- 1Molecular Plant Biology and Biotechnology Division, Central Institute of Medicinal and Aromatic Plants Research Centre, Bangalore, India
- 2Plant Genetic Engineering Laboratory, Department of Biotechnology, Bharathiar University, Coimbatore, India
Epigenetic silencing is a natural phenomenon in which the expression of genes is regulated through modifications of DNA, RNA, or histone proteins. It is a mechanism for defending host genomes against the effects of transposable elements and viral infection, and acts as a modulator of expression of duplicated gene family members and as a silencer of transgenes. A major breakthrough in understanding the mechanism of epigenetic silencing was the discovery of silencing in transgenic tobacco plants due to the interaction between two homologous promoters. The molecular mechanism of epigenetic mechanism is highly complicated and it is not completely understood yet. Two different molecular routes have been proposed for this, that is, transcriptional gene silencing, which is associated with heavy methylation of promoter regions and blocks the transcription of transgenes, and post-transcriptional gene silencing (PTGS), the basic mechanism is degradation of the cytosolic mRNA of transgenes or endogenous genes. Undesired transgene silencing is of major concern in the transgenic technologies used in crop improvement. A complete understanding of this phenomenon will be very useful for transgenic applications, where silencing of specific genes is required. The current status of epigenetic silencing in transgenic technology is discussed and summarized in this mini-review.
Introduction
Conventionally closely related species are easier to breed than inter species/genus due to compatibility issues, and this is considered a major limitation. Transgenic technologies have allowed gene transfer to completely unrelated organisms. All these advances have increased the global transgenic plant cultivation to 181 million hectares (James, 2014). Transgenic plants with stacked genes are gaining more importance lately. Here, different genes are expressed in one transgenic plant from a single transformation event, or in consecutive steps either by re-transformation or by conventional genetic crosses involving different transgenic lines expressing a single transgenic event (Dietz-Pfeilstetter, 2010). To date, diverse traits such as disease resistance, stress tolerance, nutritional improvement, and the use of plants as host systems to produce economically important molecules have been successfully proven (Ahmad et al., 2012). The purpose of gene transfer to plants in all the above cases was to achieve specific desirable traits, where lines that failed to meet expectations are discarded, so that the best performers can be propagated (Kohli et al., 2006, 2010). Initial reports of unforeseen low gene-expression levels or silencing of transgenes were considered failures. Later, those minor glitches emerged as a principal factor elucidating the role of epigenetics in this emerging technology (Meyer et al., 2013).
A major prerequisite for plant expressing a transgene is stability and segregation. Several reports have documented a deviation from the Mendelian segregation ratios in transgenic plants (Shrawat et al., 2007; Weinhold et al., 2013). This revealed the existence of hitherto unknown cellular mechanisms which regulate expression of transgenes. In the last three decades, many reports on transgene instabilities as well as the reasons behind these events were the main focus (Charrier et al., 2000; Graham et al., 2011; Stroud et al., 2013). The explanation for inactivation/silencing of transgene activity was a lack of transcription due to methylation of the promoter along with condensation of chromatin, or degradation of transcripts by different mechanism (Fagard and Vaucheret, 2000; Table 1).
Epigenetics
The British developmental biologist Conrad H. Waddington coined the term “epigenetics”. Epigenetics deals with studies related to interactions of genes and their products, which determine the phenotype of a system (Waddington, 1942). During the course of an organism’s development, cell fate is determined by genes and by other (epigenetic) factors, which underlies the notion of “epigenesis”. Modern biology has redefined as a phenomenon in which a gene’s activity is modulated by modifications of nucleic acids or the physical packaging of the chromatin in which it is embedded.
Two main classes of transgene-silencing phenomena have been reported to date. The first concerns position effects, in which the expression of a foreign gene is negatively regulated by flanking host DNA or chromosomal location (Matzke et al., 2000). The expression of a gene integrated into a region of euchromatin is also influenced by regulatory sequences of host genes (Kohli et al., 2006). Transgene integration into heterochromatic regions also leads to silencing (Grewal and Elgin, 2002).
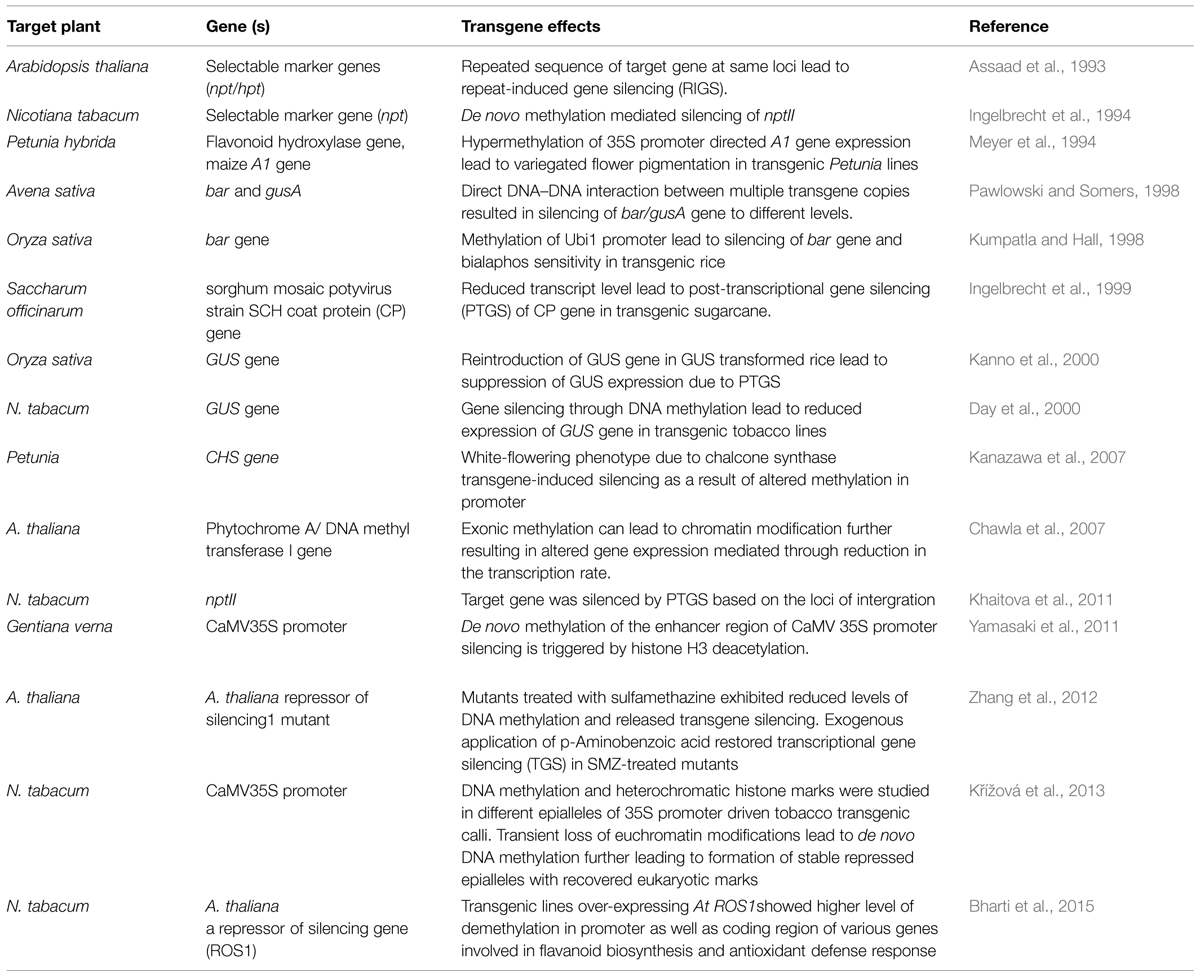
The second class of silencing phenomena is based on epigenetic regulation and is a type of inactivation mechanism that can arise when multiple copies of the same or homologous sequence are introduced in a genome. Since interactions between homologous nucleic acid sequences are responsible for these silencing, it is also called homology-dependent gene silencing (HDGS) (Meyer and Saedler, 1996). Over the years, it has become clear that HDGS occurs through distinct processes, frequent one being involvement of inverted DNA repeats (IRs) and dsRNA. T-DNA integration at the same chromosomal site leads either to ‘head-to-tail’ direct repeats (DR) or to ‘head-to-head’ or ‘tail to tail’ inverted repeats (IR). T-DNAs that are arranged as IRs are often shown to have low basal expression (Mishiba et al., 2005). IRs have the ability to interact with homologous sequences elsewhere in the genome leading to chromatin remodeling. They can also induce a sequence-specific RNA degradation process, possibly via the formation of dsRNAs (Figure 1A).
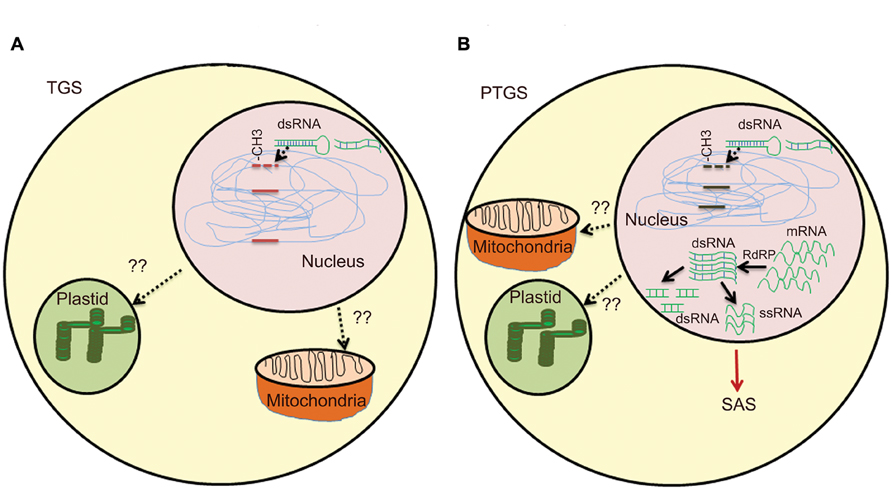
FIGURE 1. Overview of gene silencing in transgenic plants. (A) Transcriptional gene silencing (TGS)- DNA methylation induces dsRNA by endogenous gene or multiple copies of transgenes. Presence of multiple copies of transgene induces formation of dsRNA. Single copy transgene loci could also lead to formation of dsRNA due to high RNA turnover. Methylation of CG, CNG, or CNN region in promoter by different methyltransefrases that leads to TGS. Methylation in heterochromatin region also lead to TGS. T-DNA with transgene integrated as direct or inverted (IR) repeats are inactivated by DNA methylation. Cruciform structures formed by IRs act as substrate for DNA methyltransferases. (B) Post-transcriptional gene silencing (PTGS)- Methylation in coding region and high RNA turn over lead to production of dsRNA, abbrreant RNAs, cRNAs. RdRP uses these aberrant RNAs as templates and convert them into a double-stranded RNA, which is further degraded by different dsRNases yielding small dsRNAs and/or ssRNAs. The ssRNAs and/or dsRNAs act as systemic silencing signals, which are transported all over the plant and trigger PTGS in adjacent cells. SAS in mitochondria and plastids are still under study.
Homology Dependent Gene Silencing
A major breakthrough in understanding epigenetic silencing in transgenic plants was first identified in transgenic tobacco, where interaction between two homologous promoters led to DNA methylation and silencing (Matzke et al., 1989). Two types of HDGS are known based on the stage at which it occurs, called transcriptional gene silencing (TGS), which is coupled with transcription or by promoter modification, and post-transcriptional gene silencing (PTGS), which occurs after the formation of mRNA (Jauvion et al., 2012). In TGS, interacting genes that share homology in promoter regions are highly methylated. PTGS involves sequence-specific transcript turnover in the cytosol, which further requires high homology between interacting genes. Potential factors influencing HDGS are degree of homology between the transgene and endogenous gene, the complexity of the host genome, the genomic position of two transgenes, etc. A transgene locus with a complex structure with multiple scrambled T-DNAs has been reported to have strong silencing activities in tobacco, implicating transgene complexity; and vector DNA also decides the efficiency of HDGS (Fu et al., 2000). Complexity of T-DNA structure and integrated vector sequences have been shown to regulate transgene expression in grapevine (Gambino et al., 2010). An increase of endogenous transcript levels above a critical threshold induces specific degradation of homologous transcripts.
Transcriptional Gene Silencing (TGS)
Transgenes silenced at the transcriptional level acquire metastable epigenetic status that is associated with altered methylation patterns. Transgenes are frequently methylated in cytosine residues that are located within CG, CNN, or CNG sequences. De novo DNA methylation can be highly sequence-specific for a specific transgene (Matzke et al., 2007). Fungi or plants expressing foreign genes also exhibit non-symmetrical methylation leading to silencing of endogenous genes. Factors responsible for non-symmetrical methylation are still obscure. Non-symmetrical methylation patterns are aided by RNA- chromatin mechanism (McGinnis et al., 2006).
Methylation in promoter regions, histones, or in coding regions influence gene expression at both the transcriptional (Huettel et al., 2006) and post transcriptional level (Regulski et al., 2013; Tsuchiya and Eulgem, 2013). Chromatin remodeling is involved in maintenance of silenced status and also in transmission of non-symmetrical methylation patterns (Meyer, 1999). Another interesting fact about TGS in transgenic plants is the association of DNA methylation along with structural changes, as methylated and silenced transgenes were less susceptible to endonucleases, reflecting an increased level of chromatin condensation (Van Blokland et al., 1997). Hence, TGS-based silencing might also involve structural changes similar to heterochromatinization, which could be the cause of these structural changes. The responsiveness of TGS of transgenes in response to environmental change was confirmed (Meyer et al., 1992; Meyer, 2015).
Transcriptional gene silencing can be further divided into two classes:
Transcriptional cis Inactivation
In plants, transgenes integrate into the genome at random positions by illegitimate recombination; hence, copy number, their integration site, and local arrangement differ in each transformation event. Also, an inverse relation between transgene copy number and gene expression suggests that multicopy integration can lead to silencing. Integrated foreign genes can undergo TGS in cis when multicopy T-DNA is integrated at a locus adjacent to hypermethylated regions of the host genome (Mishiba et al., 2005). More rarely, single copy transgene integration at a hypomethylated locus can lead to cis inactivation (Meyer and Heidmann, 1994; Elmayan and Vaucheret, 1996). A maize A1 gene involved in floral pigmentation when overexpressed in Petunia led to silencing of A1; however, it was not silenced when Gerbera dihydroflavonol-4-reductase was over expressed in Petunia suggesting that the transgene also influenced the silencing process. Hence, some degree of difference in DNA composition of the transgene and surrounding host genomic sequences can be recognized by the cellular machinery as foreign non-compatible DNA, leading to specific methylation and silencing (Elomaa et al., 1995). It is believed that cis TGS occurs as a result of pairing between closely associated copies of transgenes or endogenous genes, which leads to the formation of secondary DNA structures which are sites for DNA methylation (Vaucheret and Fagard, 2001). Cytosine methylation at CpG and CpNpG sites of transgene and the 35S promoter were also detected in transgenic grapevine transformed with Grapevine fanleaf virus (GFLV) coat protein gene (Gambino et al., 2010).
Transcriptional Trans-Inactivation
Transcriptional gene silencing can result from unidirectional effects of one transgene on another transgene or homologous endogenous gene. A transgene can be methylated and silenced when it is crossed with a plant in which the homologous gene is in a silenced state (Meyer et al., 1993). De novo methylation of one transgene is mediated by a second transgene under control of the same promoter leading to TGS in trans (Fagard and Vaucheret, 2000). Experiments using dsRNA-containing promoter sequences initiated TGS and subsequently de novo DNA methylation of the corresponding transgene or endogenous gene, implying a role of an RNA intermediate in TGS (Meyer, 2000). Vaucheret and Fagard (2001) reported the role of different genes, including ddm1 and ddm2 in TGS in Arabidopsis transgenic lines. Yamasaki et al. (2011) reported methylation of asymmetric cytosine in the enhancer region of 35S promoter in transgenic gentian.
Post-Transcriptional Gene Silencing
Post-Transcriptional Gene silencing is a condition where transcripts do not accumulate in spite of continuous transcription (Vaucheret et al., 2001). PTGS can silence both transgenes and endogenous genes if both are homologous. An endogenous gene could be switched off, when a plant is transformed with another copy of the same gene. When genes involved in pigmentation, such as chalcone synthase A in Petunia, were overexpressed, many transgenic lines partially or completely lost activity of both transgene and endogenous gene (Napoli et al., 1990; Van der Krol et al., 1990). This was later called ‘co-suppression’, which was a result of degradation of mRNA of both transgene and endogenous gene. Analysis of degradation products in tobacco expressing β-1,3-glucanase revealed that RNAs are first cleaved by endonucleases, which are further degraded by various exonucleases (Van Eldik et al., 1998). Silencing of two endogenous genes in Arabidopsis thaliana was triggered by the antisense and hpRNA transgenes, and silencing in this case was dependent on ploidy level, as it was less pronounced in 4n compared to 2n Arabidopsis. Studies indicated that transgenes were more methylated in 4n than 2n Arabidopsis suggesting transgenes are transcriptionally repressed in 4n plants, thus resulting in reduced expression levels compared to diploid plants (Finn et al., 2011).
Transgene-induced viral resistance, recovery from infection and proteins encoded by viruses that counteract PTGS suggested it as a potential defense response to check viral infections (Brigneti et al., 1998; Kasschau and Carrington, 1998; Dalmay et al., 2000). It is speculated that the concentration of specific RNAs derived from both transgene and endogenous gene is critical to activate PTGS. dsRNAs are one of the potential candidates, as they are formed between RNAs transcribed from IR and gene homologues. dsRNA is used as a template by RNA-directed RNA polymerase (RdRP) and transcription of dsRNA by RdRP would result in antisense RNAs, which ultimately could target complementary transcripts for degradation by dsRNA-specific RNases (Bond and Baulcombe, 2015; Figure 1B).
Post-Transcriptional cis-Inactivation
Post-transcriptional gene silencing cis-inactivation is observed when foreign genes like β-Glucuronidase, neomycin phosphotransferase, etc., were driven under strong 35S promoter (Dehio and Schell, 1994; Ingelbrecht et al., 1994; Elmayan and Vaucheret, 1996). When a 35S promoter with a double enhancer was used, more transformants showed PTGS (Elmayan and Vaucheret, 1996; English et al., 1996). Initially, perceptions about PTGS were driven by higher transcript abundance above a threshold level, which ultimately triggered degradation of transgenic RNA. Later, it was found that the level of transcription was not always found to be significantly higher in silenced plants. The presence of IR at transgene locus of silenced lines was proposed to play a crucial role in cis inactivation (English et al., 1996). In same year, different models for PTGS were proposed considering RNA abundance and IRs (Baulcombe, 1996). Transgene RNA could be specifically degraded when tagged with specific molecules; these tag molecules were later named small complementary RNA (cRNA). RdRP catalyzed synthesis of cRNA using transgene RNA as template (Dougherty and Parks, 1995). They could also be internal fragments generated from transgene RNA by pairing between aberrant mRNA and normal transgene RNA due to the presence of internal sequence complementarily (Metzlaff et al., 1997). cRNA can interact with mRNA forming dsRNA, which are the target for the cellular enzymes like double-strand RNase. DNA-DNA interactions can lead to methylation, which can further interfere with transcription, ultimately producing aberrant RNA. These aberrant RNAs or higher transcript abundance were owing to the use of a strong promoter that triggered methylation of the coding sequence of the respective transgene (Wassenegger et al., 1994). Interestingly, Kanazawa et al. (2007) reported conversion of PTGS to TGS in Petunia transgenic lines as a consequence of the transgene homologous to an endogenous gene in host genome.
Post-Transcriptional Trans-Inactivation
Post-transcriptional gene silencing was originally reported as coordinated silencing of both transgenes as well as endogenous genes, which is generally termed ‘co-suppression’ (Napoli et al., 1990). Since then, several studies revealed transgenes encoding part of, or the entire transcribed sequence of, host genes have been shown to trigger co-suppression of endogenous genes (Depicker and Van Montagu, 1997). By then it was evident from studies in transgenic Petunia lines expressing a chalcone synthase, where efficiency of co-suppression correlated with the strength of the promoter, that there was an effect of transgene dose on co-suppression (Que et al., 1997). Besides, the efficiency of co-suppression is delayed when endogenous host genes are not expressed or when genes are transferred to a mutant devoid of functional gene homologues (Smith et al., 1990; Vaucheret et al., 1997). Hence, it can be concluded that co-suppression cannot be considered as the unidirectional silencing effect of transgenes, rather it is a synergistic phenomenon in which interaction or presence of host genes and transgenes aids aberrant RNA and/or cRNA leading to PTGS.
Systemic Acquired Silencing
A hallmark of PTGS in plants is that it systemically transmitted in a sequence-specific manner known as systemic acquired silencing (SAS). Remarkable and recurrent features in silencing patterns during developmental stages revealed propagation of a silencing message across the plant (Vaucheret et al., 1998; Kalantidis et al., 2008). Co-suppression of endogenous and transgenes of nitrate reductase, nitrite reductase and SAM synthase in tobacco led to chlorotic or necrotic phenotypes (Boerjan et al., 1994; Palauqui et al., 1996). The non-clonal patterns were observed in all transgenic lines silenced for a specific gene and a sequence-specific message was involved in the control of PTGS. Later, grafting experiments revealed that transgene-specific effector molecules were involved in propagation of de novo PTGS over long distances by a phenomenon called SAS (Palauqui et al., 1997). Transgenic tobacco overexpressing A. thaliana AtMYB90 involved in anthocyanin biosynthesis showed siRNA-mediated silencing as a result of SAS (Velten et al., 2012). A SAS PTGS of transgenes in N. benthamiana was initiated in localized regions of the plant when a transgene-homologous DNA was introduced (Voinnet et al., 1998). The silencing signal molecules are degraded RNA, which travels through phloem across cells through plasmodesmata (Kalantidis et al., 2008). The recipient cell can also act as a source for generating secondary signals. It has been reported that sense, antisense, and ill-defined aberrant RNAs can give rise to dsRNA which can transmit signals, ultimately leading to silencing of both transgene and endogenous gene, albeit to different levels (Figure 1B).
Small RNAs as Silencing Signals in Transgenic Plants
RNA was the driving factor for the establishment of DNA methylation patterns (Wassenegger et al., 1994) and acts a signaling agent for inducing silencing. Potato spindle tuber viroid (PSTV) in transgenic tobacco lines led to autonomous viroid RNA replication in the nucleus and induced DNA methylation in the T-DNA (Wassenegger et al., 1994). The evidence from above study clearly indicates the critical role of RNA in initiating de novo DNA methylation at homologous regions. Until then, DNA/RNA hybrids were believed to play a role in generating a target for de novo methylation. chsA co-suppression studies in Petunia led to the identification of mobile RNAs as potential candidates responsible for the induction of co-suppression (Napoli et al., 1990; Van der Krol et al., 1990). The initiation of transgene silencing has been thought to involve the generation of dsRNA. It is still under debate about factors triggering initiation of silencing even in the case of transgenes that lack unusual DNA structures.
In plants, micro RNAs (miRNAs) are produced from hairpin-like precursor RNA, which is essential for biogenesis of trans-acting siRNAs (ta-siRNAs). miRNAs are involved in regulation of gene expression by base-pairing with target RNAs further leading to their cleavage in plants. Physcomitrella patens transgenic lines expressing different levels of artificial miRNA (amiRNA) revealed transcript-dependent silencing of miRNA target. Thus, a crucial regulatory role of miRNAs might be conserved in other plants also, which are under investigation. siRNAs are another class of small RNAs that are involved in epigenetic modification (Miki and Shimamoto, 2008). Endogenous siRNAs can induce DNA methylation at CpG nucleotides leading to chromatin modification and silencing. Human H1 and Arabidopsis 7SL RNA promoters driving GUS specific short hairpin RNA resulted in the efficient silencing of GUS at both transcript and protein level, indicating a significant role of siRNAs in epigenetic regulation. However, transgenes are generally more sensitive against RNA silencing than endogenous genes in plants.
Transgene Silencing as Part of the Host Defense Mechanism?
Silencing cannot be considered as a mechanism that evolved to regulate transgene expression; it is a part of natural plant processes. TGS and PTGS can be considered as host defense responses against ‘foreign invading’ viruses. Hence, transgenes or their products can be equated to cellular invaders triggering defensive reactions leading to silencing of “trans”gene. PTGS recruits cellular components acting against foreign DNA that replicates extra-chromosomally in the nucleus, or RNA in the cytoplasm. A clear connecting link between PTGS and viral resistance was established after the discovery and characterization of various viral proteins that suppress PTGS (Kasschau and Carrington, 1998; Beclin et al., 2002). TGS may use cellular components acting against invading DNA that integrates into the genome. The involvement of DNA methylation can also be considered as a part of cellular defense mechanism against transposable elements. The probable function of dsRNA in initiating methylation can be correlated to retro-elements that produce RNAs with intricate secondary structures.
Strategies to Prevent Transgene Silencing (Depicker et al., 2005)
(1) Selection of transgenic lines with single T-DNA insert
(2) Organelle targeting/transformation
(3) Selection of favorable/unique integration sites
(4) Reactivation of silent transgenes
(5) Use of Scaffold Matrix Attachment Regions in silencing mutant host system to prevent silencing.
Concluding Remarks
The last three decades have seen immense progress and better understanding of epigenetic effects and silencing mechanisms; transgenic technologies have played a pivotal role for these achievements. Common phenomena behind different types of silencing and recent finding of involvement of siRNAs/miRNA continue to inspire efforts of scientific community to formulate comprehensive models, which also explain the silencing mechanism from an evolutionary view point. Our understanding of the influence of various factors on stability of transgene expression is improving rapidly. We cannot control or predict integration of gene into a recipient genome, nor predict the number of copies or integrity of a transgene. Hence, a comprehensive knowledge of underlying mechanisms in integration process and the influence of chromatin remodeling leading to transgene regulation are crucial. Finally, it might be useful to keep in mind that epigenetic silencing was an unexpected phenomenon; it is still hard to foresee overcoming epigenetic related silencing in transgenic system. Nevertheless, transgenic research will continue as a platform to discover new aspects of epigenetic silencing.
Author Contributions
SR and PA prepared the manuscript and RS revised it.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Ahmad, P., Ashraf, M., Younis, M., Hu, X., Kumar, A., Akram, N. A., et al. (2012). Role of transgenic plants in agriculture and biopharming. Biotechnol. Adv. 30, 524–540. doi: 10.1016/j.biotechadv.2011.09.006
Assaad, F. F., Tucker, K. L., and Signer, E. R. (1993). Epigenetic repeat-induced gene silencing (RIGS) in Arabidopsis. Plant Mol. Biol. 22, 1067–1085. doi: 10.1007/BF00028978
Baulcombe, D. C. (1996). RNA as a target and an initiator of posttranscriptional gene silencing in transgenic plants. Plant Mol. Biol. 32, 79–88. doi: 10.1007/BF00039378
Beclin, C., Boutet, S., Waterhouse, P., and Vaucheret, H. (2002). A branched pathway for transgene - induced RNA silencing in plants. Curr. Biol. 12, 684–688. doi: 10.1016/S0960-9822(02)00792-3
Bharti, P., Mahajan, M., Vishwakarma, A. K., Bhardwaj, J., and Yadav, S. K. (2015). AtROS1 overexpression provides evidence for epigenetic regulation of genes encoding enzymes of flavonoid biosynthesis and antioxidant pathways during salt stress in transgenic tobacco. J. Exp. Bot. doi: 10.1093/jxb/erv304 [Epub ahead of print].
Boerjan, W., Bauw, G., Van Montagu, M., and Inze, D. (1994). Distinct phenotypes generated by overexpression and suppression of S-adenosyl-l-methionine reveal developmental patterns of gene silencing in tobacco. Plant Cell 6, 1401–1414. doi: 10.2307/3869977
Bond, D. M., and Baulcombe, D. C. (2015). Epigenetic transitions leading to heritable, RNA-mediated de novo silencing in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 112, 917–922. doi: 10.1073/pnas.1413053112
Brigneti, G., Voinnet, O., Li, W.-X., Ding, S. W., and Baulcombe, D. C. (1998). Viral pathogenicity determinants are suppressors of transgene silencing. EMBO J. 17, 6739–6746. doi: 10.1093/emboj/17.22.6739
Charrier, B., Scollan, C., Ross, S., Zubko, E., and Meyer, P. (2000). Co-silencing of homologous transgenes in tobacco. Mol. Breed. 6, 407–419. doi: 10.1023/A:1009672714835
Chawla, R., Nicholson, S. J., Folta, K. M., and Srivastava, V. (2007). Transgene-induced silencing of Arabidopsis phytochrome A gene via exonic methylation. Plant J. 52, 1105–1118. doi: 10.1111/j.1365-313X.2007.03301.x
Dalmay, T., Hamilton, A., Mueller, E., and Baulcombe, C. D. (2000). Potato virus X amplicons in Arabidopsis mediate genetic and epigenetic gene silencing. Plant Cell 12, 369–380. doi: 10.2307/3870942
Day, C. D., Lee, E., Kobayashi, J., Holappa, L. D., Albert, H., and Ow, D. W. (2000). Transgene integration into the same chromosome location can produce alleles that express at a predictable level, or alleles that are differentially silenced. Genes Dev. 14, 2869–2880. doi: 10.1101/gad.849600
Dehio, C., and Schell, J. (1994). Identification of plant genetic loci involved in a post-transcriptional mechanism for meiotically reversible transgene silencing. Proc. Natl. Acad. Sci. U.S.A. 91, 5538–5542. doi: 10.1073/pnas.91.12.5538
Depicker, A., Sanders, M., and Meyer, P. (2005). “Transgene silencing,” in Plant epigenetics. Oxford Ames, ed. P. Meyer (Iowa: Blackwell Publishing), 1–32. doi: 10.1002/9780470988626.ch1
Depicker, A., and Van Montagu, M. (1997). Posttranscriptional gene silencing in plants. Curr. Opin. Cell Biol. 9, 373–382. doi: 10.1016/S0955-0674(97)80010-5
Dietz-Pfeilstetter, A. (2010). Stability of transgene expression as a challenge for genetic engineering. Plant Sci. 179, 164–167. doi: 10.1016/j.plantsci.2010.04.015
Dougherty, W. G., and Parks, T. D. (1995). Transgenes and gene suppression: telling us something new? Curr. Opin. Cell Biol. 7, 399–405. doi: 10.1016/0955-0674(95)80096-4
Elmayan, T., and Vaucheret, H. (1996). Expression of single copies of a strongly expressed 35S transgene can be silenced post-transcriptionally. Plant J. 9, 787–797. doi: 10.1046/j.1365-313X.1996.9060787.x
Elomaa, P., Helariutta, Y., Griesbach, J. R., Kotilainen, M., Seppanen, P., and Teeri, H. T. (1995). Transgene inactivation in Petunia hybrida is influenced by the properties of the foreign gene. Mol. Genet. Genomics 248, 649–656. doi: 10.1007/BF02191704
English, J. J., Mueller, E., and Baulcombe, D. C. (1996). Suppression of virus accumulation in transgenic plants exhibiting silencing of nuclear genes. Plant Cell 8, 179–188. doi: 10.1105/tpc.8.2.179
Fagard, M., and Vaucheret, H. (2000). (TRANS)GENE SILENCING IN PLANTS: how many mechanisms? Annu. Rev. Plant Physiol. Plant Mol. Biol. 51, 167–194. doi: 10.1146/annurev.arplant.51.1.167
Finn, E. T., Wang, L., Smolilo, D., Smith, A. N., White, R., Chaudhury, A., et al. (2011). Transgene expression and transgene-induced silencing in diploid and autotetraploid Arabidopsis. Genetics 187, 409–423. doi: 10.1534/genetics.110.124370
Fu, X., Duc, L. T., Fontana, S., Bong, B. B., Tinjuangjun, P., Sudhakar, D., et al. (2000). Linear transgene constructs lacking vector backbone sequences generate low-copy-number transgenic plants with simple integration patterns. Transgenic Res. 9, 9–11. doi: 10.1023/A:1008993730505
Gambino, G., Perrone, I., Carra, A., Chitarra, W., Boccacci, P., Torello Marinoni, D., et al. (2010). Transgene silencing in grapevines transformed with GFLV resistance genes: analysis of variable expression of transgene, siRNAs production and cytosine methylation. Transgenic Res. 19, 17–27. doi: 10.1007/s11248-009-9289-5
Graham, M. W., Mudge, S. R., Sternes, P. R., and Birch, R. G. (2011). “Understanding and avoiding transgene silencing,” in Plant Transformation Technologies, eds C. N. Stewart, A. Touraev, V. Citopvsky, and T. Tzfira (West Sussex: Blackwell Publishing). doi: 10.1002/9780470958988.ch12
Grewal, S. I. S., and Elgin, S. C. R. (2002). Heterochromatin: new possibilities for the inheritance of structure. Curr. Opin. Gen. Dev. 12, 178–187. doi: 10.1016/S0959-437X(02)00284-8
Huettel, B., Kanno, T., Daxinger, L., Aufsatz, W., Matzke, A. J. M., and Matzke, M. (2006). Endogenous targets of RNA-directed DNA methylation and Pol IV in Arabidopsis. EMBO J. 25, 2828–2836. doi: 10.1038/sj.emboj.7601150
Ingelbrecht, I., Van Houdt, H., Van Montagu, M., and Depicker, A. (1994). Post-transcriptional silencing of reporter transgenes in tobacco correlates with DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 91, 10502–10506. doi: 10.1073/pnas.91.22.10502
Ingelbrecht, L. I., Irvine, E. J., and Mirkov, T. E. (1999). Posttranscriptional gene silencing in transgenic sugarcane. dissection of homology-dependent virus resistance in a monocot that has a complex polyploid genome. Plant Physiol. 119, 1187–1197. doi: 10.1104/pp.119.4.1187
James, C. (2014). Global Status of Commercialized Biotech/GM Crops. Ithaca, NY, USA: International Service for the Acquisition of Agri-biotech Applications (ISAAA).
Jauvion, V., Rivard, M., Bouteiller, N., Elmayan, T., and Vaucheret, H. (2012). RDR2 partially antagonizes the production of RDR6-dependent siRNA in sense transgene-mediated PTGS. PLoS ONE 7:e29785. doi: 10.1371/journal.pone.0029785
Kalantidis, K., Schumacher, H. K., Alexiadis, T., and Helm, J. M. (2008). RNA silencing movement in plants. Biol. Cell 100, 13–26. doi: 10.1042/BC20070079
Kanazawa, A., O’Dell, M., and Hellens, P. R. (2007). Epigenetic inactivation of chalcone synthase-a transgene transcription in petunia leads to a reversion of the post-transcriptional gene silencing phenotype. Plant Cell Physiol. 48, 638–647. doi: 10.1093/pcp/pcm028
Kanno, T., Naito, S., and Shimamoto, G. (2000). Post-transcriptional gene silencing in cultured rice cells. Plant and Cell Physiol. 41, 321–326. doi: 10.1093/pcp/41.3.321
Kasschau, K. D., and Carrington, J. C. (1998). A counter-defensive strategy of plant viruses: suppression of posttranscriptional gene silencing. Cell 95, 461–470. doi: 10.1016/S0092-8674(00)81614-1
Khaitova, C. L., Fojtova, M., Krizova, K., Lunerova, J., Fulnecek, J., Depicker, A., et al. (2011). Paramutation of tobacco transgenes by small RNA-mediated transcriptional gene silencing. Epigenetics 6, 650–660. doi: 10.4161/epi.6.5.15764
Kohli, A., Melendi, P. G., Abranches, R., Capell, T., and Christou, P. (2006). The quest to understand the basis and mechanisms that control expression of introduced transgenes in crop plants. Plant Signal. Behav. 1, 185–195. doi: 10.4161/psb.1.4.3195
Kohli, A., Miro, B., and Twyman, R. M. (2010). “Transgene integration, expression and stability in plants: strategies for improvements,” in Transgenic Crop Plants: Principles and Development, eds C. Kole, C. H. Michler, A. G. Abbott, and T. C. Hall (Berlin: Springer-Verlag), 201–237. doi: 10.1007/978-3-642-04809-8_7
Křížová, K., Depicker, A., and Kovarik, A. (2013). Epigenetic switches of tobacco transgenes associate with transient redistribution of histone marks in callus culture. Epigenetics 8, 666–676. doi: 10.4161/epi.24613
Kumpatla, S. P., and Hall, T. C. (1998). Recurrent onset of epigenetic silencing in rice harboring a multi-copy transgene. Plant J. 14, 129–135. doi: 10.1046/j.1365-313X.1998.00097.x
Matzke, M., Kanno, T., Huettel, B., Daxinger, L., and Matzke, A. J. (2007). Targets of RNA directed DNA methylation. Curr. Opin. Plant Biol. 10, 512–519. doi: 10.1016/j.pbi.2007.06.007
Matzke, M. A., Mette, M. F., and Matzke, A. J. (2000). Transgene silencing by the host genome defense: implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol. Biol. 43, 401–415. doi: 10.1023/A:1006484806925
Matzke, M. A., Primig, M., Trnovsky, J., and Matzke, A. J. M. (1989). Reversible methylation and inactivation of marker genes in sequentially transformed tobacco plants. EMBO J. 8, 643–649.
McGinnis, K. M., Springer, C., Lin, Y., Carey, C. C., and Chandler, V. (2006). Transcriptionally silenced transgenes in maize are activated by three mutations defective in paramutation. Genetics 173, 1637–1647. doi: 10.1534/genetics.106.058669
Metzlaff, M., O’Dell, M., Cluster, P. D., and Flavell, R. B. (1997). RNA-mediated RNA degradation and chalcone synthase a silencing in petunia. Cell 88, 845–854. doi: 10.1016/S0092-8674(00)81930-3
Meyer, P. (1999). The role of chromatin remodeling in transgene silencing and plant development. In Vitro Cell. Dev. Biol. Plant 35, 29–36. doi: 10.1007/s11627-999-0006-0
Meyer, P. (2000). Transcriptional transgene silencing and chromatin components. Plant Mol. Biol. 43, 221–234. doi: 10.1023/A:1006483428789
Meyer, P. (2015). Epigenetic variation and environmental change. J. Exp. Bot. 66, 3541–3548. doi: 10.1093/jxb/eru502
Meyer, P., and Heidmann, I. (1994). Epigenetic variants of a transgenic petunia line show hypermethylation in transgene DNA: an indication for specific recognition of foreign DNA in transgenic plants. Mol. Gen. Genet. 243, 390–399. doi: 10.1007/BF00280469
Meyer, P., Heidmann, I., and Niedenhof, I. (1993). Differences in DNA-methylation are associated with a paramutation phenomenon in transgenic petunia. Plant J. 4, 89–100. doi: 10.1046/j.1365-313X.1993.04010089.x
Meyer, P., Linn, F., Heidmann, I., Heiner Meyerz, A. H., Niedenhof, I., and Saedler, H. (1992). Endogenous and environmental factors influence 35S promoter methylation of a maize A1 gene construct in transgenic petunia and its colour phenotype. Mol. Gen. Genet. 231, 345–352. doi: 10.1007/BF00292701
Meyer, P., Niedenhof, I., and Lohuis, M. T. (1994). Evidence for cytosine methylation of non-symmetrical sequences in transgenic Petunia hybrid. EMBO J. 13, 2084–2088.
Meyer, P., and Saedler, H. (1996). Homology-dependent gene silencing in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 47, 23–48. doi: 10.1146/annurev.arplant.47.1.23
Meyer, T. D., Mampaey, E., Vlemmix, M., Denil, S., Trooskens, G., Renard, J. P., et al. (2013). Quality evaluation of methyl binding domain based kits for enrichment DNA-methylation sequencing. PLoS ONE 8:e59068. doi: 10.1371/journal.pone.0059068
Miki, D., and Shimamoto, K. (2008). De novo DNA methylation induced by siRNA targeted to endogenous transcribed sequences is genespecific and OsMet1-independent in rice. Plant J. 56, 539–549. doi: 10.1111/j.1365-313X.2008.03624.x
Mishiba, K., Nishihara, M., Nakatsuka, T., Abe, Y., Hirano, H., Yokoi, T., et al. (2005). Consistent transcriptional silencing of 35S-driven transgenes in gentian. Plant J. 44, 541–556. doi: 10.1111/j.1365-313X.2005.02556.x
Napoli, C., Lemieux, C., and Jorgensen, R. (1990). Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous gene in trans. Plant Cell 2, 279–289. doi: 10.2307/3869076
Palauqui, J. C., Elmayan, T., de Borne, F. D., Crete, P., Charles, C., and Vaucheret, H. (1996). Frequencies, timing, and spatial patterns of cosuppression of nitrate reductase and nitrite reductase in transgenic tobacco plants. Plant Physiol. 112, 1447–1456.
Palauqui, J. C., Elmayan, T., Pollien, J. M., and Vaucheret, H. (1997). Systemic acquired silencing: transgene specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 16, 4738–4745. doi: 10.1093/emboj/16.15.4738
Pawlowski, P. W., and Somers, A. D. (1998). Transgenic DNA integrated into the oat genome is frequently interspersed by host DNA. Proc. Natl. Acad. Sci. U.S.A. 95, 12106–12110. doi: 10.1073/pnas.95.21.12106
Que, Q., Wang, H. Y., English, J. J., and Jorgensen, R. A. (1997). The frequency and degree of co-suppression by sense chalcone synthase transgenes are dependent on transgene promoter strength and are reduced by premature nonsense codons in the transgene coding sequence. Plant Cell 9, 1357–1368. doi: 10.1105/tpc.9.8.1357
Regulski, M., Lu, Z., Kendall, J., Donoghue, M. T., Reinders, J., Llaca, V., et al. (2013). The maize methylome influences mRNA splice sites and reveals widespread paramutation-like switches guided by small RNA. Genome Res. 23, 1651–1662. doi: 10.1101/gr.153510.112
Shrawat, A. K., Becker, D., and Lorz, H. (2007). Agrobacterium tumefaciens-mediated genetic transformation of barley (Hordeum vulgare L). Plant Sci. 172, 281–290. doi: 10.1016/j.plantsci.2006.09.005
Smith, C. J., Watson, C. F., Morris, P. C., Bird, C. R., Seymour, G. B., Gray, J. E., et al. (1990). Inheritance and effect on ripeing of antisense polygalacturonase genes in transgenic tomatoes. Plant Mol. Biol. 14, 369–379. doi: 10.1007/BF00028773
Stroud, H., Ding, B., Simon, S. A., Feng, S., Bellizzi, M., Pellegrini, M., et al. (2013). Plants regenerated from tissue culture contain stable epigenome changes in rice. Elife 2:e00354. doi: 10.7554/eLife.00354
Tsuchiya, T., and Eulgem, T. (2013). An alternative polyadenylation mechanism coopted to the Arabidopsis RPP7 gene through intronic retrotransposon domestication. Proc. Natl. Acad. Sci. U.S.A. 110, E3535–E3543. doi: 10.1073/pnas.1312545110
Van Blokland, R., Ten Lohuis, M., and Meyer, P. (1997). Condensation of chromatin in transcriptional regions of an inactivated plant transgene: evidence for an active role of transcription in gene silencing. Mol. Genet. Genomics 257, 1–13. doi: 10.1007/s004380050617
Van der Krol, A. R., Mur, L. A., Beld, M., Mol, J. N., and Stuitje, A. R. (1990). Flavonoid genes in petunia: addition of a limited number of genes copies may lead to a suppression of gene expression. Plant Cell 2, 291–299. doi: 10.1105/EPC.2.4.291
Van Eldik, G. J., Litiere, K., Jacobs, J. J., Van Montagu, M., and Cornelissen, M. (1998). Silencing of beta-1,3-glucanase genes in tobacco correlates with an increased abundance of RNA degradation intermediates. Nucleic Acids Res. 26, 5176–5181. doi: 10.1093/nar/26.22.5176
Vaucheret, H., Beclin, C., Elmayan, T., Feuerbach, F., Godon, C., Morel, J. B., et al. (1998). Transgene-induced gene silencing in plants. Plant J. 16, 651–659. doi: 10.1046/j.1365-313x.1998.00337.x
Vaucheret, H., Beclin, C., and Fagard, M. (2001). Post-transcriptional gene silencing in plants. J. Cell Sci. 114, 3083–3091.
Vaucheret, H., and Fagard, M. (2001). Transcriptional gene silencing in plants: targets, inducers and regulators. Trends Genet. 17, 29–35. doi: 10.1016/S0168-9525(00)02166-1
Vaucheret, H., Nussaume, L., Palauqui, J. C., Quillere, I., and Elmayan, T. (1997). A transcriptionally active state is required for post-transcriptional silencing (co-suppression) of nitrate reductase host genes and transgenes. Plant Cell 9, 1495–1504. doi: 10.1105/tpc.9.8.1495
Velten, J., Cakir, C., Youn, E., Chen, J., and Cazzonelli, I. C. (2012). Transgene silencing and transgene-derived siRNA production in tobacco plants homozygous for an introduced AtMYB90 construct. PLoS ONE 7:e30141. doi: 10.1371/journal.pone.0030141
Voinnet, O., Vain, P., Angell, S., and Baulcombe, D. C. (1998). Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell 95, 177–187. doi: 10.1016/S0092-8674(00)81749-3
Wassenegger, M., Heimes, S., Riedel, L., and Sanger, H. L. (1994). RNA directed de-novo methylation of genomic sequences in plants. Cell 76, 567–576. doi: 10.1016/0092-8674(94)90119-8
Weinhold, A., Kallenbach, M., and Ian Thomas Baldwin, T. I. (2013). Progressive 35S promoter methylation increases rapidly during vegetative development in transgenic Nicotiana attenuata plants. BMC Plant Biol. 13:99. doi: 10.1186/1471-2229-13–99
Yamasaki, S., Oda, M., Koizumi, N., Mitsukuri, K., Johkan, M., Nakatsuka, T., et al. (2011). De Novo Dna Methylation of the 35s Enhancer Revealed by High-Resolution Methylation Analysis. doi: 10.5511/plantbiotechnology.10.1222a
Keywords: homology-dependent gene silencing, post-transcriptional gene silencing, systematic acquired silencing, transcriptional gene silencing, transgenic plants
Citation: Rajeevkumar S, Anunanthini P and Sathishkumar R (2015) Epigenetic silencing in transgenic plants. Front. Plant Sci. 6:693. doi: 10.3389/fpls.2015.00693
Received: 14 May 2015; Accepted: 21 August 2015;
Published: 10 September 2015.
Edited by:
Clelia De-la-Peña, Centro de Investigación Científica de Yucatán, MexicoReviewed by:
Gong-yin Ye, Zhejiang University, ChinaGhulam Kadir Ahmad Parveez, Malaysian Palm Oil Board, Malaysia
Copyright © 2015 Rajeevkumar, Anunanthini and Sathishkumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ramalingam Sathishkumar, Plant Genetic Engineering Laboratory, Department of Biotechnology, Bharathiar University, Coimbatore 641 046, Tamil Nadu, India,cnNhdGhpc2hAYnVjLmVkdS5pbg==
 Sarma Rajeevkumar
Sarma Rajeevkumar Pushpanathan Anunanthini
Pushpanathan Anunanthini Ramalingam Sathishkumar
Ramalingam Sathishkumar