- 1Botanical Institute and Cluster of Excellence on Plant Sciences, University of Cologne, Cologne, Germany
- 2Lehrstuhl für Genetik, Technische Universität München, Freising, Germany
The phytoalexin camalexin and indolic glucosinolates share not only a common evolutionary origin and a tightly interconnected biosynthetic pathway, but regulatory proteins controlling the shared enzymatic steps are also modulated by the same R2R3-MYB transcription factors. The indolic phytoalexin camalexin is a crucial defense metabolite in the model plant Arabidopsis. Indolic phytoalexins and glucosinolates appear to have a common evolutionary origin and are interconnected on the biosynthetic level: a key intermediate in the biosynthesis of camalexin, indole-3-acetaldoxime (IAOx), is also required for the biosynthesis of indolic glucosinolates and is under tight control by the transcription factors MYB34, MYB51, and MYB122. The abundance of camalexin was strongly reduced in myb34/51 and myb51/122 double and in triple myb mutant, suggesting that these transcription factors are important in camalexin biosynthesis. Furthermore, expression of MYB51 and MYB122 was significantly increased by biotic and abiotic camalexin-inducing agents. Feeding of the triple myb34/51/122 mutant with IAOx or indole-3-acetonitrile largely restored camalexin biosynthesis. Conversely, tryptophan could not complement the low camalexin phenotype of this mutant, which supports a role for the three MYB factors in camalexin biosynthesis upstream of IAOx. Consistently expression of the camalexin biosynthesis genes CYP71B15/PAD3 and CYP71A13 was not negatively affected in the triple myb mutant and the MYBs could not activate pCYP71B15::uidA expression in trans-activation assays with cultured Arabidopsis cells. In conclusion, this study reveals the importance of MYB factors regulating the generation of IAOx as precursor of camalexin.
Introduction
Phytoalexins are important defense compounds produced by plants in response to infection by a large variety of microorganisms. However, the elucidation of camalexin biosynthesis benefited from the fact that abiotic elicitors like silver nitrate (AgNO3; Glawischnig et al., 2004) and UV (Müller et al., 2015) strongly induce the camalexin production. Camalexin (3-thiazol-2′-yl-indole) is an indole alkaloid phytoalexin that is specific to a group of cruciferous species including the model plant Arabidopsis thaliana, but is absent in more distantly related Brassicaceae species (Glawischnig, 2007; Rauhut and Glawischnig, 2009; Bednarek et al., 2011). The induction of camalexin biosynthesis genes is strictly localized to sites of pathogen application, as demonstrated by quantitative RT-PCR and reporter-gene analysis and there is no evidence existing for long-distance camalexin transport (Schuhegger et al., 2007). During camalexin biosynthesis, tryptophan (Trp) is converted to indole-3-acetaldoxime (IAOx; Figure 1). This step is shared with the biosynthesis of other Trp-derived metabolites and is catalyzed by two homologous cytochrome P450 enzymes, CYP79B2, and CYP79B3 (Hull et al., 2000; Mikkelsen et al., 2000; Zhao et al., 2002). The resulting IAOx is a precursor of camalexin, indolic glucosinolates (IGs) and indole-carboxylic acids (ICAs; Böttcher et al., 2014). In camalexin biosynthesis, IAOx is dehydrated to indole-3-acetonitrile (IAN) by CYP71A12 and CYP71A13 (Figure 1; Müller et al., 2015). In accordance with their specific function in phytoalexin biosynthesis, both corresponding genes are expressed at very low levels in the absence of stress and are induced by pathogen infection, application of pathogen-associated molecular patterns (PAMPs), or by AgNO3 (Nafisi et al., 2007; Millet et al., 2010). IAN is also generated during the degradation of glucobrassicin (I3M; Burow et al., 2008) and it can be converted to indole-3-carbaldehyde (ICHO) and ICA by CYP71B6 (Böttcher et al., 2014) (Figure 1). Under specific conditions IAN serves also as a precursor for IAA (Kutz et al., 2002; Park et al., 2003). However, the IAN pools seem to be strictly seperated, thus IAN from I3M breakdown cannot serve as a precursor of camalexin, but only for ICAs, as it was shown with a TALEN generated cyp71A12 cyp71a13 double knockout (Müller et al., 2015). Notably, the cyp79b2/b3 double knockout mutant cannot synthesize camalexin (Zhao et al., 2002;Glawischnig et al., 2004), but this ability was recovered in a chemical complementation strategy by feeding the mutant with camalexin precursors such as IAN and dihydrocamalexic acid (DHCA; Schuhegger et al., 2006; Nafisi et al., 2007; Böttcher et al., 2009). In camalexin biosynthesis IAN is conjugated with glutathione (Nafisi et al., 2007; Parisy et al., 2007; Böttcher et al., 2009; Su et al., 2011). From this glutathione conjugate (GS-IAN) a cysteine conjugate Cys(IAN) is formed, involving γ-Glutamyl Peptidases 1 and 3 (GGP1/3; Geu-Flores et al., 2011), which is the substrate for CYP71B15/PAD3 (Zhou et al., 1999; Schuhegger et al., 2006; Böttcher et al., 2009).
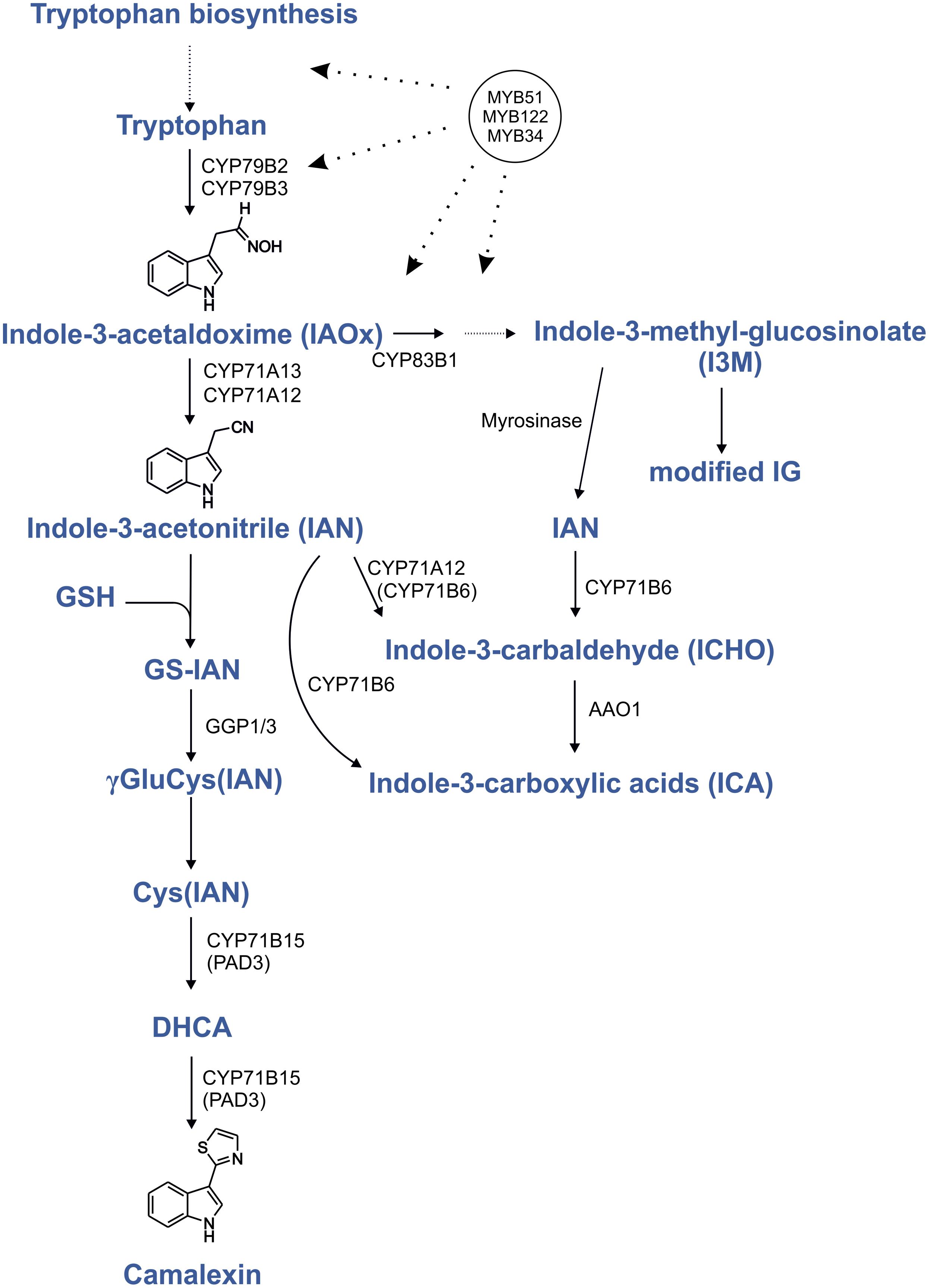
FIGURE 1. Regulation by MYB transcription factors in the camalexin biosynthesis pathway. Proven positive transcriptional regulation is shown by dotted lines with arrows. Modified from Böttcher et al. (2009), Geu-Flores et al. (2011), and Müller et al. (2015); indolic glucosinolates (IG); glutathione (GSH); dihydrocamalexic acid (DHCA).
Although the pathway leading to camalexin has been largely elucidated, its regulation remains less well understood. Perception of fungal pathogens such as Botrytis cinerea (Kliebenstein et al., 2005) and Alternaria alternata (Schuhegger et al., 2007) significantly activates camalexin production via mitogen-activated protein kinase (MAPK) cascade, which in turn phosphorylates MPK3 and MPK6 (Ren et al., 2008). Camalexin synthesis is almost completely blocked in the mpk3/6 double mutant after infection by B. cinerea (Ren et al., 2008). Mao et al. (2011) have demonstrated that the Arabidopsis transcription factor WRKY33 is a molecular target of the MPK3/6 cascade. wrky33 mutant can synthesize only very low amounts of camalexin, even in the MPK3/6 gain-of-function mutant. Furthermore, MPK4 physically interacts with MPK4 SUBSTRATE 1 (MKS1) and WRKY33 and represses WRKY33 function. Activated MPK4 phosphorylates MKS1, which in turn, releases WRKY33, which can then bind to the promoter of CYP71B15 (Qiu et al., 2008). Surprisingly, the respective wrky33 knock-out mutant contains low camalexin levels only at early stages of infection, but at later stages, contains even more camalexin than wild-type (WT) plants (Birkenbihl et al., 2012). Together, these results indicate that WRKY33 is one important regulator of camalexin, but that other regulators exist.
The transcription of NAC042 is strongly induced by AgNO3, a known inducer of camalexin biosynthesis, and the nac042 null mutant accumulates about 50% of WT camalexin levels after treatment with AgNO3 or B. cinerea (Saga et al., 2012). Furthermore, the induction of camalexin biosynthesis by acifluorfen, which generates reactive oxygen species (ROS), results in about 15% of the WT camalexin level in nac042, which highlights the key role of NAC042 in the ROS-dependent induction of camalexin biosynthesis (Saga et al., 2012).
To synthesize camalexin, it is essential that the specific genes (CYP71A12, CYP71A13, and CYP71B15) are upregulated together with the upstream Trp biosynthetic genes and CYP79B2. The known regulator of camalexin, WRKY33, binds to the promoters of CYP71B15 and CYP71A13 (Birkenbihl et al., 2012), whereas the regulators of IG biosynthesis, MYB34, MYB51, and MYB122 control genes of the shikimate pathway to Trp, i.e., anthranilate synthase α and β subunits, Trp synthases and CYP79B2 (Gigolashvili et al., 2007; Malitsky et al., 2008; Frerigmann and Gigolashvili, 2014). These MYB transcription factors thus positively regulate all the necessary steps for the production of the camalexin precursor IAOx. In addition to this intermediate, IG and camalexin biosynthesis share a glutathione conjugation step and the involvement of GGP1 (Geu-Flores et al., 2011), reflecting that camalexin biosynthesis likely has evolved from IG biosynthesis (Rauhut and Glawischnig, 2009; Bednarek et al., 2011). Therefore, MYB34, MYB51, and MYB122 possibly not only regulate the IG biosynthesis pathway, but also activate genes in the closely related camalexin biosynthesis pathway. Here we addressed the potential involvement of three MYB transcription factors in camalexin biosynthesis and show that especially MYB51 and MYB122 are involved in camalexin biosynthesis, because its synthesis is strongly reduced in corresponding double and triple mutants. Metabolite complementation of the triple myb34/51/122 mutant reveals the importance of these MYBs in the regulation of camalexin biosynthesis upstream of IAOx. Thus, camalexin and IGs not only possess a tightly interconnected biosynthetic pathway, but are at least partially regulated by the same R2R3-MYB transcription factors.
Results
Camalexin Biosynthesis Genes are Co-expressed with MYB51 and MYB122
Camalexin biosynthesis is induced locally by exposure to biotic or abiotic stresses and the genes involved in its biosynthesis are highly co-ordinately expressed. To address the role of MYB34, MYB51, and MYB122 in camalexin biosynthesis, we exploited existing co-expression databases like ATTED1 (Obayashi et al., 2009). The survey revealed that MYB51 and MYB122 are not only co-regulated with genes for Trp and IAOx biosynthesis, but also with CYP71B15/PAD3, CYP71A12, and CYP71A13 (Supplementary Tables S1 and S2). This implicates both MYB factors as good candidate regulators of camalexin biosynthesis in Arabidopsis.
MYB51 and MYB122 are Induced by Silver Nitrate and by Pathogen-Associated Molecular Pattern (PAMP) from Pythium aphanidermatum (PaNie)
To further validate the importance of R2R3-MYBs in camalexin regulation, we analyzed the induction of MYB34, MYB51, and MYB122 in response to elicitors of camalexin production. In a pilot experiment, we treated Arabidopsis Col-0 WT plants with AgNO3, a commonly used abiotic elicitor of camalexin induction, which strongly induced CYP71B15, CYP71A13, MYB122, and MYB51 (Figure 2). However, the expression of MYB34 was reduced, indicating that it plays a less important role in phytoalexin regulation.
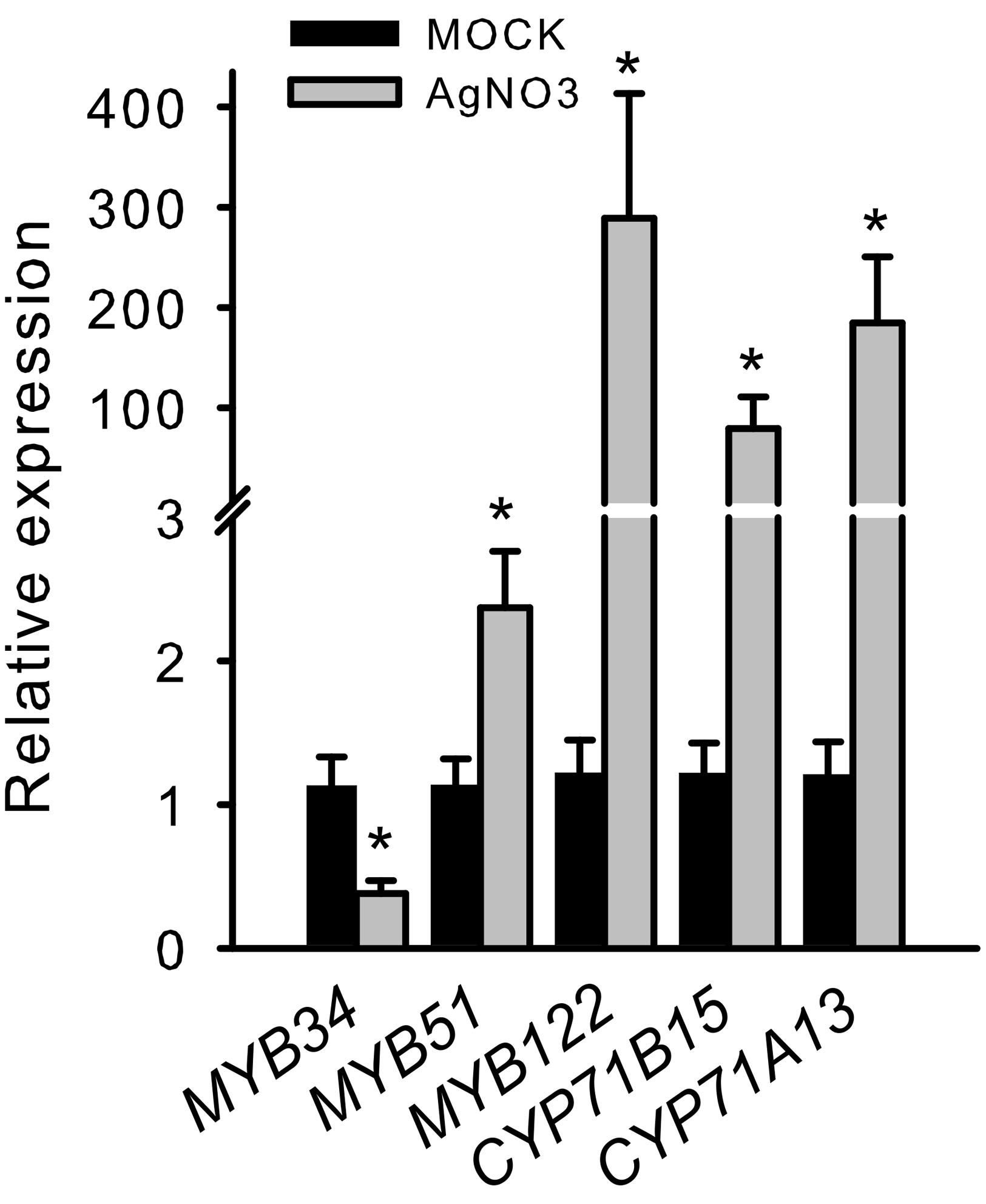
FIGURE 2. Silver nitrate induces the transcription of MYB51 and MYB122 as well as that of CYP71B15 and CYP71A13. The expression of camalexin biosynthesis genes (CYP71B15 and CYP71A13) and of MYB34, MYB51, and MYB122 upon silver nitrate (AgNO3) treatment is shown. The relative expression in Col-0 was measured in leaves of 6-week-old plants 18 h after treatment (MOCK = 1). Data are means ± SE from four independent experiments each with two to three biological replicates (n = 11). Values marked with asterisks are significantly different from those of control plants (Student’s t-test; p < 0.05).
In addition, we analyzed transgenic plants that expressed a gene encoding a Nep1-like protein from Pythium aphanidermatum (PaNie), which acts as a PAMP, under the control of an ethanol-inducible promoter (Rauhut et al., 2009). The production of this Nep1-like protein triggers the strong accumulation of camalexin 8 h following ethanol inductions (Rauhut et al., 2009). The transcription of MYB51 significantly increased 150 min after treatment, and that of CYP71B15, CYP71A13, and MYB122 after 300 min (5 h; Figure 3). Conversely, MYB34 was not induced by PaNie expression, which confirms its minor role in camalexin regulation. A similar induction pattern of the MYBs and camalexin genes was observed upon colonization with the fungus Piriformospora indica (Lahrmann et al., 2015). In addition, MYB51 transcription was also increased 40 and 88 h after infection with the necrotrophic pathogen B. cinerea, as revealed by the pMYB51::GUS reporter (Supplementary Figure S1).
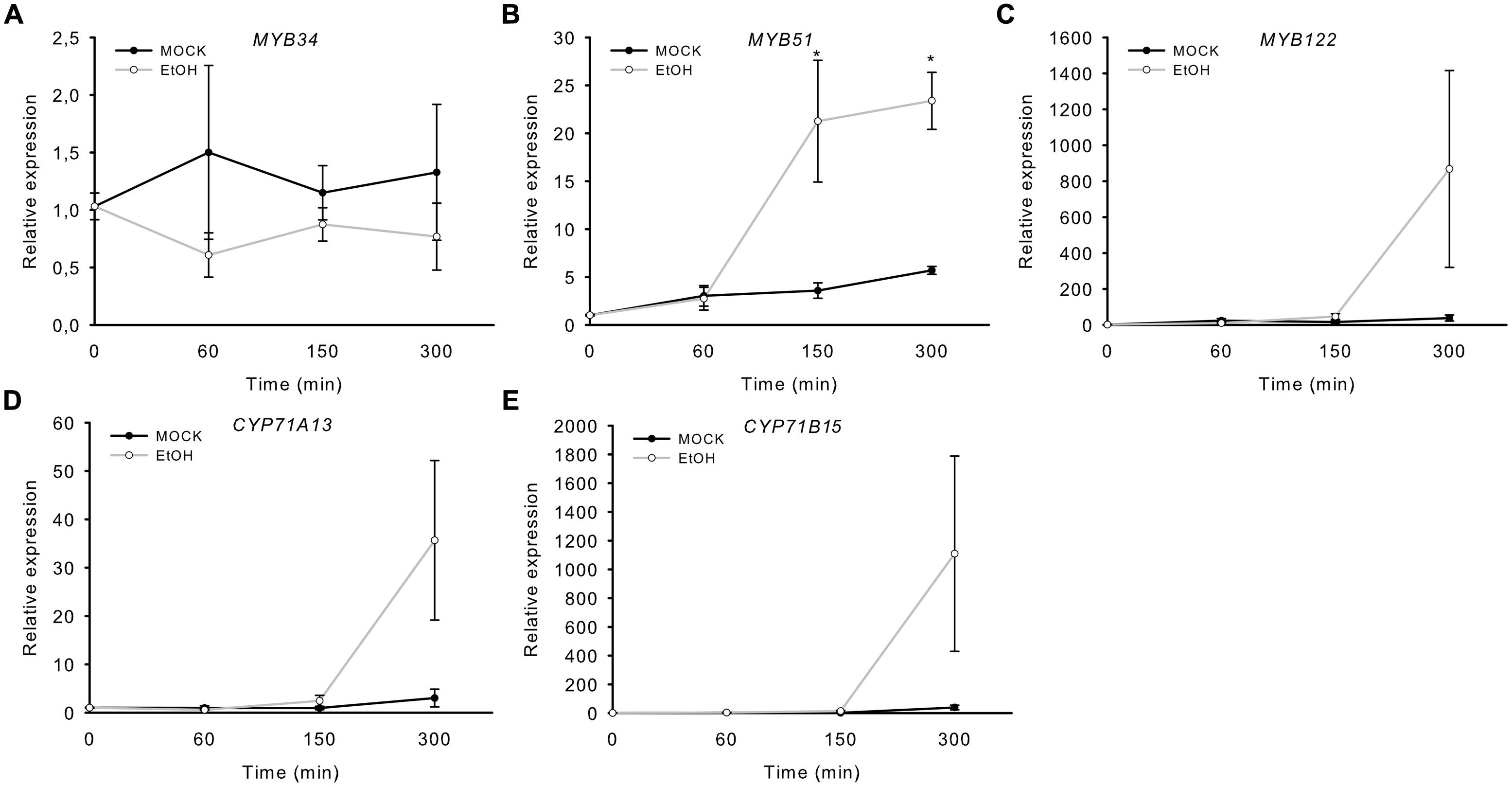
FIGURE 3. Induction of MYB51 and MYB122 in rosette leaves of Alc::PaNieDc plants. Expression of MYB34 (A), MYB51 (B), MYB122 (C), CYP71A13 (D), and the camalexin biosynthesis gene CYP71B15 (E) following AgNO3 treatment. Relative expression in pAlc::PaNieDc was measured in leaves of 6-week-old plants induced with ethanol (for 60 min, 150 min or 300 min; time point 0 = 1 min). Data are means ± SE from two independent experiments each with three biological replicates (n = 6). Values marked with asterisks are significantly different from those of control plants (Student’s t-test; p < 0.05).
Taken together, the expression patterns of MYB51 and MYB122 implicate a role in camalexin biosynthesis.
The Induction of MYB51 and MYB122 upon Wounding Coincides with that of the Camalexin Biosynthesis Gene CYP71B15
Wounding of the plant surface creates a potential entry point for invading pathogens, and plant response to injury by localized defense responses includes the induction of defense-related genes and the accumulation of anti-microbial proteins such as proteinase inhibitors, chitinase, or glucosinolates (Ryan, 1990; Chang et al., 1995; Reymond et al., 2000; Chassot et al., 2008). Especially strong wounding releases oligogalacturonides from the plant cell wall which can induce a local defense response similar to bacterial PAMPs (Denoux et al., 2008). Thus wounding of Arabidopsis leaves has been previously shown to lead to immunity to B. cinerea, because hyphal growth on wounded plants was significantly inhibited in comparison to that on unwounded controls (Chassot et al., 2008).
To address the involvement of the MYB34, MYB51, and MYB122 transcription factors in wounding response, the transcription of their respective genes was analyzed 1, 5, 10, 30, 120, and 300 min after strong wounding. Wounding of WT Arabidopsis leaves increased the transcription of MYB51 and MYB122, but not of MYB34 after 120 min of injury (Figure 4), which represented the time-point of increased expression of the camalexin biosynthesis gene CYP71B15 (Figure 3). This second induction peak of MYB51 and MYB122 transcription appears to be related to induction of camalexin biosynthesis. During the second phase of the wounding response, the transcript levels of MYB34 decreased, whereas expression of MYB51, MYB122, and CYP71B15 continued to increase and remained high even at 300 min (5 h) of treatment (Figure 4). The first peak in MYB transcription recorded after 5–10 min of injury observed in this study (Figure 3) and previously (Gigolashvili et al., 2007), was associated with an increase in IG biosynthesis.
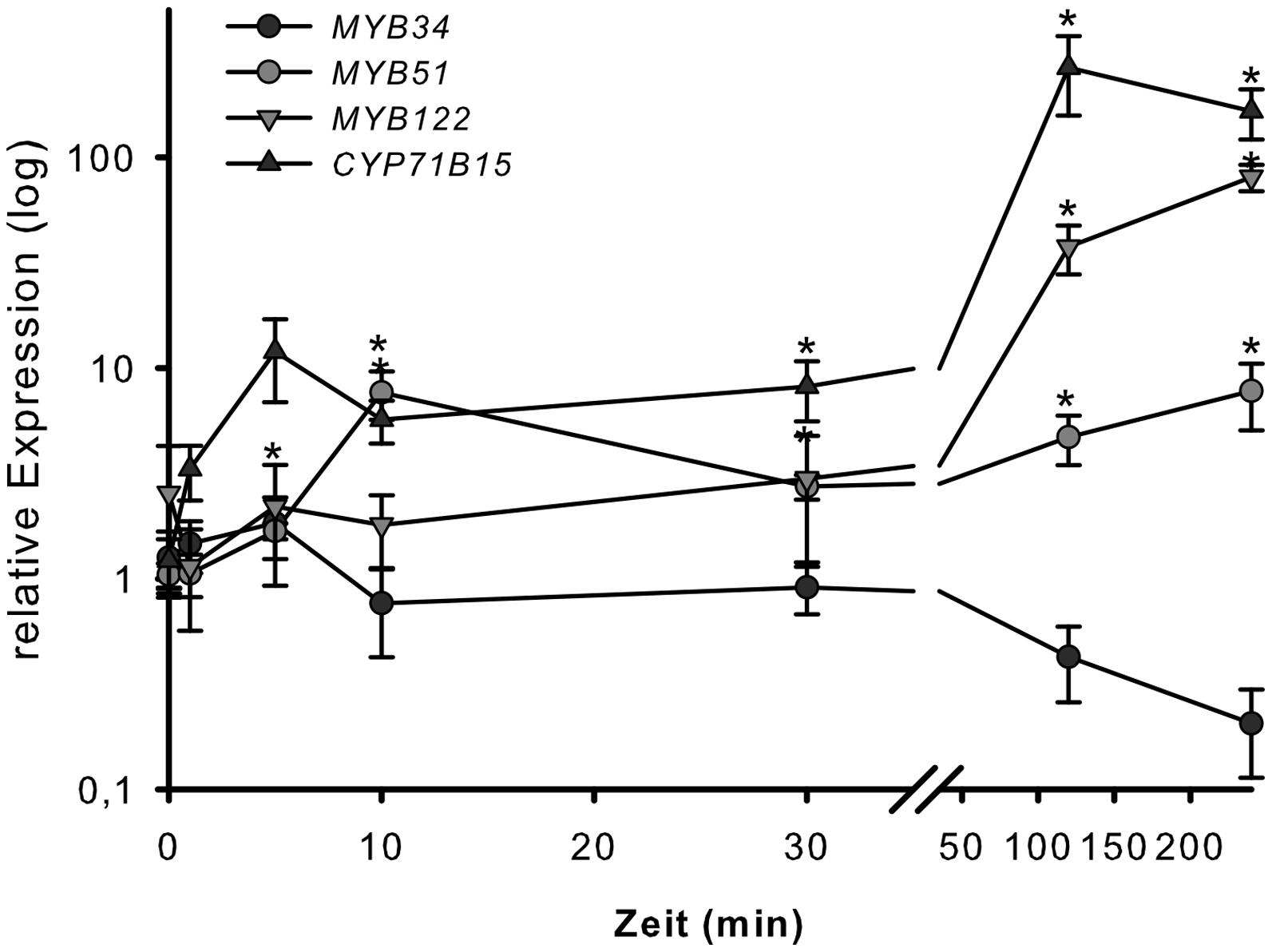
FIGURE 4. Wounding response of MYB34, MYB51, and MYB122 in leaves. Detached leaves of 6-week-old Col-0 plants grown under short day conditions were strongly wounded. Leaves were harvested after 1, 5, 10, 30, 120, and 300 min and processed for transcript analysis by qPCR. Relative transcript levels for MYB34, MYB51, MYB122, and CYP71B15 are shown for wounded vs. unwounded leaves (time-point 0 = 1 min). Data are means ± SE from three independent cultivations each with two biological replicates (n = 6). Values marked with asterisks are significantly different from the 0 time point (Student’s t-test; p < 0.05).
In order to confirm that the applied strong wounding does not resemble solely jasmonate signaling, as it is known for standard wounding application, hormone marker genes for jasmonate (VSP2), salicylic acid (PR1), and ethylene/jasmonate (PDF1.2) were checked (Supplementary Figure S2). As expected no induction, but even a repression of VSP2 was observed, while PR1 and PDF1.2 transcript levels increased similar to oligogalacturonide treatment (Denoux et al., 2008).
Together, these data suggest a role for MYB51 and MYB122 in priming camalexin biosynthesis at later stages of wounding response, to protect against biotic and abiotic stressors.
myb Mutants are Impaired in UV-Dependent Camalexin Induction
The abiotic elicitor UV can be easily applied to uniformly trigger camalexin induction in Col-0 (Müller et al., 2015 and Supplementary Figure S3B). We tested double and triple loss-of-function mutants of MYB51, MYB122, and MYB34 for their ability to synthesize camalexin after UV treatment. The camalexin content of leaves of double myb51/122, myb34/51, and triple myb34/51/122 mutants was strongly reduced after 18 h UV treatment (Figure 5), suggesting an important function for all three MYBs and especially MYB51 in camalexin accumulation. The myb34/122 double mutant showed only a minor and statistically non-significant reduction in camalexin accumulation. Camalexin levels were significantly lower in the triple myb34/51/122 mutant than in WT plants, but not in comparison to that of the myb34/51 and myb51/122 mutants. However, 24 h after UV treatment, only the myb51/122 double mutant and myb34/51/122 triple mutant contained significantly less camalexin than the WT (Supplementary Figure S3). These camalexin levels in myb mutant backgrounds confirm the importance of MYB51 and MYB122 in camalexin accumulation. The role of MYB34 appears to be minor.
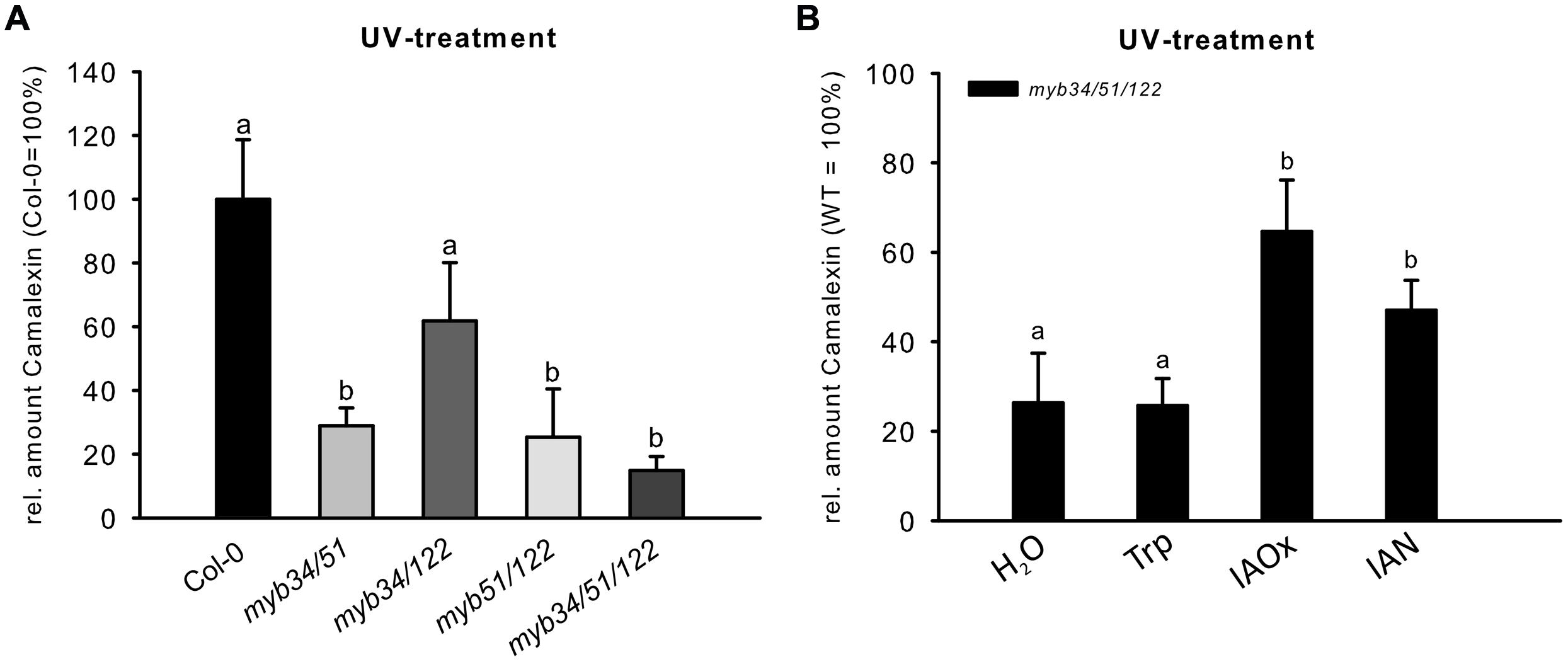
FIGURE 5. The UV-dependent induction of camalexin is impaired in multiple myb knock-out mutants. (A) The relative amount of camalexin after 18 h UV treatment in Col-0 and double and triple myb mutants (Col-0 = 100%). Data are means ± SE from two independent cultivations each with six biological replicates (n = 12). (B) The relative amount of camalexin 18 h after UV treatment in myb34/51/122 fed with H2O, or 0.25 mM Trp, IAOx or IAN (Col-0 = 100%). Data are means ± SE from three independent cultivations each with six biological replicates (n = 18). Different letters indicate significant differences at p < 0.05 (Kruskal Wallis Test, followed by a Mann Whitney U Test with Bonferroni-corrected p-values; p < 0.05).
The Camalexin Biosynthesis Genes CYP71B15 and CYP71A13 are not Downregulated in the Triple myb34/51/122 Mutant
The myb34/51/122 triple mutant is limited in the synthesis of IAOx, a precursor of IGs and camalexin and consequently, IGs (Frerigmann and Gigolashvili, 2014) and camalexin are reduced (Figure 5A). To investigate the role of MYBs on the expression of camalexin biosynthesis genes, the steady-state mRNA levels of CYP71B15 and CYP71A13 were analyzed in the triple myb34/51/122 mutant. If MYB51, MYB122, and MYB34 directly regulate camalexin biosynthesis genes, the expression of these genes should be significantly decreased in myb34/51/122, similar to that of IG biosynthesis genes.
Genes involved in IAOx biosynthesis were strongly down regulated in the myb34/51/122 mutant (Figure 6), whereas the expression of CYP71B15 and CYP71A13 either remained unchanged or increased. This increase in specific camalexin gene expression is not accompanied by higher levels of camalexin in the mutants (Supplementary Figure S3). The activity of pCYP79B2:uidA increased, whereas that of pCYP71B15:uidA was not affected by all three MYB factors, as demonstrated by co-expression via trans-activation assays with cultured Arabidopsis cells (Berger et al., 2007) (Figure 6B). Conversely, WRKY33, the transcription regulator of CYP71B15, induced pCYP71B15::uidA when co-expressed with p35S:WRKY33 in cultured cells. Thus, MYBs do not directly regulate these important camalexin biosynthesis genes downstream of IAOx.
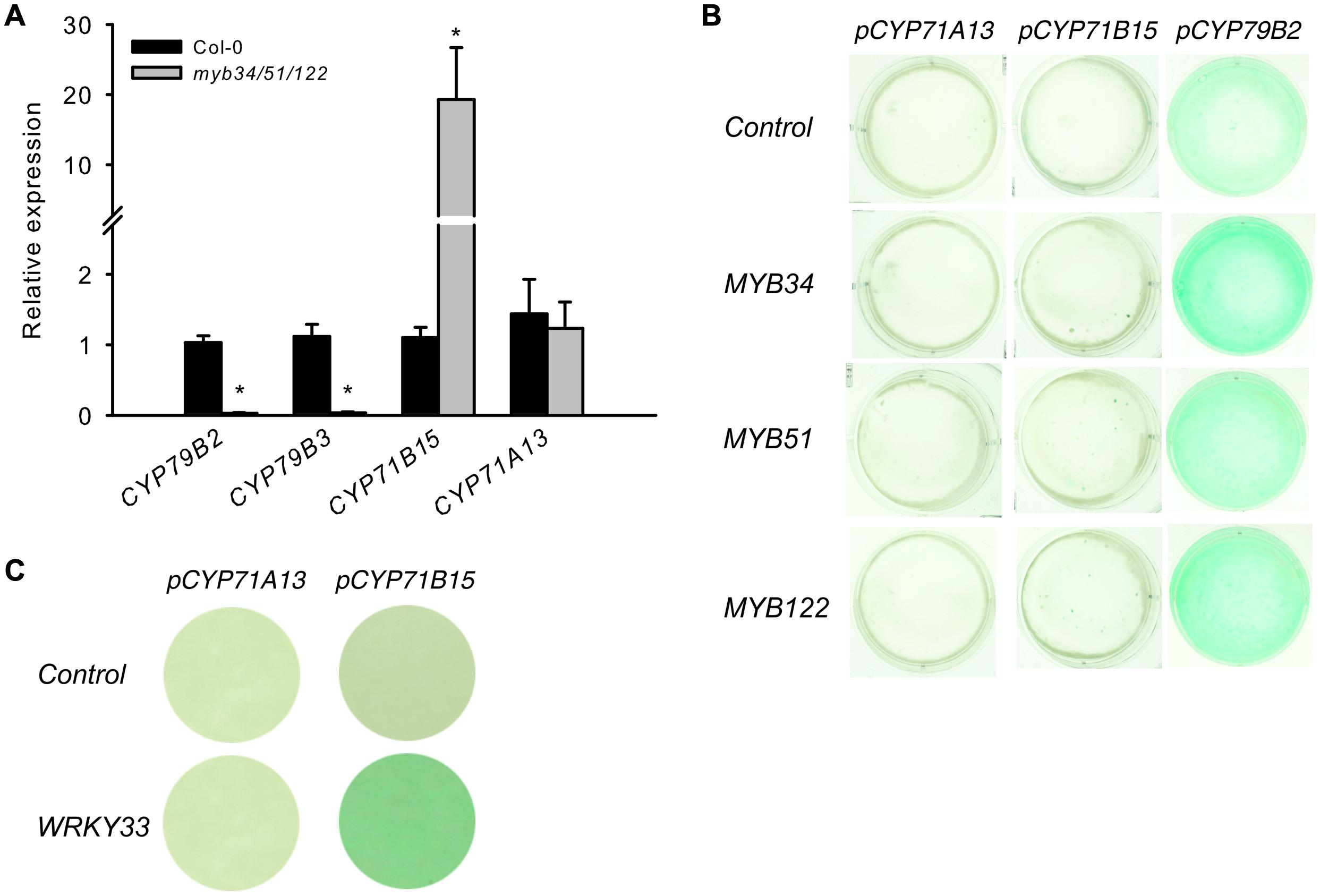
FIGURE 6. The presence of solely MYB34, MYB51, or MYB122 is not enough to activate the pCYP71B15:uidA and pCYP71A13:uidA in trans. (A) The expression of specific genes for camalexin biosynthesis (CYP71B15 and CYP71A13) and genes underlying the conversion of tryptophan to IAOx (CYP79B2 and CYP79B3) was analyzed in the myb34/51/122 mutant. Relative expression was measured in leaves of 6-week-old plants (Col-0 = 1). Data are means ± SE from three independent cultivations with three biological replicates (n = 9). Values marked with asterisks are significantly different from those of control plants (Student’s t-test; *p < 0.05). (B,C) Trans-activation with MYB34, MYB51, and MYB122 and target promoters of the camalexin biosynthesis pathway genes CYP79B2, CYP71A13, and CYP71B15. (B) The promoter–reporter constructs of pCYP71A13:uidA, pCYP71B15:uidA or pCYP79B2:uidA were co-expressed in the same cells with effector constructs p35S:MYB34, p35S:MYB51, or p35S:MYB122. The cultured A. thaliana cells were inoculated with the supervirulent Agrobacterium tumefaciens strain LBA4404.pBBR1MCS.virGN54D, containing either only the reporter construct or the reporter and effector construct in a 1:1 ratio. The GUS staining indicates trans-activation of the promoter by the effector. (C) The trans-activation potential of the p35S:WRKY33 effector toward the promoters of CYP71A13 and CYP71B15.
We also attempted to metabolically complement the low-camalexin phenotype of myb34/51/122 mutant leaves upon UV-treatment, by feeding them with IAOx, IAN or Trp. Treatment with IAOx or IAN partially restored camalexin levels in the myb34/51/122 mutant upon UV-treatment, whereas Trp feeding did not (Figure 5B). Because Trp could not complement the low-camalexin phenotype of the triple myb mutant, we conclude that the three MYB factors studied essentially regulate the synthesis of IAOx from Trp, but are not directly involved in the activation of genes downstream of IAOx.
Discussion
The camalexin biosynthetic pathway has been largely elucidated, but little is known about the regulatory components of this pathway. WRKY33 binds to the promoters of CYP71B15 and CYP71A13 to activate camalexin biosynthesis, but also other regulators have to be involved, because its loss of function leads to low camalexin levels only during early stages of pathogen infection (Birkenbihl et al., 2012). In this study, we addressed the role of the known IG regulators MYB34, MYB51, and MYB122 in the biosynthesis of camalexin in Arabidopsis. Because the camalexin and IG biosynthetic pathways have a common evolutionary origin and are tightly interconnected, these two classes of compounds might be regulated by the same set of transcription factors.
MYB51 and MYB122 are Induced by Biotic and Abiotic Triggers of Camalexin Biosynthesis
Camalexin biosynthesis is induced in plants following exposure to abiotic stresses such as heavy metal treatment or UV-C radiation or exposure to pathogens. We addressed the role of MYB34, MYB51 and MYB122 in camalexin biosynthesis by analysing their mRNA levels in plants exposed to several camalexin-inducing agents. Treatment of Arabidopsis WT plants with the abiotic elicitor AgNO3 caused a significant increase in steady-state mRNA levels of MYB122 and MYB51, but not of MYB34. Similarly, MYB122 and MYB51 were induced in transgenic plants that expressed a NEP1-like protein from PaNie under the control of an ethanol-inducible promoter (Rauhut et al., 2009), endorsing the possible role of these two transcription factors in camalexin biosynthesis. In addition, the MYB51 promoter was also induced after treatment with B. cinerea (Supplementary Figure S1). Finally, wounding of leaves, which is known to provide a protection against B. cinerea by priming camalexin production in Arabidopsis (Chassot et al., 2008), increased the expression of MYB51 and MYB122. Two induction peaks of MYB expression in response to wounding within the analyzed time scale occurred: the first peak in wounding response of MYB51 transcript level was observed after 5–10 min of injury (Gigolashvili et al., 2007) and was associated with a switch in the IG biosynthesis machinery, and the second phase of induction concerned the transcription of MYB51 and MYB122, but not MYB34, after 120 min of injury, which coincided with an strongly increased expression of the camalexin biosynthesis gene CYP71B15 (Figure 3). This second phase might therefore be related to camalexin biosynthesis. According to directed studies (Schuhegger et al., 2006; Chassot et al., 2008) and to the analysis of microarray data (see, e.g., efp browser analysis of ATH1 Affymetrix data2; Winter et al., 2007) CYP71B15 did not show strong responsiveness to wounding. In this light it was surprising that here CYP71B15 expression was induced more than 100-fold. We here applied rather harsh and extensive wounding to the tissue. Possibly severe wounding induces camalexin biosynthesis by eliciting oligogalacturonides, which originate from the plant cell wall (Denoux et al., 2008), while restricted wounding has a minor effect.
Taken together, the induced expression of MYB51 and/or MYB122 after exposure to biotic and abiotic triggers of camalexin biosynthesis [AgNO3, wounding, PAMP (PaNie) and the necrotrophic pathogen B. cinerea] suggests that the transcription factors encoded by these genes play a role in camalexin biosynthesis. Because the expression of MYB34 was not affected by the same treatments, we conclude that it is not involved in camalexin biosynthesis.
The Role of MYBs in the Regulation of IAOx – A Branch-Point in IG, Camalexin, ICA, and IAA Synthesis
The initial step of camalexin, IG, and ICA biosynthesis is the conversion of Trp to IAOx mediated by CYP79B2 and CYP79B3. The interplay between IAOx-derived metabolites was also demonstrated by the analysis of mutants deficient in IG biosynthesis. The loss of function of IG biosynthetic genes downstream of IAOx (cyp83b1/sur2, C-S lyase/sur1, and ugt74b1 null mutants) results in a strong auxin-overproducing phenotype (Barlier et al., 2000; Grubb et al., 2004; Mikkelsen et al., 2004). This is possibly due to IAOx accumulation in cells because of “biosynthetic blockage” in the IG pathway, and consequently, unspecific conversion of excess IAOx to the auxin IAA. Other metabolites such as ICA and camalexin, which can be also induced in these mutants, were not analyzed in the above-mentioned studies.
In the WT, biosynthesis of IAOx is under tight transcriptional control by MYB34, MYB51 and MYB122 transcription factors (Celenza et al., 2005; Gigolashvili et al., 2007; Frerigmann and Gigolashvili, 2014). Consequently the MYBs, especially the MYB51 and MYB122 are considered as candidates in the regulation of other Trp-derived metabolites than IGs, e.g., camalexin. We propose the following scenario for the role of MYB34, MYB51, and MYB122 in camalexin biosynthesis: they regulate genes involved in camalexin biosynthesis similar to how they regulate IG production. However, they have to act in concert with other regulators, since they are also highly expressed and regulate IG production in non-triggered tissue and would therefore lead to camalexin accumulation in healthy plants. We therefore suggest that specific signaling components exist upstream to these MYB factors, including alternative transcription factors, which activate different sets of genes for camalexin and IG biosynthesis. These different signaling components are responsive to AgNO3, PAMPs, and UV in the case of camalexin biosynthesis, and to herbivores regarding IG production. Thus, to enable camalexin biosynthesis, the MYBs and additional transcription factors are activated: MYB factors regulate IAOx biosynthesis, and alternative (unknown) regulators, together with WRKY33, control camalexin genes downstream of IAOX (e.g., CYP71B15 and CYP71A13).
The Regulation of Camalexin Biosynthesis by MYB51, MYB122, and MYB34
The analysis of camalexin accumulation in higher-order loss-of-function mutants of MYB51, MYB122, and MYB34 treated with UV revealed a strong reduction in the camalexin content of leaves of double myb51/122, myb34/51, and triple myb34/51/122 mutants (Figure 5), emphasizing the importance of MYB51 in camalexin accumulation in Arabidopsis. The role of MYB34 for camalexin induction was negligible, whereas MYB122 contributes camalexin biosynthesis, as demonstrated by the response of higher-order myb mutants after 24 h treatment with UV (Supplementary Figure S3). We propose the following explanation for the observed role of MYB122: (i) MYB122 is the lowest-expressed gene among the three MYBs (Frerigmann and Gigolashvili, 2014), therefore, the observed metabolic effects reflect its transcript abundance; (ii) transcription of MYB122 is positively regulated by MYB51, which is essential for camalexin biosynthesis. This positive correlation between MYB51 and MYB122 expression due to reciprocal regulation was previously demonstrated by the analysis of myb knock-out and overexpression plants (Frerigmann and Gigolashvili, 2014). The reciprocal activation of mRNAs of these two MYBs might play an important role in the regulation of camalexin biosynthesis.
To elucidate further the role of the MYBs, we performed a metabolic complementation experiment by feeding the UV-treated leaves of the camalexin-deficient mutant myb34/51/122 with the precursors Trp, IAOx, or IAN (Figure 5B). This experiment demonstrated that the three MYBs are essential to regulate the synthesis of IAOx from Trp during camalexin biosynthesis. However, they are not directly involved in the activation of genes downstream of IAOx, because both IAOx and IAN could complement the low camalexin phenotype of the myb34/51/122 mutant. These experiments suggest the possibility that MYB51 and MYB122 are indirectly involved in the activation of CYP71B15 or CYP71A13 by forming dynamic regulatory complexes with other transcription factors. However, even if the MYB factors interact with other transcription factors that regulate camalexin biosynthesis, they are not thought to activate CYP71B15 or CYP71A13. In support of this, qRT-PCR analysis of the triple myb34/51/122 mutant and the promoter–effector assays in cultured cells suggested that CYP71B15 and CYP71A13 are regulated independently from the MYB genes (Figure 6).
Taken together, the data substantiate the importance of three MYB factors in the regulation of camalexin biosynthesis by providing the precursor metabolite IAOx (Figure 1). There is no evidence for the direct MYB-mediated regulation of camalexin biosynthesis genes downstream of IAOx. The identification of the possible role of MYB51 and MYB122 in the activation of CYP71B15 or CYP71A13 in complex with other, yet to be identified transcription factors, is anticipated in the future.
Experimental Procedures
Arabidopsis Lines Used in this Study
The Arabidopsis loss-of-function mutants used in this study are all in the Columbia-0 (Col-0) genetic background. The T-DNA insertion mutants for MYB34, MYB51, MYB122 have been previously described and are myb34 [At5g60890; WiscDsLox424F3; (Frerigmann and Gigolashvili, 2014), myb51/hig1 (At1g18570; GK228B12; Gigolashvili et al., 2007), and myb122-2 (At1g74080; WiscDsLoxHs206_04H; Frerigmann et al., 2014). The multiple mutants were generated and described by Frerigmann and Gigolashvili (2014).
The ethanol-inducible overexpression line Alc::PaNieDc (Rauhut et al., 2009) and the pMYB51::GUS reporter line (Gigolashvili et al., 2007) were generated as described.
Biotic and Abiotic Treatments of Arabidopsis Leaves
For treatment with AgNO3, plants were grown for six weeks under short-day conditions. Pots with five plants were sprayed with AgNO3 or MOCK and harvested after 18 h in the dark [AgNO3 (5 mM AgNO3 + 0.02% Silver); MOCK (0.02% Silver)].
Expression of the NEP1-like protein in Alc::PaNieDc plants was induced by spraying with ethanol (2%) or with water for the MOCK samples. Samples were harvested at four different time points (0, 60, 150, and 300 min).
For wounding experiments, detached leaves of 6-week-old Col-0 plants were heavily crushed with forceps on the whole leaf, additionally strongly wounded with a scalpel and stored in a petri dish with wet paper tissue till harvest. After 0, 1, 5, 10, 30, 120, and 300 min leaves were frozen in liquid nitrogen and subsequent directed for RNA isolation and gene expression analysis by qRT-PCR. Wounding and storage for different time points had no effect on ACTIN2 levels.
Five-week-old plants were infected with a 6 μL droplet of B. cinerea spores (2 × 106 spores/μL in LB-media) or LB-media as MOCK. After infection, plants remained under short-day conditions but with a relative humidity of about 100%. Samples were harvested at different time points (0, 40, 88 h) and fixed immediately with ice-cold acetone. GUS staining was performed overnight at 37°C. Histochemical localisation of GUS in transgenic plants harboring the pMYB51::uidA construct was performed as described Gigolashvili et al. (2007).
UV-Treatment, Metabolite Feeding and Camalexin Measurement
For UV induction, leaves were cut at the base of the petioles and placed on wet tissue paper under a UV-lamp (Desaga UV-VIS, = 254 nm, 8 W) at a distance of 20 cm and were irradiated for 2 h (Mucha et al., 2015). Camalexin extraction and HPLC-analysis was performed essentially as previously described (Schuhegger et al., 2006). Leaves were extracted twice in 200 μl MeOH/H2O (4:1; v/v) at 65°C for 30 min. Combined extracts were centrifuged at 17,000 g for 15 min and analyzed by reverse phase HPLC (LiChroCART 250-4, RP-18, 5 μm, Merck; 1 mL⋅min-1; MeOH/H2O (1:1; v/v) for 2 min, followed by a 10 min linear gradient to 100% MeOH, followed by 3 min 100% MeOH). Camalexin was quantified using a Shimadzu F-10AXL fluorescence detector (318 nm excitation, 370 nm emission) and by UV absorption at 318 nm applying a calibration curve with authentic standard. For intermediate feeding leaves were detached at the petiole after 2 h UV treatment and incubated in 400 μl 0.25 mM precursor solution or water for an additional 16 h.
RNA Extraction and qRT-PCR
Total RNA extraction and qRT-PCR analysis were as described by Frerigmann and Gigolashvili (2014). The relative quantification of expression levels was performed using the comparative delta Ct method, and the calculated relative expression values were normalized to that of ACTIN2 and compared with the expression level in untreated WT plants (Col-0 = 1). When not specified in the figure legend, three technical replicates and three biological replicates from independently grown plants were analyzed (for primer sequences see Supplementary Table S3).
Plant Growth Conditions
Seeds of A. thaliana ecotype Col-0 and mutant lines were stratified for 2–7 days in the dark at 4°C to break seed dormancy. Plants were grown in growth cabinets with a light/dark cycle of 8 h/16 h and a day/night temperature of 21°C/18°C, 40% humidity and a mean photon flux density of 150 μmol m-2 s-1. A minimum of 100 mg rosette material was harvested from 6-week-old plants, immediately frozen in liquid nitrogen and kept at –80°C until RNA extraction or metabolite analysis.
Reporter Construction for Transient Co-transformation Experiments
The promoter regions of CYP71B15 (At3g26830; from –1,593 to +58 bp) and CYP71A13 (At1g73500; from –2,124 to +42 bp) were amplified from genomic DNA of Arabidopsis plants and cloned into the pEntry TOPO vector (Invitrogen). The construction of the CYP79B2 promoter was performed as described (Gigolashvili et al., 2007). The corresponding primer sequences are listed in Supplementary Table S4. The binary plant transformation vector pGWB3i containing an intron within the uidA gene was used to drive Agrobacterium-mediated expression of uidA from these promoters and pGWB3i was recombined with the pEntry Topo vectors containing the promoter of interest using LR reactions (Invitrogen). The final pCYP71B15::uidA, pCYP71A13::uidA and pCYP79B2::uidA clones in pGWB3i, as well as p35S::MYB34, p35S::MYB51, p35S::MYB122, and p35S:WRKY33 in pGWB2 were used to transform the supervirulent Agrobacterium tumefaciens strain LBA4404.pBBR1MCS.virGN54D as described by Berger et al. (2007).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Alexandra Chapman and Ulrike Hebbeker for practical assistance and Dr. John Chandler for critically reading the manuscript. We also cordially thank Prof. Dr. Ulf-Ingo Flügge for his continuous support over many years. This work was financially supported by the Deutsche Forschungsgemeinschaft (Project Reference Numbers: GI 824/1-1, EXC 1028, and GL346/5-1, Heisenberg fellowship to EG).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fpls.2015.00654
Footnotes
References
Barlier, I., Kowalczyk, M., Marchant, A., Ljung, K., Bhalerao, R., Bennett, M., et al. (2000). The SUR2 gene of Arabidopsis thaliana encodes the cytochrome P450 CYP83B1, a modulator of auxin homeostasis. Proc. Natl. Acad. Sci. U.S.A. 97, 14819–14824. doi: 10.1073/pnas.260502697
Bednarek, P., Piślewska-Bednarek, M., Ver Loren van Themaat, E., Maddula, R. K., Svatoš, A., and Schulze-Lefert, P. (2011). Conservation and clade-specific diversification of pathogen-inducible tryptophan and indole glucosinolate metabolism in Arabidopsis thaliana relatives. New Phytol. 192, 713–726. doi: 10.1111/j.1469-8137.2011.03824.x
Berger, B., Stracke, R., Yatusevich, R., Weisshaar, B., Flugge, U. I., and Gigolashvili, T. (2007). A simplified method for the analysis of transcription factor-promoter interactions that allows high-throughput data generation. Plant J. 50, 911–916. doi: 10.1111/j.1365-313X.2007.03088.x
Birkenbihl, R. P., Diezel, C., and Somssich, I. E. (2012). Arabidopsis WRKY33 is a key transcriptional regulator of hormonal and metabolic responses towards Botrytis cinerea infection. Plant Physiol. 159, 266–285. doi: 10.1104/pp.111.192641
Böttcher, C., Chapman, A., Fellermeier, F., Choudhary, M., Scheel, D., and Glawischnig, E. (2014). The biosynthetic pathway of indole-3-carbaldehyde and indole-3-carboxylic acid derivatives in Arabidopsis. Plant Physiol. 165, 841–853. doi: 10.1104/pp.114.235630
Böttcher, C., Westphal, L., Schmotz, C., Prade, E., Scheel, D., and Glawischnig, E. (2009). The multifunctional enzyme CYP71B15 (PHYTOALEXIN DEFICIENT3) converts cysteine-indole-3-acetonitrile to camalexin in the indole-3-acetonitrile metabolic network of Arabidopsis thaliana. Plant Cell 21, 1830–1845. doi: 10.1105/tpc.109.066670
Burow, M., Zhang, Z.-Y., Ober, J. A., Lambrix, V. M., Wittstock, U., Gershenzon, J., et al. (2008). ESP and ESM1 mediate indol-3-acetonitrile production from indol-3-ylmethyl glucosinolate in Arabidopsis. Phytochemistry 69, 663–671. doi: 10.1016/j.phytochem.2007.08.027
Celenza, J. L., Quiel, J. A., Smolen, G. A., Merrikh, H., Silvestro, A. R., Normanly, J., et al. (2005). The Arabidopsis ATR1 MYB transcription factor controls indolic glucosinolate homeostasis. Plant Physiol. 137, 253–262. doi: 10.1104/pp.104.054395
Chang, M.-M., Horovitz, D., Culley, D., and Hadwiger, L. A. (1995). Molecular cloning and characterization of a pea chitinase gene expressed in response to wounding, fungal infection and the elicitor chitosan. Plant Mol. Biol. 28, 105–111. doi: 10.1007/BF00042042
Chassot, C., Buchala, A., Schoonbeek, H., Metraux, J. P., and Lamotte, O. (2008). Wounding of Arabidopsis leaves causes a powerful but transient protection against Botrytis infection. Plant J. 55, 555–567. doi: 10.1111/j.1365-313X.2008.03540.x
Denoux, C., Galletti, R., Mammarella, N., Gopalan, S., Werck, D., De Lorenzo, G., et al. (2008). Activation of defense response pathways by OGs and Flg22 elicitors in Arabidopsis seedlings. Mol. Plant 1, 423–445. doi: 10.1093/mp/ssn019
Frerigmann, H., Berger, B., and Gigolashvili, T. (2014). bhlh05 is an interaction partner of myb51 and a novel regulator of glucosinolate biosynthesis in Arabidopsis. Plant Physiol. 166, 349–369. doi: 10.1104/pp.114.240887
Frerigmann, H., and Gigolashvili, T. (2014). MYB34, MYB51 and MYB122 distinctly regulate indolic glucosinolate biosynthesis in Arabidopsis thaliana. Mol. Plant 7, 814–828. doi: 10.1093/mp/ssu004
Geu-Flores, F., Møldrup, M. E., Böttcher, C., Olsen, C. E., Scheel, D., and Halkier, B. A. (2011). Cytosolic γ-glutamyl peptidases process glutathione conjugates in the biosynthesis of glucosinolates and camalexin in Arabidopsis. Plant Cell 23, 2456–2469. doi: 10.1105/tpc.111.083998
Gigolashvili, T., Berger, B., Mock, H. P., Müller, C., Weisshaar, B., and Flügge, U. I. (2007). The transcription factor HIG1/MYB51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J. 50, 886–901. doi: 10.1111/j.1365-313X.2007.03099.x
Glawischnig, E., Hansen, B. G., Olsen, C. E., and Halkier, B. A. (2004). Camalexin is synthesized from indole-3-acetaldoxime, a key branching point between primary and secondary metabolism in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 101, 8245–8250. doi: 10.1073/pnas.0305876101
Grubb, C. D., Zipp, B. J., Ludwig-Muller, J., Masuno, M. N., Molinski, T. F., and Abel, S. (2004). Arabidopsis glucosyltransferase UGT74B1 functions in glucosinolate biosynthesis and auxin homeostasis. Plant J. 40, 893–908. doi: 10.1111/j.1365-313X.2004.02261.x
Hull, A. K., Vij, R., and Celenza, J. L. (2000). Arabidopsis cytochrome P450s that catalyze the first step of tryptophan-dependent indole-3-acetic acid biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 97, 2379–2384. doi: 10.1073/pnas.040569997
Kliebenstein, D. J., Rowe, H. C., and Denby, K. J. (2005). Secondary metabolites influence Arabidopsis/Botrytis interactions: variation in host production and pathogen sensitivity. Plant J. 44, 25–36. doi: 10.1111/j.1365-313X.2005.02508.x
Kutz, A., Muller, A., Hennig, P., Kaiser, W. M., Piotrowski, M., and Weiler, E. W. (2002). A role for nitrilase 3 in the regulation of root morphology in sulphur-starving Arabidopsis thaliana. Plant J. 30, 95–106. doi: 10.1046/j.1365-313X.2002.01271.x
Lahrmann, U., Strehmel, N., Langen, G., Frerigmann, H., Leson, L., Ding, Y., et al. (2015). Mutualistic root endophytism is not associated with the reduction of saprotrophic traits and requires a noncompromised plant innate immunity. New Phytol. 207, 841–857. doi: 10.1111/nph.13411
Malitsky, S., Blum, E., Less, H., Venger, I., Elbaz, M., Morin, S., et al. (2008). The transcript and metabolite networks affected by the two clades of Arabidopsis glucosinolate biosynthesis regulators. Plant Physiol. 148, 2021–2049. doi: 10.1104/pp.108.124784
Mao, G., Meng, X., Liu, Y., Zheng, Z., Chen, Z., and Zhang, S. (2011). Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 23, 1639–1653. doi: 10.1105/tpc.111.084996
Mikkelsen, M. D., Hansen, C. H., Wittstock, U., and Halkier, B. A. (2000). Cytochrome P450 CYP79B2 from Arabidopsis catalyzes the conversion of tryptophan to indole-3-acetaldoxime, a precursor of indole glucosinolates and indole-3-acetic acid. J. Biol. Chem. 275, 33712–33717. doi: 10.1074/jbc.M001667200
Mikkelsen, M. D., Naur, P., and Halkier, B. A. (2004). Arabidopsis mutants in the C-S lyase of glucosinolate biosynthesis establish a critical role for indole-3-acetaldoxime in auxin homeostasis. Plant J. 37, 770–777. doi: 10.1111/j.1365-313X.2004.02002.x
Millet, Y. A., Danna, C. H., Clay, N. K., Songnuan, W., Simon, M. D., Werck-Reichhart, D., et al. (2010). Innate immune responses activated in Arabidopsis roots by microbe-associated molecular patterns. Plant Cell 22, 973–990. doi: 10.1105/tpc.109.069658
Mucha, S., Walther, D., Müller, T., Hincha, D. K., and Glawischnig, E. (2015). Substantial reprogramming of the Eutrema salsugineum (Thellungiella salsuginea) transcriptome in response to UV and silver nitrate challenge. BMC Plant Biol. 15:137. doi: 10.1186/s12870-015-0506-5
Müller, T. M., Böttcher, C., Morbitzer, R., Götz, C. C., Lehmann, J., Lahaye, T., et al. (2015). TALEN-mediated generation and metabolic analysis of camalexin-deficient cyp71a12 cyp71a13 double knockout lines. Plant Physiol. 168, 849–858. doi: 10.1104/pp.15.00481
Nafisi, M., Goregaoker, S., Botanga, C. J., Glawischnig, E., Olsen, C. E., Halkier, B. A., et al. (2007). Arabidopsis cytochrome P450 monooxygenase 71A13 catalyzes the conversion of indole-3-acetaldoxime in camalexin synthesis. Plant Cell 19, 2039–2052. doi: 10.1105/tpc.107.051383
Obayashi, T., Hayashi, S., Saeki, M., Ohta, H., and Kinoshita, K. (2009). ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 37, D987–D991. doi: 10.1093/nar/gkn807
Parisy, V., Poinssot, B., Owsianowski, L., Buchala, A., Glazebrook, J., and Mauch, F. (2007). Identification of PAD2 as a γ-glutamylcysteine synthetase highlights the importance of glutathione in disease resistance of Arabidopsis. Plant J. 49, 159–172. doi: 10.1111/j.1365-313X.2006.02938.x
Park, W. J., Kriechbaumer, V., Müller, A., Piotrowski, M., Meeley, R. B., Gierl, A., et al. (2003). The nitrilase ZmNIT2 converts indole-3-acetonitrile to indole-3-acetic acid. Plant Physiol. 133, 794–802. doi: 10.1104/pp.103.026609
Qiu, J.-L., Fiil, B. K., Petersen, K., Nielsen, H. B., Botanga, C. J., Thorgrimsen, S., et al. (2008). Arabidopsis MAP kinase 4 regulates gene expression through transcription factor release in the nucleus. EMBO J. 27, 2214–2221. doi: 10.1038/emboj.2008.147
Rauhut, T., and Glawischnig, E. (2009). Evolution of camalexin and structurally related indolic compounds. Phytochemistry 70, 1638–1644. doi: 10.1016/j.phytochem.2009.05.002
Rauhut, T., Luberacki, B., Seitz, H. U., and Glawischnig, E. (2009). Inducible expression of a Nep1-like protein serves as a model trigger system of camalexin biosynthesis. Phytochemistry 70, 185–189. doi: 10.1016/j.phytochem.2008.12.010
Ren, D., Liu, Y., Yang, K. Y., Han, L., Mao, G., Glazebrook, J., et al. (2008). A fungal-responsive MAPK cascade regulates phytoalexin biosynthesis in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 105, 5638–5643. doi: 10.1073/pnas.0711301105
Reymond, P., Weber, H., Damond, M., and Farmer, E. E. (2000). Differential gene expression in response to mechanical wounding and insect feeding in Arabidopsis. Plant Cell 12, 707–719. doi: 10.1105/tpc.12.5.707
Ryan, C. A. (1990). Protease inhibitors in plants: genes for improving defenses against insects and pathogens. Annu. Rev. Phytopathol. 28, 425–449. doi: 10.1146/annurev.py.28.090190.002233
Saga, H., Ogawa, T., Kai, K., Suzuki, H., Ogata, Y., Sakurai, N., et al. (2012). Identification and characterization of ANAC042, a transcription factor family gene involved in the regulation of camalexin biosynthesis in Arabidopsis. Mol. Plant Microbe Interact. 25, 684–696. doi: 10.1094/MPMI-09-11-0244
Schuhegger, R., Nafisi, M., Mansourova, M., Petersen, B. L., Olsen, C. E., Svatos, A., et al. (2006). CYP71B15 (PAD3) catalyzes the final step in camalexin biosynthesis. Plant Physiol. 141, 1248–1254. doi: 10.1104/pp.106.082024
Schuhegger, R., Rauhut, T., and Glawischnig, E. (2007). Regulatory variability of camalexin biosynthesis. J. Plant Physiol. 164, 636–644. doi: 10.1016/j.jplph.2006.04.012
Su, T., Xu, J., Li, Y., Lei, L., Zhao, L., Yang, H., et al. (2011). Glutathione-indole-3-acetonitrile is required for camalexin biosynthesis in Arabidopsis thaliana. Plant Cell 23, 364–380. doi: 10.1105/tpc.110.079145
Winter, D., Vinegar, B., Nahal, H., Ammar, R., Wilson, G. V., and Provart, N. J. (2007). An “electronic fluorescent pictograph” browser for exploring and analyzing large-scale biological data sets. PLoS ONE 2:e718. doi: 10.1371/journal.pone.0000718
Zhao, Y., Hull, A. K., Gupta, N. R., Goss, K. A., Alonso, J., Ecker, J. R., et al. (2002). Trp-dependent auxin biosynthesis in Arabidopsis: involvement of cytochrome P450s CYP79B2 and CYP79B3. Genes Dev. 16, 3100–3112. doi: 10.1101/gad.1035402
Keywords: camalexin biosynthesis, transcriptional regulation, MYB51, MYB122, MYB34
Citation: Frerigmann H, Glawischnig E and Gigolashvili T (2015) The role of MYB34, MYB51 and MYB122 in the regulation of camalexin biosynthesis in Arabidopsis thaliana. Front. Plant Sci. 6:654. doi: 10.3389/fpls.2015.00654
Received: 15 June 2015; Accepted: 07 August 2015;
Published: 25 August 2015.
Edited by:
Ute Roessner, The University of Melbourne, AustraliaCopyright © 2015 Frerigmann, Glawischnig and Gigolashvili. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tamara Gigolashvili, Botanical Institute and Cluster of Excellence on Plant Sciences, University of Cologne, BioCenter, D-50674 Cologne, Germany, t.gigolashvili@uni-koeln.de
 Henning Frerigmann
Henning Frerigmann Erich Glawischnig
Erich Glawischnig Tamara Gigolashvili
Tamara Gigolashvili