- 1College of Life Science, Northeast Agricultural University, Harbin, China
- 2Department of Biology, University of North Carolina, Chapel Hill, NC, USA
- 3Department of Pharmacology, University of North Carolina, Chapel Hill, NC, USA
HEXOKINASE 1 (AtHXK1) and Regulator of G-protein Signaling 1 (AtRGS1) pathways, mediate D-glucose signaling in Arabidopsis. However, it is not known the degree, if any, that these pathways overlap and how. We show modest signaling crosstalk between these pathways, albeit complex with both epistatic interactions and additive effects that may be indirect. The action of HXK1 on AtRGS1 signaling lies downstream of the primary step in G protein-mediated sugar signaling in which the WD-repeat protein, AGB1, is the propelling signaling element. RHIP1, a previously unknown protein predicted here to have a 3-stranded helical structure, interacts with both AtRGS1 and AtHXK1 in planta and is required for some glucose-regulated gene expression, providing a physical connection between these two proteins in sugar signaling. The rhip1 null mutant displays similar seedling growth phenotypes as rgs1-2 in response to glucose, further suggesting a role for RHIP1 in glucose signaling. In conclusion, glucose signaling is a complex hierarchical relationship which is specific to the target gene and sugar phenotype and suggests that there are two glycolysis-independent glucose signaling sensors: AtRGS1 and AtHXK1 that weakly communicate with each other via feed-back and feed-forward loops to fine tune the response to glucose.
Introduction
Glucose affects plant growth and development by acting as both a metabolite and a signaling molecule (Sheen, 2014). In Arabidopsis thaliana, glucose is sensed by (1) a plasma membrane G-protein-coupled pathway involving REGULATOR OF G-PROTEIN SIGNALING 1 (AtRGS1), (2) a HEXOKINASE 1 (AtHXK1) pathway, and (3) a glycolysis-dependent SNF1-RELATED KINASE 1/TARGET OF RAPAMYCIN (SnRK1/TOR) pathway. On the Arabidopsis plasma membrane, the G protein complex contains the core G-protein subunits Gα (AtGPA1), Gβ (AGB1) and Gγ (AGG1), three WNK kinases (AtWNK1, AtWNK8, and AtWNK10) and the 7-transmembrane AtRGS1 protein. AtRGS1 keeps the core G protein complex in its inactive (GDP-bound) state and physical decoupling of AtRGS1 from AtGPA1 allows this G protein to self-activate by spontaneously binding GTP. Sustained activation of glucose signaling depends, in part, on removal of the inhibiting AtRGS1 protein by endocytosis of this plasma membrane-anchored glucose sensor (Urano et al., 2012; Fu et al., 2014). The WD40 repeat subunit of the G protein AGB1 recruits the WNK kinases which phosphorylate AtRGS1 consequently triggering the endocytosis event.
AtRGS1-dependent G-protein-coupled signaling regulates many cellular biological processes, such as cell growth and proliferation (Chen et al., 2006a; Booker et al., 2010; Urano et al., 2012), abiotic stress tolerance (Chen et al., 2006b; Colaneri et al., 2014), stomatal density (Zhang et al., 2008), chloroplast development (Zhang et al., 2009), seed germination (Chen et al., 2006c), morphogenesis and development (Chen and Jones, 2004; Chen et al., 2006b). Specifically, loss of AtRGS1 confers increased growth and high glucose tolerance in Arabidopsis. Loss-of-function mutations in the WD40-repeat subunit AGB1 have the most severe phenotypes.
AtHXK1 regulates glucose-responsive gene transcription directly by forming a nuclear co-repressor complex with the vacuolar H+-ATPase B1 (VHA-B1) and the 19S regulatory particle of the proteasome subunit (RPT5B) (Cho et al., 2006). The hxk1 (aka gin2-1), vha-b1 and rpt5b null mutants display similar seedling and adult plant phenotypes (e.g., reduced growth in roots, leaves and inflorescences), and glucose response defects (namely, insensitivity to high glucose-mediated developmental arrest) (Moore et al., 2003; Cho et al., 2006).
The glycolysis-dependent SnRK1/TOR pathway contains two protein kinases SnRK1 and TOR that may function as cellular energy sensors instead of sugar sensors (Lastdrager et al., 2014; Tome et al., 2014). TOR is activated and promotes growth in response to favorable nutritional and energy conditions, while SnRK1 is stimulated upon nutrient and energy starvation conditions (Lastdrager et al., 2014; Tome et al., 2014).
Until now, the mechanisms within each of these glucose signal transduction pathways were studied independently of each other, yet, analogous to the integrative HXK/G protein mechanism for glucose sensing in yeast (Rolland et al., 2000; Kim et al., 2013), it is likely that an integration of these pathways is essential for plants to make a fast and coordinated response to extracellular and intracellular glucose conditions. In this study, we focused on functional crosstalk between AtRGS1- and AtHXK1-dependent sugar signaling (Moore et al., 2003; Chen and Jones, 2004; Urano et al., 2012). By comparing the expression of a selected set of glucose-regulated genes as well as a series of physiological sugar-related phenotypes, we determined that the two glucose sensors AtRGS1 and AtHXK1 communicate via cooperative loops in determining the final response to glucose. This occurs downstream of AtRGS1 endocytosis. RGS1-HXK1 INTERACTING PROTEIN 1 (RHIP1) serves as the physical scaffold for these two sensors.
Materials and Methods
Plant Materials
The T-DNA insertion mutant rgs1-2 was described by Chen et al. (2003), and the ethylmethane sulfonate (EMS)-mutagenized mutant gin2-1 was described by Moore et al. (2003). The 35S::RGS1-YFP over-expression line was previously described by Urano et al. (2012). hxk1-1 (SALK_034233), hxk1-2 (CS864200), hxk1-3 (CS861759), rhip1-1 (SALK_091518), and rhip1-2 (SALK_061002) mutants were obtained from the Arabidopsis Biological Resource Center (ABRC) stock. Plants homozygous for hxk1-1, hxk1-2, hxk1-3, rhip1-1, and rhip1-2 were isolated by PCR using genomic DNA with T-DNA insertion verification primers and the insertion was confirmed by sequencing. Reverse transcriptase polymerase chain reaction (PCR) was used to analyze the AtHXK1 or RHIP1 transcripts in these five mutants. The primers used are shown in Supplementary Table S1. Our analyses disproved an earlier claim by Reda (2013) that hxk1-1 (SALK_034233C) lacked expression of HXK1 (Supplementary Figure S1); hxk1-1 is not a null mutant as previously reported and the reported phenotypes of the hxk1-1 mutant should be re-interpreted. The open reading frame of AtRGS1 in pEarleyGate101 vector (C-terminal YFP-HA) was previously described by Urano et al. (2012). The 35S::AtRGS1-YFP over-expression lines in the hxk1-3, rhip1-1 or rhip1-2 mutant background were generated by Agrobacterium-mediated transformation (Bechtold et al., 1993), and T2 generation seed was used. The pCDKA;1::GUS reporter was crossed to rgs1-2 and hxk1-3 single mutant plants, and F3 and F4 generation plants were used for phenotype analysis. All mutants used are in the ecotype Columbia (Col-0) except gin2-1 which is in the Landsberg (Ler) background.
Quantitative Real-time PCR (qRT-PCR)
Quantitative real-time PCR was performed exactly as described by Grigston et al. (2008) and Urano et al. (2012). Briefly, ∼150 seeds were cultured in 75 ml 1/2 × MS liquid media with 1% sucrose at 23°C shaking (∼140 rpm) under constant low light (70 μmol s-1m-2) conditions. After 7 days, the seedlings were washed in sterilized water three times, and then starved in 1/2 × MS liquid media lacking sugar for 2 days in the dark. The seedlings were treated with fresh 1/2 × MS liquid media containing 0% (control), or 3% (w/v) D-glucose for 3 h in dark conditions, then harvested by flash freezing in liquid N2. The mRNA and cDNA were prepared with RNAeasyTM (Qiagen) and Superscript III (Invitrogen), respectively, according to the manufacturer’s instructions. Expression of TBL26, HXK1, RGS1, CA2, CAB2, DIN1, and TUB4 was analyzed by qRT-PCR with SYBRgreen (Invitrogen). The comparative CT (Threshold Cycle defined as the cycle number at which the fluorescence generated within a reaction crosses the threshold line) method described in detail in Livak and Schmittgen (2001), was performed for qRT-PCR. First, validation experiments demonstrated that the targets (TBL26, HXK1, RGS1, CA2, CAB2, DIN1) and endogenous control (TUB4) have single amplification products determined by gel electrophoresis and relatively equivalent PCR efficiencies (relative standard curve). Second, samples from each biological repeat were run in triplicate on a single plate. The CT mean and standard deviation values of the three replicate sample results were calculated. The ΔCT value was calculated by:
The standard deviation of the ΔCT was calculated by:
The ΔΔCT was calculated by:
The standard deviation of the ΔΔCT value is the same as the standard deviation of the ΔCT value. Fold-differences between test sample and control sample was calculated by 2-ΔΔCt with ΔΔCT + S and ΔΔCT - S, where S is the standard deviation of the ΔΔCT value. Fold-differences calculated using ΔΔCT method are shown as a range from 2-ΔΔCt+s to 2-ΔΔCt-s.
Plant Growth Assays
Seeds were sown onto soil and stratified at 4°C for 3 days, and then transferred to a 23°C growth chamber in 16/8 h (L/D). Two or 3-week-old seedlings were photographed. Alternatively, seeds were surface-sterilized with 70% ethanol and 95% ethanol for 10 min each, then sown onto 1/4 × MS with 0.5% phytoagar (pH adjusted to 5.75 with 5 N KOH). Seeds were stratified at 4°C in the dark for 3 days, germinated and grown horizontally at 23°C with a 16/8 h (L/D) cycle (70 μmol s-1m-2). Three days later, the seedlings were transferred to 1/4 × MS with 0.5% phytoagar (pH adjusted to 5.75 with 5 N KOH) containing 0, 0.25, 0.5, 0.75, 1, 2, or 3% (w/v) D-glucose, and grown vertically for root growth analysis. Every 24 h, the root tips were marked. The root lengths were calculated using ImageJ software. The “green seedling” assay was performed as described by Moore and coworkers (Moore et al., 2003). Briefly, surface-sterilized seeds of Col-0, rgs1-2, hxk1-3, hxk1-3/rgs1-2, and rhip1-2 mutant were sown onto 1/4 × MS with 0.5% phytoagar containing 6% (w/v) D-glucose or D-mannitol, stratified at 4°C in the dark for 3 days, germinated and grown horizontally at 23°C with continuous light (70 μmol s-1m-2) for 10 days. The average percentage of seedlings showing green cotyledons was determined. The “green seedling” assay in response to saline stress was performed as described by Colaneri et al. (2014). Briefly, surface-sterilized seeds were germinated and seedlings grown on 1/4 MS agar media supplemented with 125 mM NaCl and 0.5% D-glucose (w/v). Green seedlings per plate (36 seeds per plate) were counted 10 days after germination.
β-Glucuronidase (GUS) Staining Assay
Arabidopsis seeds were surface-sterilized and stratified as described above. Seeds germinated in 24-well plates containing 1 ml 1/2 MS liquid medium (pH adjusted to 5.75 with 5 N KOH) on a shaker (∼30 rpm) at 23°C under continuous low light (70 μmol s-1m-2). 3-day-old seedlings were then transferred to 1 ml fresh 1/2 MS liquid medium supplemented with 3% (w/v) D-glucose or D-mannitol (pH adjusted to 5.75 with 5 N KOH) for 3 more days in culture.
GUS staining was performed following the method described by Malamy and Benfey (1997). Seedlings were infiltrated in 100 mM Tris-HCl (pH 7.5) buffer containing 2.9 mg/ml NaCl, 20% (v/v) methanol, 0.001% (v/v) Triton X-100 and 0.5 mg/ml X-gluc (5-Bromo-4-chloro-3-indoxyl-beta-D-glucuronide cyclohexyl ammonium salt; Gold Biotechnology, Inc.). After incubation for 12–18 h at 37°C in dark, seedlings were cleared in 70% ethanol and photographed on a Nikon inverted microscope DIAPHOT-TMD. ImageJ software was used for analysis.
Informatic Methods
The protein sequence of RHIP1 (At4g26410) was obtained by searching against the Arabidopsis thaliana protein database supported by The Arabidopsis Information Resource (TAIR). The RHIP1 protein sequence was submitted to I-TASSER (Iterative Threading ASSEmbly Refinement) online1, to generate the predicted RHIP1 protein structure and the GO biological process of RHIP1 protein based on global and local protein similarity. PyMOL(TM) Molecular Graphics System (Version 1.7.0.0.) was used to generate the final high-quality protein structure model image based on the data of model 1 from I-TASSER. GO annotations of proteins directly interacting with RHIP1 were retrieved from TAIR GO Annotation Search2, and functional categorization by their TAIR loci for GO molecular function or GO biological process were expressed as percent of total.
AtRGS1-YFP Internalization Analysis
Fluorescence quantification for AtRGS1-YFP internalization was performed as described by Urano et al. (2012) and Fu et al. (2014). Sterilized, stratified seeds were germinated in 6-well plates containing 2 ml 1/2 × MS liquid medium (pH adjusted to 5.75 with 5 N KOH) at 23°C under darkness. Seedlings (7-day-old) were treated with 0 or 3% D-glucose (w/v) for 30 min. Hypocotyl epidermal cells located 2–4 mm below the cotyledon were imaged (Z stacks obtained) using a Zeiss LSM710 confocal laser scanning microscope equipped with a 20× Plan-NeoFluor (N.A. = 0.5) objective and a 40× C-Apochromat (N.A. = 1.20) water immersion objective. YFP fluorescence was excited by a 514 nm argon laser and detected at 526–569 nm by a photomultiplier detector. At least 10 sets of images from 5 seedlings were obtained for internalization quantification analysis by ImageJ software.
Bimolecular Fluorescence Complementation (BiFC)
Bimolecular fluorescence complementation was performed as described in Klopffleisch et al. (2011). The entire open reading frames of RHIP1 or AtHXK1 were amplified by PCR from a cDNA library made from seedlings in qRT-PCR analysis. Open reading frames were subcloned into the p-ENTR/D-TOPO vector (Invitrogen, Carlsbad, CA, USA) and recombined into the BiFC vectors pBatTL-sYFP-N and pBatTL-sYFP-C (C-terminal split-nYFP and cYFP tag, respectively), and pCL112 and pCL113 (N-terminal split-nYFP and cYFP tag, respectively) by LR recombination reaction. The open reading frame of AtRGS1 in BiFC vectors was previously described by Grigston et al. (2008). Split nYFP- and cYFP-tagged protein pairs, p19 (gene silencing suppressor), and Mt-rk (mitochondrial RFP marker, an internal transformation control) were co-expressed in 4- to 5-week-old Nicotiana benthamiana leaves by Agrobacterium tumifaciens-mediated infection. Tobacco leaf epidermal cells were imaged using a Zeiss LSM710 confocal laser scanning microscope equipped with an Apochromat 40 × water-emersion objective (N.A. = 1.2). YFP fluorescence was excited by a 514 nm argon laser and detected at 526–569 nm by a photomultiplier detector, and RFP fluorescence was excited by a diode laser and detected at 565–621 nm.
Results and Discussion
AtRGS1 and AtHXK1 Co-regulate the Expression of a Group of Glucose-regulated Genes
Glucose induces in an AtRGS1-dependent manner the expression of TRICHOMELESS 26 (TBL26, At4g01080), JACALIN-RELATED LECTIN 7 (JAL7, At1g52040), JACALIN-RELATED LECTIN23 (JAL23, At2g39330), At1g54020 (JAL gene) among ∼30 genes (Grigston et al., 2008; Urano et al., 2012). The time course and dose responsiveness for regulation of TBL26 is the best documented among the small set of genes regulated by AtRGS1. Even fewer genes to date are documented to be regulated by HXK1 (Sheen et al., 1999). The best characterized is the involvement of AtHXK1 in glucose repression of the CARBONIC ANHYDRASE 2 (CA2, At5g14740) and the photosynthetic gene CHLOROPHYL A/B-BINDING 2 (CAB2, At1g29920) (Moore et al., 2003; Cho et al., 2006). As shown in Figure 1A inset, the D-glucose induced expression of TBL26 was severely depressed in the hxk1 mutant (gin2-1 in the Ler background) with ∼80% difference between TBL26 expression in Ler and in gin2-1, suggesting an active role of AtHXK1 in glucose-induced TBL26 expression. All other studies on TBL26 were conducted using the Col ecotype. In order to exclude the influence of different ecotypes, it was necessary to generate a transcript-null allele (hxk1-3) in the Columbia ecotype (Supplementary Figure S1). Glucose-induced expression of TBL26 was impaired in seedlings lacking either AtRGS1 or AtHXK1 (Figure 1A), and this impairment was not the result of lowered expression of AtRGS1 in the hxk1-3 mutant, or of AtHXK1 in the rgs1-2 mutant (Supplementary Figure S2), suggesting an overlapping or additive function between AtRGS1 and AtHXK1 in glucose-regulated gene expression. The level of TBL26 expression in the hxk1-3 mutant was lower than in the rgs1-2 mutant (P < 0.01) but combining these alleles conferred statistically similar level as the rgs1-2 mutant indicating that the rgs1-2 allele is epistatic to the hxk1-3 allele. Note, however, that loss of both AtRGS1 and AtHXK1 did not completely eliminate glucose induction of TBL26 expression indicating that a third (glucose) input must exist. AtRGS1 slightly inhibits TBL26 expression but also positively reinforced AtHXK1 although with a different weight than inhibition.
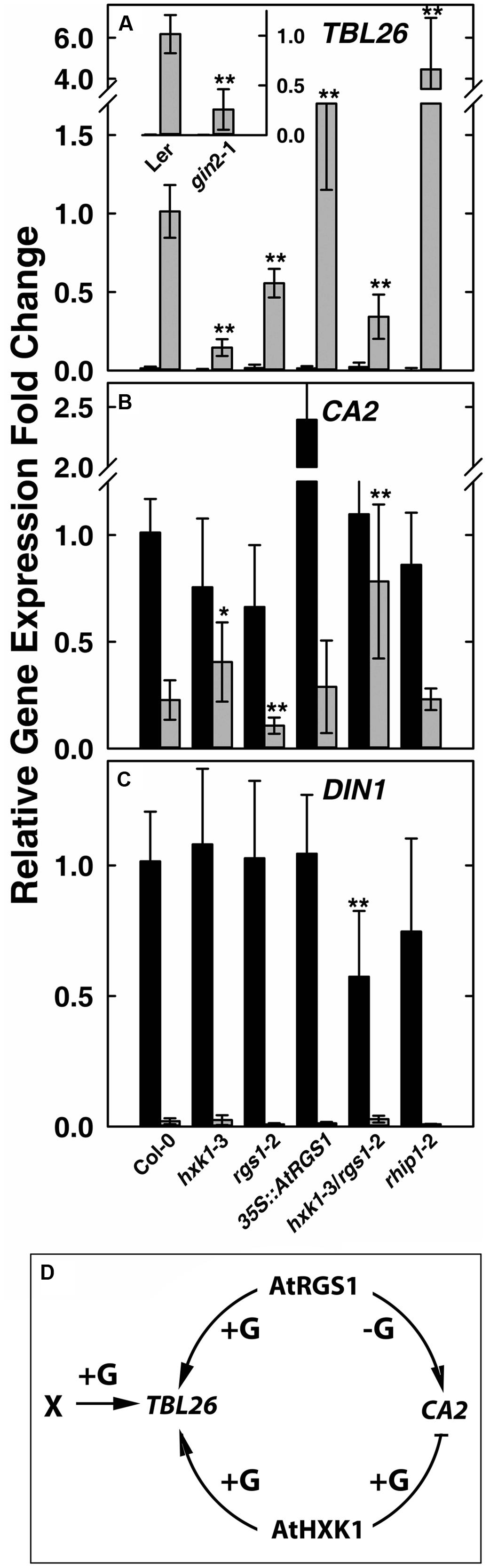
FIGURE 1. Quantitative Real-time PCR (qPCR) analysis of glucose-reporter genes TBL26, CA2 and DIN1 in Arabidopsis seedlings and the corresponding mechanism models. Seven-day-old seedlings with the indicated genotypes indicated at the bottom of panels (A–C) were starved for 2 days then treated with 1/2 × MS medium (0% D-glucose, shown by black bars) or 3%D-glucose (gray bars) for 3 h as described in Section “Materials and Methods.” Transcripts of TBL26 (A), CA2 (B), and DIN1 (C) were quantitated using qRT-PCR. Values are the means of the fold changes ± SD of 3–5 independent biological replicates. Each biological replication had at least three technical replications. ANOVA single factor analysis (α = 0.05) was conducted to compare the relative fold-change in gene expression of different plant lines with wild type control plants. ∗P < 0.05; ∗∗P < 0.01. (D) Simplest model accounting for the gene expression data based on the mutant behaviors. +G, addition of D-glucose; -G, no glucose.
Glucose represses some gene expression in an AtHXK1-dependent manner (Sheen, 1990; Koch, 1996). We next examined whether AtRGS1 influences AtHXK1-mediated gene repression of CA2 gene expression. Glucose-repression of CA2 was slightly attenuated in the hxk1 mutants (both Col and Ler alleles), consistent with previous studies (Moore et al., 2003; Cho et al., 2006). The rgs1-2 mutant displayed slightly lower CA2 gene expression with (P = 0.003236) or without (P = 0.00223) glucose treatment (Figure 1B), suggesting that AtRGS1 promotes CA2 basal gene expression. To test this hypothesis, we examined the effect of over-expression of AtRGS1 on the basal expression of CA2. The basal CA2 expression without glucose treatment was strongly increased to nearly 2.5-fold in the 35S::AtRGS1 plants (P = 1.25E-05), while the glucose-repressed level of expression was similar to wild type. The hxk1-3/rgs1-2 double mutant had statistically the same basal level of CA2 expression (P > 0.05) but the glucose repression was attenuated although statistically to no more or less degree than the in the hxk1-3 mutant (P < 0.025) (Figure 1B). The epistasis analysis did not provide strong evidence for a heirarchial role of AtRGS1 in AtHXK1 regulation of CA2 expression, however, the large effect of over-expression of AtRGS1 is suggestive of cooperation between these two pathways although with the accompanying caveats associated with interpreting gain-of-function alleles. We also examined another AtHXK1-mediated gene repression, namely CAB2, and obtained a similar result as for CA2 (Supplementary Figure S3). AtHXK1 mediates the main pathway toward CA2 repression. AtRGS1, based on the gain-of-function data, induces CA2 gene expression in the absence of glucose. An inhibitory genetic interaction by AtHXK1 on AtRGS1 fits the observed result. There is no need to implicate an independent glucose pathway since loss of both AtRGS1 and AtHXK1 statistically eliminated glucose repression of CA2 expression.
As discussed above, there are at least three sugar-sensing pathways in Arabidopsis, the two examined above and the third being the SnRK1/TOR pathway tested next. Glucose-induced repression of DARK INDUCIBLE 1 (DIN1, At4g35770) is regulated by glycolysis-dependent SnRK1(Baena-Gonzalez et al., 2007; Baena-Gonzalez and Sheen, 2008). As shown in Figure 1C, there was no obvious difference in glucose repression of DIN1 among Col-0, hxk1-3, rgs1-2, and 35S::AtRGS1 over-expression lines, suggesting no role for either HXK1 or AtRGS1 in glucose-regulated DIN1 expression. However, the hxk1-3/rgs1-2 double mutants had a lower basal level of DIN1 expression (P = 0.007405), implying redundancy or, more likely, an indirect effect on the glucose economy.
The behavior of the hxk1, rgs1, and hxk1/rgs1 mutants in glucose-regulated gene expression paints a complex relationship between two glucose sensing pathways in Arabidopsis and clearly indicate that these two pathways do not operate independently without at least modest influence on each other through direct or indirect mechanisms. The epistasis analyses suggest that AtHXK1 and AtRGS1 operate cooperatively for TBL26 and CA2 regulation (Figure 1D) and redundantly or not at all in DIN1 regulation. This unusual genetic relationship is born out in the developmental phenotypes of the mutants described below.
AtRGS1 Genetically Interacts with AtHXK1 in Modulating Early Seedling Development
The feed-back regulation of AtHXK1 and AtRGS1 on each other in glucose-regulated gene expression predicts that loss of either would confer opposite physiological phenotypes while loss of both would confer a phenotype shared by neither (e.g., an intermediate phenotype). Previous reports indicate that genetic ablation of AtHXK1 reduces growth of roots, leaves and inflorescences in the Ler ecotype (Moore et al., 2003; Cho et al., 2006), whereas, rgs1-2 null mutants have longer roots, hypocotyls, and greater rosette diameter than wild type (Chen et al., 2003; Urano et al., 2013). In order to study the different roles between AtHXK1 and AtRGS1 in glucose-regulated primary root growth, 3-day-old Arabidopsis seedlings germinated in 1/4 MS solid medium containing 0% glucose were transferred to different glucose conditions, and the lengths of primary roots on the 10th day was measured. As shown in Figure 2A, compared to wild type plants, hxk1-3 mutants had shorter roots under all glucose conditions; rgs1-2 mutants had similar root lengths to Col-0; hxk1-3/rgs1-2 double mutants had intermediate root elongation between the rgs1-2 and hxk1-3 mutants. The rgs1-2 allele rescued the hxk1-3 allele in leaf and rosette size (Figures 2B–D). Given that root elongation and leaf extension are all based on cell division in the meristem and subsequent cell expansion, the intermediate phenotype of hxk1-3/rgs1-2 double mutants imply that the two glucose sensors—AtRGS1 and AtHXK1 have opposite roles in regulation of cell proliferation. Glucose and salt stress responsiveness were also measured (Figures 2E,F). Both hxk1-3 and rgs1-2 were hyposensitive to a high dose of glucose but this hyposensitivity was not additive. The hxk1-3 allele conferred hypersensitivity to NaCl while the rgs1-2 allele and the double mutant were less sensitive. The rgs1-2 allele is epistatic to the hxk1-3 allele indicating that AtRGS1 and AtHXK1 operate in the same genetic pathway.
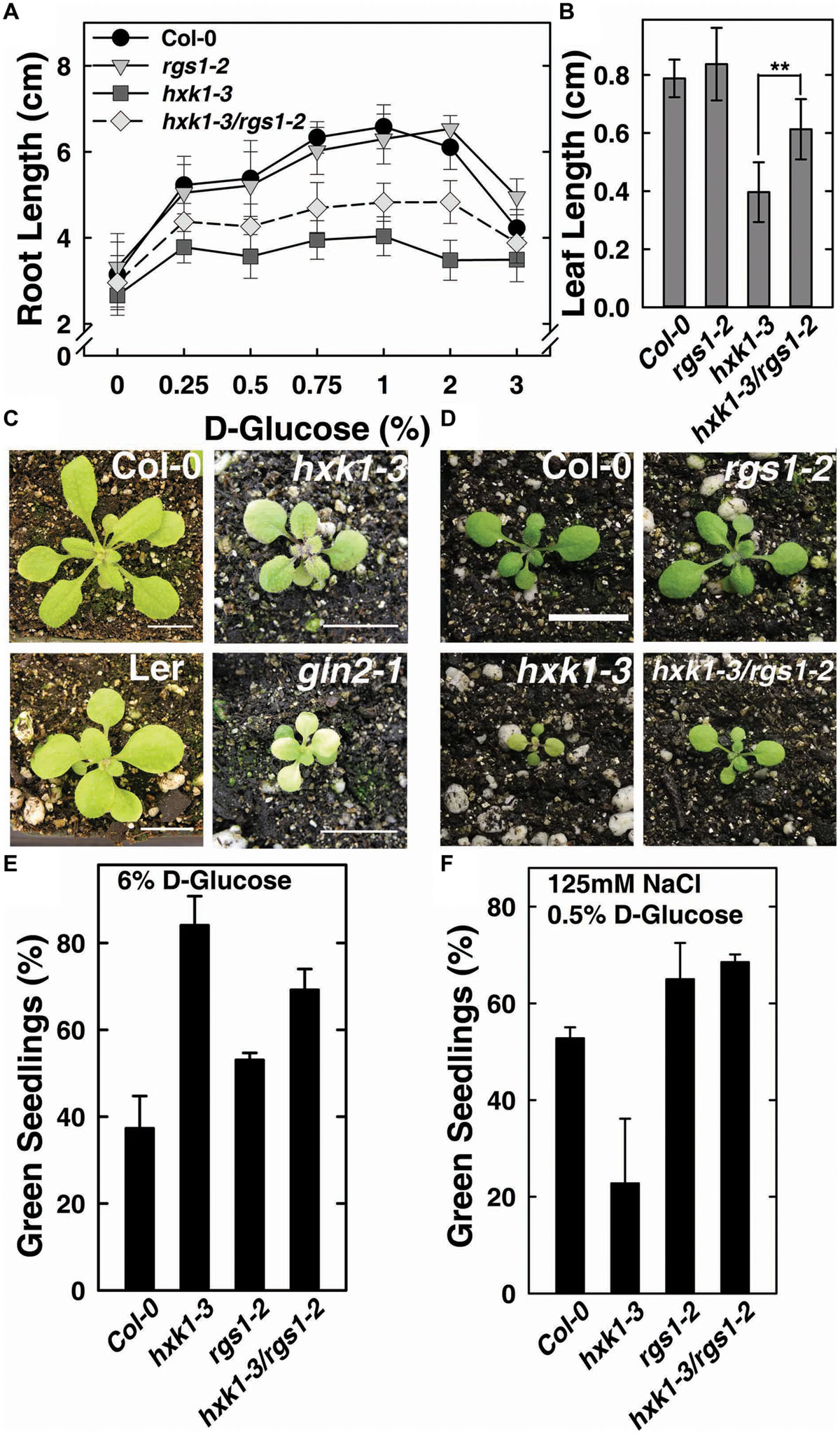
FIGURE 2. hxk1-3/rgs1-2 double mutants show intermediate growth phenotype between hxk1-3 and rgs1-2 sngle mutants. (A) Comparison of root elongation of Col-0, rgs1-2, hxk1-3, and hxk1-3/rgs1-2 mutant seedlings transferred to1/4 MS medium supplemented with 0, 0.25, 0.5, 0.75, 1, 2 or 3% D-glucose (w/v) under low intensity light (70 μmol s-1 m-2) in long-day chamber in 16/8 h (L/D). Values are means ± SD (n = 10–18). (B) Comparison of leaf size (the third leaf) of 2-week-old Col-0, rgs1-2, hxk1-3, and hxk1-3/rgs1-2 mutant seedlings grown under light (160 μmol s-1 m-2) in 16/8 h (L/D). Values are means ± SD (n = 10–12). ANOVA single factor analysis (α = 0.05) was conducted to compare the leaf size in hxk1-3and hxk1-3/rgs1-2. ∗∗P < 0.01. (C) 3-week-old Col-0, hxk1-3, Ler, and gin2-1 plants grown under high intensity light (160 μmol s-1 m-2) in 16/8 h (L/D). Scale bar = 1 cm. (D) 2-week-old Col-0, hxk1-3, rgs1-2, and hxk1-3/rgs1-2 mutant plants grown under high intensity light (160 μmol s-1 m-2) in 16/8 h (L/D). Scale bar = 1 cm. (E) Green seedling assay of the Col-0, rgs1-2, hxk1-3, and hxk1-3/rgs1-2 mutant. The assay was performed as described in Section “Materials and Methods.” The average percentage of seedlings showing green cotyledons was determined and presented with means ± SD from one representative experiment of 4 biological replications. (F) Col, rgs1-2, hxk1-3, and rgs1-2/hxk1-3 have altered responses to saline stress. The assay was performed as described in Section “Material and Methods.” Values are means ± SD from quintuplicate.
To verify the role of AtRGS1and AtHXK1 in meristematic cell activity, and to extend a previous study on the behavior of glucose sensors in sugar promoting meristem activation (Xiong et al., 2013), we compared the cell cycle activity in the primary roots of Col-0, hxk1-3 and rgs1-2 single mutant lines (Figure 3). The steady-state level of A-type CDK—CDKA;1, which is constitutively high in actively dividing cells was investigated using existing GUS reporter genes which were stably integrated into the wild-type (West et al., 2004; Skylar et al., 2011) or mutant backgrounds (this study). In order to minimize the influence of endogenous sugar from the endosperm or photosynthesis, seeds were germinated in sugar-free liquid medium, then 3-day-old seedlings were transferred to fresh liquid medium supplemented with 3% D-glucose or D-mannitol for another 3 days treatment as previously described (Zhang et al., 2010; Xiong et al., 2013). As shown in Figure 3A, D-glucose, but not D-mannitol, activated the CDKA;1 promoter in all three genotypes. Quantitative analysis showed that the rgs1-2 mutant had increased (P < 0.05) root meristem size in response to glucose as determined by GUS-staining, whereas the hxk1-3 mutant had decreased (P < 0.01) size (Figure 3B), consistent with the phenotypes we observed in glucose-promoted root growth (Figure 2A). Moreover, it indicated an overlap in function between AtRGS1 and AtHXK1 in root meristem modulation in response to glucose.
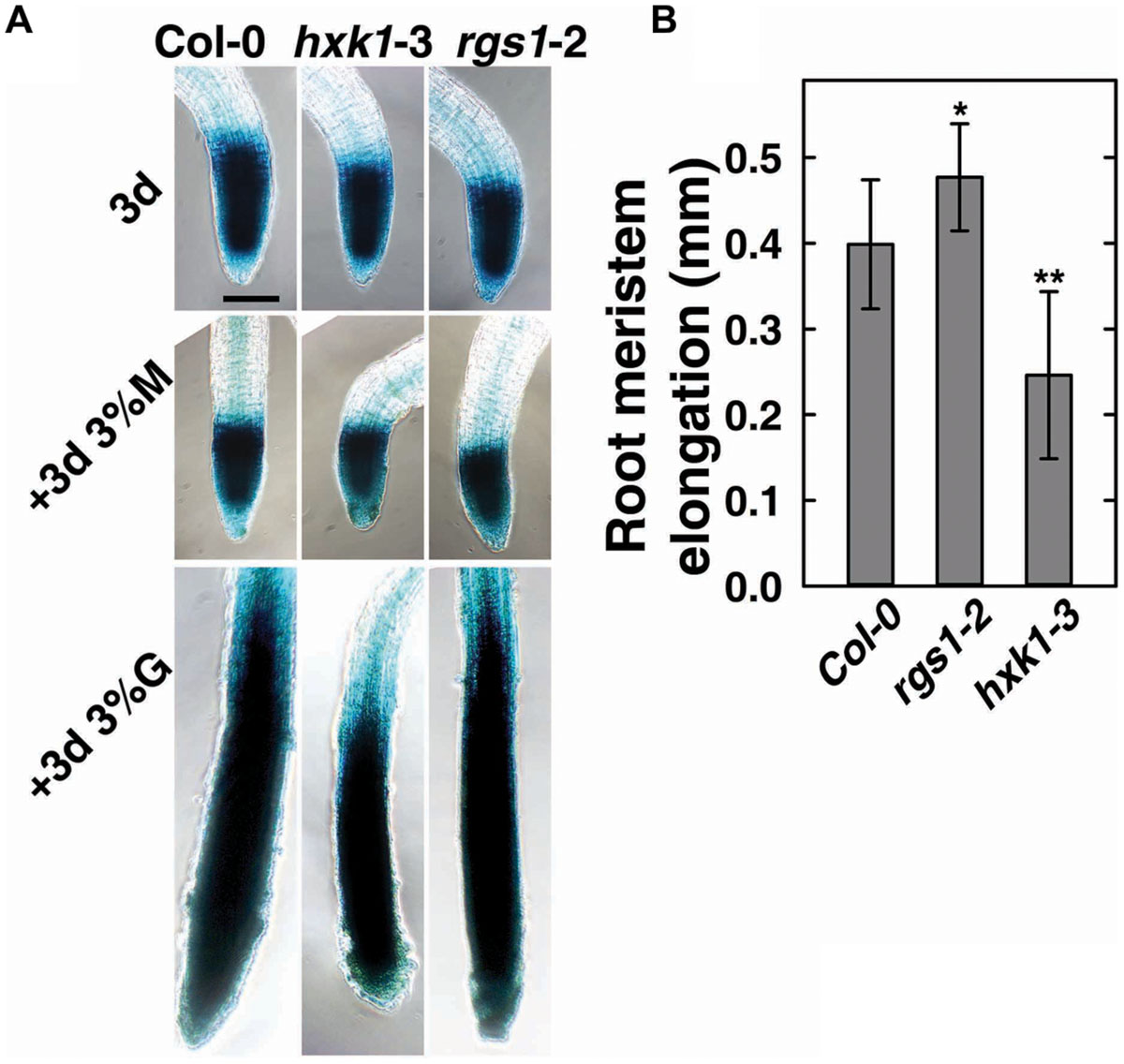
FIGURE 3. Cell cycle progress in rgs1-2 or hxk1-3 mutant showed changed response to D-glucose. (A) Meristematic activity in the primary root tip of Col-0, hxk1-3 and rgs1-2 plants as visualized by GUS-staining under the indicated sugar conditions as described in Section “Materials and Methods.” Seeds were germinated and cultured in 1/2 MS liquid medium for 3 days (3d); and then transferred to 1/2 MS liquid medium supplemented with 3% (w/v) D-glucose(+3d 3%G) or 3% (w/v) D-mannitol (+3d 3% M) for 3 more days. Scale bar = 0.1 mm. (B) The root meristem elongation as determined by the length-change of GUS-staining region in the primary root tip of Col-0, hxk1-3 and rgs1-2 seedlings. Values are means ± SD (n = 8–12) from one representative experiment of 3 biological replications. Pairwise Student’s t-test was used to compare values to the Col-0. ∗P < 0.05; ∗∗P < 0.01.
AtHXK1 Operates Downstream of Rapid Glucose-induced AtRGS1 Endocytosis
In Saccharomyces cerevisiae, glucose activation of cAMP synthesis (cAMP/PKA pathway) requires both the presence of a G-protein-coupled receptor system for extracellular glucose sensing, and phosphorylation of the sugar by hexokinases (HXKs, Hxk2/Snf1/Mig1 pathway) for cytoplasmic glucose sensing (Rolland et al., 2000). While the exact mechanism in this process is unknown, the data indicate a functional connection between G-proteins and hexokinases. To test if an analogous relationship between extracellular and cytoplasmic glucose perception exists in Arabidopsis, we quantitated glucose-induced activation of G signaling. The most rapid in vivo reporter for G protein activation currently available is glucose-induced endocytosis of AtRGS1 which occurs in minutes (Urano et al., 2012; Fu et al., 2014). Specifically, AtGPA1 (Gα) is self-activating through spontaneous binding of GTP but is held in the resting state by the inhibitory action of its physical partner, AtRGS1. Glucose relieves this inhibition by causing physical de-coupling of AtRGS1 and AtGPA1 through WNK kinase-dependent phosphorylation at the C terminus of AtRGS1 (Urano et al., 2012). Phosphorylation of AtRGS1 drives its endocytosis thus physically uncoupling AtRGS1 from AtGPA1 and allowing activation of G protein signaling (Urano et al., 2012; Fu et al., 2014). As shown in Figure 4, loss of AtHXK1 did not affect the level of glucose activated AtRGS1 re-localization, confirming that the regulatory role of AtHXK1 in AtRGS1-dependent TBL26 induction shown in Figure 1A lies downstream of the apical event in G protein coupled signaling.
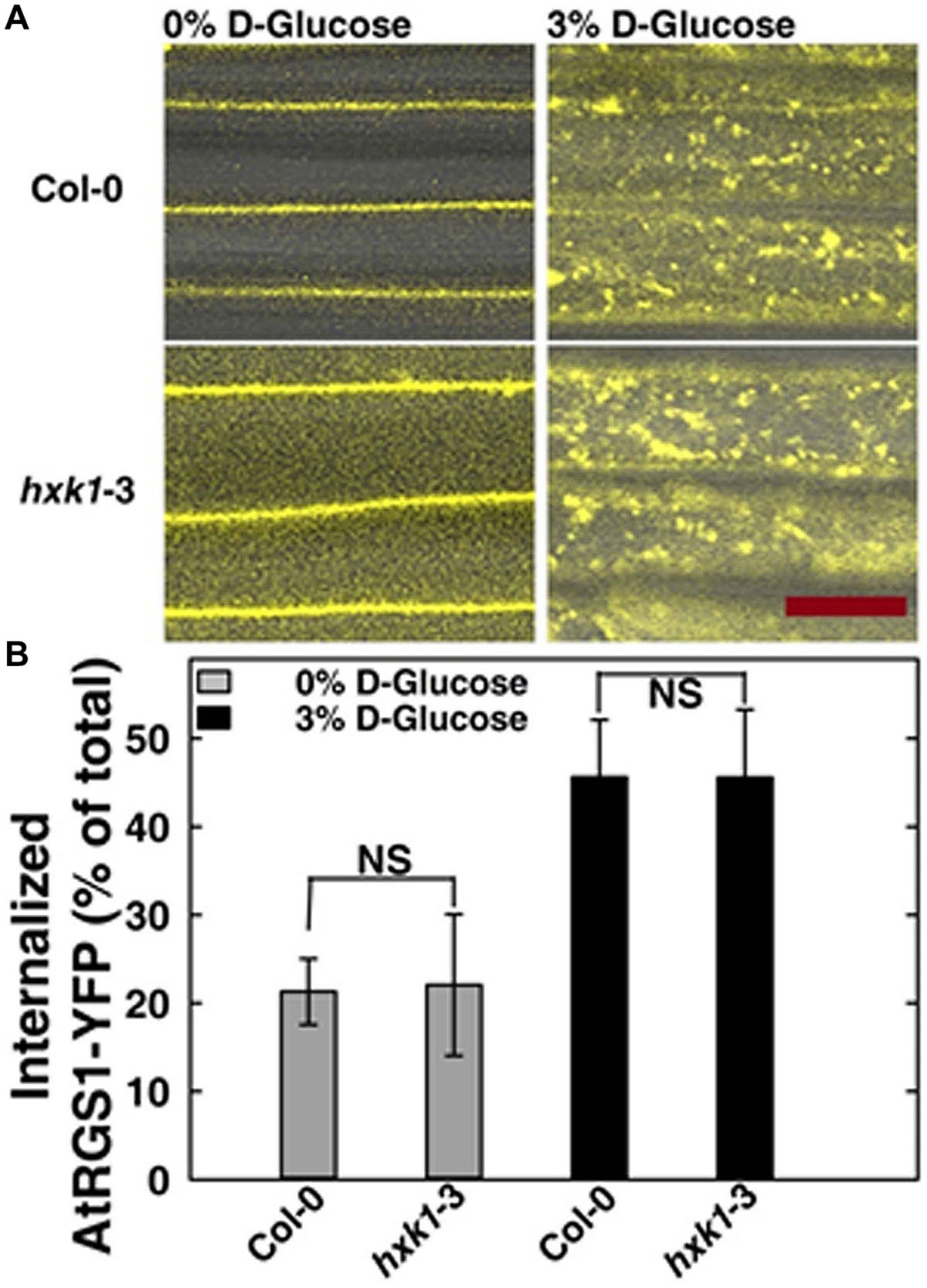
FIGURE 4. AtRGS1 internalizes in response to D-glucose even in the absence of AtHXK1. (A) Hypocotyls cells of 7-day-old Col-0 and hxk1-3 Arabidopsis seedlings stably expressing AtRGS1-YFP imaged after treatment with 3% D-glucose (w/v) for 30 min. Scale bar = 20 μm. (B) Quantization of the percentage of AtRGS1-YFP internalized in epidermal cells of Col-0 and hxk1-3 mutant seedlings hypocotyls before and after glucose stimulation. Values are percentage means ± SD, from a representative experiment (n = 10) of 3 biological replications. ANOVA single factor analysis (α = 0.05) was conducted to compare the difference between plant lines with the same glucose treatment.
RHIP1 is a Nexus in AtRGS1- and HXK1-dependent Sugar Sensing
A deep screen for Arabidopsis G-protein interacting proteins (Klopffleisch et al., 2011) yielded an uncharacterized conserved protein encoded by At4g26410 that interacts with both AtRGS1 and AtHXK1 (Supplementary Figure S4). We investigated the possibility that this protein provides a functional connection between AtHXK1 and AtRGS1 glucose signaling. We designated this protein RGS1 and HXK1 Interacting Protein 1 (RHIP1). RHIP1 contains an Uncharacterized Conserved “Protein” domain (UCP022280) with only one other protein (At2g45060) containing this domain in Arabidopsis. To our knowledge, these two proteins have not previously been identified in any study. The predicted protein structure of RHIP1 is a three-stranded helix with an N-terminal head composed of coils (Figure 5A). The predicted GO biological process of RHIP1 protein based on global and local protein similarity includes response to fungicide, receptor internalization, and regulation of receptor recycling (I-TASSER server) (Zhang, 2008; Roy et al., 2010, 2012). The top 1 enzyme homolog of RHIP1 protein in the Protein Database is the Escherichia coli histidine kinase sensor TorS sensor domain involved in signal transduction (Zhang, 2008; Moore and Hendrickson, 2009; Roy et al., 2010, 2012). RHIP1 interacts with 21 other proteins in the G protein interactome (Klopffleisch et al., 2011). Based on TAIR gene ontology annotations, 47% of these proteins are predicted to be involved in response to stimulus (Figure 5B)3. This finding prompted us to hypothesize that RHIP1 may have a role in sugar signaling through interactions with HXK1 and AtRGS1. To confirm interactions of RHIP1 with both AtRGS1 and AtHXK1 in plant cells, we used BiFC performed as described by Klopffleisch et al. (2011). As shown in Figure 6, AtRGS1 and AtHXK1 interacted with RHIP1 in vivo. Two negative and one positive control were included. The positive control was AtRGS1 and AtGPA1. The two negative controls were RHIP1-AtGPA1 and AtRGS1-AtHXK1 pairs. While in vivo interaction is confirmed, we do not reach a conclusion on the subcellular location of this interaction. Tobacco pavement cells, due to the large vacuole, are not suitable for subcellular location analysis. The dynamics of the confirmed interactions is not clear. While the data show that RHIP1 interacts with both AtRGS1 and AtHXK1, they do not show that this interaction is simultaneous; neither do the data preclude a scaffold role of RHIP1 to bring together AtRGS1 and AtHXK1. This is because that while AtRGS1 is located on the plasma membrane and the endomembrane system, AtHXK1 subcellular location is broad, including the cytoplasm, the mitochondrion and the nucleus and therefore it is plausible that RHIP1 is a protein complex scaffold.
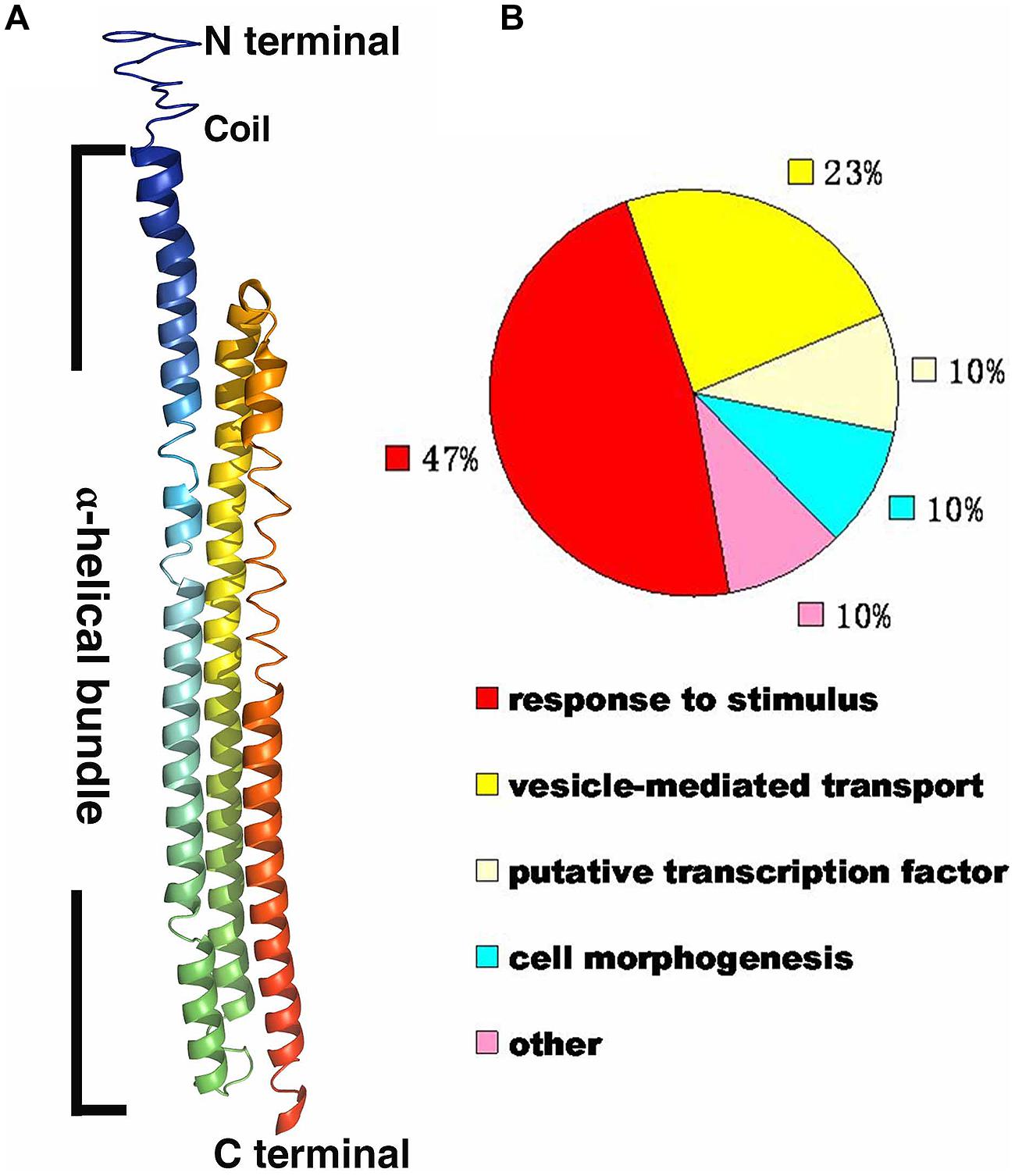
FIGURE 5. Bioinformational analysis of RHIP1 (At4g26410). (A) A modeled structure of Arabidopsis thaliana RHIP1. The structure was predicted by the I-TASSER program based on the RHIP1 protein sequence, and processed by PyMOL software. The three α-helical bundleis shown with rainbow colors, and the top is the N-terminal head composed of coils. (B) Functional analysis of 21 RHIP1 interacting proteins are annotated as “response to stimulus” (47%). These 21 proteins are AT2G32670, AT5G05760, AT1G35720, At2g26400, At2g42400, AT1G28520, AT1G20100, AT2G26300, AT4G17730, AT1G05500, AT3G26090, AT3G49290, At5g42030, AT3G60600, AT5G14240, AT5G43850, AT5G03540, AT3G59220, AT4g13640, AT5g67380, and AT4G29130. The function cluster analysis was performed based on TAIR GO annotations.
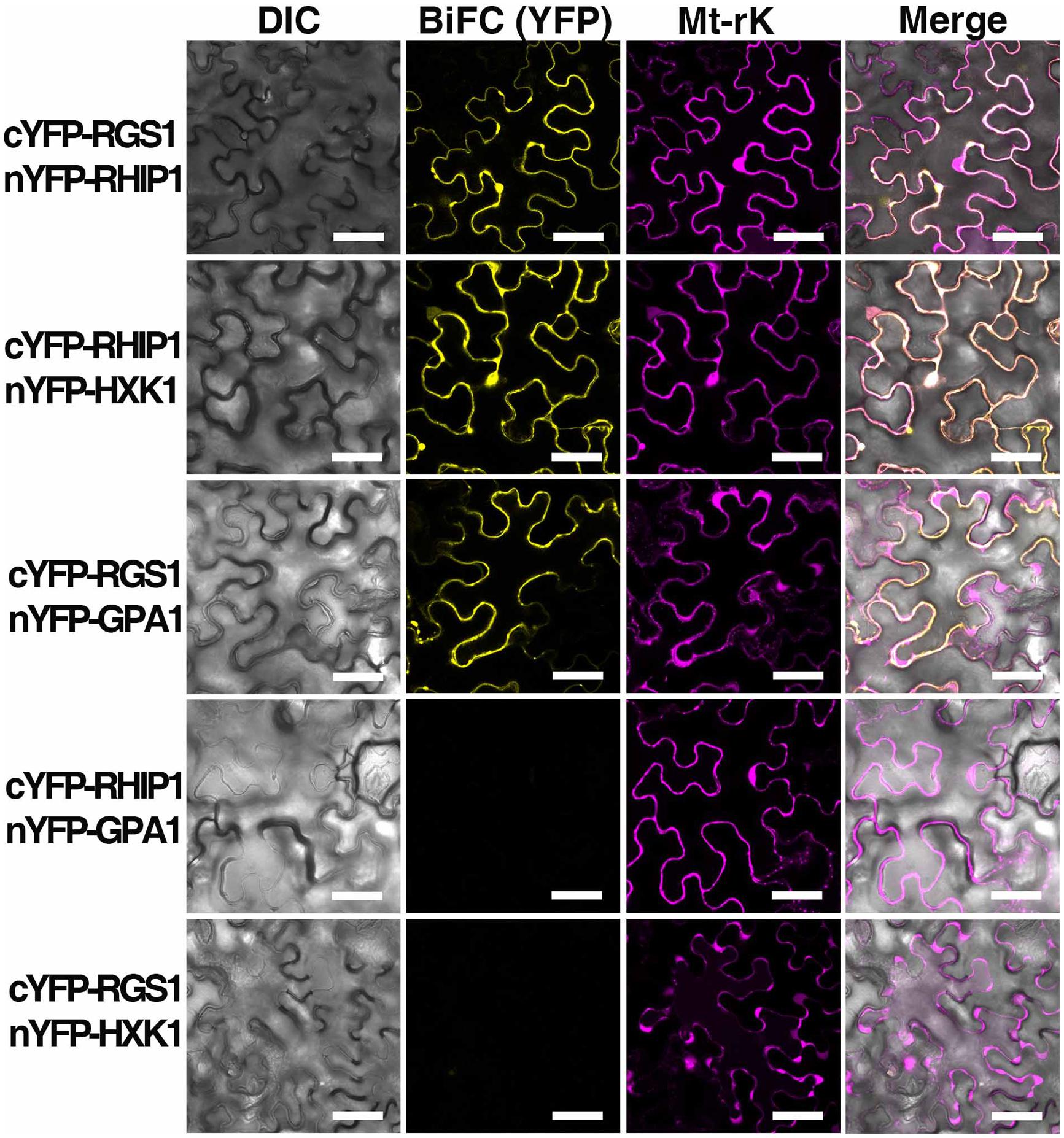
FIGURE 6. In vivo interaction between AtRGS1 and RHIP1, and AtHXK1 and RHIP1 determined by physical complementation of YFP using Bimolecular Fluorescence complementation (BiFC). The test pairs are indicated on the left. (Column 1) Differential interference contrast (DIC) images of transformed cells. (Column 2) BIFC (YFP). cYFP-tagged proteins were co-transformed with nYFP-tagged proteins into tobacco leaves as described in Section “Materials and Methods.” (Column 3) Mt-rk is an RFP mitochondria marker used for the transformation control. Fluorescence complementation of split YFP and expression of Mt-rk were observed by confocal fluorescence microscopy. (Column 4) DIC, BiFC, and Mt-rk are merged. Scale bars = 50 μm.
RHIP1 Participates in Glucose Signaling in Arabidopsis
In order to elucidate the physiological role of RHIP1 in glucose signaling, two RHIP1 T-DNA insertion mutants, rhip1-1 and rhip1-2 were generated (Supplementary Figure S5). Plants homozygous for rhip1-1 and rhip1-2 were confirmed to be transcript null. Both rhip1-1 and rhip1-2 mutants had longer roots in young seedlings and qualitatively larger inflorescence in adult plants (Figures 7A–C); these are phenotypes shared by the rgs1-2 mutant. Over-expression of RHIP1 in Col-0 resulted in defects in root elongation (Figure 7D). Acute doses of glucose (6%, w/v) cause Arabidopsis seedling development to arrest and this phenotype is attenuated in both HXK1 and RGS1 loss-of-function mutants (Moore et al., 2003; Chen and Jones, 2004). We used this standardized green seedling assay to explore the role of RHIP1. As shown in Figure 7E, rhip1-2 mutants showed less sensitivity to 6% glucose than Col-0 seedlings. These rhip1 phenotypes were not the consequences of altered transcript level for RGS1 or HXK1 (Supplementary Figure S2). As shown in Figures 1A–C, genetic ablation of RHIP1 increased glucose-induced TBL26 expression (around 4.5 fold), while there was no effect on the expression of CA2 or DIN1.
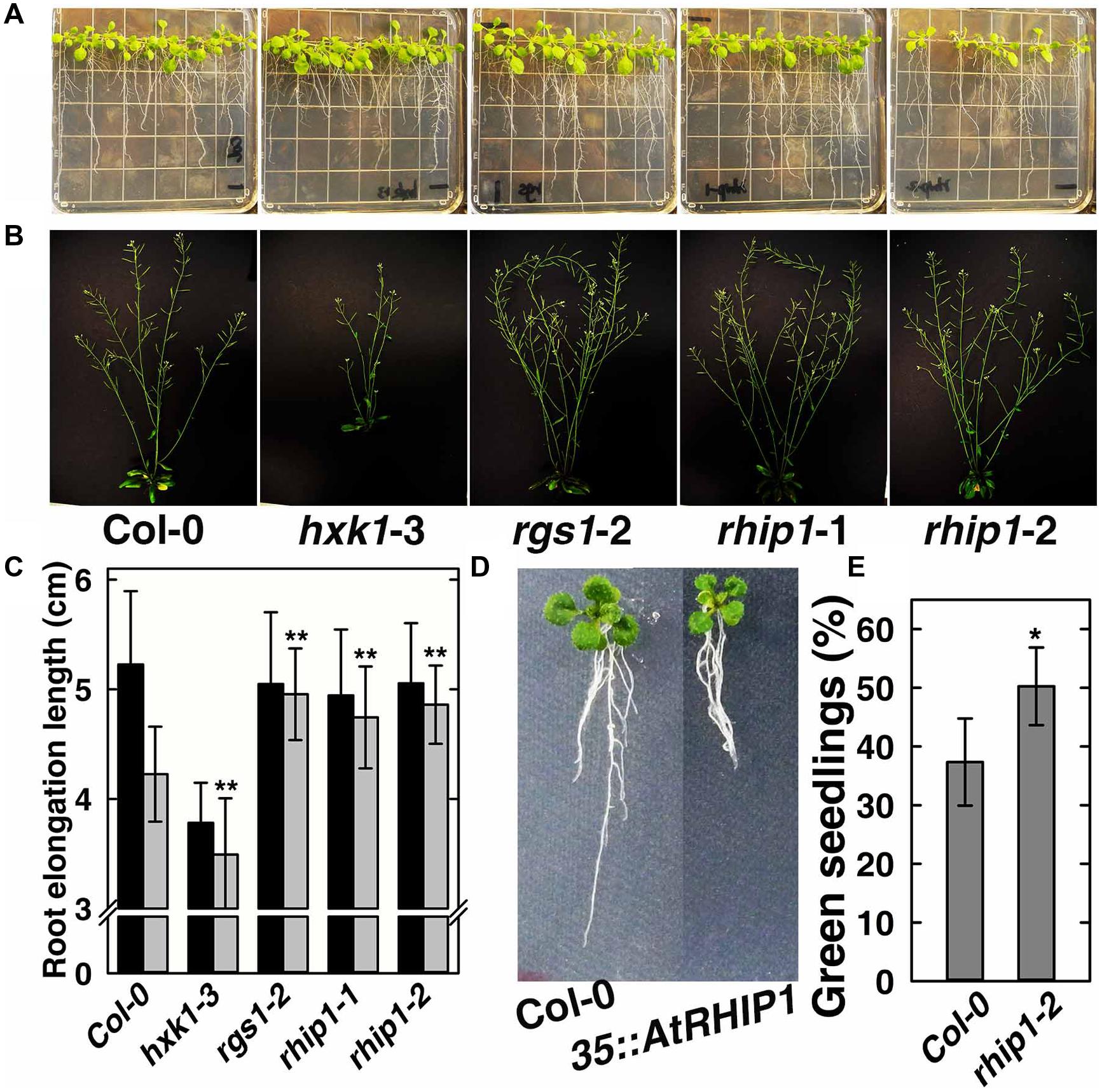
FIGURE 7. rhip1 mutants share growth phenotypes with the rgs1-2 null mutant. (A) 2-week-old Col-0, hxk1-3, rgs1-2, rhip1-1, and rhip1-2 plants grown in 1/4 × MS medium supplemented with 1% D-glucose (w/v) and grown at 200 μmol s-1 m-2 in 8/16 h (L/D). (B) 5-week-old Col-0, hxk1-3, rgs1-2, rhip1-1, and rhip1-2 plants grown at 160 μmol s-1 m-2 in 16/8 h (L/D). (C) Comparison of root elongation of Col-0, hxk1-3, rgs1-2, rhip1-1, and rhip1-2 seedlings transferred to 1/4 × MS medium supplemented with 0.25% (black) or 3% (gray) w/v D-glucose and grown at 70 μmol s-1 m-2 in 16/8 h (L/D). Values indicate means ± SD (n = 10–18) from a representative experiment. For the 3% D-glucose treatment, ANOVA single factor analysis (α = 0.05) was conducted to compare values to the Col-0. ∗∗P < 0.01. (D) 2-week-old Col-0 and 35S::YFP-RHIP1 plants grown in 1/4 × MS medium supplemented with 3% D-glucose (w/v). (E) Green seedling assay of the rhip1-2 mutant. The assay was performed as described in Section “Material and Methods.” The average percentage of seedlings showing green cotyledons was determined and presented with means ± SD from one representative experiment of 4 biological replications. ANOVA single factor analysis (α = 0.05) was conducted to compare values to the Col-0. ∗P < 0.05.
Since it is predicted that AtHXK1 operates on AtRGS1 signaling downstream of the rapid glucose-induced AtRGS1 endocytosis (Figures 1A and 4), we explored the role of their common interactor —AtRHIP1 in this process. As expected, loss of AtRHIP1 did not affect the level of glucose activated AtRGS1 re-localization (Supplementary Figure S6), consistent with the regulatory role of AtHXK1 localized downstream of the apical event in G protein coupled signaling.
In summary, a partial functional conversation occurring between AtRGS1 and AtHXK1 was shown by glucose-regulated gene expression, epistasis analyses, and protein–protein interaction: (1) AtRGS1 and AtHXK1 showed hierarchical relationships in regulation of TBL26, CA2, and CAB2 expression, suggesting collaboration between AtRGS1 and AtHXK1 in regulating gene expression. (2) hxk1 mutant exhibited the opposite root elongation and leaf expansion phenotype to the rgs1 mutants, while the double mutant showed an intermediate phenotype, implying a feed-back relationship between AtRGS1 and AtHXK1 in controlling cell activity. This was confirmed by the opposite behavior in glucose promotion of root meristem activation. The intermediate response in the double mutant was not additive since the phenotype of the single mutants were opposite. (3) Loss of AtHXK1 abrogated AtRGS1-dependent signaling as measured by TBL26 expression without changing the level of glucose activated AtRGS1 endocytosis, indicating that AtHXK1 acts downstream of AtRGS1 in sugar-activated AtRGS1 signaling. (4) AtRGS1 and AtHXK1 have a common protein interactor, RHIP1, a predicted signaling component, providing a direct nexus in AtRGS1- and AtHXK1-dependent sugar sensing. (5) Gene expression and seedling growth phenotype analyses of the rhip1 mutant confirmed a potential role in AtRGS1 and AtHXK1 signaling.
Accession Numbers
AtRGS1, At3g26090; TBL26, At4g01080; AtHXK1, At4g29130; AtCA2, At5g14740; AtTUB4, At5g44340; AtDIN1, At4g35770; RHIP1, At4g26410; CAB2, At1g29920. Mutant alleles for the genes were generated from the indicated T-DNA insertion lines: hxk1-1, SALK_034233; hxk1-2, CS864200; hxk1-3, CS861759; rhip1-1, SALK_091518C; rhip1-2, SALK_061002.
Funding
This work was supported by the National Institute of General Medical Sciences (R01GM065989), and the National Science Foundation (MCB- 0718202) to AJ. The Division of Chemical Sciences, Geosciences, and Biosciences, Office of Basic Energy Sciences of the US Department of Energy through the grant DE-FG02-05er15671 to AJ funded technical support in this study.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
J-PH thanks The University of North Carolina, where these experiments were conducted in entirety for hosting her from 2012 to 2014 as an international scholar supported by an exchange scholarship from the Chinese Scholarship Council. We thank Jing Yang for technical assistance, Alejandro Colaneri for discussions, and Dirk Inzé for providing the pCDKA;1::GUS reporter line in the Col background.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00851
Footnotes
- ^http://zhanglab.ccmb.med.umich.edu/I-TASSER/
- ^http://www.arabidopsis.org/tools/bulk/go/index.jsp
- ^http://www.arabidopsis.org/tools/bulk/go/index.jsp
References
Baena-Gonzalez, E., Rolland, F., Thevelein, J. M., and Sheen, J. (2007). A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938–942. doi: 10.1038/nature06069
Baena-Gonzalez, E., and Sheen, J. (2008). Convergent energy and stress signaling. Trends Plant Sci. 13, 474–482. doi: 10.1016/j.tplants.2008.06.006
Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris Life Sci. 316, 1194–1199.
Booker, K. S., Schwarz, J., Garrett, M. B., and Jones, A. M. (2010). Glucose attenuation of auxin-mediated bimodality in lateral root formation is partly coupled by the heterotrimeric G protein complex. PLoS ONE 5:e12833. doi: 10.1371/journal.pone.0012833
Chen, J. G., Gao, Y., and Jones, A. M. (2006a). Differential roles of Arabidopsis heterotrimeric G-protein subunits in modulating cell division in roots. Plant Physiol. 141, 887–897. doi: 10.1104/pp.106.079202
Chen, Y., Ji, F., Xie, H., and Liang, J. (2006b). Overexpression of the regulator of G-protein signalling protein enhances ABA-mediated inhibition of root elongation and drought tolerance in Arabidopsis. J. Exp. Bot. 57, 2101–2110. doi: 10.1093/jxb/erj167
Chen, Y., Ji, F., Xie, H., Liang, J., and Zhang, J. (2006c). The regulator of G-protein signaling proteins involved in sugar and abscisic acid signaling in Arabidopsis seed germination. Plant Physiol. 140, 302–310. doi: 10.1104/pp.105.069872
Chen, J. G., and Jones, A. M. (2004). AtRGS1 function in Arabidopsis thaliana. Methods Enzymol. 389, 338–350. doi: 10.1016/S0076-6879(04)89020-7
Chen, J. G., Willard, F. S., Huang, J., Liang, J., Chasse, S. A., Jones, A. M., et al. (2003). A seven-transmembrane RGS protein that modulates plant cell proliferation. Science 301, 1728–1731. doi: 10.1126/science.1087790
Cho, Y. H., Yoo, S. D., and Sheen, J. (2006). Regulatory functions of nuclear hexokinase1 complex in glucose signaling. Cell 127, 579–589. doi: 10.1016/j.cell.2006.09.028
Colaneri, A. C., Tunc-Ozdemir, M., Huang, J. P., and Jones, A. M. (2014). Growth attenuation under saline stress is mediated by the heterotrimeric G protein complex. BMC Plant Biol. 14:129. doi: 10.1186/1471-2229-14-129
Fu, Y., Lim, S., Urano, D., Tunc-Ozdemir, M., Phan, N. G., Elston, T. C., et al. (2014). Reciprocal encoding of signal intensity and duration in a glucose-sensing circuit. Cell 156, 1084–1095. doi: 10.1016/j.cell.2014.01.013
Grigston, J. C., Osuna, D., Scheible, W. R., Liu, C., Stitt, M., and Jones, A. M. (2008). D-Glucose sensing by a plasma membrane regulator of G signaling protein, AtRGS1. FEBS Lett. 582, 3577–3584. doi: 10.1016/j.febslet.2008.08.038
Kim, J. H., Roy, A., Jouandot, D., and Cho, K. H. (2013). The glucose signaling network in yeast. Biochim. Biophys. Acta 1830, 5204–5210. doi: 10.1016/j.bbagen.2013.07.025
Klopffleisch, K., Phan, N., Augustin, K., Bayne, R. S., Booker, K. S., Botella, J. R., et al. (2011). Arabidopsis G-protein interactome reveals connections to cell wall carbohydrates and morphogenesis. Mol. Syst. Biol. 7, 532. doi: 10.1038/msb.2011.66
Koch, K. E. (1996). Carbohydrate-modulated gene expression in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 509–540. doi: 10.1146/annurev.arplant.47.1.509
Lastdrager, J., Hanson, J., and Smeekens, S. (2014). Sugar signals and the control of plant growth and development. J. Exp. Bot. 65, 799–807. doi: 10.1093/jxb/ert474
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Malamy, J. E., and Benfey, P. N. (1997). Organization and cell differentiation in lateral roots of Arabidopsis thaliana. Development 124, 33–44.
Moore, B., Zhou, L., Rolland, F., Hall, Q., Cheng, W. H., Liu, Y. X., et al. (2003). Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300, 332–336. doi: 10.1126/science.1080585
Moore, J. O., and Hendrickson, W. A. (2009). Structural analysis of sensor domains from the TMAO-responsive histidine kinase receptor TorS. Structure 17, 1195–1204. doi: 10.1016/j.str.2009.07.015
Reda, M. (2013). Regulation of nitrate reduction in Arabidopsis WT and hxk1 mutant under C and N metabolites. Physiol. Plant. 149, 260–272. doi: 10.1111/ppl.12045
Rolland, F., De Winde, J. H., Lemaire, K., Boles, E., Thevelein, J. M., and Winderickx, J. (2000). Glucose-induced cAMP signalling in yeast requires both a G-protein coupled receptor system for extracellular glucose detection and a separable hexose kinase-dependent sensing process. Mol. Microbiol. 38, 348–358. doi: 10.1046/j.1365-2958.2000.02125.x
Roy, A., Kucukural, A., and Zhang, Y. (2010). I-TASSER: a unified platform for automated protein structure and function prediction. Nat. Protoc. 5, 725–738. doi: 10.1038/nprot.2010.5
Roy, A., Yang, J., and Zhang, Y. (2012). COFACTOR: an accurate comparative algorithm for structure-based protein function annotation. Nucleic Acids Res. 40, W471–W477. doi: 10.1093/nar/gks372
Sheen, J. (1990). Metabolic repression of transcription in higher plants. Plant Cell 2, 1027–1038. doi: 10.2307/3869242
Sheen, J. (2014). Master regulators in plant glucose signaling networks. J. Plant Biol. 57, 67–79. doi: 10.1007/s12374-014-0902-7
Sheen, J., Zhou, L., and Jang, J.-C. (1999). Sugars as signaling molecules. Curr. Opin. Plant Biol. 2, 410–418. doi: 10.1016/S1369-5266(99)00014-X
Skylar, A., Sung, F., Hong, F., Chory, J., and Wu, X. (2011). Metabolic sugar signal promotes Arabidopsis meristematic proliferation via G2. Dev. Biol. 351, 82–89. doi: 10.1016/j.ydbio.2010.12.019
Tome, F., Nagele, T., Adamo, M., Garg, A., Marco-Llorca, C., Nukarinen, E., et al. (2014). The low energy signaling network. Front. Plant Sci. 5:353. doi: 10.3389/fpls.2014.00353
Urano, D., Chen, J. G., Botella, J. R., and Jones, A. M. (2013). Heterotrimeric G protein signalling in the plant kingdom. Open Biol. 3, 120186. doi: 10.1098/rsob.120186
Urano, D., Phan, N., Jones, J. C., Yang, J., Huang, J., Grigston, J., et al. (2012). Endocytosis of the seven-transmembrane RGS1 protein activates G-protein-coupled signalling in Arabidopsis. Nat. Cell Biol. 14, 1079–1088. doi: 10.1038/ncb2568
West, G., Inze, D., and Beemster, G. T. (2004). Cell cycle modulation in the response of the primary root of Arabidopsis to salt stress. Plant Physiol. 135, 1050–1058. doi: 10.1104/pp.104.040022
Xiong, Y., Mccormack, M., Li, L., Hall, Q., Xiang, C., and Sheen, J. (2013). Glucose-TOR signalling reprograms the transcriptome and activates meristems. Nature 496, 181–186. doi: 10.1038/nature12030
Zhang, L., Hu, G., Cheng, Y., and Huang, J. (2008). Heterotrimeric G protein alpha and beta subunits antagonistically modulate stomatal density in Arabidopsis thaliana. Dev. Biol. 324, 68–75. doi: 10.1016/j.ydbio.2008.09.008
Zhang, L., Wei, Q., Wu, W., Cheng, Y., Hu, G., Hu, F., et al. (2009). Activation of the heterotrimeric G protein alpha-subunit GPA1 suppresses the ftsh-mediated inhibition of chloroplast development in Arabidopsis. Plant J. 58, 1041–1053. doi: 10.1111/j.1365-313X.2009.03843.x
Zhang, Y. (2008). I-TASSER server for protein 3D structure prediction. BMC Bioinformatics 9:40. doi: 10.1186/1471-2105-9-40
Keywords: plant heterotrimeric G protein, sugar signaling, Arabidopsis regulator of G-protein signaling 1 protein (AtRGS1), hexokinase 1 (HXK1)
Citation: Huang J-P, Tunc-Ozdemir M, Chang Y and Jones AM (2015) Cooperative control between AtRGS1 and AtHXK1 in a WD40-repeat protein pathway in Arabidopsis thaliana. Front. Plant Sci. 6:851. doi: 10.3389/fpls.2015.00851
Received: 15 July 2015; Accepted: 28 September 2015;
Published: 13 October 2015.
Edited by:
Christopher J. Staiger, Purdue University, USAReviewed by:
Ben Matthew Abell, Sheffield Hallam University, UKLing Zhu, The University of Texas at Austin, USA
Copyright © 2015 Huang, Tunc-Ozdemir, Chang and Jones. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Chang, Y2hhbmd5aW5nQG5lYXUuZWR1LmNu; Alan M. Jones, YWxhbl9qb25lc0B1bmMuZWR1
 Jian-Ping Huang
Jian-Ping Huang Meral Tunc-Ozdemir
Meral Tunc-Ozdemir Ying Chang
Ying Chang Alan M. Jones
Alan M. Jones