, T., and Somssich, I. E. (2007). Networks of WRKY transcription factors in defense signaling. Curr. Opin. Plant Biol. 10, 366–371. doi: 10.1016/j.pbi.2007.04.020
Feng, H., Li, X., Duan, J., Li, H., and Liang, H. (2008). Chilling tolerance of wheat seedlings is related to an enhanced alternative respiratory pathway. Crop Sci. 48, 2381–2388. doi: 10.2135/cropsci2007.04.0232
Fernández-Calvo, P., Chini, A., Fernández-Barbero, G., Chico, J. M., Gimenez-Ibanez, S., and Geerinck, J. (2011). The Arabidopsis bHLH transcription factors MYC3 and MYC4 are targets of JAZ repressors and act additively with MYC2 in the activation of jasmonate responses. Plant Cell 23, 701–715. doi: 10.1105/tpc.110.080788
Feussner, I., and Wasternack, C. (2002). The lipoxygenase pathway. Annu. Rev. Plant Biol. 53, 275–297. doi: 10.1146/annurev.arplant.53.100301.135248
Feys, B. J. F., Benedetti, C. E., Penfold, C. N., and Turner, J. G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759.
Fonseca, S., Chico, J. M., and Solano, R. (2009a). The jasmonate pathway: the ligand, the receptor and the core signalling module. Curr. Opin. Plant Biol. 12, 539–547. doi: 10.1016/j.pbi.2009.07.013
Fonseca, S., Chini, A., Hamberg, M., Adie, B., Porzel, A., Kramell, R., et al. (2009b). (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5, 344–350. doi: 10.1038/nchembio.161
Fung, R. W. M., Wang, C. Y., Smith, D. L., Gross, K. C., and Tian, M. (2004). MeSA and MeJA increase steady-state transcript levels of alternative oxidase and resistance against chilling injury in sweet peppers (Capsicum annuum L.). Plant Sci. 166, 711–719. doi: 10.1016/j.plantsci.2003.11.009
Gilmour, S. J., Fowler, S. G., and Thomashow, M. F. (2004). Arabidopsis transcriptional activators CBF1, CBF2, and CBF3 have matching functional activities. Plant Mol. Biol. 54, 767–781. doi: 10.1023/B:PLAN.0000040902.06881.d4
Gilmour, S. J., Sebolt, A. M., Salazar, M. P., Everard, J. D., and Thomashow, M. F. (2000). Overexpression of the Arabidopsis CBF3 transcriptional activator mimics multiple biochemical changes associated with cold acclimation. Plant Physiol. 24, 1854–1865. doi: 10.1104/pp.124.4.1854
Göbel, C., and Feussner, I. (2009). Methods for the analysis of oxylipins in plants. Phytochemistry 70, 1485–1503. doi: 10.1016/j.phytochem.2009.07.040
González-Aguilar, G. A., Fortizm, J., Cruz, R., Baez, R., and Wang, C. Y. (2000). Methyl jasmonate reduces chilling injury and maintains postharvest quality of mango fruit. J. Agric. Food Chem. 48, 515–519. doi: 10.1021/jf9902806
Hemsley, P. A., Hurst, C. H., Kaliyadasa, E., Lamb, R., Knight, M. R., De Cothi, E. A., et al. (2014). The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate mediator and RNA polymerase II recruitment to CBF-responsivecold-regulated genes. Plant Cell 26, 465–484. doi: 10.1105/tpc.113.117796
Howe, G. A., and Jander, G. (2008). Plant immunity to insect herbivores. Annu. Rev. Plant Biol. 59, 41–66. doi: 10.1146/annurev.arplant.59.032607.092825
Hu, Y., Jiang, L., Wang, F., and Yu, D. (2013). Jasmonate regulates the inducer of CBF expression-C-repeat binding factor/DRE binding factor1 cascade and freezingtolerance in Arabidopsis. Plant Cell 25, 2907–2924. doi: 10.1105/tpc.113.112631
IPCC (2014). “Summary for policymakers,” in Climate Change 2014: Impacts, Adaptation, and Vulnerability. Part A: Global and Sectoral Aspects. Contribution of Working Group II to the Fifth Assessment Report of the Intergovernmental Panel on Climate Change, eds C. B. Field, V. R. Barros, D. J. Dokken, K. J. Mach, M. D. Mastrandrea, T. E. Bilir, M. Chatterjee, K. L. Ebi, Y. O. Estrada, R. C. Genova, B. Girma, E. S. Kissel, A. N. Levy, S. MacCracken, P. R. Mastrandrea, and L. L. White (Cambridge, UK; New York, NY: Cambridge University Press), 1–32.
Ito, Y., Saisho, D., Nakazono, M., Tsutsumi, N., and Hirai, A. (1997). Transcript levels of tandem-arranged alternative oxidase genes in rice are increased by low temperature. Gene 203, 121–129. doi: 10.1016/S0378-1119(97)00502-7
Kim, J. H., Lee, S. Y., Oh, S. Y., Han, S. I., Park, H. G., Yoo, M. A., et al. (2004). Methyl jasmonate induces apoptosis through induction of Bax/Bcl-XS and activation of caspase-3 via ROS production in A549 cells. Oncol. Rep. 12, 1233–1238. doi: 10.3892/or.12.6.1233
Kim, Y., Park, S., Gilmour, S. J., and Thomashow, M. F. (2013). Roles of CAMTA transcription factors and salicylic acid in configuring the low-temperature transcriptome and freezing tolerance of Arabidopsis. Plant J. 75, 364–376. doi: 10.1111/tpj.12205
Knight, H., Mugford, S. G., Ulker, B., Gao, D., Thorlby, G., and Knight, M. R. (2009). Identification of SFR6, a key component in cold acclimation acting post-translationally on CBFfunction. Plant J. 58, 97–108. doi: 10.1111/j.1365-313X.2008.03763.x
Knight, H., Veale, E. L., Warren, G. J., and Knight, M. R. (1999). The sfr6 mutation in Arabidopsis suppresses low-temperature induction of genes dependent on the CRT/DRE sequence motif. Plant Cell 11, 875–886.
Koch, T., Bandemer, K., and Boland, W. (1997). Biosynthesis of cis-jasmone: a pathway for the inactivation and the disposal of the plant stress hormone jasmonic acid to the gas phase? Helv. Chim. Acta 80, 838–850. doi: 10.1002/hlca.19970800318
Koda, Y. (1997). Possible involvement of jasmonates in various morphogenic events. Physiol. Plant. 100, 639–646. doi: 10.1111/j.1399-3054.1997.tb03070.x
Koda, Y., Takahashi, K., and Kikuta, I. (1992). Potato tuber inducing activities of salicylic acid and related compounds. J. Plant Growth Regul. 11, 215–219. doi: 10.1007/BF02115480
Kosová, K., Prášil, I. T., Vítámvás, P., Dobrev, P., Motyka, V., Floková, K., et al. (2012). Complex phytohormone responses during thecold acclimation of two wheat cultivars differing in cold tolerance, winter Samanta and spring Sandra. J. Plant Physiol. 169, 567–576. doi: 10.1016/j.jplph.2011.12.013
Lee, T. M., Lur, H. S., and Chu, C. (1997). Role of abscisic acid in chilling tolerance of rice (Oryza sativa L.) seedlings: II. Modulation of free polyamine levels. Plant Sci. 126, 1–10. doi: 10.1016/S0168-9452(97)00076-9
Li, S., Zhou, X., Chen, L., Huang, W., and Yu, D. (2010). Functional characterization of Arabidopsis thaliana WRKY39 in heat stress. Mol. Cells 29, 475–483. doi: 10.1007/s10059-010-0059-2
Lorenzo, O., Chico, J. M., Sánchez-Serrano, J. J., and Solano, R. (2004). JASMONATE-INSENSITIVE1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 16, 1938–1950. doi: 10.1105/tpc.022319
Macrae, R., and Long, J. A. (2011). “Interactions between TOPLESS and histone-modifying enzymes,” in 22nd International Conference on Arabidopsis Research. (Madison, WI The North American Arabidopsis Steering Committee).
Martínez-Téllez, M. A., and Lafuente, M. T. (1993). Chilling-induced changes in phenylalanine ammonia-lyase, peroxidase, and polyphenol oxidase activities in citrus flavedo tissue. Acta Hortic. 343, 257–263. doi: 10.17660/ActaHortic.1993.343.58
Matthes, M. C., Bruce, T. J., Ton, J., Verrier, P. J., Pickett, J. A., and Napier, J. A. (2010). The transcriptome of cis-jasmone-induced resistance in Arabidopsis thaliana and its role in indirect defence. Planta 232, 1163–1180. doi: 10.1007/s00425-010-1244-4
Michelet, J. F., Olive, C., Rieux, E., Fagot, D., Simonetti, L., Galey, J. B., et al. (2012). The anti-ageing potential of a new jasmonic acid derivative (LR2412): in vitro evaluation using reconstructed epidermis Episkin™. Exp. Dermatol. 21, 398–400. doi: 10.1111/j.1600-0625.2012.01480.x
Miersch, O., Neumerkel, J., Dippe, M., Stenzel, I., and Wasternack, C. (2008). Hydroxylated jasmonates are commonly occurring metabolites of jasmonic acid and contribute to a partial switch-off in jasmonate signaling. New Phytol. 177, 114–127. doi: 10.1111/j.1469-8137.2007.02252.x
Miura, K., Lee, J., Miura, T., and Hasegawa, P. M. (2010). SIZ1 controls cell growth and plant development in Arabidopsis through salicylic acid. Plant Cell Physiol. 51, 103–113. doi: 10.1093/pcp/pcp171
Miura, K., and Ohta, M. (2010). SIZ1, a small ubiquitin-related modifier ligase, controls cold signaling through regulation of salicylic acid accumulation. J. Plant Physiol. 167, 555–560. doi: 10.1016/j.jplph.2009.11.003
Mosblech, A., Thurow, C., Gatz, C., Feussner, I., and Heilmann, I. (2011). Jasmonic acid perception by COI1 involves inositol polyphosphates in Arabidopsis thaliana. Plant J. 65, 949–957. doi: 10.1111/j.1365-313X.2011.04480.x
Murray, J. A., Jones, A., Godinm, C., and Traas, J. (2012). Systems analysis of shoot apical meristem growth and development: integrating hormonal and mechanical signaling. Plant Cell 24, 3907–3919. doi: 10.1105/tpc.112.102194
Mutlu, S., Karadaǧoǧlu, Ö., Atici, Ö., and Nalbantoǧlu, B. (2013). Protective role of salicylic acid applied before cold stress on antioxidative system and protein patterns in barley apoplast. Biol. Plant. 57, 507–513. doi: 10.1007/s10535-013-0322-4
Nishii, A., Takemura, M., Fujita, H., Shikata, M., Yokota, A., and Kohchi, T. (2000). Characterization of a novel gene encoding a putative single zinc-finger protein, ZIM, expressed during the reproductive phase in Arabidopsis thaliana. Biosci. Biotechnol. Biochem. 64, 1402–1409. doi: 10.1271/bbb.64.1402
Parthier, B. (1991). Jasmonates, new regulators of plant growth and development: many facts and few hypotheses on their actions. Botanica. Acta 104, 446–454. doi: 10.1111/j.1438-8677.1991.tb00257.x
Pauwels, L., Barbero, G. F., Geerinck, J., Tilleman, S., Grunewald, W., Pérez, A. C., et al. (2010). NINJA connects the co-repressor TOPLESS to jasmonate signalling. Nature 464, 788–791. doi: 10.1038/nature08854
Pauwels, L., and Goossens, A. (2011). The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23, 3089–3100. doi: 10.1105/tpc.111.089300
Pauwels, L., Morreel, K., De Witte, E., Lammertyn, F., Van Montagu, M., Boerjan, W., et al. (2008). Mapping methyl jasmonate-mediated transcriptional reprogramming of metabolism and cell cycle progression in cultured Arabidopsis cells. Proc. Natl. Acad. Sci. U.S.A. 105, 1380–1385. doi: 10.1073/pnas.0711203105
Peng, H. H., Shan, W., Kuang, J. F., Lu, W. J., and Chen, J. Y. (2013). Molecular characterization ofcold-responsive basic helix-loop-helix transcription factors MabHLHs that interact with MaICE1 in banana fruit. Planta 238, 937–953. doi: 10.1007/s00425-013-1944-7
Petricka, J. J., Winter, C. M., and Benfey, P. N. (2012). Control of Arabidopsis root development. Annu. Rev. Plant Biol. 63, 563–590. doi: 10.1146/annurev-arplant-042811-105501
Reinbothe, C., Springer, A., Samol, I., and Reinbothe, S. (2009). Plant oxylipins: role of jasmonic acid during programmed cell death, defence and leaf senescence. FEBS J. 276, 4666–4681. doi: 10.1111/j.1742-4658.2009.07193.x
Rotem, R., Heyfets, A., Fingrut, O., Blickstein, D., Shaklai, M., and Flescher, E. (2005). Jasmonates: novel anticancer agents acting directly and selectively on human cancer cell Mitochondria 65, 1984–1993. doi: 10.1158/0008-5472.can-04-3091
Ruelland, E., and Zachowski, A. (2010). How plants sense temperature. Environ. Exp. Bot. 69, 225–232. doi: 10.1016/j.envexpbot.2010.05.011
Sanchez-Ballesta, M. T., Lafuentea, M. T., Zacarias, L., and Granell, A. (2000). Involvement of phenylalanine ammonia-lyase in the response of Fortune mandarin fruits to cold temperature. Physiol. Plant. 108, 382–389. doi: 10.1034/j.1399-3054.2000.108004382.x
Schneider, K., Kienow, L., Schmelzer, E., Colby, T., Bartsch, M., Miersch, O., et al. (2005). A new type of peroxisomal acyl-coenzyme Asynthetasefrom Arabidopsis thaliana has the catalytic capacity to activate biosyntheticprecursors of jasmonic acid. J. Biol. Chem. 80, 13962–13972. doi: 10.1074/jbc.M413578200
Scott, I. M., Clarke, S. M., Wood, J. E., and Mur, L. A. (2004). Salicylate accumulation inhibits growth at chilling temperature in Arabidopsis. Plant Physiol. 135, 1040–1049. doi: 10.1104/pp.104.041293
Sembdner, G., and Parthier, B. (1993). The biochemistry and the physiological and molecular actions of jasmonates. Ann. Rev. Plant Physiol. Plant Mol. Biol. 44, 569–589. doi: 10.1146/annurev.pp.44.060193.003033
Seo, H. S., Song, J. T., Cheong, J. J., Lee, Y. H., Lee, Y. W., Hwang, I., et al. (2001). Jasmonic acid carboxyl methyltransferase: a key enzyme for jasmonate-regulated plant responses. Proc. Natl. Acad. Sci. U.S.A. 98, 4788–4793. doi: 10.1073/pnas.081557298
Sheard, L. B., Tan, X., Mao, H., Withers, J., Ben-Nissan, G., Hinds, T. R., et al. (2010). Jasmonate perception by inositol-phosphate-potentiated COI1-JAZ co-receptor. Nature 468, 400–405. doi: 10.1038/nature09430
Shi, Y., Tian, S., Hou, L., Huang, X., Zhang, X., Guo, H., et al. (2012). Ethylene signaling negatively regulates freezing tolerance by repressing expression of CBF and type-A ARR genes in Arabidopsis. Plant Cell 24, 2578–2595. doi: 10.1105/tpc.112.098640
Siboza, X. I., Bertling, I., and Odindo, A. O. (2014). Salicylic acid and methyl jasmonate improve chilling tolerance in cold-stored lemon fruit (Citrus limon). J. Plant Physiol. 171, 1722–1731. doi: 10.1016/j.jplph.2014.05.012
Staswick, P. E., and Tiryaki, I. (2004). The oxylipin signal jasmonic acid is activated by an enzyme that conjugates it to isoleucine in Arabidopsis. Plant Cell 16, 2117–2127. doi: 10.1105/tpc.104.023549
Swiatek, A., Van Dongen, W., Esmans, E. L., and Van-Onckelen, H. (2004). Metabolic fate of jasmonates in tobacco bright yellow-2 cells. Plant Physiol. 135, 161–172. doi: 10.1104/pp.104.040501
Tani, T., Sobajima, H., Okada, K., Chujo, T., Arimura, S., Tsutsumi, N., et al. (2008). Identification of the OsOPR7 gene encoding12-oxophytodienoate reductase involved in the biosynthesis of jasmonic acid inrice. Planta 227, 517–526. doi: 10.1007/s00425-007-0635-7
Theodoulou, F. L., Job, K., Slocombe, S. P., Footitt, S., Holdsworth, M., Baker, A., et al. (2005). Jasmonic acid levels are reduced in COMATOSE ATP-binding cassette transporter mutanC. Implications for transport of jasmonate precursors intoperoxisomes. Plant Physiol. 137, 835–840. doi: 10.1104/pp.105.059352
Thines, B., Katsir, L., Melotto, M., Niu, Y., Mandaokar, A., Liu, G., et al. (2007). JAZ repressor proteins are targets of the SCF(COI1) complex during jasmonate signalling. Nature 448, 661–665. doi: 10.1038/nature05960
Thomashow, M. F. (1999). PLANT COLD ACCLIMATION: freezing tolerance genes and regulatory mechanisms. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50, 571–599. doi: 10.1146/annurev.arplant.50.1.571
Thomashow, M. F. (2010). Molecular basis of plant cold acclimation: insights gained from studying the CBF cold response pathway. Plant Physiol. 154, 571–577. doi: 10.1104/pp.110.161794
Turner, J. G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14(Suppl.), S153–S164. doi: 10.1105/tpc.000679
Vanholme, B., Grunewald, W., Bateman, A., Kohchi, T., and Gheysen, G. (2007). The tify family previously known as ZIM. Trends Plant Sci. 12, 239–244. doi: 10.1016/j.tplants.2007.04.004
Vick, B. A., and Zimmerman, D. C. (1983). The biosynthesis of jasmonic acid: a physiological role for plant lipoxygenase. Biochem. Biophys. Res. Commun. 111, 470–477. doi: 10.1016/0006-291X(83)90330-3
Wan, S. B., Tian, L., Tian, R. R., Pan, Q. H., Zhan, J. C., Wen, P. F., et al. (2009). Involvement of phospholipase D in the low temperature acclimation induced thermotolerance in grape berry. Plant Physiol. Biochem. 47, 504–510. doi: 10.1016/j.plaphy.2008.12.010
Wang, C. Y., and Buta, G. (1994). Methyl jasmonate reduces chilling injury in Curcubita pepo through its regulation of abscisic and polyamine levels. Environ. Exp. Bot. 43, 427–432. doi: 10.1016/0098-8472(94)90025-6
Wasternack, C. (2007). Jasmonates: an update on biosynthesis, signal transduction and action in plant stress response, growth and development. Ann. Bot. 100, 681–697. doi: 10.1093/aob/mcm079
Wasternack, C., and Hause, B. (2002). Jasmonates and octadecanoids: signals in plant stress responses and development. Prog. Nucleic Acid Res. Mol. Biol. 72, 165–221. doi: 10.1016/S0079-6603(02)72070-9
Wasternack, C., and Hause, B. (2013). Jasmonates: biosynthesis, perception, signaltransduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Wathugala, D. L., Hemsley, P. A., Moffat, C. S., Cremelie, P., Knight, M. R., and Knight, H. (2012). The Mediator subunit SFR6/MED16 controls defence gene expression mediated by salicylic acid and jasmonate responsive pathways. New Phytol. 195, 217–230. doi: 10.1111/j.1469-8137.2012.04138.x
Wilen, W. R., Ewan, E. B., and Gusta, L. V. (1993). Interaction of abscisic acid and jasmonic acid on the inhibition of seed germination and the induction of freezing tolerance. Can. J. Bot. 72, 1009–1017. doi: 10.1139/b94-127
Wu, X., Shiroto, Y., Kishitani, S., Ito, Y., and Toriyama, K. (2009). Enhanced heat and drought tolerance in transgenic rice seedlings overexpressing OsWRKY11 under the control of HSP101 promoter. Plant Cell Rep. 28, 21–30. doi: 10.1007/s00299-008-0614-x
Xie, D. X., Feys, B. F., James, S., Nieto-Rostro, M., and Turner, J. G. (1998). COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. doi: 10.1126/science.280.5366.1091
Yadav, V., Mallappa, C., Gangappa, S. N., Bhatia, S., and Chattopadhyay, S. (2005). A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light-mediated photomorphogenic growth. Plant Cell 17, 1953–1966. doi: 10.1105/tpc.105.032060
Yan, J., Zhang, C., Gu, M., Bai, Z., Zhang, W., Qi, T., et al. (2009). The Arabidopsis CORONATINE INSENSITIVE1 protein is a jasmonate receptor. Plant Cell 21, 2220–2236. doi: 10.1105/tpc.109.065730
Yan, Y., Christensen, S., Isakeit, T., Engelberth, J., Meeley, R., Hayward, A., et al. (2012). Disruption of OPR7 and OPR8 reveals the versatile functions ofjasmonic acid in maize development and defense. Plant Cell 24, 1420–1436. doi: 10.1105/tpc.111.094151
Yan, Y., Stolz, S., Chételat, A., Reymond, P., Pagni, M., Dubugnon, L., et al. (2007). A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 19, 2470–2483. doi: 10.1105/tpc.107.050708
Yoshida, Y., Sano, R., Wada, T., Takabayashi, J., and Okada, K. (2009). Jasmonic acid control of GLABRA3 links inducible defense and trichome patterning in Arabidopsis. Development 136, 1039–1048. doi: 10.1242/dev.030585
Zhang, X. C., Millet, Y. A., Cheng, Z., Bush, J., and Ausubel, F. M. (2015). Jasmonate signalling in Arabidopsis involves SGT1b–HSP70–HSP90 chaperone complexes. Nat. Plants. 1:15049. doi: 10.1038/nplants.2015.49
Zhang, X., Wang, C., Zhang, Y., Sun, Y., and Mou, Z. (2012). The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24, 4294–4309. doi: 10.1105/tpc.112.103317
Zhang, Y., and Turner, J. G. (2008). Wound-induced endogenous jasmonates stunt plant growth by inhibiting mitosis. PLoS ONE 3:e3699. doi: 10.1371/journal.pone.0003699
Zhao, M. L., Wang, J. N., Shan, W., Fan, J. G., Kuang, J. F., Wu, K. Q., et al. (2013). Induction of jasmonate signalling regulators MaMYC2s and theirphysical interactions with MaICE1 in methyl jasmonate-induced chilling tolerance in banana fruit. Plant Cell Environ. 36, 30–51. doi: 10.1111/j.1365-3040.2012.02551.x
Zhu, Y., Wang, Z., Jing, Y., Wang, L., Liu, X., Liu, Y., et al. (2009). Ectopic over-expression of BhHsf1, a heat shock factor from the resurrection plant Boea hygrometrica, leads to increased thermotolerance and retarded growth in transgenic Arabidopsis and tobacco. Plant Mol. Biol. 71, 451–467. doi: 10.1007/s11103-009-9538-2
Keywords: jasmonates, abiotic stresses, heat, cold, signaling
Citation: Sharma M and Laxmi A (2016) Jasmonates: Emerging Players in Controlling Temperature Stress Tolerance. Front. Plant Sci. 6:1129. doi: 10.3389/fpls.2015.01129
Received: 15 August 2015; Accepted: 29 November 2015;
Published: 06 January 2016.
Edited by:
Maik Boehmer, University of Münster, GermanyReviewed by:
Oksoo Han, Chonnam National University, South KoreaJianye Chen, South China Agricultural University, China
Copyright © 2016 Sharma and Laxmi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ashverya Laxmi, ashverya_laxmi@nipgr.ac.in; laxmiashverya@rediffmail.com
 Manvi Sharma
Manvi Sharma