- 1Signaling Pathway Research Unit, RIKEN Center for Sustainable Resource Science, Yokohama, Japan
- 2National Key Laboratory for Plant Cell Technology, Agricultural Genetics Institute, Vietnam Academy of Agricultural Sciences, Hanoi, Vietnam
- 3Department of Biology, Lorestan University, Khorramabad, Iran
Drought causes detrimental effect to growth and productivity of many plants, including crops. NAC transcription factors have been reported to play important role in drought tolerance. In this study, we assessed the expression profiles of 19 dehydration-responsive CaNAC genes in roots and leaves of two contrasting drought-responsive chickpea varieties treated with water (control) and dehydration to examine the correlation between the differential expression levels of the CaNAC genes and the differential drought tolerability of these two cultivars. Results of real-time quantitative PCR indicated a positive relationship between the number of dehydration-inducible and -repressible CaNAC genes and drought tolerability. The higher drought-tolerant capacity of ILC482 cultivar vs. Hashem cultivar might be, at least partly, attributed to the higher number of dehydration-inducible and lower number of dehydration-repressible CaNAC genes identified in both root and leaf tissues of ILC482 than in those of Hashem. In addition, our comparative expression analysis of the selected CaNAC genes in roots and leaves of ILC482 and Hashem cultivars revealed different dehydration-responsive expression patterns, indicating that CaNAC gene expression is tissue- and genotype-specific. Furthermore, the analysis suggested that the enhanced drought tolerance of ILC482 vs. Hashem might be associated with five genes, namely CaNAC02, 04, 05, 16, and 24. CaNAC16 could be a potential candidate gene, contributing to the better drought tolerance of ILC482 vs. Hashem as a positive regulator. Conversely, CaNAC02 could be a potential negative regulator, contributing to the differential drought tolerability of these two cultivars. Thus, our results have also provided a solid foundation for selection of promising tissue-specific and/or dehydration-responsive CaNAC candidates for detailed in planta functional analyses, leading to development of transgenic chickpea varieties with improved productivity under drought.
Introduction
Drought has been considered as a major environmental constraint commonly encountered by plants, which cause significant losses to crop yield (Shao et al., 2009; Stolf-Moreira et al., 2011; Osakabe et al., 2013). Intensive research conducted in the past two decades has provided an insight into molecular mechanisms that control plant responses to drought (Shao et al., 2008; Ni et al., 2009; Hadiarto and Tran, 2011; Jogaiah et al., 2013; Albacete et al., 2014; Shanker et al., 2014). Various transcription factors (TFs) and their DNA binding sites, the so-called cis-acting elements, have been identified as molecular switches of stress-responsive gene expression (Yamaguchi-Shinozaki and Shinozaki, 2006; Tran et al., 2007). Among the TF families, the plant-specific NAC [no apical meristem (NAM), Arabidopsis transcription activation factor (ATAF), and cup-shaped cotyledon (CUC)] TF family members have been intensively studied owing to their functions in a wide range of biological processes in plants, including regulation of plant responses to environmental stimuli (Olsen et al., 2005; Tran et al., 2010; Nakashima et al., 2012; Puranik et al., 2012). Increasing number of reports have shown convincing evidence correlating drought tolerance of various plant species and expression of NAC genes (Tran et al., 2004; Hu et al., 2006; Nakashima et al., 2007; Thao et al., 2013; Thu et al., 2014a), suggesting their potential for genetic engineering of improved drought-tolerant crop varieties.
Chickpea (Cicer arietinum L.) is a nutritionally important legume crop cultivated in many countries in the Asian–African region, supplying a great source of mineral-, vitamin-, protein-, and carbohydrate-rich food for animal feeding and human consumption (Rubio, 2005; Bampidis and Christodoulou, 2011; Jukantil et al., 2012; Ngwe et al., 2012). However, drought imposes a detrimental impact on chickpea productivity worldwide, leading to a significant yield loss which has necessitated the load of chickpea research programs with the aim to develop drought-tolerant chickpea cultivars (Molina et al., 2008; Jain and Chattopadhyay, 2010; Nasr Esfahani et al., 2014). Seeing the great potential of the NAC TFs in conferring plant tolerance to drought, we recently took the advantage of the availability of the chickpea whole genomic sequence (Jain et al., 2013; Varshney et al., 2013) to identify all the CaNAC genes annotated in the chickpea genome (Ha et al., 2014). A total of 71 and 62 potential CaNAC genes was identified in the genome of the sequenced chickpea “kabuli” and “desi” cultivars, respectively (Jain et al., 2013; Varshney et al., 2013), many of which showed dehydration-responsive patterns, suggesting their involvement in regulation of drought responses in chickpea, and thus potentially playing important roles in chickpea adaptation to drought stress (Ha et al., 2014).
In this study, we further examined the functions of CaNAC genes in chickpea by comparing the expression levels of a subset of CaNAC genes in two chickpea cultivars with contrasting drought tolerance using real-time quantitative PCR (RT-qPCR) under normal and dehydration conditions. Such correlation analysis of expression levels, dehydration-responsive expression patterns and drought-tolerant degrees will enable us to identify CaNAC genes that are potentially associated with drought tolerance for in-depth in planta functional characterization prior to using them in genetic engineering for development of transgenic chickpea, as well as other crop, cultivars with superior yield under water-limited conditions.
Materials and Methods
Plant Growth, Treatments, and Collection of Tissues
Seeds of chickpea (Cicer arietinum L.) drought-sensitive Hashem and drought-tolerant ILC482 “kabuli” cultivars were received from International Center for Agricultural Research in the Dry Area (ICARDA), Syria. Hashem was developed by the Seed and Plant Improvement Institute, Karaj, Iran (Sabaghpour et al., 2005), whereas ILC482 was released by ICARDA, Syria (Singh et al., 1992). The drought-tolerant ILC482 and drought-sensitive Hashem cultivars used in this study are well-known for their contrasting drought tolerance. Their differential drought tolerability was demonstrated by the comparison of the stress tolerance index (STI), geometric mean productivity (GMP), mean productivity (MP), and harmonic mean (HM) that were determined based on their yields obtained from a field study under irrigated (well-watered) and rainfed (drought stress) conditions (Rozrokh et al., 2012, 2013). For treatments, 9-days-old chickpea seedlings grown in pots containing vermiculite under greenhouse conditions (continuous 30°C temperature, photoperiod of 12 h/12 h, 150 μmol m-2 s-1 photon flux density and 60% relative humidity) as described by Ha et al. (2014) were used. The plants were carefully removed from pots, gently washed to remove soil from roots, then subjected to either dehydration or water (control) treatments for a period of 2 and 5 h according to the methods published earlier (Tran et al., 2009). For dehydration treatment, washed plants were dried on Kim Towels (Nippon Paper Crecia Ltd.) papers, while for water treatment plants were kept in water for indicated time points. Subsequently, leaf and root samples of three biological replicates were carefully collected and frozen in liquid nitrogen for expression analysis.
RNA Isolation, DNaseI Treatment, cDNA Synthesis
Total RNA was purified from collected leaf and root samples using RNeasy Plant Mini Kit and QIAcube system (Qiagen) according to the manufacture’s instruction. Determination of RNA concentration, DNaseI digestion, and cDNA preparation for real-time quantitative PCR (RT-qPCR) were performed as previously described (Le et al., 2011a).
RT-qPCR and Statistical Analyses
Gene-specific primers, which were designed by Ha et al. (2014; Table 1), were used in the RT-qPCR analysis of 3 biological replicates to assess the expression of 19 selected dehydration-responsive CaNAC genes under various treatment conditions. Detailed information about the RT-qPCR reactions was described in (Le et al., 2011a). The RT-qPCR reactions were run using Stratagene MX3000P system (Agilent Technologies, Santa Clara, CA, USA) with the following thermal profile: 95°C for 1 min, 40 cycles at 95°C for 15 s and at 60°C for 1 min. After the last PCR cycle, the melting curves were obtained using the thermal profile of 95°C for 1 min followed by a constant increase in the temperature between 55 and 95°C. The IF4a gene, with specific RT-qPCR primers F: 5′-TGGACCAGAACACTAGGGACATT-3′ and R: 5′-AAACACGGGAAGACCCAGAA-3′, was selected as reference gene according to a report published earlier (Garg et al., 2010), and 2-ΔΔCt method was used in analysis of RT-qPCR data (Le et al., 2012). Statistical significance of the differential expression within a cultivar or between 2 cultivars under well-watered or dehydration treatment was assessed using the Student’s t-test (one tail, unpaired, equal variance). A gene was considered as dehydration-responsive if it had at least two-fold expression change (P-value < 0.05) at least at one time point under dehydration. For comparison of expression levels of CaNAC genes between drought-tolerant ILC482 and drought-sensitive Hashem, differential expression ratio with at least two-fold (P-value < 0.05) was considered as significant.
Criteria for Selection of Potential Dehydration-Responsive CaNAC Genes for In-Depth In Planta Functional Analyses and Genetic Engineering
The method was adopted from a previously published research (Thu et al., 2014b). Briefly, the selected candidate genes could be classified into two groups based on the following selection criteria. Group 1 of candidate genes are those being considered to be potential for development of improved drought-tolerant transgenic plants using overexpression approach, if they meet one of the following criteria: (i) being dehydration-inducible in tolerant cultivar vs. unchanged in sensitive cultivar and possessing higher expression levels in the tolerant cultivar under well-watered and/or dehydration conditions, (ii) showing up-regulation tendency by dehydration in both tolerant and sensitive cultivars with higher up-regulated expression change in the drought-tolerant cultivar under well-watered and/or dehydration conditions, (iii) being up-regulated in tolerant cultivar vs. unchanged in sensitive cultivar, or up-regulated/unchanged in tolerant cultivar vs. down-regulated in sensitive cultivar. Group 2 of candidate genes are those being unchanged or down-regulated by dehydration in both cultivars and showing lower expression levels in tolerant cultivar under well-watered and/or dehydration conditions. These genes could be considered for creation of improved drought-tolerant transgenic plants using gene suppression approach, such as RNA interference (RNAi).
Results
Expression Patterns of Selected CaNAC Genes in Leaves and Roots of Drought-Tolerant ILC482 Cultivar under Dehydration
The availability of natural germplasm and genetic diversity of crop varieties provides an essential key for biotechnological programs toward abiotic stress tolerance. As a means to gain a further understanding of relevant contributions of CaNAC genes to drought tolerance of chickpea and to identify candidate CaNAC genes for transgenic study, we obtained the drought-tolerant ILC482 and drought-sensitive Hashem chickpea varieties from ICARDA for comparative expression analysis of a subset of CaNAC genes. In a previous study, we found that expression of 19 of 23 CaNAC genes examined was significantly altered in leaves and roots of the drought-sensitive Hashem chickpea plants by dehydration (Ha et al., 2014), suggesting that these genes may play an important role in drought responses of chickpea. These 19 CaNAC genes, representing 26.76% (19/71 CaNAC genes identified in chickpea genome) of the CaNAC members in chickpea (Ha et al., 2014), were then selected to examine whether there is a correlation between their dehydration-responsive expression patterns in the drought-tolerant ILC482 and drought-sensitive Hashem and the differential drought tolerability of these two cultivars.
As a first step toward this objective, we determined the expression of the 19 selected CaNAC genes in the leaf and root tissues of the drought-tolerant ILC482 cultivar that was grown and subjected to dehydration treatment in parallel with the drought-sensitive Hashem cultivar. All the 19 selected CaNAC genes also displayed dehydration-responsive in ILC482 as observed in Hashem, out of which 13 and 19 genes showed altered expression in roots and leaves of ILC482, respectively, by dehydration treatment according to the pre-defined criterion (fold-change in expression ≥ 2 and P < 0.05; Figures 1 and 2). A significant overlap was observed among the dehydration-responsive CaNAC genes identified in ILC482 roots and leaves, with 10 and 1 genes being induced and repressed, respectively, in both root and leaf tissues (Figure 3).
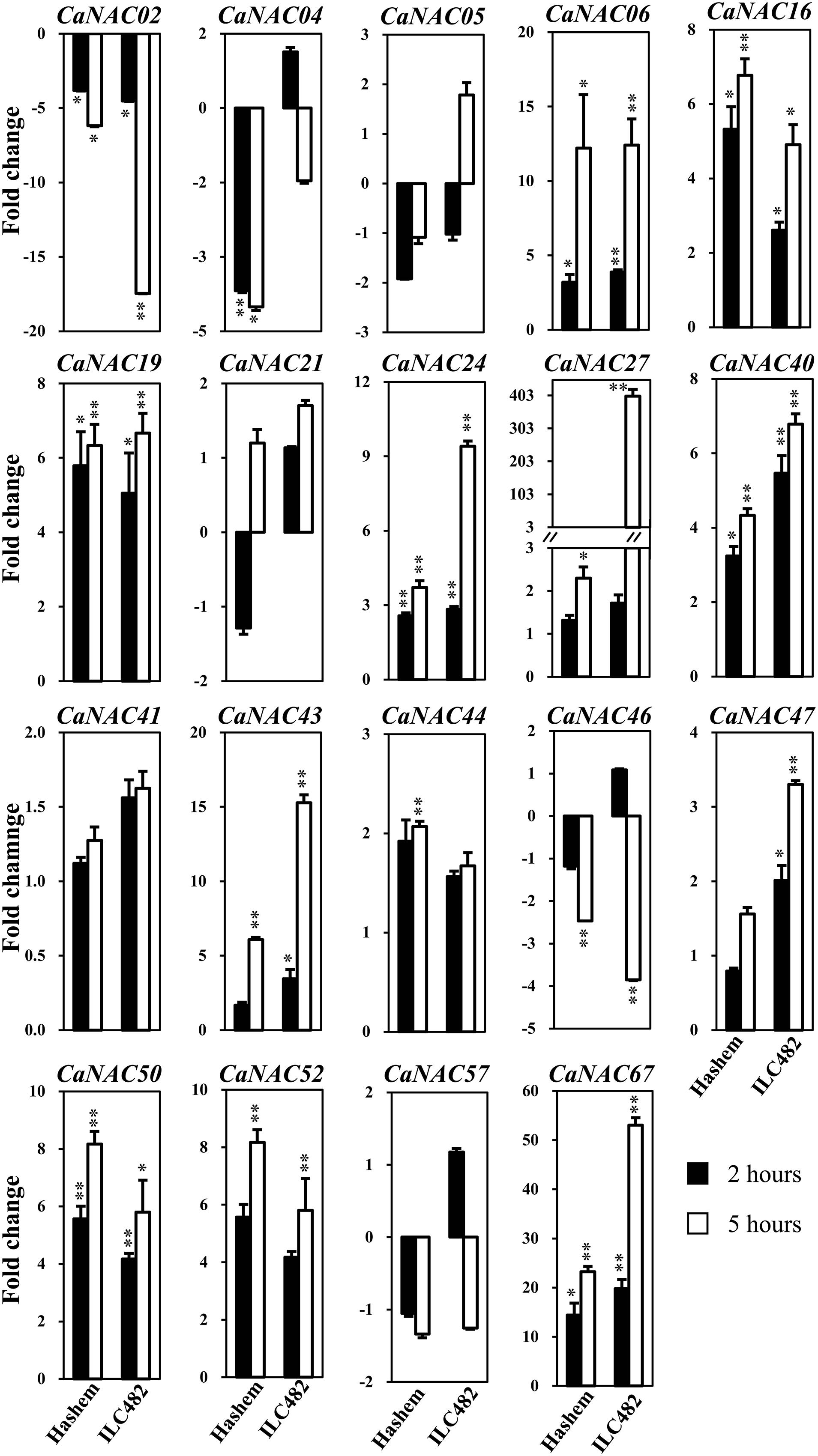
FIGURE 1. Expression of 19 selected CaNAC genes in roots of drought-tolerant ILC482 and drought-sensitive Hashem cultivars under dehydration. Expression data of the CaNAC genes in ILC482 roots were obtained by RT-qPCR of root samples treated with well-water control or dehydration for 2 or 5 h. For convenient comparison, expression data of the CaNAC genes in Hashem roots were extracted from Ha et al. (2014) and displayed. Mean relative expression levels normalized to a value of 1 in water-treated control root samples. Error bars = SE values of 3 biological replicates. Asterisks indicate significant differences as determined by a Student’s t-test (∗P < 0.05; ∗∗P < 0.01).
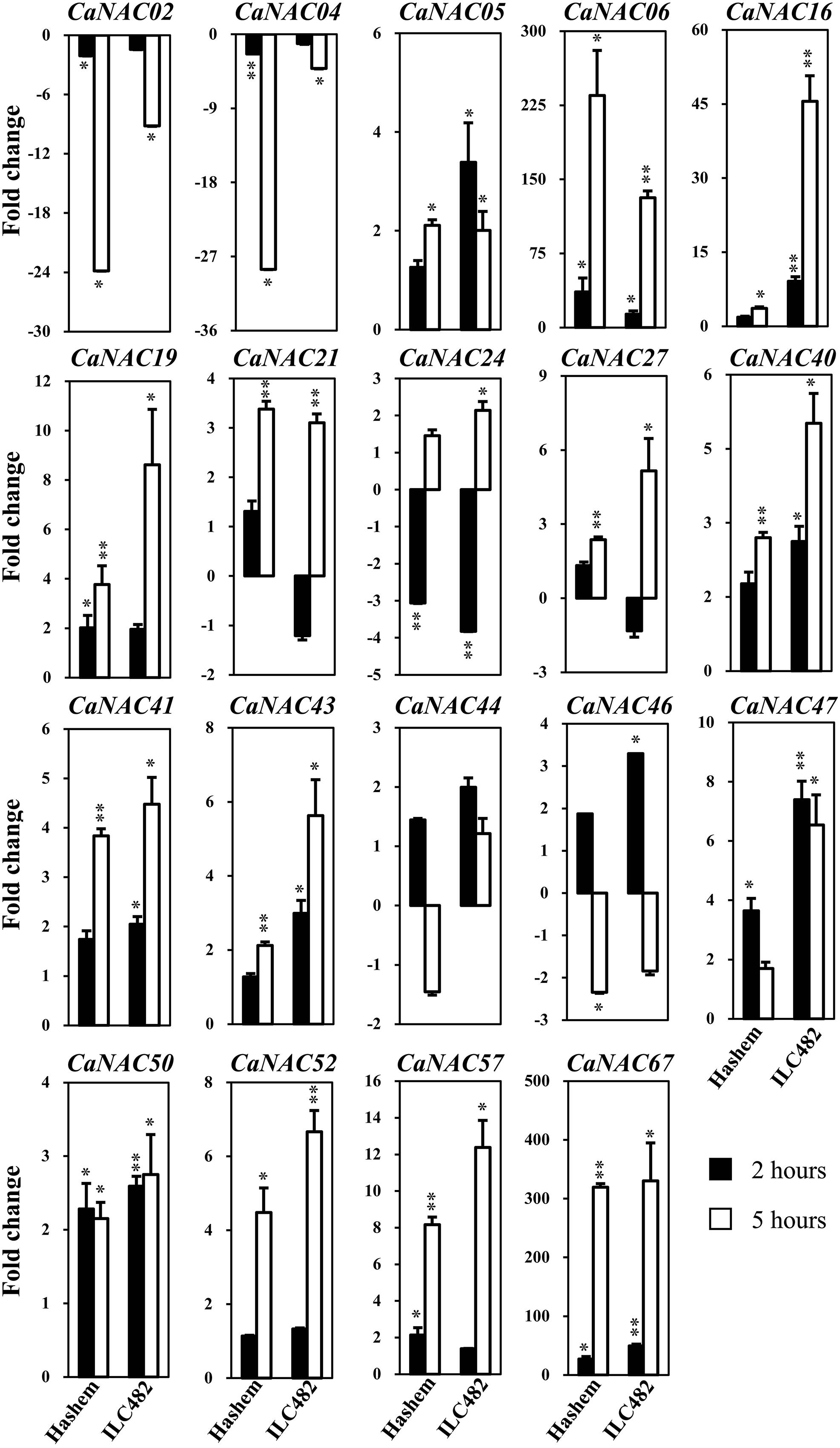
FIGURE 2. Expression of 19 selected CaNAC genes in leaves of drought-tolerant ILC482 and drought-sensitive Hashem cultivars under dehydration. Expression data of the CaNAC genes in ILC482 leaves were obtained by RT-qPCR of leaf samples treated with well-water control or dehydration for 2 or 5 h. For convenient comparison, expression data of the CaNAC genes in Hashem leaves were extracted from Ha et al. (2014) and displayed. Mean relative levels were normalized to a value of 1 in water-treated control leaf samples. Error bars = SE values of 3 biological replicates. Asterisks indicate significant differences as determined by a Student’s t-test (∗P < 0.05; ∗∗P < 0.01).
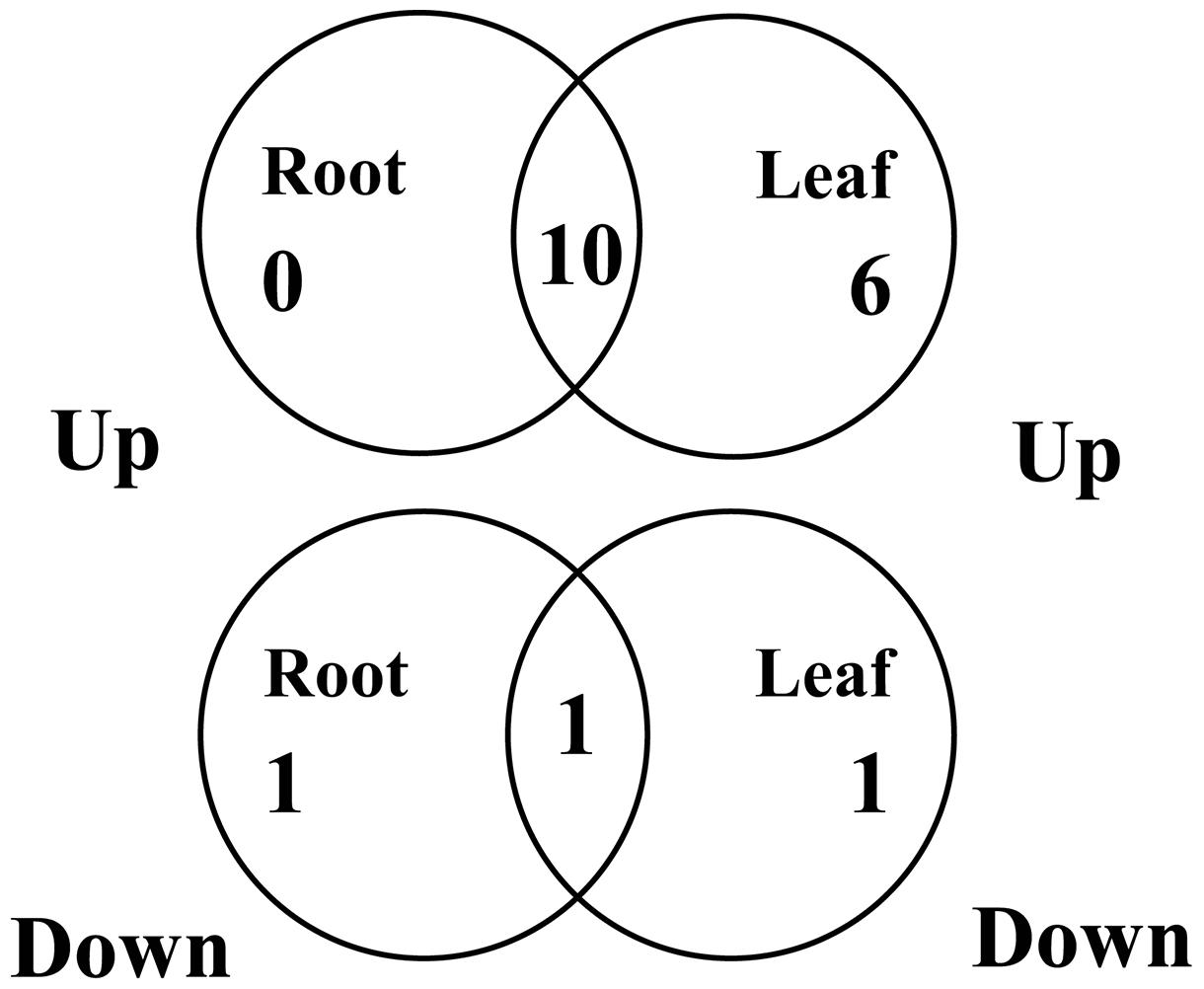
FIGURE 3. Venn diagram analysis of expression of 19 selected CaNAC genes in roots and leaves of ILC482 under dehydration. CaNAC24 was not included in the analysis because it displayed opposite expression patterns in dehydrated ILC482 leaf tissues at 2 and 5 h.
Specifically, we found 11 (CaNAC06, 16, 19, 24, 27, 40, 43, 47, 50, 52, and 67) and 17 (CaNAC05, 06, 16, 19, 21, 24, 27, 40, 41, 43, 44, 46, 47, 50, 52, 57, and 67) up-regulated CaNAC genes in dehydrated roots and leaves of ILC482, respectively, whereas 2 (CaNAC02 and 46) and 2 (CaNAC02 and 04) down-regulated CaNAC genes in the corresponding dehydrated root (Figure 1; Table 2) and leaf tissues (Figure 2; Table 3). Noticeably, CaNAC27 and CaNAC67 were the two most significantly induced genes in ILC482 roots and leaves by over 300- and 400-fold, respectively, whereas CaNAC02 was the most highly repressed gene in both roots (17.5-fold) and leaves (9.2-fold) of ILC482 after 5 h of dehydration. It is also interesting to note that CaNAC24 displayed opposite expression patterns in dehydrated ILC482 leaf tissues at 2 and 5 h, with down-regulation of 3.8-fold at 2 h but then up-regulation of 2.1-fold at 5 h of dehydration (Figure 2; Table 3). This gene was then not included in the Venn analysis to study the overlap in expression responsiveness of dehydration-responsive genes in ILC482 roots and leaves (Figure 3). In addition, CaNAC46 was noteworthy to be mentioned as its expression was repressed by 3.9-fold (at 5 h) in dehydrated ILC482 roots (Figure 1; Table 2) but induced by 3.3-fold (at 2 h) in dehydrated ILC482 leaves (Figure 2; Table 3). Such opposite dehydration-responsive expression profiles in roots and leaves indicate the diverse and tissue-specific functions of CaNAC46 in regulation of ILC482 chickpea cultivar to drought in a way that would provide the best survival of chickpea plants under water deficit conditions.
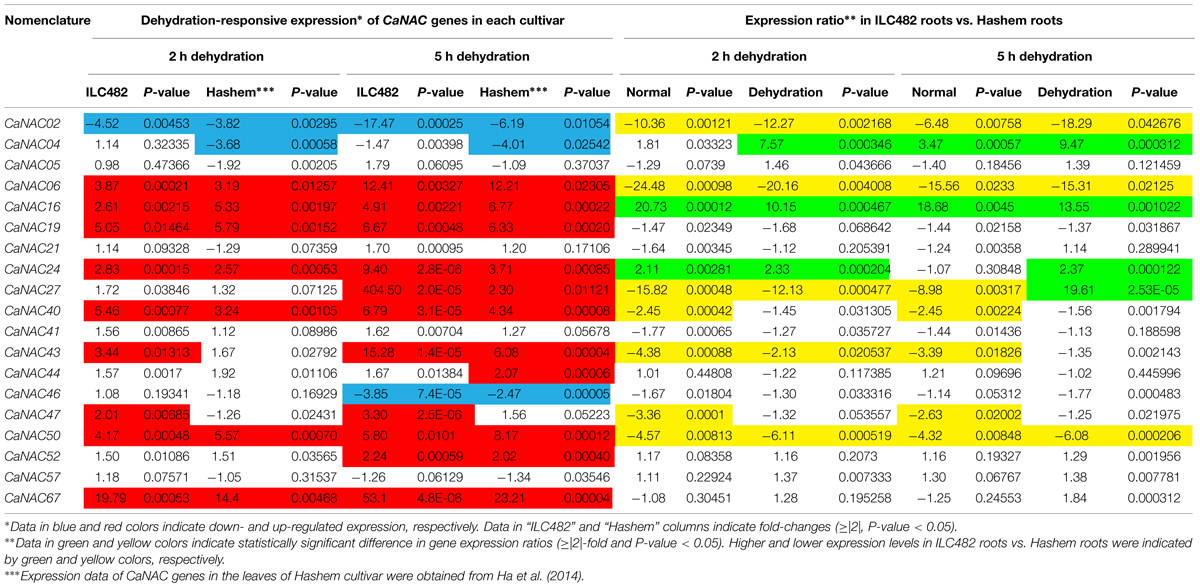
TABLE 2. Comparison of the expression levels of 19 CaNAC genes in the roots of ILC482 and Hashem cultivars under normal and dehydration conditions.
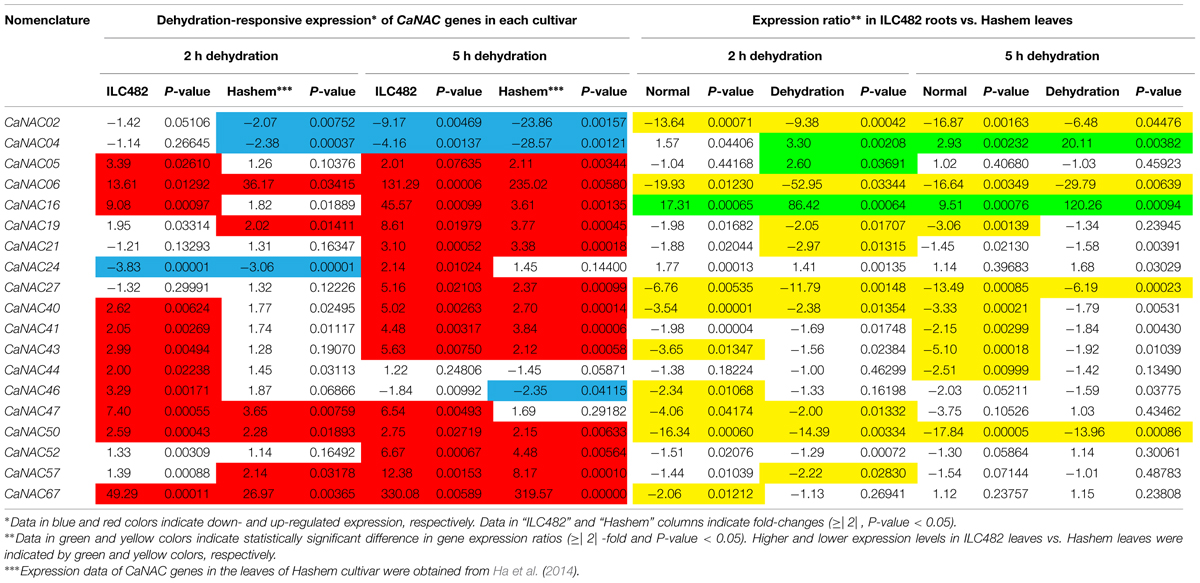
TABLE 3. Comparison of the expression levels of 19 CaNAC genes in the leaves of ILC482 and Hashem cultivars under normal and dehydration conditions.
Differential Expression of the CaNAC Genes in Roots of ILC482 and Hashem
As reported earlier by Ha et al. (2014), among the 19 tested CaNAC genes, seven (CaNAC06, 16, 19, 24, 40, 50, and 67) and two (CaNAC02 and 04) genes were up-regulated and down-regulated, respectively, in roots of Hashem cultivar by 2 h dehydration, whereas 11 (CaNAC06, 16, 19, 24, 27, 40, 43, 44, 50, 52, and 67) and 3 genes (CaNAC02, 04, and 46) were induced and repressed, respectively, in the same tissues by 5 h dehydration (Figure 1; Table 2). In comparison with drought-tolerant ILC482, our data demonstrated that more CaNAC genes were up-regulated, whereas less CaNAC genes were down-regulated by dehydration in the drought-tolerant ILC482 roots than in the drought-sensitive Hashem roots. Specifically, we detected 9 and 7 dehydration-induced, as well as 1 and 2 dehydration-repressed CaNAC genes in roots of ILC482 and Hashem, respectively, after 2 h of dehydration (Table 2). As for 5 h dehydration, we recorded the same number (11) of up-regulated CaNAC genes in roots of ILC482 and Hashem, whereas less down-regulated CaNAC genes in roots of ILC482 than in roots of Hashem (2 vs. 3; Table 2).
A comparative analysis of expression levels of the CaNAC genes in the roots of drought-tolerant ILC482 vs. those in the roots of drought-sensitive Hashem revealed that under normal conditions, 2 (CaNAC16 and 24) and 7 (CaNAC02, 06, 27, 40, 43, 47, and 50) CaNAC genes had higher and lower expression levels, respectively, in ILC482 roots than Hashem roots after 2 h water control treatment. The same 7 CaNAC genes showed lower expression levels by 5 h water treatment, while 2 CaNAC genes, namely CaNAC04 and 16, displayed higher expression levels in ILC482 roots vs. Hashem roots (Table 2). On the other hand, under dehydration conditions, 3 and 4 CaNAC genes showed higher expression levels, whereas 5 and 3 genes exhibited lower expression levels in ILC482 roots than Hashem roots after 2 and 5 h treatments, respectively (Table 2). Specifically, CaNAC04, 16, and 24 and CaNAC02, 06, 27, 43, and 50 were found to possess higher and lower expression levels, respectively, in ILC482 roots than Hashem roots after 2 h water control treatment. With regard to 5 h treatment, we recorded the same three genes CaNAC04, 16, and 24 in addition to the CaNAC27 showing higher expression levels, whereas CaNAC02, 06, and 50 displaying lower expression levels in ILC482 roots vs. Hashem roots, as in the case of 2 h dehydration treatment. With the exception of CaNAC04, which was down-regulated in Hashem roots by both 2 and 5 h dehydration treatments, CaNAC16, 24, and 27 were up-regulated by dehydration in ILC482 roots, as well as Hashem roots.
Differential Expression of the CaNAC Genes in Leaves of ILC482 and Hashem
With regard to the expression of the tested CaNAC genes in leaves, Ha et al. (2014) reported that among 19 selected CaNAC genes, 6 (CaNAC06, 19, 47, 50, 57, and 67) and 3 (CaNAC02, 04, and 24) genes showed up-regulated and down-regulated expression, respectively, in the leaves of Hashem cultivar by 2 h dehydration (Figure 2; Table 3). On the other hand, they detected more dehydration-responsive genes in 5-h-dehydrated Hashem leaves. Namely, they found 13 (CaNAC05, 06, 16, 19, 21, 27, 40, 41, 43, 50, 52, 57, and 67) and 3 genes (CaNAC02, 04, and 46) displaying up-regulated and down-regulated expression patterns, respectively, in 5-h-dehydrated Hashem leaves (Figure 2; Table 3). Similar to our observation in roots, when comparing the dehydration-regulated expression patterns of the 19 tested CaNAC genes in the leaves of ILC482 and Hashem, we found that a higher number of CaNAC genes were up-regulated, whereas a lower number of CaNAC genes were down-regulated in ILC482 leaves than in Hashem leaves by either 2 or 5 h dehydration treatment. Specifically, we recorded 11 and 15 up-regulated CaNAC genes in leaves of ILC482, while only 6 and 13 up-regulated CaNAC genes in leaves of Hashem after 2 and 5 h dehydration treatments, respectively (Table 3). As for the down-regulated CaNAC genes, we detected 1 and 2 down-regulated genes in ILC482 leaves, whereas 3 and 3 down-regulated genes in Hashem leaves after 2 and 5 h dehydration treatments, respectively (Table 3).
A comparison of the expression levels of the tested CaNAC genes in the leaves of ILC482 and Hashem revealed similar tendency as observed in the roots. Under well-watered conditions, 9 (CaNAC02, 06, 27, 40, 43, 46, 47, 50, and 67) genes showed lower expression levels, while 1 (CaNAC16) gene possessed higher transcript abundance in ILC482 leaves than Hashem leaves after 2 h water control treatment. The same number of genes (CaNAC02, 06, 19, 27, 40, 41, 43, 44, and 50) showing lower expression levels in ILC482 leaves than in Hashem leaves by 5 h water control treatment was found, whereas 2 (CaNAC04 and 16) genes were recorded with higher expression levels in the same comparison. Under dehydration conditions, 9 and 4 genes were noted to have lower expression levels in ILC482 leaves than Hashem leaves after 2 and 5 h treatments, respectively. On the other hands, 3 (CaNAC04, 05, and 16) and 2 (CaNAC04 and 16) genes showed higher transcript abundance in ILC482 leaves than Hashem leaves after 2 and 5 h treatments, respectively.
Selection of Potential CaNAC Candidate Genes for In-Depth In Planta Characterization
As a means to propose promising CaNAC candidate genes for further in-depth in planta functional analyses, which would lead to their application in generating improved drought-tolerant transgenic chickpea plants using genetic engineering, we applied the section criteria adopted from a study published previously (Thu et al., 2014b). Among the 19 CaNAC genes examined in this study, 5 genes could be suggested as top priorities for functional characterizations according to the selection criteria set in the Materials and Methods. Specifically, 3 (CaNAC04, 16, and 24) genes of Group 1 and 1 (CaNAC02) gene of Group 2 were found to be satisfied for overexpression and knock-down studies, respectively, based on the differential analysis of the root expression data. On the other hand, according to the differential analysis of the leaf expression data, 3 (CaNAC04, 05, and 16) genes and 1 (CaNAC02) gene were noted to meet the selection criteria to be classified to Groups 1 and 2, respectively.
Discussion
The plant-specific NAC TF family is one of the important TF families in plant kingdom, whose members play diverse functions during plant growth and development (Olsen et al., 2005; Tran et al., 2010; Nakashima et al., 2012; Puranik et al., 2012). The drought-related function of NAC genes was first discovered through the study of ANAC019, ANAC055, and ANAC072 in Arabidopsis (Tran et al., 2004), which then has led to many other studies in different plant species, including crops. One of the best studies that reported the potential application of NAC genes in agriculture is the work of Hu et al. (2006), who reported that transgenic rice plants overexpressing SNAC1 exhibited enhanced drought tolerance without yield penalty. Since then, an increasing number of studies, including transgenic or correlation analyses, have provided strong evidence for the correlation between NAC gene expression and drought-tolerant capacity of various crops (Nakashima et al., 2007; Zheng et al., 2009; Xue et al., 2011; Thao et al., 2013; Thu et al., 2014a; Zhu et al., 2014; Yang et al., 2015).
The root plasticity is an important root trait responding to various environmental stressors, including drought, to help plants adapt to adverse conditions. Primary root length, root biomass, and number of lateral roots are all important parameters for evaluation of drought tolerance in crops (Sharp et al., 2004; Manavalan et al., 2009; Nishiyama et al., 2011; Ha et al., 2013; Zhu et al., 2014). A recent study on SlNAC4 gene of tomato (Solanum lycopersicum) has provided convincing evidence for the regulatory function of NAC TFs in modulation of root growth under abiotic stresses. Suppression of SlNAC4 expression has resulted in hypersensitivity to drought and salt stress to SlNAC4-RNAi transgenic tomato plants, which was attributed to inhibition of root growth, as well as a decrease in water and chlorophyll contents (Zhu et al., 2014). Thus, studying expression of the CaNAC genes in roots of chickpea cultivars with contrasting drought-tolerant phenotype will enable us to determine the correlation between CaNAC gene expression and drought tolerability, which will subsequently aid us in identifying root trait-related CaNAC genes for further functional analysis. The comparative expression analysis of the 19 selected CaNAC genes has allowed us to detect a higher number of dehydration-inducible CaNAC genes (9 genes vs. 7 genes and 11 vs. 11 after 2 and 5 h dehydration treatments, respectively) and a lower number of dehydration-repressible CaNAC genes (1 gene vs. 2 genes and 2 genes vs. 3 genes after 2 and 5 h dehydration treatments, respectively) in the roots of drought-tolerant ILC482 than in the roots of drought-sensitive Hashem (Figure 1; Table 2). These findings suggested a correlation between drought tolerability of ILC482 and Hashem cultivars and the number of the dehydration-responsive CaNAC genes in their roots.
In addition, leaf-related traits, such as stomata aperture and leaf cell membrane stability, have been also well-known traits that influence drought tolerance (Kaiser, 2009; Manavalan et al., 2009; Guttikonda et al., 2014; Ha et al., 2014). Overexpression of SNAC1 gene in rice was shown to enhance stomatal closure, thereby contributing to improved drought tolerance of transgenic plants (Hu et al., 2006). This finding suggested a close association of NAC gene expression and leaf-related traits. Thus, it was also our interest to examine the correlation between drought-tolerant levels of the two contrasting chickpea cultivars and expression levels of CaNAC genes in leaf tissues under dehydration. As shown in Figure 2 and summarized in Table 3, more up-regulated CaNAC genes, whereas less down-regulated CaNAC genes were found in ILC482 leaves than in Hashem leaves. These data suggested a positive correlation between drought-tolerant degree of ILC482 and Hashem cultivars and the number of the dehydration-responsive CaNAC genes in leaves as well, which together with the results obtained in the roots (Figure 1; Table 2) firmly demonstrated this positive correlation. Taken together, the higher drought-tolerant capacity of ILC482 vs. Hashem might partly be attributed to their differential expression of the CaNAC genes in both root and leaf tissues. The more CaNAC genes are up-regulated and the less CaNAC genes down-regulated by dehydration, the higher drought-tolerant the cultivar is. In support of our results, previous studies in soybean (Glycine max) also identified positive correlation between the number of drought-inducible GmNAC genes and drought-tolerant capacity of 2 contrasting cultivars (Thao et al., 2013; Thu et al., 2014a).
From our comparative analyses of the expression of these selected 19 CaNAC genes, we also observed differential expression patterns between roots and leaves in the same cultivar, either ILC482 or Hashem, or between the same organs of the two contrasting chickpea cultivars (Tables 2 and 3). This finding suggested that the expression of CaNAC genes, at least of those examined in this study, is tissue- and genotype-dependent, which might then result in different phenotypes of different cultivars. Differential expression analyses of GmNAC genes in 3 soybean cultivars with different phenotypes also showed their tissue- and genotype-dependent expression patterns (Le et al., 2011b; Thao et al., 2013; Thu et al., 2014a,c), further supporting our observation.
One of the major aims of this study is to identify the best CaNAC candidate genes that have high potential for development of drought-tolerant chickpea cultivars by genetic engineering. On the basis of our analysis (Tables 2 and 3) and the selection criteria adopted from Thu et al. (2014b), 4 (CaNAC04, 05, 16, and 24) genes belonging to Group 1, and 1 gene (CaNAC02) classified to Group 2 could be selected for detailed in planta functional analyses in model plant systems, such as Arabidopsis, prior to using them in genetic engineering of chickpea plants or other legume crops. CaNAC04, 16, and CaNAC02 are associated with both root and leave tissues, whereas CaNAC05 and CaNAC24 are specifically associated with leaves and roots, respectively (Tables 2 and 3). All these 5 genes might potentially play important roles in conferring higher drought tolerability to ILC482 than Hashem.
Out of these 5 genes, CaNAC16 would be the best positive regulatory candidate gene as this gene was found (i) to be induced by dehydration in both roots and leaves of both ILC482 and Hashem cultivars, and (ii) to display higher expression levels in drought-tolerant ILC482 than drought-sensitive Hashem under both normal (20.73- and 18.68-fold in roots, and 17.31 and 9.51-fold in leaves at 2 and 5 h, respectively) and dehydration (10.15- and 13.55-fold in roots, and 86.42- and 120.26-fold in leaves at 2 and 5 h, respectively) conditions (Tables 2 and 3). On the other hand, CaNAC02 is a promising negative regulatory gene, as this gene was strongly down-regulated by dehydration in both roots and leaves of both 2 chickpea cultivars, and showed lower expression levels in drought-tolerant ILC482 than drought-sensitive Hashem under both normal (10.36- and 6.48-fold in roots, and 13.64- and 16.87-fold in leaves at 2 and 5 h, respectively) and dehydration (12.27- and 18.29-fold in roots, and 9.38- and 6.48-fold in leaves at 2 and 5 h, respectively) conditions (Tables 2 and 3). Taken together, CaNAC16 and CaNAC02 are highly recommended for detailed functional characterization using overexpression and knock-down approaches, respectively, with the goal to lead to their application in development of chickpea varieties with improved drought tolerance.
Author Contributions
L-SPT conceived research and wrote the manuscript. KHN, CVH, YW, UTT, and MNE performed the experiments. DVN contributed research materials.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
KN gratefully acknowledges the “International Program Associate” of Rikagaku Kenkyusho (Institute of Physical and Chemical Research, Japan) for supporting his Ph.D. study.
References
Albacete, A. A., Martinez-Andujar, C., and Perez-Alfocea, F. (2014). Hormonal and metabolic regulation of source-sink relations under salinity and drought: from plant survival to crop yield stability. Biotechnol. Adv. 32, 12–30. doi: 10.1016/j.biotechadv.2013.10.005
Bampidis, V. A., and Christodoulou, V. (2011). Chickpeas (Cicer arietinum L.) in animal nutrition: a review. Anim. Feed Sci. Technol. 168, 1–20. doi: 10.1016/j.anifeedsci.2011.04.098
Garg, R., Sahoo, A., Tyagi, A. K., and Jain, M. (2010). Validation of internal control genes for quantitative gene expression studies in chickpea (Cicer arietinum L.). Biochem. Biophys. Res. Commun. 396, 283–288. doi: 10.1016/j.bbrc.2010.04.079
Guttikonda, S. K., Valliyodan, B., Neelakandan, A. K., Tran, L. S., Kumar, R., Quach, T. N., et al. (2014). Overexpression of AtDREB1D transcription factor improves drought tolerance in soybean. Mol. Biol. Rep. 41, 7995–8008. doi: 10.1007/s11033-014-3695-3
Ha, C. V., Le, D. T., Nishiyama, R., Watanabe, Y., Tran, U. T., Dong, N. V., et al. (2013). Characterization of the newly developed soybean cultivar DT2008 in relation to the model variety W82 reveals a new genetic resource for comparative and functional genomics for improved drought tolerance. Biomed. Res. Int. 2013, 1–8. doi: 10.1155/2013/759657
Ha, C. V., Leyva-Gonzalez, M. A., Osakabe, Y., Tran, U. T., Nishiyama, R., Watanabe, Y., et al. (2014). Positive regulatory role of strigolactone in plant responses to drought and salt stress. Proc. Natl. Acad. Sci. U.S.A. 111, 851–856. doi: 10.1073/pnas.1322135111
Hadiarto, T., and Tran, L. S. (2011). Progress studies of drought-responsive genes in rice. Plant Cell Rep. 30, 297–310. doi: 10.1007/s00299-010-0956-z
Hu, H., Dai, M., Yao, J., Xiao, B., Li, X., Zhang, Q., et al. (2006). Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. U.S.A. 103, 12987–12992. doi: 10.1073/pnas.0604882103
Jain, D., and Chattopadhyay, D. (2010). Analysis of gene experession in response to water deficit of chickpea (Cicer arietinum L.) varieties differing in drought tolerance. BMC Plant Biol. 10:24. doi: 10.1186/1471-2229-10-24
Jain, M., Misra, G., Patel, R. K., Priya, P., Jhanwar, S., Khan, A. W., et al. (2013). A draft genome sequence of the pulse crop chickpea (Cicer arietinum L.). Plant J. 74, 715–729. doi: 10.1111/tpj.12173
Jogaiah, S., Govind, S. R., and Tran, L. S. (2013). Systems biology-based approaches toward understanding drought tolerance in food crops. Crit. Rev. Biotechnol. 33, 23–39. doi: 10.3109/07388551.2012.659174
Jukantil, A. K., Gaur, P. M., Gowda, C. L. L., and Chibbar, R. N. (2012). Nutritional quality and health benefits of chickpea (Cicer arietinum L.): a review. Br. J. Nutr. 108, 11–26. doi: 10.1017/S0007114512000797
Kaiser, H. (2009). The relation between stomatal aperture and gas exchange under consideration of pore geometry and diffusional resistance in the mesophyll. Plant Cell Environ. 32, 1091–1098. doi: 10.1111/j.1365-3040.2009.01990.x
Le, D. T., Aldrich, D. L., Valliyodan, B., Watanabe, Y., Van Ha, C., Nishiyama, R., et al. (2012). Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. PLoS ONE 7:e46487. doi: 10.1371/journal.pone.0046487
Le, D. T., Nishiyama, R., Watanabe, Y., Mochida, K., Yamaguchi-Shinozaki, K., Shinozaki, K., et al. (2011a). Genome-wide expression profiling of soybean two-component system genes in soybean root and shoot tissues under dehydration stress. DNA Res. 18, 17–29. doi: 10.1093/dnares/dsq032
Le, D. T., Nishiyama, R., Watanabe, Y., Mochida, K., Yamaguchi-Shinozaki, K., Shinozaki, K., et al. (2011b). Genome-wide survey and expression analysis of the plant-specific NAC transcription factor family in soybean during development and dehydration stress. DNA Res. 18, 263–276. doi: 10.1093/dnares/dsr015
Manavalan, L. P., Guttikonda, S. K., Tran, L.-S. P., and Nguyen, H. T. (2009). Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol. 50, 1260–1276. doi: 10.1093/pcp/pcp082
Molina, C., Rotter, B., Horres, R., Udupa, S. M., Besser, B., Bellarmino, L., et al. (2008). SuperSAGE: the drought stress-responsive transcriptome of chickpea roots. BMC Genomics 9:553. doi: 10.1186/1471-2164-9-553
Nakashima, K., Takasaki, H., Mizoi, J., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2012). NAC transcription factors in plant abiotic stress responses. Biochim. Biophys. Acta 1819, 97–103. doi: 10.1016/j.bbagrm.2011.10.005
Nakashima, K., Tran, L. S., Van Nguyen, D., Fujita, M., Maruyama, K., Todaka, D., et al. (2007). Functional analysis of a NAC-type transcription factor OsNAC6 involved in abiotic and biotic stress-responsive gene expression in rice. Plant J. 51, 617–630. doi: 10.1111/j.1365-313X.2007.03168.x
Nasr Esfahani, M., Sulieman, S., Schulze, J., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L. S. (2014). Mechanisms of physiological adjustment of N2 fixation in Cicer arietinum L. (chickpea) during early stages of water deficit: single or multi-factor controls. Plant J. 79, 964–980. doi: 10.1111/tpj.12599
Ngwe, T., Nukui, Y., Oyaizu, S., Takamoto, G., Koike, S., Ueda, K., et al. (2012). Bean husks as a supplemental fiber for ruminants: potential use for activation of fibrolytic rumen bacteria to improve main forage digestion. Anim. Sci. J. 83, 43–49. doi: 10.1111/j.1740-0929.2011.00916.x
Ni, F. T., Chu, L. Y., Shao, H. B., and Liu, Z. H. (2009). Gene expression and regulation of higher plants under soil water stress. Curr. Genomics 10, 269–280. doi: 10.2174/138920209788488535
Nishiyama, R., Watanabe, Y., Fujita, Y., Le, D. T., Kojima, M., Werner, T., et al. (2011). Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23, 2169–2183. doi: 10.1105/tpc.111.087395
Olsen, A. N., Ernst, H. A., Leggio, L. L., and Skriver, K. (2005). NAC transcription factors: structurally distinct, functionally diverse. Trends Plant Sci. 10, 79–87. doi: 10.1016/j.tplants.2004.12.010
Osakabe, Y., Yamaguchi-Shinozaki, K., Shinozaki, K., and Tran, L.-S. P. (2013). Sensing the environment: key roles of membrane-localized kinases in plant perception and response to abiotic stress. J. Exp. Bot. 64, 445–458. doi: 10.1093/jxb/ers354
Puranik, S., Sahu, P. P., Srivastava, P. S., and Prasad, M. (2012). NAC proteins: regulation and role in stress tolerance. Trends Plant Sci. 17, 369–381. doi: 10.1016/j.tplants.2012.02.004
Rozrokh, M., Sabaghpour, S. H., and Armin, M. (2013). Determining the best indices of drought tolerance in chickpea genotypes. Plant Ecophysiol. 4, 25–36.
Rozrokh, M., Sabaghpour, S. H., Armin, M., and Asgharipour, M. (2012). The effects of drought stress on some biochemical traits in twenty genotypes of chickpea. Eur. J. Exp. Biol. 2, 1980–1987.
Rubio, L. A. (2005). Ileal digestibility of defatted soybean, lupin and chickpea seed meals in cannulated Iberian pigs: I. Proteins. J. Sci. Food Agric. 85, 1313–1321. doi: 10.1002/Jsfa.1963
Sabaghpour, S. H., Malhotra, R. S., and Banai, T. (2005). Registration of ‘Hashem’ Kabuli chickpea. Crop Sci. 45:2651. doi: 10.2135/cropsci2004.0772
Shanker, A. K., Maheswari, M., Yadav, S. K., Desai, S., Bhanu, D., Attal, N. B., et al. (2014). Drought stress responses in crops. Funct. Integr. Genomics 14, 11–22. doi: 10.1007/s10142-013-0356-x
Shao, H. B., Chu, L. Y., Jaleel, C. A., Manivannan, P., Panneerselvam, R., and Shao, M. A. (2009). Understanding water deficit stress-induced changes in the basic metabolism of higher plants – biotechnologically and sustainably improving agriculture and the ecoenvironment in arid regions of the globe. Crit. Rev. Biotechnol. 29, 131–151. doi: 10.1080/07388550902869792
Shao, H. B., Chu, L. Y., Shao, M. A., Jaleel, C. A., and Mi, H. M. (2008). Higher plant antioxidants and redox signaling under environmental stresses. C. R. Biol. 331, 433–441. doi: 10.1016/j.crvi.2008.03.011
Sharp, R. E., Poroyko, V., Hejlek, L. G., Spollen, W. G., Springer, G. K., Bohnert, H. J., et al. (2004). Root growth maintenance during water deficits: physiology to functional genomics. J. Exp. Bot. 55, 2343–2351. doi: 10.1093/jxb/erh276
Singh, K. B., Malhotra, R. S., and Saxena, M. C. (1992). Registration of ‘ILC 482’ chickpea. Crop Sci. 32:826. doi: 10.2135/cropsci1992.0011183X003200030051x
Stolf-Moreira, R., Lemos, E. G. M., Carareto-Alves, L., Marcondes, J., Pereira, S. S., Rolla, A. A. P., et al. (2011). Transcriptional profiles of roots of different soybean genotypes subjected to drought stress. Plant Mol. Biol. Rep. 29, 19–34. doi: 10.1007/s11105-010-0203-3
Thao, N. P., Thu, N. B., Hoang, X. L., Van Ha, C., and Tran, L. S. (2013). Differential expression analysis of a subset of drought-responsive GmNAC genes in two soybean cultivars differing in drought tolerance. Int. J. Mol. Sci. 14, 23828–23841. doi: 10.3390/ijms141223828
Thu, N. B., Hoang, X. L., Doan, H., Nguyen, T. H., Bui, D., Thao, N. P., et al. (2014a). Differential expression analysis of a subset of GmNAC genes in shoots of two contrasting drought-responsive soybean cultivars DT51 and MTD720 under normal and drought conditions. Mol. Biol. Rep. 41, 5563–5569. doi: 10.1007/s11033-014-3507-9
Thu, N. B., Hoang, X. L., Nguyen, T. D. H., Thao, N. P., and Tran, L. S. (2014b). Differential expression of two-component system-related drought-responsive genes in two contrasting drought-tolerant soybean cultivars DT51 and MTD720 under well-watered and drought conditions. Plant Mol. Biol. Rep. doi: 10.1007/s11105-014-0825-y
Thu, N. B., Nguyen, Q. T., Hoang, X. L., Thao, N. P., and Tran, L. S. (2014c). Evaluation of drought tolerance of the Vietnamese soybean cultivars provides potential resources for soybean production and genetic engineering. Biomed. Res. Int. 2014:809736. doi: 10.1155/2014/809736
Tran, L. S., Nakashima, K., Sakuma, Y., Simpson, S. D., Fujita, Y., Maruyama, K., et al. (2004). Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell 16, 2481–2498. doi: 10.1105/tpc.104.022699
Tran, L. S., Nakashima, K., Shinozaki, K., and Yamaguchi-Shinozaki, K. (2007). Plant gene networks in osmotic stress response: from genes to regulatory networks. Methods Enzymol. 428, 109–128. doi: 10.1016/S0076-6879(07)28006-1
Tran, L. S., Nishiyama, R., Yamaguchi-Shinozaki, K., and Shinozaki, K. (2010). Potential utilization of NAC transcription factors to enhance abiotic stress tolerance in plants by biotechnological approach. GM Crops 1, 32–39. doi: 10.4161/gmcr.1.1.10569
Tran, L. S., Quach, T. N., Guttikonda, S. K., Aldrich, D. L., Kumar, R., Neelakandan, A., et al. (2009). Molecular characterization of stress-inducible GmNAC genes in soybean. Mol. Genet. Genomics 281, 647–664. doi: 10.1007/s00438-009-0436-8
Varshney, R. K., Song, C., Saxena, R. K., Azam, S., Yu, S., Sharpe, A. G., et al. (2013). Draft genome sequence of chickpea (Cicer arietinum) provides a resource for trait improvement. Nat. Biotechnol. 31, 240–246. doi: 10.1038/nbt.2491
Xue, G. P., Way, H. M., Richardson, T., Drenth, J., Joyce, P. A., and Mcintyre, C. L. (2011). Overexpression of TaNAC69 leads to enhanced transcript levels of stress up-regulated genes and dehydration tolerance in bread wheat. Mol. Plant 4, 697–712. doi: 10.1093/mp/ssr013
Yamaguchi-Shinozaki, K., and Shinozaki, K. (2006). Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 57, 781–803. doi: 10.1146/annurev.arplant.57.032905.105444
Yang, X., Wang, X., Ji, L., Yi, Z., Fu, C., Ran, J., et al. (2015). Overexpression of a Miscanthus lutarioriparius NAC gene MlNAC5 confers enhanced drought and cold tolerance in Arabidopsis. Plant Cell Rep. 34, 943–958. doi: 10.1007/s00299-015-1756-2
Zheng, X., Chen, B., Lu, G., and Han, B. (2009). Overexpression of a NAC transcription factor enhances rice drought and salt tolerance. Biochem. Biophys. Res. Commun. 379, 985–989. doi: 10.1016/j.bbrc.2008.12.163
Keywords: chickpea, NAC transcription factors, differential expression, differential drought tolerability, RT-qPCR
Citation: Nguyen KH, Ha CV, Watanabe Y, Tran UT, Nasr Esfahani M, Nguyen DV and Tran L-SP (2015) Correlation between differential drought tolerability of two contrasting drought-responsive chickpea cultivars and differential expression of a subset of CaNAC genes under normal and dehydration conditions. Front. Plant Sci. 6:449. doi: 10.3389/fpls.2015.00449
Received: 11 May 2015; Accepted: 01 June 2015;
Published: 19 June 2015
Edited by:
Girdhar Kumar Pandey, University of Delhi, IndiaReviewed by:
Hong-Bo Shao, Qingdao University of Science and Technology, ChinaSwati Puranik, Aberystwyth University, UK
Copyright © 2015 Nguyen, Ha, Watanabe, Tran, Nasr Esfahani, Nguyen and Tran. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lam-Son Phan Tran, Signaling Pathway Research Unit, RIKEN Center for Sustainable Resource Science, 1-7-22 Suehiro-cho, Tsurumi-ku, Yokohama 230-0045, Japan, son.tran@riken.jp
 Kien Huu Nguyen
Kien Huu Nguyen Chien Van Ha
Chien Van Ha Yasuko Watanabe1
Yasuko Watanabe1 Uyen Thi Tran
Uyen Thi Tran Maryam Nasr Esfahani
Maryam Nasr Esfahani Dong Van Nguyen
Dong Van Nguyen Lam-Son Phan Tran
Lam-Son Phan Tran