- 1State Key Laboratory of Plant Diseases and Insect Pests, Institute of Plant Protection, Chinese Academy of Agricultural Sciences, Beijing, China
- 2Department of Plant Pathology, The Ohio State University, Columbus, OH, USA
To defend against pathogens, plants have developed a sophisticated innate immunity that includes effector recognition, signal transduction, and rapid defense responses. Recent evidence has demonstrated that plants utilize the epigenetic control of gene expression to fine-tune their defense when challenged by pathogens. In this review, we highlight the current understanding of the molecular mechanisms of histone modifications (i.e., methylation, acetylation, and ubiquitination) and chromatin remodeling that contribute to plant immunity against pathogens. Functions of key histone-modifying and chromatin remodeling enzymes are discussed.
Introduction
Throughout their life cycles, plants are exposed to abiotic stresses, including temperature fluctuation and nutrition deficiency, and biotic threats, including attack by herbivores and microbial pathogens. With respect to microbial pathogens, plants are unlike animals in that they lack an adaptive immune system that produces antibodies and also lack mobile circulatory cells that detect and prevent or reduce infection. Instead, plants mainly rely on an innate immunity system to resist microbial attack. In plants, the salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) signaling pathways play pivotal roles in defending against biotrophic and necrotrophic pathogens (Pieterse et al., 2009). After detecting a pathogen, the plant activates a cascade of defense responses to establish local and systemic acquired resistance (SAR; Durrant and Dong, 2004).
Transcription of defense genes is tightly regulated by many transcription factors (TFs) that fine-tune the defense response (Thilmony et al., 2006). This requires that plants rapidly and precisely re-program gene expression. In particular, activation of an appropriate stress signaling pathway following pathogen detection is integrated in the plant cell nucleus through a set of regulatory cascades that prioritize defense over growth-related cellular functions (Moore et al., 2011). Research over the last decade has revealed that this transcriptional re-programming and regulation of defense-related genes often involves chromatin modifications and remodeling in Arabidopsis (Alvarez et al., 2010). In this review, we summarize and discuss the roles of chromatin modifications and remodeling in plant defense.
Plant Innate Immunity
Plant innate immunity is triggered by pattern recognition receptors (PRRs) located on the external cell surface. PRRs can recognize specific pathogen-/microbe-associated molecular patterns (PAMPs/MAMPs), including cell wall components, short peptides, and lipopolysaccharides derived from the pathogen, leading to PAMP/MAMP-triggered immunity (PTI/MTI), which is the first layer of defense (Boller and Felix, 2009). Several early defense responses, including the generation of reactive oxygen species (ROS), calcium flux, plant cell wall modification, and the activation of a cascade of mitogen-activated protein kinases (MAPKs), are triggered during PTI. To overcome PTI, pathogens may deliver effector proteins into host cells, resulting in effector-triggered susceptibility (ETS). An additional level of resistance associated with vigorous defense induction may occur when specific intracellular receptors/sensors called resistance (R) gene products to recognize such race-specific avirulence (avr) effectors, thereby activating effector-triggered immunity (ETI; Jones and Dangl, 2006). ETI usually triggers a localized cell death at the infection site, in a process known as the hypersensitive response (HR), which along with antimicrobial effects may restrict most pathogen growth (Caplan et al., 2008). In addition to these PTI and ETI responses after initial local infection, the uninfected portions of the plant usually develop SAR, providing resistance in distal plant tissues against subsequent pathogen challenges (Durrant and Dong, 2004; Mishina and Zeier, 2007).
Chromatin Modification in Plant Innate Immunity
Chromatin Structure and Modifications
The basic, repeated unit of chromatin is the nucleosome that contains 147 base pairs (bp) of DNA wrapped around a histone octamer, which in turn consists of two copies of the following core histones: H2A, H2B, H3, and H4 (Luger et al., 1997). The linker histone, H1, associates with DNA between two nucleosomes and participates in higher order chromatin structure formation and remodeling. Extending from the globular nucleosome core, the histone tails may harbor diverse post-translational modifications (PTMs), i.e., acetylation, methylation, phosphorylation, ubiquitination, sumoylation, carbonylation, and glycosylation. PTMs can directly affect chromatin structure or can recruit specific “readers or effectors,” thereby regulating gene expression mainly by altering nucleosome stability and positioning, which affect the accessibility for regulatory proteins or protein complexes involved in transcription, DNA replication, and repair (Kouzarides, 2007). In general, histone acetylation by histone acetyltransferases (HATs) is associated with transcriptional activation, while histone deacetylation by histone deacetylases (HDACs) is associated with transcriptional suppression (Eberharter and Becker, 2002). Depending on the context of targets, histone methylation and/or ubiquitination can either be an active or repressive marker for transcription. Generally, tri-methylations of H3K4 and H3K36 (H3K4me3 and H3K36me3) and mono-ubiquitination of H2B (H2Bub) are enriched at actively expressed genes (Xu et al., 2008; Zhang et al., 2009), H3K27me3 is associated with repressed genes, while H3K9me2 and H4K20me1 are enriched at constitutive heterochromatin and silenced transposons (Zhang et al., 2007a,b; Bernatavichute et al., 2008). In addition to histone modification, ATP-dependent chromatin-remodeling enzymes use the energy of ATP hydrolysis to remodel chromatin structure by modifying the interaction between DNA and histone to relocate or dissociate nucleosomes, move histone octamers, and catalyze the incorporation of specific histone variants. ATP-dependent chromatin-remodeling enzymes thus play crucial roles in nucleosome assembly/disassembly and allow the transcriptional machinery to access the DNA (Smith and Peterson, 2005; Clapier and Cairns, 2009).
Many studies have documented that histone modifications and ATP-dependent chromatin remodeling result in rapid, reversible, or trans-generational changes in gene expression associated with various developmental processes, such as flowering time control, cell fate determination and maintenance, and seed development. These mechanisms, however, have only recently attracted attention as potential transcriptional regulators in plant innate immunity (Table 1).

TABLE 1. Histone-modifying enzymes and chromatin-remodelling factors involved in plant responses to pathogens.
Histone Acetylation
Histone lysine acetylation is regulated by the antagonistic interactions between HATs and HDACs. Plant HDACs can be divided into four major groups or families. In addition to a plant-specific type-II HDAC (HD2) family, three other major families are designated as reduced potassium dependency 3 (RPD3), HDA1, and silence information regulator 2 (SIR2); this grouping is based on homology to yeast counterparts. Among these groups, HDA19 from Arabidopsis has been well-studied with regard to its roles in plant defense against pathogen attack. HDA19, which belongs to the RPD3 subfamily, was initially reported to be involved in the ET/JA signaling pathways of defense responses based on two lines of evidence. First, the expression of HDA19 is induced by wounding, by challenge with the pathogen Alternaria brassicicola, and by treatment with the plant hormone JA. Second, the knock-down mutant of HDA19 exhibits decreased transcription of several ET/JA pathway genes (ERF1, CHI-B, and BGL) and increased susceptibility to fungal pathogens, while overexpression results in the opposite disease phenotypes (Zhou et al., 2005). Similarly, HDA6, another Arabidopsis RPD3-type HDAC, is induced by treatments with JA and the ET precursor ACC, whereas the expression of other members of Arabidopsis RPD3-type HDACs is not inducible by these hormones (Zhou et al., 2005). In addition, HDA6 interacts with an F-box protein, coronatine insensitive 1 (COI1), which mediates JA signaling (Devoto et al., 2002). The expression of the JA-responsive genes, i.e., PDF1.2, VSP2, JIN1, and ERF1, is down-regulated in axe1-5 (HDA6 loss-of-function mutant) and HDA6-RNAi plants (Wu et al., 2008), suggesting redundant roles of HDA6 and HDA19 in plant defense against infection by necrotrophic pathogens. In addition to its role in the JA/ET defense pathway, HDA19 positively regulates SA-mediated basal defense and the expression of pathogenesis-related gene 1 (PR1) by physically interacting with WRKY38 and WRKY62 and inhibiting their transcriptional-activator activities (Kim et al., 2008). On the other hand, the basal expression of the SA-induced PR1 and PR5 is upregulated in the hda19 mutant when it is not challenged by pathogens, reflecting the negative role of HDA19 in defense responses. PR1 and PR2 are well-defined markers for SA-mediated basal and R gene-mediated defense against biotrophic pathogens (Ward et al., 1991; Rairdan and Delaney, 2002; van Loon et al., 2006). Several studies have shown that the SA-induced activation of PR1 is tightly correlated with an increase in the level of acetylated histones at the PR1 locus in Arabidopsis (Mosher et al., 2006; Koornneef et al., 2008) and tobacco (Butterbrodt et al., 2006). Additionally, HDA19 associates directly with the promoters of PR1 and PR2 and deacetylates histones at PR1 and PR2 locus. Thus, HDA19 forms a repressive chromatin environment (low histone acetylation level) under unchallenged conditions that ensures a low basal expression of defense genes as well as the proper induction of PR genes without harmful overstimulation during defense responses to pathogen attacks (Choi et al., 2012).
The HDAC proteins in the Sir2 family are NAD+-dependent HDACs that play diverse roles in a variety of physiological processes, including chromatin silencing, DNA repair, the cell cycle, and apoptosis and aging in yeast and mammalian systems (Eberharter and Becker, 2002; Yamamoto et al., 2007; Etchegaray et al., 2013). Both Arabidopsis and rice genomes contain two Sir2 family genes (Pandey et al., 2002). Knockdown of OsSRT1 by RNAi in rice plants enhances histone H3K9 acetylation on the promoters of HR-related genes, which leads to hydrogen peroxide accumulation, DNA fragmentation, and cell death, suggesting a negative role of OsSRT1 in defense (Huang et al., 2007). Highly divergent in sequence from OsSRT1, AtSRT2 is down-regulated by Pseudomonas syringae pv. tomato DC3000 (Pst DC3000) infection and negatively regulates the plant basal defense and PR1 expression, possibly by suppressing pathogen-induced expression of PAD4, EDS5, and SID2 and thereby regulating SA synthesis (Wang et al., 2010).
In addition to local resistance, SAR is also related to priming for stronger activation of various defense responses that are induced following an attack by microbial pathogens (van Hulten et al., 2006). Priming of innate immunity is correlated with chromatin modification of the promoter region of WRKY TF genes (Jaskiewicz et al., 2011) and SA- and PTI-responsive genes (Luna et al., 2012; Po-Wen et al., 2013). Researchers recently showed that repetitive abiotic stress causes the priming of PTI in Arabidopsis, leading to enhanced resistance to bacterial pathogens. This elevated defense after repeated exposure to environmental stress is compromised in the hac1 mutant, establishing a link between open chromatin configuration such as HAC1-dependent histone acetylation and primed Arabidopsis innate immunity and bacterial resistance (Singh et al., 2014a).
Histone Methylation
The Arabidopsis genome encodes 37 putative SET-domain group proteins, some of which have been experimentally demonstrated to harbor histone methyltransferase (HMT) activity (Thorstensen et al., 2011). For the removal of methyl residues from the methylated histones, the lysine-specific demethylase 1 (LSD1)-like proteins and Jomonji C-domain (JmjC) proteins are effective in histone demethylation in plants (Chen et al., 2011). Dynamic histone methylation and de-methylation are involved in many cellular processes such as gene imprinting and DNA methylation (Kohler et al., 2012), and in developmental events such as vernalization (Kim and Sung, 2014). Recent findings indicate that histone methylation contributes to plant immunity against both necrotrophic and biotrophic pathogens by affecting the expression of specific NBS-LRR proteins, WRKY family TFs, as well as TFs involved in defense signaling pathways.
The first study of immune responses involving histone methylation concerned Arabidopsis trithrox 1 (ATX1), also known as SDG27. Loss of ATX1 function affects the transcription of a subset of pathogen- and disease resistance-associated genes, including those encoding members of the TIR-NBS-LRR classes of disease resistance proteins, lectins, and heat shock proteins, as well as several WRKY family TFs (Alvarez-Venegas et al., 2006). Further findings revealed that ATX1 directly controls H3K4me3 levels at the promoter of WRKY70 and also controls the expression of WRKY70, a positive regulator of SA-mediated defense signaling against bacterial pathogens (Alvarez-Venegas et al., 2007; Saleh et al., 2008). Arabidopsis trithorax-related 7 (Atxr7), another histone H3K4 methyltransferase in the trithrox1 group, physically associates with the modifier of snc1 9 (MOS9), which is a plant-specific protein with unknown function discovered in a forward genetic screening of the snc1 mutant. Together with MOS9, Atxr7 is required for both maintaining the H3K4me3 levels at the promoter of the NBS-LRR genes Snc1 and Rpp4 and expression of these R genes residing in the RPP4 cluster (Xia et al., 2013).
In Arabidopsis, another important active signature of histone modification, H3K36 tri-methylation, is catalyzed by the SET domain group 8 (SDG8, also known as EFS, LAZ2, and Ashh2; Xu et al., 2008). SDG8 is a homolog of SET2 in yeast and ASH1 in Drosophila. Mutation in SDG8 causes pleiotropic developmental phenotypes such as early flowering time, reduced organ size, and enhanced branch shooting (Zhao et al., 2005; Dong et al., 2008; Cazzonelli et al., 2009). A recent study revealed that sdg8 mutant plants have reduced resistance to the necrotrophic fungal pathogens A. brassicicola and Botrytis cinerea, indicating that SDG8 plays a crucial role in plant defense through H3K36me3-mediated activation of a subset of genes (including ERF1, PDF1.2a, and VSP2) in the JA/ET signaling pathways (Berr et al., 2010). Another study showed that SDG8 is required for both basal and R-protein-mediated resistance and that SDG8 maintains the LAZ5 locus in a transcriptionally active state by modifying its H3K36me3 level. LAZ5 is a member of an immune receptor class involved in the detection of specific pathogens and subsequent cell death (Palma et al., 2010). In a comparative analysis of three Arabidopsis ASH1 family mutants, loss of function of ASHH2 and ASHR1 resulted in more rapid HRs to both a non-pathogenic strain (hrpA-) and a pathogenic strain (DC3000) of P. syringae. In contrast, the ashr3 mutant is more resistant to the infection than the ashr1 and ashh2 mutants. Furthermore, PR1 gene expression was highest in the ashr3 mutant, while H3K4me2 levels at the PR1 promoter region are reduced in both the ashr1 and ashh2 mutants upon infection by DC3000 (De-La-Pena et al., 2012). This result demonstrates that the ASH1 group H3K4 methyltransferases have both overlapping and distinct roles in the plant defense against pathogens.
Collectively, the active H3K4 and H3K36 methylation states, which are catalyzed by SET domain protein, have been implicated in the SA- and JA-mediated plant defense in Arabidopsis. These markers act as permissive marks for the basal expression of the defense genes or establishing the chromatin status for prompt induction when plants are challenged. In contrast, the removal of the repressive histone H3K27me3 state by the JmjC protein JMJ705 in rice also plays important roles in defense-related gene expression. When induced by a stress signal or pathogen infection, JMJ705 is involved in the methyl jasmonate-induced removal of H3K27me3 and preferential biotic stress-responsive gene activation, supporting the hypothesis that H3K27me3 maintains the resting state of defense genes under normal conditions (Li et al., 2013). FLD, a homolog of the human LSD1, was originally discovered to promote flowering time by negatively regulating the expression of flower repressor FLC (He et al., 2003; Liu et al., 2007). A forward genetic screen revealed that Arabidopsis requires FLD in order to respond to the SAR signals leading to the systemic accumulation of SA; the screen also revealed that FLD influences histone modifications at the promoters of WRKY29 and WRKY6 and thereby enables a robust activation of SA signaling in response to subsequent exposure to virulent pathogens (Singh et al., 2013, 2014b).
Histone Mono-Ubiquitination
In Arabidopsis, histone H2B mono-ubiquitination is catalyzed by the RING E3 ligases histone mono-ubqutinatio1 (HUB1) and HUB2, which participate in various developmental process such as the control of flowering time, the cell cycle, seed dormancy, and circadian clock (Xu et al., 2009; Lolas et al., 2010; Bourbousse et al., 2012). Additionally, HUB1 is a regulatory component of plant defense against necrotrophic fungal pathogens. Arabidopsis plants with mutations in the HUB1 alleles are extremely susceptible to the necrotrophic fungi B. cinerea and A. brassicicola. Consistent with the plant cell wall functioning in resistance to necrotrophic fungi by acting as a physical barrier, the thickness of epidermal cell walls is reduced in the hub1 mutant. This suggests that HUB1 may enhance defense by increasing the thickness or otherwise modifying epidermal cell walls. Interestingly, HUB1 interacts with MED21, a subunit of the Mediator complex, in regulating the function of RNA polymerase II. Arabidopsis MED21 couples critical roles in disease resistance and embryo development based on the disease susceptibility and embryo-lethal phenotypes of plant lines with reduced MED21 gene expression. Thus, MED21 together with HUB1 controls critical components involved in the regulation of defense against necrotrophic fungal pathogens, suggesting a transcriptional role of Hub1-mediated histone mono-ubiquitination in defense (Dhawan et al., 2009). In contrast, responses to the bacterial pathogen P. syringae are unaltered in hub1 plants. However, a recent report showed that both Hub1 and Hub2 regulate the expression of the R genes SNC1 and Rpp4 (Zou et al., 2014). In the auto-immunity mutant bon1, which is a negative regulator of the NB-LRR-encoding R gene SNC1 and other R-like genes (Yang and Hua, 2004; Li et al., 2007), loss of function in HUB1 or HUB2 reduces SNC1 up-regulation and suppresses the bon1 auto-immune phenotypes. Thus, HUB1 and HUB2 mediate histone 2B (H2B) mono-ubiquitination directly at the SNC1 R gene locus to regulate its expression. This is another example of how the immune response can be fine-tuned by histone modifications at an R gene locus (Zou et al., 2014).
Chromatin Remodeling
In addition to being affected by covalent histone modifications, plant defense can also be affected by chromatin-remodeling factors that regulate R gene function and specific JA or SA pathways. The Arabidopsis genome encodes more than 40 ATP-dependent chromatin-remodeling factors, which can be subdivided into at least five families based on their ATPase subunits. In the broad SWI2/SNF2 protein family, DDM1 functions antagonistically to MOS1 in regulating the expression of the R gene SNC1 (Li et al., 2010). SWR1, a component of the Arabidopsis SWR1-like complex that replaces the histone H2A with the histone variant H2A.Z, is required for maintaining the repression of SA-dependent defense genes in unstressed plants (March-Diaz et al., 2008). SWI/SNF class chromatin remodeling ATPase SPLAYED (SYD) can be directly recruited to the promoters of selected genes, i.e., PDF1.2a, VSP2, and MYC2, downstream of the JA and ET signaling pathways. Therefore, SYD is required for the expression of these genes and for resistance against the necrotrophic pathogen B. cinerea but is not required for resistance against P. syringae (Walley et al., 2008).
Concluding Remarks and Perspectives
Recent research has increased our understanding of how chromatin modifications and remodeling affect defense in the model plants Arabidopsis and rice. Based on current evidence and as summarized in Figure 1, histone modifications in plant defense responses can be grouped as follows: (1) active histone marks that establish a basal expression level of the defense genes to enable an effective induction when the plant is challenged; (2) repressive histone modifications that prevent unnecessary activation of defense-related genes under normal growth conditions; (3) histone modifications that are induced after pathogen infection and that induce or reinforce the expression of defense-related genes; and (4) histone/chromatin changes that occur in response to biotic or abiotic stresses and that can be transmitted to the next generation. In the future, a combination of new genomic and proteomic approaches should be used to identify the targets of the epigenetic-related enzymes and other factors that are involved in the regulation of plant immunity. In addition, only a few histone-modifying enzymes have been investigated. Large-scale screens and characterization of epigenetic mutants should help increase our understanding of the histone-modifying enzymes involved in the chromatin changes that occur when plants defend against pathogens. Moreover, three-dimensional structure plasticity of genomes establishes fine-tune feature in gene expression modulation rather than defined by its linear context. Emerging evidence showed that lncRNAs (long non-coding RNAs) and chromatin remodeling complexes are shaping the dynamic genome topology through chromatin loops to regulate dynamic gene expression in response to the environmental cues (Ariel et al., 2014; Jegu et al., 2014). Considering that the global genome structure is impacted in many diseases in animal systems and the participation of lncRNAs in nuclear architecture, the association between non-coding RNAs and the genome topology related to chromatin marks and organization remains an unexplored area in plant immunity.
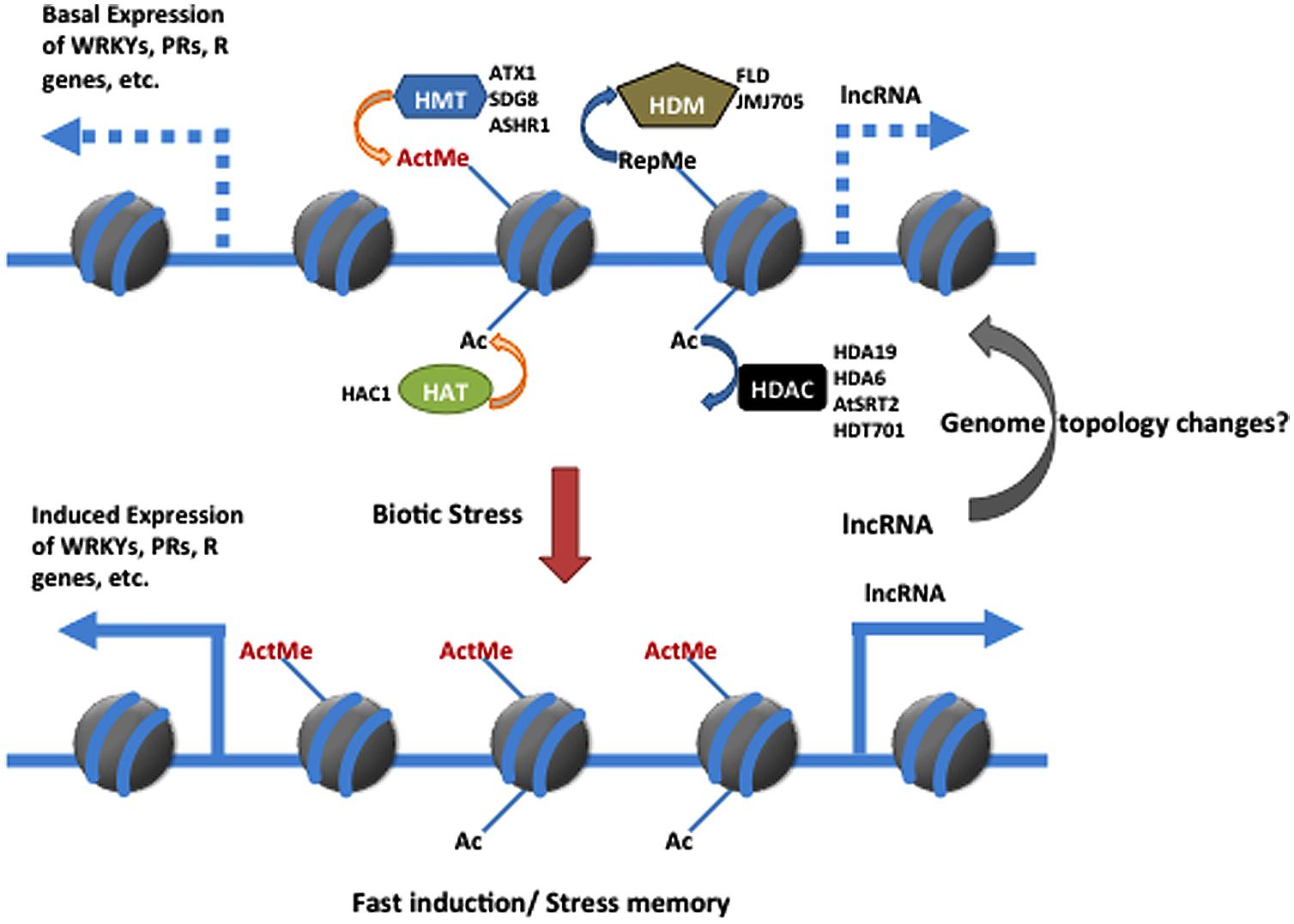
FIGURE 1. Simplified model for participation of chromatin modification in regulating plant immunity against biotic stress. Histone modification changes in defense-related gene can be achieved through methylation/demethylation and/or acetylation/deacetylation by antagonistic interaction between HMT and HDM or HAT and HDAC Each enzymes catalyzed different modification in regarding its roles in plant immunity is described in literature. The hypothetical involvement of the IncRNA in regulating the dynamin defense gene expression through the modulation of chromatin architecture is proposed as well. ActMe, active methylation marker; RepMe, Repressive methylation marker; HMT, histone methyltransferase; HDM, histone demethylase; HAC, histone acetylase; HDAC, histone de acetylase; IncRNA, long non-coding RNA; PR, pathogenesis-related; R, Resistance.
Author Contributions
BD and G-LW wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgment
This work was supported by grants from National Natural Science Foundation of China to BD (#31371928) and from National Science Foundation of USA to G-LW (IOS#1120949).
References
Alvarez, M. E., Nota, F., and Cambiagno, D. A. (2010). Epigenetic control of plant immunity. Mol. Plant Pathol. 11, 563–576. doi: 10.1111/j.1364-3703.2010.00621.x
Alvarez-Venegas, R., Abdallat, A. A., Guo, M., Alfano, J. R., and Avramova, Z. (2007). Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics 2, 106–113. doi: 10.4161/epi.2.2.4404
Alvarez-Venegas, R., Sadder, M., Hlavacka, A., Baluska, F., Xia, Y., Lu, G., et al. (2006). The Arabidopsis homolog of trithorax, ATX1, binds phosphatidylinositol 5-phosphate, and the two regulate a common set of target genes. Proc. Natl. Acad. Sci. U.S.A. 103, 6049–6054. doi: 10.1073/pnas.0600944103
Ariel, F., Jegu, T., Latrasse, D., Romero-Barrios, N., Christ, A., Benhamed, M., et al. (2014). Noncoding transcription by alternative RNA polymerases dynamically regulates an auxin-driven chromatin loop. Mol. Cell 55, 383–396. doi: 10.1016/j.molcel.2014.06.011
Bernatavichute, Y. V., Zhang, X., Cokus, S., Pellegrini, M., and Jacobsen, S. E. (2008). Genome-wide association of histone H3 lysine nine methylation with CHG DNA methylation in Arabidopsis thaliana. PLoS ONE 3:e3156. doi: 10.1371/journal.pone.0003156
Berr, A., Mccallum, E. J., Alioua, A., Heintz, D., Heitz, T., and Shen, W. H. (2010). Arabidopsis histone methyltransferase SET DOMAIN GROUP8 mediates induction of the jasmonate/ethylene pathway genes in plant defense response to necrotrophic fungi. Plant Physiol. 154, 1403–1414. doi: 10.1104/pp.110.161497
Boller, T., and Felix, G. (2009). A renaissance of elicitors: perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. doi: 10.1146/annurev.arplant.57.032905.105346
Bourbousse, C., Ahmed, I., Roudier, F., Zabulon, G., Blondet, E., Balzergue, S., et al. (2012). Histone H2B monoubiquitination facilitates the rapid modulation of gene expression during Arabidopsis photomorphogenesis. PLoS Genet. 8:e1002825. doi: 10.1371/journal.pgen.1002825
Butterbrodt, T., Thurow, C., and Gatz, C. (2006). Chromatin immunoprecipitation analysis of the tobacco PR-1a- and the truncated CaMV 35S promoter reveals differences in salicylic acid-dependent TGA factor binding and histone acetylation. Plant Mol. Biol. 61, 665–674. doi: 10.1007/s11103-006-0039-2
Caplan, J., Padmanabhan, M., and Dinesh-Kumar, S. P. (2008). Plant NB-LRR immune receptors: from recognition to transcriptional reprogramming. Cell Host Microbe 3, 126–135. doi: 10.1016/j.chom.2008.02.010
Cazzonelli, C. I., Cuttriss, A. J., Cossetto, S. B., Pye, W., Crisp, P., Whelan, J., et al. (2009). Regulation of carotenoid composition and shoot branching in Arabidopsis by a chromatin modifying histone methyltransferase, SDG8. Plant Cell 21, 39–53. doi: 10.1105/tpc.108.063131
Chen, X., Hu, Y., and Zhou, D. X. (2011). Epigenetic gene regulation by plant Jumonji group of histone demethylase. Biochim. Biophys. Acta 1809, 421–426. doi: 10.1016/j.bbagrm.2011.03.004
Choi, S. M., Song, H. R., Han, S. K., Han, M., Kim, C. Y., Park, J., et al. (2012). HDA19 is required for the repression of salicylic acid biosynthesis and salicylic acid-mediated defense responses in Arabidopsis. Plant J. 71, 135–146. doi: 10.1111/j.1365-313X.2012.04977.x
Clapier, C. R., and Cairns, B. R. (2009). The biology of chromatin remodeling complexes. Annu. Rev. Biochem. 78, 273–304. doi: 10.1146/annurev.biochem.77.062706.153223
De-La-Pena, C., Rangel-Cano, A., and Alvarez-Venegas, R. (2012). Regulation of disease-responsive genes mediated by epigenetic factors: interaction of Arabidopsis-Pseudomonas. Mol. Plant Pathol. 13, 388–398. doi: 10.1111/j.1364-3703.2011.00757.x
Devoto, A., Nieto-Rostro, M., Xie, D., Ellis, C., Harmston, R., Patrick, E., et al. (2002). COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32, 457–466. doi: 10.1046/j.1365-313X.2002.01432.x
Dhawan, R., Luo, H., Foerster, A. M., Abuqamar, S., Du, H. N., Briggs, S. D., et al. (2009). Histone Monoubiquitination1 interacts with a subunit of the mediator complex and regulates defense against necrotrophic fungal pathogens in Arabidopsis. Plant Cell 21, 1000–1019. doi: 10.1105/tpc.108.062364
Ding, B., Bellizzi Mdel, R., Ning, Y., Meyers, B. C., and Wang, G. L. (2012). HDT701, a histone H4 deacetylase, negatively regulates plant innate immunity by modulating histone H4 acetylation of defense-related genes in rice. Plant Cell 24, 3783–3794. doi: 10.1105/tpc.112.101972
Dong, G., Ma, D. P., and Li, J. (2008). The histone methyltransferase SDG8 regulates shoot branching in Arabidopsis. Biochem. Biophys. Res. Commun. 373, 659–664. doi: 10.1016/j.bbrc.2008.06.096
Durrant, W. E., and Dong, X. (2004). Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209. doi: 10.1146/annurev.phyto.42.040803.140421
Eberharter, A., and Becker, P. B. (2002). Histone acetylation: a switch between repressive and permissive chromatin. Second in review series on chromatin dynamics. EMBO Rep. 3, 224–229. doi: 10.1093/embo-reports/kvf053
Etchegaray, J. P., Zhong, L., and Mostoslavsky, R. (2013). The histone deacetylase SIRT6: at the crossroads between epigenetics, metabolism and disease. Curr. Top. Med. Chem. 13, 2991–3000. doi: 10.2174/15680266113136660213
He, Y., Michaels, S. D., and Amasino, R. M. (2003). Regulation of flowering time by histone acetylation in Arabidopsis. Science 302, 1751–1754. doi: 10.1126/science.1091109
Huang, L., Sun, Q., Qin, F., Li, C., Zhao, Y., and Zhou, D. X. (2007). Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol. 144, 1508–1519. doi: 10.1104/pp.107.099473
Jaskiewicz, M., Conrath, U., and Peterhansel, C. (2011). Chromatin modification acts as a memory for systemic acquired resistance in the plant stress response. EMBO Rep. 12, 50–55. doi: 10.1038/embor.2010.186
Jegu, T., Latrasse, D., Delarue, M., Hirt, H., Domenichini, S., Ariel, F., et al. (2014). The BAF60 subunit of the SWI/SNF chromatin-remodeling complex directly controls the formation of a gene loop at FLOWERING LOCUS C in Arabidopsis. Plant Cell 26, 538–551. doi: 10.1105/tpc.113.114454
Jones, J. D., and Dangl, J. L. (2006). The plant immune system. Nature 444, 323–329. doi: 10.1038/nature05286
Kim, D. H., and Sung, S. (2014). Genetic and epigenetic mechanisms underlying vernalization. Arabidopsis Book 12:e0171. doi: 10.1199/tab.0171
Kim, K. C., Lai, Z., Fan, B., and Chen, Z. (2008). Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 20, 2357–2371. doi: 10.1105/tpc.107.055566
Kohler, C., Wolff, P., and Spillane, C. (2012). Epigenetic mechanisms underlying genomic imprinting in plants. Annu. Rev. Plant Biol. 63, 331–352. doi: 10.1146/annurev-arplant-042811-105514
Koornneef, A., Rindermann, K., Gatz, C., and Pieterse, C. M. (2008). Histone modifications do not play a major role in salicylate-mediated suppression of jasmonate-induced PDF1.2 gene expression. Commun. Integr. Biol. 1, 143–145. doi: 10.4161/cib.1.2.6997
Kouzarides, T. (2007). Chromatin modifications and their function. Cell 128, 693–705. doi: 10.1016/j.cell.2007.02.005
Li, T., Chen, X., Zhong, X., Zhao, Y., Liu, X., Zhou, S., et al. (2013). Jumonji C domain protein JMJ705-mediated removal of histone H3 lysine 27 trimethylation is involved in defense-related gene activation in rice. Plant Cell 25, 4725–4736. doi: 10.1105/tpc.113.118802
Li, Y., Tessaro, M. J., Li, X., and Zhang, Y. (2010). Regulation of the expression of plant resistance gene SNC1 by a protein with a conserved BAT2 domain. Plant Physiol. 153, 1425–1434. doi: 10.1104/pp.110.156240
Li, Y., Yang, S., Yang, H., and Hua, J. (2007). The TIR-NB-LRR gene SNC1 is regulated at the transcript level by multiple factors. Mol. Plant Microbe Interact. 20, 1449–1456. doi: 10.1094/MPMI-20-11-1449
Liu, F., Quesada, V., Crevillen, P., Baurle, I., Swiezewski, S., and Dean, C. (2007). The Arabidopsis RNA-binding protein FCA requires a lysine-specific demethylase 1 homolog to downregulate FLC. Mol. Cell 28, 398–407. doi: 10.1016/j.molcel.2007.10.018
Lolas, I. B., Himanen, K., Gronlund, J. T., Lynggaard, C., Houben, A., Melzer, M., et al. (2010). The transcript elongation factor FACT affects Arabidopsis vegetative and reproductive development and genetically interacts with HUB1/2. Plant J. 61, 686–697. doi: 10.1111/j.1365-313X.2009.04096.x
Luger, K., Mader, A. W., Richmond, R. K., Sargent, D. F., and Richmond, T. J. (1997). Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260. doi: 10.1038/38444
Luna, E., Bruce, T. J., Roberts, M. R., Flors, V., and Ton, J. (2012). Next-generation systemic acquired resistance. Plant Physiol. 158, 844–853. doi: 10.1104/pp.111.187468
March-Diaz, R., Garcia-Dominguez, M., Lozano-Juste, J., Leon, J., Florencio, F. J., and Reyes, J. C. (2008). Histone H2A.Z and homologues of components of the SWR1 complex are required to control immunity in Arabidopsis. Plant J. 53, 475–487. doi: 10.1111/j.1365-313X.2007.03361.x
Mishina, T. E., and Zeier, J. (2007). Bacterial non-host resistance: interactions of Arabidopsis with non-adapted Pseudomonas syringae strains. Physiol. Plant. 131, 448–461. doi: 10.1111/j.1399-3054.2007.00977.x
Moore, J. W., Loake, G. J., and Spoel, S. H. (2011). Transcription dynamics in plant immunity. Plant Cell 23, 2809–2820. doi: 10.1105/tpc.111.087346
Mosher, R. A., Durrant, W. E., Wang, D., Song, J., and Dong, X. (2006). A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell 18, 1750–1765. doi: 10.1105/tpc.105.039677
Palma, K., Thorgrimsen, S., Malinovsky, F. G., Fiil, B. K., Nielsen, H. B., Brodersen, P., et al. (2010). Autoimmunity in Arabidopsis acd11 is mediated by epigenetic regulation of an immune receptor. PLoS Pathog 6:e1001137. doi: 10.1371/journal.ppat.1001137
Pandey, R., Muller, A., Napoli, C. A., Selinger, D. A., Pikaard, C. S., Richards, E. J., et al. (2002). Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 30, 5036–5055. doi: 10.1093/nar/gkf660
Pieterse, C. M., Leon-Reyes, A., Van Der Ent, S., and Van Wees, S. C. (2009). Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5, 308–316. doi: 10.1038/nchembio.164
Po-Wen, C., Singh, P., and Zimmerli, L. (2013). Priming of the Arabidopsis pattern-triggered immunity response upon infection by necrotrophic Pectobacterium carotovorum bacteria. Mol. Plant Pathol. 14, 58–70. doi: 10.1111/j.1364-3703.2012.00827.x
Rairdan, G. J., and Delaney, T. P. (2002). Role of salicylic acid and NIM1/NPR1 in race-specific resistance in Arabidopsis. Genetics 161, 803–811.
Saleh, A., Alvarez-Venegas, R., Yilmaz, M., Le, O., Hou, G., Sadder, M., et al. (2008). The highly similar Arabidopsis homologs of trithorax ATX1 and ATX2 encode proteins with divergent biochemical functions. Plant Cell 20, 568–579. doi: 10.1105/tpc.107.056614
Singh, P., Yekondi, S., Chen, P. W., Tsai, C. H., Yu, C. W., Wu, K., et al. (2014a). Environmental history modulates Arabidopsis pattern-triggered immunity in a HISTONE ACETYLTRANSFERASE1-Dependent manner. Plant Cell 26, 2676–2688. doi: 10.1105/tpc.114.123356
Singh, V., Roy, S., Singh, D., and Nandi, A. K. (2014b). Arabidopsis flowering locus D influences systemic-acquired-resistance- induced expression and histone modifications of WRKY genes. J. Biosci. 39, 119–126. doi: 10.1007/s12038-013-9407-7
Singh, V., Roy, S., Giri, M. K., Chaturvedi, R., Chowdhury, Z., Shah, J., et al. (2013). Arabidopsis thaliana FLOWERING LOCUS D is required for systemic acquired resistance. Mol. Plant Microbe Interact. 26, 1079–1088. doi: 10.1094/MPMI-04-13-0096-R
Smith, C. L., and Peterson, C. L. (2005). ATP-dependent chromatin remodeling. Curr. Top. Dev. Biol. 65, 115–148. doi: 10.1016/S0070-2153(04)65004-6
Thilmony, R., Underwood, W., and He, S. Y. (2006). Genome-wide transcriptional analysis of the Arabidopsis thaliana interaction with the plant pathogen Pseudomonas syringae pv. tomato DC3000 and the human pathogen Escherichia coli O157:H7. Plant J. 46, 34–53. doi: 10.1111/j.1365-313X.2006.02725.x
Thorstensen, T., Grini, P. E., and Aalen, R. B. (2011). SET domain proteins in plant development. Biochim. Biophys. Acta 1809, 407–420. doi: 10.1016/j.bbagrm.2011.05.008
van Hulten, M., Pelser, M., Van Loon, L. C., Pieterse, C. M., and Ton, J. (2006). Costs and benefits of priming for defense in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 5602–5607. doi: 10.1073/pnas.0510213103
van Loon, L. C., Rep, M., and Pieterse, C. M. (2006). Significance of inducible defense-related proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162. doi: 10.1146/annurev.phyto.44.070505.143425
Walley, J. W., Rowe, H. C., Xiao, Y., Chehab, E. W., Kliebenstein, D. J., Wagner, D., et al. (2008). The chromatin remodeler SPLAYED regulates specific stress signaling pathways. PLoS Pathog 4:e1000237. doi: 10.1371/journal.ppat.1000237
Wang, C., Gao, F., Wu, J., Dai, J., Wei, C., and Li, Y. (2010). Arabidopsis putative deacetylase AtSRT2 regulates basal defense by suppressing PAD4, EDS5 and SID2 expression. Plant Cell Physiol. 51, 1291–1299. doi: 10.1093/pcp/pcq087
Ward, E. R., Payne, G. B., Moyer, M. B., Williams, S. C., Dincher, S. S., Sharkey, K. C., et al. (1991). Differential Regulation of beta-1,3-Glucanase Messenger RNAs in Response to Pathogen Infection. Plant Physiol. 96, 390–397. doi: 10.1104/pp.96.2.390
Wu, K., Zhang, L., Zhou, C., Yu, C. W., and Chaikam, V. (2008). HDA6 is required for jasmonate response, senescence and flowering in Arabidopsis. J. Exp. Bot. 59, 225–234. doi: 10.1093/jxb/erm300
Xia, S., Cheng, Y. T., Huang, S., Win, J., Soards, A., Jinn, T. L., et al. (2013). Regulation of transcription of nucleotide-binding leucine-rich repeat-encoding genes SNC1 and RPP4 via H3K4 trimethylation. Plant Physiol. 162, 1694–1705. doi: 10.1104/pp.113.214551
Xu, L., Menard, R., Berr, A., Fuchs, J., Cognat, V., Meyer, D., et al. (2009). The E2 ubiquitin-conjugating enzymes, AtUBC1 and AtUBC2, play redundant roles and are involved in activation of FLC expression and repression of flowering in Arabidopsis thaliana. Plant J. 57, 279–288. doi: 10.1111/j.1365-313X.2008.03684.x
Xu, L., Zhao, Z., Dong, A., Soubigou-Taconnat, L., Renou, J. P., Steinmetz, A., et al. (2008). Di- and tri- but not monomethylation on histone H3 lysine 36 marks active transcription of genes involved in flowering time regulation and other processes in Arabidopsis thaliana. Mol. Cell. Biol. 28, 1348–1360. doi: 10.1128/MCB.01607-07
Yamamoto, H., Schoonjans, K., and Auwerx, J. (2007). Sirtuin functions in health and disease. Mol. Endocrinol. 21, 1745–1755. doi: 10.1210/me.2007-0079
Yang, S., and Hua, J. (2004). A haplotype-specific Resistance gene regulated by BONZAI1 mediates temperature-dependent growth control in Arabidopsis. Plant Cell 16, 1060–1071. doi: 10.1105/tpc.020479
Zhang, X., Bernatavichute, Y. V., Cokus, S., Pellegrini, M., and Jacobsen, S. E. (2009). Genome-wide analysis of mono-, di- and trimethylation of histone H3 lysine 4 in Arabidopsis thaliana. Genome Biol. 10:R62. doi: 10.1186/gb-2009-10-6-r62
Zhang, X., Clarenz, O., Cokus, S., Bernatavichute, Y. V., Pellegrini, M., Goodrich, J., et al. (2007a). Whole-genome analysis of histone H3 lysine 27 trimethylation in Arabidopsis. PLoS Biol. 5:e129. doi: 10.1371/journal.pbio.0050129
Zhang, X., Germann, S., Blus, B. J., Khorasanizadeh, S., Gaudin, V., and Jacobsen, S. E. (2007b). The Arabidopsis LHP1 protein colocalizes with histone H3 Lys27 trimethylation. Nat. Struct. Mol. Biol. 14, 869–871. doi: 10.1038/nsmb1283
Zhao, Z., Yu, Y., Meyer, D., Wu, C., and Shen, W. H. (2005). Prevention of early flowering by expression of FLOWERING LOCUS C requires methylation of histone H3 K36. Nat. Cell Biol. 7, 1256–1260. doi: 10.1038/ncb1329
Zhou, C., Zhang, L., Duan, J., Miki, B., and Wu, K. (2005). HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell 17, 1196–1204. doi: 10.1105/tpc.104.028514
Keywords: Chromatin, histone modification, chromatin remodeling, plant immunity
Citation: Ding B and Wang G-L (2015) Chromatin versus pathogens: the function of epigenetics in plant immunity. Front. Plant Sci. 6:675. doi: 10.3389/fpls.2015.00675
Received: 20 May 2015; Accepted: 15 August 2015;
Published: 02 September 2015.
Edited by:
Clelia De-la-Peña, Centro de Investigación Científica de Yucatán, MexicoReviewed by:
Gong-yin Ye, Zhejiang University, ChinaAna Elena Dorantes-Acosta, Universidad Veracruzana, Mexico
Federico Damian Ariel, Institute of Plant Science Paris-Saclay – Centre National de la Recherche Scientifique, France
Copyright © 2015 Ding and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guo-Liang Wang, Department of Plant Pathology, The Ohio State University, 2021 Coffey Road, Columbus, OH 43210, USA,d2FuZy42MjBAb3N1LmVkdQ==
 Bo Ding
Bo Ding Guo-Liang Wang
Guo-Liang Wang