
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Plant Sci. , 09 October 2015
Sec. Plant Pathogen Interactions
Volume 6 - 2015 | https://doi.org/10.3389/fpls.2015.00839
This article is part of the Research Topic How can secretomics help unravel the secrets of plant-microbe interactions? View all 16 articles
Botrytis cinerea is a necrotrophic fungus with high adaptability to different environments and hosts. It secretes a large number of extracellular proteins, which favor plant tissue penetration and colonization, thus contributing to virulence. Secretomics is a proteomics sub-discipline which study the secreted proteins and their secretion mechanisms, so-called secretome. By using proteomics as experimental approach, many secreted proteins by B. cinerea have been identified from in vitro experiments, and belonging to different functional categories: (i) cell wall-degrading enzymes such as pectinesterases and endo-polygalacturonases; (ii) proteases involved in host protein degradation such as an aspartic protease; (iii) proteins related to the oxidative burst such as glyoxal oxidase; (iv) proteins which may induce the plant hypersensitive response such as a cerato-platanin domain-containing protein; and (v) proteins related to production and secretion of toxins such as malate dehydrogenase. In this mini-review, we made an overview of the proteomics contribution to the study and knowledge of the B. cinerea extracellular secreted proteins based on our current work carried out from in vitro experiments, and recent published papers both in vitro and in planta studies on this fungi. We hypothesize on the putative functions of these secreted proteins, and their connection to the biology of the B. cinerea interaction with its hosts.
Phytopathogenic fungi can invade and colonize their plant host to obtain the nutrients because they are able to secrete a set of extracellular proteins and other metabolites (Gonzalez-Fernandez et al., 2010). The secretome has been recently defined as “the global group of secreted proteins into the extracellular space by a cell, tissue, cell, organ, or organism at any given time and conditions through known and unknown secretory mechanisms involving constitutive and regulated secretory organelles” (Agrawal et al., 2013). In the case of phytopatogenic fungi, the secretome consists of pathogenicity and virulence factors, which favor host tissue penetration and colonization in the susceptible plant (Girard et al., 2013). Due to the need to study these secretomes, the term secretomics emerged, and was defined as “the global study of proteins that are secreted by a cell, a tissue or an organism” (Vincent and Bedon, 2013). Many studies have been performed by using both classical approaches and modern–omic techniques through in planta experiments to unravel the plant–fungus interaction mechanisms (Allwood et al., 2008; Bhadauria et al., 2009; Tan et al., 2009; Bhadauria et al., 2010; Quirino et al., 2010; Afroz et al., 2011; Dean et al., 2012). As a proteomics sub-discipline, secretomics has contributed significantly to the study of the phytopathogenic fungus secretome by using in vitro experiments (González-Fernández and Jorrin-Novo, 2010, 2012; Girard et al., 2013; Vincent and Bedon, 2013). Botrytis cinerea Pers. Fr. (teleomorph Botryotinia fuckeliana (de Bary) Whetzel) is a necrotrophic pathogen with a wide host range, including pre- and post-harvest plant species, and it causes important economic losses in agriculture (Elad et al., 2007). The B. cinerea infection process includes host surface penetration, host tissue killing and primary lesion formation, lesion expansion, tissue maceration, and conidiation (van Kan, 2006). All these stages are mainly achieved by producing secreted proteins and other compounds, including the secretion of cell wall-degrading enzymes (CWDEs), the production of non-specific phytotoxic metabolite (botrydial and botcinolides), the boost of an oxidative burst because of reactive oxygen species (ROS) accumulation, and molecules which induce the plant hypersensitive response (HR; Williamson et al., 2007). In the last years, the use of complementary gel-based and gel-free proteomic approaches has provided important findings in the understanding of B. cinerea pathogenicity and virulence in in vitro (Gonzalez-Fernandez and Jorrin-Novo, 2012; Gonzalez-Fernandez et al., 2013; González-Fernández et al., 2014) and in planta (Shah et al., 2012) experiments.
In fungi, extracellular proteins may be secreted by both the classical pathway, via endoplasmatic reticulum and the Golgi complex, and unconventional export route non-mediated by ER-derived (Girard et al., 2013; Vincent and Bedon, 2013). The B. cinerea secretome analysis by using Fungal Secretome Database (FSD; http://fsd.snu.ac.kr/website) showed that 16% of the gene products are predicted to be secreted proteins by the canonical pathway, in which proteins have an N-terminal peptide signal (Choi et al., 2010). This percentage should be increased because it is suggested that various kinds of non-classical export pathways may exist in B. cinerea (Jain et al., 2008).
Most studies about B. cinerea secretome have been carried out through in vitro experiments, mainly because of two problems: (i) the fungal secretome is a complex analysis due to the ratio cell concentration fungus/host, and (ii) the genomic annotation quality for the two partners (Girard et al., 2013; Vincent and Bedon, 2013). To avoid the first difficulty, the in vitro experimental protocols try to simulate the in vivo conditions, where the fungus is cultured in the presence of more or less-purified fractions of its plant host (Shah et al., 2009a,b; Espino et al., 2010; Fernandez-Acero et al., 2010; González-Fernández et al., 2014). With respect to the second difficulty, it is essential that the fungal and plant genomes be sequenced in order to distinguish between fungal and plant proteins (Girard et al., 2013; Vincent and Bedon, 2013).
In the last years, great efforts have been made to explain the B. cinerea secretome complexity and versatility using secretomics from in vitro experiments. One of the first studies showed that changes during the fruit ripening process seemed to have an important role in the latent infection activation, which is probably not only dependent on changes in the pectin esterification degree of the plant cell wall (Shah et al., 2009b). By the other hand, this fungus showed significant changes in the composition and relative abundance of secreted proteins that are specific to a particular growth condition (Shah et al., 2009a,b; Espino et al., 2010; Fernandez-Acero et al., 2010; González-Fernández et al., 2014). In the presence of favorable nutrient sources, the fungus increased its protein secretion (Fernandez-Acero et al., 2010). Another key point in the establishment of a successful infection is the extracellular secreted proteins during the spore germination on the plant surface. The early secretome composition was not as variable as it could be expected, and B. cinerea seems to secrete a common set of proteins during its germination regardless of growth condition, together with a lower number of proteins that are specifics for these growth conditions (Espino et al., 2010). The aspartic protease BcAP8, a α-amylase 1, and a cerato-platanin domain-containing protein (BcPls1) were detected in the six strains. Two different pectinesterases, one endo-polygalacturonase (PG), a glucan-1,3-β-glucosidase, a glucoamylase, a carboxypeptidase S1, and a choline dehydrogenase were detected in some of the six strains (González-Fernández et al., 2014). Moreover, some hypothetical proteins, which showed differences among strains, were identified in secretome (González-Fernández et al., 2014). An example was the predicted protein BC1G_08642.1, which contains a carbohydrate-recognition domain similar to those ones included in plant and bacterial AB-toxins, glycosidases, or proteases (González-Fernández et al., 2014).
The pH was also shown to affect the B. cinerea secretome. Proteins related to proteolysis as the BcAP8, a family S53 protease, a serine-type carboxypeptidase, and a metalloprotease (Merops M35) were induced at pH 4 (similar to mature fruit environment), whereas at pH 6 (similar to leaf environment) most of the up-accumulated proteins were CWDEs (Li et al., 2012). These results were in concordance because ripe fruits generally have lower tissue pH and weakened cell walls, and they accumulate a lot of pathogenesis-related proteins for defense, so inducing protease secretion is more important than the CWDE secretion (Manteau et al., 2003). In contrast, for leaves and stem, which have higher tissue pH and harder cell walls, CWDEs are secreted in greater quantity than the proteases (Manteau et al., 2003). Therefore, pH present in each tissue could regulate the expression of secreted proteins in B. cinerea to activate the machinery required for invasion. Finally, metals present in the ambient as cooper, zinc, nickel, or cadmium, also modified the oxidoreductase production and CWDE secretion (Cherrad et al., 2012).
The comparison of these data suggests that B. cinerea secretes a common set of proteins as well as a pool of different ones, depending on the in vitro growth conditions and the strain, which were used, and making the secretome highly adaptive. Therefore, B. cinerea may greatly change the composition of the secreted protein set to satisfy the requirements of these different growing conditions. Next, we are going to discuss the importance of some B. cinerea secreted proteins, which have been identified by using a proteomic approach from in vitro experiments, and we hypothesize about their putative function in the B. cinerea interaction with its host (Table 1). All proteins, which are described in Table 1, except the pectinesterase BC1G_06840.1, were predicted to be secreted by the classical secretion pathway (Espino et al., 2010; González-Fernández et al., 2014).
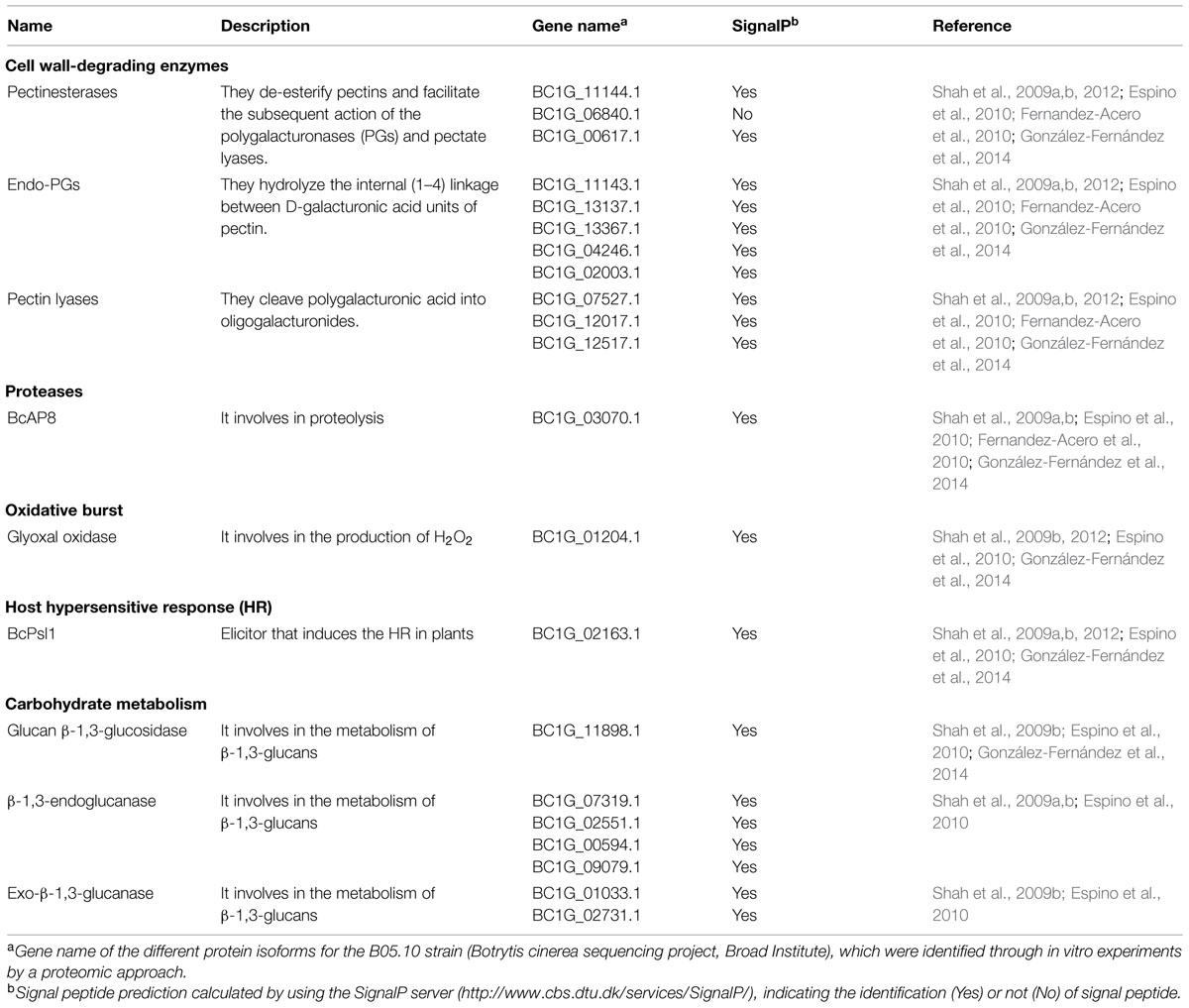
TABLE 1. Secreted proteins discussed in this mini-review, which were identified by a proteomic approach from both in vitro and in planta studies, and which may be involved in the B. cinerea–host interaction.
Several enzymes could take part in the metabolism of β-1,3-glucans that are part of the fungal cell wall. This polysaccharide is secreted by B. cinerea in high amounts to the medium for which several functions have been proposed, including extracellular energy storage, or the adhesion of the conidia to plant surface during germination (Stahmann et al., 1992). A glucan β-1,3-glucosidase, a β-1,3-endoglucanase, and an exo-β-1,3-glucanase were secreted early in B. cinerea development (Espino et al., 2010). The same β-1,3-endoglucanase and other glucanases were detected in the B. cinerea interaction with tomato (Shah et al., 2012). The degradation of β-1,3-glucans may contribute to activating the induction of the programmed cell death in plant cells by generating elicitors in the form of β-(1,3)(1,6)-oligomers (Espino et al., 2010).
Botrytis cinerea is provided with a set of CWDEs able to degrade the cell wall to allow plant tissue colonization to get nutrients (Prins et al., 2000; Shah et al., 2009b; Gonzalez-Fernandez and Jorrin-Novo, 2012). Despite having a broad spectrum of host plant species, this fungus has predilection for dicotyledonous plants with cell walls rich in pectin, which usually is highly methyl-esterified, in order to protect them from fungal PGs and pectate lyases (Prins et al., 2000). Therefore, pectin-degrading enzymes, such as pectinesterases, PGs, and pectate lyases, have a very important role in the cell wall degradation and successful fungal invasion (Kars and van Kan, 2007; Zhang and van Kan, 2013). Recently, the B. cinerea BcDW1 genome was sequenced, and the complete genes of a large set of candidate secreted CWDEs were found, among which were found 19 PGs, 15 xyloglucanases, 10 cutinases, and 9 pectin/pectate lyases (Blanco-Ulate et al., 2013). These enzymes are categorized as carbohydrate-active enzymes (CAZymes), which are proteins that degrade, modify, or create glycosidic bonds, and that are also included in diverse glycoside hydrolase (GH) families (Kubicek et al., 2014). By a genome-wide transcriptional profiling analysis, 229 potentially secreted CAZymes were expressed in three different host infected with B. cinerea (Blanco-Ulate et al., 2014). These results suggest that B. cinerea targets analogous wall polysaccharide matrix on leaves and fruit, and may selectively attack host wall polysaccharide substrates depending on the host tissue.
Two pectinesterases, the endo-PG BcPG1 and the pectin lyase A, were evidenced in the B. cinerea secretome by using a combination of 1-DE-MALDI-TOF/TOF MS/MS and label-free shotgun nUPLC–MSE techniques (González-Fernández et al., 2014), as well as in the B. cinerea interaction with tomato (Shah et al., 2012). It is known that endo-PGs show not only an elevated genetic variation, and a specialization among them, but also a potential diversification, interacting directly with host defenses (Rowe and Kliebenstein, 2007). Two endo-PGs (BcPG1 and BcPG2) out of the six previously characterized have been reported to be required for full virulence (Choquer et al., 2007; Zhang et al., 2014). For example, the BcPG1 was only detected in five of six wild-type strains (González-Fernández et al., 2014), and in the early secretome (Espino et al., 2010). Moreover, the BcPG1 was secreted by the B05.10 strain grown in glucose, tomato, or kiwifruit, although it was almost absent in strawberry (Espino et al., 2010). This enzyme exemplifies the adaptable secretome nature depending on the strain specificity by its host, and the diversity of the B. cinerea armament resulting in an over-kill strategy. Thus, some proteins, which may be required for full virulence to attack one host, may differ from others needed to invade another different host.
A high amount of proteases have been identified in the B. cinerea secretome (Shah et al., 2009a; Espino et al., 2010; Fernandez-Acero et al., 2010; González-Fernández et al., 2014). It could be explained due to their role (Espino et al., 2010): (i) generating amino acids to sustain fungal growth; (ii) contributing to cell wall softening, and, therefore, to fungal hyphal penetration; and (iii) degrading the defense proteins which plants secrete against the pathogens. The high diversity of the proteases found may also be reflecting the diverse nature of their substrate. Plant proteins have a great variety of structures by nature, and it may be needed a diverse pool of protease for its degradation (Espino et al., 2010). One of the most abundant proteins secreted by B. cinerea is the BcAP8 (Shah et al., 2009a; Espino et al., 2010; Fernandez-Acero et al., 2010; González-Fernández et al., 2014). This protease was detected in the secretome of the six wild-type strains analyzed, varying in abundance depending on the strain (González-Fernández et al., 2014). Besides, its absence in the secretome of B05.10 strain caused the reduction of up to 90% in the secreted protease activity, a rise in the intensity of high molecular weight protein bands in the 1-DE profile, and a decrease, or even the disappearance of the smallest bands (Espino et al., 2010). However, in other study, there was no changes in virulence between the Bcap8 knockout mutant and the wild-type strain B05.10 in tomato leaves and fruit (ten Have et al., 2010), and the BcAP8 was not detected in the proteomic analysis from B. cinerea–tomato interaction (Shah et al., 2012). The result indicated that BcAP8 might not be responsible for virulence in B. cinerea. The loss of BcAP8 activity in ΔBcap8 mutant may be partially compensated by expression of other genes from the Bcap family, or BcAP8 activity plays other important roles in the life cycle of the pathogen (Li et al., 2012), supporting the fact of the great adaptability of B. cinerea.
Other key mechanism in the B. cinerea invasion strategy is the active generation of an oxidative burst by the fungus during the first infection stages (van Kan, 2006; Choquer et al., 2007; Amselem et al., 2011). Several enzymes have been studied as potential ROS generators, such as a Cu–Zn superoxide dismutase (BcSOD1), whose mutation results in reduced virulence in several different hosts, and a glucose oxidase (BcGOD1; Rolke et al., 2004); however, none of them was found by proteomics (Espino et al., 2010; González-Fernández et al., 2014). There are numerous extracellular enzymes that have been previously characterized as potential ROS generators in fungi (Espino et al., 2010). An example is the glyoxal oxidase, which has proved to be an important determinant of cell morphology and virulence in B. cinerea, Ustilago maydis, or Phanerochaete chrysosporium (Soanes et al., 2008). This enzyme may also produce H2O2, together with other enzymes as superoxide dismutase (Soanes et al., 2008). Additionally, a quinoprotein glucose dehydrogenase and a cellobiose dehydrogenase were detected in the early secretome that may also generate ROS (Espino et al., 2010). The glyoxal oxidase and the cellobiose dehydrogenase were also detected in the B. cinerea interaction with tomato but only in a ripening inhibited mutant tomato (Shah et al., 2012).
Pathogens produce molecules which can activate plant defense responses so-called HR (Jones and Dangl, 2006). Unlike biotrophs, necrotrophic fungi as B. cinerea takes benefit of the HR, generating dead tissue around the infected area for rapid colonization of their hosts (Govrin and Levine, 2000; Williamson et al., 2007; Hématy et al., 2009). One protein, which may help to induce HR in plants, may be the BcPls1. This protein is a member of the hydrophobin-like cerato-platanin family, which have been reported to be a secreted protein, acting as elicitors, and, in some cases, as pathogenicity factors (Gaderer et al., 2014; Baccelli, 2015). The BcPls1 was found to have high levels of secretion in the B05.10 strain when it was grown in vitro by using different media (Shah et al., 2009a,b; Espino et al., 2010), and in planta B. cinerea–tomato interaction (Shah et al., 2012), as well as when different wild-type strains were studied (González-Fernández et al., 2014), suggesting that this protein may play an important role in host–pathogen interactions. It was required for full virulence in B. cinerea, and induced necrosis in several hosts (Frías et al., 2011). Some HR symptoms were the induction of autofluorescence and ROS, the electrolyte leakage, and the cytoplasm shrinkage (Frías et al., 2011). All these observations may imply that the cerato-platanin proteins are recognized by the immune system of the plant, and this reconnaissance induces with the programmed death of the affected cells (Frías et al., 2011).
Oxalic acid has been reported as a pathogenicity factor in B. cinerea (Lyon et al., 2007), and in the related necrotrophic fungus Sclerotinia sclerotiorum (Kim et al., 2008). Its physiological roles in pathogenesis include: (i) the enhancing of PG activity to promote cell wall degradation, (ii) suppression of the plant oxidative burst, (iii) the plant-protection enzymes inhibition, (iv) involvement in the pH signaling, (v) deregulation of stomatal guard cell closure, (vi) induction of apoptosis-like cell death, and (vii) alteration of the cellular redox status in the plant (Amselem et al., 2011). Oxalic acid secretion makes an optimal acidic environment for the pathogenicity factor expression and secretion in B. cinerea, such as CWDEs (Manteau et al., 2003), peptidases (ten Have et al., 2010), and for the phytotoxin biosynthesis (Durán-Patrón et al., 2004). Oxaloacetate is the precursor of oxalic acid. The malate dehydrogenase (MDH) catalyzes the reversible conversion of oxaloacetate and malate. A low-gene expression levels of fungal MDH, in combination with the absence of botrydial and dihydrobotrydial secretion, was found in a less virulent B. cinerea strain than in the more virulent one (Fernandez-Acero et al., 2007). Fungal MDH was also found in the mycelium proteins of six B. cinerea wild-type strains with significant quantitative differences, being higher in the strains isolated from green material (whose pH > 6; González-Fernández et al., 2014). This implies that fungal MDH plays a key role in the biosynthesis of oxalic acid to produce a more appropriate, ecological niche for the fungal pathogenic activities.
The understanding of how the B. cinerea secretome affects the interactions between this fungus and its host has become a hard task due to its adaptability at different growth conditions and plant species. B. cinerea populations manifest a significant phenotypic variability with respect to their level of aggressiveness, oxidative burst occurring during the infection, and toxin production (Prins et al., 2000; Elad et al., 2007). Considering the results from the different studies, this fungus seems to be efficiently adapted to their different host plants in terms of host preference rather than in a real host specialization (Choquer et al., 2007). Hosts and their parasites are implicated in an evolutionary fighting marked by an adaptation and a counter-adaptation of host defense and pathogen attack mechanisms. Thus, the selective influence perform by plants may affect the virulence factor evolution at population level (Choquer et al., 2007). Certain virulence factors can be important for one strain on one particular host species, but they might be dispensable on other host species, or they might be dispensable for a different strain.
Secretomics has supplied important advances in the identification of extracellular proteins, which are secreted by B. cinerea, and may be involved in the interaction with its host that result in a successful infection. So far, only the 10% of the secreted proteins, which were predicted to be involved in the classical export pathway from B. cinerea (Choi et al., 2010), has been identified by proteomic approaches (Gonzalez-Fernandez and Jorrin-Novo, 2012; González-Fernández et al., 2014). Due to its high adaptability to different hosts, more specific studies both in vitro and in planta need to be made for each host to discover the factors involved in the infection mechanisms. Finally, a very important aspect in secretomics is the knowledge of the secretion pathways (Girard et al., 2013; Vincent and Bedon, 2013). Therefore, other point for further studies may be the secretion pathway study, and the cataloging of the proteins according to their secretion mechanisms.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This publication has been supported by the Ministry of Education (Secretaría de Educación Pública, SEP) of the Federal Government of México, through the Teacher Professional Development Program (Programa para el Desarrollo Profesional Docente, para el Tipo Superior, PRODEP), and the Autonomous University of Ciudad Juárez (UACJ). We wish to thank the Spanish Ministry of Science and Innovation (BotBank Project, EUI2008-03686), the Regional Government of Andalusia (Junta de Andalucía), and the University of Córdoba (AGR-0164: Agricultural and Plant Biochemistry and Proteomics Research Group) for their previous support.
Afroz, A., Ali, A., G. M., Mir, A., and Komatsu, S. (2011). Application of proteomics to investigate stress-induced proteins for improvement in crop protection. Plant Cell Rep. 30, 745–763. doi: 10.1007/s00299-010-0982-x
Agrawal, G. K., Jwa, N. S., Lebrun, M. H., Job, D., and Rakwal, R. (2013). Plant secretome: unlocking secrets of the secreted proteins. Proteomics 10, 799–827. doi: 10.1002/pmic.200900514
Allwood, J. W., Ellis, D. I., and Goodacre, R. (2008). Metabolomic technologies and their application to the study of plants and plant-host interactions. Physiol. Plant. 132, 117–135. doi: 10.1111/j.1399-3054.2007.01001.x
Amselem, J., Cuomo, C. A., van Kan, J. A., Viaud, M., Benito, E. P., Couloux, A., et al. (2011). Genomic analysis of the necrotrophic fungal pathogens Sclerotinia sclerotiorum and Botrytis cinerea. PLoS Genet. 7:e1002230. doi: 10.1371/journal.pgen.1002230
Baccelli, I. (2015). Cerato-platanin family proteins: one function for multiple biological roles? Front. Plant Sci. 5:769. doi: 10.3389/fpls.2014.00769
Bhadauria, V., Banniza, S., Wang, L. X., Wei, Y. D., and Peng, Y. L. (2010). Proteomic studies of phytopathogenic fungi, oomycetes and their interactions with hosts. Eur. J. Plant Pathol. 126, 81–95. doi: 10.1007/s10658-009-9521-4
Bhadauria, V., Banniza, S., Wei, Y., and Peng, Y. L. (2009). Reverse genetics for functional genomics of phytopathogenic fungi and oomycetes. Comp. Funct. Genomics 2009, 11. doi: 10.1155/2009/380719
Blanco-Ulate, B., Allen, G., Powell, A. L. T., and Cantu, D. (2013). Draft genome sequence of Botrytis cinerea BcDW1, inoculum for noble rot of grape berries. Genome Announc. 1, e252-13. doi: 10.1128/genomeA.00252-13
Blanco-Ulate, B., Morales-Cruz, A., Amrine, K. C., Labavitch, J. M., Powell, A. L., and Cantu, D. (2014). Genome-wide transcriptional profiling of Botrytis cinerea genes targeting plant cell walls during infections of different hosts. Front. Plant Sci. 5:435. doi: 10.3389/fpls.2014.00435
Cherrad, S., Girard, V., Dieryckx, C., Goncalves, I. R., Dupuy, J.-W., Bonneu, M., et al. (2012). Proteomic analysis of proteins secreted by Botrytis cinerea in response to heavy metal toxicity. Metallomics 4, 835–846. doi: 10.1039/c2mt20041d
Choi, J., Park, J., Kim, D., Jung, K., Kang, S., and Lee, Y. H. (2010). Fungal secretome database: integrated platform for annotation of fungal secretomes. BMC Genomics 11:105. doi: 10.1186/1471-2164-11-105
Choquer, M., Fournier, E., Kunz, C., Levis, C., Pradier, J. M., Simon, A., et al. (2007). Botrytis cinerea virulence factors: new insights into a necrotrophic and polyphageous pathogen. FEMS Microbiol. Lett. 277, 1–10. doi: 10.1111/j.1574-6968.2007.00930.x
Dean, R., Van Kan, J. A., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Durán-Patrón, R., Cantoral, J. M., Hernández-Galán, R., Hanson, J. R., and Collado, I. G. (2004). The biodegradation of the phytotoxic metabolite botrydial by its parent organism, Botrytis cinerea. J. Chem. Res. 2004, 441–443. doi: 10.3184/0308234041423583
Elad, Y., Williamson, B., Tudzynski, P., and Delen, N. (2007). Botrytis: Biology, Pathology and Control. Dordrecht: Springer.
Espino, J. J., Gutierrez-Sanchez, G., Brito, N., Shah, P., Orlando, R., and Gonzalez, C. (2010). The Botrytis cinerea early secretome. Proteomics 10, 3020–3034. doi: 10.1002/pmic.201000037
Fernandez-Acero, F. J., Colby, T., Harzen, A., Carbu, M., Wiencke, U., Cantoral, J. M., et al. (2010). 2-DE proteomic approach to the Botrytis cinerea secretome induced with different carbon sources and plant-based elicitors. Proteomics 2010, 2270–2280. doi: 10.1002/pmic.200900408
Fernandez-Acero, F. J., Jorge, I., Calvo, E., Vallejo, I., Carbu, M., Camafeita, E., et al. (2007). Proteomic analysis of phytopathogenic fungus Botrytis cinerea as a potential tool for identifying pathogenicity factors, therapeutic targets and for basic research. Arch. Microbiol. 187, 207–215. doi: 10.1007/s00203-006-0188-3
Frías, M., González, C., and Brito, N. (2011). BcSpl1, a cerato-platanin family protein, contributes to Botrytis cinerea virulence and elicits the hypersensitive response in the host. New Phytol. 192, 483–495. doi: 10.1111/j.1469-8137.2011.03802.x
Gaderer, R., Bonazza, K., and Seidl-Seiboth, V. (2014). Cerato-platanins: a fungal protein family with intriguing properties and application potential. Appl. Microbiol. Biotechnol. 98, 4795–4803. doi: 10.1007/s00253-014-5690-y
Girard, V., Dieryckx, C., Job, C., and Job, D. (2013). Secretomes: the fungal strike force. Proteomics 13, 597–608. doi: 10.1002/pmic.201200282
Gonzalez-Fernandez, R., Aloria, K., Arizmendi, J. M., and Jorrin-Novo, J. V. (2013). Application of label-free Shotgun nUPLC-MS(E) and 2-DE approaches in the study of Botrytis cinerea mycelium. J. Proteome Res. 12, 3042–3056. doi: 10.1021/pr3010937
González-Fernández, R., Aloria, K., Valero-Galván, J., Redondo, I., Arizmendi, J. M., and Jorrín-Novo, J. V. (2014). Proteomic analysis of mycelium and secretome of different Botrytis cinerea wild-type strains. J. Proteomics 97, 195–221. doi: 10.1016/j.jprot.2013.06.022
González-Fernández, R., and Jorrin-Novo, J. V. (2010). “Proteomics of fungal plant pathogens: the case of Botrytis cinerea,” in Current Research, Technology and Education Topics in Applied Microbiology and Microbial Biotechnology, ed. A. M. Vilas (Badajoz: Formatex Research Center), 205–217.
Gonzalez-Fernandez, R., and Jorrin-Novo, J. V. (2012). Contribution of proteomics to the study of plant pathogenic fungi. J. Proteome Res. 11, 3–16. doi: 10.1021/pr200873p
Gonzalez-Fernandez, R., Prats, P., and Jorrin-Novo, J. V. (2010). Proteomics of plant pathogenic fungi. J. Biomed. Biotechnol. 2010, 36. doi: 10.1155/2010/932527
Govrin, E. M., and Levine, A. (2000). The hypersensitive response facilitates plant infection by the necrotrophic pathogen Botrytis cinerea. Curr. Biol. 10, 751–757. doi: 10.1016/S0960-9822(00)00560-1
Hématy, K., Cherk, C., and Somerville, S. (2009). Host - pathogen warfare at the plant cell wall. Curr. Opin. Plant Biol. 12, 406–413. doi: 10.1016/j.pbi.2009.06.007
Jain, P., Podila, G., and Davis, M. (2008). Comparative analysis of non-classically secreted proteins in Botrytis cinerea and symbiotic fungus Laccaria bicolor. BMC Bioinformatics 9:O3. doi: 10.1186/1471-2105-9-S10-O3
Jones, J. D. G., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kars, I., and van Kan, J. A. L. (2007). “Extracellular enzymes and metabolites involved in pathogenesis of Botrytis,” in Botrytis: Biology, Pathology and Control, eds Y. Elad, B. Williamson, P. Tudzynski, and N. Delen (Dordrecht: Springer Netherlands), 99–118.
Kim, K. S., Min, J. Y., and Dickman, M. B. (2008). Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Mol. Plant Microbe Interact. 21, 605–612. doi: 10.1094/MPMI-21-5-0605
Kubicek, C. P., Starr, T. L., and Glass, N. L. (2014). Plant cell wall-degrading enzymes and their secretion in plant-pathogenic fungi. Ann. Rev. Phytopathol. 52, 427–451. doi: 10.1146/annurev-phyto-102313-045831
Li, B., Wang, W., Zong, Y., Qin, G., and Tian, S. (2012). Exploring pathogenic mechanisms of Botrytis cinerea secretome under different ambient pH based on comparative proteomic analysis. J. Proteome Res. 11, 4249–4260. doi: 10.1021/pr300365f
Lyon, G. D., Goodman, B. A., and Williamson, B. (2007). “Botrytis cinerea perturbs redox processes as an attack strategy in plants,” in Botrytis: Biology, Pathology and Control, eds Y. Elad, B. Williamson, P. Tudzynski, and N. Delen (Dordrecht: Springer), 119–141.
Manteau, S., Abouna, S., Lambert, B., and Legendre, L. (2003). Differential regulation by ambient pH of putative virulence factor secretion by the phytopathogenic fungus Botrytis cinerea. FEMS Microbiol. Ecol. 43, 359–366. doi: 10.1111/j.1574-6941.2003.tb01076.x
Prins, T. W., Tudzynski, P., von Tiedemann, A., Tudzynski, B., ten Have, A., Hansen, M. E., et al. (2000). “Infection strategies of Botrytis cinerea and related necrotrophic pathogens,” in Fungal Pathology, ed. J. W. Kronstad (Dordrech: Kluwer Academic Publishers), 33–64.
Quirino, B. F., Candido, E. S., Campos, P. F., Franco, O. L., and Kruger, R. H. (2010). Proteomic approaches to study plant-pathogen interactions. Phytochemistry 71, 351–362. doi: 10.1016/j.phytochem.2009.11.005
Rolke, Y., Liu, S., Quidde, T., Williamson, B., Schouten, A., Weltring, K. M., et al. (2004). Functional analysis of H(2)O(2)-generating systems in Botrytis cinerea: the major Cu-Zn-superoxide dismutase (BCSOD1) contributes to virulence on French bean, whereas a glucose oxidase (BCGOD1) is dispensable. Mol. Plant Pathol. 5, 17–27. doi: 10.1111/j.1364-3703.2004.00201.x
Rowe, H. C., and Kliebenstein, D. J. (2007). Elevated genetic variation within virulence-associated Botrytis cinerea polygalacturonase loci. Mol. Plant Microbe Interact. 20, 1126–1137. doi: 10.1094/MPMI-20-9-1126
Shah, P., Atwood, J. A., Orlando, R., El Mubarek, H., Podila, G. K., and Davis, M. R. (2009a). Comparative proteomic analysis of Botrytis cinerea secretome. J. Proteome Res. 8, 1123–1130. doi: 10.1021/pr8003002
Shah, P., Gutierrez-Sanchez, G., Orlando, R., and Bergmann, C. (2009b). A proteomic study of pectin-degrading enzymes secreted by Botrytis cinerea grown in liquid culture. Proteomics 9, 3126–3135. doi: 10.1002/pmic.200800933
Shah, P., Powell, A. L., Orlando, R., Bergmann, C., and Gutierrez-Sanchez, G. (2012). Proteomic analysis of ripening tomato fruit infected by Botrytis cinerea. J. Proteome Res. 11, 2178–2192. doi: 10.1021/pr200965c
Soanes, D. M., Alam, I., Cornell, M., Wong, H. M., Hedeler, C., Paton, N. W., et al. (2008). Comparative genome analysis of filamentous fungi reveals gene family expansions associated with fungal pathogenesis. PLoS ONE 3:e2300. doi: 10.1371/journal.pone.0002300
Stahmann, K. P., Pielken, P., Schimz, K. L., and Sahm, H. (1992). Degradation of Extracellular beta-(1,3)(1,6)-d-Glucan by Botrytis cinerea. Appl. Environ. Microbiol. 58, 3347–3354.
Tan, K. C., Ipcho, S. V., Trengove, R. D., Oliver, R. P., and Solomon, P. S. (2009). Assessing the impact of transcriptomics, proteomics and metabolomics on fungal phytopathology. Mol. Plant Pathol. 10, 703–715. doi: 10.1111/j.1364-3703.2009.00565.x
ten Have, A., Espino, J. J., Dekkers, E., Van Sluyter, S. C., Brito, N., Kay, J., et al. (2010). The Botrytis cinerea aspartic proteinase family. Fungal Genet. Biol. 47, 53–65. doi: 10.1016/j.fgb.2009.10.008
van Kan, J. A. L. (2006). Licensed to kill: the lifestyle of a necrotrophic plant pathogen. Trends Plant Sci. 11, 247–253. doi: 10.1016/j.tplants.2006.03.005
Vincent, D., and Bedon, F. (2013). Secretomics of plant-fungus associations: more secrets to unravel. J. Plant Biochem. Physiol. 1, e117. doi: 10.4172/2329-9029.1000e117
Williamson, B., Tudzynski, B., Tudzynski, P., and van Kan, J. A. L. (2007). Botrytis cinerea: the cause of grey mould disease. Mol. Plant Pathol. 8, 561–580. doi: 10.1111/j.1364-3703.2007.00417.x
Zhang, L., Kars, I., Essenstam, B., Liebrand, T. W. H., Wagemakers, L., Elberse, J., et al. (2014). Fungal endopolygalacturonases are recognized as microbe-associated molecular patterns by the Arabidopsis receptor-like protein RESPONSIVENESS TO BOTRYTIS POLYGALACTURONASES1. Plant Physiol. 164, 352–364. doi: 10.1104/pp.113.230698
Keywords: Botrytis cinerea, secretomics, plant pathogenic fungi, fungal secretome, fungi–plant interactions
Citation: González-Fernández R, Valero-Galván J, Gómez-Gálvez FJ and Jorrín-Novo JV (2015) Unraveling the in vitro secretome of the phytopathogen Botrytis cinerea to understand the interaction with its hosts. Front. Plant Sci. 6:839. doi: 10.3389/fpls.2015.00839
Received: 20 May 2015; Accepted: 24 September 2015;
Published: 09 October 2015.
Edited by:
Delphine Vincent, Department of Environment and Primary Industries, AustraliaReviewed by:
Dario Cantu, University of California, Davis, USACopyright © 2015 González-Fernández, Valero-Galván, Gómez-Gálvez and Jorrín-Novo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Raquel González-Fernández, cmFxdWVsLmdvbnphbGV6QHVhY2oubXg=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.