
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 14 October 2015
Sec. Plant Genetics and Genomics
Volume 6 - 2015 | https://doi.org/10.3389/fpls.2015.00832
After yield, quality is one of the most important aspects of rice breeding. Preference for rice quality varies among cultures and regions; therefore, rice breeders have to tailor the quality according to the preferences of local consumers. Rice quality assessment requires routine chemical analysis procedures. The advancement of molecular marker technology has revolutionized the strategy in breeding programs. The availability of rice genome sequences and the use of forward and reverse genetics approaches facilitate gene discovery and the deciphering of gene functions. A well-characterized gene is the basis for the development of functional markers, which play an important role in plant genotyping and, in particular, marker-assisted breeding. In addition, functional markers offer advantages that counteract the limitations of random DNA markers. Some functional markers have been applied in marker-assisted breeding programs and have successfully improved rice quality to meet local consumers’ preferences. Although functional markers offer a plethora of advantages over random genetic markers, the development and application of functional markers should be conducted with care. The decreasing cost of sequencing will enable more functional markers for rice quality improvement to be developed, and application of these markers in rice quality breeding programs is highly anticipated.
The important attributes of rice are its cooking and eating qualities, its phytochemicals and its micronutrients. The quality of rice needs to match the preferences of local consumers in order to be acceptable. Generally, Japanese people prefer short and sticky rice, whereas Indians prefer aromatic basmati rice which elongates when cooked. Furthermore, rice quality affects the market value, given that better quality rice is able to fetch a higher premium. Indian basmati rice and Thai jasmine rice are highly priced due to their distinctive aroma when cooked. The growing income and food diversification in Asian countries such as China (Sumner et al., 2001) and some European countries (Ferrero and Nguyen, 2004) have led consumers to prefer better quality rice.
While people in some parts of the world seek a better cooking and eating quality of their rice, people in other areas seek improved nutrition. Although micronutrients are only required in small quantities, they are necessary to maintain proper bodily function. In fact, two billion people worldwide suffer from micronutrient deficiencies, particularly in vitamin A, iodine, iron (Fe), and zinc (Zn) (World Health Organization, 2007). Therefore, research has been undertaken to increase the micronutrient content in rice to avert nutrient deficiency in the human diet, especially for populations where micronutrient deficiency is prevalent. Recently, Zn-biofortified rice has been developed to avert Zn deficiency in the diet of Bangladeshi people, particularly in children (Ahmad, 2013). The International Rice Research Institute (IRRI) is expected to release Fe-rich rice by the year 2029 to alleviate Fe deficiency anemia in needy countries (David, 2014).
Due to consumers’ demand for better rice quality, rice breeders all over the world are endeavoring to develop rice varieties with improved qualities that meet local demand. The quality of a rice variety is assessed after harvesting the grains from the plant. Prior to an assessment of the acceptability of the rice variety by panelists, the quality parameters of cooking and eating quality, and phytochemical and micronutrient composition are determined by using standard procedures (Dela Cruz and Khush, 2000). Determination of the quality parameters in each individual plant is laborious and time consuming. Certain chemical analyses might require large grain samples, which can be destructive to the plant material, especially during the early stage of breeding when the breeder’s seed supply is limited.
The advent of molecular marker technology in genetic analysis has revolutionized the research on rice quality. From the time scientists first ventured into using molecular markers, from the earliest protein markers to the current DNA markers, substantial effort in molecular mapping has identified chromosome regions carrying genes of interest. Undeniably, commonly used DNA markers, such as restriction fragment length polymorphism (RFLP), randomly amplified polymorphic DNA (RAPD), and simple sequence repeats (SSRs) have contributed to the mapping and association studies that led to the discovery of genes of interest. However, these DNA markers are derived randomly from polymorphic sites of the genome, and some can be located far from the gene of interest, which might be independent from the phenotype. Functional markers (FMs, also known as perfect markers) are an alternative to random DNA markers. FMs are developed from polymorphic sites within genes that cause phenotypic trait variation (Andersen and Lübberstedt, 2003). In contrast with random DNA markers, FMs are directly linked to the allele of the trait of interest (Varshney et al., 2005). Therefore, FMs are outcompeting random DNA markers, especially in marker-assisted breeding (MAB). Thus far, numerous FMs have been developed for the breeding of quality rice (Table 1), and some of them have been applied to breeding programs that have delivered desirable quality traits unambiguously (Yi et al., 2009; Jin et al., 2010).
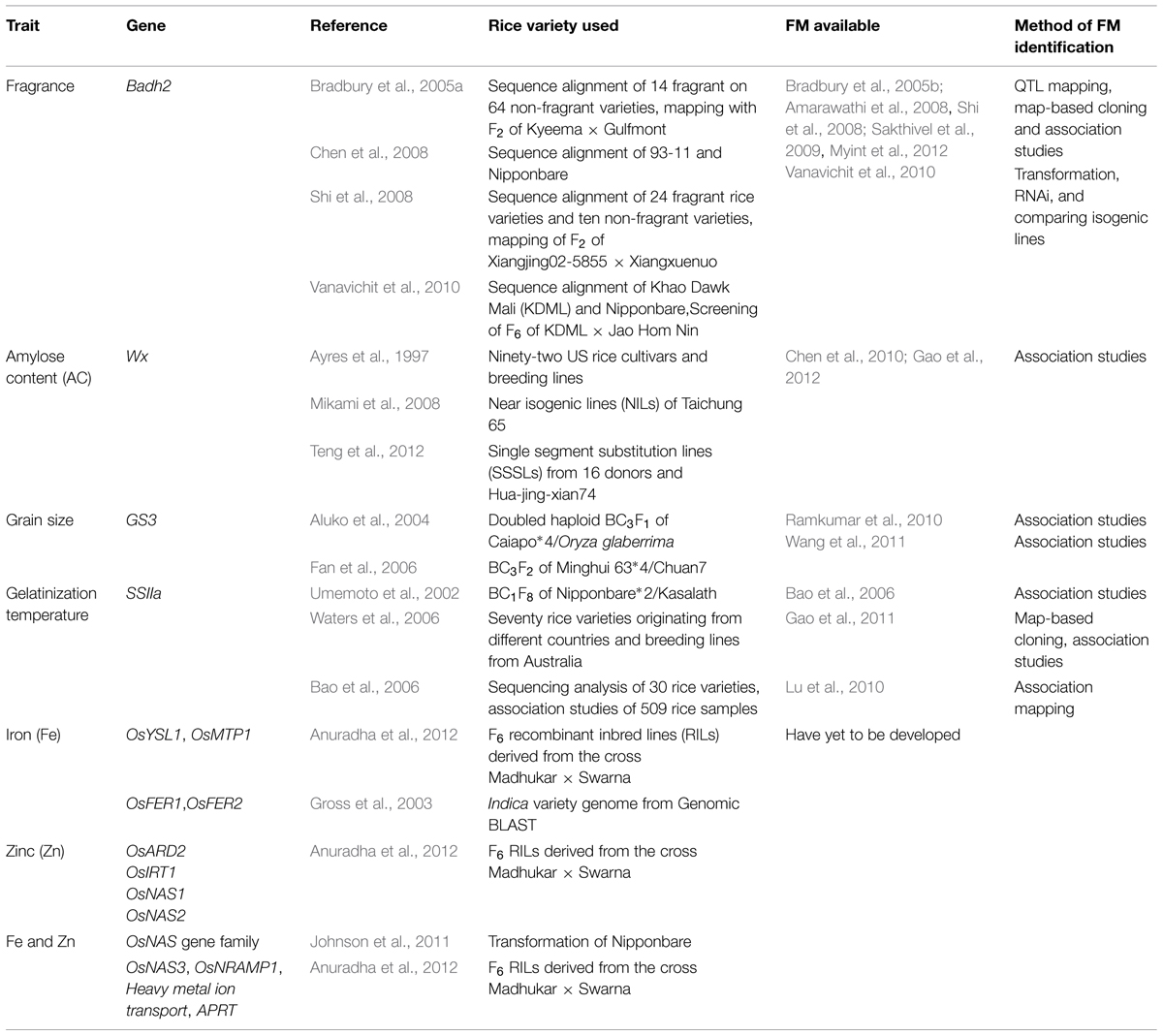
TABLE 1. Candidate genes for Functional marker (FM) development and developed FM available for quality rice breeding.
The advantage of FMs is that they can be applied to any population; random markers discovered from quantitative trait loci (QTL) mapping might be population specific. Parents from the QTL mapping have different genetic backgrounds, and might not be polymorphic when applied to other populations (Lübberstedt et al., 2005; Miklas et al., 2006; Gupta et al., 2010). In contrast, FMs can be used regardless of the genetic background of the population under study and applied to any population without revalidating markers or the QTL relationship (Gupta et al., 2010).
FMs are developed from functional gene motifs and, therefore, have complete linkage to the desired allele (Andersen and Lübberstedt, 2003). Due to the complete linkage of an FM with the target gene and the absence of recombination between the marker and the gene, the loss of information and the false selection in MAB can be prevented (Ingvardsen et al., 2008). Phenotypic validation in MAB that uses random DNA markers is essential to ensure that the target gene and marker are transferred together to the progeny; however, using FMs eliminates the need for phenotypic validation (Andersen and Lübberstedt, 2003). Therefore, FMs are more efficient than random DNA markers in MAB applications.
Another major concern in MAB is linkage drag. Random DNA markers might be located far from the target genes; therefore, when they are applied in MAB, a larger donor segment will be introgressed into the recipient parent or backcross progeny. Undesirable genes might be transferred along with the target gene, resulting in reduced performance of the phenotypic trait. To minimize linkage drag in MAB, Hospital (2001) suggested the use of flanking markers closely linked to the introgressed gene in a large population size to obtain double-recombinant genotypes. Alternatively, FMs can reduce linkage drag, particularly in foreground selection by genotyping a smaller population size (Bagge and Lübberstedt, 2008; Gupta et al., 2010).
Eating quality refers to the consumers’ sensory perception of cooked rice, which is related to characters such as flavor and texture (Hsu et al., 2014). Cooking quality refers to chemical reactions resulting from the cooking of the grain, such as gelatinization temperature (GT), kernel elongation, and water uptake (Juliano and Perez, 1984; Hsu et al., 2014). Amylose, a constituent of starch which comprises approximately 95% of the grain dry weight (Fitzgerald et al., 2009), is an important determinant of eating and cooking qualities. In addition, amylose content (AC) affects the glycemic index of a diet (Juliano and Goddard, 1986; Miller et al., 1992; Fitzgerald et al., 2011). Amylose is synthesized by granule bound starch synthase 1 (GBSSI) (Smith et al., 1997), which is encoded by the Waxy gene. At present, many Waxy alleles that correspond to different AC classes have been reported. The five common alleles are wx, Wxt, Wxg1, Wxg2, and Wxg3, which correspond to glutinous, low, intermediate, high I, and high II classes of apparent AC, respectively (Teng et al., 2012). In addition to these common alleles, a rare allele, Wxop, has been reported by Mikami et al. (2008). The identified alleles have given researchers the ability to develop FMs to develop rice varieties with desired AC by using MAB.
The Waxy gene has been reported to affect the gel consistency (GC) and GT of rice (Tan et al., 1999; Wang et al., 2007; Tian et al., 2009). Studies have confirmed that Waxy gene affects both AC and GC (Fan et al., 2005; Zhang et al., 2012). Although GT has been reported to be influenced by the Waxy gene, a major QTL corresponding to the alkali degeneration locus (alk) was found to control GT (Tian et al., 2005; Wang et al., 2007). The starch synthase IIa gene (SSIIa), located at the alk locus (Umemoto et al., 2002), is reported to have several functional single nucleotide polymorphisms (SNPs), SNP2, SNP3 (Umemoto and Aoki, 2005; Waters et al., 2006), and SNP4, (Bao et al., 2006; Waters et al., 2006) that affect GT.
Rice grain appearance is an important aspect that affects the visual preference of consumers. A major QTL on chromosome 3 has been found to be responsible for grain length (Aluko et al., 2004; Fan et al., 2006). A comparative sequencing study between short- and long-grain varieties showed that the second exon of the putative grain length gene GS3 has a nonsense mutation that is found in long-grain varieties (Fan et al., 2006). On the other hand, a loss of function mutation in GW2, a QTL located on chromosome 2, affects the grain width and weight (Song et al., 2007).
Fragrant rice varieties, such as basmati and jasmine, are of great interest to consumers due to their distinctive flavor. Researchers have identified many chemical compounds that contribute to the fragrance of fragrant rice (Yajima et al., 1979; Petrov et al., 1996). Of the identified chemical compounds, 2-acetyl-1-pyrroline (2AP) has been found to be the most significant compound in conferring fragrance to fragrant rice (Buttery et al., 1983, 1988). The elevated levels of 2AP in fragrant rice are thought to be due to a deletion within exon 7 (Bradbury et al., 2005a; Amarawathi et al., 2008) or exon 2 (Shi et al., 2008) of the gene encoding the enzyme betaine aldehyde dehydrogenase (BADH2), which is located on chromosome 8. These mutations render BADH2 non-functional, resulting in the accumulation of 2AP (Bradbury et al., 2005a, 2008). However, the deletions within exon 2 and exon 7 are likely not the only mutations for fragrance because there are varieties without them that accumulate 2AP (Fitzgerald et al., 2008). Fitzgerald et al. (2008) suggest that other mutations could influence BADH2 or that there exist other biochemical pathways, such as the one proposed by Huang et al. (2008), in addition to the pathway proposed by Bradbury et al. (2008), that lead to 2AP accumulation. Hence, the genetics and biochemical pathways of fragrance should be investigated by researchers to further understand fragrance in rice.
The micronutrients Zn and Fe are present in low quantities in rice, especially in the polished grain (Mayer et al., 2008). Therefore, biofortification strategies are undertaken to enhance the nutritional quality of rice in order to avert micronutrient deficiencies in populations for whom rice is the staple food and who have limited access to other fortified foods or micronutrient supplements (Bouis and Welch, 2010). However, biofortification in rice is no simple task. Sperotto et al. (2012) stated five constraints for concern in Fe biofortification: uptake from the soil, loading of the xylem, transport through the phloem, unloading at the base of the grain, and grain sink strength. The genetic engineering approach has reported success in increasing Zn and Fe content by overexpression of genes such as ferritin and those of the OsNAS gene family, which encode proteins that serve different purposes such as Fe accumulation or the transport of Fe ions (Johnson et al., 2011; Paul et al., 2012). Many QTLs for Zn and Fe have been reported, and candidate genes and linked markers have also been identified (Lu et al., 2008; Garcia-Oliveira et al., 2009; Sperotto et al., 2010; Anuradha et al., 2012). Based on specific QTLs, linked markers and candidate genes, the development of FMs for Zn and Fe improvement is anticipated for MAB programs.
Before the era of molecular marker technology, grain quality was evaluated on palatability, and the presence or absence of a certain trait, such as aroma. The evaluation of rice quality can also be performed visually, providing morphological data, which can then be represented by a morphological marker. Scientists investigate the proteins or enzymes underlying a specific trait, known as an allozyme marker, to discover the exact cause of the trait. Due to ambiguity and the limited information that can be extracted from enzyme analysis (Murphy et al., 1996), scientists’ attention has shifted toward DNA markers.
One classic example of DNA markers is the RFLP marker. RFLP is a hybridization based marker that utilizes restriction enzymes to cut the DNA at specific restriction sites. Single nucleotide changes, insertions or deletions cause changes in restriction sites, resulting in different molecular weight restriction fragments and variation between individuals. RFLP markers were used to map genes to chromosomes. Once a RFLP marker has been positively identified as linked to the putative gene controlling the trait under study, further investigation, such as chromosome walking, cloning, or sequencing of the gene, is undertaken. For instance, the gene controlling fragrance was initially mapped by Ahn et al. (1992) using a RFLP marker; using near isogenic lines (NILs), RFLP analysis showed that the fragrance gene (fgr) is linked to marker RG28 on chromosome 8.
The introduction of PCR-based markers such as SSR has increased scientists’ knowledge of the genetic map. The locus for a certain trait previously mapped with RFLP is saturated with SSR markers, thereby increasing proximity to the gene. In the case of fragrance, the fgr locus was mapped with SSR markers after it was discovered (Chen et al., 1997; Cho et al., 1998). Subsequently, the identified SSR markers have facilitated the development of SSR markers closely linked to the fgr, such as that developed by Garland et al. (2000) which detects changes in the mononucleotide repeat of thiamine, (T)n. This marker was unable to discriminate between genotypes using low-resolution agarose gels and was not polymorphic for some rice variety combinations; therefore, Cordeiro et al. (2002) developed another SSR marker based on the (AT)40 repeat for fragrance genotyping.
Researchers’ efforts to identify linked markers have facilitated further exploration into the genes responsible for rice quality traits. Sequencing the rice genome has also facilitated gene discovery (IRGSP, 2005; 3K RGP, 2014); now that rice genomic sequence data are available, genotype sequences of rice with and without a desirable trait can be compared, leading to discovery of the sequence underlying the trait. Using a linked SSR marker and a bacterial artificial chromosome (BAC), Bradbury et al. (2005a) identified the sequence polymorphism between fragrant and non-fragrant varieties, that is an 8-bp deletion and three SNPs and found the gene (later known as badh2) that codes for BADH2 whose functionality determines 2AP accumulation in rice. Based on sequence polymorphism and allele variation studies on different fragrant genotypes, researchers have developed FMs for use in genotyping and breeding (Bradbury et al., 2005b; Amarawathi et al., 2008; Shi et al., 2008; Sakthivel et al., 2009; Vanavichit et al., 2010; Myint et al., 2012). The progress of FM development for the example of fragrance is shown in a simplified manner in Figure 1. Some of the FMs developed by researchers for use in quality rice breeding are listed in Table 1.
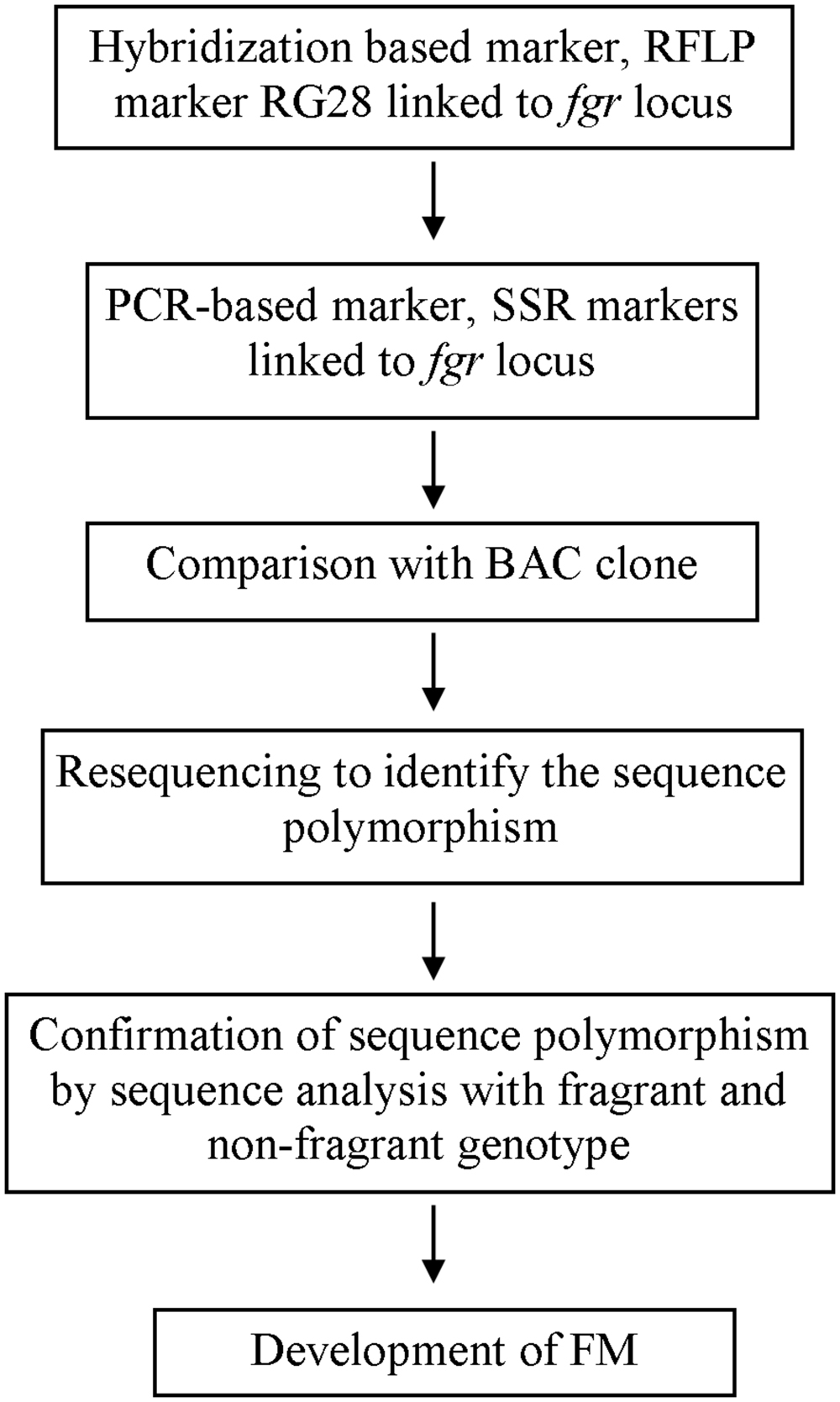
FIGURE 1. A chart of simplified progress of functional marker (FM) development for trait used as an example, fragrance, with reference to Ahn et al. (1992) and Bradbury et al. (2005a).
The development of FMs involves a series of steps (Figure 2). The initial step is discovery of the gene that controls the trait. Forward and reverse genetics approaches facilitate the identification of genes that casually affect phenotypic variation. One method of gene identification is by QTL mapping, which identifies loci that underlie the gene or genes that contribute to the trait. Family-based QTL mapping requires the development of a pedigree from crosses between different genotypes and their resulting progeny. Over the years, many family-based QTL mapping studies, especially bi-parental QTL mapping, have been conducted for rice quality traits. However, in family-based QTL mapping, the recombination events are limited to the generations of the family and therefore provide low resolution (Mitchell-Olds, 2010). To improve the resolution of QTL mapping and promote more recombination events, researchers can opt for multiple-parent advanced generation inter-cross (MAGIC). Bandillo et al. (2013) have developed a MAGIC population from half diallel-mating of eight varieties; conducted genotyping by sequencing (GBS) on 200 indica MAGIC lines and identified major genes and QTLs for many traits that influence grain quality, such as Waxy and GS3. Conversely, population-based QTL mapping and genome-wide association studies (GWAS) take into account the historical recombination events that have accumulated over thousands of generations and are, therefore, able to provide higher resolution (Mitchell-Olds, 2010). GWAS utilizes more than 100 genotypes with diverse backgrounds, which leads to a broader genetic base (Mitchell-Olds, 2010). GWAS investigates genome-wide association between SNPs and phenotypes, utilizing an array-based SNP detection platform or next generation sequencing (NGS). Chen et al. (2014) developed an array-based genotyping tool called RiceSNP50 and identified a locus in the same region as the GS3 locus. Huang et al. (2010) utilized NGS and conducted GWAS on 373 indica lines for 14 agronomic traits important to grain quality and identified major genes such as Waxy and the alk locus, which were similar to those reported by other researchers and other minor genes. Their study showed that GWAS has the potential to identify genes that contribute to natural variation of complex traits. Although the cost for this sequencing platform may be high, with time, it will be made affordable to all researchers. With the marker linked to the loci, target genes can be isolated by map-based cloning (or positional cloning), expression profiling (Duan and Sun, 2005) or transposon tagging, enabling researchers to investigate gene function.
The candidate gene approach has been used in various crop plants to identify genes that contribute to phenotypic variation. Because rice is composed mostly of starch, genes related to starch synthesis are targets of study. Tian et al. (2009) selected 18 starch synthesis-related genes and conducted an association study with AC, GC, and GT. According to Pflieger et al. (2001), genetic transformation is required to determine whether the candidate gene is the gene that causes the trait variation. Tian et al. (2009) have verified the role of each gene in the starch synthesis system by transformation. Their results suggest that selection of a single gene might be insufficient because starch synthesis-related genes cooperate with each other to form a network that determines AC, GC, and GT; therefore, modifying a single gene may alter these three properties. The verified candidate genes from this study can potentially be used in FM development.
Currently, the availability of the rice genome sequence (IRGSP, 2005; 3K RGP, 2014) facilitates gene discovery. Despite this resource, not all genes have had their functions characterized. Well-characterized gene function is a prerequisite of FM development. There are several methods by which researchers can determine a gene’s function, including genetic transformation, RNA interference (RNAi) and mutant characterization. To determine the function of the OsBADH2 gene, Niu et al. (2008) used RNAi combined with Agrobacterium tumefaciens-mediated T-DNA transfer. Their results demonstrated that down-regulated expression of the OsBADH2 gene resulted in increased 2AP, thereby validating OsBADH2 as a gene that affects fragrance in rice. Bradbury et al. (2008) proposed a pathway involving BADH2 that leads to 2AP accumulation; this pathway was supported by Chen et al. (2008), who studied it by transformation. A study by Gross et al. (2003) reported the ferritin genes OsFER1 and OsFER2. An expression profile study on OsFER1 and OsFER2 was then conducted by Stein et al. (2009), who showed that treatment with copper, excess Fe, and other metals causes differential expression of OsFER1 and OsFER2. Paul et al. (2012) showed that overexpression of theOsFER2 gene led to increased Fe and Zn levels in T3 transgenic plants.
Polymorphisms in the alleles that contribute to variation in phenotype can be in the form of SNPs, insertions/deletions (Indels) or SSRs. The relationship between the allelic polymorphism and the phenotypic variation is tested by either indirect or direct proof of allele function (Andersen and Lübberstedt, 2003). Association study is an indirect approach for proving allele function, which provides statistical proof of the relationship between the allele polymorphism and phenotype. Association studies rely on linkage disequilibrium (LD) (Andersen and Lübberstedt, 2003), which plays an important role in association studies because it affects the fine mapping of agronomically important genes. Because rice (Oryza sativa) is an autogamous species, the LD of approximately 75 kb for the indica variety is considered high; therefore it is eligible for genome-wide LD association mapping (Mather et al., 2007; Zhao et al., 2011).
Alternatively, reverse genetics approaches such as homologous recombination (HR) or targeted induced local lesions in genomes (TILLING) can be used to directly identify motif function. HR is the locus-targeted integration of alleles to produce isogenic genotypes to obtain direct proof of allele function (Andersen and Lübberstedt, 2003; Hanin and Paszkowski, 2003). Research by Terada et al. (2002) used gene targeting by HR to investigate the Waxy locus in rice with a positive/negative selection vector; these researchers obtained approximately 1% survival of transformants, suggesting that the method can be useful for gene-targeting or gene-knockout. The effects on the phenotypes of organisms generated from HR can, therefore, provide direct proof of allele function.
Targeted induced local lesions in genomes approach involves mutagenesis to create variations of mutants which are then subject to high-throughput screening for mutation discovery. By using two Nipponbare populations treated with ethyl methanesulphonate (EMS) or a combination of sodium azide andmethyl-nitrosourea (Az-MNU), Till et al. (2007) reported mutation rates of 1/294 kb and 1/265 kb, respectively. Suzuki et al. (2008) reported a mutation rate of 1/135 kb from a Taichung 65 mutant population treated with MNU, suggesting that a high mutation rate can be used to compliment other mutant resources in rice. EcoTILLING, a variant of TILLING, has been effective at revealing allele polymorphism and acts as a useful marker system for resistant genes in barley (Mejlhede et al., 2006). Recently, Tsai et al. (2011) incorporated NGS with multidimensional pooling into a TILLING protocol for identification of rare alleles. This advent in sequencing, along with researchers’ substantial efforts, will lead to the discovery of more alleles. Polymorphisms detected in mutants from TILLING or EcoTILLING provide proof of allele function; when coupled with phenotypic data, these results can facilitate the development of FMs.
Plant breeding has benefited from the advent of marker technology. The application of markers in plant breeding is known as MAB. MAB includes marker-assisted recurrent selection (MARS), marker-assisted backcrossing (MABC), and marker-assisted gene pyramiding. MABC is the most commonly used technique in rice breeding. In foreground selection of MABC, markers associated with the QTL or genes for the desired trait are used to identify plants that carry the preferred allele, allowing selection to be conducted at an early stage of the breeding program. Using an FM rather than a linked DNA marker can improve the selection precision. Because the FM is in complete linkage with the target gene, the risk of linkage drag and recombination between the marker and target gene can be minimized, thereby reducing the chance of undesirable alleles being passed down from the donor parent. Jin et al. (2010) applied FMs to select Waxy, badh2, and SSIIa genes in their backcrossing scheme and successfully improved the AC, GT, and fragrance of a maintainer line used for hybrid rice production. Study by Jin et al. (2010) showed that the availability of FMs has made possible the introgression of three traits simultaneously in a breeding program. Moreover, using FMs saves time as it circumvents the phenotypic evaluation on a limited number of seeds at every stage of breeding that would be conducted in conventional breeding.
Marker-assisted breeding is particularly useful for traits controlled by major QTLs or genes with large effects; however, it may be ineffective for traits governed by many QTLs with small effects or those influenced by the environment. Genomic selection (GS), an alternative to MAB proposed by Meuwissen et al. (2001), utilizes all marker and phenotypic data to estimate marker effects and makes predictions of which individuals would make the best parents. The genomic estimated breeding values (GEBVs) are calculated from a training population, for which both genotypic and phenotypic data have been collected and then tested on the candidate population (Chen et al., 2013). Recently, Spindel et al. (2015) attempted GS on rice for three traits: flowering time, plant height, and grain yield. Their study reported more accurate predictions of breeding line performance than pedigree data alone. With its many strengths, GS is anticipated to aid researchers in breeding for micronutrients such as Fe and Zn where many QTLs or genes are involved.
Although a plethora of developed FMs are recommended for application in quality rice breeding programs, researchers who have used them have different experiences and opinions on use. Amarawathi et al. (2008) and Sakthivel et al. (2009) reported inconsistency of the allele-specific amplification (ASA) marker system for fragrant rice genotyping developed by Bradbury et al. (2005a). Conversely, Sarhadi et al. (2011) proved the efficacy and efficiency of ASA markers in differentiating fragrant and non-fragrant rice genotypes and the genotype matches the phenotype accurately. These contrary views suggest that proper optimization of the FM assay prior to its use is essential because an optimized assay ensures reproducible results; therefore, optimization is required prior to its application in breeding programs (Poczai et al., 2013).
Another concern for the application of FMs is the pleiotropic effects of certain genes on several traits (Chen and Lübberstedt, 2010; Brenner et al., 2013). Understanding the correlation among characteristics or the pleiotropy of major genes allows breeders to decide which traits should be directly or indirectly selected or to compensate for the undesirable traits with favorable alleles (Chen and Lübberstedt, 2010). Although major genes or QTLs that influence GT and GC have been identified [SSIIa (Umemoto et al., 2002), alk2(t) (Shu et al., 2006), and qGC-6 (Su et al., 2011)], the effect of the Waxy gene on GT and GC (Lanceras et al., 2000; Bao et al., 2003; Zhang et al., 2012), has yet to be determined as pleiotropic or gene linkage. This create a challenge to breeders in selecting the traits in breeding programs (Shu et al., 2006).
Epistasis is another concern for breeders because it complicates the inheritance of quality traits. If epistatic effects of the genes are not taken into account, the associations for a single gene might be inaccurate or misleading, thereby hindering the development of FMs and causing inconsistency in FM application (Brenner et al., 2013). Epistatic effects among the QTLs controlling quality traits have been reported (Lee and Koh, 2010; Anuradha et al., 2012; Liu et al., 2013); therefore, researchers should discern the epistatic effects of the genes influencing a trait in quality rice breeding.
The main advantage of an FM is that it would have complete linkage with the desired allele; therefore, it could be applied to any population, regardless of genetic background, without having to revalidate the QTL relationship. While the above statement is technically correct, there is a subtle complication that needs to be mentioned. If a researcher selects on the basis of an FM, the possibility remains that the phenotype of interest is due to another allele that is in linkage disequilibrium with the FM. However, the effect is small. Although no large-scale assessment of linkage disequilibrium has been observed in O. sativa, the seminal work by Garris et al. (2003) indicated a linkage disequilibrium decay of 100 kb around a disease resistance locus in the aus subpopulation. More recently, linkage disequilibrium decays of 50 kb in indica, 5 kb in Oryza rufipogon (Rakshit et al., 2007), 2 Mb in indica and tropical japonica, and 500 kb in O. rufipogon have been reported by using gene-based markers and phenotypes (Caicedo et al., 2007). The physical extent of linkage disequilibrium around a gene defines the efficiency of linkage disequilibrium mapping, which is the consequence of several factors, including the degree of artificial or natural selection on the gene or region of the genome, the rate of outcrossing, recombination fraction, the age of the allele under study, chromosomal location, and the population size and structure (Garris et al., 2003).
While the costs of development and establishment and application of FMs in MAB might currently be a concern to researchers, the costs of marker development and marker genotyping are expected to drop in the near future (Lau et al., 2014). Monsanto reported that price per molecular marker decreased over sixfold from 2000 to 2006 (Eathington et al., 2007). Cost for marker discovery by sequencing technology is also expected to decrease over time (Wetterstrand, 2014). Therefore, the advancement of sequencing technology is expediting gene discovery and FM development.
There are more genes involved in eating, cooking, and the nutritional qualities of rice that have not yet been discovered. Whole-genome sequencing of rice has been conducted to identify some of these genes. The discovery of genes and gene function characterization can be conducted using the various approaches of forward and reverse genetics. FMs could revolutionize the selection strategy in quality rice breeding without linkage drag. Because the cost of marker discovery by sequencing technology is decreasing, the adoption of FMs in breeding programs, especially MAB, is greatly anticipated. We envision that the utilization of FMs will enable the incorporation of all genes for cooking, eating, and nutritional qualities into one rice genotype.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We gratefully acknowledge the financial support from the Long-term Research Grant Scheme (LRGS), the Food Security Project, and the Ministry of Education, Malaysia. This work is also supported by Geran Putra IPS, Universiti Putra Malaysia. We also thank reviewers for their critical reading and improvement of the manuscript.
Ahmad, R. (2013). Rice Revolution | Bangladesh Set to Release the World’s First zinc-Enriched Variety. Dly. Star. Available at: http://archive.thedailystar.net/beta2/news/rice-revolution/ [accessed October 15, 2014].
Ahn, S. N., Bollich, C. N., and Tanksley, S. D. (1992). RFLP tagging of a gene for aroma in rice. Theor. Appl. Genet. 84, 825–828. doi: 10.1007/BF00227391
Aluko, G., Martinez, C., Tohme, J., Castano, C., Bergman, C., and Oard, J. H. (2004). QTL mapping of grain quality traits from the interspecific cross Oryza sativa x O. glaberrima. Theor. Appl. Genet. 109, 630–639. doi: 10.1007/s00122-004-1668-y
Amarawathi, Y., Singh, R., Singh, A. K., Singh, V. P., Mohapatra, T., Sharma, T. R., et al. (2008). Mapping of quantitative trait loci for basmati quality traits in rice (Oryza sativa L.). Mol. Breed. 21, 49–65. doi: 10.1007/s11032-007-9108-8
Andersen, J. R., and Lübberstedt, T. (2003). Functional markers in plants. Trends Plant Sci. 8, 554–560. doi: 10.1016/j.tplants.2003.09.010
Anuradha, K., Agarwal, S., Rao, Y. V., Rao, K. V., Viraktamath, B. C., and Sarla, N. (2012). Mapping QTLs and candidate genes for iron and zinc concentrations in unpolished rice of Madhukar × Swarna RILs. Gene 508, 233–240. doi: 10.1016/j.gene.2012.07.054
Ayres, N. M., McClung, A. M., Larkin, P. D., Bligh, H. F. J., Jones, C. A., and Park, W. D. (1997). Microsatellites and a single-nucleotide polymorphism differentiate apparent amylose classes in an extended pedigree of US rice germ plasm. Theor. Appl. Genet. 94, 773–781. doi: 10.1007/s001220050477
Bagge, M., and Lübberstedt, T. (2008). Functional markers in wheat: technical and economic aspects. Mol. Breed. 22, 319–328. doi: 10.1007/s11032-008-9190-6
Bandillo, N., Raghavan, C., Muyco, P. A., Sevilla, M. A. L., Lobina, I. T., Dilla-Ermita, C. J., et al. (2013). Multi-parent advanced generation inter-cross (MAGIC) populations in rice: progress and potential for genetics research and breeding. Rice 6, 11. doi: 10.1186/1939-8433-6-11
Bao, J., Corke, H., He, P., and Zhu, L. (2003). Analysis of quantitative trait loci for starch properties of rice based on an RIL population. Acta Bot. Sin. 45, 986–994.
Bao, J. S., Corke, H., and Sun, M. (2006). Nucleotide diversity in starch synthase IIa and validation of single nucleotide polymorphisms in relation to starch gelatinization temperature and other physicochemical properties in rice (Oryza sativa L.). Theor. Appl. Genet. 113, 1171–1183. doi: 10.1007/s00122-006-0394-z
Bouis, H. E., and Welch, R. M. (2010). Biofortification-a sustainable agricultural strategy for reducing micronutrient malnutrition in the global south. Crop Sci. 50, S-20–S-32. doi: 10.2135/cropsci2009.09.0531
Bradbury, L. M. T., Fitzgerald, T. L., Henry, R. J., Jin, Q., and Waters, D. L. E. (2005a). The gene for fragrance in rice. Plant Biotechnol. J. 3, 363–370. doi: 10.1111/j.1467-7652.2005.00131.x
Bradbury, L. M. T., Henry, R. J., Jin, Q., Reinke, R. F., and Waters, D. L. E. (2005b). A perfect marker for fragrance genotyping in rice. Mol. Breed. 16, 279–283. doi: 10.1007/s11032-005-0776-y
Bradbury, L. M. T., Gillies, S. A., Brushett, D. J., Waters, D. L. E., and Henry, R. J. (2008). Inactivation of an aminoaldehyde dehydrogenase is responsible for fragrance in rice. Plant Mol. Biol. 68, 439–449. doi: 10.1007/s11103-008-9381-x
Brenner, E. A., Beavis, W. D., Andersen, J. R., and Lübberstedt, T. (2013). “Prospects and Limitations for development and application of functional markers in plants,” in Diagnostics in Plant Breeding, eds T. Lübberstedt and R. K. Varshney (Dordrecht: Springer), 329–348.
Buttery, R. G., Ling, L. C., Juliano, B., and Turnbaugh, J. G. (1983). Cooked rice aroma and 2-acetyl-1-pyrroline. J. Agric. Food Chem. 31, 823–826. doi: 10.1021/jf00118a036
Buttery, R. G., Turnbaugh, J. G., and Ling, L. C. (1988). Contribution of volatiles to rice aroma. J. Agric. Food Chem. 36, 1006–1009. doi: 10.1021/jf00083a025
Caicedo, A. L., Williamson, S. H., Hernandez, R. D., Boyko, A., Fledel-Alon, A., York, T. L., et al. (2007). Genome-wide patterns of nucleotide polymorphism in domesticated rice. PLoS Genet. 3:1745–1756. doi: 10.1371/journal.pgen.0030163
Chen, H., He, H., Zhou, F., Yu, H., and Deng, X. W. (2013). Development of genomics-based genotyping platforms and their applications in rice breeding. Curr. Opin. Plant Biol. 16, 247–254. doi: 10.1016/j.pbi.2013.04.002
Chen, H., Xie, W., He, H., Yu, H., Chen, W., Li, J., et al. (2014). A high-density SNP genotyping array for rice biology and molecular breeding. Mol. Plant 7, 541–553. doi: 10.1093/mp/sst135
Chen, M.-H., Fjellstrom, R. G., Christensen, E. F., and Bergman, C. J. (2010). Development of three allele-specific codominant rice Waxy gene PCR markers suitable for marker-assisted selection of amylose content and paste viscosity. Mol. Breed. 26, 513–523. doi: 10.1007/s11032-010-9419-z
Chen, S., Yang, Y., Shi, W., Ji, Q., He, F., Zhang, Z., et al. (2008). Badh2, encoding betaine aldehyde dehydrogenase, inhibits the biosynthesis of 2-acetyl-1-pyrroline, a major component in rice fragrance. Plant Cell 20, 1850–1861. doi: 10.1105/tpc.108.058917
Chen, X., Temnykh, S., Xu, Y., Cho, Y. G., and McCouch, S. R. (1997). Development of a microsatellite framework map providing genome-wide coverage in rice (Oryza sativa L.). Theor. Appl. Genet. 95, 553–567. doi: 10.1007/s001220050596
Chen, Y., and Lübberstedt, T. (2010). Molecular basis of trait correlations. Trends Plant Sci. 15, 454–461. doi: 10.1016/j.tplants.2010.05.004
Cho, Y. G., McCouch, S. R., Kuiper, M., Kang, M.-R., Pot, J., Groenen, J. T. M., et al. (1998). Integrated map of AFLP, SSLP and RFLP markers using a recombinant inbred population of rice (Oryza sativa L.). Theor. Appl. Genet. 97, 370–380. doi: 10.1007/s001220050907
Cordeiro, G. M., Christopher, M. J., Henry, R. J., and Reinke, R. F. (2002). Identification of microsatellite markers for fragrance in rice by analysis of the rice genome sequence. Mol. Breed. 9, 245–250. doi: 10.1023/A:1020350725667
David, R. B. (2014). IRRI’s Iron-rich “Miracle Rice” Ready by Year 2029: Can the Country’s Poor Wait? Available at: http://www.eaglenews.ph/ irris-iron-rich-miracle-rice-ready-by-year-2029-can-the-countrys-poor-wait/ [accessed October 15, 2014].
Dela Cruz, N., and Khush, G. S. (2000). “Rice grain quality evaluation procedures,” in Aromatic Rices, eds R. K. Singh, U. S. Singh, and G. S. Khush (New Delhi: Mohan Primlani for Oxford & IBH Publishing Co. Pvt. Ltd), 292.
Duan, M., and Sun, S. S. M. (2005). Profiling the expression of genes controlling rice grain quality. Plant Mol. Biol. 59, 165–178. doi: 10.1007/s11103-004-7507-3
Eathington, S. R., Crosbie, T. M., Edwards, M. D., Reiter, R. S., and Bull, J. K. (2007). Molecular markers in a commercial breeding program. Crop Sci. 47, S–154. doi: 10.2135/cropsci2007.04.0015IPBS
Fan, C., Xing, Y., Mao, H., Lu, T., Han, B., Xu, C., et al. (2006). GS3, a major QTL for grain length and weight and minor QTL for grain width and thickness in rice, encodes a putative transmembrane protein. Theor. Appl. Genet. 112, 1164–1171. doi: 10.1007/s00122-006-0218-1
Fan, C. C., Yu, X. Q., Xing, Y. Z., Xu, C. G., Luo, L. J., and Zhang, Q. (2005). The main effects, epistatic effects and environmental interactions of QTLs on the cooking and eating quality of rice in a doubled-haploid line population. Theor. Appl. Genet. 110, 1445–1452. doi: 10.1007/s00122-005-1975-y
Ferrero, A., and Nguyen, N. (2004). Constraints and opportunities for the sustainable development of rice-based production systems in Europe. FAO Rice Conf. 43, 12–13.
Fitzgerald, M. A., Hamilton, N. R. S., Calingacion, M. N., Verhoeven, H. A., and Butardo, V. M. (2008). Is there a second fragrance gene in rice? Plant Biotechnol. J. 6, 416–423. doi: 10.1111/j.1467-7652.2008.00327.x
Fitzgerald, M. A., McCouch, S. R., and Hall, R. D. (2009). Not just a grain of rice: the quest for quality. Trends Plant Sci. 14, 133–139. doi: 10.1016/j.tplants.2008.12.004
Fitzgerald, M. A., Rahman, S., Resurreccion, A. P., Concepcion, J., Daygon, V. D., Dipti, S. S., et al. (2011). Identification of a major genetic determinant of glycaemic index in rice. Rice 4, 66–74. doi: 10.1007/s12284-011-9073-z
Gao, L., Zhou, M., Chen, R., Gao, H., Yan, Q., Zhou, W., et al. (2012). Developing and validating the functional marker of rice Waxy gene. Rice Genomics Genet. 3, 61–65. doi: 10.5376/rgg.2012.03.0010
Gao, Z., Zeng, D., Cheng, F., Tian, Z., Guo, L., Su, Y., et al. (2011). ALK, the key gene for gelatinization temperature, is a modifier gene for gel consistency in rice. J. Integr. Plant Biol. 53, 756–765. doi: 10.1111/j.1744-7909.2011.01065.x
Garcia-Oliveira, A. L., Tan, L., Fu, Y., and Sun, C. (2009). Genetic identification of quantitative trait loci for contents of mineral nutrients in rice grain. J. Integr. Plant Biol. 51, 84–92. doi: 10.1111/j.1744-7909.2008.00730.x
Garland, S., Lewin, L., Blakeney, A., Reinke, R., and Henry, R. (2000). PCR-based molecular markers for the fragrance gene in rice (Oryza sativa L.). Theor. Appl. Genet. 101, 364–371. doi: 10.1007/s001220051492
Garris, A. J., McCouch, S. R., and Kresovich, S. (2003). Population structure and its effect on haplotype diversity and linkage disequilibrium surrounding the xa5 locus of rice (Oryza sativa L.). Genetics 165, 759–769.
Gross, J., Stein, R. J., Fett-Neto, A. G., and Fett, J. P. (2003). Iron homeostasis related genes in rice. Genet. Mol. Biol. 26, 477–497. doi: 10.1590/S1415-47572003000400012
Gupta, P. K., Kumar, J., Mir, R. R., and Kumar, A. (2010). “Marker-assisted selection as a component of conventional plant breeding,” in Plant Breeding Reviews, ed. J. Janick (Hoboken, NJ: John Wiley & Sons, Inc.), 145–217.
Hanin, M., and Paszkowski, J. (2003). Plant genome modification by homologous recombination. Curr. Opin. Plant Biol. 6, 157–162. doi: 10.1016/S1369-5266(03)00016-5
Hospital, F. (2001). Size of donor chromosome segments around introgressed loci and reduction of linkage drag in marker-assisted backcross programs. Genetics 158, 1363–1379.
Hsu, Y.-C., Tseng, M.-C., Wu, Y.-P., Lin, M.-Y., Wei, F.-J., Hwu, K.-K., et al. (2014). Genetic factors responsible for eating and cooking qualities of rice grains in a recombinant inbred population of an inter-subspecific cross. Mol. Breed. 34, 655–673. doi: 10.1007/s11032-014-0065-8
Huang, T.-C., Teng, C.-S., Chang, J.-L., Chuang, H.-S., Ho, C.-T., and Wu, M.-L. (2008). Biosynthetic mechanism of 2-acetyl-1-pyrroline and its relationship with Delta1-pyrroline-5-carboxylic acid and methylglyoxal in aromatic rice (Oryza sativa L.) callus. J. Agric. Food Chem. 56, 7399–7404. doi: 10.1021/jf8011739
Huang, X., Wei, X., Sang, T., Zhao, Q., Feng, Q., Zhao, Y., et al. (2010). Genome-wide association studies of 14 agronomic traits in rice landraces. Nat. Genet. 42, 961–967. doi: 10.1038/ng.695
Ingvardsen, C., Schejbel, B., and Lübberstedt, T. (2008). “Functional markers in resistance breeding,” in Progress in Botany, eds U. Lüttge, W. Beyschlag, and J. Murata (Berlin: Springer-Verlag), 61–87. doi: 10.1007/978-3-540-72954-9_3
IRGSP (2005). The map-based sequence of the rice genome. Nature 436, 793–800. doi: 10.1038/nature03895
Jin, L., Lu, Y., Shao, Y., Zhang, G., Xiao, P., Shen, S., et al. (2010). Molecular marker assisted selection for improvement of the eating, cooking and sensory quality of rice (Oryza sativa L.). J. Cereal Sci. 51, 159–164. doi: 10.1016/j.jcs.2009.11.007
Johnson, A. A. T., Kyriacou, B., Callahan, D. L., Carruthers, L., Stangoulis, J., Lombi, E., et al. (2011). Constitutive overexpression of the OsNAS gene family reveals single-gene strategies for effective iron- and zinc-biofortification of rice endosperm. PLoS ONE 6:e24476. doi: 10.1371/journal.pone.0024476
Juliano, B. O., and Goddard, M. S. (1986). Cause of varietal difference in insulin and glucose responses to ingested rice. Qual. Plant. Plant Foods Hum. Nutr. 36, 35–41. doi: 10.1007/BF01091751
Juliano, B. O., and Perez, C. M. (1984). Results of a collaborative test on the measurement of grain elongation of milled rice during cooking. J. Cereal Sci. 2, 281–292. doi: 10.1016/S0733-5210(84)80016-8
Lanceras, J. C., Huang, Z. L., Naivikul, O., Vanavichit, A., Ruanjaichon, V., and Tragoonrung, S. (2000). Mapping of genes for cooking and eating qualities in Thai jasmine rice (KDML105). DNA Res. 7, 93–101. doi: 10.1093/dnares/7.2.93
Lau, W. C. P., Latif, M. A., Rafii, M. Y., Ismail, M. R., and Puteh, A. (2014). Advances to improve the eating and cooking qualities of rice by marker-assisted breeding. Crit. Rev. Biotechnol. 1–12. doi: 10.3109/07388551.2014.923987 [Epub ahead of print].
Lee, J., and Koh, H. (2010). Epistatic relationships among genes related to endosperm starch synthesis in rice. Korean J. Breed. Sci. 42, 151–156.
Liu, Q., Jiang, J., Niu, F., He, Y., and Hong, D. (2013). QTL analysis for seven quality traits of RIL population in Japonica rice based on three genetic statistical models. Rice Sci. 20, 31–38. doi: 10.1016/S1672-6308(13)60105-5
Lu, K., Li, L., Zheng, X., Zhang, Z., Mou, T., and Hu, Z. (2008). Quantitative trait loci controlling Cu, Ca, Zn, Mn and Fe content in rice grains. . J. Genet. 87, 305–310. doi: 10.1007/s12041-008-0049-8
Lu, Y., Xiao, P., Shao, Y., Zhang, G., Thanyasiriwat, T., and Bao, J. (2010). Development of new markers to genotype the functional SNPs of SSIIa, a gene responsible for gelatinization temperature of rice starch. J. Cereal Sci. 52, 438–443. doi: 10.1016/j.jcs.2010.07.008
Lübberstedt, T., Zein, I., Andersen, J. R., Wenzel, G., Krützfeldt, B., Eder, J., et al. (2005). Development and application of functional markers in maize. Euphytica 146, 101–108. doi: 10.1093/dnares/dst039
Mather, K. A., Caicedo, A. L., Polato, N. R., Olsen, K. M., McCouch, S., and Purugganan, M. D. (2007). The extent of linkage disequilibrium in rice (Oryza sativa L.). Genetics 177, 2223–2232. doi: 10.1534/genetics.107.079616
Mayer, J. E., Pfeiffer, W. H., and Beyer, P. (2008). Biofortified crops to alleviate micronutrient malnutrition. Curr. Opin. Plant Biol. 11, 166–170. doi: 10.1016/j.pbi.2008.01.007
Mejlhede, N., Kyjovska, Z., Backes, G., Burhenne, K., Rasmussen, S. K., and Jahoor, A. (2006). EcoTILLING for the identification of allelic variation in the powdery mildew resistance genes mlo and Mla of barley. Plant Breed. 125, 461–467. doi: 10.1111/j.1439-0523.2006.01226.x
Meuwissen, T. H. E., Hayes, B. J., and Goddard, M. E. (2001). Prediction of total genetic value using genome-wide dense marker maps. Genetics 157, 1819–1829.
Mikami, I., Uwatoko, N., Ikeda, Y., Yamaguchi, J., Hirano, H. Y., Suzuki, Y., et al. (2008). Allelic diversification at the wx locus in landraces of Asian rice. Theor. Appl. Genet. 116, 979–989. doi: 10.1007/s00122-008-0729-z
Miklas, P. N., Kelly, J. D., Beebe, S. E., and Blair, M. W. (2006). Common bean breeding for resistance against biotic and abiotic stresses: from classical to MAS breeding. Euphytica 147, 105–131. doi: 10.1007/s10681-006-4600-5
Miller, J., Pang, E., and Bramall, L. (1992). Rice: a high or low glycemic index food? Am. J. Clin. Nutr. 56, 1034–1036.
Murphy, R. W., Jack, W., Sites, J., Buth, D. G., and Haufler, C. H. (1996). “Proteins: isozyme electrophoresis,” in Molecular Systemics, eds D. M. Hillis, C. Moritz, and B. K. Mable (Sunderland, MA: Sinauer Associates). Available at: http://www.researchgate.net/profile/Barbara_Mable/publication/49252417_Molecular_Systematics/links/0046351ade87c46b56000000.pdf [accessed June 29, 2015].
Myint, K. M., Arikit, S., Wanchana, S., Yoshihashi, T., Choowongkomon, K., and Vanavichit, A. (2012). A PCR-based marker for a locus conferring the aroma in Myanmar rice (Oryza sativa L.). Theor. Appl. Genet. 125, 887–896. doi: 10.1007/s00122-012-1880-0
Niu, X., Tang, W., Huang, W., Ren, G., Wang, Q., Luo, D., et al. (2008). RNAi-directed downregulation of OsBADH2 results in aroma (2-acetyl-1-pyrroline) production in rice (Oryza sativa L.). BMC Plant Biol. 8:100. doi: 10.1186/1471-2229-8-100
Paul, S., Ali, N., Gayen, D., Datta, S. K., and Datta, K. (2012). Molecular breeding of Osfer2 gene to increase iron nutrition in rice grain. GM Crops Food 3, 310–316. doi: 10.4161/gmcr.22104
Petrov, M., Danzart, M., Gaimpaoli, P., Faure, J., and Richard, H. (1996). Rice aroma analysis: discrimination between a scented and a non-scented rice. Sci. Aliments 16, 347–360.
Pflieger, S., Lefebvre, V., and Causse, M. (2001). The candidate gene approach in plant genetics: a review. Mol. Breed. 7, 275–291. doi: 10.1023/A:1011605013259
Poczai, P., Varga, I., Laos, M., Cseh, A., Bell, N., Valkonen, J. P., et al. (2013). Advances in plant gene-targeted and functional markers: a review. Plant Methods 9, 6. doi: 10.1186/1746-4811-9-6
Rakshit, S., Rakshit, A., Matsumura, H., Takahashi, Y., Hasegawa, Y., Ito, A., et al. (2007). Large-scale DNA polymorphism study of Oryza sativa and O. rufipogon reveals the origin and divergence of Asian rice. Theor. Appl. Genet. 114, 731–743. doi: 10.1007/s00122-006-0473-1
Ramkumar, G., Sivaranjani, A. K. P., Pandey, M. K., Sakthivel, K., Shobha Rani, N., Sudarshan, I., et al. (2010). Development of a PCR-based SNP marker system for effective selection of kernel length and kernel elongation in rice. Mol. Breed. 26, 735–740. doi: 10.1007/s11032-010-9492-3
Sakthivel, K., Shobha Rani, N., Pandey, M. K., Sivaranjani, A. K. P., Neeraja, C. N., Balachandran, S. M., et al. (2009). Development of a simple functional marker for fragrance in rice and its validation in Indian Basmati and non-Basmati fragrant rice varieties. Mol. Breed. 24, 185–190. doi: 10.1007/s11032-009-9283-x
Sarhadi, W. A., Hien, N. L., Zanjani, M., Yosofzai, W., Yoshihashi, T., and Hirata, Y. (2011). Comparative analyses for aroma and agronomic traits of native rice cultivars from Central Asia. J. Crop Sci. Biotechnol. 11, 17–22.
Shi, W., Yang, Y., Chen, S., and Xu, M. (2008). Discovery of a new fragrance allele and the development of functional markers for the breeding of fragrant rice varieties. Mol. Breed. 22, 185–192. doi: 10.1007/s11032-008-9165-7
Shu, X., Shen, S., Bao, J., Wu, D., Nakamura, Y., and Shu, Q. (2006). Molecular and biochemical analysis of the gelatinization temperature characteristics of rice (Oryza sativa L.) Starch granules. J. Cereal Sci. 44, 40–48. doi: 10.1016/j.jcs.2006.03.001
Smith, A. M., Denyer, K., and Martin, C. (1997). The synthesis of the starch granule. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 67–87. doi: 10.1146/annurev.arplant.48.1.67
Song, X.-J., Huang, W., Shi, M., Zhu, M.-Z., and Lin, H.-X. (2007). A QTL for rice grain width and weight encodes a previously unknown RING-type E3 ubiquitin ligase. Nat. Genet. 39, 623–630. doi: 10.1038/ng2014
Sperotto, R. A., Boff, T., Duarte, G. L., Santos, L. S., Grusak, M. A., and Fett, J. P. (2010). Identification of putative target genes to manipulate Fe and Zn concentrations in rice grains. J. Plant Physiol. 167, 1500–1506. doi: 10.1016/j.jplph.2010.05.003
Sperotto, R. A., Ricachenevsky, F. K., Waldow, V. D. A., and Fett, J. P. (2012). Iron biofortification in rice: it’s a long way to the top. Plant Sci. 190, 24–39. doi: 10.1016/j.plantsci.2012.03.004
Spindel, J., Begum, H., Akdemir, D., Virk, P., Collard, B., Redoña, E., et al. (2015). Genomic selection and association mapping in rice (Oryza sativa): effect of trait genetic architecture, training population composition, marker number and statistical model on accuracy of rice genomic selection in elite, tropical rice breeding lines. PLoS Genet. 11:e1004982. doi: 10.1371/journal.pgen.1004982
Stein, R. J., Ricachenevsky, F. K., and Fett, J. P. (2009). Differential regulation of the two rice ferritin genes (OsFER1 and OsFER2). Plant Sci. 177, 563–569. doi: 10.1016/j.plantsci.2009.08.001
Su, Y., Rao, Y., Hu, S., Yang, Y., Gao, Z., Zhang, G., et al. (2011). Map-based cloning proves qGC-6, a major QTL for gel consistency of japonica/indica cross, responds by Waxy in rice (Oryza sativa L.). Theor. Appl. Genet. 123, 859–867. doi: 10.1007/s00122-011-1632-6
Sumner, D. A., Rozelle, S. D., Huang, J., and Lee, H. (2001). The China Market for Rice: Current Status, Recent Trends, and Projections, with Emphasis on the Potential for Imports from the United States and Potential for External Competition with U.S. Rice. Davis, CA: A Research Study for the USA Rice Federation.
Suzuki, T., Eiguchi, M., Kumamaru, T., Satoh, H., Matsusaka, H., Moriguchi, K., et al. (2008). MNU-induced mutant pools and high performance TILLING enable finding of any gene mutation in rice. Mol. Genet. Genomics 279, 213–223. doi: 10.1007/s00438-007-0293-2
Tan, Y. F., Li, J. X., Yu, S. B., Xing, Y. Z., Xu, C. G., and Zhang, Q. (1999). The three important traits for cooking and eating quality of rice grains are controlled by a single locus in an elite rice hybrid. Shanyou 63. Theor. Appl. Genet. 99, 642–648. doi: 10.1007/s001220051279
Teng, B., Zeng, R., Wang, Y., Liu, Z., Zhang, Z., Zhu, H., et al. (2012). Detection of allelic variation at the Wx locus with single-segment substitution lines in rice (Oryza sativa L.). Mol. Breed. 30, 583–595. doi: 10.1007/s11032-011-9647-x
Terada, R., Urawa, H., Inagaki, Y., Tsugane, K., and Iida, S. (2002). Efficient gene targeting by homologous recombination in rice. Nat. Biotechnol. 20, 1030–1034. doi: 10.1038/nbt737
Tian, R., Jiang, G.-H., Shen, L.-H., Wang, L.-Q., and He, Y.-Q. (2005). Mapping quantitative trait loci underlying the cooking and eating quality of rice using a DH population. Mol. Breed. 15, 117–124. doi: 10.1007/s11032-004-3270-z
Tian, Z., Qian, Q., Liu, Q., Yan, M., Liu, X., Yan, C., et al. (2009). Allelic diversities in rice starch biosynthesis lead to a diverse array of rice eating and cooking qualities. Proc. Natl. Acad. Sci. U.S.A. 106, 21760–21765. doi: 10.1073/pnas.0912396106
Till, B. J., Cooper, J., Tai, T. H., Colowit, P., Greene, E. A., Henikoff, S., et al. (2007). Discovery of chemically induced mutations in rice by TILLING. BMC Plant Biol. 7:19. doi: 10.1186/1471-2229-7-19
Tsai, H., Howell, T., Nitcher, R., Missirian, V., Watson, B., Ngo, K. J., et al. (2011). Discovery of rare mutations in populations: TILLING by sequencing. Plant Physiol. 156, 1257–1268. doi: 10.1104/pp.110.169748
Umemoto, T., and Aoki, N. (2005). Single-nucleotide polymorphisms in rice starch synthase IIa that alter starch gelatinisation and starch association of the enzyme. Funct. Plant Biol. 32, 763–768. doi: 10.1071/FP04214
Umemoto, T., Yano, M., Satoh, H., Shomura, A., and Nakamura, Y. (2002). Mapping of a gene responsible for the difference in amylopectin structure between japonica-type and indica-type rice varieties. Theor. Appl. Genet. 104, 1–8. doi: 10.1007/s001220200000
Vanavichit, A., Tragoorung, S., Toojinda, T., Wanchana, S., and Kamolsukyunyong, W. (2010). BADH2 nucleic acids associated with grain aroma. U.S. Patent No. 6. Available at: http://www.google.com/patents/US7847083 [accessed September 21, 2014].
Varshney, R. K., Graner, A., and Sorrells, M. E. (2005). Genomics-assisted breeding for crop improvement. Trends Plant Sci. 10, 621–630. doi: 10.1016/j.tplants.2005.10.004
Wang, C., Chen, S., and Yu, S. (2011). Functional markers developed from multiple loci in GS3 for fine marker-assisted selection of grain length in rice. Theor. Appl. Genet. 122, 905–913. doi: 10.1007/s00122-010-1497-0
Wang, L. Q., Liu, W. J., Xu, Y., He, Y. Q., Luo, L. J., Xing, Y. Z., et al. (2007). Genetic basis of 17 traits and viscosity parameters characterizing the eating and cooking quality of rice grain. Theor. Appl. Genet. 115, 463–476. doi: 10.1007/s00122-007-0580-7
Waters, D. L. E., Henry, R. J., Reinke, R. F., and Fitzgerald, M. A. (2006). Gelatinization temperature of rice explained by polymorphisms in starch synthase. Plant Biotechnol. J. 4, 115–122. doi: 10.1111/j.1467-7652.2005.00162.x
Wetterstrand, K. (2014). DNA Sequencing Costs: Data from the NHGRI Genome Sequencing Program (GSP). Available at: http://www.genome.gov/sequencingcosts/ [accessed January 16, 2015].
World Health Organization (2007). Preventing and Controlling Micronutrient Deficiencies in Populations Affected by an Emergency. Geneva: Joint Statement by the World Health Organization, the World Food Programme and the United Nations Children’s Fund. Available at: http://www.who.int/nutrition/publications/WHO_WFP_UNICEFstatement.pdf [accessed March 17, 2015].
Yajima, I., Yanai, T., Nakamura, M., Sakakibara, H., and Hayashi, K. (1979). Volatile flavor components of cooked Kaorimai (scented rice. O. sativa japonica). Agric. Biol. Chem. 43, 2425–2429. doi: 10.1271/bbb1961.43.2425
Yi, M., Than, K., Vanavichit, A., Chai-arree, W., and Toojinda, T. (2009). Marker assisted backcross breeding to improve cooking quality traits in Myanmar rice cultivar Manawthukha. Field Crop. Res. 113, 178–186. doi: 10.1016/j.fcr.2009.05.006
Zhang, Z., Li, M., Fang, Y., Liu, F., Lu, Y., Meng, Q., et al. (2012). Diversification of the Waxy gene is closely related to variations in rice eating and cooking quality. Plant Mol. Biol. Report. 30, 462–469. doi: 10.1007/s11105-011-0362-x
Keywords: quantitative trait loci (QTL), DNA markers, rice quality, marker-assisted breeding (MAB), micronutrients, sequencing technology
Citation: Lau WCP, Rafii MY, Ismail MR, Puteh A, Latif MA and Ramli A (2015) Review of functional markers for improving cooking, eating, and the nutritional qualities of rice. Front. Plant Sci. 6:832. doi: 10.3389/fpls.2015.00832
Received: 12 May 2015; Accepted: 22 September 2015;
Published: 14 October 2015.
Edited by:
Richard A. Jorgensen, University of Arizona, USAReviewed by:
Ing-Feng Chang, National Taiwan University, TaiwanCopyright © 2015 Lau, Rafii, Ismail, Puteh, Latif and Ramli. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wendy C. P. Lau and Mohd Y. Rafii, Department of Crop Science, Faculty of Agriculture, Universiti Putra Malaysia, 43400 Serdang, Selangor, Malaysia, Y2h1aV9waGluZ0Bob3RtYWlsLmNvbQ==; bXJhZmlpQHVwbS5lZHUubXk=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.