- Department of Biochemistry, Cellular and Molecular Biology, University of Tennessee, Knoxville, TN, USA
The most conspicuous function of plastids is the oxygenic photosynthesis of chloroplasts, yet plastids are super-factories that produce a plethora of compounds that are indispensable for proper plant physiology and development. Given their origins as free-living prokaryotes, it is not surprising that plastids possess their own genomes whose expression is essential to plastid function. This semi-autonomous character of plastids requires the existence of sophisticated regulatory mechanisms that provide reliable communication between them and other cellular compartments. Such intracellular signaling is necessary for coordinating whole-cell responses to constantly varying environmental cues and cellular metabolic needs. This is achieved by plastids acting as receivers and transmitters of specific signals that coordinate expression of the nuclear and plastid genomes according to particular needs. In this review we will consider the so-called retrograde signaling occurring between plastids and nuclei, and between plastids and other organelles. Another important role of the plastid we will discuss is the involvement of plastid signaling in biotic and abiotic stress that, in addition to influencing retrograde signaling, has direct effects on several cellular compartments including the cell wall. We will also review recent evidence pointing to an intriguing function of chloroplasts in regulating intercellular symplasmic transport. Finally, we consider an intriguing yet less widely known aspect of plant biology, chloroplast signaling from the perspective of the entire plant. Thus, accumulating evidence highlights that chloroplasts, with their complex signaling pathways, provide a mechanism for exquisite regulation of plant development, metabolism and responses to the environment. As chloroplast processes are targeted for engineering for improved productivity the effect of such modifications on chloroplast signaling will have to be carefully considered in order to avoid unintended consequences on plant growth and development.
Introduction
According to the endosymbiotic theory plastids originated from free-living cyanobacteria that were engulfed by early eukaryotic cells. These cyanobacteria were retained by their hosts and have co-evolved with their host cells over 1.5 billion years to become an integral part of the modern plant cell (Yoon et al., 2004; Nakayama and Archibald, 2012). It is accepted that the successful stable integration occurred because of exceptional mutual benefits: the eukaryotic cell was able to establish an autotrophic lifestyle, while the engulfed cyanobacteria reached a pathogen-free asylum. Accumulating evidence suggests that this stable symbiosis between cyanobacteria and the eukaryotic cell was facilitated by infection of the latter with Chlamydiales pathogens (Ball et al., 2011, 2013).
The most conspicuous function of modern plastids is the sophisticated oxygenic photosynthesis performed by chloroplasts. However, plastids perform many other functions that are critical for proper plant development and physiology including the synthesis of amino acids, nucleotides and fatty acids, production of phytohormones, some vitamins and a multitude of secondary metabolites, as well nitrogen and sulfur assimilation. Many chloroplast secondary metabolites, besides being necessary for basic plant metabolic functions, are also important for interaction with the environment, as they function in plant defense against pathogen ingress and plant adaptation to stresses including heat, drought and high light. Thus, chloroplasts act as a hub in the cellular response to signals, generating a variety of signals that coordinate a fine-tuned and appropriate response to any given situation (Pfannschmidt and Yang, 2012).
The emerging view of the chloroplast is as a very dynamic signaling compartment. As a specific sensor of intra- and extracellular stimuli, chloroplasts constantly process and integrate a multitude of intracellular signals and pathways in order to sustain homeostasis at both the cellular and organismal levels (Figure 1A). An often over-looked aspect of cell biology is the physical interaction between organelles for coordination of signaling and metabolism (Figure 1B). This area is beginning to receive attention and we will examine these findings as they relate to chloroplasts and their roles in signaling. Given the dizzying array of signals that chloroplasts respond to and produce it is not surprising that there is considerable crosstalk between signaling pathways. This is particularly evident during responses to biotic and abiotic stress (Nakashima et al., 2014; Trotta et al., 2014; Zhou et al., 2015).
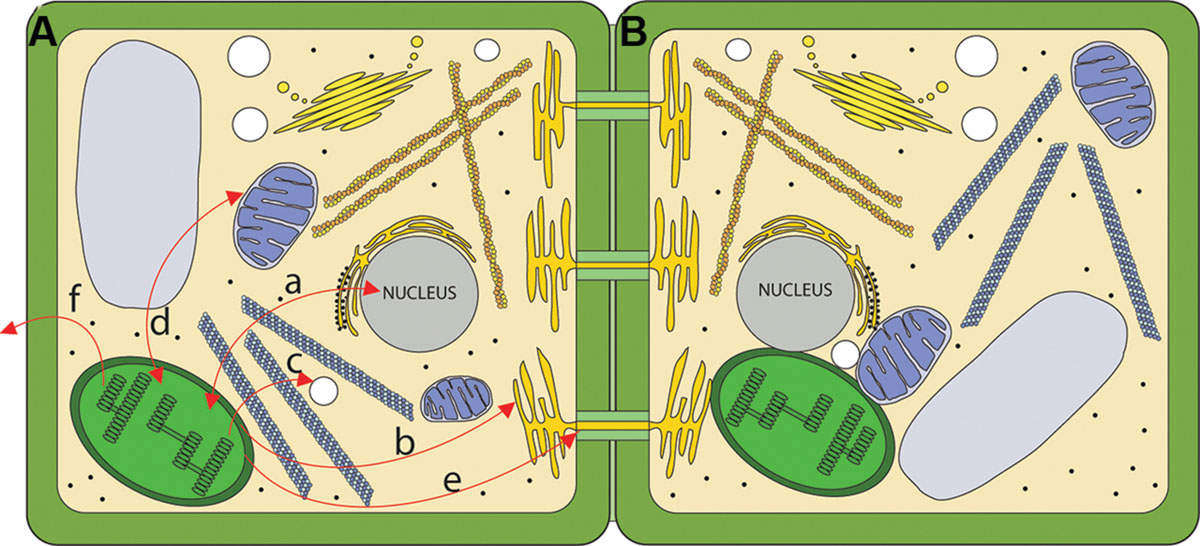
FIGURE 1. Routes for chloroplast signaling. (A) Chloroplasts generate signals that target multiple intercellular targets. (a) The majority of chloroplast proteins are encoded by the nucleus, and the import of those proteins into the chloroplast is anterograde signaling. In turn, several chloroplast products act as retrograde signals to regulate expression of nucleus-encoded genes. (b) Chloroplasts are metabolically coupled to the ER and it is likely that signals may move from the chloroplast to the ER. (c) Chloroplasts and peroxisomes are also closely associated, and numerous chloroplast products are substrates for peroxisomal pathways. (d) Mitochondria and chloroplasts are known to signal to each other. (e) Chloroplast signals regulate intercellular trafficking via plasmodesmata. It is not clear if this signaling is direct or involves retrograde signaling to the nucleus. (f) Chloroplasts produce volatile compounds that can signal to neighboring plants during pathogen attack. (B) The physical interaction between chloroplasts and various organelles may serve as a direct route for signaling.
Chloroplast signaling is not limited to exerting its effects within the cell. It is clear that chloroplast-derived signals can travel far beyond their site of production to induce changes in distal parts of the plant (e.g., Petrillo et al., 2014). Further, chloroplasts apparently regulate intercellular trafficking via the channels known as plasmodesmata. Through this action, chloroplasts could regulate almost all aspects of plant growth and development as it is becoming clear that not only do metabolites but also hormones, transcription factors and small RNA molecules use these channels for intercellular communication. Beyond whole-plant signaling, chloroplast signaling can impact entire ecosystem through production of volatile compounds.
While we focus here on the chloroplasts in mature leaves, it can be expected that other types of plastids, e.g., proplastids, etioplasts, or leucoplasts, participate in signaling networks in response to their unique developmental states and environmental conditions. Further, it is also likely that at any given time different subpopulations of chloroplasts within a cell are in various metabolic or physiologic states and are therefore likely to be involved in distinct signaling processes. Thus chloroplast signaling is complex, and dissecting crosstalk and feedback mechanisms remains a daunting task. With attempts to engineer chloroplasts for specialized or improved metabolic outputs, attention must be paid to how these adjustments may impact chloroplast behavior if unintended consequences are to be diminished. These consequences would not be limited to the chloroplasts, but could extend even to other plants cultivated in the vicinity of the engineered plants.
Chloroplasts in Intracellular Signaling
Chloroplast Signaling to the Nucleus
Over evolutionary time a significant number of the cyanobacterial genes were transferred to the host nucleus (Race et al., 1999; McFadden, 2001). These genes subsequently acquired sequences that function as transit peptides to enable import of their protein products back into plastids. The nucleus therefore exerts considerable control over chloroplast functions, and this nucleus-to-chloroplast signaling is termed anterograde signaling. Importantly, several genes encoding proteins that are integral components of photophosphorylation or photosynthesis were retained in the cyanobacterial genome (Allen, 1993; Race et al., 1999; Raven and Allen, 2003). Thus, in order to establish a stable eukaryotic plant cell it was necessary to synchronize the activities of both genomes. This has been achieved by creating a complex signaling system between the nucleus and plastids, able to transfer information and efficiently adjust gene expression in both compartments according to particular needs. Plastid derived-signals that regulate nuclear gene expression represent retrograde signaling. While anterograde signaling is well understood, it has been more challenging to unravel the molecular details of retrograde signaling.
Identification of molecules and signaling strategies underlying so-called retrograde signaling represents a long-standing quest in plant biology. Historically, Bradbeer et al. (1979) provided the first report describing the existence of communication between those organelles. They observed that the barley (Hordeum vulgare) albostrians chloroplast ribosome-deficient mutant had severely decreased chloroplast protein synthesis, in conjunction with depressed expression of nucleus-encoded chloroplast genes. This revolutionary discovery was in opposition to Ellis’ “cytoplasmic control principle” that posited control of organellar protein synthesis by cytoplasmic components (Ellis, 1977). Bradbeer’s discovery was soon confirmed by other researchers who treated young seedlings of mustard, Arabidopsis, pea or barley with lincomycin, chloramphenicol or streptomycin – inhibitors of plastid protein synthesis (Oelmuller et al., 1986; Susek et al., 1993; Yoshida et al., 1998; Sullivan and Gray, 1999). Besides the strategy of relying on systems with compromised chloroplast ribosome function, other approaches to perturbing distinct aspects of chloroplast function have also successfully interrogated chloroplast-to-nucleus signaling. The induction of carotenoid deficiency in genetic mutants or in plants treated with norflurazon (a herbicide that inhibits phytoene desaturase and blocks carotenoid synthesis), as well as in plants with compromised tetrapyrrole synthesis (resulting in accumulation of intermediates) caused suppressed expression of nucleus-encoded chloroplast genes (Johanningmeier and Howell, 1984; Mayfield and Taylor, 1984; Susek et al., 1993; Kropat et al., 1997, 2000; Strand et al., 2003; Zhang et al., 2011). These and other studies led to the realization that the plastid functional status can regulate the expression of photosynthesis associated nuclear genes (PhANGs; Surpin et al., 2002). Moreover, analysis of the Arabidopsis chloroplast ribosomal protein mutant rps1 revealed that chloroplast translational capacity is a critical factor in developing heat tolerance. This is mediated by inducing expression of the heat stress transcription factor HsfA2, a key regulator of heat tolerance (Yu et al., 2012). Therefore, the functional status of chloroplasts also regulates nuclear genes involved in heat-tolerance. The involvement of plastid translation in retrograde signaling and plant development was recently discussed in detail (Tiller and Bock, 2014).
The intense search for factors mediating this chloroplast-to-nucleus communication has identified a set of plastid metabolic intermediates. Importantly, proteins with functions in both chloroplasts and nuclei have been recently identified and have been proposed to participate in retrograde signaling. These retrograde signals are expected to work by modifying the expression of nuclear genes in order to adapt plant development and physiology to constantly changing environmental conditions. Currently two major modes of retrograde signaling are distinguished and they are involved in so-called biogenic and operational type of control. Whereas the former includes signals responsible for chloroplast and photosynthesis biogenesis, the latter act in response to changing environmental cues in fully developed chloroplasts (Pogson et al., 2008). The best-studied target of retrograde signaling is represented by PhANGs, but plastids are also involved in tuning the expression of nuclear genes involved in response to a plethora of biotic and abiotic conditions. A recent meta-analysis of microarray studies of systems where high levels of retrograde signaling were induced has identified a core module of 39 nuclear genes that were subject to regulation in response to all signals examined (Glasser et al., 2014). The genes in this group, presumably representing the core retrograde-response module, are all known to be responsive to sugar, reactive oxygen species (ROS), abscisic acid (ABA) and/or auxin signaling pathways. Thus retrograde signaling may exploit a common component of these signaling pathways to mediate changes in gene expression.
The GENOMES UNCOUPLED (GUN) Mutants – Aiming to the Nucleus with Guns
Very helpful in deciphering the retrograde signaling phenomenon were mutants isolated from genetic screens. The gun (genome uncoupled) mutants escaped the pattern of suppressed PhANG expression despite defective chloroplast physiology or inhibited biogenesis. There are numerous excellent reviews of the role of guns in retrograde signaling (Woodson and Chory, 2008, 2012; Barajas-Lopez Jde et al., 2013; Chi et al., 2013). So far, six gun mutants have been identified and they can be classified according to pathways they belong to. Whereas the gun1 mutant results from mutation in a gene encoding a chloroplast-localized pentatricopeptide repeat-containing protein (PPR), the gun2-6 mutants are associated with tetrapyrroles synthesis (Susek et al., 1993; Mochizuki et al., 2001; Larkin et al., 2003; Strand et al., 2003; Koussevitzky et al., 2007). The exact role of GUN1 in PhANG regulation is far from understood, however, it is known to act upstream of ABSCISCIC ACID INSENSITIVE 4 (ABI4), an APETALA 2-type transcription factor that binds to the ACGT motif of light- and ABA-responsive elements (Koussevitzky et al., 2007). Interestingly, the expression of ABI4 was regulated by PTM, a chloroplast PHD-type transcription factor (Sun et al., 2011). The involvement of the key enzymes of the tetrapyrrole synthesis pathway in the gun phenotype led to detailed investigations of tetrapyrroles, especially Mg-protoIX, as putative retrograde signals. However no correlation between Mg-protoIX levels and retrograde signaling could be established (Matsui et al., 2008; Moulin et al., 2008). Interestingly, the gun6 mutant identified heme as a strong candidate for mediating plastid-to-nucleus signaling (Woodson et al., 2011). Moreover, it was proposed that the impact of tetrapyrrole biosynthesis on nuclear gene expression is mediated by singlet oxygen (1O2)-induced signaling and feedback regulated 5-aminolevulinic acid (ALA) synthesis (Schlicke et al., 2014).
SAL1-PAP Chloroplast Retrograde Pathway
The detailed analysis of sal1, an Arabidopsis phosphonucleotidase mutant, has identified a known second messenger as acting in chloroplast-to-nucleus signaling. Estavillo et al. (2011) have demonstrated that the chloroplast and mitochondria-localized SAL1 phosphatase regulates the steady-state level of 3′-phosphoadenosine 5′-phosphate (PAP) by dephosphorylating it to an adenosine monophosphate (AMP). In the sal1 mutant, or in response to drought stress or high light intensity, PAP levels increased, inducing expression of ASCORBATE PEROXIDASE 2 and EARLY LIGHT INDUCIBLE PROTEIN 2, two nuclear genes whose expression is induced by high light stress (Harari-Steinberg et al., 2001; Caverzan et al., 2012). It has been proposed that PAP travels from chloroplasts to the nucleus where it regulates nuclear gene expression. Nucleus-localized exoribonucleases (XRNs) are likely targets of PAP, and by repressing their activity PAP may stimulate expression of high light and drought-responsive genes, leading to increased tolerance (Estavillo et al., 2011; Figure 2A).
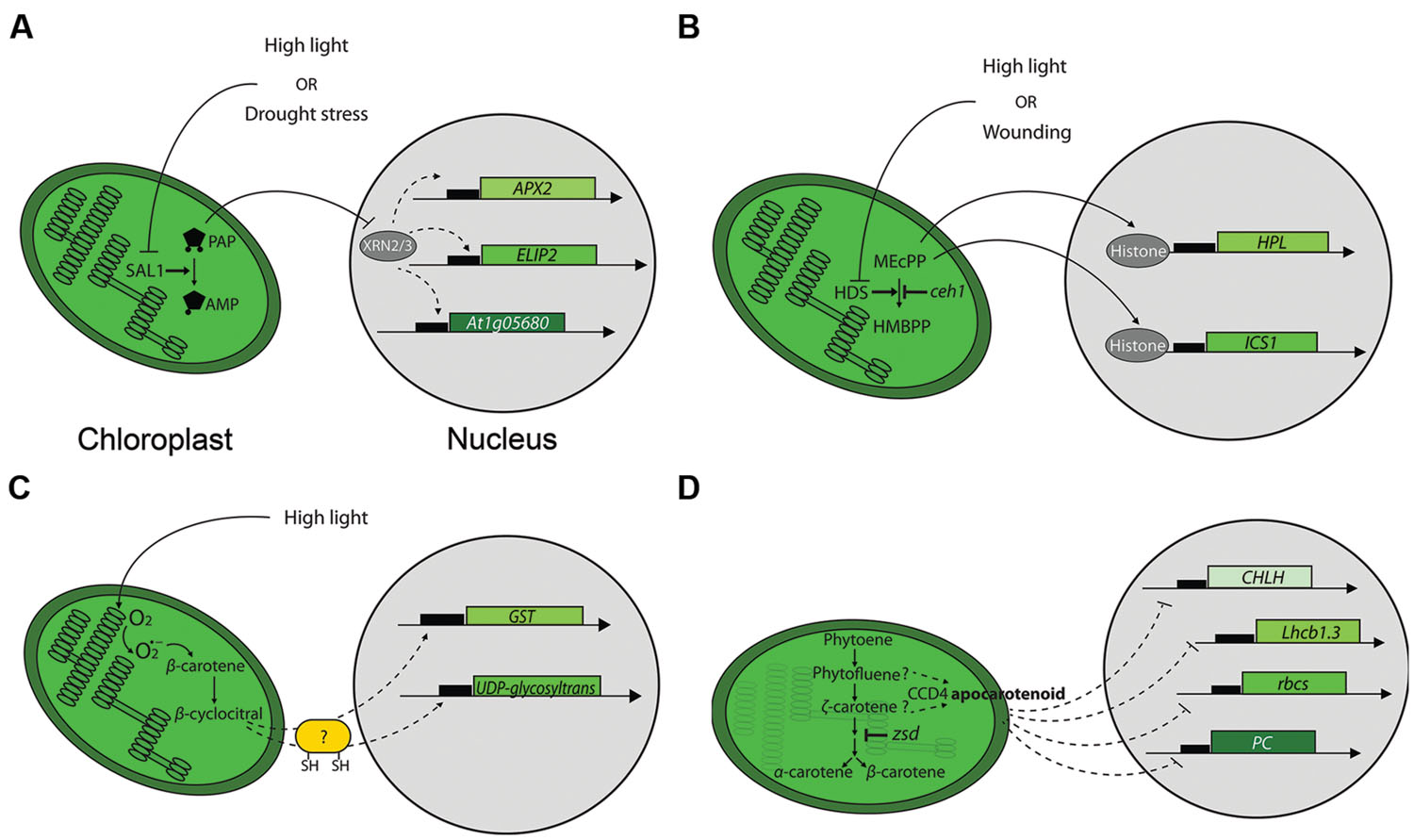
FIGURE 2. Mechanisms of chloroplast-to-nucleus signaling. (A) Retrograde signaling by PAP. High light or drought stress inhibits SAL1 phosphatase and leads to the accumulation of PAP. PAP likely inhibits specific exoribonucleases (XRNs) to modify nuclear genes expression. APX2 and ELIP2 stand for ASCORBATE PEROXIDASE 2 and EARLY LIGHT INDUCIBLE PROTEIN 2 genes, respectively. (B) Retrograde signaling by MEcPP. High light or wounding inhibits 1-hydroxy-2-methyl-2-(E)-butenyl-4-diphosphate synthase (HDS), leading to the subsequent accumulation of MEcPP. MEcPP affects nuclear gene expression via a mechanism proposed to involve chromatin remodeling by destabilizing DNA-histone interactions. HPL and ICS1 stand for HYDROPEROXIDE LYASE and ISOCHORISMATE SYNTHASE 1genes, respectively. (C) Carotenoid-derivative β-cyclocitral mediates retrograde signaling. The ROS singlet oxygen induces formation of β-cyclocitral during high light treatment. β-cyclocitral’s action on selected nuclear genes is proposed to involve proteins containing sulphydryl groups. The genes depicted are GLUTATHIONE-S-TRANSPHERASE (GST) and UDP-glycosyltransferase. (D) An unidentified apocarotenoid affects expression of nuclear genes. It is proposed that the putative signaling apocarotenoid accumulates in chloroplasts due to compromised ζ-carotene desaturase activity that results in accumulation of phytofluene and ζ-carotene, putative substrates for the carotenoid cleavage deoxygenase 4 (CCD4) enzyme that is prerequisite for the putative apocarotenoid synthesis. CHLH, Lhcb1.3, rbcs and PC stand for genes encoding the subunit H of the Mg-chelatase complex, light-harvesting complex 1.3 isoform, the Rubisco small subunit and plastocyanin, respectively.
Methylerythritol (MEcPP) Retrograde Pathway
Isoprenoid metabolism is a major biosynthetic pathway in plants (Cordoba et al., 2009). The Arabidopsis constitutively expressing HPL (ceh1) mutant displays enhanced expression of hydroperoxide lyase (HPL), a stress-inducible nuclear gene encoding a plastid-localized protein of the oxylipin pathway. The ceh1 mutation disrupted a plastid-localized enzyme (HDS) that catalyzes conversion of methylerythritol cyclodiphosphate (MEcPP) to hydroxymethylbutenyl diphosphate (HMBPP; Xiao et al., 2012). The absence of CEH1 led to accumulation of MEcPP and induced the expression of a subset of stress-associated genes, including ISOCHORISMATE SYNTHASE 1 (a key plastidial enzyme in the salicylic acid (SA)-biosynthetic pathway) and HPL, but not ALLEN OXIDE SYNTHASE [AOS, encoding a plastid-localized protein of the jasmonic acid (JA)-biosynthetic pathway]. HDS-depleted plants with increased levels of MEcPP accumulated SA and displayed increased resistance to infection by biotrophic pathogens. Importantly, elevated MEcPP levels and increased expression of HPL are observed upon both wounding and high light treatment, demonstrating involvement of MEcPP in a retrograde pathway involved in abiotic stresses (operational control) distinct from the gun signaling pathway. Therefore MEcPP is a retrograde signal inducing targeted stress responses. The proposed mechanism of MEcPP action involves direct modification of chromatin remodeling by disruption of DNA-histone interactions (Xiao et al., 2012; Figure 2B).
Derivatives of Carotenoids as Signaling Molecules
Carotenoids are tetraterpenoid products of the isoprenoid biosynthetic pathway that also generates ABA and strigolactones (Ruyter-Spira et al., 2013; Giuliano, 2014). Carotenoids are constituents of the light harvesting complexes where they serve as accessory pigments to extend the absorption spectra of the chlorophylls, and they have critical protective roles as scavengers of singlet oxygen (1O2) generated by the PSII reaction center (Telfer, 2014). Carotenoid derivatives have recently been proposed to act as chloroplast-generated signaling molecules that link chloroplast activity and nuclear gene expression (Ramel et al., 2012; Avendano-Vazquez et al., 2014; Van Norman et al., 2014).
β-cyclocitral is a product of singlet oxygen-induced β-carotene oxidation. This volatile molecule contains an α,β-unsaturated carbonyl, designating it as a reactive electrophile species (RES). β-cyclocitral’s accumulation in Arabidopsis leaves during high-light stress correlated with accumulation of singlet oxygen, supporting the notion of β-cyclocitral as an oxidation product of β-carotene (Ramel et al., 2012). Consistent with this, transcriptome analysis by DNA microarrays revealed that about 80% of β-cyclocitral-induced or repressed genes are also responsive to singlet oxygen overproduction in the Arabidopsis fluorescent (flu) mutant (op den Camp et al., 2003). Using qRT-PCR Ramel and coworkers demonstrated that all singlet oxygen marker genes tested are also induced by β-cyclocitral. Importantly, this effect seemed to be specific to β-cyclocitral as the overlap between gene expression changes induced by β-cyclocitral and other RES like methyl vinyl ketone or malondialdehyde (MDA) was smaller. Among the genes most strongly induced by β-cyclocitral were 10 glutathione-S-transferase (GST) genes and 12 UDP-glycosyltransferases (Figure 2C). Both groups of genes are involved in detoxification processes conferring tolerance to singlet oxygen in Chlaydomonas reinhardtii (Ledford et al., 2007). The protective effect of β-cyclocitral resulted in better PSII quantum efficiency and lower lipid peroxidation under high light stress (Ramel et al., 2012). Finally, the current model proposes β-cyclocitral as a stress molecule generated in chloroplasts under photooxidative stress that reprograms gene expression leading to stress acclimation. Thus, in addition to their roles in light harvesting and as antioxidants, carotenoids can also act as signaling molecules. The exact mechanism of β-cyclocitral action or its receptor in the nucleus are unknown, however, it is proposed that as an electrophile with a α,β-unsaturated carbonyl group it could react with electron donors such as proteins containing sulphydryl groups (Ramel et al., 2012, 2013). Very recently, elevated levels of β-cyclocitral were reported in Arabidopsis plastoglobule kinase mutants that have defective plastoglobule metabolism (Lundquist et al., 2013). The abc1k1 abc1k3 double mutant shows rapid chlorosis under high light stress, confirming β-cyclocitral’s, role in mediating stress responses.
In addition to β-cyclocitral, apocarotenoids are also potential chloroplast retrograde signaling components. Analysis of the Arabidopsis ζ-carotene desaturase mutant (zds/clb5/spc1/pde181) displaying arrested chloroplast biogenesis at a very early stage of development led to the conclusion that the accumulation of an uncharacterized apocarotenoid can act as a retrograde signal (Avendano-Vazquez et al., 2014). This apocarotenoid is likely generated by the activity of the carotenoid cleavage deoxygenase 4 (CCD4) enzyme on ζ-carotene (Figure 2D). Accumulation of this putative cleavage product was shown to modulate expression of many nuclear genes required for leaf development leading to a severe phenotype that included arrested chloroplast development and leaves with defective adaxial-abaxial patterning. Since neither ROS nor ABA nor strigolactone signaling pathways were responsible for the observed phenotypes, it was concluded that the putative phytofluene or ζ-carotene –derived apocaroteniod is part of a novel retrograde signaling pathway. Interestingly, the observed defects were restricted to primary leaves, underscoring the differences in developmental regulation between plastids in different organs (Avendano-Vazquez et al., 2014).
A third carotenoid derivative is implicated in plastid retrograde signaling, this time with respect to lateral root (LR) development (Van Norman et al., 2014). Reduced LR formation in Arabidopsis seedlings treated with norflurazon was observed and further investigation indicated that a β-carotene derivative is required for prebranch site formation (Van Norman et al., 2014). Extensive genetic analyses ruled out ABA and strigolactone as the carotene-derived signaling molecule involved in LR formation. Additionally, treatment of Arabidopsis seedlings with D15, a candidate inhibitor of carotenoid cleavage at the 9,10 position, resulted in a highly significant decrease in LR capacity, suggesting that this unknown apocarotenoid is likely cleaved at this position. Interestingly, even though reduced LR capacity and small albino shoots were observed in carotenoid biosynthesis mutants and in plants treated with norflurazon, plants treated with D15 had green shoots of comparable size to wild type. This demonstrates that the involvement of carotenoids in LR formation is separate from their photoprotective function (Van Norman et al., 2014). Intriguingly, carotenoid biosynthesis was found to occur in differentiated parts of the root at some distance from the oscillating zone where prebranch sites and eventually LRs formed. Thus the non-cell autonomous function of (apo)carotenoids seems to be required for development of LRs (Van Norman et al., 2014).
Chloroplast Proteins as Retrograde Signals
It is a broadly accepted paradigm that most nucleus-encoded chloroplast proteins reach this compartment due to the transit peptides located at their N-termini. Cleavage of the signal peptides after entry in to the chloroplast generates the functional chloroplast proteins. However, accumulating data show that some chloroplast proteins also act in the nucleus. While it is tempting to speculate that this is attributable to two-way protein movement between chloroplasts and cytoplasm, this has not been unambiguously demonstrated. However, a physiological function for this chloroplast-nucleus dual localization is apparent and it seems to be indispensable for proper plant response to pathogen attack and abiotic stress, and emphasizes the function of chloroplasts as signaling compartments.
One such protein, PTM (PHD type transcription factor with transmembrane domains) was shown to provide a physical link in signaling between chloroplasts and nucleus to regulate gene expression (Sun et al., 2011). This membrane-bound transcription factor (MTF) is localized to chloroplast outer envelope by four transmembrane domains at its C-terminus (Figure 3). The N-terminus of PTM contains a DNA-binding homeodomain box, a different transcription factors (DDT) domain and a plant homeodomain (PHD). Interestingly, a shorter variant of this protein, lacking the transmembrane domains, was detected in nuclear fractions. Notably, increased amounts of the shorter PTM variant were detected upon treatment with either norflurazon or lincomycin, and on exposure to high light. Through the application of protease inhibitors, it was demonstrated that the shorter form of PTM was the result of serine protease activity (Sun et al., 2011; Adam, 2015). According to the proposed model, chloroplast signals induce the intramembrane proteolytic cleavage of full length PTM, producing a soluble shorter variant (∼58 kDa) that is released to the cytoplasm and finally transclocates to the nucleus where it binds, through its PHD domain, to the ABI4 promoter to induce ABI4 expression. ABI4, in turn binds to the Lhcb promoter, close to the CUF1 element and precludes binding of G-box-binding factors required for the expression of Lhcb and other PhANGs. This model explains the gun phenotype observed in ptm and abi4 mutants. Moreover, the amount of processed PTM declined in the gun1 mutant, suggesting a complex regulatory network.
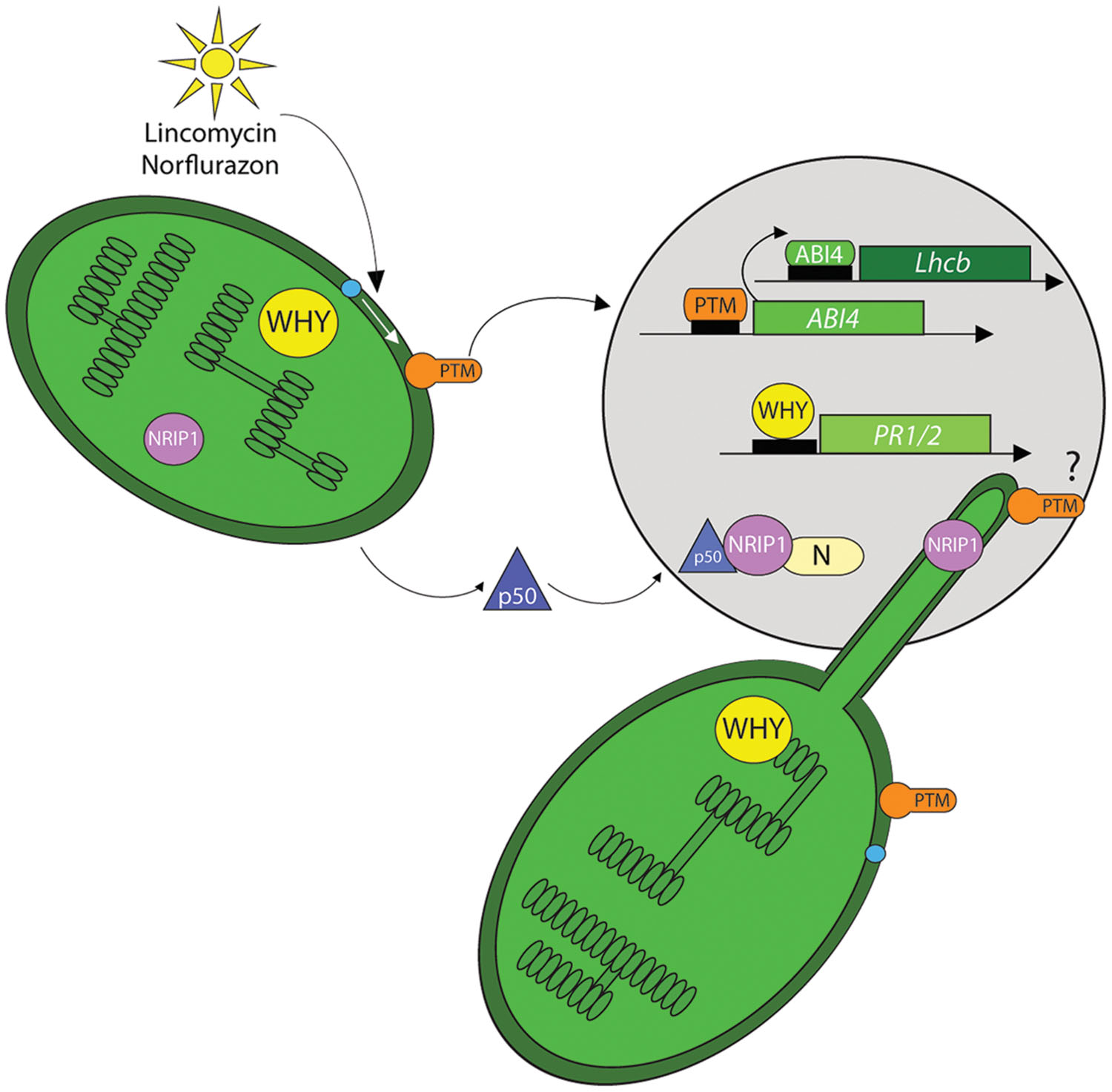
FIGURE 3. Chloroplast proteins as retrograde signals. A few chloroplast proteins have been implicated in directly modulating nuclear gene expression by their nuclear localization. These proteins may transit the cytoplasm by an unknown mechanism. Alternatively, it has also been proposed that they may move through stromules to enter the nucleus. High light, lincomycin or norflurazon treatments induce a serine protease-dependent (blue dot) proteolytic-cleavage of the PTM, a chloroplast envelope-bound plant homeodomain (PHD) transcription factor. The cleavage product is found in the nucleus where it binds to promoter region of the ABI4 transcription factor. ABI4 in turn associates with the regulatory sequences of the Lhcb genes and prevents their transcription. The chloroplast protein Whirly1 also localizes to the nucleus and this is correlated with increased expression of PATHOGENESIS RELATED GENE 1 and 2 (PR1/2). Upon TMV infection the chloroplast-localized NRIP1 is also detected in the nucleus where it interacts with the helicase domain of the TMV replicase (p50). Finally a trimeric complex of p50, NRIP1 and the N protein is localized to nucleus to provide resistance against the virus. It is suggested that NRIP1 may use stromules to translocate from the chloroplast to the nucleus.
WHIRLY1 provides another example of a chloroplast protein with a role in the nucleus. A WHIRLY1 fusion protein expressed in the plastid genome of tobacco, localized to both plastids and nuclei, and the two subpopulations were the same molecular size (Isemer et al., 2012). As a consequence WHIRLY1-regulated PR (PATHOGENESIS RELATED) genes were upregulated under normal growth conditions (Figure 3). WHIRLY1 is part of the transcriptionally active chromosome in plastids, and interestingly, another component of this complex, pTAC12 (HEMERA), also showed dual localization to chloroplasts and nuclei with unchanged molecular mass, excluding any proteolytic modification (Chen et al., 2010). However, unlike in the case of WHIRLY1, the dual localization of HEMERA in a single cell has not yet been demonstrated. Such a demonstration is important for ruling out the possibility that dual localization could be caused by fluctuating distribution resulting from specific cell types or developmental stages. It has been proposed that WHIRLY1 conveys information about the chloroplast redox state to the nucleus, and SA regulates this communication (Foyer et al., 2014). The proposed mechanism of WHIRLY1 action would be similar to that of NPR1 (See below). The mechanism and consequences of the proposed translocation of NRIP1 (another chloroplast-localized protein) to the nucleus is discussed later.
There are a few hypotheses that aim to explain the nuclear localization of chloroplast-targeted proteins and they invoke mechanisms enabling translocation of chloroplast-localized proteins to nuclei. One of them proposes permeabilization of the chloroplast outer membrane by an unknown mechanism. Another posits that stress-induced modification of chloroplasts results in the formation of stromules that contact nuclei could facilitate direct trafficking of chloroplast proteins (Caplan et al., 2008). In agreement with the former, a recent communication described GFP-fusion protein leakage from functional chloroplasts upon pathogen attack (Kwon et al., 2013). Interestingly, this occurrence was shown to be dependent on ROS. Evidence for the latter hypothesis of stromule involvement in chloroplast protein translocation is now being reported (see Stromules below). Regardless of the mechanism governing the distribution of chloroplast proteins to the cytoplasm and/or nucleus, this phenomenon may represent an important pathway for direct communication between chloroplasts and nuclei, and provides important insights in understanding the molecular basis of retrograde signaling.
Chloroplasts and Inter-Organellar Signaling
It has long been appreciated that there is metabolic crosstalk between organelles. Metabolic pathways often involve multiple organelles and metabolic intermediates may be transported across membranes by diffusion or by specialized transporters in an energy-dependent manner after traversing the cytosol. However, there is emerging evidence that direct physical contact between organelles may provide a major route for metabolic exchange (Figure 4). Such direct contact would also provide routes for inter-organelle signaling, although evidence for this is still limited. Thus, chloroplasts, besides producing signals that may travel long distances from plastids to their targets, may also communicate directly with other organelles through physical contacts. Here we consider evidence for chloroplast-organelle contacts and possible roles for these contacts in signaling.
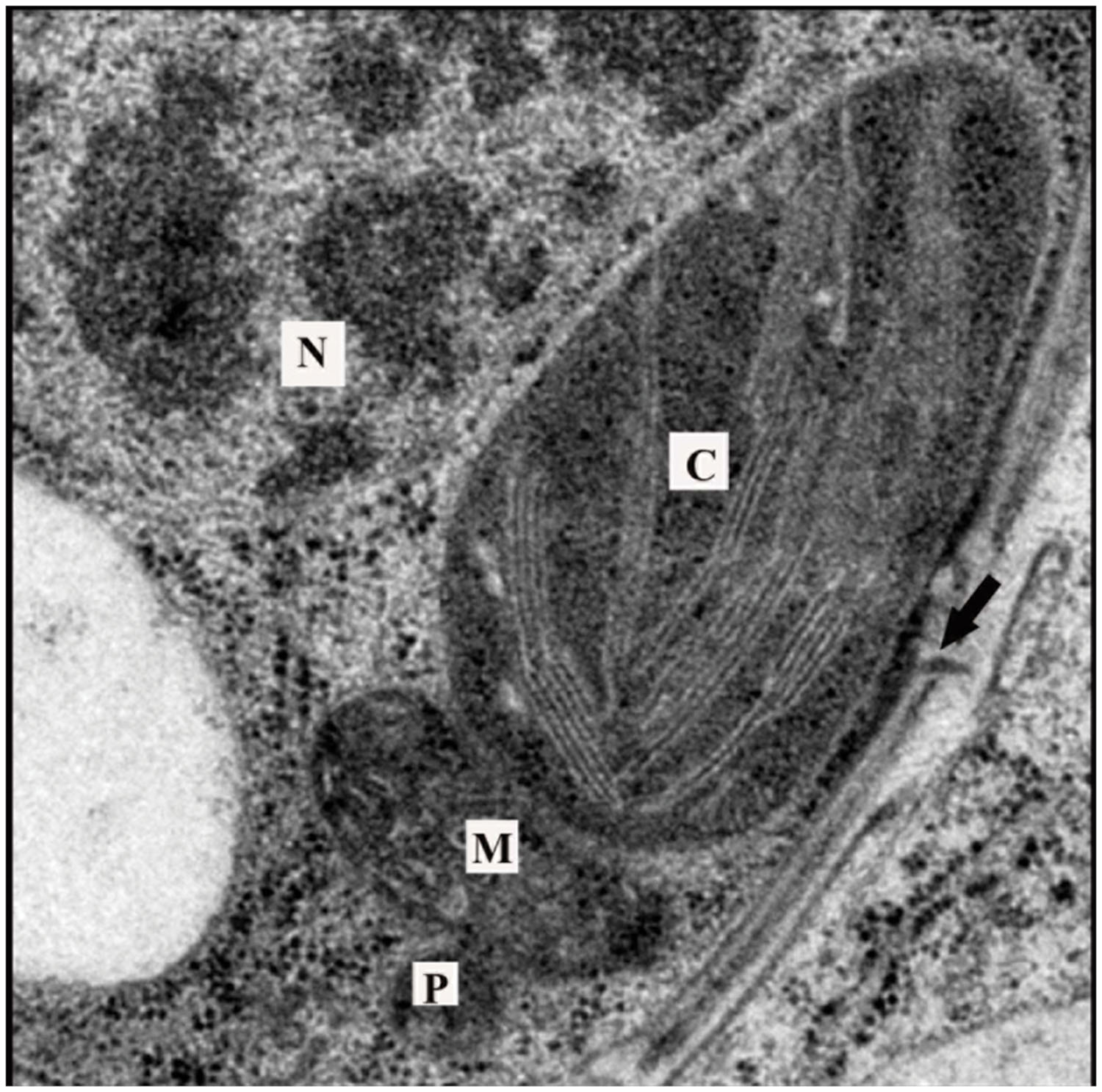
FIGURE 4. Arrangement of organelles in a leaf cell. Transmission electron microscopy images often reveal chloroplasts in close proximity with peroxisomes and mitochondria. Chloroplasts can also be observed near the nucleus and cell wall. Note the presence of a plasmodesma in the cell wall (arrow). Such arrangements of organelles would minimize distances that signals must traverse to arrive at their target. C, chloroplast; M, mitochondria; P, peroxisome; N, nucleus.
Stromules for Plastid-to-Plastid and Plastid-to-Nucleus Signaling
Rediscovered by Kohler et al. (1997), the observation of tubular protrusions from plastids changed our thinking about chloroplasts and possible pathways for signaling (Kohler et al., 1997). These so-called stromules are stroma-filled tubules enclosed by the inner and outer plastid envelope membranes, and are 0.4–0.8 μm in diameter and of variable length typically up to 65 μm (Gray et al., 2001). They are more abundant in non-green plastids than in chloroplasts. Stromules are distinct from the chloroplast protrusions (CPs) that form during stress (Holzinger et al., 2007a,b), and that are involved in the sequestration of Rubisco from the rest of the chloroplast body (Yamane et al., 2012).
Stromule formation is dependent on both intrinsic and extrinsic factors. The size of plastid, plastid identity, state of differentiation and density of plastids all determine stromule formation (Waters et al., 2004). The actin cytoskeleton has also been reported to be important for stromule formation and movement (Kwok and Hanson, 2003; Kwok and Hanson, 2004b; Gunning, 2005; Holzinger et al., 2007b), and the myosin XI motor is required for stromule formation in Nicotiana benthamiana chloroplasts (Natesan et al., 2009; Sattarzadeh et al., 2009). In contrast, it has recently been reported that isolated chloroplasts can form stromules (Brunkard et al., 2015b). Stromule formation is also temperature sensitive, and temperatures around 20°C appear optimal for stromule formation (Holzinger et al., 2007a), while lower temperatures inhibit their formation (Gray et al., 2012) and higher temperatures induce CP formation (Holzinger et al., 2007a). The light-dependence of stromule formation is somewhat controversial. There have been reports that light is not required for stromule formation (Kwok and Hanson, 2003; Gray et al., 2012), but recent findings demonstrate increased formation of stromules from Arabidopsis mesophyll chloroplasts during the day as compared to the night (Brunkard et al., 2015b). It is possible the differing results can be explained by differences in plant growth conditions and the types of plastids examined in each experiment. Besides light and temperature the hormone ABA, likely generated in response to environmental stresses, also induces stromule formation (Gray et al., 2012). Consistent with this, salt and osmotic stress also increased the fraction of stromule-bearing chloroplasts.
The most obvious consequences of stromules are significant enlargement of both plastid envelope surface and plastid volume. These modified plastid properties could affect the rate of plastid import and export, and also cause changes in plastid compartmentalization, respectively. Consistent with these presumed functions, it was shown that chlorophyll and thylakoid membranes are absent from stromules (Kohler et al., 1997; Holzinger et al., 2007a; Newell et al., 2012). The trafficking of plastid genomes or genetic material via stromules has also been ruled out (Newell et al., 2012). On the other hand proteins like GFP, aspartate aminotransferase and Rubisco complexes of a molecular weight around 550 kDa have been localized within stromules (Kwok and Hanson, 2004a). Moreover, interconnections between individual plastids via stromules have been reported, and photobleaching experiments and the use of photoconvertible proteins have demonstrated the transfer of proteins between them (Kohler et al., 2000; Hanson and Sattarzadeh, 2011, 2013, but see Schattat et al., 2012, 2015). Given that connections between plastids are rare, the biological significance of possible plastid-to-plastid trafficking remains unclear (Hanson and Sattarzadeh, 2013).
Stromules are involved in metabolic responses to stress, namely chloroplast autophagy in response to nutrient starvation (Ishida et al., 2008). Through observations of fluorescently tagged proteins and the Arabidopsis atg5 autophagy mutant, it was demonstrated that plastid stromal proteins can be remobilized to the vacuole via the ATG-dependent authophagic pathway, without destroying the chloroplast. Similar observations have been reported from rice (Izumi et al., 2015). According to the proposed model stressed-induced autophagy sequesters stromules by forming an isolation membrane that eventually clips off a given stromule and its stromal contents, and then transports the cargo to the vacuole for degradation (Ishida et al., 2008). Interestingly, both stromules and protrusions were identified in potato tuber amyloplasts (Borucki et al., 2015). These protrusions, unlike stromules, are likely involved in starch accumulation in the parenchyma storage cells. The involvement of protrusions in accumulation of starch had been previously demonstrated (Langeveld et al., 2000). Thus stromules are involved in metabolism.
Plastids and stromules have been repeatedly observed in close proximity to other organelles including mitochondria, other plastids, ER, plasma membrane and nuclei. From the perspective of signaling, however, the most interesting seems to be the distinct distribution of plastids around nuclei, including concentration of stromules around and, most intriguingly, inside the nucleus (Collings et al., 2000; Kwok and Hanson, 2004c). Clusters of plastids with long stromules of 20–30 μm localized around the nucleus were observed in N. tabacum petioles of cotyledons (Kwok and Hanson, 2004c), as well as in the lower part of the hypocotyl where the plastids were preferentially arranged around the nucleus with long stromules of up to 100 μm extending to the cell periphery (Natesan et al., 2005). Such concentration of plastids around nucleus was also observed in petal cells and in shoot meristems (Kohler and Hanson, 2000; Kwok and Hanson, 2004c). Stromules have also been observed lying in grooves and invaginations of the nuclear membranes in tobacco epidermal cells (Kwok and Hanson, 2004c). Such direct connections were proposed to increase efficiency in plastid-nucleus communication. Stromules have also been implicated in chloroplast-to-ER signaling (Schattat et al., 2011a,b), and in intercellular signaling (Kwok and Hanson, 2004c) but this has not been explicitly tested or proven. These suggestions are based on interaction between stromules and ER or the plasma membrane, respectively, and further characterization of these interactions is warranted.
Stromules and stromule-nucleus contacts may have important roles in host–pathogen interactions. The induction of stromules and remobilization of chloroplasts to surround nuclei was observed in N. benthamiana leaves in response to infiltration with GV3101, common lab strain of Agrobacterium tumefaciens (Erickson et al., 2014). In addition, starch accumulated in GV3101-treated leaves and the levels of soluble sugars also increased. However, another lab strain LBA4404 did not produce these effects. The introduction of the trans zeatin synthase (tsz) gene from the GV3101 plasmid into LBA4404 led to the induction of stromules and other cellular changes typically observed on GV3101 infiltration. Indeed, direct application of cytokinin to leaves produced phenotypes similar to those obtained with GV3101. Thus, cytokinin may mediate the production of stromules and chloroplast movements during some plant–pathogen interactions.
The question of why stromule induction and chloroplast-nucleus associations occur during plant–pathogen interactions has recently been addressed. Nicotiana N RECEPTOR INTERACTING PROTEIN (NRIP)1 is a chloroplast rhodanese sulphurtransferase that is required for effector triggered immunity (ETI) against TMV mediated by the N innate immune receptor (Caplan et al., 2008). In the presence of the viral p50 helicase effector NRIP1 was recruited to the cytoplasm and nucleus where it formed a protein complex with the TIR domain of the N immune receptor (Figure 3). The interaction with N in the nucleus and cytoplasm is necessary for ETI. These results raised the intriguing questions of whether NRIP1was remobilized from the chloroplast and how this could be accomplished. Recent data suggest that stromules may provide a route for NRIP to traffic from the chloroplast to the nucleus. Co-expression of N and p50 induced formation of stroumles during HR-PCD associated with this interaction (Caplan et al., 2015; Figure 5). Similarly, stromule induction was observed during ETI initiated in response to bacterial pathogens, and on treatment with the defense-related signaling molecules SA and the ROS H2O2. Through correlative EM-fluorescence microscopy, the authors provide convincing evidence that stromules contact the nucleus during N-mediated defense. In elegant experiments that fuse a nuclear exclusion sequence (NES) to NRIP, the authors provide quantitative evidence that NRIP1 is indeed trafficked from the chloroplast to the nucleus. H2O2 also traffics to the nucleus via stromules. In plants overexpressing chloroplast outer membrane protein CHLOROPLAST UNUSUAL POSITIONING 1 (CHUP1) stromule formation is abolished, suggesting that this membrane plays a role in stromule formation. This was confirmed in chup mutants and knockdown plants, where constitutive stromule formation was observed. The importance of stromules to HR-PCD and the defense response was underscored by the accelerated HR-PCD observed in those plants. These results are suggestive of a role for stromules in intracellular trafficking and possibly signaling during plant–pathogen interactions, and these possibilities warrant further examination. It will be exciting to test whether plastids and/or stromules are able to create hemifusion membranes with the nuclear membrane and whether other signaling molecules or metabolites use this route for chloroplast signaling.
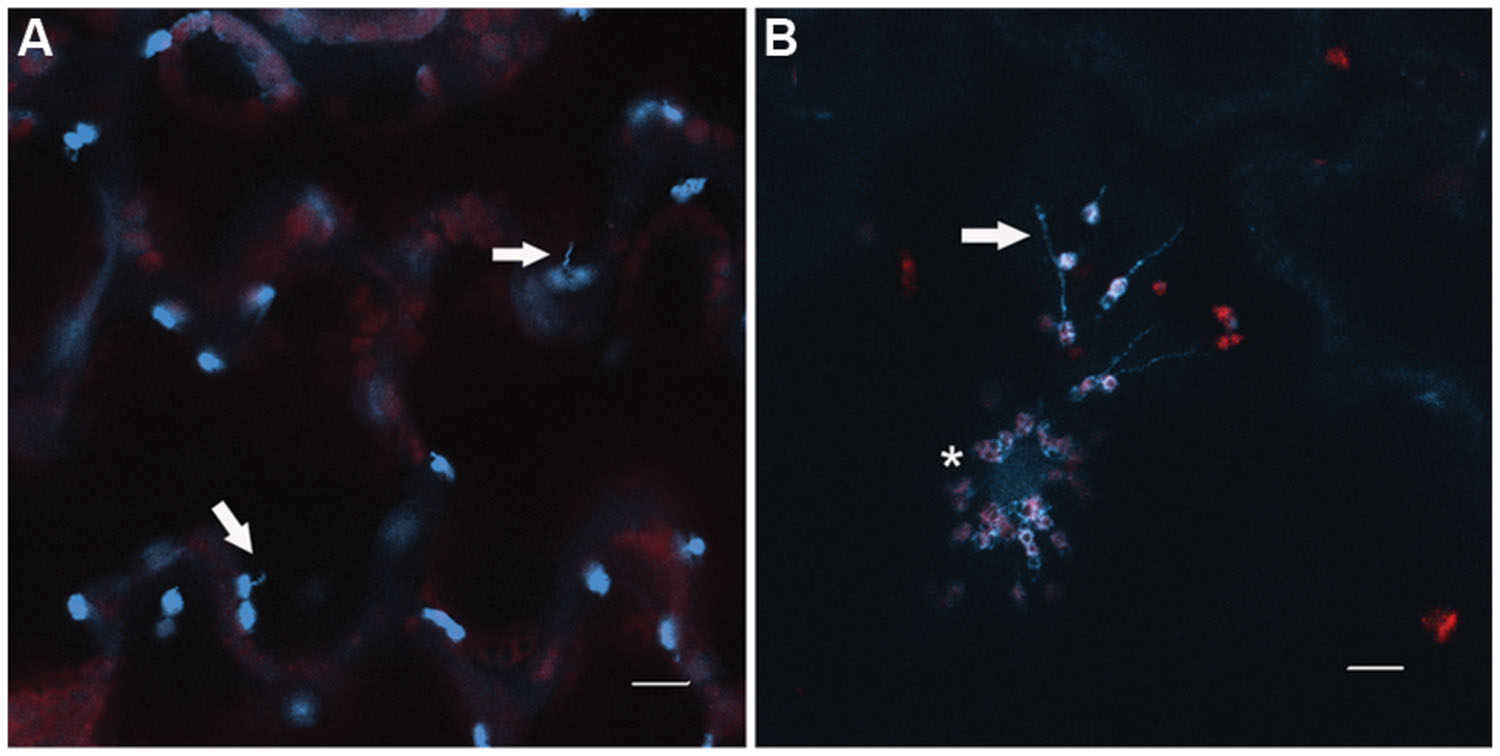
FIGURE 5. Chloroplast behavior during defense. (A) Stromules (arrows) are observed intermittently from chloroplasts in the epidermis of Nicotiana benthamiana leaves. (B) Upon infection with Tobacco mosaic virus, chloroplasts cluster around the nucleus (asterisk), and stromule formation is induced (arrow). The chloroplasts shown are expressing NRIP1-CFP (Caplan et al., 2008, 2015). Note the NRIP1-CFP signal detected in the nucleus in TMV-infected chloroplasts, indicative of the translocation of NRIP1 from the chloroplasts to the nucleus. Images were collected on a Zeiss LSM 710 confocal laser scanning microscope and single focal plane images are shown. Scale bar is 10 μm.
A separate study raises the possibility that stromules may also function in intercellular and intracellular trafficking of pathogens. The chloroplast-localized chaperone heat shock cognate 70 kDa protein (cpHsc70-1) was identified as interacting with the AbMV movement protein (MP; Krenz et al., 2010). In uninfected tissues cpHsc70-1-YFP homooligomerization was demonstrated by bimolecular fluorescence complementation (BiFC). cpHsc70-1-YFP oligomers localized to chloroplasts and near the cell periphery. Intriguingly, cpHsc70-1-YFP localized to punctate structures in chloroplasts and filaments that stretched like a “string of pearls” between chloroplasts or to the cell periphery in AbMV-infected leaves. Reduced levels of the Hsc70 ortholog in N. benthamiana by virus induced gene silencing (VIGS) led to decreased intercellular trafficking of AbMV while viral replication was unaffected, suggesting that the chloroplast chaperone and stromules may have a role in viral trafficking that is independent of viral replication and accumulation (Krenz et al., 2010). Indeed, it has been proposed that the Hsc-70 chaperone in stromules may facilitate viral transport from chloroplasts to the cell periphery and plasmodesmata, from where intercellular spread could occur (Krenz et al., 2012). AbMV is a geminivirus that replicates in the plant nucleus, but AbMV is also found in plastids (Groning et al., 1987); therefore stromule-nucleus contacts could also provide a route for viral trafficking to nuclei in newly infected cells.
Another interesting question regarding the involvement of stromules in chloroplast signaling is when during evolution would such an innovation have arisen. The R gene RESISTANCE TO POWDERY MILDEW (RPW)8.2 confers broad resistance to fungi that cause powdery mildew. RPW8.2 localizes to the extrahaustorial membrane (EHM) during fungal penetration of the host cell (Wang et al., 2009). Interestingly, RPW8.2 mutants have recently been reported to localize to an unknown membrane that appears to surround stromules (Wang et al., 2013a). This membrane has been termed the peristromule membrane (PSM) and it is proposed that they may share some as-yet-unknown characteristics with the EHM. Some RPW8.2 mutants localize to both the EHM and PSM, and while others localize to the nucleus. Notably, invasion by fungal haustoria induced the formation of stromules that purportedly connect plastids with haustoria. The authors speculate that stromules may represent an ancient interface for host–pathogen interactions. However, wild-type RPW8.2 was not observed labeling the PSM, and the authors speculate that this may be due to rapid cycling of RPW8.2 on and of the PSM (Wang et al., 2013a).
Together, these studies suggest that stromules could function in signaling in mediating responses to both biotic and abiotic stress, however, this remains to be demonstrated. Understanding the cellular functions of stromules will no doubt reveal intriguing aspects of plant cell biology and possibly identify new targets for engineering plants with modified responses to biotic and abiotic stresses.
Chloroplast-Endoplasmic Reticulum Contacts: Lipid Signaling
A conundrum in plant biology is that the enzymes for a given metabolic pathway are often found in different subcellular compartments. One striking example of this is the biogenesis of complex lipids in some plants including Arabidopsis. In these plants fatty acids synthesized de novo in the chloroplast by the prokaryotic pathway may be exported to the ER where they are assembled into lipids by the eukaryotic pathway before import back into chloroplasts (Benning, 2009). Despite concerted efforts to identify candidate transporters that would allow the substrates to translocate from the chloroplast to the ER, these transporters have not yet been discovered (Wang and Benning, 2012). This raises the intriguing possibility that metabolites may traffic directly from the chloroplast to the ER through membrane interactions. Moreover, direct physical contact sites between chloroplasts and ER, named plastid-associated membranes (PLAM), have long been reported by transmission electron microscopy in many plant and algal species (Cran and Dyer, 1973; Crotty and Ledbetter, 1973; Renaudin and Capdepon, 1977; McLean et al., 1988). More recently, confocal microscopy has also suggested chloroplast-ER membrane contact sites (MCS; Andersson et al., 2007; Tan et al., 2011). Indeed, these MCS are likely held together through strong protein–protein interactions since the ER remained associated with chloroplasts even after application of forces of 400 pN (Andersson et al., 2007). Once synthesized in the ER glycerolipids have to be imported into the chloroplasts. Mathematical modeling supports the import of diacylglycerol (DAG) from the ER to chloroplasts (Marechal and Bastien, 2014). Genetic analyses have identified the TGD complex as essential for import of ‘eukaryotic’ precursors into chloroplasts (Boudiere et al., 2014). TGD1-3 constitute a bacterial ABC transporter, while TGD4 apparently forms a β-barrel that localizes to both chloroplasts and ER (Wang and Benning, 2012). Thus transporters are indeed important mediators of lipid trafficking.
The DellaPena group has proposed an elegant hypothesis to explain chloroplast-ER membrane continuity (Mehrshahi et al., 2014). This group has developed a transorganellar assay to test whether non-polar metabolites, exemplified by tocopherols (vitamin E), located in the plastid envelope could directly access the lumen of the ER and modulate enzymes located there (Mehrshahi et al., 2013). To do this, tocopherol cyclase (TC) that is normally a chloroplast resident protein was retargeted to the ER in the background of the vte1 mutant that does not make TC and tocopherols (Porfirova et al., 2002). Excitingly, this ER-localized TC complemented the vte1 mutation and tocopherol levels were restored to almost wild-type levels in the rescued lines (Mehrshahi et al., 2013). This study revealed similar access to chloroplast-localized substrates for ER-localized γ-tocopherol methyltransferase (γTMT) and α-carotene ε-ring hydroxylase LUTEIN DEFICIENT1 (LUT1). Based on their successful transorganellar complementation assay, the authors propose that an exchange of non-polar metabolites between plastids and endoplasmic reticulum occurs, most likely due to direct contacts between those organelles at PLAM sites according to the membrane hemifusion model (Mehrshahi et al., 2014). The hemifusion model postulates that the fused membrane of the ER and chloroplast outer envelope would consist of the inner leaflet of the ER and the plastid outer envelope membranes, two envelopes with similar compositions of non-polar metabolites (Mehrshahi et al., 2014). A membrane of this nature would create an easy path for chloroplast-to-ER signaling by non-polar metabolites.
Chloroplast-Peroxisome Contacts
Peroxisomes are dynamic membrane-bound organelles of remarkable metabolic plasticity. They are found in all eukaryotic cells, and in plant cells are usually found in close association with mitochondria and chloroplasts (Figure 3). Peroxisomes are able to adjust their complement of enzymes in response to changes in environmental and developmental signals (reviewed in Goto-Yamada et al., 2015; Sandalio and Romero-Puertas, 2015). Peroxisomes were first recognized for their action in scavenging H2O2, however, it is clear that peroxisomes have much more extensive roles in cellular metabolism (Sandalio and Romero-Puertas, 2015). A subset of these reactions highlights the intimate metabolic coupling between chloroplasts and peroxisomes.
The photorespiratory pathway is the conversion of phosphoglycolate to CO2 and 3PGA, via a complex series of reactions that takes place across three separate subcellular compartments: chloroplasts, peroxisomes, and mitochondria (Foyer et al., 2009). Severe photorespiratory conditions initiate ROS-dependent lipid peroxidation in the chloroplast that leads to activation of the lipoxygenase (LOX)-mediated reaction, which is one of the starting points in the oxylipin metabolic pathway. LOX also initiates the synthesis of many cell constituents and signaling molecules, including jasmonates via oxo-phytodienoic acid (OPDA; Weber et al., 1997; Montillet et al., 2004). OPDA is then transported into the peroxisomes where two rounds of beta-oxidation serve to modify the fatty-acid side-chain of the ring. It is believed that the transport of OPDA is carried out by both active and passive transport (Leon, 2013).
Recent work has revealed that in addition to close metabolic coupling, peroxisomes and chloroplasts physically interact with each other in a light-dependent manner. Using the newly developed femtosecond laser technology and confocal laser scanning microscopy, Oikawa and colleagues demonstrated that peroxisomes adopted an elliptical shape to increase their surface area and more tightly adhered to chloroplasts in light (Oikawa et al., 2015). A force of 61 fN nm-2 was needed to disrupt the chloroplast–peroxisome interaction in the light compared to 23 fN nm-2 required to so do in the dark. Interestingly, these changes in peroxisome shape and location depended on photosynthesis, but were independent of photorespiration or the activity of photoreceptors, and the actin cytoskeleton negatively regulated the interaction between the chloroplasts and peroxisomes (Oikawa et al., 2015). The chloroplast–peroxisome physical interaction is consistent with reports from other systems that suggest direct interaction between organelles is necessary for metabolite exchange (Binns et al., 2006; de Brito and Scorrano, 2008). Thus the trafficking of compounds like OPDA during JA synthesis could potentially contribute to signaling between plastids and peroxisomes.
The Arabidopsis snowy cotyledon3 (sco3-1) mutant also provides support for signaling between chloroplasts and peroxisomes. In seedlings, the sco3-1 mutation interrupted chloroplast biogenesis, decreased chlorophyll accumulation, and disrupted thylakoids and, as a consequence, photosynthesis (Albrecht et al., 2010). This mutation also resulted in photoinhibition in mature leaves under high CO2 concentrations. It is quite interesting that the SCO3 protein is initially targeted to peroxisomes. Also interesting is that loss of SCO3 function led to cytoskeletal defects, specifically affecting microtubules. Thus, both the cytoskeleton and peroxisomes are necessary for normal chloroplast development. Further investigation of SCO3 function will illuminate the process of communication between chloroplasts and peroxisomes.
Chloroplast-Mitochondrion Contacts
Like chloroplasts, mitochondria are the end products of an endosymbiotic event, and they have also retained a portion of their ancestral genome (Woodson and Chory, 2008). Mitochondria-to-nucleus retrograde signaling is critical for coordinating expression of nuclear genes encoding mitochondrial proteins with expression of the mitochondrial genome (Woodson and Chory, 2008; Rhoads, 2011). Given their central roles in energy capture and utilization it is perhaps not surprising that chloroplasts and mitochondria exchange metabolites. Chloroplasts and mitochondria are also coupled by cellular redox status.
Mutant analyses have shed light on chloroplast-mitochondrion signaling. The expression of alternative oxidase (AOX), a nucleus-encoded mitochondrial gene, seems to be regulated by chloroplasts as increased expression of Arabidopsis and soybean AOX has been observed upon high light treatment (Finnegan et al., 1997; Blanco et al., 2014). Intriguingly, the white leaves of the chloroplast ribosome-deficient barley mutant albostrians as well as photo-bleached leaves of wild type obtained upon treatment with norflurazon, displayed elevated levels of mitochondrial DNA and transcripts (Hedtke et al., 1999). Consistent with this observation, recent analyses of leaves with green/white variegation in 12 ornamental plants confirmed that chloroplast dysfunction leads to increased levels of mitochondrial DNA in white sectors (Toshoji et al., 2012). Communication between chloroplasts and mitochondria seems to be bidirectional as mutation of genes encoding mitochondrial proteins can have profound effects on chloroplasts (e.g., Xu et al., 2008; Burch-Smith et al., 2011).
Several possible routes for communication between chloroplasts and mitochondria are proposed. There is coordinated expression of nuclear genes encoding chloroplast and mitochondria proteins, and it is therefore likely that some of the chloroplast signaling to mitochondria is accomplished by modulating nuclear gene expression. Indeed, expression of the MITOCHONDRIAL DYSFUNCTION STIMULON (MDS) suite of mitochondria-associated genes was regulated in response to chloroplast perturbations that included increased ROS and NO production (Ng et al., 2014). These MDS genes carry a common regulatory motif in their promoters that mediates their induction in response to mitochondrial retrograde signals (De Clercq et al., 2013; Ng et al., 2013). Moreover, it was demonstrated that Arabidopsis ABI4 regulates both Lhcb and AOX1A genes, providing a molecular link for nucleus-coordinated chloroplast-mitochondria communication (Koussevitzky et al., 2007; Giraud et al., 2009). Analysis of the Arabidopsis regulator of alternative oxidase 1 (rao1) mutant deficient in a nucleus-localized cyclin-dependent kinase E1 (CDKE1) that is a prerequisite for AOX induction, indicates it is another nucleus-localized sensor integrating mitochondrial and chloroplast retrograde signals (Blanco et al., 2014). Unlike wild type plants the rao1(cdke1) mutant was unable to induce AOX1A expression upon application of antimycin A or DCMU (that specifically targets chloroplasts). Further, the mutant displayed the gun phenotype upon induction of redox stress originating specifically from chloroplast photoelectron transport.
Another possible mechanism for chloroplast-to-mitochondrion signaling could involve dual targeting of proteins. The localization of proteins to multiple subcellular compartments is an ancient feature of land plants that can be observed in Physcomitrella patens (Xu et al., 2013b) and diatoms (Gile et al., 2015). In Arabidopsis, over 100 proteins are targeted to both chloroplasts and mitochondria (Carrie and Whelan, 2013). It is tempting to propose that, analogous to the situation where proteins have been shown to localize to both chloroplasts and nuclei, proteins may move from the chloroplast to the mitochondria. The regulated translocation of proteins from chloroplasts to mitochondria to modulate mitochondrial gene expression would mediate chloroplast signaling. Such translocation would be made much easier by direct contact between chloroplasts and mitochondria.
Third, there may be direct communication by physical interaction. In leaves, chloroplasts, mitochondria and peroxisomes have often been observed in close association, consistent with metabolic exchange among these organelles (Figure 4). It seems that the formation of this tri-organellar unit is regulated, with the chloroplast-peroxisome association being established first and then recruiting mitochondria (Oikawa et al., 2015). Application of the femtosecond laser pulses to chloroplast-mitochondria complexes in a variety of tissues should illuminate the biophysical characteristics of their association.
In all these instances of organelle interactions, it is apparent that more detailed analysis at the level of resolution provided by electron microscopy is needed. Such studies should incorporate state-of-the-art fixation techniques like tandem high-pressure freezing and freeze substitution in order to minimize artifact formation and to maintain intact the presumably delicate membrane extensions and contact sites. These techniques are becoming easier and less time-consuming (McDonald, 1999, 2014) and can easily be adopted for plant cell biology (Bobik et al., 2014). When coupled with fluorescence microscopy, this will be a powerful approach for ultrastructural interrogation, as exemplified by recent work from Caplan et al. (2015).
Hormones and Reactive Molecules as Chloroplast Signals
As sessile organisms plants have evolved to cope with extreme environmental conditions and fluctuations. Some of the most common environmental challenges to plant survival include drought, flooding resulting in reduced oxygen availability, and temperature extremes. In addition, the photosynthetic machinery is sensitive to excessive light and such exposure results in oxidative stress at the cellular level. In addition to these abiotic stresses, pathogens pose a constant threat of disease. Plants have therefore evolved a complex suite of responses that are exquisitely fine-tuned to allow them to cope with these stresses. It is widely recognized that chloroplasts both sense and respond to environmental conditions. Indeed, the chloroplast-generated hormones SA, JA and ABA and other secondary messengers including ROS and reactive nitrogen species (RNS) as well as redox signals are critical components of the plant stress response. Therefore chloroplast signaling is indispensable for plant survival of abiotic and biotic stress. The roles of all these chloroplast-associated molecules in coping with biotic and abiotic stress have been extensively examined and several excellent reviews are available (Miller et al., 2008; Padmanabhan and Dinesh-Kumar, 2010; Kangasjarvi et al., 2012; Pfannschmidt and Yang, 2012; Trotta et al., 2014).
Salicylic Acid
Salicylic acid is best known for its role plant–pathogen interactions and particularly in plant defense. However, SA also has roles in plant developmental processes including germination, root and shoot growth and senescence, and also functions in abiotic stress responses (Rivas-San Vicente and Plasencia, 2011). A phenolic compound, SA is largely the product of the isochorismate pathway in the chloroplasts. Pathogen infection induces the production of SA by chloroplasts mainly through the action of the chloroplast-localized ISOCHORISMATE SYNTHASE (ICS)1 enzyme although there may be some contribution by ICS2 (Wildermuth et al., 2001; Garcion et al., 2008). Analysis of the ics1 ics2 double mutant has revealed that there are other cellular sources of SA (Garcion et al., 2008), most likely via phenylpropanoid metabolism in the cytoplasm (Vlot et al., 2009). SA conjugates with glucose or a methyl side-group are the commonly active forms of SA. Indeed, methyl salicylate is a critical mediator of systemic acquired resistance (SAR; Park et al., 2007), and it may also function in ecological defense signaling when it becomes airborne (Shulaev et al., 1997).
Many SA-mediated defense responses rely on the action of the transcriptional activator NON-EXPRESSOR OF PR1 (NPR1; Fu and Dong, 2013). NPR1 interacts with TGA transcription factors, and it is believed to act with them as co-activators of defense gene expression (Zhang et al., 1999; Zhou et al., 2000; Yan and Dong, 2014). The action of NPR1 is dependent on the cellular redox state. In the absence of SA, NPR1 oligomerizes in the cytoplasm, but on perception of SA, NPR1 is reduced and the oligomer disassembles into monomers that then relocate to the nucleus to modulate gene expression (Kinkema et al., 2000; Mou et al., 2003). The redox state of NPR1 is mediated by the glutathione and thioredoxin redox systems (Mhamdi et al., 2010; Han et al., 2013). In the nucleus, NPR3 and 4, two other SA-binding proteins (SABPs) that are closely related to NPR1, modulate NPR1’s activity (Fu et al., 2012). It should be noted that there are also NPR1-independent pathways that mediate SA signaling (Rairdan et al., 2001; Shah, 2003; Uquillas et al., 2004; Blanco et al., 2005). Downstream of NPR1 the SA signaling pathway is well understood (Spoel et al., 2003; Fu and Dong, 2013).
Salicylic acid is important for basal defense as well as for ETI (Alazem and Lin, 2015), and application of SA or overexpression of its biosynthesis genes leads to increased pathogen resistance (Malamy et al., 1990; Ryals et al., 1996; Mur et al., 2008; Shah, 2009; Coll et al., 2011). Chloroplast Ca2+ signals are induced in a stress-specific manner, and this response is mediated by the calcium-sensing receptor (CAS; Nomura et al., 2012). CAS mediates the Ca2+ signals in ETI and in response to the presence of highly conserved pathogen associate molecular patterns (PAMPS). CAS, and thus CA2+, regulates chloroplast SA biosynthesis and plants depleted of CAS failed to induce SA production in response to pathogen infection. In addition, expression of several nuclear defense-related genes was shown to be dependent on CAS, and the pattern of gene regulation was most similar to that observed in response to 1O2.
Interestingly, exogenous application of SA induces closure of plasmodesmata (Wang et al., 2013b). This is mediated by PLASMODESMATA LOCALIZED PROTEIN (PDLP)5, and likely involves the action of a callose synthase (Lee et al., 2011; Wang et al., 2013b). Consistent with these findings, Arabidopsis pdlp5 mutants have decreased resistance to bacterial pathogens. These results clearly demonstrate crosstalk between SA signaling and plasmodesmata, and illustrate how chloroplast signals can act to regulate intercellular trafficking via plasmodesmata (discussed below). It will be interesting to see if similar mechanisms are deployed for viral resistance since viruses use plasmodesmata for intercellular trafficking.
There has been some controversy over the SA-binding properties of NPR1. Binding assays using recombinant GST-NPR1 and tritiated-SA ([3H]-SA) suggested that NPR1 did not bind SA (Fu et al., 2012). In contrast, equilibrium dialysis experiments revealed that NPR1 bound SA with a KD similar to that observed for other receptor-ligand interactions for plant hormones (Wu et al., 2012). Recent results confirm that SA can indeed bind NPR1 (Manohar et al., 2014). However, it is clear that NPR1 is not the only protein that binds SA. The metabolic enzymes catalase (Chen et al., 1993), ascorbate peroxidase (Durner and Klessig, 1995) and methyl salicylate esterase (SABP2; Forouhar et al., 2005) have been shown to bind SA. Recent work reveals that SA potentially has numerous targets in the cell including, not unexpectedly, numerous chloroplast proteins.
Many of these SABPs have been identified by high-throughput approaches (Moreau et al., 2013; Manohar et al., 2014). By probing Arabidopsis protein microarrays with 4-azido SA (azSA), an SA analog, the Popescu lab identified numerous chloroplast-localized proteins with roles in photosynthesis and oxidative phosphorylation as proteins interacting with AzSA (Moreau et al., 2013). Two other interesting candidate SABPs were also identified: thimet metalloendopeptidase At5g65620 (TOP1) and its homolog encoded by At5g10540 (TOP2), both of unknown cellular function. TOP1-GFP fusions localized to chloroplasts and TOP2 is likely cytosolic. Interestingly, top1 top2 double mutants had compromised ETI and PCD to bacterial pathogens. Further, TOP1 and TOP2 dimerize in an SA- and redox-dependent manner (Westlake et al., 2015). However, TOP1 and TOP2 have distinct responses to the reductant DTT, suggesting they have different activities in planta. A role for TOP1 and TOP2 in the oxidative stress response was demonstrated by treating various top mutants with methyl viologen, a potent inducer of oxidative stress, but this function is likely restricted to early in plant development. Two additional high throughput screens have recently identified another 100 candidate SABPs (Manohar et al., 2014). Of these, nine were already known SABPs and the SA binding of nine of new candidate SABPs was verified. Notably, four of the new SABPs have roles in redox regulation, reiterating the interaction between these two pathways.
Jasmonates
Jasmonic acid is a lipid-derived hormone that is perhaps best known for its roles in insect herbivory and wounding, but also has roles in plant growth and development (Leon, 2013). The term jasmonates refers to a group of compounds that are derived from linoleic acid. JA is synthesized via the octadecanoid pathway and JA synthesis is initiated in the chloroplasts but is completed in the peroxisome. JA is then derivatized to yield a diverse array of metabolites that have different functions, ranging from storage to inactivation (reviewed in Leon, 2013; Wasternack and Hause, 2013). The most active of these compounds is a JA conjugate with isoleucine, (+)-7-iso-JA-Ile (Wasternack, 2014). JA biosynthesis and signaling pathways have been elucidated (Turner et al., 2002; Antico et al., 2012; Wasternack and Hause, 2013; Zhu, 2014). JA signaling in stress is closely liked with that of another hormone, ethylene (Kunkel and Brooks, 2002). Interestingly SA and JA/ET signaling are often antagonistic to each other (Robert-Seilaniantz et al., 2011). This highlights the crosstalk between chloroplast signals.
The final chloroplastic intermediate in JA-biosysnthesis is cis-(+)-12-oxophytodienoic acid (OPDA). OPDA is then translocated to the peroxisome, where its hydrocarbon chain is shortened by β-oxidation. It is not clear how OPDA is transported to the peroxisomes, but the process must be tightly regulated, given that OPDA itself is able to act as a signaling molecule. Indeed, OPDA has been implicated in tendril coiling (Blechert, 1999); in Arabidopsis seed germination (Dave et al., 2011); in tomato embryo development (Goetz et al., 2012), and fertility in P. patens (Stumpe et al., 2010). OPDA is also known to be an important signal for defense (Stintzi et al., 2001; Scalschi et al., 2015). Interestingly, P. patens does not make JA but instead uses OPDA for defense (Stumpe et al., 2010; Ponce De Leon et al., 2012). However, SA is used as a defense signal in this moss (Ponce De Leon et al., 2012). In the liverwort Marchantia polymorpha wounding induces OPDA production, and exogenous application limited M. polymorpha growth (Yamamoto et al., 2015). As in P. patens, JA was not detected in M. polymorpha. This finding was supported by OPDA regulating growth of M. polymorpha while JA could not. Thus, the production of lipid-derived signals by the chloroplast is an ancient feature of plants, and the production of JA may have developed more recently.
Crosstalk of RNA Silencing and Chloroplast Hormones
There is also crosstalk between the chloroplast-derived phytohormones and the RNAi machinery of plant cells. Several studies have reported increased expression of RNA-DEPENDENT RNA POLYMERASE1 (RDR1) on exogenous application of SA (Xie et al., 2001; Yu et al., 2003; Hunter et al., 2013), JA (Pandey and Baldwin, 2007) and ABA (Hunter et al., 2013). RDR1 is known to have a role in antiviral RNA silencing, in the production and amplification of virus-derived siRNAs (Donaire et al., 2008; Wang et al., 2010). JA interaction with SA is mostly antagonistic, and there are few genes whose expression is induced by both hormones (Pieterse et al., 2009). The finding RDR1 can be induced by multiple hormones suggests that there is crosstalk between hormones and the RNA silencing machinery to mediate stress responses. However, RDR1 does not seem to have a role in drought resistance (Hunter et al., 2013), although other components of the RNAi components have been demonstrated to function in stress tolerance (Earley et al., 2010; Li et al., 2012; Westwood et al., 2013). Together, these findings extend the role of chloroplast signaling in the stress response.
Abscisic Acid
Abscisic acid is one of the most important hormones mediating plant biotic and abiotic stress responses. ABA also has major roles in various plant physiological processes including stomatal movement and seed dormancy (Wensuo and Zhang, 2008; Rodriguez-Gacio Mdel et al., 2009; Kim et al., 2010). ABA is a sesquiterpenoid that is produced by the methylerythritol phosphate (MEP) pathway in plastids that produces carotenoids (Finkelstein, 2013). ABA levels in a cell are the result of both its synthesis and catabolism. Interestingly, several subcellular compartments are involved in ABA metabolism (Xu et al., 2013a). All the steps of the de novo ABA biosynthetic pathway occur in plastids except for the last two, which occur in the cytosol. The first committed step in ABA synthesis is the cleavage of the carotenoid xanthopyhll by 9-cis-epoxycarotenoid dioxygenase (NCED) to produce the C15 compound xanthoxin, that is transported from the plastids into the cytosol by an unknown mechanism, where it will be converted into ABA (Finkelstein, 2013). Once synthesized, ABA is transported from the sites of synthesis to sites action via the xylem and phloem and is thus made available to both roots and shoots. ABA catabolism occurs via one of two pathways: oxidation or conjugation to glucose to produce a glucosyl ester ABA-GE. ABA-GE is stored in the vacuole or ER until it mobilized under stress conditions. Because of its central roles in modulating responses to various stresses, ABA signaling is an attractive target for engineering plants with increased tolerance to those stresses.
The mechanisms governing perception of, and signaling by, ABA are being discovered (Yoshida et al., 2015). Members of the PYR/PYL/RCAR family of soluble proteins are ABA receptors (Ma et al., 2009; Park et al., 2009). This protein family has 14 members, almost all of which appear capable of forming an ABA-receptor complex that is able to activate the transcription of ABA-responsive genes (Kline et al., 2010). Downstream of the receptors, several kinases, including calcium dependent kinases (CDPKs), and phosphatases mediate ABA signaling, culminating in changes in nuclear gene expression (Finkelstein, 2013; Yoshida et al., 2015). The 26S proteasome is also important in mediating ABA signaling (Ludwikow, 2015).
Abscisic acid signaling is particularly important during drought, salinity and cold stress. During pathogen infection, ABA signaling is antagonistic to JA/Et signaling (Soosaar et al., 2005); and it can also antagonize SA signaling (Alazem and Lin, 2015). Indeed, crosstalk between SA, JA, and ABA signaling pathways during pathogen defense is well documented (Tuteja, 2007; Flors et al., 2008; Cao et al., 2011) and underscores the role of chloroplasts in integrating inputs for plant survival. Importantly, the H subunit of Mg chelatase was shown to be an ABA receptor, and moreover, the observations that ABA can repress Lhcb expression and the gun phenotype of the abi4 mutant link this hormone to retrograde signaling (Shen et al., 2006; Koussevitzky et al., 2007).
Reactive Oxygen Species
Reactive oxygen species are formed by the reduction of molecular oxygen and the term ROS includes superoxide (O2⋅-), hydroxyl, alkoxyl (.RO), and peroxyl radicals as well as non-radical molecules like hydrogen peroxide (H2O2) and singlet oxygen (1O2). It is well established that while large amounts of ROS are damaging, small amounts act as signaling molecules (Miller et al., 2008; Gill and Tuteja, 2010). In an effort to avoid the toxicity of ROS, plants have evolved multiple antioxidant systems. It is clear then that ROS signaling is complex and is often the outcome of the balance between production and scavenging.
In a typical plant cell ROS may be generated by a variety of subcellular compartments including chloroplasts, peroxisomes, mitochondria and the apoplast. In a photosynthesizing leaf most ROS is the product of the chloroplasts and peroxisomes with smaller contributions from mitochondria (Foyer and Noctor, 2005). During photosynthesis, in the reaction center of photosystem II (PSII), excited triplet center chlorophyll P680 interacts with oxygen to generate 1O2. The acceptor side of PSI produces superoxide and hydrogen peroxide as electrons are transferred from reduced ferredoxin to molecular oxygen. Photorespiration in the peroxisomes to recycle Rubisco that has reacted with oxygen is a major source of H2O2 (Yoshida and Noguchi, 2011).
Plastid-generated ROS molecules have been shown to act as signals that modulate expression of nuclear genes. Different ROS molecules have been shown to induce expression of distinct suites of genes (Desikan et al., 2001; Vandenabeele et al., 2004; Vanderauwera et al., 2005; Laloi et al., 2007; Li et al., 2011; Balazadeh et al., 2012; Mor et al., 2014). This has made it possible to draw distinctions between the various ROS initiating responses to different stresses, (e.g., Gadjev et al., 2006). By virtue of their nature, ROS are known to interact with a variety of biological molecules. For example, as a strong electrophile, 1O2 can react spontaneously with many classes of biological molecules including proteins, lipids and nucleic acids. However, the question remains whether they act directly or indirectly through modification of other biomolecules.
Hydrogen peroxide is the least reactive ROS, and at high light intensities up to 5% of chloroplast-generated H2O2 has been detected outside the organelle (Mubarakshina et al., 2010). H2O2 may exit the chloroplasts via aquaporins (Bienert et al., 2007). Recently, H2O2 has also been shown to relocate from chloroplasts to the nucleus via stromules (Caplan et al., 2015). Indeed, chloroplast ROS production induces stromule formation (Brunkard et al., 2015b). These findings suggest that H2O2 may act directly on its targets to regulate their expression or behavior.
In contrast, 1O2 is highly reactive, and, 1O2 is involved in signaling pathways leading to cell death or to acclimation (Wagner et al., 2004; Ledford et al., 2007). Yet, because of its very high reactivity and therefore short lifetime in vivo, 1O2 is not considered to be a molecule directly involved in chloroplast-to-nucleus signaling. Studies in the Arabidopsis fluorescent (flu) mutant have shed light on 1O2 signaling (Kim and Apel, 2013). The flu mutant accumulates protochlorophyllide and when plants are exposed to light they generate large amounts of 1O2 (Meskauskiene et al., 2001). Increased 1O2 production in flu chloroplasts was associated with induction of stress responses including dramatic changes in nuclear gene expression and enhanced accumulation of the stress hormones, SA, Et, and the oxylipins OPDA and JA (op den Camp et al., 2003). Interestingly, these 1O2-induced changes were dependent on the chloroplastic EXECUTER1 (EX1) and EX2 proteins (Kim et al., 2012). To date, two direct protein targets of 1O2 have been identified: one is β-carotene and the other is the D1 protein of PSII (Kim and Apel, 2013). Reaction of 1O2 with β-carotene produces β-cyclocitral, shown to be involved in retrograde signaling (Ramel et al., 2012; see above). D1 is potent scavenger of 1O2, and this activity leads to the destruction of D1 by protease activity. However, to date no biological activity for these D1 fragments in plants has been reported.
Plants possess several enzymes including catalase and ascorbate peroxidase that detoxify ROS (Miller et al., 2008). In addition plants have evolved antioxidant systems that allow them to not only cope with ROS production during stress but also transduce ROS signals. Such systems include tocopherols (vitamin E), ascorbate (vitamin C), and glutathione (a tripeptide thiol) and they all associate with chloroplasts. Glutathione is the most important antioxidant in plants (Zechmann, 2014) and it can act on ROS directly or through the ascorbate-glutathione system. Glutathione synthesis is initiated in chloroplasts (Wachter et al., 2005). The importance of glutathione highlights the importance of thiol-disulfide reactive proteins as antioxidants (Foyer and Noctor, 2003). Proteins including thioredxoxins, glutaredoxins, glutathione peroxidases are found in chloroplasts as well as other subcellular compartments, and these proteins convey the redox state of chloroplasts to the rest of the plant cell. The involvement of redox signaling in mediating plant stress responses are extensively documented (Foyer et al., 2009).
Nitric Oxide and Reactive Nitrogen Species
Nitric oxide (NO) is a small gaseous molecule whose role in signaling in plant and non-plant systems is well established. While NO is an important second messenger in plants, major aspects of NO synthesis and action remain undiscovered. NO has critical roles in normal plant physiology including seed dormancy, germination, the floral transition, and during stress responses (Wendehenne et al., 2004; Moreau et al., 2008; Sanz et al., 2015). In plants NO is synthesized in chloroplasts (Mandal et al., 2012; Tewari et al., 2013), and synthesis is closely linked to lipid metabolism, specifically that of oleic acid (Mandal et al., 2012).
In animals NO is synthesized by NITRIC OXIDE SYNTHASEs (NOSs) that oxidize L-arginine to NO (Stuehr, 2004). Curiously, NOS-like activity has long been described in plants but the identity of this enzyme remains elusive (Besson-Bard et al., 2008). In plants, nitrate reductase can reduce nitrate to NO (Desikan et al., 2002). Another enzyme implicated in NO synthesis is AtNOA1 (formerly AtNOS1). However, AtNOA1 is a GTPase that binds ribosomes, and it lacks NOS activity (Moreau et al., 2008). Nonetheless, atnoa1 mutants displayed reduce levels of NO (Guo et al., 2003).
One of the primary mechanisms through which NO exerts its effects is by direct regulation of protein activity through nitrosylation of cysteine residues in a redox-dependent manner (Mengel et al., 2013). Nitration of tyrosine residues also occurs, and it is mediated by the peroxynitrite (ONOO-) RNS formed on reaction of NO with O2 (Corpas et al., 2013). Tyrosine nitration is viewed as a hallmark of plants under a variety of biotic and abiotic stresses (Corpas et al., 2013). While proteomic analyses have identified only relatively few nitrosylated proteins in Arabidopsis (Lindermayr et al., 2005; Romero-Puertas et al., 2008; Ortega-Galisteo et al., 2012; Camejo et al., 2013; Puyaubert et al., 2014), a recent computational approach identified more than 16, 000 potential protein target for nitrosylation (Chaki et al., 2014). Refined versions of this program should be very useful in streamlining investigation of NO function and effects in signaling.
There is substantial crosstalk between ROS and RNS signaling pathways (Lounifi et al., 2013). Indeed, even small amount of NO had profound effects on chloroplast PET and redox state (Vladkova et al., 2011; Misra et al., 2014). It is therefore not surprising that NO is important for plant defense (Zaninotto et al., 2006; Bellin et al., 2013). Further, NO signaling interacts with SA and JA signaling pathways (Wendehenne et al., 2004; Zhou et al., 2015). NO production has been proposed as a potential target for engineering plant fitness (Foresi et al., 2015). However, this approach should be carefully considered given the potentially deleterious effects of runaway NO production and the extensive crosstalk between NO and other stress signaling pathways.
Chloroplasts as Targets of Pathogen Effectors
The previous sections have highlighted the role of several chloroplast products in stress tolerance and plant defense. That chloroplasts have critical roles in pathogen defense is supported by the identification of numerous chloroplast proteins as direct targets of pathogen effectors (Table 1). Several of these targets are components of PET, suggesting that photosynthesis and chloroplast function are important for defense against pathogens. A series of experiments aimed at addressing the role of chloroplasts beyond phytohormone production in plant defense has been reported. VIGS of two genes encoding components of the PSII oxygen-evolving complex, PsbQ and PsbO, resulted in a three to four-fold increase in the number of Turnip mosaic virus (TuMV) infection foci (Manfre et al., 2011). A similar increase was observed when the chloroplast protease FtsH or Rubisco were depleted by VIGS (Manfre et al., 2011). These findings were further supported by the observation that treatment with lincomycin, and low-light conditions led to increased rates of TuMV infection. Importantly, it was demonstrated that these effects were independent of SA, as lincomycin-treated plants induced SA production to the same levels on TuMV infection as observed in non-treated controls. Given that TuMV replicates in association with the chloroplast outer envelope (Wei et al., 2010), it is perhaps not surprising that perturbing chloroplast function results in increased viral infection. However, chloroplast proteins are targets for other viruses, e.g., Tobacco mosaic virus (TMV), which are not known to replicate in association with chloroplast membranes. Interestingly, PsbO has been shown to interact the helicase domain of the TMV replicase, and VIGS of PsbO lead to a 10-fold increase in TMV infection in N. benthamiana (Abbink et al., 2002).
Chloroplasts and Intercellular Signaling
The engulfment of cyanobacteria and subsequent evolution of the cellulosic cell wall surrounding the new plant cell likely created a physical barrier that hampered cell-to-cell communication and further development of multicellular, three-dimensional plants. It is important to highlight that modern chloroplasts are a sister group to the filament and heterocyst-forming cyanobacteria Nostoc and Anabaena (Falcon et al., 2010). Intercellular movement of molecules between the cells of Anabaena cylindrica filaments has been demonstrated (Mullineaux et al., 2008). Heterocysts are specialized cells that fix N2 and lack O2-evolving PSII and typically they have thick cell walls with limited permeability to gases. Interestingly, heterocysts are connected to vegetative cells by microplasmodesmata (Falcon et al., 2010; Padmanabhan and Dinesh-Kumar, 2010). Thus, we propose that chloroplasts, as the successors of these ancient cyanobacteria, participate in regulation of the cell-to-cell connectivity of modern plant cells by regulating plasmodesmata biogenesis in contemporary plants. This hypothesis is supported by a number of reports showing that mutants with defective chloroplasts display aberrant trafficking and altered expression of cell wall-related genes (see below). The relationship between chloroplasts and plasmodesmata and thus cell wall becomes more palpable when one considers sugar transport from source to sink tissues through plasmodesmata.
Chloroplast Signaling and Symplasmic Transport
Communication between plant cells is enhanced by the presence of plasmodesmata, specialized channels traversing the cell wall between adjacent cells. Even though discovered more than 100 years ago, the structure and regulation of plasmodesmata are poorly understood. In contrast, recent advances in plant biology and genetics have helped reveal much about their function. Plasmodesmata are indispensable for proper plant development as they provide a route for exchange of metabolites including water, ions and photoassimilates, as well as information encoded by hormones, nucleic acids and proteins including transcription factors (reviewed in Gallagher et al., 2014; Brunkard et al., 2015a; Heinlein, 2015; Jackson, 2015). Plant viruses have evolved to take advantage of the intercellular connectivity afforded by plasmodesmata, and viruses use these channels for cell-to-cell spread during infections of their plant hosts. Recent studies on grafted plants resulted in a tantalizing hypothesis that plasmodesmata facilitate the movement of the entire chloroplast genomes between neighboring cells (Stegemann and Bock, 2009; Stegemann et al., 2012; Thyssen et al., 2012). However, the dimensions of plasmodesmata would constrain movement of intact organelles, and whether the translocated genomes use intact chloroplasts as vehicles or are transported through plasmodesmata as “naked” but specifically folded DNA remains to be elucidated.
Interestingly, genetic screens aimed at isolating genes regulating plasmodesmata have identified genes involved in the biogenesis and/or functioning of chloroplasts. The first such reported mutant was maize sucrose export defective1 (sxd1; Russin et al., 1996). The sxd1 mutants failed to export photosynthate from sites of photosynthesis and have reduced intercellular transport due to accumulation of callose at plasmodesmata located at the bundle-sheath and vascular parenchyma interfaces (Botha et al., 2000). Callose is a polymer composed of β-1,3-linked glucose residues and callose accumulation at the necks of plasmodesmata is a major strategy for limiting plasmodesmal trafficking (De Storme and Geelen, 2014). Notably, despite accumulating sugars and starch in source tissues, sxd1 does not repress photosynthesis (Provencher et al., 2001). SXD1 is the maize ortholog of Arabidopsis VTE1, encoding a chloroplast TC required for the production of the antioxidant vitamin E (Provencher et al., 2001; Porfirova et al., 2002). The sxd1 mutant provided the first clue that chloroplast redox state may influence plasmodesmata. However, under optimal growth conditions vte1 mutants were indistinguishable from wild-type plants, although they were more sensitive to photooxidative stress (Porfirova et al., 2002). Arabidopsis mutants with defects in tocopherol metabolism should be carefully examined for plasmodesmata-related changes in intercellular trafficking.
A genetic screen for Arabidopsis thaliana mutants with altered plasmodesmal function identified the gfp arrested trafficking (gat) mutants (Benitez-Alfonso et al., 2009). In these mutants GFP synthesized in the companion cells of the phloem failed to move via the plasmodesmata into the surrounding tissues as occurred in wild-type tissues; that is, gat mutants have decreased intercellular trafficking. The gat1, 2, 4, and 5 mutations are all seedling lethal, with development ceasing about 10 days after germination. GAT1 encodes a plastid-localized thioredoxin-m3 (TRX-m3), and gat1 roots accumulate higher levels of ROS than do wild-type roots. In addition, callose was found to accumulate at plasmodesmata in gat1 roots. Similar observations have been made for the other gat mutants (Benitez-Alfonso and Jackson, 2009). Notably, approximately five percent of plasmodesmata in gat1 seedling roots are occluded by electron-dense material (Benitez-Alfonso et al., 2009). Overexpression of GAT1 results in the reciprocal phenotype of increased intercellular transport. Thus, like SXD1/VTE1, GAT1 likely functions in redox homeostasis involving the chloroplasts and perturbation of plastid redox state results in altered plasmodesmata. In addition, it has been proposed that altered metabolic flux in the gat1 mutant may alter the redox state of TRX-m3 and ultimately influence plasmodesmata function (Benitez-Alfonso et al., 2009).
The Zambryski lab conducted a separate screen for Arabidopsis mutants with altered plasmodesmata-mediated intercellular trafficking (Kim et al., 2002; Burch-Smith and Zambryski, 2012). Several increased size exclusion limit (ise) mutants were identified by screening embryonically lethal mutants and monitoring their ability to traffic fluorescent dyes between cells. ISE1 and ISE2 have been mapped and cloned (Kobayashi et al., 2007; Stonebloom et al., 2009). In addition to increased plasmodesmal trafficking ise1 and ise2 embryos also contain increased numbers of plasmodesmata with multiple branches (Burch-Smith and Zambryski, 2010). Interestingly, nuclear ISE2 encodes a chloroplast-localized DEVH-type RNA helicase, (Burch-Smith et al., 2011) while ISE1 encodes a mitochondrial DEAD-box RNA helicase (Stonebloom et al., 2009). Further analyses in Arabidopsis and N. benthamiana revealed that loss of ISE1 and/or ISE2 leads to defective chloroplasts and ultimately leaves become yellow (Burch-Smith and Zambryski, 2010; Burch-Smith et al., 2011; Stonebloom et al., 2012). Gene expression analyses on ise1 and ise2 embryos by tiling microarrays revealed that the largest class of nuclear genes affected in both mutants, representing ∼20% of the total, encode chloroplast-localized proteins (Burch-Smith et al., 2011). Importantly, genes encoding products for chlorophyll biosynthesis, photosynthetic light-harvesting reactions, and the Calvin cycle for carbon fixation were all affected in both mutants. Thus, the overlapping plasmodesmatal phenotypes of the ise1 and ise2 mutants are likely due to defective chloroplasts.
The changes in plasmodesmata and intercellular trafficking are likely mediated by altered redox signaling from chloroplasts and mitochondria. Using redox-sensitive fluorescent reporters the redox state of chloroplasts depleted of ISE1 or ISE2 was measured (Stonebloom et al., 2012). Significantly, chloroplasts in ISE1- or ISE2-silenced N. benthamiana leaves were more reduced than in non-silenced control leaves. These results are consistent those from the gat1 mutant (Benitez-Alfonso et al., 2009). Thus, oxidized chloroplasts are associated with decreased intercellular trafficking while reduced chloroplasts increase plasmodesmata-mediated trafficking. Further experiments focused on identifying the redox pathways and signal transduction involved in regulating plasmodesmata will yield important insight into how the physiological state of the chloroplasts is communicated to the rest of the plant.
Redox Signals Travel between Cells
Recently the involvement of chloroplasts in remote control of nucleus-localized alternative splicing was reported. Specifically, the light-regulated redox state of plastoquinone is the source of a chloroplast-generated signal able to control alternative splicing (Petrillo et al., 2014). In the presence of a reduced or oxidized plastoquinone pool, the alternative splicing of selected nuclear genes is promoted or inhibited, respectively. Importantly, even though photosynthetic tissues generated it, the putative signal is able to travel to roots to control alternative splicing. Thus chloroplast redox signals can act non-cell-autonomously. The redox pathway leading to defective alternative splicing has not been identified; however, the redox-regulated protein kinases of chloroplasts are proposed to contribute to the signaling machinery through a process of phosphorylation of some other proteins (Petrillo et al., 2014). This result is consistent with the observation that an Arabidopsis plasma-membrane-localized thioredoxin, Trx h9 moved intercellularly (Meng et al., 2010), presumably via plasmodesmata. The potential for intercellular trafficking by redox signals greatly extends the reach of chloroplasts in signaling.
The question remains: how could chloroplasts regulate plasmodesmata and intercellular trafficking? It is possible that as-yet-unidentified chloroplasts signals could directly target plasmodesmata. Indeed, chloroplasts are often observed in the vicinity of plasmodesmata, and regardless of the reasons for this close proximity, this means signals would not have to travel across long distances to act (Figure 4). Studies in the algae Chara australis also suggest a relationship between chloroplast and cell wall structure (Foissner et al., 2015). It is also possible that chloroplast-to-nucelus retrograde signaling may be involved in regulating events at the cell wall. Indeed, disruption of chloroplast function often results in significant changes of expression of genes involved in cell wall synthesis and modification (Burch-Smith et al., 2011). Similarly, expression of genes with functions related to the cell wall was down regulated in plants accumulating β-cyclocitral upon high light treatment (Ramel et al., 2012). It is also perhaps significant that three of the 39 genes identified as the core response module of chloroplast retrograde signaling encode proteins involved in synthesizing or modifying cell walls (Glasser et al., 2014). Future experiments should determine the pathways used for chloroplast-to-plasmodesmata signaling.
Chloroplast Signaling in Ecosystem Functioning – the Power of Scent
In addition to being the center for production of oxygen and sugars, chloroplasts are also the source of plethora of secondary metabolites. Chloroplast-produced biogenic volatile organic compounds (BVOCs) seem to represent a fascinating signaling system that enables a plant to communicate with not only itself (e.g., propagation of a systemic response), and with the entire ecosystem that includes plant–plant, plant–animal/insect interaction, but could potentially also impact atmospheric chemistry (Atkinson, 2000; Seybold, 2006; Heil and Silva Bueno, 2007; Heil, 2010).
Volatiles directly produced in chloroplasts include isoprene, monoterpenes, diterpenes, hemiterpenes and volatile carotenoid derivatives, all synthesized by the MEP pathway. Also generated by the chloroplasts are green leaf volatiles (GLVs), synthesized in the LOX-pathway that consists of C6 and C9 aldehydes, alcohols and their acetate esters, that are responsible for the ‘mowed grass odor’ (Dudareva et al., 2013). These volatiles have important physiological roles, for example, stabilization of thylakoid membranes, reduction of ROS, as well as an ecological function, emphasizing an often underestimated contribution of chloroplasts/plastids to fine regulation of the ecosystem (Velikova et al., 2012). It is estimated that each year about 500 Tg C of isoprene is emitted to the atmosphere. This significant amount has some consequences as the highly reactive isoprene molecule can oxidize OH groups to yield peroxyl radicals that in turn can convert NO into NO2, producing O3 (Thompson, 1992; Carter, 1994; Atkinson, 2000). Therefore isoprene as a reactive molecule plays a crucial role in establishing the content of atmospheric greenhouse gasses and pollutants (ozone, methane, secondary organic aerosols, etc.; Harrison et al., 2013).
Chloroplast-produced BVOCs have other ecological functions that are a consequence of their signaling propensity. One of these is the so-called priming effect that leads to enhanced and more effective defense response to pathogen or insect attack in plants pretreated with volatiles (Engelberth, 2004). Although they are released continuously, there is enhanced emission of volatiles upon biotic or abiotic stresses including drought, high light, parasite or herbivore activity (wounding), resulting in so-called HIPVs (herbivore induced plant volatiles) emissions that include monoterpenes and GLVs (Brilli et al., 2009; Bonaventure and Baldwin, 2010; Dicke and Baldwin, 2010; Niinemets, 2010, 2013; War et al., 2011; Heil, 2014). Leaves of attacked plants emit volatiles that can prime defense responses in intact leaves of the same plant as well as on neighboring plants. Moreover, as demonstrated in maize, the intact plants exposed to HIPVs (termed receiver plants) are able to store the information conveyed by volatiles and recall it upon herbivore attack to activate defense genes encoding proteinase inhibitors (Ali et al., 2013).
The molecular mechanism of volatile signaling is far from understood, however, an attractive scenario of volatile-driven epigenetic modification through DNA methylation has been demonstrated as a very reliable strategy for priming defense induction. Moreover, since the JA-signaling pathway is triggered upon herbivore attack (wounding), it is expected that induced JA formation as a downstream effect would be relevant to the HIPV-primed defense responses (Ali et al., 2013). However, some of the most important questions concern the perception and the very first steps in volatiles signaling in receiver plants. To this end it is proposed that volatiles, as hydrophobic compounds, could diffuse through the outer, lipophilic leaf layer (cuticle) or enter through stomata (Baldwin et al., 2006). Indeed, strong membrane depolarization as well as calcium influx were observed upon treatment of tomato plants with GLVs, two phenomena often associated with a signal transduction cascade in other systems (Zebelo et al., 2012). Rearrangements of cytoskeleton and expression of selected genes encoding, among others, protein kinases and transcription factors as well as changes in stomatal aperture were observed upon treatment with the monoterpenes camphor and menthol (Kriegs et al., 2010). However, the concentrations used in the assay are inconsistent with physiological levels, thus the observed effects are questionable. Elucidating the impact of BVOCs on gene expression and impacts on general cell biology are a likely future direction of this field.
Another intriguing aspect of chloroplast originating volatiles is that their release from attacked plants can serve as attractants of the offending herbivore’s natural biological enemies, a phenomenon known as tritrophic chemical communication or tritrophic interaction (Heil, 2008; Mooney et al., 2012; Scala, 2013). Importantly, some volatiles also have allelopathic and insecticidal properties that could be deterrents for oviposition or can attract pollinators and/or seeds dispersers (Schulz et al., 2007; McCallum et al., 2011; Rodriguez et al., 2011; Reis et al., 2014). Together, these reports shed new light on chloroplasts as factories that generate volatile compounds that are important not only from the perspective of a single plant, but also the entire ecosystem. As new strategies for chloroplast biotechnology are deployed, it will be important to examine their effects on these volatiles so that susceptibility to herbivore predation and pathogen ingress are not inadvertently introduced through unintended modification of volatile production and perception.
Conclusion and Future Directions
The era of transcriptomics initiated by the microarray platform and continued by Next-generation sequencing technology, together with metabolomics and proteomic approach provided by mass-spectrometry, established new possibilities and new directions in addressing questions in biology. Inevitably, due to these two strategies contemporary plant biology is progressing toward a more holistic view, which translates into studying not a single protein per se, but a single protein in the context of an entire cell, tissue, organ, individual plant and ultimately the entire ecosystem biology. Current progress in studying chloroplast biology shows their function goes far beyond photosynthesis and includes all aspects of plant biology, making these organelles an integral part and a full-fledged player in the cell. Understanding the signaling pathways chloroplasts and other plastids use to communicate with the rest of the plant cell under normal and stress conditions will provide new ideas for developing powerful strategies for engineering improved plants. Moreover, a thorough knowledge of chloroplast signaling will also allow the design of rational approaches that could minimize if not avoid unintended crosstalk and undesirable outcomes.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We apologize to colleagues whose work we could not cite due to space limitations. We thank Dr. Elena Ganusova for helpful suggestions during manuscript preparation. This work was supported by University of Tennessee start-up funds and NSF award #1456761 to TB-S.
References
Abbink, T. E., Peart, J. R., Mos, T. N., Baulcombe, D. C., Bol, J. F., and Linthorst, H. J. (2002). Silencing of a gene encoding a protein component of the oxygen-evolving complex of photosystem II enhances virus replication in plants. Virology 295, 307–319. doi: 10.1006/viro.2002.1332
Adam, Z. (2015). Plastid intramembrane proteolysis. Biochim. Biophys. Acta 1847, 910–914. doi: 10.1016/j.bbabio.2014.12.006
Alazem, M., and Lin, N. S. (2015). Roles of plant hormones in the regulation of host-virus interactions. Mol. Plant Pathol. 16, 529–540. doi: 10.1111/mpp.12204
Albrecht, V., Simkova, K., Carrie, C., Delannoy, E., Giraud, E., Whelan, J., et al. (2010). The cytoskeleton and the peroxisomal-targeted snowy cotyledon3 protein are required for chloroplast development in Arabidopsis. Plant Cell 22, 3423–3438. doi: 10.1105/tpc.110.074781
Ali, M., Sugimoto, K., Ramadan, A., and Arimura, G. (2013). Memory of plant communications for priming anti-herbivore responses. Sci. Rep. 3, 1872. doi: 10.1038/srep01872
Allen, J. F. (1993). Control of gene expression by redox potential and the requirement for chloroplast and mitochondrial genomes. J. Theor. Biol. 165, 609–631. doi: 10.1006/jtbi.1993.1210
Andersson, M. X., Goksor, M., and Sandelius, A. S. (2007). Membrane contact sites: physical attachment between chloroplasts and endoplasmic reticulum revealed by optical manipulation. Plant Signal. Behav. 2, 185–187. doi: 10.4161/psb.2.3.3973
Antico, C. J., Colon, C., Banks, T., and Ramonell, K. M. (2012). Insights into the role of jasmonic acid-mediated defenses against necrotrophic and biotrophic fungal pathogens. Front. Biol. 7:48–56. doi: 10.1007/s11515-011-1171-1
Atkinson, R. (2000). Atmospheric chemistry of VOCs and NO. Atmos. Environ. 34, 2063–2101. doi: 10.1016/S1352-2310(99)00460-4
Avendano-Vazquez, A. O., Cordoba, E., Llamas, E., San Roman, C., Nisar, N., De La Torre, S., et al. (2014). An uncharacterized apocarotenoid-derived signal generated in zeta-carotene desaturase mutants regulates leaf development and the expression of chloroplast and nuclear genes in Arabidopsis. Plant Cell 26, 2524–2537. doi: 10.1105/tpc.114.123349
Balasubramaniam, M., Kim, B. S., Hutchens-Williams, H. M., and Loesch-Fries, L. S. (2014). The photosystem II oxygen-evolving complex protein PsbP interacts with the coat protein of Alfalfa mosaic virus and inhibits virus replication. Mol. Plant Microbe Interact. 27, 1107–1118. doi: 10.1094/MPMI-02-14-0035-R
Balazadeh, S., Jaspert, N., Arif, M., Mueller-Roeber, B., and Maurino, V. G. (2012). Expression of ROS-responsive genes and transcription factors after metabolic formation of H(2)O(2) in chloroplasts. Front. Plant Sci. 3:234. doi: 10.3389/fpls.2012.00234
Baldwin, I. T., Halitschke, R., Paschold, A., Von Dahl, C. C., and Preston, C. A. (2006). Volatile signaling in plant-plant interactions: “talking trees” in the genomics era. Science 311, 812–815. doi: 10.1126/science.1118446
Ball, S., Colleoni, C., Cenci, U., Raj, J. N., and Tirtiaux, C. (2011). The evolution of glycogen and starch metabolism in eukaryotes gives molecular clues to understand the establishment of plastid endosymbiosis. J. Exp. Bot. 62, 1775–1801. doi: 10.1093/jxb/erq411
Ball, S. G., Subtil, A., Bhattacharya, D., Moustafa, A., Weber, A. P., Gehre, L., et al. (2013). Metabolic effectors secreted by bacterial pathogens: essential facilitators of plastid endosymbiosis? Plant Cell 25, 7–21. doi: 10.1105/tpc.112.101329
Barajas-Lopez Jde, D., Blanco, N. E., and Strand, A. (2013). Plastid-to-nucleus communication, signals controlling the running of the plant cell. Biochim. Biophys. Acta 1833, 425–437. doi: 10.1016/j.bbamcr.2012.06.020
Bellin, D., Asai, S., Delledonne, M., and Yoshioka, H. (2013). Nitric oxide as a mediator for defense responses. Mol. Plant Microbe Interact. 26, 271–277. doi: 10.1094/MPMI-09-12-0214-CR
Benitez-Alfonso, Y., Cilia, M., San Roman, A., Thomas, C., Maule, A., Hearn, S., et al. (2009). Control of Arabidopsis meristem development by thioredoxin-dependent regulation of intercellular transport. Proc. Natl. Acad. Sci. U.S.A. 106, 3615–3620. doi: 10.1073/pnas.0808717106
Benitez-Alfonso, Y., and Jackson, D. (2009). Redox homeostasis regulates plasmodesmal communication in Arabidopsis meristems. Plant Signal. Behav. 4, 655–659. doi: 10.1073/pnas.0808717106
Benning, C. (2009). Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu. Rev. Cell Dev. Biol. 25, 71–91. doi: 10.1146/annurev.cellbio.042308.113414
Besson-Bard, A., Courtois, C., Gauthier, A., Dahan, J., Dobrowolska, G., Jeandroz, S., et al. (2008). Nitric oxide in plants: production and cross-talk with Ca2+ signaling. Mol. Plant 1, 218–228. doi: 10.1093/mp/ssm016
Bhat, S., Folimonova, S. Y., Cole, A. B., Ballard, K. D., Lei, Z., Watson, B. S., et al. (2013). Influence of host chloroplast proteins on Tobacco mosaic virus accumulation and intercellular movement. Plant Physiol. 161, 134–147. doi: 10.1104/pp.112.207860
Bienert, G. P., Moller, A. L., Kristiansen, K. A., Schulz, A., Moller, I. M., Schjoerring, J. K., et al. (2007). Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 282, 1183–1192. doi: 10.1074/jbc.M603761200
Binns, D., Januszewski, T., Chen, Y., Hill, J., Markin, V. S., Zhao, Y., et al. (2006). An intimate collaboration between peroxisomes and lipid bodies. J. Cell Biol. 173, 719–731. doi: 10.1083/jcb.200511125
Blanco, F., Garreton, V., Frey, N., Dominguez, C., Perez-Acle, T., Van Der Straeten, D., et al. (2005). Identification of NPR1-dependent and independent genes early induced by salicylic acid treatment in Arabidopsis. Plant Mol. Biol. 59, 927–944. doi: 10.1007/s11103-005-2227-x
Blanco, N. E., Guinea-Diaz, M., Whelan, J., and Strand, A. (2014). Interaction between plastid and mitochondrial retrograde signalling pathways during changes to plastid redox status. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130231. doi: 10.1098/rstb.2013.0231
Blechert, S. (1999). Structure-activity analyses reveal the existence of two separate groups of active octadecanoids in elicitation of the tendril-coiling response of Bryonia dioica Jacq. Planta 207, 470–479. doi: 10.1007/s004250050506
Bobik, K., Dunlap, J. R., and Burch-Smith, T. M. (2014). Tandem high-pressure freezing and quick freeze substitution of plant tissues for transmission electron microscopy. J. Vis. Exp. e51844. doi: 10.3791/51844
Bonaventure, G., and Baldwin, I. T. (2010). Transduction of wound and herbivory signals in plastids. Commun. Integr. Biol. 3, 313–317. doi: 10.4161/cib.3.4.11834
Borucki, W., Bederska, M., and Sujkowska-Rybkowska, M. (2015). Visualisation of plastid outgrowths in potato (Solanum tuberosum L.) tubers by carboxyfluorescein diacetate staining. Plant Cell Rep. 34, 853–860. doi: 10.1007/s00299-015-1748-2
Botha, C. E. J., Cross, R. H. M., Van Bel, A. J. E., and Peter, C. I. (2000). Phloem loading in the sucrose-export-defective (SXD-1) mutant maize is limited by callose deposition at plasmodesmata in bundle sheath-vascular parenchyma interface. Protoplasma 214, 65–72. doi: 10.1007/BF02524263
Boudiere, L., Michaud, M., Petroutsos, D., Rebeille, F., Falconet, D., Bastien, O., et al. (2014). Glycerolipids in photosynthesis: composition, synthesis and trafficking. Biochim. Biophys. Acta 1837, 470–480. doi: 10.1016/j.bbabio.2013.09.007
Bradbeer, J. W., Atkinson, Y. E., Borner, T., and Hagemann, R. (1979). Cytoplasmic synthesis of plastid polypeptides may be controlled by plastid-synthesized RNA. Nature 279, 816–817. doi: 10.1038/279816a0
Brilli, F., Ciccioli, P., Frattoni, M., Prestininzi, M., Spanedda, A. F., and Loreto, F. (2009). Constitutive and herbivore-induced monoterpenes emitted by Populus x euroamericana leaves are key volatiles that orient Chrysomela populi beetles. Plant Cell Environ. 32, 542–552. doi: 10.1111/j.1365-3040.2009.01948.x
Brunkard, J. O., Runkel, A. M., and Zambryski, P. C. (2015a). The cytosol must flow: intercellular transport through plasmodesmata. Curr. Opin. Cell Biol. 35, 13–20. doi: 10.1016/j.ceb.2015.03.003
Brunkard, J. O., Runkel, A. M., and Zambryski, P. C. (2015b). Chloroplasts extend stromules independently and in response to internal redox signals. Proc. Natl. Acad. Sci. U.S.A. 112, 10044–10049. doi: 10.1073/pnas.1511570112
Burch-Smith, T. M., Brunkard, J. O., Choi, Y. G., and Zambryski, P. C. (2011). Organelle-nucleus cross-talk regulates plant intercellular communication via plasmodesmata. Proc. Natl. Acad. Sci. U.S.A. 108, E1451–E1460. doi: 10.1073/pnas.1117226108
Burch-Smith, T. M., and Zambryski, P. C. (2010). Loss of INCREASED SIZE EXCLUSION LIMIT (ISE)1 or ISE2 increases the formation of secondary plasmodesmata. Curr. Biol. 20, 989–993. doi: 10.1016/j.cub.2010.03.064
Burch-Smith, T. M., and Zambryski, P. C. (2012). Plasmodesmata paradigm shift: regulation from without versus within. Annu. Rev. Plant Biol. 63, 239–260. doi: 10.1146/annurev-arplant-042811-105453
Camejo, D., Romero-Puertas Mdel, C., Rodriguez-Serrano, M., Sandalio, L. M., Lazaro, J. J., Jimenez, A., et al. (2013). Salinity-induced changes in S-nitrosylation of pea mitochondrial proteins. J. Proteomics 79, 87–99. doi: 10.1016/j.jprot.2012.12.003
Cao, F. Y., Yoshioka, K., and Desveaux, D. (2011). The roles of ABA in plant-pathogen interactions. J. Plant Res. 124, 489–499. doi: 10.1007/s10265-011-0409-y
Caplan, J. L., Kumar, A. S., Park, E., Padmanabhan, M. S., Hoban, K., Modla, S., et al. (2015). Chloroplast stromules function during innate immunity. Dev. Cell 34, 45–57. doi: 10.1016/j.devcel.2015.05.011
Caplan, J. L., Mamillapalli, P., Burch-Smith, T. M., Czymmek, K., and Dinesh-Kumar, S. P. (2008). Chloroplastic protein NRIP1 mediates innate immune receptor recognition of a viral effector. Cell 132, 449–462. doi: 10.1016/j.cell.2007.12.031
Carrie, C., and Whelan, J. (2013). Widespread dual targeting of proteins in land plants: when, where, how and why. Plant Signal. Behav. 8, e25034. doi: 10.4161/psb.25034
Carter, W. P. L. (1994). Development of ozone reactivity scales for volatile organic compounds. J. Air Waste Manag. Assoc. 44, 881–899. doi: 10.1080/1073161X.1994.10467290
Caverzan, A., Passaia, G., Rosa, S. B., Ribeiro, C. W., Lazzarotto, F., and Margis-Pinheiro, M. (2012). Plant responses to stresses: role of ascorbate peroxidase in the antioxidant protection. Genet. Mol. Biol. 35, 1011–1019. doi: 10.1590/S1415-47572012000600016
Chaki, M., Kovacs, I., Spannagl, M., and Lindermayr, C. (2014). Computational prediction of candidate proteins for S-nitrosylation in Arabidopsis thaliana. PLoS ONE 9:e110232. doi: 10.1371/journal.pone.0110232
Chen, M., Galvao, R. M., Li, M., Burger, B., Bugea, J., Bolado, J., et al. (2010). Arabidopsis HEMERA/pTAC12 initiates photomorphogenesis by phytochromes. Cell 141, 1230–1240. doi: 10.1016/j.cell.2010.05.007
Chen, Z., Ricigliano, J. W., and Klessig, D. F. (1993). Purification and characterization of a soluble salicylic acid-binding protein from tobacco. Proc. Natl. Acad. Sci. U.S.A. 90, 9533–9537. doi: 10.1073/pnas.90.20.9533
Cheng, Y. Q., Liu, Z. M., Xu, J., Zhou, T., Wang, M., Chen, Y. T., et al. (2008). HC-Pro protein of sugar cane mosaic virus interacts specifically with maize ferredoxin-5 in vitro and in planta. J. Gen. Virol. 89, 2046–2054. doi: 10.1099/vir.0.2008/001271-0
Chi, W., Sun, X., and Zhang, L. (2013). Intracellular signaling from plastid to nucleus. Annu. Rev. Plant Biol. 64, 559–582. doi: 10.1146/annurev-arplant-050312-120147
Coll, N. S., Epple, P., and Dangl, J. L. (2011). Programmed cell death in the plant immune system. Cell Death Differ. 18, 1247–1256. doi: 10.1038/cdd.2011.37
Collings, D. A., Carter, C. N., Rink, J. C., Scott, A. C., Wyatt, S. E., and Allen, N. S. (2000). Plant nuclei can contain extensive grooves and invaginations. Plant Cell 12, 2425–2440. doi: 10.1105/tpc.12.12.2425
Cordoba, E., Salmi, M., and Leon, P. (2009). Unravelling the regulatory mechanisms that modulate the MEP pathway in higher plants. J. Exp. Bot. 60, 2933–2943. doi: 10.1093/jxb/erp190
Corpas, F. J., Palma, J. M., Del Rio, L. A., and Barroso, J. B. (2013). Protein tyrosine nitration in higher plants grown under natural and stress conditions. Front. Plant Sci. 4:29. doi: 10.3389/fpls.2013.00029
Cran, D. G., and Dyer, A. F. (1973). Membrane continuity ans associations in the fern, Dryopteris borreri. Protoplasma 76, 103–108. doi: 10.1007/BF01279676
Crotty, W. J., and Ledbetter, M. C. (1973). Membrane continuities involving chloroplasts and other organelles in plant cells. Science 182, 839–841. doi: 10.1126/science.182.4114.839
Dave, A., Hernandez, M. L., He, Z., Andriotis, V. M., Vaistij, F. E., Larson, T. R., et al. (2011). 12-oxo-phytodienoic acid accumulation during seed development represses seed germination in Arabidopsis. Plant Cell 23, 583–599. doi: 10.1105/tpc.110.081489
de Brito, O. M., and Scorrano, L. (2008). Mitofusin 2 tethers endoplasmic reticulum to mitochondria. Nature 456, 605–610. doi: 10.1038/nature07534
De Clercq, I., Vermeirssen, V., Van Aken, O., Vandepoele, K., Murcha, M. W., Law, S. R., et al. (2013). The membrane-bound NAC transcription factor ANAC013 functions in mitochondrial retrograde regulation of the oxidative stress response in Arabidopsis. Plant Cell 25, 3472–3490. doi: 10.1105/tpc.113.117168
Desikan, R., A-H-Mackerness, S., Hancock, J. T., and Neill, S. J. (2001). Regulation of the Arabidopsis transcriptome by oxidative stress. Plant Physiol. 127, 159–172. doi: 10.1104/pp.127.1.159
Desikan, R., Griffiths, R., Hancock, J., and Neill, S. (2002). A new role for an old enzyme: nitrate reductase-mediated nitric oxide generation is required for abscisic acid-induced stomatal closure in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 99, 16314–16318. doi: 10.1073/pnas.252461999
De Storme, N., and Geelen, D. (2014). Callose homeostasis at plasmodesmata: molecular regulators and developmental relevance. Front. Plant Sci. 5:138. doi: 10.3389/fpls.2014.00138
Dicke, M., and Baldwin, I. T. (2010). The evolutionary context for herbivore-induced plant volatiles: beyond the ‘cry for help’. Trends Plant Sci. 15, 167–175. doi: 10.1016/j.tplants.2009.12.002
Donaire, L., Barajas, D., Martinez-Garcia, B., Martinez-Priego, L., Pagan, I., and Llave, C. (2008). Structural and genetic requirements for the biogenesis of tobacco rattle virus-derived small interfering RNAs. J. Virol. 82, 5167–5177. doi: 10.1128/JVI.00272-08
Dudareva, N., Klempien, A., Muhlemann, J. K., and Kaplan, I. (2013). Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 198, 16–32. doi: 10.1111/nph.12145
Durner, J., and Klessig, D. F. (1995). Inhibition of ascorbate peroxidase by salicylic acid and 2,6-dichloroisonicotinic acid, two inducers of plant defense responses. Proc. Natl. Acad. Sci. U.S.A. 92, 11312–11316. doi: 10.1073/pnas.92.24.11312
Earley, K., Smith, M., Weber, R., Gregory, B., and Poethig, R. (2010). An endogenous F-box protein regulates ARGONAUTE1 in Arabidopsis thaliana. Silence 1, 15. doi: 10.1186/1758-907X-1-15
Ellis, R. J. (1977). Protein synthesis by isolated chloroplasts. Biochim. Biophys. Acta 463, 185–215. doi: 10.1016/0304-4173(77)90008-8
Engelberth, J. (2004). Airborne signals prime plants against insect herbivore attack. Proc. Natl. Acad. Sci. U.S.A. 101, 1781–1785. doi: 10.1073/pnas.0308037100
Erickson, J. L., Ziegler, J., Guevara, D., Abel, S., Klosgen, R. B., Mathur, J., et al. (2014). Agrobacterium-derived cytokinin influences plastid morphology and starch accumulation in Nicotiana benthamiana during transient assays. BMC Plant Biol. 14:127. doi: 10.1186/1471-2229-14-127
Estavillo, G. M., Crisp, P. A., Pornsiriwong, W., Wirtz, M., Collinge, D., Carrie, C., et al. (2011). Evidence for a SAL1-PAP chloroplast retrograde pathway that functions in drought and high light signaling in Arabidopsis. Plant Cell 23, 3992–4012. doi: 10.1105/tpc.111.091033
Falcon, L. I., Magallon, S., and Castillo, A. (2010). Dating the cyanobacterial ancestor of the chloroplast. ISME J. 4, 777–783. doi: 10.1038/ismej.2010.2
Finkelstein, R. (2013). Abscisic acid synthesis and response. Arabidopsis Book 11, e0166. doi: 10.1199/tab.0166
Finnegan, P. M., Whelan, J., Millar, A. H., Zhang, Q., Smith, M. K., Wiskich, J. T., et al. (1997). Differential expression of the multigene family encoding the soybean mitochondrial alternative oxidase. Plant Physiol. 114, 455–466. doi: 10.1104/pp.114.2.455
Flors, V., Ton, J., Van Doorn, R., Jakab, G., Garcia-Agustin, P., and Mauch-Mani, B. (2008). Interplay between JA, SA and ABA signalling during basal and induced resistance against Pseudomonas syringae and Alternaria brassicicola. Plant J. 54, 81–92. doi: 10.1111/j.1365-313X.2007.03397.x
Foissner, I., Sommer, A., and Hoeftberger, M. (2015). Photosynthesis-dependent formation of convoluted plasma membrane domains in Chara internodal cells is independent of chloroplast position. Protoplasma 252, 1085–1096. doi: 10.1007/s00709-014-0742-9
Foresi, N., Mayta, M. L., Lodeyro, A. F., Scuffi, D., Correa-Aragunde, N., Garcia-Mata, C., et al. (2015). Expression of the tetrahydrofolate-dependent nitric oxide synthase from the green alga Ostreococcus tauri increases tolerance to abiotic stresses and influences stomatal development in Arabidopsis. Plant J. 82, 806–821. doi: 10.1111/tpj.12852
Forouhar, F., Yang, Y., Kumar, D., Chen, Y., Fridman, E., Park, S. W., et al. (2005). Structural and biochemical studies identify tobacco SABP2 as a methyl salicylate esterase and implicate it in plant innate immunity. Proc. Natl. Acad. Sci. U.S.A. 102, 1773–1778. doi: 10.1073/pnas.0409227102
Foyer, C. H., Bloom, A. J., Queval, G., and Noctor, G. (2009). Photorespiratory metabolism: genes, mutants, energetics, and redox signaling. Annu. Rev. Plant Biol. 60, 455–484. doi: 10.1146/annurev.arplant.043008.091948
Foyer, C. H., Karpinska, B., and Krupinska, K. (2014). The functions of WHIRLY1 and REDOX-RESPONSIVE TRANSCRIPTION FACTOR 1 in cross tolerance responses in plants: a hypothesis. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130226. doi: 10.1098/rstb.2013.0226
Foyer, C. H., and Noctor, G. (2003). Redox sensing snd signalling associated with reactive oxygen in chloroplasts, peroxisomes and mitochondria. Physiol. Plant. 119, 355–364. doi: 10.1034/j.1399-3054.2003.00223.x
Foyer, C. H., and Noctor, G. (2005). Redox homeostasis and antioxidant signaling: a metabolic interface between stress perception and physiological responses. Plant Cell 17, 1866–1875. doi: 10.1105/tpc.105.033589
Fu, Z. Q., and Dong, X. (2013). Systemic acquired resistance: turning local infection into global defense. Annu. Rev. Plant Biol. 64, 839–863. doi: 10.1146/annurev-arplant-042811-105606
Fu, Z. Q., Guo, M., Jeong, B. R., Tian, F., Elthon, T. E., Cerny, R. L., et al. (2007). A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature 447, 284–288. doi: 10.1038/nature05737
Fu, Z. Q., Yan, S., Saleh, A., Wang, W., Ruble, J., Oka, N., et al. (2012). NPR3 and NPR4 are receptors for the immune signal salicylic acid in plants. Nature 486, 228–232. doi: 10.1038/nature11162
Gadjev, I., Vanderauwera, S., Gechev, T. S., Laloi, C., Minkov, I. N., Shulaev, V., et al. (2006). Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 141, 436–445. doi: 10.1104/pp.106.078717
Gallagher, K. L., Sozzani, R., and Lee, C. M. (2014). Intercellular protein movement: deciphering the language of development. Annu. Rev. Cell Dev. Biol. 30, 207–233. doi: 10.1146/annurev-cellbio-100913-012915
Garcion, C., Lohmann, A., Lamodiere, E., Catinot, J., Buchala, A., Doermann, P., et al. (2008). Characterization and biological function of the ISOCHORISMATE SYNTHASE2 gene of Arabidopsis. Plant Physiol. 147, 1279–1287. doi: 10.1104/pp.108.119420
Gile, G. H., Moog, D., Slamovits, C. H., Maier, U. G., and Archibald, J. M. (2015). Dual organellar targeting of aminoacyl-tRNA synthetases in diatoms and cryptophytes. Genome Biol. Evol. 7, 1728–1742. doi: 10.1093/gbe/evv095
Gill, S. S., and Tuteja, N. (2010). Reactive oxygen species and antioxidant machinery in abiotic stress tolerance in crop plants. Plant Physiol. Biochem. 48, 909–930. doi: 10.1016/j.plaphy.2010.08.016
Giraud, E., Van Aken, O., Ho, L. H., and Whelan, J. (2009). The transcription factor ABI4 is a regulator of mitochondrial retrograde expression of ALTERNATIVE OXIDASE1a. Plant Physiol. 150, 1286–1296. doi: 10.1104/pp.109.139782
Giuliano, G. (2014). Plant carotenoids: genomics meets multi-gene engineering. Curr. Opin. Plant Biol. 19, 111–117. doi: 10.1016/j.pbi.2014.05.006
Glasser, C., Haberer, G., Finkemeier, I., Pfannschmidt, T., Kleine, T., Leister, D., et al. (2014). Meta-analysis of retrograde signaling in Arabidopsis thaliana reveals a core module of genes embedded in complex cellular signaling networks. Mol. Plant 7, 1167–1190. doi: 10.1093/mp/ssu042
Goetz, S., Hellwege, A., Stenzel, I., Kutter, C., Hauptmann, V., Forner, S., et al. (2012). Role of cis-12-oxo-phytodienoic acid in tomato embryo development. Plant Physiol. 158, 1715–1727. doi: 10.1104/pp.111.192658
Goto-Yamada, S., Mano, S., Yamada, K., Oikawa, K., Hosokawa, Y., Hara-Nishimura, I., et al. (2015). Dynamics of the light-dependent transition of plant peroxisomes. Plant Cell Physiol. 56, 1264–1271. doi: 10.1093/pcp/pcv081
Gray, J. C., Hansen, M. R., Shaw, D. J., Graham, K., Dale, R., Smallman, P., et al. (2012). Plastid stromules are induced by stress treatments acting through abscisic acid. Plant J. 69, 387–398. doi: 10.1111/j.1365-313X.2011.04800.x
Gray, J. C., Sullivan, J. A., Hibberd, J. M., and Hansen, M. R. (2001). Stromules: mobile protusions and interconnections between plastids. Plant Biol. (Stuttg.) 3, 223–233. doi: 10.1055/s-2001-15204
Groning, B. R., Abouzid, A., and Jeske, H. (1987). Single-stranded DNA from Abutilon mosaic virus is present in the plastids of infected Abutilon sellovianum. Proc. Natl. Acad. Sci. U.S.A. 84, 8996–9000. doi: 10.1073/pnas.84.24.8996
Gunning, B. E. (2005). Plastid stromules: video microscopy of their outgrowth, retraction, tensioning, anchoring, branching, bridging, and tip-shedding. Protoplasma 225, 33–42. doi: 10.1007/s00709-004-0073-3
Guo, F. Q., Okamoto, M., and Crawford, N. M. (2003). Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science 302, 100–103. doi: 10.1126/science.1086770
Han, Y., Mhamdi, A., Chaouch, S., and Noctor, G. (2013). Regulation of basal and oxidative stress-triggered jasmonic acid-related gene expression by glutathione. Plant Cell Environ. 36, 1135–1146. doi: 10.1111/pce.12048
Hanson, M. R., and Sattarzadeh, A. (2011). Stromules: recent insights into a long neglected feature of plastid morphology and function. Plant Physiol. 155, 1486–1492. doi: 10.1104/pp.110.170852
Hanson, M. R., and Sattarzadeh, A. (2013). Trafficking of proteins through plastid stromules. Plant Cell 25, 2774–2782. doi: 10.1105/tpc.113.112870
Harari-Steinberg, O., Ohad, I., and Chamovitz, D. A. (2001). Dissection of the light signal transduction pathways regulating the two early light-induced protein genes in Arabidopsis. Plant Physiol. 127, 986–997. doi: 10.1104/pp.010270
Harrison, S. P., Morfopoulos, C., Dani, K. G., Prentice, I. C., Arneth, A., Atwell, B. J., et al. (2013). Volatile isoprenoid emissions from plastid to planet. New Phytol. 197, 49–57. doi: 10.1111/nph.12021
Hedtke, B., Wagner, I., Borner, T., and Hess, W. R. (1999). Inter-organellar crosstalk in higher plants: impaired chloroplast development affects mitochondrial gene and transcript levels. Plant J. 19, 635–643. doi: 10.1046/j.1365-313x.1999.00554.x
Heil, M. (2008). Indirect defence via tritrophic interactions. New Phytol. 178, 41–61. doi: 10.1111/j.1469-8137.2007.02330.x
Heil, M. (2010). Explaining evolution of plant communication by airborne signals. Trends Ecol. Evol. 25, 137–144. doi: 10.1016/j.tree.2009.09.010
Heil, M. (2014). Herbivore-induced plant volatiles: targets, perception and unanswered questions. New Phytol. 204, 297–306. doi: 10.1111/nph.12977
Heil, M., and Silva Bueno, J. C. (2007). Within-plant signaling by volatiles leads to induction and priming of an indirect plant defense in nature. Proc. Natl. Acad. Sci. U.S.A. 104, 5467–5472. doi: 10.1073/pnas.0610266104
Heinlein, M. (2015). Plasmodesmata: channels for viruses on the move. Methods Mol. Biol. 1217, 25–52. doi: 10.1007/978-1-4939-1523-1_2
Holzinger, A., Buchner, O., Lutz, C., and Hanson, M. R. (2007a). Temperature-sensitive formation of chloroplast protrusions and stromules in mesophyll cells of Arabidopsis thaliana. Protoplasma 230, 23–30. doi: 10.1007/s00709-006-0222-y
Holzinger, A., Wasteneys, G. O., and Lutz, C. (2007b). Investigating cytoskeletal function in chloroplast protrusion formation in the arctic-alpine plant Oxyria digyna. Plant Biol. (Stuttg.) 9, 400–410. doi: 10.1055/s-2006-924727
Hunter, L. J., Westwood, J. H., Heath, G., Macaulay, K., Smith, A. G., Macfarlane, S. A., et al. (2013). Regulation of RNA-dependent RNA polymerase 1 and isochorismate synthase gene expression in Arabidopsis. PLoS ONE 8:e66530. doi: 10.1371/journal.pone.0066530
Isemer, R., Mulisch, M., Schafer, A., Kirchner, S., Koop, H. U., and Krupinska, K. (2012). Recombinant Whirly1 translocates from transplastomic chloroplasts to the nucleus. FEBS Lett. 586, 85–88. doi: 10.1016/j.febslet.2011.11.029
Ishida, H., Yoshimoto, K., Izumi, M., Reisen, D., Yano, Y., Makino, A., et al. (2008). Mobilization of rubisco and stroma-localized fluorescent proteins of chloroplasts to the vacuole by an ATG gene-dependent autophagic process. Plant Physiol. 148, 142–155. doi: 10.1104/pp.108.122770
Izumi, M., Hidema, J., Wada, S., Kondo, E., Kurusu, T., Kuchitsu, K., et al. (2015). Establishment of monitoring methods for autophagy in rice reveals autophagic recycling of chloroplasts and root plastids during energy limitation. Plant Physiol. 167, 1307–1320. doi: 10.1104/pp.114.254078
Jackson, D. (2015). Plasmodesmata spread their influence. F1000Prime Rep. 7, 25. doi: 10.12703/P7-25
Jang, C., Seo, E. Y., Nam, J., Bae, H., Gim, Y. G., Kim, H. G., et al. (2013). Insights into Alternanthera mosaic virus TGB3 functions: interactions with Nicotiana benthamiana PsbO correlate with chloroplast vesiculation and veinal necrosis caused by TGB3 over-expression. Front. Plant Sci. 4:5. doi: 10.3389/fpls.2013.00005
Jelenska, J., Van Hal, J. A., and Greenberg, J. T. (2010). Pseudomonas syringae hijacks plant stress chaperone machinery for virulence. Proc. Natl. Acad. Sci. U.S.A. 107, 13177–13182. doi: 10.1073/pnas.0910943107
Jelenska, J., Yao, N., Vinatzer, B. A., Wright, C. M., Brodsky, J. L., and Greenberg, J. T. (2007). A J domain virulence effector of Pseudomonas syringae remodels host chloroplasts and suppresses defenses. Curr. Biol. 17, 499–508. doi: 10.1016/j.cub.2007.02.028
Jimenez, I., Lopez, L., Alamillo, J. M., Valli, A., and Garcia, J. A. (2006). Identification of a plum pox virus CI-interacting protein from chloroplast that has a negative effect in virus infection. Mol. Plant Microbe Interact. 19, 350–358. doi: 10.1094/MPMI-19-0350
Jin, Y., Ma, D., Dong, J., Li, D., Deng, C., Jin, J., et al. (2007). The HC-pro protein of Potato virus Y interacts with NtMinD of tobacco. Mol. Plant Microbe Interact. 20, 1505–1511. doi: 10.1094/MPMI-20-12-1505
Johanningmeier, U., and Howell, S. H. (1984). Regulation of light-harvesting chlorophyll-binding protein mRNA accumulation in Chlamydomonas reinhardi. Possible involvement of chlorophyll synthesis precursors. J. Biol. Chem. 259, 13541–13549.
Kangasjarvi, S., Neukermans, J., Li, S., Aro, E. M., and Noctor, G. (2012). Photosynthesis, photorespiration, and light signalling in defence responses. J. Exp. Bot. 63, 1619–1636. doi: 10.1093/jxb/err402
Kim, C., and Apel, K. (2013). Singlet oxygen-mediated signaling in plants: moving from flu to wild type reveals an increasing complexity. Photosynth. Res. 116, 455–464. doi: 10.1007/s11120-013-9876-4
Kim, C., Meskauskiene, R., Zhang, S., Lee, K. P., Lakshmanan Ashok, M., Blajecka, K., et al. (2012). Chloroplasts of Arabidopsis are the source and a primary target of a plant-specific programmed cell death signaling pathway. Plant Cell 24, 3026–3039. doi: 10.1105/tpc.112.100479
Kim, I., Hempel, F. D., Sha, K., Pfluger, J., and Zambryski, P. C. (2002). Identification of a developmental transition in plasmodesmatal function during embryogenesis in Arabidopsis thaliana. Development 129, 1261–1272.
Kim, T. H., Bohmer, M., Hu, H., Nishimura, N., and Schroeder, J. I. (2010). Guard cell signal transduction network: advances in understanding abscisic acid, CO2, and Ca2+ signaling. Annu. Rev. Plant Biol. 61, 561–591. doi: 10.1146/annurev-arplant-042809-112226
Kinkema, M., Fan, W., and Dong, X. (2000). Nuclear localization of NPR1 is required for activation of PR gene expression. Plant Cell 12, 2339–2350. doi: 10.2307/3871233
Kline, K. G., Sussman, M. R., and Jones, A. M. (2010). Abscisic acid receptors. Plant Physiol. 154, 479–482. doi: 10.1104/pp.110.160846
Kobayashi, K., Otegui, M. S., Krishnakumar, S., Mindrinos, M., and Zambryski, P. (2007). INCREASED SIZE EXCLUSION LIMIT 2 encodes a putative DEVH box RNA helicase involved in plasmodesmata function during Arabidopsis embryogenesis. Plant Cell 19, 1885–1897. doi: 10.1105/tpc.106.045666
Kohler, R. H., Cao, J., Zipfel, W. R., Webb, W. W., and Hanson, M. R. (1997). Exchange of protein molecules through connections between higher plant plastids. Science 276, 2039–2042. doi: 10.1126/science.276.5321.2039
Kohler, R. H., and Hanson, M. R. (2000). Plastid tubules of higher plants are tissue-specific and developmentally regulated. J. Cell Sci. 113(Pt 1), 81–89.
Kohler, R. H., Schwille, P., Webb, W. W., and Hanson, M. R. (2000). Active protein transport through plastid tubules: velocity quantified by fluorescence correlation spectroscopy. J. Cell Sci. 113(Pt 22), 3921–3930.
Koussevitzky, S., Nott, A., Mockler, T. C., Hong, F., Sachetto-Martins, G., Surpin, M., et al. (2007). Signals from chloroplasts converge to regulate nuclear gene expression. Science 316, 715–719. doi: 10.1126/science.%201140516
Krenz, B., Jeske, H., and Kleinow, T. (2012). The induction of stromule formation by a plant DNA-virus in epidermal leaf tissues suggests a novel intra– and intercellular macromolecular trafficking route. Front. Plant Sci. 3:291. doi: 10.3389/fpls.2012.00291
Krenz, B., Windeisen, V., Wege, C., Jeske, H., and Kleinow, T. (2010). A plastid-targeted heat shock cognate 70kDa protein interacts with the Abutilon mosaic virus movement protein. Virology 401, 6–17. doi: 10.1016/j.virol.2010.02.011
Kriegs, B., Jansen, M., Hahn, K., Peisker, H., Samajova, O., Beck, M., et al. (2010). Cyclic monoterpene mediated modulations of Arabidopsis thaliana phenotype: effects on the cytoskeleton and on the expression of selected genes. Plant Signal. Behav. 5, 832–838. doi: 10.4161/psb.5.7.12032
Kropat, J., Oster, U., Rudiger, W., and Beck, C. F. (1997). Chlorophyll precursors are signals of chloroplast origin involved in light induction of nuclear heat-shock genes. Proc. Natl. Acad. Sci. U.S.A. 94, 14168–14172. doi: 10.1073/pnas.94.25.14168
Kropat, J., Oster, U., Rudiger, W., and Beck, C. F. (2000). Chloroplast signalling in the light induction of nuclear HSP70 genes requires the accumulation of chlorophyll precursors and their accessibility to cytoplasm/nucleus. Plant J. 24, 523–531. doi: 10.1046/j.1365-313x.2000.00898.x
Kunkel, B. N., and Brooks, D. M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. doi: 10.1016/S1369-5266(02)00275-3
Kwok, E. Y., and Hanson, M. R. (2003). Microfilaments and microtubules control the morphology and movement of non-green plastids and stromules in Nicotiana tabacum. Plant J. 35, 16–26. doi: 10.1046/j.1365-313X.2003.01777.x
Kwok, E. Y., and Hanson, M. R. (2004a). GFP-labelled Rubisco and aspartate aminotransferase are present in plastid stromules and traffic between plastids. J. Exp. Bot. 55, 595–604. doi: 10.1093/jxb/erh062
Kwok, E. Y., and Hanson, M. R. (2004b). In vivo analysis of interactions between GFP-labeled microfilaments and plastid stromules. BMC Plant Biol. 4:2. doi: 10.1186/1471-2229-4-2
Kwok, E. Y., and Hanson, M. R. (2004c). Plastids and stromules interact with the nucleus and cell membrane in vascular plants. Plant Cell Rep. 23, 188–195. doi: 10.1007/s00299-004-0824-9
Kwon, K. C., Verma, D., Jin, S., Singh, N. D., and Daniell, H. (2013). Release of proteins from intact chloroplasts induced by reactive oxygen species during biotic and abiotic stress. PLoS ONE 8:e67106. doi: 10.1371/journal.pone.0067106
Laloi, C., Stachowiak, M., Pers-Kamczyc, E., Warzych, E., Murgia, I., and Apel, K. (2007). Cross-talk between singlet oxygen- and hydrogen peroxide-dependent signaling of stress responses in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 104, 672–677. doi: 10.1073/pnas.0609063103
Langeveld, S. M., Van Wijk, R., Stuurman, N., Kijne, J. W., and De Pater, S. (2000). B-type granule containing protrusions and interconnections between amyloplasts in developing wheat endosperm revealed by transmission electron microscopy and GFP expression. J. Exp. Bot. 51, 1357–1361. doi: 10.1093/jexbot/51.349.1357
Larkin, R. M., Alonso, J. M., Ecker, J. R., and Chory, J. (2003). GUN4, a regulator of chlorophyll synthesis and intracellular signaling. Science 299, 902–906. doi: 10.1126/science.1079978
Ledford, H. K., Chin, B. L., and Niyogi, K. K. (2007). Acclimation to singlet oxygen stress in Chlamydomonas reinhardtii. Eukaryot. Cell 6, 919–930. doi: 10.1128/EC.00207-06
Lee, J. Y., Wang, X., Cui, W., Sager, R., Modla, S., Czymmek, K., et al. (2011). A plasmodesmata-localized protein mediates crosstalk between cell-to-cell communication and innate immunity in Arabidopsis. Plant Cell 23, 3353–3373. doi: 10.1105/tpc.111.087742
Leon, J. (2013). Role of plant peroxisomes in the production of jasmonic acid-based signals. Subcell. Biochem. 69, 299–313. doi: 10.1007/978-94-007-6889-5_16
Li, G., Froehlich, J. E., Elowsky, C., Msanne, J., Ostosh, A. C., Zhang, C., et al. (2014). Distinct Pseudomonas type-III effectors use a cleavable transit peptide to target chloroplasts. Plant J. 77, 310–321. doi: 10.1111/tpj.12396
Li, H., Ma, D., Jin, Y., Tu, Y., Liu, L., Leng, C., et al. (2015). Helper component-proteinase enhances the activity of 1-deoxy–xylulose-5-phosphate synthase and promotes the biosynthesis of plastidic isoprenoids in Potato virus Y-infected tobacco. Plant Cell Environ. doi: 10.1111/pce.12526 [Epub ahead of print].
Li, T., Li, H., Zhang, Y. X., and Liu, J. Y. (2011). Identification and analysis of seven H(2)O(2)-responsive miRNAs and 32 new miRNAs in the seedlings of rice (Oryza sativa L. ssp. indica). Nucleic Acids Res. 39, 2821–2833. doi: 10.1093/nar/gkq1047
Li, W., Cui, X., Meng, Z., Huang, X., Xie, Q., Wu, H., et al. (2012). Transcriptional regulation of Arabidopsis MIR168a and argonaute1 homeostasis in abscisic acid and abiotic stress responses. Plant Physiol. 158, 1279–1292. doi: 10.1104/pp.111.188789
Lin, L., Luo, Z., Yan, F., Lu, Y., Zheng, H., and Chen, J. (2011). Interaction between potyvirus P3 and ribulose-1,5-bisphosphate carboxylase/oxygenase (RubisCO) of host plants. Virus Genes 43, 90–92. doi: 10.1007/s11262-011-0596-6
Lindermayr, C., Saalbach, G., and Durner, J. (2005). Proteomic identification of S-nitrosylated proteins in Arabidopsis. Plant Physiol. 137, 921–930. doi: 10.1104/pp.104.058719
Lounifi, I., Arc, E., Molassiotis, A., Job, D., Rajjou, L., and Tanou, G. (2013). Interplay between protein carbonylation and nitrosylation in plants. Proteomics 13, 568–578. doi: 10.1002/pmic.201200304
Ludwikow, A. (2015). Targeting proteins for proteasomal degradation-a new function of Arabidopsis ABI1 protein phosphatase 2C. Front. Plant Sci. 6:310. doi: 10.3389/fpls.2015.00310
Lundquist, P. K., Poliakov, A., Giacomelli, L., Friso, G., Appel, M., Mcquinn, R. P., et al. (2013). Loss of plastoglobule kinases ABC1K1 and ABC1K3 causes conditional degreening, modified prenyl-lipids, and recruitment of the jasmonic acid pathway. Plant Cell 25, 1818–1839. doi: 10.1105/tpc.113.111120
Ma, Y., Szostkiewicz, I., Korte, A., Moes, D., Yang, Y., Christmann, A., et al. (2009). Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 324, 1064–1068. doi: 10.1126/science.1172408
Malamy, J., Carr, J. P., Klessig, D. F., and Raskin, I. (1990). Salicylic Acid: a likely endogenous signal in the resistance response of tobacco to viral infection. Science 250, 1002–1004. doi: 10.1126/science.250.4983.1002
Mandal, M. K., Chandra-Shekara, A. C., Jeong, R. D., Yu, K., Zhu, S., Chanda, B., et al. (2012). Oleic acid-dependent modulation of NITRIC OXIDE ASSOCIATED1 protein levels regulates nitric oxide-mediated defense signaling in Arabidopsis. Plant Cell 24, 1654–1674. doi: 10.1105/tpc.112.096768
Manfre, A., Glenn, M., Nunez, A., Moreau, R. A., and Dardick, C. (2011). Light quantity and photosystem function mediate host susceptibility to Turnip mosaic virus via a salicylic acid-independent mechanism. Mol. Plant Microbe Interact. 24, 315–327. doi: 10.1094/MPMI-08-10-0191
Manohar, M., Tian, M., Moreau, M., Park, S. W., Choi, H. W., Fei, Z., et al. (2014). Identification of multiple salicylic acid-binding proteins using two high throughput screens. Front. Plant Sci. 5:777. doi: 10.3389/fpls.2014.00777
Marechal, E., and Bastien, O. (2014). Modeling of regulatory loops controlling galactolipid biosynthesis in the inner envelope membrane of chloroplasts. J. Theor. Biol. 361, 1–13. doi: 10.1016/j.jtbi.2014.07.013
Matsui, A., Ishida, J., Morosawa, T., Mochizuki, Y., Kaminuma, E., Endo, T. A., et al. (2008). Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 49, 1135–1149. doi: 10.1093/pcp/pcn101
Mayfield, S. P., and Taylor, W. C. (1984). Carotenoid-deficient maize seedlings fail to accumulate light-harvesting chlorophyll a/b binding protein (LHCP) mRNA. Eur. J. Biochem. 144, 79–84. doi: 10.1111/j.1432-1033.1984.tb08433.x
McCallum, E. J., Cunningham, J. P., Lucker, J., Zalucki, M. P., De Voss, J. J., and Botella, J. R. (2011). Increased plant volatile production affects oviposition, but not larval development, in the moth Helicoverpa armigera. J. Exp. Biol. 214, 3672–3677. doi: 10.1242/jeb.059923
McDonald, K. (1999). High-pressure freezing for preservation of high resolution fine structure and antigenicity for immunolabeling. Methods Mol. Biol. 117, 77–97. doi: 10.1385/1-59259-201-5:77
McDonald, K. L. (2014). Out with the old and in with the new: rapid specimen preparation procedures for electron microscopy of sectioned biological material. Protoplasma 251, 429–448. doi: 10.1007/s00709-013-0575-y
McFadden, G. I. (2001). Chloroplast origin and integration. Plant Physiol. 125, 50–53. doi: 10.1104/pp.125.1.50
McLean, B., Whatley, J. M., and Juniper, B. E. (1988). Continuity of chloroplast and endoplasmic reticulum membranes in Chara and Equisetum. New Phytol. 109, 59–65. doi: 10.1111/j.1469-8137.1988.tb00219.x
Mehrshahi, P., Johnny, C., and Dellapenna, D. (2014). Redefining the metabolic continuity of chloroplasts and ER. Trends Plant Sci. 19, 501–507. doi: 10.1016/j.tplants.2014.02.013
Mehrshahi, P., Stefano, G., Andaloro, J. M., Brandizzi, F., Froehlich, J. E., and Dellapenna, D. (2013). Transorganellar complementation redefines the biochemical continuity of endoplasmic reticulum and chloroplasts. Proc. Natl. Acad. Sci. U.S.A. 110, 12126–12131. doi: 10.1073/pnas.1306331110
Meng, L., Wong, J. H., Feldman, L. J., Lemaux, P. G., and Buchanan, B. B. (2010). A membrane-associated thioredoxin required for plant growth moves from cell to cell, suggestive of a role in intercellular communication. Proc. Natl. Acad. Sci. U.S.A. 107, 3900–3905. doi: 10.1073/pnas.0913759107
Mengel, A., Chaki, M., Shekariesfahlan, A., and Lindermayr, C. (2013). Effect of nitric oxide on gene transcription – S-nitrosylation of nuclear proteins. Front. Plant Sci. 4:293. doi: 10.3389/fpls.2013.00293
Meskauskiene, R., Nater, M., Goslings, D., Kessler, F., Op Den Camp, R., and Apel, K. (2001). FLU: a negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 98, 12826–12831. doi: 10.1073/pnas.221252798
Mhamdi, A., Hager, J., Chaouch, S., Queval, G., Han, Y., Taconnat, L., et al. (2010). Arabidopsis GLUTATHIONE REDUCTASE1 plays a crucial role in leaf responses to intracellular hydrogen peroxide and in ensuring appropriate gene expression through both salicylic acid and jasmonic acid signaling pathways. Plant Physiol. 153, 1144–1160. doi: 10.1104/pp.110.153767
Miller, G., Shulaev, V., and Mittler, R. (2008). Reactive oxygen signaling and abiotic stress. Physiol. Plant. 133, 481–489. doi: 10.1111/j.1399-3054.2008.01090.x
Misra, A. N., Vladkova, R., Singh, R., Misra, M., Dobrikova, A. G., and Apostolova, E. L. (2014). Action and target sites of nitric oxide in chloroplasts. Nitric Oxide 39, 35–45. doi: 10.1016/j.niox.2014.04.003
Mochizuki, N., Brusslan, J. A., Larkin, R., Nagatani, A., and Chory, J. (2001). Arabidopsis genomes uncoupled 5 (GUN5) mutant reveals the involvement of Mg-chelatase H subunit in plastid-to-nucleus signal transduction. Proc. Natl. Acad. Sci. U.S.A. 98, 2053–2058. doi: 10.1073/pnas.98.4.2053
Montillet, J. L., Cacas, J. L., Garnier, L., Montane, M. H., Douki, T., Bessoule, J. J., et al. (2004). The upstream oxylipin profile of Arabidopsis thaliana: a tool to scan for oxidative stresses. Plant J. 40, 439–451. doi: 10.1111/j.1365-313X.2004.02223.x
Mooney, K. A., Pratt, R. T., and Singer, M. S. (2012). The tri-trophic interactions hypothesis: interactive effects of host plant quality, diet breadth and natural enemies on herbivores. PLoS ONE 7:e34403. doi: 10.1371/journal.pone.0034403
Mor, A., Koh, E., Weiner, L., Rosenwasser, S., Sibony-Benyamini, H., and Fluhr, R. (2014). Singlet oxygen signatures are detected independent of light or chloroplasts in response to multiple stresses. Plant Physiol. 165, 249–261. doi: 10.1104/pp.114.236380
Moreau, M., Lee, G. I., Wang, Y., Crane, B. R., and Klessig, D. F. (2008). AtNOS/AtNOA1 is a functional Arabidopsis thaliana cGTPase and not a nitric-oxide synthase. J. Biol. Chem. 283, 32957–32967. doi: 10.1074/jbc.M804838200
Moreau, M., Westlake, T., Zampogna, G., Popescu, G., Tian, M., Noutsos, C., et al. (2013). The Arabidopsis oligopeptidases TOP1 and TOP2 are salicylic acid targets that modulate SA-mediated signaling and the immune response. Plant J. 76, 603–614. doi: 10.1111/tpj.12320
Mou, Z., Fan, W., and Dong, X. (2003). Inducers of plant systemic acquired resistance regulate NPR1 function through redox changes. Cell 113, 935–944. doi: 10.1016/S0092-8674(03)00429-X
Moulin, M., Mccormac, A. C., Terry, M. J., and Smith, A. G. (2008). Tetrapyrrole profiling in Arabidopsis seedlings reveals that retrograde plastid nuclear signaling is not due to Mg-protoporphyrin IX accumulation. Proc. Natl. Acad. Sci. U.S.A. 105, 15178–15183. doi: 10.1073/pnas.0803054105
Mubarakshina, M. M., Ivanov, B. N., Naydov, I. A., Hillier, W., Badger, M. R., and Krieger-Liszkay, A. (2010). Production and diffusion of chloroplastic H2O2 and its implication to signalling. J. Exp. Bot. 61, 3577–3587. doi: 10.1093/jxb/erq171
Mullineaux, C. W., Mariscal, V., Nenninger, A., Khanum, H., Herrero, A., Flores, E., et al. (2008). Mechanism of intercellular molecular exchange in heterocyst-forming cyanobacteria. EMBO J. 27, 1299–1308. doi: 10.1038/emboj.2008.66
Mur, L. A., Laarhoven, L. J., Harren, F. J., Hall, M. A., and Smith, A. R. (2008). Nitric oxide interacts with salicylate to regulate biphasic ethylene production during the hypersensitive response. Plant Physiol. 148, 1537–1546. doi: 10.1104/pp.108.124404
Nakashima, K., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2014). The transcriptional regulatory network in the drought response and its crosstalk in abiotic stress responses including drought, cold, and heat. Front. Plant Sci. 5:170. doi: 10.3389/fpls.2014.00170
Nakayama, T., and Archibald, J. M. (2012). Evolving a photosynthetic organelle. BMC Biol. 10:35. doi: 10.1186/1741-7007-10-35
Natesan, S. K., Sullivan, J. A., and Gray, J. C. (2005). Stromules: a characteristic cell-specific feature of plastid morphology. J. Exp. Bot. 56, 787–797. doi: 10.1093/jxb/eri088
Natesan, S. K., Sullivan, J. A., and Gray, J. C. (2009). Myosin XI is required for actin-associated movement of plastid stromules. Mol. Plant 2, 1262–1272. doi: 10.1093/mp/ssp078
Newell, C. A., Natesan, S. K., Sullivan, J. A., Jouhet, J., Kavanagh, T. A., and Gray, J. C. (2012). Exclusion of plastid nucleoids and ribosomes from stromules in tobacco and Arabidopsis. Plant J. 69, 399–410. doi: 10.1111/j.1365-313X.2011.04798.x
Ng, S., De Clercq, I., Van Aken, O., Law, S. R., Ivanova, A., Willems, P., et al. (2014). Anterograde and retrograde regulation of nuclear genes encoding mitochondrial proteins during growth, development, and stress. Mol. Plant 7, 1075–1093. doi: 10.1093/mp/ssu037
Ng, S., Ivanova, A., Duncan, O., Law, S. R., Van Aken, O., De Clercq, I., et al. (2013). A membrane-bound NAC transcription factor, ANAC017, mediates mitochondrial retrograde signaling in Arabidopsis. Plant Cell 25, 3450–3471. doi: 10.1105/tpc.113.113985
Niinemets, U. (2010). The emission factor of volatile isoprenoids: stress, acclimation, and developmental responses. Biogeosciences 7, 2203–2223. doi: 10.5194/bg-7-2203-2010
Niinemets, U. (2013). Quantitative patterns between plant volatile emissions induced by biotic stresses and the degree of damage. Front. Plant Sci. 4:262. doi: 10.3389/fpls.2013.00262
Nomura, H., Komori, T., Uemura, S., Kanda, Y., Shimotani, K., Nakai, K., et al. (2012). Chloroplast-mediated activation of plant immune signalling in Arabidopsis. Nat. Commun. 3, 926. doi: 10.1038/ncomms1926
Oelmuller, R., Levitan, I., Bergfeld, R., Rajasekhar, V. K., and Mohr, H. (1986). Expression of nuclear genes as affected by treatments acting on the plastids. Planta 168, 482–492. doi: 10.1007/BF00392267
Oikawa, K., Matsunaga, S., Mano, S., Kondo, M., Yamada, K., Hayashi, M., et al. (2015). Physical interaction between peroxisomes and chloroplasts elucidated by in situ laser analysis. Nat. Plants 1, 15035. doi: 10.1038/NPLANTS.2015.35
op den Camp, R. G., Przybyla, D., Ochsenbein, C., Laloi, C., Kim, C., Danon, A., et al. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15, 2320–2332. doi: 10.1105/tpc.014662
Ortega-Galisteo, A. P., Rodriguez-Serrano, M., Pazmino, D. M., Gupta, D. K., Sandalio, L. M., and Romero-Puertas, M. C. (2012). S-Nitrosylated proteins in pea (Pisum sativum L.) leaf peroxisomes: changes under abiotic stress. J. Exp. Bot. 63, 2089–2103. doi: 10.1093/jxb/err414
Padmanabhan, M. S., and Dinesh-Kumar, S. P. (2010). All hands on deck-the role of chloroplasts, endoplasmic reticulum, and the nucleus in driving plant innate immunity. Mol. Plant Microbe Interact. 23, 1368–1380. doi: 10.1094/MPMI-05-10-0113
Pandey, S. P., and Baldwin, I. T. (2007). RNA-directed RNA polymerase 1 (RdR1) mediates the resistance of Nicotiana attenuata to herbivore attack in nature. Plant J. 50, 40–53. doi: 10.1111/j.1365-313X.2007.03030.x
Park, S. W., Kaimoyo, E., Kumar, D., Mosher, S., and Klessig, D. F. (2007). Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318, 113–116. doi: 10.1126/science.1147113
Park, S. Y., Fung, P., Nishimura, N., Jensen, D. R., Fujii, H., Zhao, Y., et al. (2009). Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science 324, 1068–1071. doi: 10.1126/science.1173041
Petre, B., Saunders, D. G., Sklenar, J., Lorrain, C., Win, J., Duplessis, S., et al. (2015). Candidate effector proteins of the rust pathogen Melampsora larici-populina target diverse plant cell compartments. Mol. Plant Microbe Interact. 28, 689–700. doi: 10.1094/MPMI-01-15-0003-R
Petrillo, E., Godoy Herz, M. A., Fuchs, A., Reifer, D., Fuller, J., Yanovsky, M. J., et al. (2014). A chloroplast retrograde signal regulates nuclear alternative splicing. Science 344, 427–430. doi: 10.1126/science.1250322
Pfannschmidt, T., and Yang, C. (2012). The hidden function of photosynthesis: a sensing system for environmental conditions that regulates plant acclimation responses. Protoplasma 249(Suppl. 2), S125–S136. doi: 10.1007/s00709-012-0398-2
Pieterse, C. M., Leon-Reyes, A., Van Der Ent, S., and Van Wees, S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi: 10.1038/nchembio.164
Pogson, B. J., Woo, N. S., Forster, B., and Small, I. D. (2008). Plastid signalling to the nucleus and beyond. Trends Plant Sci. 13, 602–609. doi: 10.1016/j.tplants.2008.08.008
Ponce De Leon, I., Schmelz, E. A., Gaggero, C., Castro, A., Alvarez, A., and Montesano, M. (2012). Physcomitrella patens activates reinforcement of the cell wall, programmed cell death and accumulation of evolutionary conserved defence signals, such as salicylic acid and 12-oxo-phytodienoic acid, but not jasmonic acid, upon Botrytis cinerea infection. Mol. Plant Pathol. 13, 960–974. doi: 10.1111/j.1364-3703.2012.00806.x
Porfirova, S., Bergmuller, E., Tropf, S., Lemke, R., and Dormann, P. (2002). Isolation of an Arabidopsis mutant lacking vitamin E and identification of a cyclase essential for all tocopherol biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 99, 12495–12500. doi: 10.1073/pnas.182330899
Provencher, L. M., Miao, L., Sinha, N., and Lucas, W. J. (2001). Sucrose export defective1 encodes a novel protein implicated in chloroplast-to-nucleus signaling. Plant Cell 13, 1127–1141. doi: 10.1105/tpc.13.5.1127
Puyaubert, J., Fares, A., Reze, N., Peltier, J. B., and Baudouin, E. (2014). Identification of endogenously S-nitrosylated proteins in Arabidopsis plantlets: effect of cold stress on cysteine nitrosylation level. Plant Sci. 21, 150–156. doi: 10.1016/j.plantsci.2013.10.014
Race, H. L., Herrmann, R. G., and Martin, W. (1999). Why have organelles retained genomes? Trends Genet. 15, 364–370. doi: 10.1016/S0168-9525(99)01766-7
Rairdan, G. J., Donofrio, N. M., and Delaney, T. P. (2001). Salicylic acid and NIM1/NPR1-independent gene induction by incompatible Peronospora parasitica in Arabidopsis. Mol. Plant Microbe Interact. 14, 1235–1246. doi: 10.1094/MPMI.2001.14.10.1235
Ramel, F., Birtic, S., Ginies, C., Soubigou-Taconnat, L., Triantaphylides, C., and Havaux, M. (2012). Carotenoid oxidation products are stress signals that mediate gene responses to singlet oxygen in plants. Proc. Natl. Acad. Sci. U.S.A. 109, 5535–5540. doi: 10.1073/pnas.1115982109
Ramel, F., Mialoundama, A. S., and Havaux, M. (2013). Nonenzymic carotenoid oxidation and photooxidative stress signalling in plants. J. Exp. Bot. 64, 799–805. doi: 10.1093/jxb/ers223
Raven, J. A., and Allen, J. F. (2003). Genomics and chloroplast evolution: what did cyanobacteria do for plants? Genome Biol. 4, 209. doi: 10.1186/gb-2003-4-3-209
Reis, S. L., Mantello, A. G., Rossete, E. A. G., Cardoso, A. M., and Beleboni, R. O. (2014). Insecticidal and repellent activity of typical monoterpenes from plant essential oils against Callosobruchus maculatus. BMC Proc. 8:115. doi: 10.1186/1753-6561-8-S4-P115
Renaudin, S., and Capdepon, M. (1977). Association of the endoplasmic reticulum and the plastids in Tozzia aplina L. scale leaves. J. Ultrastruct. Res. 61, 303–308. doi: 10.1016/S0022-5320(77)80055-5
Rhoads, D. M. (2011). “Plant mitochondrial retrograde regulation,” in Plant Mitochondria, ed. F. Kempken (New York, NY: Springer), 411–437. doi: 10.1007/978-0-387-89781-3_16
Rivas-San Vicente, M., and Plasencia, J. (2011). Salicylic acid beyond defence: its role in plant growth and development. J. Exp. Bot. 62, 3321–3338. doi: 10.1093/jxb/err031
Robert-Seilaniantz, A., Grant, M., and Jones, J. D. (2011). Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu. Rev. Phytopathol. 49, 317–343. doi: 10.1146/annurev-phyto-073009-114447
Rodriguez, A., San Andres, V., Cervera, M., Redondo, A., Alquezar, B., Shimada, T., et al. (2011). The monoterpene limonene in orange peels attracts pests and microorganisms. Plant Signal. Behav. 6, 1820–1823. doi: 10.4161/psb.6.11.16980
Rodriguez-Gacio Mdel, C., Matilla-Vazquez, M. A., and Matilla, A. J. (2009). Seed dormancy and ABA signaling: the breakthrough goes on. Plant Signal. Behav. 4, 1035–1049. doi: 10.4161/psb.4.11.9902
Rodriguez-Herva, J. J., Gonzalez-Melendi, P., Cuartas-Lanza, R., Antunez-Lamas, M., Rio-Alvarez, I., Li, Z., et al. (2012). A bacterial cysteine protease effector protein interferes with photosynthesis to suppress plant innate immune responses. Cell. Microbiol. 14, 669–681. doi: 10.1111/j.1462-5822.2012.01749.x
Romero-Puertas, M. C., Campostrini, N., Matte, A., Righetti, P. G., Perazzolli, M., Zolla, L., et al. (2008). Proteomic analysis of S-nitrosylated proteins in Arabidopsis thaliana undergoing hypersensitive response. Proteomics 8, 1459–1469. doi: 10.1002/pmic.200700536
Russin, W. A., Evert, R. F., Vanderveer, P. J., Sharkey, T. D., and Briggs, S. P. (1996). Modification of a specific class of plasmodesmata and loss of sucrose export ability in the sucrose export defective1 maize mutant. Plant Cell 8, 645–658. doi: 10.1105/tpc.8.4.645
Ruyter-Spira, C., Al-Babili, S., Van Der Krol, S., and Bouwmeester, H. (2013). The biology of strigolactones. Trends Plant Sci. 18, 72–83. doi: 10.1016/j.tplants.2012.10.003
Ryals, J. A., Neuenschwander, U. H., Willits, M. G., Molina, A., Steiner, H. Y., and Hunt, M. D. (1996). Systemic acquired resistance. Plant Cell 8, 1809–1819. doi: 10.1105/tpc.8.10.1809
Sandalio, L. M., and Romero-Puertas, M. C. (2015). Peroxisomes sense and respond to environmental cues by regulating ROS and RNS signalling networks. Ann. Bot. doi: 10.1093/aob/mcv074 [Epub ahead of print].
Sanz, L., Albertos, P., Mateos, I., Sanchez-Vicente, I., Lechon, T., Fernandez-Marcos, M., et al. (2015). Nitric oxide (NO) and phytohormones crosstalk during early plant development. J. Exp. Biol. 66, 2857–2868. doi: 10.1093/jxb/erv213
Sattarzadeh, A., Krahmer, J., Germain, A. D., and Hanson, M. R. (2009). A myosin XI tail domain homologous to the yeast myosin vacuole-binding domain interacts with plastids and stromules in Nicotiana benthamiana. Mol. Plant 2, 1351–1358. doi: 10.1093/mp/ssp094
Scala, A. (2013). Green leaf volatiles: a plant’s multifunctional weapon against herbivores and pathogens. Int. J. Mol. Sci. 14, 17781–17811. doi: 10.3390/ijms140917781
Scalschi, L., Sanmartin, M., Camanes, G., Troncho, P., Sanchez-Serrano, J. J., Garcia-Agustin, P., et al. (2015). Silencing of OPR3 in tomato reveals the role of OPDA in callose deposition during the activation of defense responses against Botrytis cinerea. Plant J. 81, 304–315. doi: 10.1111/tpj.12728
Schattat, M., Barton, K., Baudisch, B., Klosgen, R. B., and Mathur, J. (2011a). Plastid stromule branching coincides with contiguous endoplasmic reticulum dynamics. Plant Physiol. 155, 1667–1677. doi: 10.1104/pp.110.170480
Schattat, M., Barton, K., and Mathur, J. (2011b). Correlated behavior implicates stromules in increasing the interactive surface between plastids and ER tubules. Plant Signal. Behav. 6, 715–718. doi: 10.4161/psb.6.5.15085
Schattat, M. H., Barton, K. A., and Mathur, J. (2015). The myth of interconnected plastids and related phenomena. Protoplasma 252, 359–371. doi: 10.1007/s00709-014-0666-4
Schattat, M. H., Griffiths, S., Mathur, N., Barton, K., Wozny, M. R., Dunn, N., et al. (2012). Differential coloring reveals that plastids do not form networks for exchanging macromolecules. Plant Cell 24, 1465–1477. doi: 10.1105/tpc.111.095398
Schlicke, H., Hartwig, A. S., Firtzlaff, V., Richter, A. S., Glasser, C., Maier, K., et al. (2014). Induced deactivation of genes encoding chlorophyll biosynthesis enzymes disentangles tetrapyrrole-mediated retrograde signaling. Mol. Plant 7, 1211–1227. doi: 10.1093/mp/ssu034
Schulz, M., Kussmann, P., Knop, M., Kriegs, B., Gresens, F., Eichert, T., et al. (2007). Allelopathic monoterpenes interfere with Arabidopsis thaliana cuticular waxes and enhance transpiration. Plant Signal. Behav. 2, 231–239. doi: 10.4161/psb.2.4.4469
Seybold, S. J. (2006). Pine monoterpenes and pine bark beetles: a marriage of convenience for defense and chemical communications. Phytochem. Rev. 5, 143–178. doi: 10.1007/s11101-006-9002-8
Shah, J. (2003). The salicylic acid loop in plant defense. Curr. Opin. Plant Biol. 6, 365–371. doi: 10.1016/S1369-5266(03)00058-X
Shah, J. (2009). Plants under attack: systemic signals in defence. Curr. Opin. Plant Biol. 12, 459–464. doi: 10.1016/j.pbi.2009.05.011
Shen, Y. Y., Wang, X. F., Wu, F. Q., Du, S. Y., Cao, Z., Shang, Y., et al. (2006). The Mg-chelatase H subunit is an abscisic acid receptor. Nature 443, 823–826. doi: 10.1038/nature05176
Shi, Y., Chen, J., Hong, X., Chen, J., and Adams, M. J. (2007). A potyvirus P1 protein interacts with the rieske Fe/S protein of its host. Mol. Plant Pathol. 8, 785–790. doi: 10.1111/j.1364-3703.2007.00426.x
Shulaev, V., Silverman, P., and Raskin, I. (1997). Airborne signalling by methyl salicylate in plant pathogen resistance. Nature 385, 718–721. doi: 10.1038/385718a0
Soosaar, J. L., Burch-Smith, T. M., and Dinesh-Kumar, S. P. (2005). Mechanisms of plant resistance to viruses. Nat. Rev. Microbiol. 3, 789–798. doi: 10.1038/nrmicro1239
Spoel, S. H., Koornneef, A., Claessens, S. M., Korzelius, J. P., Van Pelt, J. A., Mueller, M. J., et al. (2003). NPR1 modulates cross-talk between salicylate– and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell 15, 760–770. doi: 10.1105/tpc.009159
Stegemann, S., and Bock, R. (2009). Exchange of genetic material between cells in plant tissue grafts. Science 324, 649–651. doi: 10.1126/science.1170397
Stegemann, S., Keuthe, M., Greiner, S., and Bock, R. (2012). Horizontal transfer of chloroplast genomes between plant species. Proc. Natl. Acad. Sci. U.S.A. 109, 2434–2438. doi: 10.1073/pnas.1114076109
Stintzi, A., Weber, H., Reymond, P., Browse, J., and Farmer, E. E. (2001). Plant defense in the absence of jasmonic acid: the role of cyclopentenones. Proc. Natl. Acad. Sci. U.S.A. 98, 12837–12842. doi: 10.1073/pnas.211311098
Stonebloom, S., Brunkard, J. O., Cheung, A. C., Jiang, K., Feldman, L., and Zambryski, P. (2012). Redox states of plastids and mitochondria differentially regulate intercellular transport via plasmodesmata. Plant Physiol. 158, 190–199. doi: 10.1104/pp.111.186130
Stonebloom, S., Burch-Smith, T., Kim, I., Meinke, D., Mindrinos, M., and Zambryski, P. (2009). Loss of the plant DEAD-box protein ISE1 leads to defective mitochondria and increased cell-to-cell transport via plasmodesmata. Proc. Natl. Acad. Sci. U.S.A. 106, 17229–17234. doi: 10.1073/pnas.0909229106
Strand, A., Asami, T., Alonso, J., Ecker, J. R., and Chory, J. (2003). Chloroplast to nucleus communication triggered by accumulation of Mg-protoporphyrinIX. Nature 421, 79–83. doi: 10.1038/nature01204
Stuehr, D. J. (2004). Enzymes of the L-arginine to nitric oxide pathway. J. Nutr. 134, 2748S–2751S; discussion 2765S–2767S.
Stumpe, M., Gobel, C., Faltin, B., Beike, A. K., Hause, B., Himmelsbach, K., et al. (2010). The moss Physcomitrella patens contains cyclopentenones but no jasmonates: mutations in allene oxide cyclase lead to reduced fertility and altered sporophyte morphology. New Phytol. 188, 740–749. doi: 10.1111/j.1469-8137.2010.03406.x
Sullivan, J. A., and Gray, J. C. (1999). Plastid translation is required for the expression of nuclear photosynthesis genes in the dark and in roots of the pea lip1 mutant. Plant Cell 11, 901–910. doi: 10.2307/3870823
Sun, X., Feng, P., Xu, X., Guo, H., Ma, J., Chi, W., et al. (2011). A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat. Commun. 2, 477. doi: 10.1038/ncomms1486
Surpin, M., Larkin, R. M., and Chory, J. (2002). Signal transduction between the chloroplast and the nucleus. Plant Cell 14(Suppl.), S327–S338.
Susek, R. E., Ausubel, F. M., and Chory, J. (1993). Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74, 787–799. doi: 10.1016/0092-8674(93)90459-4
Tan, X., Wang, Q., Tian, B., Zhang, H., Lu, D., and Zhou, J. (2011). A Brassica napus lipase locates at the membrane contact sites involved in chloroplast development. PLoS ONE 6:e26831. doi: 10.1371/journal.pone.0026831
Telfer, A. (2014). Singlet oxygen production by PSII under light stress: mechanism, detection and the protective role of beta-carotene. Plant Cell Physiol. 55, 1216–1223. doi: 10.1093/pcp/pcu040
Tewari, R. K., Prommer, J., and Watanabe, M. (2013). Endogenous nitric oxide generation in protoplast chloroplasts. Plant Cell Rep. 32, 31–44. doi: 10.1007/s00299-012-1338-5
Thompson, A. M. (1992). The oxidizing capacity of the earth’s atmosphere: probable past and future changes. Science 256, 1157–1165. doi: 10.1126/science.256.5060.1157
Thyssen, G., Svab, Z., and Maliga, P. (2012). Cell-to-cell movement of plastids in plants. Proc. Natl. Acad. Sci. U.S.A. 109, 2439–2443. doi: 10.1073/pnas.1114297109
Tiller, N., and Bock, R. (2014). The translational apparatus of plastids and its role in plant development. Mol. Plant 7, 1105–1120. doi: 10.1093/mp/ssu022
Toshoji, H., Katsumata, T., Takusagawa, M., Yusa, Y., and Sakai, A. (2012). Effects of chloroplast dysfunction on mitochondria: white sectors in variegated leaves have higher mitochondrial DNA levels and lower dark respiration rates than green sectors. Protoplasma 249, 805–817. doi: 10.1007/s00709-011-0325-y
Trotta, A., Rahikainen, M., Konert, G., Finazzi, G., and Kangasjarvi, S. (2014). Signalling crosstalk in light stress and immune reactions in plants. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130235. doi: 10.1098/rstb.2013.0235
Turner, J. G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14(Suppl.), S153–S164.
Tuteja, N. (2007). Abscisic Acid and abiotic stress signaling. Plant Signal. Behav. 2, 135–138. doi: 10.4161/psb.2.3.4156
Uquillas, C., Letelier, I., Blanco, F., Jordana, X., and Holuigue, L. (2004). NPR1-independent activation of immediate early salicylic acid-responsive genes in Arabidopsis. Mol. Plant Microbe Interact. 17, 34–42. doi: 10.1094/MPMI.2004.17.1.34
Vandenabeele, S., Vanderauwera, S., Vuylsteke, M., Rombauts, S., Langebartels, C., Seidlitz, H. K., et al. (2004). Catalase deficiency drastically affects gene expression induced by high light in Arabidopsis thaliana. Plant J. 39, 45–58. doi: 10.1111/j.1365-313X.2004.02105.x
Vanderauwera, S., Zimmermann, P., Rombauts, S., Vandenabeele, S., Langebartels, C., Gruissem, W., et al. (2005). Genome-wide analysis of hydrogen peroxide-regulated gene expression in Arabidopsis reveals a high light-induced transcriptional cluster involved in anthocyanin biosynthesis. Plant Physiol. 139, 806–821. doi: 10.1104/pp.105.065896
Van Norman, J. M., Zhang, J., Cazzonelli, C. I., Pogson, B. J., Harrison, P. J., Bugg, T. D., et al. (2014). Periodic root branching in Arabidopsis requires synthesis of an uncharacterized carotenoid derivative. Proc. Natl. Acad. Sci. U.S.A. 111, E1300–E1309. doi: 10.1073/pnas.1403016111
Velikova, V., Sharkey, T. D., and Loreto, F. (2012). Stabilization of thylakoid membranes in isoprene-emitting plants reduces formation of reactive oxygen species. Plant Signal. Behav. 7, 139–141. doi: 10.4161/psb.7.1.18521
Vladkova, R., Dobrikova, A. G., Singh, R., Misra, A. N., and Apostolova, E. (2011). Photoelectron transport ability of chloroplast thylakoid membranes treated with NO donor SNP: changes in flash oxygen evolution and chlorophyll fluorescence. Nitric Oxide 24, 84–90. doi: 10.1016/j.niox.2010.12.003
Vlot, A. C., Dempsey, D. A., and Klessig, D. F. (2009). Salicylic Acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47, 177–206. doi: 10.1146/annurev.phyto.050908.135202
Wachter, A., Wolf, S., Steininger, H., Bogs, J., and Rausch, T. (2005). Differential targeting of GSH1 and GSH2 is achieved by multiple transcription initiation: implications for the compartmentation of glutathione biosynthesis in the Brassicaceae. Plant J. 41, 15–30. doi: 10.1111/j.1365-313X.2004.02269.x
Wagner, D., Przybyla, D., Op Den Camp, R., Kim, C., Landgraf, F., Lee, K. P., et al. (2004). The genetic basis of singlet oxygen-induced stress responses of Arabidopsis thaliana. Science 306, 1183–1185. doi: 10.1126/science.1103178
Wang, W., Wen, Y., Berkey, R., and Xiao, S. (2009). Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal Haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 21, 2898–2913. doi: 10.1105/tpc.109.067587
Wang, W., Zhang, Y., Wen, Y., Berkey, R., Ma, X., Pan, Z., et al. (2013a). A comprehensive mutational analysis of the Arabidopsis resistance protein RPW8.2 reveals key amino acids for defense activation and protein targeting. Plant Cell 25, 4242–4261. doi: 10.1105/tpc.113.117226
Wang, X., Sager, R., Cui, W., Zhang, C., Lu, H., and Lee, J. Y. (2013b). Salicylic acid regulates Plasmodesmata closure during innate immune responses in Arabidopsis. Plant Cell 25, 2315–2329. doi: 10.1105/tpc.113.110676
Wang, X. B., Wu, Q., Ito, T., Cillo, F., Li, W. X., Chen, X., et al. (2010). RNAi-mediated viral immunity requires amplification of virus-derived siRNAs in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 107, 484–489. doi: 10.1073/pnas.0904086107
Wang, Z., and Benning, C. (2012). Chloroplast lipid synthesis and lipid trafficking through ER-plastid membrane contact sites. Biochem. Soc. Trans. 40, 457–463. doi: 10.1042/BST20110752
War, A. R., Sharma, H. C., Paulraj, M. G., War, M. Y., and Ignacimuthu, S. (2011). Herbivore induced plant volatiles: their role in plant defense for pest management. Plant Signal. Behav. 6, 1973–1978. doi: 10.4161/psb.6.12.18053
Wasternack, C. (2014). Perception, signaling and cross-talk of jasmonates and the seminal contributions of the Daoxin Xie’s lab and the Chuanyou Li’s lab. Plant Cell Rep. 33, 707–718. doi: 10.1007/s00299-014-1608-5
Wasternack, C., and Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 111, 1021–1058. doi: 10.1093/aob/mct067
Waters, M. T., Fray, R. G., and Pyke, K. A. (2004). Stromule formation is dependent upon plastid size, plastid differentiation status and the density of plastids within the cell. Plant J. 39, 655–667. doi: 10.1111/j.1365-313X.2004.02164.x
Weber, H., Vick, B. A., and Farmer, E. E. (1997). Dinor-oxo-phytodienoic acid: a new hexadecanoid signal in the jasmonate family. Proc. Natl. Acad. Sci. U.S.A. 94, 10473–10478. doi: 10.1073/pnas.94.19.10473
Wei, T., Huang, T. S., Mcneil, J., Laliberte, J. F., Hong, J., Nelson, R. S., et al. (2010). Sequential recruitment of the endoplasmic reticulum and chloroplasts for plant potyvirus replication. J. Virol. 84, 799–809. doi: 10.1128/JVI.01824-09
Wendehenne, D., Durner, J., and Klessig, D. F. (2004). Nitric oxide: a new player in plant signalling and defence responses. Curr. Opin. Plant Biol. 7, 449–455. doi: 10.1016/j.pbi.2004.04.002
Wensuo, J., and Zhang, J. (2008). Stomatal movements and long-distabce signaling in plants. Plant Signal. Behav. 3, 772–777. doi: 10.4161/psb.3.10.6294
Westlake, T. J., Ricci, W. A., Popescu, G. V., and Popescu, S. C. (2015). Dimerization and thiol sensitivity of the salicylic acid binding thimet oligopeptidases TOP1 and TOP2 define their functions in redox-sensitive cellular pathways. Front. Plant Sci. 6:327. doi: 10.3389/fpls.2015.00327
Westwood, J. H., Mccann, L., Naish, M., Dixon, H., Murphy, A. M., Stancombe, M. A., et al. (2013). A viral RNA silencing suppressor interferes with abscisic acid-mediated signalling and induces drought tolerance in Arabidopsis thaliana. Mol. Plant Pathol. 14, 158–170. doi: 10.1111/j.1364-3703.2012.00840.x
Wildermuth, M. C., Dewdney, J., Wu, G., and Ausubel, F. M. (2001). Isochorismate synthase is required to synthesize salicylic acid for plant defence. Nature 414, 562–565. doi: 10.1038/35107108
Woodson, J. D., and Chory, J. (2008). Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 9, 383–395. doi: 10.1038/nrg2348
Woodson, J. D., and Chory, J. (2012). Organelle signaling: how stressed chloroplasts communicate with the nucleus. Curr. Biol. 22, R690–R692. doi: 10.1016/j.cub.2012.07.028
Woodson, J. D., Perez-Ruiz, J. M., and Chory, J. (2011). Heme synthesis by plastid ferrochelatase I regulates nuclear gene expression in plants. Curr. Biol. 21, 897–903. doi: 10.1016/j.cub.2011.04.004
Wu, Y., Zhang, D., Chu, J. Y., Boyle, P., Wang, Y., Brindle, I. D., et al. (2012). The Arabidopsis NPR1 protein is a receptor for the plant defense hormone salicylic acid. Cell Rep. 1, 639–647. doi: 10.1016/j.celrep.2012.05.008
Xiao, Y., Savchenko, T., Baidoo, E. E., Chehab, W. E., Hayden, D. M., Tolstikov, V., et al. (2012). Retrograde signaling by the plastidial metabolite MEcPP regulates expression of nuclear stress-response genes. Cell 149, 1525–1535. doi: 10.1016/j.cell.2012.04.038
Xie, Z., Fan, B., Chen, C., and Chen, Z. (2001). An important role of an inducible RNA-dependent RNA polymerase in plant antiviral defense. Proc. Natl. Acad. Sci. U.S.A. 98, 6516–6521. doi: 10.1073/pnas.111440998
Xu, C., Moellering, E. R., Fan, J. B., and Benning, C. (2008). Mutation of a mitochondrial outer membrane protein affects chloroplast lipid biosynthesis. Plant J. 54, 163–175. doi: 10.1111/j.1365-313X.2008.03417.x
Xu, L., Carrie, C., Law, S. R., Murcha, M. W., and Whelan, J. (2013a). Acquisition, conservation, and loss of dual-targeted proteins in land plants. Plant Physiol. 161, 644–662. doi: 10.1104/pp.112.210997
Xu, L., Law, S. R., Murcha, M. W., Whelan, J., and Carrie, C. (2013b). The dual targeting ability of type II NAD(P)H dehydrogenases arose early in land plant evolution. BMC Plant Biol. 13:100. doi: 10.1186/1471-2229-13-100
Yamamoto, Y., Ohshika, J., Takahashi, T., Ishizaki, K., Kohchi, T., Matusuura, H., et al. (2015). Functional analysis of allene oxide cyclase, MpAOC, in the liverwort Marchantia polymorpha. Phytochemistry 116, 48–56. doi: 10.1016/j.phytochem.2015.03.008
Yamane, K., Mitsuya, S., Taniguchi, M., and Miyake, H. (2012). Salt-induced chloroplast protrusion is the process of exclusion of ribulose-1,5-bisphosphate carboxylase/oxygenase from chloroplasts into cytoplasm in leaves of rice. Plant Cell Environ. 35, 1663–1671. doi: 10.1111/j.1365-3040.2012.02516.x
Yan, S., and Dong, X. (2014). Perception of the plant immune signal salicylic acid. Curr. Opin. Plant Biol. 20, 64–68. doi: 10.1016/j.pbi.2014.04.006
Yoon, H. S., Hackett, J. D., Ciniglia, C., Pinto, G., and Bhattacharya, D. (2004). A molecular timeline for the origin of photosynthetic eukaryotes. Mol. Biol. Evol. 21, 809–818. doi: 10.1093/molbev/msh075
Yoshida, K., and Noguchi, K. (2011). “Interaction between chloroplasts and mitochondria: activity, function and regulation of the mitochondrial respiratory system during photosynthesis,” in Plant Mitochondria, ed. F. Kempken (New York, NY: Springer), 383–409. doi: 10.1007/978-0-387-89781-3_15
Yoshida, R., Sato, T., Kanno, A., and Kameya, T. (1998). Streptomycin mimics the cool temperature response in rice plants. J. Exp. Bot. 49, 221–227. doi: 10.1093/jxb/49.319.221
Yoshida, T., Mogami, J., and Yamaguchi-Shinozaki, K. (2015). Omics approaches toward defining the comprehensive abscisic acid signaling network in plants. Plant Cell Physiol. 56, 1043–1052. doi: 10.1093/pcp/pcv060
Yu, D., Fan, B., Macfarlane, S. A., and Chen, Z. (2003). Analysis of the involvement of an inducible Arabidopsis RNA-dependent RNA polymerase in antiviral defense. Mol. Plant Microbe Interact. 16, 206–216. doi: 10.1094/MPMI.2003.16.3.206
Yu, H. D., Yang, X. F., Chen, S. T., Wang, Y. T., Li, J. K., Shen, Q., et al. (2012). Downregulation of chloroplast RPS1 negatively modulates nuclear heat-responsive expression of HsfA2 and its target genes in Arabidopsis. PLoS Genet 8:e1002669. doi: 10.1371/journal.pgen.1002669
Zaninotto, F., La Camera, S., Polverari, A., and Delledonne, M. (2006). Cross talk between reactive nitrogen and oxygen species during the hypersensitive disease resistance response. Plant Physiol. 141, 379–383. doi: 10.1104/pp.106.078857
Zebelo, S. A., Matsui, K., Ozawa, R., and Maffei, M. E. (2012). Plasma membrane potential depolarization and cytosolic calcium flux are early events involved in tomato (Solanum lycopersicon) plant-to-plant communication. Plant Sci. 196, 93–100. doi: 10.1016/j.plantsci.2012.08.006
Zechmann, B. (2014). Compartment-specific importance of glutathione during abiotic and biotic stress. Front. Plant Sci. 5:566. doi: 10.3389/fpls.2014.00566
Zhang, Y., Fan, W., Kinkema, M., Li, X., and Dong, X. (1999). Interaction of NPR1 with basic leucine zipper protein transcription factors that bind sequences required for salicylic acid induction of the PR-1 gene. Proc. Natl. Acad. Sci. U.S.A. 96, 6523–6528. doi: 10.1073/pnas.96.11.6523
Zhang, Z. W., Yuan, S., Feng, H., Xu, F., Cheng, J., Shang, J., et al. (2011). Transient accumulation of Mg-protoporphyrin IX regulates expression of PhANGs – New evidence for the signaling role of tetrapyrroles in mature Arabidopsis plants. J. Plant Physiol. 168, 714–721. doi: 10.1016/j.jplph.2010.10.016
Zhao, J., Liu, Q., Zhang, H., Jia, Q., Hong, Y., and Liu, Y. (2013). The rubisco small subunit is involved in tobamovirus movement and Tm-2(2)-mediated extreme resistance. Plant Physiol. 161, 374–383. doi: 10.1104/pp.112.209213
Zhou, J., Jia, F., Shao, S., Zhang, H., Li, G., Xia, X., et al. (2015). Involvement of nitric oxide in the jasmonate-dependent basal defense against root-knot nematode in tomato plants. Front. Plant Sci. 6:193. doi: 10.3389/fpls.2015.00193
Zhou, J. M., Trifa, Y., Silva, H., Pontier, D., Lam, E., Shah, J., et al. (2000). NPR1 differentially interacts with members of the TGA/OBF family of transcription factors that bind an element of the PR-1 gene required for induction by salicylic acid. Mol. Plant Microbe Interact. 13, 191–202. doi: 10.1094/MPMI.2000.13.2.191
Keywords: retrograde signaling, plastid signaling, redox, phytohormones, plasmodesmata, cell wall, stromules, stress responses
Citation: Bobik K and Burch-Smith TM (2015) Chloroplast signaling within, between and beyond cells. Front. Plant Sci. 6:781. doi: 10.3389/fpls.2015.00781
Received: 31 July 2015; Accepted: 10 September 2015;
Published: 06 October 2015.
Edited by:
Steven James Burgess, University of Cambridge, UKReviewed by:
Taras P. Pasternak, Albert-Ludwigs-Universität Freiburg, GermanyThomas Pfannschmidt, Joseph Fourier University, France
Copyright © 2015 Bobik and Burch-Smith. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tessa M. Burch-Smith, Department of Biochemistry, Cellular and Molecular Biology, University of Tennessee, 1414 Cumberland Avenue, M407 Walters Life Science, Knoxville, TN 37932, USA, dGJ1cmNoc21AdXRrLmVkdQ==
 Krzysztof Bobik
Krzysztof Bobik Tessa M. Burch-Smith
Tessa M. Burch-Smith