- 1Department of Plant Pathology, Kansas State University, Manhattan, KS, USA
- 2Temasek Life Sciences Laboratory, National University of Singapore, Singapore, Singapore
- 3Department of Plant Pathology, University of Florida, Gainesville, FL, USA
Transcription activator-like (TAL) effectors are bacterial type III secretion proteins that function as transcription factors in plants during Xanthomonas/plant interactions, conditioning either host susceptibility and/or host resistance. Three types of TAL effector associated resistance (R) genes have been characterized—recessive, dominant non-transcriptional, and dominant TAL effector-dependent transcriptional based resistance. Here, we discuss the last type of R genes, whose functions are dependent on direct TAL effector binding to discrete effector binding elements in the promoters. Only five of the so-called executor R genes have been cloned, and commonalities are not clear. We have placed the protein products in two groups for conceptual purposes. Group 1 consists solely of the protein from pepper, BS3, which is predicted to have catalytic function on the basis of homology to a large conserved protein family. Group 2 consists of BS4C-R, XA27, XA10, and XA23, all of which are relatively short proteins from pepper or rice with multiple potential transmembrane domains. Group 2 members have low sequence similarity to proteins of unknown function in closely related species. Firm predictions await further experimentation on these interesting new members to the R gene repertoire, which have potential broad application in new strategies for disease resistance.
Introduction
Xanthomonas infects monocotyledonous and dicotyledonous plant species, and the pathogenicity of many species depends in part on the effector proteins secreted by a type III secretion (T3S) system (Leyns et al., 1984; Tampakaki et al., 2004). The transcription activator-like (TAL) effector family is a distinct family of type III effectors, which includes members with cognate susceptibility (S) and/or resistance (R) genes. TAL effectors function as host gene specific transcription factors that can target both S and R genes, leading to enhanced expression and consequential phenotypic effects (Gu et al., 2005; Yang et al., 2006; Kay et al., 2007; Römer et al., 2007). Susceptibility (S) genes are genes with TAL effector-dependent expression and have measurable effects on disease symptoms (Yang et al., 2006; Boch et al., 2014). TAL effector genes are limited to members of the genus Xanthomonas and Ralstonia (Hopkins et al., 1992; De Feyter et al., 1993; Salanoubat et al., 2002; Heuer et al., 2007). The genes are ubiquitous in some species and have apparent critical functions in a number of diseases (Yang and White, 2004; Cernadas et al., 2014; Cohn et al., 2014; Hu et al., 2014; Schwartz et al., 2015).
Three types of TAL effector associated R genes have been reported-recessive, dominant non-transcriptional (classical) and dominant TAL effector-dependent transcriptional based resistance. TAL effector-dependent recessive resistance occurs in rice lines with DNA polymorphisms in S gene effector binding elements and will not be discussed in detail here (Hutin et al., 2015). Dominant non-transcriptional based resistance is represented solely by the NBS-LRR resistance gene from tomato, Bs4, which was identified as the cognate R gene to the TAL effector gene avrBsP/avrBs4 (Bonas et al., 1993; Schornack et al., 2004). However, a transcriptionally functional TAL effector is not required for Bs4 resistance elicitation as truncated versions of the cognate avirulence gene also trigger resistance. Here, we discuss the third type, namely, TAL effector-dependent R genes that are both direct targets of TAL effectors in the host and identified as R genes. The genes have been referred to as terminator or, here, executor R (E) genes (Bogdanove et al., 2010; Tian et al., 2014). E gene expression, like Avr/R gene interactions, is associated with hypersensitive response (HR) on the respective host plants and restricts pathogen growth at the site of infection. Five E genes and the cognate TAL effector genes have been cloned, including Xa27, Bs3, Bs4C-R, Xa10, and Xa23 (Gu et al., 2005; Römer et al., 2007; Strauss et al., 2012; Tian et al., 2014; Wang et al., 2015). The TAL effector AvrXa7 may target an as yet uncharacterized E gene Xa7 due to the requirements for the effector nuclear localization signals (NLSs) and the transcription acidic activator domain in Xa7-dependent resistance (Hopkins et al., 1992; Yang et al., 2000).
E Gene Variation is in the Promoter
E genes are unique in the panoply of R genes in that specificity is not in the R gene coding sequence but in the expression of the R gene in the presence of the effector (Gu et al., 2005; Römer et al., 2007). The TAL effector, itself, contains two notable regions—the central repetitive region and a C-terminal region with NLS motifs and a potent transcription activation domain (AD). The NLS and AD were shown to be required for E gene function in the case of Bs3, Xa10, Xa7, and Xa27 (Van den Ackerveken et al., 1996; Zhu et al., 1998, 1999; Yang et al., 2000; Szurek et al., 2001). TAL effector specificity is determined by the central repetitive region (Herbers et al., 1992; Yang and Gabriel, 1995; Yang and White, 2004; Yang et al., 2005), and is the structural basis for the TAL effector code, where each repeat specifies the probability of accommodating individual nucleotides (Boch et al., 2009; Moscou and Bogdanove, 2009; Deng et al., 2012; Gao et al., 2012; Mak et al., 2012). The repetitive domain consists of 33–35 amino acid repeats that are polymorphic at amino acid residues 12 and 13, which are referred to as the repeat-variable di-residues (RVDs), each of which can be represented by amino acid residue 13 and corresponds to one DNA base in the effector binding element. Proximal to the N-terminal portion of the repetitive domain are non-canonical repeats that mediate pairing with an initial 5′ thymine (Boch et al., 2009; Moscou and Bogdanove, 2009). E gene expression occurs upon cognate effector binding to a compatible effector binding element in the respective promoter (Figure 1A). The known E genes, with the exception of Xa10, have dominant and recessive alleles that differ in DNA sequence polymorphisms in the promoter region (Figure 1B). AvrBs3, for example, fails to induce Bs3-E, an allele of Bs3 with a 13-bp insertion in the effector binding element in the promoter (Figure 1B, iv; Römer et al., 2007, 2009b; Kay et al., 2009). E genes, S genes, and TAL effector genes, therefore, reflect selective pressures in the evolution of the host and pathogen interaction. In this regard, it is important to note that naturally occurring TAL effectors are not necessarily optimized for the cognate promoters simply in terms of the binding requirements. Natural TAL effector configurations may reflect adaptive responses to other factors, including the level of target gene expression and frequency of binding sites within a genome.
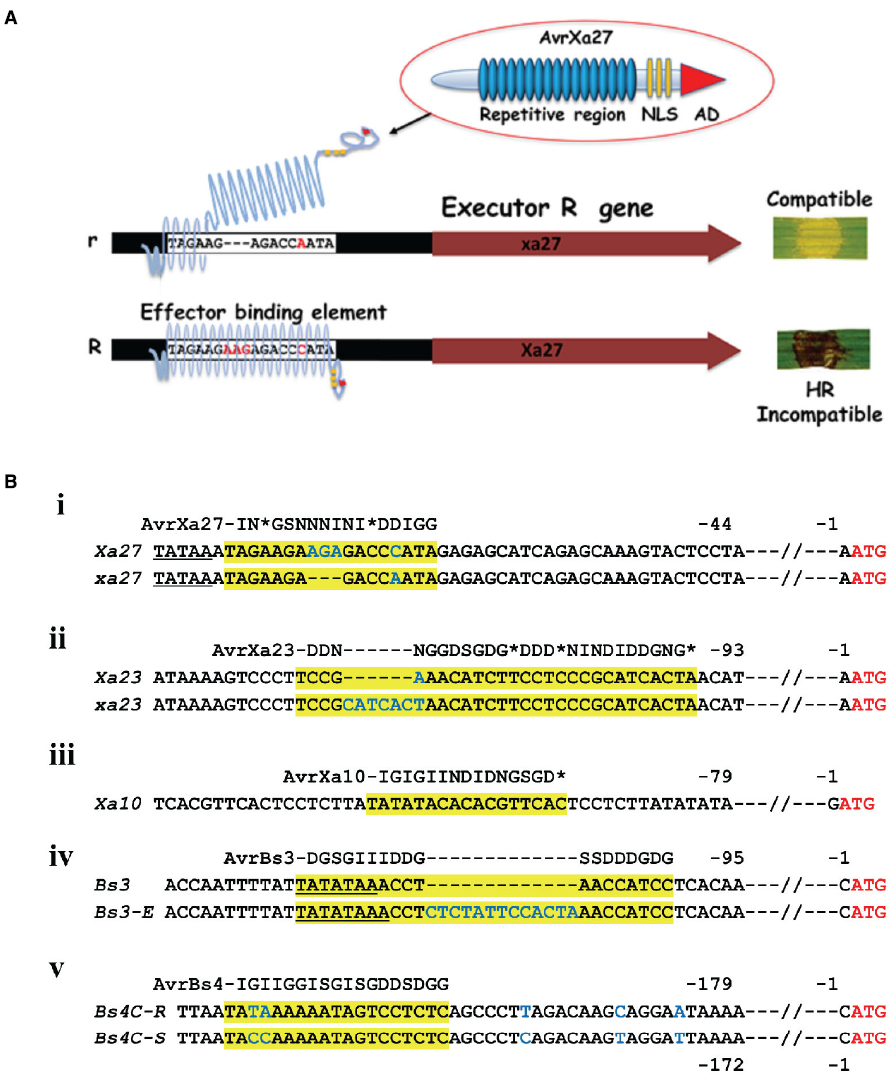
Figure 1. TAL effector and E gene interactions. (A) Schematic of the interaction between AvrXa27 and Xa27. Lower case r indicates the ineffective allele that lacks the AvrXa27 effector binding element. The r allele is missing three nucleotides and different in one nucleotide in the effector binding element of Xa27 and does not permit binding of AvrXa7 leading to a compatible interaction (Römer et al., 2010). R indicates the dominant and functional allele of Xa27. Xa27 expression leads to a resistance response and HR on leaves, indicated by the dark discoloring of the inoculation site (here, on a rice leaf). NLS, nuclear localization signal. AD, transcription activation domain of TAL effector. (B) Promoters of E genes and polymorphisms in dominant and recessive alleles. (i) Sequence alignment of a part of the promoters of Xa27 from the rice cultivar IRBB27 (gi 66735941 gb AY986491.1) and xa27 from the rice cultivar IR24 (gi 66735943 gb AY986492.1). (ii) Sequence of a part of the promoter of rice Xa10 from the rice cultivar IRBB10 (gi|448280729|gb|JX025645.1|). (iii) A part of the promoters of Xa23 from the rice cultivar CBB23 (gi|721363841|gb|KP123634.1|) and xa23 from the rice cultivar JG30 (gi|721363854|gb|KP123635.1|). (iv) A part of the promoters of Bs3 from Capsicum annuum L. cultivar ECW-30R (gi|158851516|gb|EU078684.1) and Bs3-E from C. annuum L. cultivar ECW (gi|158851512|gb|EU078683.1|). (v) A part of the promoters of Bs4C-R from pubescens cultivar PI 235047 (gi|414148024|gb|JX944826.1|) and Bs4C-S from Capsicum pubescens cultivar PI 585270 (gi|414148026|gb|JX944827.1|). The ATG start codon in each case is displayed in red letters. Nucleotides that are identical between the alleles are displayed as black letters. Predicted TATA boxes are underlined. Effector binding elements are highlighted in yellow with blue letters indicating differences between alleles. The TAL effectors are represented by the repeat regions using a single letter represents each RVD (I-NI; G-NG or HG; S-NS; D-HD or ND, *-N*, N-NN). * Represents no amino acid residue at what would otherwise be position 13.
E Proteins are not Homologs of Classical R Proteins
E proteins are not related on the basis of sequence to any other type of R protein. In fact, the proteins, with the exception of the recently reported XA10 and XA23, share no sequence identity with each other. Conceptually, the E genes and their products can be divided into two groups. Group 1 consists of proteins that likely have a function in plant development or physiology and whose function has been hijacked by host adaptation to disease. Group 1 consists solely of BS3, which is a member of a conserved family of proteins known as flavin mono-oxygenases (FMO) and, more specifically, a subclass of FMOs known variously as YUCCA or FLOOZY (Figure 2A; Römer et al., 2007; Exposito-Rodriguez et al., 2011; Zhao, 2014). Group 2 members, of which there are four, are relatively short proteins that have multiple hydrophobic potential membrane spanning domains (Figure 2B). The proteins share no sequence relatedness with proteins of known function and the relatively few related coding sequences occur within close relatives. One related sequence outside the Solanaceae, from grapevine, was reported for Bs3C-R. Several of the E proteins may have structural similarities. XA27 and XA10 are predicted or have been shown to localize to host cellular membranes and XA10, more specifically, has been shown to localize to the endoplasmic reticulum (ER; Wu et al., 2008; Tian et al., 2014). Prediction software also indicates that BS4C-R may be localized to the ER (Nakai and Horton, 1999; Strauss et al., 2012). It is tempting to speculate that BS3 requires catalytic activity for the R gene response and the group 2 proteins function as R proteins due to their interaction with host organelles. However, whether the predicted catalytic functions of BS3 are required for the R gene response has not been reported, and future analysis of the mechanism-of-action for the respective proteins may indicate some common feature.
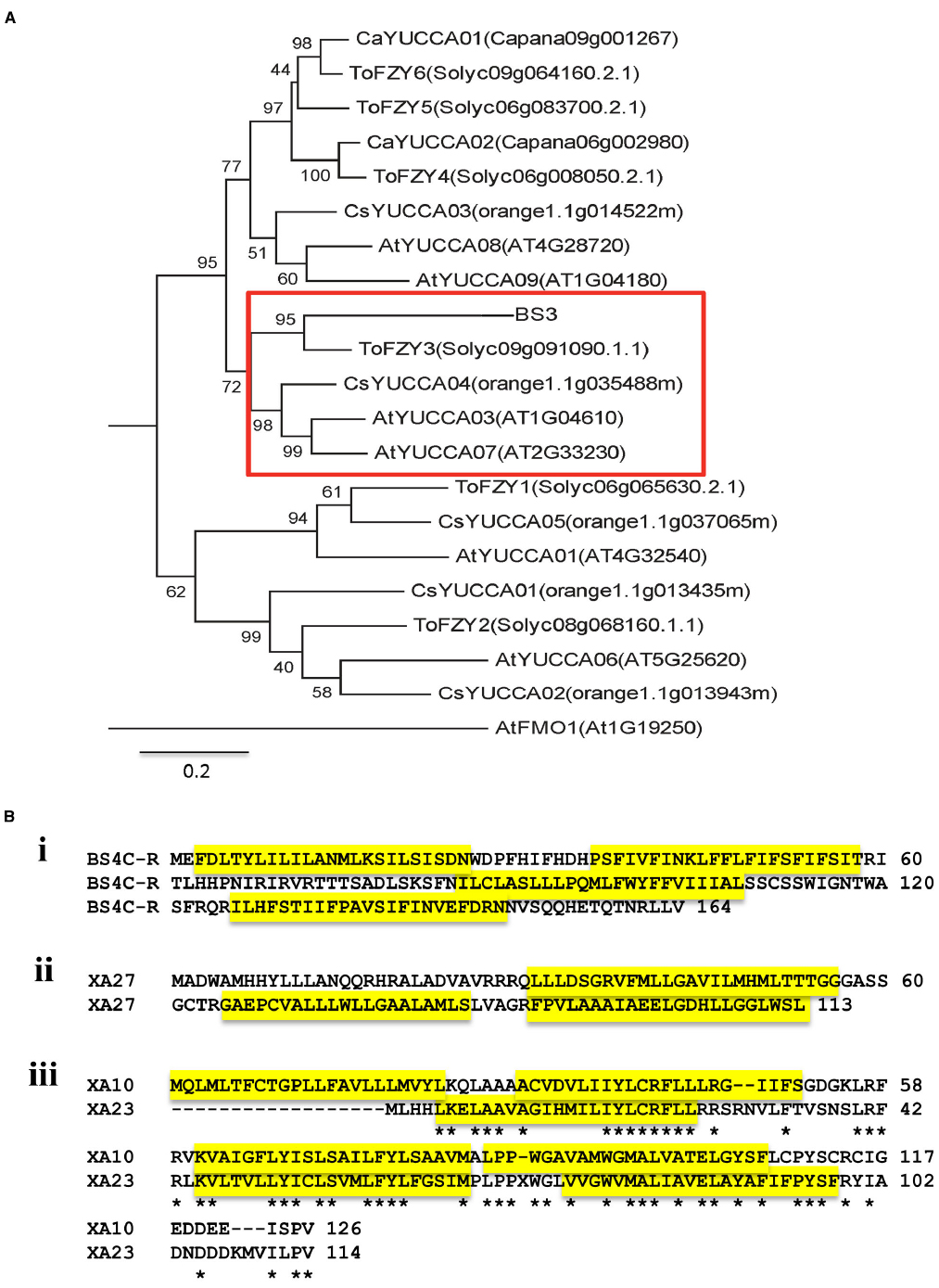
Figure 2. The known E proteins. (A) A phylogenetic tree of BS3 related proteins. Proteins from closely related YUCCA proteins of C. annuum L. (Ca), A. thaliana (At), Tomato (To), Citrus sinensis (Cs) and the BS3 protein (Capana02g001306) were aligned. Names of proteins are given with the Phytozome ID or Pepper Genome Database ID (in parentheses). A monophyletic group that contains the predicted BS3 protein and tomato YUCCA-like proteins is boxed in red. Sequences were aligned with the online ClustalW server (http://www.ch.embnet.org/software/ClustalW.html) using the default values. MEGA6.0 was used for generating a tree on the basis of ClustalW output. Phylogenetic calculations are based on the maximum likelihood method, and Bootstrap analysis was used to evaluate the reliability of the nodes of the phylogenetic trees. Bootstrap values are based on 1000 replications. The branch lengths of the tree are proportional to divergence. The 0.1 scale represents 10% change. (B) Structural predictions for group 2 E proteins. (i) Bs4C-R; (ii) XA27; (iii) XA10 and XA23. Alignment of XA10 and XA23 was conducted using the online program ClustalW2 using the default parameters (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Transmembrane helices predicted by the SOSUI program (http://bp.nuap.nagoya-u.ac.jp/sosui/sosui_submit.html) are highlighted in yellow. * Represents no amino acid residue at what would otherwise be position 13.
E Genes in Bacterial Spot Disease on Pepper
E genes for groups 1 and 2 have been cloned from pepper. The group 1 Bs3 is recognized by both TAL effectors AvrBs3 and AvrHah from the pathogens Xanthomonas campestris pv. vesicatoria and Xanthomonas gardneri, respectively, both causal organisms of bacterial spot disease of pepper and tomato (Bonas et al., 1998; Schornack et al., 2008). The gene product BS3 is a 342 amino acid protein with a high degree of relatedness with FMOs (Römer et al., 2007; Schornack et al., 2008). FMO proteins are a family of enzymes functioning in all phyla (van Berkel et al., 2006), and play roles in pathogen defense, auxin biosynthesis and metabolism of glucosinolates (Bartsch et al., 2006; Koch et al., 2006; Mishina and Zeier, 2006; Schlaich, 2007). As noted earlier, BS3 falls in a phylogenetic clade consisting of YUCCA and ToFZY members (Figure 2A; Römer et al., 2007). The most closely related proteins to BS3 have been demonstrated to be involved in auxin biosynthesis and a variety of developmental and physiological responses (Exposito-Rodriguez et al., 2011; Stepanova et al., 2011; Lee et al., 2012; Hentrich et al., 2013; Zhao, 2014). YUCCA/FLOOZY members catalyze a key intermediate in the plant pathway from indole-3-pyruvate (IPA) into indole-3-acetic acid (IAA) through oxidative decarboxylation reaction (Kim et al., 2011; Stepanova et al., 2011; Dai et al., 2013; Hentrich et al., 2013; Zhao, 2014). A homolog from tomato, ToFZY, also functions in auxin biosynthesis (Exposito-Rodriguez et al., 2011). A more distant relative of unknown enzymatic activity, AtFMO1, plays a role in systemic acquired resistance (Mishina and Zeier, 2006).
Bs4C-R encodes a member of our group 2 E proteins and is expressed in the presence of the TAL effector AvrBs4 (Strauss et al., 2012). Bs4C-R is the only E gene isolated on the basis of differential expression between resistant and susceptible cultivars and not the typical gene mapping strategy. A two-nucleotide polymorphism in the region of the effector binding element of a susceptible allele Bs4C-S leads to the failure of induction of an AvrBs4-dependent HR (Figure 1B, v; Strauss et al., 2012). Both the dominant and recessive alleles encode functionally competent proteins as constitutive expression of either Bs4C-R or Bs4C-S triggered HR Nicotiana benthamiana in leaves (Strauss et al., 2012).
E Genes in Bacterial Blight Disease of Rice
The E genes of rice are all included in our group 2 and provide resistance to bacterial blight disease. Bacterial blight of rice is caused by Xanthomonas oryzae pv. oryzae, and TAL effectors are major avirulence factors for X. oryzae pv. oryzae when the cognate E genes are present in host plants (Mew, 1987; White and Yang, 2009). Three pairs of TAL effectors and cognate E genes have been cloned from rice—AvrXa27/Xa27, AvrXa10/Xa10, and AvrXa23/Xa23. No cognate S genes or virulence effects for the TAL effectors of AvrXa10, AvrXa23, or AvrXa27 in compatible host cultivars have been reported, despite the presence of AvrXa27 and AvrXa23 in many extant strains of X. oryzae pv. oryzae (Gu et al., 2004; Wang et al., 2014).
The Xa27 product is a protein of 113 amino acids without any clear homologs based on sequence similarity in plants other than rice and several related species of the Oryza genus (Gu et al., 2005; Bimolata et al., 2013). The resistance conferred by Xa27 is affected by developmental stage, increasing with the age of the plants and reaching maximum resistance at 5 weeks. Moreover, Xa27 showed a dosage effect in the cultivar CO39 genetic background (Gu et al., 2004). At least two transmembrane α-helix domains were predicted, depending on the prediction software (Gu et al., 2005). Here, we show three based on the SOSUI program (Figure 2B, Hirokawa et al., 1998). Further experimentation has shown that the protein XA27 localizes to cytoplasmic membrane, and some protein appears in the apoplast after plasmolysis (Wu et al., 2008). Localization is dependent on the N-terminal signal anchor-like sequence, which is also essential for resistance to X. oryzae pv. oryzae (Wu et al., 2008). The protein itself appears to be toxic as gene transfer to compatible rice lines occurs with a reduced efficiency. Nevertheless recombinant lines were recovered, demonstrating that the AvrXa27-dependency of the resistance is indeed linked to the Xa27 locus (Gu et al., 2005). At the same time, lines were obtained that had elevated expression of Xa27 and displayed defense reactions, including thickened vascular elements, even in the absence of bacterial inoculation (Gu et al., 2005). The effector binding element is located immediately downstream of the predicted TATA box, and the recessive allele xa27 in the susceptible rice cultivar IR24 encodes the same protein but has a three-nucleotide deletion and one nucleotide difference in comparison to Xa27 (Figure 1B, i; Römer et al., 2009a). DNA sequence alignment of Xa27 alleles from 27 lines representing four Oryza species revealed that a Xa27-related coding sequence was indeed present in all of the lines. However, only the IRBB27 allele appears to possess the necessary effector binding element for AvrXa27 (Bimolata et al., 2013). A synthetic TAL effector directed at the recessive allele in IR24 induced a resistance reaction, indicating the product of the recessive allele could function similarly to Xa27, if expressed (Li et al., 2013).
Xa10 encodes a 126-amino acid protein, containing four potential transmembrane helices (Tian et al., 2014). A consensus effector binding element is present in the promoter region of Xa10 (Figure 1B, iii). Xa10 differs from Xa27 and Bs4C-R in sequence and by the lack of a nearly identical coding sequence in susceptible plant lines. At the same time, related sequences are found in other lines, including Xa23 (Wang et al., 2015). Ectopic and weak expression of Xa10 in rice causes a lesion mimic-like phenotype, while transient expression of Xa10 in N. benthamiana and rice induced HR in plants (Tian et al., 2014). Under the appropriate promoter, Xa10 also induced programmed cell death (PCD) in mammalian HeLa cells (Tian et al., 2014). In both rice and N. benthamiana cells, hydrogen peroxide, swelling and degradation were detected in chloroplasts. Degradation of mitochondria was also observed, supporting the model that XA10 functions as a general inducer of PCD in plant and animal cells (Tian et al., 2014). Further functional characterization revealed that XA10 forms hexamers, localizes on the ER membrane of plant and HeLa cells, and mediates Ca2+ depletion, which is consistent with some processes of PCD (Pinton et al., 2008; Williams et al., 2014).
The E gene Xa23 encodes a 113-amino acid protein that shares approximately 50% amino acid sequence identity and 64% nucleotide sequence similarity with XA10 and Xa10, respectively (Wang et al., 2015). An identical recessive allele is present on the basis of the coding region, and characterization of the effector binding element of AvrXa23 revealed a 7-bp polymorphism accounts for the failure of xa23 induction in the recessive rice varieties (Figure 1B, ii). The susceptible cultivar JG30, with xa23, became resistant to PXO99A harboring a designed TAL effector specifically targeting the xa23 promoter region including the 7-bp polymorphism (Wang et al., 2015). Moreover, Agrobacterium-mediated transient transformation of Xa23 indicated that, like Xa10, Xa23 induced an HR in N. benthamiana, and also induced an HR in tomato (Wang et al., 2015). Both XA10 and XA23 have a motif of unknown function that is comprised of five acidic amino acid residues (EDDEE and DNDDD, respectively) at the C-termini (Tian et al., 2014). Alteration of the so-called ED motif in XA10 abolished HR activity (Tian et al., 2014).
Prospects for E Genes in Disease Control
The question arises whether, as dominant major genes for resistance, the genes are effective in control of the respective diseases. In rice, only Xa10 has been deployed in field conditions and is effective against a few extant races of the pathogens (Vera-Cruz et al., 2000; Mishra et al., 2013). Xa27 has been introduced into breeding programs (Luo et al., 2012; Luo and Yin, 2013). Xa23 and Xa7 are also in the process of introduction into various breeding programs (Perez et al., 2008; Huang et al., 2012). But how durable is E gene mediated resistance? Bacteria can rapidly evolve to avoid R gene recognition through avirulence gene loss under high selection pressure for virulence (Koskiniemi et al., 2012). More specifically, TAL effectors appear to reflect exquisitely the selective forces of evolution in the form of the repetitive domain. Deletion of repeats in AvrBs3, for example, resulted in the loss of the induction of Bs3 (Römer et al., 2007). Deployment of an E gene that targets a critical TAL effector for virulence has been proposed as an approach to make adaptation less likely as the pathogen would have to maintain virulence in addition to losing Xa7 recognition. Indeed, Xa7, which is triggered by the major TAL effector AvrXa7, was found to be durable in field tests in the Philippines (Vera-Cruz et al., 2000). Field isolated strains that arose showed loss of the ability to induce resistance and were weakly virulent presumably due to associated mutations in avrXa7 (Vera-Cruz et al., 2000). However, in vitro rearrangements and in vivo selection for loss of AvrXa7-mediated resistance produced gene variants that maintained strain virulence and avoided Xa7-mediated resistance (Yang et al., 2005). Furthermore, a number of extant TAL effectors target the same S gene as AvrXa7, namely OsSWEET14, without activating Xa7-dependent resistance (Antony et al., 2010; Yu et al., 2011; Streubel et al., 2013). Strains can also acquire other major TAL effectors that target alternative paralogs of that S gene (Yang et al., 2006; Streubel et al., 2013; Zhou et al., 2015).
In India, field strain surveys have found a diversity of strains, many without AvrXa7 activity, indicating that any benefits in the deployment of Xa7 would be short-lived (Mishra et al., 2013). Thus, broad application of a single E gene like Xa7 in some environments appears to be limited. At the same time, local conditions, such as in the Philippine tests, may limit the invasion of a particular TAL effector gene into extant pathogen populations and deployment may be both broad and durable (Vera-Cruz et al., 2000). Xa27 and Xa23 are interesting from the perspective that the cognate avirulence genes are present in many strains and, therefore, broadly effective (Gu et al., 2004; Wang et al., 2014). In contrast to AvrXa7, no cognate S genes have been reported for AvrXa27 or AvrXa23, so we can speculate that these effectors may provide some fitness to the pathogen which has not been detected in laboratory or greenhouse assays so far. AvrBs3 has a phenotypic effect for strains of Xanthomonas euvesicatoria harboring the gene; a fitness benefit for the effector has been observed (Marois et al., 2002; Wichmann and Bergelson, 2004), and Bs3 is effective for many pepper strains of X. euvesicatoria. In addition, Bs3 is also effective against the emerging pepper pathogen X. gardneri, which harbors the TAL effector AvrHah1 (Schornack et al., 2008; Schwartz et al., 2015).
E Genes and New Strategies for Resistance
Despite possible shortcomings of endogenous E genes, E genes hold great potential for breeding broadly and durably resistant crop varieties. Specifically for TAL effector associated diseases, E genes can be constructed with so-called super-promoters, consisting of multiple effector binding sites, each recognizing specific corresponding TAL effectors that are expressed in the pathogen populations (Römer et al., 2009a; Hummel et al., 2012; Zeng et al., 2015). Xa27 was fused to a super promoter including binding sites for three TAL effectors from X. oryzae pv. oryzae and three from the bacterial leaf streak pathogen X. oryzae pv. oryzicola. The plants were resistant to several X. oryzae pv. oryzae and X. oryzae pv. oryzicola strains that were originally compatible on wild type homozygous Xa27 plants (Hummel et al., 2012). Similarly, transgenic rice lines containing Xa10E5 with binding elements to five TAL effectors proved to be resistant to 27 of the 28 selected X. oryzae pv. oryzae strains gathered from 11 countries (Zeng et al., 2015). Judicial choices of the effector binding sites for TAL effectors in extant populations may provide resilient barriers to TAL effector associated diseases. However, due to the risk that an added effector binding element might coincidently contains a cis regulatory element which could induce the E gene expression in response to particular stimuli and cause cell death without challenge of TAL effectors, such amended promoters should be tested thoroughly before deployment (Hummel et al., 2012). Another approach is to engineer an E gene to be under the control of a different type of pathogen inducible promoter. For example, expression of Xa27 under the control of an disease inducible or defense gene promoter, in this case, the rice PR1 promoter, which is induced by both compatible and incompatible bacteria, conferred broad resistance to X. oryzae pv. oryzae strains (Gu et al., 2005). This strategy need not be limited to Xanthomonas related diseases.
Conclusion
Thirteen R genes have been cloned for resistance to Xanthomonas diseases—all coming from rice, pepper, or tomato. Four, in addition to Bs4, are representatives of the two major classes of R genes, the receptor linked kinases (RLK) and nucleotide binding site leucine rich repeat (NBS-LRR) genes which are represented by Xa21 (RLK, rice), Xa26 (RLK, rice), Xa1 (NBS-LRR, rice), and Bs2 (NBS-LRR, pepper; Yoshimura et al., 1998; Tai et al., 1999; Zhang and Wang, 2013). Three cloned genes are recessive genes from rice and, although not discussed here, can be considered cases of loss of susceptibility (White and Yang, 2009). The five E genes and the protein products bear little or no resemblance to the other R genes, or, for that matter, other common defense response components. E genes, at least phenotypically, trigger host responses, in particular the HR, similarly to some other R gene mediated resistances. Whether the E proteins intersect other R protein mediated resistance pathways in plants remains unknown. Evidence for XA10 indicates that the protein activates PCD, possibly through ER-stress in light of the association with the ER (Williams et al., 2014). Further research into E gene functions should enhance their utility for new resistance strategies as well as improve our understanding on plant defense and PCD pathways.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank the members of the Yin laboratory for reviewing the manuscript. JZ and FW are supported by funds from National Science Foundation research award 1238189. ZY is supported by the National Research Foundation and Office of the Prime Minister Competitive Research Programme Singapore award NRF-CRP7-2010-02.
References
Antony, G., Zhou, J., Huang, S., Li, T., Liu, B., White, F., et al. (2010). Rice xa13 recessive resistance to bacterial blight is defeated by induction of the disease susceptibility gene Os11N3. Plant Cell 22, 3864–3876. doi: 10.1105/tpc.110.078964
Bartsch, M., Gobbato, E., Bednarek, P., Debey, S., Schultze, J. L., Bautor, J., et al. (2006). Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the nudix hydrolase NUDT7. Plant Cell 18, 1038–1051. doi: 10.1105/tpc.105.039982
Bimolata, W., Kumar, A., Sundaram, R. M., Laha, G. S., Qureshi, I. A., Reddy, G. A., et al. (2013). Analysis of nucleotide diversity among alleles of the major bacterial blight resistance gene Xa27 in cultivars of rice (Oryza sativa) and its wild relatives. Planta 238, 293–305. doi: 10.1007/s00425-013-1891-3
Boch, J., Bonas, U., and Lahaye, T. (2014). TAL effectors—pathogen strategies and plant resistance engineering. New Phytol. 204, 823–832. doi: 10.1111/nph.13015
Boch, J., Scholze, H., Schornack, S., Landgraf, A., Hahn, S., Kay, S., et al. (2009). Breaking the code of DNA binding specificity of TAL-type III effectors. Science 326, 1509–1512. doi: 10.1126/science.1178811
Bogdanove, A. J., Schornack, S., and Lahaye, T. (2010). TAL effectors: finding plant genes for disease and defense. Curr. Opin. Plant Biol. 13, 394–401. doi: 10.1016/j.pbi.2010.04.010
Bonas, U., Conradsstrauch, J., and Balbo, I. (1993). Resistance in tomato to Xanthomonas campestris pv vesicatoria is determined by alleles of the pepper-specific avirulence gene avrBs3. Mol. Gen. Genet. 238, 261–269.
Bonas, U., Stall, R. E., and Staskawicz, B. (1998). Genetic and structural characterization of the avirulence gene avrBs3 from Xanthomonas campestris pv. vesicatoria. Mol. Gen. Genet. 218, 127–136. doi: 10.1007/BF00330575
Cernadas, R. A., Doyle, E. L., Nino-Liu, D. O., Wilkins, K. E., Bancroft, T., Wang, L., et al. (2014). Code-assisted discovery of TAL effector targets in bacterial leaf streak of rice reveals contrast with bacterial blight and a novel susceptibility gene. PLoS Pathog. 10:e1003972. doi: 10.1371/journal.ppat.1003972
Cohn, M., Bart, R. S., Shybut, M., Dahlbeck, D., Gomez, M., Morbitzer, R., et al. (2014). Xanthomonas axonopodis virulence is promoted by a transcription activator-like effector mediated induction of a SWEET sugar transporter in cassava. Mol. Plant Microbe Interact. 27, 1186–1198. doi: 10.1094/MPMI-06-14-0161-R
Dai, X., Mashiguchi, K., Chen, Q., Kasahara, H., Kamiya, Y., Ojha, S., et al. (2013). The biochemical mechanism of auxin biosynthesis by an Arabidopsis YUCCA flavin-containing monooxygenase. J. Biol. Chem. 288, 1448–1457. doi: 10.1074/jbc.M112.424077
De Feyter, R., Yang, Y., and Gabriel, D. W. (1993). Gene-for-genes interactions between cotton R genes and Xanthomonas campestris pv. malvacearum avr genes. Mol. Plant Microbe Interact. 6, 225. doi: 10.1094/MPMI-6-225
Deng, D., Yan, C., Pan, X., Mahfouz, M., Wang, J., Zhu, J., et al. (2012). Structural basis for sequence-specific recognition of DNA by TAL effectors. Science 335, 720–723. doi: 10.1126/science.1215670
Exposito-Rodriguez, M., Borges, A. A., Borges-Perez, A., and Perez, J. A. (2011). Gene structure and spatiotemporal expression profile of tomato genes encoding YUCCA-like flavin monooxygenases: the ToFZY gene family. Plant Physiol. Biochem. 49, 782–791. doi: 10.1016/j.plaphy.2011.02.022
Gao, H., Wu, X., Chai, J., and Han, Z. (2012). Crystal structure of a TALE protein reveals an extended N-terminal DNA binding region. Cell Res. 22, 1716–1720. doi: 10.1038/cr.2012.156
Gu, K., Tian, D., Yang, F., Wu, L., Sreekala, C., Wang, D., et al. (2004). High-resolution genetic mapping of Xa27(t), a new bacterial blight resistance gene in rice, Oryza sativa L. Theor. Appl. Genet. 108, 800–807. doi: 10.1007/s00122-003-1491-x
Gu, K. Y., Yang, B., Tian, D. S., Wu, L. F., Wang, D. J., Sreekala, C., et al. (2005). R gene expression induced by a type-III effector triggers disease resistance in rice. Nature 435, 1122–1125. doi: 10.1038/nature03630
Hentrich, M., Sanchez-Parra, B., Perez Alonso, M., Carrasco Loba, V., Carrillo, L., Vicente-Carbajosa, J., et al. (2013). YUCCA8 and YUCCA9 overexpression reveals a link between auxin signaling and lignification through the induction of ethylene biosynthesis. Plant Signal. Behav. 8, e26363–e26363. doi: 10.4161/psb.26363
Herbers, K., Conrads-Strauch, J., and Bonas, U. (1992). Race-specificity of plant resistance to bacterial spot disease determined by repetitive motifs in a bacterial avirulence protein. Nature 356, 172–174. doi: 10.1038/356172a0
Heuer, H., Yin, Y., Xue, Q., Smalla, K., and Guo, J. (2007). Repeat domain diversity of avrBs3-like genes in Ralstonia solanacearum strains and association with host preferences in the field. Appl. Environ. Microbiol. 73, 4379. doi: 10.1128/AEM.00367-07
Hirokawa, T., Boon-Chieng, S., and Mitaku, S. (1998). SOSUI: classification and secondary structure prediction system for membrane proteins. Bioinformatics 14, 378–379. doi: 10.1093/bioinformatics/14.4.378
Hopkins, C. M., White, F. F., Choi, S. H., Guo, A., and Leach, J. E. (1992). Identification of a family of avirulence genes from Xanthomonas oryzae pv oryzae. Mol. Plant Microbe Interact. 5, 451–459. doi: 10.1094/MPMI-5-451
Huang, B., Xu, J. Y., Hou, M. S., Ali, J., and Mou, T. M. (2012). Introgression of bacterial blight resistance genes Xa7, Xa21, Xa22 and Xa23 into hybrid rice restorer lines by molecular marker-assisted selection. Euphytica 187, 449–459. doi: 10.1007/s10681-012-0758-1
Hu, Y., Zhang, J., Jia, H., Sosso, D., Li, T., Frommer, W. B., et al. (2014). Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. U.S.A. 111, E521–E529. doi: 10.1073/pnas.1313271111
Hummel, A. W., Doyle, E. L., and Bogdanove, A. J. (2012). Addition of transcription activator-like effector binding sites to a pathogen strain-specific rice bacterial blight resistance gene makes it effective against additional strains and against bacterial leaf streak. New Phytol. 195, 883–893. doi: 10.1111/j.1469-8137.2012.04216.x
Hutin, M., Pérez-Quintero, A., Lopez, C., and Szurek, B. (2015). MorTAL Kombat: the story of defense against TAL effectors through loss-of-susceptibility. Front. Microbiol. 6:535. doi: 10.3389/fpls.2015.00535
Kay, S., Hahn, S., Marois, E., Hause, G., and Bonas, U. (2007). A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 318, 648–651. doi: 10.1126/science.1144956
Kay, S., Hahn, S., Marois, E., Wieduwild, R., and Bonas, U. (2009). Detailed analysis of the DNA recognition motifs of the Xanthomonas type III effectors AvrBs3 and AvrBs3 delta rep16. Plant J. 59, 859–871. doi: 10.1111/j.1365-313X.2009.03922.x
Kim, J. I., Murphy, A. S., Baek, D., Lee, S., Yun, D., Bressan, R. A., et al. (2011). YUCCA6 over-expression demonstrates auxin function in delaying leaf senescence in Arabidopsis thaliana. J. Exp. Bot. 62, 3981–3992. doi: 10.1093/jxb/err094
Koch, M., Vorwerk, S., Masur, C., Sharifi-Sirchi, G., Olivieri, N., and Schlaich, N. L. (2006). A role for a flavin-containing mono-oxygenase in resistance against microbial pathogens in Arabidopsis. Plant J. 47, 629–639. doi: 10.1111/j.1365-313X.2006.02813.x
Koskiniemi, S., Sun, S., Berg, O. G., and Andersson, D. I. (2012). Selection-driven gene loss in bacteria. PLoS Genet. 8:e1002787. doi: 10.1371/journal.pgen.1002787
Lee, M., Jung, J., Han, D., Seo, P. J., Park, W. J., and Park, C. (2012). Activation of a flavin monooxygenase gene YUCCA7 enhances drought resistance in Arabidopsis. Planta 235, 923–938. doi: 10.1007/s00425-011-1552-3
Leyns, F., De Cleene, M., Swings, J., and De Ley, J. (1984). The host range of the genus Xanthomonas. Bot. Rev. 50, 308–356. doi: 10.1007/BF02862635
Li, T., Huang, S., Zhou, J., and Yang, B. (2013). Designer TAL effectors induce disease susceptibility and resistance to Xanthomonas oryzae pv. oryzae in rice. Mol. Plant 6, 781–789. doi: 10.1093/mp/sst034
Luo, Y., Sangha, J. S., Wang, S., Li, Z., Yang, J., and Yin, Z. (2012). Marker-assisted breeding of Xa4, Xa21 and Xa27 in the restorer lines of hybrid rice for broad-spectrum and enhanced disease resistance to bacterial blight. Mol. Breed. 30, 1601–1610. doi: 10.1007/s11032-012-9742-7
Luo, Y., and Yin, Z. (2013). Marker-assisted breeding of Thai fragrance rice for semi-dwarf phenotype, submergence tolerance and disease resistance to rice blast and bacterial blight. Mol. Breed. 32, 709–721. doi: 10.1007/s11032-013-9904-2
Mak, A. N., Bradley, P., Cernadas, R. A., Bogdanove, A. J., and Stoddard, B. L. (2012). The crystal structure of TAL effector PthXo1 bound to its DNA target. Science 335, 716–719. doi: 10.1126/science.1216211
Marois, E., Van den Ackerveken, G., and Bonas, U. (2002). The Xanthomonas type III effector protein AvrBs3 modulates plant gene expression and induces cell hypertrophy in the susceptible host. Mol. Plant Microbe Interact. 15, 637–646. doi: 10.1094/MPMI.2002.15.7.637
Mew, T. W. (1987). Current status and future prospects of research on bacterial blight of rice. Ann. Rev. Phytopathol. 25, 559–382. doi: 10.1146/annurev.py.25.090187.002043
Mishina, T. E., and Zeier, J. (2006). The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 141, 1666–1675. doi: 10.1104/pp.106.081257
Mishra, D., Vishnupriya, M. R., Anil, M. G., Konda, K., Raj, Y., and Sonti, R. V. (2013). Pathotype and genetic diversity amongst Indian isolates of Xanthomonas oryzae pv. oryzae. PLoS ONE 8:e81996. doi: 10.1371/journal.pone.0081996
Moscou, M. J., and Bogdanove, A. J. (2009). A simple cipher governs DNA recognition by TAL effectors. Science 326, 1501–1501. doi: 10.1126/science.1178817
Nakai, K., and Horton, P. (1999). PSORT: a program for detecting the sorting signals of proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–35. doi: 10.1016/s0968-0004(98)01336-x
Perez, L. M., Redoña, E. D., Mendioro, M. S., Vera Cruz, C. M., and Leung, H. (2008). Introgression of Xa4, Xa7 and Xa21 for resistance to bacterial blight in thermosensitive genetic male sterile rice (Oryza sativa L.) for the development of two-line hybrids. Euphytica 164, 627–636. doi: 10.1007/s10681-008-9653-1
Pinton, P., Giorgi, C., Siviero, R., Zecchini, E., and Rizzuto, R. (2008). Calcium and apoptosis: ER-mitochondria Ca2+ transfer in the control of apoptosis. Oncogene 27, 6407–6418. doi: 10.1038/onc.2008.308
Römer, P., Hahn, S., Jordan, T., Strauss, T., Bonas, U., and Lahaye, T. (2007). Plant pathogen recognition mediated by promoter activation of the pepper Bs3 resistance gene. Science 318, 645–648. doi: 10.1126/science.1144958
Römer, P., Recht, S., and Lahaye, T. (2009a). A single plant resistance gene promoter engineered to recognize multiple TAL effectors from disparate pathogens. Proc. Natl. Acad. Sci. U.S.A. 106, 20526–20531. doi: 10.1073/pnas.0908812106
Römer, P., Strauss, T., Hahn, S., Scholze, H., Morbitzer, R., Grau, J., et al. (2009b). Recognition of AvrBs3-like proteins is mediated by specific binding to promoters of matching pepper Bs3 alleles. Plant Physiol. 150, 1697–1712. doi: 10.1104/pp.109.139931
Römer, P., Recht, S., Strauss, T., Elsaesser, J., Schornack, S., Boch, J., et al. (2010). Promoter elements of rice susceptibility genes are bound and activated by specific TAL effectors from the bacterial blight pathogen Xanthomonas oryzae pv. oryzae. New Phytol. 187, 1048–1057. doi: 10.1111/j.1469-8137.2010.03217.x
Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. 415, 497–502. doi: 10.1038/415497a
Schlaich, N. L. (2007). Flavin-containing monooxygenases in plants: looking beyond detox. Trends Plant Sci. 12, 412–418. doi: 10.1016/j.tplants.2007.08.009
Schornack, S., Ballvora, A., Gurlebeck, D., Peart, J., Ganal, M., Baker, B., et al. (2004). The tomato resistance protein Bs4 is a predicted non-nuclear TIR-NB-LRR protein that mediates defense responses to severely truncated derivatives of AvrBs4 and overexpressed AvrBs3. Plant J. 37, 46–60. doi: 10.1046/j.1365-313X.2003.01937.x
Schornack, S., Minsavage, G. V., Stall, R. E., Jones, J. B., and Lahaye, T. (2008). Characterization of AvrHah1, a novel AvrBs3-like effector from Xanthomonas gardneri with virulence and avirulence activity. New Phytol. 179, 546–556. doi: 10.1111/j.1469-8137.2008.02487.x
Schwartz, A. R., Neha, P. N., Sujan, T. S., Mark, W. M., José, P. J., Joaquim, M. J., et al. (2015). Phylogenomics of Xanthomonas field strains infecting pepper and tomato reveals diversity in effector repertoires and identifies determinants of host specificity. Front. Microbiol. 6:535. doi: 10.3389/fmicb.2015.00535
Stepanova, A. N., Yun, J., Robles, L. M., Novak, O., He, W., Guo, H., et al. (2011). The Arabidopsis YUCCA1 flavin monooxygenase functions in the indole-3-pyruvic acid branch of auxin biosynthesis. Plant Cell 23, 3961–3973. doi: 10.1105/tpc.111.088047
Strauss, T., van Poecke, R. M. P., Strauss, A., Römer, P., Minsavage, G. V., Singh, S., et al. (2012). RNA-seq pinpoints a Xanthomonas TAL-effector activated resistance gene in a large-crop genome. Proc. Natl. Acad. Sci. U.S.A. 109, 19480–19485. doi: 10.1073/pnas.1212415109
Streubel, J., Pesce, C., Hutin, M., Koebnik, R., Boch, J., and Szurek, B. (2013). Five phylogenetically close rice SWEET genes confer TAL effector-mediated susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 200, 808–819. doi: 10.1111/nph.12411
Szurek, B., Marois, E., Bonas, U., and Van den Ackerveken, G. (2001). Eukaryotic features of the Xanthomonas type III effector AvrBs3: protein domains involved in transcriptional activation and the interaction with nuclear import receptors from pepper. Plant J. 26, 523–534. doi: 10.1046/j.0960-7412.2001.01046.x
Tai, T. H., Dahlbeck, D., Clark, E. T., Gajiwala, P., Pasion, R., Whalen, M. C., et al. (1999). Expression of the Bs2 pepper gene confers resistance to bacterial spot disease in tomato. Proc. Natl. Acad. Sci. U.S.A. 96, 14153–14158. doi: 10.1073/pnas.96.24.14153
Tampakaki, A. P., Fadouloglou, V. E., Gazi, A. D., Panopoulos, N. J., and Kokkinidis, M. (2004). Conserved features of type III secretion. Cell. Microbiol. 6, 805–816. doi: 10.1111/j.1462-5822.2004.00432.x
Tian, D., Wang, J., Zeng, X., Gu, K., Qiu, C., Yang, X., et al. (2014). The rice TAL effector-dependent resistance protein XA10 triggers cell death and calcium depletion in the endoplasmic reticulum. Plant Cell 26, 497–515. doi: 10.1105/tpc.113.119255
van Berkel, W. J. H., Kamerbeek, N. M., and Fraaije, M. W. (2006). Flavoprotein monooxygenases, a diverse class of oxidative biocatalysts. J. Biotechnol. 124, 670–689. doi: 10.1016/j.jbiotec.2006.03.044
Van den Ackerveken, G., Marois, E., and Bonas, U. (1996). Recognition of the bacterial avirulence protein AvrBs3 occurs inside the host plant cell. Cell 87, 1307–1316. doi: 10.1016/S0092-8674(00)81825-5
Vera-Cruz, C. M. V., Bai, J. F., Ona, I., Leung, H., Nelson, R. J., Mew, T. W., et al. (2000). Predicting durability of a disease resistance gene based on an assessment of the fitness loss and epidemiological consequences of avirulence gene mutation. Proc. Natl. Acad. Sci. U.S.A. 97, 13500–13505. doi: 10.1073/pnas.250271997
Wang, C., Qin, T., Yu, H., Zhang, X., Che, J., Gao, Y., et al. (2014). The broad bacterial blight resistance of rice line CBB23 is triggered by a novel transcription activator-like (TAL) effector of Xanthomonas oryzae pv. oryzae. Mol. Plant Pathol. 15, 333–341. doi: 10.1111/mpp.12092
Wang, C., Zhang, X., Fan, Y., Gao, Y., Zhu, Q., Zheng, C., et al. (2015). XA23 is an executor R protein and confers broad-spectrum disease resistance in rice. Mol. Plant Pathol. 8, 290–302. doi: 10.1016/j.molp.2014.10.010
Wichmann, G., and Bergelson, J. (2004). Effector genes of Xanthomonas axonopodis pv. vesicatoria promote transmission and enhance other fitness traits in the field. Genetics 166, 693–706. doi: 10.1534/genetics.166.2.693
White, F. F., and Yang, B. (2009). Host and pathogen factors controlling the rice Xanthomonas oryzae interaction. Plant Physiol. 150, 1677–1686. doi: 10.1104/pp.109.139360
Williams, B., Verchot, J., and Dickman, M. B. (2014). When supply does not meet demand-ER stress and plant programmed cell death. Front. Plant Sci. 5:211. doi: 10.3389/fpls.2014.00211
Wu, L., Goh, M. L., Sreekala, C., and Yin, Z. (2008). XA27 depends on an amino-terminal signal-anchor-like sequence to localize to the apoplast for resistance to Xanthomonas oryzae pv oryzae. Plant Physiol. 148, 1497–1509. doi: 10.1104/pp.108.123356
Yang, Y., and Gabriel, D. W. (1995). Intragenic recombination of a single plant pathogen gene provides a mechanism for the evolution of new host specificities. J. Bacteriol. 177, 4963–4968.
Yang, B., Sugio, A., and White, F. F. (2005). Avoidance of host recognition by alterations in the repetitive and C-terminal regions of AvrXa7, a type III effector of Xanthomonas oryzae pv. oryzae. Mol. Plant Microbe Interact. 18, 142–149. doi: 10.1094/MPMI-18-0142
Yang, B., Sugio, A., and White, F. F. (2006). Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. Proc. Natl. Acad. Sci. U.S.A. 103, 10503–10508. doi: 10.1073/pnas.0604088103
Yang, B., and White, F. F. (2004). Diverse members of the AvrBs3/PthA family of type III effectors are major virulence determinants in bacterial blight disease of rice. Mol. Plant Microbe Interact. 17, 1192–1200. doi: 10.1094/MPMI.2004.17.11.1192
Yang, B., Zhu, W. G., Johnson, L. B., and White, F. F. (2000). The virulence factor AvrXa7 of Xanthomonas oryzae pv. oryzae is a type III secretion pathway-dependent nuclear-localized double-stranded DNA-binding protein. Proc. Natl. Acad. Sci. U.S.A. 97, 9807–9812. doi: 10.1073/pnas.170286897
Yoshimura, S., Yamanouchi, U., Katayose, Y., Toki, S., Wang, Z. X., Kono, I., et al. (1998). Expression of Xa1, a bacterial blight-resistance gene in rice, is induced by bacterial inoculation. Proc. Natl. Acad. Sci. U.S.A. 95, 1663. doi: 10.1073/pnas.95.4.1663
Yu, Y., Streubel, J., Balzergue, S., Champion, A., Boch, J., Koebnik, R., et al. (2011). Colonization of rice leaf blades by an African strain of Xanthomonas oryzae pv. oryzae depends on a new TAL effector that induces the rice nodulin-3 Os11N3 gene. Molecular Plant Microbe Interact. 24, 1102–1113. doi: 10.1094/MPMI-11-10-0254
Zeng, X., Tian, D., Gu, K., Zhou, Z., Yang, X., Luo, Y., et al. (2015). Genetic engineering of the Xa10 promoter for broad-spectrum and durable resistance to Xanthomonas oryzae pv. oryzae. Plant Biotechnol. J. doi: 10.1111/pbi.12342 [Epub ahead of print].
Zhang, H., and Wang, S. (2013). Rice versus Xanthomonas oryzae pv. oryzae: a unique pathosystem. Curr. Opin. Plant Biol. 16, 188–195. doi: 10.1016/j.pbi.2013.02.008
Zhou, J., Peng, Z., Long, J., Sosso, D., Liu, B., Eom, J. S., et al. (2015). Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 82, 632–643. doi: 10.1111/tpj.12838
Zhu, W., Yang, B., Wills, N., Johnson, L. B., and White, F. F. (1999). The C terminus of AvrXa10 can be replaced by the transcriptional activation domain of VP16 from the herpes simplex virus. Plant Cell 11, 1665–1674. doi: 10.1105/tpc.11.9.1665
Keywords: TAL effectors, R gene, Xanthomonas
Citation: Zhang J, Yin Z and White F (2015) TAL effectors and the executor R genes. Front. Plant Sci. 6:641. doi: 10.3389/fpls.2015.00641
Received: 07 May 2015; Accepted: 02 August 2015;
Published: 20 August 2015.
Edited by:
Thomas Lahaye, Ludwig Maximilian University of Munich, GermanyReviewed by:
Brian Staskawicz, University of California, Berkeley, USAAdam Bogdanove, Cornell University, USA
Tom Schreiber, Martin Luther University of Halle-Wittenberg, Germany
Copyright © 2015 Zhang, Yin and White. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Junli Zhang, Department of Plant Pathology, Kansas State University, 4024 Throckmorton Plant Sciences Center, Manhattan, KS 66506, USA,eXVhbnl1YW4xMjU0M0BnbWFpbC5jb20=
 Junli Zhang
Junli Zhang